Abstract
This study investigated the effects of the Streptococcus agalactiae antagonizing probiotics Bacillus cereus NY5 and Bacillus subtilis as feed additives for Nile tilapia in terms of growth performance, intestinal health and resistance to S. agalactiae. A total of 720 apparently healthy juvenile Nile tilapia (0.20 ± 0.05 g) were randomly divided into 4 equal groups with 3 replicates for each group. Fish were fed a basal diet (control check group, CK group) supplemented with B. subtilis (1 × 108 CFU/g feed, BS group), B. cereus NY5 (1 × 108 CFU/g feed, BC group), and B. subtilis + B. cereus NY5 (0.5 × 108 CFU/g feed of each probiotic, BS + BC group) for 6 wk, and the probiotic supplementation groups were then fed the basal diet for 1 wk to investigate the gut microbial community. The results of this study showed that BS + BC and BC treatments significantly increased weight gain (WG), feed conversion ratio (FCR) and S. agalactiae resistance in Nile tilapia (P < 0.05). Gut microvilli length and density and c-type lysozyme (lyzc) gene expression were significantly increased by probiotic supplementation (P < 0.05). The results of high-throughput sequencing showed that the B. cereus NY5 and B. subtilis + B. cereus NY5-supplemented feed resulted in a significant improvement in tilapia autochthonous gut bacterial communities and had a stimulation effect on a variety of potential probiotics after 6 wk of feeding. After cessation of probiotic administration for 1 wk, the gut bacteria of the fish in the BS + BC and BC groups had minor changes and maintained a stable state. Consequently, it was inferred that, as a feed supplement, B. cereus NY5 and the mixture of B. subtilis and B. cereus NY5 at 1 × 108 CFU/g feed were able to promote growth and disease resistance, which may be associated with the supplement's effects on gut immune status, intestinal morphology, and intestinal microbial community composition.
Keywords: Tilapia, Bacillus subtilis, Bacillus cereus, Intestinal microbiota, Probiotic
1. Introduction
Tilapia is one of the most important species of farmed fish of freshwater aquaculture in China. In recent years, disease caused by Streptococcus agalactiae has become a major challenge for the culture of tilapia, resulting in massive losses for tilapia farmers all over the world (Amal and Saad, 2011). Probiotics (especially antagonistic probiotics) can reduce pathogenic bacteria by competitive exclusion, provide nutrients and enzymes to promote host growth, enhance the immune response by immune stimulation, and do not cause secondary pollution problems. In view of this, obtaining S. agalactiae antagonizing probiotics suitable for tilapia culture is of great practical significance for improving the resistance of tilapia and reducing the use of antibiotics.
Consumption of probiotics is an effective and attractive way to modulate the intestinal microbial composition and to maintain and promote host health (FAO/WHO, 2001). The major mechanisms of action of probiotics include enhancement of epithelial barrier function, improved adhesion to intestinal cells and pathogen inhibition by occupying adhesion sites, production of antibacterial substances, and regulation of the immune function (Rijkers et al., 2010). Through the above mechanisms, the purpose of regulating intestinal microbes and inhibiting the growth of pathogens is achieved (Almada et al., 2015). Recently, the application of beneficial bacteria in the form of probiotics has been demonstrated to be useful in aquaculture (Pérez-Sánchez et al., 2014). The main probiotic microorganisms used in aquaculture include species belonging to the lactic acid bacteria (LAB) (Beck et al., 2015, Liu et al., 2016a, Liu et al., 2017a) and Bacillus spp. (Chai et al., 2016, Giatsis et al., 2016, He et al., 2013, Sun et al., 2011).
According to the results of previous studies, due to the beneficial properties of enhancing the immune system, competitive exclusion, and producing antibacterial substances, some Bacillus spp. (including Bacillus subtilis) have been frequently used as probiotics in aquaculture (Aly et al., 2008a, Liu et al., 2010, Tseng et al., 2009). Bacillus subtilis probiotic candidates stimulated immune responses both locally and systemically in tilapia (Galagarza et al., 2018) and effectively enhanced the growth performance and disease resistance of Nile tilapia (Liu et al., 2017b). In addition, both B. subtilis and Bacillus cereus promote intestinal colonization and improve survival rate (SR) without negatively influencing feed intake, total biomass, gross revenue, partial operating costs and net revenue of tilapia (Nilton and Daniele, 2015). As a feed additive, the skin beneficial bacterial composition is promoted by the use of Bacillus spp., which can outcompete intestinal pathogenic bacteria (El-Rhman et al., 2009). Moreover, B. subtilis boosted the growth and vitality of beneficial LAB in the intestine of hosts (Hoa et al., 2000). Bacillus could prove to be effective for integrated prevention and control of streptococcosis infections (Widanarni and Tanbiyaskur, 2015).
In this study, B. cereus was extracted from healthy Nile tilapia feces (Liu et al., 2016a), and B. subtilis was isolated from a mixed-species Bacillus spp. probiotic that was used for the regulation of aquaculture water quality in our lab (Liu et al., 2015). Our previous study indicated that both strains were effective at antagonizing S. agalactiae in vitro (Liu et al., 2015, Lu et al., 2016), but it is not clear whether they have a probiotic effect in vivo. This study assessed the effects of the potential probiotic strains with special antagonistic activity in Nile tilapia. Comprehensive evaluation of the effects on growth, survival, immunity, disease resistance and the intestinal microbiota provides a solid theoretical basis for subsequent commercialization and application of the potential probiotic strains.
2. Materials and methods
All animal work in this paper was conducted according to relevant national and international guidelines. All animal care and experimental procedures were approved by the Committee on Animal Care and Use and the Committee on the Ethic of Animal Experiments of Chinese Academy of Fishery Sciences.
2.1. Bacteria and feed preparation
The probiotic B. cereus NY5 and B. subtilis were identified based on morphological, physiological, and biochemical characteristics, as well as 16S rRNA gene sequencing. The bacteria were frozen in 50% glycerol and stored at −80 °C. The antagonistic experiments showed significant inhibition zones to S. agalactiae in vitro. Luria Broth (LB) liquid medium (OXOID), which had been centrifuged at 5,000 × g (Beckman Coulter, AK, USA) for 4 min, was used to culture Bacillus strains for 24 h at a temperature of 37 °C. Distilled water was used to wash the pellets twice, and these pellets were then lyophilized and suspended in phosphate-buffered saline (PBS) (1.8 mmol/L KH2PO4, 10.1 mmol/L NaH2PO4, 2.7 mmol/L KCl and 137 mmol/L NaCl, pH 7.4). A spread plate technique was used to determine viable cells according to the cell concentrations measured at OD600, which was linearly proportional to the number of viable cells in the suspension. All cell suspension OD600 values were adjusted to an adequate value (CFU/mL) for further experiments.
The formulation and the main ingredients of the basal diet are shown in Table 1. The experimental tilapia were given the probiotics, which were added in the basal diet at a dosage of 1 × 108 CFU/g, as described in a previously conducted experiment using Bacillus spp. in tilapia culture (Aly et al., 2008a, Wang et al., 2017). The tilapia fed a pure basal diet were considered the experimental control check (CK) group. There were 3 kinds of diets prepared for the experiment: the basal diet with 1 × 108 CFU/g of B. subtilis (BS group), the basal diet with 1 × 108 CFU/g of B. cereus NY5 (BC group), or the basal diet with both 0.5 × 108 CFU/g of B. subtilis and B. cereus NY5 (BS + BC group). The preparation of these experimental diets followed the same process described in our previous study (Xia et al., 2018). Briefly, powdered dietary ingredients were thoroughly mixed by hand, then blended with oil and water, and suitable Bacillus spp. cells were added to form a soft dough. To ensure the viability of the added probiotics, the diets were freshly prepared every day and kept sealed in plastic bags that were stored at 4 °C.
Table 1.
Formulation and calculated chemical compositions of the basal diet.
| Item | Content, % |
|---|---|
| Ingredients | |
| Fish meal | 48 |
| Soybean meal | 22 |
| Wheat flour | 25 |
| Adhesives | 0.2 |
| Soybean oil | 2.0 |
| Ca(H2PO4)2 | 2.0 |
| Vitamin C phosphate ester | 0.1 |
| Choline chloride (50%) | 0.3 |
| Vitamin premix1 | 0.2 |
| Mineral premix2 | 0.2 |
| Calculated chemical compositions | |
| Crude protein | 42.0 |
| Crude lipid | 7.3 |
| Ash | 9.5 |
| Crude fibre | 3.1 |
| N free extract | 27.9 |
One kilogram of vitamin premix contained the following: thiamine, 0.438 g; riboflavin, 0.632 g; pyridoxine·HCl, 0.908 g; D-pantothenic acid, 1.724 g; nicotinic acid, 4.583 g; biotin, 0.211 g; folic acid, 0.549 g; vitamin B12, 0.001 g; inositol, 21.053 g; menadione sodium bisulfite, 0.889 g; retinyl acetate, 0.677 g; cholecalciferol, 0.116 g; DL-α-tocopherol-acetate, 12.632 g.
One kilogram of mineral premix contained the following: CoCl2·6H2O, 0.074 g; CuSO4·5H2O, 2.5 g; FeSO4·7H2O, 73.2 g; NaCl, 40.0 g; MgSO4·7H2O, 284.0 g; MnSO4·H2O, 6.50 g; KI, 0.68 g; Na2SeO3, 0.10 g; ZnSO4·7H2O, 131.93 g; cellulose, 501.09 g.
2.2. Fish and rearing conditions
All experimental juvenile tilapia were provided by the Gaoyao Fish Farm of the Pearl River Fisheries Research Institute (Guangzhou, China), and ethyl 3-aminobenzoate methanesulfonate (MS-222) was used to anaesthetize the fish when necessary. Healthy juvenile tilapia were selected and acclimated in 750-L tanks for 2 wk from 28 to 29 °C under laboratory conditions and were fed the basal diet. Thereafter, fish that were eating normally, disease-free, non-injured and smaller-sized (0.20 ± 0.05 g) were randomly distributed into twelve 50-L tanks on a random basis with 60 tilapia in each tank and 3 replicates for each treatment. The adopted feeding cycle was determined on the basis of previous reports (Wang et al., 2017, Xia et al., 2018). Every day for the next 6 wk, the tilapia were fed twice (at 09:00 and 16:00) until they were apparently satiated. Then, the basal diet was used to feed all of the tilapia for another week. The conditions of culture were identical to the description of Xia et al. (2018), and a 50% water exchange was carried out daily.
2.3. Growth performance and sampling
After the 6-wk rearing period, tilapia were processed after 24 h of starvation and exposure to an excessive dosage 200 mg/L of MS-222 and were then counted and weighed. Samples were collected and processed as described in our previous study (Xia et al., 2018). The midguts (approximately 1 cm) of 6 fish or 3 fish in each group were used for intestinal mucosal morphology investigation and reverse transcription-polymerase chain reaction (RT-PCR). The midguts of 3 fish in each group were randomly selected for gut bacteria detection by high-throughput sequencing. The calculations of various indicators, including the feed conversion ratio (FCR), weight gain (WG) and the SR, were conducted as previously described (Ran et al., 2015).
2.4. Genomic DNA isolation
DNA was extracted from fish midgut samples using the DNeasy Blood & Tissue Kit (Qiagen, Venlo, The Netherlands) according to the manufacturer's protocol, with some modifications (Giatsis et al., 2016).
2.5. Illumina high-throughput sequencing of barcoded 16S rRNA genes
Illumina MiSeq library preparation and next-generation sequencing were conducted at GENEWIZ, Inc. (Suzhou, China). A Qubit 2.0 Fluorometer (Invitrogen, Carlsbad, CA, USA) was used to quantify all of the DNA samples. Then, a MetaV Library Preparation kit (GENEWIZ, Inc., South Plainfield, NJ, USA) was used to generate amplicons with 30 to 50 ng of DNA. The selection of prokaryotic 16S rDNA's V3 and V4 hypervariable regions was performed to generate amplicons and conduct the ensuing taxonomic analysis (Lu et al., 2016). Subsequent processing and analysis methods were based on our published protocols (Xia et al., 2018).
2.6. Changes in gut microbiota after reverting to the basal diet for 1 wk
Sampling of the tilapia mid-intestine was conducted at each treatment (which contained 3 tilapia) 7 d after the fish were cut off the probiotic supply to assess the persistence of the intestinal probiotics via high-throughput sequencing analyses.
2.7. Intestinal histology
After 6 wk, transmission electron microscope (TEM) imaging and scanning electron microscopy (SEM) imaging were conducted on the mid-intestines of the 6 tilapia in each treatment group following the same operation as our recent study (Xia et al., 2018).
2.8. Intestinal c-type lysozyme gene expression
After 6 wk, tilapia intestinal tissues were harvested for determination of c-type lysozyme (lyzc) expression. Protocols for total RNA extraction of the mid-intestine, RT-PCR and quantitative polymerase chain reaction (qPCR) were consistent with our previous study (Xia et al., 2018). The target and reference genes used in this study were based on published information (Qiang et al., 2016). The following oligonucleotides were used for lyzc amplification: F-5′-AAGGGAAGCAGCAGCAGTTGTG-3′ and R-5′-CGTCCATGCCGTTAGCCTTGAG-3′. The gene encoding β-actin was chosen as the internal standard with the following primers: F-5′-CAGGGAGAAGATGACCCAGA-3′ and R-5′-CAGGGCATAACCCTAGTAGA-3′.
2.9. Challenge with S. agalactiae
Six weeks later, 25 fish from each replicate (including the CK group) were injected intraperitoneally with 20 μL of pathogenic S. agalactiae (WC1535) (median lethal dose [LD50] = 1 × 105 CFU/mL), which was provided by our lab (Key Laboratory of Tropical & Subtropical Fishery Resource Application & Cultivation, Pearl River Fisheries Research Institute of Chinese Academy of Fishery Sciences [CAFS]) (Xia et al., 2018). This strain was incubated at 37 °C for 48 h under anaerobic conditions in Brain Heart Infusion (BHI) media. The cultures were centrifuged (Beckman Coulter, AK, USA) at 2,000 × g for 5 min. The pellets were washed twice with PBS (130 mmol/L NaCl, 10 mmol/L NaH2PO4, pH 7.2). The number of bacterial cells in each suspension was determined by turbidimetry. Injected fish were monitored for clinical signs, postmortem lesions and daily mortalities for 2 wk.
2.10. Statistical analysis
Average values ± standard deviation (SD) were used to express the experimental results. Data were analyzed by one-way analysis of variance (ANOVA) with a Duncan's multiple range test. All analyses were conducted with SPSS 17.0 (SPSS, Inc.), and differences at P < 0.05 were considered significant.
3. Results
3.1. Growth performance and survival
After the 6-wk feeding trial, there were greater WG values in the BS + BC and BC groups compared with the CK group (P < 0.05) (Table 2). The FCR in the BC and BS + BC groups were lower compared to the CK group (P < 0.05). No significant differences were identified for WG and FCR between the CK and BS groups (P > 0.05). In addition, the tilapia in each group barely showed any significant differences in SR (P > 0.05).
Table 2.
Growth performance of tilapia fed diets supplemented with Bacillus subtilis and/or Bacillus cereus for 6 wk.1
| Item | Groups2 |
|||
|---|---|---|---|---|
| CK | BS | BS + BC | BC | |
| IBW, g | 0.20 ± 0.02 | 0.20 ± 0.02 | 0.19 ± 0.02 | 0.20 ± 0.02 |
| WG, % | 2,595.69 ± 229.91a | 2,772.65 ± 229.21a | 3,527.77 ± 359.08b | 3,532.20 ± 403.43b |
| FCR | 1.39 ± 0.05b | 1.33 ± 0.07b | 1.07 ± 0.04a | 1.04 ± 0.02a |
| SR, % | 94.07 ± 6.12 | 91.11 ± 2.22 | 89.63 ± 2.31 | 93.33 ± 2.94 |
IBW = initial body weight; WG = weight gain; FCR = feed conversion ratio; SR = survival rate.
a, bWithin a row, means with different letter superscripts are significantly different (P < 0.05).
Data represent means ± standard deviation (n = 6 fish).
CK group: the basal diet; BS group: the basal diet with 1 × 108 CFU/g of B. subtilis; BS + BC group: the basal diet with both 0.5 × 108 CFU/g of B. subtilis and B. cereus NY5; BC group: the basal diet with 1 × 108 CFU/g of B. cereus NY5.
3.2. Intestinal histology
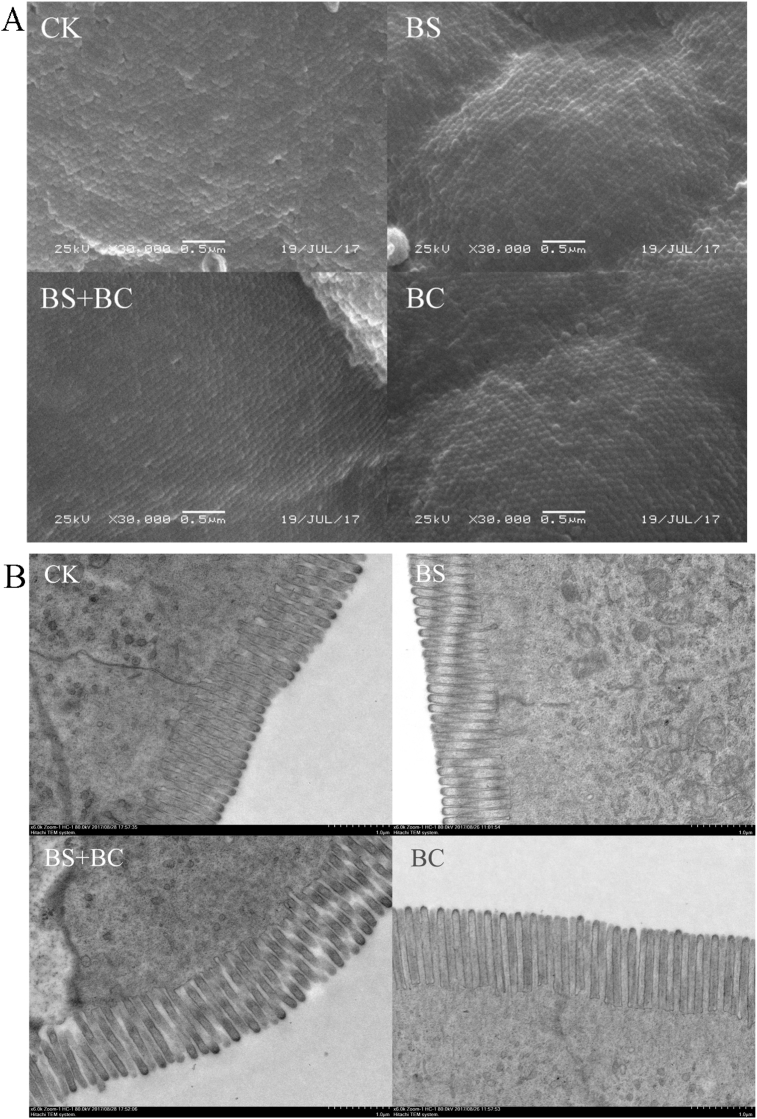
Compared with the CK group, the tilapia fed probiotic-supplemented feed had longer and denser intestinal microvilli (P < 0.05) (Fig. 1 and Table 3).
Fig. 1.
Electron microscope images of the gut microvilli. (A) Scanning electron microscopy (SEM) images for microvilli density; (B) transmission electron microscope (TEM) images of the microvilli length. CK group: the basal diet; BS group: the basal diet with 1 × 108 CFU/g of Bacillus subtilis; BS + BC group: the basal diet with both 0.5 × 108 CFU/g of B. subtilis and Bacillus cereus NY5; BC group: the basal diet with 1 × 108 CFU/g of B. cereus NY5.
Table 3.
Intestinal microvilli density and length of Nile tilapia fed diets supplemented with Bacillus subtilis or/and Bacillus cereus for 6 wk.1
| Item | Groups2 |
|||
|---|---|---|---|---|
| CK | BS | BS + BC | BC | |
| Density, count/μm2 | 85.66 ± 7.45a | 140.11 ± 13.19bc | 128.82 ± 7.83b | 142.76 ± 12.70c |
| Length, μm | 0.91 ± 0.06a | 1.03 ± 0.03c | 1.00 ± 0.03b | 1.00 ± 0.03b |
a, b, cWithin a row, means with different letter superscripts are significantly different (P < 0.05).
Data represent means ± standard deviation (n = 6 fish).
CK group: the basal diet; BS group: the basal diet with 1 × 108 CFU/g of B. subtilis; BS + BC group: the basal diet with both 0.5 × 108 CFU/g of B. subtilis and B. cereus NY5; BC group: the basal diet with 1 × 108 CFU/g of B. cereus NY5.
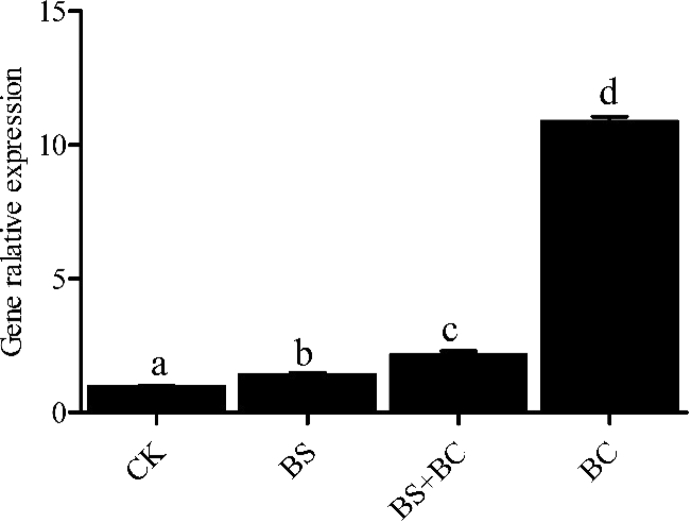
3.3. Expression of intestinal c-type lysozyme gene
As shown in Fig. 2, the expression of intestinal lyzc in the probiotic-fed groups was higher compared with the CK group (P < 0.05), and the BC group had the highest level of lyzc gene expression.
Fig. 2.
Levels of c-type lysozyme (lyzc) mRNA in the intestine of tilapia after 6 wk of feeding (n = 3). Data represent means ± SD. CK group: the basal diet; BS group: the basal diet with 1 × 108 CFU/g of Bacillus subtilis; BS + BC group: the basal diet with both 0.5 × 108 CFU/g of B. subtilis and Bacillus cereus NY5; BC group: the basal diet with 1 × 108 CFU/g of B. cereus NY5. Means sharing a common superscript letter were not significantly different (P > 0.05).
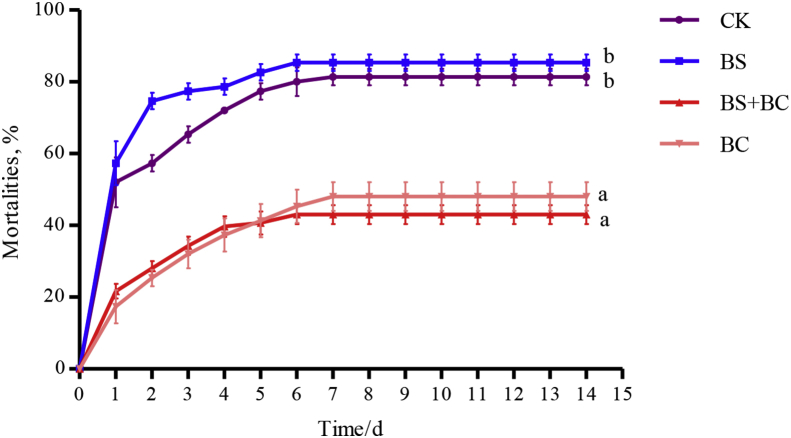
3.4. Challenge test
Fig. 3 presents the effects of probiotics on WC1535 resistance in tilapia. According to the results, each group had the highest daily mortality rate on the first day. Cumulative mortality was significantly lower in the BC (48%) and BS + BC (43%) groups compared to the CK (81.33%) and BS (85.33%) groups (P < 0.05).
Fig. 3.
Mortalities (means ± SD) of tilapia challenged by injection of Streptococcus agalactiae after being administered either the basal diet or the basal diet supplemented with probiotics for 6 wk. Data represent means ± SD. CK group: the basal diet; BS group: the basal diet with 1 × 108 CFU/g of Bacillus subtilis; BS + BC group: the basal diet with both 0.5 × 108 CFU/g of B. subtilis and Bacillus cereus NY5; BC group: the basal diet with 1 × 108 CFU/g of B. cereus NY5. Different letters indicate significant differences (P < 0.05).
3.5. High-throughput sequencing analysis
A tilapia gut microbiome analysis was conducted using high-throughput 16S rDNA deep sequencing technology (V3 and V4 regions). A total of 1,474,410 unique reads (average length 444 bp) were produced from 2,040,419 raw reads, and 189 operational taxonomic units (OTU) were identified from the gut DNA samples. The evaluation of the sample relationships was conducted on the basis of the different OTU phylogenetic diversities. As a result, the sequencing coverage (> 0.90) was sufficient for all the treatments, which indicated the representativeness of the sample OTU.
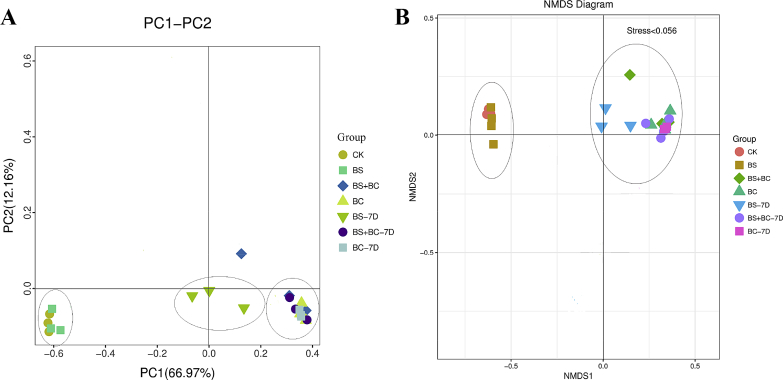
Analyses were conducted on the indices of bacterial diversity and richness from the proportion of the OTU for the calculation of each experimental group's bacterial diversity (Table 4). The BC group had a lower species richness (Chao1 and ace) in comparison with the CK group (P < 0.05). Upon cessation of probiotics for 1 wk, compared with the BS group, the BS group after cessation of probiotic consumption for 1 wk (BS-7D group) had higher Shannon and Simpson indices (P < 0.05). In comparison with the BS + BC group, the BS + BC group after cessation of probiotic consumption for 1 wk (BS + BC-7D group) had lower ace and Chao1 indices (P < 0.05). However, there was no significant difference in bacterial richness or diversity between the BC group and the BC group after cessation of probiotic consumption for 1 wk (BC-7D group) (P > 0.05). The replicates in the BS and CK groups were closely clustered, while clearly separated from those in the other groups, which all clustered closely in the principal component analysis (PCoA) plot and non-metric multidimensional scaling (NDMS) diagram (Fig. 4).
Table 4.
Richness and diversity statistics of the samples.1
| Group2 | Ace | Chao1 | Shannon | Simpson | Good's_coverage |
|---|---|---|---|---|---|
| CK | 54.89 ± 5.26bc | 57.29 ± 3.71c | 1.18 ± 0.16ab | 0.32 ± 0.08a | 0.95 ± 0.02 |
| BS | 59.61 ± 5.84c | 58.83 ± 5.71c | 1.85 ± 0.47ab | 0.45 ± 0.11a | 0.96 ± 0.01 |
| BS + BC | 67.58 ± 8.85c | 69.17 ± 6.93c | 1.94 ± 0.27b | 0.50 ± 0.08a | 0.95 ± 0.03 |
| BC | 37.06 ± 3.74a | 36.44 ± 3.09ab | 1.16 ± 0.18a | 0.31 ± 0.07a | 0.98 ± 0.02 |
| BS-7D | 63.13 ± 5.94c | 62.78 ± 4.88c | 2.85 ± 0.29c | 0.74 ± 0.07b | 0.95 ± 0.05 |
| BS + BC-7D | 48.29 ± 3.46b | 47.72 ± 4.44b | 1.74 ± 0.30ab | 0.46 ± 0.06a | 0.97 ± 0.01 |
| BC-7D | 29.29 ± 2.64a | 27.67 ± 3.88a | 1.47 ± 0.13ab | 0.40 ± 0.03a | 0.95 ± 0.05 |
a, b, cWithin a column, means with different letter superscripts are significantly different (P < 0.05).
Data represent means ± standard deviation (n = 3 fish).
CK group: the basal diet; BS group: the basal diet with 1 × 108 CFU/g of Bacillus subtilis; BS + BC group: the basal diet with both 0.5 × 108 CFU/g of B. subtilis and Bacillus cereus NY5; BC group: the basal diet with 1 × 108 CFU/g of B. cereus NY5; BS-7D group: the BS group after cessation of probiotic consumption for 1 wk; BS + BC-7D group: the BS + BC group after cessation of probiotic consumption for 1 wk; BC-7D group: the BC group after cessation of probiotic consumption for 1 wk.
Fig. 4.
Comparison of autochthonous intestinal microbiota composition between fish fed different diets. (A) Principal component (PC) analysis; (B) Non-metric multidimensional scaling (NMDS) diagram. CK group: the basal diet; BS group: the basal diet with 1 × 108 CFU/g of Bacillus subtilis; BS + BC group: the basal diet with both 0.5 × 108 CFU/g of B. subtilis and Bacillus cereus NY5; BC group: the basal diet with 1 × 108 CFU/g of B. cereus NY5; BS-7D group: the BS group after cessation of probiotic consumption for 1 wk; BS + BC-7D group: the BS + BC group after cessation of probiotic consumption for 1 wk; BC-7D group: the BC group after cessation of probiotic consumption for 1 wk.
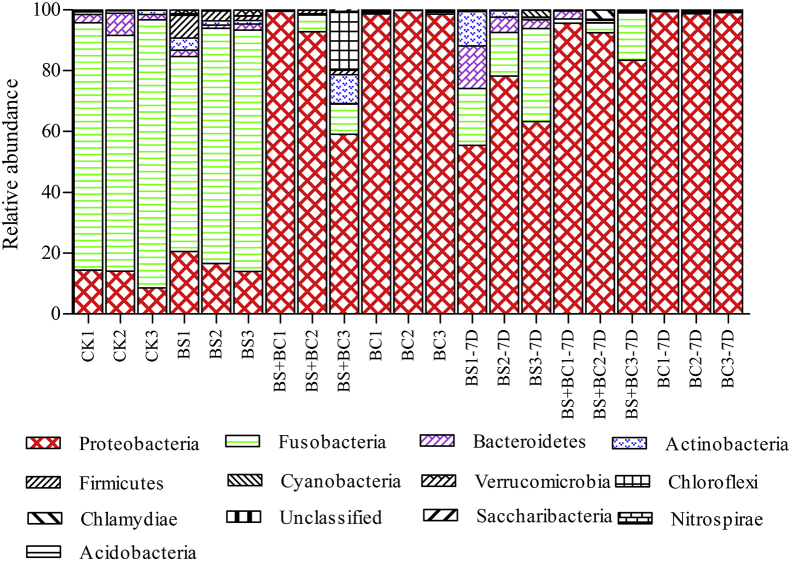
The composition of major bacteria at the phylum level in the intestines of fish fed different diets is shown in Fig. 5. In general, the most abundant phylum in the BS + BC and BC groups (accounting for 83.78% and 99.02% of 16S reads, respectively) was Proteobacteria, whereas the CK and BS groups were dominated by Fusobacteria (82.35% and 73.59%, respectively). The average amount of Proteobacteria and Bacteroidetes in the CK group (12.37% and 3.92%, respectively), Proteobacteria, Bacteroidetes, Actinobacteria, Firmicutes and Verrucomicrobia in the BS group (17.05%, 1.73%, 2.16%, 4.05% and 1.78%, respectively), and Fusobacteria, Actinobacteria and Chloroflexi in the BS + BC group (5.18%, 3.21% and 6.40%, respectively) were all above 1%. Saccharibacteria and Cyanobacteria were also present with relative abundances below 1% in the samples. Seven days after the probiotic supply was ceased, the Proteobacteria reads in BS group increased from 17.05% to 65.61%, while Fusobacteria reads dropped from 73.59% to 21.22%. The Fusobacteria and Proteobacteria reads in the BS + BC-7D (90.52% and 6.71%) and BC-7D groups (99.1% and 0.21%) were similar to those in the BS + BC and BC groups, which were 83.78% and 5.18% in the BS + BC group and 99.02% and 0.32% in the BC group, respectively.
Fig. 5.
Taxonomic distribution of gut samples at the phylum level. CK group: the basal diet; BS group: the basal diet with 1 × 108 CFU/g of Bacillus subtilis; BS + BC group: the basal diet with both 0.5 × 108 CFU/g of B. subtilis and Bacillus cereus NY5; BC group: the basal diet with 1 × 108 CFU/g of B. cereus NY5; BS-7D group: the BS group after cessation of probiotic consumption for 1 wk; BS + BC-7D group: the BS + BC group after cessation of probiotic consumption for 1 wk; BC-7D group: the BC group after cessation of probiotic consumption for 1 wk.
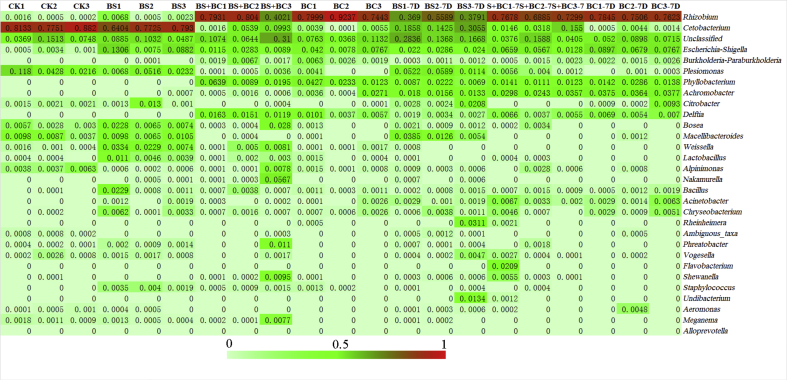
Rhizobium was found at a significant greater proportion (58.78% and 82.26%, respectively) in the BS + BC and BC groups and was also detected in the intestines of tilapia of BS and CK groups, yet it was higher in the former 2 groups (Fig. 6). Cetobacterium was the predominant genus (accounting for 82.35% and 73.53% of reads) in CK and BS groups; however, it was found much less frequently in the BS + BC and BC groups (4.18% in BS + BC group and 0.32% in BC group). Reads of Phyllobacterium, one of the major genera that accounted for 3.52% in BS + BC group and 2.61% in BC group, yet there was no Phyllobacterium detected in the 3 replicates of either BS or CK group. Plesiomonas (accounting for 6.08% and 2.72% of the reads in CK and BS groups) was present in the CK and BS groups; however, this genus was found at lower levels in the BS + BC and BC groups (0.4% in BS + BC group and 0.14% in BC group). Escherichia-Shigella (accounting for 0.16% of the reads) was detected in the CK group, and with probiotic treatment a larger amount of Escherichia-Shigella was detected (7.54% in BS group, 2.37% in BS + BC group and 4.22% in BC group). However, there were no significant differences in Phyllobacterium, Plesiomonas and Escherichia-Shigella after ANOVA of every group, due to high individual variability (Appendix Fig. 1). After the cessation of probiotic supplementation plus 7 d of basal diet feeding, reads of Rhizobium in the BS-7D group increased on average from 0.32% (BS) to 43.57%, but Cetobacterium dropped from 73.53% (BS) to 21.13%. However, Rhizobium and Cetobacterium in the BS + BC and BC groups both had smaller changes after the cessation of probiotic supplementation.
Fig. 6.
Heatmap of the 30 most predominant genus among all gut samples. CK group: the basal diet; BS group: the basal diet with 1 × 108 CFU/g of Bacillus subtilis; BS + BC group: the basal diet with both 0.5 × 108 CFU/g of B. subtilis and Bacillus cereus NY5; BC group: the basal diet with 1 × 108 CFU/g of B. cereus NY5; BS-7D group: the BS group after cessation of probiotic consumption for 1 wk; BS + BC-7D group: the BS + BC group after cessation of probiotic consumption for 1 wk; BC-7D group: the BC group after cessation of probiotic consumption for 1 wk.
At the species level, Bacillus licheniformis, Bacteroides sp., Rhizobium radiobacter, Ambiguous taxa, and Unclassified were the main components. Further, a larger fraction of R. radiobacter, which belong to the α-Proteobacteria, was detected in the BC and BS + BC groups than in the CK group (Appendix Fig. 2).
3.6. Statistical analysis of metagenomic profiles
The linear discriminate analysis (LDA) effect size (LEfSe), which is a statistical instrument developed for locating biomarkers in the metagenome, was used with default parameters for identifying possible discriminating taxa among groups. In this research, statistical analysis was only conducted on a phylum-to-genus level.
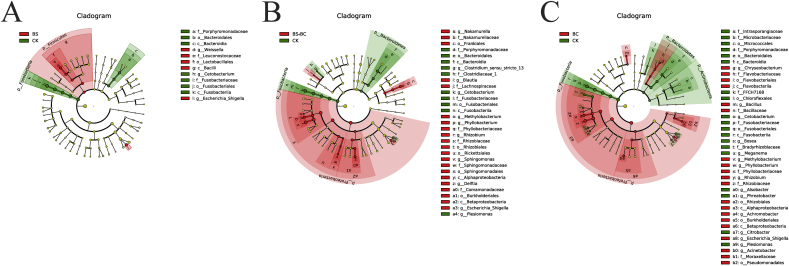
In total, 14 distinguishing taxa were detected between the groups of BS and CK with LDA scores both over 3 (Fig. 7A). One phylum (Firmicutes), 1 class (Bacilli), 1 order (Lactobacillales), 1 family and 2 genera were statistically higher in B. subtilis-treated fish. However, in the CK group, there was 1 phylum (Fusobacteria), 2 classes (Fusobacteria and Bacteroidia), 2 orders (Fusobacteriales, and Bacteroidales), 2 families and 1 genus that were enriched. The cladogram shows the 34 distinguishing taxa from the CK and BS + BC groups (LDA score > 3) (Fig. 7B). Compared to the CK group, 1 phylum (Proteobacteria), 2 classes (α- and β-Proteobacteria), 5 orders (Franklales, Rickettsiales, Sphingomonadales Burkholderiales and Rhizobiales), 6 families and 8 genera all had larger amounts in the BS + BC group. However, the amounts of 2 phyla (Fusobacteria and Bacteroidetes), 2 classes (Bacteroidia and Fusobacteria), 2 orders (Bacteroidales and Fusobacteriales), 3 families and 3 genera were greater in the CK group. In total, there were 42 obviously distinguishing taxa between the CK and BC groups (Fig. 7C). On the phylum level, the BC group clearly had a greater number of Proteobacteria compared to the CK group. In addition, in comparison, the CK group had enriched reads of Fusobacteria, Actinobacteria and Bacteroidetes.
Fig. 7.
Cladograms indicating the polygenetic distribution of bacterial lineages associated with different groups of samples. CK group: the basal diet; BS group: the basal diet with 1 × 108 CFU/g of Bacillus subtilis; BS + BC group: the basal diet with both 0.5 × 108 CFU/g of B. subtilis and Bacillus cereus NY5; BC group: the basal diet with 1 × 108 CFU/g of B. cereus NY5. Indicators between the CK and probiotic-fed groups with an linear discriminate analysis (LDA) score larger than 3, with red circles representing those bacteria richer in CK group samples, and green circles representing those richer in samples of BS (A), BS + BC (B) and BC (C) groups, respectively.
The BS group had richer Escherichia-Shigella and Weissella reads compared to the CK group, while reads from Cetobacterium were obviously richer in the CK group in comparison with the BS group at the genus level. The reads of Methylobacterium, Phyllobacterium, Rhizobium and Escherichia-Shigella were enriched in the BS + BC and BC groups compared to the CK group. However, the CK group had more enriched Plesiomonas and Cetobacterium reads compared to the BS + BC and BC groups. In comparison with the CK group, the BC group had more prevalent Bacillus sp., and the most enriched Bacillus OTU (54 OTU) shared a sequence identity of 100% with B. licheniformis.
As shown in Appendix Fig. 3, after cessation of probiotic administration for 1 wk, Weissella, Cetobacterium and Bosea were enriched in the BS group compared to the BS-7D group, but Phyllobacterium, Rhizobium and Achromobacter were significantly enriched in the BS-7D group compared to the BS group, which was under successive probiotic feeding. Reads of Achromobacter were enriched in the BS + BC-7D and BC-7D groups compared to the BS + BC and BC groups, respectively. However, Nakamurella was enriched in the BC + BC group compared to the BS + BC-7D group.
4. Discussion
The growth and feed utilization of juvenile Nile tilapia was improved by supplementation of combined B. subtilis and B. cereus or B. cereus by itself. Similar to the results of this study, tilapia growth performance has been improved by the use of Lactobacillus plantarum and B. subtilis or a mixture of Saccharomyces cerevisiae, L. plantarum and B. subtilis (Essa et al., 2010). After feeding for 4 to 8 wk, the weight gain of Nile tilapia was significantly increased in the Bacillus pumilus and the commercial probiotic product Organic Green (Hangpoong Industry Co. Ltd, Korea) supplementation groups in comparison to the CK group (Aly et al., 2008b). In addition, when the fish feed, which contained 40% or 27% crude protein, was supplemented with combined or single probiotics, it was more conducive to improving WG and FCR in comparison to the basal diet (Lara-Flores et al., 2010). After a 10-wk feeding trial, growth rather than FCR of tilapia was promoted by dietary B. licheniformis (Han et al., 2015). However, there were no positive effects of some probiotic strains on Nile tilapia growth performance in other studies. After a 21-d growth trial, the growth performance of tilapia in a B. subtilis strain-amended diet group was similar to that of the control group (Addo et al., 2017a). According to an observation of tilapia fed with B. subtilis- and Previda-supplemented feed for 8 wk, the growth performance of these fish did not significantly change (Addo et al., 2017b). Bacillus amyloliquefaciens at a concentration of 1 × 104 CFU/g was not significantly effective in improving tilapia growth performance after 30 d of feeding, but it promoted the growth of fish at the end of 60 d (Reda and Selim, 2015). The different antibiosis activities of probiotics, together with the different interactions among the intestinal beneficial bacteria, probiotics, feed, host and research conditions have led to the observed distinguishing effects of probiotics on the growth performance of Nile tilapia. These differences can consequently influence probiotic effects on growth performance in other studies. In this study, the single B. subtilis diet had no effect on tilapia WG and FCR. However, BS + BC and BC treatments positively affected the WG and FCR of these fish.
It has been reported that a multispecies (Pediococcus acidilactici, Enterococcus faecium, B. subtilis and Lactobacillus reuteri) probiotic-amended diet significantly increased tilapia mid-intestinal microvilli density in 8 wk (Pirarat et al., 2011). The numerical increases of microvilli length and perimeter ratio (PR, internal perimeter of the intestine lumen to external perimeter of the intestine ratio) suggest that intestinal morphology is improved by probiotic administration (Pirarat et al., 2011). The improved length and density of the mid-intestinal microvilli may indicate the increased intestinal absorptive surface area from the application of probiotics (Standen et al., 2016). Further, the higher microvilli density may be more conducive to the enhancement of tilapia resistance to possible pathogens by reducing the extent to which the interenterocyte junctions are exposed. In this research, supplementation with probiotics significantly increased microvillus length and density in the mid-intestine, which was consistent with previously obtained results regarding tilapia (Pirarat et al., 2011). These results indicate the improvement effect of B. subtilis and B. cereus on the health condition of the intestines. The increased length and density of mid-intestinal microvilli may be one of the reasons for enhanced resistance to disease, utilization of feed and performance in growth of tilapia within the BS + BC and BC groups in this study.
This study showed a higher lyzc gene expression in the mid-intestines of tilapia fed a probiotic-supplemented diet than those in the control group. Previous studies showed that the c-type lysozymes of tilapia were effective in lysing both Gram-negative and Gram-positive strains (Gao et al., 2012). The tilapia could recognize the bacterial challenges, generate an acute stress, and enhance body immunity by secreting specific proteins such as lysozymes (Gao et al., 2012). Lysozymes can break down the cytoderm of bacteria by hydrolyzing the chemical bonds between N-acetylglucosamine and N-acetylmuramic acid. Therefore, lysozymes are capable of lysing specific Gram-positive and even several Gram-negative strains (Alexander and Ingram, 1992). Previously, a study demonstrated the significant effect of dietary Bacillus spp. on increasing lysozyme levels in Nile tilapia, which resulted in a high rate of survival after challenge with Edwardsiella tarda (Taoka et al., 2006). Changes in the leukocyte population in the process of immune response development can be reflected in variations of lysozyme levels. A higher level of lysozymes detected in the probiotic-fed tilapia may serve as an indicator showing their improved immunity (Saurabh and Sahoo, 2008). In a previous study, enhanced nonspecific defense systems and enhanced resistance ability to Streptococcus iniae by feeding dietary B. licheniformis-supplemented feed were reported in Nile tilapia (Han et al., 2015). In this research, similar results were observed in tilapia fed with combined B. subtilis and B. cereus or B. cereus alone when challenged with S. agalactiae.
Strong adherence to the inner facets of the intestines (mucus and mucosa), which has been shown to be of great importance for beneficial activities, is one criterion for probiotic strain selection (Boyle et al., 2006, Martinez et al., 2015). As suggested in other studies, probiotics that combined unstably with the gut mucosal zone may release rapidly when suspended, bringing about dysbiosis of intestinal strains as well as ensuing disorders, and finally leading to an increased chance of infection (Liu et al., 2016b). In this research, during probiotic administration, the probiotics were not located in the gastrointestinal mucosal zone of tilapia, which indicated that these probiotics could not adhere to the intestinal mucosa and proliferate. This finding showed the challenge of successfully colonizing the fish gut with a probiotic strain. The persistence of probiotics in the gut is species-specific (Standen et al., 2015), and the dosage and duration of supplementation and the selection of probiotic strain(s) might influence colonization success, while the persistence of the probiotic might also depend on the developmental state of the animal (Pérez et al., 2010, Ramos et al., 2013, Gerritsen et al., 2011). However, the gut microbiota developed significant differences between treatments (except between the BS and CK groups), and the composition of the gut microbiota remained stable 1 wk after cessation of the administration of the probiotics, which means that discontinuation of probiotic administration (except for the BS group) did not cause significant fluctuations in gut microbes. In this study, potential probiotics, such as Rhizobium sp., which belong to the phylum Proteobacteria, were the major strains detected in tilapia intestines from the BS + BC and BC groups but were rarely found (<1%) in the CK and BS groups. Generally, many enzymes with pectolytic and cellulolytic activities were produced by Rhizobium bacteria and have the ability to hydrolyze the glycosidic skeleton in the cytoderm of plant cells (Huang et al., 2012, Robledo et al., 2008). Rhizobium live as endosymbionts in some phytophagous insect intestines and are helpful with the synthesis of nitrogen-containing substances that are generally insufficient in the food of the host insects (Russell et al., 2009). In this study, the coenzyme Q10 producing bacteria R. radiobacter was the most common Rhizobium species and may function to improve immunity (Al-Hasso, 2001, Wu et al., 2005). Changes in the autochthonous gut bacterial community and the increase in some beneficial bacteria in the intestines may be related to the improved resistance to diseases, utilization of feed and performance in growth of tilapia within the BS + BC and BC groups.
The use of combined B. subtilis and B. cereus or B. cereus by itself decreased the distribution of Plesiomonas and Cetobacterium and increased the distribution of Phyllobacterium and Escherichia-Shigella in the tilapia intestine. Operational taxonomic units of Plesiomonas and Cetobacterium were found in the intestinal tract of tilapia, forming the core microbiome, which were quite stable and resistant in response to dietary treatments (Adeoye et al., 2016). The genus Plesiomonas was also found within the intestines of tilapia that were cultured in earthen ponds (Pakingking et al., 2015), and it is considered to be a conditional pathogen in aquaculture systems. Plesiomonas is a pathogen for goldfish (Zhang et al., 2015) and grass carp (Hu et al., 2014). Plesiomonas is more prevalent in sick gibel carp than in healthy gibel carp, acting as a conditional pathogen in gibel carp (She et al., 2017). One possible way to use probiotics and prebiotics to reduce infection of hosts is by restoring intestinal microbial diversity (She et al., 2017, van Nood et al., 2013). Cetobacterium was the most abundant genus (82.15%) in control fish intestines, and this genus has previously been isolated from other fish intestines (including tilapia) (Adeoye et al., 2016, Li et al., 2015, Standen et al., 2015). Cetobacterium was the most dominant species in both sick fish that were infected by Cyprinid herpesvirus 2 (CyHV-2) and in healthy fish (Pakingking et al., 2015). Phyllobacterium sp. is a slow-growing, N-2-fixing bacterium (Rojas et al., 2001) that has the ability to degrade organic matter and remove nitrogen and phosphorus from sludge particles (Zuo et al., 2015). Phyllobacterium bacteria were specific to the intestines of both the BS + BC and BC groups in this study. Escherichia-Shigella is a common pathogen of aquatic animals and can cause disease, such as diarrhea, ascites and sepsis, in aquatic animals (Sun et al., 2012). Although there is no significant difference between the groups due to the existence of individual differences, the impact of Escherichia-Shigella in this study needs to be further explored. The metagenomics and metatranscriptomics of the intestinal bacteria should be investigated in future studies to reveal the functionality and contribution to improved immunity and growth performance of the tilapia fed the combination of B. subtilis and B. cereus or B. cereus alone.
5. Conclusions
The B. subtilis and B. cereus- or B. cereus-supplemented fish feeds at 1 × 108 CFU/g are capable of promoting growth, improving feed utilization and morphology of the intestine, enhancing intestinal lyzc expression and disease resistance, and altering the intestinal microbiota composition of tilapia. In future studies, further investigations are required to determine the effects of the probiotic-induced gastrointestinal microbiota on immune responses and growth performance in tilapia. Interesting new lines of research may someday arise from gnotobiotic fish, microbial engineering and metabonomics.
Conflict of interests
We declare that we have no financial and personal relationships with other people or organizations that can inappropriately influence our work, there is no professional or other personal interest of any nature or kind in any product, service and/or company that could be construed as influencing the content of this paper.
Acknowledgements
This work was supported by the China Agricultural Research System (CARS-46) and Science and Technology Program of Guangzhou, China (grant No.: 201707010312). We would like to thank Professor Xiaoying Hu at the South China Botanical Garden, Chinese Academy of Sciences, for the intestinal electron microscopy work in this study.
Footnotes
Peer review under responsibility of Chinese Association of Animal Science and Veterinary Medicine.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.aninu.2019.07.002.
Contributor Information
Maixin Lu, Email: mx-lu@163.com.
Gang Chen, Email: cheng@gdou.edu.cn.
Appendix A. Supplementary data
The following is the supplementary data to this article:
References
- Addo S., Carrias A.A., Williams M.A., Liles M.R., Terhune J.S., Davis D.A. Effects of Bacillus subtilis strains on growth, immune parameters, and Streptococcus iniae susceptibility in Nile Tilapia, Oreochromis niloticus. J World Aquacult Soc. 2017;48(2):257–267. doi: 10.1111/jwas.12380. [DOI] [Google Scholar]
- Addo S., Carrias A.A., Williams M.A., Liles M.R., Terhune J.S., Davis D.A. Effects of Bacillus subtilis strains and the prebiotic Previda on growth, immune parameters and susceptibility to Aeromonas hydrophila infection in Nile tilapia, Oreochromis niloticus. Aquacult Res. 2017;48(9):4798–4810. doi: 10.1111/are.13300. [DOI] [Google Scholar]
- Adeoye A.A., Yomla R., Jaramillo-Torres A., Rodiles A., Merrifield D.L., Davies S.J. Combined effects of exogenous enzymes and probiotic on Nile tilapia (Oreochromis niloticus) growth, intestinal morphology and microbiome. Aquaculture. 2016;463:61–70. doi: 10.1016/j.aquaculture.2016.05.028. [DOI] [Google Scholar]
- Al-Hasso S. Coenzyme Q10: a review. Hosp Pharm. 2001;36(1):51–55. doi: 10.1177/001857870103600107. [DOI] [Google Scholar]
- Alexander J.B., Ingram G.A. Noncellular nonspecific defence mechanisms of fish. Annu Rev Fish Dis. 1992;2:249–279. doi: 10.1016/0959-8030(92)90066-7. [DOI] [Google Scholar]
- Almada C., de Almada C.N., Martinea R.C.R., Sant'Ana A.S. Characterization of the intestinal microbiota and its interaction with probiotics and health impacts. Appl Microbiol Biotechnol. 2015;99(10):4175–4199. doi: 10.1007/s00253-015-6582-5. [DOI] [PubMed] [Google Scholar]
- Aly S.M., Abdel-Galil Ahmed Y., Abdel-Aziz Ghareeb A., Mohamed M.F. Studies on Bacillus subtilis and Lactobacillus acidophilus, as potential probiotics, on the immune response and resistance of Tilapia nilotica (Oreochromis niloticus) to challenge infections. Fish Shellfish Immunol. 2008;25(1–2):128–136. doi: 10.1016/j.fsi.2008.03.013. [DOI] [PubMed] [Google Scholar]
- Aly S.M., Mohamed M.F., John G. Effect of probiotics on the survival, growth and challenge infection in Tilapia nilotica (Oreochromis niloticus) Aquacult Res. 2008;39(6):647–656. doi: 10.1111/j.1365-2109.2008.01932.x. [DOI] [Google Scholar]
- Amal M.N.A., Saad M.Z. Streptococcosis in tilapia (Oreochromis niloticus): a review. Pertanika J Trop Agric Sci. 2011;34(2):195–206. [Google Scholar]
- Beck B.R., Kim D., Jeon J., Lee S.M., Kim H.K., Kim O.J. The effects of combined dietary probiotics Lactococcus lactis BFE920 and Lactobacillus plantarum FGL0001 on innate immunity and disease resistance in olive flounder (Paralichthys olivaceus) Fish Shellfish Immunol. 2015;42(1):177–183. doi: 10.1016/j.fsi.2014.10.035. [DOI] [PubMed] [Google Scholar]
- Boyle R.J., Robins-Browne R.M., Tang M.L. Probiotic use in clinical practice: what are the risks? Am J Clin Nutr. 2006;83(6):1256–1264. doi: 10.1093/ajcn/83.6.1256. [DOI] [PubMed] [Google Scholar]
- Chai P.C., Song X.L., Chen G.F., Xu H., Huang J. Dietary supplementation of probiotic Bacillus PC465 isolated from the gut of Fenneropenaeus chinensis improves the health status and resistance of Litopenaeus vannamei against white spot syndrome virus. Fish Shellfish Immunol. 2016;54:602–611. doi: 10.1016/j.fsi.2016.05.011. [DOI] [PubMed] [Google Scholar]
- El-Rhman A.M.A., Khattab Y.A., Shalaby A.M. Micrococcus luteus and Pseudomonas species as probiotics for promoting the growth performance and health of Nile tilapia, Oreochromis niloticus. Fish Shellfish Immunol. 2009;27(2):175–180. doi: 10.1016/j.fsi.2009.03.020. [DOI] [PubMed] [Google Scholar]
- Essa M.A., El-Serafy S.S., El-Ezabi M.M., Daboor S.M., Esmael N.A., Lall S.P. Effect of different dietary probiotics on growth, feed utilization and digestive enzymes activities of Nile tilapia, Oreochromis niloticus. J Arabian Aquacult Soc. 2010;5(2):143–161. [Google Scholar]
- FAO/WHO . 1-4 October 2001. Health and nutritional properties of probiotics in food including powder milk with liver lactic acid bacteria. Report of a Joint FAO/WHO Expert Consultation on Evaluation of Health and Nutritional Properties of Probiotics in Food Including Powder Milk with Live Lactic Acid Bacteria, Córdoba. [Google Scholar]
- Galagarza O.A., Smith S.A., Drahos D.J., Eifert J.D., Williams R.C., Kuhn D.D. Modulation of innate immunity in Nile tilapia (Oreochromis niloticus) by dietary supplementation of Bacillus subtilis endospores. Fish Shellfish Immunol. 2018;83:171–179. doi: 10.1016/j.fsi.2018.08.062. [DOI] [PubMed] [Google Scholar]
- Gao F.Y., Qu L., Yu S.G., Ye X., Tian Y.Y., Zhang L.L. Identification and expression analysis of three c-type lysozymes in Oreochromis aureus. Fish Shellfish Immunol. 2012;32(5) doi: 10.1016/j.fsi.2012.01.031. 779–488. [DOI] [PubMed] [Google Scholar]
- Gerritsen J., Smidt H., Rijkers G.T., de Vos W.M. Intestinal microbiota in human health and disease: the impact of probiotics. Genes Nutr. 2011;6:209–240. doi: 10.1007/s12263-011-0229-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giatsis C., Sipkema D., Ramiro-Garcia J., Bacanu G.M., Abernathy J., Verreth J. Probiotic legacy effects on gut microbial assembly in tilapia larvae. Sci Rep. 2016;6:33965. doi: 10.1038/srep33965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han B., Long W.Q., He J.Y., Liu Y.J., Si Y.Q., Tian L.X. Effects of dietary Bacillus licheniformis on growth performance, immunological parameters, intestinal morphology and resistance of juvenile Nile tilapia (Oreochromis niloticus) to challenge infections. Fish Shellfish Immunol. 2015;46(2):225–231. doi: 10.1016/j.fsi.2015.06.018. [DOI] [PubMed] [Google Scholar]
- He S.X., Zhang Y., Xu L., Yang Y.L., Marubashi T., Zhou Z.G. Effects of dietary Bacillus subtilis C-3102 on the production, intestinal cytokine expression and autochthonous bacteria of hybrid tilapia Oreochromis niloticus♀×Oreochromis aureus♂. Aquaculture. 2013;412–413:125–130. doi: 10.1016/j.aquaculture.2013.06.028. [DOI] [Google Scholar]
- Hoa N.T., Baccigalupi L., Huxham A., Smertenko A., Van P.H., Ammendola S. Characterization of Bacillus species used for oral bacteriotherapy and bacterioprophylaxis of gastrointestinal disorders. Appl Environ Microbiol. 2000;66(12):5241–5247. doi: 10.1128/AEM.66.12.5241-5247.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Q.D., Lin Q., Shi C.B., Fu X.Z., Li N.Q., Liu L.H. Isolation and identification of a pathogenic Plesiominas shigelloids from diseased grass carp. Acta Microbiol Sin. 2014;54:229–235. [in Chinese] [PubMed] [Google Scholar]
- Huang S.W., Sheng P., Zhang H.Y. Isolation and identification of cellulolytic bacteria from the gut of Holotrichia parallela larvae (Coleoptera: Scarabaeidae) Int J Mol Sci. 2012;13(3):2563–2577. doi: 10.3390/ijms13032563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lara-Flores M., Olivera-Castillo L., Olvera-Novoa M.A. Effect of the inclusion of a bacterial mix (Streptococcus faecium and Lactobacillus acidophilus), and the yeast (Saccharomyces cerevisiae) on growth, feed utilization and intestinal enzymatic activity of Nile tilapia (Oreochromis niloticus) Int J Fish Aquacult. 2010;2(4):93–101. [Google Scholar]
- Li T.T., Long M., Gatesoupe F.J., Zhang Q.Q., Li A.H., Gong X.N. Comparative analysis of the intestinal bacterial communities in different species of carp by pyrosequencing. Microb Ecol. 2015;69(1):25–36. doi: 10.1007/s00248-014-0480-8. [DOI] [PubMed] [Google Scholar]
- Liu G.B., Wang M., Lu M.X., Ke X.L., Liu Z.G., Zhu H.P. Application of Bacillus subtilis in comprehensive prevention and control of streptococcus disease of tilapia. Guangdong Agr Sci. 2015;42:123–129. [in Chinese] [Google Scholar]
- Liu G.B., Wang M., Lu M.X., Ke X.L., Liu Z.G., Zhu H.P. Identification of a denitrifying Bacillus strain with an antagonistic effect on Streptococcus agalactiae isolated from tilapia. J Fish Sci China. 2016;23:207–217. doi: 10.3724/SP.J.1118.2016.15095. (in Chinese) [DOI] [Google Scholar]
- Liu H.T., Wang S.F., Cai Y., Guo X.H., Cao Z.J., Zhang Y.Z. Dietary administration of Bacillus subtilis HAINUP40 enhances growth, digestive enzyme activities, innate immune responses and disease resistance of tilapia, Oreochromis niloticus. Fish Shellfish Immunol. 2017;60:326–333. doi: 10.1016/j.fsi.2016.12.003. [DOI] [PubMed] [Google Scholar]
- Liu K.F., Chiu C.H., Shiu Y.L., Cheng W., Liu C.H. Effects of the probiotic, Bacillus subtilis E20, on the survival, development, stress tolerance, and immune status of white shrimp, Litopenaeus vannamei larvae. Fish Shellfish Immunol. 2010;28(5–6):837–844. doi: 10.1016/j.fsi.2010.01.012. [DOI] [PubMed] [Google Scholar]
- Liu W.S., Wang W.W., Ran C., He S.X., Yang Y.L., Zhou Z.G. Effects of dietary scFOS and lactobacilli on survival, growth, and disease resistance of hybrid tilapia. Aquaculture. 2017;470:50–55. doi: 10.1016/j.aquaculture.2016.12.013. [DOI] [Google Scholar]
- Liu Z., Liu W.S., Ran C., Hu J., Zhou Z.G. Abrupt suspension of probiotics administration may increase host pathogen susceptibility by inducing gut dysbiosis. Sci Rep. 2016;6:23214. doi: 10.1038/srep23214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y.Y., Chen J., Zheng J.Y., Hu G.Y., Wang J.J., Huang C.L. Mucosal adherent bacterial dysbiosis in patients with colorectal adenomas. Sci Rep. 2016;6:26337. doi: 10.1038/srep26337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez R.C.R., Bedani R., Saad S.M.I. Scientific evidence for health effects attributed to the consumption of probiotics and prebiotics: an update for current perspectives and future challenges. Br J Nutr. 2015;114(12):1993–2015. doi: 10.1017/S0007114515003864. [DOI] [PubMed] [Google Scholar]
- Nilton G.M., Daniele M.A. Quantification of intestinal bacteria, operating cost and performance of fingerlings Nile tilapia subjected to probiotics. Lat Am J Aquat Res. 2015;43(2):367–373. doi: 10.3856/vol43-issue2-fulltext-13. [DOI] [Google Scholar]
- Pakingking R., Palma P., Usero R. Quantitative and qualitative analyses of the bacterial microbiota of tilapia (Oreochromis niloticus) cultured in earthen ponds in the Philippines. World J Microbiol Biotechnol. 2015;31(2):265–275. doi: 10.1007/s11274-014-1758-1. [DOI] [PubMed] [Google Scholar]
- Pérez T., Balcázar J.L., Ruiz-Zarzuela I., Halaihel N., Vendrell D., de Blas I. Host-microbiota interactions within the fish intestinal ecosystem. Mucosal Immunol. 2010;3:355–360. doi: 10.1038/mi.2010.12. [DOI] [PubMed] [Google Scholar]
- Pérez-Sánchez T., Ruiz-Zarzuela I., de Blas I., Balcázar J.L. Probiotics in aquaculture: a current assessment. Rev Aquacult. 2014;6(3):133–146. doi: 10.1111/raq.12033. [DOI] [Google Scholar]
- Pirarat N., Pinpimai K., Endo M., Katagiri T., Ponpornpisit A., Chansue N. Modulation of intestinal morphology and immunity in nile tilapia (Oreochromis niloticus) by Lactobacillus rhamnosus GG. Res Vet Sci. 2011;91(3):92–97. doi: 10.1016/j.rvsc.2011.02.014. [DOI] [PubMed] [Google Scholar]
- Qiang J., He J., Yang H., Xu P., Michael Habte-Tsion H., Ma X.Y. The changes in cortisol and expression of immune genes of GIFT tilapia Oreochromis niloticus (L.) at different rearing densities under Streptococcus iniae infection. Aquacult Int. 2016;24:1365–1378. doi: 10.1007/s10499-016-9995-y. [DOI] [Google Scholar]
- Ramos M., Weber B., Gonçalves J.F., Santos G.A., Rema P., Ozório R.O.A. Dietary probiotic supplementation modulated gut microbiota and improved growth of juvenile rainbow trout (Oncorhynchus mykiss) Comp Biochem Physiol. 2013;166:302–307. doi: 10.1016/j.cbpa.2013.06.025. [DOI] [PubMed] [Google Scholar]
- Ran C., Huang L., Liu Z., Xu L., Yang Y., Tacon P. A comparison of the beneficial effects of live and heat-inactivated baker's yeast on Nile Tilapia: suggestions on the role and function of the secretory metabolites released from the yeast. PLoS One. 2015;10(12):e0145448. doi: 10.1371/journal.pone.0145448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reda R.M., Selim K.M. Evaluation of Bacillus amyloliquefaciens on the growth performance, intestinal morphology, hematology and body composition of Nile tilapia, Oreochromis niloticus. Aquacult Int. 2015;23(1):203–217. doi: 10.1007/s10499-014-9809-z. [DOI] [Google Scholar]
- Rijkers G.T., Bengmark S., Enck P., Haller D., Herz U., Kalliomaki M. Guidance for substantiating the evidence for beneficial effects of probiotics: current status and recommendations for future research. J Nutr. 2010;140(3):671S–676S. doi: 10.3945/jn.109.113779. [DOI] [PubMed] [Google Scholar]
- Robledo M., Jimenez-Zurdo J.I., Velazquez E., Trujillo M.E., Zurdo-Pineiro J.L., Ramirez- Bahena M.H. Rhizobium cellulase CelC2 is essential for primary symbiotic infection of legume host roots. Proc Natl Acad Sci USA. 2008;105(19):7064–7069. doi: 10.1073/pnas.0802547105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas A., Holguin G., Glick B.R., Bashan Y. Synergism between Phyllobacterium sp (N-2-fixer) and Bacillus licheniformis (P-solubilizer), both from a semiarid mangrove rhizosphere. FEMS Microbiol Ecol. 2001;35(2):181–187. doi: 10.1111/j.1574-6941.2001.tb00802.x. [DOI] [PubMed] [Google Scholar]
- Russell J.A., Moreau C.S., Goldman-Huertas B., Fujiwara M., Lohman D.J., Pierce N.E. Bacterial gut symbionts are tightly linked with the evolution of herbivory in ants. Proc Natl Acad Sci USA. 2009;106(50):21236–21241. doi: 10.1073/pnas.0907926106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saurabh S., Sahoo P.K. Lysozyme: an important defence molecule of fish innate immune system. Aquacult Res. 2008;39(3):223–239. doi: 10.1111/j.1365-2109.2007.01883.x. [DOI] [Google Scholar]
- She R., Li T.T., Luo D., Li J.B., Yin L.Y., Li H. Changes in the intestinal microbiota of gibel carp (Carassius gibelio) associated with Cyprinid herpesvirus 2 (CyHV-2) infection. Curr Microbiol. 2017;74(10):1130–1136. doi: 10.1007/s00284-017-1294-y. [DOI] [PubMed] [Google Scholar]
- Standen B.T., Peggs D.L., Rawling M.D., Foey A., Davies S.J., Santos G.A. Dietary administration of a commercial mixed-species probiotic improves growth performance and modulates the intestinal immunity of tilapia, Oreochromis niloticus. Fish Shellfish Immunol. 2016;49:427–435. doi: 10.1016/j.fsi.2015.11.037. [DOI] [PubMed] [Google Scholar]
- Standen B.T., Rodiles A., Peggs D., Davies S., Santos G., Merrifield D. Modulation of the intestinal microbiota and morphology of tilapia Oreochromis niloticus, following the application of a multi-species probiotic. Appl Microbiol Biotechnol. 2015;99(20):8403–8417. doi: 10.1007/s00253-015-6702-2. [DOI] [PubMed] [Google Scholar]
- Sun Y.Z., Yang H.L., Ma R.L., Song K., Lin W.Y. Molecular analysis of autochthonous microbiota along the digestive tract of juvenile grouper Epinephelus coioides following probiotic Bacillus pumilus administration. J Appl Microbiol. 2011;110(4):1093–1103. doi: 10.1111/j.1365-2672.2011.04967.x. [DOI] [PubMed] [Google Scholar]
- Sun Z.K., Zhang W., Lu B., Zheng P., Wang Y.B., Ding P. Quantitative PCR detection method about viable but nonculturable state ShigeUa dysenteriae in water. J Environ Health. 2012;29(7):590–592. [Google Scholar]
- Taoka Y., Maeda H., Jo J.Y., Kim S.M., Park S.I., Yoshikawa T. Use of live and dead probiotic cells in tilapia Oreochromis niloticus. Fish Sci. 2006;72(4):755–766. doi: 10.1111/j.1444-2906.2006.01215.x. [DOI] [Google Scholar]
- Tseng D.Y., Ho P.L., Huang S.Y., Cheng S.C., Shiu Y.L., Chiu C.S. Enhancement of immunity and disease resistance in the white shrimp, Litopenaeus vannamei, by the probiotic, Bacillus subtilis E20. Fish Shellfish Immunol. 2009;26(2):339–344. doi: 10.1016/j.fsi.2008.12.003. [DOI] [PubMed] [Google Scholar]
- van Nood E., Vrieze A., Nieuwdorp M., Fuentes S., Zoetendal E.G., de Vos W.M. Duodenal infusion of donor feces for recurrent Clostridium difficile. N Engl J Med. 2013;368(5):407–415. doi: 10.1056/NEJMoa1205037. [DOI] [PubMed] [Google Scholar]
- Wang M., Liu G.B., Lu M.X., Ke X.L., Liu Z.G., Gao F.Y. Effect of Bacillus cereus as a water or feed additive on the gut microbiota and immunological parameters of Nile tilapia. Aquacult Res. 2017;48:3163–3173. doi: 10.1111/are.13146. [DOI] [Google Scholar]
- Widanarni, Tanbiyaskur Application of probiotic, prebiotic and synbiotic for the control of streptococcosis in tilapia Oreochromis niloticus. Pak J Biol Sci. 2015;18(2):59–66. doi: 10.3923/pjbs.2015.59.66. [DOI] [PubMed] [Google Scholar]
- Wu Z.F., Du G.C., Chen J. Effects of culture conditions on coenzyme Q10 production by Rhizobium radiobacter by metabolic flux analysis. Acta Microbiol Sin. 2005;45:231–235. doi: 10.3321/j.issn:0001-6209.2005.02.016. (in Chinese) [DOI] [PubMed] [Google Scholar]
- Xia Y., Lu M.X., Chen G., Cao J.M., Gao F.Y., Wang M. Effects of dietary Lactobacillus rhamnosus JCM1136 and Lactococcus lactis subsp. lactis JCM5805 on the growth, intestinal microbiota, morphology, immune response and disease resistance of juvenile Nile tilapia, Oreochromis niloticus. Fish Shellfish Immunol. 2018;76:368–379. doi: 10.1016/j.fsi.2018.03.020. [DOI] [PubMed] [Google Scholar]
- Zhang P., Zhu A., Hu X., Lan Y., Li X., Shen X.J. Isolation, identification and antibiotic susceptibility testing of Plesiomonas shigelloids from goldfish. Fish Sci. 2015;34:375–379. doi: 10.16378/j.cnki.1003-1111.2015.06.007. [in Chinese] [DOI] [Google Scholar]
- Zuo J.L., Jiang L.M., Wang W., Liu Z.W., Li B., Tang H.Q. PCR-DGGE Analysis of granular sludge during different culture periods with low DO. China Water Wastewater. 2015;31:77–81. [in Chinese] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.