Identifying and understanding the active microbial communities and interactions involved in plant residue utilization are key questions to elucidate the transformation of soil organic matter (SOM) in agricultural ecosystems. Microbial community composition responds strongly to management, but little is known about specific microbial groups involved in plant residue utilization and, consequently, microbial functions under different methods of fertilization. We combined DNA stable-isotope (13C) probing and high-throughput sequencing to identify active fungal and bacterial groups degrading residues in soils after 3 years of mineral fertilization with and without manure. Manuring changed the active microbial composition and complexified microbial interactions involved in residue C flow. Most fungal genera, especially Chaetomium, Staphylotrichum, Penicillium, and Aspergillus, responded to residue addition faster in soils that historically had received manure. We generated a valuable library of microorganisms involved in plant residue utilization for future targeted research to exploit specific functions of microbial groups in organic matter utilization and C sequestration.
KEYWORDS: fertilization, 13C-labeled rice residue, DNA stable-isotope probing, high-throughput sequencing, active microbes
ABSTRACT
Decomposition of crop residues in soil is mediated by microorganisms whose activities vary with fertilization. The complexity of active microorganisms and their interactions utilizing residues is impossible to disentangle without isotope applications. Thus, 13C-labeled rice residues were employed, and DNA stable-isotope probing (DNA-SIP) combined with high-throughput sequencing was applied to identify microbes active in assimilating residue carbon (C). Manure addition strongly modified microbial community compositions involved in the C flow from rice residues. Relative abundances of the bacterial genus Lysobacter and fungal genus Syncephalis were increased, but abundances of the bacterial genus Streptomyces and fungal genus Trichoderma were decreased in soils receiving mineral fertilizers plus manure (NPKM) compared to levels in soils receiving only mineral fertilizers (NPK). Microbes involved in the flow of residue C formed a more complex network in NPKM than in NPK soils because of the necessity to decompose more diverse organic compounds. The fungal species (Jugulospora rotula and Emericellopsis terricola in NPK and NPKM soils, respectively) were identified as keystone species in the network and may significantly contribute to residue C decomposition. Most of the fungal genera in NPKM soils, especially Chaetomium, Staphylotrichum, Penicillium, and Aspergillus, responded faster to residue addition than those in NPK soils. This is connected with the changes in the composition of the rice residue during degradation and with fungal adaptation (abundance and activity) to continuous manure input. Our findings provide fundamental information about the roles of key microbial groups in residue decomposition and offer important cues on manipulating the soil microbiome for residue utilization and C sequestration in soil.
IMPORTANCE Identifying and understanding the active microbial communities and interactions involved in plant residue utilization are key questions to elucidate the transformation of soil organic matter (SOM) in agricultural ecosystems. Microbial community composition responds strongly to management, but little is known about specific microbial groups involved in plant residue utilization and, consequently, microbial functions under different methods of fertilization. We combined DNA stable-isotope (13C) probing and high-throughput sequencing to identify active fungal and bacterial groups degrading residues in soils after 3 years of mineral fertilization with and without manure. Manuring changed the active microbial composition and complexified microbial interactions involved in residue C flow. Most fungal genera, especially Chaetomium, Staphylotrichum, Penicillium, and Aspergillus, responded to residue addition faster in soils that historically had received manure. We generated a valuable library of microorganisms involved in plant residue utilization for future targeted research to exploit specific functions of microbial groups in organic matter utilization and C sequestration.
INTRODUCTION
The importance of microbial residues for soil organic matter (SOM) formation has been recognized for over 2 decades (1, 2), indicating the crucial roles of microbes in sustainability and soil fertility. Therefore, dominant microbes involved in decomposition play important roles for biologically driven carbon (C) in C flows. The development of molecular biology techniques has greatly advanced our understanding of microorganisms that mediate the decomposition of plant residue in soil (3–6). To sequester more C in soil during plant residue decomposition, we need to consider the C pathways from the residue into the soil organic matter through microbial processing (7). Therefore, precise identification of the microbial groups involved in C utilization and sequestration is the key to exploring the functional roles of microbes in improving soil fertility.
Approaches using stable-isotope probing (SIP) provide novel insights into C flows in soil microbial communities. The identity of microorganisms has been linked with their activities and functions using SIP, for example, in tracing the fate of low-molecular-weight organic substrates, such as glucose (8–12), methanol (13, 14), and propionate (15). A few studies considering more complex organic compounds focused on root exudates (16–20) or high-molecular-weight organic substrates, such as cellulose (21, 22). A number of studies used SIP to characterize specific microbial groups utilizing plant residues differing in qualities or types, but all of these studies concentrated on only a single soil condition (23–29). Limited information is available to address whether the active microbiome involved in residue decomposition will be affected by specific fertilization practices although the total soil microbial community has been widely reported to be changed (30, 31).
In agricultural soils, changes in the C availability due to fertilization, such as mineral or organic fertilization, always alter microbial community structure (32, 33), which further leads to alterations in the enzyme activities related to C and N cycling (34–38). The application of organic fertilizers (i.e., cattle manure and compost) consistently resulted in higher levels of cultivable microorganisms in soil and of enzyme activities and thus can be used as an environmentally friendly and rapid measure for restoring degraded cropland (39). The organic amendments also significantly impact the decomposition rate of crop residues (40). So far, no attempt has been made to evaluate the effects of mineral and organic fertilizers on microbial groups decomposing plant residues. Considering that the degradation of plant residues is a microbe-mediated process and that fertilization selects for specifically adapted microbial communities, it is reasonable to propose that distinct active microbial communities responding to residue decomposition can exist in soils experiencing contrasting fertilization regimes. Mineral, in contrast to organic, inputs alter the general balance of soil stoichiometry, thereby resulting in adaptative selection of a resident population of oligotrophic taxa (high C use efficiency, slow growing) instead of more copiotrophic taxa (low C use efficiency, fast growing). Wang et al. (41) reported that the application of organic fertilizers increased the abundance of generally copiotrophic bacterial groups, while application of mineral fertilizers increased the abundance of oligotrophic groups. Thus, we hypothesized that there will be a shift in dominance and response strategies of specific microbial groups involved in residue assimilation in soils after manuring compared to these strategies after only mineral fertilization. To clarify this, soils were collected from a field experiment with different 3-year fertilization regimes, including fertilization with solely mineral fertilizers (NPK) and fertilization with mineral fertilizers plus manure (NPKM), and a microcosm experiment with 13C-labeled rice residues was conducted coupled with DNA-SIP technology.
RESULTS
Rice residue (0.05 g rice residue/g dry weight soil [dws]) was employed to conduct microcosm experiments. A similar amount of about 0.04 to 0.05 g of residue/g soil (dry weight) was also employed in previous SIP studies investigating microbial communities involved in residue assimilation (26–28, 42). The choice of an incubation duration of 60 days was based on a previous study showing that the cumulative CO2 efflux at the first 60 days was nearly 70% of the total 160 days’ cumulative CO2 efflux after the addition of rice residues at 25°C (43). Our data (see Fig. S1 in the supplemental material) also showed that the CO2 efflux rates were relatively stable during 30 to 60 days after the residue amendment. In addition, microbial communities are mainly affected by the addition of residue or carbon substrates (such as glucose or cellulose) during the first 7 to 31 days of incubation (26, 28, 44, 45). In this study, due to the disturbance of soil during the rice residue amendment, soils were allowed to equilibrate for 7 days prior to the first sampling. Thus, days 7, 15, 30, and 60 after rice residue addition were selected as sampling points.
Microbial communities utilizing residue-derived C.
The CO2 efflux rates and the abundances of total soil bacteria and fungi were significantly increased after the rice residue addition (Fig. S1). The dynamics of microbial abundances were similar in soils with and without manure addition (NPK plus residue and NPKM plus residue) (Fig. S1). According to the relative abundance distribution of the fungal (internal transcribed spacer [ITS]) and bacterial (16S) genes in the density gradient (Fig. S2), the fungal and bacterial DNAs from microorganisms utilizing 13C-labeled residues were clearly separated in the density gradient from 1.720 to 1.730 g ml−1 and from 1.730 to 1.740 g ml−1, respectively. The 13C-labeled DNA from the NPK plus 13C-labeled residue and NPKM plus 13C-labeled residue soil samples and the corresponding fractions from the NPK plus 12C-labeled residue and NPKM plus 12C-labeled residue soils were isolated for use in sequencing of the fungal ITS1 and bacterial 16S rRNA genes (V4-V5 regions).
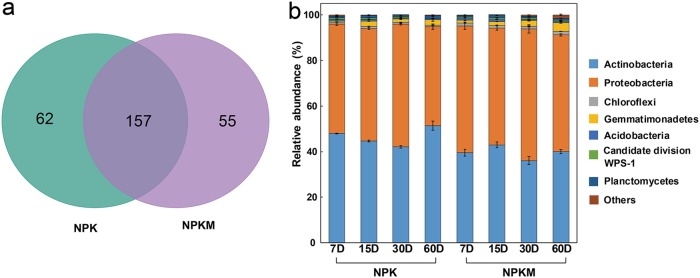
The operational taxonomic units (OTUs) were identified as being significantly labeled based on log2 fold change analysis. In total, of 219 and 212 bacterial OTUs were 13C labeled in the NPK and NPKM soils, respectively (Fig. 1a), whereas 157 of the OTUs were shared in soils with NPK and NPKM fertilization. Proteobacteria accounted for 46.5% and 39.6% of the total 13C-labeled bacterial communities in the NPK and NPKM soils, respectively, while Actinobacteria accounted for 48.6% and 54% (Fig. 1b). This indicates that Proteobacteria and Actinobacteria dominated the rice residue decomposition. The relative abundance of Proteobacteria was higher in NPKM soil than in NPK soil, whereas the relative abundance of Actinobacteria had the opposite trend. Besides these phyla, five other phyla, namely, Chloroflexi, Gemmatimonadetes, Acidobacteria, Planctomycetes, and candidate division WPS-1, also participated in residue decomposition (Fig. 1b).
FIG 1.
Effects of fertilization on the number (a) and relative abundance at the phylum level (b) of the 13C-labeled bacterial OTUs. Sampling was performed on the indicated days (D; x axis) after the addition of the rice residues. NPK, soil treated with mineral-only fertilizers; NPKM, soil treated with mineral fertilizers plus manure.
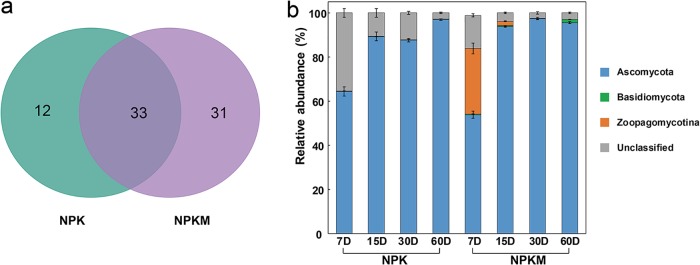
The number of 13C-enriched fungal OTUs was less than that of bacterial OTUs, with 45 fungal OTUs 13C labeled in NPK soil and 64 13C labeled in NPKM soil (Fig. 2a). In NPK soil, residue-derived C was assimilated mainly by Ascomycota and Basidiomycota (Fig. 2b). The relative abundance of 13C-labeled Ascomycota gradually increased with time, from 64.4% on day 7 to 97% on day 60. In NPKM soil, in addition to the members of Ascomycota and Basidiomycota, Zoopagomycotina was also responsible for the residue C decomposition and utilization. The relative abundance of Zoopagomycotina gradually decreased from 29.6% on day 7 to 1.92% on day 15, and it was absent on day 30. In contrast, the relative abundance of Ascomycota gradually increased from 53.8% on day 7 to 95.6% on day 60 (Fig. 2b).
FIG 2.
Effects of fertilization regimes on the number (a) and relative abundance at the phylum level (b) of the 13C-labeled fungal OTUs. Sampling was performed on the indicated days (D; x axis) after the addition of the rice residues. NPK, soil treated with mineral-only fertilizers; NPKM, soil treated with mineral fertilizers plus manure.
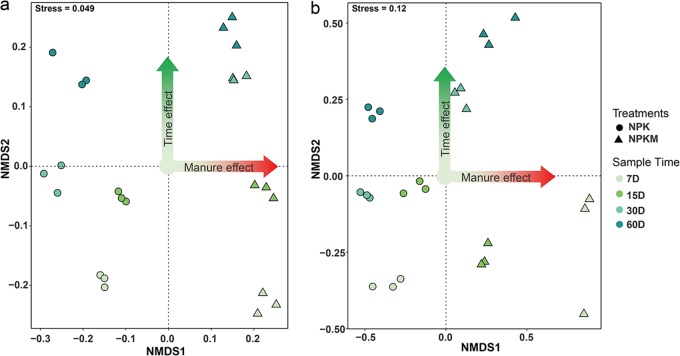
Nonmetric multidimensional scaling (NMDS) ordination of the 13C-labeled bacterial (stress value of 0.049) and fungal (stress value of 0.12) communities varied between the NPK and NPKM treatments on the horizontal axis and also clustered according to the different times on the vertical axis (Fig. 3). In addition, adonis analysis also showed that the 13C-labeled bacterial and fungal communities were significantly changed by fertilization and sampling time (Table S1). Therefore, in the following analyses, we focused mainly on the microbes that played dominant roles in the C flow depending on fertilization and on their dynamics based on the analysis of response strategies.
FIG 3.
NMDS analyses of the 13C-labeled bacterial communities (a) and fungal communities (b) involved in the utilization of rice residues. Sampling was performed on the indicated days (D) after the addition of the rice residues. NPK, soil treated with mineral-only fertilizers; NPKM, soil treated with mineral fertilizers plus manure.
Dominant microbes involved in the flow of residue-derived C.
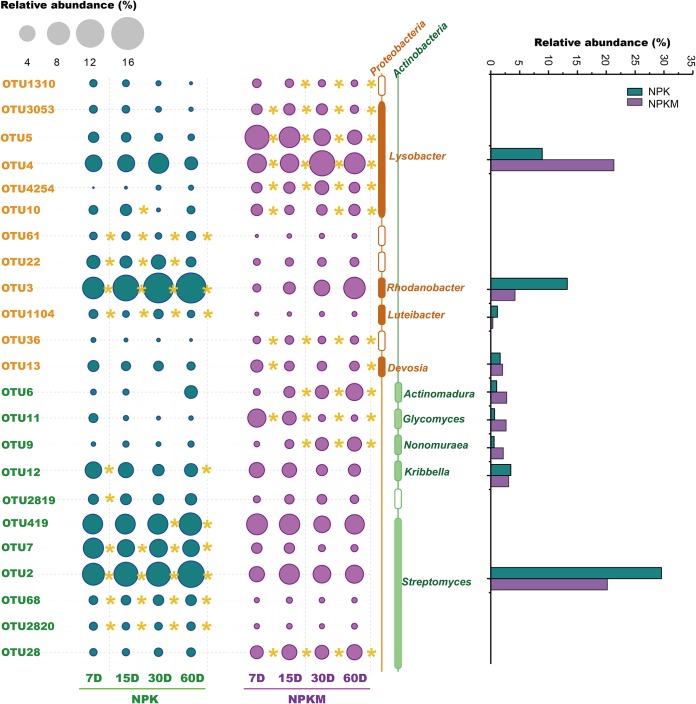
OTUs with relative abundances higher than 1% were identified as the dominant microbes. The dominant bacterial OTUs were mainly Proteobacteria and Actinobacteria, and the annotated OTUs belonged mainly to Lysobacter, Devosia, Actinomadura, Glycomyces, Nonomuraea, Rhodanobacter, Luteibacter, Kribbella, and Streptomyces (Fig. 4). The relative abundances of Lysobacter, Devosia, Actinomadura, Glycomyces, and Nonomuraea were higher in NPKM soil than in NPK soil, whereas the relative abundances of Rhodanobacter, Luteibacter, Kribbella, and Streptomyces showed the opposite trend.
FIG 4.
Dominant OTUs (relative abundance higher than 1%) in the bacterial communities involved in the C flows from rice residues. The size of the circle represents the relative abundance of the OTU. The color of the circle represents the fertilization treatment, as indicated. Sampling was performed on the indicated days (D) after the addition of the rice residues. The yellow asterisk (*) to right of a circle indicates that the relative abundance with the respective treatment was higher than that with the other treatment. Taxa are indicated to the right of the plot. The histogram in the right panel represents the average relative abundances of the dominant genera at the four sampling time points.
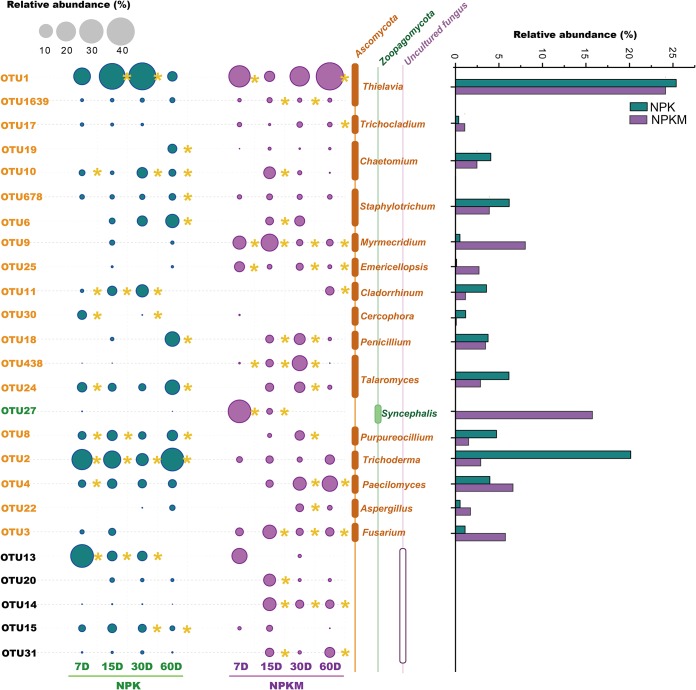
The dominant OTUs of fungi in the NPK and NPKM soils were Ascomycota and Zoopagomycotina, and most were classified into the genera Aspergillus, Thielavia, Trichocladium, Chaetomium, Emericellopsis, Cladorrhinum, Cercophora, Purpureocillium, Trichoderma, Fusarium, and Myrmecridium (Fig. 5). Other identified OTUs were classified into the genera Penicillium, Aspergillus, Talaromyces, and Staphylotrichum of the Eurotiomycetes. OTU27, identified on day 7 as accounting for approximately 29.6% of the total labeled fungal sequences in NPKM soil, was identified as a member of Syncephalis; this OTU was entirely absent from NPK soils. Compared to levels with NPK, NPKM decreased the relative abundances of Trichoderma, Purpureocillium, Talaromyces, Cercophora, Cladorrhinum, Chaetomium, Staphylotrichum, and Thielavia and increased the relative abundances of Fusarium, Aspergillus, Paecilomyces, Emericellopsis, Myrmecridium, Trichocladium, and Syncephalis.
FIG 5.
Dominant OTUs (relative abundance higher than 1%) in the fungal communities involved in the C flow from rice residues. The size of the circle represents the relative abundance of the OTU. The color of the circle represents the fertilization treatment, as indicated. Sampling was performed on the indicated days (D) after the addition of the rice residues. The yellow asterisk (*) to right of a circle indicates that the relative abundance with the respective treatment was higher than that with the other treatment. Taxa are indicated to the right of the plot. The histogram in the right panel represents the average relative abundances of the dominant genera at the four sampling time points.
Microbial networks involved in the C flow from rice residues.
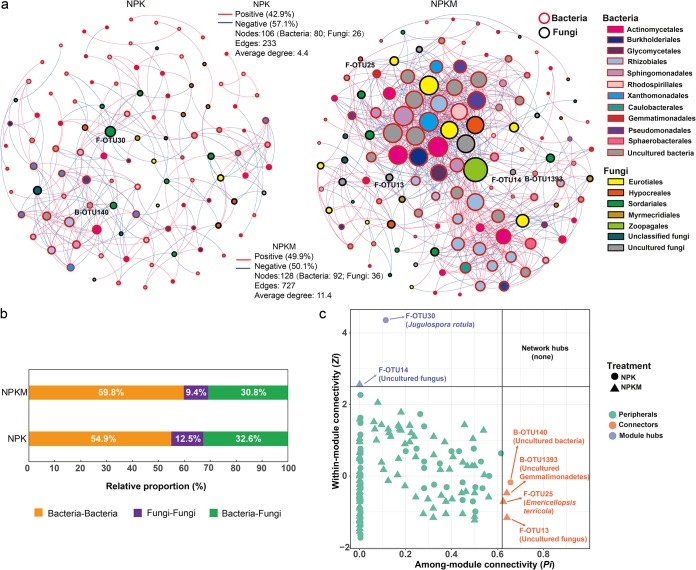
The microbes that played key roles in the flow of residue C were explored using microbial molecular ecology networks (Fig. 6). The bacterial and fungal OTUs with average relative abundances higher than 0.1% were selected for network construction by random matrix theory (RMT). There were 106 nodes in the NPK soil network, including 80 bacterial nodes and 26 fungal nodes. There were 128 nodes in the NPKM soil network, including 92 bacterial nodes and 36 fungal nodes. The average degree of the NPK network was 4.40, whereas that of the NPKM network was 11.4 (Table S2), indicating that the 3-year manure application increased the complexity of the microbial network related to the utilization of residue-derived C.
FIG 6.
Molecular ecology network analyses of microbes involved in the C flow from rice residues in soils in relation to fertilization regimes. Labeled OTUs observed at four sampling time points from each fertilization treatment were pooled to construct networks that represent random matrix theory cooccurrence models. (a) Networks in soils treated with NPK or NPKM. The circles represent nodes. Circles with red edges represent bacterial (B) OTUs, whereas circles with black edges represent fungal (F) OTUs. Lines connecting two nodes represent the significant correlations between OTUs. Red lines represent significant positive correlations, and blue lines represent significant negative correlations. (b) Proportion of intrakingdom edges (edges between bacterial OTU and bacterial OTU or between fungal OTU and fungal OTU) and interkingdom edges (edges between bacterial OTU and fungal OTU) in the networks of NPK- and NPKM-fertilized soils. (c) The distribution of Zi and Pi values for the OTUs established from their module-based topological roles. The threshold values of Zi and Pi for categorizing OTUs were 2.5 and 0.62, respectively (74, 99). Circles, NPK treatment; triangles, NPKM treatment.
Two key network nodes were found in the NPK soil (Fig. 6 and Table S3): the fungal OTU30, affiliated with Ascomycota, acted as a module hub, and uncultured bacteria, OTU140, acted as an important connector between modules. Four key network nodes were present in the NPKM soil, including a module hub and three connectors. Specifically, the fungal OTU25 (Ascomycota), unclassified fungus (OTU13), and the bacterial OTU1393 affiliated with Gemmatimonadetes acted as connectors in the network; a module hub was found to be the unclassified fungus OTU14. The average relative abundances of the two fungal OTUs that served as module hubs in NPK and NPKM soils were 2.3% and 5.81%, respectively (Table S3). However, the average relative abundances of bacterial module connectors in both NPK and NPKM soils were less than 1%. The fungal OTU30 that acted as the module hub in NPK soil was identified as Jugulospora rotula, and the fungal OTU25 that acted as a connector in the NPKM soil was classified as Emericellopsis terricola.
Response strategies of dominant microbes to residue amendment.
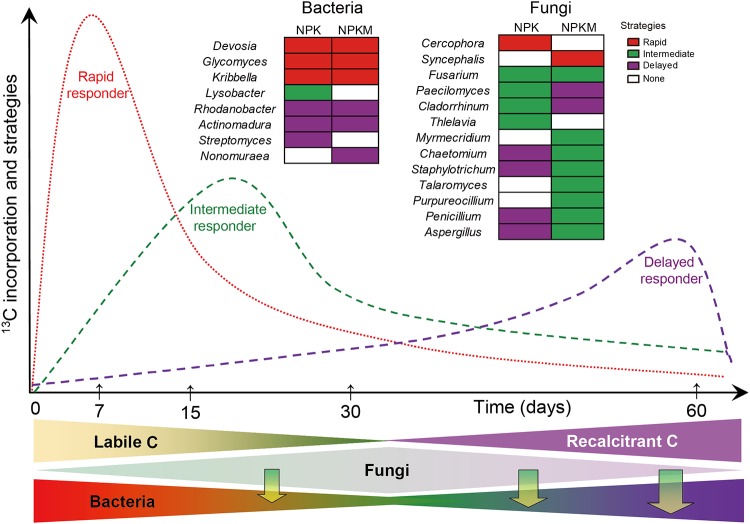
In order to assess the temporal dynamics of the microorganisms that use the residue-derived C, the abundant genera (Fig. 3 and 4) having significant changes between any two sampling points were selected. We assumed that the relative abundance changes between sampling points reflect microbial activities. Response strategies were assigned according to the time point at which the relative abundance of the genus was maximal. Genera that displayed highest relative abundances at the outset (7 days after residue addition) were referred to as rapid responders; those that displayed highest relative abundances at intermediate sampling time points (between 15 days and 30 days) were referred to as intermediate responders; those that displayed highest relative abundances at 60 days after residue addition were referred to as delayed responders. In NPK- and NPKM-fertilized soils, the response rates of the genera Devosia, Glycomyces, and Kribbella were the highest, whereas those of Rhodanobacter and Actinomadura were much lower, as were those of Streptomyces in NPK soil and Nonomuraea in NPKM soil. In contrast to the similar response strategies of bacteria in NPK and NPKM soils, fungi behaved quite diversely (Fig. 7). Only one genus, Fusarium, had the same response rates in the NPK and NPKM soils. In NPK soil, the genus Cercophora was the most rapid to respond to the addition of residue; Thielavia, Cladorrhinum, and Paecilomyces had lower response rates, and Staphylotrichum, Chaetomium, Penicillium, and Aspergillus, the delayed responders, were the slowest. In NPKM soil, Syncephalis responded rapidly while Myrmecridium, Chaetomium, Staphylotrichum, Talaromyces, Purpureocillium, Penicillium, and Aspergillus responded at intermediate levels, and Paecilomyces and Cladorrhinum exhibited delayed responses.
FIG 7.
Conceptual diagram of microbial groups responsible for utilization and decomposition of rice residues in soils in relation to fertilization regimes. NPK- mineral fertilizers, NPKM- mineral fertilizers combined with manure. Line colors correspond to the colors assigned to the response strategies for rice residue utilization, as indicated on the right of the figure. Rapid responders, genera that displayed highest relative abundance at the outset (7 days after residue addition); intermediate responders, genera that displayed highest relative abundance at intermediate sample time points (between 15 and 30 days after residue addition); delayed responders, genera that displayed highest relative abundance at 60 days after residue addition. The lower part of the figure reflects C availability and the predominance of microbial groups. The vertical arrows represent the interactions between microbial groups in residue utilization (e.g., some carbon resources are provided by fungi to bacteria). Over time, the proportion of labile C in rice residue decreases while the proportion of recalcitrant C increases gradually. Most bacteria have a limited capacity to decompose recalcitrant components at later time points. Intermediate or delayed fungal responders can provide bacteria with resources that they cannot directly make from residues themselves, such as intermediate decomposition products (see text for details).
DISCUSSION
Fertilization changed the composition of microorganisms utilizing rice residues.
Fertilization regimes significantly changed the microbial community composition involved in rice residue assimilation (Fig. 3 to 5). The changes in the composition of the residue-assimilating microbial community are affected by the differences in nutrient content between NPK and NPKM soils (28, 46) as well as by organic matter availability. The modification of the microbial community by fertilization can be direct (addition of microorganisms living in manure) (47) and indirect, including the following methods: (i) by improving the pH buffering capacity through manure application (48, 49), (ii) by addition of diverse available organic compounds or nutrients with manure, (iii) by modification of C:N:P ratios, resulting in a change in the stoichiometric ratio of food resources for microorganisms (50, 51), and (iv) by increasing and stabilizing soil aggregates, resulting in modification of microbial habitats (40, 52).
In both NPK and NPKM soils, the rice residue facilitated the growth of specific microbial groups, such as members of the Proteobacteria, Actinobacteria, and Ascomycota (Fig. 1 and 2). The bacterial genera Lysobacter and Streptomyces, which have been described to be involved in the breakdown of potato tissue, wheat straw, rice straw, and corn cellulose (26, 29, 53, 54), are also the dominant bacteria utilizing the rice residue (Fig. 4). Streptomyces is commonly used in industrial applications to produce laccase and xylanase, using rice or wheat straw as substrates (55, 56). Microbial laccase production is usually related to the ability of the microbe to degrade lignin (57), and lignin is also the most difficult decomposable straw component. The higher abundance of Streptomyces in NPK than in NPKM soil may be related to the higher content of recalcitrant components in NPK soil resulting from the 3-year application of mineral-only fertilizers (58).
Most of the fungal groups were associated with the breakdown of cellulose (Fig. 5) as they can produce a variety of cellulolytic enzymes. Fungal genera Trichocladium, Thielavia, Chaetomium, and Aspergillus can produce cellulase or xylanase (59–63). Chaetomium and Fusarium have been identified in previous SIP studies as fungal genera that can utilize crop straw or cellulose (21, 22, 64). The application of manure increased the relative abundance of Syncephalis, whereas the relative abundance of Trichoderma was reduced. Trichoderma possesses a well-developed cellulase system containing many endoglucanases and two exoglucanases (65, 66). Various organic substances, such as monosaccharides, polysaccharides, and chitin have been shown to induce enzyme secretion (67), which may indicate that Trichoderma is more competitive for rice residue than other genera. Trichoderma has a capacity to mobilize and take up nutrients superior to that of many other soil microbes, especially under oligotrophic conditions, making it more efficient and competitive (68). Thus, the manure application in NPKM soil during the 3 years reduced Trichoderma development.
Fertilization changed the cooccurrence patterns of microbes driving C flow from rice residues.
Network analyses provide a tool for identifying cooccurrence patterns and reflecting community organization (69–71) of microbes involved in the 13C-labeled residue C flow. Most previous studies have explored microbial molecular ecological networks in individual kingdoms, such as only fungi or only bacteria, but few showed the cooccurrence patterns between these groups. As microbial groups are not existing in isolation but potentially share niches in a given environment, network analyses were thus conducted on both bacteria and fungi (Fig. 6a). Linkages in microbial molecular networks can represent covariation between microbes or niche sharing (72). As our analysis might not have enough data points to construct a reliable network at each sample time, the network analyses were based on the combination of the samples across all sampling times for each fertilization regime. Pooling four sampling time points from each fertilization treatment in the network analysis can not only increase sensitivity for cooccurrence events but also show the cooccurrence patterns of microbes driving C flow from rice residues across the time points in each treated soil. Even though the microbial network will also be changed because of cross-feeding, the cross-feeding (microbes feeding on labeled microbial residues or microbial metabolites) also represents a type of C flow from rice residue through the microbial community. It is likely that the 13C label was turned over as the experiment progressed and that a second generation of microbial cells became labeled not only from 13C-labeled rice residue but also from 13C-labeled microbial metabolites, residues, and necromass of r-strategists that were formed earlier. Hence, it follows that our DNA-SIP data enabled the probing of C flow from rice residue into microbial communities in soils. Consequently, species that tended to negative correlations in the networks (Fig. 6a) indicate potential reuse of the microbial residue by some other microbes or potential competition in using rice residue. Conversely, any two species with a positive correlation reflect the potential cooperative or syntrophic in residue C utilization (Fig. 6a).
The network in NPKM soil was much more complex than that in NPK soil, with higher numbers of nodes, edges, average clustering coefficient, and average degree of the cooccurrences (Fig. 6a; see also Table S2 in the supplemental material). The complexity the of network is related to the changes in environmental factors, such as soil pH and C availability (31). An increased C input increases the functional complexity of soil microbial networks (31, 73). Three-year manure application fosters greater intermicrobial correlation and/or establishes more diverse shared niches, which represents a fundamental difference between NPKM and NPK soil. Keystone taxa acting as hubs or connectors in the network play critical roles in maintaining a network structure for other taxa (74). Fungal species, Jugulospora rotula in the NPK soil and Emericellopsis terricola in NPKM soil, were identified as keystone species (Fig. 6c; Table S3). Emericellopsis terricola was proved to have strong cellulase, protease, and laminarinase activities (75), which might be an essential factor for organizing microorganisms to cooperate in residue C utilization and may lead to a positive effect on the residue decomposition in NPKM soil. These keystone species, which are recognized as initiating components in networks, can also selectively alter microbial composition (76). Identifying these keystone groups is critical for predictive understanding of the subsequent potential for microbially mediated C sequestration when rice residue is returned to the soil. To better understand the role of these organisms in cooccurrence networks, uncultured keystone species should be the focus of future work.
Bacteria and fungi connected to each other in the network, and the organic amendment changed the ratio of the edges between bacteria and fungi to the total edges (Fig. 6b). In general, recalcitrant organic compounds in the residues (such as lignin and cellulose) are degraded mainly by fungi, and the released water-soluble substances can be utilized also by bacteria (77). Thus, fungi provide bacteria with resources that they cannot directly make from residues themselves, such as intermediate decomposition products. The 3-year input of manure increased C sources and their availability in the soil, including some soluble organics, which reduced the dependence of bacteria on fungi (78). In the NPK soil, which did not receive manure during the 3 years, bacteria strengthened their connection with fungi to get more soluble organics from residue decomposition. Thus, compared to the ratio in the NPK soil, NPKM fertilization decreased the ratio of edges between fungi and bacteria to the total edges (Fig. 6b). Therefore, the results provide some evidence that the cooccurrence patterns between the microbes are impacted by 3-year manure amendment. However, network cooccurrence patterns are ultimately based on statistical correlations, and they must be interpreted with caution as correlation does not directly demonstrate real microbial interactions. More solid experimental evidence, including more reasonable inference methods or promising culturing approaches, is still needed to explore complex ecological relationships under natural conditions and assess the effect of keystone species.
Response strategies of microbes by utilization of rice residues depend on fertilization.
Plant residues contain complex organic compounds, such as water-soluble substances (low-molecular-weight organic substrates), polymeric carbohydrates (hemicellulose and cellulose), lignin, and phenolics (79–81). Each component can be decomposed or utilized mainly by specific taxonomic groups of microorganisms. Thus, rice residue degradation could be linked to a continuous change of microbial beta diversity over time (Fig. 3). The dynamics of microbial community structure was often related to the chemical composition of straw (82, 83). Decomposition of easily degradable components can stimulate the growth of early r-strategists (84), and as the availability of resources decreases, the relative abundances of microorganisms within this group will gradually decrease, while the late K-strategist groups will gradually come to dominate (85). Thus, decomposition of labile components during the first week leads to the rapid growth of certain groups of fungi and bacteria, such as the bacterial genera Devosia, Glycomyces, and Kribbella, and then the relative abundances of these groups decrease as the labile organic components are gradually consumed (Fig. 7). The relative abundances of Rhodanobacter, Actinomadura, Streptomyces, and Nonomuraea increased gradually as these bacteria could decompose recalcitrant components at later time points (Fig. 7). However, since the classification of the response strategies was at the genus level, little can be known about whether these members were involved directly in rice residue degradation or only indirectly by cross-feeding on the intermediate or metabolic products and necromass of the r-strategists.
Bacterial groups had similar response strategies in the two fertilized soils by assimilation of rice residue-derived C. The pure-culture methods have shown that bacteria could not grow on cellulose or crop residues unless they were cultured together with fungi (86). Thus, bacteria rely on fungi and their oxidative enzymes when substrates that are difficult to decompose are used. Consequently, the oxidative exoenzymes secreted by fungi [e.g., (per)oxidases] degrade the recalcitrant components into low-molecular-weight organic substrates that can be further utilized by bacteria (Fig. 7). Thus, no significant differences in the response strategies of the bacteria were found between the two fertilization regimes. However, the response strategies of the fungal genera were quite different between NPK and NPKM soils, which may be due to changes in the composition of the rice residue during the degradation (86).
Conclusions.
The DNA-SIP approach enabled the identification of the key bacterial and fungal taxa of microbes involved in rice residue decomposition and utilization in soils under NPK and NPKM fertilization. The C from the rice residues was predominantly incorporated into bacterial phyla of Proteobacteria and Actinobacteria and the fungal phylum Ascomycota. Fertilization regimes had strong effects on microbial community composition, microbial response strategies, and microbial interactions involved in the C flow from rice residues. Since complete residue degradation involves multiple enzymatic steps that are conducted by many community members, our community-based analyses detected the microbial groups using the residues as well as the secondary products (metabolites) of initial residue cleavage. Bacterial response strategies for assimilating residues were independent of fertilization regimes. However, most of the fungal genera in soil receiving manure responded faster to residue addition than those in soil receiving solely mineral fertilizers. This is connected with the changes in the composition of the rice residue during degradation and with fungal adaptation (abundance and activity) to continuous manure input. This is confirmed by the more complex microbial network involved in the flow of rice residue-derived C in NPKM than that in NPK soils. Presumably, fungi release enzymes decomposing recalcitrant organic compounds in the residues (e.g., cellulose and lignin), and bacteria subsequently use the available soluble compounds produced by fungal exoenzymes. Future studies should be designed to discriminate the cross-feeding C flow between the species to understand mechanistic interactions of soil microbes that are directly and indirectly involved in residue-derived C utilization at any time point.
MATERIALS AND METHODS
Rice residue labeling.
Rice seeds (Oryza sativa cv. Zhendao11) were sterilized by exposure to 30% hydrogen peroxide for 30 min. Following a washing step, the rice seeds were cultured in the dark until germination. The seedlings were transferred into a transparent air-tight chamber and grown under hydroponic conditions with Hogland nutrient solution. The chamber was continually injected with 13CO2 (99 atom%; Cambridge Isotope Laboratories, Inc.) at a concentration of 400 μl liter−1. Additionally, before 13CO2 injection, high-purity air, which contained N2 (70%, vol/vol) and O2 (30%, vol/vol), was used to flush the chamber to remove the 12CO2. The environmental parameters of the illumination incubator were set as a 12-h photoperiod, temperature of 30°C day/20°C night, and relative humidity of 70% day/80% night. After 30 days of labeling, the top parts of the rice seedlings (including leaves and stems) were harvested and had a 13C content of 46 to 48 atom%. The rice residue was oven dried to constant weight at 65°C for further use.
Soil microcosm setup.
Soils used for this study were collected from an experimental farm in Rugao, Jiangsu, China (120°51′E, 32°00′N). Rice (Oryza sativa cv. Zhendao11) was rotated annually with winter wheat (Triticum aestivum cv. Yangmai16). Soil samples were collected from a short-term (3-year) experimental site that included treatment with only mineral fertilizers (NPK) and treatment with mineral fertilizers plus manure (NPKM). The NPK-treated soil annually received 440 kg N ha−1 applied as urea (240 kg N ha−1 for the rice season and 200 kg N ha−1 for the wheat season), whereas the NPKM-treated soil similarly received a total of 440 kg N ha−1 in both seasons, of which 80% was from urea and 20% was from 7.41 tons ha−1 compost of pig manure. Both the NPK and NPKM treatments were applied annually with 128 kg P ha−1 and 116 kg K ha−1 as calcium superphosphate and potassium chloride, respectively. Each treatment was applied to three plots, which served as three replicates. Soils were collected from the top 0 to 20 cm after rice harvest in October 2015. Three soil samples for each fertilization treatment were obtained from the corresponding plots. Each soil sample consisted of 10 soil cores that were then sieved through 2-mm-pore-size sieves. The NPK-treated soil had a pH of 6.34, 20.34 g SOM kg−1, 1.22 g total N kg−1, 12.97 mg Olsen-P kg−1, and 92.00 mg NH4OAc-K kg−1; the NPKM-treated soil had a pH of 6.68, 22.52 g SOM kg−1, 1.38 g total N kg−1, 29.36 mg Olsen-P kg−1, and 85.67 mg NH4OAc-K kg−1.
Each soil sample was subjected to three microcosm treatments, including a negative control (soil without rice residue), positive control (soil with 12C-labeled rice residue), and 13C treatment (soil with 13C-labeled rice residue), and each microcosm treatment group contained six replicates. Each replicate comprised a microcosm of 10 g (oven-dried basis) of soil in a 125-ml hermetically sealed serum bottle. In total, 36 microcosms [(2 fertilization treatments × 3 replicates) × (3 microcosm treatments × 2 replicates)] were employed in the incubation experiment. All microcosms were subjected to a 14-day preincubation with soil water content adjusted to 60% of the soil maximum water-holding capacity and with an incubation temperature of 25°C before the beginning of the experiments. This preincubation adjusted all of the treatments to the environmental conditions used in the incubation stage to make the rice residue addition the unique variable in the microcosm incubations. After preincubation, 0.05 g of 12C-labeled rice residue or 13C-labeled rice residue (5 mg g−1 dws) was thoroughly amended into the positive control or 13C-treatment soils, respectively. For each microcosm treatment, one was used for the measurement of microbial respiration, and the remaining one was used for sampling for DNA isolation. Sterile water was supplied to each microcosm to adjust the soil moisture content to 60% of the soil maximum water-holding capacity. All soil microcosms were incubated at 25°C in the dark for 60 days, with daily adjustment of the soil water content. Soil samples used for DNA extraction were collected after 7, 15, 30, and 60 days of incubation.
Measurements of soil respiration.
The measurement of microbial respiration was conducted at days 1, 7, 15, 30, and 60 after the addition of rice residue. The vials were closed with an air-tight butyl rubber stopper and incubated for 12 h at 25°C. Afterwards, the headspace CO2 concentration was analyzed by a gas chromatograph with a thermal conductivity detector operating at 60°C (Agilent 7890; Agilent, Santa Clara, CA). Separation was performed using a 177/149-mm (80/100 mesh) Chromosorb 102 column (Advanced Minerals, Santa Barbara, CA) at 40°C. The temperature of the injecting port was 100°C. The carrier gas (H2) flow rate was 80 ml min−1.
DNA extraction and gradient centrifugation.
Soil DNA was extracted by a FastDNA spin kit for soil (MP Biomedicals, Cleveland, OH) according to the manufacturer’s protocol. A NanoDrop ND-2000 UV-visible light (UV-Vis) spectrophotometer (Thermo Scientific, Wilmington, DE) was used to measure the concentration and quality of extracted soil DNA. DNA density gradient centrifugation and fractionation were performed according to the methods of Kong et al. (87). Briefly, 3 μg of DNA was added into 1.85 g ml−1 of CsCl gradient buffer (0.1 M Tris-HCl, 0.1 M KCl, 1 mM EDTA, pH 8.0) with an initial CsCl buoyant density of 1.725 g ml−1, which was prepared by adjusting the refractive index to 1.4025 with an AR200 digital hand-held refractometer (Reichert, Inc., Buffalo, NY). Density gradient centrifugation was performed in a 5.1-ml Quick-Seal polyallomer ultracentrifugation tube (Beckman Coulter, Palo Alto, CA) in a VTi 65.2 rotor (Beckman Coulter), which was subjected to centrifugation at 177,000 × g (45,000 rpm) for 44 h at 20°C (Optima-XPN-100; Beckman Coulter, USA). Centrifuged gradients were fractionated into 14 equal volumes (∼340 μl) by displacing the gradient medium with sterile water at the top of the tube using a syringe pump ( pump LSP01-1A; Longer, China). Then, 50-μl aliquots were used to measure the refractive index to determine the buoyant density of each collected fraction. Fractionated DNA was precipitated from CsCl by addition of 500 μl of precipitating agent (30% polyethylene glycol [PEG] 6000 and 1.6 M NaCl), incubation for 1 h at 37°C, and then two washes with 70% ethanol. The samples were then dissolved in 30 μl of Tris-EDTA buffer.
Real-time PCR quantification.
Quantitative real-time PCR was performed on an ABI 7500 real-time PCR system (Applied Biosystems, USA) to determine the abundances of bacterial 16S rRNA genes and fungal ITS1 using the primer pairs 338F/518R (338F, 5′-ACTCCTACGGGAGGCAGCAG-3′; 518R, 5′-ATTACCGCGGCTGCTGG-3′) (88) and ITS1/5.8s (ITS1, 5′-TCCGTAGGTGAACCTGCGG-3′; 5.8s, 5′-CGCTGCGTTCTTCATCG-3′) (89, 90), respectively, in the total soil DNA and fractionated DNA. Each reaction was performed in a total volume of 25 μl containing 12.5 μl of SYBR Premix Ex Taq (TaKaRa Biotechnology, Shiga, Japan), 0.5 μl of each primer (10 μM), 0.5 μl of ROX (6-carboxy-X-rhodamine) reference dye II (50×), 1 μl of DNA template (2 to 20 ng), and 10.5 μl of sterile water. Amplification conditions were as follows: 95°C for 5 min, 40 cycles of 15 s at 95°C and 34 s at 64°C, with a final temperature increase to 95°C for 15 s. Data were collected after each annealing step. One PCR product for each bacterial 16S rRNA gene and fungal ITS1 was gel purified using an Axygen PCR purification kit (Axygen Bio, USA), and the fragments were cloned separately into a pMD19-T vector (TaKaRa cloning kit). Then, ligated plasmid was transformed into competent DH5α Escherichia coli cells (TaKaRa). The reamplification and sequencing of white-positive clones using the vector-specific primers M13f/M13r were used to identify the correct insertion of the DNA fragments. Plasmids with correct inserts were extracted using an Axygen Plasmid Extraction kit (Axygen Bio, USA) for use as plasmid standards. A 10-fold dilution series of the plasmid standards was used to generate the standard curves. The PCR efficiency and correlation coefficient (R2) values for the standard curves were 95.68% and 0.9977, respectively, for 16S rRNA genes and 96.11% and 0.9978, respectively, for ITS1. The specificity of the amplification products was checked using melt curve analysis at the end of each PCR run and was then confirmed by standard agarose gel electrophoresis.
High-throughput sequencing analysis.
Based on the relative abundance distribution patterns of the bacterial 16S rRNA genes and fungal ITS1 in 14 CsCl gradients, heavy fractions of the 13C-treatment samples and the corresponding fractions from the positive controls were chosen for sequencing. In total, 24 DNA samples (2 soils × 3 replicates × 4 sampling time points) were collected from the 13C-treatment heavy fractions, and 24 DNA samples (2 soils × 3 replicates × 4 sampling time points) were collected from the corresponding positive-control fractions. Sequencing of the V4-V5 regions from bacterial 16S rRNA genes (91) and of the fungal ITS1 (92) was conducted using an Illumina MiSeq System (Roche, Switzerland) at the Nanjing Genepioneer company (Nanjing, China). The raw data were processed using the Quantitative Insights into Microbial Ecology (QIIME) toolkit (93) and UPARSE standard pipeline (94). Reads with a quality score of less than 20 and length of less than 220 bp were discarded. Paired reads were merged using the fastq_mergepairs module of USEARCH (94, 95); pairs with mismatches in the overlap were eliminated, and only sequences of >220 bp in length were included in subsequent analyses. Then, singletons were discarded using UPARSE. In addition, the SILVA reference database was used to identify and remove putative chimeras. Taxonomic assignment was accomplished by clustering the sequences into operational taxonomic units (OTUs) at a 97% similarity level. Each sample was rarefied to the same number of reads (18,000 16S rRNA reads and 20,000 ITS1 reads) for downstream analyses using Mothur software. One representative sequence from each OTU was selected using the UPARSE pipeline, and in total 5,029 bacterial OTUs and 1,864 fungal OTUs were obtained in the OTU table (see Table S4 in the supplemental material). Each representative sequence was assigned to a Ribosomal Database Project (RDP) classifier for identification using a threshold of 0.8.
Statistical analysis.
The OTUs involved in the decomposition and assimilation of 13C-labeled residue were filtered by comparing their relative abundances in the heavy fraction of 13C-treatment samples to those in the positive-control samples using R software with the DESeq2 package (96, 97). Unfortunately, the 16S rRNA sequencing data for one replicate in the NPKM positive control at 30 days was observed to be of much lower quality; thus, the mean of the NPKM positive control at 30 days was calculated based on the remaining two replicates for this analysis. Based on the result that the atomic percentage of 13C in the rice residue in the present study was lower than 50%, both the padj (false-discovery rate [FDR] adjusted P value) and the log2 fold change value were strictly controlled in subsequent analyses. The 13C-labeled OTUs were defined as the OTUs with padj values lower than 0.01 and log2 fold change values higher than 1. Nonmetric multidimensional scaling (NMDS) based on Bray-Curtis distances of all labeled OTUs and permutational multivariate analysis of variance (PERMANOVA) were performed using R (version 2.15.0) with the Vegan package. 13C-labeled OTUs with relative abundances higher than 1% were defined as the dominant OTUs involved in 13C-labeled residue decomposition and assimilation. Only genera that passed our filters for significant changes over time by analysis of variance (ANOVA) were included the assessment of response strategies for rice residue assimilation. Labeled OTUs observed in four sampling time points from each fertilization treatment were pooled to assess the roles of the microbes in driving the flow of rice residue-derived carbon using molecular ecological networks. The labeled OTUs with average relative abundances greater than 0.1% were selected to construct molecular ecological networks using the random matrix theory (RMT) (98), and the networks were visualized by Gephi (0.9.2). The threshold values of Zi (within-module connectivity) and Pi (among-module connectivity) for categorizing OTUs in networks are 2.5 and 0.62, respectively (74, 99). Furthermore, three categories that are organized as module hubs (Zi > 2.5), connectors (Pi > 0.62), and network hubs (Zi > 2.5 and Pi > 0.62) in the networks are defined as keystone microbes in the decomposition and assimilation of 13C-labeled rice residue. Origin, version 8.0, was used to draw the histogram; variance analysis was conducted using SPSS software (SPSS, version 16.0, for Windows; IBM Corp., Armonk, NY), and a P value of < 0.05 was defined as statistically significant.
Data availability.
The sequences obtained in this study have been submitted to NCBI Sequence Read Archive (SRA) under the accession numbers PRJNA520803 and PRJNA518924.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by National Natural Science Foundation of China (41977080), and the publication was prepared with the support of the RUDN University Program 5-100.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Huang Y, Eglinton G, Van der Hage E, Boon J, Bol R, Ineson P. 1998. Dissolved organic matter and its parent organic matter in grass upland soil horizons studied by analytical pyrolysis techniques. Eur J Soil Sci 49:1–15. doi: 10.1046/j.1365-2389.1998.00141.x. [DOI] [Google Scholar]
- 2.Gleixner G, Bol R, Balesdent J. 1999. Molecular insight into soil carbon turnover. Rapid Commun Mass Spectrom 13:1278–1283. doi:. [DOI] [PubMed] [Google Scholar]
- 3.Allison SD, Lu Y, Weihe C, Goulden ML, Martiny AC, Treseder KK, Martiny JB. 2013. Microbial abundance and composition influence litter decomposition response to environmental change. Ecology 94:714–725. doi: 10.1890/12-1243.1. [DOI] [PubMed] [Google Scholar]
- 4.Nuccio EE, Hodge A, Pett‐Ridge J, Herman DJ, Weber PK, Firestone MK. 2013. An arbuscular mycorrhizal fungus significantly modifies the soil bacterial community and nitrogen cycling during litter decomposition. Environ Microbiol 15:1870–1881. doi: 10.1111/1462-2920.12081. [DOI] [PubMed] [Google Scholar]
- 5.De Graaff MA, Classen AT, Castro HF, Schadt CW. 2010. Labile soil carbon inputs mediate the soil microbial community composition and plant residue decomposition rates. New Phytol 188:1055–1064. doi: 10.1111/j.1469-8137.2010.03427.x. [DOI] [PubMed] [Google Scholar]
- 6.Aneja MK, Sharma S, Munch JC, Schloter M. 2004. RNA fingerprinting—a new method to screen for differences in plant litter degrading microbial communities. J Microbiol Methods 59:223–231. doi: 10.1016/j.mimet.2004.07.005. [DOI] [PubMed] [Google Scholar]
- 7.Prescott CE. 2010. Litter decomposition: what controls it and how can we alter it to sequester more carbon in forest soils? Biogeochemistry 101:133–149. doi: 10.1007/s10533-010-9439-0. [DOI] [Google Scholar]
- 8.Padmanabhan P, Padmanabhan S, DeRito C, Gray A, Gannon D, Snape JR, Tsai CS, Park W, Jeon C, Madsen EL. 2003. Respiration of 13C-labeled substrates added to soil in the field and subsequent 16S rRNA gene analysis of 13C-labeled soil DNA. Appl Environ Microbiol 69:1614–1622. doi: 10.1128/aem.69.3.1614-1622.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kong Y, Zhu C, Ruan Y, Luo G, Wang M, Ling N, Shen Q, Guo S. 2018. Are the microbial communities involved in glucose assimilation in paddy soils treated with different fertilization regimes for three years similar? J Soils Sediments 18:2476–2490. doi: 10.1007/s11368-018-1961-z. [DOI] [Google Scholar]
- 10.Zhang H, Ding W, Luo J, Bolan N, Yu H. 2015. The dynamics of glucose-derived 13C incorporation into aggregates of a sandy loam soil following two-decade compost or inorganic fertilizer amendments. Soil Tillage Res 148:14–19. doi: 10.1016/j.still.2014.11.010. [DOI] [Google Scholar]
- 11.Dungait JAJ, Kemmitt SJ, Michallon L, Guo S, Wen Q, Brookes PC, Evershed RP. 2011. Variable responses of the soil microbial biomass to trace concentrations of 13C-labelled glucose, using 13C-PLFA analysis. Eur J Soil Sci 62:117–126. doi: 10.1111/j.1365-2389.2010.01321.x. [DOI] [Google Scholar]
- 12.Lemanski K, Scheu S. 2014. Incorporation of 13C labelled glucose into soil microorganisms of grassland: effects of fertilizer addition and plant functional group composition. Soil Biol Biochem 69:38–45. doi: 10.1016/j.soilbio.2013.10.034. [DOI] [Google Scholar]
- 13.Radajewski S, Webster G, Reay DS, Morris SA, Ineson P, Nedwell DB, Prosser JI, Murrell JC. 2002. Identification of active methylotroph populations in an acidic forest soil by stable-isotope probing. Microbiology 148:2331–2342. doi: 10.1099/00221287-148-8-2331. [DOI] [PubMed] [Google Scholar]
- 14.Ginige MP, Hugenholtz P, Daims H, Wagner M, Keller J, Blackall LL. 2004. Use of stable-isotope probing, full-cycle rRNA analysis, and fluorescence in situ hybridization-microautoradiography to study a methanol-fed denitrifying microbial community. Appl Environ Microbiol 70:588–596. doi: 10.1128/aem.70.1.588-596.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lueders T, Pommerenke B, Friedrich MW. 2004. Stable-isotope probing of microorganisms thriving at thermodynamic limits: syntrophic propionate oxidation in flooded soil. Appl Environ Microbiol 70:5778–5786. doi: 10.1128/AEM.70.10.5778-5786.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ai C, Liang G, Sun J, Wang X, He P, Zhou W, He X. 2015. Reduced dependence of rhizosphere microbiome on plant-derived carbon in 32-year long-term inorganic and organic fertilized soils. Soil Biol Biochem 80:70–78. doi: 10.1016/j.soilbio.2014.09.028. [DOI] [Google Scholar]
- 17.Wang J, Chapman SJ, Yao H. 2016. Incorporation of 13C-labelled rice rhizodeposition into soil microbial communities under different fertilizer applications. Appl Soil Ecol 101:11–19. doi: 10.1016/j.apsoil.2016.01.010. [DOI] [Google Scholar]
- 18.Yuan H, Zhu Z, Liu S, Ge T, Jing H, Li B, Liu Q, Lynn TM, Wu J, Kuzyakov Y. 2016. Microbial utilization of rice root exudates: 13C labeling and PLFA composition. Biol Fertil Soils 52:615–627. doi: 10.1007/s00374-016-1101-0. [DOI] [Google Scholar]
- 19.Hannula SE, Boschker HTS, de Boer W, van Veen JA. 2012. 13C pulse-labeling assessment of the community structure of active fungi in the rhizosphere of a genetically starch-modified potato (Solanum tuberosum) cultivar and its parental isoline. New Phytol 194:784–799. doi: 10.1111/j.1469-8137.2012.04089.x. [DOI] [PubMed] [Google Scholar]
- 20.Oburger E, Gruber B, Wanek W, Watzinger A, Stanetty C, Schindlegger Y, Hann S, Schenkeveld WDC, Kraemer SM, Puschenreiter M. 2016. Microbial decomposition of 13C-labeled phytosiderophores in the rhizosphere of wheat: mineralization dynamics and key microbial groups involved. Soil Biol Biochem 98:196–207. doi: 10.1016/j.soilbio.2016.04.014. [DOI] [Google Scholar]
- 21.Stursová M, Zifčáková L, Leigh MB, Burgess R, Baldrian P. 2012. Cellulose utilization in forest litter and soil: identification of bacterial and fungal decomposers. FEMS Microbiol Ecol 80:735–746. doi: 10.1111/j.1574-6941.2012.01343.x. [DOI] [PubMed] [Google Scholar]
- 22.Eichorst SA, Kuske CR. 2012. Identification of cellulose-responsive bacterial and fungal communities in geographically and edaphically different soils by using stable isotope probing. Appl Environ Microbiol 78:2316–2327. doi: 10.1128/AEM.07313-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Williams MA, Myrold DD, Bottomley PJ. 2006. Distribution and fate of 13C-labeled root and straw residues from ryegrass and crimson clover in soil under western Oregon field conditions. Biol Fertil Soils 42:523–531. doi: 10.1007/s00374-005-0046-5. [DOI] [Google Scholar]
- 24.Williams MA, Myrold DD, Bottomley PJ. 2006. Carbon flow from 13C-labeled straw and root residues into the phospholipid fatty acids of a soil microbial community under field conditions. Soil Biol Biochem 38:759–768. doi: 10.1016/j.soilbio.2005.07.001. [DOI] [Google Scholar]
- 25.Shrestha M, Shrestha PM, Conrad R. 2011. Bacterial and archaeal communities involved in the in situ degradation of 13C-labelled straw in the rice rhizosphere. Environ Microbiol Rep 3:587–596. doi: 10.1111/j.1758-2229.2011.00267.x. [DOI] [PubMed] [Google Scholar]
- 26.Fan F, Yin C, Tang Y, Li Z, Song A, Wakelin SA, Zou J, Liang Y. 2014. Probing potential microbial coupling of carbon and nitrogen cycling during decomposition of maize residue by 13C-DNA-SIP. Soil Biol Biochem 70:12–21. doi: 10.1016/j.soilbio.2013.12.002. [DOI] [Google Scholar]
- 27.Lee CG, Watanabe T, Sato Y, Murase J, Asakawa S, Kimura M. 2011. Bacterial populations assimilating carbon from 13C-labeled plant residue in soil: analysis by a DNA-SIP approach. Soil Biol Biochem 43:814–822. doi: 10.1016/j.soilbio.2010.12.016. [DOI] [Google Scholar]
- 28.Bernard L, Mougel C, Maron P-A, Nowak V, Lévêque J, Henault C, Haichar FEZ, Berge O, Marol C, Balesdent J, Gibiat F, Lemanceau P, Ranjard L. 2007. Dynamics and identification of soil microbial populations actively assimilating carbon from 13C-labelled wheat residue as estimated by DNA- and RNA-SIP techniques. Environ Microbiol 9:752–764. doi: 10.1111/j.1462-2920.2006.01197.x. [DOI] [PubMed] [Google Scholar]
- 29.Semenov AV, Pereira e Silva MC, Szturc-Koestsier AE, Schmitt H, Falcão Salles J, van Elsas JD. 2012. Impact of incorporated fresh 13C potato tissues on the bacterial and fungal community composition of soil. Soil Biol Biochem 49:88–95. doi: 10.1016/j.soilbio.2012.02.016. [DOI] [Google Scholar]
- 30.Sun HY, Deng SP, Raun WR. 2004. Bacterial community structure and diversity in a century-old manure-treated agroecosystem. Appl Environ Microbiol 70:5868–5874. doi: 10.1128/AEM.70.10.5868-5874.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ling N, Zhu C, Xue C, Chen H, Duan Y, Peng C, Guo S, Shen Q. 2016. Insight into how organic amendments can shape the soil microbiome in long-term field experiments as revealed by network analysis. Soil Biol Biochem 99:137–149. doi: 10.1016/j.soilbio.2016.05.005. [DOI] [Google Scholar]
- 32.Su J-Q, Ding L-J, Xue K, Yao H-Y, Quensen J, Bai S-J, Wei W-X, Wu J-S, Zhou J, Tiedje JM, Zhu Y-G. 2015. Long-term balanced fertilization increases the soil microbial functional diversity in a phosphorus-limited paddy soil. Mol Ecol 24:136–150. doi: 10.1111/mec.13010. [DOI] [PubMed] [Google Scholar]
- 33.Hartmann M, Frey B, Mayer J, Mäder P, Widmer F. 2015. Distinct soil microbial diversity under long-term organic and conventional farming. ISME J 9:1177–1194. doi: 10.1038/ismej.2014.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sinsabaugh RL, Lauber CL, Weintraub MN, Ahmed B, Allison SD, Crenshaw C, Contosta AR, Cusack D, Frey S, Gallo ME, Gartner TB, Hobbie SE, Holland K, Keeler BL, Powers JS, Stursova M, Takacs-Vesbach C, Waldrop MP, Wallenstein MD, Zak DR, Zeglin LH. 2008. Stoichiometry of soil enzyme activity at global scale. Ecol Lett 11:1252–1264. doi: 10.1111/j.1461-0248.2008.01245.x. [DOI] [PubMed] [Google Scholar]
- 35.Peacock AD, Mullen MD, Ringelberg DB, Tyler DD, Hedrick DB, Gale PM, White DC. 2001. Soil microbial community responses to dairy manure or ammonium nitrate applications. Soil Biol Biochem 33:1011–1019. doi: 10.1016/S0038-0717(01)00004-9. [DOI] [Google Scholar]
- 36.Enwall K, Nyberg K, Bertilsson S, Cederlund H, Stenström J, Hallin S. 2007. Long-term impact of fertilization on activity and composition of bacterial communities and metabolic guilds in agricultural soil. Soil Biol Biochem 39:106–115. doi: 10.1016/j.soilbio.2006.06.015. [DOI] [Google Scholar]
- 37.Chen C, Zhang J, Lu M, Qin C, Chen Y, Yang L, Huang Q, Wang J, Shen Z, Shen Q. 2016. Microbial communities of an arable soil treated for 8 years with organic and inorganic fertilizers. Biol Fertil Soils 52:455–467. doi: 10.1007/s00374-016-1089-5. [DOI] [Google Scholar]
- 38.Schlesinger WH. 2000. Carbon sequestration in soils: some cautions amidst optimism. Agric Ecosyst Environ 82:121–127. doi: 10.1016/S0167-8809(00)00221-8. [DOI] [Google Scholar]
- 39.Zhen Z, Liu H, Wang N, Guo L, Meng J, Ding N, Wu G, Jiang G. 2014. Effects of manure compost application on soil microbial community diversity and soil microenvironments in a temperate cropland in China. PLoS One 9:e108555. doi: 10.1371/journal.pone.0108555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Majumder B, Kuzyakov Y. 2010. Effect of fertilization on decomposition of 14C labelled plant residues and their incorporation into soil aggregates. Soil Tillage Res 109:94–102. doi: 10.1016/j.still.2010.05.003. [DOI] [Google Scholar]
- 41.Wang J, Xue C, Song Y, Wang L, Huang Q, Shen Q. 2016. Wheat and rice growth stages and fertilization regimes alter soil bacterial community structure, but not diversity Front Microbiol 7:1207. doi: 10.3389/fmicb.2016.01207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Blaud A, Lerch TZ, Chevallier T, Nunan N, Chenu C, Brauman A. 2012. Dynamics of bacterial communities in relation to soil aggregate formation during the decomposition of 13C-labelled rice straw. Appl Soil Ecol 53:1–9. doi: 10.1016/j.apsoil.2011.11.005. [DOI] [Google Scholar]
- 43.Devêvre OC, Horwáth WR. 2000. Decomposition of rice straw and microbial carbon use efficiency under different soil temperatures and moistures. Soil Biol Biochem 32:1773–1785. doi: 10.1016/S0038-0717(00)00096-1. [DOI] [Google Scholar]
- 44.McMahon SK, Williams MA, Bottomley PJ, Myrold DD. 2005. Dynamics of microbial communities during decomposition of carbon-13 labeled ryegrass fractions in soil. Soil Sci Soc Am J 69:1238–1247. doi: 10.2136/sssaj2004.0289. [DOI] [Google Scholar]
- 45.Schutter M, Dick R. 2001. Shifts in substrate utilization potential and structure of soil microbial communities in response to carbon substrates. Soil Biol Biochem 33:1481–1491. doi: 10.1016/S0038-0717(01)00057-8. [DOI] [Google Scholar]
- 46.Neumann D, Heuer A, Hemkemeyer M, Martens R, Tebbe CC. 2014. Importance of soil organic matter for the diversity of microorganisms involved in the degradation of organic pollutants. ISME J 8:1289–1300. doi: 10.1038/ismej.2013.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fuchs JG. 2010. Interactions between beneficial and harmful microorganisms: from the composting process to compost application, p 213–229. In Insam H, Franke-Whittle I, Goberna M (ed), Microbes at work. Springer, Berlin, Germany. [Google Scholar]
- 48.Zhang Y, Shen H, He X, Thomas BW, Lupwayi NZ, Hao X, Thomas MC, Shi X. 2017. Fertilization shapes bacterial community structure by alteration of soil pH. Front Microbiol 8:1325. doi: 10.3389/fmicb.2017.01325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rousk J, Brookes PC, Bååth E. 2010. The microbial PLFA composition as affected by pH in an arable soil. Soil Biol Biochem 42:516–520. doi: 10.1016/j.soilbio.2009.11.026. [DOI] [Google Scholar]
- 50.Qaswar M, Ahmed W, Jing H, Hongzhu F, Xiaojun S, Xianjun J, Kailou L, Yongmei X, Zhongqun H, Asghar W, Shah A, Zhang H. 2019. Soil carbon (C), nitrogen (N) and phosphorus (P) stoichiometry drives phosphorus lability in paddy soil under long-term fertilization: a fractionation and path analysis study. PLoS One 14:e0218195. doi: 10.1371/journal.pone.0218195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sileshi GW, Nhamo N, Mafongoya PL, Tanimu J. 2017. Stoichiometry of animal manure and implications for nutrient cycling and agriculture in sub-Saharan Africa. Nutr Cycling Agroecosyst 107:91–105. doi: 10.1007/s10705-016-9817-7. [DOI] [Google Scholar]
- 52.Liang Q, Chen H, Gong Y, Yang H, Fan M, Kuzyakov Y. 2014. Effects of 15 years of manure and inorganic fertilizers on enzyme activities in particle-size fractions in a North China Plain soil. Eur J Soil Biol 60:112–119. doi: 10.1016/j.ejsobi.2013.11.009. [DOI] [Google Scholar]
- 53.Schellenberger S, Kolb S, Drake HL. 2010. Metabolic responses of novel cellulolytic and saccharolytic agricultural soil Bacteria to oxygen. Environ Microbiol 12:845–861. doi: 10.1111/j.1462-2920.2009.02128.x. [DOI] [PubMed] [Google Scholar]
- 54.España M, Rasche F, Kandeler E, Brune T, Rodriguez B, Bending GD, Cadisch G. 2011. Identification of active bacteria involved in decomposition of complex maize and soybean residues in a tropical Vertisol using N-DNA stable isotope probing. Pedobiologia 54:187–193. doi: 10.1016/j.pedobi.2011.03.001. [DOI] [Google Scholar]
- 55.Berrocal M, Ball AS, Huerta S, Barrasa JM, Hernández M, Pérez-Leblic MI, Arias ME. 2000. Biological upgrading of wheat straw through solid-state fermentation with Streptomyces cyaneus. Appl Microbiol Biotechnol 54:764–771. doi: 10.1007/s002530000454. [DOI] [PubMed] [Google Scholar]
- 56.Niladevi KN, Sukumaran RK, Prema P. 2007. Utilization of rice straw for laccase production by Streptomyces psammoticus in solid-state fermentation. J Ind Microbiol Biotechnol 34:665–674. doi: 10.1007/s10295-007-0239-z. [DOI] [PubMed] [Google Scholar]
- 57.Zavarzina AG, Semenova TA, Belova OV, Lisov AV, Leontievskii AA, Ivanova A. 2018. Laccase production and humic acids decomposition by microscopic soil Fungi. Microbiology 87:308–316. doi: 10.1134/S0026261718030153. [DOI] [Google Scholar]
- 58.Xun W, Huang T, Zhao J, Ran W, Wang B, Shen Q, Zhang R. 2015. Environmental conditions rather than microbial inoculum composition determine the bacterial composition, microbial biomass and enzymatic activity of reconstructed soil microbial communities. Soil Biol Biochem 90:10–18. doi: 10.1016/j.soilbio.2015.07.018. [DOI] [Google Scholar]
- 59.Shah AR, Madamwar D. 2005. Xylanase production by a newly isolated Aspergillus foetidus strain and its characterization. Process Biochem 40:1763–1771. doi: 10.1016/j.procbio.2004.06.041. [DOI] [Google Scholar]
- 60.Katapodis P, Christakopoulou V, Kekos D, Christakopoulos P. 2007. Optimization of xylanase production by Chaetomium thermophilum in wheat straw using response surface methodology. Biochem Eng J 35:136–141. doi: 10.1016/j.bej.2007.01.007. [DOI] [Google Scholar]
- 61.Singh A, Singh N, Bishnoi NR. 2009. Production of cellulases by Aspergillus heteromorphus from wheat straw under submerged fermentation. Int J Civil Environ Eng 1:23–26. [Google Scholar]
- 62.Kapoor N, Tyagi M, Kumar H, Arya A, Siddiqui MA, Amir A, Malik AS. 2010. Production of cellulase enzyme by Chaetomium sp. using wheat straw in solid state fermentation. Res J Microbiol 5:1199–1206. doi: 10.3923/jm.2010.1199.1206. [DOI] [Google Scholar]
- 63.Berka RM, Grigoriev IV, Otillar R, Salamov A, Grimwood J, Reid I, Ishmael N, John T, Darmond C, Moisan M-C, Henrissat B, Coutinho PM, Lombard V, Natvig DO, Lindquist E, Schmutz J, Lucas S, Harris P, Powlowski J, Bellemare A, Taylor D, Butler G, de Vries RP, Allijn IE, van den Brink J, Ushinsky S, Storms R, Powell AJ, Paulsen IT, Elbourne LDH, Baker SE, Magnuson J, Laboissiere S, Clutterbuck AJ, Martinez D, Wogulis M, de Leon AL, Rey MW, Tsang A. 2011. Comparative genomic analysis of the thermophilic biomass-degrading fungi Myceliophthora thermophila and Thielavia terrestris. Nat Biotechnol 29:922–927. doi: 10.1038/nbt.1976. [DOI] [PubMed] [Google Scholar]
- 64.Kramer S, Dibbern D, Moll J, Huenninghaus M, Koller R, Krueger D, Marhan S, Urich T, Wubet T, Bonkowski M, Buscot F, Lueders T, Kandeler E. 2016. Resource partitioning between bacteria, fungi, and protists in the detritusphere of an agricultural soil. Front Microbiol 7:1524. doi: 10.3389/fmicb.2016.01524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Medve J. 1997. Cellulose hydrolysis by Trichoderma reesei cellulases: studies on adsorption, sugar production and synergism of cellobiohydrolase I, II and endoglucanase II. PhD dissertation Lund University, Lund, Sweden. [Google Scholar]
- 66.Jorgensen H, Morkeberg A, Krogh KBR, Olsson L. 2005. Production of cellulases and hemicellulases by three Penicillium species: effect of substrate and evaluation of cellulase adsorption by capillary electrophoresis. Enzyme Microb Technol 36:42–48. doi: 10.1016/j.enzmictec.2004.03.023. [DOI] [Google Scholar]
- 67.Mach RL, Peterbauer CK, Payer K, Jaksits S, Woo SL, Zeilinger S, Kullnig CM, Lorito M, Kubicek CP. 1999. Expression of two major chitinase genes of Trichoderma atroviride (T. harzianum P1) is triggered by different regulatory signals. Appl Environ Microbiol 65:1858–1863. doi: 10.1128/AEM.65.5.1858-1863.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Benítez T, Rincón AM, Limón AM, Codón MC, Antonio C. 2004. Biocontrol mechanisms of Trichoderma strains. Int Microbiol 7:249–260. [PubMed] [Google Scholar]
- 69.Faust K, Raes J. 2012. Microbial interactions: from networks to models. Nat Rev Microbiol 10:538–550. doi: 10.1038/nrmicro2832. [DOI] [PubMed] [Google Scholar]
- 70.Hallam SJ, McCutcheon JP. 2015. Microbes don’t play solitaire: how cooperation trumps isolation in the microbial world. Environ Microbiol Rep 7:26–28. doi: 10.1111/1758-2229.12248. [DOI] [PubMed] [Google Scholar]
- 71.Ren D, Madsen JS, Sørensen SJ, Burmølle M. 2015. High prevalence of biofilm synergy among bacterial soil isolates in cocultures indicates bacterial interspecific cooperation. ISME J 9:81–89. doi: 10.1038/ismej.2014.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Berry D, Widder S. 2014. Deciphering microbial interactions and detecting keystone species with co-occurrence networks. Front Microbiol 5:219. doi: 10.3389/fmicb.2014.00219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhou J, Deng Y, Luo F, He Z, Yang Y. 2011. Phylogenetic molecular ecological network of soil microbial communities in response to elevated CO2. mBio 2:e00122-11. doi: 10.1128/mBio.00122-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Olesen JM, Bascompte J, Dupont YL, Jordano P. 2007. The modularity of pollination networks. Proc Natl Acad Sci U S A 104:19891–19896. doi: 10.1073/pnas.0706375104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zuccaro A, Summerbell RC, Gams W, Schroers HJ, Mitchell JI. 2004. A new Acremonium species associated with Fucus spp., and its affinity with a phylogenetically distinct marine Emericellopsis clade. Stud Mycol 50:283–297. [Google Scholar]
- 76.Banerjee S, Schlaeppi K, van der Heijden M. 2018. Keystone taxa as drivers of microbiome structure and functioning. Nat Rev Microbiol 16:567–576. doi: 10.1038/s41579-018-0024-1. [DOI] [PubMed] [Google Scholar]
- 77.Boer W, Folman LB, Summerbell RC, Boddy L. 2005. Living in a fungal world: impact of fungi on soil bacterial niche development. FEMS Microbiol Rev 29:795–811. doi: 10.1016/j.femsre.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 78.Zhang B, Zhang J, Liu Y, Shi P, Wei G. 2018. Co-occurrence patterns of soybean rhizosphere microbiome at a continental scale. Soil Biol Biochem 118:178–186. doi: 10.1016/j.soilbio.2017.12.011. [DOI] [Google Scholar]
- 79.Heal OW, Anderson JM, Swift MJ. 1997. Plant litter quality and decomposition: an historical overview, p 3–30. In Cadisch G, Giller KE (ed), Driven by nature: plant litter quality and decomposition. CAB International, Wallingford, United Kingdom. [Google Scholar]
- 80.Puget P, Drinkwater LE. 2001. Short-term dynamics of root- and shoot-derived carbon from a leguminous green manure. Soil Sci Soc Am J 65:771–779. doi: 10.2136/sssaj2001.653771x. [DOI] [Google Scholar]
- 81.Wardle DA, Bonner KI, Barker GM. 2002. Linkages between plant litter decomposition, litter quality, and vegetation responses to herbivores. Funct Ecol 16:585–595. doi: 10.1046/j.1365-2435.2002.00659.x. [DOI] [Google Scholar]
- 82.Rui J, Peng J, Lu Y. 2009. Succession of bacterial populations during plant residue decomposition in rice field soil. Appl Environ Microbiol 75:4879–4886. doi: 10.1128/AEM.00702-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Marschner P, Umar S, Baumann K. 2011. The microbial community composition changes rapidly in the early stages of decomposition of wheat residue. Soil Biol Biochem 43:445–451. doi: 10.1016/j.soilbio.2010.11.015. [DOI] [Google Scholar]
- 84.Kuzyakov Y. 2010. Priming effects: interactions between living and dead organic matter. Soil Biol Biochem 42:1363–1371. doi: 10.1016/j.soilbio.2010.04.003. [DOI] [Google Scholar]
- 85.Fontaine S, Mariotti A, Abbadie L. 2003. The priming effect of organic matter: a question of microbial competition? Soil Biol Biochem 35:837–843. doi: 10.1016/S0038-0717(03)00123-8. [DOI] [Google Scholar]
- 86.Romaní AM, Fischer H, Mille-Lindblom C, Tranvik LJ. 2006. Interactions of bacteria and fungi on decomposing litter: differential extracellular enzyme activities. Ecology 87:2559–2569.2.0.co;2] doi: 10.1890/0012-9658(2006)87[2559:IOBAFO]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 87.Kong Y, Ling N, Xue C, Chen H, Ruan Y, Guo J, Zhu C, Wang M, Shen Q, Guo S. 2019. Long‐term fertilization regimes change soil nitrification potential by impacting active autotrophic ammonia oxidizers and nitrite oxidizers as assessed by DNA stable isotope probing. Environ Microbiol 21:1224–1240. doi: 10.1111/1462-2920.14553. [DOI] [PubMed] [Google Scholar]
- 88.Polz MF, Cavanaugh CM. 1998. Bias in template-to-product ratios in multitemplate PCR. Appl Environ Microbiol 64:3724–3730. doi: 10.1128/AEM.64.10.3724-3730.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gardes M, Bruns TD. 1993. ITS primers with enhanced specificity for basidiomycetes—application to the identification of mycorrhizae and rusts. Mol Ecol 2:113–118. doi: 10.1111/j.1365-294x.1993.tb00005.x. [DOI] [PubMed] [Google Scholar]
- 90.Vilgalys R, Hester M. 1990. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J Bacteriol 172:4238–4246. doi: 10.1128/jb.172.8.4238-4246.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Stubner S. 2002. Enumeration of 16S rDNA of Desulfotomaculum lineage 1 in rice field soil by real-time PCR with SybrGreen detection. J Microbiol Methods 50:155–164. doi: 10.1016/S0167-7012(02)00024-6. [DOI] [PubMed] [Google Scholar]
- 92.White TJ, Bruns T, Lee S, Taylor J. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics, p 315–322. In Innis MA, Gelfand DH, Srinksky JJ, White TJ (ed), PCR protocols: a guide to methods and applications. Academic Press, San Diego, CA. [Google Scholar]
- 93.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Edgar RC. 2013. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods 10:996–998. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 95.Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 96.McMurdie PJ, Holmes S. 2014. Waste not, want not: why rarefying microbiome data is inadmissible. PLoS Comput Biol 10:e1003531. doi: 10.1371/journal.pcbi.1003531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pepe-Ranney C, Campbell AN, Koechli CN, Berthrong S, Buckley DH. 2016. Unearthing the Ecology of soil microorganisms using a high resolution DNA-SIP approach to explore cellulose and xylose metabolism in soil. Front Microbiol 7:703. doi: 10.3389/fmicb.2016.00703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Deng Y, Jiang Y-H, Yang Y, He Z, Luo F, Zhou J. 2012. Molecular ecological network analyses. BMC Bioinformatics 13:113. doi: 10.1186/1471-2105-13-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Guimerà R, Nunes Amaral LA. 2005. Functional cartography of complex metabolic networks. Nature 433:895–900. doi: 10.1038/nature03288. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequences obtained in this study have been submitted to NCBI Sequence Read Archive (SRA) under the accession numbers PRJNA520803 and PRJNA518924.