Abstract
Root hairs play important roles for the acquisition of nutrients, microbe interaction and plant anchorage. In addition, root hairs provide an excellent model system to study cell patterning, differentiation and growth. Arabidopsis root hairs have been thoroughly studied to understand how plants regulate cell fate and growth in response to environmental signals. Accumulating evidence suggests that a multi-layered gene regulatory network is the molecular secret to enable the flexible and adequate response to multiple signals. In this review, we describe the key transcriptional regulators controlling cell fate and/or cell growth of root hairs. We also discuss how plants integrate phytohormonal and environmental signals, such as auxin, ethylene and phosphate availability, and modulate the level of these transcriptional regulators to tune root hair development.
Keywords: Root hair, Cell fate, Cell growth, Transcription factor, Gene regulatory network
Introduction
Root hairs form by tube-like outgrowth of root epidermal cells and they contribute to efficient acquisition of nutrients, microbe interactions and plant anchorage by expanding root surface area. Since root hair development is strongly influenced by surrounding conditions, this regulation is thought to be one of the important mechanisms of environmental adaptation in plants. Indeed, multiple environmental factors, such as availability of phosphorus, nitrogen, iron, magnesium and zinc, as well as phytohormones such as auxin, ethylene and abscisic acid (ABA) alter both cell fate and growth of root hairs (Figs. 1, 2) (Grierson et al. 2014). Root hair growth, for instance, is enhanced under low phosphate conditions but suppressed under high phosphate conditions. Due to these properties, root hairs provide a model system suitable to study cell differentiation and growth in response to environmental signals. In the past decade, various key transcription factors required for optimal root hair formation have been isolated and characterized. Accumulating evidence suggests that a gene regulatory network consisting of these transcription factors has an important role for their dynamic and robust responses. In this review we summarize the transcriptional regulation of root hair development and discuss how plants achieve the optimal response to adapt to environmental stimuli.
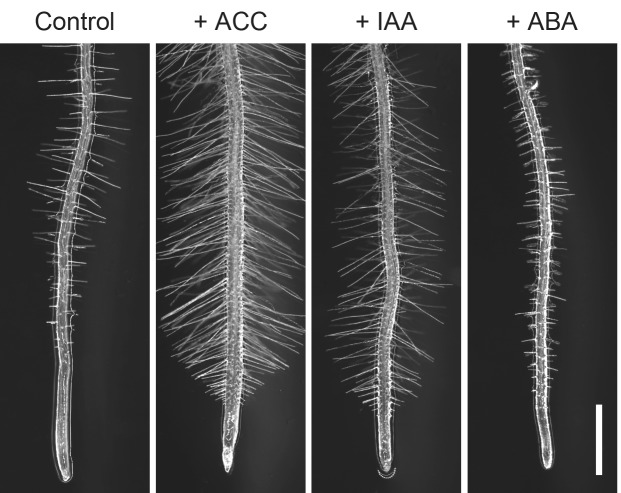
Fig. 1.
Root hair development is regulated by phytohormones. Arabidopsis seedlings grown on half strength Murashige-Skoog (MS) media with or without phytohormones. Control plants are grown on half strength MS medium without phytohormones. Root hairs are patterned and formed in accordance with the developmental program. Ethylene (100 nM 1-aminocyclopropane-1-1carboxylic acid, ACC) produces ectopic root hairs from non-hair cell files and enhances root hair growth. Auxin (10 nM indole acetic acid, IAA) enhances root hair growth without affecting the cell fate. 50 µM abscisic acid (ABA) inhibits root hair growth. Scale bar = 1 mm
Fig. 2.
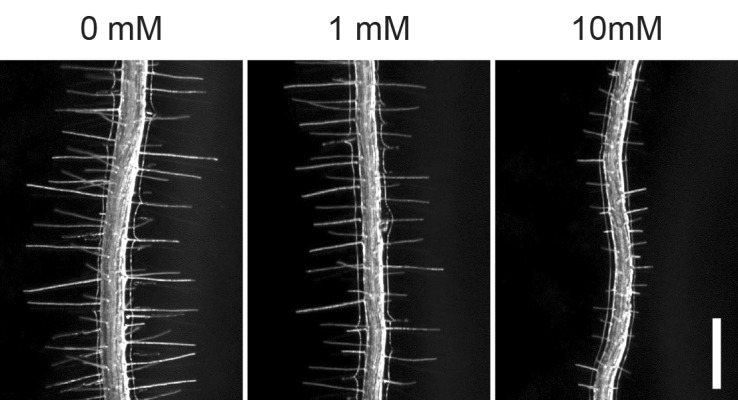
Root hair development is affected by environmental conditions. Phosphate concentration in the media affects root hair development. Scale bar = 500 µm
Developmental regulation of root hairs
Cell fate specification
During early root development, epidermal cells differentiate into either trichoblasts, i.e., root hair-forming cells or atrichoblasts, i.e., non-hair forming cells. In general, plants develop root hairs in one of the following three patterns (Datta et al. 2011). In plants that undergo type I patterning, such as rice, all epidermal cells have the potential to differentiate into root hairs and they develop root hairs in a random manner. For plants that exhibit type II patterning, including Brachypodium, root hair fate is specified by asymmetric cell division, thus smaller daughter cells gain root hair identity while larger daughter cells become non-hair cells. For plants that undergo type III patterning, such as Arabidopsis, hair cell identity is determined by the position of cells. In these plant species, epidermal cells with two underlying cortical cells become hair cells while those with only one underlying cortical cell become non-hair cells, resulting in the formation of hair cell files and non-hair cell files (Datta et al. 2011).
In the past decade, molecular mechanisms that specify cell fate of root hair have been extensively studied in Arabidopsis. It is now evident that a group of transcription factors have a key role in cell fate determination. In non-hair cells, a R2R3-type MYB transcription factor WERWOLF (WER) (Lee and Schiefelbein 1999), a basic helix-loop-helix (bHLH)-type transcription factor GLABLA3 (GL3) (Bernhardt et al. 2003) or its homolog ENHANCER OF GLABLA3 (EGL3) (Bernhardt et al. 2003), and a WD repeat protein TRANSPARENT TESTA GLABLA1 (TTG1) (Galway et al. 1994) form a protein complex and enhance the expression of GLABLA2 (GL2) which functions as a negative regulator of root hair formation (Masucci et al. 1996; Di Cristina et al. 1996). Lack of GL2 function causes ectopic root hair formation from non-hair cell files. Interestingly, the ectopic root hairs in gl2 mutants still keep non-hair cell properties such as early vacuolation and longer cell length (Masucci et al. 1996). From this perspective, GL2 seems to promote non-hair cell morphology rather than “cell fate”. To specify root hair identity, a mobile R3-type MYB transcription factor, CAPRICE (CPC) plays a key role (Wada et al. 1997). CPC is transcriptionally induced by the WER-GL3-TTG1 complex in non-hair cells and its protein moves to neighboring cells to induce root hairs. In these hair forming cells, CPC forms a protein complex with GL3 (or EGL3) and TTG1. In addition, TRIPTYCON (TRY) (Schellmann et al. 2002) and ENHANCER OF TRY AND CPC 1 (ETC1) (Simon et al. 2007), two homologs of CPC, function in a partially redundant manner with CPC (Bruex et al. 2012; Grierson et al. 2014; Ishida et al. 2008; Salazar-Henao et al. 2016). ETC1, however, does not show cell-to-cell movement like CPC, instead ETC1 moves from sub-epidermal cells to root hair cells (Rishmawi et al. 2018; Tominaga-Wada et al. 2017). ETC1 may have a different role from CPC on root hair development. In any case, CPC and its homologs are essential for the induction of a bHLH transcription factor ROOT HAIR DEFECTIVE 6 (RHD6) which plays key roles in the determination of root hair identity (Fig. 3a) (Masucci and Schiefelbein 1994; Menand et al. 2007).
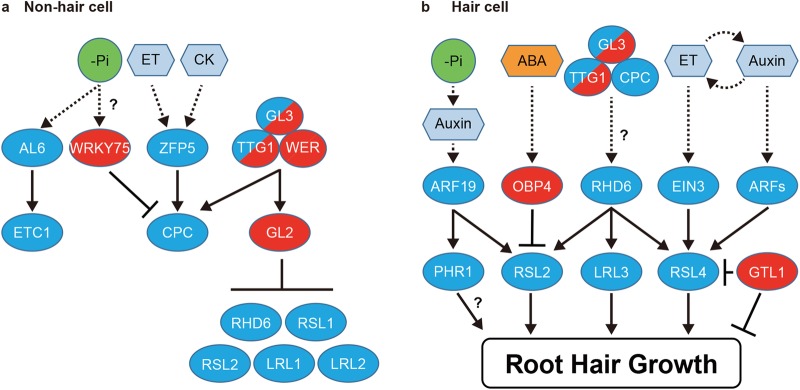
Fig. 3.
Gene regulatory network to control root hair development. Root hair development is a flexible response to developmental and environmental signals due to the gene network consisting of transcription factors. Exogenous and endogenous signals are transduced by distinct regulators. a In non-hair cells, a protein complex consisting of WER, GL3 and TTG1 induces GL2 expression. GL2 suppresses a set of transcription factors to prevent root hair formation. In addition, the WER-GL3-TTG1 complex induces CPC which moves to a hair cell. Although ZFP5 is dominantly expressed in hair cells and protein mobility is not characterized, ZFP5 is categorized in non-hair cell because ZFP5 directly regulates CPC. For the same reason, WRKY75 and AL6 are categorized in non-hair cell. b In hair cells, exogenous and endogenous signals are integrated into RSL2, RSL4 and LRL3 to change cell growth. Whether PHR1 regulates root hair growth directly or indirectly is not clear. GTL1 suppresses hair growth both directly and indirectly. Transcriptional activation and repression are indicated by arrows and blunted lines, respectively. The proteins marked in blue and red show positive and negative, respectively, regulators for root hair development. The circles containing blue and red show that they work both positively and negatively for root hair development. For two protein complexes of WER-GL3-TTG1 and CPC-GL3-TTG1, redundant proteins are not shown in the model. Note that protein mobility is not reflected in this model
Growth of root hair cells
After cell fate specification, the developmental program proceeds cell autonomously and hair-forming cells undergo extensive cell expansion to reach their maximum size. In young root hair cells, RHD6 and its homolog RHD6-LIKE1 (RSL1) promote transcription of genes encoding other bHLH transcription factors RSL2, RSL4 and Lotus japonicus ROOTHAIRLESS-LIKE 3 (LRL3) (Fig. 3a) (Karas et al. 2009; Masucci and Schiefelbein 1994; Menand et al. 2007; Yi et al. 2010). Among these bHLH homologs, RSL4 is a key regulator of root hair growth because constitutively expressed RSL4 proteins, using a CaMV 35S promoter, are sufficient to promote root hair growth (Yi et al. 2010). Transcriptome analyses of knock-out, overexpression and inducible lines of RSL4 identified very few transcription factors as downstream targets of RSL4, suggesting that RSL4 functions at the bottom of a gene regulatory network underling root hair growth (Mangano et al. 2017; Vijayakumar et al. 2016; Yi et al. 2010). Recent molecular genetic studies have demonstrated that RSL4 promotes transcription of ROOT HAIR SPECIFIC (RHS) genes that are predominantly expressed in root hairs (Hwang et al. 2017; Kim et al. 2006; Won et al. 2009). The promoter sequence of RHS genes contains a cis-element named Root Hair Element (RHE) and RSL4 binds this sequence to promote their expression in root hairs. Some RHS proteins, as well as other indirect targets of RSL4, are implicated in cell wall biogenesis and remodeling, ROS production or vesicle trafficking (Kim et al. 2006; Won et al. 2009), highlighting the key cellular processes central for root hair growth downstream of RSL4 regulation.
In addition to RSL4, RSL2 promotes root hair growth since the rsl2 single mutation causes shorter root hairs and the rsl2 rsl4 double mutation results in complete hairlessness (Mangano et al. 2018; Yi et al. 2010). Unlike RSL4, however, overexpression of RSL2 does not enhance root hair growth, indicating that RSL2 alone is not sufficient to promote hair growth (Yi et al. 2010). LRL3 also likely promotes root hair growth downstream of RHD6. Although lrl3 single mutant does not show root hair phenotypes, the lrl1 lrl3 or lrl2 lrl3 double mutants develop shorter root hairs (Karas et al. 2009). Unlike LRL3, expression of LRL1 and LRL2 does not require CPC, CPC-like MYBs, RHD6 and RSL1 (Karas et al. 2009). Thus, LRL1 and LRL2 might function in a pathway parallel to the CPC-RHD6 pathway. Further elucidation of LRL downstream targets will be key to work out how these pathways regulate hair growth coordinately.
Several recent studies have also demonstrated that root hair growth is actively repressed by transcriptional regulation. A trihelix transcription factor GT2-LIKE1 (GTL1) was initially isolated as a repressor of trichome cell growth (Breuer et al. 2009, 2012). Although the single gtl1 mutant does not show phenotypes in root hairs, the double mutation in GTL1 and its closest homolog DF1 prolongs elongation of root hairs (Shibata et al. 2018). Furthermore, root hair specific overexpression of GTL1 or DF1, using an EXPANSIN A7 promoter, strongly suppresses root hair growth, indicating that both GTL1 and DF1 act as negative regulators of root hair growth (Shibata et al. 2018). Both GTL1 and DF1 bind the RSL4 promoter region and directly suppress its expression. Interestingly, GTL1, in addition, suppresses expression of many RSL4 target genes as well as its own expression (Shibata et al. 2018). Conversely, RSL4 can induce GTL1 expression, although probably in an indirect manner (Shibata et al. 2018; Vijayakumar et al. 2016) and it can promote its own expression (Hwang et al. 2017). Therefore, RSL4 and GTL1 form a core transcriptional module to regulate each other and their common downstream targets (Fig. 4). Although the physiological role of this module has not been clarified, the RSL4-GTL1 module may provide robustness and/or flexibility to root hair growth so that they can respond to multiple environmental stimuli adequately (Shibata et al. 2018).
Fig. 4.
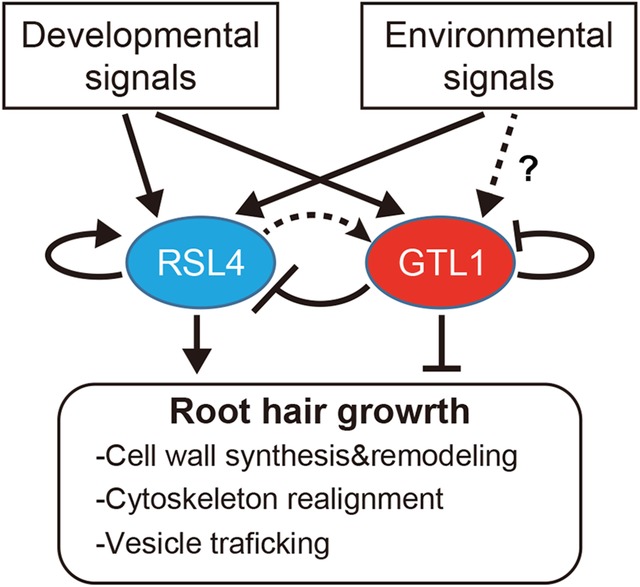
The RSL4-GTL1 module in the gene regulatory network for root hair development. Both environmental signals and developmental signals affect RSL4 expression. Whether environmental signals affect GTL1 expression is not clear but GTL1 is induced by programed developmental signals. RSL4 promotes expression of its own, GTL1 and genes involved in root hair growth. GTL1 counteracts with RSL4 by suppressing expression of RSL4, expression of its own and its downstream targets. This RSL4-GTL1 module might function as a modulator that integrates developmental signals and environmental signals to coordinate root hair growth. Transcriptional activation and repression are indicated by arrows and blunted lines, respectively
In addition to GTL1 and DF1, a DOF-type transcription factor OBF BINDING PROTEIN 4 (OBP4) is characterized as a negative regulator of root hair growth since induction of OBP4 inhibits root hair growth (Rymen et al. 2017). Unlike GTL1 and DF1, OBP4 represses RSL2 expression and does not affect RSL4 expression, suggesting that plants have at least two transcriptional pathways to repress root hair growth (Fig. 3a). OBP4 is implicated in ABA response (Rymen et al. 2017) and we will discuss further mechanistic details on this in the next section. LRL4 and LRL5 are also reported as a negative regulator of root hair growth since their ectopic overexpression, using a CaMV 35S promoter, causes short root hairs (Breuninger et al. 2016; Bruex et al. 2012). It is interesting that LRL4 and LRL5 have negative roles on root hair growth even though other bHLH transcription factors act as positive regulators.
In non-hair cells, GL2 suppresses expression of RHD6, RSL1, RSL2, LRL1 and LRL2 to prevent ectopic outgrowth of root hairs (Fig. 3b). Ectopic expression of LRL1 or LRL2 using a GL2 promoter causes ectopic hair formation, suggesting that LRL1 and LRL2 regulate both root hair growth and cell fate determination (Fig. 3b) (Lin et al. 2015).
Physiological and environmental modulation of root hair growth
Previous studies have revealed that phytohormones such as auxin, ethylene, cytokinin, strigolactone and jasmonic acid promote root hair growth while ABA and brassinosteroid inhibit root hair growth (Fig. 1) (Grierson et al. 2014; Lee and Cho 2013). Among these phytohormones, auxin and ethylene synergistically regulate root hair growth (Pitts et al. 1998), and a transcriptome analysis uncovered that almost 90% of genes are commonly upregulated by auxin and ethylene (Bruex et al. 2012). In addition, strigolactone-, jasmonic acid- and brassinosteroid-mediated signaling pathways eventually integrate with auxin- and/or ethylene-mediated pathways to coordinate root hair growth (Lee and Cho 2013). Root hair growth is also strongly influenced by nutrient availability in soil and the effect of inorganic phosphate (Pi) is best characterized to date (Fig. 2) (Salazar-Henao et al. 2016). In this section we describe the key direct transcriptional link that translates these hormonal and environmental responses into root hair growth.
Auxin
Auxin is a well-known phytohormone that massively enhances root hair growth (Fig. 1). Defects in auxin biosynthesis or signaling in mutants such as tryptophan aminotransferase of arabidopsis 1 (taa1), transport inhibitor response 1 (tir1) and auxin resistant 1 (axr1-3) cause hairless or short hair phenotypes, clearly showing that auxin is essential for root hair formation (Velasquez et al. 2016). Auxin does not confer hair cell identity to root epidermal cells because exogenous auxin treatment does not cause ectopic root hair formation. Auxin induces RSL4 expression, and several activator AUXIN RESPONSE FACTORs, such as ARF5, ARF7, ARF8 and ARF19, bind the RSL4 promoter to induce its expression (Mangano et al. 2017). A recent study, in addition, demonstrated that auxin also upregulates RSL2 expression through ARF19 although whether ARF19 directly binds to the promoter sequence of RSL2 is not yet established (Bhosale et al. 2018). Choi et al. (2018) further reported that root hair specific overexpression of repressor ARFs, i.e., ARF1-4, 9–11 and 16, using an EXPANSIN7 promoter inhibits root hair growth. These results suggest that auxin can repress root hair growth through these repressive ARFs and this response is transduced locally within root hairs (Choi et al. 2018). Membrane-anchored MYB (maMYB) is also an auxin responsive transcription factor because expression of maMYB is elevated by exogenous auxin treatment (Slabaugh et al. 2011). The knock-down mutation of maMYB by RNAi reduces root hair length, suggesting that maMYB functions in auxin-driven root hair growth. The maMYB RNAi mutants, however, still respond to auxin treatment and promote root hair growth, indicating that maMYB is not the sole regulator of auxin-dependent root hair growth (Slabaugh et al. 2011). How endoplasmic reticulum-localized maMYB functions in auxin signaling is not known and future studies should uncover further mechanistic details.
Ethylene
Ethylene is another phytohormone that enhances root hair growth (Fig. 1). Different from auxin, ethylene can also induce ectopic root hairs from non-hair cell files. Interestingly, application of ethylene or its precursor, 1-aminocyclopropane-1-1carboxylic acid (ACC), induces RSL4 expression (Fig. 3a) (Zhang et al. 2016). ETHYLENE INSENSITIVE 3 (EIN3) and its homolog EIN3-LIKE1 (EIL1) are key transcription factors to activate ethylene signaling. Both EIN3 and EIL1 are required for RSL4 induction by ethylene (Feng et al. 2017). EIN3 binds to the EIN3-binding site within the RSL4 promoter and directly regulates RSL4 expression (Feng et al. 2017). Intriguingly, EIN3 physically interacts with RHD6 and they seem to promote RSL4 expression synergistically in the presence of ethylene (Feng et al. 2017).
A C2H2 transcription factor ZINC FINGER PROTEIN 5 (ZFP5) also participates in the regulation of ethylene-dependent root hair growth. Intriguingly, the zfp5-4 loss-of-function mutation blocks only root hair growth and not ectopic root hair formation after ACC treatment (An et al. 2012). Thus, ZFP5 seems to have a specific role to elongate root hairs in response to ethylene.
Cytokinin
Cytokinin promotes root hair elongation and this regulation is independent from auxin or ethylene (Zhang et al. 2016). ZFP5 expression is elevated by cytokinin and the zfp5-4 mutant is insensitive to cytokinin for the hair growth phenotype (An et al. 2012), indicating that ZFP5 is an essential factor for the cytokinin-mediated root hair growth. ZFP5 directly activates CPC expression (An et al. 2012), thus ZFP5 appears to control root hair growth through CPC. Since ZFP5 does not affect root hair fate, it is also possible that ZFP5 regulates other factors that play more specific roles in hair growth.
Abscisic acid
ABA has a negative effect on root hair growth (Schnall and Quatrano 1992) (Fig. 1). ABA induces expression of OBP4, which subsequently represses root hair growth (Rymen et al. 2017). Moreover, OBP4 directly suppresses RSL2 expression. Thus, OBP4 inhibits root hair growth through RSL2 in response to ABA (Rymen et al. 2017).
Phosphate availability
Pi availability strongly influences root hair growth (Fig. 2). Regarding transcriptional regulations of Pi starvation, two MYB transcription factors, PHOSPHATE STARVATION RESPONSE 1 (PHR1) and its functionally redundant homolog PHR1-LIKE1 (PHL1) play a critical role on Pi starvation responses (Bustos et al. 2010; Rubio et al. 2001). Accordingly, the phr1 phl1 double mutants are insensitive to Pi starvation since they do not show enhancement of root hair development under Pi limiting conditions (Bustos et al. 2010). The overexpression line of PHR1 has much longer root hairs than those of wild type under Pi starvation condition (Bustos et al. 2010), indicating that PHR1 positively regulates root hair growth. Although PHR1 expression does not strongly respond to Pi status (Rubio et al. 2001) and the activity of PHR1 is primarily regulated by SUMOylation (Miura et al. 2005) and inhibitory binding protein SPX1 and SPX2 (Puga et al. 2014), a recent study showed that ARF7 and ARF19 promote PHR1 expression in roots (Huang et al. 2018). Thus, PHR1 may act downstream of auxin signaling. Auxin also acts systemically to transduce the Pi starvation signal since a recently proposed model suggests that Pi deficiency in soil is recognized at a root tip where auxin synthesis is enhanced by upregulation of TAA1 (Bhosale et al. 2018). Newly synthesized auxin is then transferred from the root tip to root hair cells by an auxin influx transporter AUXIN RESISTANT 1 (AUX1). After root hair cells uptake auxin, they induce cell elongation through transcriptional and non-transcriptional manners (Bhosale et al. 2018; Dindas et al. 2018).
Pi deficiency, in addition, affects fate of root hair cells and most likely ethylene, induced by Pi starvation, causes ectopic root hair formation. It is shown that Pi starvation increases the levels of the EIN3 protein which can promote root hair growth by direct transcriptional enhancement of root hair specific genes (Song et al. 2016). In addition, ALFIN-LIKE 6 (AL6) would be a key regulator of root hair formation for Pi starvation response. A homeodomain protein AL6 was isolated by forward genetic screening based on defects in root hair elongation under Pi starvation condition. AL6 induces root hair growth through activating ETC1 expression (Chandrika et al. 2013). Although cell fate of root hairs has not been evaluated in detail, AL6 can regulate cell fate of root hairs though ETC1. Another potential regulator is WRKY75 which is upregulated in response to Pi starvation (Devaiah et al. 2007). Surprisingly, however, WRKY75 is a negative regulator of root hair fate since it directly suppresses CPC expression (Devaiah et al. 2007). Thus, WRKY75 likely prevents root hair formation under low phosphate conditions. Although this seems paradoxical, having a negative regulator might be required for a precise response to rapidly changing environmental conditions.
How plants transduce high Pi stress is less understood but bHLH32 suppresses root hair development to reduce Pi uptake (Chen et al. 2007). Accordingly, the bhlh32 mutant does not inhibit root hair growth under high Pi condition and this phenotype is accompanied by up-regulation of several Pi starvation-induced genes (Chen et al. 2007).
While Pi response has mostly been studied using high or low Pi medium, a new study was conducted using a micro-device, called Dual-flow-RootChip, that allows flow of different liquids on two sides of roots (Stanley et al. 2018). When an Arabidopsis root containing RSL4-GFP was exposed to high and low Pi medium, surprisingly the RSL4-GFP signal was enhanced on the high Pi side, resulting in the production of longer root hairs only on the high Pi side (Stanley et al. 2018). This striking observation demonstrates that plants’ response to local Pi signals is different from their systemic response and plants induce root hair growth when they face high Pi locally to promote efficient Pi uptake.
Conclusions and perspectives
In this review we highlighted the key transcriptional regulations that link developmental and/or environmental inputs to root hair identity and growth. Various exogenous cues affect root hair growth and it is becoming increasingly evident that many of these upstream cues are integrated to auxin- or ethylene-mediated signaling pathways. Central regulators acting downstream of these signaling cascades are the two bHLH transcription factors RSL2 and RSL4 that function as a hub to regulate root hair growth in response to environmental signals. For cell fate specification, two MYB transcription factors CPC and ETC1 appear to function as a hub to integrate environmental and developmental cues. Another emerging characteristic of the root hair gene regulatory network is the intimate transcriptional interaction between these positive root hair regulators and negative regulators such as GTL1 and OBP4. In particular, GTL1 and RSL4 form a mutually regulating transcriptional module, and we predict that GTL1 functions as a stabilizer in this module to minimize the fluctuation of RSL4 expression under the changing environment. Although many core regulators of root hair development have been identified (Table 1), many of their transcriptional interactions remain unknown. Further elucidation of these interactions will reveal the gene regulatory network of root hair development and should facilitate better understanding of how plants regulate cell fate and growth in response to environmental signals.
Table 1.
Transcription factors involved in root hair development
| Gene name | AGI | Type | Function | References |
|---|---|---|---|---|
| CAPRICE (CPC) | At2g46410 | MYB | Cell fate | Wada et al. (1997) |
| TRIPTYCHON (TRY) | At5g53200 | MYB | Cell fate | Schellmann et al. (2002) |
| ENHANCER OF TRY AND CPC (ETC1) | At1g01380 | MYB | Cell fate | Simon et al. (2007) |
| MYB23 (MYB23) | At5g40330 | MYB | Cell fate | Kang et al. (2009) |
| WEREWOLF (WER) | At5g14750 | MYB | Cell fate | Lee and Schiefelbein (1999) |
| ENHANCER OF GLABRA3 (EGL3) | At1g63650 | bHLH | Cell fate | Bernhardt et al. (2003) |
| GLABRA3 (GL3) | At5g41315 | bHLH | Cell fate | Bernhardt et al. (2003) |
| TRANSPARENT TESTA GLABRA2 (TTG2) | At2g37260 | WRKY | Cell fate | Johnson et al. (2002), Ishida et al. (2007) |
| GLABRA (GL2) | At1g79840 | HD-ZIP | Cell fate | Masucci et al. (1996), Di Cristina et al. (1996), Lin et al. (2015) |
| ZINC FINGER PROTEIN 5 (ZFP5) | At1g10480 | C2H2 | Hormone response | An et al. (2012) |
| ETHYLENE INSENSITIVE 3 (EIN3) | At3g20770 | EIL | Hormone response | Song et al. (2016), Feng et al. (2017) |
| EIN3-LIKE1 (EIL1) | At2g27050 | EIL | Hormone response | Song et al. (2016), Feng et al. (2017) |
| AUXIN REPONSE FACTORs (ARFs) | – | ARF | Hormone response | Mangano et al. (2017), Bhosale et al. (2018), Choi et al. (2018) |
| PHOSPHATE STARVATION RESPONSE 1 (PHR1) | At4g28610 | MYB | Phosphate response | Bustos et al. (2010) |
| PHR1-LIKE1 (PHL1) | At5g29000 | MYB | Phosphate response | Bustos et al. (2010) |
| ALFIN-LIKE6 (AL6) | At2g02470 | Homeodomain protein | Phosphate response | Chandria et al. (2013) |
| WRKY75 | At5g13080 | WRKY | Phosphate response | Devaiah et al. (2007), Rishmawi et al. (2014) |
| BASIC LOOP HELIX LOOP 32 (BHLH32) | At3g25710 | bHLH | Phosphate response | Chen et al. (2007) |
| ROOT HAIR DEFECTIVE6 (RHD6) | At1g66470 | bHLH | Hair growth | Masucci and Schiefelbein (1994), Menand et al. (2007) |
| RHD SIX-LIKE 1 (RSL1) | At5g37800 | bHLH | Hair growth | Menand et al. (2007), Yi et al. (2010) |
| RHD SIX-LIKE 2 (RSL2) | At4g33880 | bHLH | Hair growth | Yi et al. (2010), Rymen et al. (2017) |
| RHD SIX-LIKE 3 (RSL3) | At2g14760 | bHLH | Hair growth | Bruex et al. (2012), Pires et al. (2013) |
| RHD SIX-LIKE 4 (RSL4) | At1g27740 | bHLH | Hair growth | Yi et al. (2010) and etc |
| RHD SIX-LIKE 5 (RSL5) | At5g43175 | bHLH | Hair growth | Pires et al. (2013) |
| Lotus japonicus ROOT HAIR LESS-LIKE 1 (LRL1) | At2g24260 | bHLH | Cell fate & Hair growth | Karas et al. (2009), Lin et al. (2015), Breuninger et al. (2016) |
| Lotus japonicus ROOT HAIR LESS-LIKE 2 (LRL2) | At4g30980 | bHLH | Cell fate & Hair growth | Karas et al. (2009), Lin et al. (2015), Breuninger et al. (2016) |
| Lotus japonicus ROOT HAIR LESS-LIKE 3 (LRL3) | At5g58010 | bHLH | Hair growth | Karas et al. (2009), Lin et al. (2015), Breuninger et al. (2016) |
| Lotus japonicus ROOT HAIR LESS-LIKE 4 (LRL4) | At1g03040 | bHLH | Hair growth | Bruex et al. (2012), Breuninger et al. (2016) |
| Lotus japonicus ROOT HAIR LESS-LIKE 5 (LRL5) | At4g02590 | bHLH | Hair growth | Bruex et al. (2012), Breuninger et al. (2016) |
| MEMBRANE ANCHORED MYB (maMYB) | At5g45420 | MYB | Hair growth | Slabaugh et al. (2011) |
| HOMEODOMAIN GLABROUS 11 (HDG11) | At1g73360 | HD-ZIP | Hair growth | Xu et al. (2014) |
| OBF BINDING PROTEIN 4 (OBP4) | At5g60850 | Dof | Hair growth | Rymen et al. (2017) |
| GT2-LIKE1 (GTL1) | At1g33240 | Trihelix | Hair growth | Shibata et al. (2018) |
| DF1 | At1g76880 | Trihelix | Hair growth | Shibata et al. (2018) |
Acknowledgements
We thank Dr. Kiminori Toyooka, Dr. Mayuko Sato and Ms. Noriko Takeda for their technical support in root hair imaging. This work was supported by grants from the Ministry of Education, Culture, Sports and Technology of Japan to M.S. (16J07464) and to K.S. (15H05961, 17H03704.)
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- An L, Zhou Z, Sun L, et al. A zinc finger protein gene ZFP5 integrates phytohormone signaling to control root hair development in Arabidopsis. Plant J. 2012;72:474–490. doi: 10.1111/j.1365-313X.2012.05094.x. [DOI] [PubMed] [Google Scholar]
- Bernhardt C, Lee MM, Gonzalez A, et al. The bHLH genes GLABRA3 (GL3) andENHANCER OF GLABRA3 (EGL3) specify epidermal cell fate in the Arabidopsis root. Development. 2003;130:6431–6439. doi: 10.1242/dev.00880. [DOI] [PubMed] [Google Scholar]
- Bhosale R, Giri J, Pandey BK, et al. A mechanistic framework for auxin dependent Arabidopsis root hair elongation to low external phosphate. Nat Commun. 2018;9:1409. doi: 10.1038/s41467-018-03851-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breuer C, Kawamura A, Ichikawa T, et al. The trihelix transcription factor GTL1 regulates ploidy-dependent cell growth in the Arabidopsis trichome. Plant Cell. 2009;21:2307–2322. doi: 10.1105/tpc.109.068387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breuer C, Morohashi K, Kawamura A, et al. Transcriptional repression of the APC/C activator CCS52A1 promotes active termination of cell growth. EMBO J. 2012;31:4488–4501. doi: 10.1038/emboj.2012.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breuninger H, Thamm A, Streubel S, et al. Diversification of a transcription factor family led to the evolution of antagonistically acting genetic regulators of root hair growth. Cur Biol. 2016;26:1622–1628. doi: 10.1016/j.cub.2016.04.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruex A, Kainkaryam RM, Wieckowski Y, et al. A gene regulatory network for root epidermis cell differentiation in Arabidopsis. PLoS Genet. 2012;8:e1002446. doi: 10.1371/journal.pgen.1002446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustos R, Castrillo G, Linhares F, et al. A central regulatory system largely controls transcriptional activation and repression responses to phosphate starvation in Arabidopsis. PLoS Genet. 2010;6:e1001102. doi: 10.1371/journal.pgen.1001102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrika NNP, Sundaravelpandian K, Yu S-M, Schmidt W. ALFIN-LIKE 6 is involved in root hair elongation during phosphate deficiency in Arabidopsis. New Phytol. 2013;198:709–720. doi: 10.1111/nph.12194. [DOI] [PubMed] [Google Scholar]
- Chen Z-H, Nimmo GA, Jenkins GI, Nimmo HG. BHLH32 modulates several biochemical and morphological processes that respond to Pi starvation in Arabidopsis. Biochem J. 2007;405:191–198. doi: 10.1042/BJ20070102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi H-S, Seo M, Cho H-T. Two TPL-binding motifs of ARF2 are involved in repression of auxin responses. Front Plant Sci. 2018;9:372. doi: 10.3389/fpls.2018.00372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta S, Kim CM, Pernas M, et al. Root hairs: development, growth and evolution at the plant-soil interface. Plant Soil. 2011;346:1–14. doi: 10.1007/s11104-011-0845-4. [DOI] [Google Scholar]
- Devaiah BN, Karthikeyan AS, Raghothama KG. WRKY75 transcription factor is a modulator of phosphate acquisition and root development in Arabidopsis. Plant Physiol. 2007;143:1789–1801. doi: 10.1104/pp.106.093971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Cristina M, Sessa G, Dolan L, et al. The Arabidopsis Athb-10 (GLABRA2) is an HD-Zip protein required for regulation of root hair development. Plant J. 1996;10:393–402. doi: 10.1046/j.1365-313X.1996.10030393.x. [DOI] [PubMed] [Google Scholar]
- Dindas J, Scherzer S, Roelfsema MRG, et al. AUX1-mediated root hair auxin influx governs SCF TIR1/AFB -type Ca 2 + signaling. Nat Commun. 2018;9:1174. doi: 10.1038/s41467-018-03582-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Xu P, Li B, et al. Ethylene promotes root hair growth through coordinated EIN3/EIL1 and RHD6/RSL1 activity in Arabidopsis. Proc Natl Acad Sci U S A. 2017;114:13834–13839. doi: 10.1073/pnas.1711723115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galway ME, Masucci JD, Lloyd AM, et al. The TTG gene is required to specify epidermal cell fate and cell patterning in the Arabidopsis root. Dev Biol. 1994;166:740–754. doi: 10.1006/dbio.1994.1352. [DOI] [PubMed] [Google Scholar]
- Grierson C, Nielsen E, Ketelaarc T, Schiefelbein J. Root hairs. Arabidopsis Book. 2014 doi: 10.1199/tab.0172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang K-L, Ma G-J, Zhang M-L, et al. The ARF7 and ARF19 Transcription factors positively regulate PHOSPHATE STARVATION RESPONSE1 in Arabidopsis roots. Plant Physiol. 2018;178:413–427. doi: 10.1104/pp.17.01713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang Y, Choi H-S, Cho H-M, Cho H-T. Tracheophytes contain conserved orthologs of a basic helix-loop-helix transcription factor that modulate ROOT HAIR SPECIFIC genes. Plant Cell. 2017;29:39–53. doi: 10.1105/tpc.16.00732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida T, Hattori S, Sano R, et al. Arabidopsis TRANSPARENT TESTA GLABRA2 is directly regulated by R2R3 MYB transcription factors and is involved in regulation of GLABRA2 transcription in epidermal differentiation. Plant Cell. 2007;19:2531–2543. doi: 10.1105/tpc.107.052274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida T, Kurata T, Okada K, Wada T. A genetic regulatory network in the development of trichomes and root hairs. Annu Rev Plant Biol. 2008;59:365–386. doi: 10.1146/annurev.arplant.59.032607.092949. [DOI] [PubMed] [Google Scholar]
- Johnson CS, Kolevski B, Smyth DR. TRANSPARENT TESTA GLABRA2, a trichome and seed coat development gene of Arabidopsis, encodes a WRKY transcription factor. Plant Cell. 2002;14:1359–1375. doi: 10.1105/tpc.001404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang YH, Kirik V, Hulskamp M, Nam KH, Hagely K, Lee MM, Schiefelbein J. The MYB23 gene provides a positive feedback loop for cell fate specification in the Arabidopsis root epidermis. Plant Cell. 2009;21:1080–1094. doi: 10.1105/tpc.108.063180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karas B, Amyot L, Johansen C, et al. Conservation of Lotus and Arabidopsis basic helix-loop-helix proteins reveals new players in root hair development. Plant Physiol. 2009;151:1175–1185. doi: 10.1104/pp.109.143867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DW, Lee SH, Choi S-B, et al. Functional conservation of a root hair cell-specific cis-element in angiosperms with different root hair distribution patterns. Plant Cell. 2006;18:2958–2970. doi: 10.1105/tpc.106.045229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RD-W, Cho H-T. Auxin, the organizer of the hormonal/environmental signals for root hair growth. Front Plant Sci. 2013 doi: 10.3389/fpls.2013.00448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MM, Schiefelbein J. WEREWOLF, a MYB-related protein in Arabidopsis, is a position-dependent regulator of epidermal cell patterning. Cell. 1999;99:473–483. doi: 10.1016/S0092-8674(00)81536-6. [DOI] [PubMed] [Google Scholar]
- Lin Q, Ohashi Y, Kato M, et al. GLABRA2 directly suppresses basic helix-loop-helix transcription factor genes with diverse functions in root hair development. Plant Cell. 2015;27:2894–2906. doi: 10.1105/tpc.15.00607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangano S, Denita-Juarez SP, Choi H-S, et al. Molecular link between auxin and ROS-mediated polar growth. Proc Natl Acad Sci USA. 2017;114:5289–5294. doi: 10.1073/pnas.1701536114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangano S, Denita-Juarez SP, Marzol E, et al. High auxin and high phosphate impact on RSL2 expression and ROS-homeostasis linked to root hair growth in Arabidopsis thaliana. Front Plant Sci. 2018 doi: 10.3389/fpls.2018.01164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masucci JD, Schiefelbein JW. The rhd6 mutation of Arabidopsis thaliana alters root-hair initiation through an auxin- and ethylene-associated process. Plant Physiol. 1994;106:1335–1346. doi: 10.1104/pp.106.4.1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masucci JD, Rerie WG, Foreman DR, et al. The homeobox gene GLABRA2 is required for position-dependent cell differentiation in the root epidermis of Arabidopsis thaliana. Development. 1996;122:1253–1260. doi: 10.1242/dev.122.4.1253. [DOI] [PubMed] [Google Scholar]
- Menand B, Yi K, Jouannic S, et al. An ancient mechanism controls the development of cells with a rooting function in land plants. Science. 2007;316:1477–1480. doi: 10.1126/science.1142618. [DOI] [PubMed] [Google Scholar]
- Miura K, Rus A, Sharkhuu A, et al. The Arabidopsis SUMO E3 ligase SIZ1 controls phosphate deficiency responses. Proc Natl Acad Sci USA. 2005;102:7760–7765. doi: 10.1073/pnas.0500778102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pires ND, Yi K, Breuninger H, et al. Recruitment and remodeling of an ancient gene regulatory network during land plant evolution. Proc Natl Acad Sci USA. 2013;110:9571–9576. doi: 10.1073/pnas.1305457110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitts RJ, Cernac A, Estelle M. Auxin and ethylene promote root hair elongation in Arabidopsis. Plant J. 1998;16:553–560. doi: 10.1046/j.1365-313x.1998.00321.x. [DOI] [PubMed] [Google Scholar]
- Puga MI, Mateos I, Charukesi R, et al. SPX1 is a phosphate-dependent inhibitor of Phosphate Starvation Response 1 in Arabidopsis. Proc Natl Acad Sci USA. 2014;111:14947–14952. doi: 10.1073/pnas.1404654111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rishmawi L, Wolff H, Schrader A, Hülskamp M. Sub-epidermal expression of ENHANCER OF TRIPTYCHON AND CAPRICE1 and its role in root hair formation upon Pi starvation. Front Plant Sci. 2018 doi: 10.3389/fpls.2018.01411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio V, Linhares F, Solano R, et al. A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev. 2001;15:2122–2133. doi: 10.1101/gad.204401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rymen B, Kawamura A, Schaefer S, et al. ABA suppresses root hair growth via the OBP4 transcriptional-regulator repression of the RSL2 promoter. Plant Physiol. 2017;173:1750–1762. doi: 10.1104/pp.16.01945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salazar-Henao JE, Vélez-Bermúdez IC, Schmidt W. The regulation and plasticity of root hair patterning and morphogenesis. Development. 2016;143:1848–1858. doi: 10.1242/dev.132845. [DOI] [PubMed] [Google Scholar]
- Schellmann S, Schnittger A, Kirik V, et al. TRIPTYCHON and CAPRICE mediate lateral inhibition during trichome and root hair patterning in Arabidopsis. EMBO J. 2002;21:5036–5046. doi: 10.1093/emboj/cdf524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnall JA, Quatrano RS. Abscisic acid elicits the water-stress response in root hairs of Arabidopsis thaliana. Plant Physiol. 1992;100:216–218. doi: 10.1104/pp.100.1.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata M, Breuer C, Kawamura A, et al. GTL1 and DF1 regulate root hair growth through transcriptional repression of ROOT HAIR DEFECTIVE 6-LIKE 4 in Arabidopsis. Development. 2018;145:dev159707. doi: 10.1242/dev.159707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon M, Lee MM, Lin Y, et al. Distinct and overlapping roles of single-repeat MYB genes in root epidermal patterning. Dev Biol. 2007;311:566–578. doi: 10.1016/j.ydbio.2007.09.001. [DOI] [PubMed] [Google Scholar]
- Slabaugh E, Held M, Brandizzi F. Control of root hair development in Arabidopsis thaliana by an endoplasmic reticulum anchored member of the R2R3-MYB transcription factor family. Plant J. 2011;67:395–405. doi: 10.1111/j.1365-313X.2011.04602.x. [DOI] [PubMed] [Google Scholar]
- Song L, Yu H, Dong J, et al. The molecular mechanism of ethylene-mediated root hair development induced by phosphate starvation. PLoS Genet. 2016 doi: 10.1371/journal.pgen.1006194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley CE, Shrivastava J, Brugman R, et al. Dual-flow-RootChip reveals local adaptations of roots towards environmental asymmetry at the physiological and genetic levels. New Phytol. 2018;217:1357–1369. doi: 10.1111/nph.14887. [DOI] [PubMed] [Google Scholar]
- Tominaga-Wada R, Kurata T, Wada T. Localization of ENHANCER OF TRY AND CPC1 protein in Arabidopsis root epidermis. J Plant Physiol. 2017;214:48–52. doi: 10.1016/j.jplph.2017.04.001. [DOI] [PubMed] [Google Scholar]
- Velasquez SM, Barbez E, Kleine-Vehn J, Estevez JM. Auxin and cellular elongation. Plant Physiol. 2016;170:1206–1215. doi: 10.1104/pp.15.01863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijayakumar P, Datta S, Dolan L. ROOT HAIR DEFECTIVE SIX-LIKE4 (RSL4) promotes root hair elongation by transcriptionally regulating the expression of genes required for cell growth. New Phytol. 2016;212:944–953. doi: 10.1111/nph.14095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada T, Tachibana T, Shimura Y, Okada K. Epidermal cell differentiation in Arabidopsis determined by a Myb homolog. CPC Sci. 1997;277:1113–1116. doi: 10.1126/science.277.5329.1113. [DOI] [PubMed] [Google Scholar]
- Won S-K, Lee Y-J, Lee H-Y, et al. cis-element- and transcriptome-based screening of root hair-specific genes and their functional characterization in Arabidopsis. Plant Physiol. 2009;150:1459–1473. doi: 10.1104/pp.109.140905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P, Cai X-T, Wang Y, Xing L, Chen Q, Xiang C-B. HDG11 upregulates cell-wall-loosening protein genes to promote root elongation in Arabidopsis. J Exp Bot. 2014;65:4285–4295. doi: 10.1093/jxb/eru202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi K, Menand B, Bell E, Dolan L. A basic helix-loop-helix transcription factor controls cell growth and size in root hairs. Nat Genet. 2010;42:264–267. doi: 10.1038/ng.529. [DOI] [PubMed] [Google Scholar]
- Zhang S, Huang L, Yan A, et al. Multiple phytohormones promote root hair elongation by regulating a similar set of genes in the root epidermis in Arabidopsis. J Exp Bot. 2016;67:6363–6372. doi: 10.1093/jxb/erw400. [DOI] [PMC free article] [PubMed] [Google Scholar]