In societies that have food choices, conscious consumers demand natural solutions to keep their food healthy and fresh during storage, simultaneously reducing food waste. The use of “good bacteria” to protect food against spoilage organisms has a long, successful history, even though the molecular mechanisms are not fully understood. In this study, we show that the depletion of free manganese is a major bioprotective mechanism of lactobacilli in dairy products. High manganese uptake and intracellular storage provide a link to the distinct, nonenzymatic, manganese-catalyzed oxidative stress defense mechanism, previously described for certain lactobacilli. The evaluation of representative Lactobacillus species in our study identifies multiple relevant species groups for fungal growth inhibition via manganese depletion. Hence, through the natural mechanism of nutrient depletion, the use of dedicated bioprotective lactobacilli constitutes an attractive alternative to artificial preservation.
KEYWORDS: lactic acid bacteria, bioprotection, food spoilage, manganese starvation, genome editing, Lactobacillus
ABSTRACT
A prominent feature of lactic acid bacteria (LAB) is their ability to inhibit growth of spoilage organisms in food, but hitherto research efforts to establish the mechanisms underlying bioactivity focused on the production of antimicrobial compounds by LAB. We show, in this study, that competitive exclusion, i.e., competition for a limited resource by different organisms, is a major mechanism of fungal growth inhibition by lactobacilli in fermented dairy products. The depletion of the essential trace element manganese by two Lactobacillus species was uncovered as the main mechanism for growth inhibition of dairy spoilage yeast and molds. A manganese transporter (MntH1), representing one of the highest expressed gene products in both lactobacilli, facilitates the exhaustive manganese scavenging. Expression of the mntH1 gene was found to be strain dependent, affected by species coculturing and the growth phase. Further, deletion of the mntH1 gene in one of the strains resulted in a loss of bioactivity, proving this gene to be important for manganese depletion. The presence of an mntH gene displayed a distinct phylogenetic pattern within the Lactobacillus genus. Moreover, assaying the bioprotective ability in fermented milk of selected lactobacilli from 10 major phylogenetic groups identified a correlation between the presence of mntH and bioprotective activity. Thus, manganese scavenging emerges as a common trait within the Lactobacillus genus, but differences in expression result in some strains showing more bioprotective effect than others. In summary, competitive exclusion through ion depletion is herein reported as a novel mechanism in LAB to delay the growth of spoilage contaminants in dairy products.
IMPORTANCE In societies that have food choices, conscious consumers demand natural solutions to keep their food healthy and fresh during storage, simultaneously reducing food waste. The use of “good bacteria” to protect food against spoilage organisms has a long, successful history, even though the molecular mechanisms are not fully understood. In this study, we show that the depletion of free manganese is a major bioprotective mechanism of lactobacilli in dairy products. High manganese uptake and intracellular storage provide a link to the distinct, nonenzymatic, manganese-catalyzed oxidative stress defense mechanism, previously described for certain lactobacilli. The evaluation of representative Lactobacillus species in our study identifies multiple relevant species groups for fungal growth inhibition via manganese depletion. Hence, through the natural mechanism of nutrient depletion, the use of dedicated bioprotective lactobacilli constitutes an attractive alternative to artificial preservation.
INTRODUCTION
Growing demand for healthy and fresh foods without added artificial preservatives is increasing the need for natural, microbial solutions. Among microbes, lactic acid bacteria (LAB) have a long history in protecting food from spoilage, and, today, bioprotective bacteria that specifically inhibit the growth of spoilage organisms have emerged as an alternative way of keeping food fresh for longer periods of time (1).
More than 200 species are covered by the exceptionally diverse genus Lactobacillus, with other genera such as Pediococcus phylogenetically intermixed (2). Believed to originally be free living, lactobacilli today also include numerous host-adapted species, with habitats in livestock feed, plants, animals, humans, and fermented foods (3). Several species have a generally recognized as safe (GRAS) status and are applied within the dairy industry for the production of fermented milk products, and certain members are widely applied as probiotics; among these are especially Lactobacillus rhamnosus and Lactobacillus paracasei (4, 5). These two species display a more nomadic lifestyle capable of colonizing the human and animal gastrointestinal tract while also displaying efficient growth in milk.
In dairy products, spoilage by mold and yeast cells is one of the major problems that reduces shelf life (6). As a nutrient-rich environment, yogurt is inherently susceptible to microbial spoilage, although certain characteristics narrow the number of potential detrimental organisms. The innate carbon source is lactose; free amino acids are scarce and are instead concentrated in caseins, which require proteolysis for liberation, and the starter cultures responsible for conversion of milk to yogurt lower the pH to around 4.5. The typically observed spoilage organisms consist of various yeasts such as Debaryomyces hansenii, Saccharomyces cerevisiae, and several others, in addition to molds, especially of the Penicillium genus (6).
In the past decade, the bioprotective potential of LAB has spurred considerable efforts in the scientific community to identify new strains with bioprotective properties from various food sources (7–9), as well as attempts to elucidate the mechanisms behind the observed bioactivity (10–12). Numerous metabolites produced by LAB have been identified as having antifungal and antibacterial activities (13), although in situ concentrations are typically significantly below MICs (14). Meanwhile, other mechanisms have, until now, been largely unexplored.
Competitive exclusion is a widespread phenomenon in nature and includes the competition for nutrients such as a carbon source (15, 16); for a physical space, such as in the gastrointestinal tract (17); as well as for essential ions. The concept of nutritional immunity in the human body (18) is an example of competition for essential ions, reinforced by the various intricate means of iron scavenging through the action of specific siderophores present in bacterial pathogens (19).
Manganese (Mn) is an essential trace element that is a key cofactor in all kingdoms of life, making it important for the growth of bacteria, yeast, and mold (20). The two major manganese uptake systems in LAB are the NRAMP-type transporter MntH and the ABC transporter SitABC. While the ABC transporter is mainly active at neutral pH, the proton-driven symporter MntH is the major transport system under acidic conditions (21).
In Escherichia coli, manganese is a critical cofactor for the superoxide dismutase (SOD) protein (22). Aerotolerant lactic acid bacteria generally lack an active SOD gene and cope with reactive oxygen species through an alternative mechanism, which has mainly been studied in Lactobacillus plantarum (23). In this bacterium, intracellular manganese(II), accumulated in excessively high concentrations of up to 25 mM manganese, scavenges oxygen radicals (O2−) as effectively as SOD (23). This defense mechanism has been shown for other lactobacilli species (e.g., Lactobacillus casei and Lactobacillus fermentum) but is not present in all members of the genus (e.g., Lactobacillus bulgaricus and Lactobacillus acidophilus) (23). The manganese(II) in L. plantarum primarily associates with a large complex of nondialyzable polyphosphate-protein aggregates (24). The nonenzymatic manganese protection against superoxide in vivo was described in the early 1980s (23), but the mechanism was shown only recently in vitro (25). So far, reports of the mechanism in other bacteria are limited, although, in some organisms, it could serve as auxiliary protection (26). High intracellular manganese(II) concentration has, for example, previously been associated with other stress defense mechanisms, such as radiation resistance (27).
In this study, we show that the competition for manganese is a major limiting factor for the growth of spoilage organisms in yogurt containing a bioprotective culture of L. paracasei and L. rhamnosus. Furthermore, we investigated the bioprotective potential of the Lactobacillus genus based on this novel competition mechanism.
RESULTS
Manganese is the limiting factor for yeast growth in yogurt containing a bioprotective culture.
To analyze the activity of the bioprotective culture in yogurt in an easy and high-throughput manner, we developed an assay to determine the bioactivity based on yeast growth measured by absorbance at 600 nm. For this, we centrifuged the yogurt and filtered the supernatant to obtain a clear aqueous phase (AQ) of the yogurt. The AQ was inoculated with yeast cells, and the yeast growth was detected by absorbance measurement in a microplate reader after several days of incubation at 17°C. Analysis of D. hansenii growth by enumeration of cells was comparable in yogurt and AQ. The AQ of the reference (REF AQ) reached slightly lower cell counts, possibly due to a smaller amount of proteins in the AQ (Fig. S1). The yeast D. hansenii was chosen as the reference organism, as it is a common spoilage organism and is sensitive to bioprotective cultures in yogurt (12).
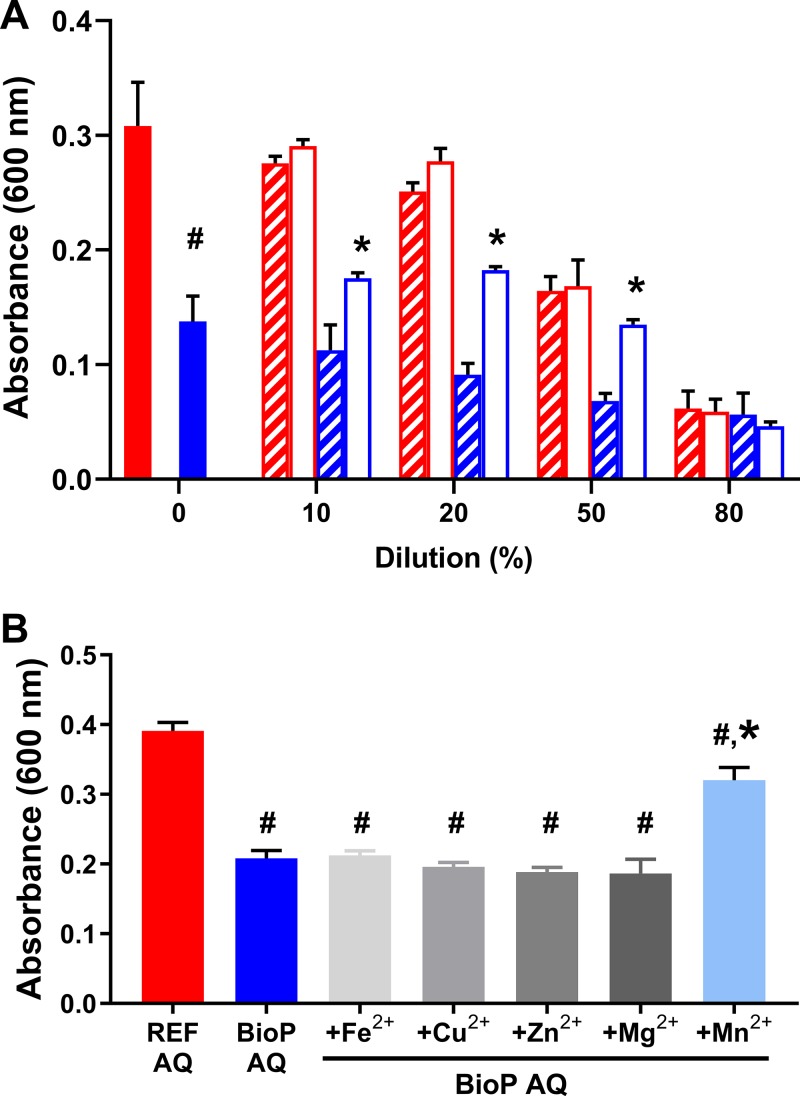
Here, the yogurt bioprotective culture consists of an L. rhamnosus strain and an L. paracasei strain, collectively designated BioP. A significant difference between D. hansenii growth in AQ of yogurt with (BioP AQ) and without (REF AQ) bioprotective culture was detected (P < 0.0001; n = 6, t = 9.292, df = 10; Student's t test, two tailed) (Fig. 1). Moreover, we noticed a difference in the bioactivity upon 10 to 50% (not 80%) dilution of BioP AQ with either tap water or MilliQ water (P < 0.0001; n = 3), while similar growth reduction was seen when diluting the REF AQ, i.e., there was no significant difference between D. hansenii growth levels in REF AQ when diluted 10 to 80% with tap water or MilliQ water (P > 0.05, n = 3; df = 32; two-way analysis of variance [ANOVA]; Fig. 1A). The decreased yeast growth in MilliQ-diluted samples suggests a dilution of a bioactive compound, but the results using tap water point to a lack of minerals in the BioP sample that can be replenished by tap water. To test which factor could be limiting, we added different metals to BioP AQ which could be present in tap water but filtered out in MilliQ water. In our assay, the added metal ion concentrations were in the same range as those present in milk (28, 29). Of all tested metals, only the addition of manganese resulted in the growth of D. hansenii that was comparable to growth in the REF AQ (Fig. 1B). In particular, the growth in BioP AQ (n = 5) and BioP AQ supplemented with iron, copper, zinc, or magnesium ions (n = 2 to 5) was not significantly different (P > 0.05), whereas the growth in BioP AQ and BioP AQ supplemented with manganese ions (n = 4) was significantly different (P < 0.0001; df = 19; one-way ANOVA). This indicates the availability of manganese ions as the sole limiting factor in the BioP AQ. We investigated the minimal concentration needed to promote growth of two different D. hansenii strains in BioP AQ (Fig. S2) and in a chemically defined medium (Fig. S3), and we identified a threshold of ∼0.01 mg/liter under both conditions. Furthermore, we measured the manganese concentration in the AQs, which was 0.03 mg/liter in REF AQ and below the quantification limit of 0.003 mg/liter in BioP AQ.
FIG 1.
Growth of D. hansenii in aqueous phase (AQ) of reference (REF) yogurt (red bars) and bioprotective strains (BioP) containing yogurt (blue bars). (A) The REF AQ and BioP AQ were diluted with tap water (clear bars) or MilliQ water (striped bars). Growth of D. hansenii was measured at 600 nm after 4 days of incubation at 17°C. (B) Effect of complementation BioP AQ with metals found in milk on the growth of D. hansenii (light blue bars). Growth was measured after 5 days of incubation at 17°C and compared to REF AQ without any addition of metal ions. Metal concentrations were used as found in milk (0.3 mg/liter Fe2+, 0.1 mg/liter Cu2+, 4.2 mg/liter Zn2+, 60 mg/liter Mg2+, and 0.03 mg/liter Mn2+). Mean and standard deviation of three replicates are indicated by the bars and error bars. Hashtags (#) indicate data being statistically significantly different (P < 0.0001) from REF AQ (A, Student's t test; B, one-way ANOVA), and asterisks (*) indicate data being different from BioP AQ diluted with MilliQ water (A, two-way ANOVA) or undiluted/nonsupplemented BioP AQ (B, one-way ANOVA).
Inhibition of different yeast and mold strains.
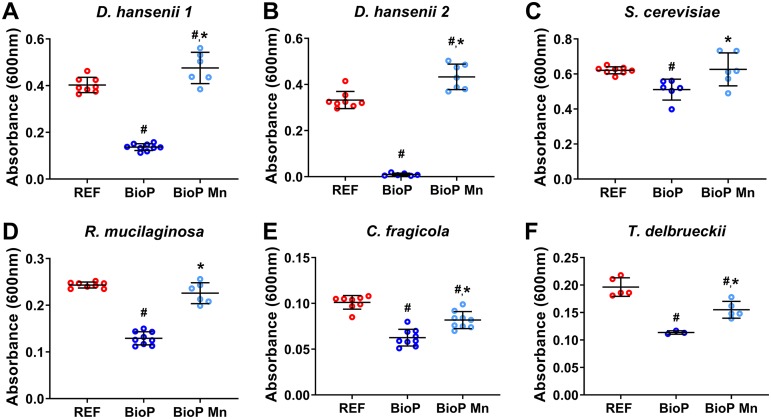
After investigating the effect of manganese on the growth of D. hansenii in BioP AQ, we analyzed the impact of manganese on different yeast strains. All yeast strains comprise food isolates that are relevant for food spoilage. The growth of all tested yeast strains was inhibited in the BioP AQ compared to REF AQ (#, P < 0.0001; n = 3 to 9; df = 10 to 23; one-way ANOVA in Fig. 2). The addition of manganese ions fully restored the growth of S. cerevisiae and Rhodotorula mucilaginosa in BioP AQ. In particular, no significant difference was detected between REF AQ and BioP AQ supplemented with manganese ions regarding the growth of R. mucilaginosa (P = 0.0990) and S. cerevisiae (P = 0.9860). In contrast, the growth of Cryptococcus fragicola, Torulaspora delbrueckii, and the two D. hansenii strains was significantly different in BioP AQ supplemented with manganese ions in comparison with REF AQ (#, P < 0.05 in Fig. 2). Nevertheless, significant differences were also observed between growth levels of these spoilage yeasts in BioP AQ with and without added manganese ions added (*, P < 0.05 in Fig. 2), supporting the view that the lack of free manganese ions in BioP AQ underlies the majority of its bioactivity. D. hansenii 1 growth was not significantly different between REF AQ and REF AQ with manganese addition (P = 0.1112) or REF AQ and BioP AQ both supplemented with manganese (P = 0.4442) (Fig. S4). We also tested the D. hansenii type strain (ATCC 36239), but the experiments with this strain were hampered due to its inability to grow in REF AQ. This exemplifies the need for relevant food isolates, as many of the strains intended for academic use have lost phenotypic traits that are key for industrial applications.
FIG 2.
Yeast growth in BioP AQ without (dark blue) and with (light blue) the addition of 0.6 mg/liter manganese and in REF AQ (red) after 5 days of incubation at 17°C. The following yeasts were tested: D. hansenii strain 1 (A), D. hansenii strain 2 (B), S. cerevisiae (C), R. mucilaginosa (D), C. fragicola (E), and T. delbrueckii (F). Individual data points for the 6 to 9 replicates are shown along with indications of mean ± standard deviation. Hashtags (#) indicate data being statistically significantly different (P < 0.05) from REF AQ, and asterisks (*) indicate data being different from nonsupplemented BioP AQ (one-way ANOVA).
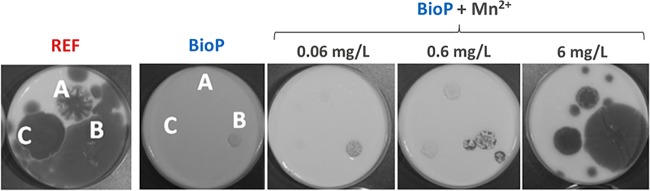
After showing the effect of limited manganese availability on yeast growth, we tested the effect of manganese on the growth of three different spoilage molds from the family Penicillium. These strains display various levels of sensitivity to bioprotective cultures in yogurt. Mold spores were spotted on the top of agar-solidified yogurt with and without bioprotective culture, and the plates were incubated for 8 days at room temperature. The sample with the bioprotective culture showed clear inhibition of the mold growth compared to the reference. The addition of increasing manganese concentrations of up to 6 mg/liter correlated positively with mold growth (Fig. 3).
FIG 3.
Growth of 3 different molds: P. brevicompactum (A), P. crustosum (B), and P. solitum (C) on plates prepared from milk fermented with starter culture alone (REF) or both starter and bioprotective culture (BioP). Different manganese concentrations were added as indicated. The spoilage molds were added in concentrations of 500 spores/spot. The plates were incubated at 22°C for 8 days.
Identification of bioprotective-relevant genes by transcriptomics.
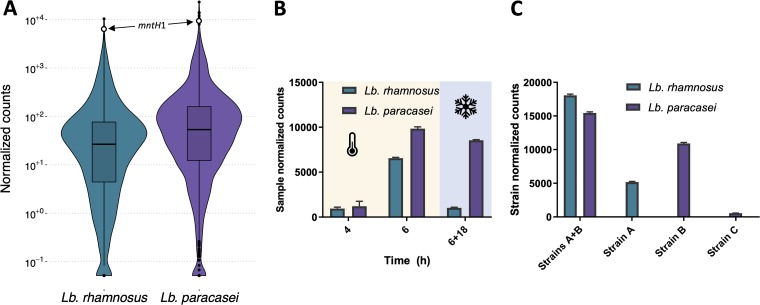
As different mechanisms of the observed manganese depletion could be hypothesized, for instance, through sequestration or uptake, we performed global gene expression profiles of the two Lactobacillus strains with bioprotective properties to examine the underlying mechanism. Experiments were performed in milk, but the starter culture required for yogurt production was omitted, as the relative abundance of BioP strain transcripts was comparatively marginal in yogurt. The depicted gene count distribution (Fig. 4A) of the cocultured BioP strains provides an example of the large count range that is typical of RNA sequencing (RNA-seq) data (30), here more than 5 orders of magnitude. For both strains, among the most expressed genes was an mntH manganese transporter-encoding gene (mntH1), remarkably displaying fifth- and seventh-highest read counts of all genes in L. rhamnosus and L. paracasei, respectively. mntH1 expression was magnitudes above the median (>100-fold) and putatively constitutes up to 1.8% of all transcripts. Such high expression, as judged from gene counts, of a manganese transporter gene is atypical. Other genes among the top 10 expressed relate to well-established highly expressed functions such as glycolysis or translation. Apart from the specified mntH1 gene, the genomes of L. rhamnosus and L. paracasei harbor one and two additional mntH homologs (mntH2 and mntH3), respectively. The two specified mntH1 genes are orthologs and are, in both species, the most distal mntH gene to the origin of replication in the clockwise direction. For the additional mntH paralogs, the expression is more than 200-fold lower; hence, their products are unlikely to contribute significantly to manganese uptake. Additionally, both genomes harbor the ABC manganese transport system (sitABC) with 20- and 10-fold lower-expression than the key mntH1 gene, respectively, in L. rhamnosus and L. paracasei (Table S1).
FIG 4.
Overall and mntH1-specific gene count distribution. (A) Sample-normalized gene count distribution of BioP L. rhamnosus and L. paracasei during coculture after 6 h milk fermentation. A white circle denotes the mntH1 count level. (B) Temporal, sample-normalized mntH1 count levels of BioP strains during 37°C milk fermentation (4 h, 6 h), and after 6 h of 37°C milk fermentation and 18 h of 7°C storage (6 + 18 h). (C) Normalized mntH1 count levels after 6 h of coculture (strains A and B) and individual culture (strain A, strain B, and strain C) in milk. Normalization is here performed individually for each strain, also for coculture. Included are the BioP strains (strains A and B) and an L. paracasei strain with a lower level of bioprotective activity (strain C).
The temporal mntH1 expression (Fig. 4B) revealed a marked increase from 4 to 6 h of milk fermentation (false-discovery rate [FDR] < 10−37) and with the expression remaining high during cold storage (400 to 500 times above median transcript levels), which indicates continued manganese uptake capacity, even during product shelf life. This correlates with an increase in bioactivity during storage (Fig. S5). Apart from temporal expression changes, mntH1 expression also seems to be a function of strain genotype and coculturing (Fig. 4C). Culturing strains individually led to lower mntH1 expression for both Lactobacillus strains than coculturing. Apart from coculturing, the effect of genotype on mntH1 expression seems to be correlated with bioprotective activity. L. paracasei strain C, for instance, was included based on its lower bioprotective activity (data not shown) and, in fact, displays a markedly lower mntH1 expression level (20-fold) than strain B, the corresponding BioP L. paracasei (Fig. 4C). In yogurt, two starter culture species (Streptococcus thermophilus and Lactobacillus delbrueckii) are ordinarily present. Of the two, only S. thermophilus also harbors an mntH gene. The relative expression of this gene in a representative strain is around 25-fold lower than the BioP strains (data not shown) and thereby comparable to L. paracasei strain C.
MntH1 is essential for bioprotective activity.
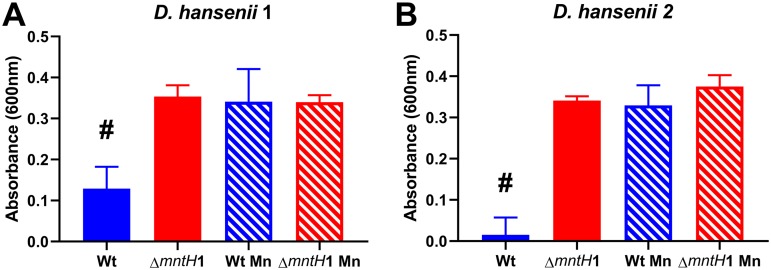
As the expression data provided strong indications of the importance of MntH1 for manganese uptake and the associated bioprotective phenotype, we sought to verify this through deletion of the mntH1 gene in the BioP L. paracasei strain B using a homologous recombination strategy coupled with CRISPR-Cas9-mediated mutant selection (Fig. S6). The bioprotective ability of the ΔmntH1 mutant was evaluated by assessing yeast growth in the AQ of milk fermented by individual strains (Fig. 5).
FIG 5.
Effect of mntH1 deletion on yeast growth inhibition. Yeast growth in AQ of milk fermented by wild-type L. paracasei strain B (WT) and ΔmntH1 L. paracasei strain B, without (full bars) and with 0.6 mg/liter manganese addition (dashed bars). Two yeast, D. hansenii 1 (A) and D. hansenii 2 (B), were included, and the growth was measured after 5 days of incubation at 17°C. Means and standard deviations of two biological replicates are indicated by bars and error bars. Hashtags (#) indicate data being statistically significantly different (P < 0.0001) from WT with manganese (Student's t test).
While the milk fermentation profile was identical for the wild-type (WT) BioP strain B and its ΔmntH1 mutant (Fig. S7), yeast growth was only inhibited in the AQ of milk fermented by the WT BioP strain B and not by the corresponding ΔmntH1 mutant. The addition of 0.6 mg/liter manganese resulted in restored yeast growth for WT BioP strain B, comparable to the growth in the AQ of milk fermented by the corresponding ΔmntH1 mutant with and without manganese addition. Complementation of the ΔmntH1 mutant with a plasmid containing the mntH1 gene under its own promoter fully restored bioactivity, while an empty plasmid control did not (Fig. S8). These results provide further evidence that mntH1 is responsible for the bioprotective phenotype. Culturing strains individually for a longer time in this experiment provides the strains with an increased potential for manganese uptake, compared to the coculture with starter culture normally performed for yogurt production. Even so, yeast inhibition by manganese uptake is insufficient in the ΔmntH strain to inhibit the growth of D. hansenii, despite the existence of two alternative manganese transporters within the genome.
Correlation between bioprotective activity and mntH phylogenetic distribution.
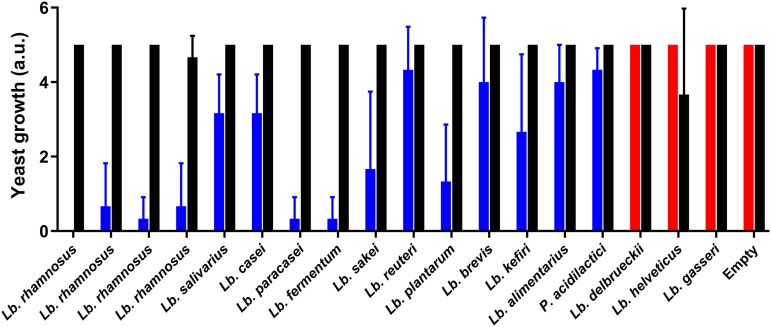
As the MntH1 transporter seems to be heavily involved in the increased uptake of manganese in certain lactobacilli, we investigated the presence of any mntH homologs in different species of the genus Lactobacillus (Fig. S9). In total, 18 strains were selected from 15 species, covering the major phylogenetic groups of the Lactobacillus sensu lato species tree (3). Of these, 15 strains harbored an mntH homolog, while 3 strains did not. In brief, a phylogenetic basis seems to exist for the distribution of an mntH gene among the lactobacilli. For the phylogenetic group containing the dairy-associated species, L. delbrueckii subsp. bulgaricus, Lactobacillus gasseri, and Lactobacillus helveticus, a homolog is absent. In contrast, in representatives of the nine other studied groups, it is present. The correlation between mntH presence and yeast inhibition was subsequently investigated experimentally. We performed milk acidification with a starter culture in the presence of the different lactobacilli. The fermented milk was divided into two samples, and 6 mg/liter manganese was added to one sample, while the other sample was left unchanged. A bioactivity assay following D. hansenii growth in both conditions was performed (Fig. 6). The majority of the tested strains containing an mntH gene were bioactive, at various levels, while those without the gene were not bioactive. The presence of the manganese ABC transporter did not correlate better with activity, as it is only present in L. rhamnosus, L. casei, L. paracasei, and L. plantarum. The addition of manganese restored the growth of D. hansenii completely in all samples, proving this metal to be the limiting factor.
FIG 6.
Growth scores of D. hansenii after 4 days of incubation at 17°C in fermented milk with a strain containing (blue) and not containing (red) an mntH gene. The addition of 6 mg/liter manganese restored the yeast growth in all cases (black). The average and standard deviation of two biological independent experiments are shown (n = 2).
DISCUSSION
In this study, we found that the ability of diverse lactobacilli to scavenge manganese results in inhibited growth of yeast and mold in fermented milk. Several studies have previously investigated the ability of lactobacilli to inhibit fungi in various foods, and the usage of lactobacilli for this purpose in commercial applications is gaining considerable interest. Thus far, bacterial production of inhibitory metabolites has been the focus of most studies, but the identified compounds are mostly found in concentrations markedly below the corresponding MIC of the metabolite (14). Our study shows that competitive exclusion is a major mechanism employed by many of lactobacilli to inhibit fungal growth in fermented milk. As a mechanism to avoid unwanted fungal growth, competitive exclusion is known from the concept of “nutritional immunity” in the human body, where depletion of iron and zinc in the respective parts of the body results in inhibition of pathogenic fungal and bacterial growth (18). In the context of lactic acid bacteria, examples of competitive exclusion of nutrients are scarce. A study by Honoré et al. (16) found that L. paracasei-mediated consumption of glucose and certain amino acids in a chemically defined medium correlated with reduced mold growth on the spent culture medium.
Here, the reduction of available manganese was proven as an effective inhibition mechanism against all the tested yeasts and molds. Yet two yeast strains, T. delbrueckii and C. fragicola, only displayed partially restored growth upon manganese addition (Fig. 2), indicating that other mechanisms, such as higher acid production or other antimicrobial compounds, could be involved as well (31). In milk, manganese is one of the essential trace metals with the lowest concentration (28, 29), and, consequently, the bacterial consumption required to reach a growth inhibitory concentration is low. Unlike the addition of other ions, the addition of 0.03 mg/liter manganese restored D. hansenii growth. This correlates well with the determined minimal manganese concentration threshold for D. hansenii growth of ∼0.01 mg/liter. In contrast, the addition of a higher concentration was needed when the assay was performed in yogurt (6 mg/liter) than during its aqueous phase (Fig. 3; Fig. S1). This can be attributed to viable bacteria present in the yogurt capable of taking up additional manganese during storage. Transcriptional profiling of BioP strains also identified high mntH1 gene expression during storage (Fig. 4B), indicating continued transport activity in this period and thereby a potential for further bacterial manganese uptake during storage.
In the BioP strains, the expression level of the mntH1 gene is exceptionally high, even surpassing most glycolytic genes. In comparison, an S. thermophilus starter culture strain and the less yeast growth-inhibiting L. paracasei strain C display a 25-fold-lower mntH1 expression level. Consequently, an excessively high mntH expression level seems to be crucial for high bioprotective activity. The mntH1 expression level of the BioP strains depends, however, on the stage of fermentation (Fig. 4B). A significant temporal increase in expression is observed, which could be an effect of a lowered manganese concentration caused by increases in cell quantity. This finding is in line with the manganese concentration-dependent mntH regulation mediated by MntR, previously observed in Bacillus subtilis (32). For the stationary-phase samples taken after 18 h of growth at 7°C, strain divergence in expression is observed, with expression remaining high in the L. paracasei strain B and less so for the L. rhamnosus strain A, possibly signifying species-dependent activity during cold storage. The level of mntH1 expression was observed to increase during coculture of BioP strains compared to individual growth, possibly revealing a mutualistic or commensalistic relationship between the two organisms.
Deleting the highly transcribed mntH1 gene in L. paracasei strain B proved this particular MntH1 transporter to be essential for the bioprotective phenotype. As the mechanism of bioprotection occurs by manganese scavenging, it follows that MntH1 is also the transporter responsible for manganese depletion. Although two additional MntH transporters and an ABC transport system (sitABC) exist in L. paracasei, there is no apparent sufficient regulatory response toward sustaining manganese uptake levels in the ΔmntH1 mutant. Either the transport kinetics or the underlying transcriptional regulation of the alternative transport systems are inadequate to achieve the required manganese uptake level for inhibition of yeast growth. The relevance of the other transport systems could, however, be a function of manganese concentration and pH. The ABC transporter type has, for instance, been shown to display improved activity at neutral or alkaline pH, while MntH displayed improved activity in an acidic medium (21, 33). Previous studies of MntH in lactobacilli have provided ambiguous findings. In L. casei, deletion of two mntH transporters was required for reducing intracellular concentrations of manganese (34), while in another L. casei study, despite deletions in both mntH and sitABC, the growth rate was unaltered (35). The low manganese concentration tested in the former and latter study were, however, set at 0.1 mg/liter and 0.08 mg/liter, respectively, thus around 3 times higher than the concentration in milk. Possibly, these are too-high concentrations for a significant effect of mntH1 deletion to materialize. Alternatively, significant strain variation in MntH1 activity exists, as already identified among the Lactobacillus strains A, B, and C examined in this study.
Variation in MntH1 activity could also explain our observation that not all selected lactobacilli (Fig. 6) harboring an mntH gene were equally proficient at inhibiting the growth of D. hansenii. Therefore, the presence of an mntH gene per se does not ensure good inhibition potential of spoilage organisms; rather, the expression level could be decisive. In the current experimental setting, the ability to take up manganese also requires the ability to grow in milk.
The main purpose of excessive manganese uptake appears to be the requirement for high intracellular manganese concentrations toward superoxide stress protection. As a simple and more efficient alternative exists in the manganese superoxide dismutase protein, additional reasons for high intracellular manganese concentrations may exist. It could be speculated whether excessive manganese scavenging as a competitive exclusion mechanism in itself could provide a fitness advantage in natural settings and thereby partially explain this characteristic among many lactobacilli.
In conclusion, a bacterial mechanism, originally identified as providing protection against oxidative stress, can, from this study, be coupled to facilitate another general bacterial mechanism, that of competitive exclusion. To our knowledge, such coupling in bacteria is without prior example. Moreover, it was shown that the principle of competitive exclusion, through the action of bioprotective strains, can be harnessed toward inhibiting the growth of unwanted spoilage organisms in a food source. The bioprotective activity of a strain in the present context, yogurt, can be defined as the sum of its ability to grow in milk, grow at the given fermentation temperature, and, not least, the expression of an mntH manganese transporter gene. Without the production of an antibiotic or other small molecule, competitive exclusion constitutes a mechanism that should be difficult for spoilage organisms to overcome by spontaneous mutation, making it an ideal mechanism for protection of food from a consumer, commercial, and regulatory perspective.
MATERIALS AND METHODS
Strains and growth conditions.
Strains used in this study are shown in Table 1. The yeast strains were streaked on yeast extract glucose chloramphenicol (YGC) plates (1 g/liter yeast extract [Merck], 20 g/liter d-glucose [Merck], and 0.1 g/liter chloramphenicol [Sigma-Aldrich]) and grown for 48 h at 25°C. Single colonies were inoculated to 25 ml YG media (1 g/liter yeast extract and 20 g/liter d-glucose) and grown overnight at 25°C with shaking. Afterward, glycerol was added to a final concentration of 15% (wt/vol), and the yeast cells were stored at −80°C until further use.
TABLE 1.
Strains used in this study
| Strain type and name | Comments/accession no. | Source |
|---|---|---|
| Yeasts | ||
| Torulaspora delbrueckii | Chr. Hansen A/S | |
| Cryptococcus fragicola | Chr. Hansen A/S | |
| Saccharomyces cerevisiae | Chr. Hansen A/S | |
| Debaryomyces hansenii strain 1 | Chr. Hansen A/S | |
| Debaryomyces hansenii strain 2 | Chr. Hansen A/S | |
| Rhodotorula mucilaginosa | Chr. Hansen A/S | |
| Molds | ||
| Penicillium brevicompactum | Chr. Hansen A/S | |
| Penicillium crustosum | Chr. Hansen A/S | |
| Penicillium solitum | Chr. Hansen A/S | |
| Lactobacilli | ||
| L. rhamnosus (strain A) | Part of BioP culture | Chr. Hansen A/S |
| L. paracasei (strain B) | Part of BioP culture | Chr. Hansen A/S |
| L. paracasei (strain C) | Chr. Hansen A/S | |
| L. rhamnosus (strain 1) | Chr. Hansen A/S | |
| L. rhamnosus (strain 2) | Chr. Hansen A/S | |
| L. rhamnosus (strain 3) | Chr. Hansen A/S | |
| L. rhamnosus (strain 4) | AZCQ00000000.1 | LMG 6400 |
| L. salivarius | Chr. Hansen A/S | |
| L. casei | Chr. Hansen A/S | |
| L. paracasei | Chr. Hansen A/S | |
| L. fermentum | Chr. Hansen A/S | |
| L. sakei | AZDN00000000 | LMG 9468 |
| L. reuteri | AZDD00000000 | LMG 9213 |
| L. plantarum | Chr. Hansen A/S | |
| L. brevis | Chr. Hansen A/S | |
| L. kefiri | AYYV00000000 | DSM 20587 |
| L. alimentarius | Chr. Hansen A/S | |
| Pediococcus acidilactici | AEEG00000000 | NCFB 2767 |
| L. delbrueckii subsp. bulgaricus | mntH not present | Chr. Hansen A/S |
| L. helveticus | mntH not present | Chr. Hansen A/S |
| L. gasseri | mntH not present | DSM 20243 |
| ΔmntH1 mutant | L. paracasei strain B ΔmntH1 | This study |
The bacterial strains were streaked on MRS (Sigma-Aldrich) plates and incubated anaerobically at 37°C for 48 h. Single colonies were inoculated into 3 ml MRS medium and grown overnight at 37°C. Ten microliters of the overnight culture were added to 2 ml heat-treated milk containing 0.02% (vol/wt) commercial starter culture and a pH indicator. The acidification was performed at 43°C and stopped after ∼6 h when a pH of 4.5 was reached. The fermented milk was kept in the refrigerator upon further use.
Yogurt production.
Reduced-fat (1.5% wt/vol) homogenized milk was heat treated at 90 ± 1°C for 20 min and cooled immediately. A commercial starter culture (S. thermophilus and L. delbrueckii subsp. bulgaricus) was inoculated at 0.02% (vol/wt) in 3-liter buckets. One bucket was inoculated with bioprotective culture (+BioP) in a total concentration of 100 U/ton, and one bucket was used as a reference (REF) and only inoculated with the starter culture. All buckets were incubated in a water bath at 43°C and fermented at these conditions until a pH of 4.60 was reached. At this time point, the fermented milk products were divided into 200-ml bottles and cooled down.
Aqueous phase production and manganese concentration determination.
The fermented milk product was centrifuged (10 min at 5,000 rpm), and the supernatant (AQ) was sterile filtered. The manganese concentration of the AQ was determined by inductively coupled plasma mass spectrometry at Eurofins Steins Laboratorium A/S.
Assay to detect bioactivity in AQ against yeasts.
The AQ of reference and BioP were transferred to a sterile 96-well plate (150 μl in each well), and dilutions were performed or different concentrations of metals were added (6 mg/liter to 6 ng/liter of manganese, 0.3 mg/liter of iron, 0.1 mg/liter of copper, 4.2 mg/liter of zinc, and 60 mg/liter magnesium).
Washed L. paracasei (wild-type strain B and ΔmntH1) cultures were added to 2 ml milk and grown for ∼18 h at 37°C until pH 4.5 was reached. The fermented milk was centrifuged (10 min at 5,000 rpm), and the supernatant (AQ) was transferred to a sterile 96-well plate (150 μl in each well).
The different yeasts (Table 1) were taken from the glycerol stock and diluted in MilliQ H2O to inoculate with ∼20 cells per well. The plates were incubated at 17°C for several days, and the yeast growth was determined by measuring the absorbance at 600 nm.
Assay to detect bioactivity of lactobacilli against yeast in fermented milk.
Lactobacilli were selected from the Chr. Hansen A/S strain collection. If available, species type strains were selected, while strains from the remaining species were selected based on the availability of the corresponding genome sequence in an otherwise unbiased selection process. One hundred fifty microliters of the fermented milk were transferred to individual wells in a 96-well plate. Manganese (6 mg/liter) was added to half of the samples, and the wells were inoculated with about 20 cells of the respective yeast. After 4 days of incubation at 17°C, a dilution row was spotted on selective YGC agar plates to analyze the yeast growth. In some cases, the growth was enumerated by optical inspection, where a value of 0 was given for no growth, a value of 1 for 1 to 2 colonies, a value of 2 for 2 to 10 colonies, a value of 3 for 10 to 30, a value of 4 for 30 to 70, and a value of 5 for confluent growth.
Assay to detect bioactivity in yogurt against molds.
The fermented milk products were divided into 200-ml bottles and cooled down. Different manganese concentrations (6 ng/liter to 6 mg/liter) were added to fermented milk products with BioP. All the fermented milk samples were heated to a temperature of 40°C and supplemented with 40 ml of a 5% sterile agar solution that had been melted and cooled down to 60°C. This solution of fermented milk and agar was then poured into sterile petri dishes and the plates were dried in an LAF bench for 30 min. Three target contaminants, Penicillium brevicompactum, Penicillium crustosum, and Penicillium solitum, were added in concentrations of 500 spores/spot. The plates were incubated at 22 ± 1°C for 8 days before assessment of mold growth.
Isolation and processing of RNA.
Initially, three independent precultures of each strain were cultivated anaerobically overnight at 37°C. Cultures were washed twice in 0.9% saline solution and inoculated in milk, which, in this case, was skim milk powder reconstituted (9.5%) in distilled water. Strain A was inoculated to an optical density at 600 nm (OD600) corresponding to 0.1, strain B to 0.5, strain C to 0.3, and the combined BioP culture to 0.1. The differences in inoculation level were chosen based on preliminary acidification rates and a growth rate proxy, with an inverse relationship between acidification rate and selected inoculation level. Prior to inoculation, lactic acid was added to reduce pH to 5.5 to simulate the acidification normally provided by the starter culture. Milk fermentation was performed at 37°C in 2-ml deep-well plates covered by foil, and proceeded either for 4 h and 6 h before harvest or for 6 h at 37°C and followed by 18 h at 7°C. Growth was performed in triplicate samples for each strain, and not all strains were subjected to all conditions. Cell harvest was performed by mixing a 1:2 volume of milk fermentate (2 ml) to RNAprotect bacteria reagent (Qiagen) (4 ml) followed by a procedure for separation of cells from milk fermentate, adapted from Derzelle et al. (36). A 2-ml volume of 1 M sodium citrate solution and 0.78 ml saline solution (0.145 M sodium chloride, 0.016 M sodium β-glycerophosphate, 0.1% Tween 80, pH 7) were added and, after 5 min, centrifuged at 10,000 × g for 2 min. The resulting cell pellet was washed with cold phosphate buffer (5 mM sodium phosphate, 1 mM EDTA, pH 7) and centrifuged as before, and the supernatant was discarded and the cell pellet was frozen at −80°C. The cell pellet was dissolved in Tris-EDTA buffer containing lysozyme (15 mg/ml), proteinase K (1.3 mg/ml), and mutanolysin (50 U) and was shaken at 1,400 rpm for 10 min at 37°C. The subsequent RNA extraction procedure was performed with the RNeasy protect bacteria minikit (Qiagen) per the manufacturer’s instructions, including removal of DNA with DNase I. The quality of total RNA was evaluated using a Bioanalyzer 2100 (Agilent). Depletion of rRNA, library preparation, and sequencing (Illumina 50-bp single-end sequencing) was performed at Keygene NV, The Netherlands.
RNA-seq data analysis.
Obtained raw reads, 15 to 45 million per sample, were trimmed with Trimmomatic (37) using default parameters, and mapping was performed using CLC Genomics Workbench v10 (Qiagen) with the following parameters: mismatch cost, 2; insertion cost, 3; deletion cost, 3; length fraction, 0.8; similarity fraction, 0.9; and strand-specific local alignment and maximum number of hits for a read of 10. For reads with 2 to 10 hits, reads were randomly assigned a gene. For BioP culture samples, mapping was performed for both genomes simultaneously and to the relevant individual genome for strain A, B, and C samples. Strain C reads were mapped to the strain B genome, as they are identical species. Following mapping, total gene counts were extracted. Within the SARTools (38) framework, the DESeq2 R package (39) was applied for normalization of sample gene counts using the median ratio method and subsequent estimation of differential expression. For comparison of mntH gene counts, it was necessary to compare gene counts between samples of different species that harbor distinct genes and therefore cannot immediately be normalized together using DESeq2. Therefore, an additional normalization between sample types containing different species compositions (BioP, strain A, strain B, or strain C) was performed on within-species normalized gene counts applying transcripts per kilobase million (TPM) normalization. For BioP culture samples, additional TPM normalization was performed for the sample as a whole (sample normalized), but also individually for strain A and strain B (strain normalized). This gives different gene count distributions, as more counts were associated overall with strain B over strain A for the 6-h samples.
Generation of the ΔmntH1 mutant.
Clean deletion of the entire ΔmntH1 in L. paracasei strain B was performed by a combination of homologous recombination and CRISPR-Cas9 counter selection (Text S1), as previously described (40). Briefly, 500-bp homology arms flanking mntH1 were employed for homologous recombination, while a targeting CRISPR-Cas9 nuclease was utilized for selection of ΔmntH1 deletion mutants. Transformation of L. paracasei was adapted from Song et al. (41), and to improve transformation efficiency, a procedure for obtaining nonmethylated plasmids was employed (42). Once a correct mutant was isolated and sequence verified, the plasmids were cured through nonselective growth cycles. Oligonucleotides and plasmids used in this study are shown in Table 2 and 3, respectively.
TABLE 2.
Oligonucleotides used in this study
| Name | Sequence |
|---|---|
| oJV2 | GGCCGCATGTTTTGGGACCATTCAAAACAGCATAGCTCTAAAACTTGTTCTGATCGTAGCTATCCTCGAT |
| oJV3 | CTGTAATTTGTTTAATTGCCATTTCAATT |
| oJV4 | GGAACTACAAAATAAATTATAAGGAGGC |
| oJV5 | AATAACTCTCCCCTTTCG |
| oJV6 | TTTGTTTGAATCTTTGGCC |
| oJV7 | TTTTGCTCACATGTTCTTTC |
| oJV8 | CTGCTTTTTGGCTATCAATC |
| oJV9 | CCGCTTCGGTTTGAGCTTTGAT |
| oJV10 | TGACATGCTGCTTTTTGGCCAT |
| oJV11 | AGCGGCTTTACTAGGGAAACTGA |
| oEFB0072 | GGAGCTGTAATATAAAAACCTTCTTC |
| oEFB0106 | AAGCTTTCTTTGAACCAAAATTAG |
| oEFB0107 | GAAGAAGGTTTTTATATTACAGCTCCGTGACTTTTTAACAATAACGGCAATTC |
| oEFB0108 | CTAATTTTGGTTCAAAGAAAGCTTTCATTGTTCGTCAACATCTGCCTTCG |
| oEFB0055 | CAACATCTTCGCTGCAAAGC |
| oEFB0056 | CTCTATTCAGGAATTGTCAG |
TABLE 3.
Plasmids used in this study
| Plasmid | Description | Resistance toa: | Source or reference |
|---|---|---|---|
| pCB578 | E. coli-lactobacilli shuttle vector containing SpCas9, tracrRNA, and a repeat-spacer-repeat array | Erm | 40 |
| pJV114 | Shuttle vector with pCB578 base containing a spacer targeting mntH1 | Erm | This work |
| pCB591 | E. coli-lactobacilli shuttle vector for homologous recombination template cloning | Amp (E. coli) Cm (Lactobacillus) | 40 |
| pJV88 | Shuttle vector with pCB591 base containing the mntH1 gene and 500-bp homology arms on either side | Amp (E. coli) Cm (Lactobacillus) | This work |
| pJV91 | Shuttle vector with pCB591 base containing 500-bp homology arms on either side of mntH1 that serves as a recombination template to generate a clean deletion | Amp (E. coli) Cm (Lactobacillus) | This work |
| pNZ8148 | Broad-host-range LAB expression vector containing NisA promoter and pepN terminator | Cm | Mobitech |
| pEFB021 | pNZ8148 in which NisA promoter has been replaced by the 207-bp native mntH1 promoter followed by mntH1 | Cm | This work |
Erm, erythromycin; Amp, ampicillin; Cm, chloramphenicol.
Statistical analysis.
Statistical analyses were conducted using Graph Pad Prism 8.2.0 (GraphPad Software, Inc., San Diego, CA). When applying one- or two-way ANOVA analyses, multiple comparisons were corrected for using Tukey’s statistical hypothesis testing. All relevant statistical information is reported along with P and n values.
Data availability.
Transcriptomic data are available from the NCBI Sequence Read Archive (SRA) repository under BioProject accession no. PRJNA602536. Strains will be available for research purposes upon request.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge our colleagues in the Discovery function of the R&D organization as well as the Dairy Bioprotection and Production Development departments in Chr. Hansen A/S for valuable discussions and sharing of results. We acknowledge Sonali Sirdesai from the University of Groningen and Morten Adler Hedegaard from the University of Copenhagen for assisting during the initial stages of this project. We acknowledge Keygene NV for method optimization and valuable discussions.
This work was supported in part by the National Science Foundation (MCB-1413044 to C.L.B.).
Certain authors are employed at Chr. Hansen A/S, a company that develops and commercializes bioprotective cultures.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Leyva Salas M, Mounier J, Valence F, Coton M, Thierry A, Coton E. 2017. Antifungal microbial agents for food biopreservation—a review. Microorganisms 5:37. doi: 10.3390/microorganisms5030037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Salvetti E, Harris HMB, Felis GE, O'Toole PW. 2018. Comparative genomics of the genus Lactobacillus reveals robust phylogroups that provide the basis for reclassification. Appl Environ Microbiol 84:e00993-18. doi: 10.1128/AEM.00993-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Duar RM, Lin XB, Zheng J, Martino ME, Grenier T, Pérez-Muñoz ME, Leulier F, Gänzle M, Walter J. 2017. Lifestyles in transition: evolution and natural history of the genus Lactobacillus. FEMS Microbiol Rev 41:S27–S48. doi: 10.1093/femsre/fux030. [DOI] [PubMed] [Google Scholar]
- 4.Smokvina T, Wels M, Polka J, Chervaux C, Brisse S, Boekhorst J, van Hylckama Vlieg JET, Siezen RJ. 2013. Lactobacillus paracasei comparative genomics: towards species pan-genome definition and exploitation of diversity. PLoS One 8:e68731. doi: 10.1371/journal.pone.0068731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Douillard FP, Ribbera A, Kant R, Pietilä TE, Järvinen HM, Messing M, Randazzo CL, Paulin L, Laine P, Ritari J, Caggia C, Lähteinen T, Brouns SJJ, Satokari R, von Ossowski I, Reunanen J, Palva A, de Vos WM. 2013. Comparative genomic and functional analysis of 100 Lactobacillus rhamnosus strains and their comparison with strain GG. PLoS Genet 9:e1003683. doi: 10.1371/journal.pgen.1003683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garnier L, Valence F, Mounier J, Garnier L, Valence F, Mounier J. 2017. Diversity and control of spoilage fungi in dairy products: an update. Microorganisms 5:42. doi: 10.3390/microorganisms5030042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Magnusson J, Ström K, Roos S, Sjögren J, Schnürer J. 2003. Broad and complex antifungal activity among environmental isolates of lactic acid bacteria. FEMS Microbiol Lett 219:129–135. doi: 10.1016/S0378-1097(02)01207-7. [DOI] [PubMed] [Google Scholar]
- 8.Bulgasem BY, Lani MN, Hassan Z, Yusoff WMW, Fnaish SG. 2016. Antifungal activity of lactic acid bacteria strains isolated from natural honey against pathogenic Candida Species. Mycobiology 44:302–309. doi: 10.5941/MYCO.2016.44.4.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wakil SM, Laba SA, Fasiku SA. 2014. Isolation and identification of antimicrobial-producing lactic acid bacteria from fermented cucumber. Afr J Biotechnol 13:2556–2564. doi: 10.5897/AJB2014.13704. [DOI] [Google Scholar]
- 10.Sjögren J, Magnusson J, Broberg A, Schnürer J, Kenne L. 2003. Antifungal 3-hydroxy fatty acids from Lactobacillus plantarum MiLAB 14. Appl Environ Microbiol 69:7554–7557. doi: 10.1128/aem.69.12.7554-7557.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schnürer J, Magnusson J. 2005. Antifungal lactic acid bacteria as biopreservatives. Trends Food Sci Technol 16:70–78. doi: 10.1016/j.tifs.2004.02.014. [DOI] [Google Scholar]
- 12.McNair LKF, Siedler S, Vinther JMO, Hansen AM, Neves AR, Garrigues C, Jäger AK, Franzyk H, Staerk D. 2018. Identification and characterization of a new antifungal peptide in fermented milk product containing bioprotective Lactobacillus cultures. FEMS Yeast Res 18:foy094. doi: 10.1093/femsyr/foy094. [DOI] [PubMed] [Google Scholar]
- 13.Crowley S, Mahony J, van Sinderen D. 2013. Current perspectives on antifungal lactic acid bacteria as natural bio-preservatives. Trends Food Sci Technol 33:93–109. doi: 10.1016/j.tifs.2013.07.004. [DOI] [Google Scholar]
- 14.Siedler S, Balti R, Neves AR. 2019. Bioprotective mechanisms of lactic acid bacteria against fungal spoilage of food. Curr Opin Biotechnol 56:138–146. doi: 10.1016/j.copbio.2018.11.015. [DOI] [PubMed] [Google Scholar]
- 15.Haidar R, Fermaud M, Calvo-Garrido C, Roudet J, Deschamps A. 2016. Modes of action for biological control of Botrytis cinerea by antagonistic bacteria. Phytopathol Mediterr 55:301–322. [Google Scholar]
- 16.Honoré AH, Aunsbjerg SD, Ebrahimi P, Thorsen M, Benfeldt C, Knøchel S, Skov T. 2016. Metabolic footprinting for investigation of antifungal properties of Lactobacillus paracasei. Anal Bioanal Chem 408:83–96. doi: 10.1007/s00216-015-9103-6. [DOI] [PubMed] [Google Scholar]
- 17.Golowczyc MA, Mobili P, Garrote GL, Abraham AG, De Antoni GL. 2007. Protective action of Lactobacillus kefir carrying S-layer protein against Salmonella enterica serovar Enteritidis. Int J Food Microbiol 118:264–273. doi: 10.1016/j.ijfoodmicro.2007.07.042. [DOI] [PubMed] [Google Scholar]
- 18.Gerwien F, Skrahina V, Kasper L, Hube B, Brunke S. 2018. Metals in fungal virulence. FEMS Microbiol Rev 42. doi: 10.1093/femsre/fux050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wandersman C, Delepelaire P. 2004. Bacterial iron sources: from siderophores to hemophores. Annu Rev Microbiol 58:611–647. doi: 10.1146/annurev.micro.58.030603.123811. [DOI] [PubMed] [Google Scholar]
- 20.Jensen AN, Jensen LT. 2014. Manganese transport, trafficking and function in invertebrates, 1–33. In Costa L, Aschner M (ed), Manganese in health and disease. The Royal Society of Chemistry, Cambridge, MA. [Google Scholar]
- 21.Porcheron G, Garénaux A, Proulx J, Sabri M, Dozois CM. 2013. Iron, copper, zinc, and manganese transport and regulation in pathogenic Enterobacteria: correlations between strains, site of infection and the relative importance of the different metal transport systems for virulence. Front Cell Infect Microbiol 3:90. doi: 10.3389/fcimb.2013.00090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takeda Y, Avila H. 1986. Structure and gene expression of the E. coli Mn-superoxide dismutase gene. Nucleic Acids Res 14:4577–4589. doi: 10.1093/nar/14.11.4577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Archibald FS, Fridovich I. 1981. Manganese, superoxide dismutase, and oxygen tolerance in some lactic acid bacteria. J Bacteriol 146:928–936. doi: 10.1128/JB.146.3.928-936.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Archibald FS, Fridovich I. 1982. Investigations of the state of the manganese in Lactobacillus plantarum. Arch Biochem Biophys 215:589–596. doi: 10.1016/0003-9861(82)90120-5. [DOI] [PubMed] [Google Scholar]
- 25.Barnese K, Gralla EB, Valentine JS, Cabelli DE. 2012. Biologically relevant mechanism for catalytic superoxide removal by simple manganese compounds. Proc Natl Acad Sci U S A 109:6892–6897. doi: 10.1073/pnas.1203051109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Culotta VC, Daly MJ. 2013. Manganese complexes: diverse metabolic routes to oxidative stress resistance in prokaryotes and yeast. Antioxid Redox Signal 19:933–944. doi: 10.1089/ars.2012.5093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun H, Xu G, Zhan H, Chen H, Sun Z, Tian B, Hua Y. 2010. Identification and evaluation of the role of the manganese efflux protein in Deinococcus radiodurans. BMC Microbiol 10:319. doi: 10.1186/1471-2180-10-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nantapo CTW, Muchenje V. 2013. Winter and spring variation in daily milk yield and mineral composition of Jersey, Friesian cows and their crosses under a pasture-based dairy system. S Afr J Anim Sci 43:17. doi: 10.4314/sajas.v43i5.3. [DOI] [Google Scholar]
- 29.Miller GD, Jarvis JK, McBean LD, National Dairy Council . 2007. Handbook of dairy foods and nutrition, 3rd ed CRC Press, Boca Raton, FL. [Google Scholar]
- 30.Dillies M-A, Rau A, Aubert J, Hennequet-Antier C, Jeanmougin M, Servant N, Keime C, Marot G, Castel D, Estelle J, Guernec G, Jagla B, Jouneau L, Laloë D, Le Gall C, Schaëffer B, Le Crom S, Guedj M, Jaffrézic F, French StatOmique Consortium . 2013. A comprehensive evaluation of normalization methods for Illumina high-throughput RNA sequencing data analysis. Brief Bioinform 14:671–683. doi: 10.1093/bib/bbs046. [DOI] [PubMed] [Google Scholar]
- 31.Lindgren SE, Dobrogosz WJ. 1990. Antagonistic activities of lactic acid bacteria in food and feed fermentations. FEMS Microbiol Rev 7:149–163. doi: 10.1111/j.1574-6968.1990.tb04885.x. [DOI] [PubMed] [Google Scholar]
- 32.Que Q, Helmann JD. 2000. Manganese homeostasis in Bacillus subtilis is regulated by MntR, a bifunctional regulator related to the diphtheria toxin repressor family of proteins. Mol Microbiol 35:1454–1468. doi: 10.1046/j.1365-2958.2000.01811.x. [DOI] [PubMed] [Google Scholar]
- 33.Kehres DG, Janakiraman A, Slauch JM, Maguire ME. 2002. Regulation of Salmonella enterica serovar Typhimurium mntH transcription by H2O2, Fe2+, and Mn2+. J Bacteriol 184:3151–3158. doi: 10.1128/jb.184.12.3151-3158.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Serata M, Yasuda E, Sako T. 2018. Effect of superoxide dismutase and manganese on superoxide tolerance in Lactobacillus casei strain Shirota and analysis of multiple manganese transporters. Biosci Microbiota Food Health 37:31–38. doi: 10.12938/bmfh.17-018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Groot MNN, Klaassens E, de Vos WM, Delcour J, Hols P, Kleerebezem M. 2005. Genome-based in silico detection of putative manganese transport systems in Lactobacillus plantarum and their genetic analysis. Microbiology 151:1229–1238. doi: 10.1099/mic.0.27375-0. [DOI] [PubMed] [Google Scholar]
- 36.Derzelle S, Bolotin A, Mistou M-Y, Rul F. 2005. Proteome analysis of Streptococcus thermophilus grown in milk reveals pyruvate formate-lyase as the major upregulated protein. Appl Environ Microbiol 71:8597–8605. doi: 10.1128/AEM.71.12.8597-8605.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Varet H, Brillet-Guéguen L, Coppée J-Y, Dillies M-A. 2016. SARTools: a DESeq2- and EdgeR-based R pipeline for comprehensive differential analysis of RNA-seq data. PLoS One 11:e0157022. doi: 10.1371/journal.pone.0157022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Leenay RT, Vento JM, Shah M, Martino ME, Leulier F, Beisel CL. 2019. Genome editing with CRISPR-Cas9 in Lactobacillus plantarum revealed that editing outcomes can vary across strains and between methods. Biotechnol J 14:1700583. doi: 10.1002/biot.201700583. [DOI] [PubMed] [Google Scholar]
- 41.Song X, Huang H, Xiong Z, Ai L, Yang S. 2017. CRISPR-Cas9D10A nickase-assisted genome editing in Lactobacillus casei. Appl Environ Microbiol 83:e01259-17. doi: 10.1128/AEM.01259-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang G, Wang W, Deng A, Sun Z, Zhang Y, Liang Y, Che Y, Wen T. 2012. A mimicking-of-DNA-methylation-patterns pipeline for overcoming the restriction barrier of bacteria. PLoS Genet 8:e1002987. doi: 10.1371/journal.pgen.1002987. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Transcriptomic data are available from the NCBI Sequence Read Archive (SRA) repository under BioProject accession no. PRJNA602536. Strains will be available for research purposes upon request.