Solventogenic clostridia are anaerobic bacteria that can produce butanol, ethanol, and acetone, which can be used as biofuels or building block chemicals. Here, we show that AdhR, a σ54-dependent transcriptional activator, senses the intracellular redox status and controls alcohol synthesis in Clostridium beijerinckii. AdhR provides a new example of a GAF domain coordinating a mononuclear non-heme iron to sense and transduce the redox signal. Our study reveals a previously unrecognized functional role of σ54 in control of cellular redox balance and provides new insights into redox signaling and regulation in clostridia. Our results reveal AdhR as a novel engineering target for improving solvent production by C. beijerinckii and other solventogenic clostridia.
KEYWORDS: enhancer binding protein, σ54, clostridia, GAF domain, redox homeostasis
ABSTRACT
The AdhR regulatory protein is an activator of σ54-dependent transcription of adhA1 and adhA2 genes, which are required for alcohol synthesis in Clostridium beijerinckii. Here, we identified the signal perceived by AdhR and determined the regulatory mechanism of AdhR activity. By assaying the activity of AdhR in N-terminally truncated forms, a negative control mechanism of AdhR activity was identified in which the central AAA+ domain is subject to repression by the N-terminal GAF and PAS domains. Binding of Fe2+ to the GAF domain was found to relieve intramolecular repression and stimulate the ATPase activity of AdhR, allowing the AdhR to activate transcription. This control mechanism enables AdhR to regulate transcription of adhA1 and adhA2 in response to cellular redox status. The mutants deficient in AdhR or σ54 showed large shifts in intracellular redox state indicated by the NADH/NAD+ ratio under conditions of increased electron availability or oxidative stress. We demonstrated that the Fe2+-activated transcriptional regulator AdhR and σ54 control alcohol synthesis to maintain redox homeostasis in clostridial cells. Expression of N-terminally truncated forms of AdhR resulted in improved solvent production by C. beijerinckii.
IMPORTANCE Solventogenic clostridia are anaerobic bacteria that can produce butanol, ethanol, and acetone, which can be used as biofuels or building block chemicals. Here, we show that AdhR, a σ54-dependent transcriptional activator, senses the intracellular redox status and controls alcohol synthesis in Clostridium beijerinckii. AdhR provides a new example of a GAF domain coordinating a mononuclear non-heme iron to sense and transduce the redox signal. Our study reveals a previously unrecognized functional role of σ54 in control of cellular redox balance and provides new insights into redox signaling and regulation in clostridia. Our results reveal AdhR as a novel engineering target for improving solvent production by C. beijerinckii and other solventogenic clostridia.
INTRODUCTION
Clostridia are Gram-positive obligate anaerobes important in human health and biotechnological applications (1, 2). The genus includes several well-known human pathogens, as well as many species capable of producing chemicals and fuels through fermentation (3–5). Among Clostridium spp., Clostridium beijerinckii is one of the best-studied species and has been extensively used for production of solvents (acetone, butanol, and ethanol) (6, 7).
As anaerobes, clostridia maintain the cellular redox balance mainly through the reactions of central metabolism. The reducing equivalents are usually generated from sugar catabolism through glycolysis and reoxidized via the formation of fermentation products, particularly the synthesis of alcohols (8, 9). Disturbance of the redox balance in clostridial cells, such as by oxidative stress or increased electron availability, can severely affect the metabolism and even damage essential cellular components (10, 11). The redox-sensing regulator Rex has been found to modulate fermentation product formation and oxidative stress tolerance in Clostridium acetobutylicum (12). However, little is known about the regulatory mechanisms that maintain redox balance in other clostridia, such as C. beijerinckii. In our previous study, the sigma factor σ54 (SigL) and the transcriptional activator AdhR were found to control alcohol synthesis in C. beijerinckii (13). σ54 is required for transcription of adhA1 and adhA2 genes, encoding alcohol dehydrogenases (ADHs). Both ADHs, which could work together or individually (14), are responsible for NAD(P)H-dependent synthesis of butanol and ethanol (13). AdhR has been identified as an activator of σ54-dependent transcription of adhA1 and adhA2 (13). However, the environmental signals perceived by AdhR are unknown.
AdhR has a modular domain architecture characteristic of bacterial enhancer binding proteins (bEBPs) for σ54-dependent transcription (15). The central domain of AdhR is an AAA+ (ATPase associated with various cellular activities) domain (Fig. 1A), which would be responsible for ATP hydrolysis to drive the formation of the transcriptionally competent open complex. The C-terminal domain contains a helix-turn-helix motif that directs the binding of AdhR to three sites of upstream activator sequences (UAS) located upstream of the adhA1 and adhA2 promoters (13). Moreover, AdhR contains a GAF domain (named for cyclic GMP-specific and stimulated phosphodiesterases, Anabaena adenylate cyclases, and Escherichia coli FhlA) and a PAS (Per, ARNT, and Sim) domain at the N terminus (Fig. 1A). The GAF and PAS domains are often found in signaling proteins (16, 17). Activation of σ54-dependent transcription is usually controlled by sensory modules of bEBPs (18). It remains to be explored how AdhR responds to environmental signals and modulates its activity.
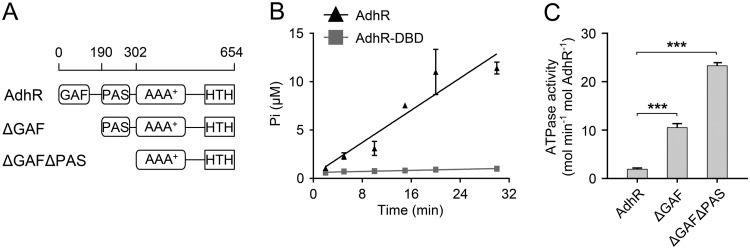
FIG 1.
N-terminal domains repress the ATPase activity of AdhR. (A) Schematic representation of the domains of AdhR and its truncated derivatives AdhRΔGAF and AdhRΔGAFΔPAS. (B) Assay of ATPase activities of AdhR determined by measuring the released Pi. (C) ATPase activities of AdhR and its truncated derivatives. Data shown in all figures are means ± the SD (n = 3 independent experiments). Statistical analysis was performed with the unpaired two-tailed Student t test (***, P < 0.001).
In this study, we constructed N-terminally truncated forms of AdhR and characterized the activity of AdhR by assaying the ATPase activity in vitro and determining the transcriptional activation of adhA1 and adhA2 genes in C. beijerinckii. We show that the N-terminal GAF and PAS domains negatively regulate the activity of the central AAA+ domain in AdhR. Binding of ferrous iron to the GAF domain was found to relieve intramolecular repression and stimulate the activity of AdhR. Furthermore, we disturbed the cellular redox balance by formate supplementation or limited exposure to O2 and rigorously analyzed its influences on transcriptional activation by AdhR, intracellular redox homeostasis, and oxidative stress tolerance in C. beijerinckii strains. The effect of truncation of AdhR on fermentation product formation was also studied. Our results demonstrate that the AdhR senses the intracellular redox state and controls alcohol synthesis to maintain redox homeostasis in C. beijerinckii.
RESULTS
N-terminal domains negatively regulate the activity of the central AAA+ domain in AdhR.
To ascertain the ability of AdhR to hydrolyze ATP, we performed ATPase activity assays using the full-length AdhR protein from C. beijerinckii, which was overexpressed in E. coli with the N-terminal hexahistidine and glutathione S-transferase (GST) tags. After protein purification, the His6 and GST tags were removed by tobacco etch virus (TEV) protease. The results clearly confirm that AdhR possesses an intrinsic ATPase activity (Fig. 1B). As a negative control, the DNA-binding domain (DBD) of AdhR was used, and no ATPase activity was observed (Fig. 1B). To test whether the GAF and PAS domains modulate the activity of AdhR, we constructed N-terminally truncated forms of AdhR protein that lacked the GAF domain or GAF and PAS domains. The purified proteins were used in the ATPase activity assays. Deletion of the GAF domain (AdhRΔGAF) or both the GAF and the PAS domains (AdhRΔGAFΔPAS) increased the ATPase activity of AdhR more than 5- and 10-fold, respectively (Fig. 1C). This result suggests that the N-terminal GAF and PAS domains repress the ATPase activity of the central AAA+ domain in the absence of the binding ligand.
Binding of ferrous iron stimulates the activity of AdhR.
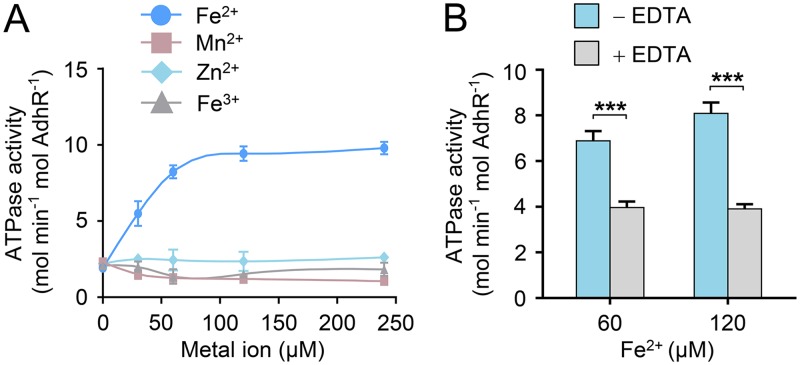
The GAF and PAS domains are known for their capacity to bind a chemically diverse range of small-molecule ligands and cofactors, including heme, flavin adenine dinucleotide (FAD), and iron-sulfur clusters (19, 20). However, the GAF and PAS domains of AdhR do not contain the conserved amino acid residues required for binding of heme, FAD, or Fe-S clusters. These domains lack transmembrane regions, suggesting that AdhR senses intracellular signals. Previous studies have shown that the N-terminal GAF domain of the nitric oxide (NO) sensor NorR contains a non-heme iron center for ligand binding (21, 22). Thus, we attempted the reconstitution of AdhR protein with ferrous iron in the absence of oxygen. After reconstitution with Fe2+, the AdhR protein showed increased ATPase activity (Fig. 2A). A 5-fold increase in AdhR activity was observed in the presence of excess Fe2+ for reconstitution. Reconstitution with other metal ions, such as Mn2+, Zn2+, or Fe3+, was unable to improve the ATPase activity of AdhR (Fig. 2A). Treatment of the reconstituted AdhR-Fe(II) with EDTA resulted in a decrease in the ATPase activity (Fig. 2B). Therefore, binding of Fe2+ leads to enhanced ATPase activity of AdhR. We then determined the content of iron bound by AdhR. AdhR purified from overexpressing cells under aerobic conditions contained about 0.2 ferrous iron ion per monomer. Ferric iron ions were not detectable. After anaerobic reconstitution and purification to remove free iron, AdhR contained 0.9 ferrous iron ion per monomer, suggesting that AdhR coordinates a mononuclear iron atom per monomer.
FIG 2.
Binding of Fe2+ ions stimulates the activity of AdhR. (A) ATPase activities of AdhR after reconstitution with increasing amounts of Fe2+, Mn2+, Zn2+, and Fe3+. (B) Effect of EDTA treatment on ATPase activities of reconstituted AdhR-Fe(II). AdhR was reconstituted with 60 and 120 μM Fe2+, respectively, and then subjected to EDTA treatment. Data shown in all figures are means ± the SD (n = 3 independent experiments). Statistical analysis was performed with the unpaired two-tailed Student t test (***, P < 0.001).
Binding of Fe2+ to the GAF domain relieves intramolecular repression of AdhR.
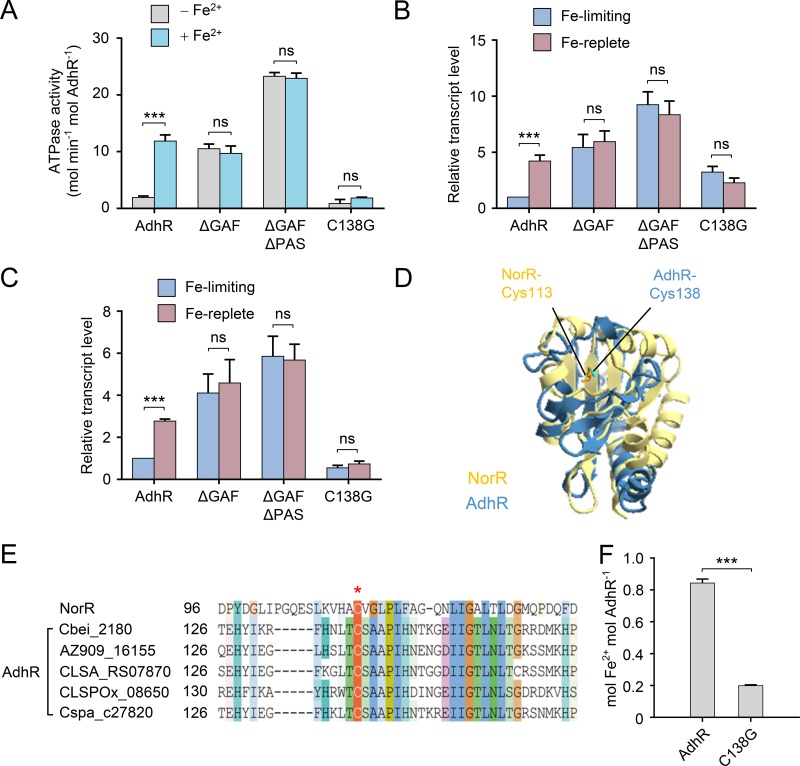
To test whether the GAF and PAS domains of AdhR are involved in Fe2+ binding, we investigated the effect of deletion of the N-terminal domains on signal-dependent activities of AdhR. The N-terminally truncated AdhR proteins were anaerobically reconstituted with Fe2+, and their ATPase activities were assayed. The AdhRΔGAF and AdhRΔGAFΔPAS proteins exhibited high and Fe2+-independent ATPase activities (Fig. 3A). The in vivo activity of AdhR was assessed by determining the transcriptional activation of adhA1 and adhA2 genes. A plasmid coding for full-length or truncated AdhR proteins was introduced into the adhR-inactivated mutant (adhR mutant). C. beijerinckii strains were grown anaerobically under iron-limited and iron-replete conditions, and the transcript levels of adhA1 and adhA2 were determined by using quantitative real-time PCR (qRT-PCR). The adhR mutant carrying the full-length AdhR-encoded plasmid showed 4- and 3-fold higher transcript levels of adhA1 and adhA2, respectively, under iron-replete conditions than under iron-limited conditions (Fig. 3B and C). Expression of truncated forms of AdhR in the adhR mutant, which lack the GAF domain or both the GAF and the PAS domains, resulted in constitutive transcriptional activation of adhA1 and adhA2 by AdhR, irrespective of iron levels in the culture. These results demonstrate that the GAF domain is critical for Fe2+ binding. The PAS domain may also play an important role in signal transduction, because AdhRΔGAFΔPAS exhibited higher activity in vitro and in vivo than did AdhRΔGAF (Fig. 3A to C).
FIG 3.
Binding of Fe2+ to the GAF domain relieves intramolecular repression of AdhR. (A) ATPase activities of AdhR and its derivatives with or without Fe2+ reconstitution. (B and C) Activation of adhA1 (B) and adhA2 (C) transcription by plasmids expressing AdhR or its derivatives in the adhR mutant. The transcript levels of adhA genes are normalized to the gene expression in the strain carrying wild-type AdhR and grown under iron-limiting conditions. (D) Structural model of the GAF domain of AdhR. The structure model of the AdhR GAF domain (blue) is overlaid with that of the GAF domain of E. coli NorR (yellow), showing the location of the conserved Cys138 of AdhR and Cys113 of NorR. (E) Sequence alignment of AdhR proteins in Clostridium spp. The Cys138 residue is strictly conserved in AdhR proteins. (F) Iron content of AdhR and AdhR-C138G after reconstitution and purification to remove free iron. The data are expressed as Fe2+ ions per monomer. Data shown in all figures are means ± the SD (n = 3 independent experiments). Statistical analysis was performed with the unpaired two-tailed Student t test (***, P < 0.001).
To provide further evidence that the GAF domain binds Fe2+, we replaced the cysteine residue at position 138 in the GAF domain with glycine (AdhR-C138G) based on comparison of the structural models of the GAF domain of AdhR and NorR (Fig. 3D). The Cys138 of AdhR is in the same location in the structure as the Cys113 of E. coli NorR, which has been reported to be involved in iron binding to NorR (22). The Cys138 residue is strictly conserved in AdhR proteins from Clostridium species (Fig. 3E). The mutated protein AdhR-C138G contained a substantially lower amount of Fe2+ ions after reconstitution than the wild-type AdhR (Fig. 3F). The mutation prevented an Fe2+-dependent increase in the ATPase activity of AdhR (Fig. 3A). Moreover, the AdhR-C138G was unable to activate transcription of adhA1 and adhA2 in response to environmental iron levels (Fig. 3B and C). Thus, these results indicate that the GAF domain is responsible for Fe2+ binding. Binding of Fe2+ relieves the repression of N-terminal domains on the central AAA+ domain.
Transcriptional activation by AdhR responds to cellular redox status.
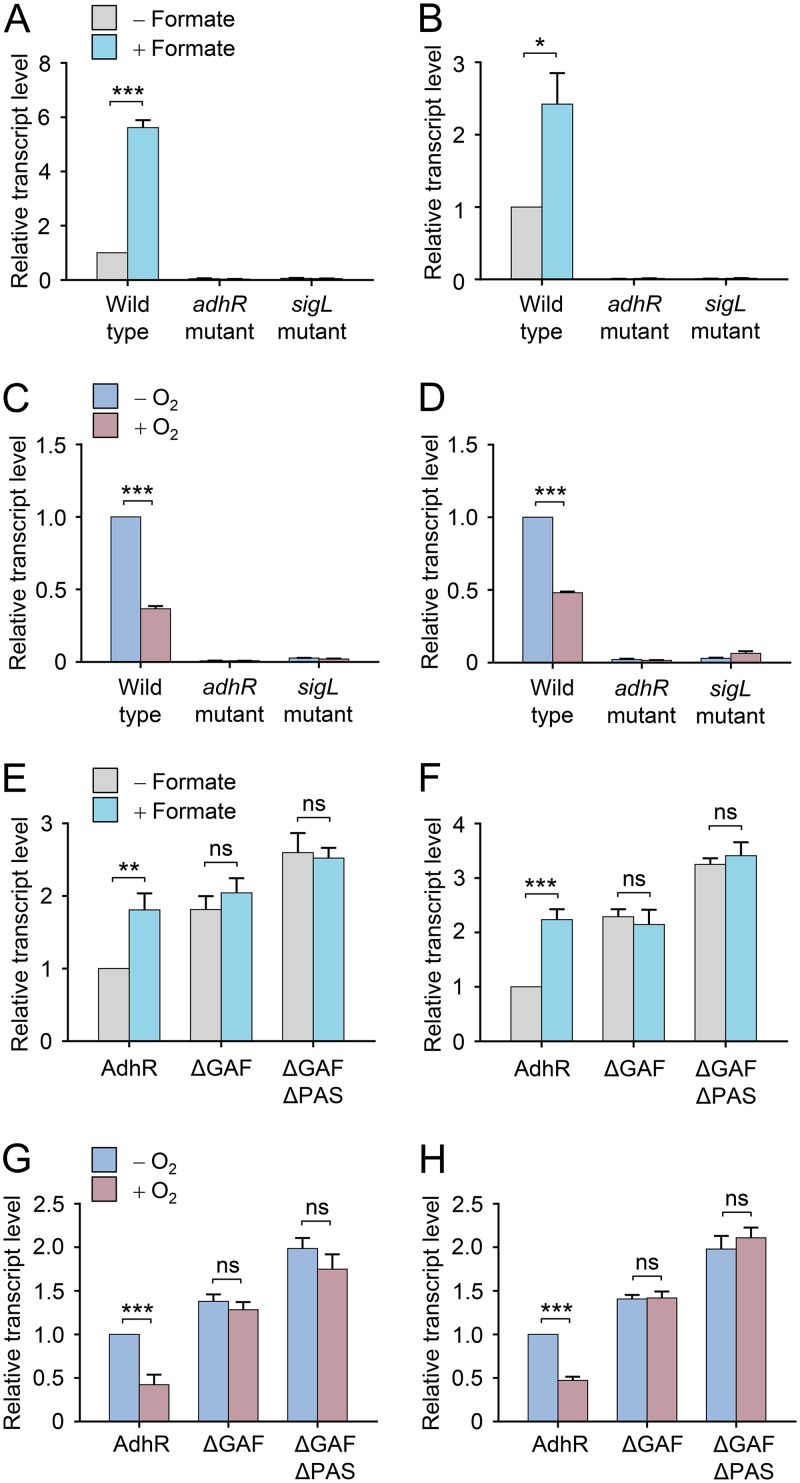
Given that the AdhR activity is dependent on binding of Fe2+, we hypothesized that AdhR can sense the redox status in the cell. To test this, we altered the cellular redox balance by adding formate to cultures or exposing cells to O2 and studied their effects on transcription of adhA1 and adhA2 genes in C. beijerinckii. Supplementation of cultures with formate has been shown to provide a surplus of electrons for cells (23, 24), whereas limited exposure to O2 causes oxidative stress to clostridial cells (10, 25). We observed that the transcript levels of adhA1 and adhA2 in the wild-type C. beijerinckii were significantly upregulated by formate supplementation and downregulated by limited exposure to O2 (Fig. 4A to D). Expression of these genes in both the adhR mutant and sigL-inactivated mutant (sigL mutant) was maintained at very low levels under all the tested conditions (Fig. 4A to D). Expression of GAF domain-truncated AdhR in the adhR mutant resulted in constitutive transcriptional activation of adhA1 and adhA2 by AdhR, regardless of the presence or absence of formate in the culture (Fig. 4E and F). The transcript levels of adhA1 and adhA2 genes in cells expressing the full-length AdhR protein were decreased by more than 50% after 10 min of exposure to O2 (Fig. 4G and H). In contrast, the expression of adhA1 and adhA2 was not downregulated by 10 min of O2 exposure in cells expressing the GAF-domain-truncated form of AdhR. These results indicate that AdhR perceives the redox state using the iron-binding GAF domain and regulates the transcription of adhA1 and adhA2 genes in response to changes in cellular redox status.
FIG 4.
Transcriptional activation by AdhR responds to cellular redox status. (A, B) Effect of formate supplementation on transcription of adhA1 (A) and adhA2 (B) genes in C. beijerinckii wild type and adhR and sigL mutants. The transcript levels of adhA1 were normalized to the gene expression in the wild type grown in the absence of formate. (C and D) Effect of exposure to O2 on adhA1 (C) and adhA2 (D) transcription in the wild type and in adhR and sigL mutants. Cells were exposed to O2 for 10 min. Data were normalized to the value in the wild type grown in the absence of O2. (E and F) Effect of formate supplementation on activation of adhA1 (E) and adhA2 (F) transcription by plasmids expressing AdhR or its truncated derivatives in the adhR mutant. Data were normalized to the value in the strain expressing full-length AdhR protein and grown in the absence of formate. (G and H) Effect of exposure to O2 on activation of adhA1 (G) and adhA2 (H) transcription by plasmids expressing AdhR or its truncated derivatives in the adhR mutant. Data were normalized to the value in the strain expressing full-length AdhR protein and grown in the absence of O2. Data shown in all figures are means ± the SD (n = 3 independent experiments). Statistical analysis was performed with the unpaired two-tailed Student t test (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
AdhR and σ54 maintain cellular redox homeostasis.
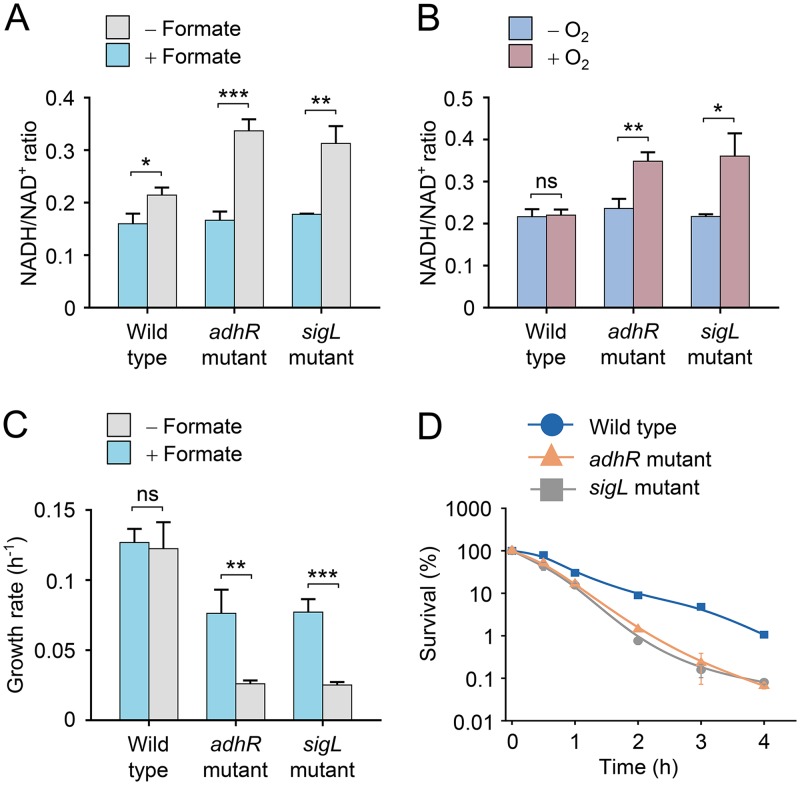
To test whether AdhR and σ54 play a role in maintaining cellular redox homeostasis in C. beijerinckii, we compared the effects of formate supplementation and limited exposure to O2 on the intracellular concentration ratio of NADH and NAD+ between the wild type and the adhR and sigL mutants. The NADH/NAD+ ratios were similar in C. beijerinckii wild type and adhR and sigL mutants without formate supplementation and O2 exposure (Fig. 5A and B). Formate supplementation resulted in a 2-fold increase in NADH/NAD+ ratio in the adhR and sigL mutants, in contrast to a slight increase in NADH/NAD+ ratio in the wild type (Fig. 5A). This result is probably due to the decrease in NAD(P)H oxidation through alcohol synthesis in the adhR and sigL mutants. An increase in NADH/NAD+ ratio was also observed in the adhR or sigL mutants after 30 min of exposure to O2, whereas the NADH/NAD+ ratio in the wild type was not changed significantly under the same condition (Fig. 5B). Thus, the adhR and sigL mutants exhibited larger shifts in the NADH/NAD+ ratio than did the wild type in response to disturbances of redox balance, indicating that AdhR and σ54 play a key role in maintaining cellular redox homeostasis.
FIG 5.
AdhR and SigL maintain cellular redox homeostasis. (A) Effect of formate supplementation on intracellular NADH/NAD+ concentration ratio in C. beijerinckii wild type and adhR and sigL mutants. NADH and NAD+ concentrations were determined at an OD600 of ∼2.0. (B) Effect of exposure to O2 on intracellular NADH/NAD+ concentration ratio in wild type and adhR and sigL mutants. Cells were exposed to O2 for 30 min. NADH and NAD+ concentrations were determined at an OD600 of ∼1.0. (C) Effect of formate supplementation on cell growth rate of wild type and adhR and sigL mutants during late-exponential growth phase. (D) Survival of wild type and adhR and sigL mutants after exposure to O2. The number of surviving cells was normalized to the value for the non-stressed wild type (time zero). Data shown in all figures are means ± the SD (n = 3 independent experiments). Statistical analysis was performed with the unpaired two-tailed Student t test (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
We studied the physiological effects of the redox vulnerability in the adhR and sigL mutants. We observed that the inhibitory effect of formate supplementation on the growth rate of the adhR and sigL mutants was more profound than that of the wild type, particularly during the late-exponential growth phase (Fig. 5C). The adhR and sigL mutants were more susceptible to O2 killing than the wild type. After 2 h of exposure to O2, 10% of the wild-type cells were viable, whereas the survival rate of the adhR and sigL mutants declined to 1% (Fig. 5D), indicating that deficiency of AdhR and σ54 impairs the ability of cells to cope with oxidative stress. This result may be explained by inhibition of glycolytic activity and NADH production by a high NADH/NAD+ ratio in the mutants (26, 27). Therefore, AdhR and σ54 are required for generating the redox states that are optimized for cell growth and oxidative stress response.
Deletion of regulatory domains of AdhR resulted in improved solvent production.
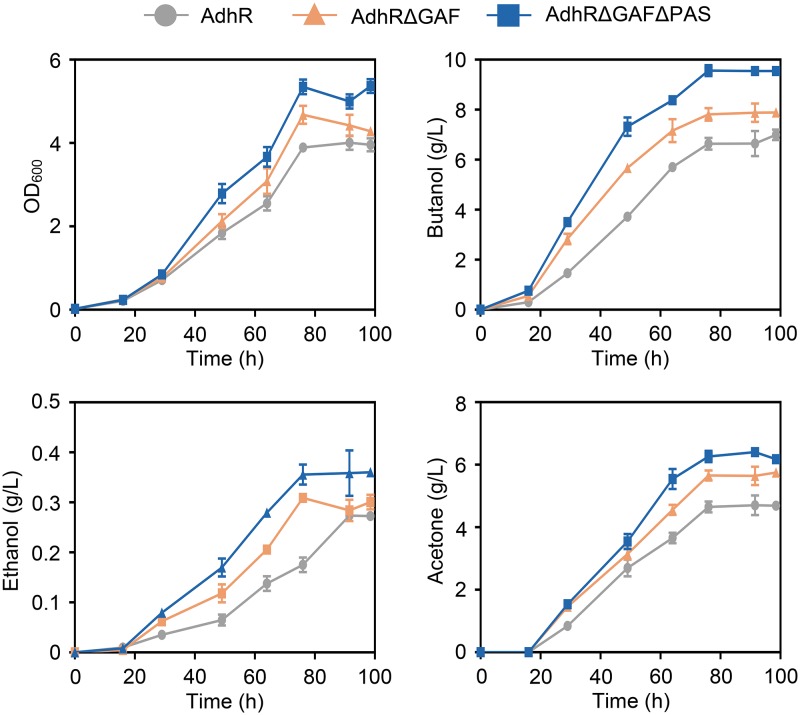
We tested the cell growth of and fermentation product formation by C. beijerinckii adhR mutant strains carrying a plasmid coding for full-length or N-terminally truncated AdhR proteins. The strains expressing truncated forms of AdhR showed higher rates of cell growth than the strain expressing full-length AdhR (Fig. 6). Synthesis of butanol and ethanol was enhanced by removal of the GAF domain or both the GAF and the PAS domains from AdhR. The production of total solvents (butanol, ethanol, and acetone) by strains expressing AdhRΔGAF and AdhRΔGAFΔPAS was increased by 16 and 34%, respectively, compared to the strain expressing full-length AdhR (Fig. 6). Compared to C. beijerinckii wild-type and adhR mutant strains carrying an empty-vector plasmid, the total solvent production by the adhR mutant expressing AdhRΔGAFΔPAS was increased by 17 and 84%, respectively. Therefore, expression of N-terminally truncated and constitutively active forms of AdhR resulted in improved solvent production by C. beijerinckii.
FIG 6.
Cell growth of and fermentation product formation by adhR mutant strains carrying plasmids expressing full-length or N-terminally truncated AdhR proteins. The strains were grown in P2 medium containing 1 g/liter yeast extract and 4 g/liter CaCO3. Cell growth was monitored spectrophotometrically at 600 nm (OD600). The concentrations of butanol, ethanol, and acetone in the medium were measured during the cultivation. Data shown in all panels are means ± the SD (n = 3 independent experiments).
DISCUSSION
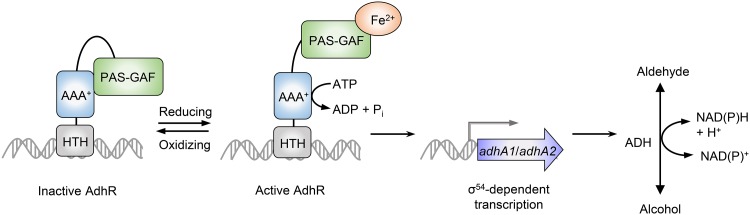
The AdhR regulatory protein is an activator of σ54-dependent transcription of adhA1 and adhA2 genes required for primary alcohol synthesis in C. beijerinckii. In this study, we identified the signal perceived by AdhR and determined the regulatory mechanism of AdhR activity. A negative control mechanism of AdhR activity was unraveled, in which the central AAA+ domain is subject to repression by the N-terminal GAF and PAS domains (Fig. 7). Binding of Fe2+ to the GAF domain was found to relieve the repression on the AAA+ domain, allowing the AdhR to hydrolyze ATP and activate the transcription of adhA1 and adhA2. This control mechanism enables AdhR to respond to changes in cellular redox status. The mutants deficient in AdhR or σ54 exhibited large shifts in intracellular redox state indicated by the NADH/NAD+ ratio under conditions of increased electron availability or oxidative stress. We propose that under anaerobic conditions with normal iron levels, the Fe2+-binding AdhR activates the σ54-dependent transcription of adhA1 and adhA2, leading to ADH-catalyzed conversion of aldehydes to alcohols concomitant with NAD(P)H oxidation (Fig. 7). When a surplus of electrons is provided for cells (e.g., by supplementing cultures with formate), the AdhR activity is increased, leading to enhanced alcohol synthesis. When cells are subjected to oxidative stress or iron starvation, repression of the AAA+ domain by the GAF and PAS domains in AdhR results in reduced NAD(P)H oxidation through ADH. Thus, the redox-sensing transcriptional activator AdhR and σ54 control alcohol synthesis to achieve redox homeostasis in clostridial cells.
FIG 7.
Schematic of the proposed mechanism for transcriptional activation by AdhR. Under oxidative-stress or iron-limited conditions, the AAA+ domain is subject to repression by the GAF and PAS domains in AdhR. Under anaerobic and iron-replete conditions, binding of Fe2+ to the GAF domain relieves the repression on the AAA+ domain. AdhR is then able to hydrolyze ATP, leading to the σ54-dependent transcription of adhA1 and adhA2, encoding ADHs that catalyze the NAD(P)H-dependent conversion of aldehydes to alcohols.
Diverse signal transduction pathways involving GAF domains have been described (17, 20). For example, the NorR protein from E. coli uses the GAF domain containing a non-heme iron center for NO sensing (21). The GAF domain in the AirS protein from Staphylococcus aureus binds an iron-sulfur cluster for sensing O2 (28), and the DosS protein from Mycobacterium tuberculosis uses the heme-binding GAF domain for redox sensing (29). However, to our knowledge, AdhR provides a new example of a GAF domain coordinating a mononuclear non-heme iron to sense and transduce the redox signal. The Cys138 in the GAF domain was identified as a key residue for iron binding, and the C138G substitution blocked the AdhR protein in the inactive state. Another regulatory domain in AdhR is the PAS domain, which is also often found in signaling proteins and can perform a variety of functions, including signal transfer and directly sensing perceived stimuli (19). It remains unknown how the PAS domain transduces the redox signal from the N-terminal GAF domain to the central AAA+ domain in the AdhR protein. The possibility that the PAS domain may also perceive another signal via ligand binding cannot be excluded. Further studies are needed to reveal the exact mechanism by which the GAF and PAS domains negatively regulate the AAA+ domain.
σ54 has been identified as a sigma factor for transcription of genes involved in various cellular processes, such as nitrogen assimilation and fixation, nitric oxide detoxification, and phage shock response (18, 30, 31). Our study reveals a previously unrecognized functional role of σ54 in control of cellular redox balance. Due to the requirement of AdhR, σ54-dependent transcription of adhA1 and adhA2 is tightly regulated. AdhR and σ54 may allow a rapid cellular response to changes in redox state or in iron availability. We observed that after wild-type C. beijerinckii was exposed to O2, the transcript levels of adhA1 and adhA2 were reduced markedly within 10 min, and the intracellular NADH/NAD+ ratio was not changed after 30 min. This implies that AdhR and σ54 respond more rapidly to O2 exposure than the NAD(H)-sensing transcriptional repressor Rex. Regulation of adhA1 and adhA2 by AdhR and σ54 could enable cells to rapidly stop synthesizing alcohols. After the intracellular NADH/NAD+ ratio starts to decline, the Rex targets, including many genes involved in fermentation and hydrogen production, are repressed, thereby providing more reducing power for removal of O2 (12).
Supplementation of formate and glycerol to cultures has been used to enhance butanol production in C. beijerinckii (32, 33). However, the underlying molecular mechanism remains unknown. Genetic engineering has been applied to enhance solvent production by C. beijerinckii, but most of the attempts did not result in a desired solvent producer (34, 35). We demonstrated that expression of N-terminally truncated and constitutively active forms of AdhR increased alcohol production by upregulating adhA1 and adhA2 in C. beijerinckii. The strains expressing AdhRΔGAF and AdhRΔGAFΔPAS also exhibited enhanced acetone synthesis, which may be due to indirect regulation of the genes involved in acetone synthesis (36). Combination of AdhR truncation and SigL overexpression could lead to a further increase in solvent production by C. beijerinckii (13).
MATERIALS AND METHODS
Strains and culture conditions.
C. beijerinckii NCIMB 8052 wild-type and mutant strains used in this study are listed in Table 1 . C. beijerinckii strains were precultured anaerobically on clostridial growth medium (37) to exponential growth phase. Erythromycin (30 μg/ml) or spectinomycin (350 μg/ml) was added when needed. The cultures were started at an optical density at 600 nm (OD600) of 0.02 and performed at 37°C in 60 ml of P2 minimal medium, which contains (per liter) 0.5 g of KH2PO4, 0.5 g of K2HPO4, 0.01 g of NaCl, 0.2 g of MgSO4⋅7H2O, 0.01 g of MnSO4⋅H2O, 0.01 g of FeSO4⋅7H2O, 1 mg of p-aminobenzoic acid, 1 mg of vitamin B1, 0.01 mg of biotin, 2.2 g of CH3COONH4, and 60 g of glucose (38). For iron-limited cultures, the initial concentration of FeSO4⋅7H2O in the medium was 3 μM (39). For formate supplementation experiments, 1 g/liter sodium formate and 1 g/liter yeast extract were added to P2 minimal medium. For oxygen exposure experiments, cells were grown anaerobically in P2 minimal medium to an OD600 of ∼1.0. Then, 10 ml of the culture was shifted to aerobic conditions by shaking in ambient air at 200 rpm in 100-ml flasks (25). Samples were removed at various time points after the shift, and the number of surviving cells was determined as described previously (25).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Genotype | Source or reference |
|---|---|---|
| C. beijerinckii strains | ||
| 8052 | Wild type | NCMIB |
| sigL mutant | sigL (Cbei_0595)::intron | 13 |
| adhR mutant | adhR (Cbei_2180)::intron | 13 |
| Wild-type control | Wild type/pXY1 | This study |
| adhR mutant control | adhR mutant/pXY1 | This study |
| AdhR | adhR::intron/pXY1-adhR | This study |
| AdhRΔGAF | adhR::intron/pXY1-adhRΔGAF | This study |
| AdhRΔGAFΔPAS | adhR::intron/pXY1-adhRΔGAFΔPAS | This study |
| AdhR-C138G | adhR::intron/pXY1-adhR-C138G | This study |
| Plasmids | ||
| pXY1 | Ampr Specr; pcb102, ColE1 origin | 41 |
| pXY1-adhR | Ampr Specr; pcb102; Pptb adhR | This study |
| pXY1-adhRΔGAF | Ampr Specr; pcb102; Pptb adhRΔGAF1–189 | This study |
| pXY1-adhRΔGAFΔPAS | Ampr Specr; pcb102; Pptb adhRΔGAFΔPAS1–301 | This study |
| pXY1-adhR-C138G | Ampr Specr; pcb102; Pptb adhR-C138G | This study |
| pTolo | Kanr; poly-His tag; GST tag; TEV protease sites PT7 | Tolo Bio |
| pET28a-adhR-DBD | Kanr; PT7 adhR-DBD | 13 |
| pTolo-adhR | Kanr; PT7 adhR | This study |
| pTolo-adhRΔGAF | Kanr; PT7 adhRΔGAF1–189 | This study |
| pTolo-adhRΔGAFΔPAS | Kanr; PT7 adhRΔGAFΔPAS1–301 | This study |
| pTolo-adhR-C138G | Kanr; PT7 adhR-C138G | This study |
Plasmid construction.
For genetic complementation experiments, the adhR gene was PCR amplified from C. beijerinckii NCIMB 8052 genomic DNA and cloned into the pXY1 vector under the control of the constitutive Pptb promoter, which is localized 116 bp upstream of the respective start codons (40). Plasmids pXY1-adhRΔGAF and pXY1-adhRΔGAFΔPAS (Table 1), which code for N-terminally truncated forms of AdhR that lacked the GAF domain or both GAF and PAS domains, were constructed with PCR using pXY1-adhR as the template. Plasmid pXY1-adhR-C138G was constructed with two steps of PCR using the mutagenic primers and flanking primers (Table 2), which codes for an AdhR derivative where a cysteine residue was replaced by a glycine. The plasmids pXY1-adhRΔGAF, pXY1-adhRΔGAFΔPAS, and pXY1-adhR-C138G were sequenced to exclude unwanted mutations in the adhR gene and individually electroporated into the adhR-inactivated mutant (Table 1).
TABLE 2.
Primers used in this study
| Primer | Sequence (5′–3ʹ) | Plasmid(s) and/or use |
|---|---|---|
| AdhR-F1 | GTAAAAGGGAGTGTCGAGGATCCATGGAGAATAAAGAACTATTAATTG | pXY1-adhR |
| AdhR-R1 | CTGAGAGTGCACCATATGTCGACTTATTTATGTTTTTTTAATTTTAGG | pXY1-adhR, pXY1-adhRΔGAF, pXY1-adhRΔGAFΔPAS, pXY1-adhR-C138G |
| ΔGAF-F1 | GTAAAAGGGAGTGTCGAGGATCCATGATATTGAATCAAACCTATAACTATATGG | pXY1-adhRΔGAF |
| ΔGAFΔPAS-F1 | GTAAAAGGGAGTGTCGAGGATCCATGGGAAAAGACAACAAAAATGCAAC | pXY1-adhRΔGAFΔPAS |
| C138G-F | AAAAGGTTTCACAATTTAACAGGCTCTGCTGCACCTATTCATAATAC | pXY1-adhR-C138G, pTolo-adhR-C138G |
| C138G-R | GTATTATGAATAGGTGCAGCAGAGCCTGTTAAATTGTGAAACCTTTT | pXY1-adhR-C138G, pTolo-adhR-C138G |
| AdhR-F2 | AGAGAGGGATCCGATGGAGAATAAAGAACTATTAATTG | pTolo-adhR |
| AdhR-R2 | AGAGAGGCGGCCGCTTATTTATGTTTTTTTAATTTTAGG | pTolo-adhR, pTolo-adhRΔGAF, pTolo-adhRΔGAFΔPAS, pTolo-adhR-C138G |
| ΔGAF-F2 | AGAGAGGGATCCGATATTGAATCAAACCTATAACTATATGG | pTolo-adhRΔGAF |
| ΔGAFΔPAS-F2 | AGAGAGGGATCCGGGAAAAGACAACAAAAATGCAAC | pTolo-adhRΔGAFΔPAS |
| 16s-F | GAAGAATACCAGTGGCGAAGGC | Internal control of qRT-PCR |
| 16s-R | ATTCATCGTTTACGGCGTGGAC | Internal control of qRT-PCR |
| Cbei_2181-F | GAGATATTGCGAGAGCCTTA | qRT-PCR of adhA1 |
| Cbei_2181-R | ATACTGCGTTATGAGCGATA | qRT-PCR of adhA1 |
| Cbei_1722-F | CTGATTGGATAGTTGCTA | qRT-PCR of adhA2 |
| Cbei_1722-R | CACTTGTTGATGGAATAG | qRT-PCR of adhA2 |
RNA isolation and real-time PCR analysis.
Cells were harvested at mid-exponential growth phase (OD600 of ∼2.0, except for oxygen-exposed cells that were harvested at an OD600 of ∼1.0), frozen immediately in liquid nitrogen, and ground into powder. RNA was isolated using TRIzol (Invitrogen) according to the manufacturer’s instructions. Contaminant DNA was removed by DNase I (TaKaRa) digestion. RNA (1 μg) was transcribed into cDNA with random primers using the ReverTraPlus kit from Toyobo. The product was quantified via real-time PCR using the CFX96 thermal cycler (Bio-Rad). The reaction mixture (20 μl) contained Power SYBR green PCR master mix (Bio-Rad) and 0.4 μM concentrations of gene-specific primers (as shown in Table 2). The PCR parameters were 1 cycle of 95°C for 2 min, followed by 40 cycles of 95°C for 20 s, 60°C for 20 s, and 72°C for 15 s. The accuracy of the PCR product was checked by melting curve analysis. The 16S rRNA gene was used as the internal standard (41).
Protein purification.
The DNA fragments coding for full-length AdhR, for N-terminally truncated forms of AdhR (AdhRΔGAF and AdhRΔGAFΔPAS), and for a mutated derivative of AdhR (AdhR-C138G) were PCR-amplified from C. beijerinckii NCIMB 8052 genomic DNA. The PCR products were cloned into the expression vector pTolo (Tolo Biotech) by a One-Step Seamless Assembly Super kit (Paisiwen Co., Ltd., China). The resulting plasmids pTolo-adhR, pTolo-adhRΔGAF, pTolo-adhRΔGAFΔPAS, and pTolo-adhR-C138G were used to produce the respective proteins with N-terminal TEV cleavable hexahistidine and glutathione S-transferase (GST) tags. E. coli BL21Rosetta(DE3) (Novagen) was transformed with individual expression plasmid. Protein overexpression and purification were performed as described previously (42). The hexahistidine and GST tags were removed by TEV cleavage for 14 h at 4°C. The purities of proteins were checked based on SDS-PAGE analysis.
Protein reconstitution.
Reconstitution of AdhR protein with ferrous iron was performed anaerobically in a glove box (<1 ppm oxygen) according to D’Autréaux et al., with modifications (21). Purified AdhR protein was degassed and transferred to the glove box. AdhR (30 μM) was incubated for 10 min with 2.5 mM dithiothreitol in 500 μl of 50 mM Tris-HCl buffer (pH 8.0) containing 150 mM NaCl. After the addition of 240 μM Fe(NH4)2(SO4)2, the reconstitution mixture was incubated for 2 h at room temperature to obtain AdhR-Fe(II). Excess Fe2+ was removed by using PD-10 desalting column (GE Healthcare). To test whether the AdhR derivatives can bind Fe2+, AdhR was replaced by AdhRΔGAF, AdhRΔGAFΔPAS, or AdhR-C138G in the reconstitution mixture. To examine the influence of metal chelators on the reconstituted protein, AdhR-Fe(II) (30 μM) was treated with 5 mM EDTA disodium. To test whether AdhR can bind other metal ions such as Mn2+, Fe(NH4)2(SO4)2 was replaced by MnSO4 in the reconstitution mixture. Reconstitution with Zn2+ or Fe3+ was conducted aerobically by incubating 30 μM AdhR with 30, 60, 120, or 240 μM ZnCl2 or FeCl3; the FeCl3 was prepared in 2.5 mM sodium citrate (pH 7.0). Ferrous and ferric iron contents in the reconstituted AdhR protein were measured using a QuantiChrom DIFE-250 kit (BioAssay Systems).
ATPase assay.
ATPase activity was determined using an ATPase/GTPase activity assay kit (Sigma-Aldrich). Briefly, the reaction mixture (150 μl) contained 50 mM Tris buffer (pH 8.0), 20 mM MgCl2, 50 mM KCl, 50 mM NaCl, 1 mM ATP, and 150 nM purified AdhR, AdhRΔGAF, AdhRΔGAFΔPAS, AdhR-C138G, or reconstituted AdhR. After anaerobic incubation at 37°C for 10 min, the formed inorganic phosphate was measured by malachite green colorimetric method (43).
Metabolite analysis.
For analysis of extracellular metabolites, culture samples were centrifuged for 10 min at 4°C and 15,000 × g to remove the cells. Acetone, Butanol, and ethanol were detected by gas chromatography as described previously (44). Intracellular NADH and NAD+ were extracted and assayed as described previously (12).
Bioinformatics.
A homology model of the GAF domain of AdhR was built using I-TASSER (45) and the N-terminal domain of E. coli DhaR (PDB ID 4LRX) as a template. RaptorX (46) was used to overlay the structural models of the GAF domain of AdhR and NorR proteins.
Statistical analysis.
Unless noted otherwise, data are presented as the means ± the standard deviations (SD) of n independent experiments. The levels of significance were calculated with unpaired two-tailed Student t test using GraphPad Prism 7.0.
ACKNOWLEDGMENTS
This study was supported by the National Natural Science Foundation of China (31630003, 31900044, 31925001, and 31921006) and the Chinese Academy of Sciences (XDB27020201).
The authors declare that they have no conflicts of interest.
REFERENCES
- 1.Al-Hinai MA, Jones SW, Papoutsakis ET. 2015. The Clostridium sporulation programs: diversity and preservation of endospore differentiation. Microbiol Mol Biol Rev 79:19–37. doi: 10.1128/MMBR.00025-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Durre P. 2014. Physiology and sporulation in Clostridium. Microbiol Spectr 2Tbs-0010–2012. doi: 10.1128/microbiolspec.TBS-0010-2012. [DOI] [PubMed] [Google Scholar]
- 3.Ren C, Wen Z, Xu Y, Jiang W, Gu Y. 2016. Clostridia: a flexible microbial platform for the production of alcohols. Curr Opin Chem Biol 35:65–72. doi: 10.1016/j.cbpa.2016.08.024. [DOI] [PubMed] [Google Scholar]
- 4.Tracy BP, Jones SW, Fast AG, Indurthi DC, Papoutsakis ET. 2012. Clostridia: the importance of their exceptional substrate and metabolite diversity for biofuel and biorefinery applications. Curr Opin Biotechnol 23:364–381. doi: 10.1016/j.copbio.2011.10.008. [DOI] [PubMed] [Google Scholar]
- 5.Durre P. 2007. Biobutanol: an attractive biofuel. Biotechnol J 2:1525–1534. doi: 10.1002/biot.200700168. [DOI] [PubMed] [Google Scholar]
- 6.Lee SY, Park JH, Jang SH, Nielsen LK, Kim J, Jung KS. 2008. Fermentative butanol production by clostridia. Biotechnol Bioeng 101:209–228. doi: 10.1002/bit.22003. [DOI] [PubMed] [Google Scholar]
- 7.Ezeji T, Milne C, Price ND, Blaschek HP. 2010. Achievements and perspectives to overcome the poor solvent resistance in acetone and butanol-producing microorganisms. Appl Microbiol Biotechnol 85:1697–1712. doi: 10.1007/s00253-009-2390-0. [DOI] [PubMed] [Google Scholar]
- 8.Lutke-Eversloh T, Bahl H. 2011. Metabolic engineering of Clostridium acetobutylicum: recent advances to improve butanol production. Curr Opin Biotechnol 22:634–647. doi: 10.1016/j.copbio.2011.01.011. [DOI] [PubMed] [Google Scholar]
- 9.Milne CB, Eddy JA, Raju R, Ardekani S, Kim PJ, Senger RS, Jin YS, Blaschek HP, Price ND. 2011. Metabolic network reconstruction and genome-scale model of butanol-producing strain Clostridium beijerinckii NCIMB 8052. BMC Syst Biol 5:130. doi: 10.1186/1752-0509-5-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Riebe O, Fischer RJ, Wampler DA, Kurtz DM Jr, Bahl H. 2009. Pathway for H2O2 and O2 detoxification in Clostridium acetobutylicum. Microbiology 155:16–24. doi: 10.1099/mic.0.022756-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Girbal L, Soucaille P. 1998. Regulation of solvent production in Clostridium acetobutylicum. Trends Biotechnol 16:11–16. doi: 10.1016/S0167-7799(97)01141-4. [DOI] [Google Scholar]
- 12.Zhang L, Nie X, Ravcheev DA, Rodionov DA, Sheng J, Gu Y, Yang S, Jiang W, Yang C. 2014. Redox-responsive repressor Rex modulates alcohol production and oxidative stress tolerance in Clostridium acetobutylicum. J Bacteriol 196:3949–3963. doi: 10.1128/JB.02037-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang B, Nie X, Gu Y, Jiang W, Yang C. 2019. Control of solvent production by sigma-54 factor and the transcriptional activator AdhR in Clostridium beijerinckii. Microb Biotechnol doi: 10.1111/1751-7915.13505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Y, Li XZ, Mao YJ, Blaschek HP. 2012. Genome-wide dynamic transcriptional profiling in Clostridium beijerinckii NCIMB 8052 using single-nucleotide resolution RNA-Seq. BMC Genomics 13:102. doi: 10.1186/1471-2164-13-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Studholme DJ, Dixon R. 2003. Domain architectures of σ54-dependent transcriptional activators. J Bacteriol 185:1757–1767. doi: 10.1128/jb.185.6.1757-1767.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Taylor BL, Zhulin IB. 1999. PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol Mol Biol Rev 63:479–506. doi: 10.1128/MMBR.63.2.479-506.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aravind L, Ponting CP. 1997. The GAF domain: an evolutionary link between diverse phototransducing proteins. Trends Biochem Sci 22:458–459. doi: 10.1016/s0968-0004(97)01148-1. [DOI] [PubMed] [Google Scholar]
- 18.Bush M, Dixon R. 2012. The role of bacterial enhancer binding proteins as specialized activators of σ54-dependent transcription. Microbiol Mol Biol Rev 76:497–529. doi: 10.1128/MMBR.00006-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Henry JT, Crosson S. 2011. Ligand-binding PAS domains in a genomic, cellular, and structural context. Annu Rev Microbiol 65:261–286. doi: 10.1146/annurev-micro-121809-151631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zoraghi R, Corbin JD, Francis SH. 2004. Properties and functions of GAF domains in cyclic nucleotide phosphodiesterases and other proteins. Mol Pharmacol 65:267–278. doi: 10.1124/mol.65.2.267. [DOI] [PubMed] [Google Scholar]
- 21.D’Autréaux B, Tucker NP, Dixon R, Spiro S. 2005. A non-haem iron centre in the transcription factor NorR senses nitric oxide. Nature 437:769–772. doi: 10.1038/nature03953. [DOI] [PubMed] [Google Scholar]
- 22.Tucker NP, D’Autréaux B, Yousafzai FK, Fairhurst SA, Spiro S, Dixon R. 2008. Analysis of the nitric oxide-sensing non-heme iron center in the NorR regulatory protein. J Biol Chem 283–918. doi: 10.1074/jbc.M705850200. [DOI] [PubMed] [Google Scholar]
- 23.Ma C, Ou J, Xu N, Fierst JL, Yang ST, Liu X. 2015. Rebalancing redox to improve biobutanol production by Clostridium tyrobutyricum. Bioengineering (Basel) 3:E2. doi: 10.3390/bioengineering3010002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Berrios-Rivera SJ, Bennett GN, San KY. 2002. Metabolic engineering of Escherichia coli: increase of NADH availability by overexpressing an NAD+-dependent formate dehydrogenase. Metab Eng 4:217–229. doi: 10.1006/mben.2002.0227. [DOI] [PubMed] [Google Scholar]
- 25.Hillmann F, Fischer RJ, Saint-Prix F, Girbal L, Bahl H. 2008. PerR acts as a switch for oxygen tolerance in the strict anaerobe Clostridium acetobutylicum. Mol Microbiol 68:848–860. doi: 10.1111/j.1365-2958.2008.06192.x. [DOI] [PubMed] [Google Scholar]
- 26.Garrett RH, Grisham CM. 2010. Glycolysis, p 535–562. In Lockwood L, Kiselica S (ed), Biochemistry, 4th ed Brooks/Cole, Boston, MA. [Google Scholar]
- 27.Richter O, Betz A, Giersch C. 1975. The response of oscillating glycolysis to perturbations in the NADH/NAD system: a comparison between experiments and a computer model. Biosystems 7:137–146. doi: 10.1016/0303-2647(75)90051-9. [DOI] [PubMed] [Google Scholar]
- 28.Sun F, Ji QJ, Jones MB, Deng X, Liang HH, Frank B, Telser J, Peterson SN, Bae T, He C. 2012. AirSR, a [2Fe-2S] cluster-containing two-component system, mediates global oxygen sensing and redox signaling in Staphylococcus aureus. J Am Chem Soc 134:305–314. doi: 10.1021/ja2071835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kumar A, Toledo JC, Patel RP, Lancaster JR, Steyn A. 2007. Mycobacterium tuberculosis DosS is a redox sensor and DosT is a hypoxia sensor. Proc Natl Acad Sci U S A 104:11568–11573. doi: 10.1073/pnas.0705054104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reitzer L, Schneider BL. 2001. Metabolic context and possible physiological themes of σ54-dependent genes in Escherichia coli. Microbiol Mol Biol Rev 65:422–444. doi: 10.1128/MMBR.65.3.422-444.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Joly N, Engl C, Jovanovic G, Huvet M, Toni T, Sheng X, Stumpf MP, Buck M. 2010. Managing membrane stress: the phage shock protein (Psp) response, from molecular mechanisms to physiology. FEMS Microbiol Rev 34:797–827. doi: 10.1111/j.1574-6976.2010.00240.x. [DOI] [PubMed] [Google Scholar]
- 32.Cho DH, Shin SJ, Kim YH. 2012. Effects of acetic and formic acid on ABE production by Clostridium acetobutylicum and Clostridium beijerinckii. Biotechnol Bioprocess Eng 17:270–275. doi: 10.1007/s12257-011-0498-4. [DOI] [Google Scholar]
- 33.Ujor V, Agu CV, Gopalan V, Ezeji TC. 2014. Glycerol supplementation of the growth medium enhances in situ detoxification of furfural by Clostridium beijerinckii during butanol fermentation. Appl Microbiol Biotechnol 98:6511–6521. doi: 10.1007/s00253-014-5802-8. [DOI] [PubMed] [Google Scholar]
- 34.Lu C, Yu L, Varghese S, Yu M, Yang ST. 2017. Enhanced robustness in acetone-butanol-ethanol fermentation with engineered Clostridium beijerinckii overexpressing adhE2 and ctfAB. Bioresour Technol 243:1000–1008. doi: 10.1016/j.biortech.2017.07.043. [DOI] [PubMed] [Google Scholar]
- 35.Wen Z, Minton NP, Zhang Y, Li Q, Liu J, Jiang Y, Yang S. 2017. Enhanced solvent production by metabolic engineering of a twin-clostridial consortium. Metab Eng 39:38–48. doi: 10.1016/j.ymben.2016.10.013. [DOI] [PubMed] [Google Scholar]
- 36.Tummala SB, Junne SG, Papoutsakis ET. 2003. Antisense RNA downregulation of coenzyme A transferase combined with alcohol-aldehyde dehydrogenase overexpression leads to predominantly alcohologenic Clostridium acetobutylicum fermentations. J Bacteriol 185:3644–3653. doi: 10.1128/jb.185.12.3644-3653.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wiesenborn DP, Rudolph FB, Papoutsakis ET. 1989. Coenzyme A transferase from Clostridium acetobutylicum ATCC 824 and its role in the uptake of acids. Appl Environ Microbiol 55:323–329. doi: 10.1128/AEM.55.2.323-329.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baer SH, Blaschek HP, Smith TL. 1987. Effect of butanol challenge and temperature on lipid composition and membrane fluidity of butanol-tolerant Clostridium acetobutylicum. Appl Environ Microbiol 53:2854–2861. doi: 10.1128/AEM.53.12.2854-2861.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bahl H, Gottwald M, Kuhn A, Rale V, Andersch W, Gottschalk G. 1986. Nutritional factors affecting the ratio of solvents produced by Clostridium acetobutylicum. Appl Environ Microbiol 52:169–172. doi: 10.1128/AEM.52.1.169-172.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tummala SB, Welker NE, Papoutsakis ET. 1999. Development and characterization of a gene expression reporter system for Clostridium acetobutylicum ATCC 824. Appl Environ Microbiol 65:3793–3799. doi: 10.1128/AEM.65.9.3793-3799.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sun Z, Chen Y, Yang C, Yang S, Gu Y, Jiang W. 2015. A novel three-component system-based regulatory model for d-xylose sensing and transport in Clostridium beijerinckii. Mol Microbiol 95:576–589. doi: 10.1111/mmi.12894. [DOI] [PubMed] [Google Scholar]
- 42.Nie X, Yang B, Zhang L, Gu Y, Yang S, Jiang W, Yang C. 2016. PTS regulation domain-containing transcriptional activator CelR and sigma factor σ54 control cellobiose utilization in Clostridium acetobutylicum. Mol Microbiol 100:289–302. doi: 10.1111/mmi.13316. [DOI] [PubMed] [Google Scholar]
- 43.Henkel RD, VandeBerg JL, Walsh RA. 1988. A microassay for ATPase. Anal Biochem 169:312–318. doi: 10.1016/0003-2697(88)90290-4. [DOI] [PubMed] [Google Scholar]
- 44.Liu L, Zhang L, Tang W, Gu Y, Hua Q, Yang S, Jiang W, Yang C. 2012. Phosphoketolase pathway for xylose catabolism in Clostridium acetobutylicum revealed by 13C metabolic flux analysis. J Bacteriol 194:5413–5422. doi: 10.1128/JB.00713-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang J, Yan R, Roy A, Xu D, Poisson J, Zhang Y. 2015. The I-TASSER Suite: protein structure and function prediction. Nat Methods 12:7–8. doi: 10.1038/nmeth.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kallberg M, Wang H, Wang S, Peng J, Wang Z, Lu H, Xu J. 2012. Template-based protein structure modeling using the RaptorX web server. Nat Protoc 7:1511–1522. doi: 10.1038/nprot.2012.085. [DOI] [PMC free article] [PubMed] [Google Scholar]