Abstract
Nearly 500 basidiomycetous yeast species were accepted in the latest edition of The Yeasts: A Taxonomic Study published in 2011. However, this number presents only the tip of the iceberg of yeast species diversity in nature. Possibly more than 99 % of yeast species, as is true for many groups of fungi, are yet unknown and await discovery. Over the past two decades nearly 200 unidentified isolates were obtained during a series of environmental surveys of yeasts in phyllosphere and soils, mainly from China. Among these isolates, 107 new species were identified based on the phylogenetic analyses of nuclear ribosomal DNA (rDNA) [D1/D2 domains of the large subunit (LSU), the small subunit (SSU), and the internal transcribed spacer region including the 5.8S rDNA (ITS)] and protein-coding genes [both subunits of DNA polymerase II (RPB1 and RPB2), the translation elongation factor 1-α (TEF1) and the mitochondrial gene cytochrome b (CYTB)], and physiological comparisons. Forty-six of these belong to 16 genera in the Tremellomycetes (Agaricomycotina). The other 61 are distributed in 26 genera in the Pucciniomycotina. Here we circumscribe eight new genera, three new families and two new orders based on the multi-locus phylogenetic analyses combined with the clustering optimisation analysis and the predicted similarity thresholds for yeasts and filamentous fungal delimitation at genus and higher ranks. Additionally, as a result of these analyses, three new combinations are proposed and 66 taxa are validated.
Key words: Basidiomycetous yeasts, Molecular phylogeny, Species diversity, Taxonomy
Taxonomic novelties: New orders: Heitmaniales Q.M. Wang & F.Y. Bai, Rosettozymales Q.M. Wang & F.Y. Bai
New families: Heitmaniaceae Q.M. Wang & F.Y. Bai, Jianyuniaceae Q.M. Wang & F.Y. Bai, Rosettozymaceae Q.M. Wang & F.Y. Bai
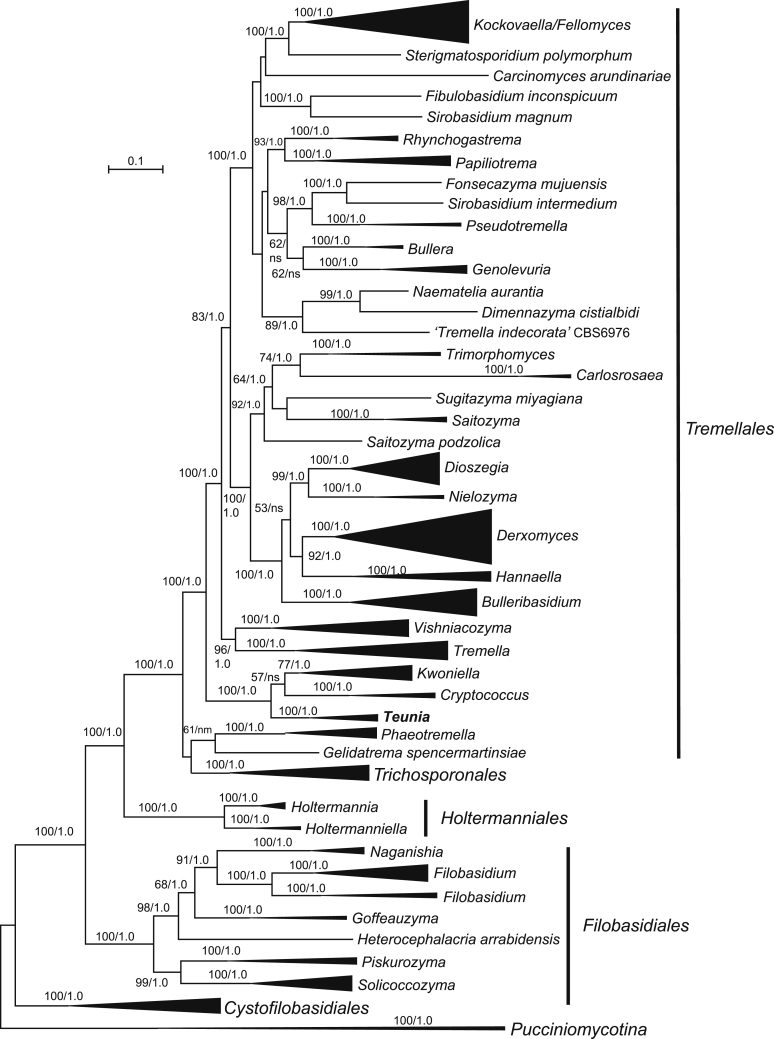
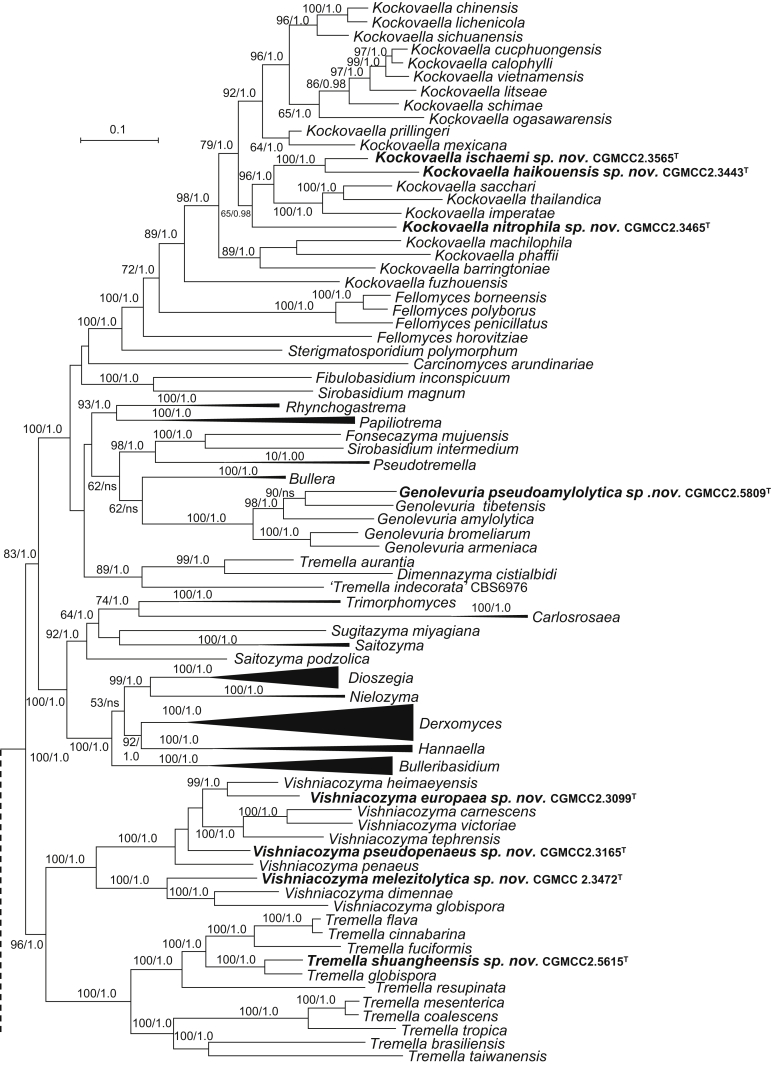
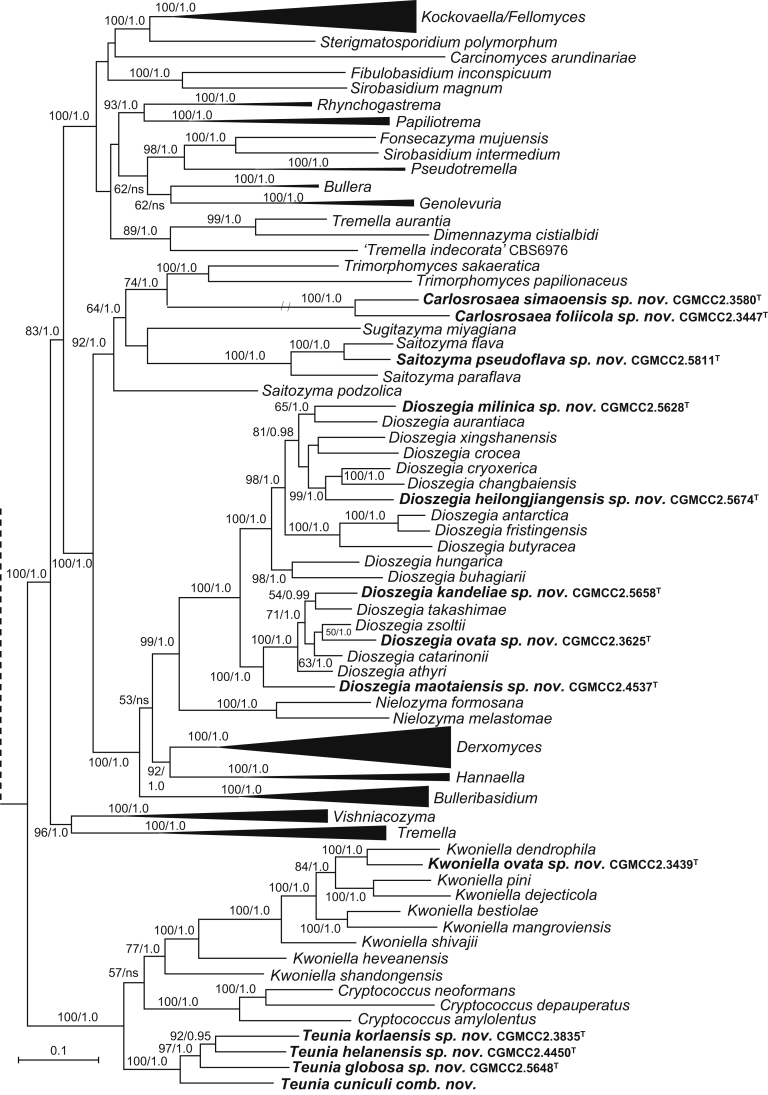
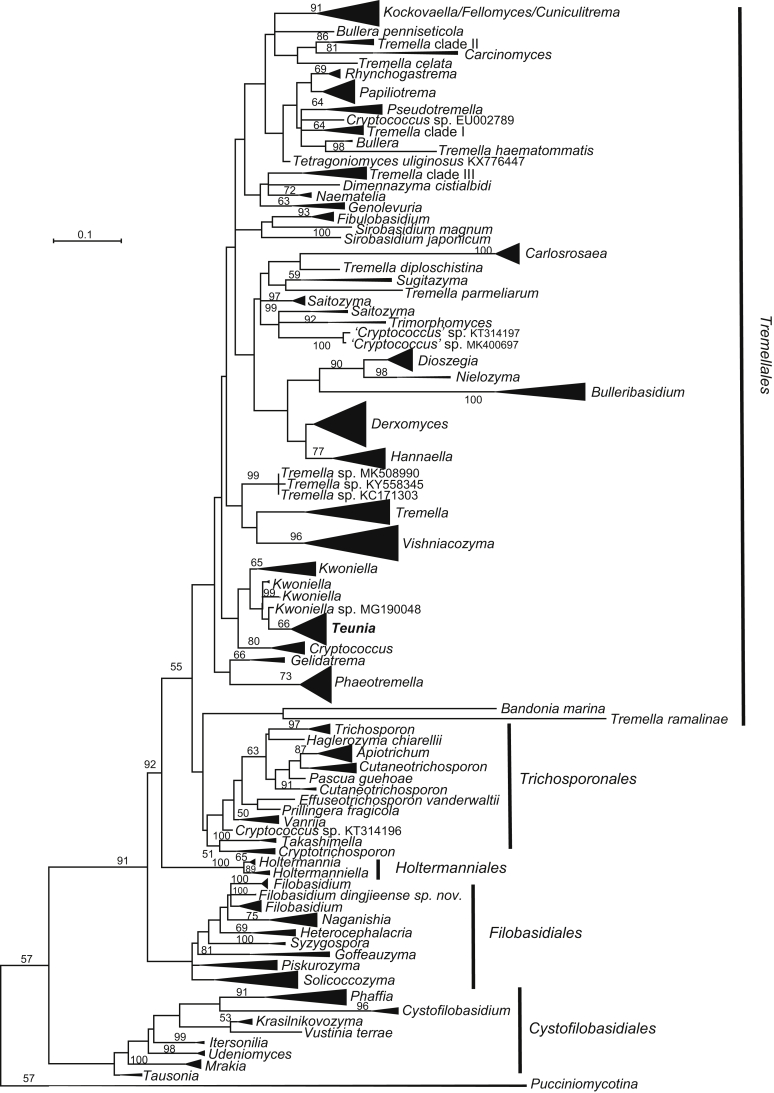
New genera: Begerowomyces Q.M. Wang & F.Y. Bai, Boekhoutia Q.M. Wang & F.Y. Bai, Meniscomyces Q.M. Wang & F.Y. Bai, Pseudosterigmatospora Q.M. Wang & F.Y. Bai, Robertozyma Q.M. Wang & F.Y. Bai, Rosettozyma Q.M. Wang & F.Y. Bai, Sterigmatospora Q.M. Wang & F.Y. Bai, Teunia Q.M. Wang & F.Y. Bai
New species: Begerowomyces foliicola Q.M. Wang, F.Y. Bai & A.H. Li; Bensingtonia pseudorectispora Q.M. Wang, F.Y. Bai & A.H. Li; Bensingtonia wuzhishanensis Q.M. Wang, F.Y. Bai & A.H. Li; Boekhoutia sterigmata Q.M. Wang, F.Y. Bai & A.H. Li; Bulleribasidium cremeum Q.M. Wang, F.Y. Bai & A.H. Li; Bulleribasidium elongatum Q.M. Wang, F.Y. Bai & A.H. Li; Bulleribasidium phyllophilum Q.M. Wang, F.Y. Bai & A.H. Li; Bulleribasidium phyllostachydis Q.M. Wang, F.Y. Bai & A.H. Li; Bulleribasidium pseudopanici Q.M. Wang, F.Y. Bai & A.H. Li; Carlosrosaea foliicola Q.M. Wang, F.Y. Bai & A.H. Li; Carlosrosaea simaoensis Q.M. Wang, F.Y. Bai & A.H. Li; Chrysozyma cylindrica Q.M. Wang, F.Y. Bai & A.H. Li; Chrysozyma flava Q.M. Wang, F.Y. Bai & A.H. Li; Chrysozyma fusiformis Q.M. Wang, F.Y. Bai & A.H. Li; Chrysozyma iridis Q.M. Wang, F.Y. Bai & A.H. Li; Chrysozyma pseudogriseoflava Q.M. Wang, F.Y. Bai & A.H. Li; Chrysozyma rhododendri Q.M. Wang, F.Y. Bai & A.H. Li; Chrysozyma sambuci Q.M. Wang, F.Y. Bai & A.H. Li; Chrysozyma sorbariae Q.M. Wang, F.Y. Bai & A.H. Li; Colacogloea aletridis Q.M. Wang, F.Y. Bai & A.H. Li; Colacogloea hydrangeae Q.M. Wang, F.Y. Bai & A.H. Li; Colacogloea rhododendri Q.M. Wang, F.Y. Bai & A.H. Li; Cystobasidium raffinophilum Q.M. Wang, F.Y. Bai & A.H. Li; Cystobasidium terricola Q.M. Wang, F.Y. Bai & A.H. Li; Derxomyces bifurcus Q.M. Wang, F.Y. Bai & A.H. Li; Derxomyces elongatus Q.M. Wang, F.Y. Bai & A.H. Li; Derxomyces longicylindricus Q.M. Wang, F.Y. Bai & A.H. Li; Derxomyces longiovatus Q.M. Wang, F.Y. Bai & A.H. Li; Derxomyces melastomatis Q.M. Wang, F.Y. Bai & A.H. Li; Derxomyces napiformis Q.M. Wang, F.Y. Bai & A.H. Li; Derxomyces ovatus Q.M. Wang, F.Y. Bai & A.H. Li; Derxomyces polymorphus Q.M. Wang, F.Y. Bai & A.H. Li; Derxomyces pseudoboekhoutii Q.M. Wang, F.Y. Bai & A.H. Li; Derxomyces pseudoyunnanensis Q.M. Wang, F.Y. Bai & A.H. Li; Derxomyces taiwanicus Q.M. Wang, F.Y. Bai & A.H. Li; Derxomyces xingshanicus Q.M. Wang, F.Y. Bai & A.H. Li; Dioszegia heilongjiangensis Q.M. Wang, F.Y. Bai & A.H. Li; Dioszegia kandeliae Q.M. Wang, F.Y. Bai, L.D. Guo & A.H. Li; Dioszegia maotaiensis Q.M. Wang, F.Y. Bai & A.H. Li; Dioszegia milinica Q.M. Wang, F.Y. Bai & A.H. Li; Dioszegia ovata Q.M. Wang, F.Y. Bai & A.H. Li; Filobasidium dingjieense Q.M. Wang, F.Y. Bai & A.H. Li; Filobasidium globosum Q.M. Wang, F.Y. Bai & A.H. Li; Filobasidium mali Q.M. Wang, F.Y. Bai & A.H. Li; Filobasidium mucilaginum Q.M. Wang, F.Y. Bai & A.H. Li; Genolevuria pseudoamylolytica Q.M. Wang, F.Y. Bai & A.H. Li; Heitmania cylindrica Q.M. Wang, F.Y. Bai & A.H. Li; Heitmania tridentata Q.M. Wang, F.Y. Bai & A.H. Li; Holtermannia saccardoi Q.M. Wang, F.Y. Bai & A.H. Li; Kockovaella haikouensis Q.M. Wang, F.Y. Bai & A.H. Li; Kockovaella ischaemi Q.M. Wang, F.Y. Bai & A.H. Li; Kockovaella nitrophila Q.M. Wang, F.Y. Bai & A.H. Li; Kondoa arboricola Q.M. Wang, F.Y. Bai & A.H. Li; Kondoa chamaenerii Q.M. Wang, F.Y. Bai & A.H. Li; Kondoa cylindrica Q.M. Wang, F.Y. Bai & A.H. Li; Kondoa daliangziensis Q.M. Wang, F.Y. Bai & A.H. Li; Kondoa foliicola Q.M. Wang, F.Y. Bai & A.H. Li; Kondoa lulangica Q.M. Wang, F.Y. Bai & A.H. Li; Kondoa myxariophila Q.M. Wang, F.Y. Bai & A.H. Li; Kondoa rhododendri Q.M. Wang, F.Y. Bai & A.H. Li; Kondoa ribitophobia Q.M. Wang, F.Y. Bai & A.H. Li; Kwoniella ovata Q.M. Wang, F.Y. Bai & A.H. Li; Meniscomyces layueensis Q.M. Wang, F.Y. Bai & A.H. Li; Microbotryozyma swertiae Q.M. Wang, F.Y. Bai & A.H. Li; Microsporomyces ellipsoideus Q.M. Wang, F.Y. Bai & A.H. Li; Microsporomyces pseudomagnisporus Q.M. Wang, F.Y. Bai & A.H. Li; Microsporomyces rubellus Q.M. Wang, F.Y. Bai & A.H. Li; Oberwinklerozyma dicranopteridis Q.M. Wang, F.Y. Bai & A.H. Li; Oberwinklerozyma nepetae Q.M. Wang, F.Y. Bai & A.H. Li; Phaeotremella lactea Q.M. Wang, F.Y. Bai & A.H. Li; Phaeotremella ovata Q.M. Wang, F.Y. Bai & A.H. Li; Phaffia aurantiaca Q.M. Wang, F.Y. Bai & A.H. Li; Phyllozyma aceris Q.M. Wang, F.Y. Bai & A.H. Li; Phyllozyma jiayinensis Q.M. Wang, F.Y. Bai & A.H. Li; Pseudobensingtonia fusiformis Q.M. Wang, F.Y. Bai & A.H. Li; Pseudohyphozyma hydrangeae Q.M. Wang, F.Y. Bai & A.H. Li; Pseudohyphozyma lulangensis Q.M. Wang, F.Y. Bai & A.H. Li; Pseudosterigmatospora motuoensis Q.M. Wang, F.Y. Bai & A.H. Li; Rhodosporidiobolus fuzhouensis Q.M. Wang, F.Y. Bai & A.H. Li; Rhodosporidiobolus jianfalingensis Q.M. Wang, F.Y. Bai & A.H. Li; Rhodosporidiobolus platycladi Q.M. Wang, F.Y. Bai & A.H. Li; Robertozyma ningxiaensis Q.M. Wang, F.Y. Bai & A.H. Li; Rosettozyma cystopteridis Q.M. Wang, F.Y. Bai & A.H. Li; Rosettozyma motuoensis Q.M. Wang, F.Y. Bai & A.H. Li; Rosettozyma petaloides Q.M. Wang, F.Y. Bai & A.H. Li; Ruinenia bangxiensis Q.M. Wang, F.Y. Bai & A.H. Li; Ruinenia fanjingshanensis Q.M. Wang, F.Y. Bai & A.H. Li; Ruinenia lunata Q.M. Wang, F.Y. Bai & A.H. Li; Saitozyma pseudoflava Q.M. Wang, F.Y. Bai & A.H. Li; Sakaguchia melibiophila M. Groenew., Q.M. Wang & F.Y. Bai; Slooffia globosa Q.M. Wang, F.Y. Bai & A.H. Li; Solicoccozyma gelidoterrea Q.M. Wang, F.Y. Bai & A.H. Li; Sporobolomyces cellobiolyticus Q.M. Wang, F.Y. Bai & A.H. Li; Sporobolomyces ellipsoideus Q.M. Wang, F.Y. Bai & A.H. Li; Sporobolomyces primogenomicus Q.M. Wang & F.Y. Bai; Sporobolomyces reniformis Q.M. Wang, F.Y. Bai & A.H. Li; Sterigmatospora layueensis Q.M. Wang, F.Y. Bai & A.H. Li; Symmetrospora rhododendri Q.M. Wang, F.Y. Bai & A.H. Li; Teunia betulae K. Sylvester, Q.M. Wang & Hittinger ex Q.M. Wang, F.Y. Bai & A.H. Li; Teunia globosa Q.M. Wang, F.Y. Bai & A.H. Li; Teunia helanensis Q.M. Wang, F.Y. Bai & A.H. Li; Teunia korlaensis Q.M. Wang, F.Y. Bai & A.H. Li; Teunia tronadorensis V. de Garcia, Zalar, Brizzio, Gunde-Cim. & van Brook ex Q.M. Wang, F.Y. Bai & A.H. Li; Tremella shuangheensis Q.M. Wang, F.Y. Bai & A.H. Li; Vishniacozyma europaea Q.M. Wang, F.Y. Bai & A.H. Li; Vishniacozyma melezitolytica Q.M. Wang, F.Y. Bai & A.H. Li; Vishniacozyma pseudopenaeus Q.M. Wang, F.Y. Bai & A.H. Li; Yamadamyces terricola Q.M. Wang, F.Y. Bai & A.H. Li; Yurkovia longicylindrica Q.M. Wang, F.Y. Bai & A.H. Li
New combinations: Colacogloea subericola (Belloch, Villa-Carv., Á;lv.-Rodríg. & Coque) Q.M. Wang, & F.Y. Bai; Symmetrospora oryzicola (Nakase & M. Suzuki) Q.M. Wang & F.Y. Bai; Teunia cuniculi (K.S. Shin & Y.H. Park) Q.M. Wang, F.Y. Bai & A.H. Li
New validations: Apiotrichum xylopini S.O. Suh, C.F. Lee, Gujjari & J.J. Zhou ex Kachalkin, Yurkov & Boekhout; Bannozyma arctica Vishniac & M. Takash. ex Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; Bulleribasidium panici Fungsin, M. Takash. & Nakase ex Xin Zhan Liu, F.Y. Bai, M. Groenew. & Boekhout; Bulleribasidium siamense Fungsin, M. Takash. & Nakase ex Q.M. Wang, F.Y. Bai, Boekhout & Nakase; Carcinomyces arundinariae Fungsin, M. Takash. & Nakase ex Yurkov; Cystobasidium alpinum Turchetti, Selbmann, Onofri & Buzzini; Cystobasidium portillonense Laich, Vaca & R. Chávez ex Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; Derxomyces cylindricus F.Y. Bai, Q.M. Wang & M. Takash. ex F.Y. Bai & Q.M. Wang; Derxomyces hubeiensis F.Y. Bai, Q.M. Wang & M. Takash. ex F.Y. Bai & Q.M. Wang; Derxomyces nakasei F.Y. Bai, Q.M. Wang & M. Takash. ex F.Y. Bai & Q.M. Wang; Dioszegia zsoltii F.Y. Bai, M. Takash. & Nakase; Genolevuria bromeliarum Landell & P. Valente ex Xin Zhan Liu, F.Y. Bai, M. Groenew. & Boekhout; Glaciozyma Turchetti, Connell, Thomas-Hall & Boekhout ex M. Groenew. & Q.M. Wang; Glaciozyma antarctica (Fell, Statzell, I.L. Hunter & Phaff) M. Groenew. & Q.M. Wang; Glaciozyma martinii Turchetti, Connell, Thomas-Hall & Boekhout; Glaciozyma watsonii Turchetti, Connell, Thomas-Hall & Boekhout; Kockovaella mexicana Lopandić, O. Molnár & Prillinger ex Xin Zhan Liu, F.Y. Bai, M. Groenew. & Boekhout; Kondoa thailandica Fungsin, Hamam. & Nakase ex Q.M. Wang, M. Groenew., F.Y. Bai & Boekhout; Kwoniella newhampshirensis K. Sylvester, Q.M. Wang & C.T. Hittinger; Kwoniella shandongensis R. Chen, Y.M. Jiang & S.C. Wei ex M. Groenew. & Q.M. Wang; Leucosporidium creatinivorum (Golubev) M. Groenew. & Q.M. Wang; Leucosporidium fragarium (J.A. Barnett & Buhagiar) M. Groenew. & Q.M. Wang; Leucosporidium intermedium (Nakase & M. Suzuki) M. Groenew. & Q.M. Wang; Leucosporidium muscorum (Di Menna) M. Groenew. & Q.M. Wang; Leucosporidium yakuticum (Golubev) M. Groenew. & Q.M. Wang; Naganishia onofrii Turchetti, Selbmann & Zucconi ex Yurkov; Naganishia vaughanmartiniae Turchetti, Blanchette & Arenz ex Yurkov; Nielozyma Xin Zhan Liu, F.Y. Bai, M. Groenew. & Boekhout; Nielozyma formosana Nakase, Tsuzuki, F.L. Lee & M. Takash. ex Xin Zhan Liu, F.Y. Bai, M. Groenew. & Boekhout; Nielozyma melastomatis Nakase, Tsuzuki, F.L. Lee & M. Takash. ex Xin Zhan Liu, F.Y. Bai, M. Groenew. & Boekhout; Oberwinklerozyma silvestris Golubev & Scorzetti ex Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; Oberwinklerozyma straminea Golubev & Scorzetti ex Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; Papiliotrema aspenensis (Ferreira-Paim, et al.) Xin Zhan Liu, F.Y. Bai, M. Groenew. & Boekhout; Papiliotrema baii Yurkov, M.A. Guerreiro & Á;. Fonseca ex Yurkov; Papiliotrema frias V. de García, Zalar, Brizzio, Gunde-Cim. & Van Broock ex Yurkov; Papiliotrema hoabinhensis D.T. Luong, M. Takash., Ty, Dung & Nakase ex Yurkov; Papiliotrema japonica J.P. Samp., Fonseca & Fell ex Xin Zhan Liu, F.Y. Bai, M. Groenew. & Boekhout; Papiliotrema terrestris Crestani, Landell, Faganello, Vainstein, Vishniac & P. Valente ex Xin Zhan Liu, F.Y. Bai, M. Groenew. & Boekhout; Papiliotrema wisconsinensis K. Sylvester, Q.M. Wang & Hittinger ex Xin Zhan Liu, F.Y. Bai, M. Groenew. & Boekhout; Piskurozyma fildesensis T.T. Zhang & Li Y. Yu ex Yurkov; Piskurozyma taiwanensis Nakase, Tsuzuki & M. Takash. ex Xin Zhan Liu, F.Y. Bai, M. Groenew. & Boekhout; Pseudoleucosporidium V. de García, et al. ex M. Groenew. & Q.M. Wang; Pseudoleucosporidium fasciculatum (Babeva & Lisichk.) M. Groenew. & Q.M. Wang; Pseudotremella lacticolour Satoh & Makimura ex Yurkov; Rhynchogastrema complexa (Landell, et al.) Xin Zhan Liu, F.Y. Bai, M. Groenew., Boekhout & Yurkov; Rhynchogastrema fermentans (C.F. Lee) Xin Zhan Liu, F.Y. Bai, M. Groenew., Boekhout & Yurkov; Rhynchogastrema glucofermentans (S.O. Suh & M. Blackw.) Xin Zhan Liu, F.Y. Bai, M. Groenew., Boekhout & Yurkov; Rhynchogastrema nanyangensis F.L. Hui & Q.H. Niu ex Xin Zhan Liu, F.Y. Bai, M. Groenew., Boekhout & Yurkov; Rhynchogastrema tunnelae (Boekhout, Fell, Scorzetti & Theelen) Xin Zhan Liu, F.Y. Bai, M. Groenew., Boekhout & Yurkov; Rhynchogastrema visegradensis (G. Péter & Dlauchy) Xin Zhan Liu, F.Y. Bai, M. Groenew., Boekhout &Yurkov; Ruinenia diospyri Nakase, Tsuzuki, F.L. Lee, Jindam. & M. Takash. ex Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; Ruinenia pyrrosiae Nakase, Tsuzuki, F.L. Lee, Jindam. & M. Takash. ex Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; Saitozyma ninhbinhensis (D.T. Luong, M. Takash., Dung & Nakase)Yurkov; Saitozyma paraflava Golubev & J.P. Samp. ex Xin Zhan Liu; F.Y. Bai, M. Groenew. & Boekhout; Tremella basidiomaticola Xin Zhan Liu & F.Y. Bai; Trimorphomyces sakaeraticus Fungsin, M. Takash. & Nakase ex Xin Zhan Liu, F.Y. Bai, M. Groenew. & Boekhout; Vanrija meifongana C.F. Lee ex Kachalkin Yurkov & Boekhout; Vanrija nantouana C.F. Lee ex Kachalkin Yurkov & Boekhout; Vanrija thermophila Vogelmann, S. Chaves & C. Hertel ex Kachalkin Yurkov & Boekhout; Vishniacozyma foliicola Q.M. Wang & F.Y. Bai ex Yurkov; Vishniacozyma heimaeyensis Vishniac ex Xin Zhan Liu, F.Y. Bai, M. Groenew. & Boekhout; Vishniacozyma psychrotolerans V. de García, Zalar, Brizzio, Gunde-Cim. & Van Broock ex Yurkov; Vishniacozyma taibaiensis Q.M. Wang & F.Y. Bai ex Yurkov; Vishniacozyma tephrensis Vishniac ex Xin Zhan Liu, F.Y. Bai, M. Groenew. & Boekhout; Yamadamyces Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; Yamadamyces rosulatus Golubev & Scorzetti ex Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout
Introduction
Basidiomycetous yeasts are fungi that can be characterisedcharacterised by unicellular growth for all or the majority of their life cycles (Boekhout et al. 2011). These occur in all three subphyla of Basidiomycota, namely Agaricomycotina, Pucciniomycotina and Ustilaginomycotina (Bauer et al., 2006, Hibbett et al., 2007, Boekhout et al., 2011). Two hundred and twenty-four basidiomycetous yeast species belonging to 39 genera were included in the fourth edition of The Yeasts, a Taxonomic Study (Kurtzman & Fell 1998). That number more than doubled in the next twelve years to 463 species distributed in 62 genera in the fifth edition (Kurtzman et al. 2011). This increase in new species and genera has largely been driven by the adoption of ribosomal DNA (rDNA) gene sequence analyses to yeast identification (Nakase, 2000, Fell et al., 2000, Scorzetti et al., 2002) and the availability of databases containing sequence data of the D1/D2 domains of the large subunit of rDNA (LSU rDNA) and the ITS (including 5.8S) region of rDNA of most of the known basidiomycetous yeast species (Fell et al., 2000, Scorzetti et al., 2002). These molecular taxonomic studies deeply improved our understanding of the phylogenetic relationships, systematics and ecology of basidiomycetous yeasts (Kurtzman & Fell 2006). However, these studies also demonstrated that many genera of basidiomycetous yeasts are polyphyletic (Aime et al., 2006, Boekhout et al., 2011). Recently, an updated taxonomic system of basidiomycetous yeasts was proposed and all polyphyletic genera were revised (Wang et al., 2014, Wang et al., 2015a, Wang et al., 2015b, Wang et al., 2015c, Liu et al., 2015a, Liu et al., 2015b, Wang and Wang, 2015). Vu et al. (2016) indicated that the above revision of basidiomycetous yeasts was a significant improvement in the generic taxonomy, although in a few cases the generic boundaries may still be too broadly defined.
It seems clear that there are still many gaps in our understanding of the yeast phylogeny and diversity. Mycologists have estimated that ca. 1 % fungal species have been described (Hawksworth, 1991, Hawksworth, 2001, Blackwell, 2011, Hawksworth and Lücking, 2017). Similar estimates exist for yeasts, indicating that ca. 12 000 undescribed yeast species await discovery (Lachance 2006), and there is ample evidence that many of these may reside in forests (Fonseca and Inácio, 2006, Morais et al., 2006, Nakase et al., 2006). For example, more than 100 unknown yeast species in forests of Thailand have not yet been described (Nakase et al. 2006).
During a survey of the basidiomycetous yeast diversity in forests, mostly in China, more than 1 000 isolates including 180 strains representing potential novel species were isolated and examined over the past 20 years. In this study, 107 new basidiomycetous yeasts species in Agaricomycotina and Pucciniomycotina are described based on phylogenetic analyses of multiple loci: three nuclear rDNA genes--the small subunit rDNA (SSU), the D1/D2 domains of the large subunit rDNA (LSU), and the internal transcribed spacer including the 5.8S rDNA (ITS)--and four protein coding genes--the largest subunit of RNA polymerase II (RPB1), the second largest subunit of RNA polymerase II (RPB2), translation elongation factor 1-α (TEF1) and the mitochondrial gene cytochrome b (CYTB), and on phenotypic properties. Based on these results, eight new genera, three new families and two new orders are proposed.
Materials and methods
Strains and phenotypic characterisation
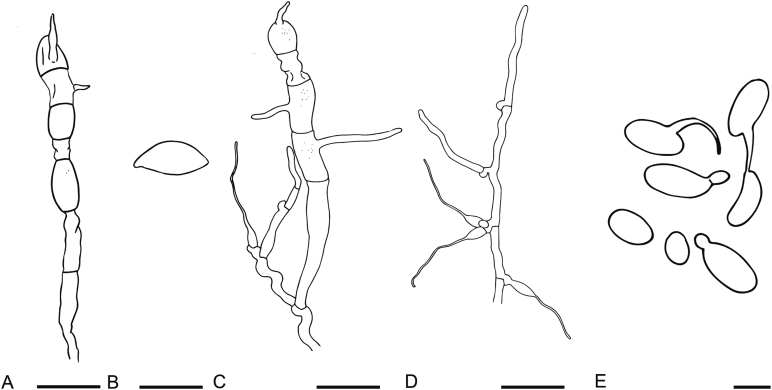
The strains studied are listed in Table 1. Strains were isolated from plant leaves by using the ballistoconidia-fall method as described by Nakase & Takashima (1993). Strains were isolated from soil by an enrichment method: one gram of each sample was placed into 10 ml Yeast Malt (YM, 0.3 % yeast extract, 0.3 % malt extract, 0.5 % peptone, 1 % glucose, Difco) broth containing 200ug/ml chloramphenicol in 15-ml conical tubes and cultured 3–7 d at 17 °C. Then enrichment samples were diluted to 1∗10-3 or 1∗10-4 and 200 μL of each dilution was plated on potato dextrose agar (PDA, 20 % potato infusion, 2 % glucose, 2 % agar, Difco) plates at 17 °C for 3–5 d to culture and isolate yeast strains. Morphological, physiological and biochemical characteristics were examined according to standard methods (Kurtzman et al. 2011). The potential sexual cycles of all new species were investigated using YM, PDA, V8 (10 % V8 juice, 2 % agar) and corn meal agar (CM, 5 % infusion corn meal, 1.5 % agar, Difco). A loopful of cells of each test strain is mixed on an agar plate incubated at 17 °C for one or two months. The cultures were examined with a microscope for the presence of filaments and sexual structures every two weeks. The ballistoconidium-forming activity of all new species was observed by the inverted-plate method (do Carmo-Sousa & Phaff 1962) using CM agar at 17 °C. After 3 to 14 d, the glass slide containing the discharged spores was removed for examination under the microscope.
Table 1.
List of yeasts employed and GenBank numbers determined in this study.
| Speceis | Strain | Date | Location | Source | 18S+ITS+D1/D2 | RPB1 | RPB2 | TEF1 | CYTB |
|---|---|---|---|---|---|---|---|---|---|
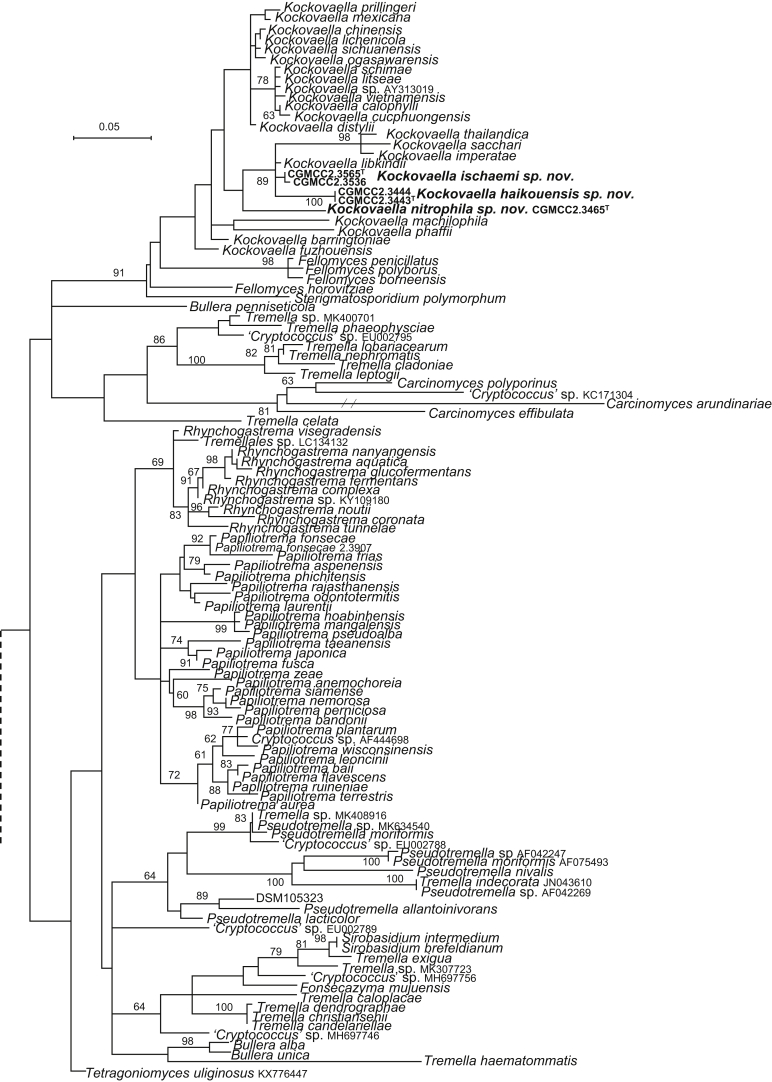
| Kockovaella haikouensis sp. nov. | CGMCC 2.3443T = HKX2 = CBS 15478 | November 14, 2006 | Haikou county, Hainan province, China | phylloplane | MK050274 | MK849163 | MK849301 | MK849032 | MK848902 |
| CGMCC 2.3444 = KX4 | November 14, 2006 | Haikou county, Hainan province, China | phylloplane | MK050275 | – | – | – | – | |
| K. ischaemi sp. nov. | CGMCC 2.3565T = JH5.17 = CBS 15500 | November 15, 2006 | Jinghong, Yunnan province, China | leaf of Ischaemum sp. | MK050276 | MK849185 | MK849323 | – | – |
| CGMCC 2.3536 = JF5.5-2 = CBS 15496 | November 15, 2006 | Jianfaling, Hainan province, China | phylloplane | MK050277 | MK849182 | MK849320 | – | – | |
| K. nitrophila sp. nov. | CGMCC 2.3465T = WZS12.1 = CBS 15487 | November 16, 2006 | Wuzhishan mountain, Hainan province, China | phylloplane | MK050278 | MK849173 | – | MK849043 | MK848913 |
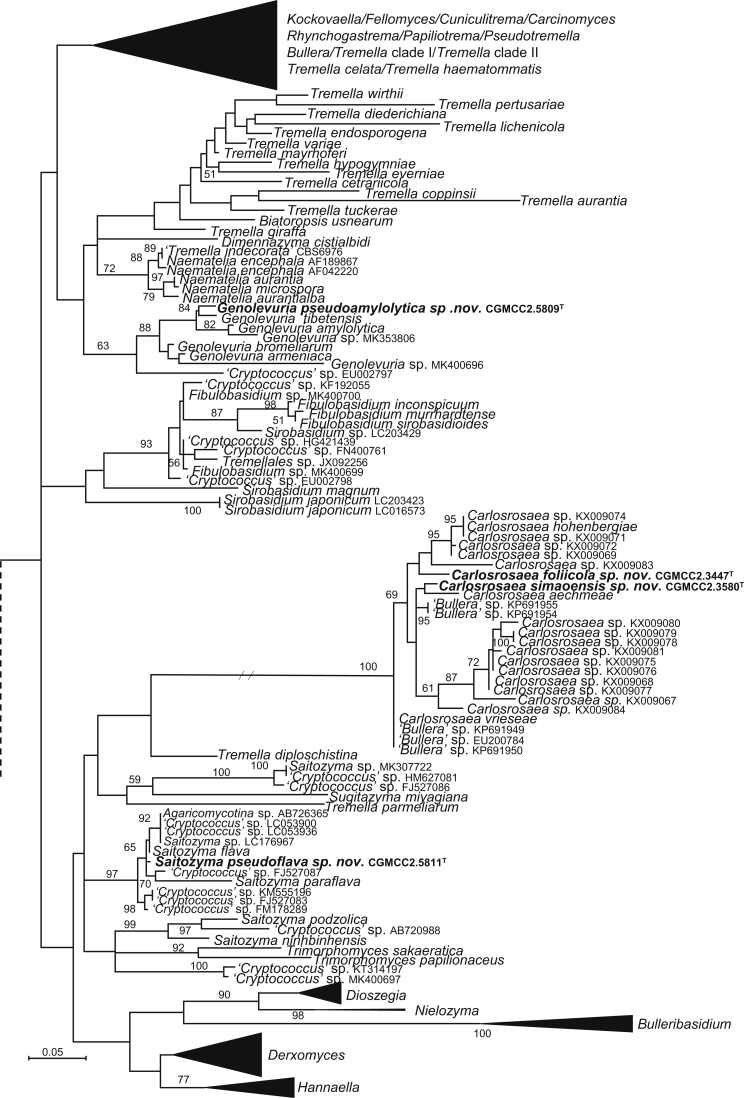
| Genolevuria pseudoamylolytica sp. nov. | CGMCC 2.5809T = HLJ1B6 = CBS 13955 | August 23, 2014 | Daliangzi river national forest park, Heilongjiang province, China | phylloplane | MK050279 | MK849257 | MK849394 | MK849118 | – |
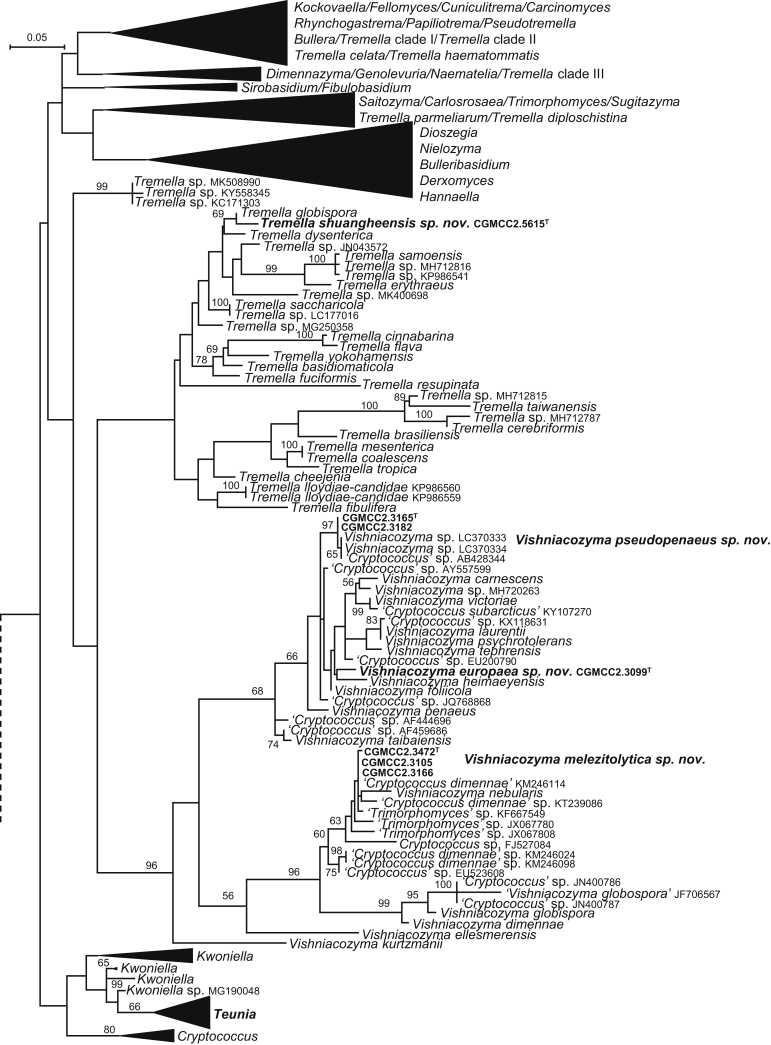
| Vishniacozyma europaea sp. nov. | CGMCC 2.3099T = G7.1-2 = CBS 15464 | September 20, 2005 | Germany | phylloplane | MK050335 | MK849148 | – | MK849018 | MK848890 |
| V. pseudopenaeus sp. nov. | CGMCC 2.3165T = G7.20 = CBS 15472 | September 20, 2005 | Germany | phylloplane | MK050333 | MK849155 | – | MK849025 | MK848897 |
| CGMCC 2.3182 = G7.14 | September 20, 2005 | Germany | phylloplane | MK050334 | MK849158 | – | MK849028 | MK848898 | |
| CBS 8412 | 1996 | Netherlands | brine bath in cheese factory | AY250757/CBS Database | – | – | – | – | |
| CBS 9328 | April 15, 1995 | Carara, Costa Rica | soil | CBS Database | – | – | – | – | |
| V. melezitolytica sp. nov. | CGMCC 2.3472T = H5A3 = CBS 15490 | April 16, 2007 | Hebei province, China | phylloplane | MK050330 | MK849177 | MK849315 | MK849046 | – |
| CGMCC 2.3105 = G18.1 = CBS 15467 | September 20, 2005 | Germany | phylloplane | MK050331 | – | – | – | – | |
| CGMCC 2.3166 = G18.11 | September 20, 2005 | Germany | phylloplane | MK050332 | MK849156 | MK849295 | MK849026 | – | |
| Saitozyma pseudoflava sp. nov. | CGMCC 2.5811T = XZ200A1 = CBS 15576 | September 22, 2014 | Tibet, China | phylloplane | MK050284 | MK849251 | MK849387 | MK849114 | MK848987 |
| Carlosrosaea foliicola sp. nov. | CGMCC 2.3447T = WZS29.4 = CBS 15481 | November 6, 2006 | Wuzhishan mountain, Hainan province, China | phylloplane | MK050282 | MK849166 | MK849304 | – | MK848905 |
| C. simaoensis sp. nov. | CGMCC 2.3580T = SM8.1 = CBS 15503 | November 14, 2006 | Simao county, Yunnan province, China | phylloplane | MK050283 | MK849188 | MK849326 | MK849056 | MK848924 |
| Tremella shuangheensis sp. nov. | CGMCC 2.5615T = SH58A1 = CBS 15561 | August 20, 2015 | Shuanghe county, Heilongjiang province, China | phylloplane | MK050285 | MK849223 | MK849362 | MK849087 | MK848956 |
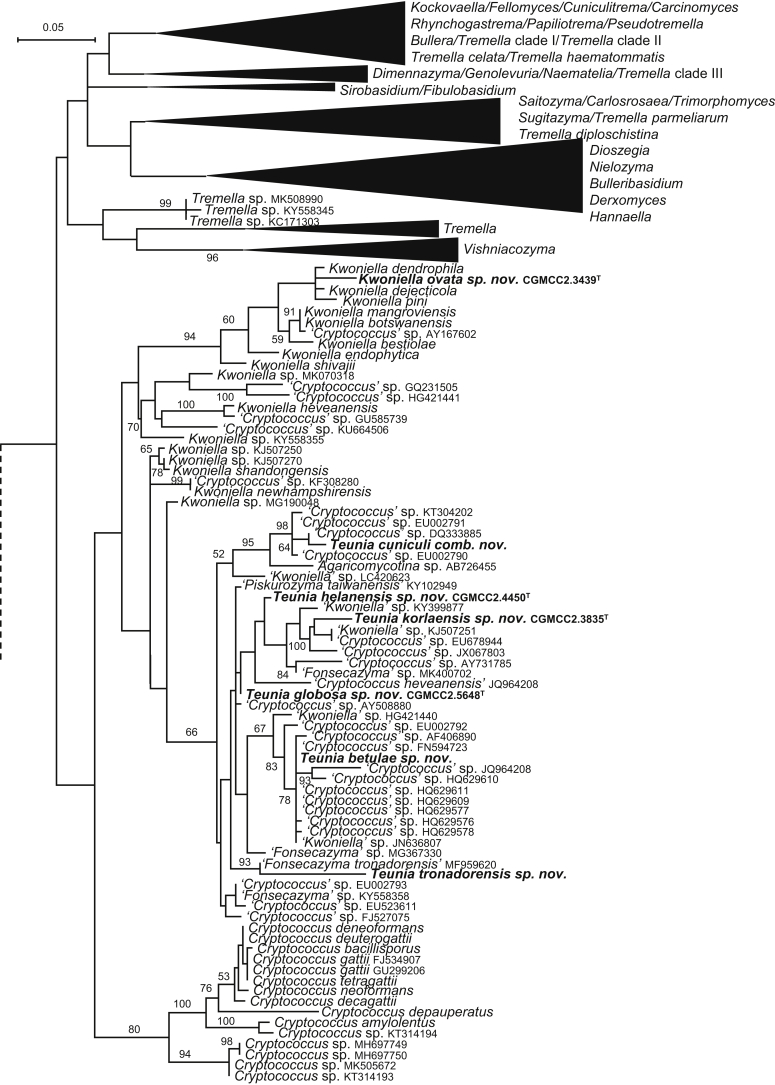
| Kwoniella ovata sp. nov. | CGMCC 2.3439T = H1C1 = CBS 15475 | November 6, 2006 | Hebei province, China | phylloplane | MK050289 | MK849160 | MK849298 | MK849030 | MK848899 |
| Teunia korlaensis sp. nov. | CGMCC 2.3835T = 141.19 = CBS 15653 | February 21, 2008 | Kuerlei county, Xinjiang province, China | soil | MK050286 | MK849194 | MK849332 | – | MK848929 |
| T. helanensis sp. nov. | CGMCC 2.4450T = HLS02-1-5 = CBS 12498 | August 21, 2009 | Helanshan mountain, Ningxia province, China | soil | MK050287 | MK849208 | MK849347 | MK849074 | MK848942 |
| T. globosa sp. nov. | CGMCC 2.5648T = GPS23.2A6 = CBS 15566 | September 22, 2015 | Lulang county, Tibet, China | phylloplane | MK050288 | MK849235 | MK849374 | MK849100 | – |
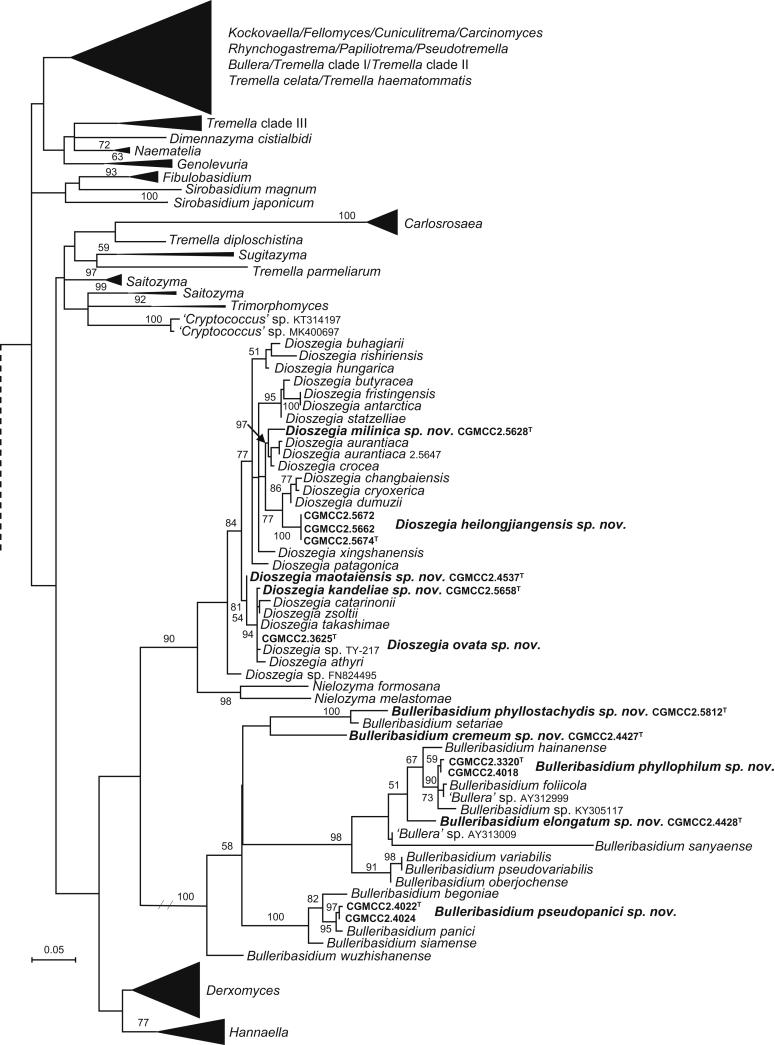
| Dioszegia milinica sp. nov. | CGMCC 2.5628T = GPS21.3B8 = CBS 15563 | September 21, 2015 | Milin county, Tibet, China | phylloplane | MK050290 | MK849231 | MK849371 | MK849097 | MK848966 |
| D. heilongjiangensis sp. nov. | CGMCC 2.5674T = HLJ13.24 = CBS 13957 | August 28, 2014 | Chelu county, Heilongjiang province, China | phylloplane | MK050291 | MK849245 | MK849382 | MK849109 | MK848981 |
| CGMCC 2.5662 = HLJ41A9 = CBS 13966 | August 26, 2014 | Wuyiling natural reserve, Heilongjiang province, China | phylloplane | MK050292 | MK849243 | MK849380 | MK849106 | MK848978 | |
| CGMCC 2.5672 = HLJ41A9B | August 26, 2014 | Wuyiling natural reserve, Heilongjiang province, China | phylloplane | MK050293 | – | – | – | – | |
| D. ovata sp. nov. | CGMCC 2.3625T = HBX1.27 = CBS 15657 | November 24, 2006 | Bangxi county, Hainan province, China | phylloplane | MK050294 | MK849190 | MK849328 | – | MK848926 |
| TY-217 | 2003 | Thailand | phylloplane | AY313036/AY313018 | – | – | – | – | |
| D. maotaiensis sp. nov. | CGMCC 2.4537T = GZMT3A9 = CBS 15516 | March 8, 2012 | Maotai county, Guizhou province, China | phylloplane | MK050295 | MK849210 | MK849350 | MK849076 | MK848945 |
| D. kandeliae sp. nov. | CGMCC 2.5658T = 224191 = CBS 13951 | April 15, 2014 | Beilunhekou natural reserve, Guangxi province, China | leaf of Kandelia candel | MK050296 | MK849241 | MK849378 | MK849104 | MK848976 |
| Bulleribasidium pseudopanici sp. nov. | CGMCC 2.4024T = WZS17.20 = CBS 15510 | November 22, 2006 | Wuzhishan mountain, Hainan province, China | phylloplane | MK050323 | MK849197 | MK849336 | MK849062 | MK848932 |
| CGMCC 2.4022 = WZS29.3 | November 16, 2006 | Wuzhishan mountain, Hainan province, China | phylloplane | MK050324 | MK849196 | MK849335 | MK849061 | – | |
| B. cremeum sp. nov. | CGMCC 2.4427T = TW1.1F-025 = CBS 12487 | August 18, 2009 | Taiwan, China | phylloplane | MK050325 | MK849198 | MK849337 | MK849064 | MK848933 |
| B. phyllostachydis sp. nov. | CGMCC 2.5812T = XZ139E1 = CBS 15575 | September 20, 2014 | Motuo, Tibet, China | leaf of Phyllostachys sp. | MK050327 | MK849261 | MK849398 | – | MK848993 |
| B. elongatum sp. nov. | CGMCC 2.4428T = TW1.1F-019 = CBS 12489 | August 18, 2009 | Taiwan, China | phylloplane | MK050326 | MK849199 | MK849338 | MK849065 | MK848934 |
| B. phyllophilum sp. nov. | CGMCC 2.3320T = HBX2.8 = CBS 15474 | November 24, 2006 | Bangxi county, Hainan province, China | phylloplane | MK050328 | MK849159 | MK849297 | MK849029 | – |
| CGMCC 2.4018 = HBX1.23 | November 24, 2006 | Bangxi county, Hainan province, China | phylloplane | MK050329 | MK849195 | MK849334 | MK849060 | MK848931 | |
| TY-199 | 2003 | Thailand | phylloplane | AY313030 | – | – | – | – | |
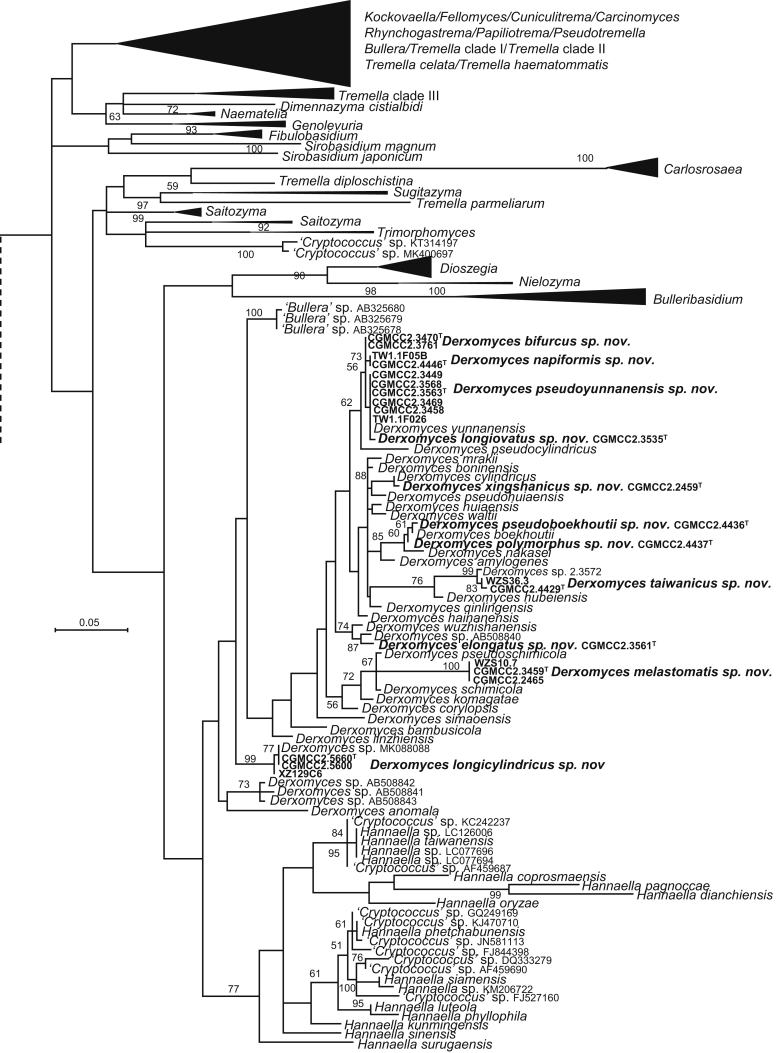
| Derxomyces pseudoboekhoutii sp. nov. | CGMCC 2.4436T = FJYZ12-8 = CBS 12493 | August 18, 2011 | Fuzhou county, Fujian province, China | phylloplane | MK050310 | MK849202 | MK849341 | MK849068 | MK848937 |
| D. polymorphus sp. nov. | CGMCC 2.4437T = FJYZ12-13 = CBS 15512 | August 18, 2011 | Fuzhou county, Fujian province, China | phylloplane | MK050309 | MK849203 | MK849342 | MK849069 | MK848938 |
| D. xingshanicus sp. nov. | CGMCC 2.2459T = HX16.1 = CBS 15445 | July 7, 2003 | Xingshan county, Hubei province, China | phylloplane | MK050308 | MK849128 | MK849269 | MK849000 | MK848873 |
| D. pseudoyunnanensis sp. nov. | CGMCC 2.3563T = SM37E2 = CBS 15499 | November 10, 2006 | Simao county, Yunnan province, China | phylloplane | MK050313 | MK849184 | MK849322 | MK849052 | MK848921 |
| CGMCC 2.3469 = WZS29.1B | November 16, 2006 | Wuzhishan mountain, Hainan province, China | phylloplane | MK050316 | MK849175 | MK849313 | MK849044 | MK848914 | |
| CGMCC 2.3568 = SM37.6 = CBS 15501 | November 14, 2006 | Simao county, Yunnan province, China | phylloplane | MK050314 | MK849186 | MK849324 | MK849053 | MK848922 | |
| CGMCC 2.3449 = WZS29.18 | November 16, 2006 | Wuzhishan mountain, Hainan province, China | phylloplane | MK050317 | – | – | – | – | |
| CGMCC 2.3458 = WZS29.1 = CBS 15484 | November 16, 2006 | Wuzhishan mountain, Hainan province, China | phylloplane | MK050315 | MK849169 | MK849307 | MK849037 | MK848907 | |
| TW1.1F026 | August 18, 2009 | Taiwan, China | phylloplane | MK050318 | – | – | – | – | |
| D. longiovatus sp. nov. | CGMCC 2.3535T = SM35.4 = CBS 15659 | November 10, 2006 | Simao county, Yunnan province, China | phylloplane | MK050312 | MK849181 | MK849319 | MK849050 | MK848919 |
| D. napiformis sp. nov. | CGMCC 2.4446T = TW1.1F028 = CBS 15748 | August 18, 2009 | Taiwan, China | phylloplane | MK050321 | MK849207 | MK849346 | MK849073 | MK848941 |
| TW1.1F05B | August 18, 2009 | Taiwan, China | phylloplane | MK050322 | – | – | – | – | |
| D. bifurcus sp. nov. | CGMCC 2.3470T = SM37.5 = CBS 15489 | November 16, 2006 | Simao county, Yunnan province, China | phylloplane | MK050319 | MK849176 | MK849314 | MK849045 | MK848915 |
| CGMCC 2.3761 = SM37.15 = CBS 15508 | October 16, 2007 | Simao county, Yunnan province, China | phylloplane | MK050320 | – | – | – | – | |
| D. elongatus sp. nov. | CGMCC 2.3561T = SM32.1 = CBS 15498 | November 10, 2006 | Simao county, Yunnan province, China | phylloplane | MK050311 | MK849183 | MK849321 | MK849051 | MK848920 |
| D. melastomatis sp. nov. | CGMCC 2.3459T = WZS19.7 = CBS 15485 | November 16, 2006 | Wuzhishan mountain, Hainan province, China | leaf of Melastoma candidum | MK050305 | MK849170 | MK849308 | MK849038 | MK848908 |
| CGMCC 2.2465 = HX7.3 | October 13, 2002 | Xingshan county, Hubei Province, China | leaf of Stephanandra chinensis | MK050306 | – | – | – | – | |
| WZS10.7 | November 15, 2006 | Wuzhishan mountain, Hainan province, China | phylloplane | MK050307 | – | – | – | – | |
| D. taiwanicus sp. nov. | CGMCC 2.4429T = TW3.1C-02 = CBS 12490 | August 18, 2009 | Taiwan, China | phylloplane | MK050303 | MK849200 | MK849339 | MK849066 | MK848935 |
| WZS36.3 | November 17, 2006 | Wuzhishan mountain, Hainan province, China | phylloplane | MK050304 | – | – | – | – | |
| D. ovatus sp. nov. | CGMCC 2.3572T = SM32.2 = CBS 15654 | November 10, 2006 | Simao county, Yunnan province, China | phylloplane | MK050302 | MK849187 | MK849325 | MK849055 | MK848923 |
| D. longicylindricus sp. nov. | CGMCC 2.5660T = XZ132E37A = CBS 13979 | September 21, 2014 | Beibeng county, Motuo, Tibet, China | phylloplane | MK050300 | MK849242 | MK849379 | MK849105 | MK848977 |
| CGMCC 2.5813 = XZ129C6A | September 20, 2014 | Motuo, Tibet, China | leaf of Nepeta sp. | MK050301 | – | – | – | – | |
| 5600 | September 21, 2014 | Beibeng county, Motuo, Tibet, China | phylloplane | MK088088 | MK849216 | MK849355 | MK849082 | MK848950 | |
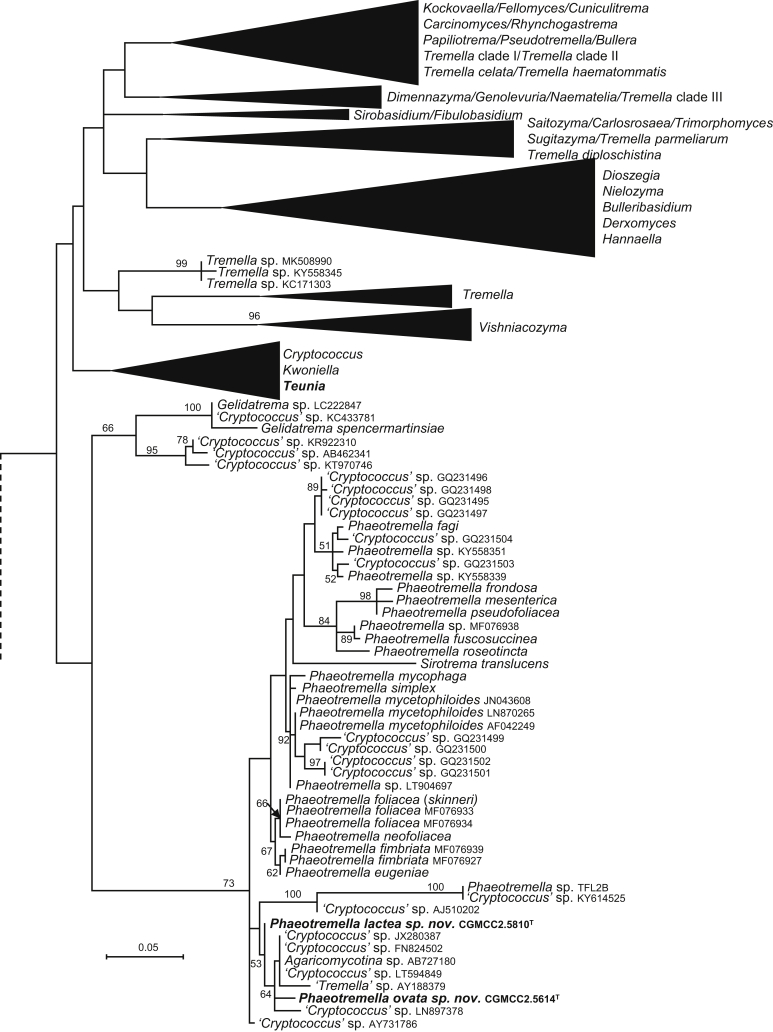
| Phaeotremella lactea sp. nov. | CGMCC 2.5810T = GPS20.4A1B = CBS 15574 | September 21, 2015 | Milin county, Tibet, China | phylloplane | MK050280 | MK849250 | – | – | MK848986 |
| P. ovata sp. nov. | CGMCC 2.5614T = NW9D3 = CBS 15756 | August 20, 2015 | Nanwenghe, Heilongjiang province, China | phylloplane | MK050281 | MK849222 | MK849361 | – | MK848949 |
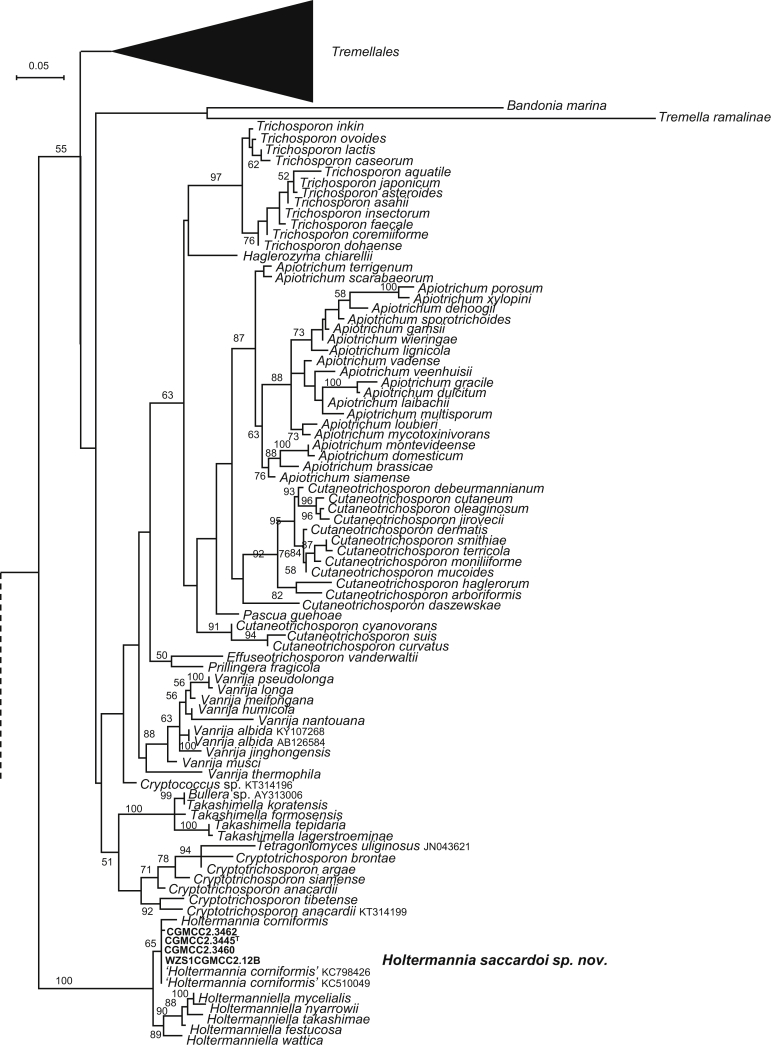
| Holtermannia saccardoi sp. nov. | CGMCC 2.3445T = SM37.10 = CBS 15479 | November 6, 2006 | Simao county, Yunnan province, China | phylloplane | MK050336 | MK849164 | MK849302 | MK849033 | MK848903 |
| CGMCC 2.3460 = SM6.3 | November 6, 2006 | Simao county, Yunnan province, China | leaf of Arisaema yunnanense | MK050337 | MK849171 | MK849309 | MK849039 | MK848909 | |
| CGMCC 2.3462 = SM32.11 | November 6, 2006 | Simao county, Yunnan province, China | phylloplane | MK050338 | – | MK849310 | MK849040 | MK848910 | |
| WZS12.12B | November 16, 2006 | Wuzhishan mountain, Hainan province, China | phylloplane | MK050339 | – | – | – | – | |
| Solicoccozyma gelidoterrea sp. nov. | CGMCC 2.5814T = HFB003-3 = CBS 15580 | August 15, 2015 | Daxinganling, China | soil | MK050340 | MK849252 | MK849388 | – | – |
| CGMCC 2.4893 = LZ3.17.4 | October 12, 2012 | China | soil | MK050341 | MK849215 | MK849354 | MK849081 | MK848948 | |
| DBVPG10727 | 2017 | Alps, Dolomites, Livigno, Italy | bark of spruce | MK070335/MK070317 | – | – | – | – | |
| CBS 9627 | November, 1981 | Colorado, Longs Peak, Rocky Mountain National Park, USA | soil | KY105431/KY109663 | – | – | – | – | |
| CBS 9287 | n/a | Providenya, Russia | soil | MK397489 | – | – | – | – | |
| Filobasidium dingjieense sp. nov. | CGMCC 2.5649T = GPS3.2A5 = CBS 15567 | September 12, 2015 | Dingjie county, Tibet, China | phylloplane | MK050342 | MK849236 | MK849375 | – | MK848971 |
| GPS23.2A5 | September 22, 2015 | Lulang county, Tibet, China | phylloplane | MK050343 | – | – | – | ||
| F. globosum sp. nov. | CGMCC 2.5680T = HLJ8A3 = CBS 15658 | August 25, 2014 | Yichun county, Heilongjiang province, China | phylloplane | MK050344 | MN014083 | MN014090 | MN014092 | MN014078 |
| CGMCC 2.5656 = HLJ8A3B | August 25, 2014 | Yichun county, Heilongjiang province, China | phylloplane | MK050345 | MK849240 | MK849377 | – | MK848975 | |
| F. mali sp. nov. | CGMCC 2.4012T = KTAPG4-11.46 = CBS 15651 | August 20, 2008 | Qufu county, Shandong province, China | leaf of apple (Malus pumila) | MK050346 | MK849333 | – | – | MK848930 |
| CGMCC 2.4052 = KTAPG1-11.63 | August 20, 2008 | Tai’an county, Shandong province, China | leaf of apple (Malus pumila) | MK050347 | – | – | – | – | |
| CGMCC 2.3464 = WZS19.13 | November 16, 2006 | Wuzhishan mountain, Hainan province, China | leaf of Melastoma candidum | MK050348 | MK849172 | MK849312 | MK849042 | MK848912 | |
| KTAPG4-11.64 | August 20, 2008 | Qufu county, Shandong province, China | leaf of apple (Malus pumila) | GQ181171 | – | – | – | – | |
| 4QVF20 = CBS 10181 | June, 1998 | Arrabida Natural Park, Portugal | Leaf of Quercus faginea | EU002869/EU002805 | – | – | – | – | |
| F. mucilaginum sp. nov. | CGMCC 2.3463T = SY2.1 = CBS 15486 | November 16, 2006 | Sanya county, Hainan province, China | phylloplane | MK050349 | – | MK849311 | MK849041 | MK848911 |
| Phaffia aurantiaca sp. nov. | CGMCC 2.5601T = GPS23.2A4 = CBS 15548 | September 22, 2015 | Lulang county, Tibet, China | phylloplane | MK050350 | MN014085 | MN014089 | MN014091 | MN014077 |
| Kondoa cylindrica sp. nov. | CGMCC 2.3102T = G6.1-1 = CBS 15466 | September 20, 2005 | Germany | phylloplane | MK050351 | MK849150 | MK849290 | MK849020 | MK848892 |
| CGMCC 2.3103 = G4.22A | September 20, 2005 | Germany | phylloplane | MK050352 | MK849151 | MK849291 | MK849021 | MK848893 | |
| CGMCC 2.3175 = G4.22B | September 20, 2005 | Germany | phylloplane | MK050353 | MK849157 | MK849296 | MK849027 | – | |
| PYCC 5566 | 1998 | Sesimbra, Portugal | basidiocarp of Myxarium nucleatum | AF444672/AF444766 | – | – | – | – | |
| K. chamaenerii sp. nov. | CGMCC 2.2652T = XJ8A5 = CBS 15453 | July 6, 2004 | Bujin county, Xinjiang province, China | leaf of Chamaenerion angustifolium | MK050354 | MK849135 | MK849275 | MK849005 | MK848878 |
| CGMCC 2.2760 = XJ10A7 | July 6, 2004 | Bujin county, Xinjiang province, China | leaf of Cotoneaster melanocarpus | MK050355 | – | MK849278 | MK849007 | MK848880 | |
| K. foliicola sp. nov. | CGMCC 2.3100T = G9.1 = CBS 15465 | September 20, 2005 | Germany | phylloplane | MK050356 | MK849262 | MK849399 | MK849120 | MK848994 |
| K. arboricola sp. nov. | CGMCC 2.2621T = XZ12B5 = CBS 15452 | September 21, 2004 | Bomi county, Tibet, China | leaf of arbor | MK050357 | MK849134 | MK849274 | – | – |
| CGMCC 2.4886 = LWL4.17.24 | October 12, 2012 | China | soil | MK050358 | MK849214 | MK849353 | – | – | |
| K. lulangica sp. nov. | CGMCC 2.2762T = XZ36D1 = CBS 15456 | September 21, 2004 | Lulang county, Tibet, China | phylloplane | MK050359 | MK849138 | MK849279 | MK849008 | MK848881 |
| K. rhododendri sp. nov. | CGMCC 2.2763T = XZ27E3 = CBS 15457 | September 21, 2004 | Bomi county, Tibet, China | leaf of Rhododendron triflorum | MK050360 | MK849139 | MK849280 | MK849009 | MK848882 |
| K. daliangziensis sp. nov. | CGMCC 2.5610T = HLJ22A8 = CBS 13974 | August 28, 2014 | Daliangzi river national forest park, Heilongjiang province, China | phylloplane | MK050361 | MK849220 | MK849359 | MK849085 | MK848954 |
| HLJ14.20B = CBS 15577 | August 20, 2014 | Chelu county, Heilongjiang province, China | phylloplane | MK050362 | MK849256 | MK849393 | MK849117 | MK848990 | |
| K. ribitophobia sp. nov. | CGMCC 2.4441T = TW2.1E-016 = CBS 12496 | August 17, 2009 | Taiwan, China | phylloplane | MK050363 | MK849204 | MK849343 | MK849070 | MK848939 |
| CGMCC 2.4875 = HZZ9D.2 | October 12, 2012 | Houzhenzi, Shaaxi province, China | phylloplane | MK050364 | MK849213 | MK849352 | MK849080 | – | |
| K. myxariophila sp. nov. | CGMCC 2.3106 = G18.2-2 = CBS 15468 | September 20, 2005 | Germany | phylloplane | MK050365 | MK849152 | MK849292 | MK849022 | MK848894 |
| AS483 = CBS 11525 | November, 2008 | Graubuenden Alp Flix, Switzerland | flower of Dianthus superbus | MN175324/FN428954 | – | – | – | – | |
| PYCC 5509T = CBS 8379 = ZP 337 | 1992 | Portugal | basidiocarps of Myxarium nucleatum | AF444596/AF189904 | – | – | – | – | |
| PYCC 8354 = ZP 338 | 1992 | Portugal | basidiocarps of Myxarium nucleatum | MN175325 | – | – | – | – | |
| PYCC 8305 = ZP 352 | 1996 | Portugal | basidiocarps of Myxarium nucleatum | MN175326 | – | – | – | – | |
| Bensingtonia wuzhishanensis sp. nov. | CGMCC 2.3569T = WZS33.18 = CBS 15661 | November 14, 2006 | Wuzhishan mountain, Hainan province, China | phylloplane | MK050366 | – | – | MK849054 | – |
| B. pseudorectispora sp. nov. | CGMCC 2.5677T = XZ154D5 = CBS 15750 | September 21, 2014 | Bomi, Tibet, China | phylloplane | MK050367 | MK849247 | MK849384 | MK849111 | MK848983 |
| Pseudobensingtonia fusiformis sp. nov. | CGMCC 2.5823T = XZ152E3A = CBS 15647 | September 21, 2014 | Bomi, Tibet, China | phylloplane | MK050370 | MK849123 | MK849265 | MK848997 | MK848870 |
| CGMCC 2.5815 = XZ152E3 = CBS 15592 | September 21, 2014 | Bomi, Tibet, China | phylloplane | MK050368 | MK849149 | MK849289 | MK849019 | MK848891 | |
| XZ152B1 = CBS 15663 | September 21, 2014 | Bomi, Tibet, China | phylloplane | MK050369 | – | – | – | – | |
| Boekhoutia sterigmata sp. nov. | CGMCC 2.4539T = FJS3F22 = CBS 15553 | October 29, 2011 | Fanjingshan Mountain, Guizhou province, China | phylloplane | MK050371 | MK849211 | – | MK849078 | MK848946 |
| Ruinenia fanjingshanensis sp. nov. | CGMCC 2.4542T = FJS6C7 = CBS 15745 | October 29, 2011 | Fanjingshan Mountain, Guizhou province, China | phylloplane | MK050372 | MK849211 | MK849267 | MK849078 | MK848946 |
| R. bangxiensis sp. nov. | CGMCC 2.3454T = HBX1.0 = CBS 10819 | November 24, 2006 | Bangxi county, Hainan province, China | phylloplane | MK050373 | MK849167 | MK849305 | MK849035 | – |
| ST-153 | February 3, 2001 | Ban Paeng Distric, Nakhon Phanom Province, Thailand | phylloplane | MN194597/DQ404467 | – | – | – | – | |
| R. lunata sp. nov. | CGMCC 2.4426T = TW 2.1E-028 = CBS 12525 | August 17, 2009 | Taiwan, China | phylloplane | KP020113 | – | MN014088 | MN014094 | MN014079 |
| TW2.1E-05B | August 18, 2009 | Taiwan, China | phylloplane | KP020110 | – | – | MK849063 | – | |
| Sterigmatospora layueensis sp. nov. | CGMCC 2.5817T = XZ100A2B = CBS 15649 | September 18, 2014 | Layue county, Tibet, China | phylloplane | MK050375 | MK849259 | MK849396 | MK849119 | – |
| Pseudosterigmatospora motuoensis sp. nov. | CGMCC 2.5816T = XZ119B3 = CBS 15591 | September 18, 2014 | Motuo, Tibet, China | leaf of Achyrospermum wallichianum | MK050374 | MK849253 | MK849389 | MK849115 | MK848988 |
| Phyllozyma jiayinensis sp. nov. | CGMCC 2.5669T = HLJ25.21 = CBS 13975 | August 25, 2014 | Qingshan county, Jiayin, Heilongjiang province, China | phylloplane | MK050376 | – | – | MK849108 | MK848980 |
| P. aceris sp. nov. | CGMCC 2.2662T = XZ17B1 = CBS 15773 | September 21, 2004 | Bomi county, Tibet, China | leaf of Acer caudatum | MK050377 | MK849136 | MK849276 | MK849006 | MK848879 |
| CGMCC 2.2617 = XZ14B2 | September 21, 2004 | Bomi county, Tibet, China | leaf of bamboo | MK050378 | MK849132 | – | MK849003 | – | |
| Meniscomyces layueensis sp. nov. | CGMCC 2.5818T = XZ100 = CBS 15747 | September 18, 2014 | Layue county,Tibet, China | phylloplane | MK050379 | MK849248 | MK849385 | MK849112 | MK848984 |
| CGMCC 2.5681 = XZ100A2 | September 18, 2014 | Layue county,Tibet, China | phylloplane | MK050380 | – | – | – | – | |
| Sakaguchia melibiophila sp. nov. | CBS 5143T = JCM 8162 = CGMCC 2.4235 = IGC 5612 | n/a | The Netherlands | bronchial secretion | KJ778625/KJ708453/KJ708356 | KJ708079 | KJ708268 | KJ707858 | KJ707732 |
| Microsporomyces pseudomagnisporus sp. nov. | CGMCC 2.4538T = FJS25C3 = CBS 15746 | October 29, 2011 | Fanjingshan Mountain, Guizhou province, China | phylloplane | MK050384 | MK849125 | MK849351 | MK849077 | – |
| M. rubellus sp. nov. | CGMCC 2.4444T = TW1.3F-017 = CBS 15622 | August 18, 2009 | Taiwan, China | phylloplane | MK050385 | MK849205 | MK849344 | MK849071 | – |
| CGMCC 2.4445 = TW1.3F-026 = CBS 12526 | August 18, 2009 | Taiwan, China | phylloplane | MK050386 | MK849206 | MK849345 | MK849072 | MK848940 | |
| M. ellipsoideus sp. nov. | CGMCC 2.5664T = XZ137E4 = CBS 16020 | September 20, 2014 | Motuo county, Tibet, China | phylloplane | MK050387 | MK849244 | MK849381 | MK849107 | MK848979 |
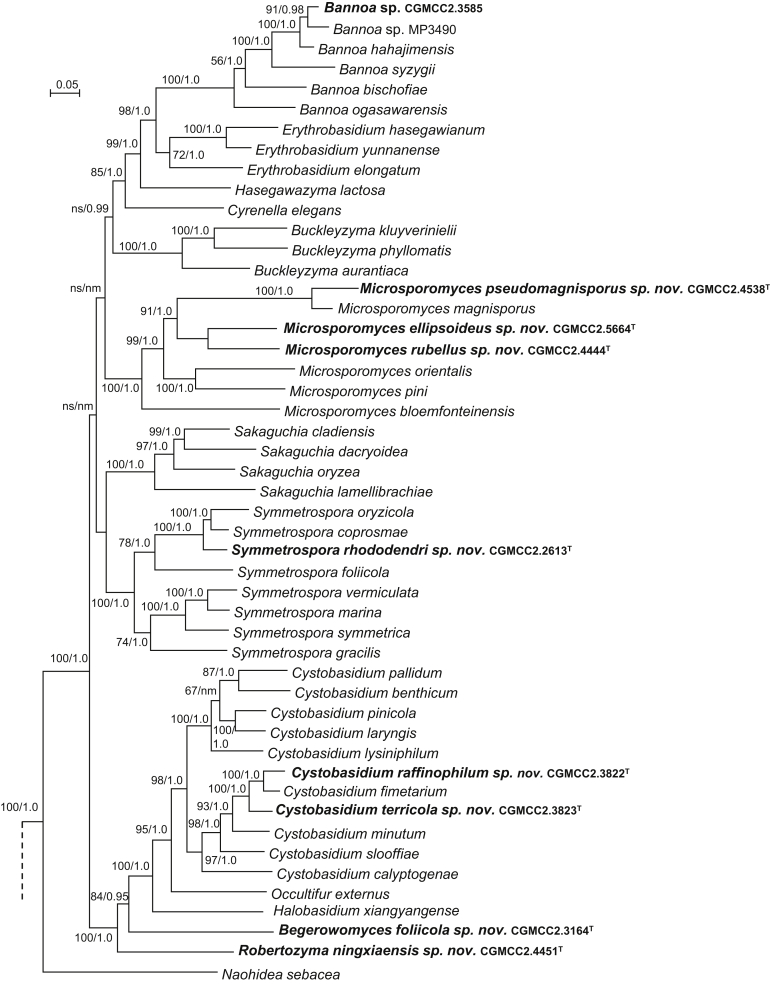
| Symmetrospora rhododendri sp. nov. | CGMCC 2.2613T = XZ49DX = CBS 15447 | September 21, 2004 | Lulang county, Tibet, China | leaf of Rhododendron sp. | MK050388 | MK849130 | MK849271 | MK849001 | – |
| Cystobasidium raffinophilum sp. nov. | CGMCC 2.3822T = 141.4 = CBS 15509 | July 6, 2007 | Yecheng county, Xinjiang province, China | soil | MK050389 | MK849191 | MK849329 | MK849058 | MK848927 |
| C. terricola sp. nov. | CGMCC 2.3823T = 140.23 = CBS 15650 | July 6, 2007 | Yecheng county, Xinjiang province, China | soil | MK050390 | MK849192 | MK849330 | MK849059 | MK848928 |
| CGMCC 2.3824 = 141.8 | July 6, 2007 | Yecheng county, Xinjiang province, China | soil | MK050391 | MK849193 | MK849331 | – | – | |
| Robertozyma ningxiaensis sp. nov. | CGMCC 2.4451T = HLS10.23 = CBS 12499 | August 21, 2009 | Helanshan mountain, Ningxia province, China | soil | MK050392 | – | MK849348 | – | MK848943 |
| CGMCC 2.4452 = HLS14.23 | August 21, 2009 | Helanshan mountain, Ningxia province, China | soil | MK050393 | MK849209 | MK849349 | MK849075 | MK848944 | |
| Begerowomyces foliicola sp. nov. | CGMCC 2.3164T = G7.4 = CBS 15655 | September 20, 2005 | Germany | phylloplane | MK050394 | MK849154 | MK849294 | MK849024 | MK848896 |
| Rosettozyma petaloides sp. nov. | CGMCC 2.3446T = WZS29.14 = CBS 15480 | November 6, 2006 | Wuzhishan mountain, Hainan province, China | phylloplane | MK050395 | MK849165 | MK849303 | MK849034 | MK848904 |
| CGMCC 2.3466 = WZS9.2 = CBS 15488 | November 16, 2006 | Wuzhishan mountain, Hainan province, China | phylloplane | MK050396 | MK849174 | – | – | – | |
| CGMCC 2.3461 = WZS29.15 | November 6, 2006 | Wuzhishan mountain, Hainan province, China | phylloplane | MK050397 | – | – | – | – | |
| R. cystopteridis sp. nov. | CGMCC 2.2615T = XZ16E1 = CBS 15448 | September 21, 2004 | Bomi county, Tibet, China | leaf of Cystopteris moupinensis | MK050398 | MK849131 | MK849272 | MK849002 | MK848876 |
| CGMCC 2.2619 = XZ5B2 = CBS 15451 | September 21, 2004 | Bomi county, Tibet, China | leaf of Rhododendron phaeochrysum | MK050399 | – | – | – | MK848877 | |
| R. motuoensis sp. nov. | CGMCC 2.5819T = XZ118E6 = CBS 15588 | September 19, 2014 | Motuo, Tibet, China | phylloplane | MK050400 | MK849260 | MK849397 | – | MK848991 |
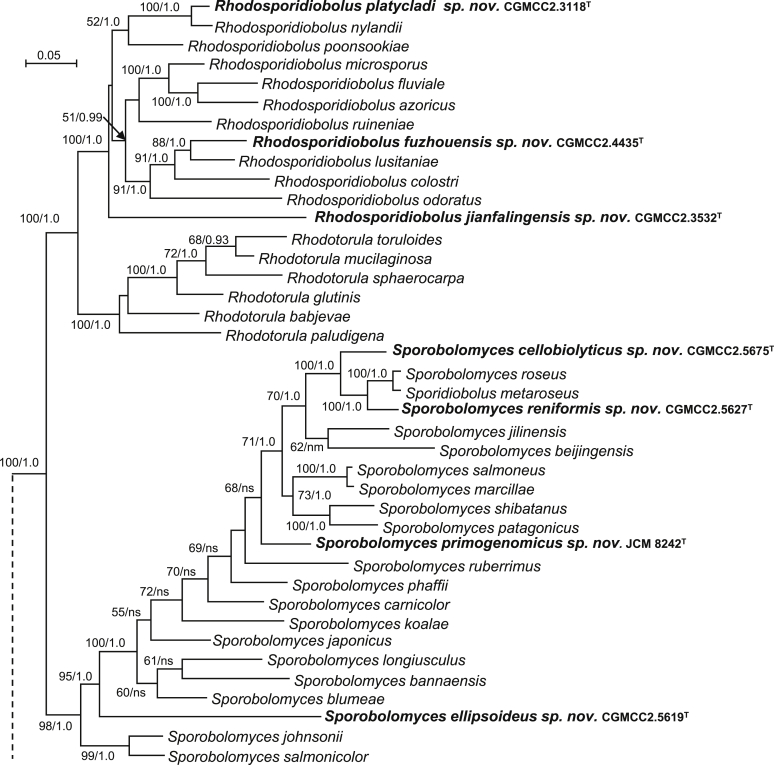
| Rhodosporidiobolus platycladi sp. nov. | CGMCC 2.3118T = BJ6-3 = CBS 15469 | March 27, 2006 | Beijing, China | leaf of Platycladus sp. | MK050401 | MK849153 | MK849293 | MK849023 | MK848895 |
| R. jianfalingensis sp. nov. | CGMCC 2.3532T = JF25.7-1 = CBS 15494 | May 10, 2007 | Jianfaling, Hainan province, China | phylloplane | MK050402 | MK849179 | MK849317 | MK849048 | MK848917 |
| CGMCC 2.3531 = JF25.7-2 | May 10, 2007 | Jianfaling, Hainan province, China | phylloplane | MK050403 | MK849178 | MK849316 | MK849047 | MK848916 | |
| R. fuzhouensis sp. nov. | CGMCC 2.4435T = FJYZ2-6 = CBS 12492 | August 18, 2011 | Fuzhou county, Fujian province, China | phylloplane | MK050404 | MK849201 | MK849340 | MK849067 | MK848936 |
| CGMCC 2.4442 = TW4.3F1 | August 18, 2009 | Taiwan, China | phylloplane | MK050405 | – | – | – | – | |
| CGMCC 2.2286 = CBS 9205 | January 1, 2001 | Xishuang Banna, Yunnan province, China | leaf of Ficus sp. | KY105509/KY109744/MN180193 | MN180194 | MN180195 | MN180197 | MN180196 | |
| Sporobolomyces cellobiolyticus sp. nov. | CGMCC 2.5675T = HLJ33B4 = CBS 13964 | August 26, 2014 | Wuyiling natural reserve, Heilongjiang province, China | phylloplane | MK050406 | MK849246 | MK849383 | MK849110 | MK848982 |
| CGMCC 2.5687 = HLJ32B2 = CBS 13963 | August 25, 2014 | Chelu county, Heilongjiang province, China | phylloplane | MK050407 | MK849249 | MK849386 | MK849113 | MK848985 | |
| MCA 3774 | n/a | Alaska, Siberia and Newfoundland, Canada | phylloplane | JN942193/JN940715 | – | – | – | – | |
| MCA 3785 | n/a | Alaska, Siberia and Newfoundland, Canada | phylloplane | JN942199/JN940720 | – | – | – | – | |
| S. reniformis sp. nov. | CGMCC 2.5627T = GPS21.2C2 = CBS 15562 | September 21, 2015 | Milin county, Tibet, China | phylloplane | MK050408 | MK849230 | MK849370 | MK849096 | MK848965 |
| S. ellipsoideus sp. nov. | CGMCC 2.5619T = GPS21.5C1 = CBS 15590 | September 21, 2015 | Milin county, Tibet, China | phylloplane | MK050409 | MK849225 | MK849364 | MK849088 | MK848957 |
| CGMCC 2.5620 = GPS23.3A5 | September 22, 2015 | Lulang county, Tibet, China | phylloplane | MK050410 | – | – | – | – | |
| CGMCC 2.5621 = GPS20.1B3 | September 21, 2015 | Milin county, Tibet, China | phylloplane | MK050411 | MK849227 | – | MK849090 | MK848959 | |
| CGMCC 2.5622 = GPS20.1A4 | September 21, 2015 | Milin county, Tibet, China | phylloplane | MK050412 | MK849228 | MK849366 | MK849091 | MK848960 | |
| CGMCC 2.5624 = GPS20.1H2 | September 21, 2015 | Milin county, Tibet, China | phylloplane | MK050413 | – | – | MK849093 | MK848962 | |
| CGMCC 2.5625 = GPS22.1B3 | September 21, 2015 | Milin county, Tibet, China | phylloplane | MK050414 | MK849229 | MK849368 | MK849094 | MK848963 | |
| CGMCC 2.5626 = GPS20.8C1 | September 21, 2015 | Milin county, Tibet, China | phylloplane | MK050415 | – | MK849369 | MK849095 | MK848964 | |
| CGMCC 2.5631 = GPS20.8C10 | September 21, 2015 | Milin county, Tibet, China | phylloplane | MK050416 | MK849233 | – | MK849099 | MK848969 | |
| CBS 2642 | n/a | UK | milk | KY105474/KY109710 | – | – | – | – | |
| S. primogenomicus sp. nov. | JCM 8242T = IAM13481 = CBS 15935 | 1983 | Kanto region, Japan | a leaf of willow | MK050417/MK050418/MK050419 | MK849124 | MK849266 | MK848998 | MK848872 |
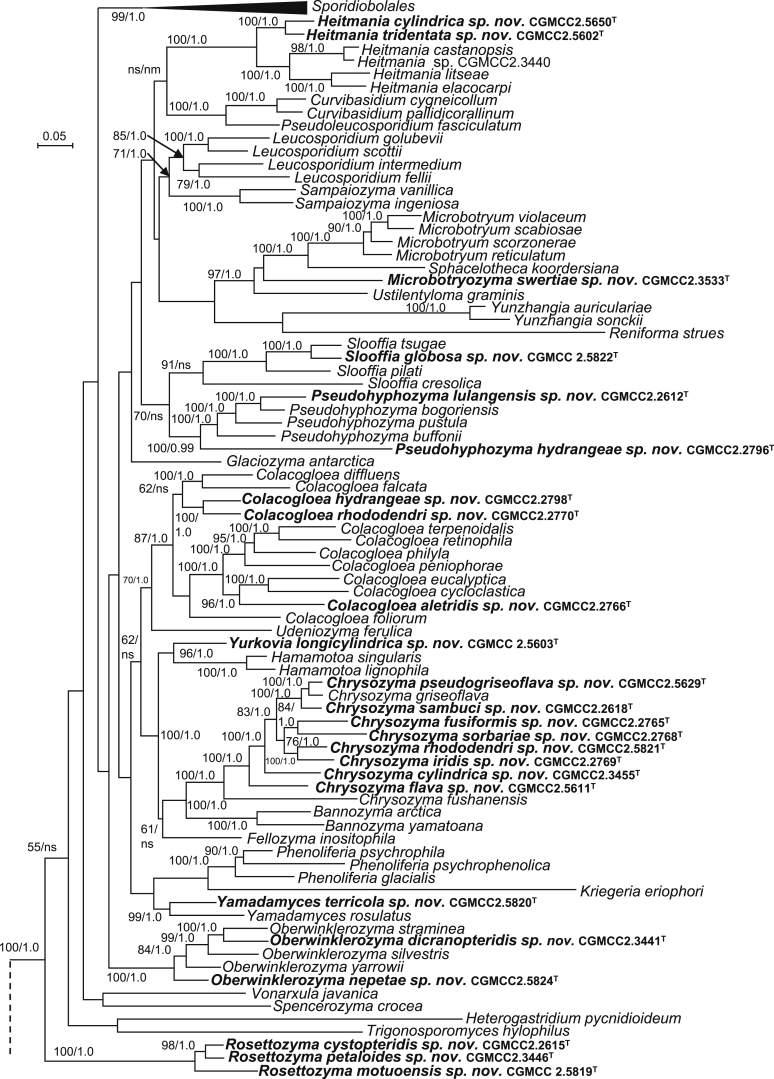
| Heitmania tridentata sp. nov. | CGMCC 2.5602T = GPS20.16B3 = CBS 15549 | September 21, 2015 | Milin county, Tibet, China | phylloplane | MK050420 | MK849217 | MK849356 | MK849083 | MK848951 |
| H. cylindrica sp. nov. | CGMCC 2.5650T = GPS20.2C8 = CBS 15568 | September 20, 2015 | Milin county, Tibet, China | phylloplane | MK050421 | MK849237 | MK849376 | MK849101 | MK848972 |
| Heitmania sp. | CGMCC 2.3440 = SM35.2A | November 10, 2006 | Simao county, Yunnan province, China | phylloplane | MK050422 | MK849161 | MK849299 | MK849031 | MK848900 |
| Heitmania sp. | CGMCC 2.3624 = SM35.2B | November 10, 2006 | Simao county, Yunnan province, China | phylloplane | MK050423 | MK849189 | MK849327 | MK849057 | MK848925 |
| Microbotryozyma swertiae sp. nov. | CGMCC 2.3533T = ZXS7.7 = CBS 15495 | May 10, 2007 | Chuxiong county, Yunnan province, China | leaf of Swertia yunnanensis | MK050424 | MK849180 | MK849318 | MK849049 | MK848918 |
| Yamadamyces terricola sp. nov. | CGMCC 2.5820T = 03-1 = CBS 15572 | August 15, 2015 | Daxinganling, China | soil | MK050425 | MK849127 | MK849268 | MK848999 | MK848874 |
| Oberwinklerozyma dicranopteridis sp. nov. | CGMCC 2.3441T = SM10.2 = CBS 15476 | November 6, 2006 | Simao county, Yunnan province, China | leaf of Dicranopteris dichotoma | MK050426 | MK849162 | MK849300 | – | MK848901 |
| O. nepetae sp. nov. | CGMCC 2.5824T = XZ129C7 = CBS 15579 | September 20, 2014 | Motuo, Tibet, China | leaf of Nepeta sp. | MK050427 | MK849254 | MK849391 | – | MK848992 |
| Chrysozyma pseudogriseoflava sp. nov. | CGMCC 2.5629T = GPS21.6B3 = CBS 15564 | September 21, 2015 | Milin county, Tibet, China | phylloplane | MK050428 | MK849232 | MK849372 | MK849098 | MK848967 |
| CGMCC 2.5646 = GPS22.3A2 | September 22, 2015 | Lulang county, Tibet, China | phylloplane | MK050430 | MK849234 | MK849373 | – | MK848970 | |
| GPS20.6D2 | September 21, 2015 | Milin county, Tibet, China | phylloplane | MK050429 | – | – | – | – | |
| C. sambuci sp. nov. | CGMCC 2.2618T = XZ13C5 = CBS 15450 | September 21, 2004 | Bomi county, Tibet, China | leaf of Sambucus williamsii | MK050431 | MK849133 | MK849273 | MK849004 | – |
| CGMCC 2.2755 = XZ13B7 | September 21, 2004 | Bomi county, Tibet, China | leaf of Sambucus williamsii | MK050432 | MK849137 | MK849277 | – | – | |
| C. rhododendri sp. nov. | CGMCC 2.5821T = XZ160D3 = CBS 15583 | September 21, 2014 | Tibet, China | leaf of Rhododendron sp. | MK050433 | MK849263 | MK849400 | MK849121 | MK848995 |
| C. iridis sp. nov. | CGMCC 2.2769T = XZ8B3 = CBS 15461 | September 21, 2004 | Bomi county, Tibet, China | leaf of Iris forrestii | MK050434 | MK849144 | MK849285 | MK849013 | MK848886 |
| C. sorbariae sp. nov. | CGMCC 2.2768T = XZ9D1 = CBS 15460 | September 21, 2004 | Bomi county, Tibet, China | leaf of Sorbaria arborea | MK050435 | MK849143 | MK849284 | MK849012 | MK848885 |
| CGMCC 2.2767 = XZ11B4 | September 21, 2004 | Bomi county, Tibet, China | leaf of Acer caudatum | MK050436 | MK849142 | MK849283 | – | MK848884 | |
| C. fusiformis sp. nov. | CGMCC 2.2765T = XZ33C2 = CBS 15458 | September 21, 2004 | Lulang county, Tibet, China | phylloplane | MK050437 | MK849140 | MK849281 | MK849010 | MK848883 |
| CGMCC 2.2764 = XZ33Z1 | September 21, 2004 | Lulang county, Tibet, China | phylloplane | MK050438 | – | – | – | – | |
| C. cylindrica sp. nov. | CGMCC 2.3455T = WZS29.2 = CBS 15482 | November 6, 2006 | Wuzhishan mountain, Hainan province, China | phylloplane | MK050439 | MK849168 | MK849306 | MK849036 | MK848906 |
| C. flava sp. nov. | CGMCC 2.5611T = GPS20.4A1 = CBS 15552 | September 21, 2015 | Milin county, Tibet, China | phylloplane | MK050440 | MK849221 | MK849360 | MK849086 | MK848955 |
| Yurkovia longicylindrica sp. nov. | CGMCC 2.5603T = GPS20.2C3 = CBS 15550 | September 21, 2015 | Milin county, Tibet, China | phylloplane | MK050441 | MK849218 | MK849357 | MK849084 | MK848952 |
| Pseudohyphozyma lulangensis sp. nov. | CGMCC 2.2612T = XZ50B2 = CBS 15446 | September 21, 2004 | Lulang county, Tibet, China | phylloplane | MK050442 | MK849129 | MK849270 | – | MK848875 |
| P. hydrangeae sp. nov. | CGMCC 2.2796T = XZ46A1 = CBS 15462 | September 21, 2004 | Lulang county, Tibet, China | leaf of Hydrangea heteromalla | MK050443 | MK849126 | MK849287 | MK849015 | MK848888 |
| CGMCC 2.2797 = XZ46C5 | September 21, 2004 | Lulang county, Tibet, China | leaf of Hydrangea heteromalla | MK050444 | MK849146 | MK849288 | MK849016 | – | |
| CGMCC 2.5607 = GPS20.2D2 | September 21, 2015 | Milin county, Tibet, China | phylloplane | MK050445 | MK849219 | MK849358 | – | MK848953 | |
| CGMCC 2.5618 = GPS23.3C2 | September 22, 2015 | Lulang county, Tibet, China | phylloplane | MK050446 | MK849224 | MK849363 | – | – | |
| CGMCC 2.5623 = GPS23.3D3 | September 22, 2015 | Lulang county, Tibet, China | phylloplane | MK050447 | – | MK849367 | MK849092 | MK848961 | |
| GPS23.3D2 | September 22, 2015 | Lulang county, Tibet, China | phylloplane | MK050448 | – | – | – | – | |
| Slooffia globosa sp. nov. | CGMCC 2.5822T = 4-6 = CBS 15573 | August 15, 2015 | Daxinganling, China | soil | MK050449 | MK849255 | MK849392 | MK849116 | MK848989 |
| Colacogloea aletridis sp. nov. | CGMCC 2.2766T = XZ31A1 = CBS 15459 | April 4, 2005 | Bomi county, Tibet, China | leaf of Aletris pauciflora | MK050450 | MK849141 | MK849282 | MK849011 | – |
| C. hydrangeae sp. nov. | CGMCC 2.2798T = XZ46B3 = CBS 15463 | April 11, 2005 | Lulang county, Tibet, China | leaf of Hydrangea heteromalla | MK050451 | MK849147 | – | MK849017 | MK848889 |
| C. rhododendri sp. nov. | CGMCC 2.2770T = XZ10F1 = CBS 15652 | April 4, 2005 | Bomi county, Tibet, China | leaf of Rhododendron lulangense | MK050452 | MK849145 | MK849286 | MK849014 | MK848887 |
| CGMCC 2.5651 = GPS20.5C1 | September 21, 2015 | Milin county, Tibet, China | phylloplane | MK050457 | MK849238 | – | MK849102 | MK848973 | |
| CGMCC 2.5652 = GPS20.5D6 | September 21, 2015 | Milin county, Tibet, China | phylloplane | MK050456 | MK849239 | – | MK849103 | MK848974 | |
| GPS20.5C5 | September 21, 2015 | Milin county, Tibet, China | phylloplane | MK050455 | – | – | – | – | |
| GPS20.5C3 | September 21, 2015 | Milin county, Tibet, China | phylloplane | MK050453 | – | – | – | – | |
| GPS20.5D1 | September 21, 2015 | Milin county, Tibet, China | phylloplane | MK050454 | – | – | – | – |
DNA extraction and ribosomal DNA sequencing
Nuclear DNA was extracted using the method described previously by Wang & Bai (2008). The ITS (including 5.8S rDNA) region and LSU rDNA D1/D2 domains were sequenced using the methods described previously (Wang & Bai 2004). The small subunit (SSU) rDNA sequences were determined according to Wang et al. (2003). The CYTB sequences were performed as described by Wang & Bai (2008). The three nuclear protein-coding genes, RPB1, RPB2 and TEF1, were obtained using methods described previously (Wang et al. 2014). GenBank accession numbers for all sequences determined in this study are listed in Table 1.
Sequences were aligned with the MAFFT program (Standley 2013) using the G-INS-i algorithm and minor gaps in all alignments were manually deleted. The most appropriate model of DNA substitution was searched with Modeltest version 3.04 (Posada & Crandall 1998) using the Akaike information criterion (AIC). The model GTR + I + G was selected for Maximum likelihood (ML) and Bayesian inference (BI) analyses. ML analysis was conducted using RAxML-HPC 7.2.8 (Stamatakis 2006) with 1 000 bootstrap replicates. BI analysis was conducted using MrBayes 3.1.2 (Ronquist et al. 2012) with 10 000 000 generations using the parameter settings described previously (Wang et al. 2015a). A bootstrap percentage (BP) of ≥70 % or a Bayesian posterior probability (PP) of ≥0.9 was considered as significantly supported in all constructed trees in this study. The alignments and trees were deposited in TreeBASE (www.treebase.org, Nos. 24640–24646).
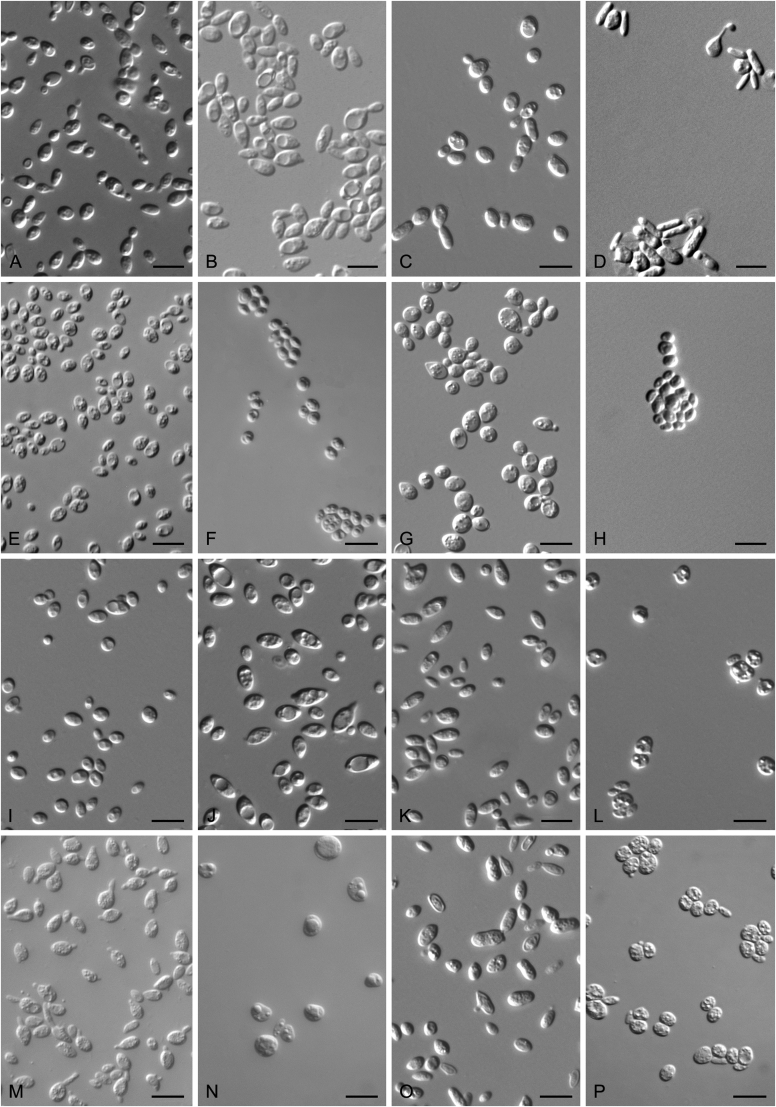
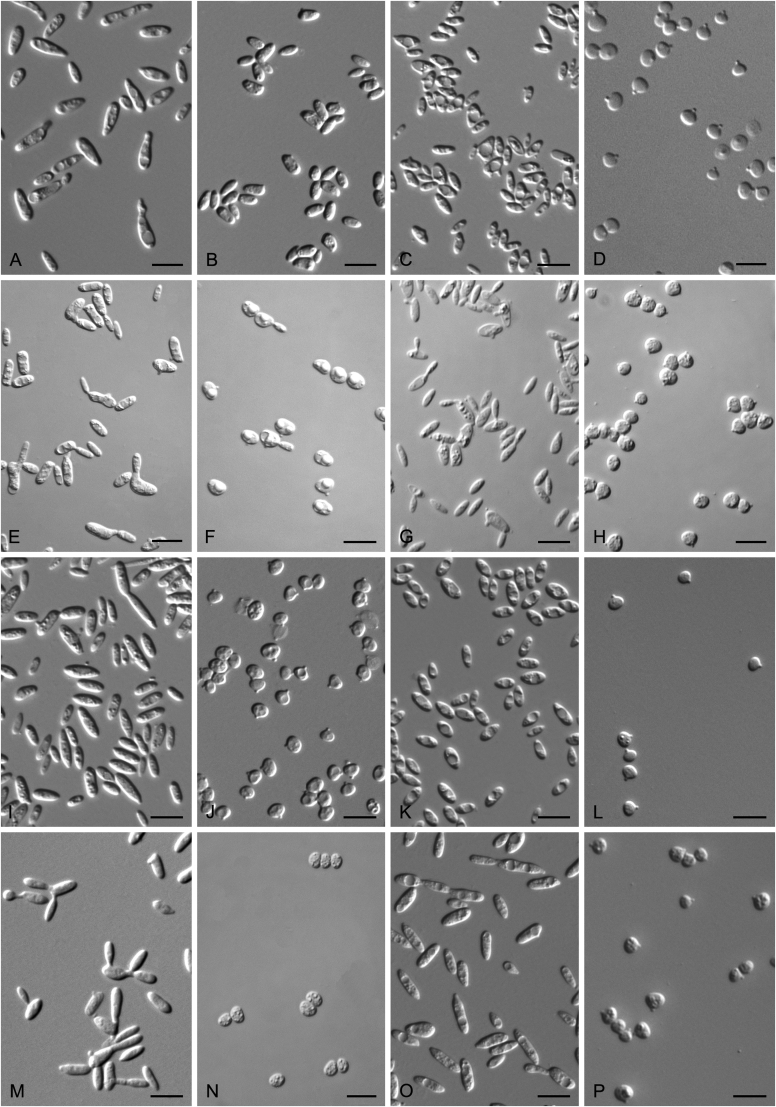
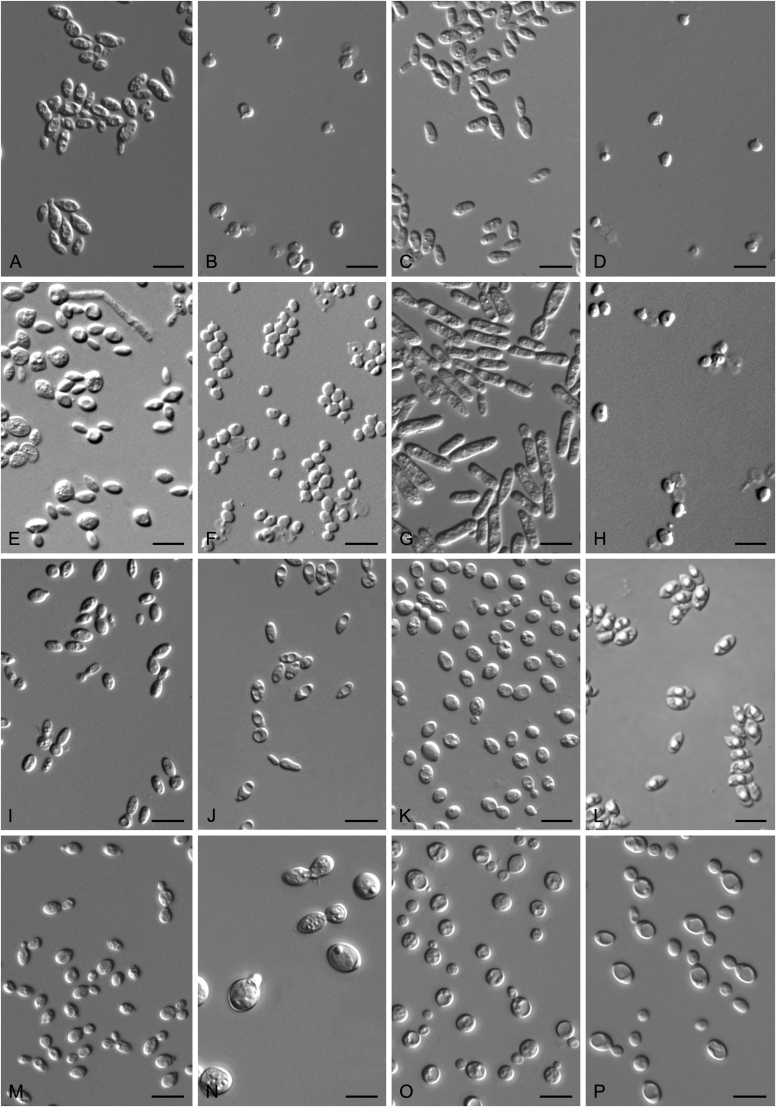
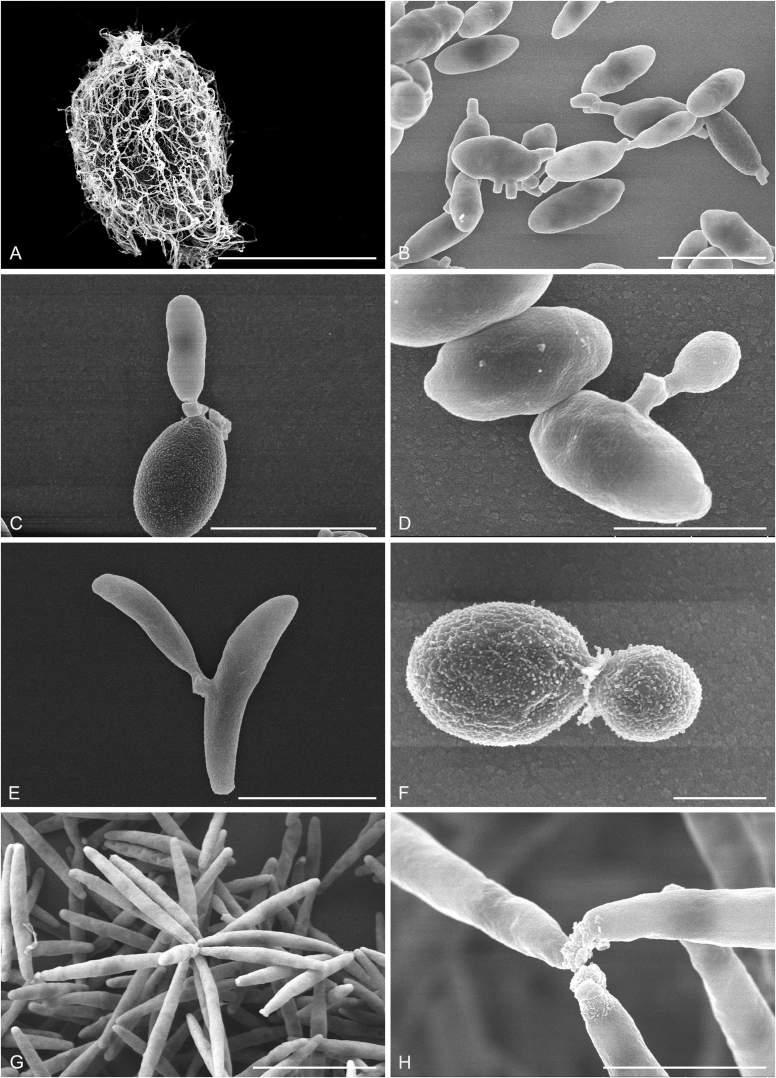
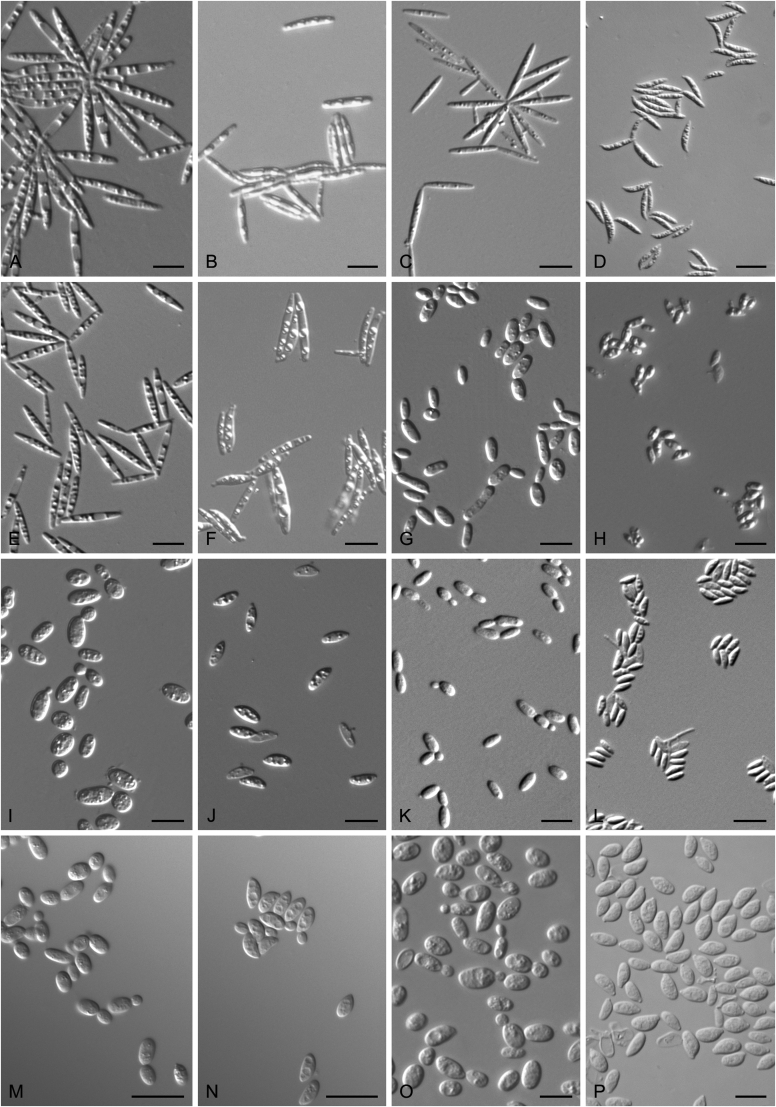
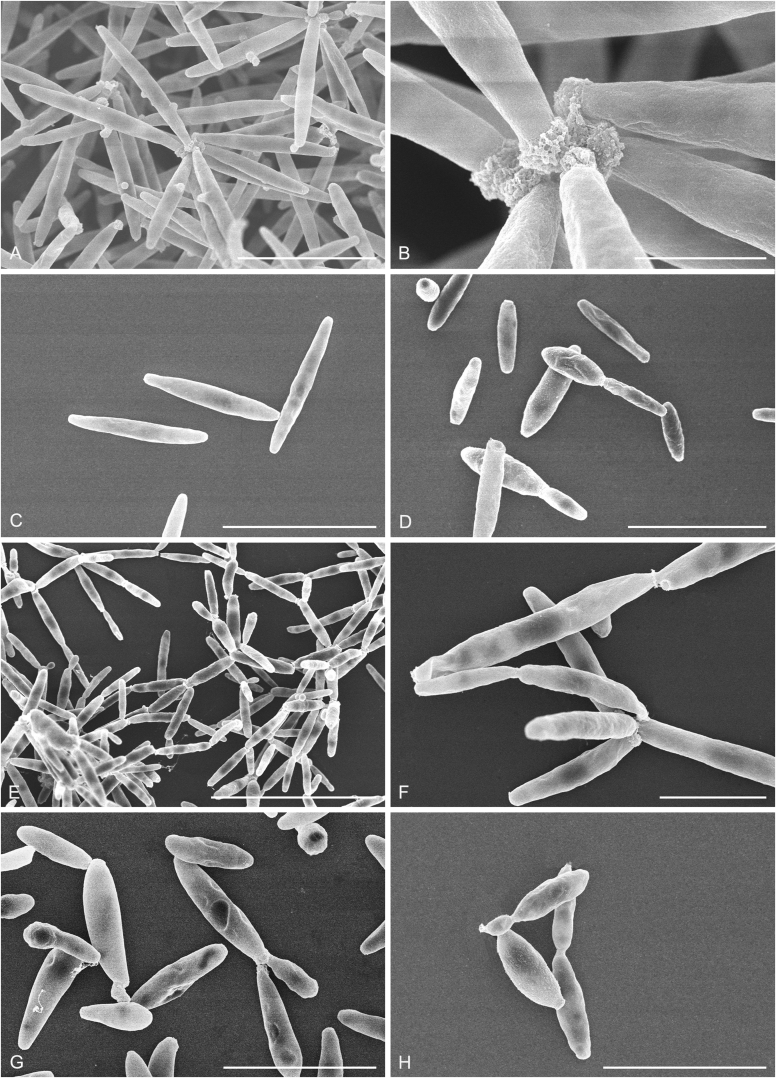
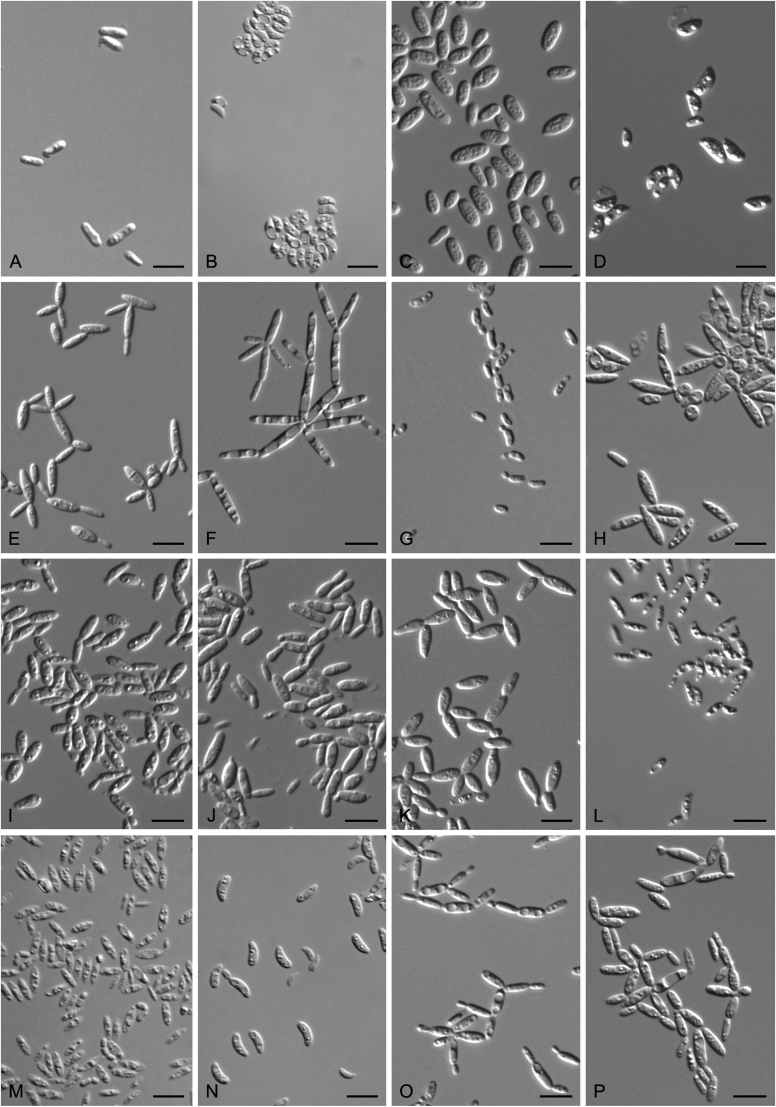
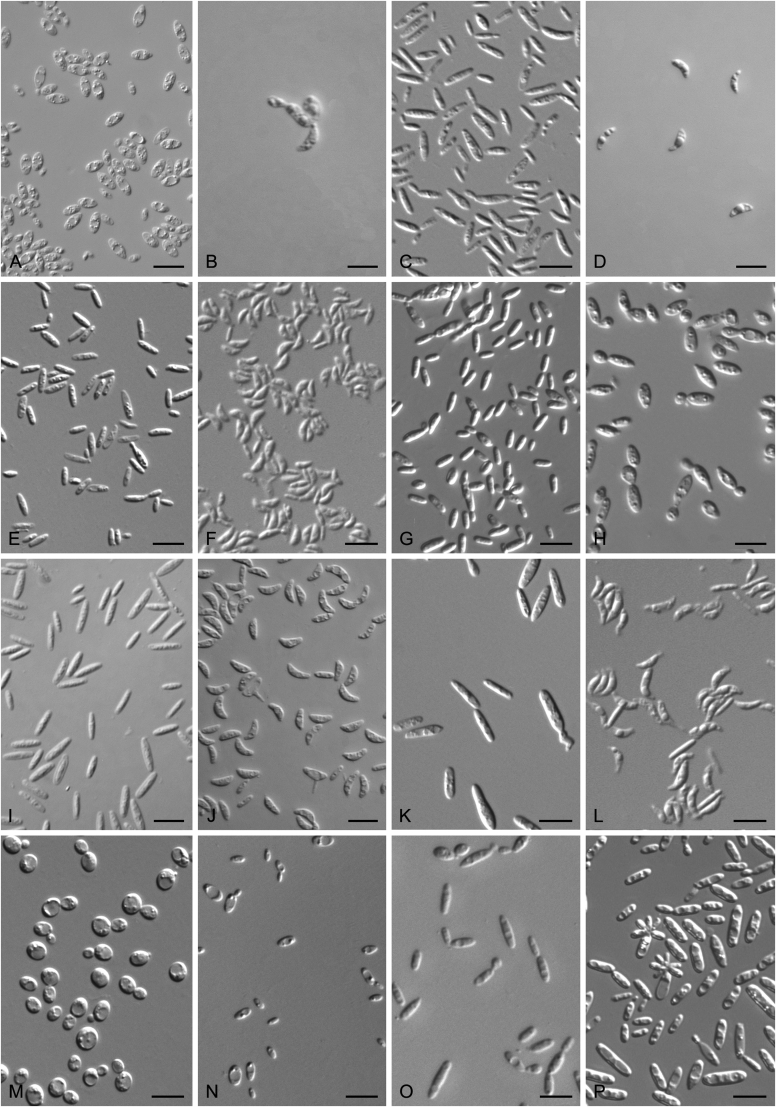
New species catalogised
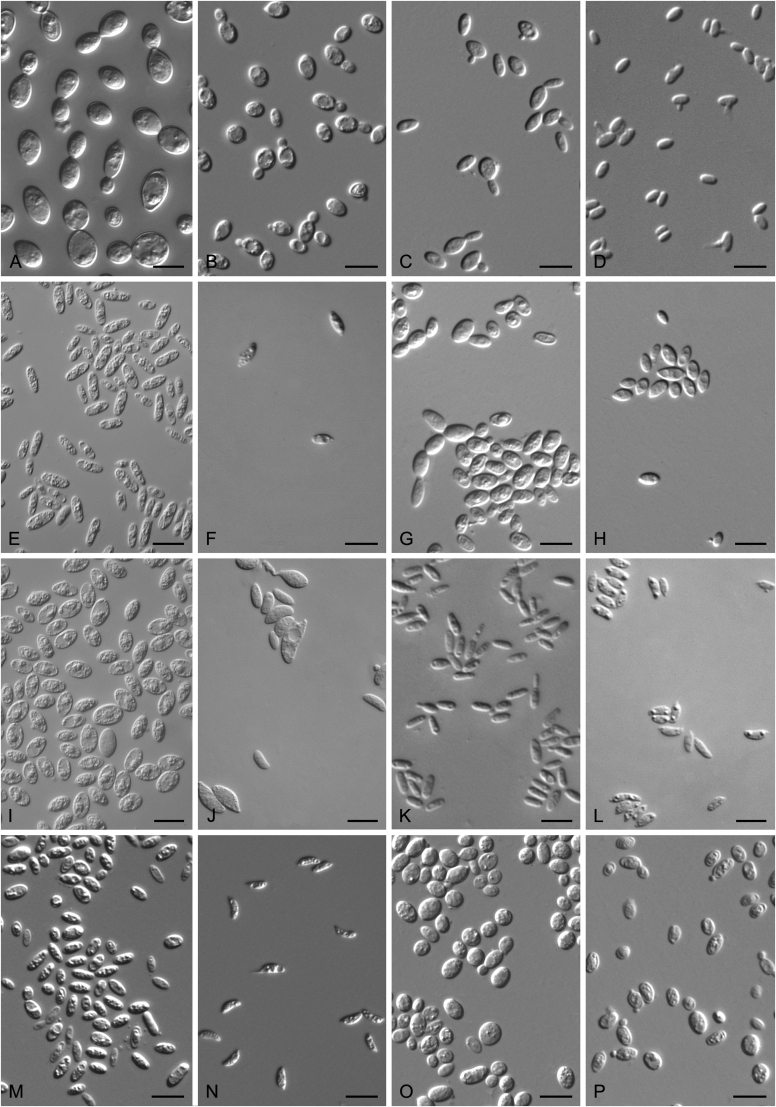
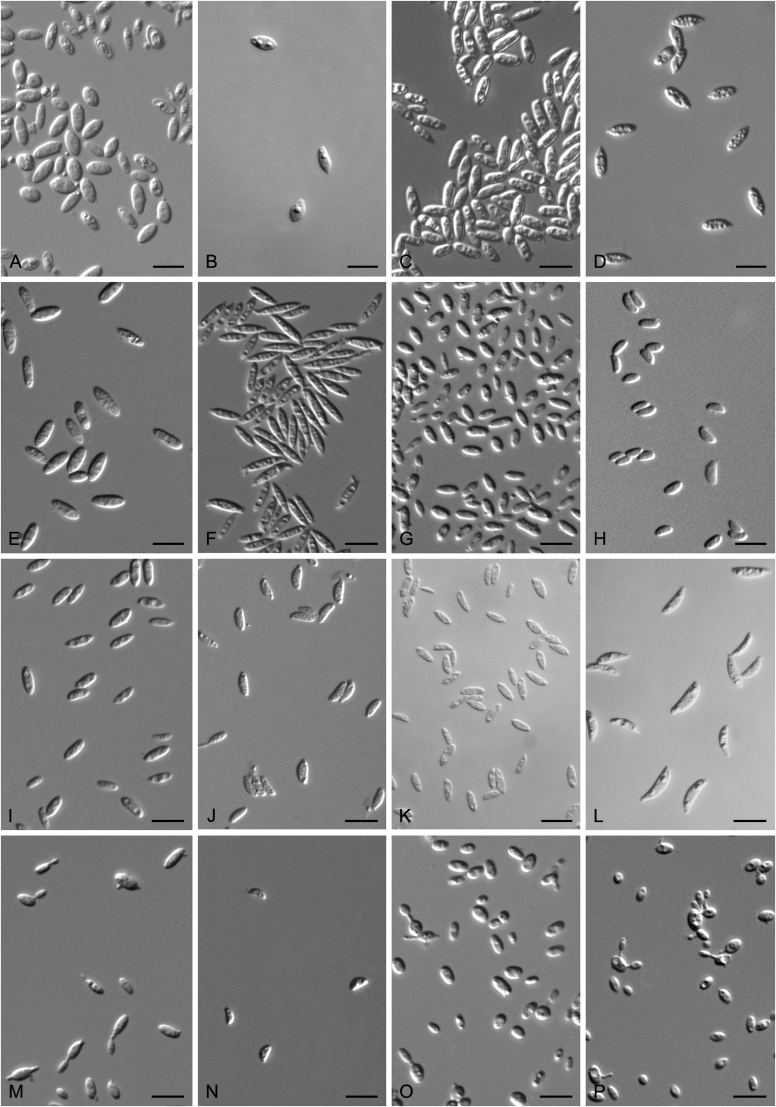
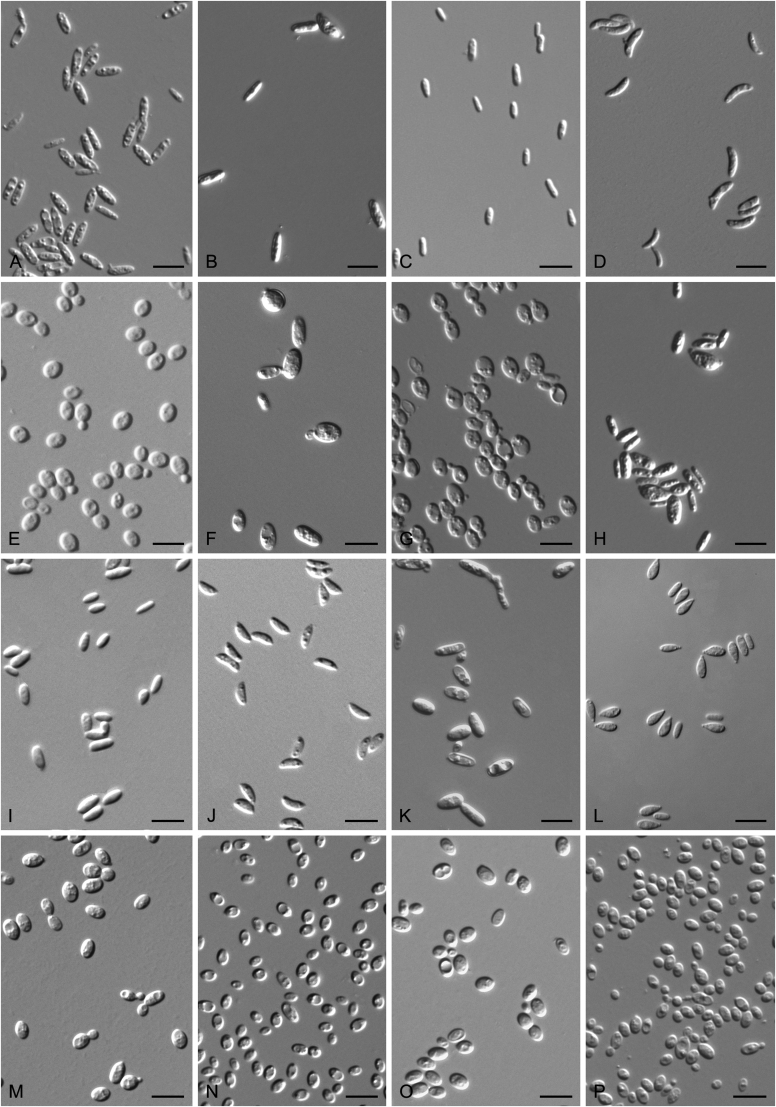
Accurate identification of known yeast species and rapid detection of new species are currently possible because of the availability of ITS and D1/D2 sequence databases for most of the known yeasts (Kurtzman and Robnett, 1998, Fell et al., 2000, Scorzetti et al., 2002, Boekhout et al., 2011, Kurtzman, 2011, Liu et al., 2015a, Wang et al., 2015a, Vu et al., 2016). Recently The Yeasts Trust announced, a new yeasts database (Boekhout et al. 2016, http://theyeasts.org/) which provides the most up-to-date and accurate taxonomic information including DNA sequences and phenotypic characteristics on all published yeasts. Vu et al. (2016) recommended that the similarity thresholds to discriminate a yeast species were 1.59 % (or 0.79 % using ex-type strains only) and 0.49 % for ITS and D1/D2, respectively, based on the barcode data of ca. 9 000 yeast strains, which are in agreement with previous studies (Kurtzman and Robnett, 1998, Fell et al., 2000, Scorzetti et al., 2002) that indicated sequence diversity among conspecific strains is less than 1 % in either the ITS or D1/D2 regions (Kurtzman and Fell, 2006, Kurtzman, 2014, Kurtzman, 2015, Kurtzman et al., 2015). However, delineation of species using single region sequence is not always reliable for yeasts, especially for basidiomycetous yeasts, because different lineages may vary in their rates of nucleotide substitution for the diagnostic gene being used (Fell et al., 2000, Scorzetti et al., 2002). Thus, a combined sequence analysis of the D1/D2 domains and ITS region is recommended for species identification by Scorzetti et al. (2002) and Kurtzman & Fell (2006). Consequently, sequence analyses of both D1/D2 and ITS were used to differentiate the potentially new species and their closely related species in this study. In order to improve the species delimitation, a case-by-case pairwise similarity approach was also provided here. We compared the sequence similarity and nucleotide variations in the ITS and D1/D2 regions among yeast genera containing more than two species in Agaricomycotina and Pucciniomycotina using the EMBOSS water alignment tool (http://www.ebi.ac.uk/Tools/psa/emboss_water/nucleotide.html; Madeira et al. 2019). The script, namely EMBOSS_water.py, was used to run the local alignment for the calculation of the sequence similarities and nucleotide variation including substitutions and deletions. All comparisons of sequence similarities were done with the type strains of the mentioned species in this study. It must be emphasised that diagnostic phenotypical features, especially physiological properties, were used to distinguish the new species from that previously described.
New generic and higher ranks circumscriptions
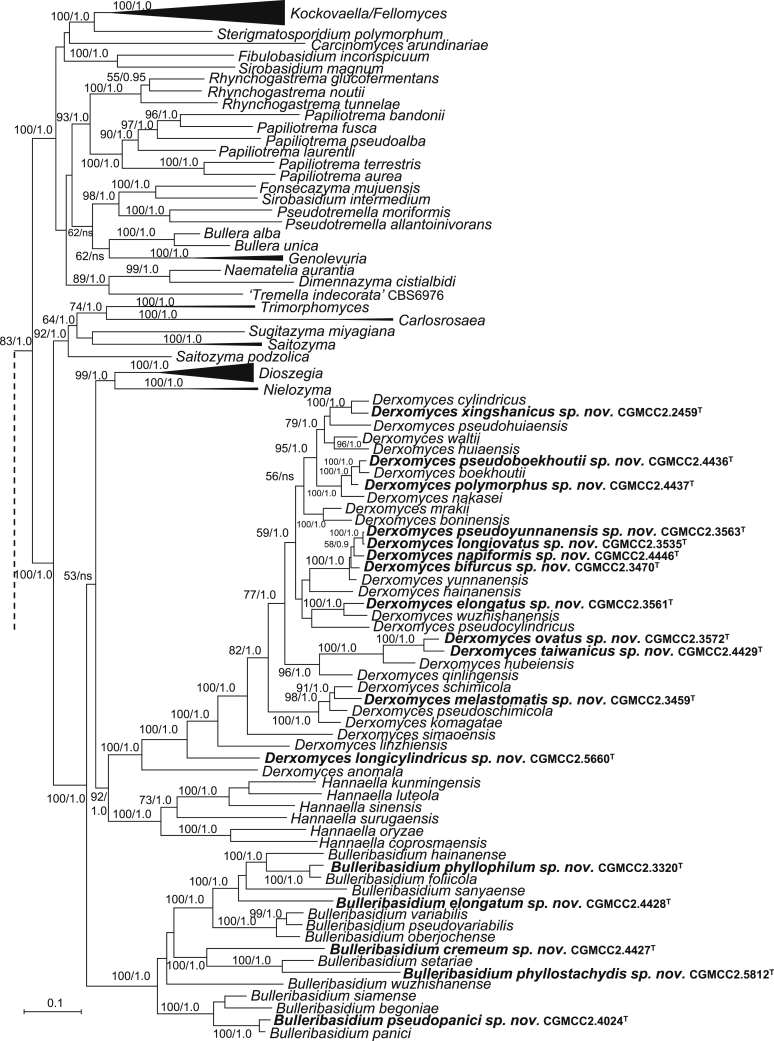
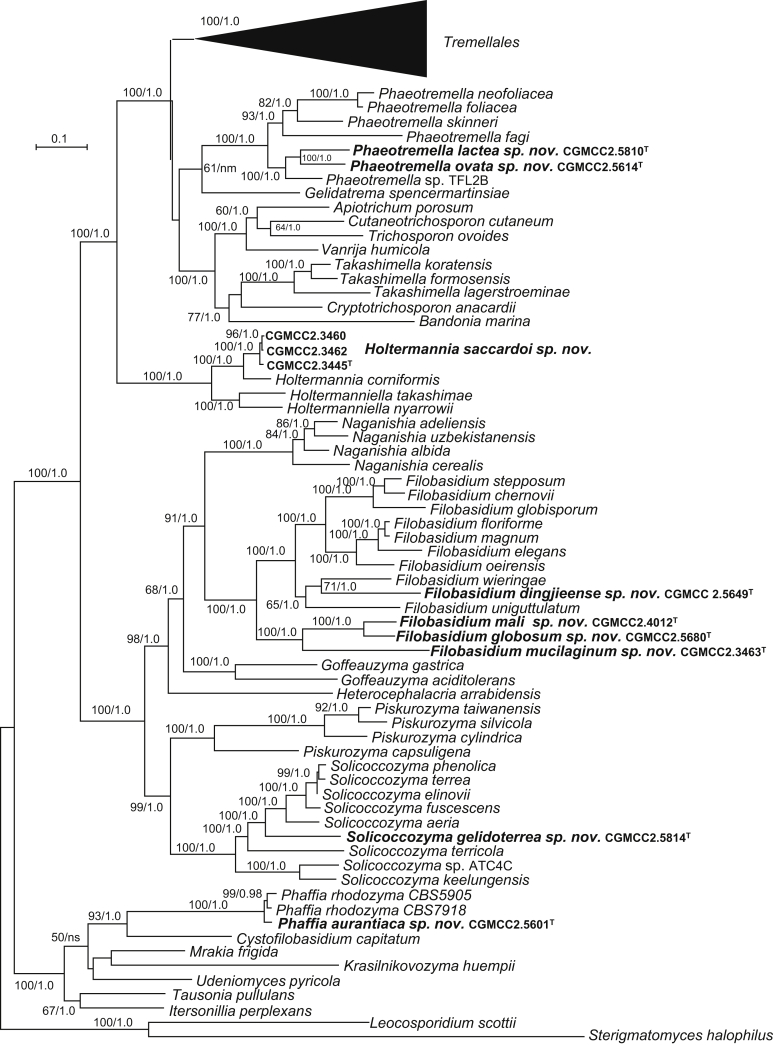
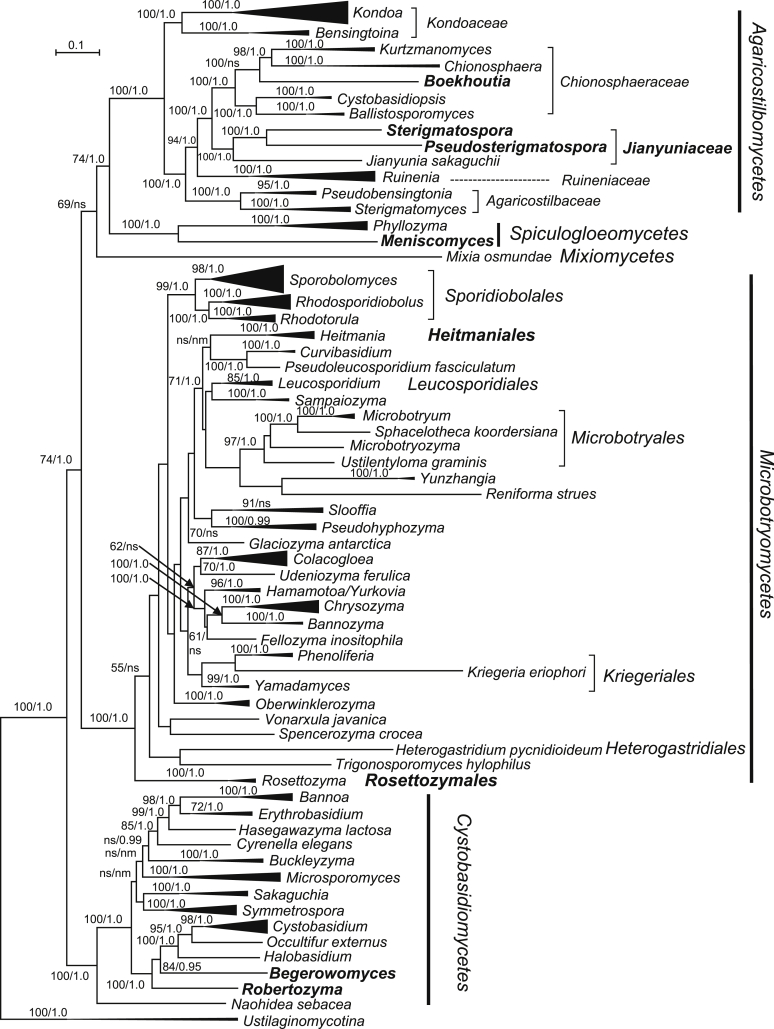
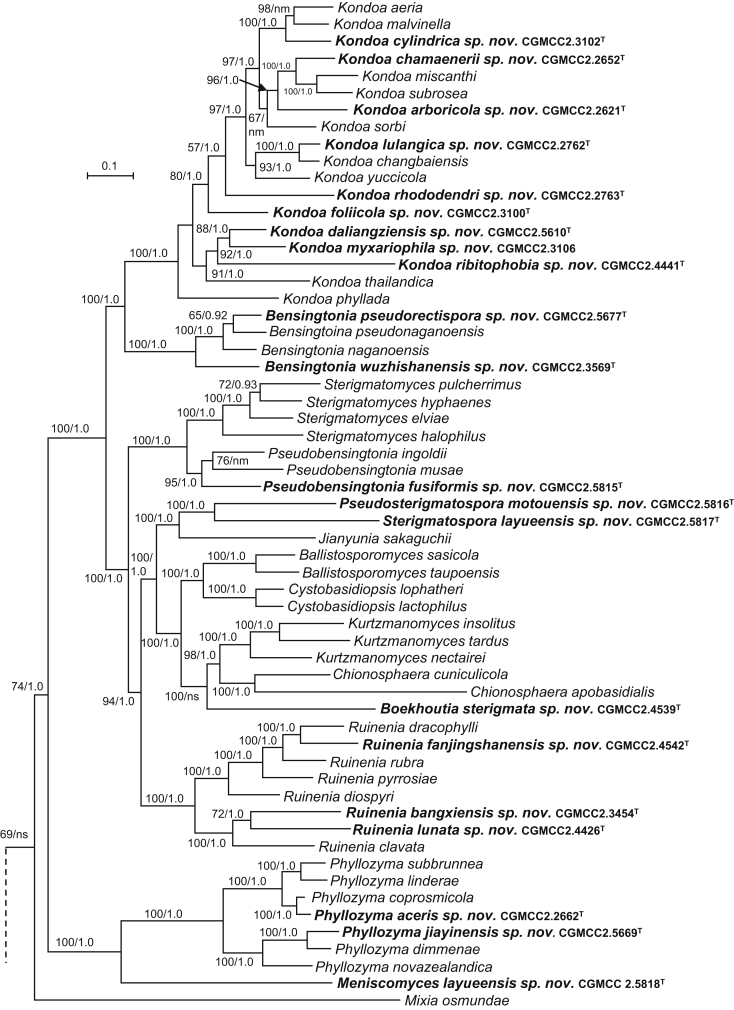
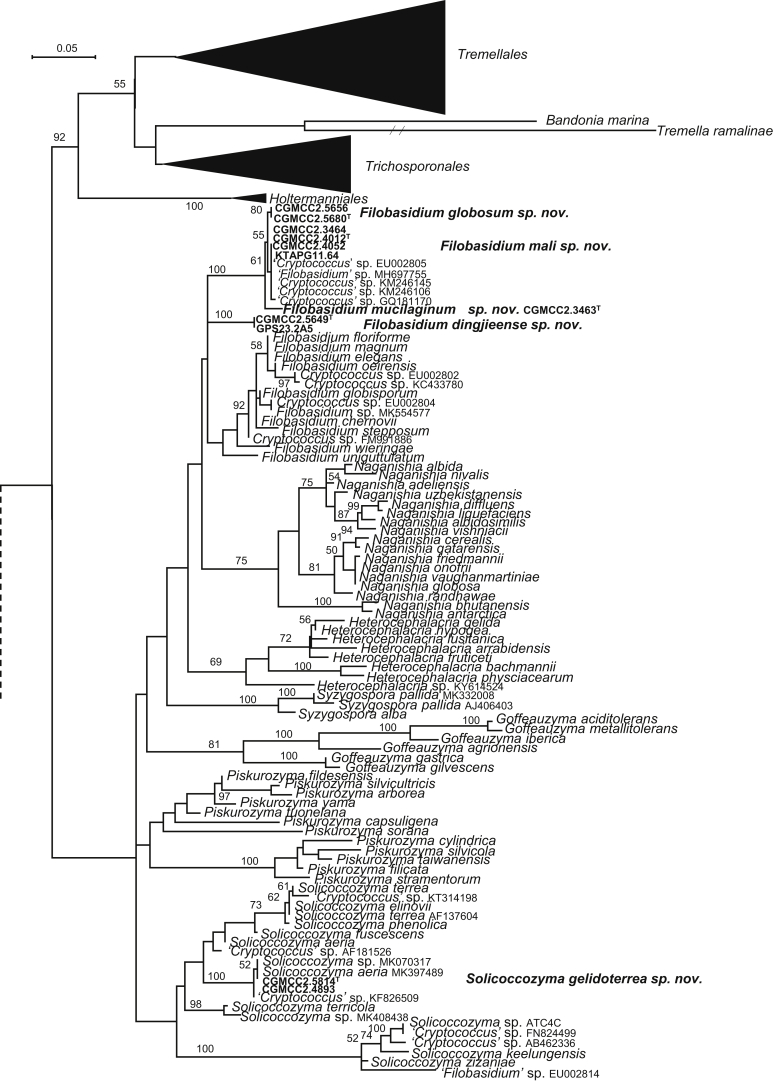
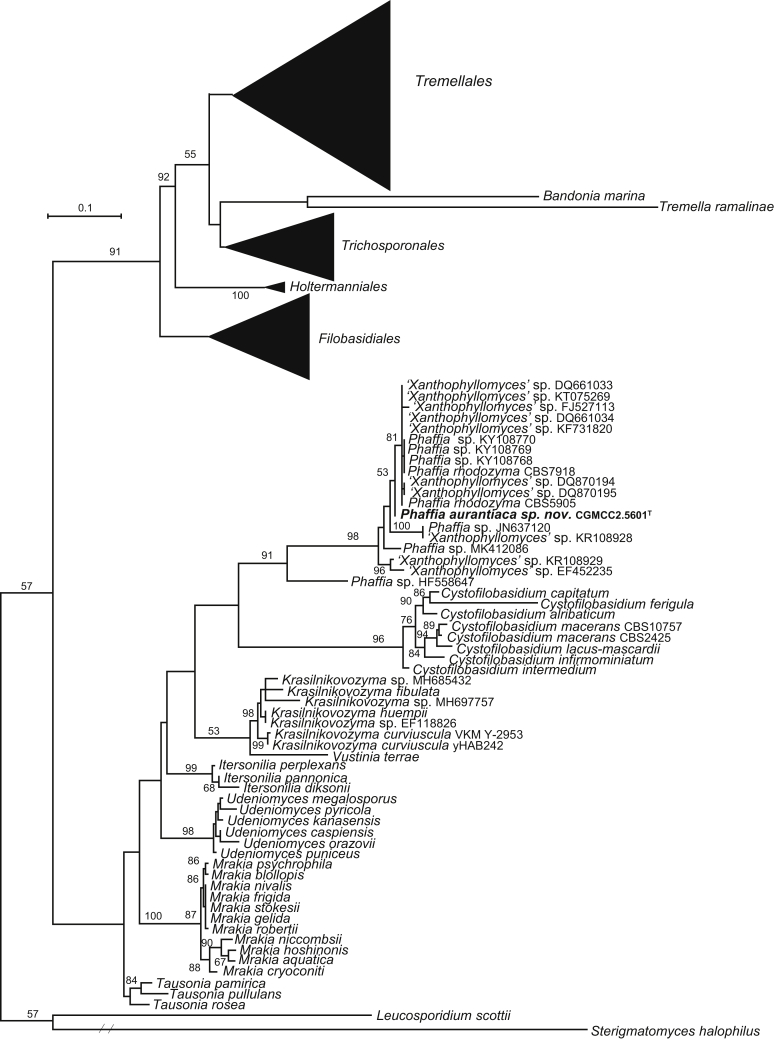
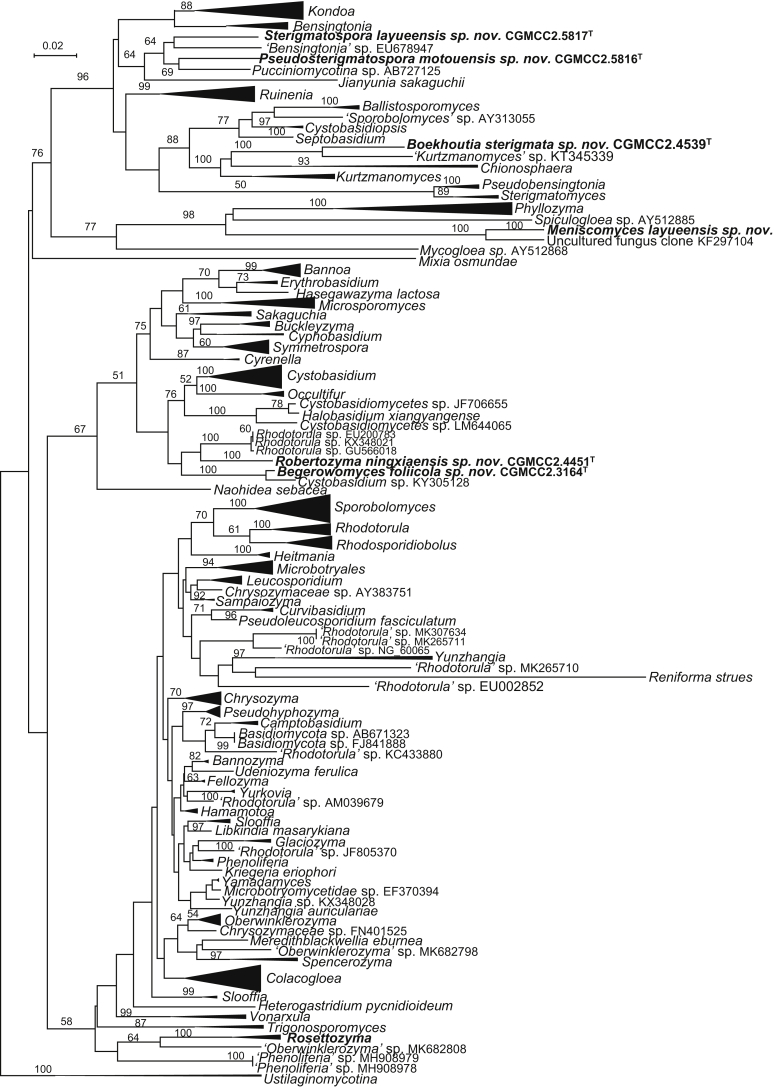
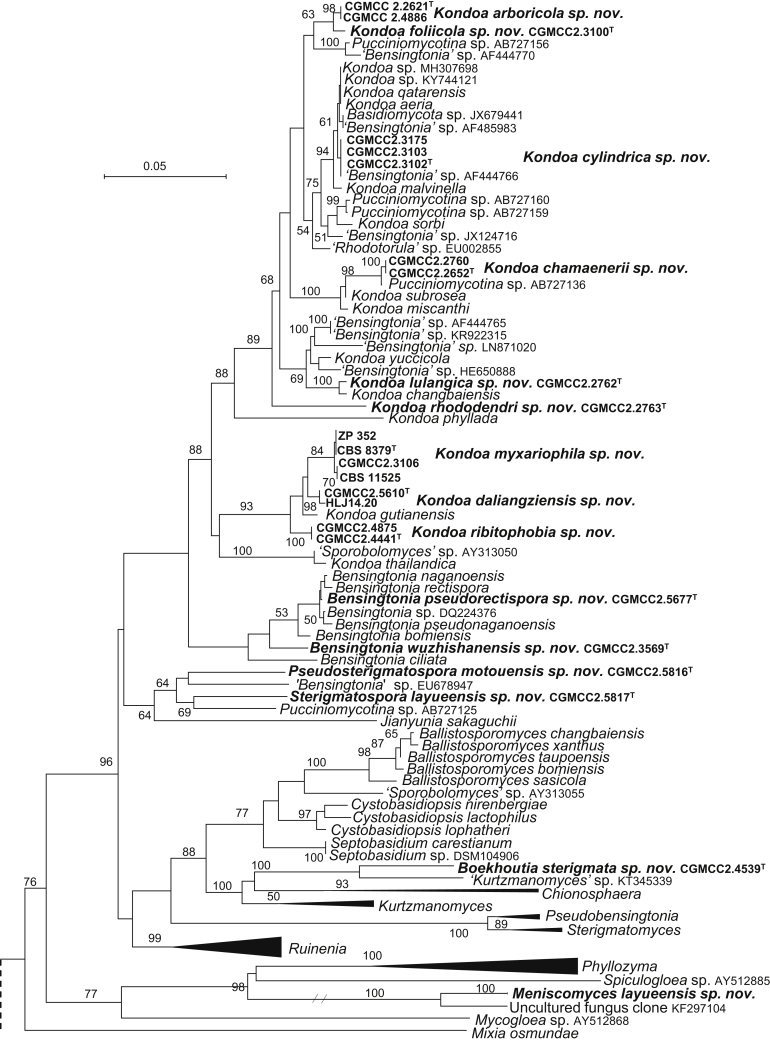
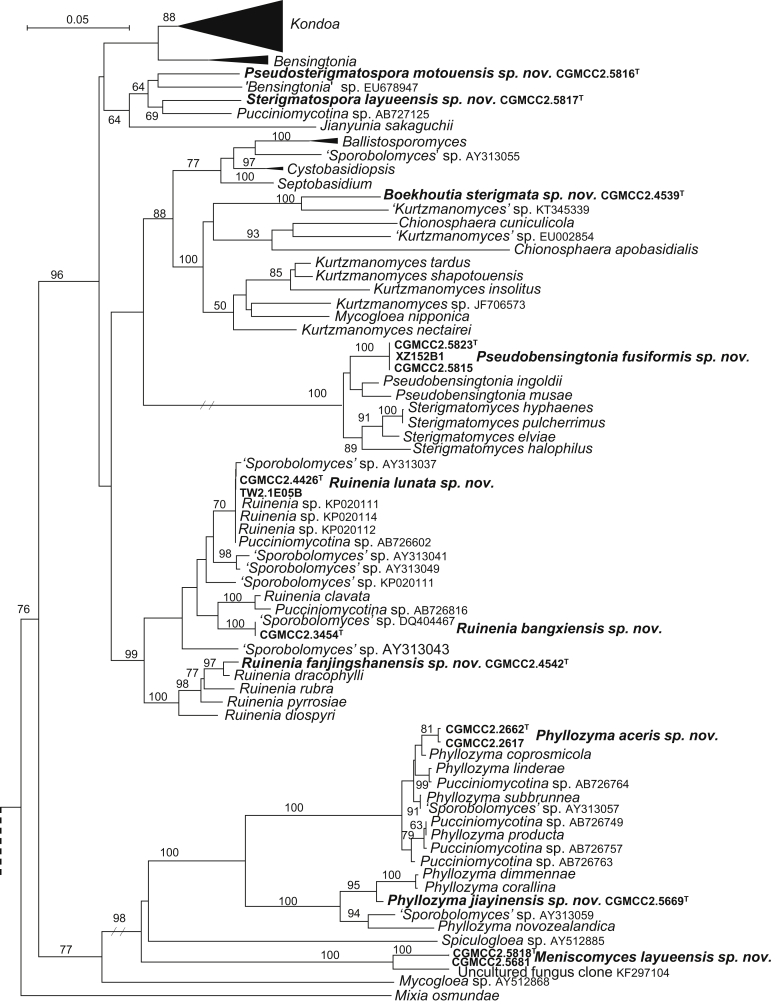
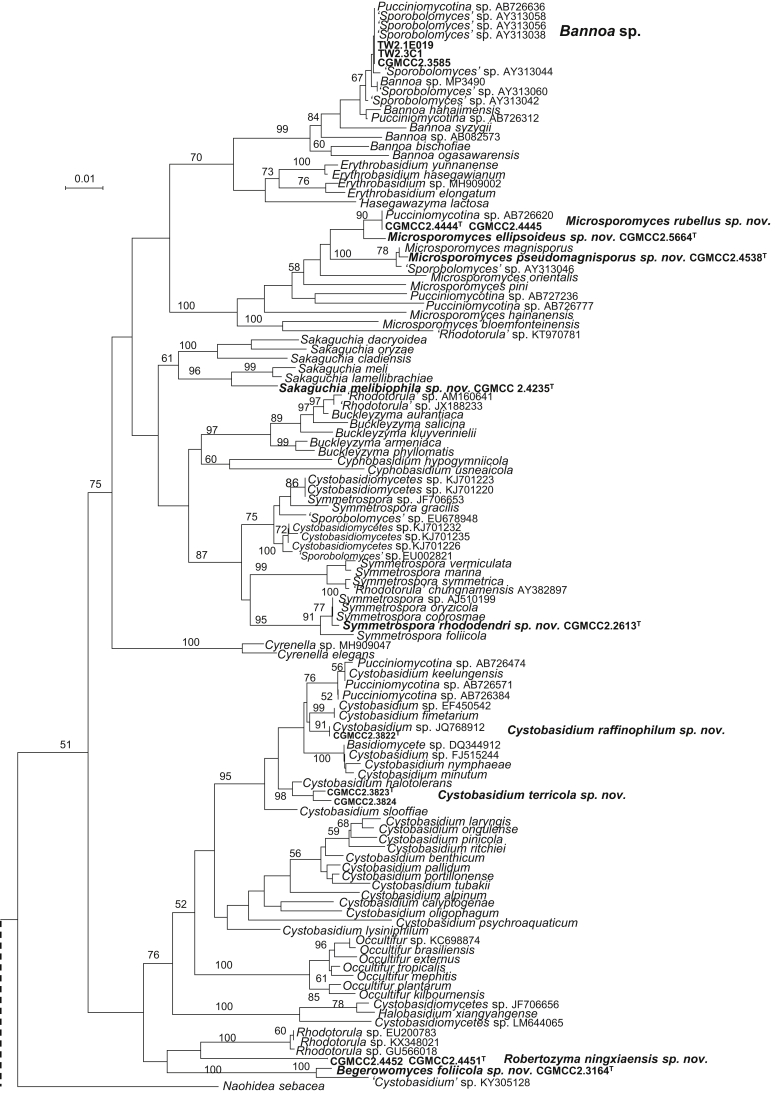
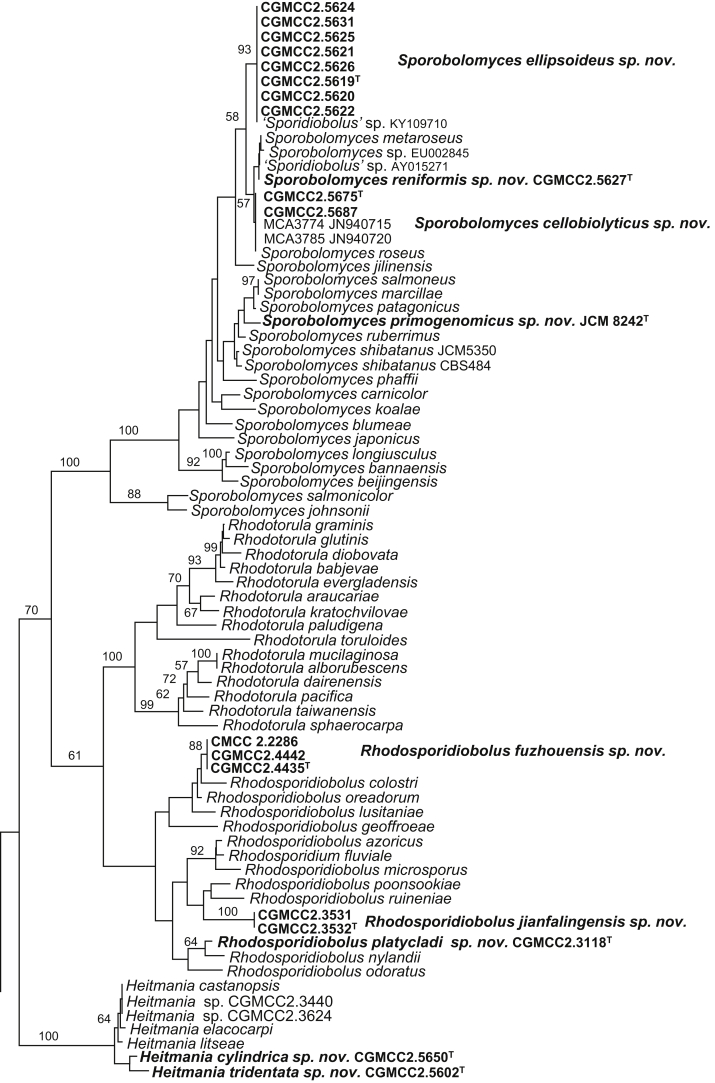
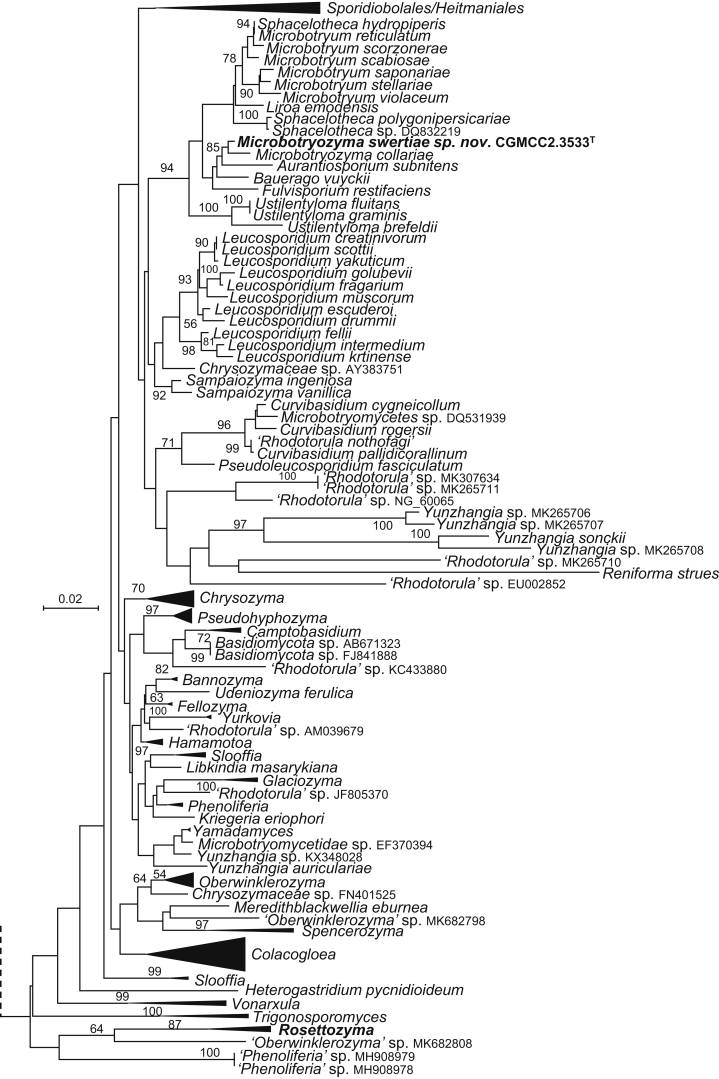
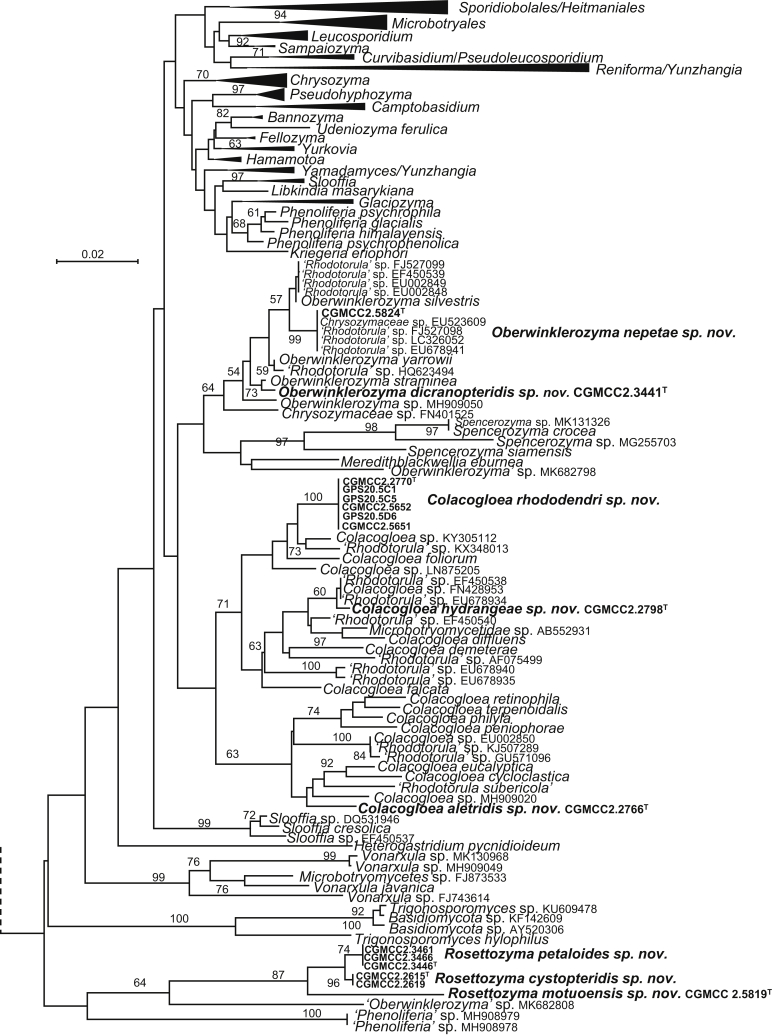
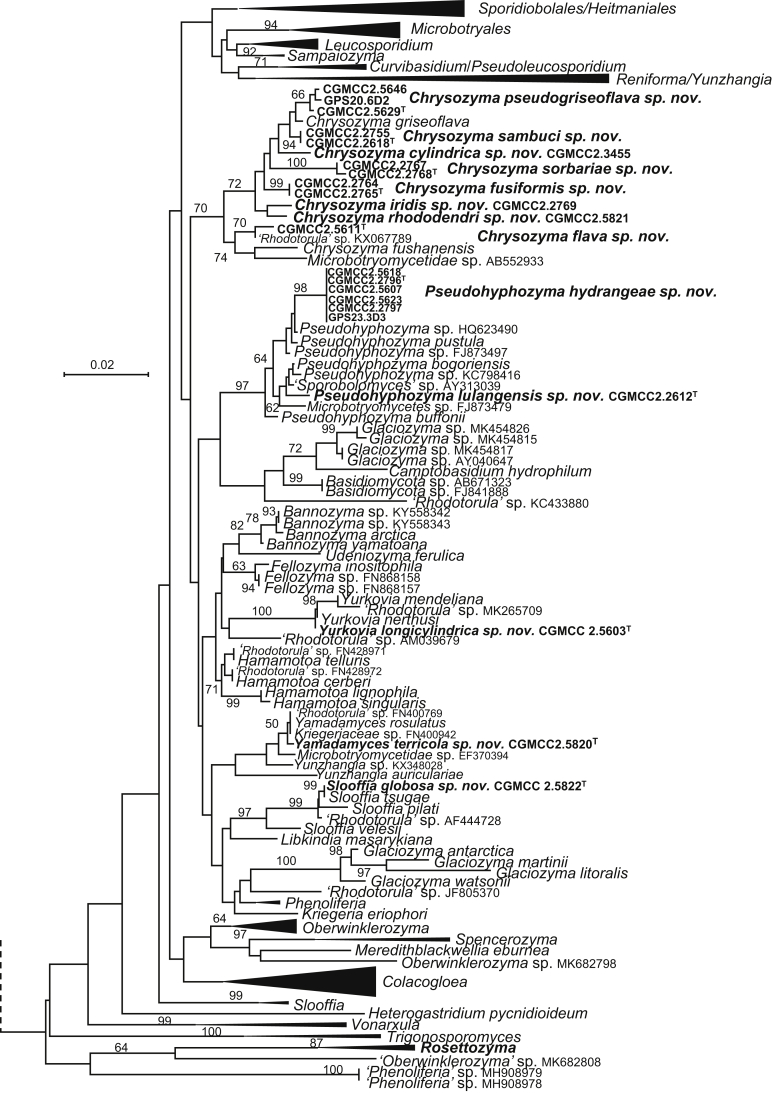
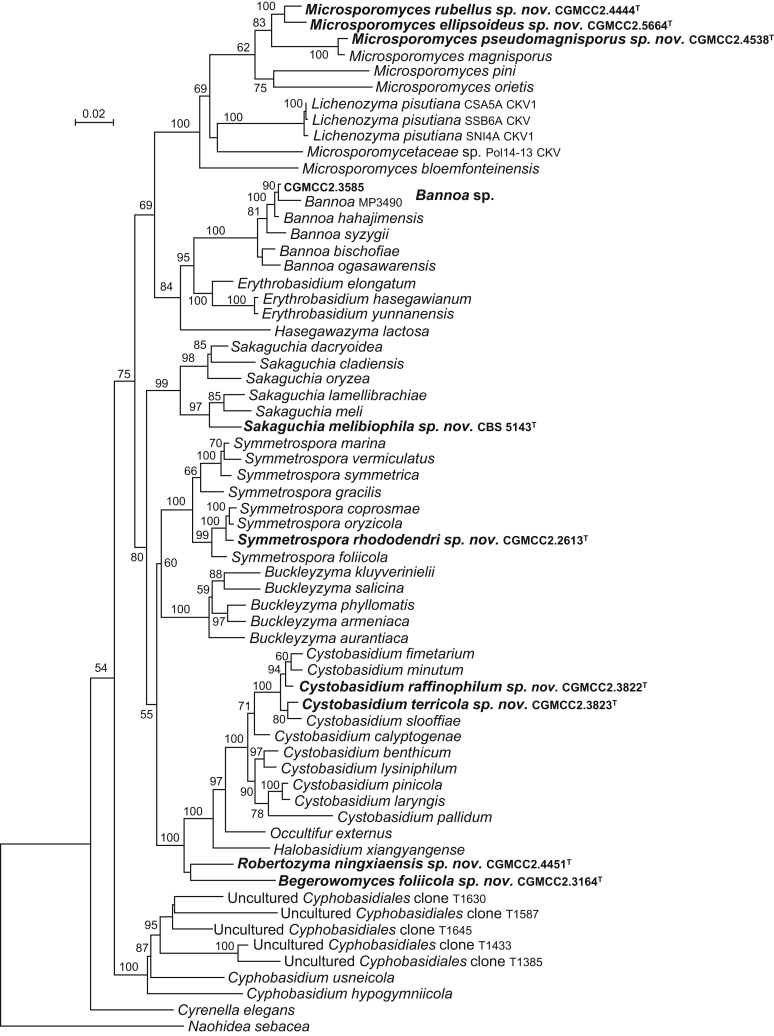
The circumscriptions of genera and higher ranks in the current study were performed mainly based on the multi-locus phylogenetic analyses used in previous studies (Wang et al., 2015a, Wang et al., 2015b, Wang et al., 2015c). The clustering optimisation analysis was done using the OPTSIL software (Göker et al. 2009) to yield non-hierarchical clusterings at generic levels by a given reference threshold, which had been employed in Liu et al. (2015b) and Wang et al. (2015b). The taxonomic thresholds predicted by Vu et al. (2016) to discriminate current yeast genera were 96.31 % for ITS and 97.11 % for D1/D2. Recently, the taxonomic thresholds predicted for filamentous fungal delimitation at the genus, family, order and class levels, recommended by Vu et al. (2019), were 94.3 %, 88.5 %, 81.2 % and 80.9 % for ITS, and 98.2 %, 96.2 %, 94.7 % and 92.7 % for D1/D2. The above taxonomic thresholds were considered, but not followed strictly, for circumscriptions of new genera and higher ranks in this study. Phenotypic differences were also discussed in the new generic circumscriptions.
Results and discussion
Diversity of phylloplane and soils yeasts
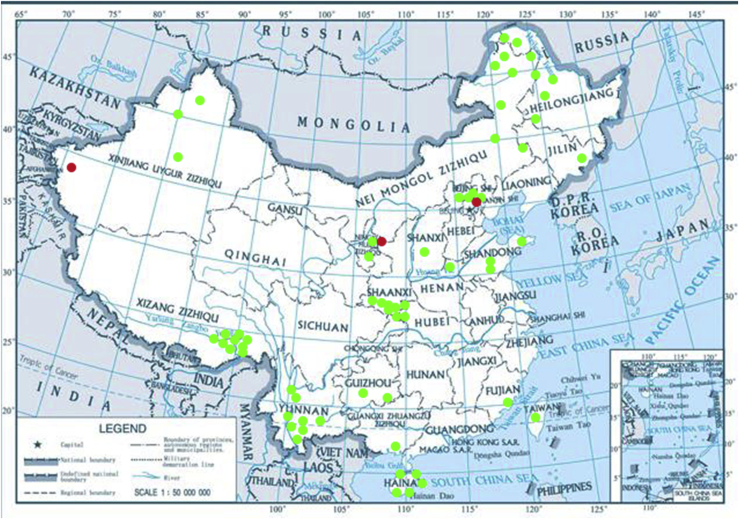
More than 1 000 plant leaves and 20 soil samples have been collected from 67 counties of 20 provinces in China (Table 1, Table 2, Fig. 1) during the past 20 years. About 1 440 strains isolated from those samples have been identified by ITS and D1/D2 sequences. Among them 180 strains belonging to Ustilaginomycotina were not considered in this study. The other 1 260 strains belonging to Agaricomycotina and Pucciniomycotina were distributed in 58 genera, e.i. Ballistosporomyces, Bannoa, Bannozyma, Bensingtonia, Buckleyzyma, Bullera, Bulleribasidium, Chrysozyma, Colacogloea, Cryptococcus, Cryptotrichosporon, Curvibasidium, Cutaneotrichosporon, Cystobasidiopsis, Cystobasidium, Cystofilobasidium, Derxomyces, Dioszegia, Erythrobasidium, Fellozyma, Fibulobasidium, Filobasidium, Genolevuria, Hannaella, Holtermannia, Holtermanniella, Itersonilia, Kockovaella, Kondoa, Kwoniella, Leucosporidium, Microbotryum, Microsporomyces, Mrakia, Naganishia, Naohidea, Oberwinklerozyma, Papiliotrema, Phaeotremella, Phyllozyma, Piskurozyma, Pseudobensingtonia, Pseudohyphozyma, Rhodosporidiobolus, Rhodotorula, Ruinenia, Saitozyma, Slooffia, Solicoccozyma, Sporobolomyces, Symmetrospora, Takashimella, Tausonia, Tremella, Trimorphomyces, Udeniomyces, Vishniacozyma and Yamadamyces, and represent 199 known species (Table 2) as well as 101 undescribed species (Table 1).
Table 2.
List of known yeasts species in China.
| Taxa | Present in number of samples | Resources | Location∗ | |
|---|---|---|---|---|
| Tremellomycetes | ||||
| Tremellales | ||||
| Bulleraceae | ||||
| Bullera alba | 54 | Phylloplane | 1; 3; 5; 13; 14; 15; 17; 19; 20; 21; 25; 26; 27; 29; 33; 34; 38; 39; 40; 41; 42; 43; 49; 50; 54; 66∗ | |
| B. penniseticola | 1 | Phylloplane | 52; | |
| Genolevuria amylolytica | 2 | Phylloplane, 1 | Soil, 1 | 2; 67; |
| G. tibetensis | 7 | Phylloplane, 6 | Soil, 1 | 2; 44; 42; 44; |
| Bulleribasidiacea | ||||
| Bulleribasidium foliicola | 10 | Phylloplane | 8; 12; 10; | |
| B. hainanense | 2 | Phylloplane | 10; 12; | |
| B. oberjochense | 3 | Phylloplane | 18; 44; | |
| B. panici | 3 | Phylloplane | 12; | |
| B. pseudovariabilis | 14 | Phylloplane | 9;12; 24; 25; 32; | |
| B. sanyaense | 3 | Phylloplane | 11; 10; | |
| B. setariae | 3 | Phylloplane | 12; 36; 44; | |
| B. variabilis | 31 | Phylloplane | 12; 25; 36; 44; 54; | |
| B. wuzhishanense | 1 | Phylloplane | 12; | |
| Derxomyces anomalus | 1 | Phylloplane | 40; 41; | |
| D. boekhoutii | 5 | Phylloplane | 4; 12; | |
| D. boninensis | 6 | Phylloplane | 10; 12; 24; 32; | |
| D. cuulongensis | 4 | Phylloplane | 44; | |
| D. cylindricus | 3 | Phylloplane | 44; | |
| D. hainanensis | 4 | Phylloplane | 12; | |
| D. hubeiensis | 4 | Phylloplane | 12; 24; 36; | |
| D. komagatae | 1 | Phylloplane | 25; | |
| D. linzhiensis | 5 | Phylloplane | 41; 44; | |
| D. mrakii | 55 | Phylloplane | 4;10;11;12; 24; 29; 32; 31; 36; 54; 55; | |
| D. nakasei | 10 | Phylloplane | 12; 24; 32; | |
| D. pseudocylindrica | 4 | Phylloplane | 12; | |
| D. pseudohuiaensis | 8 | Phylloplane | 24; 28; 31; | |
| D. pseudoschimicola | 29 | Phylloplane | 4; 10; 12; 24; 32; 36; | |
| D. qinlingensis | 2 | Phylloplane | 28; 30; | |
| D. simaoensis | 1 | Phylloplane | 54; | |
| D. waltii | 6 | Phylloplane | 12; 25; | |
| D. wuzhishanensis | 3 | Phylloplane | 12; 44; | |
| D. yunnanensis | 7 | Phylloplane | 36; 40,41; 44; 54; | |
| Dioszegia athyrium | 1 | Phylloplane | 25; | |
| D. aurantiaca | 50 | Phylloplane | 7; 12; 16; 24; 25; 27; 35; 38; 40; 41; 42; 44; 55; 67; | |
| D. butyracea | 1 | Phylloplane | 27; | |
| D. changbaiensis | 4 | Phylloplane | 25; 54; | |
| D. cream | 4 | Phylloplane | 25; 31; 45; | |
| D. fristingensis | 6 | Phylloplane | 35; 37; 42; 45; | |
| D. hungarica | 8 | Phylloplane | 3; 24; 25; 35; | |
| D. statzelliae | 1 | Phylloplane | 31; | |
| D. takashimae | 1 | Phylloplane | 8; | |
| D. xingshanensis | 2 | Phylloplane | 32; | |
| D. zsoltii | 21 | Phylloplane | 1; 3; 4; 13; 24; 25; 31;35; 51; 55; | |
| Hannaella coprosmae | 7 | Phylloplane | 25; | |
| H. kunmingensis | 2 | Phylloplane | 50; | |
| H. luteola | 18 | Phylloplane | 4; 8; 10; 12; 36; 44; 50; 51; 54; | |
| H. oryzae | 20 | Phylloplane | 1; 4; 10; 11; 25; 36; 45; 52;54; 55; | |
| H. sinensis | 25 | Phylloplane | 3;13; 8;10;11; 25; 31; 50; 51; 52; 54; | |
| H. zeae | 1 | Phylloplane | 51; | |
| H. phyllophila | 3 | Phylloplane | 67; | |
| Vishniacozyma carnescens | 5 | Phylloplane, 3 | Soil, 2 | 13; 26; 32; 35; 48; |
| V. dimennae | 1 | Phylloplane | 13; | |
| V. foliicola | 3 | Phylloplane | Soil | 2; |
| V. globispora | 1 | Phylloplane | 27; | |
| V. heimaeyensis | 1 | Soil | 48; | |
| V. taibaiensis | 2 | Phylloplane | 8;32; | |
| V. tephrensis | 2 | Phylloplane | 13; 26 | |
| V. victoriae | 13 | Phylloplane,11 | Soil, 2 | 2; 23; 24;32; 42; 45; 48; 50; 52; 67; |
| Cryptococcaceae | ||||
| Kwoniella dendrophila | 1 | Phylloplane | 13; | |
| K. dejecticola | 1 | Phylloplane | 26; | |
| Cuniculitremaceae | ||||
| Kockovaella imperatae | 1 | Phylloplane | 51; | |
| K. mexicanus | 3 | Phylloplane | 9; 51; | |
| K. sacchari | 2 | Phylloplane | 12; 51; | |
| K. schimae | 1 | Phylloplane | 51; | |
| K. sichuanensis | 1 | Phylloplane | 12; | |
| Phaeotremellaceae | ||||
| Papiliotrema aureus | 1 | Phylloplane | 4; | |
| P. flavescens | 3 | Phylloplane | 44; 52; 54; | |
| P. fonsecae | 2 | Soil | 48; | |
| P. fuscus | 1 | Phylloplane | 44; | |
| P. laurentii | 4 | Phylloplane, 1 | Soil, 3 | 31; 48; |
| Phaeotremella skinneri | 2 | Soil | 2; | |
| Sirobasidiaceae | ||||
| Fibulobasidium inconspicuum | 2 | Soil | 2; | |
| F. murrhardtense | 1 | Soil | 2; | |
| Naemateliaceae | ||||
| Tremella indecorata | 1 | Soil | 2; | |
| Trimorphmycetaceae | ||||
| Saitozyma ninhbinhensis | 1 | Phylloplane | 54; | |
| S. podzolica | 5 | Phylloplane | 36; 54; | |
| Trimorphomyces papilionaceus | 2 | Phylloplane | 12; | |
| Trichosporonales | ||||
| Tetragoniomycetaceae | ||||
| Cryptotrichosporon anacardii | 1 | Phylloplane | 67; | |
| C. tibetense | 3 | Phylloplane | 38; | |
| Takashimella formosensis | 1 | Phylloplane | 44; | |
| T. koratensis | 1 | Phylloplane | 54; | |
| Trichosporonaceae | ||||
| Cutaneotrichosporon arboriformis | 1 | Phylloplane | 36; | |
| C. moniliiforme | 2 | Phylloplane, 1 | Soil, 1 | 2; 40,41; |
| Holtermanniales | ||||
| Holtermannia corniformis | 2 | Phylloplane | 12; 54; | |
| Holtermanniella festucosa | 1 | Soil | 2; | |
| H. nyarrowii | 2 | Phylloplane | 54; | |
| H. takashimae | 1 | Phylloplane | 44; | |
| H. wattica | 7 | Phylloplane, 6 | Soil, 1 | 2; 67; |
| Filobasidiales | ||||
| Filobasidiaceae | ||||
| Filobasidium chernovii | 7 | Phylloplane, 6 | Soil, 1 | 2; 11; 22; 67; |
| F. elegans | 1 | Phylloplane | 12; | |
| F. magnum | 16 | Phylloplane, 8 | Soil, 8 | 2; 13; 22; 26; 32; 36; 48; 52; 67; |
| F. oeirense | 1 | Phylloplane | 45; | |
| F. wieringae | 1 | Phylloplane | 52; | |
| Naganishia adeliensis | 4 | Soil | 48; | |
| N. albida | 10 | Phylloplane, 3 | Soil, 7 | 26; 48; |
| N. albidosimilis | 1 | Soil | 48; | |
| N. antarctica | 1 | Soil | 48; | |
| N. diffluens | 2 | Phylloplane | 42; | |
| N. liquefaciens | 1 | Phylloplane | 32; | |
| N. uzbekistanensis | 3 | Phylloplane | Soil | 4; 48; |
| N. vishniacii | 1 | Soil | 48; | |
| Piskurozymaceae | ||||
| Piskurozyma cylindricus | 2 | Phylloplane | 67; | |
| P. filicatus | 1 | Soil | 2; | |
| Solicoccozyma terreus | 3 | Phylloplane, 1 | Soil, 2 | 2; 44; |
| S. terricola | 1 | Soil | 2; | |
| Cystofilobasidiales | ||||
| Cystofilobasidiaceae | ||||
| Cystofilobasidium capitatum | 4 | Soil | 2; 26 | |
| Itersonilia pannonica | 11 | Phylloplane | 23; 24; 24; 35; 45 38; 53; 67; | |
| I. perplexans | 10 | Phylloplane | 25; 23; 32; 38; 40; 41; 54; 55; | |
| Mrakiaceae | ||||
| Mrakia aquatica | 1 | Phylloplane | 67; | |
| M. blollopis | 1 | Phylloplane | 2; | |
| M. cryoconiti | 1 | Soil | 42; | |
| M. robertii | 1 | Soil | 2; | |
| Tausonia pullulans | 1 | Soil | 2; | |
| Udeniomyces kanasensis | 5 | Phylloplane | 45; 46; | |
| U. pseudopyricola | 26 | Phylloplane, 25 | Soil, 1 | 4; 6; 25; 27; 32; 42; 48; 54; 55; 67; |
| U. puniceus | 2 | Phylloplane | 27; 45; | |
| U. pyricola | 9 | Phylloplane | 4; 23; 24; 31; 54; | |
| Agaricostibomycetes | ||||
| Agaricostibales | ||||
| Agaricostilbaceae | ||||
| Pseudobensingtonia musae | 2 | Phylloplane | 12; 38; | |
| Chionosphaeraceae | ||||
| Ballistosporomyces bomiensis | 2 | Phylloplane | 38; | |
| B. changbaiensis | 2 | Phylloplane | 25; | |
| B. taupoensis | 3 | Phylloplane | 25; | |
| B. xanthus | 6 | Phylloplane | 25; | |
| Cystobasidiopsis lactophilus | 1 | Phylloplane | 44; | |
| C. lophatheri | 1 | Phylloplane | 37; | |
| Kondoaceae | ||||
| Kondoa changbaiensis | 9 | Phylloplane | 25; 67; | |
| K. phyllada | 2 | Phylloplane | 1; 44; | |
| K. sorbi | 3 | Phylloplane | 25; | |
| K. subrosea | 2 | Phylloplane | 45; | |
| K. thailandica | 3 | Phylloplane | 36; 44; 38; | |
| K. yuccicola | 3 | Phylloplane | 45; 67; | |
| Bensingtonia bomiensis | 1 | Phylloplane | 38; | |
| B. naganoensis | 6 | Phylloplane | 25; 55; | |
| B. pseudonaganoensis | 21 | Phylloplane | 12; 24; 25; 32; 38; 67; | |
| B. rectispora | 4 | Phylloplane | 41; | |
| Ruineniaceae | ||||
| Ruinenia clavata | 1 | Phylloplane | 25; | |
| R. diospyroris | 5 | Phylloplane | 36; | |
| Spiculogoeales | ||||
| Phyllozyma linderae | 2 | Phylloplane | 25; | |
| P. subbrunnea | 1 | Phylloplane | 25; | |
| P. coprosmicola | 2 | Phylloplane | 25; 67; | |
| P. dimmenae | 1 | Phylloplane | 25; | |
| Cystobasidiomycetes | ||||
| Cystobasidiales | ||||
| Cystobasidium calyptogenae | 2 | Phylloplane | 44; | |
| C. fimetarium | 1 | Soil | 48; | |
| C. lysinophilum | 1 | Soil | 2; | |
| C. minutum | 3 | Soil | 48; | |
| C. slooffiae | 1 | Soil | 48; | |
| C. pinicola | 1 | Phylloplane | 26; | |
| Erythrobasidiales | ||||
| Bannoa hahajimensis | 4 | Phylloplane | 36; 44; | |
| B. ogasawarensis | 13 | Phylloplane | 4; 10; 12; 25; 36; | |
| B. syzygii | 2 | Phylloplane | 25; 42; | |
| Bannozyma arctica | 3 | Phylloplane | 32; | |
| B. yamatoana | 19 | Phylloplane | 12; 23; 24; 25; 44; 54; 55; 67; | |
| Erythrobasidium hasegawianum | 4 | Phylloplane | 25; | |
| Naohidaeales | ||||
| Naohidea sebacea | 1 | Phylloplane | 16; 32; | |
| Buckeyzymaceae | ||||
| Buckleyzyma aurantiaca | 1 | Soil | 2; | |
| B. salicina | 1 | Phylloplane | 45; | |
| Symmetrosporaceae | ||||
| Symmetrospora coprosmae | 9 | Phylloplane | 25; 27; 45; 50; 67; | |
| S. oryzicola | 6 | Phylloplane | 1; 25; 31; 32; | |
| S. symmetrica | 1 | Phylloplane | 1; | |
| Microsporomycetaceae | ||||
| Microsporomyces magnisporus | 6 | Phylloplane | 36; | |
| Microbotryomycetes | ||||
| Microbotryales | ||||
| Microbotryum reticulatum | 1 | Phylloplane | 54; | |
| Sporidiobolales | ||||
| Rhodosporidium babjevae | 1 | Phylloplane | 57; | |
| Rhodosporidiobolus colostri | 2 | Phylloplane | Soil | 2; 67; |
| R. fluviale | 2 | Phylloplane | 12; 67; | |
| R. lusitaniae | 6 | Phylloplane, 4 | Soil, 2 | 4; 26; 36; 42; |
| R. microsporus | 1 | Phylloplane | 10; | |
| R. nylandii | 1 | Phylloplane | 10; | |
| R. odoratus | 32 | Phylloplane | 1; 8; 10; 12; 25; 26; 31; 32; 38; 40; 41; 42; 44; 54; 66; 67; | |
| R. poonsookiae | 1 | Phylloplane | 51; | |
| R. ruineniae | 3 | Phylloplane | 25; 36; 66 | |
| Rhodotorula glutinis | 1 | Phylloplane | 55; | |
| R. graminis | 1 | Phylloplane | 44; | |
| R. kratochvilovae | 1 | Soil | 48; | |
| R. mucilaginosa | 2 | Phylloplane | 26; 42; | |
| R. paludigena | 1 | Phylloplane | 44; | |
| Sporobolomyces bannaensis | 1 | Phylloplane | 12; | |
| S. beijingensis | 25 | Phylloplane | 1; 3; 18; 19; 22; 38; 56; 66 | |
| S. bischofiae | 1 | Phylloplane | 44; | |
| S. carnicolor | 18 | Phylloplane | 4; 13; 8; 10; 11; 12; 36; 44; 54; | |
| S. japonicus | 3 | Phylloplane | 11; 12; 35; | |
| S. jilinensis | 20 | Phylloplane | 18; 19; 25; 56; 59; 60; 61; 63; 65 | |
| S. phaffii | 7 | Phylloplane | 3; 25; 24; 27; | |
| S. roseus | 31 | Phylloplane, 29 | Soil, 2 | 1; 25; 26; 27; 34; 45; 48; |
| S. ruberrimus | 10 | Phylloplane | 18; 19; 25; 56; 57; 58; 60 | |
| S. salmonicolor | 6 | Phylloplane | 18; 19; 57; 67; | |
| S. shibatanus | 11 | Phylloplane | 3; 8; 25; 36; 51; 56; 66; | |
| Kriegeriales | ||||
| Yamadamyces rosulatus | 1 | Soil | 2; 67; | |
| Leucosporidiales | ||||
| Leucosporidium fellii | 1 | Phylloplane | 67; | |
| L. scottii | 1 | Soil | 2; 67; | |
| Colacogloeaceae | ||||
| Colacogloea diffluens | 1 | Phylloplane | 54; | |
| C. falcata | 3 | Phylloplane | 40; 41; 67; | |
| C. foliorum | 1 | Phylloplane | 67; | |
| Chrysozymaceae | ||||
| Chrysozyma griseoflava | 20 | Phylloplane | 12; 24; 25; 31; 44; 54; 67; | |
| Fellozyma inositophila | 4 | Phylloplane | 35; 32; 55; 67; | |
| incertae sedis | ||||
| Curvibasidium cygneicollum | 8 | Phylloplane | 25; 26; 38; 55; | |
| Slooffia tsugae | 3 | Phylloplane, 2 | Soil, 1 | 2; 12; |
| Oberwinklerozyma yarrowii | 2 | Phylloplane | 24; 54; | |
| Pseudohyphozyma bogoriensis | 1 | Phylloplane | 67; | |
| P. buffonii | 1 | Phylloplane | 40; 41; | |
| P. pustula | 1 | Phylloplane | 67; |
Note: ∗ 1: Baihua mountain, Beijing; 2: Mentougou, Beijing; 3: Songshan mountain, Beijing; 4: Fuzhou county, Fujian province; 5: Beilunhekou natural reserve, Guangxi province; 6: Fanjingshan Mountain, Guizhou province; 7: Maotai county, Guizhou province; 8: Bangxi county, Hainan province; 9: Haikou county, Hainan province; 10: Jianfaling, Hainan province; 11: Sanya county, Hainan province; 12: Wuzhishan mountain, Hainan province; 13: Yesanpo county, Hebei province; 14: Chelu county, Heilongjiang province; 15: Daliangzi river national forest park, Heilongjiang province; 16: Heihe county, Heilongjiang province; 17: Jiayin county, Heilongjiang province; 18: Nanwenghe, Heilongjiang province; 19: Shuanghe county, Heilongjiang province; 20: Wuyiling natural reserve, Heilongjiang province; 21: Yichun county, Heilongjiang province; 22: Hongqiqu county, Henan province; 23: Shennongjia, Hubei province; 24: Xingshan county, Hubei province; 25: Changbai Mountain, Jilin province; 26: Helanshan mountain, Ningxia province; 27: Liupan mountain, Ningxia province; 28: Fuping county, Shaaxi province; 29: Houzhenzi county, Shaaxi province; 30: Qinling Mountain, Shaaxi province; 31: Taibai County, Shaaxi province; 32: Taibai mountain, Shaaxi province; 33: Qufu county, Shandong province; 34: Tai’an county, Shandong province; 35: Taigu county, Shaxi province; 36: Taizhong county, Taiwan province; 37: Bayi county, Tibet; 38: Bomi county, Tibet; 39: Dingjie county, Tibet; 40: Layue county, Tibet; 41: Linzhi county, Tibet; 42: Lulang county, Tibet, 43: Milin county, Tibet; 44: Motuo county, Tibet; 45: unknown location, Xinjiang province; 46: Kanas Lake, Xinjiang province; 47: Kuerlei county, Xinjiang province; 48: Yecheng county, Xinjiang province; 49: Chuxiong county, Yunnan province; 50: Dali county, Yunnan province; 51: Jinghong county, Yunnan province; 52: Kunming county, Yunnan province; 53: Lijiang county, Yunnan province; 54: Simao county, Yunnan province; 55: Zixi montain, Yunnan province; 56: Tahe, Heilongjiang province; 57: Huzhong, Heilongjiang province; 58: Bailudao, Neimonggu province; 59: Dalinuoer, Neimonggu province; 60: Eerguna, Neimonggu province; 61: Hanma, Neimonggu province; 62: Honghuaerji, Neimonggu province; 63: Huihe, Neimonggu province; 64: Saihanwula, Neimonggu province; 65: Tumuji, Neimonggu province; 66: Yantai, Shandong province; 67: unknown location, Tibet.
Fig. 1.
Localisation of sampling sites in China. Red cycles represent soil origin, green cycles represent plant origin.
Among known species, 170 species belonging to 52 genera were isolated from surfaces of plant leaves commonly referred to as phylloplane (Fonseca and Inácio, 2006, Morais et al., 2006, Nakase et al., 2006, Kemler et al., 2017, Limtong and Nasanit, 2017). A total of 42 species belonging to 24 genera were isolated from soils (Table 2). The difference of species diversity between soils and leaves were not analysed in this study because soils and plants were not always collected simultaneously. Most species isolated from soils were previously reported among species occurring in soils by Botha, 2006, Botha, 2011, Yurkov et al., 2016, Yurkov, 2017 and Groenewald et al. (2018), such as Vishniacozyma victoriae, Naganishia adeliensis, Tausonia pullulans, Holtermanniella wattica, Cystobasidium minutum and Cutaneotrichosporon moniliiforme (Table 2). Among species isolated from soils in China, a few have been reported from habitats other than soils, for example, Fibulobasidium inconspicuum from leaves in a river (Sampaio et al. 2002), Genolevuria tibetensis from leaves (Wang et al. 2007) and Yamadamyces rosulatus from dead pine needle (Golubev & Scorzetti 2010).
Among the 101 undescribed species, some are represented by one or only a few isolates. It is difficult to determine if these are rare species or simply undersampled. We continuously collected samples from different locations in China over the past 20 years and some places were revisited many times, such as Milin, Lulang and Bomi counties in Tibet (Table 1). However, a number of single strain species isolated in 2004 were never isolated again despite resampling from the same locations in 2014 and 2015 (Table 1). Phenotypic for these seemingly rare species (Table S1) indicated that most of them grow at low temperature, which may result in slow-growing and competitive disadvantage to other dominating species in a microbial community. In contrast, some known species are frequently isolated from the same or different locations in China, such as Bullera alba isolated from 26 counties, Dioszegia aurantiaca from 14 locations and Rhodosporidiobolus odoratus from 16 locations (Table 2); these commonly isolated species all grow well at room temperature.
Species-by-species pairwise similarity comparison in basidiomycetous yeast genera
Over the past decades, the number of yeast species has increased from 700 (Kurtzman & Fell 1998) to 2 000 (Vu et al. 2016), which benefits from the application of DNA sequence analysis for identification of yeast species (Kurtzman et al. 2015). Relying on results from mating experiments and pairwise DNA-DNA hybridisation values for several ascomycetous genera and species, Kurtzman & Robnett (1998) suggested that different species are likely to show greater than 1 % substitutions in nucleotide sequences of the D1/D2 domains in pairwise comparisons and strains with less three nucleotides differences are likely to be either conspecific or sister species. Fell et al. (2000) observed that when a sufficient number of strains has been studied, different species of basidiomycetous yeasts differed in two or more nucleotides in the D1/D2 domains. In the same time, the authors pointed to several conflicts between taxonomic assignments and pairwise sequence comparisons. Specifically, strains of different species in both Agaricomycotina and Pucciniomycotina sometimes shared identical D1/D2 sequences but showed distinct sequences of the ITS region (Fell et al. 2000). The follow-up study performed by Scorzetti et al. (2002) did not find a common similarity threshold for basidiomycetous yeasts in both D1/D2 and ITS regions and suggested that both gene regions are necessary for a reliable species delimitation. Importantly, sequence variability patterns in these two gene regions dependent on a phylogenetic lineage. While ITS is often more variable than D1/D2 domains of the LSU, the situation was opposite in Trichosporonales and among the members of the Aerius clade of Filobasidiales (Scorzetti et al. 2002). Sequence heterogeneity among sexually compatible strains of teleomorphic species exceeded 1 % in a few genera. Despite distant evolutionary relationships between ascomycetous and basidiomycetous yeasts, the “1 % threshold” was used as an argument to delimit species in the latter group. Even in ascomycetes, this cut-off value is not uniformly applied to all genera (e.g. Clavispora, Metschnikowia, Ogataea). Nevertheless, this threshold was repeatedly used in the taxonomic literature (e.g. Kurtzman and Fell, 2006, Kurtzman, 2014, Kurtzman, 2015, Kurtzman et al., 2015). Results of studies performed by Kurtzman and Robnett, 1998, Fell et al., 2000 and Scorzetti et al. (2002) were recently revised by Vu et al. (2016). The authors observed similar taxonomic threshold, 98.41 % (or 99.21 % using ex-type strains only) for ITS and 99.51 % for LSU, considering all species recognised as yeasts.
The above two threshold values have been calculated for all yeast species and strains. A case-by-case sequences similarity analysis for each genus should be more helpful than those general values to delimit yeast species. In the present work, the sequence similarities and nucleotide variation in the ITS region and D1/D2 domains among all yeast genera that contain more than two species were determined from local alignments for 40 genera of Agaricomycotina and 30 genera of Pucciniomycotina (Table 3 and Tables S2.1–S2.70). In agreement with previous observation, sequence variability in the ITS region was, in general, greater than in D1/D2 domains for most, but not all, studied yeast genera (Table 3). All species in Holtermanniella displayed larger variability in the D1/D2 domains (11–20 nt difference) than in the ITS region (3–7 nt difference). Sequence heterogeneity among species in the following four genera, Solicoccozyma and Naganishia in the Filobasidiales, Trichosporon and Apiotrichum in Trichosporonales, did not show a stable pattern. For example, type strain of Solicoccozyma terrea differed from Solicoccozyma fuscescens by 10 nt in D1/D2 domains, and three nt in ITS region, whereas the latter species differed from Solicoccozyma aeria at eight D1/D2 positions and 13 ITS positions. Naganishia liquefaciens and Naganishia albidisimilis had identical ITS sequences but showed eight D1/D2 nucleotide differences, whereas Naganishia onofrii and Naganishia vaughanmartiniae had identical D1/D2 sequences and 17 mismatches in ITS region. Apiotrichum laibachii and Apiotrichum multisporum shared identical ITS sequences and seven differences in the D1/D2 domains. In the same time sequences of Apiotrichum scarabaeorum and Apiotrichum terrigenum differed by five and 16 nucleotides in the D1/D2 domains and ITS region, respectively. Similarly, Trichosporon asahii differed from Trichosporon coremiiforme by two nucleotides in ITS region and eight nucleotide positions in D1/D2 domains, whereas the latter species differed from Trichosporon dohaense by only one nucleotide substitution in the D1/D2 domains and nine positions in ITS.
Table 3.
Number of nucleotide variation and sequence similarities in the D1/D2 domain and ITS region among the type strains of species in the 70 genera.
| Lineage/Genus | D1/D2 | ITS |
|---|---|---|
| Agaricomycotina | ||
| Tremellomycetes | ||
| Trichosporonales | ||
| Apiotrichum | 2-66 (99.7-89.5 %) | 1-62 (99.8-88.5 %) |
| Cryptotrichosporon | 8-29 (98.5-95.7 %) | 39-109 (92.7-78.6 %) |
| Cutaneotrichosporon | 2-43 (99.7-93.1 %) | 4-103 (99.2-78.4 %) |
| Takashimella | 1-16 (99.8-96.9 %) | 5-37 (98.9-91.9 %) |
| Trichosporon | 1-49 (99.8-92.2 %) | 0-82 (100.0-85.3 %) |
| Vanrija | 11-123 (97.5-77.6 %) | 11-122 (97.7-76.4 %) |
| Holtermanniales | ||
| Holtermanniella | 11-20 (98.3-96.7 %) | 3-7 (99.3-98.4 %) |
| Cystofilobasidiales | ||
| Cystofilobasidium | 10-54 (98.3-90.7 %) | 18-98 (97.1-84.0 %) |
| Mrakia | 1-19 (99.8-97.0 %) | 8-65 (98.7-90.2 %) |
| Itersonilia | 7-30 (98.8-94.9 %) | 18-30 (97.1-95.0 %) |
| Krasilnikovozyma | 2-13 (99.6-97.5 %) | 2-83 (97.7-87.7 %) |
| Tausonia | 17-29 (97.3-95.3 %) | 84-140 (86.5-80.1 %) |
| Udeniomyces | 4-11 (99.4-98.3 %) | 38-144 (94.4-78.6 %) |
| Filobasidiales | ||
| Filobasidium | 0-21 (100.0-96.7 %) | 4-106 (99.3-84.2 %) |
| Goffeauzyma | 3-51 (99.5-90.8 %) | 3-195 (99.5-70.4 %) |
| Heterocephalacria | 5-131 (99.1-77.1 %) | 17-165 (96.2-73.8 %) |
| Naganishia | 0-47 (100.0-92.4 %) | 1-71 (99.8-89.1 %) |
| Piskurozyma | 7-71 (98.8-88.3 %) | 11-195 (98.2-73.0 %) |
| Solicoccozyma | 3-48 (99.5-92.3 %) | 3-144 (99.5-78.8 %) |
| Tremellales | ||
| Bullera | 8-45 (98.7-92.9 %) | 15-183 (97.3-71.0 %) |
| Bulleribasidium | 1-91 (99.8-84.2 %) | 8-165 (98.1-70.8 %) |
| Carcinomyces | 42-72 (91.9-86.6 %) | 117-177 (75.1-66.2 %) |
| Carlosrosaea | 14-16 (97.4-97.1 %) | 63-84 (88.0-82.8 %) |
| Cryptococcus | 0-31 (100-94.8 %) | 1-33 (99.8-93.6 %) |
| Derxomyces | 3-42 (99.5-93.5 %) | 17-206 (96.4-66.6 %) |
| Dioszegia | 3-29 (99.5-95.1 %) | 6-87 (98.7-81.8 %) |
| Fellomyces | 4-39 (99.4-93.8 %) | 10-94 (98.1-83.3 %) |
| Fibulobasidium | 2-6 (99.6-98.9 %) | 44/11 (91.4 %) |
| Genolevuria | 7-17 (98.8-96.7 %) | 23-101 (95.4-80.8 %) |
| Hannaella | 6-59 (99.0-89.6 %) | 7-85 (98.4-83.4 %) |
| Kockovaella | 2-39 (99.7-93.8 %) | 4-85 (99.2-85.0 %) |
| Kwoniella | 0-42 (100.0-93.3 %) | 6-128 (98.9-77.3 %) |
| Naematelia | 1-13 (99.8-97.9 %) | 5-26 (99.0-94.3 %) |
| Papiliotrema | 2-51 (99.6-91.7 %) | 3-127 (99.5-78.1 %) |
| Phaeotremella | 1-28 (99.8-95.3 %) | 4-89 (99.2-83.5 %) |
| Pseudotremella | 34-51 (94.4-91.9 %) | 86-181 (84.6-70.6 %) |
| Rhynchogastrema | 1-19 (99.8.0-97.0 %) | 5-42 (99-91.5 %) |
| Saitozyma | 18-64 (97.0-89.4 %) | 29-92 (94.1-81.9 %) |
| Tremella | 6-104 (99.0-88.6 %) | 0-184 (100.0-66.9 %) |
| Vishniacozyma | 7-64 (98.8-90.1 %) | 6-162 (98.7-73.8 %) |
| Pucciniomycotina | ||
| Agaricostilbomycetes | ||
| Ballistosporomyces | 2-34 (99.7 %-97.2 %) | 17-53 (97.1-91.2 %) |
| Bensingtonia | 8-52 (98.7-91.8 %) | 52-235 (92.1-67.9 %) |
| Cystobasidiopsis | 17-24 (97.2-96.1 %) | 81-101 (86.9-82.0 %) |
| Kondoa | 2-90 (99.7-83.8 %) | 29-55 (95.5-67.7 %) |
| Kurtzmanomyces | 10-61 (98.4-90.4 %) | 83-179 (87.2-74.2 %) |
| Ruinenia | 13-76 (97.8-88.3 %) | 30-134 (94.7-78.6 %) |
| Sterigmatomyces | 12-33 (97.9-94.7 %) | 47-111 (92.2-81.8 %) |
| Spiculogloeomycetes | ||
| Phyllozyma | 3-91 (99.5-85.7 %) | 8-143 (98.4-72.6 %) |
| Cystobasidiomycetes | ||
| Bannoa | 11-21 (98.3-96.8 %) | 27-35 (95.5-94 %) |
| Buckleyzyma | 4-21 (99.4-96.7 %) | 28-76 (87.8-95.3 %) |
| Cystobasidium | 3-44 (99.5-92.1 %) | 6-134 (98.9-78.0 %) |
| Erythrobasidium | 8-24 (98.7-96.0 %) | 6-75 (99.0-88.3 %) |
| Microsporomyces | 29-70 (94.3-86.1 %) | 73-126 (86.1-77.7 %) |
| Occultifur | 6-16 (99.0-97.3 %) | 13-28 (97.4-94.9 %) |
| Sakaguchia | 7-68 (98.7-87.7 %) | 28-87 (95.2-85.3 %) |
| Symmetrospora | 2-34 (99.7-94.6 %) | 12-54 (97.9-91.1 %) |
| Microbotryomycetes | ||
| Colacogloea | 14-61 (97.7-90.2 %) | 52-162 (92.2-74.0 %) |
| Curvibasidium | 3-7 (99.5-98.9 %) | 8-11 (98.7-98.2 %) |
| Glaciozyma | 7-20 (98.9-96.4 %) | 54-99 (91.3-84.2 %) |
| Hamamotoa | 1-8 (99.8-98.7 %) | 18-95 (97.2-85.5 %) |
| Heitmania | 2 (97.7 %) | 33-62 (94.6-89.9 %) |
| Leucosporidium | 1-27 (99.8-95.3 %) | 4-92 (99.3-84.7 %) |
| Oberwinklerozyma | 3-10 (99.4-98.1 %) | 42-49 (92.6-91.1 %) |
| Phenoliferia | 5-14 (99.1-97.5 %) | 3-28 (99.5-95.4 %) |
| Pseudohyphozyma | 3-8 (99.5-98.6 %) | 58-88 (90.8-86.6 %) |
| Rhodosporidiobolus | 4-41 (99.2-93.1 %) | 10-89 (98.1-85.2 %) |
| Rhodotorula | 0-45 (100.0-92.6 %) | 1-79 (99.8-85.9 %) |
| Slooffia | 7-49 (98.9-92.1 %) | 102-159 (85.1-76.9 %) |
| Spencerozyma | 4-42 (99.3-93.0 %) | 59-254 (91.7-67.5 %) |
| Sporobolomyces | 5-49 (100.0-91.9 %) | 3-94 (99.8–83.7 %) |
Our pairwise similarity comparison results indicated that a few well recognised species have less than 1 % nucleotide variation in both ITS region and D1/D2 domains, which in agreement with results by Scorzetti et al. (2002). However, these species can be separated by other taxonomic characters and multi-locus sequences analyses (MLS). For example, Rhodotorula glutinis and Rhodotorula graminis (D1/D2: 1, ITS: 2 Table S2.67) were distinguished by physiological properties (Sampaio 2011a) and on the basis of DNA-DNA hybridisation experiments (Kurtzman & Fell 1991). Recently, a MLS approach combining with the analysis of genes comprising mating locus was used to delimit species in the Papiliotrema flavescens/Papiliotrema terrestris species complex (Yurkov et al. 2015a), Cryptococcus gattii/Cryptococcus neoformans species complex (Hagen et al. 2015) and Cryptococcus amylolentus species complex (Passer et al. 2019), all of which showed less than 1 % ITS and D1/D2 sequences divergence (Tables S2. 24 and S2.34). Thus, it is important to keep in mind that delimitation of closely related species which have less than 1 % sequence heterogeneity in both D1/D2 and ITS regions requires additional analyses and more robust data such as detailed physiological characterisation, mating experiments, multi-locus analyses and even whole-genome comparisons. Delimitation of closely related species in genera with a few known species is, thus, extremely difficult in spite of the lack of data for analyses.
New taxa delineation and phylogenetic placement
The sequences of the D1/D2 and ITS regions for the 199 strains (Table 1) including 11 isolates from Germany deposited in the China General Microbiological Culture Collection Center (CGMCC) and 16 strains from Japan, Thailand, Portugal, Italy, USA, DSMZ and CBS collections employed in this study were determined. The SSU region of 138 strains representing at least one strain of each potentially new species were sequenced. A total of 142 RPB1, 137 RPB2, 126 TEF1 and 126 CYTB new sequences were generated (Table 1). The D1/D2 and ITS sequences for each strain were blasted against the GenBank database using the BLASTn tool to search for their closely related described species. Sequences of their close relatives and other phylogenetic important taxa were retrieved from GenBank (Table S3). In order to show the phylogenetic positions of these undescribed strains, multi-loci phylogenetic trees were constructed from two datasets, the combined 5.8 S, D1/D2 and SSU dataset and the combined 5.8S, D1/D2, SSU, RPB1, RPB2, TEF1 and CYTB dataset. The phylogenetic trees (Fig. 2, Fig. 4 and S1, S2) drawn from the seven-genes and three rDNA datasets were used to determine the phylogenetic positions for each new species. The trees (Fig. 3, Fig. 5) constructed from the D1/D2 dataset were used to calculate the similarity between the new species and their closely related described species as the D1/D2 sequences are available for all known species employed here, which is not the case for the ITS and SSU sequences.
Fig. 2.
Phylogenetic tree inferred using the combined sequences of RPB1, RPB2, TEF1, CYTB, SSU rDNA, LSU rDNA D1/D2 domains and 5.8S rDNA, depicting the phylogenetic positions of new taxa (in bold) within Tremellomycetes (Agaricomycotina). The tree backbone was constructed using maximum likelihood analysis. Bootstrap percentages of maximum likelihood analysis over 50 % from 1 000 bootstrap replicates and posterior probabilities of Bayesian inference above 0.9 are shown respectively from left to right on the deep and major branches. Bar = 0.05 substitutions per nucleotide position. Note: ns, not supported (BP < 50 % or PP < 0.9); nm, not monophyletic. The new taxa are in bold.
Fig. 4.
Phylogenetic tree inferred using the combined sequences of RPB1, RPB2, TEF1, CYTB, SSU rDNA, LSU rDNA D1/D2 domains and 5.8S rDNA, depicting the phylogenetic positions of new taxa (in bold) within Pucciniomycotina. The tree backbone was constructed using maximum likelihood analysis. Bootstrap percentages of maximum likelihood analysis over 50 % from 1 000 bootstrap replicates and posterior probabilities of Bayesian inference above 0.9 are shown respectively from left to right on the deep and major branches. Bar = 0.2 substitutions per nucleotide position. Note: ns, not supported (BP < 50 % or PP < 0.9); nm, not monophyletic. The new taxa are in bold.
Fig. 3.
Phylogeny of new yeast species in the Tremellomycetes (Agaricomycotina) inferred from the sequences of the LSU rDNA D1/D2 domains by maximum likelihood analysis and over 50 % from 1 000 bootstrap replicates is shown. Tree topology was backbone-constrained with the well-supported (>80 %) bipartitions of the topology of the seven-genes tree. Bar = 0.1 substitutions per nucleotide position.
Fig. 5.
Phylogeny of new yeast species in the Pucciniomycotina inferred from the sequences of the LSU rDNA D1/D2 domains by maximum likelihood analysis and over 50 % from 1 000 bootstrap replicates is shown. Tree topology was backbone-constrained with the well-supported (>80 %) bipartitions of the topology of the seven-genes tree. Bar = 0.1 substitutions per nucleotide position.
One hundred and seven new species were delimitated from the 199 strains using the species identification benchmarks suggested by Fell et al., 2000, Scorzetti et al., 2002, Kurtzman and Fell, 2006, Kurtzman, 2014, Kurtzman, 2015 and Kurtzman et al. (2015) as well as the taxonomic thresholds of yeast species recommended by Vu et al. (2016) and phenotypical features (Kurtzman et al. 2011). Forty-three new species occur in 15 genera in the Tremellomycetes (Agaricomycotina) and 52 new species distribute in 20 genera in the Pucciniomycotina (Fig. 2, Fig. 3, Fig. 4, Fig. 5, Fig. 6 and S1–S6, Table 1). However, none of these known genera appears as an obvious candidate to accommodate the other 12 new species. Therefore, eight new genera, named as Begerowomyces, Boekhoutia, Meniscomyces, Pseudosterigmatospora, Robertozyma, Rosettozyma, Sterigmatospora and Teunia, are proposed to accommodate these 12 species.
Fig. 6.
Phylogenetic tree inferred using the combined sequences of SSU rDNA, LSU rDNA D1/D2 domains and ITS region (including 5.8S rDNA), depicting the phylogenetic positions of Lichenozyma and new taxa (in bold) within Cystobasidiomycetes (Pucciniomycotina). The tree was constructed using maximum likelihood analysis and over 50 % from 1 000 bootstrap replicates is shown. Bar = 0.02 substitutions per nucleotide position.
The novel genus Teunia, located in the Cryptococcaceae (Tremellales, Tremellomycetes, Agaricomycotina), was clustered with the genera Cryptococcus and Kwoniella with 96–100 % bootstrap and 1.0 posterior probability supports in seven-genes and rDNA phylogeny (Figs 2A and S1A). However, those three genera can be separated by the clustering optimisation analysis (Table S4). Three species, namely Cryptococcus cuniculi, Fonsecazyma tronadorensis and Fonsecazyma betulae, were classified in this new genus (Fig. 2, Fig. 3G and S1C). The phylogenetic position and composition of this clade have been changing during the last decade. The oldest known species Cr. cuniculi has affinity with the erroneously identified as Cryptococcus heveanensis strain CBS 8976 in the Kwoniella clade that was described by Shin et al. (2006). Later, Boekhout et al., 2011, de Garcia et al., 2012 and Weiss et al. (2014) also indicated that Cr. cuniculi belonged to the Kwoniella clade. de Garcia et al. (2012) described another species Cryptococcus tronadorensis in this clade resolved in a LSU-based phylogenetic analysis. However, a constrained with the seven-genes topology LSU phylogenetic analysis performed by Liu et al. (2015b) showed that Cr. cuniculi was placed in the Tremella clade I (Millanes et al. 2011) and not close to the Kwoniella clade, so that this species left unclassified as Cr. cuniculi pro tem. It is important to document that the phylogenetic analysis was inconsistant with the previous results obtained by Shin et al., 2006, Boekhout et al., 2011, de Garcia et al., 2012 and Weiss et al. (2014) indicating that Cr. cuniculi was most likely a member of Cryptococcaceae. Furthermore, the two closely related species Cr. cuniculi and Cr. tronadorensis were placed in two different clades (Liu et al. 2015b). The latter species was clustered with a good support with Cryptococcus mujuensis and Kwoniella betulae. It is important to note that K. betulae was described as a species of the genus Kwoniella based on its close phylogenetic relatedness to the erroneously identified as Cr. heveanensis strain CBS 8976. Because K. betulae was not related to other species of Kwoniella and Cr. mujuensis and Cr. tronadorensis were distantly related to the genus Cryptococcus, a new genus Fonsecazyma was proposed to accommodate Fo. mujuensis, the type species of Fonsecazyma, Fo. tronadorensis and Fo. betulae (Liu et al. 2015b). The type species of Fonsecazyma was included in the seven-genes phylogeny as a single-species lineage closely related to Sirobasidium intermedium (Liu et al. 2015a) which was also in agreement with the original paper (Shin et al. 2006). This was one of a few important conflicts between constrained LSU and seven-genes analyses. The decision to propose a new genus for this clade was supported by the results of the constrained LSU analysis which also demonstrated that the Fonsecazyma clade contained three potential new species isolated but not described in earlier studies performed by Inácio (2003). It was important to name this clade so that provisionally named as “Cryptococcus” new species would be properly placed and not mistaken with either Kwoniella or Cryptococcus (Liu et al. 2015b).
The phylogenetic analyses of the three datasets in this study also supported that Fo. mujuensis has affinity with Si. intermedium instead of the Kwoniella clade (Fig. 2, Fig. 3A and S1A). Fo. tronadorensis was originally described as Cr. tronadorensis and related to the Kwoniella clade (de Garcia et al. 2012). Fo. betulae was originally described as K. betulae (Sylvester et al. 2015). The analyses in this study showed that Fo. tronadorensis, Fo. betulae, Cr. cuniculi and three newly described species formed a well supported clade closely related to Cryptococcus and Kwoniella, but still separated from them (Fig. 2, Fig. 3G and S1C), which is in agreement with the results of de Garcia et al. (2012) and Sylvester et al. (2015). The question is why Fo. tronadorensis and Fo. betulae clustered with Fo. mujuensis instead of Cr. cuniculi in the D1/D2 tree from Liu et al. (2015b). After double-checking the D1/D2 alignment used in Liu et al. (2015b), we found out that the D1/D2 sequences of Fo. mujuensis and Cr. cuniculi were swapped with each other. We also checked the placement of other species in the D1/D2 tree from Liu et al. (2015b). We have not found other mistakes in that tree, which indicated that the D1/D2 dataset is reliable except for the sequence swap between Fo. mujuensis and Cr. cuniculi. Therefore, Fo. tronadorensis, Fo. betulae and Cr. cuniculi were combined or validated in the new genus Teunia in this study (see Taxonomy section).
The novel genera Begerowomyces and Robertozyma, represented by CGMCC 2.4451 and CGMCC 2.3164, respectively, were closely related to Occultifur, Cystobasidium and two monophyletic genera, Queiroziella and Halobasidium, described by Crous et al. (2018) and Guo et al. (2019), respectively, as two separated branches in the Cystobasidiomycetes (Figs 4A and S2A), which indicated that they did not belong to the genera Occultifur, Cystobasidium, Queiroziella or Halobasidium. The BLASTn results showed that CGMCC 2.3164 and CGMCC 2.4451 had less than 90–93 % and 87–91 % (with 91 % coverage) similarities with other genera in Cystobasidiomycetes, such as Occultifur, Cystobasidium and Symmetrospora, in the D1/D2 and ITS regions, respectively. The sequence similarities between CGMCC 2.4451 and CGMCC 2.3164 were 93.6 % and 88.2 % in the D1/D2 and ITS regions, respectively. The comparison of the sequence similarities between CGMCC 2.4451, CGMCC 2.3164 and other genera in the Cystobasidiomycetes indicated that CGMCC 2.4451 and CGMCC 2.3164 should represent two novel genera according to the yeast genera thresholds predicted by Vu et al. (2016).
The novel genera Pseudosterigmatospora and Sterigmatospora, represented by CGMCC 2.5817 and CGMCC 2.5816, respectively, were located in the Agaricostilbomycetes and closely related to Jianyunia sakaguchii (Figs 4A and S2A). BLASTn searches of the D1/D2 sequences showed that CGMCC 2.5817 and CGMCC 2.5816 had the highest match with Bensingtonia rectispora with less than 90 % coverage and 90 % similarity. CGMCC 2.5817 had the highest match with species of Ballistosporomyces with 50 % coverage and less than 91 % similarity when using ITS sequences as query. However, CGMCC 2.5816 was more related to species of Ruinenia with less than 50 % coverage and 90 % similarity. CGMCC 2.5817 and CGMCC 2.5816 have 91.8 % and 65.5 % similarities in the D1/D2 and ITS regions, respectively. The low similarities in the above analyses indicated that CGMCC 2.5817 and CGMCC 2.5816 were separated and did not belong to any existing genus in the Agaricostilbomycetes. The phylogenetic analysis based on the combined three rDNA loci and seven-genes datasets showed that CGMCC 2.5817, CGMCC 2.5816 and J. sakaguchii formed a clade separated from other families in the Agaricostilbales. Because only J. sakaguchii occured in the new “Agaricostilbales family 2”, recognised by the nested analyses of the GMYC approach, this new family was not proposed by Wang et al. (2015b). It is now appropriate to proposed the “Agaricostilbales family 2” as a new family, named as Jianyuniaceae, in this study with two novel genera Pseudosterigmatospora and Sterigmatospora included in this clade.
The novel genus Boekhoutia, represented by CGMCC 2.4539, was closely related to Kurtzmanomyces and Chionosphaera. A BLASTn search of the D1/D2 sequence of CGMCC 2.4539 revealed that the closest match was Kurtzmanomyces shapotouensis with 99 % coverage and 87 % similarity. However, the closest matches using the ITS sequence were the species of Cystobasidiopsis wih 71 % coverage and less than 82 % similarity. The results of the BLASTn searches indicated that the phylogenetic position of CGMCC 2.4539 is unclear. In order to clarify its position, the phylogenetic analyses were performed based on different datasets using different algorithms (Fig. 4, Fig. 5A and S2A). CGMCC 2.4539 was located in the family Chionosphaeraceae as an isolated branch and loosely related to the genera Chionosphaera and Kurtzmanomyces without support in the tree from the single D1/D2 dataset (Fig. 5A). However, CGMCC 2.4539 formed a clade with Cystobasidiopsis in the ITS tree with 86 % bootstrap support (data not shown). CGMCC 2.4539 clustered with Kurtzmanomyces without support as a separated long branch in the tree of the combined three rDNA dataset (Fig. S2A), and located in a separated bottom branch from Kurtzmanomyces and Chionosphaera in the tree of the seven-genes dataset (Fig. 4A). The above analyses indicated that placing CGMCC 2.4539 into Chionosphaera and Kurtzmanomyces is arbitrary. Therefore, a new genus, Boekhoutia is proposed to accommodate this strain.
The novel genus Meniscomyces, represented by CGMCC 2.5818, was located in the Spiculogloeomycetes (Figs 4A and S2A). The ITS and D1/D2 sequences of CGMCC 2.5818 are very divergent from other yeast species. A BLASTn search of the D1/D2 sequences revealed that CGMCC 2.5818 matched with Phyllozyma with less than 70 % coverage and 82–83 % similarity, and genera, such as Tremella, in the Tremellomycetes (Agaricomycotina) with 79–81 % similarity. Only the 5.8S region of the ITS sequence of CGMCC 2.5818 matched with some taxa in the Pucciniomycotina and Agaricomycotina, such as Crustoderma and Tygervalleyomyces, using ITS sequences as the query. A BLASTn search of the SSU sequences showed that CGMCC 2.5818 matched to the taxa in Pucciniomycotina, with Phyllozyma as the best match with 89 % similarity. The phylogenetic analyses based on different datasets (Fig. 4, Fig. 5 and S2) showed that CGMCC 2.5818 is related to Phyllozyma, Mycogloea sp. TUB FO40962 and Spiculogloea sp. TUB RB1040 in the Spiculogloeomycetes, with Spiculogloea sp. TUB RB1040 as its closes relative (Fig. 5). Because sequences of only a few Spiculogloea and Mycogloea species are available at present, it is difficult to elucidate the higher taxonomic position of CGMCC 2.5818. Consequently, CGMCC 2.5818 was placed in a new genus Meniscomyces (see Taxonomy section), which is temporarily treated as ‘incertae sedis’ in the Spiculogloeomycetes.
The novel genus Rosettozyma, represented by the groups of CGMCC 2.2615, CGMCC 2.3466 and CGMCC 2.5819, located in a separated clade at the bottom of the tree, is separated from all known orders and other taxa in the Microbotryomycetes (Figs 4A and S2A). A BLASTn search of the D1/D2 and ITS sequences revealed that these three groups matched to the genera in the Microbotryomycetes, such as Rhodotorula, Chrysozyma, Oberwinklerozyma, Phenoliferia, Vonarxula and Yunzhangia, with 86–89 % and 84–94 % similarities (42–59 % coverage), respectively, which are below the fungal order thresholds of 94.7 % for D1/D2 and 81.2 % for ITS, recommended by Vu et al. (2016). The phylogenetic analysis and the comparison of predicted taxonomic thresholds indicated that the CGMCC 2.2615, CGMCC 2.3466 and CGMCC 2.5819 groups could represent a new order. Therefore, Rosettozyma, Rosettozymaceae and Rosettozymales are proposed (see Taxonomy section).
The genus Heitmania belongs to the Microbotryomycetes, but no higher categories were assigned to place this genus in although it represents an isolated clade in the Microbotryomycetes (Liu et al. 2017). Two new species of Heitmania are proposed in this study that represented a subclade that was separated from the already described species in the trees constructed from the different datasets (Figs 4A and S2A). The phylogenetic analysis based on the increased number of sampled species showed that this genus was more related to the order Sporidiobolales than the other taxa in the Microbotryomycetes in the tree of the three rDNA loci dataset (Fig. S2), but located in a separated branch from other existing orders in the Microbotryomycetes in the tree of the seven-genes dataset (Fig. 4) agreeing with the result from Liu et al. (2017). The genus Heitmania had a less than 93 % similarity with other taxa in the Microbotryomycetes in the D1/D2 domains and 82–88 % (60–78 % coverage) in the ITS region. The above data indicated that a new order could be circumscribed to accommodate the genus Heitmania. Therefore, Heitmaniaceae and Heitmaniales are proposed in the Taxonomy section.
Some novel species described latter were represented by a single strain or a few of isolates. In order to find potentially conspecific strains of different origin for those new species, we used the ITS and D1/D2 sequences of those species to blast the similar sequences against GenBank, The Yeasts Trust database or MycoBank (Robert et al. 2005, http://www.mycobank.org/). Sixty identical or similar sequences, which are from 46 unpublished strains and 14 uncultured fungus clones, were added in the new species delimitation below.
New species identification in the Tremellomycetes (Agaricomycotina)
Kockovaella (Cuniculitremaceae, Tremellales)
Five strains, isolated from Yunnan and Hainan provinces, South China, were located in the Kockovaella clade as three separate groups that were also separated from other species of Kockovaella (Fig. 2, Fig. 3B and S1B). Groups CGMCC 2.3443 and CGMCC 2.3536, both containing two strains, clustered together in the tree constructed by the seven-genes and three rDNA loci datasets (Figs 2B and S1B) and were most closely related to Kockovaella libkindii in the tree drawn by the D1/D2 dataset (Fig. 3B). Strains in the CGMCC 2.3443 group have identical ITS and D1/D2 sequences, which indicated that they are conspecific. Strains in the CGMCC 2.3536 group, also with identical ITS and D1/D2 sequences, differed from the CGMCC 2.3443 group by 12 nucleotides (nt) (∼2 %) substitutions in the D1/D2 domains and 16 nt (∼3.2 %) mismatches (including substitutions and deletions) in the ITS regions. These two groups differed from Koc. libkindii by three nt (∼0.5 %) and 11–17 nt (∼2.2–3.4 %) mismatches in the D1/D2 and ITS regions, respectively. Strain CGMCC 2.3465 was placed in the Kockovaella clade (Fig. 2B) as a separated branch at the bottom of the clade. It differed from other Kockovaella species by more than 3 % and 7 % mismatches in the D1/D2 and ITS regions, respectively.
The above sequence comparisons indicated that the five novel strains represent three novel species in the genus Kockovaella.
Genolevuria (Bulleraceae, Tremellales)
CGMCC 2.5809 has a close relationship with Genolevuria amylolytica and Genolevuria tibetensis (Fig. 2, Fig. 3C and S1B). They differed from each other by 10–15 nt (∼2–3 %) substitutions and ∼10 % mismatches in the D1/D2 and ITS regions, respectively. Therefore, CGMCC 2.5809 is proposed as a new species in the genus Genolevuria.
Vishniacozyma (Bulleraceae, Tremellales)
Six strains formed three groups, represented by CGMCC 2.3099, CGMCC 2.3472 and CGMCC 2.3165, in the Vishniacozyma clade (Figs 2B and S1B). Group CGMCC 2.3472, consisting of three strains, possessed similar sequences with one nt and five nt difference in the D1/D2 and ITS regions, respectively, which indicated they were conspecific. An isolate IA19 (KM246197/KM246114) named as ‘Cryptococcus dimennae’ in the GenBank database had identical or similar D1/D2 sequences (zero to one nt difference) with the CGMCC 2.3472 group, however, there were nine to ten nt (∼1.9–2.0 %) differences in the ITS regions, which indicated that the isolate IA19 may represent a different taxon and is not conspecific to the strains of the CGMCC 2.3472 group. The CGMCC 2.3472 group was closely related to Vishniacozyma nebularis, Vishniacozyma dimennae and Vishniacozyma globispora (Fig. 2, Fig. 3F and S1F), which differed from the three known species by 9–28 nt (∼1.5–4 %) substitutions in the D1/D2 domains and by more than 9 % mismatches in ITS regions. More than seven nt (∼1.1 %) D1/D2 sequence difference were observed between group CGMCC 2.3472 and other eight undescribed or erroneously identified strains (Fig. S1F), which indicated that those strains represent different species from group CGMCC 2.3472. Group CGMCC 2.3099 was more closely related to Vishniacozyma foliicola and Vishniacozyma heimaeyensis (Figs 2B and S1F). They differed from V. foliicola and V. heimaeyensis by seven to ten nt (∼1.1–1.6 %) substitutions in the D1/D2 domains and by 15–18 nt (∼3 %) mismatches in the ITS region. The two strains in the CGMCC 2.3165 group had identical sequences in the ITS and D1/D2 regions. They also had the same ITS sequences as ‘Cryptococcus’ sp. SJ8L03 (FJ153171) and SJ8L02 (FJ153172), and a similar ITS (four nt difference) and D1/D2 sequences (one nt difference) as ‘Cryptococcus’ sp. KY763 (AB428345/AB428344). A Blast search against The Yeasts Trust database (or MycoBank) showed that CBS 8412 isolated from food in the Netherlands and CBS 9328 isolated from soil in Costa Rica have similar D1/D2 sequences (99.68–99.84 %) and ITS sequences (99.45 %) with CGMCC 2.3165 group. The above analysis indicated that they should be conspecific. The CGMCC 2.3165 group differed from CGMCC 2.3099, V. foliicola and V. heimaeyensis by 8–11 nt (∼1.3–1.8 %) mismatches in the D1/D2 domains, and by more than 5 % nucleotide divergence in the ITS region.
Based on the above sequence comparisons, those six strains should represent three novel species in the genus of Vishniacozyma.
Carlosrosaea (Trimorphomycetaceae, Tremellales)
The genus Carlosrosaea was circumscribed with a single species Carlosrosaea vrieseae (Liu et al. 2015b). Recently, Felix et al. (2017) described two novel species, namely Carlosrosaea hohenbergiae and Carlosrosaea aechmeae. Strains from all three species were isolated from bromeliads in Brazil (Landell et al., 2015, Felix et al., 2017). Two Chinese isolates, CGMCC 2.3580 isolated from Yunnan province and CGMCC 2.3447 isolated from Hainan province, were placed in the genus Carlosrosaea with an affinity to Ca. vrieseae based on phylogenetic analysis of the sequences of the ITS and D1/D2 regions (Fig. 3C). They differed from Ca. vrieseae by 11–13 nt (∼1.8–2.2 %) substitutions in the D1/D2 domains, and by more than 9 % mismatches in the ITS regions. CGMCC 2.3580 and CGMCC 2.3447 differed from each other by 12 nt (∼2 %) substitutions and more than 12 % mismatches in the D1/D2 and ITS regions, respectively.
The above analyses indicated that the two novel strains represent two undescribed Carlosrosaea species.
Note: An uncultured fungal clone 2170_736 (KP891580) from Scolytus multistriatus, Sweden, has an identical ITS sequence with CGMCC 2.3447, which indicates that the species represented by CGMCC 2.3447 is also distributed outside of China.
Saitozyma (Trimorphomycetaceae, Tremellales)
CGMCC 2.5811, located in the genus Saitozyma was closely related to Saitozyma flava in the tree obtained from the combined seven-genes dataset (Fig. 2C). Although they differed from each other by only two nt in the D1/D2 domains, there were 14 nt (∼3 %) differences in the ITS region, which indicated that CGMCC 2.5811 was not conspecific to Sa. flava. More than seven nt D1/D2 heterogeneity (∼1.1 %) were observed between CGMCC 2.5811, Saitozyma paraflava and the potential new species from Thailand and Japan (Fig. 3C). The above analyses indicated that CGMCC 2.5811 represented a novel Saitozyma species.
Note: The monophyly of Saitozyma was not supported by this study (Fig. 2C), which need more robust data and species to confirm.
Tremella (Tremellaceae, Tremellales)
CGMCC 2.5615 had the closest relationship with Tremella globispora (Fig. 2, Fig. 3F). They differed from each other by seven nt (∼1.1 %) substitutions in the D1/D2 domains and 17 nt (∼3 %) mismatches in the ITS region. Thus, it should be proposed as a new species in Tremella.
Kwoniella (Cryptococcaceae, Tremellales)
CGMCC 2.3439 was placed in the Kwoniella clade with an affinity to Kwoniella endophytica, Kwoniella botswanensis, Kwoniella mangrovensis, Kwoniella pini, Kwoniella dejecticola, Kwoniella dendrophila and Kwoniella shivajii in the trees from the D1/D2 as well as the three rDNA and seven-genes datasets (Fig. 2, Fig. 3G and S1C). It differed from those six species by 11–18 nt (∼2–3 %) substitutions in the D1/D2 domains and 7 % mismatches in the ITS region. The analysis of the ITS and D1/D2 sequences indicated that CGMCC 2.3439 belongs to a novel species within Kwoniella.
Teunia (Cryptococcaceae, Tremellales)
CGMCC 2.3835, CGMCC 2.4450 and CGMCC 2.5648 formed a separate branch in the tree of the three rDNA and seven-genes datasets, and were located in the genus Teunia, a clade newly named in this study (Figs 2C and S1C). CGMCC 2.5648 was closely related to the misidentified strain ‘Piskurozyma taiwanensis’ CBS 9926 (KY102949/KY107271) and ‘Cryptococcus’ sp. F6 (AY518273/AY508880) with two nt differences in the D1/D2 domains and 12–17 nt (∼2.1–3 %) mismatches in the ITS regions, which indicated that they probably belong to different species. CGMCC 2.3835 and CGMCC 2.4450 differed from their closest undescribed or erroneously identified strains, ‘Fonsecazyma’ sp. 21S4 (MK400702),‘Kwoniella’ sp. PY016 (KY399877), ‘Kwoniella’ sp. HB31-3 (KJ507251), ‘Cryptococcus’ sp. BI226 (EU678944), ‘Cryptococcus’ sp. SAP963.4 (JX067803), ‘Cryptococcus’ sp. RT 1.5.17 (AY731785) and ‘Cryptococcus heveanensis’ YM25139 (JQ964208) (Fig. 3G), by 8–13 nt (∼1.3–2 %) substitutions in the D1/D2 domains.
The above sequence comparisons proved that the three new strains belong to three novel species in the genus Teunia.
Note: Based on the D1/D2 sequences comparisons, more than 30 undescribed or erroneously identified strains may represent more than 20 species in Teunia clade (Fig. 3G), which need to be clarified in the future because the ITS sequences are not available at present.
Dioszegia (Bulleribasidiaceae, Tremellales)
Seven strains with orange-coloured colonies distributed in five groups loctated in the Dioszegia clade (Fig. 2, Fig. 3D and S1C). Group CGMCC 2.5628 was closely related to Dioszegia crocea and Dioszegia aurantiaca and differed from them by 12–13 nt (∼1.9–2.1 %) in the D1/D2 domains and 8–11 nt (∼1.7–2.3 %) mismatches in the ITS region. Three strains in group CGMCC 2.5674 had identical sequence in the ITS and D1/D2 regions. They differed from Dioszegia cryoxerica and Dioszegia changbaiensis by 13–16 nt (∼2.1–2.6 %) in the D1/D2 domains and about 5 % mismatches in the ITS region. Groups CGMCC 2.3625, CGMCC 2.5658 and CGMCC 2.4537 formed three separate branches, clustering with Dioszegia athyrii, Dioszegia catarinoi, Dioszegia takashimae and Di. zsoltii. Group CGMCC 2.3625 and an unpublished strain, TY-217 (AY313036/AY313018) possessed similar sequences with only one and three nt differences in the D1/D2 and ITS regions, respectively, which indicated that they are conspecific. Group CGMCC 2.3625 differed from these four known species (Fig. 3D) by zero to five nt and more than 21 nt (∼4 %) mismatches in the D1/D2 and ITS regions, respectively. Group CGMCC 2.5658 differed from these four known species by one to five nt in D1/D2 domains and 11–15 nt (∼2.1–3 %) mismatches in the ITS regions. Group CGMCC 2.4537 differed from them by five to ten nt (∼0.8–1.6 %) and more than 33 nt (∼6 %) mismatches in the ITS and D1/D2 regions, respectively. An uncultured fungal clone CMH458 (KF800549) from indoor air in Kansas City, Missouri, USA had an identical ITS sequence as CGMCC 2.4537, which indicated that this species may be common in different locations.
According to the above sequence analyses five novel species in the genus Dioszegia are proposed in the Taxonomy section.
Bulleribasidium (Bulleribasidiaceae, Tremellales)
Seven strains formed five groups in the genus Bulleribasidium, represented by CGMCC 2.4024, CGMCC 2.4427, CGMCC 2.5812, CGMCC 2.4428 and CGMCC 2.3320 (Fig. 2, Fig. 3D and S1D). One nt difference was found in the D1/D2 domains between two strains in the CGMCC 2.4024 group. This group differed from its closest relative Bulleribasidium panici by three to four nt (∼0.6 %) substitutions in the D1/D2 domains and nine nt (∼1.8 %) mismatches in the ITS regions. Groups CGMCC 2.4427 and CGMCC 2.5812 were most closely related to Bulleribasidium setariae. CGMCC 2.5812 differed from Bu. setariae by nine nt (∼1.5 %) and 16 nt (∼2.6 %) mismatches in the D1/D2 and ITS regions, respectively. CGMCC 2.4427 had 45 nt (∼7.4 %) differences in the D1/D2 domains and ∼19 % in the ITS regions from Bu. setariae. Groups CGMCC 2.3320 and CGMCC 2.4428 had an affinity to Bulleribasidium foliicola. Group CGMCC 2.3320 consisted of two strains with similar D1/D2 (one nt difference) and ITS sequences (three nt differences). A Blast search against The Yeasts Trust database showed that two strains, BSB09 (KY305125) isolated from bromeliad, Brazil and TY-199 (AY313030) found in the phyllosphere of Thailand have 99.5–99.8 % sequences similarity with the CGMCC 2.3320 group in the ITS region, which indicated that they are conspecific. They differed from Bu. foliicola by three to four nt (∼0.6 %) in the D1/D2 domains and nine to ten (∼1.7–1.9 %) mismatches in the ITS regions. Group CGMCC 2.4428 had a greater sequence disparity with Bu. foliicola with 19 nt (∼3.1 %) difference in the D1/D2 domains and more than 11 % in the ITS region.
The above sequence comparisons indicated that these seven novel strains represent five undescribed species of Bulleribasidium.
Derxomyces (Bulleribasidiaceae, Tremellales)
Twenty three strains separated in twelve groups located in the Derxomyces clade (Fig. 2, Fig. 3E and S1D). Three strains in group CGMCC 2.5660 differed from each other by one nt in both the D1/D2 and ITS regions. This group differed from its closest relative Derxomyces linzhiensis by 23 nt (∼4 %) in the D1/D2 domains and more than 9 % mismatches in the ITS regions. Groups CGMCC 2.3572 and CGMCC 2.4429 were closely related to Derxomyces hubeiensis (Figs 2D and S1D). Strains in the CGMCC 2.4429 group had similar sequences with three nt differences in the ITS region, which indicated that they are conspecific. They differed from group CGMCC 2.3572 by two to four nt in the D1/D2 domains and 14–17 nt (∼2.7–3.3 %) mismatches in the ITS regions. These two groups differed from De. hubeiensis by 13–15 nt (∼2.1–2.4 %) in the D1/D2 domains and more than 6 % mismatches in the ITS regions. Group CGMCC 2.3459 contained three strains with identical ITS and D1/D2 sequences and were closely related to Derxomyces schimicola and Derxomyces pseudoschimicola. The former differed from the known two species by six to seven nt (∼1.0 %) in the D1/D2 domains and 8–18 nt (∼1.4–3.0 %) mismatches in the ITS regions. Group CGMCC 2.2459 was closely related to Derxomyces cylindricus, and differed from it by four nt (∼0.6 %) and 11 nt (∼2.2 %) mismatches in the D1/D2 and ITS regions, respectively. Groups CGMCC 2.4436 and CGMCC 2.4437 were most closely related to Derxomyces boekhoutii (Fig. S1D). These two groups differed from each other by three nt (∼0.5 %) and 18 nt (∼3.5 %) mismatches in the D1/D2 and ITS regions, respectively. Group CGMCC 2.4436 differed from De. boekhoutii by three nt (∼0.5 %) and 12 nt (∼2.4 %) mismatches in the D1/D2 and ITS regions, respectively. Group CGMCC 2.4437 and De. boekhoutii had two nt and 16 nt (∼3.1 %) differences in the D1/D2 and ITS regions, respectively. Group CGMCC 2.3561 differed from Derxomyces wuzhishanensis by seven nt (∼1.1 %) and 18 nt (∼3.5 %) mismatches in the D1/D2 and ITS regions, respectively.
Four groups represented by CGMCC 2.3535, CGMCC 2.3563, CGMCC 2.3470 and CGMCC 2.4446 were closely related to Derxomyces yunnanensis. Group CGMCC 2.3535 and De. yunnanensis had identical D1/D2 sequences, and 10 nt (∼1.9 %) differences in the ITS region, which indicated that they represent different taxa. Group CGMCC 2.3563 contained six strains that do not have more than three nt differences in both the ITS and D1/D2 regions, which indicated that they are conspecific. Group CGMCC 2.3470 comprised two strains with two nt differences in the ITS regions. The two strains in group CGMCC 2.4446 possessed identical sequences. Groups CGMCC 2.4446 and CGMCC 2.3470 differed by one nt and 13 nt (∼2.5 %) mismatches in the D1/D2 and ITS regions, respectively. These four groups and De. yunnanensis differed from one another by zero to four nt in the D1/D2 domains and 9–23 nt (∼1.8–4.5 %) mismatches in the ITS region.
Based on the above sequence comparisons twelve novel species of Derxomyces are proposed in the Taxonomy section.
Note: An uncultured fungal isolate, OTU 265 (KT328670) from coffee leaf infected by an rust fungus (Hemileia vastatrix), Finca Don Julio, USA, has one nt difference with CGMCC 2.4446 in the ITS region, which indicated that this species may be also found in the USA.
Phaeotremella (Phaeotremellaceae, Tremellales)
The BLASTn searches of the D1/D2 and ITS regions revealed that the strains CGMCC 2.5810 and CGMCC 2.5614 belonged to Phaeotremella, with the best matches Phaeotremella foliacea CBS 5029 (previously named as Phaeotremella skinneri, Spirin et al. 2018) and five ‘Cryptococcus’ spp. with 98.6–98.2 % similarity in the D1/D2 domains and 94 % similarity in the ITS regions. CGMCC 2.5810 and CGMCC 2.5614 formed a subclade with the unpublished strains TFL2B (MG909557/KY614525), ‘Tremella’ sp. H-080.13 (AY188379) and ‘Cryptococcus’ sp. CBS11775 (LT904718/FN824502) in the tree of the D1/D2 dataset (Fig. 3H). CGMCC 2.5614 differed from these unpublished strains by five to ten nt substitutions in the D1/D2 domains and by 15–17 nt (∼3–3.4 %) mismatches in the ITS regions, which indicated that they belong to different species. CGMCC 2.5810 and CGMCC 2.5614 differed from TFL2B by 11–12 nt in the D1/D2 domains and more than 6 % mismatches in the ITS regions. CGMCC 2.5614 differed from Pha. foliacea (Pha. skinneri) and Pha. foliacea (voucher Miettinen 14610) by 33–37 nt (∼6.6–7.4 %) mismatches in the ITS regions and 15–16 nt substitutions (∼2.4–2.6 %) in the D1/D2 domains. CGMCC 2.5810 differed from Pha. foliacea (Pha. skinneri), Pha. foliacea (voucher Miettinen 14610) and ‘Cryptococcus’ sp. GT-388 (HQ890369) by 8–10 nt (∼1.3–1.6 %) in the D1/D2 domains, and more than 5 % mismatches in the ITS regions.
The above data indicated that CGMCC 2.5810 and CGMCC 2.5614 represent two novel species of Phaeotremella.
Note: An uncultured Tremellales clone, 5_D20 (HQ211529) from Arctic soil, Canada has 99.4 % ITS sequence similarity with CGMCC 2.5810 by blast search against the MycoBank database, which indicated that this species may be also found in other locations than China.
Holtermannia (Holtermanniaceae, Holtermanniales)
Four isolates, CGMCC 2.3445, CGMCC 2.3460, CGMCC 2.3462 and WZS12.12B, isolated from plant leaves collected in Yunnan province clustered in the genus Holtermannia (Wuczkowski et al. 2011) in the tree obtained from the three rDNA and seven-genes datasets (Figs 2E and S1E). Similar sequences were found between these four strains with no more than 3 nt differences in the ITS and D1/D2 regions. They differed from Holtermannia corniformis by five to six nt (∼1 %) substitutions in the D1/D2 domains and 23–25 nt (∼4 %) mismatches in the ITS region, which indicated that they represent a new species of Holtermannia.
Note: An uncultured endophytic fungal clone, WFc36 (KF709568) isolated from Warburgia ugandensis in Austria has two nt difference with group CGMCC 2.3445 in the ITS region, which indicated that this new species should be found outside of China. Two strains 'Holtermannia corniformis' MB128 (KC798426) and JM11 (KC510049), have identical or very similar D1/D2 sequences, which indicated that they may be conspecific.
Solicoccozyma (Piskurozymaceae, Filobasidiales)
CGMCC 2.5814 and CGMCC 2.4893 had ten nt constituting indels in the ITS regions and identical sequence in the D1/D2 domains. Similar sequences with zero to one nt difference in the D1/D2 and ITS regions were found between CGMCC 2.5814 and ‘Solicoccozyma aeria’ CBS 9627 (KY105431/KY109663) and RUB096 (MK397489), Solicoccozyma sp. DBVPG10727 (MK070335/MK070317) and ‘Cryptococcus’ sp. CRUB 2005 (KF826509). So these six strains were considered to be conspecific. They differed from the type strain of Solicoccozyma aeria by 19 nt (∼3 %) in the D1/D2 domains, and more than 4 % mismatches in the ITS regions. Therefore, a novle species of Solicoccozyma is proposed to accommodate them (Fig. 2, Fig. 3J).
Note: The uncultured eukaryote clone LTSP_EUKA_P5P23 (FJ554237) collected from the long-term soil productivity (LTSP) in Skulow Lake in Canada, had the same ITS sequence as CGMCC 2.4893. CGMCC 2.5814 and seven uncultured clones, clone 81a17 (EU554946) and clone 54a11(EU554878) collected from soil of nptII transformed poplar plantation in Canada, clone LTSP_EUKA_P4M17 (FJ553878) from LTSP from Skulow Lake in Canada, clone BF-OTU106(AM901762) from house dust in Finland, clone C4 6B (GU366710) from temperate forest soil in USA, clone 3200K2 (KF617524) from Picea mariana forest soil mineral horizon in Bonanza Creek LTER, Alaska, USA, and clone N131(JF300706) from boreal forest soil in Sweden, contain identical or only one nt difference in the ITS regions. Based on the comparison of environmental DNA sequences, the novel species (see Taxonomy section) is commonly and abundantly found in diverse locations.
Filobasidium (Filobasidiaceae, Filobasidiales)
CGMCC 2.5649 and GPS23.2A5 have identical sequences and formed a separate branch in the Filobasidium (Fig. 2, Fig. 3J and S1E). They differed from other Filobasidium species by 19 nt (∼3 %) in the D1/D2 domains and more than 11 % mismatches in the ITS region, which indicated that they represent a novel species in Filobasidium.
Seven isolates, CGMCC 2.3463, CGMCC 2.3464, CGMCC 2.4012, CGMCC 2.4052, CGMCC 2.5680, CGMCC 2.5656 and KTAPG1-11.64, formed a separate subclade distingushied from the other Filobasidium species (Fig. 2, Fig. 3J). CGMCC 2.3464 differed from CGMCC 2.4012, CGMCC 2.4052 and KTAPG1-11.64 by two nt in the D1/D2 domains and five nt in the ITS regions. CGMCC 2.5656 and CGMCC 2.5680 with identical ITS and D1/D2 sequences differed from the above three strains by one to two nt in the D1/D2 domains, and by 29–33 nt (∼5 %) mismatches in the ITS regions. CGMCC 2.3463 differed from the above two groups, represented by CGMCC 2.4012 and CGMCC 2.5680, by five to six nt substitutions in the D1/D2 domains and more than 16 % mismatches in the ITS regions. The ITS and D1/D2 sequence comparisons indicated that these seven strains could be classified into three distinct species. Therefore, three novel species are proposed to accommodate the groups CGMCC 2.5680, CGMCC 2.4012 and CGMCC 2.3463.
Note: Three strains, ‘Cryptococcus’ sp. SC15d50p10-8 (HQ631032) isolated from Saccharum officinarum in USA, O382A (JX394019) isolated from tree hollows in Brazil and CBS 10181 (EU002869/EU002805) isolated in Portugal, differ from group CGMCC 2.4012 by one to three nt in ITS region or two nt in D1/D2 domains, which indicated that they are conspecific. Seven strains, Filobasidium sp. KBP Y-5548 (MH697755/MH697755) isolated from Plumeria obtusa in Vietnam, ‘Cryptococcus’ sp. MG34 (KM246229/KM246145), IA06 (KM246189/KM246106) isolated from coffee in Brazil, 11-1115 (KM986117/KM206723) isolated from Hungary, Filobasidium sp. HB22-2 (KJ507269) isolated from flower of Caragana sinica in South Korea, ‘Cryptococcus’ sp. LB17_3 (KJ159043) isolated from the leaf-cutting ant in Brazil and GY2L20 (FJ527080) from Taiwan, China, have one to two nt difference with group CGMCC 2.4012 in the D1/D2 domain. However, the former four strains differ from group CGMCC 2.4012 by 19–37 nt (∼3–5 %) mismatches in the ITS region, which indicated that they belong to different species. The taxonomic position of the latter three strains will be fixed in future because their ITS sequences are not available at present.
Phaffia (Mrakiaceae, Cystofilobasidiales)
Strain CGMCC 2.5601 was placed in the genus Phaffia (Fig. 2, Fig. 3K and S1E). It differed from Phaffia rhodozyma by three nt substitutions in the D1/D2 domains and 7 % mismatches in the ITS regions. Although more than four potentially new species in this genus should be described (David-Palma et al. 2014, Fig. S3), only one species, P. rhodozyma, is accepted in this genus (Liu et al. 2015b). Therefore, the second Phaffia species is proposed to accommodate CGMCC 2.5601 as a novel species to improve the species diversity in Phaffia.
New species identification in the Agaricostilbomycetes (Pucciniomycotina)
Kondoa (Kondoaceae, Agaricostilbales)
Fifteen strains, representing ten candidate novel species, were placed in the Kondoa clade (Fig. 4, Fig. 5B and S2B). Group CGMCC 2.3102, containing three strains with one nt substitution in the D1/D2 domains, had identical ITS sequences with strain PYCC 5566 (AF444672) and one nt D1/D2 sequences difference (AF444766), which indicated that they were conspecific. This group differed from its closest relative Kondoa aeria by two nt and 41 nt (∼6 %) mismatches in the D1/D2 and ITS regions, respectively. Group CGMCC 2.2652, consisting of two strains with identical sequences, was closely related to Kondoa subrosea and differed from it by ten nt (∼1.6 %) and 50 nt (∼8 %) mismatches in the D1/D2 and ITS regions, respectively. Groups CGMCC 2.2621, including two strains with identical sequences, and CGMCC 2.3100 were closely related to Kondoa sorbi (Figs 4B and S2B). These two groups differed from Kon. sorbi by 25 nt (∼4 %) in the D1/D2 domains and greater than 122 nt (∼18 %) mismatches in the ITS region. Groups CGMCC 2.3100 and CGMCC 2.2621 showed five nt differences in the D1/D2 domains and more than 12 % differences in the ITS region. Group CGMCC 2.2762 showed high affinity to Kondoa changbaiensis and differed from it by three nt substitutions and 32 nt (∼5 %) mismatches in the D1/D2 and ITS region, respectively.
Groups CGMCC 2.4441, including two strains with two nt differences in the ITS regions, CGMCC 2.5610, containing two strains with one nt substitution in the D1/D2 domains, and CGMCC 2.3106 were closely related to Kondoa gutianensis. These three groups differed from Kon. gutianensis by 9–15 nt (∼1.4–2.4 %) in the D1/D2 domains and more than 75 nt (∼12 %) mismatches in the ITS region. CGMCC 2.3106 and the unpublished strain Kondoa sp. AS483 (FN428954) isolated from flower of Dianthus superbus had identical D1/D2 sequences and three nt differences in the ITS region, which indicated that they are conspecific. An uncultured corn field bulk soil clone 09D70C34 (HG937064) collected from Göttingen, Lower Saxony, Germany, and CGMCC 2.3106 had three nt differences in the ITS regions, which indicated that this candidate novel species occurs in the soil environment. Similar sequences were found between CGMCC 2.3106, CBS 8379 and the unnamed strain, Kondoa sp. ZP 352 (AY512854) with one nt substitutions in the D1/D2 domains. CGMCC 2.3106 differed from CBS 8379 (AF444596), ZP 352 (MN175326) and ZP 338 (MN175325) by two nt substitutions and five indels in the ITS region, which indicated that they may be conspecific.
Group CGMCC 2.2763 was placed in a separate branch in the trees obtained from the rDNA and seven-genes datasets (Figs 4B and S2B). This group differed from other species of Kondoa by ∼20 % mismatches in the D1/D2 domains and with even greater diversity in the ITS regions.
The above phylogenetic analysis indicated that these fifteen strains represent nine novel species in Kondoa.
Bensingtonia (Kondoaceae, Agaricostilbales)
Strains CGMCC 2.5677 and CGMCC 2.3569 were placed in two separate branches in the genus Bensingtonia (Figs 4B and S2B). CGMCC 2.5677 was closely related to Bensingtonia naganoensis and Bensingtonia pseudonaganoensis, and differed from them by four to seven nt substitutions in the D1/D2 domains and 52–60 nt (∼8–9 %) mismatches in the ITS regions. CGMCC 2.3569 had affinity to Bensingtonia bomiensis and Bensingtonia pseudonaganoensis (Fig. 5B). 20 nt (∼3.2 %) differences in the D1/D2 domains and 93–101 nt (∼14–15 %) differences in the ITS regions were observed between them.
Based on the analysis of the ITS and D1/D2 sequences two novel species of Bensingtonia, are proposed to accommodate CGMCC 2.3569 and CGMCC 2.5677.
Pseudobensingtonia (Agaricostilbaceae, Agaricostilbales)
CGMCC 2.5815, CGMCC 2.5823 and XZ152B1 have identical sequences and were placed in the Pseudobensingtonia clade (Fig. 4, Fig. 5C and S2B). They differed from Pseudobensingtonia ingoldii and Pseudobensingtonia musae by 23–24 nt (∼4 %) in the D1/D2 domains and 87–94 nt (∼14–15 %) mismatches in the ITS regions. Therefore, a new species of Pseudobensingtonia is proposed to accommodate these three strains.
Ruinenia (Ruineniaceae, Agaricostilbales)
Four strains were placed in three separate branches in the Ruinenia clade (Fig. 4, Fig. 5C and S2B). Group CGMCC 2.4426 contained two strains with identical D1/D2 and ITS sequences. This group differed from the undescribed strains, ‘Sporobolomyces’ sp. TY-139 (AY313063/AY313037), ‘Ruinenia’ sp. TW 1.1F038 (KP020109), ‘Ruinenia’ sp. TW2.1E-026 (KP020111), ‘Ruinenia’ sp. TW2.1E-041 (KP020112) and ‘Ruinenia’ sp. TW 2.1E012 (KP020114), by 6–9 (∼1–1.4 %) mismatches in the ITS regions, which indicated that these unpublished strains may represent different species. Group CGMCC 2.4426 was closely related to Ruinenia clavata (Figs 4B and S2B) and differed from Ru. clavata by more than 34 nt (∼5.5 %) and 90 nt (∼14.5 %) mismatches in the D1/D2 and ITS regions, respectively. Group CGMCC 2.3454 and Ruinenia clavata formed a subclade (Fig. 5C). They differed from each other by 31 nt (∼5 %) in the D1/D2 domains and 111 nt (∼17 %) in the ITS regions. Group CGMCC 2.4542 differed from its closest relative Ruinenia dracophylli by 13nt (∼2 %) and 27 nt (∼4 %) mismatches in the D1/D2 and ITS regions, respectively.
Based on the above phylogenetic analysis three novel Ruinenia species are proposed.
Boekhoutia (Chionosphaeraceae, Agaricostilbales)
The circumscription of new genera in the above section showed that strain CGMCC 2.4539 represents a novel genus, Boekhoutia. Consequently, a new species name for CGMCC 2.4539 is proposed in the Taxonomy section.
Note: The unpublished strain Kurtzmanomyces sp. YM25263 (KT345339) isolated from Yunnan province, China, was closely related to CGMCC 2.4539 in the tree of the D1/D2 dataset (Fig. 5A), which indicated that this strain represents a new member of Boekhoutia.
Sterigmatospora (Jianyuniaceae, Agaricostilbales)
Strain CGMCC 2.5817 has been proposed in the above circumscription of new genera to represent the new genus Sterigmatospora. Therefore, a novel species name is proposed to accommodate this strain.
Note: Strain RP146, namely Pucciniomycotina sp. (AB727125), isolated from Japan (Takashima et al. 2012) clustered with CGMCC 2.5817 in the tree of the D1/D2 dataset (Fig. 5A), which indicated that this strain represents a member of Sterigmatospora.
Pseudosterigmatospora (Jianyuniaceae, Agaricostilbales)
CGMCC 2.5816 represented the new monotypic genus Pseudosterigmatospora, that was closely related to Sterigmatospora in the new circumscribed family Jianyuniaceae (Figs 4B and S2B). A new species name is proposed for CGMCC 2.5816.
Note: ‘Bensingtonia’ sp. BI183 (EU678947), an unpublished species isolated from Brazil, was closely related to CGMCC 2.5816 in the tree drawn from the D1/D2 dataset (Fig. 5A).
New species identification in the Spiculogloeomycetes (Pucciniomycotina)
Phyllozyma (Spiculogloeaceae, Spiculogloeales)
CGMCC 2.5669 was closely related to Phyllozyma corallina and Phyllozyma dimennae (Fig. 4, Fig. 5C and S2B), and differed from them by 17–18 nt (∼3 %) substitutions in the D1/D2 domains, and 41–47 nt (∼6.2–7.2 %) mismatches in the ITS region. Strains CGMCC 2.2662 and CGMCC 2.2617 had two nt differences in the ITS region and differed from their closest relative Phyllozyma coprosmicola by seven (∼1.1 %) and eight nt (∼1.5 %) substitutions in the D1/D2 and ITS regions, respectively.
The above data indicated that these three strains represented two new species in Phyllozyma.
Meniscomyces (incertae sedis, Spiculogloeomycetes)
CGMCC 2.5818 and CGMCC 2.5681 have identical D1/D2 and ITS sequences. They belong to the new genus Meniscomyces (Figs 4B and S2B). A new species name is proposed to accommodate these two strains.
Note: An uncultured fungal clone, 103 NA2 P31 C4 (KF297104) from a soil sample in Ellef Ringnes Island, Canada, was closely related to CGMCC 2.5681 and CGMCC 2.5818 in the tree drawn from the D1/D2 dataset (Fig. 5A), which indicated that other Meniscomyces species may occur in nature.
New species identification in the Cystobasidiomycetes (Pucciniomycotina)
Sakaguchia (Sakaguchiaceae, incertae sedis)
CGMCC 2.4235 (= JCM 8162 = CBS 5143) was named as Rhodotorula araucariae in the chemotaxonomic studies of basidiomycetous yeasts (Sugiyama et al., 1985, Hamamoto et al., 1986a, Hamamoto et al., 1986b). The result from Gadanho & Sampaio (2002) indicated that IGC 5612 (= CBS 5143) was closely related to Sakaguchia dacryoides rather than Rhodotorula and Rhodosporidium. Our analysis also supported CGMCC 2.4235 was located in the Sakaguchia clade (Fig. 5, Fig. 6 and S4), which differed from its closest relatives, Sakaguchia lamellibrachii and Sakaguchia meli, by 12–20 nt (∼2.2–4 %) and 27–33 nt (∼4–6 %) mismatches in the D1/D2 and ITS regions, respectively. Thus, a new species of Sakaguchia is proposed to accommodate it in the Taxonomy section.
Note: Two strains, ‘Sakaguchia lamellibrachiae’ MTW10.1 (LC435582) isolated from water in Thailand and ‘Rhodotorula’ sp. GY28L06 (FJ527100) isolated from plant in Taiwan, China, have one to two nt D1/D2 sequences differences from CGMCC 2.4235, which indicated they may be conspecific.
Symmetrospora (Symmetrosporaceae, incertae sedis)
CGMCC 2.2613 was found to be closely related to Symmetrospora coprosmae and Symmetrospora oryzicola (Figs 4C and S2C), and differed from them by two to three nt substitutions in the D1/D2 domains and eight to nine nt (∼1.3–1.5 %) mismatches in the ITS regions, which indicated that CGMCC 2.2613 represent a different species.
Microsporomyces (Mycrosporomycetaceae, incertae sedis)
Strain CGMCC 2.4538 and its closest relative Microsporomyces magnisporus differed from each other by one nt in their D1/D2 domains. However, 11 nt (∼2 %) mismatches were found in the ITS region, which indicated that they belong to different species. Strains CGMCC 2.4444, CGMCC 2.4445 and CGMCC 2.5664 were closely related to Microsporomyces orientalis (Fig. 5D). The former two strains with identical sequences differed from Mi. orientalis by 18 nt (∼3.2 %) and 92 nt (∼15 %) mismatches in the D1/D2 and ITS regions, respectively. The latter strain differed from Mi. orientalis by 17 nt (∼3 %) in the D1/D2 domains and 81 nt (∼14 %) mismatches in the ITS region. CGMCC 2.4444 and CGMCC CGMCC 2.5664 differed from each other by 6 nt (∼1 %) substitutions and 41 nt (∼6.8 %) mismatches in the D1/D2 and ITS regions, respectively.
Based on the above sequence comparisons three novel species are proposed to accommodate groups CGMCC 2. 4538, CGMCC 2. 5664 and CGMCC 2. 4444.
Note: Group CGMCC 2.4444 and the published strain NIP038 (AB726620) contained the same sequences in the D1/D2 domains, which indicated that they may be conspecific.
Cystobasidium (Cystobasidiaceae, Cystobasidiales)
CGMCC 2.3822, CGMCC 2.3823 and CGMCC 2.3824 were placed in the Cystobasidium clade (Fig. 4, Fig. 5D and S2C) and were closely related to Cystobasidium fimetarium and Cystobasidium minutum. CGMCC 2.3822 differed from Cy. fimetarium and Cy. minutum by 8–11 nt (∼1.3–1.8 %) and 9–12 nt (∼1.5–2 %) mismatches in the D1/D2 and ITS regions, respectively. CGMCC 2.3823 and CGMCC 2.3824, having two nt differences in the ITS regions, differed from Cy. fimetarium and Cy. minutum by 9–14 nt (∼1.5–2.4 %) and 16 nt (∼2.6 %) mismatches in the D1/D2 and ITS regions, respectivley. CGMCC 2.3823 and CGMCC 2.3824 were closely related to Cystobasidium halotolerans in the D1/D tree (Fig. 5D); they differed from Cy. halotolerans by three to four nt in the ITS region, however, by 19–26 nt mismatches in the D1/D2 domain.
The above data indicated that these strains represented two novel species of Cystobasidium.
Note: CGMCC 2.3822 and the published strain TP-Snow-Y153 (JQ768912) had three nt differences in the D1/D2 domains, which indicated that they may be conspecific.
Robertozyma (incertae sedis, Cystobasidiales)
CGMCC 2.4451 and CGMCC 2.4452 had identical D1/D2 and ITS sequences. They belong to the newly described genus Robertozyma (Fig. 4, Fig. 5D and S2C). A new species is proposed to accommodate these two strains.
Begerowomyces (incertae sedis, Cystobasidiales)
The genus Begerowomyces, represented by CGMCC 2.3164, is proposed in this study (Fig. 4, Fig. 5D and S2C). Consequently, a new species name is proposed later.
Note: ‘Cystobasidium’ sp. BSB307 (KY305128) isolated from Brazil clustered with CGMCC 2.3164, which indicated that this strain should represent a new taxon in Begerowomyces.
Lichenozyma and Halobasidium (incertae sedis)
The genus Lichenozyma has been proposed to accommodate yeasts isolated from and detected in the lichen Cladonia samples (Černajová & Škaloud, 2019). Interestingly, these yeasts have been reported as common inhabitants of lichens in Europe and USA (Spribille et al., 2016, Cernajova and Skaloud, 2019). Some yeasts detected as a part of symbiotic three-partner system in the cortex of ascomycete macrolichens by Spribille et al. (2016). Although Spribille et al. (2016) suggested in their highly cited paper that these yeasts are possibly unculturable lichen associates, seven living axenic cultures have been obtained from air-dried Cladonia thali by Černajová & Škaloud (2019).
Phylogenetically Lichenozyma has been placed inside the genus Microsporomyces (Černajová & Škaloud, 2019). It is important to note that although, the dataset used by Černajová & Škaloud (2019) was largely based on the seven-genes (Wang et al., 2015a, Wang et al., 2015b), all yeasts from lichens in the analysis contained predominantly ITS sequences. Only four isolates contained all three rDNA loci, ITS, LSU and SSU. No sequence of protein-coding genes has been obtained for the genus Lichenozyma or other lichenicolous fungi, including yeasts from ascomycete macrolichens (Spribille et al. 2016) and Cyphobasidium (Millanes et al. 2016). As the result, phylogenetic analyses inferred with Bayesian Inference and Maximum Likelihood algorithms gave different topologies and several lineages identified previously identified by Wang et al., 2015a, Wang et al., 2015b has not been resolved by Černajová & Škaloud (2019). The subsequent analysis of the three rDNA loci (consisting mainly of ITS sequences) suggested that the genus Microsporomyces is polyphyletic and supported the erection of the Lichenozyma. The fact that monophyly of the Microsporomyces clade received no statistical support (both BI and ML) can be explained with very poor taxon sampling in both genus Microsporomyces and outgroups.
We included available LSU, SSU and ITS sequences of Lichenozyma pisutiana in our combined three rDNA loci, combined ITS and LSU and LSU datasets. Our analyses demonstrated that Lichenozyma pisutiana was placed inside the genus Microsporomyces with high statistical support. The genus Microsporomyces was resolved with high statistical support in the combined three rDNA loci and combined ITS (ML: 100 %) and LSU (ML: 99 %) trees (Figs 6 and S5, S6). Our analyses do not support a separate phylogenetic position of the genus Lichenozyma. Therefore, the Lichenozyma species should be transferred into Microsporomyces as synonym.
The genus Halobasidium has been proposed to accommodate a single yeast isolate from pickling sauce for a traditional high-salt fermented food in China (Guo et al. 2019). Although the presented phylogenetic tree clearly showed a separated phylogenetic position of the Halobasidium xiangyangense, the tree is very poor in terms of taxon sampling. The analysis did not include 9 out of 18 Cystobasidium species (C. alpinum, C. fimetarium, C. halotolerans, C. iriomotense, C. keelungensis, C. oligophagum, C. ongulense, C. portillonense, and C. tubaki), including the type species of the genus C. fimetarium, Occultifur mephitis, and numerous sequences representing yet undescribed yeast species in Cystobasidiales. Among sequences representing potential new taxa in Cystobasidiaceae, Cystobasidiomycetes sp. DSM 28479 (NCBI Taxonomy ID 1524830) and JS-40 (NCBI Taxonomy ID 1082630) showed 99 and 97 % (LSU) and 97 and 95 % (ITS) similarity to Halobasidium xiangyangense, respectively. These yeasts are likely to represent closely related species (DSM 28479) or conspecific isolates (JS-40). It is important to note that Cystobasidiomycetes sp. DSM 28479 (cited as Rhodotorula sp. MB27) has been found to be the closest outgroup to the Cystobasidium - Occultifur clade by Yurkov et al. (2015b). Phylogenetic placement presented by Guo et al. (2019) contradicts larger phylogenetic analyses published by Yurkov et al. (2015b), who showed that LSU and combined rDNA phylogenies are able to resolve genera Cystobasidium, Occultifur and “Rhodotorula” sp. MB27.
Phylogenetic analysis performed in the present study showed that LSU alone is not sufficient to resolve genera in Cystobasidiaceae, including Cystobasidium, Halobasidium, Occultifur, Queiroziella and newly proposed Begerowomyces and Robertozyma (Fig. S5). Combined phylogenetic analyses of the rDNA cistron and the seven-genes analysis resolved this genus with good statistical support (Fig. 4A). The constrained LSU analysis confirmed that the two aforementioned Cystobasidiomycetes belong to the genus Halobasidium (Figs 5D and S5).
With these examples we would like to show that good taxon sampling is essential for phylogenetic studies and taxonomy of basidiomycetous yeasts. A particular attention should be given to newly erected monotypic yeast genera, which should be preferably circumscribed using multi-gene phylogenies, as in the case of recent descriptions of genera Heitmania, Libkindia and Yurkovia (Liu et al., 2017, Mašínová et al., 2017).
New species identification in the Microbotryomycetes (Pucciniomycotina)
Rhodosporidiobolus (Sporidiobolaceae, Sporidiobolales)
Three groups, represented by CGMCC 2.3532, CGMCC 2.4435 and CGMCC 2.3118, were located in the Rhodosporidiobolus clade (Fig. 4, Fig. 5E and S2D). Group CGMCC 2.3532, containing two strains with identical sequences, clustered with Rhodosporidiobolus poonsookiae and Rhodosporidiobolus ruineniae without support in the tree obtained from the D1/D2 dataset (Fig. 5E), but it was located in a different place in the trees of the three rDNA loci and seven-genes datasets (Figs 4D and S2D). The BLASTn searches of the ITS and D1/D2 indicated that CGMCC 2.3532 are more related to Rh. ruineniae than other species. This group differed from Rh. ruineniae by 16 nt (∼3 %) in the D1/D2 domains and 32 nt (∼5 %) mismatches in the ITS region. Group CGMCC 2.4435, consisting of three strains with one nt difference in the D1/D2 domains, differed from its closest relative, Rhodosporidiobolus lusitaniae by eight nt (∼1.3 %) and 22 nt (∼4 %) mismatches in the D1/D2 and ITS regions, respectively. Group CGMCC 2.3118 was most closely related to Rhodosporidiobolus nylandii and differed from it by five nt and 14 nt (∼3 %) mismatches in the D1/D2 and ITS regions, respectively.
The sequence comparisons showed that these three groups represent three distinct novel species in Rhodosporidiobolus.
Note: Three unpublished strains ‘Sporobolomyces’ sp. Vega180 (EU002899), ‘Sporobolomyces’ sp. Vega122 (EU009966) and Rhodosporidiobolus sp. Vega175 (MG471376) isolated from Coffea in Puerto Rico, USA, and ‘Rhodosporidiobolus odoratus’ AUMC 10780 (KY495748) isolated from fresh guava juice collected from shops in Assiut city, Egypt, had one to two nt difference from CGMCC 2.3118 in the ITS region, which indicated that they are conspecific. ‘Sporidiobolus’ sp. ST-88 (DQ404450) and ‘Sporidiobolus’ sp. ST-90 (DQ404451) differed from CGMCC 2.3532 group by three to five nt in the D1/D2 domains, however, the ITS sequences of those two strains are not available. Therefore, the taxonomic positions of those two strains were not delineated.
Sporobolomyces (Sporidiobolaceae, Sporidiobolales)
Twelve strains forming four groups clustered in the genus Sporobolomyces based on the sequence analysis of the seven genes and rDNA loci datasets (Fig. 4, Fig. 5E and S2D). CGMCC 2.5675, CGMCC 2.5687 and two published strains, ‘Sporobolomyces aff. jilinensis’ MCA 3774 (JN942193/JN940715) and MCA 3785 (JN942199/JN940720) had identical D1/D2 and ITS sequences. They differed from Sporobolomyces jilinensis by 12 nt (∼2 %) and 10 nt (∼1.7 %) mismatches in the D1/D2 and ITS regions, respectively. Groups CGMCC 2.5627 represented by a single strain. Group CGMCC 2.5619 contained eight Chinese strains and one strain from UK (CBS 2642) with identical D1/D2 sequences and three nt ITS sequences difference, which indicated that they are conspecific. Groups CGMCC 2.5619 and CGMCC 2.5627 were most closely related to Sporidiobolus metaroseus and Sporobolomyces roseus (Fig. 5E). Group CGMCC 2.5627 differed from these two species by two to three nt in the D1/D2 domains and nine nt (∼1.5 %) in the ITS regions. Group CGMCC 2.5619 differed from them by five to six nt (∼1 %) in the D1/D2 domains and 19 nt (∼3 %) in the ITS regions. The two new groups differed from each other by four nt and 18 nt (∼3 %) in the D1/D2 and ITS regions, respectively. Based on the above sequence analyses three novel species of Sporobolomyces, are proposed to accommodate the groups CGMCC 2. 5675, CGMCC 2. 5627 and CGMCC 2. 5619.
The placement of IAM 13481 was not stable in the trees from the three datasets (Fig. 4, Fig. 5E and S2D). The BLASTn searches of the ITS and D1/D2 sequences showed that IAM 13481 had the highest match with Sporobolomyces ruberrimus and differed from it by 12 nt (∼2 %) in both the D1/D2 and ITS regions. Originally IAM 13481 (= YK 419) was designated as Sporobolomyces roseus (Yamazaki & Komagata, 1983). Valério et al. (2008) indicated that this strain was incorrectly named and did not belong to Sp. roseus. Since then this strain was treated as an unnamed taxon of Sporobolomyces (Valério et al. 2008). However, the genome of this strain (http://genome.jgi.doe.gov/pages/search-for-genes.jsf?organism=Sporo1b) had been sequenced by the Joint Genome Institute (http://www.jgi.doe.gov) ten years ago, which was the first Pucciniomycotina species with a genome sequence. After the genome was released, the genetic and genomic studies of degrading mycotoxin and mating type genes in the basidiomycetous yeasts based on this strain have been reported (Coelho et al., 2008, Coelho et al., 2011, Ianiri et al., 2013, Ianiri et al., 2016). Unfortunately, a formal name has been unavailable for this strain until now, therefore, a new species name of Sporobolomyces is proposed to accommodate it.
Heitmania (Heitmaniaceae, Heitmaniales)
Two isolates, CGMCC 2.5602 and CGMCC 2.5650, formed a subclade in Heitmania (Figs 4D and S2D). These two strains differed from each other by four nt substitutions and 40 nt (∼6 %) mismatches in the D1/D2 and ITS regions, respectively. CGMCC 2.5602 differed from the other Heitmania species, Heitmania litseae, Heitmania elacocarpi and Heitmania castanopsis, by six to eight nt (∼1.0–1.4 %) substitutions and 86 nt (∼14 %) mismatches in the D1/D2 and ITS regions, respectively. The differences between CGMCC 2.5650 and the other three known Heitmania species ranged between four to six nt in the D1/D2 domains and were greater than 15 % in the ITS regions. Two novel species are suggested to accommodate these two strains.
Microbotryozyma (Ustilentylomataceae, Microbotryales)
The genus Microbotryozyma contains a single species, namely Microbotryozyma collariae, and was located in the family Ustilentylomataceae (Fig. 4, Fig. 5F and S2E). Strain CGMCC 2.3533 differed from Mi. collariae by six nt (∼1 %) and 57nt (∼11 %) in the D1/D2 and ITS regions, respectively. Therefore, a novel species is suggested to accommodate this strain.
Yamadamyces (Kriegeriaceae, Kriegeriales)
CGMCC 2.5820 clustered with the monotypic genus Yamadamyces in the trees obtained from the seven-genes as well as three rDNA loci datsets (Figs 4E and S2E). One nt difference in the D1/D2 domains and 56 nt (∼10 %) mismatches in the ITS regions were found between CGMCC 2.5820 and Yamadamyces rosulatus, which indicated that CGMCC 2.5820 could represent a novel Yamadamyces species .
Oberwinklerozyma (incertae sedis)
Groups CGMCC 2.3441 and CGMCC 2.5824 were located in the Oberwinklerozyma clade (Fig. 4, Fig. 5G and S2E). CGMCC 2.5824 and four unpublished strains labeled as ‘Rhodotorula’ sp. n-w29 (LC326052) and ‘Rhodotorula’ sp. BI157 (EU678941), Chrysozymaceae sp. SJ13L05 (EU523609/FJ153202) and ‘Rhodotorula’ sp. GY23L16 (HQ623608/FJ527098) contained identical sequences in the D1/D2 domains, and the latter two strains had two to three nt substitutions with CGMCC 2.5824 in the ITS regions, which indicated that they are conspecific. They differed from Oberwinklerozyma yarrowii by seven nt (∼1.2 %) and more than 40 nt (∼6 %) mismatches in the D1/D2 and ITS regions, respectively. CGMCC 2.3441 occupied a separated bottom branch in the Oberwinklerozyma clade. It differed from the other Oberwinklerozyma species by more than six nt (∼1 %) in the D1/D2 domains and 68 nt (∼11 %) mismatches in the ITS regions, respectively.
The phylogenetic analysis showed that these strains represented two novel species in Oberwinklerozyma.
Chrysozyma (Chrysozymaceae, incertae sedis)
The genus Chrysozyma, containing the two species Chrysozyma griseoflava and Chrysozyma fushanensis, was recently proposed (Wang et al. 2015b) based on the phylogenetic analysis of seven genes. Thirteen strains formed eight groups and were all closely related to Ch. griseoflava (Fig. 4, Fig. 5H and S2E). Group CGMCC 2.5629 consisted of three strains that had one nt difference in the D1/D2 domains. They differed from Ch. griseoflava by seven nt (∼1.1 %) and nine nt (∼1.3 %) substitutions in the D1/D2 and ITS regions, respectively. Groups CGMCC 2.2618 and CGMCC 2.2765, both containing two strains with identical sequences, differed from Ch. griseoflava by three to nine nt (∼0.5–1.5 %) and 20–54 nt (∼3–8 %) mismatches in the D1/D2 and ITS regions, respectively. Two strains in the CGMCC 2.2768 group had similar sequences with two nt and four nt in the D1/D2 and ITS regions, respectively. Six nt differences in the D1/D2 domains and 64 nt (∼9 %) in the ITS regions were found between the CGMCC 2.2768 group and Ch. griseoflava. Groups CGMCC 2.5821, CGMCC 2.2769 and CGMCC 2.3455, all represented by only a single strain, differed from Ch. griseoflava by two to four nt in the D1/D2 domains and 71–75 nt (∼10–11 %) mismatches in the ITS regions. Group CGMCC 2.5611 and ‘Rhodotorula’ sp. DSM 101778 (KX067789) published by Prior et al. (2017), had three nt differences in the ITS regions, which indicated that they are conspecific. This group differed from Ch. griseoflava by 11 nt (∼1.8 %) and 84 nt (∼14 %) mismatches in the D1/D2 and ITS regions, respectively.
The above sequence comparisons indicated that these eight groups represent eight novel taxa in Chrysozyma.
Yurkovia (Chrysozymaceae, incertae sedis)
Analysis of the sequences of the ITS region and D1/D2 domains suggested that CGMCC 2.5603 has affinity to the genus Yurkovia (Mašínová et al. 2017) (Fig. 4, Fig. 5H and S2E). Four nt and 40 nt (∼7 %) differences in the D1/D2 and ITS regions, respectively, were observed between CGMCC 2.5603 and Yurkovia mendeliana. CGMCC 2.5603 had identical sequences as Yurkovia nerthusi in the D1/D2 domains, however, they differed from each other by 39 (∼6 %) mismatches in the ITS region, which indicated that CGMCC 2.5603 represents a novel species in Yurkovia.
Pseudohyphozyma (incertae sedis)
The seven strains CGMCC 2.2612, CGMCC 2.2796, CGMCC 2.2797, CGMCC 2.5607, CGMCC 2.5618, CGMCC 2.5623 and GPS23.3D2 were located in the Pseudohyphozyma clade, forming two groups (Fig. 4, Fig. 5H and S2E). Group CGMCC 2.2612 and Pseudohyphozyma bogoriensis differed from each other by four nt and 22 nt (∼4 %) mismatches in the D1/D2 and ITS regions, respectively. Group CGMCC 2.2796 contained six strains with one nt difference in the ITS regions and differed from its closest relative Pseudohyphozyma pustula by five nt and 22nt (∼4 %) mismatches in the D1/D2 and ITS regions, respectively.
The above sequence comparisons indicated that these seven strains represent two novel species in Pseudohyphozyma.
Slooffia (incertae sedis)
Strain CGMCC 2.5822 was placed in the genus Sloofia with 100 % bootstrap support (Fig. 4, Fig. 5H and S2E). It had identical D1/D2 sequences with Slooffia tsugae and 37 nt (∼5 %) differences in the ITS region, which indicated that CGMCC 2.5822 represents a different species.
Colacogloea (Colacogloeaceae, incertae sedis)
Eight strains formed three groups in the Colacogloea clade (Fig. 4, Fig. 5G and S2E). Group CGMCC 2.2766 differed from its closest relative Colacogloea cycloclastica by 32 nt (∼5 %) in the D1/D2 domains and more than 13 % mismatches in the ITS region. Groups CGMCC 2.2798 and CGMCC 2.2770, consisting of six strains with one nt and two nt difference in the D1/D2 and ITS regions, respectively, were closely related to Colacogloea diffluens. The two groups differed from Co. diffluens by 22–26 nt (∼3.6–4.3 %) in the D1/D2 domains, and more than 14 % mismatches in the ITS regions. Groups CGMCC 2.2798 and CGMCC 2.2770 differed from each other by 26 nt (∼4.3 %) substitutions and 8.8 % mismatches in the D1/D2 and ITS regions, respectively.
The sequence comparisons indicated that these eight strains represent three novel species in Colacogloea.
Rosettozyma (Rosettozymaceae, Rosettozymales)
Six strains, separated in three groups were located in the newly circumscribed genus Rosettozyma (Fig. 4, Fig. 5G and S2E). CGMCC 2.3446, CGMCC 2.3461 and CGMCC 2.3466 had one nt difference in the D1/D2 domains and shared the same ITS sequences, which indicated that they are conspecific. CGMCC 2.2615 and CGMCC 2.2619 had one and three nt differences in the D1/D2 and ITS regions, respectively, which indicated that they belong to the same species. Group CGMCC 2.2615 differed from the CGMCC 2.3446 group by four to six nt and 12–14 nt (∼2–2.3 %) mismatches in the D1/D2 and ITS regions, respectively. CGMCC 2.5819 differed from groups CGMCC 2.2615 and CGMCC 2.3446 by 28–30 nt (∼4.7–5 %) in the D1/D2 domains and 30–43 nt (∼5–7.2 %) mismatches in the ITS regions.
The above data indicated that these six strains represent three novel Rosettozyma species.
Taxonomy
New taxa in Tremellomycetes (Agaricomycotina)
Kockovaella haikouensis Q.M. Wang, F.Y. Bai & A.H. Li sp. nov. MycoBank MB828736. Fig. 7A, B.
Fig. 7.
Vegetative cells grown in YM broth for 5 d at 17 °C and ballistoconidia produced on corn meal agar after 7 d at 17 °C. (A, B) Koc. haikouensis CGMCC 2.3443T; (C, D) Koc. ischaemi CGMCC 2.3565T; (E) Koc. nitrophila CGMCC 2.3465T; (F) G. pseudoamylolytica CGMCC 2.5809T; (G) Tr. shuangheensis CGMCC 2.5615T; (H) V. melezitolyticum CGMCC 2.3472T; (I) V. pseudopenaeus CGMCC 2.3165T; (J) V. europaea CGMCC 2.3099T; (K) Ca. follicola CGMCC 2.3447T; (L) Ca. simaoensis CGMCC 2.3580T; (M) Kwoni. ovata CGMCC 2.3439T; (N) Te. helanensis CGMCC 2.4450T; (O) Te. globosa CGMCC 2.5648T; (P) Te. korlaensis CGMCC 2.3835T. Bars = 10 μm.
Etymology: the specific epithet haikouensis refers to the geographic origin of the type strain, Haikou county, Hainan.
Culture characteristics: In YM broth, after 7 d at 17 °C, cells are ellipsoidal or ovoid, 1.8–3.5 × 2.5–5.0 μm and single, budding is polar (Fig. 7A), a sediment is formed. After 1 mo at 17 °C, a ring and sediment are present. On YM agar, after 1 mo at 17 °C, the streak culture is yellowish-cream, butyrous, smooth and glistening. The margin is entire. In Dalmau plate culture on CM, pseudohyphae are not formed. Sexual structures are not observed on YM, PDA, V8 and CM agar. Ballistoconidia are ellipsoidal or some what kidney-shaped, 3.3–5.0 × 5.0–8.3 μm (Fig. 7B).
Physiological and biochemical characteristics: Glucose fermentation is absent. Glucose, galactose, sucrose, maltose, cellobiose, trehalose, lactose, melibiose, raffinose, melezitose, soluble starch (variable), D-xylose (variable), L-arabinose (variable), D-ribose, D-glucosamine, N-Acetyl-D-glucosmine, ethanol (variable), glycerol (variable), erythritol (variable), ribitol (variable), galactitol, D-mannitol, methyl α-D-glucoside, salicin, DL-lactate(variable), succinate (variable) are assimilated as sole carbon sources. L-sorbose, inulin, D-arabinose, L-rhamnose, methanol, D-glucitol, citrate, myo-inositol and hexdecane are not assimilated. Ammonium sulfate, L-lysine (variable), ethylamine hydrochloride (delayed) and cadaverine dihydrochloride are assimilated as sole nitrogen sources. Potassium nitrate and sodium nitrite are not assimilated. Maximum growth temperature is 30 °C. Growth in vitamin-free medium is positive. Starch-like substances are not produced. Growth on 50 % (w/w) glucose-yeast extract agar is negative. Urease activity is positive. Diazonium Blue B reaction is positive. The major ubiquinone is Q-10.
Physiologically, Koc. haikouensis differs from the closely related species Koc. ischaemi in its inability to assimilate inulin, D-arabinose, L-rhamnose and sodium nitrite and its ability to assimilate ethylamine (Table S1.1).
Typus: China, Haikou county, Hainan province, obtained from a leaf of an unidentified plant, Nov. 2006, Q.-M. Wang (holotype CGMCC 2.3443T preserved in a metabolically inactive state, ex-type CBS 15478 = HKX2).
Kockovaella ischaemi Q.M. Wang, F.Y. Bai & A.H. Li sp. nov. MycoBank MB828738. Fig. 7C, D.
Etymology: the specific epithet ischaemi refers to Ischaemum, the plant genus from which the type strain was isolated.
Culture characteristics: In YM broth, after 7 d at 17 °C, cells are ellipsoidal or ovoid, 2.0–3.8 × 2.3–6.2 μm and single or pairs, budding is polar (Fig. 7C), blastoconidia are produced on short stalk-like conidiophores, a sediment is formed. After 1 mo at 17 °C, a ring and sediment are present. On YM agar, after 1 mo at 17 °C, the streak culture is yellowish-cream, butyrous, smooth and glistening. The margin is entire. In Dalmau plate culture on corn meal agar, pseudohyphae are not formed. Sexual structures are not observed on YM, PDA, V8 and CM agar. Ballistoconidia are ellipsoidal or some what kidney-shaped, 2.0–3.7 × 4.2–6.7 μm (Fig. 7D).
Physiological and biochemical characteristics: Glucose fermentation is absent. Glucose, galactose, sucrose, maltose, cellobiose, trehalose, lactose, melibiose, raffinose, melezitose, inulin, soluble starch, D-xylose, L-arabinose, D-arabinose (weak), D-ribose, L-rhamnose, D-glucosamine, N-Acetyl-D-glucosamine, glycerol (variable), ribitol (variable), galactitol, D-mannitol, Methyl-α-D-glucoside (variable), salicin (weak), succinate (weak), citrate (variable) and myo-Inositol (variable) are assimilated as sole carbon sources. L-sorbose, methanol, ethanol, erythritol, D-glucitol, DL-lactate and hexadecane are not assimilated. Ammonium sulfate, sodium nitrite and cadaverine dihydrochloride are assimilated as sole nitrogen sources. Potassium nitrate, L-lysine and ethylamine hydrochloride are not assimilated. Maximum growth temperature is 30 °C. Growth in vitamin-free medium is positive. Starch-like substances are not produced. Growth on 50 % (w/w) glucose-yeast extract agar is negative. Urease activity is positive. Diazonium Blue B reaction is positive. The major ubiquinone is Q-10.
Physiologically, Koc. ischaemi differs from the closely related species Koc. haikouensis in its inability to assimilate ethylamine and its ability to assimilate inulin, D-arabinose, L-rhamnose and sodium nitrite (Table S1.1).
Typus: China, Jinghong, Yunnan province, obtained from a leaf of Ischaemum sp., Nov. 2006, Q.-M. Wang (holotype CGMCC 2.3565T preserved in a metabolically inactive state, ex-type CBS 15500 = JH5.17)
Kockovaella nitrophila Q.M. Wang, F.Y. Bai & A.H. Li sp. nov. MycoBank MB828739. Fig. 7E.
Etymology: the specific epithet nitrophila refers to the physiological character of assimilating nitrate.
Culture characteristics: In YM broth, after 7 d at 17 °C, cells are subglobosal and ellipsoidal, 2.4–4.4 × 3.7–4.5 μm and single, budding is polar (Fig. 7E), a sediment is formed. After 1 mo at 17 °C, a sediment is present. On YM agar, after 1 mo at 17 °C, the streak culture is creamish white, butyrous, smooth. The margin is entire. In Dalmau plate culture on corn meal agar, pseudohyphae are not formed. Sexual structures are not observed on YM, PDA, V8 and CM agar. Ballistoconidia are not produced.
Physiological and biochemical characteristics: Glucose fermentation is absent. Glucose, galactose (weak), sucrose, maltose, trehalose, melibiose (weak), raffinose, melezitose, inulin, D-xylose (weak), L-arabinose (weak), D-arabinose (weak), D-ribose (weak) and DL-lactate (weak) are assimilated as sole carbon sources. L-sorbose, cellobiose, lactose, soluble starch, L-rhamnose, D-glucosamine, N-Acetyl-D-glucosamine, methanol, ethanol, glycerol, erythritol, ribitol, galactitol, D-mannitol, D-glucitol, Methyl-α-D-glucoside, salicin, succinate, citrate, myo-inositol and hexadecane are not assimilated. Ammonium sulfate, potassium nitrate, L-lysine (weak), ethylamine hydrochloride (weak) and cadaverine dihydrochloride (weak) are assimilated as sole nitrogen sources. Sodium nitrite is not assimilated. Maximum growth temperature is 37 °C. Growth in vitamin-free medium is positive (weak). Starch-like substances are not produced. Growth on 50 % (w/w) glucose-yeast extract agar is negative. Urease activity is positive. Diazonium Blue B reaction is positive. The major ubiquinone is Q-10.
Physiologically, Koc. nitrophila differs from its five closely related species, Koc. ischaemi, Koc. haikouensis, Koc. sacchari, Koc. thailandica and Koc. imperatae, in its inability to assimilate cellobiose, melibiose, D-glucosamine, N-Acetyl-D-glucosamine and D-mannitol and its ability to assimilate potassium nitrate (Table S1.1).
Typus: China, Wuzhishan mountain, Hainan province, obtained from a leaf of an unidentified plant, Nov. 2006, Q.-M. Wang (holotype CGMCC 2.3465T preserved in a metabolically inactive state, ex-type CBS 15487 = WZS12.1).
Genolevuria pseudoamylolytica Q.M. Wang, F.Y. Bai & A.H. Li sp. nov. MycoBank MB828740. Fig. 7F.
Etymology: the specific epithet pseudoamylolytica refers to the similar colony morphology to that of Genolevuria amylolytica.
Culture characteristics: In YM broth, after 7 d at 17 °C, cells are ellipsoidal and subglobosal, 2.9–5.2 × 3.3–7.7 μm and single, budding is polar (Fig. 7F), a sediment is formed. After 1 mo at 17 °C, a ring and sediment are present. On YM agar, after 1 mo at 17 °C, the streak culture is cream, mucoid, smooth and glistening. The margin is entire. In Dalmau plate culture on corn meal agar, pseudohyphae are not formed. Sexual structures are not observed on YM, PDA, V8 and CM agar. Ballistoconidia are not produced.
Physiological and biochemical characteristics: Glucose fermentation is absent. Glucose, galactose, L-sorbose, sucrose, maltose, cellobiose, trehalose, lactose, melibiose, raffinose, melezitose, inulin (weak), soluble starch, D-xylose (weak), L-arabinose, D-arabinose, D-ribose, L-rhamnose (weak), D-glucosamine, N-Acetyl-D-glucosamine, D-mannitol (weak), D-glucitol (weak), Methyl-α-D-glucoside and salicin are assimilated as sole carbon sources. Methanol, ethanol, glycerol, erythritol, ribitol, galactitol, DL-lactate, succinate, citrate, myo-inositol and hexadecane are not assimilated. Ammonium sulfate, potassium nitrate, sodium nitrite, L-lysine, ethylamine hydrochloride and cadaverine dihydrochloride are assimilated as sole nitrogen sources. Maximum growth temperature is 28 °C. Growth in vitamin-free medium is negative. Starch-like substances are not produced. Growth on 50 % (w/w) glucose-yeast extract agar is negative. Urease activity is positive. Diazonium Blue B reaction is positive.
Physiologically, G. pseudoamylolytica differs from the two closely related species, G. amylolytica and G. tibetensis, in its inability to assimilate ribitol and succinate and the ability to assimilate L-sorbose and potassium nitrate (Table S1.2).
Typus: China, Daliangzi river national forest park, Heilongjiang province, obtained from a leaf of an unidentified plant, Aug. 2014, Q.-M. Wang (holotype CGMCC 2.5809T preserved in a metabolically inactive state, ex-type CBS 13955 = HLJ1B6).
Tremella shuangheensis Q.M. Wang, F.Y. Bai & A.H. Li sp. nov. MycoBank MB828741. Fig. 7G.
Etymology: the specific epithet shuangheensis refers to the geographic origin of the type strain, Shuanghe county, Heilongjiang.
Culture characteristics: In YM broth, after 7 d at 17 °C, cells are subglobosal and ellipsoidal, 3.2–4.6 × 4.0–5.5 μm and single, budding is polar (Fig. 7G), a sediment is present. After 1 mo at 17 °C, a ring and sediment are present. On YM agar, after 1 mo at 17 °C, the streak culture is yellowish-cream, mucoid, smooth and glistening. The margin is entire. In Dalmau plate culture on corn meal agar, pseudohyphae are not formed. Sexual structures are not observed on YM, PDA, V8 and CM agar. Ballistoconidia are not produced.
Physiological and biochemical characteristics: Glucose fermentation is absent. Glucose, galactose (delayed and weak), L-sorbose (weak), sucrose, maltose, cellobiose, trehalose, lactose (delayed), melibiose (delayed), melezitose (delayed and weak), inulin (delayed), soluble starch (delayed and weak), D-xylose, L-arabinose (weak), D-arabinose (delayed and weak), D-ribose (delayed and weak), L-rhamnose (delayed and weak), D-glucosamine (delayed and weak), N-Acetyl-D-glucosamine (delayed and weak), ethanol (delayed and weak), glycerol, erythritol, ribitol, galactitol, D-mannitol (delayed and weak), D-glucitol (delayed and weak), Methyl-α-D-glucoside (delayed and weak), salicin (delayed and weak), D-gluconate (delayed and weak), DL-lactate (delayed and weak), succinate (delayed and weak) and myo-inositol (delayed and weak) are assimilated as sole carbon sources. Raffinose, methanol, citrate and hexadecane are not assimilated. Ammonium sulfate, L-lysine, ethylamine hydrochloride and cadaverine dihydrochloride are assimilated. Potassium nitrate and sodium nitrite are not assimilated. Maximum growth temperature is 28 °C. Growth in vitamin-free medium is delayed. Starch-like substances are not produced. Growth on 50 % (w/w) glucose-yeast extract agar is negative. Urease activity is positive. Diazonium Blue B reaction is positive.
Physiologically, T. shuangheensis differs from the closely related species T. globispora in its ability to assimilate lactose, melibiose, inulin and the inability to assimilate citrate (Table S1.3).
Typus: China, Shuanghe county, Heilongjiang province, obtained from a leaf of an unidentified plant, Aug. 2015, Q.-M. Wang (holotype CGMCC 2.5615T preserved in a metabolically inactive state, ex-type CBS 15561 = SH58A1).
Vishniacozyma melezitolytica Q.M. Wang, F.Y. Bai & A.H. Li sp. nov. MycoBank MB828742. Fig. 7H.
Etymology: the specific epithet melezitolytica refers to the physiological character of assimilating melezitose.
Culture characteristics: In YM broth, after 7 d at 17 °C, cells are ellipsoidal, 2.6–5.0 × 3.9–6.1 μm and single, budding is polar (Fig. 7H), a sediment is formed. After 1 mo at 17 °C, a pellicle and a sediment are present. On YM agar, after 1 mo at 17 °C, the streak culture is brownish-cream, butyrous, glistening and smooth. The margin is entire. In Dalmau plate culture on corn meal agar, pseudohyphae are not formed. Sexual structures are not observed on YM, PDA, V8 and CM agar. Ballistoconidia are not produced.
Physiological and biochemical characteristics: Glucose fermentation is absent. Glucose, galactose, L-sorbose (variable), sucrose, maltose, cellobiose, trehalose, lactose, raffinose, melezitose, inulin (variable), D-xylose, L-arabinose, D-arabinose (variable), D-ribose (variable), L-rhamnose, N-Acetyl-D-glucosamine (variable), D-glucosamine (variable), ethanol, glycerol, ribitol (variable), galactitol (variable), D-mannitol, D-glucitol (variable), Methyl-α-D-glucoside (variable), salicin (weak), succinate (variable) and myo-inositol (variable) are assimilated as sole carbon sources. Melibiose, soluble starch, methanol, erythritol, D-gluconate, DL-lactate, citrate and hexadecane are not assimilated. Ammonium sulfate, potassium nitrate (variable), L-lysine, ethylamine hydrochloride (variable) and cadaverine dihydrochloride (variable) are assimilated as sole nitrogen sources. Sodium nitrite is not assimilated. Maximum growth temperature is 30 °C. Growth in vitamin-free medium is variable. Starch-like substances are not produced. Growth on 50 % (w/w) glucose-yeast extract agar is negative. Urease activity is positive. Diazonium Blue B reaction is positive.
Physiologically, V. melezitolytica differs from the closely related species V. dimennae and V. globispora in its inability to assimilate DL-lactate and citrate and its ability to assimilate melezitose (Table S1.4).
Typus: China, Hebei province, obtained from a leaf of an unidentified plant, Apr. 2007, Q.-M. Wang (holotype CGMCC 2.3472T preserved in a metabolically inactive state, ex-type CBS 15490 = H5A3).
Vishniacozyma pseudopenaeus Q.M. Wang, F.Y. Bai & A.H. Li sp. nov. MycoBank MB828743. Fig. 7I.
Etymology: the specific epithet pseudopenaeus refers to the similar colony morphology and physiological characteristics to that of Vishniacozyma penaeus.
Culture characteristics: In YM broth, after 7 d at 17 °C, cells are subsphaeroidal and ellipsoidal, 2.6–3.5 × 2.8–5.0 μm and single, budding is polar (Fig. 7I), a sediment is formed. After 1 mo at 17 °C, a pellicle and a sediment are present. On YM agar, after 1 mo at 17 °C, the streak culture is pale grayish-cream, mucoid, smooth and glistening. The margin is entire. In Dalmau plate culture on corn meal agar, pseudohyphae are not formed. Sexual structures are not observed on YM, PDA, V8 and CM agar. Ballistoconidia are not produced.
Physiological and biochemical characteristics: Glucose fermentation is absent. Glucose, galactose, L-sorbose, sucrose, maltose, cellobiose, trehalose, lactose, melibiose, raffinose, melezitose, soluble starch (varialbe), D-xylose, L-arabinose, D-arabinose, D-ribose, L-rhamnose, D-glucosamine, D-gluconate, ethanol (varialbe), glycerol, erythritol (varialbe), ribitol, galactitol, D-mannitol, D-glucitol, Methyl-α-D-glucoside, salicin, , DL-lactate (varialbe), succinate (weak), citrate and myo-inositol are assimilated as sole carbon sources. Inulin, methanol and hexadecane are not assimilated. Ammonium sulfate, potassium nitrate (variable), L-lysine, ethylamine hydrochloride (weak) and cadaverine dihydrochloride (variable) are assimilated as sole nitrogen sources. Sodium nitrite are not assimilated as sole nitrogen sources. Maximum growth temperature is 32 °C. Growth in vitamin-free medium is positive. Starch-like substances are produced or not. Growth on 50 % (w/w) glucose-yeast extract agar is negative. Urease activity is positive. Diazonium Blue B reaction is positive.
Physiologically, V. pseudopenaeus differs from the closely related species V. penaeus in its ability to grow in vitamin-free medium, however, the latter does not grow in vitamin-free medium (Table S1.4).
Typus: Germany, obtained from a leaf of an unidentified plant, Sep. 2005 (holotype CGMCC 2.3165T preserved in a metabolically inactive state, ex-type CBS 15472 = G7.20).
Vishniacozyma europaea Q.M. Wang, F.Y. Bai & A.H. Li sp. nov. MycoBank MB828744. Fig. 7J.
Etymology: the specific epithet europaea refers to the geographic origin of the type strain, Europe.
Culture characteristics: In YM broth, after 7 d at 17 °C, cells are subglobosal and ellipsoidal, 2.4–4.8 × 3.0–9.6 μm and single, budding is polar (Fig. 7J), a sediment is present. After 1 mo at 17 °C, a ring and sediment are present. On YM agar, after 1 mo at 17 °C, the streak culture is yellowish cream, butyrous, smooth. The margin is entire. In Dalmau plate culture on corn meal agar, pseudohyphae are not formed. Sexual structures are not observed on YM, PDA, V8 and CM agar. Ballistoconidia are not produced.
Physiological and biochemical characteristics: Glucose fermentation is absent. Glucose, galactose, sucrose, maltose, cellobiose, trehalose, lactose, melibiose, raffinose, melezitose, soluble starch, D-xylose, L-arabinose, D-arabinose, D-ribose, L-rhamnose, D-glucosamine, ethanol (delayed and weak), glycerol (delayed and weak), erythritol, ribitol, galactitol, D-mannitol, D-glucitol, Methyl-α-D-glucoside, salicin, succinate, citrate (weak) and myo-inositol are assimilated as sole carbon sources. L-sorbose, inulin, methanol, DL-lactate and hexadecane are not assimilated. Ammonium sulfate, potassium nitrate, sodium nitrite, L-lysine, ethylamine hydrochloride (delayed and weak) and cadaverine dihydrochloride (delayed and weak) are assimilated as sole nitrogen sources. Maximum growth temperature is 23 °C. Growth in vitamin-free medium is positive. Starch-like substances are not produced. Growth on 50 % (w/w) glucose-yeast extract agar is negative. Urease activity is positive. Diazonium Blue B reaction is positive.
Physiologically, V. europaea differs from the closely related species V. foliicola in its inability to produce starch-like substances and its ability to assimilate soluble starch and potassium nitrate (Table S1.4).
Typus: Germany, obtained from a leaf of an unidentified plant, Sep. 2005 (holotype CGMCC 2.3099T preserved in a metabolically inactive state, ex-type CBS 15464 = G7.1-2).
Carlosrosaea foliicola Q.M. Wang, F.Y. Bai & A.H. Li sp. nov. MycoBank MB828745. Fig. 7K.
Etymology: the specific epithet foliicola refers to the substrate origin of the type strain, leaves.
Culture characteristics: In YM broth, after 7 d at 17 °C, cells are ovoid and ellipsoidal, 1.7–4.0 × 2.5–5.8 μm and single, budding is polar (Fig. 7K), a sediment is formed. After 1 mo at 17 °C, a part ring and sediment are present. On YM agar, after 1 mo at 17 °C, the streak culture is white-cream, butyrous, smooth and glistening. The margin is entire. In Dalmau plate culture on corn meal agar, pseudohyphae are formed. Sexual structures are not observed on YM, PDA, V8 and CM agar. Ballistoconidia are not produced.
Physiological and biochemical characteristics: Glucose fermentation is absent. Glucose, galactose, sucrose, maltose, cellobiose, trehalose, lactose, melibiose, raffinose, melezitose, soluble starch, D-xylose, L-arabinose, D-arabinose (delayed and weak), D-ribose, L-rhamnose (delayed and weak), D-glucosamine, ethanol, glycerol, erythritol, ribitol (delayed and weak), galactitol (delayed and weak), D-mannitol, D-glucitol, Methyl-α-D-glucoside, salicin, DL-lactate, succinate (delayed and weak), citrate (delayed and weak) and myo-inositol (weak) are assimilated as sole carbon sources. L-sorbose, inulin, methanol and hexadecane are not assimilated. Ammonium sulfate is assimilated as sole nitrogen sources. Potassium nitrate, L-lysine, ethylamine hydrochloride and cadaverine dihydrochloride are not assimilated. Maximum growth temperature is 30 °C. Growth in vitamin-free medium is positive. Starch-like substances are not produced. Growth on 50 % (w/w) glucose-yeast extract agar is negative. Urease activity is positive. Diazonium Blue B reaction is positive.
Physiologically, Ca. foliicola differs from the closely related species Ca. simaoensis in its ability to assimilate erythritol (Table S1.5).
Typus: China, Wuzhishan mountain, Hainan province, obtained from a leaf of an unidentified plant, Nov. 2006, Q.-M. Wang (holotype CGMCC 2.3447T preserved in a metabolically inactive state, ex-type CBS 15481 = WZS29.4).
Carlosrosaea simaoensis Q.M. Wang, F.Y. Bai & A.H. Li sp. nov. MycoBank MB828746. Fig. 7L.
Etymology: the specific epithet simaoensis refers to the geographic origin of the type strain, Simao county, Yunnan.
Culture characteristics: In YM broth, after 7 d at 17 °C, cells are ellipsoidal, 2.0–2.6 × 3.3–4.2 μm and single, budding is polar (Fig. 7L), a sediment is present. After 1 mo at 17 °C, a ring and sediment are present. On YM agar, after 1 mo at 17 °C, the streak culture is white-cream, butyrous, smooth and glossy. The margin is entire. In Dalmau plate culture on corn meal agar, pseudohyphae are not formed. Sexual structures are not observed on YM, PDA, V8 and CM agar. Ballistoconidia are not produced.
Physiological and biochemical characteristics: Glucose fermentation is absent. Glucose, galactose, sucrose, maltose, cellobiose, trehalose, lactose, melibiose, raffinose, melezitose, soluble starch, D-xylose, L-arabinose, D-arabinose (delayed and weak), D-ribose, L-rhamnose (delayed and weak), D-glucosamine, ethanol, glycerol, ribitol, galactitol (delayed and weak), D-mannitol, D-glucitol, Methyl-α-D-glucoside, salicin, DL-lactate (delayed and weak), succinate (weak), citrate and myo-inositol (delayed and weak) are assimilated as sole carbon sources. L-sorbose, inulin, methanol, erythritol and hexadecane are not assimilated. Ammonium sulfate is assimilated as sole nitrogen sources. Potassium nitrate, L-lysine, ethylamine hydrochloride and cadaverine dihydrochloride are not assimilated. Maximum growth temperature is 28 °C. Growth in vitamin-free medium is positive. Starch-like substances are not produced. Growth on 50 % (w/w) glucose-yeast extract agar is negative. Urease activity is positive. Diazonium Blue B reaction is positive.
Physiologically, Ca. foliicola and Ca. simaoensis, and their three closely related species, Ca. vrieseae, Ca. hohenbergiae and Ca. aechmeae, can be distinguished from each other by the ability to assimilate inulin, erythritol, L-lysine and cadaverine and form starch like compounds (Table S1.5).
Typus: China, Simao county, Yunnan province, obtained from a leaf of an unidentified plant, Nov. 2006, Q.-M. Wang (holotype CGMCC 2.3580T preserved in a metabolically inactive state, ex-type CBS 15503 = SM8.1).
Kwoniella ovata Q.M. Wang, F.Y. Bai & A.H. Li sp. nov. MycoBank MB828747. Fig. 7M.
Etymology: the specific epithet ovata refers to the ovoid cell morphology of the type strain.
Culture characteristics: In YM broth, after 7 d at 17 °C, cells are ovoid and ellipsoidal, 4.2–6.8 × 5.2–7.9 μm and single, budding is polar (Fig. 7M), a sediment is formed. After 1 mo at 17 °C, a ring and sediment are present. On YM agar, after 1 mo at 17 °C, the streak culture is tannish-white, butyrous, smooth and glossy. The margin is entire. In Dalmau plate culture on corn meal agar, pseudohyphae are not formed. Sexual structures are not observed on YM, PDA, V8 and CM agar. Ballistoconidia are not produced.
Physiological and biochemical characteristics: Glucose fermentation is absent. Glucose, galactose, sucrose, maltose, cellobiose, trehalose, lactose, raffinose, melezitose, soluble starch, D-xylose, L-arabinose, D-arabinose, D-ribose, L-rhamnose, ethanol, glycerol, ribitol (delayed and weak), galactitol, D-mannitol, D-glucitol, Methyl-α-D-glucoside (weak), succinate and myo-inositol (weak) are assimilated as sole carbon sources. L-sorbose, melibiose, inulin, D-glucosamine, methanol, erythritol, salicin, DL-lactate, citrate and hexadecane are not assimilated. Ammonium sulfate, sodium nitrite and L-lysine (weak) are assimilated as sole nitrogen sources. Potassium nitrate, ethylamine hydrochloride and cadaverine dihydrochloride are not assimilated. Maximum growth temperature is 37 °C. Growth in vitamin-free medium is negative. Starch-like substances are not produced. Growth on 50 % (w/w) glucose-yeast extract agar is negative. Urease activity is positive. Diazonium Blue B reaction is positive.
Physiologically, Kwon. ovata differs from its closely related species Kwon. pini and Kwon. dejecticola in its ability to grow at 37 °C (Table S1.6).
Typus: China, Hebei province, obtained from a leaf of an unidentified plant, Nov. 2006, Q.-M. Wang (holotype CGMCC 2.3439T preserved in a metabolically inactive state, ex-type CBS 15475 = H1C1).
Teunia Q.M. Wang & F.Y. Bai gen. nov. MycoBank MB828751.
Etymology: the genus is named in honour of Dr. Teun Boekhout for his contributions to yeast taxonomy.
This genus is proposed for the clade represented by Cryptococcus cuniculi, which clustered with Fonsecazyma tronadorensis (Cryptococcus tronadorensis), Fonsecazyma betulae (Kwoniella betulae) and three new species represented by CGMCC 2.4450, CGMCC 2.5648 and CGMCC 2.3835, respectively. Member of the Cryptococcaceae (Tremellales). The genus is mainly circumscribed by the phylogenetic analysis of the seven genes dataset, in which it occurred as a well supported clade within Cryptococcaceae (Fig. 2).
Sexual reproduction not known. Colonies cream to yellow, butyrous to mucoid. Budding cells present. Pseudohyphae and hyphae are not produced. Ballistoconidia are not formed.
Type species: Teunia korlaensis Q.M. Wang, F.Y. Bai & A.H. Li.
New species and combinations for Teunia
Teunia betulae K. Sylvester, Q.M. Wang & Hittinger ex Q.M. Wang, F.Y. Bai & A.H. Li, sp. nov. MycoBank MB828752.
For description see FEMS Yeast Res. 15: 7 (2015).
Holotype: NRRL Y-63732 (preserved in a metabolically inactive state).
Synonym: Kwoniella betulae K. Sylvester et al., FEMS Yeast Res. 15: 7 (2015), nom. inval., Art. 40.7 (Shenzhen).
= Fonsecazyma betulae K. Sylvester, Q.M. Wang & Hittinger ex Yurkov, Kachalkin & Boekhout, Stud. Mycol. 81: 129 (2015), nom. inval., Art. 40.7 (Shenzhen).
Teunia cuniculi (K.S. Shin & Y.H. Park) Q.M. Wang, F.Y. Bai & A.H. Li, comb. nov. MycoBank MB828753.
Basionym: Cryptococcus cuniculi K.S. Shin & Y.H. Park, Int. J. Syst. Evol. Microbiol. 56: 2243 (2006).
Teunia tronadorensis V. de García, Zalar, Brizzio, Gunde-Cim. & van Brook ex Q.M. Wang, F.Y. Bai & A.H. Li, sp. nov. MycoBank MB828754.
For description see FEMS Microbiol. Ecol. 82(2): 536 (2012).
Holotype: CRUB 1299 (preserved in a metabolically inactive state).
Synonym: Cryptococcus tronadorensis V. de García et al., FEMS Microbiol. Ecol. 82(2): 536 (2012), nom. inval., Art. 40.7 (Shenzhen).
= Fonsecazyma tronadorensis V. de García, Zalar, Brizzio, Gunde-Cim. & van Brook ex Yurkov, Stud. Mycol. 81: 129 (2015), nom. inval., Art. 40.7 (Shenzhen).
Teunia helanensis Q.M. Wang, F.Y. Bai & A.H. Li sp. nov. MycoBank MB828755. Fig. 7N.
Etymology: the specific epithet helanensis refers to the geographic origin of the type strain, Helanshan mountain, Ningxia.
Culture characteristics: In YM broth, after 7 d at 17 °C, cells are ovoid, subglobosal and ellipsoidal, 3.0–4.7 × 4.1–6.6 μm and single, budding is polar (Fig. 7N), a sediment is present. After 1 mo at 17 °C, a ring and sediment are present. On YM agar, after 1 mo at 17 °C, the streak culture is yellowish-cream, mucoid, smooth and glossy. The margin is entire. In Dalmau plate culture on corn meal agar, pseudohyphae are not formed. Sexual structures are not observed on YM, PDA, V8 and CM agar. Ballistoconidia are not produced.
Physiological and biochemical characteristics: Glucose fermentation is absent. Glucose, galactose (delayed), maltose, cellobiose, trehalose, lactose, soluble starch (delayed), D-xylose, L-arabinose (delayed), D-arabinose (delayed), D-ribose (delayed and weak), L-rhamnose, D-glucosamine (delayed), ethanol (delayed), glycerol (delayed), galactitol, D-mannitol, D-glucitol, salicin and succinate are assimilated as sole carbon sources. L-sorbose, sucrose, melibiose, raffinose, melezitose, inulin, methanol, erythritol, ribitol, Methyl-α-D-glucoside, DL-lactate, citrate, myo-inositol and hexadecane are not assimilated. Ammonium sulfate and L-lysine are assimilated as sole nitrogen sources. Potassium nitrate, sodium nitrite, ethylamine hydrochloride and cadaverine dihydrochloride are not assimilated. Maximum growth temperature is 28 °C. Growth in vitamin-free medium is negative. Starch-like substances are not produced. Growth on 50 % (w/w) glucose-yeast extract agar is negative. Urease activity is positive. Diazonium Blue B reaction is positive.
Physiologically, Te. helanensis differs from the closely related species Te. korlaensis in its inability to assimilate sucrose and its ability to assimilate soluble starch, D-arabinose, L-rhamnose, ethanol, erythritol, D-glucitol, succinate and L-lysine (Table S1.7).
Typus: China, Helanshan mountain, Ningxia province, obtained from soil, Aug. 2009, P.J. Han (holotype CGMCC 2.4450T preserved in a metabolically inactive state, ex-type CBS 12498 = HLS02-1-5).
Teunia globosa Q.M. Wang, F.Y. Bai & A.H. Li sp. nov. MycoBank MB828756. Fig. 7O.
Etymology: the specific epithet globosa refers to the globosal vegetative cells of the type strain.
Culture characteristics: In YM broth, after 7 d at 17 °C, cells are globosal, 4.5–8.0 × 5.1–8.0 μm and single, budding is polar (Fig. 7O), a sediment is present. After 1 mo at 17 °C, a ring and sediment are present. On YM agar, after 1 mo at 17 °C, the streak culture is yellowish cream, butyrous, smooth and partly wrinkled, semi-glossy. The margin is entire. In Dalmau plate culture on corn meal agar, pseudohyphae are not formed. Sexual structures are not observed on YM, PDA, V8 and CM agar. Ballistoconidia are not produced.
Physiological and biochemical characteristics: Glucose fermentation is absent. Glucose, galactose, sucrose, maltose, cellobiose, trehalose, lactose (weak), melezitose, inulin (delayed and weak), soluble starch, D-xylose (delayed and weak), D-ribose (delayed and weak), L-rhamnose (delayed and weak), D-glucosamine (delayed and weak), N-Acetyl-D-glucosamine (weak), ethanol, D-mannitol, salicin, succinate (delayed and weak) and myo-inositol are assimilated as sole carbon sources. L-sorbose, melibiose, raffinose, L-arabinose, D-arabinose, methanol, glycerol, erythritol, ribitol, galactitol, D-glucitol, Methyl-α-D-glucoside, D-gluconate, DL-lactate, citrate and hexadecane are not assimilated. Ammonium sulfate, L-lysine, ethylamine hydrochloride (delayed and weak) and cadaverine dihydrochloride are assimilated as sole nitrogen sources. Potassium nitrate and sodium nitrite are not assimilated. Maximum growth temperature is 22 °C. Growth in vitamin-free medium is negative. Starch-like substances are not produced. Growth on 50 % (w/w) glucose-yeast extract agar is negative. Urease activity is positive. Diazonium Blue B reaction is positive.
Physiologically, Te. globosa differs from the closely related species Te. betulae in its inability to assimilate L-arabinose and its ability to assimilate ethanol (Table S1.7).
Typus: China, Lulang county, Tibet, obtained from a leaf of an unidentified plant, Sep. 2015, Q.-M. Wang (holotype CGMCC 2.5648T preserved in a metabolically inactive state, ex-type CBS 15566 = GPS23.2A6).
Teunia korlaensis Q.M. Wang, F.Y. Bai & A.H. Li sp. nov. MycoBank MB828757. Fig. 7P.
Etymology: the specific epithet korlaensis refers to the geographic origin of the type strain, Korla county, Xinjiang.
Culture characteristics: In YM broth, after 7 d at 17 °C, cells are subglobosal to globosal, 3.8–5.1 × 4.3–5.9 μm and single, budding is polar (Fig. 7P), a sediment is formed. After 1 mo at 17 °C, a part ring and sediment are present. On YM agar, after 1 mo at 17 °C, the streak culture is cream, butyrous, smooth and semi-glossy. The margin is entire. In Dalmau plate culture on corn meal agar, pseudohyphae are not formed. Sexual structures are not observed on YM, PDA, V8 and CM agar. Ballistoconidia are not produced.
Physiological and biochemical characteristics: Glucose fermentation is absent. Glucose, galactose, sucrose, maltose, cellobiose, trehalose, lactose (weak), melezitose (weak), inulin (weak), D-xylose (weak), L-arabinose (weak), D-ribose (delayed and weak), L-rhamnose (weak), galactitol, D-mannitol and salicin (weak) are assimilated as sole carbon sources. L-sorbose, melibiose, raffinose, soluble starch, D-arabinose, D-glucosamine, N-Acetyl-D-glucosamine, methanol, ethanol, glycerol, ribitol, erythritol, D-glucitol, Methyl-α-D-glucoside, D-gluconate, DL-lactate, succinate, citrate, myo-inositol and hexadecane are not assimilated. Ammonium sulfate, ethylamine hydrochloride (weak) and cadaverine dihydrochloride (weak) are assimilated as sole nitrogen sources. Potassium nitrate, sodium nitrite and L-lysine are not assimilated. Maximum growth temperature is 30 °C. Growth in vitamin-free medium is negative. Starch-like substances are not produced. Growth on 50 % (w/w) glucose-yeast extract agar is negative. Urease activity is positive. Diazonium Blue B reaction is positive.
Physiologically, Te. korlaensis differs from the closely related species Te. helanensis in its inability to assimilate soluble starch, D-arabinose, L-rhamnose, ethanol, erythritol, D-glucitol, succinate and L-lysine and its ability to assimilate sucrose (Table S1.7).
Typus: China, Korla county, Xinjiang province, obtained from soil, Feb. 2008, Q.-M. Wang (holotype CGMCC 2.3835T preserved in a metabolically inactive state, ex-type CBS 15653 = 141.19).
Saitozyma pseudoflava Q.M. Wang, F.Y. Bai & A.H. Li sp. nov. MycoBank MB828758. Fig. 8A.
Fig. 8.
Vegetative cells grown in YM broth for 5 d at 17 °C and ballistoconidia produced on corn meal agar after 7 d at 17 °C. (A) Sa. pseudoflava CGMCC 2.5811T; (B) Di. milinica CGMCC 2.5628T; (C, D) Di. heilongjiangensis CGMCC 2.5674T; (E, F) Di. ovata CGMCC 2.3625T; (G, H) Di. maotaiensis CGMCC 2.4537T; (I) Di. kandeliae CGMCC 2.5658T; (J) Bu. phyllostachydis CGMCC 2.5812T; (K, L) Bu. cremeum CGMCC 2.4427T; (M, N) Bu. pseudopanici CGMCC 2.4024T; (O, P) Bu. phyllophilum CGMCC 2.3320T. Bars = 10 μm.
Etymology: the specific epithet pseudoflava refers to the similar colony morphology to that of Saitozyma flava.
Culture characteristics: In YM broth, after 7 d at 17 °C, cells are subglobosal and ovoid, 3.2–4.3 × 5.2–6.8 μm and single, budding is polar (Fig. 8A), a sediment is formed. After 1 mo at 17 °C, a pellicle and sediment are present. On YM agar, after 1 mo at 17 °C, the streak culture is yellowish-cream, butyrous, smooth and glossy. The margin is entire. In Dalmau plate culture on corn meal agar, pseudohyphae are not formed. Sexual structures are not observed on YM, PDA, V8 and CM agar. Ballistoconidia are not produced.
Physiological and biochemical characteristics: Glucose fermentation is absent. Glucose, galactose, sucrose, maltose, cellobiose, melibiose (weak), raffinose, melezitose, inulin (weak), D-xylose, L-arabinose, D-arabinose (weak), D-ribose, L-rhamnose (delayed and weak), D-glucosamine (delayed and weak), N-Acetyl-D-glucosamine (delayed and weak), ribitol (delayed and weak), galactitol (delayed and weak), D-mannitol (delayed and weak), D-glucitol, Methyl-α-D-glucoside, salicin (delayed and weak), D-gluconate (delayed and weak) and myo-inositol are assimilated as sole carbon sources. L-sorbose, trehalose, lactose, soluble starch, methanol, ethanol, glycerol, erythritol, DL-lactate, succinate, citrate and hexadecane are not assimilated. Ammonium sulfate, L-lysine and ethylamine hydrochloride (delayed and weak) are assimilated as sole nitrogen sources. Potassium nitrate, sodium nitrite and cadaverine dihydrochloride are not assimilated. Maximum growth temperature is 32 °C. Growth in vitamin-free medium is negative. Starch-like substances are not produced. Growth on 50 % (w/w) glucose-yeast extract agar is negative. Urease activity is positive. Diazonium Blue B reaction is positive.
Physiologically, Sa. pseudoflava differs from its closely related species Sa. paraflava and Sa. flava in its inability to assimilate cellobiose, trehalose, soluble starch, DL-lactate, succinate and citrate (Table S1.8).
Typus: China, Tibet, obtained from a leaf of an unidentified plant, Sep. 2014, Q.-M. Wang (holotype CGMCC 2.5811T preserved in a metabolically inactive state, ex-type CBS 15576 = XZ200A1).
Dioszegia milinica Q.M. Wang, F.Y. Bai & A.H. Li sp. nov. MycoBank MB828759. Fig. 8B.
Etymology: the specific epithet milinica refers to the geographic origin of the type strain, Milin county, Tibet.
Culture characteristics: In YM broth, after 7 d at 17 °C, cells are ovoid and ellipsoidal, 2.9–6.4 × 5.0–10.3 μm and single, budding is polar (Fig. 8B), a sediment is formed. After 1 mo at 17 °C, a ring and sediment are present. On YM agar, after 1 mo at 17 °C, the streak culture is orange, butyrous, smooth and glossy. The margin is entire. In Dalmau plate culture on corn meal agar, pseudohyphae are formed. Sexual structures are not observed on YM, PDA, V8 and CM agar. Ballistoconidia are not produced.
Physiological and biochemical characteristics: Glucose fermentation is absent. Glucose, galactose, sucrose, maltose, cellobiose, trehalose, melibiose, raffinose, melezitose, inulin, soluble starch (delayed and weak), D-xylose, L-arabinose, D-arabinose, L-rhamnose (delayed and weak), D-glucosamine (delayed and weak), galactitol, D-glucitol, succinate and citrate are assimilated as sole carbon sources. L-sorbose, lactose, D-ribose, N-Acetyl-D-glucosamine, methanol, ethanol, glycerol, erythritol, ribitol, D-mannitol, Methyl-α-D-glucoside, salicin, DL-lactate, myo-inositol and hexadecane are not assimilated. Ammonium sulfate, potassium nitrate (weak), L-lysine (weak) and ethylamine hydrochloride are assimilated as sole nitrogen sources. Sodium nitrite and cadaverine dihydrochloride are not assimilated. Maximum growth temperature is 23 °C. Growth in vitamin-free medium is positive. Starch-like substances are produced. Growth on 50 % (w/w) glucose-yeast extract agar is negative. Urease activity is positive. Diazonium Blue B reaction is positive.
Physiologically, Di. milinica differs from the closely related species Di. aurantiaca in its inability to assimilate D-ribose, N-Acetyl-D-glucosamine, glycerol, erythritol, ribitol, D-mannitol, Methyl-α-D-glucoside, salicin, DL-lactate and sodium nitrite and its ability to assimilate inulin and ethylamine (Table S1.9).
Typus: China, Milin county, Tibet, obtained from a leaf of an unidentified plant, Sep. 2015, Q.-M. Wang (holotype CGMCC 2.5628T preserved in a metabolically inactive state, ex-type CBS 15563 = GPS21.3B8).
Dioszegia heilongjiangensis Q.M. Wang, F.Y. Bai & A.H. Li sp. nov. MycoBank MB828760. Fig. 8C, D.
Etymology: the specific epithet heilongjiangensis refers to the geographic origin of the type strain, Heilongjiang province.
Culture characteristics: In YM broth, after 7 d at 17 °C, cells are subglobosal and ellipsoidal, 3.2–5.0 × 4.5–7.3 μm and single, budding is polar (Fig. 8C), a sediment is formed. After 1 mo at 17 °C, a ring and sediment are present. On YM agar, after 1 mo at 17 °C, the streak culture is yellowish to light orange, butyrous, smooth and partly wrinkled. The margin is entire. In Dalmau plate culture on corn meal agar, pseudohyphae are not formed. Sexual structures are not observed on YM, PDA, V8 and CM agar. Ballistoconidia are subglobosal to napiform, 4.0–5.0 × 5.0–6.0 μm (Fig. 8D).
Physiological and biochemical characteristics: Glucose fermentation is absent. Glucose, galactose, sucrose (weak), maltose (weak), cellobiose (weak), trehalose (weak), melibiose, raffinose, melezitose, inulin (weak), D-xylose (delayed), L-arabinose, D-arabinose (delayed and weak), galactitol (weak), D-glucitol, salicin (weak) and succinate are assimilated as sole carbon sources. L-sorbose, lactose, soluble starch, D-ribose, L-rhamnose, D-glucosamine, N-Acetyl-D-glucosamine, methanol, ethanol, glycerol, erythritol, ribitol, D-mannitol, Methyl-α-D-glucoside, DL-lactate, citrate, myo-inositol and hexadecane are not assimilated. Ammonium sulfate, L-lysine and cadaverine dihydrochloride are assimilated as sole nitrogen sources. Potassium nitrate, sodium nitrite and ethylamine hydrochloride are not assimilated. Maximum growth temperature is 26–27 °C. Growth in vitamin-free medium is negative. Starch-like substances are produced. Growth on 50 % (w/w) glucose-yeast extract agar is negative. Urease activity is positive. Diazonium Blue B reaction is positive.
Physiologically, Di. heilongjiangensis differs from the closely related species Di. changbaiensis and Di. cryoxerica in its inability to assimilate D-ribose, L-rhamnose and D-mannitol and its ability to grow in vitamin-free medium (Table S1.9).
Typus: China, Chelu county, Heilongjiang province, obtained from a leaf of an unidentified plant, Aug. 2014, Q.-M. Wang (holotype CGMCC 2.5674T preserved in a metabolically inactive state, ex-type CBS 13957 = HLJ13.24).
Dioszegia ovata Q.M. Wang, F.Y. Bai & A.H. Li sp. nov. MycoBank MB828761. Fig. 8E, F.
Etymology: the specific epithet ovata refers to the ovoid cell morphology of the type strain.
Culture characteristics: In YM broth, after 7 d at 17 °C, cells are ovoid and ellipsoidal, 2.3–4.6 × 3.8–7.7 μm and single, budding is polar (Fig. 8E), a sediment is formed. After 1 mo at 17 °C, a ring and sediment are present. On YM agar, after 1 mo at 17 °C, the streak culture is pink to orange, butyrous, smooth. The margin is entire. In Dalmau plate culture on corn meal agar, pseudohyphae are not formed. Sexual structures are not observed on YM, PDA, V8 and CM agar. Ballistoconidia are globosal and subglobosal to napiform, 3.1–6.2 × 3.8–6.9 μm (Fig. 8F).
Physiological and biochemical characteristics: Glucose fermentation is absent. Glucose, galactose, L-sorbose (delayed), sucrose, maltose, cellobiose, trehalose, lactose (delayed), melibiose, raffinose, melezitose, soluble starch, D-xylose, L-arabinose, D-arabinose, D-ribose, L-rhamnose, D-glucosamine (delayed and weak), galactitol, D-mannitol, Methyl-α-D-glucoside, salicin (weak) and succinate (delayed and weak) are assimilated as sole carbon sources. Inulin, methanol, ethanol, glycerol, erythritol, ribitol, D-glucitol, DL-lactate, citrate, myo-inositol and hexadecane are not assimilated. Ammonium sulfate and potassium nitrate (delayed and weak) are assimilated as sole nitrogen sources. Sodium nitrite, L-lysine, ethylamine hydrochloride and cadaverine dihydrochloride are not assimilated. Maximum growth temperature is 32 °C. Growth in vitamin-free medium is positive. Starch-like substances are produced. Growth on 50 % (w/w) glucose-yeast extract agar is negative. Urease activity is positive. Diazonium Blue B reaction is positive.
Physiologically, Di. ovata and the closely related species Di. maotaiensis, Di. kandeliae, Di. zsoltii, Di. catarinoi, Di. takashimae and Di. athyrii can be distinguished from one another. Di. ovata differs from the other six species in its ability to grow at 32 °C (Table S1.9).
Typus: China, Bangxi county, Hainan province, obtained from a leaf of an unidentified plant, Nov. 2006, Q.-M. Wang (holotype CGMCC 2.3625T preserved in a metabolically inactive state, ex-type CBS 15657 = HBX1.27).
Dioszegia maotaiensis Q.M. Wang, F.Y. Bai & A.H. Li sp. nov. MycoBank MB828762. Fig. 8G, H.
Etymology: the specific epithet maotaiensis refers to the geographic origin of the type strain, Maotai county, Guizhou.
Culture characteristics: In YM broth, after 7 d at 17 °C, cells are ovoid and ellipsoidal, 3.6–5.2 × 4.5–6.2 μm and single, budding is polar (Fig. 8G), a sediment is formed. After 1 mo at 17 °C, a ring and a sediment are present. On YM agar, after 1 mo at 17 °C, the streak culture is orange, butyrous, smooth and glossy. The margin is entire. In Dalmau plate culture on corn meal agar, pseudohyphae are not formed. Sexual structures are not observed on YM, PDA, V8 and CM agar. Ballistoconidia are are subglobosal to ellipsoidal, 2.4–3.5 × 3.5–5.3 μm (Fig. 8H).
Physiological and biochemical characteristics: Glucose fermentation is absent. Glucose, galactose, sucrose, maltose, cellobiose, trehalose, melibiose, raffinose, melezitose, inulin, soluble starch (delayed and weak), D-xylose, L-arabinose, D-arabinose, L-rhamnose, succinate (delayed and weak) and citrate (delayed and weak) are assimilated as sole carbon sources. L-sorbose, lactose, D-ribose, D-glucosamine, N-Acetyl-D-glucosamine, methanol, ethanol, glycerol, erythritol, ribitol, galactitol, D-mannitol, D-glucitol, Methyl-α-D-glucoside, salicin, DL-lactate, myo-inositol and hexadecane are not assimilated. Ammonium sulfate and ethylamine hydrochloride are assimilated as sole nitrogen sources. Potassium nitrate, sodium nitrite, L-lysine and cadaverine dihydrochloride are not assimilated. Maximum growth temperature is 28 °C. Growth in vitamin-free medium is positive. Starch-like substances are produced. Growth on 50 % (w/w) glucose-yeast extract agar is negative. Urease activity is positive. Diazonium Blue B reaction is positive.
Physiologically, Di. maotaiensis and the closely related species Di. ovata, Di. kandeliae, Di. zsoltii, Di. catarinoi, Di. takashimae and Di. athyrii can be distinguished from one another. Di. maotaiensis and Di. ovata differ from the other five species in their ability to grow in vitamin-free medium (Table S1.9).
Typus: China, Maotai county, Guizhou province, obtained from a leaf of an unidentified plant, Mar. 2012, Q.-M. Wang (holotype CGMCC 2.4537T preserved in a metabolically inactive state, ex-type CBS 15516 = GZMT3A9).
Dioszegia kandeliae Q.M. Wang, F.Y. Bai, L.D. Guo & A.H. Li sp. nov. MycoBank MB828763. Fig. 8I.
Etymology: the specific epithet kandeliae refers to Kandelia, the plant genus from which the type strain was isolated.
Culture characteristics: In YM broth, after 7 d at 17 °C, cells are ellipsoidal to subglobosal, 2.5–4.2 × 3.2–5.5 μm and single, budding is polar (Fig. 8I), a ring and a sediment are formed. After 1 mo at 17 °C, a ring and sediment are present. On YM agar, after 1 mo at 17 °C, the streak culture is orange-red, butyrous, smooth and glossy. The margin is entire. In Dalmau plate culture on corn meal agar, pseudohyphae are not formed. Sexual structures are not observed on YM, PDA, V8 and CM agar. Ballistoconidia are not produced.
Physiological and biochemical characteristics: Glucose fermentation is absent. Glucose, galactose, L-sorbose, sucrose, maltose, cellobiose, trehalose, lactose, melibiose, melezitose, inulin (weak), soluble starch (delayed and weak), D-xylose (delayed and weak), L-arabinose (delayed and weak), D-glucosamine (delayed and weak), N-Acetyl-D-glucosamine (delayed and weak), ethanol (delayed and weak), glycerol (delayed and weak), ribitol (delayed and weak) and D-glucitol are assimilated as sole carbon sources. Raffinose, D-arabinose, D-ribose, L-rhamnose, methanol, erythritol, galactitol, D-mannitol, Methyl-α-D-glucoside, salicin, DL-lactate, succinate, citrate, myo-inositol and hexadecane are not assimilated. Ammonium sulfate, potassium nitrate, ethylamine hydrochloride and cadaverine dihydrochloride are assimilated as sole nitrogen sources. Sodium nitrite and L-lysine are not assimilated. Maximum growth temperature is 30 °C. Growth in vitamin-free medium is negative. Starch-like substances are produced. Growth on 50 % (w/w) glucose-yeast extract agar is negative. Urease activity is positive. Diazonium Blue B reaction is positive.
Physiologically, Di. kandeliae and the closely related species Di. ovata, Di. maotaiensis, Di. zsoltii, Di. catarinoi, Di. takashimae and Di. athyrii can be distinguished from one another. Di. kandeliae differs from the other six species in its inability to assimilate raffinose and L-rhamnose (Table S1.9).
Typus: China, Beilunhekou natural reserve, Guangxi province, obtained from a leaf of Kandelia candel, Apr. 2014, L.-D. Guo (holotype CGMCC 2.5658T preserved in a metabolically inactive state, ex-type CBS 13951 = 224191).
Bulleribasidium phyllostachydis Q.M. Wang, F.Y. Bai & A.H. Li sp. nov. MycoBank MB828765. Fig. 8J.
Etymology: the specific epithet phyllostachydis refers to Phyllostachys, the plant genus from which the type strain was isolated.
Culture characteristics: In YM broth, after 7 d at 17 °C, cells are subglobosal, ovoid and ellipsoidal, 2.6–4.8 × 3.7–11.3 μm and single, budding is polar (Fig. 8J), a sediment is formed. After 1 mo at 17 °C, a pellicle and sediment are present. On YM agar, after 1 mo at 17 °C, the streak culture is yellow, butyrous, smooth and glistening. The margin is entire. In Dalmau plate culture on corn meal agar, pseudohyphae are formed. Sexual structures are not observed on YM, PDA, V8 and CM agar. Ballistoconidia are not produced.
Physiological and biochemical characteristics: Glucose fermentation is absent. Glucose, galactose, sucrose, cellobiose, trehalose, melibiose, raffinose, melezitose, D-xylose, L-arabinose, D-arabinose, D-ribose, L-rhamnose, D-glucosamine (weak), N-Acetyl-D-glucosamine (weak), galactitol, D-mannitol, D-glucitol, Methyl-α-D-glucoside (weak), salicin (weak) and D-gluconate are assimilated as sole carbon sources. L-sorbose, maltose, lactose, inulin, soluble starch, methanol, ethanol, glycerol, erythritol, ribitol, DL-lactate, succinate, citrate, myo-inositol and hexadecane are not assimilated. Ammonium sulfate and L-lysine (delayed and weak) are assimilated as sole nitrogen sources. Potassium nitrate, sodium nitrite, ethylamine hydrochloride and cadaverine dihydrochloride are not assimilated. Maximum growth temperature is 28 °C. Growth in vitamin-free medium is negative. Starch-like substances are not produced. Growth on 50 % (w/w) glucose-yeast extract agar is negative. Urease activity is positive. Diazonium Blue B reaction is positive.
Physiologically, Bu. phyllostachydis differs from its closely related species Bu. setariae in its inability to assimilate maltose, inulin, DL-lactate, succinate and citrate (Table S1.10).
Typus: China, Motuo county, Tibet, obtained from a leaf of Phyllostachys sp., Sep. 2014, Q.-M. Wang (holotype CGMCC 2.5812T preserved in a metabolically inactive state, ex-type CBS 15575 = XZ139E1).
Bulleribasidium cremeum Q.M. Wang, F.Y. Bai & A.H. Li sp. nov. MycoBank MB828766. Fig. 8K, L.
Etymology: the specific epithet cremeum refers to the pale-cream colony morphology.
Culture characteristics: In YM broth, after 7 d at 17 °C, cells are ovoid and ellipsoidal, 1.7–4.8 × 4.5–8.7 μm and single, budding is polar (Fig. 8K), a sediment is present. After 1 mo at 17 °C, a ring and sediment are present. On YM agar, after 1 mo at 17 °C, the streak culture is pale-cream, butyrous, smooth and semi-glossy. The margin is entire. In Dalmau plate culture on corn meal agar, pseudohyphae are formed. Sexual structures are not observed on YM, PDA, V8 and CM agar. Ballistoconidia are ellipsoidal to napiform, 3.3–6.7 × 4.0–6.7 μm (Fig. 8L).
Physiological and biochemical characteristics: Glucose fermentation is absent. Glucose, galactose, sucrose, maltose, cellobiose (delayed and weak), trehalose, melibiose, raffinose, melezitose, D-xylose (delayed and weak), L-arabinose (delayed and weak), D-arabinose, salicin (weak) and succinate are assimilated as sole carbon sources. L-sorbose, lactose, inulin, soluble starch, D-ribose, L-rhamnose, D-glucosamine, methanol, ethanol, glycerol, erythritol, ribitol, galactitol, D-mannitol, D-glucitol, Methyl-α-D-glucoside, DL-lactate, citrate, myo-inositol and hexadecane are not assimilated. Ammonium sulfate and L-lysine are assimilated as sole nitrogen sources. Potassium nitrate, sodium nitrite, ethylamine hydrochloride and cadaverine dihydrochloride are not assimilated. Maximum growth temperature is 28 °C. Growth in vitamin-free medium is negative. Starch-like substances are not produced. Growth on 50 % (w/w) glucose-yeast extract agar is negative. Urease activity is positive. Diazonium Blue B reaction is positive.
Physiologically, Bu. cremeum differs from its closely related species, Bu. phyllostachydis, Bu. wuzhishanense and Bu. setariae, in its inability to assimilate galactitol, D-mannitol and Methyl-α-D-glucoside (Table S1.10).
Typus: China, Taiwan province, obtained from a leaf of an unidentified plant, Aug. 2009, Q.-M. Wang (holotype CGMCC 2.4427T preserved in a metabolically inactive state, ex-type CBS 12487 = TW1.1F-025).
Bulleribasidium pseudopanici Q.M. Wang, F.Y. Bai & A.H. Li sp. nov. MycoBank MB828767. Fig. 8M, N.
Etymology: the specific epithet pseudopanici refers to the similar colony morphology to that of Bulleribasidium panici.
Culture characteristics: In YM broth, after 7 d at 17 °C, cells are ovoid, 2.3–5.0 × 3.8–7.6 μm and single, budding is polar (Fig. 8M), a sediment is formed. After 1 mo at 17 °C, a part ring and a sediment are present. On YM agar, after 1 mo at 17 °C, the streak culture is yellowish-cream, butyrous, slightly wrinkled and dull. The margin is entire. In Dalmau plate culture on corn meal agar, pseudohyphae are formed. Sexual structures are not observed on YM, PDA, V8 and CM agar. Ballistoconidia are subglobosal or ellipsoidal, 4.4–7.4 × 5.9–7.4 μm (Fig. 8N).
Physiological and biochemical characteristics: Glucose fermentation is absent. Glucose, galactose, sucrose, maltose, cellobiose, trehalose, melibiose, raffinose, melezitose, D-xylose, L-arabinose, D-arabinose, D-ribose, L-rhamnose, galactitol, D-mannitol, D-glucitol (variable), Methyl-α-D-glucoside, salicin and myo-inositol are assimilated as sole carbon sources. L-sorbose, lactose, inulin, soluble starch, D-glucosamine, N-Acetyl-D-glucosamine, methanol, ethanol, glycerol, erythritol, ribitol, D-gluconate, DL-lactate, succinate, citrate and hexadecane are not assimilated. Ammonium sulfate, L-lysine, ethylamine hydrochloride, cadaverine dihydrochloride are assimilated as sole nitrogen sources. Potassium nitrate and sodium nitrite are not assimilated. Maximum growth temperature is 28 °C. Growth in vitamin-free medium is negative. Starch-like substances are not produced. Growth on 50 % (w/w) glucose-yeast extract agar is negative. Urease activity is positive. Diazonium Blue B reaction is positive.
Physiologically, Bu. pseudopanici differs from its closely related species Bu. panici in its inability to assimilate L-sorbose, soluble starch, D-glucosamine, erythritol, ribitol, D-gluconate, DL-lactate and succinate and its ability to form starch like compounds (Table S1.10).
Typus: China, Wuzhishan mountain, Hainan province, obtained from a leaf of an unidentified plant, Nov. 2006, Q.-M. Wang (holotype CGMCC 2.4024T preserved in a metabolically inactive state, ex-type CBS 15510 = WZS17.20).
Bulleribasidium phyllophilum Q.M. Wang, F.Y. Bai & A.H. Li sp. nov. MycoBank MB828768. Fig. 8O, P.
Etymology: the specific epithet phyllophilum refers to leaves, the substrate origin of the type strain.
Culture characteristics: In YM broth, after 7 d at 17 °C, cells are ovoid and ellipsoidal, 2.0–4.0 × 4.0–9.3 μm and single, budding is polar (Fig. 8O), a sediment is present. After 1 mo at 17 °C, a ring and sediment are present. On YM agar, after 1 mo at 17 °C, the streak culture is prey-cream, butyrous, smooth and semi-glossy. The margin is entire. In Dalmau plate culture on corn meal agar, pseudohyphae are formed. Sexual structures are not observed on YM, PDA, V8 and CM agar. Ballistoconidia are ellipsoidal, subglobosal to napiform, 3.8–6.2 × 4.6–6.2 μm (Fig. 8P).
Physiological and biochemical characteristics: Glucose fermentation is absent. Glucose, galactose, sucrose, maltose, cellobiose, trehalose, melibiose, raffinose, melezitose, inulin (variable), soluble starch (variable), D-xylose, L-arabinose, D-arabinose, L-rhamnose, D-glucosamine (weak), N-Acetyl-D-glucosamine (variable), galactitol, D-mannitol, D-glucitol (variable), Methyl-α-D-glucoside (delayed and weak) and myo-inositol (variable) are assimilated as sole carbon sources. L-sorbose, lactose, D-ribose, methanol, ethanol, glycerol, erythritol, ribitol, salicin, DL-lactate, succinate, citrate and hexadecane are not assimilated. Ammonium sulfate, potassium nitrate (variable), L-lysine (variable) and ethylamine hydrochloride (variable) are assimilatedas sole nitrogen sources. Sodium nitrite and cadaverine dihydrochloride are not assimilated. Maximum growth temperature is 28 °C. Growth in vitamin-free medium is positive. Starch-like substances are not produced. Growth on 50 % (w/w) glucose-yeast extract agar is negative. Urease activity is positive. Diazonium Blue B reaction is positive.
Physiologically, Bu. phyllophilum and its closely related species Bu. foliicola cannot be distinguished from each other. The former did not grow at 30 °C, but the latter grew weak (Table S1.10).
Typus: China, Bangxi county, Hainan province, obtained from a leaf of an unidentified plant, Nov. 2006, Q.-M. Wang (holotype CGMCC 2.3320T preserved in a metabolically inactive state, ex-type CBS 15474 = HBX2.8).
Bulleribasidium elongatum Q.M. Wang, F.Y. Bai & A.H. Li sp. nov. MycoBank MB828769. Fig. 9A.
Fig. 9.
Vegetative cells grown in YM broth for 5 d at 17 °C and ballistoconidia produced on corn meal agar after 7 d at 17 °C. (A) Bu. elongatum CGMCC 2.4428T; (B) De. pseudoboekhoutii CGMCC 2.4436T; (C, D) De. polymorphus CGMCC 2.4437T; (E, F) De. xingshaicus CGMCC 2.2459T; (G, H) De. pseudoyunnanensis CGMCC 2.3563T; (I, J) De. longiovatus CGMCC 2.3535T; (K, L) De. napiformis CGMCC 2.4446T; (M, N) De. bifurcus CGMCC 2.3470T; (O, P) De. elongatus CGMCC 2.3561T. Bars = 10 μm.
Etymology: the specific epithet elongatum refers to the elongate vegetative cells of the type strain.
Culture characteristics: In YM broth, after 7 d at 17 °C, cells are ellipsoidal and cylindrical, 2.7–4.1 × 6.8–12.5 μm and single, budding is polar (Fig. 9A), a sediment is present. On YM agar, after 1 mo at 17 °C, the streak culture is cream, butyrous, wrinkled and dull. The margin is entire. In Dalmau plate culture on corn meal agar, pseudohyphae are formed. Sexual structures are not observed on YM, PDA, V8 and CM agar. Ballistoconidia are not produced.
Physiological and biochemical characteristics: Glucose fermentation is absent. Glucose, galactose, sucrose, maltose, cellobiose, trehalose, melibiose, raffinose, melezitose, D-xylose, L-arabinose (delayed and weak), D-arabinose (delayed and weak), D-ribose (delayed and weak), L-rhamnose (delayed and weak), D-glucosamine (delayed and weak), ribitol (delayed and weak) and galactitol are assimilated as sole carbon sources. L-sorbose, lactose, inulin, soluble starch, methanol, ethanol, glycerol, erythritol, D-mannitol, D-glucitol, Methyl-α-D-glucoside, salicin, DL-lactate, succinate, citrate, myo-inositol and hexadecane are not assimilated. Ammonium sulfate, sodium nitrite (delayed and weak), L-lysine, ethylamine hydrochloride and cadaverine dihydrochloride are assimilated as sole nitrogen sources. Potassium nitrate is not assimilated. Maximum growth temperature is 28 °C. Growth in vitamin-free medium is negative. Starch-like substances are not produced. Growth on 50 % (w/w) glucose-yeast extract agar is negative. Urease activity is positive. Diazonium Blue B reaction is positive.
Physiologically, Bu. elongatum differs from its closely related species, Bu. phyllophilum, Bu. foliicola and Bu. hainanense, in its inability to assimilate D-mannitol and its ability to assimilate cadaverine (Table S1.10).
Typus: China, Taiwan province, obtained from a leaf of an unidentified plant, Aug. 2009, Q.-M. Wang (holotype CGMCC 2.4428T preserved in a metabolically inactive state, ex-type CBS 12489 = TW1.1F-019).
Derxomyces pseudoboekhoutii Q.M. Wang, F.Y. Bai & A.H. Li sp. nov. MycoBank MB828770. Fig. 9B.
Etymology: the specific epithet pseudoboekhoutii refers to the similar colony morphology to that of Derxomyces boekhoutii.
Culture characteristics: In YM broth, after 7 d at 17 °C, cells are ellipsoidal or ovoid, 2.5–3.8 × 5.0–7.5 μm and single, budding is polar (Fig. 9B), a sediment is present. After 1 mo at 17 °C, a ring and a sediment are present. On YM agar, after 1 mo at 17 °C, the streak culture is cream, butyrous and semi-glossy. The margin is entire. In Dalmau plate culture on corn meal agar, pseudohyphae are not formed. Sexual structures are not observed on YM, PDA, V8 and CM agar. Ballistoconidia are not produced.
Physiological and biochemical characteristics: Glucose fermentation is absent. Glucose, galactose, sucrose, maltose, cellobiose, trehalose, melibiose, raffinose, melezitose, inulin, D-xylose, L-arabinose, D-arabinose, D-ribose (delayed), galactitol (weak), D-mannitol (delayed and weak) and Methyl-α-D-glucoside are assimilated as sole carbon sources. L-sorbose, lactose, soluble starch, L-rhamnose, D-glucosamine, N-Acetyl-D-glucosamine, methanol, ethanol, glycerol, erythritol, ribitol, D-glucitol, salicin, DL-lactate, succinate, citrate, myo-inositol and hexadecane are not assimilated. Ammonium sulfate, potassium nitrate, L-lysine, ethylamine hydrochloride (delayed and weak), cadaverine dihydrochloride (delayed and weak) are assimilated as sole nitrogen sources. Sodium nitrite is not assimilated. Maximum growth temperature is 25 °C. Growth in vitamin-free medium is negative. Starch-like substances are not produced. Growth on 50 % (w/w) glucose-yeast extract agar is negative. Urease activity is positive. Diazonium Blue B reaction is positive.
Physiologically, De. pseudoboekhoutii differs from the closely related species De. boekhoutii in its inability to assimilate soluble starch and grow in vitamin-free medium and its ability to assimilate D-arabinose and D-ribose (Table S1.11).
Typus: China, Fuzhou county, Fujian province, obtained from a leaf of an unidentified plant, Aug. 2011, Q.-M. Wang (holotype CGMCC 2.4436T preserved in a metabolically inactive state, ex-type CBS 12493 = FJYZ12-8).
Derxomyces polymorphus Q.M. Wang, F.Y. Bai & A.H. Li sp. nov. MycoBank MB828771. Fig. 9C, D.
Etymology: the specific epithet polymorphus refers to the variable vegetative cell morphology of the type strain.
Culture characteristics: In YM broth, after 7 d at 17 °C, cells are ovoid to fusiform, 2.0–4.8 × 4.7–8.0 μm and single, budding is polar (Fig. 9C), a sediment is present. After 1 mo at 17 °C, a ring and a sediment are present. On YM agar, after 1 mo at 17 °C, the streak culture is yellowish-cream, smooth and dull. In Dalmau plate culture on corn meal agar, pseudohyphae are not formed. Sexual structures are not observed on YM, PDA, V8 and CM agar. Ballistoconidia are subglobosal to napiform, 3.0–4.3 × 4.3–5.7 μm (Fig. 9D).
Physiological and biochemical characteristics: Glucose fermentation is absent. Glucose, galactose, sucrose, maltose, cellobiose, trehalose, melibiose, raffinose, melezitose, inulin (weak), soluble starch, D-xylose, L-rhamnose (weak), galactitol, D-glucitol, salicin (weak) and succinate are assimilated as sole carbon sources. L-sorbose, lactose, L-arabinose, D-arabinose, D-ribose, D-glucosamine, N-Acetyl-D-glucosamine, methanol, ethanol, glycerol, erythritol, ribitol, D-mannitol, Methyl-α-D-glucoside, DL-lactate, citrate, myo-inositol and hexadecane are not assimilated. Ammonium sulfate, potassium nitrate (weak), L-lysine, ethylamine hydrochloride, cadaverine dihydrochloride are assimilated as sole nitrogen sources. Sodium nitrite is not assimilated. Maximum growth temperature is 27–28 °C. Growth in vitamin-free medium is weak. Starch-like substances are produced. Growth on 50 % (w/w) glucose-yeast extract agar is negative. Urease activity is positive. Diazonium Blue B reaction is positive.
Physiologically, De. polymorphus differs from the closely related species De. nakasei in its inability to assimilate L-sorbose, L-arabinose, D-arabinose, D-ribose, D-glucosamine, erythritol, D-mannitol, Methyl-α-D-glucoside, DL-lactate and myo-inositol (Table S1.11).
Typus: China, Fuzhou county, Fujian province, obtained from a leaf of an unidentified plant, Aug. 2011, Q.-M. Wang (holotype CGMCC 2.4437T preserved in a metabolically inactive state, ex-type CBS 15512 = FJYZ12-13).
Derxomyces xingshanicus Q.M. Wang, F.Y. Bai & A.H. Li sp. nov. MycoBank MB828772. Fig. 9E, F.
Etymology: the specific epithet xingshanicus refers to the geographic origin of the type strain, Xingshan county, Hubei.
Culture characteristics: In YM broth, after 7 d at 17 °C, cells are ellipsoidal and cylindrical, 2.0–5.0 × 5.5–11.2 μm and single, budding is polar (Fig. 9E), a sediment is present. After 1 mo at 17 °C, a ring and a sediment are present. On YM agar, after 1 mo at 17 °C, the streak culture is yellow, butyrous, smooth and semi-glossy. The margin is entire. In Dalmau plate culture on corn meal agar, pseudohyphae and hyphae are formed. Sexual structures are not observed on YM, PDA, V8 and CM agar. Ballistoconidia are are ellipsoidal to napiform, 3.0–6.2 × 5.5–8.0 μm (Fig. 9F).
Physiological and biochemical characteristics: Glucose fermentation is absent. Glucose, galactose, sucrose, maltose, cellobiose, trehalose, melibiose, raffinose, melezitose, inulin (weak), soluble starch (weak), D-xylose, L-arabinose, D-arabinose, D-ribose, L-rhamnose, D-glucosamine (weak), erythritol, galactitol, D-mannitol, D-glucitol, Methyl-α-D-glucoside, salicin, DL-lactate, succinate (weak) and myo-inositol are assimilated as sole carbon sources. L-sorbose, lactose, methanol, ethanol, glycerol, ribitol, citrate and hexadecane are not assimilated. Ammonium sulfate, L-lysine, ethylamine hydrochloride and cadaverine dihydrochloride are assimilated as sole nitrogen sources. Potassium nitrate and sodium nitrite are not assimilated. Maximum growth temperature is 28 °C. Growth in vitamin-free medium is negative. Starch-like substances are produced. Growth on 50 % (w/w) glucose-yeast extract agar is negative. Urease activity is positive. Diazonium Blue B reaction is positive.
Physiologically, De. xingshanicus differs from the closely related species De. cylindricus in its inability to assimilate L-sorbose, ribitol and sodium nitrite and its ability to assimilate erythritol (Table S1.11).
Typus: China, Xingshan county, Hubei province, obtained from a leaf of an unidentified plant, Jul. 2003, Q.-M. Wang (holotype CGMCC 2.2459T preserved in a metabolically inactive state, ex-type CBS 15445 = HX16.1).
Derxomyces pseudoyunnanensis Q.M. Wang, F.Y. Bai & A.H. Li sp. nov. MycoBank MB828773. Fig. 9G, H.
Etymology: the specific epithet pseudoyunnanensis refers to the similar colony morphology to that of Derxomyces yunnanensis.
Culture characteristics: In YM broth, after 7 d at 17 °C, cells are ovoid and ellipsoidal, 1.5–4.3 × 5.7–10.0 μm and single, budding is polar (Fig. 9G), a sediment is formed. After 1 mo at 17 °C, a pellicle and sediment are present. On YM agar, after 1 mo at 17 °C, the streak culture is cream, butyrous, wrinkled and dull. The margin is entire. In Dalmau plate culture on corn meal agar, pseudohyphae are formed. Sexual structures are not observed on YM, PDA, V8 and CM agar. Ballistoconidia are globose and subglobosal to napiform, 3.6–4.4 × 3.6–5.1 μm (Fig. 9H).
Physiological and biochemical characteristics: Glucose fermentation is absent. Glucose, galactose, L-sorbose (variable), sucrose, maltose, cellobiose, trehalose, melibiose, raffinose, melezitose, inulin, soluble starch (variable), D-xylose, L-arabinose (variable), D-arabinose, D-ribose (variable), L-rhamnose, galactitol (variable), D-mannitol (variable), D-glucitol (variable), Methyl-α-D-glucoside (variable), salicin (variable) and myo-inositol (weak) are assimilated as sole carbon sources. Lactose, D-glucosamine, N-Acetyl-D-glucosamine, methanol, ethanol, glycerol, erythritol, ribitol, DL-lactate, succinate, citrate and hexadecane are not assimilated. Ammonium sulfate, potassium nitrate (variable), L-lysine, ethylamine hydrochloride, cadaverine dihydrochloride are assimilated as sole nitrogen sources. Sodium nitrite is not assimilated. Maximum growth temperature is 28 °C. Growth in vitamin-free medium is variable. Starch-like substances are not produced. Growth on 50 % (w/w) glucose-yeast extract agar is negative. Urease activity is positive. Diazonium Blue B reaction is positive.
Physiologically, De. pseudoyunnanensis can not be distinguished from its close relative De. longiovatus (Table S1.11).
Typus: China, Simao county, Yunnan province, obtained from a leaf of an unidentified plant, Nov. 2006, Q.-M. Wang (holotype CGMCC 2.3563T preserved in a metabolically inactive state, ex-type CBS 15499 = SM37E2).
Derxomyces longiovatus Q.M. Wang, F.Y. Bai & A.H. Li sp. nov. MycoBank MB828774. Fig. 9I, J.
Etymology: the specific epithet longiovatus refers to the long ovoid vegetative cells of the type strain.
Culture characteristics: In YM broth, after 7 d at 17 °C, cells are long ovoid, cylindrical and ellipsoidal, 1.8–3.7 × 3.9–13.8 μm and single, budding is polar (Fig. 9I), a sediment is formed. After 1 mo at 17 °C, a pellicle and sediment are present. On YM agar, after 1 mo at 17 °C, the streak culture is yellowish-cream, butyrous, dull. The margin is entire or eroded. In Dalmau plate culture on corn meal agar, pseudohyphae and hyphaeare formed. Sexual structures are not observed on YM, PDA, V8 and CM agar. Ballistoconidia are subglobosal to napiform, 3.2–4.5 × 4.8–6.5 μm (Fig. 9J).
Physiological and biochemical characteristics: Glucose fermentation is absent. Glucose, galactose, sucrose, maltose, cellobiose (delayed and weak), trehalose, melibiose, raffinose, melezitose, inulin, soluble starch, D-xylose, L-arabinose, D-arabinose, L-rhamnose (delayed and weak), salicin (delayed and weak) and myo-inositol (weak) are assimilated as sole carbon sources. L-sorbose, lactose, D-ribose, D-glucosamine, N-Acetyl-D-glucosamine, methanol, ethanol, glycerol, erythritol, ribitol, galactitol, D-mannitol, D-glucitol, Methyl-α-D-glucoside, DL-lactate, succinate, citrate and hexadecane are not assimilated. Ammonium sulfate, potassium nitrate, L-lysine, ethylamine hydrochloride, cadaverine dihydrochloride are assimilatedas sole nitrogen sources. Sodium nitrite is not assimilated. Maximum growth temperature is 28 °C. Growth in vitamin-free medium is negative. Starch-like substances are not produced. Growth on 50 % (w/w) glucose-yeast extract agar is negative. Urease activity is positive. Diazonium Blue B reaction is positive.
Physiologically, De. longiovatus and its closely related species De. pseudoyunnanensis as well as De. yunnanensis are very similar. The two new species are not distinguishable, they differ from De. yunnanensis in its ability to assimilate inulin (Table S1.11).
Typus: China, Simao county, Yunnan province, obtained from a leaf of an unidentified plant, Nov. 2006, Q.-M. Wang (holotype CGMCC 2.3535T preserved in a metabolically inactive state, ex-type CBS 15659 = SM35.4).
Derxomyces napiformis Q.M. Wang, F.Y. Bai & A.H. Li sp. nov. MycoBank MB828775. Fig. 9K, L.
Etymology: the specific epithet napiformis refers to the napiform ballistoconidia of the type strain.
Culture characteristics: In YM broth, after 7 d at 17 °C, cells are ellipsoidal to ovoid, 1.5–4.3 × 5.0–8.6 μm and single, budding is polar (Fig. 9K), a sediment is formed. After 1 mo at 17 °C, a ring and sediment are present. On YM agar, after 1 mo at 17 °C, the streak culture is yellowish-cream, butyrous, slightly wrinkled and dull. The margin is entire. In Dalmau plate culture on corn meal agar, pseudohyphae are formed. Sexual structures are not observed on YM, PDA, V8 and CM agar. Ballistoconidia are ellipsoidal to napiform, 2.9–3.6 × 4.2–4.6 μm (Fig. 9L).
Physiological and biochemical characteristics: Glucose fermentation is absent. Glucose, galactose, sucrose, maltose, cellobiose, trehalose, melibiose, raffinose, melezitose, D-xylose, L-arabinose, D-arabinose, L-rhamnose, Methyl-α-D-glucoside, succinate and myo-inositol are assimilated as sole carbon sources. L-sorbose, lactose, inulin, soluble starch, D-ribose, D-glucosamine, methanol, ethanol, glycerol, erythritol, ribitol, galactitol, D-mannitol, D-glucitol, salicin, DL-lactate, citrate and hexadecane are not assimilated. Ammonium sulfate, L-lysine, ethylamine hydrochloride and cadaverine dihydrochloride are assimilated as sole nitrogen sources. Potassium nitrate and sodium nitrite are not assimilated. Maximum growth temperature is 28 °C. Growth in vitamin-free medium is negative. Starch-like substances are not produced. Growth on 50 % (w/w) glucose-yeast extract agar is negative. Urease activity is positive. Diazonium Blue B reaction is positive.
Physiologically, De. napiformis differs from its closely related species De. bifurcus in its inability to assimilate inulin, D-ribose and potassium nitrate and its ability to assimilate Methyl-α-D-glucoside, succinate and myo-inositol (Table S1.11).
Typus: China, Taiwan province, obtained from a leaf of an unidentified plant, Aug. 2009, Q.-M. Wang (holotype CGMCC 2.4446T preserved in a metabolically inactive state, ex-type CBS 15748 = TW1.1F028).
Derxomyces bifurcus Q.M. Wang, F.Y. Bai & A.H. Li sp. nov. MycoBank MB828776. Fig. 9M, N.
Etymology: the specific epithet bifurcus refers to the vegetative cells producing bifurcate budding of the type strain.
Culture characteristics: In YM broth, after 7 d at 17 °C, cells are cylindrical and ellipsoidal, 1.5–2.8 × 5.0–8.3 μm and single, budding is bifurcate or multi-polar (Fig. 9M), a sediment is formed. After 1 mo at 17 °C, a ring and sediment are present. On YM agar, after 1 mo at 17 °C, the streak culture is yellowish-cream, butyrous, wrinkled and dull. The margin is entire or eroded. In Dalmau plate culture on corn meal agar, pseudohyphae are formed. Sexual structures are not observed on YM, PDA, V8 and CM agar. Ballistoconidia are are ellipsoidal to napiform, 3.0–4.0 × 5.0–6.6 μm (Fig. 9N).
Physiological and biochemical characteristics: Glucose fermentation is absent. Glucose, galactose, sucrose, maltose, cellobiose, trehalose, melibiose, raffinose, melezitose, inulin, soluble starch (weak), D-xylose, L-arabinose, D-arabinose, D-ribose and L-rhamnose are assimilated as sole carbon sources. L-sorbose, lactose, D-glucosamine, N-Acetyl-D-glucosamine, methanol, ethanol, glycerol, erythritol, ribitol, galactitol, D-mannitol, D-glucitol, Methyl-α-D-glucoside, salicin, DL-lactate, succinate, citrate, myo-inositol and hexadecane are not assimilated. Ammonium sulfate, potassium nitrate, L-lysine, ethylamine hydrochloride and cadaverine dihydrochloride are assimilated as sole nitrogen sources. Sodium nitrite is not assimilated. Maximum growth temperature is 28 °C. Growth in vitamin-free medium is negative. Starch-like substances are not produced. Growth on 50 % (w/w) glucose-yeast extract agar is negative. Urease activity is positive. Diazonium Blue B reaction is positive.
Physiologically, De. bifurcus differs from its closely related species De. napiformis in its inability to assimilate Methyl-α-D-glucoside, succinate and myo-inositol and its ability to assimilate inulin, D-ribose and potassium nitrate (Table S1.11).
Typus: China, Simao county, Yunnan province, obtained from a leaf of an unidentified plant, Nov. 2006, Q.-M. Wang (holotype CGMCC 2.3470T preserved in a metabolically inactive state, ex-type CBS 15489 = SM37.5).
Derxomyces elongatus Q.M. Wang, F.Y. Bai & A.H. Li sp. nov. MycoBank MB828777. Fig. 9O, P.
Etymology: the specific epithet elongatus refers to the elongate vegetative cells of the type strain.
Culture characteristics: In YM broth, after 7 d at 17 °C, cells are cylindrical and long ellipsoidal, 3.1–6.0 × 6.1–16.7 μm and single, budding is polar (Fig. 9O), a sediment is formed. After 1 mo at 17 °C, a ring and sediment are present. On YM agar, after 1 mo at 17 °C, the streak culture is cream, butyrous, slight wrinkled and dull. The margin is entire. In Dalmau plate culture on corn meal agar, pseudohyphae are formed. Sexual structures are not observed on YM, PDA, V8 and CM agar. Ballistoconidia are globosal and subglobosal to napiform, 3.3–4.0 × 3.3–5.1 μm (Fig. 9P).
Physiological and biochemical characteristics: Glucose fermentation is absent. Glucose, galactose, sucrose, maltose, cellobiose, trehalose, melibiose, raffinose, melezitose, inulin, soluble starch (delayed), D-xylose, L-arabinose, D-arabinose, D-ribose, L-rhamnose, D-glucosamine, N-Acetyl-D-glucosamine, ethanol, glycerol (delayed and weak), galactitol, D-mannitol, D-glucitol, Methyl-α-D-glucoside, succinate and citrate are assimilated as sole carbon sources. L-sorbose, lactose, methanol, erythritol, ribitol, salicin, DL-lactate, myo-inositol and hexadecane are not assimilated. Ammonium sulfate, potassium nitrate, L-lysine, ethylamine hydrochloride and cadaverine dihydrochloride are assimilated as sole nitrogen sources. Sodium nitrite is not assimilated. Maximum growth temperature is 28 °C. Growth in vitamin-free medium is negative. Starch-like substances are not produced. Growth on 50 % (w/w) glucose-yeast extract agar is negative. Urease activity is positive. Diazonium Blue B reaction is positive.
Physiologically, De. elongatus differs from the closely related species De. wuzhishanensis in its inability to grow in vitamin-free medium and its ability to assimilate D-glucosamine, D-mannitol, citrate, potassium nitrate, ethylamine and cadaverine (Table S1.11).
Typus: China, Simao county, Yunnan province, obtained from a leaf of an unidentified plant, Nov. 2006, Q.-M. Wang (holotype CGMCC 2.3561T preserved in a metabolically inactive state, ex-type CBS 15498 = SM32.1).
Derxomyces melastomatis Q.M. Wang, F.Y. Bai & A.H. Li sp. nov. MycoBank MB828778. Fig. 10A, B.
Fig. 10.
Vegetative cells grown in YM broth for 5 d at 17 °C and ballistoconidia produced on corn meal agar after 7 d at 17 °C. (A, B) De. melastomatis CGMCC 2.3459T; (C, D) De. taiwanicus CGMCC 2.4429T; (E, F) De. ovatus CGMCC 2.3572T; (G, H) De. longicylindricus CGMCC 2.5660T; ; (I) Pha. lactea CGMCC 2.5810T; (J) Pha. ovata CGMCC 2.5614T; (K, L) Ho. saccardoi CGMCC 2.3445T; (M) So. gelidoterrea CGMCC 2.5814T; (N) Fi. dingjieense CGMCC 2.5649T, (O) Fi. globosum CGMCC 2.5680T; (P) Fi. mali CGMCC 2.4012T. Bars = 10 μm.
Etymology: the specific epithet melastomatis refers to Melastoma, the plant genus from which the type strain was isolated.
Culture characteristics: In YM broth, after 7 d at 17 °C, cells are ovoid and ellipsoidal, 2.3–4.0 × 4.7–8.2 μm and single, budding is polar (Fig. 10A), a sediment is present. After 1 mo at 17 °C, a ring and a sediment are present. On YM agar, after 1 mo at 17 °C, the streak culture is yellowish-cream, butyrous, smooth and dull. The margin is entire. In Dalmau plate culture on corn meal agar, pseudohyphae are not formed. Sexual structures are not observed on YM, PDA, V8 and CM agar. Ballistoconidia are are ellipsoidal to napiform, 2.7–4.0 × 2.9–5.3 μm (Fig. 10B).
Physiological and biochemical characteristics: Glucose fermentation is absent. Glucose, galactose, sucrose, maltose, cellobiose, trehalose, melibiose, raffinose (weak), melezitose, inulin, soluble starch (weak), D-xylose, L-arabinose, D-arabinose (weak), D-ribose (weak), L-rhamnose, galactitol, D-mannitol (weak), D-glucitol, Methyl-α-D-glucoside, salicin (weak), succinate and myo-inositol are assimilated as sole carbon sources. L-sorbose, lactose, D-glucosamine, N-Acetyl-D-glucosamine, methanol, ethanol, glycerol, erythritol, ribitol, DL-lactate, citrate and hexadecane are not assimilated. Ammonium sulfate, L-lysine (weak), ethylamine hydrochloride and cadaverine dihydrochloride are assimilated as sole nitrogen sources. Potassium nitrate and sodium nitrite are not assimilated. Maximum growth temperature is 28 °C. Growth in vitamin-free medium is negative. Starch-like substances are not produced. Growth on 50 % (w/w) glucose-yeast extract agar is negative. Urease activity is positive. Diazonium Blue B reaction is positive.
Physiologically, De. melastomatis differs from the closely related species De. komagatae, De. schimicola and De. pseudoschimicola in its ability to assimilate inulin (Table S1.11).
Typus: China, Wuzhishan mountain, Hainan province, obtained from a leaf Melastoma candidum, Nov. 2006, Q.-M. Wang (holotype CGMCC 2.3459T preserved in a metabolically inactive state, ex-type CBS 15485 = WZS19.7).
Derxomyces taiwanicus Q.M. Wang, F.Y. Bai & A.H. Li sp. nov. MycoBank MB828779. Fig. 10C, D.
Etymology: the specific epithet taiwanicus refers to the geographic origin of the type strain, Taiwan.
Culture characteristics: In YM broth, after 7 d at 17 °C, cells are ellipsoidal and cylindrical, 3.0–3.7 × 4.4–8.2 μm and single, budding is polar (Fig. 10C), a sediment is present. After 1 mo at 17 °C, a ring and a sediment are present. On YM agar, after 1 mo at 17 °C, the streak culture is pale-yellow, butyrous, wrinkled and dull. The margin is entire. In Dalmau plate culture on corn meal agar, pseudohyphae are formed. Sexual structures are not observed on YM, PDA, V8 and CM agar. Ballistoconidia are are ellipsoidal to napiform, 2.9–4.3 × 3.0–4.3 μm (Fig. 10D).
Physiological and biochemical characteristics: Glucose fermentation is absent. Glucose, galactose, L-sorbose (delayed and weak), sucrose, maltose, cellobiose, trehalose, melibiose, raffinose, melezitose, D-xylose, L-arabinose, D-arabinose (delayed and weak), D-ribose (delayed and weak), L-rhamnose, ribitol (delayed and weak), galactitol (delayed and weak), D-mannitol, D-glucitol (delayed and weak), Methyl-α-D-glucoside, salicin (delayed and weak) and succinate are assimilated as sole carbon sources. Lactose, inulin, soluble starch, D-glucosamine, methanol, ethanol, glycerol, erythritol, DL-lactate, citrate, myo-inositol and hexadecane are not assimilated. Ammonium sulfate and L-lysine are assimilated as sole nitrogen sources. Potassium nitrate, sodium nitrite, ethylamine hydrochloride and cadaverine dihydrochloride are not assimilated. Maximum growth temperature is 28 °C. Growth in vitamin-free medium is negative. Starch-like substances are not produced. Growth on 50 % (w/w) glucose-yeast extract agar is negative. Urease activity is positive. Diazonium Blue B reaction is positive.
Physiologically, De. taiwanicus differs from the closely related species De. ovatus in its inability to assimilate myo-inositol (Table S1.11).
Typus: China, Taiwan province, obtained from a leaf of an unidentified plant, Aug. 2009, Q.-M. Wang (holotype CGMCC 2.4429T preserved in a metabolically inactive state, ex-type CBS 12490 = TW3.1C-02).
Derxomyces ovatus Q.M. Wang, F.Y. Bai & A.H. Li sp. nov. MycoBank MB828780. Fig. 10E, F.
Etymology: the specific epithet ovatus refers to the ovoid vegetative cells of the type strain.
Culture characteristics: In YM broth, after 7 d at 17 °C, cells are ovoid or ellipsoidal, 2.0–5.4 × 3.8–7.7 μm and single, budding is polar (Fig. 10E), a sediment is present. After 1 mo at 17 °C, a ring and a sediment are present. On YM agar, after 1 mo at 17 °C, the streak culture is yellow, butyrous, smooth and dull. The margin is entire. In Dalmau plate culture on corn meal agar, pseudohyphae are formed. Sexual structures are not observed on YM, PDA, V8 and CM agar. Ballistoconidia are ellipsoidal to napiform, 1.8–3.6 × 3.0–4.5 μm (Fig. 10F).
Physiological and biochemical characteristics: Glucose fermentation is absent. Glucose, galactose, L-sorbose (delayed and weak), sucrose, maltose, cellobiose, trehalose, melibiose, raffinose, melezitose, inulin (delayed and weak), soluble starch (weak), D-xylose, L-arabinose, L-rhamnose, ethanol (delayed and weak), galactitol, D-mannitol, D-glucitol, Methyl-α-D-glucoside, salicin (delayed and weak), succinate and myo-inositol are assimilated as sole carbon sources. Lactose, D-arabinose, D-ribose, D-glucosamine, N-Acetyl-D-glucosamine, methanol, glycerol, erythritol, ribitol, DL-lactate, citrate and hexadecane are not assimilated. Ammonium sulfate, potassium nitrate (delayed and weak) and L-lysine are assimilated as sole nitrogen sources. Sodium nitrite, ethylamine hydrochloride and cadaverine dihydrochloride are not assimilated. Maximum growth temperature is 28 °C. Growth in vitamin-free medium is netative. Starch-like substances are not produced. Growth on 50 % (w/w) glucose-yeast extract agar is negative. Urease activity is positive. Diazonium Blue B reaction is positive.
Physiologically, De. ovatus differs from the closely related species De. taiwanicus. in its ability to assimilate myo-inositol (Table S1.11).
Typus: China, Simao county, Yunnan province, obtained from a leaf of an unidentified plant, Nov. 2006, Q.-M. Wang (holotype CGMCC 2.3572T preserved in a metabolically inactive state, ex-type CBS 15654 = SM32.2).
Derxomyces longicylindricus Q.M. Wang, F.Y. Bai & A.H. Li sp. nov. MycoBank MB828781. Fig. 10G, H.
Etymology: the specific epithet longicylindricus refers to the long cylindrical vegetative cells of the type strain.
Culture characteristics: In YM broth, after 7 d at 17 °C, cells are long cylindrical, 2.9–5.0 × 7.1–22 μm and single, budding is polar (Fig. 10G), a sediment is present. After 1 mo at 17 °C, a ring and a sediment are present. On YM agar, after 1 mo at 17 °C, the streak culture is yellowish-cream, butyrous, wrinkled and dull. The margin is entire. In Dalmau plate culture on corn meal agar, pseudohyphae are not formed. Sexual structures are not observed on YM, PDA, V8 and CM agar. Ballistoconidia are ellipsoidal to napiform, 2.4–-4.2 × 3.6–6.0 μm (Fig. 10H).
Physiological and biochemical characteristics: Glucose fermentation is absent. Glucose, galactose, L-sorbose (weak), sucrose, maltose, cellobiose (weak), trehalose, melibiose, raffinose, melezitose, inulin, soluble starch (weak), D-xylose, L-arabinose, L-rhamnose, D-glucitol (delayed and weak), Methyl-α-D-glucoside and succinate (delayed and weak) are assimilated as sole carbon sources. Lactose, D-ribose, D-arabinose, D-glucosamine, N-Acetyl-D-glucosamine, methanol, ethanol, glycerol, erythritol, ribitol, galactitol, D-mannitol, salicin, DL-lactate, citrate, myo-inositol and hexadecane are not assimilated. Ammonium sulfate, L-lysine, ethylamine hydrochloride and cadaverine dihydrochloride are assimilated as sole nitrogen sources. Potassium nitrate and sodium nitrite are not assimilated. Maximum growth temperature is 28 °C. Growth in vitamin-free medium is negative. Starch-like substances are not produced. Growth on 50 % (w/w) glucose-yeast extract agar is negative. Urease activity is positive. Diazonium Blue B reaction is positive.
Physiologically, De. longicylindricus differs from the closely related species De. linzhiensis in its inability to assimilate D-arabinose, galactitol, D-mannitol and cadaverine and its ability to assimilate L-rhamnose, L-lysine and ethylamine (Table S1.11).
Typus: China, Beibeng county, Motuo, Tibet, obtained from a leaf of an unidentified plant, Sep. 2014, Q.-M. Wang (holotype CGMCC 2.5660T preserved in a metabolically inactive state, ex-type CBS 13979 = XZ132E37A).
Phaeotremella lactea Q.M. Wang, F.Y. Bai & A.H. Li sp. nov. MycoBank MB828782. Fig. 10I.
Etymology: the specific epithet lactea refers to the colony colour of this species.
Culture characteristics: In YM broth, after 7 d at 17 °C, cells are ellipsoidal, 2.7–4.0 × 4.4–6.6 μm and single, budding is polar (Fig. 10I), a sediment is present. After 1 mo at 17 °C, a pellicle and sediment are present. On YM agar, after 1 mo at 17 °C, the streak culture is cream, butyrous, smooth and glossy. The margin is entire. In Dalmau plate culture on corn meal agar, pseudohyphae are not formed. Sexual structures are not observed on YM, PDA, V8 and CM agar. Ballistoconidia are not produced.
Physiological and biochemical characteristics: Glucose fermentation is absent. Glucose, galactose, sucrose, maltose, cellobiose, trehalose, lactose, melibiose, raffinose, melezitose, inulin, D-xylose, L-arabinose, D-arabinose, D-ribose, L-rhamnose, D-glucosamine, ribitol, D-mannitol, D-glucitol, salicin, D-gluconate, succinate and myo-inositol are assimilated as sole carbon sources. L-sorbose, soluble starch, N-Acetyl-D-glucosamine, methanol, ethanol, glycerol, erythritol, galactitol, Methyl-α-D-glucoside, DL-lactate, citrate and hexadecane are not assimilated. Ammonium sulfate, potassium nitrate (weak), sodium nitrite (weak), L-lysine (weak) and ethylamine hydrochloride (weak) are assimilated as sole nitrogen sources. Cadaverine dihydrochloride is not assimilated. Maximum growth temperature is 28 °C. Growth in vitamin-free medium is positive (weak). Starch-like substances are not produced. Growth on 50 % (w/w) glucose-yeast extract agar is negative. Urease activity is positive. Diazonium Blue B reaction is positive.
Physiologically, Pha. lactea differs from the closely related species Pha. ovata in its inability to assimilate soluble starch, N-Acetyl-D-glucosamine, galactitol, Methyl-α-D-glucoside and cadaverine and its ability to assimilate raffinose, succinate and myo-inositol (Table S1.12).
Typus: China, Milin county, Tibet, obtained from a leaf of an unidentified plant, Sep. 2015, Q.-M. Wang (holotype CGMCC 2.5810T preserved in a metabolically inactive state, ex-type CBS 15574 = GPS20.4A1B).
Phaeotremella ovata Q.M. Wang, F.Y. Bai & A.H. Li sp. nov. MycoBank MB828783. Fig. 10J.
Etymology: the specific epithet ovata refers to the ovoid vegetative cells of the type strain.
Culture characteristics: In YM broth, after 7 d at 17 °C, cells are ovoid and fusiform, 2.0–3.4 × 4.8–8.2 μm and single, budding is polar (Fig. 10J), a sediment is formed. After 1 mo at 17 °C, a pellicle and sediment are present. On YM agar, after 1 mo at 17 °C, the streak culture is yellow, butyrous, smooth. The margin is entire. In Dalmau plate culture on corn meal agar, pseudohyphae are not formed. Sexual structures are not observed on YM, PDA, V8 and CM agar. Ballistoconidia are not produced.
Physiological and biochemical characteristics: Glucose fermentation is absent. Glucose, galactose, sucrose, maltose, cellobiose, trehalose, lactose, melibiose, melezitose, inulin, soluble starch, D-xylose, L-arabinose, D-arabinose, D-ribose, L-rhamnose, D-glucosamine, N-Acetyl-D-glucosamine, ribitol, galactitol, D-mannitol, D-glucitol, Methyl-α-D-glucoside, salicin and D-gluconate are assimilated as sole carbon sources. L-sorbose, raffinose, methanol, ethanol, glycerol, erythritol, DL-lactate, succinate, citrate, myo-inositol and hexadecane are not assimilated. Ammonium sulfate, potassium nitrate, L-lysine, ethylamine hydrochloride and cadaverine dihydrochloride are assimilated as sole nitrogen sources. Sodium nitrite is not assimilated. Maximum growth temperature is 28 °C. Growth in vitamin-free medium is positive. Starch-like substances are not produced. Growth on 50 % (w/w) glucose-yeast extract agar is negative. Urease activity is positive. Diazonium Blue B reaction is positive.
Physiologically, Pha. ovata differs from its closely related species Pha. lactea in its inability to assimilate raffinose, succinate and myo-inositol and its ability to assimilate soluble starch, N-Acetyl-D-glucosamine, Methyl-α-D-glucoside and cadaverine (Table S1.12).
Typus: China, Nanwenghe, Heilongjiang province, obtained from a leaf of an unidentified plant, Aug. 2015, Q.-M. Wang (holotype CGMCC 2.5614T preserved in a metabolically inactive state, ex-type CBS 15756 = NW9D3).
Holtermannia Sacc. & Traverso, Syll. Fung. 19: 871. 1910. emend. Q.M. Wang, F.Y. Bai & A.H. Li.
Type species: Holtermannia pinguis (Holterm.) Sacc. & Traverso.
This genus is emended to include Holtermannia corniformis and six other sexual species (Kobayasi 1937), and one newly described anamorphic species Holtermannia saccardoi (Figs 2E and S1E).
Sexual reproduction observed in most species. For teleomorphic taxa, the corniform basidiocarps are narrowly clavate and often slightly compressed. The basidiocarps are simple or infrequently branched. The tertiary hyphae have clamp connections (Bandoni et al. 2011). Colonies whitish to cream, mucoid. Budding cells present. Ballistoconidia formed or not.
Holtermannia saccardoi Q.M. Wang, F.Y. Bai & A.H. Li sp. nov. MycoBank MB828784. Fig. 10K, L.
Etymology: the specific epithet saccardoi named in honour of P.A. Saccardo for his proposal of the genus Holtermannia.
Culture characteristics: In YM broth, after 7 d at 17 °C, cells are globosal, ovoid and ellipsoidal, 3.1–5.8 × 3.6–6.4 μm and single, budding is polar (Fig. 10K), a sediment is formed. After 1 mo at 17 °C, a ring and a sediment are present. On YM agar, after 1 mo at 17 °C, the streak culture is cream, mucoid, smooth and shiny. The margin is entire. In Dalmau plate culture on corn meal agar, pseudohyphae are not formed. Sexual structures are not observed on YM, PDA, V8 and CM agar. Ballistoconidia are ellipsoidal, 4.1–5.9 × 7.4–9.1 μm (Fig. 10L).
Physiological and biochemical characteristics: Glucose fermentation is absent. Glucose, galactose (variable), sucrose, maltose, cellobiose, trehalose, lactose (variable), melibiose, raffinose, melezitose, soluble starch, D-xylose, L-arabinose, D-arabinose, D-ribose, L-rhamnose, ethanol, glycerol, erythritol, ribitol, galactitol, D-mannitol, D-glucitol, Methyl-α-D-glucoside, salicin, DL-lactate (variable), succinate (weak), citrate (variable) and myo-inositol are assimilated as sole carbon sources. L-sorbose, inulin, D-glucosamine, methanol and hexadecane are not assimilated. Ammonium sulfate, potassium nitrate, L-lysine and cadaverine dihydrochloride (variable) are assimilated as sole nitrogen sources. Sodium nitrite and ethylamine hydrochloride are not assimilated. Maximum growth temperature is 30 °C. Growth in vitamin-free medium is positive. Starch-like substances are not produced. Growth on 50 % (w/w) glucose-yeast extract agar is negative. Urease activity is positive. Diazonium Blue B reaction is positive.
Physiologically, Ho. saccardoi differs from its closely related species Ho. corniformis in its inability to assimilate L-sorbose and its ability to assimilate melibiose, raffinose, erythritol and potassium nitrate (Table S1.13).
Typus: China, Simao county, Yunnan province, obtained from a leaf of an unidentified plant, Nov. 2006, Q.-M. Wang (holotype CGMCC 2.3445T preserved in a metabolically inactive state, ex-type CBS 15479 = SM37.10).
Solicoccozyma gelidoterrea Q.M. Wang, F.Y. Bai & A.H. Li sp. nov. MycoBank MB828785. Fig. 10M.
Etymology: the specific epithet gelidoterrea refers to the cold environments origin of all strains used in this study.
Culture characteristics: In YM broth, after 7 d at 17 °C, cells are ellipsoidal and ovoid, 3.3–4.8 × 4.1–5.5 μm and single, budding is polar (Fig. 10M), a sediment is formed. After 1 mo at 17 °C, a ring and a sediment are present. On YM agar, after 1 mo at 17 °C, the streak culture is cream, butyrous, smooth and glistening. The margin is entire. In Dalmau plate culture on corn meal agar, pseudohyphae are not formed. Sexual structures are not observed on YM, PDA, V8 and CM agar. Ballistoconidia are not produced.
Physiological and biochemical characteristics: Glucose fermentation is absent. Glucose, galactose, L-sorbose, sucrose, maltose, cellobiose, trehalose (variable), lactose, melibiose (variable), raffinose, melezitose, inulin, D-xylose, L-arabinose, D-arabinose, D-ribose, L-rhamnose, D-glucosamine, N-Acetyl-D-glucosamine, ethanol, ribitol, galactitol, glycerol (variable), D-mannitol, D-glucitol, Methyl-α-D-glucoside, salicin, D-gluconate and myo-inositol are assimilated as sole carbon sources. Soluble starch, methanol, erythritol, DL-lactate, succinate, citrate and hexadecane are not assimilated. Ammonium sulfate, potassium nitrate, sodium nitrite, L-lysine, ethylamine hydrochloride and cadaverine dihydrochloride are assimilated as sole nitrogen sources. Maximum growth temperature is 32 °C. Growth in vitamin-free medium is positive. Starch-like substances are not produced. Growth on 50 % (w/w) glucose-yeast extract agar is negative. Urease activity is positive. Diazonium Blue B reaction is positive.
Physiologically, So. gelidoterrea differs from its four closely related species, So. aeria, So. terrea, So. phenolica and So. fuscescens, in its inability to assimilate succinate and its ability to assimilate inulin (Table S1.14).
Typus: China, Daxinganling, obtained from soil, Aug. 2015, Q.-M. Wang (holotype CGMCC 2.5814T preserved in a metabolically inactive state, ex-type CBS 15580 = HFB003-3).
Filobasidium dingjieense Q.M. Wang, F.Y. Bai & A.H. Li sp. nov. MycoBank MB828786. Fig. 10N.
Etymology: the specific epithet dingjieenese refers to the geographic origin of the type strain, Dingjie county, Tibet.
Culture characteristics: In YM broth, after 7 d at 17 °C, cells are globosal and ellipsoidal, 6.8–10.6 × 6.9–10.6 μm and single, budding is polar (Fig. 10N), a sediment is present. After 1 mo at 17 °C, a ring and a sediment are present. On YM agar, after 1 mo at 17 °C, the streak culture is gray-cream, mucoid, smooth and glossy. The margin is entire. In Dalmau plate culture on corn meal agar, pseudohyphae are formed. Sexual structures are not observed on YM, PDA, V8 and CM agar. Ballistoconidia are not produced.
Physiological and biochemical characteristics: Glucose fermentation is absent. Glucose, galactose (delayed and weak), sucrose, maltose, cellobiose, trehalose, melezitose, D-xylose, L-arabinose, ethanol (delayed and weak), glycerol (delayed and weak), Methyl-α-D-glucoside (weak), succinate, citrate and myo-inositol are assimilated as sole carbon sources. L-sorbose, lactose, melibiose, raffinose, inulin, soluble starch, D-arabinose, D-ribose, L-rhamnose, D-glucosamine, N-Acetyl-D-glucosamine, methanol, erythritol, ribitol, galactitol, D-mannitol, D-glucitol, salicin, DL-lactate and hexadecane are not assimilated. Ammonium sulfate, potassium nitrate, sodium nitrite, ethylamine hydrochloride (delayed and weak) and cadaverine dihydrochloride (delayed and weak) are assimilated as sole nitrogen sources. L-lysine is not assimilated. Maximum growth temperature is 19 °C. Growth in vitamin-free medium is positive. Starch-like substances are not produced. Growth on 50 % (w/w) glucose-yeast extract agar is negative. Urease activity is positive. Diazonium Blue B reaction is positive.
Physiologically, Fi. dingjieense differs from its closely related species Fi. uniguttulatum in its inability to assimilate raffinose, L-rhamnose, N-Acetyl-D-glucosamine, ribitol, D-mannitol, D-glucitol, salicin, hexadecane and L-lysine and its ability to assimilate cellobiose, potassium nitrate and sodium nitrite (Table S1.15).
Typus: China, Dingjie county, Tibet, obtained from a leaf of an unidentified plant, Sep. 2015, Q.-M. Wang (holotype CGMCC 2.5649T preserved in a metabolically inactive state, ex-type CBS 15567 = GPS3.2A5).
Filobasidium globosum Q.M. Wang, F.Y. Bai & A.H. Li sp. nov. MycoBank MB828788. Fig. 10O.
Etymology: the specific epithet globosum refers to the globosal vegetative cells of the type strain.
Culture characteristics: In YM broth, after 7 d at 17 °C, cells are globosal, 2.7–6.7 × 2.7–6.7 μm and single, budding is polar (Fig. 10O), a sediment is present. After 1 mo at 17 °C, a ring and a sediment are present. On YM agar, after 1 mo at 17 °C, the streak culture is gray-cream, mucoid, smooth and shiny. The margin is entire. In Dalmau plate culture on corn meal agar, pseudohyphae are not formed. Sexual structures are not observed on YM, PDA, V8 and CM agar. Ballistoconidia are not produced.
Physiological and biochemical characteristics: Glucose fermentation is absent. Glucose, galactose, sucrose, maltose, cellobiose, trehalose, lactose, melibiose, raffinose, melezitose, inulin, D-xylose (delayed and weak), L-arabinose, L-rhamnose (delayed and weak), D-mannitol, Methyl-α-D-glucoside (weak), succinate (weak) and myo-inositol (weak) are assimilated as sole carbon sources. L-sorbose, soluble starch, D-arabinose, D-ribose, D-glucosamine, N-Acetyl-D-glucosamine, methanol, ethanol, glycerol, erythritol, ribitol, galactitol, D-glucitol, salicin, DL-lactate, citrate and hexadecane are not assimilated. Ammonium sulfate, potassium nitrate, L-lysine, ethylamine hydrochloride and cadaverine dihydrochloride are assimilated as sole nitrogen sources. Sodium nitrite is not assimilated. Maximum growth temperature is 28 °C. Growth in vitamin-free medium is positive. Starch-like substances are not produced. Growth on 50 % (w/w) glucose-yeast extract agar is negative. Urease activity is positive. Diazonium Blue B reaction is positive.
Physiologically, Fi. globosum differs from its closely related species Fi. mali in its inability to assimilate ribitol, galactitol, salicin and ethylamine and its ability to assimilate lactose and grow in vitamin-free medium (Table S1.15).
Typus: China, Yichun county, Heilongjiang province, obtained from a leaf of an unidentified plant, Aug. 2014, Q.-M. Wang (holotype CGMCC 2.5680T preserved in a metabolically inactive state, ex-type CBS 15658 = HLJ8A3).
Filobasidium mali Q.M. Wang, F.Y. Bai & A.H. Li sp. nov. MycoBank MB828789. Fig. 10, Fig. 11A.
Fig. 11.
SEM image of vegetative cells grown in YM broth for 5 d at 17 °C. (A) Fi. mali CGMCC 2.4012T, Bars = 4 μm; (B) Boe. sterigmata CGMCC 2.4539T, Bars = 5 μm; (C) St. layueensis CGMCC 2.5817T, Bars = 5 μm; (D) Pse. motuoensis CGMCC 2.5816T, Bars = 2 μm; (E) Me. layueensis CGMCC 2.5818T, Bars = 5 μm; (F) Beg. foliicola CGMCC 2.3164T, Bars = 1 μm; (G, H) Ros. petaloides CGMCC 2.3446T, G Bars = 10 μm, H Bars = 3 μm.
Etymology: the specific epithet mali refers to the substrate origin of the type strain, Malus.
Culture characteristics: In YM broth, after 7 d at 17 °C, cells are subglobosal and ellipsoidal, 3.0–4.6 × 3.0–7.7 μm and single, budding is polar (Fig. 10P), a sediment is present. After 1 mo at 17 °C, a ring and a sediment are present. On YM agar, after 1 mo at 17 °C, the streak culture is gray-cream, mucoid, smooth and shiny. The margin is entire. In Dalmau plate culture on corn meal agar, pseudohyphae are not formed. Sexual structures are not observed on YM, PDA, V8 and CM agar. Ballistoconidia are not produced.
Physiological and biochemical characteristics: Glucose fermentation is absent. Glucose, Galactose, L-sorbose, sucrose, maltose, cellobiose (or weak), trehalose, melibiose (or weak), raffinose (or weak), melezitose (or weak), D-xylose (or delayed and weak), L-arabinose (or weak), L-rhamnose (or delayed and weak), ethanol (or weak), D-mannitol, ribitol, galactitol, Methyl-α-D-glucoside (or weak), salicin (or weak), D-Gluconate (weak), succinate (delayed and weak) and myo-inositol (delayed and weak) are assimilated as sole carbon sources. Lactose (variable), inulin, soluble starch, D-arabinose (variable), D-ribose, D-glucosamine, N-Acetyl-D-glucosamine, methanol, glycerol, D-glucitol (variable), erythritol, DL-lactate, citrate and hexadecane are not assimilated. Ammonium sulfate, potassium nitrate (weak), L-lysine, and cadaverine dihydrochloride are assimilated as sole nitrogen sources. Sodium nitrite and ethylamine hydrochloride (variable) are not assimilated. Maximum growth temperature is 32 °C. Growth in vitamin-free medium is negative. Starch-like substances are produced or not. Growth on 50 % (w/w) glucose-yeast extract agar is negative. Urease activity is positive. Diazonium Blue B reaction is positive.
Physiologically, Fi. mali differs from its closely related species Fi. globosum in its inability to grow in vitamin-free medium and its ability to assimilate ribitol, galactitol and salicin (Table S1.15).
Typus: China, Tai’an county, Shandong province, obtained from isolated from apple, Aug. 2008, Q.-M. Wang (holotype CGMCC 2.4012T preserved in a metabolically inactive state, ex-type CBS 15651 = KTAPG4-11.64).
Filobasidium mucilaginum Q.M. Wang, F.Y. Bai & A.H. Li sp. nov. MycoBank MB828790. Fig. 12A.
Fig. 12.
Vegetative cells grown in YM broth for 5 d at 17 °C and ballistoconidia produced on corn meal agar after 7 d at 17 °C. (A) Fi. mucilaginum CGMCC 2.3463T; (B) Pha. aurantiaca CGMCC 2.5601T; (C, D) Kon. cylindrica CGMCC 2.3102T; (E, F) Kon. chamaenerii CGMCC 2.2652T; (G, H) Kon. foliicola CGMCC 2.3100T; (I, J) Kon. arboricola CGMCC 2.2621T; (K, L) Kon. lulangica CGMCC 2.2762T; (M, N) Kon. daliangziensis CGMCC 2.5610T, (O) Kon. ribitophobia CGMCC 2.4441T; (P) Kon. myxariophila CBS 8379T. Bars = 10 μm.
Etymology: the specific epithet mucilaginum refers to the mucoid colony morphology of the type strain.
Culture characteristics: In YM broth, after 7 d at 17 °C, cells are subglobosal and ellipsoidal, 3.8–8.1 × 3.8–8.8 μm and single, budding is polar (Fig. 12A), a sediment is present. After 1 mo at 17 °C, a ring and a sediment are present. On YM agar, after 1 mo at 17 °C, the streak culture is gray-cream, mucoid, smooth and shiny. The margin is entire. In Dalmau plate culture on corn meal agar, pseudohyphae are not formed. Sexual structures are not observed on YM, PDA, V8 and CM agar. Ballistoconidia are not produced.
Physiological and biochemical characteristics: Glucose fermentation is absent. Glucose, galactose, sucrose, maltose, cellobiose, trehalose, lactose (delayed and weak), melibiose (delayed and weak), raffinose, melezitose, soluble starch (weak), D-xylose, L-arabinose (delayed and weak), D-arabinose (delayed and weak), ethanol (delayed and weak), glycerol (delayed and weak), erythritol (delayed and weak), ribitol (delayed and weak), galactitol, D-mannitol, D-glucitol, Methyl-α-D-glucoside, salicin, succinate (delayed and weak) and myo-inositol (delayed and weak) are assimilated as sole carbon sources. L-sorbose, inulin, D-ribose, L-rhamnose, D-glucosamine, methanol, DL-lactate, citrate and hexadecane are not assimilated. Ammonium sulfate and potassium nitrate (delayed and weak) are assimilated as sole nitrogen sources. Sodium nitrite, L-lysine, ethylamine hydrochloride and cadaverine dihydrochloride are not assimilated. Maximum growth temperature is 28 °C. Growth in vitamin-free medium is positive. Starch-like substances are not produced. Growth on 50 % (w/w) glucose-yeast extract agar is negative. Urease activity is positive. Diazonium Blue B reaction is positive.
Physiologically, Fi. mucilaginum differs from its closely related species Fi. globosum and Fi. mali in its inability to assimilate Methyl-α-D-glucoside and its ability to assimilate L-sorbose and D-glucitol (Table S1.15).
Typus: China, Sanya county, Hainan province, obtained from a leaf of an unidentified plant, Nov. 2006, Q.-M. Wang (holotype CGMCC 2.3463T preserved in a metabolically inactive state, ex-type CBS 15486 = SY2.1).
Phaffia aurantiaca Q.M. Wang, F.Y. Bai & A.H. Li sp. nov. MycoBank MB828791. Fig. 12B.
Etymology: the specific epithet aurantiaca refers to the orange colony colour of the type strain.
Culture characteristics: In YM broth, after 7 d at 17 °C, cells are ovoid and ellipsoidal, 3.4–6.4 × 5.2–8.9 μm and single, budding is polar (Fig. 12B), a sediment is present. On YM agar, after 1 mo at 17 °C, the streak culture is orange, butyrous, smooth and glossy. The margin is entire. In Dalmau plate culture on corn meal agar, pseudohyphae are not formed. Sexual structures are not observed on YM, PDA, V8 and CM agar. Ballistoconidia are not produced.
Physiological and biochemical characteristics: Glucose fermentation is absent. Glucose, galactose, sucrose, maltose, cellobiose, trehalose, lactose, melibiose, raffinose, melezitose, D-xylose (delayed and weak), L-arabinose, D-ribose, ethanol, glycerol, erythritol, ribitol (delayed and weak), galactitol (delayed and weak), D-mannitol, D-glucitol, Methyl-α-D-glucoside (delayed and weak), salicin (delayed and weak), DL-lactate and succinate are assimilated as sole carbon sources. L-sorbose, inulin, soluble starch, D-arabinose, L-rhamnose, D-glucosamine, N-Acetyl-D-glucosamine, methanol, citrate, myo-inositol and hexadecane are not assimilated. Ammonium sulfate, L-lysine, ethylamine hydrochloride and cadaverine dihydrochloride are assimilated as sole nitrogen sources. Potassium nitrate and sodium nitrite are not assimilated. Maximum growth temperature is 23 °C. Growth in vitamin-free medium is negative. Starch-like substances are produced. Growth on 50 % (w/w) glucose-yeast extract agar is negative. Urease activity is positive. Diazonium Blue B reaction is positive.
Physiologically, Pha. aurantiaca differs from its closely related species Pha. rhodozyma in its inability to assimilate soluble starch and its ability to assimilate galactose, lactose, melibiose, erythritol and ethylamine (Table S1.16).
Typus: China, Lulang county, Tibet, obtained from a leaf of an unidentified plant, Sep. 2015, Q.-M. Wang (holotype CGMCC 2.5601T preserved in a metabolically inactive state, ex-type CBS 15548 = GPS23.2A4).
New taxa in Agaricostilbomycetes (Pucciniomycotina)
Kondoa cylindrica Q.M. Wang, F.Y. Bai & A.H. Li sp. nov. MycoBank MB828792. Fig. 12C, D.
Etymology: the specific epithet cylindrica refers to the cylindrical ballistoconidia of the type strain.
Culture characteristics: In YM broth, after 7 d at 17 °C, cells are ellipsoidal and cylindrical, 2.4–4.8 × 4.8–8.5 μm and single, budding is polar (Fig. 12C), a sediment is formed. After 1 mo at 17 °C, a ring and a sediment are present. On YM agar, after 1 mo at 17 °C, the streak culture is pale orange, butyrous, smooth and semi-glossy. The margin is entire. In Dalmau plate culture on corn meal agar, pseudohyphae are not formed. Sexual structures are not observed on YM, PDA, V8 and CM agar. Ballistoconidia are cylindrical, 2.1–2.9 × 4.3–5.7 μm (Fig. 12D).
Physiological and biochemical characteristics: Glucose fermentation is absent. Glucose, sucrose, maltose, cellobiose (variable), trehalose, raffinose (variable), melezitose (variable), soluble starch, D-xylose (variable), L-arabinose (variable), D-ribose (variable), L-rhamnose, ethanol (variable), glycerol, erythritol (variable), ribitol (variable), galactitol (variable), D-mannitol, D-glucitol, Methyl-α-D-glucoside (variable), salicin (variable), succinate (delayed and weak) and citrate (variable) are assimilated as sole carbon sources. Galactose, L-sorbose, lactose, melibiose, inulin, D-arabinose, D-glucosamine, N-Acetyl-D-glucosamine, methanol, D-gluconate, DL-lactate, myo-inositol and hexadecane are not assimilated. Ammonium sulfate, potassium nitrate (variable) and cadaverine dihydrochloride (variable) are assimilated as sole nitrogen sources. Sodium nitrite, L-lysine and ethylamine hydrochloride are not assimilated. Maximum growth temperature is 22–25 °C. Growth in vitamin-free medium is positive. Starch-like substances are not produced. Growth on 50 % (w/w) glucose-yeast extract agar is negative. Urease activity is positive. Diazonium Blue B reaction is positive.
Physiologically, Kon. cylindrica differs from its closely related species Kon. aeria and Kon. malvinella in its inability to assimilate DL-lactate and its ability to grow in vitamin-free medium (Table S1.17).
Typus: Germany, obtained from a leaf of an unidentified plant, Sep. 2005 (holotype CGMCC 2.3102T preserved in a metabolically inactive state, ex-type CBS 15466 = G6.1-1).
Kondoa chamaenerii Q.M. Wang, F.Y. Bai & A.H. Li sp. nov. MycoBank MB828793. Fig. 12E, F.
Etymology: the specific epithet chamaenerii refers to Chamaenerion, the plant genus from which the type strain was isolated.
Culture characteristics: In YM broth, after 7 d at 17 °C, cells are cylindrical, 2.6–4.3 × 5.7–10.0 μm and single, budding is polar (Fig. 12E), a sediment is present. On YM agar, after 1 mo at 17 °C, the streak culture is pinkish-cream, butyrous, smooth and glossy. The margin is entire. In Dalmau plate culture on corn meal agar, pseudohyphae are not formed. Sexual structures are not observed on YM, PDA, V8 and CM agar. Ballistoconidia are long ellipsoidal, 2.9–4.3 × 7.1–10.0 μm (Fig. 12F).
Physiological and biochemical characteristics: Glucose fermentation is absent. Glucose, galactose (variable), L-sorbose (variable), sucrose, maltose, cellobiose (variable), trehalose, lactose (variable), raffinose (variable), inulin (weak), soluble starch (variable), glycerol, ribitol (delayed and weak), mannitol (delayed and weak) and D-glucitol (variable) are assimilated as sole carbon sources. Melibiose, melezitose, D-xylose, L-arabinose, D-arabinose, D-ribose, L-rhamnose, D-glucosamine, methanol, ethanol, erythritol, galactitol, D-Methyl-α-D-glucoside, salicin, DL-lactate, succinate, citrate, myo-inositol and hexadecane are not assimilated. Ammonium sulfate and potassium nitrate are assimilated as sole nitrogen sources. Sodium nitrite, L-lysine, ethylamine hydrochloride and cadaverine dihydrochloride are not assimilated. Maximum growth temperature is 26–27 °C. Growth in vitamin-free medium is negative. Starch-like substances are not produced. Growth on 50 % (w/w) glucose-yeast extract agar is negative. Urease activity is positive. Diazonium Blue B reaction is positive.
Physiologically, Kon. chamaenerii. differs from its closely related species Kon. subrosea and Kon. miscanthi in its inability to assimilate succinate (Table S1.17).
Typus: China, Bujin county, Xinjiang province, obtained from a leaf of Chamaenerion angustifolium, Jul. 2004, F.-Y. Bai (holotype CGMCC 2.2652T preserved in a metabolically inactive state, ex-type CBS 15453 = XJ8A5).
Kondoa foliicola Q.M. Wang, F.Y. Bai & A.H. Li sp. nov. MycoBank MB828794. Fig. 12G, H.
Etymology: the specific epithet foliicola refers to the substrate origin of the type strain, leaves.
Culture characteristics: In YM broth, after 7 d at 17 °C, cells are ellipsoidal and somewhat ovoid, 3.1–5.4 × 5.1–7.8 μm and single, budding is polar (Fig. 12G), a sediment is formed. After 1 mo at 17 °C, an incomplete ring and sediment are present. On YM agar, after 1 mo at 17 °C, the streak culture is pale-yellow, butyrous, dull. The margin is entire. In Dalmau plate culture on corn meal agar, pseudohyphae are not formed. Sexual structures are not observed on YM, PDA, V8 and CM agar. Ballistoconidia are ellipsoidal or ovoid, 2.5–4.0 × 3.8–8.8 μm (Fig. 12H).
Physiological and biochemical characteristics: Glucose fermentation is absent. Glucose, sucrose, cellobiose, trehalose, lactose, raffinose, melezitose, soluble starch, D-xylose, L-arabinose, D-arabinose, glucosamine, glycerol, ribitol and D-mannitol are assimilated as sole carbon sources. Galactose, L-sorbose, maltose, melibiose, inulin, D-ribose, L-rhamnose, D-N-methanol, ethanol, erythritol, galactitol, D-glucitol, Methyl-α-D-glucoside, salicin, DL-lactate, succinate, citrate, myo-inositol and hexadecane are not assimilated. Ammonium sulfate, potassium nitrate (weak), L-lysine and ethylamine hydrochloride (delayed and weak) are assimilated as sole nitrogen sources. Sodium nitrite and cadaverine dihydrochloride are not assimilated. Maximum growth temperature is 26–27 °C. Growth in vitamin-free medium is negative. Starch-like substances are produced. Growth on 50 % (w/w) glucose-yeast extract agar is negative. Urease activity is positive. Diazonium Blue B reaction is positive.
Physiologically, Kon. foliicola differs from its closely related species Kon. arboricola in its inability to assimilate maltose, grow in vitamin-free medium and produce starch like compounds and its ability to assimilate melezitose, D-arabinose and D-glucosamine (Table S1.17).
Typus: Germany, obtained from a leaf of an unidentified plant, Sep. 2005 (holotype CGMCC 2.3100T preserved in a metabolically inactive state, ex-type CBS 15465 = G9.1).
Kondoa arboricola Q.M. Wang, F.Y. Bai & A.H. Li sp. nov. MycoBank MB828795. Fig. 12I, J.
Etymology: the specific epithet arboricola refers to the substrate origin of the type strain, tree.
Culture characteristics: In YM broth, after 7 d at 17 °C, cells are ellipsoidal, 2.9–5.0 × 7.1–10.0 μm and single, budding is polar (Fig. 12I), a sediment is present. On YM agar, after 1 mo at 17 °C, the streak culture is yellowish cream, butyrous, smooth and semi-glossy. The margin is entire. In Dalmau plate culture on corn meal agar, pseudohyphae are not formed. Sexual structures are not observed on YM, PDA, V8 and CM agar. Ballistoconidia are allantoid or reniform, 3.0–5.7 × 7.0–15.7 μm (Fig. 12J).
Physiological and biochemical characteristics: Glucose fermentation is absent. Glucose, L-sorbose (variable), sucrose (variable), maltose, cellobiose, trehalose, lactose (delayed and weak), raffinose, inulin (variable), soluble starch (variable), D-xylose (variable), L-arabinose (variable), ethanol (variable), glycerol, ribitol (variable), D-mannitol (variable), D-glucitol (variable), DL-lactate (variable) and succinate (variable) are assimilated as sole carbon sources. Galactose, melibiose, melezitose, D-arabinose, D-ribose, L-rhamnose, D-glucosamine, methanol, erythritol, galactitol, Methyl-α-D-glucoside, salicin, citrate, myo-inositol and hexadecane are not assimilated. Ammonium sulfate, potassium nitrate (weak), L-lysine (weak) and ethylamine hydrochloride are assimilated as sole nitrogen sources. Sodium nitrite and cadaverine dihydrochloride are not assimilated. Maximum growth temperature is 26–27 °C. Growth in vitamin-free medium is positive. Starch-like substances are not produced. Growth on 50 % (w/w) glucose-yeast extract agar is negative. Urease activity is positive. Diazonium Blue B reaction is positive.
Physiologically, Kon. arboricola differs from its closely related species Kon. foliicola in its inability to assimilate melezitose, D-arabinose and D-glucosamine and its ability to assimilate maltose, grow in vitamin-free medium and produce starch like compounds (Table S1.17).
Typus: China, Bomi county, Tibet, obtained from a leaf of tree, Sep. 2004, F.-Y. Bai (holotype CGMCC 2.2621T preserved in a metabolically inactive state, ex-type CBS 15452 = XZ12B5).
Kondoa lulangica Q.M. Wang, F.Y. Bai & A.H. Li sp. nov. MycoBank MB828796. Fig. 12K, L.
Etymology: the specific epithet lulangica refers to the geographic origin of the type strain, Lulang county, Tibet.
Culture characteristics: In YM broth, after 7 d at 17 °C, cells are ellipsoidal and cylindrical, 2.4–3.8 × 5.0–7.6 μm and single, budding is polar (Fig. 12K), a sediment is formed. After 1 mo at 17 °C, a sediment is present. On YM agar, after 1 mo at 17 °C, the streak culture is pale pink, butyrous, smooth and glistening. The margin is entire. In Dalmau plate culture on corn meal agar, pseudohyphae are not formed. Sexual structures are not observed on YM, PDA, V8 and CM agar. Ballistoconidia are allantoid or reniform, 2.6–2.9 × 5.7–8.6 μm (Fig. 12L).
Physiological and biochemical characteristics: Glucose fermentation is absent. Glucose, sucrose (delayed), maltose, trehalose (delayed and weak), melezitose (delayed and weak), soluble starch (weak), glycerol, erythritol (delayed), D-mannitol, D-glucitol and Methyl-α-D-glucoside are assimilated as sole carbon sources. Galactose, L-sorbose, cellobiose, lactose, melibiose, raffinose, inulin, D-xylose, L-arabinose, D-arabinose, D-ribose, L-rhamnose, D-glucosamine, methanol, ethanol, ribitol, galactitol, salicin, DL-lactate, succinate, citrate, myo-inositol and hexadecane are not assimilated. Ammonium sulfate, potassium nitrate and ethylamine hydrochloride are assimilated as sole nitrogen sources. Sodium nitrite, L-lysine and cadaverine dihydrochloride are not assimilated. Maximum growth temperature is 24 °C. Growth in vitamin-free medium is positive. Starch-like substances are not produced. Growth on 50 % (w/w) glucose-yeast extract agar is negative. Urease activity is positive. Diazonium Blue B reaction is positive.
Physiologically, Kon. lulangica differs from its closely related species Kon. changbaiensis in its inability to assimilate cellobiose, raffinose and ribitol and its ability to assimilate erythritol, Methyl-α-D-glucoside and grow in vitamin-free medium (Table S1.17).
Typus: China, Lulang county, Tibet, obtained from a leaf of an unidentified plant, Sep. 2004, F.-Y. Bai (holotype CGMCC 2.2762T preserved in a metabolically inactive state, ex-type CBS 15456 = XZ36D1).
Kondoa daliangziensis Q.M. Wang, F.Y. Bai & A.H. Li sp. nov. MycoBank MB832014. Fig. 12M, N.
Etymology: the specific epithet daliangziensis refers to the geographic origin of the type strain, Daliangzi River National Forest Park, Heilongjiang.
Culture characteristics: In YM broth, after 7 d at 17 °C, cells are cylindrical and ellipsoidal, 2.7–4.4 × 4.3–8.4 μm and single, budding is polar (Fig. 12M), a sediment is formed. After 1 mo at 17 °C, a ring and sediment are present. On YM agar, after 1 mo at 17 °C, the streak culture is pale orange, butyrous, smooth and glistening. The margin is entire. In Dalmau plate culture on corn meal agar, pseudohyphae are not formed. Sexual structures are not observed on YM, PDA, V8 and CM agar. Ballistoconidia are allantoid or reniform, 2.6–3.1 × 7.1–8.6 μm (Fig. 12N).
Physiological and biochemical characteristics: Glucose fermentation is absent. Glucose, sucrose (variable), maltose, cellobiose, trehalose, melezitose, L-arabinose (variable), ethanol (variable), glycerol, ribitol (variable), D-mannitol (variable), D-glucitol (variable), salicin (variable) and DL-lactate (variable) are assimilated as sole carbon sources. Galactose, L-sorbose, lactose, melibiose, raffinose, inulin, soluble starch, D-xylose, D-arabinose, D-ribose, L-rhamnose, D-glucosamine, N-Acetyl-D-glucosamine, methanol, erythritol, galactitol, Methyl-α-D-glucoside, succinate, citrate, myo-inositol and hexadecane are not assimilated. Ammonium sulfate, potassium nitrate, L-lysine, ethylamine hydrochloride (weak) and cadaverine dihydrochloride (weak) are assimilated as sole nitrogen sources. Sodium nitrite is not assimilated. Maximum growth temperature is 22–23 °C. Growth in vitamin-free medium is variable. Starch-like substances are not produced. Growth on 50 % (w/w) glucose-yeast extract agar is negative. Urease activity is positive. Diazonium Blue B reaction is positive.
Physiologically, Kon. daliangziensis and Kon. ribitophobia are difficult to distinguish from each other. The latter can grow at 25 °C, but the former does not. Kon. daliangziensis differs from Kon. gutianensis in its inability to assimilate galactose and inulin and its ability to assimilate L-lysine (Table S1.17).
Typus: China, Daliangzi river national forest park, Heilongjiang province, obtained from a leaf of an unidentified plant, Aug. 2014, Q.-M. Wang (holotype CGMCC 2.5610T preserved in a metabolically inactive state, ex-type CBS 13974 = HLJ22A8).
Kondoa ribitophobia Q.M. Wang, F.Y. Bai & A.H. Li sp. nov. MycoBank MB828798. Fig. 12O.
Etymology: the specific epithet ribitophobia refers to the physiological character of not assimilating ribitol.
Culture characteristics: In YM broth, after 7 d at 17 °C, cells are globosal, oval and ellipsoidal, 3.3–4.9 × 4.5–8.3 μm and single, budding is polar (Fig. 12O), a sediment is formed. After 1 mo at 17 °C, a pellicle and a sediment are present. On YM agar, after 1 mo at 17 °C, the streak culture is pale yellow, butyrous, smooth and glossy. The margin is entire. In Dalmau plate culture on corn meal agar, pseudohyphae are not formed. Sexual structures are not observed on YM, PDA, V8 and CM agar. Ballistoconidia are not produced.
Physiological and biochemical characteristics: Glucose fermentation is absent. Glucose, galactose (variable), L-sorbose (variable), sucrose, maltose, cellobiose (variable), trehalose, melezitose, inulin (variable), L-arabinose (variable), L-rhamnose (variable), ethanol (variable), glycerol, D-mannitol (delayed and weak), D-glucitol (variable), Methyl-α-D-glucoside (variable), salicin (weak) and succinate (variable) are assimilated as sole carbon sources. Lactose, melibiose, raffinose, soluble starch, D-xylose, D-arabinose, D-ribose, D-glucosamine, N-Acetyl-D-glucosamine, methanol, erythritol, ribitol, galactitol, DL-lactate, citrate, myo-inositol and hexadecane are not assimilated. Ammonium sulfate, potassium nitrate (weak), L-lysine (variable), ethylamine hydrochloride (variable) and cadaverine dihydrochloride (variable) are assimilated as sole nitrogen sources. Sodium nitrite is not assimilated. Maximum growth temperature is 26–27 °C. Growth in vitamin-free medium is variable. Starch-like substances are not produced. Growth on 50 % (w/w) glucose-yeast extract agar is negative. Urease activity is positive. Diazonium Blue B reaction is positive.
Physiologically, Kon. ribitophobia differs from its closely related species Kon. gutianensis in its inability to assimilate ribitol (Table S1.17).
Typus: China, Taiwan province, obtained from a leaf of an unidentified plant, Aug. 2009, Q.-M. Wang (holotype CGMCC 2.4441T preserved in a metabolically inactive state, ex-type CBS 12496 = TW2.1E-016).
Kondoa myxariophila J.P. Sampaio, Q.M. Wang & F.Y. Bai sp. nov. MycoBank MB828799. Fig. 12, Fig. 13.
Fig. 13.
Vegetative cells, ballistoconidia and the sexual stage of Kon. myxariophila CBS 8379T. (A) Basidia; (B) Basidiospores; (C, D) Haustorial branches; (E) Ballistoconidia. Bars = 10 μm.
Etymology: the specific epithet myxariophila refers to the association of the novel taxon with the fruiting bodies of Myxarium nucleatum (Auriculariales).
Sexual characteristics: The sexual stage is observed PDA and MYP plates incubated at 20 °C for 8–12 wk and occurs in individual strains in the absence of mating. Hyphae are 3–5 μm in diameter and have clamp connections. Basidia are cylindrical, transversely-septate, usually four-celled and measure 40–60 × 7.5–5 μm (Fig. 13A, C). Basidiospores are formed at the end of basidial sterigmata, measuring 10–5 μm in length. Basidiospores are oval, measure 11–9 × 7–5 μm (Fig. 13B), are forcefully ejected (ballistospores) and germinate by budding. Haustorial branches are conspicuously formed and occur laterally on hyphae (Fig. 13C, D).
Culture characteristics: In YM broth, after 7 d at 17 °C, cells are ellipsoidal to ovoid, measure 3–4 × 4–6 μm and occur single or in pairs and budding is polar (Fig. 12P). A sediment is formed. After 1 mo at 17 °C, a pellicle and a sediment are present. On YM agar, after 1 mo at 17 °C, the streak culture is pale yellow, butyrous, semi-glossy and smooth. The margin is entire. In Dalmau plate culture on corn meal agar, pseudohyphae are not formed. Ballistoconidia can be produced in solid medium (CMA) but are rare and measure 4–5 × 5–8 μm (Fig. 13E).
Physiological and biochemical characteristics: Glucose fermentation is absent. Glucose, sucrose, maltose, trehalose, melibiose (variable), cellobiose (variable), raffinose (variable), melezitose, soluble starch, D-xylose, L-arabinose (delayed and weak), D-arabinose (delayed and weak), D-ribose (variable), L-rhamnose (delayed and weak), D-glucosamine (variable), glycerol (delayed and weak), ribitol (variable), salicin (variable), D-mannitol (delayed and weak), D-glucitol (delayed and weak), succinate (delayed and weak) and citrate (weak) are assimilated as sole carbon sources. Galactose, L-sorbose, lactose, inulin, methanol, ethanol, erythritol, galactitol, Methyl-α-D-glucoside, DL-lactate, myo-inositol and hexadecane are not assimilated. Ammonium sulfate, potassium nitrate (variable), sodium nitrite (variable), ethylamine hydrochloride (delayed and weak) and cadaverine dihydrochloride (delayed and weak) are assimilated as sole nitrogen sources. L-lysine is not assimilated. Maximum growth temperature is 22–25 °C. Growth in vitamin-free medium is positive. Starch-like substances are not produced. Growth on 50 % (w/w) glucose-yeast extract agar is negative. Urease activity is positive. Diazonium Blue B reaction is positive.
Physiologically, Kon. myxariophila differs from its closest relatives, Kon. daliangziensis, Kon. ribitolophobia and Kon. gutianensis, in its inability to assimilate L-lysine and its ability to assimilate soluble starch and D-xylose (Table S1.17).
Typus: Portugal, Sesimbra, obtained from the fruiting body of Myxarium nucleatum (Auriculariales), Nov. 1992, J.P. Sampaio (holotype PYCC 5509T preserved in a metabolically inactive state, ex-type CBS 8379 = ZP 337).
Note: Besides several sexual strains isolated with the ballitoconidium-fall method from basidiocarps of Myxarium nucleatum in Portugal (PYCC 5509 = ZP 337; PYCC 8354 = ZP 338; and PYCC 8305 = ZP 352) in 1992 and 1996, another strain was isolated from the leaf of an unidentified plant, collected in Germany in September 2005 (CGMCC 2.3106 = CBS 15468). Although a sexual stage has not been reported for the culture isolated in Germany, these four strains have similar ITS sequences. Therefore, Kon. myxariophila appears to be capable to engage in mycoparasitism because it produces haustorial branches and is ecologically associated with other fungi. Nevertheless, the mycoparasitic strategy might be combined with a saprobe lifestyle in the phylloplane since Kon. myxariophila is also able to produce ballistoconidia and is also found in association with plant leafs. Similarly to the other two sexual species in the genus, Kon. aeria and Kon. malvinella, Kon. myxariophila does not produce teliospores, produces transversely-septate basidia and its basidiospores are forcefully discharged (ballistospores).
Kondoa rhododendri Q.M. Wang, F.Y. Bai & A.H. Li sp. nov. MycoBank MB828800. Fig. 14A, B.
Fig. 14.
Vegetative cells grown in YM broth for 5 d at 17 °C and ballistoconidia produced on corn meal agar after 7 d at 17 °C. (A, B) Kon. rhododendri CGMCC 2.3463T; (C, D) Ben. wuzhishanensis CGMCC 2.3569T; (E) Ben. pseudorectispora CGMCC 2.5677T; (F) Ps. fusiformis CGMCC 2.5823T; (G, H) Ru. fanjingshanensis CGMCC 2.4542T; (I, J) Ru. bangxiensis CGMCC 2.3454T; (K, L) Ru. lunata CGMCC 2.4426T; (M, N) Boe. sterigmata CGMCC 2.4539T, (O) St. layueensis CGMCC 2.5817T; (P) Pse. motuoensis CGMCC 2.5816T. Bars = 10 μm.
Etymology: the specific epithet rhododendri refers to Rhododendron, the plant genus from which the type strain was isolated.
Culture characteristics: In YM broth, after 7 d at 17 °C, cells are ovoid, ellipsoidal and cylindrical, 2.7–4.8 × 4.5–9.5 μm and single, budding is polar (Fig. 14A), a sediment is formed. After 1 mo at 17 °C, a sediment is present. On YM agar, after 1 mo at 17 °C, the streak culture is pinkish cream, butyrous, smooth and glossy. The margin is entire. In Dalmau plate culture on corn meal agar, pseudohyphae are not formed. Sexual structures are not observed on YM, PDA, V8 and CM agar. Ballistoconidia are long ellipsoidal or ovoid, 3.0–4.3 × 7.9–10.0 μm (Fig. 14B).
Physiological and biochemical characteristics: Glucose fermentation is absent. Glucose, galactose (delayed), L-sorbose (delayed), sucrose, maltose, cellobiose (delayed and weak), trehalose, melezitose (delayed), inulin (weak), D-xylose (delayed), L-arabinose (delayed and weak), D-ribose (delayed and weak), ethanol (weak), glycerol (delayed), ribitol, galactitol (delayed and weak), D-mannitol and D-glucitol are assimilated as sole carbon sources. Lactose, melibiose, raffinose, soluble starch, D-arabinose, L-rhamnose, D-glucosamine, methanol, erythritol, Methyl-α-D-glucoside, salicin, DL-lactate, succinate, citrate, myo-inositol and hexadecane are not assimilated. Ammonium sulfate, potassium nitrate, L-lysine and ethylamine hydrochloride are assimilated as sole nitrogen sources. Sodium nitrite and cadaverine dihydrochloride are not assimilated. Maximum growth temperature is 25 ΊC. Growth in vitamin-free medium is positive. Starch-like substances are not produced. Growth on 50 % (w/w) glucose-yeast extract agar is negative. Urease activity is positive. Diazonium Blue B reaction is positive.
Physiologically, Kon. rhododendri differs well from other Kondoa species in its assimilation of carbon and nitrogen sources (Table S1.17).
Typus: China, Bomi county, Tibet, obtained from a leaf of Rhododendron triflorum, Sep. 2004, F.-Y. Bai (holotype CGMCC 2.2763T preserved in a metabolically inactive state, ex-type CBS 15457 = XZ27E3).
Bensingtonia wuzhishanensis Q.M. Wang, F.Y. Bai & A.H. Li sp. nov. MycoBank MB828801. Fig. 14C, D.
Etymology: the specific epithet wuzhishanensis refers to the geographic origin of the type strain, Wuzhishan mountain, Hainan.
Culture characteristics: In YM broth, after 7 d at 17 °C, cells are cylindrical or fusiform, 3.4–4.0 × 7.6–10.0 μm and single, budding is polar (Fig. 14C), a sediment is present. On YM agar, after 1 mo at 17 °C, the streak culture is ivory to cream, mucoid, smooth and glistening. The margin is entire. In Dalmau plate culture on corn meal agar, pseudohyphae are not formed. Sexual structures are not observed on YM, PDA, V8 and CM agar. Ballistoconidia are cylindrical, 2.9–3.7 × 7.4–10.0 μm (Fig. 14D).
Physiological and biochemical characteristics: Glucose fermentation is absent. Glucose, galactose, sucrose, maltose, cellobiose, trehalose, lactose (delayed and weak), melibiose, raffinose, melezitose, soluble starch, D-xylose, L-arabinose (delayed and weak), D-arabinose, D-ribose, L-rhamnose (weak), D-glucosamine (delayed and weak) , ethanol, glycerol (weak), erythritol, ribitol, galactitol, D-mannitol, D-glucitol, Methyl-α-D-glucoside, salicin, DL-lactate, succinate (delayed and weak) and citrate (weak) are assimilated as sole carbon sources. L-sorbose, inulin, methanol, myo-inositol and hexadecane are not assimilated. Ammonium sulfate, potassium nitrate, sodium nitrite (delayed and weak), L-lysine, ethylamine hydrochloride (delayed and weak) and cadaverine dihydrochloride (delayed and weak) are assimilated as sole nitrogen sources. Maximum growth temperature is 26–27 °C. Growth in vitamin-free medium is positive. Starch-like substances are not produced. Growth on 50 % (w/w) glucose-yeast extract agar is negative. Urease activity is positive. Diazonium Blue B reaction is positive.
Physiologically, Be. wuzhishanensis differs from its closely related species, Be. pseudorectispora, Be. bomiensis, Be. naganoensis, Be. pseudonaganoensis and Be. rectispora, in its ability to assimilate D-ribose, ethanol and erythritol (Table S1.18).
Typus: China, Wuzhishan mountain, Hainan province, obtained from a leaf of an unidentified plant, Nov. 2006, Q.-M. Wang (holotype CGMCC 2.3569T preserved in a metabolically inactive state, ex-type CBS 15661 = WZS33.18).
Bensingtonia pseudorectispora Q.M. Wang, F.Y. Bai & A.H. Li sp. nov. MycoBank MB828802. Fig. 14E.
Etymology: the specific epithet pseudorectispora refers to the similar colony morphology to that of Bensingtonia rectispora.
Culture characteristics: In YM broth, after 7 d at 17 °C, cells are ellipsoidal and cylindrical, 2.8–3.2 × 7.2–10.3 μm and single, budding is polar (Fig. 14E), a sediment is present. After 1 mo at 17 °C, a ring and a sediment are present. On YM agar, after 1 mo at 17 °C, the streak culture is pink red, butyrous, wrinkled and dull. The margin is entire. In Dalmau plate culture on corn meal agar, pseudohyphae are not formed. Sexual structures are not observed on YM, PDA, V8 and CM agar. Ballistoconidia are not produced.
Physiological and biochemical characteristics: Glucose fermentation is absent. Glucose, maltose, melezitose, D-mannitol and salicin are assimilated as sole carbon sources. Galactose, L-sorbose, sucrose, cellobiose, trehalose, lactose, melibiose, raffinose, inulin, soluble starch, D-xylose, L-arabinose, D-arabinose, D-ribose, L-rhamnose, D-glucosamine, N-Acetyl-D-glucosamine, methanol, ethanol, glycerol, erythritol, ribitol, galactitol, D-glucitol, Methyl-α-D-glucoside, DL-lactate, succinate, citrate, myo-inositol and hexadecane are not assimilated. Ammonium sulfate, potassium nitrate, L-lysine, ethylamine hydrochloride and cadaverine dihydrochloride are assimilated as sole nitrogen sources. Sodium nitrite is not assimilated. Maximum growth temperature is 22–23 °C. Growth in vitamin-free medium negative. Starch-like substances are not produced. Growth on 50 % (w/w) glucose-yeast extract agar is negative. Urease activity is positive. Diazonium Blue B reaction is positive.
Physiologically, Be. pseudorectispora differs from its closely related species Be. rectispora in its inability to assimilate sucrose, trehalose and glycerol and its ability to assimilate salicin and ethylamine (Table S1.18).
Typus: China, Bomi, Tibet, obtained from a leaf of an unidentified plant, Sep. 2014, Q.-M. Wang (holotype CGMCC 2.5677T preserved in a metabolically inactive state, ex-type CBS 15750 = XZ154D5).
Pseudobensingtonia fusiformis Q.M. Wang, F.Y. Bai & A.H. Li sp. nov. MycoBank MB828803. Fig. 14F.
Etymology: the specific epithet fusiformis refers to the fusiform vegetative cells of the type strain.
Culture characteristics: In YM broth, after 7 d at 17 °C, cells are cylindrical, ellipsoidal and fusiform, 7.6–13.3 × 2.2–3.6 μm and single, budding is polar (Fig. 14F), a sediment is present. On YM agar, after 1 mo at 17 °C, the streak culture is yellow, butyrous, wrinkled and dull. The margin is entire. In Dalmau plate culture on corn meal agar, pseudohyphae are not formed. Sexual structures are not observed on YM, PDA, V8 and CM agar. Ballistoconidia are not produced.
Physiological and biochemical characteristics: Glucose fermentation is absent. Glucose, L-sorbose, sucrose, cellobiose, trehalose, lactose, raffinose, inulin, D-xylose, L-arabinose (variable), D-ribose (weak), ethanol (variable), glycerol, erythritol, ribitol, D-mannitol, D-glucitol, D-gluconate and succinate are assimilated as sole carbon sources. Galactose, maltose, melibiose, melezitose, soluble starch, D-arabinose, L-rhamnose, D-glucosamine, N-Acetyl-D-glucosamine, methanol, galactitol, Methyl-α-D-glucoside, salicin, DL-lactate, citrate, myo-inositol and hexadecane are not assimilated. Ammonium sulfate, L-lysine, ethylamine hydrochloride and cadaverine dihydrochloride are assimilated as sole nitrogen sources. Potassium nitrate and sodium nitrite are not assimilated. Maximum growth temperature is 23 °C. Growth in vitamin-free medium is negative. Starch-like substances are not produced. Growth on 50 % (w/w) glucose-yeast extract agar is negative. Urease activity is positive. Diazonium Blue B reaction is positive.
Physiologically, Ps. fusiformis differs from its closely related species Ps. ingoldii and Ps. musae in its inability to assimilate citrate and its ability to assimilate inulin (Table S1.19).
Typus: China, Bomi, Tibet, obtained from a leaf of an unidentified plant, Sep. 2014, Q.-M. Wang (holotype CGMCC 2.5823T preserved in a metabolically inactive state, ex-type CBS 15647 = XZ152EA3).
Ruinenia fanjingshanensis Q.M. Wang, F.Y. Bai & A.H. Li sp. nov. MycoBank MB828804. Fig. 14G, H.
Etymology: the specific epithet fanjingshanensis refers to the geographic origin of the type strain, Fanjingshan Mountain, Guizhou.
Culture characteristics: In YM broth, after 7 d at 17 °C, cells are ellipsoidal and cylindrical, 2.1–3.6 × 5.0–7.9 μm and single, budding is polar (Fig. 14G), a sediment is present. After 1 mo at 17 °C, a ring and a sediment are present. On YM agar, after 1 mo at 17 °C, the streak culture is pink-red, butyrous, wrinkled and dull. The margin is entire. In Dalmau plate culture on corn meal agar, pseudohyphae are not formed. Sexual structures are not observed on YM, PDA, V8 and CM agar. Ballistoconidia are allantoid or reniform, 2.1–3.6 × 5.0–7.9 μm (Fig. 14H).
Physiological and biochemical characteristics: Glucose fermentation is absent. Glucose, maltose, trehalose, melibiose, raffinose, inulin, soluble starch (weak), ribitol and D-mannitol are assimilated as sole carbon sources. Galactose, L-sorbose, sucrose, cellobiose, lactose, melezitose, D-xylose, L-arabinose, D-arabinose, D-ribose, L-rhamnose, D-glucosamine, N-Acetyl-D-glucosamine, methanol, ethanol, glycerol, erythritol, galactitol, D-glucitol, Methyl-α-D-glucoside, salicin, D-gluconate, DL-lactate, succinate, citrate, myo-inositol and hexadecane are not assimilated. Ammonium sulfate, ethylamine hydrochloride and cadaverine dihydrochloride are assimilated as sole nitrogen sources. Potassium nitrate, sodium nitrite and L-lysine are not assimilated. Maximum growth temperature is 21 °C. Growth in vitamin-free medium is negative Starch-like substances are not produced. Growth on 50 % (w/w) glucose-yeast extract agar is negative. Urease activity is positive. Diazonium Blue B reaction is positive.
Physiologically, Ru. fanjingshanensis differs from its closely related species Ru. dracophylli in its inability to assimilate L-sorbose, sucrose, maltose, cellobiose, melezitose, glycerol, ribitol, galactitol, D-mannitol, D-glucitol, salicin and succinate and its ability to assimilate trehalose, inulin, ethylamine and cadaverine (Table S1.20).
Typus: China, Fanjingshan Mountain, Guizhou province, obtained from a leaf of an unidentified plant, Oct. 2011, Q.-M. Wang (holotype CGMCC 2.4542T preserved in a metabolically inactive state, ex-type CBS 15745 = FJS6C7).
Ruinenia bangxiensis Q.M. Wang, F.Y. Bai & A.H. Li sp. nov. MycoBank MB828805. Fig. 14I, J.
Etymology: the specific epithet bangxiensis refers to the geographic origin of the type strain, Bangxi county, Hainan.
Culture characteristics: In YM broth, after 7 d at 17 °C, cells are ellipsoidal and cylindrical, 2.2–3.7 × 6.4–10.5 μm and single, budding is polar (Fig. 14I), a sediment is present. After 1 mo at 17 °C, a ring and a sediment are present. On YM agar, after 1 mo at 17 °C, the streak culture is pinkish-orange, butyrous, wrinkled and dull. The margin is entire. In Dalmau plate culture on corn meal agar, pseudohyphae are not formed. Sexual structures are not observed on YM, PDA, V8 and CM agar. Ballistoconidia are allantoid or reniform, 2.4–2.9 × 5.3–7.3 μm (Fig. 14J).
Physiological and biochemical characteristics: Glucose fermentation is absent. Glucose, sucrose, maltose, cellobiose, trehalose, melibiose, raffinose, melezitose, inulin (variable), soluble starch (weak), D-xylose (weak), L-arabinose (delayed and weak), ethanol (variable), ribitol (variable), D-glucitol (variable), succinate (variable), and D-mannitol are assimilated as sole carbon sources. Galactose, L-sorbose, lactose, D-arabinose, D-ribose, L-rhamnose, D-glucosamine, methanol, glycerol, erythritol, galactitol, Methyl-α-D-glucoside, salicin, DL-lactate, citrate, myo-inositol and hexadecane are not assimilated. Ammonium sulfate, potassium nitrate and L-lysine (variable) are assimilated as sole nitrogen sources. Sodium nitrite, ethylamine hydrochloride and cadaverine dihydrochloride are not assimilated. Maximum growth temperature is 25 °C. Growth in vitamin-free medium is positive. Starch-like substances are not produced. Growth on 50 % (w/w) glucose-yeast extract agar is negative. Urease activity is positive. Diazonium Blue B reaction is positive.
Physiologically, Ru. bangxiensis differs from its closely related species Ru. clavata in its inability to assimilate D-ribose and cadaverine and its ability to assimilate potassium nitrate (Table S1.20).
Typus: China, Bangxi county, Hainan province, obtained from a leaf of an unidentified plant, Nov. 2006, Q.-M. Wang (holotype CGMCC 2.3454T preserved in a metabolically inactive state, ex-type CBS 10819 = HBX1.0).
Ruinenia lunata Q.M. Wang, F.Y. Bai & A.H. Li sp. nov. MycoBank MB828806. Fig. 14K, L.
Etymology: the specific epithet lunata refers to the falcate ballistoconidia of the type strain.
Culture characteristics: In YM broth, after 7 d at 17 °C, cells are ellipsoidal to falcate, 1.8–3.5 × 5.0–9.0 μm and single, budding is polar (Fig. 14K), a sediment is formed. After 1 mo at 17 °C, a ring and sediment are present. On YM agar, after 1 mo at 17 °C, the streak culture is orange-red, butyrous, smooth. The margin is entire. In Dalmau plate culture on corn meal agar, pseudohyphae are not formed. Sexual structures are not observed on YM, PDA, V8 and CM agar. Ballistoconidia are reniform to falcate, 3.0–6.5 × 6.0–13.0 μm (Fig. 14L).
Physiological and biochemical characteristics: Glucose fermentation is absent. Glucose, sucrose, maltose, cellobiose (variable), trehalose, melibiose, raffinose, melezitose, ribitol (delayed), D-mannitol (delayed) and D-glucitol (delayedand weak) are assimilated as sole carbon sources. Galactose, L-sorbose, lactose, inulin, soluble starch, L-rhamnose, D-xylose, L-arabinose, D-arabinose, D-ribose, D-glucosamine, methyl α-D-glucoside, methanol, ethanol, erythritol, galactitol, glycerol, DL-lactic acid, critic acid, salicin, succinic acid, inositol and hexdecane are not assimilated. Ammonium sulfate and ethylamine hydrochloride (variable) are assimilated as sole nitrogen sources. L-lysine, sodium nitrite, potassium nitrate and cadaverine dihydrochloride are not assimilated. Maximum growth temperature is 22 °C. Growth in vitamin-free medium is negative. Starch-like substances are not produced. Growth on 50 % (w/w) glucose-yeast extract agar is negative. Urease activity is positive. Diazonium Blue B reaction is positive.
Physiologically, Ru. lunata differs from its closely related species Ru. bangxiensis and Ru. clavata in its inability to assimilate soluble starch and D-xylose and grow at 25 °C (Table S1.20).
Typus: China, Taiwan province, obtained from a leaf of an unidentified plant, Aug. 2009, Q.-M. Wang (holotype CGMCC 2.4426T preserved in a metabolically inactive state, ex-type CBS 12525 = TW 2.1E-028).
Boekhoutia Q.M. Wang & F.Y. Bai gen. nov. MycoBank MB828807.
Etymology: the genus is named in honour of Dr. Teun Boekhout for his research contributions to yeast taxonomy.
This genus is proposed for the branch represented by strain CGMCC 2.4539, which formed a separate clade from Kurtzmanomyces. Member of the Chionosphaeraceae (Agaricostilbales). The genus is mainly circumscribed by the phylogenetic analysis of the seven genes dataset, in which it occurred as a separate branch within Chionosphaeraceae (Fig. 4A).
Sexual reproduction not known. Colonies orange red, butyrous. Budding cells present and blastoconidia produced at the end of a stalk-like conidiophore. Conidiophore single or multiple, usually multifurcate. Pseudohyphae and hyphae not produced. Ballistoconidia formed.
Type species: Boekhoutia sterigmata Q.M. Wang, F.Y. Bai & A.H. Li.
Note: Boekhoutia and its close relative Kurtzmanomyces can produce stalk-like conidiophores, the former usually produces multifurcate conidiophores; each conidiophore of the latter can produce sequential multiple blastoconidia (Sampaio 2011b). Boekhoutia does not assimilate ethanol and D-mannitol, whereas all species of Kurtzmanomyces assimilate these two carbon sources.
Boekhoutia sterigmata Q.M. Wang, F.Y. Bai & A.H. Li sp. nov. MycoBank MB828808. Fig. 11, Fig. 14M, N.
Etymology: the specific epithet sterigmata refers to the vegetative cells producing conidia on stalk-like conidiophores in the type strain.
Culture characteristics: In YM broth, after 7 d at 17 °C, cells are ellipsoidal and cylindrical, 2.8–3.2 × 7.2–10.3 μm and single, budding is polar (Fig. 14M), a sediment is present. One or more conidia are produced on each stalk-like conidiophore. Conidiophore is single or multiple, usually multifurcate. After 1 mo at 17 °C, a ring and a sediment are present. On YM agar, after 1 mo at 17 °C, the streak culture is deep pink red, butyrous, wrinkled and dull. The margin is entire. In Dalmau plate culture on corn meal agar, pseudohyphae are not formed. Sexual structures are not observed on YM, PDA, V8 and CM agar. Ballistoconidia are allantoid or reniform, 2.6–3.2 × 3.8–5.8 μm (Fig. 14N).
Physiological and biochemical characteristics: Glucose fermentation is absent. Glucose, galactose, sucrose, maltose (delayed and weak), cellobiose, trehalose, melezitose and inulin are assimilated as sole carbon sources. L-sorbose, lactose, melibiose, raffinose, soluble starch, D-xylose, L-arabinose, D-arabinose, D-ribose, L-rhamnose, D-glucosamine, N-Acetyl-D-glucosamine, methanol, ethanol, glycerol, erythritol, ribitol, galactitol, D-mannitol, D-glucitol, Methyl-α-D-glucoside, salicin, DL-lactate, succinate, citrate, myo-inositol and hexadecane are not assimilated. Ammonium sulfate, L-lysine, ethylamine hydrochloride and cadaverine dihydrochloride are assimilated as sole nitrogen sources. Potassium nitrate and sodium nitrite are not assimilated. Maximum growth temperature is 23 °C. Growth in vitamin-free medium is negative. Starch-like substances are not produced. Growth on 50 % (w/w) glucose-yeast extract agar is negative. Urease activity is positive. Diazonium Blue B reaction is positive.
Typus: China, Fanjingshan Mountain, Guizhou province, obtained from a leaf of an unidentified plant, Oct. 2011, Q.-M. Wang (holotype CGMCC 2.4539T preserved in a metabolically inactive state, ex-type CBS 15553 = FJS3F22).
Jianyuniaceae Q.M. Wang & F.Y. Bai fam. nov. MycoBank MB828809.
Member of the Agaricostilbales (Agaricostilbomycetes). The diagnosis of the family Jianyuniaceae is based on the the genus Jianyunia. The nomenclature of the family is based on the genus Jianyunia.
Type genus: Jianyunia Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout.
Genera accepted: Jianyunia Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, Sterigmatospora Q.M. Wang & F.Y. Bai, Pseudosterigmatospora Q.M. Wang & F.Y. Bai.
Sterigmatospora Q.M. Wang & F.Y. Bai gen. nov. MycoBank MB828810.
Etymology: the genus is named based on the morphology of the vegetative cells, which produce conidia on stalk-like conidiophores.
This genus is proposed for the branch represented by strain CGMCC 2.5817, which formed a separate clade. Member of the Jianyuniaceae (Agaricostilbales). The genus is mainly circumscribed by the phylogenetic analysis of the seven genes dataset, in which it occurred as a separate branch within Jianyuniaceae (Fig. 4A).
Sexual reproduction not known. Colonies cream, butyrous. Budding cells present and blastoconidia produced on stalk-like conidiophores. Conidiophore single or multiple, usually cluster on cells. Pseudohyphae and hyphae not produced. Ballistoconidia not formed.
Type species: Sterigmatospora layueensis Q.M. Wang, F.Y. Bai & A.H. Li.
Note: Sterigmatospora and Pseudosterigmatospora can produce stalk-like conidiophores, the former usually produces cluster of conidiophores from one site on cells, the latter can form bifurcate or trifurcate conidophores. They are also distinguished by some physiological characteristics (Table S1.21), such as assimilation of raffinose and growth in vitamin-free medium.
Sterigmatospora layueensis Q.M. Wang, F.Y. Bai & A.H. Li sp. nov. MycoBank MB828811. Fig. 11, Fig. 14O.
Etymology: the specific epithet layueensis refers to the geographic origin of the type strain, Layue county, Tibet.
Culture characteristics: In YM broth, after 7 d at 17 °C, cells are ovoid and ellipsoidal, 2.8–3.5 × 3.8–5.9 μm and single, budding is polar (Fig. 14O), a sediment is present. One or more conidia are produced on each stalk-like conidiophore. Conidiophore is single or multiple, usually cluster on cells. After 1 mo at 17 °C, a sediment is present. On YM agar, after 1 mo at 17 °C, the streak culture is pale yellow, butyrous, smooth and glossy. The margin is entire. In Dalmau plate culture on corn meal agar, pseudohyphae are not formed. Sexual structures are not observed on YM, PDA, V8 and CM agar. Ballistoconidia are not produced.
Physiological and biochemical characteristics: Glucose fermentation is absent. Glucose, L-sorbose, sucrose, maltose, cellobiose, trehalose, raffinose, melezitose, D-xylose (variable), L-arabinose (variable), ribitol (variable), D-mannitol, D-glucitol, Methyl-α-D-glucoside and salicin (variable) are assimilated as sole carbon sources. Galactose, lactose, melibiose, inulin, soluble starch, D-arabinose, D-ribose, L-rhamnose, D-glucosamine, N-Acetyl-D-glucosamine, methanol, ethanol, glycerol, erythritol, galactitol, DL-lactate, succinate, citrate, myo-inositol and hexadecane are not assimilated. Ammonium sulfate, potassium nitrate, L-lysine, ethylamine hydrochloride and cadaverine dihydrochloride are assimilated as sole nitrogen sources. Sodium nitrite is not assimilated. Maximum growth temperature is 20 °C. Growth in vitamin-free medium is positive. Starch-like substances are not produced. Growth on 50 % (w/w) glucose-yeast extract agar is negative. Urease activity is positive. Diazonium Blue B reaction is positive.
Typus: China, Layue county, Tibet, obtained from a leaf of an unidentified plant, Sep. 2014, Q.-M. Wang (holotype CGMCC 2.5817T preserved in a metabolically inactive state, ex-type CBS 15649 = XZ100A2B).
Pseudosterigmatospora Q.M. Wang & F.Y. Bai gen. nov. MycoBank MB828812.
Etymology: the genus is named because of a similar morphology as present in the genus Sterigmatospora.
This genus is proposed for the branch represented by strain CGMCC 2.5816, which formed a separate clade. Member of the Jianyuniaceae (Agaricostilbales). The genus is mainly circumscribed by the phylogenetic analysis of the seven genes dataset, in which it occurred as a separate branch within Jianyuniaceae (Fig. 4A).
Sexual reproduction not known. Colonies white to cream, butyrous. Budding cells present and blastoconidia produced on stalk-like conidiophores. Conidiophores single or multiple, usually bifurcate, somewhat trifurcate. Pseudohyphae and hyphae not produced. Ballistoconidia not formed.
Type species: Pseudosterigmatospora motuoensis Q.M. Wang, F.Y. Bai & A.H. Li.
Pseudosterigmatospora motuoensis Q.M. Wang, F.Y. Bai & A.H. Li sp. nov. MycoBank MB832545. Fig. 11, Fig. 14P.
Etymology: the specific epithet motuoensis refers to the geographic origin of the type strain, Motuo, Tibet.
Culture characteristics: In YM broth, after 7 d at 17 °C, cells are ovoid and ellipsoidal, 2.2–3.0 × 3.7–5.3 μm and single, budding is polar (Fig. 14P), a sediment is present. After 1 mo at 17 °C, a ring and a sediment are present. One or more conidia are produced on each stalk-like conidiophore. Conidiophore is single or multiple, usually bifurcate, somewhat trifurcate. On YM agar, after 1 mo at 17 °C, the streak culture is yellowish-cream, butyrous, smooth and glossy. The margin is entire. In Dalmau plate culture on corn meal agar, pseudohyphae are not formed. Sexual structures are not observed on YM, PDA, V8 and CM agar. Ballistoconidia are not produced.
Physiological and biochemical characteristics: Glucose fermentation is absent. Glucose, galactose (delayed and weak), L-sorbose (delayed and weak), sucrose, maltose (delayed and weak), trehalose, melezitose, ethanol, D-mannitol, D-glucitol and salicin (delayed and weak) are assimilated as sole carbon sources. Cellobiose, lactose, melibiose, raffinose, inulin, soluble starch, D-xylose, L-arabinose, D-arabinose, D-ribose, L-rhamnose, D-glucosamine, N-Acetyl-D-glucosamine, methanol, glycerol, erythritol, ribitol, galactitol, Methyl-α-D-glucoside, D-gluconate, DL-lactate, succinate, citrate, myo-inositol and hexadecane are not assimilated. Ammonium sulfate, potassium nitrate, sodium nitrite (delayed and weak), L-lysine, ethylamine hydrochloride and cadaverine dihydrochloride (delayed and weak) are assimilated as sole nitrogen sources. Maximum growth temperature is 26–27 °C. Growth in vitamin-free medium is negative. Starch-like substances are not produced. Growth on 50 % (w/w) glucose-yeast extract agar is negative. Urease activity is positive. Diazonium Blue B reaction is positive.
Typus: China, Motuo, Tibet, obtained from a leaf of Achyrospermum wallichianum, Sep. 2014, Q.-M. Wang (holotype CGMCC 2.5816T preserved in a metabolically inactive state, ex-type CBS 15591 = XZ119B3).
New taxa in Spiculogloeomycetes (Pucciniomycotina)
Phyllozyma aceris Q.M. Wang, F.Y. Bai & A.H. Li sp. nov. MycoBank MB828813. Fig. 15A, B.
Fig. 15.
Vegetative cells grown in YM broth for 5 d at 17 °C and ballistoconidia produced on corn meal agar after 7 d at 17 °C. (A, B) Phy. aceris CGMCC 2.2662T; (C) Phy. jiayinensis CGMCC 2.5669T; (D) Me. layueensis CGMCC 2.5818T; (E) Sa. melibiophila CBS 5143T; (F) Mi. ellipsoideus CGMCC 2.5664T; (G, H) Mi. rubellus CGMCC 2.4444T; (I, J) Mi. pseudomagnisporus CGMCC 2.4538T; (K, L) Sy. rhododendri CGMCC 2.2613T; (M) Cy. raffinophilum CGMCC 2.3822T; (N) Cy. terricola CGMCC 2.3823T, (O) Do. ningxiaensis CGMCC 2.4451T; (P) Beg. foliicola CGMCC 2.3164T. Bars = 10 μm.
Etymology: the specific epithet aceris refers to Acer, the plant genus from which the type strain was isolated.
Culture characteristics: In YM broth, after 7 d at 17 °C, cells are cylindrical, 2.6–3.5 × 5.5–8.9 μm and single, budding is polar (Fig. 15A), a sediment is present. After 1 mo at 17 °C, a pellicle and sediment are present. On YM agar, after 1 mo at 17 °C, the streak culture is pink-orange, butyrous, smooth and glossy. The margin is entire. In Dalmau plate culture on corn meal agar, pseudohyphae are not formed. Sexual structures are not observed on YM, PDA, V8 and CM agar. Ballistoconidia are allantoid, falcate or cylindrical, 2.5–3.7 × 10.0–13.3 μm (Fig. 15B).
Physiological and biochemical characteristics: Glucose fermentation is absent. Glucose, sucrose, trehalose, raffinose, inulin (delayed and weak), ribitol (delayed and weak), D-mannitol, D-glucitol (weak) and succinate (weak) are assimilated as sole carbon sources. Galactose, L-sorbose, maltose, cellobiose, lactose, melibiose, melezitose, soluble starch, D-xylose, L-arabinose, D-arabinose, D-ribose, L-rhamnose, D-glucosamine, N-Acetyl-D-glucosamine, methanol, ethanol, glycerol, erythritol, galactitol, Methyl-α-D-glucoside, salicin, DL-lactate, citrate, myo-inositol and hexadecane are not assimilated. Ammonium sulfate and potassium nitrate are assimilated as sole nitrogen sources. Sodium nitrite, L-lysine, ethylamine hydrochloride and cadaverine dihydrochloride are not assimilated. Maximum growth temperature is 20 °C. Growth in vitamin-free medium is negative. Starch-like substances are not produced. Growth on 50 % (w/w) glucose-yeast extract agar is negative. Urease activity is positive. Diazonium Blue B reaction is positive.
Physiologically, Phy. aceris differs from its closely related species Phy. coprosmicola in its inability to assimilate glycerol, D-gluconate, DL-lactate and sodium nitrite (Table S1.22).
Typus: China, Bomi county, Tibet, obtained from a leaf of Acer caudatum, Sep. 2004, F.-Y. Bai (holotype CGMCC 2. 2662T preserved in a metabolically inactive state, ex-type CBS 15773 = XZ17B1).
Phyllozyma jiayinensis Q.M. Wang, F.Y. Bai & A.H. Li sp. nov. MycoBank MB828814. Fig. 15C.
Etymology: the specific epithet jiayinensis refers to the geographic origin of the type strain, Jiayin, Heilongjiang.
Culture characteristics: In YM broth, after 7 d at 17 °C, cells are cylindrical, 1.4–2.0 × 3.2–7.3 μm and single, budding is polar (Fig. 15C), a sediment is present. After 1 mo at 17 °C, a sediment is present. On YM agar, after 1 mo at 17 °C, the streak culture is cream, butyrous, smooth and somewhat wrinkled and glossy. The margin is entire. In Dalmau plate culture on corn meal agar, pseudohyphae are not formed. Sexual structures are not observed on YM, PDA, V8 and CM agar. Ballistoconidia are not produced.
Physiological and biochemical characteristics: Glucose fermentation is absent. Glucose, trehalose, D-mannitol, D-glucitol (delayed), D-gluconate (weak) and DL-lactate (weak) are assimilated as sole carbon sources. Galactose, L-sorbose, sucrose, maltose, cellobiose, lactose, melibiose, raffinose, melezitose, inulin, soluble starch, D-xylose, L-arabinose, D-arabinose, D-ribose, L-rhamnose, D-glucosamine, N-Acetyl-D-glucosamine, methanol, ethanol, glycerol, erythritol, ribitol, galactitol, salicin, citrate, succinate, myo-inositol and hexadecane are not assimilated. Ammonium sulfate and potassium nitrate (delayed and weak) are assimilated as sole nitrogen sources. Sodium nitrite, L-lysine, ethylamine hydrochloride and cadaverine dihydrochloride are not assimilated. Maximum growth temperature is 26–27 °C. Growth in vitamin-free medium is negative. Starch-like substances are not produced. Growth on 50 % (w/w) glucose-yeast extract agar is negative. Urease activity is positive. Diazonium Blue B reaction is positive.
Physiologically, Phy. jiayinensis and its closely related species Phy. dimennae and Phy. corallina are distinguishable from one another by assimilation of sucrose, D-xylose, glycerol, ribitol, D-glucitol, Methyl-α-D-glucoside, DL-lactate, succinate and sodium nitrite (Table S1.22).
Typus: China, Qingshan county, Jiayin, Heilongjiang province, obtained from a leaf of an unidentified plant, Aug. 2014, Q.-M. Wang (holotype CGMCC 2.5669T preserved in a metabolically inactive state, ex-type CBS 13975 = HLJ25.21).
Meniscomyces Q.M. Wang & F.Y. Bai gen. nov. MycoBank MB828815.
Etymology: the genus is named after the lunately shaped vegetative cells.
This genus is proposed for the branch represented by strain CGMCC 2.5818T, which formed a separate clade. Member of the Spiculogloeomycetes. The genus is mainly circumscribed by the phylogenetic analysis of the seven genes dataset, in which it occurred as a separate branch within the Spiculogloeomycetes (Fig. 4A).
Sexual reproduction not known. Colonies cream, butyrous. Budding cells present. Cells special, lunate, allantoid and falcate, which differs from the cell morphology of other taxa in Spiculogloeomycetes (Pucciniomycotina). Pseudohyphae and hyphae not produced. Ballistoconidia not formed.
Type species: Meniscomyces layueensis Q.M. Wang, F.Y. Bai & A.H. Li.
Meniscomyces layueensis Q.M. Wang, F.Y. Bai & A.H. Li sp. nov. MycoBank MB828816. Fig. 11, Fig. 15D.
Etymology: the specific epithet layueensis refers to the geographic origin of the type strain, Layue county, Tibet.
Culture characteristics: In YM broth, after 7 d at 17 °C, cells are lunate, allantoids and falcate, 1.4–2.6 × 7.1–10.0 μm and single, budding is polar (Fig. 15D), a sediment is present. After 1 mo at 17 °C, a sediment is present. On YM agar, after 1 mo at 17 °C, the streak culture is cream, butyrous, smooth and glossy. The margin is entire. In Dalmau plate culture on corn meal agar, pseudohyphae are not formed. Sexual structures are not observed on YM, PDA, V8 and CM agar. Ballistoconidia are not produced.
Physiological and biochemical characteristics: Glucose fermentation is absent. Glucose, sucrose, maltose, cellobiose (variable), trehalose, melezitose and succinate are assimilated as sole carbon sources. Galactose, L-sorbose, lactose, melibiose, raffinose, inulin, soluble starch, D-xylose, L-arabinose, D-arabinose, D-ribose, L-rhamnose, D-glucosamine, N-Acetyl-D-glucosamine, methanol, ethanol, glycerol, erythritol, ribitol, galactitol, D-mannitol, D-glucitol, Methyl-α-D-glucoside, salicin, D-gluconate, DL-lactate, citrate, myo-inositol and hexadecane are not assimilated. Ammonium sulfate, potassium nitrate, L-lysine and ethylamine hydrochloride (weak) are assimilated as sole nitrogen sources. Sodium nitrite and cadaverine dihydrochloride are not assimilated. Maximum growth temperature is 23 °C. Growth in vitamin-free medium is negative. Starch-like substances are not produced. Growth on 50 % (w/w) glucose-yeast extract agar is negative. Urease activity is positive. Diazonium Blue B reaction is positive.
Typus: China, Layue county, Tibet, obtained from a leaf of an unidentified plant, Sep. 2014, Q.-M. Wang (holotype CGMCC 2.5818T preserved in a metabolically inactive state, ex-type CBS 15747 = XZ100).
New taxa in Cystobasidiomycetes (Pucciniomycotina)
Sakaguchia melibiophila M. Groenew., Q.M. Wang, & F.Y. Bai sp. nov. MycoBank MB828817. Fig. 15E
Etymology: the specific epithet melibiophila refers to the physiological character of assimilating melibiose.
Culture characteristics: In YM broth, after 7 d at 17 °C, cells are ellipsoidal, 2.5–4.4 × 3.8–5.6 μm and single, budding is polar (Fig. 15E), a sediment is formed. After 1 mo at 17 °C, a sediment is present. On YM agar, after 1 mo at 17 °C, the streak culture is orange-red, butyrous, smooth. The margin is entire. In Dalmau plate culture on corn meal agar, pseudohyphae are not formed. Sexual structures are not observed on YM, PDA, V8 and CM agar. Ballistoconidia are not produced.
Physiological and biochemical characteristics: Glucose fermentation is absent. Glucose, galactose, L-sorbose, cellobiose, trehalose, melibiose (delayed), D-xylose, L-arabinose, D-arabinose (delayed), D-ribose (delayed), ethanol, glycerol, ribitol, D-mannitol, salicin, D-glucitol, D-gluconate DL-lactate, succinate, citrate, myo-Inositol are assimilated as sole carbon sources. Sucrose, maltose, lactose, raffinose, melezitose, inulin, soluble starch, L-rhamnose, D-glucosamine, methanol, erythritol, galactitol and Methyl-α-D-glucoside are not assimilated. Potassium nitrate, sodium nitrite, L-lysine, ethylamine hydrochloride and cadaverine dihydrochlorideare assimilated as sole nitrogen sources. Maximum growth temperature is 35 °C. Growth in vitamin-free medium is negative. Starch-like substances are not produced. Growth on 50 % (w/w) glucose-yeast extract agar is negative. Urease activity is positive. Diazonium Blue B reaction is positive.
Physiologically, Sa. melibiophila differs from its closely related species Sa. lamellibrachiae and Sa. meli in its ability to assimilate cellobiose, melibiose, ribitol, and nitrate (Table S1.23).
Typus: Netherlands, obtained from bronchial secretion, J. Swieringa (holotype CBS 5143T preserved in a metabolically inactive state, ex-type JCM 8162 = CGMCC 2.4235).
Microsporomyces ellipsoideus Q.M. Wang, F.Y. Bai & A.H. Li sp. nov. MycoBank MB828818. Fig. 15F.
Etymology: the specific epithet ellipsoideus refers to the ellipsoidal vegetative cells of the type strain.
Culture characteristics: In YM broth, after 7 d at 17 °C, cells are ellipsoidal or cylindrical, 6.0–7.5 × 9.0–14.5 μm and single, budding is polar (Fig. 15F), a sediment is formed. After 1 mo at 17 °C, a ring and sediment are present. On YM agar, after 1 mo at 17 °C, the streak culture is brownish-orange, butyrous, smooth. The margin is entire. In Dalmau plate culture on corn meal agar, pseudohyphae are not formed. Sexual structures are not observed on YM, PDA, V8 and CM agar. Ballistoconidia are not produced.
Physiological and biochemical characteristics: Glucose fermentation is absent. Glucose, galactose, sucrose, maltose, trehalose, melibiose (weak), raffinose, soluble starch, glycerol, D-glucitol, Methyl-α-D-glucoside, salicin and succinate (delayed and weak) are assimilated as sole carbon sources. L-sorbose, cellobiose, lactose, melezitose, inulin, D-xylose, L-arabinose, D-arabinose, D-ribose, L-rhamnose, D-glucosamine, methanol, ethanol, erythritol, ribitol, galactitol, D-mannitol, DL-lactate, citrate, myo-inositol and hexadecane are not assimilated. Ammonium sulfate, potassium nitrate and L-lysine (weak) are assimilated as sole nitrogen sources. Sodium nitrite, ethylamine hydrochloride and cadaverine dihydrochloride are not assimilated. Maximum growth temperature is 26–27 °C. Growth in vitamin-free medium is negative. Starch-like substances are not produced. Growth on 50 % (w/w) glucose-yeast extract agar is negative. Urease activity is positive. Diazonium Blue B reaction is positive.
Physiologically, Mi. ellipsoideus differs from its closely related species Mi. rubellus in its inability to assimilate melezitose, ribitol and galactitol and its ability to soluble starch and Methyl-α-D-glucoside (Table S1.24).
Typus: China, Motuo county, Tibet, obtained from a leaf of an unidentified plant, Sep. 2014, Q.-M. Wang (holotype CGMCC 2.5664T preserved in a metabolically inactive state, ex-type CBS 16020 = XZ137E4).
Microsporomyces rubellus Q.M. Wang, F.Y. Bai & A.H. Li sp. nov. MycoBank MB828819. Fig. 15G, H.
Etymology: the specific epithet rubellus refers to the pale red colony colour of this species.
Culture characteristics: In YM broth, after 7 d at 17 °C, cells are ovoid and ellipsoidal, 3.8–6.2 × 5.1–8.1 μm and single, budding is polar (Fig. 15G), a sediment is formed. After 1 mo at 17 °C, a ring and a sediment are present. On YM agar, after 1 mo at 17 °C, the streak culture is pale-red, butyrous, smooth and semi-glossy. The margin is entire. In Dalmau plate culture on corn meal agar, pseudohyphae are not formed. Sexual structures are not observed on YM, PDA, V8 and CM agar. Ballistoconidia are allantoid, reniform or cylindrical, 2.1–5.7 × 5.0–11.4 μm (Fig. 15H).
Physiological and biochemical characteristics: Glucose fermentation is absent. Glucose, galactose, sucrose, maltose, trehalose, melibiose, raffinose, melezitose, glycerol (delayed and weak), ribitol, galactitol, D-mannitol (delayed and weak), D-glucitol (weak), salicin (variable) and DL-lactate (variable) are assimilated as sole carbon sources. L-sorbose, cellobiose, lactose, inulin, soluble starch, D-xylose, L-arabinose, D-arabinose, D-ribose, L-rhamnose, D-glucosamine, methanol, ethanol, erythritol, Methyl-α-D-glucoside, succinate, citrate, myo-inositol and hexadecane are not assimilated. Ammonium sulfate, potassium nitrate (variable) and L-lysine (variable) are assimilated as sole nitrogen sources. Sodium nitrite, ethylamine hydrochloride and cadaverine dihydrochloride are not assimilated. Maximum growth temperature is 32 °C. Growth in vitamin-free medium is negative. Starch-like substances are not produced. Growth on 50 % (w/w) glucose-yeast extract agar is negative. Urease activity is positive. Diazonium Blue B reaction is positive.
Physiologically, Mi. rubellus differs from its three closely related species Mi. ellipsoideus in its inability to assimilate soluble starch and Methyl-α-D-glucoside and its ability to assimilate melezitose, ribitol and galactitol (Table S1.24).
Typus: China, Taiwan province, obtained from a leaf of an unidentified plant, Aug. 2009, Q.-M. Wang (holotype CGMCC 2.4444T preserved in a metabolically inactive state, ex-type CBS 15622 = TW1.3F-017).
Microsporomyces pseudomagnisporus Q.M. Wang, F.Y. Bai & A.H. Li sp. nov. MycoBank MB828820. Fig. 15I, J.
Etymology: the specific epithet pseudomagnisporus refers to the similar colony morphology to that of Microsporomyces magnisporus.
Culture characteristics: In YM broth, after 7 d at 17 °C, cells are cylindrical, 2.0–3.0 × 4.0–8.0 μm and single, budding is polar (Fig. 15I), a sediment is formed. After 1 mo at 17 °C, a part ring and sediment are present. On YM agar, after 1 mo at 17 °C, the streak culture is orange, butyrous, wrinkled and semi-glossy. The margin is entire. In Dalmau plate culture on corn meal agar, pseudohyphae are not formed. Sexual structures are not observed on YM, PDA, V8 and CM agar. Ballistoconidia are allantoid or reniform, 2.5–3.3 × 5.8–8.3 μm (Fig. 15J).
Physiological and biochemical characteristics: Glucose fermentation is absent. Glucose, galactose, L-sorbose, sucrose, trehalose (weak), melibiose (weak), raffinose (weak), melezitose (weak), inulin (delayed), D-arabinose (weak), ethanol, ribitol (weak), D-mannitol (weak), D-glucitol (weak), Methyl-α-D-glucoside (weak) and succinate (weak) are assimilated as sole carbon sources. Maltose, cellobiose, lactose, soluble starch, D-xylose, L-arabinose, D-ribose, L-rhamnose, D-glucosamine, N-Acetyl-D-glucosamine, methanol, glycerol, erythritol, galactitol, salicin, DL-lactate, citrate, myo-inositol and hexadecane are not assimilated. Ammonium sulfate, potassium nitrate, L-lysine, ethylamine hydrochloride and cadaverine dihydrochloride are assimilated as sole nitrogen sources. Sodium nitrite is not assimilated. Maximum growth temperature is 19 °C. Growth in vitamin-free medium is postive. Starch-like substances are not produced. Growth on 50 % (w/w) glucose-yeast extract agar is negative. Urease activity is positive. Diazonium Blue B reaction is positive.
Physiologically, Mi. pseudomagnisporus differs from its closely related species Mi. magnisporus in its inability to assimilate maltose, soluble starch, N-Acetyl-D-glucosamine, DL-lactate, citrate and sodium nitrite and its ability to assimilate inulin, ethanol, L-lysine, ethylamine and cadaverine (Table S1.24).
Typus: China, Fanjingshan Mountain, Guizhou province, obtained from a leaf of an unidentified plant, Oct. 2011, Q.-M. Wang (holotype CGMCC 2.4538T preserved in a metabolically inactive state, ex-type CBS 15746 = FJS25C3).
Symmetrospora rhododendri Q.M. Wang, F.Y. Bai & A.H. Li sp. nov. MycoBank MB828821. Fig. 15K, L.
Etymology: the specific epithet rhododendri refers to Rhododendron, the plant genus from which the type strain was isolated.
Culture characteristics: In YM broth, after 7 d at 17 °C, cells are cylindrical and ellipsoidal, 3.4–4.7 × 6.6–9.4 μm and single, budding is polar (Fig. 15K), a sediment is formed. After 1 mo at 17 °C, a part ring and sediment are present. On YM agar, after 1 mo at 17 °C, the streak culture is pinkish orange, butyrous, slight wrinkled and glossy. The margin is entire. In Dalmau plate culture on corn meal agar, pseudohyphae are not formed. Sexual structures are not observed on YM, PDA, V8 and CM agar. Ballistoconidia are ellipsoidal to long ovoid, 1.7–3.6 × 5.0–7.1 μm (Fig. 15L).
Physiological and biochemical characteristics: Glucose fermentation is absent. Glucose, galactose, L-sorbose (delayed), sucrose, trehalose, melibiose, L-arabinose, glycerol, ribitol (weak), D-mannitol, D-glucitol and succinate (weak) are assimilated as sole carbon sources. Maltose, cellobiose, lactose, melezitose, raffinose, inulin, soluble starch, D-xylose, D-arabinose, D-ribose, L-rhamnose, D-glucosamine, N-Acetyl-D-glucosamine, methanol, ethanol, erythritol, galactitol, Methyl-α-D-glucoside, salicin, DL-lactate, citrate, myo-inositol and hexadecane are not assimilated. Ammonium sulfate, potassium nitrate and L-lysine are assimilated as sole nitrogen sources. Sodium nitrite, ethylamine hydrochloride and cadaverine dihydrochloride are not assimilated. Maximum growth temperature is 25 °C. Growth in vitamin-free medium is positive. Starch-like substances are not produced. Growth on 50 % (w/w) glucose-yeast extract agar is negative. Urease activity is positive. Diazonium Blue B reaction is positive.
Physiologically, Sy. rhododendri differs from its closely related species Sy. coprosmae and Sy. oryzicola in its inability to assimilate L-sorbose, cellobiose, melezitose, D-arabinose, D-ribose, Methyl-α-D-glucoside, salicin and DL-lactate and its ability to assimilate lactose, inulin, nitrate and cadaverine (Table S1.25).
Typus: China, Lulang county, Tibet, obtained from a leaf of Rhododendron sp., Sep. 2004, F.-Y. Bai (holotype CGMCC 2.2613T preserved in a metabolically inactive state, ex-type CBS 15447 = XZ49DX).
New combinations for Symmetrospora
Symmetrospora oryzicola (Nakase & M. Suzuki) Q.M. Wang & F.Y. Bai, com. nov. MycoBank MB832091.
Basionym: Sporobolomyces oryzicola Nakase & M. Suzuki, J. Gen. Appl. Microbiol., 32(2): 152 (1986).
Cystobasidium raffinophilum Q.M. Wang, F.Y. Bai & A.H. Li sp. nov. MycoBank MB828822. Fig. 15M.
Etymology: the specific epithet raffinophilum refers to the ability to assimilate raffinose.
Culture characteristics: In YM broth, after 7 d at 17 °C, cells are ellipsoidal, 3.1–5.0 × 4.5–6.8 μm and single, budding is polar (Fig. 15M), a sediment is present. After 1 mo at 17 °C, a ring and sediment are present. On YM agar, after 1 mo at 17 °C, the streak culture is orange-pink, mucoid, smooth and glistening. The margin is entire. In Dalmau plate culture on corn meal agar, pseudohyphae are not formed. Sexual structures are not observed on YM, PDA, V8 and CM agar. Ballistoconidia are not produced.
Physiological and biochemical characteristics: Glucose fermentation is absent. Glucose, galactose (delayed), sucrose, cellobiose, trehalose, raffinose, melezitose, inulin (weak), D-xylose, L-arabinose, D-arabinose, D-ribose (delayed and weak), ethanol, glycerol, ribitol (delayed and weak), galactitol, D-mannitol, succinate (weak) and citrate (weak) are assimilated as sole carbon sources. L-sorbose, maltose, lactose, melibiose, soluble starch, L-rhamnose, D-glucosamine, N-Acetyl-D-glucosamine, methanol, erythritol, D-glucitol, Methyl-α-D-glucoside, salicin, DL-lactate, myo-inositol and hexadecane are not assimilated. Ammonium sulfate and potassium nitrate are assimilated as sole nitrogen sources. Sodium nitrite, L-lysine, ethylamine hydrochloride and cadaverine dihydrochloride are not assimilated. Maximum growth temperature is 26–27 °C. Growth in vitamin-free medium is negative. Starch-like substances are not produced. Growth on 50 % (w/w) glucose-yeast extract agar is negative. Urease activity is positive. Diazonium Blue B reaction is positive.
Physiologically, Cy. raffinophilum differs from its closely related species Cy. fimetarium in its inability to assimilate lactose, salicin, DL-lactate and its ability to assimilate galactose, raffinose, melezitose, galactitol and potassium nitrate (Table S1.26).
Typus: China, Yecheng county, Xinjiang province, obtained from soil, Jul. 2007, Q.-M. Wang (holotype CGMCC 2.3822T preserved in a metabolically inactive state, ex-type CBS 15509 = 141.4).
Cystobasidium terricola Q.M. Wang, F.Y. Bai & A.H. Li sp. nov. MycoBank MB828823. Fig. 15N.
Etymology: the specific epithet terricola refers to the origin of the substrate of the type strain, soil.
Culture characteristics: In YM broth, after 7 d at 17 °C, cells are ovoid and ellipsoidal, 2.1–4.2 × 2.8–5.7 μm and single, budding is polar (Fig. 15N), a sediment is present. After 1 mo at 17 °C, a ring and sediment are present. On YM agar, after 1 mo at 17 °C, the streak culture is pink-red, mucoid, smooth and glistening. The margin is entire. In Dalmau plate culture on corn meal agar, pseudohyphae are not formed. Sexual structures are not observed on YM, PDA, V8 and CM agar. Ballistoconidia are not produced.
Physiological and biochemical characteristics: Glucose fermentation is absent. Glucose, galactose (variable), L-sorbose (variable), sucrose, cellobiose, trehalose, lactose, raffinose (variable), melezitose, D-xylose, L-arabinose, D-arabinose (delayed and weak), D-ribose, ethanol, glycerol, ribitol, D-mannitol, D-glucitol, salicin, DL-lactate (delayed and weak), succinate and citrate (variable) are assimilated as sole carbon sources. Maltose, melibiose, inulin, soluble starch, L-rhamnose, D-glucosamine, methanol, erythritol, galactitol, Methyl-α-D-glucoside, myo-inositol and hexadecane are not assimilated. Ammonium sulfate, potassium nitrate (delayed and weak), sodium nitrite (delayed and weak), L-lysine (delayed and weak), ethylamine hydrochloride (delayed and weak) and cadaverine dihydrochloride (delayed and weak) are assimilated as sole nitrogen sources. Maximum growth temperature is 35 °C. Growth in vitamin-free medium is weak. Starch-like substances are not produced. Growth on 50 % (w/w) glucose-yeast extract agar is negative. Urease activity is positive. Diazonium Blue B reaction is positive.
Physiologically, Cy. terricola and its three closely related species, Cy. raffinophilum, Cy. minutum and Cy. fimetarium, are distinguishable by the assimilation of L-sorbose, galactose, lactose, raffinose, melezitose, galactitol, D-glucitol, salicin, DL-lactate and potassium nitrate (Table S1.26).
Typus: China, Yecheng county, Xinjiang province, obtained from soil, Jul. 2007, Q.-M. Wang (holotype CGMCC 2.3823T preserved in a metabolically inactive state, ex-type CBS 15650 = 140.23).
Robertozyma Q.M. Wang & F.Y. Bai gen. nov. MycoBank MB828824.
Etymology: the genus is named in honour of Dr. V. Robert for his contributions to the yeast taxonomy.
This genus is proposed for the branch represented by strain CGMCC 2.4451 which formed a separate clade. Member of the Cystobasidiales (Cystobasidiomycetes). The genus is mainly circumscribed by the phylogenetic analysis of the seven genes dataset, in which it occurred as a separate branch within the Cystobasidiales (Fig. 4).
Sexual reproduction not known. Colonies orange, butyrous. Budding cells present. Pseudohyphae and hyphae not produced. Ballistoconidia not formed.
Type species: Robertozyma ningxiaensis Q.M. Wang, F.Y. Bai & A.H. Li.
Note: Robertozyma and its closely related genera, Begerowomyces and Halobasidium, have a similar colony morphology, however, they can be distinguished by some physiological characters (Table S1.27). Robertozyma does not assimilate sucrose, melezitose, D-xylose and ethanol, whereas species of Begerowomyces and Halobasidium can use them. Begerowomyces species assimilate erythritol and galactitol, whereas species of Robertozyma and Halobasidium do not assimilate these two carbon resources.
Robertozyma ningxiaensis Q.M. Wang, F.Y. Bai & A.H. Li sp. nov. MycoBank MB828826. Fig. 15O.
Etymology: the specific epithet ningxiaensis refers to the geographic origin of the type strain, Ningxia province, China.
Culture characteristics: In YM broth, after 7 d at 17 °C, cells are oval and ellipsoidal, 3.2–4.5 × 3.9–6.8 μm and single, budding is polar (Fig. 15O), a sediment is present. After 1 mo at 17 °C, a ring and sediment are present. On YM agar, after 1 mo at 17 °C, the streak culture is orange red, butyrous, smooth and glossy. The margin is entire. In Dalmau plate culture on corn meal agar, pseudohyphae are not formed. Sexual structures are not observed on YM, PDA, V8 and CM agar. Ballistoconidia are not produced.
Physiological and biochemical characteristics: Glucose fermentation is absent. Glucose, galactose (delayed and weak), trehalose, glycerol, D-mannitol, salicin (delayed and weak) and succinate are assimilated as sole carbon sources. L-sorbose, sucrose, maltose, cellobiose, lactose, melibiose, raffinose, melezitose, inulin, soluble starch, D-xylose, L-arabinose, D-arabinose, D-ribose, L-rhamnose, D-glucosamine, methanol, ethanol, erythritol, ribitol, galactitol, D-glucitol, Methyl-α-D-glucoside, D-gluconate, DL-lactate, citrate, myo-inositol and hexadecane are not assimilated. Ammonium sulfate, L-lysine and ethylamine hydrochloride are assimilated as sole nitrogen sources. Potassium nitrate, sodium nitrite and cadaverine dihydrochloride are not assimilated. Maximum growth temperature is 22–23 °C. Growth in vitamin-free medium is negative. Starch-like substances are not produced. Growth on 50 % (w/w) glucose-yeast extract agar is negative. Urease activity is positive. Diazonium Blue B reaction is positive.
Typus: China, Helanshan mountain, Ningxia province, obtained from soil, Aug. 2009, P.J. Han (holotype CGMCC 2.4451T preserved in a metabolically inactive state, ex-type CBS 12499 = HLS10.23).
Begerowomyces Q.M. Wang & F.Y. Bai gen. nov. MycoBank MB828827.
Etymology: the genus is named in honour of Dr. Dominik Begerow for his contributions to yeast taxonomy and his proposal of the order Cystobasidiales.
This genus is proposed for the branch represented by strain CGMCC 2.3164, which formed a separate clade. Member of the Cystobasidiales (Cystobasidiomycetes). The genus is mainly circumscribed by the phylogenetic analysis of the seven genes dataset, in which it occurred as a separate branch within the Cystobasidiales (Fig. 4).
Sexual reproduction not known. Colonies yellow, butyrous. Budding cells present. Pseudohyphae and hyphae not produced. Ballistoconidia not formed.
Type species: Begerowomyces foliicola Q.M. Wang, F.Y. Bai & A.H. Li.
Begerowomyces foliicola Q.M. Wang, F.Y. Bai & A.H. Li sp. nov. MycoBank MB828828. Fig. 11, Fig. 15P.
Etymology: the specific epithet foliicola refers to the type strain isolated from a leaf.
Culture characteristics: In YM broth, after 7 d at 17 °C, cells are ovoid and ellipsoidal, 2.6–3.9 × 2.7–6.0 μm and single, budding is polar (Fig. 15P), a sediment is formed. After 1 mo at 17 °C, a pellicle and sediment are present. On YM agar, after 1 mo at 17 °C, the streak culture is yellowish cream, butyrous, wrinkled and smooth. The margin is entire. In Dalmau plate culture on corn meal agar, pseudohyphae are not formed. Sexual structures are not observed on YM, PDA, V8 and CM agar. Ballistoconidia are not produced.
Physiological and biochemical characteristics: Glucose fermentation is absent. Glucose, galactose (delayed and weak), sucrose (delayed), maltose (delayed and weak), cellobiose (delayed and weak), trehalose (delayed and weak), melezitose, inulin (delayed and weak), D-xylose, L-arabinose (delayed and weak), ethanol, erythritol (delayed), ribitol, galactitol, D-mannitol (delayed), D-glucitol (delayed) and succinate (delayed) are assimilated as sole carbon sources. L-sorbose, lactose, melibiose, raffinose, soluble starch, D-arabinose, D-ribose, L-rhamnose, D-glucosamine, N-Acetyl-D-glucosamine, methanol, glycerol, Methyl-α-D-glucoside, salicin, DL-lactate, citrate, myo-inositol and hexadecane are not assimilated. Ammonium sulfate, potassium nitrate (delayed and weak), L-lysine and ethylamine hydrochloride are assimilated as sole nitrogen sources. Sodium nitrite and cadaverine dihydrochloride are not assimilated. Maximum growth temperature is 26–27 °C. Growth in vitamin-free medium is positive. Starch-like substances are not produced. Growth on 50 % (w/w) glucose-yeast extract agar is negative. Urease activity is positive. Diazonium Blue B reaction is positive.
Typus: Germany, obtained from a leaf of an unidentified plant, Sep. 2005 (holotype CGMCC 2.3164T preserved in a metabolically inactive state, ex-type CBS 15655 = G7.4).
New taxa in Microbotryomycetes (Pucciniomycotina)
Rosettozymales Q.M. Wang & F.Y. Bai ord. nov. MycoBank MB828829.
Member of the Microbotryomycetes. The diagnosis of the order Rosettozymales is based on the the genus Rosettozyma. The nomenclature of the order is based on the genus Rosettozyma.
Type family: Rosettozymaceae Q.M. Wang & F.Y. Bai.
Rosettozymaceae Q.M. Wang & F.Y. Bai fam. nov. MycoBank MB828830.
Member of the Rosettozymales (Microbotryomycetes). The diagnosis of the family Rosettozymaceae is based on the the genus Rosettozyma. The nomenclature of the family is based on the genus Rosettozyma.
Type genus: Rosettozyma Q.M. Wang & F.Y. Bai.
Genus accepted: Rosettozyma Q.M. Wang & F.Y. Bai.
Rosettozyma Q.M. Wang & F.Y. Bai gen. nov. MycoBank MB828831.
Etymology: the genus is named based on the morphology of the vegetative cells forming a rosette.
This genus is proposed for the clade represented by CGMCC 2.3446, which formed a separate clade from other orders and taxa in the Microbotryomycetes. Member of Microbotryomycetes. The genus is mainly circumscribed by the phylogenetic analysis of the seven genes dataset, in which it occurred as a separate clade within the Microbotryomycetes (Fig. 4).
Sexual reproduction not known. Colonies white, butyrous. Budding cells present and always form rosette-like clusters. Pseudohyphae and hyphae not produced. Ballistoconidia formed.
Type species: Rosettozyma petaloides Q.M. Wang, F.Y. Bai & A.H. Li.
Note: Except the genus Rosettozyma, species in Yamadamyces and Meredithblackwellia also form rosette-like cell clusters (Golubev and Scorzetti, 2010, Toome et al., 2013).
Rosettozyma petaloides Q.M. Wang, F.Y. Bai & A.H. Li sp. nov. MycoBank MB828832. Fig. 11, Fig. 16A, B.
Fig. 16.
Vegetative cells grown in YM broth for 5 d at 17 °C and ballistoconidia produced on corn meal agar after 7 d at 17 °C. (A, B) Ros. petaloides CGMCC 2.3446T; (C, D) Ros. cystopteridis CGMCC 2.2615T; (E, F) Ros. motuoensis CGMCC 2.5819T; (G, H) Sp. cellobiolyticus CGMCC 2.5675T; (I, J) Sp. reniformis CGMCC 2.5627T; (K, L) Sp. ellipsoideus CGMCC 2.5619T; (M, N) Sp. primogenomicus IAM13481T; (O, P) Rh. platycladi CGMCC 2.3118T. Bars = 10 μm.
Etymology: the specific epithet petaloides refers to the vegetative cells forming a petale morphology of the type strain.
Culture characteristics: In YM broth, after 7 d at 17 °C, cells are elongate fusiform, either singly or in rosettes, 2.2–3.2 × 9.8–18.7 μm, budding is polar (Fig. 16A), a sediment is formed. After 1 mo at 17 °C, a pellicle and a sediment are present. On YM agar, after 1 mo at 17 °C, the streak culture is whitish to cream, butyrous, slightly wrinkled and semi-glossy. The margin is entire. In Dalmau plate culture on corn meal agar, pseudohyphae are not formed. Sexual structures are not observed on YM, PDA, V8 and CM agar. Ballistoconidia are cylindrical or falcate, 1.3–1.6 × 9.3–12.0 μm (Fig. 16B).
Physiological and biochemical characteristics: Glucose fermentation is absent. Glucose, sucrose, maltose, cellobiose (variable), trehalose, lactose (variable), raffinose (variable), melezitose, D-xylose, L-arabinose, D-arabinose (variable), D-ribose (variable), ethanol (variable), glycerol, ribitol (variable), D-mannitol, D-glucitol, Methyl-α-D-glucoside, salicin (delayed and weak), DL-lactate (variable), succinate (delayed and weak) and citrate (delayed and weak) are assimilated as sole carbon sources. Galactose, L-sorbose, melibiose, inulin, soluble starch, L-rhamnose, D-glucosamine, methanol, erythritol, galactitol, myo-inositol and hexadecane are not assimilated. Ammonium sulfate, potassium nitrate (variable), sodium nitrite (variable), L-lysine (variable), ethylamine hydrochloride (delayed and weak) and cadaverine dihydrochloride (delayed and weak) are assimilated as sole nitrogen sources. Maximum growth temperature is 26–27 °C. Growth in vitamin-free medium is delayed and weak. Starch-like substances are not produced. Growth on 50 % (w/w) glucose-yeast extract agar is negative. Urease activity is positive. Diazonium Blue B reaction is positive.
Physiologically, Ro. petaloides and its two closely related species, Ro. cystopteridis and Ro. motuoensis, can be distinguished from one another by the assimilation of D-xylose, L-arabinose, D-arabinose, glycerol and succinate. Ro. petaloides differs from the other species in its ability to assimilate D-xylose and glycerol (Table S1.28).
Typus: China, Wuzhishan mountain, Hainan province, obtained from a leaf of an unidentified plant, Nov. 2006, Q.-M. Wang (holotype CGMCC 2.3446T preserved in a metabolically inactive state, ex-type CBS 15480 = WZS29.14).
Rosettozyma cystopteridis Q.M. Wang, F.Y. Bai & A.H. Li sp. nov. MycoBank MB828833. Fig. 16, Fig. 17A, B.
Fig. 17.
SEM image of vegetative cells grown in YM broth for 5 d at 17 °C. (A, B) Ros. cystopteridis CGMCC 2.2615T, A Bars = 10 μm, B Bars = 2 μm; (C) Ros. motuoensis CGMCC 2.5819T, Bars = 10 μm; (D) He. tridentata CGMCC 2.5602T, Bars = 10 μm; (E, F) He. cylindrica CGMCC 2.5650T, E Bars = 20 μm, F Bars = 5 μm; (G) Ya. terricola CGMCC 2.5820T, Bars = 10 μm; (H) Ch. rhododendri CGMCC 2.5821T, Bars = 5 μm.
Etymology: the specific epithet cystopteridis refers to Cystopteris, the plant genus from which the type strain was isolated.
Culture characteristics: In YM broth, after 7 d at 17 °C, cells are ellipsoidal, either singly or in rosettes, 2.2–2.8 × 11.4–20.3 μm, budding is polar (Fig. 16C), a sediment is formed. After 1 mo at 17 °C, a pellicle and a sediment are present. On YM agar, after 1 mo at 17 °C, the streak culture is whitish to cream, butyrous, slightly wrinkle, semi-glistening. The margin is entire. In Dalmau plate culture on corn meal agar, pseudohyphae are not formed. Sexual structures are not observed on YM, PDA, V8 and CM agar. Ballistoconidia are allantoid or falcate, 1.7–2.8 × 7.7–15.4 μm (Fig. 16D).
Physiological and biochemical characteristics: Glucose fermentation is absent. Glucose, sucrose, maltose, trehalose, melezitose, L-arabinose (variable), D-arabinose, ethanol, erythritol (variable), D-mannitol, D-glucitol, Methyl-α-D-glucoside (variable) and salicin are assimilated as sole carbon sources. Galactose, L-sorbose, cellobiose, lactose, melibiose, raffinose, inulin, soluble starch, D-xylose, D-ribose, L-rhamnose, D-glucosamine, methanol, glycerol, ribitol, galactitol, DL-lactate, succinate, citrate, myo-inositol and hexadecane are not assimilated. Ammonium sulfate, potassium nitrate, L-lysine, ethylamine hydrochloride and cadaverine dihydrochloride are assimilated as sole nitrogen sources. Sodium nitrite is not assimilated. Maximum growth temperature is 27 °C. Growth in vitamin-free medium is negative. Starch-like substances are not produced. Growth on 50 % (w/w) glucose-yeast extract agar is negative. Urease activity is positive. Diazonium Blue B reaction is positive.
Physiologically, Ro. cystopteridis and its two closely related species, Ro. petaloides and Ro. motuoensis, can be distinguished from one another by the assimilation of D-xylose, L-arabinose, D-arabinose, glycerol and succinate. Ro. cystopteridis differs from Ro. petaloides in its inability to assimilate D-xylose and glycerol. Ro. cystopteridis differs from Ro. motuoensis in its inability to assimilate succinate and its ability to assimilate D-arabinose (Table S1.28).
Typus: China, Bomi county, Tibet, obtained from a leaf of Cystopteris moupinensis, Sep. 2004, F.-Y. Bai (holotype CGMCC 2.2615T preserved in a metabolically inactive state, ex-type CBS 15448 = XZ16E1).
Rosettozyma motuoensis Q.M. Wang, F.Y. Bai & A.H. Li sp. nov. MycoBank MB828834. Fig. 16, Fig. 17C.
Etymology: the specific epithet motuoensis refers to the geographic origin of the type strain, Motuo, Tibet.
Culture characteristics: In YM broth, after 7 d at 17 °C, cells are ellipsoidal, either singly or in rosettes, 1.5–2.5 × 12.5–20.0 μm, budding is polar (Fig. 16E), a sediment is formed. After 1 mo at 17 °C, a pellicle and a sediment are present. On YM agar, after 1 mo at 17 °C, the streak culture is white, butyrous, smooth, semi-glistening. The margin is entire. In Dalmau plate culture on corn meal agar, pseudohyphae are not formed. Sexual structures are not observed on YM, PDA, V8 and CM agar. Ballistoconidia are allantoid or falcate, 1.4–2.3 × 11.7–21.0 μm (Fig. 16F).
Physiological and biochemical characteristics: Glucose fermentation is absent. Glucose, sucrose, maltose, trehalose, melezitose, ethanol, D-mannitol, D-glucitol, Methyl-α-D-glucoside and succinate are assimilated as sole carbon sources. Galactose, L-sorbose, cellobiose, lactose, melibiose, raffinose, inulin, soluble starch, D-xylose, L-arabinose, D-arabinose, D-ribose, L-rhamnose, D-glucosamine, N-Acetyl-D-glucosamine, methanol, glycerol, erythritol, ribitol, galactitol, salicin, D-gluconate, DL-lactate, citrate, myo-inositol and hexadecane are not assimilated. Ammonium sulfate, potassium nitrate, L-lysine and ethylamine hydrochloride are assimilated as sole nitrogen sources. Sodium nitrite and cadaverine dihydrochloride are not assimilated. Maximum growth temperature is 22–23 °C. Growth in vitamin-free medium is negative. Starch-like substances are not produced. Growth on 50 % (w/w) glucose-yeast extract agar is negative. Urease activity is positive. Diazonium Blue B reaction is positive.
Physiologically, Ro. motuoensis and their two closely related species, Ro. petaloides and Ro. cystopteridis, can be distinguished from one another by the assimilation of D-xylose, L-arabinose, D-arabinose, glycerol and succinate. Ro. motuoensis differs from Ro. petaloides in its inability to assimilate D-xylose, L-arabinose and glycerol and its ability to assimilate succinate. Ro. motuoensis differs from Ro. cystopteridis in its inability to assimilate D-arabinose and its ability to assimilate succinate (Table S1.28).
Typus: China, Motuo, Tibet, obtained from a leaf of an unidentified plant, Sep. 2014, Q.-M. Wang (holotype CGMCC 2.5819T preserved in a metabolically inactive state, ex-type CBS 15588 = XZ118E6).
Sporobolomyces cellobiolyticus Q.M. Wang, F.Y. Bai & A.H. Li sp. nov. MycoBank MB828835. Fig. 16G, H.
Etymology: the specific epithet cellobiolyticus refers to the physiological character of assimilating cellobiose.
Culture characteristics: In YM broth, after 7 d at 17 °C, cells are ovoid, ellipsoidal and cylindrical, 2.6–4.8 × 5.6–12.0 μm and single, budding is polar (Fig. 16G), a sediment is formed. On YM agar, after 1 mo at 17 °C, the streak culture is orange, butyrous, smooth. The margin is entire. In Dalmau plate culture on corn meal agar, pseudohyphae are not formed. Sexual structures are not observed on YM, PDA, V8 and CM agar. Ballistoconidia are ellipsoidal or reniform, 1.9–3.2 × 5.1–7.1 μm (Fig. 16H).
Physiological and biochemical characteristics: Glucose fermentation is absent. Glucose, galactose (variable), L-sorbose (variable), sucrose, maltose, cellobiose (delayed), trehalose, raffinose (delayed), melezitose, inulin, D-ribose (variable), ethanol (variable), glycerol (variable), ribitol (variable), D-mannitol (variable), D-glucitol, Methyl-α-D-glucoside, salicin (variable), DL-lactate (variable) and succinate (variable) are assimilated as sole carbon sources. Lactose, melibiose, soluble starch, D-xylose, L-arabinose, D-arabinose, L-rhamnose, D-glucosamine, N-Acetyl-D-glucosamine, methanol, erythritol, galactitol, D-gluconate, citrate, myo-inositol and hexadecane are not assimilated. Ammonium sulfate, potassium nitrate (variable), sodium nitrite (variable), L-lysine, ethylamine hydrochloride (variable) and cadaverine dihydrochloride (variable) are assimilated as sole nitrogen sources. Maximum growth temperature is 26–27 °C. Growth in vitamin-free medium is positive. Starch-like substances are not produced. Growth on 50 % (w/w) glucose-yeast extract agar is negative. Urease activity is positive. Diazonium Blue B reaction is positive.
Physiologically, Sp. cellobiolyticus differs from its closely related species Sp. jilinensis in its inability to assimilate soluble starch and D-xylose and its ability to assimilate cellobiose and inulin (Table S1.29).
Typus: China, Wuyiling natural reserve, Heilongjiang province, obtained from a leaf of an unidentified plant, Sep. 2014, Q.-M. Wang (holotype CGMCC 2.5675T preserved in a metabolically inactive state, ex-type CBS 13964 = HLJ33B4).
Sporobolomyces reniformis Q.M. Wang, F.Y. Bai & A.H. Li sp. nov. MycoBank MB828836. Fig. 16I, J.
Etymology: the specific epithet reniformis refers to the reniform ballistoconidia.
Culture characteristics: In YM broth, after 7 d at 17 °C, cells are ellipsoidal to ovoid, 3.8–5.7 × 5.8–10.7 μm and single, budding is polar (Fig. 16I), a sediment is formed. On YM agar, after 1 mo at 17 °C, the streak culture is orange red, butyrous, smooth and glistening. The margin is entire. In Dalmau plate culture on corn meal agar, pseudohyphae are not formed. Sexual structures are not observed on YM, PDA, V8 and CM agar. Ballistoconidia are ellipsoidal or reniform, 2.8–3.8 × 7.5–10.0 μm (Fig. 16J).
Physiological and biochemical characteristics: Glucose fermentation is absent. Glucose, L-sorbose, sucrose, maltose, trehalose, raffinose, ethanol (delayed and weak) and DL-lactate are assimilated as sole carbon sources. Galactose, cellobiose, lactose, melibiose, melezitose, inulin, soluble starch, D-xylose, L-arabinose, D-arabinose, D-ribose, L-rhamnose, D-glucosamine, N-Acetyl-D-glucosamine, methanol, glycerol, erythritol, ribitol, galactitol, D-mannitol, D-glucitol, Methyl-α-D-glucoside, salicin, D-gluconate, succinate, citrate, myo-inositol and hexadecane are not assimilated. Ammonium sulfate, and cadaverine dihydrochloride (delayed and weak) are assimilated as sole nitrogen sources. Potassium nitrate, sodium nitrite, L-lysine and ethylamine hydrochloride are not assimilated. Maximum growth temperature is 30 °C. Growth in vitamin-free medium is positive. Starch-like substances are not produced. Growth on 50 % (w/w) glucose-yeast extract agar is negative. Urease activity is positive. Diazonium Blue B reaction is positive.
Physiologically, Sp. reniformis differs from its closely related species Sp. ellipsoideus in its inability to assimilate melezitose, D-mannitol and D-glucitol (Table S1.29).
Typus: China, Milin county, Tibet, obtained from a leaf of an unidentified plant, Sep. 2015, Q.-M. Wang (holotype CGMCC 2.5627T preserved in a metabolically inactive state, ex-type CBS 15562 = GPS21.2C2).
Sporobolomyces ellipsoideus Q.M. Wang, F.Y. Bai & A.H. Li sp. nov. MycoBank MB828837. Fig. 16K, L.
Etymology: the specific epithet ellipsoideus refers to the ellipsoidal cell morphology.
Culture characteristics: In YM broth, after 7 d at 17 °C, cells are cylindrical and ellipsoidal, 2.1–2.9 × 3.6–8.8 μm and single, budding is polar (Fig. 16K), a sediment is formed. After 1 mo at 17 °C, a ring and sediment are present. On YM agar, after 1 mo at 17 °C, the streak culture is orange, butyrous, smooth and glossy. The margin is entire. In Dalmau plate culture on corn meal agar, pseudohyphae are not formed. Sexual structures are not observed on YM, PDA, V8 and CM agar. Ballistoconidia are ellipsoidal, allantoid or reniform, 1.2–2.5 × 5.0–7.1 μm (Fig. 16L).
Physiological and biochemical characteristics: Glucose fermentation is absent. Glucose, galactose (variable), L-sorbose (variable), sucrose, maltose, cellobiose (variable), trehalose, lactose (variable), raffinose (weak), melezitose, inulin (variable), soluble starch (variable), D-ribose (variable), L-arabinose (variable), D-arabinose (variable), L-rhamnose (variable), D-glucosamine (variable), ethanol (variable), glycerol (variable), ribitol (variable), D-mannitol, D-glucitol, Methyl-α-D-glucoside (variable), DL-lactate (variable), succinate (variable), citrate (variable) and salicin (variable) are assimilated as sole carbon sources. Melibiose, D-xylose, N-Acetyl-D-glucosamine, D-gluconate, methanol, erythritol, galactitol, myo-inositol and hexadecane are not assimilated. Ammonium sulfate, potassium nitrate, Sodium nitrite (variable), L-lysine, ethylamine hydrochloride (variable) and cadaverine dihydrochloride are assimilated as sole nitrogen sources. Maximum growth temperature is 26–27 °C. Growth in vitamin-free medium is positive. Starch-like substances are not produced. Growth on 50 % (w/w) glucose-yeast extract agar is negative. Urease activity is positive. Diazonium Blue B reaction is positive.
Physiologically, Sp. ellipsoideus differs from its closely related species Sp. reniformis in its ability to assimilate melezitose, D-mannitol and D-glucitol (Table S1.29).
Typus: China, Milin county, Tibet, obtained from a leaf of an unidentified plant, Sep. 2015, Q.-M. Wang (holotype CGMCC 2.5619T preserved in a metabolically inactive state, ex-type CBS 15590 = GPS21.5C1).
Sporobolomyces primogenomicus Q.M. Wang & F.Y. Bai sp. nov. MycoBank MB828838. Fig. 16M, N.
Etymology: the specific epithet primogenomicus refers to the fact that the type strain was the first sequenced genome in the Pucciniomycotina.
Culture characteristics: In YM broth, after 7 d at 17 °C, cells are ellipsoidal, 2.0–3.8 × 3.0–5.6 μm and single, budding is polar (Fig. 16M), a sediment is formed. After 1 mo at 17 °C, a ring and sediment are present. On YM agar, after 1 mo at 17 °C, the streak culture is red, butyrous, shiny. The margin is entire. In Dalmau plate culture on corn meal agar, pseudohyphae are not formed. Sexual structures are not observed on YM, PDA, V8 and CM agar. Ballistoconidia are allantoid or reniform, 2.0–2.7 × 3.3–5.8 μm (Fig. 16N).
Physiological and biochemical characteristics: Glucose, galactose, L-sorbose, sucrose, maltose, cellobiose, trehalose, raffinose, melezitose, soluble starch, D-xylose, L-arabinose, D-arabinose, D-ribose, glycerol, ribitol, D-mannitol, D-glucitol, Methyl-α-D-glucoside, salicin, DL-lactate, succinate and citrate (delayed) are assimilated as sole carbon sources. Lactose, melibiose, inulin, L-rhamnose, erythritol, galactitol and myo-inositol are not assimilated. Potassium nitrate (weak) is assimilated as sole nitrogen sources.
Physiologically, Sp. primogenomicus differs from its closely related species Sp. ruberrimus in its ability to assimilate L-sorbose, soluble starch, D-xylose, L-arabinose, D-arabinose, D-ribose, ribitol, D-glucitol, Methyl-α-D-glucoside and DL-lactate (Table S1.29). The data of carbon assimilation were collected from Yamazaki & Komagata (1983).
Typus: Japan, Kanto region, obtained from a willow leaf, 1983, M. Yoshizawa (holotype JCM 8242T preserved in a metabolically inactive state, ex-type CBS 15935 = IAM13481).
Acception of Sporobolomyces shibatanus in the genus Sporobolomyces
Sporobolomyces shibatanus (Okun.) Verona & Cif., Atti Ist. Bot. R. Univ. Pavia, 3. Ser. 10: 246. 1939.
Synonym: Sporidiobolus pararoseus Fell & Tallman, Curr. Microbiol. 5: 80. 1981.
Note: Sporobolomyces shibatanus was omitted from the list of accepted species of Sporobolomyces in our previous study (Wang et al. 2015b), deleted by accident in the final version. Sporobolomyces shibatanus is the anamorph of Sporidiobolus pararoseus (Sampaio 2011c), but the former was published earlier than the latter. Thus, Sporidiobolus pararoseus should be considered as a synonym of Sporobolomyces shibatanus at present.
Rhodosporidiobolus platycladi Q.M. Wang, F.Y. Bai & A.H. Li sp. nov. MycoBank MB828839. Fig. 16O, P.
Etymology: the specific epithet platycladi refers to Platycladus, the plant genus from which the type strain was isolated.
Culture characteristics: In YM broth, after 7 d at 17 °C, cells are ellipsoidal and ovoid, 4.0–6.2 × 5.5–9.7 μm and single, budding is polar (Fig. 16O), a sediment is present. After 1 mo at 17 °C, a ring and sediment are present. On YM agar, after 1 mo at 17 °C, the streak culture is pale cream, butyrous, smooth and glistening. The margin is entire. In Dalmau plate culture on corn meal agar, pseudohyphae are not formed. Sexual structures are not observed on YM, PDA, V8 and CM agar. Ballistoconidia are ellipsoidal or reniform, 3.1–5.0 × 7.0–10.0 μm (Fig. 16P).
Physiological and biochemical characteristics: Glucose fermentation is absent. Glucose, L-sorbose (weak), sucrose, maltose, cellobiose, trehalose, raffinose, melezitose, D-xylose, L-arabinose, glycerol, ribitol, D-mannitol, D-glucitol, Methyl-α-D-glucoside (weak) and salicin (weak) are assimilated as sole carbon sources. Galactose, lactose, melibiose, inulin, soluble starch, D-arabinose, D-ribose, L-rhamnose, D-glucosamine, N-Acetyl-D-glucosamine, methanol, ethanol, erythritol, galactitol, DL-lactate, succinate, citrate, myo-inositol and hexadecane are not assimilated. Ammonium sulfate, potassium nitrate, sodium nitrite (weak), L-lysine, ethylamine hydrochloride and cadaverine dihydrochloride (weak) are assimilated as sole nitrogen sources. Maximum growth temperature is 32 °C. Growth in vitamin-free medium is positive. Starch-like substances are not produced. Growth on 50 % (w/w) glucose-yeast extract agar is negative. Urease activity is positive. Diazonium Blue B reaction is positive.
Physiologically, Rh. platycladi differs from its closely related species Rh. nylandii in its inability to assimilate soluble starch, D-arabinose, D-ribose, ethanol and succinate and its ability to assimilate D-xylose, L-arabinose and L-lysine (Table S1.30).
Typus: China, Beijing, obtained from a leaf of Platycladus orientalis, Mar. 2006, S.-A. Wang (holotype CGMCC 2.3118T preserved in a metabolically inactive state, ex-type CBS 15469 = BJ6-3).
Rhodosporidiobolus jianfalingensis Q.M. Wang, F.Y. Bai & A.H. Li sp. nov. MycoBank MB828840. Fig. 18A, B.
Fig. 18.
Vegetative cells grown in YM broth for 5 d at 17 °C and ballistoconidia produced on corn meal agar after 7 d at 17 °C. (A, B) Rh. jianfalingensis CGMCC 2.3532T; (C, D) Rh. fuzhouensis CGMCC 2.4435T; (E) He. tridentata CGMCC 2.5602T; (F) He. cylindrica CGMCC 2.5650T; (G) Mic. swertiae CGMCC 2.3533T; (H) Ya. terricola CGMCC 2.5820T; (I) O. nepetae CGMCC 2.5824T; (J) O. dicranopteridis CGMCC 2.3441T; (K, L) Ch. pseudogriseoflava CGMCC 2.5629T; (M, N) Ch. sambuci CGMCC 2.2618T; (O) Ch. iridis CGMCC 2.2769T; (P) Ch. rhododendri CGMCC 2.5821T. Bars = 10 μm.
Etymology: the specific epithet jianfalingensis refers to the geographic origin of the type strain, Jianfaling, Hainan.
Culture characteristics: In YM broth, after 7 d at 17 °C, cells are cylindrical, 1.4–2.9 × 4.3–10.0 μm and single, budding is polar (Fig. 18A), a sediment is present. After 1 mo at 17 °C, a sediment is present. On YM agar, after 1 mo at 17 °C, the streak culture is pale cream, butyrous, slightly wrinkled and glossy. The margin is entire or eroded. In Dalmau plate culture on corn meal agar, pseudohyphae are not formed. Sexual structures are not observed on YM, PDA, V8 and CM agar. Ballistoconidia are allantoid or reniform, 2.1–2.9 × 5.0–7.1 μm (Fig. 18B).
Physiological and biochemical characteristics: Glucose fermentation is absent. Glucose, galactose, sucrose, maltose, cellobiose, trehalose, lactose (weak), melibiose (weak), raffinose, melezitose, soluble starch, D-xylose, L-arabinose, D-arabinose (weak), D-ribose (weak), L-rhamnose, D-glucosamine (weak), Methyl-α-D-glucoside, salicin, succinate (weak) and citrate (weak) are assimilated as sole carbon sources. L-sorbose, inulin, methanol, ethanol, glycerol, erythritol, ribitol, galactitol, D-mannitol, D-glucitol, DL-lactate, myo-inositol and hexadecane are not assimilated. Ammonium sulfate, potassium nitrate, sodium nitrite and L-lysine are assimilated as sole nitrogen sources. Ethylamine hydrochloride and cadaverine dihydrochloride are not assimilated. Maximum growth temperature is 25 °C. Growth in vitamin-free medium is positive. Starch-like substances are not produced. Growth on 50 % (w/w) glucose-yeast extract agar is negative. Urease activity is positive. Diazonium Blue B reaction is positive.
Physiologically, Rh. jianfalingensis differs from its closely related four species, Rh. platycladi, Rh. nylandii, Rh. odoratus and Rh. ruineniae, in its inability to assimilate L-sorbose, glycerol, ribitol, D-mannitol and D-glucitol (Table S1.30).
Typus: China, Jianfaling, Hainan province, obtained from a leaf of an unidentified plant, Nov. 2006, Q.-M. Wang (holotype CGMCC 2.3532T preserved in a metabolically inactive state, ex-type CBS 15494 = JF25.7-1).
Rhodosporidiobolus fuzhouensis Q.M. Wang, F.Y. Bai & A.H. Li sp. nov. MycoBank MB828841. Fig. 18C, D.
Etymology: the specific epithet fuzhouensis refers to the geographic origin of the type strain, Fuzhou county, Fujian.
Culture characteristics: In YM broth, after 7 d at 17 °C, cells are ellipsoidal and cylindrical, 3.2–5.0 × 6.1–11.0 μm and single, budding is polar (Fig. 18C), a sediment is formed. After 1 mo at 17 °C, a pellicle and a sediment are present. On YM agar, after 1 mo at 17 °C, the streak culture is pink orange, butyrous, smooth and glossy. The margin is entire. In Dalmau plate culture on corn meal agar, pseudohyphae are not formed. Sexual structures are not observed on YM, PDA, V8 and CM agar. Ballistoconidia are allantoid or reniform, 2.8–4.9 × 6.0–9.2 μm (Fig. 18D).
Physiological and biochemical characteristics: Glucose fermentation is absent. Glucose, galactose (delayed), L-sorbose, trehalose, D-xylose (variable), D-ribose (variable), ethanol, ribitol (variable), D-mannitol (variable), D-glucitol, salicin (variable) and succinate (variable) are assimilated as sole carbon sources. Sucrose, maltose, cellobiose, lactose, melibiose, raffinose, melezitose, inulin, soluble starch, L-arabinose, D-arabinose, L-rhamnose, D-glucosamine, methanol, glycerol, erythritol, galactitol, Methyl-α-D-glucoside, gluconate, DL-lactate, citrate, myo-inositol and hexadecane are not assimilated. Ammonium sulfate and L-lysine (delayed and weak) are assimilated as sole nitrogen sources. Potassium nitrate, sodium nitrite, ethylamine hydrochloride and cadaverine dihydrochloride are not assimilated. Maximum growth temperature is 26–27 °C. Growth in vitamin-free medium is positive. Starch-like substances are not produced. Growth on 50 % (w/w) glucose-yeast extract agar is negative. Urease activity is positive. Diazonium Blue B reaction is positive.
Physiologically, Rh. fuzhouensis differs from its closely related species Rh. lusitaniae in its inability to assimilate galactitol, citrate, potassium nitrate and sodium nitrite (Table S1.30).
Typus: China, Jinghong, Yunnan province, obtained from a leaf of an unidentified plant, Aug. 2011, Q.-M. Wang (holotype CGMCC 2.4435T preserved in a metabolically inactive state, ex-type CBS 12492 = FJYZ2-6).
Heitmaniales Q.M. Wang & F.Y. Bai ord. nov. MycoBank MB828842.
Member of the Microbotryomycetes. The diagnosis of the order Heitmaniales is based on the the genus Heitmania. The nomenclature of the order is based on the genus Heitmania.
Type family: Heitmaniaceae Q.M. Wang & F.Y. Bai.
Heitmaniaceae Q.M. Wang & F.Y. Bai fam. nov. MycoBank MB828843.
Member of the Microbotryomycetes. The diagnosis of the family Heitmaniaceae is based on the the genus Heitmania. The nomenclature of the family is based on the genus Heitmania.
Type genus: Heitmania X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, Index Fungorum 381: 1 (2018).
Genus accepted: Heitmania X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, Index Fungorum 381: 1 (2018).
Synonyms: Heitmania X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, Int. J. Syst. Evol. Microbiol. 67: 4538 (2017), nom. inval., Art. 40.1 (Shenzhen).
Heitmania tridentata Q.M. Wang, F.Y. Bai & A.H. Li sp. nov. MycoBank MB828844. Fig. 17, Fig. 18E.
Etymology: the specific epithet tridentata refers to the vegetative cell morphology of the type strain.
Culture characteristics: In YM broth, after 7 d at 17 °C, cells are cylindrical and ellipsoidal, 2.6–3.4 × 5.9–12.0 μm and single, budding is polar, usually tridentate (Fig. 18E), a sediment is formed. After 1 mo at 17 °C, a part ring and sediment are present. On YM agar, after 1 mo at 17 °C, the streak culture is cream, butyrous, smooth and glossy. The margin is entire. In Dalmau plate culture on corn meal agar, pseudohyphae are not formed. Sexual structures are not observed on YM, PDA, V8 and CM agar. Ballistoconidia are not produced.
Physiological and biochemical characteristics: Glucose fermentation is absent. Glucose, maltose, trehalose, ethanol and D-mannitol (weak) are assimilated as sole carbon sources. Galactose, L-sorbose, sucrose, cellobiose, lactose, melibiose, raffinose, melezitose, inulin, soluble starch, D-xylose, L-arabinose, D-arabinose, D-ribose, L-rhamnose, D-glucosamine, N-Acetyl-D-glucosamine, methanol, glycerol, erythritol, ribitol, galactitol, D-glucitol, Methyl-α-D-glucoside, salicin, DL-lactate, succinate, citrate, myo-inositol and hexadecane are not assimilated. Ammonium sulfate, potassium nitrate, L-lysine and cadaverine dihydrochloride (weak) are assimilated as sole nitrogen sources. Sodium nitrite and ethylamine hydrochloride are not assimilated. Maximum growth temperature is 22–23 °C. Growth in vitamin-free medium is negative. Starch-like substances are not produced. Growth on 50 % (w/w) glucose-yeast extract agar is negative. Urease activity is positive. Diazonium Blue B reaction is positive.
Physiologically, He. tridentata differs from its closely related species He. cylindrica in its inability to assimilate melezitose, glycerol, D-glucitol, succinate and ethylamine (Table S1.31).
Typus: China, Milin county, Tibet, obtained from a leaf of an unidentified plant, Sep. 2015, Q.-M. Wang (holotype CGMCC 2.5602T preserved in a metabolically inactive state, ex-type CBS 15549 = GPS20.16B3).
Heitmania cylindrica Q.M. Wang, F.Y. Bai & A.H. Li sp. nov. MycoBank MB828845. Fig. 17, Fig. 18F.
Etymology: the specific epithet cylindrica refers to the vegetative cell morphology of the type strain.
Culture characteristics: In YM broth, after 7 d at 17 °C, cells are elongate cylindrical, 2.5–3.4 × 9.9–16.3 μm and single, budding is polar (Fig. 18F), a sediment is formed. After 1 mo at 17 °C, a ring and sediment are present. On YM agar, after 1 mo at 17 °C, the streak culture is cream, butyrous, smooth and glossy. The margin is entire. In Dalmau plate culture on corn meal agar, pseudohyphae are not formed. Sexual structures are not observed on YM, PDA, V8 and CM agar. Ballistoconidia are not produced.
Physiological and biochemical characteristics: Glucose fermentation is absent. Glucose, sucrose (delayed and weak), maltose (delayed), trehalose (delayed), melezitose (delayed), ethanol, glycerol, D-mannitol, D-glucitol and succinate are assimilated as sole carbon sources. Galactose, L-sorbose, cellobiose, lactose, melibiose, raffinose, inulin, soluble starch, D-xylose, L-arabinose, D-arabinose, D-ribose, L-rhamnose, D-glucosamine, N-Acetyl-D-glucosamine, methanol, erythritol, ribitol, galactitol, Methyl-α-D-glucoside, salicin, DL-lactate, citrate, myo-inositol and hexadecane are not assimilated. Ammonium sulfate, potassium nitrate (delayed and weak), L-lysine and ethylamine hydrochloride are assimilated as sole nitrogen sources. Sodium nitrite and cadaverine dihydrochloride are not assimilated. Maximum growth temperature is 22–23 °C. Growth in vitamin-free medium is negative. Starch-like substances are not produced. Growth on 50 % (w/w) glucose-yeast extract agar is negative. Urease activity is positive. Diazonium Blue B reaction is positive.
Physiologically, He. cylindrica differs from its closely related species He. tridentata in its ability to assimilate melezitose, glycerol, D-glucitol, succinate and ethylamine (Table S1.31).
Typus: China, Milin county, Tibet, obtained from a leaf of an unidentified plant, Sep. 2015, Q.-M. Wang (holotype CGMCC 2.5650T preserved in a metabolically inactive state, ex-type CBS 15568 = GPS20.2C8).
Microbotryozyma swertiae Q.M. Wang, F.Y. Bai & A.H. Li sp. nov. MycoBank MB828846. Fig. 18G.
Etymology: the specific epithet swertiae refers to Swertia, the plant genus from which the type strain was isolated.
Culture characteristics: In YM broth, after 7 d at 17 °C, cells are cylindrical and lunate, 1.7–2.5 × 3.9–5.6 μm and single, budding is polar (Fig. 18G), a sediment is present. On YM agar, after 1 mo at 17 °C, the streak culture is cream, butyrous, smooth and glossy. The margin is entire. In Dalmau plate culture on corn meal agar, pseudohyphae are not formed. Sexual structures are not observed on YM, PDA, V8 and CM agar. Ballistoconidia are not produced.
Physiological and biochemical characteristics: Glucose fermentation is absent. Glucose, L-sorbose (delayed), sucrose, maltose, cellobiose, trehalose, lactose, melezitose, D-xylose, D-ribose (delayed and weak), glycerol, ribitol (delayed and weak), D-mannitol, D-glucitol, Methyl-α-D-glucoside, salicin and succinate (delayed and weak) are assimilated as sole carbon sources. Galactose, melibiose, raffinose, inulin, soluble starch, L-arabinose, D-arabinose, L-rhamnose, D-glucosamine, methanol, ethanol, erythritol, galactitol, D-gluconate, DL-lactate, citrate, myo-inositol and hexadecane are not assimilated. Ammonium sulfate, potassium nitrate and ethylamine hydrochloride are assimilatedas sole nitrogen sources. Sodium nitrite, L-lysine and cadaverine dihydrochloride are not assimilated. Maximum growth temperature is 26–27 °C. Growth in vitamin-free medium is negative. Starch-like substances are not produced. Growth on 50 % (w/w) glucose-yeast extract agar is negative. Urease activity is positive. Diazonium Blue B reaction is positive.
Physiologically, Mic. swertiae differs from its closely related species Mic. collariae in its inability to assimilate D-gluconate, DL-lactate and sodium nitrite (Table S1.32).
Typus: China, Chuxiong county, Yunnan province, obtained from a leaf of Swertia yunnanensis, Nov. 2006, Q.-M. Wang (holotype CGMCC 2.3533T preserved in a metabolically inactive state, ex-type CBS 15495 = ZXS7.7).
Yamadamyces terricola Q.M. Wang, F.Y. Bai & A.H. Li sp. nov. MycoBank MB828847. Fig. 17, Fig. 18H.
Etymology: the specific epithet terricola refers to the substrate from which the type strain was isolated, soil.
Culture characteristics: In YM broth, after 7 d at 17 °C, cells are fusiform, 2.5–3.4 × 6.0–11.8 μm and single, budding is polar (Fig. 18H), a sediment is formed. After 1 mo at 17 °C, a ring and a sediment are present. On YM agar, after 1 mo at 17 °C, the streak culture is cream, butyrous, smooth and glossy. The margin is entire. In Dalmau plate culture on corn meal agar, pseudohyphae are not formed. Sexual structures are not observed on YM, PDA, V8 and CM agar. Ballistoconidia are not produced.
Physiological and biochemical characteristics: Glucose fermentation is absent. Glucose, sucrose, maltose, trehalose, melezitose, D-glucosamine (delayed and weak), ethanol, glycerol (weak), ribitol, D-mannitol, D-glucitol and succinate (delayed and weak) are assimilated as sole carbon sources. Galactose, L-sorbose, cellobiose, lactose, melibiose, raffinose, inulin, soluble starch, D-xylose, L-arabinose, D-arabinose, D-ribose, L-rhamnose, N-Acetyl-D-glucosamine, methanol, erythritol, galactitol, Methyl-α-D-glucoside, salicin, DL-lactate, citrate, myo-inositol and hexadecane are not assimilated. Ammonium sulfate, L-lysine (weak), ethylamine hydrochloride and cadaverine dihydrochloride are assimilated as sole nitrogen sources. Potassium nitrate and sodium nitrite are not assimilated. Maximum growth temperature is 26–27 °C. Growth in vitamin-free medium is postive. Starch-like substances are not produced. Growth on 50 % (w/w) glucose-yeast extract agar is negative. Urease activity is positive. Diazonium Blue B reaction is positive.
Physiologically, Ya. terricola differs from its closely related species Ya. rosulatus in its inability to assimilate cellobiose, L-rhamnose, N-Acetyl-D-glucosamine, salicin, D-gluconate, DL-lactate, citrate, myo-inositol, potassium nitrate and sodium nitrite and its ability to growin vitamin-free medium (Table S1.32).
Typus: China, Daxinganling, obtained from soil, Aug. 2015, Q.-M. Wang (holotype CGMCC 2.5820T preserved in a metabolically inactive state, ex-type CBS 15572 = 03-1).
Note: The genus Yamadamyces was invalidly published because its type species was based on an invalid name (Art. 40.1, ICN Shenzhen Code), thus it was validated in the Validated Taxa section.
Oberwinklerozyma nepetae Q.M. Wang, F.Y. Bai & A.H. Li sp. nov. MycoBank MB828848. Fig. 18I.
Etymology: the specific epithet nepetae refers to Nepeta, the plant genus from which the type strain was isolated.
Culture characteristics: In YM broth, after 7 d at 17 °C, cells are cylindrical, ellipsoidal and ovoid, 2.7–3.2 × 6.4–8.9 μm and single, budding is polar (Fig. 18I), a sediment is formed. After 1 mo at 17 °C, a ring and sediment are present. On YM agar, after 1 mo at 17 °C, the streak culture is white cream, butyrous, smooth and glistening. The margin is entire. In Dalmau plate culture on corn meal agar, pseudohyphae are not formed. Sexual structures are not observed on YM, PDA, V8 and CM agar. Ballistoconidia are not produced.
Physiological and biochemical characteristics: Glucose fermentation is absent. Glucose, L-sorbose, sucrose, maltose, cellobiose, trehalose, melibiose, raffinose, melezitose, D-mannitol, D-glucitol, Methyl-α-D-glucoside and salicin are assimilated as sole carbon sources. Galactose, lactose, inulin, soluble starch, D-xylose, L-arabinose, D-arabinose, D-ribose, L-rhamnose, D-glucosamine, N-Acetyl-D-glucosamine, methanol, ethanol, glycerol, erythritol, ribitol, galactitol, DL-lactate, succinate, citrate, myo-inositol and hexadecane are not assimilated. Ammonium sulfate, potassium nitrate, sodium nitrite, L-lysine, ethylamine hydrochloride and cadaverine dihydrochloride are assimilated as sole nitrogen sources. Maximum growth temperature is 26–27 °C. Growth in vitamin-free medium is negative. Starch-like substances are not produced. Growth on 50 % (w/w) glucose-yeast extract agar is negative. Urease activity is positive. Diazonium Blue B reaction is positive.
Physiologically, O. nepetae differs from its closely related species O. yarrowii and O. silvestris in its inability to assimilate galactose, D-glucosamine, ethanol, glycerol, DL-lactate, succinate, citrate and myo-inositol (Table S1.33).
Typus: China, Motuo, Tibet, obtained from a leaf of Nepeta sp., Sep. 2014, Q.-M. Wang (holotype CGMCC 2.5824T preserved in a metabolically inactive state, ex-type CBS 15579 = XZ129C7).
Oberwinklerozyma dicranopteridis Q.M. Wang, F.Y. Bai & A.H. Li sp. nov. MycoBank MB828849. Fig. 18J.
Etymology: the specific epithet dicranopteridis refers to Dicranopteris, the plant genus from which the type strain was isolated.
Culture characteristics: In YM broth, after 7 d at 17 °C, cells are ellipsoidal and cylindrical, 2.2–3.7 × 6.4–10.5 μm and single, budding is polar (Fig. 18J), a sediment is formed. After 1 mo at 17 °C, a ring and a sediment are present. On YM agar, after 1 mo at 17 °C, the streak culture is cream, butyrous, smooth and glossy. The margin is entire. In Dalmau plate culture on corn meal agar, pseudohyphae are not formed. Sexual structures are not observed on YM, PDA, V8 and CM agar. Ballistoconidia are not produced.
Physiological and biochemical characteristics: Glucose fermentation is absent. Glucose, galactose, L-sorbose, sucrose, maltose, cellobiose, trehalose, lactose, raffinose, melezitose, D-xylose, L-arabinose (delayed and weak), D-arabinose, D-glucosamine, ethanol, glycerol, ribitol, galactitol, D-mannitol, D-glucitol, Methyl-α-D-glucoside, salicin, DL-lactate (delayed and weak), succinate, citrate (delayed and weak) and myo-inositol are assimilated as sole carbon sources. Melibiose, inulin, soluble starch, D-ribose, L-rhamnose, methanol, erythritol and hexadecane are not assimilated. Ammonium sulfate, potassium nitrate, sodium nitrite, L-lysine, ethylamine hydrochloride and cadaverine dihydrochlorideare assimilated as sole nitrogen sources. Maximum growth temperature is 26–27 °C. Growth in vitamin-free medium is positive. Starch-like substances are not produced. Growth on 50 % (w/w) glucose-yeast extract agar is negative. Urease activity is positive. Diazonium Blue B reaction is positive.
Physiologically, O. dicranopteridis differs from its closely related species O. straminea in its ability to assimilate cellobiose, lactose, D-arabinose, galactitol, Methyl-α-D-glucoside and salicin (Table S1.33).
Typus: China, Simao county, Yunnan province, obtained from a leaf of Dicranopteris dichotoma, Nov. 2006, Q.-M. Wang (holotype CGMCC 2.3441T preserved in a metabolically inactive state, ex-type CBS 15476 = SM10.2).
Chrysozyma pseudogriseoflava Q.M. Wang, F.Y. Bai & A.H. Li sp. nov. MycoBank MB828850. Fig. 18K, L.
Etymology: the specific epithet pseudogriseoflava refers to the similar colony morphology to that of Chrysozyma griseoflava.
Culture characteristics: In YM broth, after 7 d at 17 °C, cells are cylindrical, ellipsoidal to fusiform, 3.3–4.7 × 6.9–9.7 μm and single, budding is polar (Fig. 18K), a sediment is formed. After 1 mo at 17 °C, a ring and sediment are present. On YM agar, after 1 mo at 17 °C, the streak culture is yellowish-cream, butyrous, smooth, dull and partly wrinkled. The margin is entire. In Dalmau plate culture on corn meal agar, pseudohyphae are not formed. Sexual structures are not observed on YM, PDA, V8 and CM agar. Ballistoconidia are allantoid or cylindrical, 2.3–3.1 × 4.6–7.7 μm (Fig. 18L).
Physiological and biochemical characteristics: Glucose fermentation is absent. Glucose, sucrose, maltose, cellobiose, trehalose, raffinose, melezitose, ethanol and DL-lactate are assimilated as sole carbon sources. Galactose, L-sorbose, lactose, melibiose, inulin, soluble starch, D-xylose, L-arabinose, D-arabinose, D-ribose, L-rhamnose, D-glucosamine, N-Acetyl-D-glucosamine, methanol, glycerol, erythritol, ribitol, galactitol, D-mannitol, D-glucitol, Methyl-α-D-glucoside, salicin, succinate, citrate, myo-inositol and hexadecane are not assimilated. Ammonium sulfate, potassium nitrate, sodium nitrite, L-lysine, ethylamine hydrochloride and cadaverine dihydrochloride (weak) are assimilated as sole nitrogen sources. Maximum growth temperature is 22–23 °C. Growth in vitamin-free medium is negative. Starch-like substances are not produced. Growth on 50 % (w/w) glucose-yeast extract agar is negative. Urease activity is positive. Diazonium Blue B reaction is positive.
Physiologically, Ch. pseudogriseoflava differs from its closely related species Ch. griseoflava in its inability to assimilate galactose, soluble starch, D-xylose, D-arabinose, glycerol, ribitol, D-glucitol, salicin and citrate and its ability to assimilate raffinose, DL-lactate and L-lysine (Table S1.34).
Typus: China, Milin county, Tibet, obtained from a leaf of an unidentified plant, Sep. 2015, Q.-M. Wang (holotype CGMCC 2.5629T preserved in a metabolically inactive state, ex-type CBS 15564 = GPS21.6B3).
Chrysozyma sambuci Q.M. Wang, F.Y. Bai & A.H. Li sp. nov. MycoBank MB828851. Fig. 18M, N.
Etymology: the specific epithet sambuci refers to Sambucus, the plant genus from which the type strain was isolated.
Culture characteristics: In YM broth, after 7 d at 17 °C, cells are long ellipsoidal and cylindrical, 2.4–4.0 × 7.2–13.5 μm and single, budding is polar (Fig. 18M), a sediment is present. After 1 mo at 17 °C, a ring and sediment are present. On YM agar, after 1 mo at 17 °C, the streak culture is cream, butyrous, smooth and dull. The margin is entire. In Dalmau plate culture on corn meal agar, pseudohyphae are not formed. Sexual structures are not observed on YM, PDA, V8 and CM agar. Ballistoconidia are allantoid or reniform, 2.2–2.9 × 5.9–8.8 μm (Fig. 18N).
Physiological and biochemical characteristics: Glucose fermentation is absent. Glucose, sucrose, maltose, cellobiose, trehalose, melezitose, inulin (variable), soluble starch (variable), D-arabinose (variable), ethanol (delayed and weak), D-mannitol (variable), D-glucitol (variable) and salicin (variable) are assimilated as sole carbon sources. Galactose, L-sorbose, lactose, melibiose, raffinose, D-xylose, L-arabinose, D-ribose, L-rhamnose, D-glucosamine, N-Acetyl-D-glucosamine, methanol, glycerol, erythritol, ribitol, galactitol, Methyl-α-D-glucoside, DL-lactate, succinate, citrate, myo-inositol and hexadecane are not assimilated. Ammonium sulfate, potassium nitrate, L-lysine, ethylamine hydrochloride (weak) and cadaverine dihydrochloride (weak) are assimilated as sole nitrogen sources. Sodium nitrite is not assimilated. Maximum growth temperature is 23–24 °C. Growth in vitamin-free medium is negative. Starch-like substances are not produced. Growth on 50 % (w/w) glucose-yeast extract agar is negative. Urease activity is positive. Diazonium Blue B reaction is positive.
Physiologically, Ch. sambuci and its closely related species Ch. milinensis and Ch. griseoflava can be distinguished from one another by the assimilation of galactose, raffinose, D-xylose, glycerol, DL-lactate, citrate, sodium nitrite and L-lysine (Table S1.34).
Typus: China, Bomi county, Tibet, obtained from a leaf of Sambucus williamsii, Sep. 2004, F.-Y. Bai (holotype CGMCC 2.2618T preserved in a metabolically inactive state, ex-type CBS 15450 = XZ13C5).
Chrysozyma iridis Q.M. Wang, F.Y. Bai & A.H. Li sp. nov. MycoBank MB828852. Fig. 18O.
Etymology: the specific epithet iridis refers to Iris, the plant genus from which the type strain was isolated.
Culture characteristics: In YM broth, after 7 d at 17 °C, cells are cylindrical, 2.8–3.2 × 7.2–10.3 μm and single, budding is polar (Fig. 18O), a sediment is present. After 1 mo at 17 °C, a ring and sediment are present. On YM agar, after 1 mo at 17 °C, the streak culture is cream, butyrous, smooth and glossy. The margin is entire. In Dalmau plate culture on corn meal agar, pseudohyphae are not formed. Sexual structures are not observed on YM, PDA, V8 and CM agar. Ballistoconidia are not produced.
Physiological and biochemical characteristics: Glucose fermentation is absent. Glucose, galactose (weak), sucrose, maltose, cellobiose, trehalose, melezitose, inulin (weak), D-glucitol (delayed and weak), D-mannitol and salicin (delayed) are assimilated as sole carbon sources. L-sorbose, lactose, melibiose, raffinose, soluble starch, D-xylose, L-arabinose, D-arabinose, D-ribose, L-rhamnose, D-glucosamine, methanol, ethanol, glycerol, erythritol, ribitol, galactitol, Methyl-α-D-glucoside, DL-lactate, succinate, citrate, myo-inositol and hexadecane are not assimilated. Ammonium sulfate, potassium nitrate and ethylamine hydrochloride are assimilated as sole nitrogen sources. Sodium nitrite, L-lysine and cadaverine dihydrochloride are not assimilated. Maximum growth temperature is 28 °C. Growth in vitamin-free medium is positive. Starch-like substances are not produced. Growth on 50 % (w/w) glucose-yeast extract agar is negative. Urease activity is positive. Diazonium Blue B reaction is positive.
Physiologically, Ch. iridis differs from its closely related species Ch. rhododendri in its inability to assimilate raffinose, D-xylose, L-arabinose, ethanol and Methyl-α-D-glucoside (Table S1.34).
Typus: China, Bomi county, Tibet, obtained from a leaf of Iris forrestii, Sep. 2004, F.-Y. Bai (holotype CGMCC 2.2769T preserved in a metabolically inactive state, ex-type CBS 15461 = XZ8B3).
Chrysozyma rhododendri Q.M. Wang, F.Y. Bai & A.H. Li sp. nov. MycoBank MB828853. Fig. 17, Fig. 18P.
Etymology: the specific epithet rhododendri refers to Rhododendron, the plant genus from which the type strain was isolated.
Culture characteristics: In YM broth, after 7 d at 17 °C, cells are cylindrical to long ellipsoidal, 1.9–3.7 × 7.5–12.5 μm and single, budding is polar (Fig. 18P), a sediment is present. After 1 mo at 17 °C, a ring and sediment are present. On YM agar, after 1 mo at 17 °C, the streak culture is cream, mucoid, smooth and glossy. The margin is entire. In Dalmau plate culture on corn meal agar, pseudohyphae are not formed. Sexual structures are not observed on YM, PDA, V8 and CM agar. Ballistoconidia are not produced.
Physiological and biochemical characteristics: Glucose fermentation is absent. Glucose, sucrose, maltose, cellobiose, trehalose, raffinose, melezitose, D-xylose, L-arabinose, ethanol, D-mannitol, D-glucitol, Methyl-α-D-glucoside and salicin are assimilated as sole carbon sources. Galactose, L-sorbose, lactose, melibiose, inulin, soluble starch, D-arabinose, D-ribose, L-rhamnose, D-glucosamine, N-Acetyl-D-glucosamine, methanol, glycerol, erythritol, ribitol, galactitol, DL-lactate, succinate, citrate, myo-inositol and hexadecane are not assimilated. Ammonium sulfate, potassium nitrate, sodium nitrite, L-lysine, ethylamine hydrochloride and cadaverine dihydrochloride are assimilated as sole nitrogen sources. Maximum growth temperature is 22–23 °C. Growth in vitamin-free medium is positive. Starch-like substances are not produced. Growth on 50 % (w/w) glucose-yeast extract agar is negative. Urease activity is positive. Diazonium Blue B reaction is positive.
Physiologically, Ch. rhododendri differs from its closely related species Ch. iridis in its ability to assimilate raffinose, D-xylose, L-arabinose, ethanol and Methyl-α-D-glucoside (Table S1.34).
Typus: China, Tibet, obtained from a leaf of Rhododendron sp., Sep. 2014, Q.-M. Wang (holotype CGMCC 2.5821T preserved in a metabolically inactive state, ex-type CBS 15583 = XZ160D3).
Chrysozyma fusiformis Q.M. Wang, F.Y. Bai & A.H. Li sp. nov. MycoBank MB828854. Fig. 19A, B.
Fig. 19.
Vegetative cells grown in YM broth for 5 d at 17 °C and ballistoconidia produced on corn meal agar after 7 d at 17 °C. (A, B) Ch. fusiformis CGMCC 2.2765T; (C, D) Ch. sorbariae CGMCC 2.2768T; (E, F) Ch. cylindrica CGMCC 2.3455T; (G) Ch. flava CGMCC 2.5611T; (H) Pseu. hydrangea CGMCC 2.2796T; (I, J) Pseu. lulangensis CGMCC 2.2612T; (K, L) Yu. longicylindrica CGMCC 2.5603T; (M) Sl. globosa CGMCC 2.5822T; (n) Co. aletridis CGMCC 2.2766T; (O) Co. hydrangeae CGMCC 2.2798T; (P) Co. rhododendri CGMCC 2.2770T. Bars = 10 μm.
Etymology: the specific epithet fusiformis refers to the fusiform vegetative cells of the type strain.
Culture characteristics: In YM broth, after 7 d at 17 °C, cells are ellipsoidal to fusiform, 3.0–4.6 × 4.7–8.2 μm and single, budding is polar (Fig. 19A), a sediment is present. After 1 mo at 17 °C, a ring and sediment are present. On YM agar, after 1 mo at 17 °C, the streak culture is cream, butyrous, smooth and dull surface. The margin is entire. In Dalmau plate culture on corn meal agar, pseudohyphae are not formed. Sexual structures are not observed on YM, PDA, V8 and CM agar. Ballistoconidia are ellipsoidal orallantoid, 2.9–4.3 × 7.1–11.4 μm (Fig. 19B).
Physiological and biochemical characteristics: Glucose fermentation is absent. Glucose, sucrose, maltose, cellobiose, trehalose, melibiose, melezitose, ethanol, D-mannitol and succinate (delayed and weak) are assimilated as sole carbon sources. Galactose, L-sorbose, lactose, raffinose, inulin, soluble starch, D-xylose, L-arabinose, D-arabinose, D-ribose, L-rhamnose, D-glucosamine, methanol, glycerol, erythritol, ribitol, galactitol, D-glucitol, Methyl-α-D-glucoside, salicin, DL-lactate, citrate, myo-inositol and hexadecane are not assimilated. Ammonium sulfate, potassium nitrate, sodium nitrite, L-lysine, ethylamine hydrochloride and cadaverine dihydrochloride are assimilated as sole nitrogen sources. Maximum growth temperature is 24 °C. Growth in vitamin-free medium is negative. Starch-like substances are not produced. Growth on 50 % (w/w) glucose-yeast extract agar is negative. Urease activity is positive. Diazonium Blue B reaction is positive.
Physiologically, Ch. fusiformis differs well from other Chrysozyma species in its assimilation of carbon and nitrogen sources (Table S1.34).
Typus: China, Lulang county, Tibet, obtained from a leaf of an unidentified plant, Sep. 2004, F.-Y. Bai (holotype CGMCC 2.2765T preserved in a metabolically inactive state, ex-type CBS 15458 = XZ33C2).
Chrysozyma sorbariae Q.M. Wang, F.Y. Bai & A.H. Li sp. nov. MycoBank MB828855. Fig. 19C, D.
Etymology: the specific epithet sorbariae refers to Sorbaria, the plant genus from which the type strain was isolated.
Culture characteristics: In YM broth, after 7 d at 17 °C, cells are elongate ellipsoidal and cylindrical, 1.7–2.7 × 5.8–10.4 μm and single, budding is polar (Fig. 19C), a sediment is present. On YM agar, after 1 mo at 17 °C, the streak culture is cream, butyrous, smooth and semi-gloosy. The margin is entire. In Dalmau plate culture on corn meal agar, pseudohyphae are not formed. Sexual structures are not observed on YM, PDA, V8 and CM agar. Ballistoconidia are allantoid or falcate, 2.1–2.9 × 6.4–7.9 μm (Fig. 19D).
Physiological and biochemical characteristics: Glucose fermentation is absent. Glucose, sucrose, maltose, trehalose, melezitose, inulin (delayed and weak), D-mannitol and D-glucitol are assimilated as sole carbon sources. Galactose, L-sorbose, cellobiose, lactose, melibiose, raffinose, soluble starch, D-xylose, L-arabinose, D-arabinose, D-ribose, L-rhamnose, D-glucosamine, methanol, ethanol, glycerol, erythritol, ribitol, galactitol, Methyl-α-D-glucoside, salicin, DL-lactate, succinate, citrate, myo-inositol and hexadecane are not assimilated. Ammonium sulfate, potassium nitrate (weak) and sodium nitrite are assimilated as sole nitrogen sources. L-lysine, ethylamine hydrochloride and cadaverine dihydrochloride are not assimilated. Maximum growth temperature is 26–27 ΊC. Growth in vitamin-free medium is variable. Starch-like substances are not produced. Growth on 50 % (w/w) glucose-yeast extract agar is negative. Urease activity is positive. Diazonium Blue B reaction is positive.
Physiologically, Ch. sorbariae differs well from other Chrysozyma species in its assimilation of carbon and nitrogen sources (Table S1.34).
Typus: China, Bomi county, Tibet, obtained from a leaf of Sorbaria arboricola, Sep. 2004, F.-Y. Bai (holotype CGMCC 2.2768T preserved in a metabolically inactive state, ex-type CBS 15460 = XZ9D1).
Chrysozyma cylindrica Q.M. Wang, F.Y. Bai & A.H. Li sp. nov. MycoBank MB828856. Fig. 19E, F.
Etymology: the specific epithet cylindrica refers to the cylindrical vegetative cells of the type strain.
Culture characteristics: In YM broth, after 7 d at 17 °C, cells are cylindrical, 2.2–3.2 × 3.9–10.0 μm and single, budding is polar (Fig. 19E), a sediment is formed. After 1 mo at 17 °C, a ring and a sediment are present. On YM agar, after 1 mo at 17 °C, the streak culture is yellowish-cream, butyrous, smooth and semi-glossy. The margin is entire. In Dalmau plate culture on corn meal agar, pseudohyphae are formed. Sexual structures are not observed on YM, PDA, V8 and CM agar. Ballistoconidia are allantoid or reniform, 1.5–2.5 × 3.8–6.3 μm (Fig. 19F).
Physiological and biochemical characteristics: Glucose fermentation is absent. Glucose, galactose (delay and weak), sucrose, trehalose (delay), melezitose, D-mannitol and D-glucitol are assimilated as sole carbon sources. L-sorbose, maltose, cellobiose, lactose, melibiose, raffinose, inulin, soluble starch, D-xylose, L-arabinose, D-arabinose, D-ribose, L-rhamnose, D-glucosamine, N-Acetyl-D-glucosamine, methanol, ethanol, glycerol, erythritol, ribitol, galactitol, Methyl-α-D-glucoside, salicin, DL-lactate, succinate, citrate, myo-inositol and hexadecane are not assimilated. Ammonium sulfate, potassium nitrate (weak), ethylamine hydrochloride and cadaverine dihydrochloride are assimilated as sole nitrogen sources. Sodium nitrite and L-lysine are not assimilated. Maximum growth temperature is 22–23 °C. Growth in vitamin-free medium is positive. Starch-like substances are not produced. Growth on 50 % (w/w) glucose-yeast extract agar is negative. Urease activity is positive. Diazonium Blue B reaction is positive.
Physiologically, Ch. cylindrica differs well from other Chrysozyma species in its assimilation of carbon and nitrogen sources (Table S1.34).
Typus: China, Wuzhishan mountain, Hainan province, obtained from a leaf of an unidentified plant, Nov. 2006, Q.-M. Wang (holotype CGMCC 2.3455T preserved in a metabolically inactive state, ex-type CBS 15482 = WZS29.2).
Chrysozyma flava Q.M. Wang, F.Y. Bai & A.H. Li sp. nov. MycoBank MB828857. Fig. 19G.
Etymology: the specific epithet flava refers to the yellow colony colour of the type strain.
Culture characteristics: In YM broth, after 7 d at 17 °C, cells are ellipsoidal or to cylindrical, 2.1–3.1 × 4.0–10.8 μm and single, budding is polar (Fig. 19G), a sediment is formed. After 1 mo at 17 °C, a ring and a sediment are present. On YM agar, after 1 mo at 17 °C, the streak culture is yellow, butyrous, smooth and glossy. The margin is entire. In Dalmau plate culture on corn meal agar, pseudohyphae are not formed. Sexual structures are not observed on YM, PDA, V8 and CM agar. Ballistoconidia are not produced.
Physiological and biochemical characteristics: Glucose fermentation is absent. Glucose, sucrose, maltose, cellobiose, trehalose, melezitose, ethanol, D-mannitol and D-gluconate (weak) are assimilated as sole carbon sources. Galactose, L-sorbose, lactose, melibiose, raffinose, inulin, soluble starch, D-xylose, L-arabinose, D-arabinose, D-ribose, L-rhamnose, D-glucosamine, N-Acetyl-D-glucosamine, D-glucitol, Methyl-α-D-glucoside methanol, glycerol, erythritol, ribitol, galactitol, salicin, DL-lactate, succinate, citrate, myo-inositol and hexadecane are not assimilated. Ammonium sulfate is assimilated as sole nitrogen sources. Potassium nitrate, sodium nitrite, L-lysine, ethylamine hydrochloride and cadaverine dihydrochloride are not assimilated. Maximum growth temperature is 26–27 °C. Growth in vitamin-free medium is negative. Starch-like substances are not produced. Growth on 50 % (w/w) glucose-yeast extract agar is negative. Urease activity is positive. Diazonium Blue B reaction is positive.
Physiologically, Ch. flava differs well from other Chrysozyma species in its assimilation of carbon and nitrogen sources (Table S1.34).
Typus: China, Milin county, Tibet, obtained from a leaf of an unidentified plant, Sep. 2015, Q.-M. Wang (holotype CGMCC 2.5611T preserved in a metabolically inactive state, ex-type CBS 15552 = GPS20.4A1).
Pseudohyphozyma hydrangeae Q.M. Wang, F.Y. Bai & A.H. Li sp. nov. MycoBank MB828858. Fig. 19H.
Etymology: the specific epithet hydrangeae refers to Hydrangea, the plant genus from which the type strain was isolated.
Culture characteristics: In YM broth, after 7 d at 17 °C, cells are cylindrical and ellipsoidal, 3.0–4.3 × 5.8–9.1 μm and single, budding is polar (Fig. 19H), a sediment is present. After 1 mo at 17 °C, a ring and a sediment are present. On YM agar, after 1 mo at 17 °C, the streak culture is cream, butyrous, smooth and glossy. The margin is entire. In Dalmau plate culture on corn meal agar, pseudohyphae are not formed. Sexual structures are not observed on YM, PDA, V8 and CM agar. Ballistoconidia are not produced.
Physiological and biochemical characteristics: Glucose fermentation is absent. Glucose, maltose, cellobiose, trehalose, melezitose, inulin (variable), soluble starch (variable), D-xylose (variable), L-arabinose (variable), D-arabinose (variable), ethanol, ribitol, D-mannitol, D-glucitol and succinate (variable) are assimilated as sole carbon sources. Galactose, L-sorbose, sucrose, lactose, melibiose, raffinose, D-ribose, L-rhamnose, D-glucosamine, N-Acetyl-D-glucosamine, methanol, glycerol, erythritol, galactitol, Methyl-α-D-glucoside, salicin, DL-lactate, citrate, myo-inositol and hexadecane are not assimilated. Ammonium sulfate, potassium nitrate, L-lysine, ethylamine hydrochloride and cadaverine dihydrochloride are assimilated as sole nitrogen sources. Sodium nitrite is not assimilated. Maximum growth temperature is 29 °C. Growth in vitamin-free medium is variable. Starch-like substances are not produced. Growth on 50 % (w/w) glucose-yeast extract agar is negative. Urease activity is positive. Diazonium Blue B reaction is positive.
Physiologically, Ps. hydrangeae and its four closely related species, Ps. lulangensis, Ps. bogoriensis, Ps. pustula and Ps. buffonii, can be distinguished from one another by the assimilation of galactose, L-sorbose, melezitose, glycerol, salicin, citrate, potassium nitrate and sodium nitrite (Table S1.35).
Typus: China, Lulang county, Tibet, obtained from a leaf of Hydrangea heteromalla, Sep. 2004, F.-Y. Bai (holotype CGMCC 2.2796T preserved in a metabolically inactive state, ex-type CBS 15462 = XZ46A1).
Pseudohyphozyma lulangensis Q.M. Wang, F.Y. Bai & A.H. Li sp. nov. MycoBank MB828859. Fig. 19I, J.
Etymology: the specific epithet lulangensis refers to the geographic origin of the type strain, Lulang county, Tibet.
Culture characteristics: In YM broth, after 7 d at 17 °C, cells are cylindrical, 3.0–4.0 × 8.4–11.1 μm and single, budding is polar (Fig. 19I), a sediment is present. After 1 mo at 17 °C, a ring and a sediment are present. On YM agar, after 1 mo at 17 °C, the streak culture is white cream, butyrous, smooth and glossy. The margin is entire. In Dalmau plate culture on corn meal agar, pseudohyphae are not formed. Sexual structures are not observed on YM, PDA, V8 and CM agar. Ballistoconidia are allantoid or reniform, 1.9–2.7 × 5.1–8.3 μm (Fig. 19J).
Physiological and biochemical characteristics: Glucose fermentation is absent. Glucose, maltose (weak), cellobiose, trehalose, D-xylose (delayed), L-arabinose, D-arabinose (delayed and weak), D-ribose (delayed), D-glucosamine, ethanol, ribitol (delayed), D-mannitol, D-glucitol (delayed) and salicin are assimilated as sole carbon sources. Galactose, L-sorbose, sucrose, lactose, melibiose, raffinose, melezitose, inulin, soluble starch, L-rhamnose, methanol, glycerol, erythritol, galactitol, Methyl-α-D-glucoside, DL-lactate, succinate, citrate, myo-inositol and hexadecane are not assimilated. Ammonium sulfate, L-lysine, ethylamine hydrochloride and cadaverine dihydrochloride are assimilated as sole nitrogen sources. Potassium nitrate and sodium nitrite are not assimilated. Maximum growth temperature is 23 °C. Growth in vitamin-free medium is positive. Starch-like substances are not produced. Growth on 50 % (w/w) glucose-yeast extract agar is negative. Urease activity is positive. Diazonium Blue B reaction is positive.
Physiologically, Ps. lulangensis differs from its closely related species Ps. bogoriensis in its inability to assimilate galactose, L-sorbose, soluble starch, glycerol and succinate and its ability to grow in vitamin-free medium (Table S1.35).
Typus: China, Lulang county, Tibet, obtained from a leaf of an unidentified plant, Sep. 2004, F.-Y. Bai (holotype CGMCC 2.2612T preserved in a metabolically inactive state, ex-type CBS 15446 = XZ50B2).
Yurkovia longicylindrica Q.M. Wang, F.Y. Bai & A.H. Li sp. nov. MycoBank MB828860. Fig. 19K, L.
Etymology: the specific epithet longicylindrica refers to the elongate cylindrical cells of the type strain.
Culture characteristics: In YM broth, after 7 d at 17 °C, cells are elongate cylindrical, 2.5–4.5 × 7.5–15.9 μm and single, budding is polar (Fig. 19K), a sediment is present. After 1 mo at 17 °C, a ring and a sediment are present. On YM agar, after 1 mo at 17 °C, the streak culture is cream, butyrous, wrinkled and dull. The margin is entire. In Dalmau plate culture on corn meal agar, pseudohyphae are not formed. Sexual structures are not observed on YM, PDA, V8 and CM agar. Ballistoconidia are allantoid, falcate or cylindrical, 1.4–2.6 × 7.1–12.9 μm (Fig. 19L).
Physiological and biochemical characteristics: Glucose fermentation is absent. Glucose, galactose, sucrose, trehalose, melibiose, melezitose, inulin, soluble starch (delayed and weak), D-arabinose, ethanol, ribitol, D-mannitol and D-glucitol are assimilated as sole carbon sources. L-sorbose, maltose, cellobiose, lactose, raffinose, D-xylose, L-arabinose, D-ribose, L-rhamnose, D-glucosamine, N-Acetyl-D-glucosamine, methanol, glycerol, erythritol, galactitol, Methyl-α-D-glucoside, salicin, DL-lactate, succinate, citrate, myo-inositol and hexadecane are not assimilated. Ammonium sulfate, potassium nitrate (weak), sodium nitrite, L-lysine, ethylamine hydrochloride and cadaverine dihydrochloride are assimilated as sole nitrogen sources. Maximum growth temperature is 22–23 °C. Growth in vitamin-free medium is positive. Starch-like substances are not produced. Growth on 50 % (w/w) glucose-yeast extract agar is negative. Urease activity is positive. Diazonium Blue B reaction is positive.
Physiologically, Yu. longicylindrica differs from its closely related species Yu. mendeliana and Yu. nerthusi in its inability to assimilate L-sorbose, maltose, L-arabinose, glycerol and succinate and its ability to assimilate melibiose (Table S1.36).
Typus: China, Milin county, Tibet, obtained from a leaf of an unidentified plant, Sep. 2015, Q.-M. Wang (holotype CGMCC 2.5603T preserved in a metabolically inactive state, ex-type CBS 15550 = GPS20.2C3).
Slooffia globosa Q.M. Wang, F.Y. Bai & A.H. Li sp. nov. MycoBank MB828861. Fig. 19M.
Etymology: the specific epithet globosa refers to the globosal vegetative cells of the type strain.
Culture characteristics: In YM broth, after 7 d at 17 °C, cells are globosal, 4.1–5.9 × 4.8–5.9 μm and single, budding is polar (Fig. 19M), a sediment is present. After 1 mo at 17 °C, a ring and a sediment are present. On YM agar, after 1 mo at 17 °C, the streak culture is yellowish cream, butyrous, slightly wrinkled and glossy. The margin is entire. In Dalmau plate culture on corn meal agar, pseudohyphae are not formed. Sexual structures are not observed on YM, PDA, V8 and CM agar. Ballistoconidia are not produced.
Physiological and biochemical characteristics: Glucose fermentation is absent. Glucose, sucrose, maltose, cellobiose, trehalose, lactose (weak), melezitose (delayed and weak), ethanol, glycerol, D-mannitol, Methyl-α-D-glucoside (delayed and weak) and D-gluconate (weak) are assimilated as sole carbon sources. Galactose, L-sorbose, melibiose, raffinose, inulin, soluble starch, D-xylose, L-arabinose, D-arabinose, D-ribose, L-rhamnose, D-glucosamine, N-Acetyl-D-glucosamine, methanol, erythritol, ribitol, galactitol, D-glucitol, salicin, succinate, DL-lactate, citrate, myo-inositol and hexadecane are not assimilated. Ammonium sulfate, potassium nitrate, L-lysine (weak), ethylamine hydrochloride and cadaverine dihydrochlorideare (delayed and weak) are assimilated as sole nitrogen sources. Sodium nitrite is not assimilated. Maximum growth temperature is 26–27 °C. Growth in vitamin-free medium is negative. Starch-like substances are not produced. Growth on 50 % (w/w) glucose-yeast extract agar is negative. Urease activity is positive. Diazonium Blue B reaction is positive.
Physiologically, Sl. globosa differs from its closely related species Sl. tsugae in its inability to assimilate L-sorbose, D-xylose, D-glucitol, salicin, DL-lactate, succinate, citrate and sodium nitrite (Table S1.37).
Typus: China, Daxinganling, obtained from soil, Aug. 2015, Q.-M. Wang (holotype CGMCC 2.5822T preserved in a metabolically inactive state, ex-type CBS 15573 = 4–6).
Colacogloea aletridis Q.M. Wang, F.Y. Bai & A.H. Li sp. nov. MycoBank MB828862. Fig. 19N.
Etymology: the specific epithet aletridis refers to Aletris, the plant genus from which the type strain was isolated.
Culture characteristics: In YM broth, after 7 d at 17 °C, cells are ellipsoidal and ovoid, 2.0–3.8 × 3.0–7.6 μm and single, budding is polar (Fig. 19N), a sediment is formed. After 1 mo at 17 °C, a ring and a sediment are present. On YM agar, after 1 mo at 17 °C, the streak culture is cream, butyrous, smooth and glistening. The margin is entire. In Dalmau plate culture on corn meal agar, pseudohyphae are not formed. Sexual structures are not observed on YM, PDA, V8 and CM agar. Ballistoconidia are not produced.
Physiological and biochemical characteristics: Glucose fermentation is absent. Glucose, sucrose, maltose (weak), trehalose, melezitose, ethanol, ribitol (delayed), D-mannitol (weak), D-glucitol (weak) and Methyl-α-D-glucoside (weak) are assimilated as sole carbon sources. Galactose, L-sorbose, cellobiose, lactose, melibiose, raffinose, inulin, soluble starch, D-xylose, L-arabinose, D-arabinose, D-ribose, L-rhamnose, D-glucosamine, methanol, glycerol, erythritol, galactitol, salicin, DL-lactate, succinate, citrate, myo-inositol and hexadecane are not assimilated. Ammonium sulfate, potassium nitrate, sodium nitrite, L-lysine, ethylamine hydrochloride and cadaverine dihydrochloride are assimilated as sole nitrogen sources. Maximum growth temperature is 30 ΊC. Growth in vitamin-free medium is positive. Starch-like substances are not produced. Growth on 50 % (w/w) glucose-yeast extract agar is negative. Urease activity is positive. Diazonium Blue B reaction is positive.
Physiologically, Co. aletridis differ well from other Colacogloe species in its assimilation of carbon and nitrogen sources (Table S1.38).
Typus: China, Bomi county, Tibet, obtained from a leaf of Aletris pauciflora, Sep. 2004, F.-Y. Bai (holotype CGMCC 2.2766T preserved in a metabolically inactive state, ex-type CBS 15459 = XZ31A1).
Colacogloea hydrangeae Q.M. Wang, F.Y. Bai & A.H. Li sp. nov. MycoBank MB828863. Fig. 19O.
Etymology: the specific epithet hydrangeae refers to Hydrangea, the plant genus from which the type strain was isolated.
Culture characteristics: In YM broth, after 7 d at 17 °C, cells are cylindrical and ellipsoidal, 2.7–4.1 × 5.7–10.9 μm and single, budding is polar (Fig. 19O), a sediment is present. On YM agar, after 1 mo at 17 °C, the streak culture is yellowish cream, butyrous, smooth with partly wrinkled, glossy. The margin is entire. In Dalmau plate culture on corn meal agar, pseudohyphae are not formed. Sexual structures are not observed on YM, PDA, V8 and CM agar. Ballistoconidia are not produced.
Physiological and biochemical characteristics: Glucose fermentation is absent. Glucose, sucrose, maltose, trehalose, melezitose (delayed), D-glucosamine, ethanol, ribitol (delayed), D-mannitol, D-glucitol and salicin are assimilated as sole carbon sources. Galactose, L-sorbose, cellobiose, lactose, raffinose, melibiose, inulin, soluble starch, D-xylose, L-arabinose, D-arabinose, D-ribose, L-rhamnose, methanol, glycerol, erythritol, galactitol, Methyl-α-D-glucoside, DL-lactate, succinate, citrate, myo-inositol and hexadecane are not assimilated. Ammonium sulfate, potassium nitrate, sodium nitrite, L-lysine and ethylamine hydrochloride are assimilated as sole nitrogen sources. Cadaverine dihydrochlorideare is not assimilated. Maximum growth temperature is 28 °C. Growth in vitamin-free medium is weak. Starch-like substances are not produced. Growth on 50 % (w/w) glucose-yeast extract agar is negative. Urease activity is positive. Diazonium Blue B reaction is positive.
Physiologically, Co. hydrangeae differs from its closely related species Co. rhododendri in its inability to assimilate glycerol and its ability to assimilate salicin (Table S1.38).
Typus: China, Lulang county, Tibet, obtained from a leaf of Hydrangea heteromalla, Sep. 2004, Q.-M. Wang (holotype CGMCC 2.2798T preserved in a metabolically inactive state, ex-type CBS 15463 = XZ46B3).
Colacogloea rhododendri Q.M. Wang, F.Y. Bai & A.H. Li sp. nov. MycoBank MB828864. Fig. 19P.
Etymology: the specific epithet rhododendri refers to Rhododendron, the plant genus from which the type strain was isolated.
Culture characteristics: In YM broth, after 7 d at 17 °C, cells are cylindrical, 1.0–3.8 × 4.3–15.0 μm and single, budding is polar (Fig. 19P), a sediment is present. On YM agar, after 1 mo at 17 °C, the streak culture is prey-cream, butyrous, wrinkled and dull. The margin is entire or eroded. In Dalmau plate culture on corn meal agar, pseudohyphae are not formed. Sexual structures are not observed on YM, PDA, V8 and CM agar. Ballistoconidia are not produced.
Physiological and biochemical characteristics: Glucose fermentation is absent. Glucose, galactose (variable), sucrose, maltose, cellobiose (variable), trehalose, melezitose, inulin (variable), D-glucosamine (weak), N-Acetyl-D-glucosamine (weak), ethanol, glycerol (delayed), ribitol (variable), D-mannitol, D-glucitol (variable) and D-gluconate are assimilated as sole carbon sources. L-sorbose, lactose, melibiose, raffinose, soluble starch, D-xylose, L-arabinose, D-arabinose, D-ribose, L-rhamnose, methanol, erythritol, galactitol, Methyl-α-D-glucoside, salicin, DL-lactate, succinate, citrate, myo-inositol and hexadecane are not assimilated. Ammonium sulfate, potassium nitrate (weak), sodium nitrite (variable), L-lysine, ethylamine hydrochloride and cadaverine dihydrochlorideare (variable) are assimilated as sole nitrogen sources. Maximum growth temperature is 26–27 °C. Growth in vitamin-free medium is positive. Starch-like substances are not produced. Growth on 50 % (w/w) glucose-yeast extract agar is negative. Urease activity is positive. Diazonium Blue B reaction is positive.
Physiologically, Co. rhododendri differs from its closely related species Co. hydrangeae in its inability to assimilate salicin and its ability to assimilate glycerol (Table S1.38).
Typus: China, Bomi county, Tibet, obtained from a leaf of Rhododendron lulangense, Sep. 2004, F.-Y. Bai (holotype CGMCC 2.2770T preserved in a metabolically inactive state, ex-type CBS 15652 = XZ10F1).
New combination for Colacogloea
Colacogloea subericola (Belloch, Villa-Carv., Álv.-Rodríg. & Coque) Q.M. Wang & F.Y. Bai com. nov. MycoBank MB832093.
Basionym: Rhodotorula subericola Belloch, Villa-Carv., Álv.-Rodríg. & Coque, Int. J. Syst. Evol. Microbiol. 57(7): 1670 (2007).
Validated Taxa
Usually the type culture of a new yeast species should be conserved in two or more collections when it was described. Thus, two or more collection numbers of type culture were always listed for new species by many yeast taxonomists but often without explicitely indicating the holotype, which, however, resulted in numerous invalidly described species according the Art. 40.7 of the Shenzhen Code (Turland et al. 2018) during the last ten years. In order to avoid this embarrassing situation, 70 invalidly described taxa were validated here.
Apiotrichum xylopini S.O. Suh, C.F. Lee, Gujjari & J.J. Zhou ex Kachalkin, Yurkov & Boekhout, sp. nov. MycoBank MB831708.
For description see Int. J. Syst. Evol. Microbiol. 61(10): 2540 (2011).
Holotype: CBS 11841 (preserved in a metabolically inactive state).
Synonyms: Trichosporon xylopini S.O. Suh, C.F. Lee, Gujjari & J.J. Zhou, Int. J. Syst. Evol. Microbiol. 61(10): 2540 (2011), nom. inval., Art. 40.7 (Shenzhen).
= Apiotrichum xylopini S.O. Suh, C.F. Lee, Gujjari & J.J. Zhou ex Kachalkin, Yurkov & Boekhout, Stud. Mycol. 81: 142 (2015), nom. inval., Art. 40.7 (Shenzhen).
Bannozyma arctica Vishniac & M. Takash. ex Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, sp. nov. MycoBank MB831713.
For description see Int. J. Syst. Evol. Microbiol. 60(5): 1217 (2010).
Holotype: CBS 9278 (preserved in a metabolically inactive state).
Synonyms: Rhodotorula arctica Vishniac & M. Takash., Int. J. Syst. Evol. Microbiol. 60(5): 1217 (2010), nom. inval., Art. 40.7 (Shenzhen).
= Bannozyma arctica Vishniac & M. Takash. ex Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, Stud. Mycol. 81: 183 (2015), nom. inval., Art. 40.7 (Shenzhen).
Bulleribasidium panici Fungsin, M. Takash. & Nakase ex Xin Zhan Liu, F.Y. Bai, M. Groenew. & Boekhout, sp. nov. MycoBank MB831675.
For description see Microbiol. Culture Coll. 19(1): 27 (2003).
Holotype: JCM 11819 (preserved in a metabolically inactive state).
Synonyms: Bullera panici Fungsin et al., Microbiol. Culture Coll. 19(1): 27 (2003), nom. inval., Art. 40.7 (Shenzhen).
= Bulleribasidium panici Fungsin, M. Takash. & Nakase ex Xin Zhan Liu, F.Y. Bai, M. Groenew. & Boekhout, Stud. Mycol. 81: 123 (2015), nom. inval., Art. 40.7 (Shenzhen).
Bulleribasidium siamense Fungsin, M. Takash. & Nakase ex Q.M. Wang, F.Y. Bai, Boekhout & Nakase, sp. nov. MycoBank MB831676.
For description see Microbiol. Culture Coll. 19(1): 29 (2003).
Holotype: JCM 11820 (preserved in a metabolically inactive state).
Synonyms: Bullera siamensis Fungsin et al., Microbiol. Culture Coll. 19(1): 29 (2003), nom. inval., Art. 40.6 (Shenzhen).
= Mingxiaea siamensis Fungsin, M. Takash. & Nakase ex Q.M. Wang, F.Y. Bai, Boekhout & Nakase, Int. J. Syst. Evol. Microbiol. 61: 214 (2011), nom. inval., Art. 40.6 (Shenzhen).
= Bulleribasidium siamense Fungsin, M. Takash. & Nakase ex Xin ZhanLiu, F.Y. Bai, M. Groenew. & Boekhout, Stud. Mycol. 81: 123 (2015), Nom. inval., Art. 40.6 (Shenzhen).
Carcinomyces arundinariae Fungsin, M. Takash. & Nakase ex Yurkov, sp. nov. MycoBank MB831698.
For description see Microbiol. Culture Coll. 18(2): 86 (2002).
Holotype: JCM 11818 (preserved in a metabolically inactive state).
Synonyms: Bullera arundinariae Fungsin, M. Takash. & Nakase, in Fungsin et al., Microbiol. Culture Coll. 18(2): 86 (2002), nom. inval., Art. 40.6 (Shenzhen).
= Carcinomyces arundinariae Fungsin, M. Takash. & Nakase ex Yurkov, Stud. Mycol. 81: 133 (2015), nom. inval., Art. 40.6 (Shenzhen).
Cystobasidium alpinum Turchetti, Selbmann, Onofri & Buzzini, sp. nov. MycoBank MB831749.
For description see Life 8 (2, no 9): 10 (2018).
Holotype: CBS 14809 (preserved in a metabolically inactive state).
Synonyms: Cystobasidium alpinum Turchetti, Selbmann, Onofri & Buzzini, Life 8 (2, no 9): 10 (2018), nom. inval., Art. 40.7 (Shenzhen).
Cystobasidium portillonense (Laich, Vaca & R. Chávez) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB831741.
Basionym: Rhodotorula portillonensis Laich, Vaca & R. Chávez, Index Fungorum 361: 1 (2018).
Synonyms: Rhodotorula portillonensis Laich, Vaca & R. Chávez, Int. J. Syst. Evol. Microbiol. 63(10): 3889 (2013), nom. inval., Art. 40.7 (Shenzhen).
= Cystobasidium portillonense Laich, Vaca & R. Chávez ex Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, Stud. Mycol. 81: 173 (2015), nom. inval., Art. 40.7 (Shenzhen).
Derxomyces cylindricus F.Y. Bai, Q.M. Wang & M. Takash. ex F.Y. Bai & Q.M. Wang, sp. nov. MycoBank MB831863.
For description see Int. J. Syst. Evol. Microbiol. 54(5): 1879 (2004).
Holotype: CGMCC AS 2.2308 (preserved in a metabolically inactive state).
Synonyms: Bullera cylindrica F.Y. Bai, Q.M. Wang & M. Takash., Int. J. Syst. Evol. Microbiol. 54(5): 1879 (2004), nom. inval., Art. 40.7 (Shenzhen).
= Derxomyces cylindrica F.Y. Bai, Q.M. Wang & M. Takash. ex F.Y. Bai & Q.M. Wang, FEMS Yeast Res. 8(5): 804 (2008), nom. inval., Art. 40.7 (Shenzhen).
Derxomyces hubeiensis F.Y. Bai, Q.M. Wang & M. Takash. ex F.Y. Bai & Q.M. Wang, sp. nov. MycoBank MB831864.
For description see Int. J. Syst. Evol. Microbiol. 54(5): 1880 (2004).
Holotype: CGMCC AS 2.2466 (preserved in a metabolically inactive state).
Synonyms: Bullera hubeiensis F.Y. Bai, Q.M. Wang & M. Takash., Int. J. Syst. Evol. Microbiol. 54(5): 1880 (2004), nom. inval., Art. 40.7 (Shenzhen).
= Derxomyces hubeiensis F.Y. Bai, Q.M. Wang & M. Takash. ex F.Y. Bai & Q.M. Wang, FEMS Yeast Res. 8(5): 805 (2008), nom. inval., Art. 40.7 (Shenzhen).
Derxomyces nakasei F.Y. Bai, Q.M. Wang & M. Takash. ex F.Y. Bai & Q.M. Wang, sp. nov. MycoBank MB831865.
For description see Int. J. Syst. Evol. Microbiol. 54(5): 1880 (2004).
Holotype: CGMCC AS 2.2435 (preserved in a metabolically inactive state).
Synonyms: Bullera nakasei F.Y. Bai, Q.M. Wang & M. Takash., Int. J. Syst. Evol. Microbiol. 54(5): 1880 (2004), nom. inval., Art. 40.7 (Shenzhen).
= Derxomyces nakasei F.Y. Bai, Q.M. Wang & M. Takash. ex F.Y. Bai & Q.M. Wang, FEMS Yeast Res. 8(5): 805 (2008), nom. inval., Art. 40.7 (Shenzhen).
Dioszegia zsoltii F.Y. Bai, M. Takash. & Nakase, sp. nov. MycoBank MB831868.
For description see J. Gen. Appl. Microbiol., 48(1): 21 (2002).
Holotype: CGMCC AS 2.2089 (preserved in a metabolically inactive state).
Synonyms: Dioszegia zsoltii F.Y. Bai, M. Takash. & Nakase, J. Gen. Appl. Microbiol., 48(1): 21 (2002), nom. inval., Art. 40.7 (Shenzhen).
= Dioszegia yunnanensis F.Y. Bai, M. Takash. & Nakase, J. Gen. Appl. Microbiol., 48(1): 22 (2002), nom. inval., Art. 40.7 (Shenzhen).
Genolevuria bromeliarum Landell & P. Valente ex Xin Zhan Liu, F.Y. Bai, M. Groenew. & Boekhout, sp. nov. MycoBank MB831695.
For description see Int. J. Syst. Evol. Microbiol. 59(4): 911 (2009).
Holotype: CBS 10424 (preserved in a metabolically inactive state).
Synonyms: Cryptococcus bromeliarum Landell & P. Valente, Int. J. Syst. Evol. Microbiol. 59(4): 911 (2009), nom. inval., Art. 40.7 (Shenzhen).
= Genolevuria bromeliarum Landell & P. Valente ex Xin Zhan Liu, F.Y. Bai, M. Groenew. & Boekhout, Stud. Mycol. 81: 129 (2015), nom. inval., Art. 40.7 (Shenzhen).
Glaciozyma Turchetti, Connell, Thomas-Hall & Boekhout ex M. Groenew. & Q.M. Wang, gen. nov. MycoBank MB831869.
For description see Extremophiles 15 (5): 579 (2011).
Type species:Glaciozyma antarctica (Fell, Statzell, I.L. Hunter & Phaff) M. Groenew. & Q.M. Wang.
Synonym: Glaciozyma Turchetti, Connell, Thomas-Hall & Boekhout, Extremophiles 15 (5): 579 (2011), nom. inval., Art. 40.1, see Arts 6.3, 12.1 (Melbourne).
Glaciozyma antarctica (Fell, Statzell, I.L. Hunter & Phaff) M. Groenew. & Q.M. Wang, comb. nov. MycoBank MB831870.
Basionym: Leucosporidium antarcticum Fell, Statzell, I.L. Hunter & Phaff, Antonie van Leeuwenhoek 35 (4): 447 (1970).
Synonym: Glaciozyma antarctica (Fell, Statzell, I.L. Hunter & Phaff) Turchetti, Connell, Thomas-Hall & Boekhout, Extremophiles 15 (5): 579 (2011), nom. inval., Art. 41.5, see Note 1 (Shenzhen).
Glaciozyma martinii Turchetti, Connell, Thomas-Hall & Boekhout, sp. nov. MycoBank MB831872.
For description see Extremophiles 15 (5): 579 (2011).
Holotype: CBS 10620 (preserved in a metabolically inactive state).
Synonym: Glaciozyma martinii Turchetti, Connell, Thomas-Hall & Boekhout, Extremophiles 15 (5): 579 (2011), nom. inval., Arts 35.1, 40.7 (Shenzhen).
Glaciozyma watsonii Turchetti, Connell, Thomas-Hall & Boekhout, sp. nov. MycoBank MB831873.
For description see Extremophiles 15 (5): 582 (2011).
Holotype: CBS 10986 (preserved in a metabolically inactive state).
Synonym: Glaciozyma watsonii Thomas-Hall, Connell, Boekhout & Turchetti, Extremophiles 15 (5): 582 (2011), nom. inval., Arts 35.1, 40.7 (Shenzhen).
Kockovaella mexicana Lopandić, O. Molnár & Prillinger ex Xin Zhan Liu, F.Y. Bai, M. Groenew. & Boekhout, sp. nov. MycoBank MB831697.
For description see Microbiol. Res. 160(1): 8 (2005).
Holotype: CBS 8279 (preserved in a metabolically inactive state).
Synonyms: Fellomyces mexicanus Lopandić et al., Microbiol. Res. 160(1): 8 (2005), nom. inval., Art. 40.7 (Shenzhen).
= Kockovaella mexicana Lopandić, O. Molnár & Prillinger ex Xin Zhan Liu, F.Y. Bai, M. Groenew. & Boekhout, Stud. Mycol. 81: 131 (2015), nom. inval., Art. 40.7 (Shenzhen).
Kondoa thailandica Fungsin, Hamam. & Nakase ex Q.M. Wang, M. Groenew., F.Y. Bai & Boekhout, sp. nov. MycoBank MB831742.
For description see Int. J. Syst. Evol. Microbiol. 51(3): 1210 (2001).
Holotype: JCM 10651 (preserved in a metabolically inactive state).
Synonyms: Bensingtonia thailandica Fungsin, Hamam. & Nakase, Int. J. Syst. Evol. Microbiol. 51(3): 1210 (2001), nom. inval., Art. 40.7 (Shenzhen).
= Kondoa thailandica Fungsin, Hamam. & Nakase ex Q.M. Wang, M. Groenew., F.Y. Bai & Boekhout, Stud. Mycol. 81: 171 (2015), nom. inval., Art. 40.7 (Shenzhen).
Kwoniella newhampshirensis K. Sylvester, Q.M. Wang & Hittinger, sp. nov. MycoBank MB828749.
For description see FEMS Yeast Research 15: 7 (2015).
Holotype: NRRL Y-63731 (preserved in a metabolically inactive state).
Synonyms: Kwoniella newhampshirensis K. Sylvester et al., FEMS Yeast Res. 15: 7 (2015), nom. inval., Art. 40.7 (Shenzhen).
Kwoniella shandongensis R. Chen, Yuan M. Jiang & S.C. Wei ex M. Groenew. & Q.M. Wang, sp. nov. MycoBank MB828750.
For description see Int. J. Syst. Evol. Microbiol. 62: 2775 (2012).
Holotype: CGMCC 2.04458 (preserved in a metabolically inactive state).
Synonyms: Kwoniella shandongensis Chen et al., Int. J. Syst. Evol. Microbiol. 62: 2775 (2012), nom. inval., Art. 40.7 (Shenzhen).
Leucosporidium creatinivorum (Golubev) M. Groenew. & Q.M. Wang, comb. nov. MycoBank MB831751.
Basionym: Rhodotorula creatinivora Golubev, Mikol. Fitopatol. 32(3): 8 (1998), as ‘creatinovora’.
Synonyms: Leucosporidiella creatinivora (Golubev) J.P. Samp., Mycol. Progr. 2(1): 66 (2003).
= Leucosporidium creatinivorum (Golubev) V. de García et al., FEMS Yeast Research 15 (4): 9 (2015), nom. inval., Art. 41.5 (Shenzhen).
Leucosporidium fragarium (J.A. Barnett & Buhagiar) M. Groenew. & Q.M. Wang, comb. nov. MycoBank MB831752.
Basionym: Torulopsis fragaria J.A. Barnett & Buhagiar, J. Gen. Microbiol. 67(2): 237 (1971).
Synonyms: Leucosporidiella fragaria (J.A. Barnett & Buhagiar) J.P. Samp., Mycol. Progr. 2(1): 66 (2003).
= Leucosporidium fragarium (J.A. Barnett & Buhagiar) V. de García et al., FEMS Yeast Res. 15: 9 (2015), nom. inval., Art. 41.5 (Shenzhen).
Leucosporidium intermedium (Nakase & M. Suzuki) M. Groenew. & Q.M. Wang, comb. nov. MycoBank MB831754.
Basionym: Bullera intermedia Nakase & M. Suzuki, J. Gen. Appl. Microbiol. 32(2): 150 (1986).
Synonyms: Sporobolomyces intermedius (Nakase & M. Suzuki) Nakase & M. Suzuki, J. Gen. Appl. Microbiol. 33(2): 193 (1987).
= Bensingtonia intermedia (Nakase & M. Suzuki) Nakase & Boekhout, J. Gen. Appl. Microbiol. 34(3): 435 (1988).
= Leucosporidium intermedium (Nakase & M. Suzuki) V. de García et al., FEMS Yeast Res. 15: 9 (2015), nom. inval., Art. 41.5 (Shenzhen).
Leucosporidium muscorum (Di Menna) M. Groenew. & Q.M. Wang, comb. nov. MycoBank MB831755.
Basionym: Candida muscorum Di Menna, J. Gen. Microbiol. 18: 269 (1958).
Synonyms: Leucosporidiella muscorum (Di Menna) J.P. Samp., Mycol. Progr. 2(1): 66 (2003).
= Leucosporidium muscorum (Di Menna) V. de García et al., FEMS Yeast Res. 15: 9 (2015), nom. inval., Art. 41.5 (Shenzhen).
Leucosporidium yakuticum (Golubev) M. Groenew. & Q.M. Wang, comb. nov. MycoBank MB831756.
Basionym: Rhodotorula yakutica Golubev, Mikol. Fitopatol. 32(3): 9 (1998).
Synonyms: Leucosporidiella yakutica (Golubev) J.P. Samp., Mycol. Progr. 2(1): 66 (2003).
= Leucosporidium yakuticum (Golubev) V. de García et al., FEMS Yeast Res. 15: 9 (2015), nom. inval., Art. 41.5 (Shenzhen).
Naganishia onofrii Turchetti, Selbmann & Zucconi ex Yurkov, sp. nov. MycoBank MB831673.
For description see Extremophiles 19: 157 (2015).
Holotype: CBS 13732 (preserved in a metabolically inactive state).
Synonyms: Cryptococcus onofrii Turchetti et al., Extremophiles 19: 157 (2015), nom. inval., Art. 40.7 (Shenzhen).
= Naganishia onofrii Turchetti, Selbmann & Zucconi ex Yurkov, Stud. Mycol. 81: 119 (2015), nom. inval., Art. 41.5 (Shenzhen).
Naganishia vaughanmartiniae Turchetti, Blanchette & Arenz ex Yurkov, sp. nov. MycoBank MB831674.
For description see Extremophiles 19: 157 (2015).
Holotype: CBS 13731 (preserved in a metabolically inactive state).
Synonyms: Cryptococcus vaughanmartiniae Turchetti et al., Extremophiles 19: 157 (2015), nom. inval., Art. 40.7 (Shenzhen).
= Naganishia vaughanmartiniae Turchetti, Blanchette & Arenz. ex Yurkov, Stud. Mycol. 81: 119 (2015), nom. inval., Art. 40.7 (Shenzhen).
Nielozyma Xin Zhan Liu, F.Y. Bai, M. Groenew. & Boekhout, gen. nov. MycoBank MB831677.
For description see Stud. Mycol. 81: 123 (2015).
Type species: Nielozyma melastomatis Nakase, Tsuzuki, F.L. Lee & M. Takash. ex Xin Zhan Liu, F.Y. Bai, M. Groenew. & Boekhout.
Synonym: Nielozyma Xin Zhan Liu, F.Y. Bai, M. Groenew. & Boekhout, Stud. Mycol. 81: 123 (2015), nom. inval., Art. 40.1 (Shenzhen).
Nielozyma formosana Nakase, Tsuzuki, F.L. Lee & M. Takash. ex Xin Zhan Liu, F.Y. Bai, M. Groenew. & Boekhout, sp. nov. MycoBank MB831678.
For description see Syst. Appl. Microbiol. 27(5): 562 (2004).
Holotype: JCM 12154 (preserved in a metabolically inactive state).
Synonyms: Bullera formosana Nakase et al., Syst. Appl. Microbiol. 27(5): 562 (2004), nom. inval., Art. 40.6 (Shenzhen).
= Nielozyma formosana Nakase, Tsuzuki, F.L. Lee & M. Takash. ex Xin Zhan Liu, F.Y. Bai, M. Groenew. & Boekhout, Stud. Mycol. 81: 123 (2015), nom. inval., Art. 40.6 (Shenzhen).
Nielozyma melastomatis Nakase, Tsuzuki, F.L. Lee & M. Takash. ex Xin Zhan Liu, F.Y. Bai, M. Groenew. & Boekhout, sp. nov. MycoBank MB831679.
For description see Syst. Appl. Microbiol. 27(5): 560 (2004).
Holotype: JCM 12153 (preserved in a metabolically inactive state).
Synonyms: Bullera melastomatis Nakase et al., Syst. Appl. Microbiol. 27(5): 560 (2004), as ‘melastomae’, nom. inval., Art. 40.6 (Shenzhen).
= Nielozyma melastomatis Nakase, Tsuzuki, F.L. Lee & M. Takash. ex Xin Zhan Liu, F.Y. Bai, M. Groenew. & Boekhout, Stud. Mycol. 81: 123 (2015), as ‘melastomae’, nom. inval., Art. 40.6 (Shenzhen).
Oberwinklerozyma silvestris Golubev & Scorzetti ex Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, sp. nov. MycoBank MB831743.
For description see Int. J. Syst. Evol. Microbiol. 60(10): 2504 (2010).
Holotype: CBS 11420 (preserved in a metabolically inactive state).
Synonyms: Rhodotorula silvestris Golubev & Scorzetti, Int. J. Syst. Evol. Microbiol. 60(10): 2504 (2010), nom. inval., Art. 40.7 (Shenzhen).
= Oberwinklerozyma silvestris Golubev & Scorzetti ex Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, Stud. Mycol. 81: 185 (2015), nom. inval., Art. 40.7 (Shenzhen).
Oberwinklerozyma straminea Golubev & Scorzetti ex Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, sp. nov. MycoBank MB831744.
For description see Int. J. Syst. Evol. Microbiol. 60(10): 2505 (2010).
Holotype: CBS 10976 (preserved in a metabolically inactive state).
Synonyms: Rhodotorula straminea Golubev & Scorzetti, Int. J. Syst. Evol. Microbiol. 60(10): 2505 (2010), nom. inval., Art. 40.7 (Shenzhen).
= Oberwinklerozyma straminea Golubev & Scorzetti ex Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, Stud. Mycol. 81: 185 (2015), nom. inval., Art. 40.7 (Shenzhen).
Papiliotrema aspenensis (Ferreira-Paim et al.) Xin Zhan Liu, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB831707.
Basionym: Cryptococcus aspenensis Ferreira-Paim et al., PLoS ONE 9(9): e108633, 10 (2014).
Synonym: Papiliotrema aspenensis (Ferreira-Paim, et al.) Xin Zhan Liu, F.Y. Bai, M. Groenew. & Boekhout, Stud. Mycol. 81: 126 (2015), nom. inval., Art. 41.5 (Shenzhen).
Papiliotrema baii Yurkov, M.A. Guerreiro & Á. Fonseca ex Yurkov, sp. nov. MycoBank MB831705.
For description see PLoS ONE 10(4): e0126996, 15 (2015).
Holotype: PYCC 6352 (preserved in a metabolically inactive state).
Synonyms: Cryptococcus baii Yurkov, M.A. Guerreiro & Á. Fonseca, in Yurkov et al., PLoS ONE 10(4): e0126996, 15 (2015), nom. inval., Art. 40.7 (Shenzhen).
= Papiliotrema baii Yurkov, M.A. Guerreiro & Á. Fonseca ex Yurkov, Stud. Mycol. 81: 126 (2015), nom. inval., Art. 40.7 (Shenzhen).
Papiliotrema frias V. de García, Zalar, Brizzio, Gunde-Cim. & van Broock ex Yurkov, sp. nov. MycoBank MB831685.
For description see FEMS Microbiology Ecology 82(2): 537 (2012).
Holotype: EXF-5992 (preserved in a metabolically inactive state).
Synonyms: Cryptococcus frias V. de García et al., FEMS Microbiol. Ecol. 82(2): 537 (2012), nom. inval., Art. 40.7 (Shenzhen).
= Papiliotrema frias V. de García, Zalar, Brizzio, Gunde-Cim. & van Broock ex Yurkov, Stud. Mycol. 81: 126 (2015), nom. inval., Art. 40.7 (Shenzhen).
Papiliotrema hoabinhensis D.T. Luong, M. Takash., Ty, Dung & Nakase ex Yurkov, sp. nov. MycoBank MB831686.
For description see J. Gen. Appl. Microbiol. 51(6): 340 (2005).
Holotype: JCM 10835 (preserved in a metabolically inactive state).
Synonyms: Bullera hoabinhensis D.T. Luong et al., J. Gen. Appl. Microbiol. 51(6): 340 (2005), nom. inval., Art. 40.7 (Shenzhen).
= Papiliotrema hoabinhensis D.T. Luong, M. Takash., Ty, Dung & Nakase ex Yurkov, Stud. Mycol. 81: 126 (2015), nom. inval., Art. 40.7 (Shenzhen).
Papiliotrema japonica J.P. Samp., Fonseca & Fell ex Xin Zhan Liu, F.Y. Bai, M. Groenew. & Boekhout, sp. nov. MycoBank MB831687.
For description see Int. J. Syst. Evol. Microbiol. 54(3): 990 (2004).
Holotype: CBS 2013 (preserved in a metabolically inactive state).
Synonyms: Bullera japonica J.P. Samp. et al., Int. J. Syst. Evol. Microbiol. 54(3): 990 (2004), nom. inval., Art. 40.6 (Shenzhen).
= Papiliotrema japonica J.P. Samp., Fonseca & Fell ex Xin Zhan Liu, F.Y. Bai, M. Groenew. & Boekhout, Stud. Mycol. 81: 126 (2015), nom. inval., Art. 40.6 (Shenzhen).
Papiliotrema terrestris Crestani, Landell, Faganello, Vainstein, Vishniac & P. Valente ex Xin Zhan Liu, F.Y. Bai, M. Groenew. & Boekhout, sp. nov. MycoBank MB831688.
For description see Int. J. Syst. Evol. Microbiol. 59(3): 635 (2009).
Holotype: CBS 10810 (preserved in a metabolically inactive state).
Synonyms: Cryptococcus terrestris Crestani et al., Int. J. Syst. Evol. Microbiol. 59(3): 635 (2009), nom. inval., Art. 40.7 (Shenzhen).
= Papiliotrema terrestris Crestani, Landell, Faganello, Vainstein, Vishniac & P. Valente ex Xin Zhan Liu, F.Y. Bai, M. Groenew. & Boekhout, Stud. Mycol. 81: 121 (2015), nom. inval., Art. 40.7 (Shenzhen).
Papiliotrema wisconsinensis K. Sylvester, Q.M. Wang & Hittinger ex Xin Zhan Liu, F.Y. Bai, M. Groenew. & Boekhout, sp. nov. MycoBank MB831712.
For description see FEMS Yeast Res. 15(3): 7 (2015).
Holotype: CBS 13895 (preserved in a metabolically inactive state).
Synonyms: Cryptococcus wisconsinensis K. Sylvester, Q.M. Wang & Hittinger, FEMS Yeast Res. 15(3): 7 (2015), nom. inval., Art. 40.7 (Shenzhen).
= Papiliotrema wisconsinensis K. Sylvester, Q.M. Wang & Hittinger ex Xin Zhan Liu, F.Y. Bai, M. Groenew. & Boekhout, Stud. Mycol. 81: 127 (2015), nom. inval., Art. 40.7 (Shenzhen).
Piskurozyma fildesensis T.T. Zhang & Li Y. Yu ex Yurkov, sp. nov. MycoBank MB831672.
For description see Int. J. Syst. Evol. Microbiol. 64(2): 676 (2013).
Holotype: CBS 12705 (preserved in a metabolically inactive state).
Synonyms: Cryptococcus fildesensis T.T. Zhang & Li Y. Yu, in Zhang et al., Int. J. Syst. Evol. Microbiol. 64(2): 676 (2013), nom. inval., Art. 40.7 (Shenzhen).
= Piskurozyma fildesensis T.T. Zhang & Li Y. Yu ex Yurkov, Stud. Mycol. 81: 121 (2015), nom. inval., Art. 40.7 (Shenzhen).
Piskurozyma taiwanensis Nakase, Tsuzuki & M. Takash. ex Xin Zhan Liu, F.Y. Bai, M. Groenew. & Boekhout, sp. nov. MycoBank MB831670.
For description see J. Gen. Appl. Microbiol. 48(6): 349 (2002).
Holotype: JCM 11143 (preserved in a metabolically inactive state).
Synonyms: Bullera taiwanensis Nakase et al., J. Gen. Appl. Microbiol. 48(6): 349 (2002), nom. inval., Art. 40.7 (Shenzhen).
= Cryptococcus taiwanensis Nakase, Tsuzuki & M. Takash. ex Golubev, in Golubev & Tomashevskaya, Mikrobiologiya 79 (3): 408 (2010), nom. inval., Art. 40.7 (Shenzhen).
= Piskurozyma taiwanensis Nakase, Tsuzuki & M. Takash. ex Xin Zhan Liu, F.Y. Bai, M. Groenew. & Boekhout, Stud. Mycol. 81: 121 (2015), nom. inval., Art. 40.7 (Shenzhen).
Pseudoleucosporidium V. de García et al. ex M. Groenew. & Q.M. Wang, gen. nov. MycoBank MB831877.
For description see FEMS Yeast Res. 15: 11 (2015).
Type species:Pseudoleucosporidium fasciculatum (Babeva & Lisichk.) M. Groenew. & Q.M. Wang.
Synonyms: Pseudoleucosporidium V. de García et al., FEMS Yeast Research 15 (4): 11 (2015), nom. inval., Art. 40.1 (Shenzhen).
Pseudoleucosporidium fasciculatum (Babeva & Lisichk.) M. Groenew. & Q.M. Wang, comb. nov. MycoBank MB831878.
Holotype: VKM Y-2869 (preserved in a metabolically inactive state).
Basionym: Leucosporidium fasciculatum Babeva & Lisichk., Mikrobiologiya 69(6): 801 (2000).
Synonym: Pseudoleucosporidium fasciculatum (Babeva & Lisichk.) V. de García, et al., FEMS Yeast Res. 15: 11 (2015), nom. inval., Art 41.5 (Shenzhen).
Pseudotremella lacticolour Satoh & Makimura ex Yurkov, sp. nov. MycoBank MB831696.
For description see Antonie van Leeuwenhoek 104(1): 90 (2013).
Holotype: JCM 15449 (preserved in a metabolically inactive state).
Synonyms: Cryptococcus lacticolour Satoh & Makimura, Antonie van Leeuwenhoek 104(1): 90 (2013), nom. inval., Art. 40.7 (Shenzhen).
= Pseudotremella lacticolour Satoh & Makimura ex Yurkov, Stud. Mycol. 81: 130 (2015) nom. inval., Art. 40.7 (Shenzhen).
Rhynchogastrema complexa (Landell et al.) Xin Zhan Liu, F.Y. Bai, M. Groenew., Boekhout & Yurkov, comb. nov. MycoBank MB831689.
Basionym: Bandoniozyma complexa Landell, et al., in Valente et al., PLoS ONE 7(10): e46060, 9 (2012).
Synonym: Rhynchogastrema complexa (Landell, et al.) Xin Zhan Liu, F.Y. Bai, M. Groenew., Boekhout & Yurkov, Stud. Mycol. 81: 127 (2015), nom. inval., Art. 41.5 (Shenzhen).
Rhynchogastrema fermentans (C.F. Lee) Xin Zhan Liu, F.Y. Bai, M. Groenew., Boekhout & Yurkov, comb. nov. MycoBank MB831690.
Basionym: Bandoniozyma fermentans C.F. Lee, in Valente et al., PLoS ONE 7(10): e46060, 9 (2012).
Synonym: Rhynchogastrema fermentans (C.F. Lee) Xin Zhan Liu, F.Y. Bai, M. Groenew., Boekhout & Yurkov, Stud. Mycol. 81: 127 (2015), nom. inval., Art. 41.5 (Shenzhen).
Rhynchogastrema glucofermentans (S.O. Suh & M. Blackw.) Xin Zhan Liu, F.Y. Bai, M. Groenew., Boekhout & Yurkov, comb. nov. MycoBank MB831691.
Basionym: Bandoniozyma glucofermentans S.O. Suh & M. Blackw., in Valente et al., PLoS ONE 7(10): e46060, 9 (2012).
Synonym: Rhynchogastrema glucofermentans (S.O. Suh & M. Blackw.) Xin Zhan Liu, F.Y. Bai, M. Groenew., Boekhout & Yurkov, Stud. Mycol. 81: 127 (2015), nom. inval., Art. 41.5 (Shenzhen).
Rhynchogastrema nanyangensis F.L. Hui & Q.H. Niu ex Xin Zhan Liu, F.Y. Bai, M. Groenew., Boekhout & Yurkov, sp. nov. MycoBank MB831692.
For description see Curr. Microbiol. 65(5): 619 (2012).
Holotype: CBS 12474 (preserved in a metabolically inactive state).
Synonyms: Cryptococcus nanyangensis F.L. Hui & Q.H. Niu, in Hui et al., Curr. Microbiol. 65(5): 619 (2012), nom. inval., Art. 40.7 (Shenzhen).
= Rhynchogastrema nanyangensis F.L. Hui & Q.H. Niu ex Xin Zhan Liu, F.Y. Bai, M. Groenew., Boekhout & Yurkov, Stud. Mycol. 81: 127 (2015), nom. inval., Art. 40.7 (Shenzhen).
Rhynchogastrema tunnelae (Boekhout, Fell, Scorzetti & Theelen) Xin Zhan Liu, F.Y. Bai, M. Groenew., Boekhout & Yurkov, comb. nov. MycoBank MB831693.
Basionym: Bandoniozyma tunnelae Boekhout, Fell, Scorzetti & Theelen, in Valente et al., PLoS ONE 7(10): e46060, 9 (2012).
Synonym: Rhynchogastrema tunnelae (Boekhout, Fell, Scorzetti & Theelen) Xin Zhan Liu, F.Y. Bai, M. Groenew., Boekhout & Yurkov, Stud. Mycol. 81: 128 (2015), nom. inval., Art. 41.5 (Shenzhen).
Rhynchogastrema visegradensis (G. Péter & Dlauchy) Xin Zhan Liu, F.Y. Bai, M. Groenew., Boekhout & Yurkov, comb. nov. MycoBank MB831694.
Basionym: Bandoniozyma visegradensis G. Péter & Dlauchy, in Valente et al., PLoS ONE 7(10): e46060, 10 (2012).
Synonym: Rhynchogastrema visegradensis (G. Péter & Dlauchy) Xin Zhan Liu, F.Y. Bai, M. Groenew., Boekhout & Yurkov, Stud. Mycol. 81: 128 (2015), nom. inval., Art. 41.5 (Shenzhen).
Ruinenia diospyri Nakase, Tsuzuki, F.L. Lee, Jindam. & M. Takash. ex Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, sp. nov. MycoBank MB831745.
For description see J. Gen. Appl. Microbiol. 51(5): 280 (2005).
Holotype: JCM 12157 (preserved in a metabolically inactive state).
Synonyms: Sporobolomyces diospyri Nakase, Tsuzuki, F.L. Lee, Jindam. & M. Takash. J. Gen. Appl. Microbiol. 51(5): 280 (2005), as ‘diospyroris’, nom. inval., Art. 40.7 (Shenzhen).
= Ruinenia diospyri Nakase, Tsuzuki, F.L. Lee, Jindam. & M. Takash. ex Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, Stud. Mycol. 81: 171 (2015), as ‘diospyroris‘, nom. inval., Art. 40.7 (Shenzhen).
Ruinenia pyrrosiae Nakase, Tsuzuki, F.L. Lee, Jindam. & M. Takash. ex Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, sp. nov. MycoBank MB831746.
For description see J. Gen. Appl. Microbiol. 51(5): 284 (2005).
Holotype: JCM 12159 (preserved in a metabolically inactive state).
Synonyms: Sporobolomyces pyrrosiae Nakase, et al., J. Gen. Appl. Microbiol. 51(5): 284 (2005), nom. inval., Art. 40.7 (Shenzhen).
= Ruinenia pyrrosiae Nakase, Tsuzuki, F.L. Lee, Jindam. & M. Takash. ex Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, Stud. Mycol. 81: 171 (2015), nom. inval., Art. 40.7 (Shenzhen).
Saitozyma ninhbinhensis (D.T. Luong, M. Takash., Dung & Nakase) Yurkov, comb. nov. MycoBank MB831700.
For description see J. Gen. Appl. (Special Issue) Biotechnol.: 36 (2002).
Holotype: VTCC 10184 (preserved in a metabolically inactive state).
Basionym: Bullera ninhbinhensis D.T. Luong, M. Takash., Ty, Dung & Nakase, Journal of Genetics and Applications (Special Issue) Biotechnology: 36 (2002).
Synonyms: Saitozyma ninhbinhensis D.T. Luong, M. Takash., Ty, Dung & Nakase ex Yurkov, Stud. Mycol. 81: 134 (2015), nom. inval., Art. 41.5 (Shenzhen).
Saitozyma paraflava Golubev & J.P. Samp. ex Xin Zhan Liu, F.Y. Bai, M. Groenew. & Boekhout, sp. nov. MycoBank MB831704.
For description see J. Gen. Appl. Microbiol. 50(2): 68 (2004).
Holotype: VKM Y-2923 (preserved in a metabolically inactive state).
Synonyms: Cryptococcus paraflavus Golubev & J.P. Samp., in Golubev et al., J. Gen. Appl. Microbiol. 50(2): 68 (2004), nom. inval., Art. 40.6 (Shenzhen).
= Saitozyma paraflava Golubev & J.P. Samp. ex Xin Zhan Liu, F.Y. Bai, M. Groenew. & Boekhout, Stud. Mycol. 81: 134 (2015), nom. inval., Art. 40.6 (Shenzhen).
Tremella basidiomaticola Xin Zhan Liu & F.Y. Bai, sp. nov. MycoBank MB831876.
For description see Mycokeys 47: 80 (2019).
Holotype: CGMCC 2.5724 (preserved in a metabolically inactive state).
Synonym: Tremella basidiomaticola Xin Zhan Liu & F.Y. Bai, Mycokeys 47: 80 (2019), nom. inval., Art. 40.8 (Shenzhen).
Trimorphomyces sakaeraticus Fungsin, M. Takash. & Nakase ex Xin Zhan Liu, F.Y. Bai, M. Groenew. & Boekhout, sp. nov. MycoBank MB831699.
For description see Microbiol. Culture Coll. 19(1): 37 (2003).
Holotype: JCM 11900 (preserved in a metabolically inactive state).
Synonyms: Bullera sakaeratica Fungsin, M. Takash. & Nakase, Microbiol. Culture Coll. 19(1): 37 (2003), nom. inval., Art. 40.7 (Shenzhen).
= Trimorphomyces sakaeraticus Fungsin, M. Takash. & Nakase ex Xin Zhan Liu, F.Y. Bai, M. Groenew. & Boekhout, Stud. Mycol. 81: 134 (2015), nom. inval., Art. 40.7 (Shenzhen).
Vanrija meifongana C.F. Lee ex Kachalkin, Yurkov & Boekhout, sp. nov. MycoBank MB831709.
For description see Antonie van Leeuwenhoek 99(3): 647 (2011).
Holotype: CBS 11424 (preserved in a metabolically inactive state).
Synonyms: Asterotremella meifongana C.F. Lee, in Liu et al., Antonie van Leeuwenhoek 99(3): 647 (2011), nom. inval., Art. 40.7 (Shenzhen).
= Vanrija meifongana C.F. Lee ex Kachalkin, Yurkov & Boekhout, Stud. Mycol. 81: 142 (2015), nom. inval., Art. 40.7 (Shenzhen).
Vanrija nantouana C.F. Lee ex Kachalkin, Yurkov & Boekhout, sp. nov. MycoBank MB831710.
For description see Antonie van Leeuwenhoek 99(3): 648 (2011).
Holotype: CBS 10890 (preserved in a metabolically inactive state).
Synonyms: Asterotremella nantouana C.F. Lee, Antonie van Leeuwenhoek 99(3): 648 (2011), nom. inval., Art. 40.7 (Shenzhen).
= Vanrija nantouana C.F. Lee ex Kachalkin, Yurkov & Boekhout, Stud. Mycol. 81: 142 (2015), nom. inval., Art. 40.7 (Shenzhen).
Vanrija thermophila Vogelmann, S. Chaves & C. Hertel ex Kachalkin, Yurkov & Boekhout, sp. nov. MycoBank MB831711.
For description see Int. J. Syst. Evol. Microbiol. 62(7): 1719 (2012).
Holotype: CBS 10687 (preserved in a metabolically inactive state).
Synonyms: Cryptococcus thermophilus Vogelmann, S. Chaves & C. Hertel, Int. J. Syst. Evol. Microbiol. 62(7): 1719 (2012), nom. inval., Art. 40.7 (Shenzhen).
= Vanrija thermophila Vogelmann, S. Chaves & C. Hertel ex Kachalkin, Yurkov & Boekhout, Stud. Mycol. 81: 142 (2015), nom. inval., Art. 40.7 (Shenzhen).
Vishniacozyma foliicola Q.M. Wang & F.Y. Bai ex Yurkov, sp. nov. MycoBank MB831680.
For description see J. Gen. Appl. Microbiol. 57(5): 287 (2011).
Holotype: CGMCC AS 2.2471 (preserved in a metabolically inactive state).
Synonyms: Cryptococcus foliicola Q.M. Wang & F.Y. Bai, J. Gen. Appl. Microbiol. 57(5): 287 (2011), nom. inval., Art. 40.7 (Shenzhen).
= Vishniacozyma foliicola Q.M. Wang & F.Y. Bai ex Yurkov, Stud. Mycol. 81: 124 (2015), nom. inval., Art. 40.7 (Shenzhen).
Vishniacozyma heimaeyensis Vishniac ex Xin Zhan Liu, F.Y. Bai, M. Groenew. & Boekhout, sp. nov. MycoBank MB831682.
For description see Canad. J. Microbiol. 48(5): 464 (2002).
Holotype: CBS 8933 (preserved in a metabolically inactive state).
Synonyms: Cryptococcus heimaeyensis Vishniac, Canad. J. Microbiol. 48(5): 464 (2002), nom. inval., Art. 40.6 (Shenzhen).
= Vishniacozyma heimaeyensis Vishniac ex Xin Zhan Liu, F.Y. Bai, M. Groenew. & Boekhout, Stud. Mycol. 81: 124 (2015), nom. inval., Art. 40.6 (Shenzhen).
Vishniacozyma psychrotolerans V. de García, Zalar, Brizzio, Gunde-Cim. & Van Broock ex Yurkov, sp. nov. MycoBank MB831684.
For description see FEMS Microbiology Ecology 82(2): 535 (2012).
Holotype: EXF-7039 (preserved in a metabolically inactive state).
Synonyms: Cryptococcus psychrotolerans V. de García, Zalar, Brizzio, Gunde-Cim. & Van Broock, FEMS Microbiol. Ecol. 82(2): 535 (2012), nom. inval., Art. 40.7 (Shenzhen).
= Vishniacozyma psychrotolerans V. de García, Zalar, Brizzio, Gunde-Cim. & Van Broock ex Yurkov, Stud. Mycol. 81: 124 (2015), nom. inval., Art. 40.7 (Shenzhen).
Vishniacozyma taibaiensis Q.M. Wang & F.Y. Bai ex Yurkov, sp. nov. MycoBank MB831681.
For description see J. Gen. Appl. Microbiol. 57(5): 288 (2011).
Holotype: CGMCC AS 2.2444 (preserved in a metabolically inactive state).
Synonyms: Cryptococcus taibaiensis Q.M. Wang & F.Y. Bai, J. Gen. Appl. Microbiol. 57(5): 288 (2011), nom. inval., Art. 40.7 (Shenzhen).
= Vishniacozyma taibaiensis Q.M. Wang & F.Y. Bai ex Yurkov, Stud. Mycol. 81: 124 (2015), nom. inval., Art. 40.7 (Shenzhen).
Vishniacozyma tephrensis Vishniac ex Xin Zhan Liu, F.Y. Bai, M. Groenew. & Boekhout, sp. nov. MycoBank MB831683.
For description see Canad. J. Microbiol. 48(5): 466 (2002).
Holotype: CBS 8935 (preserved in a metabolically inactive state).
Synonyms: Cryptococcus tephrensis Vishniac, Canad. J. Microbiol. 48(5): 466 (2002), nom. inval., Art. 40.6 (Shenzhen).
= Vishniacozyma tephrensis Vishniac ex Xin Zhan Liu, F.Y. Bai, M. Groenew. & Boekhout, Stud. Mycol. 81: 124 (2015), nom. inval., Art. 40.6 (Shenzhen).
Yamadamyces Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, gen. nov. MycoBank MB831747.
For description see Stud. Mycol. 81: 178 (2015).
Type species:Yamadamyces rosulatus Golubev & Scorzetti ex Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout.
Synonym: Yamadamyces Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, Stud. Mycol. 81: 178 (2015), nom. inval., Art. 40.1 (Shenzhen).
Yamadamyces rosulatus Golubev & Scorzetti ex Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, sp. nov. MycoBank MB831748.
For description see Int. J. Syst. Evol. Microbiol. 60(10): 2503 (2010).
Holotype: CBS 10977 (preserved in a metabolically inactive state).
Synonym: Rhodotorula rosulata Golubev & Scorzetti, Int. J. Syst. Evol. Microbiol. 60(10): 2503 (2010), nom. inval., Art. 40.7 (Shenzhen).
= Yamadamyces rosulatus Golubev & Scorzetti ex Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, Stud. Mycol. 81: 178 (2015), nom. inval., Art. 40.7 (Shenzhen).
Contributions
F.-Y.B. and Q.-M.W. conceived and designed the project. Q.-M.W., F.-Y.B., P.-J.H. and L.-D. G. performed sampling and yeast isolation. A.-H. Li, F.-X.Y. and Q.-M.W. performed phenotypic characterisation and analysed the molecular data. L.K. run the emboss water analysis. A.Y. analysed the D1/D2 data. K.B. registered the taxa in MycoBank and handled the invalid taxonomic names. Q.-M.W., M.G. and F.-Y.B. wrote the paper. Q.-M.W., M.C.A., A.Y. and K.B. revised the paper. A.Y., M.T., J.P.S., B.F., S.J., M.C.A., B.T., J.I. supported the sequences and physiological data or strains generated and conserved in their laboratory.
Acknowledgements
We thank Prof. Jian-Yun Zhuang for his advice on nomenclatural matters. We thank Dr. Alexander Idnurm for his kindly providing the sequences and informations of strain IAM13481 and his critical comments for this manuscript, Dr. Aleksey Kachalkin for his sharing the physilogical data of strain KBP Y-5548 and Masako Takashima for her sharing the physilogical data of strain TY-217. We also thank Ana Pontes and Cláudia Carvalho for editing illustrations of Kondoa myxariophila and for ITS sequencing, respectively. This study was supported by grants No. 31570016 from the National Natural Science Foundation of China (NSFC) and national project on scientific groundwork No. 2014FY210400 from the Ministry of Science and Technology of China. The authors are solely responsible for the content of this work.
Footnotes
Peer review under responsibility of Westerdijk Fungal Biodiversity Institute.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.simyco.2020.01.002.
Contributor Information
Q.-M. Wang, Email: wangqm@im.ac.cn.
F.-Y. Bai, Email: baify@im.ac.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Fig. S1. Phylogenetic tree inferred using the combined sequences of SSU rDNA, LSU rDNA D1/D2 domains and 5.8S rDNA, depicting the phylogenetic positions of new taxa (in bold) within Tremellomycetes (Agaricomycotina). The tree backbone was constructed using maximum likelihood analysis. Bootstrap percentages of maximum likelihood analysis over 50 % from 1 000 bootstrap replicates and posterior probabilities of Bayesian inference above 0.9 are shown respectively from left to right on the deep and major branches. Bar = 0.1 substitutions per nucleotide position. Note: ns, not supported (BP < 50 % or PP < 0.9); nm, not monophyletic. The new taxa are in bold. Fig. S2. Phylogenetic tree inferred using the combined sequences of SSU rDNA, LSU rDNA D1/D2 domains and 5.8S rDNA, depicting the phylogenetic positions of new taxa (in bold) within Pucciniomycotina. The tree backbone was constructed using maximum likelihood analysis. Bootstrap percentages of maximum likelihood analysis over 50 % from 1 000 bootstrap replicates and posterior probabilities of Bayesian inference above 0.9 are shown respectively from left to right on the deep and major branches. Bar = 0.1 substitutions per nucleotide position. Note: ns, not supported (BP < 50 % or PP < 0.9); nm, not monophyletic. The new taxa are in bold. Fig. S3. Phylogenetic tree inferred using the ITS (including 5.8S rDNA), depicting the phylogenetic positions of Phaffia species (new taxa in bold). The tree was constructed using maximum likelihood analysis and over 50 % from 1 000 bootstrap replicates is shown. Bar = 0.2 substitutions per nucleotide position. Fig. S4. Phylogenetic tree inferred using the combined sequences of RPB1, RPB2, TEF1, CYTB, SSU rDNA, LSU rDNA D1/D2 domains and 5.8S rDNA, depicting the phylogenetic positions of Sakaguchia melibiophila and other new taxa (in bold) within Cystobasidiomycetes (Pucciniomycotina). The tree was constructed using maximum likelihood analysis and over 50 % from 1 000 bootstrap replicates is shown. Bar = 0.05 substitutions per nucleotide position. Fig. S5. Phylogenetic tree inferred using the sequences of LSU rDNA D1/D2 domains, depicting the phylogenetic positions of Lichenozyma and new taxa (in bold) within Cystobasidiomycetes (Pucciniomycotina). The tree was constructed using maximum likelihood analysis and over 50 % from 1 000 bootstrap replicates is shown. Bar = 0.05 substitutions per nucleotide position. Fig. S6. Phylogenetic tree inferred using the combined LSU rDNA D1/D2 domains and ITS (including 5.8S rDNA), depicting the phylogenetic positions of Lichenozyma and new taxa (in bold) within Cystobasidiomycetes (Pucciniomycotina). The tree was constructed using maximum likelihood analysis and over 50 % from 1 000 bootstrap replicates is shown. Bar = 0.05 substitutions per nucleotide position.
Table S1Physiological and biochemical characteristics of new species and their closest relatives.
Multimedia component 33
Table S2. Number of nucleotide variation and sequence similarities in the D1/D2 domain and ITS region among species in each of 70 genera.
List of yeasts and sequence numbers retrieved from GenBank.
The results of clustering optimization analysis.
The results of clustering optimization analysis.
References
- Aime M.C., Matheny P.B., Henk D.A., et al. An overview of the higher-level classification of Pucciniomycotina based on combined analyses of nuclear large and small subunit rDNA sequences. Mycologia. 2006;98:896–905. doi: 10.3852/mycologia.98.6.896. [DOI] [PubMed] [Google Scholar]
- Bandoni R.J., Teun Boekhout T., Sampaio J.P. In: The Yeasts, A Taxonomic Study. 5th edn. Kurtzman C.P., Fell J.W., Boekhout T., editors. Elsevier; Netherland: 2011. Holtermannia Saccardo & Traverso (1910) pp. 1467–1470. [Google Scholar]
- Bauer R., Begerow D., Sampaio J.P., et al. The simple–septate basidiomycetes: a synopsis. Mycological progress. 2006;5:41–66. [Google Scholar]
- Blackwell M. The Fungi: 1, 2, 3 ... 5.1 million species? American Journal of Botany. 2011;98:426–438. doi: 10.3732/ajb.1000298. [DOI] [PubMed] [Google Scholar]
- Boekhout T., Bai F.Y., Daniel H.-M., et al. 2016. The Yeasts Trust Database.http://theyeasts.org/ [Google Scholar]
- Boekhout T., Fonseca A., Sampaio J.P., et al. In: The Yeasts, A Taxonomic Study. 5th edn. Kurtzman C.P., Fell J.W., Boekhout T., editors. Elsevier; Netherland: 2011. Discussion of teleomorphic and anamorphic basidiomycetous yeasts; pp. 1339–1372. [Google Scholar]
- Botha A. In: Biodiversity and ecophysiology of yeasts. Lachance M.A., Kurtzman C.P., Fell J.W., editors. Springer; Germany: 2006. Yeasts in Soil; pp. 221–240. [Google Scholar]
- Botha A. The importance and ecology of yeasts in soil. Soil Biology and Biochemistry. 2011;43:1–8. [Google Scholar]
- Černajová I., Škaloud P. The first survey of Cystobasidiomycete yeasts in the lichen genus Cladonia; with the description of Lichenozyma pisutiana gen. nov., sp. nov. Fungal Biology. 2019;123:625–637. doi: 10.1016/j.funbio.2019.05.006. [DOI] [PubMed] [Google Scholar]
- Coelho M.A., Gonçalves P., Sampaio J.P. Evidence for maintenance of sex determinants but not of sexual stages in red yeasts, a group of early diverged basidiomycetes. BMC Evolutionary Biology. 2011;11:249. doi: 10.1186/1471-2148-11-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coelho M.A., Rosa A., Rodrigues N., et al. Identification of mating type genes in the bipolar basidiomycetous yeast Rhodosporidium toruloides: first insight into the MAT locus structure of the Sporidiobolales. Eukaryot Cell. 2008;7:1053–1061. doi: 10.1128/EC.00025-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous P.W., Wingfield M.J., Burgess T.I., et al. Fungal Planet description sheets: 716–784. Persoonia. 2018;40:240–393. doi: 10.3767/persoonia.2018.40.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David-Palma M., Libkind D., Sampaio J.P. Global distribution, diversity hot spots and niche transitions of an astaxanthin–producing eukaryotic microbe. Molecular Ecology. 2014;23:921–932. doi: 10.1111/mec.12642. [DOI] [PubMed] [Google Scholar]
- de Garcia V., Zalar P., Brizzio S., et al. Cryptococcus species (Tremellales) from glacial biomes in the southern (Patagonia) and northern (Svalbard) hemispheres. FEMS Microbiolology Ecology. 2012;82:523–539. doi: 10.1111/j.1574-6941.2012.01465.x. [DOI] [PubMed] [Google Scholar]
- do Carmo-Sousa L., Phaff H.J. An improved method for the detection of spore discharge in the Sporobolomycetaceae. Journal of Bacteriology. 1962;83:434–435. doi: 10.1128/jb.83.2.434-435.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felix C.R., Navarro H.M.C., Paulino G.V.B., et al. Carlosrosaea hohenbergiae sp. nov. and Carlosrosaea aechmeae sp. nov., two tremellaceous yeasts isolated from bromeliads in north–eastern Brazil. International Journal of Systematic and Evolutionary Microbiology. 2017;67:1752–1757. doi: 10.1099/ijsem.0.001856. [DOI] [PubMed] [Google Scholar]
- Fell J.W., Boekhout T., Fonseca A., et al. Biodiversity and systematics of basidiomycetous yeasts as determined by large-subunit rDNA D1/D2 domain sequence analysis. International Journal of Systematic and Evolutionary Microbiology. 2000;50:1351–1371. doi: 10.1099/00207713-50-3-1351. [DOI] [PubMed] [Google Scholar]
- Fonseca F., Inácio J. In: Biodiversity and ecophysiology of yeasts. Lachance M.A., Kurtzman C.P., Fell J.W., editors. Springer; Germany: 2006. Phylloplane yeasts; pp. 263–302. [Google Scholar]
- Gadanho M., Sampaio J.P. Polyphasic taxonomy of the basidiomycetous yeast genus Rhodotorula: Rh. glutinis sensu stricto and Rh. dairenensis comb. nov. FEMS Yeast Research. 2002;2:47–58. doi: 10.1111/j.1567-1364.2002.tb00068.x. [DOI] [PubMed] [Google Scholar]
- Göker M., García-Blázquez G., Voglmayr H., et al. Molecular taxonomy of phytopathogenic fungi: a case study in Peronospora. PLoS One. 2009;4 doi: 10.1371/journal.pone.0006319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golubev W.I., Scorzetti G. Rhodotorula rosulata sp. nov., Rhodotorula silvestris sp. nov. and Rhodotorula straminea sp. nov., novel myo-inositol-assimilating yeast species in the Microbotryomycetes. International Journal of Systematic and Evolutionary Microbiology. 2010;60:2501–2506. doi: 10.1099/ijs.0.016303-0. [DOI] [PubMed] [Google Scholar]
- Groenewald M., Lombard L., de Vries M., et al. Diversity of yeast species from Dutch garden soil and the six novel Ascomycetes. FEMS Yeast Research. 2018;18:1–14. doi: 10.1093/femsyr/foy076. foy076. [DOI] [PubMed] [Google Scholar]
- Guo Z., Wang Y., Hou Q., et al. Halobasidium xiangyangense gen. nov., sp. nov., a new xylose–utilizing yeast in the family Cystobasidiaceae, isolated from the pickling sauce used to make Datoucai, a high–salt fermented food. International Journal of Systematic and Evolutionary Microbiology. 2019;69:139–145. doi: 10.1099/ijsem.0.003119. [DOI] [PubMed] [Google Scholar]
- Hagen F., Khayhan K., Theelen B., et al. Recognition of seven species in the Cryptococcus gattii/Cryptococcus neoformans species complex. Fungal Genetics and Biology. 2015;78:16–48. doi: 10.1016/j.fgb.2015.02.009. [DOI] [PubMed] [Google Scholar]
- Hamamoto M., Sugiyama J., Komagata K. DNA base composition of strains in the genera Rhodosporidium, Cystofilobasidium, and Rhodotorula determined by reversed-phase high-performance liquid chromatography. The Journal of General and Applied Microbiology. 1986;32:215–223. [Google Scholar]
- Hamamoto M., Sugiyama J., Goto S., et al. Numerical taxonomy based on the electrophoretic mobility of enzymes in the genera Rhodosporidium, Cystofilobasidium, and Rhodotorula. The Journal of General and Applied Microbiology. 1986;32:89–99. [Google Scholar]
- Hawksworth D.L. The fungal dimension of biodiversity: magnitude, significance, and conservation. Mycology Research. 1991;95:641–655. [Google Scholar]
- Hawksworth D.L. The magnitude of fungal diversity: the 1.5 million species estimate revisited. Mycology Research. 2001;105:1422–1432. [Google Scholar]
- Hawksworth D.L., Lücking R. Fungal diversity revisited: 2.2 to 3.8 million species. Microbiology Spectrum. 2017;5:1–2. doi: 10.1128/microbiolspec.funk-0052-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibbett D.S., Binder M., Bischoff J.F., et al. A higher–level phylogenetic classification of the Fungi. Mycological research. 2007;111:509–547. doi: 10.1016/j.mycres.2007.03.004. [DOI] [PubMed] [Google Scholar]
- Ianiri G., Idnurm A., Castoria R. Transcriptomic responses of the basidiomycete yeast Sporobolomyces sp. to the mycotoxin patulin. BMC Genomics. 2016;17:210. doi: 10.1186/s12864-016-2550-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ianiri G., Idnurm A., Wright S.A., et al. Searching for genes responsible for patulin degradation in a biocontrol yeast provides insight into the basis for resistance to this mycotoxin. Applied and Environmental Microbiology. 2013;79:3101–3115. doi: 10.1128/AEM.03851-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inácio J. Faculdade de Ciências e Tecnologia da Universidade Nova de Lisboa; Portugal: 2003. Ocorrência e Diversidade de Leveduras no Filoplano de Plantas Seleccionadas do Parque Natural da Arrábida. PhD Thesis. (in Portuguese) [Google Scholar]
- Kemler M., Witfeld F., Begerow D., et al. In: Yeasts in natural ecosystems: diversity. Buzzini P., Lachance M.A., Yurkov A., editors. Springer; Cham: 2017. Phylloplane yeasts in temperate climates; pp. 171–198. [Google Scholar]
- Kobayasi Y. On the genus Holtermannia of Tremellaceae. Science Reports of the Tokyo Bunrika Daigaku Section B. 1937;3:75–81. [Google Scholar]
- Kurtzman C.P. In: The Yeasts, A Taxonomic Study. 5th edn. Kurtzman C.P., Fell J.W., Boekhout T., editors. Elsevier; Netherland: 2011. Discussion of teleomorphic and anamorphic ascomycetous yeasts and yeast-like taxa; pp. 293–307. [Google Scholar]
- Kurtzman C.P. Use of gene sequence analyses and genome comparisons for yeast systematics. International Journal of Systematic and Evolutionary Microbiology. 2014;64:325–332. doi: 10.1099/ijs.0.054197-0. [DOI] [PubMed] [Google Scholar]
- Kurtzman C.P. Identification of food and beverage spoilage yeasts from DNA sequence analyses. International Journal of Food Microbiology. 2015;213:71–78. doi: 10.1016/j.ijfoodmicro.2015.05.023. [DOI] [PubMed] [Google Scholar]
- Kurtzman C.P., Fell J.W. Molecular relatedness between the basidiomycetous yeasts Sporidiobolus ruinenii and Sporobolomyces coprophilus. Mycologia. 1991;83:107–110. [Google Scholar]
- Kurtzman C.P., Fell J.W. 4th edn. Elsevier; Netherland: 1998. The Yeasts, A Taxonomic Study. [Google Scholar]
- Kurtzman C.P., Fell J.W. In: Biodiversity and ecophysiology of yeasts. Lachance M.A., Kurtzman C.P., Fell J.W., editors. Springer; Gemany: 2006. Yeast systematics and phylogeny–implications of molecular identification methods for studies in ecology; pp. 11–30. [Google Scholar]
- Kurtzman C.P., Fell J.W., Boekhout T., et al. In: The Yeasts, A Taxonomic Study. 5th edn. Kurtzman C.P., Fell J.W., Boekhout T., editors. Elsevier; Netherland: 2011. Methods for isolation, phenotypic characterization and maintenance of yeasts; pp. 87–110. [Google Scholar]
- Kurtzman C.P., Mateo R.Q., Kolecka A., et al. Advances in yeast systematics and phylogeny and their use as predictors of biotechnologically important metabolic pathways. FEMS Yeast Research. 2015;15:1–17. doi: 10.1093/femsyr/fov050. pii: fov050. [DOI] [PubMed] [Google Scholar]
- Kurtzman C.P., Robnett C.J. Identification and phylogeny ascomycetous yeasts from analysis of nuclear large subunit (26S) ribosomal DNA partial sequences. Antonie Van Leeuwenhoek. 1998;73:331–371. doi: 10.1023/a:1001761008817. [DOI] [PubMed] [Google Scholar]
- Lachance M.A. In: Biodiversity and ecophysiology of yeasts. Lachance M.A., Kurtzman C.P., Fell J.W., editors. Springer; Germany: 2006. Yeast Biodiversity: How Many and How Much? pp. 1–10. [Google Scholar]
- Landell M.F., Brandão L.R., Safar S.V., et al. Buller avrieseae sp. nov., a tremellaceous yeast species isolated from bromeliads. International Journal of Systematic and Evolutionary Microbiology. 2015;65:2466–2471. doi: 10.1099/ijs.0.000285. [DOI] [PubMed] [Google Scholar]
- Limtong S., Nasanit R. In: Yeasts in natural ecosystems: diversity. Buzzini P., Lachance M.A., Yurkov A., editors. Springer; Cham: 2017. Phylloplane yeasts in tropical climates; pp. 199–224. [Google Scholar]
- Liu X.Z., Groenewald M., Boekhout T., et al. Heitmania gen. nov., a new yeast genus in Microbotryomycetes, and three novel species: Heitmania litseae sp. nov., Heitmania castanopsis sp. nov. and Heitmania elacocarpi sp. nov. International Journal of Systematic and Evolutionary Microbiology. 2017;67:4534–4540. doi: 10.1099/ijsem.0.002323. [DOI] [PubMed] [Google Scholar]
- Liu X.Z., Wang Q.M., Theelen B., et al. Phylogeny of tremellomycetous yeasts and related dimorphic and filamentous basidiomycetes reconstructed from multiple gene sequence analyses. Studies in Mycology. 2015;81:1–26. doi: 10.1016/j.simyco.2015.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X.Z., Wang Q.M., Göker M., et al. Towards an integrated phylogenetic classification of the Tremellomycetes. Studies in Mycology. 2015;81:85–147. doi: 10.1016/j.simyco.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madeira F., Park Y.M., Lee J., et al. The EMBL–EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Research. 2019:1–6. doi: 10.1093/nar/gkz268. pii: gkz268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mašínová T., Pontes A., Carvalho C., et al. Libkindia masarykiana gen. et sp. nov., Yurkovia mendeliana gen. et sp. nov. and Leucosporidium krtinense f.a. sp. nov., isolated from temperate forest soils. International Journal of Systematic and Evolutionary Microbiology. 2017;67:902–908. doi: 10.1099/ijsem.0.001707. [DOI] [PubMed] [Google Scholar]
- Millanes A.M., Diederich P., Ekman S., et al. Phylogeny and character evolution in the jelly fungi (Tremellomycetes, Basidiomycota, Fungi) Molecular Phylogenetics and Evolution. 2011;61:12–28. doi: 10.1016/j.ympev.2011.05.014. [DOI] [PubMed] [Google Scholar]
- Millanes A.M., Diederich P., Wedin M. Cyphobasidium gen. nov., a new lichen-inhabiting lineage in the Cystobasidiomycetes (Pucciniomycotina, Basidiomycota, Fungi) Fungal Biology. 2016;120:1468–1477. doi: 10.1016/j.funbio.2015.12.003. [DOI] [PubMed] [Google Scholar]
- Morais P.B., Pagnocca F.C., Rosa C.A. In: Biodiversity and ecophysiology of yeasts. Lachance M.A., Kurtzman C.P., Fell J.W., editors. Springer; Germany: 2006. Yeast communities in tropical rain forests in Brazil and other South American ecosystem; pp. 461–484. [Google Scholar]
- Nakase T. Expanding world of ballistosporous yeasts: Distribution in the phyllosphere, systematics and phylogeny. The Journal of General and Applied Microbiology. 2000;46:189–216. doi: 10.2323/jgam.46.189. [DOI] [PubMed] [Google Scholar]
- Nakase T., Jindamorakot S., Am-in S., et al. In: Biodiversity and ecophysiology of yeasts. Lachance M.A., Kurtzman C.P., Fell J.W., editors. Springer; Germany: 2006. Yeast biodiversity in tropical forests of Asia; pp. 441–460. [Google Scholar]
- Nakase T., Takashima M. A simple procedure for the high frequency isolation of new taxa of ballistosporous yeasts living on the surfaces of plants. RIKEN Review. 1993;3:33–34. [Google Scholar]
- Passer A.R., Coelho M.A., Billmyre R.B., et al. Genetic and genomic analyses reveal boundaries between species closely related to Cryptococcus Pathogens. MBio. 2019;10 doi: 10.1128/mBio.00764-19. pii: e00764-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posada D., Crandall K.A. Modeltest: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- Prior R., Mittelbach M., Begerow D. Impact of three different fungicides on fungal epi- and endophytic communities of common bean (Phaseolus vulgaris) and broad bean (Viciafaba) Journal of Environmental Science and Health Part B–Pesticides Food Contaminants and Agricultural Wastes. 2017;52:376–386. doi: 10.1080/03601234.2017.1292093. [DOI] [PubMed] [Google Scholar]
- Robert V., Stegehuis G., Stalpers J. 2005. The MycoBank engine and related databases.http://www.mycobank.org [Google Scholar]
- Ronquist F., Teslenko M., van der Mark P., et al. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampaio J.P. In: The Yeasts, A Taxonomic Study. 5th edn. Kurtzman C.P., Fell J.W., Boekhout T., editors. Elsevier; Amsterdam: 2011. Rhodotorula Harrison (1928) pp. 1873–1927. [Google Scholar]
- Sampaio J.P. In: The Yeasts, A Taxonomic Study. 5th edn. Kurtzman C.P., Fell J.W., Boekhout T., editors. Elsevier; Amsterdam: 2011. Kurtzmanomyces Y. Yamada, M. Itoh, Kawasai, Banno & Nakase (1998) pp. 1795–1800. [Google Scholar]
- Sampaio J.P. In: The Yeasts, A Taxonomic Study. 5th edn. Kurtzman C.P., Fell J.W., Boekhout T., editors. Elsevier; Netherland: 2011. Sporidiobolus Nyland (1949) pp. 1549–1561. [Google Scholar]
- Sampaio J.P., Weiß M., Gadanho M., et al. New taxa in the Tremellales: Bulleribasidium oberjochense gen. et sp. nov., Papiliotrema bandonii gen. et sp. nov. and Fibulobasidium murrhardtense sp. nov. Mycologia. 2002;94:873–887. [PubMed] [Google Scholar]
- Scorzetti G., Fell J.W., Fonseca A., et al. Systematics of basidiomycetous yeasts: a comparison of large subunit D1/D2 and internal transcribed spacer rDNA regions. FEMS Yeast Research. 2002;2:495–517. doi: 10.1111/j.1567-1364.2002.tb00117.x. [DOI] [PubMed] [Google Scholar]
- Shin K.S., Oh H.M., Park Y.H., et al. Cryptococcus mujuensis sp. nov. and Cryptococcus cuniculi sp. nov., basidiomycetous yeasts isolated from wild rabbit faeces. International Journal of Systematic and Evolutionary Microbiology. 2006;56:2241–2244. doi: 10.1099/ijs.0.64353-0. [DOI] [PubMed] [Google Scholar]
- Spirin V., Malysheva V., Yurkov A., et al. Studies in the Phaeotremella foliacea group (Tremellomycetes, Basidiomycota) Mycological Progress. 2018;17:451–466. [Google Scholar]
- Spribille T., Tuovinen V., Resl P., et al. Basidiomycete yeasts in the cortex of ascomycete macrolichens. Science. 2016;353:488–492. doi: 10.1126/science.aaf8287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Standley K. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama J., Fukagawa M., Chiu S., et al. Cellular carbohydrate composition, DNA base composition, ubiquinone systems, and diazonium blue B colour test in the genera Rhodosporidium, Leucosporidium, Rhodotorula and related basidiomycetous yeasts. The Journal of General and Applied Microbiology. 1985;31:519–550. [Google Scholar]
- Sylvester K., Wang Q.M., James B., et al. Temperature and host preferences drive the diversification of Saccharomyces and other yeasts: a survey and the discovery of eight new yeast species. FEMS Yeast Research. 2015;15:1–16. doi: 10.1093/femsyr/fov002. pii: fov002. [DOI] [PubMed] [Google Scholar]
- Takashima M., Sugita T., Van B.H., et al. Taxonomic richness of yeasts in Japan within subtropical and cool temperate areas. PLoS One. 2012;7 doi: 10.1371/journal.pone.0050784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toome M., Roberson R.W., Aime M.C. Meredithblackwellia eburnea gen. et sp. nov., Kriegeriaceae fam. nov. and Kriegeriales ord. nov. --toward resolving higher-level classification in Microbotryomycetes. Mycologia. 2013;105:486–495. doi: 10.3852/12-251. [DOI] [PubMed] [Google Scholar]
- Turland N.J., Wiersema J.H., Barrie F.R., et al. 2018. International Code of Nomenclature for Algae, Fungi, and Plants (Shenzhen Code) [DOI] [Google Scholar]
- Valério E., Gadanho M., Sampaio J.P. Reappraisal of the Sporobolomyces roseus species complex and Sporidiobolus metaroseus sp. nov. International Journal of Systematic and Evolutionary Microbiology. 2008;58:736–741. doi: 10.1099/ijs.0.65580-0. [DOI] [PubMed] [Google Scholar]
- Vu D., Groenewald M., de Vries M., et al. Large-scale generation and analysis of filamentous fungal DNA barcodes boosts coverage for kingdom fungi and reveals thresholds for fungal species and higher taxon delimitation. Studies in Mycology. 2019;92:1–20. doi: 10.1016/j.simyco.2018.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vu D., Groenewald M., Szöke S., et al. DNA barcoding analysis of more than 9000 yeast isolates contributes to quantitative thresholds for yeast species and genera delimitation. Studies in Mycology. 2016;85:91–105. doi: 10.1016/j.simyco.2016.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q.M., Bai F.Y. Four new yeast species of the genus Sporobolomyces from plant leaves. FEMS Yeast Research. 2004;4:579–586. doi: 10.1016/j.femsyr.2003.11.002. [DOI] [PubMed] [Google Scholar]
- Wang Q.M., Bai F.Y. Molecular phylogeny of basidiomycetous yeasts in the Cryptococcus luteolus lineage (Tremellales) based on nuclear rDNA and mitochondrial cytochrome b gene sequence analyses: proposal of Derxomyces gen. nov. and Hannaella gen. nov., and eight novel Derxomyces species. FEMS Yeast Research. 2008;8:799–814. doi: 10.1111/j.1567-1364.2008.00403.x. [DOI] [PubMed] [Google Scholar]
- Wang Q.M., Bai F.Y., Zhao J.H., et al. Bensingtonia changbaiensis sp. nov. and Bensingtonia sorbi sp. nov., novel ballistoconidium-forming yeast species from plant leaves. International Journal of Systematic and Evolutionary Microbiology. 2003;53:2085–2089. doi: 10.1099/ijs.0.02736-0. [DOI] [PubMed] [Google Scholar]
- Wang Q.M., Groenewald M., Takashima M., et al. Phylogeny of yeasts and related filamentous fungi within Pucciniomycotina determined from multigene sequence analyses. Studies in Mycology. 2015;81:27–54. doi: 10.1016/j.simyco.2015.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q.M., Groenewald M., Göker M., et al. Phylogenetic classification of yeasts and related taxa within Pucciniomycotina. Studies in Mycology. 2015;81:149–189. doi: 10.1016/j.simyco.2015.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q.M., Begerow D., Groenewald M., et al. Multigene phylogeny and taxonomic revision of yeasts and related fungi in the Ustilaginomycotina. Studies in Mycology. 2015;81:55–83. doi: 10.1016/j.simyco.2015.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q.M., Theelen B., Groenewald M., et al. Moniliellomycetes and Malasseziomycetes, two new classes in Ustilaginomycotina. Persoonia. 2014;33:41–47. doi: 10.3767/003158514X682313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Wang Q.M. Molecular phylogenetic analysis of ballistoconidium– forming yeasts in Trichosporonales (Tremellomycetes): A proposal for Takashimella gen. nov.and Cryptotrichosporon tibetense sp. nov. PLoS One. 2015;10 doi: 10.1371/journal.pone.0132653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q.M., Wang S.A., Jia J.H., et al. Cryptococcus tibetensis sp. nov., a novel basidiomycetous anamorphic yeast species isolated from plant leaves. The Journal of General and Applied Microbiology. 2007;53:281–285. doi: 10.2323/jgam.53.281. [DOI] [PubMed] [Google Scholar]
- Weiss M., Bauer R., Sampaio J.P., et al. In: Systematics and Evolution, The Mycota VII Part A. McLaughlin D.J., Spatafora J.W., editors. Springer; Germany: 2014. Tremellomycetes and related groups; pp. 349–350. [Google Scholar]
- Wuczkowski M., Passoth V., Turchetti B., et al. Holtermanniella takashimae sp. nov., Holtermanniella gen. nov. and proposal of the order Holtermanniales to accommodate Tremellomycetous yeasts of the Holtermanniales clade. Internatonal Journal of Systematic and Evolutionary Microbiology. 2011;61:680–689. doi: 10.1099/ijs.0.019737-0. [DOI] [PubMed] [Google Scholar]
- Yamazaki M., Komagata K. An electrophoretic comparison of enzymes of ballistosporogenous yeasts. The Journal of General and Applied Microbiology. 1983;29:115–143. [Google Scholar]
- Yurkov A. In: Yeasts in natural ecosystems: diversity. Buzzini P., Lachance M.A., Yurkov A., editors. Springer; Cham: 2017. Yeasts in forest soils; pp. 87–116. [Google Scholar]
- Yurkov A., Guerreiro M.A., Sharma L., et al. Multigene assessment of the species boundaries and sexual status of the basidiomycetous yeasts Cryptococcus flavescens and C. terrestris (Tremellales) PLoS ONE. 2015;10:1–24. doi: 10.1371/journal.pone.0120400. e0120400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurkov A.M., Kachalkin A.V., Daniel H.M., et al. Two yeast species Cystobasidium psychroaquaticum f.a. sp. nov. and Cystobasidium rietchieii f.a. sp. nov. isolated from natural environments, and the transfer of Rhodotorula minuta clade members to the genus Cystobasidium. Antonie Van Leeuwenhoek. 2015;107:173–185. doi: 10.1007/s10482-014-0315-0. [DOI] [PubMed] [Google Scholar]
- Yurkov A.M., Wehde T., Federici J., et al. Yeast diversity and species recovery rates from beech forest soils. Mycological Progress. 2016;15:845–859. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Phylogenetic tree inferred using the combined sequences of SSU rDNA, LSU rDNA D1/D2 domains and 5.8S rDNA, depicting the phylogenetic positions of new taxa (in bold) within Tremellomycetes (Agaricomycotina). The tree backbone was constructed using maximum likelihood analysis. Bootstrap percentages of maximum likelihood analysis over 50 % from 1 000 bootstrap replicates and posterior probabilities of Bayesian inference above 0.9 are shown respectively from left to right on the deep and major branches. Bar = 0.1 substitutions per nucleotide position. Note: ns, not supported (BP < 50 % or PP < 0.9); nm, not monophyletic. The new taxa are in bold. Fig. S2. Phylogenetic tree inferred using the combined sequences of SSU rDNA, LSU rDNA D1/D2 domains and 5.8S rDNA, depicting the phylogenetic positions of new taxa (in bold) within Pucciniomycotina. The tree backbone was constructed using maximum likelihood analysis. Bootstrap percentages of maximum likelihood analysis over 50 % from 1 000 bootstrap replicates and posterior probabilities of Bayesian inference above 0.9 are shown respectively from left to right on the deep and major branches. Bar = 0.1 substitutions per nucleotide position. Note: ns, not supported (BP < 50 % or PP < 0.9); nm, not monophyletic. The new taxa are in bold. Fig. S3. Phylogenetic tree inferred using the ITS (including 5.8S rDNA), depicting the phylogenetic positions of Phaffia species (new taxa in bold). The tree was constructed using maximum likelihood analysis and over 50 % from 1 000 bootstrap replicates is shown. Bar = 0.2 substitutions per nucleotide position. Fig. S4. Phylogenetic tree inferred using the combined sequences of RPB1, RPB2, TEF1, CYTB, SSU rDNA, LSU rDNA D1/D2 domains and 5.8S rDNA, depicting the phylogenetic positions of Sakaguchia melibiophila and other new taxa (in bold) within Cystobasidiomycetes (Pucciniomycotina). The tree was constructed using maximum likelihood analysis and over 50 % from 1 000 bootstrap replicates is shown. Bar = 0.05 substitutions per nucleotide position. Fig. S5. Phylogenetic tree inferred using the sequences of LSU rDNA D1/D2 domains, depicting the phylogenetic positions of Lichenozyma and new taxa (in bold) within Cystobasidiomycetes (Pucciniomycotina). The tree was constructed using maximum likelihood analysis and over 50 % from 1 000 bootstrap replicates is shown. Bar = 0.05 substitutions per nucleotide position. Fig. S6. Phylogenetic tree inferred using the combined LSU rDNA D1/D2 domains and ITS (including 5.8S rDNA), depicting the phylogenetic positions of Lichenozyma and new taxa (in bold) within Cystobasidiomycetes (Pucciniomycotina). The tree was constructed using maximum likelihood analysis and over 50 % from 1 000 bootstrap replicates is shown. Bar = 0.05 substitutions per nucleotide position.
Multimedia component 33
List of yeasts and sequence numbers retrieved from GenBank.
The results of clustering optimization analysis.
The results of clustering optimization analysis.