Abstract
Background
CDKN2A and TP53 mutations are recurrent events in melanoma, occurring in 13.3% and 15.1% of cases respectively and are associated with poorer outcomes. It is unclear what effect CDKN2A and TP53 mutations have on the clinical outcomes of patients treated with checkpoint inhibitors.
Methods
All patients with cutaneous melanoma or melanoma of unknown primary who received checkpoint inhibitor therapy and underwent genomic profiling with the 50-gene Mayo Clinic solid tumor targeted cancer gene panel were included. Patients were stratified according to the presence or absence of mutations in BRAF, NRAS, CDKN2A, and TP53. Patients without mutations in any of these genes were termed quadruple wild type (QuadWT). Clinical outcomes including median time to progression (TTP), median overall survival (OS), 6-month and 12-month OS, 6-month and 12-month without progression, ORR and disease control rate (DCR) were analyzed according to the mutational status of CDKN2A, TP53 and QuadWT.
Results
A total of 102 patients were included in this study of which 14 had mutations of CDKN2A (CDKN2Amut), 21 had TP53 mutations (TP53mut), and 12 were QuadWT. TP53mut, CDKN2Amut and QuadWT mutational status did not impact clinical outcomes including median TTP, median OS, 6-month and 12-month OS, 6-month and 12-month without progression, ORR and DCR. There was a trend towards improved median TTP and DCR in CDKN2Amut cohort and a trend towards worsened median TTP in the QuadWT cohort.
Conclusion
Cell cycle regulators such as TP53 and CDKN2A do not appear to significantly alter clinical outcomes when immune checkpoint inhibitors are used.
Introduction
Activating mutations of BRAF and NRAS are the most common mutations observed in melanoma. They are present in approximately 51–63% and 26–28% respectively of the molecular mutations in melanomas [1,2]. Accordingly, the prognostic implications of these mutations are well characterized [3]. Both BRAF mutations (BRAFmut) and NRAS mutations (NRASmut) have been shown to be early events that occur in benign and pre-invasive lesions and are not sufficient to induce carcinogenesis [4,5]. Rather, an accumulation of additional pathogenic mutations is required for pre-malignant BRAFmut or NRASmut lesions to progress to invasive melanoma [4].
Mitogenic driver mutations such as BRAF and NRAS induce senescence in premalignant disease and require secondary mutations in cell cycle control genes to convert BRAF and NRAS aberrations into oncogenes [4,6–9]. Loss of function mutations of CDKN2A and TP53 are two significant genomic alterations that allow oncogene driven melanocytes to overcome senescence and evade apoptosis [10,11]. Given the importance of TP53 and CDKN2A mutations in the pathogenesis of invasive melanoma it is understandable that both of these mutations are common mutations in melanoma. According to The Cancer Genome Atlas (TCGA) data genomic alterations of TP53 and CDKN2A are found in 15.1% and 13.3% of melanomas respectively [2], and are are frequently co-mutated with BRAF and NRAS mutations. When CDKN2A mutations are present they are found to be co-mutated with BRAF, NRAS and non-NRAS/BRAF mutations at rates of 33.3% - 67.4%, 23.9% - 40.7% and 8.7% - 29.9% respectively [2,12]. Similarly TP53 is co-mutated with BRAF, NRAS and non-BRAF/NRAS mutations with frequencies of 33.1% - 73.9%, 17.4% - 35.7% and 8.7% - 32.2% respectively. CDKN2A and TP53 mutations were present together in 5.5% - 8.3% of cases. BRAF, NRAS, CDKN2A and TP53 mutations were absent in 8.3% - 32.2% of cases.
Historically both TP53 and CDKN2A mutations are associated with a poor prognosis in melanoma patients. Several studies have shown that patients with TP53 or CDKN2A mutations have a shorter expected survival [13–15]. This occurs, at least in part, because TP53 and CDKN2A mutated tumors are more resistant to chemotherapy [13]. Several preclinical studies have demonstrated that TP53 and CDKN2A mutations lead to a loss of normal cell cycle regulation which in turn causes malignant cells to develop chemoresistance [16,17]. This paradigm of poor outcomes and chemoresistance is pervasive and has been demonstrated in multiple other malignancies. This is further supported by the observation that the use of cyclin-dependent kinase (CDK) inhibitors can enhance responsiveness to chemotherapy in tumors with loss of P16INK4a [CDKN2A loss of function mutation] [18,19]. However, it is not clear that this paradigm remains true in melanoma with the era of checkpoint inhibitors.
Neither BRAF nor NRAS mutations are thought to directly impact the efficacy of immunotherapy; however, previous studies have demonstrated nuances in response rates with checkpoint inhibitors according to genotypes. Douglas et al were the first to report the influence of NRASmut on immunotherapy outcomes and concluded that individuals harboring NRASmut had improved response rates, clinical benefit and progression free survival [20]. However, this study included all subtypes of melanoma and also only contained a small cohort of patients who received programmed death-1 (PD-1) inhibitors. Kim et al subsequently published a study assessing the effects of TP53 and non-V600 BRAF mutations (BRAFnon-v600) on clinical outcomes of cutaneous melanomas [21]. Neither TP53 nor BRAFnon-v600 mutations were associated with overall survival (OS) with ipilumumab treatment. There is a paucity of literature discussing the clinical outcomes of patients with CDKN2A and TP53 mutations since the introduction of immune checkpoint inhibitors. Herein we report the effect of TP53 and CDKN2A mutations on the response to immune checkpoint inhibitors, including PD1 inhibitors, in patients with advanced cutaneous melanoma and melanoma of unknown primary.
Results
Mutational status and patient characteristics
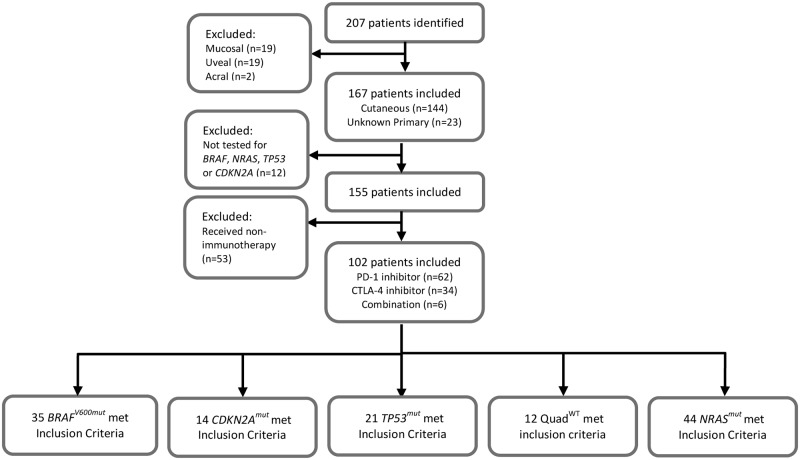
A total of 207 melanoma patients had genomic profiling using our in house 50 gene panel, of which 102 patients met the inclusion criteria for this analysis (Fig 1). Genomic profiling was performed between March 1, 2014 and October 1, 2016. Clinical data were collected between January 1, 1990 and April 7, 2017. Of the 102 patients evaluated 14 (13.7%) patients were identified to have CDKN2Amut, 21 (20.6%) had TP53mut, and 12 (11.8%) were QuadWT; the genotypes of CDKN2Amut, TP53mut and QuadWT patients are displayed in S1 Fig. The patient characteristics for this cohort of patients are summarized in Table 1.
Fig 1. Flow diagram of patient selection.
Table 1. Patient characteristics stratified by TP53 and CDKN2A mutations.
| TP53mut | TP53WT | P Value | CDKN2Amut | CDKN2AWT | P Value | QuadWT | Not Quad WT | P Value | |
|---|---|---|---|---|---|---|---|---|---|
| (N = 21) | (N = 81) | (N = 14) | (N = 88) | (N = 12) | (N = 90) | ||||
| Age at diagnosis | 0.83 a | 0.50 a | 0.15a | ||||||
| Median | 60.7 | 62.3 | 63.9 | 61.2 | 71.9 | 60.8 | |||
| Range | (32.4–82.2) | (22.8–91.0) | (31.6–88.5) | (22.8–91.0) | (41.3–77.3) | (22.8–91.0) | |||
| Gender | 0.74 b | 0.87 b | 0.67b | ||||||
| Male | 14 (66.7%) | 57 (70.4%) | 10 (71.4%) | 61 (69.3%) | 9 (75.0%) | 62 (68.9%) | |||
| Female | 7 (33.3%) | 24 (29.6%) | 4 (28.6%) | 27 (30.7%) | 3 (25.0%) | 28 (31.1%) | |||
| Ethnicity | 0.05 b | 0.69 b | 0.71b | ||||||
| Caucasian | 20 (95.2%) | 81 (100.0%) | 14 (100.0%) | 87 (98.9%) | 12 (100.0%) | 89 (98.9%) | |||
| Hispanic | 1 (4.8%) | 0 (0%) | 0 (0%) | 1 (1.1%) | 0 (0%) | 1 (1.1%) | |||
| Sites of Disease | |||||||||
| CNS | 3 (16.7%) | 20 (26.7%) | 0.38 b | 3 (23.1%) | 20 (25.0%) | 0.88 b | 4 (36.4%) | 19 (23.2%) | 0.34b |
| Liver | 5 (27.8%) | 19 (25.3%) | 0.83 b | 5 (38.5%) | 19 (23.8%) | 0.26 b | 5 (45.5%) | 19 (23.2%) | 0.11b |
| Lung | 10 (55.6%) | 37 (49.3%) | 0.64 b | 8 (61.5%) | 39 (48.8%) | 0.39 b | 9 (81.8%) | 38 (46.3%) | 0.03b |
| Adrenal | 1 (5.6%) | 6 (8.0%) | 0.72 b | 2 (15.4%) | 5 (6.3%) | 0.25 b | 1 (9.1%) | 6 (7.3%) | 0.83b |
| Bone | 4 (22.2%) | 16 (21.3%) | 0.93 b | 0 (0%) | 20 (25.0%) | 0.04 b | 2 (18.2%) | 18 (22.0%) | 0.78b |
| Skin | 2 (11.1%) | 16 (21.3%) | 0.32 b | 0 (0%) | 18 (22.5%) | 0.06 b | 1 (9.1%) | 17 (20.7%) | 0.36b |
| Lymph Node | 8 (44.4%) | 33 (44.0%) | 0.97 b | 7 (53.8%) | 34 (42.5%) | 0.44 b | 6 (54.5%) | 35 (42.7%) | 0.46b |
| Other | 4 (25.0%) | 20 (29.0%) | 0.75 b | 6 (50.0%) | 18 (24.7%) | 0.07 b | 1 (10.0%) | 23 (30.7%) | 0.17b |
| Melanoma Subtype | 0.10 b | 0.80 b | 0.41b | ||||||
| Cutaneous | 15 (71.4%) | 70 (86.4%) | 12 (85.7%) | 73 (83.0%) | 11 (91.7%) | 74 (82.2%) | |||
| Unknown Primary | 6 (28.6%) | 11 (13.6%) | 2 (14.3%) | 15 (17.0%) | 1 (8.3%) | 16 (17.8%) | |||
| Metastases | 0.32 b | 0.81 b | 0.95b | ||||||
| Yes | 18 (85.7%) | 75 (92.6%) | 13 (92.9%) | 80 (90.9%) | 11 (91.7%) | 82 (91.1%) | |||
| No | 3 (14.3%) | 6 (7.4%) | 1 (7.1%) | 8 (9.1%) | 1 (8.3%) | 8 (8.9%) | |||
| Name of Therapy | 0.14 b | 0.68 b | 0.03b | ||||||
| Pembrolizumab | 7 (33.3%) | 46 (56.8%) | 8 (57.1%) | 45 (51.1%) | 7 (58.3%) | 46 (51.1%) | |||
| Nivolumab | 1 (4.8%) | 5 (6.2%) | 0 (0%) | 6 (6.8%) | 3 (25.0%) | 3 (3.3%) | |||
| Ipilimumab | 11 (52.4%) | 23 (28.4%) | 5 (35.7%) | 29 (33.0%) | 2 (16.7%) | 32 (35.6%) | |||
| Nivolumab/Ipilimumab | 2 (9.5%) | 3 (3.7%) | 0 (0%) | 5 (5.7%) | 0 (0%) | 5 (5.6%) | |||
| Other therapy | 0 (0%) | 4 (4.9%) | 1 (7.1%) | 3 (3.4%) | 0 (0%) | 4 (4.4%) | |||
| Other Therapy Name | - | - | - | ||||||
| Ipilimumab/Dabrafenib | 0 (0%) | 1 (25.0%) | 0 (0.0%) | 1 (33.3%) | 0 (0%) | 1 (25.0%) | |||
| Ipilimumab/Dacarbazine | 0 (0%) | 1 (25.0%) | 1 (100.0%) | 0 (0%) | 0 (0%) | 1 (25.0%) | |||
| Pembrolizumab/Indoximod | 0 (0%) | 2 (50.0%) | 0 (0.0%) | 2 (66.7%) | 0 (0%) | 2 (50.0%) | |||
| Lines of Therapy | 0.84 b | 0.43 b | 0.87b | ||||||
| 1 | 18 (85.7%) | 68 (84.0%) | 12 (85.7%) | 74 (84.1%) | 11 (91.7%) | 75 (83.3%) | |||
| 2 | 3 (14.3%) | 10 (12.3%) | 1 (7.1%) | 12 (13.6%) | 1 (8.3%) | 12 (13.3%) | |||
| 3 | 0 (0%) | 1 (1.2%) | 0 (0%) | 1 (1.1%) | 0 (0%) | 1 (1.1%) | |||
| 4 | 0 (0%) | 2 (2.5%) | 1 (7.1%) | 1 (1.1%) | 0 (0%) | 2 (2.2%) | |||
| LDH elevated c | 0.45 b | 0.06 b | 0.09b | ||||||
| Yes | 2 (10.5%) | 16 (21.6%) | 1 (8.3%) | 17 (21.0%) | 4 (33.3%) | 14 (17.3%) | |||
| No | 13 (68.4%) | 40 (54.1%) | 5 (41.7%) | 48 (59.3%) | 8 (66.7%) | 45 (55.6%) | |||
| Not tested | 4 (21.1%) | 18 (24.3%) | 6 (50.0%) | 16 (19.8%) | 0 (0%) | 22 (27.2%) | |||
| Number of metastatic sites | 0.79 b | 0.38 a | 0.35 | ||||||
| Median Number of Sites | 1.0 | 2.0 | 0.48a | 2.0 | 1.5 | 2.0 | 2.0 | 0.32a | |
| Range | 0–5.0 | 0–5.0 | 0–5.0 | 0–5.0 | 0–4.0 | 0–5.0 | |||
| Response Rate (ORR) d | 0.30 b | 0.54 b | 0.73b | ||||||
| ORR | 9 (47.4%) | 23 (34.3%) | 5 (45.5%) | 27 (36.0%) | 5 (41.7%) | 27 (36.5%) | |||
| Disease Control Rate (DCR) d | 0.58 b | 0.15 b | 0.86b | ||||||
| DCR | 11 (57.9%) | 34 (50.7%) | 8 (72.7%) | 37 (49.3%) | 6 (50.0%) | 39 (52.7%) | |||
|
Duration of Immunotherapy (months) e |
0.87 a | 0.50 a | 0.54a | ||||||
| Median | 2 | 3 | 4.0 | 3.0 | 3.0 | 2.0 | |||
| Range | 1.0–9.0 | 0–13.0 | 0–9.0 | 0–13.0 | 1.0–9.0 | 0–13.0 |
a Wilcoxon rank-sum test;
b Chi square test;
c 9 subjects missing LDH data;
d 16 subjects missing response data;
e 24 subjects with incomplete duration data;
Overall response rate (ORR) = complete response + partial response; disease control rate (DCR) = complete response + partial response + stable disease; TP53mut: TP53 pathogenic mutation; TP53WT: TP53 wild type; CDKN2Amut: CDKN2A pathogenic mutation; CDKN2AWT: CDKN2A wild type; QuadWT: Quadruple wild type; TTP: Time to progression
Of the 102 patients included in the analysis 93 patients had metastatic disease. Metastases were present in 92.9% of CDKN2Amut, 85.7% of TP53mut and 91.7% of QuadWT patients. The presence of these mutations did not affect the sites of disease with exception of a lack of bone metastases in CDKN2Amut patients and increased lung metastases in the QuadWT cohort. Cutaneous melanoma was by far the most common subtype of melanoma, while melanoma of unknown primary was far less common with the latter comprising 14.3%, 28.6% and 8.3% in CDKN2Amut, TP53mut and QuadWT patients, respectively. In the CDKN2Amut and QuadWT cohorts PD-1 inhibitors were the most commonly used agents representing 57.1% and 83.3% of immunotherapies respectively, while in the TP53mut cohort the most common immunotherapy was ipilumumab (CTLA-4 inhibitor) with 52.4% of patients receiving the CTLA-4 inhibitor. Combination immunotherapies were more commonly used in the TP53mut cohort as compared to the CDKN2Amut or QuadWT cohorts. The demographics of the entire cohort were predominantly Caucasian and male.
Time to progression outcomes
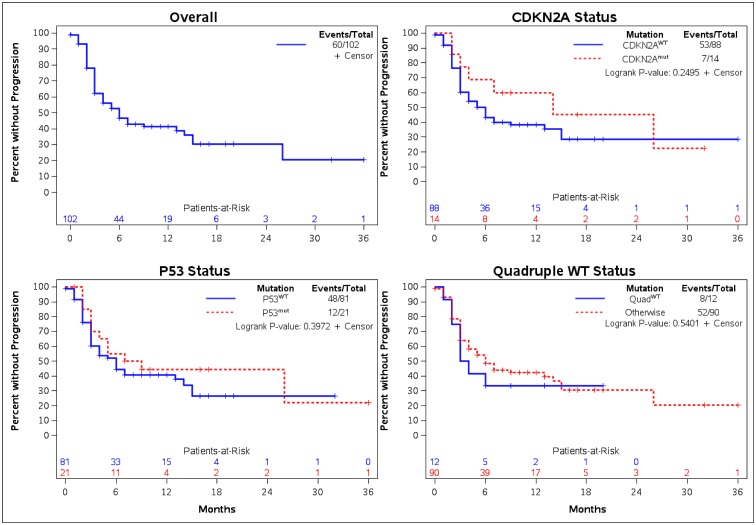
There were no statistically significant differences in TTP identified between the various mutational cohorts (Fig 2). The median TTP for CDKN2Amut and CDKN2AWT were 14.0 months (95% CI: 3.0 months–NE) and 6.0 months (95% CI: 3.0–9.0 months) respectively. The median TTP for TP53mut and TP53WT were 8.0 (95% CI: 3.0 months–NE) and 6.0 months (95% CI: 3.0–13.0 months) respectively. Those with QuadWT had a TTP of 3.5 months (95% CI: 2.0 months–NE) versus 6.0 months (95% CI: 4.0–14.0 months) in those that did not have quadruple wild type. All trends were preserved for TTP in the CDKN2A, TP53 and QuadWT cohorts at 6 and 12-month intervals (Table 2). The proportion of patients without progression at 12-months for CDKN2Amut and CDKN2AWT patients were 60.0% (95% CI: 28.5–81.2%) and 38.3% (95% CI: 27.4–49.0%) respectively. For TP53mut and TP53WT the percentage of patients without progression at 12-months were 44.4% (95% CI: 22.5–64.4%) and 40.7% (95% CI: 29.2–51.9%) respectively. The percentage of patients with QuadWT mutational status without progression at 12-months were 33.3% (95% CI: 10.3–58.8%) versus 42.3% (95% CI: 31.2–53.1%) in those without QuadWT status. Fig 2 displays TTP graphs for CDKN2A, TP53 and QuadWT cohorts.
Fig 2. Time-to-progression by mutation status.
Table 2. Time to progression by mutation.
| Mutation | Event/Total | Median Months (95% CI)KM | w/o progression (%) at 6-Months | w/o progression (%) at 12-Months |
|---|---|---|---|---|
| (95% CI)KM | (95% CI)KM | |||
| TP53mut | 12/21 | 8.0 (3.0-NE) | 55.0 (31.3–73.5%) | 44.4 (22.5–64.4%) |
| TP53WT | 48/81 | 6.0 (3.0–13.0) | 44.2 (32.6–55.2%) | 40.7 (29.2–51.9%) |
| CDKN2Amut | 7/14 | 14.0 (3.0-NE) | 68.6 (35.9–87.0%) | 60.0 (28.5–81.2%) |
| CDKN2AWT | 53/88 | 6.0 (3.0–9.0) | 43.2 (32.2–53.7%) | 38.3 (27.4–49.0%) |
| QuadWT | 8/12 | 3.5 (2.0-NE) | 33.3 (10.3–58.8%) | 33.3 (10.3–58.8%) |
| Otherwise | 52/90 | 6.0 (4.0–14.0) | 48.5 (37.3–58.8%) | 42.3 (31.2–53.1%) |
| Overall TTP: | 60/102 | 6.0 (4.0–13.0) | 46.6 (36.2–56.3%) | 41.2 (30.9–51.2%) |
CI: Confidence interval; KM: Kaplan-Meier estimate; NE: Not estimable; TP53mut: TP53 pathogenic mutation; TP53WT: TP53 wild type; CDKN2Amut: CDKN2A pathogenic mutation; CDKN2AWT: CDKN2A wild type; QuadWT: Quadruple wild type; w/o: Without
Overall survival outcomes
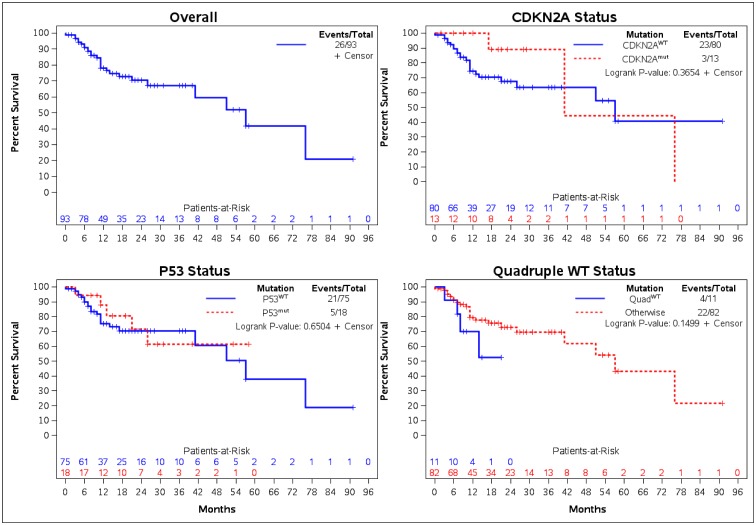
There were no statistically significant differences in OS between the various mutational cohorts (Fig 3). The median OS for CDKN2Amut and CDKN2AWT patients were 41.0 months (95% CI: 17.0–76.0 months) and 57.0 months (95% CI: 26.0 months–NE) respectively. For those with TP53mut and TP53WT mutational status the median OS were NE (95% CI: 21.0 months–NE) and 57.0 months (95% CI: 41.0 months–NE) respectively. The median OS for QuadWT cohort was NE (95% CI: 7.0 months–NE) and those without QuadWT had a median OS of 57.0 months (95% CI: 41.0 months–NE). The proportion of patients alive at 12 months with CDKN2Amut and CDKN2AWT mutational status were 100% (95% CI: 100.0–100.0%) and 74.5% (95% CI: 61.8–83.5%) respectively. The percentage of TP53mut and TP53WT patients alive at 12 months were 87.7% (95% CI: 58.8–96.8%) and 75.4% (95% CI: 62.2–84.6%) respectively. The proportion of patients with 12-month OS in the QuadWT cohort was 70.1% (95% CI: 32.3–89.5%) versus those without QuadWT was 79.5% (95% CI: 67.6% - 87.4%). The OS outcomes are also shown in Table 3.
Fig 3. Overall survival by mutation status.
Table 3. Overall survival from metastatic diagnosis by mutation.
| Mutation | Event/Total | Median Months (95% CI)KM | Survival (%) at 6-Months | Survival (%) at 12-Months |
|---|---|---|---|---|
| (95% CI)KM | (95% CI)KM | |||
| TP53mut | 5/18 | NE (21.0-NE) | 94.4 (66.6–99.2%) | 87.7 (58.8–96.8%) |
| TP53WT | 21/75 | 57.0 (41.0-NE) | 90.0 (80.2–95.1%) | 75.4 (62.2–84.6%) |
| CDKN2Amut | 3/13 | 41.0 (17.0–76.0) | 100.0 (100.0–100.0%) | 100.0 (100.0–100.0%) |
| CDKN2AWT | 23/80 | 57.0 (26.0-NE) | 89.5 (80.1–94.6%) | 74.5 (61.8–83.5%) |
| QuadWT | 4/11 | NE (7.0-NE) | 90.9 (50.8–98.7%) | 70.1 (32.3–89.5%) |
| Otherwise | 22/82 | 57.0 (41.0-NE) | 90.9 (81.9–95.6%) | 79.5 (67.6–87.4%) |
| Overall Survival: | 26/93 | 57.0 (41.0-NE) | 91.0 (82.7–95.4%) | 78.2 (67.0–85.9%) |
CI: Confidence interval; KM: Kaplan-Meier estimate; NE: Not estimable; TP53mut: TP53 pathogenic mutation; TP53WT: TP53 wild type; CDKN2Amut: CDKN2A pathogenic mutation; CDKN2AWT: CDKN2A wild type; QuadWT: Quadruple wild type
Response to immunotherapy
There was no statistically significant difference in overall response rate (ORR) or disease control rate (DCR) between the different mutational cohorts as shown in Table 1. The ORR for CDKN2Amut and CDKN2AWT patients were 45.5% and 36.0% respectively (p-value = 0.54), while the DCR for CDKN2Amut and CDKN2AWT were 72.7% and 49.3% respectively (p-value = 0.15). The ORR for TP53mut and TP53WT patients were 47.4% and 34.3% respectively (p-value = 0.30), while the DCR for TP53mut and TP53WT were 57.9% and 50.7% respectively (p-value = 0.58). The ORR for QuadWT and non-QuadWT patients were 41.7% and 36.5% respectively (p-value = 0.73). The DCR for QuadWT patients was 50.0% versus those without quadruple wild type who had a DCR of 52.7% (p-value = 0.86).
Discussion
Despite the negative prognostic significance typically ascribed to loss of TP53 in malignancies, the data from this study demonstrates no adverse prognostic or predictive significance for mutations of TP53 or CDKN2A in melanoma patients treated with immune checkpoint inhibitor therapy. The lack of deleterious effect from mutations in genes controlling cell cycle regulators is further supported by consistency across mutational cohorts in regards to TTP, 12-month OS, DCR and ORR. While not statistically significant, CDKN2Amut patients appeared to have a trend towards improved DCR and TTP. However, a larger cohort would be needed to investigate whether clinical outcomes are truly enhanced in patients with CDKN2Amut.
The devaluation of cell cycle regulators with immune checkpoint inhibitor therapy is likely explained by the mechanism in which cytotoxic T cells induce cell death. Chemotherapy primarily induces apoptosis in malignant cells via cellular stress and the intrinsic caspase pathway. For instance, many chemotherapy treatments will induce DNA damage, which will in turn signal cell cycle regulators such as TP53 and CDKN2A to activate the intrinsic caspase pathway to induce apoptosis [22–24]. The lethality of immune checkpoint inhibitors is derived primarily from the activation of cytotoxic T cells, which induce apoptosis through granzyme [25]. Granzyme is a serine protease that enters the cytoplasm via perforin and directly activates the caspase pathway independent of cell cycle regulators and induces apoptosis. Additionally, activation of the adaptive immune system will also initiate the extrinsic caspase pathway via death ligands such as tumor necrosis factor (TNF) super family and FasL. Therefore, the cytotoxic effects activated by the adaptive immune system do not appear to be driven by the internal machinery of the cell cycle and its regulators. Rather, T cell recognition of tumor cells via tumor epitopes and immune activating markers that initiates the introduction of granzyme are the more relevant drivers for checkpoint inhibitors. The clinical findings from this small retrospective study support the preclinical rationale that immune checkpoints are not adversely affected by the absence of cell cycle regulators. In addition, these findings are further supported by a previous study that did not show an adverse impact of TP53 mutations on clinical outcomes when patients were treated with ipilimumab [21].
It is more difficult to interpret the findings of the QuadWT cohort given that this cohort includes a diverse collection of mutations. A number of pathogenic mutations were identified in the QuadWT group including: KIT (n = 3), BRAFnon-V600 (n = 2), APC (n = 1), CTNNB1 (n = 1), HRAS (n = 1) and STK11 (n = 1). Similar to the CDKN2Amut and TP53mut cohorts there was no statistically significant difference in clinical outcomes observed in this study. However, there was a trend towards worsened median TTP in this patient cohort (3.5 months vs 6.0 months). However, other clinical outcomes including ORR, DCR and 12-month OS were similar between QuadWT cohort and the non-QuadWT cohorts. Given the small size of this cohort (n = 12) and heterogeneous genotype of this cohort conclusions cannot be drawn.
The small size of the study cohort and the retrospective nature of this study are limitations of this exploratory study. Because of the limited sample size the effect of co-mutations on clinical outcomes could not be analyzed with this cohort of patients. Additionally there was heterogeneous use of PD-1 inhibitors and CTLA-4 inhibitors between mutational cohorts. For instance, the TP53mut cohort and non-QuadWT cohort both had a higher proportion of ipilimumab use. Given that PD-1 inhibitors are known to have higher response rates and improved clinical outcomes compared to CTLA-4 inhibitors this may have underestimated the benefit of checkpoint inhibitors in these cohorts. However, despite the difference in treatment modalities there appeared to be consistency across clinical outcomes with similar OS, TTP, ORR and DCR results. Additionally, the TP53mut and non-QuadWT cohorts did not appear to fare any worse despite the higher utilization of ipilimumab.
This exploratory study suggests that immune checkpoint inhibitors are able to function at least as well in the presence of CDKN2A or TP53 pathogenic mutations. The lack of clear driver mutations such as CDKN2A, TP53, NRAS or BRAFV600 mutations (QuadWT) also did not seem to significantly impact clinical outcomes in this cohort of patients. While the role for CDKN2A and TP53 are integral to oncogenesis of melanoma and escape from senescence these mutations do not appear to have a significant deleterious effect on prognosis when immune checkpoint inhibitors are used for therapy.
Given the increasing frequency with which large gene mutation panels are being ordered by practicing clinicians, it is necessary to analyze the significance of common mutations in a given cancer type in order to both focus the clinician on relevant findings, and help them ignore irrelevant ones. Researchers with access to large databases of clinical and genomic findings should systematically analyze the association between common genetic events and clinical outcomes. As in this case, such retrospective studies are exploratory and can help guide larger prospective studies.
Methods
Study population/study design
This is a retrospective study which was approved by Mayo Clinic IRB(16–005168). No consent was needed as information was obtain anonymously. This study was conducted in accordance with principles for human experimentation as defined in the Declaration of Helsinki and International Conference on Harmonization Good Clinical Practice guidelines. No participating physicians have conflicts of interest to declare. The Mayo Clinic IRB waived the requirement for informed consent since the data was analyzed anonymously. Patients were identified from all three Mayo Clinic campuses (Minnesota, Arizona and Florida). Patients with a diagnosis of metastatic or unresectable cutaneous melanoma or melanoma of unknown primary whose tumors were analyzed with our 50 gene Solid Tumor Targeted Cancer Gene Panel were included. Patients who received an immune checkpoint inhibitor at any point during their treatment course were included. However, data associated with the first immunotherapy regimen and overall patient outcomes were evaluated for this analysis. Response to targeted therapy, chemotherapy and subsequent lines of immunotherapy treatments were collected but are not the focus of this study. This study allowed for treatment with cytotoxic T-lymphocyte associated protein 4 (CTLA-4) inhibitors, PD-1 inhibitors, or combinations that included either a PD-1 inhibitor or CTLA-4 inhibitor.
The objective of this study is to investigate the impact of the presence of CDKN2A mutations (CDKN2Amut), TP53 mutations (TP53mut) and quadruple wild type (QuadWT) mutational status on clinical outcomes in patients who received immune checkpoint inhibitors. Patients who did not carry TP53, CDKN2A, NRAS or BRAFv600 mutations were termed QuadWT. The primary endpoint measured was median time-to-progression (TTP) with secondary endpoints including the percentage of participants without progression at 6 and 12 months, median overall survival (OS), OS at 6 and 12 months, disease control rate (DCR) and overall response rate (ORR) to immunotherapy. Response rates were assessed using available CT or MRI imaging and their associated reports. Calculations were based on the best overall response using the immune related response criteria (irRC) and were categorized as complete response (CR), partial response (PR), stable disease (SD) and progressive disease (PD). Pathologic tumor characteristics, patient demographic and clinical details were also collected by chart review.
Genomic profiling
The Solid Tumor Targeted Cancer Gene Panel is a 50 gene panel that evaluated the following genes: ABL1, AKT1, ALK, APC, ATM, BRAF, CDH11, CKDN2A, CSF1R, CTNNB1, EGFR, ERBB2, ERBB4, EZH2, FBXW7, FGFR1, FGFR2, FGFR3, FLT3, GNA11, GNAQ, GNAS, HNF1A, HRAS, IDH1, IDH2, JAK2, JAK3, KDR, KIT, KRAS, MET, MLH1, MPL, NOTCH1, NPM1, NRAS, PDGFRA, PIK3CA, PTEN, PTPN11, RB1, RET, SMAD4, SMARCB1, SMO, SRC, STK11, TP53 and VHL. This is a laboratory-developed test using Research Use Only reagents. Extracted DNA from the clinical specimen is fragmented, adapter ligated, and a sequence library of fragments is prepared using a custom capture hybridization method. Individual patient samples are indexed for identification and the library is sequenced on an Illumina platform. Sequence data are processed through the Mayo Clinic Clinical Genome Sequencing Lab bioinformatics pipeline and a variant call file is generated for final analysis and reporting(Unpublished Mayo method). This testing is clinically available through Mayo Clinic.
Statistics
Patient characteristics were compared between mutation statuses (TP53mut versus TP53 wild type [TP53WT], CDKN2Amut versus CDKN2A wild type [CDKN2AWT] and QuadWT versus non-QuadWT). Wilcoxon rank-sum compared non-normally distributed continuous data and chi-square tests compared categorical data. Nonparametric survival analysis was used to model TTP and OS. TTP was defined as the time from first line immunotherapy date until date of progression. A patient’s progression time was censored if they received subsequent treatment, were lost to follow-up, or death occurred before known progression. OS was defined as the time from metastatic diagnosis date until date of death. Survival time was censored when patients were lost to follow-up. Kaplan-Meier (KM) method was used to estimate event rates, median time and 95% confidence intervals. Median TTP and OS estimates were not estimable (NE) where rates were greater than 50% at the last time point in the cohort. Log-rank test was used to compare TTP and OS event rates between mutation statuses. P values ≤ .05 were considered statistically significant. Analyses were performed in SAS Statistical Software 9.4 (SAS Institute, Cary, NC).
Supporting information
(DOCX)
Data Availability
All relevant data is available within the Dryad Repository at DOI: 10.5061/dryad.m0cfxpp0g.
Funding Statement
The author(s) received no specific funding for this work.
References
- 1.Carlino MS, Haydu LE, Kakavand H, Menzies AM, Hamilton AL, Yu B, et al. Correlation of BRAF and NRAS mutation status with outcome, site of distant metastasis and response to chemotherapy in metastatic melanoma. Br J Cancer 2014; 111 (2):292–299. 10.1038/bjc.2014.287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang T, Dutton-Regester K, Brown KM, Hayward NK. The genomic landscape of cutaneous melanoma. Pigment Cell Melanoma Res 2016; 29 (3):266–283. 10.1111/pcmr.12459 [DOI] [PubMed] [Google Scholar]
- 3.Jakob JA, Bassett RL, Ng CS, Curry JL, Joseph RW, Alvarado GC, et al. NRAS mutation status is an independent prognostic factor in metastatic melanoma. Cancer 2012; 118 (16):4014–4023. 10.1002/cncr.26724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shain AH, Yeh I, Kovalyshyn I, Sriharan A, Talevich E, Gagnon A, et al. The Genetic Evolution of Melanoma from Precursor Lesions. N Engl J Med 2015; 373 (20):1926–1936. 10.1056/NEJMoa1502583 [DOI] [PubMed] [Google Scholar]
- 5.Pollock PM, Harper UL, Hansen KS, Yudt LM, Stark M, Robbins CM, et al. High frequency of BRAF mutations in nevi. Nat Genet 2003; 33 (1):19–20. 10.1038/ng1054 [DOI] [PubMed] [Google Scholar]
- 6.Viros A, Sanchez-Laorden B, Pedersen M, Furney SJ, Rae J, Hogan K, et al. Ultraviolet radiation accelerates BRAF-driven melanomagenesis by targeting TP53. Nature 2014; 511 (7510):478–482. 10.1038/nature13298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dovey M, White RM, Zon LI. Oncogenic NRAS cooperates with p53 loss to generate melanoma in zebrafish. Zebrafish 2009; 6 (4):397–404. 10.1089/zeb.2009.0606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goel VK, Ibrahim N, Jiang G, Singhal M, Fee S, Flotte T, et al. Melanocytic nevus-like hyperplasia and melanoma in transgenic BRAFV600E mice. Oncogene 2009; 28 (23):2289–2298. 10.1038/onc.2009.95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu H, McDaid R, Lee J, Possik P, Li L, Kumar SM, et al. The role of BRAF mutation and p53 inactivation during transformation of a subpopulation of primary human melanocytes. Am J Pathol 2009; 174 (6):2367–2377. 10.2353/ajpath.2009.081057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bennett DC. Genetics of melanoma progression: the rise and fall of cell senescence. Pigment Cell Melanoma Res 2016; 29 (2):122–140. 10.1111/pcmr.12422 [DOI] [PubMed] [Google Scholar]
- 11.Kato S, Lippman SM, Flaherty KT, Kurzrock R. The Conundrum of Genetic "Drivers" in Benign Conditions. J Natl Cancer Inst 2016; 108 (8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hodis E, Watson IR, Kryukov GV, Arold ST, Imielinski M, Theurillat JP, et al. A landscape of driver mutations in melanoma. Cell 2012; 150 (2):251–263. 10.1016/j.cell.2012.06.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Busch C, Geisler J, Knappskog S, Lillehaug JR, Lonning PE. Alterations in the p53 Pathway and p16INK4a Expression Predict Overall Survival in Metastatic Melanoma Patients Treated with Dacarbazine. J Invest Dermatol 2010; 130 (10):2514–2516. 10.1038/jid.2010.138 [DOI] [PubMed] [Google Scholar]
- 14.Grafstrom E, Egyhazi S, Ringborg U, Hansson J, Platz A. Biallelic deletions in INK4 in cutaneous melanoma are common and associated with decreased survival. Clinical Cancer Research 2005; 11 (8):2991–2997. 10.1158/1078-0432.CCR-04-1731 [DOI] [PubMed] [Google Scholar]
- 15.Rothberg BEG, Berger AJ, Molinaro AM, Subtil A, Krauthammer MO, Camp RL, et al. Melanoma Prognostic Model Using Tissue Microarrays and Genetic Algorithms. J Clin Oncol 2009; 27 (34):5772–5780. 10.1200/JCO.2009.22.8239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gallagher SJ, Thompson JF, Indsto J, Scurr LL, Lett M, Gao BF, et al. p16INK4a expression and absence of activated B-RAF are independent predictors of chemosensitivity in melanoma tumors. Neoplasia 2008; 10 (11):1231–1239. 10.1593/neo.08702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rudolf K, Cervinka M, Rudolf E. Dual inhibition of topoisomerases enhances apoptosis in melanoma cells. Neoplasma 2010; 57 (4):316–324. 10.4149/neo_2010_04_316 [DOI] [PubMed] [Google Scholar]
- 18.Zhang J, Zhou L, Zhao S, Dicker DT, El-Deiry WS. The CDK4/6 inhibitor palbociclib synergizes with irinotecan to promote colorectal cancer cell death under hypoxia. Cell Cycle 2017; 16 (12):1193–1200. 10.1080/15384101.2017.1320005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iwadate Y, Mochizuki S, Fujimoto S, Namba H, Sakiyama S, Tagawa M, et al. Alteration of CDKN2/p16 in human astrocytic tumors is related with increased susceptibility to antimetabolite anticancer agents. Int J Oncol 2000; 17 (3):501–505. 10.3892/ijo.17.3.501 [DOI] [PubMed] [Google Scholar]
- 20.Johnson DB, Lovly CM, Flavin M, Panageas KS, Ayers GD, Zhao Z, et al. Impact of NRAS mutations for patients with advanced melanoma treated with immune therapies. Cancer Immunol Res 2015; 3 (3):288–295. 10.1158/2326-6066.CIR-14-0207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim DW, Haydu LE, Joon AY, Bassett RL Jr., Siroy AE, Tetzlaff MT, et al. Clinicopathological features and clinical outcomes associated with TP53 and BRAF(N)(on-)(V)(600) mutations in cutaneous melanoma patients. Cancer 2017; 123 (8):1372–1381. 10.1002/cncr.30463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dixon H, Norbury CJ. Therapeutic exploitation of checkpoint defects in cancer cells lacking p53 function. Cell Cycle 2002; 1 (6):362–368. 10.4161/cc.1.6.257 [DOI] [PubMed] [Google Scholar]
- 23.Vousden KH, Lu X. Live or let die: the cell’s response to p53. Nat Rev Cancer 2002; 2 (8):594–604. 10.1038/nrc864 [DOI] [PubMed] [Google Scholar]
- 24.Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature 2000; 408 (6810):307–310. 10.1038/35042675 [DOI] [PubMed] [Google Scholar]
- 25.Cullen SP, Martin SJ. Mechanisms of granule-dependent killing. Cell Death Differ 2008; 15 (2):251–262. 10.1038/sj.cdd.4402244 [DOI] [PubMed] [Google Scholar]