Abstract
Background
Red oak pollen is an important cause of allergic respiratory disease and it is widely distributed in North America and central Europe. To date, however, red oak pollen allergens have not been identified. Here, we describe the allergenic protein profile from red oak pollen.
Methods
Total proteins were extracted from red oak pollen using a modified phenolic extraction method, and, subsequently, proteins were separated by two-dimensional gel electrophoresis (2DE) for both total protein stain (Coomassie Blue) and immunoblotting. A pool of 8 sera from red oak sensitive patients was used to analyze blotted proteins. Protein spots were analyzed by Mass Spectrometry.
Results
Electrophoretic pattern of total soluble proteins showed higher intensity bands in the regions of 26–40 and 47–52 kDa. Two dimensional immunoblots using pool sera from patients revealed four allergenic proteins spots with molecular masses in the range from 50 to 55 kDa. Mass spectrometry analysis identified 8 proteins including Enolase 1 and Enolase 1 chloroplastic, Xylose isomerase (X1 isoform), mitochondrial Aldehyde dehydrogenase, UTP-Glusose-1-phosphate uridylyltransferase, Betaxylosidase/alpha-l-arabinofuranosidase and alpha- and beta subunits of ATP synthase.
Conclusions
This study has identified for first time 8 IgE binding proteins from red oak pollen. These findings will pave the way towards the development of new diagnostic and therapeutic modalities for red oak allergy.
Keywords: Immunoproteomics, Mass spectrometry, Pollen allergy, Red oak, Two-dimensional gel electrophoresis
Abbreviations: 2-DE, Two-dimensional electrophoresis; AIT, Allergy immunotherapy; BSA, Bovine serum albumin; CHAPS, (3-(3-Cholamidopropyl)dimethylammonio)-1-propanesulfonate); DTT, Dithiothreitol; ED, Emergency department; IEF, Isoelectric focusing; IPG, Immobilized pH gradient; LC, Liquid chromatography; MS, Mass spectrometry; MS/MS, Tandem mass spectrometry; PBS, Phosphate-buffered saline; PMSF, Phenyl methyl sulfonyl fluoride; PVDF, Polyvinylidene difluoride; Q-TOF, Quadrupole Time-of-Flight; SDS, Sodium dodecyl sulfate
Introduction
Oak pollen allergy has a remarkable clinical impact all over the world, and there is plenty of evidence indicating that the prevalence of respiratory allergic reactions induced by oak pollens is increasing. A study conducted in 10 large Canadian cities, found that daily oak tree pollen concentrations were associated within 2.32% of daily asthma hospitalization.1 Similarly, in the USA, the oak pollen was associated with 21,200 asthma emergency department (ED) visits in the northeastern, southeastern, and midwestern states in 2010, with costs valued at $10.4 million.2 In the UK oak pollen is as abundant as birch, and oak trees are found abundantly in Europe,3 where oak has been highlighted as a major tree allergen by the European Academy of Allergy and Clinical Immunology (EAACI).4 Climate change is expected to lengthen and intensify the pollen seasons of a series of allergenic plant taxa.5 It has been predicted that climate change may increase oak pollen season length and asthma ED visits by 5% on average in 2050 in the USA.2,6 In Europe high seasonal patterns of oak pollen are found from Spain to Sweden, although oak pollen seasons varied highly between countries.7
There are 500–600 oak species worldwide and many of them have been associated with allergic disease.8 However, in only 3 oak subspecies pollen allergens have been identified: white oak (Quercus alba), red oak (Q. rubra) and Mongolian oak (Q. mongolica).9 White oak allergens include Que a 1, Ca2+ binding protein and profilin, while Mongolian oak pollen produces Que m 1.9, 10, 11 A sequence Bet v1-like allergen was isolated from red oak and deposited in the UniProt database (Entry identifier:H9NJ54). However, oak allergens have only been partly characterized,12 and there are no publications regarding red oak allergens. This tree is wildly distributed in Central and North America, and Western and Central Europe.13, 14, 15 Thus, it is important to identify allergens produced by oak in order to improve the diagnosis and treatment of allergic patients, in particular, for those residing in complex exposure areas where cross-reactive allergens as Bet v1-like proteins do not guarantee a specific diagnosis.
In the present study, we have investigated the allergenic protein profile of red oak pollen using a discovery immunoproteomics approach.
Methods
Patient selection
Eight polysensitized allergic patients to red oak (Q. rubra) pollen and 4 healthy control subjects were recruited from the outpatient allergy clinic. None of these patients were taking antihistamines, corticosteroids, or specific allergen immunotherapy at the time of the study. Skin prick tests were performed on the forearm to the most common aeroallergens including red oak, white oak, Dermatophagoides pteronyssinus, grass mix, dog hair, feather mix, cat fur, cockroach, weed mix, tree mix, birch, mesquite, Populus alba, cedar, privet, Arizona cypress, alder black, Populus tremuloides, western juniperus, and eucalyptus (Hollister Stier, Elkhart, IN, USA). A skin reaction characterized by ≥ 3 mm wheal diameter within 20 min indicated that the subject was positive. Histamine-induced reaction was used as positive control, and saline solution was used as negative control; 3 out of the 8 asthmatics were also allergic to white oak and the remaining 5 patients were skin prick test positive to other allergens including cat and Dermatophagoides pteronyssinus (Supplementary table 1). After skin prick test, peripheral blood was extracted, and anticoagulated whole blood samples were used to estimate the number of leukocytes by electrical impedance using a cell counter (Sysmex XP-300, Sysmex Corporation, Kobe, Japan), and the percentage of eosinophils was estimated by direct observation through Wright's stain blood preparations, whereas serum was collected from coagulated whole blood samples and stored at −80 °C until use. The protocol was approved by the human ethics and research committees at the National Institute of Respiratory Diseases, Mexico. The study was conducted under the ethical principles of the 1975 Declaration of Helsinki (as revised in 1983), and it was consistent with Good Clinical Practice Guidelines.
Pollen collection and pollen protein extraction
Fresh inflorescences were collected from oak trees (May 2016) in Xalisco, Nayarit, Mexico, and identified as Quercus rubra L. with the assistance of a biologist and a forest engineer. Once dried, anthers were obtained from the inflorescence by passing through a 2 mm mesh, and pollen grains were separated using two standard test sieves (250 and 63 μm, VWR International, Radnor, Pennsylvania. USA). Pollen grain morphology was observed with an optical microscope equipped with camera and micrometer (Lumenera Corporation, Ottawa, Ontario, Canada). Total soluble pollen proteins were extracted using the protocol reported by Faurobert et al.,16 with minor modifications. Briefly, 100 mg of pollen were mixed with 3 mL of SDS-Extraction buffer (30% sucrose, 2% SDS, 0.1 M Tris-HCl, 2% 2-Mercaptoethanol and 1 mM PMSF) and the solution was incubated at 4 °C with shaking for 10 min. One volume of phenol (equilibrated with 10 mM Tris-HCl) was added to the above solution, mixed, and centrifuged for 10 min at 5500×g and 4 °C. The phenolic phase was recovered in a new tube and mixed with 0.1 M ammonium acetate and incubated overnight at −20 °C. After 20 min centrifugation at 20,000×g and 4 °C, the pellet was washed 3 times with cold acetone plus 2 times with 80% acetone prior to vacuum drying the pellet (Vacufuge plus, Eppendorf, Hamburg, Germany). The pellet was suspended in 400 μL of rehydration buffer [8 M urea, 2% CHAPS, 20 mM DTT, 0.002% Bromophenol blue, 0.5% IPG buffer pH 3–10 (Bio-Rad. Hercules, California, USA)]. Total protein concentration was determined by the Bradford method using bovine serum albumin (BSA) as standard.
One and two-dimensional electrophoresis
Pollen proteins were separated by SDS-PAGE in the Mini-PROTEAN Tetra Cell (Bio-Rad). Gels (13% polyacrylamide) were run under reducing-denaturing conditions. To obtain the one-dimensional electrophoretic profile, total soluble pollen proteins (30 μg) were separated in a multi-channel SDS-PAGE gel and stained with Coomassie blue.
For a two-dimensional electrophoretic separation (2-DE), pollen proteins (2 mg) were loaded onto 13 cm linear pH 3–10 IPG (immobilized pH gradient) strips (GE Healthcare, Piscataway, New Jersey, USA). Passive rehydration was carried out at room temperature during 14 h. Isoelectric focusing (IEF) was conducted at 50 mA per IPG strip and 20 °C in an Ettan IPGphor system 3 (GE Healthcare). The IEF conditions were: (1) Constant 500 V until 0.5 kVh, (2) 4000 V linear gradient until 0.8 kVh, (3) 8000 V linear gradient until 11.3 kVh, and (4) Constant 8000 V until 4.4 kVh. After IEF, the strips were equilibrated by shaking for 15 min in SDS equilibration buffer (6 M urea, 30% glycerol, 2% SDS, 50 mM Tris-HCl buffer pH 8.8, 0.002% bromophenol blue) the first time containing 1% dithiothreitol and the second time, equilibration was performed in the same solution but containing 2.5% iodoacetamide instead of dithiothreitol. Equilibrated strips were transferred to a vertical SDS-polyacrylamide gel, sealed with agarose and the second dimension was performed using the Hoeffer SE 600 Ruby system (GE Healthcare). Two independent preparative 2-DE gels were run. One for total protein (Coomassie blue) stain that was scanned with the Typhoon FLA 9500 laser scanner (GE Healthcare). Whereas pollen proteins from a second gel were electro-transferred onto polyvinylidene difluoride (PVDF) sheets (Immun-Blot, Bio-Rad) by using a Trans-Blot SD Semi-Dry Electrophoretic Transfer Cell (Bio-Rad). After electro-transference, membranes were rinsed in Phosphate-buffered saline (PBS) solution pH 7.5 and non-specific binding was blocked by incubating for 2 h in sodium azide-free BSA solution (5%).
Immunodetection analysis
For immunodetection, PVDF membranes were incubated overnight at 4 °C with pooled sera diluted to 1:50 in PBS. After washing (3 times for 5 min with PBS-Tween 20, plus one wash with PBS), the membranes were incubated for 1 h with a mouse monoclonal (B3102E8) horseradish peroxidase-conjugated anti-human IgE-Fc antibody (ABCAM Laboratories, Cambridge, Massachusetts, USA) diluted to 1 μg/mL in PBS. Membranes were washed again. The enzyme-substrate reaction was performed by incubating the membrane in peroxidase substrate for enhanced chemiluminescence by Clarity Western ECL Substrate (Bio-Rad) and imaging was performed with a ChemiDoc MP Imaging System (Bio-Rad) and the software Image Lab v5.1 (Bio-Rad). Acquisition and exposition parameters were automatically optimized by the software.
In-gel protein digestion and liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis
Comparisons of the IgE-reactive spots from the immunoblots to the Coomassie stained 2-DE (Preparative gels) were performed and the protein spots were excised from preparative gels, reduced with 10 mM DTT in 25 mM ammonium bicarbonate followed by protein alkylation with 55 mM iodoacetamide. Protein digestion was carried out overnight at 37 °C with sequencing grade trypsin (Promega, Madison, WI, USA). Tryptic peptide separation was performed using the 1290 Infinity LC System (Agilent Technologies, Santa Clara, California, USA) equipped with an analytical column ZORBAX 300SB-C8 (5 μm × 2.1 mm x 150 mm, Agilent Technologies), and MS/MS analysis was performed by a 6530 Accurate-Mass Quadrupole Time-of-Flight (Q-TOF) LC/MS system (Agilent Technologies) as previously reported by Morales-Amparano et al.17
Protein identification
MS/MS raw data files (.d files) were processed in the Spectrum Mill MS Proteomics Workbench server (Agilent Technologies) to obtain .mzXML files which were transformed into .mgf files in the MSConvert program (available at http://proteowizard.sourceforge.net/). Proteins were then identified using the .mgf files and the MASCOT search engine (http://www.matrixscience.com). Searches were conducted against the Viridiplantae subset of the NCBInr protein database (6,686,534 sequences, August 2018). Trypsin was used as the specific protease, allowing one missed cleavage. Mass error tolerance for precursor and fragment ions was set to 20 ppm and 0.1 Da, respectively. Carbamidomethyl cysteine was set as fixed modification and oxidation of methionine was specified as variable modification. Identifications were considered successful when significant MASCOT scores (≥30) were obtained, indicating the identity or extensive homology at p < 0.05.
Results
Patients
Eight allergic patients to red oak pollen and 4 healthy non-atopic subjects participated in the study (clinical features are in Table 1). The two groups were almost the same age (45 ± 3.5 years vs 42 ± 3.2 years, respectively). IgE levels in allergic patients were higher when compared with healthy controls (median 375 [range: 84–1630 U/dL vs median 58 [range: 30–82) p < 0.05. Similarly, eosinophil numbers were greater in the asthmatic group as compared with healthy controls (median 350 [range: 180–700 U/dL vs median 103 [range: 61–145]) p < 0.05.
Table 1.
Clinical characteristics of allergic and control subjects.
| Allergic patients | Control subjects | |
|---|---|---|
| No. subjects | 8 | 4 |
| Age (years) | 45 ± 3.5 | 42 ± 3.2 |
| Females | 4 | 3 |
| Males | 4 | 1 |
| FEV1% | 86% (range 81–91) | 102% (range 98–110)∗ |
| Total IgE (U/dL) | 375 (84–1630) | 58 (30–82)∗ |
| Eosinophils (cells/mm3) | 350 (180–700) | 103 (61–145)∗ |
| Atopy | atopic | no |
| Asthma | 2 | 0 |
| Asthma + Allergic rhinitis | 3 | 0 |
| Allergic rhinitis | 3 | 0 |
FEV1 = Forced expiratory volume at the end of the 1st second.
∗ = P < 0.05
One-dimensional electrophoresis
Protein extraction from red oak pollen (100 mg starting material) yielded around 20 mg of protein/g of starting material consistently. Electrophoretic pattern of total soluble proteins shows bands in the molecular mass range of 10–100 kDa. Higher intensity bands are observed in the regions of about 25–40 and 40–60 kDa (Fig. 1).
Fig. 1.
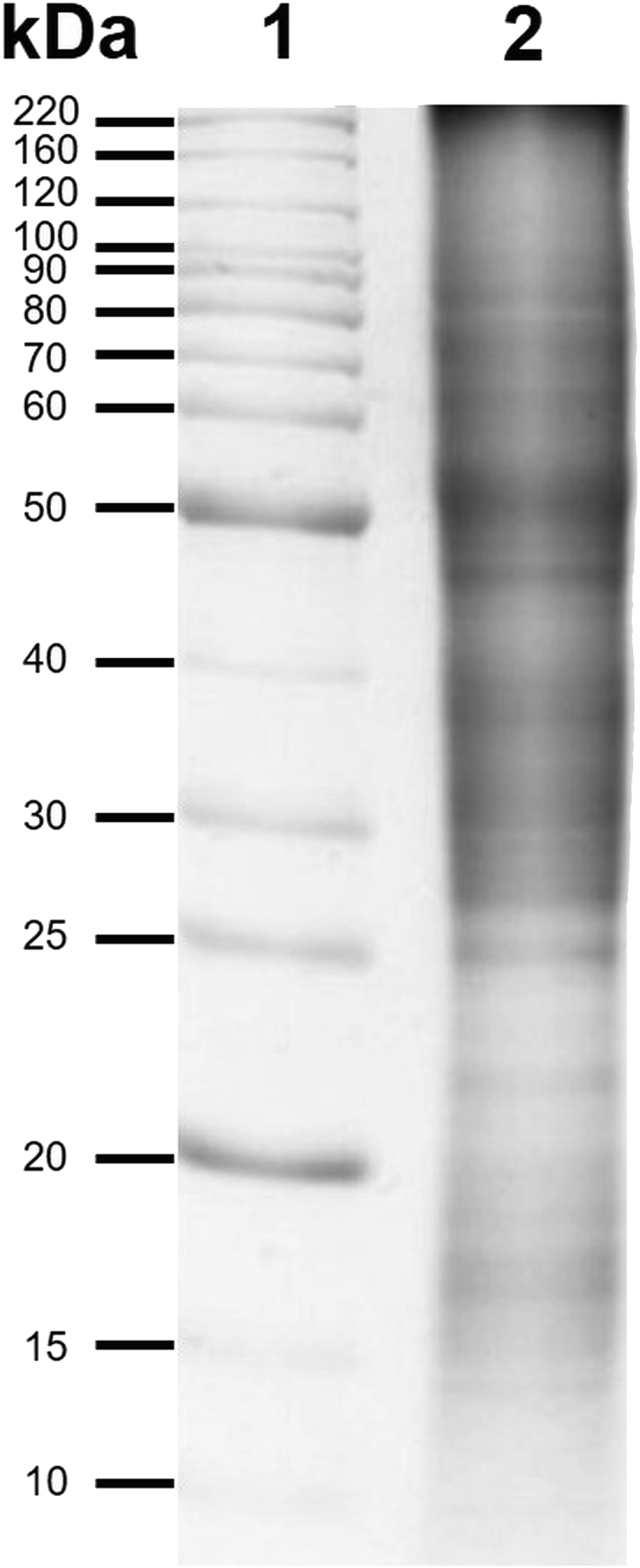
One-dimensional total soluble protein profile of Quercus rubra pollen. Lane 1, Molecular weight marker. Lane 2. Total soluble proteins (30 μg)
Two-dimensional profile of total pollen proteins and two-dimensional immunoblot
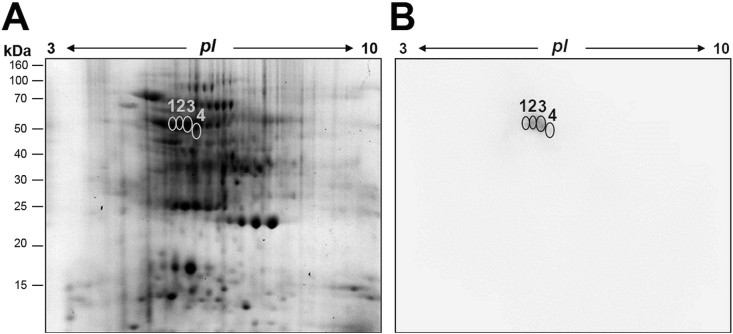
Analysis of red oak pollen extracts resolved into 400 protein spots approximately upon 2-DE analysis (Fig. 2A). Interestingly, two-dimensional immunoblots using pool sera from allergic patients revealed 4 allergenic protein spots with molecular masses in the range from 50 to 55 kDa and isoelectric points in the range from 5.5 to 6.2 (Fig. 2B). Moreover, no IgE binding reactivity was detected with the sera from healthy control subjects (data not shown).
Fig. 2.
2-DE and Immunoblotting using pollen proteins from Quercus rubra. A) 2-DE of total soluble proteins stained with Coomassie blue. B) Immunoreactive protein spots detected with pooled sera from Quercus rubra allergic patients using 2-DE immunoblotting (2 mg of protein, pH 3–10)
Protein identification by mass spectrometry
Homology database search with the Mascot search engine allowed identifying 8 allergenic proteins including alpha and beta subunits of ATP synthase, Xylose isomerase (X1 isoform), mitochondrial aldehyde dehydrogenase,UTP-Glucose-1-phosphate uridylyltransferase, beta xylosidase/alpha-l-arabinofuranosidase 1, and both enolase 1 and a chloroplastic enolase (Table 2). Some IgE binding proteins showed 2- or more proteoforms: ATP synthase subunit alpha, mitochondrial (n = 2), ATP synthase subunit beta, mitochondrial (3), X1isoformxyloseisomerase(n = 4), enolase 1, chloroplastic-like (n = 2), enolase 1 (n = 2) and aldehyde dehydrogenase (n = 2) (Table 2 and Supplementary Table 2). In contrast, the sera from healthy volunteers did not exhibit any IgE binding peptide (data not shown). And we did not identify any Bet v 1-like protein, which is usually found in allergenic fagales trees.
Table 2.
Allergenic proteins identified in Quercus rubra pollen.
| Spota | Protein | Organism | Functional category | Accession numberb | Exper. Mr/pIc |
Theo. Mr/pId |
PM/SCe | Scoref |
|---|---|---|---|---|---|---|---|---|
| 1 | ATP synthase subunit beta | Quercus suber | ATP biosynthetic process | POF09998.1 | 52.8/5.66 | 83.3/5.93 | 13/20% | 556 |
| 1 | ATP synthase subunit alpha | Oenothera biennis | ATP biosynthetic process | P05492.1 | 52.8/5.66 | 55.8/6.23 | 2/6% | 90 |
| 1 | Xyloseisomeraseisoform X1 | Quercus suber | Carbohydrate metabolism | XP_023880138.1 | 52.8/5.66 | 57.8/5.65 | 3/7% | 89 |
| 1 | Enolase 1, chloroplastic-like | Quercus suber | Glycolytic process | XP_023876868.1 | 52.8/5.66 | 51.6/5.72 | 3/8% | 81 |
| 2 | ATP synthase subunit beta, | Quercus suber | ATP biosynthetic process | XP_023880780.1 | 52.4/5.85 | 59.6/5.80 | 10/31% | 384 |
| 2 | Xyloseisomeraseisoform X1 | Quercus suber | Carbohydrate metabolism | XP_023880138.1 | 52.4/5.85 | 57.8/5.65 | 4/9% | 233 |
| 2 | ATP synthase subunit alpha | Oenothera biennis | ATP biosynthetic process | P05492.1 | 52.4/5.85 | 55.8/6.23 | 4/10% | 194 |
| 2 | Enolase 1, chloroplastic-like | Quercus suber | Glycolytic process | XP_023876868.1 | 52.4/5.85 | 51.6/5.72 | 4/13% | 58 |
| 3 | Enolase 1 | Quercus suber | Glycolytic process | XP_023928354.1 | 53.2/6.04 | 48.2/5.64 | 9/31% | 363 |
| 3 | Xyloseisomeraseisoform X1 | Quercus suber | Carbohydrate metabolism | XP_023880138.1 | 53.2/6.04 | 57.8/5.65 | 5/13% | 347 |
| 3 | ATP synthase subunit alpha | Oenothera biennis | ATP biosynthetic process | P05492.1 | 53.2/6.04 | 55.8/6.23 | 2/6% | 174 |
| 3 | Aldehyde dehydrogenase | Quercus suber | Detoxification | XP_023903360.1 | 53.2/6.04 | 54.2/5.71 | 3/8% | 124 |
| 4 | Enolase 1 | Quercus suber | Glycolytic process | XP_023928354.1 | 50.9/6.15 | 48.2/5.64 | 10/33% | 497 |
| 4 | Xyloseisomeraseisoform X1 | Quercus suber | Carbohydrate metabolism | XP_023880138.1 | 50.9/6.15 | 57.8/5.65 | 4/9% | 252 |
| 4 | Aldehyde dehydrogenase | Quercus suber | Detoxification | XP_023903360.1 | 50.9/6.15 | 54.2/5.71 | 3/8% | 154 |
| 4 | UTP--glucose-1-phosphate uridylyltransferase | Quercus suber | UDP-glucose metabolic process | XP_023870341.1 | 50.9/6.15 | 52.2/6.14 | 3/9% | 148 |
| 4 | Beta-xylosidase/alpha-l-arabinofuranosidase 1-like | Quercus suber | Carbohydrate metabolism | XP_023884910.1 | 50.9/6.15 | 90.1/5.97 | 3/5% | 131 |
Spot numbers as indicated in Fig. 2.
Accession numbers according to NCBInrdatabse.
Experimental molecular mass (kDa) and pI.
Theoretical mass (kDa) and pI of identified proteins retrieved from NCBInr database.
Number of peptides matched/protein sequence coverage.
Mascot score reported after database search, score >47 indicate identity or extensive homology at p < 0.05
Discussion
This is the first study to identify allergens from red oak pollen by using 2-DE followed by mass spectrometry.
Allergies, including pollen allergies, are recognized by the World Allergy Organization (WAO) as a global public health issue. Oak pollen allergy is of clinical relevance in both the American and European continents. Patients suffering from allergic rhinitis caused by red oak are usually treated with inhaled antihistamines and inhaled steroids. While these medications control patient's symptoms, they do not cure the disease. Allergy immunotherapy (AIT) is a causal treatment targeting the underlying allergic disease, affecting immunological mechanisms and resulting in the induction of immunological tolerance leading to sustained symptom relief and prevention of disease progression. Indeed, the AIT not only averts allergy symptoms, it also prevents both the development of asthma and new allergies in children. To date, the allergens from red oak remain unknown. Thus, it is important to recognize the allergens from this pollen tree, as they could be used for diagnostics and AIT.
In the present study, 2 enolases were identified in red oak pollen; enolase-1 and enolase chloroplastic-like, both of which were originally identified as orthologous of Quercus suber enolases. These proteins are essential glycolytic metalloenzymes, which catalyzed the conversion of 2-phosphoglycerate to phosphoenolpyruvate during glycolysis.18 Enolases also play a part in growth control and hypoxia tolerance responses. Indeed, enolase is an allergenic protein which has been identified in other plants including Ailanthus altissima, Ligustrum, Cynodon dactylon (Bermuda grass), Ambrosia, latex (Hev b 9) and coconut.19, 20, 21, 22, 23, 24, 25, 26 Interestingly, both enolases share a 68% of sequence identity, but not a single peptide identified by mass spectrometry. Enolase 1 exhibited high identity with enolases from other plant allergenic sources including Bermuda grass (88.31%), ambrosia (90.99%) and latex (Hebv 9, 91.46%). Enolases have also been reported as allergens from fungi and seafood.27 Enolases identified here share between 67 and 69% sequence identity with fully recognized fungal and seafood allergenic enolases (Rho m 1, Cur l 2, Cla h 6, Alt a 6, Asp f 22, Pen c 22, Sal s 2). In all, this finding suggests that enolases from red oak pollen may play a major role in specific allergic reactions.
Xylose isomerase is a critical enzyme in xylose metabolism which is vital for the transformation of glucose and xylose into fructose and xylulose, respectively. X1 isoform of the Xylose isomerase was found as a novel allergen in red oak pollen. This functional protein was reported as allergenic in the pollen of the Phoenix sylvestris palm tree (Arecaceae).28 Quercus rubra Xylose isomerase isoform X1 exhibited high sequence identity (78.5%) with Phoenix sylvestris homologous allergenic protein. This high sequence homology may represent a conserved allergenicity of the protein even in taxonomically distant trees. Several proteins involved in carbohydrate metabolism were identified as IgE binding allergens in this study. UTP-Glucose-1-phosphate uridylyltransferase is a key protein in glycogenesis and cell wall formation by UDP-glucose from glucose-1-phosphate and UTP. Our results showed one IgE-binding protein spot which corresponded to UTP-Glucose-1-phosphate uridylyltransferases, as similarly reported in fruit latex29 and short ragweed.30 Beta xylosidase/alpha-l-arabinofuranosidase was another enzyme we identified in red oak pollen. This protein has been reported as an allergen in Aspergillus niger.31 Beta xylosidase participates in the degradation of cell membrane hemicelluloses during pollen tube formation and has never been described as allergenic in pollen, therefore, this may be the first report of a likely role of this enzyme as an IgE recognized plant protein.
Mitochondrial proteins were also identified in immunodetected protein spots, including the alpha and beta subunits of ATP synthase, proteins that are widely recognized as pollen allergens.21,26,28,32 Likewise, aldehyde dehydrogenase family 2 member B7 was also identified in immunoreactive spots. Aldehyde dehydrogenase is a major allergen in the fungi Alternaria alternata (Alt a 10) and Cladosporium herbarum (Cla h 10), in the Asian lady beetle Harmonia axyridis (Har a 2), and the mite Tyrophagus putrescentiae (Tyr p 35).33, 34, 35 Quercus rubra Aldehyde dehydrogenase showed 51–53% sequence identity with fungal (Alternaria and Cladosporium) and mite (Tyrophagus) allergenic aldehyde dehydrogenases, but low sequence identity with allergenic insect (Harmonia) aldehyde dehydrogenases (26%). It should be noted that the allergens detected are present in several spots in the gel slab. However, this is common in most 2-DE map analyses: such spots likely represent post-translational modifications, including proteolytic cleavages.
The novel finding that red oak produces 8 allergenic proteins may have clinical implications for sensitized patients. For example, the development of recombinant proteins could have potential applications in diagnosis and therapy. Indeed, recombinant allergen molecules exhibit the structural characteristics of the natural allergens in as much as they could be used in skin prick tests, in vitro measurements of specific IgE and basophil activation test. Moreover, hypoallergenic derivatives could also be developed for application in allergy immunotherapy; hypoallergenic recombinant variants preserve T-cell epitopes while removing B-cell epitopes, which offers several advantages over the natural forms as they seek to ensure better standardization and eliminate unwanted side effects.
Our study has some potential limitations. First, we did not analyze the samples individually for each patient of the cohort, which could have provided some additional information about the grade and frequency of the specific recognition of these red oak allergens. For example, 3 out 8 red oak sensitive subjects had also positive skin test reaction to white oak, suggesting a co-sensitization that could have been further investigated with the individual profile of these patients. Secondly, although we have identified a relatively high amount of novel aeroallergens, no validation studies were undertaken yet to investigate their potential recognition in a larger cohort of patients. To uncover red oak allergens, we rather undertook a discovery proteomics approach by optimizing protein identification and placing more effort on analyzing a reduced number of samples, while validation-based approach optimizes the proteomics methods to achieve the highest sensitivity and throughput for large number of samples.36
Conclusions
In summary, we have identified 8 IgE-reactive proteins from red oak pollen including Enolase 1 and Enolase 1 chloroplastic, Xylose isomerase (X1 isoform), mitochondrial Aldehyde dehydrogenase, UTP-Glusose-1-phosphate uridylyltransferase, beta xylosidase/alpha-l-arabinofuranosidase and alpha- and beta subunits of ATP synthase. These findings will pave the way towards the development of new diagnostic and therapeutic modalities for red oak allergy.
Declaration of Competing Interest
The authors declare that they have no competing interests.
Acknowledgments
Thanks to CONACYT-Mexico Grant-251744-Infrastructure; Plataforma Analítica Institucional-CIAD, A.C. (Project PAI-10363); and to the Alexander Von Humboldt Foundation, Germany.
Footnotes
Full list of author information is available at the end of the article
Supplementary data to this article can be found online at https://doi.org/10.1016/j.waojou.2020.100111.
Contributor Information
José Ángel Huerta-Ocampo, Email: jose.huerta@ciad.mx.
Luis M. Terán, Email: lmteran@iner.gob.mx.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Dales R.E., Cakmak S., Judek S., Coates F. Tree pollen and hospitalization for asthma in urban Canada. Int Arch Allergy Immunol. 2008;146(3):241–247. doi: 10.1159/000116360. [DOI] [PubMed] [Google Scholar]
- 2.Anenberg S.C., Weinberger K.R., Roman H. Impacts of oak pollen on allergic asthma in the United States and potential influence of future climate change. GeoHealth. 2017;1(3):80–92. doi: 10.1002/2017GH000055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Skjøth C.A., Baker P., Sadyś M., Adams-Groom B. Pollen from alder (Alnus sp.), birch (Betula sp.) and oak (Quercus sp.) in the UK originate from small woodlands. Urban Clim. 2015;14:414–428. [Google Scholar]
- 4.Ferreira F., Gadea Maier Y., Wallner M. Tree pollen allergens. In: Akdis C.A., Agache I., editors. Global Atlas of Allergy. European Academy of Allergy and Clinical Immunology; Zurich, Switzerland: 2014. pp. 18–21. [Google Scholar]
- 5.Ziska L.H., Makra L., Harry S.K. Temperature-related changes in airborne allergenic pollen abundance and seasonality across the northern hemisphere: a retrospective data analysis. Lancet Planet Health. 2019;3(3):e124–e131. doi: 10.1016/S2542-5196(19)30015-4. [DOI] [PubMed] [Google Scholar]
- 6.Zhang Y., Bielory L., Mi Z., Cai T., Robock A., Georgopoulos P. Allergenic pollen season variations in the past two decades under changing climate in the United States. Global Change Biol. 2015;21(4):1581–1589. doi: 10.1111/gcb.12755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grundström M., Adams-Groom B., Pashley C.H. Oak pollen seasonality and severity across Europe and modelling the season start using a generalized phenological model. Sci Total Environ. 2019;663:527–536. doi: 10.1016/j.scitotenv.2019.01.212. [DOI] [PubMed] [Google Scholar]
- 8.Lewis W.H., Vinay P., Zenger V.E. Johns Hopkins University Press; Baltimore, United States: 1983. Airborne and Allergenic Pollen of North America. [Google Scholar]
- 9.Lee J.Y., Yang M., Jeong K.Y. Characterization of a major allergen from Mongolian oak, Quercus mongolica, a dominant species of oak in Korea. Int Arch Allergy Immunol. 2017;174(2):77–85. doi: 10.1159/000481092. [DOI] [PubMed] [Google Scholar]
- 10.Ipsen H., Hansen O.C. The NH2-terminal amino acid sequence of the immunochemically partial identical major allergens of alder (Alnus glutinosa) Aln g I, birch (Betula verrucosa) Bet v I, hornbeam (Carpinus betulus) Car b I and oak (Quercus alba) Que a I pollens. Mol Immunol. 1991;28(11):1279–1288. doi: 10.1016/0161-5890(91)90015-c. [DOI] [PubMed] [Google Scholar]
- 11.Movérare R., Everberg H., Carlsson R. Purification and characterization of the major oak pollen allergen Que a 1 for component-resolved diagnostics using ImmunoCAP®. Int Arch Allergy Immunol. 2008;146(3):203–211. doi: 10.1159/000115888. [DOI] [PubMed] [Google Scholar]
- 12.Egger C., Focke M., Bircher A.J. The allergen profile of beech and oak pollen. Clin Exp Allergy. 2008;38(10):1688–1696. doi: 10.1111/j.1365-2222.2008.03092.x. [DOI] [PubMed] [Google Scholar]
- 13.Torres-Miranda A., Luna-Vega I., Oyama K. Conservation biogeography of red oaks (Quercus, section Lobatae) in Mexico and Central America. Am J Bot. 2011;98(2):290–305. doi: 10.3732/ajb.1000218. [DOI] [PubMed] [Google Scholar]
- 14.Sander I.L. Northern red oak (Quercus rubra L.) In: Burns R.M., Honkala B.H., editors. Silvics of North America: Volume 2. Hardwoods. vol. 2. United States Department of Agriculture; Washington, DC: 1990. pp. 727–733. [Google Scholar]
- 15.Woziwoda B., Kopec D., Witkowski J. The negative impact of intentionally introduced Quercus rubra L. on a forest community. Acta Soc Bot Pol. 2014;83(1) [Google Scholar]
- 16.Faurobert M., Pelpoir E., Chaïb J. Phenol extraction of proteins for proteomic studies of recalcitrant plant tissues. In: Thiellement H., Zivy M., Damerval C., Méchin V., editors. Plant Proteomics: Methods and Protocols. Humana Press; Totowa, NJ: 2007. pp. 9–14. [DOI] [PubMed] [Google Scholar]
- 17.Morales-Amparano M.B., Ramos-Clamont Montfort G., Baqueiro-Peña I., Robles-Burgueño MdR., Vázquez-Moreno L., Huerta-Ocampo J.Á. Proteomic response of Saccharomyces boulardii to simulated gastrointestinal conditions and encapsulation. Food Sci Biotechnol. 2019;28(3):831–840. doi: 10.1007/s10068-018-0508-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Didiasova M., Schaefer L., Wygrecka M. When place matters: shuttling of enolase-1 across cellular compartments. Front Cell Dev Biol. 2019;7(61) doi: 10.3389/fcell.2019.00061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wagner S., Breiteneder H., Simon-Nobbe B. Hev b 9, an enolase and a new cross-reactive allergen from Hevea latex and molds. Eur J Biochem. 2000;267(24):7006–7014. doi: 10.1046/j.1432-1327.2000.01801.x. [DOI] [PubMed] [Google Scholar]
- 20.Mousavi F., Majd A., Shahali Y., Ghahremaninejad F., Shokouhi Shoormasti R., Pourpak Z. Immunoproteomics of tree of heaven (Ailanthus atltissima) pollen allergens. J Proteom. 2017;154:94–101. doi: 10.1016/j.jprot.2016.12.013. [DOI] [PubMed] [Google Scholar]
- 21.Mani B.M., Huerta-Ocampo J.A., Garcia-Sanchez J.R., Barrera-Pacheco A., de la Rosa A.P.B., Teran L.M. Identification of Ligustrum lucidum pollen allergens using a proteomics approach. Biochem Biophys Res Commun. 2015;468(4):788–792. doi: 10.1016/j.bbrc.2015.11.033. [DOI] [PubMed] [Google Scholar]
- 22.Shi H., Ye T., Zhong B., Liu X., Chan Z. Comparative proteomic and metabolomic analyses reveal mechanisms of improved cold stress tolerance in bermudagrass (Cynodon dactylon (L.) Pers.) by exogenous calcium. J Integr Plant Biol. 2014;56(11):1064–1079. doi: 10.1111/jipb.12167. [DOI] [PubMed] [Google Scholar]
- 23.Wu W.-S., McClain K.L. DNA polymorphisms and mutations of the tumor necrosis factor-α(TNF-α) promoter in Langerhans cell histiocytosis (LCH) J Interferon Cytokine Res. 1997;17(10):631–635. doi: 10.1089/jir.1997.17.631. [DOI] [PubMed] [Google Scholar]
- 24.Barton J.S., Schomacker R. Comparative protein profiles of the Ambrosia plants. Biochim Biophys Acta Protein Proteonomics. 2017;1865(6):633–639. doi: 10.1016/j.bbapap.2017.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Troncoso-Ponce M.A., Rivoal J., Dorion S. Molecular and biochemical characterization of the sunflower (Helianthus annuus L.) cytosolic and plastidial enolases in relation to seed development. Plant Sci. 2018;272:117–130. doi: 10.1016/j.plantsci.2018.04.007. [DOI] [PubMed] [Google Scholar]
- 26.Saha B., Sircar G., Pandey N., Gupta Bhattacharya S. Mining novel allergens from coconut pollen employing manual de novo sequencing and homology-driven proteomics. J Proteome Res. 2015;14(11):4823–4833. doi: 10.1021/acs.jproteome.5b00657. [DOI] [PubMed] [Google Scholar]
- 27.Ruethers T., Taki A.C., Nugraha R. Variability of allergens in commercial fish extracts for skin prick testing. Allergy. 2019;74(7):1352–1363. doi: 10.1111/all.13748. [DOI] [PubMed] [Google Scholar]
- 28.Saha B., Bhattacharya S.G. Charting novel allergens from date palm pollen (Phoenix sylvestris) using homology driven proteomics. J Proteom. 2017;165:1–10. doi: 10.1016/j.jprot.2017.05.021. [DOI] [PubMed] [Google Scholar]
- 29.Conti A., Giuffrida M.G., Hoffmann-Sommergruber K. Identification of latex UDP glucose pyrophosphorylase (Hev b UDPGP) as a novel cause of latex fruit allergy syndrome. Eur Ann Allergy Clin Immunol. 2007;39(4):116–118. [PubMed] [Google Scholar]
- 30.Smiljanic K., Apostolovic D., Trifunovic S. Subpollen particles are rich carriers of major short ragweed allergens and NADH dehydrogenases: quantitative proteomic and allergomic study. Clin Exp Allergy. 2017;47(6):815–828. doi: 10.1111/cea.12874. [DOI] [PubMed] [Google Scholar]
- 31.Sander I., Raulf-Heimsoth M., Siethoff C., Lohaus C., Meyer H.E., Baur X. Allergy to Aspergillus-derived enzymes in the baking industry: identification of β-xylosidase from Aspergillus niger as a new allergen (Asp n 14) J Allergy Clin Immunol. 1998;102(2):256–264. doi: 10.1016/s0091-6749(98)70109-5. [DOI] [PubMed] [Google Scholar]
- 32.Ghosal K., Saha B., Gupta Bhattacharya S. Clinical and immuno-proteomic approach on Lantana camara pollen allergy—a major health hazard. Allergy Asthma Clin Immunol. 2016;12(1):33. doi: 10.1186/s13223-016-0135-z. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 33.Achatz G., Oberkofler H., Lechenauer E. Molecular cloning of major and minor allergens of Alternaria alternata and Cladosporium herbarum. Mol Immunol. 1995;32(3):213–227. doi: 10.1016/0161-5890(94)00108-d. [DOI] [PubMed] [Google Scholar]
- 34.Nakazawa T., Satinover S.M., Naccara L. Asian ladybugs Harmonia axyridis: a new seasonal indoor allergen. J Allergy Clin Immunol. 2007;119(2):421–427. doi: 10.1016/j.jaci.2006.11.633. [DOI] [PubMed] [Google Scholar]
- 35.Cui Y., Yu L., Teng F. Transcriptomic/proteomic identification of allergens in the mite Tyrophagus putrescentiae. Allergy. 2016;71(11):1635–1639. doi: 10.1111/all.12999. [DOI] [PubMed] [Google Scholar]
- 36.Parker C.E., Borchers C.H. Mass spectrometry based biomarker discovery, verification, and validation — quality assurance and control of protein biomarker assays. Mol Oncol. 2014;8(4):840–858. doi: 10.1016/j.molonc.2014.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.