Abstract
The human long- and middle-wavelength sensitive cone opsin genes exhibit an extraordinary degree of haplotype diversity that results from recombination mechanisms that have intermixed the genes. As a first step in expression, genes—including the protein coding exons and intervening introns—are transcribed. Next, transcripts are spliced to remove the introns and join the exons to generate a mature message that codes for the protein. Important information necessary for splicing is contained within exons, and is overlaid by the protein code. Intermixing the long- and middle-wavelength sensitive cone opsin genes has disrupted the splicing code, leading to exclusion of some exons from the mature message and is associated with several vision disorders including nearsightedness, cone dystrophy, and color vision deficiencies.
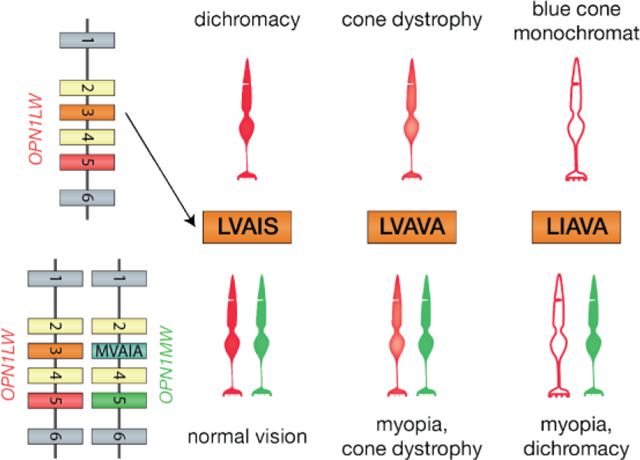
Graphical Abstract
Introduction
Humans with normal color vision have three types of cone photoreceptor that are classed according to their relative spectral sensitivities as short- (S), middle- (M) and long- (L) wavelength sensitive. Neural comparisons between their quantal catches give rise to six main hue percepts – red, green, blue, yellow, black and white – and combinations thereof yield more than a million distinguishable colors [1–3]. The color palette is greatly reduced for individuals with inherited red-green color vision deficiencies. Rearrangements and mutations of genes encoding the protein component (opsin) of cone photopigments are the primary cause of inherited color vision deficiencies (dichromacies and anomalous trichromacies) [4–6]. Except at very low light levels when rods are active—all vision, not just color vision—is based on cones [2]. Here we briefly review the role of genetic variation in the cone opsin genes in inherited color vision deficiencies, including recent discoveries of mutations that cause defective splicing and result in several vision disorders including cone dysfunction, nearsightedness (myopia), cone dystrophy, blue cone monochromacy, and dichromacy.
Genetics of Common Inherited Color Vision Deficiencies
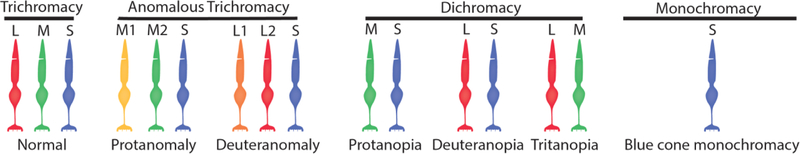
Inherited color vision deficiencies are categorized according to the number of functional cone types contributing to vision (Figure 1). All are caused by mutations that alter the complement of functional cone opsins expressed. The L, M and S-cone opsin genes, designated OPN1LW, OPN1MW, and OPN1SW, respectively, have similar organizations. Inherited color vision deficiencies associated with mutations in OPN1SW are rare and will not be discussed here. OPN1LW and OPN1MW are on the X-chromosome. The number of X-chromosomes per cell is two for females and one for males, but female cells—including L- and M-cones—express genes from one of the two X-chromosomes.
Figure 1.
Complement of cones types in normal color vision and inherited color vision deficiency. Protanopia and protanomaly lack contribution from L-cones, deuteranopia and deuteranomaly lack contributions for M-cones, tritanopia lacks contributions from S-cones, and blue cone monochromacy lacks contribution from L- and M-cones.
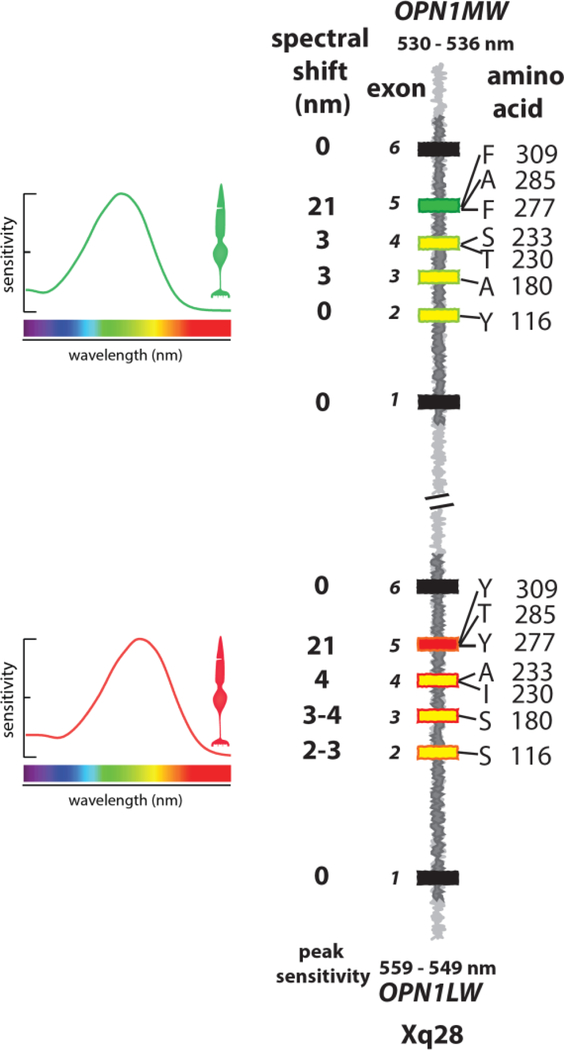
Trichromatic color vision appeared recently in mammalian evolution, and is found only in primates. In Old World Primates, including humans, OPN1LW and OPN1MW gene expression is segregated to separate cone populations, allowing males and females alike to have trichromatic color vision. In non-human Old World Primates, L- and M-cone photopigments are stereotyped within a species, differing at ~18 amino acid positions, seven of which shift the spectra (Figure 2). Presumably differences not involved in spectral tuning were necessary to preserve correct splicing or to stabilize the opsins after they diverged.
Figure 2.
Organization of OPN1LW and OPN1MW and spectral tuning of L- and M-cones. OPN1LW and OPN1MW lie in tandem on the X-chromosome at Xq28 and are nearly identical in sequence. Exons are black, yellow, green and red boxes. Exons 1 and 6 are invariant (black boxes), exon 5 (red and green boxes) encodes amino acids that tune the spectra and define whether the encoded pigment is of the L-class or M-class. Exons 2, 3 and 4 specify amino acids that tune the spectra but do not define whether the encoded pigment is of the L- versus M-class. These are indicated using the single letter amino acid code followed by the codon/amino acid position. In this illustration, the amino acids encoded by exons 2, 3 and 4 that shift the spectrum toward the long-wavelength (magnitudes given in figure) are shown in OPN1LW (yellow boxes with red outlines) and those that shift the spectrum to the short-wavelengths are shown in OPN1MW (yellow boxes with green outlines). Depending on the combination of amino acids encoded by exons 2–4 L-cone peak sensitivity ranges from 549 to 559 nanometers (nm). Depending on exon 3 and 4, M-cone peak sensitivity ranges from 530 to 536 nm. The single letter amino acid code is A=alanine, F=phenylalanine, G=glycine, I=isoleucine, L=leucine, P=proline, Q=glutamine, R=arginine, S=serine, T=threonine, Y=tyrosine.
Unlike in non-human Old World Primates, human OPN1LW and OPN1MW genes vary in copy number and exhibit genetic variability because recombination mechanisms intermix the genes and redistribute them to form new gene arrays that underlie inherited red-green color vision deficiencies. Relaxation of selection against inherited red-green color vision deficiencies presumably allowed opsin gene arrays underlying these defects to propagate, as evidenced by the 15% prevalence of female carriers of inherited red-green color vision defects in present day populations [6].
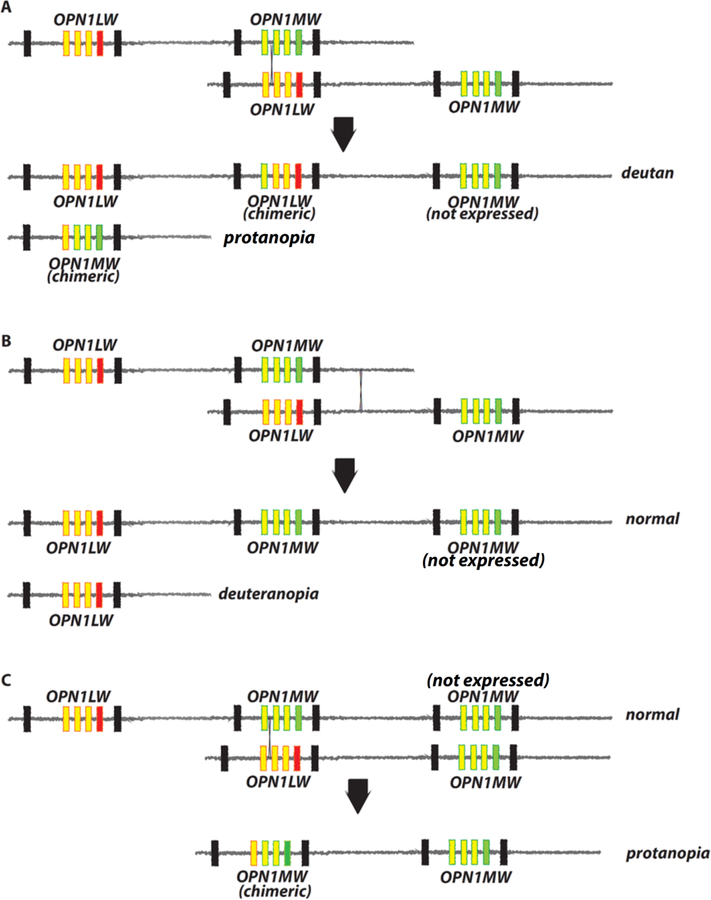
Figure 3 illustrates recombination events responsible for generating opsin gene arrays that cause inherited red-green color vision defects. In females, exchange of genetic material mediated by a cross-over between an OPN1LW gene and an OPN1MW gene on different X-chromosomes produces two new arrays that both contain intermixed, chimeric genes. The example in Figure 3A shows recombination between two arrays that each have one OPN1LW and one OPN1MW gene. The genes from the parental chromosomes are redistributed, creating a new array with three genes, and another with one gene. Both arrays have chimeric genes. The chimeric gene in the three-gene array derives exon 5 from the parental OPN1LW gene, and it encodes an L-class pigment. Only the leftmost two genes in the array are expressed, so the array underlies deutan-type color vision. Whether the array underlies dichromacy or anomalous trichromacy depends on whether spectral tuning sites in exons 2, 3 and 4 differ between the two L pigments. The one-gene array carries a chimeric gene that encodes an M-class pigment because it derives exon 5 from a parental OPN1LW gene, and it will confer protanopia.
Figure 3.
Recombination intermixes the OPN1LW and OPN1MW gene sequences, generates variability in the number of opsin genes on the X-chromosome, and causes red-green color vision defects. (A)-(C) The top two parental X-chromosome opsin gene arrays exchange genetic material at the location indicated by the thin black X. All parental arrays confer normal color vision. Parental OPN1LW genes have a red box for exon 5 and yellow boxes with red outlines for exons 2–4. Parental OPN1MW genes have a green box for exon 5 and yellow boxes with green outlines for exons 2–4. Below the black arrows in (A)-(C) are the recombination products and the type of color vision they confer. The parental gene of origin for exons 2–4 in chimeric genes are indicated by the color of the outline (red for OPN1LW, green for OPN1MW origin), and by the color of the box for exon 5 (red for OPN1LW, green for OPN1MW).
The region between the genes is nearly identical to the region following the last gene in the array. Recombination between these two regions (Figure 3B) does not intermix OPN1LW and OPN1MW, but does redistribute the genes such that one new array carries one gene, which encodes an L pigment and will confer dichromacy (deuteranopia). The other carries OPN1LW followed by two OPN1MW genes and will confer normal color vision.
Variation in the number of opsin genes on the X-chromosome in present day populations allows recombination between the middle gene in a three gene array and the first gene in a two gene array as illustrated in Figure 3C. This recombination gives rise to a new array in the which the first gene is chimeric followed by an OPN1MW gene. The chimeric gene derives exon 5 from a parental OPN1MW, and thus encodes an M class pigment. The array underlies a protan color vision deficiency, dichromacy or anomalous trichromacy, depending on whether spectral tuning sites encoded by exon 3 and 4 differ in the two genes. The other product of the recombination illustrated in Figure 3C is the same as that underlying deutan color vision shown in Figure 3A.
Because expression of opsin genes from the two X-chromosomes is segregated to separate cone populations, a female will have an inherited red-green color vision defect only if both of her X-chromosomes carry the same category of inherited red-green defect —protan or deutan. If one X-chromosome carries a protan defect and the other a deutan defect, she will have L- and M-cones and normal color vision.
Intermixing OPN1LW and OPN1MW Causes Inherited Color Vision Defects, Cone Dysfunction, and High-Myopia.
Variability in the nucleotide sequence (haplotype diversity) of exons 2 – 4 of OPN1LW and OPN1MW has been evaluated with regard to selective pressure for red-green color vision [7–10]. However, cones provide more than color vision. Importantly, they mediate high acuity spatial vision and achromatic black-white vision [11], as was recently illustrated when color percepts elicited by adaptive optics-mediated micro-stimulation of individual cones, revealed that the majority of L- and M-cones elicit the percept of white [2]. A small subset of L- and M-cones in the vicinity of S-cones elicited red, green or blue color percepts [12]. Thus, most L- and M-cones mediate high-acuity achromatic spatial vision. After birth, human eyes undergo emmetropization, a controlled axial elongation, governed by a feedback mechanism in which L-and M-cones play a critical role so that the eye stops growing when the length is optimally matched to the power of the optical components for high acuity vision. Nearsightedness (myopia) results if the eye grows too long.
An exon 3 haplotype was recently identified as the cause of Bornholm Eye Disease a syndrome characterized by high-myopia, and inherited red-green color vision defects [13,14]. The haplotype is abbreviated LVAVA for the amino acids at the polymorphic positions encoded by exon 3 which are Leucine 153, Valine 171, Alanine 174, Valine 178, and Alanine 180. Bornholm Eye Disease was genetically linked to the first officially designated high-myopia gene, MYP1, located at Xq28, the same location as OPN1LW and OPN1MW [15]. The LVAVA haplotype has been demonstrated independently by several labs as a cause of high-myopia [13,16–18].
Initially it was assumed that if OPN1LW/OPN1MW were responsible for Bornholm Eye Disease, all affected individuals would have the same color vision phenotype. However, they do not because the role of the cone opsin genes in myopia is independent of their role in color vision. That is, the color vision phenotype in Bornholm Eye Disease individuals with a deutan defect is accounted for by their opsin gene arrays in which the first two genes encode L-class pigments such as the deutan array in Figure 3A. They have high-myopia because one of the first two genes has the LVAVA haplotype, which causes a splicing defect and produces cones with dramatically reduced amounts of photopigment [14]. These patients have two submosaics of L-cones, one expressing the LVAVA opsin and another expressing a normal OPN1LW haplotype and this appears to interfere with emmetropization. We hypothesized that the associated high-myopia is the result of an erroneous contrast signal initiated at the level of the cones [14] due to dramatically different amounts of photopigment in the two subpopulations of cones. Similarly, the protan color viison in the second Bornholm Eye Disease family is accounted for by their opsin gene array having two OPN1MW genes such as the protanomalous array in Figure 3C, and they have high-myopia because one of the OPN1MW genes specifies the LVAVA haplotype, the other specifies a normal variant. Cones expressing a gene encoding the LVAVA haplotype are associated with cone dystrophy [19,20].
The first OPN1LW/OPN1MW exon 3 haplotype associated with cone dysfunction was abbreviated “LIAVA” and differs from LVAVA in encoding isoleucine instead of valine at positon 171. This haplotype causes a splicing defect and appears to give rise to “empty cones” which are stable over many years [19–21]. When a normal haplotype is expressed in a separate population of cones, the LIAVA haplotype is associated with high-myopia particularly when the larger fraction of L-/M-cones are those expressing the LIAVA haplotype [17], and with dichromacy. LIAVA is also associated with blue cone monochromacy when all but the S-cones express an LIAVA opsin gene haplotype.
Intermixing the OPN1LW and OPN1MW Genes Disrupts the Splicing Code
The LIAVA- and LVAVA-exon 3 haplotypes fail to be recognized during pre-messenger RNA (pre-mRNA) splicing [22,23]. Genes are initially transcribed in their entirety to generate pre-mRNA. Splicing removes the introns from the pre-mRNA and joins the exons to form the mature mRNA, which carries a contiguous protein coding sequence. When the splicing apparatus fails to recognize an exon, it is excluded from the mature mRNA [14,22,23], which disrupts the protein coding sequence. Opsin mRNA lacking exon 3 likely is targeted for destruction and not translated into protein.
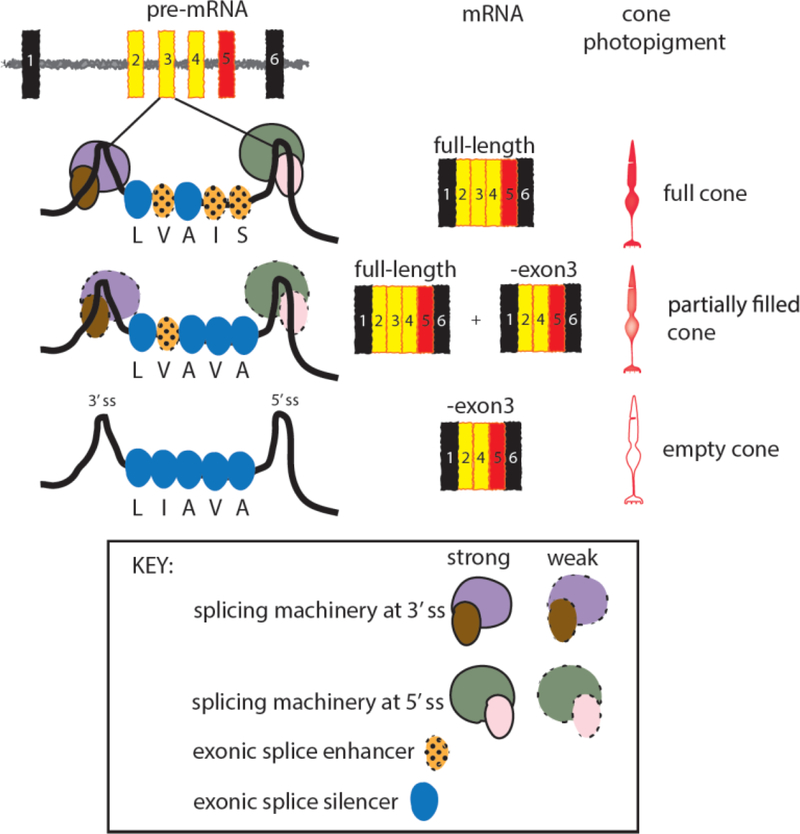
The 3’ and 5’ consensus splice sites at the intron/exon junctions in pre-mRNA contain insufficient information for correct splicing [24]. Additional information is provided by 6–8 nucleotide sequences that serve as enhancers of splicing (Figure 4). When located in exons, these serve a dual role, providing information needed for splicing and for making a functional protein. Intermixing the OPN1LW and OPN1MW exon 3 sequences has created haplotypes that disrupt the splicing code. For example, the LVAIS haplotype is normal-splicing haplotype, not associated with vision disorders. It contains sufficient information for exon 3 to be included in the mRNA always [23]. Cones expressing this haplotype contain the maximum amount of photopigment. Splicing enhancers in LVAIS associated with I178 and S180 are disrupted in LVAVA, and nucleotide changes associated with V178 and A180 in LVAVA introduce splicing silencers [25]. Splicing silencers are also short nucleotide sequences and, when in exons, they promote exon skipping. The splicing apparatus does not always recognize the LVAVA-exon 3, and produces a mixture of exon 3-skipped and full-length mRNA (Figure 4). Functional photopigment is made only from the full length mRNA so cones expressing the LVAVA haplotype have a reduced amount of photopigment. Splicing enhancers associated with V171, I178 and S180 in LVAIS are all disrupted by nucleotides associated with I171, V178 and A180 in LIAVA, and silencers are created at all three locations in LIAVA [25]. LIAVA-exon 3 is excluded from the mRNA most, if not all of the time, and thus, the LIAVA haplotype gives rise to empty cones. It is not known whether exon 3-skipping is caused by disruption of enhancers or activation of silencers, or some combination of the two.
Figure 4.
Model for how nucleotide sequence variability in exon 3 alters splicing and the amount of photopigment in cones. The most common human OPN1LW exon 3 haplotype is abbreviated “LVAIS” for the amino acids specified at the five exon 3-encoded polymorphic amino acid positions (see Figure 2 single letter amino acid code). Nucleotide triplets specify the amino acid code, hexamers and octamers specify the splicing code and, within exons, can act as enhancers to promote exon inclusion in mRNA or act as silencers to inhibit exon inclusion. Components of the splicing machinery bind to enhancers and interact with the RNA-Protein complexes bound to the 5’ and 3’ splice sites at the intron/exon junctions and assist in including the exon in the mRNA. Exonic splicing silencers can block the binding of proteins to enhancers or inhibit assembly of the splicing machinery, promoting exon exclusion. The LVAIS haplotype includes exon 3 most, perhaps all, of the time. LIAVA does not include exon 3 a detectable amount. LVAVA includes exon 3 some of the time. Based on observations in humans the amount of photopigment in the cone correlates to the amount of full length mRNA [14].
Potential for Splicing Differences to Alter Photopigment Optical Density in Cones.
In anomalous trichromacy (Figure 1), color vision is usually mediated by a difference in peak sensitivity of the underlying L or M-cones; but can be mediated by two cone populations that have the same peak sensitivity but different optical densities [26]. The discovery of exon 3-skipping haplotypes of OPN1LW and OPN1MW provides a potential mechanism for generating optical density differences that could underlie anomalous trichromacy.
Conclusions
For most of the 30 years since the molecular genetics of inherited color vision deficiencies was elucidated, ideas about evolutionary advantages or disadvantages of cone opsin gene variation, including comparisons between humans and non-human Old World Primates, have considered only the potential effects on color vision and protein structure and function.
The human OPN1LW and OPN1MW genes have been intermixing over the course of human evolution, presumably due to reduced selection against disorders associated with recombinant arrays. Compared to other Old World Primates, human innovations make us less dependent on normal color vision and non-myopic vision for survival. The recent discovery that intermixing the opsin gene sequences produces haplotypes that cause splicing defects which lead to a much broader spectrum of vision disorders, not just inherited red-green color vision defects, is a revelation that is expected to ultimately have a significant impact on our understanding of cone-based vision disorders and on the development of treatments for them.
Highlights Normal color vision requires short-, medium- and long-wavelength sensitive cones.
Inherited color vision deficiency reduces the number of cone types
Cones are responsible for color vision and high acuity spatial vision
L- and M-cones play a critical role eye growth
Intermixing OPN1LW and OPN1MW genes causes splicing errors and vision defects
Acknowledgements:
M. Neitz is the Ray H. Hill Endowed Professor, J. Neitz is the Bishop Professor. This work was supported by unrestricted funds from Research to Prevent Blindness to the Department of Ophthalmology at the University of Washington, by NIH grants R01 EY0028118 and P30 EY001730. S. Patterson is supported by NEI T32 EY007031.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests:
Maureen Neitz and Jay Neitz have significant financial interest in the company Waveshift and its subsidiaries, SightGlass and Visu, and receive royalties from the University of Washington for intellectual property, including patents, licensed to these companies.
Sara Patterson: none
References
Referenced papers have been labeled as:
* of special interested
** of outstanding interest
- 1.Schmidt BP, Neitz M, Neitz J: The neurobiological explanation for color appearance and hue perception. Journal of the Optical Society of America (2014) 31(3):A195–A207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sabesan R, Schmidt BP, Tuten WS, Roorda A: The elementary representation of spatial and color vision in the human retina. Science Advances (2016) 2(e1600797. [DOI] [PMC free article] [PubMed] [Google Scholar]; ** Microstimulation of individual cones using adaptive optics and eye tracking measured, for the first time, the color perceptions elicited by stimulating individual cones of known spectral type in the human retina. The results suggests that the nervous system encodes high-resolution achromatic information and lower-resolution color signals in separate pathways that emerge as early as the first synapse.
- 3.Schmidt BP, Touch P, Neitz M, Neitz J: Circuitry to explain how the relative number of l and m cones shapes color experience. J Vis (2016) 16(8):18. [DOI] [PMC free article] [PubMed] [Google Scholar]; ** Psychophysical tests were performed to measure the wavelengths of unique yellow and unique green, and the relative ratio of L to M cones estimated. The authors evaluated biologically motivated models to explain how color appearance could be influenced by L:M cone ratio and an environmental normalization process. The results suggest that post-retinal processes may not be necessary to explain unique hues. This is counter to the traditional view.
- 4.Nathans J: The evolution and physiology of human color vision: Insights from molecular genetic studies of visual pigments. Neuron (1999) 24(299–312. [DOI] [PubMed] [Google Scholar]
- 5.Jacobs GH: Evolution of colour vision in mammals. Philosophical transactions of the Royal Society of London Series B, Biological sciences (2009) 364(1531):2957–2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Neitz J, Neitz M: The genetics of normal and defective color vision. Vision Research (2011) 51(7):633–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Winderickx J, Battisti L, Hibibya Y, Motulsky AG, Deeb SS: Haplotype diversity in the human red and green opsin genes: Evidence for frequent sequence exchange in exon 3. Human Molecular Genetics (1993) 2(1413–1421. [DOI] [PubMed] [Google Scholar]
- 8.Sharpe LT, Stockman A, Jägle H, Knau H, Klausen G, Reitner A, Nathans J: Red, green, and red-green hybrid pigments in the human retina: Correlations between deduced protein sequences and psychophysically measured spectral sensitivities. Journal of Neuroscience (1998) 18(10053–10069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Verrelli BC, Tishkoff SA: Signatures of selection and gene conversion associated with human color vision variation. American Journal of Human Genetics (2004) 75(363–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Verrelli BC, Lewis CMJ, Stone AC, Perry GH: Different selective pressures shape the molecual evolution of color vision in chimpanzee and human populations. Mol Biol Evol (2008) 25(12):2735–2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Neitz J, Neitz M: Evolution of the circuitry for conscious color vision in primates. Eye (London, England) (2017) 31(2):286–300. [DOI] [PMC free article] [PubMed] [Google Scholar]; ** examines evidence from experiments in combination with consideration of constraints from evolution to gain a better understand the neural machinery responsible for the hues, red, green blue and yellow, and how they are separated from black and white.
- 12.Schmidt BP, Sabesan R, Tuten WS, Neitz J, Roorda A: Sensations from a single m-cone depend on the activity of surrounding s-cones. Scientific reports (2018) 8(1):8561. [DOI] [PMC free article] [PubMed] [Google Scholar]; ** Using adaptive optics and eye tracking, the authors performed microstimulation of indivdiual cones and found that the majority of L and M cones elicted the sensation of white, while a minority elicted hue sensations. This is a direct demonstration that the majority of L and M cones are resonsible for spatial acuity, not color perception.
- 13.McClements M, Davies WI, Michaelides M, Young T, Neitz M, MacLaren RE, Moore AT, Hunt DM: Variations in opsin coding sequences cause x-linked cone dysfunction syndrome with myopia and dichromacy. Invest Ophthalmol Vis Sci (2013) 54(2):1361–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Greenwald S, Kuchenbecker JA, Rowlan JS, Neitz J, Neitz M: Role of a dual splicing and amino acid code in myopia, cone dysfunction and cone dystrophy associated with l/m opsin interchange mutations. TVST (2017) 6(3):2. [DOI] [PMC free article] [PubMed] [Google Scholar]; ** Genetically modified mice, human electrophysiology, and in vitro assays were used to demonstrate that the LIAVA and LVAVA human opsin gene haplotypes do not exert their deleterious effects at the level of protein structure and function, but rather at the level of splicing.
- 15.Young TL, Deeb SS, Ronan SM, Dewan AT, Alvear AB, Scavello GS, Paluru PC, Brott MS, Hayashi T, Holleschau AM, Benegas N et al. : X-linked high myopia associated with cone dysfunction. Archives of Ophthalmology (2004) 122(6):897–908. [DOI] [PubMed] [Google Scholar]
- 16.Li J, Gao B, Guan L, Xiao X, Zhang J, Li S, Jiang H, Jia X, Yang J, Guo X, Yin Y et al. : Unique variants in opn1lw cause both syndromic and nonsyndromic x-linked high myopia mapped to myp1. Invest Ophthalmol Vis Sci (2015) 56(6):4150–4155. [DOI] [PubMed] [Google Scholar]; * Genetic analysis of subjects with LVAVA opsin gene haplotypes shows that LVAVA causes myopia with and without other vision problems. This is an illustration at the effects of exon-skipping exon 3 haploytpes on regulating eye growth is independent of the cone opsin genes’ role in color vision.
- 17.Buena-Atienza E, Ruther K, Baumann B, Bergholz R, Birch D, De Baere E, Dollfus H, Greally MT, Gustavsson P, Hamel CP, Heckenlively JR et al. : De novo intrachromosomal gene conversion from opn1mw to opn1lw in the male germline results in blue cone monochromacy. Scientific reports (2016) 6(28253. [DOI] [PMC free article] [PubMed] [Google Scholar]; * The authors identified de novo gene conversion events generating LVAVA and LIAVA exon 3 haplotypes that cause exon skipping.
- 18.Orosz O, Rajta I, Vajas A, Takacs L, Csutak A, Fodor M, Kolozsvari B, Resch M, Senyi K, Lesch B, Szabo V et al. : Myopia and late-onset progressive cone dystrophy associate to lvava/mvava exon 3 interchange haplotypes of opsin genes on chromosome x. Invest Ophthalmol Vis Sci (2017) 58(3):1834–1842. [DOI] [PubMed] [Google Scholar]; * Studies of human subjects demonstrates the LVAVA and MVAVA exon 3 haplotypes cause both myopia and cone dystrophy.
- 19.Patterson EJ, Kalitzeos A, Kasilian M, Gardner JC, Neitz J, Hardcastle AJ, Neitz M, Carroll J, Michaelides M: Residual cone structure in patients with x-linked cone opsin mutations. Invest Ophthalmol Vis Sci (2018) 59(10):4238–4248. [DOI] [PMC free article] [PubMed] [Google Scholar]; ** High resolution adaptive optics and optical coherence tomography imaging of the eyes of living humans with LIAVA and LVAVA cone opsin gene haplotypes demonstrates the effects of these haplotypes on cone structure and viability over time.
- 20.Patterson EJ, Wilk M, Langlo CS, Kasilian M, Ring M, Hufnagel RB, Dubis AM, Tee JJ, Kalitzeos A, Gardner JC, Ahmed ZM et al. : Cone photoreceptor structure in patients with x-linked cone dysfunction and red-green color vision deficiency. Invest Ophthalmol Vis Sci (2016) 57(8):3853–3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carroll J, Dubra A, Gardner JC, Mizrahi-Meissonnier L, Cooper RF, Dubis AM, Nordgren R, Genead M, Connor TB Jr., Stepien KE, Sharon D et al. : The effect of cone opsin mutations on retinal structure and the integrity of the photoreceptor mosaic. Invest Ophthalmol Vis Sci (2012) 53(13):8006–8015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gardner JC, Liew G, Quan YH, Ermetal B, Ueyama H, Davidson AE, Schwarz N, Kanuga N, Chana R, Maher ER, Webster AR et al. : Three different cone opsin gene array mutational mechanisms with genotype-phenotype correlation and functional investigation of cone opsin variants. Human mutation (2014) 35(11):1354–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ueyama H, Muraki-Oda S, Yamade S, Tanabe S, Yamashita T, Shichida Y, Ogita H: Unique haplotype in exon 3 of cone opsin mrna affects splicing of its precursor, leading to congenital color vision defect. Biochem Biophys Res Commun (2012) 424(1):152–157. [DOI] [PubMed] [Google Scholar]
- 24.Fairbrother WG, Yeh RF, Sharp PA, Burge CB: Predictive identification of exonic splicing enhancers in human genes. Science (2002) 297(5583):1007–1013. [DOI] [PubMed] [Google Scholar]
- 25.Desmet FO, Hamroun D, Lalande M, Collod-Beroud G, Claustres M, Beroud C: Human splicing finder: An online bioinformatics tool to predict splicing signals. Nucleic Acids Res (2009) 37(9):e67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Neitz J, Neitz M, He JC, Shevell SK: Trichromatic color vision with only two spectrally distinct photopigments. Nature Neuroscience (1999) 2(884–888. [DOI] [PubMed] [Google Scholar]