Human cytomegalovirus (HCMV) is an opportunistic herpesvirus that is asymptomatic for healthy individuals but that can lead to severe pathology in patients with congenital infections and immunosuppressed patients. Thus, it is important to understand the modulation of the immune response by HCMV, which is understudied in the context of endogenous MHC class II regulation. Using Kasumi-3 cells as a myeloid progenitor cell model endogenously expressing MHC class II (HLA-DR), this study shows that HCMV decreases the expression of HLA-DR in infected cells by reducing the transcription of HLA-DR transcripts early during infection independently of the expression of previously implicated genes. This is an important finding, as it highlights a mechanism of immune evasion utilized by HCMV to decrease the expression of MHC class II in a relevant cell system that endogenously expresses the MHC class II complex.
KEYWORDS: cytomegalovirus, Kasumi-3, MHC class II, myeloid
ABSTRACT
Human cytomegalovirus (HCMV) is a ubiquitous pathogen that encodes many proteins to modulate the host immune response. Extensive efforts have led to the elucidation of multiple strategies employed by HCMV to effectively block NK cell targeting of virus-infected cells and the major histocompatibility complex (MHC) class I-primed CD8+ T cell response. However, viral regulation of the MHC class II-mediated CD4+ T cell response is understudied in endogenous MHC class II-expressing cells, largely because the popular cell culture systems utilized for studying HCMV do not endogenously express MHC class II. Of the many cell types infected by HCMV in the host, myeloid cells, such as monocytes, are of particular importance due to their role in latency and subsequent dissemination throughout the host. We investigated the impact of HCMV infection on MHC class II in Kasumi-3 cells, a myeloid-progenitor cell line that endogenously expresses the MHC class II gene, HLA-DR. We observed a significant reduction in the expression of surface and total HLA-DR at 72 h postinfection (hpi) and 120 hpi in infected cells. The decrease in HLA-DR expression was independent of the expression of previously described viral genes that regulate the MHC class II complex or the unique short (US) region of HCMV, a region expressing many immunomodulatory genes. The altered surface level of HLA-DR was not a result of increased endocytosis and degradation but was a result of a reduction in HLA-DR transcripts due to a decrease in the expression of the class II transactivator (CIITA).
IMPORTANCE Human cytomegalovirus (HCMV) is an opportunistic herpesvirus that is asymptomatic for healthy individuals but that can lead to severe pathology in patients with congenital infections and immunosuppressed patients. Thus, it is important to understand the modulation of the immune response by HCMV, which is understudied in the context of endogenous MHC class II regulation. Using Kasumi-3 cells as a myeloid progenitor cell model endogenously expressing MHC class II (HLA-DR), this study shows that HCMV decreases the expression of HLA-DR in infected cells by reducing the transcription of HLA-DR transcripts early during infection independently of the expression of previously implicated genes. This is an important finding, as it highlights a mechanism of immune evasion utilized by HCMV to decrease the expression of MHC class II in a relevant cell system that endogenously expresses the MHC class II complex.
INTRODUCTION
Human cytomegalovirus (HCMV) is a clinically significant herpesvirus that drastically alters the host cell during its protracted replication cycle. Among the well-characterized alterations is a striking change to the cellular proteome and a remodeling of the protein composition of the plasma membrane (1). The plasma membrane composition can be altered by multiple mechanisms, which include modifying the kinetics of endocytosis, recycling, and lysosomal degradation of target proteins, altering their trafficking and localization away from the plasma membrane and changing the rate of synthesis at the transcriptional and translational levels.
Immune proteins constitute a significant portion of the plasma membrane proteins regulated by HCMV. Examples of a few of the many immune proteins whose surface expression is regulated by this virus include MICA (major histocompatibility complex class I polypeptide-related sequence A), MICB, tumor necrosis factor alpha (TNF-α), TNF-related apoptosis-inducing ligand receptor 1 (TRAIL-R1), TRAIL-R2, and the UL16 binding proteins 1, 2, 3, and 6 (2–6). Perhaps the best-studied example is major histocompatibility complex (MHC) class I, as HCMV utilizes several well-described strategies to ensure reduced MHC class I antigen presentation (7–10). Modulating the presentation of MHC class II, which presents both exogenous and endogenous antigens, would also be beneficial for HCMV infection. The pool of MHC class II-presented antigens could include antigens derived from viral proteins expressed within infected cells (reviewed in reference 11). A CD4+ T cell response to endogenous antigen has been demonstrated for many different viruses (reviewed in reference 12). HCMV-infected cells can present endogenous antigen to prime the CD4+ T cell response (13). CD4+ T cells specific for both murine cytomegalovirus (MCMV) and HCMV express granzyme B and possess cytolytic activity to control infection (14–16). Therefore, controlling the action of CD4+ T cells would be advantageous for HCMV infection, specifically, to block the endogenous presentation of viral proteins.
The MHC class II pathway involves the synthesis of MHC class II proteins, their association with the invariant chain (Ii) within the endoplasmic reticulum, trafficking of the complex through the Golgi apparatus to the MHC class II loading compartment (MIIC), and loading of the peptide with help from accessory factors in endosomal MIICs (reviewed in reference 17). These mature MHC class II molecules move to the plasma membrane to present peptide to CD4+ T cells. Pathogens employ various mechanisms to block MHC class II presentation at different steps of the pathway. These strategies targeting MHC class II and accessory factors can be broadly divided into three categories: (i) synthesis, assembly, and loading, (ii) altered trafficking and localization, and (iii) targeted degradation. Blocking the assembly of MHC class II components, for example, interfering with the interaction between the invariant chain and the MHC class II alpha and beta chains, is an attractive strategy. In fact, US3-mediated inhibition of Ii-MHC class II complex formation has been reported for HCMV (18). Altered localization of MHC class II can be achieved by rerouting MHC class II to endosomes or lysosomes, where the complex is degraded or retained in these vesicles away from the plasma membrane. HCMV-infected cells have been shown to contain MHC class II retained within perinuclear vesicles in the cell, and this aberrant localization has been observed in fibroblasts, myeloid cells, and endothelial cells (19–21). Additionally, US2 directly targets MHC class II for degradation by binding and degrading the alpha chain of human leukocyte antigen DR (HLA-DR) and HLA-DM within HCMV-infected glioblastoma U373 cells (13). For MCMV, M78 binds to MHC class II and degrades it in the lysosomes within infected myeloid cells (22).
In addition to these mechanisms, viruses can use cytokine-mediated effects to regulate MHC class II. MHC class II is induced by interferon gamma (IFN-γ), and blocking IFN-γ signal transduction is an effective approach for preventing induced MHC class II production. Accordingly, HCMV blocks IFN-γ-induced surface MHC class II in infected endothelial and glioblastoma cells by degrading Jak1 to prevent IFN-γ signal transduction (19, 23). MCMV exerts a similar effect on IFN-γ-driven MHC class II expression (24). While IFN-γ induces MHC class II synthesis, interleukin-10 (IL-10), an immunosuppressive cytokine, decreases MHC class II via multiple mechanisms. HCMV encodes an IL-10 homologue, UL111A, that has an immunomodulatory function on responder cells and decreases MHC class II during latent infections (25, 26). Additionally, HCMV can induce cellular IL-10 to achieve a similar function (27), and MCMV utilizes IL-10 signaling for viral persistence in the salivary gland (28). Thus, blocking antigen presentation by MHC class II molecules is a strategy employed by cytomegaloviruses to promote infection.
HCMV can infect many different cell types, such as fibroblasts, epithelial cells, and myeloid cells. HCMV infections in vitro have classically been studied in fibroblasts because of the robust lytic replication of the virus in this cell type. However, the dissemination of the virus in vivo occurs primarily through cells of the myeloid lineage. The virus infects myeloid progenitor cells and undergoes latency. Upon reactivation of the virus, myeloid cells are integral for viral spread throughout the host. Given their importance for viral spread and their ability to act as antigen-presenting cells, it is critical to understand how HCMV manipulates myeloid cells to its advantage during infection. The use of primary cells as an HCMV infection model has been challenging due to the fact that these cells obtained ex vivo have considerable donor variability in terms of infectivity and are not amenable for techniques that require teasing out specific viral proteins and mechanisms. However, HCMV can infect several cell lines of the myeloid lineage that mimic primary cells with regard to many aspects of HCMV infection and thus have a great potential to be experimental models for studying HCMV infection in vitro (29–32). One of these cell lines, Kasumi-3, expresses appreciable levels of MHC class II HLA-DR, and we took advantage of this line to investigate how HCMV controls endogenously expressed MHC class II complexes. These cells are a clonal myeloid progenitor cell line derived from a myeloperoxidase-negative leukemia patient, express surface CD34 and MHC class II, and serve as a model of latency and reactivation (29, 30, 33). Previous work implicating viral genes for MHC class II regulation has involved IFN-γ stimulation to drive MHC class II expression in nonmyeloid cells. The impact of HCMV infection in myeloid progenitor cells for MHC class II has been understudied. Using Kasumi-3 cells enables us to understand the regulation of endogenous MHC class II in infected myeloid cells without differentiation or activation of the cells. We found that HCMV does downregulate the surface levels of MHC class II but that this decrease is independent of the mechanisms reported for regulating MHC class II under either induced or overexpressed conditions. We observed that surface MHC class II molecules are endocytosed and degraded at the same rate in uninfected and HCMV-infected cells, indicating that HCMV does not promote the degradation of surface MHC class II molecules. Rather, our results show that HCMV repression of MHC class II primarily occurs at the transcriptional level as a result of the downregulation of class II transactivator (CIITA) expression.
RESULTS
HCMV reduces surface and total levels of MHC class II.
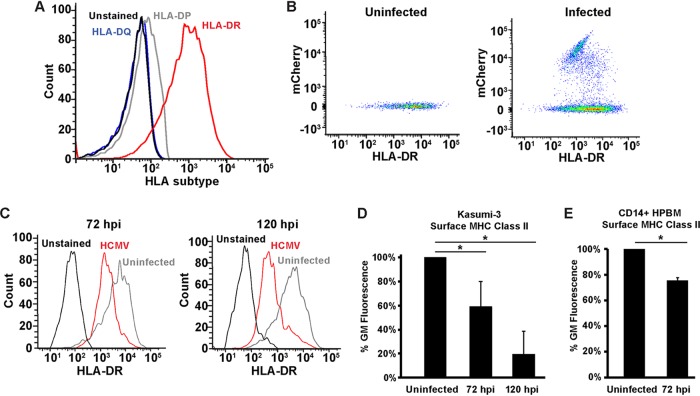
To determine a suitable model for addressing the mechanism of MHC class II downregulation in an endogenously expressing system, we checked for the surface levels of the MHC class II human leukocyte antigen DR (HLA-DR) isotype in three different myeloid cell lines, Kasumi-3, KG1, and THP-1. While both KG1 and Kasumi-3 cells were positive for surface HLA-DR, very little was detected in undifferentiated THP-1 cells (data not shown). Since we found the Kasumi-3 cells to be more amenable to infection and could attain a higher percentage of infected cells, we chose them as our model for addressing the mechanism of MHC class II regulation by HCMV. We next investigated the surface levels of other MHC class II HLAs on Kasumi-3 cells. We detected no HLA-DQ, as the cell population stained for HLA-DQ was indistinguishable from unstained cells (Fig. 1A). While there was very little staining for HLA-DP, the predominant HLA subtype detected on Kasumi-3 cells was HLA-DR (Fig. 1A). Thus, we used HLA-DR for our subsequent experiments to investigate how HCMV regulates MHC class II surface levels.
FIG 1.
HCMV downregulates surface expression of MHC class II in a myeloid progenitor cell line. (A) Flow cytometry analysis of unstained Kasumi-3 cells or cells stained for HLA-DR, HLA-DQ, and HLA-DP. (B) Flow cytometry scatter plot of HCMV-infected (72 hpi) and uninfected cells showing the relationship between mCherry (a marker of HCMV infection) and HLA-DR. (C) Histograms of HCMV-infected (mCherry) and uninfected Kasumi-3 cells stained for surface HLA-DR at 72 and 120 h postinfection. (D) Bar graph of the geometric mean (GM) fluorescence values for the samples used in the assay whose results are presented in panel C, displayed as a percentage of the value for the uninfected sample. (E) Geometric mean fluorescence values from uninfected or HCMV-infected CD14+ human peripheral blood monocytes (HPBM) at 72 hpi. Values are averages from a minimum of three independent experiments. *, P < 0.05.
Since reduced surface expression of MHC class II complexes would benefit a viral infection, we hypothesized that surface HLA-DR would decrease following infection by HCMV. To test this hypothesis, we infected Kasumi-3 cells with a virus expressing mCherry from its genome (34) to allow for identification of infected cells. Although only a minority of the population was infected, these cells could be identified as mCherry positive (Fig. 1B) and surface HLA-DR levels could be assessed on both the mCherry-positive (infected) and -negative (uninfected) populations. Gating the cells on mCherry allowed us to separate infected and uninfected cells; however, we did not characterize the infection as lytic or latent. Following HCMV infection, surface HLA-DR levels were reduced compared to those for uninfected samples at both 72 and 120 h postinfection (hpi) (Fig. 1C). Quantification using the geometric mean fluorescence showed that the levels were reduced to ∼60% at 72 h, further decreasing to ∼20% by 120 hpi (Fig. 1D). Thus, MHC class II surface expression steadily declines throughout HCMV infection of Kasumi-3 cells. To determine whether this surface downregulation also occurred in primary cells, we next infected CD14+ human peripheral blood monocytes and monitored surface HLA-DR at 72 hpi. We observed decreased surface HLA-DR (Fig. 1E), similar to what was observed in the Kasumi-3 cells.
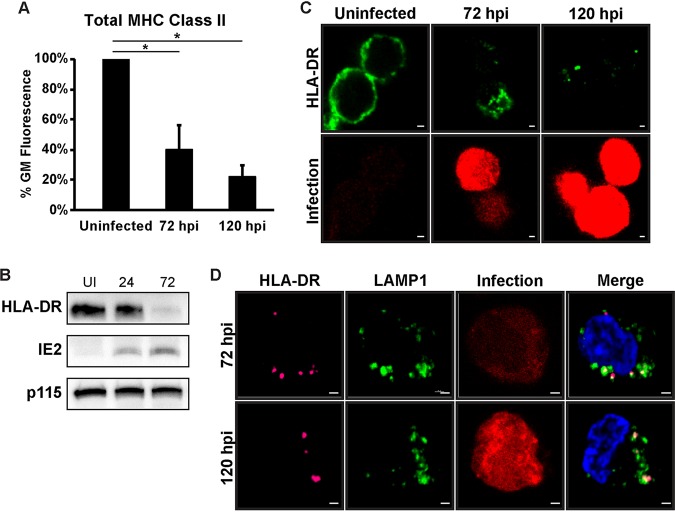
We next wondered whether the decrease in surface MHC class II correlated with a decrease in total MHC class II. Reduced surface levels could be due to intracellular sequestration of the protein, in which total MHC class II levels would remain steady. Total HLA-DR levels were measured in permeabilized Kasumi-3 cells that were uninfected or in infected cells at 72 or 120 hpi. Similar to the surface levels, total HLA-DR protein was reduced at 72 hpi and further declined by 120 hpi (Fig. 2A). The decrease in total HLA-DR protein in infected cells was confirmed by Western blot analysis (Fig. 2B). Immunofluorescence analysis of uninfected Kasumi-3 cells showed that HLA-DR localized to both the cell surface and intracellular puncta (Fig. 2C). At 72 h postinfection, some surface staining remained, but much of the HLA-DR protein localized to internal puncta. Any HLA-DR detected at 120 hpi was entirely localized to intracellular puncta, and very little HLA-DR was detected in some cells (Fig. 2C). At 72 hpi, most of the HLA-DR puncta either colocalized with or were adjacent to LAMP1 (Fig. 2D); however, some HLA-DR puncta with no correlation to LAMP1 were present. At 120 hpi, almost all HLA-DR puncta colocalized with LAMP1. These LAMP1/HLA-DR-positive puncta could represent MHC class II loading compartments or degradative lysosomes, which would be consistent with the decrease observed for both surface and total HLA-DR molecules during infection.
FIG 2.
Total MHC class II is reduced during HCMV infection. (A) Bar graph of the geometric mean fluorescence values of total HLA-DR protein in uninfected and infected samples (72 and 120 hpi). Values are a percentage of the value for the uninfected sample and are averages from at least three independent experiments. *, P < 0.05. (B) Western blot analysis of uninfected and HCMV-infected Kasumi-3 cells at 24 and 72 hpi to detect HLA-DR, IE2, and p115 (loading control) protein levels. (C) Immunofluorescence of total HLA-DR protein (green) in uninfected and HCMV-infected Kasumi-3 cells at 72 and 120 hpi. Infected cells expressed mCherry (red) from the genome as marker for infection. (D) Immunofluorescence of total HLA-DR (pink) and LAMP1 (green) in HCMV-infected (red) Kasumi-3 cells at 72 and 120 hpi. Nuclei stained with DAPI. Bars (C and D), 1 μm.
The HCMV-dependent reduction in MHC class II is independent of viral proteins previously reported to downregulate MHC class II.
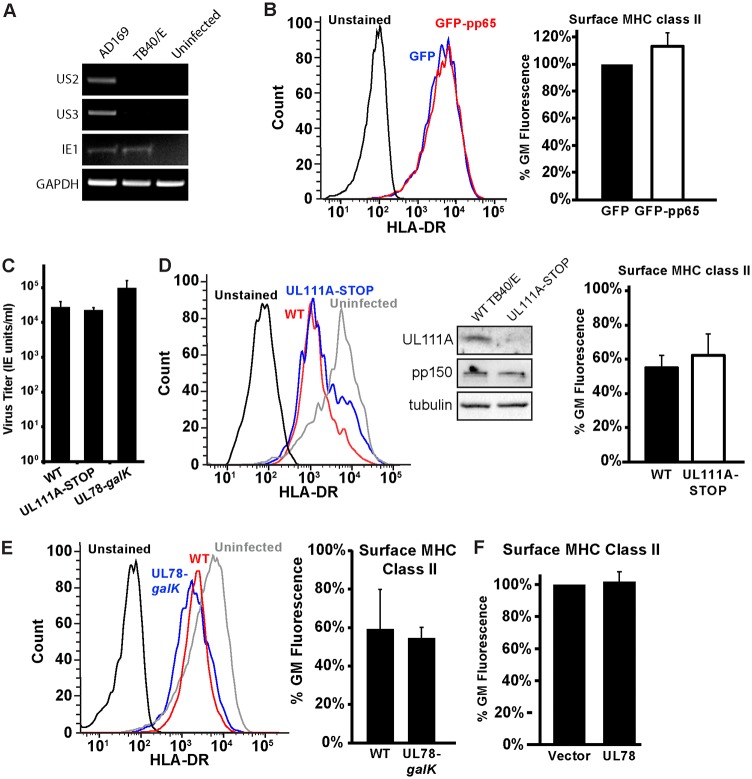
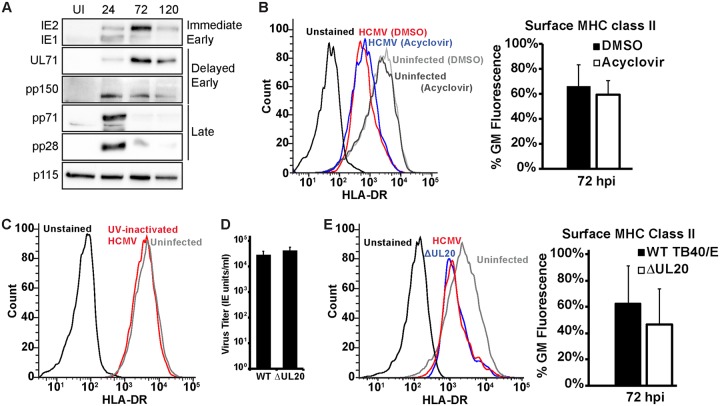
Several mechanisms for how HCMV regulates MHC class II in cell lines in which the complex is not natively expressed have been reported (13, 18, 21, 22, 25, 35). We next sought to determine which of these mechanisms might apply to the regulation of endogenously expressed MHC class II in cells of the myeloid lineage. Two HCMV proteins expressed from the unique short (US) region of the genome, US2 and US3, alter the stability, loading, and trafficking of MHC class II molecules (13, 18). However, the expression of these genes was disrupted during the generation of the bacterial artificial chromosome (BAC) used to propagate the TB40/E strain (36). As confirmation, both US2 and US3 were detected in cDNA harvested from cells infected with the AD169 strain but not in that harvested from cells infected with the TB40/E strain, utilized for the experiments described above (Fig. 3A). Thus, the MHC class II regulation that we observed is not dependent on US2 or US3.
FIG 3.
Kasumi-3 cells do not regulate MHC class II using viral proteins previously reported to decrease MHC class II expression and localization. (A) RT-PCR analysis of US2, US3, IE1, and GAPDH transcripts from uninfected fibroblasts or fibroblasts infected with the AD169 or TB40/E strains of HCMV (96 hpi). (B) (Left) Histograms of surface HLA-DR protein in Kasumi-3 cells 48 h after electroporation with either the GFP control or GFP-pp65. (Right) Quantification of the geometric mean fluorescence as a percentage of the value for the GFP control sample. (C) Infectious titers at 120 hpi of wild-type TB40/E (wild type [WT]) and the UL111A-STOP and UL78-galK viruses following infection of fibroblasts at an MOI of 3. (D) (Left) Histograms of surface HLA-DR protein in Kasumi-3 cells infected with mCherry expressing wild-type TB40/E or a virus lacking UL111A (UL111A-STOP) expression at 72 hpi. (Middle) Western blot analysis confirms the absence of the UL111A protein. (Right) Quantification of the geometric mean fluorescence as a percentage of the value for the uninfected sample. (E) (Left) Histograms of surface HLA-DR protein in Kasumi-3 cells infected with mCherry expressing wild-type TB40/E or a virus lacking UL78 expression (UL78-galK) at 72 hpi. (Right) Quantification of the geometric mean fluorescence of TB40/E or a virus lacking UL78 (UL78-galK) as a percentage of the value for the uninfected sample. (F) Graph representing the geometric mean fluorescence of surface HLA-DR protein in Kasumi-3 cells at 48 h postelectroporation with either the vector control or a plasmid expressing UL78. The histograms in panels B, D, and E are representative images from one of at least three independent experiments. The values in the bar graphs in panels B to F are percentages of the geometric mean fluorescence for uninfected or control transfected cells and are averages from at least three independent experiments.
MHC class II regulation has been reported for two other viral proteins, the tegument protein pp65 and the viral IL-10 homologue UL111A (21, 25, 35). In IFN-γ-stimulated fibroblasts, transfection of pp65 alone was sufficient to reduce surface HLA-DR levels (21). To test whether endogenously expressed MHC class II was reduced in the presence of pp65, we expressed either green fluorescent protein (GFP) or GFP-pp65 in Kasumi-3 cells and compared the expression of surface HLA-DR in the GFP-positive populations. Expression of pp65 did not reduce MHC class II surface levels (Fig. 3B). Thus, pp65 alone is not responsible for the downregulation of MHC class II during infection of Kasumi-3 cells. To test the contribution of the viral IL-10 or UL111A protein, we generated a virus in which the start codon of UL111A was replaced with a stop codon (the UL111A-STOP virus). The UL111A-STOP virus produced infectious virions at wild-type levels (Fig. 3C), and Western blot analysis confirmed that UL111A was not expressed (Fig. 3D). MHC class II surface levels were still decreased, despite the absence of UL111A (Fig. 3D). Thus, the HCMV downregulation of MHC class II is independent of the viral IL-10 protein.
A recent study using mouse cytomegalovirus demonstrated a role for the MCMV M78 protein in degrading MHC class II (22). To test whether the HCMV homologue may play a similar role, we generated a virus deficient for UL78 expression by replacing the entire UL78 open reading frame (ORF) with galK. The UL78-galK virus grew as well as and perhaps even slightly better than the wild-type virus (Fig. 3C) and was able to downregulate MHC class II with kinetics similar to those in the wild-type virus (Fig. 3E). To confirm that UL78 was not responsible for the observed MHC class II surface downregulation, we next transfected Kasumi-3 cells with a vector control or a plasmid expressing UL78. We observed no decrease in surface MHC class II in the UL78-transfected samples (Fig. 3F). Collectively these data show that UL78 is not responsible for MHC class II downregulation in Kasumi-3 cells.
Class II molecule downregulation is independent of the unique short region of the HMCV genome.
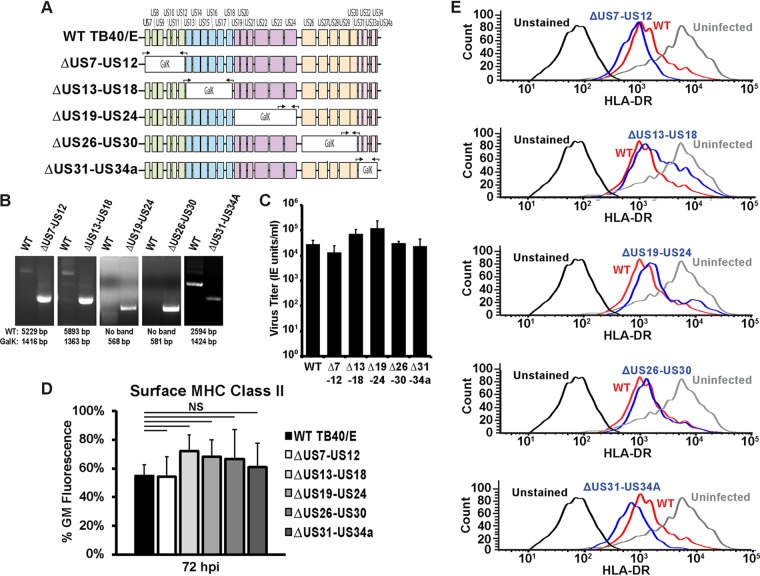
HCMV downregulates MHC class II in the myeloid lineage Kasumi-3 cell line; however, this downregulation appears to be independent of previously published mechanisms. We sought to determine the viral factors responsible for downregulating MHC class II and hypothesized that the factor(s) responsible may be expressed from the unique short region of the genome, a region that has been shown to encode several factors important for immune modulation and the regulation of MHC class I and NK cell ligands. To screen this region for class II-regulating factors, we generated five viruses in which five to six contiguous genes were replaced with galK (the ΔUS7-US12, ΔUS13-US18, ΔUS19-US24, ΔUS26-US30, and ΔUS31-US34A viruses), as depicted in the schematic in Fig. 4A. Successful replacement of the US region gene segments with galK was confirmed by PCR (Fig. 4B).
FIG 4.
The unique short region is not required for the downregulation of surface MHC class II molecules in Kasumi-3 cells. (A) Schematic of the US region of the HCMV genome showing the strategy for segmentally knocking out proteins expressed from the US region. Black arrows indicate primer sets either flanking the galK insertion region or binding within galK and a neighboring flanking region. (B) PCR analysis showing replacement of the indicated segment of the US region with galK. The expected values for wild-type and galK-containing bands are indicated below the images. (C) Infectious titers at 120 hpi of wild-type TB40/E (WT) and the US deletion mutants (indicated by Δ7-12, Δ13-18, Δ19-24, Δ26-30, and Δ31-34A) following the infection of fibroblasts at an MOI of 3. (D) Bar graph generated from the geometric mean fluorescence values of surface HLA-DR staining of Kasumi-3 cells infected with wild-type TB40/E or viruses lacking segments of the US region gene (the ΔUS7-US12, ΔUS13-US18, ΔUS19-US24, ΔUS26-US30, and ΔUS31-US34A viruses) at 72 hpi. NS, not significant. (E) Histograms from one representative experiment for the samples for which the results are shown in panel D. The values in panels C and D are averages from three independent experiments.
We were able propagate all of the US region deletion viruses, and importantly, none of the viruses exhibited a growth phenotype on fibroblasts (Fig. 4C). Analysis of surface MHC class II levels following infection with US region deletion viruses showed that all five viruses were competent for downregulating MHC class II surface expression (Fig. 4D and 4E). In cells infected with the ΔUS13-US18 virus, a slight increase in HLA-DR surface levels compared to the levels in a wild-type infection was observed; however, this increase was not statistically significant and the virus still largely retained the ability to downregulate surface MHC class II. This may indicate that a factor in this region could be a minor participant in regulating surface HLA-DR; however, this factor is clearly not solely responsible for reducing surface MHC class II. Thus, MHC class II surface levels were still reduced in the absence of each the HCMV US region proteins. This suggests that none of these proteins alone are responsible for the MHC class II downregulation, although we cannot rule out the possibility of a redundancy among other US region genes that were still expressed in each of the deletion viruses. Attempts to replace the entire US region of the genome were unsuccessful, as the virus was unable to spread following electroporation of the US region-deficient BAC.
HCMV utilizes an immediate or early protein to regulate surface MHC class II levels.
To this point, none of the viral factors investigated were responsible for the decrease in MHC class II surface protein during infection of the myeloid progenitor Kasumi-3 cell line. It is unclear which, if any, of the above-described gene products are even expressed during an infection of Kasumi-3 cells. To investigate the expression of viral genes in our Kasumi-3 cell infections, we examined the protein levels of two immediate early proteins (IE1 and IE2), two delayed early proteins (UL71 and pp150), and two late proteins (pp28 and pp71) at 24, 72, and 120 h postinfection. We found that both the immediate early and early proteins were expressed and that their expression peaked at 72 hpi, with markedly reduced levels being detected at 120 hpi (Fig. 5A). Both late proteins, which are present in the virion tegument layer, were present at 24 hpi, likely a result of incoming protein from the infections at a high multiplicity of infection (MOI). However, very little late protein was detected at 72 and 120 hpi, indicating suppressed expression of late proteins in our Kasumi-3 cell infections. This may indicate that the infected cells were shutting down viral protein expression, although we have not adequately characterized the infection as lytic or latent. At least some cells were undergoing a productive infection, as small amounts of infectious virions could be recovered (data not shown). Thus, we hypothesized that MHC class II downregulation was a result of immediate or early gene expression. To test this, we investigated whether acyclovir could prevent the decrease in surface MHC class II levels. Expression of viral late proteins is dependent upon DNA replication, which can be blocked by the addition of a replication inhibitor, such as acyclovir. Addition of acyclovir did not prevent the decrease in HLA-DR surface levels (Fig. 5B). Thus, the regulation of MHC class II is not dependent on viral DNA replication and therefore appears to be dependent on a viral factor expressed with immediate early or early kinetics.
FIG 5.
Reduced MHC class II protein requires early viral gene synthesis. (A) Western blot analysis of uninfected (UI) or HCMV-infected Kasumi-3 cells at 24, 72, and 120 hpi, showing the results for two immediate early proteins (IE1 and IE2), two delayed early proteins (UL71 and pp150), two late proteins (pp71 and pp28), and a loading control (p115). (B) (Left) Histograms of one representative experiment of surface HLA-DR staining of uninfected Kasumi-3 cells or cells infected with wild-type TB40/E and treated with DMSO or acyclovir. (Right) The bar graph shows the geometric mean fluorescence values for infected samples at 72 hpi. (C) Histograms of one representative experiment of surface HLA-DR staining at 72 hpi of Kasumi-3 cells infected with UV-inactivated virus. (D) Infectious titers at 120 hpi of wild-type TB40/E (WT) and the ΔUL20 viruses following infection of fibroblasts at an MOI of 3. (E) (Left) Histograms of one representative experiment of surface HLA-DR staining at 72 hpi of Kasumi-3 cells infected with wild-type TB40/E or a virus expressing GFP in place of UL20 (ΔUL20). (Right) Bar graph generated from geometric mean fluorescence values. The values graphed in panels A, D, and E are averages from a minimum of three independent experiments.
Our protein expression data detected the presence of tegument proteins at 24 hpi. While the levels of these proteins were nearly undetectable at 72 and 120 hpi, when the greatest reduction in surface MHC class II was observed, it is nonetheless important to rule out the possibility of a contribution of incoming virion proteins in downregulating MHC class II. This is particularly important due to the high number of virions added to the Kasumi-3 cells to initiate infection. To test whether incoming virion proteins were responsible for the MHC class II downregulation, we added UV-inactivated virus to Kasumi-3 cells and measured surface MHC class II levels. The levels of surface HLA-DR were indistinguishable from the levels on uninfected cells (Fig. 5C), indicating that incoming virion proteins were not responsible for the MHC class II downregulation. Thus, the downregulation of MHC class II appears to be mediated by a gene whose expression is not dependent on DNA replication.
One factor of interest expressed with immediate early kinetics is UL20. UL20 is a putative T cell receptor homologue that is immediately trafficked to the lysosome for degradation following expression and is not required for infection in fibroblasts (37). We hypothesized that in Kasumi-3 cells, UL20 may bind to newly synthesized MHC class II molecules and deliver them to lysosomes for degradation, thus resulting in decreased surface localization. To test this hypothesis, we generated a UL20-null virus that expresses GFP in place of UL20. UL20 has previously been reported to be dispensable for growth on fibroblasts (38), and we similarly found that infectious virion production was equivalent between the ΔUL20 and wild-type viruses (Fig. 5D). No difference in the reduction of MHC class II molecules was observed in the absence of UL20 (Fig. 5E). Thus, UL20 is not the factor responsible for reducing surface MHC class II levels.
HCMV does not alter the internalization rate of class II molecules.
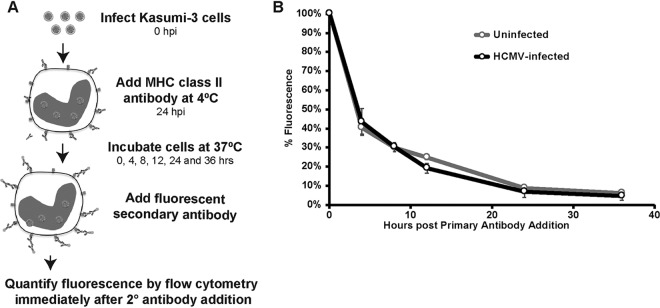
Surface expression of MHC class II is reduced by HCMV, and understanding the mechanism of this downregulation is important for identifying the viral factor mediating this effect. Potential mechanisms of downregulation include actively targeting the MHC class II molecules for internalization and degradation, preventing the proper assembly and loading of the MHC class II complex, or modulating MHC class II expression at the transcriptional or translational level. We first tested whether HCMV increases the endocytosis rate of surface MHC class II molecules (Fig. 6A). HCMV-infected Kasumi-3 cells were labeled with antibody against HLA-DR at 24 h postinfection. Secondary antibody conjugated to a fluorescent probe was then added at 0, 4, 8, 12, 24, and 36 h after the addition of primary antibody, and immunofluorescence was measured using flow cytometry. The internalization of MHC class II, as measured by the rate of fluorescence loss, was indistinguishable between uninfected and HCMV-infected cells (Fig. 6B). Thus, the difference in surface MHC class II levels is not likely due to an increase in the internalization and degradation of the complex.
FIG 6.
HCMV does not alter the rate of MHC class II internalization in Kasumi-3 cells. (A) Schematic showing the strategy for measuring the rate of MHC class II internalization. (B) Graph plotting the remaining surface HLA-DR levels in uninfected or infected samples at 0, 4, 8, 12, 24, and 36 h after the addition of primary antibody. Values are the means from three independent experiments.
HCMV reduces MHC class II transcription by preventing CIITA expression.
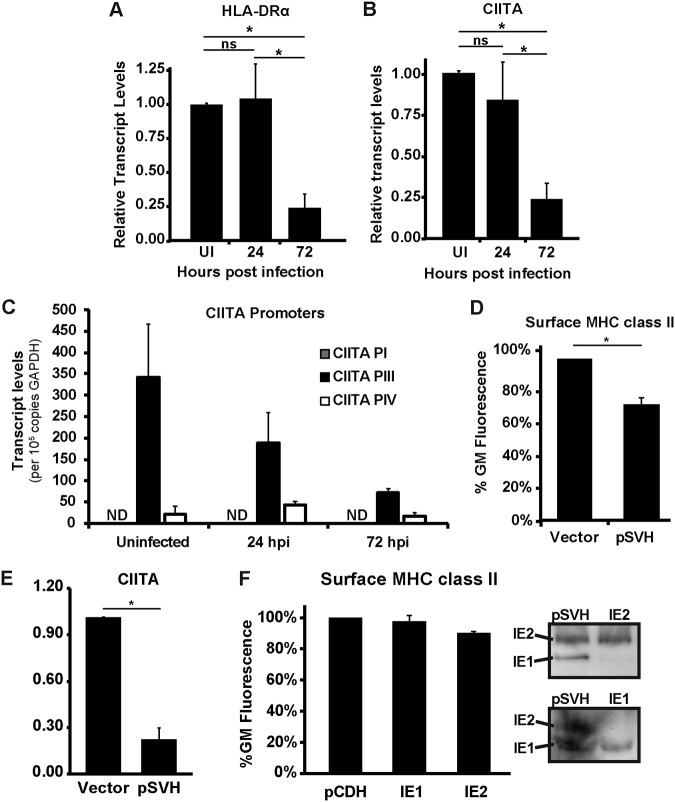
We next tested whether MHC class II was regulated at the transcriptional level. At 24 hpi, the levels of the MHC class II HLA-DRα transcript were unchanged. However, at 72 hpi, transcript levels were significantly reduced (Fig. 7A). Thus, HCMV either reduces the expression or enhances the degradation of HLA-DRα transcripts. The HLA class II promoter is dependent upon the MHC class II transactivator (CIITA). We next investigated whether HCMV also transcriptionally downregulates CIITA. We detected a small but not significant decrease in CIITA transcripts at 24 hpi, with a much greater and significant reduction in transcript levels being observed at 72 hpi (Fig. 7B). Thus, the reduction in MHC class II can be explained by the downregulation of its essential transcriptional transactivator.
FIG 7.
HCMV reduces MHC class II in Kasumi-3 cells by downregulating the expression of CIITA. (A and B) Quantitative PCR analysis of HLA-DRα (A) or CIITA (B) transcript levels in uninfected (UI) and HCMV-infected samples at 24 and 72 hpi. The values shown are relative to those for uninfected samples after normalization to the value for GAPDH. (C) Quantitative PCR analysis of CIITA transcripts derived from promoter I, III, or IV (PI, PIII, or PIV, respectively) in uninfected and HCMV-infected samples at 24 and 72 hpi. Values are the absolute numbers of transcripts per 105 copies of GAPDH. ND, not detected. (D) Quantification of the geometric mean fluorescence for surface HLA-DR in Kasumi-3 cells electroporated with a DNA control (Vector) or pSVH. (E) Quantitative PCR analysis of CIITA transcripts following electroporation of a DNA control (Vector) or pSVH into Kasumi-3 cells. (F) (Left) Quantification of the geometric mean fluorescence for surface HLA-DR in Kasumi-3 cells electroporated with the pCDH vector, pCDH-IE1, or pCDH-IE2. (Right) Western blots of the samples for which the results are shown in panels D and F, showing the expression of IE1 and IE2 after transfection of the pSVH, pCDH-IE1, and pCDH-IE2 plasmids. All values in panels A to F are averages from at least three independent experiments. *, P < 0.05; ns, not statistically significant.
CIITA contains four promoters, and to define how HCMV downregulates CIITA transcription, it is first important to understand the contribution of each of these promoters. Transcriptional activity has largely been assigned to promoters I, III, and IV. Promoter I is active in monocyte-derived dendritic cells, promoter III is active in B cells, T cells, and monocytes, and promoter IV is active in response to IFN-γ induction (39–41). We investigated which promoter(s) was active in uninfected Kasumi-3 cells and the effect of infection on promoter usage. The predominant transcripts in uninfected Kasumi-3 cells were a product of promoter III (Fig. 7C). A very small population of transcripts derived from promoter IV was also present, but no transcripts corresponding to promoter I could be detected. Thus, Kasumi-3 cells primarily utilize promoter III to endogenously express CIITA. As expected, the levels of transcripts generated from promoter III were decreased at 24 and 72 hpi, similar to what was observed for total CIITA transcript levels (Fig. 7C). The minor pool of transcripts derived from promoter IV remained relatively constant throughout the infection. Thus, the decrease in CIITA transcript levels can be attributed to either repression of promoter III transcription or a decrease in the stability of transcripts derived from promoter III.
The major immediate early (MIE) proteins are multifunctional proteins essential for HCMV infection that regulate the transcription of a diverse set of genes. We hypothesized that these genes transcriptionally repress CIITA promoter III. This would be consistent with our observation that the factor responsible for downregulation is an immediate early or early gene, since addition of acyclovir did not block the downregulation of surface HLA-DR. We first transfected Kasumi-3 cells with a plasmid that contains the entire major immediate early gene region (42) and that can express both IE1 and IE2 as well as the other gene products produced from this region. We observed reduced surface HLA-DR levels in these samples at 48 h posttransfection (Fig. 7D). The reduced surface HLA-DR correlated with a decrease in CIITA transcripts (Fig. 7E). We next wanted to determine which of the major immediate proteins was responsible for this downregulation. The major immediate protein 2 (IE2) was the more abundantly expressed MIE protein in our infections (Fig. 5A) and can act as a transcriptional repressor, which was first identified by its ability to directly bind a target sequence in its own promoter, termed the cis repression signal (43, 44). IE2 has since been shown to repress the transcription of other genes (45–48), and we hypothesized that IE2 is the MIE protein responsible for the transcriptional repression of CIITA. We next transfected Kasumi-3 cells with a plasmid that expressed IE2 alone. Interestingly, surface HLA-DR was not reduced in the presence of IE2 alone, suggesting that IE2 is insufficient by itself to mediate the transcriptional repression of CIITA and, hence, MHC class II transcription and the subsequent decrease in surface protein (Fig. 7F). IE1 was also insufficient by itself to mediate the downregulation of surface MHC class II (Fig. 7F). Western blot analysis confirmed protein expression from the transfected plasmids. Therefore, while HCMV decreases MHC class II surface expression by using the MIE region, IE1 and IE2 alone are insufficient. Thus, IE1 and IE2 either work in concert or one of the other gene products expressed from the pSVH plasmid is responsible for repressing CIITA transcript levels.
DISCUSSION
HCMV is an opportunistic pathogen that can cause disease in immunocompromised individuals and neonates. The virus has developed multiple strategies to effectively combat the host immune response, specifically, the MHC class I-driven CD8+ T cell response and NK cells. While there is some information on how HCMV controls MHC class II expression following interferon gamma induction or in overexpression systems, the regulation of endogenous MHC class II has been less well characterized. It is important to investigate MHC class II regulation under endogenous expression conditions, as both HCMV and MCMV are known to interfere with interferon gamma signal transduction (19, 24, 49). Also, the MHC class II synthesis and processing pathways can be quite distinct within different cell types (reviewed in references 17 and 50), thus highlighting the importance of investigating MHC class II regulation in cells relevant to the host infection. HCMV establishes latency in undifferentiated cells of the myeloid lineage, and Kasumi-3 cells express markers of myeloid progenitor cells, can differentiate down the myeloid pathway, and have been shown to be a suitable model for supporting cytomegalovirus latency and reactivation (29). Importantly, they robustly express HLA-DR on their surface, making them a great model for investigating the HCMV-mediated mechanism of MHC class II downregulation.
Modulation of MHC class II is functionally important for infection. CD4+ T cells are critical for resolution of persistent murine cytomegalovirus (MCMV) infection in salivary glands (51). Additionally, CD4+ T cells reactive to HCMV antigens are found within the human host (52). Blunting of the MHC class II-primed CD4+ T cell response would be advantageous to the virus since CD4+ T cells form an essential branch of the adaptive immune system for clearance of the virus. Endogenous gB can be presented by infected cells to prime CD4+ T cells (53); therefore, understanding MHC class II presentation in the context of endogenous antigen presentation in infected cells is essential. We observed a reduction in surface and total HLA-DR in infected Kasumi-3 cells over the course of infection. Interestingly, this downregulation occurred only in infected cells and was not observed in uninfected cells in the same culture. Thus, the downregulation is not mediated by a secreted factor, such as cellular IL-10. This highlights the importance of MHC class II regulation within the infected cells and serves to demonstrate that HCMV decreases HLA-DR in these cells as a means to prevent endogenous MHC class II presentation.
In our efforts to define the mechanism of the HCMV-mediated decrease in MHC class II during infection of Kasumi-3 cells, we were surprised to find that previously reported genes were not singularly responsible for this phenotype. However, rat cytomegalovirus (RCMV) downregulates MHC class II in bone marrow-derived dendritic cells independently of these previously described mechanisms as well (54). Although of interest, RCMV downregulates MHC class II by endocytic degradation and not transcription, as we found for HCMV, suggesting a divergence of the mechanism (54). In light of these results, we undertook a targeted screen of the unique short (US) region of the viral genome. HCMV encodes various proteins within the US region to block different steps of the MHC class I processing and presentation pathway and NK cell activation. We wanted to investigate whether a similar mechanism exists for MHC class II presentation, as many of the proteins encoded within this region have no known function. Surprisingly, none of the mutants in our screen blocked the ability of the virus to decrease HLA-DR surface expression. This is an important finding, since it shows that the US region does not contribute to the endogenous MHC class II regulation in Kasumi-3 cells, unlike the important role that this region plays in limiting the MHC class I pathway. However, we cannot rule out the possibility that this may occur as a concerted action of multiple genes and that deletion of these genes singularly does not block the MHC class II decrease.
Viral modulation of immune proteins can occur through a variety of mechanisms. One such strategy is the targeting of immune proteins directly for degradation. The related gammaherpesvirus, Epstein-Barr virus (EBV), utilizes BDFL3 to increase the internalization and promote the proteosomal degradation of MHC class I and class II (55). We assessed two potential HCMV-encoded proteins to determine whether they play a role in the altered trafficking of MHC class II, UL20 and UL78. UL20 is a putative T cell receptor gamma homologue expressed early in infection that is targeted to the lysosomes immediately after synthesis and that is dispensable for replication in culture (37). A recent report showed the role of M78 in MCMV infection for the regulation of MHC class II and CD4+ T cell activation (22). UL78 is the HCMV homologue of M78 that has endocytic activity and localization similar to those of M78 (56). However, the loss of either UL20 or UL78 did not rescue MHC class II surface expression. Furthermore, we found that the rates of HLA-DR internalization between infected and uninfected cells were not significantly different. Hence, HCMV utilizes an alternative strategy for downregulating surface MHC class II molecules in Kasumi-3 cells.
Another strategy utilized by viruses is to reduce the class II transactivator (CIITA), the master regulator for MHC class II synthesis and processing. CIITA is essential for MHC class II presentation because it increases the transcription of multiple factors in the pathway, including the MHC class II HLA genes (HLA-DR, HLA-DQ, and HLA-DP) and their accessory factors (HLA-DM, HLA-DO, and invariant chain) (reviewed in reference 57). Our results show that CIITA transcript levels in fact decreased during infection. Thus, by blocking CIITA, the entire MHC class II presentation pathway comes to a significant halt. This is consistent with the findings of a study showing decreased CIITA expression in mature Langerhans cells, a specialized dendritic cell, after HCMV infection (58). These results show that HCMV may utilize the repression of CIITA as a general mechanism within cells of the myeloid lineage to inhibit the MHC class II machinery.
CIITA transcription is modulated by differential promoter usage in different cell types in response to various conditions. The type I promoter is active in dendritic cells, and the type III promoter is active in B cells, monocytes, and T cells, whereas the type IV promoter is driven by interferon gamma stimulation in multiple cell types (39–41). We found promoter III-derived CIITA transcripts to be the primary form expressed in Kasumi-3 cells. Accordingly, these accounted for the downregulation during infection. While we did not find promoter IV transcripts to be significantly regulated by HCMV, this may be due to the absence of interferon gamma in our in vitro, cell culture infection system. However, HCMV may regulate these transcripts in the host, where IFN-γ may be present, and activate transcription from this promoter. Since HCMV can modulate interferon gamma signaling (19, 23), this may be one way of decreasing promoter IV CIITA transcripts. Coculturing infected Kasumi-1 cells with uninfected peripheral blood mononuclear cells resulted in an increase in HLA-DR transcript levels, illustrating the complexity of the HCMV-mediated regulation of MHC class II under different conditions (59). Importantly, our results focus on the endogenous regulation of MHC class II and show that HCMV downregulates the expression of CIITA by decreasing transcription from promoter III, resulting in decreased HLA-DRα transcripts and MHC class II surface expression.
HCMV encodes factors that have demonstrated transcriptional repressor activity. The major immediate early proteins IE1 and IE2 exercise transcriptional control over many viral and cellular genes and are essential for promoting viral replication. IE2 has transcription repressor activity, as first identified for its own promoter (44). While expression of the entire major immediate early region resulted in reduced surface MHC class II levels and CIITA transcript levels, IE1 and IE2 individually were insufficient to mediate this decrease. Thus, other products from this region are necessary to achieve the MHC class II reduction, either independently or in conjunction with IE1 or IE2. There are no known viral microRNAs reported to be expressed from the major immediate early promoter and protein region contained in pSVH. However, seven other open reading frames have been reported in this region, and six of these are classified as cytomegalovirus latency-specific transcripts and originate from either an upstream start site or the antisense strand of the MIE proteins (60). The products of several of these ORFs elicit a serological response, suggesting that they are produced in the host (61) and can be present in both a lytic infection and a latent infection (62). Of particular interest is ORF94, which is a nuclear protein that has been shown to reduce the constitutive expression of 2′,5′-oligoadenylate synthetase (63). The mechanism for decreasing CIITA promoter III transcripts certainly requires further investigation and may be direct transcriptional repression by binding the promoter or could involve inhibition by noncoding RNAs, epigenetic modification of the CIITA gene, or downregulation of cellular factors required for CIITA transcription. We have also not ruled out the possibility that the decrease in transcripts may be posttranscriptional, for example, as a result of a decrease in message stability. Although more work is required to define this mechanism, we have described the regulation of endogenous MHC class II surface expression by the major immediate early region in a relevant cell type for infection.
MATERIALS AND METHODS
Cell culture.
Normal human dermal fibroblasts (HDFs; catalog number 106-05n; Cell Applications Inc.) and normal human lung fibroblast MRC-5 cells (ATCC CCL-171) were maintained in Dulbecco’s modified Eagle’s medium (DMEM; Corning) containing 10% fetal bovine serum (FBS; HyClone), 2 mM GlutaMAX (Gibco), and 100 U/ml penicillin and 100 g/ml streptomycin (Corning). Kasumi-3 cells (ATCC CRL-2725) were maintained in RPMI 1640 medium (ATCC) supplemented as described above for DMEM, with the exception that 20% FBS was used. Human peripheral blood monocytes (catalog number 6906K-50A; Cell Applications Inc.) were maintained in the manufacturer’s supplied human blood cell culture medium. Cells were maintained at 37°C with 5% CO2.
BAC mutagenesis.
Recombinant viruses were generated using BAC mutagenesis in Escherichia coli strain SW105 and galK selection as described previously (64). Mutant viruses were generated from a TB40/E BAC expressing mCherry (34). A virus that expresses IE2-2A-EGFP (65) was utilized for the internalization studies. The parent BAC for this IE2-2A-EGFP virus, which also expressed mCherry, was a gift from Eain Murphy, and the mCherry sequence was replaced with the wild-type sequence, as described below. The primers used for recombineering for the generation of all mutant strains were as follows: UL111A-galk-For (5′-TGGGACGCGCAGTTGGGTGGCGGACTGGGGCGGCATGCTGCGGCGCCTGTTGACAATTAATCATCGGCA), UL111A-galk-Rev (5′-AAAAAAGACGATCAGGACCAGAGAGGAAGAGACCATCACCGACAGTCAGCACTGTCCTGCTCCTT), UL111A-STOP-sense (5′-GACGCGCAGTTGGGTGGCGGACTGGGGCGGCATGCTGCGGCGTAGCTGTCGGTGATGGTCTCTTCCTCTCTGGTCCTGATCGTCTTTTTT), UL111A-STOP-antisense (5′-AAAAAAGACGATCAGGACCAGAGAGGAAGAGACCATCACCGACAGCTACGCCGCAGCATGCCGCCCCAGTCCGCCACCCAACTGCGCGTC), UL78-galk-For (5′-GGAGAGGGTATATTCGTTCGGCGAGAGCGGGCGGCGGTGGTGGGTCCTGTTGACAATTAATCATCGGCA), UL78-galk-Rev (3′-GACGTGATTTATCTGCCACTTTTCTCCCCGCTGCCGTACAGCGCCGCCGC TCAGCACTGTCCTGCTCCTT), US7-US12-galk-For (5′-GGTTTATATATGACCATCCACGCTTATAACGAACCTAACAGTTTACCTGTTGACAATTAATCATCGGCA), US7-12-galk-Rev (5′-CCCATCGTCCCCCTTTCTCTATAAAACTTGCCGGGTACCTGAAGCTCAGCACTGTCCTGCTCCTT), US13-US18-galk-For (5′-GCTTCAGGTACCCGGCAAGTTTTATAGAGAAAGGGGGACGATGGGCCTGTTGACAATTAATCATCGGCA), US13-US18-galk-Rev (5′-GTAACCGGGTGCTGATAAGACGGACTGTTTCATCGACGCCTACCTTCAGCACTGTCCTGCTCCTT), US19-US24-galk-For (5′-TCACGAGTGTGGTCAAACCGTGGCGGCACCCTGTATCCGACCCGTCCTGTTGACAATTAATCATCGGCA), US19-US24-galk-Rev (5′-GGTGACGGTGTAGCGTTGCTTTCTCTGTATTTGGCTCGGCTTCTGTCAGCACTGTCCTGCTCCTT), US26-US30-galk-For (5′-CTCTCAGCCGGACAACCGGCGTCACTGACAGAAGCCGAGCCAAATCCTGTTGACAATTAATCATCGGCA), US26-US30-galk-Rev (5′-GTCTCACCGATGAGACACCGACCGCACTCGAGAGTAAAGACAAATTCAGCACTGTCCTGCTCCTT), US31-US34A-galk-For (5′-TGTCTCATCGGTGAGACGAGGCCGCCGCCCGACAAGTTCGATCTCCCTGTTGACAATTAATCATCGGCA), US31-US34A-galk-Rev (5′-TCGGCATCTTTGTCAATAAGACGCACGCCGCCGTGACCCATACCGTCAGCACTGTCCTGCTCCTT), UL20-galk-For (5′-CGGTCTTTATATATACAAACGCCGTTATGCTCAGTGTCCGGCAAGCCTGTTGACAATTAATCATCGGCA), UL20-galk-Rev (5′-TGCCCAGTGGTAATTCAGCATCACCAGCATAGCATGTATCCCGAGTCAGCACTGTCCTGCTCCTT), UL20-STOPGFP-For (5′-CGGTCTTTATATATACAAACGCCGTTATGCTCAGTGTCCGGCAAGATGGTGAGCAAGGGCGAG), UL20-STOPGFP-Rev (5′-TGCCCAGTGGTAATTCAGCATCACCAGCATAGCATGTATCCCGAGTTACTTGTACAGCTCGTC), mCherry-restore-galk-For (5′-TGTATTTGTGACTATACTATGTGCAGTCGTGTGTCGATGTTCCTATTGGGCCTGTTGACAATTAATCATCGGCA), mCherry-restore-galk-Rev (5′-GATGTCTTCCTGCGTCCCACCATTCTTTATACCTCCTACATTCACACCCTTTCAGCACTGTCCTGCTCCTT), mCherry-restore-For (5′-TGTATTTGTGACTATACTATGTGCAGTCGTGTGTCGATGTTCCTATTGGGAAGGGTGTGAATGTAGGAGGTATAAAGAATGGTGGGACGCAGGAAGACATC), mCherry-restore-Rev (5′-GATGTCTTCCTGCGTCCCACCATTCTTTATACCTCCTACATTCACACCCTTCCCAATAGGAACATCGACACACGACTGCACATAGTATAGTCACAAATACA).
BAC sequences were PCR amplified from purified BAC DNA and verified by Sanger sequencing (Genewiz). The primers used for verifying insertion at the desired sites were US7-US12-galk-For (5′-AACGAACCTAACAGTTTA), US7-12-galk-Rev (5′-TGACCATGTTTAAAATCACGC), US13-US18-galk-For (5′-AGAAAGGGGGACGATGGG), US13-US18-galk-Rev (5′-CATGACGACGGGTCGGATACA), galk-237.2-For (5′-TCCAGCAGCCAGCCCTGC), US19-US24-galk-Rev (5′-TAGGTCGGTTCTCAGACCGAG), US26-US30-galk-Rev (5′-GTACACCACAGGTTGTGCGAG), US31-US34A-galk-For (5′-CCCGACAAGTTCGATCTC), and US31-US34A-galk-Rev (5′-CCCACATCCCACCGCCTTTTA).
HCMV infections and virus titrations.
All cytomegaloviruses stocks were generated from BACs. These included AD169, TB40/E, and the TB40/E derivatives, generated as described above. Virus stocks were propagated by electroporating purified BAC DNA into MRC-5 cells according to previously published protocols (66) and subsequently freezing the infected cells as passage 0 (P0) stocks. These stocks were added to MRC-5 cells in roller bottles (Greiner) to generate P1 virus stocks. The virions produced in roller bottles were concentrated by ultracentrifugation on a 20% sorbitol cushion at 20,000 rpm for 1 h at 20°C in a Beckman SW32 rotor. HCMV stocks were titrated by serial dilution on MRC-5 cells and quantified by the immunological detection of immediate early proteins as previously described (67), using an antibody that detects the HCMV major immediate early proteins (68). Images of stained monolayers were acquired on a Nikon Eclipse Ti inverted microscope, and fluorescent nuclei were quantified using NIS Elements software.
Kasumi-3 cells were infected as previously described, with some modification (29). Briefly, cells were cultured in X-Vivo medium (Lonza) for 48 h prior to infection. Cells were infected with TB40/E-mCherry wild-type or mutant viruses (as described above) in X-Vivo medium with sufficient virus to result in infection of 10 to 20% of the cells (MOI, 10 to 40) and subjected to a spin at 1,000 × g for 30 min at room temperature. The cells were incubated overnight at 37°C, washed twice with 1× phosphate-buffered saline (PBS) on the following day, and resuspended in fresh RPMI (with 20% FBS) following the final wash. Infected cells were cultured in RPMI (with 20% FBS) at 37°C until analysis. Where indicated, acyclovir (EMD Millipore) or dimethyl sulfoxide (DMSO; Fisher) was added at the time of infection at a concentration of 100 μg/ml, and the medium was supplemented with acyclovir or DMSO following washing and resuspension of the cells at 24 h. For UV inactivation, virus was placed under a Mineralight ultraviolet lamp (shortwave UV at 254 nm) for 30 min at a distance of 10 cm. UV inactivation was confirmed by a lack of mCherry-positive cells upon infection. For infection of primary monocytes, cells were resuspended in X-Vivo medium, infected at an MOI of 40, and subjected to a spin at 400 × g for 30 min at room temperature. The cells were incubated overnight at 37°C and were washed twice with blood culture medium on the following day. Infected cells were cultured in blood culture medium at 37°C until analysis.
Single-step growth curve analysis of all BAC-generated mutant HCMVs was performed on HDFs infected at an MOI of 3. Virus was added to the HDFs and allowed to incubate for 3 h at 37°C. The cells were washed twice with 1× PBS and the medium was replaced with fresh growth medium. Virus was harvested at 120 h postinfection by scraping the cells in the medium, sonicating with 1-s pulses 10 times, vortexing for 15 s, and centrifuging at 13,000 rpm for 10 min at 4°C. The supernatants were collected, aliquoted, flash frozen in liquid nitrogen, and stored at −80°C until analysis. For analysis, the samples were titrated by serial dilution and quantified as described above.
Flow cytometry.
Kasumi-3 cells were harvested, washed with cold wash buffer (2% bovine serum albumin [BSA] in 1× PBS with 1 mM EDTA and 0.01% sodium azide), and blocked with human TruStain FcX for 10 min at room temperature, followed by incubation with antibodies against HLA-DR-allophycocyanin (clone LN3; Thermo Fisher Scientific), HLA-DP (clone B7/21; Abcam), and anti-HLA-DQ-fluorescein isothiocyanate (clone Tü169; BioLegend) on ice for 30 min. For unconjugated primary antibodies, the cells were pelleted and incubated with secondary antibody (Thermo Fisher Scientific) on ice for 30 min. After antibody staining, the cells were washed, resuspended in cold wash buffer, and kept on ice until data acquisition on an LSR II flow cytometer (BD Biosciences). At the time points indicated above, live cells were identified using Sytox blue dead cell stain (Thermo Fisher Scientific) or Zombie UV fixable dye (BioLegend), and infected cells were selected by gating for mCherry. For total HLA-DR staining, cells were fixed and permeabilized using a fix/permeabilization kit (BD Biosciences) and staining was performed as described above. Data analysis for the samples was performed with FlowJo software (TreeStar), and the geometric mean fluorescence intensity was calculated for HLA-DR on gated cell populations.
Western blotting.
Kasumi-3 cells were infected with a virus lacking both US23 and US24, both of which augment virus growth when individually disrupted by transposon mutagenesis (69). Lacking these genes allowed the virus to grow to higher titers on fibroblasts (titers 100- to 1,000-fold higher than those of wild-type viruses) to obtain a population of nearly completely infected cells (>95%). At 24 and 72 h postinfection, uninfected and infected cells were harvested in radioimmunoprecipitation assay (RIPA) buffer (1% NP-40, 1% sodium deoxycholate, 0.1% sodium dodecyl sulfate [SDS], 0.15 M NaCl, 10 mM sodium phosphate [pH 7.2], 2 mM EDTA, 1 mM dithiothreitol, 50 mM NaF, 1 mM phenylmethylsulfonyl fluoride, 1 mM aprotinin, 0.2 mM Na3VO4, 1 μg/ml leupeptin). The cells were pelleted, resuspended in RIPA buffer, and stored at −80°C. Samples were analyzed by SDS-polyacrylamide gel electrophoresis and immunoblotting on a polyvinylidene difluoride membrane blocked with 5% milk Tris-buffered saline (with 0.1% Tween 20). The following primary antibodies were used: anti-HLA-DR (clone TAL 1B5; Novus Biologicals), anti-exon2/3 against cytomegalovirus immediate early genes (a gift from Jim Alwine [70]), anti-p115 (Proteintech), anti-pp71 (clone 2H10-9; kindly provided by John Purdy [71]), anti-pp28 (clone 5C3; Virusys), anti-pp150 (clone 36-14; the clone was a gift from David Spector and was originally generated by Bill Britt), and anti-UL71 (clone 1G, generated by Neil Christensen). The horseradish peroxidase-conjugated secondary antibodies were from GE Healthcare. Antibody dilutions were in accordance with the manufacturers’ instructions. Blots were developed with the SuperSignal West Pico Plus chemiluminescent substrate (Thermo Fisher Scientific).
Immunofluorescence microscopy and imaging.
Kasumi-3 cells were harvested, washed twice with RPMI (with 20% FBS), and pelleted by centrifugation at 100 × g for 8 min. The cell pellets were resuspended in 100 μl of RPMI (with 20% FBS) and added to poly-d-lysine coverslips (Electron Microscopy Sciences). The cells were allowed to attach for 10 min, followed by centrifugation at 1,000 × g for 10 min and incubation for 1 h at room temperature. Coverslips with adhered cells were fixed by adding 100 μl of 4% paraformaldehyde directly to the cells and medium for 15 min at room temperature. The cells were blocked in PBS containing 10% human serum, 0.5% Tween 20, and 5% glycine. Triton X-100 (0.1%) was added for permeabilization. Primary and secondary antibodies were diluted in blocking buffer. The primary antibody for HLA-DR was clone L243 (BioLegend), and that for LAMP1 was clone D2D11 (Cell Signaling Technologies). Alexa Fluor 488 and Alexa Fluor 647 were used as the secondary antibodies (Thermo Fisher Scientific). Coverslips were mounted with ProLong Diamond antifade mountant with DAPI (4′,6-diamidino-2-phenylindole; Thermo Fisher Scientific). Images were taken on a C2+ confocal microscope system (Nikon). Images were minimally processed using NIS Elements software. The images shown are a single slice of a z-stack.
Reverse transcriptase and quantitative PCR.
Kasumi-3 cells were infected as described above for Western blot analysis. Uninfected and infected cells were harvested at 24 and 72 hpi by washing with 1× PBS and resuspending in lysis buffer supplied in an RNeasy minikit (Qiagen) for RNA extraction, which was subsequently performed following the manufacturer’s protocol. The RNA concentration was quantified using a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific). cDNA was synthesized from RNA using SuperScript first-strand synthesis system for reverse transcription-PCR (RT-PCR) (Thermo Fisher Scientific) following the manufacturer’s protocol. Samples were cycled as follows on a StepOnePlus real-time PCR system (Thermo Fisher Scientific): 50°C for 2 min, 95°C for 10 min, and 95°C for 15 s, 60°C for 60 s, and 55°C for 30 s for 40 cycles. The primers used for quantitative PCR were as follows: HLA-DR-For (5′-CGAGTTCTATCTGAATCCTG-3′), HLA-DR-Rev (5′-CTGGAGGTACATTGGTGA-3′), pan-CIITA-For (5′-AGCCTTTCAAAGCCAAGTCC-3′), pan-CIITA-Rev (5′-TTGTTCTCACTCAGCGCATC-3′), CIITA-PI-For (5′-GGAGACCTGGATTTGGCCCT-3′), CIITA-PIII-For (5′-GGGGAAGCTGAGGGCACG-3′), CIITA-PIV-For (5′-GCGGCCCCAGAGCTGG-3′), CIITA-PI-PIII-PIV-Rev (5′-GAAGCTCCAGGTAGCCACCTTCTA-3′), GAPDH-For (5′-ACCCACTCCTCCACCTTTGAC-3′), GAPDH-Rev (5′-CTGTTGCTGTAGCCAAATTCGT-3′), US2-For (5′-ATGAACAATCTCTGGAAA-3′), US2-Rev (5′-GATTTGAAACCAGGGATG-3′), US3-For (5′-GACCATCAACTGGTACCT-3′), US3-Rev (5′-AATAAATCGCAGACGGGC-3′), IE1-For (5′-TTCCCAGAATTGGCCGAA-3′), and IE1-Rev (5′-TCTGTTTGACGAGTTCTGCCA-3′). The data were analyzed using the delta threshold cycle method with GAPDH (glyceraldehyde-3-phosphate dehydrogenase) as a normalization control. To calculate absolute transcript numbers, standard curves for each primer set were generated by use of a dilution series (1 ng to 1 fg). Samples from RT-PCR were run on a 4% agarose gel for visualization.
Amaxa transfection and cell sorting.
Kasumi-3 cells (1 million or 2 million cells per transfection) were centrifuged at 100 × g for 10 min, resuspended in 100 μl of Cell Line Nucleofector solution R (Lonza), and mixed with 1 or 2 μg plasmid DNA. The cell suspension and reagent mixture were added to Nucleofector cuvettes, and electroporation was performed using the program V-001 in a Nucleofector 2b device. Prewarmed RPMI medium (with 20% FBS) was added to the electroporated cells, and the cells were incubated at 37°C until they were harvested for flow cytometry analysis. Electroporated cells were identified by the presence of GFP. The nonfluorescent pSVH and vector control plasmid were cotransfected with 0.2 μg pMAX-GFP to allow for the detection of transfected cells. The plasmid DNA used was from the plasmids pEGFPC1 (Clontech), pEGFPC1-pp65 and pSVH (both of which were gifts from Jim Alwine), pCDH (puro2AGFP), pCDH-IE1 (puro2AGFP), pCDH-IE2 (puro2AGFP), and pMAX-GFP (Lonza). pCDH (puro2AGFP) was generated by stitching a T2A-enhanced green fluorescent protein (EGFP) sequence to the 3′ end of puromycin. The EGFP sequence was amplified from pEGFPN2 with a primer that introduced an N-terminal T2A sequence downstream of 12 bp homologous to the 3′ end of puromycin. Puromycin was amplified from pCDH-CMV-MCS-E1-Puro (Systems Biosciences) with primers that introduced C-terminal homology to 15 bp at the 5′ end of the T2A-EGFP PCR product. The T2A-EGFP and puromycin PCR products were stitched together in a PCR that introduced an XhoI site 3′ to the EGFP sequence. The resulting insert was digested with BsiWI and XhoI and ligated into pCDH-CMV-MCS-E1-Puro that had been digested with BsiWI and SalI. pCDH-IE2 (puro2AGFP) was generated by amplifying the IE2 sequence from pIE86 with primers containing XbaI and EcoRI and ligating the resulting insert into pCDH (puro2AGFP) that had been cut with XbaI and EcoRI. The ORF of IE1 flanked by XbaI and EcoRI sites was purchased from Integrated DNA Technologies and inserted into pCDH (puro2AGFP) that had been cut with XbaI and EcoRI. pCDH-UL78 (puro2AGFP) was generated by amplifying the UL78 sequence from HCMV BAC (TB40/E) DNA with primers containing BamHI and EcoRI and ligated into pCDH (puro2AGFP) that had been cut with BamHI and EcoRI. To generate RNA for cDNA synthesis and subsequent quantitative PCR analysis, Kasumi-3 cells were electroporated with pSVH or pCDH and cotransfected with pMax-GFP, as described above. GFP-positive cells were sorted using a FACSAria SORP cell sorter (BD Biosciences) at 48 h posttransfection, and RNA was isolated immediately after sorting.
Internalization assay.
For the HLA-DR internalization assay, TB40/E IE2-2A-EGFP virus was used to infect Kasumi-3 cells as described above. Infected and uninfected cells were harvested at 24 h postinfection, incubated with unconjugated anti-HLA-DR (clone L243; BioLegend) for 30 min on ice, washed with cold, sterile wash buffer (2% BSA in 1× PBS), and resuspended in RPMI (with 20% FBS). The cells were incubated at 37°C and harvested at 0 h, 4 h, 8 h, 12 h, 24 h, and 36 h after primary antibody labeling. Cells were stained with anti-mouse immunoglobulin-Alexa Fluor 647 (Thermo Fisher Scientific), and data acquisition was performed on an LSR II flow cytometer (BD Biosciences). Data analysis was performed using FlowJo software (TreeStar), and the geometric mean fluorescence intensity was calculated for HLA-DR.
ACKNOWLEDGMENTS
We thank Alexandria Ostman and Natalie Buchkovich for technical assistance in generating reagents and performing experiments. We thank Chris Norbury and John Wills for helpful discussions and suggestions on preparing the manuscript and Christine O’Connor for advice on working with the Kasumi-3 cell infection system. We especially thank Neil Christensen for generating the UL71 antibody. We also thank Nate Sheaffer and Joseph Bednarczyk from the Penn State College of Medicine’s Flow Cytometry Core for assistance with flow cytometry analysis and cell sorting.
The work and the authors were supported by NIH grant R01AI130156 to N.J.B.
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
REFERENCES
- 1.Weekes M, Tomasec P, Huttlin E, Fielding C, Nusinow D, Stanton R, Wang E, Aicheler R, Murrell I, Wilkinson G, Lehner P, Gygi S. 2014. Quantitative temporal viromics: an approach to investigate host-pathogen interaction. Cell 157:1460–1472. doi: 10.1016/j.cell.2014.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fielding CA, Aicheler R, Stanton RJ, Wang ECY, Han S, Seirafian S, Davies J, McSharry BP, Weekes MP, Antrobus PR, Prod'homme V, Blanchet FP, Sugrue D, Cuff S, Roberts D, Davison AJ, Lehner PJ, Wilkinson GWG, Tomasec P. 2014. Two novel human cytomegalovirus NK cell evasion functions target MICA for lysosomal degradation. PLoS Pathog 10:e1004058. doi: 10.1371/journal.ppat.1004058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fielding CA, Weekes MP, Nobre LV, Ruckova E, Wilkie GS, Paulo JA, Chang C, Suárez NM, Davies JA, Antrobus R, Stanton RJ, Aicheler RJ, Nichols H, Vojtesek B, Trowsdale J, Davison AJ, Gygi SP, Tomasec P, Lehner PJ, Wilkinson GW. 2017. Control of immune ligands by members of a cytomegalovirus gene expansion suppresses natural killer cell activation. Elife 6:e22206. doi: 10.7554/eLife.22206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baillie J, Sahlender DA, Sinclair JH. 2003. Human cytomegalovirus infection inhibits tumor necrosis factor alpha (TNF-alpha) signaling by targeting the 55-kilodalton TNF-alpha receptor. J Virol 77:7007–7016. doi: 10.1128/jvi.77.12.7007-7016.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dunn C, Chalupny NJ, Sutherland CL, Dosch S, Sivakumar PV, Johnson DC, Cosman D. 2003. Human cytomegalovirus glycoprotein UL16 causes intracellular sequestration of NKG2D ligands, protecting against natural killer cell cytotoxicity. J Exp Med 197:1427–1439. doi: 10.1084/jem.20022059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith W, Tomasec P, Aicheler R, Loewendorf A, Nemčovičová I, Wang EC, Stanton RJ, Macauley M, Norris P, Willen L, Ruckova E, Nomoto A, Schneider P, Hahn G, Zajonc DM, Ware CF, Wilkinson GW, Benedict CA. 2013. Human cytomegalovirus glycoprotein UL141 targets the TRAIL death receptors to thwart host innate antiviral defenses. Cell Host Microbe 13:324–335. doi: 10.1016/j.chom.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ahn K, Gruhler A, Galocha B, Jones TR, Wiertz EJ, Ploegh HL, Peterson PA, Yang Y, Früh K. 1997. The ER-luminal domain of the HCMV glycoprotein US6 inhibits peptide translocation by TAP. Immunity 6:613–621. doi: 10.1016/s1074-7613(00)80349-0. [DOI] [PubMed] [Google Scholar]
- 8.Wiertz EJ, Jones TR, Sun L, Bogyo M, Geuze HJ, Ploegh HL. 1996. The human cytomegalovirus US11 gene product dislocates MHC class I heavy chains from the endoplasmic reticulum to the cytosol. Cell 84:769–779. doi: 10.1016/s0092-8674(00)81054-5. [DOI] [PubMed] [Google Scholar]
- 9.Jones TR, Wiertz EJ, Sun L, Fish KN, Nelson JA, Ploegh HL. 1996. Human cytomegalovirus US3 impairs transport and maturation of major histocompatibility complex class I heavy chains. Proc Natl Acad Sci U S A 93:11327–11333. doi: 10.1073/pnas.93.21.11327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jones TR, Sun L. 1997. Human cytomegalovirus US2 destabilizes major histocompatibility complex class I heavy chains. J Virol 71:2970–2979. doi: 10.1128/JVI.71.4.2970-2979.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ganesan A, Eisenlohr L. 2017. The elucidation of non-classical MHC class II antigen processing through the study of viral antigens. Curr Opin Virol 22:71–76. doi: 10.1016/j.coviro.2016.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Juno JA, van Bockel D, Kent SJ, Kelleher AD, Zaunders JJ, Munier CM. 2017. Cytotoxic CD4 T cells—friend or foe during viral infection? Front Immunol 8:19. doi: 10.3389/fimmu.2017.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tomazin R, Boname J, Hegde NR, Lewinsohn DM, Altschuler Y, Jones TR, Cresswell P, Nelson JA, Riddell SR, Johnson DC. 1999. Cytomegalovirus US2 destroys two components of the MHC class II pathway, preventing recognition by CD4+ T cells. Nat Med 5:1039–1043. doi: 10.1038/12478. [DOI] [PubMed] [Google Scholar]
- 14.Verma S, Weiskopf D, Gupta A, McDonald B, Peters B, Sette A, Benedict CA. 2016. Cytomegalovirus-specific CD4 T cells are cytolytic and mediate vaccine protection. J Virol 90:650–658. doi: 10.1128/JVI.02123-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Leeuwen EM, Remmerswaal EB, Heemskerk MH, ten Berge IJ, van Lier RA. 2006. Strong selection of virus-specific cytotoxic CD4+ T-cell clones during primary human cytomegalovirus infection. Blood 108:3121–3127. doi: 10.1182/blood-2006-03-006809. [DOI] [PubMed] [Google Scholar]
- 16.Casazza JP, Betts MR, Price DA, Precopio ML, Ruff LE, Brenchley JM, Hill BJ, Roederer M, Douek DC, Koup RA. 2006. Acquisition of direct antiviral effector functions by CMV-specific CD4+ T lymphocytes with cellular maturation. J Exp Med 203:2865–2877. doi: 10.1084/jem.20052246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Unanue ER, Turk V, Neefjes J. 2016. Variations in MHC class II antigen processing and presentation in health and disease. Annu Rev Immunol 34:265–297. doi: 10.1146/annurev-immunol-041015-055420. [DOI] [PubMed] [Google Scholar]
- 18.Hegde NR, Tomazin RA, Wisner TW, Dunn C, Boname JM, Lewinsohn DM, Johnson DC. 2002. Inhibition of HLA-DR assembly, transport, and loading by human cytomegalovirus glycoprotein US3: a novel mechanism for evading major histocompatibility complex class II antigen presentation. J Virol 76:10929–10941. doi: 10.1128/jvi.76.21.10929-10941.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller DM, Rahill BM, Boss JM, Lairmore MD, Durbin JE, Waldman JW, Sedmak DD. 1998. Human cytomegalovirus inhibits major histocompatibility complex class II expression by disruption of the Jak/Stat pathway. J Exp Med 187:675–683. doi: 10.1084/jem.187.5.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Slobedman B, Mocarski E, Arvin A, Mellins E, Abendroth A. 2002. Latent cytomegalovirus down-regulates major histocompatibility complex class II expression on myeloid progenitors. Blood 100:2867–2873. doi: 10.1182/blood.V100.8.2867. [DOI] [PubMed] [Google Scholar]
- 21.Odeberg J, Plachter B, Brandén L, Söderberg-Nauclér C. 2003. Human cytomegalovirus protein pp65 mediates accumulation of HLA-DR in lysosomes and destruction of the HLA-DR alpha-chain. Blood 101:4870–4877. doi: 10.1182/blood-2002-05-1504. [DOI] [PubMed] [Google Scholar]
- 22.Yunis J, Farrell HE, Bruce K, Lawler C, Sidenius S, Wyer O, Davis-Poynter N, Stevenson PG. 2018. Murine cytomegalovirus degrades MHC class II to colonize the salivary glands. PLoS Pathog 14:e1006905. doi: 10.1371/journal.ppat.1006905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Le Roy E, Mühlethaler-Mottet A, Davrinche C, Mach B, Davignon JL. 1999. Escape of human cytomegalovirus from HLA-DR-restricted CD4(+) T-cell response is mediated by repression of gamma interferon-induced class II transactivator expression. J Virol 73:6582–6589. doi: 10.1128/JVI.73.8.6582-6589.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heise MT, Connick M, Virgin HW. 1998. Murine cytomegalovirus inhibits interferon gamma-induced antigen presentation to CD4 T cells by macrophages via regulation of expression of major histocompatibility complex class II-associated genes. J Exp Med 187:1037–1046. doi: 10.1084/jem.187.7.1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jenkins C, Garcia W, Godwin MJ, Spencer JV, Stern JL, Abendroth A, Slobedman B. 2008. Immunomodulatory properties of a viral homolog of human interleukin-10 expressed by human cytomegalovirus during the latent phase of infection. J Virol 82:3736–3750. doi: 10.1128/JVI.02173-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cheung AK, Gottlieb DJ, Plachter B, Pepperl-Klindworth S, Avdic S, Cunningham AL, Abendroth A, Slobedman B. 2009. The role of the human cytomegalovirus UL111A gene in down-regulating CD4+ T-cell recognition of latently infected cells: implications for virus elimination during latency. Blood 114:4128–4137. doi: 10.1182/blood-2008-12-197111. [DOI] [PubMed] [Google Scholar]
- 27.Poole E, Lau JC, Sinclair J. 2015. Latent infection of myeloid progenitors by human cytomegalovirus protects cells from FAS-mediated apoptosis through the cellular IL-10/PEA-15 pathway. J Gen Virol 96:2355–2359. doi: 10.1099/vir.0.000180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Humphreys IR, de Trez C, Kinkade A, Benedict CA, Croft M, Ware CF. 2007. Cytomegalovirus exploits IL-10-mediated immune regulation in the salivary glands. J Exp Med 204:1217–1225. doi: 10.1084/jem.20062424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O’Connor C, Murphy E. 2012. A myeloid progenitor cell line capable of supporting human cytomegalovirus latency and reactivation, resulting in infectious progeny. J Virol 86:9854–9865. doi: 10.1128/JVI.01278-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Albright E, Kalejta R. 2013. Myeloblastic cell lines mimic some but not all aspects of human cytomegalovirus experimental latency defined in primary CD34(+) cell populations. J Virol 87:9802–9812. doi: 10.1128/JVI.01436-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Poole E, Huang CJZ, Forbester J, Shnayder M, Nachshon A, Kweider B, Basaj A, Smith D, Jackson SE, Liu B, Shih J, Kiskin FN, Roche K, Murphy E, Wills MR, Morrell NW, Dougan G, Stern-Ginossar N, Rana AA, Sinclair J. 2019. An iPSC-derived myeloid lineage model of herpes virus latency and reactivation. Front Microbiol 10:2233. doi: 10.3389/fmicb.2019.02233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saffert RT, Kalejta RF. 2007. Human cytomegalovirus gene expression is silenced by Daxx-mediated intrinsic immune defense in model latent infections established in vitro. J Virol 81:9109–9120. doi: 10.1128/JVI.00827-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Forte E, Swaminathan S, Schroeder MW, Kim JY, Terhune SS, Hummel M. 2018. Tumor necrosis factor alpha induces reactivation of human cytomegalovirus independently of myeloid cell differentiation following posttranscriptional establishment of latency. mBio 9:e01560-18. doi: 10.1128/mBio.01560-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.O'Connor CM, Shenk T. 2011. Human cytomegalovirus pUS27 G protein-coupled receptor homologue is required for efficient spread by the extracellular route but not for direct cell-to-cell spread. J Virol 85:3700–3707. doi: 10.1128/JVI.02442-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Redpath S, Angulo A, Gascoigne NR, Ghazal P. 1999. Murine cytomegalovirus infection down-regulates MHC class II expression on macrophages by induction of IL-10. J Immunol 162:6701–6707. [PubMed] [Google Scholar]
- 36.Sinzger C, Hahn G, Digel M, Katona R, Sampaio KL, Messerle M, Hengel H, Koszinowski U, Brune W, Adler B. 2008. Cloning and sequencing of a highly productive, endotheliotropic virus strain derived from human cytomegalovirus TB40/E. J Gen Virol 89:359–368. doi: 10.1099/vir.0.83286-0. [DOI] [PubMed] [Google Scholar]
- 37.Jelcic I, Reichel J, Schlude C, Treutler E, Sinzger C, Steinle A. 2011. The polymorphic HCMV glycoprotein UL20 is targeted for lysosomal degradation by multiple cytoplasmic dileucine motifs. Traffic 12:1444–1456. doi: 10.1111/j.1600-0854.2011.01236.x. [DOI] [PubMed] [Google Scholar]
- 38.Fehr AR, Yu D. 2010. Human cytomegalovirus gene UL21a encodes a short-lived cytoplasmic protein and facilitates virus replication in fibroblasts. J Virol 84:291–302. doi: 10.1128/JVI.01116-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Muhlethaler-Mottet A, Otten LA, Steimle V, Mach B. 1997. Expression of MHC class II molecules in different cellular and functional compartments is controlled by differential usage of multiple promoters of the transactivator CIITA. EMBO J 16:2851–2860. doi: 10.1093/emboj/16.10.2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van der Stoep N, Quinten E, Marcondes Rezende M, van den Elsen PJ. 2004. E47, IRF-4, and PU.1 synergize to induce B-cell-specific activation of the class II transactivator promoter III (CIITA-PIII). Blood 104:2849–2857. doi: 10.1182/blood-2004-03-0790. [DOI] [PubMed] [Google Scholar]
- 41.Landmann S, Mühlethaler-Mottet A, Bernasconi L, Suter T, Waldburger JM, Masternak K, Arrighi JF, Hauser C, Fontana A, Reith W. 2001. Maturation of dendritic cells is accompanied by rapid transcriptional silencing of class II transactivator (CIITA) expression. J Exp Med 194:379–391. doi: 10.1084/jem.194.4.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stenberg RM, Fortney J, Barlow SW, Magrane BP, Nelson JA, Ghazal P. 1990. Promoter-specific trans activation and repression by human cytomegalovirus immediate-early proteins involves common and unique protein domains. J Virol 64:1556–1565. doi: 10.1128/JVI.64.4.1556-1565.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lang D, Stamminger T. 1993. The 86-kilodalton IE-2 protein of human cytomegalovirus is a sequence-specific DNA-binding protein that interacts directly with the negative autoregulatory response element located near the cap site of the IE-1/2 enhancer-promoter. J Virol 67:323–331. doi: 10.1128/JVI.67.1.323-331.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cherrington JM, Khoury EL, Mocarski ES. 1991. Human cytomegalovirus ie2 negatively regulates alpha gene expression via a short target sequence near the transcription start site. J Virol 65:887–896. doi: 10.1128/JVI.65.2.887-896.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang YC, Huang CF, Tung SF, Lin YS. 2000. Competition with TATA box-binding protein for binding to the TATA box implicated in human cytomegalovirus IE2-mediated transcriptional repression of cellular promoters. DNA Cell Biol 19:613–619. doi: 10.1089/104454900750019371. [DOI] [PubMed] [Google Scholar]
- 46.Tsai HL, Kou GH, Chen SC, Wu CW, Lin YS. 1996. Human cytomegalovirus immediate-early protein IE2 tethers a transcriptional repression domain to p53. J Biol Chem 271:3534–3540. doi: 10.1074/jbc.271.7.3534. [DOI] [PubMed] [Google Scholar]
- 47.Tsai HL, Kou GH, Tang FM, Wu CW, Lin YS. 1997. Negative regulation of a heterologous promoter by human cytomegalovirus immediate-early protein IE2. Virology 238:372–379. doi: 10.1006/viro.1997.8855. [DOI] [PubMed] [Google Scholar]
- 48.Shlapobersky M, Sanders R, Clark C, Spector DH. 2006. Repression of HMGA2 gene expression by human cytomegalovirus involves the IE2 86-kilodalton protein and is necessary for efficient viral replication and inhibition of cyclin A transcription. J Virol 80:9951–9961. doi: 10.1128/JVI.01300-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Feng L, Sheng J, Vu GP, Liu Y, Foo C, Wu S, Trang P, Paliza-Carre M, Ran Y, Yang X, Sun X, Deng Z, Zhou T, Lu S, Li H, Liu F. 2018. Human cytomegalovirus UL23 inhibits transcription of interferon-γ stimulated genes and blocks antiviral interferon-γ responses by interacting with human N-myc interactor protein. PLoS Pathog 14:e1006867. doi: 10.1371/journal.ppat.1006867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Roche PA, Furuta K. 2015. The ins and outs of MHC class II-mediated antigen processing and presentation. Nat Rev Immunol 15:203–216. doi: 10.1038/nri3818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jonjić S, Mutter W, Weiland F, Reddehase MJ, Koszinowski UH. 1989. Site-restricted persistent cytomegalovirus infection after selective long-term depletion of CD4+ T lymphocytes. J Exp Med 169:1199–1212. doi: 10.1084/jem.169.4.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lachmann R, Bajwa M, Vita S, Smith H, Cheek E, Akbar A, Kern F. 2012. Polyfunctional T cells accumulate in large human cytomegalovirus-specific T cell responses. J Virol 86:1001–1009. doi: 10.1128/JVI.00873-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hegde NR, Dunn C, Lewinsohn DM, Jarvis MA, Nelson JA, Johnson DC. 2005. Endogenous human cytomegalovirus gB is presented efficiently by MHC class II molecules to CD4+ CTL. J Exp Med 202:1109–1119. doi: 10.1084/jem.20050162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Baca Jones CC, Kreklywich CN, Messaoudi I, Vomaske J, McCartney E, Orloff SL, Nelson JA, Streblow DN. 2009. Rat cytomegalovirus infection depletes MHC II in bone marrow derived dendritic cells. Virology 388:78–90. doi: 10.1016/j.virol.2009.02.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Quinn LL, Williams LR, White C, Forrest C, Zuo J, Rowe M. 2016. The missing link in Epstein-Barr virus immune evasion: the BDLF3 gene induces ubiquitination and downregulation of major histocompatibility complex class I (MHC-I) and MHC-II. J Virol 90:356–367. doi: 10.1128/JVI.02183-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wagner S, Arnold F, Wu Z, Schubert A, Walliser C, Tadagaki K, Jockers R, Mertens T, Michel D. 2012. The 7-transmembrane protein homologue UL78 of the human cytomegalovirus forms oligomers and traffics between the plasma membrane and different intracellular compartments. Arch Virol 157:935–949. doi: 10.1007/s00705-012-1246-6. [DOI] [PubMed] [Google Scholar]
- 57.LeibundGut-Landmann S, Waldburger JM, Krawczyk M, Otten LA, Suter T, Fontana A, Acha-Orbea H, Reith W. 2004. Mini-review: specificity and expression of CIITA, the master regulator of MHC class II genes. Eur J Immunol 34:1513–1525. doi: 10.1002/eji.200424964. [DOI] [PubMed] [Google Scholar]
- 58.Lee AW, Wang N, Hornell TM, Harding JJ, Deshpande C, Hertel L, Lacaille V, Pashine A, Macaubas C, Mocarski ES, Mellins ED. 2011. Human cytomegalovirus decreases constitutive transcription of MHC class II genes in mature Langerhans cells by reducing CIITA transcript levels. Mol Immunol 48:1160–1167. doi: 10.1016/j.molimm.2011.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Koldehoff M, Lindemann M, Ross SR, Elmaagacli AH. 2018. Cytomegalovirus induces HLA-class-II-restricted alloreactivity in an acute myeloid leukemia cell line. PLoS One 13:e0191482. doi: 10.1371/journal.pone.0191482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kondo K, Kaneshima H, Mocarski ES. 1994. Human cytomegalovirus latent infection of granulocyte-macrophage progenitors. Proc Natl Acad Sci U S A 91:11879–11883. doi: 10.1073/pnas.91.25.11879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Landini MP, Lazzarotto T, Xu J, Geballe AP, Mocarski ES. 2000. Humoral immune response to proteins of human cytomegalovirus latency-associated transcripts. Biol Blood Marrow Transplant 6:100–108. doi: 10.1016/s1083-8791(00)70072-3. [DOI] [PubMed] [Google Scholar]
- 62.Lunetta JM, Wiedeman JA. 2000. Latency-associated sense transcripts are expressed during in vitro human cytomegalovirus productive infection. Virology 278:467–476. doi: 10.1006/viro.2000.0666. [DOI] [PubMed] [Google Scholar]
- 63.Tan JC, Avdic S, Cao JZ, Mocarski ES, White KL, Abendroth A, Slobedman B. 2011. Inhibition of 2′,5′-oligoadenylate synthetase expression and function by the human cytomegalovirus ORF94 gene product. J Virol 85:5696–5700. doi: 10.1128/JVI.02463-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Warming S, Costantino N, Court DL, Jenkins NA, Copeland NG. 2005. Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Res 33:e36. doi: 10.1093/nar/gni035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Roche KL, Nukui M, Krishna BA, O'Connor CM, Murphy EA. 2018. Selective 4-thiouracil labeling of RNA transcripts within latently infected cells after infection with human cytomegalovirus expressing functional uracil phosphoribosyltransferase. J Virol 92:e00880-18. doi: 10.1128/JVI.00880-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Spector DJ, Yetming K. 2010. UL84-independent replication of human cytomegalovirus strain TB40/E. Virology 407:171–177. doi: 10.1016/j.virol.2010.08.029. [DOI] [PubMed] [Google Scholar]
- 67.Britt WJ. 2010. Human cytomegalovirus: propagation, quantification, and storage. Curr Protoc Microbiol Chapter 14:Unit 14E.3. doi: 10.1002/9780471729259.mc14e03s18. [DOI] [PubMed] [Google Scholar]
- 68.Desai D, Lauver M, Ostman A, Cruz L, Ferguson K, Jin G, Roper B, Brosius D, Lukacher A, Amin S, Buchkovich N. 2019. Inhibition of diverse opportunistic viruses by structurally optimized retrograde trafficking inhibitors. Bioorg Med Chem 27:1795–1803. doi: 10.1016/j.bmc.2019.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yu D, Silva MC, Shenk T. 2003. Functional map of human cytomegalovirus AD169 defined by global mutational analysis. Proc Natl Acad Sci U S A 100:12396–12401. doi: 10.1073/pnas.1635160100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Harel NY, Alwine JC. 1998. Phosphorylation of the human cytomegalovirus 86-kilodalton immediate-early protein IE2. J Virol 72:5481–5492. doi: 10.1128/JVI.72.7.5481-5492.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kalejta RF, Bechtel JT, Shenk T. 2003. Human cytomegalovirus pp71 stimulates cell cycle progression by inducing the proteasome-dependent degradation of the retinoblastoma family of tumor suppressors. Mol Cell Biol 23:1885–1895. doi: 10.1128/mcb.23.6.1885-1895.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]