Most microbes de novo synthesize compatible solutes for adaptation to salt stress or fluctuating salinity environments. However, to date, one of the core questions involved in these physiological processes, i.e., the regulation of salt-induced compatible solute biosynthesis, is still not well understood. Here, this issue was systematically investigated by employing the model freshwater cyanobacterium Synechococcus elongatus PCC 7942. A novel mechanism for cyanobacterial adaption to salt stress and fluctuating salinity, i.e., the ion-induced synergistic modulation of key synthesizing and degrading enzymes of compatible solutes, is proposed. Because the ion-induced activation/inhibition of enzymes is a fast and efficient process, it may represent a common strategy of microbes for adaptation to environments with fluctuating salinity.
KEYWORDS: cyanobacteria, salt stress, salt acclimation, compatible solute
ABSTRACT
Salinity is one of the most important abiotic factors in various natural habitats of microbes. Cyanobacteria are the most widely distributed family of photosynthetic microorganisms in environments with fluctuating salinity. In response to salt stress, many cyanobacteria de novo synthesize compatible solutes to maintain osmotic balance in the cell. However, the regulation of intracellular accumulation of these compounds is still not well understood. The freshwater cyanobacterium Synechococcus elongatus PCC 7942 (Syn7942) exclusively accumulates sucrose as a compatible solute upon salt stress and is thus an ideal model microorganism for studying the metabolism of compatible solute dynamics. Here, we focused on elucidating the regulatory mechanisms involved in salt-induced sucrose accumulation in Syn7942. Using a series of physiological and biochemical experiments, we showed that the ionic effect of salt stress plays an important role in inducing sucrose synthesis, whereby elevated ion concentration directly activates the sucrose-synthesizing enzyme sucrose-phosphate synthase and simultaneously inhibits the sucrose-degrading enzyme invertase, resulting in a rapid sucrose accumulation. Thus, we propose a novel mechanism for cyanobacterial adaption to salt stress and fluctuating salinity, i.e., the ion-induced synergistic modulation of the enzymes synthesizing and degrading compatible solutes. These findings greatly enhance our current understanding of microbial adaptation to salt.
IMPORTANCE Most microbes de novo synthesize compatible solutes for adaptation to salt stress or fluctuating salinity environments. However, to date, one of the core questions involved in these physiological processes, i.e., the regulation of salt-induced compatible solute biosynthesis, is still not well understood. Here, this issue was systematically investigated by employing the model freshwater cyanobacterium Synechococcus elongatus PCC 7942. A novel mechanism for cyanobacterial adaption to salt stress and fluctuating salinity, i.e., the ion-induced synergistic modulation of key synthesizing and degrading enzymes of compatible solutes, is proposed. Because the ion-induced activation/inhibition of enzymes is a fast and efficient process, it may represent a common strategy of microbes for adaptation to environments with fluctuating salinity.
INTRODUCTION
Salinity is one of the most important abiotic factors in various natural habitats of microbes. It highly fluctuates in association with geographical (e.g., water merging in estuaries and tidal fluxes) and climatic (e.g., rainfall and desiccation) changes and factitious activities (e.g., discharge of sewage). Salt stress usually challenges living cells via its two associated effects, i.e., high ionic strength and high external osmotic pressure. The former leads to an influx of inorganic ions (e.g., Na+ and Cl–), some of which are toxic to cell metabolism (1, 2). The latter causes a direct loss of intracellular water, which in turn reduces cell turgor and inhibits cell growth. To cope with these challenges, microbes have developed two strategies, the “salt-in” and “salt-out” strategies (3). Some archaea and a few halophilic bacteria adopt the “salt-in” strategy. High concentrations of inorganic salts (generally K+ and Cl–, up to 2 to 3 M) are accumulated in the cytoplasm and required for the growth and structural stability of cells. The intracellular enzymatic machineries of these microorganisms maintain proper conformation and activity at near-saturating salt environments. Under low salt concentrations, most proteins become denatured (3–7). For most microbes, including cyanobacteria, the “salt-out” strategy is adopted. These microorganisms maintain a relatively low intracellular ion concentration by actively pumping out inorganic ions and accumulate high levels of compatible solutes, which are small water-soluble organic compounds such as sucrose, glycerol, glycine betaine, ectoine, etc., by either de novo biosynthesis or uptake from the environment to ensure osmotic balance (6, 8, 9).
Microbes dynamically maintain intracellular levels of compatible solutes as stimulus-induced intermediates. When the environmental salinity abruptly or gradually decreases, the cell secretes or degrades these compounds for a rapid or gradual decrease of their levels. This response is physiologically important for microbes, enabling adaptation to fluctuating salinity. However, the majority of previous studies on the regulation of compatible solute metabolism focused on the expression and enzyme activity of key synthesizing genes/enzymes (10–17). Much less attention has been paid to the degradation of compatible solutes. Only a few enzymes participating in the degradation of compatible solutes, such as sucrose and glucosylglycerol (GG), have been identified (18–21). The regulation and the role of degrading actions in compatible solute metabolism, especially from the point of view of the synthesis/degradation balance, are rarely examined (22) and require further study. Their delineation will enable a deep understanding of the mechanisms of microbial salt acclimation.
Cyanobacteria are a large group of microorganisms adopting the “salt-out” strategy for salt acclimation. These bacteria can be found in almost all light-exposed environments and play an important role in the global substance and energy cycling because of their oxygenic photosynthetic capabilities (23). A rough correlation between the salt tolerance of and the compatible solutes produced by cyanobacteria is found (24–27). Species with low salt tolerance (up to 0.7 M NaCl, equivalent to 4.1% salinity) synthesize sucrose and/or trehalose, and species with moderate salt tolerance (up to 1.8 M NaCl) mainly accumulate GG and sometimes glucosylglycerate. Highly salt-tolerant cyanobacteria (up to 3.0 M NaCl) mainly accumulate glycine betaine and glutamate betaine. Recently, a novel compatible solute, homoserine betaine, was identified from the oceanic cyanobacterium Trichodesmium erythraeum (28). Among these compatible solutes, some have great potential for industrial or biotechnological applications, such as food additives, human nutrients, and moisturizing agents (29). Direct photosynthetic production of sucrose by cyanobacteria is also considered a potential strategy to provide abundant sugar feedstock for biorefineries (30).
The molecular basis of sucrose metabolism in cyanobacteria has been clearly elucidated. Whereas sucrose-phosphate synthase (SPS) and sucrose-phosphate phosphatase (SPP) catalyze sucrose synthesis from fructose-6-phosphate and UDP-glucose, three enzymes, i.e., alkaline/neutral invertase (19, 22), sucrose synthase (18), and amylosucrase (20), are able to catalyze sucrose degradation. Recently, cyanobacteria have been extensively metabolically engineered for photosynthetic production of sucrose. The overexpression of key enzymes and/or sugar transporters, inactivation of competing pathways, and rerouting of carbon flux in cellular metabolism have greatly improved their sucrose productivity (30–34). For example, overexpressing the native sps gene in Synechocystis sp. PCC 6803 (Syn6803) and Synechococcus elongatus PCC 7942 (here referred to as Syn7942) improved the intracellular sucrose accumulation by approximately 3- and 2-fold, respectively (32, 33). The introduction of Escherichia coli proton/sucrose symporter CscB into S. elongatus strains resulted in a successful export of sucrose from the cytoplasm to the medium, channeling up to 80% of the fixed CO2 into sucrose production (30, 31). The enhancement of glycogen synthesis and inactivation of invertase also effectively improved sucrose production in Syn7942 and Syn6803, respectively (22, 34).
Compared to the well-characterized genetic background and the great progress in metabolic engineering, the inducing and regulatory mechanisms of salt-induced sucrose biosynthesis in cyanobacteria are poorly understood. A few core questions about this physiological process still must be clearly answered. What is the core inducing factor of sucrose synthesis? At which level(s) is sucrose synthesis regulated? How is sucrose synthesis/degradation dynamically regulated? Finally, what is the sensory/signaling mechanism for cyanobacterial acclimation to fluctuating salinity? Previous studies on the regulation of salt-induced GG synthesis in Syn6803 (a moderately salt-tolerant cyanobacterium) provide us valuable clues. GG metabolism was found to be regulated at the levels of both gene expression and biochemical regulation (13, 15, 21). In the present study, we systematically addressed these questions using the model freshwater cyanobacterium Syn7942 (synthesizing sucrose as the only compatible solute upon salt stress) on multiple levels (transcriptional, translational, and biochemical). The present study provides evidence that the ion-induced enzymatic balance of compatible solute turnover represents an important regulatory mechanism.
RESULTS
Ionic compounds are more effective than osmotic compounds in the induction of sucrose synthesis.
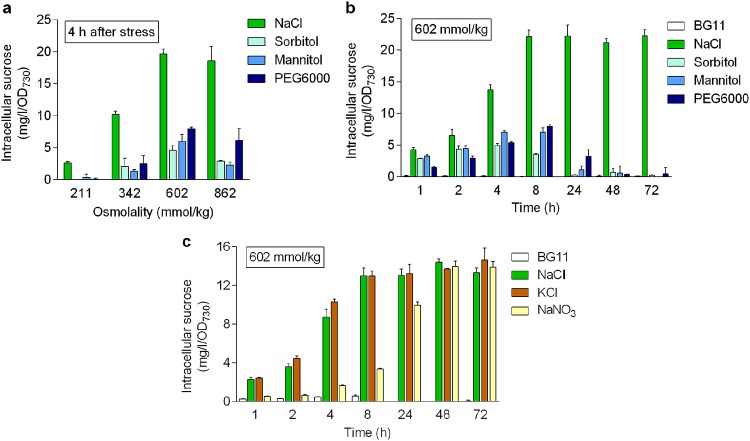
Salt stress causes two basic types of stresses to the living cell, i.e., ionic stress and osmotic stress. To obtain more information about the effects of these stresses on the induction of sucrose synthesis, NaCl and three commonly used osmotic compounds (sorbitol, mannitol, and PEG6000) were used to challenge Syn7942 cells under equal osmolality conditions (see Table S1 in the supplemental material). We first analyzed sucrose production after a short-term (4 h) stress (Fig. 1a). All osmotic compounds induced sucrose synthesis, and the induction was osmolality dependent within a certain range (≤602 mmol/kg), similar to the effect of NaCl. However, the inductive efficiency of these compounds was markedly lower than that of NaCl. Sucrose accumulation in the sorbitol-, mannitol-, and PEG6000-stressed cells was only 0 to 23%, 12 to 31%, and 4 to 40% of that in the NaCl-stressed cells, respectively. The long-term effects of these compounds were examined in a time course manner (up to 72 h) at 602 mmol/kg (Fig. 1b). Differences in sucrose accumulation were apparent for the two types of compounds. The sucrose content in the NaCl-stressed cells rapidly increased after stress and achieved the highest level (22.1 mg/liter/OD730 [optical density at 730 nm]) within 8 h, which was subsequently maintained. In contrast, the intracellular sucrose levels induced by sorbitol, mannitol, and PEG6000 increased slowly; the maximum amounts were only 5.0 to 8.0 mg/liter/OD730, which corresponded to 22.6 to 36.2% of the level induced by iso-osmolar NaCl. After achieving the highest level at 4 h (sorbitol) or 8 h (mannitol and PEG6000), the sucrose amount turned to decline. At 24 h (sorbitol) or 48 h (mannitol and PEG6000), only traces of sucrose were detected. Thus, the patterns of sucrose accumulation caused by NaCl and the osmotic compounds were clearly different. Whereas NaCl induced a relatively high and long-term production of sucrose in Syn7942, sorbitol, mannitol, or PEG6000 only induced a relatively low and transient sucrose accumulation. These results suggest that ionic compounds may be more effective than osmotic compounds in the induction of sucrose production.
FIG 1.
Sucrose accumulation in Syn7942 cells challenged by different stresses. (a) Cells were stressed by NaCl, sorbitol, mannitol, and PEG6000 under different osmolality conditions for 4 h. (b) Cells were stressed by NaCl, sorbitol, mannitol, and PEG6000 under the same osmolality (602 mmol/kg) for different times. (c) Cells were stressed by NaCl, KCl, and NaNO3 under the same osmolality (602 mmol/kg) for different times. The data are presented as means from at least three independent replicates (n = 3 in panels a and c; n = 4 in panel b) with the standard deviations.
To verify this hypothesis, the effects of two additional inorganic salts, KCl and NaNO3, on the induction of sucrose synthesis were examined and compared to that of NaCl (Fig. 1c). Iso-osmolar KCl exhibited an effect almost identical to that of NaCl. The intracellular sucrose rapidly increased after KCl stress and attained the steady level at 8 h (13.0 mg/liter/OD730). After 72 h of stress, a similar level of intracellular sucrose (14.6 mg/liter/OD730) was still maintained. For NaNO3, although the accumulation of sucrose was not as pronounced in the first 24 h compared to NaCl and KCl, a similar amount of sucrose was attained in the fully acclimated cells (at 48 and 72 h). Thus, all three inorganic salts exhibited effective induction of sucrose synthesis in Syn7942. In addition to sucrose accumulation, cell growth of Syn7942 cells under different salt stresses was also examined to evaluate the possible toxicity of high concentrations of the involved ions (Fig. S1). At a concentration of 300 mM (corresponding to an osmolality of 600 mmol/kg) or below, the KCl- or NaNO3-stressed cells showed growth similar to that of the NaCl-stressed cells, indicating that NaCl, KCl, and NaNO3 at the same concentrations have similar effects on the growth of Syn7942.
Sucrose synthesis in Syn7942 is regulated on multiple levels of sps/SPS.
SPS is the key enzyme catalyzing the rate-limiting step of sucrose synthesis in cyanobacteria (32, 33). In Syn7942, this enzyme is encoded as a fusion protein containing both SPS and SPP domains by Synpcc7942_0808 (referred to as sps7942) (35). To unravel the regulatory mechanisms of salt-induced sucrose synthesis in cyanobacteria, the expression of sps7942 and the biochemical aspects of sucrose production by Syn7942 in response to different stresses were analyzed.
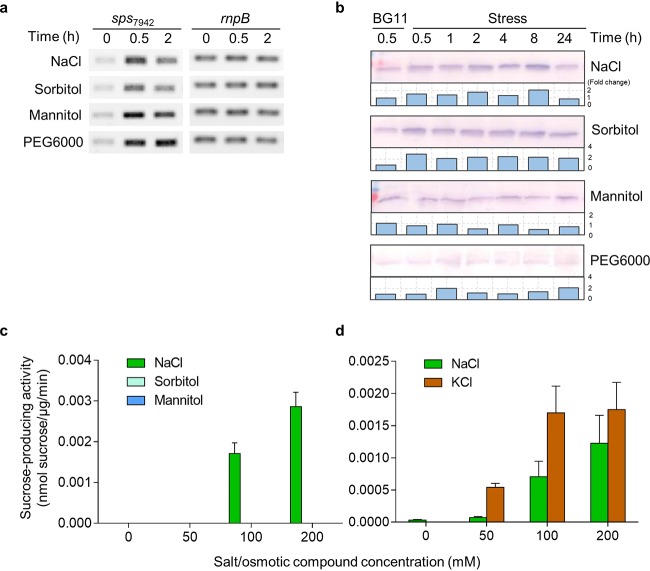
The transcription of sps7942 was first examined in cells stressed by 300 mM NaCl (causing an osmolality of 602 mmol/kg) (Fig. 2a). A basic level of transcription of sps7942 was observed under the nonstress condition (0 h). After addition of NaCl, the transcription of sps7942 was upregulated. The mRNA levels increased by approximately 4.3- and 2.9-fold at 0.5 and 2 h, respectively. The sps7942 transcription was also analyzed in cells stressed by sorbitol, mannitol, and PEG6000. In all cases, an increase of the sps7942 mRNA level was observed in the tested period, despite a certain reduction of this level after 0.5 h in the presence of sorbitol.
FIG 2.
Effects of ionic and osmotic compounds on the expression of sps7942 (a and b) and the sucrose-producing activity (c and d) in Syn7942. In panels a and b, Syn7942 cells were stressed by NaCl, sorbitol, mannitol, and PEG6000 at 602 mmol/kg for the indicated times, and the amounts of sps7942 transcript (a) and SPS7942 protein (b) were analyzed by RT-PCR and immunoblotting, respectively. In panels c and d, the sucrose-producing activity of Syn7942 was analyzed with extracts of Syn7942 cells grown under standard conditions. Different concentrations of NaCl, KCl, sorbitol, and mannitol were included in the enzymatic reactions. The produced sucrose was determined by ion-exchange chromatography (d) or HPLC (c). The data are presented as means from three independent replicates with the standard deviations.
The expression of sps7942 was further examined by immunoblotting (Fig. 2b). Consistent with the transcription analysis, a basic level of SPS7942 protein was detected under the nonstress condition. The amount of SPS7942 mildly increased after addition of NaCl, achieving the highest level at 8 h (∼2-fold), and then returned to the initial level at 24 h. The sorbitol stress also resulted in an immediate increase of SPS7942, by ∼3-fold at 0.5 h. The SPS7942 level remained constant in the following 24 h. In the case of mannitol and PEG6000 stresses, the compounds did not appear to clearly affect the SPS7942 levels within the tested period. Hence, the notable increase in sps7942 mRNA levels was not observed on the protein level. The transcription and translation of sps7942 under the tested stresses were not always causally correlated.
In the above-described experiments, the slight increase of SPS7942 level under NaCl stress seemed not sufficient to fully explain the rapid sucrose accumulation, and the similar increase of SPS7942 amount under NaCl and sorbitol stresses also could not explain the different profiles of sucrose accumulation. Therefore, the biochemical aspects of sucrose production were further examined. Sorbitol and mannitol are small molecules compared to PEG6000, and they might flow into the cell following the concentration gradient during stress. The subsequent results confirmed this hypothesis, i.e., increasing amounts of sorbitol and mannitol in Syn7942 cells were detected after applying sorbitol or mannitol stress (Fig. S2). To determine whether the influx of ions (e.g., Na+ and Cl–) during salt stress or small osmotic compounds (e.g., sorbitol and mannitol) during osmotic stress into the cell affects sucrose production, the sucrose-producing activity of Syn7942 was analyzed in vitro using cell extracts of Syn7942 grown in the standard BG11 medium. Different concentrations of inorganic salts and osmotic compounds were used in the assay reactions (Fig. 2c). Under the control condition (0 mM), the sucrose-producing activity was under the detection limit and could not be detected. The activity was notably induced by the addition of NaCl at concentrations greater than 50 mM and exhibited a concentration-dependent profile. The activity at 100 and 200 mM NaCl rose to 1.7 × 10−3 and 2.9 × 10−3 nmol sucrose/μg/min, respectively (Fig. 2c). In contrast, the presence of isomolar concentrations of sorbitol and mannitol did not induce any sucrose synthesis, indicating that these small compounds did not directly stimulate the sucrose-producing enzyme activity. To evaluate whether the activation of sucrose-producing activity is specific to NaCl, the effect of KCl was also examined (Fig. 2d). Similar to NaCl, KCl exhibited a concentration-dependent induction of sucrose-producing activity. At all tested concentrations (50 to 200 mM), isomolar KCl appeared to be more effective than NaCl. The sucrose-producing activity induced by KCl was 7.7- and 2.3-fold that induced by NaCl at 50 and 100 mM, respectively. At the concentration of 200 mM, the activity caused by KCl was still 43% higher than that of NaCl. These observations strongly suggested that sucrose production in Syn7942 is mainly regulated on the biochemical level and that such biochemical regulation is mediated by the inorganic ions from the inorganic salts. To further explore the regulatory mechanisms, detailed analyses of the enzymes involved in the synthesizing and degrading aspects of sucrose metabolism were performed.
SPS7942 possesses a bifunctional SPS/SPP activity.
In many cyanobacteria, SPS and SPP are encoded by separate genes or are expressed as (i) a fused protein of a functional SPS domain and a nonfunctional SPP domain and (ii) a separate functional SPP (16, 35, 36). However, the genomes of some Synechococcus species (e.g., Syn7942, S. elongatus PCC 6301, and S. elongatus UTEX 2973) only encode fused forms of SPS and SPP, suggesting the bifunctional activity of these proteins. Recently, Martínez-Noël and colleagues provided the first evidence supporting the dual biochemical activity of SPS7942 using native and recombinant proteins (35). In the present study, this was further verified by a comprehensive analysis of the wild-type SPS7942 and its single amino acid-substituted variants, before the detailed enzymatic analyses of SPS7942 were performed.
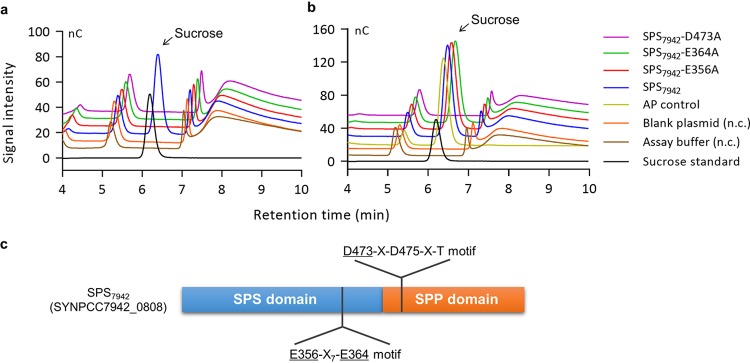
We heterologously expressed SPS7942 in E. coli and purified the recombinant protein. The activity of SPS7942 was examined using two sets of substrates, fructose-6-phosphate and UDP-glucose (SPS substrates) and sucrose-6-phosphate (SPP substrate). Compared to the negative controls, SPS7942 was found to catalyze the apparent formation of sucrose from both sets of substrates (Fig. 3a and b), agreeing well with the bifunctional activity of this protein.
FIG 3.
Analysis of the SPS and SPP activities of SPS7942 and its variants (a and b) and schematic illustration of the functional architecture and conserved motifs of SPS7942 (c). In panel a, fructose-6-phosphate and UDP-glucose were used as the substrates (5 mM each) to determine the bifunctional activity of SPS7942. In panel b, sucrose-6-phosphate (5 mM) was used to examine the SPP activity of SPS7942. All assays were performed in the presence of 200 mM NaCl. The generated sucrose was examined by ion-exchange chromatography. The assay buffer and empty plasmid pRSFDuet-1 were used as negative controls (n.c.). AP (alkaline phosphatase; NEB, Ipswich, MA) was used as a positive control. In panel c, the underlined amino acids were replaced.
E-X7-E and D-X-D-X-T are the highly conserved motifs of SPS and SPP, respectively, and they play important roles in enzymatic catalysis (37–39). The structural analyses of SPS and SPP indicated that these motifs participate in the formation of active sites of the enzymes (39, 40). Whereas the first Glu residue of E-X7-E may function as the nucleophile and the second Glu may function as the general acid/base catalyst (38), the invariant first Asp of D-X-D-X-T works as a functional nucleophile initiating the phosphohydrolysis of sucrose-6-phosphate (39). Therefore, the corresponding residues (E356, E364, and D473) in the E-X7-E and D-X-D-X-T motifs of SPS7942 were analyzed with regard to the protein activity (Fig. 3c). Three variants (E356A, E364A, and D473A) of SPS7942 were constructed and purified. Although the substitutions in the SPS domain (E356A and E364A) disrupted sucrose synthesis only from fructose-6-phosphate and UDP-glucose, the substitution in the SPP domain (D473A) inhibited sucrose synthesis from fructose-6-phosphate and UDP-glucose, as well as from sucrose-6-phosphate (Fig. 3a and b). Therefore, the glutamate residues at positions 356 and 364 (E356 and E364) are important for the SPS activity, and the aspartate residue at position 473 (D473) is necessary for the SPP activity. Together, these results confirmed that SPS7942 is a bifunctional enzyme.
The activity of SPS7942 is activated at the SPS domain in an ion-induced manner.
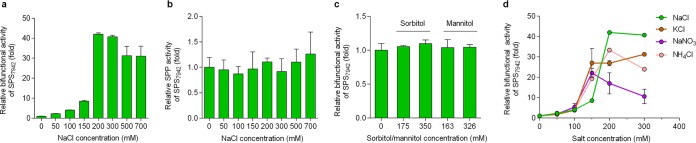
To further analyze the enzymatic regulation of sucrose synthesis, the effects of inorganic ions on the activity of purified SPS7942 were investigated. In line with the increased sucrose-producing activity in the cyanobacterial cell extract, the addition of NaCl also induced the bifunctional activity of SPS7942 (Fig. 4a). In the presence of NaCl concentrations below 150 mM, the bifunctional activity was slightly increased. At 200 mM or higher concentrations, the activity was dramatically stimulated, by approximately 30- to 40-fold. However, when the SPP activity was analyzed using sucrose-6-phosphate as the substrate, the activity was almost unaffected by the addition of NaCl at all tested concentrations (Fig. 4b), indicating that the activity of the SPS domain, rather than the SPP domain, was activated. In parallel, the effects of the osmotic compounds sorbitol and mannitol on SPS7942 activity were analyzed (Fig. 4c). The bifunctional activity was almost unaffected regardless of the presence or absence of sorbitol and mannitol, indicating that these compounds do not directly modulate SPS7942 activity. This agrees well with the results of the sucrose-producing activity when using Syn7942 cell extracts (Fig. 2c). Thus, it becomes clear that the activation of SPS7942 activity is realized in an ion-induced manner and occurs at the SPS domain.
FIG 4.
Activating effect of ionic and osmotic compounds on the enzyme activity of SPS7942. (a, c, and d) Fructose-6-phosphate and UDP-glucose were used as substrates (2 mM each) to determine the bifunctional activity of SPS7942. (b) Sucrose-6-phosphate (2 mM) was used to examine the SPP activity of SPS7942. Different concentrations of NaCl, KCl, NaNO3, NH4Cl, sorbitol, and mannitol were included in reaction mixtures, as indicated. The data are presented as means from independent replicates (n = 3 in panels a and d; n = 4 in panels b and c) with the standard deviations; the data of KCl and NH4Cl in panel d are presented as means from two independent replicates with the standard deviations. In each panel, all obtained values at different concentrations were normalized by comparing them to the value at 0 mM.
To investigate whether such activation exhibits an ion preference, three additional inorganic salts (i.e., KCl, NaNO3, and NH4Cl) were analyzed in parallel with NaCl. Similar to NaCl, all of these salts markedly activated SPS7942. The enzyme activity was increased by approximately 22- to 33-fold at concentrations of 150 or 200 mM of these compounds (Fig. 4d).
The Syn7942 invertase is regulated in a manner contrary to SPS7942.
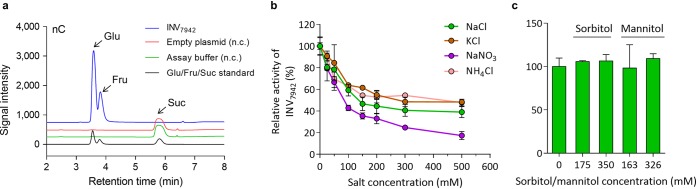
The accumulation of sucrose in cyanobacteria is an outcome of sucrose synthesis and degradation under salt stress. Hence, the sucrose catabolism of Syn7942 was also analyzed in the present study. To this end, the enzyme(s) responsible for sucrose degradation was first identified. A search of the Syn7942 genome for sequences encoding the reported cyanobacterial enzymes for sucrose catabolism, e.g., alkaline/neutral invertase (19, 22), sucrose synthase (18), and amylosucrase (20), was performed. Only one invertase homolog was identified in this cyanobacterium, encoded by Synpcc7942_0397. The deduced protein sequence shares 36 to 42% identity with the biochemically identified invertases of Synechocystis sp. PCC 6803 (Sll0626) and Anabaena sp. PCC 7120 (InvA-Alr1521 and InvB-Alr0819). To characterize the biochemical properties of the enzyme, an N-terminal His-tagged invertase of Syn7942 (termed INV7942) was produced and purified in E. coli. As predicted, the protein efficiently catalyzed the hydrolysis of sucrose to fructose and glucose (Fig. 5a). Trehalose, raffinose, and stachyose were also used as substrates, but no noticeable activities toward these oligosaccharides were detected (data not shown).
FIG 5.
Characterization of the invertase activity of INV7942 (a) and the effect of ionic (b) and osmotic (c) compounds on enzyme activity. All assays were performed using sucrose (Suc, 2 mM) as a substrate. In panel a, the products glucose (Glu) and fructose (Fru) were examined by ion-exchange chromatography. The assay buffer and the empty plasmid pRSFDuet-1 were used as negative controls (n.c.). In panels b and c, different concentrations of NaCl, KCl, NaNO3, NH4Cl, sorbitol, or mannitol were included in reaction mixtures, as indicated. The measured activities were normalized by comparing them with the starting values (at 0 mM). The data are presented as means from at least three independent replicates (n = 3 in panel b; n = 4 in panel c) with the standard deviations.
To analyze the impact of ionic compounds on INV7942 activity, increasing concentrations of NaCl, KCl, NaNO3, and NH4Cl were included in the assay buffer (Fig. 5b). The activity of INV7942 was quickly reduced to approximately 50% at 150 mM NaCl and slowly decreased to 39% at 500 mM NaCl. In the case of KCl, NaNO3, and NH4Cl, a strong inhibition was also observed at 0 to 150 mM, which was followed by a sustained inhibition at 150 to 500 mM. Similar to SPS7942, the INV7942 activity was almost unaffected by the addition of sorbitol or mannitol (Fig. 5c). Taken together, these data indicated that the activity of INV7942 is regulated in an ion-dependent manner, with the inhibition contrasting with SPS7942 activation.
DISCUSSION
Syn7942 is a freshwater cyanobacterium synthesizing sucrose as the only compatible solute for adaption to salt stress. Early studies have shown that this cyanobacterium is able to tolerate maximally 0.5 M NaCl (32, 41), a salinity (2.9%) lower than that of seawater (3.5%). Due to the influences of geographical, climatic, and factitious factors, one can image that this cyanobacterium often encounters fluctuating salinity in its aquatic habitats. It is of interest to understand how the salt-induced sucrose synthesis of this cyanobacterium is dynamically regulated according to the environmental salinity.
Induction of compatible solute synthesis.
Salt stress and osmotic stress have often been used as synonymous terms in the early studies of microbial responses and adaption to high-salinity environments. However, there is increasing evidence that the ionic and osmotic challenges involved in salt stress exhibit clearly different effects on the genetics, biochemical processes, and physiology of microbes. For example, salt (NaCl) and osmotic (sorbitol) stresses induce different sets of genes and cause differential changes in the cytoplasmic volume and different levels of damage to the photosynthetic machinery in the cyanobacteria Syn6803 and/or Syn7942 (2, 42, 43), indicating that the two stresses are recognized as different stimuli by these microorganisms. Therefore, before delving deep into the regulatory mechanisms of salt-induced sucrose synthesis in Syn7942, we first determined the accumulating profiles of intracellular sucrose under the two types of stresses. To this end, three commonly used osmotic compounds (sorbitol, mannitol, and PEG6000) and three inorganic salts (NaCl, KCl, and NaNO3) were used to apply exogenous stresses. Different profiles of sucrose accumulation in Syn7942 were observed. Whereas the ionic compounds exhibited a notable and relatively long-term inducing effect on sucrose accumulation, the osmotic compounds had only a minor and relatively short-term effect (Fig. 1a and b). A similar phenomenon was observed by Marin et al. in the study of the synthesis of GG, the main compatible solute in the intermediate salt-tolerant cyanobacteria (44). Elevated NaCl levels induced a proportional increase of GG content in Syn6803 cells, but sorbitol and maltose only caused a weak GG accumulation, close to the background level. Based on these observations, we propose that, in general, the ionic compounds could more effectively induce compatible solute synthesis than the osmotic compounds in cyanobacteria. However, these results did not provide us enough information to figure out the “real” or key factor that induces compatible solute synthesis. Further analyses on the levels of gene expression and biochemical regulation were thus performed.
Regulation of compatible solute synthesis.
Compared to the well-characterized synthetic pathways and the expanding metabolic engineering of cyanobacteria for target compound production (28, 31, 32, 45, 46), the regulatory mechanisms of compatible solute synthesis in cyanobacteria still need to be further studied. The existing data indicate that such regulation could proceed on multiple levels, i.e., the expression of key genes and the modulation of key enzymes (14, 15, 47, 48). However, the importance of the two strategies in a specific microorganism may be different. In Syn6803 and Anabaena sp. PCC 7120, in which sucrose is used as a secondary compatible solute or plays an additional role in N2 fixation (14, 49), the regulation of sucrose synthesis is accomplished mainly by the salt-induced stimulation of sps expression (12, 14, 47, 48). With regard to GG synthesis in Syn6803, it is regulated via a combination of alteration of ggpS (the GG-phosphate synthase gene) expression and biochemical regulation of GGPS activity, with the latter playing a key role in the rapid early adaptation of cells to salt stress environments (15).
In the present study, the transcription, translation, and enzyme activity of sps7942/SPS7942 were systematically investigated, and the regulation at all these levels was observed. While the transcription of sps7942 was notably upregulated shortly after salt (NaCl) stress (0.5 h) (Fig. 2a), the amount of SPS7942 was only slightly affected and slowly increased after NaCl stress (within 24 h), i.e., the fast induction of sps7942 transcription was not directly relayed on the protein level. According to a previous study, immediately after a salt shock of 684 mM NaCl, the protein synthesis in Syn6803 in almost completely blocked (50). Therefore, the nonsynchronous regulation of sps7942 transcription and translation in Syn7942 might be caused by the fast and serious inhibition of protein synthesis machinery after a salt shock. The salt-induced sucrose synthesis in Syn7942 is a rapid physiological process. It could reach a steady level within 3 h of salt stress, without a lag phase (51). The slow and slight increase of SPS7942 levels seems insufficient to fully explain the rapid increase of sucrose accumulation. Hence, additional efficient strategies may be used by Syn7942. NaCl-mediated modulation of SPS activity has been demonstrated in Synechococcus sp. PCC 6301, a species closely related to Syn7942 (48). However, this phenomenon was observed using cyanobacterial cell extracts rather than purified SPS; the question of whether NaCl activates SPS directly or indirectly still remains unanswered. Data presented in the present study clearly indicated that regardless of whether the cell extracts of Syn7942 or the purified protein was used, the enzyme activity depends on the high NaCl content in the assay reaction (Fig. 2 and 4), thus confirming the direct activation of SPS7942 by NaCl. The activation occurs at the SPS domain, since the SPP activity was not affected at all by increased salt concentrations (Fig. 4b). Furthermore, this activation seems not to be restricted to NaCl. Other selected inorganic salts also showed marked stimulation of enzyme activity (Fig. 4d). We conclude from these observations that the salt-induced sucrose synthesis in Syn7942 is mainly regulated by the ion-mediated activation of the SPS domain activity of the bifunctional SPS7942.
In the physiological experiment, it is interesting to see that the osmotic compounds (sorbitol, mannitol, and PEG6000) also caused low levels of sucrose accumulation in Syn7942 at the early stage of salt acclimation (Fig. 1a and b). We suppose that the osmotic stress caused by these compounds led to an increase of intracellular ion concentration by shrinking cells (43), and the elevated ion concentration in turn induced gentle sucrose synthesis. Further investigations on changes in the cell volume and intracellular ion concentration of the stressed Syn7942 cells will provide more information.
Although the regulation modes of sucrose synthesis in Syn7942 and GG synthesis in Syn6803 are similar, there is an obvious difference in enzyme modulation. While GGPS can be activated in cell extracts but not purified protein, the activity of SPS7942 can be activated in either cell extracts or purified protein (Fig. 2 and 4). Hagemann et al. proposed that a factor inhibiting GGPS might be present in the cell extracts of Syn6803 and is lost during biochemical purification of GGPS, with the purified protein retaining an active state in the in vitro assays (13). In the present study, the inhibiting factor might not have been removed during purification and the purified SPS7942 remained inactive in the absence of salts. It is also possible that SPS7942-inhibiting factors do not exist and that the ion-mediated modulation of SPS7942 uses a mechanism distinct from that of GGPS. Further studies of the molecular basis of salt-induced activation of SPS7942 will help elucidate the activation or inhibition-activation mechanism.
Recycling of compatible solute.
Compatible solutes are temporary metabolites involved in the stress response. Hence, the recycling of compatible solutes obviously makes important cellular or physiological sense for microbes from at least two perspectives: first, to finely regulate their intracellular contents in response to the extracellular salt stress, and second, to avoid a net loss of carbon and energy. Three sucrose breakdown enzymes have been identified in cyanobacteria: (i) sucrose synthase, which uses (A/U)DP to reversibly split sucrose into (A/U)DP-glucose and fructose; (ii) amylosucrase, which catalyzes the splitting of sucrose into free fructose and glucose that is transferred to maltooligosaccharides; and (iii) invertase, which hydrolyzes sucrose directly into glucose and fructose (18–20). Whereas sucrose synthase is mainly found in N2-fixing filamentous cyanobacteria and amylosucrase is only present in a few cyanobacteria, invertase is widespread among both unicellular and N2-fixing cyanobacteria species and represents the main degrading pathway for intracellular sucrose balancing. In the present study, the invertase encoded by Synpcc7942_0397 was identified as the only enzyme catalyzing sucrose breakdown in Syn7942. The purified enzyme efficiently hydrolyzed sucrose to glucose and fructose. More interestingly, the activity of this enzyme was sensitive to elevated ion concentrations in the assay buffer. This response was in contrast to that of SPS7942 (activated by ions). Such synergistic enzyme modulation of SPS7942 and INV7942 enables the cell to meet the biochemical requirements for the dynamic balance of intracellular sucrose.
Enzymatic balance of compatible solute.
Based on the information presented above, we demonstrate that the regulation of salt-induced sucrose accumulation in Syn7942 occurs on multiple levels, including gene expression and biochemical regulation, and that the biochemical modulation mediated by inorganic ions plays an important role in this process. In Syn7942, the sucrose-synthesizing gene sps7942 is constitutively expressed. A basic amount of SPS7942 is maintained by the cell (Fig. 2). Under the nonstress condition, the SPS7942 protein may retain an inactive state, and INV7942 functions with a high activity. As a result, the dynamics of sucrose metabolism tends to be in the direction of degradation. Only traces of sucrose could be detected in cyanobacterial cells (Fig. 1b and c). Under the salt stress (e.g., NaCl) condition, the inorganic ions (Na+ and Cl–) rapidly diffuse into cyanobacterial cells following the salt gradient, leading to an increase of intracellular ion concentrations. To ensure a fast response to salt stress, the activity of the existing SPS7942 is directly activated by the elevated ion concentrations and the activity of INV7942 is depressed. Thus, the dynamics of sucrose metabolism tends to be in the direction of sucrose synthesis. A fast accumulation of intracellular sucrose is thereby observed (Fig. 1). During further salt acclimation (a few hours after salt stress), the intracellular content of inorganic ions may be reduced by actively pumping out the ions (7), and a new balance of intracellular sucrose is achieved by reducing SPS7942 activity and enhancing INV7942 activity. The gentle upregulation of sps7942 expression during this time (Fig. 2a and b) also favors the maintenance of a steadily high level of intracellular sucrose. A similar regulatory effect of inorganic ions has also been reported for GG metabolism in Syn6803. Whereas GGPS, the key enzyme catalyzing GG synthesis, is biochemically activated by inorganic ions, GghA, the GG-degrading enzyme, is simultaneously inhibited, which results in a dynamic regulation of the intracellular GG level (17, 21). In the natural environment, the salinity highly fluctuates. A fast and fine regulation of intracellular compatible solute is needed for microbe acclimation to such fluctuating environments. The ion-induced modulation of the synthesizing and degrading enzymes is an efficient way to accomplish this response and thus may be the predominant manner by which cyanobacteria, and possibly other microbes, regulate intracellular compatible solutes.
With regard to the photosynthetic production of sucrose by cyanobacteria, the highest level of sucrose productivity was achieved in Syn7942 and S. elongatus UTEX 2973 (a strain highly similar to Syn7942) (30, 31). In both strains, the sucrose transporter CscB was introduced. Our results provide useful information for guiding further biotechnological investigations in these strains. Because the biochemical modulation of key enzymes seems to play the predominant role in the regulation of sucrose metabolism in Syn7942 (and probably also in UTEX 2973), we suppose that further engineering to improve the expression level of SPS would have limited effects for the further improvement of sucrose productivity. More efforts might be put into protein engineering of SPS and invertase after further explorations of the ion-mediated modulating mechanisms of these enzymes, in combination with the search for more efficient sucrose exporters and the development of efficient sucrose-utilizing strategies (e.g., cocultivation with heterotrophic microorganisms) (33, 52).
MATERIALS AND METHODS
Microbial strains and cultivation conditions.
For standard cultivation, Syn7942 cells were grown in 100 ml of BG11 medium (53) in 250-ml flasks, on a horizontal rotary shaker at 150 rpm and 30°C under constant white-light illumination (30 μmol/m2/s). The starting cell density of cyanobacterial cultures was set to an OD730 of 0.08. Cell growth was monitored by measuring the OD730. To perform stress experiments, cyanobacterial cells pregrown in the BG11 medium were transferred into 100 ml of BG11 medium supplemented with different concentrations of ionic salts (NaCl, KCl, NaNO3, or NH4Cl) or osmotic compounds (sorbitol, mannitol, or PEG6000) in the 250-ml flasks described above. E. coli strain DH5α was used for routine cloning purposes, and E. coli strain BL21(DE3) (Transgen, Beijing, China) was used for the overproduction of target proteins.
Determination of sucrose in Syn7942 cells or enzymatic reactions.
For sucrose determination, 1 ml of Syn7942 culture grown in the flasks on a shaker under different conditions (see above) was sampled at designated time points. Cells were harvested by centrifugation at 10,000 × g for 2 min and treated as described previously (34). The amount of sucrose in the resulting liquid samples or enzymatic reactions was analyzed by ion-exchange chromatography as described previously (34) or using a sucrose/d-glucose assay kit (Megazyme, Bray, Ireland) according to the manufacturer’s instructions.
Heterologous expression and purification of SPS7942 and invertase.
The coding sequences of the sps7942 (Synpcc7942_0808) and invertase (Synpcc7942_0397) genes were amplified from the total DNA of Syn7942 using the primer pairs 0808-F/0808-R and 0397-F/0397-R, respectively (see Table S2 in the supplemental material), via PCR. The resulting fragments were cloned into the pRSFDuet-1 vector (Novagen, Merck, Darmstadt, Germany) by using the BamHI and NotI sites, generating the recombinant expression plasmids pRSFDuet-0808 and pRSFDuet-0397, respectively, which were then used to transform E. coli strain BL21(DE3). The sequences of both cloned PCR products were confirmed by sequencing.
For the overexpression of N-terminal His-tagged SPS7942 and INV7942, the transformed E. coli cells were grown in a modified lysogeny broth (LB) medium (NaCl-free) containing 37.5 μg/ml kanamycin at 37°C. At an OD600 of ∼0.6, cells were induced by 0.2 mM IPTG (isopropyl-β-d-thiogalactopyranoside) overnight at 18°C (for SPS7942) or 25°C (for INV7942). After induction, cells were harvested by centrifugation and disrupted by sonication in prechilled lysis buffer (50 mM KH2PO4-K2HPO4 buffer [pH 7.4], 5 mM imidazole, 0.1% Tween 20, and 5 mM β-mercaptoethanol). The lysates were centrifuged at 20,000 × g for 50 min, and the resulting supernatants were filtered through 0.22-μm-pore polyethersulfone membranes and then loaded onto a Ni Sepharose 6 FF column (GE Healthcare, Chicago, IL) for affinity chromatography. Eluted factions were further loaded onto Superdex 200 10/300 GL column (GE Healthcare) equilibrated with 50 mM KH2PO4-K2HPO4 buffer (pH 7.4) with (for SPS7942) or without (for INV7942) 5 mM MgCl2. The protein concentration in the fractions was determined by the Bradford method (54), and the purity of target proteins was examined by SDS-PAGE.
To obtain SPS7942 variants, the designed mutations were introduced into pRSFDuet-0808 by site-directed mutagenesis. The primers used for this purpose, i.e., E356A-F/E356A-R, E364A-F/E364A-R, and D473A-F/D473A-R, are listed in Table S2. All generated constructs were confirmed by DNA sequencing. The expression and purification of SPS7942 variants were performed using the procedure described above.
RNA isolation, reverse transcription, and semiquantitative analysis.
To determine gene transcription, Syn7942 cells were inoculated into 100 ml of BG11 medium supplemented with different stress compounds in the 250-ml flasks with a starting OD730 of around 1.0. At designated time points, cells from 15-ml cultures were harvested by centrifugation at 4°C and frozen immediately by liquid nitrogen. Cells were broken by grinding in a mortar for 10 min. Proper amounts of liquid nitrogen were supplemented into the mortar to keep cells frozen during grinding. RNA was extracted by using a MiniBEST plant RNA extraction kit (TaKaRa Bio, Shiga, Japan) and treated with DNase I (TaKaRa Bio). Reverse transcription of RNA was performed as previously described (55). Equal amounts of cDNA were used for further DNA amplification by PCR by using the primer pairs RT-sps-F/RT-sps-R and RT-rnpB-F/RT-rnpB-R (Table S2). The resulting products were visualized by electrophoresis. Semiquantitative analysis was performed with windows covering all signals for each sample using the software ImageJ (56). The data obtained were normalized using the rnpB signal.
Immunodetection of SPS7942.
Rabbit polyclonal antibodies against SPS7942 were provided by Yangkai Duan of our lab and used for immunoblotting. Syn7942 cells grown under different stresses were harvested by centrifugation and resuspended in 50 mM Tris-HCl buffer (pH 7.4). An appropriate amount of quartz sand was added to the cell suspension, and the cells were disrupted by vortex mixing. After centrifugation, the protein concentration in the supernatant was determined by the Bradford method (54). Then, 100 μg of total protein was subjected to SDS-PAGE, and the immunoblotting analysis was performed according to a previously described protocol (57). The semiquantification of SPS7942 hybridization signals was accomplished using the software ImageJ (56).
Enzymatic assay with Syn7942 cell extracts.
Syn7942 cells grown under standard conditions were harvested by centrifugation, resuspended in 50 mM HEPES buffer (pH 8.0), and disrupted by quartz sand as described above. The obtained cell extracts were then used in enzymatic assays. The reaction mixtures contained 20 mM HEPES (pH 8.0), 10 mM MgCl2, 20 mM UDP-glucose, 20 mM fructose-6-phosphate, the appropriate volume of cell extract (for a final total protein concentration of 1 mg/ml), and different concentrations of ionic or osmotic compounds, as indicated. After incubation at 30°C for 12 h, the reactions were terminated by boiling, and the generated sucrose was analyzed by ion-exchange chromatography or high-pressure liquid chromatography (HPLC) by using an Agilent 1260 Infinity II HPLC system equipped with a refractive index detector (Agilent Technologies, Inc., La Jolla, CA) and a Shodex Asahipak NH2P-50 4E analytical column (Showa Denko, Japan); the column was equilibrated and eluted with 75% acetonitrile at a flow rate of 1.0 ml/min (column temperature, 30°C).
Enzymatic assay with purified SPS7942 and INV7942.
Fructose-6-phosphate (Solarbio, Beijing, China), UDP-glucose (Solarbio), and sucrose-6-phosphate (Sigma-Aldrich, St. Louis, MO) were used as the substrates to determine the SPS and/or SPP activities of SPS7942; sucrose was used to determine the invertase activity of INV7942. For the assay involving SPS7942, 20 mM HEPES buffer (pH 8.0) containing 5 mM MgCl2 and different concentrations of ionic compounds or osmotic compounds was used. For the assay involving INV7942, 20 mM HEPES buffer (pH 8.0) containing different concentrations of ionic compounds or osmotic compounds was used. All enzymatic reactions were performed with incubation at 30°C and terminated by heating at 85°C for 5 min. The enzymatic rates were calculated based on reactions in which less than 20% of the substrate was consumed and normalized to controls for comparison. The reaction product, i.e., sucrose or glucose, was quantified by using the sucrose/d-glucose assay kit.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge financial support from the National Science Fund for Distinguished Young Scholars of China (31525002), the National Science Foundation of China Project (31872622), the Shandong Key Basic Research Project (ZR2017ZB0211), and the Shandong Taishan Scholarship.
We thank Martin Hagemann of Rostock University for helpful discussion. We thank Huiyuan Gao and Shijie Zhao of Shandong Agricultural University for assistance with measuring osmolality.
Y.L. and M.Z. performed most of the research and analyzed data. M.W., W.Z., and C.Q. performed part of the research and analyzed data. Y.L. and Q.L. designed research, discussed results, and wrote the manuscript. X.L. designed research, discussed results, and revised the manuscript. All authors read and approved the final manuscript.
All authors declare that no conflicting interests are present with respect to publication of the manuscript.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Allakhverdiev SI, Nishiyama Y, Suzuki I, Tasaka Y, Murata N. 1999. Genetic engineering of the unsaturation of fatty acids in membrane lipids alters the tolerance of Synechocystis to salt stress. Proc Natl Acad Sci U S A 96:5862–5867. doi: 10.1073/pnas.96.10.5862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allakhverdiev SI, Sakamoto A, Nishiyama Y, Inaba M, Murata N. 2000. Ionic and osmotic effects of NaCl-induced inactivation of photosystems I and II in Synechococcus sp. Plant Physiol 123:1047–1056. doi: 10.1104/pp.123.3.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Oren A. 2008. Microbial life at high salt concentrations: phylogenetic and metabolic diversity. Saline Systems 4:2. doi: 10.1186/1746-1448-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oren A. 1986. Intracellular salt concentrations of the anaerobic halophilic eubacteria Haloanaerobium praevalens and Halobacteroides halobius. Can J Microbiol 32:4–9. doi: 10.1139/m86-002. [DOI] [Google Scholar]
- 5.Ventosa A, Nieto JJ, Oren A. 1998. Biology of moderately halophilic aerobic bacteria. Microbiol Mol Biol Rev 62:504–544. doi: 10.1128/MMBR.62.2.504-544.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Galinski EA, Trüper HG. 1994. Microbial behavior in salt-stressed ecosystems. FEMS Microbiol Rev 15:95–108. doi: 10.1111/j.1574-6976.1994.tb00128.x. [DOI] [Google Scholar]
- 7.Hagemann M. 2011. Molecular biology of cyanobacterial salt acclimation. FEMS Microbiol Rev 35:87–123. doi: 10.1111/j.1574-6976.2010.00234.x. [DOI] [PubMed] [Google Scholar]
- 8.Robert MF. 2005. Organic compatible solutes of halotolerant and halophilic microorganisms. Saline Syst 1:5. doi: 10.1186/1746-1448-1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kempf B, Bremer E. 1998. Uptake and synthesis of compatible solutes as microbial stress responses to high-osmolality environments. Arch Microbiol 170:319–330. doi: 10.1007/s002030050649. [DOI] [PubMed] [Google Scholar]
- 10.Hagemann M, Schoor A, Erdmann N. 1996. NaCl acts as a direct modulator in the salt adaptive response: salt-dependent activation of glucosylglycerol synthesis in vivo and in vitro. J Plant Physiol 149:746–752. doi: 10.1016/S0176-1617(96)80101-5. [DOI] [Google Scholar]
- 11.Hagemann M, Schoor A, Jeanjean R, Zuther E, Joset F. 1997. The stpA gene form Synechocystis sp. strain PCC 6803 encodes the glucosylglycerol-phosphate phosphatase involved in cyanobacterial osmotic response to salt shock. J Bacteriol 179:1727–1733. doi: 10.1128/jb.179.5.1727-1733.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lunn JE, Price GD, Furbank RT. 1999. Cloning and expression of a prokaryotic sucrose-phosphate synthase gene from the cyanobacterium Synechocystis sp. PCC 6803. Plant Mol Biol 40:297–305. doi: 10.1023/a:1006130802706. [DOI] [PubMed] [Google Scholar]
- 13.Hagemann M, Effmert U, Kerstan T, Schoor A, Erdmann N. 2001. Biochemical characterization of glucosylglycerol-phosphate synthase of Synechocystis sp. strain PCC 6803: comparison of crude, purified, and recombinant enzymes. Curr Microbiol 43:278–283. doi: 10.1007/s002840010301. [DOI] [PubMed] [Google Scholar]
- 14.Desplats P, Folco E, Salerno GL. 2005. Sucrose may play an additional role to that of an osmolyte in Synechocystis sp. PCC 6803 salt-shocked cells. Plant Physiol Biochem 43:133–138. doi: 10.1016/j.plaphy.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 15.Marin K, Huckauf J, Fulda S, Hagemann M. 2002. Salt-dependent expression of glucosylglycerol-phosphate synthase, involved in osmolyte synthesis in the cyanobacterium Synechocystis sp. strain PCC 6803. J Bacteriol 184:2870–2877. doi: 10.1128/jb.184.11.2870-2877.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cumino AC, Perez-Cenci M, Giarrocco LE, Salerno GL. 2010. The proteins involved in sucrose synthesis in the marine cyanobacterium Synechococcus sp. PCC 7002 are encoded by two genes transcribed from a gene cluster. FEBS Lett 584:4655–4660. doi: 10.1016/j.febslet.2010.10.040. [DOI] [PubMed] [Google Scholar]
- 17.Novak JF, Stirnberg M, Roenneke B, Marin K. 2011. A novel mechanism of osmosensing, a salt-dependent protein-nucleic acid interaction in the cyanobacterium Synechocystis species PCC 6803. J Biol Chem 286:3235–3241. doi: 10.1074/jbc.M110.157032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Curatti L, Porchia AC, Herrera-Estrella L, Salerno GL. 2000. A prokaryotic sucrose synthase gene (susA) isolated from a filamentous nitrogen-fixing cyanobacterium encodes a protein similar to those of plants. Planta 211:729–735. doi: 10.1007/s004250000343. [DOI] [PubMed] [Google Scholar]
- 19.Vargas W, Cumino A, Salerno GL. 2003. Cyanobacterial alkaline/neutral invertases. Origin of sucrose hydrolysis in the plant cytosol? Planta 216:951–960. doi: 10.1007/s00425-002-0943-x. [DOI] [PubMed] [Google Scholar]
- 20.Perez-Cenci M, Salerno GL. 2014. Functional characterization of Synechococcus amylosucrase and fructokinase encoding genes discovers two novel actors on the stage of cyanobacterial sucrose metabolism. Plant Sci 224:95–102. doi: 10.1016/j.plantsci.2014.04.003. [DOI] [PubMed] [Google Scholar]
- 21.Kirsch F, Pade N, Klähn S, Hess WR, Hagemann M. 2017. The glucosylglycerol-degrading enzyme GghA is involved in acclimation to fluctuating salinities by the cyanobacterium Synechocystis sp. strain PCC 6803. Microbiology 163:1319–1328. doi: 10.1099/mic.0.000518. [DOI] [PubMed] [Google Scholar]
- 22.Kirsch F, Luo Q, Lu X, Hagemann M. 2018. Inactivation of invertase enhances sucrose production in the cyanobacterium Synechocystis sp. PCC 6803. Microbiology 164:1220–1228. doi: 10.1099/mic.0.000708. [DOI] [PubMed] [Google Scholar]
- 23.Scanlan DJ, Ostrowski M, Mazard S, Dufresne A, Garczarek L, Hess WR, Post AF, Hagemann M, Paulsen I, Partensky F. 2009. Ecological genomics of marine picocyanobacteria. Microbiol Mol Biol Rev 73:249–299. doi: 10.1128/MMBR.00035-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reed RH, Richardson DL, Warr SRC, Stewart W. 1984. Carbohydrate accumulation and osmotic stress in cyanobacteria. J Gen Microbiol 130:1–4. doi: 10.1099/00221287-130-1-1. [DOI] [Google Scholar]
- 25.Reed RH, Borowitzka LJ, Mackay MA, Chudek JA, Foster R, Warr SRC, Moore DJ, Stewart W. 1986. Organic solute accumulation in osmotically stressed cyanobacteria. FEMS Microbiol Lett 39:51–56. doi: 10.1111/j.1574-6968.1986.tb01842.x. [DOI] [Google Scholar]
- 26.Stal LJ, Reed RH. 1987. Low-molecular mass carbohydrate accumulation in cyanobacteria from a marine microbial mat in response to salt. FEMS Microbiol Ecol 45:305–312. doi: 10.1111/j.1574-6968.1987.tb02381.x. [DOI] [Google Scholar]
- 27.Klähn S, Steglich C, Hess WR, Hagemann M. 2010. Glucosylglycerate: a secondary compatible solute common to marine cyanobacteria from nitrogen-poor environments. Environ Microbiol 12:83–94. doi: 10.1111/j.1462-2920.2009.02045.x. [DOI] [PubMed] [Google Scholar]
- 28.Pade N, Michalik D, Ruth W, Belkin N, Hess WR, Berman-Frank I, Hagemann M. 2016. Trimethylated homoserine functions as the major compatible solute in the globally significant oceanic cyanobacterium Trichodesmium. Proc Natl Acad Sci U S A 113:13191–13196. doi: 10.1073/pnas.1611666113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tan X, Luo Q, Lu X. 2016. Biosynthesis, biotechnological production, and applications of glucosylglycerols. Appl Microbiol Biotechnol 100:6131–6139. doi: 10.1007/s00253-016-7608-3. [DOI] [PubMed] [Google Scholar]
- 30.Song K, Tan X, Liang Y, Lu X. 2016. The potential of Synechococcus elongatus UTEX 2973 for sugar feedstock production. Appl Microbiol Biotechnol 100:7865–7875. doi: 10.1007/s00253-016-7510-z. [DOI] [PubMed] [Google Scholar]
- 31.Ducat DC, Avelar-Rivas JA, Way JC, Silver PA. 2012. Rerouting carbon flux to enhance photosynthetic productivity. Appl Environ Microbiol 78:2660–2668. doi: 10.1128/AEM.07901-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Du W, Liang F, Duan Y, Tan X, Lu X. 2013. Exploring the photosynthetic production capacity of sucrose by cyanobacteria. Metab Eng 19:17–25. doi: 10.1016/j.ymben.2013.05.001. [DOI] [PubMed] [Google Scholar]
- 33.Duan Y, Luo Q, Liang F, Lu X. 2016. Sucrose secreted by the engineered cyanobacterium and its fermentability. J Ocean Univ China 15:890–896. doi: 10.1007/s11802-016-3007-8. [DOI] [Google Scholar]
- 34.Qiao C, Duan Y, Zhang M, Hagemann M, Luo Q, Lu X. 2017. Effects of reduced and enhanced glycogen pools on salt-induced sucrose production in a sucrose-secreting strain of Synechococcus elongatus PCC 7942. Appl Environ Microbiol 84:e02023-17. doi: 10.1128/AEM.02023-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martínez-Noël GMA, Cumino AC, Kolman MDLA, Salerno GL. 2013. First evidence of sucrose biosynthesis by single cyanobacterial bimodular proteins. FEBS Lett 587:1669–1674. doi: 10.1016/j.febslet.2013.04.012. [DOI] [PubMed] [Google Scholar]
- 36.Lunn JE. 2002. Evolution of sucrose synthesis. Plant Physiol 128:1490–1500. doi: 10.1104/pp.010898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cumino A, Curatti L, Giarrocco L, Salerno GL. 2002. Sucrose metabolism: Anabaena sucrose-phosphate synthase and sucrose-phosphate phosphatase define minimal functional domains shuffled during evolution. FEBS Lett 517:19–23. doi: 10.1016/S0014-5793(02)02516-4. [DOI] [PubMed] [Google Scholar]
- 38.Cid E, Gomis RR, Geremia RA, Guinovart JJ, Ferrer JC. 2000. Identification of two essential glutamic acid residues in glycogen synthase. J Biol Chem 275:33614–33621. doi: 10.1074/jbc.M005358200. [DOI] [PubMed] [Google Scholar]
- 39.Fieulaine S, Lunn JE, Borel F, Ferrer JL. 2005. The structure of a cyanobacterial sucrose-phosphatase reveals the sugar tongs that release free sucrose in the cell. Plant Cell 17:2049–2058. doi: 10.1105/tpc.105.031229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chua TK, Bujnicki JM, Tan TC, Huynh F, Patel BK, Sivaraman J. 2008. The structure of sucrose phosphate synthase from Halothermothrix orenii reveals its mechanism of action and binding mode. Plant Cell 20:1059–1072. doi: 10.1105/tpc.107.051193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ladas NP, Papageorgiou GC. 2000. Cell turgor: a critical factor for the proliferation of cyanobacteria at unfavorable salinity. Photosynth Res 65:155–164. doi: 10.1023/A:1006423221150. [DOI] [PubMed] [Google Scholar]
- 42.Kanesaki Y, Suzuki I, Allakhverdiev SI, Mikami K, Murata N. 2002. Salt stress and hyperosmotic stress regulate the expression of different sets of genes in Synechocystis sp. PCC 6803. Biochem Biophys Res Commun 290:339–348. doi: 10.1006/bbrc.2001.6201. [DOI] [PubMed] [Google Scholar]
- 43.Allakhverdiev SI, Sakamoto A, Nishiyama Y, Murata N. 2000. Inactivation of photosystems I and II in response to osmotic stress in Synechococcus: contribution of water channels. Plant Physiol 122:1201–1208. doi: 10.1104/pp.122.4.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Marin K, Stirnberg M, Eisenhut M, Kramer R, Hagemann M. 2006. Osmotic stress in Synechocystis sp. PCC 6803: low tolerance towards nonionic osmotic stress results from lacking activation of glucosylglycerol accumulation. Microbiology 152:2023–2030. doi: 10.1099/mic.0.28771-0. [DOI] [PubMed] [Google Scholar]
- 45.Klähn S, Hagemann M. 2011. Compatible solute biosynthesis in cyanobacteria. Environ Microbiol 13:551–562. doi: 10.1111/j.1462-2920.2010.02366.x. [DOI] [PubMed] [Google Scholar]
- 46.Tan X, Du W, Lu X. 2015. Photosynthetic and extracellular production of glucosylglycerol by genetically engineered and gel-encapsulated cyanobacteria. Appl Microbiol Biotechnol 99:2147–2154. doi: 10.1007/s00253-014-6273-7. [DOI] [PubMed] [Google Scholar]
- 47.Salerno GL, Porchia AC, Vargas WA, Abdian PL. 2004. Fructose-containing oligosaccharides: novel compatible solutes in Anabaena cells exposed to salt stress. Plant Sci 167:1003–1008. doi: 10.1016/j.plantsci.2004.05.029. [DOI] [Google Scholar]
- 48.Hagemann M, Marin K. 1999. Salt-induced sucrose accumulation is mediated by sucrose-phosphate-synthase in cyanobacteria. J Plant Physiol 155:424–430. doi: 10.1016/S0176-1617(99)80126-6. [DOI] [Google Scholar]
- 49.Cumino AC, Marcozzi C, Barreiro R, Salerno GL. 2007. Carbon cycling in Anabaena sp. PCC 7120: sucrose synthesis in the heterocysts and possible role in nitrogen fixation. Plant Physiol 143:1385–1397. doi: 10.1104/pp.106.091736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hagemann M, Wölfel L, Krüger B. 1990. Alterations of protein synthesis in the cyanobacterium Synechocystis sp. PCC 6803 after a salt shock. J Gen Microbiol 136:1393–1399. doi: 10.1099/00221287-136-7-1393. [DOI] [Google Scholar]
- 51.Fulda S, Huckauf J, Schoor A, Hagemann M. 1999. Analysis of stress responses in the cyanobacterial strains Synechococcus sp. PCC 7942, Synechocystis sp. PCC 6803, and Synechococcus sp. PCC 7418: osmolyte accumulation and stress protein synthesis. J Plant Physiol 154:240–249. doi: 10.1016/S0176-1617(99)80215-6. [DOI] [Google Scholar]
- 52.Weiss TL, Young EJ, Ducat DC. 2017. A synthetic, light-driven consortium of cyanobacteria and heterotrophic bacteria enables stable polyhydroxybutyrate production. Metab Eng 44:236–245. doi: 10.1016/j.ymben.2017.10.009. [DOI] [PubMed] [Google Scholar]
- 53.Castenholz RW. 1988. Culturing methods for cyanobacteria. Methods Enzymol 167:68–93. doi: 10.1016/0076-6879(88)67006-6. [DOI] [Google Scholar]
- 54.Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 55.Qiao C, Zhang M, Luo Q, Lu X. 2019. Identification of two two-component signal transduction mutants with enhanced sucrose biosynthesis in Synechococcus elongatus PCC 7942. J Basic Microbiol 59:465–476. doi: 10.1002/jobm.201800676. [DOI] [PubMed] [Google Scholar]
- 56.Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gao Z, Zhao H, Li Z, Tan X, Lu X. 2012. Photosynthetic production of ethanol from carbon dioxide in genetically engineered cyanobacteria. Energ Environ Sci 5:9857–9865. doi: 10.1039/C2EE22675H. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.