Abstract
Diazotrophic microorganisms regulate marine productivity by alleviating nitrogen limitation. However, we know little about the identity and activity of diazotrophs in deep-sea sediments, a habitat covering nearly two-thirds of the planet. Here, we identify candidate diazotrophs from Pacific Ocean sediments collected at 2893 m water depth using 15N-DNA stable isotope probing and a novel pipeline for nifH sequence analysis. Together, these approaches detect an unexpectedly diverse assemblage of active diazotrophs, including members of the Acidobacteria, Firmicutes, Nitrospirae, Gammaproteobacteria, and Deltaproteobacteria. Deltaproteobacteria, predominately members of the Desulfobacterales and Desulfuromonadales, are the most abundant diazotrophs detected, and display the most microdiversity of associated nifH sequences. Some of the detected lineages, including those within the Acidobacteria, have not previously been shown to fix nitrogen. The diazotrophs appear catabolically diverse, with the potential for using oxygen, nitrogen, iron, sulfur, and carbon as terminal electron acceptors. Therefore, benthic diazotrophy may persist throughout a range of geochemical conditions and provide a stable source of fixed nitrogen over geologic timescales. Our results suggest that nitrogen-fixing communities in deep-sea sediments are phylogenetically and catabolically diverse, and open a new line of inquiry into the ecology and biogeochemical impacts of deep-sea microorganisms.
Subject terms: Microbial ecology, Biogeochemistry, Microbial biooceanography, Stable isotope analysis, Environmental microbiology
Introduction
Nitrogen is an essential element for life and microorganisms play a critical role in its bioavailability. Through the enzymatic reduction of dinitrogen gas to ammonia, nitrogen-fixing organisms (i.e., diazotrophs) produce the Earth’s largest natural source of bioavailable nitrogen [1]. In particular, marine diazotrophs supply nearly one-half of the global fixed nitrogen demand [2] and their activity often regulates marine primary productivity [3–5]. However, despite its well-documented ecological and biogeochemical importance in the pelagic euphotic zone [6], and its continued investigation in shallow marine sediments [7–10], nitrogen fixation in deep marine sediments (>200 m water depth) remains relatively unexplored. This may be a significant oversight, given that deep-sea sediments cover nearly two-thirds of Earth’s surface, have high microbial densities (up to 1000× greater than surface waters) [11], and can have relatively low concentrations of bioavailable nitrogen at the surface (<25 μM) [12].
Recently, nitrogen fixation has been detected or implicated in several geochemically anomalous deep-sea habitats, including methane seeps [12–15], hydrothermal vents [16], whale falls [12], and oxygen minimum zones [17, 18]. The emerging perspective from nifH sequencing [19], inhibition experiments [20], and geochemical correlation analyses [12, 15, 17, 18] suggests that the diazotrophs across these sites are phylogenetically and catabolically diverse. However, only two deep-sea taxa have been directly identified as diazotrophs to date, the methanogenic Methanocaldococcus sp. FS406-22 isolated from hydrothermal vent fluid [21] and the methanotrophic ANME-2 archaea found at methane seeps [22]. Even less is known about the identity of deep-sea diazotrophs outside these geochemically anomalous regions, despite diverse nifH sequences [19] and bulk 15N2 assimilation [12] suggesting their presence and activity. Identifying the organisms that fix nitrogen in widely representative deep-sea sediments would likely reveal novel nitrogen-fixing taxa, prescribe ecosystem function to uncultivated lineages, and help evaluate the biogeochemical impacts of and controls on diazotrophy in the greater marine benthos.
In the present study, we investigate the identity and activity of diazotrophs within deep-sea sediment collected at the distal end of Monterey Canyon, CA, USA (0–3 and 9–12 cm below seafloor [cmbsf]; 2893 m water depth). Active diazotrophy was previously demonstrated within this sediment via 15N2 tracer assays [12], however, the responsible organisms were unidentified. Here, we identify the active diazotrophs using a combination of density-gradient 15N-DNA stable isotope probing (15N-SIP) and a novel nifH amplicon analysis pipeline. We investigate sediment incubated with a headspace of either argon or methane to examine the effect of methane on diazotroph community composition and activity. From our integrative approach, we identify diazotrophs with broad phylogenetic and catabolic diversity, bearing implications for the resiliency and biogeochemical impacts of deep-sea diazotrophy through time.
Materials and methods
Sample collection and 15N2 sediment microcosm incubations
Sediment pushcores from Monterey Canyon, CA, USA were collected in October 2010 on the R/V Western Flyer using ROV Doc Ricketts (Fig. 1a). The sampling site was located at 2893 m water depth and showed no visible signs of physical or geochemical anomalies. The site was 28 m from a previous whale fall (deposited ~10 years prior to sampling). 15N2 incubations from the 0–3 and 9–12 cmbsf horizons were conducted with sediment mixed 1:1 with argon-sparged 0.2 μm-filtered bottom water in 60 ml serum bottles amended with argon or methane headspaces and 15N2 gas (Sigma-Isotec lot #SZ1694). For details on sample collection, sediment geochemistry, 15N2 incubation set up and subsampling, including an assessment of the negligible 15N-contaminants in the 15N2 gas, and bulk 15N-incorporation over time, see Dekas et al. [12].
Fig. 1. Sampling location and previously measured N2 fixation in the sediment core used in this study.
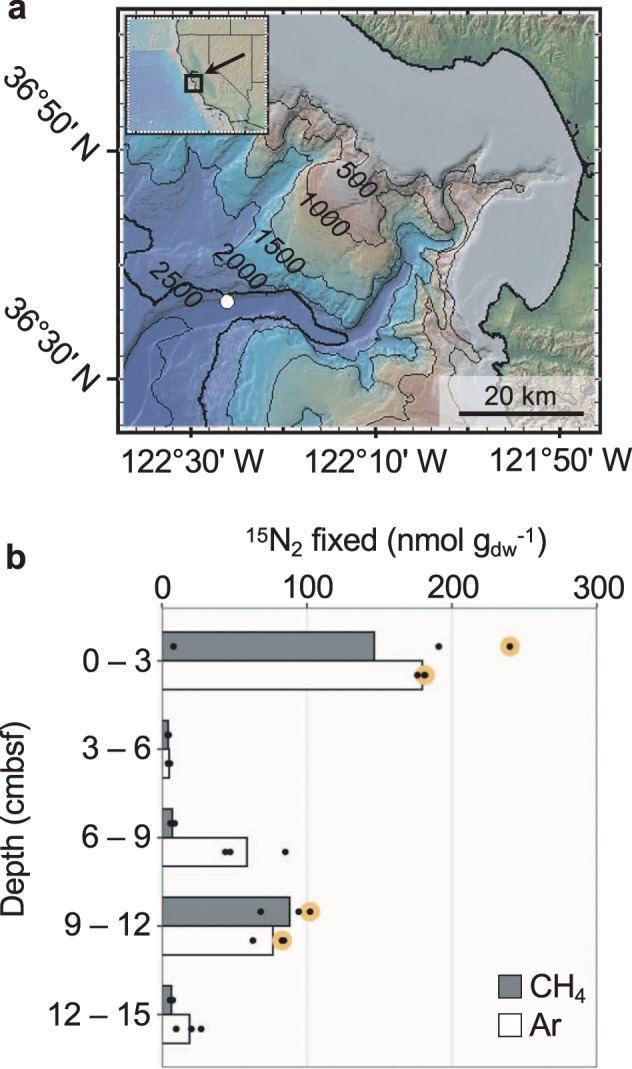
a Map of sampling site at Monterey Canyon, California, USA. Sampled location marked with a white circle. Contour lines show 500 m depth intervals. b 15N2 assimilation in sediment microcosms measured using isotope ratio mass spectrometry (reproduced from Dekas et al. [12]). Microcosm headspace gas is indicated. Each circle represents a biological replicate, with bars indicating the average 15N2 assimilation. Large, yellow circles indicate the replicate bottle used for 15N-SIP and nifH analyses.
DNA extractions
DNA was extracted in duplicate from raw (unincubated) and 15N2-incubated (2-month time point) sediment using a PowerSoil DNA Isolation Kit (MoBio Laboratories, Carlsbad, CA, USA) following manufacturer protocol with the following modifications. Samples were initially centrifuged at 10 000 rcf for 30 s and supernatant removed. After addition of Solution C1, bead beat tubes were heated at 65 °C for 10 min, briefly vortexing after 5 min. Cells were lysed by bead-beating at 5.5 m s−1 for 45 s using a FastPrep-24 5 G sample homogenizer (MP Biomedicals, Santa Ana, CA, USA). Duplicate DNA extracts were combined and stored at −80 °C.
Density-gradient stable isotope probing
Density-gradient separation of DNA was performed according to Buckley et al. [23] with the exception of excluding the secondary bis-benzimide CsCl gradient [24]. In summary, DNA fragments >4 kb were selected using a BluePippin platform (Sage Science, Beverly, MA, USA) and added to gradient buffer (15 mM Tris-HCl, pH 8.0; 15 mM EDTA; 15 mM KCl) containing 1.762 g ml−1 CsCl in 4.7 ml polypropylene tubes. Samples were ultracentrifuged to isopycnic equilibrium at 164 000 rcf for 66 h at 20 °C in a TLA-110 fixed angle rotor (Beckman Coulter, Brea, CA, USA). Following centrifugation, tubes were fractionated from bottom to top in 100 μl increments via displacement by Milli-Q water. Fraction densities were calculated using an AR200 Digital Refractometer (Reichert Technologies, Depew, NY, USA). Fractions were then desalted using an Ampure XP bead clean-up kit (Beckman Coulter) and stored at −80 °C.
16S rRNA gene and nifH amplification and sequencing
The 16S rRNA gene was amplified from all density fractions as well as unfractionated DNA extract (subsampled prior to size selection). PCR was performed in duplicate 25 μl reactions containing: 1 × Quantabio 5Prime HotMasterMix (Quantabio, Beverly, MA, USA), 0.2 μM each of V4/V5 515F-Y and 926R primers [25], 1 μl of DNA template, and 13 μl of molecular-grade water. Primer sequences were modified to include an Illumina overhang adapter. Negative controls and a 16S rRNA mock community (as in Parada et al. [25]) were included. Initial denaturation was performed at 95 °C for 180 s, followed by 30 cycles of 95 °C for 45 s, 50 °C for 45 s, and 68 °C for 90 s, and a final elongation step at 68 °C for 5 min. Oligonucleotide barcodes were added during a second PCR with the following conditions: 95 °C for 180 s, 8 cycles of 95 °C for 30 s, 55 °C for 30 s, and 72 °C for 30 s, and a final elongation step at 72 °C for 5 min. No amplification was observed in the negative controls in a 2% agarose gel. PCR duplicates received the same oligonucleotide barcodes and were pooled after the second PCR.
The nifH gene was amplified from unfractionated DNA in duplicate 25 μl PCR reactions containing: 2.5 μl 10 × ExTaq Buffer (+Mg2+), 0.5 μl 10 mM dNTPs (Takara Bio USA, Mountain View, CA, USA), 0.5 μl 2.5 mg/ml bovine serum albumin (New England BioLabs, Ipswich, MA, USA), 0.3 μl ExTaq DNA polymerase Hot-Start version (Takara Bio USA), 0.5 μl each of forward and reverse nifH primers (10 μM) described in Mehta et al. [16], 1 μl of DNA template, and 19.2 μl of molecular-grade water. Primer sequences were modified to include an Illumina overhang adapter. One negative control was included using 1 μl of molecular-grade water as template. Initial denaturation was performed at 95 °C for 120 s, followed by 35 cycles of 95 °C for 30 s, 55 °C for 30 s, and 72 °C for 45 s, and a final elongation step at 72 °C for 5 min. Oligonucleotide barcodes were added during a second PCR and duplicates were pooled, as described above.
Barcoded PCR products were purified using an Ampure XP bead clean-up kit (Beckman Coulter) at 0.7 × bead solution, pooled in equal concentrations, purified again at 0.7 × bead solution, and sent to the UC Davis DNA Technologies Core Facility (Davis, CA, USA) for 2 × 250 bp paired-end sequencing on an Illumina MiSeq platform.
Quality filtering and inferring sequence variants
16S rRNA and nifH primer sequences were removed from demultiplexed fastq files using cutadapt (v.1.13; [26]). The following filtering and processing steps were performed using DADA2 (v.1.4.0; [27]). Reads were trimmed to 220 bp and reads containing ambiguous bases or >2 expected sequencing errors were removed. Quality-filtered reads were pooled, amplicon sequence variants (ASVs) inferred, paired-end reads merged, and chimeric sequences removed. Merged paired-end sequences were filtered, keeping those 368–378 bp for 16S rRNA gene and 330–370 bp for nifH. The nifH ASVs that did not align to the target region were manually discarded. Taxonomy was assigned to 16S rRNA sequences via alignment to SILVA Train Set v132. Singleton, unclassified, and eukaryotic sequences were pruned.
15N-incorporator identification
16S rRNA gene sequences labeled with 15N were identified using the multiple window high-resolution DNA-SIP (MW-HR-SIP) method from the HTSSIP software package (v.1.4.0; [28]) in R (v.3.4.0; [29]). The HTSSIP package implements DESeq2 [30] to test if a given ASV is significantly enriched in abundance in a given buoyant density range between control (here, the unincubated sediment) and treatment (here, the sediment incubated with 15N2). Data were pre-filtered to prune ASVs that: (1) differed in abundance by 10× between unfractionated control-treatment pairs and (2) did not appear in at least three fractions >1.69 g ml−1 in both control and treatment. These criteria filtered taxa whose abundances may have been affected due to bottle effects and/or incubation with either N2 or CH4. Moderated log2-fold-changes were calculated per taxon across four buoyant density windows: 1.7000–1.7150 g ml−1, 1.7075–1.7225 g ml−1, 1.7150–1.7300 g ml−1, and 1.7225–1.7375 g ml−1 with a null threshold of 0.25. These windows were narrow enough to detect the small expected buoyant density shifts, while containing at least 3 fractions per sample. Fold changes were tested for statistical significance using one-sided Wald tests and adjusted for multiple hypothesis testing using the Benjamini–Hochberg method to a false detection rate of 0.05.
Evaluating 15N-SIP accuracy
To evaluate SIP accuracy, the percentage of false positives as a function of 15N-enrichment of the labeled biomass was estimated using simulations in SIPSim (v.0.1; [31]). Microbial communities for simulated experiments were assembled by downloading 816 complete genomes from the NCBI RefSeq database. Percent taxa shared and percent of rank abundances permuted between control and treatment communities were set to match the values from the sample incubated with argon from the 9–12 cmbsf horizon analyzed here. Default SIPSim settings were used, with the exception of simulating 1000 fragments per genome. Simulated OTU tables were then filtered and analyzed following the same pipeline used for the real samples.
Atom percent of 15N-labeled biomass was estimated to be 40.4 at% 15N by mass balance, using the previous measurement of 0.422 as the total at% 15N of the N pool in this sediment [12] and assuming that: (1) 98.7% of total organic N was from dead biomass and (2) 10% of the community was 15N-labeled [32]. Given the potential error in these assumptions, specificity estimates for simulated communities with an at% labeling within a factor of two of our 15N-labeled biomass estimate (i.e., 20–50 at% 15N) were averaged together for the final specificity estimate.
Closest characterized relatives of 15N-incorporators
Closest characterized relatives of 15N-incorporators were identified via BLAST search using the NCBI 16S ribosomal RNA sequences database. Complete or near-complete 16S rRNA gene sequences were downloaded, aligned using MAFFT (v.7.055b; [33]), and maximum likelihood phylogenetic trees inferred using RAxML (v.7.7.2; [34]) with the GTR + G + I evolutionary model and 100 bootstrap replicates. One 16S rRNA gene copy from Methanocaldococcus jannaschii was used as the outgroup (GenBank accession no. NR_113292).
Inferring nifH host identity
A reference nifH database was curated from the NCBI Nucleotide database that contained all sequences annotated as “nifH”, “nitrogenase reductase”, “nitrogenase iron protein”, “anfH”, or “vnfH” (n = 83 318 as of August 28, 2018). The database was filtered to remove sequences that were: (1) from unidentified organisms, (2) <200 bp or >1 kb in length, and (3) not flanked by start and stop codons. The filtered nifH database (n = 6040 sequences) was aligned using MAFFT (v.7.055b), adjusting for sequence direction, and then a phylogenetic tree was inferred using RAxML (v.8.2.12) with the GTR + CAT evolutionary model and 100 bootstrap replicates.
SEPP (v.4.3.5; [35]) was used to insert the nifH ASVs into the full-length nifH reference alignment and reference tree. After placement on the reference tree, the taxonomic classifications for reference sequences that were within a patristic distance <0.48 (i.e., sum of branch lengths; k) to each ASV were obtained using myTAI (v.0.8.0; [36]). Reference sequences within this threshold were 81% (±2.6%) identical to amplicon sequences (min. 71% sequence identity) (Supplementary Fig. S1). The host identity for each nifH ASV was then inferred based on these references’ conserved taxonomic ranks. ASVs separated by patristic distances >0.48 to their nearest neighbors remained unclassified. The k value was set empirically to the value that maximized the number of annotated ASVs (Supplementary Fig. S2). This balances: (1) between being too restrictive and failing to identify reference sequences near an ASV (<<k) and (2) overexploring tree space and including reference sequences from different taxonomic lineages that display patterns of vertical inheritance (>>k). All trees were visualized using the Interactive Tree of Life [37].
The sequence placements for the 42 nifH ASVs with k > 0.48 were inspected manually. Nine of these ASVs were found to display clustering characteristics consistent with a valid taxonomic signature. Eight unidentified nifH ASVs clustered with the single Acidobacteria nifH sequence available from GenBank, but were not assigned to the Acidobacteria due to their patristic distance to Desulfovibrio nifH sequences being within the selected k threshold (kAc-Dv = 0.40; k = 0.48). However, the failure to classify is likely a function of the poor representation of Acidobacteria nifH sequences in the database (n = 1); if more reference sequences were available, these ASVs may have been assigned to Acidobacteria hosts. Conversely, one ASV clustered within the Betaproteobacteria, but its patristic distance to its nearest neighbor was greater than our selected annotation threshold, so it remained unclassified (knear = 0.49; k = 0.48). Thus, while these ASVs were not annotated under the optimized k threshold, they are considered to belong to the lineages in their respective clusters.
Results and discussion
Identity and relative abundance of 15N-incorporators
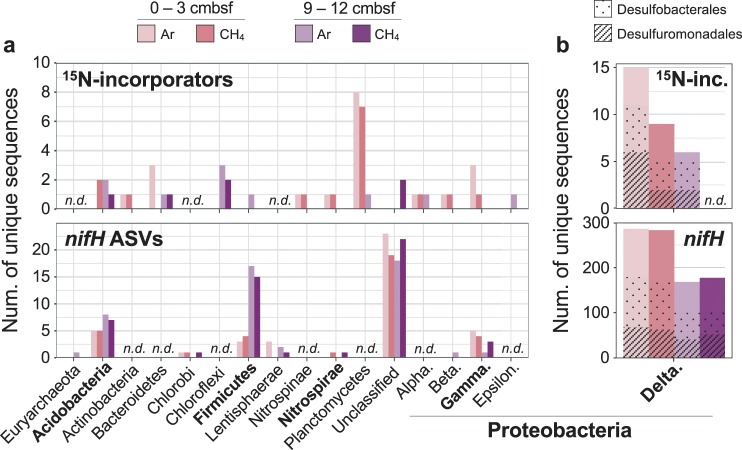
We used 15N-SIP to identify organisms that assimilated 15N2 in our sediment incubations. 15N-SIP links taxonomic identity to active function without requiring cultivation or lineage-specific hypotheses [38], thereby increasing the potential to observe a broad community of diazotrophs, including novel taxa [24, 39]. We identified a total of 61 taxa (unique 16S rRNA gene ASVs) as having incorporated 15N into their DNA across both sediment depths and headspace compositions (Fig. 2). We hereafter refer to these taxa as 15N-incorporators. Deltaproteobacteria composed 21 of the 61 15N-incorporators, with the Desulfobacterales and Desulfuromonadales each containing eight. Outside the Deltaproteobacteria, the 15N-incorporators were phylogenetically diverse, falling within the Acidobacteria, Actinobacteria, Bacteroidetes, Chloroflexi, Firmicutes, Nitrospinae, Nitrospirae, Planctomycetes, and Proteobacteria (Alpha-, Beta-, Gamma-, and Epsilon-).
Fig. 2. Taxonomic diversity of 15N-incorporators and nifH-containing taxa identified via 15N-SIP and DNA sequencing, respectively.
a Taxa listed by class for Proteobacteria and phylum for all other groups. b Deltaproteobacteria 15N-incorporators and nifH ASVs shown separately to accommodate for different vertical axis scales. The fraction of sequences identified as Desulfobacterales (dots), Desulfuromonadales (stripes), and other Deltaproteobacteria (no pattern) is additionally indicated. n.d. = not detected.
In the 0–3 cmbsf sediment, we identified 34 and 25 taxa as 15N-incorporators after incubation with headspaces of argon or methane, respectively (Fig. 2). Seventeen of these were identified as incorporators in both incubations and belonged to the Nitrospinae (n = 1), Nitrospirae (n = 1), Planctomycetes (n = 4), Betaproteobacteria (n = 1), Gammaproteobacteria (n = 1), and Deltaproteobacteria (n = 9) (Supplementary Fig. S3). The taxa identified as 15N-incorporators from the Ar incubation at this depth composed 8% of the incubated community’s 16S rRNA gene reads (Fig. 3a). In the corresponding raw (unincubated) sediment, the same taxa composed 3% of reads, indicating their approximate in situ relative abundance. The taxa identified as 15N-incorporators from the CH4 incubation at this depth accounted for 5% and 2% of the incubated and raw communities, respectively (Fig. 3b).
Fig. 3. Relative abundance of 15N-incorporators in the incubation in which they were identified, as well as the corresponding raw (unincubated) sediment.
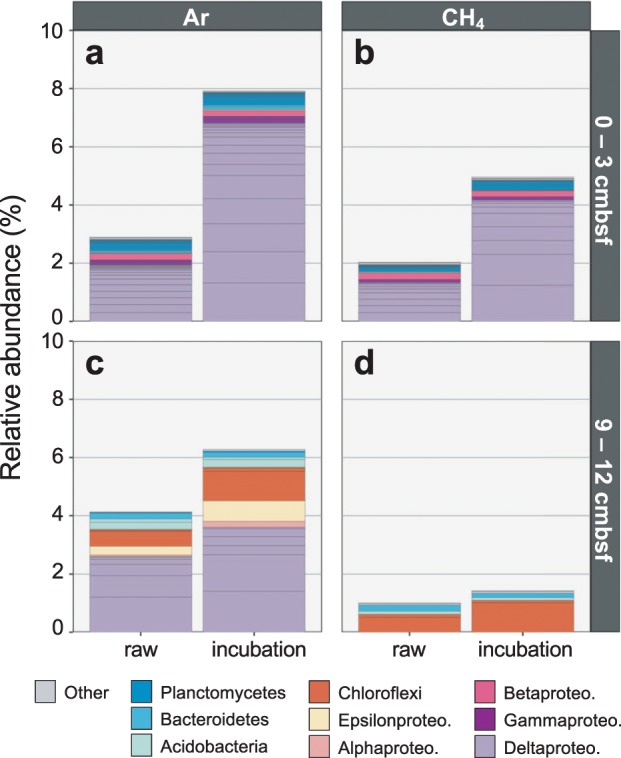
Relative abundances shown for 0–3 cmbsf samples incubated with argon (a) or methane (b) and 9–12 cmbsf samples incubated with argon (c) or methane (d). Lineages that accounted for >0.1% of 16S rRNA gene reads in at least one sample are colored by class (for Proteobacteria only) or phylum. Low abundance lineages (i.e., <0.1% of reads in each sample) are grouped as ‘Other’. Internal bar lines show relative abundances of individual ASVs.
Fewer taxa were identified as 15N-incorporators in the 9–12 cmbsf incubations, consistent with the lower detected rates of N2 fixation in that sediment horizon (Fig. 1b). From these incubations, we identified 16 and 6 taxa as 15N-incorporators after incubation with Ar or CH4, respectively (Fig. 2). These incubations shared three 15N-incorporators, one each from the Acidobacteria, Bacteroidetes, and Chloroflexi (Supplementary Fig. S3). The 15N-incorporators from the Ar incubation at this depth accounted for 6% of the incubated community’s 16S rRNA gene reads, and the same taxa composed 4% of reads in the corresponding raw community (Fig. 3c). The taxa identified as 15N-incorporators from the CH4 incubation at this depth composed 1% of reads from both the incubated and raw samples (Fig. 3d). The results from both sediment depths therefore indicate that active diazotrophs represent a relatively low percentage of the in situ microbial communities (<5%).
15N-SIP accuracy depends upon a suite of parameters, including experimental design, number of isotopically-enriched taxa, sequencing depth, and computational method of incorporator detection [31]. Using a recently developed software package to quantify SIP accuracy [31], we estimate our 15N-SIP analysis has a false positive detection rate of 18% (±16%) (see “Methods”; Supplementary Fig. S4). However, three additional lines of evidence support the conclusion that 15N-labeled taxa are identified accurately: the list of 15N-incorporators was not predicted by (1) ASV rank abundance, (2) lineages with the most ASVs, nor (3) ASV abundance ratios between control and treatment unfractionated samples (Supplementary Fig. S5). It is also important to note the possibility of false negatives. For instance, we did not identify Deltaproteobacteria 15N-incorporators in the 9–12 cmbsf incubation with CH4 (Figs. 2b and 3d). Although this could indicate a change in community dynamics with methane, their lack of detection may be an artifact introduced during data pre-filtering to minimize false positives (Supplemental Fig. S4).
Separately, cross-feeding of 15N-labeled substrates between diazotrophic and non-diazotrophic organisms can inflate 15N-incorporator diversity. While previous nanoscale secondary ion mass spectrometry analyses of individual cells from deep-sea methane seep sediments incubated with 15N2 did not find significant cross-feeding after six months of incubation [14, 22], a length three times longer than the experiments here, it still remains a potential cause of 15N-labeling, especially in the case of physically associated cells. To minimize the impact of these potential sources of error, we supplemented the 15N-SIP results with a molecular analysis of nifH.
Inferred diversity and identity of nifH-containing taxa
To survey the diversity of taxa potentially capable of fixing nitrogen, which includes those not active under the incubation conditions, we performed amplicon sequencing of the nifH gene. We recovered a total of 62 748 reads after quality-filtering, comprising 1026 unique ASVs across all raw and incubated sediment samples from both depths. Of these ASVs, 434 were bona fide nifH sequences (hereafter referred to as nifH ASVs) and 592 were homologs of nifH. This high-throughput approach combined with greater sequence resolution recovered nearly an order of magnitude more unique nifH sequences than previous deep-sea studies [12, 19], implying a greater diazotroph diversity in the benthos than previously known.
In addition, to corroborate the 15N-SIP results with an independent assessment of diazotroph identity, we applied a novel pipeline to infer taxonomic identity from the nifH amplicon sequences. Our approach adopts a phylogenetic framework whereby amplicons are inserted into a reference tree of curated nifH sequences. Then, the identities of the amplicons’ source organisms are inferred based on the neighboring sequences according to empirically-defined parameters (see Methods). When amplicons are located near reference sequences that differ in taxonomic origin, source organism identity is not inferred. This approach therefore minimizes errors in host identification due to horizontal gene transfer. We hereafter refer to the inferred hosts as nifH-containing taxa. Following this approach, we were able to propose identities at the phylum rank for 392 of the 434 recovered nifH sequences.
Consistent with the 15N-SIP results, we identified a phylogenetically diverse assemblage of nifH-containing taxa across all samples, with most belonging to the Deltaproteobacteria (84%; 331 of 392 ASVs) (Fig. 2). The nifH ASVs from inferred Deltaproteobacteria hosts dominated the relative abundance in each sample as well, comprising 81% of nifH reads from the raw 0–3 cmbsf horizon and 79% from the 9–12 cmbsf horizon (Supplementary Fig. S6) Of the inferred Deltaproteobacterial nifH ASVs, 38% were inferred to belong to the order Desulfuromonadales, 23% to the Desulfobacterales, 3% to the Desulfovibrionales, and 37% to undetermined orders. Within the Desulfuromonadales, most ASVs fell into one of three nifH clusters: those most closely related to the nifH sequence from Desulfuromusa kysingii (78–84% nucleotide sequence id), to four nifH sequences from the genus Desulfuromonas (81–89% nuc. id), or to two nifH sequences within the genus Pelobacter (81–87% nuc. id). These three nifH clusters accounted for ~70% of all nifH reads from the raw sediments and 26–45% from the incubated sediments. These findings suggest that, while we inferred diverse nifH-containing taxa overall, a few clusters of nifH-containing taxa from the Deltaproteobacteria composed the majority of the in situ diazotroph community.
Outside the Deltaproteobacteria, we recovered many lower abundance nifH sequences from phylogenetically diverse inferred taxa, including the Euryarchaeota (n = 2 ASVs), Acidobacteria (n = 8 ASVs), Bacteroidetes (n = 1 ASV), Chlorobi (n = 1 ASV), Firmicutes (n = 31 ASVs), Lentisphaerae (n = 7 ASVs), Nitrospirae (n = 2 ASVs), Betaproteobacteria (n = 1 ASV), and Gammaproteobacteria (n = 7 ASVs) (Fig. 4). Those inferred to be from the Acidobacteria and Firmicutes were most abundant, and respectively accounted for 6% and 10% of nifH reads in the 9–12 cmbsf incubations (Supplementary Fig. S6). The nifH ASVs from the Euryarchaeota, Bacteroidetes, Chlorobi, Lentisphaerae, Nitrospirae, and Gammaproteobacteria each accounted for 0.03–3% of nifH reads in each sample where detected. With the exception of the Euryarchaeota (found in only 9–12 cmbsf), we recovered nifH ASVs from each lineage in both 0–3 and 9–12 cmbsf horizons (Fig. 2a). These highly diverse, lower abundance nifH sequences suggest a taxonomically diverse assemblage of nifH-containing taxa in the deep-sea benthos.
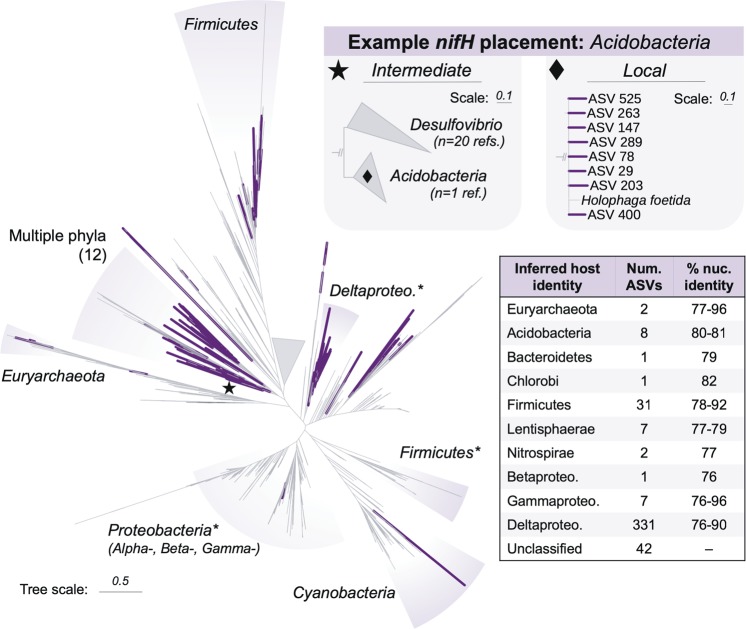
Fig. 4. Diversity of recovered nifH sequences.
Bold branches show nifH ASVs (n = 434) placed at their maximum likelihood positions on the reference nifH tree (n = 6040). Shaded regions indicate taxonomic identities of select clades for reference. Gray wedge shows collapsed branches of nifH homologs; the full tree including these sequences can be found in Supplementary Fig. S7. Reference sequences on terminal branches with lengths >1.0 were pruned for clarity (n = 34). Phyla within clade labeled “Multiple phyla”: Acidobacteria, Actinobacteria, Bacteroidetes, Chlorobi, Chloroflexi, Cyanobacteria, Elusimicrobia, Lentisphaerae, Nitrospirae, Proteobacteria, Spirochaetes, and Verrucomicrobia. Star and diamond insets display example nifH ASV placements at finer phylogenetic scales. ‘% nuc. identity’ in table shows range of percent nucleotide identities between nifH ASVs and closest references for each taxonomic group. *clades which include one or two nifH sequences from other phyla (clades without asterisks contain sequences from one phylum).
Comparison of 15N-SIP and nifH results to identify ‘candidate diazotrophs’
We combine the 15N-SIP and nifH analyses to present the most convincing cases for diazotrophy, and refer to those taxa as ‘candidate diazotrophs’ if supported by both lines of evidence. Both the 15N-SIP and nifH results indicate a phylogenetically diverse community of diazotrophs, including the Acidobacteria, Firmicutes, Nitrospirae, Gammaproteobacteria, and Deltaproteobacteria (Fig. 2). Both datasets also suggest that Deltaproteobacteria are the most abundant and diverse group of diazotrophs in both the 0–3 and 9–12 cmbsf sediment horizons. In particular, our combined results suggest that Deltaproteobacteria similar to Pelobacter carbinolicus and Desulfuromonas acetoxidans fixed nitrogen, since: (1) we identify as 15N-incorporators 16S rRNA gene ASVs that share >96% nucleotide sequence identity (V4/V5 region) with these two organisms, (2) both organisms contain copies of nifH in their reference genomes, and (3) we recovered nifH ASVs that clustered with those sequences in all incubations where they were identified as 15N-incorporators (81% nifH nuc. id for D. acetoxidans and 81–89% nifH nuc. id for P. carbinolicus). These findings provide the first anabolic evidence of active diazotrophy in these two lineages, extending previous molecular studies that implicated them as diazotrophs in both shallow- and deep-sea sediments [18–20, 40, 41].
Although the detected lineages within the Acidobacteria, Firmicutes, Nitrospirae, and Gammaproteobacteria each compose <1% of 16S rRNA reads in each sample (Fig. 3), their detection indicates diverse diazotrophs beyond the Deltaproteobacteria, and in some cases reveals novel diazotrophs. To our knowledge, this is the first evidence suggesting Acidobacteria fix nitrogen in the environment. Diazotrophy within the Acidobacteria is supported here by repeated detection of 15N incorporation into Acidobacterial 16S rRNA gene ASVs (n = 4) and multiple nifH ASVs (n = 8) (Fig. 2a; Supplementary Fig. S8). The low sequence identity in 16S rRNA gene amplicons between the Acidobacteria ASVs and their closest characterized relatives (88–91% nuc. id) suggests these candidate diazotrophs represent novel lineages within this phylum. Currently, a single published Acidobacteria genome, Holophaga foetida [42], contains the full genetic complement required to fix nitrogen (nifHDKENB), but the organism’s ability to fix nitrogen was not assayed at the time of isolation [43]. Although Acidobacteria are widespread and can be abundant in sediments (>10% of the bacterial community) [44–47], their ecological functions in marine ecosystems remain largely unexplored.
While the results of the 15N-SIP and nifH analyses are consistent, 11 taxa were implicated as diazotrophs in only one of the two datasets. We cannot exclude the possibility that these taxa are false positives in a single dataset, and therefore do not refer to them as ‘candidate diazotrophs’ here. However, they may indeed be diazotrophs. Instances where nifH amplicons were recovered from a group for which no 15N-incorporators were identified (e.g., Euryarchaeota) may be due to 15N-SIP data filtering (see Methods) or may indicate diazotrophic organisms that were inactive or were active but did not fix nitrogen during incubation. Instances where 15N-incorporators were identified but lack accompanying nifH ASVs (e.g., Planctomycetes) may indicate nifH primer bias, insufficient sequencing depth, and/or a lack of diazotrophic cultured representatives with available nifH sequence data to aid identification. These cases should be further investigated by targeted approaches to assess diaozotrophy, such as fluorescence in situ hybridization coupled to nanoscale secondary ion mass spectrometry.
In particular, the Planctomycetes present a compelling case for further investigation. The Planctomycetes are a cosmopolitan phylum that lack nitrogen-fixing representatives, though recent metagenomic evidence from the ocean surface indicates that some members contain the full complement of genes required to fix nitrogen [48]. Our 15N-SIP results suggest novel members of benthic Planctomycetes are active diazotrophs (n = 12; 82–94% 16S rRNA nuc. id to closest characterized relatives) (Fig. 2a; Supplementary Fig. S8). Additional work is needed to confirm diazotrophic function within the Planctomycetes and help evaluate their ecological role and biogeochemical significance in the marine environment.
Indications of diverse catabolisms fueling nitrogen fixation
Previous work has demonstrated that nitrogen fixation at deep-sea methane seeps is dependent on methane and methane oxidizers are the dominant diazotrophs [12, 14, 15, 22]. In the sediments investigated here, 15N2 assimilation was not dependent on methane (Fig. 1b), suggesting other catabolisms support diazotrophy outside of seeps [12]. Indeed, inspection of the metabolisms associated with each candidate diazotroph’s closest characterized relative revealed diverse potential catabolisms (Fig. 5). We detected five candidate diazotrophs within the Deltaproteobacteria whose closest characterized relatives were either obligate or facultative sulfate reducers (95–100% 16S rRNA nuc. id; Fig. 5). This is consistent with previous work demonstrating that sulfate reduction is an important catabolism fueling diazotrophy in shallow marine sediments [7, 9, 20, 49–53]. In addition, closest cultured relatives from 10 of the other candidate diazotrophs have previously been shown to use elemental sulfur as a terminal electron acceptor (91–97% 16S rRNA nuc. id; Fig. 5), further underscoring the likely importance of sulfur metabolism to diazotrophy.
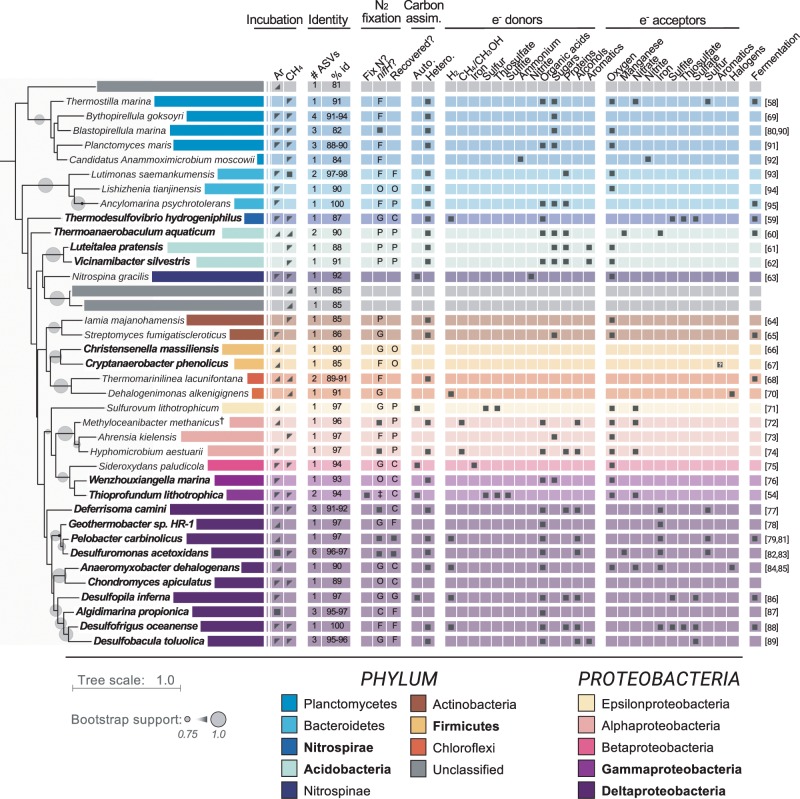
Fig. 5. Metabolic profiles of the 15N-incorporators’ closest characterized relatives [54, 58–95].
Empty cells indicate metabolisms not tested or not found to support growth. Bold taxon names indicate 15N-incorporators within phyla or classes (Proteobacteria only) for which nifH sequences were also detected. Incubation column: Indicates the headspace (‘Ar’ or ‘CH4′) and sediment horizon (0–3 cmbsf, shaded top left corner; 9–12 cmbsf, shaded bottom right corner; both, full shaded square) for which taxa were identified as 15N-incorporators. Identity column: ‘# ASVs’ indicates number of ASVs identified as 15N-incorporators that shared the same closest relative; ‘% id’ indicates percent sequence identity to closest relative across the amplified V4/V5 16S rRNA gene region (~370 bp). N2 fixation column: ‘Fix N?’ indicates if closest relative previously shown to fix nitrogen (dark square); ‘nifH?’ indicates if relative contains a copy of nifH in its genome (dark square) or the relative’s lowest taxonomic rank that contains an organism with a copy of nifH (P phylum, C class, O Order, F Family, G Genus); ‘Recovered?’ indicates if one of the recovered nifH ASV’s closest nifH reference sequence was the copy from that relative (dark square) or the lowest common taxonomic rank shared with that relative. Tree rooted to 16S rRNA gene from Methanocaldococcus jannaschii (GenBank accession no. NR_113292). †: 15N-incorporator shared equal sequence identity to M. methanicus and M. superfactus. The 16S rRNA gene sequence from M. methanicus was used for tree construction and the metabolic profiles for both organisms are shown. ‡: nifH sequence not available at the time of analysis.
The closest cultured relatives of fifteen of the candidate diazotrophs within the Acidobacteria and Deltaproteobacteria can use iron as a terminal electron acceptor (90–100% 16S rRNA nuc. id; Fig. 5). This is consistent with recent findings in sediments from the Mauritanian oxygen-deficient zone (47–1108 m water depth), where ferrous iron porewater concentrations, nifH gene abundances from Pelobacter carbinolicus (an iron- and sulfur-reducer), and nitrogenase activity were found to co-occur [18]. Diazotrophy coupled to iron reduction may therefore be widespread in benthic habitats.
In addition, we recovered six ASVs spanning Acidobacteria, Nitrospirae, and Deltaproteobacteria whose closest characterized relatives can ferment diverse organic substrates (87–100% 16S rRNA nuc. id) (Fig. 5). This is consistent with previous work suggesting that fermentation may support diazotrophy in salt marsh sediments [52]. Interestingly, it suggests that deep-sea nitrogen fixation may be ecologically linked to the remineralization of complex organic matter at both the initial organic hydrolysis (mediated by fermenters) and complete oxidation stages (e.g., oxidation of acetate to CO2 by some sulfate reducers).
Strikingly, we identified two candidate diazotrophs from the Gammaproteobacteria whose closest cultured relative, isolated from a deep-sea hydrothermal vent chimney [54], is a facultative anaerobe capable of both denitrification and nitrogen fixation (94% 16S rRNA nuc. id) (Fig. 5). We identified two additional 15N-incorporators from the Alphaproteobacteria and Epsilonproteobacteria whose closest cultured relatives are also facultative denitrifiers (97% 16S rRNA nuc. id) (Fig. 5). It is likely that these organisms temporally separate nitrogen fixation and denitrification depending on nitrogen availability. These organisms may therefore alternate between being fixed nitrogen sources and sinks, as has been previously suggested [55, 56], potentially contributing to changes in sediment net fixed nitrogen fluxes [8].
In contrast to methane seep sediments, we have little evidence suggesting that methane oxidation was coupled to nitrogen fixation in these incubations. Although we detected two candidate diazotrophs whose closest relatives can oxidize methane (96–97% 16S rRNA nuc. id), they were identified in the Ar-amended incubations, suggesting they used alternative electron donors during incubation (Fig. 5). Furthermore, the potential catabolisms coupled to nitrogen fixation when methane was added was not overall different from those when methane was not added (Fig. 5). Therefore, while methane cycling coupled to nitrogen fixation in situ remains a possibility, our findings highlight the potential for methane-independent diazotrophy outside of seeps.
Inferring catabolic potential based on comparisons to similar cultured organisms is not definitive, as phylogenetically similar organisms can be metabolically dissimilar. However, other methods to link metabolisms such as culturing and assembling genomes from metagenomes generally assess only a small subset of the community, which makes these approaches insufficient to obtain a community-level perspective. This is particularly true in deep-sea sediments, where the slow growth of microorganisms renders them difficult to obtain in pure culture, and the high intra-species genetic variability presents challenges for assembling high-quality genomes from metagenomes. Therefore, examining the metabolisms of the closest cultured relatives to 15N-incorporators, although an indirect assessment of metabolic capacity, provides a broad overview of the diazotrophic community’s potential ecology, which can both inform the design of future studies and aid in their interpretation.
Implications for deep-sea diazotrophy
Taken together, our findings imply a large diazotrophic niche space in the marine benthos, with the potential for diazotrophic activity across a range of redox and geochemical conditions. Since the samples investigated here were collected at a water depth approaching the average ocean depth and had total organic matter contents (1.2–1.4 wt%) similar to those from continental margins (~1.0 wt%) [57], these diazotrophs may represent those from continental margins more broadly. If indeed widespread, these lineages could impact rates of climatically important metabolisms on the seafloor (e.g., methanogenesis/methanotrophy, organic matter remineralization) by ameliorating ecosystem-level nitrogen limitation. Furthermore, the potential catabolic diversity in the diazotrophic assemblage suggests that benthic diazotrophy may be a stable source of fixed nitrogen despite changing environmental conditions. The catabolic diversity implicated here contrasts with the current understanding of diazotrophs at methane seeps, where nitrogen fixation is methane-dependent and thought to be mediated exclusively by methane-oxidizing archaea and/or methane-dependent sulfate-reducing bacteria [13–15, 18, 22]. Thus, widely distributed, low abundance diazotrophs may serve as a ‘seed bank’ from which certain diazotrophic groups can proliferate if and when environmental conditions select for them, for example, as methane seeps, whale falls, and oxygen minimum zones develop. This taxonomic and metabolic flexibility in the diazotroph assemblage could therefore provide a robust source of fixed nitrogen across vast spatial and temporal scales.
Conclusions
Our 15N-SIP and nifH sequencing analyses together suggest that phylogenetically diverse organisms fix nitrogen in deep-sea sediment. The results from both techniques suggest that, on account of their diversity and relative abundance, Deltaproteobacteria are important diazotrophs in this habitat, particularly those from the taxonomic orders Desulfobacterales and Desulfuromonadales. Our findings also suggest that deep-sea sediments host novel diazotrophs of unanticipated phylogenetic breadth, as both datasets detected the Acidobacteria, Firmicutes, Nitrospirae, and Gammaproteobacteria, in addition to the Deltaproteobacteria. The candidate diazotrophs’ closest cultured relatives can use many terminal electron acceptors, including oxygen, nitrate, iron, sulfur, sulfate, and organic compounds, making it likely that deep-sea diazotrophy is coupled to multiple biogeochemical cycles. These findings have broad biogeochemical implications in considering both modern and ancient environments and highlight the need for additional analyses exploring the diversity and activity of diazotrophs in widely representative marine sediments.
Supplementary information
Acknowledgements
We thank all members of the Dekas Laboratory for valuable discussions and feedback, and thank Spencer Debenport (Buckley Laboratory) for assistance with 15N-SIP sample processing and analysis. We thank Victoria Orphan, and the crew and science party of R/V Western Flyer DR204–208 (Monterey Bay Aquarium Research Institute), including Shana Goffredi, Bob Vrijenhoek, Julio Harvey and Lonny Lundsten, for providing access to the samples and assisting with sample collection (funded by NSF MCB-0348492). Funding for this research was provided by the Center for Dark Energy Biosphere Investigations (Research Grant to AED), and the National Science Foundation (OCE-1634297 to AED and a Graduate Research Fellowship to BJK).
Data availability
All 16S rRNA gene and nifH sequence data have been deposited in the GenBank, EMBL, and DDBJ databases under BioProject number PRJEB32101.
Code availability
Code to reproduce both 15N-SIP and nifH analyses are available on github (https://github.com/BKapili/SIP_2019).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Bennett J. Kapili, Email: kapili@stanford.edu
Anne E. Dekas, Email: dekas@stanford.edu
Supplementary information
The online version of this article (10.1038/s41396-019-0584-8) contains supplementary material, which is available to authorized users.
References
- 1.Gruber N, Galloway JN. An Earth-system perspective of the global nitrogen cycle. Nature. 2008;451:293–6. doi: 10.1038/nature06592. [DOI] [PubMed] [Google Scholar]
- 2.Galloway JN, Dentener FJ, Capone DG, Boyer EW, Howarth RW, Seitzinger SP, et al. Nitrogen cycles: past, present, and future. Biogeochemistry. 2004;70:153–226. doi: 10.1007/s10533-004-0370-0. [DOI] [Google Scholar]
- 3.Ryther JH, Dunstan WM. Nitrogen phosphorus and eutrophication in the coastal marine environment. Science. 1971;171:1008–13. doi: 10.1126/science.171.3975.1008. [DOI] [PubMed] [Google Scholar]
- 4.Howarth RW. Nutrient limitation of net primary production in marine ecosystems. Annu Rev Ecol Syst. 1988;19:89–110. doi: 10.1146/annurev.es.19.110188.000513. [DOI] [Google Scholar]
- 5.Elser JJ, Bracken MES, Cleland EE, Gruner DS, Harpole WS, Hillebrand H, et al. Global analysis of nitrogen and phosphorus limitation of primary producers in freshwater, marine and terrestrial ecosystems. Ecol Lett. 2007;10:1135–42. doi: 10.1111/j.1461-0248.2007.01113.x. [DOI] [PubMed] [Google Scholar]
- 6.Sohm JA, Webb EA, Capone DG. Emerging patterns of marine nitrogen fixation. Nat Rev Microbiol. 2011;9:499–508. doi: 10.1038/nrmicro2594. [DOI] [PubMed] [Google Scholar]
- 7.Burns JA, Zehr JP, Capone DG. Nitrogen-fixing phylotypes of Chesapeake Bay and Neuse River estuary sediments. Microb Ecol. 2002;44:336–43. doi: 10.1007/s00248-002-1000-9. [DOI] [PubMed] [Google Scholar]
- 8.Fulweiler RW, Nixon SW, Buckley BA, Granger SL. Reversal of the net dinitrogen gas flux in coastal marine sediments. Nature. 2007;448:180–2. doi: 10.1038/nature05963. [DOI] [PubMed] [Google Scholar]
- 9.Bertics VJ, Löscher CR, Salonen I, Dale AW, Gier J, Schmitz RA, et al. Occurrence of benthic microbial nitrogen fixation coupled to sulfate reduction in the seasonally hypoxic Eckernförde Bay, Baltic Sea. Biogeosciences. 2013;10:1243–58. doi: 10.5194/bg-10-1243-2013. [DOI] [Google Scholar]
- 10.Brown SM, Jenkins BD. Profiling gene expression to distinguish the likely active diazotrophs from a sea of genetic potential in marine sediments. Environ Microbiol. 2014;16:3128–42. doi: 10.1111/1462-2920.12403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Orcutt BN, Sylvan JB, Knab NJ, Edwards KJ. Microbial ecology of the dark ocean above, at, and below the seafloor. Microbiol Mol Biol Rev. 2011;75:361–422. doi: 10.1128/MMBR.00039-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dekas AE, Fike DA, Chadwick GL, Green-Saxena A, Fortney J, Connon SA, et al. Widespread nitrogen fixation in sediments from diverse deep-sea sites of elevated carbon loading. Environ Microbiol. 2018;20:4281–96. doi: 10.1111/1462-2920.14342. [DOI] [PubMed] [Google Scholar]
- 13.Pernthaler A, Dekas AE, Brown CT, Goffredi SK, Embaye T, Orphan VJ. Diverse syntrophic partnerships from deep-sea methane vents revealed by direct cell capture and metagenomics. Proc Natl Acad Sci USA. 2008;105:7052–7. doi: 10.1073/pnas.0711303105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dekas AE, Chadwick GL, Bowles MW, Joye SB, Orphan VJ. Spatial distribution of nitrogen fixation in methane seep sediment and the role of the ANME archaea. Environ Microbiol. 2014;16:3012–29. doi: 10.1111/1462-2920.12247. [DOI] [PubMed] [Google Scholar]
- 15.Dekas AE, Connon SA, Chadwick GL, Trembath-Reichert E, Orphan VJ. Activity and interactions of methane seep microorganisms assessed by parallel transcription and FISH-NanoSIMS analyses. ISME J. 2016;10:678–92. doi: 10.1038/ismej.2015.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mehta MP, Butterfield DA, Baross JA. Phylogenetic diversity of nitrogenase (nifH) genes in deep-sea and hydrothermal vent environments of the Juan de Fuca Ridge. Appl Environ Microbiol. 2003;69:960–70. doi: 10.1128/AEM.69.2.960-970.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gier J, Sommer S, Löscher CR, Dale AW, Schmitz RA, Treude T. Nitrogen fixation in sediments along a depth transect through the Peruvian oxygen minimum zone. Biogeosciences. 2016;13:4065–80. doi: 10.5194/bg-13-4065-2016. [DOI] [Google Scholar]
- 18.Gier J, Löscher CR, Dale AW, Sommer S, Lomnitz U, Treude T. Benthic dinitrogen fixation traversing the oxygen minimum zone off Mauritania (NW Africa) Front Mar Sci. 2017;4:1–16. doi: 10.3389/fmars.2017.00390. [DOI] [Google Scholar]
- 19.Dang H, Yang J, Li J, Luan X, Zhang Y, Gu G, et al. Environment-dependent distribution of the sediment nifH-harboring microbiota in the northern South China Sea. Appl Environ Microbiol. 2013;79:121–32. doi: 10.1128/AEM.01889-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bertics VJ, Sohm JA, Treude T, Chow C-ET, Capone DG, Fuhrman JA, et al. Burrowing deeper into benthic nitrogen cycling: the impact of bioturbation on nitrogen fixation coupled to sulfate reduction. Mar Ecol Prog Ser. 2010;409:1–15. doi: 10.3354/meps08639. [DOI] [Google Scholar]
- 21.Mehta MP, Baross JA. Nitrogen fixation at 92°C by a hydrothermal vent archaeon. Science. 2006;314:1783–6. doi: 10.1126/science.1134772. [DOI] [PubMed] [Google Scholar]
- 22.Dekas AE, Poretsky RS, Orphan VJ. Deep-sea archaea fix and share nitrogen in methane-consuming microbial consortia. Science. 2009;326:422–6. doi: 10.1126/science.1178223. [DOI] [PubMed] [Google Scholar]
- 23.Buckley DH, Huangyutitham V, Hsu S-F, Nelson TA. Stable isotope probing with 15N achieved by disentangling the effects of genome G+C content and isotope enrichment on DNA density. Appl Environ Microbiol. 2007;73:3189–95. doi: 10.1128/AEM.02609-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pepe-Ranney C, Koechli C, Potrafka R, Andam C, Eggleston E, Garcia-Pichel F, et al. Non-cyanobacterial diazotrophs mediate dinitrogen fixation in biological soil crusts during early crust formation. ISME J. 2016;10:287–98. doi: 10.1038/ismej.2015.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parada AE, Needham DM, Fuhrman JA. Every base matters: assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ Microbiol. 2016;18:1403–14. doi: 10.1111/1462-2920.13023. [DOI] [PubMed] [Google Scholar]
- 26.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011;17:10–2. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 27.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJ, Holmes SP. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13:581–3. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Youngblut ND, Barnett SE, Buckley DH. HTSSIP: an R package for analysis of high throughput sequencing data from nucleic acid stable isotope probing (SIP) experiments. PLoS ONE. 2018;13:e0189616. doi: 10.1371/journal.pone.0189616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.R Core Team. R: a language and environment for statistical computing. Vienna, Austria: R Core Team; 2017.
- 30.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Youngblut ND, Barnett SE, Buckley DH. SIPSim: a modeling toolkit to predict accuracy and aid design of DNA-SIP experiments. Front Microbiol. 2018;9:1–16. doi: 10.3389/fmicb.2018.00570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Capone DG. Benthic nitrogen fixation. In: Blackburn TH, Sørensen J, editors. Nitrogen cycling in coastal marine environments. John Wiley & Sons; 1988. p. 85–123.
- 33.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–80. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–3. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mirarab S, Nguyen N, Warnow T. SEPP: SATé-enabled phylogenetic placement. Pacific Symp Biocomput. 2012;17:247–58. [DOI] [PubMed]
- 36.Drost H-G, Gabel A, Liu J, Quint M, Grosse I. myTAI: evolutionary transcriptomics with R. Bioinformatics. 2018;34:1589–90. doi: 10.1093/bioinformatics/btx835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Letunic I, Bork P. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016;44:242–5. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cadisch G, Espana M, Causey R, Richter M, Shaw E, Morgan JAW, et al. Technical considerations for the use of 15N-DNA stable-isotope probing for functional microbial activity in soils. Rapid Commun Mass Spectrom. 2005;19:1424–8. doi: 10.1002/rcm.1908. [DOI] [PubMed] [Google Scholar]
- 39.Buckley DH, Huangyutitham V, Hsu SF, Nelson TA. Stable isotope probing with 15N2 reveals novel noncultivated diazotrophs in soil. Appl Environ Microbiol. 2007;73:3196–204. doi: 10.1128/AEM.02610-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou H, Dang H, Klotz MG. Environmental conditions outweigh geographical contiguity in determining the similarity of nifH-harboring microbial communities in sediments of two disconnected marginal seas. Front Microbiol. 2016;7(July):1–13. doi: 10.3389/fmicb.2016.01111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fulweiler RW, Brown SM, Nixon SW, Jenkins BD. Evidence and a conceptual model for the co-occurrence of nitrogen fixation and denitrification in heterotrophic marine sediments. Mar Ecol Prog Ser. 2013;482:57–68. doi: 10.3354/meps10240. [DOI] [Google Scholar]
- 42.Anderson I, Held B, Lapidus A, Nolan M, Lucas S, Tice H, et al. Genome sequence of the homoacetogenic bacterium Holophaga foetida type strain (TMBS4T) Stand Genom Sci. 2012;6:174–84. doi: 10.4056/sigs.2746047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liesack W, Bak F, Kreft J-U, Stackebrandt E. Holophaga foetida gen. nov., sp. nov., a new, homoacetogenic bacterium degrading methoxylated aromatic compounds. Arch Microbiol. 1994;162:85–90. doi: 10.1007/BF00264378. [DOI] [PubMed] [Google Scholar]
- 44.Polymenakou PN, Bertilsson S, Tselepides A, Stephanou EG. Bacterial community composition in different sediments from the Eastern Mediterranean Sea: a comparison of four 16S ribosomal DNA clone libraries. Microb Ecol. 2005;50:447–62. doi: 10.1007/s00248-005-0005-6. [DOI] [PubMed] [Google Scholar]
- 45.Schauer R, Bienhold C, Ramette A, Harder J. Bacterial diversity and biogeography in deep-sea surface sediments of the South Atlantic Ocean. ISME J. 2009;4:159–70. doi: 10.1038/ismej.2009.106. [DOI] [PubMed] [Google Scholar]
- 46.Bienhold C, Boetius A, Ramette A. The energy – diversity relationship of complex bacterial communities in Arctic deep-sea sediments. ISME J. 2012;6:724–32. doi: 10.1038/ismej.2011.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhu D, Tanabe S-H, Yang C, Zhang W, Sun J. Bacterial community composition of South China Sea sediments through pyrosequencing-based analysis of 16S rRNA genes. PLoS One. 2013;8:1–9. doi: 10.1371/journal.pone.0078501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Delmont TO, Quince C, Shaiber A, Esen ÖC, Lee STM, Rappé MS, et al. Nitrogen-fixing populations of Planctomycetes and Proteobacteria are abundant in surface ocean metagenomes. Nat Microbiol. 2018;3:804–13. doi: 10.1038/s41564-018-0176-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Capone DG. Nitrogen fixation (acetylene reduction) by rhizosphere sediments of the eelgrass Zostera marina. Mar Ecol Prog Ser. 1982;10:67–75. doi: 10.3354/meps010067. [DOI] [Google Scholar]
- 50.Capone DG, Oremland R, Taylor BF. Significance of N2 fixation to the production of Thalassia testudinum. In: CICAR-II Symposium. Caracas, Venezuela; 1976.
- 51.Nedwell DB, Abdul Aziz SA. Heterotrophic nitrogen fixation in an intertidal saltmarsh sediment. Estuar Coast Mar Sci. 1980;10:699–702. doi: 10.1016/S0302-3524(80)80097-1. [DOI] [Google Scholar]
- 52.Gandy EL, Yoch DC. Relationship between nitrogen-fixing sulfate reducers and fermenters in salt marsh sediments and roots of Spartina alterniflora. Appl Environ Microbiol. 1988;54:2031–6. doi: 10.1128/AEM.54.8.2031-2036.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McGlathery KJ, Risgaard-Petersen N, Christensen PB. Temporal and spatial variation in nitrogen fixation activity in the eelgrass Zostera marina rhizosphere. Mar Ecol Prog Ser. 1998;168:245–58. doi: 10.3354/meps168245. [DOI] [Google Scholar]
- 54.Takai K, Miyazaki M, Hirayama H, Nakagawa S, Querellou J, Godfroy A. Isolation and physiological characterization of two novel, piezophilic, thermophilic chemolithoautotrophs from a deep-sea hydrothermal vent chimney. Environ Microbiol. 2009;11:1983–97. doi: 10.1111/j.1462-2920.2009.01921.x. [DOI] [PubMed] [Google Scholar]
- 55.Neyra CA, Dobereiner J, Lalande R, Knowles R. Denitrification by N2-fixing Spirillum lipoferum. Can J Microbiol. 1977;23:300–5. doi: 10.1139/m77-044. [DOI] [PubMed] [Google Scholar]
- 56.Newell SE, Pritchard KR, Foster SQ, Fulweiler RW. Molecular evidence for sediment nitrogen fixation in a temperate New England estuary. PeerJ. 2016;4:e1615. doi: 10.7717/peerj.1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Emerson S, Hedges JI. Processes controlling the organic carbon content of open ocean sediments. Paleoceanography. 1988;3:621–34. doi: 10.1029/PA003i005p00621. [DOI] [Google Scholar]
- 58.Slobodkina GB, Panteleeva AN, Beskorovaynaya DA, Bonch-Osmolovskaya EA, Slobodkina AI. Thermostilla marina gen. nov., sp. nov., a thermophilic, facultatively anaerobic planctomycete isolated from a shallow submarine hydrothermal vent. Int J Syst Evol Microbiol. 2016;66:633–8. doi: 10.1099/ijsem.0.000767. [DOI] [PubMed] [Google Scholar]
- 59.Haouari O, Fardeau M-L, Cayol J-L, Fauque G, Casiot C, Elbaz-Poulichet F, et al. Thermodesulfovibrio hydrogeniphilus sp. nov., a new thermophilic sulphate-reducing bacterium isolated from a Tunisian hot spring. Syst Appl Microbiol. 2008;31:38–42. doi: 10.1016/j.syapm.2007.12.002. [DOI] [PubMed] [Google Scholar]
- 60.Losey NA, Stevenson BS, Busse HJ, Damsté JSS, Rijpstra WIC, Rudd S, et al. Thermoanaerobaculum aquaticum gen. nov., sp. nov., the first cultivated member of Acidobacteria subdivision 23, isolated from a hot spring. Int J Syst Evol Microbiol. 2013;63:4149–57. doi: 10.1099/ijs.0.051425-0. [DOI] [PubMed] [Google Scholar]
- 61.Vieira S, Luckner M, Wanner G, Overmann J. Luteitalea pratensis gen. nov., sp. nov. a new member of subdivision 6 Acidobacteria isolated from temperate grassland soil. Int J Syst Evol Microbiol. 2017;67:1408–14. doi: 10.1099/ijsem.0.001827. [DOI] [PubMed] [Google Scholar]
- 62.Huber KJ, Geppert AM, Wanner G, Fösel BU, Wüst PK, Overmann J. The first representative of the globally widespread subdivision 6 Acidobacteria, Vicinamibacter silvestris gen. nov., sp. nov., isolated from subtropical savannah soil. Int J Syst Evol Microbiol. 2016;66:2971–9. doi: 10.1099/ijsem.0.001131. [DOI] [PubMed] [Google Scholar]
- 63.Watson SW, Waterbury BJ. Characteristics of two marine nitrite oxidizing bacteria, Nitrospina gracilis nov. gen. nov. sp. and Nitrococcus mobilis nov. gen. nov. sp. Arch Mikrobiol. 1971;77:203–30. doi: 10.1007/BF00408114. [DOI] [Google Scholar]
- 64.Kurahashi M, Fukunaga Y, Sakiyama Y, Harayama S, Yokota A. Iamia majanohamensis gen. nov., sp. nov., an actinobacterium isolated from sea cucumber Holothuria edulis, and proposal of Iamiaceae fam. nov. Int J Syst Evol Microbiol. 2009;59:869–73. doi: 10.1099/ijs.0.005611-0. [DOI] [PubMed] [Google Scholar]
- 65.Goodfellow M, Williams ST, Alderson G. Transfer of Chainia species to the genus Streptomyces with emended description of species. Syst Appl Microbiol. 1986;8:55–60. doi: 10.1016/S0723-2020(86)80148-5. [DOI] [Google Scholar]
- 66.Ndongo S, Dubourg G, Khelaifia S, Fournier PE, Raoult D. Christensenella timonensis, a new bacterial species isolated from the human gut. N. Microbes N. Infect. 2016;13:32–3. doi: 10.1016/j.nmni.2016.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Juteau P, Côté V, Duckett MF, Beaudet R, Lépine F, Villemur R, et al. Cryptanaerobacter phenolicus gen. nov., sp. nov., an anaerobe that transforms phenol into benzoate via 4-hydroxybenzoate. Int J Syst Evol Microbiol. 2005;55:245–50. doi: 10.1099/ijs.0.02914-0. [DOI] [PubMed] [Google Scholar]
- 68.Nunoura T, Hirai M, Miyazaki M, Kazama H, Makita H, Hirayama H, et al. Isolation and characterization of a thermophilic, obligately anaerobic and heterotrophic marine Chloroflexi bacterium from a Chloroflexi-dominated microbial community associated with a Japanese shallow hydrothermal system, and proposal for Thermomarinilinea lacunofontalis gen. nov., sp. nov. Microbes Environ. 2013;28:228–35. doi: 10.1264/jsme2.ME12193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Storesund JE, Øvreås L. Diversity of Planctomycetes in iron-hydroxide deposits from the Arctic Mid Ocean Ridge (AMOR) and description of Bythopirellula goksoyri gen. nov., sp. nov., a novel Planctomycete from deep sea iron-hydroxide deposits. Antonie Van Leeuwenhoek. 2013;104:569–84. doi: 10.1007/s10482-013-0019-x. [DOI] [PubMed] [Google Scholar]
- 70.Bowman KS, Nobre MF, da Costa MS, Rainey FA, Moe WM. Dehalogenimonas alkenigignens sp. nov., a chlorinated-alkane-dehalogenating bacterium isolated from groundwater. Int J Syst Evol Microbiol. 2012;63:1492–8. doi: 10.1099/ijs.0.045054-0. [DOI] [PubMed] [Google Scholar]
- 71.Inagaki F, Takai K, Nealson KH, Horikoshi K. Sulfurovum lithotrophicum gen. nov., sp. nov., a novel sulfur-oxidizing chemolithoautotroph within the ε-Proteobacteria isolated from Okinawa Trough hydrothermal sediments. Int J Syst Evol Microbiol. 2004;54:1477–82. doi: 10.1099/ijs.0.03042-0. [DOI] [PubMed] [Google Scholar]
- 72.Vekeman B, Kerckhof FM, Cremers G, de Vos P, Vandamme P, Boon N, et al. New Methyloceanibacter diversity from North Sea sediments includes methanotroph containing solely the soluble methane monooxygenase. Environ Microbiol. 2016;18:4523–36. doi: 10.1111/1462-2920.13485. [DOI] [PubMed] [Google Scholar]
- 73.Rüger H-J, Höfle MG. Marine star-shaped-aggregate-forming bacteria: Agrobacterium atlanticum sp. nov.; Agrobacterium meteori sp. nov.; Agrobacterium ferrugineum sp. nov., nom. rev.; Agrobacterium gelatinovorum sp. nov., nom. rev.; and Agrobacterium stellulatum sp. nov., nom. rev. Int J Syst Bacteriol. 1992;42:133–43. doi: 10.1099/00207713-42-1-133. [DOI] [PubMed] [Google Scholar]
- 74.Hirsch P. Genus Hyphomicrobium Stutzer and Hartleb 1898, 76AL. In: Staley JT, Bryant MP, Pfennig N, Holt JG, (editors.) Bergey’s manual of systematic bacteriology. 3rd ed. Baltimore, Maryland: Williams & Wilkins; 1989. p. 1895–904.
- 75.Weiss JV, Rentz JA, Plaia T, Neubauer SC, Merrill-Floyd M, Lilburn T, et al. Characterization of neutrophilic Fe(II)-oxidizing bacteria isolated from the rhizosphere of wetland plants and description of Ferritrophicum radicicola gen. nov. sp. nov., and Sideroxydans paludicola sp. nov. Geomicrobiol J. 2007;24:559–70. doi: 10.1080/01490450701670152. [DOI] [Google Scholar]
- 76.Wang G, Tang M, Li T, Dai S, Wu H, Chen C, et al. Wenzhouxiangella marina gen. nov, sp. nov, a marine bacterium from the culture broth of Picochlorum sp. 122, and proposal of Wenzhouxiangellaceae fam. nov. in the order Chromatiales. Antonie Van Leeuwenhoek. 2015;107:1625–32. doi: 10.1007/s10482-015-0458-7. [DOI] [PubMed] [Google Scholar]
- 77.Slobodkina GB, Reysenbach AL, Panteleeva AN, Kostrikina NA, Wagner ID, Bonch-Osmolovskaya EA, et al. Deferrisoma camini gen. nov., sp. nov., a moderately thermophilic, dissimilatory iron(III)-reducing bacterium from a deep-sea hydrothermal vent that forms a distinct phylogenetic branch in the Deltaproteobacteria. Int J Syst Evol Microbiol. 2012;62:2463–8. doi: 10.1099/ijs.0.038372-0. [DOI] [PubMed] [Google Scholar]
- 78.Emerson D. Potential for iron-reduction and iron-cycling in iron oxyhydroxide-rich microbial mats at Loihi Seamount. Geomicrobiol J. 2009;26:639–47. doi: 10.1080/01490450903269985. [DOI] [Google Scholar]
- 79.Schink B. Fermentation of 2,3-butanediol by Pelobacter carbinolicus sp. nov., and evidence for propionate formation from C2 compounds. Arch Microbiol. 1984;137:33–41.
- 80.Schlesner H. Pirella marina sp. nov., a budding, peptidoglycan-less bacterium from brackish water. Syst Appl Microbiol. 1986;8:177–80. doi: 10.1016/S0723-2020(86)80073-X. [DOI] [Google Scholar]
- 81.Lovley DR, Phillips EJP, Lonergan DJ, Widman PK. Fe(III) and S0 reduction by Pelobacter carbinolicus. Appl Environ Microbiol. 1995;61:2132–8. [DOI] [PMC free article] [PubMed]
- 82.Pfennig N, Biebl H. Desulfuromonas acetoxidans gen. nov. and sp. nov., a new anaerobic, sulfur-reducing, acetate-oxidizing bacterium. Arch Microbiol. 1976;110:3–12. doi: 10.1007/BF00416962. [DOI] [PubMed] [Google Scholar]
- 83.Roden EE, Lovley DR. Dissimilatory Fe(III) reduction by the marine microorganism Desulfuromonas acetoxidans. Appl Environ Microbiol. 1993;59:734–42. doi: 10.1128/AEM.59.3.734-742.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sanford RA, Cole JR, Tiedje JM. Characterization and description of Anaeromyxobacter dehalogenans gen. nov., sp. nov., an aryl-halorespiring facultative anaerobic myxobacterium. Appl Environ Microbiol. 2002;68:893–900. doi: 10.1128/AEM.68.2.893-900.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Treude N, Rosencrantz D, Liesack W, Schnell S. Strain FAc12, a dissimilatory iron-reducing member of the Anaeromyxobacter subgroup of Myxococcales. FEMS Microbiol Ecol. 2003;44:261–9. doi: 10.1016/S0168-6496(03)00048-5. [DOI] [PubMed] [Google Scholar]
- 86.Gittel A, Seidel M, Kuever J, Galushko AS, Cypionka H, Könneke M. Desulfopila inferna sp. nov., a sulfate-reducing bacterium isolated from the subsurface of a tidal sand-flat. Int J Syst Evol Microbiol. 2010;60:1626–30. doi: 10.1099/ijs.0.015644-0. [DOI] [PubMed] [Google Scholar]
- 87.Kendall MM, Liu Y, Boone DR. Butyrate- and propionate-degrading syntrophs from permanently cold marine sediments in Skan Bay, Alaska, and description of Algorimarina butyrica gen. nov., sp. nov. FEMS Microbiol Lett. 2006;262:107–14. doi: 10.1111/j.1574-6968.2006.00380.x. [DOI] [PubMed] [Google Scholar]
- 88.Knoblauch C, Sahm K, Jørgensen BB. Psychrophilic sulfate-reducing bacteria isolated from permanently cold Arctic marine sediments: description of Desulfofrigus oceanense gen. nov., sp. nov., Desulfofrigus fragile sp. nov., Desulfofaba gelida gen. nov., sp. nov., Desulfotalea psychrophila gen. nov., sp. nov. and Desulfotalea arctica sp. nov. Int J Syst Bacteriol. 1999;49:1631–43. doi: 10.1099/00207713-49-4-1631. [DOI] [PubMed] [Google Scholar]
- 89.Rabus R, Nordhaus R, Ludwig W, Widdel F. Complete oxidation of toluene under strictly anoxic conditions by a new sulfate-reducing bacterium. Appl Environ Microbiol. 1993;59:1444–51. doi: 10.1128/AEM.59.5.1444-1451.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Schlesner H, Rensmann C, Tindall BJ, Gade D, Rabus R, Pfeiffer S, et al. Taxonomic heterogeneity within the Planctomycetales as derived by DNA-DNA hybridization, description of Rhodopirellula baltica gen. nov., sp. nov., transfer of Pirellula marina to the genus Blastopirellula gen. nov. as Blastopirellula marina comb. nov. and emended description of the genus Pirellula. Int J Syst Evol Microbiol. 2004;54:1567–80. doi: 10.1099/ijs.0.63113-0. [DOI] [PubMed] [Google Scholar]
- 91.Bauld J, Staley JT. Planctomyces maris sp. nov.: a marine isolate of the Planctomyces-Blastocaulis group of budding bacteria. J Gen Microbiol. 1976;97:45–55. doi: 10.1099/00221287-97-1-45. [DOI] [Google Scholar]
- 92.Khramenkov SV, Kozlov MN, Kevbrina MV, Dorofeev AG, Kazakova EA, Grachev VA, et al. A novel bacterium carrying out anaerobic ammonium oxidation in a reactor for biological treatment of the filtrate of wastewater fermented sludge. Microbiology. 2013;82:628–36. doi: 10.1134/S002626171305007X. [DOI] [PubMed] [Google Scholar]
- 93.Yoon J-H, Kang S-J, Jung Y-T, Oh T-K. Aestuariicola saemankumensis gen. nov., sp. nov., a member of the family Flavobacteriaceae, isolated from tidal flat sediment. Int J Syst Evol Microbiol. 2008;58:2126–31. doi: 10.1099/ijs.0.65717-0. [DOI] [PubMed] [Google Scholar]
- 94.Chen LP, Xu HY, Fu SZ, Fan HX, Zhou YG, Liu ZP. Lishizhenia tianjinensis sp. nov., isolated from coastal seawater. Int J Syst Evol Microbiol. 2009;59:2400–3. doi: 10.1099/ijs.0.008524-0. [DOI] [PubMed] [Google Scholar]
- 95.Jia C, Cui H-c, Han Y-q, Fu T-y, Du R, Wang X-l, et al. Ancylomarina psychrotolerans sp. nov., isolated from sediments of Fildes Peninsula and emended the description of genus Ancylomarina. Antonie Van Leeuwenhoek. 2018;111:1183–9. doi: 10.1007/s10482-018-1025-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All 16S rRNA gene and nifH sequence data have been deposited in the GenBank, EMBL, and DDBJ databases under BioProject number PRJEB32101.
Code to reproduce both 15N-SIP and nifH analyses are available on github (https://github.com/BKapili/SIP_2019).