Abstract
The last few decades have been marked by a rapid genetic improvement in chicken growth rates. The modern-day chicken is more efficient in converting feed into muscle mass than their predecessors. This enhanced efficiency emanates from better nutrient digestion, absorption, and metabolism. The gut has therefore become a research focus especially after the ban on the use of antibiotics as growth promoters (AGP) in poultry. In pursuance of better gut health in the post-AGP era, many different strategies are being continuously sought and tested. The gut is inhabited by more than 900 bacterial species along with fungi and archaea, and they play an important role to maintain a conducive milieu for the host. A beneficial shift in the microbial ecosystem of the chicken can be promoted by many dietary and non-dietary interventions, however, diet is ranked as one of the most important and potent regulators of gut microbiota composition. Therefore, the constituents of the diet warrant special attention in the modulation of the gut ecosystem. Among dietary constituents, fiber possesses a significant ability to modulate the microbiota. In this review, we will highlight the importance of fiber in poultry nutrition and will also discuss the effects of fiber on gut microbiota and its resultant ramifications on the liver and brain.
Keywords: Poultry, Microbiota, Gut health, Antibiotic alternatives
1. Introduction
The continuous scrutinization and improvement in genetic programs over the last few decades have made the chicken an ever more efficient converter of feed into muscle mass than its predecessors. Concomitantly, the nutrient requirements of the chicken have also been better estimated so as to achieve the maximum genetic potential in minimum possible time. Thus, the present-day chicken strains come with the ability to digest feed more efficiently and absorb the nutrients needed for maximum growth potential (Clavijo and Flórez, 2017). The growth rate and efficiency depend on a number of factors among which diet type and nutrient density are considered extremely important. Over the years, antimicrobial growth promoters (AGP; antibiotics supplemented at sub-therapeutic levels) have been used in broiler diets with the intent of preserving gut health thus optimizing growth performance. However, consumer's concern about antibiotic resistance resulted in the complete ban on AGP in the European Union (Regulation 1831/2003/EC) followed by other countries. This specific measure to completely ban the use of AGP raised the risks of bacterial diseases and dysbiosis, thereby compromising the performance of the birds (Paiva and McElroy, 2014). The challenge of maintaining gut health is still evolving as many alternatives to AGP (pre-, pro-biotics, organic acids, essential oils, etc.) yield variable results (Gadde et al., 2017). The key to preserving gut health is to understand the complex interplay between the host and its intestinal microbiome. This information thus helps to strike a balance between beneficial and pathogenic microbiota residing inside the gut. Any disturbance to this balance can initiate a cascade of reactions leading towards the inflammation of the gut and can jeopardize the whole process of digestion, absorption, and metabolism of the nutrients in the host.
There are innumerable factors including diet, breed type, housing environment, hatchery conditions, breeder chicken health, etc., which can affect the gut microbiome (Shang et al., 2018). Of these, diet is the principal environmental factor that can directly influence the nature of microbiota in the host (Yadav and Jha, 2019). The diet is a mixture of energy and protein, carbohydrates, fats, vitamins and minerals sources. Some of these sources brings a varying amount and type of fiber into the diet which exerts a direct effect on the gut microbiome (Apajalahti et al., 2004, Pan and Yu, 2014). Here, we will focus on the role of fiber in poultry diets and how it can influence the gut microbiota in chickens.
2. Fiber and non-starch polysaccharides
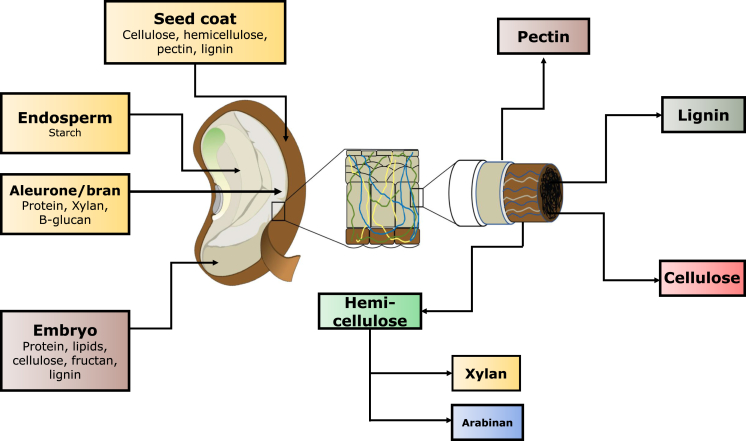
Dietary fiber polysaccharides and oligosaccharides are composed of relatively small numbers of monosaccharides e.g. glucose, galactose, arabinose, xylose, fructose, mannose, rhamnose fucose and some of their uronic acid forms (Fig. 1). However, a large number of combinations are possible between these simple monosaccharides, some forming the backbone of the oligosaccharide with varying linkages (e.g. β1-4) and others forming side chains at various points on the backbone (e.g. 02, 03, or more than one side chain per backbone linkage). Due to this flexibility, dietary fiber is one of the most diverse groups of molecules found in nature (Hamaker and Tuncil, 2014). Dietary fiber can be broken down only by enzymes that pair appropriately with specific sugar composition, linkages and chain length (Hamaker and Tuncil, 2014). Based on chemistry and accessibility of specific dietary fiber to particular microbial groups and consortia, dietary fiber presents a huge potential to modulate gut microbiota.
Fig. 1.
Distribution of dietary fiber components in grain.
Conventionally, dietary fiber was considered a diluent of poultry diet with a negative link to voluntary feed intake and nutrient digestibility (Mateos et al., 2012, Jha et al., 2019). Therefore, diets were formulated with crude fiber not exceeding 3%, especially for young broiler chickens. However, dietary fiber has also been suggested to significantly increase the gizzard weight (27.9 vs. 30.1 g/kg) (Hetland et al., 2005, González-Alvarado et al., 2007, Hetland and Svihus, 2007), amylase activity (178 vs. 261 U/g jejunal DM) and bile acid (14.7 vs. 21.9 jejunum mg/g) (Hetland et al., 2005; Svihus, 2011) in broiler chickens. Further, fiber sources such as hulls and wood shavings can decrease the pH content of the gizzard by the magnitude of 0.2 to 1.2 units (Jiménez-Moreno et al., 2009, Senkoylu et al., 2009). Increased gizzard volume and longer retention time allows for more HCl secretion along with stimulative effect of gizzard activity on acid secretion (Svihus, 2011). All these factors, in toto, can promote nutrient digestibility and growth performance along with improved intestinal health (Jha and Berrocoso, 2016). In many countries, corn-soybean meal-based diets are commonly used in the poultry industry as this nutritional regimen is considered highly digestible. However, in some countries, locally grown cereal such as wheat, barley, triticale in combination with their co-products are also used in poultry diets. Likewise, protein sources other than soybean meal (SBM) e.g. lupin, rapeseed meal, sunflower meal, canola meal are also used to fully or partially replace SBM in poultry diets (Jezierny et al., 2010). Notably, most cereal grains and protein sources contain fiber, of which, the most interesting fraction is the soluble form of fiber (Mateos et al., 2013). The major non-starch polysaccharides (NSP) fractions present in cereals include arabinoxylan and β-glucan. The contents may vary depending upon plant species and tissue of the grains (Saulnier et al., 2007, Izydorczyk and Dexter, 2008). Wheat, triticale, and corn are reported to be rich in arabinoxylan and barley and oats contain high β-glucan (Knudsen, 2014). Arabinoxylan is the major polymer in the cell wall of most cereals and with different structural features as demonstrated by varying arabinose-to-xylose ratio that can vary in the whole grain from 0.48 in barley to 0.74 in corn and in flour from 0.53 in wheat to 1.06 in corn. The concentration of β-glucan in particular varies from very little in corn (0.1%), to intermediate in wheat, rye and triticale (0.7% to 1.7%) (Knudsen, 2014). However, the proportion of NSP in protein feedstuffs varies considerably because the protein feedstuffs belong to different botanical families (Knudsen, 2014). Further, they have a diverse composition of the individual tissues making up the seeds (Caffall and Mohnen, 2009, Pustjens et al., 2013). Thus, arabinan (peas and rapeseed), arabinogalactan (soybeans and rapeseed) and galactans (lupine) are present either free or linked to rhamnogalacturonan (Knudsen, 2014) in protein feedstuffs.
3. Chicken microbiome functionality
The gastrointestinal tract (GIT) of the newly hatched chick is generally not sterile rather microbiota already reside there transmitted by different routes inter alia vertical transmission from mother in the oviduct (Gantois et al., 2009), from the environment through the pores on the eggshell (Roto et al., 2016). Finally, the hatchery and transportation vehicle, as well as the farm provides other sources of microbiota to colonize the chick's gut (Pedroso et al., 2005). Among other factors modulating the intestinal microbiota, maternal antibodies supplied through the yolk can protect against harmful bacteria during the early phase of life. In this way, maternal antibodies modulate chicken intestinal microbiota and thus the immune system (Cebra, 1999). This argument is supported by the differential development of the immune system in Germ-free animals pointing out that microbiota have a role to play in immuno-competence development (Williams, 2014). The intestinal microbiota diversity increases during the first weeks of life (Ballou et al., 2016) but the individual variation in microbiota composition decreases as the chicken grows older.
The GIT of chickens is enriched with complex microbial communities including bacteria, fungi, archaea, protozoa, and viruses but are dominated by bacteria (Wei et al., 2013). The host accommodates and forms a symbiotic relationship with the resident bacteria (Neish, 2009). This tolerance is manifested by suppression of the host immune response towards the microbiome. The gut microbiota constitutes a protective layer by associating themselves with the intestinal epithelial surface of enterocyte (Yegani and Korver, 2008). In this way, they shield the host from colonization by pathogenic bacteria. These bacteria constitute a commensal relationship with the host driving nutrients and aiding in digestion of a portion of the indigestible fraction of the host's diet. The gut epithelium is covered by a mucous layer that separates bacteria from the mucosa (Biasato et al., 2019). A well-structured and intact mucus layer is an integral component of defense used by the host against microbial invasion and infection. In this regard, the gut microbiota and diet are considered extremely important to maintain a normal structure and production of the intestinal mucus (Jha et al., 2019). Despite being a barrier against the gut microbiota, normal functional development of mucus cannot be ensured without the presence of bacteria (Schroeder, 2019). Mucins are the main components of mucus layer and are secreted by goblet cells of the GIT epithelium. Their protein backbone is highly glycosylated with carbohydrate chains of different monosaccharides thus allowing histological differentiation of mucin into neutral and acidic mucins with the latter further subdivided into sulfated and sialylated mucin types (Biasato et al., 2019). Bacterial colonization and proliferation have been suggested to alter the gut mucin composition by synthesizing mucin-specific glycosidases, glycosulfatases and proteases (Forder et al., 2007). In one of the initial studies, Forder et al. (2007) demonstrated that microbiota can influence small intestine goblet cell mucus composition and these changes occurred from 3 to 4 d post hatch. Although, the total number of goblet cells containing acidic mucin was not affected by bacterial colonization but mucin composition was altered with a decrease in sulfated mucin and an increase in sialylated mucin. Further, they noticed that sulfated mucin did not alter in low bacterial load chickens during the first week post hatch. They postulated that retention of sulfated mucin during post hatch development may be indicative of immature gut outlining the effect of bacteria on mucin production and overall gut maturity (Forder et al., 2007). Thus, diet and its components especially fiber fits perfectly well in this complex relationship of gut microbiota and mucosal barrier since ingested nutrients play important role in the development and functionality of the GIT. Therefore, an altered gut microbiota emanating from dietary changes especially as a result of feeding a low fiber ration can severely damage the mucus layer and enhance susceptibility to gut inflammation and infection by pathogenic bacteria.
This commensalism comes with a cost-to-benefit ratio for the host. These microbial communities competitively exclude the pathogenic bacteria from attachment to the epithelial surface of the gut (Dibner and Richards, 2005), stimulate and regulate the host immune system and contribute to host nutrition. The greater susceptibility of germ-free animals to pathogenic bacteria infection compared with their conventionally-raised counterparts indicates that the beneficial microbiota of the host reinforces the host's immune system (Pan and Yu, 2014). This implies that the intestinal immune system is more robust due to the presence of beneficial bacteria, and hence is more able to secrete immunoglobulin A (IgA) which binds to bacterial epitopes and helps in regulating microbial composition (Mitchell and Moretó, 2006, Suzuki and Nakajima, 2014). Most importantly, the commensal microbiota strengthens the barrier (mucus layer, epithelial monolayer, and intestinal immune cells) function, thereby keeping a check on pathogenic microbe invasion (Shakouri et al., 2009, Oakley and Kogut, 2016).
The commensal, beneficial bacteria attack the indigestible fraction of the diet, in particular, NSP, thereby yielding fermentation products on which the other members of the gut ecosystem thrive and produce short-chain fatty acids (SCFA) which in turn, are utilized by the host (Borda-Molina et al., 2018). Towards the distal segment of the gut i.e. ceca, the microbiota yields SCFA but in different proportions to that produced in the small intestine, and some vitamins e.g. vitamin K and B complex by fermenting the indigestible fraction of the host diet (Dibner and Richards, 2005). The most abundant SCFA are propionate, butyrate, and acetate (Tazoe et al., 2008). The SCFA are a source of energy for the host and aids in the proliferation of enterocytes and enhances the absorptive area of the gut (Dibner and Richards, 2005). SCFA are rapidly absorbed by the intestinal epithelial cells and regulate a number of cellular functions including gene expression, chemotaxis, differentiation and apoptosis (Corrêa-Oliveira et al., 2016). They also reduce the pH at the site of production and expedite the nutrient absorption. These SCFA can influence energy metabolism and regulate host feed intake through their interaction with free fatty acid receptors (FFA2,3) (Sleeth et al., 2010). Among SCFA, butyrate has received the most interest because of its anti-inflammatory properties and also as a source of energy for enterocytes (Canani et al., 2011). It has been reported that the butyrate protects the host against the inflammatory response. Importantly, intestinal macrophages and dendritic cells respond to the presence of butyrate through niacin receptor GPR109a, thereby increasing the production of IL-10, up-regulating the retinaldehyde dehydrogenase enzyme and enhancing Treg cell differentiation (Singh et al., 2014).
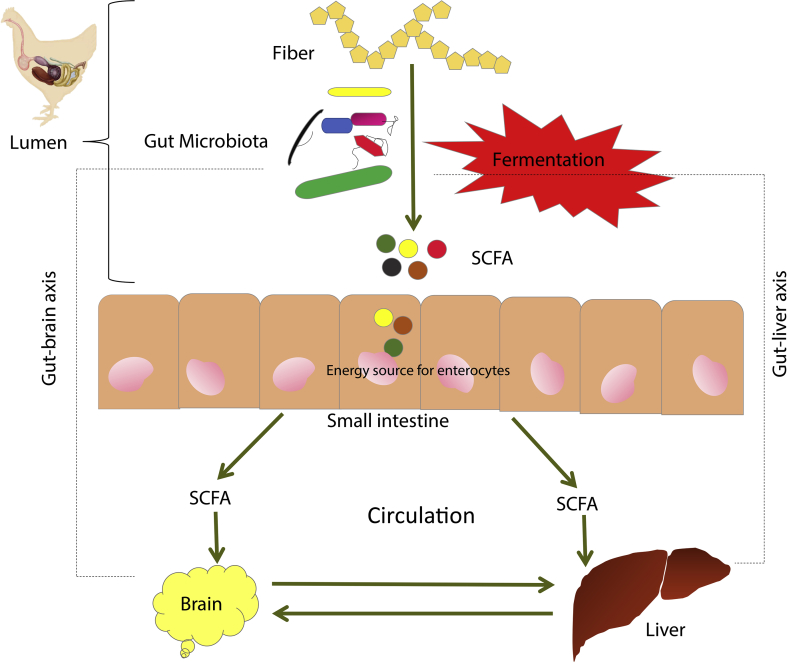
It has been postulated that the gut microbiota modulates the host physiology via the gut–brain axis, a bi-directional communication system based on neural, endocrine and immunological mechanisms (Fig. 2). Although this phenomenon has been vigorously investigated in mammals, not much work has been conducted on birds. A study on rodents demonstrated a shift in behavioral patterns in addition to variation in brain-derived neurotropic factors (BDNF) in different regions of the brain in response to perturbation in gut microbiota (Bercik et al., 2011). A transient perturbation of the microbiota increased hippocampal BDNF and exploratory behavior. These changes were reversed upon the normalization of microbiota after the withdrawal of antimicrobials. These findings suggest that there is a strong communicational link between the brain and the resident microbiota. Interestingly, these changes were not initiated by an intestinal inflammatory response, specific enteric neurotransmitters or the autonomic nervous system. It was suggested that specific bacterial products acted directly on the central nervous system, the authors implicated butyrate as a potential candidate (Bercik et al., 2011). It was also suggested that the immune system takes into account many cellular components of the microbiota. This is how a complex gut–brain axis operates, developing dynamic interactions between the host's innate and adaptive immune systems and specific microbial species which are assumed to demonstrate probiotic effects (Jarchum and Pamer, 2011).
Fig. 2.
Role of fiber in gut-liver and gut brain axis.
A recent study by Calefi et al. (2016) supported this argument in poultry. They found that Clostridium perfringens, heat stress, and thioglycolate produced a different behavioral response and affected c-fos expression in the paraventricular nucleus of the hypothalamus, nucleus taenia of the amygdala, medial preoptic area and globus pallidus brain nuclei of chickens. The authors reported that in any case, this phenomenon clearly demonstrates increased neural activity in the analyzed brain area. It is quite obvious from these studies that the right balance of the microbiota inside the gut is of paramount importance in order to maintain a vibrant immune system which can be helpful to exploit the maximum potential of the chickens.
From the physiological point of view, the gut–liver axis is likely the most important connection between the resident microbiota and extra-intestinal organs (Fig. 2). It represents a close functional and bidirectional communication between the intestine and the liver. It is well known that the liver is continually exposed not only to the products of digestion and absorption but also to the gut-derived factors including bacteria and bacterial components such as lipopolysaccharide (LPS). The venous system of the portal circulation directs the gut-liver axis and implies the close anatomical and functional interaction of the gut and the liver (O'hara et al., 2017). Physiologically, this link establishes itself as an important operative unit that helps protect the host against potentially harmful substances from the gut by reacting to and neutralizing many potential toxins and/or immune-active compounds, thereby maintaining the homeostasis of the immune system (Ponziani et al., 2018). The portal vein is the direct venous outflow from the intestine. In situations where the intestinal barrier is damaged followed by increased permeability, the liver becomes increasingly susceptible to numerous toxic factors originating from the intestine and gut microbiota. Some of these toxic components are termed as pathogen-associated molecular patterns (PAMP) and damage-associated molecular patterns (DAMP); both can result in hepatic injury. PAMP can directly attack hepatocytes or cells of the hepatic innate immune system e.g. Kupffer cells or stellate cells. The activated hepatic immune system initiates pro-inflammatory pathways and can also influence anti-viral and anti-apoptotic pathways in hepatocytes. These effects can be both detrimental (activation of the immune response, release of pro-inflammatory cytokines) and beneficial (cytoprotection and regeneration of hepatocytes). Briefly, the bacterial products after reaching the liver stimulate pattern recognition receptors (PRR) such as membrane-bound Toll-like receptors (TLR) and cytoplasmic nucleotide-binding oligomerization domain-like receptors (NLR). Toll-like receptors are usually expressed on sentinel cell-like macrophages and dendritic cells but TLR have also been identified in response to LPS on hepatic non-immune cell e.g. stellate cells and endothelial cells (Chesta et al., 1993, Crispe, 2009). TLR4 starts the innate immune response in recognition of the presence of LPS via co-receptors CD14 or myeloid differential protein-2 (MD-2) (Guo and Friedman, 2010, Takeuchi and Akira, 2010). Downstream TLR4 signaling may be myeloid differentiation factor 88 (MyD88)-dependent or -independent (Mandrekar and Szabo, 2009). The MyD88 dependent pathway initiates the nuclear translocation of nuclear factor kappa-B (NF-κB), a heterodimeric transcription factor expressed in all cell types. This, in turn, initiates the release of proinflammatory cytokines tumor necrosis factor alpha (TNFα), interleukin 6 (IL-6), IL-1β (Akira et al., 2006). The MyD88-independent pathway induces the phosphorylation of IL regulatory factor 3 which induce the type-1 interferon (Seki and Schnabl, 2012). On the other hand, activation of NLR results in the recruitment and activation of more inflammatory cells by activation of IL-1β and IL-18 (Martinon et al., 2002, Agostini et al., 2004). An already existing intestinal dysbiosis would further aggravate this ecosystem (Carotti et al., 2015).
The gut microbiota also modulates bile acid (BA) pool size and composition, modifying its signaling properties and subsequent action on BA receptors (Ridlon et al., 2014, Ridlon and Bajaj, 2015, Arab et al., 2017). In addition to their role in lipid absorption, BA regulate the intestinal microbiome (van Best et al., 2015) through farnesoid-X receptors (FXR)-induced production of antimicrobial peptides e.g. angiogenin 1 and RNase family member 4 (Inagaki et al., 2006, Parséus et al., 2017). FXR activation has been linked with inhibition of inflammation and improved gut integrity by reducing the bacterial translocation (Gadaleta et al., 2011). This phenomenon keeps a check on bacterial overgrowth and preserves epithelial cell integrity (Cariou and Staels, 2006). In animal models, using germ-free and FXR-deficient mice, it has been noted that the gut microbiota influences BA profile and FXR signaling (Arab et al., 2017, Staley et al., 2017). It is important to mention that the liver not only receives microbial input but conversely influences the intestinal microbes via bile acid and IgA antibody production and secretion into the intestine via the bile duct, thus playing an important regulatory role in the control of microbial populations (Brandl et al., 2017).
4. Microbial modulation through fiber
Many strategies can be employed to maintain eubiosis in the gut, among these, supplementation of fiber sources in the form of prebiotics has received much attention after phasing out of AGP in poultry feed. There are many possible explanations about the positive influence of prebiotics on the performance of the birds. However, it is generally believed that prebiotics are utilized by beneficial bacteria e.g. Lactobacillus, Bifidobacterium, thus promoting their growth and presence in the gut. These bacteria protect the host by strictly controlling the propagation of pathogenic bacteria (Pourabedin and Zhao, 2015, Yadav et al., 2016).
The variation in the chemical structure of the fiber is bound to affect its utilization by resident bacteria as specific enzymes are required for specific fiber types. This implies that the gut microbiota has a wide-ranging ability to break down and utilize these complex molecules. This ability is evident from the analysis of the gene content of the bacteria that encode carbohydrate-active enzyme (CAZymes) for cleaving linkage type and associated protein i.e. carbohydrate-linkage protein and transporters (Hamaker and Tuncil, 2014). It is noteworthy that bacteria degrade carbohydrate-based substrates in a highly competitive milieu, their ability to compete depends upon the presence of specific and relevant CAZyme encoding genes. Successful bacteria compete based on their ability to produce a specific set of enzymes that have a higher binding efficiency and rate of reaction towards the fiber substrate, coupled with the ability to capture the released product(s). This is often backed by the potential to colonize around fiber particles with greater tenacity than potential competitors (Hamaker and Tuncil, 2014).
Interestingly, the site and rate of fermentation of different carbohydrates also vary with their respective degree of polymerization (DP), the higher DP tends to be fermented in the distal part of the intestine (Henningsson et al., 2002). It is also evident that the DP and solubility of oligosaccharides are two important factors for the production of SCFA. Generally, low DP oligosaccharides favor butyric acid production whereas high DP oligosaccharides have the propensity to produce propionic acid (Nilsson and Nyman, 2005).
In general, dietary fiber alters the niche environment in the gut by providing substrates and binding platforms for microbial growth, allowing microbial species that are able to utilize these substrates to thrive (Deehan et al., 2017). Together, the gut microbiome harbors 130 glycoside hydrolase, 22 polysaccharide lyase, and 16 carbohydrate esterase families, which provide the microbiome with the flexibility to switch between different energy sources of fiber depending upon availability (Flint et al., 2012). The impact of dietary fiber on microbiota composition displays several consistent characteristics (Makki et al., 2018). Firstly, the observed shifts induced by non-digestible carbohydrates irrespective of whether they are categorized as prebiotics or not, are restricted to a limited number of taxa. Secondly, the magnitude of the changes can be substantial, with specific species constituting more than 30% of the total sequences obtained by amplicon sequencing of the fecal microbiota. Thirdly, the response of the microbiota to dietary fiber is highly individualized (Davis et al., 2011, Walker et al., 2011). The individual animal may lack some “keystone” species or strains that possess a specific set of enzymes to degrade a specific substrate (Zhao et al., 2018).
In return for these benefits to the host, microbes do tax the host in terms of creating a demand for energy and protein. They can also catabolize bile acids and thus reduce fat digestibility (Gaskins et al., 2002). Clearly, these processes increase the demand for energy and other essential nutrients from the host and any deficiency can lead to a reduction in performance.
Now-a-days, next generation sequencing has contributed immensely towards the characterization of microbial communities. The respective studies employ the amplification of small subunit of 16S ribosomal gene of bacteria and archea, 18S rRNA gene of eukaryotic species and nuclear ribosomal internal transcribed spacer region of fungi (Meyer et al., 2010). Thus, it helps in deep characterization of microbial communities along with quantification of relative abundance of different organisms. Interestingly, bacterial 16S rRNA tool has been in use in different field of studies, however, the first study characterizing the chicken microbiota was published in 2011 (Danzeisen et al., 2011). The 16S rRNA gene comprise 9 hypervariable regions but V1–V3, V3–V4, V4–V5, V1, V3 and V4 have been covered in characterizing chicken gut microbiota. Further, there are various sequencing technologies (Roche 454 pyrosequencing, Illumina MiSeq, HiSeq and Ion PGS system) with their respective pros and cons, however, can be effectively used for characterization of microbiota. Then bioinformatic processing is carried out by open sources platform such as QIIME and Mothur that in turn access public data base e.g. GreenGenes, the ribosomal database project and SILVA (DeSantis et al., 2006, Caporaso et al., 2010) to perform taxonomic assignment. Algorithms such as PICRUSt and Tax4Fun can be used to predict the metabolic functions based on the taxonomic identities from 16S rRNA gene sequencing (Langille et al., 2013, Aßhauer et al., 2015). To catalog the gene functions or analysis of individual genome, metagenomic or metatransciptomic approaches in which gene or transcripts respectively are directly sequenced with no PCR are useful in getting information on community structure, diversity and metabolic functions or gene expressions (Zinicola et al., 2015). Bioinformatic analysis of such dataset are more complex than 16S amplicon data and involve a sequence assembler such as Velvet (CLC workbench, Newbler version 3.0, Biospace) or MG-RAST. Basic Local Alignment Search Tool (BLAST) is used to assign bacterial taxa and functional groups, and gene functions may be analyzed by using Kyoto Encyclopedia of Genes and Genome (KEGG) or Cluster of Orthologous Genes (COG). These functional prediction processes produce interesting information but extreme care shall be exercised in drawing strong conclusions especially in avian species since their deviating organism may exhibit different functions and association between microorganisms and host (Borda-Molina et al., 2018).
It has been estimated that more than 900 species are harbored by chick GIT that are involved in many important body functions ranging from digestion of feed, breakdown of toxins and stimulation of immune system (Apajalahti et al., 2004). The upper GIT of chickens especially crop is inhabited by Lactobacillus sp. and Bifidobacterium sp. along with some members of Enterobacteriaceae family (Witzig et al., 2015). It is suggested that Lactobacilli is present in high abundance in proventriculus and gizzard as well. Ileum is the main site of nutrient absorption and is characterized by high abundance of Lactobacillus sp. and less abundance of butyrate producing bacteria e.g. Clostridium, Streptococcus, and Enterococcus. The cecum is the section of GIT where maximum fermentation takes place and feed resident time is also high compared with other segments of the GIT i.e. 12 to 20 h vs. 2.5 h for the rest of the GIT (Singh et al., 2012). Clostridiaceae, Bacteroidaceae, Lactobacillaceae and butyrate producers like Lachnospiraceae are the most abundant families present in cecum. In addition, many bacteria which are still to be characterized have also been found in cecum. Importantly, ceca demonstrate extreme diversity with 50 genera detected at the time of hatch to 200 at d 42. For more detailed information about use of high throughput sequencing application in chicken studies, readers are referred to an excellent review paper (Borda-Molina et al., 2018).
5. Dietary fiber for improved gut health in chickens
Gut health is relatively a broad term involving physiological, microbiological and physical functions that works in consonance to maintain intestinal homeostasis (Kogut, 2019). Nutrient digestion and absorption (Kiarie et al., 2013), barrier function, effective immune system (Wigley, 2013) and a neuroendocrine organ of the body (Cani and Knauf, 2016) are some of the most important attributes of the gut. The gut microbiota, through evolution with the host, exert considerable influence on each component that aides in maintaining intestinal homeostasis. Keeping in view these important functions of gut microbiota, it is quite evident that favorable microbiota can exert beneficial effects on the host including better growth performance and immune status, whereas hostile microbiota can lead to enteric disease that can not only reduce the growth performance but also increase the mortality of the flock (Kogut, 2019).
Although many non-AGP products can be employed to achieve improved gut health in chickens, the diet is the chief regulator of the chicken gut microbiome and should be considered much more seriously. As discussed earlier, dietary fiber has the potential to shift the microbial population in the gut, and thus some fiber sources could be a suitable option, e.g. wheat bran . It is a readily available by-product of flour milling in most countries and serves as a rich source of (in)soluble fiber (Vermeulen et al., 2018), and only a small proportion is soluble (Maes and Delcour, 2001, Maes and Delcour, 2002). The resident bacteria attach themselves to the insoluble polysaccharide segments forming a colony around fiber particles. Several studies suggest that structural dissimilarities exist between fiber-attached bacterial community and free-living bacteria. The former possesses a greater enzymatic capacity. Heavy colonization of fiber particles by bacteria can be considered as a hub of microbial processes and leads to leaching out of solute from the aggregate (Simon et al., 2002). These bacterial communities are dependent on the type of substrate. Though no such evidence exists in poultry, ruminal bacteria form a kind of biofilm around the fiber particles and exhibit higher fibrolytic activity than the free-living bacteria (Michalet-Doreau et al., 2001, Shinkai and Kobayashi, 2007). In a recent publication, it was highlighted that the inclusion of wheat bran in chicken diets could lead to more elaborate cross-feeding among bacteria strengthening the beneficial microbial population (Vermeulen et al., 2018).
Fiber particle size and structure are 2 important factors that should be considered before the inclusion of any fiber source in the chicken diet. Smaller particle size in the case of wheat bran has been linked with rapid fermentation and SCFA production (Vermeulen et al., 2017). It is noteworthy that fiber particle reduction does not alter the chemical composition of the fiber but may enhance the water-extractable GIT fraction (Jacobs et al., 2016). It may also result in the particle being small enough to pass through the sieves at the entrance of the caeca, and thus they can be more effectively fermented. These observations were ratified by Vermeulen et al. (2018) who demonstrated that wheat bran successfully increased the Lactobacillus and Bifidobacterium, genera which are known to produce lactate. At the same time, Lachnospiracaea were considerably enriched on reduced particle-sized wheat bran. Importantly, many members of Lachnospiracaea family are known users of lactate, producing butyrate in the process (Belenguer et al., 2006, De Maesschalck et al., 2015). The fermentation products of fiber can generate prebiotic effects as was suggested by Ribeiro et al. (2018). They noted that the supplementation of exogenous xylanase in wheat and corn-based diets decreased the viscosity of the digesta by degrading the water-soluble arabinoxylan and generated arabinoxylo-oligosaccharides (AXOS) and xylo-oligosaccharides (XOS). The XOS and AXOS increased the number of Solirubrobacter and Bifidobacterium genera. Such a shift in gut microbiota is accompanied by greater production of lactate (Kabel et al., 2002).
6. Future perspective
It is encouraging to note that a lot of work is being carried out to understand the dynamics of the microbial ecosystem of the gut and how it is linked with gut health. In this regard, many feed additives are being tested for their potential to modulate the intestinal microbiome. However, the inherent dietary macro-ingredients of the diet are the chief regulator of the gut ecosystem and require much greater attention. The bulk of the work has been and is still being done on the role of fiber in human gut microbial modulation nutrition, this trend is relatively new in animal nutrition and has accelerated only as a result of the decision by many countries to phase out AGP in response to consumers' concern. The work in poultry has mainly highlighted the effect of fiber on gut microbial shift and its functional outcome i.e. quantification of SCFA and other metabolic ramifications. More efforts are needed to understand fiber structure in general and how it and bacteria interact in a highly competitive environment. Thus, a holistic framework including physical form, type, and amount of fiber duly observing its concomitant influence on the liver, brain and gut health is required. Such work can lead us to unravel the consistent effects of fiber in bringing a desirable shift in the intestinal microbiome for improved gut health in chickens.
Conflict of interest
We declare that we have no financial and personal relationships with other people or organizations that can inappropriately influence our work, there is no professional or other personal interest of any nature or kind in any product, service and/or company that could be construed as influencing the content of this paper.
Acknowledgements
The authors wish to extend their gratitude to Dr. Mike R. Bedford (AB Vista Feed ingredients Ltd. Marlborough, UK) for his invaluable suggestions in preparing this manuscript.
Footnotes
Peer review under responsibility of Chinese Association of Animal Science and Veterinary Medicine.
References
- Agostini L., Martinon F., Burns K., McDermott M.F., Hawkins P.N., Tschopp J. NALP3 forms an IL-1β-processing inflammasome with increased activity in Muckle-Wells autoinflammatory disorder. Immunity. 2004;20:319–325. doi: 10.1016/s1074-7613(04)00046-9. [DOI] [PubMed] [Google Scholar]
- Akira S., Uematsu S., Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- Apajalahti J., Kettunen A., Graham H. Characteristics of the gastrointestinal microbial communities, with special reference to the chicken. World's Poult Sci J. 2004;60:223–232. [Google Scholar]
- Arab J.P., Karpen S.J., Dawson P.A., Arrese M., Trauner M. Bile acids and nonalcoholic fatty liver disease: molecular insights and therapeutic perspectives. Hepatology. 2017;65:350–362. doi: 10.1002/hep.28709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aßhauer K.P., Wemheuer B., Daniel R., Meinicke P. Tax4Fun: predicting functional profiles from metagenomic 16S rRNA data. Bioinformatics. 2015;31:2882–2884. doi: 10.1093/bioinformatics/btv287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballou A.L., Ali R.A., Mendoza M.A., Ellis J., Hassan H.M., Croom W. Development of the chick microbiome: how early exposure influences future microbial diversity. Front Vet Sci. 2016;3:2. doi: 10.3389/fvets.2016.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belenguer A., Duncan S.H., Calder A.G., Holtrop G., Louis P., Lobley G.E. Two routes of metabolic cross-feeding between Bifidobacterium adolescentis and butyrate-producing anaerobes from the human gut. Appl Environ Microbiol. 2006;72:3593–3599. doi: 10.1128/AEM.72.5.3593-3599.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bercik P., Denou E., Collins J., Jackson W., Lu J., Jury J. The intestinal microbiota affect central levels of brain-derived neurotropic factor and behavior in mice. Gastroenterology. 2011;141:599–609. doi: 10.1053/j.gastro.2011.04.052. e3. [DOI] [PubMed] [Google Scholar]
- Biasato I., Ferrocino I., Grego E., Dabbou S., Gai F., Gasco L. Gut microbiota and mucin composition in female broiler chickens fed diets including yellow mealworm (Tenebrio molitor, L.) Animals. 2019;9:213. doi: 10.3390/ani9050213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borda-Molina D., Seifert J., Camarinha-Silva A. Current perspectives of the chicken gastrointestinal tract and its microbiome. Comput Struct Biotechnol J. 2018;16:131–139. doi: 10.1016/j.csbj.2018.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandl K., Kumar V., Eckmann L. Gut-liver axis at the frontier of host-microbial interactions. Am J Physiol-Gastrl. 2017;312:G413–G419. doi: 10.1152/ajpgi.00361.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caffall K.H., Mohnen D. The structure, function, and biosynthesis of plant cell wall pectic polysaccharides. Carbohydr Res. 2009;344:1879–1900. doi: 10.1016/j.carres.2009.05.021. [DOI] [PubMed] [Google Scholar]
- Calefi A.S., da Silva Fonseca J.G., Cohn D.W.H., Honda B.T.B., Costola-de-Souza C., Tsugiyama L.E. The gut-brain axis interactions during heat stress and avian necrotic enteritis. Poult Sci. 2016;95:1005–1014. doi: 10.3382/ps/pew021. [DOI] [PubMed] [Google Scholar]
- Canani R.B., Di Costanzo M., Leone L., Pedata M., Meli R., Calignano A. Potential beneficial effects of butyrate in intestinal and extraintestinal diseases. World J Gastroenterol. 2011;17:1519. doi: 10.3748/wjg.v17.i12.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cani P.D., Knauf C. How gut microbes talk to organs: the role of endocrine and nervous routes. Mol Metab. 2016;5:743–752. doi: 10.1016/j.molmet.2016.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso J.G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F.D., Costello E.K. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cariou B., Staels B. The expanding role of the bile acid receptor FXR in the small intestine. J Hepatol. 2006;44:1213–1215. doi: 10.1016/j.jhep.2006.03.006. [DOI] [PubMed] [Google Scholar]
- Carotti S., Guarino M.P.L., Vespasiani-Gentilucci U., Morini S. Starring role of toll-like receptor-4 activation in the gut-liver axis. World J Gastroenterol. 2015;6:99. doi: 10.4291/wjgp.v6.i4.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cebra J.J. Influences of microbiota on intestinal immune system development. Am J Clin Nutr. 1999;69 doi: 10.1093/ajcn/69.5.1046s. 1046s-51s. [DOI] [PubMed] [Google Scholar]
- Chesta J., Defilippi C., Defilippi C. Abnormalities in proximal small bowel motility in patients with cirrhosis. Hepatology. 1993;17:828–832. [PubMed] [Google Scholar]
- Clavijo V., Flórez M.J.V. The gastrointestinal microbiome and its association with the control of pathogens in broiler chicken production: a review. Poult Sci. 2017;97:1006–1021. doi: 10.3382/ps/pex359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrêa-Oliveira R., Fachi J.L., Vieira A., Sato F.T., Vinolo M.A.R. Regulation of immune cell function by short-chain fatty acids. Clin Transl Immunol. 2016;5:e73. doi: 10.1038/cti.2016.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crispe I.N. The liver as a lymphoid organ. Annu Rev Immunol. 2009;27:147–163. doi: 10.1146/annurev.immunol.021908.132629. [DOI] [PubMed] [Google Scholar]
- Danzeisen J.L., Kim H.B., Isaacson R.E., Tu Z.J., Johnson T.J. Modulations of the chicken cecal microbiome and metagenome in response to anticoccidial and growth promoter treatment. PLoS One. 2011;6 doi: 10.1371/journal.pone.0027949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis L.M., Martínez I., Walter J., Goin C., Hutkins R.W. Barcoded pyrosequencing reveals that consumption of galactooligosaccharides results in a highly specific bifidogenic response in humans. PLoS One. 2011;6 doi: 10.1371/journal.pone.0025200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Maesschalck C., Eeckhaut V., Maertens L., De Lange L., Marchal L., Nezer C. Effects of xylo-oligosaccharides on broiler chicken performance and microbiota. Appl Environ Microbiol. 2015;81:5880–5888. doi: 10.1128/AEM.01616-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deehan E.C., Duar R.M., Armet A.M., Perez-Munoz M.E., Jin M., Walter J. Modulation of the gastrointestinal microbiome with nondigestible fermentable carbohydrates to improve human health. Microbiol Spectr. 2017;5 doi: 10.1128/microbiolspec.BAD-0019-2017. [DOI] [PubMed] [Google Scholar]
- DeSantis T.Z., Hugenholtz P., Larsen N., Rojas M., Brodie E.L., Keller K. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dibner J., Richards J. Antibiotic growth promoters in agriculture: history and mode of action. Poult Sci. 2005;84:634–643. doi: 10.1093/ps/84.4.634. [DOI] [PubMed] [Google Scholar]
- Flint H.J., Scott K.P., Duncan S.H., Louis P., Forano E. Microbial degradation of complex carbohydrates in the gut. Gut Microb. 2012;3:289–306. doi: 10.4161/gmic.19897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forder R., Howarth G., Tivey D., Hughes R. Bacterial modulation of small intestinal goblet cells and mucin composition during early posthatch development of Poultry1. Poult Sci. 2007;86:2396–2403. doi: 10.3382/ps.2007-00222. [DOI] [PubMed] [Google Scholar]
- Gadaleta R.M., Van Erpecum K.J., Oldenburg B., Willemsen E.C., Renooij W., Murzilli S. Farnesoid X receptor activation inhibits inflammation and preserves the intestinal barrier in inflammatory bowel disease. Gut. 2011;60:463–472. doi: 10.1136/gut.2010.212159. [DOI] [PubMed] [Google Scholar]
- Gadde U., Kim W., Oh S., Lillehoj H.S. Alternatives to antibiotics for maximizing growth performance and feed efficiency in poultry: a review. Anim Health Res Rev. 2017;18:26–45. doi: 10.1017/S1466252316000207. [DOI] [PubMed] [Google Scholar]
- Gantois I., Ducatelle R., Pasmans F., Haesebrouck F., Gast R., Humphrey T.J. Mechanisms of egg contamination by Salmonella Enteritidis. FEMS Microbiol Rev. 2009;33:718–738. doi: 10.1111/j.1574-6976.2008.00161.x. [DOI] [PubMed] [Google Scholar]
- Gaskins H., Collier C., Anderson D. Antibiotics as growth promotants: mode of action. Anim Biotechnol. 2002;13:29–42. doi: 10.1081/ABIO-120005768. [DOI] [PubMed] [Google Scholar]
- González-Alvarado J., Jiménez-Moreno E., Lázaro R., Mateos G. Effect of type of cereal, heat processing of the cereal, and inclusion of fiber in the diet on productive performance and digestive traits of broilers. Poult Sci. 2007;86:1705–1715. doi: 10.1093/ps/86.8.1705. [DOI] [PubMed] [Google Scholar]
- Guo J., Friedman S.L. Toll-like receptor 4 signaling in liver injury and hepatic fibrogenesis. Fibrogenesis Tissue Repair. 2010;3:21. doi: 10.1186/1755-1536-3-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamaker B.R., Tuncil Y.E. A perspective on the complexity of dietary fiber structures and their potential effect on the gut microbiota. J Mol Biol. 2014;426:3838–3850. doi: 10.1016/j.jmb.2014.07.028. [DOI] [PubMed] [Google Scholar]
- Henningsson AsM., Björck I.M., Nyman E.M.G. Combinations of indigestible carbohydrates affect short-chain fatty acid formation in the hindgut of rats. J Nutr. 2002;132:3098–3104. doi: 10.1093/jn/131.10.3098. [DOI] [PubMed] [Google Scholar]
- Hetland H., Svihus B. Inclusion of dust bathing materials affects nutrient digestion and gut physiology of layers. J Appl Poult Res. 2007;16:22–26. [Google Scholar]
- Hetland H., Svihus B., Choct M. Role of insoluble fiber on gizzard activity in layers. J Appl Poult Res. 2005;14:38–46. [Google Scholar]
- Inagaki T., Moschetta A., Lee Y.-K., Peng L., Zhao G., Downes M. Regulation of antibacterial defense in the small intestine by the nuclear bile acid receptor. Proc Natl Acad Sci USA. 2006;103:3920–3925. doi: 10.1073/pnas.0509592103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izydorczyk M., Dexter J. Barley β-glucans and arabinoxylans: molecular structure, physicochemical properties, and uses in food products–a Review. Food Res Int. 2008;41:850–868. [Google Scholar]
- Jacobs P.J., Bogaerts S., Hemdane S., Delcour J.A., Courtin C.M. Impact of wheat bran hydration properties as affected by toasting and degree of milling on optimal dough development in bread making. J Agric Food Chem. 2016;64:3636–3644. doi: 10.1021/acs.jafc.5b05958. [DOI] [PubMed] [Google Scholar]
- Jarchum I., Pamer E.G. Regulation of innate and adaptive immunity by the commensal microbiota. Curr Opin Immunol. 2011;23:353–360. doi: 10.1016/j.coi.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jezierny D., Mosenthin R., Bauer E. The use of grain legumes as a protein source in pig nutrition: a review. Anim Feed Sci Technol. 2010;157:111–128. [Google Scholar]
- Jha R., Berrocoso J.F. Dietary fiber and protein fermentation in the intestine of swine and their interactive effects on gut health and on the environment: a review. Anim Feed Sci Technol. 2016;212:18–26. [Google Scholar]
- Jha R., Fouhse J.M., Tiwari U.P., Li L., Willing B.P. Dietary fiber and intestinal health of monogastric animals. Front Vet Sci. 2019;6 doi: 10.3389/fvets.2019.00048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez-Moreno E., González-Alvarado J., Lázaro R., Mateos G. Effects of type of cereal, heat processing of the cereal, and fiber inclusion in the diet on gizzard pH and nutrient utilization in broilers at different ages. Poult Sci. 2009;88:1925–1933. doi: 10.3382/ps.2009-00193. [DOI] [PubMed] [Google Scholar]
- Kabel M.A., Kortenoeven L., Schols H.A., Voragen A.G. In vitro fermentability of differently substituted xylo-oligosaccharides. J Agric Food Chem. 2002;50:6205–6210. doi: 10.1021/jf020220r. [DOI] [PubMed] [Google Scholar]
- Kiarie E., Romero L.F., Nyachoti C.M. The role of added feed enzymes in promoting gut health in swine and poultry. Nutr Res Rev. 2013;26:71–88. doi: 10.1017/S0954422413000048. [DOI] [PubMed] [Google Scholar]
- Knudsen K.E.B. Fiber and nonstarch polysaccharide content and variation in common crops used in broiler diets. Poult Sci. 2014;93:2380–2393. doi: 10.3382/ps.2014-03902. [DOI] [PubMed] [Google Scholar]
- Kogut M.H. The effect of microbiome modulation on the intestinal health of poultry. Anim Feed Sci Technol. 2019;250:32–40. [Google Scholar]
- Langille M.G., Zaneveld J., Caporaso J.G., McDonald D., Knights D., Reyes J.A. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol. 2013;31:814. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maes C., Delcour J. Alkaline hydrogen peroxide extraction of wheat bran non-starch polysaccharides. J Cereal Sci. 2001;34:29–35. [Google Scholar]
- Maes C., Delcour J. Structural characterisation of water-extractable and water-unextractable arabinoxylans in wheat bran. J Cereal Sci. 2002;35:315–326. [Google Scholar]
- Makki K., Deehan E.C., Walter J., Bäckhed F. The impact of dietary fiber on gut microbiota in host health and disease. Cell Hotspot Microb. 2018;23:705–715. doi: 10.1016/j.chom.2018.05.012. [DOI] [PubMed] [Google Scholar]
- Mandrekar P., Szabo G. Signalling pathways in alcohol-induced liver inflammation. J Hepatol. 2009;50:1258–1266. doi: 10.1016/j.jhep.2009.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinon F., Burns K., Tschopp J. The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-β. Mol Cell. 2002;10:417–426. doi: 10.1016/s1097-2765(02)00599-3. [DOI] [PubMed] [Google Scholar]
- Mateos G., Guzman P., Saldana B., Bonilla A.P., Lazar R., Jimenez-Moreno E. Proc. European Symposium of Poultry Nutrition Potsdam. 2013. Relevance of dietary fiber in poultry feeding. [Google Scholar]
- Mateos G., Jiménez-Moreno E., Serrano M., Lázaro R. Poultry response to high levels of dietary fiber sources varying in physical and chemical characteristics. J Appl Poult Res. 2012;21:156–174. [Google Scholar]
- Meyer A., Todt C., Mikkelsen N.T., Lieb B. Fast evolving 18S rRNA sequences from Solenogastres (Mollusca) resist standard PCR amplification and give new insights into mollusk substitution rate heterogeneity. BMC Evol Biol. 2010;10:70. doi: 10.1186/1471-2148-10-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalet-Doreau B., Fernandez I., Peyron C., Millet L., Fonty G. Fibrolytic activities and cellulolytic bacterial community structure in the solid and liquid phases of rumen contents. Reprod Nutr Dev. 2001;41:187–194. doi: 10.1051/rnd:2001122. [DOI] [PubMed] [Google Scholar]
- Mitchell M., Moretó M. Absorptive function of the small intestine: adaptations meeting demand. Avian Gut Funct Health Dis. 2006:43–63. [Google Scholar]
- Neish A.S. Microbes in gastrointestinal health and disease. Gastroenterology. 2009;136:65–80. doi: 10.1053/j.gastro.2008.10.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson U., Nyman M. Short-chain fatty acid formation in the hindgut of rats fed oligosaccharides varying in monomeric composition, degree of polymerisation and solubility. Br J Nutr. 2005;94:705–713. doi: 10.1079/bjn20051531. [DOI] [PubMed] [Google Scholar]
- O’hara S.P., Karlsen T.H., LaRusso N.F. Cholangiocytes and the environment in primary sclerosing cholangitis: where is the link? Gut. 2017;66:1873–1877. doi: 10.1136/gutjnl-2017-314249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakley B.B., Kogut M.H. Spatial and temporal changes in the broiler chicken cecal and fecal microbiomes and correlations of bacterial taxa with cytokine gene expression. Front Vet Sci. 2016;3:11. doi: 10.3389/fvets.2016.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paiva D., McElroy A. Necrotic enteritis: applications for the poultry industry. J Appl Poult Res. 2014;23(3):557–566. [Google Scholar]
- Pan D., Yu Z. Intestinal microbiome of poultry and its interaction with host and diet. Gut Microb. 2014;5:108–119. doi: 10.4161/gmic.26945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parséus A., Sommer N., Sommer F., Caesar R., Molinaro A., Ståhlman M. Microbiota-induced obesity requires farnesoid X receptor. Gut. 2017;66:429–437. doi: 10.1136/gutjnl-2015-310283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedroso A., Menten J.F.M., Lambais M.R. The structure of bacterial community in the intestines of newly hatched chicks. J Appl Poult Res. 2005;14:232–237. [Google Scholar]
- Ponziani F.R., Zocco M.A., Cerrito L., Gasbarrini A., Pompili M. Bacterial translocation in patients with liver cirrhosis: physiology, clinical consequences, and practical implications. Expert Rev Gastroenterol Hepatol. 2018;12:641–656. doi: 10.1080/17474124.2018.1481747. [DOI] [PubMed] [Google Scholar]
- Pourabedin M., Zhao X. Prebiotics and gut microbiota in chickens. FEMS Microbiol Lett. 2015;362:fnv122. doi: 10.1093/femsle/fnv122. [DOI] [PubMed] [Google Scholar]
- Pustjens A.M., Schols H.A., Kabel M.A., Gruppen H. Characterisation of cell wall polysaccharides from rapeseed (Brassica napus) meal. Carbohydr Polym. 2013;98:1650–1656. doi: 10.1016/j.carbpol.2013.07.059. [DOI] [PubMed] [Google Scholar]
- Ribeiro T., Cardoso V., Ferreira L., Lordelo M., Coelho E., Moreira A. Xylo-oligosaccharides display a prebiotic activity when used to supplement wheat or corn-based diets for broilers. Poult Sci. 2018;97:4330–4341. doi: 10.3382/ps/pey336. [DOI] [PubMed] [Google Scholar]
- Ridlon J.M., Bajaj J.S. The human gut sterolbiome: bile acid-microbiome endocrine aspects and therapeutics. Acta Pharm Sin B. 2015;5:99–105. doi: 10.1016/j.apsb.2015.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridlon J.M., Kang D.J., Hylemon P.B., Bajaj J.S. Bile acids and the gut microbiome. Curr Opin Gastroenterol. 2014;30:332. doi: 10.1097/MOG.0000000000000057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roto S.M., Kwon Y.M., Ricke S.C. Applications of in ovo technique for the optimal development of the gastrointestinal tract and the potential influence on the establishment of its microbiome in poultry. Front Vet Sci. 2016;3:63. doi: 10.3389/fvets.2016.00063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saulnier L., Sado P.-E., Branlard G., Charmet G., Guillon F. Wheat arabinoxylans: exploiting variation in amount and composition to develop enhanced varieties. J Cereal Sci. 2007;46:261–281. [Google Scholar]
- Schroeder B.O. Fight them or feed them: how the intestinal mucus layer manages the gut microbiota. Gastroenterol Rep. 2019;7:3–12. doi: 10.1093/gastro/goy052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki E., Schnabl B. Role of innate immunity and the microbiota in liver fibrosis: crosstalk between the liver and gut. J Physiol. 2012;590:447–458. doi: 10.1113/jphysiol.2011.219691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senkoylu N., Samli H.E., Akyurek H., Okur A.A., Kanter M. Effects of whole wheat with or without xylanase supplementation on performance of layers and digestive organ development. Ital J Anim Sci. 2009;8:155–163. [Google Scholar]
- Shakouri M., Iji P., Mikkelsen L., Cowieson A. Intestinal function and gut microflora of broiler chickens as influenced by cereal grains and microbial enzyme supplementation. J Anim Physiol Anim Nutr. 2009;93:647–658. doi: 10.1111/j.1439-0396.2008.00852.x. [DOI] [PubMed] [Google Scholar]
- Shang Y., Kumar S., Oakley B., Kim W.K. Chicken gut microbiota: importance and detection technology. Front Vet Sci. 2018;5:254. doi: 10.3389/fvets.2018.00254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinkai T., Kobayashi Y. Localization of ruminal cellulolytic bacteria on plant fibrous materials as determined by fluorescence in situ hybridization and real-time PCR. Appl Environ Microbiol. 2007;73:1646–1652. doi: 10.1128/AEM.01896-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon M., Grossart H.-P., Schweitzer B., Ploug H. Microbial ecology of organic aggregates in aquatic ecosystems. Aquat Microb Ecol. 2002;28:175–211. [Google Scholar]
- Singh K., Shah T., Deshpande S., Jakhesara S., Koringa P., Rank D. High through put 16S rRNA gene-based pyrosequencing analysis of the fecal microbiota of high FCR and low FCR broiler growers. Mol Biol Rep. 2012;39:10595–10602. doi: 10.1007/s11033-012-1947-7. [DOI] [PubMed] [Google Scholar]
- Singh K.M., Shah T.M., Reddy B., Deshpande S., Rank D., Joshi C. Taxonomic and gene-centric metagenomics of the fecal microbiome of low and high feed conversion ratio (FCR) broilers. J Appl Genet. 2014;55:145–154. doi: 10.1007/s13353-013-0179-4. [DOI] [PubMed] [Google Scholar]
- Sleeth M.L., Thompson E.L., Ford H.E., Zac-Varghese S.E., Frost G. Free fatty acid receptor 2 and nutrient sensing: a proposed role for fibre, fermentable carbohydrates and short-chain fatty acids in appetite regulation. Nutr Res Rev. 2010;23:135–145. doi: 10.1017/S0954422410000089. [DOI] [PubMed] [Google Scholar]
- Staley C., Weingarden A.R., Khoruts A., Sadowsky M.J. Interaction of gut microbiota with bile acid metabolism and its influence on disease states. Appl Microbiol Biotechnol. 2017;101:47–64. doi: 10.1007/s00253-016-8006-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K., Nakajima A. New aspects of IgA synthesis in the gut. Int Immunol. 2014;26:489–494. doi: 10.1093/intimm/dxu059. [DOI] [PubMed] [Google Scholar]
- Svihus B. The gizzard: function, influence of diet structure and effects on nutrient availability. World's Poult Sci J. 2011;67:207–224. [Google Scholar]
- Takeuchi O., Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140:805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- Tazoe H., Otomo Y., Kaji I., Tanaka R., Karaki S., Kuwahara A. Roles of short-chain fatty acids receptors, GPR41 and GPR43 on colonic functions. J Physiol Pharmacol. 2008;59:251–262. [PubMed] [Google Scholar]
- van Best N., Jansen P.L., Rensen S.S. The gut microbiota of nonalcoholic fatty liver disease: current methods and their interpretation. Hepatol Int. 2015;9:406–415. doi: 10.1007/s12072-015-9640-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeulen K., Verspreet J., Courtin C., Haesebrouck F., Ducatelle R., Van Immerseel F. Reduced particle size wheat bran is butyrogenic and lowers Salmonella colonization, when added to poultry feed. Vet Mircobiol. 2017;198:64–71. doi: 10.1016/j.vetmic.2016.12.009. [DOI] [PubMed] [Google Scholar]
- Vermeulen K., Verspreet J., Courtin C.M., Haesebrouck F., Baeyen S., Haegeman A. Reduced-particle-size wheat bran is efficiently colonized by a lactic acid-producing community and reduces levels of Enterobacteriaceae in the cecal microbiota of broilers. Appl Environ Microbiol. 2018;84 doi: 10.1128/AEM.01343-18. e01343-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker A.W., Ince J., Duncan S.H., Webster L.M., Holtrop G., Ze X. Dominant and diet-responsive groups of bacteria within the human colonic microbiota. ISME J. 2011;5:220. doi: 10.1038/ismej.2010.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei S., Morrison M., Yu Z. Bacterial census of poultry intestinal microbiome. Poult Sci. 2013;92:671–683. doi: 10.3382/ps.2012-02822. [DOI] [PubMed] [Google Scholar]
- Wigley P. Immunity to bacterial infection in the chicken. Dev Comp Immunol. 2013;41:413–417. doi: 10.1016/j.dci.2013.04.008. [DOI] [PubMed] [Google Scholar]
- Williams S.C. Gnotobiotics. Proc Natl Acad Sci USA. 2014;111:1661. doi: 10.1073/pnas.1324049111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witzig M., da Silva A.C., Green-Engert R., Hoelzle K., Zeller E., Seifert J. Spatial variation of the gut microbiota in broiler chickens as affected by dietary available phosphorus and assessed by T-RFLP analysis and 454 pyrosequencing. PLoS One. 2015;10 doi: 10.1371/journal.pone.0143442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav A.S., Kolluri G., Gopi M., Karthik K., Singh Y. Exploring alternatives to antibiotics as health promoting agents in poultry-A review. J Exp Biol. 2016;4:3S. [Google Scholar]
- Yadav S., Jha R. Strategies to modulate the intestinal microbiota and their effects on nutrient utilization, performance, and health of poultry. J Anim Sci Biotechnol. 2019;10:2. doi: 10.1186/s40104-018-0310-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yegani M., Korver D. Factors affecting intestinal health in poultry. Poult Sci. 2008;87:2052–2063. doi: 10.3382/ps.2008-00091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L., Zhang F., Ding X., Wu G., Lam Y.Y., Wang X. Gut bacteria selectively promoted by dietary fibers alleviate type 2 diabetes. Science. 2018;359:1151–1156. doi: 10.1126/science.aao5774. [DOI] [PubMed] [Google Scholar]
- Zinicola M., Higgins H., Lima S., Machado V., Guard C., Bicalho R. Shotgun metagenomic sequencing reveals functional genes and microbiome associated with bovine digital dermatitis. PLoS One. 2015;10 doi: 10.1371/journal.pone.0133674. [DOI] [PMC free article] [PubMed] [Google Scholar]