Abstract
Long interspersed element-1 (LINE-1 or L1) is the only autonomous retrotransposon active in human cells. Different host factors have been shown to influence L1 mobility however, systematic analyses of these factors are limited. Here, we developed a high-throughput microscopy-based retrotransposition assay that identified the Double-Stranded Break (DSB) repair and Fanconi Anemia factors active in the S/G2 phase as potent inhibitors and regulators of L1 activity. In particular BRCA1, an E3 ubiquitin ligase with a key role in several DNA repair pathways, directly affects L1 retrotransposition frequency and structure and also plays a distinct role in controlling L1 ORF2 protein translation through L1 mRNA binding. These results suggest the existence of a “battleground” at the DNA replication fork between HR factors and L1 retrotransposons, and revealing a potential role for L1 in the genotypic evolution of tumors characterized by BRCA1 and HR repair deficiencies.
Keywords: BRCA1, Homologous Recombination (HR), LINE-1 (L1), retrotransposon, siRNA screen
Introduction
Retrotransposons replicate through RNA intermediates that are reverse transcribed and inserted at new genomic loci. LINE-1 (Long Interspersed element 1 or L1), the only active, autonomous, non-LTR transposable element in human cells, constitutes ~21% of the human genome and has had a substantial impact on the evolution of mammals1–3. The canonical, full-length L1 element is ~6 kb long and consists of a 5′ UTR containing an internal promoter, two open reading frames (ORF1 and ORF2) and a 3′ UTR. ORF1p encodes a RNA-binding protein and exhibits chaperone activity4; ORF2p encodes a protein with endonuclease and reverse transcriptase activities5,6. The mobility of the L1 retrotransposon is dependent on its transcription and the subsequent translation of its encoded proteins, therefore both cytoplasmic and nuclear events are critical for L1 retrotransposition. Interestingly, translation of the second open reading frame of L1 which encodes ORF2p, is highly regulated in the cytoplasm of human cells7. An understudied and largely unknown unconventional termination/re-initiation mechanism has been proposed to explain the regulation of ORF2p translation8.
L1 ribonucleoprotein (RNP) complexes are able to enter the nucleus mainly during mitosis7 and, based on timing experiments, the subsequent steps in retrotransposition are thought to occur mainly during S phase. The nuclear L1 mRNA is reverse transcribed and inserted in a new genomic locus through a partially understood molecular mechanism known as target-primed reverse transcription (TPRT)6,9–13. Several proteins have been shown to inhibit L1 retrotransposition to protect the host cell from potentially genotoxic effects of L1 mobilization14. Several DNA repair factors have been linked to L1 retrotransposition15–21 but the relationship between DNA repair and L1 retrotranspositionremains uncertain and a molecular model that mechanistically explains roles of damage repair factors on retrotransposition is still lacking.
In order to gain a more systematic understanding of the cellular regulators of L1 retrotransposition, we undertook an arrayed, genome-wide siRNA knockdown screen utilizing a high-throughput microscopy-based readout. Our screen reveals that HR factors, and in particular the BRCA1 protein, have strong inhibitory effects on L1 retrotransposition. Our results suggest that BRCA1-mediated HR repair competes with TPRT at pre-existing or L1-created DNA breaks, and that resected DNA ends and protected replication forks represent potential structures incompatible with completion of TPRT. In the cytoplasm, BRCA1 suppresses ORF2 translation through association with its mRNA.
These findings pave the way to a better understanding of L1 regulation in normal human cells as well as in cancers often characterized by deficiency of DNA repair factors such as BRCA1 and Fanconi anemia proteins.
Results
Whole-genome siRNA screen using image-based retrotransposition assays
To study cellular factors that regulate L1 retrotransposition, we developed and optimized a high-throughput microscopy and siRNA-based retrotransposition assay in HeLa M2 cells22 expressing a well-established GFP-AI reporter23,24 (Figure 1A–B). DAPI and GFP signals were quantified algorithmically using optimized parameters (Supplementary Figure 1E and Methods). We ensured that L1 retrotransposition did not have a toxic effect on cells for the duration of our assay (Supplementary Figure 1B) and siRNAs that displayed excessive cell death were discarded from the analysis (see Methods and Supplementary Notes).
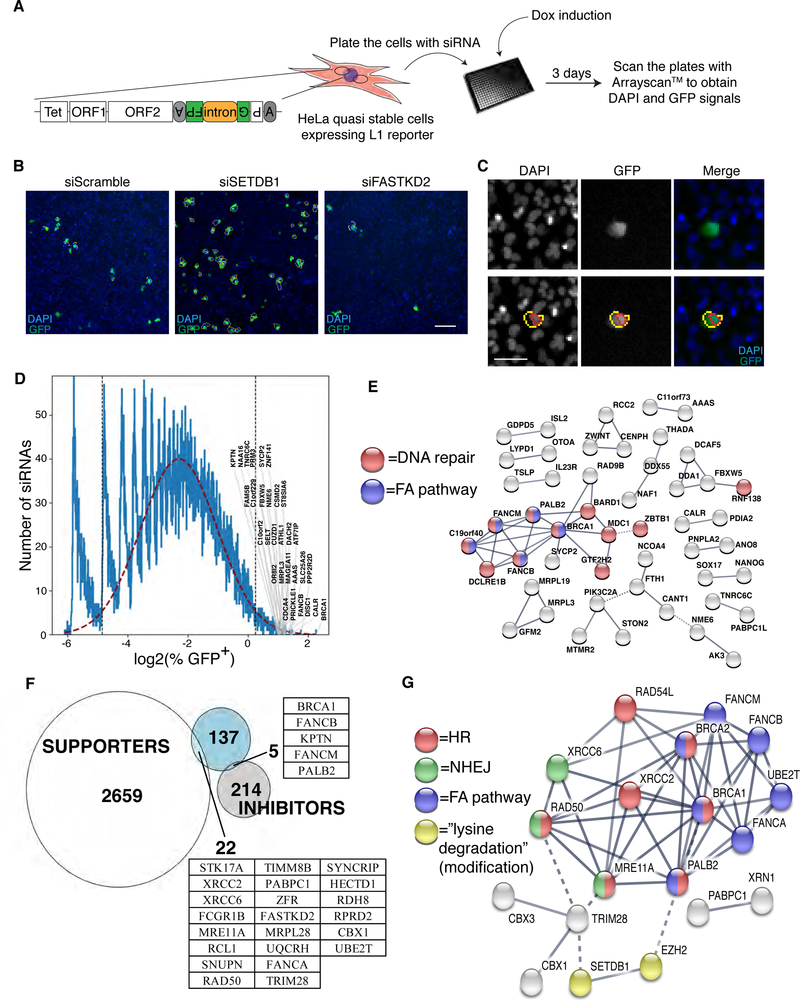
Figure 1. A whole-genome siRNA knockdown screen for L1 regulators.
A. A schematic the screen workflow. B. Examples of images obtained with Arrayscan™. siScramble knockdown represents the baseline L1 retrotransposition rate. Positive (e.g. siSETDB1) and negative (e.g. siFASTKD2) controls are shown as examples Scale bar: 100μm. C. Images were quantified using the Arrayscan™ built-in software. DAPI was used to detect and identify each nucleus (red circle around nuclei); GFP signal (yellow circle around cell) was subsequently measured for each cell. (larger images are presented in Supplementary Figure 1E). Scale bar: 50μm D. Histogram of the percentage of GFP+ cells in log2 scale, for the whole screen. The red dashed line represents a Gaussian curve fitted to the histogram. The vertical dotted lines delimit the 95% of the fitted Gaussian curve. Representative genes are specified for siRNAs that induced an increase of % GFP+ (inhibitor of L1 retrotransposition). E. Cluster analysis (STRING, high fidelity) of the L1 inhibitors obtained from the identification of outliers in panel D. High fidelity siRNAs targets are shown connected by lines, with line thickness proportional to the confidence of the interaction. Unconnected siRNA targets are not shown. siRNA targets involved in the FA or DNA repair pathways are colored blue or red, respectively. F. Proportional Venn diagram of the genes identified in our screen (white and gray; Gaussian curve analysis) and in 21 (blue). The genes in common between the two screens are specified in the tables. G. Cluster analysis of the genes affecting L1 retrotransposition identified in both 21 and our screen (from both the Gaussian curve and “hard threshold” analyses; Supplementary Table 1). Nodes are colored according to their KEGG pathway and connected by lines of thickness proportional to the confidence of the interaction.
Analysis using a “hard threshold” based on the robust Z-score of the percentage of GFP+ cells (Z ≤ –0.8 for supporters and Z ≥ 3 for inhibitors), identified 790 inhibitors,such as SETDB1 (Figure 1B), that when depleted, increase L1 retrotransposition, and 1133 supporters,such as FASTKD2 (Figure 1B), that when depleted decrease L1 retrotransposition (Supplementary Table 1). GO term analysis of the inhibitors showed significant enrichment of genes involved in RNA binding, cell cycle and DNA repair (Supplementary Figure 2A); whereas supporters significantly clustered in GO classes such as mediator complex, THO complex, helicase and lysosome (Supplementary Figure 2B). We also analyzed the data by fitting a Gaussian curve to the distribution of %GFP+ cells (Figure 1D) and identifying “outliers” from 95% of the curve. This analysis (Figure 1D) identified 220 “inhibitors” and 2681 “supporters” of L1 retrotransposition (Supplementary Table 1). Cluster analysis of L1 inhibitors (STRING25) clearly identified “Fanconi anemia pathway” (KEGG, hsa03460) and “DNA repair” (UniProt keyword enrichment, KW-0234) as the two most highly enriched clusters of proteins, with false discovery rates (FDRs) of 0.0242 and 0.0093 respectively (Figure 1E). We found many enriched clusters and GO classes among the “supporters” (Supplementary Table 2 and Supplementary Figure 2B).
We also compared our screen to a previously published21 whole genome CRISPR screen that identified 164 regulators of L1 retrotransposition using HeLa and K-562 cells (111 regulators in HeLa cells and 142 regulators in K562; 89 in common). The values obtained for L1 retrotransposition (“combo CaSTLE score” for Liu et al.21 and “% GFP+” for our screen) were mostly uncorrelated (Figure 1F and Supplementary Figure 1F). The few genes overlapping between the two screens cluster into distinct KEGG pathways: homologous recombination (HR) (FDR=5.66e-11), Fanconi anemia pathway (FDR=1.33e-10), Non-Homologous end-joining (FDR=2.97e-05), “Lysine degradation” (meaning lysyl side chain modification; FDR=0.0411) (Figure 1G). This analysis clearly confirmed that several genes, including BRCA1, involved in DNA damage repair and more specifically in DSB repair, strongly inhibited L1 retrotransposition (Figure 1D–G, Figure 2D, and Supplementary Figure 2A). We therefore validated and investigated the relevance of these genes on L1 activity.
Figure 2. Secondary validations of the DNA repair factors.
A. Exon-junction qPCR analysis (green bars) and Arrayscan analysis (red bars) (n=2 independent samples, error= s.d.) B. BRCA1 was knocked out in HCT116 cells. A BRCA1 expressing vector was transfected and retrotransposition (% GFP positive cells) quantified. Recoded (ORFeus) or non-recoded (L1rp) L1 GFP-AI reporters were used. Expressed plasmids are indicated on the X axis, blue bars indicate wildtype HCT116 cells, red bars, BRCA1 deficient HCT116 cells. (n=2 independent samples, error= s.d.) C. Factors from the three major DSB repair pathways (HR, NHEJ, MMEJ) are grouped and their effect on L1 retrotransposition shown in a bar plot. The red bars indicate HR core factors (n=2 independent samples, error= s.d.). D. Canonical FA factors and FA-like factors plotted for their effect on L1 retrotransposition. The factors within each group were ordered by their fold change compared to siScramble control. (n=2 independent samples). E. Immunoblot of iPOND inputs and immunoprecipitations (IP) of replication forks from ORFeus or L1rp expressing HeLa-M2 cells. The blotted proteins are indicated at left of each panel. Cell treatments: #1= BrdU for 10 minutes prior to fixation and CLICK reaction; #2= BrdU for 10 minutes prior to fixation but no CLICK reaction; 3#= BrdU for 10 minutes, prior to washing and incubation for 1h in media with thymidine 10μM before fixation and CLICK reaction. If two panels are presented they represent different “exposures” of the membrane. Quantification of ORF1p bands after iPOND are reported. Right panel: immunoblot of the indicated proteins using a prefixation aliquot of cells used for iPOND and expressing ORFeus or L1rp. Uncropped blot images are presented in Supplementary Data 1.
Secondary validations of DNA repair genes
The enrichment of DNA damage proteins among the genes with the strongest effect on L1 retrotransposition in our screen (Figure 1E) led us to focus on the DNA damage repair cluster also identified by Liu et al.21 (Figure 1G) and by Ardeljan et al. (co-submitted) who paradoxically identified this same cluster of genes as synthetic lethal in p53– L1-expressing cells. In order to understand which specific DSB repair pathways restrict L1 retrotransposition, we curated a list of 176 genes (Supplementary Table 3) with the following GO term annotations: Fanconi Anemia (FA), Homologous recombination (HR), non-homologous end joining (NHEJ) and micro-homology mediated end joining (MMEJ). We performed siRNA knockdown of these 176 selected genes in a 96 well format (Supplementary Figure 3A, Supplementary Table 3) that enabled us to perform an exon junction RT-qPCR assay to detect intron-less GFP (from cells that had undergone retrotransposition) as an additional validation of retrotransposition at the genomic level (Supplementary Figure 3A–B). We observed a good correlation between the results obtained in the 96 well setting versus the results in our whole genome screen (Pearson’s r = 0.57) with, as expected, a significant decrease in noise in the 96 well format (Supplementary Figure 2C–D). Our GFP-AI exon-junction qPCR measurements are in agreement with the results obtained in the screen and in our 96 well validation (Supplementary Figure 3C, correlation coefficient=0.48). Considering both measurements (96-well secondary screen and exon junction qPCR), we set a threshold of more than 1.8 (log2 fold change of the percentage of GFP+ cells) for inhibitors and less than −1 (log2 fold change of the percentage of GFP+ cells) for supporters (Supplementary Figure 3C, horizontal lines). 42 genes behaved as strong inhibitors of L1 retrotransposition (Figure 2A) and 32 genes behaved as strong supporters (Supplementary Table 3).
Among the inhibitors, BRCA1 showed the highest inhibition of L1 retrotransposition. Corroborating a potentially important role of BRCA1 and the HR repair pathway in L1 retrotransposition, we also identified members of the FA/BRCA1 pathway as strong L1 retrotransposition inhibitors (Figure 2D). Depletion of BRCA1, as part of the ATM pathway, was previously noted to increase LRE3 retrotransposon activity in HCT116 colorectal carcinoma cells16. Overexpression of BRCA1 restored L1 retrotransposition frequency to wild-type levels in BRCA1 knockout cells (Figure 2B). Analysis of genes involved in HR, NHEJ and MMEJ revealed that both HR core proteins26,27 (Figure 2C, red bars) and MMEJ had a bias towards factors that inhibit L1 retrotransposition whereas NHEJ factors showed a more ambiguous behavior (Figure 2C).
Overall these data show that the DNA damage response mediated by BRCA1 and other HR and FA factors strongly inhibits L1 retrotransposition, pointing to replication forks and sites of replication stress as preferred sites for L1 activity and potentially for its inhibition.
Fanconi Anemia factors limit L1 retrotransposition during replication
Our whole-genome screen identified a cluster of L1 inhibitors within the FA pathway28 that recognize and repair lesions at stalled replication forks. Increased L1 activity has also been reported in cells depleted of SLX4/FANCP and FANCD2, triggering an interferon response20. Interestingly, most FA factors behave as inhibitors of L1 activity excepting FANCJ/BRIP1, which acts as a supporter (Figure 2D). Measurements of retrotransposition using a luciferase-AI reporter, also showed that FANCJ behaves differently than other FANC proteins. FANCM, FANCB and FAAP24 (FA Core Complex Associated Protein 24) depletion together with BRCA1 knockdown showed a synergistic effect, suggesting possible parallel pathways of L1 inhibition for BRCA1 and the Fanconi complex (see section below).
To show that L1 proteins are recruited to DNA replication forks as suggested by our previous work7 and corroborated by functional interactions with FA and HR factors reported here, we performed iPOND (isolation of proteins on nascent DNA)29 coupled to immunoblotting using HeLa-M2 cells expressing recoded (ORFeus) and non-recoded (L1rp) L1. iPOND analysis showed that ORF1p encoded by ORFeus or L1rp was enriched on DNA replication forks (Figure 2E). Not surprisingly, when considering the much lower abundance of ORF2p compared to ORF1p and the sensitivity of immunoblot analysis, we could not detect ORF2p. Moreover, ORF2p is a relatively large protein (160KDa) and thus less amenable to analysis after fixation and reverse crosslinking, steps required to perform iPOND (note absence of ORF2p in control fixed and reverse crosslinked iPOND INPUT samples compared to the same cells prior to fixation). Similar results were obtained using a reverse transcriptase (RT) mutant recoded L1 (ORF2p D702Y) (Supplementary Figure 3D).
Overall these data support a model in which replication forks are exploited by L1 as a retrotransposition target. This model is supported by the involvement of FA factors in L1 retrotransposition and by our previously published data7 demonstrating that L1 retrotransposition mainly occurs during S phase, when HR is active, and that L1 proteins interact with stalled replication forks on which HR and FA factors are localized to repair DNA.
Evidence that LINE-1 both creates and uses stalled replication forks to retrotranspose
Because of the central role of FA factors in protecting replication forks arrested by DNA damage sites30 and sites of “replication stress”28,31, we focused on elucidating the role of L1 retrotransposition in potentially exploiting or creating stalled replication forks e.g. upon endonuclease cutting (Figure 3F). We designed experiments to distinguish between these two non-mutually exclusive possibilities: (i) measuring retrotransposition after treatment with aphidicolin (APH), a DNA polymerase inhibitor that creates stalled replication forks and (ii) evaluating the impact of L1 expression on the cell cycle. Due to the strong inhibitory effect exerted by BRCA1 on L1 retrotransposition and the known role of BRCA1 on cell cycle, we also analyzed the cell cycle distribution of cells depleted of BRCA1 and expressing L1.
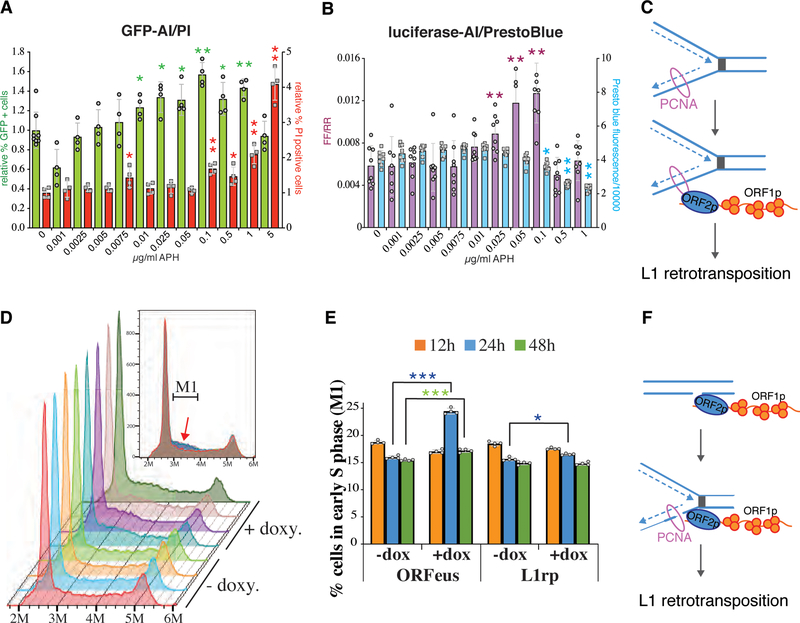
Figure 3. Evidence that L1 uses and induces stalled replication forks.
A. L1 retrotransposition (green bars) measured with a GFP-AI reporter in HeLa-M2 cells treated with the indicated concentrations of Aphidicolin. Cell death was internally measured using propidium iodide (PI) uptake. Signal was compensated for GFP overlap (red bars) (n=4 independent samples, error= s.d., two tailed distribution homoscedastic T-test, *p≤0.05, **≤0.01). B. Luciferase-AI measurements of L1 retrotransposition normalized by Renilla activity (FF/RR, purple bars) in HeLa-M2 cells treated with the indicated amount of APH. Cell viability was measured using PrestoBlue (blue bars) performed before FF and RR measurements (n=4 independent samples, error= s.d., two tailed distribution homoscedastic T-test, *p≤0.05, **≤0.01). C. Scheme of the mode of L1 recruitment on replication fork supported by data presented in A and B (L1 uses the replication fork). D. Cell cycle distribution of HeLa-M2 cells transfected with a pCEP-puro plasmid for doxycycline inducible expression of L1 ORFeus. Three independent measurements were conducted for cells treated (+doxy.) or untreated (−doxy.) with 1μg/ml doxycycline for 24h. The inset shows an overlay of all the profiles with the M1 gate used for quantification of cells in early S phase presented in E. The red arrow indicates the “hump” of cells accumulating in early S phase in cells expressing L1 (+doxy.). E. Percentage of cells in early S phase (M1 gate shown in D) for HeLa-M2 treated or not treated with 1μg/ml doxycycline for the indicated times to induce expression of recoded L1 (ORFeus) or non-recoded L1 (L1rp) (n=4 independent samples, error= s.d., two tailed distribution homoscedastic T-test, *p≤0.05, **p≤0.01) F. Scheme of the modes of L1 recruitment to replication forks supported by data presented in D and E (L1 induces stall in the replication fork by nicking the DNA).
We measured L1 retrotransposition efficiency in cells treated with different concentrations of APH, expecting no increase in retrotransposition if L1 interacts with the replication fork only when the fork stalls or collapses because of collision with a DNA nick created by retrotransposing L1 (Figure 3F). We measured retrotransposition with both GFP-AI and luciferase-AI (luc-AI) reporters with internal controls for cell death and cell viability using propidium iodide staining (PI) for GFP-AI measurements and PrestoBlue analysis for luc-AI measurements. We observed that L1 insertions increased as cells accumulate in early S phase induced by the APH treatments (Figure 3A, B and Supplementary Figure 4A,B). As a low concentration of APH slows and stalls replication fork progression, while treatment with more APH induces cell death (Figure 3A and B), this result points to a possible recruitment of L1 to stalledreplication forks, where retrotransposition may occur at high frequency (see Discussion) (Figure 3C).
We also measured the distribution across the cell cycle stages of cells expressing ORFeus or L1rp. Overexpression of L1, especially using the codon optimized and high expressing ORFeus, causes a clear accumulation of cells in early S phase, reminiscent of APH treatment (Figure 3D–E and Supplementary Figure 5A 24h). This effect is transient (Figure 3E), presumably because prolonged stalling in S phase caused by high expression of L1 triggers apoptosis and cell death similar to what is observed after treatment with high concentrations of APH (Figure 3A and B). As expected, cell toxicity induced by L1 expression is rescued, at least partially, by an inactivating mutation in the endonuclease domain of ORF2p (EN-) (Supplementary Figure 5B). This cell toxicity also explains the absence of early S phase accumulation by cells depleted of BRCA1 (Supplementary Figure 5A). These observations suggest that L1 is not only able to use stalled replication forks as a target but can also create them via ORF2p endonuclease mediated nicking of DNA (see Supplementary Note).
BRCA1 depletion increases endonuclease-dependent and -independent L1 retrotransposition
The highest increase of L1 retrotransposition in response to gene knockdown was observed upon BRCA1 depletion (Figure 1D and Figure 2A,C,D). We confirmed the repressive function of BRCA1 on L1 retrotransposition using the p53 mutant and BRCA1-null ovarian carcinoma cell line UWB1.289 and its isogenic cell line with re-established expression of wild type BRCA1 (UWB1.289+BRCA1)32. We used an “all-in-one” plasmid expressing rtTA and recoded L1 retrotransposition reporter based on firefly (ORFeus-luc-AI) that expresses renilla luciferase from a bidirectional doxycycline responsive CMV promoter that also drives the ORFeus-luc-AI reporter (Figure 4A). The use of this reporter in retrotransposition assays clearly showed that, in the BRCA1+ cell line (shaded red), L1 retrotransposition was strongly inhibited compared to retrotransposition observed in the BRCA1 mutant cell line (Figure 4B).
Figure 4. Re-expression of wild type BRCA1 represses L1 retrotransposition in UWB1.289 ovarian carcinoma cells.
A. Scheme of the “all-in-one” Renilla-ORFeus-luciferase-AI retrotransposition reporter constitutively expressing the rtTA trans-activator. B. L1 retrotransposition measurements as ratio of Firefly (FF) over Renilla (RR) signals from UWB1.289 BRCA1 null cells (UWB) and UWB1.289 cells stably expressing a functional BRCA1 (UWB+BRCA1) transfected with the plasmid depicted in A or with a plasmid expressing RR alone. Cell were treated with 0.125 or 0.250μg/ml doxycycline for 48h before measurement of FF and RR (n=16 independent samples, two tailed distribution homoscedastic T-test, ***p≤0.005, ****p≤0.001). The median, interquartile range and 95% confidence intervals are represented within the density plots (violin plots)
It was previously shown that L1 is able to retrotranspose at pre-existing DNA damage sites and that these events are independent of L1 endonuclease (EN) activity18,33. Because BRCA1 depletion impairs DNA damage repair function and therefore increases the number of unresolved DNA damage sites34,35, we measured the EN-independent activity of L1 in BRCA1 depleted cells. Due to the rarity of endonuclease-independent retrotransposition events, we utilized a retrotransposition assay based on drug selection (Blasticidin resistance selected over 10 days) instead of GFP fluorescence detection, to quantify L1 insertions. We also used the more sensitive luciferase-AI reporter, growing cells expressing the reporter for 5 days in 3 cm wells (instead of 2 days in 96 well plates as for Figure 3B, Figure 5F and Figure 6A). As expected, BRCA1 depletion, as well as depletion of a centrally important FANC protein (FANCD2), induced an increase of L1 EN-dependent retrotransposition (Figure 5A). Knockdown of BRCA1, BRCA2 and FANCD2 also increased EN-independent retrotransposition (Figure 5B), demonstrating a role of the HR/FANC pathway on both EN-dependent and -independent retrotransposition. These results were recapitulated using the luciferase-AI reporter (Figure 5C). Using the reverse transcriptase (RT) inhibitor lamivudine/3TC, we confirmed that the EN dependent and independent retrotransposition observed in HeLa-M2 cells depends on RT activity (Supplementary Figure 6A).
Figure 5. HR/FA pathways repress L1 EN-dependent and independent retrotransposition through BRCA1-mediated DNA resection.
A. Retrotransposition assays using a L1 wild type -BSD-AI reporter. Clones of HeLa cells after Blasticidin selection are stained with crystal violet. The number of cells plated per well and the transfected siRNA are indicated. B. Same experiment shown in A but using an endonuclease dead (EN-) L1. Schematic representation of each of the L1-BSD-AI reporter plasmids used are shown at the bottom of each corresponding panel. C. L1 Retrotransposition measurements using a Firefly-AI reporter also expressing Renilla for normalization (Figure 4A, without the rtTA). HeLa-M2 cells, expressing wild type or H230A lacking endonuclease activity (L1 EN-) L1 luciferase-AI reporters, were plated in 3 cm wells and induced with doxycycline for 5 days. Retrotransposition activity is reported as the ratio (FF/RR)*1000 (n=4 independent samples, error= s.d., two tailed distribution homoscedastic T-test, *p≤0.05, **p≤0.01, ***p≤0.001, blue*=significantly decreased compared to siCtrl, red*=significantly increased compared to siCtrl). (qPCR measurements are presented in Supplementary Figure 6B). D. BRCA1 and CtIP knockdowns promote, while 53BP1 knockdown inhibits resection at a DNA break. Resection can still occur when BRCA1 and its antagonist 53BP1 are both absent. E. Single and double knockdown of the indicated proteins in HeLa-M2 cells plated in 6-well plates. Fold change of L1 retrotransposition was calculated by normalizing %GFP upon knockdown to %GFP of scrambled siRNA control treated cells (n=2 independent samples, error= s.d.). F. Retrotransposition measurements upon knockdown of the same factors depleted in E. but using a pCEP-puro ORFeus-luciferase-AI and Renilla reporter episomally maintained in HeLa-M2 cells plated in 96 well plates. (n=8 independent samples, error= s.d., two tailed distribution homoscedastic T-test, ***p≤0.001, blue*=significantly decreased compared to siCtrl, red*=significantly increased compared to siCtrl).
Figure 6. Mechanism of BRCA1 mediated repression of L1 retrotransposition.
A. Left panel: Epistasis interaction ratios for factors involved in fork protection (shaded green) or fork reversal/degradation (shaded blue). ZRANB3 and DNA2 bars are colored in green stripes as they may play also a role in fork protection though they are normally classified as promoters of fork degradation. Epistasis ratio=[(FF/RR)value for siBRCA1+siFactorX] / [(FF/RR)value for siFactorX]. The two controls are represented by: (i) the ratio of (FF/RR) after siCtrl+siBRCA1 treatment, divided by the (FF/RR) after siCtrl treatment (siCtrl+siCtrl) (gray bar; full L1 de-repression mediated by BRCA1); (ii) the ratio of (FF/RR) after siBRCA1+siBRCA1 treatment divided by (FF/RR) after siCtrl+siBRCA1 treatment (dark green bar; full epistasis interaction). Right panel: retrotransposition measured as FF/RR values after treatment with the indicated siRNAs . (n=8 independent samples, error= s.d., two tailed distribution homoscedastic T-test, *p≤0.05, **p≤0.01, ***p≤0.001, blue*=significantly decreased compared to siCtrl or to siBRCA1, red*=significantly increased compared to siCtrl or to siBRCA1) B. Model of the modes of inhibition of L1 retrotransposition by mechanisms of fork protection and fork reversal. C. Diagram depicting the procedure used to recover and analyze de novo L1 insertions. D. Pie charts showing the percentages of the different L1 insertion types detected in the control (scrambled siRNA) and BRCA1 deficient HeLa cells (siBRCA1). E. Proposed model for target site deletions in BRCA1 deficient cells. Genomic DNA is shown in blue; L1 mRNA is shown as dashed lines; L1 cDNA is shown in orange. Sequences between the two nicks induced by L1 are shown in red for target site duplications (TSD) and purple for target site deletions.
Surprisingly, and in contrast with measurements based on the GFP-AI reporter, we observed that knockdown of BRCA2 induced a paradoxical decrease in L1 EN-dependent retrotransposition (Figure 5A and 5C) while it increased EN-independent L1 activity (Figure 5B and 5C). Moreover, the BSD-AI reporter revealed a smaller effect of BRCA1 knockdown on L1 retrotransposition in this longer-term assay compared to the three-day readout employed with GFP-AI (Figure 2A–D and 1D). These observations suggest that elevated L1 retrotransposition is detrimental to long-term growth of cells deficient in HR/FA pathways as also reported by Ardeljan et al. (see Discussion).
BRCA1-dependent HR pathway restricts L1 retrotransposition through resection
One of the common features of HR and MMEJ is thatboth require resected DNA end structures. Because of the crucial role of BRCA1 in this process, we hypothesized that resection may act as a physical barrier to L1 retrotransposition. GFP-AI (assay performed in 6 well plates for 3 days; Figure 5E) and luciferase-AI (assay performed in 96 well plates for 2 days; Figure 5F) reporters confirmed that CtIP, a known promoter of DNA resection36–38 like BRCA1, is a retrotransposition inhibitor while 53BP1, which inhibits end resection39–42, is a supporter. Moreover, the increased level of retrotransposition measured upon BRCA1 depletion returned to basal levels upon rescuing resection by knocking down both 53BP1 and BRCA1 (Figure 5E–F). Similar effects on retrotransposition were observed using L1rp (the native L1 sequence) instead of recoded L1 (ORFeus) (Supplementary Figure 7). This evidence strongly points to a pivotal role of DNA end resection mediated by CtIP, synergized by BRCA138 and inhibited by 53BP1, in the repression of L1 retrotransposition.
Fork protection inhibits L1 retrotransposition while fork reversal permits L1 activity
We showed that BRCA1 as well as key factors in strand coating and resection inhibit retrotransposition and other data supporting the importance of stalled replication forks in retrotransposition. We therefore wondered whether fork protection or fork reversal and degradation play roles in regulating LINE-1 retrotransposition. Toexplore functional interactions between L1 and fork protection, reversal and degradation we conducted a series of single and double siRNA knockdowns to identify epistasis interactions with BRCA1 mediated L1 inhibition using the luc-AI reporter. We depleted these factors by co-transfecting cells with siRNA against a target of interest together with siRNA control (siCtrl), or together with siRNA against BRCA1 and measured retrotransposition after 48hrs of ORFeus-luc-AI expression (Figure 6A and Supplementary Figure 8). As in the case of 53BP1, we sought factors that, in the context of BRCA1 depletion (L1 de-repression), could rescue retrotransposition to control levels (“alleviating” epistasis interactions43) (Figure 5E–F). We argue that such factors play key roles in inhibiting L1 through a BRCA1-dependent mechanism. To identify factors necessary for BRCA1 mediated inhibition of L1, we analyzed epistasis interactions of BRCA1 with known fork protection and fork degradation factors (epistasis ratio in Figure 6A calculated from values presented in Supplementary Figure 8, see Methods). A ratio equal to 1 indicates complete abrogation of BRCA1-mediated increase of LINE-1 retrotransposition (Figure 6A, second bar in green), thus values of 1 or close to 1, indicate a critical role for the considered factor in the BRCA1-mediated suppression of LINE-1 (see Methods).
This analysis clearly showed that most factors involved in fork protection are needed for BRCA1-mediated repression. BRCA2 knockdown, a key factor in fork protection and Rad51 recruitment, almost completely restored L1 retrotransposition to control levels in cells lacking BRCA1, an outcome that we call “alleviation” of BRCA1-mediated L1 inhibition (in Figure 6A an epistasis ratio almost as low as 1; in Supplementary Figure 8, compare first bar (siCtrl+siCtrl) to 14th bar (siBRCA1+siBRCA1) and 15th bar (siBRCA1+siBRCA2)). This observation indicates that BRCA2 is a key player in inhibition of L1 mediated by BRCA1. In line with this evidence, the single stranded DNA (ssDNA)-coating proteins RPA1 and Rad51 similarly play important roles in repressing L1. Interestingly, between the two proteins known to repress exonuclease activity, only deletion of BOD1L, that promotes Rad51 loading44, and not depletion of ABRO1, independent from Rad5145, is able to significantly alleviate BRCA1-mediated L1 inhibition. This confirms that RPA1 and subsequent Rad51 coating of DNA are both essential for L1 inhibition by BRCA1. Surprisingly, DNA2 and ZRANB3 behave like fork protection factors despite their canonical classification as fork reversal/degradation factors (Figure 6A). A mechanism that protects the replication fork through CtIP cutting of the reversed fork and partial DNA degradation by DNA2 has been proposed38 and may explain our result. If DNA2 with CtIP plays a BRCA1-dependent role in fork protection, depletion of BRCA1 and CtIP together with DNA2 should reverse the alleviation of the BRCA1-mediated effect observed upon DNA2 knockdown (Figure 6A right panels). We indeed found that, in the absence of BRCA1, CtIP and DNA2 (siBRCA1+siCtIP+siDNA2), retrotransposition is identical to that observed for cells treated only with siBRCA1 (Figure 6A right panel). These data are in line with the model proposed by Przetocka et al.38 and suggest that L1 is able to retrotranspose at reversed replication forks left “deprotected” in the absence of BRCA1 and DNA2 (model Figure 6B).
The ratio reported in Figure 6A for ZRANB3 is explained by the fact that depletion of ZRANB3 itself induces an increase in L1 retrotransposition and depletion of both BRCA1 and ZRANB3 has the same effect as knocking down only BRCA1 (Supplementary Figure 8C). This intriguing result suggests a complex functional interaction between the annealing helicase and fork remodeler46 ZRANB3, L1 and BRCA1 (see Discussion).
BRCA1 depletion induces formation of target site deletions upon L1 retrotransposition
To understand the molecular mechanism of BRCA1-mediated inhibition on L1 TPRT specifically, we sequenced de novo L1 insertions and their flanking regions in cells depleted of BRCA1 using a modified recovery assay18 (Figure 6C). We evaluated the sequence at which the first nick occurred, the length of the inserted polyA, the length of the L1 insertions and the type of L1 insertions (inversion at 5’ end versus canonical) (Supplementary Table 4). The majority of L1 insertions were characterized by a L1 A/T rich target site (TTTTAA) demonstrating that most L1 insertions were mediated by ORF2 endonuclease activity. No significant differences between control cells and cells depleted of BRCA1 were observed in the length of inserted L1 or polyA (Supplementary Figure 9B–C) and L1 insertions were not enriched at common fragile sites (CFSs)47,48 as defined by 47(Supplementary Figure 9A). We observed a striking increase in the formation of target site deletions relative to target site duplications upon BRCA1 knockdown (Figure 6D). During canonical L1 TPRT, the second nick is usually “downstream” of the first nick, whereas for the less common target site deletions, the second DNA nick is inferred to occur upstream of the first nick (Figure 6E)13,49,50. The reason for the biased preference for “downstream” second nicking during L1 TPRT is not known, but BRCA1 seems to be a strong determinant of this bias.
L1 insertions were also sequenced from cells depleted of FANCM (Figure 2D, 6A, S8 and Supplementary Table 3). No notable differences with control cells were found, further corroborating that FA factors inhibit L1 through a different mechanism from that used by BRCA1 (see below and Supplementary Table 4).
BRCA1 inhibits ORF2 translation
During our study of L1 in BRCA1-depleted cells, we were surprised to observe an additional distinct layer of regulation of L1 induced by BRCA1. Specifically,BRCA1 depletion induced anincrease in L1 ORF2p for both ORFeus and L1rp (non-recoded L1) while ORF1p levels were not affected (Figure 7A and Supplementary Figure 10A, B). We excluded an effect of BRCA1 on ORF2p degradation by comparing the stability of ORF2 protein over time (Supplementary Figure 10B–C). A small increase in L1 mRNA could also be measured upon depletion of BRCA1 (Supplementary Figure 10D) but because ORF1p and ORF2p are translated from a bicistronic mRNA, we hypothesized an effect of BRCA1 depletion on ORF2p translation. Immunoblot analyses of cells depleted of BRCA1, 53BP1 and FANCM proteins revealed that ORF2p levels increased specifically in BRCA1-depleted cells while ORF1p remained unaltered, suggesting that this translational effect was independent of the HR repair pathway (Figure 7A). With a similar approach used in Figure 2B, we rescued the increase of ORF2p due to CRISPR-Cas9 knockout of BRCA1, overexpressing a gRNA-resistant BRCA1 (Figure 7B). Immunofluorescence staining showed a clear increase in ORF2p signal upon knocking down BRCA1 compared to control siRNA treated cells (Figure 7C). BRCA1 expression changes during the cell cycle decreasing in G1 and increasing in S, G2 and M phase (cyclebase 3.0)51 as we confirmed using sorted HeLa S.FUCCI cells (Figure 7D). In line with the inhibitory effect of BRCA1 on ORF2p translation (Figure 7A–B), we observed that cells expressing low levels of BRCA1 protein (most likely in G1 phase) were more likely to express ORF2p, whereas cells displaying high levels of BRCA1p did not usually express ORF2p (Figure 7E–F; p-value: 1.7 × 10−9).
Figure 7. BRCA1 inhibits ORF2 translation in the cytoplasm.
A. Immunoblot of ORF1 and ORF2-FLAG proteins from control HeLa cells (siCtrl) or cells treated with the indicated siRNAs and expressing L1 ORFeus. Doxycycline (DOX) is used to induce the expression of L1; tubulin used as loading control. B. Left panel: Immunoblot of HeLa cells expressing recoded L1 and BRCA1 knocked out using the indicated amounts of CRSIPR/Cas9 lentivirus. An empty plasmid (-BRCA1) or a plasmid encoding BRCA1 (+BRCA1) was also transfected and L1 expression induced for 48h Right panel: quantification of ORF2p normalized to tubulin. C. Immunofluorescence of HeLa cells expressing recoded L1 and treated for 24 hrs with control siRNA (siScramble) or siRNA against BRCA1 (siBRCA1). Scale bar: 100μm D. BRCA1 mRNA quantification in different phases of the cell cycle using sorted FUCCI cells. (n=3 independent samples, error= s.d., two tailed distribution homoscedastic T-test). Quantification (E) and imaging (F) of ORF2p in cells expressing L1 and displaying low or high BRCA1 (n=3 independent samples, error= s.d., two tailed distribution homoscedastic T-test) F. L1 with halo-tagged ORF2p detected with JF657 dye (green). BRCA1 protein (red) detected with antibody D-9. Red arrows indicate cells with low BRCA1; blue arrows indicate cells with higher BRCA1 expression. Scale bar: 50μm G. L1 mRNA bound to BRCA1 measured by BRCA1 IP and qPCR compared to a standard curve of known concentrations of L1 DNA. (n=3 independent samples, error= s.d.) H. left panel: Cell cycle stages displayed by HeLa S.FUCCI cells. Right panel: Stacked bar plots showing the percentages of cells starting to express ORF2p in different cell cycle stages for control (CTR) or siBRCA1 treated cells. The total number of quantified cells is reported on the side of the bars. Uncropped blot images for panels A and B are shown in Supplementary Data 1.
We hypothesized that BRCA1 may inhibit ORF2 translation through direct binding to L1 mRNA in the cytoplasm as such a function has been previously attributed to BRCA1 for several mRNAs and proteins52. To evaluate a possible physical interaction between BRCA1 and L1 mRNA, we performed immunoprecipitation of BRCA1 protein followed by RT-qPCR of L1 mRNA. We observed a >5 fold enrichment of L1 mRNA binding to BRCA1 compared to an IgG control (Figure 7G). Because BRCA1 expression is regulated during the cell cycle (Figure 7D), we measured the timing of ORF2 protein translation and expression during the cell cycle to interrogate a possible role of BRCA1 in this regulation. Using live cell imaging of HeLa S.FUCCI cells expressing an inducible ORF2-Halo tagged LINE-17, we observed that ORF2 protein starts to be detectable during G1 phase, when BRCA1 transcripts are low (Figure 7H, Supplementary Movie 1). However, cells depleted of BRCA1 showed an increased fraction of cells that express ORF2p in cell cycle stages other than G1 (Figure 7H, Supplementary Movie 2) suggesting that BRCA1 inhibits ORF2p translation mainly in S/G2/M phases when BRCA1 is highly expressed. These data suggest that BRCA1 regulates L1 ORF2p translation through binding to its mRNA, possibly in a cell cycle regulated fashion.
Overall, our results demonstrate several lines of defense deployed by BRCA1 against L1 retrotransposon activity (Figure 8). These lines of regulation of L1 activity may be misregulated in certain BRCA1 deficient cancers, highlighting a possible role of LINE-1 in the progression of tumors such as breast and ovarian cancers.
Figure 8. Model of BRCA1 inhibition of LINE-1 retrotransposition.
Model of mechanisms by which BRCA1 inhibits LINE-1 activity. In the nucleus, L1 interacts (or creates) stalled replication forks but its retrotransposition activity is usually inhibited by fork protection processes such as coating of ssDNA by RPA1. In the presence of BRCA1, double strand breaks created, for example, by collapse of replication forks at sites of nicked DNA, are resected and immediately coated by RPA proteins that limit L1 retrotransposition and bias the second cut of L1 toward DNA downstream of the first nick. This process will therefore create canonical target site duplications (TSDs) flanking new insertions. In cells depleted of BRCA1, resection is impaired and L1 is able to efficiently retrotranspose, cutting DNA downstream or upstream of the first nick and creating target site duplications or target site deletions. Cytoplasmic BRCA1 interacts with L1 mRNA and inhibits ORF2p translation and formation of functional L1 RNPs.
Discussion
HR factors restrict L1 retrotransposition
We have previously demonstrated that L1 retrotransposition is biased towards the S phase and that ORF2p binds replication fork components (PCNA and MCM proteins)12. Here we present a whole-genome siRNA screen to identify L1 retrotransposition factors. Consistent with our prior results, we show that Fanconi anemia proteins involved in replication fork stability, BRCA1 and homology mediated repair pathways, active during the S/G2 phase, functionally compete with L1 retrotransposition. We observed that knockdown of HR components, and to a lesser extent MMEJ factors, enhance L1 retrotransposition frequency, implying that these pathways interfere with retrotransposition. The mechanistic similarities between HR and MMEJ include the following features: 1) both pathways dominate during the S/G2 phase53 when L1 activity peaks7 2) both pathways require resected DNA end structures to perform strand invasion, homology search and subsequent recombination. We thus postulate that DNA end resection complexes act as a physical barrier to L1 retrotransposition by inhibiting TPRT. Also, during TPRT, in BRCA1 knockdown cells, L1 appears to choose between a downstream or upstream DNA second nick without bias. This suggests that BRCA1 and associated proteins may sterically block the formation of the second DNA nick upstream of the first nick during L1 retrotransposition, perhaps by recruiting ssDNA coating proteins RPA1 and Rad51 that render the DNA upstream the first nick inaccessible to the EN domain of ORF2p (model in Figure 8). Alternatively, the DNA ends created by L1 ORF2p endonuclease activity are differentially resected by endonucleases such Mre11 (3’ to 5’), EXO1 or DNA2 (5’ to 3’), and may be responsible for the formation of target site duplications or target site deletions at completion of TPRT.
This observation connects up-regulation of L1 to cancers characterized by mis-regulated HR pathways such as mutated of BRCA1 and/or BRCA2 proteins, known to drive several cancers, including breast and ovarian cancer. It is therefore possible that, in an environment of mis-regulated HR, the expression of L1 and its activity may increase genome instability, accelerating tumor progression or the development of resistant clones upon treatment. Further studies are necessary to evaluate the role of L1 expression on tumor progression in tumors with a compromised HR background and to evaluate the possible use of L1 expression and activity as a biomarker for specific stages of tumor progression or potentially as a therapeutic target.
Synthetic lethality and retrotransposition
Our results and those of21 are consistent in that retrotransposition activity is enhanced by knockdown or knockout of BRCA1 and FA factors (Figure 1E and 1G). However, Ardeljan et al. (co-submitted) paradoxically observe exactly the same sets of genes as synthetic lethal with LINE-1 overexpression. How can we resolve this apparent paradox? We note that our group and Liu et al.21 both used a short-term readout of retrotransposition efficiency based on GFP (3 and 10 days respectively) whereas Ardeljan et al. examined cultures after 27 days of selection. In all three cases, experiments were done in p53 negative cells with retrotransposon overexpression. Knockdown or knockout of these genes may have two effects: a large short term increase in retrotransposition and a long-term synthetic fitness defects. Consistent with this interpretation, BRCA1 and BRCA2 knockdown in a longer-term drug resistance-based selection revealed a more modest enhancement of retrotransposition and, in the case of BRCA2 knockdown, shows an opposite effect on retrotransposition (Figure 5A–B). We also observe that simultaneous BRCA1 depletion and expression of L1 confers a strong fitness defect (Supplementary Figure 5B) suggesting that sustained L1 expression and activity in conditions of impaired DNA damage repair has dire consequences for genome stability. Under normal circumstances these consequences lead to cell toxicity and death but in a tumorigenic context, could trigger clonal selection and tumor progression towards more aggressive and/or therapy-refractory cancers.
L1 ORF2p translation and BRCA1
Translational control of ORF2 remains a completely unknown process, and here we observed an inhibitory effect on ORF2 protein level by BRCA1 in the cytoplasm. This effect is likely RNA sequence independent, as the effect of BRCA1 depletion affects ORF2 levels in both L1RP and ORFeus expressing cells. Binding of BRCA1 between ORF1p and ORF2p might impede the “sliding” of the ribosomes toward the ORF2p translation start site, regulating its specification. Inhibition of ORF2p translation may depend on the L1 polyA tail. PABPC1, a polyA binding protein and L1 interactor54 mayexplain how BRCA1 can recognize and bind L1 mRNA. In addition, we observed that ORF2p expression mirrors the abundance of BRCA1 during the cell cycle; in G1 phase when BRCA1 protein is lower, ORF2p is more likely to be translated (Figure 7). We also observed a mild change in cell cycle distribution of ORF2 expression when BRCA1 is depleted, suggesting that BRCA1 repression of ORF2p translation may be cell cycle linked. The small effect induced by BRCA1 depletion may reflect incomplete BRCA1 depletion or the fact that BRCA1 is probably not the only ORF2p translation regulator.
Methods
Cell Culture and Cell lines
HeLa M2 cells (a gift from Gerald Schumann, Paul Ehrlich Institute) were cultured in DMEM supplemented with 10% FBS (Gemini, prod. number 100–106) and 1 mM L-glutamine (ThermoFisher/Life Technologies, prod. number 25030–081). HCT116+ch3 cells (an extra copy of chromosome 3 corrects a mismatch repair deficiency) were cultured in DMEM/F12, HEPES (ThermoFisher Scientific, catalog number 11330032) supplemented with 10% FBS and 400 μg/ml G418. UWB1.289 and UWB1.289+BRCA1 cells were a gift from Dr. A. Sfeir (NYU Langone Health). Cells were cultured in 50% ATCC-formulated RPMI 1640 medium-50% MEGM (Mammary Epithelial Growth Medium from Clonetics/Lonza; MEGM Bullet Kit; CC-3150) supplemented with SingleQuots additives (no gentamycin or amphotericin B). The medium was supplemented with 3% fetal bovine serum (FBS). UWB1.289+BRCA1 were cultured as UWB1.289 with the exception of adding 200μg/ml geneticin/G418 selection in the culture media during cell line maintenance. Transfection of the all-in-one ORFeus-luc-AI reporter was done by plating 0.01×10^6 cells/well in 96 well plates the day before transfection. 10μl transfection mix composed of 0.2μg of DNA, 0.4μl of P3000 reagent and 0.15μl of lipofectamine 3000 (ThermoFisher) in Opti-MEM, was directly added to the 50μl of medium in each well. After 6h, the media with transfection mix was aspirated and cells were left recovering over-night in complete media with doxycycline and without selection. After 48h firefly and renilla luminescence were measured using DualGlo kit (Promega) and a SynergyH1 plate reader (BioTek) with 1mm read height, 1 s integration time and autoscale gain option.
All cells were routinely tested for mycoplasma by PCR detection of conditioned media.
Plasmids and DNA constructs
pLD401 (ORFeus with 3×Flag ORF2) and pMT302 (L1rp with 3×Flag ORF2 and L1 5’UTR) plasmids were previously described and characterized in 23. pEA79 (untagged ORFeus with a GFP-AI cassette) was constructed by subcloning ORFeus under a Tet inducible promoter into a pCEP-puro vector 7. pSS141 (pCEP ORFeus BSD-AI) and pSS207 (pCEP ORFeus-endo-dead BSD-AI) were constructed by replacing the GFP-AI cassette with BSD-AI. pSS131 (BSD-AI rescue construct) was constructed by replacing the L1PA1 in pBS-L1PA1CH_blast rescue (Addgene #69611) with tet-ORFeus and the backbone was replaced with pCEP-puro sequences. pPM404 (renillla-ORFeus-luc-AI reporter) was assembled in a pCEP-puro backbone adding ,by “Gibson deletion” cloning55, the Renilla-CMV dual promoter from pYX50(=pPM97) provided by W. An56(South Dakota State University), to PM255, a derivative of pLD401 with the luciferase-AI cassette inserted in the BstZ17I site in the 3’UTR of L1 ORFeus. PM415, Renilla-ORFeus-luc-AI reporter, was constructed using Gibson cloning and single strand oligos55 to integrate a FspAI and AscI fragment from ORFeus-EN-(pLD62423) into FspAI-AscI cut PM404. The all-in-one Renilla-ORFeus-luc-AI reporter (pPM417) was constructed adding a CMV-rtTA -SV40pA fragment from pTET-ON advanced (Clontech) into the EcoRI site of pPM404. A list of the constructs used in this study is presented in Supplementary Table 5.
Primary siRNA screen
The high-throughput LINE-1 retrotransposition screen utilizes HeLa M2 cells22 (which constitutively express the rtTA/“Tet-ON” system) expressing a well-established GFP-AI reporter (Figure 1A). A synthetic L1 (ORFeus) retrotransposon was expressed from a Doxycycline-inducible (Tet) promoter with its 3’UTR bearing an antisense GFP disrupted by an inverted intron (GFP-AI). This reporter design, extensively evaluated and used in previous studies23,24, allows quantification of retrotransposition by measuring the percentage of GFP positive cells (Figure 1A). We cloned the L1 reporter in the pCEP-puro vector for episomal expression, thus allowing the maintenance of a “quasi-stable” cell line under puromycin selection23. Cells carrying the L1 reporter were plated on 384 well plates with arrayed siRNA pools to knockdown each individual human gene. 3 days after knock-down (Supplementary Figure 1A), the cells were stained with DAPI and the total number of cells and the number of green cells quantified by high content microscopy (Figure 1A).
Secondary validation screen
The settings of the 96 well validation followed the same procedures as the whole genome screen. We calculated the fold change of %GFP compared to siScramble for each knockdown.
Exon-junction qPCR
After Arrayscan VTI analysis exon junction qPCR was performed, lysing the cells in the same wells by freezing and thawing in TE buffer. Cell lysates were then treated with 0.3μg/μl proteinase K (Invitrogen, prod. number 25530049) for 1 h at 37˚C and ~50ng DNA was used for qPCR reaction. GFP and the control gene RPL21 were amplified using primers and probes reported in Supplementary Table 6.
Taqman probes were designed spanning the GFP exon junction (these anneal only to the intron-less [i.e. post RNA-splicing] GFP region). The probe sequences were designed as previously described in 57. The GFP expression was quantified by normalizing to the internal control, RPL21, using custom TaqMan FAM/HEX-labeled probes. Probes were synthesized by Integrated DNA Technologies (IDT).
We used LightCycler® 480 Probes Master (Roche, prod. number 04707494001) for the qPCR reaction.
CRISPR and BRCA1 rescue experiment
A gRNA (GGAGCCCACTTCATTAGTAC) targeting BRCA1 was cloned into lentiCRISPR v2 (Addgene #52961) following Golden Gate cloning protocol 58. Lentivirus carrying CRISPR-Cas9 constructs were produced using HEK293T cells with the following plasmids: pPM333/pSS172 (pMD2.G, Addgene #12259), pPM332/pSS173 (psPAX2, Addgene #12260), pSS164 (CRISPR Cas9 with BRCA1 gRNA). These three plasmids were co-transfected with Lipofectamine 2000 in 10cm plates. Viral supernatants were collected and filtered with 0.45μm filters. HCT116+ch3 or HeLa-M2 cells (plated the previous day) were then infected with the collected lentivirus using 8μg/ml Hexadimethrine bromide (polybrene). Infected cells were selected by 0.8μg/ml (HCT116 cells) or 1μg/ml (HeLa cells) puromycin for > 3 d and used as a pool. Infection was optimized at and MOI of ~0.3. A BRCA1 rescue construct (pSS146) was built by cloning the BRCA1 cDNA into the ORFeus GFP-AI reporter plasmid (pEA79). Cells expressing integrated Cas9 and gRNA were than transfected with L1 reporter (with or without BRCA1 cDNA) to measure the retrotransposition efficiency. Experiments that showed rescue of ORF2p protein upon BRCA1 knockout and overexpression (pSS146) (Figure 7B) were conducted in a similar manner except cells were then lysed and analyzed by immunoblotting.
Retrotransposition assay in 6 well plates
HeLa-M2 cells were plated into 6 well plates at 0.3 million cells per well. The next day, cells were transfected with L1 reporter construct using FuGENE-HD Transfection Reagent (Promega, prod number E2311) following the standard protocol. 24 h after transfection, quasi-stable cells carrying the reporter plasmids were selected with 1μg/ml puromycin for > 5 d. Doxycycline (1μg/ml) was added to induce L1 expression. After 3 d, the percentage of GFP positive cells was measured by flow cytometry (FACS buffer: 1% FBS, 1mM EDTA and 100U/ml of Penicillin-Streptomycin) and dead cells were excluded from the analysis by 5μg/ml propidium iodate staining (ThermoFisher Scientific, prod. number P1304MP).
Retrotransposition assay using L1 luciferase-AI reporter
Measurement of retrotransposition using the Renilla-ORFeus-luc-AI reporter (pPM404) were performed in 96 well plates or 6 well plates as specified in the text. In both procedures a quasi-stable cell line of HeLa-M2 cells episomally maintaining the reporter plasmid ,was generated by transfecting pPM404 in HeLa-M2 cells then maintained in media with 1μg/ml puromycin for more than 5 days. 0.005×10^6 cells were plated in each well of a 96 well plate or 0.2×10^6 cells in 6 well plates. Knockdown of specific genes was performed in 96 wells transfecting 2pmoles (0.2μl of a 10μM siRNA solution) of 3 siRNAs pooled together, with 0.3μl of RNAiMAX (ThermoFisher) in 10μl of OptiMEM/well. Transfection in 6 well plates was performed using 75pmoles (7.5μl of a 10μM siRNA solution), 7.5μl of RNAiMAX in 50μl of OptiMEM. The day following siRNA transfection the expression of the reported was induced with 0.25μg/ml doxycycline for 48h (96 well plates) or 5 days (6 well plates). PrestoBlue viability assay was performed adding a volume of reagent equal to 10% of the media into each well. After 30 minutes fluorescence was measure (ex./em.= 560/590). Following PrestoBlue assay, DualGlow reagents (Promega) were directly added to the media following manufacturer specifications. For assays conducted in 6 well plates, cells were trypsinized and collected in 1.5ml tubes and lysed with Reporter Lysis Buffer (1X; Promega E397A) supplemented with protease inhibitors. Cells in lysis buffer were subject to 2 cycles of freezing in LN2 and thawing at 55°C for 30 sec. The lysates were then spinned down at max speed (> 20000 rcf) for 10 minutes at 4°C and the supernatant collected. 2, 10 and 50μl (to ensure linearity) of this cleared lysate was then mixed with DualGlow reagents for Firefly (FF) and Renilla (RR) measurements. L1 retrotransposition rate was expressed as the ration of (FF/RR)*1000.
Knockdown experiment and Aphidicolin treatment
All our knockdown experiments were conducted using a pool of 3 siRNAs against the considered target to minimize off target effects and increase specificity of knockdown. We used highly validated (in GPRM259,KB60 ,TC3261, HCT11662, HEK29363 and HeLa64 cells) ThermoFisher Silencer Select siRNAs guaranteed to produce at least a 70% reduction of the target gene (our unpublished validations and qPCR quantifications presented in Supplementary Figure 6B–C support this claim). HeLa cells were plated into 6 well plates with 0.3 million cells per well. The next day we transfected the cells with 7.5μl 10μM siRNA pools (3 siRNAs against same gene) using 100μl OPTIMEM (Gibco, prod. number 31985070) and 7.5μl Lipofectamine® RNAiMAX Transfection Reagent (Life Technologies, prod. number 13778030). The next day, cells were replated into 6 well plates with 0.1 million cells per well. After 24 h, we added 1μg/ml doxycycline and various concentrations of Aphidicolin to the cells. After 24 h, we washed off Aphidicolin 3 times with PBS and cultured the cells in Doxycycline for another 2 d. After 2 d, we measured the retrotransposition efficiency by flow cytometry as described above. For a typical knockdown experiment, we add the doxycycline the next day after transfection and measure the retrotransposition by flow cytometry after 3 d.
Experiments assessing the effect of APH treatment on L1 retrotransposition using the luc-AI reporter were conducted in 96 well plates using HeLa-M2 pPM404 “quasi-stable” cell lines treated as for the experiments that used the GFP-AI reporter and measuring viability by PrestoBlue, FF and RR 48h after APH treatment.
Analysis of epistasis interaction with BRCA1 knock down
To identify factors necessary for BRCA1 mediated inhibition of L1 we analyzed epistasis ratios obtained by dividing the Firefly/Renilla (FF/RR) values after co-transfection with siRNAs against BRCA1 (siBRCA1) and the considered factor (double knock-down), by the FF/RR values from treatments with siRNA against the considered factor and siCtrl (single knock down) (epistasis ratio in Figure 6A calculated from values presented in Supplementary Figure 8 and summarized in Supplementary Table 7). The ratio of the siCtrl-siBRCA1 treatment values divided by siCtrl-siCtrl treatment values (Figure 6A, first bar in gray) provide the “full de-inhibition” induced by BRCA1 depletion (neutral interaction); on the contrary, a ratio equal to 1 will indicate that depletion of the considered factor completely abrogates the increase of LINE-1 retrotransposition upon depletion of BRCA1, thus indicating a critical role for the considered factor in the BRCA1-mediated suppression of LINE-1. This latter situation can be exemplified by the depletion of BRCA1 itself (Figure 6A, second bar in green) that is, needless to say, essential for the BRCA1-mediated repression of LINE-1. It is important to note that, including different denominators (FF/RR from the single knock down) in the calculation of the epistasis ratio excludes effects on L1 retrotransposition induced by parallel pathways not involving BRCA1 (absence of epistasis interaction or neutral interactions).
BSD-AI experiment
HeLa cells were first transfected with pSS141 (ORFeus BSD-AI) or pSS207 (endonuclease dead ORFeus BSD-AI) and quasi-stable cell lines were selected after 5 d of 1 μg/ml puromycin selection. We then performed knockdown with siRNA as described above. 24 h after knockdown, doxycycline was added for 3 d. We then plated 0.1 and 0.5 million cells for pSS141; 0.5 million and 5 million cells for pSS207 into 10cm plates under 15μg/ml Blasticidin selection. Cells were stained with crystal violet after 10 d for imaging.
L1 insertion rescue
Quasi-stable cell line expressing pSS131 (ORFeus L1 rescue construct) was maintained under puromycin (1 μg/ml) selection for at least 5 d. We then performed knockdown experiment following the procedures described above. After 3 d of 1 μg/ml doxycycline treatment, cells were plated 1:10, 1:100 and 1:1000 in 10cm plates and kept under 15μg/ml Blasticidin selection. Single clones were hand-picked under microscope and then expanded. Each of the clones was expanded to reach confluency in a 6 cm plate. 3–5 million cells were then harvested and genomic DNA was isolated using QIAamp DNA mini kit (QIAGEN, prod. number 51304). The following L1 recovery steps were adapted from 19,49 with modifications. 50μg of purified DNA was used for the following steps. 1) Genomic DNA was digested with 200U HindIII-HF (New England BioLabs, prod. number R3104S) at 37°C overnight. 2) The restriction enzyme was heat inactivated at 65C for 20min, followed by a self-ligation using 1200U T4 DNA ligase (New England BioLabs, prod. number M0202S) in 1ml total volume at room temperature overnight. 3) We then purified the DNA using Amicon Ultra 0.5ml, 50K (Millipore Sigma, prod number UFC505096) by centrifuging for 20 m at 14,000g and columns were washed with 500μl H2O twice. 4) We concentrated the DNA using ethanol precipitation into 5μl volume and pulled 5 samples into one transformation mix. 5) We then transformed the DNA into ElectroMAX™ DH10B™ T1 R cells (Life Technologies, prod. number 12033–015) using electroporation and recovered the colonies on LB plates supplemented with 100μg/ml Blasticidin. 6) Each of the clones were then Sanger sequenced (Genewiz) using the primers reported in Supplementary Table 6.
Immunoblotting
HeLa M2 cells were lysed in SKL Triton lysis buffer (50 mM Hepes pH7.5, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 10% glycerol, 1% Triton X-100, 25 mM NaF, 10 μM ZnCl2) supplemented with protease and phosphatase inhibitors (Complete-EDTA free, Roche/Sigma prod. number 11873580001; 1 mM PMSF and 1 mM NaVO4). NuPage 4XLDS sample buffer (ThermoFisher Scientific, prod. number NP0007) supplemented with 1.43M β-mercaptoethanol was added to the samples to reach a 1× dilution (350 mM β-mercaptoethanol final concentration) before gel electrophoresis performed using 4–12% Bis-Tris gels (ThermoFisher Scientific, prod. number WG1402BOX). Proteins were transferred on Immobilon-FL membrane (Millipore, prod. number IPFL00010), blocked for 1 hr with blocking buffer (LiCOR prod. number 927–40000):TBS buffer (50 mM Tris Base, 154 mM NaCl) 1:1 and then incubated with primary antibodies solubilized in LiCOR blocking buffer: TBS-Tween (0.1% Tween in TBS buffer) 1:1. Secondary goat antibodies conjugated to IRDye680 (anti-rabbit) or IRDye800 (anti-mouse) dyes (LiCOR prod. number 926–32210 and 926–68071), were used for detection of the specific bands on an Odyssey CLx scanner (LiCOR). 4H1 mouse monoclonal antibody targeting amino acids 35 to 44 of human ORF1p 7,23,65 (Millipore, cat. number MABC1152) was used at 1:15000 dilution. ORF2-Flag were detected using FLAG-M2 antibody (Sigma, cat. number F1804) at 1:1000 dilution. Anti-α/β tubulin (Cell Signaling #2148) at 1:1000 dilution was used as loading control. Other antibodies used for immunoblot analysis in this manuscript are reported below:
Anti-BRCA1 MS13, Abcam ab16781, dilution 1:200; anti-Cas9, OriGene TA190309, dilution 1:500; anti-Cdt1, Abcam ab202067, dilution 1:1000; anti-p53 FL-393, Santa Cruz sc-6243, dilution 1:200.
Immunofluorescence
Immunofluorescence was performed as previously described 7. We used the mouse BRCA1 [D9] antibody (Santa Cruz cat. sc-6954) at 1:100 dilution and FLAG-M2 antibody (Sigma, cat. number F1804) at 1:1000 dilution for staining. Halotag7-ORF2p was detected using Halo tag ligand JF646 66 as previously described 7.
iPOND
iPOND IP was performed as previously described29. Briefly, HeLa-M2 cells expressing recoded (LD401) or non-recoded (MT302) L1 from a pCEP-puro based plasmid were grown in 15cm plates. When 95% confluent, the plates were incubated for 10 minutes with 10μM EdU at 37°C 5% CO2. Cells in 2/3 of the plates (conditions #1 and #2) were then fixed with 1% formaldehyde in PBS for 20 minutes, while 1/3 of the plates (condition #3) were washed once in media supplemented with 10μM thymidine and then incubated for an additional 1h with complete media with 10μM thymidine. After 1h these plates also were fixed with 1% formaldehyde for 20 minutes. After fixation formaldehyde was quenched with 0.0625M (final concentration) glycine added directly to the media. Cells were washed in cold PBS and scraped in 50ml tubes. After 2 PBS washes, cells were permeabilized for 30 minutes in 0.25% Triton-X100 (in PBS) and then washed in 0.5% BSA in PBS. Click reaction was performed incubating the cells for 2h in a solution of 10mM sodium ascorbate, 2mM CuSO4 and 10μM biotin azide (in PBS) (2×10^7 cells/ml of click reaction). Biotin azise was omitted form condition #2. Cells were then washed in PBS+0.5% BSA, lysed in 1% SDS, 50mM Tris-HCl pH8 (lysis buffer) and sonicated with 5 pulses (20”on, 40” off) at 30% amplitude) of sonication in ice. Lysed were clarified spinning at 20.000×g (max speed). The supernatant was then diluted 1:1 in PBS and 25μl of Dynabeads MyOne Streptavidin T1 (Invitrogen, 65601) added to each IP solution. 15μl of solution for each condition (INPUTs) were collected before adding beads and stored at 4°C. After an over-night incubation at 4°C the beads were washed once in lysis buffer, once in 1M NaCl and twice in lysis buffer. The immunoprecipitated complexes were eluted adding 30μl of 1X LDS supplemented with β-mercaptoethanol (350 mM β-mercaptoethanol final concentration) directly to the washed beads and 4X LDS + β-mercaptoethanol to the INPUT solutions to reach a 1X dilution. Crosslinking was reversed incubating the beads and the INPUTs in LDS at 95°C for 25 minutes. Samples were then centrifuged at maximum speed and supernatant stored at −20°C until immunoblotting analysis was performed. 5μl INPUT and 20μl of IP samples were loaded on a 4–12% Bis-Tris NuPage gel. Proteins were transferred onto Immobilon-FL (Millipore) membrane, blocked with LiCOR blocking buffer (1:1 in TBS) and blotted with the indicated antibodies: anti-PCNA, Abcam ab18197, 1:5000 dilution; anti H3, Abcam ab1791, 1:10.000 dilution.
IP-RT-qPCR and qPCR
HeLa-M2 cells expressing ORFeus (pLD401) from one 15 cm plate per IP were collected by trypsinization and lysed in SKL Triton lysis buffer (50 mM Hepes pH7.5, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 10% glycerol, 1% Triton X-100, 25 mM NaF, 10 μM ZnCl2) supplemented with 1 mM DTT, 400 μM Ribonucleoside Vanadyl Complex (VRC) (NEB, prod. number S1402), 400U/(ml of buffer) of RNASEOUT (Thermo, prod. number 10777019), protein inhibitor and phosphatase inhibitor (Complete-EDTA free, Roche/Sigma prod. number 11873580001; 1 mM PMSF and 1 mM NaVO4). The cells-lysate was centrifuged for 15 min. at 16,000 rcf at 4°C. Immunoprecipitations were conducted overnight at 4°C using mouse 4H1 ORF1p antibodies, mouse BRCA1 [D9] Santa Cruz cat. sc-6954) or control IgG from normal mouse (Santa Cruz sc-2025) coupled to magnetic beads (Dynabeads Antibody Coupling kit, Life Technologies, prod. number 14311D). Beads were washed five times in Triton buffer and resuspended in 100 μl of Triton buffer containing 30 μg of proteinase K (Invitrogen, prod. number 25530049). The mixture was incubated at 55°C for 30 min. 1ml of Trizol (Life Technologies, prod. number 15596026) was directly added to the beads mixture and mRNA was purified using RNA clean-up and concentration columns (Norgen Biotek, prod. number 23600). cDNA was generated from RNA using the USB First-Strand cDNA synthesis kit for Real-Time PCR (Affimetrix, prod. number 75780). q-PCR was performed using a standard curve of pLD401 plasmid 23. Each q-PCR reaction contained 2.5μl of Sybr Green mastermix 2X (Roche, LightCycler 480 SYBR Green I Master, prod. number 04887352001), 25nl of forward primer (100 μM), 25nl of reverse primer (100 μM), 500nl of cDNA for IP and water to 5μl (final volume). qPCR was performed using a Light Cycler 480 (Roche) with standard conditions. The primers used for qPCR are reported in Supplementary Table 6.
Live-cell imaging
Hela S.FUCCI cells expressing rtTA and stably expressing a pCEP-puro plasmid expressing ORFeus L1 with Halotag7-ORF2p and untagged ORF1p under a Tet inducible promoter 7 were plated in 24 well plates at a density of 0.025×10^6 cells/well in 500ml of media. The next day 500μl of fresh media containing doxycycline 2μg/ml and Halo ligand JF646 200nM was added to the cells. After 4hr cells were moved to an EVOS-FL auto on stage incubator (Thermo-Fisher) for image collection every 30 m for 48hr. Bright field was used for autofocus and GFP, RFP and cy5 cubes were used to collect fluorescence images of about 30 beacons per experiment.
Supplementary Material
Supplementary Information
Supplementary Information including Supplementary Note and 10 Supplementary Figures
Supplementary Table 6
Oligos and primers used in this study
Supplementary Table 5
List and description of DNA constructs used in this study
Supplementary Table 4
L1 insertion sequences recovered in control cells and cells depleted of BRCA1 or FANCM
Supplementary Table 7 Summary table of the epistasis analysis reported in Supplementary Figure 8
Cells are colored red if depletion of the considered gene induced a significant increase of L1 retrotransposition compared to siCtrl (in single knockdowns) or siBRCA1 (in double knockdowns). Cells are colored blue if the depletion of the considered gene induced a significant decrease of L1 retrotransposition compared to siCtrl (in single knockdowns) or siBRCA1 (in double knockdowns). The number of + indicate the magnitude of decrease or increase. X means that depletion of the considered gene did not induce any significant alteration in L1 retrotransposition compared to siCtrl (in single KDs) or siBRCA1 (in double KDs) treatments.
Supplementary Table 3
log fold changes of the DNA repair factors in 96-well validation screen
Supplementary Table 2
Enrichment analysis results for the L1 supporters identified in our screen
Supplementary Table 1
Raw data and hit lists for the genome-wide siRNA knockdown screen
Supplementary Video 2 Live-cell imaging of FUCCI cells
expressing ORF2 in S/G2. FUCCI cells expressing L1 were imaged every 30 min for 48 h. Geminin and Cdt1 peptides are visualized in green and red, respectively. Merged channels, ORF2p (cy5 channel) and bright field are shown as a movie. The merged channel (left panel) shows two cells in the center of the field starting to express ORF2p in S/G2 phase (green nuclei).
Supplementary Video 1 Live-cell imaging of FUCCI cells expressing ORF2 in G1
FUCCI cells expressing L1 were imaged every 30 min for 48 h. Geminin and Cdt1 peptides are visualized in green and red, respectively. Merged channels, ORF2p (cy5 channel) and bright field are shown as a movie. The merged channel (left panel) shows a cluster of cells in the center of the field starting to express ORF2p in G1 phase (red nuclei).
ACKNOWLEDGMENTS
We thank Tony Huang and Kathleen H. Burns for helpful discussions and comments on the manuscript. This work was supported by NIH grants P50GM107632 to J.D.B. and P01AG051449 to John Sedivy.
Footnotes
STATISTICS
Two-tailed unpaired homoscedastic T-test was applied to assess statistical significance. The P values intervals were reported in the corresponding figure legends.
REPORTING SUMMARY STATEMENT
Further information on experimental design is available in the Nature Research Reporting Summary linked to this article.
DATA AVAILABILITY STATEMENT
All the raw data of the primary and secondary screens are provided as supplemental tables.
COMPETING INTERESTS STATEMENT
The authors declare that they have no competing interests.
Supplementary Data 1
Unprocessed western blots for Figs. 2 and 7 and Supplementary Figs. 3 and 10
References:
- 1.Burns KH & Boeke JD Human transposon tectonics. Cell 149, 740–752 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang CRL, Burns KH & Boeke JD Active Transposition in Genomes. Annual Review of Genetics 46, 651–675 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sassaman DM et al. Many human L1 elements are capable of retrotransposition. Nature Genetics (1997). [DOI] [PubMed] [Google Scholar]
- 4.Martin SL & Bushman FD Nucleic acid chaperone activity of the ORF1 protein from the mouse LINE-1 retrotransposon. Molecular and cellular biology 21, 467–475 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cost GJ, Feng Q, Jacquier A & Boeke JD Human L1 element target-primed reverse transcription in vitro. The EMBO Journal 21, 5899–5910 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feng Q, Moran JV, Kazazian HH & Boeke JD Human L1 Retrotransposon Encodes a Conserved Endonuclease Required for Retrotransposition. Cell 87, 905–916 (1996). [DOI] [PubMed] [Google Scholar]
- 7.Mita P. et al. LINE-1 protein localization and functional dynamics during the cell cycle. eLife 7, 210–210 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alisch RS, Garcia-Perez JL, Muotri AR, Gage FH & Moran JV Unconventional translation of mammalian LINE-1 retrotransposons. Genes & development 20, 210–224 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luan DD, Korman MH, Jakubczak JL & Eickbush TH Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: A mechanism for non-LTR retrotransposition. Cell 72, 595–605 (1993). [DOI] [PubMed] [Google Scholar]
- 10.Jurka J & Klonowski P. Integration of Retroposable Elements in Mammals: Selection of Target Sites. Journal of Molecular Evolution (1996). [DOI] [PubMed] [Google Scholar]
- 11.Gilbert N, Lutz S, Morrish TA & Moran JV Multiple fates of L1 retrotransposition intermediates in cultured human cells. Molecular and cellular biology 25, 7780–7795 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moran JV et al. High frequency retrotransposition in cultured mammalian cells. Cell 87, 917–927 (1996). [DOI] [PubMed] [Google Scholar]
- 13.Symer DE et al. Human l1 retrotransposition is associated with genetic instability in vivo. Cell 110, 327–338 (2002). [DOI] [PubMed] [Google Scholar]
- 14.Goodier JL Restricting retrotransposons: a review. Mobile DNA 7, 16–16 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gasior SL, Wakeman TP, Xu B & Deininger PL The Human LINE-1 Retrotransposon Creates DNA Double-strand Breaks. Journal of molecular biology 357, 1383–1393 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coufal NG et al. Ataxia telangiectasia mutated (ATM) modulates long interspersed element-1 (L1) retrotransposition in human neural stem cells. Proceedings of the National Academy of Sciences of the United States of America 108, 20382–20387 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Suzuki J. et al. Genetic Evidence That the Non-Homologous End-Joining Repair Pathway Is Involved in LINE Retrotransposition. PLoS genetics 5, e1000461–e1000461 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morrish TA et al. DNA repair mediated by endonuclease-independent LINE-1 retrotransposition. Nature Genetics 31, 159–165 (2002). [DOI] [PubMed] [Google Scholar]
- 19.Servant G. et al. The Nucleotide Excision Repair Pathway Limits L1 Retrotransposition. Genetics 205, 139–153 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brégnard C. et al. Upregulated LINE-1 Activity in the Fanconi Anemia Cancer Susceptibility Syndrome Leads to Spontaneous Pro-inflammatory Cytokine Production. EBioMedicine 8, 184–194 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu N. et al. Selective silencing of euchromatic L1s revealed by genome-wide screens for L1 regulators. Nature (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hampf M & Gossen M. Promoter Crosstalk Effects on Gene Expression. Journal of Molecular Biology (2007). [DOI] [PubMed] [Google Scholar]
- 23.Taylor MS et al. Affinity Proteomics Reveals Human Host Factors Implicated in Discrete Stages of LINE-1 Retrotransposition. Cell 155, 1034–1048 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.An W. et al. Characterization of a synthetic human LINE-1 retrotransposon ORFeus -Hs. Mobile DNA 2, 1–1 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Szklarczyk D. et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 47, D607–D613 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krejci L, Altmannova V, Spirek M & Zhao X. Homologous recombination and its regulation. (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li X & Heyer W-D Homologous recombination in DNA repair and DNA damage tolerance. Cell Research (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ceccaldi R, Sarangi P & D’Andrea AD The Fanconi anaemia pathway: new players and new functions. Nature Reviews Molecular Cell … 17, 337–349 (2016). [DOI] [PubMed] [Google Scholar]
- 29.Dungrawala H & Cortez D. Purification of proteins on newly synthesized DNA using iPOND. Methods Mol Biol 1228, 123–31 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Michl J, Zimmer J & Tarsounas M. Interplay between Fanconi anemia and homologous recombination pathways in genome integrity. The EMBO Journal 35, 909923 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schlacher K, Wu H & Jasin M. A distinct replication fork protection pathway connects Fanconi anemia tumor suppressors to RAD51-BRCA½. Cancer Cell 22, 10616 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.DelloRusso C. et al. Functional characterization of a novel BRCA1-null ovarian cancer cell line in response to ionizing radiation. Mol Cancer Res 5, 35–45 (2007). [DOI] [PubMed] [Google Scholar]
- 33.Morrish TA et al. Endonuclease-independent LINE-1 retrotransposition at mammalian telomeres. Nature 446, 208–212 (2007). [DOI] [PubMed] [Google Scholar]
- 34.Ruffner H, Joazeiro CA, Hemmati D, Hunter T & Verma IM Cancer-predisposing mutations within the RING domain of BRCA1: loss of ubiquitin protein ligase activity and protection from radiation hypersensitivity. Proceedings of the National Academy of Sciences 98, 5134–5139 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shen SX et al. A targeted disruption of the murine Brca1 gene causes gamma-irradiation hypersensitivity and genetic instability. Oncogene 17, 3115–3124 (1998). [DOI] [PubMed] [Google Scholar]
- 36.Huertas P & Jackson SP Human CtIP mediates cell cycle control of DNA end resection and double strand break repair. J Biol Chem 284, 9558–65 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.You Z & Bailis JM DNA damage and decisions: CtIP coordinates DNA repair and cell cycle checkpoints. Trends Cell Biol 20, 402–9 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Przetocka S. et al. CtIP-Mediated Fork Protection Synergizes with BRCA1 to Suppress Genomic Instability upon DNA Replication Stress. Mol Cell 72, 568–582e6 (2018). [DOI] [PubMed] [Google Scholar]
- 39.Her J, Ray C, Altshuler J, Zheng H & Bunting SF 53BP1 Mediates ATR-Chk1 Signaling and Protects Replication Forks under Conditions of Replication Stress. Mol Cell Biol 38(2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Villa M, Bonetti D, Carraro M & Longhese MP Rad9/53BP1 protects stalled replication forks from degradation in Mec1/ATR-defective cells. EMBO Rep 19, 351–367 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bunting SF et al. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell 141, 243–254 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Polato F. et al. CtIP-mediated resection is essential for viability and can operate independently of BRCA1. The Journal of experimental medicine 211, 1027–1036 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Beyer A, Bandyopadhyay S & Ideker T. Integrating physical and genetic maps: from genomes to interaction networks. Nat Rev Genet 8, 699–710 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Higgs MR et al. BOD1L Is Required to Suppress Deleterious Resection of Stressed Replication Forks. Mol Cell 59, 462–77 (2015). [DOI] [PubMed] [Google Scholar]
- 45.Xu S. et al. Abro1 maintains genome stability and limits replication stress by protecting replication fork stability. Genes Dev 31, 1469–1482 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Poole LA & Cortez D. Functions of SMARCAL1, ZRANB3, and HLTF in maintaining genome stability. Crit Rev Biochem Mol Biol 52, 696–714 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fungtammasan A, Walsh E, Chiaromonte F, Eckert KA & Makova KD A genome-wide analysis of common fragile sites: What features determine chromosomal instability in the human genome? Genome research 22, 993–1005 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Arlt MF et al. BRCA1 is required for common-fragile-site stability via its G2/M checkpoint function. Molecular and cellular biology 24, 6701–6709 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gilbert N, Lutz-Prigge S & Moran JV Genomic deletions created upon LINE-1 retrotransposition. Cell 110, 315–325 (2002). [DOI] [PubMed] [Google Scholar]
- 50.Jensen S, Gassama MP & Heidmann T. Retransposition of the Drosophilia LINE I Element Can Induce Deletion in the Target DNA: A Simple Model Also Accounting for the Variability of the Normally Observed Target Site Duplications. Biochemical and biophysical research communications 202, 111–119 (1994). [DOI] [PubMed] [Google Scholar]
- 51.Santos A, Wernersson R & Jensen LJ Cyclebase 3.0: a multi-organism database on cell-cycle regulation and phenotypes. Nucleic Acids Res 43, D1140–4 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dacheux E. et al. BRCA1-Dependent Translational Regulation in Breast Cancer Cells. PloS one 8, e67313–e67313 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mao Z, Bozzella M, Seluanov A & Gorbunova V. DNA repair by nonhomologous end joining and homologous recombination during cell cycle in human cells. Cell Cycle 7, 2902–2906 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dai L, Taylor MS, O’Donnell KA & Boeke JD Poly(A) Binding Protein C1 Is Essential for Efficient L1 Retrotransposition and Affects L1 RNP Formation. Molecular and Cellular Biology (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods references:
- 55.Kalva S, Boeke JD & Mita P. Gibson Deletion: a novel application of isothermal in vitro recombination. Biol Proced Online 20, 2 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xie Y, Rosser JM, Thompson TL, Boeke JD & An W. Characterization of L1 retrotransposition with high-throughput dual-luciferase assays. Nucleic Acids Res 39, e16 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xie Y. et al. Cell division promotes efficient retrotransposition in a stable L1 reporter cell line. Mobile DNA 4, 10–10 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sanjana NE, Shalem O & Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nature Methods 11, 783–784 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hasson SA et al. High-content genome-wide RNAi screens identify regulators of parkin upstream of mitophagy. Nature 504, 291–5 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pasetto M. et al. Whole-genome RNAi screen highlights components of the endoplasmic reticulum/Golgi as a source of resistance to immunotoxin-mediated cytotoxicity. Proc Natl Acad Sci U S A 112, E1135–42 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Grohar PJ et al. Identification of an inhibitor of the EWS-FLI1 oncogenic transcription factor by high-throughput screening. J Natl Cancer Inst 103, 962–78 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vassilev A. et al. Identification of genes that are essential to restrict genome duplication to once per cell division. Oncotarget 7, 34956–76 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xiao S. et al. Genome-scale RNA interference screen identifies antizyme 1 (OAZ1) as a target for improvement of recombinant protein production in mammalian cells. Biotechnol Bioeng 113, 2403–15 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sivan G, Ormanoglu P, Buehler EC, Martin SE & Moss B. Identification of Restriction Factors by Human Genome-Wide RNA Interference Screening of Viral Host Range Mutants Exemplified by Discovery of SAMD9 and WDR6 as Inhibitors of the Vaccinia Virus K1L-C7L- Mutant. MBio 6, e01122 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rodić N. et al. Long interspersed element-1 protein expression is a hallmark of many human cancers. The American journal of pathology 184, 1280–1286 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Grimm JB et al. A general method to improve fluorophores for live-cell and single-molecule microscopy. Nature Methods (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information
Supplementary Information including Supplementary Note and 10 Supplementary Figures
Supplementary Table 6
Oligos and primers used in this study
Supplementary Table 5
List and description of DNA constructs used in this study
Supplementary Table 4
L1 insertion sequences recovered in control cells and cells depleted of BRCA1 or FANCM
Supplementary Table 7 Summary table of the epistasis analysis reported in Supplementary Figure 8
Cells are colored red if depletion of the considered gene induced a significant increase of L1 retrotransposition compared to siCtrl (in single knockdowns) or siBRCA1 (in double knockdowns). Cells are colored blue if the depletion of the considered gene induced a significant decrease of L1 retrotransposition compared to siCtrl (in single knockdowns) or siBRCA1 (in double knockdowns). The number of + indicate the magnitude of decrease or increase. X means that depletion of the considered gene did not induce any significant alteration in L1 retrotransposition compared to siCtrl (in single KDs) or siBRCA1 (in double KDs) treatments.
Supplementary Table 3
log fold changes of the DNA repair factors in 96-well validation screen
Supplementary Table 2
Enrichment analysis results for the L1 supporters identified in our screen
Supplementary Table 1
Raw data and hit lists for the genome-wide siRNA knockdown screen
Supplementary Video 2 Live-cell imaging of FUCCI cells
expressing ORF2 in S/G2. FUCCI cells expressing L1 were imaged every 30 min for 48 h. Geminin and Cdt1 peptides are visualized in green and red, respectively. Merged channels, ORF2p (cy5 channel) and bright field are shown as a movie. The merged channel (left panel) shows two cells in the center of the field starting to express ORF2p in S/G2 phase (green nuclei).
Supplementary Video 1 Live-cell imaging of FUCCI cells expressing ORF2 in G1
FUCCI cells expressing L1 were imaged every 30 min for 48 h. Geminin and Cdt1 peptides are visualized in green and red, respectively. Merged channels, ORF2p (cy5 channel) and bright field are shown as a movie. The merged channel (left panel) shows a cluster of cells in the center of the field starting to express ORF2p in G1 phase (red nuclei).