Abstract
Background
Identification of biomarkers for acute myeloid leukemia (AML) is important for treating this malignancy. Recent studies have reported that microRNAs (miRNAs) are stably detectable in the blood/plasma and can be used as biomarkers for various types of cancer including AML. The aim of this study was to analyze miR‐223 level in serum as a potential indicator for AML diagnosis and prognosis prediction.
Methods
Quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) was used to detect the levels of miR‐223 in the serum samples from 131 patients and 70 healthy individuals.
Results
The results revealed that serum miR‐223 was underexpressed in AML patients, particularly those in intermediate and unfavorable cytogenetic risk groups. Further analysis revealed that serum miR‐223 could yield a receiver operating characteristic (ROC) area under the curve (AUC) of 0.849 with 83.2% sensitivity and 81.4% specificity. Moreover, a significant increase in serum miR‐223 level was observed in AML subjects after their treatment. Reduced serum miR‐223 level was highly associated with aggressive clinical variables and shorter survival of patients. Furthermore, miR‐223 expression was identified to be an independent prognostic predictor of worse overall survival.
Conclusion
In conclusion, miR‐223 may be a reliable diagnostic and prognostic biomarker for AML.
Keywords: acute myeloid leukemia, biomarker, miR‐223, serum
1. INTRODUCTION
Acute myeloid leukemia (AML), the most frequent type of acute leukemia in adults, is characterized by an accumulation and differentiation arrest of myeloid blasts in the bone marrow and blood.1, 2 According to cytogenetic information, patients diagnosed with AML are classified into three risk‐based subgroups: favorable, intermediate, and poor.3 Though there has been a significant improvement in the 5‐year survival rate of AML patients over the past years, the prognosis of elderly patients and relapsed AML remains dismal.4, 5 To date, the clinical outcome and treatment response of AML patients are still difficult to predict. It is therefore necessary to identify an effective and sensitive marker for the diagnosis and prognosis of this malignancy.
MicroRNAs (miRNAs) are a class of single strand, small, and non‐coding RNAs with highly conserved sequences about 18‐25 nucleotides in length. MiRNAs are known to play an important role in multiple biological processes, such as cell growth, differentiation, apoptosis, and tumor angiogenesis.6, 7 Accumulating studies have demonstrated that aberrant miRNAs expression contributes to cancer initiation and progression, suggesting their important roles in tumorigenesis as oncogenes or suppressors.8, 9 Recently, increasing evidence has shown miRNAs are stably detected in the plasma and sera of tumor‐bearing cases and used as non‐invasive indicators for the diagnosis of specific cancers. In the research of AML, upregulation of miR‐155,10 miR‐210,11 and miR‐126‐5p12 was associated with poor clinical outcome of patients with AML, while high miR‐370,13 miR‐328,14 and miR‐20415 expression at diagnosed AML predicted a good prognosis.
Recently, the role of miR‐223 in AML had been analyzed by several researches. A study by Xiao et al revealed an inverse correlation between miR‐223 expression and FBXW7 expression. Elevated miR‐223 levels significantly inhibited cell motility and proliferation, as well as stimulated cell apoptosis in AML cell lines.16 Pulikkan et al reported miR‐223 levels were reduced in different AML subtypes, and ectopic miR‐223 expression suppressed tumorigenesis by regulating G1/S cell cycle phase transition by degrading E2F1.17 In addition, Gentner and colleagues classified AML cases into four genetic risk groups and found patients with adverse prognosis exhibited lower miR‐223 levels than those with favorable risk group.18 In the present study, we aimed to detect the expression of serum miR‐223 in AML and investigate the relationship of its expression with aggressive variables and clinical prognosis.
2. MATERIALS AND METHODS
2.1. Patient samples
This research was approved by the Ethics Committee of our hospital, and all participants signed the informed consent form before serum sample collection. All specimens were handled and made anonymous according to the ethical and legal standards.
This study included a case group of 131 subjects diagnosed with AML. The patient group comprised 80 men and 51 women, aged 41 to 73 years. The diagnosis of AML patients was according to the French‐American‐British (FAB) and World Health Organization criteria combined to immunophenotyping and cytogenetic analysis. The control group enrolled 70 healthy individuals with no clinical symptoms of cancer or other disease. Summary of patients’ characteristics is listed in Table 1.
Table 1.
Clinical characteristics of 131 patients and serum miR‐223 expression in acute myeloid leukemia cases
| Clinical feature | Patients (n = 131) | Low miR‐223 (n = 69) | High miR‐223 (n = 62) | P value |
|---|---|---|---|---|
| Age (years) | ||||
| <60 | 85 | 44 | 41 | NS |
| ≥60 | 46 | 25 | 21 | |
| Gender | ||||
| Man | 80 | 42 | 38 | NS |
| Woman | 51 | 27 | 24 | |
| Blast in BM | ||||
| <50% | 54 | 21 | 33 | .0081 |
| ≥50% | 77 | 48 | 29 | |
| WBC (×109/L) | ||||
| <10 | 52 | 25 | 27 | NS |
| ≥10 | 79 | 44 | 35 | |
| PLT (×109/L) | ||||
| <50 | 66 | 32 | 34 | NS |
| ≥50 | 65 | 37 | 28 | |
| Cytogenetics | ||||
| Favorable | 52 | 16 | 36 | <.0001 |
| Intermediate | 61 | 38 | 23 | |
| Unfavorable | 18 | 15 | 3 | |
| Complete remission | ||||
| Yes | 55 | 20 | 35 | .0015 |
| No | 76 | 49 | 27 | |
Abbreviations: BM, bone marrow; NS, no significant; PLT, platelet; WBC, white blood cells.
Blood samples were obtained from all participants early in the morning. The samples were centrifuged at 1600 g for 10 minutes, then the supernatant was transferred to 1.5‐mL tubes and stored at −80°C until further processing.
2.2. RNA isolation and quantitative real‐time polymerase chain reaction (qRT‐PCR)
Total RNA was isolated from serum using a miRVana PARIS Kit (Ambion). RNA concentration was determined with a NanoDrop ND‐1000 spectrophotometer (NanoDrop Technologies). About 1 μg (10 µL) RNA was used in reverse transcription. Reverse transcription was performed with the TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems). PCR reaction for quantification of serum miR‐223 was carried out in triplicate using the SYBR Green PCR Master Mix (Applied Biosystems) as described by the manufacturer. The primers for real‐time PCR were designed as follows: miR‐223 F: 5′‐ GCGTGTATTTGACAAGCTGAG TT ‐3′; miR‐223 R: 5′‐ GTGTCAGTTTGTCAAATACCC CA ‐3′ (Invitrogen). The ABI Prism 7900 HT Sequence detection system (Applied Biosystems) was used to perform real‐time PCR reactions. During the RNA purification step, the same amount mentioned above of cel‐miR‐39 spike‐in control was added (Qiagen) according to the provider recommendations and previous publication.19 The relative levels of miR‐223 expression in serum were normalized against cel‐miR‐39 using the 2−ΔΔCt method. The primer sequences are listed in Table 2.
Table 2.
Primers used in Quantitative reverse transcription‐polymerase chain reaction analysis
| Primer Sequences (written 5′ to 3′) | |
|---|---|
| miR‐223, forward | GCGGCGGTGTCAGTTTGTC |
| miR‐223, reverse | GTGCAGGGTCCGAGGT |
| cel‐miR‐39, forward | CAGAGTCACCGGGTGTAAAT |
| cel‐miR‐39, reverse | CCAGTGCGTGTCGTGGAGTC |
2.3. Statistical analysis
All statistical analyses were conducted using Prism 5.0 software (GraphPad Software Inc). The Mann‐Whitney U test or Kruskal‐Wallis test was used to assess the differences in serum miR‐223 levels between groups. The Pearson's Chi‐square test was used to analyze the association between serum miR‐223 levels and clinicopathological variables. Area under receiver operating characteristic (ROC) curve (AUC) was generated to evaluate the diagnostic accuracy of serum miR‐223 for AML diagnosis. A Kaplan‐Meier survival analysis was done between the serum miR‐223 levels and survival time.
Cox proportional hazards regression analysis was performed for univariate and multivariate analyses of prognostic values. Overall survival (OS) was defined as the time from diagnosis until death. Relapse‐free survival (RFS) was defined as the time from diagnosis until induction failure or relapse. Complete remission (CR) was defined by 5% or fewer blast cells in the bone marrow and normalization of the peripheral blood counts after one course of induction chemotherapy. A P value <.05 was considered significant.
3. RESULTS
3.1. Down‐regulation of miR‐223 in AML patients
The miR‐223 expression was measured in serum samples from all the participants by using qRT‐PCR. Compared to healthy controls, we found that serum miR‐223 expression was significantly decreased in AML samples. In addition, miR‐223 levels in favorable cytogenetic risk group were frequently higher those in intermediate/unfavorable cytogenetic risk groups (Figure 1).
Figure 1.
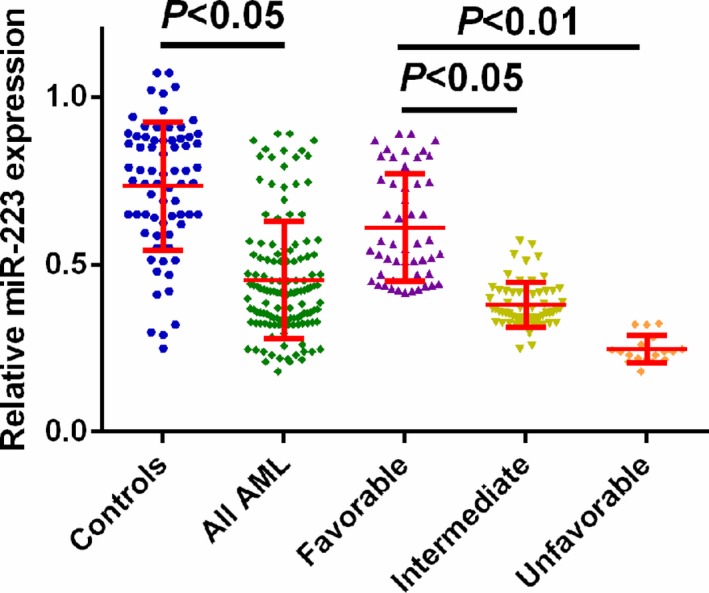
Measurement of miR‐223 expression by using Quantitative reverse transcription‐polymerase chain reaction. MiR‐223 expression was significantly decreased in acute myeloid leukemia cases compared with normal controls (P < .05). MiR‐223 levels in favorable cytogenetic risk group were frequently higher those in intermediate/unfavorable cytogenetic risk groups (P < .05)
Next, we evaluated the diagnostic power of serum miR‐223 in AML. The ROC curve analysis demonstrated the AUC was 0.849, and the sensitivity and specificity were 83.2% and 81.4%, respectively. Thus, serum miR‐223 showed good performance to discriminate AML patients from controls (Figure 2).
Figure 2.
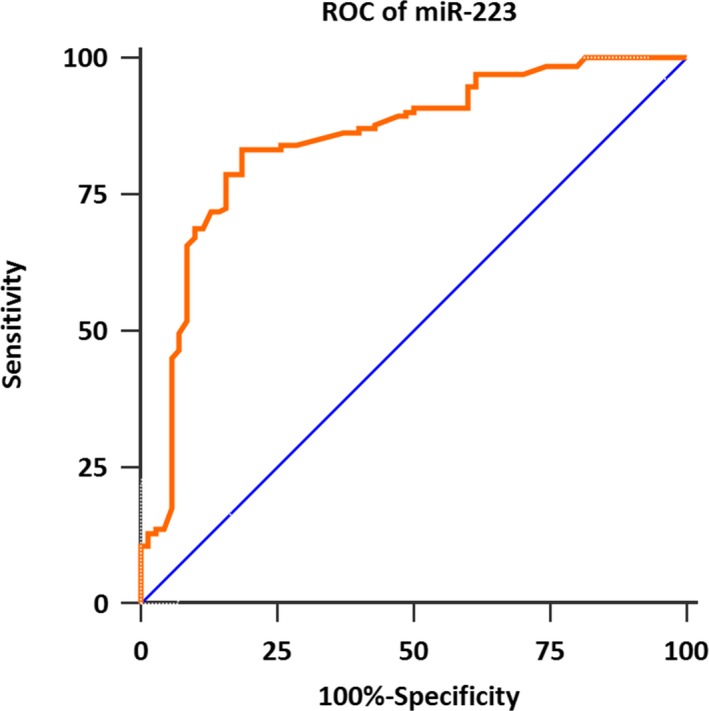
The receiver operating characteristic curve of the diagnostic power of serum miR‐223 in acute myeloid leukemia. Serum miR‐223 yielded an area under the curve value of 0.849, with 83.2% sensitivity, and 81.4% specificity in identifying acute myeloid leukemia from normal controls
3.2. Comparison of miR‐223 levels in preoperative and postoperative serum samples
We next compared the miR‐223 levels in paired blood samples from all cases before and after the induction chemotherapy. Among all 131 subjects, 55 cases achieved a CR while 76 patients failed to achieve a CR. For AML cases in CR, the levels of miR‐223 in postoperative blood samples were greatly increased compared with those in preoperative samples (P < .01, Figure 3A). Likewise, miR‐223 expression in AML patients without achievement of CR was also significantly elevated after treatment (P < .05, Figure 3B).
Figure 3.
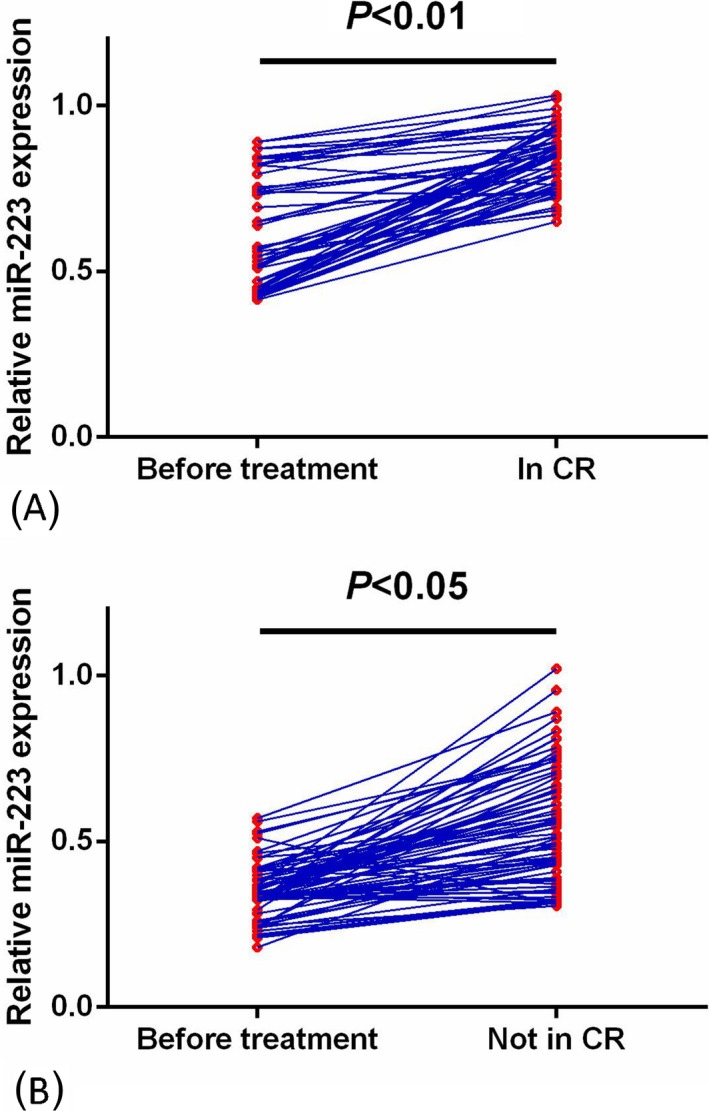
Comparison of miR‐223 levels in preoperative and postoperative. A, Changes in serum miR‐223 levels in acute myeloid leukemia (AML) subjects achieving Complete remission (CR) before and after treatment. The levels of miR‐223 in postoperative blood samples were greatly increased compared with those in preoperative samples (P < .01). B, Changes in serum miR‐223 levels in AML subjects who did not achieve CR before and after treatment. MiR‐223 expression in AML patients without achievement of CR was also significantly elevated after treatment (P < .01)
3.3. Low miR‐223 expression is associated with aggressive clinical findings in AML
We analyzed the correlation between miR‐223 expression and clinical variables. All AML cases were assigned to the low expression group (n = 69) and the high expression group (n = 62) based on the median expression level of miR‐223. The high miR‐223 expression was significantly more common in AML patients with achievement of CR than those without. Also, low miR‐223 expression was closely associated with cytogenetics and blasts in bone marrow (all P < .05, Table 1). However, no significant association was found between miR‐223 expression and age, gender, white blood cells, or platelets (all P > .05, Table 1).
3.4. Low miR‐223 expression predicted poor prognosis of AML
In the 5‐year’ follow‐up, the correlation between miR‐223 expression and prognosis was analyzed using Kaplan‐Meier method plus log‐rank test. The results showed OS/RFS of AML patients with low miR‐223 expression was significantly shorter than those with high expression (both P < .05, Figure 4A,B).
Figure 4.
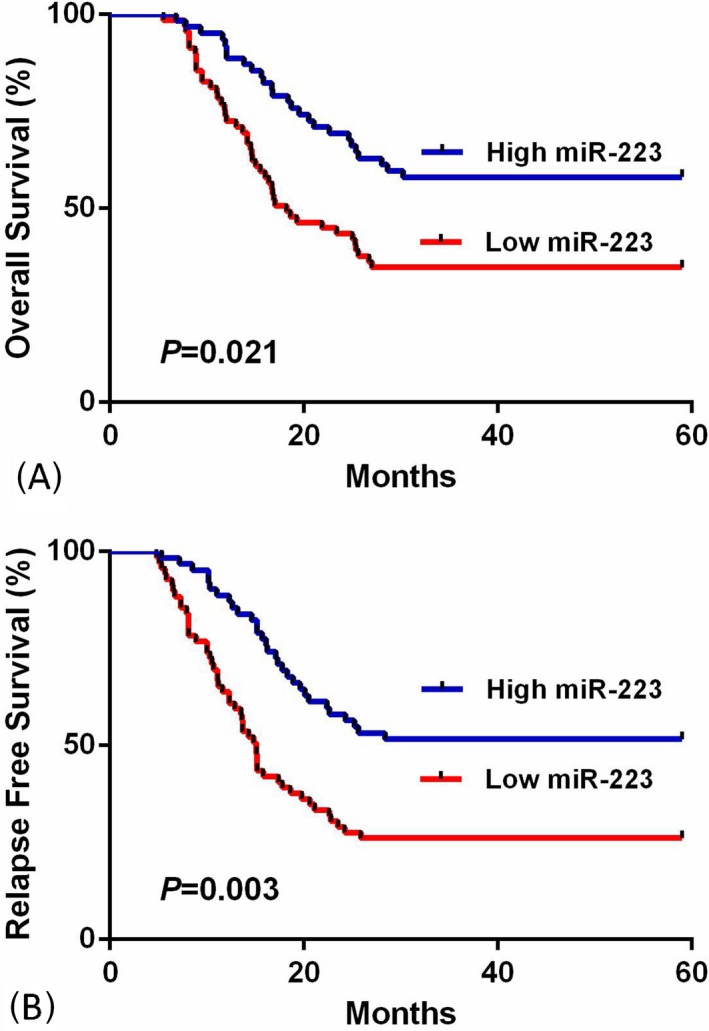
The correlation between miR‐223 expression and prognosis of acute myeloid leukemia (AML). A, Overall survival rates of AML patients with high serum miR‐223 levels were significantly longer (P < .05). B, Relapse‐free survival rates of AML patients with high serum miR‐223 levels were significantly longer (P < .05)
In the Cox proportional hazard model, univariate analysis showed that serum miR‐223 expression (RR 3.54; 95% CI, 1.47‐5.79; P = .016), cytogenetics (RR 5.12; 95% CI, 1.82‐8.62; P = .004), and blasts in bone marrow (RR 2.37; 95% CI, 1.28‐3.46; P = .027) were independent prognostic indicators for predicting worse OS in AML patients. Furthermore, multivariate analysis revealed that serum miR‐223 expression (RR 4.25; 95% CI, 1.29‐7.38; P = .011), cytogenetics (RR 5.84; 95% CI, 2.15‐9.82; P = .001), and blasts in bone marrow (RR 2.76; 95% CI, 1.37‐4.32; P = .023) were independent predictors of OS in AML cases (Table 3).
Table 3.
Univariate and multivariate analysis of overall survival in 131 acute myeloid leukemia patients
| Findings | Univariate analysis | Multivariate analysis | ||
|---|---|---|---|---|
| RR (95% CI) | P | RR (95% CI) | P | |
| MiR‐223 in serum | 3.54 (1.47‐5.79) | .016 | 4.25 (1.29‐7.38) | .011 |
| Cytogenetics | 5.12 (1.82‐8.62) | .004 | 5.84 (2.15‐9.82) | .001 |
| Blast in BM | 2.37 (1.28‐3.46) | .027 | 2.76 (1.37‐4.32) | .023 |
4. DISCUSSION
This study explored the clinical utility of serum miR‐223 to serve as a potential non‐invasive prognostic biomarker in AML patients. First, serum miR‐223 expression was significantly reduced in AML patients, particularly in intermediate and unfavorable cytogenetic risk groups. Second, ROC analysis showed serum miR‐223 could discriminate AML subjects from controls. Third, miR‐223 levels in AML patients were greatly upregulated after the induction chemotherapy, particularly in those cases achieving a CR. Fourth, low miR‐223 expression was strongly associated with worse clinical measures and poorer survival. Finally, miR‐223 was confirmed to be an independent prognostic factor for AML patients. Our findings were in line with previously published data, suggesting miR‐223 exerted a tumor suppressive function in AML.16, 17, 18 It was demonstrated that miR‐223 was downregulated in AML patients compared with healthy subjects. Moreover, miR‐223 was shown to inhibit cell proliferation and enhance cell apoptosis in AML cell lines.16 Another research has also showed that there were significantly higher miR‐223 levels in patients with favorable prognosis, whereas patients with low miR‐223 expression levels were associated with worse outcome, by measuring miR‐223 expression in blasts from 115 AML patients. Furthermore, miR‐223 was hierarchically expressed in AML subpopulations, with lower expression in leukemic stem cell–containing fractions.18
More interestingly, miR‐223 was also reported to act as a tumor suppressor in various types of cancer by accumulating evidence. Kurozumi et al found ITGA3/ITGB1 was directly targeted by miR‐223, and enhanced miR‐223 expression greatly suppressed prostate cell proliferation, migration, and invasion.20 In nasopharyngeal carcinoma, Yang et al showed the levels of miR‐223 were reduced in both NPC cell lines and the plasma of nasopharyngeal carcinoma patients, and exogenous miR‐223 greatly inhibited cell proliferation, invasion, and tumor growth by regulating MAFB.21 Tang and colleagues revealed miR‐223 expression was decreased not only in cervical cancer cell lines but also in cancerous tissues; in vitro analysis showed miR‐223 upregulation significantly restrained cervical cancer cell metastasis and repressed the epithelial‐mesenchymal transition.22 In gallbladder cancer, reduced miR‐223 expression was found both in gallbladder cancer cell lines and tissues. miR‐223 overexpression or STMN1 inhibition significantly suppressed cancer cell viability and enhanced gallbladder cancer sensitivity to docetaxel in vitro and in vivo, and vice versa.23 Compared with normal controls and chronic liver disease patients, patients with hepatocellular carcinoma exhibited significantly lower miR‐223 expression, indicating this miRNA had potential as an oncogene in hepatocellular carcinoma.24 The miR‐223 levels were lower in breast cancer tissues and cells than those in normal tissues and cells, due to the fact that enforced miR‐223 expression exhibited multiple anti‐oncogenic effects in BC cells through silencing STIM1 expression.25
In contrast, several studies had found high miR‐223 expression in some cancers. In gastric cancer, upregulation of miR‐223 more frequently occurred in tissues, cisplatin‐resistant cells, and metastatic cells of gastric cancer, and high miR‐223 expression markedly increased cancer cell metastasis, as well as stimulated cisplatin‐induced apoptosis of resistant cells. Moreover, ROC analysis showed circulating miR‐223 could be used as an indicator to discriminate gastric cancer from controls. Both EPB41L3 and FBXW7 were identified as the downstream target genes.26, 27, 28 Also, Liu et al showed miR‐223 expression was remarkably increased in colon cancer tissues and inversely correlated with p120 expression. miR‐223 overexpression increased c‐Myc, cyclinD1, MMP7 protein levels which are related to cell proliferation and invasion, suggesting miR‐223 played an oncogenic role in colon cancer.29 In non‐small cell lung cancer (NSCLC), high miR‐223 expression was observed in sputum or blood samples of NSCLC patients, and ROC curve analysis revealed serum miR‐223 had high predictive accuracy in early‐stage NSCLC detection.30, 31 The role of miR‐223 in human tumorigenesis seemed to be a double‐edged sword and its function might depend on tumor progression.
Daschkey et al has reported higher miR‐223 levels in core‐binding factor infant AML.32 Interestingly, Eyholzer et al highlighted in their analysis miR‐223 levels in AML M2 samples comparable to that in monocytes and significantly less than that found in granulocytes.33 Considering the role of miR‐223 as regulator of monocytic and granulocytic differentiation in AML cells and considering that lentiviral miR‐223 overexpression enhances myeloid differentiation in vitro,34, 35 it is plausible that AMLs with a favorable prognosis have retained more differentiation capacity than poor prognosis AMLs.
In summary, we provided convincing evidence that serum miR‐223 levels were significantly decreased in AML patients, and reduced serum miR‐223 could distinguish AML from normal controls and predicted poor patient survival. Based on these findings, miR‐223 may be a novel biomarker for AML diagnosis and prognosis.
AUTHORS' CONTRIBUTION
GY, ZY, and HH carried out the studies, participated in collecting data, and drafted the manuscript. ZZ, YC, and XL performed the statistical analysis and participated in its design. RL, QW, JL, and DX helped to draft the manuscript and participated in the data analysis and study design. All authors read and approved the final manuscript.
ACKNOWLEDGMENTS
The present study was funded by the National Natural Science Foundation of China (grant no.81870128); Clinical Research Program of Nanfang Hospital, Southern Medical University (grant no. 2018CR028); Science and Technology Planning Project of Guangdong (2014A020212185).
Yu G, Yin Z, He H, et al Low serum miR‐223 expression predicts poor outcome in patients with acute myeloid leukemia. J Clin Lab Anal. 2020;34:e23096 10.1002/jcla.23096
REFERENCES
- 1. Löwenberg B. Acute myeloid leukemia: the challenge of capturing disease variety. Hematology. 2008;2008(1):1‐11. [DOI] [PubMed] [Google Scholar]
- 2. Tenen DG. Disruption of differentiation in human cancer: AML shows the way. Nat Rev Cancer. 2003;3:89‐101. [DOI] [PubMed] [Google Scholar]
- 3. Gregory TK, Wald D, Chen Y, Vermaat JM, Xiong Y, Tse W. Molecular prognostic markers for adult acute myeloid leukemia with normal cytogenetics. J Hematol Oncol. 2009;2:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Breems DA, Van Putten WL, Huijgens PC, et al. Prognostic index for adult patients with acute myeloid leukemia in first relapse. J Clin Oncol. 2005;23:1969‐1978. [DOI] [PubMed] [Google Scholar]
- 5. Creutzig U, Zimmermann M, Ritter J, et al. Treatment strategies and long‐term results in paediatric patients treated in four consecutive AML‐BFM trials. Leukemia. 2005;19:2030‐2042. [DOI] [PubMed] [Google Scholar]
- 6. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281‐297. [DOI] [PubMed] [Google Scholar]
- 7. Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215‐233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Esquela‐Kerscher A, Slack FJ. Oncomirs ‐ microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259‐269. [DOI] [PubMed] [Google Scholar]
- 9. Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834‐838. [DOI] [PubMed] [Google Scholar]
- 10. Xu LH, Guo Y, Cen JN, et al. Overexpressed miR‐155 is associated with initial presentation and poor outcome in Chinese pediatric acute myeloid leukemia. Eur Rev Med Pharmacol Sci. 2015;19:4841‐4850. [PubMed] [Google Scholar]
- 11. Tang X, Chen L, Yan X, Li Y, Xiong Y, Zhou X. Overexpression of miR‐210 is associated with poor prognosis of acute myeloid leukemia. Med Sci Monit. 2015;21:3427‐3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Shibayama Y, Kondo T, Ohya H, Fujisawa S, Teshima T, Iseki K. Upregulation of microRNA‐126‐5p is associated with drug resistance to cytarabine and poor prognosis in AML patients. Oncol Rep. 2015;33:2176‐2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lin X, Wang Z, Wang Y, Feng W. Serum MicroRNA‐370 as a potential diagnostic and prognostic biomarker for pediatric acute myeloid leukemia. Int J Clin Exp Pathol. 2015;8:14658‐14666. [PMC free article] [PubMed] [Google Scholar]
- 14. Liu L, Chen R, Zhang Y, Fan W, Xiao F, Yan X. Low expression of circulating microRNA‐328 is associated with poor prognosis in patients with acute myeloid leukemia. Diagn Pathol. 2015;10:109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Butrym A, Rybka J, Baczyńska D, Tukiendorf A, Kuliczkowski K, Mazur G. Low expression of microRNA‐204 (miR‐204) is associated with poor clinical outcome of acute myeloid leukemia (AML) patients. J Exp Clin Cancer Res. 2015;34:68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Xiao Y, Su C, Deng T. MiR‐223 decreases cell proliferation and enhances cell apoptosis in acute myeloid leukemia via targeting FBXW7. Oncol Lett. 2016;12:3531‐3536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pulikkan JA, Dengler V, Peramangalam PS, et al. Cell‐cycle regulator E2F1 and microRNA‐223 comprise an autoregulatory negative feedback loop in acute myeloid leukemia. Blood. 2010;115:1768‐1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gentner B, Pochert N, Rouhi A, et al. MicroRNA‐223 dose levels fine tune proliferation and differentiation in human cord blood progenitors and acute myeloid leukemia. Exp Hematol. 2015;43(10): 858‐868.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Enderle D, Spiel A, Coticchia CM, et al. Characterization of RNA from exosomes and other extracellular vesicles isolated by a novel spin column‐based method. PLoS ONE. 2015;10(8):e0136133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kurozumi A, Goto Y, Matsushita R, et al. Tumor‐suppressive microRNA‐223 inhibits cancer cell migration and invasion by targeting ITGA3/ITGB1 signaling in prostate cancer. Cancer Sci. 2016;107:84‐94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yang W, Lan X, Li D, Li T, Lu S. MiR‐223 targeting MAFB suppresses proliferation and migration of nasopharyngeal carcinoma cells. BMC Cancer. 2015;15:461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tang Y, Wang Y, Chen Q, Qiu N, Zhao Y, You X. MiR‐223 inhibited cell metastasis of human cervical cancer by modulating epithelial‐mesenchymal transition. Int J Clin Exp Pathol. 2015;8:11224‐11229. [PMC free article] [PubMed] [Google Scholar]
- 23. Lu W, Hu Y, Ma Q, et al. MiR‐223 increases gallbladder cancer cell sensitivity to docetaxel by downregulating STMN1. Oncotarget. 2016;7:62364‐62376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bhattacharya S, Steele R, Shrivastava S, Chakraborty S, Di Bisceglie AM, Ray RB. Serum miR‐30e and miR‐223 as novel noninvasive biomarkers for hepatocellular carcinoma. Am J Pathol. 2016;186:242‐247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yang Y, Jiang Z, Ma N, et al. MicroRNA‐223 targeting STIM1 inhibits the biological behavior of breast cancer. Cell Physiol Biochem. 2018;45:856‐866. [DOI] [PubMed] [Google Scholar]
- 26. Li X, Zhang Y, Zhang H, et al. MiRNA‐223 promotes gastric cancer invasion and metastasis by targeting tumor suppressor EPB41L3. Mol Cancer Res. 2011;9:824‐833. [DOI] [PubMed] [Google Scholar]
- 27. Zhou X, Jin W, Jia H, Yan J, Zhang G. MiR‐223 promotes the cisplatin resistance of human gastric cancer cells via regulating cell cycle by targeting FBXW7. J Exp Clin Cancer Res. 2015;34:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhou X, Ji G, Chen H, Jin W, Yin C, Zhang G. Clinical role of circulating miR‐223 as a novel biomarker in early diagnosis of cancer patients. Int J Clin Exp Med. 2015;8:16890‐16898. [PMC free article] [PubMed] [Google Scholar]
- 29. Liu L, Zhang C, Li X, et al. MiR‐223 promotes colon cancer by directly targeting p120 catenin. Oncotarget. 2017;8:63764‐63779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bagheri A, Khorram Khorshid HR, Mowla SJ, et al. Altered miR‐223 expression in sputum for diagnosis of non‐small cell lung cancer. Avicenna J Med Biotechnol. 2017;9:189‐195. [PMC free article] [PubMed] [Google Scholar]
- 31. Zhang H, Mao F, Shen T, et al. Plasma miR‐145, miR‐20a, miR‐21 and miR‐223 as novel biomarkers for screening early‐stage non‐small cell lung cancer. Oncol Lett. 2017;13:669‐676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Daschkey S, Röttgers S, Giri A, et al. MicroRNAs distinguish cytogenetic subgroups in pediatric AML and contribute to complex regulatory networks in AML‐relevant pathways. PLoS ONE. 2013;8:e56334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Eyholzer M, Schmid S, Schardt JA, Haefliger S, Mueller BU, Pabst T. Complexity of miR‐223 regulation by CEBPA in human AML. Leuk Res. 2010;34:672‐676. [DOI] [PubMed] [Google Scholar]
- 34. Fazi F, Rosa A, Fatica A, et al. A minicircuitry comprised of microRNA‐223 and transcription factors NFI‐A and C/EBPalpha regulates human granulopoiesis. Cell. 2005;123:819‐831. [DOI] [PubMed] [Google Scholar]
- 35. Fazi F, Racanicchi S, Zardo G, et al. Epigenetic silencing of the myelopoiesis regulator microRNA‐223 by the AML1/ETO oncoprotein. Cancer Cell. 2007;12:457‐466. [DOI] [PubMed] [Google Scholar]


