Abstract
Esophageal squamous cell carcinoma (ESCC) is one of the most prevalent cancers worldwide. Recent studies have shown that cancer stem cells (CSCs) are present in ESCC, are thought to lead to aggressive tumor behavior and the prognosis. The CXC chemokine receptor 4 (CXCR4), is regarded as a putative CSCs marker in various malignancies. Here, we demonstrate that CXCR4 played a key role in ESCC progression and CXCR4 positive ESCC cells possessed stem-like properties. Furthermore, the anti-malarial agent chloroquine (CQ) targeted CXCR4-positive ESCC cells via STAT3 pathway. Therefore, CQ with anti-CSCs effects may be an effective adjunct to current ESCC chemotherapy regimens.
Keywords: esophageal squamous cell carcinoma (ESCC), cancer stem cells (CSCs), CXCR4, chloroquine (CQ), STAT3
Introduction
Esophageal cancer is the fifth and eighth most frequent cause of cancer-related death in male and female worldwide, respectively (1). Esophageal squamous cell carcinoma (ESCC) is the predominant histopathological subtype and comprises up to 90% of esophageal cancer cases worldwide. The incidence of ESCC varies widely by region. The highest prevalence of ESCC was reported in Southern and Eastern Africa and Eastern Asia, particularly in Linzhou of China, however, in Western and Middle Africa and Central America, the lowest prevalence was estimated (2). The prognosis for ESCC remains poor largely due to late diagnosis and propensity for metastasis. Despite significant advances in diagnostic techniques and therapeutic approaches, the overall 5-year survival rate ranges from 15 to 25% (3–5). Recent studies have shown that many malignancies contain a cell subpopulation known as cancer stem cells (CSCs), which are thought to cause aggressive tumor behavior and therapy resistance and have high tumor-initiating capacity, thereby leading to recurrence, metastasis, and insufficient response to conventional therapies (6). Therefore, CSCs markers that are enriched in ESCC must be determined to isolate cells with stem-like characteristics, improving the prognosis of ESCC patients.
The CXC chemokine receptor 4 (CXCR4), a G-protein-coupled receptor (GPCR), also known as a co-receptor for HIV-1 and HIV-2 (7, 8), is activated exclusively by chemokine CXCL12 (9). Although initial studies were centered on the participation of CXCR4 in HIV infection of T cells, its connection to cancer became a hot research topic. CXCR4 was overexpressed in many types of human cancers, and its expression was correlated with angiogenesis, metastasis, and prognosis (10–14). Cancer cells are thought to hijack the CXCR4/CXCL12 axis to establish distant metastasis. The abrogation of the CXCR4/CXCL12 axis results in reduced metastatic burden in a variety of mouse models of cancer. Increasing evidence suggested that CXCR4 could be regarded as a putative CSCs marker in various malignancies, including glioma, non-small cell lung cancer, prostate cancer, renal cell carcinoma, and breast cancer (15, 16). CXCR4 expression in CSCs confers increased invasiveness and metastatic potential as well as improved self-renewal and survival capacity (17). The anti-malarial agent chloroquine (CQ) has been suggested to have an impact on the cancer stem-like cells via inhibition of autophagy (18, 19). Recently, It has been reported that CQ might be a novel antagonist to CXCR4 (20). Balic et al. demonstrated that CQ preferentially targeted CSCs via inhibition of CXCR4 and hedgehog signaling in pancreatic cancer, which acted through autophagy-independent mode of action (21).
In this study, we confirmed that CXCR4 was significantly up-regulated in ESCC specimens and overall survival was significantly shorter for patients with CXCR4 overexpression, suggesting that CXCR4 play a key role in ESCC progression. CXCR4+ ESCC cells exhibited high expression of stemness-related genes, had higher metastatic ability, and more resistant to anti-cancer drugs, indicating that CXCR4+ cells may possess some characteristics of CSCs. Furthermore, CQ targeted CXCR4-positive ESCC cells via STAT3 pathway, independent of autophagy.
Materials and Methods
Cell Lines and Cell Culture
The human ESCC cell line (EC109) were maintained in RPMI 1640 medium containing 10% fetal bovine serum supplemented with 100 U/ml penicillin G and 100 μg/ml streptomycin in humidified atmosphere at 37°C, 5% CO2.
Tissue Specimens and Clinicopathological Characteristics
A total of 121 ESCC tissues and 91 neighboring non-cancerous tissues were collected immediately following surgical resection of ESCC patients at the First Affiliated Hospital of Zhengzhou University (Zhengzhou, China). Ethical approval from the First Affiliated Hospital of Zhengzhou University Research Ethics Committee and patient written informed consent were obtained. Histological examination was carried out by pathologists. None of the patients received any anti-tumor therapy prior to the surgical resection.
RNA Isolation, Reverse Transcription, and Quantitative Reverse Transcription Polymerase Chain Reaction (qRT-PCR)
Total RNA was isolated using RNAiso Plus (Takara, Japan) and reverse-transcribed using PrimeScript™ II 1st Strand cDNA Synthesis Kit (Takara, Japan) according to the manufacturer's instructions as described earlier (22). QRT-PCR was performed on Agilent Mx3005P system using FastStart Essential DNA Green Master (Roche, USA). Human GAPDH were used as an internal control. All reactions were performed in triplicate. The data were analyzed by 2ΔΔCt method. All the primers used in this research are in Table 1.
Table 1.
Primer sequences are shown for all genes tested.
| Gene name | Sense sequence | Anti-sense sequence | Product size (bp) |
|---|---|---|---|
| GAPDH | GCACCGTCAAGGCTGAGAAC | TGGTGAAGACGCCAGTGGA | 138 |
| CXCR4 | ACTACACCGAGGAAATGGGCT | CCCACAATGCCAGTTAAGAAGA | 133 |
| OCT4 | CCCCTGGTGCCGTGAAG | GCAAATTGCTCGAGTTCTTTCTG | 97 |
| NANOG | CAAAGGCAAACAACCCACTT | TCTGCTGGAGGCTGAGGTAT | 158 |
| SOX9 | AGCGAACGCACATCAAGAC | CTGTAGGCGATCTGTTGGGG | 85 |
| LIN28 | CGGGCATCTGTAAGTGGTTC | CAGACCCTTGGCTGACTTCT | 191 |
| MMP9 | TGTACCGCTATGGTTACACTCG | GGCAGGGACAGTTGCTTCT | 97 |
| MMP15 | AGGTCCATGCCGAGAACTG | GTCTCTTCGTCGAGCACACC | 156 |
| ATG7 | ATGATCCCTGTAACTTAGCCCA | CACGGAAGCAAACAACTTCAAC | 114 |
| BECN1 | CCATGCAGGTGAGCTTCGT | GAATCTGCGAGAGACACCATC | 215 |
Transwell Migration and Invasion Assay
The migration and invasion of ESCC cells were examine using 24-well Transwell chambers (8-μm pore size polycarbonate membrane, Corning, USA). Harvested cells (2 × 104) were resuspended in 100 μl serum-free RMPI-1640 medium and placed in the upper compartment of the chamber. A total of 500 μl of RMPI-1640 medium containing 10% fetal bovine serum was used as a source of chemoattractant and was added in the bottom compartment of the chamber. After 24 h, the cells that had migrated to the basal side of the membrane were stained with crystal violet (0.005%, sigma, USA) and quantified by counting 5 independent symmetrical visual fields under the microscope. All experiments were repeated three times independently.
Immunohistochemistry
Formalin-fixed, paraffin-embedded sections (3 mm) were deparaffinized in xylene, rehydrated with an alcohol gradient, and washed briefly in tap water. Endogenous peroxidase was blocked by with methanol containing 0.3% hydrogen peroxide (H2O2) for 30 min. To retrieve antigenicity, sections were boiled in 10 mM citrate buffer (pH 5.8) for 30 min in a microwave. Next, sections were incubated with goat serum diluted in PBS (pH 7.4) for 30 min at 22°C. Subsequently, sections were incubated at 4°C overnight with primary antibody specific for CXCR4 diluted at 1:200 (Abcam, Cambridge, UK). After incubation with horseradish peroxidase-conjugated secondary antibody for 1 h at 37°C, sections were treated with substrate 3,3-Diaminobenzidine (DAB) and counterstained with hematoxylin and visualized under a microscope (Leica, USA).
Separation of CXCR4+ Cells by Magnetic Cell Sorting (MACS)
EC109 cells were harvested, prepared into single cell suspension and counted. CXCR4+ cells were separated according to the manufacturer's instructions. <2 × 107 cells were suspended in 200 μl PEB buffer. Twenty microliter of CD184 (CXCR4)-APC was added, mixed and incubated for 10 min in the refrigerator (2–8°C). The cells were washed and resuspended in 160 μl PEB buffer. Forty microliter of Anti-APC MicroBeads was added, mixed well and incubated for 15 min in the refrigerator (2–8°C). Then cells were washed, resuspended in 1,000 μl PB buffer and proceeded to magnetic separation. The flow-through was unlabeled cells (CXCR4− cells). MACS column was removed from MACS separator and the magnetically labeled cells (CXCR+ cells) were obtained.
Mouse Experiment
Animal protocols were approved by the Animal Care and Use Committee of the First Affiliated Hospital of Zhengzhou University. Female BALB/c-nu mice (15–18 g, 5–6 weeks old) were obtained from the animal facility (Beijing Vital River Laboratory Animal Technology Co., Ltd.). ESCC cells (1 × 106) were injected subcutaneously into the left flank of recipient BALB/c-nu mice. Tumor bearing mice were treated with CQ (50 mg/kg, once daily I.P. for 18 days). The control mice received vehicle (PBS) alone. Tumor growth was monitored using a caliper and tumor volume was calculated according to the formula: (L × W × W)/2.
Statistics
Statistical analyses were performed with SPSS 17.0. All data were presented as means ± SD of experiments performed in triplicate. Data were tested for normal distribution and Student's t-test, ANOVAs, or non-parametric Mann-Whitney U/Wilcoxon tests were applied. A P < 0.05 was accepted as statistically significant.
Results
CXCR4 Expression Was Correlated With Tumor Invasion and Survival
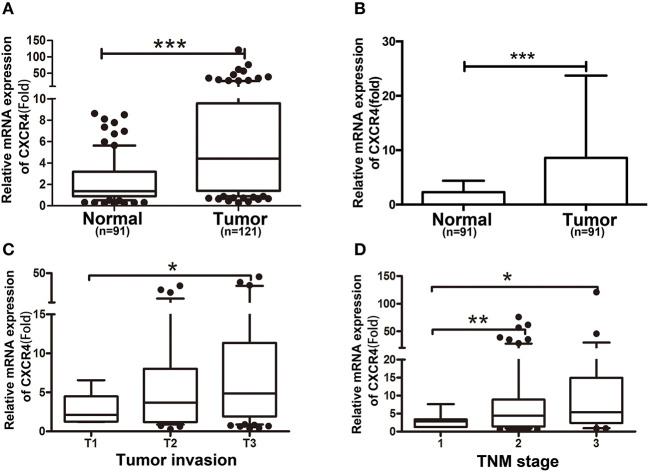
We first detected CXCR4 mRNA expression in human ESCC specimens and neighboring non-cancerous specimens. We found CXCR4 was significantly up-regulated in ESCC specimens, compared with non-cancerous specimens (Figure 1A). Further, 91 paired ESCC specimens and the corresponding neighboring non-cancerous specimens were analyzed, showing significant up-regulation of CXCR4 expression in ESCC specimens (Figure 1B). Moreover, CXCR4 expression was significantly correlated with tumor invasion and stage (Figures 1C,D). No correlation was observed between CXCR4 expression and patient's age, sex, lymph node metastasis and differentiation. Then we performed immunohistochemistry (IHC) on a paraffin-embedded human ESCC tissues. We found that CXCR4 protein expression was significantly upregulated in ESCC compared with the adjacent non-cancerous tissues (Figure 2A). Further, Kaplan-Meier analysis showed that overall survival was significantly shorter for patients with CXCR4 overexpression (Figure 2B). These data suggested that CXCR4 might play an oncogenic role in human ESCC tumorigenesis and development.
Figure 1.
CXCR4 expression is increased in ESCC tissues. (A) The expression of CXCR4 in neighboring non-cancerous tissues (Normal, n = 91) and ESCC tissues (Tumor, n = 121) was determined by real time PCR. (B) Expression of CXCR4 in 91 representative carcinoma tissues and their corresponding non-cancerous tissues from the same patients were analyzed side by side for comparison. The correlation of NEDD9 expression with clinicopathological characteristics such as tumor invasion (C) and TNM stage (D) was analyzed. *P < 0.05, **P < 0.01, and ***P < 0.001.
Figure 2.
Kaplan-Meier survival analysis in patients with ESCC. (A) Representative immunohistochemical staining of CXCR4 expression in ESCC tissues and their corresponding non-cancerous tissues (magnification, ×100). (B) Kaplan-Meier analysis indicating the correlation of CXCR4 overexpression with poorer overall survival of ESCC patients.
CXCR4 Positive ESCC Cells Possessed Stem-Like Properties
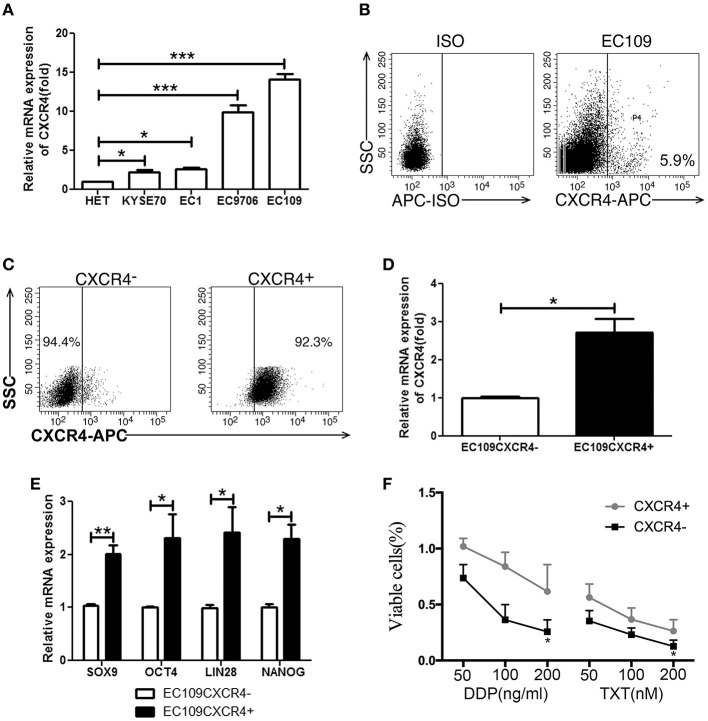
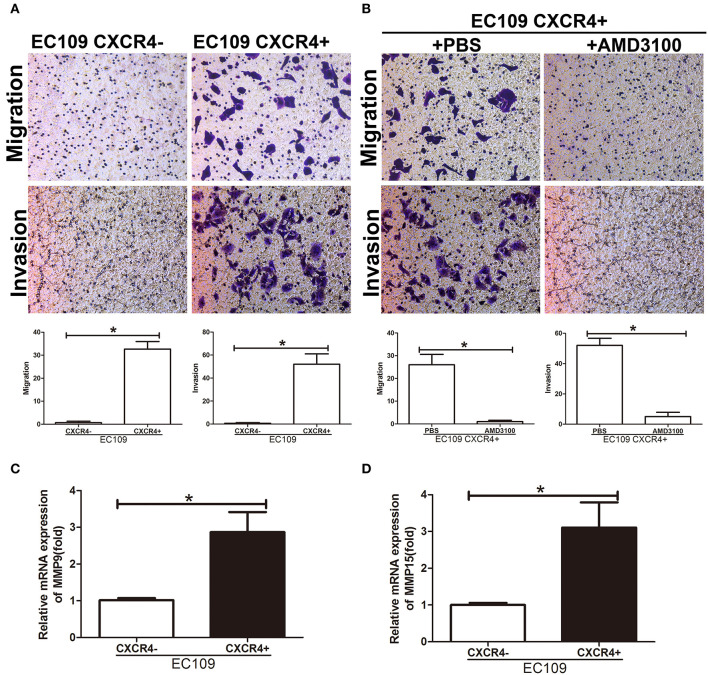
We next analyzed the CXCR4 expression in ESCC cell lines and human esophageal epithelial cell line Het-1a. As expected, we got the same result that CXCR4 was overexpressed in ESCC cell lines compared with human esophageal epithelial cell line Het-1a (Figure 3A, Supplementary Figure S1). CXCR4 expression was highest in EC109 cells and detected by flow cytometry (Figure 3B). To investigate the role of CXCR4 in ESCC, we isolated CXCR4 positive and negative cells in ESCC cell line EC109 by MACS. The purity of the two sorted subpopulations was 92.3 and 94.4%, respectively (Figure 3C). QPCR was used to confirm the expression of CXCR4 in the two subpopulations (Figure 3D). To assess whether the CXCR4 positive cells have the features of the tumor-initiating cells, we compared the mRNA expression levels of stemness-associated markers such as NANOG, OCT4, LIN28, and SOX9 between CXCR4 positive and negative cells by quantitative real-time PCR. The high expression levels of NANOG, OCT4, LIN28, and SOX9 were observed in the CXCR4 positive cells, compared with the CXCR4 negative cells (Figure 3E). Since CSCs have been demonstrated to be more resistant to chemo- and radiation therapy and thus might contribute to drug resistance and tumor recurrence, we analyzed the sensitivity of CXCR4 positive and negative cells toward cisplatin (DDP) and docetaxel (TXT). The survival rates of CXCR4 positive cells were higher under the treatment of DDP and TXT, compared with CXCR4 negative cells (Figure 3F). Further, we analyzed the migration and invasion ability in the two subpopulations. CXCR4 positive cells showed stronger migration and invasion ability (Figure 4A). AMD3100, a CXCR4 antagonist, significantly inhibit migration and invasion ability of CXCR4 positive cells (Figure 4B). And CXCR4 positive cells increased expression of MMP9 and MMP15 (Figures 4C,D), which are known to play an important role in extracellular matrix remodeling during the process of tumor invasion and metastasis. These data suggested that CXCR4 had a functional role in the maintenance of stemness of ESCC cells.
Figure 3.
CXCR4 positive cells possessed stem-like properties. (A) The CXCR4 expression was detected by real time PCR in 1 immortalized esophageal cell line (Het-1a) and 4 ESCC cell lines. (B) The expression analysis of CXCR4 was detected by flow cytometry. (C) The purity of sorted EC109 cells with or without CXCR4 expression. (D) The expression analysis of CXCR4 was detected by real time PCR. (E) The expression analysis of stemness-related transcription factors (SOX9, OCT4, LIN28, and NANOG) was detected by real time PCR. (F) Cell survival rate was tested by CCK-8 method. *P < 0.05, **P < 0.01, and ***P < 0.001.
Figure 4.
CXCR4 promoted cell migration and invasion. (A,B) Migration and invasion assays were performed using Transwell chambers. (C) The expression analysis of MMP9 was detected by real time PCR. (D) The expression analysis of MMP15 was detected by real time PCR. *P < 0.05.
CQ Targeted CXCR4-Positive ESCC Cells via STAT3
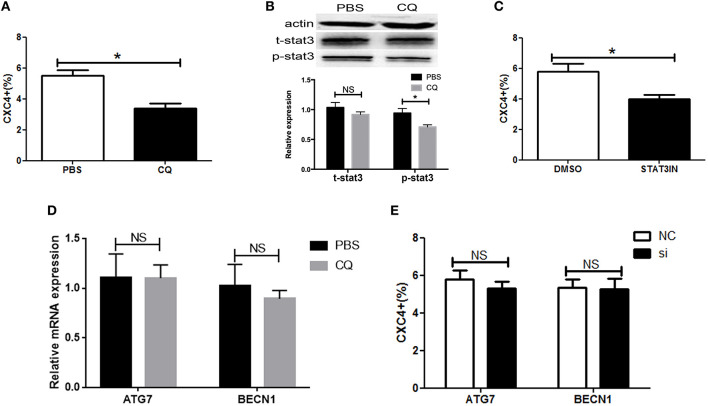
Recently, CQ had been demonstrated to function as an inhibitor of CXCR4 signaling and have an impact on the cancer stem-like phenotype (20, 21, 23, 24). We aimed to assess the effect of CQ on CXCR4 expression in ESCC cells. We found CQ could reduce the amount of cells with CXCR4 expression (Figure 5A). Next, we dissected the molecular mechanisms by which CQ inhibited CXCR4 expression. Signal transducer and activator of transcription 3 (STAT3) plays a key role in the tumorigenesis and cancer stem cells (25, 26). We observed that CQ inhibited STAT3 in ESCC cells (Figure 5B). Further, we found that CXCR4 expression was decreased by the STAT3 inhibitor (Figure 5C). CQ is an anti-malarial drug known to inhibit autophagy by disrupting lysosomal stability and function. We found that CQ had no effect on the expression of ATG7 and BECN1, known as key molecules in autophagy pathway (Figure 5D). Autophagy inhibition with ATG7 or BECN1 siRNAs did not affect CXCR4 expression (Figure 5E). These data suggested that CQ targeted CXCR4-positive ESCC cells via STAT3 pathway, independent of autophagy.
Figure 5.
Chloroquine targeted CXCR4-positive ESCC cells via STAT3. (A) The expression of CXCR4 was analyzed by flow cytometry. (B) The activity of pSTAT3 and tSTAT3 were measured by western blotting in ESCC cells with or without CQ treatment (5 μM). (C) The expression of CXCR4 was analyzed by flow cytometry in ESCC cells with or without STAT3 inhibitor (S3I-201, Sigma, USA) treatment. (D) The expression analysis of ATG7 and BECN1 was detected by real time PCR in ESCC cells with or without CQ treatment (5 μM). (E) The expression of CXCR4 was analyzed by flow cytometry in ESCC cells after ATG7 or BECN1 knockdown. *P < 0.05.
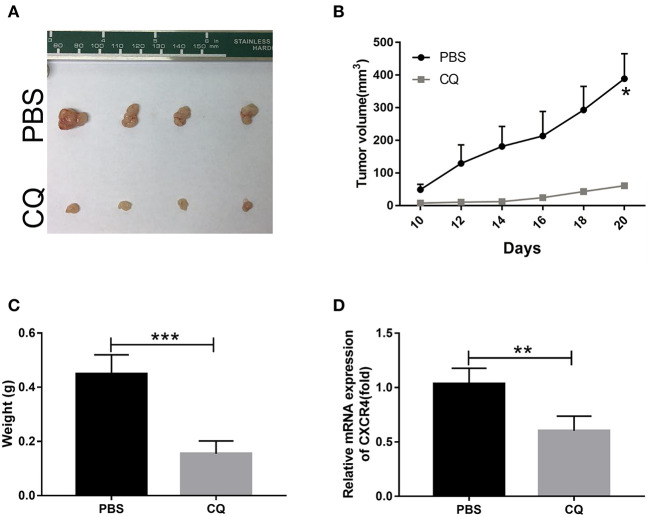
CQ Inhibited Tumor Growth in vivo
The most important property of CSCs is their ability to efficiently propagate tumors in vivo that recapitulate the parental tumors. To examine the role of CQ on the tumor initiating capability, BALB/c nude mice were transplanted with ESCC EC109 cells and treated with or without CQ. We found that animals with CQ treatment markedly delayed the tumor progression relative to the control group (Figures 6A,B). CXCR4 and stemness-related transcription factors showed a significant decrease in animals with CQ treatment (Figures 6C,D). Taken together, these data demonstrate that CQ inhibits the tumorigenic capacity of ESCC cells in vivo.
Figure 6.
Chloroquine inhibited tumor growth in vivo. (A) Images of the xenograft tumors formed in nude mice. (B) Tumor volume was measured every second day and expressed in mm3. (C) Weights of xenograft tumors were summarized. (D) The expression of CXCR4 was confirmed. *P < 0.05, **P < 0.01, and ***P < 0.001.
Discussion
CXCR4, also known as cluster of differentiation 184 (CD184) or fusin, one of the most well-studied chemokine receptors due to its earlier found role as a coreceptor for HIV entry (8), is widely expressed throughout the human body during embryonic development and adult life, with uniquely high-expression levels in the hematopoietic system (27). Recent studies confirmed a wide expression of CXCR4 in the tumors with a consequent implication of this receptor in a panel of pathological processes such as cell proliferation, migration, metastasis and cancer emergence and progression. Importantly, the CXCR4 has emerged as a drug target for its crucial role in promoting and maintaining cancer stem cells (CSCs). Kimura et al. found CXCR4 was associated with cell proliferation and tumorigenicity, concluding that CXCR4 is the marker of synovial sarcoma-initiating cells, a new biomarker for prognosis and a new potential therapeutic target (28). Sun et al. demonstrated that CD133+CXCR4+ primary endometrial cancer cells grew faster, exhibited high expression of stemness-related genes, produced more spheres, had higher clonogenic ability, and more resistant to anti-cancer drugs than other subpopulations, indicating that CD133+CXCR4+ cells may possess some characteristics of CSCs in primary endometrial cancer (29). CXCR4 increases the growth and sphere formation efficiency of hypoxic breast cancer side population (SP) cells by c-Jun/ABCG2 pathway (30).
In human esophageal cancer, CXCR4 overexpression promoted cell invasion in vitro and tumor growth in vivo and indicated worse survival outcome (31–33). Consistent with previous studies, we found that CXCR4 was significantly up-regulated in ESCC tissues, and correlated with tumor invasion and survival. Increasing evidence suggests that CXCR4 is potentially the CSCs marker of malignant tumors including synovial sarcoma (28), endometrial cancer (29), lung cancer (34), and so on. To investigate the role of CXCR4 in ESCC, we isolated CXCR4 positive and negative cells by MACS. We showed that CXCR4 positive cells overexpressed stem-related genes, more resistant to chemotherapy, and possessed stronger invasion ability, suggesting CXCR4 positive cells displayed stem-like properties.
Recently, the CXCL12/CXCR4 signaling pathway has emerged as a potential therapeutic target for human tumors because of its critical role in promoting and maintaining CSCs (35). Over the past few years, a number of inhibitors of CXCL12/CXCR4 have been identified and currently are in different development stages as potential agents for the treatment of cancers. So far, CXCR4 antagonists are developed by a number of programs, including five major classes: (1) small modified peptides, such as T140, (2) small-molecules, such as AMD3100, (3) antibodies, such as MDX-1338/BMS 93656, (4) modified agonists and antagonists for CXCL12 such as CTCE-9908, and (5) microRNAs, such as miR-302a (35).
CQ, a well-known antimalarial, has been administered to patients for over 60 years and the safety profiles are well-established. In malaria infection, CQ inhibits heme polymerase activity in Plasmodium parasites, thereby leading to the buildup of free heme, a substance toxic to the parasite (36). CQ has recently been reported to inhibit self-renewal of CSCs and essentially abrogate their in-vivo tumorigenicity. Application of CQ for cancer treatment is an example of drug re-purposing. Sotelo et al. observed that addition of CQ to standard therapy leads to a significant prolongation of survival in patients with glioblastoma (37). Until recently, the generally accepted antitumor mechanisms of CQ was inhibition of autophagy (38). Nearly simultaneously, it has been demonstrated that tumor-suppressing effects of CQ are independent from its autophagy-inhibiting activities (39, 40). Choi et al. demonstrated that CQ regulates the CSCs in triple negative breast cancer through altering Jak2-STAT3 signaling pathway and DNMT1 expression in addition to autophagy inhibition (24). It was reported that CQ targeted pancreatic cancer stem cells via inhibition of CXCR4 and hedgehog signaling, and its inhibitory effect was not related to inhibition of autophagy (21). We observed that CQ targeted CXCR4-positive ESCC cells, independent of autophagy. This is in line with recent data that CQ can act independently of its effect on autophagy.
In summary, our study demonstrates that CXCR4 is involved in the maintenance of stemness of ESCC cells and plays an oncogenic role in human ESCC. CQ targets CXCR4-positive ESCC cells via STAT3 pathway, independent of autophagy. Therefore, CQ with anti-CSCs effects, warrants further clinical evaluation as a targeted therapy, especially in combination therapy with chemotherapy, for the comprehensive treatment of ESCC.
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.
Ethics Statement
The studies involving human participants were reviewed and approved by the First Affiliated Hospital of Zhengzhou University Research Ethics Committee. The patients/participants provided their written informed consent to participate in this study. This animal study was reviewed and approved by the Animal Care and Use Committee of the First Affiliated Hospital of Zhengzhou University. Written informed consent was obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article.
Author Contributions
YZ and DY contributed conception and design of the study. DY, DZ, XS, SL, AL, DW, GQ, YP, YQ, and XC contributed to the experiments and data acquisition. FW, RC, SZ, and LW performed the statistical analysis. DY wrote the first draft of the manuscript. YZ revised the manuscript. All authors contributed to manuscript revision, read, and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This study was supported by grants from the National Natural Science Foundation of China (Nos. 81602599, 81771781, and U1804281) and the National Key Research and Development Program of China (No. 2016YFC1303501).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2020.00311/full#supplementary-material
The CXCR4 expression was analyzed by flow cytometer. Representative flow image is presented.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. (2015) 65:87–108. 10.3322/caac.21262 [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. (2011) 61:69–90. 10.3322/caac.20107 [DOI] [PubMed] [Google Scholar]
- 3.Lin EW, Karakasheva TA, Hicks PD, Bass AJ, Rustgi AK. The tumor microenvironment in esophageal cancer. Oncogene. (2016). 10.1038/onc.2016.34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zeng H, Zheng R, Zhang S, Zuo T, Xia C, Zou X, et al. Esophageal cancer statistics in China, 2011: estimates based on 177 cancer registries. Thorac Cancer. (2016) 7:232–7. 10.1111/1759-7714.12322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Qiao Y, Zhang C, Li A, Wang D, Luo Z, Ping Y, et al. IL6 derived from cancer-associated fibroblasts promotes chemoresistance via CXCR7 in esophageal squamous cell carcinoma. Oncogene. (2017). 10.1038/onc.2017.387 [DOI] [PubMed] [Google Scholar]
- 6.Alison MR, Lin WR, Lim SM, Nicholson LJ. Cancer stem cells: in the line of fire. Cancer Treat Rev. (2012) 38:589–98. 10.1016/j.ctrv.2012.03.003 [DOI] [PubMed] [Google Scholar]
- 7.Endres MJ, Clapham PR, Marsh M, Ahuja M, Turner JD, McKnight A, et al. CD4-independent infection by HIV-2 is mediated by fusin/CXCR4. Cell. (1996) 87:745–56. 10.1016/S0092-8674(00)81393-8 [DOI] [PubMed] [Google Scholar]
- 8.Feng Y, Broder CC, Kennedy PE, Berger EA. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science. (1996) 272:872–7. 10.1126/science.272.5263.872 [DOI] [PubMed] [Google Scholar]
- 9.Bleul CC, Farzan M, Choe H, Parolin C, Clark-Lewis I, Sodroski J, et al. The lymphocyte chemoattractant SDF-1 is a ligand for LESTR/fusin and blocks HIV-1 entry. Nature. (1996) 382:829–33. 10.1038/382829a0 [DOI] [PubMed] [Google Scholar]
- 10.Yang P, Liang SX, Huang WH, Zhang HW, Li XL, Xie LH, et al. Aberrant expression of CXCR4 significantly contributes to metastasis and predicts poor clinical outcome in breast cancer. Curr Mol Med. (2014) 14:174–84. 10.2174/1566524013666131121115656 [DOI] [PubMed] [Google Scholar]
- 11.Sun X, Charbonneau C, Wei L, Yang W, Chen Q, Terek RM. CXCR4-targeted therapy inhibits VEGF expression and chondrosarcoma angiogenesis and metastasis. Mol Cancer Ther. (2013) 12:1163–70. 10.1158/1535-7163.MCT-12-1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sobolik T, Su YJ, Wells S, Ayers GD, Cook RS, Richmond A. CXCR4 drives the metastatic phenotype in breast cancer through induction of CXCR2 and activation of MEK and PI3K pathways. Mol Biol Cell. (2014) 25:566–82. 10.1091/mbc.e13-07-0360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lv B, Yang X, Lv S, Wang L, Fan K, Shi R, et al. CXCR4 signaling induced epithelial-mesenchymal transition by PI3K/AKT and ERK pathways in glioblastoma. Mol Neurobiol. (2014) 52:1263–8. 10.1007/s12035-014-8935-y [DOI] [PubMed] [Google Scholar]
- 14.Gockel I, Schimanski CC, Heinrich C, Wehler T, Frerichs K, Drescher D, et al. Expression of chemokine receptor CXCR4 in esophageal squamous cell and adenocarcinoma. BMC Cancer. (2006) 6:290. 10.1186/1471-2407-6-290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gassenmaier M, Chen D, Buchner A, Henkel L, Schiemann M, Mack B, et al. CXC chemokine receptor 4 is essential for maintenance of renal cell carcinoma-initiating cells and predicts metastasis. Stem Cells. (2013) 31:1467–76. 10.1002/stem.1407 [DOI] [PubMed] [Google Scholar]
- 16.Jung MJ, Rho JK, Kim YM, Jung JE, Jin YB, Ko YG, et al. Upregulation of CXCR4 is functionally crucial for maintenance of stemness in drug-resistant non-small cell lung cancer cells. Oncogene. (2013) 32:209–21. 10.1038/onc.2012.37 [DOI] [PubMed] [Google Scholar]
- 17.Gatti M, Pattarozzi A, Bajetto A, Wurth R, Daga A, Fiaschi P, et al. Inhibition of CXCL12/CXCR4 autocrine/paracrine loop reduces viability of human glioblastoma stem-like cells affecting self-renewal activity. Toxicology. (2013) 314:209–20. 10.1016/j.tox.2013.10.003 [DOI] [PubMed] [Google Scholar]
- 18.Yang S, Wang X, Contino G, Liesa M, Sahin E, Ying H, et al. Pancreatic cancers require autophagy for tumor growth. Genes Dev. (2011) 25:717–29. 10.1101/gad.2016111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Firat E, Weyerbrock A, Gaedicke S, Grosu AL, Niedermann G. Chloroquine or chloroquine-PI3K/Akt pathway inhibitor combinations strongly promote gamma-irradiation-induced cell death in primary stem-like glioma cells. PLoS ONE. (2012) 7:e47357. 10.1371/journal.pone.0047357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim J, Yip ML, Shen X, Li H, Hsin LY, Labarge S, et al. Identification of anti-malarial compounds as novel antagonists to chemokine receptor CXCR4 in pancreatic cancer cells. PLoS ONE. (2012) 7:e31004. 10.1371/journal.pone.0031004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Balic A, Sorensen MD, Trabulo SM, Sainz B, Jr, Cioffi M, et al. Chloroquine targets pancreatic cancer stem cells via inhibition of CXCR4 and hedgehog signaling. Mol Cancer Ther. (2014) 13:1758–71. 10.1158/1535-7163.MCT-13-0948 [DOI] [PubMed] [Google Scholar]
- 22.Yue D, Fan Q, Chen X, Li F, Wang L, Huang L, et al. Epigenetic inactivation of SPINT2 is associated with tumor suppressive function in esophageal squamous cell carcinoma. Exp Cell Res. (2014) 322:149–58. 10.1016/j.yexcr.2013.11.009 [DOI] [PubMed] [Google Scholar]
- 23.Yu F, Li J, Xie Y, Sleightholm RL, Oupicky D. Polymeric chloroquine as an inhibitor of cancer cell migration and experimental lung metastasis. J Control Release. (2016) 244 (Pt B):347–56. 10.1016/j.jconrel.2016.07.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Choi DS, Blanco E, Kim YS, Rodriguez AA, Zhao H, Huang TH, et al. Chloroquine eliminates cancer stem cells through deregulation of Jak2 and DNMT1. Stem Cells. (2014) 32:2309–23. 10.1002/stem.1746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kryczek I, Lin Y, Nagarsheth N, Peng D, Zhao L, Zhao E, et al. IL-22(+)CD4(+) T cells promote colorectal cancer stemness via STAT3 transcription factor activation and induction of the methyltransferase DOT1L. Immunity. (2014) 40:772–84. 10.1016/j.immuni.2014.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Agarwal S, Lakoma A, Chen Z, Hicks J, Metelitsa LS, Kim ES, et al. G-CSF promotes neuroblastoma tumorigenicity and metastasis via STAT3-dependent cancer stem cell activation. Cancer Res. (2015) 75:2566–79. 10.1158/0008-5472.CAN-14-2946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kircher M, Herhaus P, Schottelius M, Buck AK, Werner RA, Wester HJ, et al. CXCR4-directed theranostics in oncology and inflammation. Ann Nucl Med. (2018) 32:503–11. 10.1007/s12149-018-1290-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kimura T, Wang L, Tabu K, Tsuda M, Tanino M, Maekawa A, et al. Identification and analysis of CXCR4-positive synovial sarcoma-initiating cells. Oncogene. (2016) 35:3932–43. 10.1038/onc.2015.461 [DOI] [PubMed] [Google Scholar]
- 29.Sun Y, Yoshida T, Okabe M, Zhou K, Wang F, Soko C, et al. Isolation of stem-like cancer cells in primary endometrial cancer using cell surface markers CD133 and CXCR4. Transl Oncol. (2017) 10:976–87. 10.1016/j.tranon.2017.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.He C, Zhang H, Wang B, He J, Ge G. SDF-1/CXCR4 axis promotes the growth and sphere formation of hypoxic breast cancer SP cells by c-Jun/ABCG2 pathway. Biochem Biophys Res Commun. (2018) 505:593–99. 10.1016/j.bbrc.2018.09.130 [DOI] [PubMed] [Google Scholar]
- 31.Guo J, Yu X, Gu J, Lin Z, Zhao G, Xu F, et al. Regulation of CXCR4/AKT-signaling-induced cell invasion and tumor metastasis by RhoA, Rac-1, and Cdc42 in human esophageal cancer. Tumour Biol. (2016) 37:6371–8. 10.1007/s13277-015-4504-x [DOI] [PubMed] [Google Scholar]
- 32.Wu J, Wu X, Liang W, Chen C, Zheng L, An H. Clinicopathological and prognostic significance of chemokine receptor CXCR4 overexpression in patients with esophageal cancer: a meta-analysis. Tumour Biol. (2014) 35:3709–15. 10.1007/s13277-013-1490-8 [DOI] [PubMed] [Google Scholar]
- 33.Lu CL, Guo J, Gu J, Ge D, Hou YY, Lin ZW, et al. CXCR4 heterogeneous expression in esophageal squamous cell cancer and stronger metastatic potential with CXCR4-positive cancer cells. Dis Esophagus. (2014) 27:294–302. 10.1111/dote.12100 [DOI] [PubMed] [Google Scholar]
- 34.Chen WL, Huang AF, Huang SM, Ho CL, Chang YL, Chan JY. CD164 promotes lung tumor-initiating cells with stem cell activity and determines tumor growth and drug resistance via Akt/mTOR signaling. Oncotarget. (2017) 8:54115–35. 10.18632/oncotarget.11132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou W, Guo S, Liu M, Burow ME, Wang G. Targeting CXCL12/CXCR4 axis in tumor immunotherapy. Curr Med Chem. (2017) 26:3026–41. 10.2174/0929867324666170830111531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pelt J, Busatto S, Ferrari M, Thompson EA, Mody K, Wolfram J. Chloroquine and nanoparticle drug delivery: a promising combination. Pharmacol Ther. (2018) 191:43–9. 10.1016/j.pharmthera.2018.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sotelo J, Briceno E, Lopez-Gonzalez MA. Adding chloroquine to conventional treatment for glioblastoma multiforme: a randomized, double-blind, placebo-controlled trial. Ann Int Med. (2006) 144:337–43. 10.7326/0003-4819-144-5-200603070-00008 [DOI] [PubMed] [Google Scholar]
- 38.Cai Y, Cai J, Ma Q, Xu Y, Zou J, Xu L, et al. Chloroquine affects autophagy to achieve an anticancer effect in EC109 esophageal carcinoma cells in vitro. Oncol Lett. (2018) 15:1143–8. 10.3892/ol.2017.7415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eng CH, Wang Z, Tkach D, Toral-Barza L, Ugwonali S, Liu S, et al. Macroautophagy is dispensable for growth of KRAS mutant tumors and chloroquine efficacy. Proc Natl Acad Sci USA. (2016) 113:182–7. 10.1073/pnas.1515617113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.King MA, Ganley IG, Flemington V. Inhibition of cholesterol metabolism underlies synergy between mTOR pathway inhibition and chloroquine in bladder cancer cells. Oncogene. (2016) 35:4518–28. 10.1038/onc.2015.511 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The CXCR4 expression was analyzed by flow cytometer. Representative flow image is presented.
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.