Bats are natural hosts of many FiVs, from which diverse FiVs were serologically or virologically detected in Africa, Europe, and East Asia. Recently, very divergent FiVs were identified in the Chinese bat population, but their antigenic relationship with other known FiVs remains unknown. Here, we conducted serological characterization and investigation of Chinese indigenous FiVs and prototypes of other viruses in bats. Results indicated that Chinese indigenous FiVs are antigenically distant to other FiVs, and infection of novel or multiple FiVs occurred in Chinese bats, with FiV DH04 or an antigenically related one being the most widely and the most highly prevalent. Additionally, besides Rousettus leschenaultii and Eonycteris spelaea bats, the insectivorous Myotis horsfieldii and M. schreibersii bats are highly preferential hosts of FiVs. Seroreactive and viral metagenomic results indicated that more as yet unknown bat-borne FiVs circulate in Southern China, and to uncover them further, investigation and timely surveillance is needed.
KEYWORDS: filovirus, bats, cross-antigenicity, ELISA, genetic diversity
ABSTRACT
Southern China is a hot spot of emerging infectious diseases, in which diverse species of bats dwell, a large group of flying mammals considered natural reservoirs for zoonotic viruses. Recently, divergent filoviruses (FiVs) have been identified in bats within this region, which pose a potential risk to public health, but the true infection situation in bats remains largely unclear. Here, 689 archived bat serum samples were analyzed by enzyme-linked immunosorbent assay (ELISA), Western blotting, and neutralization assay to investigate the seroprevalence and cross-reactivity of four divergent FiVs and two other viruses (rabies virus and Tuhoko pararubulavirus 1) of different families within the order Mononegavirales. Results showed no cross-antigenicity between FiVs and other mononegaviruses but different cross-reactivity among the FiVs themselves. The total FiV seroreactive rate was 36.3% (250/689), with infection by the indigenous Chinese FiV DH04 or an antigenically related one being the most widely and the most highly prevalent. Further viral metagenomic analysis of fruit bat tissues also identified the gene sequence of a novel FiV. These results indicate the likely prevalence of other so far unidentified FiVs within the Chinese bat population, with frugivorous Rousettus leschenaultii and Eonycteris spelaea bats and insectivorous Myotis horsfieldii and Miniopterus schreibersii bats being their major reservoirs.
IMPORTANCE Bats are natural hosts of many FiVs, from which diverse FiVs were serologically or virologically detected in Africa, Europe, and East Asia. Recently, very divergent FiVs were identified in the Chinese bat population, but their antigenic relationship with other known FiVs remains unknown. Here, we conducted serological characterization and investigation of Chinese indigenous FiVs and prototypes of other viruses in bats. Results indicated that Chinese indigenous FiVs are antigenically distant to other FiVs, and infection of novel or multiple FiVs occurred in Chinese bats, with FiV DH04 or an antigenically related one being the most widely and the most highly prevalent. Additionally, besides Rousettus leschenaultii and Eonycteris spelaea bats, the insectivorous Myotis horsfieldii and M. schreibersii bats are highly preferential hosts of FiVs. Seroreactive and viral metagenomic results indicated that more as yet unknown bat-borne FiVs circulate in Southern China, and to uncover them further, investigation and timely surveillance is needed.
INTRODUCTION
Filoviruses (FiVs) are a group of baculiform viruses within the family Filoviridae featuring some notorious members responsible for highly contagious human hemorrhagic diseases, such as Ebola and Marburg virus diseases with fatality rates ranging from 25 to 90% (1). In addition to the 3 established genera of the family that exclusively infect mammals, Ebolavirus, Marburgvirus, and Cuevavirus (2), the FiVs have recently been expanded by the addition of two new genera discovered in fish, Striavirus and Thamnovirus with Xilang striavirus and Huangjiao thamnovirus as their respective unique species (2, 3). The genus Ebolavirus is composed of five species, with each containing only one member as follows: Zaire ebolavirus (Ebola virus, EBOV), Sudan ebolavirus (Sudan virus, SUDV), Bundibugyo ebolavirus (Bundibugyo virus, BDBV), Taï Forest ebolavirus (Taï Forest virus, TAFV), and Reston ebolavirus (Reston virus, RESTV) (2). Genus Marburgvirus contains a single species, Marburg marburgvirus with two members, Marburg virus (MARV) and Ravn virus (RAVV), which are approximately 21% divergent from each other at the nucleotide level (2). Genus Cuevavirus consists of a single species, Lloviu cuevavirus (Lloviu virus, LLOV) (2).
Bats have been closely associated with FiVs; e.g., the cave-roosting Rousettus aegyptiacus bats are considered to be natural reservoir hosts for MARV (4–6), cumulative evidence based on serological and virological detections has indicated that various bat species could play a role in the ecological circle of ebolaviruses (5, 7, 8), and the LLOV genome has been detected in Miniopterus schreibersii bats in Spain and Hungary (9, 10). In addition, a number of novel FiVs have been discovered in bats in China and Africa and are divergent enough to be candidates for new species or even genera (11–16). Particularly, in China, bat-borne FiVs such as FiV DH04 (DH04) and Měnglà virus (MLAV), show great genetic diversity and are quite distinct from other currently known FiVs (11–13). They exhibit broad cell tropisms (12, 13), indicative of a complicated infection situation in bats and of the high zoonotic potential of FiVs in China. Serological methods based on nucleoprotein (NP) have been widely applied to investigate the seroprevalence of FiVs in bats and have revealed the possibility of an increased number of bat species acting as reservoirs (17–22). However, such serological data can be challenged as to interpretation, mainly due to unidentified cross-antigenicity between different FiV NPs. To address this problem, we conducted a serological investigation of FiVs in bats, revealing the antigenic relationships between FiVs and two other mononegaviruses, Tuhoko pararubulavirus 1 (TUHV) and rabies virus (RABV), and the extensive infection of Chinese bat populations with diverse filoviruses.
RESULTS
Recombinant NP-based Western blotting analysis of cross-antigenicity of FiVs, TUHV, and RABV.
As shown in Fig. 1A, all mononegaviruses formed 3 phylogroups, corresponding to the families of Filoviridae, Paramyxoviridae, and Rhabdoviridae, with very low amino acid identities between them (≤12.3%). Five filoviruses clustered within 3 clades, with one consisting of EBOV and RESTV with the highest mutual amino acid identity (54.5%) and belonging to the genus Ebolavirus. Chinese isolate DH04 formed a unique clade sharing 32.9 to 35.9% identities with ebolaviruses. The third clade included MARV and another Chinese FiV (MLAV) with the 2nd highest mutual similarity (45.9%) but with ≤28.1% similarity with other FiVs. Western blotting (WB) analyses (Fig. 1B) showed that mouse anti-EBOV-NP hyperimmune serum had a weak cross-reactivity with RESTV and MARV; anti-RESTV-NP strongly reacted with EBOV but not with others; anti-FiV_DH04-NP and anti-RABV-CVS11-NP reacted only with their own antigen; and anti-MARV-NP showed a weak cross-reactivity with MLAV. In addition, anti-TUHV-NP also showed very weak cross-reactivity with MARV but not with the others. This indicates that some FiVs are minimally related antigenically to other mononegaviruses (e.g., paramyxovirus and rhabdovirus). However, within the family Filoviridae, varying cross-antigenicity between viruses was detected, while very weak or nonexistent between clades; however, stronger cross-reactivity was found within the EBOV clade.
FIG 1.
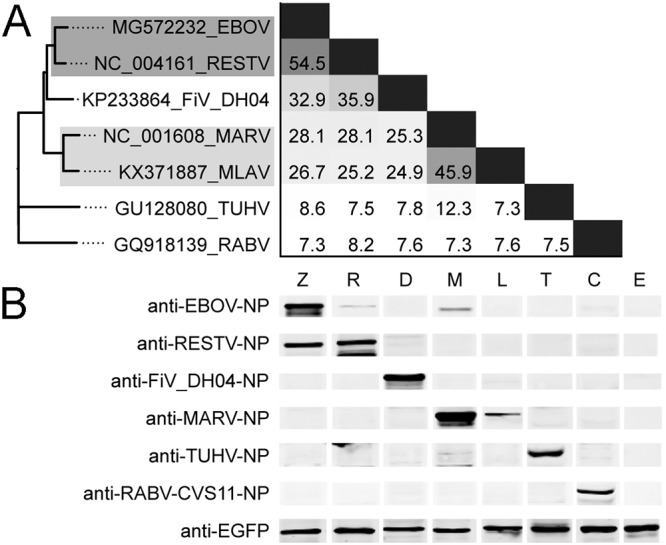
The cross-antigenicity characterization of the five FiVs, TUHV, and RABV. (A) Phylogeny and pairwise comparison of the 340-aa NP C-terminal sequences. Viruses in phylogenetic tree showing significant cross-reactivity are in grayed boxes with the depth of gray representing the reactive strength. (B) WB analysis of eukaryotically expressed NPs of the 7 viruses using 6 antiserum samples. Lanes were taken from different gels. Different loading amounts of these NPs were determined using ImageJ to ensure the same amounts of the target proteins. Z, EBOV (loading amount, 52.3 μg); R, RESTV (59.5 μg); D, FiV DH04 (95.55 μg); M, MARV (60.8 μg); L, MLAV (51.8 μg); T, TUHV (48.6 μg); C, RABV CVS11 (60.9 μg); E, EGFP (11.0 μg).
Serological investigation of FiVs in bat serum.
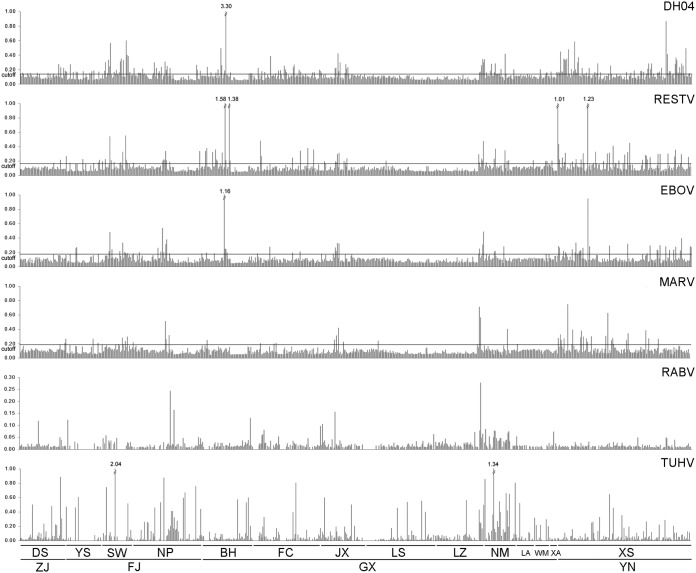
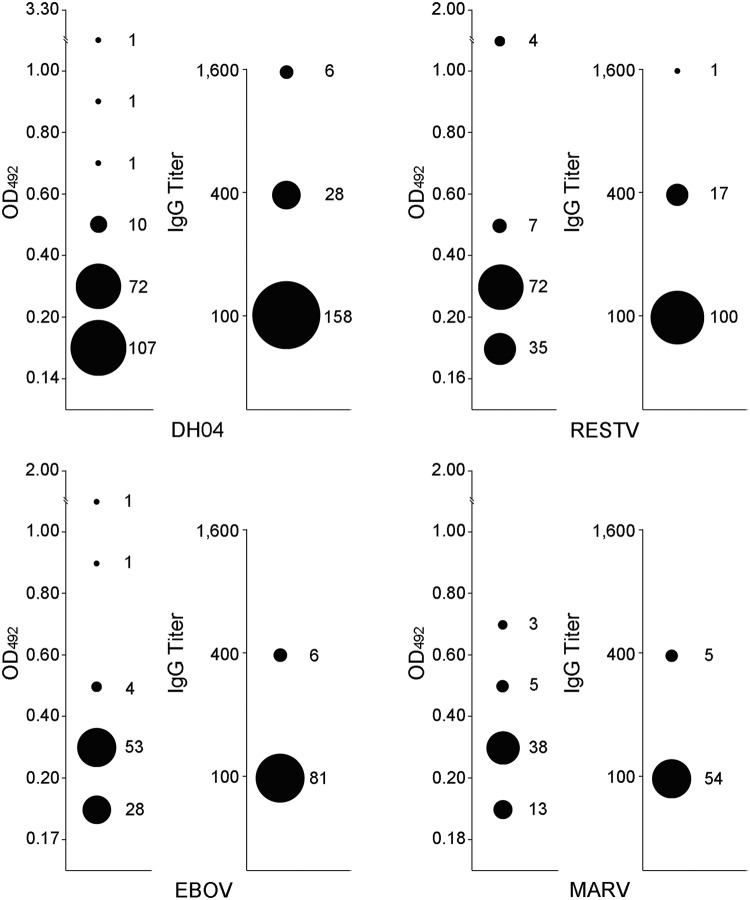
Results of enzyme-linked immunosorbent assay (ELISA) reactions of bat serum samples with DH04, RESTV, EBOV, MARV, RABV, and TUHV are shown in Fig. 2 with the reactivity of 151 being further confirmed by WB analyses against NPs of DH04, RESTV, EBOV, and MARV (Fig. 3). The optical density at 492 nm (OD492) reading ranges of WB-validated serum samples were 0.05 to 0.87, 0.05 to 1.23, 0.05 to 0.95, and 0.04 to 0.75, respectively, against the 4 viruses. WB results showed that 81, 53, 45, and 39 serum samples were reactive to DH04, RESTV, EBOV, and MARV, respectively (Fig. 3). As previously described, ELISA and WB results were combined to determine the ELISA cutoff values (23). By this method, 250 of 689 (36.3%) bat serum samples tested showed reactivity to one or more of the 4 FiVs with DH04 showing the highest reactive rate (27.9%, P < 0.001), followed by RESTV (17.1%), EBOV (12.6%), and MARV (8.6%) (Table 1). Among the reactive serum samples, most had OD492 readings of <0.4, with only 1, 4, and 1 showing OD492 readings of >1.0 against DH04, RESTV, and EBOV, respectively (Fig. 2 and 4). Their titers were determined by 4-fold dilutions, and results showed that most had titers of 100, with only 6 and 1 serum samples showing titers of 1,600 against DH01 and RESTV, respectively (Fig. 4).
FIG 2.
The OD492 readings (y axis) of bat sera against the rNPs of DH04, RESTV, EBOV, MARV, RABV, and TUHV. Each bar on the x axis represents 1 serum sample. OD492 readings of >1.0 are labeled above the bar. ZJ, Zhejiang; FJ, Fujian; GX, Guangxi; YN, Yunnan; DS, Daishan; YS, Yanshi; SW, Shawu; NP, Nanping; BH, Beihai; FC, Fangchenggang; JX, Jingxi; LS, Lingshan; LZ, Longzhou; NM, Ningming; LA, Long’an; WM, Wuming; XA, Xing’an; XS, Xishuangbanna.
FIG 3.
WB analyses of 151 bat serum samples. Four lanes from left to right are, respectively, DH04, RESTV, EBOV, and MARV. The sample code is shown above the WB picture. The value under each lane is the OD492 reading.
TABLE 1.
Specimen details and results of ELISAs
| Province | Location | Bat species | Yr | Dieta | Serum no. | ELISA P (%)b
|
||||
|---|---|---|---|---|---|---|---|---|---|---|
| FiV | DH04 | RESTV | EBOV | MARV | ||||||
| Yunnan | Xishuangbanna | R. leschenaultii | 2012 | F | 143 | 87 (60.8) | 66 (46.2) | 43 (30.1) | 33 (23.1) | 23 (16.1) |
| Guangxi | Lingshan | S. kuhlii | 2015 | I | 14 | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| M. australis | 2015 | I | 15 | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | ||
| S. kuhlii | 2016 | I | 43 | 2 (4.7) | 0 (0.0) | 1 (2.3) | 0 (0.0) | 1 (2.3) | ||
| Subtotal | 72 | 2 (2.8) | 0 (0.0) | 1 (1.4) | 0 (0.0) | 1 (1.4) | ||||
| Long'an | H. larvatus | 2015 | I | 19 | 2 (10.5) | 1 (5.3) | 0 (0.0) | 0 (0.0) | 1 (5.3) | |
| Wuming | H. larvatus | 2015 | I | 15 | 2 (13.3) | 2 (13.3) | 1 (6.7) | 0 (0.0) | 1 (6.7) | |
| Xing'an | H. larvatus | 2015 | I | 9 | 6 (66.7) | 3 (33.3) | 3 (33.3) | 2 (22.2) | 0 (0.0) | |
| Beihai | S. kuhlii | 2015 | I | 50 | 21 (42.0) | 10 (20.0) | 13 (26.0) | 7 (14.0) | 2 (4.0) | |
| Fangchenggang | H. larvatus | 2016 | I | 28 | 9 (32.1) | 8 (28.6) | 3 (10.7) | 2 (7.1) | 2 (7.1) | |
| Hipposideros pomona | 2016 | I | 1 | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | ||
| R. affinis | 2016 | I | 40 | 14 (35.0) | 12 (30.0) | 4 (10.0) | 2 (5.0) | 1 (2.5) | ||
| Subtotal | 69 | 23 (33.3) | 20 (29.0) | 7 (10.1) | 4 (5.8) | 3 (4.3) | ||||
| Jingxi | Aselliscus stoliczkanus | 2016 | I | 2 | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | |
| Hipposideros turpis | 2016 | I | 8 | 5 (62.5) | 4 (50.0) | 1 (12.5) | 1 (12.5) | 0 (0.0) | ||
| H. armiger | 2016 | I | 15 | 8 (53.0) | 5 (33.3) | 5 (33.3) | 6 (40.0) | 3 (20.0) | ||
| H. larvatus | 2016 | I | 8 | 4 (50.0) | 4 (50.0) | 3 (37.5) | 1 (12.5) | 1 (12.5) | ||
| H. pomona | 2016 | I | 4 | 1 (25.0) | 1 (25.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | ||
| Rhinolophus pearsonii | 2016 | I | 2 | 1 (50.0) | 0 (0.0) | 0 (0.0) | 1 (50.0) | 0 (0.0) | ||
| Rhinolophus thomasi | 2016 | I | 7 | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | ||
| Subtotal | 46 | 19 (41.3) | 15 (32.6) | 11 (23.9) | 9 (20.0) | 4 (8.7) | ||||
| Ningming | M. schreibersii | 2016 | I | 33 | 20 (60.6) | 17 (51.5) | 11 (33.3) | 9 (27.3) | 5 (15.2) | |
| Longzhou | T. melanopogon | 2016 | I | 32 | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | |
| H. armiger | 2016 | I | 17 | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | ||
| Subtotal | 49 | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | ||||
| Fujian | Yanshi | R. sinicus | 2016 | I | 33 | 11 (33.3) | 11 (33.3) | 7 (21.2) | 4 (12.1) | 3 (9.1) |
| Nanping | H. armiger | 2016 | I | 11 | 2 (18.2) | 2 (18.2) | 0 (0.0) | 0 (0.0) | 0 (0.0) | |
| R. sinicus | 2016 | I | 1 | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | ||
| R. affinis | 2016 | I | 61 | 19 (31.1) | 17 (27.9) | 8 (13.1) | 9 (14.8) | 5 (8.2) | ||
| Subtotal | 73 | 21 (28.8) | 19 (26.0) | 8 (11.0) | 14 (19.2) | 6 (8.2) | ||||
| Shawu | R. affinis | 2016 | I | 8 | 5 (62.5) | 3 (37.5) | 4 (50.0) | 1 (12.5) | 2 (25.0) | |
| M. horsfieldii | 2016 | I | 23 | 18 (78.3) | 14 (60.9) | 9 (39.1) | 8 (34.8) | 8 (34.8) | ||
| Subtotal | 31 | 23 (74.2) | 17 (54.8) | 13 (41.9) | 9 (29.0) | 10 (32.3) | ||||
| Zhejiang | Daishan | R. ferrumequinum | 2016 | I | 47 | 13 (27.7) | 13 (27.7) | 2 (4.3) | 1 (2.1) | 2 (4.3) |
| Totals | 689 | 250 (36.3) | 192 (27.9) | 118 (17.1) | 87 (12.6) | 59 (8.6) | ||||
F, frugivorous; I, insectivorous.
P (%), number reactive (percentage). The column FiV indicates the general reactive rate of the 4 FiVs.
FIG 4.
Distribution of the OD492 readings and the antibody titers of bat serum reactive to DH04, RESTV, EBOV, and MARV. Each virus has two panels with the left one showing the OD492 readings at a dilution of 1:100 and the right showing the antibody titers. The size of the filled circle represents the number of serum samples at the same OD492 reading level or titer.
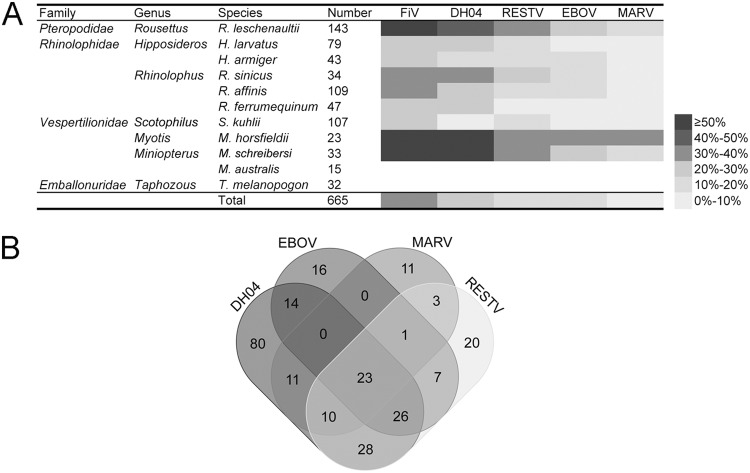
In terms of bat species, as shown in Fig. 5, the Rousettus leschenaultii, Myotis horsfieldii, and M. schreibersii bats showed the highest total FiV seroreactivity (60.8%, 78.3%, and 60.6%, respectively) (P < 0.001), followed by bats in the Hipposideros larvatus, Hipposideros armiger, Rhinolophus sinicus, Rhinolophus affinis, Rhinolophus ferrumequinum, and Scotophilus kuhlii species with seroreactivity of 21.5 to 34.9%. Miniopterus australis and Taphozous melanopogon bats anywhere were negative for all 4 FiVs. Single- or multiple-reactivity of these serum samples to the 4 FiVs are summarized in Fig. 5B, with 80, 20, 16, and 11 serum samples showing single-reactivity to DH04, RESTV, MARV, and EBOV, respectively. Interestingly, while EBOV/MARV and DH04/EBOV/MARV cross-reactions were not observed, 3 serum samples showed reactivity to MARV/RESTV, 14 to DH04/EBOV, 11 to DH04/MARV and 28 to DH04/RESTV. Of note is that 23 serum samples reacted with all 4 FiVs.
FIG 5.
FiV seroreactivity in bat serum samples. (A) The seroreactivity of different FiVs varied in bat species. Bat species with <10 were not included in the statistical analysis. The column FiV indicates the total seroreactivity of the 4 viruses. (B) The Venn diagram shows single- or multiple-reactivity of the reactive serum sample.
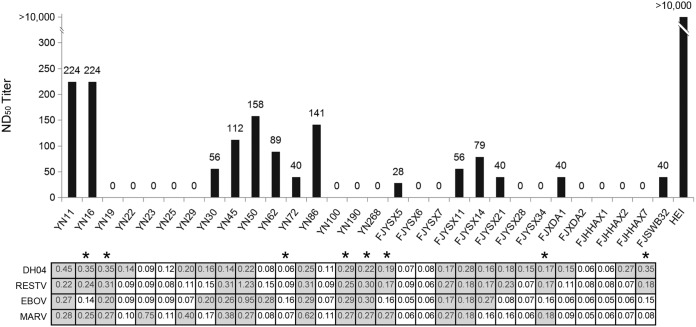
Thirty serum samples available in sufficient volume, including 12 reactive to EBOV by ELISA, were further tested to determine their neutralizing antibodies against EBOV transcription-competent virus-like particles (trVLPs). Compared to the 50% neutralizing dose (ND50) of the refined control hyperimmunized equine immunoglobulin (HEI) (>10,000), 14 serum samples had neutralizing antibody titers ranging between 28 and 224 (Fig. 6), showing 73.3% (22/30) coincidence with ELISA results. Among the 8 serum samples showing discrepancy between neutralization assay (NA) and ELISA results, 5 ELISA-negative serum samples showed neutralizing capacity with titers between 28 and 224, while the remaining 3 ELISA-positive serum samples against EBOV did not neutralize trVLPs.
FIG 6.
EBOV trVLP-based NA analyses of the 30 bat serum samples with HEI as positive control. The ELISA results are shown with OD492 reading > cutoff value highlighted in gray. Serum samples not showing coincidence between NA and ELISA are identified with a star.
Viral metagenomics.
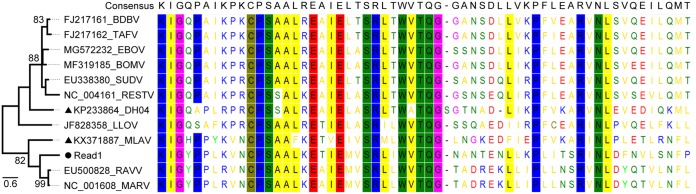
A total of 57 adult R. leschenaultii bats were sampled during January 2019 and subjected to viral metagenomic analysis. The Illumina sequencing generated ∼2.0 Gb of high-quality data with an average length of 180 bp. After annotation, 2 reads that were exactly the same were related to FiVs, corresponding to the L gene. They shared the highest 71.7% amino acid identity with MARV, followed by 58.3% with MLAV and the lowest 45.0% with DH04. The phylogenetic tree showed that the sequence was placed between MLAV and MARV, indicative of a remote relationship with currently known FiVs (Fig. 7). The multiple sequence alignment of reads with other representatives revealed consistency in conserved motifs and sites (Fig. 7), which ensured that the short reads truly originated from FiVs. To confirm the presence of FiV in these samples, a previously validated nested reverse transcriptase PCR (RT-PCR) method (11, 12) was employed to screen all organ samples, and no positive results were obtained.
FIG 7.
Phylogeny and multiple amino acid sequence alignment of FiV-like reads generated by viral metagenomic analysis of fruit bats (filled circle) with other representatives of known FiVs, including two newly identified Chinese FiVs (filled triangles). Conserved motifs or sites are shadowed in different colors.
DISCUSSION
As one of the most abundant, diverse, and widely distributed mammals, bats have been confidently linked to many emerging zoonotic viruses, including Hendra virus, Nipah virus, severe acute respiratory syndrome (SARS) and swine acute diarrhea syndrome (SADS)-related coronaviruses, with frequent high-consequence spillover to humans and other animals (24, 25). Bats are also deeply involved in the ecology of FiVs (5), but their roles to maintain the circulation of FiVs remain largely unknown and, therefore, need further investigation. In addition, the recent discovery of novel FiVs in bats, especially in different bat species in China (11–14), has added more mystery to these viruses and has driven numerous efforts to investigate the background of FiVs in bats (15, 16). Such work is critical to prevent the outbreak of, and to control, FiV-associated zoonotic diseases. Field studies have uncovered FiV dynamics in many bat populations, with increasing numbers of viral RNA detection cases being reported, e.g., EBOV RNA positivity in Hypsignathus monstrosus, Epomops franqueti, and Myonycteris torquata bats in central Africa (7), MARV RNA in R. aegyptiacus, Miniopterus inflatus, and Rhinolophus eloquens bats in Gabon and D. R. Congo (26, 27), LLOV RNA in M. schreibersii bats in Spain and Hungary (9, 10), RESTV RNA in M. schreibersii, M. australis, Cynopterus brachyotis, and Chaerephon plicata bats in Philippines (8), MLAV and other novel FiV RNA in R. leschenaultii and Eonycteris spelaea bats in China (11–13), and Bombali virus RNA in Chaerephon pumilus and Mops condylurus bats in Sierra Leone, Kenya, and Guinea (14–16). These viral nucleic acid data have extensively increased our knowledge of the genetic diversity, distribution, and host range of FiVs.
Serological epidemiology is another important aspect to uncover the present and historical situation of bat-borne FiVs and is critical for estimating the potential risk of these viruses to public health. Compared to RNA detection, the serological investigation will reveal more insights into FiV infection in bats and its distribution (5). However, serological investigations face some challenges, especially in regard to widely used methods based on the NP due to the uncharacterized cross-antigenicity between FiVs. In addition, a small region in the middle of FiV NP sequences shows certain homology with NPs of paramyxoviruses or even with rhabdoviruses (28), which could introduce false-positive serological results. In the present investigation, the involvement of NP in this cross-antigenicity was identified by WB using a combination of eukaryotically expressed recombinant NPs (rNPs) of four FiVs, TUHV and RABV, and their NP-specific antiserum. TUHV is a new bat-originated paramyxovirus detected in Southern China, and RABV also showed seroprevalence in Chinese bat populations (29, 30). Based on the identity comparison and WB analysis of the 340 amino acids (aa) at the C terminus of DH04 NP and the counterparts of EBOV, RESTV, MARV, MLAV, TUHV, and RABV (Fig. 1), these FiVs are genetically distant and antigenically unrelated to paramyxoviruses and rhabdoviruses, demonstrating that such a serological investigation of FiVs based on NP shows high specificity among the mononegaviruses, a conclusion confirmed by the distinct profiles of the ELISA readings of the FiVs, TUHV and RABV (Fig. 2). However, among the FiVs, strong cross-reactivity was shown to occur between ebolaviruses, i.e., EBOV/RESTV, and only very weak one-way cross-reactivity between EBOV/MARV. We did not generate antiserum against MLAV, thereby preventing a reciprocal cross-reactivity identification of MLAV and other FiVs, but the one-way cross-reactivity and 45.9% amino acid identity between MARV/MLAV indicates that they are genetically close and antigenically related to each other. Phylogeny based on the L protein has also shown that MLAV is relatively close to MARV (13). DH04 showed no cross-reactivity with other FiVs, which, together with its distant phylogenetic position among the other FiVs, indicates that it is genetically distant and antigenically unrelated to other FiVs. These cross-antigenic traits between FiVs based on NP show some consistency with cross-reactivity characterization using bat convalescent-phase serum generated by prime-boost immunization of cultivatable FiVs (31).
The profiling of cross-reactivity among the four FiVs revealed that reactive serum samples could be classified into four groups. Firstly, as an indigenous FiV, DH04 was first reported in R. leschenaultii bats in Dehong, western Yunnan (11), and later in E. spelaea bats in Jinghong and Mengla, southern Yunnan (12), regions >350 km apart (Fig. 8). Our ELISA results also showed that 27.9% (192/689) of bat serum samples react with DH04, with 11.6% (80/689) of serum samples from R. leschenaultii bats in Yunnan, H. larvatus and H. armiger bats in Guangxi, R. ferrumequinum and R. affinis bats in Zhejiang, and M. horsfieldii and M. schreibersii bats in Fujian exclusively reacting with DH04. The RNA and serological evidence indicates that DH04 or DH04-like FiVs possess a wide distribution in Chinese bat populations. Secondly, there were 11 serum samples exclusively reacting with MARV, e.g., an R. leschenaultii bat serum YN23 had the highest and exclusive OD492 reading of 0.75 to MARV (Fig. 3) and also exclusively and strongly reacted with MARV by WB (Fig. 3), suggesting that these bats were infected by another Chinese indigenous MLAV, or similar ones, rather than the African MARV, and that the distribution of these MLAV-like viruses are more restricted than DH04. Thirdly, there were also 16 and 20 serum samples exclusively reacting with EBOV and RESTV, respectively, even one showing an OD492 reading as high as 1.38 against RESTV (Fig. 2), while 7 serum samples cross-reacted with them (Fig. 5B). As RESTV is another Eastern Asian FiV that was serologically and/or virologically detected in bats in China (20), Bangladesh (21), and Philippines (8, 18), this portion of the serum samples possibly reflected infection of RESTV-like viruses. Finally, multiple-reactivity was widely observed in serum samples here, with 23 cross-reactive with all 4 viruses (Fig. 5B), thereby making the OD492 reading profiles of the 4 FiVs seemingly similar (Fig. 2), which undoubtedly indicates a complex infection involving novel and/or multiple FiVs in bats. This conclusion can be proven by Yang et al. (12), who has revealed much diversity of FiV RNAs in bats, with one bat even infected by four viruses, and by our viral metagenomic data that also indicated the signal of novel FiVs in bats. Unfortunately, our viral metagenomic data here revealed very limited reads related to FiVs, which was not confirmed using RT-PCR detection, probably due to incompatibility of RT-PCR primer pairs to novel FiV or low replication of this virus in bats at capture. The neutralization tests were performed to validate the ELISA and WB, and results did not show good coincidence, which may be ascribed to cross-reactive and potent neutralizing antibodies of unidentified bat FiVs. In support of this suggestion, it is known that antibodies from human survivors of Ebola or Marburg viral infections exhibit cross-reactive and potent neutralizing responses to FiVs other than the original infecting species (32, 33), which suggests that these new bat-borne FiVs may be of scientific and medical value for the development of therapeutics and vaccines.
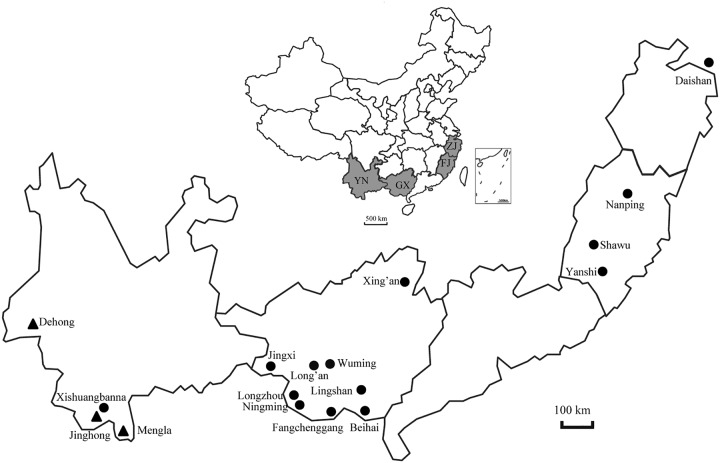
FIG 8.
Locations of bat serum samples. The black-filled circles indicate the location of bat serum samples, and the black-filled triangles indicate the locations where the bats were positive for FiVs by RNA detection. YN, Yunnan province; GX, Guangxi Zhuang autonomous region; FJ, Fujian province; ZJ, Zhejiang province.
Due to a lack of specific positive serum samples against the 4 viruses, we cannot assess the sensitivity of the ELISA established here. However, serological results here showed that FiV infections in Chinese bats are highly prevalent, particularly in R. leschenaultii, M. horsfieldii, and M. schreibersii bats. Such high seroreactive rates were also observed in Africa, e.g., as high as 43.8% of Zambian Rousettus sp. were seroreactive to MARV using NP- and glycoprotein (GP)-based ELISA (22), 36.4% of Ghanaian bat serum samples reacted with EBOV NPs (19), which adds evidence that bats in the old world are natural hosts for FiVs, though some viruses have not yet been isolated from those creatures. Besides the frugivorous R. leschenaultii and E. spelaea bats, from which the FiV RNA was detected (11, 12), our results also indicated that the insectivorous M. horsfieldii, M. schreibersii, Hipposideros spp., and Rhinolophus spp. bats are likely new hosts of FiVs in China. Three of them, except for Hipposideros spp. were also included in the FiV host spectrum in Africa and Europe, e.g., Rhinolophus spp. showed RNA- and antibody-positive rates in Africa (4, 27, 34) while M. schreibersii bats in Europe were positive for LLOV RNA (9, 10). Of note is that these reactive serum samples showed relatively low titers in ELISA (≤1,600) and NA (≤224) results, which was also widely observed in the bat serum samples of other countries, such as Ghana (≤800) (19), Gabon (≤1,600) (34), and Philippines (≤1,280) (18). This could be ascribed to the following: (i) bats have special immune systems against FiVs that do not produce high levels of antibodies; (ii) the antigenic structure of FiV N protein might fold mistakenly in vitro and cannot be efficiently bound by bat antibodies; (iii) those bats were infected by FiVs other than those used here, but antigenically related to the viruses used here, making detection methods based on certain viruses not 100% recognizing and binding antibodies triggered by other FiVs in bats.
Altogether, the present study, along with the identification of genetic diversities in previous studies (11, 12), has indicated that there are more unknown bat-borne FiVs circulating in Southern China, and to uncover them further, investigation and timely surveillance is needed.
MATERIALS AND METHODS
Ethics statement.
The procedures for sampling bats in this study were reviewed and approved by the Administrative Committee on Animal Welfare of the Institute of Military Veterinary Medicine, Academy of Military Medical Sciences, China (Laboratory Animal Care and Use Committee Authorization, permit number JSY-DW-2015-01).
Sample information.
The 689 serum samples used in this study were archived samples stored at −80°C following collection between 2012 and 2016 in the Yunnan, Guangxi, Fujian, and Zhejiang provinces, China, and were used to investigate group A rotaviruses and hantaviruses in our previous studies (23, 35). Bat species were identified as described previously, covering 16 species within 8 genera and 4 families (23) as follows: Rhinolophidae (n = 336), Vespertilionidae (n = 178), Emballonuridae (n = 32), and Pteropodidae (n = 143). Serum details are shown in Table 1 and Fig. 8. In addition, 57 adult R. leschenaultii bats were captured with nets at an orchard in Dehong prefecture of Yunnan province during January 2019, where FiV DH04 was identified from the same bat species in 2015 (11). All trapped bats were dead at collection every morning, and their rectums with the contents, livers, kidneys, and lungs were immediately collected at the local CDC and shipped to the laboratory on dry ice, where they were stored at −80°C.
Expression of recombinant NP and generation of mouse anti-rNP-specific hyperimmune serum.
The NP sequence (340 aa at C terminus) available for DH04 (GenBank accession number KP233864) was amplified from a plasmid preserved in the laboratory using Hi-Fidelity polymerase (Tiangen), and its counterparts in EBOV (GenBank accession number MG572232), RESTV (GenBank accession number NC_004161), MARV (GenBank accession number NC_001608), LLOV (GenBank accession number NC_016144), and TUHV (GenBank accession number GU128080) were amplified from preserved plasmids or chemically synthesized. The expression and purification of prokaryotic rNPs and preparation of mouse-specific hyperimmune serum were as previously described (23). Briefly, the NP gene fragments were subcloned into pET-28a(+) with a His tag at the C terminus and transfected Escherichia coli strain BL21 (Tiangen). After induction with 0.5 mM isopropyl-β-d- thiogalactopyranoside (IPTG), the rNPs were purified and quantified by Ni-NTA His bind resin (Novagen) and a BCA protein assay kit (CWBio). Specific hyperimmune serum was prepared by injecting 4-week-old female Kunming mice intramuscularly with purified rNPs. Attempts to produce the rNP of LLOV failed, even with codon optimization, change of bacterial host, adjustment of inducement temperature, and truncation of the NP; hence, LLOV was not included in the experiments. Eukaryotic enhanced green fluorescent protein (EGFP)-tagged NP C-terminal fragments of EBOV (predicted molecular weight [MW], 64.9 kDa), RESTV (110.8 kDa), MARV (61.8 kDa), DH04 (67.23 kDa), TUHV (43.9 kDa), and MLAV (GenBank accession number KX371887; 75.4 kDa) were expressed as described previously (23). In addition, the entire NP of RABV CVS11 strain (GenBank accession number GQ918139) was prokaryotically and eukaryotically expressed, with mouse-specific hyperimmune serum prepared as before.
Cross-antigenicity analysis by WB and serological assay of bat serum by ELISA and WB.
The details of cross-antigenicity characterization, WB, and ELISA have been previously described (23). Six mouse-specific hyperimmune serum samples were used to detect the 7 eukaryotically expressed NPs by WB. Eukaryotically expressed NPs were separated by SDS-PAGE and transferred onto nitrocellulose blotting membranes (GE Healthcare), blocked with 5% skimmed milk (Promega), then incubated with mouse anti-NP serum at a 1:300 dilution. After washing with phosphate-buffered saline with Tween 20 (PBST) 3×, the membranes were incubated with donkey anti-mouse IgG 1:1,000 for 50 min, and then visualized using an Odyssey imager (LI-COR Biosciences). All bat serum samples were tested by ELISA against prokaryotic rNPs of DH04, RESTV, EBOV, and MARV. Briefly, 96-well microplates (Corning) were coated with purified prokaryotic rNPs at 4°C overnight and blocked with 5% skimmed milk at 37°C for 1 h. Then, 100-fold phosphate-buffered saline (PBS)-diluted serum samples were added to the wells and incubated at 37°C for 1 h followed by the addition of horseradish peroxidase (HRP)-conjugated goat anti-bat IgG (Bethyl) at a 1:20,000 dilution. After incubation at 37°C for 5 min, freshly prepared o-phenylenediamine (OPD) substrate solution (Sigma) was added to each well for 5 min for color reaction and then stopped by addition of 2 M sulfuric acid. The OD492 values were immediately read and blanked by the OD630 value using a multimode microplate reader (Infinite 200 Pro; Tecan). Due to lack of specific positive bat serum against the 4 viruses, the cutoff value of the ELISA could not be determined by traditional methods. Instead, the ELISA OD492 readings of 151 bat serum samples were compared with their corresponding WB results (23), with the cutoff being established as the lowest reading at which 95% of the WB results were positive. In addition, the 518 bat serum samples available in sufficient quantity were tested by ELISA against RABV and TUHV rNPs, but due to the lack of sufficient serum, the cutoff values of these were not determined.
Neutralization assay.
Reactivity of the EBOV serum was further confirmed by neutralization assay (NA) with EBOV transcription-competent virus-like particles (trVLPs). Cells infected with trVPs continuously express the EBOV proteins responsible for genome replication and transcription, thereby permitting safe performance of NA under biosafety level 2 conditions (36). To generate trVLPs, 293T cells were transfected with plasmids bearing NP, VP35, VP30, L, vRNA-RLuc, or T7, using TransIT-LT1 transfection reagent (Mirus Bio). At 96 h posttransfection, the resulting trVLPs were harvested and diluted to 100 TCID50 (50% tissue culture infective dose)/50 μl, followed by reaction of 2-fold dilutions (1:20 to 1:2,560) with bat serum for 1 h at 37°C and addition to 293T cells transfected by 4 plasmids bearing NP, VP35, VP30, L, or Tim1. Luciferase activity was measured 72 h postincubation in a GloMax 20/20 single tube luminometer (Promega) using the Renilla-Glo luciferase assay system (Promega). Antiserum titers (ND50) were calculated by the Kärber method and expressed as dilution endpoints. A refined hyperimmunized equine immunoglobulin (HEI) against EBOV virus-like particles (VLPs) was used as a positive NA control (37).
Viral metagenomic analysis and RT-PCR screening.
Organ samples of 57 fruit bats were pooled and subjected to viral metagenomic analysis as per our published method (38, 39). All sequences generated in one lane by the Illumina HiSeq 4000 platform were subjected to host genome removal and virus annotation. To detect FiVs, all organ samples were subjected to RNA extraction using the RNeasy minikit (Qiagen). Reverse transcription was conducted with the 1st cDNA synthesis kit (TaKaRa) according to the manufacturer’s protocol. The cDNA was amplified using our published nested PCR method, which was successfully used to detect DH04 and MLAV in bats (11, 12). The FiV-like reads revealed by high-throughput sequencing (HTS) were compared with the counterparts of other FiVs, and their preliminary phylogeny was implemented using MEGA v.6.06 under the maximum likelihood method with 100 bootstrap replications.
Statistical analyses.
To compare the differences of the OD492 readings of bat serum to DH04, RESTV, EBOV, and MARV, normal distribution tests were conducted separately. The serum titers were not normally distributed, so the differences between them were compared using a Wilcoxon rank sum test. All data processes were conducted using Statistical Analysis System (SAS) v.9.2.
Data availability.
The Illumina-generated raw data in this study have been deposited in the NCBI Short Read Archive (SRA) under BioProject number PRJNA597258.
ACKNOWLEDGMENTS
We are grateful to Jonathan S. Towner at the Centers for Disease Control and Prevention, Atlanta, Georgia, USA, for critical readings of the manuscript.
The study was supported by the National Natural Science Foundation of China (31572529 and 31502077) and the National Key Basic Research and Development Program of China (2016YFC1200100).
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
We declare no conflict of interest.
REFERENCES
- 1.Feldmann H, Sanchez A, Geisbert TW. 2013. Filoviridae: Marburg and Ebola virus, p 923–956. In Knipe DM, Howley PM (ed), Fields virology, 6th ed, vol 1 Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 2.International Committee on Taxonomy of Viruses. 2019. Virus taxonomy: 2018b release. https://talk.ictvonline.org/taxonomy.
- 3.Shi M, Lin X-D, Chen X, Tian J-H, Chen L-J, Li K, Wang W, Eden J-S, Shen J-J, Liu L, Holmes EC, Zhang Y-Z. 2018. The evolutionary history of vertebrate RNA viruses. Nature 556:197–202. doi: 10.1038/s41586-018-0012-7. [DOI] [PubMed] [Google Scholar]
- 4.Towner JS, Amman BR, Sealy TK, Carroll SAR, Comer JA, Kemp A, Swanepoel R, Paddock CD, Balinandi S, Khristova ML, Formenty PBH, Albarino CG, Miller DM, Reed ZD, Kayiwa JT, Mills JN, Cannon DL, Greer PW, Byaruhanga E, Farnon EC, Atimnedi P, Okware S, Katongole-Mbidde E, Downing R, Tappero JW, Zaki SR, Ksiazek TG, Nichol ST, Rollin PE. 2009. Isolation of genetically diverse Marburg viruses from Egyptian fruit bats. PLoS Pathog 5:e1000536. doi: 10.1371/journal.ppat.1000536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Olival JK, Hayman TD. 2014. Filoviruses in bats: current knowledge and future directions. Viruses 6:1759–1788. doi: 10.3390/v6041759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Amman BR, Carroll SA, Reed ZD, Sealy TK, Balinandi S, Swanepoel R, Kemp A, Erickson BR, Comer JA, Campbell S, Cannon DL, Khristova ML, Atimnedi P, Paddock CD, Crockett RJK, Flietstra TD, Warfield KL, Unfer R, Katongole-Mbidde E, Downing R, Tappero JW, Zaki SR, Rollin PE, Ksiazek TG, Nichol ST, Towner JS. 2012. Seasonal pulses of Marburg virus circulation in juvenile Rousettus aegyptiacus bats coincide with periods of increased risk of human infection. PLoS Pathog 8:e100287. doi: 10.1371/journal.ppat.1002877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leroy EM, Kumulungui B, Pourrut X, Rouquet P, Hassanin A, Yaba P, Délicat A, Paweska JT, Gonzalez J-P, Swanepoel R. 2005. Fruit bats as reservoirs of Ebola virus. Nature 438:575–576. doi: 10.1038/438575a. [DOI] [PubMed] [Google Scholar]
- 8.Jayme SI, Field HE, de Jong C, Olival KJ, Marsh G, Tagtag AM, Hughes T, Bucad AC, Barr J, Azul RR, Retes LM, Foord A, Yu M, Cruz MS, Santos IJ, Lim TMS, Benigno CC, Epstein JH, Wang L-F, Daszak P, Newman SH. 2015. Molecular evidence of Ebola Reston virus infection in Philippine bats. Virol J 12:107. doi: 10.1186/s12985-015-0331-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Negredo A, Palacios G, Vazquez-Moron S, Gonzalez F, Dopazo H, Molero F, Juste J, Quetglas J, Savji N, Martınez MC, Herrera JE, Pizarro M, Hutchison SK, Echevarrıa JE, Lipkin WI, Tenorio A. 2011. Discovery of an ebolavirus-like filovirus in Europe. PLoS Pathog 7:e1002304. doi: 10.1371/journal.ppat.1002304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kemenesi G, Kurucz K, Dallos B, Zana B, Földes F, Boldogh S, Görföl T, Carroll MW, Jakab F. 2018. Re-emergence of Lloviu virus in Miniopterus schreibersii bats, Hungary, 2016. Emerg Microbes Infect 7:66. doi: 10.1038/s41426-018-0067-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.He B, Feng Y, Zhang H, Xu L, Yang W, Zhang Y, Li X, Tu C. 2015. Filovirus RNA in fruit bats, China. Emerg Infect Dis 21:1675–1677. doi: 10.3201/eid2109.150260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang X-L, Zhang Y-Z, Jiang R-D, Guo H, Zhang W, Li B, Wang N, Wang L, Waruhiu C, Zhou J-H, Li S-Y, Daszak P, Wang L-F, Shi Z-L. 2017. Genetically diverse filoviruses in Rousettus and Eonycteris spp. bats, China, 2009 and 2015. Emerg Infect Dis 23:482–486. doi: 10.3201/eid2303.161119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang X-L, Tan CW, Anderson DE, Jiang R-D, Li B, Zhang W, Zhu Y, Lim XF, Zhou P, Liu X-L, Guan W, Zhang L, Li S-Y, Zhang Y-Z, Wang L-F, Shi Z-L. 2019. Characterization of a filovirus (Měnglà virus) from Rousettus bats in China. Nat Microbiol 4:390–395. doi: 10.1038/s41564-018-0328-y. [DOI] [PubMed] [Google Scholar]
- 14.Goldstein T, Anthony SJ, Gbakima A, Bird BH, Bangura J, Tremeau-Bravard A, Belaganahalli MN, Wells HL, Dhanota JK, Liang E, Grodus M, Jangra RK, DeJesus VA, Lasso G, Smith BR, Jambai A, Kamara BO, Kamara S, Bangura W, Monagin C, Shapira S, Johnson CK, Saylors K, Rubin EM, Chandran K, Lipkin WI, Mazet J. 2018. The discovery of Bombali virus adds further support for bats as hosts of ebolaviruses. Nat Microbiol 3:1084–1089. doi: 10.1038/s41564-018-0227-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Forbes KM, Webala PW, Jääskeläinen AJ, Abdurahman S, Ogola J, Masika MM, Kivistö I, Alburkat H, Plyusnin I, Levanov L, Korhonen EM, Huhtamo E, Mwaengo D, Smura T, Mirazimi A, Anzala O, Vapalahti O, Sironen T. 2019. Bombali virus in Mops condylurus bat, Kenya. Emerg Infect Dis 25:955–957. doi: 10.3201/eid2505.181666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karan LS, Makenov MT, Korneev MG, Sacko N, Boumbaly S, Yakovlev SA, Kourouma K, Bayandin RB, Gladysheva AV, Shipovalov AV, Yurganova IA, Grigorieva YE, Fedorova MV, Scherbakova SA, Kutyrev VV, Agafonov AP, Maksyutov RA, Shipulin GA, Maleev VV, Boiro M, Akimkin VG, Popova AY. 2019. Bombali virus in Mops condylurus bats, Guinea. Emerg Infect Dis 25:1774–1775. doi: 10.3201/eid2509.190581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hayman DTS, Emmerich P, Yu M, Wang L-F, Suu-Ire R, Fooks AR, Cunningham AA, Wood J. 2010. Long-term survival of an urban fruit bat seropositive for Ebola and Lagos bat viruses. PLoS One 5:e11978. doi: 10.1371/journal.pone.0011978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Taniguchi S, Watanabe S, Masangkay JS, Omatsu T, Ikegami T, Alviola P, Ueda N, Iha K, Fujii H, Ishii Y, Mizutani T, Fukushi S, Saijo M, Kurane I, Kyuwa S, Akashi H, Yoshikawa Y, Morikawa S. 2011. Reston ebolavirus antibodies in bats, the Philippines. Emerg Infect Dis 17:1559–1560. doi: 10.3201/eid1708.101693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hayman DTS, Yu M, Crameri G, Wang L-F, Suu-Ire R, Wood JLN, Cunningham AA. 2012. Ebola virus antibodies in fruit bats, Ghana, West Africa. Emerg Infect Dis 18:1207–1209. doi: 10.3201/eid1807.111654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yuan J, Zhang Y, Li J, Zhang Y, Wang L-F, Shi Z. 2012. Serological evidence of ebolavirus infection in bats, China. Virol J 9:236. doi: 10.1186/1743-422X-9-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Olival KJ, Islam A, Yu M, Anthony SJ, Epstein JH, Khan SA, Khan SU, Crameri G, Wang L-F, Lipkin WI, Luby SP, Daszak P. 2013. Ebola virus antibodies in fruit bats, Bangladesh. Emerg Infect Dis 19:270–273. doi: 10.3201/eid1902.120524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Changula K, Kajihara M, Mori-Kajihara A, Eto Y, Miyamoto H, Yoshida R, Shigeno A, Hang'ombe B, Qiu Y, Mwizabi D, Squarre D, Ndebe J, Ogawa H, Harima H, Simulundu E, Moonga L, Kapila P, Furuyama W, Kondoh T, Sato M, Takadate Y, Kaneko C, Nakao R, Mukonka V, Mweene A, Takada A. 2018. Seroprevalence of filovirus infection of Rousettus aegyptiacus bats in Zambia. J Infect Dis 218:S312–S317. doi: 10.1093/infdis/jiy266. [DOI] [PubMed] [Google Scholar]
- 23.Xu L, Wu J, Li Q, Wei Y, Tan Z, Cai J, Guo H, Yang L, Huang X, Chen J, Zhang F, He B, Tu C. 2019. Seroprevalence, cross antigenicity and circulation sphere of bat-borne hantaviruses revealed by serological and antigenic analyses. PLoS Pathog 15:e1007545. doi: 10.1371/journal.ppat.1007545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moratelli R, Calisher CH. 2015. Bats and zoonotic viruses: can we confidently link bats with emerging deadly viruses? Mem Inst Oswaldo Cruz 110:1–22. doi: 10.1590/0074-02760150048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou P, Fan H, Lan T, Yang X-L, Shi W-F, Zhang W, Zhu Y, Zhang Y-W, Xie Q-M, Mani S, Zheng X-S, Li B, Li J-M, Guo H, Pei G-Q, An X-P, Chen J-W, Zhou L, Mai K-J, Wu Z-X, Li D, Anderson DE, Zhang L-B, Li S-Y, Mi Z-Q, He T-T, Cong F, Guo P-J, Huang R, Luo Y, Liu X-L, Chen J, Huang Y, Sun Q, Zhang X-L-L, Wang Y-Y, Xing S-Z, Chen Y-S, Sun Y, Li J, Daszak P, Wang L-F, Shi Z-L, Tong Y-G, Ma J-Y. 2018. Fatal swine acute diarrhoea syndrome caused by an HKU2-related coronavirus of bat origin. Nature 556:255–258. doi: 10.1038/s41586-018-0010-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Towner JS, Pourrut X, Albarino CG, Nkogue CN, Bird BH, Grard G, Ksiazek TG, Gonzalez J-P, Nichol ST, Leroy EM. 2007. Marburg virus infection detected in a common African bat. PLoS One 2:e764. doi: 10.1371/journal.pone.0000764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Swanepoel R, Smit SB, Rollin PE, Formenty P, Leman PA, Kemp A, Burt FJ, Grobbelaar AA, Croft J, Bausch DG, Zeller H, Leirs H, Braack LEO, Libande ML, Zaki S, Nichol ST, Ksiaze TG, Paweska JT, on behalf of the International Scientific and Technical Committee for Marburg Hemorrhagic Fever Control in the Democratic Republic of the Congo. 2007. Studies of reservoir hosts for Marburg virus. Emerg Infect Dis 13:1847–1851. doi: 10.3201/eid1312.071115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sanchez A, Kiley MP, Klenk H-D, Feldmann H. 1992. Sequence analysis of the Marburg virus nucleoprotein gene: comparison to Ebola virus and other non-segmented negative-strand RNA viruses. J Gen Virol 73:347–357. doi: 10.1099/0022-1317-73-2-347. [DOI] [PubMed] [Google Scholar]
- 29.Lau SKP, Woo PCY, Wong BHL, Wong AYP, Tsoi H-W, Wang M, Lee P, Xu H, Poon RWS, Guo R, Li KSM, Chan K-H, Zheng B-J, Yuen K-Y. 2010. Identification and complete genome analysis of three novel paramyxoviruses, Tuhoko virus 1, 2 and 3, in fruit bats from China. Virology 404:106–116. doi: 10.1016/j.virol.2010.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jiang Y, Wang L, Lu Z, Xuan H, Han X, Xia X, Zhao F, Tu C. 2010. Seroprevalence of rabies virus antibodies in bats from Southern China. Vector Borne Zoonotic Dis 10:177–181. doi: 10.1089/vbz.2008.0212. [DOI] [PubMed] [Google Scholar]
- 31.Schuh AJ, Amman BR, Sealy TS, Flietstra TD, Guito JC, Nichol ST, Towner JS. 2019. Comparative analysis of serologic cross-reactivity using convalescent sera from filovirus-experimentally infected fruit bats. Sci Rep 9:6707. doi: 10.1038/s41598-019-43156-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Flyak AI, Shen X, Murin CD, Turner HL, David JA, Fusco ML, Lampley R, Kose N, Ilinykh PA, Kuzmina N, Branchizio A, King H, Brown L, Bryan C, Davidson E, Doranz BJ, Slaughter JC, Sapparapu G, Klages C, Ksiazek TG, Saphire EO, Ward AB, Bukreyev A, Crowe JE. 2016. Cross-reactive and potent neutralizing antibody responses in human survivors of natural ebolavirus infection. Cell 164:392–405. doi: 10.1016/j.cell.2015.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Natesan M, Jensen SM, Keasey SL, Kamata T, Kuehne AI, Stonier SW, Lutwama JJ, Lobel L, Dye JM, Ulrich RG. 2016. Human survivors of disease outbreaks caused by Ebola or Marburg virus exhibit cross-reactive and long-lived antibody responses. Clin Vaccine Immunol 23:717–724. doi: 10.1128/CVI.00107-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pourrut X, Souris M, Towner JS, Rollin PE, Nichol ST, Gonzalez J-P, Leroy E. 2009. Large serological survey showing cocirculation of Ebola and Marburg viruses in Gabonese bat populations, and a high seroprevalence of both viruses in Rousettus aegyptiacus. BMC Infect Dis 9:159. doi: 10.1186/1471-2334-9-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.He B, Huang X, Zhang F, Tan W, Matthijnssens J, Qin S, Xu L, Zhao Z, En YL, Wang Q, Hu T, Bao X, Wu J, Tu C. 2017. Group A rotaviruses in Chinese bats: genetic composition, serology, and evidence for bat-to-human transmission and reassortment. J Virol 91:e02493-16. doi: 10.1128/JVI.02493-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Watt A, Moukambi F, Banadyga L, Groseth A, Callison J, Herwig A, Ebihara H, Feldmann H, Hoenen T. 2014. A novel life cycle modeling system for Ebola virus shows a genome length-dependent role of VP24 in virus infectivity. J Virol 88:10511–10524. doi: 10.1128/JVI.01272-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zheng X, Wong G, Zhao Y, Wang H, He S, Bi Y, Chen W, Jin H, Gai W, Chu D, Cao Z, Wang C, Fan Q, Chi H, Gao Y, Wang T, Feng N, Yan F, Huang G, Zheng Y, Li N, Li Y, Qian J, Zou Y, Kobinger G, Gao GF, Qiu X, Yang S, Xia X. 2016. Treatment with hyperimmune equine immunoglobulin or immunoglobulin fragments completely protects rodents from Ebola virus infection. Sci Rep 6:24179. doi: 10.1038/srep24179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.He B, Li Z, Yang F, Zheng J, Feng Y, Guo H, Li Y, Wang Y, Su N, Zhang F, Fan Q, Tu C. 2013. Virome profiling of bats from Myanmar by metagenomic analysis of tissue samples reveals more novel mammalian viruses. PLoS One 8:e61950. doi: 10.1371/journal.pone.0061950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tan Z, Yu H, Xu L, Zhao Z, Zhang P, Qu Y, He B, Tu C. 2019. Virome profiling of rodents in Xinjiang Uygur autonomous region, China: isolation and characterization of a new strain of Wenzhou virus. Virology 529:122–134. doi: 10.1016/j.virol.2019.01.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The Illumina-generated raw data in this study have been deposited in the NCBI Short Read Archive (SRA) under BioProject number PRJNA597258.