The interaction between the immune system and the nervous system has been at the center of multiple research studies in recent years. Whereas the role played by cytokines as neuronal mediators is no longer contested, the mechanisms by which cytokines modulate pain processing remain to be elucidated. In this study, we have analyzed the involvement of granulocyte-macrophage colony stimulating factor (GM-CSF) in nociceptor activation in male and female mice.
Keywords: chronic pain, GM-CSF, neuroimmune interaction
Abstract
The interaction between the immune system and the nervous system has been at the center of multiple research studies in recent years. Whereas the role played by cytokines as neuronal mediators is no longer contested, the mechanisms by which cytokines modulate pain processing remain to be elucidated. In this study, we have analyzed the involvement of granulocyte-macrophage colony stimulating factor (GM-CSF) in nociceptor activation in male and female mice. Previous studies have suggested GM-CSF might directly activate neurons. However, here we established the absence of a functional GM-CSF receptor in murine nociceptors, and suggest an indirect mechanism of action, via immune cells. We report that GM-CSF applied directly to magnetically purified nociceptors does not induce any transcriptional changes in nociceptive genes. In contrast, conditioned medium from GM-CSF-treated murine macrophages was able to drive nociceptor transcription. We also found that conditioned medium from nociceptors treated with the well established pain mediator, nerve growth factor, could also modify macrophage gene transcription, providing further evidence for a bidirectional crosstalk.
SIGNIFICANCE STATEMENT The interaction of the immune system and the nervous system is known to play an important role in the development and maintenance of chronic pain disorders. Elucidating the mechanisms of these interactions is an important step toward understanding, and therefore treating, chronic pain disorders. This study provides evidence for a two-way crosstalk between macrophages and nociceptors in the peripheral nervous system, which may contribute to the sensitization of nociceptors by cytokines in pain development.
Introduction
Chronic pain is a debilitating condition affecting large numbers of people (Phillips, 2009), with the prevalence in Europe estimated to be ∼20% (Breivik et al., 2006). More surprising perhaps is that >50% of those suffering do not respond or get effective relief with current treatments (Nicol et al., 2018). Over the last decade, considerable advances have been made toward understanding the neurobiological mechanisms underlying chronic pain, with several promising trials of new classes of drug (Brown et al., 2012; Ford, 2012; Schwertner et al., 2013).
Substantial evidence has been presented to suggest that the interaction between neurons and immune cells can result in pain-related conditions stemming from the activation of nociceptors by immune system mediators (Marchand et al., 2005; Cook et al., 2018; Hore and Denk, 2019). Cytokines are also potent neuromodulators that are capable of activation and sensitization of nociceptors (Moalem and Tracey, 2006; Scholz and Woolf, 2007). One such mediator that we have chosen to investigate in this study is granulocyte-macrophage colony stimulating factor (GM-CSF).
GM-CSF has been shown to act as a proinflammatory cytokine (Hamilton, 2008). GM-CSF can enhance antigen presentation and drive macrophages into a proinflammatory phenotype that produces inflammatory cytokines such as TNF, IL-6, IL-1β, and CCL17 (Cook et al., 2004; Fleetwood et al., 2007; Hamilton, 2008; Metcalf, 2008; Achuthan et al., 2016; Wicks and Roberts, 2016). GM-CSF signaling requires the presence of the GM-CSF receptor (CSF2R), a heterodimer made up of a low-affinity ligand binding α chain (CSF2Rα) and the signal transducing β chain (CSF2Rβ) in a ternary complex (Hamilton, 2008; Hansen et al., 2008; Broughton et al., 2016). Downstream signaling of GM-CSF involves the Ras/MAPK pathway as well as the JAK/STAT pathway (Hansen et al., 2008; Broughton et al., 2016).
Within the CNS, GM-CSF has been shown to play a neuroinflammatory role by activating microglia (Parajul et al., 2012; Nicol et al., 2018). The expression of GM-CSFR has also been shown to be increased in infiltrating macrophages and in microglia-like cells in human spinal cord of patients with multiple sclerosis (Donatien et al., 2018). Inhibition of GM-CSF signaling was found to attenuate arthritic pain (Cook et al., 2012). Additionally, silencing GM-CSF and the gene for its receptor resulted in analgesic effects in models of bone cancer and inflammatory pain (Schweizerhof et al., 2009; Cook et al., 2013). Functional studies have shown that injection of GM-CSF into the paw of laboratory animals produces pain-related behavior (Schweizerhof et al., 2009; Achuthan et al., 2016).
However, the pathways and mechanisms behind GM-CSF mediated pain remain elusive (Wicks and Roberts, 2016). There have been claims that the receptor for GM-CSF is expressed in the peripheral nervous system, suggesting that GM-CSF could directly activate nociceptors and thereby drive pain and hyperalgesia (Schweizerhof et al., 2009; Bali et al., 2013). However, multiple recent high-throughput RNA sequencing studies suggest that neurons in the dorsal root ganglion (DRG) express the CSF2Rα transcript at very low levels but do not express any CSF2Rβ (Thakur et al., 2014; Flegel et al., 2015; Lopes et al., 2017; Zeisel et al., 2018). Because both receptor subunits are needed for GM-CSF signaling, these datasets suggest that any effect of GM-CSF on neurons would have to be indirect, i.e., via another cell type. Many immune cells found in neuronal tissues do express appropriate receptors. Many studies of GM-CSF have to date studied systems containing multiple cell types, making it difficult to identify direct versus indirect effects.
This study addresses this discrepancy and seeks to elucidate the mechanism behind the activation of nociceptors by GM-CSF. It demonstrates that GM-CSF can exert an indirect effect on nociceptors via macrophages. We show that pain-related genes are transcriptionally upregulated by conditioned media from bone marrow-derived macrophages (BMDMs) treated in vitro with GM-CSF. Hence, although GM-CSF may be incapable of directly activating nociceptors, it can do so indirectly, and contribute to the algesic effects of GM-CSF.
Materials and Methods
Animals.
For most experiments, adult female C57BL/6J mice 6–8 weeks of age, weighing ∼20–25 g were ordered from Envigo. The animals were housed with a 12 h light/dark cycle with lights on between 7:00 A.M. and 7:00 P.M. and unrestricted access to food and water. Animals were housed in groups of 4–8 and cared for in accordance to the United Kingdom Animals Scientific Procedures Act (1986).
In some experiments, adult male and female C57BL/6J mice from the Walter and Eliza Hall Institute were used. Nav1.8-cre Csf2rbfl/fl mice were generated by crossing the Csf2rbfl/fl mouse (Croxford et al., 2015) with the Nav1.8-cre mouse (gift from J. N. Wood, Institute for Biomedical Research, University College London, London; described by Stirling et al., 2005), i.e., mice with any GM-CSFR expression deleted in Nav1.8+ neurons. Where appropriate, experiments were approved by The University of Melbourne Animal Ethics Committee.
Isolation of DRGs and their dissociation by magnetic separation.
Adult female C57BL/6J mice were killed with an overdose of pentobarbital and death confirmed by decapitation. The DRG were taken from all vertebral levels as previously described (Malin et al., 2007). DRG were washed in F12 medium and then dissociated by enzymatic digestion, followed by gentle mechanical dissociation (Thakur et al., 2014). The single-cell suspension was exposed to a biotinylated non-neuronal antibody mixture (Miltenyi MACS Neuron Isolation Kit), followed by antibiotin microbeads (Miltenyi MACS Neuron Isolation Kit). Cells were then run through a LD exclusion column and placed in a QuadroMACS separator (Miltenyi Biotec) so that only neuronal cells were eluted (>95% pure neuronal cells generated). Neurons were then plated on Matrigel-coated coverslips and cultured for 48 h (5% CO2, 95% O2, at 37°C) in medium with different stimuli as discussed in the following sections on cell culture. For the initial set of experiments, magnetically-activated cell sorting (MACS) nociceptor cultures were prepared in parallel to traditional whole DRG cultures. These were treated for 48 h with either mouse GM-CSF (2 μg/ml; Peprotech) or, as a positive control, mouse 2.5S nerve growth factor (NGF; 10 ng/ml; Alomone Labs).
BMDM isolation and cell culture.
Adult female C57BL/6J mice were killed with pentobarbital and death confirmed by decapitation. The lower body was sterilized with 70% ethanol. The skin, muscles and fat surrounding femur, tibia, and fibula were removed, and the bones collected in cold DMEM. The bones were flushed with 5–10 ml of cold PBS and the cells collected, resuspended and plated in DMEM containing 10% FBS, 1% penicillin–streptomycin (Sigma-Aldrich) and macrophage-CSF (M-CSF; CSF-1; PeproTech). Cultures were maintained for 1 week at 37°C (5% CO2/95% O2). Once confluent, cells were incubated with non-enzymatic cell dissociation buffer (Millipore) at 37°C for 10 min, scraped carefully and re-plated at a density of 30,000–50,000 cells per well in DMEM containing M-CSF. Twenty-four hours later, the medium was replaced with M-CSF-free medium and cells were treated with either GM-CSF (2 μg/ml) or LPS (100 ng/ml) for 48 h.
Cross stimulation of nociceptor and BMDM cultures.
To look for indirect effects of mediators on pure nociceptors and BMDMs, MACS-sorted neurons and BMDMs were cultured for 48 h with either media alone, GM-CSF, or, as a positive control, NGF (for neurons) or LPS (for BMDMs). Forty-eight hours later, fresh cultures of MACS-sorted neurons and BMDMs were plated, as described. Supernatants from the neurons treated for 48 h were added to the fresh BMDM cultures, and similarly supernatants from the BMDMs treated for 48 h were added to the fresh neuron cultures. Supernatants were centrifuged to remove any cells and then 1 ml was added to the respective wells. These were further cultured for 24 h, following which cells were taken for RNA extraction and gene expression analysis.
RNA extraction and TaqMan qPCR array cards.
In each of the experiments, cells were lysed and RNA was extracted from cultured whole DRG and MACS-sorted DRG samples using the RNeasy microkit (Qiagen) following the manufacturer's protocol with some minor modifications. RNA integrity was assessed on the Agilent 2100 Bioanalyzer Pico Chip (Agilent). The RNA integrity number (RIN) for each of the samples used was >8. Samples with a RIN of <8 were not used for qPCR analysis. Following RNA extraction, the samples were amplified and reverse transcribed using the Repli-g WTA single-cell amplification kit (Qiagen). The cDNA was used for gene expression analysis by using the TaqMan custom-made microfluidic array cards (ThermoFisher). These custom-made cards were designed in-house and contained primers and probes to detect 45 test genes as well as three housekeeping genes for reference [18S, GAPDH, and Ywhaz (B2M in macrophage card)]. Three types of cards were used in this study. The first card, used to look for differences between whole DRG and MACS-sorted samples, contained probe sets for a mixture of neuronal and non-neuronal genes known to be present in the DRG that can be activated by NGF and other mediators. These include genes such as TRPVI and TRPA1, ion channels widely expressed on neuronal cells known to be involved in nociception (Caterina and Julius, 2001; Bevan et al., 2014; Huang et al., 2017). In addition, the array card contained probe sets for some cytokine and chemokine genes. The second card contained probe sets for genes that are known to be specifically involved in axotomy and pain-related behavior. These included neuropeptides, such as galanin and neuropeptide Y, known for their role in nociception (Kerr et al., 2000; Brothers and Wahlestedt, 2010), proteins such as annexin 1 and ADAM8 known for their role in modulating inflammatory pain (Schlomann et al., 2000; Chen et al., 2014) in addition to other markers associated with pain such as CSF-1, BDNF, and NGF. Finally, the third card contained probe sets for genes that are present in macrophages. They include canonical inflammatory mediators such as IL6, TNF, and CCL17 (Laskin, 2009). The transcripts measured by each card are given in Table 1.
Table 1.
Genes probe sets present on qPCR array cards
| A | B | C | |||
|---|---|---|---|---|---|
| Adcyap1 | Tac1 | Gapdh | Sfpq | Arg1 | Il4ra |
| Atf3 | Trpa1 | Ywhaz | Scn10a | B2m | Il6 |
| Bdnf | Trpv1 | Hbb | Calca | Gapdh | Irf4 |
| Cacna2d1 | Gapdh | Fabp7 | Hoxb5 | Ccl17 | Irf5 |
| Calca | Ywhaz | Sox10 | Kcnt1 | Ccl22 | Mertk |
| Ccl2 | Il6st | CCL21b | Scn4a | Ccl24 | Mmp9 |
| Nos1 | Ccl4 | Csf1 | Prdm12 | Ccr2 | Nfkbiz |
| Vgf | Il6 | Il34 | Gamt | Ccr6 | Nos2 |
| Gal | Il11 | Gap43 | Prmt8 | Cd19 | Ppard |
| Gch1 | Stat3 | Gal | Ngf | Fcgr1 | Pparg |
| 18S | Tnf | 18S | Areg | 18S | Ptgs2 |
| Ngf | Tlr4 | Bdnf | Il6 | Chil3 | Retnlb |
| Ngfr | Il1b | Sema6a | Vgf | Cybb | Sbno2 |
| Npy | Ccl3 | Npy | Dpysl5 | Foxp3 | Socs1 |
| Ntrk1 | Ccl5 | Nts | Jak2 | Gata3 | Socs2 |
| Ntrk2 | Cxcl12 | Npy2r | Srrm4 | Gata6 | Socs3 |
| Ntrk3 | Il18 | Star | Camk1 | Ido1 | Sox10 |
| Oprm1 | Areg | Adam8 | Usp18 | Ifng | Stat1 |
| P2rx3 | Csf1 | Casp3 | Ntrk1 | Il10 | Stat6 |
| P2rx4 | Csf3 | Atf3 | Ucn | Il12a | Tbx21 |
| Il6ra | Csf2ra | Cacna2d1 | Jun | Il1b | Dpysl5 |
| Scn10a | Ccl20 | P2rx3 | Anxa1 | Il22 | Tgfb2 |
| Scn11a | Il17a | Kcnmb1 | Ngfr | Il27 | Tnf |
| Scn9a | Ereg | Dnm3 | Tnfsf12 | Il4 | Nfil3 |
A, Genes represented on a DRG card. B, Genes represented on an axotomy card. C, Genes represented on a macrophage card.
Each cDNA sample was quantified using a Qubit BR ssDNA assay kit and diluted in PCR grade water to a final concentration of 6 ng/μl. This was added to TaqMan Universal 2x Master mix (ThermoFisher) to achieve a final volume of 100 μl. TaqMan array cards were run on a 7900HT Fast Real-Time PCR system (Applied Biosystems) and gene expression calculated using the ddCT method (normalizing each sample to the average of the three housekeeping genes and then to their respective internal controls, usually the unstimulated/untreated samples). Samples with cycling thresholds of 40 in the unstimulated conditions were not included in the analysis.
Measurement of [Ca2+]i in DRG neuron.
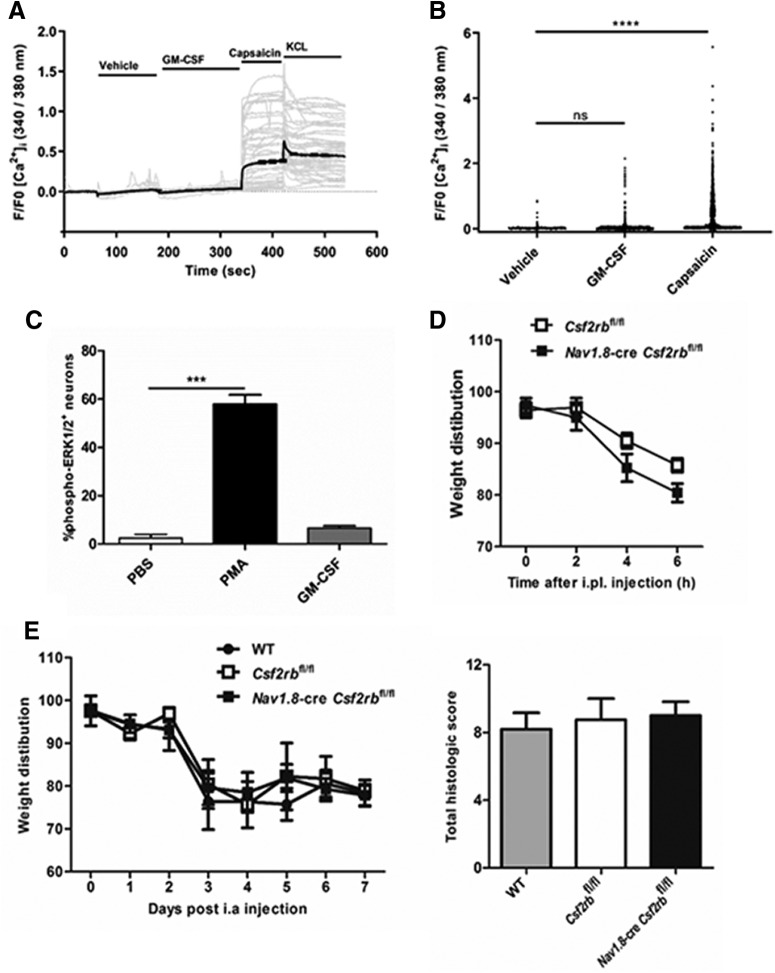
Mouse DRG neurons were dissociated from whole DRGs as previously described (Rajasekhar et al., 2015) and plated onto coverslips coated with poly-l-lysine and 100 μg/ml laminin. The DRG neurons were maintained in DMEM containing antibiotic-antimitotic, 10% FBS, and N-1 supplement at 37°C (5% CO2/95% O2) for 24 h. The DRG neurons were loaded with Fura-2/AM ester (5 μm, 45 min, 37°C) in calcium assay buffer (10 mm HEPES, 0.5% BSA, 10 mm[scap] [scap]d[scap]-glucose, 2.2 mm CaCl2·6H2O, 2.6 mm KCl, 150 mm NaCl) containing 4 mm probenecid and 0.05% pluronic F127. Cells were washed and incubated in calcium assay buffer for 30 min before imaging. Cells were observed using a Leica DMI-6000B microscope with an HC PLAN APO 0.4 numerical aperture 10× objective and maintained at 37°C. Images were collected at 1 s intervals (excitation: 340 nm/380 nm; emission: 530 nm). Cells were challenged sequentially with vehicle, GM-CSF (200 ng/ml), capsaicin (0.5 μm; TRPV1 agonist). KCl (50 mm) in calcium assay buffer containing probenecid, was applied at the end of the experiment to obtain maximal [Ca2+]i].
Results are expressed as the 340/380 nm fluorescence emission ratio, which is proportional to changes in [Ca2+]i. Data are presented as F/F0, where F is the measured fluorescence intensity and F0 is the basal fluorescence. All F/F0 values have been subtracted by 1. In each experiment two technical replicates were included with 68–559 neurons recorded in each repeat. The experiment was repeated three times (n = 3) with equivalent results. A response was deemed positive if it was ≥10% above baseline. Results were excluded from the analysis if they showed a fluctuating calcium response before addition of GM-CSF or did not show pronounced reversibility (>50%) from the peak response to GM-CSF application and did not respond to KCL addition. This constituted <1% of DRG neurons studied.
Detection of ERK1/2 and STAT5 activation in neurons stimulated with GM-CSF.
The dissociated DRG neurons plated onto coverslips, as described above for measurement of [Ca2+] (Rajasekhar et al., 2015), were also used for the detection of ERK1/2 and STAT5 activation following GM-CSF stimulation. Following a 24 h culture in DMEM containing antibiotic-antimitotic, 10% FBS, and N-1 supplement at 37°C (5% CO2/95% O2), the neurons were serum-starved overnight (17–18 h) by incubating them in DMEM supplemented with 0.1% (w/v) BSA, 100 U/ml penicillin, 100 mg/ml streptomycin, and 1% (v/v) N1 in a humidified incubator at 37°C (95% O2, 5% for CO2). Subsequently, neurons were stimulated for 15 min with PBS, GM-CSF (200 ng/ml) or PMA (2 μm; Sigma-Aldrich). Cells were then washed in ice-cold PBS and fixed in 4% paraformaldehyde in PBS for 20 min at room temperature. After three washes with PBS, cells were blocked and permeabilized by incubating with PBS supplemented with 0.01% Triton-X, 5% heat-inactivated FBS, and 5% goat serum for 60 min. Neurons were washed (3× PBS), then stained overnight with mouse anti-mouse NeuN mAb (clone A60; Millipore) in combination with either rabbit anti-mouse phospho-p44/42 MAPK (Erk1/2) (Thr202/Tyr204) (197G2) mAb (Cell Signaling Technology) or rabbit anti-mouse phospho-STAT5 (Y694) (D47E7) XP mAb (Cell Signaling Technology); all primary antibodies were diluted in PBS with 5% FBS and 0.01% Triton-X. Following washing (3× PBS), neurons were incubated with goat anti-rabbit IgG (H+L) antibody, AlexaFluor 568 conjugate (ThermoFisher) and goat anti-mouse IgG (H+L) antibody, Alexa Fluor488 conjugate (ThermoFisher). Neurons were washed (3× PBS), then stained with DAPI (1 μg/ml, 5 min; EMD Millipore). In all experiments, secondary antibody only and single primary antibody controls were included to check for nonspecific secondary binding and bleed-through of fluorochromes, respectively.
Images were obtained with a Zeiss Axioskop 2 at 10× magnification and captured by a Zeiss AxioCam MRm. Each condition included two technical replicates and five images were taken from each replicate. Quantification of positive cells was performed with ImageJ software. For neurons, only NeuN-positive cells were included in the analysis. To determine when cells were positive a lower threshold for staining intensity in the green channel (AlexaFluor 488) was set based on the PBS-treated control cells. Cells with fluorescence intensities above this threshold were regarded as positive. A mean of positive cells across the 10 images from each condition was calculated. Three separate experiments were performed.
GM-CSF-induced inflammatory pain.
Inflammatory pain was induced by a single intraplantar injection (10 μl) of GM-CSF (50 ng/paw; R&D Systems) into the left hind footpad (Achuthan et al., 2016; Cook and Hamilton, 2018).
mBSA/GM-CSF-induced arthritis.
Monoarticular arthritis was induced by an intraarticular injection of methylated BSA (mBSA; 100 μg in 10 μl) into the right knee on Day 0, and saline into the left knee, followed by a subcutaneous injection of GM-CSF (600 ng) into the scruff of the neck on Days 0–2, as before (Achuthan et al., 2016; Cook and Hamilton, 2018). Mice were killed (Day 7) and knee joints were removed, fixed, decalcified, and paraffin embedded (Achuthan et al., 2016; Cook and Hamilton, 2018). Frontal sections (7 μm) were stained with H&E and cellular infiltration, synovitis, pannus formation, cartilage damage, and bone erosions were each scored separately from 0 (normal) to 5 (severe) as described previously (Achuthan et al., 2016; Cook and Hamilton, 2018); these scores were then added to give the total histologic score for each mouse.
Assessment of pain-related behaviors.
As an indicator of pain, the differential weight distribution over a 3 s period between the inflamed paw or limb relative to the non-inflamed paw or limb was measured using the incapacitance meter (IITC Life Science). This technique has been validated for measurement of both paw and arthritic knee pain (Achuthan et al., 2016; Cook and Hamilton, 2018). Mice were acclimatized to the incapacitance meter on at least 3 separate days before the commencement of the experiment. Three measurements were taken for each time point and averaged.
Experimental design and statistical analysis.
All data are expressed as mean ± SEM, except where stated as median. Statistical analyses were performed using SPSS v23 (IBM). Kruskal–Wallis nonparametric independent samples tests were used for analysis of Figures 1, 3, and 4. The samples were corrected for multiple testing using the Bonferroni correction. For calcium imaging in Figure 2, GM-CSF activation of neurons and histology, a one-way ANOVA was used, and for pain readings, a two-way ANOVA was used, with either a Bonferroni or Tukey post hoc test. A p value <0.05 was considered significantly different to the null hypothesis of no difference at the 95% confidence level.
Figure 1.

GM-CSF causes dysregulation of genes in mixed DRG cultures but not in purified neuronal cultures. A, Heatmap representing the transcriptional changes in a panel of genes (Table 2) was assessed in mixed DRG cultures and pure MACS sorted neuronal cultures from C57BL/6J mice following treatment with GM-CSF (2 μg/ml) for 48 h and NGF (10 ng/ml). Each column represents average data of n = 8 independent experiments. Each individual experiment contained pooled cells from two mice. B, Genes showing a twofold or greater change in expression changes following GM-CSF treatment in unsorted DRG cell cultures as compared with purified neurons. Each dot represents a separate gene which is an average of n = 8 experiments. Dotted line represents untreated control. Solid line represents mean of each group. Kruskal–Wallis test was conducted to identify genes that were significantly modulated after treatment with GM-CSF in mixed DRG cultures (highlighted black dots). The results were corrected for multiple comparisons using the Bonferroni correction. None of the genes from purified neuronal cultures reached statistical significance after GM-CSF treatment. *adjusted p < 0.05; *genes significantly different from untreated control. +adjusted p < 0.05 and ++adjusted p < 0.01; +genes significantly different between whole DRG and purified neurons.
Figure 3.
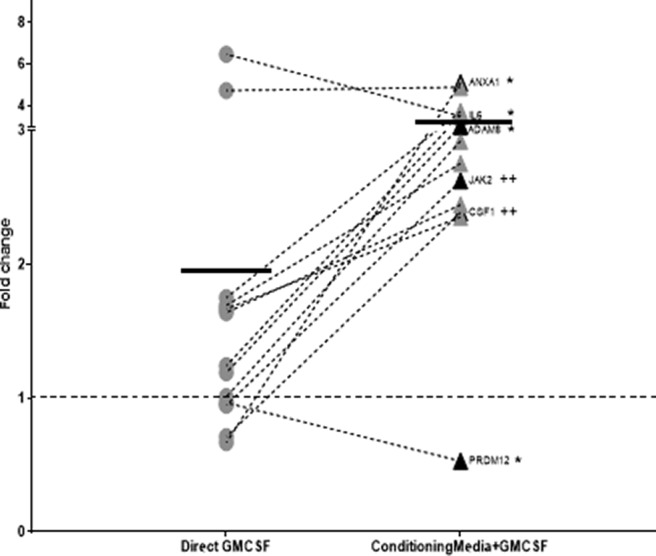
Nociceptor gene expression can be indirectly modulated by conditioning media from GM-CSF stimulated BMDMs. Genes dysregulated by twofold or more from nociceptors that received conditioning medium from GM-CSF (2 μg/ml) treated BMDMs. Each dot represents a separate gene which is an average of n = 10 individual experiments. Kruskal–Wallis test was conducted to identify genes that were significantly modulated after treatment with the conditioning medium (highlighted black dots). The results were corrected for multiple comparisons using the Bonferroni correction. None of the genes from purified neuronal cultures that were treated directly with GM-CSF (2 μg/ml) reached statistical significance. Samples with cycling thresholds of 40 in the unstimulated conditions were not included in the analysis. Dotted line represents untreated control. Solid line represents mean of each group. No significant changes were seen with untreated conditioning media control from BMDMs on neuronal cultures. *adjusted p < 0.05; *genes significantly different from untreated control. ++adjusted p < 0.01; +genes significantly different between direct GM-CSF stimulation and conditioning media with GM-CSF.
Figure 4.
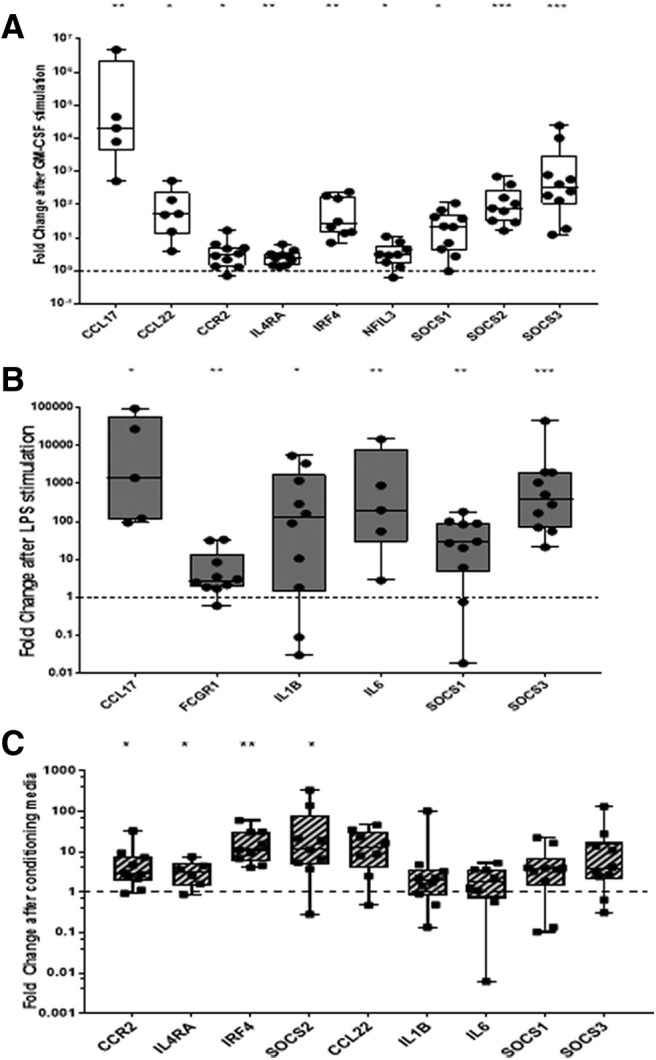
Macrophage gene expression can be indirectly modulated by NGF stimulated nociceptors. BMDMs were treated with (A) GM-CSF, (B) LPS, and (C) conditioning medium from NGF-stimulated nociceptors for 48 h (see Materials and Methods). The fold-change in the expression of dysregulated genes on a macrophage card (Table 1, column C) is depicted on a log10 scale. Only significantly dysregulated genes are depicted in A and B. Each dot represents a separate experiment (n = 10). Kruskal–Wallis test was conducted to identify genes that were significantly modulated after treatment. The results were corrected for multiple comparisons using the Bonferroni correction. Dotted line represents untreated controls where conditioning media from untreated nociceptors was applied to BMDMs. No significant changes were seen in BMDM cultures treated with conditioning medium from untreated nociceptors as a control. Box-and-whisker plots showing maximum to minimum range. Samples with cycling thresholds of 40 in the unstimulated conditions were not included in the analysis. *adjusted p < 0.05, **adjusted p < 0.01, ***adjusted p < 0.001.
Figure 2.
GM-CSF does not directly activate neurons in vitro and in vivo. A, B, Time course and peak Ca2+ responses in mixed DRG cultures in response to vehicle, GM-CSF (200 ng/ml), capsaicin (0.5 μm), and KCl (50 mm; only A), respectively. A, Gray lines, Individual traces from 50 random cells; black lines, mean response; (B) n = 1767 neurons (pooled data from two independent experiments). C, Percentage of DRG neurons positive for phospho-ERK1/2 following stimulation with PBS, PMA, or GM-CSF (200 ng/ml) for 15 min. Three independent experiments were performed. D, E, Pain development (incapacitance meter, ratio of weight bearing on injected relative to non-injected knee/hindpaw, a value <100 indicates pain) was measured following (D) intra-planatar (i.pl.) injection of GM-CSF (20 ng) in Csf2rbfl/fl and Nav1.8-cre Csf2rbfl/fl mice (n = 5–8 mice/group); and (E) mBSA/GM-CSF arthritis [mBSA intra-articular (i.a.) (Day 0); GM-CSF or saline subcutaneously (Days 0–2)] induction in WT, Csf2rbfl/fl, and Nav1.8-cre Csf2rbfl/fl mice (n = 4–7 mice/group). Arthritis (histology, Day 7) was also assessed in E. C–E, Data are expressed as mean ± SEM. For B and C, a one-way ANOVA was used. ***p < 0.001, ****p < 0.0001.
Results
The literature around the involvement of GM-CSF in chronic and neuropathic pain remains sparse. However, even within this limited literature there is little consensus on the possible mechanisms behind the actions of GM-CSF in pain. To clarify, we have undertaken a number of experiments, as follows in the next sections.
GM-CSF does not modulate gene expression in purified neurons from mouse DRG
Previous studies have reported that GM-CSF can act directly on nociceptive neurons, and as a result, cause hyperalgesia (Schweizerhof et al., 2009; Parajul et al., 2012). Here, we began by addressing the discrepancy in the literature on the mode of action of GM-CSF by using MACS to enrich for small and medium diameter neurons (which are nearly all nociceptors) from mouse DRG. Thakur et al. (2014) showed that dissociated DRG preparations that are commonly used for analysis actually contain predominantly non-neuronal cells. In contrast, they showed, that following MACS isolation, a culture of 95% pure nociceptors can be produced from adult mouse DRG. Large diameter neurons (>30 μm), which are lost during MACS, are largely non-nociceptive (Dubin and Patapoutian, 2010), and hence their absence is an asset rather than a disadvantage when studying the role of GM-CSF is nociception and peripheral sensitization.
Parallel cultures of cells from adult mouse DRG were set up using either the traditional dissociation technique to prepare the mixed (i.e., unsorted) cultures and purified cultures (i.e., sorted) from adult mouse DRG obtained after MACS. For these sets of experiments, 48 genes that are known to be expressed in the DRG, including some internal housekeeping controls (GAPDH, 18S and YWhAZ), were developed into a Taqman qPCR array card (Thermofisher). This card was used as a screening tool to help provide an indication toward specific pathways or areas of interest to be investigated further. The list of genes present on the card is provided in Table 1, column A.
Figure 1A is a heatmap that shows the gene expression changes in mixed DRG cultures and pure neurons following GM-CSF and NGF treatment in the panel tested. It is evident that GM-CSF had an overall greater impact in mixed cultures as compared with pure neuronal cultures. Only 6% of the genes (n = 2/34, namely CSF2RA and IL6) showing a >2-fold average increase in expression level following GM-CSF treatment in the purified neuronal cultures and none of the differences reached statistical significance.
However, when GM-CSF was applied to the mixed DRG cultures, 44% of the genes (n = 15/34) showed a twofold or more average increase in gene expression, and four of these were found to reach statistical significance with an average increase in expression of 2.3-fold. Figure 1B shows the significantly altered genes (black dots) along with those showing a >2-fold increase in expression. The overall average increase in gene expression in the mixed cultures with GM-CSF stimulation was 3.9-fold, whereas purified cultures following GM-CSF stimulation showed an average of 1.6-fold increase.
As a positive control, we applied NGF instead of GM-CSF to the mixed and purified cultures and found, as expected, a significantly increased expression of 12 and 5 genes, respectively. Fifty percent of the genes showed a twofold or greater average increase in expression in the mixed DRG cultures, whereas ∼32% of the genes in purified cultures showed a two-fold or more average increase in expression. The average fold increase of the significant genes was 5.8- and 2.5-fold in the mixed and purified cell cultures, respectively (data not shown).
These results suggest that GM-CSF is incapable of driving direct transcriptional changes in neuronal genes in nociceptors. However, changes in neuronal genes in the mixed cultures following GM-CSF treatment indicate that it might be having an indirect effect on nociceptors via satellite cells or other non-neuronal cell types that make up the majority of the cells in the DRG, and indeed in the mixed DRG cultures. To obtain supporting evidence for the proposal that GM-CSF is incapable of directly stimulating nociceptor transcription, we reviewed recent publications that have made use of RNA sequencing to examine gene expression in mouse and human DRG (Tables 2, 3; Thakur et al., 2014; Flegel et al., 2015; Lopes et al., 2017; Ray et al., 2018, 2019; Zeisel et al., 2018). The Table compares the expression of the two GM-CSF receptor chains to several control transcripts: Calca, one of the most highly expressed genes in DRG; TrpV1 and Nav1.8, which are well expressed in nociceptive neurons; and Dnmt3a, which is very lowly expressed (Saunders et al., 2018). It is evident the two transcripts coding for the receptor chains of the GM-CSF receptor, namely CSF2Rα and CSF2Rβ, are expressed at levels below our negative control transcript in the DRG, the CSF2Rβ gene, in particular, appears to be undetectable, even by a technique as sensitive as RNA-seq. In whole human tibial nerve, mRNA for both receptors can be detected at higher levels, presumably because of a contribution from non-neuronal cells (Ray et al., 2019).
Table 2.
GM-CSF receptor subunit expression in neurons by RNA sequencing[1]
| Bulk-sequencing | |||||||
|---|---|---|---|---|---|---|---|
| Expression Units | Mouse tissue |
Human tissue |
|||||
|
Thakur et al., 2014; MACS-sorted nociceptors |
Lopes et al., 2017; MACS-sorted nociceptors after nerve injury |
Lopes et al., 2017; FACS-sorted nociceptors |
Flegel et al., 2015; Whole DRG |
Ray et al., 2018; Whole DRG |
Ray et al., 2019; Human tibial nerve |
||
| FPKM | FPKM | FPKM | FPKM | TPM | TPM | ||
| Csf2ra | 4 | 3 | 2 | 0 | 0 | CSF2RA | 15 |
| Csf2rb | 1 | 0 | 0 | 1 | 1 | CSF2RB | 6 |
| Calca | 912 | 3987 | 10287 | 313 | 1701 | CD40 | 49 |
| TrpV1 | 58 | 154 | 112 | 48 | 73 | TRPV1 | 7 |
| Dnmt3a | 4 | 2 | 2 | 5 | 4 | UCHL1 | 92 |
Expression values derived from publicly available bulk RNA-sequencing datasets. Data for Csf2ra and Csf2rb are provided along with the following control/comparison genes: Calca, which is one of the most highly expressed genes in DRG; TrpV1, which is well expressed in nociceptive neurons; Dnmt3a, which is very lowly expressed if at all in neurons (Saunders et al., 2018); Nav1.8; CD40, a myeloid cell marker; and Uchl1, the gene coding for a protein which is highly expressed in nerve fibers,. FPKM, Fragments per kilobase per million mapped reads; TPM, transcripts per million.
Table 3.
Single-cell Sequencing of mouse DRG (Zeisel et al; mousebrain.org): Trinarization scores
| Csf2ra | Csf2rb | Calca | TrpV1 | Dnmt3a | Nav1.8 | |
|---|---|---|---|---|---|---|
| Peptidergic (TrpM8), DRG | 0.18 | 0 | 0.39 | 2.21 | 0.36 | 0 |
| Peptidergic (TrpM8), DRG | 0.27 | 0 | 0.32 | 0.67 | 0.11 | 0.08 |
| Peptidergic (TrpM8), DRG | 0.11 | 0 | 4.04 | 0.31 | 0.22 | 0 |
| Peptidergic (PEP1.2), DRG | 0.2 | 0 | 11.3 | 3.07 | 0.1 | 0.19 |
| Peptidergic (PEP1.3), DRG | 0.13 | 0 | 43.4 | 2.68 | 0.15 | 1.56 |
| Peptidergic (PEP1.1), DRG | 0.19 | 0 | 37.3 | 1.02 | 0.13 | 1.16 |
| Peptidergic (PEP1.4), DRG | 0.19 | 0 | 52.3 | 3.26 | 0.32 | 2.51 |
| Peptidergic (PEP2), DRG | 0.12 | 0 | 61.6 | 0.56 | 0.24 | 3.38 |
| Neurofilament (NF2/3), DRG | 0 | 0 | 0.64 | 0 | 0.19 | 0.61 |
| Neurofilament (NF4/5), DRG | 0.11 | 0 | 0.07 | 0.05 | 0.35 | 0.04 |
| Neurofilament (NF1), DRG | 0.08 | 0 | 0.07 | 0 | 0.13 | 0.03 |
| Non-peptidergic (TH), DRG | 0.18 | 0 | 0.17 | 0.01 | 0.35 | 1.08 |
| Non-peptidergic (NP1.1), DRG | 0.15 | 0 | 6.38 | 0.06 | 0.33 | 3.71 |
| Non-peptidergic (NP1.2), DRG | 0.22 | 0 | 3.23 | 0.05 | 0.27 | 5.28 |
| Non-peptidergic (NP2.1), DRG | 0.24 | 0 | 11.1 | 0.04 | 0.38 | 5.47 |
| Non-peptidergic (NP2.2), DRG | 0.18 | 0 | 34.5 | 0.73 | 0.27 | 4.99 |
| Non-peptidergic (NP3), DRG | 0.26 | 0 | 0.74 | 1.95 | 0.26 | 4 |
Expression values derived from publicly available B single-cell RNA-sequencing datasets. Data for Csf2ra and Csf2rb are provided along with the following control/comparison genes: Calca, which is one of the most highly expressed genes in DRG; TrpV1, which is well expressed in nociceptive neurons; Dnmt3a, which is very lowly expressed if at all in neurons (Saunders et al., 2018); Nav1.8; CD40, a myeloid cell marker; and Uchl1, the gene coding for a protein which is highly expressed in nerve fibers.
GM-CSF does not directly activate neurons in vitro and in vivo
To support the above gene expression data, suggesting an indirect effect of GM-CSF on neurons, we monitored some signaling pathways in cultured DRG neurons. We were unable to observe any GM-CSF-stimulated elevation in intracellular Ca2+ levels (Fig. 2A,B) or ERK1/2 phosphorylation (Fig. 2C) compared with our positive controls, namely capsaicin and PMA, respectively. We were also unable to detect STAT5 phosphorylation following GM-CSF stimulation in these neurons, unlike in murine macrophages grown from bone marrow cells in GM-CSF (Fleetwood et al., 2007; data not shown).
Table 3 indicates that Nav1.8+ neurons do not express the Csf2rb gene and therefore cannot express a functional GM-CSFR. To demonstrate in vivo that GM-CSF-induced pain development is not due to GM-CSF receptor signaling via Nav1.8+ neuronal cells (that is, the majority of nociceptors), Nav1.8-cre Csf2rbfl/fl mice were generated by crossing the Csf2rbfl/fl mouse (Croxford et al., 2015) with the Nav1.8-cre mouse (Stirling et al., 2005), these mice will lack any functional GM-CSF receptors that may possibility be expressed in Nav1.8+ neurons. GM-CSF-induced inflammatory pain and GM-CSF-induced arthritic pain were then initiated, and pain development measured by a change in weight distribution (using the well validated incapacitance meter method; Achuthan et al., 2016; Cook et al., 2018). Following intraplantar injection of GM-CSF, pain was evident in Csf2rbfl/fl control and also in Nav1.8-cre Csf2rbfl/fl mice (Fig. 2D). Similarly, following induction of mBSA/GM-CSF arthritis, similar pain development was evident in WT, Csf2rbfl/fl control and Nav1.8-cre Csf2rbfl/fl mice from Day 3 onward (Fig. 2E); all three strains developed a similar degree of arthritis (at Day 7, as judged by histology; Fig. 2E). Together, these in vitro and in vivo data do not support a direct action of GM-CSF on neurons consistent with a lack of GM-CSF receptor gene expression in neurons.
Nociceptor gene expression can be indirectly modulated by GM-CSF stimulated BMDMs
As mentioned, based on these data, we hypothesized that GM-CSF might be having an indirect effect on nociceptors via non-neuronal cells that are present within the DRG and in the periphery at a site of injury. Macrophages are one cell type present in the DRG and known to be responsive to GM-CSF as well as being a potential source of pain mediators (Cook et al., 2018; Hore and Denk, 2019). To look for potential indirect effects of GM-CSF, supernatants from GM-CSF-stimulated BMDM cultures were added to sorted neuronal cultures to test whether these BMDMs are capable of producing mediators that can elicit transcriptional changes in neurons. Because our overall aim was to look at the mechanism of GM-CSF action in pain, a second TaqMan card containing probe sets for genes that are known to be involved in axotomy and pain-related behavior was used (Table 1, column B).
Once again, direct treatment of purified nociceptors with GM-CSF did not cause any significant dysregulation in the genes present on this array card (Fig. 3). Conditioning medium from unstimulated BMDMs had no significant impact on neuronal gene transcription (data not shown). Following treatment with conditioning medium from GM-CSF treated BMDMs, 31% of the genes tested showed twofold or more average increase in gene expression, calculated by normalizing the transcriptional changes to neuronal cultures that received supernatants from unstimulated BMDMs. Six genes were found to be significantly dysregulated following indirect stimulation with GM-CSF. These were ADAM8 (3-fold increase), ANXA1 (5-fold increase), IL6 (3.5-fold increase), PRDM12 (0.5-fold decrease), CSF-1 (2.4-fold increase), and JAK2 (2.6-fold increase). In addition to the genes that reached statistical significance, there were several other changes in known pain-related genes, such as TNFSF12 (3.6-fold increase), USP18 (5-fold), GAL (2.9-fold), NGF (2.4-fold), and NPY (2.4-fold), which showed increased expression following indirect activation using GM-CSF treated conditioning medium, but which did not reach statistical significance (Fig. 3).
Macrophage gene expression can be indirectly modulated by NGF stimulated nociceptors
We investigated next the possibility of cross talk between stimulated nociceptors and macrophages. Although there is growing evidence to support the view that stimulated immune cells can communicate with neurons (Watkins and Maier, 2002; Marchand et al., 2005; Scholz and Woolf, 2007; Sorge et al., 2015; Hore and Denk, 2019), which is supported by the data in Figure 3, the literature on the ability of stimulated neurons to communicate with immune cells is more limited (McMahon et al., 2015). To examine this possibility, we used a similar strategy to that used in Figure 3 to explore whether nociceptors that had been treated with NGF were capable of producing mediators that could modulate macrophage gene expression. A third TaqMan card containing 48 genes, of which 29 genes are known to be expressed in macrophages at levels which depend on their functional state (Murray et al., 2014) was used (Table 1, column C).
As positive controls, we found that GM-CSF (Fig. 4A) and LPS (4B) stimulation of BMDMs, as expected, had large impacts on gene transcription. GM-CSF treatment led to 55% of the genes having a twofold or more increase in expression; of these, nine were found to be statistically significant after correcting for multiple testing. They were Ccl17, Ccl22, Ccr2, Il4ra, Irf4, Nfil3, Socs1, Socs2, and Socs3 (Fig. 4A). Additionally, cytokine genes such as Il6, Il1b, and Il27 were also found to be upregulated, although without reaching statistical significance. Stimulation of BMDMs with LPS led to 72% of the genes having a twofold or more increase in expression and, of these, six reached statistical significance, namely, Ccl17, Fcgr1, Il1b, Il6, Socs1, and Socs3 (Fig. 4B).
Conditioning medium from unstimulated neurons had no impact on BMDM gene transcription (data not shown). Conditioning medium from NGF treated nociceptors caused a twofold or more increase in 69% of the genes. Although only four genes reached statistical significance, namely CCR2, IL4Ra, IRF4, and SOCS2 (Fig. 4C). There were several other genes, namely CCL22, IL1b, IL6, SOCS1, and SOCS3, that showed a trend toward increased expression following treatment with NGF-stimulated conditioning medium (Fig. 4C). It should be noted that BMDMs do not express the receptors for NGF (TRKA and p75; e.g., see RNA-seq data by Ostuni et al., 2013; Piccolo et al., 2017; Hill et al., 2018), demonstrating that NGF-stimulated neurons can produce mediators capable of activating macrophages.
Discussion
In this present study we provide evidence that GM-CSF does not directly activate nociceptors but suggest that GM-CSF acts via macrophages to produce mediators that interact with nociceptors. We provide evidence for a bidirectional cross talk between neurons and macrophages.
Previous studies have suggested that GM-CSF can act on and stimulate sensory neurons. Bali et al. (2013) suggested that GM-CSF brought about transcriptional regulation of several pain genes in sensory neurons in a model of cancer pain, an observation replicated by Schweizerhof et al. (2009) and F. Zhang et al. (2019). Donatien et al., 2018 report that GM-CSF can enhance capsaicin-induced calcium influx in DRG neurons, although not directly induce calcium influx. However, these studies did not separate neuronal cells from non-neuronal cells within the DRG and hence it is difficult to attribute these changes specifically to sensory neurons. In contrast, other recent publications (Lopes et al., 2017; Zeisel et al., 2018) making use of RNA-sequencing to look for transcriptional changes in a cell-specific manner have indicated the absence of the GM-CSFRβ chain on nociceptors, indicating alternate mechanisms of action. In this context, a TrkA inhibitor was able to reduce the GM-CSF enhanced capsaicin-induced calcium influx response, suggesting that GM-CSF may be acting indirectly via NGF (Donatien et al., 2018).
Therefore, we looked for changes caused by stimulating purified nociceptors with GM-CSF and found no significant transcriptional changes. Also, even if there was some expression of the GM-CSF receptor on neurons, deleting the Csf2rβ subunit in Nav1.8+ neurons (i.e., most nociceptors) in vivo showed no effect on the generation of GM-CSF-driven inflammatory and arthritic pain, suggesting that GM-CSF does not act directly via nociceptors. It has been reported that low and high threshold Aβ fibers respond to GM-CSF (Schweizerhof et al., 2009). Based on our findings, we consider that these responses are possibly indirect although further studies are needed to address this issue. Overall, our results lead us to hypothesize that the reported effects of GM-CSF on DRGs (Schweizerhof et al., 2009; Bali et al., 2013) were predominantly due to the ability of GM-CSF to activate non-neuronal cells associated with nociceptors, likely in the peripheral nerve itself or during myeloid cell infiltration into the DRG. These non-neuronal cells might then indirectly bring about transcriptional changes in nociceptors associated with pain/hyperalgesia.
Macrophages are one of the most commonly studied cell type in the pain field due to their involvement in the pathogenesis of various neuropathies (Lu and Richardson, 1993). H. Zhang et al. (2016) showed that recruitment of macrophages to the DRG was important for inducing and maintaining chemotherapy-induced peripheral neuropathy, an observation in accordance with several other studies showing increased myeloid cells in the DRG following peripheral injury (Fenzi et al., 2001; Hu and McLachlan, 2002; Hu and McLachlan, 2003). Furthermore, Shepherd et al. (2018) showed that the angiotensin II receptor (AT2R) antagonist reduces neuropathic pain by blocking the downstream signaling of AT2R in infiltrating peripheral macrophages, as sensory neurons lack expression of this receptor. Blocking of macrophage activation using TLR antagonists (Jurga et al., 2018) and inhibitors of p38 MAPK/MMP9 (Mika et al., 2007; Hutchinson et al., 2008), PI3K and NF-kB (Popiolek-Barczyk et al., 2015) has analgesic effects in various models of neuropathic pain, consistent with our proposed mechanism of action.
We therefore analyzed whether factors from stimulated macrophages can bring about transcriptional changes in nociceptors that mimic injured or activated nociceptors. We found that supernatants from GM-CSF stimulated macrophages upregulated several neuronal genes, namely ADAM8, ANXA1, IL6, CSF-1, and JAK2, which are also significantly upregulated following injury (Pei et al., 2011; Chen et al., 2014; Guan et al., 2016; Diaz-delCastillo et al., 2018; Tang et al., 2018). Supernatants from GM-CSF stimulated macrophages were found to significantly downregulate expression of PRDM12, an important nociceptor gene (Desiderio et al., 2019). There is evidence to suggest that, following injury, activated monocytes from the spleen and lymph nodes infiltrate into the site of injury as well as the associated DRG (Hu and McLachlan, 2002). It is expected that inflammatory cytokines from these immune cells can then impact the neurons by affecting their firing rates and causing changes in gene expression (Ohtori et al., 2004; Ozaktay et al., 2006).
Of the mediators that were upregulated in our experimental set up, CSF1 was of particular interest from the perspective of nerve injury. The role of microglia in chronic pain is well established, with various proposed mechanisms to drive microglial activation and central sensitization in a variety of pains states (Calvo and Bennett, 2012; Denk et al., 2016; Fernandez-Zafra et al., 2019). It has been demonstrated that peripheral nerve injury induces the production of CSF-1 in neurons, which then recruit spinal cord microglia to proliferate (Guan et al., 2016). The presence of large numbers of activated microglia is responsible for further activation of spinal neurons and maintenance of neuropathic pain through the release of inflammatory and neuropathic mediators (Kawasaki et al., 2008; Zhao et al., 2017). The release of CSF-1 from nociceptors raises the possibility of bidirectional cross talk with nociceptors further recruiting and stimulating macrophages in a positive feedback loop. Therefore, we looked for transcriptional changes in macrophages following treatment with conditioning media from stimulated neurons.
Analysis of macrophages at a site of nerve injury has shown them to be predominantly anti-inflammatory in nature and involved in regeneration and recovery of the nerve (Gaudet et al., 2011; Ydens et al., 2012). Interestingly, macrophages stimulated with supernatants from NGF treated neurons led to an upregulation of cytokine and chemokine receptors (IL4Ra and CCR2) and transcription factors (SOCS2 and IRF4). Because NGF by itself is incapable of directly activating macrophages (Ostuni et al., 2013; Piccolo et al., 2017; Hill et al., 2018), it can be assumed that the transcriptional changes in macrophages were due to mediators being released by these stimulated nociceptors. Furthermore, these transcriptional changes were distinct from those following direct stimulation with LPS or GM-CSF, suggesting a distinct mechanism of action. We found that NGF stimulated nociceptors upregulate the expression of inflammatory mediators and chemokines, such as IL-1β, IL6, and CCL22, which have the potential to activate and recruit macrophages.
Here we, like many others, have used in vitro dissociated DRG cultures to study nociceptive processes. However, unlike nearly all previous studies, we use highly purified neurons in the culture. This allows us to disambiguate direct versus indirect effects of applied agents; a key advantage and main point of this study. The disadvantage being that the cellular properties inevitably change somewhat over time in culture as seen by transcriptional profiling of such cultures (Thakur et al., 2014; Wangzhou et al., 2019; Lopes et al., 2017). Some of the emergent changes suggest that cultured nociceptors take on a “neuropathic” phenotype (Wangzhou et al., 2019) and so one caveat of the current work is that, inevitably, the neurons we studied are not in their native state.
One of the problems we faced during these experiments was the intra-group variability observed in the transcriptional analysis. Variability in transcriptional analysis is a common phenomenon (Raser and O'Shea, 2005; Volfson et al., 2006) since transcription is not a continuous process, but rather a discontinuous one that takes place in “bursts” and “pulses”. Hence differences in the expression levels of lowly and highly expressed genes can be observed even in the absence of any stimulus leading to the observed variability (Chubb and Liverpool, 2010). In this study, we have made use of stringent statistical tests to cover the inherent intra-group variability and hence identify transcripts that are genuinely dysregulated because of the treatments.
It is important to note in this context, that although nociceptor transcriptional change is very common in persistent pain states, nociceptors can be activated and sensitized without transcriptional change (Wu et al., 2001; J. M. Zhang and Strong, 2008). But transcriptional change in nociceptors, when it does occur, can lead to changes in the sensitivity and activity of these neurons and is thereby an important regulator of nociceptor function. In the current experiment we looked for acute effects of GM-CSF on calcium signaling in purified nociceptors but did not observe any of these non-transcriptional actions. Others who have seen non-transcriptional effects of GM-CSF on cultured neurons have used mixed cultures containing a variety of cell types which may allow for indirect activation of nociceptors via non-neuronal cells (Schweizerhof et al., 2009; Bali et al., 2013; Donatien et al., 2018). Indeed, in those experiments, the non-transcriptional effects of GM-CSF were blocked by trkA inhibitors, suggesting the release of secondary mediators.
In conclusion, the findings in this study highlight the need to dissect the mechanisms of action of cytokines at a cell-type-specific level, with a view to developing more targeted therapies and interventions to treat pain. Our findings support the concept that immune cells and neurons at the site of nerve injury are engaged in a loop that involves crosstalk between them. More specifically, proinflammatory mediators and cytokines released from GM-CSF stimulated monocytes or macrophages act on neurons, which in turn release neurotransmitters that can further activate these immune cells. The net effect is likely to be peripheral sensitization and consequent chronic pain.
Footnotes
This work was supported by the Wellcome trust Senior Investigator Award to S.B.M., GlaxoSmith-Kline, and by Grant 1043147 from the National Health and Medical Research Council of Australia. We thank Dr. Franziska Denk for advice on accessing publicly available RNA-seq data. Julia E. Smith is an employee of GSK.
J.E.S. is an employee and shareholder of GSK. The remaining authors declare no competing financial interests.
References
- Achuthan A, Cook AD, Lee MC, Saleh R, Khiew HW, Chang MW, Louis C, Fleetwood AJ, Lacey DC, Christensen AD, Frye AT, Lam PY, Kusano H, Nomura K, Steiner N, Förster I, Nutt SL, Olshansky M, Turner SJ, Hamilton JA (2016) Granulocyte macrophage colony-stimulating factor induces CCL17 production via IRF4 to mediate inflammation. J Clin Investig 126:3453–3466. 10.1172/JCI87828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bali KK, Venkataramani V, Satagopam VP, Gupta P, Schneider R, Kuner R (2013) Transcriptional mechanisms underlying sensitization of peripheral sensory neurons by granulocyte-/granulocyte-macrophage colony stimulating factors. Mol Pain 9:48. 10.1186/1744-8069-9-48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevan S, Quallo T, Andersson DA (2014) Trpv1. In Mammalian transient receptor potential (TRP) cation channels, pp 207–245. Berlin; Heidelberg: Springer. [Google Scholar]
- Breivik H, Collett B, Ventafridda V, Cohen R, Gallacher D (2006) Survey of chronic pain in Europe: prevalence, impact on daily life, and treatment. Eur J Pain 10:287–333. [DOI] [PubMed] [Google Scholar]
- Brothers SP, Wahlestedt C (2010) Therapeutic potential of neuropeptide Y (NPY) receptor ligands. EMBO Mol Med 2:429–439. 10.1002/emmm.201000100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broughton SE, Hercus TR, Nero TL, Dottore M, McClure BJ, Dhagat U, Taing H, Gorman MA, King-Scott J, Lopez AF, Parker MW (2016) Conformational changes in the GM-CSF receptor suggest a molecular mechanism for affinity conversion and receptor signaling. Structure 24:1271–1281. 10.1016/j.str.2016.05.017 [DOI] [PubMed] [Google Scholar]
- Brown MT, Murphy FT, Radin DM, Davignon I, Smith MD, West CR (2012) Tanezumab reduces osteoarthritic knee pain: results of a randomized, double-blind, placebo-controlled phase III trial. J Pain 13:790–798. 10.1016/j.jpain.2012.05.006 [DOI] [PubMed] [Google Scholar]
- Calvo M, Bennett DL (2012) The mechanisms of microgliosis and pain following peripheral nerve injury. Exp Neurol 234:271–282. 10.1016/j.expneurol.2011.08.018 [DOI] [PubMed] [Google Scholar]
- Caterina MJ, Julius D (2001) The vanilloid receptor: a molecular gateway to the pain pathway. Ann Rev Neurosci 24:487–517. 10.1146/annurev.neuro.24.1.487 [DOI] [PubMed] [Google Scholar]
- Chen L, Lv F, Pei L (2014) Annexin 1: a glucocorticoid-inducible protein that modulates inflammatory pain. Eur J Pain 18:338–347. 10.1002/j.1532-2149.2013.00373.x [DOI] [PubMed] [Google Scholar]
- Chubb JR, Liverpool TB (2010) Bursts and pulses: insights from single cell studies into transcriptional mechanisms. Curr Opin Genet Dev 20:478–484. 10.1016/j.gde.2010.06.009 [DOI] [PubMed] [Google Scholar]
- Cook AD, Hamilton JA (2018) Investigational therapies targeting the granulocyte macrophage colony-stimulating factor receptor-α in rheumatoid arthritis: focus on mavrilimumab. Ther Adv Musculoskelet Dis 10:29–38. 10.1177/1759720X17752036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook AD, Braine EL, Hamilton JA (2004) Stimulus-dependent requirement for granulocyte-macrophage colony-stimulating factor in inflammation. J Immunol 173:4643–4651. 10.4049/jimmunol.173.7.4643 [DOI] [PubMed] [Google Scholar]
- Cook AD, Pobjoy J, Steidl S, Dürr M, Braine EL, Turner AL, Lacey DC, Hamilton JA (2012) Granulocyte-macrophage colony-stimulating factor is a key mediator in experimental osteoarthritis pain and disease development. Arthritis Res Ther 14:R199. 10.1186/ar4037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook AD, Pobjoy J, Sarros S, Steidl S, Dürr M, Lacey DC, Hamilton JA (2013) Granulocyte-macrophage colony-stimulating factor is a key mediator in inflammatory and arthritic pain. Ann Rheum Dis 72:265–270. 10.1136/annrheumdis-2012-201703 [DOI] [PubMed] [Google Scholar]
- Cook AD, Christensen AD, Tewari D, McMahon SB, Hamilton JA (2018) Immune cytokines and their receptors in inflammatory pain. Trends Immunol 39:240–255. 10.1016/j.it.2017.12.003 [DOI] [PubMed] [Google Scholar]
- Croxford AL, Lanzinger M, Hartmann FJ, Schreiner B, Mair F, Pelczar P, Clausen BE, Jung S, Greter M, Becher B (2015) The cytokine GM-CSF drives the inflammatory signature of CCR2+ monocytes and licenses autoimmunity. Immunity 43:502–514. 10.1016/j.immuni.2015.08.010 [DOI] [PubMed] [Google Scholar]
- Denk F, Crow M, Didangelos A, Lopes DM, McMahon SB (2016) Persistent alterations in microglial enhancers in a model of chronic pain. Cell Rep 15:1771–1781. 10.1016/j.celrep.2016.04.063 [DOI] [PubMed] [Google Scholar]
- Desiderio S, Vermeiren S, Van Campenhout C, Kricha S, Malki E, Richts S, Fletcher EV, Vanwelden T, Schmidt BZ, Henningfeld KA, Pieler T, Woods CG, Nagy V, Verfaillie C, Bellefroid EJ (2019) Prdm12 directs nociceptive sensory neuron development by regulating the expression of the NGF receptor TrkA. Cell Rep 26:3522–3536.e5. 10.1016/j.celrep.2019.02.097 [DOI] [PubMed] [Google Scholar]
- Diaz-delCastillo M, Woldbye DP, Heegaard AM (2018) Neuropeptide Y and its involvement in chronic pain. Neuroscience 387:162–169. 10.1016/j.neuroscience.2017.08.050 [DOI] [PubMed] [Google Scholar]
- Donatien P, Anand U, Yiangou Y, Sinisi M, Fox M, MacQuillan A, Quick T, Korchev YE, Anand P (2018) Granulocyte-macrophage colony-stimulating factor receptor expression in clinical pain disorder tissues and role in neuronal sensitization. Pain Rep 3:e676. 10.1097/PR9.0000000000000676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubin AE, Patapoutian A (2010) Nociceptors: the sensors of the pain pathway. J Clin Investig 120:3760–3772. 10.1172/JCI42843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenzi F, Benedetti MD, Moretto G, Rizzuto N (2001) Glial cell and macrophage reactions in rat spinal ganglion after peripheral nerve lesions: an immunocytochemical and morphometric study. Arch Ital Biol 139:357–365. [PubMed] [Google Scholar]
- Fernandez-Zafra T, Gao T, Jurczak A, Sandor K, Hore Z, Agalave NM, Su J, Estelius J, Lampa J, Hokfelt T, Wiesenfeld-Hallin Z, Xu X, Denk F, Svensson CI (2019) Exploring the transcriptome of resident spinal microglia after collagen antibody-induced arthritis. Pain 160:224–236. 10.1097/j.pain.0000000000001394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleetwood AJ, Lawrence T, Hamilton JA, Cook AD (2007) Granulocyte-macrophage colony-stimulating factor (CSF) and macrophage CSF-dependent macrophage phenotypes display differences in cytokine profiles and transcription factor activities: implications for CSF blockade in inflammation. J Immunol 178:5245–5252. 10.4049/jimmunol.178.8.5245 [DOI] [PubMed] [Google Scholar]
- Flegel C, Schöbel N, Altmüller J, Becker C, Tannapfel A, Hatt H, Gisselmann G (2015) RNA-seq analysis of human trigeminal and dorsal root ganglia with a focus on chemoreceptors. PLoS One 10:e0128951. 10.1371/journal.pone.0128951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford AP. (2012) In pursuit of P2X3 antagonists: novel therapeutics for chronic pain and afferent sensitization. Purinergic Signal 8:3–26. 10.1007/s11302-011-9271-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudet AD, Popovich PG, Ramer MS (2011) Wallerian degeneration: gaining perspective on inflammatory events after peripheral nerve injury. J Neuroinflammation 8:110. 10.1186/1742-2094-8-110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Z, Kuhn JA, Wang X, Colquitt B, Solorzano C, Vaman S, Guan AK, Evans-Reinsch Z, Braz J, Devor M, Abboud-Werner SL, Lanier LL, Lomvardas S, Basbaum AI (2016) Injured sensory neuron-derived CSF1 induces microglial proliferation and DAP12-dependent pain. Nat Neurosci 19:94–101. 10.1038/nn.4189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton JA. (2008) Colony-stimulating factors in inflammation and autoimmunity. Nat Rev Immunol 8:533–544. 10.1038/nri2356 [DOI] [PubMed] [Google Scholar]
- Hansen G, Hercus TR, McClure BJ, Stomski FC, Dottore M, Powell J, Ramshaw H, Woodcock JM, Xu Y, Guthridge M, McKinstry WJ, Lopez AF, Parker MW (2008) The structure of the GM-CSF receptor complex reveals a distinct mode of cytokine receptor activation. Cell 134:496–507. 10.1016/j.cell.2008.05.053 [DOI] [PubMed] [Google Scholar]
- Hill DA, Lim HW, Kim YH, Ho WY, Foong YH, Nelson VL, Nguyen HCB, Chegireddy K, Kim J, Habertheuer A, Vallabhajosyula P, Kambayashi T, Won KJ, Lazar MA (2018) Distinct macrophage populations direct inflammatory versus physiological changes in adipose tissue. Proc Natl Acad Sci U S A 115:E5096–E5105. 10.1073/pnas.1802611115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hore Z, Denk F (2019) Neuroimmune interactions in chronic pain–an interdisciplinary perspective. Brain Behav Immun 79:56–62. 10.1016/j.bbi.2019.04.033 [DOI] [PubMed] [Google Scholar]
- Huang K, Bian D, Jiang B, Zhai Q, Gao N, Wang R (2017) TRPA1 contributed to the neuropathic pain induced by docetaxel treatment. Cell Biochem Funct 35:141–143. 10.1002/cbf.3258 [DOI] [PubMed] [Google Scholar]
- Hu P, McLachlan EM (2002) Macrophage and lymphocyte invasion of dorsal root ganglia after peripheral nerve lesions in the rat. Neuroscience 112:23–38. 10.1016/S0306-4522(02)00065-9 [DOI] [PubMed] [Google Scholar]
- Hu P, McLachlan EM (2003) Distinct functional types of macrophage in dorsal root ganglia and spinal nerves proximal to sciatic and spinal nerve transections in the rat. Exp Neurol 184:590–605. 10.1016/S0014-4886(03)00307-8 [DOI] [PubMed] [Google Scholar]
- Hutchinson MR, Coats BD, Lewis SS, Zhang Y, Sprunger DB, Rezvani N, Baker EM, Jekich BM, Wieseler JL, Somogyi AA, Martin D, Poole S, Judd CM, Maier SF, Watkins LR (2008) Proinflammatory cytokines oppose opioid-induced acute and chronic analgesia. Brain Behav Immun 22:1178–1189. 10.1016/j.bbi.2008.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurga AM, Rojewska E, Makuch W, Mika J (2018) Lipopolysaccharide from rhodobacter sphaeroides (TLR4 antagonist) attenuates hypersensitivity and modulates nociceptive factors. Pharm Biol 56:275–286. 10.1080/13880209.2018.1457061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki Y, Zhang L, Cheng JK, Ji RR (2008) Cytokine mechanisms of central sensitization: distinct and overlapping role of interleukin-1β, interleukin-6, and tumor necrosis factor-α in regulating synaptic and neuronal activity in the superficial spinal cord. J Neurosci 28:5189–5194. 10.1523/JNEUROSCI.3338-07.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr BJ, Cafferty WB, Gupta YK, Bacon A, Wynick D, McMahon SB, Thompson SW (2000) Galanin knockout mice reveal nociceptive deficits following peripheral nerve injury. Eur J Neurosci 12:793–802. 10.1046/j.1460-9568.2000.00967.x [DOI] [PubMed] [Google Scholar]
- Laskin DL. (2009) Macrophages and inflammatory mediators in chemical toxicity: a battle of forces. Chem Res Toxicol 22:1376–1385. 10.1021/tx900086v [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes DM, Denk F, McMahon SB (2017) The molecular fingerprint of dorsal root and trigeminal ganglion neurons. Front Mol Neurosci 10:304. 10.3389/fnmol.2017.00304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Richardson PM (1993) Responses of macrophages in rat dorsal root ganglia following peripheral nerve injury. J Neurocytol 22:334–341. 10.1007/BF01195557 [DOI] [PubMed] [Google Scholar]
- Malin SA, Davis BM, Molliver DC (2007) Production of dissociated sensory neuron cultures and considerations for their use in studying neuronal function and plasticity. Nat Protoc 2:152–160. 10.1038/nprot.2006.461 [DOI] [PubMed] [Google Scholar]
- Marchand F, Perretti M, McMahon SB (2005) Role of the immune system in chronic pain. Nat Rev Neurosci 6:521–532. 10.1038/nrn1700 [DOI] [PubMed] [Google Scholar]
- McMahon SB, La Russa F, Bennett DL (2015) Crosstalk between the nociceptive and immune systems in host defence and disease. Nat Rev Neurosci 16:389–402. 10.1038/nrn3946 [DOI] [PubMed] [Google Scholar]
- Metcalf D. (2008) Hematopoietic cytokines. Blood 111:485–491. 10.1182/blood-2007-03-079681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mika J, Osikowicz M, Makuch W, Przewlocka B (2007) Minocycline and pentoxifylline attenuate allodynia and hyperalgesia and potentiate the effects of morphine in rat and mouse models of neuropathic pain. Eur J Pharmacol 560:142–149. 10.1016/j.ejphar.2007.01.013 [DOI] [PubMed] [Google Scholar]
- Moalem G, Tracey DJ (2006) Immune and inflammatory mechanisms in neuropathic pain. Brain Res Rev 51:240–264. 10.1016/j.brainresrev.2005.11.004 [DOI] [PubMed] [Google Scholar]
- Murray PJ, Allen JE, Biswas SK, Fisher EA, Gilroy DW, Goerdt S, Gordon S, Hamilton JA, Ivashkiv LB, Lawrence T, Locati M, Mantovani A, Martinez FO, Mege JL, Mosser DM, Natoli G, Saeij JP, Schultze JL, Shirey KA, Sica A, et al. (2014) Macrophage activation and polarization: nomenclature and experimental guidelines. Immunity 41:14–20. 10.1016/j.immuni.2014.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicol LSC, Thornton P, Hatcher JP, Glover CP, Webster CI, Burrell M, Hammett K, Jones CA, Sleeman MA, Billinton A, Chessell I (2018) Central inhibition of granulocyte-macrophage colony-stimulating factor is analgesic in experimental neuropathic pain. Pain 159:550–559. 10.1097/j.pain.0000000000001130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtori S, Takahashi K, Moriya H, Myers RR (2004) TNF-α and TNF-α receptor type 1 upregulation in glia and neurons after peripheral nerve injury: studies in murine DRG and spinal cord. Spine 29:1082–1088. 10.1097/00007632-200405150-00006 [DOI] [PubMed] [Google Scholar]
- Ostuni R, Piccolo V, Barozzi I, Polletti S, Termanini A, Bonifacio S, Curina A, Prosperini E, Ghisletti S, Natoli G (2013) Latent enhancers activated by stimulation in differentiated cells. Cell 152:157–171. 10.1016/j.cell.2012.12.018 [DOI] [PubMed] [Google Scholar]
- Özaktay AC, Kallakuri S, Takebayashi T, Cavanaugh JM, Asik I, DeLeo JA, Weinstein JN (2006) Effects of interleukin-1 beta, interleukin-6, and tumor necrosis factor on sensitivity of dorsal root ganglion and peripheral receptive fields in rats. Eur Spine J 15:1529–1537. 10.1007/s00586-005-0058-8 [DOI] [PubMed] [Google Scholar]
- Parajul B, Sonobe Y, Kawanokuchi J, Doi Y, Noda M, Takeuchi H, Mizuno T, Suzumura A (2012) GM-CSF increases LPS-induced production of proinflammatory mediators via upregulation of TLR4 and CD14 in murine microglia. J Neuroinflammation 9:268. 10.1186/1742-2094-9-268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei L, Zhang J, Zhao F, Su T, Wei H, Tian J, Li M, Shi J (2011) Annexin 1 exerts anti-nociceptive effects after peripheral inflammatory pain through formyl-peptide-receptor-like 1 in rat dorsal root ganglion. Br J Anaesth 107:948–958. 10.1093/bja/aer299 [DOI] [PubMed] [Google Scholar]
- Phillips CJ. (2009) The cost and burden of chronic pain. Rev Pain 3:2–5. 10.1177/204946370900300102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccolo V, Curina A, Genua M, Ghisletti S, Simonatto M, Sabò A, Amati B, Ostuni R, Natoli G (2017) Opposing macrophage polarization programs show extensive epigenomic and transcriptional cross-talk. Nat Immunol 18:530–540. 10.1038/ni.3710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popiolek-Barczyk K, Kolosowska N, Piotrowska A, Makuch W, Rojewska E, Jurga AM, Pilat D, Mika J (2015) Parthenolide relieves pain and promotes M2 microglia/macrophage polarization in rat model of neuropathy. Neural Plast 2015:676473. 10.1155/2015/676473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajasekhar P, Poole DP, Liedtke W, Bunnett NW, Veldhuis NA (2015) P2Y1 receptor activation of the TRPV4 ion channel enhances purinergic signaling in satellite glial cells. J Biol Chem 290:29051–29062. 10.1074/jbc.M115.689729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raser JM, O'Shea EK (2005) Noise in gene expression: origins, consequences, and control. Science 309:2010–2013. 10.1126/science.1105891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray P, Torck A, Quigley L, Wangzhou A, Neiman M, Rao C, Lam T, Kim JY, Kim TH, Zhang MQ, Dussor G, Price TJ (2018) Comparative transcriptome profiling of the human and mouse dorsal root ganglia: an RNA-seq-based resource for pain and sensory neuroscience research. Pain 159:1325–1345. 10.1097/j.pain.0000000000001217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray PR, Khan J, Wangzhou A, Tavares-Ferreira D, Akopian AN, Dussor G, Price TJ (2019) Transcriptome analysis of the human tibial nerve identifies sexually dimorphic expression of genes involved in pain, inflammation and neuro-immunity. Front Mol Neurosci 12:37. 10.3389/fnmol.2019.00037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders J, Hore Z, Gentry C, McMahon S, Denk F (2018) Negative evidence for a functional role of neuronal DNMT3a in persistent pain. Front Mol Neurosci 11:332. 10.3389/fnmol.2018.00332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlomann U, Rathke-Hartlieb S, Yamamoto S, Jockusch H, Bartsch JW (2000) Tumor necrosis factor α induces a metalloprotease-disintegrin, ADAM8 (CD 156): implications for neuron–glia interactions during neurodegeneration. J Neurosci 20:7964–7971. 10.1523/JNEUROSCI.20-21-07964.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholz J, Woolf CJ (2007) The neuropathic pain triad: neurons, immune cells and glia. Nat Neurosci 10:1361–1368. 10.1038/nn1992 [DOI] [PubMed] [Google Scholar]
- Schweizerhof M, Stösser S, Kurejova M, Njoo C, Gangadharan V, Agarwal N, Schmelz M, Bali KK, Michalski CW, Brugger S, Dickenson A, Simone DA, Kuner R (2009) Hematopoietic colony-stimulating factors mediate tumor-nerve interactions and bone cancer pain. Nat Med 15:802–807. 10.1038/nm.1976 [DOI] [PubMed] [Google Scholar]
- Schwertner A, Conceição dos Santos CC, Costa GD, Deitos A, de Souza A, de Souza ICC, Torres IL, da Cunha Filho JSL, Caumo W (2013) Efficacy of melatonin in the treatment of endometriosis: a phase II, randomized, double-blind, placebo-controlled trial. Pain 154:874–881. 10.1016/j.pain.2013.02.025 [DOI] [PubMed] [Google Scholar]
- Shepherd AJ, Mickle AD, Golden JP, Mack MR, Halabi CM, de Kloet AD, Samineni VK, Kim BS, Krause EG, Gereau RW 4th, Mohapatra DP (2018) Macrophage angiotensin II type 2 receptor triggers neuropathic pain. Proc Natl Acad Sci U S A 115:E8057–E8066. 10.1073/pnas.1721815115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorge RE, Mapplebeck JC, Rosen S, Beggs S, Taves S, Alexander JK, Martin LJ, Austin JS, Sotocinal SG, Chen D, Yang M, Shi XQ, Huang H, Pillon NJ, Bilan PJ, Tu Y, Klip A, Ji RR, Zhang J, Salter MW, Mogil JS (2015) Different immune cells mediate mechanical pain hypersensitivity in male and female mice. Nat Neurosci 18:1081–1083. 10.1038/nn.4053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirling LC, Forlani G, Baker MD, Wood JN, Matthews EA, Dickenson AH, Nassar MA (2005) Nociceptor-specific gene deletion using heterozygous NaV1. 8-cre recombinase mice. Pain 113:27–36. 10.1016/j.pain.2004.08.015 [DOI] [PubMed] [Google Scholar]
- Tang Y, Liu L, Xu D, Zhang W, Zhang Y, Zhou J, Huang W (2018) Interaction between astrocytic colony stimulating factor and its receptor on microglia mediates central sensitization and behavioral hypersensitivity in chronic post ischemic pain model. Brain Behav Immun 68:248–260. 10.1016/j.bbi.2017.10.023 [DOI] [PubMed] [Google Scholar]
- Thakur M, Crow M, Richards N, Davey GI, Levine E, Kelleher JH, Agley CC, Denk F, Harridge SD, McMahon SB (2014) Defining the nociceptor transcriptome. Front Mol Neurosci 7:87. 10.3389/fnmol.2014.00087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volfson D, Marciniak J, Blake WJ, Ostroff N, Tsimring LS, Hasty J (2006) Origins of extrinsic variability in eukaryotic gene expression. Nature 439:861–864. 10.1038/nature04281 [DOI] [PubMed] [Google Scholar]
- Wangzhou A, McIlvried LA, Paige C, Barragan-Iglesias P, Guzman CA, Dussor G, Ray PR, Gereau RW, Price TJ (2019) Transcriptomic analysis of native versus cultured human and mouse dorsal root ganglia focused on pharmacological targets. BioRxiv. doi: 10.1101/766865 10.1101/766865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins LR, Maier SF (2002) Beyond neurons: evidence that immune and glial cells contribute to pathological pain states. Physiol Rev 82:981–1011. 10.1152/physrev.00011.2002 [DOI] [PubMed] [Google Scholar]
- Wicks IP, Roberts AW (2016) Targeting GM-CSF in inflammatory diseases. Nat Rev Rheumatol 12:37–48. 10.1038/nrrheum.2015.161 [DOI] [PubMed] [Google Scholar]
- Wu G, Ringkamp M, Hartke TV, Murinson BB, Campbell JN, Griffin JW, Meyer RA (2001) Early onset of spontaneous activity in uninjured C-fiber nociceptors after injury to neighboring nerve fibers. J Neurosci 21:RC140. 10.1523/JNEUROSCI.21-08-j0002.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ydens E, Cauwels A, Asselbergh B, Goethals S, Peeraer L, Lornet G, Almeida-Souza L, Van Ginderachter JA, Timmerman V, Janssens S (2012) Acute injury in the peripheral nervous system triggers an alternative macrophage response. J Neuroinflammation 9:176. 10.1186/1742-2094-9-176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeisel A, Hochgerner H, Lönnerberg P, Johnsson A, Memic F, van der Zwan J, Häring M, Braun E, Borm LE, La Manno G, Codeluppi S, Furlan A, Lee K, Skene N, Harris KD, Hjerling-Leffler J, Arenas E, Ernfors P, Marklund U, Linnarsson S (2018) Molecular architecture of the mouse nervous system. Cell 174:999–1014.e22. 10.1016/j.cell.2018.06.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JM, Strong JA (2008) Recent evidence for activity-dependent initiation of sympathetic sprouting and neuropathic pain. Sheng Li Xue Bao 60:617–627. [PubMed] [Google Scholar]
- Zhang F, Wang Y, Liu Y, Han H, Zhang D, Fan X, Du X, Gamper N, Zhang H (2019) Transcriptional regulation of voltage-gated sodium channels contributes to GM-CSF induced pain. J Neurosci 39:5222–5233. 10.1523/JNEUROSCI.2204-18.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Li Y, de Carvalho-Barbosa M, Kavelaars A, Heijnen CJ, Albrecht PJ, Dougherty PM (2016) Dorsal root ganglion infiltration by macrophages contributes to paclitaxel chemotherapy-induced peripheral neuropathy. J Pain 17:775–786. 10.1016/j.jpain.2016.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Alam A, Chen Q, Eusman MA, Pal A, Eguchi S, Wu L, Ma D (2017) The role of microglia in the pathobiology of neuropathic pain development: what do we know? Br J Anaesth 118:504–516. 10.1093/bja/aex006 [DOI] [PubMed] [Google Scholar]