Abstract
Osteoporosis is characterized by the reduction of bone mineral density and deterioration of bone quality which leads to high risk of fractures. Some microRNAs (miRNAs) have been confirmed as potential modulators of osteoblast differentiation to maintain bone mass maintenance. We aimed to clarify whether miR-122 could regulate osteoblast differentiation in ovariectomized rats with osteoporosis. miR-122 was upregulated and Purkinje cell protein 4 (PCP4) was downregulated in ovariectomized rats. PCP4 was identified as a target of miR-122 by dual-luciferase reporter gene assay. We transfected isolated osteoblasts from ovariectomized rats with miR-122 mimic or inhibitor or PCP4 overexpression vectors. Proliferation and differentiation of osteoblasts were repressed by the overexpression of miR-122 but enhanced by overexpression of PCP4. miR-122 could induce the activation of the c-Jun NH2-terminal kinase (JNK) signaling pathway, while PCP4 blocked this pathway. Rescue experiments further demonstrated that the inhibiting effects of miR-122 on osteoblast differentiation could be compensated by activation of the PCP4 or inhibition of JNK signaling pathway. Collectively, our data imply that miR-122 inhibits osteoblast proliferation and differentiation in rats with osteoporosis, highlighting a novel therapeutic target for osteoporotic patients.
Keywords: osteoporosis, microRNA-122, PCP4, JNK signaling pathway, osteoblasts, proliferation
Introduction
Osteoporosis is a bone metabolic disease characterized by a decline in bone mass accompanied by deterioration of bone tissue and microstructure, as well as bone mineral density degradation.1 At present, there is a consensus opinion regarding the causative factors of osteoporosis, in which an imbalance between bone formation and resorption ultimately results in bone degeneration and a greater susceptibility to fractures.2 Epidemiology shows that women have a considerably higher incidence of osteoporosis than men. This phenomenon has led to speculation implicating the sharp decline in estrogen production among menopausal females as a factor in the incidence of osteoporosis, whereas males are naturally spared this hormone-deficiency syndrome.3 Current treatment protocols for osteoporosis include bisphosphonates, teriparatide, and selective estrogen receptor modulators, as well as denosumab.4 Although these osteoporosis medicals are effective in restoring bone strength, they inadvertently decrease bone strain, and generally exert inadequate effects on the prevention of hip and non-vertebral fractures.5 Recently, microRNAs (miRNAs) have been identified as key regulators in bone formation, contributing to normal remodeling, as well as degeneration in conditions such as osteoporosis.6 Therefore, given the incomplete adequacy of current treatments, it seems important to elucidate further the potential of miRNAs to augment current therapeutic strategies for osteoporosis.
Increasing evidence has suggested that miR-122 is a robust biomarker for osteoporosis diagnosis and prognosis. The ectopic expression of miR-122a in serum samples from osteoporotic patients was reported by Seeliger et al.7 More recently, plasma hsa-miR-122-5p has been proposed as a diagnostic biomarker for osteoporosis.8 However, the mechanism by which these miRNAs participate in osteoporosis remains largely unknown. Another previous study revealed that miR-122-5p negatively regulates the expression of T-box brain 1, thus influencing the differentiation of mouse bone mesenchymal stem cells (bMSCs) in neuron-like cells.9 Located in chromosome 11q12, the gene for Purkinje cell protein 4 (PCP4) belongs to a family of proteins associated with calcium transduction signals, which is involved not only in synaptic transmission and calcium homeostasis but also in morphogenesis, cell-cell interactions, and Purkinje cell formation.9,10 Importantly, the Gene Expression Omnibus (GEO): GSE63009 dataset indicated differentially expressed genes (DEGs) in bisphosphonates-treated osteoclastic precursor cells, whereas untreated cells showed abnormal expression of PCP4 during osteoblast differentiation. Bisphosphonates including alendronate or risedronate exhibited an anabolic effect on the activity of osteoblasts in culture.11 Other expression databases including TargetScan, microRNA, and miRWalk have suggested that PCP4 is a possible target of miR-122. Hence, we speculated that miR-122 might participate in the pathology of osteoporosis by targeting PCP4 in osteoblasts.
The involvement of c-Jun N-terminal kinases (JNKs) in cell-cycle regulatory processes such as apoptosis and cellular stress has been previously associated with the regulation of osteogenesis.2 The ovariectomized rat model of osteoporosis has proven useful in the preclinical investigation of osteoporotic therapies.12 Based on the biological characteristics of miR-122, PCP4, and the JNK signaling pathway, the current study aimed to identify whether miR-122 could affect the proliferation and differentiation of osteoblasts in ovariectomized Sprague-Dawley (SD) rats through regulation of PCP4 and the JNK signaling pathway, in an attempt to provide a novel molecular target for osteoporosis treatment.
Results
The Potential Significance of PCP4 and miR-122 in Osteoporosis
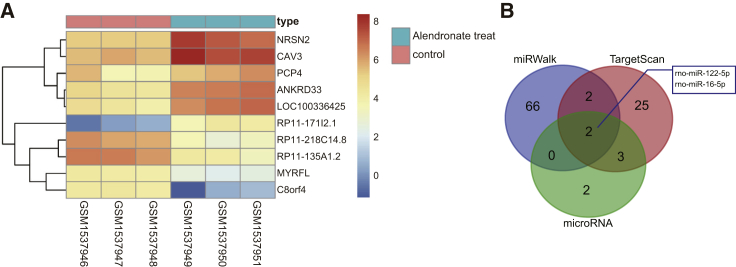
After retrieval in the GEO database, DEGs in osteoclastic precursor cells treated with or without alendronate were screened from the GEO: GSE63009 dataset.13 A total of 211 DEGs were screened out with p <0.05 and |LogFoldChange| > 2.0 as criteria. Hence, a heatmap of the top 10 DEGs was subsequently plotted (Figure 1A), wherein the top three DEGs with largest fold change were Neurensin 2 (NRSN2), Caveolin 3 (CAV3), and PCP4. Through literature review, we were aware of few investigations about the functions of NRSN2 and CAV3 in osteoporosis perspective, while PCP4 has been reported to promote osteoblast differentiation.10 Thus, in the present study, we considered PCP4 to be a key gene in osteoporosis. For the purposes of further exhaustive analysis regarding the molecular mechanism by which PCP4 is implicated in osteoporosis, 32, 7, and 288 miRNAs predicted to be capable of regulating PCP4 of SD rats were identified using TargetScan (Table S1), microRNA (Table S2), and miRWalk (Table S3). We then plotted a Venn diagram of the top 70 miRNAs from each website (Figure 1B) to find the intersection miRNAs. This analysis showed that rno-miR-122-5p and rno-miR-16-5p were predicted in all three databases to bind to PCP4. In addition, a previous report has shown that miR-122-5p was upregulated in the event of fractures secondary to osteoporosis.14 Another earlier study concluded that increased expression of PCP4 could activate the Ca2+/calmodulin-dependent protein kinase (CaMK) signaling pathway,15 a regulator of the JNK signaling pathway,15, 16, 17 and indeed the inhibition of JNK has been applied in osteoporosis therapy.18,19 Hence, we speculate that miR-122 and PCP4 may be involved in osteoporosis and JNK signaling pathway is likely associated with the role of PCP4 in osteoporosis.
Figure 1.
PCP4 Expression Is Increased in Osteoporosis, and May Be a Target Gene of miR-122-5p
(A) The heatmap of the top 10 DEGs in osteoclastic precursor cells treated with or without alendronate in the GEO: GSE63009 microarray dataset searched from GEO database, where the x axis represents the sample number and the y axis is reflective of the DEG. Histogram in the upper right is the color gradation in which every rectangle refers to the expression of one sample, with red indicating high expression and blue indicating low expression. (B) Predicted miRNAs that could target PCP4 obtained from TargetScan, microRNA, and miRWalk databases. miR-122-5p, microRNA-122-5p; PCP4, Purkinje cell protein 4; JNK, c-Jun NH2-terminal kinase; DEGs, differentially expressed genes.
PCP4 Has Low Expression but miR-122 Has High Expression in Our Rat Model of Osteoporosis
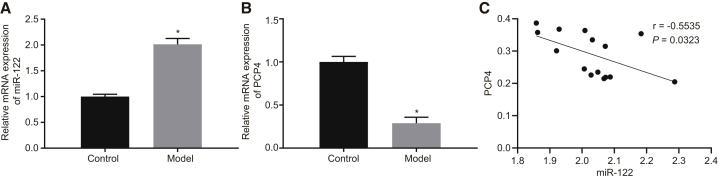
In this study, we induced osteoporosis by ovariectomy in SD rats. We measured the bone mineral density (BMD) in the sham-operated and ovariectomized rats to confirm the successful induction of osteoporosis. We found that the BMD was significantly lower in the ovariectomized SD rats than in the sham-operated SD rats (p < 0.05; Table 1), suggesting that the rat model of osteoporosis was successfully developed in this study. Subsequently, we measured the miR-122 expression and mRNA expression of PCP4 in the bone tissues from the sham-operated and ovariectomized rats by qRT-PCR. This analysis revealed decreased mRNA expression of PCP4 and increased expression of miR-122 in the bone tissues of ovariectomized rats relative to the sham-operated rats (Figures 2A and 2B). The expression of miR-122 correlated negatively with PCP4 expression in the bone tissue of the ovariectomized rats (Figure 2C), which followed the predicted expression pattern from microarray-based gene-expression analysis.
Table 1.
Bone Mineral Density of SD Rats in the Sham Group and the Experimental Group
| Normal SD Rat Tibia | Ovariectomy SD Rat Tibia | p | |
|---|---|---|---|
| Bone mineral density (mg/cm2) | 197.93 ± 6.34 | 185.52 ± 4.34 | < 0.001 |
SD, Spague-Dawlay.
Figure 2.
miR-122 Is Overexpressed while PCP4 Is Reduced in the Bone Tissues of Ovariectomized Rats
(A) The miR-122 expression in the bone tissues of sham-operated and ovariectomized rats determined by qRT-PCR. (B) The mRNA expression of PCP4 in the bone tissues of sham-operated and ovariectomized rats determined by qRT-PCR. (C) The correlation between miR-122 and PCP4 in bone tissue of ovariectomized rats analyzed by Pearson’s correlation analysis. *p < 0.05 versus the control group; statistical data were measurement data, described as mean ± standard deviation; the independent sample t test was used for the comparison between the two groups; and the repeated-measurement analysis of variance was applied for the comparison of data at different time points, followed by Bonferroni post hoc test. miR-122-5p, microRNA-122-5p; PCP4, Purkinje cell protein 4.
PCP4 Is a Target Gene of miR-122
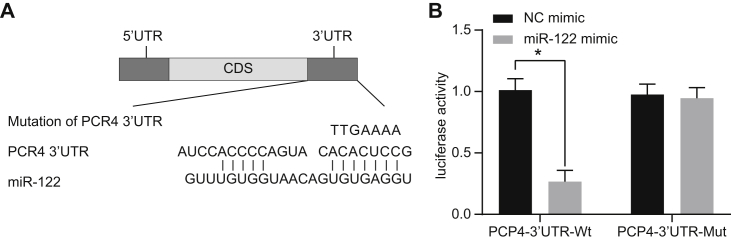
The TargetScan website was employed to aid in the detection of the binding sites of miR-122 on the PCP4 3′ untranslated regions (UTRs) of SD rats (Figure 3A). A dual luciferase reporter gene assay provided validation of predictions, suggesting that PCP4 was indeed a target of miR-122 (Figure 3B). Compared with the negative control (NC) mimic group, luciferase activity of PCP4-3′ UTR-WT (wild-type) in the miR-122 mimic group was significantly lower (p < 0.05), but there was little difference in luciferase activity of PCP4-3′ UTR-Mut (mutant) between the miR-122 mimic and NC mimic groups (p > 0.05). These findings constitute evidence that miR-122 binds to PCP4 3′ UTR and can indeed downregulate its expression.
Figure 3.
PCP4 Is Verified to Be a Target Gene of miR-122
(A) Binding regions between PCP4 3′ UTR and miR-122 sequence. (B) Luciferase activity of the PCP4-WT and PCP4-Mut after transfection of miR-122 mimic or NC mimic. Statistical data were measurement data, which were described as mean ± standard deviation; the independent sample t test was used for the comparison between the two groups; the one-way analysis of variance with Tukey’s post hoc test was used to analyze multiple groups; and *p < 0.05 versus the NC mimic group. miR-122, microRNA-122; PCP4, Purkinje cell protein 4; 3′ UTR, 3′ untranslated region; WT, wild-type; Mut, mutation.
PCP4 Overexpression Vector Was Successfully Constructed
To investigate the role of PCP4 in osteoporosis, we constructed a PCP4 overexpression plasmid. After endonuclease cleavage of the constructed PCP4 overexpression plasmid, we conducted agarose gel electrophoresis. As displayed in Figure S1A, two bands of pcDNA3.1 (+) plasmid and PCP4 overexpression plasmid (198 bp in size) presented after endonuclease cleavage. After the PCP4 band was excised, the DNA was harvested and verified by sequencing to identify whether the PCP4 overexpression vector was successfully constructed. The correctly sequenced results in Figure S1B exhibited that PCP4 overexpression vector was indeed successfully constructed without mutation.
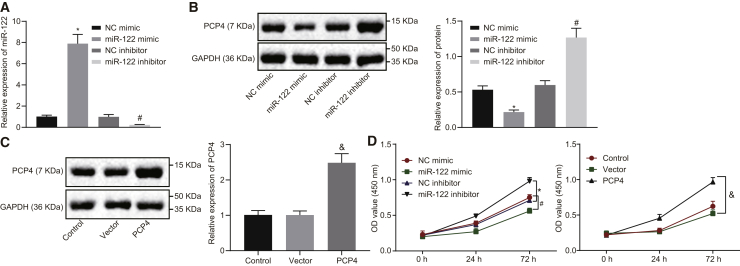
Overexpression of miR-122 Inhibits PCP4 Expression
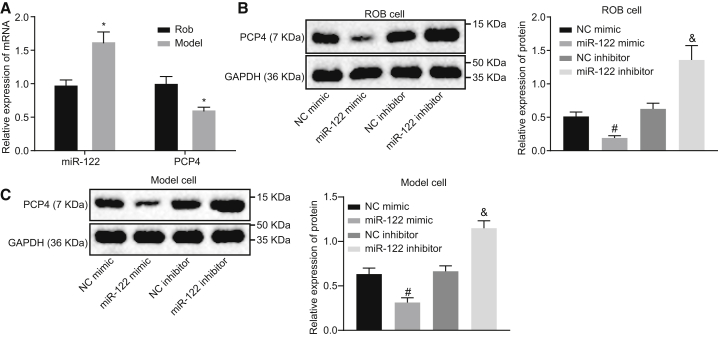
The expression of miR-122, as well as the mRNA expression of PCP4, was determined by qRT-PCR in the commercial rat osteoblast cell line ROB and also in the osteoblasts isolated from the ovariectomized rats. This analysis showed that miR-122 expression was appreciably elevated and PCP4 expression markedly reduced in the osteoblasts isolated from the ovariectomized rats relative to ROB cells (Figure 4A), which further supports a role for miR-122 and PCP4 in osteoporosis. Next, the ROB cells and primary cultured osteoblasts from the ovariectomized rats were transfected with miR-122 mimic or inhibitor to determine the regulatory effect of miR-122 on PCP4. As measured by western blot analysis (Figures 4B and 4C), the PCP4 protein expression was significantly inhibited in ROB cells and primary cultured osteoblasts from the ovariectomized rats following miR-122 mimic transfection when compared with the NC mimic transfection, whereas we saw enhanced PCP4 expression after the miR-122 inhibitor transfection in comparison with the NC inhibitor transfection. Based on the aforementioned results, we conclude that miR-122 can potently suppress PCP4 expression and may exert a regulatory role in the biological functions associated with osteoblasts.
Figure 4.
Elevated miR-122 Inhibits PCP4 Expression
(A) The expression of miR-122 and PCP4 mRNA expression in commercial rat osteoblast cell line ROB and osteoblasts isolated from ovariectomized rats measured by qRT-PCR. (B) The protein expression of PCP4 in ROB cells transfected with miR-122 mimic or inhibitor measured by western blot assay. (C) The protein expression of PCP4 in isolated osteoblasts transfected with miR-122 mimic or inhibitor measured by western blot assay. *p < 0.05 versus the ROB cells; #p the NC mimic group; &p < 0.05 versus the NC inhibitor group; statistical data were measurement data, described as mean ± standard deviation; the independent sample t test was used for the comparison between the two groups; and the one-way analysis of variance with Tukey’s post hoc test was used to analyze data among multiple groups. miR-122, microRNA-122; PCP4, Purkinje cell protein 4; NC, negative control; qRT-PCR, quantitative reverse transcriptase polymerase chain reaction.
Elevated miR-122 Expression Decreases Proliferation of Osteoblasts from Ovariectomized Rats
The primary osteoblasts of the ovariectomized rats were transfected with miR-122 mimic, miR-122 inhibitor, NC mimic, or NC inhibitor to evaluate the effects of miR-122 on PCP4 expression. The results of qRT-PCR (Figure 5A) exhibited that the expression of miR-122 was enhanced after miR-122 mimic transfection, while that of miR-122 was suppressed by transfection with miR-122 inhibitor (p < 0.05). Western blot analysis showed that the protein expression of PCP4 was lowered by miR-122 mimic transfection but restored by miR-122 inhibitor transfection (Figure 5B). Moreover, the primary osteoblasts of the ovariectomized rats were transfected with PCP4 overexpression vector or vector pcDNA3.1 (+). The expression of PCP4 in the PCP4 treatment group was significantly higher than that in the vector group (p < 0.05; Figure 5C). A cell counting kit-8 (CCK-8) assay was applied in order to determine the cell viability after transfection. The results collected indicated reduced cell viability induced by miR-122 mimic transfection. This analysis revealed elevated viability of the cells transfected with miR-122 inhibitor (p < 0.05). Additionally, the viability was increased by PCP4 overexpression (p < 0.05; Figure 5D). These findings suggested that elevated expression of miR-122 can act to impair the survival, while PCP4 overexpression enhanced survival of osteoblasts.
Figure 5.
Enhanced miR-122 Expression Downregulates Proliferation of Osteoblasts Isolated from Ovariectomized Rats
(A) The transfection efficiency of miR-122 mimic or inhibitor or PCP4 overexpression vector in osteoblasts isolated from ovariectomized rats tested by qRT-PCR. (B) The protein expression of PCP4 in osteoblasts isolated from ovariectomized rats in response to upregulation or downregulation of miR-122 measured by western blot analysis. (C) The protein expression of PCP4 in osteoblasts isolated from ovariectomized rats in response to transfection with PCP4 overexpression vector measured by western blot analysis. (D) Viability of osteoblasts isolated from ovariectomized rats in response to upregulation or downregulation of miR-122 or PCP4 overexpression assessed by CCK-8; *p < 0.05 versus the NC mimic group; #p < 0.05 versus the NC inhibitor group; &p < 0.05 versus the vector group; statistical data were measurement data, described as mean ± standard deviation; the independent sample t test was used for the comparison between the two groups; the one-way analysis of variance with Tukey’s post hoc test was used to analyze data among multiple groups; and the repeated-measurement analysis of variance with Bonferroni post hoc test was applied for the comparison of data at different time points. miR-122, microRNA-122; PCP4, Purkinje cell protein 4; NC, negative control.
miR-122 Downregulation or PCP4 Upregulation Promotes Osteoblast Differentiation from Ovariectomized Rats
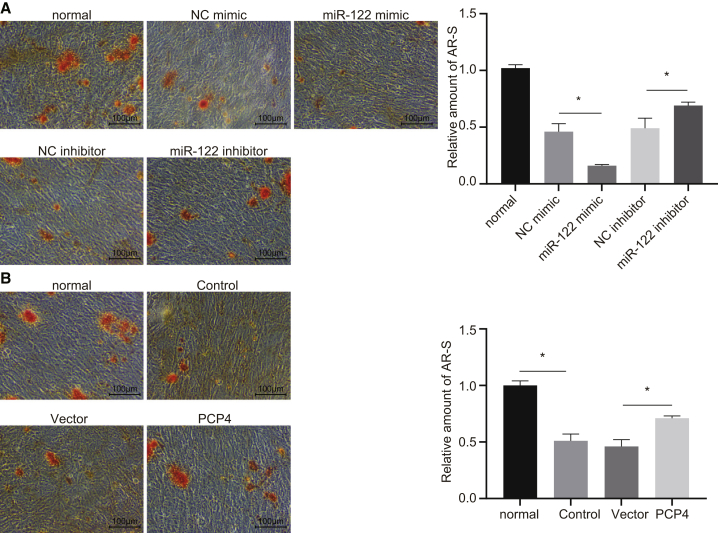
Moreover, we performed Alizarin red S staining to determine the differentiation of osteoblasts derived from the ovariectomized rats. We found decreased staining intensity of the Alizarin red S and lower calcium deposition with forced expression of miR-122, whereas miR-122 inhibition elevated the staining intensity and calcium deposition (Figure 6A). The overexpression of PCP4 was likewise found to augment Alizarin red S staining intensity and calcium deposition (Figure 6B).
Figure 6.
miR-122 Represses but PCP4 Promotes Differentiation of Osteoblasts Isolated from Ovariectomized Rats
(A) Representative microphotographs (×100, scale bar represents 100 μm) and the relative amount of Alizarin red S-stained osteoblasts isolated from ovariectomized rats in response to elevation or inhibition of miR-122 (×100), scale bar represents 100 μm. (B) Representative micrographs (×100, scale bar represents 100 μm) and relative amount of Alizarin red S-stained osteoblasts in response to overexpression of PCP4; *p < 0.05 &p < 0.05 versus the control group. The results were normalized to that in osteoblasts isolated from sham-operated rats. Statistical data were measurement data, described as mean ± standard deviation; the independent sample t test was used for the comparison between the two groups; and the one-way analysis of variance with Tukey’s post hoc test was used to analyze data among multiple groups. miR-122, microRNA-122; PCP4, Purkinje cell protein 4; NC, negative control.
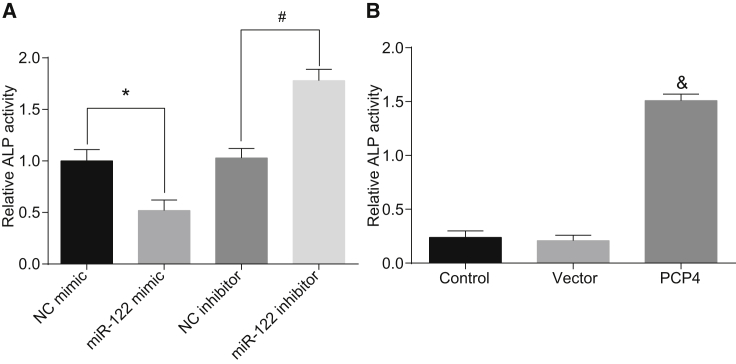
The activity of alkaline phosphatase (ALP) is one of the most important indexes to judge the differentiation and maturation of osteoblasts.20 We used a microplate reader to examine the ALP activity in the present study, finding that ALP activity was significantly decreased in the miR-122 mimic group compared with the NC mimic group. In addition, ALP activity was enhanced in the miR-122 inhibitor group compared with the NC inhibitor group (p < 0.05; Figure 7A). ALP activity was also enhanced when PCP4 was overexpressed (p < 0.05; Figure 7B). Based on the above results, we conclude that inhibited miR-122 or overexpressed PCP4 promotes the differentiation of osteoblasts derived from the ovariectomized rats.
Figure 7.
miR-122 Inhibits but PCP4 Promotes ALP Activity in Osteoblasts Isolated from Ovariectomized Rats
(A) Activity of ALP in osteoblasts isolated from ovariectomized rats after overexpressing or inhibiting miR-122. (B) Activity of ALP in osteoblasts isolated from ovariectomized rats after overexpressing PCP4. *p < 0.05 versus the NC mimic group; #p < 0.05 versus the NC inhibitor group; &p < 0.05 versus the control group; statistical data were measurement data, described as mean ± standard deviation; and the one-way analysis of variance with Tukey’s post hoc test was used to analyze data among multiple groups. miR-122, microRNA-122; PCP4, Purkinje cell protein 4; NC, negative control; ALP, alkaline phosphatase.
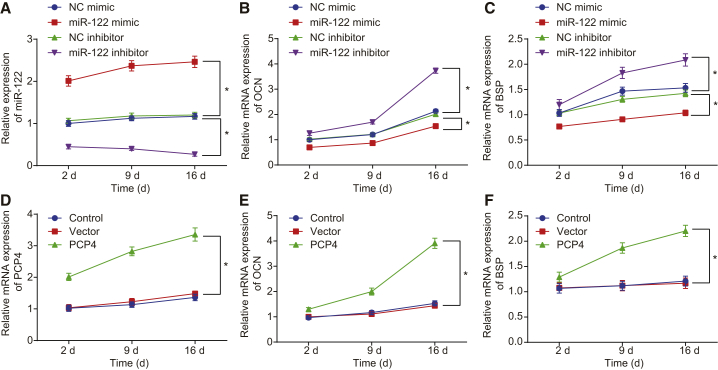
miR-122 Downregulation or PCP4 Upregulation Elevates Expression of Osteocalcin (OCN) and Bone Sialoprotein (BSP)
OCN and BSP are critical molecular markers for revealing osteoblast differentiation, which can be used to judge the maturation of osteoblasts.21 We determined the expression of OCN and BSP using qRT-PCR at the 2nd, 9th, and 16th days after cell transfection to test the effects of miR-122 and PCP4 on osteoblast differentiation. The expression of miR-122 was higher, while that of OCN and BSP was significantly lower at each time point in the miR-122 mimic group than in the NC mimic group (p < 0.05). Meanwhile, the miR-122 inhibitor group demonstrated a lower miR-122 expression and a higher expression of OCN and BSP relative to the NC inhibitor group at each time point (p < 0.05; Figures 8A–8C). The expression of PCP4, OCN, and BSP increased in the osteoblasts overexpressing PCP4 at each time point (p < 0.05; Figures 8D–8F). Hence, inhibited miR-122 or overexpressed PCP4 promotes the differentiation of osteoblasts from the ovariectomized rats by upregulating OCN and BSP.
Figure 8.
miR-122 Inhibits but PCP4 Upregulates Expression of OCN and BSP
The expression of miR-122, PCP4, and osteoblast differentiation markers OCN and BSP in osteoblasts isolated from ovariectomized rats was determined using qRT-PCR at the 2nd, 9th, and 16th days after cell transfection. A, The expression of miR-122 in osteocytes of osteoporotic rats after transfection of miR-122 mimic or miR-122 inhibitor measured by qRT-PCR. B, The expression of OCN in osteocytes of osteoporotic rats after transfection of miR-122 mimic or miR-122 inhibitor measured by qRT-PCR. C, The expression of BSP in osteocytes of osteoporotic rats after transfection of miR-122 mimic or miR-122 inhibitor measured by qRT-PCR. D, The expression of PCP4 in osteocytes of osteoporotic rats after overexpression of PCP4 measured by qRT-PCR. E, The expression of OCN in osteocytes of osteoporotic rats after overexpression of PCP4. F, The expression of BSP in osteocytes of osteoporotic rats after overexpression of PCP4. Statistical data were measurement data, described as mean ± standard deviation; the one-way analysis of variance with Tukey’s post hoc test was used to analyze data among multiple groups; and the repeated-measurement analysis of variance with Bonferroni post hoc test was applied for the comparison of data at different time points. *p < 0.05 versus the NC mimic, NC inhibitor and vector groups. miR-122, microRNA-122; PCP4, Purkinje cell protein 4; OCN, osteocalcin; BSP, bone sialoprotein; NC, negative control.
miR-122 Elevation Downregulates but PCP4 Overexpression Upregulates ALP and Col-I
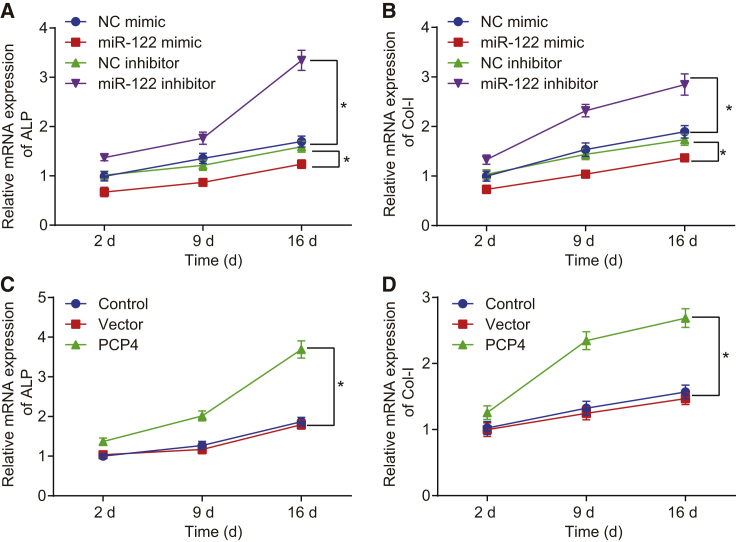
The osteoblast phenotypic markers ALP and Col-I were assessed by means of qRT-PCR at the 2nd, 9th, and 16th days after cell transfection, which revealed that mRNA expression of ALP and Col-I decreased in the miR-122 mimic group when compared with the NC mimic group (p < 0.05). In comparison with the NC inhibitor group, the miR-122 inhibitor group showed raised mRNA expression of ALP and Col-I (p < 0.05; Figure 9A). PCP4 overexpression contributed to the elevated mRNA expression of ALP and Col-I (p < 0.05; Figure 9B). These findings indicate that upregulated miR-122 decreased ALP and Col-I, thus inhibiting osteoblast differentiation, while PCP4 exhibited a promoting effect on osteoblast differentiation.
Figure 9.
miR-122 Represses and PCP4 Elevates the Expression of ALP and Col-I
The osteoblast markers ALP and Col-I in osteoblasts isolated from ovariectomized rats were determined by means of qRT-PCR at the 2nd, 9th, and 16th days after cell transfection. A, The expression of ALP in osteocytes of osteoporotic rats after transfection of miR-122 mimic or miR-122 inhibitor measured by qRT-PCR. B, The expression of Co1-I in osteocytes of osteoporotic rats after transfection of miR-122 mimic or miR-122 inhibitor measured by qRT-PCR. C, The expression of ALP in osteocytes of osteoporotic rats after overexpression of PCP4 measured by qRT-PCR. D, The expression of Co1-I in osteocytes of osteoporotic rats after overexpression of PCP4. *p < 0.05. Statistical data were measurement data, described as mean ± standard deviation; the independent sample t test was used for the comparison between the two groups; the one-way analysis of variance with Tukey’s post hoc test was used to analyze data among multiple groups; and the repeated-measurement analysis of variance with Bonferroni post hoc test was applied for the comparison of data at different time points. miR-122, microRNA-122; PCP4, Purkinje cell protein 4; NC, negative control; ALP, alkaline phosphatase; Col-I, collagen type I.
miR-122 Elevation Enhances but PCP4 Overexpression Blocks the JNK Signaling Pathway
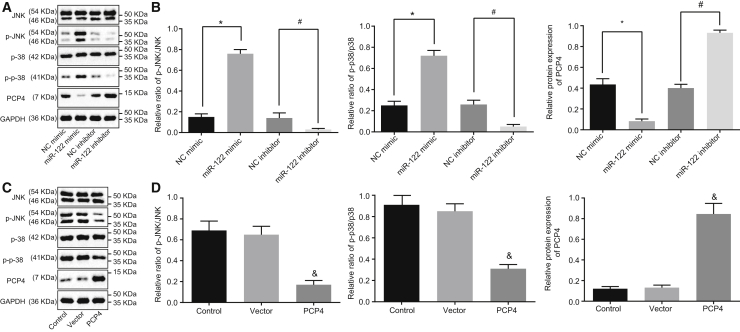
To explore further the molecular mechanisms of PCP4 in osteoporosis, we used western blot analysis to measure changes in the JNK signaling pathway-related proteins upon altering miR-122 and PCP4 expression in osteoblasts from ovariectomized rats. The results (Figures 10A and 10B) demonstrated that forced or inhibited miR-122 expression and PCP4 overexpression had no effects on the protein expression of JNK and p38 (p > 0.05). However, the PCP4 protein expression was diminished while the extents of JNK and p38 phosphorylation were enhanced when miR-122 was upregulated in osteoblasts (p < 0.05). In comparison with the NC inhibitor group, the miR-122 inhibitor group had normalized PCP4 protein expression, but the degree of JNK and p38 phosphorylation was reduced (p < 0.05). The PCP4 group showed upregulated protein expression of PCP4 and decreased JNK and p38 phosphorylation versus the vector control group (p < 0.05). Taken together, we find that inhibited miR-122 or overexpressed PCP4 blocks activation of the JNK signaling pathway.
Figure 10.
miR-122 Increases and PCP4 Decreases the Extents of JNK and p38 Phosphorylation
The protein expression of PCP4, JNK, p38 and the extents of JNK, and p38 phosphorylation in osteoblasts isolated from ovariectomized rats were measured by western blot assay after cell transfection. A, The band diagram of protein expression in osteocytes of osteoporotic rats after overexpressing or inhibiting miR122 measured by Western blot analysis. B, The gray diagram of p-JNK/JNK, p-p38/p38 and PCP4 in osteocytes of osteoporotic rats after overexpressing or inhibiting miR-122 measured by Western blot analysis. C, The band diagram of protein expression in osteocytes of osteoporotic rats after overexpressing PCP4 measured by Western blot analysis. D, The gray diagram of p-JNK/JNK, p-p38/p38 and PCP4 in osteocytes of osteoporotic rats after overexpressing PCP4 measured by Western blot analysis. *p < 0.05 versus the NC mimic group; #p < 0.05 versus the NC inhibitor group; &p < 0.05 versus the control group; statistical data were measurement data, described as mean ± standard deviation; the independent sample t test was used for the comparison between the two groups; and the one-way analysis of variance with Tukey’s post hoc test was used to analyze data among multiple groups. miR-122, microRNA-122; PCP4, Purkinje cell protein 4; NC, negative control; JNK, c-Jun NH2-terminal kinase.
miR-122 Suppresses Osteoblast Differentiation via Activating the JNK Signaling Pathway
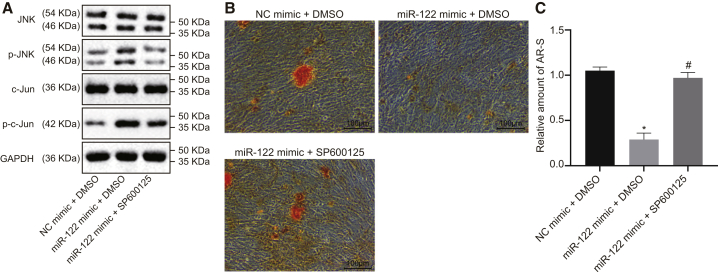
SP600125 was found to be a specific inhibitor of the JNK signaling pathway.22 Osteoblasts from the ovariectomized rats were treated with SP600125 or dimethyl sulfoxide (DMSO) in the presence of miR-122 mimic to explore the implications of this pathway in the osteoblast differentiation. Western blot analysis revealed that the extents of JNK and c-Jun phosphorylation in the miR-122 mimic + DMSO group were appreciably higher than those in the NC mimic + DMSO group, and those in the miR-122 mimic + SP600125 group were noticeably lower than those in the miR-122 mimic + DMSO group (Figure 11A). The results of the Alizarin red S staining showed that the Alizarin red S staining intensity and calcium deposition in the miR-122 mimic + DMSO group were reduced relative to those in the NC mimic + DMSO group. Compared with the miR-122 mimic + DMSO group, the Alizarin red S staining intensity and calcium deposition in the miR-122 mimic + SP600125 group were increased (Figure 11B). Together, these data demonstrate further that the stimulative effect of miR-122 overexpression on osteoblast differentiation depends on the activation of JNK signaling pathway.
Figure 11.
The Inhibitor of the JNK Signaling Pathway SP600125 Reverses the Inhibiting Effect of miR-122 on Osteoblast Differentiation
Osteoblasts isolated from ovariectomized rats were treated with DMSO or SP600125 in the presence of miR-122 mimic. (A) The protein expressions of JNK and c-Jun as well as the extents of JNK and p38 phosphorylation measured by western blot assay. (B) Representative images of Alizarin red S-stained osteoblasts (×100). * p < 0.05 vs. that of cells treated by NC mimic + DMSO, # p < 0.05 vs. that of cells treated by miR-122 mimic + DMSO. Scale bar represents 100 μm. (C) Relative amount of Alizarin red S-stained osteoblasts. Statistical data were measurement data, described as mean ± standard deviation; the independent sample t test was used for the comparison between the two groups; and the one-way analysis of variance with Tukey’s post hoc test was used to analyze data among multiple groups. miR-122, microRNA-122; JNK, c-Jun NH2-terminal kinase; p-JNK, phosphorylated JNK; p-c-Jun; CCK-8, cell counting kit-8.
Effect of miR-122 on Osteoblast Proliferation and Differentiation Can Be Compensated by PCP4
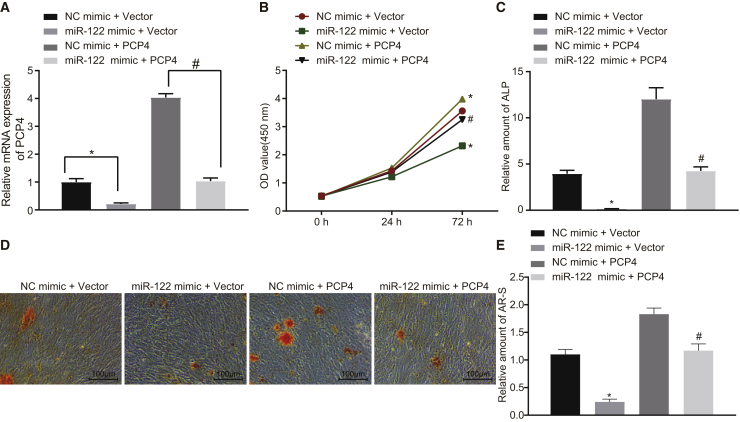
As depicted in Figure 12, when compared with the NC + vector group, osteoblast viability (Figures 12A and 12B) and ALP activity (Figure 12C) were markedly inhibited and the number of Alizarin red S-stained osteoblasts (Figures 12D and 12E) was reduced in the miR-122 mimic + vector group (p < 0.05). However, we saw potentiated osteoblast viability and ALP activity, as well as more numerous Alizarin red S-stained osteoblasts and enhanced calcium deposition in the NC + PCP4 group (p < 0.05), which was consistent with our earlier results. However, osteoblast viability and ALP activity, Alizarin red S-stained osteoblasts and calcium deposition were increased in the miR-122 mimic + PCP4 group compared with the miR-122 mimic + vector group (p < 0.05). Collectively, our data demonstrate that the suppression of osteoblast proliferation and differentiation mediated by miR-122 was rescued by restoration of PCP4.
Figure 12.
Effect of miR-122 on Osteoblast Proliferation and Differentiation Can Be Reversed by PCP4
Osteoblasts isolated from ovariectomized rats were transfected with miR-122 mimic, NC mimic, PCP4 overexpression vector, or vector pcDNA3.1 (+). (A) The expression of PCP4 determined by qRT-PCR. (B) OD value at 0, 24, and 72 h showing osteoblast proliferation assessed by CCK-8. (C) The activity of ALP in osteoblasts. (D) Representative images of Alizarin red S-stained osteoblasts (× 100), scale bar represents 100 μm. (E) Amount of Alizarin red S-stained osteoblasts relative to the NC + vector group; *p < 0.05 versus the NC + vector group; #, p < 0.05 versus the miR-122 mimic + vector group; statistical data were measurement data, described as mean ± standard deviation; the independent sample t test was used for the comparison between the two groups; the one-way analysis of variance with Tukey’s post hoc test was used to analyze data among multiple groups; and the repeated-measurement analysis of variance with Bonferroni post hoc test was applied for the comparison of data at different time points. miR-122, microRNA-122; PCP4, Purkinje cell protein 4; NC, negative control; OD, optical density; ALP, alkaline phosphatase; AR-S, Alizarin red S stained osteoblasts.
Discussion
The role of miRNAs in the incidence and regulation of diverse diseases has recently received extensive attention. From the perspective of osteoporosis, miRNAs play central roles in the regulation of bone remodeling.23 The present study provides evidence indicating that miR-122 targets and negatively regulates PCP4 in a model of osteoporosis. Xiao et al.10 demonstrated in their earlier study that PCP4 mRNA expression was increased upon osteoplastic differentiation of bone marrow mesenchymal stem cells (BMSCs), which further substantiates PCP4’s involvement in osteogenic differentiation of BMSCs. In the present analysis, qRT-PCR and western blot assay methods presented evidence that PCP4 decreases, while the expression of miR-122 increases in osteoblasts derived from ovariectomized rats with osteoporosis. Our findings consequently suggest that the overexpression of miR-122 inhibits PCP4 and activates the JNK signaling pathway. Furthermore, miR-122 exhibits inhibitory effects on osteoblast proliferation and differentiation via reducing PCP4 expression and activating the JNK signaling pathway.
Our study provides evidence that miR-122 expression is increased in ovariectomized SD rats with osteoporosis, and that this overexpression suppressed proliferation and differentiation of the osteoblasts. Yang et al.9 asserted that miR-122-5p may act to regulate the differentiation of BMSCs by directly or indirectly controlling the expression of neuronal markers. In addition, miR-122 has been validated to aid in the regulation of osseointegration by targeting OPN.24 Notably, treatment with BMSCs-derived exosomal miR-122-5p relieved osteonecrosis of the femoral head by facilitating proliferation and differentiation of osteoblasts.25 Furthermore, we confirmed in the present study that PCP4 is a target of miR-122. Although it is previously reported that PCP4 stimulation during osteoblast differentiation enhances deposition of calcium and formation of mineral nodules, the functions of PCP4 in relation to osteogenic differentiation and bone formation currently remain unclear,10 thus highlighting the need for further exploration of this topic. In our study, we found PCP4 to promote osteoblast proliferation and differentiation in SD rats with osteoporosis. Relatively few studies have previously confirmed such an interaction of miR-122 and PCP4, but our results identify their binding relationship and indicate them to be important modulators of osteoblast proliferation and differentiation, with a definite potential in osteoporosis therapy.
Our study also showed that miR-122 activates the JNK signaling pathway by enhancing the extents of JNK and p38 phosphorylation. Besides, the JNK signaling pathway inhibitor was found to restore the osteoblast differentiation that had been lowered by miR-122 mimic. In line with our present findings, a low dose of bisphenol A was capable of promoting the transcript level of miR-122 and the extent of JNK phosphorylation in rat liver, implying a positive correlation between miR-122 and the JNK signaling pathway.26 Consistent with that result, forced expression of miR-122 induced subsequent activation of JNK signaling pathway by enhancing JNK phosphorylation.27 JNK is a critical stress response kinase, which activates in various pathological and physiological cellular processes.28 Increasing evidence has indicated that p38 has powerful effects in the regulating bone development and maintenance. More specifically, early osteoblast differentiation is regulated by p38a, while late osteoblast maturation is controlled by p38b.29 Guo et al.30 provided evidence verifying promotion of apoptosis of osteoblasts and repression of osteoblast differentiation by interleukin-1α (IL-1α) through the activation of the JNK and the p38MAPK pathways. Hisae et al.31 concluded that treatment with p-cresyl sulfate promoted the dysfunction of osteoblasts via the activation of JNK/p38 pathways. Considering that miR-22 downregulation may increase PCP4 expression, there may thus exist a potential therapeutic channel through repressing the JNK signaling pathway. Indeed, we found that PCP4 overexpression blocked the JNK signaling pathway by suppressing JNK and p38 phosphorylation.
OCN and BSP are osteoblast differentiation markers, and ALP and Col-I are osteoblast phenotypic markers. In this study we demonstrated that overexpression of miR-122 resulted in the downregulation of OCN, BSP, ALP, and Col-I, by which miR-122 inhibits osteoblast proliferation and differentiation in osteoporosis. Wang et al.32 have asserted that ALP was the earliest marker of BMSC differentiation into osteoblasts, such that ALP activity could represent a biomarker for the functional state of osteoblasts. Col-I activity is considered to be a maturity indicator for osteoblasts, while the metabolism of Col-I is characteristic of bone with higher sensitivity, specificity, and stability.33 OCN is a marker of osteogenic maturation, being a bone-specific protein synthesized in osteoblasts.34 In addition, BSP is regarded as a matrix-associated signal able to promote osteoblast differentiation, thereby leading to elevated mineralized matrix production.35 Thus, based on our findings, we propose a potential osteoporosis therapy through the maintenance and upregulation of OCN, BSP, ALP, and Col-I.
In conclusion, our key findings provide evidence that miR-122 inhibits osteoblast proliferation and differentiation in ovariectomized rats with osteoporosis through activation of the PCP4-dependent JNK signaling pathway. These findings may provide a basis for novel therapeutic avenues for osteoporosis therapies. However, because this study is based on a SD rat model, we remain uncertain about overdose or side effects occurring in practical clinical application, a matter that must be settled before ever proceeding to translational studies.
Materials and Methods
Ethical Statements
All procedures of this study were performed in strict accordance with the recommendations of the Guide for the Care and Use of Laboratory Animals provided by the National Institutes of Health. The protocols of the study were approved by the Institutional Animal Care and Use Committee of Changzheng Hospital, Second Military Medical University.
Microarray-Based Gene-Expression Analysis
Microarray data related to osteoporosis were investigated with “osteoporosis” used as the key word in the GEO database (https://www.ncbi.nlm.nih.gov/geo/), among which GSE63009 was selected for subsequent analysis. GSE63009 is generally employed during the gene screening process where changes in osteoclastic precursor cells are monitored after treatment with a common therapeutic drug for osteoporosis such as alendronate or risedronate. We included three blank controls and three bisphosphonate-treated samples in this dataset.13 The Limma package in R language was applied to standardize microarray data for screening DEGs under specific conditions (p < 0.05 and |LogFoldChange| > 2.0). After plotting a heatmap of DEGs the miRNAs capable of regulating DEGs were predicted by three different miRNA-mRNA relationship prediction websites as follows: TargetScan (http://www.targetscan.org/vert_71/), microRNA (http://34.236.212.39/microrna/getGeneForm.do), and miRWalk (http://mirwalk.umm.uni-heidelberg.de/). The differences between the respective miRNAs were then subsequently predicted in concordance with various other websites, after which the online analysis tool was applied to calculate and construct custom Venn diagrams of the overlaps (http://bioinformatics.psb.ugent.be/webtools/Venn/).
Model Establishment
45 20-week-old female SD rats (weighing 263–303 g) were used in the study, of which 30 rats were subjected to oophorectomy and the remaining 15 rats were sham-operated. The oophorectomized rats were intraperitoneally injected with 1% pentobarbital at a dose of 0.4–0.5 mL/100 g prior to the operation. Then their fallopian tubes were ligated with a suture. After surgical incision, the fallopian tubes of rats in the sham group were not ligated, but were rather touched with the operating forceps, followed by excision of a small block of adipose tissue. After the operation, the SD rats underwent recovery in a well-lit sterile incubator until they had awoken. If rats showed any signs of postoperative pain within a 6 h period, they were treated with xylazine (0.5 mg/100 g subcutaneously [s.c.]). Rat behavior and life signs were observed and recorded at regular intervals after surgery. If any test subject appeared excessively weak to sustain the osteoporosis model, they were euthanized. The osteoporosis model was successfully established 3 months after the bilateral tubal ligation and ovariectomy. 15 successfully modeled rats were then randomly selected and treated with daily injections of 0.2 mL miR-122 inhibitor (10 μl/mL) via the tail vein for 3 consecutive weeks (from the first to third day of a week).36
BMD measurement was determined using a dual energy X-ray absorptiometry (DEXA; QDR-4500A, Hologic, Waltham, MA, USA) at the tibia site. The scanner was programmed using its animal scanning mode with the appropriate parameters of voltage (140/100 kV), current (2.5 mA), and scan speed (4.8 s/cm). After scanning, the scan analysis interface was opened for BMD measurement.
Isolation and Identification of the Primary Cultured Osteoblasts
Osteoblasts were extracted through the application of trypsin-collagenase digestion and tissue culture methods. After euthanasia, the rats were disinfected with 75% ethanol for 5 min. The tibia was sheared with periosteum and associated blood removed. Phosphate buffered saline (PBS) was used to rinse the tibia until it appeared white and transparent. Finally, the tibia was sheared into pieces (about 1 × 1 mm) using ophthalmic scissors. The bone pieces were treated with 0.25% trypsin until they turned white, when their internal fibroblasts were cleared. The samples were then detached using 1% type-II collagenase at 200 r/min at 37°C for 60 min in an air bath. The collected supernatant was then detached and centrifuged in order to obtain the white precipitate, which was incubated with low glucose Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal bovine serum (FBS). Once confluence reached approximately 80%, cells were passaged.
The collected osteoblasts were purified using the differential adhesion method. After a 10 min culture, the cell suspension was transferred into a second culture flask. After another 10 min culture period, the cell suspension was then placed into the third flask where fibroblasts were adherent to the wells, yielding the high-purity osteoblast suspension. Finally, osteoblasts at passage 4 were selected for the experiments described as follows.
Osteoblasts were observed under a microscope to identify cellular morphology and were stained with the ALP kit for identification. The cells isolated above were lysed with Trizol, and the supernatant was then collected for identification purposes, whereupon the ALP activity was detected based on the instructions of the ALP Assay Kit (Nanjing Jiancheng Bioengineering Institute, Nanjing, Jiangsu, China). The commercial rat osteoblast cell line ROB (adult) was purchased from Cell Applications (R406-05a; Cell Applications, San Diego, CA, USA).
Cell Transfection
The osteoblasts isolated from ovariectomized rats were untransfected or transfected with NC mimic, miR-122 mimic, miR-122 inhibitor, NC inhibitor, vector pcDNA3.1 (+), or PCP4 overexpression vector. Osteoblasts isolated from sham-operated rats were employed as normal control material. Osteoblasts from the ovariectomized rats were treated with SP600125 or DMSO in addition to transfection with miR-122 mimic or NC mimic.
Construction of PCP4 Overexpression Vector
The coding sequences of PCP4 in rats were examined in National Center for Biotechnology Information Nucleotide and amplified by high fidelity enzyme (Takara, Tokyo, Japan) with the primer sequences: 5′-CCCTCGAGGGATGAGTGAGAGACAAAGTGCTGGAG-3′ (xho1) and 5′-GGGGTACCCCCTAGGACTGTGATCCTGCCTTTTTCT-3′ (Kpn1). First, Xho1 (Thermo Fisher Scientific, Sunnyvale, CA, USA) and Kpn1 (Thermo Fisher Scientific, Sunnyvale, CA, USA) were used for the endonuclease cleavage of the pcDNA3.1 (+) plasmids and recovery fragments of quantitative real-time PCR. The fragments were ligated with T4 DNA ligase (Thermo Fisher Scientific, Sunnyvale, CA, USA) overnight at 4°C post-purification. The next day, the fragments were transformed to Escherichia coli DH5α competent cells and cultured in an Amp Resistant LB culture dish at 37°C overnight. The following day, the positive clones from the colony PCR were selected. The qualified positive samples, following detection, were sequenced by Sangon Biotech (Shanghai, China) to ascertain whether they were unmutated. The plasmids were then extracted using an endotoxin-free plasmid extraction kit (Omega, Norcross, GA, USA) and stored for further use.
CCK-8 Assay
The isolated cells were detached using 0.25% trypsin and seeded into 96-well plates at a density of 1 × 104 cells/well, with five replicates prepared for each sample. Transfection was conducted using the Lipofectamine 2000 kit (11668019, Invitrogen, Carlsbad, CA, USA) with NC mimic, miR-122 mimic, NC inhibitor, and miR-122 inhibitor (synthesized by Shanghai GenePharma, Shanghai, China), vector and PCP4 overexpression vector. The untransfected cells were employed as control material. After 6 h of transfection, the medium was renewed. Next, 10 μL CCK-8 reagent (40203ES60, Shanghai Yeasen Biotechnology, Shanghai, China) was added to the plates at 0, 24, and 72 h of culturing. The optical density (OD) values at each time point were subsequently measured at 450 nm using a microplate reader (MK3, Thermo Fisher Scientific, Sunnyvale, CA, USA).
Alizarin Red S Staining
The cells were seeded into a 12-well plate at a density of 5 × 103 per well. When a confluence of 80% was reached, the cells were then untransfected or transfected with NC mimic, miR-122 mimic, NC inhibitor, miR-122 inhibitor, vector, and PCP4 overexpression vector. After removal of the original medium, the cells were rinsed three times with PBS and fixed with 4% paraformaldehyde for 15 min. The cells were then subjected to Alizarin red S staining at 37°C for 30 min. After staining, the Alizarin red S-stained cells were eluted with cetylpyridinium chloride, and the OD value was detected on a spectrophotometer at 570 nm.
ALP Staining
The primary cultured osteoblasts were seeded into a 24-well plate at a density of 1 × 104 cells/well and cultured with 2 mL induction medium per well. The cells were subsequently transfected upon attaining a confluence of 80%. Three replicates were prepared for each group, with addition of culture medium containing 0.5% DMSO serving as the NC. The cells in each well were then lysed with 600 μL PBS containing 0.1% Triton at 4°C for 12 h. Finally, the ALP kit (Nanjing Jiancheng Bioengineering Institute, Nanjing, Jiangsu, China) was used to assess the ALP activity of the cells.
Dual-Luciferase Reporter Gene Assay
The bioinformatics prediction website, microRNA.org, or TargetScan software was used to analyze the target genes of miR-122, among which the PCP4 gene, known to be closely linked with cell differentiation, was set as the object of study. WT and Mut primers were designed for the predicted targeted fragments of PCP4 3′ UTR and synthesized by Sangon Biotech (Shanghai, China). The pMIR-Report luciferase vector was cleaved using the restriction endonucleases Hind III/Pme I into fragments, among which the large ones were harvested by electrophoresis. The predicted targeted fragments of PCP4-3′ UTR-WT and PCP4 3′ UTR-Mut were ligated into the luciferase vector with ligase 4 using the two sites Hind III/Pme I at both ends, forming PCP4 3′ UTR-WT-Luc plasmid and PCP4 3′ UTR-Mut-Luc plasmid. PCP4 3′ UTR-WT-Luc plasmid and PCP4 3′ UTR-Mut-Luc plasmid were co-transfected respectively with NC mimic and miR-122 mimic into the 293T cells. Finally, a Firefly Luciferase Reporter Gene Assay Kit (RG005, Beyotime Institute of Biotechnology, Shanghai, China) and a microplate reader (MK3, Thermo Fisher Scientific, Sunnyvale, CA, USA) were used to assess the luciferase activity at 560 nm.
Extraction, Reverse Transcription, and Quantitation of RNA
Total RNA was extracted by Trizol (Takara, Tokyo, Japan) from the cells at the 2nd, 9th, and 16th days of culture. The concentration, purity and integrity of the total RNA were measured by means of a spectrophotometric assay and agarose gel electrophoresis. A VeriQuest SYBRTM Green One-Step qRT-PCR Master Mix Kit (75705200RXN, Sigma-Aldrich Chemical Company, St. Louis, MO, USA) was applied for further experimentation using 1 μL total RNA from cells in each group.
The quantitative real-time PCR conditions applied were as follows: synthesis at 50°C for 10 min and at 95°C for 10 min in 1 cycle; denaturation at 95°C for 15 s and annealing at 60°C for 30 s in 35–45 cycles. The reaction systems were as follows: 25 μL VerQuest SYBR Green One-Step qRT-PCR Master Mix (2×); 0.5 μL VeriQuest (100×) RT Enzyme Mix for SYBR Green Assay; 2.5 μL forward primer (10 μM); 2.5 μL reverse primer (10 μM); 1 μL Template RNA, and brought to 50 μL with RNase Free, diethylpyrocarbonate-treated water. The primers (Table 2) were all synthesized by the Beijing Genomics Institute company (Beijing, China). GAPDH served as the internal reference for mRNAs and U6 served as the internal reference for miR-122. Finally, quantitative analysis was performed by 2-ΔΔCT assay.
Table 2.
Primers Sequences for Reverse Transcription Quantitative Polymerase Chain Reaction
| Targeted Genes | Forward Primer (5′-3′) | Reverse Primer (5′-3′) |
|---|---|---|
| ALP | 5′-CCTAGACACAAGCACTCCCACTA-3′ | 5′-GTCAGTCAGGTTGTTCCGATTC-3′ |
| OCN | 5′-GACCCTCTCTCTGCTCACTCTG-3′ | 5′-CACCTTACTGCCCTCCTGCTT-3′ |
| Col-I | 5′-TCTGACTGGAAGAGCGGAGAG-3′ | 5′-GAGTGGGGAACACACAGGTCT-3′ |
| miR-122 | 5′-GAGTGTGACAATGGTGTTTGG-3′ | 5′-CCCAGTTATGGCCGTTTA-3′ |
| GAPDH | 5′-CGGCAAGTTCAACGGCACAGTCAAGG-3′ | 5′-ACGACATACTCAGCACCAGCATCACC-3′ |
| U6 | 5′-CTCGCTTCGGCAGCACA-3′ | 5′-GTGCAGGGTCCGAGGT-3′ |
| PCP4 | 5′-GGCAGAAGAAGGTCCAAGAA-3′ | 5′-TCTGAACTGAGACTGAATGGC-3′ |
| BSP | 5′-ACGCTGGAAAGTTGGAGTTAG-3′ | 5′-CCTCCTCTTCCTCTTCCTCTT-3′ |
ALP, alkaline phosphatase; Col-I, collagen type I; OCN, osteocalcin; miR-122, microRNA-122; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; PCP4, Purkinje cell protein 4; BSP, bone sialoprotein.
Western Blot Analysis
Appropriate amount of phenylmethanesulfonyl fluoride was mixed with radio-immunoprecipitation assay lysis buffer (P0013B, Beyotime Institute of Biotechnology, Shanghai, China) to a final concentration of 1 mM. The cells were centrifuged at 450 g for 5 min after 24 h culture, and subsequently resuspended and centrifuged again. The supernatant was resuspended with the lysis buffer mixture at a ratio of 1 mL per 107 cells, followed by ice bathing at 4°C for 5 min. The supernatant (namely the total cell protein) was then obtained after a 10 min centrifugation at 12,000 rpm. A total of 30 μg cellular protein was subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto a polyvinylidene fluoride membrane via a wet transfer method. After this, the membranes were blocked in 5% skim milk powder for 2.5 h, then immunoblotted with the following primary rabbit antibodies to PCP4 (ab197377), JNK (ab179461), and p-JNK (ab124956), c-Jun (ab32137), p-c-Jun (ab32385), glyceraldehyde-3-phosphate dehydrogenase (GAPDH; ab181602), p38 (ab170099), and p-p38 (ab47363). All antibodies were purchased from Abcam (Cambridge, UK). Next, the membrane was probed with the corresponding secondary rabbit antibody labeled by horseradish peroxidase (HRP) for 2 h. The proteins were visualized using enhanced chemiluminescence (ECL) reagent. Here, the equal volumes of the ECL solutions A and B were mixed and added into the membrane. After the membrane was completely infiltrated, the excess liquid was discarded, and the membrane was wrapped with a fresh plastic film. In a darkened room (avoiding direct red lighting), the film was cut and transferred into a dark box, which was then pressed in the membrane for exposure for 1 min. After that, the membrane was developed for approximately 1 min and fixed for 3–5 min. Finally, the software Quanity One was used to analyze the gray value of the protein bands.
Statistical Analysis
SPSS21.0 statistical software (IBM, Armonk, NY, USA) was used to analyze the data and calculate the average value and standard deviation. All experiments were repeated at least 3 times. Data between two groups were analyzed by t test; data comparison among multiple groups by one-way analysis of variance (ANOVA). p < 0.05 was considered to be reflective of statistically significant difference; while p < 0.01 was indicative of extreme statistically significant difference.
Author Contributions
Y.-C.M. and T.L. designed the study. H.J., J.Y., and X.M. collated the data, carried out data analyses, and produced the initial draft of the manuscript. R.G. and X.-H.Z. contributed to drafting the manuscript. All authors have read and approved the final submitted manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
The authors would like to extend their sincere gratitude to the reviewers.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2019.11.038.
Supplemental Information
References
- 1.Zhang X., Zhu Y., Zhang C., Liu J., Sun T., Li D., Na Q., Xian C.J., Wang L., Teng Z. miR-542-3p prevents ovariectomy-induced osteoporosis in rats via targeting SFRP1. J. Cell. Physiol. 2018;233:6798–6806. doi: 10.1002/jcp.26430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Song N., Wang Z.M., He L.J., Xu Y., Ren Y.L. Estradiol-enhanced osteogenesis of rat bone marrow stromal cells is associated with the JNK pathway. Mol. Med. Rep. 2017;16:8589–8594. doi: 10.3892/mmr.2017.7699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Johnston B.D., Ward W.E. The ovariectomized rat as a model for studying alveolar bone loss in postmenopausal women. BioMed Res. Int. 2015;2015:635023. doi: 10.1155/2015/635023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Khan M., Cheung A.M., Khan A.A. Drug-Related Adverse Events of Osteoporosis Therapy. Endocrinol. Metab. Clin. North Am. 2017;46:181–192. doi: 10.1016/j.ecl.2016.09.009. [DOI] [PubMed] [Google Scholar]
- 5.Sugiyama T., Kim Y.T., Oda H. Osteoporosis therapy: a novel insight from natural homeostatic system in the skeleton. Osteoporos. Int. 2015;26:443–447. doi: 10.1007/s00198-014-2923-y. [DOI] [PubMed] [Google Scholar]
- 6.van Wijnen A.J., van de Peppel J., van Leeuwen J.P., Lian J.B., Stein G.S., Westendorf J.J., Oursler M.J., Im H.J., Taipaleenmäki H., Hesse E. MicroRNA functions in osteogenesis and dysfunctions in osteoporosis. Curr. Osteoporos. Rep. 2013;11:72–82. doi: 10.1007/s11914-013-0143-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Seeliger C., Karpinski K., Haug A.T., Vester H., Schmitt A., Bauer J.S., van Griensven M. Five freely circulating miRNAs and bone tissue miRNAs are associated with osteoporotic fractures. J. Bone Miner. Res. 2014;29:1718–1728. doi: 10.1002/jbmr.2175. [DOI] [PubMed] [Google Scholar]
- 8.Mandourah A.Y., Ranganath L., Barraclough R., Vinjamuri S., Hof R.V., Hamill S., Czanner G., Dera A.A., Wang D., Barraclough D.L. Circulating microRNAs as potential diagnostic biomarkers for osteoporosis. Sci. Rep. 2018;8:8421. doi: 10.1038/s41598-018-26525-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang Y., Shen Z., Sun W., Gao S., Li Y., Guo Y. The role of miR-122-5p in negatively regulating T-box brain 1 expression on the differentiation of mouse bone mesenchymal stem cells. Neuroreport. 2017;28:367–374. doi: 10.1097/WNR.0000000000000752. [DOI] [PubMed] [Google Scholar]
- 10.Xiao J., Wu Y., Chen R., Lin Y., Wu L., Tian W., Liu L. Expression of Pcp4 gene during osteogenic differentiation of bone marrow mesenchymal stem cells in vitro. Mol. Cell. Biochem. 2008;309:143–150. doi: 10.1007/s11010-007-9652-x. [DOI] [PubMed] [Google Scholar]
- 11.Im G.I., Qureshi S.A., Kenney J., Rubash H.E., Shanbhag A.S. Osteoblast proliferation and maturation by bisphosphonates. Biomaterials. 2004;25:4105–4115. doi: 10.1016/j.biomaterials.2003.11.024. [DOI] [PubMed] [Google Scholar]
- 12.Mathavan N., Turunen M.J., Tägil M., Isaksson H. Characterising bone material composition and structure in the ovariectomized (OVX) rat model of osteoporosis. Calcif. Tissue Int. 2015;97:134–144. doi: 10.1007/s00223-015-9991-7. [DOI] [PubMed] [Google Scholar]
- 13.Yuen T., Stachnik A., Iqbal J., Sgobba M., Gupta Y., Lu P., Colaianni G., Ji Y., Zhu L.L., Kim S.M. Bisphosphonates inactivate human EGFRs to exert antitumor actions. Proc. Natl. Acad. Sci. USA. 2014;111:17989–17994. doi: 10.1073/pnas.1421410111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Panach L., Mifsut D., Tarín J.J., Cano A., García-Pérez M.A. Serum Circulating MicroRNAs as Biomarkers of Osteoporotic Fracture. Calcif. Tissue Int. 2015;97:495–505. doi: 10.1007/s00223-015-0036-z. [DOI] [PubMed] [Google Scholar]
- 15.Zohn I.E., Yu H., Li X., Cox A.D., Earp H.S. Angiotensin II stimulates calcium-dependent activation of c-Jun N-terminal kinase. Mol. Cell. Biol. 1995;15:6160–6168. doi: 10.1128/mcb.15.11.6160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mishra J.P., Mishra S., Gee K., Kumar A. Differential involvement of calmodulin-dependent protein kinase II-activated AP-1 and c-Jun N-terminal kinase-activated EGR-1 signaling pathways in tumor necrosis factor-alpha and lipopolysaccharide-induced CD44 expression in human monocytic cells. J. Biol. Chem. 2005;280:26825–26837. doi: 10.1074/jbc.M500244200. [DOI] [PubMed] [Google Scholar]
- 17.Chase R., Tolloczko B. Synaptic innervation of the giant cerebral neuron in sated and hungry snails. J. Comp. Neurol. 1992;318:93–102. doi: 10.1002/cne.903180107. [DOI] [PubMed] [Google Scholar]
- 18.Ma J., Ma Y., Liu X., Chen S., Liu C., Qin A., Fan S. Gambogic acid inhibits osteoclast formation and ovariectomy-induced osteoporosis by suppressing the JNK, p38 and Akt signalling pathways. Biochem. J. 2015;469:399–408. doi: 10.1042/BJ20150151. [DOI] [PubMed] [Google Scholar]
- 19.Lu S.Y., Wang C.Y., Jin Y., Meng Q., Liu Q., Liu Z.H., Liu K.X., Sun H.J., Liu M.Z. The osteogenesis-promoting effects of alpha-lipoic acid against glucocorticoid-induced osteoporosis through the NOX4, NF-kappaB, JNK and PI3K/AKT pathways. Sci. Rep. 2017;7:3331. doi: 10.1038/s41598-017-03187-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jo S., Han J., Lee Y.L., Yoon S., Lee J., Wang S.E., Kim T.H. Regulation of osteoblasts by alkaline phosphatase in ankylosing spondylitis. Int. J. Rheum. Dis. 2019;22:252–261. doi: 10.1111/1756-185X.13419. [DOI] [PubMed] [Google Scholar]
- 21.Zhou F., Ma D., Ouyang M., Liu G., Zhu T., Yang Y. Repair mechanism of mesenchymal stem cells derived from nasal mucosa in orbital fracture. Am. J. Transl. Res. 2018;10:1722–1729. [PMC free article] [PubMed] [Google Scholar]
- 22.Yoon S.P., Kim J. Exogenous CGRP upregulates profibrogenic growth factors through PKC/JNK signaling pathway in kidney proximal tubular cells. Cell Biol. Toxicol. 2018;34:251–262. doi: 10.1007/s10565-017-9399-4. [DOI] [PubMed] [Google Scholar]
- 23.Taipaleenmäki H. Regulation of Bone Metabolism by microRNAs. Curr. Osteoporos. Rep. 2018;16:1–12. doi: 10.1007/s11914-018-0417-0. [DOI] [PubMed] [Google Scholar]
- 24.Sartori E.M., Magro-Filho O., Silveira Mendonça D.B., Li X., Fu J., Mendonça G. Modulation of Micro RNA Expression and Osteoblast Differentiation by Nanotopography. Int. J. Oral Maxillofac. Implants. 2018;33:269–280. doi: 10.11607/jomi.5372. [DOI] [PubMed] [Google Scholar]
- 25.Liao W., Ning Y., Xu H.J., Zou W.Z., Hu J., Liu X.Z., Yang Y., Li Z.H. BMSC-derived exosomes carrying microRNA-122-5p promote proliferation of osteoblasts in osteonecrosis of the femoral head. Clin. Sci. (Lond.) 2019;133:1955–1975. doi: 10.1042/CS20181064. [DOI] [PubMed] [Google Scholar]
- 26.Vahdati Hassani F., Mehri S., Abnous K., Birner-Gruenberger R., Hosseinzadeh H. Protective effect of crocin on BPA-induced liver toxicity in rats through inhibition of oxidative stress and downregulation of MAPK and MAPKAP signaling pathway and miRNA-122 expression. Food Chem. Toxicol. 2017;107(Pt A):395–405. doi: 10.1016/j.fct.2017.07.007. [DOI] [PubMed] [Google Scholar]
- 27.Wang H., Wei S. Tanshinol relieves lipopolysaccharide-induced inflammatory injury of HaCaT cells via down-regulation of microRNA-122. Phytother. Res. 2019;33:910–918. doi: 10.1002/ptr.6283. [DOI] [PubMed] [Google Scholar]
- 28.Gao X., Wu X., Yan J., Zhang J., Zhao W., DeMarco D., Zhang Y., Bakhos M., Mignery G., Sun J. Transcriptional regulation of stress kinase JNK2 in pro-arrhythmic CaMKIIδ expression in the aged atrium. Cardiovasc. Res. 2018;114:737–746. doi: 10.1093/cvr/cvy011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thouverey C., Caverzasio J. Focus on the p38 MAPK signaling pathway in bone development and maintenance. Bonekey Rep. 2015;4:711. doi: 10.1038/bonekey.2015.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guo C., Yang X.G., Wang F., Ma X.Y. IL-1α induces apoptosis and inhibits the osteoblast differentiation of MC3T3-E1 cells through the JNK and p38 MAPK pathways. Int. J. Mol. Med. 2016;38:319–327. doi: 10.3892/ijmm.2016.2606. [DOI] [PubMed] [Google Scholar]
- 31.Tanaka H., Iwasaki Y., Yamato H., Mori Y., Komaba H., Watanabe H., Maruyama T., Fukagawa M. p-Cresyl sulfate induces osteoblast dysfunction through activating JNK and p38 MAPK pathways. Bone. 2013;56:347–354. doi: 10.1016/j.bone.2013.07.002. [DOI] [PubMed] [Google Scholar]
- 32.Wang Y., Luo S., Zhang D., Qu X., Tan Y. Sika pilose antler type I collagen promotes BMSC differentiation via the ERK1/2 and p38-MAPK signal pathways. Pharm. Biol. 2017;55:2196–2204. doi: 10.1080/13880209.2017.1397177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dawson-Hughes B., Looker A.C., Tosteson A.N., Johansson H., Kanis J.A., Melton L.J., 3rd The potential impact of the National Osteoporosis Foundation guidance on treatment eligibility in the USA: an update in NHANES 2005-2008. Osteoporos. Int. 2012;23:811–820. doi: 10.1007/s00198-011-1694-y. [DOI] [PubMed] [Google Scholar]
- 34.Nakamura A., Dohi Y., Akahane M., Ohgushi H., Nakajima H., Funaoka H., Takakura Y. Osteocalcin secretion as an early marker of in vitro osteogenic differentiation of rat mesenchymal stem cells. Tissue Eng. Part C Methods. 2009;15:169–180. doi: 10.1089/ten.tec.2007.0334. [DOI] [PubMed] [Google Scholar]
- 35.Gordon J.A., Tye C.E., Sampaio A.V., Underhill T.M., Hunter G.K., Goldberg H.A. Bone sialoprotein expression enhances osteoblast differentiation and matrix mineralization in vitro. Bone. 2007;41:462–473. doi: 10.1016/j.bone.2007.04.191. [DOI] [PubMed] [Google Scholar]
- 36.Yang S., Wang Y., Gao H., Wang B. MicroRNA-30a-3p overexpression improves sepsis-induced cell apoptosis in vitro and in vivo via the PTEN/PI3K/AKT signaling pathway. Exp. Ther. Med. 2018;15:2081–2087. doi: 10.3892/etm.2017.5644. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.