Abstract
Prion diseases exhibit different disease phenotypes in their natural hosts and when transmitted to rodents, and this variability is regarded as indicative of prion strain diversity. Phenotypic characterization of scrapie strains in sheep can be attempted by histological, immunohistochemical and biochemical approaches, but it is widely considered that strain confirmation and characterization requires rodent bioassay. Examples of scrapie strains obtained from original sheep isolates by serial passage in mice include ME7, 79A, 22A and 87V. In order to address aspects of prion strain stability across the species barrier, we transmitted the above murine strains to sheep of different breeds and susceptible Prnp genotypes. The experiment included 40 sheep dosed by the oral route alone and 36 sheep challenged by combined subcutaneous and intracerebral routes. Overall, the combined route produced higher attack rates (~100%) than the oral route (~50%) and 2–4 times shorter incubation periods. Uniquely, 87V given orally was unable to infect any sheep. Overall, scrapie strains adapted and cloned in mice produce distinct but variable disease phenotypes in sheep depending on breed or Prnp genotype. Further re-isolation experiments in mice are in progress in order to determine whether the original cloned murine disease phenotype will reemerge.
Keywords: biological properties, interspecies transmission, prion protein, Scrapie diversity, species barrier, strain typing
Introduction
Scrapie is a chronic, progressive and invariably fatal neurodegenerative disorder of sheep and goats. It is the archetype of the transmissible spongiform encephalopathies (TSEs) that naturally affect several mammalian species. Other naturally-occurring TSEs include chronic wasting disease in cervids, transmissible mink encephalopathy and bovine spongiform encephalopathy (BSE). They belong to a group of misfolding protein disorders, also known as prion diseases, whose pathogenesis primarily involves the conversion of a host-encoded glycoprotein, termed cellular prion protein (PrPc), into a misfolded form usually termed diseased-associated PrP (PrPd), PrP scrapie (PrPSc) or protease-resistant PrP (PrPres).1 The phenomenon of PrPc conversion is initiated by exposure to exogenous prions followed by an autocatalytic process that results in the deposition of PrPd/PrPres in different tissues and viscera. The accumulation of PrPd in the ovine brain is associated with progressive neurodegeneration characterized by variable early astrocytosis, spongiform change, loss of synaptic proteins, microglial activation and abnormal neuromodulary responses at clinical stages of the disease.2
The significance of different lesions in TSEs can be difficult to establish as they may vary markedly from case to case depending on the source (strain)3-6 or on host factors like the Prnp genotype.7-9 Vacuolar lesion profiles in sheep7,10 were insufficiently reproducible when individuals experimentally challenged with a single scrapie isolate were compared.10 Instead, the PrPd profiling and the epitope mapping approaches proved to be more reliable.11 The full3-5 or short6 PrPd profiles in clinically-affected sheep with different TSE strains show that the relative magnitude of different morphological types of PrPd found in specific cells and brain areas are characteristic of particular strains. The recent demonstration of different PrPd profiles in different European sheep populations of the same PrP genetic background refutes the proposition that differences in the Prnp genotype are the sole determinate of the PrPd profile.12 The epitope mapping approach using immunohistochemistry (IHC) revealed differences between strains like BSE and CH1641 in comparison with scrapie.13
Experimentally, scrapie can be transmitted to a number of species, including livestock and laboratory animals. Cross-species transmission, if successful, is generally characterized by prolonged incubation periods, which is interpreted as the species barrier phenomenon.14-16 Nevertheless the “species-barrier” is abrogated with the adaptation of the strain to the new host after 2–3 passages.17 Characterization of sheep TSE strains, also known as strain typing, has been historically performed by serial passage at limiting dilution of individual or pooled sheep scrapie brains in rodents.14,18-20 Strain typing metrics consist of incubation period (survival times), vacuolar lesion profile, PrPd immunohistochemistry and the molecular PrPres profile. Serial passages of different natural sheep scrapie sources showed distinct disease phenotypes, and perpetuation of the same biological properties upon serial passages in mouse lines of the same genetic background. These data suggested that mice could be used for TSE strain characterization.21 Subsequently, the mouse bioassay tool was able to discriminate between sheep scrapie and cattle BSE and similarities in the rodent transmission phenotype suggested that variant Creutzfeldt-Jakob disease in humans originated from cattle BSE sources.22,23 This multiplicity of different disease phenotypes and inferred strains in mice with identical genetic background24 has been difficult to accommodate within the protein-only hypothesis as few stable conformational variants of abnormal PrP have been identified.25
Around 20 murine scrapie-adapted strains have been identified following initial transmission from natural sheep or goat scrapie infected brains.24,26 Nevertheless the relationship between these mouse strains and natural strains in the sheep population remains obscure. It remains possible that murine scrapie strains isolated from sheep sources are novel strains that emerge during the process of inter-species limiting dilution serial passage and the significance of murine strains for field strain classification or discrimination between natural scrapie strains therefore remains untested. If, within a particular species, there are several determinants (or factors) influencing and controlling prion adaptation, in cross-species transmission they may be maximized and result in an unpredictable outcomes. Documented scenarios16 indicate that some species are refractory to specific prions. However, the single documented attempt to transmit the murine ME7 strain into sheep was successful27 in 2 out of 25 Cheviot sheep when inoculated with 5ml of a 10% brain homogenate supernatant by the sub-cutaneous route. Despite no strain typing tools were available at the time histopathological examination of the sheep brains confirmed that spongiform lesions were characteristic of scrapie. The present paper describes the phenotype of the disease in sheep after inoculation with cloned ME7, 79A, 22A and 87V murine strains. The limitations and relevance of biological cloning after interspecies transmission in an attempt to characterize strains are discussed.
Results
Attack Rates and Survival Times
Table 1 gives an overview of the experimental design for both single oral and combined challenges, and summarizes the results on attack rates (AR) and survival times (ST). Six sheep, which died of intercurrent disease, have not been included in the AR calculations: i) in the ME7 oral experiment, one VRQ/VRQ (1/5) and one ARQ/ARQ (1/5) Cheviot sheep that died at 844 and 838 d post-inoculation (dpi), respectively; ii) in the 79A combined experiment, one VRQ/VRQ and one ARQ/ARQ Cheviot sheep that died at 52dpi and 48 dpi, respectively; iii) two VRQ/VRQ Cheviot sheep in the 22A oral and combined experiments, that died at 1624dpi and 208dpi, respectively.
Table 1. Summary of the experimental design, attack rates (AR), survival times (ST) and global immunohistochemical results on the central nervous system (CNS), peripheral nervous system (PNS), gut-associated lymphoid tissues (GALT) and non-GALT (nGALT).
| Strain | Host | Route | Recipients | AR | ST | CNS | PNS | GALT | nGALT |
|---|---|---|---|---|---|---|---|---|---|
| ME7 | Suff AA | Oral | 3 | 2/3R | 763, 1209 | 8.4 | 4.6 | 0.4 | 1.8 |
| Combined | 3 | 3/3R | 365 ± 256 | 7.6 | 3.2 | 12 | 3.8 | ||
| Chev VV | Oral | 5 | 4/4 | 1146 ± 193 | 11.6 | 6.6 | 9.1 | 35.7 | |
| Combined | 3 | 3/3 | 659 ± 100 | 9.1 | 3.1 | 4.7 | 19.2 | ||
| Chev AA | Oral | 5ID1 | 4/4 | 1388 ± 12 | 11 | 5 | 2.5 | 10.2 | |
| Combined | 3 | 3/3 | 624 ± 60 | 5.7 | 0.9 | 0 | 2.2 | ||
| 79A | Suff AA | Oral | 3 | 0/3R, * | - | - | - | - | - |
| Combined | 3 | 3/3R | 653 ± 335 | 7.7 | 1 | 0 | 0.4 | ||
| Chev VV | Oral | 3 | 1/3 | 481 | 6.9 | 3.1 | 1 | 5.4 | |
| Combined | 3ID1 | 2/2 | 671, 743 | 8.8 | 0.7 | 4.1 | 12.4 | ||
| Chev AA | Oral | 3 | 1/3* | 2450 | 7.5 | 0.5 | 0 | 1.7 | |
| Combined | 3ID1 | 2/2 | 647, 1043 | 5.5 | 0.2 | 0 | 1.7 | ||
| 22A | Suff AA | Oral | 3 | 2/3* | 2274* | 0.5 | 0 | 0 | 0 |
| Combined | 3 | 3/3 | 592 ± 575 | 7.3 | 1.3 | 3.7 | 16.4 | ||
| Chev VV | Oral | 3ID2 | 1/2 | 1865 | 12.3 | 2.1 | 1.3 | 14.4 | |
| Combined | 3ID1 | 2/2 | 259, 479 | 8.9 | 1.5 | 3.1 | 18.3 | ||
| Chev AA | Oral | 3 | 3/3 | 2107 ± 296 | 6.3 | 1 | 4 | 13.5 | |
| Combined | 3 | 3/3 | 687 ± 270 | 7.2 | 0.4 | 4.9 | 16.9 | ||
| 87V | Suff AA | Oral | 3 | 0/3 | - | - | - | - | - |
| Combined | 3 | 3/3R | 991 ± 607 | 9.1 | 0 | 0 | 0 | ||
| Chev VV | Oral | 3 | 0/3 | - | - | - | - | - | |
| Combined | 3 | 3/3 | 687 ± 156 | 14.4 | 0 | 0 | 0 | ||
| Chev AA | Oral | 3 | 0.3 | - | - | - | - | - | |
| Combined | 3 | 3/3 | 509 ± 85 | 10.1 | 0 | 0 | 0 | ||
Standard deviations are given for the ST (in days) to highlight individual variation. Scores for the different compartments indicate the sum of the averaged magnitude of PrPd in each individual tissue or area contained in each compartment. The combined route (in italic) includes the oral, sub-cutaneous and intracerebral routes. All groups, except for the oral ME7 Cheviot sheep, which had a total of five VRQ and five ARQ sheep, initially contained a total of three sheep. However, some sheep died of intercurrent deaths before the first positive animal of that group did and are excluded from the table. The superscript (ID) followed by a number in the column of recipients indicates the number of sheep that died with intercurrent deaths. The superscript R indicates that one animal in that group was polymorphic at codon M112T (R). An asterix (*) designates culled at the end of the experiment. Survival times (in days), which are defined as the interval of days between the inoculation date and the post-mortem, are only given for sheep that were demonstrably infected, and are given as individual values rather than as an average if groups are smaller than three sheep.
Oral infections with ME7, 79A and 22A were successful in 10/11 (91%), 2/9 (22%) and 6/9 (67%) sheep, respectively. The 87V strain did not produce disease by the oral route (0/9). In contrast, 100% AR (9/9) were obtained for the ME7 and 87V infections, and 78% (7/9) and 89% (8/9) for the 79A and 22A strains, respectively, when the combined routes were used. Ataxia was the most frequent clinical sign. Occasional signs included weight loss and pruritus. Dysphagia and behavioral problems were rarely observed.
Overall, sheep developed clinical signs with significantly shorter incubation periods when infected by the combined routes. For the oral ME7 infection, Cheviot VRQ/VRQ (4/4) and ARQ/ARQ (4/4) sheep died with averaged ST of 1146 ± 193dpi and 1388 ± 12dpi, and only two of the three Suffolk sheep developed terminal disease at 763dpi and 1209dpi (the sheep that did not acquire infection -culled at 2465dpi- was methionine/threonine (MT) heterozygote at codon 112). In the combined counterpart, all Suffolk (3/3), VRQ/VRQ (3/3) and ARQ/ARQ (3/3) Cheviot sheep developed terminal disease at 365dpi ± 256, 659dpi ± 100 and 624dpi ± 60, respectively. The longest and shortest ST were found in the single ARQ/ARQ (2450dpi) and VRQ/VRQ (481dpi) Cheviot sheep after oral 79A. Although no evidence of infection was found in the three ARQ/ARQ Suffolk sheep when inoculated orally with 79A (culled at 2457dpi), all of this breed and genotype combination succumbed to terminal disease when challenged by the combined routes (446dpi, 473dpi and 1039dpi). Similarly, 79A combined challenge VRQ/VRQ (2/2) Cheviot sheep developed terminal disease at 671dpi and 743dpi whereas the 79A combined challenge ARQ/ARQ (2/2) Cheviot sheep did so with incubation periods of 647dpi and 1043dpi. In the 22A oral experiment, ARQ/ARQ Cheviot sheep succumbed to terminal end point at 1870dpi, 2012dpi and 2439dpi whereas 374dpi, 843dpi and 843dpi were obtained when challenged by the combined routes. A single Cheviot sheep of the VRQ/VRQ genotype orally challenged with 22A died with clinical disease after 1865dpi whereas two Suffolk sheep, which were culled at the termination of the experiment (2274dpi), showed pathological features consistent with preclinical infection. In the 22A combined challenge, the two VRQ/VRQ Cheviot sheep died with 259dpi and 479dpi, whereas all three Suffolk sheep developed PrPd deposition in their brain at intercurrent death at 87dpi, terminal scrapie at 471dpi and at cull at 1218dpi. For the 87V strain, 100% (9/9) of sheep succumbed to disease when the combined route was used with variable survival times: Cheviot ARQ/ARQ sheep (452dpi, 467dpi and 607dpi) and VRQ/VRQ (544dpi, 663dpi and 854dpi) sheep, and ARQ/ARQ Suffolk sheep (567dpi, 719dpi and 1686dpi).
Accumulation of PrPd in Peripheral Organs
None of the 87V infections resulted in detectable PrPd accumulation in peripheral tissues. Sheep infected with ME7 and 22A showed widespread PrPd accumulation in the LRS and PNS compartments (Table 1), while those infected with 79A showed lesser amounts of PrPd in the same compartments (with the exception of the single orally-infected VRQ/VRQ sheep), which appeared to correlate with the low AR after oral infection. Overall, the combined route gave more widespread PrPd accumulation than the oral route alone. The VRQ/VRQ Cheviot sheep had the most generalized accumulation of PrPd in peripheral tissues regardless of the murine strain, followed by the ARQ/ARQ Suffolk sheep, while the ARQ/ARQ Cheviot sheep showed the least involvement of peripheral tissues (Table 1). Overall, there was a correlation between the GALT and non-GALT compartments: when one was positive usually the other also was, and when one is little positive or negative the other was too. In contrast no such correlation was found when the LRS and PNS compartments were compared, at least for ARQ/ARQ sheep infected with 22A or 79A.
Accumulation of PrPd in the CNS
No correlation was found between the PrPd CNS magnitude or its topography, and the severity of clinical signs or incubation period. Cheviot VRQ/VRQ sheep had the highest magnitude of PrPd in the CNS and peripheral organs but they were not necessarily the ones with the most severe clinical signs (data not shown) or the longest ST (Table 1). In general sheep with widespread distribution of PrPd in LRS and PNS showed high magnitudes of CNS PrPd.
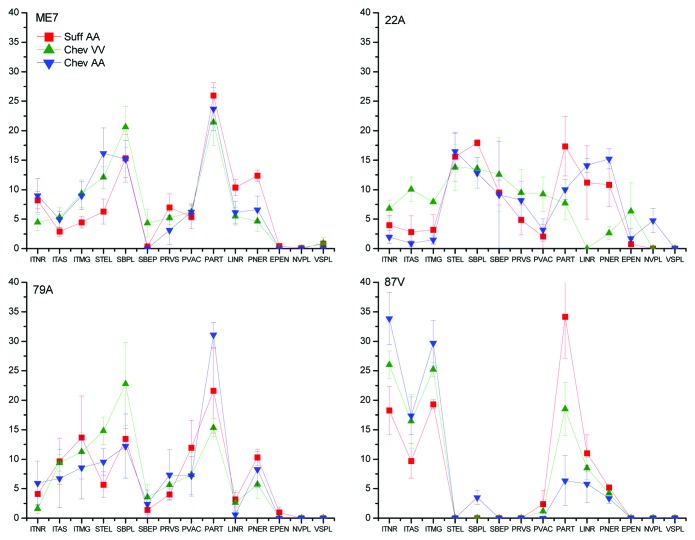
The patterns of PrPd accumulation in the telencephalon (Fig. 1, Table 2) or in the entire brain (Fig. S1) were distinct between strains. However, within a single strain the breed, PrnP genotype, route of inoculation also affected the pathological phenotype. Some PrPd types were more frequent in a particular breed or genotype, and were independent of the strain. For example, Cheviot sheep had higher magnitudes of intraglial types than did Suffolk sheep whereas the neuropil-associated (NRPL) PrPd types predominated in Suffolk sheep (Fig. One and Table 2). The influence of the route of inoculation was evident in ARQ/ARQ Cheviot sheep (Fig. S1), while the disease phenotype of the 87V strain appeared less susceptible to host factors. The 87V strain was characterized by abundant intracellular forms of PrPd and the lack of or minimal astrocyte-associated (GLAS) PrPd forms, and this profile was consistent across breed and genotypes. ME7 infected sheep displayed low amounts of truncated intracellular PrPd forms and abundant NRPL-associated types (Figs. One and 2). However, modulation of the PrPd profile was influenced by the breed. Cheviot sheep displayed high stellate and intraglial levels of PrPd, whereas Suffolk sheep revealed high NRPL-associated PrPd types. In the 22A infections, the predominant pattern was glia-associated extracellular PrPd (Fig. 1). However, the PrPd phenotypes also appeared to be modulated by the Prnp genotype: both ARQ/ARQ Cheviot and Suffolk sheep displayed similar profiles, which were very different from those of VRQ/VRQ Cheviot sheep (Fig. S1). Higher levels of intraglial PrPd types compared with intraneuronal accumulations were characteristic of 79A infections (Fig. 1). Nevertheless, modulation of the PrPd pattern was observed according to the route of inoculation in Cheviot ARQ/ARQ sheep, which displayed unusually high intraneuronal types when orally-dosed with 79A and lower intraglial types when inoculated by the combined route (Fig. S1).
Figure 1. Influence of the host breed and genotype on the PrPd profiles when inoculated with ME7, 79A, 22A and 87V by the combined route. The x axis of the above graph refers to different morphological types of PrPd labeling identified by immunohistochemistry. PrPd types are: ITNR, intraneuronal; ITAS, intrastrocytic; ITMG, intramicroglial; STEL, stellate; SBPL, subpial; SBEP, subependymal; PRVS, perivascular; PVAC, perivacuolar; PART, fine particulate-coalescing; LINR, linear; PNER, perineuronal; EPEN, ependymal; NVPL, non vascular plaques; VASC, vascular plaques.
Table 2. Summary of the PrPd cell distribution according to strain and host.
| Strain | Host | ITNR | ITGL | GLAS | NRPL | EPEN | NVPL | VSPL |
|---|---|---|---|---|---|---|---|---|
| ME7 | Suff AA | 23 ± 2 | 10 ± 2 | 19 ± 0 | 44 ± 5 | 1 ± 2 | 0.3 ± 0 | 2 ± 2 |
| Chev VV | 14 ± 9 | 23 ± 5 | 29 ± 8 | 31 ± 18 | 0.0 | 0.0 | 2 ± 2 | |
| Chev AA | 23 ± 16 | 18 ± 4 | 23 ± 2 | 35 ± 21 | 0.0 | 0.0 | 0.0 | |
| 79A | Suff AA | 12 | 32 | 21 | 35 | 3 | 0.0 | 0.0 |
| Chev VV | 4 ± 3 | 37 ± 10 | 35 ± 13 | 25 ± 0 | 0.0 | 0.0 | 0.0 | |
| Chev AA | 18 ± 15 | 23 ± 20 | 21 ± 23 | 37 ± 13 | 0.0 | 0.0 | 0.0 | |
| 22A | Suff AA | 22 | 23 | 23 | 29 | 2 | 0.0 | 0.0 |
| ChevVV | 19 ± 6 | 26 ± 8 | 32 ± 1 | 10 ± 5 | 13 ± 19 | 0.0 | 0.0 | |
| Chev AA | 5 ± 5 | 4 ± 2 | 34 ± 12 | 44 ± 15 | 4 ± 6 | 11 ± 15 | 0.0 | |
| 87A | Suff AA | 44 | 29 | 1 | 26 | 0.0 | 0.0 | 0.0 |
| Chev VV | 45 | 36 | 0.5 | 18 | 0.0 | 0.0 | 0.0 | |
| Chev AA | 52 | 37 | 1 | 10 | 0.0 | 0.0 | 0.0 |
The scores shown are average short PrPd profiles grouped according to the host and to the strain but not the route. The intraneuronal (ITNR) score (in percentage) is the amount of PrPd found within the perikarya of neurons relative to that found elsewhere. The intraglial (ITGL) type includes the intra-astrocytic and intra-microglial. The glia-associated PrPd (GLAS) includes the stellate, subpial, subependymal, perivascular and perivacuolar PrPd types. The neuropil-associated PrPd includes the particulate, linear and perineuronal. Plaque-like PrPd was scored as associated with plaques (vascular plaques [VSPL]) or not (non-vascular plaques [NVPL]). The host was either Suffolk (Suff) or Cheviot (Chev) sheep and of ARQ/ARQ (AA) or VRQ/VRQ (VV) Prnp genotype
Although there were some similarities in the PrPd profiles of ME7 and 79A strains (Fig. 1), Suffolk sheep with 79A displayed high intraglial and low linear PrPd types, but the reverse was the case for the ME7 strain. Coalescing plaque-like types (NVPL) were present in the white matter of 22A inoculated sheep (Fig. 2) but these were absent following ME7 infection. In contrast, ME7-infected VRQ/VRQ Cheviot and ARQ/ARQ Suffolk sheep displayed vascular plaques (VSPL, Figure 1 and Table 2). Suffolk sheep displayed high intraglial PrPd and weak stellate PrPd when infected with 79A (Fig. 2), which was in contrast with the patterns observed after 22A infection.
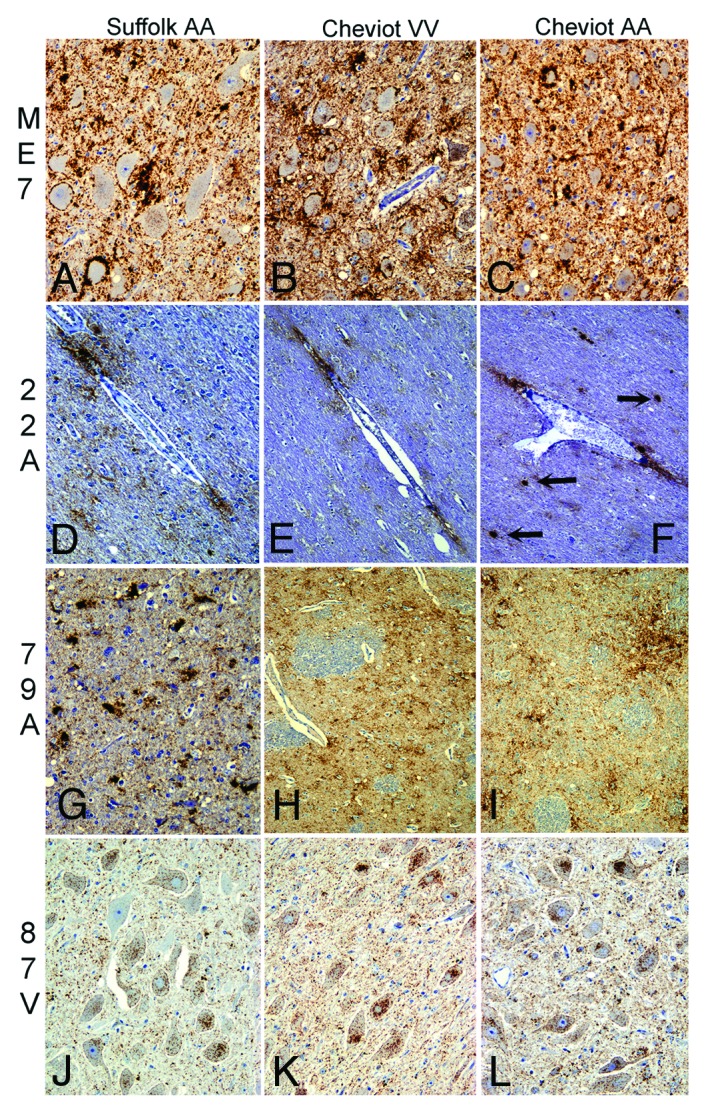
Figure 2. Main PrPd features in brains of sheep infected with ME7, 79A, 22A, or 87V. In ME7 infections (A-C), neuropil-associated PrPd types predominated, like-wise perineuronal, coalescing or diffuse punctate. A prominent characteristic of the 22A agent in sheep (D-F) was the glia-associated PrPd types like the perivascular and perivacuolar in the white matter with some plaque-like depositions (arrows) in particular in Cheviots of the ARQ/ARQ genotype. 79A infections (G-I) resulted in a combination of intraglial-coalescing pattern (G) with stellate-like labeling (H, I). Sheep infected with 87V (J-L) unequivocally showed most PrPd associated with neurons or glial cells. Immunohistochemistry for PrPd using R145 antibody. Magnifications: x10 (D-F, H and I); x20 (A-C, G, J-L).
IHC results on epitope mapping indicated that all infected sheep behave like scrapie cases with the exception of 87V-infected Cheviot sheep, which showed truncation sites similar to those of sheep infected with BSE or CH1641 (Fig. 3).
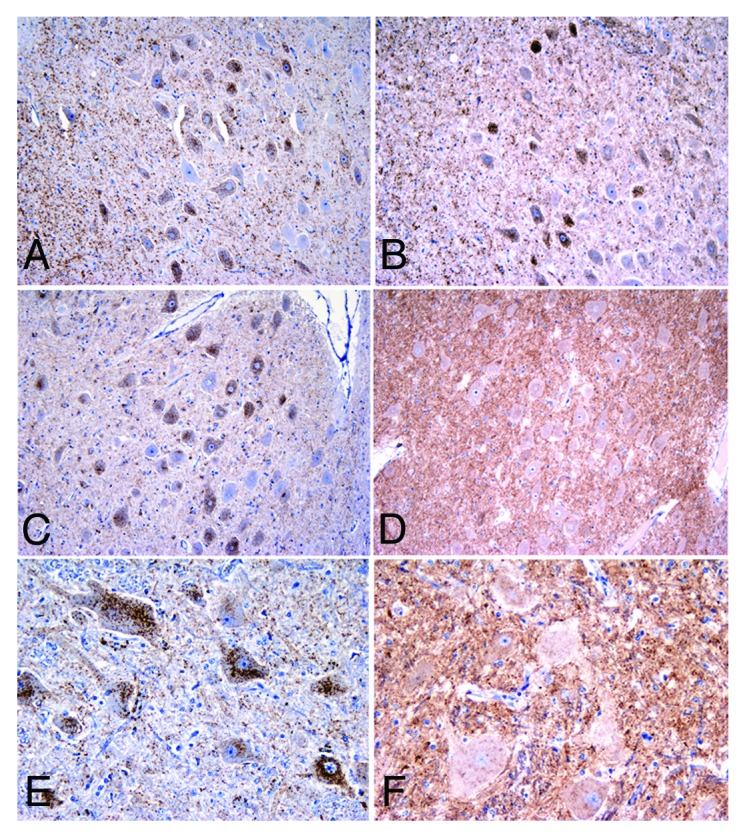
Figure 3. Epitope mapping results for the 87V infections in the dorsal motor nucleus of the vagus nerve of sheep. Note that Suffolk ARQ/ARQ sheep displayed intraneuronal immunolabelling with C-terminal antibodies such as Bar224 (A) and N-terminal antibodies such as P4 (B). Instead, Cheviot VRQ/VRQ (C, D) and ARQ/ARQ (E, F) sheep displayed the intraneuronal type with Bar224 (C, E) but not with P4 (D, F), a feature that is characteristic of strains like BSE and CH1641 when experimentally passaged in sheep. Immunohistochemistry for PrPd using Bar224 or P4 antibodies. Magnifications: x10 (A-D), x20 (E, F).
Glycoform Ratio of Sheep PrPres Originated from Murine Strains
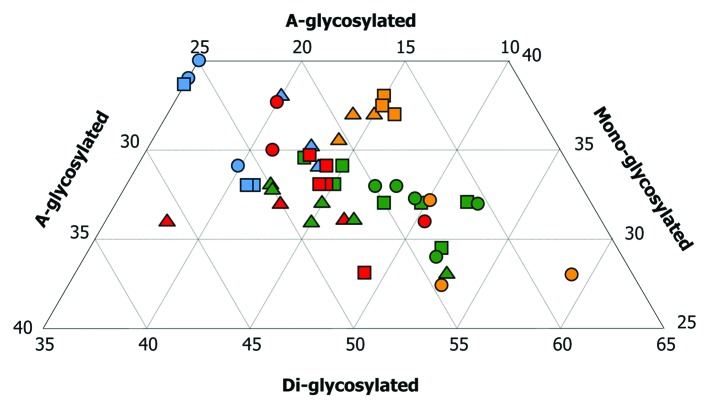
An overview of the glycoprofiles of PrPres is shown in Figure 4. For sheep infected with ME7 or 22A the di-glycosylated band was the most prominent. Sheep infected with 79A displayed a different glycoprofile, with little di-glycosylated PrPres and higher percentages of the mono-glycosylated band. 87V infection in Cheviot sheep produced prominent mono-glycosylated bands, similar to 79A-infected sheep, but with lesser and lower a-glycosylated bands.
Figure 4. Triplot analyses from brains of sheep infected with cloned murine strains. Colors denote strain whereas symbol denotes breed and genotype. This plot shows total number of animals infected either by a single oral dose or a combined challenge. Eighteen sheep infected with ME7 (green) gave a positive result in Western-blot. Nine did with 79A (blue). Eleven sheep infected with 22A (red) gave a positive result in WB. All nine sheep infected with 87V (yellow) displayed strong signals of PrPres in their brains. Note that the PrPres associated with ovine ME7 showed higher amounts of the di-glycosylated band, followed by intermediate amounts of the mono-glycosylated band and low amounts of the a-glycosylated band. The characteristics of PrPres in ovine 22A infections are similar to ME7 ones but with a relatively lower and higher amount of the di- and a-glycosylated bands, respectively. The mono- and the a-glycosylated bands of PrPres are significantly higher after 79A infections. This pattern was similar to that observed for the PrPres associated with the 87V strain in Cheviot sheep of the VRQ/VRQ (triangles) and ARQ/ARQ (squares) genotypes, but not in the Suffolk sheep of the ARQ/ARQ genotype (circles), in which, the di-glycosylated band predominated.
Discussion
The species barrier is known to play a major role in interspecies transmissions of scrapie resulting in prolongation of ST and incomplete AR at first passage. Potentially, the species barrier may predispose toward the emergence of new strains. The present series of experiments demonstrated successful transmissions of cloned scrapie strains ME7, 79A, 22A and 87V from mice to their putative natural sheep host. Sheep were most susceptible to the ME7 strain regardless of the route of challenge but they were completely or significantly resistant to murine 87V and 79A infection by the oral route. Nevertheless, the species barrier to 87V and 79A was overcome when routes other than the oral were used. Another factor contributing to prolonged ST in ARQ/ARQ Suffolk sheep was the MT polymorphism at codon 112.32,33 Polymorphic sheep inoculated by the combined route (with ME7, 79A or 87V) displayed prolonged ST (608dpi, 1039dpi and 1686dpi, respectively) when compared with the non-polymorphic ones and these polymorphic sheep contribute to the high standard deviations of ST in Suffolk sheep compared with Cheviot sheep. The two polymorphic sheep inoculated with ME7 or 79A by the oral route (Table 1) did not succumb to disease. The ST was substantially reduced (by 1/3 or ¼ when compared with the oral route) for all strains administered by intracerebral challenge. Interestingly, those strains that transmitted best by the oral route (ME7 and 22A) were also those that replicated most efficiently in peripheral tissues.
Variation in pathological phenotype, that is, the pattern of histological lesions and PrPd accumulation in scrapie affected mice, is related to the interaction between the host (genotype, age, sex, breed) and the agent strain (passage history, dose and route of inoculation, donor genotype and organ used for preparing inoculum).20 The present study used the PrPd phenotype to monitor interactions that might occur between the pathogen and the host. The 87V strain displayed the most consistent profiles across sheep of different breeds and genotypes suggesting that it is highly stable upon inter-species transmission. Using epitope mapping, the 87V strain transmitted to Cheviot sheep showed N-terminal PrP truncation characteristics similar to BSE- or CH1641. All other strain-host combinations behaved like classical scrapie. The 87V strain suffers frequently from a strain breakdown upon transmissions in mice and is considered unstable in this species,28 but the significance of the emergence of a BSE-like or CH1641-like strain in the 87V challenged sheep is unclear. According to previous cross-species experiments,14-16,35 passage of this sheep strain variant back into their original hosts, in this case the VM mouse line, should allow the properties of 87V to re-emerge. The experiment challenge of the above sheep 87V strain back into VM mice is still in progress and it therefore remains to be determined whether a new strain has been established.36
According to the protein-only hypothesis, the properties of any given prion isolate are determined, at least in part, by the sequence of PrP encoded within both the inoculated recipient and donor animals. Intra-species transmission of natural scrapie shows a strong interaction between the amino acid sequence of the sheep donor and of the recipient.37 The current results may suggest an influence of the genetic background of the donor and of the recipient. Only the Prnpaa allele, used for the ME7 and 79A transmissions, but not the Prnpbb allele used for the 87V and 22 transmissions, might have a disease effect as the former resulted in significant variation in disease phenotype although not necessarily with shorter ST. Moreover, the Prnp genotype of the recipient seems to influence both the nature and magnitude of the PrPd profile regardless of the scrapie strain/mouse line. VRQ/VRQ sheep displayed the highest magnitudes of PrPd in the CNS and LRS tissues with similar incubation periods (or even shorter) than ARQ/ARQ sheep did. Nevertheless, the PrPd accumulated in brains from VRQ/VRQ sheep is usually associated with glial cells, both intra- and extracellular whereas that observed in brains from ARQ/ARQ Suffolk sheep is more associated with neurites (Table 2).
Prion strain biology is hampered by the absence of specific molecular or structural characteristics that may be used to define strains. When a new disease phenotype is seen following passage, it has been difficult therefore to differentiate between host and strain effects, and in particular to know whether there has been strain mutation or adaptation or whether the new host has selected from a mixture of pre-existing strains in the donor.38-40 Our results confirm that not all the pathological features of cloned murine strains are transferred to the sheep host, in which phenotypic variability appears to be dependant of host factors such as Prnp genotype or breed. Although not definitive as the inocula were cloned this phenotypic variability would not appear to be due to the existence of a mixture of strains in the infectious material given to sheep.
Our studies show several examples of this PrPd profile shift. Amyloid plaques are abundant and characteristic of murine 87V, occur inconsistently in ME7 and 22A infections, and are absent in mice challenged with 79A.41 However, plaques were absent in 79A and 87V infected sheep brains and only vascular plaques were occasionally present in ME7 infections of VRQ/VRQ Cheviot and ARQ/ARQ Suffolk sheep, while non-vascular plaques were found in Cheviot ARQ/ARQ sheep infected with 22A. Thus it would appear that the host can have both qualitative and quantitative influence of some PrPd types. In contrast, some disease characteristics that were intrinsic to each cloned scrapie strain were apparently transferred to sheep. Thus, different sheep PrP proteins arising from different Prnp haplotypes did not necessarily alter the specific brain PrPd profile or LRS phenotype. By IHC, for instance particulate labeling types predominated in 87V and ME7; the perineuronal types in 79A infections, and high frequencies of the perineuronal and linear types in 22A infections regardless of the genetic background of the host. In addition, some tissue tropisms appeared common to both murine and ovine hosts: the 87V agent did not replicate in the periphery of any host and the 79A only did exceptionally. Similarly, the Western-blot analyses revealed distinct patterns of PrPres glycoform ratio which showed better correlation with strain rather than sheep breeds or genotypes. Therefore it seems reasonable to suggest that some features of the disease phenotype might be strain-encoded and propagated in the new host.
As shown in Table 2, different sheep scrapie isolates showed different proportions of PrPd types. Thus, 22A in sheep displayed the highest glia-associated PrPd depositions, the 87V the intracellular ones, the ME7 the neuropil one and the 79A the intraglial. As suggested for other sheep TSEs27,42 these accumulation patterns may reflect strain specific differences in the propagation efficiency in different cell types, and differences in PrPd trafficking and processing pathways.
These results show that i) some properties of cloned strains are lost upon interspecies transmissions resulting in new characteristics in the new host, ii) some strain properties remain distinct and consistent on transmission from mice to sheep of different breeds and genotypes, but ii) a selective process operates among sheep of different Prnp genotypes or breed; each likely to favor the selection of different cellular tropisms and processing pathways for PrPd when compared with the original cloned strain. Without further data on the re-isolation of the sheep adapted strains back into mice it is not yet possible to know whether new strains have been created from the original cloned murine strains by interspecies passage. However, the profound phenotypic changes found on passage of the cloned murine scrapie strains into sheep combined with the marked phenotypic changes associated with different sheep host factors suggest that greater caution needs to be exercised in the use of rodents to bioassay strains in natural hosts.
Materials and Methods
Selection of Cloned Mouse-adapted Scrapie Strains and Transmission to Sheep
Cloned murine scrapie strains are defined by their i) relative incubation periods in different inbred mouse lines ii) vacuolar lesion profiles produced in the brain of infected mice, and iii) stability upon serial passages in mice in terms of reproducing such properties. We selected the murine scrapie strains ME7, 79A, 22A and 87V for transmission into sheep based on their differences in origin and distinct stability and pathology.14,28,29 Briefly, the ME7 strain originated from a natural scrapie isolate in Suffolk sheep following passage into C57BL mouse lines. The 79A strain originated from serial passages of the “drowsy” experimental scrapie in goats to C57BL mice. The 22A strain, from Cheviot sheep experimentally infected with SSBP/1 transmitted into VM mouse line, and the 87V, from natural scrapie in crossbred Cheviot and Border Leicester sheep also into VM mice.
A total of 220 mice per strain were inoculated intracerebrally with 20μl of a 10% brain homogenate at 8 weeks of age in order to generate enough inoculum to perform the sheep transmissions. Following previous experiments17 mouse inoculations with the ME7 or the 79A strain were performed in mice homozygous for the Prnpaa (formerly known as s7) whereas those with the 22A or the 87V strain used mice homozygous for the Prnpbb allele (formerly known as p7) of the Prnp (Sinc) gene. Mice were left to develop clinical disease and killed at terminal end-point. Mouse brains were removed immediately after death, pooled as a 20% homogenate and kept at –80°C until required. Representative brains not used for the inocula were examined by histology and IHC to confirm that specific strain characteristics were maintained in this further mouse serial passage (data not shown).
A total of 24 Suffolk and 52 Cheviot 3–5 mo-old sheep were challenged. Sheep were homozygous for either the VRQ or ARQ Prnp allele (Table 1). Each sheep received either a single oral dose of 25ml of 20% suspension of mouse brain infected with the appropriate strain of scrapie or a combination of that oral dose and additional injections of 1ml and 2ml of a 10−1 dilution of the mouse brain homogenates by the intracerebral and subcutaneous routes, respectively. Five additional sheep of different Prnp genotypes were introduced as environmental controls. Sheep inoculated with different mouse strains were kept in separate boxes in order to avoid cross-contamination. Monitoring for clinical signs of scrapie was performed daily in order to detect signs of i) locomotor dysfunction/ataxia, ii) behavioral changes, iii) dysphagia, iv) pruritus, and v) weight loss. These clinical abnormalities were scored from 0 to 3 (data not shown). Clinical end point was reached when any one of those five groups of signs was given a score of 3 or when a combined score of 8 was reached. At that point, sheep were killed by an intravenous overdose of barbiturates and necropsies performed.
All animal procedures complied with the Animals (Scientific Procedures) Act 1986 and were approved by the ethics committee at Moredun Research Institute.
At necropsy, brains were removed rapidly from the skull and sliced sagittally; one hemi-brain was immersed in 10% buffered formalin for 8–12 d and processed for morphological studies, and the other hemi-brain was rapidly frozen and stored at -80°C until required for biochemical examinations. A total of 38 tissue samples representative of different areas of the central nervous system (CNS), nerves and ganglia of the peripheral nervous system (PNS), gut-associated lymphoid tissues (GALT) and secondary lymphoid organs (LRS) were taken. All formalin-fixed samples were processed for paraffin-wax embedding according to standard procedures. For PrPd IHC, sections (4 μm) were mounted on glass microscope slides and subjected to immunolabelling as described previously.5 Briefly, antigen retrieval included immersion in 98% formic acid for 15 min at 24°C, followed by autoclaving in 0.2% citrate buffer (pH 6.2) at 121°C for 30 min. Tissues were incubated overnight at 24°C with monoclonal antibody R145 (diluted 1:4000). The IHC procedure was completed by an immunoperoxidase method (Vector-elite ABC kit; Vector Laboratories, Peterborough, UK) with diaminobenzidine (DAB, Sigma-Aldrich Company Ltd, Gillingham, UK) as a substrate, and sections were finally counterstained with Mayer’s hematoxylin. For the epitope mapping approach sections were incubated with primary antibody P4 (dilution 1:12000) as described previously.11 Sections from positive-control and negative-control tissue blocks were included in each IHC run to ensure consistency in the sensitivity and specificity of the IHC procedure, respectively.
Assessment of PrPd Detected by IHC
The PNS structures examined included nerve cell ganglia (trigeminal, nodose, stellate, sympathetic chain and cranial mesenteric ganglion), plexi of the enteric nervous system (ENS) at several gut segments (jejunum, distal ileum and colon) and peripheral nerves (vagus and sciatic). Intra- and extra-cellular PrPd deposits in those structures were subjectively scored from 0 to 3. The GALT comprised the Peyer’s patches at jejunal, ileal and colon intestinal segments, and the recto-anal mucosal associated lymphoid tissue (RAMALT). The LRS tissues comprised structures enclosed in the head (palatine tonsil and retropharyngeal and prescapular lymph nodes), in the gut (the proximal and the distal jejunal lymph nodes) and in the periphery (the spleen and the popliteal lymph node). The degree of involvement of the GALT and LRS tissues was determined as a factor of the proportion of lymphoid follicles with PrPd accumulation, and of the magnitude and distribution of the PrPd immunolabelling in positive follicles as previously detailed.30
The morphometric examination of the sheep brain was based on the short PrPd profiling in several divisions of the telencephalon.6 The PrPd types subjectively scored from 0 (absence) to 3 (severe) included intracellular and extracellular types, ependymal, non-vascular plaques and vascular plaques. Average values for each PrPd type in the telencephalon were converted into percentage figures of the total PrPd magnitude, and its graphical representation constituted the “short PrPd profile.”6 In addition morphological PrPd types were subjectively scored from 0 to 3 in the seven brain areas and selected spinal cord segments, so that for each animal, the total magnitude of PrPd was expressed as the sum of all CNS topographical individual scores.
Western Blotting (WB)
Samples of caudal medulla were analyzed by WB with monoclonal antibody SAF84 (R-biopharm, Darmstadt, Germany) as described previously.31 Briefly, samples were homogenized at 20% (w/v) in lysis buffer and frozen at -20°C overnight. Lysates were further diluted to 10% in lysis buffer before centrifugation at 100 × g for 5 min and then the supernatant was aspirated to new tubes. 200 μl of lysate were treated with 20% sarkosyl for 20 min and then digested with 50 μg/ml proteinase K solution for 1 h, both at 37°C and with agitation. Digestion was terminated by adding 1 mM Pefabloc SC (Roche Diagnostics, Burgess Hill, West Sussex, UK). Samples were then centrifuged at 20,000 × g for 1 h at 4°C, the supernatants discarded and the pellets resuspended in 45 μl 2 × SB (Invitrogen, Paisley, UK) containing 5 μl of 10 × sample reducing agent (Invitrogen, Paisley, UK). Samples were heated at 100°C for 10 min and once cooled pulsed for 5 sec at 5000rpm. SDS-PAGE was performed on 10 μl of sample on 4–12% Bis-Tris NuPAGE gels (Invitrogen, Paisley, UK) at 150 V for 1 h. Proteins were electrotransferred onto Hybond P PVDF membrane (GE Healthcare, Chalfont St Giles, Buckinghamshire, UK) at 30 V for 1 h. Non-specific antigen binding on the membrane was blocked by soaking in 2% non-fat milk in trizma buffered saline (TBS) with 0.1% Tween20 (Sigma Chemical Company, Poole, Dorset, UK) and probed with SAF 84. Signal was detected using Super Signal West Dura Chemiluminescent Substrate (Pierce, Rockford, IL, USA) and a Kodak IS440 image station (Labtech International Ltd., Lewes, UK). The relative intensities of the di-, mono- and aglycosyl bands of PrPres were determined by scanning the western blot image using Kodak 1D Image Analysis Software.
Supplementary Material
Glossary
Abbreviations:
- ME7
79A, 22A, 87V, cloned murine strains
- PrP
Prion protein
- Prnp
Prion protein gene
- TSEs
Transmissible spongiform encephalopathies
- BSE
Bovine spongiform encephalopathy
- PrPc
Cellular prion protein
- PrPd
Diseased-associated PrP
- PrPSc
PrP scrapie
- PrPres
Protease-resistant PrP
- IHC
Immunohistochemistry
- CH1641
Experimental sheep scrapie strain
- VRQ
Valine, Arginine and Glutamine at codons 136, 154 and 171, respectively of the PrP gene
- ARQ
Alanine, Arginine and Glutamine at codons 136, 154 and 171, respectively of the PrP gene
- C57Bl and VM
mouse lines
- CNS
central nervous system
- PNS
peripheral nervous system
- GALT
gut-associated lymphoid tissues
- non-GALT
secondary lymphoid organs not associated with the gut
- ENS
enteric nervous system
- LRS
lymphoreticular system
- WB
western-blotting
- AR
attack rates
- ST
survival times
- dpi
days post inoculation
- M112T or R
methionine and thiamine at codon 112 of the prion protein gene
- ID
intercurrent death
- ITNR
intraneuronal
- ITGL
intraglial
- GLAS
glia-associated PrPd
- NRPL
neuropil-associated PrPd
- VSPL
vascular plaques
- NVPL
non-vascular plaques
- Suff
Suffolk breed sheep
- Chev
Cheviot breed sheep
- ITAS
intrastrocytic
- ITMG
intramicroglial
- STEL
stellate
- SBPL
subpial
- SBEP
subependymal
- PRVS
perivascular
- PVAC
perivacuolar
- PART
fine particulate-coalescing
- LINR
linear
- PNER
perineuronal
- EPEN
ependymal
AUTHOR:
Please cite reference 34 in the text. Citations for references 28-31 appear to be out of order and can be corrected upon inclusion of the reference 34 citation.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
The authors greatly appreciate the help provided by Prof. Nora Hunter (Roslin Institute and Royal (Dick) School of Veterinary Studies) with the supply of the cloned murine strains used to generate the inocula. The excellent technical support in immunohistochemistry from Mrs. Lynne Algar, Ms. Ann Dunachie and Ms. Maria Oliva. Finally, the authors thank Dr. James Hope’s critical reading of the manuscript.
References
- 1.Prusiner SB, Bolton DC, Groth DF, Bowman KA, Cochran SP, McKinley MP. . Further purification and characterization of scrapie prions. Biochemistry 1982; 21:6942 - 50; http://dx.doi.org/ 10.1021/bi00269a050; PMID: 6818988 [DOI] [PubMed] [Google Scholar]
- 2.Sisó S, González L, Blanco R, Chianini F, Reid HW, Jeffrey M, et al. . Neuropathological changes correlate temporally but not spatially with selected neuromodulatory responses in natural scrapie. Neuropathol Appl Neurobiol 2011; 37:484 - 99; http://dx.doi.org/ 10.1111/j.1365-2990.2010.01152.x; PMID: 21114681 [DOI] [PubMed] [Google Scholar]
- 3.González L, Martin S, Jeffrey M. . Distinct profiles of PrP(d) immunoreactivity in the brain of scrapie- and BSE-infected sheep: implications for differential cell targeting and PrP processing. J Gen Virol 2003; 84:1339 - 50; http://dx.doi.org/ 10.1099/vir.0.18800-0; PMID: 12692301 [DOI] [PubMed] [Google Scholar]
- 4.González L, Martin S, Houston FE, Hunter N, Reid HW, Bellworthy SJ, et al. . Phenotype of disease-associated PrP accumulation in the brain of bovine spongiform encephalopathy experimentally infected sheep. J Gen Virol 2005; 86:827 - 38; http://dx.doi.org/ 10.1099/vir.0.80299-0; PMID: 15722546 [DOI] [PubMed] [Google Scholar]
- 5.González L, Martin S, Begara-McGorum I, Hunter N, Houston F, Simmons M, et al. . Effects of agent strain and host genotype on PrP accumulation in the brain of sheep naturally and experimentally affected with scrapie. J Comp Pathol 2002; 126:17 - 29; http://dx.doi.org/ 10.1053/jcpa.2001.0516; PMID: 11814318 [DOI] [PubMed] [Google Scholar]
- 6.Sisó S, Jeffrey M, Martin S, Chianini F, Dagleish MP, González L. . Characterization of strains of ovine transmissible spongiform encephalopathy with a short PrPd profiling method. J Comp Pathol 2010; 142:300 - 10; http://dx.doi.org/ 10.1016/j.jcpa.2009.12.003; PMID: 20153480 [DOI] [PubMed] [Google Scholar]
- 7.Ligios C, Jeffrey M, Ryder SJ, Bellworthy SJ, Simmons MM. . Distinction of scrapie phenotypes in sheep by lesion profiling. J Comp Pathol 2002; 127:45 - 57; http://dx.doi.org/ 10.1053/jcpa.2002.0589; PMID: 12354545 [DOI] [PubMed] [Google Scholar]
- 8.Spiropoulos J, Casalone C, Caramelli M, Simmons MM. . Immunohistochemistry for PrPSc in natural scrapie reveals patterns which are associated with the PrP genotype. Neuropathol Appl Neurobiol 2007; 33:398 - 409; http://dx.doi.org/ 10.1111/j.1365-2990.2007.00800.x; PMID: 17617872 [DOI] [PubMed] [Google Scholar]
- 9.Ersdal C, Ulvund MJ, Espenes A, Benestad SL, Sarradin P, Landsverk T. . Mapping PrPSc propagation in experimental and natural scrapie in sheep with different PrP genotypes. Vet Pathol 2005; 42:258 - 74; http://dx.doi.org/ 10.1354/vp.42-3-258; PMID: 15872372 [DOI] [PubMed] [Google Scholar]
- 10.Begara-McGorum I, González L, Simmons M, Hunter N, Houston F, Jeffrey M. . Vacuolar lesion profile in sheep scrapie: factors influencing its variation and relationship to disease-specific PrP accumulation. J Comp Pathol 2002; 127:59 - 68; http://dx.doi.org/ 10.1053/jcpa.2002.0558; PMID: 12354546 [DOI] [PubMed] [Google Scholar]
- 11.Jeffrey M, Martin S, González L, Ryder SJ, Bellworthy SJ, Jackman R. . Differential diagnosis of infections with the bovine spongiform encephalopathy (BSE) and scrapie agents in sheep. J Comp Pathol 2001; 125:271 - 84; http://dx.doi.org/ 10.1053/jcpa.2001.0499; PMID: 11798244 [DOI] [PubMed] [Google Scholar]
- 12.González L, Sisó S, Monleón E, Casalone C, van Keulen LJ, Balkema-Buschmann A, et al. . Variability in disease phenotypes within a single PRNP genotype suggests the existence of multiple natural sheep scrapie strains within Europe. J Gen Virol 2010; 91:2630 - 41; http://dx.doi.org/ 10.1099/vir.0.022574-0; PMID: 20538906 [DOI] [PubMed] [Google Scholar]
- 13.Jeffrey M, González L, Chong A, Foster J, Goldmann W, Hunter N, et al. . Ovine infection with the agents of scrapie (CH1641 isolate) and bovine spongiform encephalopathy: immunochemical similarities can be resolved by immunohistochemistry. J Comp Pathol 2006; 134:17 - 29; http://dx.doi.org/ 10.1016/j.jcpa.2005.06.005; PMID: 16324707 [DOI] [PubMed] [Google Scholar]
- 14.Dickinson AG. Scrapie in sheep and goats. In: Kimberlin RH, ed. Slow Virus Diseases of Animals and Man. Amsterdam, North-Holland, 1976: 209-39. [Google Scholar]
- 15.Kimberlin RH. Early events in the pathogenesis of scrapie in mice: biological and biochemical studies. In: Prusiner SB and Hadlow WJ. Slow Transmissible Diseases of the Nervous System. Vol. 2. New York: Academic Press, 1979:33-54. [Google Scholar]
- 16.Béringue V, Vilotte JL, Laude H. . Prion agent diversity and species barrier. Vet Res 2008; 39:47; http://dx.doi.org/ 10.1051/vetres:2008024; PMID: 18519020 [DOI] [PubMed] [Google Scholar]
- 17.Bruce ME, McConnell I, Fraser H, Dickinson AG. . The disease characteristics of different strains of scrapie in Sinc congenic mouse lines: implications for the nature of the agent and host control of pathogenesis. J Gen Virol 1991; 72:595 - 603; http://dx.doi.org/ 10.1099/0022-1317-72-3-595; PMID: 1672371 [DOI] [PubMed] [Google Scholar]
- 18.Fraser H, Dickinson AG. . The sequential development of the brain lesion of scrapie in three strains of mice. J Comp Pathol 1968; 78:301 - 11; http://dx.doi.org/ 10.1016/0021-9975(68)90006-6; PMID: 4970192 [DOI] [PubMed] [Google Scholar]
- 19.Fraser H, Dickinson AG. . Scrapie in mice. Agent-strain differences in the distribution and intensity of grey matter vacuolation. J Comp Pathol 1973; 83:29 - 40; http://dx.doi.org/ 10.1016/0021-9975(73)90024-8; PMID: 4199908 [DOI] [PubMed] [Google Scholar]
- 20.Fraser H. The pathology of natural and experimental scrapie. In: Kimberlin RH, ed. Slow Virus Diseases of Animals and Man. Amsterdam, North-Holland, 1976:267-303. [Google Scholar]
- 21.Bruce ME, Fraser H, McBride PA, Scott JR, Dickinson AG. The basis of strain variation in scrapie. In: Prusiner SB, Collinge J, Powell J and Anderton B, eds. Prion Diseases of Humans and Animals. Chichester: Ellis Horwood, 1992:497-508. [Google Scholar]
- 22.Bruce ME, Will RG, Ironside JW, McConnell I, Drummond D, Suttie A, et al. . Transmissions to mice indicate that ‘new variant’ CJD is caused by the BSE agent. Nature 1997; 389:498 - 501; http://dx.doi.org/ 10.1038/39057; PMID: 9333239 [DOI] [PubMed] [Google Scholar]
- 23.Bruce ME, Boyle A, Cousens S, McConnell I, Foster J, Goldmann W, et al. . Strain characterization of natural sheep scrapie and comparison with BSE. J Gen Virol 2002; 83:695 - 704; PMID: 11842264 [DOI] [PubMed] [Google Scholar]
- 24.Bruce ME. . TSE strain variation. Br Med Bull 2003; 66:99 - 108; http://dx.doi.org/ 10.1093/bmb/66.1.99; PMID: 14522852 [DOI] [PubMed] [Google Scholar]
- 25.Prusiner SB. . Novel proteinaceous infectious particles cause scrapie. Science 1982; 216:136 - 44; http://dx.doi.org/ 10.1126/science.6801762; PMID: 6801762 [DOI] [PubMed] [Google Scholar]
- 26.Vorberg I, Priola SA. . Molecular basis of scrapie strain glycoform variation. J Biol Chem 2002; 277:36775 - 81; http://dx.doi.org/ 10.1074/jbc.M206865200; PMID: 12138171 [DOI] [PubMed] [Google Scholar]
- 27.Zlotnik I, Rennie JC. . Experimental transmission of mouse passaged scrapie to goats, sheep, rats and hamsters. J Comp Pathol 1965; 75:147 - 57; http://dx.doi.org/ 10.1016/0021-9975(65)90005-8; PMID: 14319384 [DOI] [PubMed] [Google Scholar]
- 28.Bruce ME, Dickinson AG. Biological stability of different classes of scrapie agent. In: Slow transmissible diseases of the nervous system. Prusiner SB and Hadlow WJ, eds. II Vol. London: Academic Press, Inc, 1979:71-86. [Google Scholar]
- 29.Outram GW. The pathogenesis of scrapie in mice. In: Kimberlin RH, ed. Slow Virus Diseases of Animals and Man. Amsterdam, North-Holland, 1976: 325-57. [Google Scholar]
- 30.Martin S, González L, Chong A, Houston FE, Hunter N, Jeffrey M. . Immunohistochemical characteristics of disease-associated PrP are not altered by host genotype or route of inoculation following infection of sheep with bovine spongiform encephalopathy. J Gen Virol 2005; 86:839 - 48; http://dx.doi.org/ 10.1099/vir.0.80364-0; PMID: 15722547 [DOI] [PubMed] [Google Scholar]
- 31.Dagleish MP, Hamilton S, González L, Eaton SL, Steele P, Finlayson J, et al. . Digestion and transportation of bovine spongiform encephalopathy-derived prion protein in the sheep intestine. J Gen Virol 2010; 91:3116 - 23; http://dx.doi.org/ 10.1099/vir.0.025049-0; PMID: 20826616 [DOI] [PubMed] [Google Scholar]
- 32.Ikeda T, Horiuchi M, Ishiguro N, Muramatsu Y, Kai-Uwe GD, Shinagawa M. . Amino acid polymorphisms of PrP with reference to onset of scrapie in Suffolk and Corriedale sheep in Japan. J Gen Virol 1995; 76:2577 - 81; http://dx.doi.org/ 10.1099/0022-1317-76-10-2577; PMID: 7595361 [DOI] [PubMed] [Google Scholar]
- 33.Laegreid WW, Clawson ML, Heaton MP, Green BT, O’Rourke KI, Knowles DP. . Scrapie resistance in ARQ sheep. J Virol 2008; 82:10318 - 20; http://dx.doi.org/ 10.1128/JVI.00710-08; PMID: 18632863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kimberlin RH, Cole S, Walker CA. . Temporary and permanent modifications to a single strain of mouse scrapie on transmission to rats and hamsters. J Gen Virol 1987; 68:1875 - 81; http://dx.doi.org/ 10.1099/0022-1317-68-7-1875; PMID: 3110370 [DOI] [PubMed] [Google Scholar]
- 35.Kimberlin RH, Walker CA, Fraser H. . The genomic identity of different strains of mouse scrapie is expressed in hamsters and preserved on reisolation in mice. J Gen Virol 1989; 70:2017 - 25; http://dx.doi.org/ 10.1099/0022-1317-70-8-2017; PMID: 2504883 [DOI] [PubMed] [Google Scholar]
- 36.Scott MR, Groth D, Tatzelt J, Torchia M, Tremblay P, DeArmond SJ, et al. . Propagation of prion strains through specific conformers of the prion protein. J Virol 1997; 71:9032 - 44; PMID: 9371560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.González L, Nonno R, Chianini F, Goldmann W, Sisó S, Di Bari M, et al. . Cross-genotype transmission of scrapie strains results in the emergence of strain variants. Prion 2011; 5:Supplement 102 [Google Scholar]
- 38.Pattison IH, Gordon WS, Millson GC. . Experimental production of scrapie in goats. J Comp Pathol 1959; 69:300 - 12; PMID: 14430941 [DOI] [PubMed] [Google Scholar]
- 39.Pattison IH, Millson GC. . Scrapie produced experimentally in goats with special reference to the clinical syndrome. J Comp Pathol 1961; 71:101 - 9; PMID: 13733383 [DOI] [PubMed] [Google Scholar]
- 40.Thackray AM, Hopkins L, Lockey R, Spiropoulos J, Bujdoso R. . Emergence of multiple prion strains from single isolates of ovine scrapie. J Gen Virol 2011; 92:1482 - 91; http://dx.doi.org/ 10.1099/vir.0.028886-0; PMID: 21270287 [DOI] [PubMed] [Google Scholar]
- 41.Bruce ME, Dickinson AG, Fraser H. . Cerebral amyloidosis in scrapie in the mouse: effect of agent strain and mouse genotype. Neuropathol Appl Neurobiol 1976; 2:471 - 8; http://dx.doi.org/ 10.1111/j.1365-2990.1976.tb00521.x [DOI] [Google Scholar]
- 42.Jeffrey M, McGovern G, Sisó S, González L. . Cellular and sub-cellular pathology of animal prion diseases: relationship between morphological changes, accumulation of abnormal prion protein and clinical disease. Acta Neuropathol 2011; 121:113 - 34; http://dx.doi.org/ 10.1007/s00401-010-0700-3; PMID: 20532540 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.