Abstract
MicroRNAs (miRNAs) are small RNAs that guide Argonaute (AGO) proteins to specific target messenger RNAs (mRNAs) to repress their translation and stability. Canonically, miRNA targeting is reliant on base pairing of the seed region, nucleotides (nts) 2–7, of the miRNA to sites in mRNA 3’UTRs. Recently, the 3’ half of the miRNA has gained attention for newly appreciated roles in regulating target specificity and regulation. Additionally, the extent of pairing to the miRNA 3’-end can influence the stability of the miRNA itself. These findings highlight the importance of sequences beyond the seed in controlling the function and existence of miRNAs.
Keywords: microRNA, miRNA targeting, seed pairing, miRNA family
Target recognition and regulation by miRNAs
Since their discovery in the early 1990s [1,2], thousands of microRNAs (miRNAs) have been identified across the plant and animal kingdoms [3]. There is now evidence that miRNAs impact every major biological pathway by regulating the expression of substantial fractions of protein-coding genes [4,5]. Given this omnipresent role in gene regulation, it is not surprising that misregulation of individual miRNAs can have dire consequences, contributing to a variety of diseases and afflictions in humans [6].
Within the miRNA induced silencing complex (miRISC) (see Glossary), the ~22 nucleotide (nt) miRNA recruits Argonaute (AGO) to specific target sites via base-pairing interactions [4,7]. Perfect base-pairing of the miRNA with its target site, which is common in plants but rare in animals, results in endonucleolytic cleavage by AGO of the target RNA. Animal miRNAs typically form a partial duplex with their target site, which prevents cleavage and instead relies on AGO co-factors to regulate target expression through translational repression and mRNA destabilization [4,7]. Pairing of nucleotides (nts) 2–7 of the miRNA, called the seed, to its target site has generally been considered the minimal element needed to engage a target mRNA [4]. Indeed, structural studies have shown that only sequences within the seed of the AGO-bound miRNA are available for initial pairing to a target site [8–10]. Additionally, single molecule studies have demonstrated the importance of the seed in stable target site engagement [11–13]. Interestingly, once miRISC binds target RNA, AGO can undergo a conformational change that allows for extended seed pairing and exposes part of the miRNA 3’ region (nts 13–16) for additional interactions with the target [14]. A recent systematic investigation of pairing interactions between a miRNA and target corroborates a sequential recognition model where the miRNA seed binds first and then nts in the 3’ half are able to bind the target site [15]. The relevance of pairing to miRNA 3’-end sequences has been demonstrated in several new studies showing that it can impact the specificity of targeting, the regulatory mechanism, and the stability of the miRNA itself.
Same seed, different targets
Given its importance in many established miRNA-target interactions, seed pairing potential is the foundation of popular target prediction programs [4]. As such, members of a miRNA family that share seed sequences are typically assigned to the same target sites. However, there is mounting in vivo evidence that pairing interactions beyond the seed can lead to non-overlapping target profiles for individual miRNAs in a family. On a genome-wide scale, crosslinking and immunoprecipitation with sequencing (CLIP-seq) methods have been used to identify endogenous AGO-bound target sites. In some of these studies, the target sequence became ligated to the presumptive targeting miRNA, producing miRNA-target chimeric reads [16–19]. These types of sequencing reads allow for the unambiguous identification of which specific miRNA recruited AGO to a particular target site. Many chimeras contained target sequences that could pair to the seed of the attached miRNA, reaffirming the prevalence of this pairing motif in endogenous miRNA-target interactions [16–19]. Contrary to expectation though, miRNAs within a family did not always produce chimeras with the same target sites, suggesting that sequences beyond the seed can contribute to target recognition in vivo [18,19].
The examination of miRNA-target chimeras generated from CLIP-based studies in human cell culture, mouse brain and whole larval stage C. elegans revealed that individual miRNA family members, often called sisters, can exhibit biased target interactions [18,19]. With identical seed sequences, miRNA sisters apparently rely on their potential for unique 3’-end pairing interactions to engage some targets. For sister-specific target sites in both the mammalian and worm studies, the chimera-forming miRNA had a more favorable predicted binding affinity than that of its sisters [18,19]. Consistent with non-identical target preferences, sister miRNAs were also shown to differ in their regulatory capacity for targets with 3’-pairing interactions that favored one of the family members. In mammalian cells, reporters with sister-specific target sites were generally repressed more potently by the chimera-forming miRNA versus other family members upon transfection into the culture system [18].
Taking advantage of a well-established let-7 miRNA target in Caenorhabditis elegans, Broughton et al., further demonstrated the importance of 3’-end pairing interactions for specific and functional targeting in vivo [19]. The lin-41 3’UTR contains two let-7 target sites that lack perfect seed pairing (one involves a G-U pair and the other forces a target nucleotide bulge) but support perfect pairing to nucleotides 11–19 of the let-7 miRNA (Fig 1A) [20,21]. Loss of let-7 results in misregulation of lin-41 and lethality, despite the expression of sister miRNAs that apparently cannot compensate [19–21]. The wild-type lin-41 3’UTR only formed chimeras with let-7 miRNA, but this specificity was found to be transferrable when 3’-end pairing was designed to favor a sister [19]. CRISPR/Cas9-based genome editing was used to swap each let-7 target site for a site that had formed chimeras exclusively with its sister miRNA, miR-48. Importantly, these new lin-41 3’UTR sites supported canonical seed pairing with any of the let-7 family members but were predicted to bind more extensively to the 3’-end of miR-48 (Fig 1B). Worms with the edited lin-41 3’UTR were found now to depend on miR-48, but not let-7, for regulation of this gene and, ultimately, viability (Fig 1B) [19]. This study confirmed that pairing to sequences beyond the seed can confer specific and functional miRNA target interactions in vivo.
Figure 1:

Auxiliary pairing of miRNA 3’-end sequences can overcome seed imperfections and confer target specificity to miRNA sisters. A) In C. elegans, the lin-41 3’UTR contains two let-7 miRNA target sites that each feature extensive complementarity to the 3’-half of let-7 and imperfect seed-pairing potential: Site 1 forces a target nt bulge and Site 2 includes an unfavorable G:U base pair (pairing to the miRNA seed, nts 2–7, is shaded gray). While let-7 family members, such as miR-48, can support the same seed-pairing architecture, only let-7 has sufficient 3’-end pairing capacity to regulate lin-41, allowing for normal worm development; loss of lin-41 regulation by let-7 results in lethality (depicted by skull and crossbones) because the let-7 sisters cannot compensate. B) Exchange of the let-7 sites for sequences predicted to correct the seed imperfections but pair more favorably to the 3’-end of miR-48 transfers regulation of lin-41 from let-7 to miR-48. Sites 1’ and 2’ are duplications of a sequence in the dot-1.1 3’UTR that only formed chimeras with miR-48 [19]. C) The inclusion of pairing to nt 8 (shaded yellow) in this context, provides a seed architecture that allows regulation by let-7 or miR-48, regardless of 3’-pairing capacity [22]. Sites 1” and 2” are duplications of the sequence in (B) except for the substitution of U for C to enable canonical pairing to the G at the 8th position in let-7 and miR-48.
While imperfect pairing to the miRNA seed can be compensated by extended pairing interactions with the 3’-portion of the miRNA, even seemingly perfect seed matches can depend on additional pairing [4]. One explanation for this phenomenon is that sequences immediately adjacent to the seed can influence targeting. In fact, there is a hierarchy of seed pairing architecture wherein targets that pair to miRNA nts 2–7 alone are generally less repressed than those that include pairing to the 8th position [4]. New work from Brancati and Grosshans shows that the ability to pair with nt 8 can also influence the specificity of miRNA target interactions [22]. Using the same C. elegans model described above, these authors demonstrated that perfect pairing of the lin-41 target sites to nts 2–8 of let-7 permits regulation by other family members, regardless of differences in potential 3’-pairing interactions (Fig 1C). However, seed pairing at nts 2–7 with a G:U wobble pair at position 8 was sufficient to reinstate dependence on pairing to miRNA 3’-end sequences (Fig 1B). In some cases, bias for targeting by a specific sister was sensitive to the expression levels of other family members [22]. This work highlights the importance of considering overall miRNA-target pairing architecture as well as miRNA abundance in understanding target recognition and regulation in vivo.
Target sites within coding regions supersede canonical recognition and regulation
Many of the miRISC binding sites identified through CLIP-based studies include the expected features: seed-pairing capacity, 3’UTR residence, and an association with target mRNA destabilization (Fig 2) [16–19,23–26]. However, in all of these reports sizeable fractions of AGO-bound sites were also detected in protein-coding sequences (CDSs), often lacking in complementary seed motifs or clear effects on target mRNA regulation [16–19,23–26]. While specific examples of functional CDS-located miRNA target sites have emerged [27–29], in general this position in the mRNA has been regarded as suboptimal for eliciting a regulatory outcome because stable miRISC association would be thwarted by translating ribosomes [30]. Thus, the relevance of the thousands of CDS miRNA target sites, including some that are highly reproducible across biological replicates, has been an outstanding question.
Figure 2:

The structure and position of miRNA-target interactions impose different regulatory outcomes. Canonical miRNA target sites reside in 3’UTRs, require seed-pairing, and rely on the AGO co-factor GW182/ TNRC6 to recruit deadenylases and other factors that act to destabilize and repress translation initiation of the target mRNA (top) [4,7]. The example is a 3’UTR site engineered to limit pairing to the miR-20a seed region [31]. A new class of target sites are located in coding sequences (CDSs), lack seed complementarity and, instead, offer extensive pairing to miRNA 3’-ends; these types of sites seem to block translation elongation independently of GW182/ TNRC6, resulting in reduced protein but not mRNA levels of the target (bottom) [31]. The example is the CDS site in exon 2 of DAPK3 mRNA paired to miR-20a [31].
In recent work, Zhang et al. provide compelling evidence that some CDS miRNA target sites actually comprise a new category of recognition elements [31]. Multiple independent studies have identified a miR-20a target site in the second exon of DAPK (a p53-activating kinase) [16,23,26,32]. Curiously, this site lacks seed pairing and, except for a G:U wobble at position 6 and a bulged C at position 12, is perfectly complementary to miR-20a nts 5–23 (Fig 2). Within the CDS context, this pairing architecture sufficed for target regulation, but it lost functionality when inserted into in the 3’UTR [31]. Additional examples of CDS sites with weak seed and extensive 3’-end pairing interactions for a different miRNA were also shown to mediate target regulation when placed in coding but not untranslated regions, leading the authors to define these as a novel class of miRNA recognition elements (MREs) [31].
Not only are the pairing architecture and location of MREs in this class unusual, the regulatory mechanism is also atypical. These MREs apparently repress translation without triggering mRNA destabilization (Fig 2) [31]. In contrast to miRNA seed-dependent interactions in 3’UTRs, seedless sites in CDSs interfere with translation through a mechanism that does not rely on the AGO co-factor GW182/TNRC6 (Fig 2) [31]. The GW182/TNRC6 protein is instrumental in recruiting deadenylation factors to initiate mRNA decay of canonical targets [4,7]. Its absence in complexes that regulate CDS targets may explain their lack of influence on mRNA levels. The stark contrasts between canonical 3’UTR target sites and the newly described CDS sites that depend more on 3’-pairing interactions highlight wide gaps in our understanding of functional miRNA targeting rules.
Seeding miRNA decay with 3’-end pairing
While miRNA pairing potential can greatly influence the specificity and mechanism of target regulation, it can also impact the fate of the miRNA. Following initial studies in Drosophila and mammalian cells [33,34], numerous examples of Target RNA-Directed MicroRNA Degradation (TDMD) have emerged [35]. In this pathway, seed along with extensive pairing to the 3’-half of the miRNA to a target site can trigger decay of the miRNA itself. While the factors and mechanisms that sense this pairing structure and elicit destruction of the miRNA are yet to be fully revealed, TDMD often is associated with non-templated nt additions, called tailing, and trimming at the miRNA 3’-end [35]. Since efficient TDMD seems to require an unusually high degree of pairing to nts in the 3’-half of the miRNA [33,36], how often this pathway functions in vivo has been an open question. Nonetheless, intriguing examples of viruses expressing transcripts that trigger TDMD of host miRNAs established that this is a biologically relevant mechanism for regulating gene expression [34,37–39].
In just this past year, several examples of host-mediated TDMD of endogenous miRNAs have come to light [40–42]. In one study, a conserved block of sequence containing a highly complementary site to miR-29b was shown to regulate spatial expression of this miRNA (Fig. 3) [40]. This element is present within a long non-coding RNA (lncRNA) called libra or the 3’UTR of the Nrep mRNA throughout vertebrate evolution. In mice and zebrafish, high expression of these transcripts in the cerebellum leads to TDMD of miR-29b in this brain region [40]. Importantly, loss of this regulatory mechanism resulted in striking behavioral defects, including impaired motor functions in mice and aberrant exploratory and anxiety-like behaviors in zebrafish [40].
Figure 3:
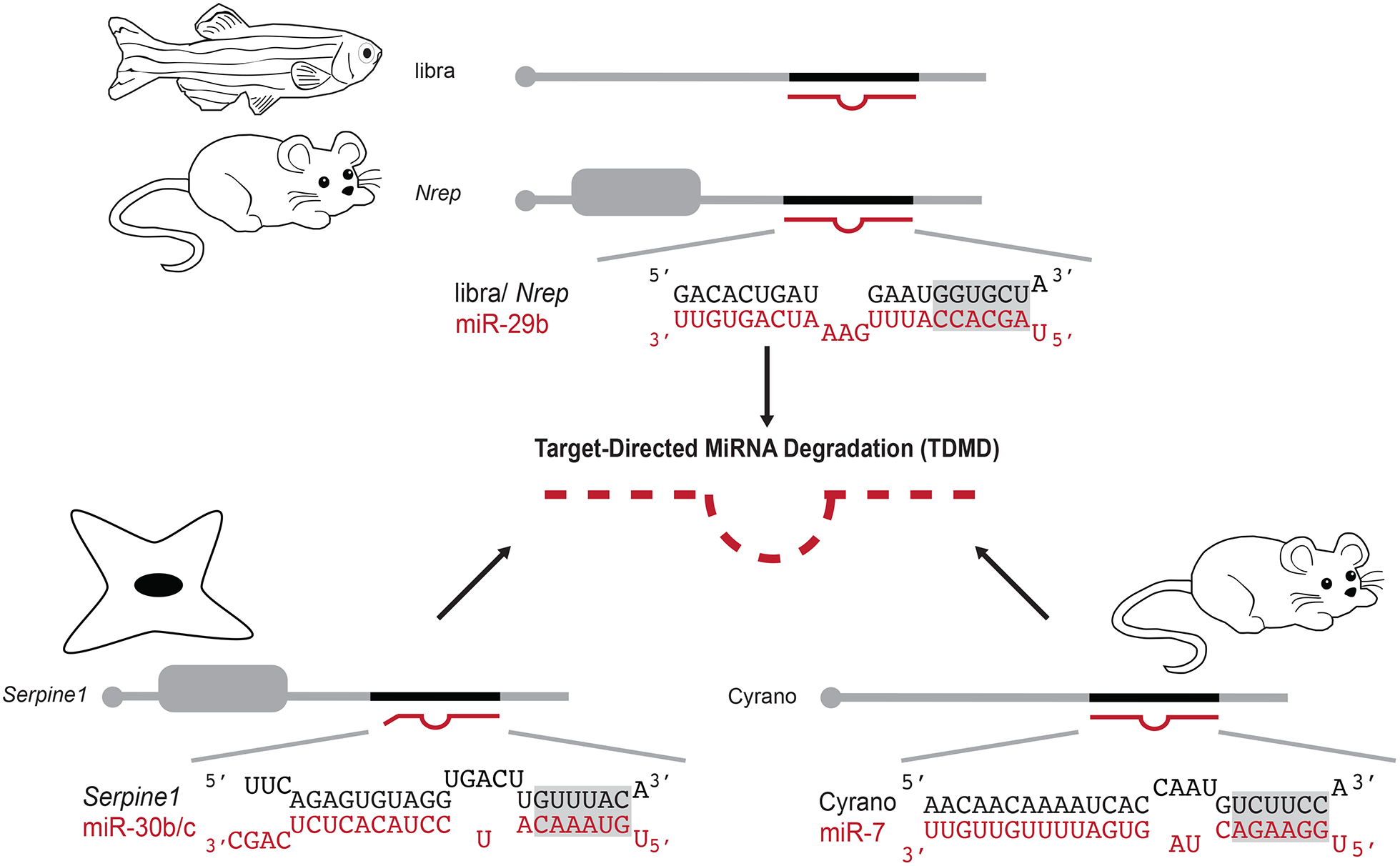
Extensive pairing between a miRNA and target can induce Target-Directed miRNA Degradation (TDMD). TDMD of miR-29b can be triggered by pairing to a conserved region in the zebrafish lncRNA, libra, or the mouse Nrep 3’UTR (top) [40]. A site in the 3’UTR of Serpine1 induces TDMD of miR-30b/c in mouse fibroblasts (bottom left) [42].
In mice, pairing of miR-7 to a site in the lncRNA Cyrano results in rapid decay of the miRNA through TDMD [41].
One advantage of using TDMD to regulate miRNA expression is that it can be selective for miRNA family members with differing degrees of 3’-end pairing interactions. Indeed, the extended complementarity of miR-29b designates this miRNA, but not its sisters, for TDMD through the pathway described above [40]. Likewise, the ability of a site in the Serpine1 3’UTR to form 10 contiguous pairs with miR-30b/c triggered TDMD of those sisters but not the less complementary miR-30a/d/e family members in mouse fibroblasts (Fig 3) [42]. When this site was removed from the Serpine1 3’UTR by CRISPR/Cas9, levels of miR-30b/c increased, which impacted the degree and specificity of targeting by these miRNAs. Loss of Serpine1-mediated TDMD led to cell cycle and stress response defects, suggesting that this pathway plays a critical role in the regulation of miR-30b/c activity [42].
While Ghini et al. suggested that Serpine1 may be just one of over a thousand endogenous TDMD trigger mRNAs, non-coding RNAs provide another source of potential targets for controlling miRNA stability through this pathway [42]. The Cyrano lncRNA, which includes a highly complementary miR-7 site, is broadly conserved across vertebrates (Fig 3) [43]. While knockdown of Cyrano in zebrafish resulted in neurodevelopmental defects [43], mouse knockouts of Cyrano appear normal [41]. However, loss of Cyrano in mouse brain tissue led to increased miR-7 levels, which was associated with a general derepression of its targets [41]. While the importance of miR-7 TDMD by Cyrano in mammals awaits further studies, the conservation of this non-coding RNA and its potency in triggering miR-7 decay make it an intriguing model.
Concluding remarks and future perspectives
While the seed is long-recognized and well-supported as a critical element in miRNA targeting [4,7], there is a growing appreciation that sequences in the 3’-half of the miRNA have roles to play as well. Members of a miRNA family can recognize unique targets depending on seed region architecture along with the potential for 3’-pairing interactions. In parallel, specific sisters can be subjected to TDMD via differences in their 3’-end regions. Further elucidation of the pairing rules that govern selective targeting and TDMD will be needed to realize how widespread these events are (see Outstanding Questions). The demonstration of a new class of miRNA targets that depend on 3’-end interactions, but not seed pairing, suggests there is still much to be learned about how the miRNA complex engages targets in vivo. Furthermore, the existence of targets that lack seed complementarity, are located in CDSs, and exclusively undergo translational repression could mean that the extent of gene regulation by the miRNA pathway may be farther-reaching than previously considered. With limited sequence content of only ~22 nts, it is now becoming clear that each nucleotide in a miRNA contributes to an overall pairing architecture that can influence recognition and regulation of the target, as well as stability of the miRNA.
Acknowledgements
We thank members of the Pasquinelli lab for suggestions and critical reading of the manuscript. Support for this work was from the UCSD Cellular and Molecular Genetics Training Program through an institutional grant from the National Institute of General Medicine (T32 GM007240) and a National Science Foundation Graduate Research Fellowship (DGE-1650112) to L.B.C. and grants from the National Institute of Aging (R01 AG056562) and the National Institute of General Medicine (R35 GM127012) to A.E.P.
Glossary
- CLIP-seq
crosslinking and immunoprecipitation with sequencing (CLIP-seq) is a technique used to isolate and sequence RNA bound to a specific protein. This method has been used to identify miRNAs and target sites bound by AGO
- Chimeric Reads
contiguous sequences from CLIP-seq-based assays that contain two independently derived RNA sequences that became ligated during library preparation. The chimeric reads in AGO CLIP-seq datasets represent a miRNA associated with a specific target site. The name refers to different entities brought together into one being, as in the Chimera in Greek mythology composed of a lion’s head, goat’s body and a serpent’s tail
- CRISPR/Cas9
a genome editing method that uses guide RNAs to target CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) associated nuclease (Cas9) to specific DNA sequences for endonucleolytic cleavage. Repair of the cut DNA can be engineered to incorporate any new sequence of interest. This method is based on a natural Prokaryotic defense system against bacteriophage that involves the integration of foreign DNA sequences between CRISPR segments in a bacterial genome
- GW182/TNRC6
alternative names for a miRISC factor that bridges AGO to proteins that promote deadenylation and translational repression of bound targets. GW182 refers to the molecular weight and glycine/tryptophan repeats that characterize this protein. TNRC6 stands for Trinucleotide repeat containing gene 6
- miRISC
miRNA induced silencing complex (miRISC). A complex consisting of a mature miRNA, AGO, and potentially other proteins that function in target regulation
- miRNA family
a group of miRNAs that share a seed but differ to varying degrees in the rest of their sequence
- miRNA Recognition Element (MRE)
miRNA binding site, the region in a target RNA that can base pair to a miRNA
- Seed
nucleotides (nts) 2–7 counting from the 5’end of a miRNA. Canonical targeting involves perfect pairing of the miRNA seed to target sequences
- Sisters
members of the same miRNA family
- Target RNA-Directed miRNA Degradation (TDMD)
degradation of a miRNA caused by binding to a highly complementary target RNA sequence
- Tailing
the addition of non-templated nucleotides to the 3’-end of an RNA, usually by addition of uridines and/or adenosines
References
- 1.Lee RC et al. (1993) The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75, 843–54 [DOI] [PubMed] [Google Scholar]
- 2.Wightman B et al. (1993) Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 75, 855–862 [DOI] [PubMed] [Google Scholar]
- 3.Kozomara A et al. (2018) miRBase: from microRNA sequences to function. Nucleic Acids Res. DOI: 10.1093/nar/gky1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartel DP (2018) Metazoan MicroRNAs. Cell 173, 20–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu H et al. (2018) Small but powerful: function of microRNAs in plant development. Plant Cell Rep. 37, 515–528 [DOI] [PubMed] [Google Scholar]
- 6.Paul P et al. (2018) Interplay between miRNAs and human diseases. J. Cell. Physiol 233, 2007–2018 [DOI] [PubMed] [Google Scholar]
- 7.Gebert LFR and MacRae IJ (2018) Regulation of microRNA function in animals. Nat. Rev. Mol. Cell Biol DOI: 10.1038/s41580-018-0045-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elkayam E et al. (2012) The structure of human argonaute-2 in complex with miR-20a. Cell 150, 100–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nakanishi K et al. (2012) Structure of yeast Argonaute with guide RNA. Nature 486, 368–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schirle NT and MacRae IJ (2012) The Crystal Structure of Human Argonaute2. Science 336, 1037–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chandradoss SD et al. (2015) A Dynamic Search Process Underlies MicroRNA Targeting. Cell 162, 96–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jo MH et al. (2015) Human Argonaute 2 Has Diverse Reaction Pathways on Target RNAs. Mol. Cell 59, 117–124 [DOI] [PubMed] [Google Scholar]
- 13.Salomon WE et al. (2015) Single-Molecule Imaging Reveals that Argonaute Reshapes the Binding Properties of Its Nucleic Acid Guides. Cell 162, 84–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schirle NT et al. (2014) Structural basis for microRNA targeting. Science 346, 608–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yan Y et al. (2018) The sequence features that define efficient and specific hAGO2-dependent miRNA silencing guides. Nucleic Acids Res. 46, 8181–8196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Helwak A et al. (2013) Mapping the human miRNA interactome by CLASH reveals frequent noncanonical binding. Cell 153, 654–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grosswendt S et al. (2014) Unambiguous Identification of miRNA: Target site interactions by different types of ligation reactions. Mol. Cell 54, 1042–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moore MJ et al. (2015) MiRNA-target chimeras reveal miRNA 3′-end pairing as a major determinant of Argonaute target specificity. Nat. Commun 6, 1–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Broughton JP et al. (2016) Pairing beyond the Seed Supports MicroRNA Targeting Specificity. Mol. Cell 64, 320–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reinhart BJ et al. (2000) The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 403, 901–906 [DOI] [PubMed] [Google Scholar]
- 21.Slack FJ et al. (2000) The lin-41 RBCC Gene Acts in the C. elegans Heterochronic Pathway between the let-7 Regulatory RNA and the LIN-29 Transcription Factor. Molecular 5, 659–669 [DOI] [PubMed] [Google Scholar]
- 22.Brancati G and Großhans H (2018) An interplay of miRNA abundance and target site architecture determines miRNA activity and specificity. Nucleic Acids Res. 46, 3259–3269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chi SW et al. (2009) Argonaute HITS-CLIP decodes microRNA – mRNA interaction maps. Nature 460, 479–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hafner M et al. (2010) Transcriptome-wide Identification of RNA-Binding Protein and MicroRNA Target Sites by PAR-CLIP. Cell 141, 129–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zisoulis DG et al. (2010) Comprehensive discovery of endogenous Argonaute binding sites in Caenorhabditis elegans. Nat. Struct. Mol. Biol 17, 173–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xue Y et al. (2013) Direct conversion of fibroblasts to neurons by reprogramming PTB-regulated MicroRNA circuits. Cell 152, 82–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Duursma AM et al. (2008) miR-148 targets human DNMT3b protein coding region. RNA 14, 872–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tay Y et al. (2008) MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature 455, 1124–1128 [DOI] [PubMed] [Google Scholar]
- 29.Hausser J et al. (2013) Analysis of CDS-located miRNA target sites suggests that they can effectively inhibit translation. Genome Res. 23, 604–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gu S et al. (2009) Biological basis for restriction of microRNA targets to the 3′ untranslated region in mammalian mRNAs. Nat. Struct. Mol. Biol 16, 144–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang K et al. (2018) A novel class of microRNA-recognition elements that function only within open reading frames. Nat. Struct. Mol. Biol DOI: 10.1038/s41594-018-0136-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cai Z et al. (2015) Oncogenic miR-17/20a forms a positive feed-forward loop with the p53 kinase DAPK3 to promote tumorigenesis. J. Biol. Chem 290, 19967–19975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ameres SL et al. (2010) Target RNA-directed trimming and tailing of small silencing RNAs. Science 328, 1534–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cazalla D et al. (2010) Down-regulation of a host microRNA by a Herpesvirus saimiri noncoding RNA. Science 328, 1563–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fuchs Wightman F et al. (2018) Target RNAs strike back on microRNAs. Front. Genet 9, 435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baccarini A et al. (2011) Kinetic analysis reveals the fate of a MicroRNA following target regulation in mammalian cells. Curr. Biol 21, 369–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Libri V et al. (2012) Murine cytomegalovirus encodes a miR-27 inhibitor disguised as a target. Proc. Natl. Acad. Sci 109, 279–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marcinowski L et al. (2012) Degradation of cellular mir-27 by a novel, highly abundant viral transcript is important for efficient virus replication in vivo. PLoS Pathog. 8, e1002510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee S et al. (2013) Selective degradation of host MicroRNAs by an intergenic HCMV noncoding RNA accelerates virus production. Cell Host Microbe 13, 678–690 [DOI] [PubMed] [Google Scholar]
- 40.Bitetti A et al. (2018) MicroRNA degradation by a conserved target RNA regulates animal behavior. Nat. Struct. Mol. Biol 25, 244–251 [DOI] [PubMed] [Google Scholar]
- 41.Kleaveland B et al. (2018) A Network of Noncoding Regulatory RNAs Acts in the Mammalian Brain. Cell 174, 350–362.e17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ghini F et al. (2018) Endogenous transcripts control miRNA levels and activity in mammalian cells by target-directed miRNA degradation. Nat. Commun 9, 3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ulitsky I et al. (2011) Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell 147, 1537–50 [DOI] [PMC free article] [PubMed] [Google Scholar]


