Abstract
Nucleic acid-based diagnostic tests often require isolation and concentration of nucleic acids from biological samples. Commercial purification kits are difficult to use in low-resource settings because of their cost and insufficient laboratory infrastructure. Several recent approaches based on the use of magnetic beads offer a potential solution but remain limited to small volume samples. We have developed a simple and low-cost nucleic acid extraction method suitable for isolation and concentration of nucleic acids from small or large sample volumes. The method uses magnetic beads, a transfer pipette, steel wool, and an external magnet to implement high-gradient magnetic separation (HGMS) to retain nucleic acid-magnetic bead complexes within the device’s steel wool matrix for subsequent processing steps. We demonstrate the method’s utility by extracting tuberculosis DNA from both sputum and urine, two typical large volume sample matrices (5–200 mL), using guanidine-based extraction chemistry. Our HGMS-enabled extraction method is statistically indistinguishable from commercial extraction kits when detecting a spiked 123-base DNA sequence. For our HGMS-enabled extraction method, we obtained extraction efficiencies for sputum and urine of approximately 10 and 90%, whereas commercial kits obtained 10–17 and 70–96%, respectively. We also used this method previously in a blinded sample preparation comparison study published by Beall et al., 2019. Our manual extraction method is insensitive to high flow rates and sample viscosity, with capture of ∼100% for flow rates up to 45 mL/min and viscosities up to 55 cP, possibly making it suitable for a wide variety of sample volumes and types and point-of-care users. This HGMS-enabled extraction method provides a robust instrument-free method for magnetic bead-based nucleic acid extraction, potentially suitable for field implementation of nucleic acid testing.
Keywords: sample preparation, low-resource, nucleic acid extraction, high-gradient magnetic separation, magnetic bead separation, qPCR, tuberculosis
1. Introduction
Tuberculosis (TB) remains a global challenge with an estimated 10 million infections and 1.3 million deaths per annum worldwide.1 Diagnosis of active, transmissible infections remains a significant public health challenge, particularly in low-resource settings.2,3 Sputum is the standard patient sample used in both traditional microscopic inspection2 and nucleic acid-based tests such as GeneXpert.4 However, some patient populations, including children and HIV-positive individuals,5 have difficulty producing sputum. Therefore, there is interest in assessing other potential sample types. Urine, in particular, is easily and noninvasively obtained. Unfortunately, the highly dilute number of targeted biomarkers available for detection and the presence of inhibitors of downstream detection methods limit its utility in nucleic acid-based testing. One fundamental limitation to improved diagnostic sensitivity remains the development of sample extraction and concentration methods, which convert dilute biomarkers into pure, inhibitor-free samples appropriate for downstream detection.
Magnetic beads have become a valuable tool in laboratory separations because of their low cost, tunable surface chemistries, and the use of magnets for isolation using simple processing steps. Because of this, magnetic bead biomarker isolation assays have been applied to liquid sample handling and processing in point-of-care biomarker detection assays.6−9 Furthermore, magnetic beads are easily added to a variety of liquid samples, providing a high surface area for specific chemical capture of biomarkers, which can then be recollected to purify and concentrate biomarkers of interest.10
One of the critical features in magnetic bead-based extraction methods is the efficient transfer of magnetic beads from one processing solution to the next. The most common approach for magnetic bead processing is difficult to implement in low-resource settings. This approach uses stationary magnets to apply a magnetic field across a sample volume to physically separate magnetic beads and bound analyte(s) from solution. After separation, the fluid phase is removed, and subsequent processing solutions are added; after removing the magnetic field and mixing the beads into the new solution, the separation process is repeated. Although this method can be automated using robotics, without robotics, sample processing using this method is labor-intensive, requires careful removal of solutions by pipetting, and is inefficient for processing large volume samples.
Alternatively, magnetic beads can be moved between different processing solutions while the liquid solutions remain stationary. The advantage of this approach is that it decreases the number of manual steps, eliminates the requirement of liquid handling, and allows for simple automation of bead manipulation.7,11−13 The challenge with this approach, however, is maximizing the applied magnetic force on the beads, which is needed to efficiently transfer magnetic beads across fluid interfaces between processing solutions while minimizing bead loss. Despite this shortcoming, at least one sample-to-answer system based on this approach is commercially available, the Cobas Liat polymerase chain reaction (PCR) System (Roche Molecular Diagnostics, Pleasanton, CA).
The third potential handling method for magnetic bead processing is based on a flow-through design. In this approach, a moving suspension of magnetic beads flows through a magnetic field, and beads are collected on the inside wall of the tube. Successive processing solutions are then applied by flowing through the tube. However, unless the fluid is moved at a very low flow rate, the drag forces of the fluid flow dominate over the magnetic forces on the beads, resulting in low bead capture and retention. Whereas this has the potential for processing large sample volumes, there is a trade-off between processing speed and bead retention. Implementing this approach requires the incorporation of a highly robust magnetic bead capture and retention method beyond that achievable with just an external magnet.
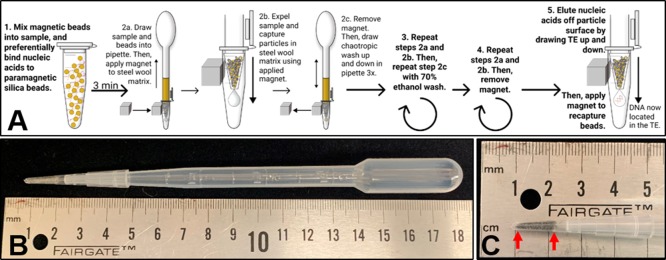
In a previous nucleic acid extraction study, we used a transfer pipette as a way to efficiently contain and mix lyophilized DNA-binding reagents and magnetic beads with patient urine samples before automated extraction of TB DNA.13 Wanting to further expand the use of the transfer pipette for simple and contained sample handling, we sought to develop a flow-through magnetic bead separation platform by incorporating a highly efficient method to retain the paramagnetic beads and their surface-bound nucleic acids within the transfer pipette. This report describes the incorporation of a ferromagnetic matrix, such as steel wool, into the flow path of the magnetic bead suspension and the use of an external magnet to capture paramagnetic beads on the surface of the matrix (Figure 1). Once the magnetic bead-biomarker complex is magnetically separated from the fluid suspension, subsequent processing steps are performed using the transfer pipette squeeze bulb to expose prealiquotted processing liquids to the bead surface by flow-through fluid exchange, ultimately yielding a purified and concentrated biomarker.
Figure 1.
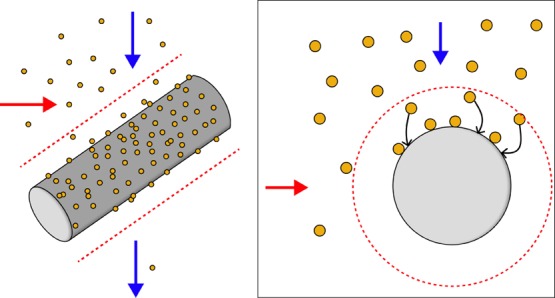
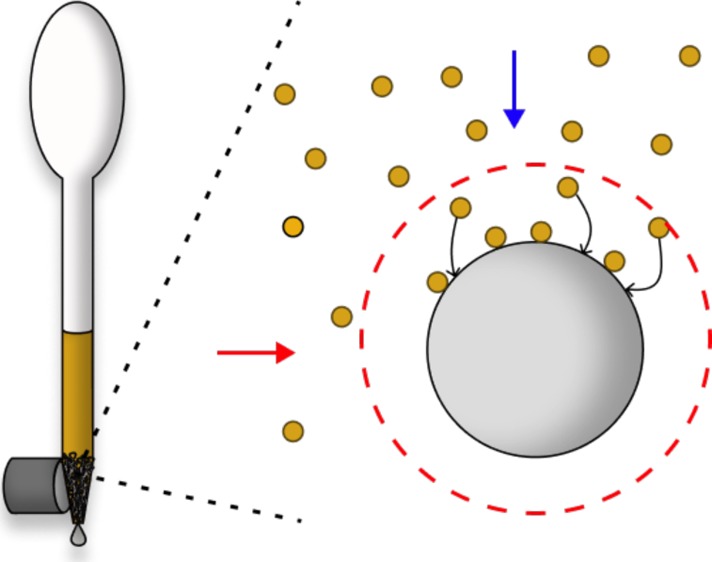
Simplified schematic showing the capture of paramagnetic beads using HGMS by a single matrix wire. An external magnetic field is applied (red horizontal arrow) to magnetize a ferromagnetic matrix. When a paramagnetic bead suspension flows through a magnetized matrix (blue vertical arrow), the high-gradient magnetic field around the wire strands in the matrix (red dashed line) creates a magnetic force on the beads that dominate over the viscous drag force in the region near the matrix, favoring bead capture on the wire surface.
As early as the 1930s,14 the use of a ferromagnetic matrix for paramagnetic particle capture from a flowing fluid stream is described in the literature as high-gradient magnetic separation (HGMS).15 This process and theory were implemented in the 1970s for mining and sewage applications as a means to capture weakly paramagnetic materials, such as CuO, from a flowing mine slurry or to remove paramagnetic contaminants from flowing sewage or water supplies.15−19 Previous reports using HGMS for biological applications focused on direct separation of erythrocytes from whole blood by chemically converting the hemoglobin in the cell from a diamagnetic to paramagnetic state.20,21 Using columns of loosely packed steel wool, the paramagnetic cells were efficiently captured in the matrix in the presence of an externally applied magnetic field. Miltenyi et al. then went on to modify the steel wool/immunospecific magnetic particle separation system22 to develop magnetic activated cell sorting (MACS) in the 1990s.23 These reusable columns were first composed of steel wool,23,24 which was then followed by stacked magnetic spheres with samples passing between them.25 Miltenyi’s MACS is the most prevalent use of HGMS in biological systems.
The method described in this paper was used previously and applied in a blinded sample preparation study sponsored by the Bill and Melinda Gates Foundation. In that study, Beall et al.26 compared our HGMS-enabled method to five other commercial extraction methods for total nucleic acid purification from sputum, whole blood, and stool. However, because the methods were not reported due to conflicts with commercial proprietary interests of the commercial participants, we describe here the details of the methods we used in this comparison study.
As described in this report, the HGMS phenomenon is robust, and for a variety of flows produced, the capture of paramagnetic beads in the steel wool matrix is consistently near 100%. The approach also offers the advantage of magnetic purification of biomarkers with minimal external contamination during processing. The device performance and the low cost of materials suggest that the HGMS phenomenon may be a promising approach for efficiently extracting nucleic acids from patient samples in low-resource settings.
2. Materials and Methods
2.1. Preparation of Synthetic Biological Samples
Synthetic sputum was prepared using a protocol generously provided by the nonprofit global health organization PATH (Seattle, WA): 114 mM NaCl, 33 mg/mL bovine serum albumin, 3.6 mg/mL phosphatidylcholine (Millipore Sigma, P3556), 4.7 mM CaCl2, 47 mg/mL mucin from porcine stomach (Millipore Sigma, M2378), 6 mg/mL salmon sperm DNA (Millipore Sigma, D1626), and 2 mM sodium azide. These concentrations are based on sputum component concentrations as determined by Sanders et al.27 The ingredients were mixed with a stir bar at 4 °C overnight and then stored at 4 °C. Deidentified, disease-negative, residual urine samples were obtained from the Vanderbilt University Molecular Infectious Disease Laboratory. An exemption from Institutional Review Board oversight was granted by the Vanderbilt University Institutional Review Board for use of these samples. Fifteen milliliters of each sample was pooled, pipetted into 1 mL aliquots, and stored at −80 °C. The urine samples were pooled to minimize variability of PCR inhibitors expected among individual urine samples. These samples were thawed and allowed to equilibrate to room temperature before spiking with DNA. Synthetic sputum was stored at 4 °C and warmed to room temperature with gentle mixing before spiking with DNA. One hundred microliters of synthetic sputum or pooled urine was spiked with 5 μL containing a total of 5 × 106 copies of a synthetic 123-base DNA oligomer of the IS6110 gene found in Mycobacterium tuberculosis (Integrated DNA Technologies, Coralville, IA). IS6110 is a variably repeating DNA insertion element found in M. tuberculosis and is used as a specific diagnostic marker for infection.5 The 123-mer sequence of the IS6110 insertion sequence was previously reported by Ogusku and Salem.28
2.2. Extraction Chemistry for Biological Samples
The steps for the extraction procedure are illustrated schematically in Figure 2A. A DNA-spiked sample was combined with 300 μL of binding buffer [4 M guanidine thiocyanate, 10 mM Tris HCl (pH 8), 1 mM ethylenediamine tetraacetic acid (EDTA, pH 8), and 0.5% Triton X-100], 300 μL of isopropanol, and 3 μL of β-mercaptoethanol in a 1.5 or 2 mL Eppendorf tube and mixed through inversion. For urine samples only, 5.6 μg of poly-A carrier RNA (Qiagen, 1017647) was also added into the extraction mixture. Then, 2 mg (50 μL) of MyOne Silane Dynabeads (Thermo Fisher, 37002D) was added to the sputum or urine solution, mixed through inversion, and incubated at room temperature for 3 min, with inversions every minute to maintain bead suspension. A 200 μL pipette tip (Fisher Scientific, 02-707-505) containing 17 ± 1 mg of alloy 434 stainless steel wool (Lustersheen-online.com, SKU16162) was affixed to the end of a 3.2 mL transfer pipette (Fisher Scientific, 13-711-9D). The very bottom of the 200 μL tip was trimmed to remove the void space located below the steel wool capture matrix (Figure 2B and C). The solution was then drawn up and down using the squeeze bulb of the transfer pipette. Once well-mixed and drawn into the pipette bulb, a magnet (K&J Magnetics, B666-N52) was applied to the steel wool matrix through the wall of the sample tube. As the bead solution was dispensed back into the original sample tube, the beads were captured in the magnetized matrix. Flow-through was discarded, and the magnet was removed. Next, the beads were washed in the transfer pipette by passing 1.5 mL of chaotropic wash [84% ethanol, 640 mM guanidine thiocyanate, 1.6 mM Tris HCl (pH 8), and 160 mM EDTA (pH 8)] up and down through the pipette three times. The total volume was then drawn into the transfer pipette, and the beads were magnetically captured through the wall of a 2 mL Eppendorf tube as previously described. Flow-through was discarded, and the magnet was removed. The previous step was repeated with 1.5 mL of 70% ethanol wash, and the flow-through was discarded. The pipettes were then allowed to sit upright in a clean Eppendorf tube for 1–2 min to allow any residual rinse liquid to pool into the pipette tip, which was then expelled while maintaining bead capture in the steel wool matrix with the externally applied magnet. In addition, for sputum samples only, an additional rinse step was incorporated before eluting the nucleic acids off the beads. To perform this rinse, the magnet was applied to the matrix through the tube wall, and 100 μL of 10 mM Tris-HCl and 1 mM EDTA (TE) buffer, pH 8 was gently drawn up and then immediately expelled to rinse the beads without disturbing them. After rinsing, the magnet was removed, and 50 μL of TE, pH 8 was gently drawn up and down for approximately 1 min to elute the nucleic acids (approximately 20 times). The solution was then drawn into the pipette, and the beads were captured in the matrix using the magnet placed along the outside of a clean collection tube, into which the eluate was saved; real-time PCR (qPCR) was performed on the final sample eluate. Any beads that may have passed into the eluate were left in the sample and not removed. A single user processed triplicate samples in parallel in approximately 10–15 min. The experiment was also performed in triplicate samples with no spiked IS6110 DNA, and no DNA was detected in these samples.
Figure 2.
Design and detailed workflow of the HGMS-enabled steel wool extraction device. (A) Five processing steps of HGMS-enabled nucleic acid extraction. DNA is adsorbed to silica-coated paramagnetic beads using guanidine-based chemistry. Once bound, the solution is drawn into the transfer pipette, and an external magnet is applied to capture the magnetic beads in the steel wool matrix tip as the fluid flows through. The magnet is then removed to allow for release of the beads, which washes the bead surface and the bound DNA. The magnet is then reapplied to recapture magnetic beads after each wash step. In the final step, the nucleic acids are eluted off the beads while the magnetic beads are retained in the steel wool. (B) HGMS-enabled steel wool separator. (C) Steel wool capture matrix. The matrix is placed between the two graduations on the pipette tip (indicated by red arrows), and the excess is removed from the bottom.
Commercial extractions were also performed for comparison using the DNeasy Blood and Tissue Kit (Qiagen, 69504) for synthetic sputum and the QIAamp Viral RNA Mini Kit for urine (Qiagen, 52904) as per the manufacturer’s protocol. Both of these kits use guanidine-based chemistry to bind nucleic acids to the silica centrifuge columns. Qiagen recommends that the QIAamp Viral RNA Mini Kit be used for DNA extraction from urine because the included lysis buffer, AVL (Qiagen, 19073), is optimized to inactivate the PCR inhibitors found in urine, unlike buffers found in their other commercially available kits. Extraction of both sputum and urine were also performed as per the manufacturer’s protocol, with the Chargeswitch gDNA Mini Tissue Kit (Thermo Fisher, CS11204), which is a magnetic bead-based extraction kit that uses guanidine-free extraction chemistry.
2.3. Determination of Extraction Efficiency Using qPCR
To determine the efficiency of the nucleic acid extractions, qPCR was performed using the Quanta UltraPlex 1-Step ToughMix (4×) (VWR, 10804-944) according to the manufacturer’s protocol. We used this kit to most closely match the published methods performed in the study by Beall et al.26 The 123-base IS6110 sequence from M. tuberculosis was amplified using the forward primer 5′-CCTGCGAGCGTAGGCGT-3′, reverse primer 5′CTCGTCCAGCGCCGCTT-3′, and probe 5′-/56-FAM/CGACACATA/ZEN/GGTGAGGTCTGCTAC/3IABkFQ/-3′. All primers and probes were synthesized by Integrated DNA Technologies (Coralville, IA). The 20-mer primers in the study by Ogusku and Salem28 were modified by trimming the 3′-ends to make 17-mers, which prevented the potential formation of primer dimers or hairpins.
The concentration of the primers and probes in the final PCR reaction was 600 nM of forward and reverse primers and 400 nM of probe. To the 1× master mix, 5 μL of the extracted sample was added for a total reaction volume of 20 μL. The samples were run according to the manufacturer’s protocol; for the initial step, the samples were held at 50 °C for 10 min, followed by a hold at 95 °C for 3 min. The samples were then cycled from 95 °C for 30 s to 60 °C for 60 s for a total of 45 cycles in a Rotor-Gene Q thermal cycler (Qiagen, Germantown, MD). In addition to a no-template negative control, an IS6110 standard curve using 5 × 106 copies/reaction diluted 10-fold sequentially to 5 × 103 copies/reaction was included for conversion of Ct values to copies/reaction. Control samples containing 5 × 106 copies of DNA in a total of 50 μL of TE were used to quantify the DNA eluate in a “perfect extraction” with 100% recovery and detection, which was used to calculate the percentage extraction efficiency using postextraction qPCR. Exact volumes of the final eluates were measured for accurate calculation of DNA recovery before performing PCR, because it was not uncommon for a few microliters of the eluate to remain in the steel wool matrix after expulsion because of liquid surface tension. The PCR efficiencies of the standards, controls, and extracted samples were calculated using LinRegPCR software (available at http://linregpcr.nl), setting the fluorescence threshold above the fluorescence values of the negatives,29,30 and a statistical comparison was performed.
2.4. Baseline Bead Capture Protocol for Aqueous Solutions
A total of 800 μg (20 μL) of MyOne Silane Dynabeads (Thermo Fisher, 37002D) was mixed into 3 mL of 1× phosphate-buffered saline (PBS) + 0.1% Tween-20 (final bead concentration of 267 μg/mL) and gently vortexed to combine. The solution was drawn into a 3.2 mL transfer pipette (Fisher Scientific, 13-711-9D), and 35 mg of Grade 000 steel wool (Global Material Technologies, Inc., Buffalo Grove, IL) was placed in a 200 μL pipette tip (Fisher Scientific, 02-707-505) at an approximate packing density of 0.91 g/mm3. The very bottom of the 200 μL tip was trimmed to remove the void space located below the steel wool matrix (Figure 2C), and then the tip was affixed to the end of the transfer pipette (Figure 2B). With a magnet (K&J Magnetics, DCX0) beside the steel wool matrix, the bead solution was expelled (Figure S1, Video S1). Unless otherwise noted, flow rates less than 20 mL/min were used to process the samples.
To estimate the quantity of beads captured in the steel wool matrix, the beads in the flow-through were concentrated using centrifugation at 3086g for 5 min and resuspended in an appropriate volume of PBS-T. This volume was variable and was selected based on the quantity of beads in the sample. This was done to make certain the absorbance was within the linear range of the device and ensure accuracy of the measurement. Absorbance at 700 nm was measured in at least triplicate on a Nanodrop Spectrophotometer ND-1000, and the quantity of beads was calculated using a standard curve (Figure S2). The quantity of beads captured was estimated by subtracting the quantity of beads located in the flow-through from the quantity of starting beads.
2.5. Effect of Steel Wool Matrix on Paramagnetic Bead Capture
The effect of steel wool quantity on capture was measured by including increasing quantities from 0 to 35 mg of Grade 000 steel wool placed in the pipette tip and performing the protocol for bead capture. Packing density was maintained as described across all steel wool masses. The effect of steel wool grade and alloy on capture was measured by incorporating 35 mg of steel wool of Grade 0000 (Lustersheen-online.com, SKU16246214), 000, 1, and 3 (Red Devil, Inc., 3332), steel wool alloys 434 (Lustersheen-online.com, SKU16162) and 316L (Lustersheen-online.com, SKU162751), aluminum (Global Material Technologies, Inc., 166510-P) or copper (Rogue River Tools, 731847303978) wool in the pipette tip, or no matrix at all. Packing density was maintained across the different grades and materials.
2.6. Effect of Magnet Properties on Paramagnetic Bead Capture
The effect of magnet surface field on capture was investigated by measuring the capture with cylinder magnets 3/4” (1.91 cm) in diameter with surface fields increasing from 548 to 6180 Gauss (K&J Magnetics, DC01, DCH1, DC2, DC3, DC4, DC8, and DCX0) and performing the capture protocol outlined. The effect of magnet size on capture was also investigated by incorporating magnets increasing in size from 1/8″ cube (0.32 cm) to 1″ cube (2.54 cm), with a constant surface field of 6451 Gauss (K&J Magnetics, B222-N52, B333-N52, B444-N52, B666-N52, B888-N52, BCCC-N52, and BX0X0X0-N52) and performing the capture protocol outlined above.
2.7. Effect of Flow Rate and Sample Viscosity on Paramagnetic Bead Capture
The effect of flow rate on paramagnetic bead capture was measured using a stock solution of Dynabeads at a concentration of 267 μg/mL in 1× PBS + 0.1% Tween-20. This solution was placed into a 60 mL syringe and loaded onto a syringe pump (RK TCI-II Syringe Pump). A 35 mg Grade 000 steel wool pellet was placed at the end of the pipette tip and trimmed as specified. This end was sealed onto a flexible tube affixed to the syringe. A total volume of 3 mL of the bead solution was passed through the steel wool with a magnet present (K&J Magnetics, DCX0). The uncaptured beads were centrifuged and measured as previously outlined. In between each trial, the solution in the syringe was mixed through inversion to maintain a well-distributed solution. For flows greater than 20 mL/min, the syringe was operated manually, and the flow rate was estimated with a stopwatch.
Two types of viscous solutions were measured. First, glycerol solutions of 0 to 80% (v/v) in 1× PBS + 0.1% Tween-20 were prepared. Second, a diluent for sputum was also prepared as outlined by Creecy et al.:31 4 M guanidine thiocyanate, 25 mM sodium citrate, 4.9% Triton X-100, and 0.2% sodium dodecyl sulfate. The diluted sputum was prepared by mixing synthetic sputum in equal parts with the guanidine diluent on a Fisher Vortex Genie 2 at speed 4 for 10 min. The viscosities of these solutions were measured using a total volume of 500 μL of well-mixed, room-temperature solution using a Brookfield DV-II+ Pro Viscometer (Middleboro, MD). Spindle 40Z was used for the measurement. Measurements were taken at a strain rate of 37.5 s–1 after increasing the strain rate from a minimum speed. This was important because of the non-Newtonian, shear-thinning rheology of the sputum. The setup was allowed to settle for a minimum of two to three full spindle rotations before a measurement was recorded. Measurements were performed in triplicate.
The effect of viscosity on capture was measured using 800 μg of MyOne Silane Dynabeads placed into 2.5 mL of each of the viscous solutions and vortexed to thoroughly combine. The solution was drawn into the transfer pipette, and 35 mg of Grade 000 steel wool was placed directly into the end of the transfer pipette. With the magnet beside the steel wool matrix, the bead-containing solution was expelled through the pipette tip. To measure the concentration of uncaptured beads, the Dynabeads contained in the flow-through were centrifuged at 3082g for a minimum of 5 min. More viscous solutions were diluted with PBS-T to reduce the solution viscosity and then centrifuged until the beads pelleted. The pelleted beads were washed in 1× PBS + 0.1% Tween-20 and then resuspended in an appropriate volume of PBS-T as discussed to ensure accuracy of the spectrographic reading. The bead concentration was measured in triplicate using absorbance at 700 nm as previously outlined here.
2.8. Statistical Analyses
All statistical analyses were performed using MATLAB. One-way analysis of variance was used to determine the statistical significance for data containing three or more groups. Unpaired t-tests were performed for two group comparisons. Statistical significance was defined as p < 0.05. Tukey’s range test was used to determine which data points were statistically significant relative to the rest of the data. Experimental variation was compared to the baseline method described above.
All samples and standards were analyzed in triplicate PCR reactions, except the analysis of one QIAamp Viral RNA Mini Kit urine extraction; one of the PCR replicates was identified to be an outlier and removed. This value was greater than 2 standard deviations outside of the mean, which averaged all PCR values for each experimental condition (triplicate extractions with triplicate reactions for each sample, nine reactions total). This criterion was applied to all samples, and this was the only identified outlier across all conditions in this study and is likely due to a pipetting error.
3. Results
3.1. M. tuberculosis IS6110 DNA Extraction from Sputum and Urine
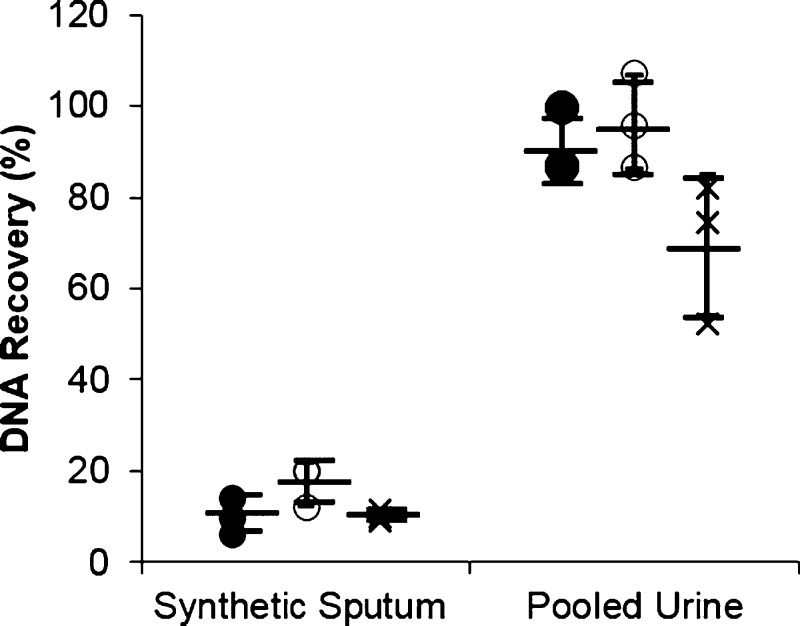
For synthetic sputum samples, the HGMS-enabled extraction method recovered a total of 10.2 ± 4.03% of spiked DNA, the commercially available Qiagen DNeasy Blood and Tissue kit recovered 17.3 ± 4.65%, and the Chargeswitch gDNA Mini kit recovered 10.1 ± 1.12% (Figure 3). Extractions were more efficient for urine samples, with the HGMS-enabled extraction yielding 91.2 ± 7.46% of spiked DNA, the commercial Qiagen QIAamp Viral RNA Mini kit recovering 96.5 ± 10.46% of spiked DNA, and the commercial Chargeswitch gDNA Mini kit recovering 69.5 ± 15.4% (Figure 3). For all sample types and methods, there is some variation in the extraction efficiency, but the three methods were not statistically different. Using LinRegPCR for comparison, the HGMS-enabled extraction of sputum was approximately 10% lower than controls. A260/A280 readings for sample purity of the HGMS-enabled extraction measured at 1.86, suggesting that the PCR inhibitor(s) reducing the reaction efficiency are not likely a contaminating protein32 (Figure S3). Although not measured, likely inhibitor candidates are low concentrations of salts, metals, or alcohols introduced during processing.
Figure 3.
HGMS-enabled extraction performed similarly to commercial kits. Extraction of 5 × 106 copies of a 123-base IS6110 TB DNA sequence using HGMS-enabled extraction (●), commercially available Qiagen kits (○), and Chargeswitch kit (×) is not statistically different from one another for both synthetic sputum and pooled urine. (mean ± s.d.), n ≥ 3.
3.2. Effect of Physical Properties of the Steel Wool Matrix on Paramagnetic Bead Capture
The mass and magnetic susceptibility of the separation matrix used in the pipette tip affected the capture of paramagnetic beads. As shown in Figure 4A, as the amount of steel wool was increased from 0 mg to 10 ± 1 mg, the capture efficiency of 1 μm Dynabeads increased from 15.8 ± 5.27 to 99.2 ± 1.41%, with steel wool masses greater than 10 mg also capturing ∼99% of beads. Tukey’s range test identified 0 and 5 mg of steel wool as significantly less than the higher densities tested, suggesting that for this design, as long as the matrix is significantly magnetized and the packing density is maintained, the amount of matrix needed to capture a majority of the beads is quite small (Video S2).
Figure 4.
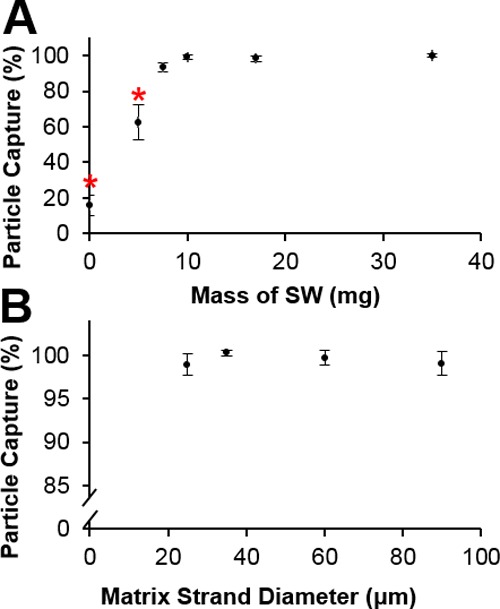
Effect of the ferromagnetic matrix mass and strand diameter on the capture of beads. (A) Steel wool matrix mass ≤5 mg captured statistically fewer beads. (B) Steel wool matrix strand diameter does not have an effect on the paramagnetic bead capture efficiency (mean ± s.d.), n ≥ 3. * indicates statistical significance at p < 0.05.
Within the range of available materials, the diameter of the steel wool matrix had essentially no effect on bead capture. Steel wool Grades 0000, 000, 1, and 3, which correspond to strand diameters of approximately 25, 35, 60, and 90 μm, performed similarly and had capture efficiencies of at least 98% (Figure 4B).
Because the different alloy compositions of the matrix can have different magnetic properties, we also looked at the effect of the alloy on capture (Figure 5). The magnetic susceptibilities (χ) of the materials were varied, allowing for evaluation of their effect on capture; diamagnetic (χ < 0), paramagnetic (χ > 0), and ferromagnetic (χ > 1) materials were tested.33 In the presence of an externally applied magnetic field, a weak magnetic dipole is induced in the opposite direction of the field for diamagnetic materials and weakly in the same direction for paramagnetic materials. For diamagnetic materials, copper and water (corresponding to no matrix), capture efficiencies are 41.8 ± 8.19 and 22.7 ± 8.44%, respectively. For the paramagnetic materials, aluminum and 316L stainless steel, paramagnetic bead capture was 51.7 ± 4.44 and 47.4 ± 8.38%, respectively. In contrast, for ferromagnetic materials, which form a strong magnetic dipole parallel to the applied external field, the capture efficiencies for 434 stainless steel and low-carbon steel are 100.4 ± 0.082 and 99.9 ± 0.071%, respectively. The capture in paramagnetic and diamagnetic matrices is statistically different from that in ferromagnetic matrices. When no magnet is applied, the capture efficiencies of each matrix are ∼10%, with the exception of no matrix and low carbon steel. The negative values are most likely due to measurement error, because the solutions measured were at the edge of the linear measurement range of the spectrophotometer. The capture values greater than 100% for 434 stainless steel are due to the concentration of beads in the centrifuged flow-through being lower than the detection limit of the spectrophotometer.
Figure 5.
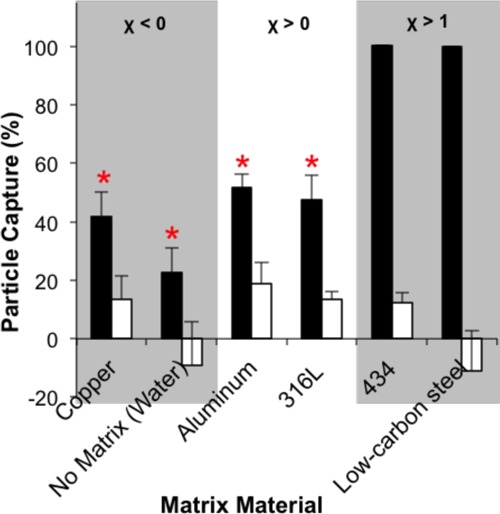
Effect of matrix magnetic susceptibility (χ) on capture. Ferromagnetic materials (χ > 1) captured more magnetic beads than paramagnetic (χ > 0) and diamagnetic (χ < 0) materials. Black bars—with applied magnetic field. White bars—no magnetic field present (mean ± s.d.), n ≥ 3. * indicates statistical significance at p < 0.05.
3.3. Effect of Magnet Properties on Paramagnetic Bead Capture
As shown in Figure 6A, the capture efficiency increased with increasing surface field, with 548 G (Gauss) and 1701 G having capture efficiencies of 76.5 ± 13.1 and 88.6 ± 6.61%, respectively. Capture for the remaining magnets plateaued at ∼99%. Only the smallest surface field examined, 548 G, exhibited a statistically reduced capture efficiency. This suggests that as long as the magnetic surface field is greater than ∼1700 G, the capture efficiency will be nearly 100%. A close inspection reveals a minor decrease in the bead capture efficiency from ∼99 to ∼94% when using the 3309 G magnet; as theorized by Himmelblau,34 the region of attractive magnetic force temporarily falls when the applied magnetic field is greater than the field needed to reach saturation. We hypothesize that this value is near the maximum saturation point of the ferromagnetic matrix. This effect is overcome when the applied field is sufficiently large. Further studies are needed to validate this hypothesis.
Figure 6.
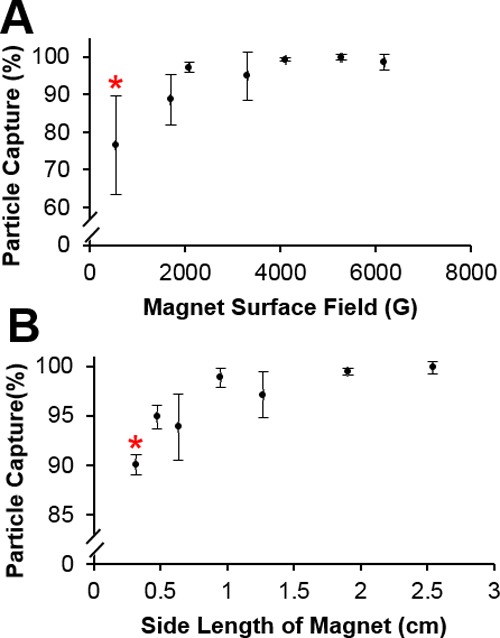
Effect of magnet surface field and magnet dimensions on bead capture. (A) Smaller statistical percentage of paramagnetic beads were captured using a 548 G magnet. (B) Smallest cube magnet with a side length of 0.32 cm captured statistically fewer beads because of decreased effective trapping length (mean ± s.d.), n ≥ 3. * indicates statistical significance at p < 0.05.
The different cube magnets used (Figure 6B) had their surface field held constant while the side length of the cube magnet varied. With increasing magnet size, the capture efficiencies increased from 90.0 ± 1.03% for the smallest 0.32 cm magnet up to 99.9 ± 0.58% for the largest 2.54 cm magnet, with the smallest magnet of 0.32 cm capturing statistically fewer beads than the rest. This difference is likely due to the reduction in the effective trapping length of the steel wool, because in this case, the magnet dimension is smaller than the size of the steel wool matrix. However, it is important to note that the capture efficiency only varies between the highest and lowest by 10%. While statistically significant, the difference may not be enough to warrant the use of a larger and more expensive magnet, assuming a strong enough surface field is applied.
3.4. Effect of Sample Viscosity and Flow Rate on Paramagnetic Bead Capture
We also assessed how the flow rate and sample viscosity influenced paramagnetic bead capture (Figure 7). Capture measurements were performed for samples passed through the HGMS-enabled separator with and without a steel wool matrix (Figure S1). Samples applied to the HGMS-enabled device with a steel wool capture matrix at flow rates from 1 ± 0 to 46.5 ± 5.01 mL/min had capture efficiencies of at least 96%, and 60.1 ± 3.01 mL/min captured 89.2 ± 1.07%. Even though this is only a ∼10% reduction, it is statistically significant (Figure 7A). For samples passed through without a matrix, capture efficiencies were measured for flows only up to 20 mL/min, with capture efficiency rapidly declining with flow rate from ∼100% bead capture at 1 mL/min to only ∼25% capture at 20 mL/min. The capture efficiency was statistically reduced for flow rates of at least 10 mL/min.
Figure 7.
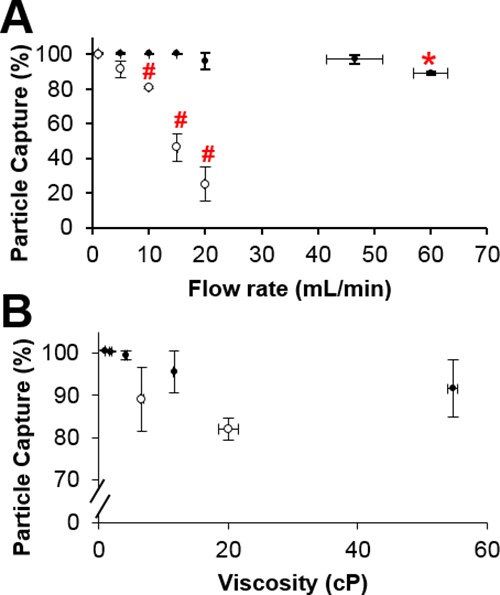
Effect of sample properties on bead capture. (A) Increasing the flow rate did not statistically reduce bead capture in a 35 mg steel wool matrix (●) until reaching 60 mL/min, but was significantly decreased at 10 mL/min without any matrix present (○). (B) Sample viscosity up to 55 cP did not reduce paramagnetic bead capture from either glycerol solutions (●) or synthetic sputum (○) (mean ± s.d.) n ≥ 3. * and # indicate statistical significance at p < 0.05.
Biological samples often differ in rheological properties that may influence the capture performance of the HGMS-enabled system, because viscous drag forces on beads are much higher than the magnetic force, even when viscosity limits the sample flow rates. As seen in Figure 7B, for the v/v % glycerol solutions prepared, the viscosities increased with increasing percentage of glycerol, measuring from 0.98 ± 0.06 to 54.7 ± 0.76 cP. The capture efficiency decreased by ∼9% with increasing glycerol percentage from 100 ± 0.12 to 91.7 ± 6.78%, but this was not statistically different even though there is a statistically significant difference in the sample viscosities. We also tested this using synthetic sputum with shear-thinning rheologic behavior. This was compared to bead capture from liquefied sputum in Figure 7B because it is common for sputum samples to undergo liquefaction and decontamination in protocols, such as those used for TB diagnosis. The measured viscosities of the synthetic and liquefied sputum were 20.0 ± 1.59 and 6.66 ± 0.22 cP. There was no statistically significant difference between the capture efficiencies, with both near 85% capture.
4. Discussion
The method reported here was used in a blinded sample preparation study sponsored by the Bill and Melinda Gates Foundation published by Beall et al.26 This publication compared the sensitivity, specificity, and outcome features for stool, sputum, and whole blood for the HGMS-enabled method described in this report against five other commercial extraction methods. In parallel to this effort, The Gates Foundation also commissioned a study comparing eight nucleic acid amplification technologies suitable for low-resource settings.35 In both of these studies, blinded samples were supplied to each of the study participants. For the extraction comparison, each sample matrix was spiked with a high, medium, or low amount of microbe appropriate to the matrix. In performing the experiments reported in Figure 3, we aimed to replicate the protocols and use the same processing reagents whenever possible as used by Beall et al. In the study by Beall et al., we are coded as “Developer E″ and ranked third overall. The reported sensitivity and specificity for our extraction of chemically inactivated M. tuberculosis DNA from the patient sputum were 76 and 100%, respectively. Urine was not part of this comparison study. Other aspects of the six test systems, such as the number of steps, time for extraction, and ASSUR criteria are reported in the study by Beall et al.26
In this report, we focus on presenting the details of our HGMS-enabled method and a comparison to several commercially available manual kits. We found that in addition to performing well in comparison to automated systems reported by Beall et al., our approach also performed well in comparison to other manual commercial kits. Figure 3 shows the percentage recovery of the HGMS-based method compared to both a silica column and another magnetic bead-based method, and the recovery from sputum and urine for the three systems is similar.
We have also previously reported sputum and urine extraction results using a magnetic bead-based system both manually and semiautomatically. In these previous reports, the extraction processing was performed using a system of prearrayed solutions held stationary in a small diameter tube by surface tension forces, and biomarkers bound to magnetic beads were passed through consecutive solutions to perform the processing steps. With respect to sputum, this automated method was used to extract TB DNA from chemically inactivated M. tuberculosis in synthetic sputum in the study by Creecy et al., and the extracted TB DNA was amplified by both PCR and isothermal amplification.31,36 In both studies, clinically relevant concentrations of TB mycobacteria were detectable by DNA amplification, but extraction efficiencies were not calculable.
In previous urine extraction studies described in the study by Bordelon et al., using the tube extraction system manually, we extracted approximately 46% of DNA from urine, which is less than the 91.2% found in this report.37 Whereas here we report results from smaller volumes, our prior work used shorter binding times, fewer beads, and did not incorporate the addition of carrier RNA.38 When carrier RNA was removed from the extraction protocols for both the HGMS-enabled extraction and Qiagen kit in this report, extraction efficiencies were reduced to 59.5 ± 7.59 and 88.2 ± 7.45%, respectively. This was a statistically significant reduction for the HGMS-enabled extraction method, whereas the difference of the Qiagen extracts was not statistically significant (Figure S4). This is consistent with previous reports. Carrier RNA was not tested with the Chargeswitch kit.39
A key difference between the Beall et al. study and the extraction studies reported here is the length of the DNA extraction targets. The Beall et al. studies were performed with whole pathogens, and the manual studies reported here used only a 123-base DNA fragment. We believe that the extraction efficiencies for a range of DNA fragment sizes would be similar to those reported here, provided the DNA fragments were greater than 100 bp in length. This is based on our previous work37 and others40,41 showing that it is difficult to efficiently extract and recover short DNA fragments using chaotrope/silica chemistries. For example, Oreskovic et al. compared two published DNA extraction methods and three commercial kits to their hybridization-capture system for extraction of single-stranded IS6110 TB DNA (25–150 base). They found that their 1.5 h protocol performed most consistently for all methods across different fragment sizes, consistently isolating 73 to 84% of fragments.41 The other method using Q Sepharose only performed well for larger fragments (63–75%) and was significantly reduced for 25-base fragments (9%).41 We also believe that the extraction efficiencies reported for the HGMS-enabled extractions are not dependent on the spiked DNA concentration. Based on what we have observed previously, if the bead concentration is sufficient to bind available nucleic acids, the extraction efficiency is preserved.37
There are two elements of our HGMS-enabled protocol that are critical for optimum performance. Our protocol used a 1–2 min gravity-driven pooling step to remove residual liquid from the pipette tip before eluting the nucleic acids, and this was found to be critical for DNA detection. Though the beads themselves have a low carry over, the surface tension in the capture matrix has potential to retain liquid. Specifically, this step removed approximately 100 μL of the residual ethanol wash from the sample chamber. Ethanol contained in this wash is known to inhibit PCR and other downstream detection methods. Therefore, ethanol removal is critical. Second, we found that it was important to include a rapid 100 μL TE rinse for sputum samples and have seen this to be beneficial under specific conditions in a prior work.8 We found that while this rinse can lead to some nucleic acid loss (data not shown), this rinse helps remove excess PCR contaminants (e.g., residual proteins, salts, and alcohols) and allows for improved detection. The total rinse time appears to be critical too. If the rinse time is too quick, the beads may be disrupted, resulting in DNA elution and reduced recovery. In addition, contaminants may remain in the system, inhibiting the PCR. If the rinse is too slow, DNA can be partially eluted and recovery is reduced (data not shown). Use of alternative processing chemistries or DNA detection strategies may allow for the elimination of these two steps.
Our HGMS-enabled magnetic separation system, combined with chaotrope/silica binding chemistry, resulted in a flow-through design for magnetic bead extraction of nucleic acids that performed as well as available commercial kits. A set of optimal physical features related to maximizing HGMS capture capabilities were identified, such as a high magnetic susceptibility steel wool matrix with a mass of at least 7.5 mg, packed at a density of 0.91 g/mm3 under a 6180 G field (Figure 4). These consistently captured ∼100% of paramagnetic beads. Variation around these values did not significantly reduce the capture. For example, the same percentage capture is achieved with a 35 mg matrix in an externally applied field of at least ∼1700 G (Figure 6). The HGMS-enabled system can also capture essentially all beads from solution flows up to 45 mL/min and viscosities up to 55 cP (Figure 7). Our methods characterized the effect of each variable individually; however, the interplay between multiple variables will need to be further elucidated for final optimization for potential applications.
Although previous studies were limited in their scope of investigation, there are some clear differences between our system and previously described HGMS systems. For example, the packing density of 0.91 mg/mm3 or ∼11.5% packing volume used in these experiments is higher than those in the previous work, where HGMS was used to efficiently isolate cells with columns typically packed at 2–5% of the total volume.21−23 Owen et al. reported capture of up to 92% of paramagnetic erythrocytes using 1 g of magnetic stainless steel wool at flow rates less than 2 mL/min in an applied magnetic field of 33,000 G.42 Molday and Molday reported 96 ± 2% capture of red blood cells using immunospecific magnetic nanoparticles at 2 mL/min in a 7500 G field.22 Miltenyi et al. only looked at relative proportions of cell mixtures before and after MACS, but demonstrated that two different cell populations could be efficiently separated, with the retained cell type generally making up <2% of the cells that were not captured in the MACS matrix. Miltenyi et al. used flow rates between 0.16 and 8 mL/min, but most often less than 2 mL/min.23 Despite these differences, these systems demonstrate capture efficiencies similar to the ones reported here, albeit at lower flow rates, greater applied fields, and with more steel wool than used in our system.
As far as we can tell, there have been no other studies specifically looking at the effect of sample viscosity on biological applications, because prior studies used aqueous solutions, such as saline.21,23 Our viscosity results are consistent with the industrial application of mineral particle separation by Dobby et al., which demonstrated that viscosity plays a minor role in capture.43
Prior work performed limited studies looking at the effect of magnet surface field, typically only looking at two different fields.21,44 For example, Owen et al. investigated the effect of field strength on capture, demonstrating that capture efficiencies are proportional to the magnetic moment of the beads captured divided by the flow rate through the magnetic column.21 Our data are consistent with Owen’s findings, in that capture efficiency decreased with decreasing magnetic field. In addition, assuming that the magnetic moment of the magnetic beads is constant and uniform, the flow-rate data collected is consistent with Owen’s findings, with capture efficiency decreasing with increasing flow rate; this is obvious when there is no ferromagnetic matrix present in the path of fluid flow, and it is more subtly observed at high flow rates with the presence of ferromagnetic matrix (Figure 7A). These findings are consistent with the available theory.19,43 We also note that the prior work fully immersed and maximally saturated the entire steel wool matrix in the applied magnetic field but did not assess the effect of magnet size specifically on a single system. This is likely because it was much more common to use electromagnets for HGMS systems when first implemented,21,22 with an exception to this being Miltenyi.23
The differences in the capture effect with changes in matrix magnetic susceptibility suggest that the feature dominating paramagnetic bead capture is the matrix itself. However, the fact that there is a difference in capture efficiency between the two diamagnetic materials, copper (χ = −9.36 × 10–6) and water/no matrix (χ = −9.05 × 10–6),33 suggests that just the presence of a metal matrix, even when traditionally identified as “nonmagnetic,” has an impact on capture. We and others have observed that the micron-scale paramagnetic beads can form chains while in a magnetic field45,46 and can form complex, magnetically anisotropic structures that could potentially further contribute to increasing bead capture. The importance of the chaining phenomenon is unclear. Further, the flow likely remains laminar for all of the flows achievable in this system. Treating a single strand of the 35 μm steel wool matrix wire as a cylinder sitting perpendicular to the path of the fluid flow at a flow rate of 15 mL/min (fluid velocity of 7.29 cm/s) yields a cylinder Reynolds number (Re) of 2.58 in the laminar range.47
In general, manual extraction methods are not as desirable as automated systems, which require less training and are more reproducible. However, when the processing instruments and/or training are unavailable, simple manual methods may fill a critical gap. In this report, we describe the details of an HGMS-based manual approach. This manual system performs similar to other manual systems, has some attractive features for low-resource use, particularly, in the areas of cost and robust operation, and has the potential for further improvement to make it even more suitable for low-resource settings. One of the attractive features of HGMS-enabled extraction is that its robust performance is less dependent on user training. In our experience, the range of flow rates produced by different users expelling the transfer pipette is no greater than 20 mL/min (data not shown), and the capture of magnetic beads in the steel wool matrix is not affected by a range of flows up to 45 mL/min (Figure 7). For highly viscous samples, the maximum flow rate of a liquid sample through the device primarily depends on viscous shear forces within the magnetic matrix material. Therefore, variations in flow rates produced by a user squeezing the pipette bulb as quickly as possible are limited. The two dominant forces acting in a magnetic bead in a high-gradient magnetic separator are the hydrodynamic drag force and the magnet force; all other forces are negligible.19 For samples with viscosities near that of water, the hydrodynamic drag force has the potential to overcome the magnetophoretic force at sufficiently high flow rates, reducing the capture efficiency of beads from solution.48 It is our conclusion that HGMS-enabled capture demonstrated a robustness against potential operator variability, which could occur from variations in squeezing pressure and the resulting flow produced from different users and samples with a range of viscosities.
In addition, though the applied magnetic field needs to encompass the steel wool matrix for bead capture, a user can see the matrix inside of the pipette tip (Figure 2C), and the magnet is selected to be the same size or larger than the steel wool matrix. Because a surface field of only ∼1700 G is needed to efficiently capture particles (Figure 6A), the magnet does not need to be held in a precise location and can be a distance away from the steel wool matrix, with the condition that the magnet has a strong enough surface field. In the DNA extractions, the magnet was applied through an Eppendorf tube wall with the pipette tip resting inside, and the steel wool separator was able to efficiently capture magnetic particles repeatedly during each wash step. While robust, it is still possible for a user to make an error. If a mistake is made and magnetic beads are lost, which would be visible due to the brown color of the magnetic beads, it is quick and easy to redraw the solution into the transfer pipette and repeat the magnetic bead capture. Repeating this step to overcome a processing error without loss is another major advantage of HGMS-enabled extraction.
The HGMS separator described here could potentially be used for sample preparation in a low-resource setting, where the cost of a point-of-care assay is a significant contributor to use and adoption. With the steel wool and transfer pipettes costing just pennies, the most expensive components of the assay described are the magnetic beads at $3.30/assay at the time of publication. This large number of beads was kept constant because we sought to describe the methods that closely match the method used by Beall et al.,26 which were designed to maximize performance, rather than minimize the cost. Further assay optimization could reduce the cost, and it is believed that the resulting extraction kit will likely cost under $1.00 per test. Quantitative PCR was used as the readout and was not inhibited by the extraction methods. We expect methods such as loop-mediated isothermal amplification or other nucleic acid amplification methods in development, with lateral flow assay readout, would also perform well and might serve as a more suitable detection method in resource-limited settings.
Another important consideration for the low-resource use is the identification of less hazardous extraction chemistry. There are alternative methods to guanidinium-based methods, such as the Chargeswitch system (Thermo Fisher). Claremont BioSolutions has also developed an extraction method that does not use hazardous extraction chemistry.49 A compelling future direction is the testing of the Chargeswitch chemistry using the HGMS format. This potentially could eliminate the critical timing step currently required for maximum recovery from sputum. Improved reagent storage could also be achieved. For example, the transfer pipette bulb allows for future lyophilization of reagents within the extraction device,13 further simplifying the assay protocol, and reagent packing; this would also allow for inclusion of poly-A carrier RNA, which helped significantly improve the extraction efficiency of DNA from urine. Other liquid-phase processing solutions and containers could be packaged and could arrive in a disposable, single-use “kit” format, with all components prealiquotted or individually wrapped. The user would simply remove the assay components from their packaging and quickly start performing extraction(s) without any preprocessing steps. The use of a single inlet and outlet of the transfer pipette, rather than separate entry/exit ports in flow-through systems, yields additional mixing potential that could improve the biomarker yield.
The use of a larger sample can increase the sensitivity of low-resource detection methods.13,50 This is simple to do with HGMS-enabled extraction. Pipettes with different sized bulbs or syringes could be easily interchanged for application-specific design, with larger, more dilute samples using larger volume actuators. Whereas we only processed 100 μL for these proof-of-principle studies, we demonstrate that larger sample volumes, up to 3 mL, can be processed using the HGMS-enabled transfer pipette system. Further, it is the maximum number of beads that can be captured in a matrix that limits a HGMS’s performance, not the volume in which these beads are suspended; this was demonstrated by Miltenyi23 and in industrial applications as discussed. The large number of beads captured in our baseline design suggests that the small amount of steel wool used in these studies is still well below the load-limiting capacity. This implicates potential for very large volumes or dilution, though overzealous dilution can reduce bead-biomarker interactions necessary for DNA isolation.
With modifications, this approach can be adapted for other sample types. In the study by Beall et al., the methods described here were successfully modified with a proteinase K treatment for successful extraction from whole blood and stool. This shows that the HGMS-enabled extraction can be adapted for other sample types at clinically relevant concentrations. Further optimization may be required, however. For example, we tried to adapt our previous sputum protocol,31,36 but the high detergent concentration in the lysis/binding buffer resulted in significant bubble formation while using the HGMS-enabled device. In addition, we hypothesize that our sensitivity for M. tuberculosis detection was only 76% because our guanidine buffer did not lyse all spiked M. tuberculosis cells, which are known to be difficult to chemically lyse without the use of organic solvents.51 These examples demonstrate the need for buffers tailored to the extraction system design as well as the application.
Additional processing strategies are possible with this approach and have not been explored. The initial capture of biomarkers onto the magnetic beads is an area where we speculate this HGMS-based approach may improve the performance. In this alternate approach, magnetic beads could be transiently captured within the steel wool and the entire sample passed through it multiple times to accelerate the binding of the nucleic acid biomarkers to the surface of the magnetic beads. This is likely to have advantages over other bead-sample mixing strategies in that the extraction efficiency can be maintained while the bead number and initial binding times are reduced. This could also be implemented during the processing steps. Instead of releasing and recapturing the beads in the steel wool matrix during each step of the extraction processes, the magnet could be applied throughout. However, if the beads are nonuniformly distributed throughout the steel wool matrix, there is the possibility that not all beads will receive sufficient contact with the processing solutions. Another future direction is to incorporate this processing method into a simple instrument to reduce the number of manual steps and operational errors.
This study did not look at the effect of bead magnetic susceptibility or size, which would also influence the capture efficiency of beads in the steel wool matrix. The size of the beads used, if at all,20,21,42 across previous studies was also smaller than 200 nm,22,23 giving the beads a smaller magnetic susceptibility than the 1 μm beads used in our study because of their reduced iron content. Nevertheless, the use of smaller beads would increase the surface area available for capture, potentially reducing the cost of the system while maintaining the capture efficiency.
In summary, the proposed HGMS-enabled nucleic acid extraction method is an effective alternative for magnetic bead-based sample processing that is as efficient as gold standard commercially available systems but also inexpensive, rapid, and simple. We have shown that the device offers advantages over existing magnetic extraction methods, including magnetic separation of beads from viscous and large-volume samples without the use of specialized laboratory equipment, making the approach potentially useful in resource-limited applications. With changes in the surface chemistry of the beads, we expect that this robust HGMS-enabled system can be applied to extraction and purification of other biomarkers of interest.
Acknowledgments
The authors would like to thank Jami Hamman and Christia M. Victoriano for their efforts in assisting with data collection. They would also like to thank John Albert Rector IV for assistance in collecting the supporting information.
Glossary
Abbreviations
- HGMS
high-gradient magnetic separation
- TB
tuberculosis
- qPCR
real-time polymerase chain reaction
- χ
magnetic susceptibility
- Re
Reynolds number
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsami.9b21564.
Figure S1-S4: Overview of paramagnetic bead separation from a flowing fluid stream; Nanodrop Spectrophotometer standard curve for bead quantitation; A260/A280 measurements for sample purity of DNA extracted samples; Effect of carrier RNA on DNA extraction from urine (PDF)
Video S1: Paramagnetic bead separation from a flowing fluid stream using high-gradient magnetic separation (MP4)
Video S2: Paramagnetic bead separation from a flowing fluid stream using a stationary magnet (MP4)
Author Contributions
S.I.P. gathered and analyzed the experimental data and wrote the manuscript. S.I.P., M.L., K.A.R., N.M.A., and M.E.P. developed extraction protocols. P.K.R. helped to draft the manuscript. A.E.W. and C.W. performed experimental procedures and measurements under the supervision of S.I.P. S.I.P. and F.R.H. designed the study. F.R.H. edited the manuscript.
This work was funded in part by the Bill and Melinda Gates Foundation, OPP1172605, and the National Institutes of Health, R01 AI135937.
The authors declare the following competing financial interest(s): Vanderbilt University has filed a patent describing the technology discussed in this publication.
Supplementary Material
References
- Organization W. H.Global Tuberculosis Report 2018, 2018.
- Cambanis A.; Ramsay A.; Wirkom V.; Tata E.; Cuevas L. E. Investing time in microscopy: an opportunity to optimise smear-based case detection of tuberculosis. Int. J. Tuberc. Lung Dis. 2007, 11, 40–5. [PubMed] [Google Scholar]
- Hänscheid T. The future looks bright: low-cost fluorescent microscopes for detection of Mycobacterium tuberculosis and Coccidiae. Trans. R. Soc. Trop. Med. Hyg. 2008, 102, 520–521. 10.1016/j.trstmh.2008.02.020. [DOI] [PubMed] [Google Scholar]
- WHO . Policy Statement: Automated Real-Time Nucleic Acid Amplification Technology for Rapid and Simultaneous Detection of Tuberculosis and Rifampicin Resistance: Xpert MTB/RIF System, 2011. [PubMed]
- Green C.; Huggett J. F.; Talbot E.; Mwaba P.; Reither K.; Zumla A. I. Rapid diagnosis of tuberculosis through the detection of mycobacterial DNA in urine by nucleic acid amplification methods. Lancet Infect. Dis. 2009, 9, 505–511. 10.1016/s1473-3099(09)70149-5. [DOI] [PubMed] [Google Scholar]
- Crowe S.; Turnbull S.; Oelrichs R.; Dunne A. Monitoring of Human Immunodeficiency Virus Infection in Resource-Contrained Countries. Clin. Infect. Dis. 2003, 37, S25–S35. 10.1086/375369. [DOI] [PubMed] [Google Scholar]
- Sur K.; McFall S. M.; Yeh E. T.; Jangam S. R.; Hayden M. A.; Stroupe S. D.; Kelso D. M. Immiscible phase nucleic acid purification eliminates PCR inhibitors with a single pass of paramagnetic particles through a hydrophobic liquid. J. Mol. Diagn. 2010, 12, 620–628. 10.2353/jmoldx.2010.090190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordelon H.; Adams N. M.; Klemm A. S.; Russ P. K.; Williams J. V.; Talbot H. K.; Wright D. W.; Haselton F. R. Development of a low-resource RNA extraction cassette based on surface tension valves. ACS Appl. Mater. Interfaces 2011, 3, 2161–2168. 10.1021/am2004009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russ P. K.; Karhade A. V.; Bitting A. L.; Doyle A.; Solinas F.; Wright D. W.; Haselton F. R. A Prototype Biomarker Detector Combining Biomarker Extraction and Fixed Temperature PCR. J. Lab. Autom. 2016, 21, 590–598. 10.1177/2211068216634072. [DOI] [PubMed] [Google Scholar]
- Aguilar-Arteaga K.; Rodriguez J. A.; Barrado E. Magnetic solids in analytical chemistry: a review. Anal. Chim. Acta 2010, 674, 157–165. 10.1016/j.aca.2010.06.043. [DOI] [PubMed] [Google Scholar]
- Long Z.; Shetty A. M.; Solomon M. J.; Larson R. G. Fundamentals of magnet-actuated droplet manipulation on an open hydrophobic surface. Lab Chip 2009, 9, 1567–1575. 10.1039/b819818g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry S. M.; Pezzi H. M.; LaVanway A. J.; Guckenberger D. J.; Anderson M. A.; Beebe D. J. AirJump: Using Interfaces to Instantly Perform Simultaneous Extractions. ACS Appl. Mater. Interfaces 2016, 8, 15040–15045. 10.1021/acsami.6b02555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordelon H.; Ricks K. M.; Pask M. E.; Russ P. K.; Solinas F.; Baglia M. L.; Short P. A.; Nel A.; Blackburn J.; Dheda K.; Zamudio C.; Cáceres T.; Wright D. W.; Haselton F. R.; Pettit A. C. Design and use of mouse control DNA for DNA biomarker extraction and PCR detection from urine: Application for transrenal Mycobacterium tuberculosis DNA detection. J. Microbiol. Methods 2017, 136, 65–70. 10.1016/j.mimet.2017.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frantz S. G.Magnetic Separator. U.S. Patent 2,074,085A, 1937.
- Kolm H. H.; Oberteuffer J. A.; Kelland D. R.. High-Gradient Magnetic Separation; Scientific American, 1975; pp 45–54. [Google Scholar]
- Oberteuffer J. High Gradient Magnetic Separation. IEEE Trans. Magn. 1973, 9, 303–306. 10.1109/tmag.1973.1067673. [DOI] [Google Scholar]
- Mitchell R.; Bitton G.; Oberteuffer J. A. High Gradient Magnetic Filtration of Magnetic and Non-Magnetic Contaminants from Water. Separ. Purif. Methods 1975, 4, 267–303. 10.1080/03602547508066042. [DOI] [Google Scholar]
- Oberteuffer J.; Wechsler I.; Marston P.; Mcnallan M. High Gradient Magnetic Filtration of Steel Mill Process and Waste-Waters. IEEE Trans. Magn. 1975, 11, 1591–1593. 10.1109/tmag.1975.1058803. [DOI] [Google Scholar]
- Gerber R.; Birss R. R.. High Gradient Magnetic Separation; Research Studies Press, 1983. [Google Scholar]
- Melville D.; Paul F.; Roath S. Direct magnetic separation of red cells from whole blood. Nature 1975, 255, 706. 10.1038/255706a0. [DOI] [PubMed] [Google Scholar]
- Owen C. S. High gradient magnetic capture of red blood cells. J. Appl. Phys. 1982, 53, 3884–3887. 10.1063/1.331089. [DOI] [Google Scholar]
- Molday R. S.; Molday L. L. Separation of cells labeled with immunospecific iron dextran microspheres using high gradient magnetic chromatography. FEBS Lett. 1984, 170, 232–238. 10.1016/0014-5793(84)81319-8. [DOI] [PubMed] [Google Scholar]
- Miltenyi S.; Müller W.; Weichel W.; Radbruch A. High Gradient Magnetic Cell Separation With MACS. Cytometry 1990, 11, 231–238. 10.1002/cyto.990110203. [DOI] [PubMed] [Google Scholar]
- Miltenyi S.; Radbruch A.; Weichel W.; Muller W.; Gottlinger C.; Meyer K. L.. Metal matricies for use in high gradient magnetic separation of biological materials and method for coating the same. U.S. Patent 5,385,707A, 1995.
- Miltenyi S.Magnetic Separation Apparatus. U.S. Patent 5,795,470A, 1998.
- Beall S. G.; Cantera J.; Diaz M. H.; Winchell J. M.; Lillis L.; White H.; Kalnoky M.; Gallarda J.; Boyle D. S. Performance and workflow assessment of six nucleic acid extraction technologies for use in resource limited settings. PLoS One 2019, 14, e0215753 10.1371/journal.pone.0215753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders N. N.; Van Rompaey E.; De Smedt S. C.; Demeester J. Structural Alterations of Gene Complexes by Cystic Fibrosis. Am. J. Respir. Crit. Care Med. 2001, 164, 486–493. 10.1164/ajrccm.164.3.2011041. [DOI] [PubMed] [Google Scholar]
- Ogusku M. M.; Salem J. I. Analysis of different primers used in the PCR method: diagnosis of tuberculosis in the state of Amazons, Brazil. J. Bras. Pneumol. 2004, 30, 433–439. 10.1590/s1806-37132004000400008. [DOI] [Google Scholar]
- Ramakers C.; Ruijter J. M.; Deprez R. H. L.; Moorman A. F. M. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci. Lett. 2003, 339, 62–66. 10.1016/s0304-3940(02)01423-4. [DOI] [PubMed] [Google Scholar]
- Ruijter J. M.; Ramakers C.; Hoogaars W. M. H.; Karlen Y.; Bakker O.; van den Hoff M. J. B.; Moorman A. F. M. Amplification efficiency: linking baselike and bias in the analysis of quantitative PCR data. Nucleic Acids Res. 2009, 37, e45 10.1093/nar/gkp045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creecy A.; Russ P. K.; Solinas F.; Wright D. W.; Haselton F. R. Tuberculosis Biomarker Extraction and Isothermal Amplification in an Integrated Diagnostic Device. PLoS One 2015, 10, e0130260 10.1371/journal.pone.0130260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrell R. E., Jr., RNA Methodologies: Laboratory Guide for Isolation and Characterization, 5th ed.; Academic Press, 2017. [Google Scholar]
- Schenck J. F. The role of magnetic susceptibility in magnetic resonance imaging: MRI magnetic compatibility of the first and second kinds. Med. Phys. 1996, 23, 815–850. 10.1118/1.597854. [DOI] [PubMed] [Google Scholar]
- Himmelblau D. A.Observation and Modeling of Paramagnetic Particle Entrapment in a Magnetic Field; Massachusettes Institute of Technology, 1973. [Google Scholar]
- Cantera J. L.; White H.; Diaz M. H.; Beall S. G.; Winchell J. M.; Lillis L.; Kalnoky M.; Gallarda J.; Boyle D. S. Assessment of eight nucleic acid amplification technologies for potential use to detect infectious agents in low-resource settings. PLoS One 2019, 14, e0215756 10.1371/journal.pone.0215756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitting A. L.; Bordelon H.; Baglia M. L.; Davis K. M.; Creecy A. E.; Short P. A.; Albert L. E.; Karhade A. V.; Wright D. W.; Haselton F. R.; Adams N. M. Automated Device for Asynchronous Extraction of RNA, DNA, or Protein Biomarkers from Surrogate Patient Samples. J. Lab. Autom. 2016, 21, 732–742. 10.1177/2211068215596139. [DOI] [PubMed] [Google Scholar]
- Bordelon H.; Russ P. K.; Wright D. W.; Haselton F. R. A magnetic bead-based method for concentrating DNA from human urine for downstream detection. PLoS One 2013, 8, e68369 10.1371/journal.pone.0068369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishore R.; Reef Hardy W.; Anderson V. J.; Sanchez N. A.; Buoncristiani M. R. Optimization of DNA extraction from low-yield and degraded samples using the BioRobot EZ1 and BioRobot M48. J. Forensic Sci. 2006, 51, 1055–1061. 10.1111/j.1556-4029.2006.00204.x. [DOI] [PubMed] [Google Scholar]
- He H.; Li R.; Chen Y.; Pan P.; Tong W.; Dong X.; Chen Y.; Yu D. Integrated DNA and RNA extraction using magnetic beads from viral pathogens causing acute respiratory infections. Sci. Rep. 2017, 7, 45199. 10.1038/srep45199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook L.; Starr K.; Boonyaratanakornkit J.; Hayden R.; Sam S. S.; Caliendo A. M. Does Size Matter? Comparison of Extraction Yields for Different-Sized DNA Fragments by Seven Different Routine and Four New Circulating Cell-Free Extraction Methods. J. Clin. Microbiol. 2018, 56, e01061 10.1128/jcm.01061-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oreskovic A.; Brault N. D.; Panpradist N.; Lai J. J.; Lutz B. R. Analytical Comparison of Methods for Extraction of Short Cell-Free DNA from Urine. J. Mol. Diagn. 2019, 21, 1067–1078. 10.1016/j.jmoldx.2019.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen C. S. High Gradient Magnetic Separation of Erythrocytes. Biophys. J. 1978, 22, 171–178. 10.1016/s0006-3495(78)85482-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobby G.; FInch J. A. Capture of mineral particles in a high gradient magnetic field. Powder Technol. 1977, 17, 73–82. 10.1016/0032-5910(77)85044-4. [DOI] [Google Scholar]
- Maxwell E.; Kelland D. High Gradient Magnetic Separation in Coal Desulfurization. IEEE Trans. Magn. 1978, 14, 482–487. 10.1109/tmag.1978.1059826. [DOI] [Google Scholar]
- Gijs M. A. M. Magnetic bead handling on-chip: new opportunities for analytical applications. Microfluid. Nanofluidics 2004, 1, 22–40. 10.1007/s10404-004-0010-y. [DOI] [Google Scholar]
- Gao Y.; Beerens J.; van Reenen A.; Hulsen M. A.; de Jong A. M.; Prins M. W. J.; den Toonder J. M. J. Strong vortical flows generated by the collective motion of magnetic particle chains rotating in a fluid cell. Lab Chip 2015, 15, 351–360. 10.1039/c4lc01198h. [DOI] [PubMed] [Google Scholar]
- Sato M.; Kobayashi T.. A fundamental study of the flow past a cylinder using Abaqus/CFD. 3DS SIMULIA Conference, Providence, RI, 2012.
- Oberteuffer J. Magnetic Separation: A Review of Principles, Devices, and Applications. IEEE Trans. Magn. 1974, 10, 223–238. 10.1109/tmag.1974.1058315. [DOI] [Google Scholar]
- Ferguson T. M.; Weigel K. M.; Lakey Becker A.; Ontengco D.; Narita M.; Tolstorukov I.; Doebler R.; Cangelosi G. A.; Niemz A. Pilot study of a rapid and minimally instrumented sputum sample preparation method for molecular diagnosis of tuberculosis. Sci. Rep. 2016, 6, 19541. 10.1038/srep19541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis K. M.; Gibson L. E.; Haselton F. R.; Wright D. W. Simple sample processing enhances malaria rapid diagnostic test performance. Analyst 2014, 139, 3026–3031. 10.1039/c4an00338a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrakchi H.; Lanéelle M.-A.; Daffé M. Mycolic Acids: Structures, Biosynthesis, and Beyond. Chem. Biol. 2014, 21, 67–85. 10.1016/j.chembiol.2013.11.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.