Abstract
Bacterial pathogens display an impressive arsenal of molecular mechanisms that allow survival in diverse host niches. Subversion of plasma membrane and cytoskeletal functions are common themes associated to infection by both extracellular and intracellular pathogens. Moreover, intracellular pathogens modify the structure/stability of their membrane-bound compartments and escape degradation from phagocytic or autophagic pathways. Here, we review the manipulation of host membranes by Listeria monocytogenes, Francisella tularensis, Shigella flexneri and Yersinia spp. These four bacterial model pathogens exemplify generalized strategies as well as specific features observed during bacterial infection processes.
Keywords: Listeria monocytogenes, Francisella tularensis, Shigella flexneri, Yersinia pseudotuberculosis, Phagocytosis, Membrane trafficking
1. Introduction
The eukaryotic cell is a complex environment. Vital functions such as internalization of external solutes, synthesis of novel proteins or genetic information storage are compartmentalized in membrane-bound structures. From the perspective of a pathogen, the eukaryotic cell might be considered as a rich source of nutrients as well as a protected environment in which microbial replication may take place, avoiding contact with extracellular host defenses such as the complement or antibodies. Cells have adapted molecular strategies (e.g. phagocytosis or autophagy) to counteract and destroy intracellular pathogens, which in turn have developed strategies to cope with host cell defenses.
Four major bacterial pathogenesis paradigms are Listeria monocytogenes, Francisella tularensis, Shigella flexneri and Yersinia spp. Listeria monocytogenes is responsible for a food-borne disease associated with meningitis and abortions. Francisella tularensis is the agent of tularemia, responsible for ulcero-glandular and pneumonic severe infections. Shigella flexneri is the etiological agent of human bacillary dysentery. Yersinia pseudotuberculosis and Y. enterocolitica are responsible for enteritis, ileitis, and mesenteric lymphadenitis in humans, and are closely related to Y. pestis, the plague agent. Besides their clinical importance, these bacteria are useful paradigms to understand the diverse strategies used by microbial pathogens to subvert host cell functions. While Listeria and Yersinia invade cultured epithelial cells by interacting with host cell plasma membrane receptors via bacterial surface proteins, Shigella injects bacterial effectors within the host cell cytoplasm to promote internalization and Francisella induces spacious pseudopods to invade macrophages. Once inside epithelial cells, Yersinia proliferates in a membrane-bound compartment while Listeria, Shigella and Francisella disrupt their internalization vacuole and escape into the host cell cytoplasm. In vivo, Yersinia is mainly extracellular. We will review here the major interactions that occur between these important bacterial pathogens and host membranes.
2. Listeria monocytogenes
Listeria monocytogenes is a Gram-positive bacterial pathogen responsible for listeriosis, a disease causing meningitis in the new born and abortions in pregnant women. Listeria is now a classical model in the study of bacterial intracellular parasitism [1]. The pathogenic potential of Listeria is intimately associated with its capacity to traverse the intestinal, the foeto/placental and the blood/brain barriers, which requires invasion of host cells [2]. Using tissue cultured cells, it has been observed that upon internalization Listeria is able to disrupt its membrane-bound compartment and to escape to the host cytoplasm, where it proliferates [3]. Listeria is then able to avoid the autophagosomal machinery by polymerizing host cell actin, which drives bacterial cytoplasmic movement and cell-to-cell spread [4] (Fig. 1). Exceptions to this canonical intracellular cycle model have been observed in vivo, particularly during the traversal of the intestinal barrier. Indeed, in goblet cells Listeria does not escape from the internalization vacuole and is transcytosed to the lamina propia [5]. It may also persist in spacious phagosomes [6].
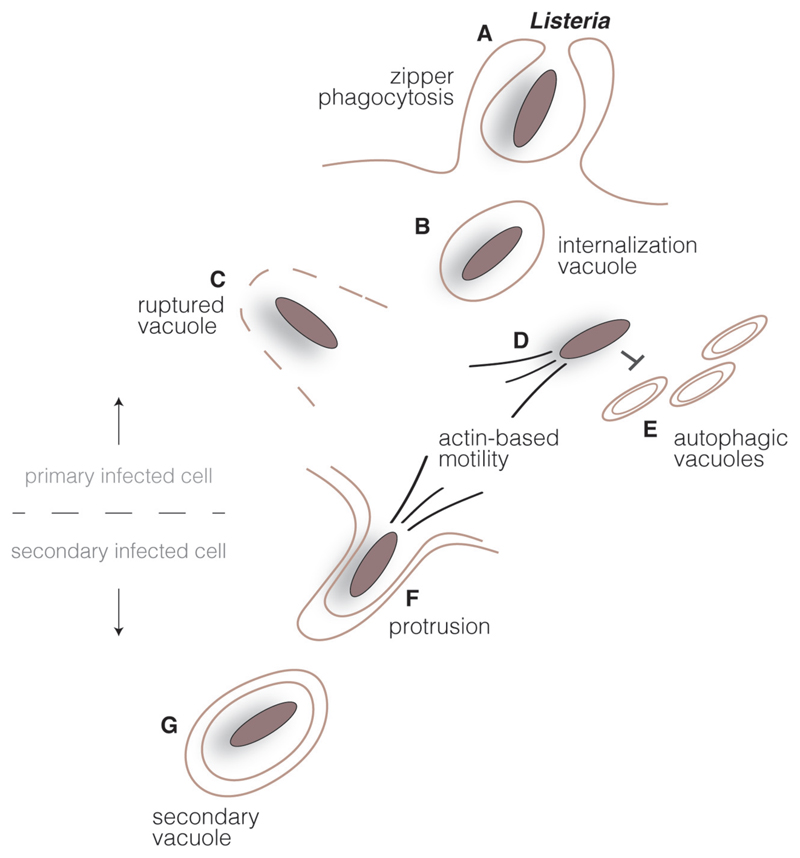
Fig. 1.
Listeria intracellular cycle. Listeria surface invasion proteins interact with host cell receptors in epithelial cells triggering bacterial engulfment (A). Listeria transiently resides in an internalization vacuole (B), but bacterial secreted toxins lyse the vacuole (C) and bacteria are released in the host cell cytoplasm. Cytoplasmic bacteria multiply and polymerize host cell actin, promoting an actin-based motility system (D) which propels the bacteria and allows escape from the autophagic pathway (E). Upon reaching the host cell plasma membrane, Listeria induces the formation of protrusions (F) and are found in double-membrane vacuoles in secondary cells (G). Disruption of this compartment by bacterial secreted toxins allows the start of new infection cycle.
2.1. Cell invasion mediated by InlA and InlB
Listeria induces its internalization within mammalian cells by using surface invasion molecules that activate host plasma membrane receptors. These proteins belong to the internalin family, whose members are characterized by the presence of N-terminal leucine-rich repeats which drive protein–protein interactions [7]. The prototype internalin (InlA) is a covalently-anchored bacterial cell wall protein that binds the cellular adherens junction molecule E-cadherin [8] and triggers Listeria internalization in polarized epithelial cells and tissues, particularly during traversal of the intestinal and the foeto/placental barriers [9,10]. InlB is a second bacterial surface molecule loosely attached to lipoteichoic acids which can interact with the host molecule Met [11], the receptor for the hepatocyte growth factor. In vitro, InlB promotes Listeria internalization in a wide variety of epithelial cells; in vivo, InlB has been shown to cooperate with InlA in crossing the foeto/placental barrier [2] (Fig. 1A).
2.1.1. InlA-invasion pathway
Lipid rafts are cholesterol-rich signaling platforms that allow the clustering of signaling proteins or lipids, and which play a central role in the infectious process of many bacterial pathogens. Interaction between InlA and E-cadherin leads to E-cadherin clustering, a process which is alleviated if lipid rafts are disorganized [12]. This event is followed by E-cadherin cytoplasmic tail phosphorylation and ubiquitylation by Src and Hakai, respectively [13], allowing clathrin recruitment via its adaptor Dab2 [14]. Clathrin is a major membrane coat implicated in the endocytosis of many surface receptors, and clathrin-mediated endocytosis is involved in the infectious process of several important viruses [15]. The study of Listeria internalization demonstrated that during bacterial entry, clathrin does not function as a classical endocytic coat but instead as a molecular hub that triggers actin reorganization at bacterial entry sites [16–18]. Indeed, clathrin coats are stabilized by phosphorylation of the clathrin heavy chain, followed by the sequential recruitment of Hip1R, actin and myosin VI [14]. PI 3-kinase activity, constitutively present in intestinal goblet cells, is also required for the InlA-dependent bacterial crossing of the intestinal barrier following the InlA/E-cadherin interaction [19] (see below for discussion on phosphoinositide metabolism).
2.1.2. InlB-invasion pathway
The second Listeria invasion protein InlB activates the receptor tyrosine kinase Met and triggers recruitment of several protein adaptors [20,21]. Cbl is a ubiquitin ligase which post-translationally modifies Met, favoring clathrin recruitment and actin reorganization during cell entry [14,22,23]. Cbl, together with the protein adaptors Shc, Gab-1 and CrkII, also contributes to the infection process by recruiting PI 3-kinase to the Listeria entry foci [24]. PI 3-kinase is a central enzyme in many signaling cascades which locally produces the phosphoinositide PI(3,4,5)P3 and promotes further recruitment of cellular effectors that link receptor activation with actin cytoskeleton rearrangements. In the InlB-invasion pathway, cholesterol-rich lipid rafts are not required for the initial Met clustering but instead, for optimal PI(3,4,5)P3 distribution within the inner leaflet of the plasma membrane, and for subsequent Rac1 recruitment [12,25]. Rac1 is a member of the Rho family of small GTPases which modulates cortical actin polymerization during Listeria entry by activating WASP-related complexes WAVE-1/-2 [26]. The Arp2/3 complex is the final component of this signaling cascade which directly promotes nucleation of actin polymerization at bacterial entry foci [27]. The Arp2/3 complex has been traditionally considered as a single molecular entity composed of 7 subunits: ARPC1/p41, ARPC2/p34, ARPC3/p21, ARPC4/p20 and ARPC5/p16 together with the Arp2 and Arp3 subunits [28]. A genome-wide siRNA screen recently determined that the subunits ARPC4/p20 and ARPC5/p16 are not strictly required to drive Listeria internalization within host cells, revealing the presence of alternative Arp2/3 complexes in mammalian cells [29]. Another cytoskeletal element implicated in Listeria infection are septins, a family of small GTPases which form non-polar filaments [30]. The septin cytoskeleton contributes to the anchorage of Met to the actin cytoskeleton [31] and different members of the septin family have been shown to modulate host cell invasion by Listeria [32–34].
As mentioned above, PI 3-kinase activity is constitutive in intestinal cells but not in the placenta, and therefore InlB plays a critical role in the traversal of the foeto/placental barrier by promoting the PI 3-kinase-dependent production of PI(3,4,5)P3 [19]. Additionally, in epithelial cells PI 4-kinase activity is necessary for efficient Listeria internalization [35–37]. PI4P production takes place downstream of the tetraspanin CD81 [38], a plasma membrane molecular hub which interacts with integrins and has been also implicated in cellular entry of Plasmodium falciparum [39] and the hepatitis C virus [40]. In vivo, it has been recently demonstrated that CD81-associated signaling inhibits T-cell anti-Listeria responses [41].
Host molecules may also counterbalance plasma membrane signaling triggered by Listeria and down-regulate infection: for example, the host phosphatidyinositol 5-phosphatase Oculocerebrorenal Syndrome of Lowe Protein (OCRL) cleaves PI(4,5)P2 and PI(3,4,5)P3, inhibiting actin polymerization and therefore bacterial entry [42].
2.2. Phagosomal membrane disruption by LLO, PlcA and PlcB
Upon cellular invasion, Listeria is located in a phagosomal compartment (Fig. 1B) that is disrupted via the lytic action of the pore-forming toxin listeriolysin O (LLO), a member of the cholesterol-dependent cytolysin (CDC) family. CDCs can mediate hemolysis in their reduced state, and the cellular enzyme γ-interferon-inducible lysosomal thiol reductase (GILT) present on lysosomes contributes to activate LLO within phagosomes [43]. An acidic pH also potentiates LLO lytic activity [44]. Insertion of LLO in the Listeria phagosome allows Ca+2 leakage which inhibits phagosomal maturation and blocks interaction with lysosomal compartments [45,46]. Rupture of the phagosomal membrane is enhanced by the two bacterial phospholipases PlcA and PlcB which display phosphatiylinositol-specific and broad-range phospolipase C activities, respectively [47] (Fig. 1C). It is important to mention that besides its critical function in disrupting phagosomal compartments, LLO released by extracellular Listeria also contributes to infection by inducing Ca+2 influx [48] that promotes bacterial internalization [49], while triggering the targeting of late endosomal compartments to the bacterial entry sites (Kühbacher, Cossart and Pizarro-Cerda, unpublished observations) as has been observed for the internalization process of parasites like Leishmania or Trypanosoma [50,51]. LLO also favors Listeria infection by influencing host protein sumoylation [52], histone post-translational modifications [53] and mitochondrial dynamics [54].
2.3. Autophagy escape mediated by ActA and InlK
After vacuolar escape, cytosolic Listeria replicate and polymerize host actin via the recruitment to bacterial poles of the Arp2/3 complex by the Listeria surface protein ActA [55,56]. ActA mimics WASP and activates actin filament barbed-end branching by the Arp2/3 complex [57]. Similar to what has been mentioned above for bacterial entry, it has been recently found that the Arp2/3 subunit ARPC5/p16 is not required for the Listeria actin-based motility, suggesting that other cellular molecules may associate with the Arp2/3 complex to modulate its function and/or localization [29]. A similar phenotype has been recently observed for the motility of the Vaccinia virus [58], indicating that the diversity of Arp2/3 complexes is more widespread than initially appreciated. The actin comet tails formed by Listeria contain more than 20 different molecules which stabilize the actin filaments or modulate force generation [59] (Fig. 1D). Actin-based motility is important not only to favor invasion of host neighboring cells (see below) but it is also critical to escape from the autophagy machinery [60–62] (Fig. 1E). Indeed, the autophagic pathway has been shown to limit Listeria cytoplasmic growth [63]. Besides ActA, the surface molecule InlK also protects Listeria from autophagic degradation by recruiting to the bacterial surface the major vault protein and therefore inhibiting autophagic recognition [64]. In addition, it has been recently shown that the ubiquitin-like protein ISG15 is induced upon Listeria infection, modifies a subset of host proteins involved in cytokine secretion and restricts Listeria proliferation both in vitro and in vivo. Interestingly, ISG15 leads to increased cytokine secretion following over-expression or infection, a phenomenon that could explain the in vivo phenotype. How ISG15 leads to fewer bacteria in vitro is still an open question [65].
2.4. Cell-to-cell spread
Initial studies suggested that actin-based motility was sufficient to induce the formation of host membrane protrusions and uptake by neighboring cells during Listeria cell-to-cell spread [66–68] (Fig. 1F). Attachment of actin tails to plasma membrane by the phosphorylated form of the actin-binding molecule ezrin has been also proposed to promote the formation of Listeria protrusions [69]. Interestingly, formins seem to catalyze the generation of unbranched actin filaments that assist the formation of Listeria protrusions initially triggered by the propulsive force generated by the Arp2/3 complex [70]. A siRNA screen has revealed a role for the CNSK1A1 kinase in actin-tail formation and for the CNSK2B in favoring cell-to-cell spread, but their precise mechanism of action remains to be identified [71].
Interestingly, the secreted molecule InlC has been shown to contribute to cell-to-cell spread by directly interacting with the host cell molecule Tuba [72]. Tuba normally controls the structure of cortical actin structures by recruiting N-WASP and by interacting with COPII components: by sequestering Tuba, InlC induces loose membrane junctions that are more permissive to Listeria protrusion formation [72,73]. More recently, it has been proposed that Listeria cell-to-cell spread exploits efferocytosis, which is a process by which dying or dead cells are removed by phagocytosis [74]. Listeria uptake in secondary-infected cells leads to bacterial sequestration within a double-membrane compartment (Fig. 1G). The phospholipases PlcA and PlcB are required for disruption of the inner membrane while LLO is involved in lysis of the outer membrane, allowing initiation of a new Listeria intracellular cycle [75].
3. Francisella tularensis
Francisella tularensis is a Gram-negative bacterium and the causative agent of the zoonotic disease tularemia. This highly infectious bacterial pathogen can infect a broad variety of animal species, from mammals to arthropods. Tularemia can be transmitted to humans in numerous ways [76] and the clinical outcome of the disease depends on the route of infection and bacterial subspecies (subsp) [77]. Four major F. tularensis subsps exist that differ in virulence and geographic distribution. They are designated subsps tularensis, holarctica, mediasiatica and novicida. The most virulent subsp tularensis, capable of causing a life-threatening disease in humans, is considered a potential bioterrorism agent [78] by the U.S. Centers for Disease Control and Prevention (CDC). Francisella is an intracellular pathogen which upon adhesion and invasion of host cells resides transiently in a phagosomal compartment; disruption of this phagosome is followed by active cytosolic multiplication. Dissemination to adjacent cells does not involve actin polymerization and relies instead in cell lysis and bacterial dissemination.
3.1. Adhesion and invasion
Francisella is able to infect and multiply in vitro within numerous cell types including hepatocytes, endothelial cells and fibroblasts, to which bacterial attachment is mediated by subsps-specific outer membrane proteins and the type IV pilus [79]. However, in vivo the Francisella main cellular niche is the macrophage, used for bacterial dissemination [80]. Different macrophage receptors involved in Francisella uptake have been identified over the past ten years, including the complement receptor CR3 (CD11b/CD18), the mannose receptor (MR), the scavenger receptor A (SRA), FcγRs and surface-exposed nucleolin [81]. The subversion of phagocytic or scavenger receptors to invade macrophages is not specific to Francisella and is common to a subset of bacterial intracellular pathogens including Mycobacterium tuberculosis [82], which do not actively promote surface cytoskeletal rearrangements and instead rely on the internalization machineries of host cells. As observed for the vast majority of bacterial intracellular pathogens, Francisella internalization is dependent on actin polymerization; remarkably, Francisella is captured by macrophages via a unique engulfment mechanism which involves the initial formation of large loops or pseudopodia [83] (Fig. 2A). As in the case of Listeria, lipid rafts are required for efficient Francisella internalization [84]. In order to avoid cellular recognition by the host immune system, Francisella harbors a peculiar lipopolysaccharide (LPS) which does not activate host Toll-like receptor 4 (TLR4); nevertheless, the bacterial lipoproteins (BLPs) do activate TLR2, the primary TLR involved in the inflammatory response to Francisella infection.
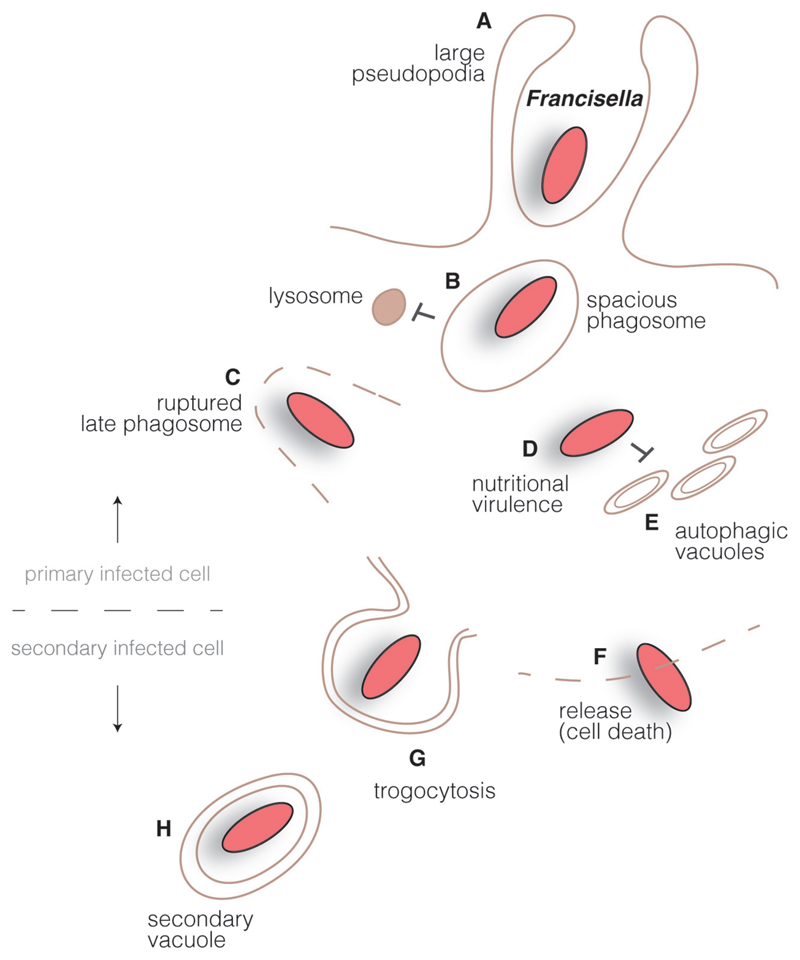
Fig. 2.
Francisella intracellular cycle. Francisella interact with innate immune receptors at the surface of macrophages in order to get internalized by large pseudopodia (A). Inside cells, bacteria reside in a large spacious phagosome that matures into a late phagocytic compartment but avoids fusion with lysosomes (B). A type VI secretion system is involved in the rupture of the bacteria-containing compartment (C) and Francisella is found in the host cell cytoplasm, where is takes advantage of available resources (‘nutritional virulence’) (D) and avoids interaction with autophagic vacuoles (E). Bacteria can be released from dying cells (F) or can be trapped by ‘trogocytosis’ in double membrane compartments that allows infection of neighboring cells (H).
3.2. Phagosomal stage
3.2.1. Spacious phagosome maturation
After engulfment by phagocytic cells, Francisella transiently resides in a spacious phagosome (Fig. 2B). This bacterial-containing compartment sequentially displays membrane markers of early (EEA1) and late endosomes/lysosomes (LAMP-1 and -2) but fails to acquire the hydrolase cathepsin D or lysosomal tracers [85]. Depending on the uptake route, the phagosomal stage may last 30 min for non-opsonized bacteria to 2–4 h for opsonized Francisella. While the importance of phagosomal acidification has been shown to be an absolute prerequisite for Listeria to escape from the phagosome as mentioned above [44], controversy exists on the importance of this process on the ability of Francisella to access to the host cytosol. Indeed, some studies indicate that compartments containing Francisella become acidified prior to phagosomal disruption [86,87] while others report that Francisella phagosomes resist acidification and acquire limited amounts of vacuolar ATPases, while still capable of accessing the cellular cytoplasm [88].
3.2.2. Inhibition of NADPH oxidase
During this transient time spent in the phagosome, Francisella must actively evade several host antimicrobial defenses, including anti-microbial peptides and reactive oxygen species (ROS) produced by the NADPH oxidase. Francisella is precisely one of the few bacteria capable of surviving within human polymorphonuclear neutrophils (PMNs), which are potent ROS producers, due to its ability to block either the assembly of the NADPH oxidase integral membrane gp91phox/p22phox components, or the phosphorylation and recruitment of the its cytosolic p47phox/p40phox subunits [89]. Francisella-infected monocytes or macrophages also fail to trigger a robust oxidative burst [90]. Francisella is additionally equipped with a series of enzymes that include superoxide dismutase, catalase and acid phosphatases that allow bacterial survival in the hostile phagosomal environment [91].
3.2.3. Phagosomal disruption by a T6SS
Bacterial secretion systems are sophisticated nano-machines that allow protein export from the bacterial cytosol toward specific external environments. Type VI secretion systems (T6SS) have been recently discovered in Gram-negative bacteria and are critical to the virulence of many pathogens such as Vibrio cholerae and Pseudomonas aeruginosa. A Francisella pathogenicity island (FPI) has been presumed to encode a T6SS apparatus, and many genes expressed from this locus or regulating expression of this locus, including IglC and mglA [92], iglI and iglJ [89] or fevR [93], were previously implicated in promoting Francisella escape from its phagosome and bacterial cytoplasmic proliferation (Fig. 2C). The atomic structure of the Francisella T6SS was recently solved by cryoelectron microscopy, demonstrating for the first time in this type of system how its two main components, IglA and IglB, are interdigitated into a single fold [94]. Moreover, it is the first T6SS which displays assembly upon KCl stimulus or intracellular residence, and its contribution to phagosomal escape and intra-macrophage replication has been confirmed [94]. However, the precise molecular contribution of the Francisella T6SS effectors to phagosomal membrane disruption, together with additional not FPI-encoded proteins also implicated in this process, remain to be identified.
3.3. Cytoplasmic life and interactions with the autophagic pathway
Once in the cytosol, Francisella adapts its nutritional needs to the available nutrient sources, a function that has been recently coined as ‘nutritional virulence’ [95] (Fig. 2D). Asparagine uptake has been identified as critical for efficient bacterial cytosolic multiplication [96]. It has been proposed that in mouse embryonic fibroblasts (MEFs), cytoplasmic Francisella stimulates an ATG5-independent autophagy process which further supply nutrients that supports bacterial growth [97]. An early report by Celli et al. indicated that, at late stages of infection in bone-marrow derived macrophages, a subset of intracellular Francisella could be found inside LC3-positive, double membrane-bound vacuoles [98]. This observation suggested that late residency within autophagic compartments constitutes an intrinsic part of the Francisella intracellular life cycle [98]. However, further studies in other cell types contradicted this notion by demonstrating that metabolically active Francisella in fact avoid autophagic capture [85] (Fig. 2E) whereas mutant bacteria displaying impaired cytosolic multiplication are captured and destroyed by autophagy. In addition, guanylate-binding proteins (GBPs) represent another cell system to attack cytosolic Francisella in order to activate the Absent in Melanoma 2 (AIM2) inflammasome, which controls Francisella replication [99].
3.4. Cell-to-cell spread
Unlike Listeria (mentioned above) or Shigella (see below), Francisella is not able of actin-based cytosolic movements. Hence, Francisella dissemination to adjacent cells was thought to occur exclusively after their release from lysed infected cells following apoptotic or pyroptotic cell death (Fig. 2F). A novel cell-to-cell dissemination mechanism was very recently described by which Francisella can also infect adjacent cells without passing through an extracellular stage: this unique process, designated trogocytosis, allows bacterial transfer from infected cells to uninfected macrophages through a transient, contact-dependent mechanism, leaving both donor and recipient cells intact and viable [100] (Fig. 2G). Therefore, Francisella is able to exploit natural host cell cytosolic exchange mechanisms to directly transfer from infected to non-infected cells.
4. Shigella flexneri
The Gram-negative pathogenic bacteria Shigella spp. cause bacillary dysentery in humans, also known as shigellosis. Shigella transmission takes place generally through the fecal-oral route by the uptake of contaminated food or water, giving rise to a disease that results in colonic epithelium destruction and elevated intestinal inflammation. Annually, about 160 million cases of shigellosis with about 1 million deaths have been estimated to occur mainly in the developing world; hence, Shigella infections constitute an important health burden, particularly for young children [101]. The rise in antibiotic resistance and the absence of an efficient vaccine against Shigella underline the urgency for the development of novel antibacterial strategies against this important pathogen [102].
Four Shigella species have been described: S. dysenteriae, S. flexneri, S. boydii and S. sonnei. The molecular basis of infection has been studied using the Shigella flexneri model (referred from now on as Shigella). Primordial for Shigella infectivity is the presence of a large virulence plasmid that encompasses the mxi-spa pathogenicity island, encoding the structural genes for a type 3 secretion system (T3SS) and about 25 effector proteins that are injected into targeted host cells upon contact with the bacterium [103,104]. The T3SS is a major virulence determinant that has been described in many pathogenic bacteria including E. coli, Salmonella and Yersina (see below). The Shigella T3SS is expressed and assembled at 37 °C together with a first wave of effector proteins, however secretion together with the expression of a second wave of effectors is only triggered upon contact with the host cell. The effectors are translocated directly from the bacterial cytoplasm into the host cytoplasm through the needle complex that connects with the bacterial translocon, made of the two proteins IpaB and IpaC spanning the host cell membrane. Subsequently, the translocated effector proteins subvert host cellular processes resulting in cytoskeletal rearrangements, bacterial invasion and the modulation of the host immune response. Injection of the bacterial effectors is a rapid process with half the pre-stored proteins translocated within 240 s [104,105].
4.1. T3SS effector injection triggers Shigella internalization
The cellular mechanisms that take place during the early contact between Shigella and the host cells have been investigated in detail over the last two decades. Recent data have highlighted the capture of the pathogen in the vicinity of host cells through cellular extensions that resemble filopodial-like structures involving a functional T3SS. The interactions between the pathogen and the filopodial tips induce their retraction to give rise to close contacts between the bacterium and the cellular surface [106]. Additionally, the bacterial translocator/effector IpaB, the bacterial factor IcsA and the host receptors CD44 and α5β1-integrin have been proposed in the establishment of early bacterial-host contacts [107–109]. Also, the presence of cholesterol in lipid-raft-like domains is required for proper translocon assembly and bacterial internalization [110,111]. Together, these interactions promote Shigella entry through a site-specific invasion process [112].
The translocon complex assembles within the host membrane and is connected with the T3SS, to allow the injection of the bacterial effectors into the host cytoplasm or in association with the host membrane complex. These effectors include IpaA, IpaB, IpaC, IpgB1, IpgB2, IpgD and VirA, all involved in the entry of Shigella through a trigger-like process [113] (Fig. 3A). IpaA has been found to interact directly with human vinculin, which would allow a link with the actin cytoskeleton [114]. More precisely, IpaA promotes the capping of F-actin barbed ends via vinculin controlling the polymerization dynamics of actin at the bacterial contact sites [114]. Also, the targeting of β1-integrin by IpaA has been suggested to indirectly stimulate the GTPase activity of the small GTPase RhoA, which would lead in turn to the loss of actin stress fibers via the ROCK/myosin-II pathway [115]. Such stress fiber disassembly may facilitate the availability of actin for the formation of membrane ruffles at the bacterial entry site (Fig. 3A). IpaC is a constituent of the T3SS translocon complex and plays a direct role in the bacterial entry process. It recruits and activates the host tyrosine kinase Src to the site of bacterial contact where it phosphorylates the actin-binding protein cortactin that implicates the Arp2/3 complex and leads to actin remodeling and polymerization [116]. In addition, the activated form of cortactin interacts also with the Src-related tyrosine kinase CrK that further boosts actin polymerization at the Shigella entry site [117].
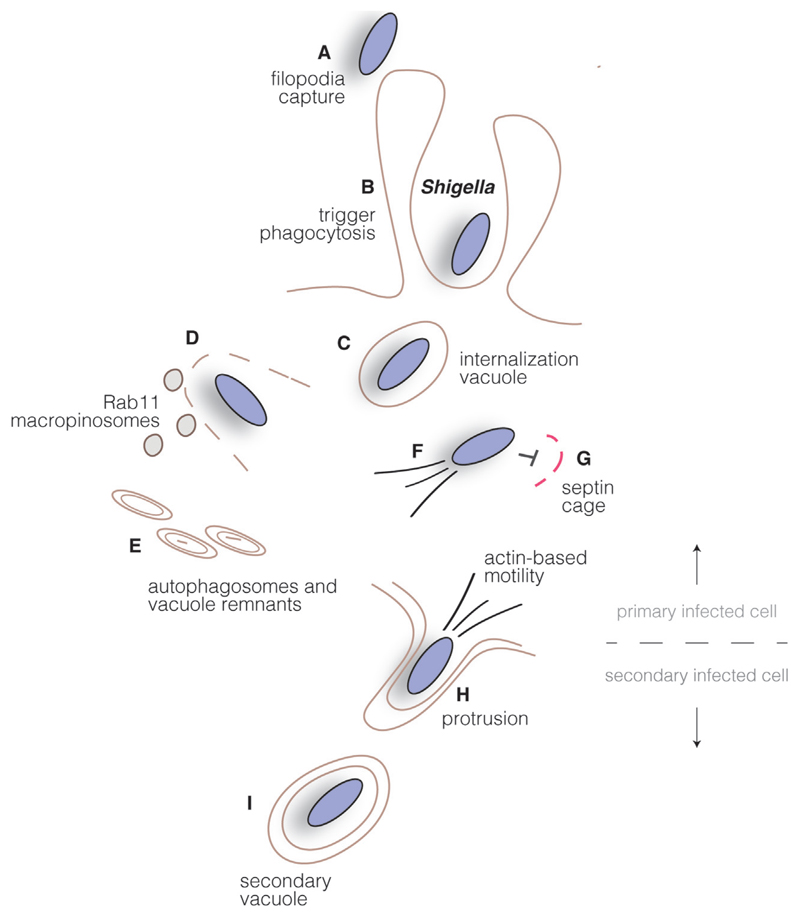
Fig. 3.
Shigella intracellular cycle. Shigella promotes its uptake into host cells by triggering plasma membrane rearrangements through injection of type III secretion system effectors in the cytosol of host cells. Bacteria are transiently found in an internalization vacuole (B), which is disrupted upon recruitment of Rab11-positive macropinosomes (C), leading the uptake of vacuole remnants by autophagosomes (D). Cytoplasmic bacteria polymerize actin and move intracellularly (E), while septin cages can also function as bacterial traps (F). Upon reaching the host cell plasma membrane, Shigella induces the formation of protrusions (G) and subsequently localize in double-membrane vacuoles in neighboring cells (H). Disruption of this compartment allows the start of a new infection cycle.
The two homologues effectors, IpgB1 and IpgB2, play a key role in Shigella entry. IpgB1 stimulates Rac and Cdc42 [118]. It binds ELMO, a protein involved in engulfment and cell motility, leading to the activation of Rac1 through the ELMO/Dock180 pathway. Consequently, it induces Arp2/3 complex-dependent membrane ruffling at the bacterial contact site [119], and it has also been revealed as a pace maker for Shigella internalization [120]. In contrast, IpgB2 binds to mDia1 and to ROCK, which leads to actin nucleation and stress fiber formation, thus mimicking RhoA [121,122]. Even though IpgD, a PI(4,5)P2 phosphatase that specifically depletes PtdIns(4,5)P2 into PtdIns(5)P, does not affect Shigella entry in general, it may weaken the connection between cortical actin and the plasma membrane to facilitate membrane extensions [15]. Another effector that has been implicated during the early entry of Shigella is VirA, which correlates with local degradation of microtubules, which indirectly affects ruffle formation through a feedback loop via RhoA and Rac1 [123–125]. This illustrates, how different enzymatic activities of the bacterial effectors lead to finely tuned spatiotemporal modulations of the host cytoskeleton at the contact site allowing the internalization of Shigella in a vacuolar compartment.
4.2. Rupture of the Shigella containing vacuole
Shigella ruptures its endocytic vacuole rapidly upon internalization to reach the host cytoplasm (Fig. 3B). The T3SS in particular, through the action of the translocon complex proteins IpaB and IpaC, has been proposed to play a crucial role in the direct destabilization of the Shigella containing vacuole: the T3SS translocon proteins IpaB and IpaC could form a pore complex in cholesterol-rich membrane domains of the vacuole, causing its disruption and subsequent bacterial escape to the cytosol [126,127]. This was substantiated through experiments that demonstrated red blood cell hemolysis via IpaB and IpaC [128]. Recently, T3SS reconstitution of the Shigella injection device in E. coli resulted in the destabilization of the vacuole containing the E. coli expressing the T3SS in macrophages and hinted at the capacity of the T3SS to disrupt endocytic vacuoles [129]. Nevertheless, it has been puzzling that other T3SS containing bacteria, including Salmonella and Yersinia remain mainly within phagolysosomes without damage arguing for a vacuolar damage mechanism that requires additional factors. For example, a non characterized region on the Shigella virulence plasmid has been implicated in the uncoupling of Shigella entry step and the vacuolar escape step [130]. Novel approaches for the tracking of vacuolar damage in single cells at high temporal resolution reveals that this process occurs in less than 10 min after bacterial entry [120,131,132]. Using this method, a new mechanism of vacuolar membrane damage caused by Shigella has been discovered, which depends on the subversion of host membrane trafficking pathways through the injection of Shigella T3SS effectors. A high-content small interfering RNA (siRNA) screen identified several host proteins that modulate S. flexneri vacuolar membrane rupture [133]. They include a network of Rab GTPases, namely Rab5 and Rab11, a component of the host recycling pathway (Fig. 3C). Depletion of Rab5 and Rab11 delays vacuolar rupture, and both GTPases are recruited to the bacterial entry site through the effector IpgD that converts of PI(4,5)P2 to PI(5)P. Also, the ipgD mutant increases the time of vacuolar escape by Shigella. Using correlative light electron microscopy (CLEM) in three dimensions, it was further shown that Rab11 was only recruited to macropinosomes in the surrounding of the bacterial containing vacuole, and that Shigella was not taken up within such spacious compartments [134]. It was therefore identified that Shigella exclusively interacts with in situ induced macropinosomes having no contact with other endomembrane compartments. The interaction between the Shigella containing vacuole and surrounding macropinosomes through membrane contacts is necessary for vacuolar rupture through a mechanism that requires further investigation [134].
4.3. Shigella lifestyle after vacuolar rupture and cell-to-cell spread
Vacuolar rupture and bacterial escape into the cytoplasm results in the generation of small vesicles derived from vacuolar membrane remnants, which are associated with poly-ubiquitinated proteins. The autophagy marker LC3 and the adaptor p62 are recruited, as well as inflammasome components and caspase-1, and the membrane remnants are targeted to autophagic degradation [120,135] (Fig. 3D). Therefore, the vacuolar rupture process itself represents a site for the establishment of a signaling platform in the host cell. Within the cytoplasm, Shigella polymerizes actin at one of its poles with the help of the outer membrane protein IcsA (or VirG). IcsA interacts with and recruits N-WASP, which in turn activates the Arp2/3 complex to form an actin comet tail [136] (Fig. 3E). Intracellular bacterial motility can reach speeds of 3–26 μm/min [136,137]. Structurally, as for Listeria, actin comet tails are composed of short, highly branched cross-linked actin filaments that can leave a long trail behind one of the bacterial poles. A subset of intracytoplasmic Shigella may be found surrounded by septin-cages that impede their mobility and interact with the host autophagy machinery [138] (Fig. 3F). As Listeria, intracellular moving Shigella make protrusions upon reaching the host cell plasma membrane (Fig. 3G) which allows them to invade neighboring cells, in which bacteria will be found in two-membrane secondary vacuoles (Fig. 3H). Rupture of these secondary vacuoles by still non-identified mechanisms initiate a new intracellular infection cycle.
5. Yersinia spp.
Yersinia spp. are Gram-negative, facultative anaerobes that belong to the Enterobacteriaceae family, for which eighteen species have been reported, including three species that display pathogenic features in humans and animals [139]. Y. pseudotuberculosis and Y. enterocolitica are enteropathogenic bacteria responsible for enteritis, ileitis, and mesenteric lymphadenitis; Y pestis, the etiological agent of bubonic plague, is still endemic in rodent populations on several continents and considered by the World Health Organization as a reemerging disease. Y. pseudotuberculosis and Y. enterocolitica (referred from now on as Yersinia) are mainly transmitted by ingestion: upon uptake of contaminated food or water, bacteria gain access to the gastrointestinal tract, reach the terminal ileum and translocate across the epithelial barrier by transcytosis through antigen-sampling M cells associated with Peyer’s patches [140]. Thereafter, Yersinia predominantly survives extracellularly (Fig. 4A), inhibiting its internalization by PMNs and monocytes (Fig. 4B).
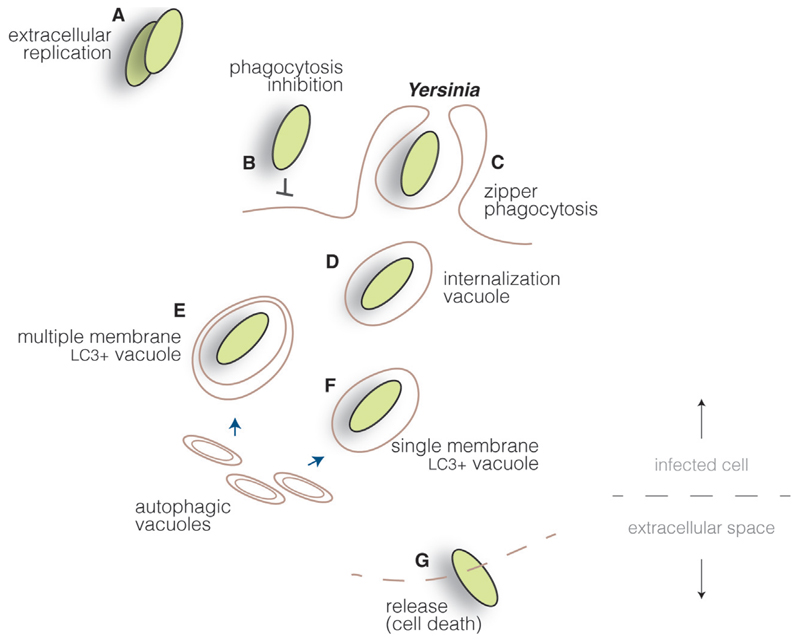
Fig. 4.
Yersinia cycle. Yersinia proliferate mainly as extracellular pathogens (A), as they actively inhibit cellular uptake by injecting type III secretion system effectors into host cells, blocking actin cytoskeleton rearrangements (B). However, through the interaction of surface invasion proteins and host cell receptors, Yersinia can induce its internalization in specific cell lines (C). Bacteria are found initially within internalization vacuoles (D) which, through interaction with autophagic vacuoles, can mature into multiple (E) or single (F) membrane compartments, depending on the cell type. Yersinia are probably released upon cell death in cells permissive for bacterial replication (G).
5.1. Facultative intracellular life
5.1.1. Cell invasion
In the gastrointestinal tract, Yersinia displays on its surface at high density the outer membrane protein (OMP) invasin [141]. Invasin is related to the intimins OMPs present in enteropathogenic and enterohemorragic Escherichia coli, and binds with high affinity to multiple members of the β1 chain integrin family on a variety of cells, including M cells [142,143]. Binding of β1 integrins by invasin promotes recruitment at bacterial entry sites of the focal adhesion kinase (FAK) and Src, which normally drive actin-dependent rearrangements required for cellular migration, but which are used by Yersinia to promote bacterial invasion [144,145]. Indeed, as in the case of Listeria infection process, the small GTPase Rac1 is subsequently recruited at Yersinia entry sites, where it induces actin polymerization via the Arp2/3 complex [146] and bacterial engulfment through zipper phagocytosis (Fig. 4C). The small GTPase Arf6 also participates in the bacterial invasion process and cooperates with Rac1 in the localized activation of PIP 5-kinase, which produces a focal increase of the phosphoinositide PI(4,5)P2 [147], probably required to recruit other actin-remodeling proteins required for Yersinia invasion. The final stages of Yersinia entry into host cells require PI 3-kinase-dependent fusion of Rab5-positive vesicles with bacterial-containing compartments that are still open to the extracellular space: the host lipid phosphatase OCRL is then recruited to these non-sealed vacuoles promoting hydrolysis of PI(4,5)P2 and vacuole scission from the plasma membrane [148] (Fig. 4D). Clathrin coats are, as for Listeria, critical for Yersinia internalization in host cells [23]. The lollipop-shaped multifunctional adhesin YadA also facilitates epithelial cell entry in the absence of invasin activity, through indirect binding of β1 integrins via engagement of extra-cellular matrix molecules [149].
5.1.2. Autophagy subversion
Yersinia actively inhibits its own internalization in PMNs and macrophages (see below). Interestingly, while Y. enterocolitica upon internalization in phagocytic cells is degraded by autophagy [150], Y. pestis replicates within intracellular compartments positive for the autophagy marker protein LC3 and which do not acidify [151]. Recently, it has been suggested that the small GTPase Rab1 plays a critical role in the inhibition of acidification by directly inhibiting the maturation of the Y. pestis-containing compartment [152]. Y. pseudotuberculosis has been also found to proliferate in LC3-positive vacuoles: in bone marrow-derived macrophages, the bacteria are detected in classical autophagosomes characterized by multiple limiting membranes which also avoid acidification [153] (Fig. 4E). Surprisingly, in epithelial HeLa cells, Y. pseudotuberculosis resides in single-membrane compartments also labelled by LC3, suggesting an autophagosomal origin [154] (Fig. 4F). The vesicle-associated membrane protein 7 (VAMP7), which belongs to the SNARE family of mediators of membrane docking and fusion, was found to regulate the association of LC3 to both single- and multiple-membrane compartments supporting Y. pseudotuberculosis replication; however, VAMP3 was shown to play a critical role in the commitment of bacteria towards single-membrane LC3-positive compartments in HeLa cells, suggesting that this SNARE functions as a molecular checkpoint for bacterial trafficking to single- or double-membrane autophagosomes [154]. The potential functional differences of these two types of autophagosomes are unknown. Interestingly, vacuolar escape for Y. enterocolitica has been described [155] but the relevance of this observation requires further investigation.
5.2. Extracellular life
As mentioned above, Yersinia spp. has developed molecular adaptations to survive within macrophages if uptake takes places. However, the first bacterial defense strategy to avoid degradation is to actively inhibit internalization within professional phagocytes. The T3SS of Yersinia, which is one of the first to have been thoroughly characterized, delivers into the cytoplasm of host cells, molecules which interfere with actin cytoskeleton rearrangements in order to inhibit phagocytosis [156]. The effector YopH is a tyrosine phosphatase that dephosphorylates critical cytoskeletal-related proteins such as FAK, p130Cas and paxillin [157], inhibiting Rac1 activation required for bacterial internalization, as mentioned above [145]. Direct manipulation of Rho GTPases is performed by YopE, which functions as a GTPase-activating protein (GAP) for Rho, Rac1 and Cdc42, completely inhibiting the rearrangement of the actin cytoskeleton [158,159]. The effector YopO/YpkA is a multidomain effector which displays a GDP-dissociation inhibitor (GDI)-like domain which sequesters the GDI-free pool of Rac1 localized at the plasma membrane [160] and which also displays a phosphatase domain which targets the vasodilator-stimulated phosphoprotein (VASP), another critical regulator of actin assembly [161]. Finally, the Y. enterocolitica-specific YopT is a cysteine protease that cleaves the carboxy-terminal prenylyl groups of Rho, Rac1 and Cdc42, disrupting their membrane localization [162].
6. Conclusions
In the course of infection, bacterial pathogens manipulate host cell membranes in exquisite ways. During internalization, the subversion of small Rho GTPases and phosphoinositide metabolism has been extensively characterized as mechanisms to control actin cytoskeleton rearrangements and vesicular traffic. Several recent findings have now refined our current view of bacterial infection strategies to harness host membranes, and clathrin stands as a novel critical regulator of actin polymerization for zippering pathogens such as Listeria and Yersinia [23]. Bacterial transcytosis has also emerged as a novel invasion mechanism for Listeria that allows translocation across the epithelial barrier [5]. It would be interesting to determine whether Yersinia, which also uses a zipper invasion mechanism based on the activation of surface host cell receptors, exploits a similar strategy during the initial steps of host infection. Another important finding concerns the co-existence of diverse Arp2/3 complexes in cells which differentially participate in Listeria entry or actin comet tail formation [29]. Similar observations have been made for viral infection [58] and focal adhesion formation [163], highlighting that cellular functions implicating the Arp2/3 complex may need to be re-investigated.
The rupture process of bacterial-containing compartments has been also recently enriched by the characterization of the Francisella T6SS, that appears essential for bacterial escape to the host cytosol [94] and by the identification of small GTPases of the Rab family which are required for Shigella cytosolic translocation [133]. In the future, it will be important to determine whether other host molecules can directly influence vacuolar membrane stability and what are the specific molecular contributions of T6SS effectors for Francisella phagosomal escape. Concerning bacterial replication within host cells, the finding that Yersinia may reside in single- or multiple-membrane compartments [154] highlights the versatility of bacterial pathogens to adapt their molecular survival strategies to their specific local infection environment. This is also exemplified by the Francisella nutritional virulence phenotype [95].
Infectious diseases are still a major public health problem in the 21st century. Profound knowledge of how host cellular components are subverted by bacterial pathogens should allow identification of host targets to be used in novel therapeutic strategies.
Acknowledgments
We apologize to colleagues whose work could not be included in this review owing to space limitations. This work was supported by the Institut Pasteur (PTR 460 and PTR 521 to JPC), L’Agence Nationale de la Recherche (ANR) (ANR-15-CE15-0017 StopBugEntry to JPC, AC, JE and FL), Institut National de la Santé et de la Recherche Médicale (PC: INSERM Unité 604), Institut National de la Recherche Agronomique (PC: INRA Unité Sous Contrat 2020), Centre National de la Recherche Scientifique AC: CNRS Unité Mixte de Recherche 8253; FL: CNRS Unité Mixte de Recherche 8204 (ERC) Advanced Grant (670823 BacCellEpi to PC) and ERC Starting Grant (26116 RuptEffects to JE). PC is an International Senior Research Scholar of the Howard Hughes Medical Institute.
Abbreviations
- ECM
extracellular matrix
- FPI
Francisella pathogenicity island
- LLO
listeriolysin O
- OMP
outer membrane protein
- PMN
polymorphonuclear neutrophils
- ROS
reactive oxygen species
- Subsp
subspecies
- T3SS
type 3 secretion system
- TLR
toll-like receptor
- YCV
Yersinia-containing vacuole
Footnotes
The authors declare no conflict of interest.
References
- [1].Cossart P. Illuminating the landscape of host-pathogen interactions with the bacterium Listeria monocytogenes. Proc Natl Acad Sci. 2011 doi: 10.1073/pnas.1112371108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Disson O, Grayo S, Huillet E, Nikitas G, Langa-Vives F, Dussurget O, et al. Conjugated action of two species-specific invasion proteins for fetoplacental listeriosis. Nature. 2008;455:1114–1118. doi: 10.1038/nature07303. [DOI] [PubMed] [Google Scholar]
- [3].Pizarro-Cerdá J, Kühbacher A, Cossart P. Entry of Listeria monocytogenes in mammalian epithelial cells: an updated view. Cold Spring Harb Perspect Med. 2012;2 doi: 10.1101/cshperspect.a010009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Gouin E, Welch MD, Cossart P. Actin-based motility of intracellular pathogens. Curr Opin Microbiol. 2005;8:35–45. doi: 10.1016/j.mib.2004.12.013. [DOI] [PubMed] [Google Scholar]
- [5].Nikitas G, Deschamps C, Disson O, Niault T, Cossart P, Lecuit M. Transcytosis of Listeria monocytogenes across the intestinal barrier upon specific targeting of goblet cell accessible E-cadherin. J Exp Med. 2011;208:2263–2277. doi: 10.1084/jem.20110560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Birmingham CL, Canadien V, Kaniuk NA, Steinberg BE, Higgins DE, Brumell JH. Listeriolysin O allows Listeria monocytogenes replication in macrophage vacuoles. Nature. 2008;451:350–354. doi: 10.1038/nature06479. [DOI] [PubMed] [Google Scholar]
- [7].Cabanes D, Dehoux P, Dussurget O, Frangeul L, Cossart P. Surface proteins and the pathogenic potential of Listeria monocytogenes. Trends Microbiol. 2002;10:238–245. doi: 10.1016/s0966-842x(02)02342-9. [DOI] [PubMed] [Google Scholar]
- [8].Mengaud J, Ohayon H, Gounon P, Mege R-M, Cossart P. E-cadherin is the receptor for internalin, a surface protein required for entry of L. monocytogenes into epithelial cells. Cell. 1996;84:923–932. doi: 10.1016/s0092-8674(00)81070-3. [DOI] [PubMed] [Google Scholar]
- [9].Lecuit M, Vandormael-Pournin S, Lefort J, Huerre M, Gounon P, Dupuy C, et al. A transgenic model for listeriosis: role of internalin in crossing the intestinal barrier. Science. 2001;292:1722–1725. doi: 10.1126/science.1059852. [DOI] [PubMed] [Google Scholar]
- [10].Lecuit M, Nelson DM, Smith SD, Khun H, Huerre M, Vacher-Lavenu M-C, et al. Targeting and crossing of the human maternofetal barrier by Listeria monocytogenes: role of internalin interaction with trophoblast E-cadherin. Proc Natl Acad Sci U S A. 2004;101:6152–6157. doi: 10.1073/pnas.0401434101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Shen Y, Naujokas M, Park M, Ireton K. InIB-dependent internalization of Listeria is mediated by the Met receptor tyrosine kinase. Cell. 2000;103:501–510. doi: 10.1016/s0092-8674(00)00141-0. [DOI] [PubMed] [Google Scholar]
- [12].Seveau S. Role of lipid rafts in E-cadherin- and HGF-R/met-mediated entry of Listeria monocytogenes into host cells. J Cell Biol. 2004;166:743–753. doi: 10.1083/jcb.200406078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Bonazzi M, Veiga E, Pizarro-Cerdá J, Cossart P. Successive post-translational modifications of E-cadherin are required for InlA-mediated internalization of Listeria monocytogenes. Cell Microbiol. 2008;10:2208–2222. doi: 10.1111/j.1462-5822.2008.01200.x. [DOI] [PubMed] [Google Scholar]
- [14].Bonazzi M, Vasudevan L, Mallet A, Sachse M, Sartori A, Prevost MC, et al. Clathrin phosphorylation is required for actin recruitment at sites of bacterial adhesion and internalization. J Cell Biol. 2011;195:525–536. doi: 10.1083/jcb.201105152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Cossart P, Helenius A. Endocytosis of Viruses and Bacteria. Cold Spring Harb Perspect Biol. 2014;6(8) doi: 10.1101/cshperspect.a016972. pii: a016972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Pizarro-Cerdá J, Bonazzi M, Cossart P. Clathrin-mediated endocytosis: what works for small, also works for big. Bioessays. 2010;32:496–504. doi: 10.1002/bies.200900172. [DOI] [PubMed] [Google Scholar]
- [17].Pizarro-Cerdá J, Cossart P. Listeria monocytogenes membrane trafficking and lifestyle: the exception or the rule? Annu Rev Cell Dev Biol. 2009;25:649–670. doi: 10.1146/annurev.cellbio.042308.113331. [DOI] [PubMed] [Google Scholar]
- [18].Bonazzi M, Kühbacher A, Toledo-Arana A, Mallet A, Vasudevan L, Pizarro-Cerdá J, et al. A common clathrin-mediated machinery co-ordinates cell-cell adhesion and bacterial internalization. Traffic. 2012;13:1653–1666. doi: 10.1111/tra.12009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Gessain G, Tsai YH, Travier L, Bonazzi M, Grayo S, Cossart P, et al. PI3-kinase activation is critical for host barrier permissiveness to Listeria monocytogenes. J Exp Med. 2015;212:165–183. doi: 10.1053/j.gastro.2009.05.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Ireton K, Payrastre B, Cossart P. The Listeria monocytogenes protein InlB is an agonist of mammalian phosphoinositide 3-kinase. J Biol Chem. 1999;274:17025–17032. doi: 10.1074/jbc.274.24.17025. [DOI] [PubMed] [Google Scholar]
- [21].Dokainish H, Gavicherla B, Shen Y, Ireton K. The carboxyl-terminal SH3 domain of the mammalian adaptor CrkII promotes internalization of Listeria monocytogenes through activation of host phosphoinositide 3-kinase. Cell Microbiol. 2007;9:2497–2516. doi: 10.1111/j.1462-5822.2007.00976.x. [DOI] [PubMed] [Google Scholar]
- [22].Veiga E, Cossart P. Listeria hijacks the clathrin-dependent endocytic machinery to invade mammalian cells. Nat Cell Biol. 2005;7:894–900. doi: 10.1038/ncb1292. [DOI] [PubMed] [Google Scholar]
- [23].Veiga E, Guttman JA, Bonazzi M, Boucrot E, Toledo-Arana A, Lin AE, et al. Invasive and adherent bacterial pathogens co-opt host clathrin for infection. Cell Host Microbe. 2007;2:340–351. doi: 10.1016/j.chom.2007.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Ireton K, Payrastre B, Chap H, Ogawa W, Sakaue H, Kasuga M, et al. A role for phosphoinositide 3-kinase in bacterial invasion. Science. 1996;274:780–782. doi: 10.1126/science.274.5288.780. [DOI] [PubMed] [Google Scholar]
- [25].Seveau S, Tham TN, Payrastre B, Hoppe AD, Swanson JA, Cossart P. A FRET analysis to unravel the role of cholesterol in Rac1 and PI 3-kinase activation in the InlB/Met signalling pathway. Cell Microbiol. 2007;9:790–803. doi: 10.1111/j.1462-5822.2006.00832.x. [DOI] [PubMed] [Google Scholar]
- [26].Bierne H, Miki H, Innocenti M, Scita G, Gertler FB, Takenawa T, et al. WASP-related proteins, Abi1 and Ena/VASP are required for Listeria invasion induced by the met receptor. J Cell Sci. 2005;118:1537–1547. doi: 10.1242/jcs.02285. [DOI] [PubMed] [Google Scholar]
- [27].Bierne H, Gouin E, Roux P, Caroni P, Yin HL, Cossart P. A role for cofilin and LIM kinase in Listeria-induced phagocytosis. J Cell Biol. 2001;155:101–112. doi: 10.1083/jcb.200104037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Welch MD, DePace AH, Verma S, Iwamatsu A, Mitchison TJ. The human Arp2/3 complex is composed of evolutionarily conserved subunits and is localized to cellular regions of dynamic actin filament assembly. J Cell Biol. 1997;138:375–384. doi: 10.1083/jcb.138.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Kühbacher A, Emmenlauer M, Rämö P, Kafai N, Dehio C, Cossart P, et al. Genome-wide siRNA screen identifies complementary signaling pathways involved in Listeria infection and reveals different actin nucleation mechanisms during Listeria cell invasion and actin comet tail formation. mBio. 2015;6:e00598–15. doi: 10.1128/mBio.00598-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Mostowy S, Cossart P. Septins: the fourth component of the cytoskeleton. Nat Rev Mol Cell Biol. 2012;13:183–194. doi: 10.1038/nrm3284. [DOI] [PubMed] [Google Scholar]
- [31].Mostowy S, Janel S, Forestier C, Roduit C, Kasas S, Pizarro-Cerdá J, et al. A role for septins in the interaction between the Listeria monocytogenes invasion protein InlB and the met receptor. Biophys J. 2011;100:1949–1959. doi: 10.1016/j.bpj.2011.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Pizarro-Cerdá J, Jonquières R, Gouin E, Vandekerckhove J, Garin J, Cossart P. Distinct protein patterns associated with Listeria monocytogenes InlA- or InlB-phagosomes. Cell Microbiol. 2002;4:101–115. doi: 10.1046/j.1462-5822.2002.00169.x. [DOI] [PubMed] [Google Scholar]
- [33].Mostowy S, Danckaert A, Tham TN, Machu C, Guadagnini S, Pizarro-Cerdá J, et al. Septin 11 restricts InlB-mediated invasion by Listeria. J Biol Chem. 2009;284:11613–11621. doi: 10.1074/jbc.M900231200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Mostowy S, Tham TN, Danckaert A, Guadagnini S, Boisson-Dupuis S, Pizarro-Cerdá J, et al. Septins regulate bacterial entry into host cells. PLoS One. 2009;4:e4196. doi: 10.1371/journal.pone.0004196.g005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Pizarro-Cerdá J, Payrastre B, Wang Y-J, Veiga E, Yin HL, Cossart P. Type II phosphatidylinositol 4-kinases promote Listeria monocytogenes entry into target cells. Cell Microbiol. 2007;9:2381–2390. doi: 10.1111/j.1462-5822.2007.00967.x. [DOI] [PubMed] [Google Scholar]
- [36].Pizarro-Cerdá J, Kühbacher A, Cossart P. Phosphoinositides and host-pathogen interactions. Biochim Biophys Acta. 2014 doi: 10.1016/j.bbalip.2014.09.011. [DOI] [PubMed] [Google Scholar]
- [37].Rämö P, Drewek A, Arrieumerlou C, Beerenwinkel N, Ben-Tekaya H, Cardel B, et al. Simultaneous analysis of large-scale RNAi screens for pathogen entry. BMC Genom. 2014;15:1162. doi: 10.1186/1471-2164-15-1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Tham TN, Gouin E, Rubinstein E, Boucheix C, Cossart P, Pizarro-Cerdá J. Tetraspanin CD81 is required for Listeria monocytogenes invasion. Infect Immun. 2010;78:204–209. doi: 10.1128/IAI.00661-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Silvie O, Rubinstein E, Franetich J-F, Prenant M, Belnoue E, Rénia L, et al. Hepatocyte CD81 is required for Plasmodium falciparum and Plasmodium yoelii sporozoite infectivity. Nat Med. 2003;9:93–96. doi: 10.1038/nm808. [DOI] [PubMed] [Google Scholar]
- [40].Kapadia SB, Barth H, Baumert T, McKeating JA, Chisari FV. Initiation of hepatitis C virus infection is dependent on cholesterol and cooperativity between CD81 and scavenger receptor B type I. J Virol. 2006;81:374–383. doi: 10.1128/JVI.01134-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Martinez del Hoyo G, Ramirez-Huesca M, Levy S, Boucheix C, Rubinstein E, Minguito de la Escalera M, et al. CD81 controls immunity to Listeria infection through Rac-dependent inhibition of proin-ammatory mediator release and activation of cytotoxic T cells. J Immunol. 2015;194:6090–6101. doi: 10.4049/jimmunol.1402957. [DOI] [PubMed] [Google Scholar]
- [42].Kühbacher A, Dambournet D, Echard A, Cossart P, Pizarro-Cerdá J. Phosphatidylinositol 5-phosphatase oculocerebrorenal syndrome of Lowe protein (OCRL) controls actin dynamics during early steps of Listeria monocytogenes infection. J Biol Chem. 2012;287:13128–13136. doi: 10.1074/jbc.M111.315788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Singh R, Jamieson A, Cresswell P. GILT is a critical host factor for Listeria monocytogenes infection. Nature. 2008;455:1244–1247. doi: 10.1038/nature07344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Beauregard KE, Lee KD, Collier RJ, Swanson JA. pH-dependent perforation of macrophage phagosomes by listeriolysin O from Listeria monocytogenes. J Exp Med. 1997;186:1159–1163. doi: 10.1084/jem.186.7.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Henry R, Shaughnessy L, Loessner MJ, Alberti-Segui C, Higgins DE, Swanson JA. Cytolysin-dependent delay of vacuole maturation in macrophages infected with Listeria monocytogenes. Cell Microbiol. 2006;8:107–119. doi: 10.1111/j.1462-5822.2005.00604.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Shaughnessy LM, Hoppe AD, Christensen KA, Swanson JA. Membrane perforations inhibit lysosome fusion by altering pH and calcium in Listeria monocytogenes vacuoles. Cell Microbiol. 2006;8:781–792. doi: 10.1111/j.1462-5822.2005.00665.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Smith GA, Marquis H, Jones S, Johnston NC, Portnoy DA, Goldfine H. The two distinct phospholipases C of Listeria monocytogenes have overlapping roles in escape from a vacuole and cell-to-cell spread. Infect Immun. 1995;63:4231–4237. doi: 10.1128/iai.63.11.4231-4237.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Dramsi S, Cossart P. Listeriolysin O-mediated calcium in-ux potentiates entry of Listeria monocytogenes into the human Hep-2 epithelial cell line. Infect Immun. 2003;71:3614–3618. doi: 10.1128/IAI.71.6.3614-3618.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Vadia S, Arnett E, Haghighat A-C, Wilson-Kubalek EM, Tweten RK, Seveau S. The pore-forming toxin listeriolysin O mediates a novel entry pathway of L. monocytogenes into human hepatocytes. PLoS Pathog. 2011;7:e1002356. doi: 10.1371/journal.ppat.1002356.g009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Forestier C-L, Machu C, Loussert C, Pescher P, Späth GF. Imaging host cell-leishmania interaction dynamics implicates parasite motility, lysosome recruitment, and host cell wounding in the infection process. Cell Host Microbe. 2011;9:319–330. doi: 10.1016/j.chom.2011.03.011. [DOI] [PubMed] [Google Scholar]
- [51].Fernandes MC, Cortez M, Flannery AR, Tam C, Mortara RA, Andrews NW. Trypanosoma cruzi subverts the sphingomyelinase-mediated plasma membrane repair pathway for cell invasion. J Exp Med. 2011;208:909–921. doi: 10.1083/jcb.140.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Ribet D, Hamon M, Gouin E, Nahori M-A, Impens F, Neyret-Kahn H, et al. Listeria monocytogenes impairs SUMOylation for efficient infection. Nature. 2010;464:1192–1195. doi: 10.1038/nature08963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Hamon MA, Batsché E, Régnault B, Tham TN, Seveau S, Muchardt C, et al. Histone modifications induced by a family of bacterial toxins. Proc Natl Acad Sci U S A. 2007;104:13467–13472. doi: 10.1073/pnas.0702729104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Stavru F, Bouillaud F, Sartori A, Ricquier D, Cossart P. Listeria monocytogenes transiently alters mitochondrial dynamics during infection. Proc Natl Acad Sci. 2011;108:3612–3617. doi: 10.1073/pnas.1100126108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Kocks C, Gouin E, Tabouret M, Berche P, Ohayon H, Cossart P. L.monocytogenes-induced actin assembly requires the actA gene product, a surface protein. Cell. 1992;68:521–531. doi: 10.1016/0092-8674(92)90188-i. [DOI] [PubMed] [Google Scholar]
- [56].Welch MD, Rosenblatt J, Skoble J, Portnoy DA, Mitchison TJ. Interaction of human Arp2/3 complex and the Listeria monocytogenes ActA protein in actin filament nucleation. Science. 1998;281:105–108. doi: 10.1126/science.281.5373.105. [DOI] [PubMed] [Google Scholar]
- [57].Boujemaa-Paterski R, Gouin E, Hansen G, Samarin S, Le Clainche C, Didry D, et al. Listeria protein ActA mimics WASp family proteins: it activates filament barbed end branching by Arp2/3 complex. Biochemistry. 2001;40:11390–11404. doi: 10.1021/bi010486b. [DOI] [PubMed] [Google Scholar]
- [58].Abella JVG, Galloni C, Pernier J, Barry DJ, Kjær S, Carlier M-F, et al. Isoform diversity in the Arp2/3 complex determines actin filament dynamics. Nat Cell Biol. 2015;18:76–86. doi: 10.1038/ncb3286. [DOI] [PubMed] [Google Scholar]
- [59].Van Troys M, Lambrechts A, David V, Demol H, Puype M, Pizarro-Cerdá J, et al. The actin propulsive machinery: the proteome of Listeria monocytogenes tails. Biochem Biophys Res Commun. 2008;375:194–199. doi: 10.1016/j.bbrc.2008.07.152. [DOI] [PubMed] [Google Scholar]
- [60].Rich KA, Burkett C, Webster P. Cytoplasmic bacteria can be targets for autophagy. Cell Microbiol. 2003;5:455–468. doi: 10.1046/j.1462-5822.2003.00292.x. [DOI] [PubMed] [Google Scholar]
- [61].Birmingham CL, Canadien V, Gouin E, Troy EB, Yoshimori T, Cossart P, et al. Listeria monocytogenes evades killing by autophagy during colonization of host cells. Autophagy. 2007;3:442–451. doi: 10.4161/auto.4450. [DOI] [PubMed] [Google Scholar]
- [62].Yoshikawa Y, Ogawa M, Hain T, Yoshida M, Fukumatsu M, Kim M, et al. Listeria monocytogenes ActA-mediated escape from autophagic recognition. Nat Cell Biol. 2009;11:1233–1240. doi: 10.1038/ncb1967. [DOI] [PubMed] [Google Scholar]
- [63].Py BF, Lipinski MM, Yuan J. Autophagy limits Listeria monocytogenes intracellular growth in the early phase of primary infection. Autophagy. 2007;3:117–125. doi: 10.4161/auto.3618. [DOI] [PubMed] [Google Scholar]
- [64].Dortet L, Mostowy S, Louaka AS, Gouin E, Nahori M-A, Wiemer EAC, et al. Recruitment of the major vault protein by InlK: a Listeria monocytogenes strategy to avoid autophagy. PLoS Pathog. 2011;7:e1002168. doi: 10.1371/journal.ppat.1002168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Radoshevich L, Impens F, Ribet D, Quereda JJ, Nam Tham T, Nahori M-A, et al. ISG15 counteracts Listeria monocytogenes infection. eLife. 2015;4 doi: 10.7554/eLife.06848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Robbins JR, Barth AI, Marquis H, de Hostos EL, Nelson WJ, Theriot JA. Listeria monocytogenes exploits normal host cell processes to spread from cell to cell. J Cell Biol. 1999;146:1333–1350. doi: 10.1083/jcb.146.6.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Monack DM, Theriot JA. Actin-based motility is sufficient for bacterial membrane protrusion formation and host cell uptake. Cell Microbiol. 2001;3:633–647. doi: 10.1046/j.1462-5822.2001.00143.x. [DOI] [PubMed] [Google Scholar]
- [68].Wang J, King JE, Goldrick M, Lowe M, Gertler FB, Roberts IS. Lamellipodin is important for cell-to-cell spread and actin-based motility in Listeria monocytogenes. Infect Immun. 2015;83:3740–3748. doi: 10.1128/IAI.00193-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Pust S, Morrison H, Wehland J, Sechi AS, Herrlich P. Listeria monocytogenes exploits ERM protein functions to efficiently spread from cell to cell. EMBO J. 2005;24:1287–1300. doi: 10.1038/sj.emboj.7600595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Fattouh R, Kwon H, Czuczman MA, Copeland JW, Pelletier L, Quinlan ME, et al. The diaphanous-related formins promote protrusion formation and cell-to-cell spread of Listeria monocytogenes. J Infect Dis. 2015;211:1185–1195. doi: 10.1093/infdis/jiu546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Chong R, Squires R, Swiss R, Agaisse H. RNAi screen reveals host cell kinases specifically involved in Listeria monocytogenes spread from cell to cell. PLoS One. 2011;6:e23399. doi: 10.1371/journal.pone.0023399.t001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Rajabian T, Gavicherla B, Heisig M, Müller-Altrock S, Goebel W, Gray-Owen SD, et al. The bacterial virulence factor InlC perturbs apical cell junctions and promotes cell-to-cell spread of Listeria. Nat Cell Biol. 2009;11:1212–1218. doi: 10.1038/ncb1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Gianfelice A, Le PHB, Rigano LA, Saila S, Dowd GC, McDivitt T, et al. Host endoplasmic reticulum COPII proteins control cell-to-cell spread of the bacterial pathogen Listeria monocytogenes. Cell Microbiol. 2015;17:876–892. doi: 10.1111/cmi.12409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Czuczman MA, Fattouh R, van Rijn JM, Canadien V, Osborne S, Muise AM, et al. Listeria monocytogenes exploits efferocytosis to promote cell-to-cell spread. Nature. 2014:1–17. doi: 10.1038/nature13168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Alberti-Segui C, Goeden KR, Higgins DE. Differential function of Listeria monocytogenes listeriolysin O and phospholipases C in vacuolar dissolution following cell-to-cell spread. Cell Microbiol. 2007;9:179–195. doi: 10.1111/j.1462-5822.2006.00780.x. [DOI] [PubMed] [Google Scholar]
- [76].Carvalho CL, de Carvalho IL, Zé-Zé L, Núncio MS, Duarte EL. Comparative immunology, microbiology and infectious diseases, comparative immunology. Microbiol Infect Dis. 2014;37:85–96. doi: 10.1016/j.cimid.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Keim P, Johansson A, Wagner DM. Molecular epidemiology, evolution, and ecology of francisella. Ann N Y Acad Sci. 2007;1105:30–66. doi: 10.1196/annals.1409.011. [DOI] [PubMed] [Google Scholar]
- [78].Dennis DT, Inglesby TV, Henderson DA, Bartlett JG, Ascher MS, Eitzen E, et al. Tularemia as a biological weapon: medical and public health management. JAMA. 2001:2763–2773. doi: 10.1001/jama.285.21.2763. [DOI] [PubMed] [Google Scholar]
- [79].Rowe HM, Huntley JF. From the outside-in: the francisella tularensis envelope and virulence. Front Cell Infect Microbiol. 2015;5:2041. doi: 10.1016/j.str.2015.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Santic M, Molmeret M, Klose KE, Abu Kwaik Y. Francisella tularensis travels a novel, twisted road within macrophages. Trends Microbiol. 2006;14:37–44. doi: 10.1016/j.tim.2005.11.008. [DOI] [PubMed] [Google Scholar]
- [81].Moreau GB, Mann BJ. Adherence and uptake of Francisellainto host cells. Virulence. 2014;4:826–832. doi: 10.4161/viru.25629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Schlesinger LS, Bellinger-Kawahara CG, Payne NR, Horwitz MA. Phagocytosis of Mycobacterium tuberculosis is mediated by human monocyte complement receptors and complement component C3. J Immunol. 1990;144:2771–2780. [PubMed] [Google Scholar]
- [83].Clemens DL, Lee BY, Horwitz MA. Francisella tularensis enters macrophages via a novel process involving pseudopod loops. Infect Immun. 2005;73:5892–5902. doi: 10.1128/IAI.73.9.5892-5902.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Tamilselvam B, Daefler S. Francisella targets cholesterol-rich host cell membrane domains for entry into macrophages. J Immunol. 2008;180:8262–8271. doi: 10.4049/jimmunol.180.12.8262. [DOI] [PubMed] [Google Scholar]
- [85].Celli J, Zahrt TC. Mechanisms of Francisella tularensis intracellular pathogenesis. Cold Spring Harb Perspect Med. 2013;3:a010314. doi: 10.1101/cshperspect.a010314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Chong A, Wehrly TD, Nair V, Fischer ER, Barker JR, Klose KE, et al. The early phagosomal stage of Francisella tularensis determines optimal phagosomal escape and Francisella pathogenicity island protein expression. Infect Immun. 2008;76:5488–5499. doi: 10.1128/IAI.00682-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Santic M, Asare R, Skrobonja I, Jones S, Abu Kwaik Y. Acquisition of the vacuolar ATPase proton pump and phagosome acidification are essential for escape of Francisella tularensis into the macrophage cytosol. Infect Immun. 2008;76:2671–2677. doi: 10.1128/IAI.00185-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Clemens DL, Lee BY, Horwitz MA. Virulent and avirulent strains of Francisella tularensis prevent acidification and maturation of their phagosomes and escape into the cytoplasm in human macrophages. Infect Immun. 2004;72:3204–3217. doi: 10.1128/IAI.72.6.3204-3217.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].McCaffrey RL, Schwartz JT, Lindemann SR, Moreland JG, Buchan BW, Jones BD, et al. Multiple mechanisms of NADPH oxidase inhibition by type A and type B Francisella tularensis. J Leukoc Biol. 2010;88:791–805. doi: 10.1189/jlb.1209811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Geier H, Celli J. Phagocytic receptors dictate phagosomal escape and intracellular proliferation of Francisella tularensis. Infect Immun. 2011;79:2204–2214. doi: 10.1128/IAI.01382-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Dieppedale J, Gesbert G, Ramond E, Chhuon C, Dubail I, Dupuis M, et al. Possible links between stress defense and the tricarboxylic acid (TCA) cycle in francisella pathogenesis. Mol Cell Proteom. 2013;12:2278–2292. doi: 10.1074/mcp.M112.024794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Santic M, Molmeret M, Klose KE, Jones S, Kwaik YA. The Francisella tularensis pathogenicity island protein IglC and its regulator MglA are essential for modulating phagosome biogenesis and subsequent bacterial escape into the cytoplasm. Cell Microbiol. 2005;7:969–979. doi: 10.1111/j.1462-5822.2005.00526.x. [DOI] [PubMed] [Google Scholar]
- [93].Brotcke A, Monack DM. Identification of fevR, a novel regulator of virulence gene expression in Francisella novicida. Infect Immun. 2008;76:3473–3480. doi: 10.1128/IAI.00430-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Clemens DL, Ge P, Lee B-Y, Horwitz MA, Zhou ZH. Atomic structure of T6SS reveals interlaced array essential to function. Cell. 2015;160:940–951. doi: 10.1016/j.cell.2015.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Barel M, Ramond E, Gesbert G, Charbit A. The complex amino acid diet of Francisella in infected macrophages. Front Cell Infect Microbiol. 2015;5:1–5. doi: 10.3389/fcimb.2015.00009/abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Gesbert G, Ramond E, Rigard M, Frapy E, Dupuis M, Dubail I, et al. Asparagine assimilation is critical for intracellular replication and dissemination of Francisella. Cell Microbiol. 2014;16:434–449. doi: 10.1111/cmi.12227. [DOI] [PubMed] [Google Scholar]
- [97].Steele S, Brunton J, Ziehr B, Taft-Benz S, Moorman N, Kawula T. Francisella tularensis harvests nutrients derived via ATG5-independent autophagy to support intracellular growth. PLoS Pathog. 2013;9:e1003562. doi: 10.1371/journal.ppat.1003562.s007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Checroun C, Wehrly TD, Fischer ER, Hayes SF, Celli J. Autophagy-mediated reentry of Francisella tularensis into the endocytic compartment after cytoplasmic replication. Proc Natl Acad Sci U S A. 2006;103:14578–14583. doi: 10.1073/pnas.0601838103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Meunier E, Wallet P, Dreier RF, Costanzo S, Anton L, Rühl S, et al. Guanylate-binding proteins promote activation of the AIM2 inflammasome during infection with Francisella novicida. Nat Publ Group. 2015;16:476–484. doi: 10.1038/ni.3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Steele S, Radlinski L, Taft-Benz S, Brunton J, Kawula TH. Trogocytosis-associated cell to cell spread of intracellular bacterial pathogens. eLife. 2016;5 doi: 10.7554/elife.10625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Kotloff KL, Winickoff JP, Ivanoff B, Clemens JD, Swerdlow DL, Sansonetti PJ, et al. Global burden of Shigella infections: implications for vaccine development and implementation of control strategies. Bull World Health Org. 1999;77:651–666. [PMC free article] [PubMed] [Google Scholar]
- [102].Phalipon A, Sansonetti PJ. Shigella’s ways of manipulating the host intestinal innate and adaptive immune system: a tool box for survival? Immunol Cell Biol. 2007;85:119–129. doi: 10.1038/sj.icb7100025. [DOI] [PubMed] [Google Scholar]
- [103].Buchrieser C, Glaser P, Rusniok C, Nedjari H, D’Hauteville H, Kunst F, et al. The virulence plasmid pWR100 and the repertoire of proteins secreted by the type III secretion apparatus of Shigella flexneri. Mol Microbiol. 2000;38:760–771. doi: 10.1046/j.1365-2958.2000.02179.x. [DOI] [PubMed] [Google Scholar]
- [104].Parsot C. Shigella type III secretion effectors: how, where, when, for what purposes? Curr Opin Microbiol. 2009;12:110–116. doi: 10.1016/j.mib.2008.12.002. [DOI] [PubMed] [Google Scholar]
- [105].Enninga J, Mounier J, Sansonetti P, Tran-Van-Nhieu G. Secretion of type III effectors into host cells in real time. Nat Meth. 2005;2:959–965. doi: 10.1038/nmeth804. [DOI] [PubMed] [Google Scholar]
- [106].Romero S, Grompone G, Carayol N, Mounier J, Guadagnini S, Prévost M-C, et al. ATP-mediated erk1/2 activation stimulates bacterial capture by filopodia, which precedes shigella invasion of epithelial cells. Cell Host Microbe. 2011;9:508–519. doi: 10.1016/j.chom.2011.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Watarai M, Funato S, Sasakawa C. Interaction of Ipa proteins of Shigella flexneri with alpha5beta1 integrin promotes entry of the bacteria into mammalian cells. J Exp Med. 1996;183:991–999. doi: 10.1084/jem.183.3.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Mounier J, Boncompain G, Senerovic L, Lagache T, Chrétien F, Perez F, et al. Shigella effector IpaB-induced cholesterol relocation disrupts the golgi complex and recycling network to inhibit host cell secretion. Cell Host Microbe. 2012;12:381–389. doi: 10.1016/j.chom.2012.07.010. [DOI] [PubMed] [Google Scholar]
- [109].Zumsteg AB, Goosmann C, Brinkmann V, Morona R, Zychlinsky A. IcsA is a Shigella flexneri adhesin regulated by the type III secretion system and required for pathogenesis. Cell Host Microbe. 2014;15:435–445. doi: 10.1016/j.chom.2014.03.001. [DOI] [PubMed] [Google Scholar]
- [110].Lafont F, Nhieu GTV, Hanada K, Sansonetti P, Goot FGVD. Initial steps of Shigella infection depend on the cholesterol/sphingolipid raft-mediated CD44-IpaB interaction. EMBO J. 2002;21:4449–4457. doi: 10.1093/emboj/cdf457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Van Der Goot FG. Rafts can trigger contact-mediated secretion of bacterial effectors via a lipid-based mechanism. J Biol Chem. 2004;279:47792–47798. doi: 10.1074/jbc.M406824200. [DOI] [PubMed] [Google Scholar]
- [112].Skoudy A, Mounier J, Aruffo A, Ohayon H, Gounon P, Sansonetti P, et al. CD44 binds to the Shigella IpaB protein and participates in bacterial invasion of epithelial cells. Cell Microbiol. 2000;2:19–33. doi: 10.1046/j.1462-5822.2000.00028.x. [DOI] [PubMed] [Google Scholar]
- [113].Cossart P, Sansonetti PJ. Bacterial invasion: the paradigms of enteroinvasive pathogens. Science. 2004;304:242–248. doi: 10.1126/science.1090124. [DOI] [PubMed] [Google Scholar]
- [114].Ramarao N, Le Clainche C, Izard T, Bourdet-Sicard R, Ageron E, Sansonetti PJ, et al. Capping of actin filaments by vinculin activated by the Shigella IpaA carboxyl-terminal domain. FEBS Lett. 2007;581:853–857. doi: 10.1016/j.febslet.2007.01.057. [DOI] [PubMed] [Google Scholar]
- [115].Demali KA, Jue AL, Burridge K. IpaA targets beta1 integrins and rho to promote actin cytoskeleton rearrangements necessary for Shigella entry. J Biol Chem. 2006;281:39534–39541. doi: 10.1074/jbc.M605939200. [DOI] [PubMed] [Google Scholar]
- [116].Mounier J, Popoff MR, Enninga J, Frame MC, Sansonetti PJ, Van Nhieu GT. The IpaC carboxyterminal effector domain mediates src-dependent actin polymerization during shigella invasion of epithelial cells. PLoS Pathog. 2009;5:e1000271. doi: 10.1371/journal.ppat.1000271.t001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Bougnères L, Girardin SE, Weed SA, Karginov AV, Olivo-Marin J-C, Parsons JT, et al. Cortactin and Crk cooperate to trigger actin polymerization during Shigella invasion of epithelial cells. J Cell Biol. 2004;166:225–235. doi: 10.1083/jcb.200402073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Ohya K, Handa Y, Ogawa M, Suzuki M, Sasakawa C. IpgB1 is a novel Shigella effector protein involved in bacterial invasion of host cells. J Biol Chem. 2005;280:24022–24034. doi: 10.1074/jbc.M502509200. [DOI] [PubMed] [Google Scholar]
- [119].Handa Y, Suzuki M, Ohya K, Iwai H, Ishijima N, Koleske AJ, et al. Shigella IpgB1 promotes bacterial entry through the ELMO–Dock180 machinery. Nat Cell Biol. 2006;9:121–128. doi: 10.1038/ncb1526. [DOI] [PubMed] [Google Scholar]
- [120].Ehsani S, Santos JC, Rodrigues CD, Henriques R, Audry L, Zimmer C, et al. Hierarchies of host factor dynamics at the entry site of Shigella flexneri during host cell invasion. Infect Immun. 2012;80:2548–2557. doi: 10.1128/IAI.06391-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Alto NM, Shao F, Lazar CS, Brost RL, Chua G, Mattoo S, et al. Identification of a bacterial type III effector family with G protein mimicry functions. Cell. 2006;124:133–145. doi: 10.1016/j.cell.2005.10.031. [DOI] [PubMed] [Google Scholar]
- [122].Orchard RC, Alto NM. Mimicking GEFs: a common theme for bacterial pathogens. Cell Microbiol. 2011;14:10–18. doi: 10.1111/j.1462-5822.2011.01703.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Ogawa M, Handa Y, Ashida H, Suzuki M, Sasakawa C. The versatility of Shigella effectors. Nat Rev Microbiol. 2008;6:11–16. doi: 10.1038/nrmicro1814. [DOI] [PubMed] [Google Scholar]
- [124].Yoshida S, Katayama E, Kuwae A, Mimuro H, Suzuki T, Sasakawa C. Shigella deliver an effector protein to trigger host microtubule destabilization, which promotes Rac1 activity and efficient bacterial internalization. EMBO J. 2002;21:2923–2935. doi: 10.1093/emboj/cdf319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Germane KL, Ohi R, Goldberg MB, Spiller BW. Structural and functional studies indicate that shigella VirA is not a protease and does not directly destabilize microtubules ‡. Biochemistry. 2008;47:10241–1024. doi: 10.1021/bi801533k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Hayward RD, Cain RJ, McGhie EJ, Phillips N, Garner MJ, Koronakis V. Cholesterol binding by the bacterial type III translocon is essential for virulence effector delivery into mammalian cells. Mol Microbiol. 2005;56:590–603. doi: 10.1111/j.1365-2958.2005.04568.x. [DOI] [PubMed] [Google Scholar]
- [127].High N, Mounier J, Prevost MC, Sansonetti PJ. IpaB of Shigella flexneri causes entry into epithelial cells and escape from the phagocytic vacuole. EMBO J. 1992;11:1991–1999. doi: 10.1002/j.1460-2075.1992.tb05253.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Blocker A, Gounon P, Larquet E, Niebuhr K, Cabiaux V, Parsot C, et al. The tripartite type III secreton of Shigella flexneri inserts IpaB and IpaC into host membranes. J Cell Biol. 1999;147:683–693. doi: 10.1083/jcb.147.3.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Du J, Reeves AZ, Klein JA, Twedt DJ, Knodler LA, Lesser CF. The type III secretion system apparatus determines the intracellular niche of bacterial pathogens. Proc Natl Acad Sci U S A. 2016:1–6. doi: 10.1073/pnas.1520699113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Paetzold S, Lourido S, Raupach B, Zychlinsky A. Shigella flexneri phagosomal escape is independent of invasion. Infect Immun. 2007;75:4826–4830. doi: 10.1128/IAI.00454-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Paz I, Sachse M, Dupont N, Mounier J, Cederfur C, Enninga J, et al. Galectin-3, a marker for vacuole lysis by invasive pathogens. Cell Microbiol. 2010;12:530–544. doi: 10.1111/j.1462-5822.2009.01415.x. [DOI] [PubMed] [Google Scholar]
- [132].Ray K, Bobard A, Danckaert A, Paz-Haftel I, Clair C, Ehsani S, et al. Tracking the dynamic interplay between bacterial and host factors during pathogen-induced vacuole rupture in real time. Cell Microbiol. 2010;12:545–556. doi: 10.1111/j.1462-5822.2010.01428.x. [DOI] [PubMed] [Google Scholar]
- [133].Mellouk N, Weiner A, Aulner N, Schmitt C, Elbaum M, Shorte SL, et al. Shigella subverts the host recycling compartment to rupture its vacuole. Cell Host Microbe. 2014;16:517–530. doi: 10.1016/j.chom.2014.09.005. [DOI] [PubMed] [Google Scholar]
- [134].Weiner A, Mellouk N, Lopez-Montero N, Chang Y-Y, Souque C, Schmitt C, et al. Macropinosomes are key players in early shigella invasion and vacuolar escape in epithelial cells. PLoS Pathog. 2016;12:e1005602. doi: 10.1371/journal.ppat.1005602.s014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Dupont N, Lacas-Gervais S, Bertout J, Paz I, Freche B, Van Nhieu GT, et al. Shigella phagocytic vacuolar membrane remnants participate in the cellular response to pathogen invasion and are regulated by autophagy. Cell Host Microbe. 2009;6:137–149. doi: 10.1016/j.chom.2009.07.005. [DOI] [PubMed] [Google Scholar]
- [136].Stevens JM, Galyov EE, Stevens MP. Actin-dependent movement of bacterial pathogens. Nat Rev Microbiol. 2006;4:91–101. doi: 10.1038/nrmicro1320. [DOI] [PubMed] [Google Scholar]
- [137].Ray K, Marteyn B, Sansonetti PJ, Tang CM. Life on the inside: the intracellular lifestyle of cytosolic bacteria. Nat Rev Microbiol. 2009;7:333–340. doi: 10.1038/nrmicro2112. [DOI] [PubMed] [Google Scholar]
- [138].Mostowy S, Bonazzi M, Hamon MA, Tham TN, Mallet A, Lelek M, et al. Entrapment of intracytosolic bacteria by septin cage-like structures. Cell Host Microbe. 2010;8:433–444. doi: 10.1016/j.chom.2010.10.009. [DOI] [PubMed] [Google Scholar]
- [139].McNally A, Thomson NR, Reuter S, Wren BW. Add, stir and reduce: Yersinia spp. as model bacteria for pathogen evolution. Nat Rev Microbiol. 2016;14:177–190. doi: 10.1038/nrmicro.2015.29. [DOI] [PubMed] [Google Scholar]
- [140].Grützkau A, Hanski C, Hahn H, Riecken EO. Involvement of M cells in the bacterial invasion of Peyer’s patches: a common mechanism shared by Yersinia enterocolitica and other enteroinvasive bacteria. Gut. 1990;31:1011–1015. doi: 10.1136/gut.31.9.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Pepe JC, Miller VL. Yersinia enterocolitica invasin: a primary role in the initiation of infection. Proc Natl Acad Sci U S A. 1993;90:6473–6477. doi: 10.1073/pnas.90.14.6473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Clark MA, Hirst BH, Jepson MA. M-cell surface beta1 integrin expression and invasin-mediated targeting of Yersinia pseudotuberculosis to mouse Peyer’s patch M cells. Infect Immun. 1998;66:1237–1243. doi: 10.1128/iai.66.3.1237-1243.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Isberg RR, Voorhis DL, Falkow S. Identification of invasin: a protein that allows enteric bacteria to penetrate cultured mammalian cells. Cell. 1987;50:769–778. doi: 10.1016/0092-8674(87)90335-7. [DOI] [PubMed] [Google Scholar]
- [144].Alrutz MA, Isberg RR. Involvement of focal adhesion kinase in invasin-mediated uptake. Proc Natl Acad Sci U S A. 1998;95:13658–13663. doi: 10.1073/pnas.95.23.13658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].Bruce-Staskal PJ, Weidow CL, Gibson JJ, Bouton AH. Cas Fak and Pyk2 function in diverse signaling cascades to promote Yersinia uptake. J Cell Sci. 2002;115:2689–2700. doi: 10.1242/jcs.115.13.2689. [DOI] [PubMed] [Google Scholar]
- [146].Alrutz MA, Srivastava A, Wong KW, D’Souza-Schorey C, Tang M, Ch’Ng LE, et al. Efficient uptake of Yersinia pseudotuberculosis via integrin receptors involves a Rac1-Arp 2/3 pathway that bypasses N-WASP function. Mol Microbiol. 2001;42: 689–703. doi: 10.1046/j.1365-2958.2001.02676.x. [DOI] [PubMed] [Google Scholar]
- [147].Wong KW, Isberg RR. Arf6 and phosphoinositol-4-phosphate-5-kinase activities permit bypass of the rac1 requirement for 1 integrin-mediated bacterial uptake. J Exp Med. 2003;198:603–614. doi: 10.1084/jem.20021363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Sarantis H, Balkin DM, De Camilli Pietro, Isberg RR, Brumell JH, Grinstein S. Yersinia entry into host cells requires rab5-dependent dephosphorylation of PI(4, 5)P2 and membrane scission. Cell Host Microbe. 2012;11:117–128. doi: 10.1016/j.chom.2012.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Mikula KM, Kolodziejczyk R, Goldman A. Yersinia infection tools-characterization of structure and function of adhesins. Front Cell Infect Microbiol. 2012;2(169) doi: 10.3389/fcimb.2012.00169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].Deuretzbacher A, Czymmeck N, Reimer R, Trülzsch K, Gaus K, Hohenberg H, et al. Beta1 integrin-dependent engulfment of Yersinia enterocolitica by macrophages is coupled to the activation of autophagy and suppressed by type III protein secretion. J Immunol. 2009;183:5847–5860. doi: 10.4049/jimmunol.0804242. [DOI] [PubMed] [Google Scholar]
- [151].Pujol C, Klein KA, Romanov GA, Palmer LE, Cirota C, Zhao Z, et al. Yersinia pestis can reside in autophagosomes and avoid xenophagy in murine macrophages by preventing vacuole acidification. Infect Immun. 2009;77:2251–2261. doi: 10.1128/IAI.00068-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Connor MG, Pulsifer AR, Price CT, Abu Kwaik Y, Lawrenz MB. Yersinia pestis requires host rab1b for survival in macrophages. PLoS Pathog. 2015;11:e1005241. doi: 10.1371/journal.ppat.1005241.s004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].Moreau K, Lacas-Gervais S, Fujita N, Sebbane F, Yoshimori T, Simonet M, et al. Autophagosomes can support Yersinia pseudotuberculosis replication in macrophages. Cell Microbiol. 2010;12:1108–1123. doi: 10.1111/j.1462-5822.2010.01456.x. [DOI] [PubMed] [Google Scholar]
- [154].Ligeon L-A, Moreau K, Barois N, Bongiovanni A, Lacorre D-A, Werkmeister E, et al. Role of VAMP3 and VAMP7 in the commitment of Yersinia pseudotuberculosisto LC3-associated pathways involving single- or double-membrane vacuoles. Autophagy. 2014;10:1588–1602. doi: 10.4161/auto.29411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Curfs JH, Meis JF, Fransen JA, van der Lee HA, Hoogkamp-Korstanje JA. Interactions of Yersinia enterocolitica with polarized human intestinal Caco-2 cells. Med Microbiol Immunol. 1995;184:123–127. doi: 10.1007/BF00224348. [DOI] [PubMed] [Google Scholar]
- [156].Cornelis GR. The Yersinia Ysc-Yop type III weaponry. Nat Rev Mol Cell Biol. 2002;3:742–754. doi: 10.1038/nrm932. [DOI] [PubMed] [Google Scholar]
- [157].Persson C, Carballeira N, Wolf-Watz H, Fallman M. The PTPase YopH inhibits uptake of Yersinia, tyrosine phosphorylation of p130Cas and FAK, and the associated accumulation of these proteins in peripheral focal adhesions. EMBO J. 1997;16:2307–2318. doi: 10.1093/emboj/16.9.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [158].Von Pawel-Rammingen U, Telepnev MV, Schmidt G, Aktories K, Wolf-Watz H, Rosqvist R. GAP activity of the Yersinia YopE cytotoxin specifically targets the Rho pathway: a mechanism for disruption of actin microfilament structure. Mol Microbiol. 2000;36:737–748. doi: 10.1046/j.1365-2958.2000.01898.x. [DOI] [PubMed] [Google Scholar]
- [159].Viboud GI, Mejia E, Bliska JB. Comparison of YopE and YopT activities in counteracting host signalling responses to Yersinia pseudotuberculosis infection. Cell Microbiol. 2006;8:1504–1515. doi: 10.1111/j.1462-5822.2006.00729.x. [DOI] [PubMed] [Google Scholar]
- [160].Groves E, Rittinger K, Amstutz M, Berry S, Holden DW, Cornelis GR, et al. Sequestering of Rac by the Yersinia effector YopO blocks Fcgamma receptor-mediated phagocytosis. J Biol Chem. 2010;285:4087–4098. doi: 10.1074/jbc.M109.071035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [161].Ke Y, Tan Y, Wei N, Yang F, Yang H, Cao S, et al. Yersinia protein kinase A phosphorylates vasodilator-stimulated phosphoprotein to modify the host cytoskeleton. Cell Microbiol. 2014;17:473–485. doi: 10.1111/cmi.12378. [DOI] [PubMed] [Google Scholar]
- [162].Shao F, Vacratsis PO, Bao Z, Bowers KE, Fierke CA, Dixon JE. Biochemical characterization of the Yersinia YopT protease: cleavage site and recognition elements in Rho GTPases. Proc Natl Acad Sci. 2003;100:904–909. doi: 10.1073/pnas.252770599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [163].Chorev DS, Moscovitz O, Geiger B, Sharon M. Regulation of focal adhesion formation by a vinculin-Arp2/3 hybrid complex. Nat Commun. 2014;5:1–11. doi: 10.1038/ncomms4758. [DOI] [PubMed] [Google Scholar]