Abstract
To explain the patchy distribution of West Nile virus (WNV), we propose that avian immunity encountered by Culex vectors regulates WNV transmission, particularly at communal bird roosts. To test this hypothesis, we selected two test sites with communally roosting American robins (Turdus migratorius) and two control sites that lacked communal roosts. The density of vector-vertebrate contacts, represented by engorged Culex pipiens, was 23-fold greater at test sites compared to control sites, and the density of blood-engorged Cx. pipiens measured in resting mosquito traps correlated positively with the presence of robins and negatively with the presence of other birds, confirming an attraction to robins for blood feeding. WNV transmission was alternately up-regulated (amplification) and down-regulated (suppression) at both test sites. At one test site, infection in resting Cx. pipiens surged from zero to 37.2 per thousand within four weeks, and robin immunity rose from 8.4% to 64% before reducing to 33%. At this site, ten potentially infectious contacts between vector and vertebrates (including nine robins and a mourning dove [Zenaida macroura]) were documented. Infectious vector-vertebrate contacts were absent from control sites. The use of infectious vector-vertebrate contacts, rather than infected mosquitoes, to evaluate a transmission focus is novel.
Keywords: Arbovirus, transmission, ecology, vector, bird, mosquito, West Nile virus, communal bird roost
INTRODUCTION
West Nile virus (WNV; Flavivirus, Flaviviridae) emerged as a pathogen of humans, wildlife, and domestic animals throughout the U.S.A. between 1999–2004 and is now endemic/enzootic throughout the country. The virus spread to Colorado late in 2002 and subsequently caused a major human and wildlife disease outbreak in 2003. In Colorado, WNV has been locally active annually since then, particularly in the months of July and August when infection rates in vector mosquitoes (Culex tarsalis and Culex pipiens) tend to peak (Fauver et al. 2016). In the early years of the invasion of WNV in North America, dead corvids (crows, jays, and magpies) became the hallmark of epizootic activity. However, other avian species that are frequently fed upon by vectors and that die infrequently as a result of infection (e.g., American robin, Turdus migratorius) are probably more important for driving amplification of the virus in the environment (Kilpatrick 2011).
Seroprevalence surveys and vector host-utilization studies, in concert with reservoir competence and relative abundance data, have implicated the house sparrow (Passer domesticus) and American robin as candidate amplifiers in rural and suburban biomes in Colorado (Kent et al. 2009, Komar et al. 2003, McKenzie and Goulet 2010). Studies in Colorado have determined that Culex vectors feed mainly on certain bird species (i.e., robins and doves), presumably due to the combined effects of evolved host preferences of mosquitoes, permissive defensive behaviors of certain birds, avian relative abundance, and avian roosting behaviors (Kent et al. 2009). In Colorado, WNV transmission peaks in late July-early August, coinciding with post-breeding dispersal and communal roosting of certain reservoir-competent passerine birds, such as American robin, American crow (Corvus brachyrhynchos), common grackle (Quiscalus quiscula), house finch (Haemorhous mexicanus) and house sparrow, and some non-passerine birds (i.e., doves, gulls, herons, egrets, pelicans, and cormorants). These nocturnal communal bird roosts may serve as WNV amplification foci. However, efforts to test this hypothesis have produced conflicting results. Some studies found a positive spatial association between communal bird roosts and WNV transmission (Kent et al. 2009, Diuk-Wasser et al. 2010, Reisen et al. 2009). Other studies found the opposite (Reisen et al. 2005, Komar et al. 2015). Critics of the communal roost amplification theory argue that the vector-to-host ratio within dense aggregations of birds is too low to sustain transmission (Janousek et al. 2014, Krebs et al. 2014). A competing hypothesis is that herd immunity among amplifier hosts regulates amplification (Kwan et al. 2012). Essentially, transmission amplifies among competent species until immunity builds up in the amplifier host population. Once immunity wanes due to population turnover, amplification may resume, resulting in patchy distribution of transmission activity over space and time. However, the solution to the puzzle of where and when birds amplify WNV remains unsolved.
In order to address the question of how WNV persists and amplifies in the environment, we propose that both the communal roost amplification and the herd immunity regulation theories are involved. Avian immunity encountered by WNV vectors feeding at communal bird roosts will drive (regulate) WNV transmission activity (i.e., amplification and suppression). To test this hypothesis in Colorado, we selected two study sites that harbored a communal passerine roost in previous years and two control sites for comparison. We measured transmission activity and interactions between vectors and amplifiers throughout the peak WNV transmission season during July and August, 2013.
MATERIALS AND METHODS
Study sites
Four mosquito collection sites were selected in suburban environments in eastern Larimer County, north-central Colorado, based on the presence or absence of nocturnal communal bird roosts. Roost Site A (40.702594, −105.003284) was located in the town of Wellington. Roost Site B (40.4166508, −105.0711679) was located in the city of Loveland. Control Site A (40.711357, −105.028882) was near Wellington, about 1.5 miles from Roost Site A. Control Site B (40.516202, −105.072094) was located in south Fort Collins, about 6.5 miles from Roost Site B.
Avian surveys
Surveys for nocturnally roosting birds were carried out by a single observer (NK) at each mosquito collection site during a ten-min period within the last half hour of daylight, once per week, for six weeks beginning the fourth week of July, 2013. All birds seen or heard entering the site, or already present at the site, were identified and counted. Birds flying over the site were noted but not considered to be roosting locally.
Mosquito sampling
Host-seeking mosquitoes were collected at each site in a single miniature CDC light trap with light bulb removed, and baited each night with approximately 2 kg solid CO2 for three consecutive days each week for eight weeks beginning the second week of July, 2013. Concurrently, resting mosquitoes were collected at each site in three CDC resting traps (BioQuip Products Inc, Rancho Dominguez, CA) for three consecutive days each week. To increase sample sizes of blood-engorged Culex sp. mosquitoes, collections of resting mosquitoes at communal bird roost sites were supplemented using an Insectazooka™ wand aspirator (BioQuip Products, Inc.) for 5 to 15 min, three to four days per week. At Roost Site A, resting mosquitoes were aspirated primarily from a 2.1 m wood security fence. At Roost Site B, resting mosquitoes were aspirated from discarded tires and wood fiber pots placed on the ground (Komar et al. 1995).
Mosquitoes collected in the field were killed by freezing and stored in 2 ml collection tubes at −80° C. Collections were sorted by date, location, collection method, and species after examination using a bifocal dissecting microscope on a custom-built refrigerated table. Species were identified using a standard identification key for North American mosquitoes (Darsie and Ward 2005). Male mosquitoes and other insects were discarded. Female mosquito pools were combined within collection week, with a cap of 50 mosquitoes per pool for non-gravid mosquitoes and 30 per pool for gravid mosquitoes. For the purposes of virus detection, small pools of resting mosquitoes were combined across collection method (i.e., resting trap and aspiration). Engorged mosquitoes with at least half of their blood meal undigested were separated and tested individually (abdomens only) to determine the identity of the blood source from extracted nucleic acid using PCR. Infection status of these individual mosquitoes was determined from testing extracted nucleic acid using RT-PCR.
Mosquitoes were pooled in polystyrene 1.8 ml grinding tubes (model MCT-200-C, Axygen Scientific, Union City, CA) along with a single copper-coated iron ball bearing (BB; Crosman Corporation, Bloomfield, NY) and 1 ml BA1 buffer (M199-Hanks’ salts with L-glutamine; 0.05 M TRIS-HCl, pH 7.5; 1% bovine serum albumin [Bovuminar Cohn Fraction V], pH 7.0; 0.35g/liter sodium bicarbonate; 100 units/ml penicillin; 100 mg/ml streptomycin; 1 mg/ml Fungizone®). Grinding tubes were placed in a cassette and vigorously shaken using a MixerMill® MM300 (Retsch-Allee 1–5, Haan, Germany) set to 25 Hz for 4 min within a Class II biosafety cabinet. After mixing, homogenates were clarified by centrifugation at 10,000 rpm for 3 min and refrigerated (short term) or frozen at −80° C (long-term) until further use.
Virus detection
Virus isolation by plaque assay and a WNV-specific real-time RT-PCR assay were used for detecting arboviruses. For plaque assay, mosquito pool supernatants were inoculated (0.1 ml) in duplicate onto a Vero cell monolayer using a 6-well culture plate (Costar Inc., Cambridge, MA) for selective isolation of arboviruses. After 1 h of incubation at 37° C (5% CO2), all plates were overlaid with 0.5% agarose containing extra antibiotics (100 units/ml penicillin, 100 μg/ml streptomycin, 50 μg/ml gentamycin, 1 mg/ml Fungizone®) and returned to the incubator. After two days, one set of plates was then overlaid again with 0.5% agarose containing neutral red stain and returned to the incubator. The duplicate set of plates was incubated an additional three days prior to adding the second overlay. After staining, both sets of plates were observed daily for viral plaque formation until the cells expired five days later.
For RT-PCR, sub-aliquots were prepared for all mosquito pool homogenates in a 96-well S-block, mixing 140 μl of each homogenate with 150 μl of extraction buffer. DNA and RNA were simultaneously extracted from mosquito homogenates in a 96-well plate format using a Qiagen Biorobot 9604 (Qiagen, Valencia, CA), according to the manufacturer’s instructions. Nucleic acids were eluted in 100 μl AVE elution buffer supplied with the Qiagen Biorobot 9604 extraction kit and stored at −20° C until use. Four wells consisting of tap water were included on each extraction plate as a control for contamination. RNA was transcribed to cDNA and used in a real-time PCR assay for detection of WNV genomic sequences as described previously (Lanciotti et al. 2000). Any positive mosquito pool (Ct ≤ 38.5) was retested with a second set of primers and probe to rule out false positive test results. For individual (blood-engorged) mosquitoes, we severed each abdomen from its respective thorax while frozen using forceps decontaminated with ethanol. The same methods were used to extract RNA from the abdomens, except that a zinc-coated BB was used in 0.5 ml PBS for homogenization with the mixer mill set to 18 cycles/sec for 2.5 min. Pools of eight RNA extracts were prepared (1 μl each). If a positive result was obtained, original RNA extracts were repeat tested individually to determine which of the eight specimens was positive. If an abdomen tested positive, legs were removed from the corresponding mosquito carcass, homogenized, and clarified. Extracted nucleic acid from the leg homogenates was tested by real-time RT-PCR to determine if the original infection detected in the abdomen was already disseminated in the mosquito. If yes, the mosquito was assumed to have been infected prior to blood-feeding and the blood considered uninfected. If no, the mosquito was assumed to have been uninfected prior to blood-feeding and the blood considered viremic.
Blood meal identification
To determine the vertebrate source of blood in the engorged abdomens of mosquitoes, extracted nucleic acid was subjected to PCR using vertebrate-degenerate primers for the mitochondrial CO1 gene, following a previously described protocol (Kent et al. 2009). Successful amplification of a DNA product was confirmed by visualization of an ethidium bromide-stained fragment approximately 648 bases in length by 2% agarose gel electrophoresis. The amplified fragment was column-purified and sequenced in both directions by the Sanger method using an ABI 3130 genetic analyzer. Vertebrate identification was accomplished by choosing the best match in the Barcode of Life Database (BOLD) (Ivanova et al. 2007) and/or GenBank.
Antibody detection
Antibodies in the blood-engorged abdomen homogenates were labeled with biotin to provide a means of virus-specific antibody detection, following the protocol described by Basile et al. (2010) with minor modifications. Briefly, 55 μl of mosquito abdomen homogenate or control media was loaded into each well of a 100,000-molecular-weight-cutoff filter plate (Acroprep 96 Omega 100K; VWR Scientific, San Francisco, CA) and supplemented with 5 μl of 5.55 mg/ml sulfo-LC-biotin (Pierce, Rockford, IL). The filter plate was incubated at room temperature for 30 min on a rotary plate shaker (Lab-Line Instruments, VWR Scientific) at 800 rpm. Biotinylated antibodies were retained in the wells and unwanted components were removed by vacuum filtration. Samples/controls were subsequently washed in the filter plate using 100 μl PBS and then re-suspended in 60 μl PBS. The entire volume (60 μl) of each sample/control was added to a low-binding 96-well plate and diluted with 60 μl of Candor Low Cross buffer (Boca Scientific, Boca Raton, FL). These samples were then tested for WNV-specific and St. Louis encephalitis virus-specific antibodies using a biotin-microsphere immunoassay (b-MIA) as previously described (Komar et al. 2015). Briefly, biotinylated antibody samples were mixed with microsphere set 132 (Radix Biosolutions, Georgetown, TX) conjugated to either West Nile viral antigen or normal control antigen (Hennessey Research, Kansas City, MO). A corresponding assay for detection of SLEV-reactive antibodies utilized microsphere set 157. The amount of binding was determined by the addition of streptavidin-phycoerythrin (Jackson Immunoresearch, West Grove, PA), with measurement of median fluorescent intensities (MFI) for each microsphere set using a BioPlex instrument (Bio-Rad Laboratories, Hercules, CA). A blood-engorged mosquito abdomen spiked with flavivirus group-reactive monoclonal antibody 6B6C-1 was used as a positive control.
Statistical methods
An Excel add-in computed point and confidence interval estimates of mosquito infection rate (i.e., infection prevalence) using data from pooled samples, where pool sizes may differ. Bias-corrected maximum likelihood methods were used to estimate infection rate and a skew-corrected score confidence interval computed (Biggerstaff 2009, https://www.cdc.gov/westnile/resourcepages/mosqsurvsoft.html, accessed 12 Mar 2018). Confidence limits for seroprevalence estimates were generated using the Wilson score method for binomial proportions (S-PLUS 6.1 Professional software, Insightful Inc., Seattle, WA). Pearson’s correlation coefficient (r) and associated p-values were calculated from scatter plots comparing density of resting mosquitoes (Cx. pipiens and Cx. tarsalis analyzed separately) and counts of communally roosting robins or counts of all other birds at each of the four study sites (Pagano and Gauvreau 1993).
RESULTS
Avian surveys
American robins were identified roosting communally at Roost Sites A and B but were infrequently detected at Control Sites A and B (Figure 1). Groups of American robins were observed flying over Control Site B and were later discovered roosting communally nearby, about 0.3 miles from Control Site B. At Roost Site A (Wellington), communal roosting of American robin was stable at a relatively low level (weekly count range 8–13, mean 11.2) throughout the study period but spiked (count = 27) for one survey in mid-August. At Roost Site B (Loveland), communal roosting increased rapidly through the end of July (reaching a high count of 111 individual robins) and then decreased rapidly in early August such that the roost site appeared abandoned by mid-August. The cause of the abandonment is unknown but may have been influenced by a construction project adjacent to the site. Control Site A had relatively high numbers of breeding house sparrows (maximum weekly count 24), barn swallows (Hirundo rustica, maximum weekly count 16), and Eurasian collared-doves (Streptopelia decaocto, maximum weekly count 4) late into the summer. Overall American robins comprised 46.3% of all birds counted (N=555) at the four sites and 72.3% of all birds counted at Roost Sites A and B (N=343). Other species of birds counted, in order of decreasing abundance, included house sparrow (13.3% of all birds), house finch (13.0%), mourning dove (Zenaida macroura, 4.9%), Eurasian collared-dove (4.7%), black-capped chickadee (Poecile atricapilla, 4.7%), barn swallow (4.5%), blue jay (Cyanocitta cristata, 2.9%), American goldfinch (Spinus tristis, 1.1%), western meadowlark (Sturnella neglecta, 0.9%), western kingbird (Tyrannus verticalis, 0.7%), blue grosbeak (Guiraca caerulea, 0.5%), red-winged blackbird (Agelaius phoeniceus, 0.4%), killdeer (Charadrius vociferus, 0.4%), and northern flicker (Colaptes auratus, 0.4%). The following species were observed only once each (0.2%): cedar waxwing (Bombycilla cedrorum), common grackle, common nighthawk (Chordeiles minor), cliff swallow (Petrochelidon pyrrhonota), downy woodpecker (Picoides pubescens), hummingbird species (Selasphorus sp.), and western tanager (Piranga ludoviciana).
Figure 1.
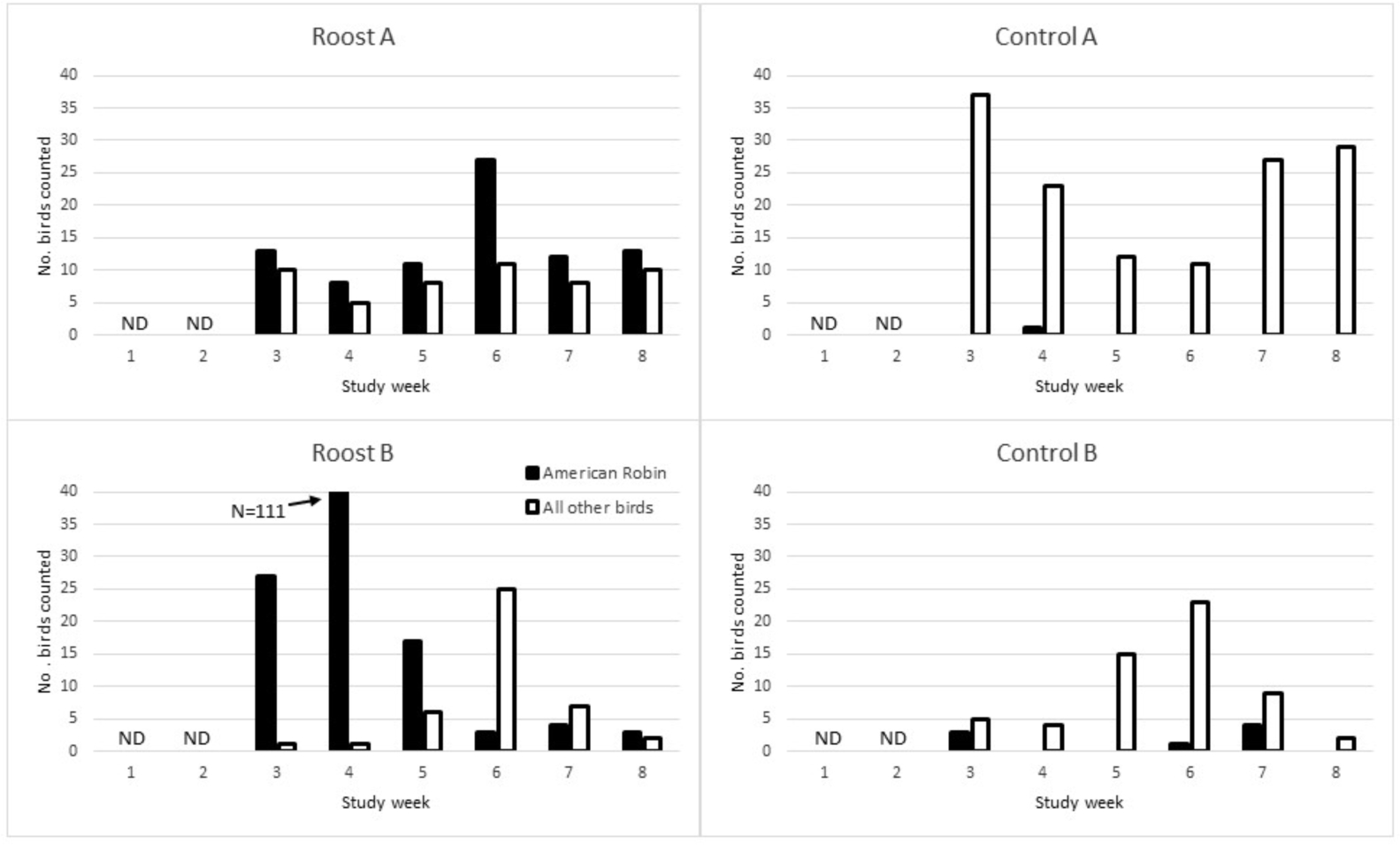
Single-observer counts of all birds present or entering study sites (Roost A, Roost B, Control A, Control B) during a ten-min period at dusk, by week, July-August, 2013. Solid bars represent counts for the American Robin, which were roosting communally at Roost Sites A and B, but were essentially absent at Control Sites A and B. Open bars represent all other bird species combined. See text for list of other bird species detected. No bird counts occurred during the first two weeks of the study. ND = no data.
Mosquito sampling
Overall, 7,772 adult female mosquitoes of 11 species were collected in July and August, 2013, of which 5,594 were host-seeking (captured in C02-baited fan traps) and 2,178 were resting (collected in resting traps and by aspiration) (Table 1). Aedes vexans was the most abundant mosquito collected in the CO2-baited traps. Cx. pipiens was the most abundant resting mosquito.
Table 1.
Adult female mosquitoes (N=7,936) collected during July-August, 2013, at four sites in Larimer County and tested (pools ≤50) for viral RNA by RT-PCR, according to collection method. Engorged mosquitoes were screened in pools of eight, and retested as individuals if the screen was positive. To calculate the infection rate, they were treated as pools of one. Other than West Nile virus (WNV), only Flanders virus was detected (in Cx. tarsalis, see text). CI, 95% confidence interval.
| Species | Collection method | Total female mosquitoes | Pools tested | WNV detections by RT-PCR | WNV I.R. per 1000 [CI] |
|---|---|---|---|---|---|
| Culex tarsalis | CO2 | 1,719 | 70 | 21 | 15.6 [9.9–23.7] |
| Culex pipiens | CO2 | 308 | 30 | 1 | 3.3 [0.2–15.9] |
| Culex restuans | CO2 | 1 | 1 | 0 | 0.0 [0.0–793.4] |
| Culex erythrothorax | CO2 | 1 | 1 | 0 | 0.0 [0.0–793.4] |
| Culiseta inornata | CO2 | 9 | 4 | 0 | 0.0 [0.0–220.1] |
| Aedes vexans | CO2 | 2,751 | 86 | 1 | 0.4 [0.02–1.75] |
| Aedes dorsalis | CO2 | 390 | 21 | 0 | 0.0 [0.0–8.7] |
| Aedes melanimon | CO2 | 182 | 9 | 0 | 0.0 [0.0–16.3] |
| Aedes trivittatus | CO2 | 205 | 19 | 0 | 0.0 [0.0–14.4] |
| Aedes nigromaculis | CO2 | 12 | 1 | 0 | 0.0 [0.0–123.2] |
| Aedes hendersoni | CO2 | 16 | 3 | 0 | 0.0 [0.0–124.1] |
Resting traps were used to monitor density of vertebrate-vector contacts over time at each site. Of 744 freshly engorged Culex mosquitoes, 99.2% were collected as resting mosquitoes and 0.8% were collected as host-seeking mosquitoes. However, many of the engorged mosquitoes from resting collections were derived from supplemental aspiration of resting mosquitoes. We relied on the density of freshly engorged mosquitoes (containing more than half undigested blood meal, indicating a recent vertebrate contact, i.e., less than two days old) in the CDC resting traps to indicate intensity of vertebrate-vector interaction at the four sites. Overall, the cumulative density of vector-vertebrate contacts was significantly greater at roost sites compared to control sites: 23-fold greater for Cx. pipiens, and five-fold greater for Cx. tarsalis (Table 2). At both roost sites, Cx. tarsalis-vertebrate contact density was high (above two contacts per trap-night) early in the study but subsequently dropped to below one contact per trap night from study weeks 4–7 at Roost Site A (Figure 2A) and weeks 4–8 at Roost Site B (Figure 2C). Culex pipiens followed a similar pattern at Roost Site A, starting off with a high contact density during mid-July but then dropping below one contact per trap night for study weeks 3–8 (Figure 2A). At Roost Site B, density of vertebrate contacts for Cx. pipiens dropped below one at week 5, staying low except for a spike up to two contacts per trap-night at week 7 (Figure 2C). At Control Sites A and B, vertebrate contacts were essentially undetectable for both Cx. pipiens and Cx. tarsalis except for the month of July at Control Site A when Cx. tarsalis vertebrate contact density reached 1.5 per trap-night in early July and slowly declined throughout the month (Figures 2B and 2D). The density of engorged mosquitoes encountered at a site was moderately correlated with the counts of roosting American robin for Cx. pipiens (r=0.574, r2=0.329, p=0.003) but not for Cx. tarsalis (r=0.089, r2=0.008, p=0.7) (Figures 3A and 3B). Correlation was weaker when plotting counts of birds of all species (Cx. pipiens: r=0.418, r2=0.1745, p=0.04; Cx. tarsalis: r=0.077, r2=0.006, p=0.7, data not shown). However, when correlation was evaluated for all birds except American robin, correlations became negative (Cx. pipiens: r=−0.425, r2=0.1805, p=0.04; Cx. tarsalis: r=−0.039, r2=0.0015, p=0.9), implying that Culex mosquitoes (especially Cx. pipiens) were attracted to the robins for blood feeding and repelled (or dispatched) by other birds (Figures 3C and 3D).
Table 2.
Cumulative density of vector-vertebrate contacts for Culex pipiens and Cx. tarsalis at each study site, as determined by the mean number of engorged mosquitoes with less than half of the blood meal digested per resting trap night, July-August, 2013.
| Cx. pipiens | Cx. tarsalis | ||||
|---|---|---|---|---|---|
| Study Site | No. resting trap-nights | No. resting engorged mosquitoes collected | Density of resting engorged mosquitoes (No. per trap night) | No. resting engorged mosquitoes collected | Density of resting engorged mosquitoes (No. per trap night) |
| Roost Site A (Wellington) | 66 | 82 | 1.2 | 101 | 1.5 |
| Control Site A (Wellington) | 53 | 6 | 0.1 | 21 | 0.4 |
| Roost Site B (Loveland) | 56 | 72 | 1.3 | 33 | 0.6 |
| Control Site B | 57 | 0 | 0.0 | 3 | 0.05 |
Figure 2.
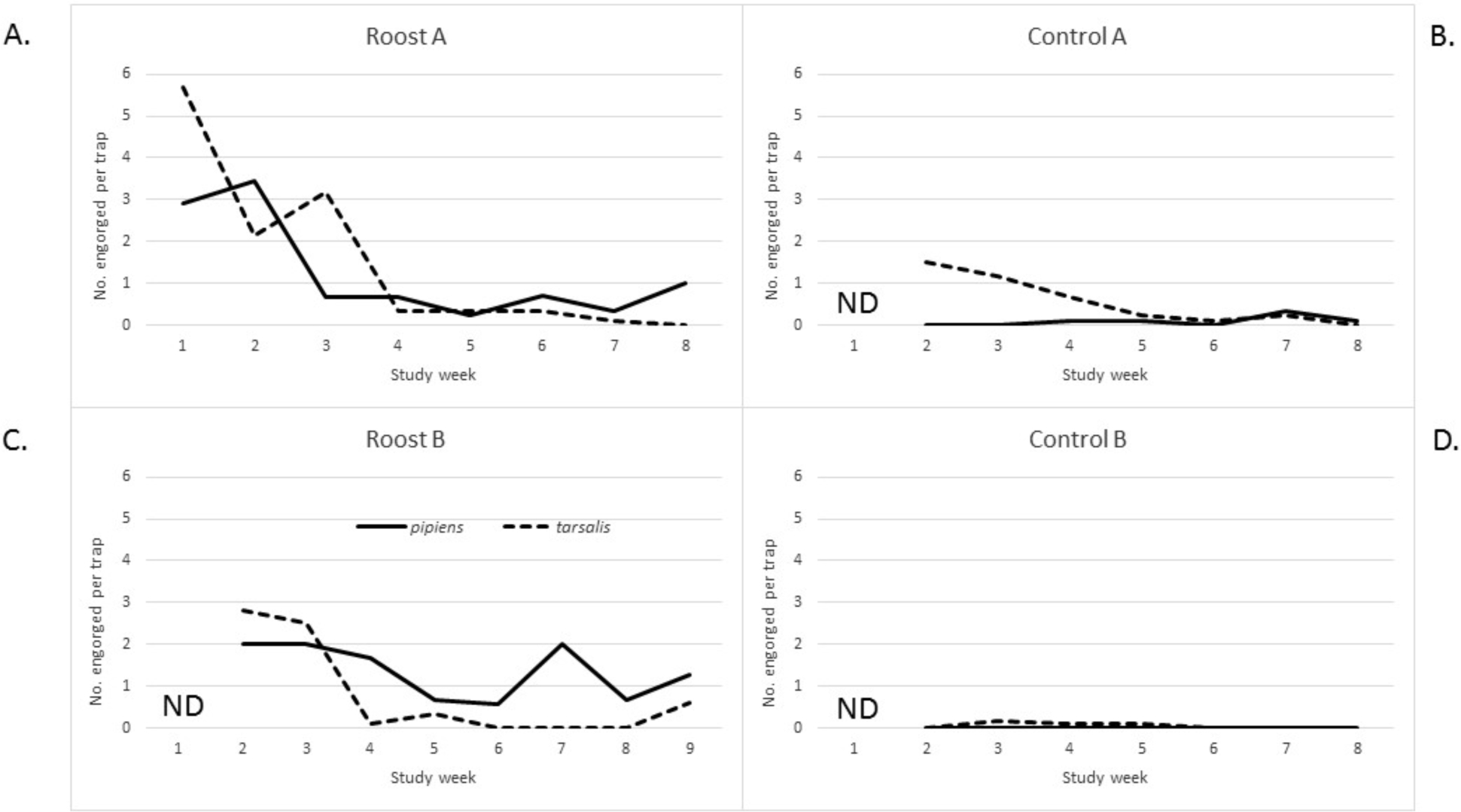
Density of vector-vertebrate contacts over time at each of the four study sites, as determined by the number of freshly engorged Culex mosquitoes (blood meal less than half digested) collected per resting-trap-night, July-August, 2013. A, Roost Site A. B, Control Site A. C, Roost Site B. D, Control Site B. Cx. pipiens represented by solid line; Cx. tarsalis represented by dashed line.
Figure 3.
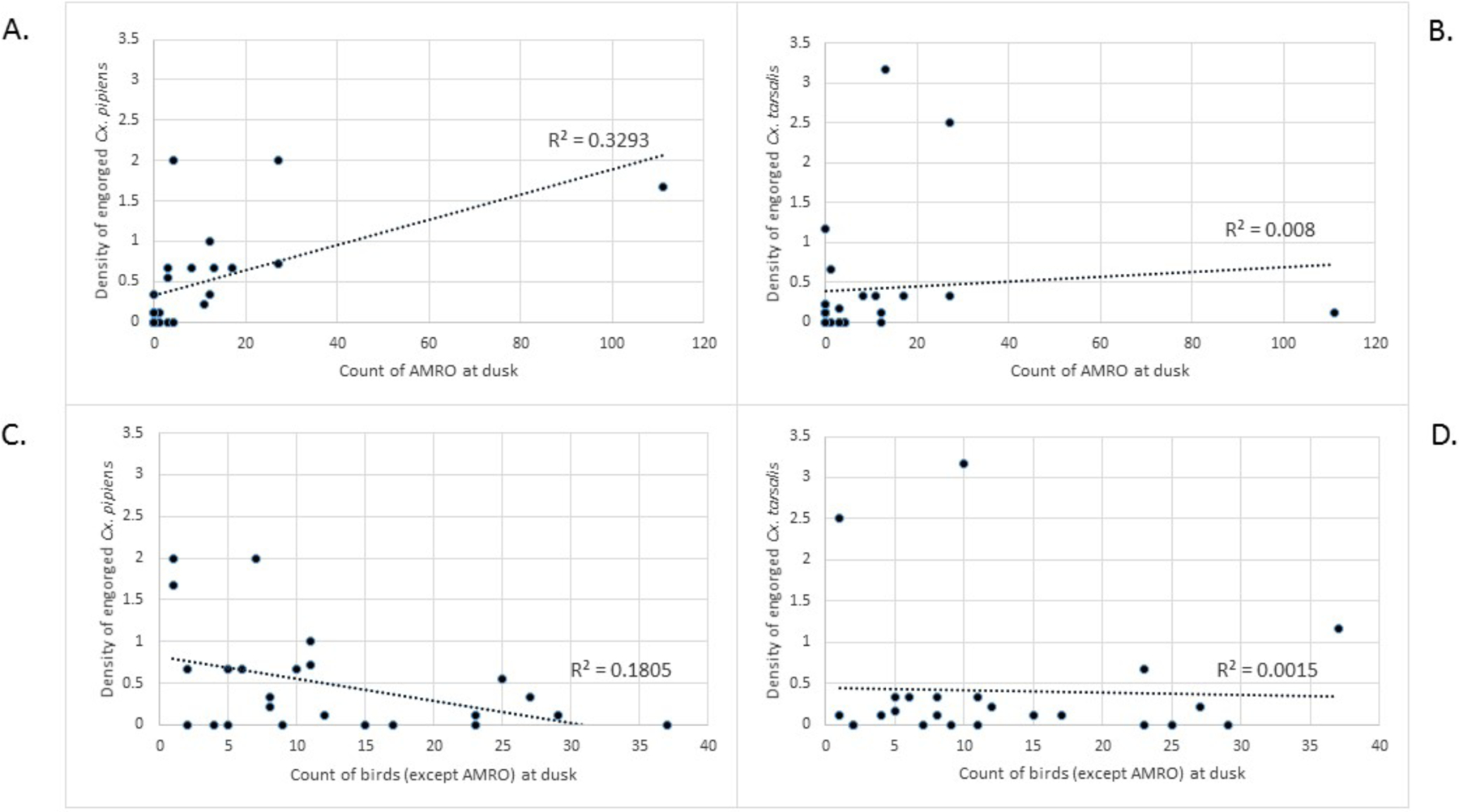
Scatter plot of vector-vertebrate contact density vs American robin (AMRO) count for Cx. pipiens (A), or Cx. tarsalis (B), and vs all other birds for Cx. pipiens (C) or Cx. tarsalis (D).
Blood meal identification
Most engorged mosquitoes came from resting collections (from resting traps and aspiration) at Roost Site A (N=70 Cx. pipiens; N=130 Cx. tarsalis) and Roost Site B (N=400 Cx. pipiens; N=116 Cx. tarsalis). Most vector-vertebrate contacts for Cx. pipiens and Cx. tarsalis involved the American robin (84% and 91%, respectively, at Roost Site A; 94% and 96%, respectively, at Roost Site B). All other species identified as vertebrate contacts for these vectors represented <3% of all contacts, except for house finch at Roost Site A, for which seven (10.0%) of the contacts with Cx. pipiens were attributed to this passerine species. House finch also accounted for three (2.3%) of the contacts for Cx. tarsalis at Roost Site A, and three (0.8%) of the contacts for Cx. pipiens only, at Roost Site B. Other avian species identified among the roost site blood meals included house sparrow (two [2.9%] of the contacts for Cx. pipiens and four [3.1%] for Cx. tarsalis at Roost Site A), common grackle (one [0.8%] of the contacts for Cx. tarsalis only, at Roost Site A), Eurasian collared-dove (three [0.8 %] of the contacts for Cx. pipiens only, at Roost Site B, mourning dove (two [0.5%] of the contacts for Cx. pipiens and one [0.9%] of the contacts for Cx. tarsalis, at Roost Site B only), mallard (Anas platyrhynchos; one [0.9%] of the contacts for Cx. tarsalis, at Roost Site B only), and finally black-capped chickadee and song sparrow (Melospiza melodia; one [0.2%] of the contacts each for Cx. pipiens, at Roost Site B only). The only vector-mammal contact identified at the roost sites was a Cx. pipiens blood meal from a horse (Caballus equinus) at Roost Site A.
Other blood meals identified at roost sites from non-vectors include, from Roost Site A: 2 cattle (Bos taurus), one horse and one house finch from Aedes melanimon; one cattle, one desert cottontail (Sylvilagus audubonii) from Ae. vexans; two cattle, two horses from Culiseta inornata; and from Roost Site B: four American robins, one red fox (Vulpes vulpes), one domestic cat (Felis cattus) from Ae. trivittatus; two humans (Homo sapiens) and one red fox from Aedes vexans.
Blood meals identified from control sites include, from Control Site A: one house sparrow from Ae. melanimon; one cattle from Ae. trivittatus; two cattle, one horse, one barn swallow, one house finch, one house sparrow from Ae. vexans; one house finch from Cs. inornata; two cattle, one house finch, one mourning dove from Cx. pipiens; four cattle, four mourning doves, three Eurasian collared-dove, one house finch, one house sparrow from Cx. tarsalis; and from Control Site B: one American robin, one domestic chicken (Gallus gallus), one horse from Cx. tarsalis; one American robin from Aedes vexans.
Antibody detection
All freshly engorged mosquitoes were tested for presence of WNV-reTableactive and SLEV-reactive antibodies. All samples were negative for SLEV antibodies. Of 13 vertebrates identified among the 738 blood meals tested, WNV-reactive antibodies were detected in blood meals from just seven host species, including American robin, house finch, mourning dove, Eurasian collared-dove, mallard, fox, and horse (Table 3). House sparrow, common grackle, black-capped chickadee, cow, human, and cat were all represented by small sample sizes (ranging from one to six) and were all negative for antibodies. WNV antibody prevalence curves by week were prepared for the American robin by selecting results from mosquitoes that had fed upon blood of the American robin. These data were further divided into a curve for mosquitoes collected at Roost Site A and those collected at Roost Site B (Figure 4). At Roost Site A, the immunity encountered by vectors feeding on robins appeared to fluctuate wildly between 0 and 67% through the eight-week sampling period. At Roost Site B, the immunity encountered increased over time from 8.5% to 64% and then decreased to 33%. The sample size of mosquitoes was lower at Roost Site A (N=177) compared to Roost Site B (N=491), and consequently the precision of the data is lower for Roost Site A.
Table 3.
Cumulative vertebrate seroprevalence for West Nile antibody, by species, as determined by microsphere immunoassay of mosquito blood meals in Northern Colorado, July-August, 2013.
| Vertebrate species | No. tested | No. WN Ab positive | % positive (95% CI) |
|---|---|---|---|
| American robin | 668 | 175 | 26.2 (23.0, 29.7) |
| House Finch | 13 | 3 | 23.1 (8.2, 50.3) |
| House Sparrow | 6 | 0 | 0.0 (0.0, 39.0) |
| Common Grackle | 1 | 0 | 0.0 (0.0, 79.4) |
| Black-capped Chickadee | 1 | 0 | 0.0 (0.0, 79.4) |
| Mallard | 1 | 1 | 100.0 (20.6, 100.0) |
| Eurasian Collared-Dove | 3 | 1 | 33.0 (6.2, 79.2) |
| Mourning Dove | 3 | 2 | 66.7 (20.8, 93.8) |
| Horse | 4 | 2 | 50.0(15.0,85.0) |
| Cow | 3 | 0 | 0.0 (0.0, 56.2) |
| Human | 2 | 0 | 0.0 (0.0, 65.8) |
| Fox | 2 | 1 | 50.0 (9.4, 90.6) |
| Cat | 1 | 0 | 0.0 (0.0, 79.4) |
| Unidentified | 30 | 5 | 16.7 (7.3, 33.6) |
Figure 4.
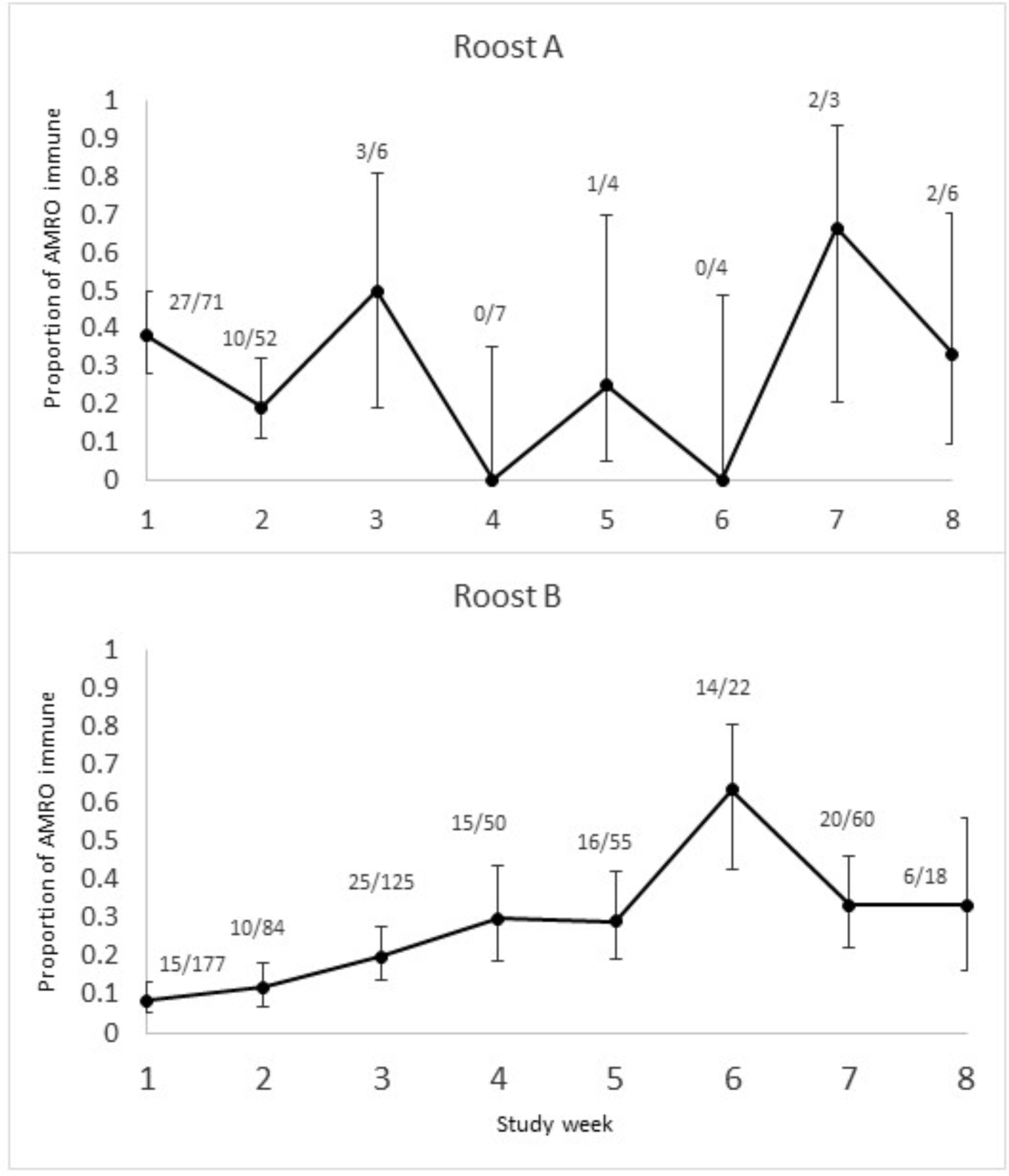
Prevalence of West Nile virus-reactive antibodies in abdomens of engorged Culex mosquitoes that had fed on blood of American robin, by week, in A. Roost Site A in Wellington, CO, and B. Roost Site B in Loveland, CO, July-August, 2013. Error bars represent 95% confidence intervals derived by the Wilson score method for binomial proportions.
Virus detection
WNV infections were detected in 42 pools of adult female mosquitoes collected from all four study sites. WNV was detected in 27 of 371 pools of Cx. tarsalis (N=2,547) and in 14 of 556 pools of Cx. pipiens (N=1,459) (Table 1). A single RT-PCR positive pool was detected among 97 pools of Aedes vexans (N=2842). Infection rates were derived for both Culex vector species by study site and collection method as a surrogate for behavior (host-seeking vs resting; 16 categories in total; Table 4). This analysis revealed that the highest detected rate (cumulative across the eight-week study) was among host-seeking Cx. tarsalis at Control Site B (25.7 per thousand), followed by host-seeking Cx. tarsalis at Control Site A (14.7 per thousand), resting Cx. pipiens at Roost Site B (14.2 per thousand), resting Cx. tarsalis at Roost Site B (11.3 per thousand), host-seeking Cx. pipiens at Control Site B (8.2 per thousand), resting Cx. tarsalis at Roost Site A (7.2 per thousand), host-seeking Cx. tarsalis at Roost Site B (4.9 per thousand), with no detected infections for the remaining nine categories. However, due to small sample sizes (ranging from N=6 for resting Cx. pipiens at Control Site B, to N=975 for resting Cx. pipiens at Roost Site B), none of the estimated infection rates were significantly different from any other among the 16 categories.
Table 4.
Cumulative WNV infection rates in adult female Cx. pipiens and Cx. tarsalis, by site and by behavior (resting vs host-seeking), July-August, 2013. Cohorts with no virus-positive pools used uncorrected methods of estimation for the point estimate and confidence interval. Units for the infection rate point estimates and confidence intervals are per 1,000 mosquitoes.
| Study Site | Cx. pipiens | Cx. tarsalis | ||
|---|---|---|---|---|
| Resting | Host-seeking | Resting | Host-seeking | |
| (CI; N) | (CI; N) | (CI; N)) | (CI; N)) | |
| Roost A | 0.0 | 0.0 | 7.2 | 0.0 |
| (Wellington) | (0.0–24.9; 140) | (0.0–54.2; 51) | (1.3–23.1; 280) | (0.00–30.2; 97) |
| Control A | 0.0 | 0.0 | 0.0 | 14.7 |
| (Wellington) | (0.0–116.0; 26) | (0.0–27.2; 105) | (0.0–22.7; 142) | (7.6–26.6; 824) |
| Roost B | 14.2 | 0.0 | 11.3 | 4.9 |
| (Loveland) | (8.0–23.5; 975) | (0.0–88.1; 31) | (3.8–26.5; 351) | (0.3–24.4; 199) |
| Control B | 0.0 | 8.2 | 0.0 | 25.7 |
| (Fort Collins) | (0.0–341.7; 6) | (0.49–41.0; 121) | (0.0–97.9; 26) | (13.1–49.0; 599) |
Collections of resting Cx. pipiens were adequately robust at Roost Site B to permit an assessment of the WNV infection rate by week across the eight weeks of the study (Figure 5). WNV was first detected in these mosquitoes during the third week of the study and peaked during study week 6 when the infection rate reached 37.2 per 1,000 mosquitoes (maximum likelihood estimate, 95% C.I. 9.3 – 108.4).
Figure 5.
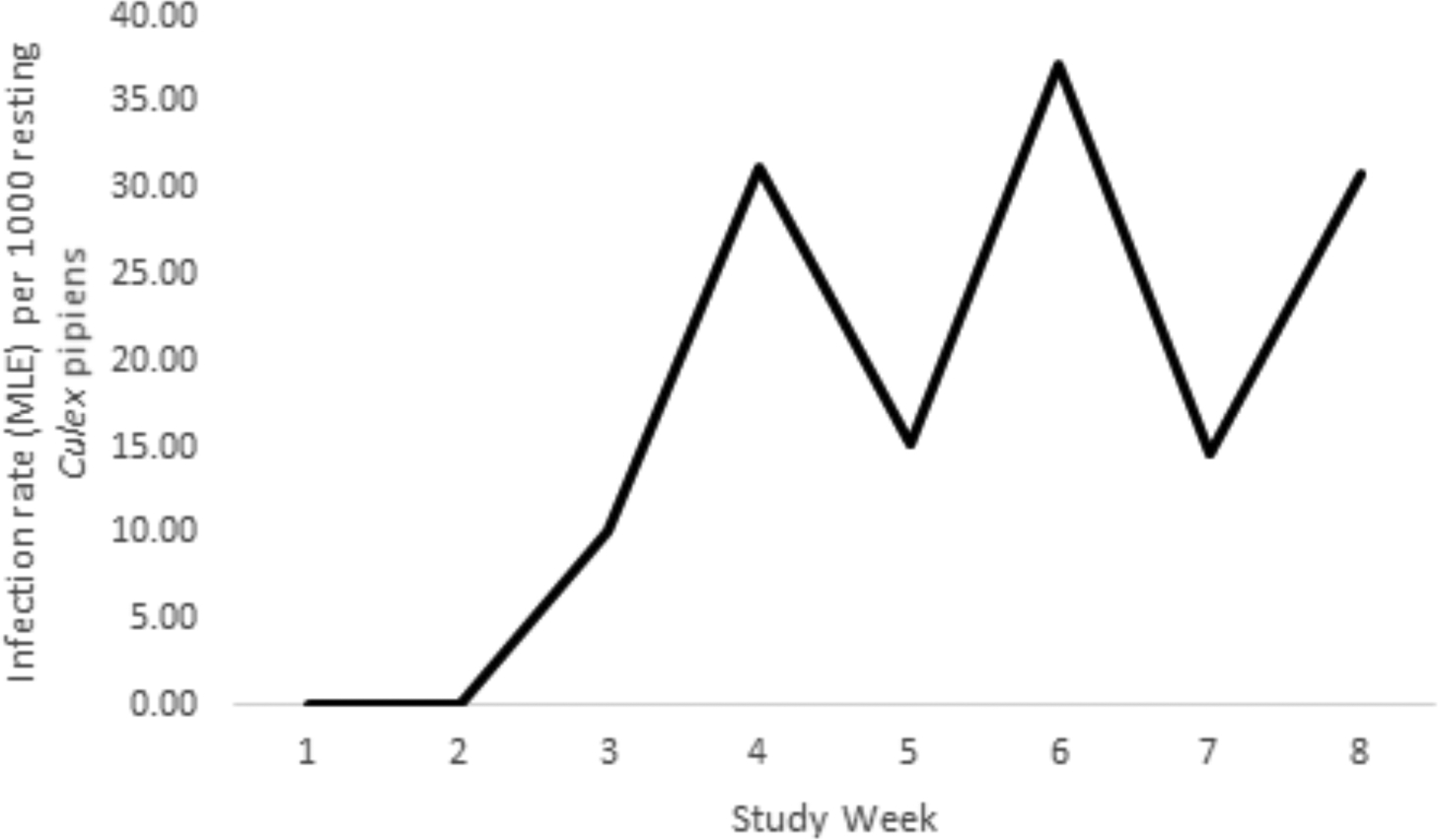
Infection rate estimates by week during July-August, 2013, among resting Culex pipiens mosquitoes at Roost Site B. The number of mosquitoes sampled per week ranged between 70 and 201.
Among the engorged mosquitoes tested individually, 11 tested positive for WNV based on detection of viral RNA in the abdomens, including one from Roost Site A, ten from Roost Site B, and none from Control Sites A and B. For each of the positive abdomens, we determined the corresponding infection status of legs from the same mosquito (Table 5). Just one of these mosquitoes had a disseminated infection (legs tested positive for WNV RNA). This presumably infectious Cx. tarsalis had engorged on robin blood at Roost Site B during study week 2, but the contact was not an example of vector-to-vertebrate transmission because the robin blood contained WNV-specific antibodies, indicating that this robin was already immune. At Roost Site A, the one infected engorged mosquito was a non-infectious Cx. tarsalis that had fed on a robin during study week 5. The robin blood was antibody-free, and if due to viremic blood, it would have been above the threshold level for infectiousness. Therefore, this vector-vertebrate contact was interpreted as a possible bird-to-vector transmission event. At Roost Site B, nine other similar contacts (gut-limited infections interpreted as possibly derived from viremic blood) yielded five possible bird-to-vector transmission events, four involving American robin with three Cx. pipiens (study weeks 3, 5 and 7, respectively) and one Cx. tarsalis (study week 2), and one involving mourning dove with Cx. pipiens (study week 5). The four unsuccessful transmission contacts involved the American robin with sub-infectious viremia (Ct>35.0) (Table 4). American robin-to-Culex transmission of WNV may be regulated by immunity levels in the robin population, leading to WNV amplification when immunity is low and WNV suppression when immunity is high (Figure 6).
Table 5.
Disseminated (leg) infection status for engorged Culex mosquitoes with WNV-infected abdomens, collected over eight weeks during July-August, 2013, at Roost Site B. If the legs are negative, it is assumed that the vertebrate host blood is the source of the infection and that the vector-host contact represents host-to-vector transmission if the viremia is infectious (Ct<35). If legs are positive, the vector was likely infectious and the contact represents vector-to-host transmission. Presence of antibody negates transmission. Ct, mean threshold cycle (<40=positive); Ab, antibody; MIA, microsphere immunoassay; tx, transmission; AMRO, American robin; MODO, mourning dove.
| Week | Vector | Abdomen Ct | Legs | Host | Ab (MIA) | Bird-to-vector tx | Vector-to-bird tx | Site |
|---|---|---|---|---|---|---|---|---|
| 1 | -- | -- | -- | -- | -- | -- | -- | |
| 2 | tarsalis | 37.0 | POS | AMRO | POS | No | No | Roost B |
| tarsalis | 25.1 | Neg | AMRO | Neg | YES | No | Roost B | |
| 3 | tarsalis | 38.0 | Neg | AMRO | Neg | No | No | Roost B |
| pipiens | 26.9 | Neg | AMRO | Neg | YES | No | Roost B | |
| 4 | -- | -- | -- | -- | -- | -- | -- | |
| 5 | tarsalis | 27.1 | Neg | AMRO | Neg | YES | No | Roost A |
| pipiens | 34.2 | Neg | AMRO | Neg | YES | No | Roost B | |
| pipiens | 34.3 | Neg | MODO | Neg | YES | No | Roost B | |
| 6 | -- | -- | -- | -- | -- | -- | -- | |
| 7 | pipiens | 36.6 | Neg | AMRO | POS | No | No | Roost B |
| pipiens | 32.6 | Neg | AMRO | Neg | YES | No | Roost B | |
| pipiens | 36.7 | Neg | AMRO | Neg | No | No | Roost B | |
| 8 | pipiens | 38.1 | Neg | AMRO | Neg | No | No | Roost B |
Figure 6.
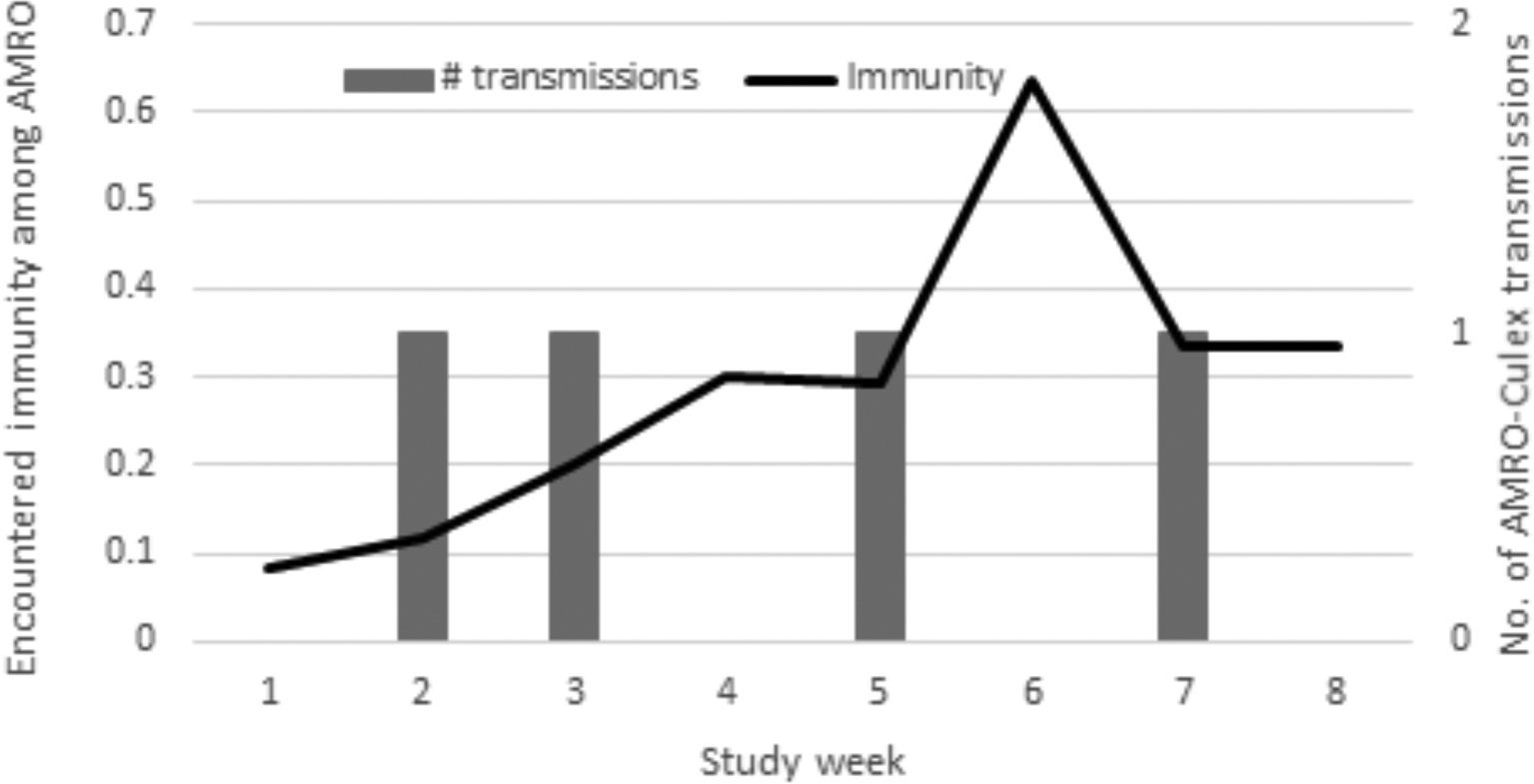
Temporal relationship between the number of putative American robin to vector transmissions detected at Roost Site B and the proportion of immune robins encountered by feeding mosquitoes. Early in July, encountered immunity is low (<20%) which allows for transmission events to occur (amplification). Transmission events detected during weeks two and three (bars) result in amplification and increasing immunity. Transmission is suppressed when immunity reaches approximately 30%, and consequently detected transmission slows, with just one event detected during weeks four and five. No transmission event is detected when immunity spikes to more than 60% in week six but continues slowly with one additional detected event in week seven when immunity returns to 30%. Immunity levels are increased by virus amplification and decreased by virus suppression coupled with either immigration of susceptible birds (e.g., influx of hatch-year birds due to reproduction) or emigration of immune birds (e.g., migration or roost site abandonment or fatal infections). AMRO = American robin.
Five isolates of Flanders virus (Rhabdoviridae) were cultured from pools of non-engorged Cx. tarsalis at Roost Site A (two pools), Control Site A (one pool), and Control Site B (two pools), but none from pools of non-engorged Cx. pipiens. Engorged mosquitoes were not tested for Flanders virus.
DISCUSSION
Outbreak investigations of WNV activity throughout the U.S.A. have recognized the focal nature of transmission of this arthropod-borne virus (Komar et al. 2005, Godsey et al. 2005, Kilpatrick et al. 2006, Hamer et al. 2011). However, the ecological basis for this focality is not well understood. This ignorance presents an important barrier to effective prediction, control, and prevention of human WNV infections. We explored the hypothesis that communal roosts of passerine birds provide an ecological context for both amplification and suppression of WNV transmission, depending on the immunity levels in the blood of the roosting birds encountered by hematophagous vectors. Field studies were carried out in Larimer County, CO, in 2013, a year when Colorado reported 90 West Nile neuroinvasive disease cases with seven deaths, and Larimer County was the most affected county with 28% of all Colorado cases (ArboNet data, CDC).
This study sought evidence that WNV transmission either increased or decreased around two communal robin roosts in Larimer County during the 2013 WNV transmission season. We demonstrated that the vectors of WNV, Cx. pipiens and Cx tarsalis, were attracted to these communal roost sites for the purpose of blood-feeding. Each blood-engorged vector captured in CDC resting traps represented a vector-vertebrate contact. The density of vector-vertebrate contacts was 23-fold greater at the two communal robin roost sites compared with two control sites for Cx. pipiens, and five-fold greater for Cx. tarsalis. The corresponding measurements taken in 2010 in suburbs of Phoenix, AZ (using sparrow and blackbird communal roosts) was 33-fold for Cx. pipiens quinquefasciatus and three-fold for Cx. tarsalis, indicating that the attraction of Culex mosquitoes to communal bird roosts for blood-feeding is not a phenomenon limited to our Colorado field sites (Komar et al. 2013).
These vector-vertebrate contacts represented blood meals derived mostly from the communally roosting robins at the sites. The identification of the vertebrate source of these blood meals confirmed that robins provided 84–96% of the blood meals between the two vector species, while just 73% of the birds counted at the two communal roost sites were robins. Thus, the proportion of blood meals taken from robins was more than expected based on the relative abundance of robins compared to all birds present. This demonstrates a preference for robin blood by these mosquitoes, a result observed previously in Colorado (Kent et al. 2009). This preference was further corroborated by noting a positive correlation between density of Cx. pipiens blood meals and the number of robins present at a site, and a negative correlation between this density and the number of birds of other species.
We hoped to detect a difference in WNV infection rates among mosquitoes at the four study sites, but small sample sizes prevented this. Fine geographic scale of the study sites, rather than insufficient collection effort, was primarily responsible for this outcome. A field site was comprised of just a few dozen trees. However, by examining the infections of individual engorged mosquitoes and identifying the vertebrate source of the imbibed blood, we were able to deduce that transmission events among vector-vertebrate contacts (i.e., infectious contacts between vertebrate hosts and mosquito vectors) had occurred from five viremic robins and one viremic mourning dove to Culex vectors at the communal roost sites, compared to zero transmission events detected at control sites. These deductions operate under the assumption that an engorged mosquito with a gut-limited infection acquired its infection from the current blood meal. However, a small percentage of these mosquitoes may be old enough to have had a previous vertebrate encounter that resulted in a non-disseminated gut infection. In vector competence experiments, Cx. pipiens often developed non-disseminated gut infections (Turell et al. 2001).
Failure to detect vector-to-vertebrate transmission events through our examination of infections in engorged mosquitoes was no surprise. This is because of the daily survival rate of mosquitoes, estimated at 90% per day (Jones et al. 2012). The 10% daily mortality rate of mosquitoes implies that roughly ten infected mosquitoes are required for one of them to survive the extrinsic incubation period and infect a new vertebrate host. In fact, we did detect one infectious vector (out of a total of 11 engorged mosquitoes that tested positive for WNV). However, this one infectious vector had fed on an immune robin, therefore resulting in a dead end for its viral load. This observation illustrates the regulatory effect of bird immunity among the population of amplifier hosts. The more immune amplifiers present, the more infected vectors are needed to successfully amplify the infection. In this way, immunity suppresses transmission and leads to herd immunity.
While we failed to detect evidence of vector-to-vertebrate transmission among the mosquitoes, we were able to indirectly observe this type of transmission by examining the change in the proportion of immune American robins encountered by mosquitoes during the course of the transmission season. At Roost Site A, the sample size of communally roosting robins was low during 2013 and perhaps consequently the proportion of immune robins encountered by mosquitoes was observed to fluctuate wildly. This fluctuation may be due to low precision of the observed data (a statistical phenomenon), or to rapidly alternating amplification and suppression (a biological phenomenon). When the ratio of vector to vertebrate amplifier host is high, amplification occurs very quickly and rapidly leads to a situation of herd immunity and suppression of transmission (Janousek et al. 2014). However, given a small population with a high rate of turnover, the departure of just a few immune hosts or the arrival of a small number of susceptible hosts can have dramatic effects on this ratio and the potential for new transmission events. At Roost Site B, the larger populations of both vectors and vertebrate hosts led to a more stable situation, with an observed bell curve of encountered immunity. The initial increase in robin immunity encountered by vectors at the start of the peak transmission season would be due to amplification. However, this increase in encountered immunity results in the suppression of transmission. With reduced transmission, the population turnover (arrival of new susceptible birds to the communal roost) results in a decrease in encountered immunity.
The observation that transmission intensity fluctuates over time on a scale of days/weeks may explain why some previous studies have failed to detect a positive association between communal roosting and mosquito infection rates (Reisen et al. 2005, Diuk-Wasser et al. 2010, Komar et al. 2015). The cited studies used cumulative mosquito infection rate as the dependent variable, which means that they measured the net effect of weekly transmission measures. To illustrate this point, consider a communal roost that amplifies for two weeks and then suppresses for six weeks. The heavier collection of mosquitoes during suppression weeks will bias the measured cumulative infection rate downward, thereby masking the amplification effect. To alleviate this problem, we suggest that mosquito infection rates are inappropriate for measuring amplification and suppression. Rather, the change in the rate over time is the important measurement, and the number of infectious vertebrate-vector contacts. We only detected six of the latter at communal roosts, but this was compared to zero at control sites, despite the detection of elevated mosquito infection rates at these control sites. This assessment of transmission events among vector-vertebrate contacts as a means of evaluating a transmission focus is novel. It is permitted by the relatively new technologies enabling detection of infection and immunity in mosquito blood meals.
The detection of high infection rates in host-seeking mosquitoes at all four study sites (test sites and controls) can be explained by mosquitoes moving through space from feeding sites, such as the communal roost sites, to appropriate breeding habitats and vice versa. Capturing these mosquitoes in baited CO2 traps indicates that host-seeking mosquitoes will take advantage of blood sources wherever they may be found, and therefore risk of WNV transmission to people exists wherever people and host-seeking Culex mosquitoes coexist. On the other hand, this risk is much lower (by multiple orders of magnitude) relative to that of American robins and certain other birds. Indeed, our related study of WNV transmission risk around communal bird roosts in metropolitan Phoenix observed a lower risk of human WNV infection near great-tailed grackle roosts. This lower risk was probably multifactorial, including a combination of explanations from grackles eating mosquitoes, transmission suppression due to herd immunity, and zooprophylaxis, among others (Komar et al 2015).
Immunity levels encountered by feeding Culex vectors regulate virus amplification and suppression (Kwan et al. 2012). We show that many of these Culex feed at communal bird roost sites. Therefore, the roost site provides an opportunity to monitor these trends and/or manipulate the regulation in a manner that reduces risk of transmission to humans. However, interpreting surveillance data collected from mosquitoes and/or birds at communal roost sites can be complicated, largely due to time delays both in the laboratory and in nature. Laboratory-based surveillance of WNV infection rates in mosquitoes will experience unavoidable delays in reporting virus detection results. Delays are caused by the time required for sorting and identifying mosquitoes (a human resource-dependent delay that depends on the availability of entomologists for working with mosquitoes). Once identified and sorted into pools, the pooled mosquitoes are homogenized in batches, then tested for viruses either by cell culture (which requires about three days of incubation prior to detection of arbovirus-induced plaques) and/or by molecular detection systems (such as real-time RT-PCR), which also takes several days to generate a confirmed result. Similarly, laboratory-based detection of avian antibodies to WNV, whether from individual mosquito abdomens or avian serum samples, suffers from a variety of confounders. First, laboratory procedures require several days for processing samples, running the test, and eventually reporting the result. Second, detectable antibodies imply a minimum delay of four days after infection during which the vertebrate host begins the physiological process of generating WNV-specific antibodies (Komar et al. 2003). Add on several more days for organizing a field-based intervention, and one is now several weeks later in the season than the transmission event that served as trigger for the intervention. If the intervention response targets a communal bird roost site, great care must be taken to avoid converting an arbovirus suppressive location into a potential for additional arbovirus amplification. This could happen, for example, if the intervention inadvertently causes numerous birds to emigrate from the roost site. In the few weeks that had passed, the immunity may have surged, converting the site into a suppressive site. The departure of immune birds reduces herd immunity. If the departing birds are replaced by new susceptible arrivals, amplification once again would be favored.
This study adds to a growing body of data and published literature that implicates post-breeding communal roosts of passerine birds (e.g., American robin) as vital to the environmental persistence of WNV. The vector control and public health communities must investigate methods to harness this relationship to the benefit of public health. However, with the complexity of virus–vector–vertebrate–environment–climate interactions, such a mandate presents a significant challenge. Ideally, this focal basis of amplification could be marshalled for early detection of WNV transmission activity. When surveillance indicators signal an impending outbreak, swift and refined interventions could target these transmission foci. However, targeting control efforts to a site that has become suppressive to transmission is counterproductive and potentially could even have the opposite effect. More efforts are needed to define these approaches and demonstrate their utility for control and prevention of WNV disease.
Acknowledgments
This study was funded by the Centers for Disease Control and Prevention. Programmatic support was provided by Harry Savage and Roger Nasci. Jason Velez and others in the cell culture facility provided technical support for cell culture-based assays. Janae Stovall and Karen Boroughs operated the DNA sequence analyzer. Brad Biggerstaff advised on biostatistics.
REFERENCES CITED
- Basile AJ, Biggerstaff BJ, Kosoy O, Junna A, Panella NA, Powers AM, Stark LM, and Nemeth N. 2010. Removal of species constraints in antibody detection. Clin. Vaccine Immunol 17: 56–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darsie RF and Ward RA. 2005. Identification and Geographical Distribution of the Mosquitoes of North America, North of Mexico. Florida: University of Florida Press. 398 pp. [Google Scholar]
- Diuk-Wasser MA, Molaei G, Simpson JE, Folsom-O’Keefe CM, Armstrong PM, and Andreadis TG. 2010. Avian communal roosts as amplification foci for West Nile virus in urban areas in northeastern United States. Am. J. Trop. Med. Hyg 82: 337–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fauver JR, Pecher L, Schurich JA, Bolling BG, Calhoon M, Grubaugh ND, Burkhalter KL, Eisen L, Andre BG, Nasci RS, LeBailly A, Ebel GD, and Moore CG. 2016. Temporal and spatial variability of entomological risk indices for West Nile virus infection in northern Colorado: 2006–2013. J. Med. Entomol 53: 425–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godsey MS Jr., Blackmore M, Panella NA, Burkhalter K, Gottfried K, Halsey L, Rutledge R, Langevin S, Gates R, Lamonte K, Lambert A, Lanciotti R, Loyless T, Stark L, Olivieri R, Conti L, and Komar N. 2005. West Nile virus epizootiology in the southeastern United States, 2001. Vector-Borne Zoonotic Dis. 5: 82–89. [DOI] [PubMed] [Google Scholar]
- Hamer GL, Chaves LF, Anderson TK, Kitron UD, Brawn JD, Ruiz MO, Loss SR, Walker ED, and Goldberg TL. 2011. Fine-scale variation in vector host use and force of infection drive localized patterns of West Nile virus transmission. PLoS One 6: e23767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova NV, Zemlak TS, Hanner RH, and Hebert PDN. 2007. Universal primer cocktails for fish DNA barcoding. Mol. Ecol. Notes 7: 544–548. [Google Scholar]
- Janousek WM, Marra PP, and Kilpatrick AM. 2014. Avian roosting behavior influences vector-host interactions for West Nile virus hosts. Parasit. Vectors 7: 399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones CE, Lounibos LP, Marra PP, and Kilpatrick AM. 2012. Rainfall influences survival of Culex pipiens (Diptera: Culicidae) in a residential neighborhood in the mid-Atlantic United States. J. Med. Entomol 49: 467–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent R, Juliusson L, Weissmann M, Evans S, and Komar N. 2009. Seasonal blood-feeding behavior of Culex tarsalis (Diptera: CulicidAe) in Weld County, Colorado, 2007. J. Med. Entomol 46: 380–390. [DOI] [PubMed] [Google Scholar]
- Kilpatrick AM, Daszak P, Jones MJ, Marra PP, and Kramer LD. 2006. Host heterogeneity dominates West Nile virus transmission. Proc. Biol. Sci 273: 2327–2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilpatrick AM 2011. Globalization, land use, and the invasion of West Nile virus. Science 334: 323–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komar N, Pollack RJ, and Spielman A. 1995. A nestable fiber pot for sampling resting mosquitoes. J. Am. Mosq. Contr. Assoc 11: 463–467. [PubMed] [Google Scholar]
- Komar N, Langevin S, Hinten S, Nemeth N, Edwards E, Hettler D, Davis B, Bowen R, and Bunning M. 2003. Experimental infection of North American birds with the New York 1999 strain of West Nile virus. Emerg. Infect. Dis 9: 311–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komar N, Panella NA, Langevin SA, Brault AC, Amador M, Edwards E, and Owen JC. 2005. Avian hosts for West Nile virus in St. Tammany Parish, Louisiana 2002. Am. J. Trop. Med. Hyg 73: 1031–1037. [PubMed] [Google Scholar]
- Komar N, Colborn JM, Horiuchi K, Delorey M, Biggerstaff B, Damian D, Smith K, and Townsend J. 2015. Reduced West Nile virus transmission around communal roosts of great-tailed grackle (Quiscalus mexicanus). Ecohealth 12: 144–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komar N, Panella NA, Young GR, Brault AC, and Levy CE. 2013. Avian hosts of West Nile virus in Arizona. Am. J. Trop. Med. Hyg 89: 474–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komar N, Panella NA, Young GR, and Basile AJ. 2015. Methods for detection of West Nile virus antibodies in mosquito blood meals. J. Am. Mosq. Contr. Assoc 31: 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs BL, Anderson TK, Goldberg TL, Hamer GL, Kitron UD, Newman CM, Ruiz MO, Walker ED, and Brawn JD. 2014. Host group formation decreases exposure to vector-borne disease: a field experiment in a ‘hotspot’ of West Nile virus transmission. Proc. R. Soc. B 281: 20141586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan JL, Kluh S, and Reisen WK. 2012. Antecedent avian immunity limits tangential transmission of West Nile virus to humans. PLoS One 7: e34127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanciotti RS, Kerst AJ, Nasci RS, Godsey MS, Mitchell CJ, Savage HM, Komar N, Panella NA, Allen BC, Volpe KE, Davis BS, and Roehrig JT. 2000. Rapid detection of West Nile virus from human clinical specimens, field-collected mosquitoes, and avian samples by a TaqMan reverse transcriptase-PCR assay. J. Clin. Microbiol 38: 4066–4071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenzie VJ and Goulet NE. 2010. Bird community composition linked to human West Nile virus cases along the Colorado Front Range. Ecohealth 7: 439–447. [DOI] [PubMed] [Google Scholar]
- Pagano M and Gauvreau K. 1993. Principles of Biostatistics. Belmont, CA: Duxbury Press; 524 pp. [Google Scholar]
- Reisen WK, Wheeler S, Armijos MV, Fang Y, Garcia S, Kelley K, and Wright S. 2009. Role of communally nesting ardeid birds in the epidemiology of West Nile virus revisited. Vector-Borne Zoonotic Dis. 9: 275–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reisen WK, Wheeler SS, Yamamoto S, Fang Y, and Garcia S. 2005. Nesting ardeid colonies are not a focus of elevated West Nile virus activity in southern California. Vector-Borne Zoonotic Dis. 5: 258–266. [DOI] [PubMed] [Google Scholar]
- Turell MJ, O’Guinn ML, Dohm DJ, and Jones JW. 2001. Vector competence of North American mosquitoes (Diptera: Culicidae) for West Nile virus. J. Med. Entomol 38: 130–134. [DOI] [PubMed] [Google Scholar]


