Diffuse brainstem gliomas, historically termed diffuse intrinsic pontine glioma (DIPG), account for approximately 75% of pediatric brainstem tumors and have a particularly poor prognosis with a median survival of only 10 months [8, 10]. Until recently, the diagnosis of DIPG was principally made by imaging, with biopsy relegated to an ancillary role owing to the delicate anatomic location [1]. However, with improved surgical techniques [4, 14] and the discovery of canonical histone H3 lysine-27-methionine (H3K27M) driver mutations, direct examination of these lesions to distinguish DIPG from radiologic mimics has reemerged as an important component of the diagnostic process [17, 19].
H3K27M mutations in DIPG result in global loss of the repressive H3K27 trimethylation (H3K27me3) through multiple mechanisms including inhibition of PRC2 methyltransferase activity and spread of H3K27me3 [5, 9, 18]. Global reduction in H3K27me3 is also observed in a subset of childhood posterior fossa (PF) ependymomas termed PF-group A ependymomas (PFA) [2, 12]. PFAs show overexpression of EZH inhibitory protein (EZHIP) in most cases, or harbor mutations in EZHIP in ~ 10% of tumors [11]. Additionally, three independent groups have demonstrated that EZHIP mimics the H3K27M “oncohistone” to cause global H3K27me3 reduction. EZHIP bears a methionine residue, similar to the H3 lysine-to-methionine (K27M) mutation, that is critical for mediating global H3K27me3 reduction [6, 7, 13]. The genomic distribution of H3K27me3 in H3K27M DIPGS and PFAs show remarkable similarities suggesting that these two tumors may be epigenetically related and share similar pathogenic mechanisms [2, 7]. Indeed, in support of this hypothesis, ~ 4% of PFAs demonstrate H3K27M mutations that are mutually exclusive from EZHIP mutations [11].
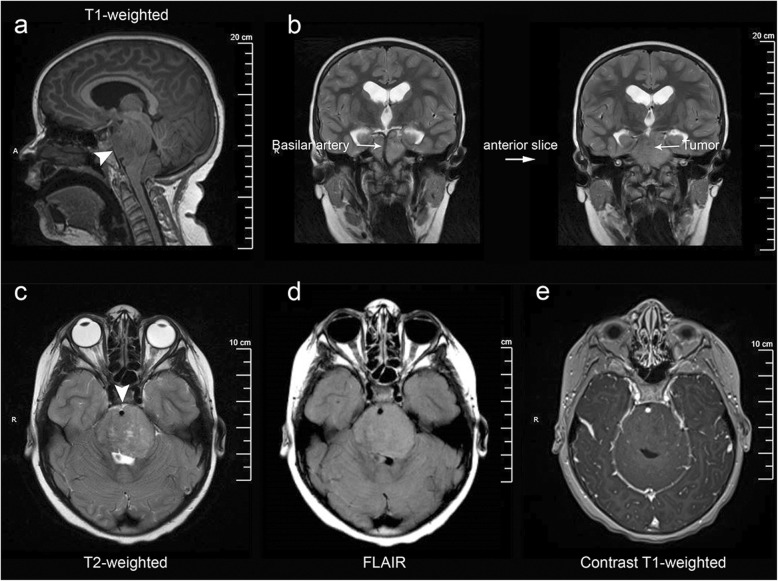
Here, we present an unusual case of a brainstem tumor with diagnostic radiographic and characteristic histopathologic features of DIPG, but demonstrating methylation features of PFA ependymoma. A 5-year-old boy presented to a local hospital with new-onset headache. Exam revealed evidence of cranial nerve deficits and cerebellar dysfunction (dysconjugate gaze and difficulty with tandem walking). MRI disclosed an infiltrating, expansile mass centered within the pons and obstructive hydrocephalus (Fig. 1a). The mass encased the basilar artery and contained a ventral exophytic component within the prepontine and suprasellar cisterns (Fig. 1b). Imaging characteristics included hyperintensity on T2 and fluid attenuated inversion recovery (FLAIR) sequences (Fig. 1c-d) and absence of enhancement following gadolinium administration (Fig. 1e). The mass did not involve the fourth ventricle or the cerebellum. Cystic change was noted. Other atypical features for DIPG, such as circumscription or dorsally exophytic growth, were not seen. The findings met clinicoradiologic criteria for DIPG [6]. Biopsy of the mass was deferred and radiation therapy was commenced.
Fig. 1.
Representative MRI characteristics. T1-weighted sagittal image showed an infiltrative mass centered within and expanding the pons (a). The exophytic portion of the tumor is seen encasing the basilar artery (b). The mass demonstrated classic MR characteristics of DIPG including increased T2 (c) and FLAIR (d) signals, and lack of post contrast enhancement (e)
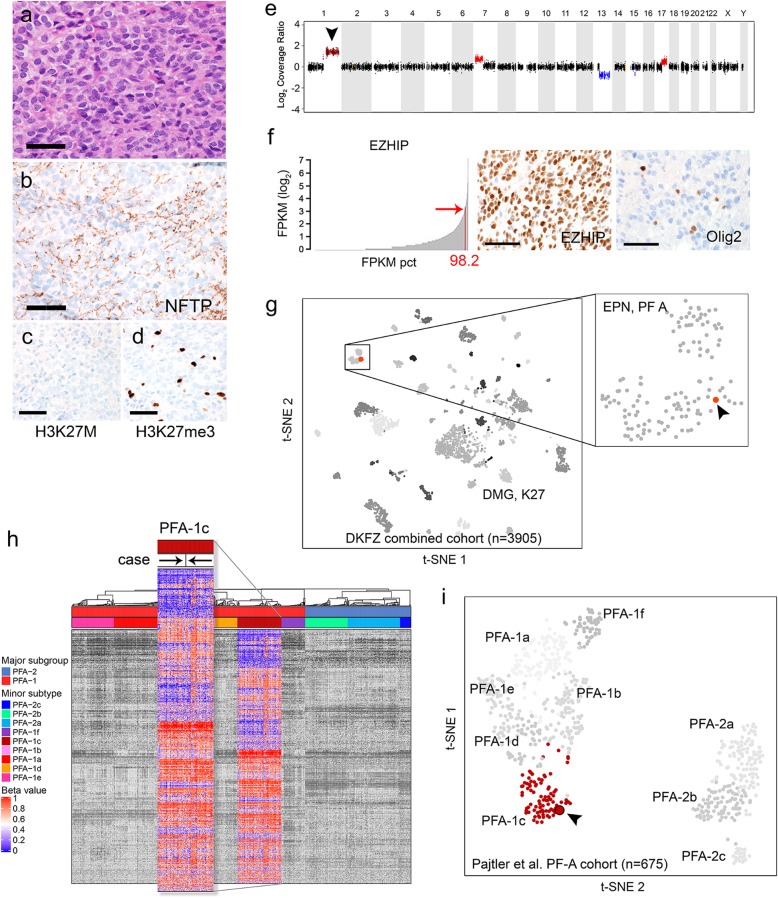
The patient was then evaluated at our institution for clinical eligibility in a trial for DIPG that prompted a tissue diagnosis. Biopsy revealed small fragments of densely cellular tumor (Fig. 2a). Scattered entrapped neurons hinted at its infiltrative nature (Supplemental Figure 1a-b). Tumor cells were positive for GFAP and neurofilament highlighted variable patterns of infiltration (Fig. 2b, Supplemental Figure 1c), with involvement of seemingly normal brain parenchyma by single tumor cells (Supplemental Figure 1e-f). No necrosis, microvascular proliferation, true ependymal or perivascular pseudorosettes were noted. Staining for H3K27M mutant protein was negative (Fig. 2c). Initial histologic and immunophenotypic findings suggested an H3-wildtype infiltrating astrocytoma consistent with DIPG. At our institution, pediatric CNS tumors frequently undergo integrative sequencing through the Michigan Oncology Sequencing Project (MI-ONCOSEQ) [16]. Tumors are assayed using whole exome and transcriptome-based techniques (see [15] for a description of the project). Sequencing revealed relatively few genomic alterations (Supplemental Table 1), but was notable for 1q gain (Fig. 2e) and confirmation of its H3-wildtype (H3F3A, HIST1H3B/C, HIST2H3A) status. RNA-seq showed overexpression of EZHIP mRNA that was confirmed by immunohistochemistry (Fig. 2f, Supplemental Figure 1d). Subsequent immunohistochemistry for H3K27me3 and Olig2 showed complete loss of nuclear expression in tumor cells (Fig. 2d, f). The relatively few genomic alterations and EZHIP overexpression in conjunction with loss of H3K27me3 in the absence of H3 mutations did not fit with classic molecular features of a H3-wildtype DIPG and prompted us to perform methylation analyses.
Fig. 2.
Histopathology and results of integrative sequencing and array-based methylation profiling. Routine H&E sections showed a high-grade cellular tumor (a). Tumor cells were seen infiltrating axons (b). H3K27M stain was negative (with appropriate control staining) and H3K27me3 showed loss with expression in admixed non-neoplastic cells (c, d). Copy number profiling through MI-ONCOSEQ mainly showed whole-arm changes including gains in 1q, 7p, 17p, and losses of 13q and partial 13p (e). Gene expression (RNA-seq) showed high expression of EZHIP (CXorf67) (f, expression is presented as log-transformed fragments per kilobase of exon model per million reads mapped, or FPKM, and shown as a percentile rank among the MI-ONCOSEQ compendium). Immunohistochemistry confirmed EZHIP protein overexpression and loss of Olig2 in tumor cells (f). Reproduction of the unsupervised clustering analysis of reference and diagnostic cohorts by Capper et al. [3] using t-Distributed Stochastic Neighbor Embedding (t-SNE), with incorporation of the presented case (g). Reproduction of the Consensus Clustering analysis by Pajtler et al. [11] and incorporation of our case showing clustering with the PFA-1c subtype (h). Heatmap representation and clustering were performed identically to the previously published methods [11]. Illustration of the previously defined [11] Consensus Clustering-based PFA major subgroups and minor subtypes (n = 675) using t-SNE dimensionality reduction (i); arrowhead denotes placement of the current case. Scale bars = 40 μm
Array-based profiling of CpG methylation in brain tumors has recently been shown to result in diagnostic refinements that are highly robust and prognostically meaningful [3]. We profiled our tumor using the Infinium MethylationEPIC BeadChip (interrogating ~ 850,000 CpG sites) in conjunction with the DKFZ Classifier tool recently implemented for CNS tumors (http://www.molecularneuropathology.org) [3]. While the methylation class most closely matched ‘ependymoma, posterior fossa group A’, the calibrated Classifier score was 0.62, below the proposed threshold of 0.9 (potential reasons are discussed below). To further assess the methylation profile of this tumor in relation to other CNS entities, we performed unsupervised clustering on the DKFZ cohort that comprises the 82 tumor methylation classes used in the Classifier (v11b4). Reproduction of the unsupervised clustering (t-SNE) demonstrated that the tumor clusters with the group ‘EPN, PF A’ (Fig. 2g). We next evaluated the tumor in relation to recently defined nine subtypes of PFA ependymoma [11]. Hierarchical clustering analysis revealed clustering within the PFA-1c subtype (Fig. 2h) and this was concordant with the results of t-SNE (Fig. 2i). While the overall Classifier score was 0.62, this may be due to the composition of PFA ependymomas in the current version of the Classifier (v11b4). The ‘EPN, PF A’ tumor class contains tumors arising solely within the fourth ventricle and/or cerebellum. Thus, the low calibrated score we encountered may reflect a potential subgroup of PFA ependymomas not yet recognized in the current implementation of the Classifier.
In summary, we present an unusual childhood brain tumor arising within the pons that met all clinical criteria for a DIPG but unexpectedly demonstrated H3K27me3 global reduction in the absence of H3.3 or H3.1 mutations, EZHIP overexpression, as well as clustering with tumors in the DNA methylation class ‘EPN, PF A’. While overall prognostic outcome has yet to be determined, a 7 month follow up MRI showed an increase in tumor size, portending an aggressive outcome. This case illustrates the significant contribution of molecular pathology to routine surgical neuropathology practice and highlights the increasingly important role of tissue procurement for genetic and epigenetic profiling of pediatric brain tumors in general. This case expands our knowledge of the epigenetic similarities including H3K27me3 global reduction between H3K27M DIPGs and PFA ependymomas [2, 7], and have biologic implications from both a neurodevelopmental perspective and in the design of targeted epigenetic therapies.
Supplementary information
Additional file 1: Supplemental Table 1. Results of relevant alterations detected through MI-ONCOSEQ integrative sequencing.
Additional file 2: Supplemental Figure 1. H&E images showing entrapped neurons within the tumor mass (a-b). Neurofilament immunohistochemistry showing infiltrative densely cellular regions adjacent to more delineated areas (a). EZHIP (CXorf67) immunohistochemistry showed increased nuclear expression in tumor cells (d) and served to highlight individual tumor cells percolating surrounding normal-appearing brain (e-f). Scale bars = 40 μm (a-b), 100 μm (c-f).
Acknowledgements
The Venneti lab is supported by grants from NINDS R01NS110572, Sidney Kimmel, St Baldrick’s, Claire McKenna, Chad Tough, Doris Duke and Sontag Foundations, Michigan Taubmann Institute and the University of Michigan Pediatric Brain Tumor Initiative. Funding CK is supported by NINDS K08-NS099427-01, the UM Department of Pediatrics Chad Carr Pediatric Brain Tumor Center, the Chad Tough Foundation, the Catching Up with Jack Foundation, the Prayers from Maria Foundation, and the U CAN-CER VIVE Foundation. The University of Michigan Peds-MiOncoSeq study was supported by grant 1UM1HG006508 from the NIH Clinical Sequencing Exploratory Research Award (private investigator: Arul Chinnaiyan, coinvestigator: Rajen Mody).
Authors’ contributions
DP conceptualized the project, drafted the manuscript, performed the bioinformatic analyses, and designed the figures. MQ, ZA, and KA performed and interpreted the methylation analysis. DH, FY and ARJ performed and interpreted IHC. RM and AC supervised the project. HJLG provided surgical care. CK supervised the project and provided clinical care. SV performed histopathologic review, conceptualized and supervised the project, and wrote the manuscript. The authors read and approved the final manuscript.
Consent for publication
Obtained.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Carl Koschmann, Email: ckoschma@med.umich.edu.
Sriram Venneti, Email: svenneti@med.umich.edu.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s40478-020-00905-w.
References
- 1.Albright AL, Packer RJ, Zimmerman R, Rorke LB, Boyett J, Hammond GD. Magnetic resonance scans should replace biopsies for the diagnosis of diffuse brain stem gliomas: a report from the Children's Cancer group. Neurosurgery. 1993;33:1026–1029. doi: 10.1227/00006123-199312000-00010. [DOI] [PubMed] [Google Scholar]
- 2.Bayliss J, Mukherjee P, Lu C, Jain SU, Chung C, Martinez D, Sabari B, Margol AS, Panwalkar P, Parolia A, et al. Lowered H3K27me3 and DNA hypomethylation define poorly prognostic pediatric posterior fossa ependymomas. Sci Transl Med. 2016;8:366ra161. doi: 10.1126/scitranslmed.aah6904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Capper D, Jones DTW, Sill M, Hovestadt V, Schrimpf D, Sturm D, Koelsche C, Sahm F, Chavez L, Reuss DE, et al (2018) DNA methylation-based classification of central nervous system tumours. Nature 555: 469–474 Doi 10.1038/nature26000 [DOI] [PMC free article] [PubMed]
- 4.Gupta N, Goumnerova LC, Manley P, Chi SN, Neuberg D, Puligandla M, Fangusaro J, Goldman S, Tomita T, Alden T, et al. Prospective feasibility and safety assessment of surgical biopsy for patients with newly diagnosed diffuse intrinsic pontine glioma. Neuro-Oncology. 2018;20:1547–1555. doi: 10.1093/neuonc/noy070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harutyunyan AS, Krug B, Chen H, Papillon-Cavanagh S, Zeinieh M, De Jay N, Deshmukh S, Chen CCL, Belle J, Mikael LG, et al. H3K27M induces defective chromatin spread of PRC2-mediated repressive H3K27me2/me3 and is essential for glioma tumorigenesis. Nat Commun. 2019;10:1262. doi: 10.1038/s41467-019-09140-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hubner JM, Muller T, Papageorgiou DN, Mauermann M, Krijgsveld J, Russell RB, Ellison DW, Pfister SM, Pajtler KW, Kool M (2019) EZHIP / CXorf67 mimics K27M mutated oncohistones and functions as an intrinsic inhibitor of PRC2 function in aggressive posterior fossa ependymoma. Neuro Oncol: Doi. 10.1093/neuonc/noz058 [DOI] [PMC free article] [PubMed]
- 7.Jain SU, Do TJ, Lund PJ, Rashoff AQ, Diehl KL, Cieslik M, Bajic A, Juretic N, Deshmukh S, Venneti S, et al (2019) PFA ependymoma-associated protein EZHIP inhibits PRC2 activity through a H3 K27M-like mechanism. Nat Commun 10: 2146 Doi 10.1038/s41467-019-09981-6 [DOI] [PMC free article] [PubMed]
- 8.Jansen MH, Veldhuijzen van Zanten SE, Sanchez Aliaga E, Heymans MW, Warmuth-Metz M, Hargrave D, van der Hoeven EJ, Gidding CE, de Bont ES, Eshghi OS, et al. Survival prediction model of children with diffuse intrinsic pontine glioma based on clinical and radiological criteria. Neuro-Oncology. 2015;17:160–166. doi: 10.1093/neuonc/nou104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lewis PW, Muller MM, Koletsky MS, Cordero F, Lin S, Banaszynski LA, Garcia BA, Muir TW, Becher OJ, Allis CD. Inhibition of PRC2 activity by a gain-of-function H3 mutation found in pediatric glioblastoma. Science. 2013;340:857–861. doi: 10.1126/science.1232245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ostrom QT, Cioffi G, Gittleman H, Patil N, Waite K, Kruchko C, Barnholtz-Sloan JS. CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the United States in 2012-2016. Neuro-Oncology. 2019;21:v1–v100. doi: 10.1093/neuonc/noz150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pajtler KW, Wen J, Sill M, Lin T, Orisme W, Tang B, Hubner JM, Ramaswamy V, Jia S, Dalton JD, et al. Molecular heterogeneity and CXorf67 alterations in posterior fossa group a (PFA) ependymomas. Acta Neuropathol. 2018;136:211–226. doi: 10.1007/s00401-018-1877-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Panwalkar P, Clark J, Ramaswamy V, Hawes D, Yang F, Dunham C, Yip S, Hukin J, Sun Y, Schipper MJ, et al. Immunohistochemical analysis of H3K27me3 demonstrates global reduction in group-a childhood posterior fossa ependymoma and is a powerful predictor of outcome. Acta Neuropathol. 2017;134:705–714. doi: 10.1007/s00401-017-1752-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Piunti A, Smith ER, Morgan MAJ, Ugarenko M, Khaltyan N, Helmin KA, Ryan CA, Murray DC, Rickels RA, Yilmaz BD et al (2019) CATACOMB: an endogenous inducible gene that antagonizes H3K27 methylation activity of Polycomb repressive complex 2 via an H3K27M-like mechanism. Sci Adv 5: eaax2887. 10.1126/sciadv.aax2887 [DOI] [PMC free article] [PubMed]
- 14.Puget S, Beccaria K, Blauwblomme T, Roujeau T, James S, Grill J, Zerah M, Varlet P, Sainte-Rose C. Biopsy in a series of 130 pediatric diffuse intrinsic Pontine gliomas. Childs Nerv Syst. 2015;31:1773–1780. doi: 10.1007/s00381-015-2832-1. [DOI] [PubMed] [Google Scholar]
- 15.Robinson DR, Wu YM, Lonigro RJ, Vats P, Cobain E, Everett J, Cao X, Rabban E, Kumar-Sinha C, Raymond V, et al. Integrative clinical genomics of metastatic cancer. Nature. 2017;548:297–303. doi: 10.1038/nature23306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roychowdhury S, Iyer MK, Robinson DR, Lonigro RJ, Wu YM, Cao X, Kalyana-Sundaram S, Sam L, Balbin OA, Quist MJ et al (2011) Personalized oncology through integrative high-throughput sequencing: a pilot study. Sci Transl med 3: 111ra121. 10.1126/scitranslmed.3003161 [DOI] [PMC free article] [PubMed]
- 17.Schwartzentruber J, Korshunov A, Liu XY, Jones DT, Pfaff E, Jacob K, Sturm D, Fontebasso AM, Quang DA, Tonjes M, et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature. 2012;482:226–231. doi: 10.1038/nature10833. [DOI] [PubMed] [Google Scholar]
- 18.Stafford JM, Lee CH, Voigt P, Descostes N, Saldana-Meyer R, Yu JR, Leroy G, Oksuz O, Chapman JR, Suarez F et al (2018) Multiple modes of PRC2 inhibition elicit global chromatin alterations in H3K27M pediatric glioma. Sci Adv 4: eaau5935. 10.1126/sciadv.aau5935 [DOI] [PMC free article] [PubMed]
- 19.Wu G, Broniscer A, McEachron TA, Lu C, Paugh BS, Becksfort J, Qu C, Ding L, Huether R, Parker M, et al. Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nat Genet. 2012;44:251–253. doi: 10.1038/ng.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Supplemental Table 1. Results of relevant alterations detected through MI-ONCOSEQ integrative sequencing.
Additional file 2: Supplemental Figure 1. H&E images showing entrapped neurons within the tumor mass (a-b). Neurofilament immunohistochemistry showing infiltrative densely cellular regions adjacent to more delineated areas (a). EZHIP (CXorf67) immunohistochemistry showed increased nuclear expression in tumor cells (d) and served to highlight individual tumor cells percolating surrounding normal-appearing brain (e-f). Scale bars = 40 μm (a-b), 100 μm (c-f).