Abstract
We report the updated classification of Inborn Errors of Immunity/Primary Immunodeficiencies, compiled by the International Union of Immunological Societies Expert Committee. This report documents the key clinical and laboratory features of 430 inborn errors of immunity, including 64 gene defects that have either been discovered in the past 2 years since the previous update (published January 2018) or were characterized earlier but have since been confirmed or expanded upon in subsequent studies. The application of next-generation sequencing continues to expedite the rapid identification of novel gene defects, rare or common; broaden the immunological and clinical phenotypes of conditions arising from known gene defects and even known variants; and implement gene-specific therapies. These advances are contributing to greater understanding of the molecular, cellular, and immunological mechanisms of disease, thereby enhancing immunological knowledge while improving the management of patients and their families. This report serves as a valuable resource for the molecular diagnosis of individuals with heritable immunological disorders and also for the scientific dissection of cellular and molecular mechanisms underlying inborn errors of immunity and related human diseases.
Keywords: IUIS, primary immune deficiency, inborn errors of immunity, immune dysregulation, autoinflammatory disorders, next-generation sequencing
Inborn errors of immunity, also referred to as primary immunodeficiencies, manifest as increased susceptibility to infectious diseases, autoimmunity, autoinflammatory diseases, allergy, and/or malignancy. These conditions are caused by monogenic germline mutations that result in loss of expression, loss-of-function (LOF; amorphic/hypomorphic), or gain-of-function (GOF; hypermorphic) of the encoded protein [1, 2]. Heterozygous lesions may underlie autosomal dominant traits by GOF, haploinsufficiency, or negative dominance. Biallelic lesions typically cause autosomal recessive traits by LOF of the encoded protein (rarely GOF), while X-linked recessive traits arise from LOF of genes on the X chromosome, either in the hemizygous state in males or in the homozygous state in females. Rare X-linked dominant traits can also arise from LOF or GOF variants. This results in aberrant immunity due to the critical roles of these proteins in the development, maintenance and function of cells of the immune system, or cells other than leukocytes that contribute to immunity, during homeostasis and in response to external (e.g., infectious agents or environmental antigens) and internal (e.g., cytokines, self-antigens and cancer cells) stimuli [3–5]. Inborn errors of immunity were traditionally considered to be rare diseases, affecting ~ 1 in 10,000 to 1 in 50,000 births. However, with ongoing discovery of novel inborn errors of immunity (Fig. 1a) and improved definition of clinical phenotypes [6–8], the collective prevalence of these conditions is more likely to be at least 1/1000–1/5000 [9]. Indeed, more common inborn errors have recently been described [10]. Regardless of their exact incidence and prevalence, inborn errors of immunity represent an unprecedented model to link defined monogenic defects with clinical phenotypes of immune dysregulation, in a broad sense of the term. As a committee, we are aware that human immunity involves cells other than circulating or tissue leukocytes and that it can be scaled up from the immune system to the whole organism. Inborn errors of immunity have unequivocally revealed non-redundant roles of single genes and their products in immune function [3, 4, 6–8], formed the basis of improved mechanism-based therapies for the immunopathology underlying many diseases [8, 11], established immunological paradigms representing the foundations of basic, clinical and translational immunology [3–5, 9, 12–14], and provided insights into the molecular pathogenesis of more common diseases [9, 15]. Clear examples of these include:
The initial description by Bruton of X-linked agammaglobulinemia (XLA) and the ability to treat this condition with antibody replacement therapy (the mainstay treatment for antibody deficiency diseases such as CVID) [16]
The discovery of mutations in BTK [12] and the subsequent development of BTK-inhibitors such as ibrutinib for the treatment of B cell malignancies [14]
Progressive CD4 T cell deficiency explains opportunistic infections secondary to HIV infection [9].
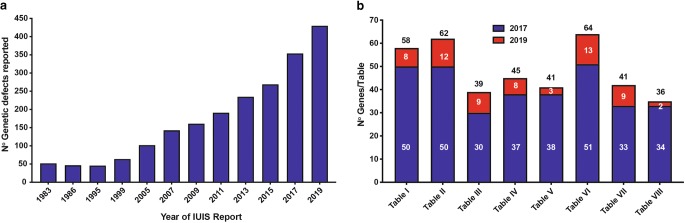
Fig. 1.
Rate of discovery of novel inborn errors of immunity: 1983–2019. a The number of genetic defects underlying monogenic immune disorders as reported by the IUIS/WHO committee in the indicated year. b The number of pathogenic gene variants listed in each table by the IUIS committee. Report published in 2017, and the number of new genes for each table contained in this report (red bars). The numbers in each column correspond to the number of genes reported in the 2017 IUIS update (blue bars) [1, 2], the number of new genes for each table contained in this report (red bars), and the total number of genes for each table. Note: only data for Tables 1, 2, 3, 4, 5, 6, 7, and 8 are shown, because Table 9 (bone marrow failure) is a new addition to the current report.
Thus, the study of inborn errors of immunity has provided profound advances in the practice of precision molecular medicine.
Since the early 1950s, when XLA was one of the first primary immune deficiencies to be described [16], clinical immunology has leveraged advances in the development of new methods to expedite the identification of defects of the immune system and the cellular, molecular, and genetic aberrations underlying these conditions. Indeed, the completion of the Human Genome Project in the early 2000s, coupled with rapid developments in next generation DNA sequencing (NGS) technologies, enabled the application of cost-effective and time-efficient sequencing of targeted gene panels, whole exomes, or whole genomes to cohorts of patients suspected of having a monogenic explanation for their disease. These platforms have led to a quantum leap in the identification and diagnosis of previously undefined genetically determined defects of the immune system (Fig. 1a, b; [6–8]).
The International Union of Immunological Societies Expert Committee of Inborn Errors of Immunity comprises pediatric and adult clinical immunologists, clinician/scientists and researchers in basic immunology from across the globe (https://iuis.org/committees/iei/). A major objective and responsibility of the committee is to provide the clinical and research communities with an update of genetic causes of immune deficiency and dysregulation. The committee has existed since 1970 and has published an updated report approximately every 2 years to inform the field of these advances (Fig. 1a). In March 2019, the committee met in New York to discuss and debate the inclusion of genetic variants published over the preceding 2 years (since June 2017) [1, 2], as well as gene mutations that had appeared in the literature earlier but, based on newly available evidence, were now substantiated (Fig. 1b).
Rather than simply including every gene variant reported, the committee applies very stringent criteria such that only those genes with convincing evidence of disease pathogenicity are classified as causes of novel inborn errors of immunity [17]. The Committee makes informed judgments for including new genetic causes of immunological conditions based on what we believe is most useful for practitioners caring for patients. Our current, and continuously evolving, practice is that criteria for inclusion can be met by several ways, for instance peer-reviewed publication of (1) multiple cases from unrelated kindreds, including detailed immunologic data, or (2) very few cases, or even a single case (see below), for whom compelling mechanistic/pathogenic data is also provided, generally from parallel studies in an animal or cell culture model.
Herein, we provide this latest update. The inborn errors of immunity are listed in 10 tables: Combined immunodeficiencies (Table 1), Combined immunodeficiencies with syndromic features (Table 2), Predominantly antibody deficiencies (Table 3), Diseases of immune dysregulation (Table 4), Congenital defects of phagocytes (Table 5), Defects in intrinsic and innate immunity (Table 6), Autoinflammatory diseases (Table 7), Complement deficiencies (Table 8), and Phenocopies of inborn errors of immunity (Table 10) (Fig. 1b). Since the last update (published January 2018) [1, 2], we have added a new table to consolidate genes that cause bone marrow failure (Table 9). Our division into phenotypes does not imply that the presentation is homogeneous. Rather, we recognize that substantial phenotypic and clinical heterogeneity exists within groups of patients with mutations in the same gene and even between individuals from the same pedigree with the identical gene mutation. To simplify the classification, each disorder has been listed only once, although distinct disorders due to mutations in the same gene, but with different modes of inheritance and pathogenic mechanisms are listed individually. Thus, several genes appear more than once in this update (some examples are listed below). Sub-divisions within each table segregate groups of disorders into coherent phenotypic sets. OMIM numbers are also provided within each table. If a OMIM number has not yet been issued for a particular genetic condition, then the number provided generally refers to the OMIM for that gene. Beneath each table, the new disorders added to this update are highlighted for easy reference.
Table 1.
Immunodeficiencies affecting cellular and humoral immunity
| Disease | Genetic defect | Inheritance | OMIM | T cells | B cells | Ig | Associated features |
|---|---|---|---|---|---|---|---|
| 1. T-B+ severe combined immune deficiency (SCID) | |||||||
| γc deficiency (common gamma chain SCID, CD132 deficiency) | IL2RG | XL | 308380 | Very low | Normal to high | Low | Low NK |
| JAK3 deficiency | JAK3 | AR | 600173 | Very low | Normal to high | Low | Low NK |
| IL7Rα deficiency | IL7R | AR | 146661 | Very low | Normal to high | Low | Normal NK |
| CD45 deficiency | PTPRC | AR | 151460 | Very low | Normal | Low | Normal γ/δ Τ cells |
| CD3δ deficiency | CD3D | AR | 186790 | Very low | Normal | Low | Normal NK, no γ/δ T cells |
| CD3ε deficiency | CD3E | AR | 186830 | Very low | Normal | Low | Normal NK, no γ/δ T cells |
| CD3ζ deficiency | CD3Z | AR | 186780 | Very low | Normal | Low | Normal NK, no γ/δ T cells |
| Coronin-1A deficiency | CORO1A | AR | 605000 | Very low | Normal | Low | Detectable thymus |
| LAT deficiency | LAT | AR | 602354 | Normal to low | Normal to low | High | Typical SCID or combined immunodeficiency, the latter with adenopathy, splenomegaly, recurrent infections, autoimmunity |
| 2. T-B- SCID | |||||||
| RAG deficiency |
RAG1 RAG2 |
AR |
179615 179616 |
Very low | Very low | Decreased | Normal NK cell number, but increased risk of graft rejection, possibly due to activated NK cells |
| DCLRE1C (Artemis) deficiency | DCLRE1C | AR | 605988 | Very low | Very low | Decreased | Normal NK cell number, but increased risk of graft rejection, possibly due to activated NK cells, radiation sensitivity |
| DNA PKcs deficiency | PRKDC | AR | 615966 | Very low | Very low | Variable | Normal NK, radiation sensitivity, microcephaly |
| Cernunnos/XLF deficiency | NHEJ1 | AR | 611290 | Very low | Very low | Decreased | Normal NK, radiation sensitivity, microcephaly |
| DNA ligase IV deficiency | LIG4 | AR | 601837 | Very low | Very low | Decreased | Normal NK, radiation sensitivity, microcephaly |
| Adenosine deaminase (ADA) deficiency | ADA | AR | 608958 | Very low | Low, decreasing | Low, decreasing | Low NK, bone defects, may have pulmonary alveolar proteinosis, cognitive defects |
| AK2 defect | AK2 | AR | 103020 | Very low | Very Low | Decreased | Reticular dysgenesis with neutropenia; deafness |
| Activated RAC2 defect | RAC2 | AD GOF | 602049 | Very low | Very Low | Low, poor specific antibody responses | Recurrent bacterial and viral infections, lymphoproliferation; neutropenia |
| 3. Combined immunodeficiency (CID), generally less profound than SCID | |||||||
| CD40 ligand (CD154) deficiency | CD40LG | XL | 308230 | Normal to low | sIgM+IgD+ naïve B cells present; IgG+, IgA+, IgE+ memory B cells absent | IgM normal or high, other Ig isotypes low | Severe and opportunistic infections, idiopathic neutropenia; hepatitis and cholangitis, Cryptosporidium infections, cholangiocarcinoma; autoimmune blood cytopenias; peripheral neuroectodermal tumors |
| CD40 deficiency | CD40 | AR | 606843 | Normal | Neutropenia, opportunistic infections, gastrointestinal and biliary tract and liver disease, Cryptosporidium infections | ||
| ICOS deficiency | ICOS | AR | 604558 | Normal | Normal | Low | Recurrent infections, autoimmunity, gastroenteritis, granulomas |
| ICOSL deficiency | ICOSLG | AR | 605717 | Low | Low | Low | Recurrent bacterial and viral infections, neutropenia |
| CD3γ deficiency | CD3G | AR | 186740 | Normal number, but low TCR expression | Normal | Normal | Immune deficiency and autoimmunity of variable severity |
| CD8 deficiency | CD8A | AR | 186910 | Absent CD8, Normal CD4 | Normal | Normal | Recurrent infections, may be asymptomatic |
| ZAP-70 deficiency (ZAP70 LOF) | ZAP70 | AR | 269840 | Low CD8 number, normal CD4 number but with poor function | Normal | Normal | May have immune dysregulation, autoimmunity |
| ZAP-70 combined hypomorphic and activating mutations | ZAP70 | AR (LOF/GOF) | 617006 | Decreased CD8, normal or decreased CD4 cells | Normal or decreased | Normal IgA, low IgM, low/normal IgG; protective Ab responses to vaccines | Severe autoimmunity (bullous pemphigoid, inflammatory colitis |
| MHC class I deficiency | TAP1 | AR | 170260 | Low CD8, normal CD4, absent MHC I on lymphocytes | Normal | Normal | Vasculitis, pyoderma gangrenosum |
| TAP2 | AR | 170261 | |||||
| TAPBP | AR | 601962 | |||||
| B2M | AR | 109700 | Sinopulmonary infections, cutaneous granulomas. Absent β2m associated proteins MHC-I, CD1a, CD1b, and CD1c | ||||
| MHC class II deficiency group A, B, C, D | CIITA | AR | 600005 | Low CD4+ T cells, reduced MHC II expression on lymphocytes | Normal | Normal to low | Failure to thrive, respiratory and gastrointestinal infections, liver/biliary tract disease |
| RFXANK | AR | 603200 | |||||
| RFX5 | AR | 601863 | |||||
| RFXAP | AR | 601861 | |||||
| IKAROS deficiency | IKZF1 | AD DN | 603023 | no memory T cells | no memory B cells | Low Ig, | recurrent sinopulmonary infections, pneumocystis early CID onset |
| DOCK8 deficiency | DOCK8 | AR | 243700 | T cell lymphopenia, reduced naïve CD8 T cells, increased exhausted CD8+ TEM cells, reduced MAIT, NKT cells, increased γδ T cells; poor proliferation; few Treg with poor function | increased total B cells, reduced memory B cells Poor peripheral B cell tolerance. | Low IgM, normal/high IgG and IgA, very high IgE, poor antibody responses | Low NK cells with poor function. Eosinophilia, recurrent infections, cutaneous viral, fungal and staphylococcal infections, severe atopy/allergic disease, cancer diathesis |
| DOCK2 deficiency | DOCK2 | AR | 603122 | Low | Normal | IgG normal or low, poor antibody responses | Early invasive herpes viral, bacterial infections, Normal NK cell number, but defective function. Poor interferon responses in hematopoietic and non-hematopoietic cells |
| Polymerase and deficiency |
POLD1 POLD2 |
AR |
174761 600815 |
Low CD4 T cells | Low B cells but normal maturation | Low igG | Recurrent respiratory tract infections, skin infections, warts and molluscum, short stature, intellectual disability |
| RHOH deficiency | RHOH | AR | 602037 | Normal, few naïve T cells, restricted repertoire, poor proliferation to CD3 | Normal | Normal | HPV infection, lung granulomas, molluscum contagiosum, lymphoma |
| STK4 deficiency | STK4 | AR | 614868 | CD4 lymphopenia, reduced naïve T cells, increased TEM and TEMRA cells, poor proliferation | Reduced memory B cells | Reduced IgM, increased IgG, IgA, IgE; impaired Ab responses | Intermittent neutropenia, bacterial, viral (HPV, EBV, molluscum), candidal infections, lymphoproliferation, autoimmune cytopenias, lymphoma, congenital heart disease |
| TCRα deficiency | TRAC | AR | 615387 | Absent TCRαβ except for a minor CD3-dim TCRαβ population; most T cells γδ; poor proliferation | Normal | Normal | Recurrent viral, bacterial, fungal infections, immune dysregulation and autoimmunity, diarrhea |
| LCK deficiency | LCK | AR | 615758 | Low CD4+, low Treg, restricted T cell repertoire, poor TCR signaling | Normal | Normal IgG and IgA, high IgM | Recurrent infections, immune dysregulation, autoimmunity |
| ITK deficiency | ITK | AR | 186973 | Progressive CD4 T cell lymphopenia; reduced T cell activation | Normal | Normal to low serum Ig | EBV associated B cell lymphoproliferation, lymphoma, immune dysregulation |
| MALT1 deficiency | MALT1 | AR | 615468 | Normal number, poor proliferation | Normal | Normal levels, poor specific antibody response | Bacterial, fungal and viral infections |
| CARD11 deficiency | CARD11 | AR LOF | 615206 | Normal number, predominantly naïve T cells, poor proliferation | Normal, transitional B cell predominance | Absent/low | Pneumocystis jirovecii pneumonia, bacterial and viral infections |
| BCL10 deficiency | BCL10 | AR | 616098 | Normal number, few memory T and Treg cells, poor antigen and anti-CD3 proliferation | Normal number, decreased memory and switched B cells | Low | Recurrent bacterial and viral infections, candidiasis, gastroenteritis |
| IL-21 deficiency | IL21 | AR | 615767 | Normal number, normal/low function | Low, decreased memory and switched B cells | Hypogammaglobulinemia, poor specific antibody responses; increased IgE | Severe early onset colitis, recurrent sinopulmonary infections |
| IL-21R deficiency | IL21R | AR | 615207 | Normal number, low cytokine production, poor antigen proliferation | Normal, decreased memory and switched B cells | Recurrent infections, Pneumocystis jiroveci, Cryptosporidium infections, liver disease | |
| OX40 deficiency | TNFRSF4 | AR | 615593 | Normal numbers, low antigen specific memory CD4+ | Normal numbers, low memory B cells | Normal | Impaired immunity to HHV8, Kaposi’s sarcoma |
| IKBKB deficiency | IKBKB | AR | 615592 | Normal number, absent Treg and γ/δ T cells, impaired TCR activation | Normal number, poor function | Low | Recurrent bacterial, viral, fungal infections, opportunistic infections |
| NIK deficiency | MAP 3 K14 | AR | 604655 | Normal number, poor proliferation to antigen | Low, low switched memory B cells | Low Ig’s | Low NK number and function, recurrent bacterial, viral and Cryptosporidium infections |
| RelB deficiency | RELB | AR | 604758 | Normal number, poor diversity, reduced proliferation to mitogens; no response to Ag | Marked increase in B cell number | Normal Ig levels but Impaired specific antibody responses | Recurrent infections |
| RelA haploinsufficiency | RELA | AD | 618287 | Normal/increased | Normal | Normal | Chronic mucocutaneous ulceration, Impaired NFkB activation; reduced production of inflammatory cytokines |
| Moesin deficiency | MSN | XL | 300988 | Normal number, defective migration, proliferation | Low number | Low Ig’s over time | Recurrent infections with bacteria, varicella, neutropenia |
| TFRC deficiency | TFRC | AR | 616740 | Normal number, poor proliferation | Normal number, low memory B cells | Low | Recurrent infections, neutropenia, thrombocytopenia |
| c-Rel deficiency | REL | AR | 164910 | Normal, decreased memory CD4, poor proliferation | Low, mostly naïve; few switched memory B cells, impaired proliferation | Low, poor specific antibody responses | Recurrent infections with bacteria, mycobacteria, salmonella and opportunistic organisms. Defective innate immunity |
| FCHO1 deficiency | FCHO1 | AR | 613437 | Low, poor proliferation | Normal number | Normal | Recurrent infections (viral, mycobacteria, bacterial, fungal), lymphoproliferation, failure to thrive, increased activation-induced T cell death, defective clathrin-mediated endocytosis |
SCID/CID spectrum: Infants with SCID who have maternal T cell engraftment may have T cells in normal numbers that do not function normally; these cells may cause autoimmune cytopenias or graft versus host disease. Hypomorphic mutations in several of the genes that cause SCID may result in Omenn syndrome (OS), or “leaky” SCID, or still less profound combined immunodeficiency (CID) phenotypes. Both OS and leaky SCID can be associated with > 300 autologous T cells/μL of peripheral blood and reduced, rather than absent, proliferative responses when compared with typical SCID caused by null mutations. A spectrum of clinical findings including typical SCID, OS, leaky SCID, CID, granulomas with T lymphopenia, autoimmunity and CD4 T lymphopenia can be found in an allelic series of RAG1/2 and other SCID-associated genes. There can be clinical overlap between some genes listed here and those listed in Table 7
Total number of disorders in Table 1: 50
Total number of mutant genes: 58
New inborn errors of immunity: 8; New inborn errors of immunity: 8; RAC2 GOF [18–21]; ICOSLG [22]; AD DN IKZF1 [23]; POLD1 [24, 25]; POLD2 [24]; RELA [26, 27]; REL [28]; FCHO1 [29]
SCID severe combined immunodeficiency, CID combined immunodeficiency, EBV Epstein-Barr virus, MHC major histocompatibility complex, HPV human papillomavirus, Treg T regulatory cell, XL X-linked inheritance, AR autosomal recessive inheritance, AD autosomal dominant inheritance, LOF loss-of-function, GOF gain-of-function
Table 2.
Combined immunodeficiencies with associated or syndromic features
| Disease | Genetic defect | Inheritance | OMIM | T cells | B cells | Ig | Associated features |
|---|---|---|---|---|---|---|---|
| 1. Immunodeficiency with congenital thrombocytopenia | |||||||
| Wiskott-Aldrich syndrome (WAS LOF) | WAS | XL | 300392 | Progressive decrease in numbers, abnormal lymphocyte responses to anti-CD3 | Normal numbers | Low IgM and antibody responses to polysaccharides, often high IgA and IgE | Thrombocytopenia with small platelets, eczema, recurrent bacterial/viral infections, bloody diarrhea, lymphoma, autoimmune disease, IgA- nephropathy. Patients with XL-thrombocytopenia have later onset of complications and more favourable life expectancy but eventually develop similar complications as observed in WAS |
| WIP deficiency | WIPF1 | AR | 602357 | Reduced, defective lymphocyte responses to anti-CD3 | Normal or low | Normal, except for high IgE | Thrombocytopenia with or without small platelets, recurrent bacterial and viral infections, eczema, bloody diarrhea; WAS protein absent |
| Arp2/3-mediated filament branching defect | ARPC1B | AR | 604223 | Normal | Normal numbers | Normal except for high IgA and IgE | Mild thrombocytopenia with normal sized platelets, recurrent invasive infections; colitis, vasculitis, autoantibodies (ANA, ANCA), eosinophilia; defective Arp2/3 filament branching |
| 2. DNA repair defects other than those listed in Table 1 | |||||||
| Ataxia-telangiectasia | ATM | AR | 607585 | Progressive decrease, poor proliferation to mitogens; may have low TRECs and T cells by newborn screening (NBS) | Normal | Often low IgA, IgE and IgG subclasses, increased IgM monomers; antibodies variably decreased | Ataxia, telangiectasia especially of sclerae; pulmonary infections; lymphoreticular and other malignancies; increased alpha fetoprotein; increased radiosensitivity, chromosomal instability and chromosomal translocations |
| Nijmegen breakage syndrome | NBS1 | AR | 602667 | Progressive decrease; may have low TRECs and T cells by NBS | Variably reduced | Often low IgA, IgE, and IgG subclasses, increased IgM; antibodies variably decreased | Microcephaly, dysmorphic facies; lymphomas and solid tumors; increased radiosensitivity;, chromosomal instability |
| Bloom syndrome | BLM | AR | 604610 | Normal | Normal | Low | Short stature, dysmorphic facies sun-sensitive erythema; marrow failure; leukemia, lymphoma; chromosomal instability |
| Immunodeficiency with centromeric instability and facial anomalies (ICF types 1, 2, 3, 4) | DNMT3B | AR | 602900 | Decreased or normal, responses to PHA may be decreased | Decreased or normal | Hypogammaglobulinemia or agammaglobulinemia, variable antibody deficiency | Facial dysmorphic features, developmental delay, macroglossia; bacterial/opportunistic infections; malabsorption; cytopenias; malignancies; multiradial configurations of chromosomes 1, 9, 16 |
| ZBTB24 | AR | 614064 | Decreased or normal | Facial dysmorphic features, macroglossia; bacterial/opportunistic infections; malabsorption; cytopenias; malignancies; multiradial configurations of chromosomes 1, 9, 16 | |||
| CDCA7 | AR | 609937 | Decreased or normal; responses to PHA may be decreased | ||||
| HELLS | AR | 603946 | Decreased or normal | ||||
| PMS2 deficiency | PMS2 | AR | 600259 | Normal | Low B cells, switched and non-switched | Low IgG and IgA, high IgM, abnormal antibody responses | Recurrent infections; café-au-lait spots; lymphoma, colorectal carcinoma, brain tumors |
| RNF168 deficiency (Radiosensitivity, Immune Deficiency, Dysmorphic features, Learning difficulties [RIDDLE] syndrome) | RNF168 | AR | 612688 | Normal | Normal | Low IgG or IgA | Short stature, mild defect of motor control to ataxia; normal intelligence to learning difficulties; mild facial dysmorphism to microcephaly; increased radiosensitivity |
| MCM4 deficiency | MCM4 | AR | 602638 | Normal | Normal | Normal | NK cells: low number and function; viral infections (EBV, HSV, VZV); short stature; B cell lymphoma; adrenal failure |
| X-linked reticulate pigmentary disorder (POLA1 deficiency) | POLA1 | XL | 301220 | Not assessed | Not assessed | Not assessed | Hyperpigmentation, characteristic facies, lung and GI involvement |
| POLE1 (Polymerase ε subunit 1) deficiency (FILS syndrome) | POLE1 | AR | 174762 | Normal; decreased T cell proliferation | Low memory B cells | Low IgG2 and IgM, lack of antibody to PPS | Recurrent respiratory infections, meningitis; facial dysmorphism, livido, short stature |
| POLE2 (Polymerase ε subunit 2) deficiency | POLE2 | AR | 602670 | Lymphopenia, lack of TRECS at NBS, absent proliferation in response to antigens | Very low | Hypogammaglobulinemia | Recurrent infections, disseminated BCG infections; autoimmunity (type 1 diabetes), hypothyroidism, facial dysmorphism |
| Ligase I deficiency | LIG1 | AR | 126391 | Lymphopenia, increased γδ T cells, decreased mitogen response | Normal | Hypogammaglobulinemia, Reduced antibody responses | Recurrent bacterial and viral infections; growth retardation; sun sensitivity, radiation sensitivity; macrocytic red blood cells |
| NSMCE3 deficiency | NSMCE3 | AR | 608243 | Decreased number, poor responses to mitogens and antigens | Normal | Normal IgG, IgA, normal to elevated IgM; decreased antibody responses to PPS | Severe lung disease (possibly viral); thymic hypoplasia; chromosomal breakage, radiation sensitivity |
| ERCC6L2 (Hebo deficiency) | ERCC6L2 | AR | 615667 | Lymphopenia | Low | Normal | Facial dysmorphism, microcephaly; bone marrow failure |
| GINS1 deficiency | GINS1 | AR | 610608 | Low or normal | Low or normal | High IgA, low IgM and IgG | Neutropenia; IUGR; NK cells very low |
| 3. Thymic defects with additional congenital anomalies | |||||||
|
DiGeorge/velocardio-facial syndrome Chromosome 22q11.2 deletion syndrome (22q11.2DS) |
Large deletion (3 Mb) typically in chromosome 22 (TBX1) | AD | 602054 | Decreased or normal, 5% have low TRECs at NBS and < 1500 CD3T cells/μL in neonatal period | Normal | Normal or decreased | Hypoparathyroidism; conotruncal cardiac malformation, velopalatal insufficiency; abnormal facies; intellectual disability |
| DiGeorge/velocardio-facial syndrome | Unknown | Sporadic | Decreased or normal | ||||
| TBX1 deficiency | TBX1 | AD | 602054 | Decreased or normal, may have low TRECs at NBS | |||
| CHARGE syndrome | CHD7 | AD | 608892 | Decreased or normal, may have low TRECs at NBS; response to PHA may be decreased | Normal | Normal or decreased | Coloboma of eye; heart anomaly; choanal atresia; intellectual disability; genital and ear anomalies, CNS malformation; some are SCID-like |
| SEMA3E | AD | 608166 | |||||
| Unknown | |||||||
| Winged helix nude FOXN1 deficiency | FOXN1 | AR | 601705 | Very low | Normal | Decreased | Severe infections; abnormal thymic epithelium, immunodeficiency; congenital alopecia, nail dystrophy; neural tube defect |
| FOXN1 haploinsufficiency | FOXN1 | AD | 600838 | Severe T cell lymphopenia at birth, normalised by adulthood | Normal/low | Not assessed | Recurrent, viral and bacterial respiratory tract infections; skin involvement (eczema, dermatitis), nail dystrophy |
| Chromosome 10p13-p14 deletion syndrome (10p13-p14DS) | Del10p13-p14 | AD | 601362 | Normal, rarely lymphopenia and decreased lymphoproliferation to mitogens and antigens; hypoplastic thymus may be present | Normal | Normal | Hypoparathyroidism; renal disease; deafness; growth retardation; facial dysmorphism; cardiac defects may be present; recurrent infections ± |
| Chromosome 11q deletion syndrome (Jacobsen syndrome) | 11q23del | AD | 147791 | Lymphopenia; low NK cells | Decreased B cells and switched memory B cells | Hypogammaglobulinemia, decreased antibody responses | Recurrent respiratory infections; multiple warts; facial dysmorphism, growth retardation |
| 4. Immuno-osseous dysplasias | |||||||
| Cartilage hair hypoplasia (CHH) | RMRP | AR | 157660 | Varies from severely decreased (SCID) to normal; impaired lymphocyte proliferation | Normal | Normal or reduced, antibodies variably decreased | Short-limbed dwarfism with metaphyseal dysostosis; sparse hair; bone marrow failure; autoimmunity; susceptibility to lymphoma and other cancers; impaired spermatogenesis; neuronal dysplasia of the intestine |
| Schimke immuno-osseous dysplasia | SMARCAL1 | AR | 606622 | Decreased | Normal | Normal | Short stature, spondiloepiphyseal dysplasia, intrauterine growth retardation; nephropathy; bacterial, viral, fungal infections; may present as SCID; bone marrow failure |
| MYSM1 deficiency | MYSM1 | AR | 612176 | T cell lymphopenia, reduced naïve T cells, low NK cells | B cell deficiency | Hypogammaglobulinemia | Short stature; recurrent infections; congenital bone marrow failure, myelodysplasia; immunodeficiency affecting B cells and granulocytes; skeletal anomalies; cataracts; developmental delay |
| MOPD1 deficiency (Roifman syndrome) | RNU4ATAC | AR | 601428 | Decreased NK cell function | Decreased total and memory B cells | Hypogammaglobulinemia, variably decreased specific antibodies | Recurrent bacterial infections; lymphadenopathy; spondyloepiphyseal dysplasia, extreme intrauterine growth retardation; retinal dystrophy; facial dysmorphism; may present with microcephaly; short stature |
| Immunoskeletal dysplasia with neurodevelopmental abnormalities (EXTL3 deficiency) | EXTL3 | AR | 617425 | Decreased | Normal | Decreased to normal | Short stature; cervical spinal stenosis, neurodevelopmental impairment; eosinophilia; may have early infant mortality |
| 5. Hyper IgE syndromes (HIES) | |||||||
| AD-HIES STAT3 deficiency (Job syndrome) | STAT3 | AD LOF (dominant negative) | 147060 | Normal overall; Th17, T follicular helper, MAIT, NKT cells decreased, Tregs may be increased; impaired responses to STAT3-activatng cytokines | Normal, reduced memory B cells, BAFF expression increased, impaired responses to STAT3-activating cytokines | Very high IgE, specific antibody production decreased | Distinctive facial features (broad nasal bridge); bacterial infections (boils, pulmonary abscesses, pneumatoceles) due to S. aureus, pulmonary aspergillus, Pneumocystis jirovecii; eczema, mucocutaneous candidiasis; hyperextensible joints, osteoporosis and bone fractures, scoliosis, retained primary teeth; coronary and cerebral aneurysms |
| IL6 receptor deficiency | IL6R | AR | 147880 | Normal/increased; normal responses to mitogens | Normal total and memory B; reduced switched memory B | Normal/low serum IgM, G, A. Very high IgE; specific antibody production low | Recurrent pyogenic infections, cold abscesses; high circulating IL-6 levels |
| IL6 signal transducer (IL6ST) deficiency | IL6ST | AR | 618523 | Decreased Th17 cells | Reduced switched and non-switched memory B cells | High IgE, specific antibody production variably affected | Bacterial infections, boils, eczema, pulmonary abscesses, pneumatoceles; bone fractures; scoliosis; retention of primary teeth; craniosynostosis |
|
ZNF341 deficiency AR-HIES |
ZNF341 | AR | 618282 | Decreased Th17 and NK cells | Normal, reduced memory B cells, impaired responses to STAT3-activaitng cytokines | High IgE and IgG, specific antibody production decreased | Phenocopy of AD-HIES; mild facial dysmorphism; early onset eczema, MCC, bacterial skin infections, abscesses, recurrent bacterial respiratory infections (S. aureus), lung abscesses and pneumatoceles; hyperextensible joints; bone fractures and retention of primary teeth |
| ERBIN deficiency | ERBB2IP | AD | 606944 | Increased circulating Treg | Normal | Moderately increased IgE | Recurrent respiratory infections, susceptibility to S. aureus, eczema; hyperextensible joints, scoliosis; arterial dilatation in some patients |
| Loeys-Dietz syndrome (TGFBR deficiency) | TGFBR1 | AD | 609192 | Normal | Normal | Elevated IgE | Recurrent respiratory infectons; eczema, food allergies; hyper-extensible joints, scoliosis, retention of primary teeths; aortic aneurisms. |
| TGFBR2 | 610168 | ||||||
| Comel-Netherton syndrome | SPINK5 | AR | 605010 | Normal | Low switched and non-switched B cells | High IgE and IgA, Antibody variably decreased | Congenital ichthyosis, bamboo hair, atopic diathesis; increased bacterial infections; failure to thrive |
| PGM3 deficiency | PGM3 | AR | 172100 | CD8 and CD4 T cells may be decreased | Low B and memory B cells | Normal or elevated IgG and IgA, most with high IgE, eosinophilia | Severe atopy; autoimmunity; bacterial and viral infections; skeletal anomalies/dysplasia: short stature, brachydactyly, dysmorphic facial features; intellectual disability and cognitive impairment, delayed CNS myelination in some affected individuals |
| CARD11 deficiency (heterozygous DN) | CARD11 | AD LOF | 617638 | Normal overall, but defective T cell activation and proliferation; skewing toward Th2 | Normal to low | High IgE, poor specific antibody production; impaired activation of both NF-κB and mTORC1 pathways | Variable atopy, eczema, food allergies, eosinophilia; cutaneous viral infections, recurrent respiratory infections; lymphoma; CID |
| 6. Defects of vitamin B12 and folate metabolism | |||||||
| Transcobalamin 2 deficiency | TCN2 | AR | 613441 | Normal | Variable | Decreased | Megaloblastic anemia, pancytopenia; if untreated (B12) for prolonged periods results in intellectual disability |
| SLC46A1/PCFT deficiency causing hereditary folate malabsorption | SLC46A1 | AR | 229050 | Variable numbers and activation profile | Variable | Decreased | Megaloblastic anemia, failure to thrive; if untreated for prolonged periods results in intellectual disability |
| Methylene-tetrahydrofolate dehydrogenase 1 (MTHFD1) deficiency | MTHFD1 | AR | 172460 | Low thymic output, normal in vitro proliferation | Low | Decreased/poor antibody responses to conjugated polysaccharide antigens | Recurrent bacterial infection, Pneumocystis jirovecii; megaloblastic anemia; failure to thrive; neutropenia; seizures, intellectual disability; folate-responsive |
| 7. Anhidrotic ectodermodysplasia with immunodeficiency (EDA-ID) | |||||||
| EDA-ID due to NEMO/IKBKG deficiency (ectodermal dysplasia, immune deficiency) | IKBKG | XL | 300248 | Normal or decreased, TCR activation impaired | Normal; Low memory and isotype switched B cells | Decreased, some with elevated IgA, IgM, poor specific antibody responses, absent antibodies to polysaccharide antigens | Anhidrotic ectodermal dysplasia (in some); various infections (bacteria, mycobacteria, viruses, fungi); colitis; conical teeth, variable defects of skin, hair and teeth; monocyte dysfunction |
| EDA-ID due to IKBA GOF mutation | NFKBIA | AD GOF | 164008 | Normal total T cells, TCR activation impaired | Normal B cell numbers, impaired BCR activation, low memory and isotype switched B cells | Decreased IgG and IgA, elevated IgM, poor specific antibody responses, absent antibody to polysaccharide antigens | Anhidrotic ectodermal dysplasia; various infections (bacteria, mycobacteria, viruses, fungi); colitis; variable defects of skin, hair and teeth; T cell and monocyte dysfunction |
| EDA-ID due to IKBKB GOF mutation | IKBKB | AD GOF | 618204 | Decreased T cells, impaired TCR activation | Normal number, poor function | Reduced | Recurrent bacterial, viral, fungal infections; variable ectodermal defects |
| 8. Calcium channel defects | |||||||
| ORAI-1 deficiency | ORAI1 | AR | 610277 | Normal, defective TCR mediated activation | Normal | Normal | Autoimmunity; EDA; non-progressive myopathy |
| STIM1 deficiency | STIM1 | AR | 605921 | ||||
| 9. Other defects | |||||||
| Purine nucleoside phosphorylase (PNP) deficiency | PNP | AR | 164050 | Progressive decrease | Normal | Normal or low | Autoimmune hemolytic anemia; neurological impairment |
| Immunodeficiency with multiple intestinal atresias | TTC7A | AR | 609332 | Variable, but sometimes absent or low TRECs at NBS; may have SCID phenotype at birth | Normal or low | Markedly decreased IgG, IgM, IgA | Bacterial (sepsis), fungal, viral infections; multiple intestinal atresias, often with intrauterine polyhydramnios and early demise |
| Tricho-Hepato-Enteric Syndrome (THES) | TTC37 | AR | 222470 | Impaired IFNγ production | Variably low numbers of switched memory B cells | Hypogammaglobulinemia, may have low antibody responses | Respiratory infections; IUGR; facial dysmorphic features, wooly hair; early onset intractable diarrhea, liver cirrhosis; platelet abnormalities |
| SKIV2L | 614602 | ||||||
| Hepatic veno-occlusive disease with immunodeficiency (VODI) | SP110 | AR | 604457 | Normal (decreased memory T cells) | Normal (decreased memory B cells) | Decreased IgG, IgA, IgM, absent germinal center and tissue plasma cells | Hepatic veno-occlusive disease; susceptibility to Pneumocystis jirovecii pneumonia, CMV, candida; thrombocytopenia; hepatosplenomegaly; cerebrospinal leukodystrophy |
| BCL11B deficiency | BCL11B | AD | 617237 | Low, poor proliferation | Normal | Normal | Congenital abnormalities, neonatal teeth, dysmorphic facies; absent corpus callosum, neurocognitive deficits |
| EPG5 deficiency (Vici syndrome) | EPG5 | AR | 615068 | Profound depletion of CD4+ cells | Defective | Decreased (particularly IgG2) | Agenesis of the corpus callosum; cataracts; cardiomyopathy; skin hypopigmentation; intellectual disability; microcephaly; recurrent infections, chronic mucocutaneous candidiasis |
| HOIL1 deficiency | RBCK1 | AR | 610924 | Normal numbers | Normal, decreased memory B cells | Poor antibody responses to polysaccharides | Bacterial infections; autoinflammation; amylopectinosis |
| HOIP deficiency | RNF31 | AR | 612487 | Normal numbers | Normal, decreased memory B cells | decreased | Bacterial infections; autoinflammation; amylopectinosis; lymphangiectasia |
| Hennekam-lymphangiectasia-lymphedema syndrome | CCBE1 | AR | 612753 | Low/variable | Low/variable | decreased | Lymphangiectasia and lymphedema with facial abnormalities and other dysmorphic features |
| FAT4 | AR | 612411 | Low/variable | Low/variable | decreased | Lymphangiectasia and lymphedema with facial abnormalities and other dysmorphic features | |
| Activating de novo mutations in nuclear factor, erythroid 2- like (NFE2L2) | NFE2L2 | AD | 617744 | Not reported | Decreased switched memory B cells | Hypogammaglobulinemia, decreased antibody responses | Recurrent respiratory and skin infections; growth retardation, developmental delay; white matter cerebral lesions; increased level of homocysteine; increased expression of stress response genes |
| STAT5b deficiency | STAT5B | AR | 245590 | Modestly decreased, reduced Treg number and function | Normal | hypergammaglobulinemia, increased IgE | Growth-hormone insensitive dwarfism; dysmorphic features; eczema; lymphocytic interstitial pneumonitis; prominent autoimmunity |
| STAT5b deficiency | STAT5B | AD (dominant negative) | 604260 | Normal | Normal | Increased IgE | Growth-failure; eczema (no immune defects compared to AR STAT5 deficiency) |
| Kabuki syndrome (type 1 and 2) | KMT2D | AD | 602113 | Normal | Normal | Low IgA and occasionally low IgG | Typical facial abnormalities, cleft or high arched palate, skeletal abnormalities, short stature; intellectual disability; congenital heart defects; recurrent infections (otitis media, pneumonia) in 50% of patients; autoimmunity may be present |
| KDM6A | XL (females may be affected) | 300128 | |||||
| KMT2A deficiency (Wiedemann-Steiner syndrome) | KMT2A | AD | 605130 | Normal | Decreased switched and non-switched memory B cells | Hypogammaglobulinemia, decreased antibody responses | Respiratory infections; short stature; hypertelorism; hairy elbows; developmental delay, intellectual disability |
Total number of disorders in Table 2: 58
Total number of mutant genes in Table 2: 62
New inborn errors of immunity: 12; LIG1 [30]; FOXN1 haploinsufficiency [31]; IL6R [32, 33]; IL6ST [34, 35]; ZNF341 [36, 37]; ERBB2IP [38]; TGFBR1 [39]; TGFBR2 [39]; AD LOF CARD11 [40, 41]; AD GOF IKBKB [42]; SKIV2L [43]; NFE2L2 [44]
Unknown cause of DiGeorge syndrome, unknown cause of CHARGE syndrome, unknown gene(s) within 10p13–14 deletion responsible for phenotype
EDA ectodermal dysplasia anhydrotic, HSV herpes simplex virus, VZV varicella zoster virus, BCG Bacillus Calmette-Guerin, NBS newborn screen, TREC T cell receptor excision circle (biomarker for low T cells used in NBS), IUGR interuterine growth retardation
Table 3.
Predominantly antibody deficiencies
| Disease | Genetic defect | Inheritance | OMIM | Ig | Associated features |
|---|---|---|---|---|---|
| 1. Severe reduction in all serum immunoglobulin isotypes with profoundly decreased or absent B cells, agammaglobulinemia | |||||
| BTK deficiency, X-linked agammaglobulinemia (XLA) | BTK | XL | 300300 | All isotypes decreased in majority of patients, some patients have detectable immunoglobulins | Severe bacterial infections, normal numbers of pro-B cells |
| μ heavy chain deficiency | IGHM | AR | 147020 | All isotypes decreased | Severe bacterial infections, normal numbers of pro-B cells |
| λ5 deficiency | IGLL1 | AR | 146770 | ||
| Igα deficiency | CD79A | AR | 112205 | ||
| Igβ deficiency | CD79B | AR | 147245 | ||
| BLNK deficiency | BLNK | AR | 604515 | ||
| p110δ deficiency | PIK3CD | AR | 602839 | Severe bacterial infections; autoimmune complications (IBD) | |
| p85 deficiency | PIK3R1 | AR | 615214 | Severe bacterial infections, cytopenias, decreased or absent pro-B cells | |
| E47 transcription factor deficiency | TCF3 | AD | 616941 | Recurrent bacterial infections | |
| TCF3 | AR | 147141 | Severe, recurrent bacterial infections, failure to thrive | ||
| SLC39A7 (ZIP7) deficiency | SLC39A7 | AR | 601416 | Early onset infections, blistering dermatosis, failure to thrive, thrombocytopenia | |
| Hoffman syndrome/TOP2B deficiency | TOP2B | AD | 126431 | Recurrent infections, facial dysmorphism, limb anomalies | |
| 2. Severe reduction in at least 2 serum immunoglobulin isotypes with normal or low number of B cells, CVID phenotype | |||||
| Common variable immune deficiency with no gene defect specified (CVID) | Unknown | Variable | Low IgG and IgA and/or IgM | Clinical phenotypes vary: most have recurrent infections, some have polyclonal lymphoproliferation, autoimmune cytopenias and/or granulomatous disease | |
| Activated p110δ syndrome (APDS) | PIK3CD GOF | AD | 615513 (APDS1) | Normal/increased IgM, reduced IgG and IgA | Severe bacterial infections; reduced memory B cells and increased transitional B cells, EBV ± CMV viremia, lymphadenopathy/splenomegaly, autoimmunity, lymphoproliferation, lymphoma |
| PIK3R1 | AD | 616005 (APDS2) | Severe bacterial infections, reduced memory B cells and increased transitional B cells, lymphadenopathy/splenomegaly, lymphoproliferation, lymphoma; developmental delay | ||
| PTEN deficiency (LOF) | PTEN | AD | 158350 | Normal/Decreased | Recurrent infections, Lymphoproliferation, Autoimmunity; developmental delay |
| CD19 deficiency | CD19 | AR | 107265 | Low IgG and IgA and/or IgM | Recurrent infections, may have glomerulonephritis (CD81 mutation abolishes expression of CD19, thereby phenocopying CD19 mutations) |
| CD81 deficiency | CD81 | AR | 186845 | Low IgG, low or normal IgA and IgM | |
| CD20 deficiency | CD20 | AR | 112210 | Low IgG, normal or elevated IgM and IgA | Recurrent infections |
| CD21 deficiency | CD21 | AR | 120650 | Low IgG, impaired anti-pneumococcal response | Recurrent infections |
| TACI deficiency# | TNFRSF13B | AR or AD | 604907 | Low IgG and IgA and/or IgM | Variable clinical expression and penetrance for monoallelic variants |
| BAFF receptor deficiency | TNFRSF13C | AR | 606269 | Low IgG and IgM, | Variable clinical expression |
| TWEAK deficiency | TNFSF12 | AD | 602695 | Low IgM and A, lack of anti-pneumococcal antibody | Pneumonia, bacterial infections, warts, thrombocytopenia. Neutropenia |
| TRNT1 deficiency | TRNT1 | AR | 612907 | B cell deficiency and hypogammaglobulinemia | congenital sideroblastic anemia, deafness, developmental delay |
| NFKB1 deficiency | NFKB1 | AD | 164011 | Normal or low IgG, IgA, IgM, low or normal B cells, low memory B cells | Recurrent sinopulmonary infections, COPD, EBV proliferation, autoimmune cytopenias, alopecia and autoimmune thyroiditis |
| NFKB2 deficiency | NFKB2 | AD | 615577 | Low serum IgG, A and M; low B cell numbers | Recurrent sinopulmonary infections, alopecia and endocrinopathies |
| IKAROS deficiency | IKZF1 | AD (haploinsufficiency) | 603023 | Low IgG, IgA, IgM, low or normal B cells; B cells and Ig levels reduce with age | Decreased pro-B cells, recurrent sinopulmonary infections; increased risk of ALL, autoimmunity, CVID phenotype |
| IRF2BP2 deficiency | IRF2BP2 | AD | 615332 | Hypogammaglobulinemia, absent IgA | Recurrent infections, possible autoimmunity and inflammatory disease |
| ATP6AP1 deficiency | ATP6AP1 | XL | 300972 | Variable immunoglobulin findings | Hepatopathy, leukopenia, low copper |
| ARHGEF1 deficiency | ARHGEF1 | AR | 618459 | Hypogammaglobulinemia; lack of antibody | Recurrent infections, bronchiectasis |
| SH3KBP1 (CIN85) deficiency | SH3KBP1 | XL | 300310 | IgM, IgG deficiency; loss of antibody | Severe bacterial infections |
| SEC61A1 deficiency | SEC61A1 | AD | 609213 | Hypogammaglobulinemia | Severe recurrent respiratory tract infections |
| RAC2 deficiency | RAC2 | AR | 602049 | Low IgG, IgA, IgM, low or normal B cells; reduced Ab responses following vaccination | Recurrent sinopulmonary infections, selective IgA deficiency; poststreptococcal glomerulonephritis; urticaria |
| Mannosyl-oligosaccharide glucosidase deficiency | MOGS | AR | 601336 | Low IgG, IgA, IgM, increased B cells; poor Ab responses following vaccination | Bacterial and viral infections; severe neurologic disease; also known as congenital disorder of glycosylation type IIb (CDG-IIb) |
| 3. Severe reduction in serum IgG and IgA with normal/elevated IgM and normal numbers of B cells, hyper IgM | |||||
| AID deficiency | AICDA | AR | 6055258 | IgG and IgA decreased, IgM increased; normal memory B cells but lacking somatic hypermutation | Bacterial infections, enlarged lymph nodes and germinal centers; autoimmunity |
| AD | 605257 | IgG absent or decreased, IgA undetected, IgM increased; normal memory B cells with intact somatic hypermutation | Bacterial infections, enlarged lymph nodes and germinal centers. Mutations uniquely localize to the nuclear export signal. | ||
| UNG deficiency | UNG | AR | 191525 | IgG and IgA decreased, IgM increased | Enlarged lymph nodes and germinal centers |
| INO80 deficiency | INO80 | AR | 610169 | IgG and IgA decreased, IgM increased | Severe bacterial infections |
| MSH6 deficiency | MSH6 | AR | 600678 | Variable IgG, defects, increased IgM in some, normal B cells, low switched memory B cells, Ig class switch recombination and somatic hypermutation defects | Family or personal history of cancer |
| 4. Isotype, light chain, or functional deficiencies with generally normal numbers of B cells | |||||
| Ig heavy chain mutations and deletions | Mutation or chromosomal deletion at 14q32 | AR | One or more IgG and/or IgA subclasses as well as IgE may be absent | May be asymptomatic | |
| Kappa chain deficiency | IGKC | AR | 147200 | All immunoglobulins have lambda light chain | Asymptomatic |
| Isolated IgG subclass deficiency | Unknown | ? | Reduction in one or more IgG subclass | Usually asymptomatic, a minority may have poor antibody response to specific antigens and recurrent viral/bacterial infections | |
| IgG subclass deficiency with IgA deficiency | Unknown | ? | Reduced IgA with decrease in one or more IgG subclass | Recurrent bacterial infections | |
| May be asymptomatic | |||||
| Selective IgA deficiency | Unknown | ? | Absent IgA with other isotypes normal, normal subclasses and specific antibodies | May be asymptomatic Bacterial infections, autoimmunity mildly increased | |
| Specific antibody deficiency with normal Ig levels and normal B cells | Unknown | ? | Normal | Reduced ability to produce antibodies to specific antigens | |
| Transient hypogammaglobulinemia of infancy | Unknown | ? | IgG and IgA decreased | Normal ability to produce antibodies to vaccine antigens, usually not associated with significant infections | |
| CARD11 GOF | CARD11 | AD GOF | 616452 | Polyclonal B cell lymphocytosis due to constitutive NF-κB activation | Splenomegaly, lymphadenopathy, poor vaccine response |
| Selective IgM deficiency | Unknown | ? | Absent serum IgM | Pneumococcal/bacterial | |
Common variable immunodeficiency disorders (CVID) include several clinical and laboratory phenotypes that may be caused by distinct genetic and/or environmental factors. Some patients with CVID and no known genetic defect have markedly reduced numbers of B cells as well as hypogammaglobulinemia. Identification of causal variants can assist in defining treatment. In addition to monogenic causes on this table, a small minority of patients with XLP (Table 4), WHIM syndrome (Table 6), ICF (Table 2), VODI (Table 2), thymoma with immunodeficiency (Good syndrome), or myelodysplasia are first seen by an immunologist because of recurrent infections, hypogammaglobulinemia, and normal or reduced numbers of B cells
Total number of disorders in Table 3: 46
Total number of mutant genes in Table 3: 39
New disorders: 9: AR PIK3CD [46–48]; AR TCF3 [49, 50]; SLC39A7 [51]; TOP2B [52]; ARHGEF1 [53]; SH3KBP1 [54]; SEC61A1 [55]; AR LOF RAC2 [56]; AD AICDA
EBV Epstein-Barr virus, COPD chronic obstructive pulmonary disease
#Heterozygous variants in TNFRSF13B have been detected in healthy individuals, thus such variants are likely to be disease-modifying rather than disease-causing
Table 4.
Diseases of immune dysregulation
| Disease | Genetic defect | Inheritance | OMIM | Circulating T cells | Circulating B cells | Functional defect | Associated features |
|---|---|---|---|---|---|---|---|
| 1. Familial hemophagocytic lymphohistiocytosis (FHL syndromes) | |||||||
| Perforin deficiency (FHL2) | PRF1 | AR | 170280 | Increased activated T cells | Normal | Decreased to absent NK and CTL activities cytotoxicity | Fever, HSM, hemophagocytic lymphohistiocytosis (HLH), cytopenias |
| UNC13D/Munc13–4 deficiency (FHL3) | UNC13D | AR | 608897 | Increased activated T cells | Normal | Decreased to absent NK and CTL activities (cytotoxicity and/or degranulation) | Fever, HSM, HLH, cytopenias, |
| Syntaxin 11 deficiency (FHL4) | STX11 | AR | 605014 | ||||
| STXBP2/Munc18–2 deficiency (FHL5) | STXBP2 | AR or AD | 601717 | ||||
| FAAP24 deficiency | FAAP24 | AR | 610884 | Increased activated T cells | Normal | Failure to kill autologous EBV transformed B cells. Normal NK cell function | EBV-driven lymphoproliferative disease |
| SLC7A7 deficiency | SLC7A7 | AR | 222700 | Normal | Normal |
Hyper-inflammatory response of macrophages Normal NK cell function |
Lysinuric protein intolerance, bleeding tendency, alveolar proteinosis |
| 2. FHL syndromes with hypopigmentation | |||||||
| Chediak-Higashi syndrome | LYST | AR | 606897 | Increased activated T cells | Normal | Decreased NK and CTL activities (cytotoxicity and/or degranulation) | Partial albinism, recurrent infections, fever, HSM, HLH, giant lysosomes, neutropenia, cytopenias, bleeding tendency, progressive neurological dysfunction |
| Griscelli syndrome, type 2 | RAB27A | AR | 603868 | Normal | Normal | Decreased NK and CTL activities (cytotoxicity and/or degranulation) | Partial albinism, fever, HSM, HLH, cytopenias |
| Hermansky-Pudlak syndrome, type 2 | AP3B1 | AR | 603401 | Normal | Normal | Decreased NK and CTL activities (cytotoxicity and/or degranulation) | Partial albinism, recurrent infections, pulmonary fibrosis, increased bleeding, neutropenia, HLH |
| Hermansky-Pudlak syndrome, type 10 | AP3D1 | AR | 617050 | Normal | Normal | Decreased NK and CTL activities (cytotoxicity and/or degranulation) | Oculocutaneous albinism, severe neutropenia, recurrent infections, seizures, hearing loss and neurodevelopmental delay |
| 3. Regulatory T cell defects | |||||||
| IPEX, immune dysregulation, polyendocrinopathy, enteropathy X-linked | FOXP3 | XL | 300292 | Normal | Normal | Lack of (and/or impaired function of) CD4+ CD25+ FOXP3+ regulatory T cells (Tregs) | Autoimmune enteropathy, early onset diabetes, thyroiditis hemolytic anemia, thrombocytopenia, eczema, elevated IgE and IgA |
| CD25 deficiency | IL2RA | AR | 147730 | Normal to decreased | Normal | No CD4 + C25+ cells with impaired function of Tregs cells | Lymphoproliferation, autoimmunity, impaired T cell proliferation in vitro |
| CD122 deficiency | IL2RB | AR | 618495 | Increased memory CD8 T cells, decreased Tregs | Increased memory B cells | Diminished IL2Rβ expression, dysregulated signaling in response to IL-2/IL-15; increased immature NK cells | Lymphoproliferation, lymphadenopathy, hepatosplenomegaly, autoimmune hemolytic anemia, dermatitis, enteropathy, hypergammaglobulinemia, recurrent viral (EBV, CMV) infections |
| CTLA4 haploinsufficiency (ALPS-V) | CTLA4 | AD | 123890 | Decreased | Decreased | Impaired function of Tregs. | Autoimmune cytopenias, enteropathy, interstitial lung disease, extra-lymphoid lymphocytic infiltration, recurrent infections |
| LRBA deficiency | LRBA | AR | 606453 | Normal or decreased CD4 numbers T cell dysregulation | Low or normal numbers of B cells | Reduced IgG and IgA in most | Recurrent infections, inflammatory bowel disease, autoimmunity |
| DEF6 deficiency | DEF6 | AR | 610094 | Mild CD4 and CD8 lymphopenia | Low or normal numbers of B cells | Impaired Treg function | Enteropathy, hepatosplenomegaly, cardiomyopathy, recurrent infections |
| STAT3 GOF mutation | STAT3 | AD GOF | 102582 | Decreased | Decreased | Enhanced STAT3 signaling, leading to increased Th17 cell differentiation, lymphoproliferation and autoimmunity. Decreased Tregs and impaired function | Lymphoproliferation, solid organ autoimmunity, recurrent infections |
| BACH2 deficiency | BACH2 | AD | 605394 | Progressive T cell lymphopenia | Impaired memory B cell development | Haploinsufficiency for a critical lineage specification transcription factor | Lymphocytic colitis, sinopulmonary infections |
| FERMT1 deficiency | FERMT1 | AR | 173650 | Normal | Normal | Intracellular accumulation of IgG, IgM, IgA, and C3 in colloid bodies under the basement membrane | Dermatosis characterized by congenital blistering, skin atrophy, photosensitivity, skin fragility, and scaling |
| 4. Autoimmunity with or without lymphoproliferation | |||||||
| APECED (APS-1), autoimmune polyendocrinopathy with candidiasis and ectodermal dystrophy | AIRE | AR or AD | 240300 | Normal | Normal | AIRE serves as check-point in the thymus for negative selection of autoreactive T cells and for generation of Tregs | Autoimmunity: hypoparathyroidism, hypothyroidism, adrenal insufficiency, diabetes, gonadal dysfunction and other endocrine abnormalities; dental enamel hypoplasia, alopecia areata enteropathy, pernicious anemia; chronic mucocutaneous candidiasis |
| ITCH deficiency | ITCH | AR | 606409 | Not assessed | Not assessed | Itch deficiency may cause immune dysregulation by affecting both anergy induction in auto-reactive effector T cells and generation of Tregs | Early-onset chronic lung disease (interstitial pneumonitis), autoimmunity (thyroiditis, type I diabetes, chronic diarrhea/enteropathy, and hepatitis), failure to thrive, developmental delay, dysmorphic facial features |
| Tripeptidyl-peptidase II deficiency | TPP2 | AR | 190470 | Decreased | Decreased | TPP2 deficiency results in premature immunosenescence and immune dysregulation | Variable lymphoproliferation, severe autoimmune cytopenias, hypergammaglobulinemia, recurrent infections |
| JAK1 GOF | JAK1 | AD GOF | 147795 | Not assessed | Not assessed | Hyperactive JAK1 | HSM, eosinophilia, eosinophilic enteritis, thyroid disease, poor growth, viral infections |
| Prolidase deficiency | PEPD | AR | 613230 | Normal | Normal | Peptidase D | Autoantibodies common, chronic skin ulcers, eczema, infections |
| 5. Immune dysregulation with colitis | |||||||
| IL-10 deficiency | IL10 | AR | 124092 | Normal | Normal | No functional IL-10 secretion | Inflammatory bowel disease (IBD), folliculitis, recurrent respiratory diseases, arthritis, |
| IL-10R deficiency | IL10RA | AR | 146933 | Normal | Normal | Leukocytes unresponsive to IL-10 | IBD, folliculitis, recurrent respiratory diseases, arthritis, lymphoma |
| IL10RB | AR | 123889 | Normal | Normal | Leukocytes unresponsive to IL-10, and IL-22, IL-26, IL-28A, IL-28B and IL-29 | ||
| NFAT5 haploinsufficiency | NFAT5 | AD | 604708 | Normal | Normal | Decreased memory B cells and plasmablasts | IBD, recurrent sinopulmonary infections |
| TGFB1 deficiency | TGFB1 | AR | 618213 | Normal | Normal | Decreased T cell proliferation in response to anti-CD3 | IBD, immunodeficiency, recurrent viral infections, microcephaly, and encephalopathy |
| RIPK1 | RIPK1 | AR | 618108 | Reduced | Normal/reduced | Reduced activation of MAPK, NFkB pathways to | Recurrent infections, early-onset IBD, progressive polyarthritis |
| 6. Autoimmune lymphoproliferative syndrome (ALPS, Canale-Smith syndrome) | |||||||
| ALPS-FAS | TNFRSF6 |
AD AR |
134637 | Increased TCR α/β+CD4−CD8− double negative (DN) T cells | Normal, low memory B cells | Apoptosis defect FAS mediated | Splenomegaly, adenopathies, autoimmune cytopenias, increased lymphoma risk, IgG and A normal or increased, elevated serum FasL, IL-10, vitamin B12 |
| ALPS-FASLG | TNFSF6 | AR | 134638 | Increased DN T cells | Normal | Apoptosis defect FASL mediated | Splenomegaly, adenopathies, autoimmune cytopenias, SLE, soluble FasL is not elevated |
| ALPS-Caspase10 | CASP10 | AD | 601762 | Increased DN T cells | Normal | Defective lymphocyte apoptosis | Adenopathies, splenomegaly, autoimmunity |
| ALPS-Caspase 8 | CASP8 | AR | 601763 | Slightly increased DN T cells | Normal | Defective lymphocyte apoptosis and activation | Adenopathies, splenomegaly, bacterial and viral infections, hypogammaglobulinemia |
| FADD deficiency | FADD | AR | 602457 | Increased DN T cells | Normal | Defective lymphocyte apoptosis | Functional hyposplenism, bacterial and viral infections, recurrent episodes of encephalopathy and liver dysfunction |
| 7. Susceptibility to EBV and lymphoproliferative conditions | |||||||
| SAP deficiency (XLP1) | SH2D1A | XL | 300490 | Normal or Increased activated T cells | Reduced Memory B cells | Reduced NK cell and CTL cytotoxic activity |
Clinical and immunologic features triggered by EBV infection: HLH, Lymphoproliferation, Aplastic anemia, Lymphoma. Hypogammaglobulinemia, Absent iNKT cells |
| XIAP deficiency (XLP2) | XIAP | XL | 300079 | Normal or Increased activated T cells; low/normal iNK T cells | Normal or reduced Memory B cells | Increased T cells susceptibility to apoptosis to CD95 and enhanced activation-induced cell death (AICD) |
EBV infection, Splenomegaly, lymphoproliferation HLH, Colitis, IBD, hepatitis Low iNKT cells |
| CD27 deficiency | CD27 | AR | 615122 | Normal | No memory B cells | hypogammaglobulinemia; poor Ab responses to some vaccines/infections | Features triggered by EBV infection, HLH, aplastic anemia, low iNKT cells, B-lymphoma |
| CD70 deficiency | CD70 | AR | 602840 | Normal number, low Treg, poor activation and function | Decreased memory B cells | hypogammaglobulinemia; poor Ab responses to some vaccines/infections | EBV susceptibility, Hodgkin lymphoma; autoimmunity in some patients |
| CTPS1 deficiency | CTPS1 | AR | 615897 | Normal to low, but reduced activation, proliferation | Decreased memory B cells | Normal/high IgG poor proliferation to antigen | Recurrent/chronic bacterial and viral infections (EBV, VZV), EBV lymphoproliferation, B cell non-Hodgkin lymphoma |
| CD137 deficiency (41BB) | TNFRSF9 | AR | 602250 | Normal | Normal | Low IgG, low IgA, poor responses to T cell-dependent and T cell independent antigens, decreased T cell proliferation, IFNγ secretion, cytotoxicity | EBV lymphoproliferation, B cell lymphoma, chronic active EBV infection |
| RASGRP1 deficiency | RASGRP1 | AR | 603962 | Poor activation, proliferation, motility. Reduced naïve T cells | Poor activation, proliferation, motility | Normal IgM, IgG, increased IgA |
Recurrent pneumonia, herpesvirus infections, EBV associated lymphoma Decreased NK cell function |
| RLTPR deficiency | CARMIL2 | AR | 610859 | Normal number, high CD4, increased naïve CD4+ and CD8+ T cells, low Treg and MAIT, poor CD28-induced function | Normal B cell numbers, reduced memory B cells | Normal to low, poor T dependent antibody response | Recurrent bacterial, fungal and mycobacterial infections, viral warts, molluscum and EBV lymphoproliferative and other malignancy, atopy |
| X-linked magnesium EBV and neoplasia (XMEN) | MAGT1 | XL | 300853 | Low CD4 Low recent thymic emigrant cels, inverted CD4/CD8 ratio, reduced MAIT cells, poor proliferation to CD3 | Normal but decreased memory B cells |
Progressive hypogammaglobulinemia Reduced NK cell and CTL cytotoxic activity due to impaired expression of NKG2D |
EBV infection, lymphoma, viral infections, respiratory and GI infections Glycosylation defects |
| PRKCD deficiency | PRKCD | AR | 615559 | Normal | Low memory B cells, high CD5 B cells | Apoptotic defect in B cells | Recurrent infections, EBV chronic infection, lymphoproliferation, SLE-like autoimmunity (nephrotic and antiphospholipid syndromes), low IgG |
Total number of disorders in Table 4: 44
Total number of mutant genes in Table 4: 45
New disorders: 8; SLC7A7 [57]; IL2RB [58, 59]; DEF6 [60]; FERMT1 [61]; TGFB1 [62]; RIPK1 [63, 64]; TNFRSF9 [46, 65, 66]; STAT5B AD DN []
FHL familial hemophagocytic lymphohistiocytosis, HLH hemophagocytic lymphohistiocytosis, HSM hepatosplenomegaly, DN double-negative, SLE systemic lupus erythematous, IBD Inflammatory bowel disease
Table 5.
Congenital defects of phagocyte number or function
| Disease | Genetic defect | Inheritance | OMIM | Affected cells | Affected function | Associated features |
|---|---|---|---|---|---|---|
| 1. Congenital neutropenias | ||||||
| Elastase deficiency (Severe congential neutropenia [SCN] 1) | ELANE | AD | 130130 | N | Myeloid differentiation |
Susceptibility to MDS/leukemia Severe congenital neutropenia or cyclic neutropenia |
| GFI 1 deficiency (SCN2) | GFI1 | AD | 600871 | N | Myeloid differentiation | B/T lymphopenia |
| HAX1 deficiency (Kostmann Disease) (SCN3) | HAX1 | AR | 605998 | N | Myeloid differentiation | Cognitive and neurological defects in patients with defects in both HAX1 isoforms, susceptibility to MDS/leukemia |
| G6PC3 deficiency (SCN4) | G6PC3 | AR | 611045 | N | Myeloid differentiation, chemotaxis, O2− production | Structural heart defects, urogenital abnormalities, inner ear deafness, and venous angiectasias of trunks and limbs |
| VPS45 deficiency (SCN5) | VPS45 | AR | 610035 | N | Myeloid differentiation, migration | Extramedullary hematopoiesis, bone marrow fibrosis, nephromegaly |
| Glycogen storage disease type 1b | G6PT1 | AR | 602671 | N + M | Myeloid differentiation, chemotaxis, O2− production | Fasting hypoglycemia, lactic acidosis, hyperlipidemia, hepatomegaly |
| X-linked neutropenia/myelodysplasia | WAS | XL GOF | 300299 | N | Differentiation, mitosis. Results from GOF mutations in GTPase binding domain of WASp | Neutropenia, myeloid maturation arrest, monocytopenia, variable lymphoid anomalies |
| P14/LAMTOR2 deficiency | LAMTOR2 | AR | 610389 | N + M | Endosomal biogenesis |
Neutropenia Hypogammaglobulinemia ↓CD8 cytotoxicity, partial albinism, growth failure |
| Barth Syndrome (3-Methylglutaconic aciduria type II) | TAZ | XL | 300394 |
N + L Mel |
Mitochondrial function | Cardiomyopathy, myopathy, growth retardation, neutropenia |
| Cohen syndrome | VPS13B | AR | 607817 | N | Myeloid differentiation | Dysmorphism, mental retardation, obesity, deafness, neutropenia |
| Clericuzio syndrome (Poikiloderma with neutropenia) | USB1 | AR | 613276 | N | Myeloid differentiation | Retinopathy, developmental delay, facial dysmorphisms, poikiloderma |
| JAGN1 deficiency | JAGN1 | AR | 616012 | N | Myeloid differentiation | Myeloid maturation arrest, osteopenia |
| 3-Methylglutaconic aciduria | CLPB | AR | 616254 | N |
Myeloid differentiation Mitochondrial protein |
Neurocognitive developmental aberrations, microcephaly, hypoglycemia, hypotonia, ataxia, seizures, cataracts, IUGR |
| G-CSF receptor deficiency | CSF3R | AR | 138971 | N | Stress granulopoiesis disturbed | |
| SMARCD2 deficiency | SMARCD2 | AR | 601736 | N | Chromatin remodeling, Myeloid differentiation and neutrophil functional defect | Neutropenia, developmental aberrations, bones, hematopoietic stem cells, myelodysplasia |
| Specific granule deficiency | CEBPE | AR | 189965 | N | Terminal maturation and global dysfunction | Neutropenia, Neutrophils with bilobed nuclei |
| Shwachman-Diamond Syndrome | SBDS | AR | 607444 | N | Neutrophil maturation, chemotaxis, ribosomal biogenesis | Pancytopenia, exocrine pancreatic insufficiency, chondrodysplasia |
| DNAJC21 | AR | 617052 | N + HSC | Pancytopenia, exocrine pancreatic insufficiency | ||
| EFL1 | AR | 617941 | N + HSC | |||
| HYOU1 deficiency | HYOU1 | AR | 601746 | N | Unfolded protein response | Hypoglycemia, inflammatory complications |
| SRP54 deficiency | SRP54 | AD | 604857 | N | Protein translocation to ER, myeloid differentiation and neutrophil functional defect | Neutropenia, exocrine pancreatic insufficiency |
| 2. Defects of motility | ||||||
| Leukocyte adhesion deficiency type 1 (LAD1) | ITGB2 | AR | 600065 |
N + M + L + NK |
Adherence, chemotaxis, endocytosis, T/NK cytotoxicity | Delayed cord separation, skin ulcers, periodontitis, leukocytosis |
| Leukocyte adhesion deficiency type 2 (LAD2) | SLC35C1 | AR | 605881 | N + M | Rolling, chemotaxis | Mild LAD type 1 features with hh-blood group, growth retardation, developmental delay |
| Leukocyte adhesion deficiency type 3 (LAD3) | FERMT3 | AR | 607901 |
N + M + L + NK |
Adherence, chemotaxis | LAD type 1 plus bleeding tendency |
| Rac2 deficiency | RAC2 | AD LOF | 608203 | N |
Adherence, chemotaxis O2− production |
Poor wound healing, leukocytosis |
| β actin deficiency | ACTB | AD | 102630 | N + M | Motility | Mental retardation, short stature |
| Localized juvenile periodontitis | FPR1 | AR | 136537 | N | Formylpeptide induced chemotaxis | Periodontitis only |
| Papillon-Lefèvre syndrome | CTSC | AR | 602365 | N + M | Chemotaxis | Periodontitis, palmoplantar hyperkeratosis in some patients |
| WDR1 deficiency | WDR1 | AR | 604734 | N | Spreading, survival, chemotaxis | Mild neutropenia, poor wound healing, severe stomatitis, neutrophil nuclei herniate |
| Cystic fibrosis | CFTR | AR | 602421 | M only | Chemotaxis | Respiratory infections, pancreatic insufficiency, elevated sweat chloride |
| Neutropenia with combined immune deficiency due to MKL1 deficiency | MKL1 | AR | 606078 | N + M + L + NK | Impaired expression of cytoskeletal genes | Mild thrombocytopenia |
| 3. Defects of respiratory burst | ||||||
| X-linked chronic granulomatous disease (CGD), gp91phox | CYBB | XL | 306400 | N + M | Killing (faulty O2− production) |
Infections, autoinflammatory phenotype, IBD McLeod phenotype in patients with deletions extending into the contiguous Kell locus |
| Autosomal recessive CGD | CYBA | AR | 608508 | Infections, autoinflammatory phenotype | ||
| CYBC1 | 618334 | |||||
| NCF1 | 608512 | |||||
| NCF2 | 608515 | |||||
| NCF4 | 613960 | |||||
| G6PD deficiency class I | G6PD | XL | 305900 | N | Reduced O2− production | Infections |
| 4. Other non-lymphoid defects | ||||||
| GATA2 deficiency | GATA2 | AD | 137295 | Monocytes + peripheral DC | Multi lineage cytopenias | Susceptibility to mycobacteria, HPV, histoplasmosis, alveolar proteinosis, MDS/AML/CMML, lymphedema |
| Pulmonary alveolar proteinosis | CSF2RA | XL (Biallelic mutations in pseudo-autosomal gene) | 300770 | Alveolar macrophages | GM-CSF signaling | Alveolar proteinosis |
| CSFR2B | AR | 614370 | ||||
Total number of disorders in Table 5: 34
Total number of mutant genes in Table 5: 41
New disorders: 3; SRP54 [67, 68]; DNAJC21 [69]; CYBC1 [70, 71]
Removed: Cyclic neutropenia was merged with elastase deficiency
MDS myelodysplastic syndrome, IUGR intrauterine growth retardation, LAD leukocyte adhesion deficiency, AML acute myelogenous leukemia, CMML chronic myelomonocytic leukemia, N neutrophil, M monocyte, MEL melanocyte, L lymphocyte, NK natural killer
Table 6.
Defects in intrinsic and innate immunity
| Disease | Genetic defect | Inheritance | OMIM | Affected cells | Affected function | Associated features |
|---|---|---|---|---|---|---|
| 1. Mendelian susceptibility to mycobacterial disease (MSMD) | ||||||
| IL-12 and IL-23 receptor β1 chain deficiency | IL12RB1 | AR | 601604 | L + NK | IFN-γ secretion | Susceptibility to mycobacteria and Salmonella |
| IL-12p40 (IL-12 and IL-23) deficiency | IL12B | AR | 161561 | M | ||
| IL-12Rβ2 deficiency | IL12RB2 | AR | 601642 | L + NK | ||
| IL-23R deficiency | IL23R | AR | 607562 | L + NK | ||
| IFN-γ receptor 1 deficiency | IFNGR1 | AR | 209950 | M + L | IFN-γ binding and signaling | |
| AD | 615978 | M + L | ||||
| IFN-γ receptor 2 deficiency | IFNGR2 | AR | 147569 | M + L | IFN-γ signaling | |
| STAT1 deficiency | STAT1 | AD LOF | 614892 | M + L | ||
| Macrophage gp91 phox deficiency | CYBB | XL | 300645 | Macrophage only | Killing (faulty O2− production) | Isolated susceptibility to mycobacteria |
| IRF8 deficiency | IRF8 | AD | 614893 | M + L | Impaired development of cDCs and Th1* cells | Susceptibility to mycobacteria |
| AR | 226990 | M | Lack of circulating monocytes and DCs, reduced NK cell numbers and function reported in some patients | Susceptibility to mycobacteria and multiple other infectious agents including EBV | ||
| SPPL2a deficiency | SPPL2A | AR | 608238 | M + L | Impaired development of cDCs and Th1* cells | Susceptibility to mycobacteria and Salmonella |
| Tyk2 deficiency | TYK2 | AR | 611521 | M + L | Impaired cellular responses to IL-10, IL-12, IL-23, and type I IFNs | Susceptibility to intracellular bacteria (mycobacteria, Salmonella), and viruses |
| P1104A TYK2 homozygosity | TYK2 | AR | 176941 | L | Impaired cellular responses to IL-23 | MSMD or tuberculosis |
| ISG15 deficiency | ISG15 | AR | 147571 | IFNγ production defect | Susceptibility to mycobacteria (BCG), brain calcification | |
| RORγt deficiency | RORC | AR | 602943 | L + NK | Lack of functional RORγT protein, IFNγ production defect, complete absence of IL-17A/F-producing T cells | Susceptibility to mycobacteria and candida |
| JAK1 deficiency | JAK1 | AR LOF | 147795 | N + L | Reduced JAK1 activation to cytokines, Reduced IFNγ production | Susceptibility to mycobacteria and viruses, urothelial carcinoma |
| 2. Epidermodysplasia verruciformis (HPV) | ||||||
| EVER1 deficiency | TMC6 | AR | 605828 | Keratinocytes | EVER1, EVER2 and CIB1 form a complex in keratinocytes | Human papillomavirus (HPV) (group B1) infections and cancer of the skin (typical EV) |
| EVER2 deficiency | TMC8 | 605829 | ||||
| CIB1 deficiency | CIB1 | 618267 | ||||
| WHIM (warts, hypogammaglobulinemia, infections, myelokathexis) syndrome | CXCR4 | AD GOF | 162643 | Leukocytes | Increased response of the CXCR4 chemokine receptor to its ligand CXCL12 (SDF-1) | Warts (HPV) infection, neutropenia, low B cell number, hypogammaglobulinemia |
| 3. Predisposition to severe viral infection | ||||||
| STAT1 deficiency | STAT1 | AR LOF | 600555 | Leukocytes and other cells |
STAT1-dependent IFN-α/β, γ and λ responses |
Severe viral infections, mycobacterial infection |
| STAT2 deficiency | STAT2 | AR | 600556 | Leukocytes and other cells |
STAT2-dependent IFN-α/β and λ response |
Severe viral infections (disseminated vaccine-strain measles) |
| IRF9 deficiency | IRF9 | AR | 147574* | Leukocytes and other cells | IRF9- and ISGF3-dependent IFN-α/β and λ responses | Severe influenza disease |
| IRF7 deficiency | IRF7 | AR | 605047 | Leukocytes, plasmacytoid dendritic cells, non-hematopoietic cells | IFN-α, β and γ production and IFN-λ production | |
| IFNAR1 deficiency | IFNAR1 | AR | 107450* | Leukocytes and other cells | IFNAR1-dependent responses to IFN-α/β | Severe disease caused by Yellow Fever vaccine and Measles vaccine |
| IFNAR2 deficiency | IFNAR2 | AR | 602376 | Broadly expressed | IFNAR2-dependent responses to IFN-α/β | Severe viral infections (disseminated vaccine-strain measles, HHV6) |
| CD16 deficiency | FCGR3A | AR | 146740 | NK cells | Altered NK cells function | Severe herpes viral infections, particularly VZV, Epstein-Barr virus (EBV), and (HPV) |
| MDA5 deficiency | IFIH1 | AR LOF | 606951 | Broadly expressed | Viral recognition and IFN induction | Rhinovirus and other RNA viruses |
| RNA polymerase III deficiency | POLR3A | AD | 614258 | Leukocytes and other cells | Impaired viral recognition and IFN induction in response to VZV or poly I:C | Severe VZV infection |
| POLR3C | AD | 617454 | ||||
| POLR3F | AD | 617455 | ||||
| 4. Herpes simplex encephalitis (HSE) | ||||||
| TLR3 deficiency | TLR3 | AD | 613002 | Central nervous system (CNS) resident cells and fibroblasts | TLR3-dependent IFN-α, β and γ response | Herpes simplex virus 1 encephalitis (incomplete clinical penetrance for all etiologies listed here); severe pulmonary influenza; VZV |
| AR | ||||||
| UNC93B1 deficiency | UNC93B1 | AR | 608204 | UNC-93B-dependent IFN-α, β and γ response | Herpes simplex virus 1 encephalitis | |
| TRAF3 deficiency | TRAF3 | AD | 601896 | TRAF3-dependent IFN-α, β and γ response | ||
| TRIF deficiency | TICAM1 | AD | 607601 | TRIF-dependent IFN-α, β and γ response | ||
| AR | ||||||
| TBK1 deficiency | TBK1 | AD | 604834 |
TBK1-dependent IFN-α, β and γ response |
||
| IRF3 deficiency | IRF3 | AD | 616532 | Low IFN-α/β production in response to HSV1 and decreased IRF3 phosphorylation | ||
| DBR1 deficiency | DBR1 | AR | 607024 | Impaired production of anti-viral IFNs | HSE of the brainstem. Other viral infections of the brainstem. | |
| 5. Predisposition to invasive fungal diseases | ||||||
| CARD9 deficiency | CARD9 | AR | 607212 | Mononuclear phagocytes | CARD9 signaling pathway | Invasive candidiasis infection, deep dermatophytoses, other invasive fungal infections |
| 6. Predisposition to mucocutaneous candidiasis | ||||||
| IL-17RA deficiency | IL17RA | AR | 605461 | Epithelial cells, fibroblasts, mononuclear phagocytes | IL-17RA signaling pathway | CMC, folliculitis |
| IL-17RC deficiency | IL17RC | AR | 610925 | IL-17RC signaling pathway | CMC | |
| IL-17F deficiency | IL17F | AD | 606496 | T cells | IL-17F-containing dimers | CMC, folliculitis |
| STAT1 GOF | STAT1 | AD GOF | 600555 | T cells, B cells, monocytes | Gain-of-function STAT1 mutations that impair the development of IL-17-producing T cells | CMC, various fungal, bacterial and viral (HSV) infections, auto-immunity (thyroiditis, diabetes, cytopenias), enteropathy |
| ACT1 deficiency | TRAF3IP2 | AR | 607043 | T cells, fibroblasts | Fibroblasts fail to respond to IL-17A and IL-17F, and their T cells to IL-17E | CMC, blepharitis, folliculitis, and macroglossia |
| 7. TLR signaling pathway deficiency with bacterial susceptibility | ||||||
| IRAK4 deficiency | IRAK4 | AR | 606883 | Lymphocytes + granulocytes+ monocytes | TIR-IRAK4 signaling pathway | Bacterial infections (pyogens) |
| MyD88 deficiency | MYD88 | AR | 602170 | Lymphocytes + granulocytes + monocytes | TIR-MyD88 signaling pathway | |
| IRAK1 deficiency | IRAK1 | XL | 300283 | Lymphocytes + granulocytes + monocytes | TIR-IRAK1 signaling pathway | Bacterial infections, X-linked MECP2 deficiency-related syndrome due to a large de novo Xq28 chromosomal deletion encompassing both MECP2 and IRAK1 |
| TIRAP deficiency | TIRAP | AR | 614382 | Lymphocytes + granulocytes + monocytes | TIRAP- signaling pathway, TLR1/2, TLR2/6, and TLR4 agonists were impaired in the fibroblasts and leukocytes | Staphylococcal disease during childhood |
| 8. Other inborn errors of immunity related to non-hematopoietic tissues | ||||||
| Isolated congenital asplenia (ICA) | RPSA | AD | 271400 | No spleen | RPSA encodes ribosomal protein SA, a component of the small subunit of the ribosome | Bacteremia (encapsulated bacteria) |
| HMOX | AR | 141250 | Macrophages | HO-1 regulates iron recycling and heme-dependent damage occurs | Hemolysis, nephritis, inflammation | |
| Trypanosomiasis | APOL1 | AD | 603743 | Somatic | Pore forming serum protein | Trypanosomiasis |
| Acute liver failure due to NBAS deficiency | NBAS | AR | 608025 | Somatic and hematopoietic | ER stress | Fever induces liver failure |
|
Acute necrotizing encephalopathy Osteopetrosis |
RANBP2 | AR | 601181 | Ubiquitous expression | Nuclear pore | Fever induces acute encephalopathy |
| CLCN7 | AR | 602727 | Osteoclasts | Secretory lysosomes | Osteopetrosis with hypocalcemia, neurologic features | |
| SNX10 | AR | 614780 | Osteopetrosis with visual impairment | |||
| OSTM1 | AR | 607649 | Osteopetrosis with hypocalcemia, neurologic features | |||
| PLEKHM1 | AR | 611466 | Osteopetrosis | |||
| TCIRG1 | AR | 604592 | Osteopetrosis with hypocalcemia | |||
| TNFRSF11A | AR | 603499 | Osteoclastogenesis | Osteopetrosis | ||
| TNFSF11 | AR | 602642 | Stromal | Osteoclastogenesis | Osteopetrosis with severe growth retardation | |
| Hidradenitis suppurativa | NCSTN | AD | 605254 | Epidermis | Notch signaling/gamma-secretase in hair follicle regulates keratinization | Verneuil’s disease/Hidradenitis suppurativa with acne |
| PSEN | AD | 613737 | Verneuil’s disease/Hidradenitis suppurative with cutaneous hyperpigmentation | |||
| PSENEN | AD | 613736 | Verneuil’s disease/Hidradenitis suppurativa | |||
| 9. Other inborn errors of immunity related to leukocytes | ||||||
| IRF4 haploinsufficiency | IRF4 | AD | 601900 | L + M | IRF4 is a pleiotropic transcription factor | Whipple’s disease |
| IL-18BP deficiency | IL18BP | AR | 604113 | Leukocytes and other cells | IL-18BP neutralizes secreted IL-18 | Fulminant viral hepatitis |
Total number of disorders in Table 6: 53
Total number of mutant genes in Table 6: 64
New genes: 13, IL12RB2 [72]; IL23R [72]; SPPL2A [73]; TYK2 P1104A allele [10]; CIB1 [74]; IRF9 [75]; IFNAR1 [76]; POLR3A [77]; POLR3C [77]; POLR3F [78]; DBR1 [79]; IRF4 [80]; IL18BP [81]
NF-κB nuclear factor kappa B, TIR Toll and Interleukin 1 receptor, IFN interferon, TLR Toll-like receptor, MDC myeloid dendritic cell, CNS central nervous system, CMC chronic mucocutaneous candidiasis, HPV human papillomavirus, VZV varicella zoster virus, EBV, Epstein-Barr virus
Table 7.
Autoinflammatory disorders
| Disease | Genetic defect | Inheritance | OMIM | T cells | B cells | Functional defect | Associated features |
|---|---|---|---|---|---|---|---|
| 1. Type 1 interferonopathies | |||||||
| STING-associated vasculopathy, infantile-onset (SAVI) | TMEM173 | AR | 612374 | Not assessed | Not assessed | STING activates both the NF-kappa-B and IRF3 transcription pathways to induce expression of IFN | Skin vasculopathy, inflammatory lung disease, systemic autoinflammation and ICC, FCL |
| ADA2 deficiency | ADA2 | AR | 607575 | Not assessed | Not assessed | ADAs deactivate extracellular adenosine and terminate signaling through adenosine receptors | Polyarteritis nodosa, childhood-onset, early-onset recurrent ischemic stroke and fever; some patients develop hypogammaglobulinemia |
| TREX1 deficiency, Aicardi-Goutieres syndrome 1 (AGS1) | TREX1 | AR | 606609 | Not assessed | Not assessed | Intracellular accumulation of abnormal ss DNA species leading to increased type I IFN production | Classical AGS, SLE, FCL |
| RNASEH2B deficiency, AGS2 | RNASEH2B | AR | 610326 | Not assessed | Not assessed | Intracellular accumulation of abnormal RNA-DNA hybrid species leading to increased type I IFN production | Classical AGS, SP |
| RNASEH2C deficiency, AGS3 | RNASEH2C | AR | 610330 | Not assessed | Not assessed | Classical AGS | |
| RNASEH2A deficiency, AGS4 | RNASEH2A | AR | 606034 | Not assessed | Not assessed | Classical AGS | |
| SAMHD1 deficiency, AGS5 | SAMHD1 | AR | 606754 | Not assessed | Not assessed | Controls dNTPs in the cytosol, failure of which leads to increased type I IFN production | Classical AGS, FCL |
| ADAR1 deficiency, AGS6 | ADAR1 | AR | 146920 | Not assessed | Not assessed | Catalyzes the deamination of adenosine to inosine in dsRNA substrates, failure of which leads to increased type I IFN production | Classical AGS, BSN, SP |
| Aicardi-Goutieres syndrome 7 (AGS7) | IFIH1 | AD GOF | 615846 | Not assessed | Not assessed | IFIH1 gene encodes a cytoplasmic viral RNA receptor that activates type I interferon signaling through the MAVS adaptor molecule | Classical AGS, SLE, SP, SMS |
| DNAse II deficiency | DNASE2 | AR | 126350 | Not assessed | Not assessed | DNAse II degrades and eliminates DNA. Loss of DNase II activity induces type I interferon signaling | AGS |
| Pediatric systemic lupus erythematosus due to DNASE1L3 deficiency | DNASE1L3 | AR | 614420 | DNASE1L3 is an endonuclease that degrades extracellular DNA. DNASE1L3 deficiency decreases clearance of apoptotic cells | Very early onset SLE, reduced complement levels, autoantibodies (dsDNA, ANCA), lupus nephritis, hypocomplementemic urticarial vasculitis syndrome | ||
| Spondyloenchondro-dysplasia with immune dysregulation (SPENCD) | ACP5 | AR | 171640 | Not assessed | Not assessed | Upregulation of IFN through mechanism possibly relating to pDCS | Short stature, SP, ICC, SLE, thrombocytopenia and autoimmune hemolytic anemia, possibly recurrent bacterial and viral infections |
| X-linked reticulate pigmentary disorder | POLA1 | XL | 301220 | Not assessed | Not assessed | POLA1 is required for synthesis of cytosolic RNA:DNA and its deficiency leads to increase production of type I interferon | Hyperpigmentation, characteristic facies, lung and GI involvement |
| USP18 deficiency | USP18 | AR | 607057 | Not assessed | Not assessed | Defective negative regulation of ISG15 leading to increased IFN | TORCH-like syndrome |
| OAS1 deficiency | OAS1 | AD GOF | 164350 | Low | Increased interferon from recognition of RNA | Pulmonary alveolar proteinosis, skin rash | |
| 2. Defects affecting the inflammasome | |||||||
| Familial Mediterranean fever | MEFV | AR LOF | 249100 | Mature granulocytes, cytokine-activated monocytes. | Increased inflammasome-mediated induction of IL1β. | Recurrent fever, serositis and inflammation responsive to colchicine. Predisposes to vasculitis and inflammatory bowel disease. | |
| AD | 134610 | Mature granulocytes, cytokine-activated monocytes. | Usually M694del variant. | ||||
| Mevalonate kinase deficiency (Hyper IgD syndrome) | MVK | AR | 260920 | Somatic and hemaotpoietic | affecting cholesterol synthesis, pathogenesis of disease unclear | Periodic fever and leukocytosis with high IgD levels | |
| Muckle-Wells syndrome | NLRP3 | AD GOF | 191900 | PMNs Monocytes | Defect in cryopyrin, involved in leukocyte apoptosis and NFkB signaling and IL-1 processing | Urticaria, SNHL, amyloidosis. | |
| Familial cold autoinflammatory syndrome 1 | AD GOF | 120100 | PMNs, monocytes | Non-pruritic urticaria, arthritis, chills, fever and leukocytosis after cold exposure. | |||
| Neonatal onset multisystem inflammatory disease (NOMID) or chronic infantile neurologic cutaneous and articular syndrome (CINCA) | AD GOF | 607115 | PMNs, chondrocytes | Neonatal onset rash, chronic meningitis, and arthropathy with fever and inflammation. | |||
| Familial cold autoinflammatory syndrome 2 | NLRP12 | AD GOF | 611762 | PMNs, monocytes | Non-pruritic urticaria, arthritis, chills, fever and leukocytosis after cold exposure. | ||
| NLRC4-MAS (macrophage activating syndrome) | NLRC4 | AD GOF | 616050 | PMNs monocytes macrophages | Gain of function mutation in NLRC4 results in elevated secretion of IL-1β and IL-18 as well as macrophage activation | Severe enterocolitis and macrophage activation syndrome | |
| Familial cold autoinflammatory syndrome 4 | 616115 | ||||||
| PLAID (PLCγ2 associated antibody deficiency and immune dysregulation) | PLCG2 | AD GOF | 614878 | B cells, NK, Mast cells | Mutations activate IL-1 pathways | Cold urticaria hypogammaglobulinemia, impaired humoral immunity, autoinflammation | |
| Familial cold autoinflammatory syndrome 3 or APLAID (c2120A > C) | 614468 | ||||||
| NLRP1 deficiency | NLRP1 | AR | 617388 | leukocytes | Systemic elevation of IL-18 and caspase 1, suggesting involvement of NLRP1 inflammasome | Dyskeratosis, autoimmunity and arthritis | |
| NLRP1 GOF | NLRP1 | AD GOF | 615225 | Keratinocytes | Increased IL1β | Palmoplantar carcinoma, corneal scarring; recurrent respiratory papillomatosis | |
| 3. Non-inflammasome-related conditions | |||||||
| TNF receptor-associated periodic syndrome (TRAPS) | TNFRSF1A | AD | 142680 | PMNs, monocytes | Mutations of 55-kD TNF receptor leading to intracellular receptor retention or diminished soluble cytokine receptor available to bind TNF | Recurrent fever, serositis, rash, and ocular or joint inflammation | |
| Pyogenic sterile arthritis, pyoderma gangrenosum, acne (PAPA) syndrome, hyperzincemia and hypercalprotectinemia | PSTPIP1 | AD | 604416 | Hematopoietic tissues, upregulated in activated T cells | Disordered actin reorganization leading to compromised physiologic signaling during inflammatory response | Destructive arthritis, inflammatory skin rash, myositis | |
| Blau syndrome | NOD2 | AD | 186580 | Monocytes | Mutations in nucleotide binding site of CARD15, possibly disrupting interactions with lipopolysaccharides and NF-kB signaling | Uveitis, granulomatous synovitis, camptodactyly, rash and cranial neuropathies, 30% develop Crohn colitis | |
| ADAM17 deficiency | ADAM17 | AR | 614328 | Leukocytes and epithelial cells | Defective TNFα production | Early onset diarrhea and skin lesions | |
| Chronic recurrent multifocal osteomyelitis and congenital dyserythropoietic anemia (Majeed syndrome) | LPIN2 | AR | 609628 | Neutrophils, bone marrow cells | Undefined | Chronic recurrent multifocal osteomyelitis, transfusion-dependent anemia, cutaneous inflammatory disorders | |
| DIRA (Deficiency of the Interleukin 1 Receptor Antagonist) | IL1RN | AR | 612852 | PMNs, Monocytes | Mutations in the IL1 receptor antagonist allow unopposed action of Interleukin 1 | Neonatal onset of sterile multifocal osteomyelitis, periostitis and pustulosis. | |
| DITRA (Deficiency of IL-36 receptor antagonist) | IL36RN | AR | 614204 | Keratinocytes, leukocytes | Mutations in IL-36RN leads to increase IL-8 production | Pustular psoriasis | |
| SLC29A3 mutation | SLC29A3 | AR | 602782 | Leukocytes, bone cells | – | Hyperpigmentation hypertrichosis, histiocytosis-lymphadenopathy plus syndrome | |
| CAMPS (CARD14 mediated psoriasis) | CARD14 | AD | 602723 | Mainly in keratinocytes | Mutations in CARD14 activate the NF-kB pathway and production of IL-8 | Psoriasis | |
| Cherubism | SH3BP2 | AD | 118400 | Stroma cells, bone cells | Hyperactived macrophage and increase NF-kB | Bone degeneration in jaws | |
| CANDLE (chronic atypical neutrophilic dermatitis with lipodystrophy) | PSMB8* | AR and AD | 256040 | Keratinocytes, B cell adipose cells | Mutations cause increased IFN signaling through an undefined mechanism | Contractures, panniculitis, ICC, fevers | |
| PSMG2 | AR | 609702 | Lymphocytes | Panniculitis, lipodystrophy, autoimmune hemolytic anemia | |||
| COPA defect | COPA | AD | 6011924 | PMN and tissue specific cells | Defective intracellular transport via the coat protein complex I (COPI) | Autoimmune inflammatory arthritis and interstitial lung disease with Th17 dysregulation and autoantibody production | |
| Otulipenia/ORAS | OTULIN | AR | 615712 | Leukocytes | Increase LUBAC induction of NF-KB activation leading to high proinflamatory cytokines levels. | Fever, diarrhea, dermatitis | |
| A20 deficiency | TNFAIP3 | AD | 616744 | Lymphocytes | Defective inhibition of NF-KB signaling pathway | Arthralgia, mucosal ulcers, ocular inflammation | |
| AP1S3 deficiency | AP1S3 | AR | 615781 | Keratinocytes | Disrupted TLR3 translocation | Pustular psoriasis | |
| ALPI deficiency | ALPI | AR | 171740 | Intestinal epithelial cells | Deficient inhibition of LPS in intestine | Inflammatory bowel disease | |
| TRIM22 | TRIM22 | AR | 606559 | Macrophages, intestinal epithelial cells | Granulomatous colitis | Inflammatory bowel disease | |
| T cell lymphoma subcutaneous panniculitis-like (TIM3 deficiency) | HAVCR2 | AR | 618398 | Leukocytes | Increased inflammasome activity due to defective checkpoint signaling | Panniculitis, HLH, polyclonal cutaneous T cell infiltrates or T cell lymphoma | |
Total number of disorders in Table 7: 45
Total number of mutant genes in Table 7: 42
New disorders: 9; DNASE2 [82]; DNASE1L3 [83–85]; OAS1 [86]; AD MEFV; NLRP1 GOF [87, 88]; ALPI [89]; TRIM22 [90]; PSMG2 [91]; HAVCR2 [92, 93]
IFN interferon, HSM hepatosplenomegaly, CSF cerebrospinal fluid, SLE systemic lupus erythematosus, TORCH toxoplasmosis, other, rubella, cytomegalovirus, and herpes infections, SNHL sensorineural hearing loss, AGS Aicardi-Goutières syndrome, BSN bilateral striatal necrosis, FCL familial chilblain lupus, ICC intracranial calcification, IFN interferon type I, pDCs plasmacytoid dendritic cells, SP spastic paraparesis, SMS Singleton-Merten syndrome, ss single-stranded DNA
*Variants in PSMB4, PSMB9, PSMA3, and POMP have been proposed to cause a similar CANDLE phenotype in compound heterozygous monogenic (PSMB4), digenic (PSMA3/PSMB8, PSMB9/PSMB4, PSMB4/PSMB8) and AD monogenic (POMP) models [94]
Table 8.
Complement deficiencies
| Disease | Genetic defect | Inheritance | Gene OMIM | Laboratory features | Associated features |
|---|---|---|---|---|---|
| Complement deficiencies | |||||
| C1q deficiency due to defects | C1QA | AR | 120550 | Absent CH50 hemolytic activity, defective activation of the classical pathway, diminished clearance of apoptotic cells | SLE, infections with encapsulated organisms |
| C1QB | AR | 120570 | |||
| C1QC | AR | 120575 | |||
| C1r deficiency | C1R | AR | 613785 | Absent CH50 hemolytic activity, defective activation of the classical pathway | SLE, infections with encapsulated organisms, Ehlers-Danlos phenotype |
| C1r Periodontal Ehlers-Danlos | C1R | AD GOF | 613785 | Normal CH50 | Hyperpigmentation, skin fragility |
| C1s deficiency | C1S | AR | 613785 | Absent CH50 hemolytic activity, defective activation of the classical pathway | SLE, infections with encapsulated organisms, Ehlers-Danlos phenotype |
| C1s Periodontal Ehlers-Danlos | C1S | AD GOF | 613785 | Normal CH50 | Hyperpigmentation, skin fragility |
| Complete C4 deficiency | C4A + C4B | AR | 120810 | Absent CH50 hemolytic activity, defective activation of the classical pathway, complete deficiency requires biallelic mutations/deletions/conversions of both C4A and C4B | SLE, infections with encapsulated organisms, partial deficiency is common (either C4A or C4B) and appears to have a modest effect on host defense |
| C2 deficiency | C2 | AR | 217000 | Absent CH50 hemolytic activity, defective activation of the classical pathway | SLE, infections with encapsulated organisms, atherosclerosis |
| C3 deficiency (LOF) | C3 | AR | 120700 | Absent CH50 and AH50 hemolytic activity, defective opsonization, defective humoral immune response | Infections, glomerulonephritis, atypical hemolytic-uremic syndrome with GOF mutations. |
| C3 GOF | C3 | AD GOF | 120700 | Increased activation of complement | Atypical hemolytic-uremic syndrome |
| C5 deficiency | C5 | AR | 120900 |
Absent CH50 and AH50 hemolytic activity Defective bactericidal activity |
Disseminated neisserial infections |
| C6 deficiency | C6 | AR | 217050 | Absent CH50 and AH50 hemolytic activity, defective bactericidal activity | |
| C7 deficiency | C7 | AR | 217070 | ||
| C8α deficiency | C8A | AR | 120950 | ||
| C8 γ deficiency | C8G | AR | 120930 | ||
| C8 β deficiency | C8B | AR | 120960 | ||
| C9 deficiency | C9 | AR | 120940 | Reduced CH50 and AP50 hemolytic activity, deficient bactericidal activity | Mild susceptibility to disseminated neisserial infections |
| MASP2 deficiency | MASP2 | AR | 605102 | Deficient activation of the lectin activation pathway | Pyogenic infections, inflammatory lung disease, autoimmunity |
| Ficolin 3 deficiency | FCN3 | AR | 604973 | Absence of complement activation by the Ficolin 3 pathway. | Respiratory infections, abscesses |
| C1 inhibitor deficiency | SERPING1 | AD | 606860 | Spontaneous activation of the complement pathway with consumption of C4/C2, spontaneous activation of the contact system with generation of bradykinin from high molecular weight kininogen | Hereditary angioedema |
| Factor B GOF | CFB | AD GOF | 612924 | Gain-of-function mutation with increased spontaneous AH50 | Atypical hemolytic-uremic syndrome |
| Factor B deficiency | CFB | AR | 615561 | Deficient activation of the alternative pathway | Infections with encapsulated organisms |
| Factor D deficiency | CFD | AR | 134350 | Absent AH50 hemolytic activity | Neisserial infections |
| Properdin deficiency | CFP | XL | 300383 | Absent AH50 hemolytic activity | Neisserial infections |
| Factor I deficiency | CFI | AR | 217030 | Spontaneous activation of the alternative complement pathway with consumption of C3 | Infections, disseminated neisserial infections, atypical Hemolytic-uremic syndrome, preeclampsia |
| Factor H deficiency | CFH | AR or AD | 134370 | Spontaneous activation of the alternative complement pathway with consumption of C3 | |
| Factor H-related protein deficiencies | CFHR1 | AR or AD | 134371, | Normal CH50, AH50, autoantibodies to Factor H., linked deletions of one or more CFHR genes leads to susceptibility autoantibody-mediated aHUS | Older onset atypical hemolytic-uremic syndrome, disseminated neisserial infections |
| CFHR2 | 600889, | ||||
| CFHR3 | 605336, | ||||
| CFHR4 | 605337, | ||||
| CFHR5 | 608593 | ||||
| Thrombomodulin deficiency | THBD | AD | 188040 | Normal CH50, AH50 | Atypical hemolytic-uremic syndrome |
| Membrane Cofactor Protein (CD46) deficiency | CD46 | AD | 120920 | Inhibitor of complement alternate pathway, decreased C3b binding | Atypical hemolytic-uremic syndrome, infections, preeclampsia |
| Membrane Attack Complex Inhibitor (CD59) deficiency | CD59 | AR | 107271 | Erythrocytes highly susceptible to complement-mediated lysis | Hemolytic anemia, polyneuropathy |
| CD55 deficiency (CHAPEL disease) | CD55 | AR | 125240 | Hyperactivation of complement on endothelium | Protein losing enteropathy, thrombosis |
Table 10.
Phenocopies of inborn errors of immunity
| Disease | Genetic defect/presumed pathogenesis | Circulating T cells | Circulating B cells | Serum Ig | Associated features/similar PID |
|---|---|---|---|---|---|
| 1. Phenocopies of inborn errors of immunity | |||||
| Associated with somatic mutations | |||||
| Autoimmune lymphoproliferative syndrome (ALPS–SFAS) | Somatic mutation in TNFRSF6 | Increased CD4−CD8−double negative (DN) αβ T cells | Normal, but increased number of CD5+ B cells | Normal or increased | Splenomegaly, lymphadenopathy, autoimmune cytopenias, Defective lymphocyte apoptosis/ALPS–FAS (=ALPS type Im) |
| RAS-associated autoimmune leukoproliferative disease (RALD) | Somatic mutation in KRAS (GOF) | Normal | B cell lymphocytosis | Normal or increased | Splenomegaly, lymphadenopathy, autoimmune cytopenias, granulocytosis, monocytosis/ALPS-like |
| RAS-associated autoimmune leukoproliferative disease (RALD) | Somatic mutation in NRAS (GOF) | Increased CD4−CD8− double negative (DN) T alpha/beta cells | Lymphocytosis | Normal or increased | Splenomegaly, lymphadenopathy, autoantibodies/ALPS-like |
| Cryopyrinopathy, (Muckle-Wells/CINCA/NOMID-like syndrome) | Somatic mutation in NLRP3 | Normal | Normal | Normal | Urticaria-like rash, arthropathy, neurological signs |
| Hypereosinophilic syndrome due to somatic mutations in STAT5b | Somatic mutation in STAT5B (GOF) | Normal | Normal | Normal | Eosinophilia, atopic dermatitis, urticarial rash, diarrhea |
| Associated with autoantibodies | |||||
| Chronic mucocutaneous candidiasis | AutoAb to IL-17 and/or IL-22 | Normal | Normal | Normal | Endocrinopathy, chronic mucocutaneous candidiasis/CMC |
| Adult-onset immunodeficiency with susceptibility to mycobacteria | AutoAb to IFNγ | Decreased naive T cells | Normal | Normal | Mycobacterial, fungal, Salmonella VZV infections/MSMD, or CID |
| Recurrent skin infection | AutoAb to IL-6 | Normal | Normal | Normal | Staphylococcal infections/STAT3 deficiency |
| Pulmonary alveolar proteinosis | AutoAb to GM-CSF | Normal | Normal | Normal | Pulmonary alveolar proteinosis, cryptococcal meningitis, disseminated nocardiosis/CSF2RA deficiency |
| Acquired angioedema | AutoAb to CI inhibitor | Normal | Normal | Normal | Angioedema/C1 INH deficiency (hereditary angioedema) |
| Atypical hemolytic uremic syndrome | AutoAb to Complement Factor H | Normal | Normal | Normal | aHUS = Spontaneous activation of the alternative complement pathway |
| Thymoma with hypogammaglobulinemia (Good syndrome) | AutoAb to various cytokines | Increased CD8+ T cells | No B cells | Decreased | Invasive bacterial, viral or opportunistic infections, autoimmunity, PRCA, lichen planus, cytopenia, colitis, chronic diarrhea |
aHUS atypical hemolytic uremic syndrome, XL X-linked inheritance, AR autosomal recessive inheritance, AD autosomal dominant inheritance, LOF loss-of-function, GOF gain-of-function, PRCA pure red cell aplasia
Total number of conditions for Table 10: 12
Table 9.
Bone marrow failure
| Disease | Genetic defect | Inheritance | Gene OMIM | T cells | B cells | Other affected cells | Associated features | Major Category | Subcategory |
|---|---|---|---|---|---|---|---|---|---|
| Bone marrow failure | |||||||||
| Fanconi anemia type A | FANCA | AR | 227650 | Normal to low | Normal to low | HSC | Normal to low NK, CNS, skeletal, skin, cardiac, GI, urogenital anomalies, increased chromosomal breakage | Bone marrow failure with immune deficiency | Fanconi Anemia |
| Fanconi anemia type B | FANCB | XLR | 300514 | ||||||
| Fanconi anemia type C | FANCC | AR | 227645 | ||||||
| Fanconi anemia type D1 | BRCA2 | AR | 605724 | ||||||
| Fanconi anemia type D2 | FANCD2 | AR | 227646 | ||||||
| Fanconi anemia type E | FANCE | AR | 600901 | ||||||
| Fanconi anemia type F | FANCF | AR | 603467 | ||||||
| Fanconi anemia type G | XRCC9 | AR | 614082 | ||||||
| Fanconi anemia type I | FANCI | AR | 609053 | ||||||
| Fanconi anemia type J | BRIP1 | AR | 609054 | ||||||
| Fanconi anemia type L | FANCL | AR | 614083 | ||||||
| Fanconi anemia type M | FANCM | AR | 618096 | ||||||
| Fanconi anemia type N | PALB2 | AR | 610832 | ||||||
| Fanconi anemia type O | RAD51C | AR | 613390 | ||||||
| Fanconi anemia type P | SLX4 | AR | 613951 | ||||||
| Fanconi anemia type Q | ERCC4 | AR | 615272 | ||||||
| Fanconi anemia type R | RAD51 | AR | 617244 | ||||||
| Fanconi anemia type S | BRCA1 | AR | 617883 | ||||||
| Fanconi anemia type T | UBE2T | AR | 616435 | ||||||
| Fanconi anemia type U | XRCC2 | AR | 617247 | ||||||
| Fanconi anemia type V | MAD2L2 | AR | 617243 | ||||||
| Fanconi anemia type W | RFWD3 | AR | 617784 | ||||||
| MIRAGE (myelodysplasia, infection, restriction of growth, adrenal hypoplasia, genital phenotypes, enteropathy) | SAMD9 | AD GOF | 617053 | Not reported | Not reported | HSC, myeloid cells | Intrauterine growth retardation, gonadal abnormalities, adrenal failure, MDS with chromosome 7 aberrations, predisposition to infections, enteropathy, absent spleen | ||
| Ataxia pancytopenia syndrome | SAMD9L | AD GOF | 611170 | Normal | Low | HSC, myeloid cells | MDS, neurological features | ||
| DKCX1 | DKC1 | XL | 305000 | Normal to low | Normal to low | HSC | Bone marrow failure, pulmonary and hepatic fibrosis, nail dystrophy, leukoplakia, reticulate skin pigmentation; microcephaly, neurodevelopmental delay | Dyskeratosis Congenita | |
| DKCA1 | TERC | AD | 127550 | ||||||
| DKCA2 | TERT | AD | 187270 | ||||||
| DKCA3 | TINF2 | AD | 604319 | ||||||
| DKCA4 | RTEL1 | AD | 616373 | ||||||
| DKCA5 | TINF2 | AD | 268130 | ||||||
| DKCA6 | ACD | AD | 616553 | ||||||
| DKCB1 | NOLA3 | AR | 224230 | ||||||
| DKCB2 | NOLA2 | AR | 613987 | ||||||
| DKCB3 | WRAP53 | AR | 613988 | ||||||
| DKCB4 | TERT | AR | 613989 | ||||||
| DKCB5 | RTEL1 | AR | 615190 | Low | Nail dystrophy, leukoplakia, bone marrow failure, severe B cell immunodeficiency, intrauterine growth retardation, growth retardation, microcephaly, cerebellar hypoplasia, and esophageal dysfunction | ||||
| DKCB6 | PARN | AR | 616353 | Normal to low | Developmental delay, microcephaly, and cerebellar hypoplasia | ||||
| DKCB7 | ACD | AR | 616553 | Normal to low | Bone marrow failure, pulmonary and hepatic fibrosis, nail dystrophy, leukoplakia, reticulate skin pigmentation; microcephaly, neurodevelopmental delay | ||||
| BMFS1 (SRP72-deficiency) | SRP72 | AD | 602122 | NA | NA | Bone marrow failure and congenital nerve deafness | |||
| BMFS2 | ERCC6L2 | AR | 615667 | NA | NA | Bone marrow failure, learning difficulties, microcephaly | |||
| BMFS5 | TP53 | AD | 618165 | NA | Low B | Erythroid hypoplasia, B cell deficiency | |||
| Coats plus syndrome | STN1 | AR | 613129 | Normal | Normal | Intrauterine growth retardation, premature aging, pancytopenia, hypocellular bone marrow, gastrointestinal hemorrhage due to vascular ectasia, intracranial calcification, abnormal telomeres | |||
| CTC1 | AR | 617053 | Not reported | Not reported | |||||
Total number of disorders in Table 9: 43
Total number of mutant genes in Table 9: 43
HSC hematopoietic stem cell, NK natural killer, CNS central nervous system, GI gastrointestinal, MDS myelodysplastic syndrome, DKCX X-inked dyskeratosis congenital, DKCA autosomal dominant dyskeratosis congenita, DKCB autosomal recessive dyskeratosis congenita, BMFS bone marrow failure syndrome
The advances in our understanding of clinical immunology continue to expand at a vast and remarkable rate, with the addition in this update of many—64, distributed across all tables (Fig. 1b)—novel genetic defects underlying inborn errors of immunity. Perhaps not surprisingly, most if not all of these new variants were identified by NGS, thus highlighting that whole exome/whole genome sequencing has become the gold standard for identifying novel pathogenic gene variants [6–8]. Indeed, since the first application of NGS to identify novel inborn errors of immunity was published in 2010 [18], ~ 45% of all currently known disease-causing variants have been discovered by whole exome/genome sequencing. Thus, a typical approach to identifying a pathogenic variant in a new patient might now consist of first sequencing a phenotype-driven panel of genes and advancing to whole exome/genome sequencing if the cause of disease remains elusive.
In this update, we increase the list of immunological diseases to 404, with 430 known genetic defects identified as causing these conditions. The unbiased application of NGS to the discovery and characterization of novel inborn errors of immunity continues to inform clinical and basic immunology. Thus, additional phenotypes have been identified for conditions resulting from variants in known and novel genes; the penetrance of genetic variants on clinical phenotypes has been shown to be highly variable; and clinical entities sharing common phenotypes have been discovered. For example, this update includes the findings that bi-allelic mutations in ZNF341 [19, 20], IL6ST (encoding gp130, a common component of the receptors for IL-6, IL-11, IL-27, LIF, OSM, CNTF) [21, 22], or IL6R [23, 24] all cause conditions that resemble autosomal dominant hyper-IgE syndrome due to dominant negative mutations in STAT3 [15]. Detailed analyses of these patients revealed a novel mechanism of regulating STAT3 signaling (via the transcription factor ZNF341) and defined the exact consequences of impaired IL-6/IL-6R/gp130 and putatively IL-11/IL-11R/gp130 signaling to the phenotype of AD-HIES.
Furthermore, key findings over the past 2 years continue to reveal that distinct mechanisms of disease (GOF, LOF, dominant negative, haploinsufficient), as well as different modes of inheritance (autosomal recessive, autosomal dominant) of variants in the same gene can cause disparate clinical conditions. This is a fascinating aspect of the genetics of human disease, and a salient reminder to be cognizant of the nature of the genetic variants identified from NGS. It is these genes that have several entries in this update. A few recent examples include:
Heterozygous variants in CARD11 [25, 26] or STAT5B [27] can be pathogenic due to negative dominance. This was potentially unexpected because autosomal recessive LOF variants in both of these genes were previously reported to cause combined immunodeficiency and severe immune dysregulation, respectively, yet heterozygous relatives of these affected individuals were healthy [28, 29].
While heterozygous dominant negative mutations in TCF3, encoding the transcription factor E47, cause B cell deficiency and agammaglobulinemia [30], nonsense mutations in TCF3 have now been identified that are pathogenic only in an autosomal recessive state, as heterozygous carriers of these particular allelic variants remained healthy [31, 32].
A heterozygous hypermorphic variant in IKBKB was found to cause a combined immunodeficiency [33] not too dissimilar to the original description of bi-allelic, recessive variants in IKBKB [34]. Similarly, bi-allelic LOF mutations in PIK3CD are now known to cause B cell deficiency and agammaglobulinemia [35–37], which is quite distinct from the immune dysregulated state of individuals with monoallelic activating PIK3CD mutations [1, 37]. This observation nicely parallels the earlier findings of either homozygous or heterozygous mutations in PIK3R1 that clinically phenocopy recessive or activating mutations in PIK3CD respectively [1, 37].
Distinct diseases can result from heterozygous mutations in IKZF1 (Ikaros): combined immunodeficiency due to dominant negative alleles [38] or CVID due to haploinsufficiency [39].
Similar to STAT1 [40], variants in RAC2 [41–45] or CARD11 [25, 26, 28] can be pathogenic either as monoallelic GOF or LOF or bi-allelic recessive LOF.
Thus, these findings have revealed the fundamental importance of elucidating the impact of a novel variant on the function of the encoded protein and thus the mechanism of pathogenicity. Furthermore, these new entries are an important reminder not to overlook the potential significance of identifying heterozygous variants in genes previously believed to cause disease only in a biallelic manner or to result in a previously defined specific clinical entity. Indeed, there are now at least 35 genes that have multiple entries in the current update, reflecting the distinct mechanisms by which variants result in or cause disease (e.g., STAT1, STAT3, NLRP1, RAC2, ZAP70, CARD11, IKBKB, WAS, JAK1, IFIH1, C3, C1R, C1S–GOF or LOF; STAT5, STAT1, CARD11, ACD, CFH, CFHR1–5, FOXN1, RAC2, TCF3, AICDA, PIK3R1, IFNGR1, TREX1, TICAM1, IRF8–AD or AR; PIK3CD–AD GOF, AR LOF; IKZF1–AD, or haploinsufficient; NLRP3—distinct disease phenotypes despite all resulting from GOF alleles).
As noted above, genetic, biochemical, and functional analyses of putative novel pathogenic variants need to meet stringent criteria to be considered for inclusion in this update [17]. These criteria can make reporting genetic findings from single cases challenging, as often the best evidence that a novel variant is disease-causing is to identify additional, similarly affected but unrelated individuals with the same variants, or functionally similar variants in the same gene. While this can be challenging, particularly in light of the rarity of individual inborn errors of immunity, robust mechanistic laboratory investigations continue to provide compelling data from single patients, with or without evidence from animal models. Specifically, homozygous LOF mutations in IRF9 [46] and IL18BP [47] were identified and rigorously characterized in single patients and found to be the molecular cause of life-threatening influenza and fulminant viral hepatitis, respectively.
The study and discovery of novel inborn errors of immunity can also enable improved patient management by implementing gene-specific targeted therapies. Thus, JAK inhibitors are being used to treat disorders of immune dysregulation resulting from GOF mutations in JAK1, STAT1 or STAT3 [11], while mTOR inhibitors such as rapamycin or PI3K p110δ-specific inhibitors have been reported for the treatment of individuals with PIK3CD GOF or PIK3R1 LOF mutations [37]. Regarding novel gene defects, immune dysregulation due to DEF6 deficiency was successfully treated with abatacept (CTLA4-Ig) [48]. This correlated with impaired CTLA4 expression and function in DEF6-deficient T cells [48] and parallels the therapeutic use of abatacept and belatacept for LRBA-deficiency and CTLA4 haploinsufficiency, both of which are characterized by reduced CTLA4 expression in affected regulatory T cells [49, 50]. From a theoretical perspective, the finding that MSMD can be caused by mutations in IL12RB2, IL23R or SPPL2A and that these mutations are associated with impaired production of IFNγ—a requisite of anti-mycobacterial immunity—implies that IFNγ administration could be therapeutically beneficial in these clinical settings [51, 52]. Similarly, recombinant IL18BP could potentially ameliorate viral-induced liver toxicity due to IL18BP deficiency [47].
The goals of the IUIS Expert Committee on Inborn Errors of Immunity are to increase awareness, facilitate recognition, promote optimal treatment, and support research in the field of disorders of immunity. Thus, this 2019 Update and the accompanying “Phenotypical IUIS Classification” publications are intended as resources for clinicians and researchers. Importantly, these tables underpin the design of panels used for targeted gene sequencing to facilitate genetic diagnoses or inborn errors. In the past 5 years, the number of gene defects underlying inborn errors of immunity has nearly doubled from ~ 250 to 430 (Fig. 1a). The human genome contains 1800–2000 genes that are known to be involved in immune responses [13]. Thus, the discovery and study of inborn errors of immunity has elegantly illustrated that > 20% of these immune genes play non-redundant roles in host defense and immune regulation. With the improved identification and phenotyping of patients with rare diseases, combined with high throughput genome sequencing, the number of genes fundamentally required for immunity will no doubt continue to increase, further revealing critical and novel roles for specific genes, molecules, pathways and cell types in immune responses, as well as mechanisms of disease pathogenesis and targets for immunotherapies. The field of inborn errors of immunity, and the global clinical and research communities, will therefore continue to provide key insights into basic and clinical immunology.
Acknowledgments
The members of the Inborn Errors of Immunity committee would like to thanks the International Union of Immunological Societies (IUIS) for funding, as well as CSL Behring, Baxalta and Shire/Takeda for providing educational grants to enable us to compile this classification update.
Compliance with Ethical Standards
Conflict of Interest
The authors declare that they have no conflict of interest.
Footnotes
The original version of this article was revised: The in-text citations and the references were mismatched.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
2/22/2020
The original version of this article unfortunately contained mistakes in reference numbers. The in-text citations and the references were mismatched. The original article has been corrected.
References
- 1.Picard C, Bobby Gaspar H, Al-Herz W, Bousfiha A, Casanova JL, Chatila T, et al. International Union of Immunological Societies: 2017 primary immunodeficiency diseases committee report on inborn errors of immunity. J Clin Immunol. 2018;38(1):96–128. doi: 10.1007/s10875-017-0464-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bousfiha A, Jeddane L, Picard C, Ailal F, Bobby Gaspar H, Al-Herz W, et al. The 2017 IUIS phenotypic classification for primary Immunodeficiencies. J Clin Immunol. 2018;38(1):129–143. doi: 10.1007/s10875-017-0465-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Casanova JL, Abel L. Human genetics of infectious diseases: unique insights into immunological redundancy. Semin Immunol. 2018;36:1–12. doi: 10.1016/j.smim.2017.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fischer A, Rausell A. What do primary immunodeficiencies tell us about the essentiality/redundancy of immune responses? Semin Immunol. 2018;36:13–16. doi: 10.1016/j.smim.2017.12.001. [DOI] [PubMed] [Google Scholar]
- 5.Zhang SY, Jouanguy E, Zhang Q, Abel L, Puel A, Casanova JL. Human inborn errors of immunity to infection affecting cells other than leukocytes: from the immune system to the whole organism. Curr Opin Immunol. 2019;59:88–100. doi: 10.1016/j.coi.2019.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bucciol G, Moens L, Bosch B, Bossuyt X, Casanova JL, Puel A, et al. Lessons learned from the study of human inborn errors of innate immunity. J Allergy Clin Immunol. 2019;143(2):507–527. doi: 10.1016/j.jaci.2018.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meyts I, Bosch B, Bolze A, Boisson B, Itan Y, Belkadi A, et al. Exome and genome sequencing for inborn errors of immunity. J Allergy Clin Immunol. 2016;138(4):957–969. doi: 10.1016/j.jaci.2016.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Picard C, Fischer A. Contribution of high-throughput DNA sequencing to the study of primary immunodeficiencies. Eur J Immunol. 2014;44(10):2854–2861. doi: 10.1002/eji.201444669. [DOI] [PubMed] [Google Scholar]
- 9.Zhang Q, Frange P, Blanche S, Casanova JL. Pathogenesis of infections in HIV-infected individuals: insights from primary immunodeficiencies. Curr Opin Immunol. 2017;48:122–133. doi: 10.1016/j.coi.2017.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kerner G, Ramirez-Alejo N, Seeleuthner Y, Yang R, Ogishi M, Cobat A, et al. Homozygosity for TYK2 P1104A underlies tuberculosis in about 1% of patients in a cohort of European ancestry. Proc Natl Acad Sci U S A. 2019;116(21):10430–10434. doi: 10.1073/pnas.1903561116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leiding JW, Forbes LR. Mechanism-based precision therapy for the treatment of primary immunodeficiency and primary Immunodysregulatory diseases. J Allergy Clin Immunol Pract. 2019;7(3):761–773. doi: 10.1016/j.jaip.2018.12.017. [DOI] [PubMed] [Google Scholar]
- 12.Conley ME, Dobbs AK, Farmer DM, Kilic S, Paris K, Grigoriadou S, et al. Primary B cell immunodeficiencies: comparisons and contrasts. Annu Rev Immunol. 2009;27:199–227. doi: 10.1146/annurev.immunol.021908.132649. [DOI] [PubMed] [Google Scholar]
- 13.Fischer A, Rausell A. Primary immunodeficiencies suggest redundancy within the human immune system. Sci Immunol. 2016;1(6). 10.1126/sciimmunol.aah5861. [DOI] [PubMed]
- 14.Gayko U, Fung M, Clow F, Sun S, Faust E, Price S, et al. Development of the Bruton's tyrosine kinase inhibitor ibrutinib for B cell malignancies. Ann N Y Acad Sci. 2015;1358:82–94. doi: 10.1111/nyas.12878. [DOI] [PubMed] [Google Scholar]
- 15.Ma CS, Tangye SG. Flow Cytometric-based analysis of defects in lymphocyte differentiation and function due to inborn errors of immunity. Front Immunol. 2019;10:2108. doi: 10.3389/fimmu.2019.02108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bruton OC. Agammaglobulinemia Pediatrics. 1952;9(6):722–728. [PubMed] [Google Scholar]
- 17.Casanova JL, Conley ME, Seligman SJ, Abel L, Notarangelo LD. Guidelines for genetic studies in single patients: lessons from primary immunodeficiencies. J Exp Med. 2014;211(11):2137–2149. doi: 10.1084/jem.20140520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Byun M, Abhyankar A, Lelarge V, Plancoulaine S, Palanduz A, Telhan L, et al. Whole-exome sequencing-based discovery of STIM1 deficiency in a child with fatal classic Kaposi sarcoma. J Exp Med. 2010;207(11):2307–2312. doi: 10.1084/jem.20101597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beziat V, Li J, Lin JX, Ma CS, Li P, Bousfiha A, et al. A recessive form of hyper-IgE syndrome by disruption of ZNF341-dependent STAT3 transcription and activity. Sci Immunol. 2018;3(24). 10.1126/sciimmunol.aat4956. [DOI] [PMC free article] [PubMed]
- 20.Frey-Jakobs S, Hartberger JM, Fliegauf M, Bossen C, Wehmeyer ML, Neubauer JC, et al. ZNF341 controls STAT3 expression and thereby immunocompetence. Sci Immunol. 2018;3(24). 10.1126/sciimmunol.aat4941. [DOI] [PMC free article] [PubMed]
- 21.Shahin T, Aschenbrenner D, Cagdas D, Bal SK, Conde CD, Garncarz W, et al. Selective loss of function variants in IL6ST cause hyper-IgE syndrome with distinct impairments of T-cell phenotype and function. Haematologica. 2019;104(3):609–621. doi: 10.3324/haematol.2018.194233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schwerd T, Twigg SRF, Aschenbrenner D, Manrique S, Miller KA, Taylor IB, et al. A biallelic mutation in IL6ST encoding the GP130 co-receptor causes immunodeficiency and craniosynostosis. J Exp Med. 2017;214(9):2547–2562. doi: 10.1084/jem.20161810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Spencer S, Kostel Bal S, Egner W, Lango Allen H, Raza SI, Ma CA, et al. Loss of the interleukin-6 receptor causes immunodeficiency, atopy, and abnormal inflammatory responses. J Exp Med. 2019;216(9):1986–1998. doi: 10.1084/jem.20190344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nahum A, Sharfe N, Broides A, Dadi H, Naghdi Z, Mandola AB, et al. Defining the biological responses of IL-6 by the study of a novel IL-6 receptor chain (IL6R) immunodeficiency. J Allergy Clin Immunol. 2019. 10.1016/j.jaci.2019.11.015. [DOI] [PubMed]
- 25.Ma CA, Stinson JR, Zhang Y, Abbott JK, Weinreich MA, Hauk PJ, et al. Germline hypomorphic CARD11 mutations in severe atopic disease. Nat Genet. 2017;49(8):1192–1201. doi: 10.1038/ng.3898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dorjbal B, Stinson JR, Ma CA, Weinreich MA, Miraghazadeh B, Hartberger JM, et al. Hypomorphic caspase activation and recruitment domain 11 (CARD11) mutations associated with diverse immunologic phenotypes with or without atopic disease. J Allergy Clin Immunol. 2019;143(4):1482–1495. doi: 10.1016/j.jaci.2018.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klammt J, Neumann D, Gevers EF, Andrew SF, Schwartz ID, Rockstroh D, et al. Dominant-negative STAT5B mutations cause growth hormone insensitivity with short stature and mild immune dysregulation. Nat Commun. 2018;9(1):2105. doi: 10.1038/s41467-018-04521-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu HY, Bauman BM, Arjunaraja S, Dorjbal B, Milner JD, Snow AL, et al. The CBM-opathies-A rapidly expanding Spectrum of human inborn errors of immunity caused by mutations in the CARD11-BCL10-MALT1 complex. Front Immunol. 2018;9:2078. doi: 10.3389/fimmu.2018.02078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nadeau K, Hwa V, Rosenfeld RG. STAT5b deficiency: an unsuspected cause of growth failure, immunodeficiency, and severe pulmonary disease. J Pediatr. 2011;158(5):701–708. doi: 10.1016/j.jpeds.2010.12.042. [DOI] [PubMed] [Google Scholar]
- 30.Boisson B, Wang YD, Bosompem A, Ma CS, Lim A, Kochetkov T, et al. A recurrent dominant negative E47 mutation causes agammaglobulinemia and BCR(−) B cells. J Clin Invest. 2013;123(11):4781–4785. doi: 10.1172/JCI71927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ben-Ali M, Yang J, Chan KW, Ben-Mustapha I, Mekki N, Benabdesselem C, et al. Homozygous transcription factor 3 gene (TCF3) mutation is associated with severe hypogammaglobulinemia and B-cell acute lymphoblastic leukemia. J Allergy Clin Immunol. 2017;140(4):1191–4 e4. doi: 10.1016/j.jaci.2017.04.037. [DOI] [PubMed] [Google Scholar]
- 32.Qureshi S, Sheikh MDA, Qamar FN. Autosomal recessive Agammaglobulinemia - first case with a novel TCF3 mutation from Pakistan. Clin Immunol. 2019;198:100–101. doi: 10.1016/j.clim.2018.07.016. [DOI] [PubMed] [Google Scholar]
- 33.Cardinez C, Miraghazadeh B, Tanita K, da Silva E, Hoshino A, Okada S, et al. Gain-of-function IKBKB mutation causes human combined immune deficiency. J Exp Med. 2018;215(11):2715–2724. doi: 10.1084/jem.20180639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pannicke U, Baumann B, Fuchs S, Henneke P, Rensing-Ehl A, Rizzi M, et al. Deficiency of innate and acquired immunity caused by an IKBKB mutation. N Engl J Med. 2013;369(26):2504–2514. doi: 10.1056/NEJMoa1309199. [DOI] [PubMed] [Google Scholar]
- 35.Sogkas G, Fedchenko M, Dhingra A, Jablonka A, Schmidt RE, Atschekzei F. Primary immunodeficiency disorder caused by phosphoinositide 3-kinase delta deficiency. J Allergy Clin Immunol. 2018;142(5):1650–1653. doi: 10.1016/j.jaci.2018.06.039. [DOI] [PubMed] [Google Scholar]
- 36.Cohen SB, Bainter W, Johnson JL, Lin TY, Wong JCY, Wallace JG, et al. Human primary immunodeficiency caused by expression of a kinase-dead p110delta mutant. J Allergy Clin Immunol. 2019;143(2):797–9 e2. doi: 10.1016/j.jaci.2018.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tangye SG, Bier J, Lau A, Nguyen T, Uzel G, Deenick EK. Immune Dysregulation and disease pathogenesis due to activating mutations in PIK3CD-the Goldilocks' effect. J Clin Immunol. 2019;39(2):148–158. doi: 10.1007/s10875-019-00612-9. [DOI] [PubMed] [Google Scholar]
- 38.Boutboul D, Kuehn HS, Van de Wyngaert Z, Niemela JE, Callebaut I, Stoddard J, et al. Dominant-negative IKZF1 mutations cause a T, B, and myeloid cell combined immunodeficiency. J Clin Invest. 2018;128(7):3071–3087. doi: 10.1172/JCI98164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kuehn HS, Boisson B, Cunningham-Rundles C, Reichenbach J, Stray-Pedersen A, Gelfand EW, et al. Loss of B cells in patients with heterozygous mutations in IKAROS. N Engl J Med. 2016;374(11):1032–1043. doi: 10.1056/NEJMoa1512234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Toubiana J, Okada S, Hiller J, Oleastro M, Lagos Gomez M, Aldave Becerra JC, et al. Heterozygous STAT1 gain-of-function mutations underlie an unexpectedly broad clinical phenotype. Blood. 2016;127(25):3154–3164. doi: 10.1182/blood-2015-11-679902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alkhairy OK, Rezaei N, Graham RR, Abolhassani H, Borte S, Hultenby K, et al. RAC2 loss-of-function mutation in 2 siblings with characteristics of common variable immunodeficiency. J Allergy Clin Immunol. 2015;135(5):1380–4 e1-5. doi: 10.1016/j.jaci.2014.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hsu AP, Donko A, Arrington ME, Swamydas M, Fink D, Das A, et al. Dominant activating RAC2 mutation with lymphopenia, immunodeficiency, and cytoskeletal defects. Blood. 2019;133(18):1977–1988. doi: 10.1182/blood-2018-11-886028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lougaris V, Chou J, Beano A, Wallace JG, Baronio M, Gazzurelli L, et al. A monoallelic activating mutation in RAC2 resulting in a combined immunodeficiency. J Allergy Clin Immunol. 2019;143(4):1649–1653. doi: 10.1016/j.jaci.2019.01.001. [DOI] [PubMed] [Google Scholar]
- 44.Sharapova SO, Haapaniemi E, Sakovich IS, Kostyuchenko LV, Donko A, Dulau-Florea A, et al. Heterozygous activating mutation in RAC2 causes infantile-onset combined immunodeficiency with susceptibility to viral infections. Clin Immunol. 2019;205:1–5. doi: 10.1016/j.clim.2019.05.003. [DOI] [PubMed] [Google Scholar]
- 45.Smits BM, Lelieveld PHC, Ververs FA, Turkenburg M, de Koning C, van Dijk M, et al. A dominant activating RAC2 variant associated with immunodeficiency and pulmonary disease. Clin Immunol. 2019;108248. 10.1016/j.clim.2019.108248. [DOI] [PubMed]
- 46.Hernandez N, Melki I, Jing H, Habib T, Huang SSY, Danielson J, et al. Life-threatening influenza pneumonitis in a child with inherited IRF9 deficiency. J Exp Med. 2018;215(10):2567–2585. doi: 10.1084/jem.20180628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Belkaya S, Michailidis E, Korol CB, Kabbani M, Cobat A, Bastard P, et al. Inherited IL-18BP deficiency in human fulminant viral hepatitis. J Exp Med. 2019;216(8):1777–1790. doi: 10.1084/jem.20190669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Serwas NK, Hoeger B, Ardy RC, Stulz SV, Sui Z, Memaran N, et al. Human DEF6 deficiency underlies an immunodeficiency syndrome with systemic autoimmunity and aberrant CTLA-4 homeostasis. Nat Commun. 2019;10(1):3106. doi: 10.1038/s41467-019-10812-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lo B, Zhang K, Lu W, Zheng L, Zhang Q, Kanellopoulou C, et al. AUTOIMMUNE DISEASE. Patients with LRBA deficiency show CTLA4 loss and immune dysregulation responsive to abatacept therapy. Science. 2015;349(6246):436–440. doi: 10.1126/science.aaa1663. [DOI] [PubMed] [Google Scholar]
- 50.Schwab C, Gabrysch A, Olbrich P, Patino V, Warnatz K, Wolff D, et al. Phenotype, penetrance, and treatment of 133 cytotoxic T-lymphocyte antigen 4-insufficient subjects. J Allergy Clin Immunol. 2018;142(6):1932–1946. doi: 10.1016/j.jaci.2018.02.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Martinez-Barricarte R, Markle JG, Ma CS, Deenick EK, Ramirez-Alejo N, Mele F, et al. Human IFN-gamma immunity to mycobacteria is governed by both IL-12 and IL-23. Sci Immunol. 2018;3(30). 10.1126/sciimmunol.aau6759. [DOI] [PMC free article] [PubMed]
- 52.Kong XF, Martinez-Barricarte R, Kennedy J, Mele F, Lazarov T, Deenick EK, et al. Disruption of an antimycobacterial circuit between dendritic and helper T cells in human SPPL2a deficiency. Nat Immunol. 2018;19(9):973–985. doi: 10.1038/s41590-018-0178-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Roussel L, Landekic M, Golizeh M, Gavino C, Zhong MC, Chen J, et al. Loss of human ICOSL results in combined immunodeficiency. J Exp Med. 2018;215(12):3151–3164. doi: 10.1084/jem.20180668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Conde CD, Petronczki OY, Baris S, Willmann KL, Girardi E, Salzer E, et al. Polymerase delta deficiency causes syndromic immunodeficiency with replicative stress. J Clin Invest. 2019;129(10):4194–4206. doi: 10.1172/JCI128903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cui Y, Keles S, Charbonnier LM, Jule AM, Henderson L, Celik SC, et al. Combined immunodeficiency due to a loss of function mutation in DNA Polymerase Delta 1. J Allergy Clin Immunol. 2019. 10.1016/j.jaci.2019.10.004. [DOI] [PMC free article] [PubMed]
- 56.Badran YR, Dedeoglu F, Leyva Castillo JM, Bainter W, Ohsumi TK, Bousvaros A, et al. Human RELA haploinsufficiency results in autosomal-dominant chronic mucocutaneous ulceration. J Exp Med. 2017;214(7):1937–1947. doi: 10.1084/jem.20160724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Comrie WA, Faruqi AJ, Price S, Zhang Y, Rao VK, Su HC, et al. RELA haploinsufficiency in CD4 lymphoproliferative disease with autoimmune cytopenias. J Allergy Clin Immunol. 2018;141(4):1507–1510. doi: 10.1016/j.jaci.2017.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Beaussant-Cohen S, Jaber F, Massaad MJ, Weeks S, Jones J, Alosaimi MF, et al. Combined immunodeficiency in a patient with c-Rel deficiency. J Allergy Clin Immunol. 2019;144(2):606–608. doi: 10.1016/j.jaci.2019.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Calzoni E, Platt CD, Keles S, Kuehn HS, Beaussant-Cohen S, Zhang Y, et al. F-BAR domain only protein 1 (FCHO1) deficiency is a novel cause of combined immune deficiency in human subjects. J Allergy Clin Immunol. 2019;143(6):2317–2321. doi: 10.1016/j.jaci.2019.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Maffucci P, Chavez J, Jurkiw TJ, O'Brien PJ, Abbott JK, Reynolds PR, et al. Biallelic mutations in DNA ligase 1 underlie a spectrum of immune deficiencies. J Clin Invest. 2018;128(12):5489–5504. doi: 10.1172/JCI99629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bosticardo M, Yamazaki Y, Cowan J, Giardino G, Corsino C, Scalia G, et al. Heterozygous FOXN1 variants cause low TRECs and severe T cell Lymphopenia, revealing a crucial role of FOXN1 in supporting early Thymopoiesis. Am J Hum Genet. 2019;105(3):549–561. doi: 10.1016/j.ajhg.2019.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lyons JJ, Liu Y, Ma CA, Yu X, O'Connell MP, Lawrence MG, et al. ERBIN deficiency links STAT3 and TGF-beta pathway defects with atopy in humans. J Exp Med. 2017;214(3):669–680. doi: 10.1084/jem.20161435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schepers D, Tortora G, Morisaki H, MacCarrick G, Lindsay M, Liang D, et al. A mutation update on the LDS-associated genes TGFB2/3 and SMAD2/3. Hum Mutat. 2018;39(5):621–634. doi: 10.1002/humu.23407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fabre A, Charroux B, Martinez-Vinson C, Roquelaure B, Odul E, Sayar E, et al. SKIV2L mutations cause syndromic diarrhea, or trichohepatoenteric syndrome. Am J Hum Genet. 2012;90(4):689–692. doi: 10.1016/j.ajhg.2012.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Huppke P, Weissbach S, Church JA, Schnur R, Krusen M, Dreha-Kulaczewski S, et al. Activating de novo mutations in NFE2L2 encoding NRF2 cause a multisystem disorder. Nat Commun. 2017;8(1):818. doi: 10.1038/s41467-017-00932-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rodriguez R, Fournier B, Cordeiro DJ, Winter S, Izawa K, Martin E, et al. Concomitant PIK3CD and TNFRSF9 deficiencies cause chronic active Epstein-Barr virus infection of T cells. J Exp Med. 2019. 10.1084/jem.20190678. [DOI] [PMC free article] [PubMed]
- 67.Anzilotti C, Swan DJ, Boisson B, Deobagkar-Lele M, Oliveira C, Chabosseau P, et al. An essential role for the Zn(2+) transporter ZIP7 in B cell development. Nat Immunol. 2019;20(3):350–361. doi: 10.1038/s41590-018-0295-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Broderick L, Yost S, Li D, McGeough MD, Booshehri LM, Guaderrama M, et al. Mutations in topoisomerase IIbeta result in a B cell immunodeficiency. Nat Commun. 2019;10(1):3644. doi: 10.1038/s41467-019-11570-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bouafia A, Lofek S, Bruneau J, Chentout L, Lamrini H, Trinquand A, et al. Loss of ARHGEF1 causes a human primary antibody deficiency. J Clin Invest. 2019;129(3):1047–1060. doi: 10.1172/JCI120572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Keller B, Shoukier M, Schulz K, Bhatt A, Heine I, Strohmeier V, et al. Germline deletion of CIN85 in humans with X chromosome-linked antibody deficiency. J Exp Med. 2018;215(5):1327–1336. doi: 10.1084/jem.20170534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schubert D, Klein MC, Hassdenteufel S, Caballero-Oteyza A, Yang L, Proietti M, et al. Plasma cell deficiency in human subjects with heterozygous mutations in Sec61 translocon alpha 1 subunit (SEC61A1) J Allergy Clin Immunol. 2018;141(4):1427–1438. doi: 10.1016/j.jaci.2017.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mauhin W, Habarou F, Gobin S, Servais A, Brassier A, Grisel C, et al. Update on Lysinuric protein intolerance, a multi-faceted disease retrospective cohort analysis from birth to adulthood. Orphanet J Rare Dis. 2017;12(1):3. doi: 10.1186/s13023-016-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fernandez IZ, Baxter RM, Garcia-Perez JE, Vendrame E, Ranganath T, Kong DS, et al. A novel human IL2RB mutation results in T and NK cell-driven immune dysregulation. J Exp Med. 2019;216(6):1255–1267. doi: 10.1084/jem.20182015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang Z, Gothe F, Pennamen P, James JR, McDonald D, Mata CP, et al. Human interleukin-2 receptor beta mutations associated with defects in immunity and peripheral tolerance. J Exp Med. 2019;216(6):1311–1327. doi: 10.1084/jem.20182304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Has C, Castiglia D, del Rio M, Diez MG, Piccinni E, Kiritsi D, et al. Kindler syndrome: extension of FERMT1 mutational spectrum and natural history. Hum Mutat. 2011;32(11):1204–1212. doi: 10.1002/humu.21576. [DOI] [PubMed] [Google Scholar]
- 76.Kotlarz D, Marquardt B, Baroy T, Lee WS, Konnikova L, Hollizeck S, et al. Human TGF-beta1 deficiency causes severe inflammatory bowel disease and encephalopathy. Nat Genet. 2018;50(3):344–348. doi: 10.1038/s41588-018-0063-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cuchet-Lourenco D, Eletto D, Wu C, Plagnol V, Papapietro O, Curtis J, et al. Biallelic RIPK1 mutations in humans cause severe immunodeficiency, arthritis, and intestinal inflammation. Science. 2018;361(6404):810–813. doi: 10.1126/science.aar2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Li Y, Fuhrer M, Bahrami E, Socha P, Klaudel-Dreszler M, Bouzidi A, et al. Human RIPK1 deficiency causes combined immunodeficiency and inflammatory bowel diseases. Proc Natl Acad Sci U S A. 2019;116(3):970–975. doi: 10.1073/pnas.1813582116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Alosaimi MF, Hoenig M, Jaber F, Platt CD, Jones J, Wallace J, et al. Immunodeficiency and EBV-induced lymphoproliferation caused by 4-1BB deficiency. J Allergy Clin Immunol. 2019;144(2):574–83 e5. doi: 10.1016/j.jaci.2019.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Somekh I, Thian M, Medgyesi D, Gulez N, Magg T, Gallon Duque A, et al. CD137 deficiency causes immune dysregulation with predisposition to lymphomagenesis. Blood. 2019. 10.1182/blood.2019000644. [DOI] [PMC free article] [PubMed]
- 81.Carapito R, Konantz M, Paillard C, Miao Z, Pichot A, Leduc MS, et al. Mutations in signal recognition particle SRP54 cause syndromic neutropenia with Shwachman-diamond-like features. J Clin Invest. 2017;127(11):4090–4103. doi: 10.1172/JCI92876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bellanne-Chantelot C, Schmaltz-Panneau B, Marty C, Fenneteau O, Callebaut I, Clauin S, et al. Mutations in the SRP54 gene cause severe congenital neutropenia as well as Shwachman-diamond-like syndrome. Blood. 2018;132(12):1318–1331. doi: 10.1182/blood-2017-12-820308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Dhanraj S, Matveev A, Li H, Lauhasurayotin S, Jardine L, Cada M, et al. Biallelic mutations in DNAJC21 cause Shwachman-diamond syndrome. Blood. 2017;129(11):1557–1562. doi: 10.1182/blood-2016-08-735431. [DOI] [PubMed] [Google Scholar]
- 84.Arnadottir GA, Norddahl GL, Gudmundsdottir S, Agustsdottir AB, Sigurdsson S, Jensson BO, et al. A homozygous loss-of-function mutation leading to CYBC1 deficiency causes chronic granulomatous disease. Nat Commun. 2018;9(1):4447. doi: 10.1038/s41467-018-06964-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Thomas DC, Charbonnier LM, Schejtman A, Aldhekri H, Coomber EL, Dufficy ER, et al. EROS/CYBC1 mutations: decreased NADPH oxidase function and chronic granulomatous disease. J Allergy Clin Immunol. 2019;143(2):782–785. doi: 10.1016/j.jaci.2018.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.de Jong SJ, Crequer A, Matos I, Hum D, Gunasekharan V, Lorenzo L, et al. The human CIB1-EVER1-EVER2 complex governs keratinocyte-intrinsic immunity to beta-papillomaviruses. J Exp Med. 2018;215(9):2289–2310. doi: 10.1084/jem.20170308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hernandez N, Bucciol G, Moens L, Le Pen J, Shahrooei M, Goudouris E, et al. Inherited IFNAR1 deficiency in otherwise healthy patients with adverse reaction to measles and yellow fever live vaccines. J Exp Med. 2019;216(9):2057–2070. doi: 10.1084/jem.20182295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ogunjimi B, Zhang SY, Sorensen KB, Skipper KA, Carter-Timofte M, Kerner G, et al. Inborn errors in RNA polymerase III underlie severe varicella zoster virus infections. J Clin Invest. 2017;127(9):3543–3556. doi: 10.1172/JCI92280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Carter-Timofte ME, Hansen AF, Mardahl M, Fribourg S, Rapaport F, Zhang SY, et al. Varicella-zoster virus CNS vasculitis and RNA polymerase III gene mutation in identical twins. Neurol Neuroimmunol Neuroinflamm. 2018;5(6):e500. doi: 10.1212/NXI.0000000000000500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhang SY, Clark NE, Freije CA, Pauwels E, Taggart AJ, Okada S, et al. Inborn errors of RNA lariat metabolism in humans with brainstem viral infection. Cell. 2018;172(5):952–965. doi: 10.1016/j.cell.2018.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Guerin A, Kerner G, Marr N, Markle JG, Fenollar F, Wong N, et al. IRF4 haploinsufficiency in a family with Whipple's disease. Elife. 2018;7. 10.7554/eLife.32340. [DOI] [PMC free article] [PubMed]
- 92.Brehm A, Liu Y, Sheikh A, Marrero B, Omoyinmi E, Zhou Q, et al. Additive loss-of-function proteasome subunit mutations in CANDLE/PRAAS patients promote type I IFN production. J Clin Invest. 2015;125(11):4196–4211. doi: 10.1172/JCI81260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Rodero MP, Tesser A, Bartok E, Rice GI, Della Mina E, Depp M, et al. Type I interferon-mediated autoinflammation due to DNase II deficiency. Nat Commun. 2017;8(1):2176. doi: 10.1038/s41467-017-01932-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Al-Mayouf SM, Sunker A, Abdwani R, Abrawi SA, Almurshedi F, Alhashmi N, et al. Loss-of-function variant in DNASE1L3 causes a familial form of systemic lupus erythematosus. Nat Genet. 2011;43(12):1186–1188. doi: 10.1038/ng.975. [DOI] [PubMed] [Google Scholar]
- 95.Ozcakar ZB, Foster J, 2nd, Diaz-Horta O, Kasapcopur O, Fan YS, Yalcinkaya F, et al. DNASE1L3 mutations in hypocomplementemic urticarial vasculitis syndrome. Arthritis Rheum. 2013;65(8):2183–2189. doi: 10.1002/art.38010. [DOI] [PubMed] [Google Scholar]
- 96.Carbonella A, Mancano G, Gremese E, Alkuraya FS, Patel N, Gurrieri F, et al. An autosomal recessive DNASE1L3-related autoimmune disease with unusual clinical presentation mimicking systemic lupus erythematosus. Lupus. 2017;26(7):768–772. doi: 10.1177/0961203316676382. [DOI] [PubMed] [Google Scholar]
- 97.Cho K, Yamada M, Agematsu K, Kanegane H, Miyake N, Ueki M, et al. Heterozygous mutations in OAS1 cause infantile-onset pulmonary alveolar Proteinosis with Hypogammaglobulinemia. Am J Hum Genet. 2018;102(3):480–486. doi: 10.1016/j.ajhg.2018.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhong FL, Mamai O, Sborgi L, Boussofara L, Hopkins R, Robinson K, et al. Germline NLRP1 mutations cause skin inflammatory and Cancer susceptibility syndromes via Inflammasome activation. Cell. 2016;167(1):187–202. doi: 10.1016/j.cell.2016.09.001. [DOI] [PubMed] [Google Scholar]
- 99.Drutman SB, Haerynck F, Zhong FL, Hum D, Hernandez NJ, Belkaya S, et al. Homozygous NLRP1 gain-of-function mutation in siblings with a syndromic form of recurrent respiratory papillomatosis. Proc Natl Acad Sci U S A. 2019;116(38):19055–19063. doi: 10.1073/pnas.1906184116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Parlato M, Charbit-Henrion F, Pan J, Romano C, Duclaux-Loras R, Le Du MH, et al. Human ALPI deficiency causes inflammatory bowel disease and highlights a key mechanism of gut homeostasis. EMBO Mol Med. 2018;10(4). 10.15252/emmm.201708483. [DOI] [PMC free article] [PubMed]
- 101.Li Q, Lee CH, Peters LA, Mastropaolo LA, Thoeni C, Elkadri A, et al. Variants in TRIM22 that affect NOD2 signaling are associated with very-early-onset inflammatory bowel disease. Gastroenterology. 2016;150(5):1196–1207. doi: 10.1053/j.gastro.2016.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.de Jesus AA, Brehm A, VanTries R, Pillet P, Parentelli AS, Montealegre Sanchez GA, et al. Novel proteasome assembly chaperone mutations in PSMG2/PAC2 cause the autoinflammatory interferonopathy CANDLE/PRAAS4. J Allergy Clin Immunol. 2019;143(5):1939–1943. doi: 10.1016/j.jaci.2018.12.1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Gayden T, Sepulveda FE, Khuong-Quang DA, Pratt J, Valera ET, Garrigue A, et al. Germline HAVCR2 mutations altering TIM-3 characterize subcutaneous panniculitis-like T cell lymphomas with hemophagocytic lymphohistiocytic syndrome. Nat Genet. 2018;50(12):1650–1657. doi: 10.1038/s41588-018-0251-4. [DOI] [PubMed] [Google Scholar]
- 104.Polprasert C, Takeuchi Y, Kakiuchi N, Yoshida K, Assanasen T, Sitthi W, et al. Frequent germline mutations of HAVCR2 in sporadic subcutaneous panniculitis-like T-cell lymphoma. Blood Adv. 2019;3(4):588–595. doi: 10.1182/bloodadvances.2018028340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kapferer-Seebacher I, Pepin M, Werner R, Aitman TJ, Nordgren A, Stoiber H, et al. Periodontal Ehlers-Danlos syndrome is caused by mutations in C1R and C1S, which encode subcomponents C1r and C1s of complement. Am J Hum Genet. 2016;99(5):1005–1014. doi: 10.1016/j.ajhg.2016.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]