Influenza viruses are highly contagious pathogens and are a major threat to human health. Vaccination remains the most effective tool to protect humans against influenza infection. However, vaccination does not always guarantee complete protection against drifted or, more noticeably, shifted influenza viruses. Although U.S. Food and Drug Administration (FDA) drugs are approved for the treatment of influenza infections, influenza viruses resistant to current FDA antivirals have been reported and continue to emerge. Therefore, there is an urgent need to find novel antivirals for the treatment of influenza viral infections in humans, a search that could be expedited by repurposing currently approved drugs. In this study, we assessed the influenza antiviral activity of 10 compounds previously shown to inhibit mammarenavirus infection. Among them, eight drugs showed antiviral activities, providing a new battery of drugs that could be used for the treatment of influenza infections.
KEYWORDS: Orthomyxovirus, influenza virus, antivirals, prophylactic, therapeutic, drug treatment, reporter genes
ABSTRACT
Influenza A (IAV) and influenza B (IBV) viruses are highly contagious pathogens that cause fatal respiratory disease every year, with high economic impact. In addition, IAV can cause pandemic infections with great consequences when new viruses are introduced into humans. In this study, we evaluated 10 previously described compounds with antiviral activity against mammarenaviruses for their ability to inhibit IAV infection using our recently described bireporter influenza A/Puerto Rico/8/34 (PR8) H1N1 (BIRFLU). Among the 10 tested compounds, eight (antimycin A [AmA], brequinar [BRQ], 6-azauridine, azaribine, pyrazofurin [PF], AVN-944, mycophenolate mofetil [MMF], and mycophenolic acid [MPA]), but not obatoclax or Osu-03012, showed potent anti-influenza virus activity under posttreatment conditions [median 50% effective concentration (EC50) = 3.80 nM to 1.73 μM; selective index SI for 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide (MTT) assay, >28.90 to 13,157.89]. AmA, 6-azauridine, azaribine, and PF also showed potent inhibitory effect in pretreatment (EC50 = 0.14 μM to 0.55 μM; SI-MTT = 70.12 to >357.14) or cotreatment (EC50 = 34.69 nM to 7.52 μM; SI-MTT = 5.24 to > 1,441.33) settings. All of the compounds tested inhibited viral genome replication and gene transcription, and none of them affected host cellular RNA polymerase II activities. The antiviral activity of the eight identified compounds against BIRFLU was further confirmed with seasonal IAVs (A/California/04/2009 H1N1 and A/Wyoming/3/2003 H3N2) and an IBV (B/Brisbane/60/2008, Victoria lineage), demonstrating their broad-spectrum prophylactic and therapeutic activity against currently circulating influenza viruses in humans. Together, our results identified a new set of antiviral compounds for the potential treatment of influenza viral infections.
IMPORTANCE Influenza viruses are highly contagious pathogens and are a major threat to human health. Vaccination remains the most effective tool to protect humans against influenza infection. However, vaccination does not always guarantee complete protection against drifted or, more noticeably, shifted influenza viruses. Although U.S. Food and Drug Administration (FDA) drugs are approved for the treatment of influenza infections, influenza viruses resistant to current FDA antivirals have been reported and continue to emerge. Therefore, there is an urgent need to find novel antivirals for the treatment of influenza viral infections in humans, a search that could be expedited by repurposing currently approved drugs. In this study, we assessed the influenza antiviral activity of 10 compounds previously shown to inhibit mammarenavirus infection. Among them, eight drugs showed antiviral activities, providing a new battery of drugs that could be used for the treatment of influenza infections.
INTRODUCTION
Influenza A (IAV) and B (IBV) viruses are members of the Orthomyxoviridae family and are responsible for severe human respiratory disease (1). The World Health Organization (WHO) estimates that about 3 to 5 million severe cases of illness and approximately 250,000 to 500,000 deaths are caused every year by seasonal influenza virus infections (1–5). IAV and IBV genomes are made of eight negative-sense, single-stranded RNA segments (1). IAVs are classified into subtypes based on the major antigenic surface glycoproteins hemagglutinin (HA; 18 subtypes) and neuraminidase (NA; 11 subtypes) (1, 6–9). IBVs have no subtype classification but are divided into two major lineages referred to as Victoria and Yamagata (1, 5, 10, 11). H1N1 and H3N2 IAVs and the two lineages of IBVs are presently circulating in the human population, causing recurrent epidemics (1, 2, 5, 10). Moreover, the impact of IAV is increased by the risk of sporadic pandemics when novel viruses are introduced into the human population (12).
Currently, NA inhibitors, including oseltamivir, zanamivir, and peramivir, matrix protein 2 (M2) inhibitors (amantadine and rimantadine), and the polymerase acid endonuclease (PA) inhibitor baloxavir marboxil (Xofluza), are approved by the Food and Drug Administration (FDA) for the treatment of influenza infections in humans (1, 13). However, 90% of the currently circulating H1N1 and H3N2 IAVs and all IBVs are resistant to M2 inhibitors (1). Moreover, NA inhibitor-resistant IAVs have also been reported (1, 14). Likewise, Xofluza-resistant viruses have also been identified (15, 16). Therefore, there is an urgent need to find novel antiviral drugs for the treatment of influenza infections. However, discovery and implementation of new antivirals is a long and complicated process that requires multiple levels of approval, including safety and effectiveness testing. Drug repurposing approaches can reduce the time and resources required to advance a candidate antiviral drug into the clinic, as available knowledge about the pharmacology and toxicology of the repurposed candidate drug can alleviate the labor and resource-intensive efforts involved in preclinical testing of newly discovered drug candidates (17, 18).
We have documented the screening of the Repurposing, Focused Rescue, and Accelerated Medchem (ReFRAME) library (19) for compounds with antiviral activity against the prototypic mammarenavirus lymphocytic choriomeningitis virus (LCMV), and identified 10 compounds with a potent and dose-dependent anti-LCMV activity (20). These include inhibitors of IMP dehydrogenase (IMPDH), dihydroorotate dehydrogenase (DHODH), and UMP synthetase (UMPS) enzymes, as well as the proviral MCL1 apoptosis regulator and the mitochondrial electron transport complex (mETC) III (19).
Since mammarenaviruses and influenza viruses are both negative-stranded RNA viruses with a segmented genome, we examined the ability of the 10 identified anti-mammarenavirus compounds to inhibit influenza viral infection, taking advantage of a novel recombinant replication-competent bireporter IAV (BIRFLU) expressing both fluorescent (Venus) and luciferase (Nano luciferase [Nluc]) reporter genes (21). We found that eight of the ten repurposing candidate compounds tested exhibited broad-spectrum and potent prophylactic and therapeutic antiviral activities against currently circulating H1N1 and H3N2 IAVs, and IBVs, suggesting the feasibility of their implementation for the treatment of influenza viral infections. Importantly, information from these studies could also provide new insights into important cellular pathways required for influenza viral infection that could be used for the identification of new targets for the efficient treatment of these important human respiratory pathogens.
RESULTS
Compound effects on IAV multiplication.
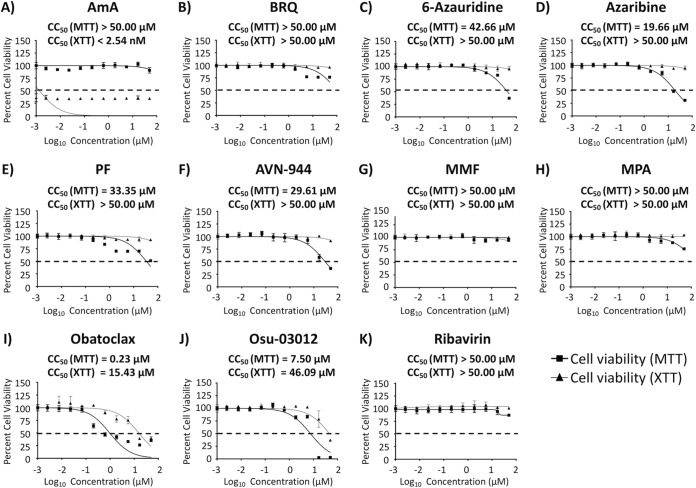
Prior to examining the effects on IAV multiplication of the selected 10 compounds (Fig. 1) with antiviral activity against mammarenavirus, we first determined the 50% cytotoxicity concentrations (CC50) of each of the compounds in Madin-Darby canine kidney (MDCK) cells using a 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide (MTT) assay for evaluation of cell viability and a 2,3-bis-(2-methoxy-4-nitro-5-sulfophenyl)-2H-tetrazolium-5-carboxanilide salt [XTT] assay for evaluation of cellular proliferation (Fig. 2 and Table 1). In the MTT assay, antimycin A (AmA), brequinar (BRQ), mycophenolate mofetil (MMF), and mycophenolic acid (MPA) were not toxic at all tested concentrations, while 6-azauridine (CC50 = 42.66 μM), azaribine (CC50 = 19.66 μM), pyrazofurin (PF) (CC50 = 33.35 μM), AVN-944 (CC50 = 29.61 μM), obatoclax (CC50 = 0.23 μM), and Osu-03012 (CC50 = 7.50 μM) had different levels of toxicity (Fig. 2 and Table 1). In the XTT assay, none of the drugs, except AmA (CC50 < 2.54 nM), obatoclax (CC50 = 15.43 μM), and Osu-03012 (CC50 = 46.09 μM), showed toxicity, even at the highest tested concentration (50 μM) (Fig. 2 and Table 1).
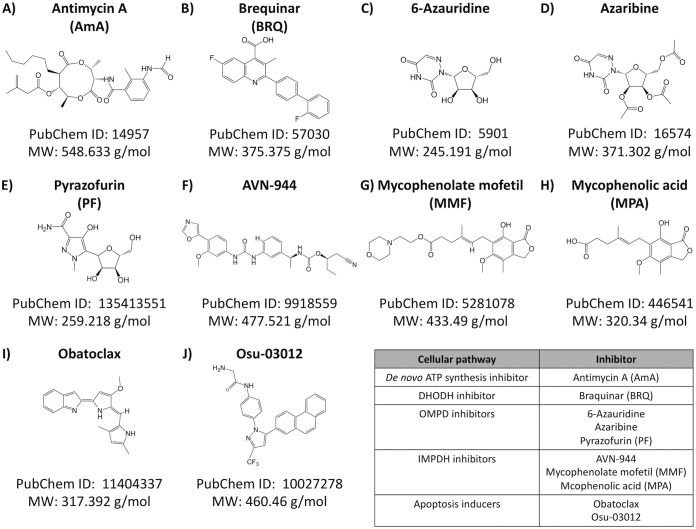
FIG 1.
Structure of the compounds. Structure, molecular weight, and compound identifier [ID] (PubChem) of each of the compounds used in this study are indicated.
FIG 2.
Cytotoxicity of the compounds. Confluent monolayers (96-well plate format; 5.0 × 104 cells/well; quadruplicates) of MDCK cells were treated with the indicated doses (3-fold serial dilutions, starting concentration of 50 μM) of each of the indicated compounds. Cell proliferation assays were performed at 48 h posttreatment, and the CC50 for each compound was calculated as indicated in Materials and Methods. Compound-treated cell viability was calculated as a percentage relative to values obtained for dimethyl sulfoxide (DMSO) vehicle-treated cells. Dotted lines indicate 50% cell viability. Data are expressed as mean and SD from three independent experiments conducted in quadruplicates.
TABLE 1.
Summary of CC50, EC50, and SI values of the compounds against BIRFLU during posttreatment in MDCK cellsa
| Compound | Activity statistics of compound against BIRFLUb
|
||||
|---|---|---|---|---|---|
| CC50 (MTT) (μM) | CC50 (XTT) | EC50 | SI (MTT) | SI (XTT) | |
| AmA | >50.00 | <2.54 nM | 3.80 nM | >13,157.89 | <0.66 |
| BRQ | >50.00 | >50.00 μM | 0.58 μM | >86.21 | >86.21 |
| 6-Azauridine | 42.66 | >50.00 μM | 0.34 μM | 125.47 | >147.06 |
| Azaribine | 19.66 | >50.00 μM | 0.29 μM | 67.79 | >172.41 |
| PF | 33.35 | >50.00 μM | 38.90 nM | 857.33 | >1,285.35 |
| AVN-944 | 29.61 | >50.00 μM | 0.21 μM | 141.00 | >238.10 |
| MMF | >50.00 | >50.00 μM | 0.77 μM | >64.94 | >64.94 |
| MPA | >50.00 | >50.00 μM | 1.73 μM | >28.90 | >28.90 |
| Obatoclax | 0.23 | 15.43 μM | 0.42 μM | 0.55 | 36.74 |
| Osu-03012 | 7.50 | 46.09 μM | 14.42 μM | 0.52 | 3.20 |
| Ribavirin | >50.00 | >50.00 μM | 9.50 μM | >5.26 | >5.26 |
BIRFLU, A/Puerto Rico/8/34 H1N1.
CC50, median 50% cytotoxicity concentration; EC50, median 50% effective concentration; SI, selective index (CC50/EC50).
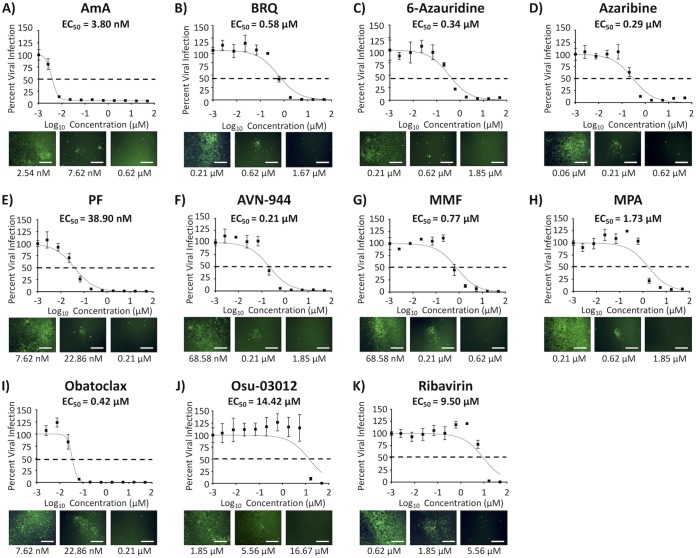
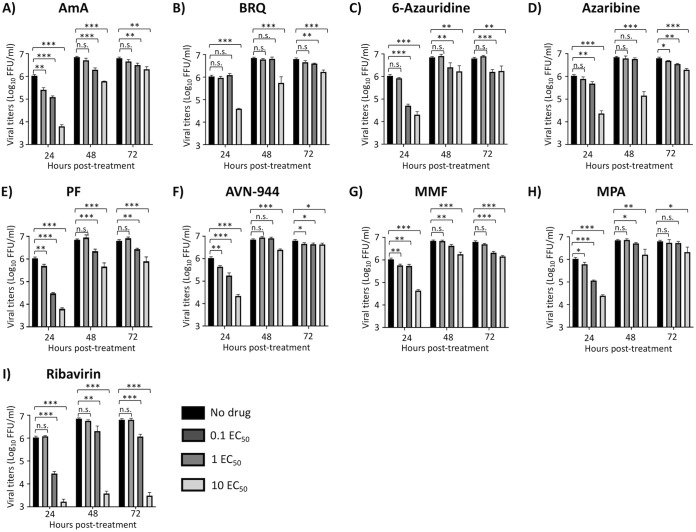
To determine the 50% effective concentration (EC50) of the compounds against IAV infection, MDCK cells were infected with 200 fluorescence-forming units (FFU) per well of our recently described bireporter influenza A/Puerto Rico/8/34 (PR8) H1N1 (BIRFLU) isolate (21). After 1 h of viral absorption, virus inoculum was replaced with infection medium containing 3-fold serial dilutions (starting concentration of 50 μM) of the 10 individual compounds (Fig. 3). Eight of the compounds tested showed a potent inhibitory effect on BIRFLU multiplication based on their EC50 and selective index (SI; CC50/EC50) values with either the MTT (SI-MTT) or the XTT (SI-XTT) assay: BRQ (EC50 = 0.58 μM; SI-MTT and SI-XTT > 86.21), 6-azauridine (EC50 = 0.34 μM; SI-MTT = 125.47 and SI-XTT > 147.06), azaribine (EC50 = 0.29 μM; SI-MTT = 67.79 and SI-XTT > 172.41), PF (EC50 = 38.9 nM; SI-MTT = 857.33 and SI-XTT > 1,285.35), AVN-944 (EC50 = 0.21 μM; SI-MTT = 141.00 and SI-XTT > 238.10), MMF (EC50 = 0.77 μM; SI-MTT and SI-XTT > 64.94), and MPA (EC50 = 1.73 μM; SI-MTT and SI-XTT > 28.90) (Fig. 3 and Table 1). AmA (EC50 = 3.80 nM) showed the most potent antiviral activity with the MTT assay (SI-MTT > 13,157.89) but the lowest SI values with the XTT assay (SI-XTT < 0.66) because of the different CC50 in the MTT and XTTs assay (Fig. 2 and 3; and Table 1). In contrast, obatoclax (EC50 = 0.42 μM; SI-MTT = 0.55 and SI-XTT = 36.74) and Osu-03012 (EC50 = 14.42 μM; SI-MTT = 0.52 and SI-XTT = 3.20) did not show potent inhibitory effect on IAV multiplication (Fig. 3 and Table 1) that could be distinguished from their inhibitory effect on cell viability (Fig. 2 and Table 1). Ribavirin, included as a control in these assays, showed a CC50 of >50 μM (Fig. 2), an EC50 of 9.5 μM (Fig. 3), and SI values of >5.26 from MTT and XTT assays (Table 1), values consistent with previous published data (21, 22). Next, we evaluated the effects of the eight compounds that showed a potent inhibitory effect on BIRFLU multiplication (AmA, 6-azauridine, azaribine, BRQ, AVN-944, MMF, MPA, and PF) on production of infectious virus progeny. The eight tested compounds exerted dose- and time-dependent inhibition of production of infectious IAV (Fig. 4). As expected, inhibition of IAV production by ribavirin was also dose dependent, as previously described (21, 22).
FIG 3.
Inhibition of BIRFLU. Confluent monolayers (96-well plate format; 5.0 × 104 cells/well; quadruplicates) of MDCK cells were infected with 200 FFU of BIRFLU. After 1 h of viral adsorption, the indicated concentrations (3-fold serial dilutions, starting concentration of 50 μM) of the different compounds or 0.1% DMSO vehicle control were added to the postinfection medium. At 48 hpi, tissue culture supernatants from infected cells were collected and used to measure Nluc expression. Images of Venus expression were taken using a fluorescence microscope. Percent viral infection and the EC50 were calculated based on Nluc expression. Dotted lines indicate 50% viral inhibition. Data are expressed as mean and SD from three independent experiments conducted in quadruplicates. Bar, 50 μm.
FIG 4.
Inhibition of viral growth kinetics. MDCK cells (24-well plate format; 2.5 × 105 cells/well; triplicates) were infected (MOI = 0.1) with BIRFLU. After 1 h of viral adsorption, infected cells were treated with the indicated 0.1, 1, and 10 EC50 of the different compounds calculated based on the results shown in Fig. 3. Tissue culture supernatants from infected cells were collected at 24, 48, and 72 hpi, and viral titers were calculated using an immunofocus assay (FFU/ml). Data are expressed as mean and SD from three independent experiments conducted in triplicates. Statistical analysis was conducted using an unpaired Student’s t test. *, P < 0.05; **, P < 0.01; ***, P < 0.001; n.s., not significant.
Compound effects on IAV infection during pretreatment and cotreatment.
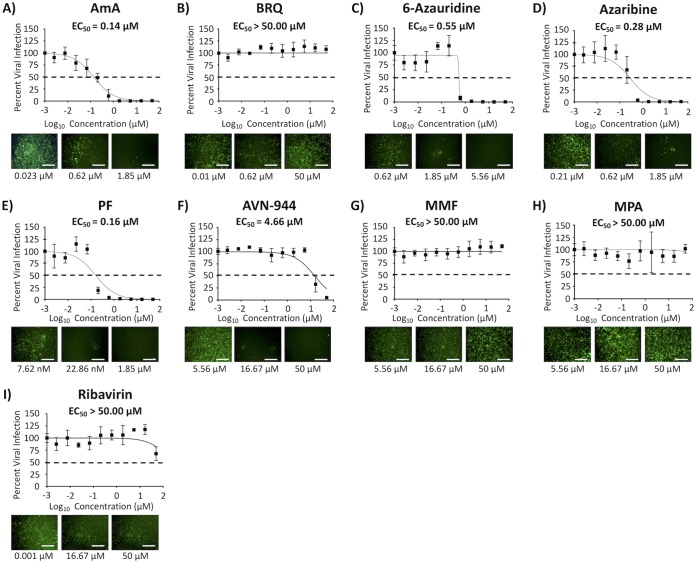
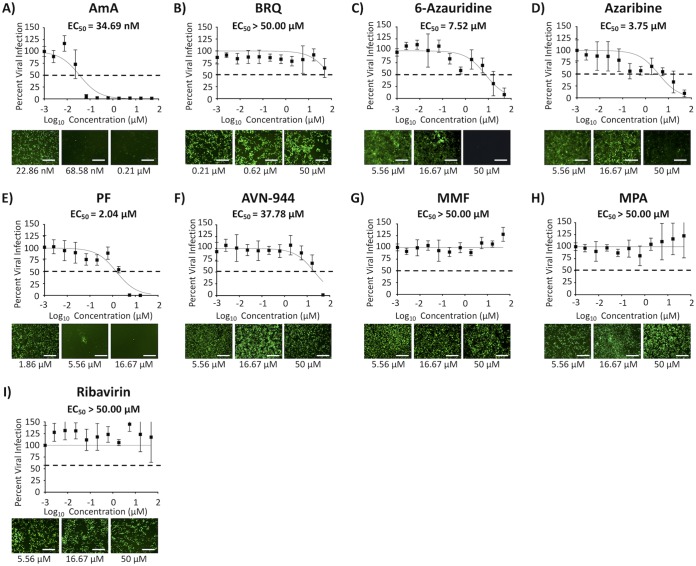
We next tested the prophylactic (pretreatment) or binding-inhibitory (cotreatment) effect of the compounds against BIRFLU (Fig. 5 and 6, respectively). EC50 and SI-MTT values of AmA and SI-MTT and SI-XTT values of 6-azauridine, azaribine, AVN-944, and PF in pretreatment or cotreatment experiments were similar to those observed in posttreatment experiments (Fig. 5 and 6, respectively, and Tables 2 and 3, respectively). AVN-944 pretreatment (Fig. 5 and Table 2) and cotreatment (Fig. 6 and Table 3) showed either significantly diminished or no antiviral activity against BIRFLU compared to that of the posttreatment experiment (SI values of posttreatment, pretreatment, and cotreatments were 141.00, 6.35, and 0.78 [SI-MTT], and >238.10, 10.72, and 1.32 [SI-XTT], respectively) (Tables 1 to 3). BRQ, MMF, and MPA did not inhibit viral infection even at the highest tested concentration (50 μM) in the pretreatment (Fig. 5) or cotreatment (Fig. 6) experiments (Tables 2 and 3, respectively). Based on the results from the cotreatment experiments (Fig. 6), we next evaluated the ability of the compounds to prevent hemagglutination or viral HA-mediated hemagglutination of turkey red blood cells (RBCs) using hemagglutination (HA) and hemagglutinin inhibition (HAI) assays, respectively (Table S1). In our HA assays, only AmA and AVN-944 were able to inhibit hemagglutination of RBCs at 500 μM and 1,000 μM, respectively, which is 10 to 20 times higher than the concentration used in our inhibition assays (50 μM). Notably, none of the compounds showed HAI activity, even at the highest tested concentration of 1,000 μM. These results suggest that the compounds are not targeting influenza viral entry and that other steps in the replication cycle of the virus might be targeted by the compounds.
FIG 5.
Prophylactic antiviral activity. Confluent monolayers (96-well plate format; 5 × 104 cells/well; quadruplicates) of MDCK cells were pretreated with the indicated 3-fold serial dilutions of the indicated compounds (starting concentration of 50 μM) or with 0.1% DMSO vehicle control for 24 h before infection with 200 FFU of BIRFLU. At 48 hpi, tissue culture supernatants from infected cells were collected and used to measure Nluc expression. Images of Venus expression were taken using a fluorescence microscope. Percent viral infection and the EC50 were calculated based on Nluc expression. Dotted lines indicate 50% viral inhibition. Data are expressed as mean and SD from three independent experiments conducted in quadruplicates. Bar, 50 μm.
FIG 6.
Inhibition of viral binding. The indicated 3-fold serial dilutions of the compounds (starting concentration of 50 μM) or 0.1% DMSO vehicle control were mixed with BIRFLU (200 FFU/well). After 1 h of incubation at room temperature, the virus-compound mixture was used to infect confluent monolayers of MDCK cells (96-well plate format; 5.0 × 104 cells/well; quadruplicates). After 1 h viral absorption, cells were washed with phosphate-buffered saline (PBS), and fresh postinfection medium was added. At 48 hpi, tissue culture supernatants from infected cells were collected and used to measure Nluc expression. Images of Venus expression were taken using a fluorescence microscope. Percent viral infection and the EC50 were calculated based on Nluc expression. Dotted lines indicate 50% viral inhibition. Data are expressed as mean and SD from three independent experiments conducted in quadruplicates. Bar, 50 μm.
TABLE 2.
Summary of CC50, EC50, and SI values of the compounds against BIRFLU during pretreatment in MDCK cellsa
| Compound | Activity statistics of compound against BIRFLUb
|
||||
|---|---|---|---|---|---|
| CC50 (MTT) (μM) | CC50 (XTT) | EC50 (μM) | SI (MTT) | SI (XTT) | |
| AmA | >50.00 | <2.54 nM | 0.14 | >357.14 | <0.02 |
| BRQ | >50.00 | >50.00 μM | >50.00 | 1.00 | 1.00 |
| 6-Azauridine | 42.66 | > 50.00 μM | 0.55 | 77.56 | >90.91 |
| Azaribine | 19.66 | >50.00 μM | 0.28 | 70.12 | >178.57 |
| PF | 33.35 | >50.00 μM | 0.16 | 208.43 | >312.50 |
| AVN-944 | 29.61 | >50.00 μM | 4.66 | 6.35 | >10.72 |
| MMF | >50.00 | >50.00 μM | >50.00 | 1.00 | 1.00 |
| MPA | >50.00 | >50.00 μM | >50.00 | 1.00 | 1.00 |
| Ribavirin | >50.00 | >50.00 μM | >50.00 | 1.00 | 1.00 |
BIRFLU, A/Puerto Rico/8/34 H1N1.
CC50, median 50% cytotoxicity concentration; EC50, median 50% effective concentration; SI, selective index (CC50/EC50).
TABLE 3.
Summary of CC50, EC50, and SI values of the compounds against BIRFLU during cotreatment in MDCK cellsa
| Compound | Activity statistics of compound against BIRFLUb
|
||||
|---|---|---|---|---|---|
| CC50 (MTT) (μM) | CC50 (XTT) | EC50 | SI (MTT) | SI (XTT) | |
| AmA | >50.00 | <2.54 nM | 34.69 nM | >1,441.33 | <0.07 |
| BRQ | >50.00 | >50.00 μM | >50.00 μM | 1.00 | 1.00 |
| 6-Azauridine | 42.66 | >50.00 μM | 7.52 μM | 5.67 | >6.65 |
| Azaribine | 19.66 | >50.00 μM | 3.75 μM | 5.24 | >13.33 |
| PF | 33.35 | >50.00 μM | 2.04 μM | 16.35 | >24.51 |
| AVN-944 | 29.61 | >50.00 μM | 37.78 μM | 0.78 | >1.32 |
| MMF | >50.00 | >50.00 μM | >50.00 μM | 1.00 | 1.00 |
| MPA | >50.00 | >50.00 μM | >50.00 μM | 1.00 | 1.00 |
| Ribavirin | >50.00 | >50.00 μM | >50.00 μM | 1.00 | 1.00 |
BIRFLU, A/Puerto Rico/8/34 H1N1.
CC50, median 50% cytotoxicity concentration; EC50, median 50% effective concentration; SI, selective index (CC50/EC50).
Compound effects on viral genome replication and gene transcription.
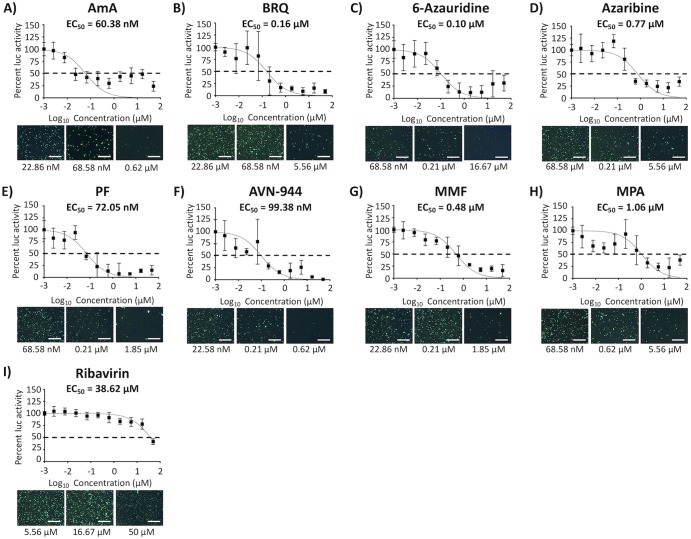
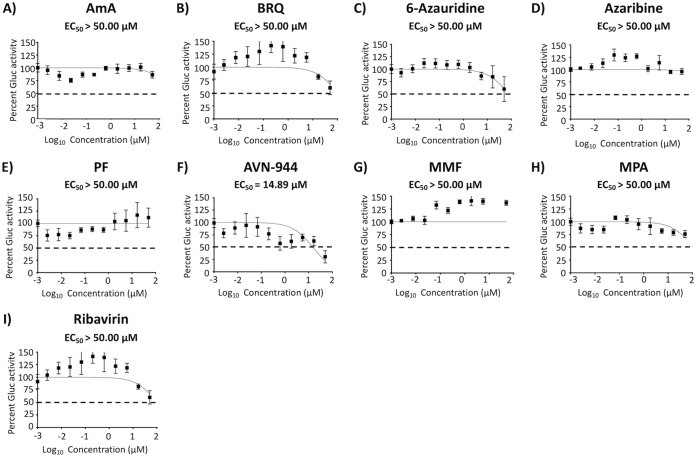
To evaluate if the compounds with antiviral activity against BIRFLU were targeting viral replication, transcription, or both, we conducted a minigenome (MG) assay (Fig. 7). All tested compounds reduced levels of MG-directed green fluorescent protein (GFP) and Gaussia luciferase (Gluc) expression (Fig. 7), suggesting that they exerted their antiviral effect via inhibition of the activity of the viral ribonucleoprotein complex (vRNP), which is responsible for directing viral RNA replication and gene transcription. Notably, none of the compounds inhibited Gluc expression levels mediated by the host cellular RNA polymerase II (Fig. 8), suggesting that a rapidly replicating RNA virus is more sensitive than the host transcriptional machinery to changes in host cell nucleotide pools.
FIG 7.
Inhibition of viral replication and transcription. Human 293T cells (96-well plate format; 5.0 × 104 cells/well; quadruplicates) were transiently transfected with 125 ng of ambisense pDZ plasmids encoding PR8 PB2, PB1, PA, and NP, together with 250 ng of hpPol-I Gluc and hpPol-I GFP vRNA-like expression plasmids and 50 ng of pCAGGS-Cluc. Cells transfected in the absence of pDZ-PB2 were used as a negative control. After 6 h, transfection medium was replaced with medium containing 3-fold serial dilutions of the indicated compounds (starting concentration of 50 μM). At 24 h posttransfection, Gluc and Cluc expression levels were determined from tissue culture supernatants from transfected cells. Transfected cells were also imaged for GFP expression using a fluorescence microscope. Cells treated with 0.1% DMSO vehicle were used as an internal control. The EC50 was calculated as a percentage relative to values obtained with DMSO vehicle-treated cells. Dotted lines indicate 50% inhibition viral replication and transcription. Data are expressed as mean and SD from three independent experiments conducted in quadruplicate. Bar, 50 μm.
FIG 8.
Inhibition of cellular host gene expression. MDCK cells (96-well plates; 5.0 × 104 cells/well; quadruplicates) were transiently transfected with 50 ng of pCAGGS-Gluc plasmid. After 6 h, transfection medium was replaced with medium containing serial 3-fold dilutions (starting concentration 50 μM) of the indicated compounds. At 24 h posttransfection (hpt), Gluc expression levels were determined from tissue culture supernatants. Cells treated with 0.1% DMSO vehicle were used as an internal control. The EC50 was calculated as a percentage relative to values obtained with DMSO vehicle-treated cells. Dotted lines indicate 50% inhibition of reporter gene expression (Gluc). Data are expressed as mean and SD from three independent experiments conducted in quadruplicates.
Effect of the compounds on seasonal H1N1 and H3N2 IAVs, and IBV.
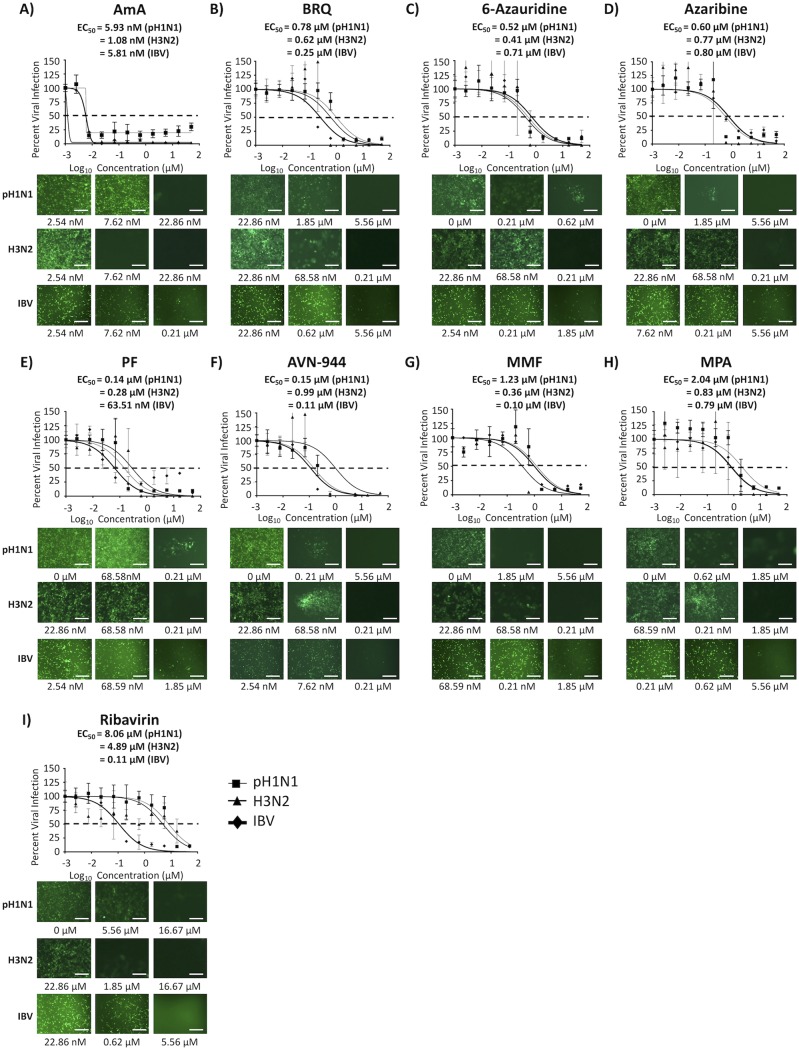
We next tested the ability of the compounds to inhibit seasonal H1N1 and H3N2 IAVs and IBV using a fluorescence-based microneutralization assay and Venus fluorescence-expressing pH1N1 and H3N2 IAVs and IBV (Fig. 9 and Table 4). The EC50 and SI values of the eight compounds with pH1N1-Venus, H3N2-Venus, and IBV-Venus (Fig. 9 and Table 4) were similar to those obtained with BIRFLU (except the SI-XTT value of AmA) (Fig. 3 and Table 1). These results indicated a broad-spectrum antiviral activity of the tested compounds against different types (IAV and IBV) and subtypes (H1N1 and H3N2) of influenza, including currently circulating human seasonal influenza viruses.
FIG 9.
Inhibition of seasonal H1N1 and H3N2 IAVs and IBV. Confluent monolayers of MDCK cells (96-well plates; 5.0 × 104 cells/well; quadruplicates) were infected with 200 FFU of the indicated Venus-expressing A/California/04/09 H1N1 (pH1N1) and A/Wyoming/3/03 H3N2 IAVs or with B/Brisbane/60/08 IBV. After 1 h of viral adsorption, the indicated concentrations (3-fold serial dilutions, starting concentration of 50 μM) of the different compounds or 0.1% DMSO vehicle control were added to the postinfection medium. Cells treated with 0.1% DMSO vehicle were used as an internal control. At 48 hpi, infected cells were evaluated for viral infection by Venus fluorescence expression using a fluorescence microscope or a fluorescent plate reader. Percent viral infection and the EC50 were calculated based on Nluc expression. Dotted lines indicate 50% viral inhibition. Data are expressed as mean and SD from three independent experiments conducted in quadruplicates. Bar, 50 μm.
TABLE 4.
Summary of CC50, EC50, and SI values of the compounds against seasonal H1N1 and H3N2 IAVs and IBV during posttreatment in MDCK cells
| Virusa | Compound | Activity statistics of compound against virusb
|
||||
|---|---|---|---|---|---|---|
| CC50 (MTT) (μM) | CC50 (XTT) | EC50 | SI (MTT) | SI (XTT) | ||
| pH1N1 | AmA | >50.00 | <2.54 nM | 5.93 nM | >8,431.70 | <0.43 |
| pH1N1 | BRQ | >50.00 | >50.00 μM | 0.78 μM | >64.10 | >64.10 |
| pH1N1 | 6-Azauridine | 42.66 | >50.00 μM | 0.52 μM | 82.04 | >96.15 |
| pH1N1 | Azaribine | 19.66 | >50.00 μM | 0.60 μM | 32.77 | >83.33 |
| pH1N1 | PF | 33.35 | >50.00 μM | 0.14 μM | 238.21 | >357.14 |
| pH1N1 | AVN-944 | 29.61 | >50.00 μM | 0.15 μM | 197.4 | >333.33 |
| pH1N1 | MMF | >50.00 | >50.00 μM | 1.23 μM | >40.65 | >40.65 |
| pH1N1 | MPA | >50.00 | >50.00 μM | 2.04 μM | >24.51 | >24.51 |
| pH1N1 | Ribavirin | >50.00 | >50.00 μM | 8.06 μM | >6.20 | >6.20 |
| H3N2 | AmA | >50.00 | <2.54 nM | 1.08 nM | >46,296.30 | <2.35 |
| H3N2 | BRQ | >50.00 | >50.00 μM | 0.62 μM | >80.65 | >80.65 |
| H3N2 | 6-Azauridine | 42.66 | >50.00 μM | 0.41 μM | 104.05 | >121.95 |
| H3N2 | Azaribine | 19.66 | >50.00 μM | 0.77 μM | 25.53 | >64.94 |
| H3N2 | PF | 33.35 | >50.00 μM | 0.28 μM | 119.11 | >178.57 |
| H3N2 | AVN-944 | 29.61 | >50.00 μM | 0.99 μM | 29.91 | >50.51 |
| H3N2 | MMF | >50.00 | >50.00 μM | 0.36 μM | 138.89 | 138.89 |
| H3N2 | MPA | >50.00 | >50.00 μM | 0.83 μM | 60.24 | 60.24 |
| H3N2 | Ribavirin | >50.00 | >50.00 μM | 4.89 μM | >10.22 | >10.22 |
| IBV | AmA | >50.00 | <2.54 nM | 5.81 nM | >8,605.85 | <0.44 |
| IBV | BRQ | >50.00 | >50.00 μM | 0.25 μM | >200.00 | >200.00 |
| IBV | 6-Azauridine | 42.66 | >50.00 μM | 0.71 μM | 60.08 | >70.42 |
| IBV | Azaribine | 19.66 | >50.00 μM | 0.80 μM | 24.57 | >62.50 |
| IBV | PF | 33.35 | >50.00 μM | 63.51 nM | 525.11 | >787.28 |
| IBV | AVN-944 | 29.61 | >50.00 μM | 0.11 μM | 269.18 | >454.54 |
| IBV | MMF | >50.00 | >50.00 μM | 0.10 μM | >500.00 | >500.00 |
| IBV | MPA | >50.00 | >50.00 μM | 0.79 μM | >63.29 | >63.29 |
| IBV | Ribavirin | >50.00 | >50.00 μM | 0.11 μM | >454.55 | >454.55 |
pH1N1, A/California/04/09 H1N1; H3N2, A/Wyoming/03/03 H3N2; IBV, B/Brisbane/60/08.
CC50, median 50% cytotoxicity concentration; EC50, median 50% effective concentration; SI, selective index (CC50/EC50).
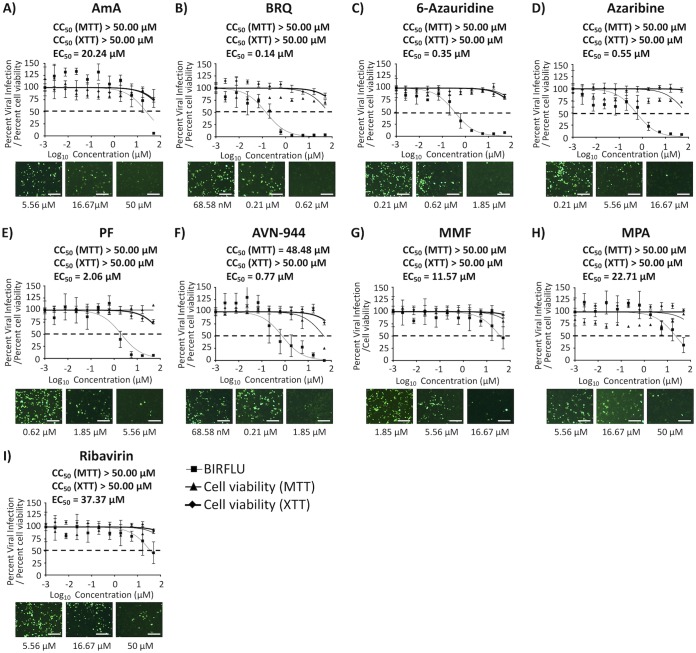
Effect of the compounds on IAV multiplication in human A549 cells.
To further examine the anti-influenza activity of these compounds for the treatment of influenza viral infections in humans, we evaluated their toxicity and antiviral activity in human alveolar A549 cells (Fig. 10). Four compounds, BRQ (Fig. 10B), 6-azauridine (Fig. 10C), azaribine (Fig. 10D), and AVN-944 (Fig. 10F), showed an EC50 of <1 μM and similar or higher SI-MTT and SI-XTT values in A549 than those observed in MDCK cells (Table 5). In contrast, AmA (Fig. 10A), PF (Fig. 10E), MMF (Fig. 10G), and MPA (Fig. 10H) showed lower SI values (except the SI-XTT of AmA) compared to those observed in MDCK cells (Table 1). Even though AmA showed alleviated SI values, it did not show any toxicity in the MTT and XTT assays (Fig. 10 and Table 5). These results support the consideration of 6-azauridine, azaribine, BRQ, and AVN-944 as repurposing candidate drugs for the treatment of influenza viral infections.
FIG 10.
Toxicity and inhibition of BIRFLU in human A549 cells. Confluent monolayers (96-well plates; 5.0 × 104 cells/well; quadruplicates) of human A549 cells were infected with 200 FFU of BIRFLU. After 1 h of viral adsorption, the indicated concentrations (3-fold serial dilutions, starting concentration of 50 μM) of the indicated compounds or 0.1% DMSO vehicle control were added to the postinfection media. At 48 hpi, tissue culture supernatants from infected A549 cells were collected and used to measure Nluc expression. Images of Venus expression were taken using a fluorescence microscope. Percent viral infection and the EC50 were calculated based on Nluc expression. Dotted lines indicate 50% viral inhibition. Data are expressed as mean and SD from three independent experiments conducted in quadruplicates. Bar, 50 μm.
TABLE 5.
Summary of CC50, EC50, and SI values of the compounds against BIRFLU during posttreatment in A549 cellsa
| Compound | Activity statistics of compound against BIRFLUb
|
||||
|---|---|---|---|---|---|
| CC50 (MTT) (μM) | CC50 (XTT) (μM) | EC50 (μM) | SI (MTT) | SI (XTT) | |
| AmA | >50.00 | >50.00 | 20.24 | >2.47 | >2.47 |
| BRQ | >50.00 | >50.00 | 0.14 | >357.14 | >357.14 |
| 6-Azauridine | >50.00 | >50.00 | 0.35 | >142.86 | >142.86 |
| Azaribine | >50.00 | >50.00 | 0.55 | >90.91 | >90.91 |
| PF | >50.00 | >50.00 | 2.06 | >24.27 | >24.27 |
| AVN-944 | 48.48 | >50.00 | 0.77 | 62.96 | 64.93 |
| MMF | >50.00 | >50.00 | 11.57 | >4.32 | >4.32 |
| MPA | >50.00 | >50.00 | 22.71 | >2.20 | >2.20 |
| Ribavirin | >50.00 | >50.00 | 37.37 | >1.33 | >1.33 |
BIRFLU, A/Puerto Rico/8/34 H1N1.
CC50, median 50% cytotoxicity concentration; EC50, median 50% effective concentration; SI, selective index (CC50/EC50).
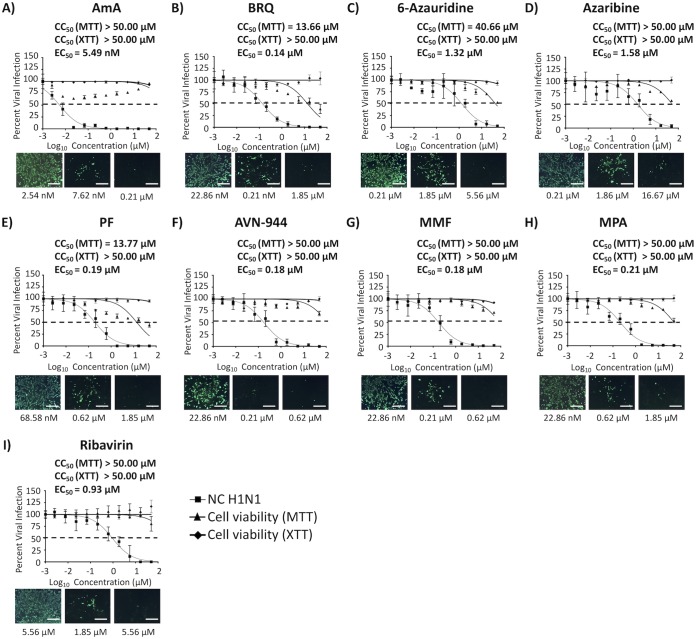
Effect of the compounds on IAV multiplication in 16HBE cells.
To examine the anti-influenza activity of these compounds in a more relevant cell substrate, we evaluated their toxicity and antiviral activity in primary human bronchial epithelial immortalized HBE (16HBE) cells with wild-type A/New Caledonia/20/1999 H1N1 (NC H1N1) (Fig. 11). Eight compounds showed potent inhibitory activity, with SI values ranging from 30.80 to 9,107.46 (Table 6). Among them, five compounds, AmA (Fig. 11A), BRQ (Fig. 11B), AVN-944 (Fig. 11F), MMF (Fig. 11G), and MPA (Fig. 11H), showed an EC50 of <1 μM and similar or higher SI values in 16HBE cells than those observed in MDCK cells (Table 6). As with A549 cells, AmA did not show any toxicity in the MTT and XTT assays and showed potent SI values in 16HBE cells (Fig. 11 and Table 6). The SI values of six compounds (i.e., those other than 6-azauridine and azaribine), were higher than those obtained with ribavirin, a well-characterized viral inhibitor in various influenza studies (23–25).
FIG 11.
Toxicity and inhibition of A/New Caledonia/20/1999 H1N1 in 16HBE cells. Confluent monolayers (96-well plate format; 5.0 × 104 cells/well; quadruplicates) of primary human bronchial epithelial (HBE) immortalized (16HBE) cells were infected with 50 PFU of A/New Caledonia/20/1999 H1N1 (NC H1N1). After 1 h of viral adsorption, the indicated concentrations (3-fold serial dilutions, starting concentration of 50 μM) of the different compounds or 0.1% DMSO vehicle control with 1% Avicel were added to the postinfection medium. At 72 hpi, cells were fixed and immunostained with anti-NP monoclonal antibody (MAb) (HB-65) and followed by fluorescein isothiocyanate (FITC)-conjugated secondary antibody (Ab). Images of FITC Ab were taken using a fluorescence microscope. Percent viral infection and the EC50 were calculated based on Nluc expression. Dotted lines indicate 50% viral inhibition. Data are expressed as mean and SD from three independent experiments conducted in quadruplicates. Bar, 50 μm.
TABLE 6.
Summary of CC50, EC50, and SI values of the compounds against NC H1N1 during posttreatment in 16HBE cellsa
| Compound | Activity statistics of compound against NC H1N1b
|
||||
|---|---|---|---|---|---|
| CC50 (MTT) (μM) | CC50 (XTT) (μM) | EC50 | SI (MTT) | SI (XTT) | |
| AmA | >50.00 | >50.00 | 5.49 nM | >9,107.46 | >9,107.46 |
| BRQ | >13.66 | >50.00 | 0.14 μM | >97.57 | >357.14 |
| 6-Azauridine | 40.66 | >50.00 | 1.32 μM | 30.80 | >37.88 |
| Azaribine | >50.00 | >50.00 | 1.58 μM | >31.65 | >31.65 |
| PF | 13.77 | >50.00 | 0.19 μM | 81.00 | >263.16 |
| AVN-944 | >50.00 | >50.00 | 0.18 μM | >277.78 | >277.78 |
| MMF | >50.00 | >50.00 | 0.18 μM | >277.78 | >277.78 |
| MPA | >50.00 | >50.00 | 0.21 μM | >238.10 | >238.10 |
| Ribavirin | >50.00 | >50.00 | 0.93 μM | >53.76 | >53.76 |
NC H1N1, A/New Caledonia/20/1999 H1N1.
CC50, median 50% cytotoxicity concentration; EC50, median 50% effective concentration; SI, selective index (CC50/EC50).
DISCUSSION
In this study, we investigated the ability of ten compounds to inhibit influenza infection (Fig. 1). The compounds were selected based on their ability to inhibit mammarenavirus (LCMV) infection (20) and on evidence that efficient multiplication of viruses with a negative-sense RNA segmented genome involves some common cellular pathways that could be targeted by existing compounds, including those targeting reactive oxygen species (ROS) production (AmA), pyrimidine synthesis (BRQ, 6-azauridine, azaribine, and PF), GMP synthesis (AVN-944, MMF, and MPA), and apoptosis (obatoclax and Osu-03012). All of the compounds, except obatoclax and Osu-03012, exhibited different levels of safety and antiviral activity against IAV infection (Fig. 2 to 4 and Table 1), with SI-MTT values ranging from 28.90 to 13,157.89 and SI-XTT values (except that of AmA) ranging from 28.90 to 1,285.35 in posttreatment settings (Table 1). We observed differences in antiviral activity when the compounds were used in pretreatment and cotreatment experiments (Fig. 5 and 6 and Tables 2 and 3). Our results indicated that compounds with antiviral activity targeted viral genome replication or gene transcription or both (Fig. 7) without affecting RNA polymerase II mediated cellular transcription (Fig. 8). Our findings support the feasibility of using these compounds for the treatment of seasonal influenza viruses, including currently circulating human H1N1 and H3N2 IAVs and IBV (Fig. 9). Comparable CC50 (MTT and XTT) and EC50 values were obtained in A549 (Fig. 10 and Table 5) and 16HBE cells (Fig. 11 and Table 6), supporting the feasibility of repurposing these drugs for the treatment of influenza viral infections in humans.
The different compounds could be grouped into five different categories based on their potential mechanism of antiviral activity. The first group included AmA, a known inhibitor of de novo ATP synthesis (26). AmA is known to inhibit the cellular mETC III, resulting in generation of ROS and suppression of production of ATP (26, 27). AmA has been described to have antiviral activity against multiple viruses, including porcine reproductive and respiratory syndrome virus (PRRSV) (28), dengue virus (DENV) (29), Venezuelan and Western equine encephalitis viruses (VEEV and WEEV, respectively), La Crosse virus (LACV), vesicular stomatitis virus (VSV), encephalomyocarditis virus (EMCV), Sendai virus (SeV), and hepatitis C virus (HCV) (26). Likewise, AmA has been described as inhibiting influenza viral budding (30) and protein expression (31). Although these previous findings imply that the antiviral effects of AmA take place only after the virus enters the cells, our results also demonstrate that AmA inhibits influenza infection in pretreated or cotreated settings (Fig. 5 and 6). Although AmA showed the most potent EC50 values in MDCK and 16HBE cells, the SI values were different in MDCK cells because of the different results with the cell viability assays (SI-MTT > 13,157.89 versus SI-XTT < 0.66) (Table 1). Although both MTT and XTT assays measure cellular proliferation (32), the MTT assay is influenced not only by the mitochondrial electron chain but also by nonmitochondrial components and processes, such as oxidoreductases, superoxides, and/or glycolysis (33). It is possible that these differences are responsible for the different CC50 values for AmA in MDCK cells in the MTT (CC50 > 50.00 μM) and XTT (CC50 < 2.54 nM) assays. Importantly, as shown in Fig. 8, AmA did not inhibit RNA polymerase II activity in human 293T cells or cellular proliferation (both MTT and XTT) in human A549 or 16HBE cells (Fig. 10 and 11, respectively, and Tables 5 and 6, respectively), suggesting the feasibility of using AmA for the treatment of influenza infections in humans. It is also possible that the different CC50 values for AmA in canine (MDCK) and human (A549 and 16HBE) cells with the XTT assay are cell specific.
The second group of compounds included BRQ, a known inhibitor of DHODH, a key enzyme of the pyrimidine biosynthesis pathway (34). Previous studies have described the antiviral effect of BRQ against yellow fever virus (YFV) (34), Kunjin virus (34), DENV (34, 35), and IAV (36). In the case of IAV, BRQ was reported to have an EC50 of 2.6 μM and an SI of >3.8 against an influenza A/WSN/33 H1N1 strain expressing Nluc in a human bronchial epithelial BEAS-2B cell line (36). These values are different than those observed in our studies with PR8 BIRFLU (EC50 = 0.58 μM and SI > 86.21 in MDCK cells; EC50 = 0.14 μM and SI > 357.14 in A549 cells), pH1N1 (EC50 = 0.78 μM and SI > 64.10 in MDCK cells), H3N2 (EC50 = 0.62 μM and SI > 80.65 in MDCK cells), and IBV (EC50 = 0.25 μM and SI > 200.00 in MDCK cells) (Table 4), which likely reflects differences in virus strains, cell types, and/or assay conditions.
The third group included the orotidine monophosphate decarboxylase (OMPD) inhibitors 6-azauridine, azaribine, and PF (34, 37–39). OMPD catalyzes key steps in pyrimidine synthesis, and, as with the DHODH inhibitors, these compounds inhibit de novo pyrimidine synthesis (34). Pyrimidine biosynthesis inhibitors have shown very limited in vivo efficacy as antiviral drugs (40, 41), likely reflecting that the pyrimidine salvage pathway can provide infected cells with levels of pyrimidine pools able to counteracting the effect of pyrimidine de novo biosynthesis inhibitors. Therefore, targeting the pyrimidine salvage pathway might enhance the antiviral effect of pyrimidine biosynthesis inhibitors in vivo. This strategy is supported by the fact that interfering with the activity of uridine-cytidine kinase 2 (UCK2), a key enzyme of the pyrimidine salvage pathway, sensitized cells to treatment with a DHODH inhibitor (42). 6-Azauridine is metabolized from azaribine and is known to inhibit a wide range of RNA viruses, including WNV (43, 44), YFV (44), Langat virus (LGTV) (44), Wesselsbron virus (WESSV) (44), Zika virus (ZIKV) (44), Usutu virus (USUV) (44), Rift Valley fever virus (RVFV) (45), and IAV (46, 47). PF was isolated from Streptomyces candidus (48) and is reported to have broad-spectrum antimicrobial activities (37, 49, 50).
The fourth group included the IMPDH inhibitors AVN-944, MMF, and MPA, which, similarly to ribavirin, inhibit the replication of DNA and RNA viruses via reduction of GTP pools in infected cells (51–54). IMPDH is a key enzyme in the de novo synthesis of GMP, and its inhibition by AVN-944, MMF, and MPA results in depleted pools of intracellular GMP (54). AVN-944 has been developed mainly for the treatment of cancer (55). It was tested in a human phase I clinical trial in doses of up to 250 mg without showing serious side effects (56). MMF, a prodrug of MPA, is an immunosuppressive agent that is commonly used as a prophylactic for allograft rejections in kidney, cardiac, or liver transplants (54, 57, 58). MMF and MPA have been described to have antiviral activity against IAV (MMF and MPA) and IBV (MPA) (51–53). MMF and MPA exhibited similar inhibitory activities, with EC50 values of 0.208 μM against B/Hong Kong/411989/2011 (53), 0.24 μM against A/WSN/1933 H1N1 (52), and 1.51 μM against A/Hong Kong/415742/2009 H1N1 (53) in the case of MPA; and 0.94 μM against A/Vietnam/1194/2004 H5N1 in the case of MMF (51).
The fifth group included the inducers of apoptosis obatoclax and Osu-03012 (59, 60). Obatoclax has been described to have antitumor activity in several hematologic malignant tumors, including leukemia and myelodysplasia (60, 61). Osu-03012 is a derivative of celecoxib (59, 62) and has been previously shown to inhibit mumps, IAV and IBV, measles, Junín virus, rubella virus, and HIV infections by stimulating autophagosome formation (62). However, we did not observe any antiviral activity against IAV in our study.
MDCK cells are the gold standard cell line to study influenza virus infection in multiple laboratories, but we also investigated the toxicity and antiviral activity of the compounds in more relevant human A549 and 16HBE cells (Fig. 10 and 11, respectively). While all of the compounds had antiviral activity against BIRFLU in human A549 and 16HBE cells, their antiviral effects in human cells were different from those observed in MDCK cells. Previous studies have already reported that influenza virus replicates to a lesser extent in A549 or 16HBE than in MDCK cells, with viral titers being lower in A549 and 16HBE than in MDCK cells (63, 64). Differences in virus replication may account for the observed differences in antiviral activity of the compounds in these three cell lines (Tables 1, 5, and 6).
In conclusion, our study has identified AmA, BRQ, 6-azauridin, azaribine, PF, AVN-944, MMF, and MPA as drugs that should be further investigated for their potential repurposing for the treatment of IAV and IBV infections. Moreover, and due to the different mechanisms of antiviral activity, it is feasible that a mixture of these compounds, alone or in combination with currently in-use influenza antivirals, could be implemented for the treatment of influenza infections. Future pharmacologic and in vivo studies will need to be undertaken to demonstrate the antiviral activity of the compounds in validated animal models for the feasibility of treating influenza viral infections. Likewise, studies aimed at identifying potential drug-resistant mutants will further support the feasibility of treating influenza viral infections with these new antiviral compounds, but since these antivirals target cellular host proteins that are important for influenza viral replication rather than the virus itself, it is unlikely that IAV and/or IBV could escape the antiviral effect of these compounds. Importantly, identification of new host factors required by these and other antivirals against influenza infection could also reveal new cellular pathways that could be targeted for the rational design of new antivirals for the treatment of influenza infections.
MATERIALS AND METHODS
Cell lines.
Madin-Darby canine kidney (MDCK) (ATCC CCL-34), primary human bronchial epithelial (HBE) immortalized (16HBE), human adenocarcinoma alveolar basal epithelial A549 (ATCC CCL-185), and human embryonic kidney 293T (ATCC CRL-11268) cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM; Mediatech, Inc.) supplemented with 5% fetal bovine serum (FBS) and 1% PSG (100 U/ml penicillin, 100 μg/ml streptomycin, and 2 mM l-glutamine) at 37°C in a 5% CO2 atmosphere (65).
Influenza viruses.
Influenza A/Puerto Rico/8/34 (PR8) H1N1 bireporter influenza A virus (BIRFLU) expressing two reporter genes (Venus and Nano luciferase [Nluc]) (21), pandemic influenza A/California/04/2009 expressing Venus (pH1N1-Venus) (66), and influenza B/Brisbane/60/2008 expressing Venus (IBV-Venus) (67) were prepared as previously described (21, 66, 67). Influenza A/Wyoming/3/2003 expressing Venus (H3N2-Venus) was generated and prepared as previously described (68). Influenza A/New Caledonia/20/1999 H1N1 (NC H1N1) was propagated in MDCK cells as previously described (23).
Compounds.
Antimycin A (AmA, catalog no. A8674; Sigma-Aldrich), 6-azauridine (catalog no. A1882; Sigma-Aldrich), azaribine (catalog no. T340057; Sigma-Aldrich), brequinar (BRQ, catalog no. SML0113; Sigma-Aldrich), AVN-944 (catalog no. A13652; AdooQ Bio), mycophenolate mofetil, (MMF, catalog no. J90063; AkSci), mycophenolic acid (MPA, catalog no. E480; AkSci), pyrazofurin (PF, catalog no. SLM1502; Sigma-Aldrich,), obatoclax (catalog no. V2454; AkSci), Osu-03012 (catalog no. Y0267; AkSci), and ribavirin (catalog no. AK-49185; Ark Pharmer) were prepared as a 10 mM stock solution in dimethyl sulfoxide (DMSO) and kept at −20°C until experimental use. Each drug was diluted in infectious DMEM supplemented with 2% FBS and 1% PSG medium at the maximum DMSO concentration of 0.1%.
Cell viability assay.
MDCK, 16HBE, and A549 cell viabilities were determined using the MTT assay (CellTiter 96 nonradioactive cell proliferation assay; Promega) and/or the XTT assay (cell viability and proliferation assay, Sigma-Aldrich) following the manufacturer’s instructions and as described previously (32). Briefly, confluent monolayers (96-well plate format; 5.0 × 104 cells/well; quadruplicates) of MDCK or A549 cells were treated with 100 μl of DMEM containing serially diluted (3-fold dilutions, starting concentration of 50 μM) compounds or with 0.1% DMSO (vehicle control). Plates were incubated at 37°C in a 5% CO2 atmosphere for 48 h. Cells and supernatants were treated with either 15 μl of dye solution for the MTT assay or 100 μl of XTT labeling reagent for the XTT assay and incubated at 37°C in a 5% CO2 atmosphere for 4 h. Next, absorbance at 570 nm of cells was measured using a VMax kinetic microplate reader (Molecular Devices, Waltham, MA). Viability of compound-treated cells was calculated as a percentage relative to values obtained with DMSO vehicle-treated cells. Nonlinear regression curves and the median cytotoxic concentration (CC50) were calculated using GraphPad Prism software version 8.0 (32).
Virus inhibition assay.
Confluent monolayers (96-plate format; 5.0 × 104 cells/well; quadruplicates) of MDCK or A549 cells were infected with 200 fluorescence-forming units (FFU)/well of BIRFLU, H3N2-Venus, pH1N1-Venus, or IBV-Venus at room temperature for 1 h. 16HBE cells were similarly infected with 50 PFU/well of NC H1N1 at room temperature for 1 h. After viral absorption, cells were washed three times with phosphate-buffered saline (PBS) and incubated with virus infection medium containing 1 μg/ml of N-tosyl-l-phenylalanine chloromethyl ketone (TPCK)-treated trypsin (Sigma). The different compounds or 0.1% DMSO vehicle control were used to treat cells either 1 h before virus infection (pretreatment), simultaneously with virus inoculum (cotreatment), or after virus absorption (posttreatment). Infected cells were incubated at 33°C for 48 h. In the case of BIRFLU, tissue culture supernatants were collected and used to measure Nluc expression using the Nano-Glo luciferase substrate (Promega) and a Lumicount luminometer. For the inhibition assay in 16HBE cells with NC H1N1, a microplaque reduction assay was used. Briefly, infected cells were incubated at 37°C for 72 h with infection medium containing the indicated concentrations and 1% Avicel. Then, cells were fixed for immunostaining with an anti-nucleoprotein (anti-NP) monoclonal antibody (MAb) (HB-65) as the primary antibody (Ab), and incubated with fluorescein isothiocyanate (FITC)-conjugated rabbit anti-mouse IgG secondary antibody (Dako) as the secondary Ab. Stained plaques were counted and the percentage of viral infection was calculated. In the cases of Venus-expressing pH1N1, H3N2, and IBV, cells were washed with PBS, and Venus expression levels were measured using a fluorescence plate reader (DTX-880; Becton, Dickinson). Percent of viral infection was calculated based on Nluc or Venus measurements. Images of immunostaining or Venus expression were obtained using a fluorescence microscope (Nikon Eclipse TE2000). Individual wells in the quadruplicates were used to calculate the average and standard deviation (SD) of viral inhibition using Microsoft Excel software. The median inhibitory concentration (EC50) was determined by a sigmoidal dose response curve (GraphPad Prism, version 8.0).
Virus growth kinetics inhibition.
Multicycle virus growth kinetic inhibitions were evaluated in confluent monolayers of MDCK cells (24-well plate format; 2.5 × 105 cells/well; triplicates) infected with BIRFLU at a multiplicity of infection (MOI) of 0.01. After 1 h of viral absorption at room temperature, cells were washed with PBS and incubated with infection medium containing the indicated concentrations (0.1, 1, and 10 EC50) of each compound. At 24, 48, and 72 h postinfection (hpi), viruses in culture supernatants were determined using an immunofocus assay (fluorescence-forming units [FFU]/ml) (24).
Hemagglutination and hemagglutinin inhibition assays.
HA (hemagglutination) and HAI (hemagglutinin inhibition) assays were used to determine the HA-neutralizing ability of the compounds (69). For both assays, the compounds were serially diluted (2-fold, starting concentration of 1,000 μM) in 96-well V-bottom plates. HA titers were determined by adding 0.5% turkey red blood cells (RBCs) to the serially diluted compounds for 30 min on ice. For the HAI assay, four hemagglutinating units (HAU) of BIRFLU were added to the serially diluted compounds and incubated for 60 min at room temperature. HAI titers were evaluated after adding 0.5% turkey RBCs to the virus-compound mixtures for 30 min on ice. The HAI titer was defined as the minimum concentration of the compound that inhibited hemagglutination.
Minigenome assay.
To analyze the ability of the compounds to inhibit influenza polymerase replication and transcription activity, we used a previously described minigenome (MG) assay (65). Briefly, human 293T cells (5.0 × 105 cells/well; 12-well plate format; quadruplicates) were transiently cotransfected in suspension, using Lipofectamine 2000 (LPF2000; Invitrogen) with 125 ng of each of the ambisense pDZ-PB2, pDZ-PB1, pDZ-PA, and pDZ-NP plasmids; 250 ng of an IAV MG viral RNA (vRNA)-like expression plasmids encoding Gaussia luciferase (Gluc) or GFP driven by a human RNA polymerase I promoter (hpPol-I Gluc and hpPol-I GFP, respectively); and 50 ng of a plasmid constitutively expressing Cypridina luciferase (Cluc) (pCAGGS-Cluc). Cells transfected in the absence of pDZ-PB2 were used as a negative control. At 6 h posttransfection (hpt), transfection medium was replaced with medium containing serially diluted compounds (3-fold dilutions, starting concentration of 50 μM) and incubated at 33°C. At 24 hpt, Gluc and Cluc expression levels were determined using the BioLux Gaussia luciferase assay kit (New England BioLabs) and the BioLux Cypridina luciferase assay kit (New England BioLabs), using a Lumicount luminometer (Packard). The mean value and SD value were calculated using Microsoft Excel software.
Inhibition of host gene expression.
To evaluate the effect of the compounds on host protein synthesis, MDCK cells (5.0 × 104 cells/well; 96-well plates; quadruplicates) were transiently transfected, using LPF2000, with 50 ng of a plasmid expressing Gluc under a polymerase II-dependent promoter (pCAGGS-Gluc). After 6 h, transfection medium was replaced with medium containing serial dilutions (3-fold dilutions, starting concentration of 50 μM) of the indicated compounds. At 24 hpt, Gluc expression levels were determined from tissue culture supernatants using the BioLux Gaussia luciferase assay kit (New England BioLabs) and a Lumicount luminometer (Packard) (65). The mean value and SD value were calculated using Microsoft Excel software.
Statistical analysis.
The unpaired Student’s t test was used to evaluate significant differences. Data of at least three independent experiments in quadruplicates are expressed as the mean ± standard deviation (SD), which were calculated using Microsoft Excel software. Values were considered statistically significant when P < 0.05 (*), P < 0.01 (***), or P < 0.001 (***), or were not significant (n.s.). All data were analyzed using Prism software version 8.0 (GraphPad Software, CA, USA). CC50 and EC50 were drawn using sigmoidal dose response curves (GraphPad Software), and the selective index (SI) of each compound was evaluated by dividing the CC50 by the EC50.
Supplementary Material
ACKNOWLEDGMENT
This research was partially funded by the New York Influenza Center of Excellence (NYICE), a member of the National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health (NIH), Department of Health and Human Services, Centers of Excellence for Influenza Research and Surveillance (CEIRS) contract no. HHSN272201400005C (NYICE) and by Department of Defense (DoD) Peer Reviewed Medical Research Program (PRMRP) grant W81XWH-18-1-0460 to L.M.-S.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Knipe DM, Howley PM, Cohen JI, Griffin DE, Lamb RA, Martin MA, Racaniello VR, Roizman B. 2013. Fields virology, 6th ed Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 2.Clark AM, DeDiego ML, Anderson CS, Wang J, Yang H, Nogales A, Martinez-Sobrido L, Zand MS, Sangster MY, Topham DJ. 2017. Antigenicity of the 2015–2016 seasonal H1N1 human influenza virus HA and NA proteins. PLoS One 12:e0188267. doi: 10.1371/journal.pone.0188267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Molinari NA, Ortega-Sanchez IR, Messonnier ML, Thompson WW, Wortley PM, Weintraub E, Bridges CB. 2007. The annual impact of seasonal influenza in the US: measuring disease burden and costs. Vaccine 25:5086–5096. doi: 10.1016/j.vaccine.2007.03.046. [DOI] [PubMed] [Google Scholar]
- 4.Federici C, Cavazza M, Costa F, Jommi C. 2018. Health care costs of influenza-related episodes in high income countries: a systematic review. PLoS One 13:e0202787. doi: 10.1371/journal.pone.0202787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barr IG, McCauley J, Cox N, Daniels R, Engelhardt OG, Fukuda K, Grohmann G, Hay A, Kelso A, Klimov A, Odagiri T, Smith D, Russell C, Tashiro M, Webby R, Wood J, Ye Z, Zhang W, Writing Committee of the World Health Organization Consultation on Northern Hemisphere Influenza Vaccine Composition for June 2009–2010 . 2010. Epidemiological, antigenic and genetic characteristics of seasonal influenza A(H1N1), A(H3N2) and B influenza viruses: basis for the WHO recommendation on the composition of influenza vaccines for use in the 2009–2010 northern hemisphere season. Vaccine 28:1156–1167. doi: 10.1016/j.vaccine.2009.11.043. [DOI] [PubMed] [Google Scholar]
- 6.Parrish CR, Murcia PR, Holmes EC. 2015. Influenza virus reservoirs and intermediate hosts: dogs, horses, and new possibilities for influenza virus exposure of humans. J Virol 89:2990–2994. doi: 10.1128/JVI.03146-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tong S, Zhu X, Li Y, Shi M, Zhang J, Bourgeois M, Yang H, Chen X, Recuenco S, Gomez J, Chen LM, Johnson A, Tao Y, Dreyfus C, Yu W, McBride R, Carney PJ, Gilbert AT, Chang J, Guo Z, Davis CT, Paulson JC, Stevens J, Rupprecht CE, Holmes EC, Wilson IA, Donis RO. 2013. New world bats harbor diverse influenza A viruses. PLoS Pathog 9:e1003657. doi: 10.1371/journal.ppat.1003657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tong S, Li Y, Rivailler P, Conrardy C, Castillo DA, Chen LM, Recuenco S, Ellison JA, Davis CT, York IA, Turmelle AS, Moran D, Rogers S, Shi M, Tao Y, Weil MR, Tang K, Rowe LA, Sammons S, Xu X, Frace M, Lindblade KA, Cox NJ, Anderson LJ, Rupprecht CE, Donis RO. 2012. A distinct lineage of influenza A virus from bats. Proc Natl Acad Sci U S A 109:4269–4274. doi: 10.1073/pnas.1116200109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Parrish CR, Kawaoka Y. 2005. The origins of new pandemic viruses: the acquisition of new host ranges by canine parvovirus and influenza A viruses. Annu Rev Microbiol 59:553–586. doi: 10.1146/annurev.micro.59.030804.121059. [DOI] [PubMed] [Google Scholar]
- 10.Nogales A, Perez DR, Santos J, Finch C, Martinez-Sobrido L. 2017. Reverse genetics of influenza B viruses. Methods Mol Biol 1602:205–238. doi: 10.1007/978-1-4939-6964-7_14. [DOI] [PubMed] [Google Scholar]
- 11.Chen R, Holmes EC. 2008. The evolutionary dynamics of human influenza B virus. J Mol Evol 66:655–663. doi: 10.1007/s00239-008-9119-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smith GJ, Vijaykrishna D, Bahl J, Lycett SJ, Worobey M, Pybus OG, Ma SK, Cheung CL, Raghwani J, Bhatt S, Peiris JS, Guan Y, Rambaut A. 2009. Origins and evolutionary genomics of the 2009 swine-origin H1N1 influenza A epidemic. Nature 459:1122–1125. doi: 10.1038/nature08182. [DOI] [PubMed] [Google Scholar]
- 13.Mifsud EJ, Hayden FG, Hurt AC. 2019. Antivirals targeting the polymerase complex of influenza viruses. Antiviral Res 169:104545. doi: 10.1016/j.antiviral.2019.104545. [DOI] [PubMed] [Google Scholar]
- 14.Earhart KC, Elsayed NM, Saad MD, Gubareva LV, Nayel A, Deyde VM, Abdelsattar A, Abdelghani AS, Boynton BR, Mansour MM, Essmat HM, Klimov A, Shuck-Lee D, Monteville MR, Tjaden JA. 2009. Oseltamivir resistance mutation N294S in human influenza A(H5N1) virus in Egypt. J Infect Public Health 2:74–80. doi: 10.1016/j.jiph.2009.04.004. [DOI] [PubMed] [Google Scholar]
- 15.Omoto S, Speranzini V, Hashimoto T, Noshi T, Yamaguchi H, Kawai M, Kawaguchi K, Uehara T, Shishido T, Naito A, Cusack S. 2018. Characterization of influenza virus variants induced by treatment with the endonuclease inhibitor baloxavir marboxil. Sci Rep 8:9633. doi: 10.1038/s41598-018-27890-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takashita E, Kawakami C, Morita H, Ogawa R, Fujisaki S, Shirakura M, Miura H, Nakamura K, Kishida N, Kuwahara T, Mitamura K, Abe T, Ichikawa M, Yamazaki M, Watanabe S, Odagiri T, Influenza Virus Surveillance Group of Japan . 2019. Detection of influenza A(H3N2) viruses exhibiting reduced susceptibility to the novel cap-dependent endonuclease inhibitor baloxavir in Japan, December 2018. Euro Surveill 24:pii=1800698 https://www.eurosurveillance.org/content/10.2807/1560-7917.ES.2019.24.3.1800698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ashburn TT, Thor KB. 2004. Drug repositioning: identifying and developing new uses for existing drugs. Nat Rev Drug Discov 3:673–683. doi: 10.1038/nrd1468. [DOI] [PubMed] [Google Scholar]
- 18.Pushpakom S, Iorio F, Eyers PA, Escott KJ, Hopper S, Wells A, Doig A, Guilliams T, Latimer J, McNamee C, Norris A, Sanseau P, Cavalla D, Pirmohamed M. 2019. Drug repurposing: progress, challenges and recommendations. Nat Rev Drug Discov 18:41–58. doi: 10.1038/nrd.2018.168. [DOI] [PubMed] [Google Scholar]
- 19.Janes J, Young ME, Chen E, Rogers NH, Burgstaller-Muehlbacher S, Hughes LD, Love MS, Hull MV, Kuhen KL, Woods AK, Joseph SB, Petrassi HM, McNamara CW, Tremblay MS, Su AI, Schultz PG, Chatterjee AK. 2018. The ReFRAME library as a comprehensive drug repurposing library and its application to the treatment of cryptosporidiosis. Proc Natl Acad Sci U S A 115:10750–10755. doi: 10.1073/pnas.1810137115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim YJ, Cubitt B, Chen E, Hull MV, Chatterjee AK, Cai Y, Kuhn JH, de la Torre JC. 2019. The ReFRAME library as a comprehensive drug repurposing library to identify mammarenavirus inhibitors. Antiviral Res 169:104558. doi: 10.1016/j.antiviral.2019.104558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nogales A, Ávila-Pérez G, Rangel-Moreno J, Chiem K, DeDiego ML, Martínez-Sobrido L. 2019. A novel fluorescent and bioluminescent bi-reporter influenza A virus (BIRFLU) to evaluate viral infections. J Virol 93:e00032-19. doi: 10.1128/JVI.00032-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Markland W, McQuaid TJ, Jain J, Kwong AD. 2000. Broad-spectrum antiviral activity of the IMP dehydrogenase inhibitor VX-497: a comparison with ribavirin and demonstration of antiviral additivity with alpha interferon. Antimicrob Agents Chemother 44:859–866. doi: 10.1128/aac.44.4.859-866.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nogales A, Baker SF, Martinez-Sobrido L. 2015. Replication-competent influenza A viruses expressing a red fluorescent protein. Virology 476:206–216. doi: 10.1016/j.virol.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nogales A, Rodríguez-Sánchez I, Monte K, Lenschow DJ, Perez DR, Martínez-Sobrido L. 2016. Replication-competent fluorescent-expressing influenza B virus. Virus Res 213:69–81. doi: 10.1016/j.virusres.2015.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pagadala NS. 2019. AZT acts as an anti-influenza nucleotide triphosphate targeting the catalytic site of A/PR/8/34/H1N1 RNA dependent RNA polymerase. J Comput Aided Mol Des 33:387–404. doi: 10.1007/s10822-019-00189-w. [DOI] [PubMed] [Google Scholar]
- 26.Raveh A, Delekta PC, Dobry CJ, Peng W, Schultz PJ, Blakely PK, Tai AW, Matainaho T, Irani DN, Sherman DH, Miller DJ. 2013. Discovery of potent broad spectrum antivirals derived from marine actinobacteria. PLoS One 8:e82318. doi: 10.1371/journal.pone.0082318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Quinlan CL, Gerencser AA, Treberg JR, Brand MD. 2011. The mechanism of superoxide production by the antimycin-inhibited mitochondrial Q-cycle. J Biol Chem 286:31361–31372. doi: 10.1074/jbc.M111.267898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karuppannan AK, Wu KX, Qiang J, Chu JJ, Kwang J. 2012. Natural compounds inhibiting the replication of porcine reproductive and respiratory syndrome virus. Antiviral Res 94:188–194. doi: 10.1016/j.antiviral.2012.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shum D, Smith JL, Hirsch AJ, Bhinder B, Radu C, Stein DA, Nelson JA, Fruh K, Djaballah H. 2010. High-content assay to identify inhibitors of dengue virus infection. Assay Drug Dev Technol 8:553–570. doi: 10.1089/adt.2010.0321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hui E-W, Nayak D. 2001. Role of ATP in influenza virus budding. Virology 290:329–341. doi: 10.1006/viro.2001.1181. [DOI] [PubMed] [Google Scholar]
- 31.Hao L, Sakurai A, Watanabe T, Sorensen E, Nidom CA, Newton MA, Ahlquist P, Kawaoka Y. 2008. Drosophila RNAi screen identifies host genes important for influenza virus replication. nature 454:890–893. doi: 10.1038/nature07151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aslantürk ÖS. 2018. In vitro cytotoxicity and cell viability assays: principles, advantages, and disadvantages, vol 2 InTech, London, UK. [Google Scholar]
- 33.Stepanenko AA, Dmitrenko VV. 2015. Pitfalls of the MTT assay: direct and off-target effects of inhibitors can result in over/underestimation of cell viability. Gene 574:193–203. doi: 10.1016/j.gene.2015.08.009. [DOI] [PubMed] [Google Scholar]
- 34.Qing M, Zou G, Wang QY, Xu HY, Dong H, Yuan Z, Shi PY. 2010. Characterization of dengue virus resistance to brequinar in cell culture. Antimicrob Agents Chemother 54:3686–3695. doi: 10.1128/AAC.00561-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang QY, Bushell S, Qing M, Xu HY, Bonavia A, Nunes S, Zhou J, Poh MK, Florez de Sessions P, Niyomrattanakit P, Dong H, Hoffmaster K, Goh A, Nilar S, Schul W, Jones S, Kramer L, Compton T, Shi PY. 2011. Inhibition of dengue virus through suppression of host pyrimidine biosynthesis. J Virol 85:6548–6556. doi: 10.1128/JVI.02510-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yan D, Weisshaar M, Lamb K, Chung HK, Lin MZ, Plemper R. 2015. Replication-competent influenza virus and respiratory syncytial virus luciferase reporter strains engineered for co-infections identify antiviral compounds in combination screens. Biochemistry 54:5589–5604. doi: 10.1021/acs.biochem.5b00623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.De Clercq E. 2016. C-nucleosides to be revisited. J Med Chem 59:2301–2311. doi: 10.1021/acs.jmedchem.5b01157. [DOI] [PubMed] [Google Scholar]
- 38.McDonald CJ. 1981. The uses of systemic chemotherapeutic agents in psoriasis. Pharmacol Ther 14:1–24. doi: 10.1016/0163-7258(81)90008-5. [DOI] [PubMed] [Google Scholar]
- 39.Slavik M. 1979. Changes in amino acid metabolism caused by 6-azauridine triacetate: relevance to cancer treatment. Cancer Treat Rep 63:1041–1044. [PubMed] [Google Scholar]
- 40.Cheung NN, Lai KK, Dai J, Kok KH, Chen H, Chan KH, Yuen KY, Kao R. 2017. Broad-spectrum inhibition of common respiratory RNA viruses by a pyrimidine synthesis inhibitor with involvement of the host antiviral response. J Gen Virol 98:946–954. doi: 10.1099/jgv.0.000758. [DOI] [PubMed] [Google Scholar]
- 41.Davis IC, Lazarowski ER, Chen FP, Hickman-Davis JM, Sullender WM, Matalon S. 2007. Post-infection A77-1726 blocks pathophysiologic sequelae of respiratory syncytial virus infection. Am J Respir Cell Mol Biol 37:379–386. doi: 10.1165/rcmb.2007-0142OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Deans RM, Morgens DW, Okesli A, Pillay S, Horlbeck MA, Kampmann M, Gilbert LA, Li A, Mateo R, Smith M, Glenn JS, Carette JE, Khosla C, Bassik MC. 2016. Parallel shRNA and CRISPR-Cas9 screens enable antiviral drug target identification. Nat Chem Biol 12:361–366. doi: 10.1038/nchembio.2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Morrey JD, Smee DF, Sidwell RW, Tseng C. 2002. Identification of active antiviral compounds against a New York isolate of West Nile virus. Antiviral Res 55:107–116. doi: 10.1016/s0166-3542(02)00013-x. [DOI] [PubMed] [Google Scholar]
- 44.Crance JM, Scaramozzino N, Jouan A, Garin D. 2003. Interferon, ribavirin, 6-azauridine and glycyrrhizin: antiviral compounds active against pathogenic flaviviruses. Antiviral Res 58:73–79. doi: 10.1016/s0166-3542(02)00185-7. [DOI] [PubMed] [Google Scholar]
- 45.Lang Y, Li Y, Jasperson D, Henningson J, Lee J, Ma J, Li Y, Duff M, Liu H, Bai D, McVey S, Richt JA, Ikegami T, Wilson WC, Ma W. 2019. Identification and evaluation of antivirals for Rift Valley fever virus. Vet Microbiol 230:110–116. doi: 10.1016/j.vetmic.2019.01.027. [DOI] [PubMed] [Google Scholar]
- 46.Noah JW, Severson W, Noah DL, Rasmussen L, White EL, Jonsson C. 2007. A cell-based luminescence assay is effective for high-throughput screening of potential influenza antivirals. Antiviral Res 73:50–59. doi: 10.1016/j.antiviral.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 47.Smee DF, Morrison AC, Barnard DL, Sidwell R. 2002. Comparison of colorimetric, fluorometric, and visual methods for determining anti-influenza (H1N1 and H3N2) virus activities and toxicities of compounds. J Virol Methods 106:71–79. doi: 10.1016/s0166-0934(02)00137-4. [DOI] [PubMed] [Google Scholar]
- 48.Gutowski GE, Sweeney MJ, DeLong DC, Hamill RL, Gerzon K, Dyke RW. 1975. Biochemistry and biological effects of the pyrazofurins (pyrazomycins): initial clinical trial. Ann N Y Acad Sci 255:544–551. doi: 10.1111/j.1749-6632.1975.tb29257.x. [DOI] [PubMed] [Google Scholar]
- 49.Kucukguzel SG, Senkardes S. 2015. Recent advances in bioactive pyrazoles. Eur J Med Chem 97:786–815. doi: 10.1016/j.ejmech.2014.11.059. [DOI] [PubMed] [Google Scholar]
- 50.Shigeta S, Konno K, Yokota T, Nakamura K, De Clercq E. 1988. Comparative activities of several nucleoside analogs against influenza A, B, and C viruses in vitro. Antimicrob Agents Chemother 32:906–911. doi: 10.1128/aac.32.6.906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cho J, Yi H, Jang EY, Lee M-S, Lee J-Y, Kang C, Lee CH, Kim K. 2017. Mycophenolic mofetil, an alternative antiviral and immunomodulator for the highly pathogenic avian influenza H5N1 virus infection. Biochem Biophys Res Commun 494:298–304. doi: 10.1016/j.bbrc.2017.10.037. [DOI] [PubMed] [Google Scholar]
- 52.Chan JF, Chan KH, Kao RY, To KK, Zheng BJ, Li CP, Li PT, Dai J, Mok FK, Chen H, Hayden FG, Yuen KY. 2013. Broad-spectrum antivirals for the emerging Middle East respiratory syndrome coronavirus. J Infect 67:606–616. doi: 10.1016/j.jinf.2013.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.To KKW, Mok K-Y, Chan ASF, Cheung NN, Wang P, Lui Y-M, Chan JFW, Chen H, Chan K-H, Kao RYT, Yuen K-Y. 2016. Mycophenolic acid, an immunomodulator, has potent and broad-spectrum in vitro antiviral activity against pandemic, seasonal and avian influenza viruses affecting humans. J Gen Virol 97:1807–1817. doi: 10.1099/jgv.0.000512. [DOI] [PubMed] [Google Scholar]
- 54.Padalko E, Verbeken E, Matthys P, Aerts JL, De Clercq E, Neyts J. 2003. Mycophenolate mofetil inhibits the development of coxsackie B3-virus-induced myocarditis in mice. BMC Microbiol 3:25–25. doi: 10.1186/1471-2180-3-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Floryk D, Thompson TC. 2008. Antiproliferative effects of AVN944, a novel inosine 5-monophosphate dehydrogenase inhibitor, in prostate cancer cells. Int J Cancer 123:2294–2302. doi: 10.1002/ijc.23788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hamilton JM, Harding MW, Genna T, Bol DK. 2009. A phase I dose-ranging study of the pharmacokinetics, pharmacodynamics, safety, and tolerability of AVN944, an IMPDH inhibitor, in healthy male volunteers. J Clin Pharmacol 49:30–38. doi: 10.1177/0091270008325149. [DOI] [PubMed] [Google Scholar]
- 57.Pan Q, de Ruiter PE, Metselaar HJ, Kwekkeboom J, de Jonge J, Tilanus HW, Janssen HL, van der Laan LJ. 2012. Mycophenolic acid augments interferon‐stimulated gene expression and inhibits hepatitis C Virus infection in vitro and in vivo. Hepatology 55:1673–1683. doi: 10.1002/hep.25562. [DOI] [PubMed] [Google Scholar]
- 58.Fulton B, Markham A. 1996. Mycophenolate mofetil: a review of its pharmacodynamic and pharmacokinetic properties and clinical efficacy in renal transplantation. Drugs 51:278–298. doi: 10.2165/00003495-199651020-00007. [DOI] [PubMed] [Google Scholar]
- 59.Booth L, Roberts JL, Cash DR, Tavallai S, Jean S, Fidanza A, Cruz ‐Luna T, Siembiba P, Cycon KA, Cornelissen CN, Dent P. 2015. GRP78/BiP/HSPA5/Dna K is a universal therapeutic target for human disease. J Cell Physiol 230:1661–1676. doi: 10.1002/jcp.24919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jimenez-Guerrero R, Gasca J, Flores ML, Perez-Valderrama B, Tejera-Parrado C, Medina R, Tortolero M, Romero F, Japon MA, Saez C. 2018. Obatoclax and paclitaxel synergistically induce apoptosis and overcome paclitaxel resistance in urothelial cancer cells. Cancers (Basel) 10:E490. doi: 10.3390/cancers10120490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schimmer AD, O’Brien S, Kantarjian H, Brandwein J, Cheson BD, Minden MD, Yee K, Ravandi F, Giles F, Schuh A, Gupta V, Andreeff M, Koller C, Chang H, Kamel-Reid S, Berger M, Viallet J, Borthakur G. 2008. A phase I study of the pan Bcl-2 family inhibitor obatoclax mesylate in patients with advanced hematologic malignancies. Clin Cancer Res 14:8295–8301. doi: 10.1158/1078-0432.CCR-08-0999. [DOI] [PubMed] [Google Scholar]
- 62.Booth L, Roberts JL, Ecroyd H, Tritsch SR, Bavari S, Reid SP, Proniuk S, Zukiwski A, Jacob A, Sepúlveda CS, Giovannoni F, García CC, Damonte E, González-Gallego J, Tuñón MJ, Dent P. 2016. AR‐12 inhibits multiple chaperones concomitant with stimulating autophagosome formation collectively preventing virus replication. J Cell Physiol 231:2286–2302. doi: 10.1002/jcp.25431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ilyushina NA, Ikizler MR, Kawaoka Y, Rudenko LG, Treanor JJ, Subbarao K, Wright PF. 2012. Comparative study of influenza virus replication in MDCK cells and in primary cells derived from adenoids and airway epithelium. J Virol 86:11725–11734. doi: 10.1128/JVI.01477-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhai W, Zhang DN, Mai C, Choy J, Jian G, Sra K, Galinski MS. 2012. Comparison of different cell substrates on the measurement of human influenza virus neutralizing antibodies. PLoS One 7:e52327. doi: 10.1371/journal.pone.0052327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nogales A, Rodriguez L, DeDiego ML, Topham DJ, Martinez-Sobrido L. 2017. Interplay of PA-X and NS1 proteins in replication and pathogenesis of a temperature-sensitive 2009 pandemic H1N1 influenza A virus. J Virol 91:e00720-17. doi: 10.1128/JVI.00720-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.DiPiazza A, Nogales A, Poulton N, Wilson PC, Martinez-Sobrido L, Sant AJ. 2017. Pandemic 2009 H1N1 influenza Venus reporter virus reveals broad diversity of MHC class II-positive antigen-bearing cells following infection in vivo. Sci Rep 7:10857. doi: 10.1038/s41598-017-11313-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Breen M, Nogales A, Baker SF, Perez DR, Martinez-Sobrido L. 2016. Replication-competent influenza A and B viruses expressing a fluorescent dynamic timer protein for In Vitro and In Vivo studies. PLoS One 11:e0147723. doi: 10.1371/journal.pone.0147723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wasik BR, Voorhees IEH, Barnard KN, Alford-Lawrence BK, Weichert WS, Hood G, Nogales A, Martínez-Sobrido L, Holmes EC, Parrish CR, Wasik BR, Voorhees IEH, Barnard KN, Alford-Lawrence BK, Weichert WS, Hood G, Nogales A, Martínez-Sobrido L, Holmes EC, Parrish CR. 2019. Influenza viruses in mice: deep sequencing analysis of serial passage and effects of sialic acid structural variation. J Virol 93:e01039-19. doi: 10.1128/JVI.01039-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nogales A, Piepenbrink MS, Wang J, Ortega S, Basu M, Fucile CF, Treanor JJ, Rosenberg AF, Zand MS, Keefer MC, Martinez-Sobrido L, Kobie JJ. 2018. A highly potent and broadly neutralizing H1 influenza-specific human monoclonal antibody. Sci Rep 8:4374. doi: 10.1038/s41598-018-22307-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.