Old World alphaviruses comprise several medically relevant viruses, including chikungunya virus and Ross River virus. Recurrent outbreaks and the lack of antivirals and vaccines demand ongoing research to fight the emergence of these infectious diseases. In this context, a thorough investigation of virus-host interactions is critical. Here, we highlight the importance of the host protein G3BP for several Old World alphaviruses. Our data strongly suggest that G3BP plays a crucial role for the activity of the viral replicase and, thus, the amplification of the viral RNA genome. To our knowledge, the present work is the first to provide a functional link between the regulation of viral polyprotein processing and RNA replication and a host factor for alphaviruses. Moreover, the results of this study raise several questions about the fundamental regulatory mechanisms that dictate the activity of the viral replicase, thereby paving the way for future studies.
KEYWORDS: virus-host interactions, polyprotein processing, RNA replication, replication complexes
ABSTRACT
We present a comprehensive overview of the dependency of several Old World alphaviruses for the host protein G3BP. Based on their replication ability in G3BP-deleted cells, Old World alphaviruses can be categorized into two groups, being either resistant or sensitive to G3BP deletion. We observed that all sensitive viruses have an Arg residue at the P4 position of the cleavage site between the nonstructural protein P1 (nsP1) and nsP2 regions of the replicase precursor polyprotein (1/2 site), while a different residue is found at this site in viruses resistant to G3BP deletion. Swapping this residue between resistant and sensitive viruses also switches the G3BP deletion sensitivity. In the absence of G3BP, chikungunya virus (CHIKV) replication is at the limit of detection. The P4 Arg-to-His substitution partially rescues this defect. The P4 residue of the 1/2 site is known to play a regulatory role during processing at this site, and we found that if processing is blocked, the influence of the P4 residue on the sensitivity to G3BP deletion is abolished. Immunofluorescence experiments with CHIKV replicase with manipulated processing indicate that the synthesis of double-stranded RNA is defective in the absence of G3BP and suggest a role of G3BP during negative-strand RNA synthesis. This study provides a functional link between the host protein G3BP and the P4 residue of the 1/2 site for viral RNA replication of Old World alphaviruses. While this suggests a link between G3BP proteins and viral replicase polyprotein processing, we propose that G3BP proteins do not have a regulatory role during polyprotein processing.
IMPORTANCE Old World alphaviruses comprise several medically relevant viruses, including chikungunya virus and Ross River virus. Recurrent outbreaks and the lack of antivirals and vaccines demand ongoing research to fight the emergence of these infectious diseases. In this context, a thorough investigation of virus-host interactions is critical. Here, we highlight the importance of the host protein G3BP for several Old World alphaviruses. Our data strongly suggest that G3BP plays a crucial role for the activity of the viral replicase and, thus, the amplification of the viral RNA genome. To our knowledge, the present work is the first to provide a functional link between the regulation of viral polyprotein processing and RNA replication and a host factor for alphaviruses. Moreover, the results of this study raise several questions about the fundamental regulatory mechanisms that dictate the activity of the viral replicase, thereby paving the way for future studies.
INTRODUCTION
The genus Alphavirus belongs to the Togaviridae family, a group of enveloped viruses with a single-stranded, positive-sense RNA genome (1). Alphaviruses are globally distributed and geographically distinguished into Old World and New World alphaviruses. Most members are mosquito borne, and many pose health threats for their vertebrate host. The virus genome contains two open reading frames (ORFs). The 5′ ORF encodes a nonstructural (ns) polyprotein (P1234) and is translated directly from the viral genomic RNA. The second ORF encodes a structural polyprotein and is translated from a subgenomic RNA, which is transcribed from a negative-strand RNA template during RNA replication. The P1234 is cleaved into four mature ns proteins (nsP1 to -4) by the nsP2 protease domain (2, 3), which belongs to the papain superfamily of cysteine proteases (4). Each nsP fulfills specific tasks during the viral life cycle, but the combined action of all four nsPs is required for formation of the viral replicase and, thus, replication of the viral RNA genome. Synthesis of the different viral RNA species is regulated by the sequential processing of the three cleavage sites within the P1234 (3, 5, 6). First, processing between nsP3 and nsP4 (3/4 site) activates RNA-dependent RNA polymerase (RdRp) activity, which resides in nsP4 (7), and the resulting P123-nsP4 replicase synthesizes predominantly full-length negative-strand RNA, complementary to genomic RNA, whereas the synthesis of positive-strand genomic and subgenomic RNAs remains inefficient. Subsequent processing between nsP1 and nsP2 (1/2 site), generating nsP1-P23-nsP4 replicase, allows the synthesis of both negative- and positive-strand RNA, but the production of genomic RNA dominates over the production of subgenomic RNA. The fully processed mature nsP1-nsP2-nsP3-nsP4 replicase produces positive-strand RNA exclusively, predominantly subgenomic RNA.
As the mature replicase no longer produces negative-strand RNA (3), its synthesis depends on the continuous production of P1234 (8). During the course of infection, with rising levels of free nsP2 protease, cleavage of P1234 occurs rapidly at the site between nsP2 and nsP3 (2/3 site) (9), leading to the shutdown of negative-strand RNA synthesis 3 to 4 h postinfection (pi) (8).
The temporal regulation of P1234 processing is thus crucial for proper replicase function (10). The P4 to P1′ residues (i.e., amino acids −4 to +1 with respect to the scissile bond) play a critical role in how efficiently the processing of the cleavage sites occurs (11). Interestingly, for Semliki Forest virus (SFV), the timeliness of P1234 processing is linked to neurovirulence (12). The processing of the 1/2 site occurs relatively slowly for SFV6 (consensus clone of L10 strain of SFV [13]), which is neurovirulent in adult mice, while SFV A774 [a consensus clone of the A7(74) strain of SFV (12)] exhibits faster 1/2 site processing and is avirulent in adult mice. For both SFV clones, the 1/2 site processing efficiency is determined by amino acid residue 534 of the ns polyprotein (P4 position of 1/2 site) and residue 1052, which resides within the nsP2 protease region (amino acids [aa] 515 of nsP2) and is functionally linked to the P4 residue. Replacement of residues 534 and 1052 of SFV A774 with those of SFV6 results in decelerated P1234 processing and increased neurovirulence (12).
In addition to the intrinsic properties of the viral replicase, virus replication also depends on the availability of specific cellular proteins. Several studies have established the role of G3BP proteins as host factors for alphavirus infections. G3BP1 and -2 (here combinedly referred to as G3BP) are homologous multifunctional RNA-binding proteins, best characterized for their stress granule-nucleating function in response to environmental stress, including virus infection (14, 15). Whereas New World alphaviruses do not interact with G3BP, with Eastern equine encephalitis virus (EEEV) being the only reported exception to this (16), Old World alphaviruses recruit G3BP to viral replication complexes (17, 18). This recruitment is mediated by FGDF motifs, located in the hypervariable domain (HVD) of nsP3, which bind to the N-terminal NTF2-like domain of G3BP (19). This interaction is also conserved within the mosquito host, in which G3BP is referred to as Rasputin (20). For some Old World alphaviruses, such as SFV and chikungunya virus (CHIKV), two FGDF motifs can be found in the HVD of nsP3. In a 3-dimensional structure of the NTF2-like domain of G3BP1 (residues 1 to 139) in complex with a 25-residue peptide of SFV nsP3, each motif binds to a separate dimer of G3BP1 (21). The two FGDF motifs thereby interlink G3BP dimers into an nsP3-G3BP polycomplex. We have previously shown that the NTF2-like domain of G3BP1 promotes clustering of SFV4 (molecular clone derived from prototype SFV [22]) replication complexes, likely via the formation of these nsP3-G3BP polycomplexes, and is sufficient to facilitate efficient replication of SFV4, suggesting a predominantly structural proviral role of G3BP for SFV4 (17). In the absence of G3BP, SFV4 is attenuated but continues to produce progeny viruses to detectable levels. CHIKV, in contrast, has a strict requirement also for the C-terminal RGG domain of G3BP1 and is unable to replicate if either the NTF2-like or RGG domain is absent. We demonstrated that nsP3-G3BP1 complexes interact with translation initiation factors and 40S ribosomal subunits and that these factors are recruited to viral replication sites, which correlates with enhanced localized translational activity. These results showed that SFV4 and CHIKV, despite being closely related, differ remarkably in their requirement for G3BP. This motivated us to extend the investigation of the role of G3BP also for other Old World alphaviruses with the aim to better understand the seemingly versatile proviral activities of this host protein.
In the present study, we test several Old World alphaviruses for their sensitivity to G3BP deletion and show that the degree of sensitivity correlates with a particular residue within the viral replicase and its regulatory role in ns polyprotein processing. While we cannot exclude a role of G3BP during this processing, our data suggest its participation in viral RNA replication.
RESULTS
Alphaviruses differ in their sensitivity to G3BP deletion.
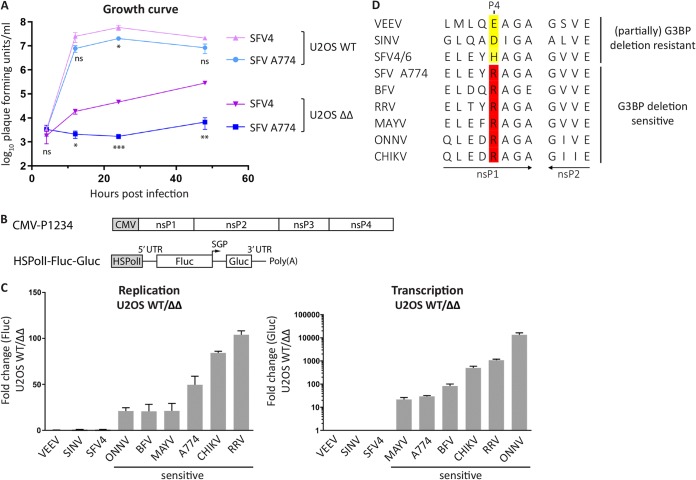
Initially, we aimed to determine if different SFV strains share a resistance to G3BP deletion as previously observed for SFV4 (17). We infected human U2OS wild-type (WT) cells or G3BP1/2 double-knockout cells (U2OS ΔΔ [15]) with either SFV4 or SFV A774 at a multiplicity of infection (MOI) of 0.1 and determined virus titers over a period of 48 hpi (Fig. 1A). In the presence of G3BP, i.e., in U2OS WT cells, the two viruses reached similar titers at every time point measured. However, in U2OS ΔΔ cells, the titers of the two viruses were significantly lower and significantly different from each other. While SFV4 was partially resistant to G3BP deletion, SFV A774 did not detectably replicate within 24 hpi, and only at 48 hpi was a very low virus titer detected.
FIG 1.
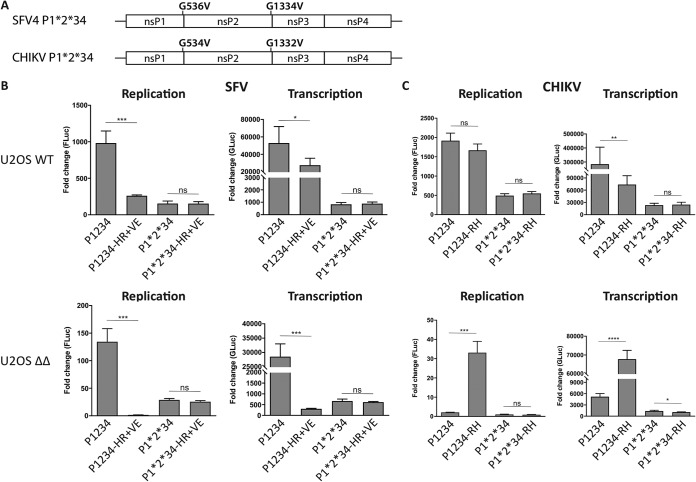
Sensitivity to G3BP deletion correlates with the presence of Arg residue at P4 position of 1/2 site. (A) U2OS WT or ΔΔ cells were infected with SFV4 or SFV A774 at an MOI of 0.1, and at indicated time points, viral titers were quantified by a plaque assay. Data are means of the results from three independent experiments. Error bars indicate the standard error of mean (SEM). SFV A774 virus titers were compared to SFV4 titers at each time point and cell line, and statistical differences were as follows: ns, P > 0.05; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001. (B) Schematic representation of trans-replication components for expression in human cells. The ns proteins are expressed from a cytomegalovirus (CMV) promoter-based plasmid. The RNA template, which is replicated in trans by the ns proteins, is expressed from a separate plasmid using the human RNA polymerase I promoter (HSPolI). Upon RNA replication, the RNA template resembles the viral RNA but encodes Fluc and Gluc reporters in place of the ns and structural proteins, respectively. SGP, subgenomic promoter; Fluc contains 77 amino acids of nsP1 on its N terminus that are not shown in the figure. UTR, untranslated region. (C) U2OS WT or ΔΔ cells were cotransfected with indicated replicase and corresponding template expression constructs. Fluc (replication) and Gluc (transcription) activities were measured 18 h posttransfection and normalized to signals obtained from control cells transfected with catalytic-inactive replicases and templates. Data are shown as the ratios of replication and transcription signals in U2OS WT to ΔΔ cells from three independent experiments. Means are shown, and error bars indicate the standard deviation (SD). (D) Sequence analysis of 1/2 site residues of replicases analyzed in panel C. Residues at the P4 position are highlighted.
The observation that SFV A774 was more sensitive than was SFV4 to G3BP deletion encouraged us to investigate the G3BP dependence of a panel of Old World alphaviruses, including Sindbis virus (SINV), SFV4, SFV A774, Barmah Forest virus (BFV), Ross River virus (RRV), Mayaro virus (MAYV), o’nyong’nyong virus (ONNV), and CHIKV; New World alphavirus Venezuelan equine encephalitis virus (VEEV) was used as a control. As these viruses have different host cell requirements and several of them are biosafety level 3 (BSL3) pathogens, we made use of a highly sensitive trans-replication system, in which replicase expression is uncoupled from viral RNA replication (23); among other things, this system allows one to circumvent differences in virus host cell ranges as well as biosafety issues. In a trans-replicase system, a reporter template, which is expressed from a separate plasmid via an RNA-polymerase I promoter, is replicated in trans and encodes firefly luciferase (Fluc) in place of the ns proteins and Gaussia luciferase (Gluc) in place of the structural proteins (Fig. 1B). Measurement of Fluc and Gluc activities is therefore representative of viral genomic RNA synthesis (replication) and subgenomic RNA synthesis (transcription), respectively (23). To examine the influence of G3BP on replicase activity of the above-mentioned alphaviruses, we transfected U2OS WT or U2OS ΔΔ cells with their respective replicases and corresponding template expression plasmids and measured replication and transcription activities 18 h posttransfection. Figure 1C shows the results for individual replicases as the signal in U2OS WT cells divided by the signal in U2OS ΔΔ cells, thus indicating the sensitivity of each replicase to G3BP deletion. As expected, neither the RNA replication nor transcription for New World alphavirus VEEV, which does not interact with G3BP (24), was affected by G3BP knockout. For most Old World alphaviruses, in contrast, replication and transcription were strongly reduced in G3BP knockout cells, despite some variation in the extent. The only exceptions were SINV and SFV4 trans-replicases, which were resistant to G3BP deletion in this system.
We searched for features that are shared among sensitive or resistant alphaviruses but different between these two groups. It came to our notice that all trans-replicases that were G3BP deletion sensitive share an arginine (Arg) residue at the P4 position of the 1/2 site of the P1234 (Fig. 1D). This residue is always altered in the resistant replicases. Consequently, we aimed to test if the identity of this residue influences G3BP sensitivity and if substitution of the P4 residue alters G3BP dependence.
SFV sensitivity to G3BP deletion is linked to the processing determinants of the 1/2 site.
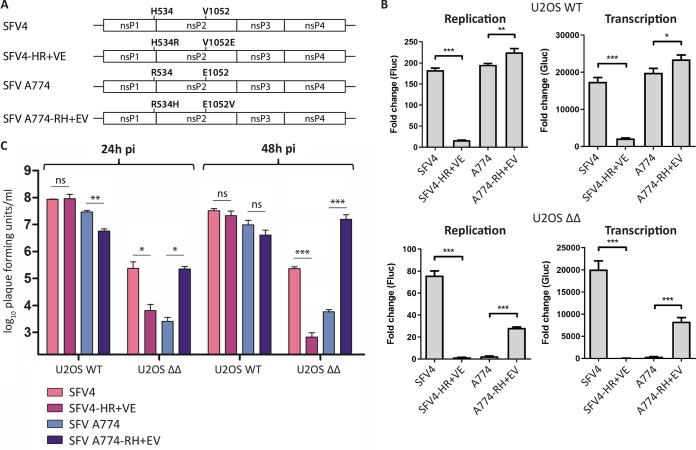
It is known that substitution of the P4 residue of the 1/2 site of SFV A774 and SFV6 (aa residue 534 of nsP1) influences processing efficiency at this site (12). This residue is functionally coupled to amino acid residue 1052 of the P1234 (aa 515 of nsP2). For SFV6, as well as for SFV4, residues 534 and 1052 of P1234 are His and Val, respectively (Fig. 2A); for SFV6, this combination is associated with slow 1/2 site processing and high virulence in adult mice. SFV A774 naturally contains an Arg residue at the P4 position of the 1/2 site and Glu at position 1052 of the P1234, which is associated with fast 1/2 site processing and an avirulent phenotype in adult mice. To determine if the identity of the P4 residue is functionally connected to G3BP sensitivity, we mutated residues 534 and 1052 of SFV4 and SFV A774, generating SFV4-HR+VE (His534Arg plus Val1052Glu) and SFV A774-RH+EV (Arg534His plus Glu1052Val), and tested the activities of the corresponding trans-replicases in the presence and absence of G3BP in comparison to their corresponding wild-type (WT) replicases (Fig. 2B). The activity of the SFV4-HR+VE trans-replicase was clearly impaired compared to that of SFV4 in U2OS WT cells, and in U2OS ΔΔ cells, replicase activity was reduced to the background level. Though an increase in sensitivity of the SFV4-HR+VE trans-replicase to G3BP deletion was evident, the results of statistical analysis did not allow us to make strong conclusions about the link of the P4 residue of SFV4 and sensitivity to G3BP deletion. On the other hand, in U2OS WT cells the trans-replicase of SFV A774-RH+EV was more active than its WT counterpart. Importantly, it showed a profound increase in replication and transcription activity in the absence of G3BP compared to that with SFV A774. Thus, resistance to G3BP deletion could to some extent be transferred from SFV4 to SFV A774 via the double substitution of Arg534His plus Glu1052Val.
FIG 2.
Substitution of the residues involved in processing of the 1/2 site alters the G3BP sensitivity of SFV trans-replicase and virus. (A) Schematic representation of substitutions examined in the context of (B) trans-replicase and (C) virus. (B) U2OS WT or ΔΔ cells were cotransfected with indicated SFV replicases and corresponding templates. Fluc (replication) and Gluc (transcription) activities were measured 20 h posttransfection, normalized to signals obtained from cells transfected with catalytic-inactive replicases, and plotted as the fold change. Data are means of the results from three independent experiments. Error bars indicate the standard deviation (SD). Statistical differences were as follows: *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001. (C) U2OS WT or ΔΔ cells were infected with indicated SFV viruses at an MOI of 0.1, and viral titers at 24 hpi and 48 hpi were quantified by plaque assay. Data are means of the results from three independent experiments. Error bars indicate the SEM. Statistical differences were as follows: ns, not significant; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001.
Next, we tested the effect of these substitutions in the context of recombinant viruses. We generated recombinant SFV4-HR+VE and SFV A774-RH+EV viruses, infected U2OS WT and ΔΔ cells, and measured the production of progeny viruses in comparison to the corresponding WT SFV4 and SFV A774 (Fig. 2C). All viruses replicated to high titers in the presence of G3BP, including the SFV4-HR+VE virus, which was severely attenuated in the trans-replicase system (Fig. 2B). This discrepancy is likely due to increased sensitivity in that system for mutations conferring polyprotein processing phenotypes (10). All viruses were attenuated in cells lacking G3BP, as expected. However, swapping of residues 534 and 1052 between SFV4 and SFV A774 had a substantial impact on the G3BP deletion sensitivity. The double substitution in SFV4-HR+VE resulted in increased G3BP sensitivity compared to that of WT SFV4, as well as a phenotype similar to that of A774. In contrast, SFV A774-RH+EV is more resistant to G3BP deletion than is WT SFV A774, and at 48 hpi, viral titers significantly exceeded those of SFV4, reaching similar levels as in U2OS WT cells. These results suggest that the identities of the SFV residues 534 and 1052 are a determining factor for G3BP sensitivity. More precisely, while the combination of His534 and Val1052 (in SFV4 and SFV A774-RH+EV) provides some resistance to G3BP deletion, Arg534 and Glu1052 (in SFV A774 and SFV4-HR+VE) are associated with increased sensitivity to G3BP deletion. It is noteworthy that the previously reported link between residues 534 and 1052 and the cleavage of the 1/2 site suggests a link between 1/2 site processing efficiency and G3BP dependence. Thus, an increase of 1/2 site processing efficiency appears to be associated with higher sensitivity to G3BP deletion, whereas a reduced processing efficiency may provide some resistance.
P4 Arg-to-His substitution rescues CHIKV replication in G3BP-deleted cells or if the nsP3-G3BP interaction is disrupted.
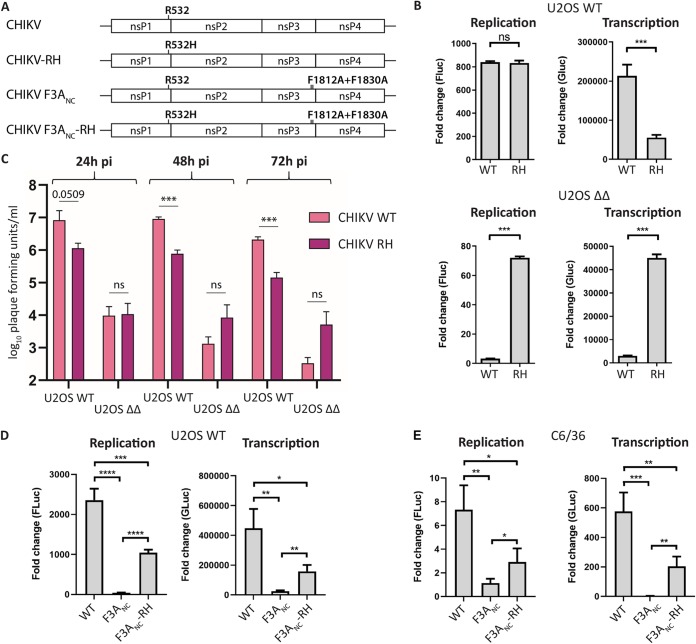
Our analysis has shown that all highly G3BP depletion-sensitive alphaviruses tested contain an Arg residue at the P4 position of the 1/2 site. In the case of SFV A774, substitution of this residue (together with residue 1052) partially overcame the G3BP dependence. To determine if this is also the case for other G3BP-dependent alphaviruses, we tested the P4 Arg-to-His substitution in the context of CHIKV. The SFV P1234 residues 534 and 1052 correspond to CHIKV P1234 residues 532 and 1050, respectively (Fig. 3A). For CHIKV, the single Arg532His substitution has been shown to have a similar effect on replicase activity in mammalian cells as the combination of Arg532His and Glu1050Val (25); the phenotypes caused by single and double mutations were also similar in context of virus, both in vitro and in vivo (26). This allowed us to exclude a role for Glu1050 and focus on the role of Arg532 (P4 residue) alone. In the presence of G3BP, the Arg532His substitution did not affect replication in the CHIKV trans-replication system, but transcriptional activity was significantly reduced, likely due to delayed polyprotein processing (Fig. 3B). Importantly, the Arg-to-His substitution rescued replicase activity in G3BP-deleted cells to a large extent, while replication and transcription activities were at the limit of detection for WT CHIKV. Similarly, infection of U2OS WT with CHIKV WT or CHIKV-RH showed that the mutant was impaired for replication in WT cells. In contrast, replication of CHIKV-RH was improved in U2OS ΔΔ cells, albeit at low levels (Fig. 3C). These results strongly support the observations made with SFV and suggest that the identity of the P4 residue of the 1/2 site is a determinant for G3BP dependence.
FIG 3.
P4 Arg-to-His substitution rescues CHIKV trans-replicase activity in the absence of the nsP3-G3BP interaction. (A) Schematic representation of amino acid substitutions in CHIKV R532H (RH), CHIKV F3ANC, and CHIKV F3ANC-RH. (B) U2OS WT or ΔΔ cells were cotransfected with WT CHIKV (WT) or CHIKV R532H (RH) replicase expression plasmids and corresponding templates. Fluc (replication) and Gluc (transcription) activities were measured 20 h posttransfection and normalized to signals obtained from control cells transfected with plasmids encoding catalytic-inactive replicases and templates and plotted as fold change. Data are means of the results from three independent experiments. Error bars indicate the standard deviation (SD). Statistical differences were as follows: ns, P > 0.05; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001. (C) U2OS WT or ΔΔ cells were infected with indicated CHIKV variants at an MOI of 0.1, and viral titers at 24 hpi, 48 hpi, and 72 hpi were quantified by plaque assay. Data are means of the results from three independent experiments. Error bars indicate the SEM. Statistical differences were as follows: ns, not significant; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001. (D and E) U2OS WT (D) or Aedes albopictus C6/36 (E) cells were cotransfected with indicated trans-replicases and templates and 18 h or 48 h posttransfection, respectively, analyzed as described for panel B. *, P ≤ 0.05, **, P ≤ 0.01, ***, P ≤ 0.001, ****, P ≤ 0.0001.
To furthermore confirm that the effect of the P4 substitution on G3BP dependence is indeed linked to the nsP3-G3BP interaction, we generated CHIKV trans-replicases with mutations in both G3BP-binding motifs of nsP3 (CHIKV F3ANC), which completely abolish the nsP3-G3BP interaction (21). This allowed us to test the role of the P4 residue in parental U2OS WT cells, in the presence and absence of G3BP binding. The results are in accordance with the observations made in U2OS ΔΔ cells, showing that in the absence of G3BP binding, CHIKV trans-replicase activity was severely impaired but could be partially rescued by the Arg-to-His substitution of the P4 residue (Fig. 3D).
This work clearly showed a link between the P4 residue of the 1/2 site and G3BP dependence using human U2OS cells. In the mosquito vector, the interaction of Old World alphavirus nsP3 and G3BP (named Rasputin) is conserved and crucial for CHIKV replication (20, 27). To investigate if the P4 residue of the 1/2 site can also modulate G3BP/Rasputin sensitivity in insect cells, we measured the activities of CHIKV trans-replicase constructs in Aedes albopictus C6/36 cells (Fig. 3E). The activity of CHIKV F3ANC trans-replicase was severely impaired compared to that of WT CHIKV. The defect of CHIKV F3ANC was, however, partially reversed by the Arg-to-His substitution of the P4 residue. Taken together, the results presented in Fig. 3 provide evidence that the severe defect of CHIKV replicase activity in the absence of G3BP/Rasputin binding in mammalian and mosquito cells can be partially overcome by a single amino acid substitution at the P4 position of the 1/2 site.
P4 Arg-to-His substitution allows for the production of infectious CHIKV particles in the absence of the nsP3-G3BP interaction.
Next, we aimed to validate that the P4 Arg-to-His substitution also rescues the production of infectious CHIKV virions in the absence of the nsP3-G3BP interaction. For this, we transfected BHK-21 cells with infectious cDNA (icDNA) clones of WT CHIKV, CHIK F3ANC, or CHIKV F3ANC-RH and measured virus rescue by an infectious center assay (ICA) (Fig. 4A). As expected, transfection of icDNA of WT CHIKV led to the appearance of a high number of plaques, whereas no plaques were detected for CHIKV F3ANC. Despite the severe defect caused by the F3ANC mutation, infectivity was rescued by the additional Arg-to-His substitution of the P4 residue (CHIKV F3ANC-RH), even though it remained ∼50-fold lower than that of WT CHIKV.
FIG 4.
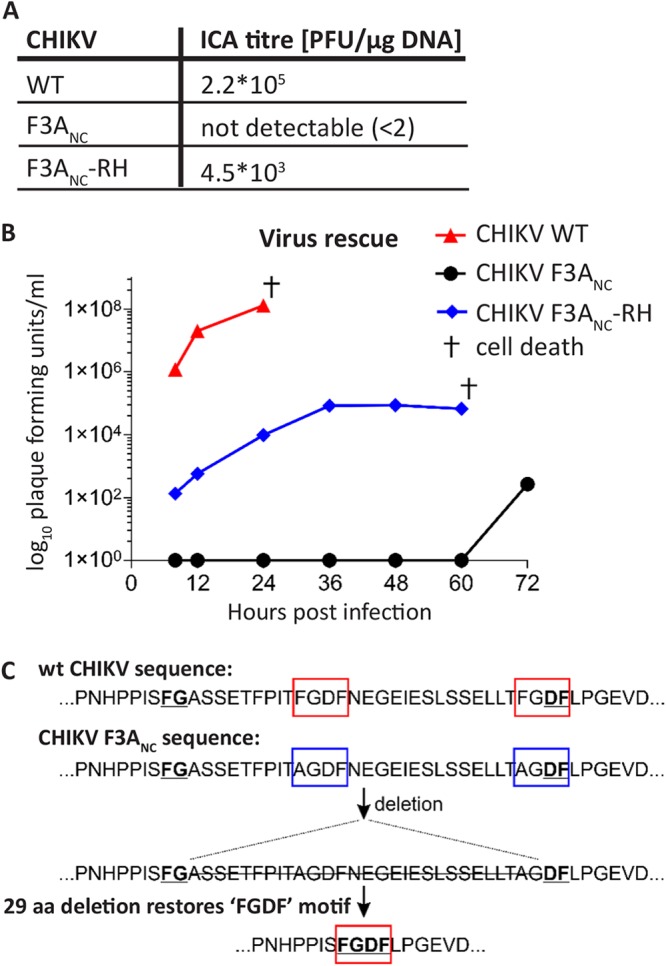
P4 Arg-to-His substitution rescues CHIKV replication in the absence of the nsP3-G3BP interaction. (A) BHK-21 cells were electroporated with infectious cDNA of WT CHIKV, the G3BP-binding mutant CHIKV F3ANC, and CHIKV F3ANC-RH, and infectivity was assessed by ICA. (B) BHK-21 cells were electroporated with infectious cDNA plasmid and viral titers quantified by plaque assay at the indicated time points. †, time point at which electroporated cells had died. (C) CHIKV F3ANC virus, rescued 72 h postelectroporation from panel B, was passaged 5 times and plaque purified, and 2 clones were subjected to sequence analysis. Both clones as well as pool of passage 5 viruses shared a mutation resulting in a restoration of an FGDF motif via a 29-amino-acid deletion, flanked by an N-terminal FG and a C-terminal DF motif (underlined).
We additionally assessed the rescue of viruses from these icDNA clones in BHK-21 cells by a plaque assay (Fig. 4B). Cells transfected with icDNA of WT CHIKV produced high titers as early as 8 h posttransfection. In contrast, CHIKV F3ANC did not produce infectious virus particles up to 60 h posttransfection. Again, this defect was partially overcome by the Arg532His substitution (CHIKV F3ANC-RH), which confirms the results of the ICA experiment. We conclude that the P4 Arg-to-His substitution provides CHIKV with partial resistance to the lack of nsP3-G3BP interaction and rescues the production of infectious virus.
Surprisingly, at 72 h posttransfection, we detected the onset of infectious virus production from CHIKV F3ANC icDNA-transfected cells (Fig. 4B), presumably due to reversion or adaptation. Therefore, we performed sequence analysis of the genomic region coding for the nsP1-nsP2 part of the ns polyprotein of 12 plaque-purified CHIKV F3ANC-derived virus isolates. Altogether, we found 30 different mutations in this region; however, none of them had an apparent connection to the 1/2 site or protease activity of nsP2. The obtained virus stock was then passaged five times, and complete ns polyprotein regions were sequenced for two plaque-purified viruses. Again, no mutation associated with the 1/2 site or nsP2 protease was found. Instead, both viruses had a mutation resulting in a 29-amino-acid deletion within the HVD of nsP3 (Fig. 4C). Sequencing of the region encoding the nsP3 HVD of the pool of passage-5 viruses revealed that this deletion was present in a large majority, if not all, virus genomes. Remarkably, the deleted region is flanked by an N-terminal FG sequence and a C-terminal DF sequence, resulting in restoration of an FGDF motif (Fig. 4C). It has been shown that a single intact FGDF/G3BP-binding motif is sufficient to allow for CHIKV replication, although it was delayed compared to WT virus with both motifs (21). Restoration of the FGDF motif is thus likely the cause for the sudden production of infectious virus, detected at 72 h posttransfection, highlighting the importance of the nsP3-G3BP interaction for CHIKV replication.
Stability of the ns polyprotein is reduced in cells lacking G3BP proteins but not by a mutation blocking the nsP3-G3BP interaction.
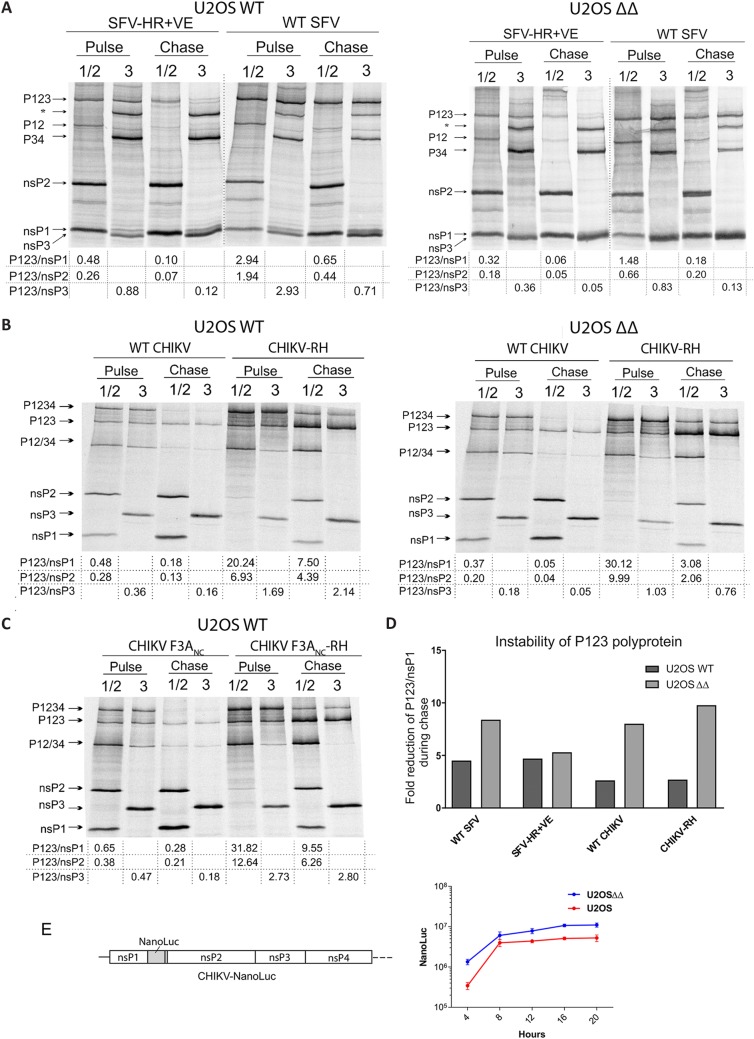
The identity of the P4 residue of the 1/2 site can influence the cleavage efficiency of the 1/2 and consequently also the 2/3 site (12). An Arg residue at this position, which, based on our results, correlates with high sensitivity to G3BP deletion, is associated with rapid cleavage of the 1/2 site. In contrast, a His residue in this position is associated with slow cleavage and correlates with partial resistance to G3BP deletion. This correlation urged us to investigate if binding of G3BP to nsP3 influences the processing of the ns polyprotein. For this, we transfected U2OS WT or U2OS ΔΔ cells with WT SFV4 or SFV4-HR+VE replicase and assessed the processing of the ns polyprotein by pulse-chase labeling (Fig. 5A). The P1234, its cleavage intermediates, and the mature nsPs were immunoprecipitated using nsP1 and nsP2 or nsP3 antibodies after a 15-min pulse labeling with [35S]methionine-cysteine or after an additional 45-min chase period in the presence of excess unlabeled methionine-cysteine and subsequently resolved by SDS-PAGE. The efficiency of P1234 processing and production of mature nsPs were assessed by analyzing the ratios of the amount of uncleaved P123 intermediate to the amount of mature nsPs in pulse samples. For technical reasons, immunoprecipitation of nsP4 could not be performed, and hence, the data do not provide conclusions about the processing at the 3/4 site. As expected, the P123/nsP ratios are consistently higher after the 15-min pulse than at the following 45-min chase. In accordance with previous observations (12), processing of SFV4-HR+VE P123 was always faster than was the processing of its WT counterpart. In a comparison of the processing of WT SFV4 ns polyprotein in U2OS WT and U2OS ΔΔ cells, the P123/nsP ratios were higher in U2OS WT cells, suggesting that G3BP proteins had a minor inhibitory effect on the processing of the SFV ns polyprotein (Fig. 5A). The P123/nsP1 and P123/nsP2 ratios of SFV4-HR+VE were, nonetheless, relatively similar in the U2OS WT and U2OS ΔΔ cells (Fig. 5A).
FIG 5.
Processing of ns polyprotein of SFV or CHIKV is not strongly influenced by G3BP. (A and B) U2OS WT and ΔΔ cells were transfected with plasmid expressing P1234 of WT SFV and SFV-HR+VE (A) or WT CHIKV and CHIKV-RH (B). (C) Additionally, U2OS WT cells were transfected with CHIKV F3ANC or CHIKV F3ANC-RH P1234 expression plasmids. At 8 h (SFV, A) or 12 h (CHIKV, B and C) posttransfection, cells were labeled with [35S]methionine-[35S]cysteine for 15 min (pulse), followed by a 45-min chase in the presence of excess unlabeled methionine and cysteine. Viral nsPs and their precursors in pulse-labeled or chased samples were then immunoprecipitated using antibodies against nsP1 and nsP2 or nsP3, separated by SDS-PAGE, and visualized using a Typhoon imager. Positions of mature nsPs and polyprotein precursors are indicated. Band intensities were quantified using the ImageQuant software and processing efficiencies calculated as ratios of P123 to nsP1, nsP2 or nsP3. Data are representative of at least two repeated experiments. *, nonspecific band. (D) Instability of P123 shown as a fold reduction of the P123/nsP1 ratio during chase. One representative image is shown. (E) Left, schematic representation of CHIKV-NanoLuc trans-replicase construct; right, comparison of CHIKV ns polyprotein expression in U2OS WT and ΔΔ cells. Cells were transfected with CHIKV-NanoLuc and incubated at 37°C for 4, 8, 12, 16, or 20 h. At these time points, cells were collected and lysed, and NanoLuc activities were determined. Data are shown as means of the results from three experiments. Error bars show the SD.
Next, we investigated if CHIKV ns polyprotein processing is influenced by G3BP proteins by repeating the above-described experiment in an analogous manner for CHIKV (Fig. 5B). Similar to the observations with SFV, P123/nsP ratios of WT CHIKV ns polyprotein were only slightly different between U2OS WT and U2OS ΔΔ cells after the pulse, and the difference became more prominent after subsequent chase period. The R532H substitution in CHIKV-RH strongly reduced processing efficiency of P123 both in U2OS WT and U2OS ΔΔ cells. Interestingly, the increased P123/nsP ratios of CHIKV-RH were observed to rather similar extents in pulsed samples, independent of whether G3BP was present or absent. Although the decrease in P123/nsP ratios in chased samples was more prominent in U2OS ΔΔ cells, we concluded that, similar to SFV, the effect of G3BP on the processing efficiency of P123 of CHIKV is detectable but is relatively minor.
Though the pulse-chase experiment supported the hypothesis that G3BP affects the processing of SFV and CHIKV ns polyproteins, the observed differences may not result directly from the lack of nsP3-G3BP interaction. We observed that the expression of nsPs was consistently higher in U2OS ΔΔ cells, possibly due to the inability of these cells to form stress granules. Increased nsP2 levels may, however, contribute to decreased stability of P123. Indeed, a comparison of P123/nsP ratios in pulse to those in chase revealed a trend that stability of P123 in U2OS ΔΔ cells was generally lower than in U2OS WT cells (Fig. 5D). To confirm that replicase protein expression levels were higher in U2OS ΔΔ than in WT U2OS cells, we constructed a CHIKV replicase containing the nanoluciferase (NanoLuc) coding region fused with nsP1 (Fig. 5E, left) in the position that was previously shown to tolerate large insertions (28) and measured its expression of NanoLuc at various times postinfection (Fig. 5E, right). The results confirmed sustained higher levels of nsP expression in the U2OS ΔΔ than in WT U2OS cells. This is consistent with a previous observation made for transient transfection of these cells lacking the ability to form stress granules (SGs) (15, 17). The introduction of RH, F3ANC, or F3ANC-RH mutations had no effect on the observed difference of expression in the U2OS ΔΔ compared to WT cells (data not shown). Based on this, we propose that the higher levels of nsP2 expressed in the U2OS ΔΔ cells were the reason for the generally lower stability of the ns polyprotein in these cells.
To address the issue of importance of nsP3-G3BP interaction for ns polyprotein processing in one and the same cell type, we performed experiment using the replicase of G3BP-binding mutants CHIKV F3ANC and CHIKV F3ANC-RH in parental U2OS WT cells. As expected, the RH mutation again prominently slowed down the processing of P123 (Fig. 5C). A comparison of the ns polyprotein processing of CHIKV F3ANC and CHIKV F3ANC-RH to that of WT CHIKV and CHIKV-RH (Fig. 5B, left), respectively, revealed that processing of CHIKV F3ANC and WT CHIKV replicases, as well as CHIKV F3ANC-RH and CHIKV-RH, occurred at very similar efficiencies. Thus, the ability of CHIKV nsP3 to bind G3BP had no detectable effect on ns polyprotein processing.
Altogether, the results of the pulse-chase experiments suggest that G3BP proteins do not have a direct prominent effect on the efficiency of the P123 processing of either SFV or CHIKV. Most importantly, the inability of CHIKV F3ANC to replicate in U2OS WT cells cannot be attributed to the increase in speed of P123 processing, as, if anything, processing of P123 of CHIKV F3ANC was somewhat slower than that of its WT counterpart (compare Fig. 5B and C).
P4 substitution does not influence sensitivity to G3BP deletion in the absence of ns polyprotein processing.
As the ns polyprotein processing efficiency remained relatively similar regardless of whether nsP3 interacted with G3BP, we considered the possibility that the P4 residue itself confers the altered sensitivity to G3BP deletion, independent of its effect on ns polyprotein processing. All alphaviruses harbor a universally conserved Gly residue at the P2 position of every processing site (11), and the Gly-to-Val substitution completely abolishes processing at the respective site (2, 29). In contrast to the 3/4 site, the processing of which is absolutely essential for the replicase activity of alphaviruses (3, 11), blocking of the cleavage of 1/2 and 2/3 sites results in infectious viruses (30) and trans-replicase constructs which are still capable of performing RNA replication and boosting of reporter gene expression (25, 31). Here, a combination of the Gly-to-Val substitutions of the P2 residues at the 1/2 and 2/3 sites was introduced into SFV4, SFV4-HR+VE, CHIKV, and CHIKV-RH ns polyproteins, and the resulting constructs are designated P1*2*34 (Fig. 6A). In both U2OS WT and U2OS ΔΔ cells, the replication and transcription activities of SFV4 P1*2*34 replicase were reduced compared to those with WT SFV4 P1234 replicase (Fig. 6B); this was expected since blocking P123 processing locks the replicase in a state which primarily produces negative-strand RNA, while the synthesis of positive-strand RNAs is defective (3). We then tested the effect of the SFV4-HR+VE substitutions in the context of active ns polyprotein processing (P1234-HR+VE) and when processing is blocked (P1*2*34-HR+VE). As seen before (Fig. 2B), enhanced ns polyprotein processing caused by the HR+VE substitutions attenuated SFV replicase activity and also increased the sensitivity of the SFV4 P1234-HR+VE trans-replicase to G3BP deletion (Fig. 6B). This effect was completely abolished for SFV P1*2*34-HR+VE trans-replicase, which in both U2OS WT and U2OS ΔΔ cells had activity similar to that of SFV P1*2*34 trans-replicase. Thus, as it could be expected, for SFV, the effect originating from blocking of P123 processing was dominant over the effect(s) caused by the P4 substitution, a finding that would suggest the importance of P123 processing for G3BP deletion resistance. On the other hand, blocking of P123 processing did not abolish the negative effect of G3BP deletion on SFV trans-replicase activity, as the levels of both replication and transcription by P1*2*34 replicases in U2OS ΔΔ cells were at least several fold lower than those in U2OS WT cells (Fig. 6B). This observation clearly indicates that G3BP acts, at least in part, through a mechanism that does not involve P123 processing.
FIG 6.
The effect of the P4 residue is abolished if P123 polyprotein processing is blocked. (A) Illustration of Gly-to-Val substitutions at the P2 positions which prevent cleavage at the respective cleavage sites. Corresponding replicases with mutations at the 1/2 and 2/3 sites are designated P1*2*34. (B and C) U2OS WT or ΔΔ cells were cotransfected with indicated SFV (B) or CHIKV (C) trans-replicases and corresponding templates. Fluc (replication) and Gluc (transcription) activities were measured at 18 h posttransfection, normalized to signals obtained from control cells transfected with catalytic-inactive replicases and corresponding templates, and plotted as the fold change. Data are the means of the results from three independent experiments. Error bars indicate the SD. Statistical differences are as follows: ns, P > 0.05; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001.
We proceeded to perform analogous experiments using CHIKV trans-replicase. In this experiment, we investigated the effect of CHIKV Arg532His substitution in the context of active ns polyprotein processing (P1234-RH) and when processing is blocked (P1*2*34-RH). In U2OS ΔΔ cells, CHIKV P1234 trans-replicase activity was strongly impaired but was partially rescued by the Arg532His substitution (P1234-RH) (Fig. 6C). If polyprotein processing was blocked (P1*2*34), the effect of the P4 mutation was again lost. Interestingly, however, for CHIKV, the replicase was locked into a G3BP deletion-sensitive phenotype. Thus, like CHIKV P1234, the CHIKV P1*2*34 trans-replicase was also severely affected by G3BP knockout, but unlike the P1234 replicase, its activity could not be rescued by the Arg532His substitution (P1*2*34-RH) (Fig. 6C).
In conclusion, the ability of the P4 residue to alter SFV or CHIKV replicase sensitivity to G3BP deletion is entirely dependent on its effect on 1/2 site processing efficiency. In the absence of ns polyprotein processing, P4 substitution has no effect on G3BP dependence. For reasons unknown, the CHIKV and SFV P1*2*34 replicases maintained the phenotype characteristic to the corresponding WT replicase, where the G3BP deletion is sensitive for CHIKV and partly G3BP deletion resistant for SFV. The fact that the G3BP deletion-sensitive phenotype of CHIKV was maintained also in the complete absence of P123 processing strongly supports our conclusion that at least for CHIKV, G3BP does not act by regulating the efficiency of P123 processing.
dsRNA synthesis is defective in G3BP-deleted cells.
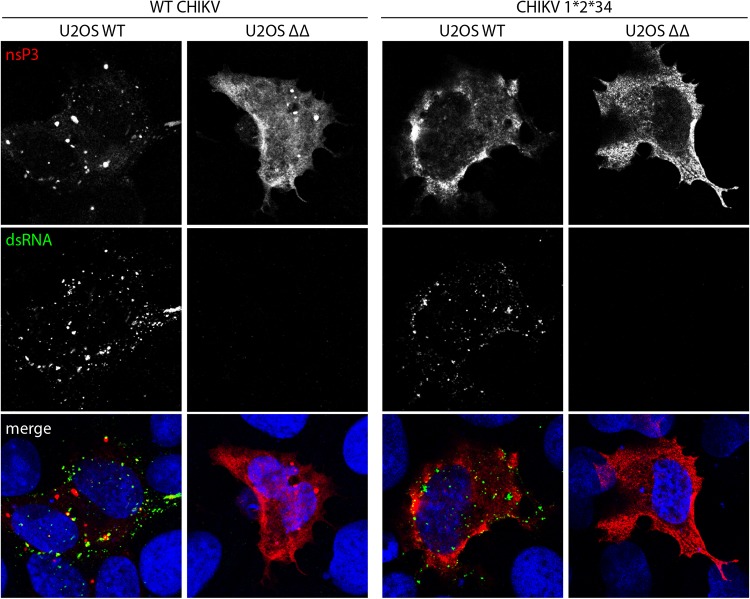
The CHIKV trans-replication data show that reporter expression is severely impaired in cells lacking G3BP, or if the nsP3-G3BP interaction is disrupted, despite the presence of replicase proteins, expressed from a separate plasmid. This suggests a possible defect in viral RNA replication/transcription that could be partially rescued by reducing 1/2 processing efficiency but not by complete block of P123 processing. However, the measurement of reporter activities does not reveal if the defect is at the step of negative-strand or positive-strand RNA synthesis. In order to address this, we transfected U2OS WT or U2OS ΔΔ cells with CHIKV replicase and template expression plasmids and stained cells for double-stranded RNA (dsRNA) and CHIKV nsP3 (Fig. 7). In U2OS WT cells, expression of the CHIKV trans-replicase components was associated with punctate nsP3 staining, often colocalizing with strong dsRNA signals, indicating active RNA replication. In contrast, transfected U2OS ΔΔ cells gave diffuse nsP3 staining, as expected from previous work (17, 32), and did not produce any detectable levels of dsRNA, suggesting a lack of negative-strand RNA synthesis. The production of negative-strand RNA is the first step of RNA replication and is performed by the P123+nsP4 replicase. By using the cleavage-deficient mutant CHIKV P1*2*34, which only allows cleavage at the 3/4 site, we were able to lock the replicase to predominantly produce negative-strand RNA. This replicase also produces strong dsRNA signals in U2OS WT cells, but no dsRNA signal was detected in U2OS ΔΔ cells. This finding is consistent with the lack of induction of reporter expression (Fig. 6C). In the context of the P1*2*34 replicase, nsP3 signals were diffuse, and a large amount was found to accumulate at the plasma membrane in both cell types, likely due to the fact that nsP3 cannot be released from the replicase complex and remains associated with the membrane-anchored P123 polyprotein.
FIG 7.
CHIKV trans-replicase fails to produce dsRNA in the absence of G3BP. U2OS WT or ΔΔ cells were cotransfected with WT CHIKV or CHIKV P1*2*34 trans-replicase plasmids. At 24 h posttransfection, cells were fixed and stained for nsP3 (red) and double-stranded RNA (dsRNA) (green). Nuclei are shown in blue. Representative images from three independent experiments are shown.
To further analyze the replication and transcription of CHIKV variants in U2OS WT or U2OS ΔΔ cells, we mock transfected or transfected cells with HSPolI-Fluc-Gluc template plasmid and either CHIKV, CHIKV-RH, CHIKV-P1*2*34, CHIKV-P1*2*34-RH, or CHIKV-GAA replicase plasmid. Total RNA was extracted and first analyzed for the presence of genomic and subgenomic RNAs by Northern blotting (Fig. 8A). The results indicate that WT CHIKV and CHIKV-RH replicate very efficiently in WT U2OS cells, and that both CHIKV P1*2*34 and CHIKV P1*2*34-RH are replication competent but at lower levels than with the fully processed replicases. In U2OS ΔΔ cells, the levels of positive-strand RNAs were much lower and could be detected only using longer exposures. The template RNA, synthesized by cellular RNA polymerase I, is also detectable under these conditions (Fig. 8A, right, line “GAA”) masking any genomic RNAs possibly made by active replicases. For these reasons, the only replicase-generated RNA detected in U2OS ΔΔ cells is subgenomic RNA generated by CHIKV-RH; these data are in perfect agreement with data from Gluc reporter expression (Fig. 6C). When blots were probed for negative-strand RNA (Fig. 8B), all replicases produced negative-strand RNA in WT U2OS cells, with higher signals seen in CHIKV P1*2*34 replicases, as expected. However, in U2OS ΔΔ cells, no negative-strand RNA was detected for any replicase. These results are consistent with those shown in Fig. 7 and suggest that G3BP plays a role at or preceding the CHIKV negative-strand RNA synthesis.
FIG 8.
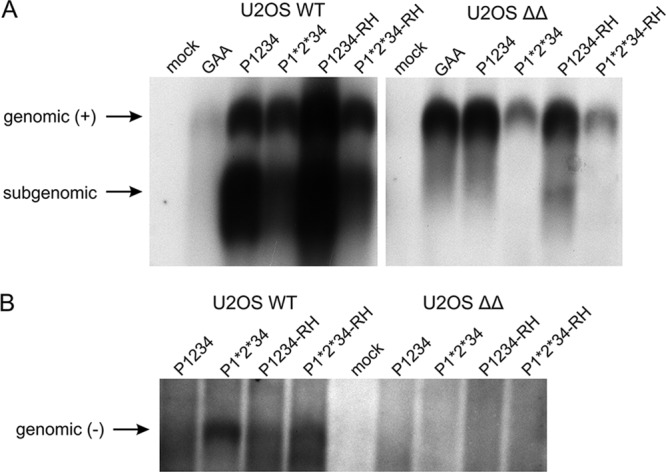
Northern blot analysis of CHIKV positive-strand (+) and negative-strand (−) RNAs in trans-replication system. U2OS WT or ΔΔ cells were cotransfected with HSPolI-Fluc-Gluc and CHIKV, CHIKV-P1*2*34, CHIKV-RH, CHIKV-P1*2*34-RH, or CHIKV-GAA; control cells were mock transfected. Samples were collected at 18 h posttransfection. (A) RNA was analyzed by Northern blotting using a probe complementary to the Gluc reporter gene to detect positive strands. Longer exposure was required for U2OS ΔΔ cells, and that gel section is thus presented as a separate panel. (B) RNA was analyzed using a probe corresponding to the Fluc reporter gene to detect negative strands. In both panels, “genomic” designates the full-length RNA of either strand, while “subgenomic” designates RNA synthesized by CHIKV replicase using the subgenomic promoter. The experiment was performed twice, with similar results, and data from one experiment are shown.
G3BP deletion or the P4 Arg-to-His substitution does not facilitate the switch from CHIKV RNA translation to RNA replication.
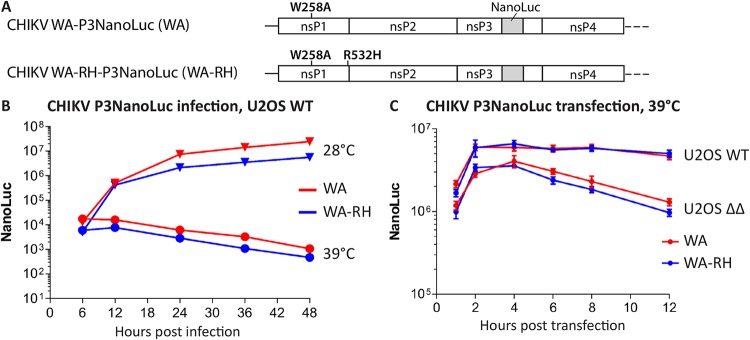
Our observations are in agreement with previous work showing that knockdown of G3BP is accompanied by inefficient production of viral negative-strand RNA (33). In that work, it was proposed that G3BP might be involved in the switch of incoming viral RNA translation to RNA replication. The inability of the viral replicase to stop translation and engage with the viral RNA template would indeed prevent the initiation of viral RNA replication and explain the observed phenotype. We aimed to investigate this hypothesis and for that purpose made use of CHIKV harboring the W258A (WA) substitution (25, 34). This substitution causes a temperature-dependent effect such that RNA replication is severely impaired at 39°C but is very similar to WT CHIKV at 28°C. Therefore, at the restrictive temperature, viral proteins are expressed exclusively from the incoming viral RNA. To assess viral protein production, we tested the effect of R532H substitution in the context of a recombinant CHIKV-WA-P3NanoLuc, encoding the NanoLuc fused to the C-terminal domain of nsP3 (34) (Fig. 9A). At the permissive temperature (28°C), active replication of CHIKV-WA-P3NanoLuc leads to increasing production of viral ns proteins in infected U2OS WT cells, which is reflected by the increasing P3NanoLuc signal (Fig. 9B). At the restrictive temperature (39°C), RNA replication does not occur, and the P3NanoLuc levels, expressed only from the incoming viral RNA, cease over time due to degradation of the reporter protein and template RNA. CHIKV-WA-RH-P3NanoLuc, containing the R532H substitution, which partially rescues CHIKV replication in the absence of G3BP, had the same phenotype at either temperature. This implies that the R532H substitution does not prolong the time for translation of the ns polyprotein and, accordingly, does not alter the switch from RNA translation to RNA replication in the presence of G3BP.
FIG 9.
G3BP deletion or the P4 Arg-to-His substitution does not prolong or increase ns protein translation. (A) Schematic representation of modifications present in the ns polyprotein region of the infectious clones CHIKV-WA-P3NanoLuc (WA) or CHIKV-WA-RH-P3NanoLuc (WA-RH). The NanoLuc luciferase sequence was inserted between the codons of amino acid residues 1716 and 1717. (B) U2OS WT cells, cultured at 28°C or 39°C, were infected with CHIKV-WA or CHIKV-WA-RH at an MOI of 1, and reporter activity was measured from lysates prepared at indicated time points. (C) U2OS WT or ΔΔ cells, cultured at 39°C, were transfected with in vitro-transcribed infectious RNA of CHIKV-WA or CHIKV-WA-RH. Reporter activity was measured from lysates prepared at the indicated time points. Data are the means of the results from three independent experiments. Error bars indicate the SD.
We speculated that if G3BP is involved in the switch from viral RNA translation to RNA replication, the expression of P3NanoLuc would be prolonged in U2OS ΔΔ cells compared to that in U2OS WT cells. We transfected U2OS WT and U2OS ΔΔ cells with in vitro-transcribed RNA of CHIKV-WA-P3NanoLuc or CHIKV-WA-RH-P3NanoLuc and compared early P3NanoLuc signals at the restrictive temperature only (Fig. 9C). Again, the two virus RNAs, with and without the R532H substitution, behaved very similarly in the two cell lines, ruling out the possibility that the partial rescue of G3BP dependence by the R532H substitution was due to a change in the switch from translation to replication. There was, however, a clear difference between the two cell lines in that P3NanoLuc levels were noticeably higher in U2OS WT cells than in U2OS ΔΔ cells. Since we previously reported that nsP3-G3BP complexes interact with 40S ribosomal subunits and translation initiation factors (17), this might imply a role of G3BP in promoting translation of the ns polyprotein from the viral genome. Alternatively, this might indicate that P3NanoLuc protein is more stable when bound to G3BP, i.e., in U2OS WT cells. Clearly, neither excessive nor prolonged production of P3NanoLuc was observed in U2OS ΔΔ cells. Thus, the results of these experiments allow us to conclude that G3BP is unlikely to facilitate the switch from CHIKV genomic RNA translation to RNA replication.
DISCUSSION
The role of the host protein G3BP as an essential proviral factor for CHIKV has been repeatedly reported (17, 20, 21, 24, 33) and is furthermore confirmed by the present study. The G3BP-binding FGDF motifs, present in the HVD of nsP3, are conserved among Old World alphaviruses (19), and here, we demonstrate that the interaction with G3BP is indeed crucial for the replication of many representative Old World alphaviruses. Our investigation also shows that Old World alphaviruses are affected quite differently by G3BP deletion, as can be seen for different strains of SFV, SFV4, and SFV A774 (Fig. 1C). Despite the common nsP3-G3BP interaction, these observations highlight major differences among these viruses and suggest versatile proviral roles for G3BP, in line with our previous work (17).
A remarkable observation in our work is that a determinant for ns polyprotein processing efficiency has an influence on G3BP sensitivity. An Arg residue at the P4 position of the 1/2 site confers fast processing at this site and is associated with increased sensitivity to G3BP deletion. On the other hand, a His residue at this position confers slow processing at the 1/2 site and is associated with decreased G3BP sensitivity. We furthermore confirm that the link of the CHIKV P4 residue to G3BP sensitivity is indeed dependent on the processing at the 1/2 site, as the effect of substitution of this residue is abolished if the processing of P123 is blocked (Fig. 6). Naturally, we speculated that this residue might mimic the function of G3BP. Indeed, the pulse-chase experiment revealed that in U2OS ΔΔ cells lacking G3BP, the stability of the CHIKV and SFV ns polyproteins is somewhat reduced (Fig. 5D). However, this effect may simply originate from higher ns polyprotein expression levels in U2OS ΔΔ cells resulting in increased levels of nsP2 protease (Fig. 5E). Consistent with this, no difference in the processing of ns polyproteins of CHIKV and CHIKV-F3ANC was detected in U2OS WT cells (Fig. 5B and C).
How does the Arg-to-His substitution of the P4 residue in cleavable P1234 overcome the strong defect caused by the abolished nsP3-G3BP interaction as seen for CHIKV? Since a His residue at the P4 position is associated with slow processing, it should lead to increased stability of the P123+nsP4 negative-strand replicase. However, since CHIKV has an Arg residue at P4, ns polyprotein processing is more efficient at the 1/2 site, and negative-strand replicase has a shorter half-life, which restricts the production of negative-strand RNA. Our data also clearly point toward a role of G3BP during negative-strand RNA synthesis (Fig. 7 and 8). In the absence of G3BP, this process might be inefficient, and an extended half-life of the P123-nsP4 replicase could increase the probability of such an inefficient process to occur. Previous work confirms that for efficient viral replication, the timeliness of processing at the 1/2 site is critical (10). Sequences at the 1/2 site that result in too-efficient cleavage by the nsP2 protease reduce infectivity. This defect can be rescued through adaptive changes in the nsP2 protease domain or the cleavage site itself, which normalize the cleavage rate at the 1/2 site. In a similar manner, a hyperactive nsP2 protease causes growth defects which can be rescued by compensatory changes in the cleavage sequence of the 1/2 site that are less efficiently recognized by nsP2.
However, preventing further processing of the CHIKV P123-nsP4 trans-replicase (P1*2*34) is not sufficient to rescue replicase activity in the absence of G3BP (Fig. 6C). In U2OS WT cells, the P1*2*34 replicase activity is attenuated compared to the WT replicase, but replication and transcription remain detectable, and synthesis of both negative- and positive-strand RNA occurs (Fig. 8). For SFV and SINV, the combination of blocked 1/2 and 2/3 sites still allows for the production of infectious progeny at 30°C (3, 5). But, despite being locked in its predominantly negative-strand-RNA-producing activity, the CHIKV P1*2*34 replicase fails to show replicase activity in the absence of G3BP, revealing that blocking of P123 processing does not have the same effect as the Arg-to-His substitution of the P4 residue. Furthermore, as the effect of Arg-to-His substitution is completely lost in the context of P1*2*34 replicase, it is also unlikely that the P4 residue itself, independent of its influence on the processing efficiency of the 1/2 site, can function to rescue replication in the absence of G3BP. Interestingly, though the Arg-to-His substitution rescues replication of CHIKV that is unable to bind G3BP, no virus rescued from icDNA of CHIKV-F3ANC has this kind of adaptive mutation. Most likely, these observations indicate the involvement of additional host component(s) that may be different even for related alphaviruses in the formation/functioning of the replicase complex. Unfortunately, information about alphavirus replicase complex biogenesis, its exact composition, as well as structural information about the replicase is still insufficiently available. The 1/2 site must be accessible for the nsP2 protease active site, but it is not known which conformational changes occur before, during, and after the cleavage. The P4 residue could influence these structural rearrangements or stabilize transition states which could alter specific functions of the replicase. Interestingly, several types of cancer are also associated with a prominent Arg-to-His mutation signature, which demonstrates that this particular mutation is frequently associated with altered protein function and/or interactions (35). Currently, we do not know if other P4 substitutions have similar effects on G3BP sensitivity as seen for Arg and His. Furthermore, the P4 residue and/or ns polyprotein processing have critical roles for several biological functions of alphaviruses. For SFV isolates, the P4 His-residue and slow processing are required for neurovirulence, and the P4 Arg-to His substitution results in a more virulent phenotype (12). Intriguingly, the situation is reversed for CHIKV, where the substitution of a natural Arg residue with His attenuates virus in vivo (26). Whether or not the interaction with G3BP or different requirements for G3BP domains (17) have any direct role for the different phenotypes of SFV strains and CHIKV mutants remains currently unknown.
Our previous work has shown that CHIKV strictly depends on the C-terminal RGG domain of G3BP, and we proposed a functional role of this domain in the recruitment of the translation initiation machinery to ensure efficient synthesis of viral proteins (17). In the present work, we applied a trans-replicase system, in which synthesis of the viral replicase proteins is guaranteed via the expression of a replication-independent plasmid. Nevertheless, replication of CHIKV was greatly diminished also in this system if the nsP3-G3BP interaction was abolished, despite the availability of replicases (Fig. 3B). This clearly indicates that G3BP has a critical proviral function, independent of translation of viral proteins. It has been suggested that G3BP acts postentry and negatively affects viral RNA synthesis if absent (33). Our experiments with temperature-sensitive CHIKV-WA suggest that it is unlikely that G3BP is involved in the switch from RNA translation to RNA replication or that the Arg-to-His substitution affects the time during which translation of ns proteins takes place (Fig. 9). Furthermore, during CHIKV infection, the replicase must be able to engage the template in cis, whereas in the trans-replication system, the template is provided in trans, but despite these differences, both systems are severely affected by the absence of G3BP (Fig. 3B and 4A and B). Together, these observations point toward a role of G3BP in the formation and/or functional activation of the early viral replicase and, hence, in negative-strand RNA synthesis. Substitution of the P4 residue of the 1/2 site overcomes the G3BP dependence, but only to some extent. In fact, in none of our experiments did the P4 substitution fully compensate for the loss of the nsP3-G3BP interaction (Fig. 3, 4, 6, and 8). It is likely, therefore, that G3BP exerts several proviral functions at different stages of the viral life cycle and that the various Old World alphaviruses depend on these functions to different extents.
In our previous work, we found that SFV tolerates G3BP knockout to some extent, whereas CHIKV is nonviable in its absence (17). The application of the trans-replication system in the present study validates this observation. In the context of the proposed role of G3BP in viral RNA replication, the question arises as to which RNA replication-related differences might confer the discrepancy of these viruses regarding G3BP sensitivity. A noteworthy difference is that the SFV replicase is capable of transcribing nonviral host cell RNA (36), a characteristic which is not shared by the CHIKV replicase (A. Merits, A. Utt, and S. Saul, unpublished data). This indicates that CHIKV has more stringent requirements for replicase activity than does SFV, likely caused by differences in the RdRp activity itself or other replicase activities which ensure that the RNA template is maintained in a state accessible for the RdRp to initiate RNA replication.
Analogous to mammalian cells, the interaction of nsP3 with Rasputin, the mosquito homolog of G3BP, is crucial for the propagation of CHIKV in mosquito cells (27). This suggests that mammalian G3BP and mosquito Rasputin are required for similar functions during the replication cycle of CHIKV. Our data support this hypothesis by showing that the CHIKV R532H substitution results in reduced G3BP sensitivity in both mammalian and mosquito cells. Future work addressing the role of Rasputin’s RGG domain and the influence of Rasputin on viral RNA replication will help to clarify which proviral functions are shared between G3BP and Rasputin. With EEEV being the only reported exception (16), New World alphaviruses do not interact with G3BP in mammalian cells and replicate efficiently in its absence (24). Consequently, it is likely that Rasputin is dispensable for the efficient replication of New World alphaviruses in the mosquito vector.
We have shown that several alphaviruses rely on G3BP as a proviral host factor but with various levels of dependence, presumably at different stages of the life cycle and targeting different domains. This suggests that the interaction with G3BP goes back to a common viral ancestor from which the functional role of G3BP diverged over time. Interestingly, G3BP-like proteins are also found in plants, such as Arabidopsis thaliana, which are bound by FGDF-like motifs present in the nuclear shuttle proteins of the begomovirus Abutilon mosaic virus and pea necrotic yellow dwarf virus (37). Consistent with this hypothesis, phylogenetic analysis suggests that the genus Alphavirus originates from an insect-borne plant virus (38).
The present work underlines the importance of regulated polyprotein processing for the Old World alphavirus replicase activity and suggests a critical role for the host factor G3BP for replicase functionality. However, our data also prompt further investigation, as the regulatory mechanisms that determine replicase activity and its interaction with viral RNA are still insufficiently understood.
MATERIALS AND METHODS
Cell lines and cell culture.
Human osteosarcoma (U2OS) cells (ATCC HTB-96) were kept in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 100 U/ml penicillin, and 100 μg/ml streptomycin. U2OS-derived double-null ΔΔG3BP1/2 knockout (KO) cells (15) were maintained in the same medium as wild-type U2OS. Baby hamster kidney (BHK-21) cells (ATCC CCL-10) were maintained in Glasgow’s modified Eagle’s medium (GMEM) supplemented with 10% FBS, 10% tryptose phosphate broth, 20 mM HEPES, 1 mM l-glutamine, 100 U/ml penicillin, and 0.1 mg/ml streptomycin. U2OS and BHK-21 cells were cultured at 37°C in 5% CO2. Aedes albopictus-derived C6/36 cells were maintained in Eagle’s minimum essential medium supplemented with 10% FBS, 100 U/ml penicillin, and 0.1 mg/ml streptomycin and cultured at 28°C.
Construction of trans-replicase plasmids.
Plasmids CMV-P1234, CMV-P1234GAA, CMV-P1RH234, and CMV-P1GV2GV34 expressing in mammalian cells CHIKV WT P1234, P1234 with GAA, P1234 with R532H, or P1234 with G534V and G1332V mutations, respectively, have been previously described (25, 28). To avoid confusion with plasmids expressing replicases of other alphaviruses, they are here designated CHIKV, CHIKV-GAA, CHIKV-RH, and CHIKV-P1*2*34, respectively. Similarly, plasmids Ubi-P1234, Ubi-P1234GAA, and Ubi-P1RH234 for the expression of CHIKV polyproteins in Aedes cells have been described (25). Plasmids for expression of replicases of SFV4, SFV A774, SINV, RRV, and MAYV have the same design as CMV-P1234 and were assembled from synthetic DNAs (GenScript, USA) and restriction fragments of corresponding infectious cDNA clones (12, 22, 39–41). Plasmids for the expression of replicases of ONNV, BFV, and VEEV were assembled from synthetic DNA fragments (GenScript). For each replicase-expressing plasmid, a negative control, harboring a GDD-to-GAA mutation in the active site of nsP4 polymerase, was obtained using PCR-based mutagenesis and subcloning. Similarly, the H534R and V1052E or G536V and G1334V mutations were introduced into the replicase of SFV4; the R534H and E1052V mutations were introduced into the replicase of SFV A774, and the mutation F3ANC was introduced into the replicase of CHIKV. In plasmids expressing CHIKV F3ANC-RH replicase, the F3ANC and RH mutations were combined. In a plasmid encoding CHIKV P1*2*34-RH cleavage site-inactivating mutations were combined with the R532H mutation, and in a plasmid encoding an SFV4 P1*2*34-HR+VE cleavage site-inactivating mutations were combined with the H534R and V1052E mutations. To obtain plasmid for quantification of CHIKV P1234 expression, a sequence encoding an enhanced green fluorescent protein (EGFP) marker inserted after codon 516 of nsP1 in CMV-P1E234-C (28) was replaced with sequence encoding nanoluciferase (NanoLuc), and the resulting plasmid was designated CHIKV-NanoLuc. The DNA fragment containing the inserted sequence was then transferred to CHIKV-RH, CHIKV F3ANC, and CHIKV F3ANC-RH, resulting in CHIKV-NanoLuc-RH, CHIKV-NanoLuc-F3ANC, and CHIKV-NanoLuc-F3ANC-RH, respectively. All of these constructs were obtained using standard cloning techniques. Cellular RNA polymerase I promoter-based plasmids for the production of a replication-competent RNA template of CHIKV in mammalian (HSPolI-Fluc-Gluc) and Aedes albopictus (AlbPolI-Fluc-Gluc) cells have also been described (23). Plasmids for the expression of replication-competent RNA templates of SFV, ONNV, SINV, RRV, BFV, and VEEV have the same design as HSPolI-Fluc-Gluc and were assembled from synthetic DNA fragments (GenScript). The sequences of all plasmids were verified using Sanger sequencing.
Viruses, their mutants, and virus infections.
SFV was rescued by transfection of BHK-21 cells with the infectious plasmid pCMV-SFV4 (42). The generation of pCMV-A774 and infectious cDNA clones for SFV4-HR+VE and SFV A774-RH+EV is described in reference 12. CHIKV was rescued from the pCMV-CHIKV-ICRES (CHIKV LR2006-OPY1) infectious clone. The generation of pCMV-CHIKV F3ANC and the icDNA clone for CHIKV-RH, a virus with an R532H mutation, are described in references 21 and 26, respectively. pCMV-CHIKV F3ANC-R532H (RH) was constructed from pCMV-CHIKV F3ANC and icDNA of CHIKV-RH using standard cloning techniques. The construction of icDNA of CHIKV-nsP1W258A-NanoLuc (here designated CHIKV-WA) has been previously described (25, 34). The NanoLuc sequence was inserted between the codons of amino acid residues 1716 and 1717 of CHIKV P1234. To obtain CHIKV-WA-RH, the R532H mutation was transferred from the icDNA clone of CHIKV-RH to that of CHIKV-nsP1W258A-NanoLuc using standard cloning techniques.
Virus titers for infection experiments on U2OS-based cell lines were determined on wild-type U2OS cells by a plaque assay. The exception was CHIKV-RH that did not form visible plaques on U2OS cells and whose titer was therefore determined using BHK-21 cells. For infections, cell monolayers were washed with phosphate-buffered saline (PBS), and virus was added in infection medium (DMEM supplemented with 0.2% bovine serum albumin (BSA), 2 mM l-glutamine, and 20 mM HEPES) with periodic shaking for 1 h at 37°C. Infectious media were then removed and cells washed with PBS before adding prewarmed complete medium. For virus growth curve experiments, cells were grown in six-well plates to ∼90% confluence, infected as described above, and overlaid with 2 ml complete medium, and samples of the supernatant were taken at different time points pi. Virus titers were then determined on BHK-21 cells by a plaque assay.
Virus titration by plaque assay.
A 10-fold serial dilution of virus suspension was prepared and used to infect monolayers of BHK-21 cells for 1 h at 37°C. Cells were washed with PBS, kept in BHK-21 medium supplemented with 0.8% (wt/vol) agarose, and incubated at 37°C. After 36 h, 10% (vol/vol) formaldehyde in PBS was added and incubated for at least 4 h at room temperature, and plaques were revealed by crystal violet staining.
Mutant virus propagation, plaque purification, RNA isolation, and sequencing.
Rescued CHIKV F3ANC virus was plaque purified directly or after 5 passages at a low MOI in BHK-21 cells. Plaque purification and a search for adaptive mutations were performed as described previously (43). Briefly, the stock of CHIKV F3ANC was used to infect confluent monolayers of BHK-21 cells. Following infection, the cells were overlaid with GMEM containing 2% fetal calf serum (FCS) and 0.9% Bacto agar and incubated at 37°C for 48 h. After, a neutral red dye in 1% agar was added, and the plates were incubated for a further 2 h. The plaques were cut out, transferred to a BHK-21 monolayer on 24-well tissue culture plates, and incubated in GMEM containing 2% FCS. Viral stocks were collected following the observation of cytopathic effects.
The presence of the adaptive mutations was verified through reverse transcription-PCR (RT-PCR) and sequencing. For these procedures, viral RNA was extracted from each stock using the RNeasy minikit (Qiagen) and then reverse transcribed using the first-strand cDNA synthesis kit (Thermo Fisher Scientific). A set of PCR fragments covering either the nsP1-nsP2 region or complete ns polyprotein region of CHIKV were obtained and sequenced. To analyze the pooled sequences, the plaque purification was omitted, and viral RNA, isolated from passage-5 stock, was used as the template for RT-PCR amplifying the region encoding the HVD of nsP3. The obtained PCR fragment was sequenced using Sanger sequencing.
Trans-replication assay.
Wild-type U2OS or U2OS ΔΔG3BP1/2 cells were grown on six-well plates to 90% confluence and cotransfected with mixtures of 1 μg of plasmid encoding replicase polyprotein and 1 μg of plasmid for expression of the corresponding RNA template using the Lipofectamine LTX reagent, according to the manufacturer’s protocols (Invitrogen). At 18 or 20 h posttransfection, cells were lysed with passive lysis buffer (Promega), and firefly luciferase (Fluc) and Gaussia luciferase (Gluc) activities were analyzed using the luciferase assay system (Promega) and native coelenterazine (Promega), respectively. Trans-replication in C6/36 cells was performed as described previously (23). Luciferase signals were measured using the GloMax SIS luminometer (Promega) or FLUOstar Omega microplate reader (BMG Labtech).
Infectious center assay.
ICA was performed as previously described (44), except that 5 μg of purified CHIKV icDNA plasmid was used per electroporation. In brief, 10-fold dilutions of electroporated BHK-21 cells were seeded in six-well plates containing subconfluent naive BHK-21 cells. After 2 h of incubation at 37°C in a 5% CO2 incubator, the cells were overlaid with 2 ml GMEM supplemented with 2% FBS containing 0.8% carboxymethyl cellulose (Sigma Life Science). Plaques were stained with crystal violet after 2 to 3 days of incubation at 37°C.
Metabolic labeling and immunoprecipitation.
Pulse-chase labeling was carried out as described previously (11), except that experiments were performed using U2OS WT and ΔΔ cells transfected with plasmids expressing SFV, SFV-HR+VE, CHIKV, CHIKV-RH, CHIKV-F3ANC, or CHIKV-F3ANC-RH ns polyproteins. Briefly, cells were transfected with 2 μg of these plasmids. At 8 (SFV) or 12 (CHIKV) h posttransfection, cells were starved in methionine- and cysteine-free medium for 30 min and then labeled with 5 μCi of a [35S]methionine-[35S]cysteine mixture (PerkinElmer) for 15 min. In the chased samples, the pulse was followed by a chase for 45 min in the presence of excess unlabeled methionine and cysteine. The cells were then lysed by boiling in 1% SDS, and samples were subjected to immunoprecipitation (IP) using antibodies against the nsPs of SFV or CHIKV (in-house) and protein A-Sepharose CL-AB (Sigma-Aldrich). The precipitated proteins were separated using SDS-PAGE and visualized using a Typhoon imager. Band intensities were quantified using the ImageQuant TL software (GE Healthcare).
Immunofluorescence and microscopy.
Cells grown on cover glasses (VWR) were fixed with 3.7% (vol/vol) formaldehyde in PBS for 15 min at room temperature, immersed in methanol for 10 min at –20°C, and blocked with 5% horse serum (Sigma) in PBS at 4°C overnight. Antibodies were diluted in blocking buffer as described below, and samples were incubated for 1 h with primary antibodies, followed by 30 min of incubation with secondary antibodies at room temperature. Cover glasses were mounted on glass slides using vinol mounting medium (45) and imaged by confocal laser scanning microscopy using a Supercontinuum confocal Leica TCS SP5 X equipped with a pulsed white-light laser and a Leica HCX PL Apo 63×/1.40 oil objective. Images were processed using Adobe Photoshop. The settings for image acquisition and adjustment were kept constant for all samples for dsRNA signals. Settings for nsP3 and GFP were varied slightly between samples to compensate for strong differences in localized signal intensities. Primary antibodies used were mouse anti-dsRNA (English and Scientific Consulting; 1:200) and rabbit antiserum against CHIKV-nsP3 (1:500 [46]). Secondary antibodies used were Alexa Fluor 488 (Molecular Probes; 1:200), Alexa Fluor 568 (Molecular Probes; 1:1,000), and Alexa Fluor 647 (Molecular Probes; 1:500).
Analysis of CHIKV P1234 expression in U2OS and ΔΔ cells.
U2OS and ΔΔ cells grown on 24-well cell culture plates (∼105 cells/well) were transfected with 0.5 μg of CHIKV-NanoLuc, CHIKV-NanoLuc-RH, CHIKV-NanoLuc-F3ANC, and CHIKV-NanoLuc-F3ANC-RH. Transfected cells were incubated at 37°C for 4, 8, 12, 16, or 20 h. At these time points, cells were collected and lysed, and reporter activities were determined as described above.
Northern blotting.
Northern blotting was performed as previously described (23). Briefly, U2OS WT and ΔΔ cells were cotransfected with HSPolI-Fluc-Gluc and CHIKV, CHIKV-RH, CHIKV-P1*2*34, CHIKV-P1*2*34-RH, or CHIKV-GAA; control cells were mock transfected. At 18 h posttransfection, total RNA was extracted using the TRIzol reagent (Life Technologies). Two micrograms of RNA was used for positive-strand detection, while 10 μg of RNA was used for negative-strand detection. RNAs were denatured, separated on a denaturing gel (1% agarose/6% formaldehyde), and transferred to a Hybond-N+ filter (GE Healthcare). Digoxigenin (DIG)-labeled RNA probe complementary to the sequence encoding the Gluc marker was used to detect positive-strand RNAs; a probe corresponding to the sequence encoding the Fluc marker was used to detect negative-strand RNAs. Filters were hybridized overnight; blots were washed and developed according to the manufacturer’s (Roche) protocols.
Experiments with CHIKV-WA and CHIKV-WA-RH.
Temperature-sensitive mutants of CHIKV, CHIKV-WA and CHIKV-WA-RH, were rescued and titers determined as described above, except that icDNAs were first linearized and in vitro transcribed using the mMessage mMachine SP6 transcription kit (Ambion), the experiment was performed at 28°C, and the extended incubation times were used. Next, U2OS cells were infected with rescued viruses at an MOI of 1 and incubated at 28°C or 39°C for 6, 12, 24, 36, or 48 h. At these time points, cells were collected and lysed, and reporter activities were determined as described above.
In the transfection experiment, U2OS WT or U2OS ΔΔ cells were transfected with 5 μg of in vitro-transcribed infectious RNAs of CHIKV-WA or CHIKV-WA-RH. Transfected cells were incubated at 39°C for 1, 2, 4, 6, 8, or 12 h. At these time points, cells were collected and lysed, and reporter activities were determined.
Statistical analysis.
All statistical analysis was performed using an unpaired, two-tailed Student t test with a 95% confidence interval.
ACKNOWLEDGMENTS
We thank Kai Rausalu and Laura Sandra Lello (University of Tartu) for help with insect cell and Northern blot experiments, and members of the G.M.M. laboratory (Karolinska Institutet) for critical readings. B.G. was supported by a studentship from Karolinska Institutet (KI Doktorander). Work in the G.M.M. laboratory is supported by project grants from the Swedish Research Council (grant 621-2014-4718) and the Swedish Cancer Foundation (grant CAN 2015–751). Work in the A.M. laboratory was supported by European Regional Development Fund through the Centre of Excellence in Molecular Cell Engineering, Estonia, 2014-2020.4.01.15-013, institutional research funding (IUT20-27) from Estonian Research Council (to A.M.), and by The Wellcome Trust (grant 200171/Z/15/Z).
The funders had no role in the study design, data collection and interpretation, or the decision to submit the work for publication.
REFERENCES
- 1.Chen R, Mukhopadhyay S, Merits A, Bolling B, Nasar F, Coffey LL, Powers A, Weaver SC, Ictv Report C . 2018. ICTV virus taxonomy profile: Togaviridae. J Gen Virol 99:761–762. doi: 10.1099/jgv.0.001072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vasiljeva L, Valmu L, Kääriäinen L, Merits A. 2001. Site-specific protease activity of the carboxyl-terminal domain of Semliki Forest virus replicase protein nsP2. J Biol Chem 276:30786–30793. doi: 10.1074/jbc.M104786200. [DOI] [PubMed] [Google Scholar]
- 3.Kim KH, Rumenapf T, Strauss EG, Strauss JH. 2004. Regulation of Semliki Forest virus RNA replication: a model for the control of alphavirus pathogenesis in invertebrate hosts. Virology 323:153–163. doi: 10.1016/j.virol.2004.03.009. [DOI] [PubMed] [Google Scholar]
- 4.Strauss EG, De Groot RJ, Levinson R, Strauss JH. 1992. Identification of the active-site residues in the Nsp2 proteinase of Sindbis virus. Virology 191:932–940. doi: 10.1016/0042-6822(92)90268-t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shirako Y, Strauss JH. 1994. Regulation of Sindbis virus RNA replication: uncleaved P123 and nsP4 function in minus-strand RNA synthesis, whereas cleaved products from P123 are required for efficient plus-strand RNA synthesis. J Virol 68:1874–1885. doi: 10.1128/JVI.68.3.1874-1885.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lemm JA, Rumenapf T, Strauss EG, Strauss JH, Rice CM. 1994. Polypeptide requirements for assembly of functional Sindbis virus replication complexes: a model for the temporal regulation of minus- and plus-strand RNA synthesis. EMBO J 13:2925–2934. doi: 10.1002/j.1460-2075.1994.tb06587.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tomar S, Hardy RW, Smith JL, Kuhn RJ. 2006. Catalytic core of alphavirus nonstructural protein nsP4 possesses terminal adenylyltransferase activity. J Virol 80:9962–9969. doi: 10.1128/JVI.01067-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sawicki DL, Sawicki SG. 1980. Short-lived minus-strand polymerase for Semliki Forest virus. J Virol 34:108–118. doi: 10.1128/JVI.34.1.108-118.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vasiljeva L, Merits A, Golubtsov A, Sizemskaja V, Kääriäinen L, Ahola T. 2003. Regulation of the sequential processing of Semliki Forest virus replicase polyprotein. J Biol Chem 278:41636–41645. doi: 10.1074/jbc.M307481200. [DOI] [PubMed] [Google Scholar]
- 10.Lulla V, Karo-Astover L, Rausalu K, Saul S, Merits A, Lulla A. 2018. Timeliness of proteolytic events is prerequisite for efficient functioning of the alphaviral replicase. J Virol 92:e00151-18. doi: 10.1128/JVI.00151-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lulla A, Lulla V, Tints K, Ahola T, Merits A. 2006. Molecular determinants of substrate specificity for Semliki Forest virus nonstructural protease. J Virol 80:5413–5422. doi: 10.1128/JVI.00229-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saul S, Ferguson M, Cordonin C, Fragkoudis R, Ool M, Tamberg N, Sherwood K, Fazakerley JK, Merits A. 2015. Differences in processing determinants of nonstructural polyprotein and in the sequence of nonstructural protein 3 affect neurovirulence of Semliki Forest virus. J Virol 89:11030–11045. doi: 10.1128/JVI.01186-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ferguson MC, Saul S, Fragkoudis R, Weisheit S, Cox J, Patabendige A, Sherwood K, Watson M, Merits A, Fazakerley JK. 2015. Ability of the encephalitic arbovirus Semliki Forest virus to cross the blood-brain barrier is determined by the charge of the E2 glycoprotein. J Virol 89:7536–7549. doi: 10.1128/JVI.03645-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tourrière H, Chebli K, Zekri L, Courselaud B, Blanchard JM, Bertrand E, Tazi J. 2003. The RasGAP-associated endoribonuclease G3BP assembles stress granules. J Cell Biol 160:823–831. doi: 10.1083/jcb.200212128. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 15.Kedersha N, Panas MD, Achorn CA, Lyons S, Tisdale S, Hickman T, Thomas M, Lieberman J, McInerney GM, Ivanov P, Anderson P. 2016. G3BP-Caprin1-USP10 complexes mediate stress granule condensation and associate with 40S subunits. J Cell Biol 212:845–860. doi: 10.1083/jcb.201508028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frolov I, Kim DY, Akhrymuk M, Mobley JA, Frolova EI. 2017. Hypervariable domain of eastern equine encephalitis virus nsP3 redundantly utilizes multiple cellular proteins for replication complex assembly. J Virol 91:e00371-17. doi: 10.1128/JVI.00371-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Götte B, Panas MD, Hellstrom K, Liu L, Samreen B, Larsson O, Ahola T, McInerney GM. 2019. Separate domains of G3BP promote efficient clustering of alphavirus replication complexes and recruitment of the translation initiation machinery. PLoS Pathog 15:e1007842. doi: 10.1371/journal.ppat.1007842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Panas MD, Varjak M, Lulla A, Eng KE, Merits A, Karlsson Hedestam GB, McInerney GM. 2012. Sequestration of G3BP coupled with efficient translation inhibits stress granules in Semliki Forest virus infection. Mol Biol Cell 23:4701–4712. doi: 10.1091/mbc.E12-08-0619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Panas MD, Schulte T, Thaa B, Sandalova T, Kedersha N, Achour A, McInerney GM. 2015. Viral and cellular proteins containing FGDF motifs bind G3BP to block stress granule formation. PLoS Pathog 11:e1004659. doi: 10.1371/journal.ppat.1004659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fros JJ, Geertsema C, Zouache K, Baggen J, Domeradzka N, van Leeuwen DM, Flipse J, Vlak JM, Failloux AB, Pijlman GP. 2015. Mosquito Rasputin interacts with chikungunya virus nsP3 and determines the infection rate in Aedes albopictus. Parasit Vectors 8:464. doi: 10.1186/s13071-015-1070-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schulte T, Liu L, Panas MD, Thaa B, Dickson N, Götte B, Achour A, McInerney GM, Schulte T, Liu L, Panas MD, Thaa B, Dickson N, Götte B, Achour A, McInerney GM. 2016. Combined structural, biochemical and cellular evidence demonstrates that both FGDF motifs in alphavirus nsP3 are required for efficient replication. Open Biol 6:160078. doi: 10.1098/rsob.160078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liljeström P, Lusa S, Huylebroeck D, Garoff H. 1991. In vitro mutagenesis of a full-length cDNA clone of Semliki Forest virus: the small 6,000-molecular-weight membrane protein modulates virus release. J Virol 65:4107–4113. doi: 10.1128/JVI.65.8.4107-4113.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Utt A, Rausalu K, Jakobson M, Männik A, Alphey L, Fragkoudis R, Merits A, Utt A, Rausalu K, Jakobson M, Männik A, Alphey L, Fragkoudis R, Merits A. 2019. Design and use of chikungunya virus replication templates utilizing mammalian and mosquito RNA polymerase I-mediated transcription. J Virol 93:e01794-19. doi: 10.1128/JVI.00794-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim DY, Reynaud JM, Rasalouskaya A, Akhrymuk I, Mobley JA, Frolov I, Frolova EI. 2016. New World and Old World alphaviruses have evolved to exploit different components of stress granules, FXR and G3BP proteins, for assembly of viral replication complexes. PLoS Pathog 12:e1005810. doi: 10.1371/journal.ppat.1005810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bartholomeeusen K, Utt A, Coppens S, Rausalu K, Vereecken K, Arien KK, Merits A. 2018. A chikungunya virus trans-replicase system reveals the importance of delayed nonstructural polyprotein processing for efficient replication complex formation in mosquito cells. J Virol 92:e00152-18. doi: 10.1128/JVI.00152-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chan YH, Teo TH, Utt A, Tan JJL, Amrun SN, Abu Bakar F, Yee WX, Becht E, Lee CYP, Lee B, Rajarethinam R, Newell E, Merits A, Carissimo G, Lum FM, Ng L. 2019. Mutating chikungunya virus non-structural protein produces potent live-attenuated vaccine candidate. EMBO Mol Med 11:e10092. doi: 10.15252/emmm.201810092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Göertz GP, Lingemann M, Geertsema C, Abma-Henkens MHC, Vogels CBF, Koenraadt CJM, van Oers MM, Pijlman GP. 2018. Conserved motifs in the hypervariable domain of chikungunya virus nsP3 required for transmission by Aedes aegypti mosquitoes. PLoS Negl Trop Dis 12:e0006958. doi: 10.1371/journal.pntd.0006958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Utt A, Quirin T, Saul S, Hellstrom K, Ahola T, Merits A. 2016. Versatile trans-replication systems for chikungunya virus allow functional analysis and tagging of every replicase protein. PLoS One 11:e0151616. doi: 10.1371/journal.pone.0151616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shirako Y, Strauss JH. 1990. Cleavage between Nsp1 and Nsp2 initiates the processing pathway of Sindbis virus nonstructural polyprotein-P123. Virology 177:54–64. doi: 10.1016/0042-6822(90)90459-5. [DOI] [PubMed] [Google Scholar]
- 30.Gorchakov R, Frolova E, Sawicki S, Atasheva S, Sawicki D, Frolov I. 2008. A new role for ns polyprotein cleavage in Sindbis virus replication. J Virol 82:6218–6231. doi: 10.1128/JVI.02624-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hellström K, Kallio K, Utt A, Quirin T, Jokitalo E, Merits A, Ahola T. 2017. Partially uncleaved alphavirus replicase forms spherule structures in the presence and absence of RNA template. J Virol 91:e00787-17. doi: 10.1128/JVI.00787-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Panas MD, Ahola T, McInerney GM. 2014. The C-terminal repeat domains of nsP3 from the Old World alphaviruses bind directly to G3BP. J Virol 88:5888–5893. doi: 10.1128/JVI.00439-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scholte FE, Tas A, Albulescu IC, Zusinaite E, Merits A, Snijder EJ, van Hemert MJ. 2015. Stress granule components G3BP1 and G3BP2 play a proviral role early in Chikungunya virus replication. J Virol 89:4457–4469. doi: 10.1128/JVI.03612-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matkovic R, Bernard E, Fontanel S, Eldin P, Chazal N, Hersi DH, Merits A, Peloponese JM, Briant L. 2018. The host DHX9 DExH-box helicase is recruited to chikungunya virus replication complexes for optimal genomic RNA translation. J Virol 93:e01764-18. doi: 10.1128/JVI.01764-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Szpiech ZA, Strauli NB, White KA, Ruiz DG, Jacobson MP, Barber DL, Hernandez RD. 2017. Prominent features of the amino acid mutation landscape in cancer. PLoS One 12:e0183273. doi: 10.1371/journal.pone.0183273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nikonov A, Molder T, Sikut R, Kiiver K, Mannik A, Toots U, Lulla A, Lulla V, Utt A, Merits A, Ustav M. 2013. RIG-I and MDA-5 detection of viral RNA-dependent RNA polymerase activity restricts positive-strand RNA virus replication. PLoS Pathog 9:e1003610. doi: 10.1371/journal.ppat.1003610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Krapp S, Greiner E, Amin B, Sonnewald U, Krenz B. 2017. The stress granule component G3BP is a novel interaction partner for the nuclear shuttle proteins of the nanovirus pea necrotic yellow dwarf virus and geminivirus abutilon mosaic virus. Virus Res 227:6–14. doi: 10.1016/j.virusres.2016.09.021. [DOI] [PubMed] [Google Scholar]
- 38.Powers AM, Brault AC, Shirako Y, Strauss EG, Kang W, Strauss JH, Weaver SC. 2001. Evolutionary relationships and systematics of the alphaviruses. J Virol 75:10118–10131. doi: 10.1128/JVI.75.21.10118-10131.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Polo JM, Davis NL, Rice CM, Huang HV, Johnston RE. 1988. Molecular analysis of Sindbis virus pathogenesis in neonatal mice by using virus recombinants constructed in vitro. J Virol 62:2124–2133. doi: 10.1128/JVI.62.6.2124-2133.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kuhn RJ, Niesters HGM, Hong Z, Strauss JH. 1991. Infectious RNA transcripts from Ross River virus cDNA clones and the construction and characterization of defined chimeras with Sindbis virus. Virology 182:430–441. doi: 10.1016/0042-6822(91)90584-X. [DOI] [PubMed] [Google Scholar]
- 41.Chuong C, Bates TA, Weger-Lucarelli J. 2019. Infectious cDNA clones of two strains of Mayaro virus for studies on viral pathogenesis and vaccine development. Virology 535:227–231. doi: 10.1016/j.virol.2019.07.013. [DOI] [PubMed] [Google Scholar]
- 42.Ulper L, Sarand I, Rausalu K, Merits A. 2008. Construction, properties, and potential application of infectious plasmids containing Semliki Forest virus full-length cDNA with an inserted intron. J Virol Methods 148:265–270. doi: 10.1016/j.jviromet.2007.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lulla V, Karo-Astover L, Rausalu K, Merits A, Lulla A. 2013. Presentation overrides specificity: probing the plasticity of alphaviral proteolytic activity through mutational analysis. J Virol 87:10207–10220. doi: 10.1128/JVI.01485-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Utt A, Das PK, Varjak M, Lulla V, Lulla A, Merits A. 2015. Mutations conferring a noncytotoxic phenotype on chikungunya virus replicons compromise enzymatic properties of nonstructural protein 2. J Virol 89:3145–3162. doi: 10.1128/JVI.03213-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fukui Y, Yumura S, Yumura TK. 1987. Agar-overlay immunofluorescence: high-resolution studies of cytoskeletal components and their changes during chemotaxis. Methods Cell Biol 28:347–356. doi: 10.1016/s0091-679x(08)61655-6. [DOI] [PubMed] [Google Scholar]
- 46.Mutso M, Morro AM, Smedberg C, Kasvandik S, Aquilimeba M, Teppor M, Tarve L, Lulla A, Lulla V, Saul S, Thaa B, McInerney GM, Merits A, Varjak M. 2018. Mutation of CD2AP and SH3KBP1 binding motif in alphavirus nsP3 hypervariable domain results in attenuated virus. Viruses 10:226. doi: 10.3390/v10050226. [DOI] [PMC free article] [PubMed] [Google Scholar]