Abstract
In nature, biological processes are regulated with precise spatial and temporal resolution at the molecular, cellular, and organismal levels. In order to perturb and manipulate these processes, optically controlled chemical tools have been developed and applied in living systems. The use of light as an external trigger provides spatial and temporal control with minimal adverse effects. Incorporation of light-responsive amino acids into proteins in cells and organisms with an expanded genetic code has enabled the precise activation/deactivation of numerous, diverse proteins, such as kinases, nucleases, proteases, and polymerases. Using unnatural amino acids to generate light-triggered proteins enables a rational engineering approach that is based on mechanistic and/or structural information. This review focuses on the most recent developments in the field, including technological advances and biological applications.
Table of Content Graphic
Introduction
In order to study and manipulate biological processes with the same precision as nature, chemical biologists have developed a number of optical tools [1]. The use of light to control protein activity provides non-invasive, precise, spatiotemporal control and allows for more acute perturbation than other methods (such as RNA interference or gene editing). Optical control of protein function in live systems has primarily been achieved through two approaches: genetic encoding of light-responsive amino acids or optogenetic methods using natural photoresponsive protein domains. Over the last two decades, more than a hundred non-canonical amino acids have been genetically encoded in a range of organisms to provide functionalities not found in the common set of 20 amino acids [2]. The incorporation of light-triggered amino acids into proteins has been used to control a wide range of biological processes in cells and animals [3], and this review highlights select examples from the past 5 years in order to demonstrate the versatility of this approach. Due to space limitations, we are not including other important methodologies, such as protein bioconjugation of photoswitchable ligands [4,5]. Purely optogenetic approaches have been extensively reviewed elsewhere [6–9].
Technical Advances in the Field
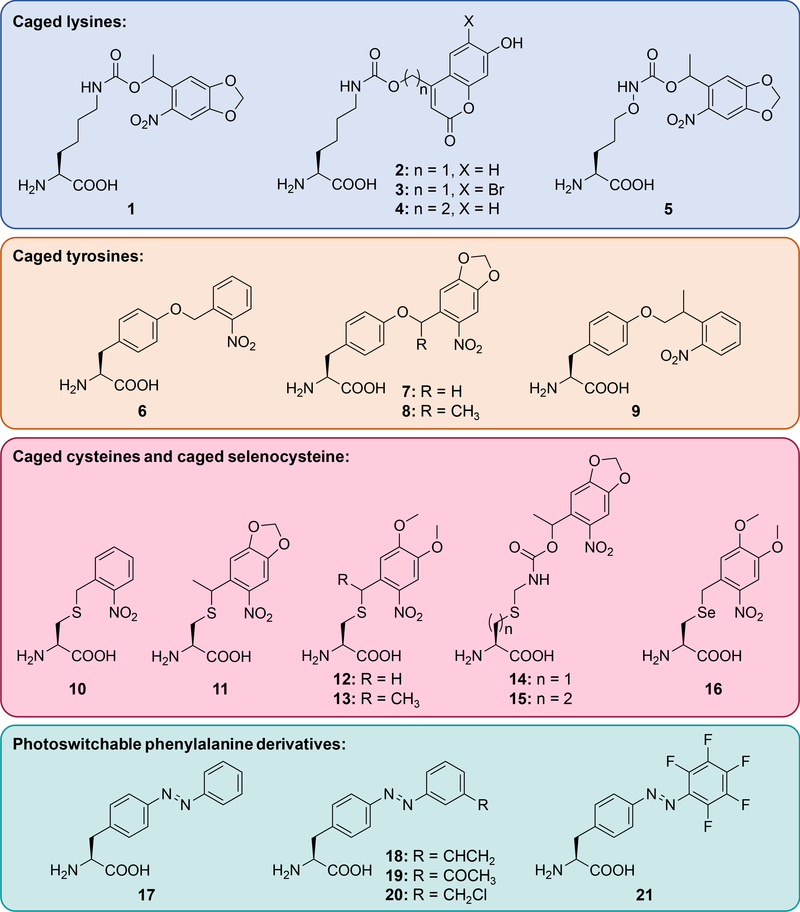
Caged lysine:
The photocaged lysine 1 (Figure 1) has been applied toward optical triggering of Cas9 nuclease [10], T7 RNA polymerase [11], Cre recombinase [12], MEK [13,14] and LCK [15] kinases, isocitrate dehydrogenase [16], and protein-protein interactions [17,18]. Optical control of lysine, which plays an essential role in enzymatic catalysis of many biological processes, has been instrumental in gaining a deeper understanding of living systems at the molecular level. This photocaged lysine utilizes 365 nm light for activation and may be incompatible with certain experiments performed in E. coli due to the abundance of nitroreductases. In order to develop a system that is compatible with a range of organisms and to provide activation with blue (405 nm) and near-IR (two-photon 760 nm) light, the coumarin-caged lysines 2 and 3 were developed [19]. Both were applied in mammalian cells for the optical control of luciferase function and of GFP folding and the different decaging wavelengths for 2 (405 nm) and 3 (760 nm) enable sequential, wavelength-selective activation. Additionally, the coumarin chromophore provides fluorescent tracking of the incorporated amino acid prior to decaging, thus 2 and 3 can act as both fluorescent and photo-activatable probes in live cells. Introduction of the additional methylene group in 4, blocks photolysis and provides a stable and small fluorophore that can be site-specifically placed into proteins. The caged lysine 2 has subsequently been applied to control MEK kinase in zebrafish embryos [14] (see Optical Control of Cell Signaling section) and DNA helicase [20] (see Optical Control of Nucleic Acid Processing section).
Figure 1. Genetically encoded, light-responsive unnatural amino acids.
These include caged lysines 1 - 5, caged tyrosines 6 - 9, caged cysteines 10 - 15, caged selenocysteine 16, and photoswitchable phenylalanine derivatives 17 - 21.
While lysine often plays an essential role in enzymatic catalysis, replacement of the ε-carbon with oxygen generates an amino-oxy functionality which can undergo bio-orthogonal oxime ligation with a ketone or aldehyde. The Virdee group generated the corresponding lysine analog 5 with a nitrobenzyl caging group to render it unreactive until UV-induced photolysis and encoded it using the same synthetase/tRNA pair engineered for 1 [21]. While incorporation efficiency was low, masking a reactive bio-orthogonal handle with a caging group may minimize off-target reactivity and may enable the encoding of other, more reactive bio-orthogonal handles.
Caged tyrosine:
Photocaged tyrosine 6 was genetically encoded a decade ago and has been applied to the optical control of several enzyme classes [22–27]; however, in order to facilitate decaging through red-shifting of the chromophore’s absorption maximum, the Deiters group developed three additional photocaged tyrosine derivatives 7 - 9 [28]. Use of a dual-luciferase reporter allowed for simultaneous assessment of incorporation and decaging efficiency. While 8 delivered the most efficient optical activation of protein function, the caged tyrosine 7 proved to be the better analog when both incorporation efficiency and decaging efficiency are considered. Thus, caged tyrosine 7 was subsequently applied to the demonstration of spatial control of luciferase activity and the efficient optical triggering of TEV protease (TEVp) activity in mammalian cells. Optical control of TEVp may enable the engineering of precise spatiotemporal activation/deactivation of a protein of interest at a desired subcellular region or protein translocation through light-triggered peptide cleavage. Furthermore, caged amino acids may be applicable to the photocontrol of other proteases [29].
Caged cysteine and caged selenocysteine:
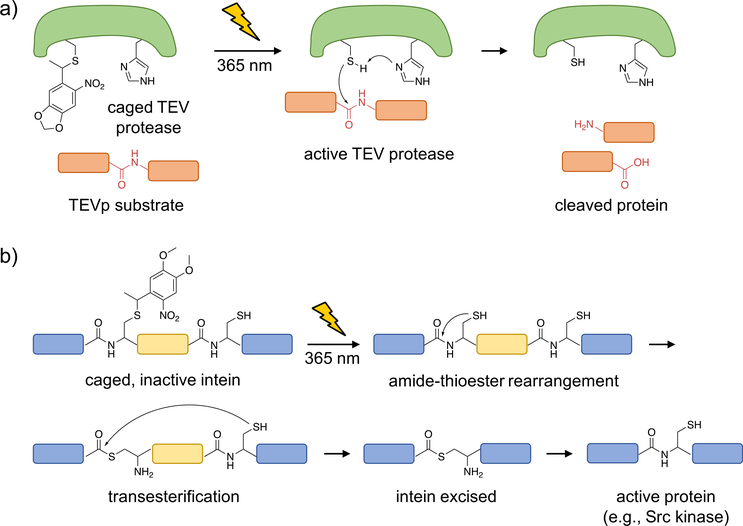
Although it is the least abundant amino acid found in proteins, cysteine plays an essential role in nucleophilic catalysis, redox signaling, metal binding, and structural support through disulfide formation [30]. In order to optically control these different functions, several caged cysteine analogs have emerged in the last few years. Schultz genetically encoded 10 in yeast using an E. coli leucyl synthetase/tRNA pair to cage the active site of caspase-3 [29]. More recently, the Chin lab developed the caged cysteine 11, which was incorporated by an engineered pyrrolysine tRNA synthetase/tRNA pair and applied in mammalian cells for the photo-activation of TEVp (Figure 2a) [31].
Figure 2. Optical control of TEV protease function and intein splicing using photocaged cysteine.
a) Upon caging of TEVp, the catalytic activity is blocked until photolysis of the caged, catalytically active cysteine, which performs a nucleophilic attack onto the protein substrate to generate cleaved fragments. b) A caged intein, indicated by the region in yellow, can be strategically placed within a protein (shown in blue; e.g., mCherry or Src kinase) such that it is misfolded and inactive prior to irradiation. Upon photo-activation, the native cysteine is generated and the intein is excised, leading to full-length, active protein (with only a cysteine scar).
Photoactivated proteins are typically generated through replacement of an essential amino acid with a caged analog. In contrast, the Ai lab developed an approach that can be utilized in cases where such a critical amino acid residue is not available, by developing a light-triggered intein [32]. Inteins are protein segments which can self-cleave and excise, thereby rejoining the cleaved N- and C-terminal fragments to form a new, truncated protein. This cleavage event often utilizes a nucleophilic cysteine residue, making it amenable to optical control through incorporation of the caged cysteines 12 and 13. As proof-of-concept, a caged Nostoc punctiforme DnaE intein (splicing occurs with a reaction half-life of one minute) was placed in the middle of the mCherry protein sequence such that an inactive, non-fluorescent protein was expressed. Following UV irradiation and protein splicing, full-length, active mCherry was generated. In a second application, the caged intein was placed into the catalytic domain of Src kinase in order to control its enzymatic activity and downstream phosphorylation (Figure 2b). One limitation of the approach is the necessity of a cysteine residue at the splice site; which, if not naturally present, will leave a scar following intein excision. In addition, optically controlled inteins have been applied to protein splicing in yeast [33] and generation of cyclic peptides in E. coli [34].
The light-activated cysteines discussed above required tRNA synthetase engineering for genetic encoding. Alternatively, the Deiters lab engineered the caged cysteine structure and developed the caged cysteine 14 and the homocysteine 15, which structurally mimic the caged lysine 1 and serve as substrates for the corresponding tRNA synthetase [35]. Not surprisingly, the homocysteine 15 was incorporated as efficiently as caged lysine 1, while incorporation of 14 was slightly less efficient. Light activation of 14 and 15 was demonstrated through optical control of Renilla luciferase. While both 14 and 15 showed an increase in luminescence following UV irradiation, the activity of the homocysteine containing enzyme was substantially lower. Thus, site-specific incorporation of homocysteine through light-activation of 15 may enable perturbation of an active site with single-atom resolution due to the introduction of an additional methylene unit.
Selenocysteine (Sec) is structurally and functionally similar to its periodic neighbor cysteine. Recently, the role of Sec in functional proteins and enzymes has received much attention [36], and clever approaches to its introduction have been developed [37–39]. The Klimasauskas group masked the nucleophilic selenium with a nitrobenzyl moiety to generate 16 and incorporated it into sfGFP [40]. Following UV irradiation, native Sec was generated and reacted with maleimide-modified biotin.
Photoswitchable amino acids:
While the amino acids described above have enabled optical control of a wide range of protein functions, they are limited to an irreversible activation event. Since many biological processes undergo cycles of activation and deactivation, tools that mimic this reversibility may be better suited for studying these systems. The photoswitchable azobenzene amino acid 17 was first genetically encoded in bacteria in 2006 and applied to controlling catabolite activator protein binding [41] and GFP fluorescence [42]. More recently, additional photoswitchable amino acids have been genetically encoded, in order to provide improved photostationary states, red-shifted wavelengths for photoisomerization, and increased modulation of protein conformation through azo-bridging. The Wang group developed three azobenzene-derived photoswitchable amino acids 18 - 20 with thiol-reactive handles and demonstrated their function in the control of calmodulin conformation [43]. Subsequently, the pentafluoro-azobenzene derivative 21 with red- shifted isomerization wavelength and improved azo-bridging efficiency was reported [44]. The Lin group synthesized and genetically encoded a full set of fluorinated azobenzene analogs with improved photoswitching properties; however, these amino acids have yet to be applied to the control of protein function [45]. Most recently, the Wang group applied 18 to the reversible control of glutamate receptors in mammalian cells without utilizing the inherent cross-linking capabilities [46]. The approaches presented here have established a foundation for obtaining reversible, spatiotemporal control of biological processes in living systems through unnatural amino acid mutagenesis; however, the field is still in its early stages and further work is underway to improve this technology.
Applications
Optical Control of Nucleic Acid Processing:
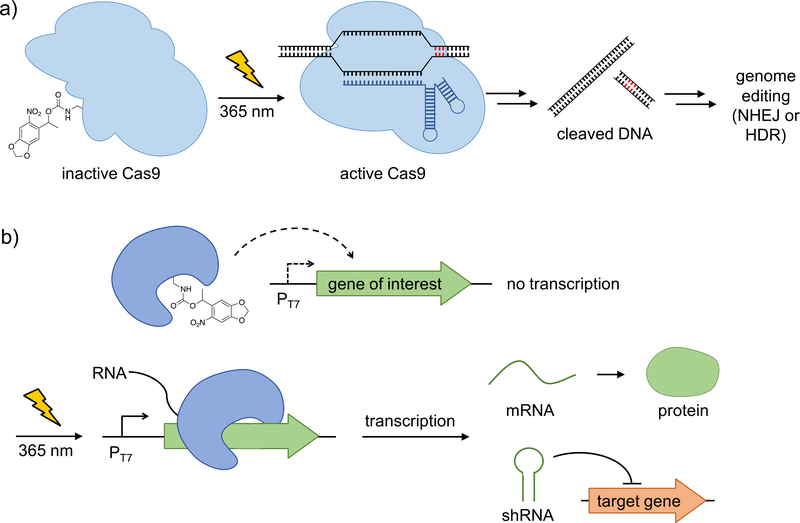
The CRISPR/Cas9 system is a highly versatile genome-editing tool that enables modification, insertion, or deletion of sequences of genomic DNA [47–49]. While natural Cas9 is constitutively active, conditional control of its function enables applications with spatio-temporal precision and may minimize off-target effects [49]. The Deiters group developed the first optically controlled CRISPR/Cas9 system by replacing a lysine residue in the HNH nuclease domain with the photocaged lysine 1, in order to prevent the conformational change necessary for nuclease activity (Figure 3a) [10]. Using a dual fluorescent reporter, which relies on the excision of mCherry and the subsequent expression of GFP, spatial and temporal control of Cas9 activity was achieved in mammalian cells. Additionally, light-triggered gene silencing of an endogenous target, the cell surface receptor CD71, was demonstrated.
Figure 3. Optical control of nucleic acid processing via caged T7 RNA polymerase and CRISPR/Cas9.
a) CRISPR/Cas9 was rendered inactive by the incorporation of the photocaged lysine 1, which blocks HNH nuclease activity. UV irradiation generates active Cas9, which results in DNA double strand cleavage and subsequent genomic editing. b) Caged T7 RNA polymerase is catalytically inactive, preventing transcription of genes under control of the T7 promoter. Upon UV-induced decaging, wild-type T7 RNA polymerase is restored and T7- promoter induced genes (e.g., mRNAs to code for protein or shRNA for gene silencing) are transcribed. Adapted with permission from Angew. Chem. Int. Ed., 55, 5394. Copyright 2016 Wiley-VCH; and J. Am. Chem. Soc., 135, 13433. Copyright 2013 American Chemical Society.
In order to expand the toolkit of optically controlled DNA-processing enzymes, the Deiters group developed a caged helicase (UvrD) [20] and a caged DNA recombinase [12] for spatiotemporal control of DNA unwinding and recombination, respectively. UvrD was rendered light responsive through the installation of the photocaged lysine 2 at a conserved lysine residue within the ATPase domain. In conjunction with optical control of kinase function [13,15], this indicates the possibility of universal photochemical triggering of ATP-dependent processes. Cre recombinase was initially photocaged at an active-site tyrosine with 6; however, in order to improve enzyme expression levels in mammalian cells, a conserved lysine residue was replaced with 1. Excellent off to on photocontrol of Cre recombinase was achieved and the potential for performing knock- in/knock-out experiments with high spatiotemporal resolution was demonstrated in developing zebrafish embryos [50].
In order to optically control transcription in mammalian cells, the Deiters group developed a photocaged T7 RNA polymerase in which an active site lysine was replaced with the caged lysine 1 (Figure 3b) [11]. This enables light-activation of a transcriptional pathway (gene of interest placed under the T7 promoter) that is orthogonal to the endogenous cellular machinery, and the caged T7 RNA polymerase was applied to the triggering of gene expression (control of EGFP mRNA as a proof-of-concept), as well as, gene silencing (control of shRNA targeting the motor protein Eg5 as a proof-of-concept) with spatial and temporal resolution.
Optical Control of Cell Signaling:
Cell signaling networks exhibit a high degree of spatial and temporal dynamics, suggesting light as a preferred external control element. Unlike genetic tools, which require days or hours to knock down or inhibit signaling proteins, light activation of caged amino acids enables one to study the acute effects of kinase function. Since the caged amino acids are genetically directed to their incorporation site in cell signaling proteins, they provide an unmatched specificity that is difficult to achieve with small molecule inhibitors.
LCK (lymphocyte-specific protein tyrosine kinase) is responsible for initiating the T-cell receptor (TCR) signaling pathway, following MHC protein recognition by the TCR. The Chin and James groups used the photocaged lysine 1 to place LCK function under optical control by following the general strategy of blocking an essential and conserved lysine residue in the ATP binding pocket [15]. Through light-triggering of kinase function, they were able to quantify the phosphorylation kinetics of LCK and identified its ability to stimulate its own activation. This work nicely showcases that acute optical activation of kinase function allows for uncovering of mechanistic details of cell signaling activity.
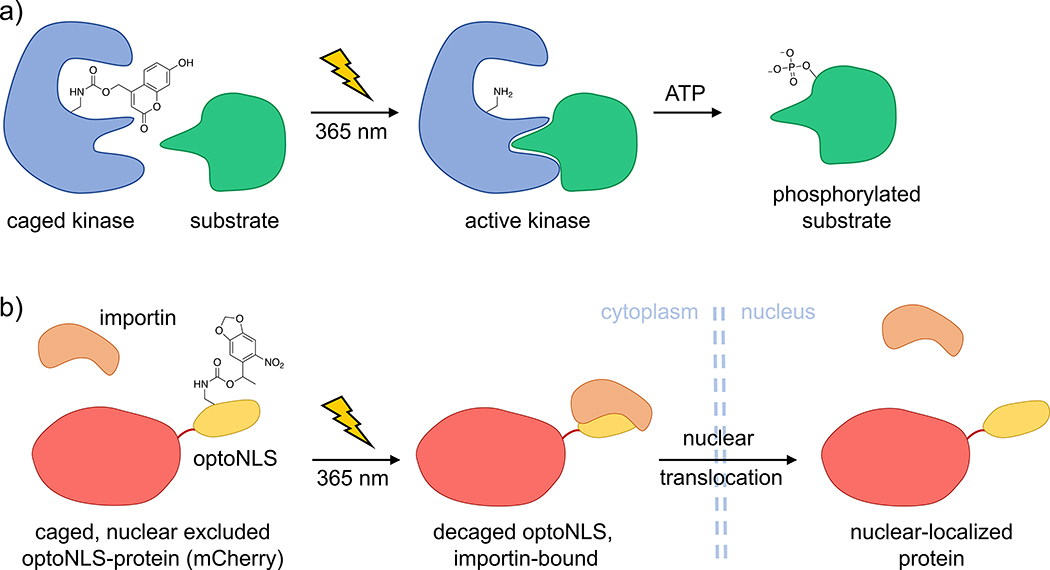
The photocaged lysine 1 has previously been utilized to render the MEK/ERK pathway light-responsive in mammalian cells [13]. More recently, MEK kinase was placed under light control using the more sensitive caged lysine 2 in zebrafish embryos (Figure 4a) [14]. Temporal control of MEK function in the developing animal revealed an essential time window in which hyperactive MEK affects dorsal/ventral patterning, a discovery that is relevant to human birth defects caused by Ras/MAPK pathway mutations. The expansion of optical control of cell signaling to multicellular model organisms will enable investigations in complex systems through more precise perturbation of enzymes/pathways required for normal/mutant embryonic development.
Figure 4. Optical control of kinase activity via genetic incorporation of the caged lysine 2 and optical control of nuclear translocation through caging of a protein-protein interaction.
a) Upon replacement of an essential lysine in the ATP-binding site of a kinase with the caged lysine 2, the enzyme is rendered catalytically inactive. Following UV irradiation, the native, active kinase is generated and is capable of phosphorylating its downstream substrate. b) Replacement of the endogenous nuclear localization sequence (NLS) in the transcription factor FOXO3 (mCherry labeled) with an optoNLS sequence from SatB1 (indicated in yellow), containing a single caged lysine residue enables optical triggering of nuclear translocation.
Control of neural circuits with light has been extensively explored via optogenetic approaches using light-responsive ion channels [6]. In addition to classical optogenetic methods, the use of unnatural amino acid mutagenesis has been utilized by the Wang group by genetically incorporating the caged cysteine 12 into an inward rectifying potassium channel in mouse neocortex tissue slices in order to obtain precise activation of neuronal suppression as measured by patch-clamp recordings [51,52].
Optical Control of Protein-Protein Interactions:
Most examples of using photocaged amino acids for optical control of protein function target enzymatic processes through caging of an essential residue within the active site. However, even a relatively small nitrobenzyl caging group can be efficiently used in the regulation of protein-protein interactions when strategically placed into the protein binding interface. The Engelke and Deiters groups replaced an important lysine residue in the nuclear localization sequence (NLS) of the transcription factor SatB1 with caged lysine 1 [17]. This led to sequestration of NLS fusion protein in the cytoplasm until UV irradiation generated the native NLS sequence, followed by translocation into the nucleus (Figure 4b). This optical NLS approach was applied to controlling FOXO3 transcription factor-DNA binding and TEVp-nuclear substrate interaction. Due to the small size of this optical NLS (20 amino acids), it can easily be appended to any protein of interest to modulate cytoplasmic to nuclear localization. One limitation of the approach is the relatively slow rate of translocation to the nucleus compared to other NLS.
A similar strategy was applied for optically controlling a virus-host protein-protein interaction using the caged lysine 1 [18]. The Chatterjee group incorporated 1 into VP1, a surface protein, of the adeno-associated virus (AAV) capsid. This blocked interaction with human heparin sulfate proteoglycan (HPSG) and prevented viral infection of HEK293T cells, until photolysis released the caging group, forming the native capsid. This approach provides an innovative tool to probe the cellular entry process of human viruses by disrupting the interactions between the virus and the host cell through introduction of photocaged residues and should be broadly applicable to viruses that utilize Lys/Arg, Cys/Ser/Thr, or Tyr as critical regulatory residues at the binding interface.
Optical Control of other Enzymatic Processes:
Isocitrate dehydrogenase (IDH) is an essential enzyme in the citric acid cycle, and has been found mutated through an active site arginine to lysine mutation in various cancers. In order to better study the effects of mutant IDH2 activation, the Chin group replaced an active site lysine with the photocaged lysine 1 to block substrate binding until UV induced decaging [16]. Upon photoactivation of IDH2, a decrease in 5-hydroxymethylcytosine was observed, validating a previously proposed sequence of events in cancer cell epigenetic modifications.
The study of pathogenic bacteria has increased over the last several years as the result of a spike in MRSA-related illnesses. Traditionally, it has been challenging to purify S. aureus toxinantitoxin proteins due to the toxicity caused in E. coli upon overexpression. The Hergenrother group developed a strategy to overcome this limitation by replacing the active site tyrosine with 6, rendering the toxin inactive until a defined activation time-point post-expression and purification [53]. This approach of expressing toxic proteins as their benign, caged precursors may constitute a general method for obtaining otherwise hard-to-isolate proteins.
Summary
Optical control of proteins in cells and organisms with an expanded genetic code has provided precise, spatiotemporal regulation of a diverse range of protein functions. These include optical control of proteolysis, genome editing, protein splicing, phosphorylation, DNA recombination, RNA polymerization, neuronal activity, and protein translocation. In many of the applications discussed above, replacement of an essential residue with a photocaged analog in either an active site or a binding interface rendered the protein inactive. In instances where an essential residue cannot be identified, incorporation of a photocaged intein into the protein may provide an alternative approach for optical control. However, requirements and kinetics of the splicing event will need to be considered in experimental designs. In recent years, photocaged amino acids with improved optical properties, e.g., red-shifted excitation maxima, have been developed, further broadening the scope of this approach.
Compared to other means to optically control protein function, the site-specific, genetic encoding of photocaged and photoswitchable amino acids in cells and animals has several unique advantages: (i) the small size of the various caging groups (~150–250 Da) results in modification of only the most essential site of the protein of interest; (ii) the location of the caged residue can often be predicted based on structural and mechanistic protein data, thereby minimizing the need for extensive trial-and-error experiments; (iii) light-triggered removal of the caging group yields the native, wild-type protein; and (iv) only the unnatural photocaged/photoswitchable amino acid needs to be synthesized and the unnaturally modified protein is generated by the biosynthetic machinery within the cell or organism. Reversible control of protein function with light-switchable amino acids is promising and improved photostationary states and methods for translating the small configurational change of a single light-switchable amino acids into large changes in protein activity and structure will further improve the applicability of these tools. For structurally complex unnatural amino acids, protein yields can (depending on the protein and the site of incorporation) be significantly reduced, leaving room for improvement of the existing tRNA/tRNA synthetase expression systems. This is particularly important for expansion of photocaged amino acid mutagenesis into multicellular model organisms such as zebrafish, fruit flies, and mice which will enable enhanced developmental studies with spatio-temporal precision in order to better understand the complex underpinnings of metazoan development.
Acknowledgement
T.C. was supported by a National Science Foundation Graduate Research Fellowship. A.D. acknowledges support from the National Institutes of Health (GM112728 and HD085206) and the National Science Foundation (CBET-1603930).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ankenbruck N, Courtney T, Naro Y, Deiters A: Optochemical Control of Biological Processes in Cells and Animals. Angewandte Chemie International Edition 2018, 57:2–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wan W, Tharp JM, Liu WR: Pyrrolysyl-tRNA synthetase: An ordinary enzyme but an outstanding genetic code expansion tool. Biochimica et Biophysica Acta (BBA) - Proteins and Proteomics 2014, 1844:1059–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baker AS, Deiters A: Optical control of protein function through unnatural amino acid mutagenesis and other optogenetic approaches. ACS Chem Biol 2014, 9:1398–1407. [DOI] [PubMed] [Google Scholar]
- 4.Tsai Y-H, Essig S, James JR, Lang K, Chin JW: Selective, rapid and optically switchable regulation of protein function in live mammalian cells. Nature chemistry 2015, 7:554–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Broichhagen J, Damijonaitis A, Levitz J, Sokol KR, Leippe P, Konrad D, Isacoff EY, Trauner D: Orthogonal Optical Control of a G Protein-Coupled Receptor with a SNAP- Tethered Photochromic Ligand. ACS Cent Sci 2015, 1:383–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deisseroth K: Optogenetics: 10 years of microbial opsins in neuroscience. Nat Neurosci 2015,18:1213–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang K, Cui B: Optogenetic control of intracellular signaling pathways. Trends in biotechnology 2015, 33:92–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weitzman M, Hahn KM: Optogenetic approaches to cell migration and beyond. Curr Opin Cell Biol 2014, 30:112–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Repina NA, Rosenbloom A, Mukherjee A, Schaffer DV, Kane RS: At Light Speed: Advances in Optogenetic Systems for Regulating Cell Signaling and Behavior. Annual Review of Chemical and Biomolecular Engineering 2017, 8:13–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hemphill J, Borchardt EK, Brown K, Asokan A, Deiters A: Optical Control of CRISPR/Cas9 Gene Editing. J Am Chem Soc 2015, 137:5642–5645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hemphill J, Chou C, Chin JW, Deiters A: Genetically encoded light-activated transcription for spatiotemporal control of gene expression and gene silencing in mammalian cells. J Am Chem Soc 2013, 135:13433–13439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luo J, Arbely E, Zhang J, Chou C, Uprety R, Chin JW, Deiters A: Genetically encoded optical activation of DNA recombination in human cells. Chem Commun (Camb) 2016, 52:8529–8532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gautier A, Deiters A, Chin JW: Light-Activated Kinases Enable Temporal Dissection of Signaling Networks in Living Cells. Journal of the American Chemical Society 2011, 133:2124–2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.**.Liu J, Hemphill J, Samanta S, Tsang M, Deiters A: Genetic Code Expansion in Zebrafish Embryos and Its Application to Optical Control of Cell Signaling. J Am Chem Soc 2017, 139:9100–9103. Genetic code expansion was applied in live zebrafish embryos in order to elucidate the temporal effects of MEK activation on development. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.*.Liaunardy-Jopeace A, Murton BL, Mahesh M, Chin JW, James JR: Encoding optical control in LCK kinase to quantitatively investigate its activity in live cells. Nat Struct Mol Biol 2017, 24:1155–1163. Optical control of LCK kinase enabled quantification of phosphorylation kinetics in live cells and provided greater mechanistic details of the role of LCK in signaling. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Walker OS, Elsasser SJ, Mahesh M, Bachman M, Balasubramanian S, Chin JW: Photoactivation of Mutant Isocitrate Dehydrogenase 2 Reveals Rapid Cancer- Associated Metabolic and Epigenetic Changes. J Am Chem Soc 2016, 138:718–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Engelke H, Chou C, Uprety R, Jess P, Deiters A: Control of protein function through optochemical translocation. ACS Synth Biol 2014, 3:731–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.*.Erickson SB, Mukherjee R, Kelemen RE, Wrobel CJ, Cao X, Chatterjee A: Precise Photoremovable Perturbation of a Virus-Host Interaction. Angew Chem Int Ed Engl 2017, 56:4234–4237. A caged lysine was incorporated into the viral capsid of an AAV protein in order to optically control virus-host interactions, thus demonstrating the ability to precisely tune viral uptake in cells. [DOI] [PubMed] [Google Scholar]
- 19.**.Luo J, Uprety R, Naro Y, Chou C, Nguyen DP, Chin JW, Deiters A: Genetically encoded optochemical probes for simultaneous fluorescence reporting and light activation of protein function with two-photon excitation. J Am Chem Soc 2014, 136:1555115558 Development of blue and near-IR activated lysine analogs provides improved photochemical properties and enables wavelength-selective protein activation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luo J, Kong M, Liu L, Samanta S, Van Houten B, Deiters A: Optical Control of DNA Helicase Function through Genetic Code Expansion. Chembiochem 2017, 18:466469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stanley M, Virdee S: Genetically Directed Production of Recombinant, Isosteric and Nonhydrolysable Ubiquitin Conjugates. Chembiochem 2016, 17:1472–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deiters A, Groff D, Ryu Y, Xie J, Schultz PG: A genetically encoded photocaged tyrosine. Angew Chem Int Ed Engl 2006, 45:2728–2731. [DOI] [PubMed] [Google Scholar]
- 23.Edwards WF, Young DD, Deiters A: Light-activated Cre recombinase as a tool for the spatial and temporal control of gene function in mammalian cells. ACS Chem Biol 2009,4:441–445. [DOI] [PubMed] [Google Scholar]
- 24.Chou C, Young DD, Deiters A: A light-activated DNA polymerase. Angew Chem Int Ed Engl 2009, 48:5950–5953. [DOI] [PubMed] [Google Scholar]
- 25.Chou C, Young DD, Deiters A: Photocaged T7 RNA Polymerase for the Light Activation of Transcription and Gene Function in Pro- and Eukaryotic Cells. Chembiochem 2010,11:972–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chou C, Deiters A: Light-Activated Gene Editing with a Photocaged Zinc-Finger Nuclease. Angew Chem Int Ed Engl 2011, 50:6839–6842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arbely E, Torres-Kolbus J, Deiters A, Chin JW: Photocontrol of tyrosine phosphorylation in mammalian cells via genetic encoding of photocaged tyrosine. J Am Chem Soc 2012, 134:11912–11915. [DOI] [PubMed] [Google Scholar]
- 28.Luo J, Torres-Kolbus J, Liu J, Deiters A: Genetic Encoding of Photocaged Tyrosines with Improved Light-Activation Properties for the Optical Control of Protease Function. ChemBioChem 2017, 18:1442–1447. [DOI] [PubMed] [Google Scholar]
- 29.Wu N, Deiters A, Cropp TA, King D, Schultz PG: A genetically encoded photocaged amino acid. J Am Chem Soc 2004, 126:14306–14307. [DOI] [PubMed] [Google Scholar]
- 30.Pace NJ, Weerapana E: Diverse functional roles of reactive cysteines. ACS Chem Biol 2013, 8:283–296. [DOI] [PubMed] [Google Scholar]
- 31.Nguyen DP, Mahesh M, Elsasser SJ, Hancock SM, Uttamapinant C, Chin JW: Genetic encoding of photocaged cysteine allows photoactivation of TEV protease in live mammalian cells. J Am Chem Soc 2014, 136:2240–2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.**.Ren W, Ji A, Ai HW: Light activation of protein splicing with a photocaged fast intein. J Am Chem Soc 2015, 137:2155–2158. An optically triggered intein was developed and applied to the control of mCherry fluorescence and Src kinase function, potentially providing a general approach to render proteins light-responsive. [DOI] [PubMed] [Google Scholar]
- 33.Tyszkiewicz AB, Muir TW: Activation of protein splicing with light in yeast. Nature Methods 2008, 5:303. [DOI] [PubMed] [Google Scholar]
- 34.Bocker JK, Friedel K, Matern JC, Bachmann AL, Mootz HD: Generation of a genetically encoded, photoactivatable intein for the controlled production of cyclic peptides. Angew Chem Int Ed Engl 2015, 54:2116–2120. [DOI] [PubMed] [Google Scholar]
- 35.Uprety R, Luo J, Liu J, Naro Y, Samanta S, Deiters A: Genetic encoding of caged cysteine and caged homocysteine in bacterial and mammalian cells. Chembiochem 2014,15:1793–1799. [DOI] [PubMed] [Google Scholar]
- 36.Mousa R, Notis Dardashti R, Metanis N: Selenium and Selenocysteine in Protein Chemistry. Angew Chem Int Ed Engl 2017, 56:15818–15827. [DOI] [PubMed] [Google Scholar]
- 37.Gupta N, DeMong LW, Banda S, Copeland PR: Reconstitution of Selenocysteine Incorporation Reveals Intrinsic Regulation by SECIS Elements. Journal of Molecular Biology 2013, 425:2415–2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mukai T, Englert M, Tripp HJ, Miller C, Ivanova NN, Rubin EM, Kyrpides NC, Soll D: Facile Recoding of Selenocysteine in Nature. Angew Chem Int Ed Engl 2016, 55:5337–5341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Metanis N, Hilvert D: Harnessing selenocysteine reactivity for oxidative protein folding. Chem Sci 2015, 6:322–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rakauskaite R, Urbanaviciute G, Ruksenaite A, Liutkeviciute Z, Juskenas R, Masevicius V, Klimasauskas S: Biosynthetic selenoproteins with genetically-encoded photocaged selenocysteines. Chem Commun (Camb) 2015, 51:8245–8248. [DOI] [PubMed] [Google Scholar]
- 41.Bose M, Groff D, Xie J, Brustad E, Schultz PG: The incorporation of a photoisomerizable amino acid into proteins in E. coli. J Am Chem Soc 2006, 128:388–389. [DOI] [PubMed] [Google Scholar]
- 42.Padilla MS, Young DD: Photosensitive GFP mutants containing an azobenzene unnatural amino acid. Bioorg Med Chem Lett 2015, 25:470–473. [DOI] [PubMed] [Google Scholar]
- 43.Hoppmann C, Lacey VK, Louie GV, Wei J, Noel JP, Wang L: Genetically encoding photoswitchable click amino acids in Escherichia coli and mammalian cells. Angew Chem Int Ed Engl 2014, 53:3932–3936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hoppmann C, Maslennikov I, Choe S, Wang L: In Situ Formation of an Azo Bridge on Proteins Controllable by Visible Light. J Am Chem Soc 2015, 137:11218–11221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.John AA, Ramil CP, Tian Y, Cheng G, Lin Q: Synthesis and Site-Specific Incorporation of Red-Shifted Azobenzene Amino Acids into Proteins. Org Lett 2015, 17:62586261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.*.Klippenstein V, Hoppmann C, Ye S, Wang L, Paoletti P: Optocontrol of glutamate receptor activity by single side-chain photoisomerization. eLife 2017, 6:e25808 A photoswitchable amino acid was genetically inserted into the NMDA receptor in mammalian cells and is the first example of reversible optical control in cells using unnatural amino acid mutagenesis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jiang F, Doudna JA: CRISPR-Cas9 Structures and Mechanisms. Annual Review of Biophysics 2017, 46:505–529. [DOI] [PubMed] [Google Scholar]
- 48.Hsu Patrick D, Lander Eric S, Zhang F: Development and Applications of CRISPR-Cas9 for Genome Engineering. Cell 157:1262–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhou W, Deiters A: Conditional Control of CRISPR/Cas9 Function. Angew Chem Int Ed Engl 2016, 55:5394–5399. [DOI] [PubMed] [Google Scholar]
- 50.Brown W, Liu J, Tsang M, Deiters A: Cell Lineage Tracing in Zebrafish Embryos with an Expanded Genetic Code. ChemBioChem 2018, 0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kang JY, Kawaguchi D, Coin I, Xiang Z, O’Leary DD, Slesinger PA, Wang L: In vivo expression of a light-activatable potassium channel using unnatural amino acids. Neuron 2013, 80:358–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kang JY, Kawaguchi D, Wang L: Optical Control of a Neuronal Protein Using a Genetically Encoded Unnatural Amino Acid in Neurons. J Vis Exp 2016:e53818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Larson AS, Hergenrother PJ: Light activation of Staphylococcus aureus toxin YoeBSa1 reveals guanosine-specific endoribonuclease activity. Biochemistry 2014, 53:188201. [DOI] [PMC free article] [PubMed] [Google Scholar]