Abstract
Poly(A) tails are non-templated additions of adenosines at the 3’ end of most eukaryotic messenger RNAs. In the nucleus, these RNAs are co-transcriptionally cleaved at a poly(A) site and then polyadenylated before being exported to the cytoplasm. In the cytoplasm, poly(A) tails play pivotal roles in the translation and stability of the mRNA. One challenge in studying poly(A) tails is that they are difficult to sequence and accurately measure. However, recent advances in sequencing technology, computational algorithms and other assays have enabled a more detailed look at poly(A) tail length genome-wide throughout many developmental stages and organisms. With the help of these advances, our understanding of poly(A) tail length has evolved over the past five years with the recognition that highly expressed genes can have short poly(A) tails and the elucidation of seemingly contradictory roles for poly(A) binding protein (PABP) in facilitating both protection and deadenylation.
Keywords: poly(A) tail, deadenylation, translation, Poly(A) Binding Protein
Poly(A) Tails are a Dynamic and Important Modification of RNA
The first reports of a repetitive poly(A) stretch found on RNA came in the early 1970s [1–3]. At the time, little was known about how or why RNAs had a poly(A) tail, but already there was speculation that it could be “a signal related most probably to translation or mRNA formation and transport” [1]. Over the following years, these predictions were validated and the dynamic regulation of poly(A) tails became more apparent. Numerous polymerases and deadenylases have been identified that are important for modulating tail length [4–6]. The poly(A) tail was beginning to reveal itself as a key player in post-transcriptional regulation, much more than just an afterthought on a messenger RNA (mRNA).
The creation of a poly(A) tail on newly-synthesized RNAs involves the cooperation of many proteins and sequence elements. Almost all metazoan mRNAs contain a polyadenylation signal (PAS) (see Glossary), which has the canonical sequence AAUAAA or a close variant. This PAS as well as a downstream GU- or U-rich sequence guide the formation of the poly(A) tail by recruiting multiple protein complexes involved in initial 3’ end processing [7,8]. Other sequence elements can modulate the efficiency or exact location of polyadenylation. The position where polyadenylation takes place is not decided by the RNA Polymerase terminating the pre-mRNA; instead, cleavage of the RNA occurs co-transcriptionally, within 10–30 nucleotides downstream of the PAS, and poly(A) polymerase (PAP) then adds the poly(A) tail. Once 11–14 adenosines have been added, nuclear poly(A) binding protein (PABPN) is able to bind the growing poly(A) tail [9]. This binding allows PAP to transition from distributive synthesis to processive synthesis, and PAP is then capable of rapidly synthesizing a poly(A) tail, which is thought to be around 200–250 nucleotides in length in metazoans [10,11]. Several polyadenylation studies have contributed to our understanding of this transition and the full-length key proteins involved, but it has been difficult to capture this initial moment of polyadenylation in the context of an intact whole organism [12,13]. Whether all transcripts are ‘fully’ polyadenylated to ~250 adenosines in all tissues is not entirely clear. For example, some genes have been found to include a poly(A) limiting element (PLE) which acts to restrict the initial length of the poly(A) tail on the pre-mRNA to less than 20 nucleotides [14]. Nearly all mRNAs undergo cleavage and polyadenylation to some extent. The known metazoan exceptions are the replication-dependent histone protein genes, which terminate in a stem-loop structure. Some non-coding RNAs, such as several long non-coding RNAs (lncRNAs) and a few small non-coding RNAs, have also been found to contain poly(A) tails [15].
Cleavage and polyadenylation are thought to be necessary for proper export of an mRNA from the nucleus into the cytoplasm. Once in the cytoplasm, the poly(A) tail is predominantly coated with the cytoplasmic poly(A) binding protein (PABPC). Little is known about the transition between PABPN and PABPC on the poly(A) tail. Though both are shuttling proteins that can move between the cytoplasm and nucleus, how and when they complete their trade-off is poorly understood. The protein landscape of a newly synthesized transcript is quite different than that of an actively translating mRNA in the cytoplasm and remodeling of many proteins must take place; for PABPN and PABPC, the first round of translation seems to promote this transformation [16,17]. The exchange between PABPN and PABPC could also be influenced by nuclear export or through passive remodeling in the cytoplasm. PABPN has not been found to have any repeating footprint pattern, but PABPC repetitively coats the poly(A) tail with a footprint of about 20–30 adenosine nucleotides [18,19]. PABPC facilitates a host of interactions important for translation and stability including binding to initiation factor eIF4G of the eIF4F cap-binding complex, as well as the translation termination factor eRF3 [20–24]. Through these interactions, PABPC and the poly(A) tail are able to synergistically promote translation. Interestingly, PABPC has been implicated in protection and stability, as well as recruitment of deadenylases- a seemingly contradictory role that has just begun to be elucidated in recent publications. The complex landscape of poly(A) tail binding interactions is further complicated by the fact that there are other factors such as La proteins that can bind to the poly(A) tail and to PABPC to potentially modulate translation [25,26]. The journey from initial biogenesis to decay contains many multifaceted relationships between the poly(A) tail and various protein factors.
In many instances, the poly(A) tail serves as a gatekeeper to protect the mRNA. Coming from the 3’ end, an enzyme looking to degrade an mRNA would have to chew through the whole poly(A) tail before affecting the protein-coding region. At the 5’ end, the mRNA is protected by its cap, and this cap needs to be specifically removed before degradation can initiate there. Additionally, deadenylation generally occurs before decapping. This positions the poly(A) tail at a key threshold in mRNA decay. Due to this role, the traditional view for many decades was that a longer tail meant more protection, greater stability, and overall a positive influence on translation [4,27,28]. Although these roles in decay are a key part of the life of a poly(A) tail, recent research has shown that this is a simplified version of a fuller story.
Quantitative Measurements of Poly(A) Tail Length
Much of the early work studying poly(A) tail length relied on reporter genes, single-gene analysis, or was conducted in a specific cellular context such as embryogenesis. Although these studies provided insight into some of the factors controlling length, we were still missing the full story of poly(A) tail length dynamics on a genome-wide scale in multiple cellular contexts. With the introduction of high throughput sequencing, many fields within RNA biology took great leaps forward in understanding transcriptome-level events. However, poly(A) sequencing was still not possible with standard RNA-seq protocols primarily due to the difficulty of reading homopolymeric sequences [29]. Current sequencers are largely unable to accurately call multiple adenosines in a row. Despite these challenges, creative approaches were developed to circumvent this issue.
Chang et al. developed a solution termed TAIL-seq that included both an experimental procedure as well as software that uses a machine learning model to accurately measure poly(A) tails (Figure 1a) [30]. In brief, their experimental method involves depletion of ribosomal RNA (rRNA), and size selection against other small noncoding RNAs (tRNA, snRNA, snoRNA, and miRNA). Since noncoding RNAs make up the vast majority of the cellular RNA, these steps allow for enrichment of the library with RNAs that are of interest. A 3’ biotin adaptor is ligated to the RNA and RNase T1 is used to partially digest the RNA selectively after guanosine, leaving the poly(A) tail intact. The ligated RNAs are isolated with streptavidin beads and gel purified before a 5’ adaptor is added. These libraries undergo paired-end sequencing on the Illumina platform (MiSeq or HiSeq instruments). The first read sequences 51 nucleotides and is used for genome mapping, while the second read of 231 nucleotides is used to determine the 3’ end sequence. The fluorescence intensity files are then reanalyzed using their Tailseeker software in order to more accurately assess the length of the poly(A) tail as well as determine any non-A residues present in the tail.
Figure 1:
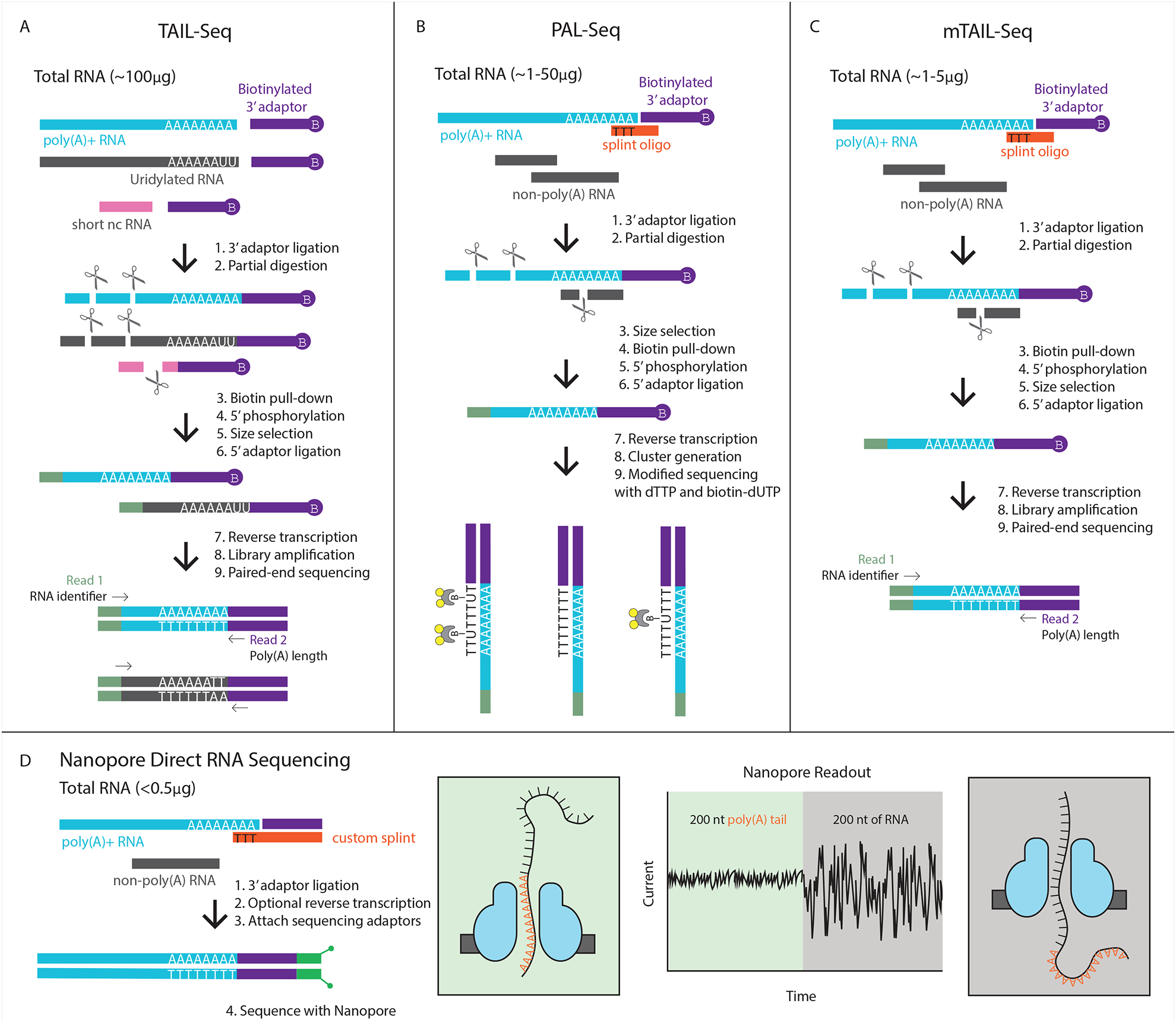
Comparison of different sequencing methods for reading poly(A) tails. A) TAIL-seq is able to capture the 3’ end of any RNA, and therefore gives a readout of both poly(A) tail length as well as other modifications such as uridylation through an innovative Tailseeker algorithm. B) PAL-seq uses a splint oligo to preferentially capture polyadenylated RNAs, thus bypassing rRNA removal. Biotin-labeled dUTP marks each cluster in proportion to the length of the tail. C) mTAIL-seq uses the splint oligo approach in order to reduce the amount of starting material needed, and uses the Tailseeker software to read poly(A) tail length. D) Nanopore technology is a new way to sequence that can be used to directly sequence RNA or cDNA with minimal library preparation needed. The nucleic acid travels through the nanopore at a constant rate; therefore, the dwell time of the poly(A) tail in the nanopore correlates to its length.
Another method was developed by Subtelny et al. in the same year, called PAL-seq (Poly(A) tail Length Sequencing) (Figure 1b) [31]. In order to enrich for polyadenylated species and remove noncoding RNAs, this protocol relies on a ligation step with a DNA splint oligo that bridges the last part of the poly(A) tail with sequence that matches the 3’ biotin adaptor, therefore preferentially adding the adaptor only to RNAs that contain a poly(A) tail. Another key difference in this protocol is the modification done during the sequencing, which is performed on the Genome Analyzer, originally made by Solexa and acquired by Illumina. During sequencing, a mixture of dTTP and biotin-conjugated dUTP is introduced, and each cluster will be marked with an amount of biotin proportional to the length of the tail. Finally, fluorophore-tagged streptavidin is introduced, which will report on the amount of biotin included in each poly(A) tract, and therefore provide a way to determine tail length.
Both of these innovative technologies have expanded our knowledge of poly(A) tail length at a genome-wide scale. Each has its own pros and cons that will vary depending on the application. The original TAIL-seq protocol requires a large amount of starting material, on the order of 100μg of RNA, but because the 3’ ligation is not biased in any way, it is possible to capture other nucleotides present in the tail such as uridylation or guanylation events. On the other hand, PAL-seq requires much less starting RNA and bypasses costly (and sometimes ineffective) rRNA depletion steps thanks to the splint ligation, but this step results in capturing tails that only have adenosines present at their most 3’ end. The final quantification of poly(A) tail length is reliant on the random distribution of biotin tagged uridine and may not be as accurate as determining the identity of each nucleotide, as in the Tailseeker program. Additionally, PAL-seq requires the user to modify the standard sequencing workflow of a Genome Analyzer.
Further advances have been made that combine key components of these two protocols. Two different labs devised methods that took advantage of the splint oligo approach during the ligation step and combined that with the powerful Tailseeker algorithm and ease of Illumina sequencing [32,33]. Since these approaches were very similar to one another, they are both termed mTAIL-seq (Figure 1C).
Although still in early stages, an exciting new avenue for poly(A) tail length measurement is through the use of Nanopore technology (Figure 1D) [34]. Nanopore still faces the same difficulty in accurately calling long stretches of homopolymeric sequence as other sequencing platforms. However, since the nucleic acid being read is pulled through the pore at a constant rate, the amount of time that the poly(A) tail spends going through the pore correlates to its length. Since Nanopore technology is able to directly sequence RNA (and cDNA with minimal library preparation) this method would bypass much of the time and cost associated with preparing a traditional library. Additionally, this can completely bypass PCR amplification, thus eliminating any potential biases introduced at that step. Although this dwell-time readout does not report on whether any other nucleotides are present in the tail besides adenosines, this method could be used to get from experimental condition to actual tail length readout in a very short amount of time. Another new option for inferring poly(A) tail length is TED-seq, which relies on precise size selection of libraries so that the tail length can be deduced by subtracting the distance from the mapped 5’ end of the read to the expected 3’ cleavage/polyadenylation site from the selected fragment size [35]. While a direct tail measurement is not possible, this allows researchers to get an idea of tail length without complicated sequencing methods.
Poly(A) Tails: Connections to Expression and Translation
For many years, it was thought that for most transcripts, a long poly(A) tail would protect the mRNA from decay and degradation. As with many areas of biology, there seems to be greater nuance involved in poly(A) tail length control than previously appreciated. One of the first clues came with the discovery that, contrary to earlier thoughts that most tails would stay long, several transcripts had much shorter poly(A) tails than expected [36,37]. These studies did not map a specific gene with an exact tail length, but instead looked at pools of RNAs that were either not captured by earlier studies that used oligo(dT) beads, or by preferentially eluting RNA off of oligo(dT) beads to produce short tailed and long tailed fractions. Some very stable transcripts such as beta actin were shown to have a short poly(A) tail of less than 30 nucleotides. Overall there were many more transcripts found to have short poly(A) tails than previously expected. This opened up several new inquiries: do certain types of genes have short poly(A) tails? How could a stable transcript have a short tail? Does this serve a biological purpose? Subsequent genome-wide studies began to address these questions. With the implementation of new sequencing methods, the landscape of poly(A) tail length became more clear. Not only were many short-tailed species present, but overall median tail lengths were seen to be in the range of 50–100 adenosines for nearly all species studied: human, Drosophila, mouse, and Caenorhabditis elegans [30,31,33]. Yeast was the only exception with a median tail length of around 33 adenosines, but this was actually not strikingly different considering yeast poly(A) tails were already known to be limited in their initial lengths to around 90 nucleotides [38]. It was becoming clear that a shorter tail is not always linked to destabilization and decay.
These median values reflected much shorter tails than expected, but there still was a broad range of sizes found in each organism. Investigating whether certain genes were enriched for short or long poly(A) tails was actually a more complex undertaking than one might expect. For one, the depth obtained in each sequencing study greatly influences the conclusions reached, especially for genes expressed at a low level. If only a handful of reads are captured for a particular gene, this median may or may not reflect the entire pool of poly(A) lengths for that gene at a given time. In order to assess the poly(A) tail length profile at a truly transcriptome-wide level, sequencing methods must capture even lowly expressed transcripts at their various tail lengths. Second, the use of words such as “short” or “long” to describe tails in the literature must be carefully considered because these words are limited to describing differences within the total length spectrum that an experiment was able to capture. What may be described as “long” in one context could perhaps fall within the “short” category according to another researcher, particularly due to recent changes and advances in the field.
By analyzing recent poly(A) datasets relative to their own median lengths, and taking into account the greatest number of reads possible for each gene, it was determined that short poly(A) tails are associated with highly expressed, well-translated genes [33]. Longer poly(A) tails are associated with transcripts of lower abundance and poor translation (Figure 2). This was a surprising finding and counters the idea that a long tail is universally better for protection and translation. These shortened tails do not appear to be on their way to decay, as they accumulate at steady state at discrete lengths. Furthermore, there was an inverse correlation between poly(A) tail length and half-life of a transcript. This is consistent with early reports which found that short poly(A) tails were associated with the most stable mRNAs in vegetatively growing Dictyostelium discoideum cells [39,40].
Figure 2:
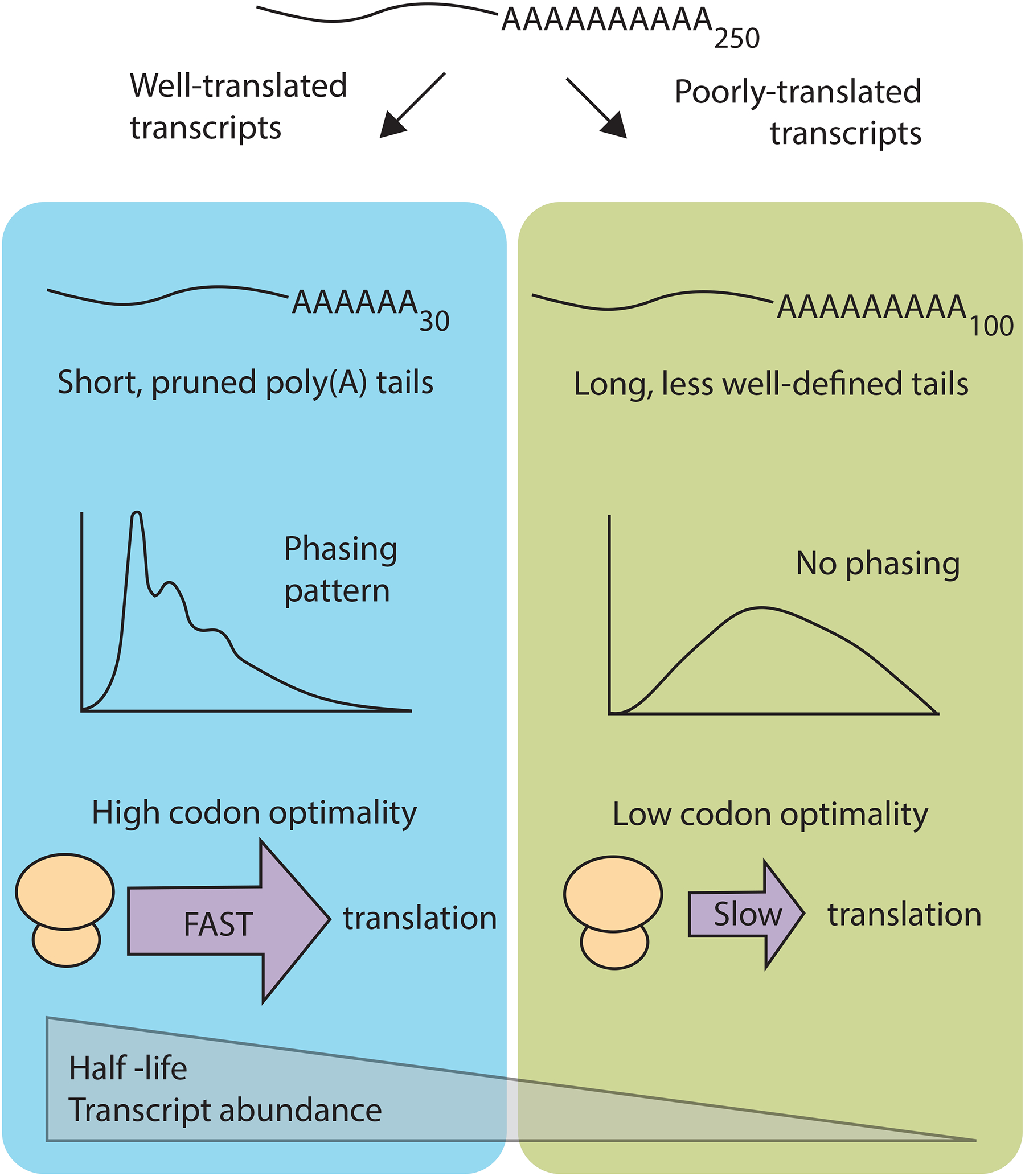
Short poly(A) tails are associated with highly expressed, well-translated transcripts. These shortened tails occur at discrete lengths that have a phasing pattern matching the footprint of serial binding of cytoplasmic poly(A) binding protein. Longer tails do not show this phasing and have less well-defined tails. In somatic cells, short tailed transcripts tend to have high codon optimality and long half lives.
Another interesting feature found in some poly(A) datasets is that there is a phasing pattern to poly(A) tail length wherein there is a greater enrichment for lengths that would be expected to occur with serial binding of PABPC [33,41]. This enrichment for tails at these footprint lengths suggests that unprotected adenosines are likely to be quickly removed. Intriguingly, this phasing distribution is only found on mRNA poly(A) tails and not on other polyadenylated but untranslated species such as lncRNAs [33]. Furthermore, phasing is seen most clearly on highly expressed, well-translated transcripts. This suggests that the shortening of poly(A) tails to discrete lengths, termed pruning, is a process linked to translational activity.
Thus far, the available sequencing datasets have measured poly(A) tail lengths only at steady state. Pulse-chase time course experiments will provide a new level of information regarding the dynamics of poly(A) tail pruning. Deadenylases may remove adenosine nucleotides from all transcripts, regardless of translation status or half-life, but are blocked from fully deadenylating certain transcripts because efficient translation has resulted in a closed-loop formation protecting the transcript. Tails might be deadenylated at the same rate until the deadenylase reaches the critical point of pruning, stopping at the most proximal PABP. However, another possibility is that a sensor recognizes high translation and recruits pruning factors. In this scenario, the tails on well-translated transcripts would be shortened faster, thus resulting in short tails primarily being detected at steady state. Future time course experiments would address these kinetic questions.
These recent studies linking short poly(A) tails with high expression and translation have all been conducted outside of the embryonic context, where it has long been known that a very different polyadenylation landscape is at play. During oocyte maturation and early embryonic development, selective cytoplasmic polyadenylation lengthens the tails of certain mRNAs which actually increases translation, thereby reactivating silenced transcripts [31,32,42–44]. Cytoplasmic polyadenylation has also been found to activate some neuronal transcripts [45]. These seem to be scenarios that are specific to a particular cellular or developmental context.
On top of length as a way to modulate the tail, the composition of poly(A) tails can be variable as well. Uridylation typically occurs on very short tails (less than 25 nucleotides) and is a way to mark a transcript for decay. PABPC binding on the tail inhibits uridylation, and miRNA targeting induces it [46–49]. Guanylation is found selectively on longer poly(A) tails and can stall deadenylation by the Ccr4-Not complex, thus delaying decay [30,50]. Cytosine addition has also been seen on poly(A) tails (though less frequently) but its biological function has not yet been characterized [30,50].
Dual Roles of Poly(A) Binding Protein
The poly(A) tail facilitates numerous interactions between mRNA and proteins. Many of these interactions occur through poly(A) binding protein, which binds the poly(A) tail with high affinity [51,52]. A definitive role for PABPC has remained obscure due to the seemingly conflicting roles it plays in gene expression. On the one hand, it is able to promote deadenylation by direct binding to deadenylase complexes Pan2-Pan3 and Ccr4-Not [53–56]. However, it is most commonly recognized as a protein that directly binds and protects the poly(A) tail from degradation [57,58]. Moreover, through its interaction with 5’ cap-binding factors as well as translation termination factor eRF3, PABPC is thought to promote translation. Nevertheless, as a protein that can recruit deadenylases, it is also involved in decay and down-regulation of genes. These pleiotropic effects have necessitated careful inquiry in order to delineate the parameters of each of these roles.
One major breakthrough came in defining the deadenylase activity of the Ccr4 and Caf1 enzymes, components of the larger Ccr4-Not complex, in the presence of PABPC. Two different groups studying yeast and human deadenylases simultaneously found that binding by PABPC (Pab1 in yeast) does not block Ccr4 activity and in fact, Ccr4 can release PABPC from the tail and continue deadenylating [41,56]. In contrast, Caf1 can only remove adenosines outside of the protective footprint of PABPC (Figure 3A). PABPC still retains a central role in recruitment though, as depletion of Pab1 resulted in much slower deadenylation rates by the Ccr4-Not complex overall [56]. These distinct functional roles suggest that the amount of PABPC on the tail could result in varying tail lengths.
Figure 3:
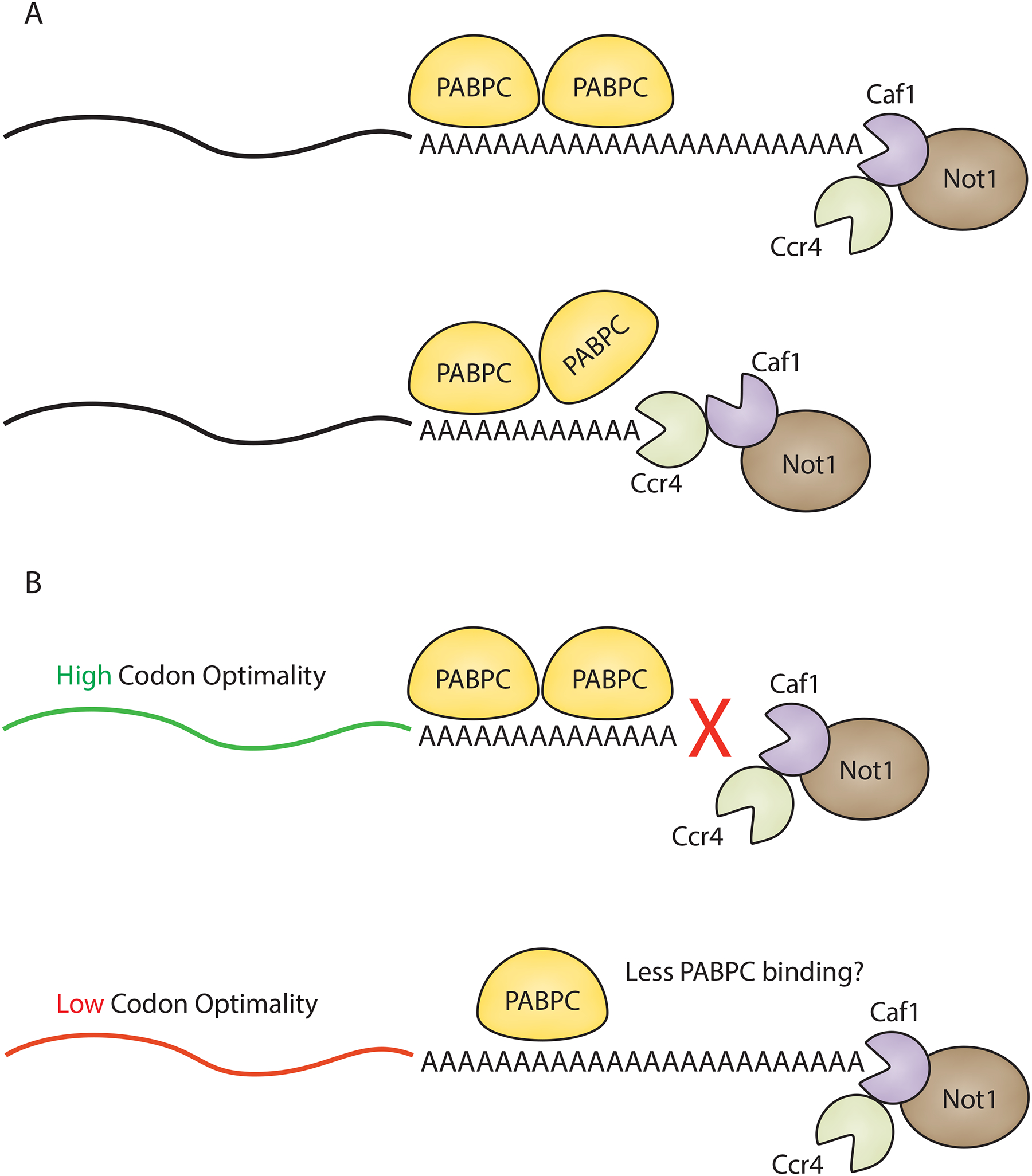
Differential activities of the Ccr4 and Caf1 deadenylases. A) Caf1 is able to deadenylate portions of the poly(A) tail that are not tightly bound by cytoplasmic poly(A) binding protein (PABPC) but will halt once it reaches PABPC. On the other hand, Ccr4 is able to displace PABPC from the poly(A) tail and continue deadenylating. Ccr4 is also able to act on poly(A) stretches that are not bound by PABPC. B) Caf1 preferentially accelerates deadenylation of low codon optimality transcripts. Ccr4 is able to act on both substrates but high codon optimality transcripts seem to rely solely on Ccr4-mediated deadenylation. This may be due to differences in PABPC occupancy on the tails of transcripts with higher (more PABPC) and lower (less PABPC) levels of optimal codons.
Interestingly, deadenylation patterns of Ccr4 and Caf1 were also shown to be influenced by codon optimality, a proxy for translation status [56,59]. Using a reporter system, transcripts with lower codon optimality were deadenylated more rapidly than counterparts with higher optimality. Additionally, depletion of Caf1 preferentially influenced the deadenylation rate of the low codon optimality reporter, suggesting that its poly(A) tail might have less PABPC bound, allowing Caf1 to be more active on lowly compared to highly translated transcripts (Figure 3B).
These studies again situate translation and poly(A) tail length in close contact with one another, with PABPC as a main player in modulating this relationship. It’s unclear how certain well-translated transcripts might end up with more PABPC coating their tail than others. The mechanistic work that has been done to characterize PABPC has revealed non-identical roles for the four RNA binding domains, and it has been suggested that the arrangement of PABPC on the tail may change in response to deadenylation [56]. Whether this shifting of PABPC influences translation or decay is not understood. Further investigation into the multi-pronged roles of PABPC in gene expression will provide answers in this area.
Concluding Remarks and Future Perspectives
Given the importance of fine-tuned gene expression across all domains of biology, it may come as no surprise that the poly(A) tail is not a passive bystander in this process. From its initial biogenesis to dynamic control in the cytoplasm and ultimately decay, the length of the poly(A) tail continues to show itself as an important player in processes as central as translation and mRNA stability. Elucidating the cause and effect of these relationships will be difficult but of ultimate importance in understanding these complex interactions (see Outstanding Questions). As these types of central questions are addressed, the role of the poly(A) tail in more specific contexts, such as mediating miRNA target regulation or consequential alterations in disease may become clearer. With the advent of new technology able to read poly(A) tail size more accurately than ever before, a more coherent and, at times, surprising picture is developing to explain how an addition present on almost all mRNAs is dynamically controlled to exert a vast influence on gene expression.
Acknowledgements
We thank Jens Lykke-Andersen and members of the Pasquinelli lab for suggestions and critical reading of the manuscript. Support for this work was from the UCSD Cellular and Molecular Genetics Training Program through an institutional grant from the National Institute of General Medicine (T32 GM007240) and an NSF Graduate Research Fellowship DGE-1650112 to A.L.N. and a grant from the NIH (R35 GM127012) to A.E.P.
Glossary
- biotin
a molecule frequently used to tag or label a nucleic acid or protein of interest. Biotin binds to streptavidin with a very high affinity, and therefore this interaction can be used to pull out your molecule of interest, separating it away from other molecules that do not have this biotin tag
- cap
the 5’ methylguanosine cap is added to RNA as part of initial processing into a mature mRNA. It protects the transcript from degradation on the 5’ side. The cap is added to the growing RNA co-transcriptionally and is important for export from the nucleus into the cytoplasm, mRNA stability and translation initiation
- deadenylases
Enzymes that catalyze removal of adenosines. There are several different known deadenylases that can act on the poly(A) tail for both pruning and decay
- distributive
referring to the activity of an enzyme, a distributive process is one in which the enzyme dissociates from its substrate frequently after a catalytic event. This is in contrast to a processive enzyme, defined below
- homopolymeric
a repeating sequence of the same nucleotide. The poly(A) tail is a homopolymeric sequence as it contains all adenosine nucleotides. Homopolymeric sequences are notoriously difficult to sequence with current technology
- Nanopore
a type of sequencing that enables minimal library preparation and long sequencing reads. Protein nanopores are embedded on a membrane and an ionic current is passed through the nanopore. When a strand of nucleic acid is going through the pore, the current changes depending on which base is going through the pore, therefore giving a readout of G, C, T or A
- poly(A) polymerase (PAP)
Canonical PAP (also called PAPα) is the enzyme responsible for adding the poly(A) tail to the newly made RNA. Other nuclear and cytoplasmic polymerases have also been discovered which have more specialized functions, different than the general function of canonical PAP
- polyadenylation signal (PAS)
a sequence element directing where cleavage and polyadenylation should occur for most Pol II transcripts. Since cleavage then occurs downstream of this element, it remains in the mature messenger RNA. The canonical sequence AAUAAA is found in most mammalian transcripts, with some minor variability. There can be greater variability in this signal sequence in other eukaryotes
- processive
a processive enzyme can catalyze multiple reactions after a single substrate-enzyme encounter. This would result in continual activity because once an enzyme is associated with the correct substrate, it can continue its activity until complete
References
- 1.Darnell JE et al. (1971) An adenylic acid-rich sequence in messenger RNA of HeLa cells and its possible relationship to reiterated sites in DNA. Proc. Natl. Acad. Sci. U. S. A 68, 1321–1325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee SY et al. (1971) A polynucleotide segment rich in adenylic acid in the rapidly-labeled polyribosomal RNA component of mouse sarcoma 180 ascites cells. Proc. Natl. Acad. Sci. U. S. A 68, 1331–1335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Edmonds M et al. (1971) Polyadenylic acid sequences in the heterogeneous nuclear RNA and rapidly-labeled polyribosomal RNA of HeLa cells: possible evidence for a precursor relationship. Proc. Natl. Acad. Sci. U. S. A 68, 1336–1340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goldstrohm AC and Wickens M (2008) Multifunctional deadenylase complexes diversify mRNA control. Nat. Rev. Mol. Cell Biol 9, 337–344 [DOI] [PubMed] [Google Scholar]
- 5.Schmidt MJ and Norbury CJ (2010) Polyadenylation and beyond: Emerging roles for noncanonical poly(A) polymerases. Wiley Interdiscip. Rev. RNA 1, 142–151 [DOI] [PubMed] [Google Scholar]
- 6.Laishram RS (2014) Poly(A) polymerase (PAP) diversity in gene expression – Star-PAP vs canonical PAP. FEBS Lett. 588, 2185–2197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Proudfoot NJ (2011) Ending the message: poly(A) signals then and now. Genes Dev. 25, 1770–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tian B and Graber JH (2012) Signals for pre-mRNA cleavage and polyadenylation. Wiley Interdiscip. Rev. RNA 3, 385–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meyer S et al. (2002) Equilibrium Studies on the Association of the Nuclear Poly(A) Binding Protein with Poly(A) of Different Lengths. Biochemistry 41, 6082–6089 [DOI] [PubMed] [Google Scholar]
- 10.Brawerman G (1981) The role of the poly(a) sequence in mammalian messenger RN. Crit. Rev. Biochem. Mol. Biol 10, 1–38 [DOI] [PubMed] [Google Scholar]
- 11.Sheiness D and Darnell JE (1973) Polyadenylic Acid Segment in mRNA Becomes Shorter with Age. Nat. New Biol 241, 265–268 [DOI] [PubMed] [Google Scholar]
- 12.Sawicki S et al. (1977) 3’ Terminal Addition to HeLa Cell Nuclear and Cytoplasmic poly(A). J. Mol. Biol 113, 219–235 [DOI] [PubMed] [Google Scholar]
- 13.Wahle E (1995) Poly(A) Tail Length Control is Caused by Termination of Processive Synthesis. J. Biol. Chem 270, 2800–2808 [DOI] [PubMed] [Google Scholar]
- 14.Gu H et al. (1999) The poly(A)-limiting element is a conserved cis-acting sequence that regulates poly(A) tail length on nuclear pre-mRNAs. Proc. Natl. Acad. Sci. U. S. A 96, 8943–8948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cai X et al. (2004) Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA 10, 1957–1966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sato H and Maquat LE (2009) Remodeling of the pioneer translation initiation complex involves translation and the karyopherin importin beta. Genes Dev. 23, 2537–2550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Singh G et al. (2015) The Clothes Make the mRNA: Past and Present Trends in mRNP Fashion. Annu. Rev. Biochem 84, 325–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith BL et al. (1997) Visualization of poly(A)-binding protein complex formation with poly(A) RNA using atomic force microscopy. J. Struct. Biol 119, 109–117 [DOI] [PubMed] [Google Scholar]
- 19.Baer BW et al. (1983) The Protein Responsible for the Repeating Strcuture of Cytoplasmic Poly(A)-Ribonucleoprotein. J. Cell Biol 96, 717–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoshino S et al. (1999) Novel function of the eukaryotic polypeptide-chain releasing factor 3 (eRF3/GSPT) in the mRNA degradation pathway. Biochemistry. (Mosc) 64, 1367–1372 [PubMed] [Google Scholar]
- 21.Hoshino S et al. (1999) The eukaryotic polypeptide chain releasing factor (eRF3/GSPT) carrying the translation termination signal to the 3’-Poly(A) tail of mRNA. Direct association of erf3/GSPT with polyadenylate-binding protein. J. Biol. Chem 274, 16677–16680 [DOI] [PubMed] [Google Scholar]
- 22.Tarun SZ et al. (1997) Translation initiation factor eIF4G mediates in vitro poly(A) tail-dependent translation. Proc. Natl. Acad. Sci. U. S. A 94, 9046–9051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jacobson A and Favreau M (1983) Possible involvement of poly(A) in protein synthesis. Nucleic Acids Res. 11, 6353–6368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wigington CP et al. (2014) Poly(A) RNA-binding proteins and polyadenosine RNA: new members and novel functions. Wiley Interdiscip. Rev. RNA 5, 601–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maraia RJ et al. (2017) The La and related RNA-binding proteins (LARPs): structures, functions, and evolving perspectives. Wiley Interdiscip. Rev. RNA 8, e1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vinayak J et al. (2018) Human La binds mRNAs through contacts to the poly(A) tail. Nucleic Acids Res. 46, 4228–4240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weill L et al. (2012) Translational control by changes in poly(A) tail length: recycling mRNAs. Nat. Struct. Mol. Biol 19, 577–585 [DOI] [PubMed] [Google Scholar]
- 28.Jalkanen AL et al. (2014) Determinants and implications of mRNA poly(A) tail size--does this protein make my tail look big? Semin. Cell Dev. Biol 34, 24–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Quail MA et al. (2012) A tale of three next generation sequencing platforms: comparison of Ion Torrent, Pacific Biosciences and Illumina MiSeq sequencers. BMC Genomics 13, 341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chang H et al. (2014) TAIL-seq: Genome-wide determination of poly(A) tail length and 3’ end modifications. Mol. Cell 53, 1044–1052 [DOI] [PubMed] [Google Scholar]
- 31.Subtelny AO et al. (2014) Poly(A)-tail profiling reveals an embryonic switch in translational control. Nature 508, 66–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lim J et al. (2016) mTAIL-seq reveals dynamic poly(A) tail regulation in oocyte-to-embryo development. Genes Dev. 30, 1671–1682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lima SA et al. (2017) Short poly(A) tails are a conserved feature of highly expressed genes. Nat. Struct. Mol. Biol 24, 1057–1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Garalde DR et al. (2018) Highly parallel direct RNA sequencing on an array of nanopores. Nat. Methods 15, 201–206 [DOI] [PubMed] [Google Scholar]
- 35.Woo YM et al. (2018) TED-Seq Identifies the Dynamics of Poly(A) Length during ER Stress. Cell Rep. 24, 3630–3641.e7 [DOI] [PubMed] [Google Scholar]
- 36.Meijer HA et al. (2007) A novel method for poly(A) fractionation reveals a large population of mRNAs with a short poly(A) tail in mammalian cells. Nucleic Acids Res. 35, e132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Choi YH and Hagedorn CH (2003) Purifying mRNAs with a high-affinity eIF4E mutant identifies the short 3’ poly(A) end phenotype. Proc. Natl. Acad. Sci. U. S. A 100, 7033–7038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brown CE and Sachs AB (1998) Poly(A) tail length control in Saccharomyces cerevisiae occurs by message-specific deadenylation. Mol. Cell. Biol 18, 6548–6559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Palatnik CM et al. (1980) Messenger RNA stability in Dictyostelium discoideum. J. Mol. Biol 141, 99–118 [DOI] [PubMed] [Google Scholar]
- 40.Palatnik CM et al. (1979) Fractionation and functional analysis of newly synthesized and decaying messenger RNAs from vegetative cells of Dictyostelium discoideum. J. Mol. Biol 128, 371–395 [DOI] [PubMed] [Google Scholar]
- 41.Yi H et al. (2018) PABP Cooperates with the CCR4-NOT Complex to Promote mRNA Deadenylation and Block Precocious Decay. Mol. Cell 70, 1081–1088.e5 [DOI] [PubMed] [Google Scholar]
- 42.Sheets MD et al. (1994) The 3’-untranslated regions of c-mos and cyclin mRNAs stimulate translation by regulating cytoplasmic polyadenylation. Genes Dev. 8, 926–938 [DOI] [PubMed] [Google Scholar]
- 43.Eichhorn SW et al. (2016) mRNA poly(A)-tail changes specified by deadenylation broadly reshape translation in Drosophila oocytes and early embryos. Elife 5, 1–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bazzini AA et al. (2016) Codon identity regulates mRNA stability and translation efficiency during the maternal-to-zygotic transition. EMBO J. 35, 2087–2103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Udagawa T et al. (2012) Bidirectional control of mRNA translation and synaptic plasticity by the cytoplasmic polyadenylation complex. Mol. Cell 47, 253–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rissland OS et al. (2007) Efficient RNA polyuridylation by noncanonical poly(A) polymerases. Mol. Cell. Biol 27, 3612–3624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rissland OS and Norbury CJ (2009) Decapping is preceded by 3′ uridylation in a novel pathway of bulk mRNA turnover. Nat. Struct. Mol. Biol 16, 616–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lim J et al. (2014) Uridylation by TUT4 and TUT7 Marks mRNA for Degradation. Cell 159, 1365–1376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Morgan M et al. (2017) mRNA 3′ uridylation and poly(A) tail length sculpt the mammalian maternal transcriptome. Nature 548, 347–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lim J et al. (2018) Mixed tailing by TENT4A and TENT4B shields mRNA from rapid deadenylation. Science 361, 701–704 [DOI] [PubMed] [Google Scholar]
- 51.Görlach M et al. (1994) The mRNA Poly(A)-Binding Protein: Localization, Abundance, and RNA-Binding Specificity. Exp. Cell Res 211, 400–407 [DOI] [PubMed] [Google Scholar]
- 52.Sachs AB et al. (1987) A Single Domain of Yeast Poly(A)-Binding Protein Is Necessary and Sufficient for RNA Binding and Cell Viability. Mol. Cell. Biol 7, 3268–3276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mangus D. a et al. (2004) Positive and Negative Regulation of Poly (A) Nuclease. Mol. Cell. Biol 24, 5521–5533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Funakoshi Y et al. (2007) Mechanism of mRNA deadenylation: evidence for a molecular interplay between translation termination factor eRF3 and mRNA deadenylases. Genes Dev. 21, 3135–3148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Uchida N et al. (2004) Identification of a Human Cytoplasmic Poly(A) Nuclease Complex Stimulated by Poly(A)-binding Protein. J. Biol. Chem 279, 1383–1391 [DOI] [PubMed] [Google Scholar]
- 56.Webster MW et al. (2018) mRNA Deadenylation Is Coupled to Translation Rates by the Differential Activities of Ccr4-Not Nucleases. Mol. Cell 70, 1089–1100.e8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bernstein P et al. (1989) The poly(A)-poly(A)-binding protein complex is a major determinant of mRNA stability in vitro. Mol. Cell. Biol 9, 659–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang Z et al. (1999) An mRNA stability complex functions with poly(A)-binding protein to stabilize mRNA in vitro. Mol. Cell. Biol 19, 4552–4560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hanson G and Coller J (2017) Codon optimality, bias and usage in translation and mRNA decay. Nat. Rev. Mol. Cell Biol 19, 20–30 [DOI] [PMC free article] [PubMed] [Google Scholar]


