Campylobacter is a leading cause of foodborne diarrheal disease worldwide. Poultry are a key source of human infections, but there are currently few effective measures against Campylobacter in poultry during production. One option to control Campylobacter may be to alter the composition of microbial communities in the avian intestines by introducing beneficial bacteria, which exclude the harmful ones. We previously described two inbred chicken lines which differ in resistance to intestinal colonization by Campylobacter. Here, we investigated the composition of the microbial communities in the gut of these lines and whether transferring gut bacteria between the resistant and susceptible lines alters their resistance to Campylobacter. No major differences in microbial populations were found, and resistance or susceptibility to colonization was not conferred by transferring gut bacteria between lines. The data suggest that gut microbiota did not play a role in resistance to Campylobacter colonization, at least in the lines used.
KEYWORDS: Campylobacter, chickens, microbiota, colonization, resistance, intestinal microbiota
ABSTRACT
Campylobacteriosis is the leading foodborne bacterial diarrheal illness in many countries, with up to 80% of human cases attributed to the avian reservoir. The only control strategies currently available are stringent on-farm biosecurity and carcass treatments. Heritable differences in the resistance of chicken lines to Campylobacter colonization have been reported and resistance-associated quantitative trait loci are emerging, although their impact on colonization appears modest. Recent studies indicated a protective role of the microbiota against colonization by Campylobacter in chickens. Furthermore, in murine models, differences in resistance to bacterial infections can be partially transferred between lines by transplantation of gut microbiota. In this study, we investigated whether heritable differences in colonization of inbred chicken lines by Campylobacter jejuni are associated with differences in cecal microbiota. We performed homologous and heterologous cecal microbiota transplants between line 61 (resistant) and line N (susceptible) by orally administering cecal contents collected from 3-week-old donors to day-of-hatch chicks. Recipient birds were challenged (day 21) with C. jejuni 11168H. In birds given homologous microbiota, the differential resistance of lines to C. jejuni colonization was reproduced. Contrary to our hypothesis, transfer of cecal microbiota from line 61 to line N significantly increased C. jejuni colonization. No significant difference in the overall composition of the cecal microbial communities of the two lines was identified, although line-specific differences for specific operational taxonomic units were identified. Our data suggest that while heritable differences in avian resistance to Campylobacter colonization exist, these are not explained by significant variation in the cecal microbiota.
IMPORTANCE Campylobacter is a leading cause of foodborne diarrheal disease worldwide. Poultry are a key source of human infections, but there are currently few effective measures against Campylobacter in poultry during production. One option to control Campylobacter may be to alter the composition of microbial communities in the avian intestines by introducing beneficial bacteria, which exclude the harmful ones. We previously described two inbred chicken lines which differ in resistance to intestinal colonization by Campylobacter. Here, we investigated the composition of the microbial communities in the gut of these lines and whether transferring gut bacteria between the resistant and susceptible lines alters their resistance to Campylobacter. No major differences in microbial populations were found, and resistance or susceptibility to colonization was not conferred by transferring gut bacteria between lines. The data suggest that gut microbiota did not play a role in resistance to Campylobacter colonization, at least in the lines used.
INTRODUCTION
Campylobacter is the main bacterial cause of zoonotic foodborne infections in many countries. In the United Kingdom, approximately 90% of human cases are caused by Campylobacter jejuni, with C. coli and other species playing a relatively minor role (1). Symptoms can range from mild gastroenteritis to severe hemorrhagic diarrhea that can last as long as 2 weeks and occasionally relapse. In addition, campylobacteriosis can involve severe sequelae, including inflammatory bowel disease and debilitating inflammatory neuropathies, such as the Guillain-Barré syndrome. Recent estimates place its economic cost at £50 million per year in the United Kingdom (2), where 63,946 laboratory-confirmed cases of human infection were recorded in 2017 (1) and 9.3 cases are predicted to be unreported for every one captured by national surveillance (3).
Poultry are an important reservoir of human campylobacteriosis, with some estimates attributing up to 80% of human infections to this source (4). The ceca are a key site of persistence of Campylobacter in chickens, where numbers of C. jejuni can reach as high as 1010 CFU/g of contents. Given such levels, contamination of carcasses with numbers of C. jejuni predicted to be adequate for human infection is challenging to prevent during the slaughter process. Control of Campylobacter relies mainly on stringent on-farm biosecurity measures and carcass treatments, including freezing, rapid surface chilling, or the application of organic acid solutions or chlorinated water, where permitted by national regulations. There are currently no effective commercial vaccines for Campylobacter in poultry, and even though some protective candidates have been described in the literature, these often confer modest protection that has proven challenging to reproduce across repeated studies and laboratories (5).
Inbred chicken lines 61 and N are known to exhibit heritable differences in resistance to colonization by several C. jejuni strains (6), and recent work using backcross and advanced intercross populations of these lines has identified quantitative trait loci (QTL) associated with this phenotype (7). Differential resistance to C. jejuni colonization has also been detected between other chicken lines and was associated with variation in cecal and systemic transcriptional responses (8–10). Heritable differences in resistance to Campylobacter also appear to exist in commercial broilers, although only 10% of the variation in Campylobacter colonization phenotype was explained by host genetics in a population studied recently (11).
In murine models, Campylobacter-induced enteritis and colonization require the prior depletion of the indigenous microbiota with antibiotics (12). This study also indicated that Enterococcus faecalis is a protective constituent of the microbiota. More recently, Clostridium cluster XI, Bifidobacterium, and Lactobacillus species were reported to be significantly enriched in mice that were protected against Campylobacter-induced colitis (13). These authors also demonstrated that oral administration of sodium deoxycholate, a secondary bile acid that is produced via the metabolism of the aforementioned bacteria, reduced enteritis. Moreover, removal of these bacterial taxa through antibiotic treatment enhanced the severity of Campylobacter-induced colitis (13).
In chickens, Han et al. demonstrated that the presence of intestinal microbiota is protective against Campylobacter by comparing colonization levels in C. jejuni-challenged birds that had naturally acquired microbiota or that had been reared under germfree conditions or treated with antibiotics (14). However, the individual components of the microbiota that were associated with the protective effect and relative role(s) of direct competition versus immune priming by the microbiota were not investigated. More recently, Connerton et al. studied the effects of Campylobacter colonization on the cecal microbiota of chickens and found that colonization by Campylobacter significantly alters the composition of the gut microbiota, with decreases in the abundance of operational taxonomic units (OTUs) in Lactobacillaceae and Clostridium cluster XIVa (15). However, they also found that the age of the bird had an effect on the composition of the microbiota and that the effect of age exceeded that of Campylobacter infection as time progressed (15).
In this study, we sought to investigate whether intestinal microbiota plays a role in the differential resistance of chicken inbred lines 61 (resistant) and N (susceptible) to colonization by C. jejuni. This involved analyzing the cecal microbiome of birds of each line at 3 weeks of age, when they are known to differ in resistance to C. jejuni challenge (6, 7), and performing homologous and heterologous microbiota transplants between the two lines. A precedent exists in the literature for transferring resistance against bacterial colonization in this way. For example, when using inbred mouse lines that differ in resistance to colonization by the murine attaching and effacing pathogen Citrobacter rodentium, reciprocal transfer of the microbiota to the heterologous line altered susceptibility to colonization (16). Furthermore, fecal microbiota transplants are now accepted treatments for acute and recurrent Clostridium difficile infections in humans, even in cases where antibiotic treatment failed (17, 18). The rationale for our study is given further impetus by the recent observation that the introduction of adult microbiota into flocks of neonatal chicks has a mild protective effect against Campylobacter colonization and altered the gut microbiome (19).
RESULTS
Reciprocal transfer of cecal microbiota from resistant or susceptible inbred lines does not confer the phenotype against C. jejuni colonization.
We hypothesized that differential resistance of inbred lines 61 and N to C. jejuni colonization is associated with variation in indigenous microbiota at a key site of persistence and that heterologous transplants of cecal microbiota would transfer susceptibility (microbiota from line N into line 61) or resistance (microbiota from line 61 into line N) to Campylobacter challenge. Cecal microbiota was pooled from five donor birds of each line at 21 days of age. Following homologous or heterologous administration of cecal contents from the donor birds to day-of-hatch recipient chicks, 4 recipient birds were sampled from each group at 1, 7, and 21 days posthatch and on day 21; the remaining 10 birds from each group were challenged with 104 CFU of C. jejuni 11168H as described in Materials and Methods. The data presented derive from a single study of this design.
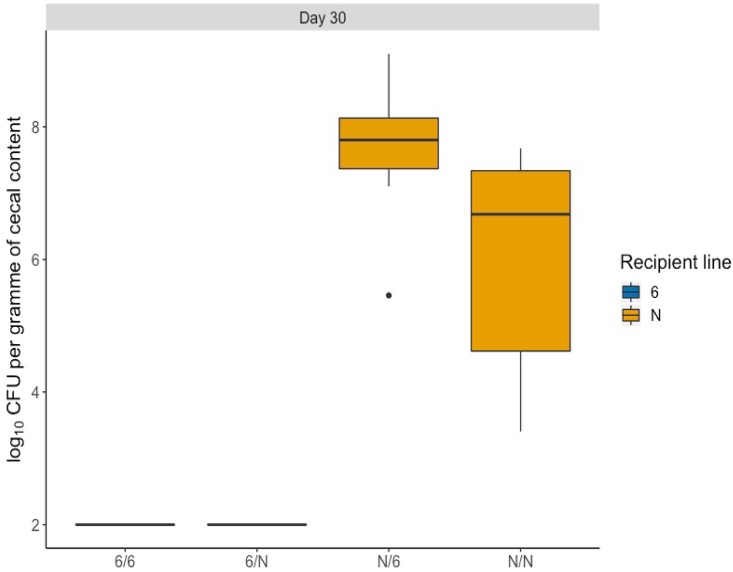
In birds of each line given homologous cecal microbiota, the previously described differences in C. jejuni colonization were reproduced (Fig. 1). A mean of 1.3 × 107 CFU/g cecal contents was detected in susceptible line N birds given line N microbiota, whereas for resistant line 61 birds given line 61 microbiota, no Campylobacter was isolated at the limit of detection by direct plating (2 log10 CFU/g). In the groups that received heterologous microbiota, none of the line 61 birds given microbiota from susceptible line N were colonized at the limit of detection (Fig. 1). In birds of line N given microbiota from the resistant line 61, a mean of 2.1 × 108 CFU/g cecal contents was detected, which represents a statistically significant increase compared to that of the birds of the same line that received homologous microbiota (P = 0.002) (Fig. 1).
FIG 1.
Transfer of cecal microbiota between inbred lines 61 and N is not protective against C. jejuni colonization. Chickens were given homologous or heterologous cecal microbiota from 3-week-old donor birds on the day of hatch and infected with 104 CFU of C. jejuni 11168H at 21 days posttransplant. Ten chickens were sampled in each group at 9 days postinfection, and significant differences were identified using a one-way two-sided ANOVA (Minitab, UK). Birds from line N that received microbiota from line 61 had a significantly higher number of cecal C. jejuni than the line N birds that received line N microbiota (P < 0.05). For groups noted on the x axis, the first letter denotes the recipient line and the second letter denotes the donor line. The boxes show the quartile 1-to-quartile 3 range, and the whiskers indicate the minimum and maximum, unless values were more than 1.5 times the interquartile range. Statistical outliers identified by R are plotted as individual solid circles; they were not excluded from the analysis.
The global composition of the cecal microbiota of donor birds varying in Campylobacter resistance is not significantly different, but line-specific OTUs exist.
From a total of 106 samples (including positive and negative controls) in this experiment, a total of 23,065,560 reads were sequenced on the Illumina MiSeq platform. After quality filtering and the chimeric read removal step, there were 9,911,881 reads from all cecal content samples that passed through the OTU classification step. The average number of reads per cecal sample was 101,620 ± 46,029. All samples were rarefied at 43,808 reads. In the mock bacterial population control sample used as the DNA extraction control, we obtained, on average, 11.15% (±7.97%) abundance of the six bacterial species, and Enterobacteriaceae were present at 31%. In the mock DNA control sample used as the control for the PCR step, we obtained, on average, 13.24% (±5.28%) abundance of the six bacterial species, and Enterobacteriaceae were present at 19%. The relative abundance of the individual bacterial species in the mock controls is given in Table S1 in the supplemental material. While we observed some differences between the observed and the expected mock community compositions, any biases are likely to be consistent across samples. Furthermore, while Listeria was underrepresented in the PCR control sample, it is not anticipated to be a major genus in the intestinal microbiota of chickens.
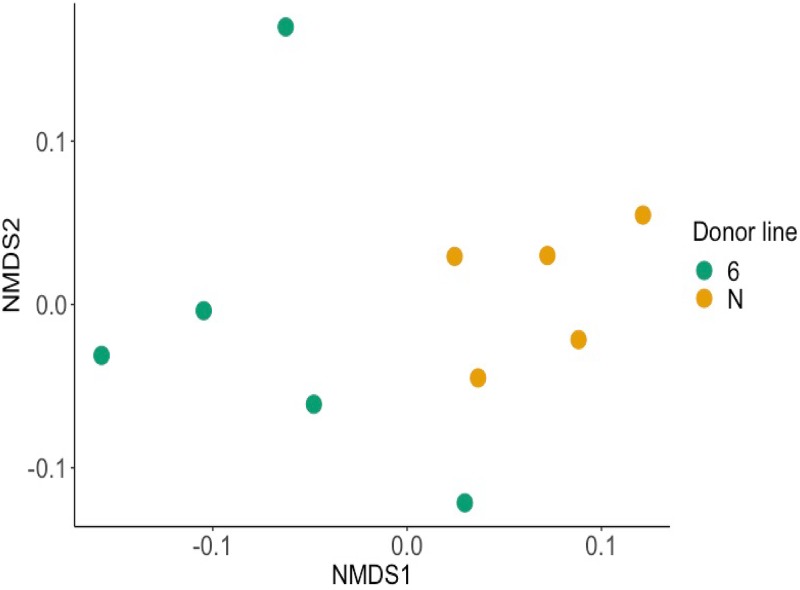
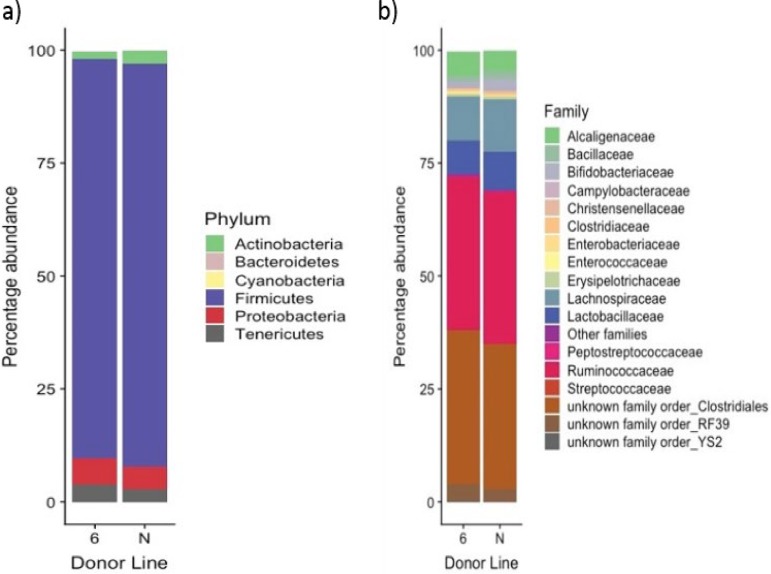
The total number of OTUs generated from the classification step was 1,297 OTUs (from all cecal and positive-control samples). The average number of OTUs from cecal samples was 662 OTUs per sample. This compares favorably to independent analysis of microbial diversity in broiler chicken ceca (20, 21). Using a nonmetric multidimensional scaling (NMDS) plot to compare the cecal microbiota obtained from donor lines 61 and N at 21 days of age, no significant difference was observed between them (P = 0.061 by the adonis test) (Fig. 2). The cecal bacterial communities of the donor birds were dominated by the phylum Firmicutes (Fig. 3). At the family level, an unknown family in the order Clostridiales and the family Ruminococcaceae dominated in both chicken lines (Fig. 3). However, a comparison of bacterial abundance between lines at the level of individual OTUs using the analysis of the composition of microbiomes (ANCOM) method revealed three significantly different OTUs between the lines from the Ruminococcaceae family (P < 0.05). One OTU in the genus Oscillospira was present at a mean of 146 ± 64 reads in cecal microbiota from resistant line 61 donor birds but was completely absent from susceptible donor birds of line N. An OTU of an unclassified genus was found to be significantly more abundant in line N, while another OTU of an unclassified genus was found to be significantly more abundant in line 61.
FIG 2.
Cecal microbial communities of donor birds of inbred lines that exhibit heritable differences in resistance to C. jejuni colonization are not significantly different. Shown is a nonmetric multidimensional scaling (NMDS) plot of the cecal microbiota from the 5 donor chickens of each line, 61 and N, at 21 days of age. While spatially the two lines clustered separately, there was no statistically significant difference between the microbiota of the two lines of chickens when investigated using the adonis test (P = 0.061).
FIG 3.
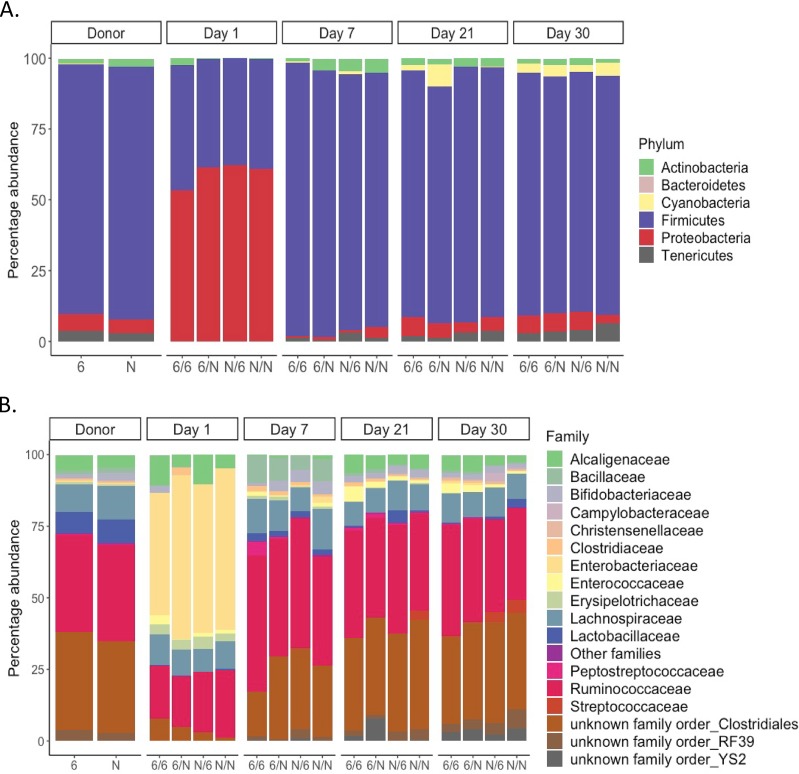
Bacterial composition of the cecal microbiota of 21-day-old donor chickens of lines 61 and N is dominated by Firmicutes at the phylum level and Ruminococcaceae and an unknown family in the order Clostridiales at the family level. Five birds were sampled in each of the donor lines. The data represent the composition of the individual samples averaged postsequencing. The overall composition of the microbiota was not significantly different between the two lines.
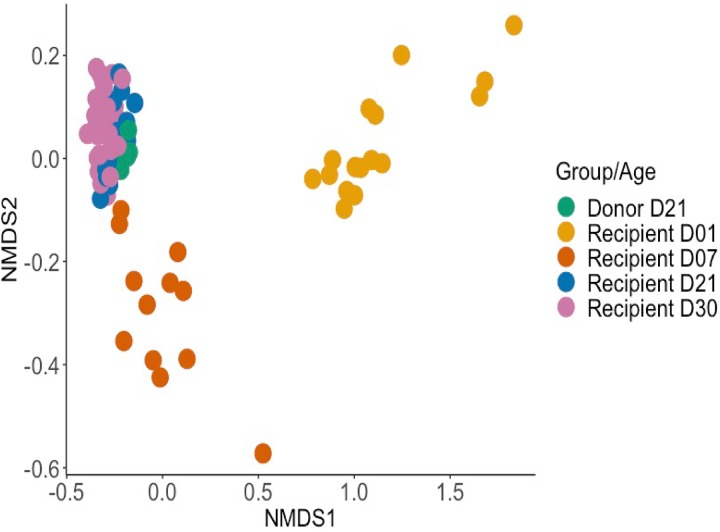
Age rather than the origin of transplanted microbiota had a dominant effect on the cecal microbial communities studied.
We next investigated whether microbiota transplants influenced the composition of the cecal microbiota over time. The NMDS plot in Fig. 4 indicates that age rather than the treatment received was the major factor that influenced the cecal microbiome. The microbiota of day-old birds clustered separately from 21- and 30-day-old birds, with the 7-day-old birds showing an intermediate clustering (P value of ≤0.001 at all time intervals studied). At family and phylum levels, no significant differences were detected between the microbial communities found in the ceca of line 61 or N birds given homologous or heterologous microbiota over time (Fig. 5). The microbiota of chickens at 1 day posttransplant had a lower diversity of bacteria and was mainly dominated by the phylum Proteobacteria (Fig. 5). At day 1 posttransplant, the microbiota clustered separately from that of donor chickens (Fig. 4), which was dominated by the Firmicutes phylum (Fig. 5). Within this phylum, the Enterobacteriaceae family dominated at 1 day posttransplant (Fig. 5). The analysis pipeline used in this study was not able to identify the bacteria to the species level; however, nucleotide sequence alignments using BLAST searches with representative sequences of this OTU indicated that the dominant bacterium at 1 day posttransplant was Escherichia coli. The intermediate phenotype of 7-day-old chickens was largely a consequence of the presence of bacteria in the Bacillaceae family (Fig. 5).
FIG 4.
Composition of the cecal microbiota of the recipients of transplants was determined primarily by the age of birds rather than the treatment received. The NMDS plot shows the clustering of cecal samples by bacterial community composition for all recipient chickens, grouped by age (12 to 16 birds were sampled at days 1, 7, and 21 and 40 birds at day 30) and by donor chickens (10 birds sampled at 21 days of age). Samples from all ages were found to cluster separately by the adonis test (P ≤ 0.001). Bray-Curtis dissimilarity values were used to calculate the dissimilarity between samples.
FIG 5.
Cecal microbiota of lines 61 and N that received homologous or heterologous microbiota transplants are not significantly different. No significant differences were detected in the average bacterial abundance at the phylum (A) or family (B) level in the cecal microbiota of inbred lines 61 and N given a homologous or heterologous microbiota transplant. Five birds were sampled per group for the donor birds, 2 to 4 birds per group at days 1, 7, and 21, and 10 birds per group at day 30. For groups noted on the x axis, the first letter denotes the recipient line and the second letter denotes the donor line.
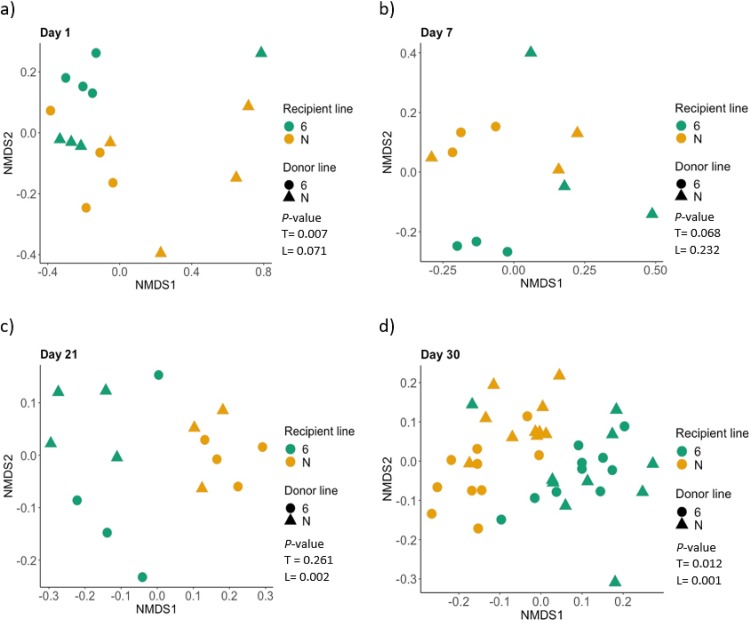
To further investigate the stability of the transplanted microbiota, we used a multivariate comparison of all the treatment groups at each time point separately to determine if the origin of the transplant or recipient line contributed significantly to the clustering of the microbiota. This analysis revealed that the donor transplant had some effect on the composition of the microbiota, but not at all time points studied (Fig. 6). Using the adonis test, the origin of transplant influenced the cecal microbiota of 1-day-old recipient chickens but the genotype of the recipient did not (P values of 0.007 and 0.071, respectively). At 7 days of age, neither transplanted bacteria nor the genotype of the recipient birds had a significant effect on the cecal microbiota of the recipient (P values of 0.068 and 0.232, respectively), possibly owing to the low number of birds sampled at each of the first three time points. After 21 days of age, the genotype of the recipient significantly affected the cecal microbiota of the recipient (P values of 0.002 and 0.001 at 21 and 30 days old, respectively). The transplanted microbiota did not affect the cecal microbiota at 21 days of age (P = 0.261) but had a significant effect in the 30-day-old recipients (P = 0.012).
FIG 6.
Cecal microbiota transplants influenced the composition of the microbiota early in the experiment, but bird line had a dominant effect with increasing age. NMDS plot of gut microbiota at each time point: day 1, top left; day 7, top right; day 21, bottom left; day 30, bottom right. P values for the effect of the transplant (T) or the bird line (L) were obtained using the adonis test and are presented in the key for each plot.
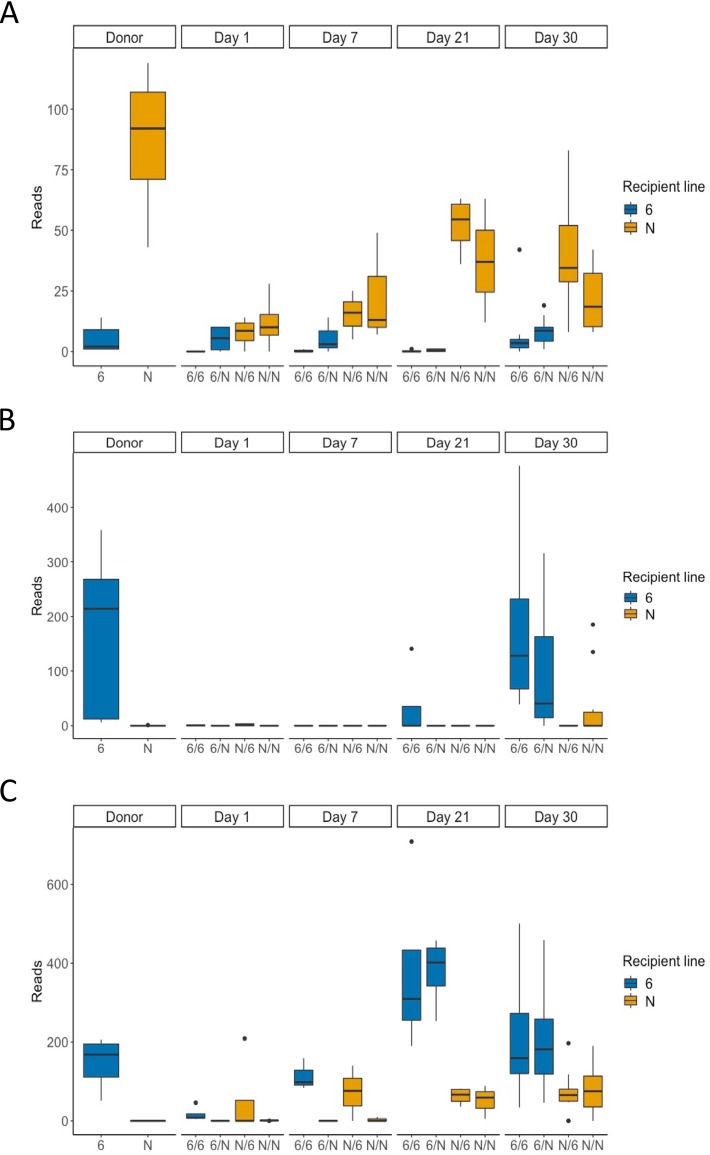
Lastly, because we observed significant effects of the transplant at some time intervals (Fig. 6) but the average bacterial abundance at phylum and family levels was not statistically different (Fig. 5), we investigated whether the microbiota transplants changed the relative abundance in the recipient birds of the same OTUs, which we identified to be significantly different in donor birds. An OTU in an unknown genus of Ruminococcaceae was more abundant in the donor birds of line N (Fig. 7A); an OTU from an unknown genus of Ruminococcaceae was significantly more abundant in the donor birds of line 61 (Fig. 7B), and an OTU in the genus Oscillospira was significantly more abundant in the donor birds of line 61 (Fig. 7C). The abundance of these OTUs per line following homologous or heterologous microbiota transplants was determined using ANCOM analysis. We found that these OTUs did not show significant differences between recipient lines or donor bacteria at 1 or 7 days posttransplant (Fig. 7). The only significant effect was that of the genotype of the recipient line at both 21 and 30 days of age (Fig. 7). This suggests that the transplanted bacteria were only able to persist in the recipient birds for a limited period of time.
FIG 7.
Abundance of specific OTUs in donor microbiota and in ceca following homologous or heterologous transplants. (A) Unknown genus in the Ruminococcaceae family. (B) A different unknown genus in the Ruminococcaceae family. (C) The genus Oscillospira. Differences were investigated using ANCOM. Significant differences were found between line 61 and N in the donor groups for all three OTUs and in the recipient birds at days 21 and 30 for the OTU shown in panel A and day 30 for the OTU shown in panel B (P < 0.05). The boxes show the quartile 1-to-quartile 3 range, and the whiskers indicate the minimum and maximum, unless values were more than 1.5 times the interquartile range. Statistical outliers identified by R are plotted as individual solid circles; they were not excluded from the analysis. For groups noted on the x axis, the first letter denotes the recipient line and the second letter denotes the donor line.
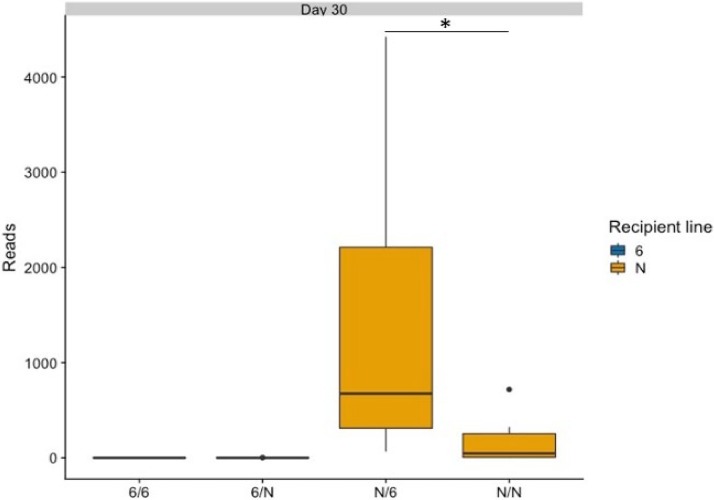
At the level of individual OTUs, we also examined the relative abundance of Campylobacter (Fig. 8). Sequence reads corresponding to Campylobacter OTUs were only detected in the susceptible line N birds, and within this line, the birds that received the heterologous microbiota transplant had significantly higher abundance of Campylobacter than the birds that received homologous microbiota (P < 0.05) (Fig. 8), consistent with the bacterial counts detected (Fig. 1).
FIG 8.
Abundance of Campylobacter detected by direct plating on mCCDA was validated by the abundance of OTUs detecting by sequencing. The graph shows the abundance of sequence reads corresponding to the OTU for the genus Campylobacter across all groups. Ten birds were sampled per group at day 30 of age. Significant differences were identified using a one-way two-sided ANOVA (Minitab, UK). Birds from line N that received microflora from line 61 had a significantly higher number of reads than the other three groups (P < 0.05). The boxes show the quartile 1-to-quartile 3 range, and the whiskers indicate the minimum and maximum, unless values were more than 1.5 times the interquartile range. Statistical outliers identified by R are plotted as individual solid circles; they were not excluded from the analysis. Only the difference between the N/6 and N/N groups is shown on the graph (*). For groups noted on the x axis, the first letter denotes the recipient line and the second letter denotes the donor line.
DISCUSSION
Control of Campylobacter infections in poultry remains challenging, and, to date, no methods for effective control at the farm level have been developed, other than the application of stringent biosecurity. Previous literature has demonstrated that the intestinal microbiota can play a role in resistance to enteric pathogens in mice (16), chickens (14, 19), and pigs (22). Consequently, we investigated the contribution of cecal microbiota to the differential resistance of inbred chicken lines 61 and N to colonization by C. jejuni, which have been demonstrated by experimental inoculation with several C. jejuni strains (6, 7). The same lines also differ in resistance to enteric carriage of Salmonella enterica serovar Typhimurium in the same direction (23), and we reasoned that differences in their microbiota may contribute to this. To this end, we performed homologous and heterologous microbiota transplants between these two lines of chickens, followed by inoculation with a dose per bird of 104 CFU of C. jejuni 11168H. Contrary to a precedent in the literature that described resistance to Citrobacter being transferable between strains of inbred mice following transfer of fecal microbiota (16), we observed a significant increase in susceptibility of line N to C. jejuni following the transfer of cecal microbiota from resistant line 61 birds (Fig. 1 and 8). The underlying basis of this effect will require repetition and further investigation.
The colonization phenotypes observed following heterologous transfer of microbiota are to be interpreted in the context of 16S ribosomal DNA amplicon analysis. This revealed no statistically significant difference in the clustering of the microbiota of the donor birds, although visually there appeared to be separation of the microbiota of the two lines by principal component analysis plots (Fig. 2). It is possible that if we had sampled more birds of each line, differences at the level of the global community, phyla, or families would have become significant. A similar separation of cecal microbial communities by the recipient line was detected (Fig. 6), which was significantly different at the last two time points, possibly owing to the higher number of birds analyzed. As we only examined C. jejuni colonization of the ceca of lines 61 and N for parity with preceding studies (6, 7), we cannot preclude the possibility that microbial transplants affected the fecal excretion of Campylobacter and bird-to-bird transmission, which was reported to be significantly impaired following fecal microbiota transplantation in a seeder-bird challenge model up to a typical slaughter age of broiler chickens (19).
It could be argued that the microbiota transplant did not successfully establish in the recipient birds, as no significant differences were observed at the level of the entire microbiome after the transplant (Fig. 5). However, when dissected across the time course, the microbiota transplant exerted a significant effect on the microbiota of the recipient birds (Fig. 6), although later in the experiment the line of recipient birds had a larger influence. RNA sequencing analysis using cecal mucosa from these two chicken lines supports the notion that bird genetics have the greatest influence on C. jejuni colonization, as we observed the largest number of differences in gene expression between uninfected birds of the two lines, with the expression of relatively few additional genes affected by Campylobacter infection (K. M. Russell, J. Smith, A. Bremner, C. Chintoan-Uta, L. Vervelde, A. Psifidi, and M. P. Stevens, submitted for publication). Previous experiments have made similar observations, with the line of chickens being described as one of the main factors that influence the intestinal microbiota (21). While we detected some significant differences in the prevalence of specific OTUs between donor birds, we could not conclusively demonstrate the early transfer of these OTUs in reciprocal transplants, although we did observe these OTUs in recipient birds were present in proportions similar to those of donor birds of the same line later following inoculation (Fig. 7). Alternatively, given the delay in observing this phenotype, these OTUs may be differentially selected from the environment by each recipient line, as with increasing age, the bird line exerted a stronger effect on the microbiota composition.
We observed that the age of birds has a large effect on the composition of the microbiota. At 1 day posthatch, irrespective of the origin and composition of donor microbiota, we observed a large population of Proteobacteria (and, more specifically, E. coli) in the ceca. By 1 week following inoculation with microbiota, Firmicutes dominate the ceca. Similar observations were reported in other microbiota studies of chickens (24, 25). It is not known what causes this proliferation of E. coli in neonatal chicks, but it may be linked to the susceptibility of neonatal chickens to colibacillosis, which is widely recognized as a key cause of mortality of chicks in hatcheries and soon after placement (26). A large influence of the age of the chickens on the composition of their microbiota was also reported in relation to colonization by C. jejuni (15) and was identified via meta-analysis of available data sets (27).
Our study determined that, at least in the case of these two particular inbred chicken lines under our experimental conditions, the microbiota does not play a major role in their differential resistance to Campylobacter colonization, and the transplantation of the microbiota from resistant to susceptible birds may not be a viable control strategy. Recent evidence in mice (28) highlights variability in the effect of the transplant when using recipient mice of different ages. Indeed, it has been reported that while fecal microbiota transfer reduced C. jejuni colonization and transmission when given to neonatal chicks, it had little impact when administration was delayed to day 7 of age (19). The observations of these authors indicate that the concept of microbiota transplantation has merit; however, while they found the microbiota of recipients to be affected by the transplant, they also observed expansion of OTUs that were not a major component of the transplanted material (e.g., Lactobacillus spp.) (19). This indicates that the transplant changes the gut environment to favor other microbes as much as transfer them directly. Such changes may account for the significant increase in C. jejuni colonization in the susceptible line following transplant of cecal contents from the resistant line. Therefore, where future studies reliably detect protective effects, they may need to consider impacts on metabolites and the mucosal immune system, not just the microbes present per se.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
C. jejuni 11168H was obtained from the National Collection of Typed Cultures and has been fully sequenced (29) and confirmed to be proficient in colonization of chickens (7). It was cultured on modified charcoal-cephoperazone-deoxycholate agar (mCCDA) (Oxoid, UK) or in Mueller-Hinton broth (MH; Oxoid) at 37°C in a microaerophilic workstation (Don Whitley Scientific, UK) in a low-oxygen atmosphere (5% O2, 5% CO2, and 90% N2). Broth cultures of Campylobacter were grown with shaking at 400 rpm.
Experimental animals.
All procedures were conducted under Home Office project license PCD70CB48, according to the requirements of the Animal (Scientific Procedures) Act 1986, with the approval of local ethical review committees. A total of 88 chickens were used in licensed procedures. Forty-four chickens of each of the inbred lines 61 (http://www.narf.ac.uk/chickens/line-6/) and N (http://www.narf.ac.uk/chickens/line-n/) were obtained on the day of hatch from the National Avian Research Facility at The Roslin Institute, a Home Office-licensed breeding establishment. Eggs were incubated and hatched under specific-pathogen-free (SPF) conditions. Animals were housed in four groups of 22 in colony cages in a single study. Groups were of mixed sex and were individually wing tagged for identification. Water and sterile irradiated feed based on vegetable protein (DBM Ltd., UK) were provided ad libitum. A further five birds of each line, reared under SPF conditions, were culled at 3 weeks of age by cervical dislocation to act as donors of cecal microbiota.
As previously described (6), chicken lines 61 and N were derived originally from White Leghorn flocks at the Avian Disease and Oncology Laboratory of the U.S. Department of Agriculture, Agricultural Research Service (the former Regional Poultry Laboratory in East Lansing, MI, USA). The lines had been maintained by random mating within the flock at the Institute for Animal Health (IAH) since 1972 (line 61) or 1982 (line N) before being transferred to the SPF unit of the National Avian Research Facility in 2013, where they have been maintained since. Inbred chicken line 61 was obtained from a White Leghorn background in 1939 and is resistant to avian leukosis virus (30). These two inbred lines have previously been reported to differ in resistance to intestinal colonization by C. jejuni when challenged on the day of hatch (6) or at 3 weeks old (7), as well as to enteric colonization by Salmonella enterica serovar Typhimurium (23).
Microbiota transplant experiment.
The donor birds were housed in separate floor pens in the SPF unit of the NARF until 3 weeks of age. At this age, they were culled by cervical dislocation, and cecal contents from the donor birds of each line were collected for separate DNA extractions to assess variability in their microbiota and for transplantation. For DNA extraction, the samples were promptly processed without freeze-thawing as described below. For transplants, the cecal contents of five birds were mixed within line in equal weight and diluted 1:6 (wt/wt) in sterile phosphate-buffered saline (PBS) to provide a mixture of sufficiently low viscosity that could be reliably administered by oral gavage using a syringe and blunt-ended needle. Homologous transplants (61 microbiota into line 61 or N microbiota into line N) and heterologous transplants (61 microbiota into line N or N microbiota into line 61) were performed by administering 100 μl of a suspension of cecal contents by oral gavage, within 30 min of collection from donors, under aerobic conditions. Four birds from each group were sampled at 1, 7, and 21 days after the microbiota transplant. At 21 days posttransplant, all remaining birds (10 per group) were inoculated with 104 CFU of C. jejuni 11168H, administered by oral gavage in a volume of 100 μl and diluted in sterile PBS. All infected birds were culled by cervical dislocation at 9 days postchallenge to enumerate cecal Campylobacter by plating 100 μl of serial 10-fold dilutions of cecal contents in PBS on mCCDA plates. At the same time, samples of cecal contents were promptly transported on ice to the laboratory for DNA isolation and analysis of the microbiota. Differences in cecal colonization by Campylobacter were investigated using a one-way, two-sided analysis of variance (ANOVA) test in Minitab (Minitab LLC, USA). P values of ≤0.05 were taken to be significant. Power analysis using measures of interanimal variance from our past research on Campylobacter vaccines, mutants, and heritable resistance indicated that 10 birds per group can detect a 2 log10 CFU/g difference with 80% power at a significance level of α = 0.05.
DNA extraction.
DNA extractions were performed using pooled contents from both ceca of each bird, with a separate extraction for each individual. Extraction was performed using a DNeasy PowerSoil kit (Qiagen, Valencia, CA, USA), with minimal delay from the time of collection and without freezing. Samples were extracted in a single batch at the earlier time points and in two batches at the last time point, with samples collected from birds that received the transplant from the same donor birds extracted in the same batch. Due to the low volume of cecal contents at 1 day posthatch, the entire ceca (tissues and contents) were used for DNA extraction. For birds of all other ages, only cecal contents were used. Cecal contents or tissues were transferred to bead-containing tubes with PowerSoil solution c1 and were heated at 65°C for 10 min. A bead-beating step was performed using a Precellys 24 homogenizer (Bertin Technologies, France) at 5,000 rpm for 45 s. After this step, DNA extraction was carried out by following the manufacturer’s protocol. A reagent-only control was produced for every DNA extraction batch. All negative-control samples returned between 22 and 810 reads per sample, whereas the cutoff of the cecal samples was 43,808 reads per sample. As such, any low-level contamination is unlikely to affect our analysis, given the high biomass of the cecal samples. DNA was also extracted from a ZymoBIOMICS Microbial Community Standard (Zymo Research, Irvine, CA, USA) in the same manner as that for cecal samples, and this was used as a mock community positive control. After DNA extraction, DNA samples were stored for up to 3 months at –80°C until sequence analysis.
Amplicon library construction and sequencing.
Barcoded primers specific to the variable 4 (V4) region of 16S bacterial ribosomal DNA were used for amplification by PCR (31). PCR was performed with Q5 high-fidelity 2× master mix (New England BioLabs, Beverly, MA, USA) with denaturation at 95°C for 2 min, followed by 30 cycles of 95°C for 20 s, 55°C for 15 s, and 72°C for 5 min, with a final extension at 72°C for 10 min. PCR amplicons were purified using the AMPure XP PCR purification system (Beckman Coulter, La Brea, CA, USA). The concentration of purified amplicons was measured using the Qubit dsDNA HS assay kit (Thermo Fisher Scientific, Hemel Hempstead, UK). Amplicons then were pooled at equimolar concentrations into a single library, whereby samples from each bird could be identified via unique barcodes. A mock DNA control sample from Zymobiotics was included as a control for the PCR step, containing bacterial DNA comprised of 25% Enterobacteriaceae and 12.5% of each of six other bacterial species. The pooled library was sequenced by paired-end 250-bp reads on the Illumina MiSeq platform (Illumina, San Diego, CA, USA) using v2 chemistry. Sequencing was carried out by Edinburgh Genomics, The University of Edinburgh.
Bioinformatic analysis.
The microbiome helper pipeline (32) was used in this study by following version 1 of the 16S Bacteria and Archaea standard operating procedure from the developer. In brief, paired-end reads were stitched with PEAR v0.9.6 (33). Stitched reads were filtered by quality score (q = 30) and length (250 bp) with the read_filter.pl command. Chimeric sequences were removed from the samples with VSEARCH v2.7.0 (34) using the RDP trainset database (35). QIIME wrapper scripts version 1.9.1 was used for OTU classification (36). SortMeRNA v2.1b (37) was used as the reference-based OTU picking method, while SUMACLUST v1 (https://git.metabarcoding.org/obitools/sumaclust/wikis/home/) was used for the de novo OTU picking method (38). Samples were rarefied by using the lowest number of reads from any sample in the analysis, excluding negative-control samples. Finally, OTU tables were generated in BIOM format for diversity analysis and abundance comparison with R version 3.4.2. Nonmetric multidimensional scaling (NMDS) plots were constructed using Bray-Curtis dissimilarity values, and statistical analyses comparing the distance of bacterial community compositions between groups were performed using the adonis function in R, which is part of the vegan package (39). Comparisons of bacterial abundance were calculated with analysis of composition of microbiomes (ANCOM) (40). Data visualization was performed with the ggplot2 package (41). P values of ≤0.05 were taken to be significant.
Accession number(s).
Sequencing reads can be accessed in the European Nucleotide Archive under accession number PRJEB35577.
Supplementary Material
ACKNOWLEDGMENTS
We gratefully acknowledge the support of the Biotechnology & Biological Sciences Research Council (Institute Strategic Programmes BBS/E/D/20002172 and BBS/E/D/30002276 and Core Capability Grant in support of the National Avian Research Facility) and Scottish Government funding via the Rural & Environmental Science and Analytical Services program of research for 2016 to 2021.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Anonymous. 2017. Zoonoses report UK 2017. Public Health England, London, United Kingdom: https://www.gov.uk/government/publications/zoonoses-uk-annual-reports. [Google Scholar]
- 2.Tam CC, O'Brien SJ. 2016. Economic cost of Campylobacter, norovirus and rotavirus disease in the United Kingdom. PLoS One 11:e0138526. doi: 10.1371/journal.pone.0138526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tam CC, Rodrigues LC, Viviani L, Dodds JP, Evans MR, Hunter PR, Gray JJ, Letley LH, Rait G, Tompkins DS, O'Brien SJ, ID2 Study Executive Committee . 2012. Longitudinal study of infectious intestinal disease in the UK (IID2 study): incidence in the community and presenting to general practice. Gut 61:69–77. doi: 10.1136/gut.2011.238386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.EFSA Panel on Biological Hazards (BIOHAZ). 2011. Scientific opinion on Campylobacter in broiler meat production: control options and performance objectives and/or targets at different stages of the food chain. EFSA J 9:2105. doi: 10.2903/j.efsa.2011.2105. [DOI] [Google Scholar]
- 5.Johnson TJ, Shank JM, Johnson JG. 2017. Current and potential treatments for reducing Campylobacter colonisation in animal hosts and disease in humans. Front Microbiol 8:487. doi: 10.3389/fmicb.2017.00487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boyd Y, Herbert EG, Marston KL, Jones MA, Barrow PA. 2005. Host genes affect intestinal colonisation of newly hatched chickens by Campylobacter jejuni. Immunogenetics 57:248–253. doi: 10.1007/s00251-005-0790-6. [DOI] [PubMed] [Google Scholar]
- 7.Psifidi A, Fife M, Howell J, Matika O, van Diemen PM, Kuo R, Smith J, Hocking PM, Salmon N, Jones MA, Hume DA, Banos G, Stevens MP, Kaiser P. 2016. The genomic architecture of resistance to Campylobacter jejuni intestinal colonisation in chickens. BMC Genomics 17:293. doi: 10.1186/s12864-016-2612-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li X, Swaggerty CL, Kogut MH, Chiang HI, Wang Y, Genovese KJ, He H, Zhou H. 2010. Gene expression profiling of the local cecal response of genetic chicken lines that differ in their susceptibility to Campylobacter jejuni colonization. PLoS One 5:e11827. doi: 10.1371/journal.pone.0011827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li XY, Swaggerty CL, Kogut MH, Chiang HI, Wang Y, Genovese KJ, He H, Pevzner IY, Zhou HJ. 2011. Caecal transcriptome analysis of colonized and non-colonized chickens within two genetic lines that differ in caecal colonization by Campylobacter jejuni. Anim Genet 42:491–500. doi: 10.1111/j.1365-2052.2010.02168.x. [DOI] [PubMed] [Google Scholar]
- 10.Li X, Swaggerty CL, Kogut MH, Chiang HI, Wang Y, Genovese KJ, He H, McCarthy FM, Burgess SC, Pevzner IY, Zhou H. 2012. Systemic response to Campylobacter jejuni infection by profiling gene transcription in the spleens of two genetic lines of chickens. Immunogenetics 64:59–69. doi: 10.1007/s00251-011-0557-1. [DOI] [PubMed] [Google Scholar]
- 11.Bailey RA, Kranis A, Psifidi A, Watson KA, Rothwell L, Hocking PM, Kaiser P, Stevens MP, Avendano S. 2018. Colonization of a commercial broiler line by Campylobacter is under limited genetic control and does not significantly impair performance or intestinal health. Poult Sci 97:4167–4176. doi: 10.3382/ps/pey295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O'Loughlin JL, Samuelson DR, Braundmeier-Fleming AG, White BA, Haldorson GJ, Stone JB, Lessmann JJ, Eucker TP, Konkel ME. 2015. The intestinal microbiota influences Campylobacter jejuni colonization and extraintestinal dissemination in mice. Appl Environ Microbiol 81:4642–4650. doi: 10.1128/AEM.00281-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun X, Winglee K, Gharaibeh RZ, Gauthier J, He Z, Tripathi P, Avram D, Bruner S, Fodor A, Jobin C. 2018. Microbiota-derived metabolic factors reduce campylobacteriosis in mice. Gastroenterology 154:1751–1763. doi: 10.1053/j.gastro.2018.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Han Z, Willer T, Pielsticker C, Rychlik I, Velge P, Kaspers B, Rautenschlein S. 2017. Influence of the gut microbiota composition on Campylobacter jejuni colonization in chickens. Infect Immun 85:e00380-17. doi: 10.1128/IAI.00380-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Connerton PL, Richards PJ, Lafontaine GM, O'Kane PM, Ghaffar N, Cummings NJ, Smith DL, Fish NM, Connerton IF. 2018. The effect of the timing of exposure to Campylobacter jejuni on the gut microbiome and inflammatory responses of broiler chickens. Microbiome 6:88. doi: 10.1186/s40168-018-0477-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Willing BP, Vacharaksa A, Croxen M, Thanachayanont T, Finlay BB. 2011. Altering host resistance to infections through microbial transplantation. PLoS One 6:e26988. doi: 10.1371/journal.pone.0026988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bakken JS, Fecal Microbiota Transplantation Workgroup, Borody T, Brandt LJ, Brill JV, Demarco DC, Franzos MA, Kelly C, Khoruts A, Louie T, Martinelli LP, Moore TA, Russell G, Surawicz C. 2011. Treating Clostridium difficile infection with faecal microbiota transplant. Clin Gastroenterol Hepatol 9:1044–1049. doi: 10.1016/j.cgh.2011.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brandt JL, Aroniadis OC, Mellow M, Kanatzar A, Kelly C, Park T, Stollman N, Rohlke F, Surawicz C. 2012. Long-term follow-up of colonoscopic faecal microbiota transplant for recurrent Clostridium difficile infection. Am J Gastroenterol 107:1079–1087. doi: 10.1038/ajg.2012.60. [DOI] [PubMed] [Google Scholar]
- 19.Gilroy R, Chaloner G, Wedley A, Lacharme-Lora L, Wigley P. 2018. Campylobacter jejuni transmission and colonisation in broiler chickens is inhibited by faecal microbiota transplantation. BioRxiv 10.1101/476119. [DOI]
- 20.Sergeant MJ, Constantinidou C, Cogan TA, Bedford MR, Penn CW, Pallen MJ. 2014. Extensive microbial and functional diversity within the chicken cecal microbiome. PLoS One 9:e91941. doi: 10.1371/journal.pone.0091941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pandit RJ, Hinsu AT, Patel NV, Koringa PG, Jakhesara SJ, Thakkar JR, Shah TM, Limon G, Psifidi A, Guitian J, Hume DA, Tomley FM, Rank DN, Raman M, Tirumurugaan KG, Blake DP, Joshi CG. 2018. Microbial diversity and community composition of caecal microbiota in commercial and indigenous Indian chickens determined using 16s rDNA amplicon sequencing. BMC Microbiome 6:115. doi: 10.1186/s40168-018-0501-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Niederwerder MC, Constance LA, Rowland RRR, Abbas W, Fernando SC, Potter ML, Sheahan MA, Burkey TE, Hesse RA, Cino-Ozuna AG. 2018. Fecal microbiota transplantation is associated with reduced morbidity and mortality in porcine circovirus associated disease. Front Microbiol 9:1631. doi: 10.3389/fmicb.2018.01631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fife MS, Howell JS, Salmon N, Hocking PM, van Diemen PM, Jones MA, Stevens MP, Kaiser P. 2011. Genome-wide SNP analysis identifies major QTL for Salmonella colonization in the chicken. Anim Genet 42:134–140. doi: 10.1111/j.1365-2052.2010.02090.x. [DOI] [PubMed] [Google Scholar]
- 24.Johnson TJ, Youmans BP, Noll S, Cardrona C, Evans NP, Karnezos P, Ngunjiri JM, Abundo MC, Lee CW. 2018. A consistent and predictable commercial broiler chicken bacterial microbiota in antibiotic-free production displays strong correlations with performance. Appl Environ Microbiol 84:e00362-18. doi: 10.1128/AEM.00362-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jurburg SD, Brouwer MSM, Ceccarelli D, van der Goot J, Jansman AJM, Bossers A. 2019. Patterns of community assembly in the developing chicken microbiome reveal rapid primary succession. Microbiologyopen 8:e00821. doi: 10.1002/mbo3.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kemmett K, Williams NJ, Chaloner S, Humphrey P, Wigley P, Humphrey T. 2014. The contribution of systemic E. coli infection to the early mortalities of commercial broiler chickens. Avian Pathol 43:37–42. doi: 10.1080/03079457.2013.866213. [DOI] [PubMed] [Google Scholar]
- 27.Kers JG, Velkers FC, Fischer EAJ, Hermes GDA, Stegeman JA, Smidt H. 2018. Host and environmental factors affecting the intestinal microbiota in chickens. Front Microbiol 9:235. doi: 10.3389/fmicb.2018.00235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Le Roy T, Debedat J, Marquet F, Da-Cunha C, Ichou F, Guerre-Millo M, Kapel N, Aron-Wisniewsky J, Clement K. 2019. Comparative evaluation of microbiota engraftment following faecal microbiota transfer in mice models: age, kinetics and microbial status matter. Front Microbiol 9:3289. doi: 10.3389/fmicb.2018.03289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pascoe B, Williams LK, Calland JK, Meric G, Hitchings MD, Dyer M, Ryder J, Shaw S, Lopes BS, Chintoan-Uta C, Allan E, Vidal A, Fearnley C, Everest P, Pachebat JA, Cogan TA, Stevens MP, Humphrey TJ, Wilkinson TS, Cody AJ, Colles FM, Jolley KA, Maiden MCJ, Strachan N, Pearson BM, Linton D, Wren BW, Parkhill J, Kelly DJ, van Vliet AHM, Forbes KJ, Sheppard SK. 2019. Domestication of Campylobacter jejuni NCTC 11168. Microb Genom 5:e000279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Waters NF. 1945. Breeding for resistance and susceptibility to avian lymphomatosis. Poult Sci 24:259–269. doi: 10.3382/ps.0240259. [DOI] [Google Scholar]
- 31.Kozich JJ, Westcott SL, Baxter TT, Highlander SK, Schloss PD. 2013. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol 79:5112–5120. doi: 10.1128/AEM.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Comeau AM, Douglas GM, Langille M. 2017. Microbiome Helper: a custom and streamlined workflow for microbiome research. mSystems 2:e00127-16. doi: 10.1128/mSystems.00127-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang J, Kobert K, Flouri T, Stamatakis A. 2014. PEAR: a fast and accurate Illumina paired-end read merger. Bioinformatics 30:614–620. doi: 10.1093/bioinformatics/btt593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rognes T, Flouri T, Nichols B, Quince C, Mahé F. 2016. VSEARCH: a versatile open source tool for metagenomics. PeerJ 4:e2584. doi: 10.7717/peerj.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cole JR, Wang Q, Fish JA, Chai B, McGarrell DM, Sun Y, Brown CT, Porras-Alfaro A, Kuske CR, Tiedje JM. 2014. Ribosomal Database Project: data and tools for high throughput rRNA analysis. Nucleic Acids Res 42:D633–D642. doi: 10.1093/nar/gkt1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kopylova E, Noé L, Touzet H. 2012. SortMeRNA: fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics 28:3211–3217. doi: 10.1093/bioinformatics/bts611. [DOI] [PubMed] [Google Scholar]
- 38.Mercier C, Boyer F, Bonin A, Coissac E. 2013. SUMATRA and SUMACLUST: fast and exact comparison and clustering of sequences. Programs Abstr SeqBio 2013 Workshop, Montpellier, France. [Google Scholar]
- 39.Dixon P. 2003. VEGAN, a package of R functions for community ecology. J Veg Sci 14:927–930. doi: 10.1111/j.1654-1103.2003.tb02228.x. [DOI] [Google Scholar]
- 40.Mandal S, Van Treuren W, White RA, Eggesbø M, Knight R, Peddada SD. 2015. Analysis of composition of microbiomes: a novel method for studying microbial composition. Microb Ecol Health Dis 26:27663. doi: 10.3402/mehd.v26.27663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wickham H. 2016. ggplot2, elegant graphics for data analysis. Springer International Publishing, New York, NY. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.