Graphical abstract
Keywords: Flavonoids, Terpenoids, Lamiaceae family, LC-MS profiling, LC-MS/MS analysis
Abstract
Dereplication of crude plant extracts through liquid chromatography-mass spectrometry is a powerful technique for the discovery of novel natural products. Unfortunately, this technique is often plagued by a low level of confidence in natural product identification. This is mainly due to the lack of extensive chromatographic and mass spectrometric optimizations that result in improper and incomplete MS/MS fragmentation data. This study proposes a solution to this problem by the optimization of chromatographic separation and mass spectrometry parameters. We report herein a direct and high-throughput strategy for natural product dereplication in five Salvia species using high-resolution ESI-QTOF-MS/MS data. In the present study, we were able to identify a total of forty-seven natural products in crude extracts of five Salvia species using MS/MS fragmentation data. In addition to dereplication of Salvia species, quantitative profiling of twenty-one bioactive constituents of the genus was also performed on an ion trap mass spectrometer. For the quantitation study, method development focused on chromatographic optimizations to achieve maximum sensitivity. The developed dereplication and quantitation strategy can be extended to develop comprehensive metabolic profiles of other plant genera and species and thus can prove useful in the field of drug discovery from plants.
Introduction
The genus Salvia is the largest genus in the family Lamiaceae (mint family) comprising about 1000 species of shrubs, annuals and perennials [1], [2]. Salvia species have been used for centuries for the treatment of various ailments. The representative plant of this genus Salvia officinalis L. is commonly used in the form of an aqueous infusion to treat cough, bronchitis, asthma and digestive disturbances [3]. The essential oils and fractionated extracts of this plant have been shown to possess cytotoxic and antiviral properties [4], [5]. In Chinese medicine, roots of Salvia miltiorrhiza (Danshen) have been long used for longevity and to treat cardiovascular problems such as hypertension, angina, myocardial infarction and ischemic stroke [6], [7]. Salvia moorcroftiana roots are used to treat cough and cold, while its seeds are used to treat diarrhea [8], [9].
The genus Salvia is rich in low molecular weight compounds such as sesquiterpenoids of germacrane, carotane, caryophyllane and guaiane classes; diterpenoids of abietane, clerodane, pimarane, labdane and other classes; sesterterpenoids of C-23 and C-25 classes; triterpenoids of ursane, oleanane, lupane and dammarane classes; phenolic acids and flavonoids [10]. Many compounds isolated from various Salvia species exhibit interesting biological activities such as antimicrobial, antiviral, anticancer and antioxidant activities. Tanshinones, the most well-known compounds first isolated from S. miltiorrhiza and other species, are known for their antioxidant [11], anti-inflammatory [12], anticancer [13], antibacterial and antiplatelet aggregation activities [14]. Salvianolic acid A and B, isolated from various Salvia species, show antioxidant and cardioprotective activities [15]. Specific compounds reported from plants included in this study have also shown promising biological activities. For example, 5-hydroxy-7,4′-dimethoxyflavone isolated from S. moorcroftiana has shown inhibitory activities against α-glucosidase [16]. Nubiol isolated from S. nubicola has been proven active against Pseudomonas aeruginosa [17]. Ethyl acetate fractions of S. Plebeia and the isolated compound 6-methoxyluteolin-7-glucoside have shown antioxidant properties [18], [19].
Natural products have always played a key role in the discovery of novel drugs. It has been estimated that about half of new drugs approved by the FDA during 1981–2014 were either natural products, their mimics or their derivatives [20]. However, the content of natural products in a plant, in qualitative and quantitative terms, varies due to several factors such as the weather. This makes the traditional isolation and characterization of natural products even more tedious and difficult than it already is. To overcome this problem, it is essential to obtain reliable metabolic profiles that represent the characteristics and pharmacologically active natural products of a plant. Metabolic profile development requires prior identification of natural products for which HPLC-MS/MS is a fast and reliable approach. It is often done without the use of chemically pure standards since the availability of a compound in question through synthesis, isolation or commercial sources is not always possible. Another problem in addition to the unavailability of pure standards is that every plant specie can have its own unique chemistry. This poses a big challenge to the development of a versatile chromatographic method that can be used to analyze a diverse range of plant extracts. Even if a method is versatile enough to effectively separate many components of a mixture, the level of certainty of natural product identification through mass spectrometry varies depending upon what information is available. Unambiguous identification is only possible when a purified standard is available. However, in the case of a large metabolomics study, it is neither economical nor practical to have a large number of purified standards available. In cases where purified standards are unavailable, it is still possible to identify natural products in a sample based on accurate mass and MS/MS fragmentation data [21]. The available information is what constitutes the so-called “identification levels” in metabolomics [22]. Unambiguous identification using a standard is termed Level 1, while Level 2 is used for identification using MS/MS fragmentation data. Since Level 1 is only achievable in a small number of cases, Level 2 is the most commonly used level of natural product identification using mass spectrometry.
We present herein a direct and high-throughput approach for the profiling of flavonoids and terpenoids in five important Salvia species along with quantitation of twenty-one bioactive principles in the same number of plants. The five Salvia species included in this study have been long used in the indigenous medicinal system of Indo-Pak region but, a comprehensive approach for the dereplication of natural products in these species has never been reported. The high-throughput screening method developed in this study is an approach that presents a clear picture of the natural product content of the studied Salvia species. The knowledge of what natural products are present in a plant can serve as a means of discovery of potential drug leads. The current study can prove useful for bioactivity-guided drug discovery from Salvia species and study various biochemical pathways in plant metabolomics.
Experimental
Chemicals and reagents
Compounds 2–3, 6–8, 11, 15 and 17–20 were purchased from Sigma-Aldrich (Riedstr. 2 D-89555, Steinheim 49 7329 970, Germany) while compounds 1, 4–5, 9–10, 12–14, 16 and 21 (Table 1) were previously isolated by our research group from various sources. All analytes including their class, formula, molecular weight and the instrument polarity used for analysis are listed in Table S1. Formic acid was used as an additive for the mobile phase and purchased from Daejung (Daejung Chemicals & Metals Co. Ltd., Korea). Methanol (MeOH) and acetonitrile (ACN) for mobile phase ware purchased from Merck (Merck KGaA, Darmstadt, Germany) and Daejung (Daejung Chemicals & Metals Co. Ltd., Korea), respectively. Type I water (ISO 3696) for the mobile phase was obtained from Barnstead™ GenPure™ ultrapure water system (Thermo Fisher Scientific Inc., USA).
Table 1.
Optimized MS/MS parameters for compounds 1–21.
| Analyte | Compound analyzed | Retention time | m/z | Ion type | Fragmentation amplitude | MRM transitions |
|---|---|---|---|---|---|---|
| 1 | Apigenin-7-O-glucoside | 3.15 | 431.3 | [M−H]− | 90 | 431.3 → 269.1, 311.2 |
| 2 | Salvianolic acid B | 4.19 | 717.6 | [M−H]− | 75 | 717.6 → 519.2, 321.1 |
| 3 | Salvianolic acid A | 4.55 | 517.1 | [M+Na]+ | 75 | 517.1 → 337.0, 319.0, 221.0 |
| 4 | (2S,3R)-Morelloflavone-7-O-rhamnopyranoside | 4.74 | 703.3 | [M+H]+ | 55 | 703.3 → 541.1, 415.1 |
| 5 | (2S,3R)-Volkensiflavone-7-O-rhamnopyranoside | 4.81 | 731.6 | [M+HCOOH-H]− | 65 | 731.6 → 514.4, 463.1, 605.3, 443.2, 569.3 |
| 6 | Luteolin | 4.83 | 285.1 | [M−H]− | 100 | 285.1 → 217.0, 175.0, 151.0 |
| 7 | Quercetin | 4.84 | 301.1 | [M−H]− | 100 | 301.1 → 179.0, 151.0 |
| 8 | Apigenin | 5.41 | 269.1 | [M−H]− | 105 | 269.1 → 149.0 |
| 9 | (2S,3R)-Morelloflavone | 5.43 | 557.1 | [M+H]+ | 80 | 557.1 → 451.1, 405.1, 431.0 |
| 10 | Naringenin | 5.49 | 271.1 | [M−H]− | 90 | 271.1 → 177.0, 151.0 |
| 11 | Diosmetin | 5.55 | 299.1 | [M−H]− | 110 | 299.1 → 284.1 |
| 12 | (2S,3R)-Volkensiflavone | 5.77 | 541.1 | [M+H]+ | 85 | 541.1 → 415.0, 389.0 |
| 13 | Chrysin | 6.31 | 253.1 | [M−H]− | 110 | 253.1 → 209.0, 181.0 |
| 14 | 3,5,7-Trimethoxyflavone | 6.5 | 313.0 | [M+H]+ | 125 | 313.0 → 298.0, 269.0 |
| 15 | Salvinorin A | 6.83 | 455.1 | [M+Na]+ | 85 | 455.1 → 238.9, 395.0 |
| 16 | 3-Methylflavone | 7.02 | 237.0 | [M+H]+ | 130 | 237.0 → 178.0, 133.0 |
| 17 | Carnosic acid | 7.19 | 331.2 | [M−H]− | 75 | 331.2 → 303.1, 285.1 |
| 18 | Carnosol | 7.2 | 329.3 | [M−H]− | 60 | 329.3 → 285.3 |
| 19 | Cryptotanshinone | 7.57 | 297.1 | [M+H]+ | 100 | 297.1 → 279.0, 251.0 |
| 20 | Tanshinone IIA | 7.97 | 295.0 | [M+H]+ | 87 | 295.0 → 277.0, 249.0 |
| 21 | Rutin | 8.06 | 611.3 | [M+H]+ | 70 | 611.3 → 471.8, 317.1 |
Instrumentation and experimental conditions
HPLC-MS/MS analysis for natural product identification was performed on Bruker maXis II™ HR-QTOF mass spectrometer (Bremen, Germany) coupled to Dionex UltiMate™ 3000 series HPLC system (Thermo Fisher Scientific, Waltham, MA, USA) fitted with a binary RS pump, column thermostat and auto-sampler. Sample chromatography was performed on Macherey-Nagel Nucleodur® C18 Gravity column (3.0 × 100 mm, 1.8 µm) kept at 40 °C. 4-µL sample was injected while the mobile phase consisted of A (0.1% formic acid in H2O) and B (0.1% formic acid in MeOH). Mobile phase flow rate was set at 0.7 mL/min using a linear gradient of A and B starting at 10% B, increased to 90% B in 5.5 min, maintained at 90% for 1.5 min, and returned to 10% B in 1 min. Total run time was 10 min including a 1-min holding time at the start and 1-min equilibration time at the end of the gradient.
Mass spectra were recorded using electrospray ionization employing the Bruker CaptiveSpray™ ion source. MS and MS/MS spectra were recorded separately both in positive and negative mode. Ion source parameters used are mentioned as follows (parameters for negative mode next to positive mode parameters): capillary voltage at 4500 V (−3500 V), end plate offset at 500 V, nebulizer gas 45.0 psi, drying gas at 12.0 L/min and drying gas temperature at 270 °C. All spectra were recorded in the mass range from m/z 100 to 2000 while the scan speed was set at 5 Hz for MS while 12 Hz for MS/MS spectra. The active exclusion feature of the instrument was used which enables the instrument to remove a precursor ion from consideration from further consideration after a set number of MS/MS spectra have been recorded for that particular precursor ion. The active exclusion number was set at 3, and the precursor reconsideration time was set at 30 s.
HPLC-MS/MS analysis for quantitation was performed on Bruker amaZon™ speed ion trap mass spectrometer (Bremen, Germany) coupled to Dionex UltiMate™ 3000 series HPLC system (Thermo Fisher Scientific, Waltham, MA, USA) fitted with a binary pump, column thermostat and auto-sampler. Chromatographic separation of analytes was achieved on the reverse-phase C-Phenyl column (Agilent ZORBAX Eclipse XDB-Phenyl 4.6 × 75 mm, 3.5 µm) kept at 40 °C. A 4-µL sample was injected while the mobile phase consisted of A (0.1% formic acid in H2O) and C (0.1% formic acid in ACN). Mobile phase flow rate was set at 0.6 mL/min using a linear gradient of A and C starting at 25% C, increased to 95% C in 6 min, maintained at 95% for 1 min, and returned to 25% C in 1 min. Total run time was 10 min including a 1-min holding time at the start and 1-min equilibration time at the end of the gradient.
Mass spectra for quantitation were recorded under positive and negative modes as appropriate for the individual analyte (Table S1). Ion source parameters were as follows: capillary voltage at 4500 V (−3500 V for negative mode), end plate offset at 500 V, nebulizer gas 35.0 psi, drying gas at 8.0 L/min and drying gas temperature at 250 °C. Mass spectra scan range was set at m/z 50 to 850 while the number of spectral averages was set at 5. Ion charge control (ICC) was used for transferring a certain number of ions to the ion trap. ICC was set at 200,000 while the accumulation time was 100 ms. Fragmentation was performed under collision-induced dissociation (CID) with a time interval of 1.0 s between MS and MS/MS while the fragmentation time was set at 20 ms. Fragmentation amplitude was optimized for each analyte to obtain the maximum abundance of fragment ions. Table 1 summarizes optimized MS/MS parameters for all analytes.
Method performance
All MS and MS/MS data was saved using both profile and line spectra to minimize the possibility of instrumental noise being mistaken as a precursor ion. For qualification of precursor ion for MS/MS analysis, isotopic pattern matching (hereto referred to as the mSigma value). Mass spectra for all samples were recorded under both ionization modes (positive and negative) to counter check the authenticity of a molecular ion peak while active exclusion was used to minimize the chance of common contaminant peaks being put under MS/MS fragmentation. Each sample was injected in triplicate. A pooled QC sample was prepared by combining all plant extracts to check the accuracy of data by comparing the identified compounds in a sample against the QC sample.
The performance of the developed quantitation method was assessed through the determination of accuracy and precision. Accuracy (% bias) and precision (% RSD) were assessed by analyzing three different QC samples with six replicates for intra-day and twelve replicates (Two days, six replicates/day) for inter-day analysis. The accuracy of analysis was calculated using the expected concentration (CE) and the mean value of measured concentration (CM) by using the following relation: Accuracy (bias, %) = [(CE − CM)/CE] × 100. Similarly, relative standard deviation, % RSD was used as an indicator of analytical precision and calculated from the standard deviation and mean value of measured concentrations by the following equation: Precision (RSD, %) = (Standard Deviation (SD)/CM) × 100. LOD and LOQ values for the analyzed compounds were calculated using the standard deviation of the response (σ) and the slopes (S) i.e. LOD = 3.3σ/S and LOQ = 10σ/S.
Method performance was further evaluated through the analysis of fortified samples prepared by spiking additional amounts of compounds 1, 2–3, 6–8, 11 and 17–19 at three levels of 100, 200 and 400 ng/mL, respectively in the original sample solutions used for analysis.
Sample preparation
Shade-dried plant material (whole plant) was crushed using a dry mill. 1 g of each plant was accurately weighed and extracted with 10 mL methanol through sonication for 20 min. Each sample was centrifuged for 15 min at 6000 rpm to settle large particles, and the supernatant was filtered through a 0.22 µm PTFE syringe-driven filter. 50 µL of the filtered extract was diluted to 1000 µL with methanol for LC-MS and LC-MS/MS analysis.
For quantitation 1 mg of each standard compound was weighed and dissolved into 1 mL methanol to prepare standard stock solutions. These solutions were diluted with 50:50 water: ACN in a serial manner to prepare eight calibrant solutions ranging from 50 to 1500 ng/mL. Analysis of plant samples was performed using diluted plant extract. 50 µL of filtered plant extract was diluted to 1500 µL with 50:50 water: ACN for LC-MS/MS analysis.
Spiked samples for method validation were prepared in a similar manner as the plant samples. 50 µL of filtered plant extract plus an amount of standard solution equivalent to spike concentrations of 50, 100 and 150 ng/mL were diluted to a final volume of 1500 µL with 50:50 water: ACN for three samples, and labelled as S1, S2 and S3, respectively.
Spiked samples for method validation were prepared in a similar manner as the plant samples. 50 µL of filtered plant extract plus an amount of standard solution equivalent to spike concentrations of 100, 200 and 400 ng/mL was diluted to a final volume of 1500 µL with 50:50 water: ACN for three samples and labelled as S1, S2 and S3, respectively.
Results and discussion
LC-MS/MS optimization
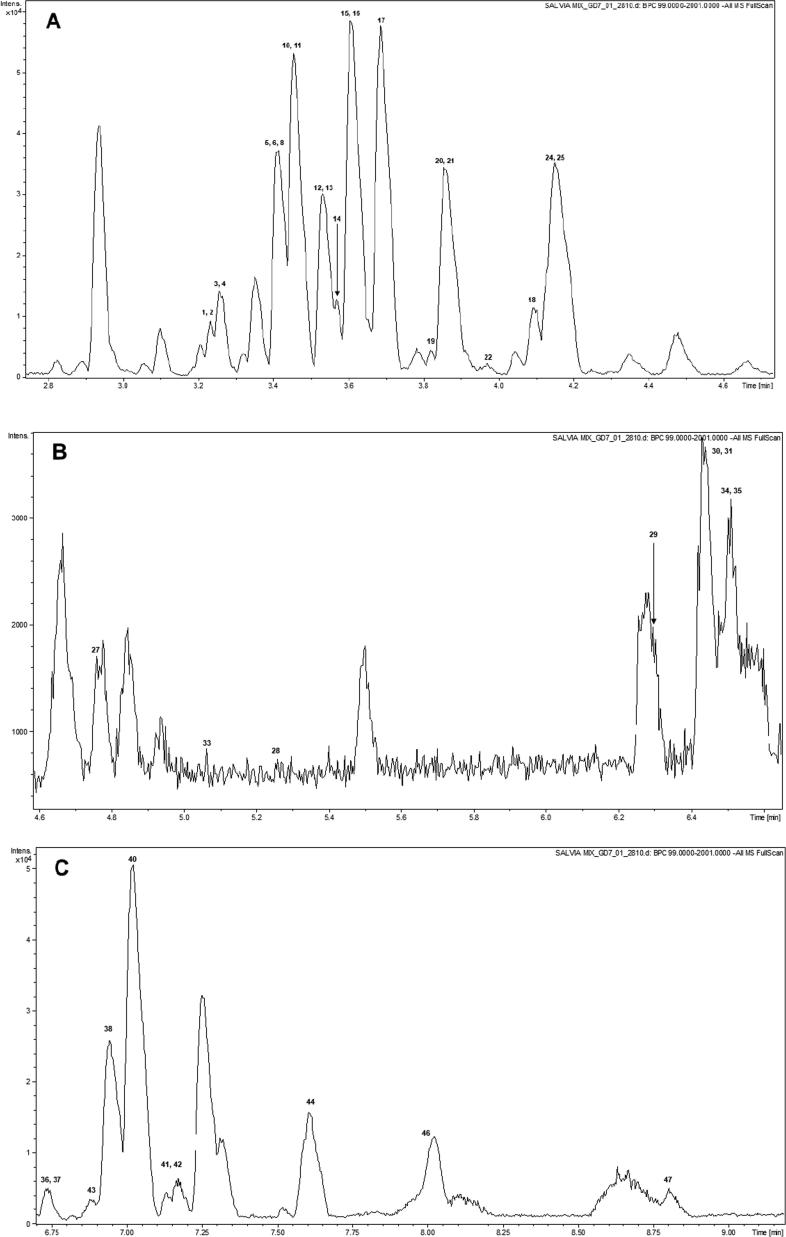
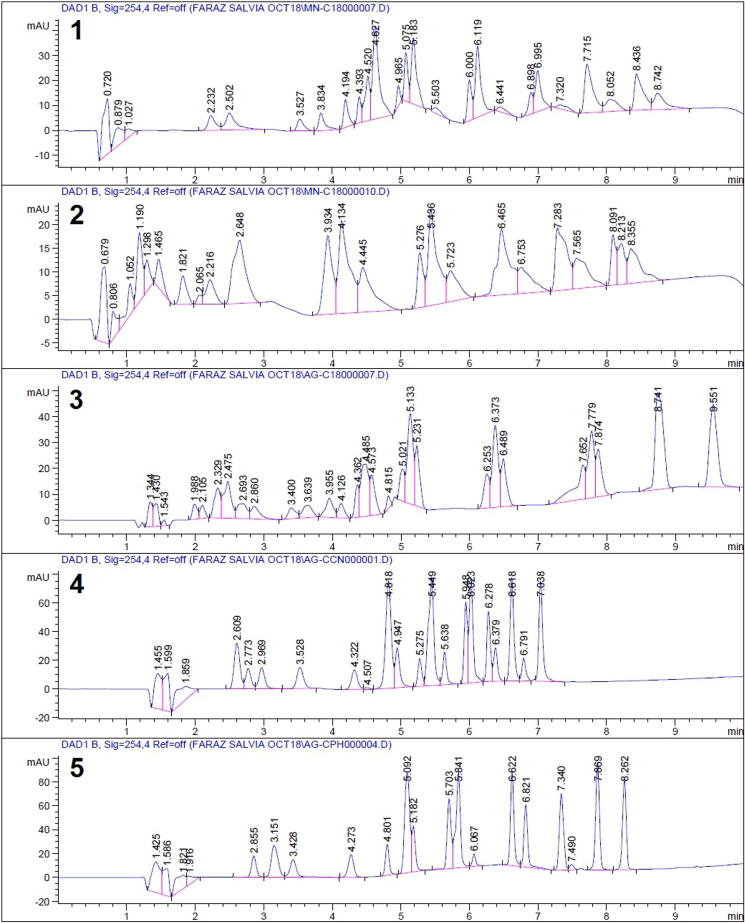
The profiling of Salvia species was performed through an untargeted metabolomics workflow. Chromatographic optimizations included the variation of the mobile phase gradient to obtain an optimum separation of visible peaks. It was found that an optimum separation was achieved on a linear gradient starting at 10% B and reaching 90% in 5.5 min. A pooled sample was prepared by mixing crude extracts of each plant. The aim of preparing a pooled sample was to optimize the chromatography on a sample as complex as possible so that the optimized method could be used effectively for samples with varied chemistry and natural product composition. Fig. 1 shows a TIC chromatogram on the pooled sample of Salvia species. The numbers on the peaks correspond to compounds numbers as mentioned in Table 3. Good separation was achieved in a total run time of 10 min with most of the peaks separated by baseline. The developed method was able to effectively analyze plant samples belonging to different species and no carryover peaks were detected in consecutive runs. In addition to the optimization of separation efficiency, we also optimized the method for MS signal intensities so that most sensitive quantitation results could be obtained. To obtain the maximum number of data points for maximum sensitivity, the scan frequency of the instrument was kept at maximum (12 Hz). To decrease the level of noise in the data, the active exclusion was used to avoid contaminant peaks (from the solvent) being put under MS/MS fragmentation. Precursor reconsideration time was set at 0.5 min after careful examination of the peak widths. This reconsideration time ensured that no precursor ions were excluded from the MS/MS analysis. To ensure that the recorded data was of high accuracy, every LC-MS/MS run was accompanied by a calibration segment at the start of the analysis. The calibration segment lasted for 0.3 min, during which the instrument was injected with sodium formate (10 mM in 1:1 water:2-propanol) at a flow rate of 3 μL/min. Calibration was performed through a comparison of obtained m/z values of sodium formate clusters with the known m/z values.
Fig. 1.
TIC chromatogram of the pooled sample from Salvia species.
Table 3.
Table of compounds detected in Salvia species (positive and negative ionization modes).
| S. No. | Compound Name | Formula | RT (min) | Ion Type | m/z Measured | m/z Calculated | Error (ppm) | mSigma | MS/MS | MSI level |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 6-Hydroxyluteolin-7-O-glucoside | C21H20O12 | 3.25 | [M−H]− | 463.0878 | 463.0882 | −0.86 | 32.9 | 301.0349 | 2 |
| 2 | Nubenoic acid | C15H20O5 | 3.25 | [M+H]+ | 281.1387 | 281.1384 | 1.07 | 49.7 | 245.1189, 203.1061, 201.0917, 187.0762 | 2 |
| [M−H]− | 279.1238 | 279.1238 | 0.00 | 29.8 | 261.1142, 235.1338, 217.1277, 202.1000 | |||||
| 3 | trans-p-Coumaric acid | C9H8O3 | 3.34 | [M−H]− | 163.0403 | 163.0401 | 1.23 | NC | NP | 2 |
| 4 | Luteolin-7-O-glucoside | C21H20O11 | 3.37 | [M−H]− | 447.0936 | 447.0933 | 0.67 | 31.4 | 285.0403 | 2 |
| 5 | 6-Methoxyluteolin-7-O-glucoside | C22H22O12 | 3.41 | [M+H]+ | 479.1184 | 479.1184 | 0.00 | 16.2 | 317.0655, 302.0419 | 2 |
| [M−H]− | 477.1033 | 477.1038 | −1.05 | 18.3 | 462.0798, 315.0508, 299.0193 | |||||
| 6 | 6-Hydroxyluteolin-7-O-glucoside | C21H20O12 | 3.42 | [M−H]− | 463.0878 | 463.0882 | −0.86 | 44.7 | 301.0358, 300.0274 | 2 |
| 7 | Plebeiolide G | C15H20O4 | 3.45 | [M+H]+ | 265.1431 | 265.1434 | −1.13 | NC | 247.1316, 229.1210, 219.1746, 183.1161 | 2 |
| 8 | Luteolin-7-O-glucuronide | C21H18O12 | 3.45 | [M−H]− | 461.0721 | 461.0725 | −0.87 | 29.7 | NF | 2 |
| 9 | Loliolide | C11H16O3 | 3.46 | [M+H]+ | 197.1171 | 197.1172 | −0.51 | 25.2 | 179.1070, 161.0969, 151.1130 | 2 |
| 10 | Rosmarinic acid | C18H16O8 | 3.49 | [M−H]− | 359.0771 | 359.0772 | −0.28 | 15.4 | 197.0454, 179.0351, 161.0244, 151.0403 | 2 |
| 11 | 8-Methoxygenistein-7-O-α-L-rhamnoside-4′-O-β-D-glucoside | C28H32O15 | 3.48 | [M+H]+ | 609.1820 | 609.1814 | 0.98 | 29.1 | 301.0707, 463.1235 | 2 |
| [M−H]− | 607.1671 | 607.1668 | 0.49 | 39 | 299.0560, 284.0309 | |||||
| 12 | Apigenin-7-O-glucoside† | C21H20O10 | 3.52 | [M−H]− | 431.0981 | 431.0984 | −0.70 | 45.7 | 269.0444 | 1 |
| 13 | Plebeiolide A | C17H24O6 | 3.56 | [M+Na]+ | 347.1475 | 347.1465 | 2.88 | 287.1292 | 2 | |
| [M+H]+ | 325.1645 | 325.1646 | −0.31 | 46.7 | 323.1846, 247.1338, 229.1227 | |||||
| [M−H]− | 323.1491 | 323.1500 | −2.79 | 25 | 281.1407, 279.1233, 237.1492 | |||||
| 14 | 1α-Hydroxy-2-oxoeudesman-3,7(11)-dien-8β,12-olide | C15H18O4 | 3.57 | [M+H]+ | 263.1278 | 263.1278 | 0.00 | 46.4 | 245.1172, 243.1013, 217.1222, 227.1054, 203.1075 | 2 |
| [M−H]− | 261.1137 | 261.1132 | 1.91 | 22.7 | 246.0899, 217.1234, 202.0996 | |||||
| 15 | Salviacoccin | C20H20O6 | 3.61 | [M+H]+ | 357.1335 | 357.1333 | 0.56 | 17.6 | 339.1235, 293.1173, 245.0804, 181.1012 | 2 |
| [M−H]− | 355.1196 | 355.1187 | 2.53 | 16.5 | 311.1276, 288.1754, 287.0645 | |||||
| 16 | Salvidivin C | C23H28O10 | 3.68 | [M−H]− | 463.1612 | 463.1610 | 0.43 | 48.3 | NF | 2 |
| 17 | Nubdienolide | C15H18O5 | 3.73 | [M−H]− | 277.1078 | 277.1081 | −1.08 | 14.4 | NF | 2 |
| 18 | Nubenolide | C15H16O4 | 4.09 | [M+H]+ | 261.1120 | 261.1121 | −0.38 | 47.7 | 243.1011, 233.1172, 217.1223, 215.1064, 189.0909 | 2 |
| [M−H]− | 259.0982 | 259.0976 | 2.32 | 15.1 | NF | |||||
| 19 | Nubatin | C17H16O7 | 3.80 | [M−H]− | 331.0824 | 331.0823 | 0.30 | 31.4 | 316.0579, 301.0358, 195.0298, 135.0461 | 2 |
| 20 | Salvitin | C16H12O6 | 3.83 | [M−H]− | 299.0561 | 299.0561 | 0.00 | 23.6 | 284.0327, 256.0352 | 2 |
| 21 | Luteolin† | C15H10O6 | 3.87 | [M+H]+ | 287.0551 | 287.0550 | 0.35 | 44.6 | NF | 1 |
| [M−H]− | 285.0405 | 285.0405 | 0.00 | 15.5 | 283.0238, 255.0291, 151.0024 | |||||
| 22 | Castanin E | C15H20O6 | 3.96 | [M−H]− | 295.1194 | 295.1187 | 2.37 | 27.6 | 233.1192 | 2 |
| 23 | Nubenone | C15H16O4 | 4.12 | [M+H]+ | 261.1124 | 261.1121 | 1.15 | 47.3 | 243.1030, 215.1063, 201.0915, 189.0917 | 2 |
| 24 | Apigenin† | C15H10O5 | 4.13 | [M+H]+ | 271.0603 | 271.0601 | 0.74 | 48.3 | NF | 1 |
| [M−H]− | 269.0452 | 269.0455 | −1.12 | 24.0 | 225.0559 | |||||
| 25 | Diosmetin† | C16H12O6 | 4.17 | [M+H]+ | 301.0707 | 301.0707 | 0.00 | 27.7 | 286.0472, 258.0531, 168.0041 | 1 |
| [M−H]− | 299.0564 | 299.0561 | 1.00 | 17.7 | 284.0331 | |||||
| 26 | Nubiol | C15H18O3 | 4.35 | [M+H]+ | 247.1328 | 247.1329 | −0.40 | 10.3 | 229.1221, 201.1275, 183.1171, 214.0987 | 2 |
| 27 | Takakin | C16H12O6 | 4.74 | [M−H]− | 299.0568 | 299.0561 | 2.34 | 36.8 | 284.0325 | 2 |
| 28 | Przewalskinone B | C16H12O5 | 5.24 | [M−H]− | 283.0614 | 283.0612 | 0.71 | 17.5 | 268.0380 | 2 |
| 29 | Salviviridinol | C21H32O4 | 6.29 | [M−H]− | 347.2224 | 347.2228 | −1.15 | NC | 345.2102, 315.1965 | 2 |
| 30 | Salvinolone | C20H26O3 | 6.42 | [M−H]− | 313.1804 | 313.1809 | −1.60 | 30.1 | 311.1679, 298.1576 | 2 |
| 31 | Santolinoic acid | C30H48O5 | 6.47 | [M−H]− | 487.3424 | 487.3429 | −1.03 | 33.0 | 469.3287, 451.2690, 443.3494, | 2 |
| 32 | Isopimara-6,8(14),15-triene | C20H30 | 6.49 | [M+H]+ | 271.2423 | 271.2420 | 1.11 | 49.4 | 215.1794, 201.1644, 229.1958, 159.1181, 189.1637, 177.1641 | 2 |
| 33 | Carnosol† | C20H26O4 | 6.50 | [M+H]+ | 331.1899 | 331.1904 | −1.51 | NC | 287.1994, 177.1642 | 1 |
| [M−H]− | 329.1758 | 329.1758 | 0.00 | 25.3 | NF | |||||
| 34 | 3β-Hydroxydehydroabietic acid | C20H28O3 | 6.54 | [M−H]− | 315.1971 | 315.1966 | 1.59 | 40.6 | 313.1781, 271.2070, 289.3319 | 2 |
| 35 | 2-(2-Acetoxypentadecyl)-6-hydroxy-4-methoxybenzoic acid | C25H40O6 | 6.55 | [M−H]- | 435.2753 | 435.2752 | 0.23 | 30.5 | 375.2545, 349.2738 | 2 |
| 36 | Nemorosin | C20H28O4 | 6.71 | [M−H]− | 331.1921 | 331.1915 | 1.81 | 27.3 | 313.1818, 287.2019, 285.1856 | 2 |
| 37 | Salvimirzacolide | C25H38O5 | 6.72 | [M−H]− | 417.2646 | 417.2646 | 0.00 | 48.5 | 373.2766, 221.1547 | 2 |
| 38 | Divinatorin A | C20H28O4 | 6.92 | [M+H]+ | 333.2051 | 333.2060 | −2.70 | NC | 331.1829, 315.1955, 287.2006, 273.1851 | 2 |
| [M−H]− | 331.1914 | 331.1915 | −0.30 | 50.0 | 329.1758, 313.1826, 287.2025, 285.1874 | |||||
| 39 | Cryptotanshinone† | C19H20O3 | 6.96 | [M+H]+ | 297.1499 | 297.1480 | 6.39 | 50.0 | 238.0761 | 1 |
| 40 | Cryptanol | C20H28O3 | 7.03 | [M−H]− | 315.1965 | 315.1966 | −0.32 | 43.2 | 299.1652, 285.1879, 243.1034 | 2 |
| 41 | 16-Hydroxy-6,7-didehydroferruginol | C20H28O2 | 7.13 | [M−H]− | 299.2017 | 299.2017 | 0.00 | 47.4 | 227.1094 | 2 |
| 42 | 19-Acetoxy-15,16-epoxy-6-hydroxy-ent-cleroda-3,13(16),14-trien-18-al | C22H32O5 | 7.16 | [M−H]− | 375.2184 | 375.2177 | 1.87 | 45.6 | 343.1920, 328.1680, 313.1448 | 2 |
| 43 | Carnosic acid† | C20H28O4 | 6.82 | [M−H]− | 331.1914 | 331.1915 | −0.30 | 49.9 | 313.1822, 329.1778, 287.2018, 285.1868 | 1 |
| 44 | Divinatorin C | C22H30O5 | 7.61 | [M−H]− | 373.2021 | 373.2020 | 0.27 | 48.0 | 373.2021, 331.1915, 313.1809, 287.2018, 285.1878 | 2 |
| 45 | Isopimara-8(14),15-diene | C20H32 | 7.71 | [M+H]+ | 273.2575 | 273.2577 | −0.73 | 48.1 | 217.1950, 203.1794, 191.1796 | 2 |
| 46 | Royleanone | C20H28O3 | 7.93 | [M−H]− | 315.1966 | 315.1966 | 0.00 | 41.4 | 299.1671, 243.1036 | 2 |
| 47 | 7α-Hydroxy-14,15-dinorlabd-8(17)-en-13-one | C18H30O2 | 8.80 | [M+H]+ | 279.2297 | 279.2319 | −7.88 | NC | 261.2232, 149.0969 | 2 |
| [M−H]− | 277.2170 | 277.2173 | −1.08 | 12.3 | 277.2170, 259.2058, 233.1543, 205.1590 |
*NC = Not calculated.
**NP = Not performed.
***NF = No fragmentation seen.
Identified using standard.
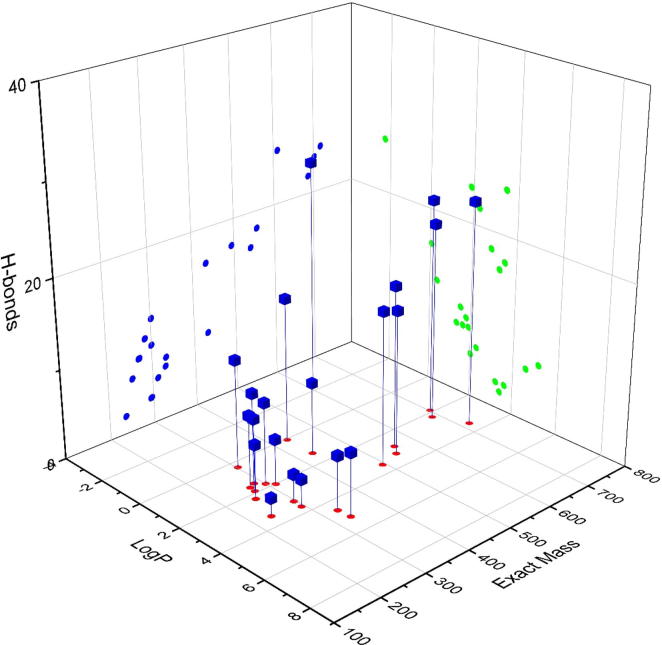
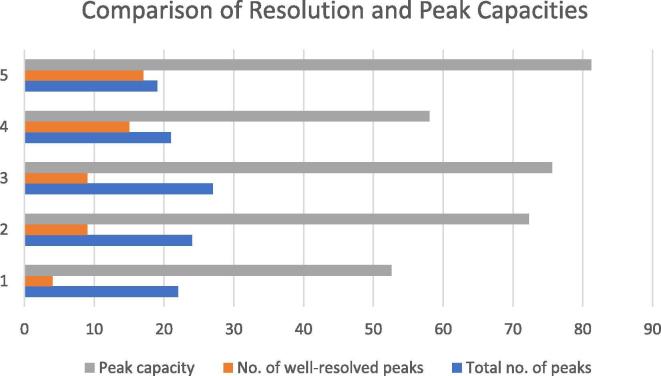
An important parameter related to the sensitivity of a quantitation method is the instrument duty cycle which is greatly reduced if many analytes elute at retention times close to each other. This results in the instrument being busy performing MS/MS fragmentation on too many ions in a tiny time window. To increase the instrument duty cycle, it was necessary to optimize the chromatographic separation in such a way that all analytes elute at retention times as much different from one another as possible. We started the optimization of chromatography by careful examination of the physicochemical characteristics of compounds 1–21. Fig. 2 shows a chemical space of compounds 1–21 using three important parameters: exact mass, LogP and number of hydrogen bonds. The use of these three parameters gave an impression of analyte polarity and affinity for stationary and mobile phases. It was found that the LogP values for most of the analytes ranged from 1.33 to 3.63 and only five compounds had LogP values out of this range. We used four different columns under various mobile phase compositions and the results were compared in terms of peak capacities and the number of well-resolved peaks. Table 2 lists the columns used along with the calculated peak capacities and the number of well-resolved peaks. A visual comparison among column peak capacities, the total number of eluted peaks and the number of well-resolved (baseline-separated) peaks can be seen in Fig. 3. Numbers on the Y-axis correspond to the serial number of experiments as listed in Table 2. Chromatographic parameters such as the mobile phase flow rate, gradient composition and column temperature were all varied and the effect of each was seen on the separation of analytes. It was observed that greater ACN percentages at the start of a chromatographic run resulted in better peak shapes but a narrow range of retention times. On the contrary, smaller percentages of ACN resulted in distorted peak shapes. Good peak shapes and optimum separation was achieved at 25% ACN at the start of the run. The optimum column temperature was found to be 40 °C and the best flow rates were 0.6–0.7 mL/min. It should be noted that Table 2 only lists the best chromatographic conditions used for each column used in method development and optimization. The obtained chromatograms are shown in Fig. 4.
Fig. 2.
Chemical space of compounds 1–21 used for quantitation.
Table 2.
Selection of optimum stationary phase.
| S. No. | Manufacturer | Column | Dimensions | Flow rate (mL/min) | Temperature (°C) | Gradient used | Well-resolved peaks | Peak capacity |
|---|---|---|---|---|---|---|---|---|
| 1 | Macherey-Nagel | Nucleodur C18 Gravity | 3 × 100 mm, 1.8 µm particle size | 0.7 | 40 | 20% C, 0–1 min; 20–95% C, 1–7 min; 95% C, 7–8 min; 95–20% C, 8–9 min; 20% C, 9–10 min. | 4 | 52.61 |
| 2 | 0.7 | 40 | 30% C, 0–1 min; 30–95% C, 1–7 min; 95% C, 7–8 min; 95–30% C, 8–9 min; 30% C, 9–10 min. | 9 | 72.28 | |||
| 3 | Agilent | Zorbax Eclipse XDB-C18 | 4.6 × 100 mm, 1.8 µm particle size | 0.6 | 40 | 30% C, 0–1 min; 30–60% C, 1–3 min; 60% C, 4 min; 60–95% C, 4–6.5 min; 95% C, 6.5–8 min; 95–30% C, 8–9 min; 30% C, 9–10 min. | 9 | 75.61 |
| 4 | Agilent | Zorbax Eclipse XDB-Phenyl | 4.6 × 75 mm, 3.5 µm particle size | 0.6 | 40 | 25% C, 0–1 min; 25–95% C, 1–7 min; 95% C, 7–8 min; 95–25% C, 8–9 min; 25% C, 9–10 min. | 17 | 81.22 |
| 5 | Agilent | Zorbax Eclipse XDB-CN | 4.6 × 75 mm, 3.5 µm particle size | 0.6 | 40 | 25% C, 0–1 min; 25–95% C, 1–7 min; 95% C, 7–8 min; 95–25% C, 8–9 min; 25% C, 9–10 min. | 15 | 58.02 |
Fig. 3.
Comparison of separation efficiencies of different columns used in quantitation method development.
Fig. 4.
HPLC-UV Chromatograms showing separation efficiencies of columns 1–5.
Based on the observations from these experiments, it was found that Agilent Zorbax Eclipse XDB-Phenyl was the best column for the analysis of compounds 1–21 in Salvia species as it gave the best peak capacities with the maximum number of well-resolved peaks with good shapes. It was concluded that the presence of phenyl rings in the stationary phase results in aromatic-aromatic interactions as all the analytes except salvinorin A (15) contained aromatic rings in their structures. This resulted in better retention and selectivity which can be spread throughout the entire length of chromatogram through the selection of a proper mobile phase gradient. It can be seen in Fig. 4(5) that all the analyte peaks are baseline-separated and have good retention time differences. This led to a method with optimum differences in retention times of all analytes.
HPLC-QTOF-MS/MS analysis was performed under both ionization modes (positive and negative) to ensure that all types of compounds can be ionized, detected and subsequently identified. To ensure optimum scan speed, all MS/MS spectra were recorded at a scan speed of 12 Hz so that as much as possible data could be recorded in a single HPLC-MS/MS run. Many natural products tend to be abundant and at several folds higher concentrations than other natural products. It was, therefore, necessary to ensure that MS peaks belonging to natural products present in smaller concentrations are not left without MS/MS fragmentation. To achieve this goal, we used the active exclusion feature of the instrument. It was found that an active exclusion number of 3 was optimum to acquire data containing a maximum number of MS/MS spectra. It is also very common to have multiple natural products of identical molecular weights in the same plant specie. To make sure that active exclusion does not bar the isomers from MS/MS to be performed on, the precursor reconsideration time feature was used and set at 30 s.
Eleven out of twenty-one analytes used in the quantitation study showed better MS and MS/MS signals in the negative mode ionization while ten were better observed under positive ionization mode. We did not use the instrument in the alternating polarity mode because that would have decreased the sensitivity of quantitation because of unnecessary polarity switches. Instead, we used a scheduled precursor list that contained information about ionization polarity, m/z values, and retention times of all analytes. This ensured that the instrument polarity was only switched at the time when a particular analyte was being eluted. The sensitivity of quantitation in an MS/MS method also depends upon the intensities of fragment ions. To improve the fragment ion intensities, fragmentation amplitude for each analyte was carefully tuned. For this, we monitored the intensities of fragment ions of each analyte at different fragmentation amplitudes. It was observed that fragmentation amplitudes between 60 and 130 % were optimum for all analytes. A summary of all optimized parameters for quantitation is shown in Table 1.
Identification and quantification of natural products
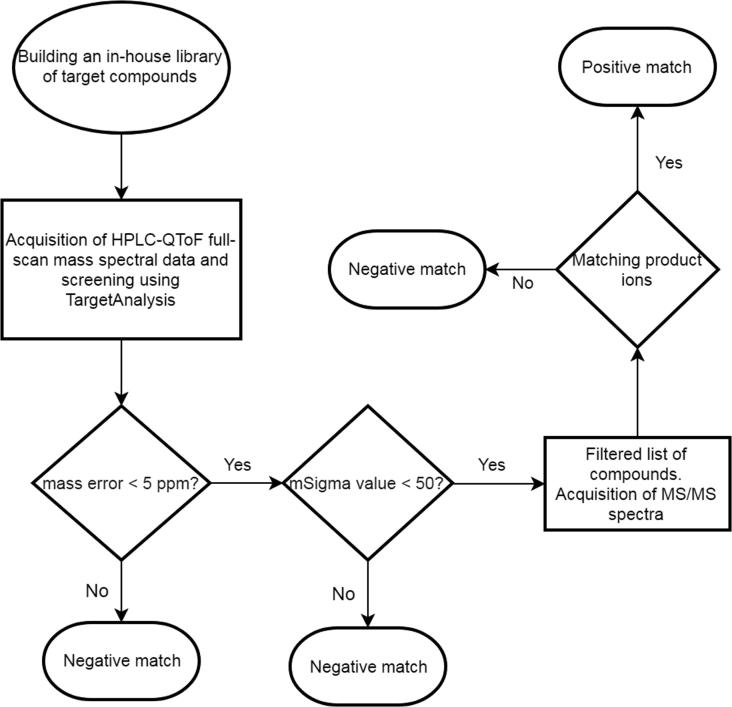
Analysis of data was performed on Bruker Compass DataAnalysis (ver. 4.4 SR1, 64-bit) and Bruker Compass TargetAnalysis (ver. 1.3). All obtained data were first subjected to noise removal using the spectral background subtraction algorithm built-in in DataAnalysis 4.4. Each data file was calibrated using sodium formate clusters m/z values in the high-precision calibration (HPC) mode. Compound identification strategy involved screening of obtained data based on accurate mass, mSigma, fragmentation pattern matching using a data post-processing routine [23]. An in-house library of compounds previously known to be isolated from the genus Salvia was prepared after an extensive literature survey and through the use of the Dictionary of Natural Products on DVD (DNP ver. 26.2). All acquired LC-MS data, after noise removal and calibration, was screened against the built library using TargetAnalysis to get a list of candidate compounds. The candidate compounds were then filtered using their mass error and mSigma values. Every m/z value in the candidate compounds list was first checked for its mass accuracy, the tolerance for which was set at 5 ppm. Every m/z value was also checked for its mSigma value which is the measure of how good an observed isotopic pattern fits onto a simulated isotopic pattern. Smaller values of mSigma indicate a good isotopic pattern match, which in turn ensures good quality of data. The tolerance for mSigma value was set at 50.
The filtered list thus obtained was used to prepare a scheduled precursor list with retention time and m/z values of all candidate compounds. The sample was rerun in the MS/MS mode and fragmentation data was acquired. MS/MS-based identification of compounds was performed using the comparison of obtained fragment m/z values with the theoretical fragmentation patterns of candidate compounds. The theoretical fragmentation patterns were generated using the Fragmentation Explorer functionality built into DataAnalysis. The workflow for compound identification is presented in Fig. 5.
Fig. 5.
A workflow of natural product identification using LC-ESI-MS/MS.
It was observed that most analytes, under the positive ionization mode, were observed as protonated molecules and as deprotonated molecules under negative ionization conditions. The most common fragments observed were neutral losses such as H2O and CO2. Loss of H2O was the predominant mode of fragmentation in positive mode followed by other modes of fragmentation. The loss of CO2 was prevalent in negative ionization mode for molecules containing carboxylic acid groups. The MS/MS fragmentation data was analyzed for both modes to identify compounds. For example, In positive ionization mode, nubenoic acid (Entry 2, Table 3) showed the successive losses to two H2O molecules to form an ion of formula C15H17O3+ which appeared at an m/z of 245.1189. In the negative mode, nubenoic acid showed the loss of a water molecule to yield an anion with the formula C15H17O4− (m/z 261.1142). The fragmentation went on to show the loss of a CO2 molecule and the loss of a CH3 radical to yield ions of formulas C14H17O2− (m/z 217.1277) and C13H14O2•− (m/z 202.1000), respectively. The fragmentation patterns of nubenoic acid in both modes are shown in Figs. S1 and S2. Loliolide (Entry 9, Table 3) appeared in positive ionization mode as a protonated ion of formula C11H17O3+ (m/z 197.1169). It showed two neutral losses: The loss of water molecule to yield the ion of formula C11H15O2+ (m/z 179.1070) and a successive loss of CO molecule to yield the ion of formula C10H15O+ (m/z 151.1130). The fragmentation of loliolide is shown in Fig. S3. Carnosol (Entry 33, Table 3) appeared in the positive ionization mode as a protonated ion of formula C20H27O4+ (m/z 331.1899). It showed the characteristic loss of CO2 molecule to yield ion of formula C19H27O2+ (m/z 287.1994). Carnosic acid (Entry 43, Table 3) appeared as a deprotonated ion (C20H27O4−, m/z 331.1914). It also showed the loss of a CO2 molecule in the negative ionization mode to yield an ion of formula C19H27O2− (m/z 287.2018). The fragmentation of carnosol and carnosic acid is shown in Figs. S4 and S5.
Flavonoids were identified by characteristic losses such as the loss of CO and the decomposition of molecules through retro-Diels-Alder (RDA) reaction. Such reactions were seen in both ionization modes. For example, apigenin (Entry 24, Table 3) was seen as a protonated ion in the positive mode (C15H11O5+, m/z 271.0603) and as a deprotonated ion in the negative mode (C15H09O5−, m/z 269.0452). RDA in the negative ionization mode resulted in the ion of formula C7H3O4− (m/z 151.0039). Fragmentation of apigenin in negative ionization mode is shown in Fig. S6. All compounds were identified using the same strategy. A complete list of compounds identified in Salvia species is shown in Table 3. The MS/MS spectra of all identified compounds alongwith the assigned fragment ion structures are provided with the supplementary data.
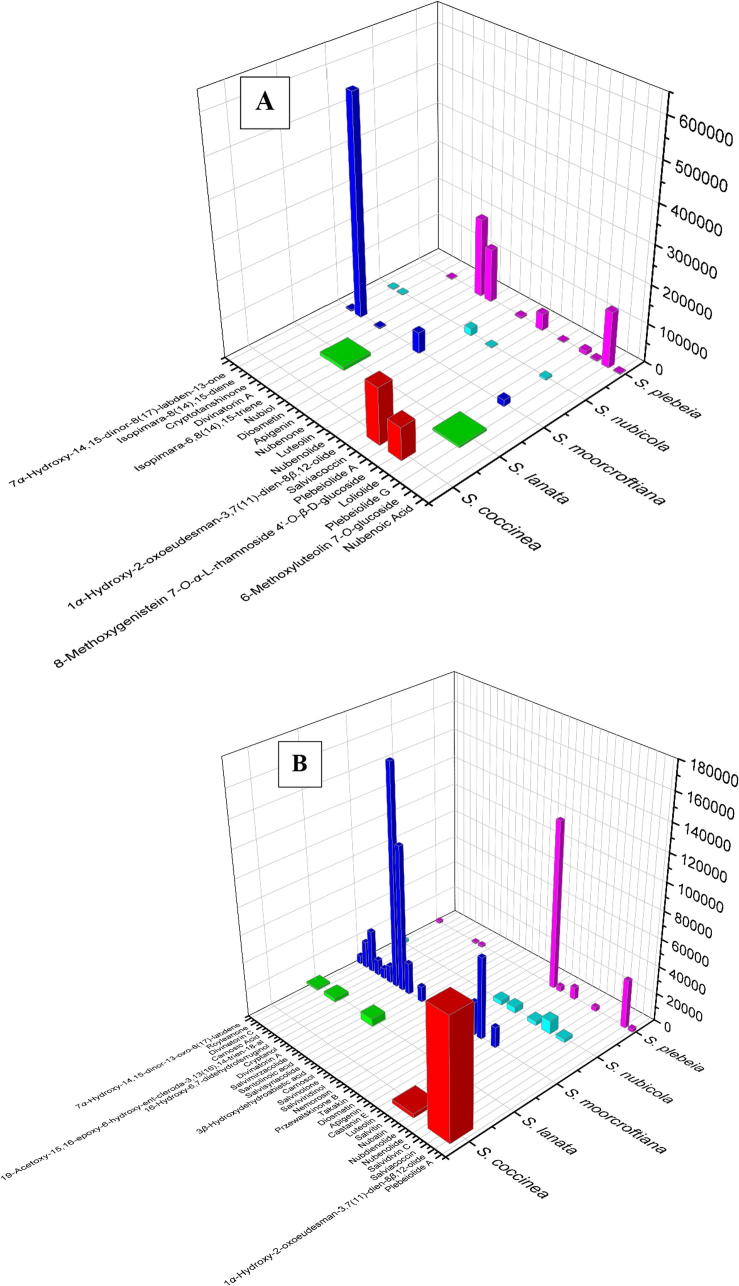
We were able to identify forty-seven compounds based on their exact masses, mSigma values and MS/MS fragmentation pattern. Twenty compounds were identified in the positive ionization mode, forty compounds were identified in the negative ionization mode and thirteen compounds were commonly identified in both modes. Based on ion intensities observed in the positive and negative ionization modes, bar graphs were constructed that show the distribution of identified compounds in five Salvia species (Fig. 6). The distribution of various identified natural products in a plant specie is a unique fingerprint that can serve to identify the plant. The graphs show that the highest concentrations of compounds showing antioxidant activities are present in the three Salvia species: moorcroftiana, nubicola and plebeia. The presence of various bioactive natural products in these Salvia species concurs with their traditional use. The results of this dereplication study can prove useful for bioactivity-guided drug discovery. It can be achieved through bioassays and dereplication of various fractions of plant extracts and then the activity of a fraction can be attributed to the presence of specific natural products in that fraction. For example, Salvia nubicola has been used traditionally in China and India to treat common cough, flu, cold, asthma and inflammatory diseases [24]. These activities concur with the fact that flavonoids and terpenoids isolated from S. plebeia show potent antiviral activities against H1N1 [25].
Fig. 6.
Distribution of identified compounds in positive mode (A) and negative mode (B).
Similar observations were also made when the developed method was applied for quantitation of analytes 1–21 in five Salvia species. It was found that the five species studies in this project contained varying amounts of flavonoids apigenin (8), diosmetin (11), luteolin (6) and quercetin (7) along with phenolic compounds salvianolic acid A (3) and B (2) and abietane diterpenoids carnosol (18) and carnosic acid (17). All these compounds are known to exhibit various biological activities. Carnosol (18) and carnosic acid (17) have been shown to possess anticancer, anti-inflammatory and antioxidant properties [26], [27], [28], [29]. In addition, flavonoids are well-known for their various bioactivities such as antioxidant, antidiabetic, anti-inflammatory, antibacterial, antifungal, antitumor and various other activities [30], [31], [32], [33]. The reported antibacterial and antifungal properties concur with the bioactivities of S. nubicola [34]. S. moorcroftiana is shown to possess anti-inflammatory activities [35]. The results of quantitation are summarized in Table S2.
Method performance and validation
Eight calibrants for each analyte were used between the concentration range of 50 ng/mL to 1500 ng/mL. Linear calibration curves were obtained with excellent correlation coefficients (≥0.9990). LOD and LOQ values were found to be between 0.48 and 0.98 ng/mL and 1.58–3.23 ng/mL, respectively. Table S3 summarizes obtained LOD and LOQ values along with calibration equations. LOD and LOQ values indicate excellent sensitivity and selectivity of the developed method. Method accuracy and precision (intraday and interday precision) were calculated using three QC levels at 175, 625 and 1100 ng/mL, respectively. The accuracy of the method was found to be > 95% in all cases while % RSD was found to be lower than 5% in all cases. The data for accuracy and bias of standard are listed in Table S4.
For validation of quantitation results, all plant samples were fortified with analytes 1–3, 6–8, 11, and 17–19 at three concentration levels: 100, 200 and 400 ng/mL. The method used for the preparation of fortified samples remained the same as unfortified samples. The fortification (spiking) of samples was done before the final dilution stage. The fortified samples were marked as S1, S2 and S3 for fortification levels of 100, 200 and 400 ng/mL, respectively. Analyses of fortified samples showed increased concentrations of analytes 1–3, 6–8, 11, and 17–19 in all samples and excellent recoveries (>95%) were observed. The results of recovery studies are summarized in Table S5.
Conclusions
The present study was focused on the development of a dereplication method for the identification of natural products in five Salvia species. A total of forty-seven compounds belonging to phenolics, flavonoids, diterpenoids and other compound families were identified. A method for quantitation was also developed for the determination of twenty-one important compounds in the five Salvia species. A major focus of the quantitation study was to develop a method that can be used to quantitate natural products in samples of varied chemistries. The chromatographic optimizations resulted in optimum differences in analyte retention times that led to excellent resolution and sensitivity of the developed method.
The developed dereplication and quantitation methods were effective in the analysis of five Salvia species with varied compositions. Due to its effectiveness, the same method or a modified version of it can be used for the dereplication of other medicinally important Salvia species. Such work is of great importance for people working in the field of bioactivity-guided drug discovery from plants. Furthermore, the distribution profiles can be used for plant raw material authentication and quality control of herbal formulations.
Compliance with Ethics Requirements
This article does not contain any studies with human or animal subjects.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgments
The authors express gratitude to Mr. Arsalan Tahir and Mr. Junaid Ul Haq for technical assistance in UHPLC-MS/MS analyses. Dr. Faraz Ul Haq would also like to acknowledge the Higher Education Commission (HEC), Pakistan for financial assistance under the Indigenous Ph.D. Fellowship Program.
Funding
This work was supported by the Organization for the Prohibition of Chemical Weapons (OPCW), The Hague, Netherlands (L/ICA/ICB/210500/17).
Footnotes
Peer review under responsibility of Cairo University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jare.2020.02.001.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- 1.Walker J.B., Sytsma K.J., Treutlein J., Wink M. Salvia (Lamiaceae) is not monophyletic: implications for the systematics, radiation, and ecological specializations of Salvia and tribe Mentheae. Am J Bot. 2004;91:1115–1125. doi: 10.3732/ajb.91.7.1115. [DOI] [PubMed] [Google Scholar]
- 2.Zhou Y., Xu G., Choi F.F.K., Ding L.S., Han Q.B., Song J.Z. Qualitative and quantitative analysis of diterpenoids in Salvia species by liquid chromatography coupled with electrospray ionization quadrupole time-of-flight tandem mass spectrometry. J Chrom A. 2009;1216:4847–4858. doi: 10.1016/j.chroma.2009.04.017. [DOI] [PubMed] [Google Scholar]
- 3.Walch S.G., Tinzoh L.N., Zimmermann B.F., Stuhlinger W., Lachenmeier D.W. Antioxidant capacity and polyphenolic composition as quality indicators for aqueous infusions of Salvia officinalis L. (sage tea) Front Pharmacol. 2011;2:79–94. doi: 10.3389/fphar.2011.00079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Loizzo M.R., Tundis R., Menichini F., Saab A.M., Statti G.A. Menichini, F Cytotoxic activity of essential oils from Labiatae and Lauraceae families against in vitro human tumor models. Anticancer Res. 2007;27:3293–3299. [PubMed] [Google Scholar]
- 5.Smidling D., Mitic-Culafic D., Vukovic-Gacic B., Simic D., Knezevic-Vukcevic J. Evaluation of antiviral activity of fractionated extracts of sage Salvia officinalis L. (Lamiaceae) Arch Biol Sci. 2008;60:421–429. [Google Scholar]
- 6.Zhou L., Zuo Z., Chow M.S. Danshen: an overview of its chemistry, pharmacology, pharmacokinetics, and clinical use. J Clin Pharmacol. 2005;45:1345–1359. doi: 10.1177/0091270005282630. [DOI] [PubMed] [Google Scholar]
- 7.Cheng T.O. Cardiovascular effects of Danshen. Int J Cardiol. 2007;121:9–22. doi: 10.1016/j.ijcard.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 8.Shinwari M.I., Khan M.A. Folk use of medicinal herbs of Margalla Hills National Park, Islamabad. J Ethnopharmacol. 2000;69:45–56. doi: 10.1016/s0378-8741(99)00135-x. [DOI] [PubMed] [Google Scholar]
- 9.Raziq A., de Verdier K. Younas, M Ethnoveterinary treatments by dromedary camel herders in the Suleiman Mountainous Region in Pakistan: an observation and questionnaire study. J Ethnobiol Ethnomed. 2010;6:16. doi: 10.1186/1746-4269-6-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu Y.B., Ni Z.Y., Shi Q.W., Dong M., Kiyota H., Gu Y.C. Constituents from Salvia species and their biological activities. Chem Rev. 2012;112:5967–6026. doi: 10.1021/cr200058f. [DOI] [PubMed] [Google Scholar]
- 11.Tao S., Zheng Y., Lau A., Jaramillo M.C., Chau B.T., Lantz R.C. Tanshinone I activates the Nrf2-dependent antioxidant response and protects against As(III)-induced lung inflammation in vitro and in vivo. Antioxid Redox Signal. 2013;19:1647–1661. doi: 10.1089/ars.2012.5117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fan G.W., Gao X.M., Wang H., Zhu Y., Zhang J., Hu L.M. The anti-inflammatory activities of Tanshinone IIA, an active component of TCM, are mediated by estrogen receptor activation and inhibition of iNOS. J Steroid Biochem Mol Biol. 2009;113:275–280. doi: 10.1016/j.jsbmb.2009.01.011. [DOI] [PubMed] [Google Scholar]
- 13.Gao H., Sun W., Zhao J., Wu X., Lu J.J., Chen X. Tanshinones and diethyl blechnics with anti-inflammatory and anti-cancer activities from Salvia miltiorrhiza Bunge (Danshen) Sci Rep. 2016;6:33720. doi: 10.1038/srep33720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maione F., De Feo V., Caiazzo E., De Martino L., Cicala C., Mascolo N. Tanshinone IIA, a major component of Salvia milthorriza Bunge, inhibits platelet activation via Erk-2 signaling pathway. J Ethnopharmacol. 2014;155:1236–1242. doi: 10.1016/j.jep.2014.07.010. [DOI] [PubMed] [Google Scholar]
- 15.Ho J.H. Hong, CY Salvianolic acids: small compounds with multiple mechanisms for cardiovascular protection. J Biomed Sci. 2011;18:30. doi: 10.1186/1423-0127-18-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khan T., Zahid M., Asim M., ul-Hussan S., Iqbal Z., Choudhary M.I. Pharmacological activities of crude acetone extract and purified constituents of Salvia moorcraftiana Wall. Phytomedicine. 2002;9:749–752. doi: 10.1078/094471102321621386. [DOI] [PubMed] [Google Scholar]
- 17.Ali M.S., Ibrahim S.A., Ahmed S. Lobkovsky, E Guaiane sesquiterpene lactones from Salvia nubicola (Lamiaceae) Chem Biodivers. 2007;4:98–104. doi: 10.1002/cbdv.200790011. [DOI] [PubMed] [Google Scholar]
- 18.Weng X.C., Wang W. Antioxidant activity of compounds isolated from Salvia plebeia. Food Chem. 2000;71:489–493. [Google Scholar]
- 19.Gu L., Weng X. Antioxidant activity and components of Salvia plebeia R. Br. — a Chinese herb. Food Chem. 2001;73:299–305. [Google Scholar]
- 20.Newman D.J., Cragg G.M. Natural products as sources of new drugs from 1981 to 2014. J Nat Prod. 2016;79:629–661. doi: 10.1021/acs.jnatprod.5b01055. [DOI] [PubMed] [Google Scholar]
- 21.Schymanski E.L., Jeon J., Gulde R., Fenner K., Ruff M., Singer H.P. Identifying small molecules via high resolution mass spectrometry: communicating confidence. Environ Sci Technol. 2014;48:2097–2098. doi: 10.1021/es5002105. [DOI] [PubMed] [Google Scholar]
- 22.Sumner L.W., Amberg A., Barrett D., Beale M.H., Beger R., Daykin C.A. Proposed minimum reporting standards for chemical analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI) Metabolomics. 2007;3:211–221. doi: 10.1007/s11306-007-0082-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rios J.J., Roca M., Perez-Galvez A. Systematic HPLC/ESI-high resolution-qTOF-MS methodology for metabolomic studies in nonfluorescent chlorophyll catabolites pathway. J Anal Methods Chem. 2015;2015:490627. doi: 10.1155/2015/490627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liang Y.Y., Wan X.H., Niu F.J., Xie S.M., Guo H., Yang Y.Y. Salvia plebeia R. Br.: an overview about its traditional uses, chemical constituents, pharmacology and modern applications. Biomed Pharmacother. 2020;121:109589. doi: 10.1016/j.biopha.2019.109589. [DOI] [PubMed] [Google Scholar]
- 25.Bang S., Quy Ha T.K., Lee C., Li W., Oh W.K. Shim, SH Antiviral activities of compounds from aerial parts of Salvia plebeia R. Br. J Ethnopharmacol. 2016;192:398–405. doi: 10.1016/j.jep.2016.09.030. [DOI] [PubMed] [Google Scholar]
- 26.Lo A.H., Liang Y.C., Lin-Shiau S.Y., Ho C.T., Lin J.K. Carnosol, an antioxidant in rosemary, suppresses inducible nitric oxide synthase through down-regulating nuclear factor-κB in mouse macrophages. Carcinogenesis. 2002;23:983–991. doi: 10.1093/carcin/23.6.983. [DOI] [PubMed] [Google Scholar]
- 27.Guerrero I.C., Andres L.S., Leon L.G., Machin R.P., Padron J.M., Luis J.G. Abietane diterpenoids from Salvia pachyphylla and S. clevelandii with cytotoxic activity against human cancer cell lines. J Nat Prod. 2006;69:1803–1805. doi: 10.1021/np060279i. [DOI] [PubMed] [Google Scholar]
- 28.Johnson J.J. Carnosol: a promising anti-cancer and anti-inflammatory agent. Cancer Lett. 2011;305:1–7. doi: 10.1016/j.canlet.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schwarz K., Ternes W. Antioxidative constituents of Rosmarinus officinalis and Salvia officinalis. Zeitschrift für Lebensmittel-Untersuchung und Forschung. 1992;195:99–103. doi: 10.1007/BF01201766. [DOI] [PubMed] [Google Scholar]
- 30.Sen P., Sahu P.K., Haldar R., Sahu K.K., Prasad P., Roy A. Apigenin naturally occurring flavonoids: occurrence and bioactivity. UK J Pharm Biosci. 2016;4:56. [Google Scholar]
- 31.Xiao J. Dietary flavonoid aglycones and their glycosides: which show better biological significance? Crit Rev Food Sci Nutr. 2017;57:1874–1905. doi: 10.1080/10408398.2015.1032400. [DOI] [PubMed] [Google Scholar]
- 32.Anand-David A.V., Arulmoli R., Parasuraman S. Overviews of biological importance of quercetin: a bioactive flavonoid. Pharmacogn Rev. 2016;10:84–89. doi: 10.4103/0973-7847.194044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rodriguez-Mateos A., Vauzour D., Krueger C.G., Shanmuganayagam D., Reed J., Calani L. Bioavailability, bioactivity and impact on health of dietary flavonoids and related compounds: an update. Arch Toxicol. 2014;88:1803–1853. doi: 10.1007/s00204-014-1330-7. [DOI] [PubMed] [Google Scholar]
- 34.Melkani A.B., Negi A., Sati S.C., Khulbe K., Dev V. Terpenoid composition and antimicrobial activity of the essential oil from Salvia nubicola Wall ex Sweet. J Essent Oil Res. 2010;22:575–577. [Google Scholar]
- 35.Hussain L., Akash H., Sajid M., Iqbal R., Irfan M., Qadir M.I. Analgesic, anti-inflammatory and antipyretic activity of Salvia moorcroftiana. Pak J Pharm Sci. 2017;30(2):481–486. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.