Key Points
Question
What is the frequency of pathogenic or likely pathogenic germline genetic variants in known cancer-susceptibility genes in a large population of patients with osteosarcoma who were unselected for family history?
Findings
In this next-generation exome sequencing study of 1244 patients with osteosarcoma, 28.0% of patients in the discovery set carried a rare pathogenic or likely pathogenic germline variant in a cancer-susceptibility gene compared with 12.1% of individuals without cancer who were comparably sequenced and 9.3% of individuals of non-Finnish European ancestry identified through the Exome Aggregation Consortium database.
Meaning
A higher than expected frequency of patients with osteosarcoma carrying a pathogenic or likely pathogenic germline variant suggests germline genetic testing may be warranted for individuals with osteosarcoma.
Abstract
Importance
Osteosarcoma, the most common malignant bone tumor in children and adolescents, occurs in a high number of cancer predisposition syndromes that are defined by highly penetrant germline mutations. The germline genetic susceptibility to osteosarcoma outside of familial cancer syndromes remains unclear.
Objective
To investigate the germline genetic architecture of 1244 patients with osteosarcoma.
Design, Setting, and Participants
Whole-exome sequencing (n = 1104) or targeted sequencing (n = 140) of the DNA of 1244 patients with osteosarcoma from 10 participating international centers or studies was conducted from April 21, 2014, to September 1, 2017. The results were compared with the DNA of 1062 individuals without cancer assembled internally from 4 participating studies who underwent comparable whole-exome sequencing and 27 173 individuals of non-Finnish European ancestry who were identified through the Exome Aggregation Consortium (ExAC) database. In the analysis, 238 high-interest cancer-susceptibility genes were assessed followed by testing of the mutational burden across 736 additional candidate genes. Principal component analyses were used to identify 732 European patients with osteosarcoma and 994 European individuals without cancer, with outliers removed for patient-control group comparisons. Patients were subsequently compared with individuals in the ExAC group. All data were analyzed from June 1, 2017, to July 1, 2019.
Main Outcomes and Measures
The frequency of rare pathogenic or likely pathogenic genetic variants.
Results
Among 1244 patients with osteosarcoma (mean [SD] age at diagnosis, 16 [8.9] years [range, 2-80 years]; 684 patients [55.0%] were male), an analysis restricted to individuals with European ancestry indicated a significantly higher pathogenic or likely pathogenic variant burden in 238 high-interest cancer-susceptibility genes among patients with osteosarcoma compared with the control group (732 vs 994, respectively; P = 1.3 × 10−18). A pathogenic or likely pathogenic cancer-susceptibility gene variant was identified in 281 of 1004 patients with osteosarcoma (28.0%), of which nearly three-quarters had a variant that mapped to an autosomal-dominant gene or a known osteosarcoma-associated cancer predisposition syndrome gene. The frequency of a pathogenic or likely pathogenic cancer-susceptibility gene variant was 128 of 1062 individuals (12.1%) in the control group and 2527 of 27 173 individuals (9.3%) in the ExAC group. A higher than expected frequency of pathogenic or likely pathogenic variants was observed in genes not previously linked to osteosarcoma (eg, CDKN2A, MEN1, VHL, POT1, APC, MSH2, and ATRX) and in the Li-Fraumeni syndrome-associated gene, TP53.
Conclusions and Relevance
In this study, approximately one-fourth of patients with osteosarcoma unselected for family history had a highly penetrant germline mutation requiring additional follow-up analysis and possible genetic counseling with cascade testing.
This next-generation exome sequencing study investigates the frequency of pathogenic or likely pathogenic germline genetic variants in known cancer-susceptibility genes among patients with osteosarcoma.
Introduction
The peak incidence of osteosarcoma (OMIM 259500) occurs during the pubertal growth spurt.1,2,3 Osteosarcoma risk factors include tall height,4,5 high birth-weight,4,5 previous radiotherapy,6 and at least 8 established cancer predisposition syndromes,7,8 including autosomal-dominant disorders (Li-Fraumeni syndrome [OMIM 151623],9,10 hereditary retinoblastoma [OMIM 180200],11,12 and Diamond-Blackfan anemia [OMIM 105650]13,14) and autosomal-recessive disorders (primarily DNA helicase disorders,15,16,17,18 such as Rothmund-Thomson syndrome [OMIM 268400], RAPADILINO syndrome [OMIM 266280], Werner syndrome [OMIM 277700], and Bloom syndrome [OMIM 210900]). Candidate gene and genome-wide association studies suggest that common single-nucleotide polymorphisms are also associated with osteosarcoma,19,20,21 affirming a complex underlying architecture for its genetic etiology but one that appears to be weighted disproportionately toward rare variants.
An earlier study reported that 4% of patients with osteosarcoma younger than 30 years with an unknown family history of cancer carried a pathogenic germline variant of TP53 (OMIM 191170) that was known to be or highly likely to be associated with Li-Fraumeni syndrome; in addition, 6% of those patients carried rare likely pathogenic TP53 variants.22 A survey of 72 candidate genes across 1162 sarcomas, including 124 osteosarcomas, observed that 217 individuals (18.7%) had a pathogenic or likely pathogenic (pathogenic/likely pathogenic) germline variant in autosomal-recessive or autosomal-dominant genes; 7% of variants were in autosomal-dominant genes.23 Previous studies estimated that approximately 8% to 10% of all children with cancer carry a pathogenic germline variant in a known cancer-susceptibility gene.24,25 The frequency of pathogenic/likely pathogenic variants in children with osteosarcoma was reported to be between 3 of 42 patients (7.1%)25 and 7 of 39 patients (17.9%).24
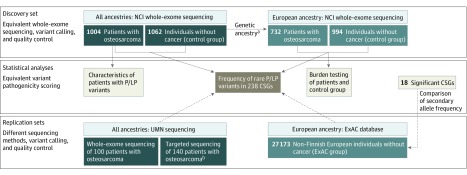
We used a 2-phase approach to evaluate rare germline variants in 1244 patients with osteosarcoma, beginning with assessment of 238 cancer-susceptibility genes followed by burden testing for an additional 736 candidate genes. We compared the frequency of pathogenic/likely pathogenic variants in patients with those of 1062 individuals without cancer (the control group), and for significant findings, with 27 173 individuals of non-Finnish European ancestry who were identified through the Exome Aggregation Consortium (ExAC) database26 (the ExAC group; Figure 1).
Figure 1. Overview of Study Samples and Design.
CSG indicates cancer-susceptibility gene; ExAC, Exome Aggregation Consortium database; NCI, National Cancer Institute; P/LP, pathogenic/likely pathogenic; and UMN, University of Minnesota.
aGenetic ancestry was determined using the available microarray from single-nucleotide polymorphism genome-wide association studies or whole-exome sequencing data from structure and principal component analyses. Individuals with more than 80% European ancestry were considered European.
bSequencing of 238 cancer-susceptibility genes.
Methods
The NCI Retrospective Study of Genetic Risk Factors for Osteosarcoma was approved by the institutional review board of the National Institutes of Health. All of the participants in the Genetic Epidemiology of Osteosarcoma study provided written informed consent, and the study was approved by the institutional review board of the University of Minnesota. The study was also approved by the respective local institutional review boards, and all participants provided written informed consent.
Study Populations
A total of 1244 patients with osteosarcoma were assembled from 10 participating centers and studies, including the National Cancer Institute retrospective Children’s Oncology Group study of genetic risk factors for osteosarcoma19 (United States); the Genetic Epidemiology of Osteosarcoma study of the Children’s Oncology Group21 (United States); the Clinica Universidad de Navarra (Pamplona, Spain); the Instituto de Oncologia Pediatrica, Grupo de Apoio ao Adolescente e a Crianca com Cancer/Universidade Federal de Sao Paulo27 (Sao Paulo, Brazil); the Childhood Cancer Survivor Study28 (United States); the National Cancer Institute Bone Disease and Injury Study of Osteosarcoma29 (United States); the Unidad Nacional de Oncologia Pediatrica30 (Guatemala City, Guatemala); the Royal National Orthopaedic Hospital NHS Trust and University College London Cancer Institute (Middlesex, United Kingdom); the Istituto Ortopedico Rizzoli (Bologna, Italy); and the Ankara Oncology Training and Research Hospital (Ankara, Turkey; eMethods and eTable 1 in the Supplement). Of those, 782 patients were previously reported in a genome-wide association study,19,27 which included 48 patients from the Instituto de Oncologia Pediatrica, Grupo de Apoio ao Adolescente e a Crianca com Cancer /Universidade Federal de Sao Paulo. A total of 462 additional patients were included, drawn from the Childhood Cancer Survivor Study, the NCI Bone Disease and Injury Study of Osteosarcoma, the Hospital Infantil Manuel De Jesus Rivera (Managua, Nicaragua), and from the Unidad Nacional de Oncologia Pediatrica. Each center provided data on patient and clinical variables, which were harmonized across studies.
A total of 1004 patients who underwent whole-exome sequencing at the National Cancer Institute were included as a primary discovery set, and 240 additional (nonoverlapping) patients with osteosarcoma21 comprised a replication set of patients who underwent whole-exome sequencing (n = 100) or targeted sequencing (n = 140) at the University of Minnesota (Figure 1; eMethods in the Supplement). Patients from the replication sets were drawn from the Genetic Epidemiology of Osteosarcoma study of the Children’s Oncology Group (United States). Neither family history nor tumor sequence data were available for the patients in this study.
The 1062 individuals without osteosarcoma who were assigned to the control group were assembled internally from 4 participating studies. This group included 994 adults of European ancestry (mean [SD] age at enrollment, 64.6 [7.2] years) who were drawn from 3 large studies: the Prostate, Lung, Colon and Ovarian Cancer Prevention clinical trial (United States),31 the American Cancer Society Cancer Prevention Study II (United States),32 and the Environment and Genes in Lung Cancer Etiology study (Italy).33 In addition, 68 individuals were enrolled from the Instituto de Oncologia Pediatrica, Grupo de Apoio ao Adolescente e a Crianca com Cancer/Universidade Federal de Sao Paulo study and were drawn from the same population as the 48 patients with osteosarcoma from Sao Paulo, Brazil (eMethods and eTable 1 in the Supplement).
The population substructure was determined for the patient group and the control group using the available single-nucleotide polymorphism microarray data or whole-exome sequencing data based on structure and principal component analyses, as previously described.19,34 Individuals with more than 80% European ancestry were considered European (Figure 1; eTable 1 in the Supplement).
The population frequency of pathogenic/likely pathogenic germline variants was estimated for 238 cancer-susceptibility genes using publicly available noncancer whole-exome sequencing data from the ExAC database.26 Variant data for each gene were analyzed for secondary comparisons with individuals in the ExAC group using similar pathogenicity scoring and in silico analysis.
Sequencing
Whole-exome sequencing was performed on a discovery set of 1004 patients and 1062 individuals in the control group using germline DNA extracted from either leukocytes or buccal samples between April 21, 2014, and July 1, 2017, at the National Cancer Institute (eMethods in the Supplement; Figure 1).34,35,36 All analyses evaluated variants with minor allele frequencies of less than 0.01 that passed quality-control filters.34,35,37 For the patient replication sets, we used buccal sample DNA to conduct whole-exome sequencing on 100 patients with osteosarcoma and targeted sequencing of 238 cancer-susceptibility genes on an additional 140 patients at the University of Minnesota from August 1, 2017, to September 1, 2018 (eMethods in the Supplement).
Genes and Variants
We assembled a set of 238 cancer-susceptibility genes, including 114 cancer-predisposing genes,38 14 genes associated with Diamond-Blackfan anemia,34,39,40,41 and 110 cancer-associated genes previously described25,42,43 or reported to have germline associations in the Catalogue of Somatic Mutations in Cancer44 (eTable 2 in the Supplement). These genes were grouped by mode of inheritance: 141 genes were autosomal-dominant, 45 were autosomal-recessive, 25 were autosomal-dominant and autosomal-recessive, 11 were X-linked, 1 was Y-linked, and 15 had de novo or unknown inheritance patterns (eTable 2 in the Supplement). An additional 736 candidate genes were evaluated, including 140 genes associated with osteosarcoma that were identified through the Human Genome Epidemiology (HuGE) phenopedia45 and manual curation of published reports and 596 genes somatically altered in pediatric bone cancers or recurrent in any pediatric cancer that were identified through the Catalogue of Somatic Mutations in Cancer44 and annotation of published osteosarcoma somatic data46,47,48,49 (eTable 3 in the Supplement).
A stepwise pipeline was constructed to evaluate each rare variant that passed quality-control filters in the genes of interest. Variants were classified as pathogenic, likely pathogenic, of uncertain significance, likely benign, or benign based on previous reports24,25 and recommendations from the American College of Medical Genetics and Genomics and the Association for Molecular Pathology50 (eMethods and eTable 4 in the Supplement). An in silico prediction algorithm was also used to further filter the variant of uncertain significance category as damaging or not damaging (eMethods in the Supplement). All pathogenic/likely pathogenic variants are summarized in eTable 5 in the Supplement.
Statistical Analyses
We analyzed the 1004 patients with osteosarcoma in the discovery set, which included 732 patients of European ancestry, with the 1062 individuals in the control group, which included 994 patients of European ancestry (Figure 1; eMethods in the Supplement). The replication set consisted of 240 patients with osteosarcoma who had germline whole-exome sequencing or targeted sequencing data available, and we performed secondary patient comparisons with individuals in the ExAC group.26
Rare-variant burden tests were conducted on the 732 European patients in the discovery set and the 994 European individuals in the control group using burden and optimal sequence kernel association tests.51 The comparisons between the patient group and the ExAC group were restricted to genes identified as substantially different between the primary discovery set of patients and the control group. Comparisons among individuals with and without pathogenic/likely pathogenic variants were performed using 2-sided χ2 or Fisher exact tests for categorical variables and Mann-Whitney U tests for continuous variables (eg, age). We used 2-sided exact binomial tests and logistic regression models to compare the frequencies of pathogenic/likely pathogenic variants between patients and individuals in the ExAC group only for the selected genes identified as substantially different between the primary discovery set of patients and individuals in the control group who had comparable whole-exome sequencing performed at the National Cancer Institute. We compared overall survival between patients carrying pathogenic/likely pathogenic variants and individuals without pathogenic/likely pathogenic variants for all cancer-susceptibility genes and the TP53 gene using adjusted Cox proportional hazards regression models and estimated hazard ratios (HRs) and 95% CIs. All data were analyzed from June 1, 2017, to July 1, 2019.
Results
Among 1244 patients with osteosarcoma, the mean (SD) age at diagnosis was 16 (8.9) years (age range, 2-80 years), and 684 patients (55.0%) were male (eTable 1 in the Supplement). Our primary analyses were based on patients and individuals in the control group with whole-exome sequencing data jointly called that yielded comparable quality-control measures and coverage (Figure 1; eFigure 1 in the Supplement).
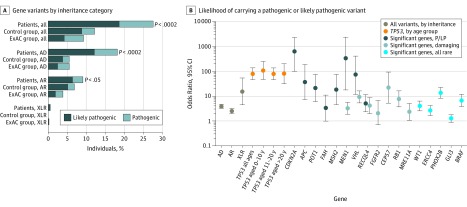
We assessed the frequency of pathogenic/likely pathogenic variants in 238 cancer-susceptibility genes in the discovery set of patients and the control group. Overall, 281 of 1004 patients with osteosarcoma (28.0%; 95% CI, 22.7%-33.2%) had a pathogenic/likely pathogenic variant in a gene of interest, which was significantly higher than the frequency observed in the control group (128 of 1062 individuals [12.1%]; 95% CI, 6.4%-17.7%; Fisher exact P = 1.3 × 10−18; Figure 2A, Figure 2B, and Figure 3; eTable 6 in the Supplement). The pathogenic/likely pathogenic frequency among European patients with osteosarcoma was also higher compared with the frequency among individuals in the ExAC group (2527 individuals [9.3%]; 95% CI, 8.2%-10.5%; Fisher exact P = 2.3 × 10−53; Figure 2A and Figure 2B; eTable 6 in the Supplement). Patients with pathogenic/likely pathogenic variants were significantly younger (mean [SD] age, 15.3 [7.2] years; age range, 2-61 years) than patients without pathogenic/likely pathogenic variants (mean [SD] age, 16.9 [10.2] years; age range, 2-80 years; Mann-Whitney U P = .02; Figure 4; eFigure 2 in the Supplement).
Figure 2. Frequency of Rare Pathogenic or Likely Pathogenic Germline Variants in Cancer-Susceptibility Genes.
A, Includes 1004 patients in the discovery set, 1062 individuals in the control group, and 27 173 individuals in the ExAC group. Genes with both AD and AR inheritance patterns and variants in genes with unknown inheritance are grouped with AD genes. B, Includes genes with a significantly higher frequency of pathogenic/likely pathogenic, damaging, or rare variants in 732 European patients compared with 27 173 individuals in the ExAC group. P values represent European patient-control group burden tests, with P < .0002 significant at the Bonferroni threshold. AD indicates autosomal-dominant genes; AR, autosomal-recessive genes; ExAC, Exome Aggregation Consortium database; P/LP, pathogenic/likely pathogenic; and XLR, X-linked or Y-linked recessive inheritance pattern genes.
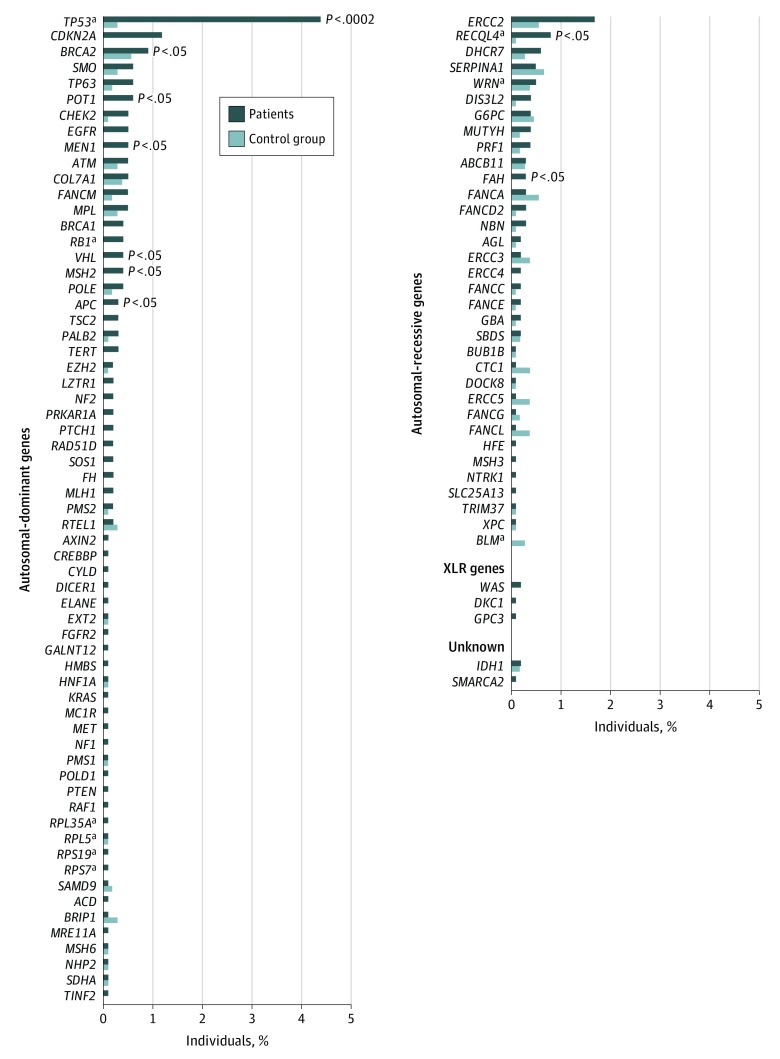
Figure 3. Frequency of Pathogenic or Likely Pathogenic Variants.
Frequency of pathogenic or likely pathogenic variants in the 1004 patients and 1062 individuals in the control group. P values represent European patient-control group burden tests, with P < .0002 significant at the Bonferroni threshold.
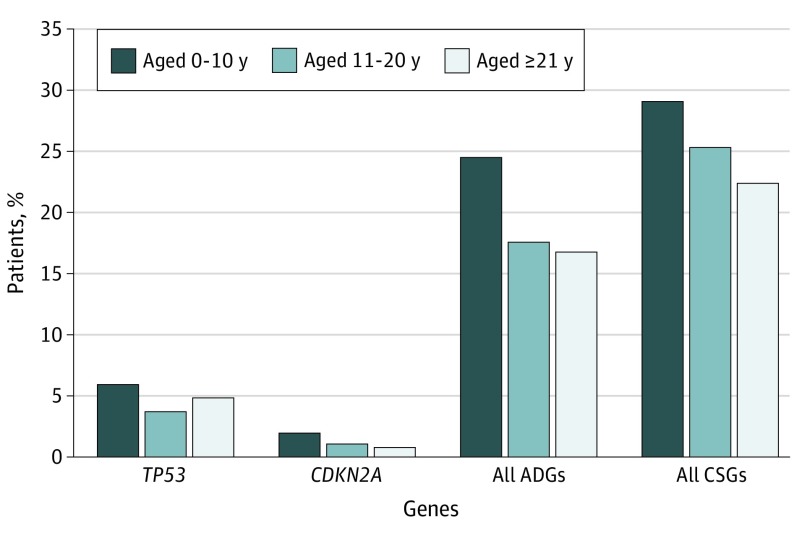
Figure 4. Frequency of Rare Pathogenic or Likely Pathogenic Variants.
All ADGs include genes TP53 and CDKN2A. A total of 151 patients were aged 0 to 10 years, 698 patients were aged 11 to 20 years, and 125 patients were aged 21 years and older. ADG indicates autosomal-dominant gene and CSG, cancer-susceptibility gene.
Among 364 patients with osteosarcoma subtype information, cancer-susceptibility genes with pathogenic/likely pathogenic variants were less common in those with surface subtypes (3 of 22 patients [13.6%]) vs conventional subtypes (104 of 342 patients [30.4%]; eTable 8 in the Supplement). A pathway enrichment analysis52,53 of the 101 cancer-susceptibility genes with pathogenic/likely pathogenic variants indicated enrichment in DNA repair pathway genes (Fisher exact P = 3.4 × 10−28; eFigure 3 and eTable 9 in the Supplement).
Autosomal-Dominant Genes
Overall, 185 of 1004 patients (18.4%; 95% CI, 12.8%-24.0%) with osteosarcoma had a pathogenic/likely pathogenic variant in an autosomal-dominant or an autosomal-dominant and autosomal-recessive cancer-susceptibility gene, whereas the variant frequency was 56 individuals (5.3%; 95% CI, 0%-11.1%) in the control group and 1494 individuals (5.5%; 95% CI, 4.3%-6.6%) in the ExAC group (Figure 2A and Figure 2B; eTable 6 in the Supplement). The highest frequency of pathogenic/likely pathogenic autosomal-dominant cancer-susceptibility gene variants was found in patients aged 0 to 10 years (37 of 151 patients [24.5%]; Mann-Whitney U P = .006; Figure 4). The 732 European patients with cancer had a higher burden of pathogenic/likely pathogenic autosomal-dominant variants than the 994 European individuals in the control group (burden P = 1.9 × 10−16). This higher burden translated to a nearly 4-fold greater risk of carrying a pathogenic/likely pathogenic variant compared with the ExAC group (odds ratio [OR], 3.9; 95% CI, 3.3-4.6).
Eighteen patients (1.8%) had more than 1 pathogenic/likely pathogenic autosomal-dominant variant compared with 4 individuals (0.4%) in the control group (Fisher exact P = .002). No significant difference was observed in overall patient survival for those carrying any pathogenic/likely pathogenic variant or an autosomal-dominant pathogenic/likely pathogenic variant compared with patients without these variants (Cox [adjusted for age, sex, and tumor location] P = .55 for all genes and P = .34 for autosomal-dominant genes) in the subset of 407 patients for whom outcome data was available.
Pathogenic/likely pathogenic variants in the TP53 gene were the most frequent of all autosomal-dominant genes (44 of 1004 total patients [4.4%]; 30 of 732 European patients [4.1%]) and substantially higher than those observed in the control group (3 of 1062 total individuals [0.3%]; 3 of 994 European individuals [0.3%]; burden P = 3.2 × 10−8) and the ExAC group (27 individuals [0.1%]; Fisher exact P = 9.0 × 10−44; Figure 2B and Figure 3; eTable 7 and eTable 10 in the Supplement). This finding is consistent with a previous study,22 which included 360 patients who were also in the current study. Analyses restricted to European patients who did not participate in the previous study found that 32 of 644 patients (5.0%) had a pathogenic TP53 variant.
All pathogenic/likely pathogenic TP53 variants were observed in patients younger than 30 years at diagnosis, with the exception of 1 patient, who was aged 39 years at diagnosis (Mann-Whitney U P = .05; eFigure 2 and eTable 8 in the Supplement). Patients aged 0 to 10 years (n = 151) had the highest estimated likelihood of carrying a TP53 pathogenic/likely pathogenic variant (OR, 108; 95% CI, 47-247; Figure 2B and Figure 4). Patients with a pathogenic/likely pathogenic TP53 variant were more likely to have osteosarcoma of the axial skeleton (χ2 P = .001), and the data suggested that patients with TP53 pathogenic/likely pathogenic variants were more likely to have metastases at diagnosis (χ2 P = .06; eTable 8 in the Supplement). In the subset of patients with outcome data, an adjusted Cox proportional hazards model indicated that patients carrying a TP53 pathogenic/likely pathogenic variant had significantly worse overall survival compared with patients without these variants (HR, 2.2; 95% CI, 1.2-4.0; Cox P = .009). These variants occurred in several functional domains, including the DNA-binding domain (subregion-based burden54 P = 1.5 × 10−6; eFigure 4A in the Supplement), which is consistent with previous studies.55,56,57,58,59
The gene CDKN2A (OMIM 600160) had the second highest frequency of pathogenic/likely pathogenic variants in the patients with osteosarcoma (12 of 1004 total patients [1.2%]; 8 of 732 European patients [1.1%]) compared with no pathogenic/likely pathogenic variants among individuals in the control group (burden P = 3.1 × 10−3) and the ExAC group (Fisher exact P = 2.2 × 10−13; Figure 2B and Figure 3; eTable 7 in the Supplement). Individuals with a CDKN2A pathogenic/likely pathogenic variant were younger (mean [SD] age, 12.9 [4.4] years) than patients without pathogenic/likely pathogenic variants (mean [SD] age, 6.9 [10.2] years; Mann-Whitney U P = .03). Notably, the youngest patients (aged 0-10 years) had the highest frequency of these variants (3 of 151 patients [2.0%]; Figure 4). The CDKN2A variants mapped to sites that were somatically mutated in bone cancers58 (eFigure 4B in the Supplement). Five additional autosomal-dominant genes (MEN1 [OMIM 613733], VHL [OMIM 608537], POT1 [OMIM 606478], APC [OMIM 611731], and MSH2 [OMIM 609309]) had a significantly higher pathogenic/likely pathogenic burden in European patients compared with European individuals in the control group (Figure 3; eTable 7 in the Supplement).
We compared the frequency of pathogenic/likely pathogenic variants among individuals in the ExAC group and observed that the risk of carrying a pathogenic/likely pathogenic variant in genes MEN1, VHL, POT1, and APC was elevated in European patients with osteosarcoma after a Bonferroni adjustment (Figure 2B; eTable 7 in the Supplement). Fifty-five additional autosomal-dominant genes had pathogenic/likely pathogenic variants in 1 or more patients (each were present in 0.1%-0.6% of patients; Figure 3; eTable 10 in the Supplement). Most of the specific variants observed in patients were absent in individuals in both the control group and the ExAC group as well as other public databases (the 1000 Genomes Project and the Exome Sequencing Project).
In addition, 316 patients (25.4%) with osteosarcoma had a rare variant of uncertain significance that was predicted to be damaging in silico in an autosomal-dominant gene, in the absence of another pathogenic/likely pathogenic autosomal-dominant variant. Altogether, 545 patients (43.8%) had at least 1 pathogenic variant, likely pathogenic variant, or variant of uncertain significance that was predicted to be damaging in silico in an autosomal-dominant gene. The European patients had more variants of uncertain significance that were predicted to be damaging in silico in the genes RB1 (OMIM 614041) and VHL (OMIM 608537) compared with individuals in the control group after adjustment for multiple testing (eTable 7 in the Supplement).
Autosomal-Recessive Genes
A total of 92 of 1004 patients (9.2%; 95% CI, 3.3%-15.1%) had a pathogenic/likely pathogenic variant in 33 autosomal-recessive genes, which is higher than that of the control group (72 of 1062 individuals [6.8%]; burden P = .03) and the ExAC group (1041 individuals [3.8%]; Fisher exact P = 2.6 × 10−13; Figure 2A and Figure 2B; eTable 6 in the Supplement). All autosomal-recessive gene variants were present as single heterozygotes, with the exception of 1 patient aged 13 years who had osteosarcoma with 2 RECQL4 (OMIM 603780) pathogenic/likely pathogenic variants; we were unable to phase the variants. The gene RECQL4 had the highest frequency of pathogenic/likely pathogenic variants in European patients with osteosarcoma (7 of 732 patients [1.0%]) compared with European individuals in the control group (1 of 994 individuals [0.1%]; burden P = .02; Figure 2B and Figure 3; eFigure 4C and eTable 7 in the Supplement). One RECQL4 variant was previously reported in a patient with Rothmund-Thomson syndrome and osteosarcoma (c.2476C>T, p.Arg826*).60 Several other autosomal-recessive genes had more pathogenic/likely pathogenic variants in patients than in the control group but were not significantly associated (Figure 2B and Figure 3; eTable 7 and eTable 10 in the Supplement).
We observed a preponderance of male patients (4 of 1004 patients [0.4% of the total patients and 0.7% of the 540 male patients in the discovery set]) who carried a pathogenic/likely pathogenic variant in an X-linked cancer-susceptibility gene (DKC1 [OMIM 300126], GPC3 [OMIM 300037], or WAS [OMIM 300392]) compared with no individuals in the control group and 7 individuals (0.03%) in the ExAC group (OR, 15.5; 95% CI, 5-53; Fisher exact P = 4.4 × 10−9; Figure 2; eTable 6 in the Supplement).
Known Osteosarcoma Syndrome Genes
Genes associated with cancer-predisposing syndromes associated with the occurrence of osteosarcoma were also associated with sporadic osteosarcoma or patients unselected for family history. We identified that 6.5% of the patients had a pathogenic/likely pathogenic variant in 1 of the following syndromic genes: RB1, RECQL4, RPL35A (OMIM 180468), RPL5 (OMIM 603634), RPS19 (OMIM 603474), RPS7 (OMIM 603658) , TP53, and WRN (OMIM 277700). A total of 2.2% of patients had a pathogenic/likely pathogenic variant in a syndromic gene without TP53. A total of 19.6% of patients had either a pathogenic/likely pathogenic variant in any autosomal-dominant gene or an osteosarcoma-associated autosomal-recessive syndrome gene.
Replication of Findings
Two independent patient data sets were used to evaluate the frequency of pathogenic/likely pathogenic variants in the cancer-susceptibility genes; set 1 comprised 100 patients with whole-exome sequencing data, and set 2 comprised 140 patients with targeted sequencing data. The overall prevalence of pathogenic/likely pathogenic variants in the replication sets (28 patients [28.0%] in set 1 and 38 patients [27.1%] in set 2; eTable 6 in the Supplement) was consistent with the carrier rates observed in our larger discovery set of 1004 patients with osteosarcoma (281 patients [28.0%]).
Of note, pathogenic/likely pathogenic variants in specific genes identified in our discovery set were also identified to have pathogenic/likely pathogenic variants in the 240 total patients in the replication sets; these genes were TP53 (13 patients [5.4%]), MEN1 (1 patient [0.4%]), MSH2 (1 patient [0.4%]), FAH (OMIM 613871; 4 patients [1.7%]), RECQL4 (8 patients [0.8%]), DKC1 (1 patient [0.4%]), and WAS (1 patient [0.4%]; eTable 10 and eTable 11 in the Supplement).
Candidate Gene Rare-Variant Burden
To explore whether unidentified germline genetic associations with osteosarcoma existed, we evaluated rare variants in 736 candidate genes, which included 140 genes previously associated with osteosarcoma and 596 somatically altered genes (eTable 3 in the Supplement).
Burden tests of in silico–predicted damaging variants (minor allele frequency ≤0.005) and all rare variants (minor allele frequency ≤0.01) did not identify an association with the evaluated genes (eTable 12 in the Supplement). One exception was observed; the gene ATRX (OMIM 300032) had a higher rare-variant burden in European patients (28 of 732 patients [3.8%]) compared with European individuals in the control group (18 of 994 individuals [1.8%]). One variant was pathogenic (c.6532C>T, p.Arg2178Trp, in 1 male patient; absent in the control group) and was previously reported to be pathogenic for alpha-thalassemia X-linked (ATR-X) intellectual disability syndrome in a patient with ATR-X syndrome who also developed osteosarcoma.61,62
Discussion
We report that 28.0% of patients with osteosarcoma had a pathogenic/likely pathogenic variant in a cancer-susceptibility gene, with 18.4% of those variants in an autosomal-dominant gene; to our knowledge, this frequency is higher than previously reported for any other pediatric cancer.23,24,25,63 The highest carrier frequency was observed in the youngest patients, with 24.5% of patients aged 0 to 10 years carrying a pathogenic/likely pathogenic variant in an autosomal-dominant gene. An additional 25.4% of the total patients had an in silico–predicted damaging variant of uncertain significance in an autosomal-dominant cancer-susceptibility gene. We confirmed previous observations of a high frequency of germline TP53 pathogenic/likely pathogenic variants in patients with osteosarcoma22,49 with double the sample. These data suggest that germline TP53 pathogenic/likely pathogenic variants are associated with a younger age at diagnosis, an axial tumor location, and worse survival.
Previous reports of smaller numbers of patients with osteosarcoma have suggested enrichment of pathogenic/likely pathogenic variants in other autosomal-dominant cancer-susceptibility genes.24,25 We similarly identified pathogenic/likely pathogenic variants in the genes RB1, APC, MSH2, and PALB2 (OMIM 610355). We additionally report that 6.5% of patients with osteosarcoma unselected for family history had a pathogenic/likely pathogenic variant in a gene associated with a cancer-predisposing syndrome that is associated with osteosarcoma.
This study identified several new candidate osteosarcoma-susceptibility genes that are worthy of additional study, including CDKN2A, MEN1, VHL, POT1, APC, MSH2, and ATRX. Notably, CDKN2A had the second highest frequency (1.2%) of pathogenic/likely pathogenic variants in patients with osteosarcoma and has not been associated with pediatric cancer; however, it has been associated with melanoma and pancreatic cancer.64,65,66 A germline variant located 150 kilobases upstream of CDKN2A has been associated with the risk of canine osteosarcoma,67 which has biologic similarity to human osteosarcoma.68 Somatic CDKN2A loss is an important somatic event in human osteosarcomas.48,49,69,70 Four of 6 of the CDKN2A pathogenic/likely pathogenic variants (p.Asp125His, p.Gly101Trp, p.Ile49Ser, and p.Ile49Thr) observed in the patients with osteosarcoma have been previously associated with a predisposition for melanoma or pancreatic cancer.71,72,73,74
The X-linked cancer-susceptibility genes have not been previously associated with osteosarcoma and were identified in both the discovery and replication patient sets. We report 2 patients with pathogenic DKC1 variants (c.1223C>T and c.-142C>G) that are known to cause dyskeratosis congenita,75,76 which is associated with a high risk of select solid tumors77,78 but has not been previously associated with osteosarcoma.79 We identified WAS loss-of-function mutations, which are associated with Wiskott-Aldrich syndrome and have previously been associated with lymphoma susceptibility but not with osteosarcoma. Our data further associate osteosarcoma with rare variants in ATRX, which has been reported to have somatic driver mutations in osteosarcoma.48 Osteosarcoma has been reported in 5 children with the rare ATR-X genetic disorder, which is associated with heterozygous pathogenic germline variants in ATRX.62,80,81 One of these previously reported patients with ATR-X syndrome developed osteosarcoma61,62 and had a worse outcome, which is comparable with the osteosarcoma patient who had the same ATRX variant.
Strengths and Limitations
A strength of our study is that, to our knowledge, the 1244 patients with osteosarcoma in our analysis represent the largest set of patients with a single solid pediatric cancer to be evaluated for cancer-susceptibility gene pathogenic variants to date and consequently provide more precise pathogenic/likely pathogenic carrier prevalence estimates. The use of internal individuals without cancer who were jointly called with the patients improved the whole-exome sequencing quality-control measures.
The limitations of our study include the inability to assess family history, the incomplete data on important clinical variables from all centers, and the use of ExAC whole-exome sequencing data, which could not be directly used for discovery analyses or burden testing owing to distinct biases associated with its accumulation of data from many sources. In addition, 284 of the 1004 patients in the discovery set were derived from the Childhood Cancer Survivor Study, which could have resulted in survival bias for this subset. Notably, patients in the Childhood Cancer Survivor Study had a lower carrier frequency of TP53 pathogenic/likely pathogenic variants compared with all other patients (2.8% vs 5.0%, respectively; Fisher exact P = .17).
Conclusions
We report that an estimated 28.0% of patients with osteosarcoma carried a rare germline pathogenic/likely pathogenic variant in a cancer-susceptibility gene, and more patients carried likely damaging variants in autosomal-dominant cancer-susceptibility genes. We confirm known associations and identify new genes that provide insight into the biology of osteosarcoma. Our findings have important implications for the genetic testing of patients, especially younger patients, who are newly diagnosed with osteosarcoma because these patients were more likely to have a potentially clinically relevant disease-associated pathogenic/likely pathogenic variant. We acknowledge that our estimates, particularly those based on in silico analyses, may be high because functional studies are required to prove pathogenicity.
Our data underscore the high frequency of potentially actionable cancer risk variants in patients with osteosarcoma, suggesting a need for further preventive and early detection strategies as well as a consideration of cascade genetic testing for the patient and the entire family.82,83,84 We note that individuals harboring Li-Fraumeni syndrome–associated TP53 mutations benefit from active screening, which could translate into improved outcomes.85,86 Further studies are needed to refine our observations and identify optimal approaches to genetic testing and counseling for patients with osteosarcoma.
eMethods. Study Population, Whole-Exome Sequencing, Pathogenicity Scoring and In Silico Analysis, and Statistical Analyses
eTable 1. Description of Participating Studies
eTable 2. Description of 238 Cancer-Susceptibility Genes Evaluated
eTable 3. Osteosarcoma Candidate Genes and Known Pediatric/Osteosarcoma Somatically Altered Genes Evaluated
eTable 4. Details of Criteria for Classification of Pathogenicity Categories
eTable 5. Details of Pathogenic and Likely Pathogenic Variants in 1004 Patients With Osteosarcoma in Discovery Set
eTable 6. Prevalence of Potentially Pathogenic Variants in 238 Cancer-Susceptibility Genes in 1244 Patients With Osteosarcoma Compared With 28 235 Individuals Without Cancer
eTable 7. Top Statistically Significant Cancer-Susceptibility Genes With a Higher Burden of Variants
eTable 8. Characteristics of Individuals With TP53 Pathogenic/Likely Pathogenic Variants and All Pathogenic/Likely Pathogenic Cancer-Susceptibility Gene Variants in 1004 Patients With Osteosarcoma in the Discovery Set
eTable 9. Pathways Significantly Enriched for 101 Cancer-Susceptibility Genes With 1 or More Pathogenic/Likely Pathogenic Variants in 1004 Patients With Osteosarcoma
eTable 10. Number of Rare Variants by Pathogenicity Score in 1004 Patients With Osteosarcoma and 1062 Individuals Without Cancer for 238 Cancer-Susceptibility Genes and Pathogenic/Likely Pathogenic Variants in 240 Patients in the Replication Set
eTable 11. Predicted Pathogenic and Likely Pathogenic Variants in 240 Patients in the Replication Set
eTable 12. Top Statistically Significant Candidate Genes With a Higher Burden of Variants
eFigure 1. Mean Whole-Exome Sequencing Coverage per Sample
eFigure 2. Frequency of Rare Pathogenic/Likely Pathogenic Variants in the TP53 Gene and the Other Cancer-Susceptibility Genes
eFigure 3. Pathway Enrichment Illustrated With a Network Analysis and Protein-Protein Interactions for 101 Cancer-Susceptibility Genes With 1 or More Pathogenic/Likely Pathogenic Variant Identified in Discovery Set of 1004 Patients With Osteosarcoma
eFigure 4. Lollipop Plots Illustrating Location of Variants Within the Specified Genes That Contain a Significantly Increased Burden (Number) of Rare Pathogenic/Likely Pathogenic Variants in Patients Compared With the Control Group by Protein/Functional Domain
eReferences
References
- 1.Ward E, DeSantis C, Robbins A, Kohler B, Jemal A. Childhood and adolescent cancer statistics, 2014. CA Cancer J Clin. 2014;64(2):83-103. doi: 10.3322/caac.21219 [DOI] [PubMed] [Google Scholar]
- 2.Mirabello L, Troisi RJ, Savage SA. Osteosarcoma incidence and survival rates from 1973 to 2004: data from the Surveillance, Epidemiology, and End Results program. Cancer. 2009;115(7):1531-1543. doi: 10.1002/cncr.24121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mirabello L, Troisi RJ, Savage SA. International osteosarcoma incidence patterns in children and adolescents, middle ages and elderly persons. Int J Cancer. 2009;125(1):229-234. doi: 10.1002/ijc.24320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mirabello L, Pfeiffer R, Murphy G, et al. Height at diagnosis and birth-weight as risk factors for osteosarcoma. Cancer Causes Control. 2011;22(6):899-908. doi: 10.1007/s10552-011-9763-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arora RS, Kontopantelis E, Alston RD, Eden TO, Geraci M, Birch JM. Relationship between height at diagnosis and bone tumours in young people: a meta-analysis. Cancer Causes Control. 2011;22(5):681-688. doi: 10.1007/s10552-011-9740-9 [DOI] [PubMed] [Google Scholar]
- 6.Ron E. Cancer risks from medical radiation. Health Phys. 2003;85(1):47-59. doi: 10.1097/00004032-200307000-00011 [DOI] [PubMed] [Google Scholar]
- 7.Calvert GT, Randall RL, Jones KB, Cannon-Albright L, Lessnick S, Schiffman JD. At-risk populations for osteosarcoma: the syndromes and beyond. Sarcoma. 2012;2012:152382. doi: 10.1155/2012/152382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gianferante DM, Mirabello L, Savage SA. Germline and somatic genetics of osteosarcoma—connecting aetiology, biology and therapy. Nat Rev Endocrinol. 2017;13(8):480-491. doi: 10.1038/nrendo.2017.16 [DOI] [PubMed] [Google Scholar]
- 9.Li FP, Fraumeni JF Jr, Mulvihill JJ, et al. A cancer family syndrome in twenty-four kindreds. Cancer Res. 1988;48(18):5358-5362. [PubMed] [Google Scholar]
- 10.Tinat J, Bougeard G, Baert-Desurmont S, et al. 2009 version of the Chompret criteria for Li Fraumeni syndrome. J Clin Oncol. 2009;27(26):e108-e109. doi: 10.1200/JCO.2009.22.7967 [DOI] [PubMed] [Google Scholar]
- 11.Lohmann D. Retinoblastoma. Adv Exp Med Biol. 2010;685:220-227. doi: 10.1007/978-1-4419-6448-9_21 [DOI] [PubMed] [Google Scholar]
- 12.Chauveinc L, Mosseri V, Quintana E, et al. Osteosarcoma following retinoblastoma: age at onset and latency period. Ophthalmic Genet. 2001;22(2):77-88. doi: 10.1076/opge.22.2.77.2228 [DOI] [PubMed] [Google Scholar]
- 13.Lipton JM, Federman N, Khabbaze Y, et al. ; Diamond-Black Anemia Registry . Osteogenic sarcoma associated with Diamond-Blackfan anemia: a report from the Diamond-Blackfan Anemia Registry. J Pediatr Hematol Oncol. 2001;23(1):39-44. doi: 10.1097/00043426-200101000-00009 [DOI] [PubMed] [Google Scholar]
- 14.Vlachos A, Rosenberg PS, Atsidaftos E, Alter BP, Lipton JM. Incidence of neoplasia in Diamond Blackfan anemia: a report from the Diamond Blackfan Anemia Registry. Blood. 2012;119(16):3815-3819. doi: 10.1182/blood-2011-08-375972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kansara M, Thomas DM. Molecular pathogenesis of osteosarcoma. DNA Cell Biol. 2007;26(1):1-18. doi: 10.1089/dna.2006.0505 [DOI] [PubMed] [Google Scholar]
- 16.Siitonen HA, Sotkasiira J, Biervliet M, et al. The mutation spectrum in RECQL4 diseases. Eur J Hum Genet. 2009;17(2):151-158. doi: 10.1038/ejhg.2008.154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ishikawa Y, Miller RW, Machinami R, Sugano H, Goto M. Atypical osteosarcomas in Werner syndrome (adult progeria). Jpn J Cancer Res. 2000;91(12):1345-1349. doi: 10.1111/j.1349-7006.2000.tb00924.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.German J. Bloom’s syndrome. XX. the first 100 cancers. Cancer Genet Cytogenet. 1997;93(1):100-106. doi: 10.1016/S0165-4608(96)00336-6 [DOI] [PubMed] [Google Scholar]
- 19.Savage SA, Mirabello L, Wang Z, et al. Genome-wide association study identifies two susceptibility loci for osteosarcoma. Nat Genet. 2013;45(7):799-803. doi: 10.1038/ng.2645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Savage SA, Mirabello L. Using epidemiology and genomics to understand osteosarcoma etiology. Sarcoma. 2011;2011:548151. doi: 10.1155/2011/548151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Musselman JR, Bergemann TL, Ross JA, et al. Case-parent analysis of variation in pubertal hormone genes and pediatric osteosarcoma: a Children’s Oncology Group (COG) study. Int J Mol Epidemiol Genet. 2012;3(4):286-293. [PMC free article] [PubMed] [Google Scholar]
- 22.Mirabello L, Yeager M, Mai PL, et al. Germline TP53 variants and susceptibility to osteosarcoma. J Natl Cancer Inst. 2015;107(7):djv101. doi: 10.1093/jnci/djv101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ballinger ML, Goode DL, Ray-Coquard I, et al. ; International Sarcoma Kindred Study . Monogenic and polygenic determinants of sarcoma risk: an international genetic study. Lancet Oncol. 2016;17(9):1261-1271. doi: 10.1016/S1470-2045(16)30147-4 [DOI] [PubMed] [Google Scholar]
- 24.Zhang J, Walsh MF, Wu G, et al. Germline mutations in predisposition genes in pediatric cancer. N Engl J Med. 2015;373(24):2336-2346. doi: 10.1056/NEJMoa1508054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grobner SN, Worst BC, Weischenfeldt J, et al. ; ICGC PedBrain-Seq Project; ICGC MMML-Seq Project . The landscape of genomic alterations across childhood cancers. Nature. 2018;555(7696):321-327. doi: 10.1038/nature25480 [DOI] [PubMed] [Google Scholar]
- 26.Exome Aggregation Consortium; Exome Aggregation Consortium . (ExAC) [online database]. https://omictools.com/exac-browser-tool. Published January 13, 2015. Accessed September 18, 2018.
- 27.Mirabello L, Koster R, Moriarity BS, et al. A genome-wide scan identifies variants in NFIB associated with metastasis in patients with osteosarcoma. Cancer Discov. 2015;5(9):920-931. doi: 10.1158/2159-8290.CD-15-0125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morton LM, Sampson JN, Armstrong GT, et al. Genome-wide association study to identify susceptibility loci that modify radiation-related risk for breast cancer after childhood cancer. J Natl Cancer Inst. 2017;109(11):djx058. doi: 10.1093/jnci/djx058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mirabello L, Yu K, Berndt SI, et al. ; National Osteosarcoma Etiology Study Group . A comprehensive candidate gene approach identifies genetic variation associated with osteosarcoma. BMC Cancer. 2011;11:209. doi: 10.1186/1471-2407-11-209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dean M, Bendfeldt G, Lou H, et al. Increased incidence and disparity of diagnosis of retinoblastoma patients in Guatemala. Cancer Lett. 2014;351(1):59-63. doi: 10.1016/j.canlet.2014.04.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prorok PC, Andriole GL, Bresalier RS, et al. ; Prostate, Lung, Colorectal and Ovarian Cancer Screening Trial Project Team . Design of the prostate, lung, colorectal and ovarian (PLCO) cancer screening trial. Control Clin Trials. 2000;21(6)(suppl):273S-309S. doi: 10.1016/S0197-2456(00)00098-2 [DOI] [PubMed] [Google Scholar]
- 32.Calle EE, Rodriguez C, Jacobs EJ, et al. The American Cancer Society Cancer Prevention Study II Nutrition Cohort: rationale, study design, and baseline characteristics. Cancer. 2002;94(9):2490-2501. doi: 10.1002/cncr.101970 [DOI] [PubMed] [Google Scholar]
- 33.Landi MT, Consonni D, Rotunno M, et al. Environment And Genetics in Lung cancer Etiology (EAGLE) study: an integrative population-based case-control study of lung cancer. BMC Public Health. 2008;8:203. doi: 10.1186/1471-2458-8-203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mirabello L, Macari ER, Jessop L, et al. Whole-exome sequencing and functional studies identify RPS29 as a novel gene mutated in multicase Diamond-Blackfan anemia families. Blood. 2014;124(1):24-32. doi: 10.1182/blood-2013-11-540278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mirabello L, Khincha PP, Ellis SR, et al. Novel and known ribosomal causes of Diamond-Blackfan anaemia identified through comprehensive genomic characterisation. J Med Genet. 2017;54(6):417-425. doi: 10.1136/jmedgenet-2016-104346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim J, Luo W, Wang M, et al. Prevalence of pathogenic/likely pathogenic variants in the 24 cancer genes of the ACMG secondary findings v2.0 list in a large cancer cohort and ethnicity-matched controls. Genome Med. 2018;10(1):99. doi: 10.1186/s13073-018-0607-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shi J, Yang XR, Ballew B, et al. ; NCI DCEG Cancer Sequencing Working Group; NCI DCEG Cancer Genomics Research Laboratory; French Familial Melanoma Study Group . Rare missense variants in POT1 predispose to familial cutaneous malignant melanoma. Nat Genet. 2014;46(5):482-486. doi: 10.1038/ng.2941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rahman N. Realizing the promise of cancer predisposition genes. Nature. 2014;505(7483):302-308. doi: 10.1038/nature12981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boria I, Garelli E, Gazda HT, et al. The ribosomal basis of Diamond-Blackfan anemia: mutation and database update. Hum Mutat. 2010;31(12):1269-1279. doi: 10.1002/humu.21383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gazda HT, Preti M, Sheen MR, et al. Frameshift mutation in p53 regulator RPL26 is associated with multiple physical abnormalities and a specific pre-ribosomal RNA processing defect in Diamond-Blackfan anemia. Hum Mutat. 2012;33(7):1037-1044. doi: 10.1002/humu.22081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sankaran VG, Ghazvinian R, Do R, et al. Exome sequencing identifies GATA1 mutations resulting in Diamond-Blackfan anemia. J Clin Invest. 2012;122(7):2439-2443. doi: 10.1172/JCI63597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ripperger T, Bielack SS, Borkhardt A, et al. Childhood cancer predisposition syndromes—a concise review and recommendations by the Cancer Predisposition Working Group of the Society for Pediatric Oncology and Hematology. Am J Med Genet A. 2017;173(4):1017-1037. doi: 10.1002/ajmg.a.38142 [DOI] [PubMed] [Google Scholar]
- 43.Futreal PA, Coin L, Marshall M, et al. A census of human cancer genes. Nat Rev Cancer. 2004;4(3):177-183. doi: 10.1038/nrc1299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Forbes SA, Beare D, Boutselakis H, et al. COSMIC: somatic cancer genetics at high-resolution. Nucleic Acids Res. 2017;45(D1):D777-D783. doi: 10.1093/nar/gkw1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yu W, Clyne M, Khoury MJ, Gwinn M. Phenopedia and genopedia: disease-centered and gene-centered views of the evolving knowledge of human genetic associations. Bioinformatics. 2010;26(1):145-146. doi: 10.1093/bioinformatics/btp618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Behjati S, Tarpey PS, Haase K, et al. Recurrent mutation of IGF signalling genes and distinct patterns of genomic rearrangement in osteosarcoma. Nat Commun. 2017;8:15936. doi: 10.1038/ncomms15936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen X, Bahrami A, Pappo A, et al. ; St Jude Children’s Research Hospital–Washington University Pediatric Cancer Genome Project . Recurrent somatic structural variations contribute to tumorigenesis in pediatric osteosarcoma. Cell Rep. 2014;7(1):104-112. doi: 10.1016/j.celrep.2014.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kovac M, Blattmann C, Ribi S, et al. Exome sequencing of osteosarcoma reveals mutation signatures reminiscent of BRCA deficiency. Nat Commun. 2015;6:8940. doi: 10.1038/ncomms9940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Perry JA, Kiezun A, Tonzi P, et al. Complementary genomic approaches highlight the PI3K/mTOR pathway as a common vulnerability in osteosarcoma. Proc Natl Acad Sci U S A. 2014;111(51):E5564-E5573. doi: 10.1073/pnas.1419260111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Richards S, Aziz N, Bale S, et al. ; ACMG Laboratory Quality Assurance Committee . Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405-424. doi: 10.1038/gim.2015.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ionita-Laza I, Lee S, Makarov V, Buxbaum JD, Lin X. Sequence kernel association tests for the combined effect of rare and common variants. Am J Hum Genet. 2013;92(6):841-853. doi: 10.1016/j.ajhg.2013.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xie C, Mao X, Huang J, et al. KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011;39(web server issue):W316-W322. doi: 10.1093/nar/gkr483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rahmati S, Abovsky M, Pastrello C, Jurisica I. pathDIP: an annotated resource for known and predicted human gene-pathway associations and pathway enrichment analysis. Nucleic Acids Res. 2017;45(D1):D419-D426. doi: 10.1093/nar/gkw1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhu B, Mirabello L, Chatterjee N. A subregion-based burden test for simultaneous identification of susceptibility loci and subregions within. Genet Epidemiol. 2018;42(7):673-683. doi: 10.1002/gepi.22134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ognjanovic S, Olivier M, Bergemann TL, Hainaut P. Sarcomas in TP53 germline mutation carriers: a review of the IARC TP53 database. Cancer. 2012;118(5):1387-1396. doi: 10.1002/cncr.26390 [DOI] [PubMed] [Google Scholar]
- 56.Olivier M, Hollstein M, Hainaut P. TP53 mutations in human cancers: origins, consequences, and clinical use. Cold Spring Harb Perspect Biol. 2010;2(1):a001008. doi: 10.1101/cshperspect.a001008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Petitjean A, Mathe E, Kato S, et al. Impact of mutant p53 functional properties on TP53 mutation patterns and tumor phenotype: lessons from recent developments in the IARC TP53 database. Hum Mutat. 2007;28(6):622-629. doi: 10.1002/humu.20495 [DOI] [PubMed] [Google Scholar]
- 58.Zhou X, Edmonson MN, Wilkinson MR, et al. Exploring genomic alteration in pediatric cancer using ProteinPaint. Nat Genet. 2016;48(1):4-6. doi: 10.1038/ng.3466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Guha T, Malkin D. Inherited TP53 Mutations and the Li-Fraumeni syndrome. Cold Spring Harb Perspect Med. 2017;7(4):a026187. doi: 10.1101/cshperspect.a026187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang LL, Gannavarapu A, Kozinetz CA, et al. Association between osteosarcoma and deleterious mutations in the RECQL4 gene in Rothmund-Thomson syndrome. J Natl Cancer Inst. 2003;95(9):669-674. doi: 10.1093/jnci/95.9.669 [DOI] [PubMed] [Google Scholar]
- 61.Badens C, Lacoste C, Philip N, et al. Mutations in PHD-like domain of the ATRX gene correlate with severe psychomotor impairment and severe urogenital abnormalities in patients with ATRX syndrome. Clin Genet. 2006;70(1):57-62. doi: 10.1111/j.1399-0004.2006.00641.x [DOI] [PubMed] [Google Scholar]
- 62.Masliah-Planchon J, Levy D, Heron D, et al. Does ATRX germline variation predispose to osteosarcoma? three additional cases of osteosarcoma in two ATR-X syndrome patients. Eur J Hum Genet. 2018;26(8):1217-1221. doi: 10.1038/s41431-018-0147-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Waszak SM, Northcott PA, Buchhalter I, et al. Spectrum and prevalence of genetic predisposition in medulloblastoma: a retrospective genetic study and prospective validation in a clinical trial cohort. Lancet Oncol. 2018;19(6):785-798. doi: 10.1016/S1470-2045(18)30242-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Berwick M, Orlow I, Hummer AJ, et al. ; GEM Study Group . The prevalence of CDKN2A germ-line mutations and relative risk for cutaneous malignant melanoma: an international population-based study. Cancer Epidemiol Biomarkers Prev. 2006;15(8):1520-1525. doi: 10.1158/1055-9965.EPI-06-0270 [DOI] [PubMed] [Google Scholar]
- 65.Nelson AA, Tsao H. Melanoma and genetics. Clin Dermatol. 2009;27(1):46-52. doi: 10.1016/j.clindermatol.2008.09.005 [DOI] [PubMed] [Google Scholar]
- 66.McWilliams RR, Wieben ED, Chaffee KG, et al. CDKN2A germline rare coding variants and risk of pancreatic cancer in minority populations. Cancer Epidemiol Biomarkers Prev. 2018;27(11):1364-1370. doi: 10.1158/1055-9965.EPI-17-1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Karlsson EK, Sigurdsson S, Ivansson E, et al. Genome-wide analyses implicate 33 loci in heritable dog osteosarcoma, including regulatory variants near CDKN2A/B. Genome Biol. 2013;14(12):R132. doi: 10.1186/gb-2013-14-12-r132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Withrow SJ, Khanna C. Bridging the gap between experimental animals and humans in osteosarcoma. Cancer Treat Res. 2009;152:439-446. doi: 10.1007/978-1-4419-0284-9_24 [DOI] [PubMed] [Google Scholar]
- 69.Mohseny AB, Tieken C, van der Velden PA, et al. Small deletions but not methylation underlie CDKN2A/p16 loss of expression in conventional osteosarcoma. Genes Chromosomes Cancer. 2010;49(12):1095-1103. doi: 10.1002/gcc.20817 [DOI] [PubMed] [Google Scholar]
- 70.Nielsen GP, Burns KL, Rosenberg AE, Louis DN. CDKN2A gene deletions and loss of p16 expression occur in osteosarcomas that lack RB alterations. Am J Pathol. 1998;153(1):159-163. doi: 10.1016/S0002-9440(10)65556-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Holland EA, Schmid H, Kefford RF, Mann GJ. CDKN2A (p16[INK4a]) and CDK4 mutation analysis in 131 Australian melanoma probands: effect of family history and multiple primary melanomas. Genes Chromosomes Cancer. 1999;25(4):339-348. doi: [DOI] [PubMed] [Google Scholar]
- 72.Lal G, Liu L, Hogg D, Lassam NJ, Redston MS, Gallinger S. Patients with both pancreatic adenocarcinoma and melanoma may harbor germline CDKN2A mutations. Genes Chromosomes Cancer. 2000;27(4):358-361. doi: [DOI] [PubMed] [Google Scholar]
- 73.Goldstein AM, Chan M, Harland M, et al. ; Lund Melanoma Study Group; Melanoma Genetics Consortium (GenoMEL) . Features associated with germline CDKN2A mutations: a GenoMEL study of melanoma-prone families from three continents. J Med Genet. 2007;44(2):99-106. doi: 10.1136/jmg.2006.043802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hussussian CJ, Struewing JP, Goldstein AM, et al. Germline p16 mutations in familial melanoma. Nat Genet. 1994;8(1):15-21. doi: 10.1038/ng0994-15 [DOI] [PubMed] [Google Scholar]
- 75.Dokal I. Dyskeratosis congenita in all its forms. Br J Haematol. 2000;110(4):768-779. doi: 10.1046/j.1365-2141.2000.02109.x [DOI] [PubMed] [Google Scholar]
- 76.Vulliamy TJ, Marrone A, Knight SW, Walne A, Mason PJ, Dokal I. Mutations in dyskeratosis congenita: their impact on telomere length and the diversity of clinical presentation. Blood. 2006;107(7):2680-2685. doi: 10.1182/blood-2005-07-2622 [DOI] [PubMed] [Google Scholar]
- 77.Alter BP, Giri N, Savage SA, Rosenberg PS. Cancer in the National Cancer Institute inherited bone marrow failure syndrome cohort after fifteen years of follow-up. Haematologica. 2018;103(1):30-39. doi: 10.3324/haematol.2017.178111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bertuch AA. The molecular genetics of the telomere biology disorders. RNA Biol. 2016;13(8):696-706. doi: 10.1080/15476286.2015.1094596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Alter BP, Giri N, Savage SA, Rosenberg PS. Cancer in dyskeratosis congenita. Blood. 2009;113(26):6549-6557. doi: 10.1182/blood-2008-12-192880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ji J, Quindipan C, Parham D, et al. Inherited germline ATRX mutation in two brothers with ATR-X syndrome and osteosarcoma. Am J Med Genet A. 2017;173(5):1390-1395. doi: 10.1002/ajmg.a.38184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Smolle MA, Heitzer E, Geigl JB, et al. A novel mutation in ATRX associated with intellectual disability, syndromic features, and osteosarcoma. Pediatr Blood Cancer. 2017;64(10):e26522. doi: 10.1002/pbc.26522 [DOI] [PubMed] [Google Scholar]
- 82.Caswell-Jin JL, Zimmer AD, Stedden W, Kingham KE, Zhou AY, Kurian AW. Cascade genetic testing of relatives for hereditary cancer risk: results of an online initiative. J Natl Cancer Inst. 2019;111(1):95-98. doi: 10.1093/jnci/djy147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Krawczak M, Cooper DN, Schmidtke J. Estimating the efficacy and efficiency of cascade genetic screening. Am J Hum Genet. 2001;69(2):361-370. doi: 10.1086/321973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Walsh MF, Kennedy J, Harlan M, et al. Germline BRCA2 mutations detected in pediatric sequencing studies impact parents’ evaluation and care. Cold Spring Harb Mol Case Stud. 2017;3(6):a001925. doi: 10.1101/mcs.a001925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bojadzieva J, Amini B, Day SF, et al. Whole body magnetic resonance imaging (WB-MRI) and brain MRI baseline surveillance in TP53 germline mutation carriers: experience from the Li-Fraumeni Syndrome Education and Early Detection (LEAD) Clinic. Fam Cancer. 2018;17(2):287-294. doi: 10.1007/s10689-017-0034-6 [DOI] [PubMed] [Google Scholar]
- 86.Ballinger ML, Best A, Mai PL, et al. Baseline surveillance in Li-Fraumeni syndrome using whole-body magnetic resonance imaging: a meta-analysis. JAMA Oncol. 2017;3(12):1634-1639. doi: 10.1001/jamaoncol.2017.1968 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
eMethods. Study Population, Whole-Exome Sequencing, Pathogenicity Scoring and In Silico Analysis, and Statistical Analyses
eTable 1. Description of Participating Studies
eTable 2. Description of 238 Cancer-Susceptibility Genes Evaluated
eTable 3. Osteosarcoma Candidate Genes and Known Pediatric/Osteosarcoma Somatically Altered Genes Evaluated
eTable 4. Details of Criteria for Classification of Pathogenicity Categories
eTable 5. Details of Pathogenic and Likely Pathogenic Variants in 1004 Patients With Osteosarcoma in Discovery Set
eTable 6. Prevalence of Potentially Pathogenic Variants in 238 Cancer-Susceptibility Genes in 1244 Patients With Osteosarcoma Compared With 28 235 Individuals Without Cancer
eTable 7. Top Statistically Significant Cancer-Susceptibility Genes With a Higher Burden of Variants
eTable 8. Characteristics of Individuals With TP53 Pathogenic/Likely Pathogenic Variants and All Pathogenic/Likely Pathogenic Cancer-Susceptibility Gene Variants in 1004 Patients With Osteosarcoma in the Discovery Set
eTable 9. Pathways Significantly Enriched for 101 Cancer-Susceptibility Genes With 1 or More Pathogenic/Likely Pathogenic Variants in 1004 Patients With Osteosarcoma
eTable 10. Number of Rare Variants by Pathogenicity Score in 1004 Patients With Osteosarcoma and 1062 Individuals Without Cancer for 238 Cancer-Susceptibility Genes and Pathogenic/Likely Pathogenic Variants in 240 Patients in the Replication Set
eTable 11. Predicted Pathogenic and Likely Pathogenic Variants in 240 Patients in the Replication Set
eTable 12. Top Statistically Significant Candidate Genes With a Higher Burden of Variants
eFigure 1. Mean Whole-Exome Sequencing Coverage per Sample
eFigure 2. Frequency of Rare Pathogenic/Likely Pathogenic Variants in the TP53 Gene and the Other Cancer-Susceptibility Genes
eFigure 3. Pathway Enrichment Illustrated With a Network Analysis and Protein-Protein Interactions for 101 Cancer-Susceptibility Genes With 1 or More Pathogenic/Likely Pathogenic Variant Identified in Discovery Set of 1004 Patients With Osteosarcoma
eFigure 4. Lollipop Plots Illustrating Location of Variants Within the Specified Genes That Contain a Significantly Increased Burden (Number) of Rare Pathogenic/Likely Pathogenic Variants in Patients Compared With the Control Group by Protein/Functional Domain
eReferences