Abstract
Cell-based therapeutic strategies afford major potential advantages in the repair of injured tendons. Generation of induced pluripotent stem cells (iPSCs) expands cell sources for “regenerative” therapy. However, its application in tendon repair is still limited and the effects remain unclear. In this study, equine tenocyte-derived iPSCs (teno-iPSCs) were generated by expressing four Yamanaka factors. Compared to parental tenocytes and bone marrow derived mesenchymal stem cells (BMSCs), the transcriptional activities of lineage-specific genes, including Mkx, Col1A2, Col14, DCN, ELN, FMOD, and TNC, were highly repressed in the resulting teno-iPSCs. Exposure to cyclic uniaxial mechanical loading increased the expression of Scx, Egr1, Col1A2, DCN, and TNC in teno-iPSCs and the expression of Scx, Egr1, DCN, and TNC in BMSCs. Reintroduction of tenogenic transcription factor Mohawk (Mkx) upregulated the expression of DCN in teno-iPSCs and the expression of Scx, Col14, and FMOD in BMSCs. Mechanical loading combined with ectopic expression of equine Mkx further enhanced the expression of Egr1, Col1A2, DCN, and TNC in teno-iPSCs and the expression of Scx, Egr1, and TNC in BMSCs. These data suggest that the repressed lineage-specific genes in the teno-iPSCs can be re-activated by mechanical loading and ectopic expression of Mkx. Our findings offer new insights into the application of iPSCs for basic and clinic research in tendon repair.
Keywords: Tenogenic differentiation, Induced pluripotent stem cell (iPSC), Bone marrow mesenchymal stem cell (BMSC), Mechanical loading, Mohawk
1. Introduction
Tendon is a unique form of connective tissue that transmits the force from muscle to bone, thus allowing joint movement efficiently (Nourissat et al., 2015). Tendon injuries are common diseases in the musculoskeletal system, affecting both human and veterinary species (Docheva et al., 2015). Due to the limited capacity for regeneration, the natural healing process for damaged tendon is slow and often insufficient (Ho et al., 2014).
Cell-based tissue engineering approaches show great promise in tendon therapy, and a large number of cell populations, including tenocytes, mesenchymal stem cells (MSCs), embryonic stem cells (ESCs) and tendon progenitor/stem cells (TPSCs), have been proposed for tendon repair and regeneration under various conditions (Gaspar et al., 2015). However, none of the tested cells is perfect. For example, although MSCs have been extensively studied for tendon repair, their proliferation and differentiation potential are affected by the age of their origin and the in vitro culture conditions (Beane et al., 2014). Moreover, the limited passage number and increasing cellular senescence during passaging also hinder their application in regenerative. medicine (Turinetto et al., 2016). TPSCs show the capacity to form a tendon-like tissue in vitro and in vivo (Bi et al., 2007; Tan et al., 2012), but they are also prone to lose their phenotype along with continuous passages (Webb et al., 2016). iPSCs derived from differentiated somatic cells expand the cell sources and hold high potential for cell therapy and tissue engineering. However, their application in tendon regeneration is still very limited and the effects remain debatable (Sun et al., 2015; Zhang et al., 2015; Czaplewski et al., 2014; Xu et al., 2013; Bavin et al., 2015). While Xu et al. reported that human iPSC-derived neural crest stem cells could promote tendon repair in a rat patellar tendon window defect model(Xu et al., 2013), the study from Bavin et al. showed that, compared to embryonic stem cells, equine fetal fibroblast-derived iPSCs had a reduced tendon differentiation capacity (Bavin et al., 2015).
Besides the cell source, various strategies, including both biological (transcription factor, growth factor and microenvironment) and biomechanical stimuli, have been applied to initiate/promote the in vitro tenogenic differentiation. For example, TGF beta signaling has been reported to be essential for tendon development (Havis et al., 2016; Pryce et al., 2009) and to promote the tendon differentiation of equine embryo-derived stem cells (Barsby and Guest, 2013). Animals with depletion of transcription factor SCX (Killian and Thomopoulos, 2016; Murchison et al., 2007; Yoshimoto et al., 2017), MKX (Ito et al., 2010; Liu et al., 2010; Onizuka et al., 2014; Suzuki et al., 2016), or Egr1 (Guerquin et al., 2013) showed aberrant and dysfunctional tendon. Ectopic expression of SCX converted human bone marrow derived mesenchymal stem cells (BMSCs) into tendon progenitor cells(Alberton et al., 2012), and in a rat patellar window injury model, the Scx-transduced TPSCs promoted tendon repair(Tan et al., 2014). Moreover, MKX over-expressed mesenchymal stem cells showed enhanced tenogenic differentiation potential through activation of the TGF beta signalling (Liu et al., 2015). It has also been reported that the cumulative mechanical loading was associated with mesenchymal stem cell-to-tenocyte differentiation(Morita et al., 2013; Morita et al., 2018) and cyclic tensile strain was able to induce the tenogenic differentiation of rat TPSCs (Xu et al., 2015). Furthermore, combination of biochemical and biomechanical cues synergistically enhanced the tenocyte/ligament cell differentiation of various types of cells (Chen et al., 2014; Chen et al., 2012; Nichols et al., 2018a; Nichols et al., 2018b).
During reprogramming process, fully differentiated somatic cells undergo significant epigenome rearrangements to re-establish the pluripotent stem cell network (Shipony et al., 2014). However, these rearrangements are insufficient to fully erase the parental epigenetic signatures, and iPSCs still retain a memory of their origin to some extent (Bar-Nur et al., 2011; Kim et al., 2010), which appears to have strong impact on their regeneration capacity (Hiler et al., 2015). In the present study, we aimed to generate tenocyte-derived iPSCs and to compare their tenogenic differentiation capacity with BMSCs by induction with mechanical loading and ectopic expression of transcription factor Mohawk. Our work should benefit both basic and clinic research on tendon repair with iPSCs.
2. Materials and methods
2.1. Isolation and culture of primary tenocytes, chondrocytes and BMSCs
Equine patellar tendon tissue or articular cartilage were surgically dissected from a two-year old horse. After thorough wash with phosphate buffered solution (PBS) containing 2× antibiotic/antimycotic solution (Gibco), the tissue was minced in basic medium (DMEM/F12 (Invitrogen) supplemented with 10% FCS (Gemini) and 1× antibiotic/ antimycotic solution), and then incubated with 2 mg/mL of collagenase D (Roche) in basic medium at 37 °C with gentle agitation. After 16 h, the digested materials were passed through a 100 μm strainer. The flow-through was then centrifuged at 200g for 10 min, and cell pellet was washed twice with medium. The cells were finally resuspended and cultured in basic medium at 37 °C under 5% CO2, and medium was changed every 2–3 days. At the confluency of 80–90%, cells were dissociated with 0.25% trypsin-EDTA, and sub-cultured at a density of 1–2 × 105 cells/cm2.
For equine BMSCs isolation, bone marrow aspirates from three horses were collected individually in ACD solution (anticoagulant citrate dextrose solution) and washed twice with PBS followed by two more washes with basic medium at 170 g for 7 min each, and then resuspended and cultured in BMSC growth medium (basic medium plus 4 ng/mL bFGF) at 37 °C, 5% CO2. After 72 h, cells were thoroughly washed with PBS, and fresh medium was added with a change of every 2–3 days. Upon reaching 80–90% confluency, cells were dissociated with 0.25% trypsin-EDTA, and further expanded at a density of 1–2 × 105 cells/cm2. BMSCs at passages 2–5 were used for experiments. Characterization of mesenchymal stem cell was carried out by flow cytometry with positive expression of CD29 (EMD Millipore, Cat# CBL481), CD44 (ThermoFisher Scientific, Cat#MA1–10229), CD90 (WSU Monoclonal Antibody Center, Item#DG2015), CD105 (Bio-Rad Laboratories, Cat#MCA1557A), MHC-I (gift from Dr. Douglas F. Antczak, Cornell University) and with negative expression of CD45 (WSU Monoclonal Antibody Center, Item#HR-DG2009), CD79 (Bio-Rad Laboratories, Cat#MCA2538A) and MHC-II (gift from Dr. Douglas F. Antczak).
2.2. Generation of iPSCs from equine tenocytes
Tenocyte-derived iPSCs were generated by using a single lentiviral stem cell cassette as previously described (Sommer et al., 2009). Briefly, pHAGE-STEMCCA lentiviruses expressing mouse Oct3/4, Sox2, Klf4, and c-Myc were produced in 293 T packaging cells, and supernatant containing the viral particles were filtered through 0.45 μm filter. Tenocytes were seeded on 35-mm culture plates at a density of 20,000 cells/cm2 the day before infection, and then incubated with viral particles for eight hours in the presence of polybrene B (8 μg/mL). Infected cells were maintained in basic medium for 30 h, and then transferred to mitomycin C inactivated MEF feeder cells in iPSC medium (DMEM containing 10% FCS, 1× NEAA, 1× L-glutamine, 1× sodium pyruvate, 0.055 mM beta-Mercaptoethanol, 1000 U/mL of LIF, and 1× antibiotic/antimycotic solution). Medium was replaced every other day. About 10–15 days individual colonies (designated as P0) were manually picked, trypsinized, and further expanded in 6-well plates pre-seeded with feeders in iPSC medium containing 10 μM of Rock inhibitor Y27632. Medium was replaced daily. When the cells reached about 80% confluence, they were mechanically dissociated, and passaged further or frozen in liquid nitrogen. At passages 3–5, cells were switched to and maintained in feeder-free StemFlex™ medium (Fisher Scientific). Cells at passages 10–25 were used in this study.
2.3. Karyotyping of teno-iPSCs
To harness metaphase cells, teno-iPSCs at 80–90% confluence were treated with 50 ng/mL nocodazole for fourhours, then collected at 150 g for 5 min after trypsin-EDTA treatment. Cells were resuspended in 3.7 mM KCl solution and incubated at 37 °C for 20 min. After two washes with fixative solution (glacial acetic acid/methanol in a ratio of 1:3), cells were spread onto microscope slides (Fisher Scientific). Chromosomes were visualized using DAPI staining under fluorescent microscope.
2.4. Immunofluorescent imaging
Fluorescence microscopy was performed as described in a previous study(Yang et al., 2012) with a few modifications. Briefly, cells seeded on glass coverslips were washed with PBS and fixed with 4% paraformaldehyde (PFA) in PBS for 20 min at room temperature. After permeabilization by incubation with 0.5% Triton X-100 in PBS for 5 min, cells were incubated with 2% bovine serum albumin (BSA) in PBS for one hour, followed by incubating overnight at 4 °C with primary antibodies: Oct3/4 (Santa Cruz Biotechnology Inc., Cat#sc-365,509), Sox2 (Cat#sc-17,320), Nanog (Cat#sc-134,218), TRA-1–81 (Invitrogen, Cat#MA1–024-D488), or Mohawk (Abcam, Cat#ab179597). Cells were stained with Alex Fluor 555-conjugated or Alex Fluor 488-conjugated 2nd antibodies (Abcam) for one hour at room temperature. Cellular DNA was stained with 4′,6-diamidino-2-phenylindole (DAPI, Molecular Probe, Eugene, OR). Fluorescence signals were detected on a Nikon Eclipse TE300 fluorescent microscope or on a Leica TCS SP5 confocal microscope.
2.5. Multi-lineage differentiation
For embryoid body formation, cells were dissociated with 0.5 mM EDTA in PBS, washed with basic media, then passed through 100 μM strainer and cultured in suspension for up to four weeks in ultra-low attachment 6-well plate (Fisher Scientific). Medium was changed every 2–3 days. Embryoid bodies were imaged on a Nikon eclipse TE300 and collected for gene expression analysis at desired time points.
For osteogenic differentiation, EDTA-dissociated cells were washed with basic medium and then seeded at a density of 1.2 × 104 cells/cm2 in 12-well tissue culture plates. After three days, the medium was switched to either basic medium (as control) or differentiation medium, which contains basic media, 100 nM dexamethasone, 10 mM β-glycerophosphate, 50 μM ascorbic acid-2 phosphate. Cells were maintained in the differentiation medium for three weeks with medium change every 2–3 days. At the end of differentiation, samples were either collected for osteogenic gene expression analysis, or subject to Alizarin Red S (Sigma) staining and imaged on a Nikon eclipse TE300.
For chondrogenic differentiation, EDTA-dissociated cells were washed with basic medium and then seeded at a density of 1.2 × 104 cells/cm2 in 12-well tissue culture plates. After three days, the medium was changed to chondrogenic medium, which was composed of basic medium, 50 μg/mL ascorbic acid, 100 nM dexamethasone, 1× ITS premix (BD Biosciences), 40 μg/mL L-proline, and 10 ng/mL human recombinant TGF-β3 (R&D systems). Cells were fed with fresh differentiation medium every 2–3 days. At 21 days, cells were collected for chondrogenic gene expression analysis, or subject to Alcian blue staining and imaged on a Nikon eclipse TE300.
For adipogenic differentiation, EDTA-dissociated cells were washed with basic medium and then seeded at a density of 1.2 × 104 cells/cm2 in 12-well tissue culture plates. After three days, cells were cultured in basic medium containing 1 μM dexamethasone, 0.5 mM 3-IBMX, 0.1 mM indomethacine and 1.7 μM insulin for 1 week with medium change every 2–3 days, then maintained in medium consisting of basic medium and 1.7 μM insulin for another two weeks. At the end of differentiation, cells were harvested for adipogenic gene expression analysis or subject to Oil-red O staining (Sigma-Aldrich).
2.6. Mechanical loading[42]
To test the effects of mechanic force on tenogenic differentiation of BMSCs and teno-iPSCs, a customized bioreactor was used to apply cyclic uniaxial sinusoidal deformations to cell-seeded poly(ɛ-caprolactone) (80 kDa; Sigma-Aldrich, St. Louis, MO) nanofibrous scaffolds during in vitro culture. The device was programmed to approximate sinusoidal waveforms equating to 3% strain amplitude (0%–6% strain) at a frequency of 1.0 Hz for 18 h. At the end of mechanical stretching, samples were either collected for RNA extraction, or fixed in 4% PFA for immunofluorescent staining.
2.7. Gene expression analysis by reverse transcription PCR (RT-PCR) or quantitative real-time PCR (qPCR)
Samples were lysed in trizol (Invitrogen), and total RNA was extracted according to the manufacturer’s instruction. One microgram of RNA was treated with RQ1 RNase-free DNase (Roche), and then used for complimentary DNA (cDNA) synthesis by using High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific). Equine specific primer pairs were designed using NCBI primer-blast or published data (Bavin et al., 2015), and the list of primer sequences can be found in Supplemental Table 1. Standard RT-PCR was conducted with 10 ng of cDNA per reaction by using Hot Start Taq 2× Master Mix (New England Biolabs, USA) on an Eppendorf Mastercycler Nexus Thermal Cycler (Fisher Scientific, USA), and qPCR was carried out with 5 ng of cDNA per reaction using SYBR Green PCR Master Mix (Fisher Scientific, USA) on an Applied Biosystems 7500 real time PCR system. All PCR reactions were performed in duplicates, and qPCR cycling was started with 95 °C for 10 min, followed by 40 cycles of 95 °C for 15s, 60 °C for 15 s, and 72 °C for 15s. At the end of the program, a melt curve was produced by taking readings every 1 °C from 65 °C to 95 °C. The reference gene GAPDH was used to normalize gene expression levels between groups by 2−ΔCt method.
2.8. Construction of equine Mohawk-expressing vectors
To clone equine Mkx gene, primers were designed based on online predicted mRNA sequence for Equus caballus mohawk homeobox (accession number: XM_014737017). Total mRNA was extracted from tendon tissue, and cDNA was synthesized as mentioned above. PCR was performed using Platinum PCR SuperMix High Fidelity (Invitrogen) with primer pairs (Forward: 5′- ACGA AGATCTATGCGGGAA GTGGGT CGGCGCGGGGCTG-3′; Reverse:5’-GAGCGTCGACGGG TTTCAGTCCTG GAATGGTTCG-3′). The PCR products were then cloned into BglII and SalI sites of eGFP-C1 vector, and further sub-cloned into pHAGE lentiviral vector. Individual clones with Mkx gene were confirmed by DNA sequencing. Of note, the accession number for predicted Mkx mRNA sequence used here is different from current updated version XM_023632370 or XM_023632373.
2.9. Mohawk short hairpin RNA (shRNA) interference
The shRNA oligonucleotides contained sense strands of equine Mkx nucleotide sequences, followed by short spacers (TTCAAGAGA), the reverse complement of the sense strands, and six thymidines as RNA polymerase III transcriptional stop signal. The detailed oligonucleotide sequences, targeting the open reading frame (shMKX) and 3′-untranslated region (shM3U) of equine Mkx gene, are shown in Supplemental table 2. The oligonucleotides were annealed and ligated into the HpaI and XhoI sites of pLB vector (Addgene plasmid#11619). All the plasmids were confirmed by sequence analysis. Replication-defective lentiviruses were produced in 293 T packaging cells, and supernatant containing viral particles were harvested at 48, 60, and 72 h and filtered through a 0.45 μm PVDF filter (Millipore, Ireland). Cells were exposed to viral supernatant at 1:1 ratio for six hours in the presence of Polybrene B (8 μg/mL). The infection efficiency was evaluated by the percentage of green fluorescent protein (GFP)-positive cells under a fluorescent microscope.
2.10. Western blot
Cells were lysed in RIPA buffer on ice, and protein concentration was quantitated using Bio-Rad Protein Assay (Bio-Rad Laboratories, USA, Cat#5000006). SDS-PAGE was carried out using a mini gel system from Bio-Rad. Proteins were transferred to PVDF membranes. After blocking with TBST (Cell Signaling Technology, USA) containing 5% nonfat dry milk (blocking buffer) for at least one hour at room temperature, the membranes were incubated at 4 °C overnight with primary antibodies at a dilution of 1:1000 in the blocking buffer, followed by incubation with horseradish peroxidase-conjugated secondary antibodies (Abcam, USA) for one hour at room temperature. After thorough wash with TBST buffer, signals on the membranes were developed with an enhanced chemiluminescent system (Pierce, USA).
2.11. Statistics
Data were expressed as means ± SD. Anova single factor test was used to determine the statistically significant differences in gene expression among different cell types. Paired student’s t-test was used to determine statistically significant differences in gene expression between the control and treated groups. Significance was shown as * (P < .05) or ** (P < .01).
3. Results
3.1. Generation and characterization of tenocyte-derived iPSCs
It has been reported that the origin of somatic cell type affects the differentiation potential of the resulting iPSCs(Polo et al., 2010). Tenocytes are the major cells found throughout the tendon structure and essential for the production of extracellular tendon matrix proteins. Cells isolated from adult equine patellar tendon expressed abundant tenocyte marker tenomodulin, which was barely shown in equine chondrocytes (Fig. 1A&B). After infection of tenocytes with a single lentiviral stem cell cassette system, visible colonies appeared at day 7, and the colonies with appropriate size were individually picked and expanded (Fig. 1C&D). Immunofluorescent staining with antibodies against Oct4, Sox2, Nanog, and TRA-1–81 confirmed the expression of pluripotency markers in the resulting iPSCs (Fig. 1E & S1). Moreover, RT-PCR with equine-specific primer pairs showed high levels of Oct4 Nanog, and REX1 in all the tested iPSC clones, which were not detected in the primary tenocyte cultures (Fig. 1F). Compared to donor tenocytes, iPSCs showed increased mRNA level of DNMT3b (Fig. 1F). Additionally, normal karyotype (n = 64, Fig. 1G) indicated the maintenance of genomic stability in the iPSCs.
Fig. 1.
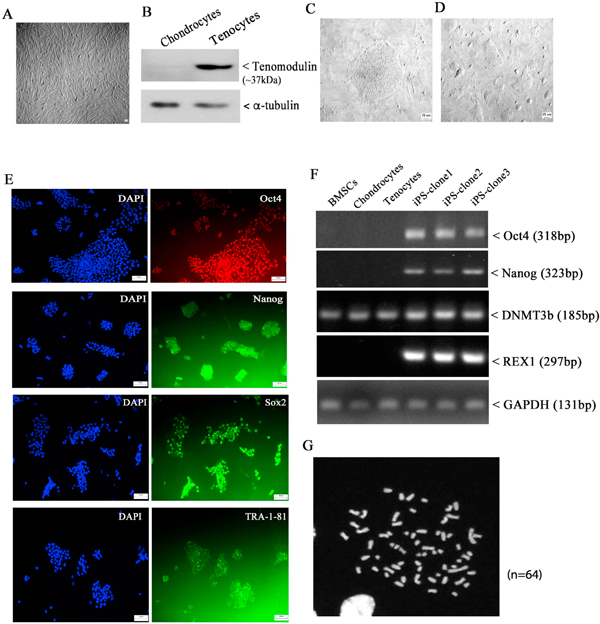
Generation of iPSCs from adult tenocytes. A: morphology of primary tenocytes (at passage 3) isolated from equine patellar tendon tissue (scale bar = 10μm); B: chondrocytes and tenocytes were lysed in RIPA buffer, and equal amounts of cell lysates were immunoblotted with antibodies against α-tubulin and tenocyte marker Tenomodulin. C: Formation of teno-iPSC colony after infection of tenocytes with a single lentiviral stem cell cassette (scale bar = 25μm); D: expansion of teno-iPSC on mouse embryonic fibroblast feeder cells (scale bar = 25μm); E: teno-iPSCs cultured in 12-well plate were fixed with PFA for immunofluorescent staining with antibodies against stem cell markers Oct4, Nanog, Sox2, or Tra-1–81 (scale bar = 25 μm). F: RT-PCR showing the expression of stem cell markers POU5F1, NANOG, DNMT3B and REX1 in teno-iPSCs; G: teno-iPSCs (passage 5) maintains normal equine karyotype (n = 64).
To determine their pluripotency, iPSCs were cultured in suspension to spontaneously form embryoid bodies (EBs, Fig. 2A), and the expression of germ layer-specific genes was measured by RT-PCR (Sheridan et al., 2012). As expected, ectodermal markers GFAP and Pax6 were activated in EBs at days of 7 and 14, which were barely detected in either donor tenocytes or the resulting iPSCs (Fig. 2B). Notably, Nestin (NES), another ectodermal marker, was expressed not only in EBs, but also in tenocytes. This is not surprising as a sub-population of nestin+ cells has been identified within the tendon cell population(Yin et al., 2016). On the other hand, the expression of endodermal marker alpha-fetoprotein (AFP) was evident in EBs at days of 21 and 28, which was negligible in tenocytes and iPSCs (Fig. 2C). When iPSCs were cultured under defined condition for adipogenesis, oil-red staining showed the formation of lipid droplet, and the expression of adipogenesis marker Leptin was detected at mRNA level (Fig. 2D&E). Culture of iPSCs with osteogenesis medium for three weeks resulted in significant calcium deposit, along with increased expression of osteocalcin (BGLAP, Fig. 2F&G). When cells were cultured in chondrogenesis medium, the production of proteoglycan proteins was visualized by positive Alcian blue staining, and the expression of Aggrecan (ACAN) was detected by RT-PCR (Fig. 2H&I). Taken together, these data confirmed that our tenocyte-derived iPSCs (teno-iPSCs) have the capacity to differentiate into other lineages.
Fig. 2.
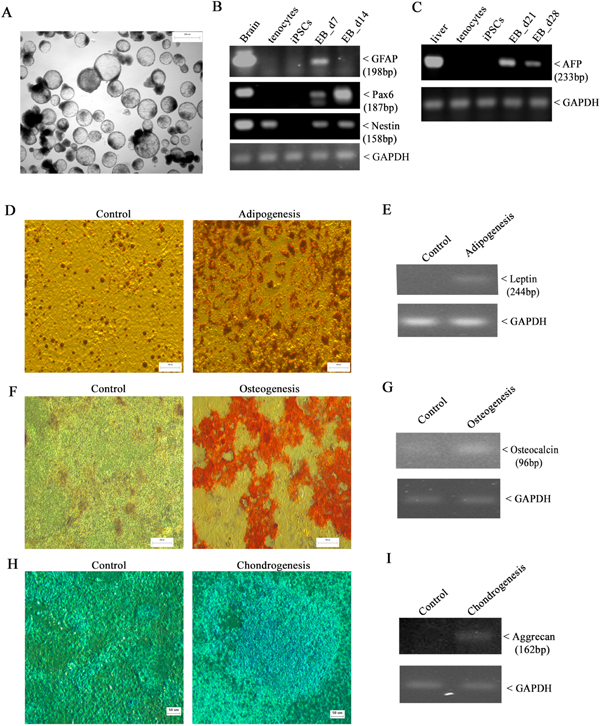
Multi-lineage differentiation capacity of teno-iPSC. A: morphology of 7-day old embryoid bodies (EBs) formed in suspension culture (scale bar = 100 μm); B: RT-PCR analysis showing the expression of ectodermal markers Glial fibrillary acidic protein (GFAP), Pax6, and Nestin (NES) in teno-iPSC-derived EBs at days 7 and 14; C: RT-PCR analysis showing the expression of endodermal marker α-fetoprotein (AFP) in teno-iPSC-derived EBs at days 21 and 28; D&E: in vitro adipogenic differentiation of teno-iPSCs. The fat droplets were displayed by oil-red staining (scale bar = 200 μm), and the induction of adipocyte marker leptin (LEP) was assessed by RT-PCR; F&G: in vitro osteogenic differentiation of teno-iPSCs. The calcium deposition was revealed by Alizarin Red S staining (scale bar = 200μm), and the expression of osteocalcin (BGLAP) was measured by RT-PCR; H&I: in vitro chondrogenic differentiation of teno-iPSCs. The production of proteoglycan proteins was shown by Alcian blue staining (scale bar = 50μm), and the induction of aggrecan (ACAN) was determined by RT-PCR. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.2. Altered tenocyte-lineage gene expression in teno-iPSCs
To determine whether our teno-iPSCs retained active expression of lineage-specific genes from donor cells, a panel of tenocyte-linked genes in teno-iPSCs and parental tenocytes were measured by qPCR. As expected, Scx, Mkx, Egr1, Col1A2, Col14, DCN, ELN, FMOD, and TNC were highly expressed in donor tenocytes (Fig. 3A–I). However, those genes were significantly suppressed in teno-iPSCs (Fig. 3A–I). Interestingly, the mRNA levels of the above genes varied among individual iPSC clones (Fig. S2), despite the fact they were derived from the same donor horse. Since the Clone 3 showed overall higher levels of tenocyte-linked genes among the three clones, it was chosen for further study. As a comparison, we also measured the tenogenic gene expression in equine BMSCs in parallel with teno-iPSCs through the study. As shown in Fig. 3D–I, except ELN, all other tested tendon-related extracellular matrix (ECM) genes, including Col1A2, Col14, DCN, FMOD, and TNC, were expressed at much lower level in teno-iPSCs than those in BMSCs. As to the tenogenesis-regulating transcription factors, the mRNA level of Scx in teno-iPSCs was about 1/10 of that in tenocytes and about 3 times of that in BMSCs (Fig. 3K–L), whereas the mRNA level of Mkx in teno-iPSCs was only about 1/100 of that in tenocytes and about 1/12 of that in BMSCs (Fig. 3K–L). For unknown reason, RT-PCR with three sets of primer pairs failed to detect TNMD expression in any type of cells used in this study. However, immunoblotting with antibodies against tenomodulin (TNMD) protein showed specific signal at expected size for cell lysates from tenocytes and BMSCs, and this signal was absent in all three tested teno-iPSC lines (Fig. 3M). Taken together, these data strongly indicate that the reprogramming process initiated by Yamanaka factors have significantly altered the tenogenic gene transcriptional activities in the resulting teno-iPSCs.
Fig. 3.
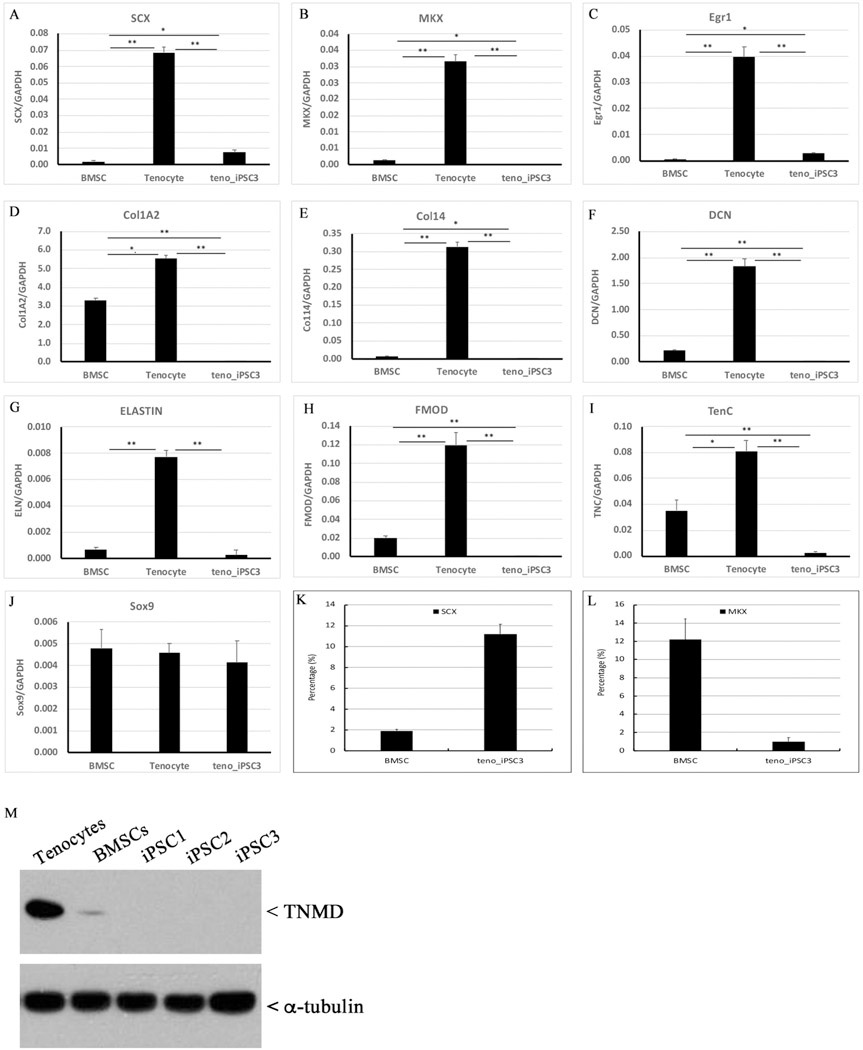
Alteration of tenocyte-linked gene expression in teno-iPSCs. A-J: cDNA samples were prepared from BMSCs, tenocytes, and teno-iPSCs, and the expression of tenocyte-linked genes, including Scx, Mkx, Egr1, Col1A2, Col14, DCN, ELN, FMOD, TNC, and Sox9, was determined by qPCR; K&L: relative transcriptional activity of Scx and Mkx in BMSCs and teno-iPSCs (normalized to tenocytes); M: Cells were lysed in RIPA buffer, and equal amounts of cell lysates were blotted with antibodies against α-tubulin and TNMD.
3.3. Regulation of tenogenic gene expression by mechanical loading
So far, there is still no well-defined condition for the tenogenic differentiation of either MSCs or ESCs. It has been recognized that mechanical force plays important roles in tendon development and that in vitro mechanical loading can potentially induce tenogenic gene expression in mesenchymal stem cells(Filomeno et al., 2012; Galloway et al., 2013; Liu et al., 2017; Song et al., 2010). Consistent with this notion, when equine BMSCs were subject to uniaxial mechanical force for 18 h, the expression of Scx, DCN, TNC, and the mechanosensitive transcription factor Egr1 was significantly increased, and the expression of Mkx, Col1A2, Col14, and ELN also trended upwards (Fig. S3). When teno-iPSCs underwent mechanical stretching, the expression of Scx, Egr1, Col1A2, DCN, and TNC was notably elevated, while the increases for Mkx and ELN were not significant (Fig. 4). Taken together, these data suggest that, like BMSCs, the tenogenic gene activities in teno-iPSCs can be regulated by mechanical loading.
Fig. 4.
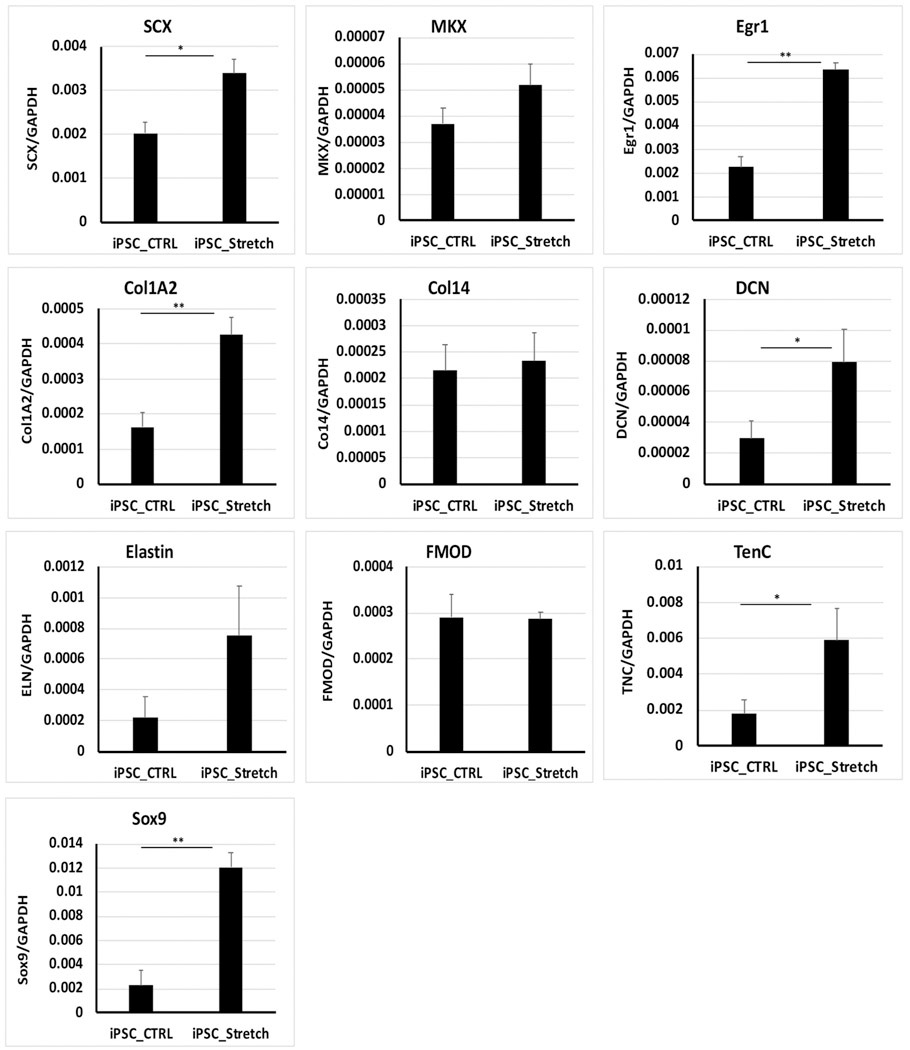
Expression of tenogenic genes in mechanical force loaded teno-iPSCs. Teno-iPSCs were seeded on vitronectin-coated PCL scaffolds for two days in basic medium, and then subject to uniaxial mechanical stretching for 18h (iPSC_Str). Cells seeded on PCL scaffolds without mechanical loading were served as control (iPSC_CTRL). Gene expression was determined by qPCR;
3.4. Mohawk regulates tenogenic gene expression in tenocytes and BMSCs
Transcription factor MKX has been reported to play critical roles in tendon development through regulating the expression of tendon extracellular matrix genes(Huang et al., 2015). In our study, knockdown of Mkx by shRNA targeting either open-reading frame (shMKX) or 3′-untranslated region (shM3U) of Mkx gene indeed decreased mRNA levels of Scx, Mkx, Egr1, Co1A2, Col14, DCN, and FMOD in either equine tenocytes or BMSCs (Fig. S4 & S5). Interestingly, knockdown of Mkx downregulated the expression of ELN in tenocyts but not in BMSCs. Moreover, in both tenocytes and BMSCs, knockdown of Mkx upregulated the expression of Sox9, a known chondrogenesis master regulator. Efficient knockdown of Mkx was also confirmed by immunoblotting of cell lysates with antibodies against MKX (Fig. S4 &S5). These data support the notion that MKX is required for the regulation of tenogenic gene expression in BMSCs and tenocytes.
3.5. Cloning of equine Mkx gene
As mentioned above, the expression of Mkx and most of the tendon-related ECM genes was highly suppressed in teno-iPSCs (Fig. 3). Although the response to mechanical loading was evident, the relative mRNA levels of the tested tenogenic genes in stimulated teno-iPSCs were still lower than those in BMSCs (Fig. 4&S3). We then asked whether overexpression of Mohawk into teno-iPSCs would boost the expression of ECM genes and facilitate their tenogenic differentiation. To this purpose, total mRNA isolated from tendon tissue was used to clone equine Mohawk gene. The PCR products were sub-cloned into mammalian expression eGFP-C1 vector or pHAGE-lentiviral vector (Fig. 5A–B). After transfection of HEK293T cells with plasmids expressing MKX tagged with or without GFP, immunoblotting of whole cell lysates with antibodies against MKX and GFP-tag confirmed the ectopic expression of MKX as well as the specificity of Mohawk antibody (Fig. 5C). Moreover, as expected, immunofluorescent staining of BMSCs infected with lentivirus expressing GFP-tagged MKX displayed apparent nuclear localization of GFP signals that were co-localized with MKX signals (Fig. 5D). Taken together, these data confirmed the success in cloning of equine Mkx gene.
Fig. 5.
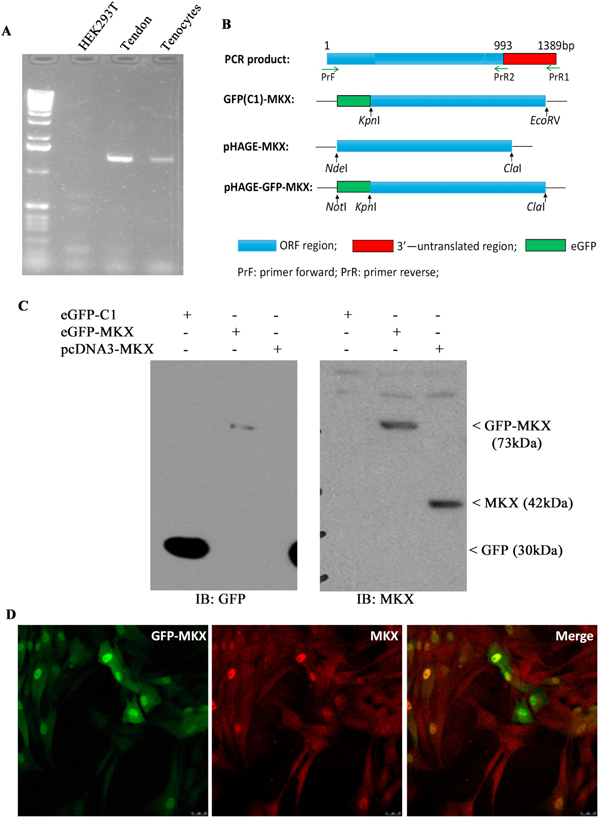
Cloning of equine Mkx gene. A) Detection of Mohawk expression by PCR in HEK293T cells, patellar tendon tissue and adult tenocytes; B) diagram for construction of MKX-expressing plasmids; C) HEK293T cells were transiently transfected with plasmids expressing GFP alone, or GFP-tagged MKX, or MKX alone for 48 h, and then lysed in RIPA buffer. Equal amounts of cell lysates was blotted for GFP and MKX; D) BMSCs were infected with pHAGE-GFP-MKX lentivirus for two days, then seeded on glass coverslips for another two days before being fixed for immunofluorescent staining with antibodies against MKX. Signals were detected on a Leica TCS SP5 confocal microscope. Scale bar =25 μm.
3.6. Regulation of tenogenic gene expression by re-introduction of Mohawk
To study the effects of MKX on tenogenesis, BMSCs were infected with MKX-expressing lentivirus for seven days, and the expression of tenogenic genes was measured. qPCR revealed a significant increase of Scx, Mkx, Col14, and FMOD (Fig. S6). Furthermore, exposure of MKX-ectopically expressed BMSCs to lenti-shMKX downregulated the expression of Scx, Egr1, Col1A2, Col14, DCN, ELN, FMOD, and TNC, which was not observed by infection with lenti-shM3U (Fig. S7). The ectopic expression of MKX and efficient knockdown of endogenous MKX by shM3U were confirmed by immunoblotting with antibodies against MKX (Fig. S7). These data not only demonstrate the biological function of the cloned equine Mkx gene, also indicate the essential role of Mohawk in regulating tenogenic gene expression in BMSCs. To determine the role of MKX on tenogenic gene expression in teno-iPSCs, cells were infected with MKX-expressing lentivirus for seven days, and the expression of tenogenesis-related genes were analyzed by qPCR. As expected, the expression of Mkx was strongly increased, and its level was comparable to that in the primary BMSCs (Fig. 6). Interestingly, ectopic expression of Mkx also resulted in significant elevation of DCN in teno-iPSCs. The expression of Scx, Egr1, Col1A2, Col14, ELN, FMOD, and TNC was increased, but not significant (Fig. 6).
Fig. 6.
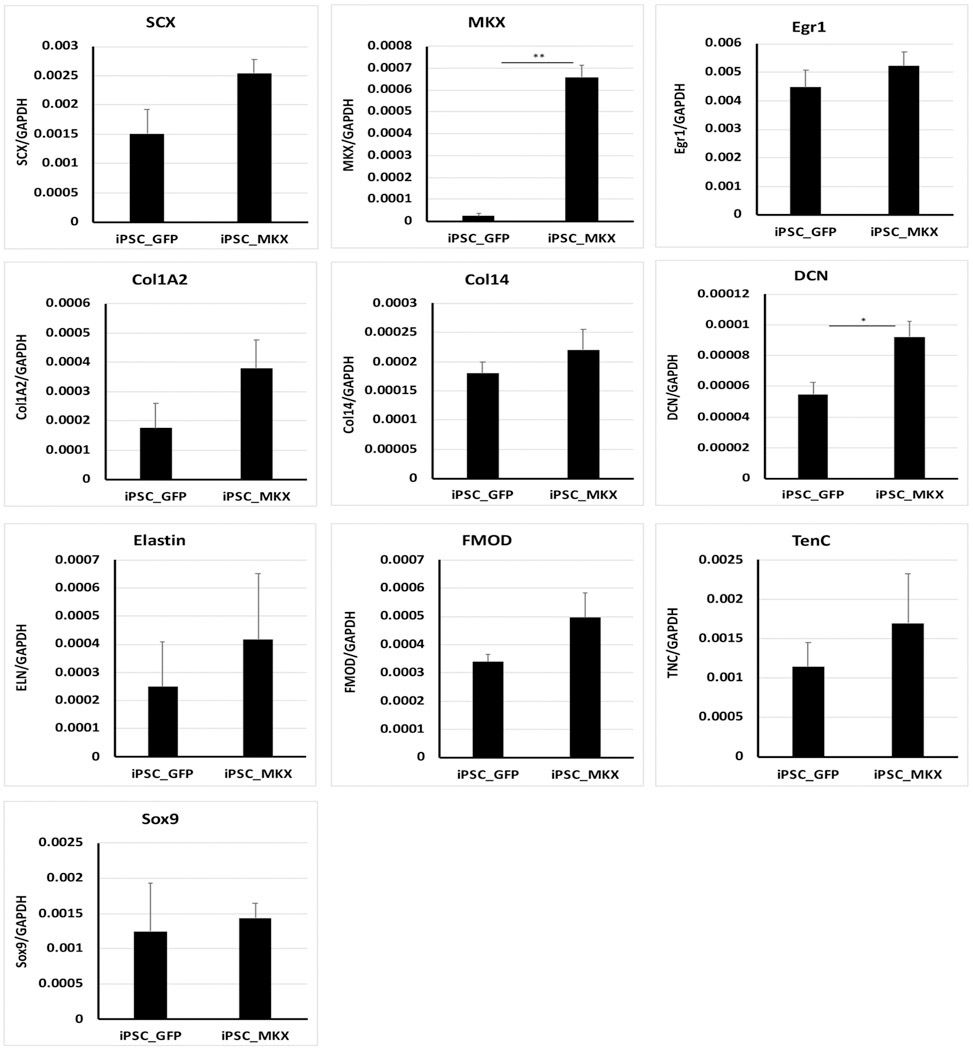
Expression of tenogenic genes in Mkx-transduced teno-iPSCs. Teno-iPSCs were infected with pHAGE-MKX (iPSC-MKX) or control pHAGE-GFP (iPSC-GFP) for seven days in basic medium, and the expression of tenocyte-linked genes was measured by qPCR.
3.7. Synergistic effects of Mohawk and mechanical loading on tenogenic gene expression
To test whether the tenogenic gene expression can be synergistically regulated by MKX and mechanical loading, MKX-expressing BMSCs and teno-iPSCs were subject to mechanical loading for 18 h, and the expression of tenogenic genes was assessed by qPCR. As expected, the mRNA levels of Egr1 in both BMSCs and teno-iPSCs were augmented upon mechanical loading. Ectopic expression of Mkx further increased the mechanical stimulation-induced expression of Scx, Egr1, Col1A2, DCN, and TNC in teno-iPSCs (Fig. 7) and that of Scx, Egr1, DCN, and TNC in BMSCs (Fig. S8). These data suggest that the combination of Mkx overexpression and mechanical stretching may have synergistic effects on facilitating the tenogenic gene expression in teno-iPSCs and BMSCs.
Fig. 7.
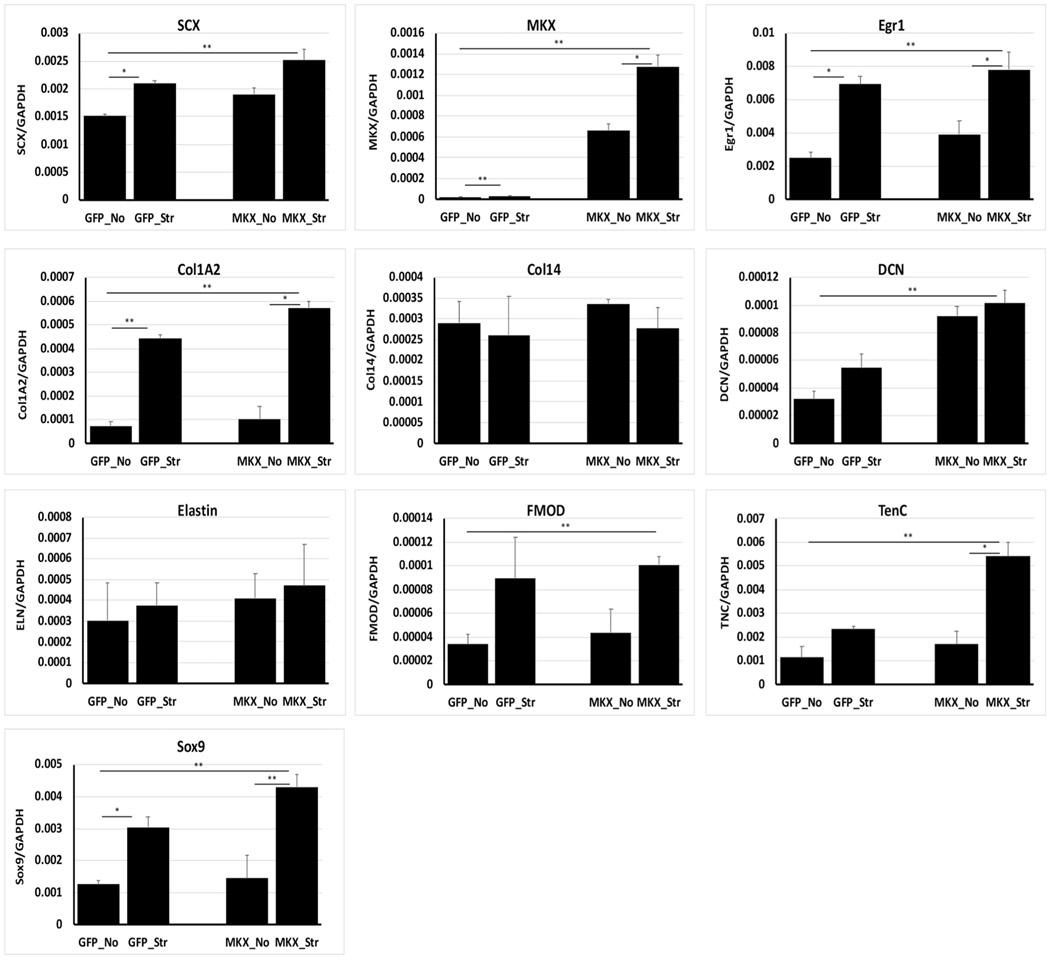
Synergistic effects of Mohawk and mechanical stretching on tenogenic gene expression in teno-iPSCs. Teno-iPSCs were infected with pHAGE-MKX or control pHAGE-GFP for four days, then split and seeded on vitronectin-coated PCL scaffolds for another two days before being subject to mechanical stretching for 18 h (GFP_Str; MKX_Str). Cells seeded on PCL scaffolds without mechanical loading were served as control (GFP_No; MKX-No). Expression of tenocyte-linked genes was determined by qPCR.
4. Discussion
In this study, by using a single lentiviral stem cell cassette expressing four Yamanaka transcription factors, we generated equine tenocyte-derived iPSCs which showed multi-lineage differentiation capacity. Compared to parental tenocytes, the resulting teno-iPSCs expressed significantly lower levels of lineage-specific genes, including Scx, Mkx, Egr1, Col1A2, Col14, DCN, ELN, FMOD, and TNC. These repressed genes, except Col14 and ELN, could be re-activated by cyclic uniaxial mechanical loading and ectopically transduced transcription factor Mkx alone or combined.
4.1. Altered lineage-specific transcriptional activity in teno-iPSCs
Although it is believed that successful reprogramming of differentiated somatic cells requires complete erasure of the parental epigenetic signature(Nashun et al., 2015), retention of epigenetic memory from the donor cells has also been reported(Bar-Nur et al., 2011; Kim et al., 2010; Phetfong et al., 2016; Roberts et al., 2017). While this might be a problem for the differentiation of iPSCs into lineages different from the donor cells, it might be advantageous for re-differentiation of iPSCs into the same lineage. Indeed, human osteoblast cells derived iPSCs were able to differentiate into osteoblast progenitors (Roberts et al., 2017). In our study, reprogramming of tenocytes with traditional Yamanaka factors significantly repressed the transcriptional activity of all the tested tenocyte-lineage specific genes, indicating the re-establishment of transcriptional network in the resulting iPSCs. Kyttaelae et al. reported that iPSC lines derived from the same donor were transcriptionally similar to each other(Kyttala et al., 2016), but our results demonstrated varied mRNA levels of parental lineage genes among the three tested teno-iPSC clones, implying that the retention degree of parental gene activities differed from one another. Kilpinen et al. suggested that many factors, such as cell type of origin, culture condition and reprogramming method, contributed to the variations between iPSC lines.(Kilpinen et al., 2017). One possible reason for the transcriptional variations among our teno-iPSC clones could be due to the heterogeneity nature of the primary tenocyte cultures, even though they were originally derived from the same donor horse. It is also noteworthy that, although teno-iPSCs displayed significant changes on the parental lineage-specific transcriptional network, they might still be inclined to re-differentiate to the same lineage than to others. Since Clone 3 showed higher level of tenogenic gene expression than other two clones, we assumed that this line might possess higher tenogenic differentiation potential. Further study will be necessary to understand whether and how variations among teno-iPSC clones affect their isogenic differentiation capacity.
4.2. Mechanical stretching alone on tenogenic differentiation
Tendon is a mechanosensitive tissue transferring force from muscle to bone. Biomechanical force plays critical roles in normal tendon development as well as in tendon repair(Galloway et al., 2013; Aspenberg, 2007; Brown et al., 2014; Snedeker and Foolen, 2017). Various types of cells and different mechanical loading parameters have been tested for mechanical force-induced tenogenic differentiation (Murchison et al., 2007; Song et al., 2010; Scott et al., 2011; Youngstrom et al., 2016; Zhang et al., 2010). Xu et al. reported that the cyclic tensile strain with different amplitudes (2%, 4%, and 8%) and frequencies (0.2 Hz, 0.5 Hz, and 1.0 Hz) had no influence on TPSC viability but showed different effects on the proliferation and the expression of tenogenic genes including Col1, TNC, TNMD, and Scx(Xu et al., 2015). Nam et al. also reported that cyclic uniaxial loading with 4% strain and 1.0 Hz frequency enhanced human BMSC proliferation, but higher strains were required for the superior expression of tenogenic genes, including COL1, COL3, TNC, Scx, DCN, and TNMD (Nam et al., 2015). In our study, the mechanical loading parameters (1.0 Hz with 0%–6% sinusoidal wave of strain) were also used in previous study (Baker et al., 2011). The cyclic uniaxial stretching upregulated the expression of Egr1 in both BMSCs and teno-iPSCs, which was expected as EGR1 is a known biomechanical sensor(Gaut et al., 2016) and thus can be served as a marker for the effective mechanical loading on the targeting cells. Furthermore, the mechanical stretching also increased the expression of Scx, DCN, and TNC in BMSCs and teno-iPSCs. This agrees with the findings in other studies where human BMSCs were subject to mechanical strain for 7 or 14 days(Xu et al., 2015; Nam et al., 2015; Qiu et al., 2016). Interestingly, mechanical loading elevated collagen I gene expression in rat TPSCs (Xu et al., 2015) but not in human BMSCs (Qiu et al., 2016). Our data revealed an apparent increase of Col1A2 expression in teno-iPSCs but not in BMSCs, indicating there might be different regulatory networks on Col1A2 gene activity between these two types of cells. In addition, a significant increase of chondrogenic marker Sox9 was detected in teno-iPSCs upon mechanical stretching (Fig. 4). Given to the low transcriptional activity of Mkx in teno-iPSCs, this result is in line with the report from another study where Mkx−/− tendon-derived cells also showed increased expression of Sox9 after mechanical stimulation.(Suzuki et al., 2016).
4.3. Transcriptional regulation on tenogenic differentiation
Mohawk is highly expressed in developing tendon and appears to be an important regulator of tenogenic differentiation, as Mkx−/− mice and Mkx−/− rats display marked tendon phenotype(Ito et al., 2010; Liu et al., 2010; Onizuka et al., 2014; Suzuki et al., 2016). However, it is unclear whether the phenotype is a direct cause from the loss of MKX-regulated ECM gene activity or an indirect cause from the change of ECM-regulating transcription factor(s) by the depletion of MKX. Although Liu et al. reported that, compared to wild littermates, there was no changes on the expression of Scx in Mkx−/− neonatal tendon cells(Liu et al., 2010), another study with a Mkx−/− mouse model showed significant increase of Scx in 8-week old Achille tendon than that in the wildtype(Pryce et al., 2009). Contradictory to the above report, the expression of Scx in the patellar tendon of 3-week old Mkx−/− rat was significantly lower than that in the wildtype (Killian and Thomopoulos, 2016). In our study, knockdown of endogenous MKX using lentiviral-shRNA downregulated the expression of Scx in both BMSCs and tenocytes but not in Mkx-transduced BMSCs, strongly suggesting that MKX regulates the expression of of Scx at the transcription level. Furthermore, the expression of Egr1, which was not explored in Mkx−/− animal models, was decreased in BMSCs and tenocytes after knockdown of MKX, indicating an interdependent transcriptional regulation network among tenogenesis-related transcription factors. Very likely, it was the loss of those key tenogenic transcription factors that caused the significant decrease of ECM gene expression, such as Col1A2, Col14, and FMOD in BMSCs, and Col1A2, Col14, DCN, ELN, and FMOD in tenocytes. Of note, reduction of FMOD expression was observed in the 3-week old Mkx−/− rat patellar tendon (Suzuki et al., 2016) but not in 8-week old Mkx−/− mouse Achille tendon (Ito et al., 2010). Additionally, compared to knockdown of Mkx, ectopic expression of Mkx seems to have less impact on the regulation of downstream ECM gene activities, except that, among the tested ECM genes, only Col14 and FMOD in BMSCs, and DCN in teno-iPSCs, were highly up-regulated after Mkx transduction. In other words, ectopic expression of Mkx alone may be insufficient to induce tenogenic gene expression in BMSCs and teno-iPSCs.
4.4. Combination of biochemical and biomechanical cues in tenogenic regulation
It has been reported overexpression Scx combined with mechanical force have a synergistic effect on promoting the commitment of human ESC-derived MSCs to tenocytes (Chen et al., 2012). Another study by Nichols et al. also showed that combination of transient Scx overexpression with cyclic strain enhanced the differentiation of murine MSC line C3H10T1/2 into ligament-like cells (Nichols et al., 2018b). In a Mkx−/− rat model, mechanical stretching enhanced the chondrogenic gene expression in tendon-derived cells(Ito et al., 2010; Suzuki et al., 2016), suggesting that MKX may play a role in regulating mechanical force-sensitive gene activities and thus affecting the cell fate. Indeed, our results demonstrated that the mechanical stimulation combined with ectopic expression of Mkx significantly increased the expression of Scx, Egr1, Col1A2, DCN, FMOD, and TNC in teno-iPSCs (Fig. 7) and that of Scx, Egr1, Col1A2, Col14, DCN, FMOD, and TNC in BMSCs (Fig. S7). On the other hand, whereas ectopic expression of Mkx did not influence the mechanical loading-induced expression of Sox9 in teno-iPSCs (Fig. 6 &7), the levels of other chondrogenic-related genes ACAN and Col2A1 were not upregulated by either ectopic expression of Mkx or mechanical loading (unpublished data). It is also worthy to mention that, as a marker for mature tenocytes, tenomodulin was not detected in cell lysates from either teno-iPSCs alone, or mechanical loading-stimulated teno-iPSCs, or Mkx-transduced teno-iPSCs. Although the underlying mechanism is unclear, the lack of tenomodulin expression could be a reflection of the persistent colony-like morphology of teno-iPSCs under the conditions used in this study (unpublished observation). Stimulation with soluble cytokines and cultivation in matrix-based environment may be necessary to overcome the challenge(Yang et al., 2017; Barsby et al., 2014). Nevertheless, our data indicate the tenogenic differentiation capacity of tenocyte-derived iPSCs. Future work will focus on investigating their potential in tendon repair and regeneration under in vivo conditions, such as the repair of collagenase-induced tendon injury in animal models.
5. Conclusion
In summary, our results demonstrated that forced reprogramming of tenocytes by Yamanaka factors repressed the lineage-specific transcriptional network in the resulting teno-iPSCs, which could be re-activated to some extent by mechanical stretching and ectopic expression of tenogenic transcription factor MKX. Our data suggest that tenocyte-derived iPSCs can be a promising cell source for basic and clinic research on tendon repair.
Supplementary Material
Acknowledgements
The authors would like to thank Dr. Filipa Pinto for assistance on the generation of teno-iPSCs, Drs. Robert L. Mauck, Spencer Szczesny and Su-chin Heo for mechanical loading experiments. Our thanks also go to Drs. Gary Althouse, Kyla Ortvet and Renata L Linardi for their help on flow cytometry assay. This work was funded by James Foundation, Applestone Farm, and partially supported by NIH/NIAMS (P30AR069619).
Abbreviations:
- BMSC
bone marrow-derived mesenchymal stem cells
- ECM
extracellular matrix
- ESC
embryonic stem cell
- FCS
fetal calf serum
- MEF
mouse embryonic fibroblasts
- RT-PCR
reverse transcription PCR
- qPCR
quantitative real time-polymerase chain reaction
- teno-iPSC
tenocyte-derived induced pluripotent stem cells
- TPSC
tendon progenitor/stem cell
Footnotes
Ethics approval
The animal care and experimental procedures were carried out in accordance with the protocol reviewed by the Institutional Animal Care and Use Committee (IACUC) at University of Pennsylvania.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.scr.2019.101489.
Competing interests
The authors declare that they have no competing interests.
References
- Alberton P, Popov C, Pragert M, Kohler J, Shukunami C, Schieker M, Docheva D, 2012. Conversion of human bone marrow-derived mesenchymal stem cells into tendon progenitor cells by ectopic expression of scleraxis. Stem Cells Dev. 21, 846–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aspenberg P, 2007. Stimulation of tendon repair: mechanical loading, GDFs and platelets. A mini-review. Int. Orthop. 31, 783–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker BM, Shah RP, Huang AH, Mauck RL, 2011. Dynamic tensile loading improves the functional properties of mesenchymal stem cell-laden nanofiber-based fibrocartilage. Tissue Eng Part A 17, 1445–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Nur O, Russ HA, Efrat S, Benvenisty N, 2011. Epigenetic memory and preferential lineage-specific differentiation in induced pluripotent stem cells derived from human pancreatic islet beta cells. Cell Stem Cell 9, 17–23. [DOI] [PubMed] [Google Scholar]
- Barsby T, Guest D, 2013. Transforming growth factor beta3 promotes tendon differentiation of equine embryo-derived stem cells. Tissue Eng Part A 19, 2156–2165. [DOI] [PubMed] [Google Scholar]
- Barsby T, Bavin EP, Guest DJ, 2014. Three-dimensional culture and transforming growth factor beta3 synergistically promote tenogenic differentiation of equine embryo-derived stem cells. Tissue Eng Part A 20, 2604–2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bavin EP, Smith O, Baird AE, Smith LC, Guest DJ, 2015. Equine induced pluripotent stem cells have a reduced tendon differentiation capacity compared to embryonic stem cells. Front Vet Sci 2, 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beane OS, Fonseca VC, Cooper LL, Koren G, Darling EM, 2014. Impact of aging on the regenerative properties of bone marrow-, muscle-, and adipose-derived mesenchymal stem/stromal cells. PLoS One 9, e115963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi Y, Ehirchiou D, Kilts TM, Inkson CA, Embree MC, Sonoyama W, Li L, Leet AI, Seo BM, Zhang L, Shi S, Young MF, 2007. Identification of tendon stem/ progenitor cells and the role of the extracellular matrix in their niche. Nat. Med. 13, 1219–1227. [DOI] [PubMed] [Google Scholar]
- Brown JP, Finley VG, Kuo CK, 2014. Embryonic mechanical and soluble cues regulate tendon progenitor cell gene expression as a function of developmental stage and anatomical origin. J. Biomech. 47, 214–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Yin Z, Chen JL, Shen WL, Liu HH, Tang QM, Fang Z, Lu LR, Ji J, Ouyang HW, 2012. Force and scleraxis synergistically promote the commitment of human ES cells derived MSCs to tenocytes. Sci. Rep. 2, 977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Yin Z, Chen JL, Liu HH, Shen WL, Fang Z, Zhu T, Ji J, Ouyang HW, Zou XH, 2014. Scleraxis-overexpressed human embryonic stem cell-derived mesenchymal stem cells for tendon tissue engineering with knitted silk-collagen scaffold. Tissue Eng Part A 20, 1583–1592. [DOI] [PubMed] [Google Scholar]
- Czaplewski SK, Tsai TL, Duenwald-Kuehl SE, Vanderby R Jr., Li WJ, 2014. Tenogenic differentiation of human induced pluripotent stem cell-derived mesenchymal stem cells dictated by properties of braided submicron fibrous scaffolds. Biomaterials 35, 6907–6917. [DOI] [PubMed] [Google Scholar]
- Docheva D, Muller SA, Majewski M, Evans CH, 2015. Biologics for tendon repair. Adv. Drug Deliv. Rev. 84, 222–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filomeno P, Dayan V, Tourino C, 2012. Stem cell research and clinical development in tendon repair. Muscles Ligaments Tendons J 2, 204–211. [PMC free article] [PubMed] [Google Scholar]
- Galloway MT, Lalley AL, Shearn JT, 2013. The role of mechanical loading in tendon development, maintenance, injury, and repair. J. Bone Joint Surg. Am. 95, 1620–1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaspar D, Spanoudes K, Holladay C, Pandit A, Zeugolis D, 2015. Progress in cell-based therapies for tendon repair. Adv. Drug Deliv. Rev. 84, 240–256. [DOI] [PubMed] [Google Scholar]
- Gaut L, Robert N, Delalande A, Bonnin MA, Pichon C, Duprez D, 2016. EGR1 regulates transcription downstream of mechanical signals during tendon formation and healing. PLoS One 11, e0166237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerquin MJ, Charvet B, Nourissat G, Havis E, Ronsin O, Bonnin MA, Ruggiu M, Olivera-Martinez I, Robert N, Lu Y, Kadler KE, Baumberger T, Doursounian L, Berenbaum F, Duprez D, 2013. Transcription factor EGR1 directs tendon differentiation and promotes tendon repair. J. Clin. Invest. 123, 3564–3576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havis E, Bonnin MA, Esteves de Lima J, Charvet B, Milet C, Duprez D, 2016. TGFbeta and FGF promote tendon progenitor fate and act downstream of muscle contraction to regulate tendon differentiation during chick limb development. Development 143, 3839–3851. [DOI] [PubMed] [Google Scholar]
- Hiler D, Chen X, Hazen J, Kupriyanov S, Carroll PA, Qu C, Xu B, Johnson D, Griffiths L, Frase S, Rodriguez AR, Martin G, Zhang J, Jeon J, Fan Y, Finkelstein D, Eisenman RN, Baldwin K, Dyer MA, 2015. Quantification of Retinogenesis in 3D cultures reveals epigenetic memory and higher efficiency in iPSCs derived from rod photoreceptors. Cell Stem Cell 17, 101–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho JO, Sawadkar P, Mudera V, 2014. A review on the use of cell therapy in the treatment of tendon disease and injuries. J Tissue Eng 5 2041731414549678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang AH, Lu HH, Schweitzer R, 2015. Molecular regulation of tendon cell fate during development. J. Orthop. Res. 33, 800–812. [DOI] [PubMed] [Google Scholar]
- Ito Y, Toriuchi N, Yoshitaka T, Ueno-Kudoh H, Sato T, Yokoyama S, Nishida K, Akimoto T, Takahashi M, Miyaki S, Asahara H, 2010. The Mohawk homeobox gene is a critical regulator of tendon differentiation. Proc. Natl. Acad. Sci. U. S. A.107, 10538–10542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killian ML, Thomopoulos S, 2016. Scleraxis is required for the development of a functional tendon enthesis. FASEB J. 30, 301–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilpinen H, Goncalves A, Leha A, Afzal V, Alasoo K, Ashford S, Bala S, Bensaddek D, Casale FP, Culley OJ, Danecek P, Faulconbridge A, Harrison PW, Kathuria A, McCarthy D, McCarthy SA, Meleckyte R, Memari Y, Moens N, Soares F, Mann A, Streeter I, Agu CA, Alderton A, Nelson R, Harper S,Patel M, White A, Patel SR, Clarke L, Halai R, Kirton CM, Kolb-Kokocinski A, Beales P, Birney E, Danovi D, Lamond AI, Ouwehand WH, Vallier L, Watt FM, Durbin R, Stegle O, Gaffney DJ, 2017. Common genetic variation drives molecular heterogeneity in human iPSCs. Nature 546, 370–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K, Doi A, Wen B, Ng K, Zhao R, Cahan P, Kim J, Aryee MJ, Ji H, Ehrlich LI, Yabuuchi A, Takeuchi A, Cunniff KC, Hongguang H, McKinney-Freeman S, Naveiras O, Yoon TJ, Irizarry RA, Jung N, Seita J, Hanna J, Murakami P, Jaenisch R, Weissleder R, Orkin SH, Weissman IL, Feinberg AP, Daley GQ, 2010. Epigenetic memory in induced pluripotent stem cells. Nature 467, 285–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyttala A, Moraghebi R, Valensisi C, Kettunen J, Andrus C, Pasumarthy KK, Nakanishi M, Nishimura K, Ohtaka M, Weltner J, Van Handel B, Parkkonen O, Sinisalo J, Jalanko A, Hawkins RD, Woods NB, Otonkoski T, Trokovic R, 2016. Genetic variability overrides the impact of parental cell type and determines iPSC differentiation potential. Stem Cell Reports 6, 200–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Watson SS, Lan Y, Keene DR, Ovitt CE, Liu H, Schweitzer R, Jiang R, 2010. The atypical homeodomain transcription factor Mohawk controls tendon morphogenesis. Mol. Cell. Biol. 30, 4797–4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Zhang C, Zhu S, Lu P, Zhu T, Gong X, Zhang Z, Hu J, Yin Z, Heng BC, Chen X, Ouyang HW, 2015. Mohawk promotes the tenogenesis of mesenchymal stem cells through activation of the TGFbeta signaling pathway. Stem Cells 33, 443–455. [DOI] [PubMed] [Google Scholar]
- Liu Y, Suen CW, Zhang JF, Li G, 2017. Current concepts on tenogenic differentiation and clinical applications. J Orthop Translat 9, 28–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita Y, Watanabe S, Ju Y, Xu B, 2013. Determination of optimal cyclic uniaxial stretches for stem cell-to-tenocyte differentiation under a wide range of mechanical stretch conditions by evaluating gene expression and protein synthesis levels. Acta Bioeng Biomech 15, 71–79. [PubMed] [Google Scholar]
- Morita Y, Yamashita T, Toku T, Ju Y, 2018. Optimization of differentiation time of mesenchymal-stem-cell to tenocyte under a cyclic stretching with a microgrooved culture membrane and selected measurement cells. Acta Bioeng Biomech 20, 3–10. [PubMed] [Google Scholar]
- Murchison ND, Price BA, Conner DA, Keene DR, Olson EN, Tabin CJ, Schweitzer R, 2007. Regulation of tendon differentiation by scleraxis distinguishes force-transmitting tendons from muscle-anchoring tendons. Development 134, 2697–2708. [DOI] [PubMed] [Google Scholar]
- Nam HY, Pingguan-Murphy B, Amir Abbas A, Mahmood Merican A, Kamarul T, 2015. The proliferation and tenogenic differentiation potential of bone marrow-derived mesenchymal stromal cell are influenced by specific uniaxial cyclic tensile loading conditions. Biomech. Model. Mechanobiol. 14, 649–663. [DOI] [PubMed] [Google Scholar]
- Nashun B, Hill PW, Hajkova P, 2015. Reprogramming of cell fate: epigenetic memory and the erasure of memories past. EMBO J. 34, 1296–1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols AEC, Settlage RE, Werre SR, Dahlgren LA, 2018a. Novel roles for scleraxis in regulating adult tenocyte function. BMC Cell Biol. 19, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols AEC, Werre SR, Dahlgren LA, 2018b. Transient Scleraxis overexpression combined with cyclic strain enhances ligament cell differentiation. Tissue Eng Part A 24, 1444–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nourissat G, Berenbaum F, Duprez D, 2015. Tendon injury: from biology to tendon repair. Nat. Rev. Rheumatol. 11, 223–233. [DOI] [PubMed] [Google Scholar]
- Onizuka N, Ito Y, Inagawa M, Nakahara H, Takada S, Lotz M, Toyama Y, Asahara H, 2014. The Mohawk homeobox transcription factor regulates the differentiation of tendons and volar plates. J. Orthop. Sci. 19, 172–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phetfong J, Supokawej A, Wattanapanitch M, Kheolamai P, Issaragrisil S, 2016. Cell type of origin influences iPSC generation and differentiation to cells of the hematoendothelial lineage. Cell Tissue Res. 365, 101–112. [DOI] [PubMed] [Google Scholar]
- Polo JM, Liu S, Figueroa ME, Kulalert W, Eminli S, Tan KY, Apostolou E, Stadtfeld M, Li Y, Shioda T, Natesan S, Wagers AJ, Melnick A, Evans T, Hochedlinger K, 2010. Cell type of origin influences the molecular and functional properties of mouse induced pluripotent stem cells. Nat. Biotechnol. 28, 848–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryce BA, Watson SS, Murchison ND, Staverosky JA, Dunker N, Schweitzer R, 2009. Recruitment and maintenance of tendon progenitors by TGFbeta signaling are essential for tendon formation. Development 136, 1351–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Y, Lei J, Koob TJ, Temenoff JS, 2016. Cyclic tension promotes fibroblastic differentiation of human MSCs cultured on collagen-fibre scaffolds. J. Tissue Eng. Regen. Med. 10, 989–999. [DOI] [PubMed] [Google Scholar]
- Roberts CL, Chen SS, Murchison AC, Ogle RA, Francis MP, Ogle RC, Sachs PC, 2017. Preferential lineage-specific differentiation of osteoblast-derived induced pluripotent stem cells into Osteoprogenitors. Stem Cells Int. 2017, 1513281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott A, Danielson P, Abraham T, Fong G, Sampaio AV, Underhill TM, 2011.Mechanical force modulates scleraxis expression in bioartificial tendons. J.Musculoskelet. Neuronal Interact. 11, 124–132. [PubMed] [Google Scholar]
- Sheridan SD, Surampudi V, Rao RR, 2012. Analysis of embryoid bodies derived from human induced pluripotent stem cells as a means to assess pluripotency. Stem Cells Int. 2012, 738910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shipony Z, Mukamel Z, Cohen NM, Landan G, Chomsky E, Zeliger SR, Fried YC, Ainbinder E, Friedman N, Tanay A, 2014. Dynamic and static maintenance of epigenetic memory in pluripotent and somatic cells. Nature 513, 115–119. [DOI] [PubMed] [Google Scholar]
- Snedeker JG, Foolen J, 2017. Tendon injury and repair - a perspective on the basic mechanisms of tendon disease and future clinical therapy. Acta Biomater. 63, 18–36. [DOI] [PubMed] [Google Scholar]
- Sommer CA, Stadtfeld M, Murphy GJ, Hochedlinger K, Kotton DN, Mostoslavsky G, 2009. Induced pluripotent stem cell generation using a single lentiviral stem cell cassette. Stem Cells 27, 543–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song G, Luo Q, Xu B, Ju Y, 2010. Mechanical stretch-induced changes in cell morphology and mRNA expression of tendon/ligament-associated genes in rat bonemarrow mesenchymal stem cells. Mol Cell Biomech 7, 165–174. [PubMed] [Google Scholar]
- Sun HB, Schaniel C, Leong DJ, Wang JH, 2015. Biology and mechano-response of tendon cells: Progress overview and perspectives. J. Orthop. Res. 33, 785–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H, Ito Y, Shinohara M, Yamashita S, Ichinose S, Kishida A, Oyaizu T,Kayama T, Nakamichi R, Koda N, Yagishita K, Lotz MK, Okawa A, Asahara H, 2016. Gene targeting of the transcription factor Mohawk in rats causes heterotopic ossification of Achilles tendon via failed tenogenesis. Proc. Natl. Acad. Sci. U. S. A. 113, 7840–7845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Q, Lui PP, Rui YF, Wong YM, 2012. Comparison of potentials of stem cells isolated from tendon and bone marrow for musculoskeletal tissue engineering. Tissue Eng Part A 18, 840–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan C, Lui PP, Lee YW, Wong YM, 2014. Scx-transduced tendon-derived stem cells (tdscs) promoted better tendon repair compared to mock-transduced cells in a rat patellar tendon window injury model. PLoS One 9, e97453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turinetto V, Vitale E, Giachino C, 2016. Senescence in human Mesenchymal stem cells: functional changes and implications in stem cell-based therapy. Int. J. Mol. Sci. 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb S, Gabrelow C, Pierce J, Gibb E, Elliott J, 2016. Retinoic acid receptor signaling preserves tendon stem cell characteristics and prevents spontaneous differentiation in vitrox. Stem Cell Res Ther 7, 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Wang Y, Liu E, Sun Y, Luo Z, Xu Z, Liu W, Zhong L, Lv Y, Wang A, Tang Z, Li S, Yang L, 2013. Human iPSC-derived neural crest stem cells promote tendon repair in a rat patellar tendon window defect model. Tissue Eng Part A 19, 2439–2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Wang Q, Li Y, Gan Y, Li P, Li S, Zhou Y, Zhou Q, 2015. Cyclic tensile strain induces Tenogenic differentiation of tendon-derived stem cells in bioreactor culture. Biomed. Res. Int. 2015, 790804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, Hu L, Chen C, Yu J, O’Connell CB, Khodjakov A, Pagano M, Dai W, 2012. BubR1 is modified by sumoylation during mitotic progression. J. Biol. Chem. 287, 4875–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang G, Rothrauff BB, Lin H, Yu S, Tuan RS, 2017. Tendon-derived extracellular matrix enhances transforming growth factor-beta3-induced Tenogenic differentiation of human adipose-derived stem cells. Tissue Eng Part A 23, 166–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Z, Hu JJ, Yang L, Zheng ZF, An CR, Wu BB, Zhang C, Shen WL, Liu HH, Chen JL, Heng BC, Guo GJ, Chen X, Ouyang HW, 2016. Single-cell analysis reveals a nestin(+) tendon stem/progenitor cell population with strong tenogenic potentiality. Sci. Adv. 2, e1600874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto Y, Takimoto A, Watanabe H, Hiraki Y, Kondoh G, Shukunami C, 2017. Scleraxis is required for maturation of tissue domains for proper integration of the musculoskeletal system. Sci. Rep. 7, 45010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youngstrom DW, LaDow JE, Barrett JG, 2016. Tenogenesis of bone marrow-, adipose-, and tendon-derived stem cells in a dynamic bioreactor. Connect. Tissue Res. 57, 454–465. [DOI] [PubMed] [Google Scholar]
- Zhang J, Pan T, Liu Y, Wang JH, 2010. Mouse treadmill running enhances tendons by expanding the pool of tendon stem cells (TSCs) and TSC-related cellular production of collagen. J. Orthop. Res. 28, 1178–1183. [DOI] [PubMed] [Google Scholar]
- Zhang C, Yuan H, Liu H, Chen X, Lu P, Zhu T, Yang L, Yin Z, Heng BC, Zhang Y, Ouyang H, 2015. Well-aligned chitosan-based ultrafine fibers committed Teno-lineage differentiation of human induced pluripotent stem cells for Achilles tendon regeneration. Biomaterials 53, 716–730. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


