Abstract
In the face of ongoing habitat fragmentation, many primate species have experienced reduced gene flow resulting in a reduction of genetic diversity, population bottlenecks, and inbreeding depression, including golden snub-nosed monkeys Rhinopithecus roxellana. Golden snub-nosed monkeys live in a multilevel society composed of several 1 male harem units that aggregate to form a cohesive breeding band, which is followed by one or more bachelor groups composed of juvenile, subadult, and adult male members. In this research, we examine the continuous landscape resistance surface, the genetic diversity and patterns of gene flow among 4 isolated breeding bands and 1 all-male band in the Qinling Mountains, China. Landscape surface modeling suggested that human activities and ecological factors severely limit the movement of individuals among breeding bands. Although these conditions are expected to result in reduced gene flow, reduced genetic diversity, and an increased opportunity for a genetic bottleneck, based on population genetic analyses of 13 microsatellite loci from 188 individuals inhabiting 4 isolated breeding bands and 1 all-male band, we found high levels of genetic diversity but low levels of genetic divergence, as well as high rates of gene flow between males residing in the all-male band and each of the 4 breeding bands. Our results indicate that the movement of bachelor males across the landscape, along with their association with several different breeding bands, appears to provide a mechanism for promoting gene flows and maintaining genetic diversity that may counteract the otherwise isolating effects of habitat fragmentation.
Keywords: gene flow; male dispersal; multilevel society; Rhinopithecus roxellana, social organization
In natural populations, genetic diversity is maintained by persistent migration, interbreeding avoidance, and genetic fusion (Orr 2005; Barrett and Schluter 2008). However, with extensive human activities, natural habitats have become fragmented into small isolated units (Pfeifer et al. 2017), resulting in the formation of insular subpopulations of individual animal species (Coltman 2005; Liu et al. 2009). Such situations restrict the movement of individuals across populations (Lande 1993), leading to the increased risk of inbreeding depression and the occurrence of genetic bottlenecks (Charlesworth and Willis 2009; Hedrick and Garcia-Dorado 2016; Trewick et al. 2017). Notable examples include the gray wolf Canis lupus (Leonard et al. 2005), the northern elephant seal Mirounga angustirostris (Weber et al. 2000), the moose Alces alces (Broders et al. 1999), and several species of nonhuman primate. At present, some 60% of the 504 living primate species are listed as vulnerable, endangered, or critically endangered and over 75% of all primate populations are in decline (Estrada et al. 2017). In most cases, population decline has resulted in a marked decrease in genetic diversity. However, there remains a small number of primate species, which despite a recent reduction in population size, maintain relatively high genetic diversity in the face of habitat fragmentation (Quéméré et al. 2010; Swedell 2011).
Several common factors associated with social, behavioral, and biological adaptability appear to enable these “resilient” species to persist and maintain genetic diversity in highly-fragmented and human-disturbed habitats (Parrish and Edelstein-Keshet 1999; Silk 2007; Arseneau et al. 2015). Among primates, a small number of taxa including hamadryas baboons Papio hamadryas, geladas Theropithecus gelada, Guinea baboons P. papio, Yunnan snub-nosed monkeys Rhinopithecus bieti, black snub-nosed monkeys R. strykeri, Guizhou snub-nosed monkeys R. brelichi, golden snub-nosed monkeys R. roxellana, and possibly humans have evolved a modular or tiered multilevel society (Dyble et al. 2016; Goffe et al. 2016; Grueter et al. 2017). A multilevel society is characterized by several independent social and breeding units nested within a larger community that can number several hundred individuals (Shultz et al. 2011; Xiang et al. 2014). As the utilization of space and resources can be optimized by the joint actions of individuals who live in groups, group members may acquire multiple benefits beyond those obtain by solitarily individuals (Macfarlan et al. 2014; Dyble et al. 2015; Dyble et al. 2016; van Cise et al. 2017). Thereby, animal species with multilevel society usually develop such flexibility to overcome ecological challenges and may avoid inbreeding risk (Kirkpatrick and Grueter 2010; Schreier and Swedell 2012; Grueter et al. 2017). However, studies on the evolutionary and social mechanisms of the maintenance of genetic diversity for multilevel society, remain limited.
Golden snub-nosed monkeys, represent an endangered species of Asian colobine or leaf-eating primate that was once widely distributed across central and southern China (Li et al. 2003). Today, however, they exist in only 3 isolated mountainous regions in central and northwest China (Minshan, Shennongjia, and Qinling Mountains; Li et al. 2002; Long and Richardson 2008; Fang et al. 2018). Golden snub-nosed monkeys exhibit a special social organization described as a modular or the multilevel society, which is composed of 4 levels: the unit, band, herd, and troop (Qi et al. 2010; Grueter et al. 2012; Qi et al. 2014; Ren et al. 2018). The most basic level of golden snub-nosed monkey social organization is formed by 1 adult male, multiple adult females, subadult females, juveniles, and infants, and is called a 1-male unit or harem (Qi et al. 2009; Grueter and van Schaik 2010; Zhao et al. 2016). Several 1-male units feed, forage, rest, and travel together and form a breeding band (Qi et al. 2009). Bachelor males including juveniles, subadults, and adults aggregate into an all-male unit (Qi et al. 2017). Several all-male units may form an all-male band that shadows multiple breeding bands (Qi et al. 2014; Qi et al. 2017). The all-male band consists of males waiting for reproductive opportunities and former breeding males whose 1-male unit has been taken over by another male (Qi et al. 2017). The breeding bands and their associated all-male band together form a herd that occasionally interacts with other independent bands or herds to form a large troop (Qi et al. 2014).
The population of golden snub-nosed monkeys inhabiting the Qinling Mountains, exploit a region of high biodiversity that has been environmentally altered by extensive human exploitation and ecological damage (Oates et al. 1994). An expanding lumber industry and the conversion of natural forest to agricultural and farmland during the last century have resulted in unprecedented reduction and fragmentation of suitable habitat for several important animal and plant species (Li et al. 2002; Long and Richardson 2008). In recent years, a large number of roads, expressways, and high-speed railways have been constructed in the area. Due to both ecological and anthropogenic resistance, populations of the giant panda Ailuropoda melanoleuca and the golden takin Budorcas taxicolor have become isolated into small and fragmented subpopulations in the Qinling Mountains (Zeng et al. 2005; Zhang et al. 2006). These same anthropogenic disturbances have affected range size, quality, and connectivity of the natural habitats for the 39 troops of snub-nosed monkeys that remain in the Qinling Mountains (Li et al. 2000). However, a recent study indicated that the genetic diversity and effective population size of R. roxellana in the Qinling Mountain has not significantly decreased (Huang et al. 2016). Two other studies have hypothesized that the transfer of adult males among different breeding bands is likely to play a critical role in promoting genetic exchanges between otherwise highly-fragmented populations (Huang et al. 2017; Qi et al. 2014), implying that the evolution and adaptive function of a multilevel society, including the existence of an all-male band, serves to provide a reservoir of genetically distinct adult males that can increase gene flow under conditions of habitat fragmentation.
In this study, we test the hypothesis that despite living in highly-fragmented habitats, the multilevel society of golden snub-nosed monkeys facilitates the movement of bachelor males across otherwise semi-isolated modular social units resulting in a system of dynamic gene exchange and the maintenance of genetic diversity. We accomplish this by presenting genetic evidence that bachelor males from different natal breeding bands commonly transfer into the all-male band, resulting in an opportunity for gene flow and high levels of genetic diversity between isolated subpopulations within this multilevel society. By linking the evolution of this specialized social system with male reproductive strategies that function to promote gene flow, this research offers new insights into the benefits of group living.
Materials and Methods
Study group and genetic sampling
Our research site is located on the northern slope of the Qinling Mountains in the Zhouzhi National Nature Reserve, China (108°14′–108°18′E, 33°45′–33°50′N, Figure 1A). The region has a temperate climate and ranges in elevation from 1,400 to 2,890 m in elevation a.s.l. The annual average temperature is 10.7°C, and the annual average rainfall is 894 mm (Li et al. 2000).
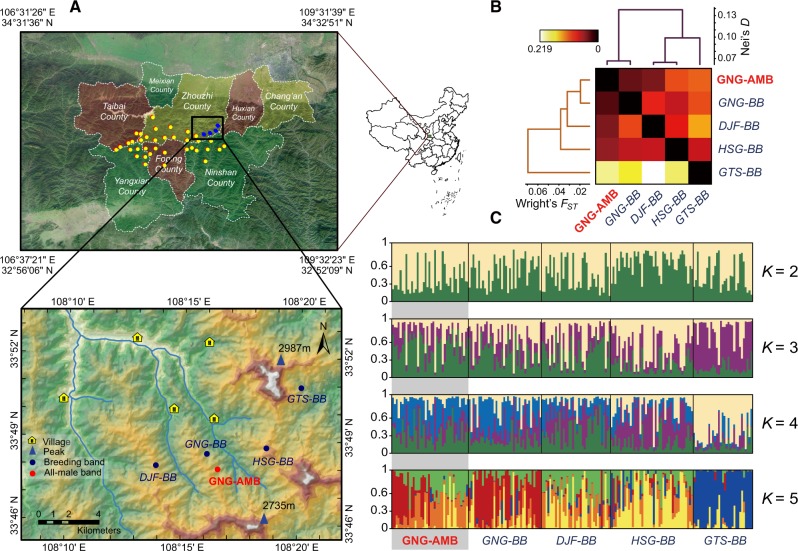
Figure 1.
(A) Study location of the 4 isolated breeding bands and the all-male band (bachelor group) of R. roxellana in the Qinling Mountains, China. (B) A heat map of genetic differentiation among bachelor groups and 4 breeding bands. Color brightness in each cell denotes the value of the pair-wise genetic differentiation index. Two clusters represent the genetic relationships among 5 bands that constructed by UPGMA. Dendrograms in the left and top were built based on Wright’s FST and Nei’s standard genetic distance, respectively, which are illustrated in the lower and upper triangular matrix of the heat map. (C) structure Bayesian clustering revealed that the 5 isolated social bands of snub-nosed monkeys had a lower divergence and high levels of admixture under fragmented habitats, which implies a potential gene flow. The clustering was built under assumption of locpriori, nonadmixture model and independent allele frequencies. BB, breeding band; AMB, all-male band.
There were 4 breeding bands and 1 all-male band in the study area during our study (Li et al. 2000). The HSG breeding band (HSG-BB) and the GTS breeding band (GTS-BB) from the East Ridge Troop, and the GNG breeding band (GNG-BB) and the DJF breeding band (DJF-BB) from the West Ridge Troop. The 2 troops are separated by the Nancha River. The GNG all-male band (GNG-AMB) principally shadowed the GNG-BB, but had occasional contact with the other 3 breeding bands. Due to the difficulty associated with following the monkeys across steep cliffs and mountainous terrain, it took 1.5 years to collect fecal and hair samples used for genetic analysis (December 2014 to March 2016).
The 4 breeding bands analyzed in this study varied in size and compositions. There were 15 one-male units in the GNG-BB, 12 one-male units in the DJF-BB, 11 one-male units in the HSG-BB and 10 one-male units in the GTS-BB. The size, age, and sex composition of each breeding bands and the all-male band are reported in Table 1. The GNG-AMB, which was composed of 12 adult, 14 subadult, and 14 juvenile males, traveled independently, but maintained a close spatial association to the GNG-BB, resulting in a high degree of social affinity and spatial overlap. The GNG herd was habituated to the presence of researchers and had been provisioned for more than 20 years. Thus, we were able to maintain close proximity to the monkeys and identify each individual based on pelage color, body size, sex, and idiosyncratic physical traits. The monkeys were categorized into 4 age/sex classes (Qi et al. 2009, 2014), including adult males and females (adult males reach maturity at 7 years of age and adult females reach maturity at 5 years of age), subadults (females are subadults at age 3–4 and males are subadults from ages 5–7 years old), juveniles (females aged from 1 to 3 years and males aged 1 to 5 years), and infants (<12 months; Qi et al. 2009).
Table 1.
Genetic diversity of 5 bands among R. roxellana multilevel society
| Band | Longitude (°E) | Latitude (°N) | Population size | Sample size | Genetic diversitya |
F IS | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Nm | PIC | Ar | HO | HE | ||||||
| GNG-AMB | 108.278 | 33.798 | 40 | 40 | 3.92 | 0.560 | 2.34 | 0.565 | 0.553 | −0.026 |
| GNG-BB | 108.270 | 33.812 | 130 | 38 | 3.54 | 0.552 | 2.32 | 0.565 | 0.545 | −0.030 |
| DJF-BB | 108.233 | 33.805 | 100 | 36 | 3.92 | 0.582 | 2.46 | 0.573 | 0.574 | −0.009 |
| HSG-BB | 108.312 | 33.813 | 90 | 43 | 3.39 | 0.506 | 2.15 | 0.513 | 0.500 | −0.034 |
| GTS-BB | 108.337 | 33.850 | 70 | 31 | 3.15 | 0.510 | 2.28 | 0.563 | 0.502 | −0.126 |
Five genetic diversity indices were presented, that is, number of alleles (Nm), polymorphism information content (PIC), allelic richness (Ar), observed heterozygosity (HO), and expected heterozygosity (HE). FIS denotes the Wright’s inbreeding coefficient.
Both fecal and hair samples were collected from all individuals in the GNG-AMB, including 12 adults, 14 subadults, and 14 juveniles, as well from 72 adults in the GNG-BB (15 males and 57 females). Because the monkeys in the remaining 3 breeding bands were not habituated, we were able only to collect fecal samples from unidentified individuals.
Fresh fecal samples were stored in DETs (20% DMSO, 0.25 M sodium-EDTA, 100 mM Tris·HCl, pH 7.5, and NaCl to saturation) solution at −20°C. A stick with adhesive tape was used to collect hair samples. It was made from an 80 cm × 6 cm wooden board covered with glue and baited with a fruit reward. In the process of grabbing the fruit, hair from the monkey’s hand and arm adhered to the glue. The collected samples were then stored in silica gel and then dried at room temperature.
Molecular methods
Follicle DNA was extracted with proteinase K digestion (Shenggon, Shanghai) in a PCR compatible buffer, whereas fecal DNA was extracted using QIAamp DNA Stool Mini Kits (Qiagen, German). All DNA samples were amplified at 19 tetra-nucleotide microsatellite loci (please see Supplementary Table S1) in an ABI Veriti Thermal Cycler with the following processes: 95°C for 5 min, followed by 30 cycles (94°C for 30 s, 55°C−60°C for 45 s, 72°C for 45 s), and 72°C for 10 min. PCR products were segregated with an ABI PRISM 3100 Genetic Analyser, and their sizes relative to the internal size standard (ROX-labeled HD400) were determined with genemapper V3.7 (Applied Biosystems). Homozygous genotypes were confirmed by 5 independent replicates, which were observed and repeated in at least 3 separate reactions (Taberlet et al. 1996). Replicates were detected by polyrelatedness V1.6 (Huang et al. 2016) and excluded from subsequent analyses. To avoid the possibility of analyzing the same individual twice, a probability of identity (PID) analysis was carried out using the software Cervus V3.0.7 (Kalinowski et al. 2010). In addition, for each locus, if the samples of missing data occupied >20% of all samples, this locus was discarded. Furthermore, to prevent genotyping errors, such as false alleles, scoring errors, and allelic dropouts, the software MICRO-CHECKER V2.2.3 (van Oosterhout et al. 2010) was employed.
Genetic diversity
Statistical analysis of genetic diversity included observed and expected heterozygosity, polymorphic information content, allelic richness, and Wright’s inbreeding coefficient at each locus for each band—and was calculated with GEnaIEx V6.5 (Peakall et al. 2003). We performed a Hardy–Weinberg equilibrium (HWE) test for each band at each locus with Fisher’s exact tests in genepop V4.3 (Rousset 2008). Significance thresholds were adjusted for multiple tests by sequential Bonferroni procedures (Rice 1989).
We employed 3 methods to test the presence of a bottleneck within each breeding band and the GNG-AMB. The first method was based on deviations of allele frequencies in the calculations of heterozygosity, where we used signed-test and 2-tailed Wilcoxon test in Bottleneck V1.2.02 (Piry et al. 1999). We considered 2 types of mutation models: 1) a 2-phase model (TPM) with 95% stepwise mutations and a variance of 12, and 2) a stepwise mutation model (SMM with iteration number set to 1,000). The second method involved calculating the GW coefficient (Garza and Williamson 2001) in arlequin V3.6 (Excoffier and Lischer 2010). Finally, we performed an effective population size change estimation, which was inferenced with Bayesian computation by the DIYABC v2.0 software (Jean-Marie et al. 2008).
Population structure
To evaluate the significance of genetic differentiation, the pairwise FST between the 4 golden snub-nosed monkey breeding bands was calculated with 100,000 permutations using arlequin V3.6 (Excoffier and Lischer 2010). Significance levels were assessed via permutation tests, where the number of steps in the Markov chain was set to 100,000, and the number of burn-in steps was set to 10,000. Nei’s standard genetic distances D (Nei 1972) were calculated in GEnalex V6.5 (Peakall et al. 2003). We performed a hierarchical cluster analysis based on genetic differentiation and genetic distance using the UPGMA method (unweighted pair group method with arithmetic mean).
In addition, the common model of isolation by distance was examined using the Mantel test implemented in the program GENALEX V6.5 (Peakall and Smouse 2012). Generally, Nei’s genetic distance and the geographic distance were used as input, and their correlation significance was assessed by conducting 1,000 permutations.
A Bayesian cluster analysis was performed using structure V2.3.4 (Pritchard et al. 2000) to examine population genetic structure. This method estimates the likelihoods of various numbers of genetically distinct groups (K) in the samples by assigning individuals into one or more groups in a manner that minimizes each group’s deviation from HWE. The program was run for K from 1 to 5 under the admixture model with correlated allele frequencies. For each run, we used 700,000 MCMC cycles, following 100,000 burn-in cycles. Ten replications were performed for each K, to test whether the number of iterations was sufficient. We set the iteration number from 700,000 MCMC cycles to 800,000, and we found that the log-likelihood value was saturated (P = 0.43, likelihood ratio test, toward the mean value of Ln-likelihood values derived from both iteration numbers of 700,000 and 800,000). Thus, our saturation analysis showed that the number of iterations was sufficient. The optimal K was estimated according to Evanno et al.’s delta K (ΔK) method (Evanno et al. 2005). The evaluation of the appropriate K is presented in the Supplementary Figure S1.
Population assignment
Population assignment was to calculate the log likelihood for each individual, with allele frequencies of the respective population (Paetkau et al. 2010). Individuals were assigned to the population with the highest likelihood, which is an effective technique to identify the natal population among individuals. We calculated the posterior probabilities for each individual that originated from the 4 different breeding bands with the Bayes equation:
where is the genotypes vector of i-th individual, and is the j-th breeding band, is the number of individuals in out of all breeding bands, and is the probability of sampling an individual with a genotypes vector equal to within . can be obtained by taking the product of the genotypic frequencies across all loci:
where is the genotype of i-th individual at j-th locus, is the frequency of in :
Where a and b are the alleles within , and , and are their frequencies in , respectively. For each individual, the breeding band with the highest posterior probability was considered to be the natal band of this individual. We used chi-squared goodness-of-fit tests to assess whether the distribution of individual origin was in accord with expectation, where the expected values were proportional to . We used the originated probability coefficient (OPC) to describe the probable percentage of individuals from different origins within a breeding band.
Gene flow among bands
Gene flow among different bands was assessed using migrate V3.1 (Beerli 2006). The amount and direction of gene flow were estimated from microsatellite genotypes by calculating mutation-scaled effective population size θ (4 times the effective population size multiplied by the mutation rate of per site in each generation), and mutation-scaled migration rate M (migration rate divided by mutation rate). Based on the continuous Brownian motion model, we implemented a pre-run with FST values to obtain the prior setting of θ and M. Five independent MCMC chains with 5,000,000 generations were used. We sampled every 100 steps under a constant mutation model, discarding the first 1,000,000 records as burn-in. The mode and 95% of the highest posterior density were estimated after checking for convergence.
Landscape resistance surface
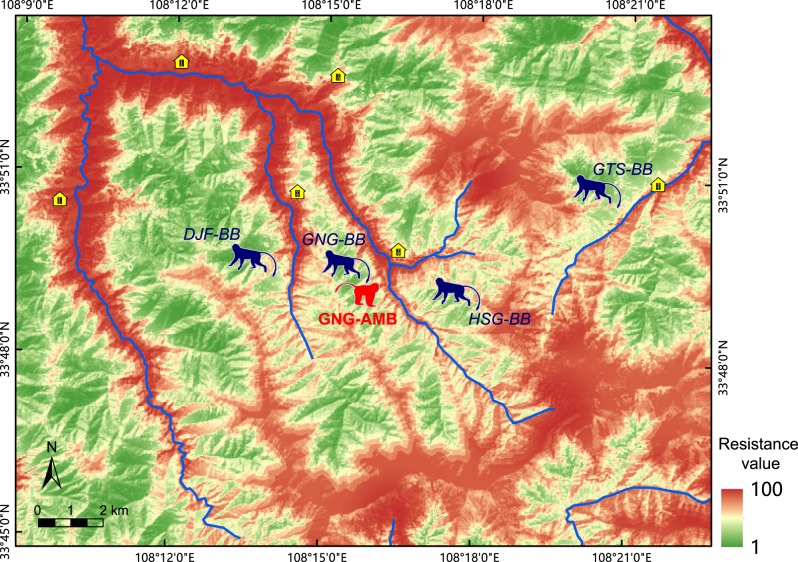
We calculated the continuous landscape resistance surface to identify the degree to which local landscape elements limited the distribution or movement of R. roxellana within the study area. We modeled landscape resistance as a function of 4 landscape variables deemed to be potentially important for the species (Li Y-L et al., unpublished data): elevation, distance to human disturbances, distance to rivers, and the enhanced vegetation index, as shown in Figure 3. Each landscape variable’s grid cell resistance value was assigned based on the frequency of species occurrence from the analysis of radio-tracking data of R. roxellana (Li Y-L et al., unpublished data; see Supplementary File S2). We assumed that all landscape variables had equal effects on the ranging and distribution of golden snub-nosed monkeys. Then the landscape resistance values were estimated as the averaged resistance values of all landscape variables, ranging from 1 (least resistance to movement) to 100 (greatest barrier to movement). All maps were processed in ArcGIS v10.2 (ESRI 2010).
Figure 3.
Satellite result of continuous landscape resistance surface. Color ranging from green and red suggests the resistance value caused by ecological factors and human activities. The red color suggests high resistance value, and green suggests that the monkeys can move freely in these areas. The result confirms that geographical factors would resist the monkeys to immigrate between breeding bands with the fragmented habitats.
Ethical standards
All research protocols reported here adhere to the regulatory requirements and were approved by the animal care committee of the Wildlife Protection Society of China (SL-2012-42). The genetic sampling received the clearance from, and complied with the protocols approved by, the specialist committee of the State Forestry Administration of China (SFA-LHXZ-2012-2788), and Chinese Academy of Science.
Results
Microsatellite dataset and genetic diversity
We analyzed 19 microsatellites from 188 golden snub-nosed monkeys residing in 4 breeding bands and 1 all-male band (sample sizes of each band are reported in Table 1). Details of these loci are presented in Supplementary Table S1. In the case of 4 loci (D19S248, D6S1036, D6S1040, and P115), samples of missing data accounted for >20% of all samples (Supplementary Table S1), and thus these loci were discarded from our analysis. The MICRO-CHECKER analysis indicated that null alleles were likely present at loci D10S676 and D7S2204 across the GNG-AMB and the DJF-BB, respectively (Supplementary Table S2). We, therefore, removed these 2 loci from subsequent analyses. As for the remaining 13 highly polymorphic loci, the largest multilocus PID was 4.34E-09, and therefore our dataset contained no identical samples. In addition, the HWE test found no evidence of linkage disequilibrium (Supplementary Table S1).
Summary statistics for the genetic diversity analyses and genetic diversity indices can be found in Table 1. Genetic variability among the 5 bands was generally moderate, with the number of alleles per locus ranging from 3.15 to 3.92 (average = 3.50). The observed heterozygosity ranged from 0.513 to 0.573, with an average of 0.5556. The expected heterozygosity (HE) ranged from 0.50 to 0.574 (average = 0.535). The polymorphism information content ranged from 0.506 to 0.582 (average = 0.542), and the allelic richness for the 5 bands ranged from 2.15 to 2.46 (average = 2.31). Values of Wright’s inbreeding coefficient (FIS) showed that the effects of inbreeding in the DJF-BB were relatively stronger than found in the other bands. In general, the effect of inbreeding among all 5 bands was weak.
Based on microsatellite data, we found no evidence that a genetic of bottlenecks had formed in any of the 4 breeding bands (Table 2; none of the sign or Wilcoxon tests suggested excess heterozygosity or deficiency in either SMM or TPM). The lowest P-value is 0.108 (Wilcoxon test with TPM model in HSG-BB). GW coefficients of all bands exceeded the empirical value of 0.68 (Garza and Williamson 2001), and the lowest GW coefficient was 0.872 ± 0.220 (HSG-BB). Inferring population history with Bayesian computation similarly indicated no evidence that a genetic bottleneck had occurred in any of the 4 breeding bands (Supplementary Table S3).
Table 2.
Bottleneck effect tests of 1 all-male band and 4 breeding bands
| Band | Sign text |
Wilcoxon test |
M ± SD | ||
|---|---|---|---|---|---|
| TPM | SMM | TPM | SMM | ||
| GNG-AMB | 0.507 | 0.505 | 0.473 | 0.580 | 0.914± 0.144 |
| GNG-BB | 0.129 | 0.141 | 0.122 | 0.153 | 0.889 ± 0.162 |
| DJF-BB | 0.560 | 0.567 | 0.170 | 0.368 | 0.876 ± 0.156 |
| HSG-BB | 0.471 | 0.470 | 0.108 | 0.318 | 0.872± 0.220 |
| GTS-BB | 0.494 | 0.521 | 0.153 | 0.188 | 0.877 ± 0.178 |
Notes: M is the Garza and Williamson’s (2001) coefficient. No results were significant.
Population structure
The matrices and dendrograms representing genetic relationships and identifying the strength of FST and D are presented in Figure 1B. The results of pairwise FST revealed relatively low genetic divergence among each of 5 bands (FST < 0.10), and the permutation test between each pair of bands (breeding and all-male) was significant (P < 0.05). Nei’s standard genetic distance ranged from 0.031 to 0.122; the largest distance was between the DJF-BB and GTS-BB. FST and Nei’s D between the GNG-AMB and GNG-BB was lower than between all other band pairs. Finally, our Mantel test analysis showed a significant correlation between geographic and genetic distance among all 5 bands (r = 902, P = 0.019; Supplementary Figure S1).
Bayesian cluster results documented high levels of admixture among the 2 putative genetic clusters, with the Evanno’s ΔK method indicating that the approximate K was maximized at K = 2 (Supplementary Figure S2). The cluster results among these 5 bands are shown in Figure 1C (results for K = 3, 4, and 5 also are shown for comparison). In addition, we found similar genetic compositions between individuals in the GNG-AMB and the GNG-BB, indicating substantive gene flow between these 2 modular social units has occurred. Although allele frequencies in the other 3 breeding bands (DJF-BB, HSG-BB, and GTS-BB) were significantly different from that of the GNG-AMB, Bayesian cluster revealed that DJF-BB, HSG-BB, DJF-BB, and GNG-AMB shared partial common genetic compositions, indicating gene flow has occurred between the all-male band and these 3 breeding bands (Figure 1C).
Population assignment
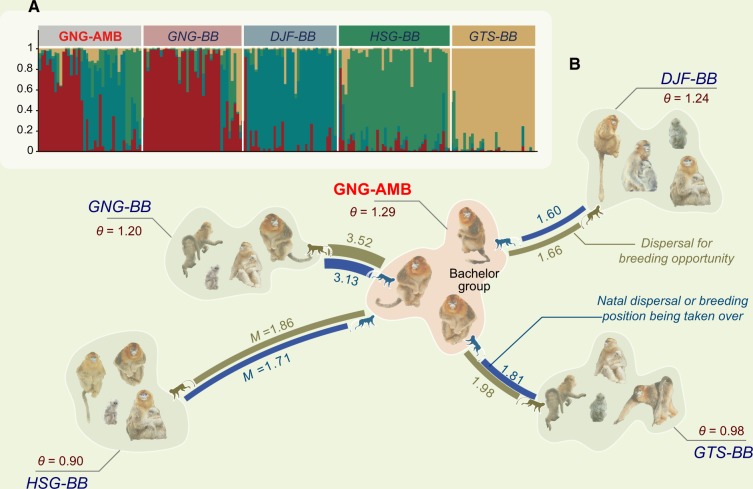
Population assignment analysis demonstrated that the members of the GNG-AMB originated unevenly from the 4 breeding bands (Figure 2A, χ2 =18.0, df = 3, P < 0.001). The OPC revealed that approximately half of the individuals came from the GNG-BB (OPCGNG-BB = 0.450), with the remaining males in the GNG-AMB originating from the DJF-BB, HSG-BB, and GTS-BB (OPCDJF-BB = 0.375 vs. OPCHSG-BB = 0.150 vs. OPCGTS-BB = 0.025).
Figure 2.
(A) The posterior probability based on population assignment reveals individual exchange among 4 different bands and the bachelor group. Each box represents a social band. Each bar within the box represents a sampled individual from the band. Genetic characters of each band were estimated by unique allele frequencies, and marked by a corresponding color. The percentage of each color within the bar denotes the posterior probability of the individual origin. (B) Gene flow among the 4 breeding bands and GNG-AMB. The levels of gene flow are represented by θ and M value estimated by migrate. θ(4Neµ): mutation-scaled effective population size. M(m/µ): mutation-scaled migration rate; where m is the migration rate and µ is the mutation rate. The blue curves show gene flow from the breeding bands to the all-male band, whereas the green curves show the opposite gene flow from the all-male band to breeding band. The widths and the numbers beside the curves denote M values.
Gene flow among bands
The migrate analysis revealed the existence of highly asymmetric gene flow between the GNG-AMB and the 4 breeding bands (θ and M are significantly >0; Figure 2B). The θ value was highest in the GNG-AMB. Allele movement between the GNG-AMB and the GNG-BB was higher than between the GNG-BB and the other 3 breeding bands. The M value is not especially noteworthy for the comparison between the GNG-AMB and the breeding bands (MGNG-BB > MGTS-BB > MHSG-BB > MDJF-BB). However, variation in the rate of gene migration from the breeding bands to the GNG-AMB was apparently greater.
Landscape resistance surface
The continuous landscape resistance surface modeling suggests that the construction of roads, farms, and villages, along with natural ecological barriers (river, high elevations) highly restrict the distribution and patterns of habitats utilization of golden snub-nosed monkeys (Figure 3). As shown in Figure 3, the habitats occupied by each of the 4 breeding bands of golden snub-nosed monkeys were highly fragmented. Recent anthropogenic changes (roads, farms, and villages) to the landscape appear to have permanently isolated the GNG-BB and the HSG-BB, and rivers and high mountains have, over some extended period of time, served as effective barriers severely limiting the ability of individuals from the GNG-BB to transfer into the DJF-BB or the GTS-BB.
Discussion
In this study, we examined genetic diversity, gene flow, and population assignment in an endangered species of nonhuman primate, the golden snub-nosed monkey, that inhabits a highly anthropogenically-disturbed and fragmented mountainous habitat in central China. We combined genetic data with continuous landscape resistance surface modeling to study the degree to which human activities and ecological factors restrict the immigration of individuals among breeding bands. In addition, we examined whether the all-male band, which represents an important component of the snub-nosed monkey multilevel social system serves to promote gene exchanges and maintain genetic diversity across otherwise isolated breeding bands.
The continuous landscape resistance surface modeling indicated that ecological/anthropogenic resistance severs as a filter limiting the ability of individuals to migrate directly between the 4 studied breeding bands, leading to a negative effect on the individual transfer among the 4 breeding bands. This is supported by satellite telemetry data of GPS collared golden snub-nosed monkeys indicating that each studied breeding band inhabited a distinct range or territory that only minimally overlapped with other breeding bands, resulting in limited opportunities for individuals to transfer (Qi et al. 2014). Moreover, human-constructed barriers such as rivers, villages, logging roads, and farmlands have limited the ability of breeding bands to freely move across this fragment landscape for decades or hundreds of years (Wang et al. 2014). We, therefore, expected this would result in low population-level genetic diversity and high population-level genetic differentiation across breeding bands (Huang et al. 2016). For example, the Xiaowangjian forest lumbering station was established in this area in 1978, with the formation of a logging road in the area occupied by the East Ridge Troop (HSG-BB and GTS-BB; Li et al. 2000). This road is frequently used by both the logging industry and local traffic, particularly farmers, and represents a barrier to the free movement of each of the 4 breeding bands. Although these barriers are not completely impenetrable, their impact may increase the costs of individual transfer among the studied breeding bands (Greenwood 1980; Ehrlen and Eriksson 2000; Bester-van der Merwe et al. 2011; Robson and Blouindemers 2013). Moreover, the topography of the Qinling Mountains, which is dominated by high altitude temperate forest habitats, and open areas of fragmented and cleared forests containing villages and agricultural fields, as well as the threat of domesticated animals such as dogs may greatly increase the risks and decrease the opportunities for successful dispersal (Wang et al. 2014). Either enhanced mortality risk reduces the occurrence of successful transfer or individual is choose not to disperse due to such cost (Lin and Batzli 2004). Thus, the expected result is reduced gene flow, reduced genetic diversity, and an increased opportunity for a genetic bottleneck.
However, we found no evidence of genetic bottlenecks, high levels of genetic diversity, low levels of genetic divergence or similar genetic backgrounds in golden snub-nosed monkeys. Population structure results showed that K = 2 was the most probable number of genetic clusters, indicating that the 5 isolated bands could be divided into 2 genetically distinct population, implying high levels of admixture and high levels of gene flow among these bands. This, along with the results of migrate V3.1 software indicated the possibility of high levels of gene flow between the all-male band and all-breeding bands. Population assignment analysis showed that the GNG-AMB was comprised of bachelor males who originated from each of 4 breeding bands, indicating the possibility of gene flow among these bands. Overall, all these evidences confirmed persistent gene flow among the breeding bands, as well as between the all-male band and each of the breeding bands.
Gene flows are likely to be the result of the movement of bachelor males among the study bands promoting genetic diversity. Mating strategies adopted by bachelor male golden snub-nosed monkeys include periodic attempts to take over a 1-male unit and usurp the leader or breeding male position, attracting adult females to leave their 1-male unit, or engaging in sneaky copulations with harem females (Patzelt et al. 2014; Qi et al. 2014; Smith et al. 2017; Qi et al. 2020). Bachelor male movements include young sexually immature males who migrated from their natal breeding band into the all-male band (primary transfer; Yao et al. 2011; Chang et al. 2014; Huang et al. 2017); fully adult males who transferred from the all-male band back into their natal breeding band or to a neighboring breeding band and successfully take over an existing 1-male unit, or sexually mature males who leave the all-male band in attempt to attract harem females to form a new 1-male unit (breeding transfer); fully adult males who formed a temporary consort relationship with 1-male unit females (temporary transfer) and older adult males who had left their previous breeding band and rejoined the all-male band after losing their residential position as an 1-male unit leader male (Grueter et al. 2017). The dynamic movements of bachelor males into and out of the all-male band and breeding bands contribute to multidirectional gene flow that serves to maintain a high level of genetic diversity otherwise across semi-isolated populations. The results support the hypothesis that bachelor males residing in an all-male band can effectively mitigate inbreeding depression across fragmented landscapes.
Supplementary Material
Acknowledgments
We thank Zhouzhi National Nature Reserve for the permission of this study. We thank Zhi-Pang Huang, Kun Bian, and Qi Li from NWU for their assistance in genetic sample. We thank Prof. Ruliang Pan, Dr. Kang Huang, and Dr. Shi-Yi Tang for their contributions to the early draft of this manuscript. We appreciate the reviewers’ advice to improve this article.
Funding
The study was funded by the National Natural Science Foundation of China (31730104 and 31622053), the Promotional project for Innovation team, the Department of Science and Technology of Shaanxi Prov. China (2018TD-017), Strategic Priority Research Program of the Chinese Academy of Sciences (XDB31020000), and the National Key Programme of Research and Development, Ministry of Science and Technology of China (2016YFC0503200).
Authors’ Contributions
X.-G.Q. conceived and designed the study. Y.-L.L. performed the experiments and wrote the article. L.W. and P.-A.G. revised the article. X.-P.Y. and J.-W.W. reanalyzed the genetic data. B.-G.L. provided the experimental materials. All authors provided input for the article and approved the final version.
Conflict of interest
The authors declare that they have no conflicts of interest to this work.
References
- Arseneau TJM, Taucher AL, van Schaik CP, Willems EP, 2015. Male monkeys fight in between-group conflicts as protective parents and reluctant recruits. Anim Behav 110:39–50. [Google Scholar]
- Barrett RDH, Schluter D, 2008. Adaptation from standing genetic variation. Trends Ecol Evol 23:38–44. [DOI] [PubMed] [Google Scholar]
- Beerli P, 2006. Comparison of Bayesian and maximum-likelihood inference of population genetic parameters. Bioinformatics 22:341–345. [DOI] [PubMed] [Google Scholar]
- Bester-van der Merwe AE, Roodt-Wilding R, Volckaert FAM, D’Amato ME, 2011. Historical isolation and hydrodynamically constrained gene flow in declining populations of the South-African abalone Haliotis midae. Conserv Genet 12:543–555. [Google Scholar]
- Broders HG, Mahoney SP, Montevecchi WA, Davidson WS, 1999. Population genetic structure and the effect of founder events on the genetic variability of moose Alces alces in Canada. Mol Ecol 8:1309–1315. [DOI] [PubMed] [Google Scholar]
- Chang ZF, Yang BH, Vigilant L, Liu ZJ, Ren BP. et al. , 2014. Evidence of male-biased dispersal in the endangered Sichuan snub-nosed monkey Rhinopithexus roxellana. Am J Primatol 76:72–83. [DOI] [PubMed] [Google Scholar]
- Charlesworth D, Willis JH, 2009. The genetics of inbreeding depression. Nat Rev Genet 10:783–796. [DOI] [PubMed] [Google Scholar]
- Coltman DW, 2005. Differentiation by dispersal. Nature 433:23–24. [DOI] [PubMed] [Google Scholar]
- Dyble M, Salali GD, Chaudhary N, Page A, Smith D. et al. , 2015. Sex equality can explain the unique social structure of hunter-gatherer bands. Science 348:796–798. [DOI] [PubMed] [Google Scholar]
- Dyble M, Thompson J, Smith D, Salali Gul D, Chaudhary N. et al. , 2016. Networks of food sharing reveal the functional significance of multilevel sociality in two hunter-gatherer groups. Curr Biol 26:2017–2021. [DOI] [PubMed] [Google Scholar]
- Ehrlen J, Eriksson O, 2000. Dispersal limitation and patch occupancy in forest herbs. Ecology 81:1667–1674. [Google Scholar]
- Estrada A, Garber PA, Rylands AB, Roos C, Fernandez-Duque E. et al. , 2017. Impending extinction crisis of the world’s primates: why primates matter. Sci Adv 3:e1600946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evanno G, Regnaut S, Goudet S, 2005. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Lischer HEL, 2010. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under linux and windows. Mol Ecol Resour 10:564–567. [DOI] [PubMed] [Google Scholar]
- Fang G, Li M, Liu XJ, Guo WJ, Jiang YT. et al. , 2018. Preliminary report on Sichuan golden snub-nosed monkeys Rhinopithecus roxellana roxellana at Laohegou Nature Reserve, Sichuan, China. Sci Rep 8:16183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garza JC, Williamson EG, 2001. Detection of reduction in population size using data from microsatellite loci. Mol Ecol 10:305–318. [DOI] [PubMed] [Google Scholar]
- Goffe AS, Zinner D, Fischer J, 2016. Sex and friendship in a multilevel society: behavioural patterns and associations between female and male guinea baboons. Behav Ecol Sociobiol 70:323–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwood PJ, 1980. Mating systems, philopatry and dispersal in birds and mammals. Anim Behav 28:1140–1162. [Google Scholar]
- Grueter CC, Chapais B, Zinner D, 2012. Evolution of multilevel social systems in nonhuman primates and humans. Int J Primatol 33:1002–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grueter CC, Qi XG, Li BG, Li M, 2017. Multilevel societies. Curr Biol 27:R984–R986. [DOI] [PubMed] [Google Scholar]
- Grueter CC, van Schaik CP, 2010. Evolutionary determinants of modular societies in colobines. Behav Ecol 21:63–71. [Google Scholar]
- Hedrick PW, Garcia-Dorado A, 2016. Understanding inbreeding depression, purging, and genetic rescue. Trends Ecol Evol 31:940–952. [DOI] [PubMed] [Google Scholar]
- Huang K, Guo ST, Cushman SA, Dunn DW, Qi XG. et al. , 2016. Population structure of the golden snub-nosed monkey Rhinopithecus roxellana in the Qinling Mountains, central China. Integr Zool 11:350–360. [DOI] [PubMed] [Google Scholar]
- Huang K, Ritland K, Dunn DW, Qi XG, Guo ST. et al. , 2016. Estimating relatedness in the presence of null alleles. Genetics 202:247–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang ZP, Bian K, Liu Y, Pan RL, Qi XG. et al. , 2017. Male dispersal pattern in golden snub-nosed monkey Rhinopithecus roxellana in Qinling Mountains and its conservation implication. Sci Rep 7:46217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jean-Marie C, Filipe S, Beaumont MA, Robert CP, Jean-Michel M. et al. , 2008. Inferring population history with DIY ABC: a user-friendly approach to Approximate Bayesian computation. Bioinformatics 24:2713–2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalinowski ST, Taper ML, Marshall TC, 2010. Revising how the computer program cervus accommodates genotyping error increases success in paternity assignment. Mol Ecol 16:1099–1106. [DOI] [PubMed] [Google Scholar]
- Kirkpatrick RC, Grueter CC, 2010. Snub-nosed monkeys: multilevel societies across varied environments. Evol Anthropol 19:98–113. [Google Scholar]
- Lande R, 1993. Risks of population extinction from demographic and environmental stochasticity and random catastrophes. Am Nat 142:911–927. [DOI] [PubMed] [Google Scholar]
- Leonard JA, Vilà C, Wayne RK, 2005. Legacy lost: genetic variability and population size of extirpated us grey wolves Canis lupus. Mol Ecol 14:9–17. [DOI] [PubMed] [Google Scholar]
- Li BG, Chen C, Ji WH, Ren BP, 2000. Seasonal home range changes of the Sichuan snub-nosed monkey Rhinopithecus roxellana in the Qinling Mountains of China. Folia Primatol 71:375–386. [DOI] [PubMed] [Google Scholar]
- Li BG, Jia ZY, Pan RL, Ren BP, 2003. Changes in distribution of the snub–nosed monkey in China In: Marsh LK, editor. Primates in Fragments: Ecology and Conservation. New York: Kluwer Academic/Plenum Press; 29–51. [Google Scholar]
- Li BG, Pan RL, Oxnard CE, 2002. Extinction of snub-nosed monkeys in China during the past 400 years. Int J Primatol 23:1227–1244. [Google Scholar]
- Lin YK, Batzli GO, 2004. Emigration to new habitats by voles: the cost of dispersal paradox. Anim Behav 68:367–372. [Google Scholar]
- Liu ZJ, Ren BP, Wu RD, Zhao L, Hao YL. et al. , 2009. The effect of landscape features on population genetic structure in Yunnan snub-nosed monkeys Rhinopithecus bieti implies an anthropogenic genetic discontinuity. Mol Ecol 18:3831–3846. [DOI] [PubMed] [Google Scholar]
- Long YC, Richardson M, 2008. Rhinopithecus Roxellana. IUCN Red List of Threatened Species 2008:e.T19596A8985735. Available from: 10.2305/IUCN.UK.2008.RLTS.T19596A8985735.en. [DOI] [Google Scholar]
- Macfarlan SJ, Walker RS, Flinn MV, Chagnon NA, 2014. Lethal coalitionary aggression and long–term alliance formation among yanomamö men. Proc Natl Acad Sci USA 111:16662–16669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M, 1972. Genetic distance between populations. Am Nat 106:283–292. [Google Scholar]
- Oates JF, Davies AG, Delson E, 1994. The diversity of living colobines In: Davies AG, Oates JF, editors. Colobine Monkeys: Their Ecology, Behaviour and Evolution. Cambridge: Cambridge University Press; 45–73. [Google Scholar]
- Orr HA, 2005. The genetic theory of adaptation: a brief history. Nat Rev Genet 6:119–127. [DOI] [PubMed] [Google Scholar]
- Paetkau D, Slade R, Burden M, Estoup A, 2010. Genetic assignment methods for the direct, real-time estimation of migration rate: a simulation-based exploration of accuracy and power. Mol Ecol 13:55–65. [DOI] [PubMed] [Google Scholar]
- Parrish JK, Edelstein-Keshet L, 1999. Complexity, pattern, and evolutionary trade-offs in animal aggregation. Science 284:99–101. [DOI] [PubMed] [Google Scholar]
- Patzelt A, Kopp GH, Ndao I, Kalbitzer U, Zinner D. et al. , 2014. Male tolerance and male-male bonds in a multilevel primate society. Proc Natl Acad Sci USA 111:14740–14745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peakall R, Ruibal M, Lindenmayer DB, Tonsor S, 2003. Spatial autocorrelation analysis offers new insights into gene flow in the Australian bush rat Rattus fuscipes. Evolution 57:1182–1195. [DOI] [PubMed] [Google Scholar]
- Peakall R, Smouse PE, 2012. GenAIEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research: an update. Bioinformatics 28:2537–2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer M, Lefebvre V, Peres CA, Banks-Leite C, Wearn OR. et al. , 2017. Creation of forest edges has a global impact on forest vertebrates. Nature 551:187–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piry S, Luikart G, Cornuet JM, 1999. Bottleneck: a computer program for detecting recent reductions in the effective size using allele frequency data. J Hered 90:502–503. [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P, 2000. Inference of population structure using multilocus genotype data. Genetics 155:945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi XG, Garber PA, Ji WH, Huang ZP, Huang K. et al. , 2014. Satellite telemetry and social modeling offer new insights into the origin of primate multilevel societies. Nat Commun 5:5296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi XG, Grueter CC, Fang G, Huang PZ, Zhang J. et al. , 2020. Multilevel societies facilitate infanticide avoidance through increased extrapair matings. Anim Behav. doi: 10.1016/j.anbehav.2019.12.014. [Google Scholar]
- Qi XG, Huang K, Fang G, Grueter CC, Dunn DW. et al. , 2017. Male cooperation for breeding opportunities contributes to the evolution of multilevel societies. Proc Biol Sci 284:20171480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi XG, Li BG, Garber PA, Ji WH, Watanabe K, 2009. Social dynamics of the golden snub-nosed monkey Rhinopithecus roxellana: female transfer and one-male unit succession. Am J Primatol 71:670–679. [DOI] [PubMed] [Google Scholar]
- Qi XG, Zhang P, Li BG, Watanabe K, 2010. The diversity of polygynous social systems among multi-level societies in non-human primates. Acta Theriol Sin 30:322–338. [Google Scholar]
- Quéméré E, Crouau-Roy B, Rabarivola C, Louis EE Jr, Chikhi L, 2010. Landscape genetics of an endangered lemur Propithecus tattersalli within its entire fragmented range. Mol Ecol 19:1606–1621. [DOI] [PubMed] [Google Scholar]
- Ren Y, Huang K, Guo ST, Pan RL, Derek DW. et al. , 2018. Kinship promotes affiliative behaviors in a monkey. Curr Zool 64:441–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice WR, 1989. Analyzing tables of statistical tests. Evolution 43:223–225. [DOI] [PubMed] [Google Scholar]
- Robson LE, Blouindemers G, 2013. Eastern hognose snakes Heterodon platirhinos avoid crossing paved roads, but not unpaved roads. Copeia 2013:507–511. [Google Scholar]
- Rousset F, 2008. Genepop’007: a complete re-implementation of the Genepop software for windows and Linux. Molecular Ecology Resources 8:103–106. [DOI] [PubMed] [Google Scholar]
- Schreier AL, Swedell L, 2012. Ecology and sociality in a multilevel society: ecological determinants of spatial cohesion in hamadryas baboons. Am J Phys Anthropol 148:580–588. [DOI] [PubMed] [Google Scholar]
- Shultz S, Opie C, Atkinson QD, 2011. Stepwise evolution of stable sociality in primates. Nature 479:219–222. [DOI] [PubMed] [Google Scholar]
- Silk JB, 2007. The adaptive value of sociality in mammalian groups. Philos Trans R Soc Lond B Biol Sci 362:539–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D, Schlaepfer P, Major K, Dyble M, Page AE. et al. , 2017. Cooperation and the evolution of hunter-gatherer storytelling. Nat Commun 8:1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swedell L, 2011. African papionins: diversity of social organization and ecological flexibility In: Campbell CJ, Fuentes A, MacKinnon KC, Bearder SK, Stumpf RM, editors. Primates in Perspective. New York: Oxford University Press; 241–277. [Google Scholar]
- Taberlet P, Griffin S, Goossens B, Questiau S, Manceau V. et al. , 1996. Reliable genotyping of samples with very low DNA quantities using pcr. Nucleic Acids Res 24:3189–3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trewick SA, Pilkington S, Shepherd LD, Gibb GC, Morgan-Richards M, 2017. Closing the gap: avian lineage splits at a young, narrow seaway imply a protracted history of mixed population response. Mol Ecol 26:5752–5772. [DOI] [PubMed] [Google Scholar]
- van Cise AM, Martien KK, Mahaffy SD, Baird RW, Webster DL. et al. , 2017. Familial social structure and socially driven genetic differentiation in Hawaiian short-finned pilot whales. Mol Ecol 26:6730–6741. [DOI] [PubMed] [Google Scholar]
- van Oosterhout C, Hutchinson WF, Wills DP, Shipley PS, 2010. Micro-checker: software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol Res 4:535–538. [Google Scholar]
- Wang CL, Wang XW, Qi XG, Guo ST, Zhao HT. et al. , 2014. Influence of human activities on the historical and current distribution of Sichuan snub-nosed monkeys in the Qinling Mountains, China. Folia Primatol 85:343–357. [DOI] [PubMed] [Google Scholar]
- Weber D, Stewart BS, Garza JC, Lehman N, 2000. An empirical genetic assessment of the severity of the northern elephant seal population bottleneck. Curr Biol 10:1287–1290. [DOI] [PubMed] [Google Scholar]
- Xiang ZF, Yang BH, Yu Y, Yao H, Grueter CC. et al. , 2014. Males collectively defend their one-male units against bachelor males in a multi-level primate society. Am J Primatol 76:609–617. [DOI] [PubMed] [Google Scholar]
- Yao H, Liu XC, Stanford C, Yang JY, Huang TP. et al. , 2011. Male dispersal in a provisioned multilevel group of Rhinopithecus roxellana in Shennongjia Nature Reserve, China. Am J Primatol 73:1280–1288. [DOI] [PubMed] [Google Scholar]
- Zeng ZG, Gong HS, Song YL, Miao T, 2005. A new distribution record of Sichuan takin Budorcas taxicolor tibetana in Qinling Mountains in Shaanxi, China. Acta Zool Sin 51:743–747. [Google Scholar]
- Zhang Z, Zhang S, Wei F, Wang H, Ming LI. et al. , 2006. Translocation and discussion on reintroduction of captive giant panda. Acta Theriol Sin 26:292–299. [Google Scholar]
- Zhao HT, Wang XW, Li JX, Zhang J, Wang CL. et al. , 2016. Postconflict behavior among Rhinopithecus roxellana leader males in the Qinling Mountains, China. Curr Zool 62:33–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.