Abstract
Background:
Germline-encoded innate immune pattern recognition receptors (PRR) are expressed at epithelial surfaces and modulate epithelial defenses. Evidence suggests that stimulation of the Toll-like receptor (TLR) family of PRR may regulate epithelial barrier integrity by upregulating tight junction (TJ) complex protein expression, but it is not known if this mechanism is utilized in esophageal epithelial cells. TJ complex proteins maintain intact barrier function, and are dysregulated in atopic disorders including eosinophilic esophagitis.
Methods:
Pattern recognition receptors expression was assessed in EoE and control primary esophageal epithelial cells, demonstrating robust expression of TLR2 and TLR3. The three‐dimensional air‐liquid interface culture (ALI) model was used to test whether TLR2 or TLR3 stimulation alters epithelial barrier function using an in vitro model of human epithelium. Transepithelial electrical resistance (TEER) and FITC‐Dextran permeability were evaluated to assess membrane permeability. ALI cultures were evaluated by histology, immunohistochemistry, Western blotting, and chromatin immunoprecipitation (ChIP).
Results:
TLR3 stimulation did not change TEER in the ALI model. TLR2 stimulation increased TEER (1.28 to 1.31-fold) and decreased paracellular permeability to FITC-Dextran, and this effect was abolished by treatment with anti-TLR2 blocking antibody. TJ complex proteins claudin-1 and zonula occludens-1 were upregulated following TLR2 stimulation, and ChIP assay demonstrated altered histone 4 acetyl binding at the TJP1 enhancer and CLDN1 enhancer and promoter following zymosan treatment, implying the occurrence of durable chromatin changes.
Conclusions:
Our findings implicate the TLR2 pathway as a potential regulator of esophageal epithelial barrier function and suggest that downstream chromatin modifications are associated with this effect.
Keywords: eosinophilic esophagitis, tight junctions, toll-like receptors, innate immunity, epithelium
Introduction:
Eosinophilic esophagitis (EoE) is an allergic disorder of the esophagus. The pathophysiology of EoE is thought to arise from the presence of food allergens in contact with the esophageal mucosa, which causes a T-helper type 2 (Th2) immune response leading to mucosal inflammation and eosinophilia, epithelial dysfunction and long-term fibrosis (1). Epithelial barrier dysfunction plays a critical role in this process because it promotes continued exposure to allergens and microbiota in the esophageal lumen, which stimulates epithelial cells and leukocytes to release chemokines and cytokines (2,3). Decreased barrier integrity in EoE has been measured by loss of mucosal impedance in vivo and transepithelial electrical resistance in vitro (4–6).
The loss of epithelial barrier integrity in EoE is attributed to the downregulation of important structural proteins in the inflamed mucosa including tight junction (TJ) proteins (2,5,7). The tight junction (TJ) complex is a dynamic structure comprised of transmembrane claudins, occludin, and cytosolic proteins (i.e., zonula occludens-1, zonula occludens-2, and zonula occludens-3), which connect the TJ to the cytoskeleton (8–10). Expression of occludin and claudin-1 are decreased in biopsy tissue of EoE patients both before and after treatment with swallowed corticosteroid (2). In addition to tight junctions, multiple structural proteins contribute to the integrity and stratification of the esophageal epithelial barrier. For example, recent descriptions of kindreds with biallelic mutations in DSG1 and DSP implicate desmosomes as a critical structural protein in the esophageal epithelium (11,12). Th2 cytokines, including IL-13 have been implicated as an important signal contributing to the downregulation of DSG1 (13), but its role in the regulation of other key TJ proteins is less clear.
Innate sensing of microbial products through epithelial pattern recognition receptors (PRR) can regulate epithelial barrier function in human nasal, intestinal and skin mucosa (14–19). The mechanisms which have been described vary across epithelial sites, and it can be hypothesized that there may be some degree of tissue specificity in the effector function of different PRR based upon their mucosal location. Several studies highlight a role for the PRR family of Toll-like receptors (TLR) as a regulator of TJ integrity in epithelial surfaces. An increasing body of evidence suggests that Toll-like receptor 2 (TLR2) signaling upregulates TJ proteins ZO-1 and claudin-1 in epithelium in the airway, intestine and skin (14–19). This mechanism is thought to be one of many host defenses maintaining balance with commensal bacteria.
In this study, our aim was to determine if the esophageal epithelium has mechanisms to modulate TJ complex protein expression based upon innate immune sensing via TLR. We examined the expression of TLR, observing robust expression of TLR2 and TLR3 in primary isolated epithelial cells. We utilized the three-dimensional, air-liquid interface (ALI) in vitro culture model to examine epithelial barrier function following stimulation of TLR2 and TLR3. TLR3 stimulation had little effect on barrier function in the ALI model, whereas TLR2 improved esophageal epithelial barrier function. We hypothesized that TLR2 stimulation was associated with upregulation of TJ proteins, resulting in improved epithelial barrier function.
Methods:
Esophageal epithelial cell culture:
Immortalized esophageal epithelial cell line (EPC2-hTERT) and isolated pprimary esophageal epithelial cells were used in this study (20–22). Patients were enrolled in the study following informed consent. This study was approved by the Children’s Hospital of Philadelphia Institutional Review Board following the US Federal Policy for Protection of Human Subjects.
Primary control patient epithelial lines were derived from esophageal biopsies of pediatric patients (under 18 years of age) during clinically-indicated endoscopic evaluations. Samples were eligible for enrollment in control group if on no treatment at the time of biopsy and found to have normal clinical endoscopy, normal esophageal histology, and no evidence of additional GI pathology. EoE cell lines were derived from pediatric patients with symptoms suggestive of EoE found to have eosinophil count >15 per high powered field on biopsy (23,24). For this study, no patients were on proton pump inhibitor, swallowed steroid or dietary therapy at the time of endoscopy. Primary cells were used between passages 2 and 4 for these experiments.
All cells were grown in keratinocyte serum-free media supplemented with bovine pituitary extract and epidermal growth factor (KSFM, Thermo-Fischer Scientific). Endotoxin-free reagents and media were used for cell culture and stimulation experiments.
Flow Cytometry:
LSR-Fortessa and FlowJo software (BD Biosciences) were used to examine TLR2 (11G7, Invitrogen mouse anti-human) and Dectin-1 (GE2, Bio-Rad mouse anti-human) expression on EPC2-hTERT cells compared to isotype controls.
Air-liquid interface (ALI) culture system:
EPC2-hTERT cells were used for ALI experiments (25). Cells were grown submerged in KSFM media on a 0.4 μm filter (Corning Life Sciences) for three days to reach confluency (protocol schematic, Figure 2A). Cultures were then switched to high-calcium concentration KSFM media (1.8 mM Ca++) for 5 days. Media was removed from the upper chamber to promote epithelial differentiation for days 7 to 10 (25).
Figure 2: TLR2 stimulation of EPC2-hTERT cells in air-liquid interface (ALI) culture results in improved barrier function.
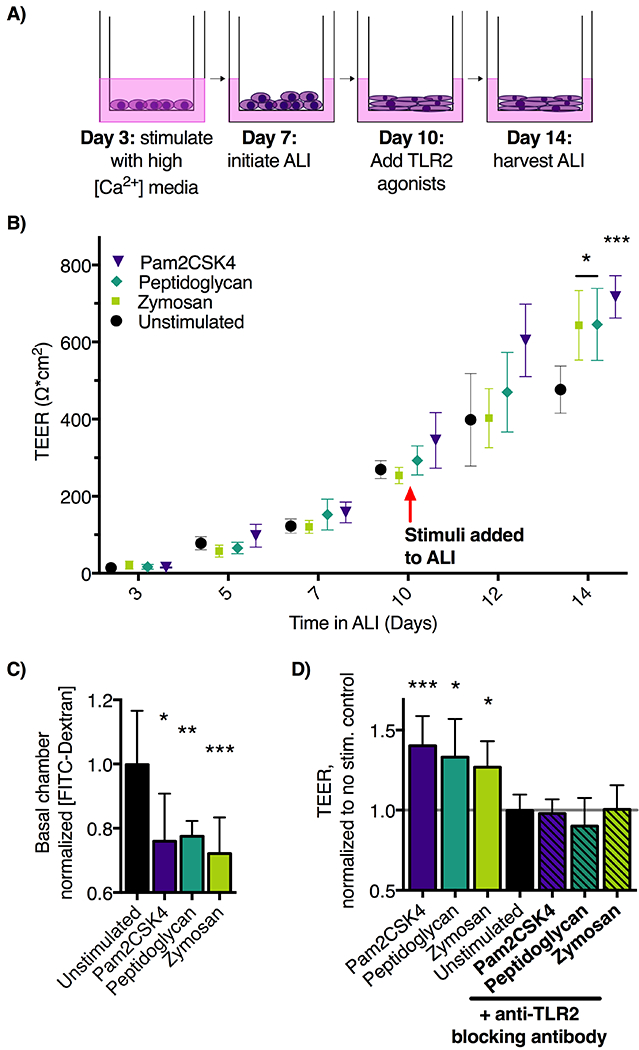
A) Schematic of ALI epithelial culture model. Culture days 1-7 allow for proliferation and initial differentiation, and days 7-10 permit terminal differentiation. Stimuli applied in the basolateral media chamber during days 10-14 allow assessment of effects on epithelial barrier function. B) ALI cultures were stimulated with TLR2 agonists zymosan (10 μg/ml), peptidoglycan (10 μg/ml), or Pam2CSK4 (10ng/mL) from days 10-14 in ALI culture. Transepithelial electrical resistance (TEER, Ω*cm2) was measured by ohmmeter and calculated by multiplying by the area of the ALI membrane. TLR2-agonist treated cultures demonstrate significantly increased day 14 TEER compared to unstimulated ALI cultures. n=10 wells per group, representative of three experiments. C) On day 14, translocation of 70 kDa FITC-Dextran across ALI membranes into the basolateral culture chamber was decreased in the zymosan and peptidoglycan treated groups (n=6 wells per group, mean ±SD. Representative of two experiments. D) Addition of pAB-hTLR2, anti-TLR2 blocking antibody to ALI cultures stimulated with 10 μg/ml of zymosan block zymosan-induced TEER increase. *p<0.05, **p<0.01, and ***p<0.001 using ANOVA followed by Dunnett’s test compared to unstimulated control throughout.
TLR stimulation:
TLR agonists were added to stimulate monolayer cultures or to ALI cultures during days 10-14 of ALI protocol. TLR2 stimulation was assessed using final concentrations of: 10 μg/ml zymosan, 10 μg/ml peptidoglycan from Staphylococcus aureus, and Pam2CSK4 (Invivogen). To assess response to TLR3 stimulation, Poly(I:C) was annealed per manufacturer’s recommendations and used at a final concentration of 10 μg/ml in ALI cultures from days 10 until 14.
TLR2 inhibition:
Rat anti-human polyclonal anti-TLR2 neutralizing antibody (Invivogen) was reconstituted and added to the basolateral chamber media (5 μg/ml final concentration, Invivogen) during ALI days 10-14 concurrent with TLR2 stimulants as described above.
Transepithlial electrical resistance (TEER):
ALI culture resistance was measured with a Millicell ERS-2 Voltohmmeter (Merck Millipore), and the TEER unit area resistance (Ω*cm2) was calculated by multiplying the measured sample resistance by the membrane area (0.33 cm2 for 24-well Millicell inserts). Only epithelial monolayers with TEER >200 ohms·cm2 on day 10 of ALI culture were used for stimulation experiments.
Transepithelial Flux Assay:
70 kDa fluorescein isothiocyanate-dextran (FITC-dextran; 3 mg/mL; Sigma) was added to the upper chamber of day 14 ALI cultures. The initial 3mg/mL upper solution was serially diluted to generate the assay standard curve. The fluorescein levels in basolateral samples were detected after 4 hours on a Spectramax M5 plate reader (Molecular Devices, San Jose USA) and normalized to the mean of the unstimulated control samples.
Histology and Immunohistochemistry:
ALI were harvested on day 14, then fixed in formalin, paraffin-embedded and serially sectioned. Slides were deparaffinized in xylene and rehydrated thru a series of ethanol washes. Histology slides were stained in hematoxylin, rinsed, then stained in eosin (Azer Scientific) on a Shandon Gemini automated stainer (ThermoFisher). Immunohistochemistry slides were incubated with E1 (Leica Biosystems) immunohistochemistry antigen retrieval solution for 20min. Claudin-1 (1:50; LS Bio LS-C415827; 1hour primary incubation) and ZO-1 (1:50; Sigma HPA001636; 1hour primary incubation) antibodies were used for staining with Bond Refine polymer staining kit on the Bond-Max automated staining system (Leica Biosystems).
All stained slides were dehydrated in ascending ethanol and xylene washes before coverslipping with cytoseal (Fisher Scientific, USA). Stained slides were digitally scanned at 20x magnification on an Aperio CS-O slide scanner (Leica Biosystems). ALI membrane thickness, number of basolateral nuclei per 100 μm, percent dilated intracellular space and percent strong 3,3′-Diaminobenzidine (DAB) staining intensity were calculated using Aperio imaging software algorithms.
Western Blot:
Day 14 ALI were washed in PBS and lysed in RIPA buffer containing protease cocktail. Bradford assay was used to normalize sample protein content prior to electrophoresis on a 4-12% Bis-tris gel, followed by transfer to nitrocellulose membranes. Mouse anti-human β-actin (1:1000, Cell-Signaling Technologies 3700S), rabbit anti-human claudin-1 (1:500, LSBio LS-C415827), and rabbit anti-human ZO-1 (1:500, Sigma HPA001636) primary antibodies and secondary HRP conjugated anti-rabbit and anti-mouse antibodies (1:2000, Cell Signaling Technologies) were used to probe the membrane. Semi-quantitative densitometry analysis was performed in NIH ImageJ software.
Quantitative real-time PCR (qRT-PCR):
We harvested RNA using Direct-zol kit with removal of DNA by column (Zymo Research). 0.2 μg of total RNA was used for reverse-transcription reactions using Advantage RT-for-PCR kit (TakaRa Bio), and the cDNA was used for qRT-PCR. Taqman gene expression assays (Applied Biosystems, human TLR1-10, CLEC7A, CD14), were normalized to the GAPDH internal control signal and custom SYBR green primer assays (primers, Table S1) were normalized to β-actin control.
Chromatin Immunoprecipitation (ChIP):
Five million cells per condition were used to perform ChIP experiments following the protocol from Upstate Biotechnology (Lake Placid, NY) with some modifications (26). EPC2-hTERT cells were harvested from monolayer after 30 minutes of stimulation, treated with 1% formaldehyde for 10 min at room temperature, sonicated and immunoprecipitated overnight at 4 °C. 5 μg of anti-H4ac (Histone 4 pan-acetyl, Merck Millipore #06-866), anti-H3K27ac (histone H3 lysine 27 acetyl Abcam ab4729), anti-trimethylated H3 lysine 4 (trimethylated H3 lysine 4 ,Active Motif 39159), and anti-GST (glutathione-S-transferase, ThermoFisher 71-7500) antibodies were used for immunoprecipitation. Antibody-bound complexes were collected, eluted and extracted as previously described (26), then RNase treated, and quantitated before analysis by qRT-PCR (primers, Table S2). The primers for the ChIP assays are listed below. ChIP DNA qRT-PCR results were normalized to GAPDH and then to anti-GST signal for each sample.
Statistical analysis:
Data were collected from a minimum of two independent experiments. Quantitative data are expressed as means ± standard deviation. Statistical analyses were performed (GraphPad Software) using one-way analysis of variance (ANOVA) to analyze multiple groups followed by Dunnett posthoc testing to compare samples against control condition. t-Test was used to compare two means to each other for analysis of ChIP data. P values ≤ 0.05 were considered significant for all analyses.
Results:
TLR2 and TLR3 are expressed in esophageal epithelial cells.
We examined gene expression of TLRs 1–10, CD14 and Dectin-1 (CLEC7A) using qRT-PCR in primary esophageal epithelial cells (Figure 1A), and in the EPC2-hTERT esophageal epithelial cell line (Figure 1B). Both primary esophageal epithelial cells and the EPC2-hTERT cell line express TLR1 through TLR6, with minimal expression of TLR7 through TLR10, CD14 and CLEC7A. As in prior studies, we observe robust mRNA expression of TLR2 and TLR3 (27–29). We did not observe significant differences in PRR expression between cell lines from EoE patients and controls (Figure 1A). TLR2 forms heterodimers with TLR6 and TLR1, both of which were also transcribed in the esophageal cells (Figure 1A & B). TLR2 protein was detected on the epithelial cell surface using flow cytometry (Figure 1C & D). We assessed levels of Dectin-1 expression on the epithelial surface (Figure 1E). Dectin-1 has been shown to have functional overlap with TLR2 in macrophages, resulting in enhanced TLR2 signaling (30–33). We observe low levels of Dectin-1 surface expression on esophageal epithelial cells (Figure 1E), confirming that esophageal epithelial cells express multiple families of PRR which may be capable of signaling crosstalk.
Figure 1: Esophageal epithelial cells express high levels of TLR2 and 3.
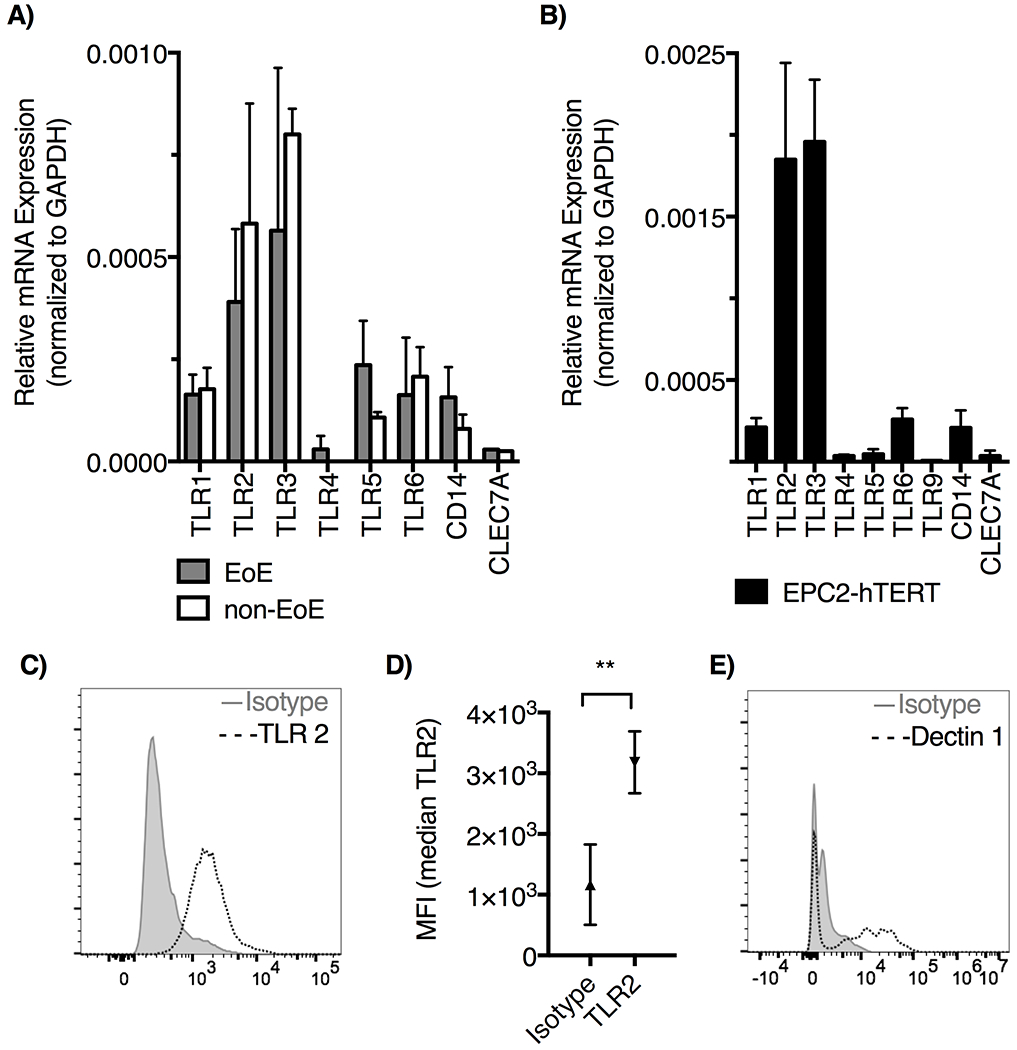
TLR expression was determined by qRT-PCR. A) Primary esophageal epithelial cells from EoE patients and controls express TLR1-6, CD14, CLEC7A (Dectin-1); mean ±SD, n=3 biologic replicates. TLR7-10 not shown, resulted as not amplified. B) TLR1-6, TLR9, CD14, and CLEC7A (Dectin1) are expressed on immortalized epithelial cell line EPC2-hTERT; mean ±SD, n=3. TLR7, 8 and 10 not amplified. C) TLR2 surface expression on EPC2-hTERT cells was assessed by flow cytometry, and D) the median fluorescence intensity is shown (**p<0.001 by t-test, n=3). E) EPC2-hTERT cells evaluated for expression of Dectin-1 by flow cytometry.
TLR2 stimulation improved epithelial barrier function in air liquid interface culture.
We next set out to test the effect of TLR2 and TLR3 stimulation on esophageal epithelial cell barrier function. These were chosen for further investigation due to their robust expression in esophageal epithelium. We used the three-dimensional air liquid interface (ALI) model and added TLR ligands to the basolateral chamber media on days 10-14 of culture (Figure 2A, schematic).
TLR3 on esophageal epithelial cells has been shown to respond to synthetic ligands and necrotic cell debris, resulting in NF-kB activation, and IL-8 and thymic stromal lymphopoietin secretion (27,34,35). We stimulated ALI with the TLR3 ligand Poly(I:C), however, we did not observe any change in the epithelial barrier function (Supplemental Figure 1). Following treatment with Poly(I:C), transepithelial electrical resistance (TEER) and FITC-Dextran permeability were unchanged compared to control untreated ALI cultures.
In contrast, stimulation of ALI cultures with TLR2 agonists resulted in a significant increase of epithelial barrier function measured by both TEER (Figure 2B) and FITC-Dextran flux assays (Figure 2C). Several TLR2 ligands were utilized in order to determine if this effect varied by targeting the TLR2/6 or TLR2/1 complex. However, increased TEER and decreased FITC-Dextran permeability was seen using irrespective of whether the ligand targeted the TLR2/1 heterodimer or TLR2/6 heterodimer. TLR2 forms a heterodimer with TLR 1 to recognize triacylated lipopeptides, and heterodimerizes with TLR6 to recognize zymosan and other diacylated lipopeptides. The average TEER was increased by 26.8% in zymosan-treated ALI, 32.2% in peptidoglycan treated ALI and 40.3% over control in Pam2CSK4 treated ALI. The FITC-Dextran assay (Fig. 2C) confirms these findings with TLR2 agonist-treated samples allowing less flux of FITC-Dextran trough the ALI membrane (Figure 2C). These results are consistent with improved barrier function in the TLR2-agonist treated samples.
Improved barrier function is TLR2-specific.
The full complement of PRR in esophageal epithelium has not been defined, and their functions are incompletely understood. For example, although zymosan is recognized by the TLR 2/6 heterodimer it can also be recognized by Dectin-1 (31–33) and potentially by epithelial-specific, innate receptors such as Ephrin A2 (36). Our data demonstrate that EPC2-hTERT cells do express low levels of Dectin-1 (Figure 1E), therefore we next set out to determine if the observed improvement in epithelial barrier function was TLR2-specific. To do so, we utilized PAb-hTLR2, which is a polyclonal rat anti-human blocking antibody specific for TLR2 (37,38). Co-incubation of ALI with PAb-hTLR2 during addition of TLR2 agonists into the culture media abrogated improvement in epithelial TEER (Figure 2E). This demonstrates a specific requirement for TLR2 to achieve improved esophageal epithelial barrier function in vitro.
TLR2 stimulation alters epithelial membrane architecture:
We next performed histology to examined epithelial architectural changes associated with zymosan and peptidoglycan treatment in ALI culture (Figure 3A). We observe the expected stratification of the epithelial membrane in unstimulated samples, with thinning of the epithelial membrane following TLR2 agonist treatment (Figure 3B, 24.8±5.6 μm untreated vs 17.6±6.5 μm zymosan-treated, p<0.001, and 18.7±3.9 peptidoglycan-treated p<0.05). The density of basolateral nuclei was decreased in TLR2 agonist treated cultures (Figure 3C). Dilated intracellular spaces are a characteristic finding of epithelial barrier dysfunction in EoE. However, there was no difference in the amount of dilated intracellular space between treated and untreated ALI cultures (Figure 3D). Dilated intracellular spaces have been shown to correlate with poor barrier function (39). Therefore, this corresponds with data obtained in our barrier function assays as we observed improved barrier function with TLR2 stimulation (40,41).
Figure 3: Zymosan stimulation of esophageal ALI alters epithelial morphology.
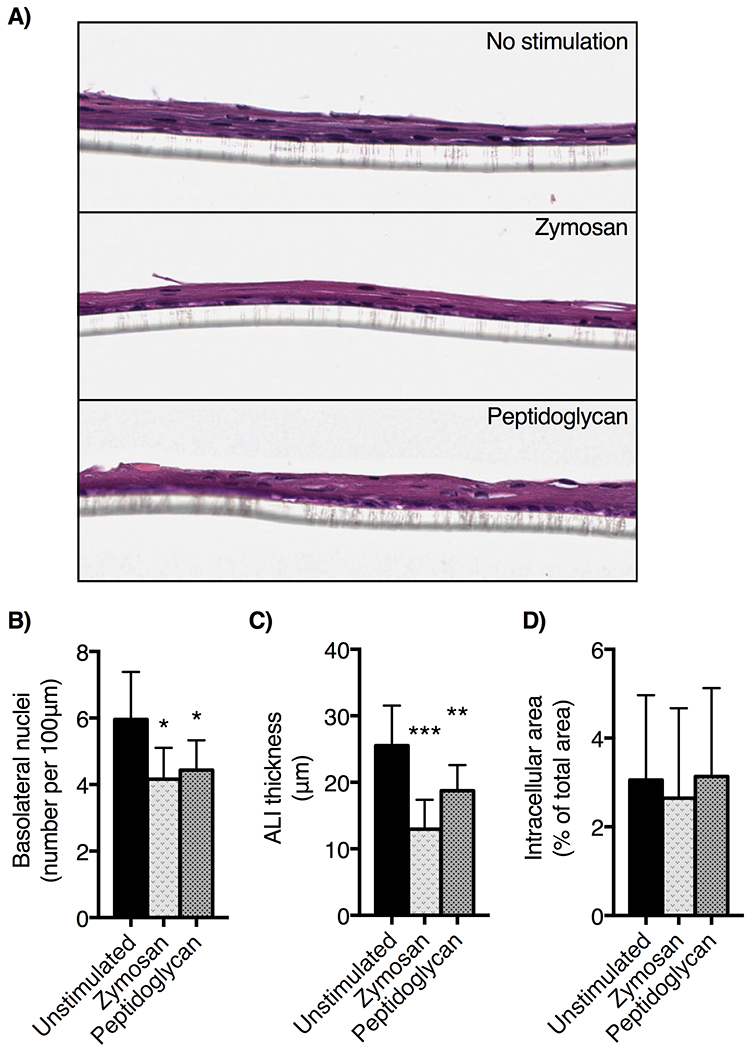
EPC2-hTERT cells grown in ALI were harvested on day 14 of culture, fixed, paraffinized and stained with hematoxylin and eosin. A) Images were captured at 20x. We quantified B) ALI thickness, C) basolateral nuclei (expressed per every 100 μm of basolateral membrane adjacent to the Transwell membrane) and D) the percent of total area dilated intracellular space quantified using Aperio software imaging algorithms (n=5 slides per condition; mean ± SD, * p<0.05, **p<0.01, *** p<0.001, ANOVA, post-hoc Dunnett test).
TLR2 stimulation upregulated TJ complex gene and protein expression:
TJ complex proteins are known to be a primary mediator of epithelial barrier integrity (42). Stimulation of TLR2 in intestinal and airway epithelium increases expression of TJ complex proteins (15,17,19), however this association has not previously been examined in the esophageal epithelium. In light of our data demonstrating improved ALI barrier function following TLR2 stimulation, we hypothesized that TLR2 stimulation may upregulate TJ complex protein expression in ALI cultures. EPC2-hTERT cells in monolayer culture were stimulated with zymosan or peptidoglycan and qRT-PCR was used to assess expression of TJ complex genes CLDN1 and TJP1 (claudin-1 and zonula occludens-1 ZO-1, Figure 4). TJP1 was significantly upregulated over 2-fold following 6 hours of TLR2 stimulation with zymosan and peptidoglycan. mRNA levels of CLDN1 rose to 2-fold over control unstimulated samples by 30 minutes of stimulation with zymosan and after 6 hours of stimulation with peptidoglycan. We did not see significant upregulation desmoglein 1 mRNA following TLR2 stimulation with either zymosan or peptidoglycan (Supplemental Figure 2), or following stimulation of TLR3 (Supplemental Figure 1C).
Figure 4: Zymosan treatment of EPC2-hTERT cells upregulates expression of cell adhesion molecules.
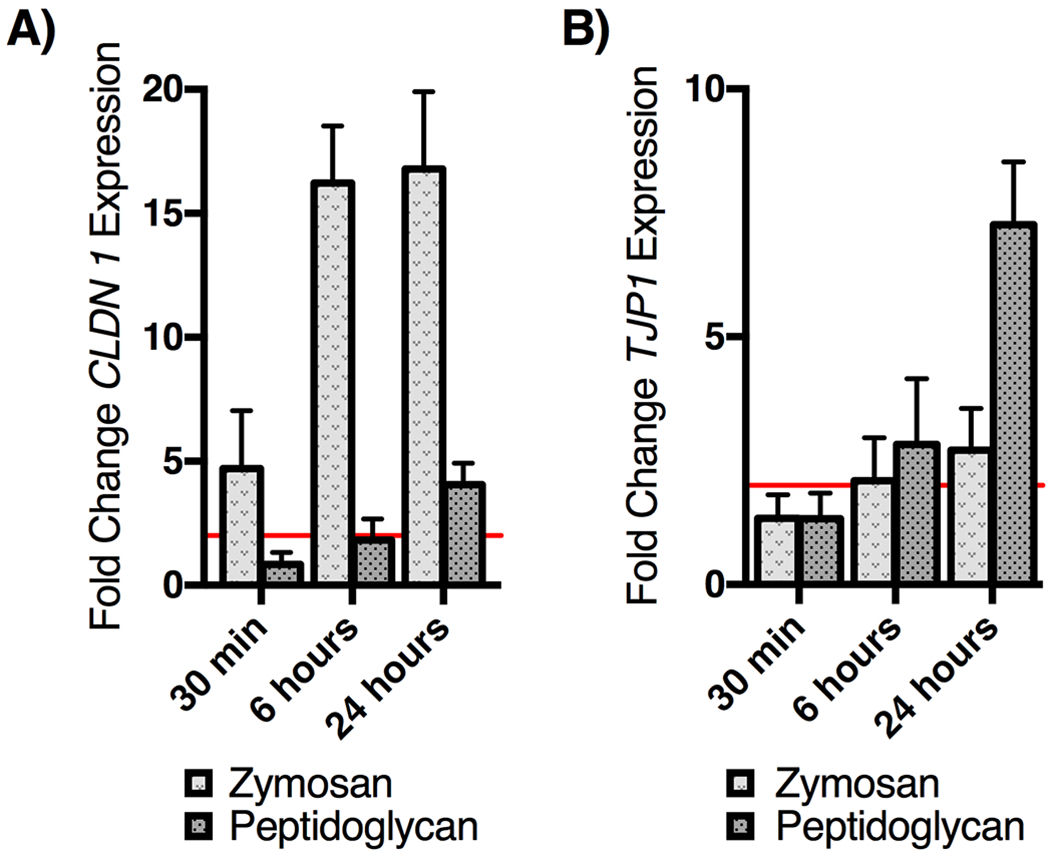
EPC2-hTERT cells stimulated with zymosan or peptidoglycan (10 μg/ml ) in culture results in greater than 2-fold upregulation of A) CLDN1, and B) TJP1 by 6 hours of stimulation. mRNA signal was normalized to β-actin and expressed as fold change compared to unstimulated control cells (n=4; mean ± SD).
To test the effect of TLR2 stimulation on TJ complex protein expression in esophageal epithelial cells, we examined claudin-1 and ZO-1 protein expression by immunohistochemistry (IHC) and western blotting (Figure 5). ALI cultures were harvested on day 14 for IHC (Figure 5A), demonstrating an increased staining intensity for claudin-1 and ZO-1 in TLR2 agonist samples. 3,3′-diaminobenzidine staining intensity was quantified in Aperio imaging software to confirm upregulation of ZO-1 and claudin-1 in TLR2 agonist treated samples (Figure 5B and 5C). We additionally observed increased expression of claudin-1 and ZO-1 protein from TLR2-stimulated ALI using western blot (Figure 5D). Semi-quantitative analysis of western blot band intensity in ImageJ software (Figure 5E) shows a mean 2.3- to 2.4-fold increase in claudin-1 and mean 2.9- to 4.2-fold increase in ZO-1 expression with stimulation. From these studies, we conclude that TLR2 stimulation is associated with increased expression of TJ proteins in the ALI model of esophageal epithelium.
Figure 5: ALI cultures treated with zymosan upregulate epithelial cell-cell adhesion molecules.
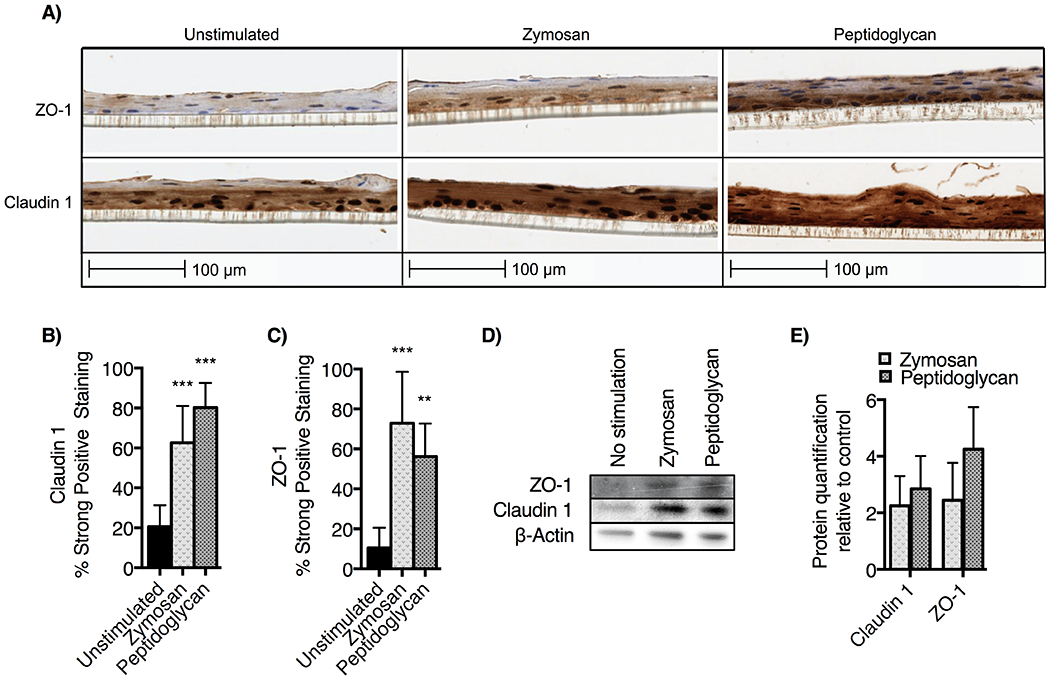
A) EPC2-hTERT ALI cultures were stimulated with zymosan and peptidoglycan then stained for immunohistochemistry with anti-claudin-1 and anti-zonula occludens-1 (ZO-1). Claudin-1 is diffusely upregulated while the pattern of ZO-1 staining is more dense in the basal layer of ALI culture. Using DAB staining quantification algorithm in Aperio, we find that B) Claudin-1 and C) ZO-1 staining are significantly upregulated (mean ± SD ** p<0.01 or *** p<0.001 ANOVA, posthoc Dunnett test compared to unstimulated control). D) Western blotting of day 14 ALI cultures confirms upregulation of claudin-1 and ZO-1, with E) densitometry normalization to beta-actin (NIH ImageJ) demonstrating 2- to 4-fold upregulation of protein following TLR2 stimulation. Results shown are representative of three experiments.
Histone modifications in zymosan-treated epithelial cells.
Modification to the chromatin regulatory landscape alters gene expression in response to changes in the cellular environment. We hypothesized TLR2 stimulation may induce regulatory histone modifications at tight junction complex gene loci (43–45). We performed chromatin immunoprecipitation (ChIP) assays of EPC2-hTERT cells following stimulation with zymosan (Figure 6) to assess for changes in the chromatin environment at the promoter and enhancer of CLDN1 and TJP1. Acetylation of histone H4 was significantly increased at the CLDN1 enhancer and promoter and decreased in the TJP1 promoter of zymosan-treated samples. This suggests changes in acetylation of histone H4 occur subsequent to TLR2 stimulation with zymosan, and may be a novel mechanism for regulation of esophageal tight junction proteins.
Figure 6: EPC2-hTERT cells stimulated with Zymosan alter chromatin at CLDN1 and TJP1 promoters and enhancers.
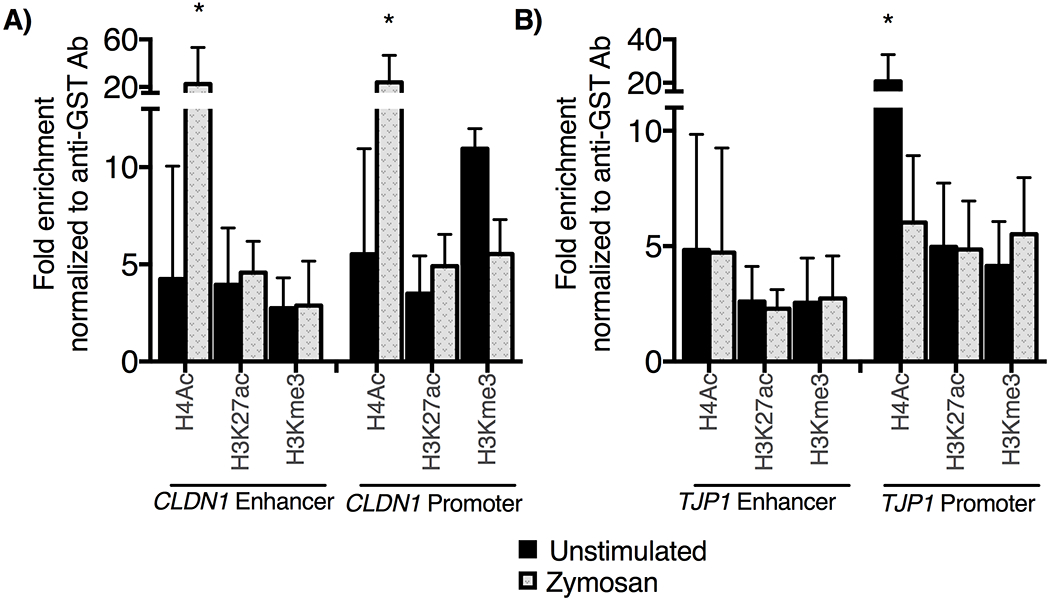
Following a 30-minute incubation with zymosan, chromatin immunoprecipitation assays were performed using antibodies specific to H4ac, H3K27ac, H3Kme3. PCR was performed to amplify immunoprecipitated DNA, and results were normalized to GAPDH and displayed as fold enrichment over anti-glutathione-S-transferase antibody. A) We observe significant enrichment of H4ac at CLDN1 enhancer and promoter locus following zymosan treatment, whereas B) there is decreased H4ac signal following zymosan treatment at the TJP1 enhancer (n=4, mean ± SD *p<0.05, t-test)
Discussion:
The pathophysiology of eosinophilic esophagitis (EoE) is not solely defined by eosinophil-predominant inflammation, but additionally by abnormal epithelial barrier morphology (39). Increased esophageal epithelial permeability is a major aspect of the pathophysiology of EoE, which perpetuates disease by allowing allergens and microbes to penetrate the mucosal barrier (4,6). Little is known about innate esophageal epithelial responses to stimuli. In this study we explore the role of TLR stimulation on epithelial barrier function utilizing the air-liquid interface model.
Our data demonstrate that esophageal epithelial cells express multiple pattern recognition receptors, including TLR1-6, Dectin-1 and CD14. Our findings demonstrate robust expression of TLR2 and TLR3 in primary epithelium isolated from patients with and without EoE. This is similar to prior findings in primary esophageal epithelial cells from pediatric patients (27). We do not observe gene expression differences in the PRRs on primary epithelial cells from EoE and control patients. In contrast, Arias et al. have recently published gene expression data from whole biopsy tissue of adults with EoE demonstrating significant upregulation of TLR1, TLR2, TLR4, and TLR9 when compared to control patient biopsy tissue (46). The overexpression of these TLR improved with six-food elimination diet (46). Our current study used isolated cells between passages 2 and 4, which may have allowed for some normalization of expression between the EoE and control cell lines in vitro. To our knowledge, there are no published data comparing expression of PRR in esophageal biopsy tissue and freshly isolated esophageal epithelial cells to determine the gene expression signal from inflammatory cells present in the biopsy transcriptome. Data from Rochman et al examining the overlap of the EoE biopsy transcriptome and esophageal-specific proteome demonstrates a functional enrichment of IL-1 cytokine pathways in EoE, further supporting a role for epithelial innate signaling in EoE (47).
In the ALI model, we have tested if stimulation of TLR2 and 3 can improve barrier function. Our data show that recognition of lipoprotein agonists by TLR2 is a mechanism that increases epithelial barrier integrity (Figure 2), with a modest increase in ALI TEER following stimulation with TLR2 ligands zymosan, peptidoglycan and Pam2CSK4. It is not known what magnitude of in vitro barrier effect corresponds to a meaningful in vivo effect in this system. In contrast, stimulation of TLR3 with Poly(I:C) resulted in no change in barrier function compared to unstimulated control ALI cultures (Supplemental Figure 1). TLR3 stimulation of esophageal epithelial cells has previously been shown to activate NF-κB, resulting in secretion of IL-8 and TSLP (34,35). Therefore, although stimulation of the TLR3 pathway does not affect barrier function in this model, this does not exclude the possibility that the TLR3 pathway engages additional epithelial innate effector mechanisms which were not examined.
The esophagus is not a sterile environment, and changes in microbiome composition have been reported in patients with reflux, Barrett’s esophagus as well as EoE patients with active inflammation (48–50). Further, tissue damage from inflammation or reflux causes release of endogenous damage-associated molecular patterns from necrotic cells which can also interact with esophageal PRR (27,34). Our findings describe one potential mechanism for esophageal barrier modulation in response to these changes. Our data shows that TLR2 stimulation increases expression of TJ-associated proteins claudin-1 and ZO-1 (Figures 4 and 5). TJ complex molecules claudin-1 and ZO-1 are upregulated within 24 hours following TLR2 stimulation (Figure 4). Our results are consistent with findings from other epithelial types, including intestinal, airway and skin keratinocytes, where TLR2 agonism has been associated with increased expression of TJ complex molecules claudin-1 and ZO-1 (14–17,19,51).
Extrapolating upon these findings, we hypothesized that signaling thru innate immune receptors may play a critical role in epithelial cell fate. We find that stimulation of TLR2 with zymosan results in significant enrichment of H4ac at both the CLDN1 promoter and enhancer (Figure 6). H4ac is an activation mark with a widespread distribution at regulatory regions. TLR2-induced chromatin remodeling could be secondary to an overall shift in the balance of histone acetyltransferases and deacetylase enzymes or more likely due to targeting of chromatin modification complexes to the regulatory regions (52). Novel therapies with histone deacetylase inhibitors could be investigated to target this pathway within the mucosal for disorders like EoE.
In this study we focused on the response of claudin-1 and ZO-1 because of their critical role in tight junction complex formation (42,53). A number epithelial proteins including desmosomes, serine peptidase inhibitors, filaggrin, and calpains are involved in maintenance of esophageal epithelial barrier integrity (13,25,54,55). We did not see alterations in the expression of desmoglein 1, however further work will be required to determine the full scope of epithelial change in response to TLR2 stimulation. We hypothesize that TLR2 stimulation induces additional alterations in esophageal epithelial cell gene expression that were not fully captured in our experiments, and future experiments characterizing these additional effects will clarify the full impact of TLR2 stimulation on the esophageal mucosa.
A TLR3 single nucleotide polymorphism (SNP) was recently linked to increase risk of developing EoE in a Spanish cohort (56). TLR2 SNPs have not been associated directly with EoE, but have been associated with closely-linked atopic diseases. Specifically, there are TLR2 SNPS which predispose to the development of asthma in Puerto Rican, Norwegian, Danish and Caucasian African populations (57–63), and development of severe atopic dermatitis in Italian and German populations (64,65). Interestingly, several of the SNPs decrease TLR2 signaling activity (57). Further studies elucidating how PRRs such as TLRs contribute to host barrier responses may help clarify mechanisms of atopic disease including EoE.
In conclusion, we have shown that esophageal epithelial cells express TLR1-6, TLR9, Dectin-1 and CD14, with membrane expression of TLR2. Stimulation of epithelial TLR2 with lipoproteins Pam2CSK4, zymosan, and peptidoglycan upregulate Claudin-1 and ZO-1, resulting in enhanced esophageal epithelial barrier function in the ALI model. This effect is TLR2-specific and abrogated by TLR2 blocking antibody. Our data highlight a potential homeostatic role of esophageal epithelial TLR2 activity in upregulating tight junction complex protein expression. This has implications for esophageal mucosal responses to infection, colonizing microbiota and endogenous DAMPs released in the context of reflux and inflammation. TLR2 agonism appears to convey a protective effect on the esophageal mucosal barrier, and, further investigation is warranted to determine if this response is intact during mucosal inflammatory diseases such as EoE.
Supplementary Material
Acknowledgements:
EPC2-hTERT cell line was the kind gift of Dr. Hiro Nakagawa. We wish to thank Drs. De’broski Herbert, Rob Rubenstein and Yann Bikard for advice regarding the ALI model and to acknowledge support from the Children’s Hospital of Philadelphia Center for Epithelial Excellence. We are grateful to Swathi Raman and the Children’s Hospital of Philadelphia Flow Cytometry and Pathology Core Facilities for technical assistance.
Funding:
MAR was funded by NIH T32-HD043021, KL2TR001879 and the ACAAI Young Faculty Award. ABM is funded by NIH K08DK106444. KES is funded by the Wallace Endowed Chair. JMS is funded by Stuart Starr Endowed Chair. JMS and ABM are funded by the Consortium of Eosinophilic Gastrointestinal Disease Researchers (CEGIR U54 AI117804) which is part of the Rare Diseases Clinical Research Network, an initiative of the Office of Rare Diseases Research, NCATS, and is funded through collaboration between NIAID, NIDDK, and NCATS and patient advocacy groups including APFED, CURED, and EFC.
Abbreviations:
- ALI
air-liquid interface
- CAPN14
calpain 14
- CLDN1
claudin-1
- EPC2-hTERT
primary human esophageal keratinocytes, EPC2 human telomerase immortalized
- GST
glutathione-S-transferase
- HAT
histone acetyl transferase
- HDAC
histone deacetylases
- KSFM
keratinocyte serum-free media
- PRR
pattern recognition receptor
- SNP
single nucleotide polymorphism
- TEER
transepithelial electrical resistance
- Th2
T-helper type 2
- TJ
tight junction
- TLR
Toll-like receptor
- USA
United States of America
- ZO-1
zonula occludens-1
Footnotes
Conflict of Interest:
The authors declare that they have no conflict of interest.
References:
- 1.Rochman M, Azouz NP, Rothenberg ME. Epithelial origin of eosinophilic esophagitis. J Allergy Clin Immunol 2018;142:10–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Simon D, Page B, Vogel M, Bussmann C, Blanchard C, Straumann A et al. Evidence of an abnormal epithelial barrier in active, untreated and corticosteroid-treated eosinophilic esophagitis. Allergy Eur J Allergy Clin Immunol 2018;73:239–247. [DOI] [PubMed] [Google Scholar]
- 3.Simon D, Straumann A, Dahinden C, Simon HU. Frequent sensitization to Candida albicans and profilins in adult eosinophilic esophagitis. Allergy Eur J Allergy Clin Immunol 2013;68:945–948. [DOI] [PubMed] [Google Scholar]
- 4.Warners MJ, van Rhijn BD, Verheij J, Smout AJPM, Bredenoord AJ. Disease activity in eosinophilic esophagitis is associated with impaired esophageal barrier integrity. Am J Physiol Liver Physiol 2017;313:G230–G238. [DOI] [PubMed] [Google Scholar]
- 5.Warners MJ, Vlieg-Boerstra BJ, Verheij J, van Hamersveld PHP, van Rhijn BD, Van Ampting MTJ et al. Esophageal and Small Intestinal Mucosal Integrity in Eosinophilic Esophagitis and Response to an Elemental Diet. Am J Gastroenterol 2017;112:1061–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Katzka DA, Ravi K, Geno DM, Smyrk TC, Iyer PG, Alexander JA et al. Endoscopic mucosal impedance measurements correlate with eosinophilia and dilation of intercellular spaces in patients with eosinophilic esophagitis. Clin Gastroenterol Hepatol 2015;13:1242–1248.e1. [DOI] [PubMed] [Google Scholar]
- 7.Katzka DA, Tadi R, Smyrk TC, Katarya E, Sharma A, Geno DM et al. Effects of Topical Steroids on Tight Junction Proteins and Spongiosis in Esophageal Epithelia of Patients With Eosinophilic Esophagitis. Clin Gastroenterol Hepatol 2014;12:1824–1829. [DOI] [PubMed] [Google Scholar]
- 8.Zeisel MB, Dhawan P, Baumert TF. Tight junction proteins in gastrointestinal and liver disease. Gut 2019;68:547–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zihni C, Mills C, Matter K, Balda MS. Tight junctions: From simple barriers to multifunctional molecular gates. Nat. Rev. Mol. Cell Biol 2016;17:564–580. [DOI] [PubMed] [Google Scholar]
- 10.Farquhar MG, Palade GE. Junctional complexes in various epithelia. J Cell Biol 1963;17:375–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Samuelov L, Sarig O, Harmon RM, Rapaport D, Ishida-Yamamoto A, Isakov O et al. Desmoglein 1 deficiency results in severe dermatitis, multiple allergies and metabolic wasting. Nat Genet 2013;45:1244–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McAleer MA, Pohler E, Smith FJD, Wilson NJ, Cole C, MacGowan S et al. Severe dermatitis, multiple allergies, and metabolic wasting syndrome caused by a novel mutation in the N-terminal plakin domain of desmoplakin. J Allergy Clin Immunol 2015;136:1268–1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sherrill JD, Kc K, Wu D, Djukic Z, Caldwell JM, Stucke EM et al. Desmoglein-1 regulates esophageal epithelial barrier function and immune responses in eosinophilic esophagitis. Mucosal Immunol 2014;7:718–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuo I-HH, Carpenter-Mendini A, Yoshida T, McGirt LY, Ivanov AI, Barnes KC et al. Activation of epidermal toll-like receptor 2 enhances tight junction function: Implications for atopic dermatitis and skin barrier repair. J Invest Dermatol 2013;133:988–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cario E, Gerken G, Podolsky DK. Toll-Like Receptor 2 Controls Mucosal Inflammation by Regulating Epithelial Barrier Function. Gastroenterology 2007;132:1359–1374. [DOI] [PubMed] [Google Scholar]
- 16.Tunis MC, Marshall JS. Toll-like receptor 2 as a regulator of oral tolerance in the gastrointestinal tract. Mediators Inflamm 2014;2014:606383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cario E, Gerken G, Podolsky DK. Toll-like receptor 2 enhances ZO-1-associated intestinal epithelial barrier integrity via protein kinase C. Gastroenterology 2004;127:224–238. [DOI] [PubMed] [Google Scholar]
- 18.Chun J, Prince A. TLR2-Induced Calpain Cleavage of Epithelial Junctional Proteins Facilitates Leukocyte Transmigration. Cell Host Microbe 2009;5:47–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ragupathy S, Esmaeili F, Paschoud S, Sublet E, Citi S, Borchard G. Toll-like receptor 2 regulates the barrier function of human bronchial epithelial monolayers through atypical protein kinase C zeta, and an increase in expression of claudin-1. Tissue Barriers 2014;2:e29166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Takaoka M, Harada H, Deramaudt TB, Oyama K, Andl CD, Johnstone CN et al. Ha-RasG12V induces senescence in primary and immortalized human esophageal keratinocytes with p53 dysfunction. Oncogene 2004;23:6760–6768. [DOI] [PubMed] [Google Scholar]
- 21.Harada H, Nakagawa H, Oyama K, Takaoka M, Andl CD, Jacobmeier B et al. Telomerase induces immortalization of human esophageal keratinocytes without p16INK4a inactivation. Mol Cancer Res 2003;1:729–738. [PubMed] [Google Scholar]
- 22.Muir AB, Lim DM, Benitez AJ, Modayur Chandramouleeswaran P, Lee AJ, Ruchelli ED et al. Esophageal epithelial and mesenchymal cross-talk leads to features of epithelial to mesenchymal transition in vitro. Exp Cell Res 2013;319:850–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lucendo AJ, Molina-Infante J, Arias Á, von Arnim U, Bredenoord AJ, Bussmann C et al. Guidelines on eosinophilic esophagitis: evidence-based statements and recommendations for diagnosis and management in children and adults. United Eur Gastroenterol J 2017;5:335–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dellon ES, Liacouras CA, Molina-Infante J, Furuta GT, Spergel JM, Zevit N et al. Updated International Consensus Diagnostic Criteria for Eosinophilic Esophagitis: Proceedings of the AGREE Conference. In: Gastroenterology. 2018: 1022–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Davis BP, Stucke EM, Khorki ME, Litosh VA, Rymer JK, Rochman M et al. Eosinophilic esophagitis–linked calpain 14 is an IL-13–induced protease that mediates esophageal epithelial barrier impairment. JCI Insight 2016;1:e86355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maurer K, Ramen S, Shi L, Song L, Sullivan KE. Rapid induction of expression by LPS is accompanied by favorable chromatin and rapid binding of c-Jun. Mol Immunol 2018;95:99–106. [DOI] [PubMed] [Google Scholar]
- 27.Lim DM, Narasimhan S, Michaylira CZ, Wang M-L. TLR3-mediated NF-{kappa}B signaling in human esophageal epithelial cells. Am J Physiol Gastrointest Liver Physiol 2009;297:G1172–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Verbeek RE, Siersema PD, Vleggaar FP, Ten Kate FJ, Posthuma G, Souza RF et al. Toll-like Receptor 2 Signalling and the Lysosomal Machinery in Barrett’s Esophagus. J Gastrointestin Liver Dis 2016;25:273–282. [DOI] [PubMed] [Google Scholar]
- 29.Huhta H, Helminen O, Lehenkari PP, Saarnio J, Karttunen TJ, Kauppila JH. Toll-like receptors 1, 2, 4 and 6 in esophageal epithelium, Barrett’s esophagus, dysplasia and adenocarcinoma. Oncotarget 2016;7:23658–23667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao G-Q, Qiu X-Y, Lin J, Li Q, Hu L-T, Wang Q et al. Co-regulation of Dectin-1 and TLR2 in inflammatory response of human corneal epithelial cells induced by Aspergillus fumigates. Int J Ophthalmol 2016;9:185–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yadav M, Schorey JS. The β-glucan receptor dectin-1 functions together with TLR2 to mediate macrophage activation by mycobacteria. Blood 2006;108:3168–3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shin DM, Yang CS, Yuk JM, Lee JY, Kim KH, Shin SJ et al. Mycobacterium abscessus activates the macrophage innate immune response via a physical and functional interaction between TLR2 and dectin-1. Cell Microbiol 2008;10:1608–1621. [DOI] [PubMed] [Google Scholar]
- 33.Ferwerda G, Meyer-Wentrup F, Kullberg BJ, Netea MG, Adema GJ. Dectin-1 synergizes with TLR2 and TLR4 for cytokine production in human primary monocytes and macrophages. Cell Microbiol 2008;10:2058–2066. [DOI] [PubMed] [Google Scholar]
- 34.Lim DM, Wang M-L. Toll-like receptor 3 signaling enables human esophageal epithelial cells to sense endogenous danger signals released by necrotic cells. Am J Physiol Gastrointest Liver Physiol 2011;301:G91–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chandramouleeswaran PM, Shen D, Lee AJ, Benitez A, Dods K, Gambanga F et al. Preferential secretion of thymic stromal lymphopoietin (TSLP) by terminally differentiated esophageal epithelial cells: Relevance to eosinophilic esophagitis (EoE). PLoS One 2016;11:e0150968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Swidergall M, Solis NV, Lionakis MS, Filler SG. EphA2 is an epithelial cell pattern recognition receptor for fungal β-glucans. Nat Microbiol 2018;3:53–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brandt KJ, Fickentscher C, Boehlen F, Kruithof EKO, De Moerloose P. NF-κB is activated from endosomal compartments in antiphospholipid antibodies-treated human monocytes. J Thromb Haemost 2014;12:779–791. [DOI] [PubMed] [Google Scholar]
- 38.Vogt LM, Meyer D, Pullens G, Faas MM, Venema K, Ramasamy U et al. Toll-Like Receptor 2 Activation by 2->1-Fructans Protects Barrier Function of T84 Human Intestinal Epithelial Cells in a Chain Length-Dependent Manner. J Nutr 2014;144:1002–1008. [DOI] [PubMed] [Google Scholar]
- 39.Collins MH, Martin LJ, Alexander ES, Todd Boyd J, Sheridan R, He H et al. Newly developed and validated eosinophilic esophagitis histology scoring system and evidence that it outperforms peak eosinophil count for disease diagnosis and monitoring. Dis Esophagus 2017;30:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kiran KC, Rothenberg ME, Sherrill JD. In vitro model for studying esophageal epithelial differentiation and allergic inflammatory responses identifies keratin involvement in eosinophilic esophagitis. PLoS One 2015;10:e0127755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Björkman EVC, Edebo A, Oltean M, Casselbrant A. Esophageal barrier function and tight junction expression in healthy subjects and patients with gastroesophageal reflux disease: Functionality of esophageal mucosa exposed to bile salt and trypsin in vitro. Scand J Gastroenterol 2013;48:1118–1126. [DOI] [PubMed] [Google Scholar]
- 42.Furuse M, Hata M, Furuse K, Yoshida Y, Haratake A, Sugitani Y et al. Claudin-based tight junctions are crucial for the mammalian epidermal barrier: a lesson from claudin-1-deficient mice. J Cell Biol 2002;156:1099–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stebe-Frick S, Ostaff MJ, Stange EF, Malek NP, Wehkamp J. Histone deacetylase-mediated regulation of the antimicrobial peptide hBD2 differs in intestinal cell lines and cultured tissue. Sci Rep 2018;8:12886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Neagos J, Standiford TJ, Newstead MW, Zeng X, Huang SK, Ballinger MN. Epigenetic Regulation of Tolerance to Toll-Like Receptor Ligands in Alveolar Epithelial Cells. Am J Respir Cell Mol Biol 2015;53:872–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Perkins DJ, Patel MC, Blanco JCG, Vogel SN. Epigenetic Mechanisms Governing Innate Inflammatory Responses. J Interferon Cytokine Res 2016;36:454–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Arias Á, Vicario M, Bernardo D, Olalla JM, Fortea M, Montalban-Arques A et al. Toll-like receptors-mediated pathways activate inflammatory responses in the esophageal mucosa of adult eosinophilic esophagitis. Clin Transl Gastroenterol 2018;9:147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rochman M, Travers J, Miracle CE, Bedard MC, Wen T, Azouz NP et al. Profound loss of esophageal tissue differentiation in patients with eosinophilic esophagitis. J Allergy Clin Immunol 2017;140:738–749.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.May M, Julian Abrams BA. Emerging Insights into the Esophageal Microbiome. Curr Treat Options Gastro 1938;16:72–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Benitez AJ, Hoffmann C, Muir AB, Dods KK, Spergel JM, Bushman FD et al. Inflammation-associated microbiota in pediatric eosinophilic esophagitis. Microbiome 2015;3:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Harris JK, Fang R, Wagner BD, Choe HN, Kelly CJ, Schroeder S et al. Esophageal microbiome in eosinophilic esophagitis. PLoS One 2015;10. doi: 10.1371/journal.pone.0128346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ren C, Zhang Q, De Haan BJ, Zhang H, Faas MM, De Vos P. Identification of TLR2/TLR6 signalling lactic acid bacteria for supporting immune regulation. Sci Rep 2016;6:34561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Allis CD, Jenuwein T. The molecular hallmarks of epigenetic control. Nat Rev Genet 2016;17:487–500. [DOI] [PubMed] [Google Scholar]
- 53.Matter K, Balda MS. Functional analysis of tight junctions. Methods 2003;30:228–234. [DOI] [PubMed] [Google Scholar]
- 54.Wu L, Tadayuki Oshima X, Li M, Tomita T, Fukui H, Watari J et al. Filaggrin and tight junction proteins are crucial for IL-13-mediated esophageal barrier dysfunction. Am J Physiol Gastrointest Liver Physiol 2018;315:341–350. [DOI] [PubMed] [Google Scholar]
- 55.Azouz NP, Ynga-Durand MA, Caldwell JM, Jain A, Rochman M, Fischesser DM et al. The antiprotease SPINK7 serves as an inhibitory checkpoint for esophageal epithelial inflammatory responses. Sci Transl Med 2018;10:82–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ávila-Castellano R, García-Lozano J-RR, Cimbollek S, Lucendo AJ, Bozada J-MM, Quiralte J et al. Genetic variations in the TLR3 locus are associated with eosinophilic esophagitis. United Eur Gastroenterol J 2018;6:349–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kormann MSD, Ferstl R, Depner M, Klopp N, Spiller S, Illig T et al. Rare TLR2 mutations reduce TLR2 receptor function and can increase atopy risk. Allergy Eur J Allergy Clin Immunol 2009;64:636–642. [DOI] [PubMed] [Google Scholar]
- 58.Smit LAM, Bongers SIM, Ruven HJT, Rijkers GT, Wouters IM, Heederik D et al. Atopy and new-onset asthma in young Danish farmers and CD14, TLR2, and TLR4 genetic polymorphisms: A nested case-control study. Clin Exp Allergy 2007;37:1602–1608. [DOI] [PubMed] [Google Scholar]
- 59.Kormann MSD, Depner M, Hartl D, Klopp N, Illig T, Adamski J et al. Toll-like receptor heterodimer variants protect from childhood asthma. J Allergy Clin Immunol 2008;122:86–92.e8. [DOI] [PubMed] [Google Scholar]
- 60.Potaczek DP, Nastalek M, Okumura K, Wojas-Pelc A, Undas A, Nishiyama C. An association of TLR2-16934A>T polymorphism and severity/phenotype of atopic dermatitis. J Eur Acad Dermatology Venereol 2011;25:715–721. [DOI] [PubMed] [Google Scholar]
- 61.Lau MYZ, Dharmage SC, Burgess JA, Win AK, Lowe AJ, Lodge C et al. The interaction between farming/rural environment and TLR2, TLR4, TLR6 and CD14 genetic polymorphisms in relation to early- and late-onset asthma. Sci Rep 2017;7:43681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Eder W, Klimecki W, Yu L, Von Mutius E, Riedler J, Braun-Fahrländer C et al. Toll-like receptor 2 as a major gene for asthma in children of European farmers. J Allergy Clin Immunol 2004;113:482–488. [DOI] [PubMed] [Google Scholar]
- 63.Tizaoui K, Kaabachi W, Hamzaoui K, Hamzaoui A. Association of single nucleotide polymorphisms in toll-like receptor genes with asthma risk: A systematic review and meta-analysis. Allergy, Asthma Immunol Res 2014;7:130–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Salpietro C, Rigoli L, Miraglia Del Giudice M, Cuppari C, Di Bella C, Salpietro A, Maiello N, La Rosa M, Marseglia GL, Leonardi S, Briuglia SCG. TLR2 and TLR4 gene polymorphisms and atopic dermatitis in Italian children: a multicenter study. 2011 [DOI] [PubMed]
- 65.Oh D-YY, Schumann RR, Hamann L, Neumann K, Worm M, Heine G. Association of the toll-like receptor 2 A-16934T promoter polymorphism with severe atopic dermatitis. Allergy Eur J Allergy Clin Immunol 2009;64:1608–1615. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


