Abstract
Background
Prolyl 4‐hydroxylase subunit alpha 1 (P4HA1) plays a critical role in modulating the extracellular matrix and promoting tumor progression in various cancers. However, the association between P4HA1 and head and neck squamous cell carcinomas (HNSCC) has not been thoroughly elucidated to date.
Methods
P4HA1 mRNA and protein expression in cancer and normal tissues were analyzed using The Cancer Genome Atlas (TCGA), Gene Expression Omnibus, and Human Protein Atlas databases. Quantitative PCR was applied to determine P4HA1 mRNA expression levels in 162 paired HNSCC and adjacent normal tissues. The cBioPortal for Cancer Genomics was utilized to explore P4HA1 genetic alterations in HNSCC. Then, KEGG analysis of P4HA1 co‐expressed genes in HNSCC was conducted using ClueGo in Cytoscape.
Results
P4HA1 mRNA and protein levels were significantly increased in HNSCC tissues compared with normal tissues. High P4HA1 expression in HNSCC tissues was significantly associated with tumor category, lymphatic metastasis and pathological stage. The area under summary receiver operating characteristic curve of TCGA and validation cohort was 0.887 and 0.883, respectively. Moreover, elevated P4HA1 expression was associated with unfavorable OS (HR: 1.728, P = .001) and RFS (HR: 2.025, P = .002) in HNSCC patients.
Conclusions
This integrated analysis provides strong evidence that increasing P4HA1 expression is significantly associated with the carcinogenesis of HNSCC. Additionally, high P4HA1 expression serves as both diagnostic biomarker and independent prognostic factor for poor OS and RFS in HNSCC patients.
Keywords: bioinformatics, carcinogenesis, diagnosis, head and neck squamous cell carcinoma, P4HA1, prognosis
1. INTRODUCTION
Head and neck cancers represent the sixth most common cancer worldwide. The vast majority (greater than 90%) are head and neck squamous cell carcinomas (HNSCC), such that the term head and neck cancer refers to cancer arising from the epithelium lining the of the upper aerodigestive tract (lip, oral cavity, pharynx, and larynx) and exhibiting microscopic evidence of squamous differentiation.1 According to the latest report of the International Agency for Research on Cancer, approximately one million new HNSCC patients were estimated to be clinically diagnosed in 2018 with greater than 542 943 deaths worldwide.2 The risk for developing HNSCC is associated with several traditional etiological factors, including cigarette smoking and alcohol abuse.3 Increasing evidence also demonstrates that infection with a high‐risk human papillomavirus (HPV) strain is associated with HNSCC and is an important favorable prognostic factor, especially for oral cavity and oropharynx cancer.4, 5 Although the recent diagnostic and therapeutic strategies have yielded some significant improvements, the 5‐year survival rate for HNSCC patients over the last decade remained at approximately 50%.6 Although multiple molecular mechanisms are associated with HNSCC initiation, growth, invasion, and metastasis, the exact pathogenesis of tumorigenesis remains unclear. Development of new technologies, such as microarray technology and next‐generation sequencing, has allowed for collection of large amounts of data to explore the key genes in the pathogenesis of HNSCC,7, 8 such as CDKN2A,9 CDH1,10 and EGFR.11 Therefore, the identification of oncogenic drivers and potential therapeutic targets is crucial for both early diagnosis and effective treatment for HNSCC.
Tumor hypoxia is an essential characteristic of the neoplastic microenvironment that may be correlated with cell proliferation, apoptosis, differentiation, vascularization/angiogenesis, genetic instability, tumor metabolism, tumor immune responses, and invasion and metastasis.12 Under hypoxic microenvironments, hypoxia inducible factor‐1 (HIF‐1) promotes extracellular matrix (ECM) remodeling by inducing prolyl 4‐hydroxylase subunit alpha 1 (P4HA1), prolyl 4‐hydroxylase subunit alpha 2 (P4HA2), and procollagen‐lysine, 2‐oxoglutarate 5‐dioxygenase 2 (PLOD2) expression, leading to changes in cancer cell morphology, adhesion and motility that enhance invasion and metastasis.13 Located at 10q22.1, P4HA1 encodes an active catalytic subunit of prolyl 4‐hydroxylase that catalyzes the formation of 4‐hydroxyproline in collagen, which is essential to the formation and stabilization of the triple helical domain of newly synthesized procollagen chains.14 P4HA1 was identified as hypoxia‐responsive gene and plays a critical role in regulating collagen biosynthesis.15 Previous evidence suggested that P4HA1 overexpression plays a critical role in cancer progression. Hu et al16 reported that high P4HA1 expression is correlated with the malignancy of gliomas and could serve as a prognostic indicator for patients with high‐grade gliomas. In human breast cancer, P4HA1 plays an essential role in enhancing invasion and metastasis and is significantly associated with decreased patient survival.17 However, until now, the association between P4HA1 and HNSCC as well as its clinical value was not clearly delineated.
In this study, we evaluated the expression of P4HA1 in HNSCC and its clinical value. In addition, we also investigated the enrichment of P4HA1 co‐expressed genes in KEGG pathways to explore its underlying mechanism in HNSCC.
2. MATERIALS AND METHODS
2.1. Bioinformatic analysis using UCSC Xena browser
P4HA1 mRNA expression and details of the clinicopathological characteristics of patients with primary HNSCC in TCGA cohort (Project Id: TCGA‐HNSC) were obtained by using the University of California Santa Cruz (UCSC) Xena browser (://xenabrowser.net/).
2.2. Comparison of P4HA1 gene expression between tumor vs non‐tumor samples using Gene Expression Omnibus database
P4HA1 mRNA expression in HNSCC samples compared with normal tissue was also analyzed using published databases (GSE6631) 18 downloaded from Gene Expression Omnibus (GEO). In the GSE6631 database, data for gene expression profiling of 22 paired HNSCC samples and corresponding adjacent normal tissues were obtained.
2.3. Specimens collection
To validate the findings of the bioinformatics analysis, 162 HNSCC tissues and their adjacent non‐tumorous tissues were collected from the Ningbo Medical Centre Lihuili Hospital and the Affiliated Tumor Hospital of Xiangya Medical School, from February 2014 to November 2018. None of the patients underwent treatment before operation. Each specimen was histopathologically confirmed by two pathologists. All specimens were preserved in RNA‐fixer Reagent (Bioteke) and stored at −80°C until further experiments. This study was approved by the Human Research Ethics Committee of Ningbo Medical Centre Lihuili Hospital and the Affiliated Tumor Hospital of Xiangya Medical School. Written informed consent was obtained from all patients.
2.4. Total RNA extraction and quantitative real‐time PCR
Total RNA was extracted from 162 paired HNSCC and normal tissues using the TRIzol reagent (Invitrogen), then reverse transcribed into cDNA by GoScript Reverse Transcription (RT) System (Promega) following the manufacturer's instructions. Real‐time quantitative reverse transcription‐polymerase chain reaction quantitative real‐time PCR (qRT‐PCR) was performed as previously described.19 The housekeeping gene glyceraldehyde 3‐phosphate dehydrogenase (GAPDH) was used as a normalize control. The primers were synthesized by Huada Biotech. The sequences of the PCR primers were as follows: 5′‐AGTACAGCGACAAAAGATCCAG‐3′ and 5′‐CTCCAACTCACTCCACTCAGTA‐3′ for P4HA1; 5′‐CCATGGAGAAGGCTGGGG‐3′, and 5′‐CAAAGTTGTCATGGATGACC‐3′ for GAPDH. The conditions of thermal cycling were as follows: 95°C at 10 minutes for a hot‐start, 45 amplification cycles at 95°C for 15 seconds, 55°C for 35 seconds, and 70°C for 30 seconds. The expression of P4HA1 was calculated using the ΔCt method. Larger ΔCt value indicates lower expression. All experiments were performed in triplicate.
2.5. Immunohistochemistry staining
P4HA1 protein expression levels in HNSCC tissues and in normal tissues were explored using immunohistochemistry (IHC) staining data from the Human Protein Atlas (HPA; ://www.proteinatlas.org/).
2.6. P4HA1 genetic alteration analysis using cBioPortal for Cancer Genomics and KEGG analysis using ClueGo in Cytoscape
P4HA1 genetic alterations in HNSCC were examined using cBioPortal for Cancer Genomics (http://www.cbioportal.org/). The associations between P4HA1 genetic alterations and overall survival (OS) as well as disease‐free survival (DFS) in HNSCC patients were assessed by generating Kaplan‐Meier survival curves. The genes co‐expressed with P4HA1 in HNSCC were defined as (|Pearson's r| ≥ .4 and |Spearman's r| ≥ .4). Then, the co‐expressed genes were loaded into ClueGo in Cytoscape for analysis of KEGG pathways. Only pathways with a P‐value ≥.05 were included.
2.7. Statistical analysis
All statistical analysis was performed using Statistical Program for Social Sciences (SPSS) 20.0 software (SPSS Inc) and R 3.1.2 software (https://www.r-project.org/), which were also used to generate figures. For comparisons of P4HA1 expression between groups, independent Student's t test and one‐way analysis of variance (one‐way ANOVA) tests were employed as appropriate. Receiver operating characteristic (ROC) analysis was used to assess the diagnostic value of P4HA1 expression for HNSCC. The cut‐off point was defined as the maximum Youden index. HNSCC patients with integrated survival data were divided into high and low P4HA1 expression groups according to the maximum Youden index based on ROC curves for death and recurrence detection in HNSCC patients. Kaplan‐Meier curves of overall survival (OS) and recurrent‐free survival (RFS) after initial therapy were generated, and log‐rank tests were performed to evaluate the difference between the survival curves. Univariate and multivariate Cox regression analyses were performed to determine the independent prognostic value of P4HA1 expression in terms of OS and RFS in HNSCC patients. P‐value <.05 was considered to be statistically significant.
3. RESULTS
3.1. P4HA1 expression is significantly elevated in HNSCC tissues
By comparing P4HA1 mRNA expression using the RNA‐Seq data of 520 HNSCC tissues and normal tissues in TCGA, we revealed that HNSCC tissues exhibited significantly elevated P4HA1 mRNA expression compared with normal tissues (P = 1.64E‐23; Figure 1A,B), consistent with our findings using GEO data (P = 6.16E‐04; Figure 1C). The qRT‐PCR analysis using the 162 paired HNSCC samples confirmed that P4HA1 mRNA expression levels were significantly upregulated in HNSCC tissues compared with adjacent normal tissues (P = 1.41E‐40, Figure 2). We further explored P4HA1 protein expression in HNSCC tissues and normal tissues using the HPA database. Immunohistochemical staining images revealed that P4HA1 exhibited high expression in HNSCC tissues (Figure 3A). In comparison, oral mucosa exhibited medium P4HA1 expression (Figure 3B).
Figure 1.
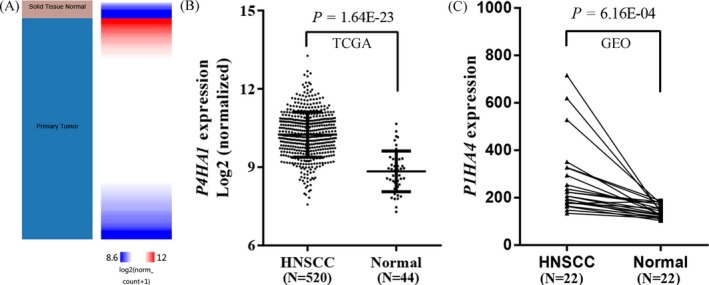
P4HA1 expression levels are significantly elevated in HNSCC tissues compared with normal tissues using public databases. A–B, Heatmap (A) and plot (B) showing P4HA1 expression in HNSCC tissue and normal tissue using TCGA database. C, P4HA1 expression in HNSCC tissue and normal tissue using GEO database. N, Sample number
Figure 2.
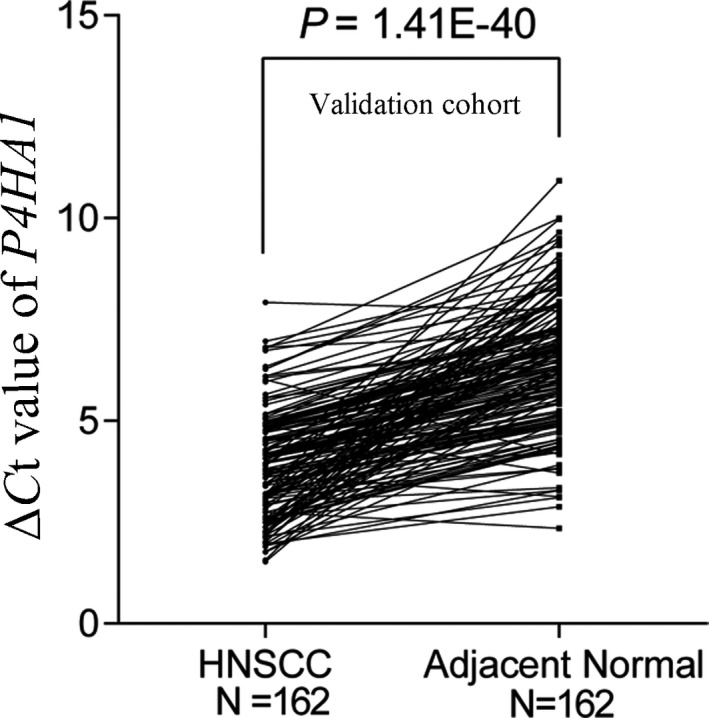
P4HA1 expression levels were significantly higher in HNSCC tissues vs paired non‐tumor tissues in our validation cohort. N, sample number
Figure 3.
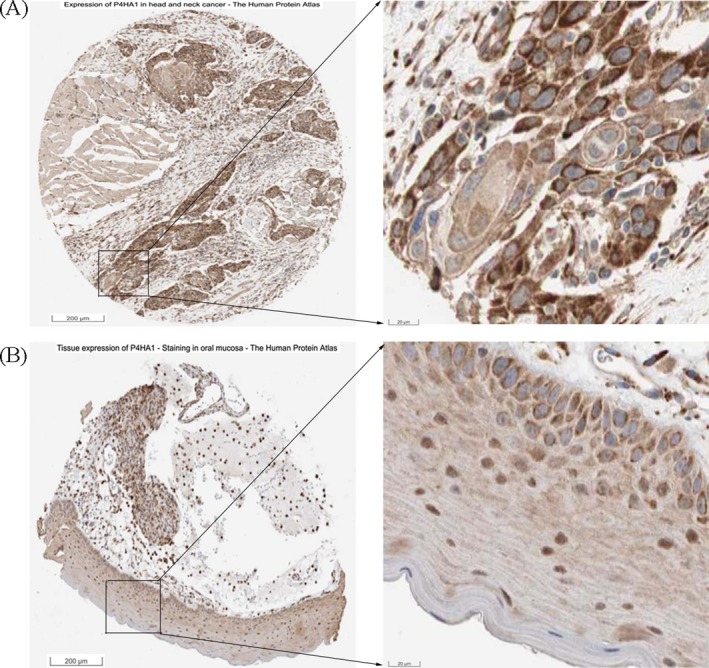
P4HA1 immumohistochemical staining images in HNSCC tissue and normal tissue. Images were obtained from Human Protein Atlas (://v18.proteinatlas.org/). A, High P4HA1 protein expression in HNSCC tissue (://www.proteinatlas.org/ENSG00000122884-P4HA1/pathology/tissue/head+and+neck+cancer#img). B, Medium P4HA1 protein expression in normal oral mucosa (https://www.proteinatlas.org/ENSG00000122884-P4HA1/tissue/oral+mucosa#img)
3.2. Association of P4HA1 expression with some clinical features of HNSCC
Then, we analyzed the association between P4HA1 mRNA expression levels and clinicopathological characteristics of patients with HNSCC. As shown in Table 1, high P4HA1 expression in HNSCC tissues was significantly associated with alcohol consumption (P = .019), tumor location (P = .017), HPV infection (P = .011), tumor category (P = .006), lymphatic metastasis (P = .006), and pathological stage (P = .002).
Table 1.
Association between P4HA1 expression and clinicopathological features of HNSCC patients
| Characteristics | N | Mean ± SD | P‐value |
|---|---|---|---|
| Gender | |||
| Female | 136 | 10.168 ± 0.877 | 0.208 |
| Male | 384 | 10.276 ± 0.858 | |
| Age | |||
| <60 y | 233 | 10.229 ± 0.827 | 0.63 |
| ≥60 y | 286 | 10.265 ± 0.894 | |
| Smoking history | |||
| No | 117 | 10.149 ± 0.894 | 0.199 |
| Yes | 391 | 10.266 ± 0.853 | |
| Alcohol history | |||
| No | 162 | 10.115 ± 0.892 | 0.019 |
| Yes | 347 | 10.306 ± 0.842 | |
| Histologic grade | |||
| G1 + 2 | 366 | 10.199 ± 0.830 | 0.084 |
| G3 + 4 | 132 | 10.350 ± 0.927 | |
| Tumor site | |||
| Oral cavity + oropharynx | 394 | 10.197 ± 0.870 | 0.017 |
| Hypopharynx + larynx | 126 | 10.408 ± 0.824 | |
| HPV status | |||
| Negative | 73 | 10.328 ± 0.724 | 0.011 |
| Positive | 38 | 9.942 ± 0.798 | |
| Tumor category | |||
| Tis/T1/T2 | 185 | 10.104 ± 0.853 | 0.006 |
| T3/T4 | 273 | 10.330 ± 0.850 | |
| Lymphatic metastasis | |||
| No | 176 | 10.111 ± 0.820 | 0.006 |
| Yes | 244 | 10.341 ± 0.862 | |
| Pathological stage | |||
| I + II | 101 | 10.021 ± 0.812 | 0.002 |
| III + IV | 347 | 10.329 ± 0.875 | |
Abbreviation: N, sample number.
3.3. Diagnostic value of P4HA1 expression for HNSCC
We examined the diagnostic value of P4HA1 expression in HNSCC using ROC curves. An area under the ROC curve (AUC) closer to 1.0 signifies that the test exhibits more perfect discrimination. The maximum Youden index was used as a cut‐off point. The result suggested that P4HA1 expression yielded an AUC of 0.887, a sensitivity of 88.8%, and a specificity of 78.1% using TCGA cohort (Figure 4A) and yielded an AUC of 0.883, a sensitivity of 78.4% and a specificity of 83.3% using our validation cohort (Figure 4B).
Figure 4.
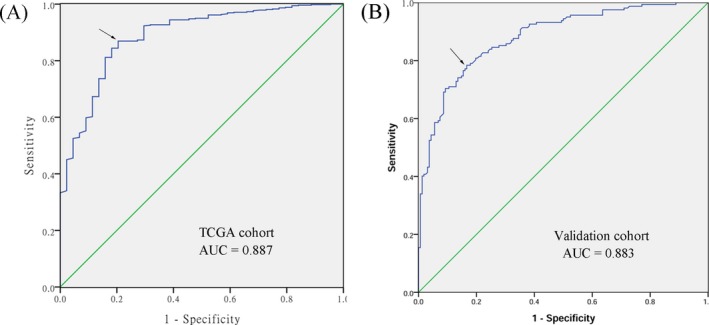
Receiver operating characteristic (ROC) curves to assess the diagnostic value of P4HA1 expression in HNSCC patients. The area under the curve (AUC) was 0.887 based on TCGA cohort. The area under the curve (AUC) was 0.883 based on our validation cohort. The arrow points to the intercept
3.4. High P4HA1 expression was an independent prognostic predictor of unfavorable OS and RFS in HNSCC patients
Using the maximum Youden index as cut‐off point (10.665), we classified 517 HNSCC patients with integrated OS data into the high P4HA1 expression group (N = 173) and low P4HA1 expression group (N = 344). Kaplan‐Meier curves and log‐rank tests revealed that high P4HA1 expression was associated with significantly worse OS in HNSCC (P = 2.07E‐5). In addition, 437 HNSCC patients with integrated RFS data were divided into high (N = 106) and low (N = 331) P4HA1 expression groups according to a cut‐off value of 10.845. Kaplan‐Meier curves and log‐rank tests revealed that HNSCC patients in the high P4HA1 expression group exhibited significantly poorer RFS (P = .002).
In univariate Cox proportional hazards analysis, the results showed that elderly (hazard ratio (HR): 1.318, 95% confidence interval (CI): 1.003‐1.731, P = .047), female (HR: 1.349, 95% CI: 1.014‐1.796, P = .04), advanced stages (HR: 1.754, 95% CI: 1.203‐2.558, P = .004), lymphatic metastasis (HR: 1.86, 95% CI: 1.343‐2.576, P = 1.86E‐04), and elevated P4HA1 expression (HR: 1.775, 95% CI: 1.358‐2.321, P = 2.68E‐05) were associated with unfavorable OS. Of note, we found that alcohol consumption (HR: 1.809, 95% CI: 1.130‐2.896, P = .014), advanced stages (HR: 2.302, 95% CI: 1.249‐4.242, P = .007), lymphatic metastasis (HR: 1.653, 95% CI: 1.062‐2.573, P = .026), and high P4HA1 expression (HR: 1.865, 95% CI: 1.249‐2.785, P = .002) were significantly associated with shorter RFS (Table 2). Multivariate Cox proportional hazard analysis was conducted to investigate the independent prognostic factors in terms of OS and RFS in HNSCC patients by adjusting only variables that exhibited significance in univariate analysis. We found that high P4HA1 expression (OS: HR: 1.728, 95% CI: 1.267‐2.357, P = .001; RFS: HR: 2.025, 95% CI: 1.296‐3.162, P = .002) was independent unfavorable prognostic factor in terms of OS and RFS in HNSCC patients (Figure 5).
Table 2.
Univariate and multivariate analysis of overall survival and recurrent‐free survival in HNSCC patients
| Characteristics | Univariate analysis | Multivariate analysis | ||||
|---|---|---|---|---|---|---|
| HR | 95% CI | P value | HR | 95% CI | P value | |
| Overall survival | ||||||
| Age (≥60 y vs <60 y) | 1.318 | 1.003‐1.731 | .047 | 1.223 | 0.891‐1.679 | .214 |
| Gender (female vs male) | 1.349 | 1.014‐1.796 | .04 | 1.372 | 0.978‐1.926 | .067 |
| Smoking history (yes vs no) | 1.123 | 0.803‐1.572 | .498 | |||
| Alcohol history (yes vs no) | 0.942 | 0.709‐1.252 | .68 | |||
| Histologic grade (G3/4 vs G1/2) | 0.867 | 0.637‐1.180 | .419 | |||
| Pathologic stage (III/IV vs I/II) | 1.754 | 1.203‐2.558 | .004 | 1.878 | 1.055‐3.345 | .032 |
| Pathologic N (N1/2/3 vs N0) | 1.86 | 1.343‐2.576 | 1.86E‐04 | 1.422 | 0.973‐2.078 | .069 |
| HPV (positive vs negative) | 0.856 | 0.420‐1.746 | .67 | |||
| P4HA1 expression (high vs low) | 1.775 | 1.358‐2.321 | 2.68E‐05 | 1.728 | 1.267‐2.357 | .001 |
| Recurrence‐free survival | ||||||
| Age (≥60 y vs <60 y) | 1.291 | 0.878‐1.899 | .194 | |||
| Gender (female vs male) | 1.118 | 0.714‐1.751 | .626 | |||
| Smoking history (yes vs no) | 0.973 | 0.626‐1.513 | .904 | |||
| Alcohol history (yes vs no) | 1.809 | 1.130‐2.896 | .014 | 1.36 | 0.827‐2.236 | .226 |
| Histologic grade (G3/4 vs G1/2) | 0.821 | 0.526‐1.281 | .384 | |||
| Pathologic stage (III/IV vs I/II) | 2.302 | 1.249‐4.242 | .007 | 1.514 | 0.721‐3.179 | .274 |
| Pathologic N (N1/2/3 vs N0) | 1.653 | 1.062‐2.573 | .026 | 1.19 | 0.709‐1.996 | .511 |
| HPV (positive vs negative) | 0.914 | 0.343‐2.438 | .858 | |||
| P4HA1 expression (high vs low) | 1.865 | 1.249‐2.785 | .002 | 2.025 | 1.296‐3.162 | .002 |
Abbreviations: CI, confidence interval; HR, hazard ratio.
Figure 5.
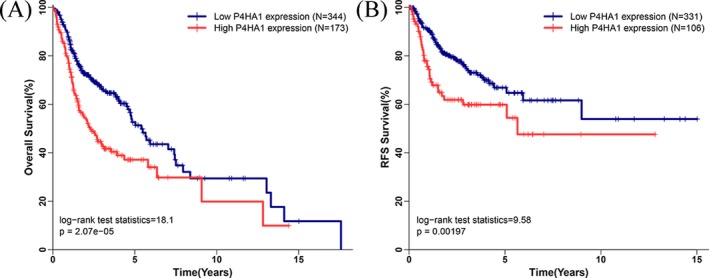
Association between P4HA1 expression and survival in HNSCC. A, High P4HA1 expression is associated with poor OS in HNSCC patients; B, High P4HA1 expression is associated with poor RFS in HNSCC patients
3.5. P4HA1 genetic alteration was associated with worse OS and DFS in HNSCC patients
The cBioPortal for Cancer Genomics was utilized to explore P4HA1 genetic alterations in HNSCC P4HA1 was only altered in 8 samples, including 504 sequenced HNSCC patients from the Cancer Genome Atlas Research (Figure 6). Then, we also evaluated the association between P4HA1 genetic alteration and survival in HNSCC patients. Survival curves indicated that HNSCC patients with P4HA1 genetic alterations exhibited significantly worse OS (log‐rank P = .025) and DFS (log‐rank P = .007).
Figure 6.
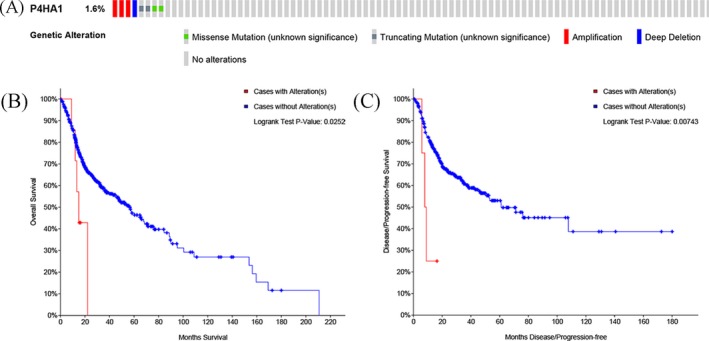
P4HA1 genetic alterations in HNSCC and its correlation with prognosis of HNSCC patients in OS and DFS. A, P4HA1 is altered in 1.6% (8/504) of sequenced HNSCC patients. B, P4HA1 genetic alterations were associated with significantly worse overall survival; C, P4HA1 genetic alterations were associated with significantly worse disease‐free survival
3.6. KEGG analysis based on P4HA1 co‐expressed genes
By data mining using cBioPortal for Cancer Genomics, we identified 282 co‐expressed genes with P4HA1 in HNSCC. To further investigate the possible signaling pathways that P4HA1 might be involved in, P4HA1 co‐expressed genes in HNSCC were subjected to KEGG pathway analysis. In HNSCC, P4HA1 co‐expressed genes were enriched in the HIF‐1 signaling pathway, focal adhesion, regulation of actin cytoskeleton, protein processing in endoplasmic reticulum, vibrio cholera infection, lysosome, lysine degradation, glycolysis/gluconeogenesis, and other types of O‐glycan biosynthesis (Figure7).
Figure 7.
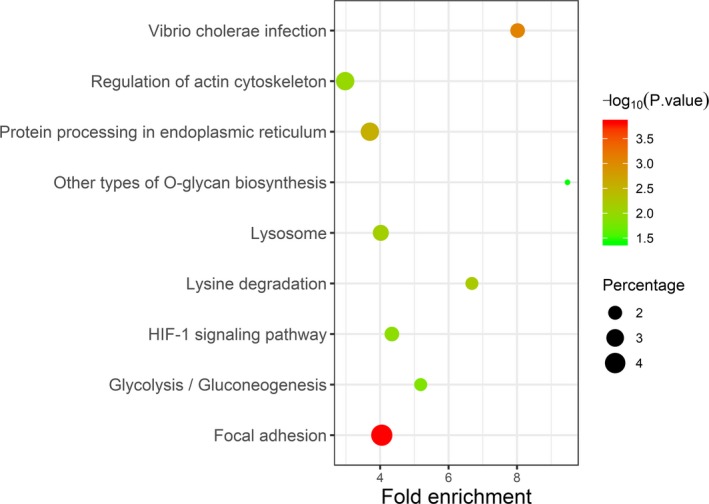
KEGG pathway analysis of the genes co‐expressed with P4HA1 in HNSCC
4. DISCUSSION
Collagens are the major structural extracellular matrix (ECM) proteins and form fibers or networks in tumor tissues to support the tumor microenvironment and play crucial roles in carcinogenesis.20, 21 P4HA1 is a key intracellular enzyme to catalyze the formation of 4‐hydroxyproline that is essential for proper three‐dimensional folding of newly synthesized procollagen chains, maintaining ECM homeostasis.14 Accumulating evidence indicates that increased P4HA1 is associated with the initiation, invasion, and metastasis of many human cancers, including hepatocellular carcinoma,22 breast cancer,17 and prostate cancer.23 However, the association of P4HA1 with HNSCC remains uninvestigated. In the present study, significantly increased P4HA1 mRNA levels were observed in HNSCC tissues compared with nontumor tissues using TCGA database, which is consistent with the analysis results of the GEO database. Furthermore, the HPA database validated that P4HA1 protein levels were elevated in HNSCC compared with surrounding normal tissue and demonstrated that P4HA1 was mainly localized to the endoplasmic reticulum and slightly localized to the mitochondria and vesicles. All these results suggested that P4HA1 plays an important role in HNSCC transformation.
The history of high alcohol consumption is a crucial factor for increased the risk of HNSCC.24 In this study, using RNA‐seq data in TCGA‐HNSC, we found that increased P4HA1 expression was significantly correlated with alcohol consumption, suggesting that alcohol might contribute to HNSCC by inducing P4HA1 expression. Accumulating evidence indicates that HPV infection is an important risk factor for HNSCC. HPV‐positive HNSCCs and HPV‐negative HNSCCs differ with respect to the molecular mechanisms underlying their oncogenic processes.4 HPV‐positive cancers are more susceptible to chemotherapy and radiation with better prognosis compared with HPV‐negative patients.25, 26 Studies have consistently demonstrated that most HNSCCs with HPV detected in the tumor are from the oral cavity and oropharynx,27 and HPV is driving the increasing incidence of oral cavity and oropharyngeal cancer over the past 30 years.28, 29 Consistent with prior reports that integration of HPV into the genome results in altered DNA copy number and mRNA transcript abundance and splicing,30 our analysis demonstrated that downregulated P4HA1 was more frequently found in tumors located in the oral cavity and oropharynx as well as in HPV‐positive patients, indicating that HPV infection contributes to HNSCC by inhibiting P4HA1 expression. Moreover, P4HA1 was overexpressed in HNSCC at advanced stages and with lymphatic metastasis compared with early stage disease and no lymphatic metastasis, suggesting the involvement of P4HA1 in the tumorigenesis and metastatic progression of HNSCC.
One of the most important issues concerning cancer patients is how to screen and diagnose at an early stage. Screening for HNSCC depends on clinical symptoms and imaging examinations (laryngoscopy, computed tomography, magnetic resonance imaging, and positron emission tomography), and a definite diagnosis depends on biopsy and histopathological examination.1 However, given the nonspecificity of symptoms in the early stage, the early detection of HNSCC remains unsatisfactory. In the present study, we constructed ROC curves and calculated the AUC to determine the diagnostic value of P4HA1 for HNSCC. The AUC value of TCGA and validation cohort was 0.887 and 0.883, respectively, signifying greater diagnostic accuracy compared with conventional cancer‐related biomarkers, such as carcinoembryonic antigen (CEA), squamous cell carcinoma antigen (SCC Ag), TPS (tissue polypeptide specific antigen), and Cyfra 21‐1.31, 32 These results suggest that P4HA1 expression levels might represent a promising diagnostic biomarker for HNSCC.
Despite current treatment regimens with curative intent, including surgery, radiotherapy and chemotherapy, local or distant recurrence rates remain high, and the 5‐year overall survival rate of HNSCC patients is less than 50%.33 Emerging therapeutic strategies, such as anti‐EGFR antibody (cetuximab) and anti‐PD‐1 antibodies (pembrolizumab and nivolumab) that have recently been approved for the treatment of advanced and metastatic HNSCC, are promising options for the management of high‐risk patients. However, predicting high‐risk HNSCC patients remains a challenge for both the clinician and the pathologist.34 Tumor diameter, lymphatic metastasis, distal metastasis, and clinical stage are vital factors affecting tumor patient outcomes;35 however, these factors are unable to absolutely justify clinical application due to heterogeneous molecular mechanisms and clinical behaviors of HNSCC.36 Therefore, reliable prognostic biomarkers are urgently needed to identify HNSCC patients at risk of disease recurrence and subsequent death. Recently, dysregulation of P4HA1 expression was reported to promote tumor progression and associated with unfavorable prognosis in various cancers, including gliomas,16 breast cancer,17 and prostate cancer.23 With regard to the findings in the present study, the log‐rank test and univariate Cox proportional hazard analysis showed that high P4HA1 expression was correlated with inferior OS and RFS of HNSCC patients, and these findings are consistent with previous report.37 Future multivariate Cox proportional hazard analysis confirmed that both elevated P4HA1 and advanced stages were dependent poor prognostic factors for OS and RFS of HNSCC patients after adjusting for age, gender, smoking behavior, alcohol consumption, and histologic grade. Taken together, the present study indicates that P4HA1 expression may be of great value for tailoring of individual therapies and risk stratification of recurrence and subsequent death, which might help these patients benefit from an intensified first‐line treatment and surveillance.
Based on large HNSCC samples in TCGA using cBioPortal for Cancer Genomics, we found that although P4HA1 genetic alteration was less frequent in HNSCC (8/504), its alteration was associated with significantly worse overall survival and disease‐free survival. Given that elevated P4HA1 was dependent poor prognostic biomarker for OS and RFS of HNSCC patients, we hypothesized that P4HA1 genetic alterations might increase its expression level, which should be confirmed in further investigations. In HNSCC, P4HA1 coexpressed genes were additionally enriched in some cancer‐related and metabolism‐related pathways, such as HIF‐1 signaling pathway, lysine degradation pathway, and gluconeogenesis pathway. These results can provide novel insight HNSCC pathogenesis. In breast cancer, HIF‐1 mediates increasing P4HA1 expression in conditions of hypoxic stress, resulting in fibrillary collagen deposition and the induction of a more invasive cell phenotype.17 However, several limitations of our study should be considered. Due to the size of sample, we did not validate the P4HA1 protein expression level in HNSCC tissues. Additionally, the role of P4HA1 in these pathways in the HNSCC is not completely clear. Thus, further studies are needed to explore underlying mechanism of P4HA1 in these pathways in the HNSCC.
5. CONCLUSIONS
This integrated bioinformatics analysis provides strong evidence that increasing P4HA1 is significantly associated with HNSCC carcinogenesis and metastasis. Additionally, high P4HA1 expression is both a diagnostic biomarker and an independent prognostic factor for poor OS and RFS in HNSCC patients.
CONFLICTS OF INTEREST
None of the authors have any commercial or other associations that might pose a conflict of interest.
ACKNOWLEDGMENTS
This work was supported by grants from the National Natural Science Foundation of China (No. 81670920), the Zhejiang Provincial Natural Science Foundation of China (Nos. LY14H160003, LY15H130003, and LY19H160090), the Medical and Health Science Research Foundation of Zhejiang Province (Nos. 2016KYB272, 2018RC063, and 2019ZD018), the Medical and Health Training Project of Zhejiang Province (Nos. 2014PYA017 and 2015RCB025), the Scientific Innovation Team Project of Ningbo (No. 2012B82019), the Ningbo Natural Science Foundation (Nos. 2013A610217, 2015A610221, 2017A610236, 2018A610363, and 2018A610361), the Huimin Technology Research and Development Projects of Ningbo (No. 2015C50026), and the Ningbo Health Branding Subject Fund (PPXK2018‐02). Thanks are due to the professional English language copy editing provided by American Journal Experts Company.
Li Q, Shen Z, Wu Z, et al. High P4HA1 expression is an independent prognostic factor for poor overall survival and recurrent‐free survival in head and neck squamous cell carcinoma. J Clin Lab Anal. 2020;34:e23107 10.1002/jcla.23107
Contributor Information
Chongchang Zhou, Email: zhou900709900709@163.com.
Huigao Liu, Email: 8658259386582593@163.com.
REFERENCES
- 1. Vigneswaran N, Williams MD. Epidemiologic trends in head and neck cancer and aids in diagnosis. Oral Maxillofac Surg Clin North Am. 2014;26(2):123‐141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394‐424. [DOI] [PubMed] [Google Scholar]
- 3. Hashibe M, Brennan P, Chuang S‐C, et al. Interaction between tobacco and alcohol use and the risk of head and neck cancer: pooled analysis in the International Head and Neck Cancer Epidemiology Consortium. Cancer Epidemiol Biomarkers Prev. 2009;18(2):541‐550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kobayashi K, Hisamatsu K, Suzui N, Hara A, Tomita H, Miyazaki T. A review of HPV‐related head and neck cancer. J Clin Med. 2018;7(9):241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lewis A, Kang R, Levine A, Maghami E. The new face of head and neck cancer: the HPV epidemic. Oncology. 2015;29(9):616‐626. [PubMed] [Google Scholar]
- 6. Marur S, Forastiere AA. Head and neck squamous cell carcinoma: update on epidemiology, diagnosis, and treatment. Mayo Clin Proc. 2016;91(3):386‐396. [DOI] [PubMed] [Google Scholar]
- 7. Sailer V, Gevensleben H, Dietrich J, et al. Clinical performance validation of PITX2 DNA methylation as prognostic biomarker in patients with head and neck squamous cell carcinoma. PLoS ONE. 2017;12(6):e0179412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhou C, Ye M, Ni S, et al. DNA methylation biomarkers for head and neck squamous cell carcinoma. Epigenetics. 2018;13(4):398‐409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhou C, Shen Z, Ye D, et al. The association and clinical significance of CDKN2A promoter methylation in head and neck squamous cell carcinoma: a meta‐analysis. Cell Physiol Biochem. 2018;50(3):868‐882. [DOI] [PubMed] [Google Scholar]
- 10. Shen Z, Zhou C, Li J, Deng H, Li Q, Wang J. The association, clinicopathological significance, and diagnostic value of CDH1 promoter methylation in head and neck squamous cell carcinoma: a meta‐analysis of 23 studies. Onco Targets Ther. 2016;9:6763‐6773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sivarajah S, Kostiuk M, Lindsay C, et al. EGFR as a biomarker of smoking status and survival in oropharyngeal squamous cell carcinoma. J Otolaryngol Head Neck Surg. 2019;48(1):1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wigerup C, Pahlman S, Bexell D. Therapeutic targeting of hypoxia and hypoxia‐inducible factors in cancer. Pharmacol Ther. 2016;164:152‐169. [DOI] [PubMed] [Google Scholar]
- 13. Gilkes DM, Bajpai S, Chaturvedi P, Wirtz D, Semenza GL. Hypoxia‐inducible factor 1 (HIF‐1) promotes extracellular matrix remodeling under hypoxic conditions by inducing P4HA1, P4HA2, and PLOD2 expression in fibroblasts. J Biol Chem. 2013;288(15):10819‐10829. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 14. Myllyharju J, Kivirikko KI. Collagens, modifying enzymes and their mutations in humans, flies and worms. Trends Genet. 2004;20(1):33‐43. [DOI] [PubMed] [Google Scholar]
- 15. Sorensen BS, Toustrup K, Horsman MR, Overgaard J, Alsner J. Identifying pH independent hypoxia induced genes in human squamous cell carcinomas in vitro. Acta Oncol. 2010;49(7):895‐905. [DOI] [PubMed] [Google Scholar]
- 16. Hu W‐M, Zhang JI, Sun S‐X, et al. Identification of P4HA1 as a prognostic biomarker for high‐grade gliomas. Pathol Res Pract. 2017;213(11):1365‐1369. [DOI] [PubMed] [Google Scholar]
- 17. Gilkes DM, Chaturvedi P, Bajpai S, et al. Collagen prolyl hydroxylases are essential for breast cancer metastasis. Can Res. 2013;73(11):3285‐3296. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18. Kuriakose MA, Chen WT, He ZM, et al. Selection and validation of differentially expressed genes in head and neck cancer. Cell Mol Life Sci. 2004;61(11):1372‐1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Shen Z, Li Q, Deng H, Lu D, Song H, Guo J. Long non‐coding RNA profiling in laryngeal squamous cell carcinoma and its clinical significance: potential biomarkers for LSCC. PLoS ONE. 2014;9(9):e108237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lu P, Weaver VM, Werb Z. The extracellular matrix: a dynamic niche in cancer progression. J Cell Biol. 2012;196(4):395‐406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Provenzano PP, Eliceiri KW, Campbell JM, Inman DR, White JG, Keely PJ. Collagen reorganization at the tumor‐stromal interface facilitates local invasion. BMC Med. 2006;4(1):38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Feng G, Shi H, Li J, et al. MiR‐30e suppresses proliferation of hepatoma cells via targeting prolyl 4‐hydroxylase subunit alpha‐1 (P4HA1) mRNA. Biochem Biophys Res Comm. 2016;472(3):516‐522. [DOI] [PubMed] [Google Scholar]
- 23. Chakravarthi BVSK, Pathi SS, Goswami MT, et al. The miR‐124‐prolyl hydroxylase P4HA1‐MMP1 axis plays a critical role in prostate cancer progression. Oncotarget. 2014;5(16):6654‐6669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sarafim‐Silva BAM, Duarte GD, Sundefeld M, Biasoli ER, Miyahara GI, Bernabe DG. Childhood trauma is predictive for clinical staging, alcohol consumption, and emotional symptoms in patients with head and neck cancer. Cancer. 2018;124(18):3684‐3692. [DOI] [PubMed] [Google Scholar]
- 25. Joseph AW, D'Souza G. Epidemiology of human papillomavirus‐related head and neck cancer. Otolaryngol Clin North Am. 2012;45(4):739‐764. [DOI] [PubMed] [Google Scholar]
- 26. Lechner M, Fenton TR. The genomics, epigenomics, and transcriptomics of HPV‐associated oropharyngeal cancer‐understanding the basis of a rapidly evolving disease. Adv Genet. 2016;93:1‐56. [DOI] [PubMed] [Google Scholar]
- 27. Sturgis EM, Cinciripini PM. Trends in head and neck cancer incidence in relation to smoking prevalence: an emerging epidemic of human papillomavirus‐associated cancers? Cancer. 2007;110(7):1429‐1435. [DOI] [PubMed] [Google Scholar]
- 28. Ernster JA, Sciotto CG, O’Brien MM, et al. Rising incidence of oropharyngeal cancer and the role of oncogenic human papilloma virus. Laryngoscope. 2007;117(12):2115‐2128. [DOI] [PubMed] [Google Scholar]
- 29. Hammarstedt L, Lindquist D, Dahlstrand H, et al. Human papillomavirus as a risk factor for the increase in incidence of tonsillar cancer. Int J Cancer. 2006;119(11):2620‐2623. [DOI] [PubMed] [Google Scholar]
- 30. Parfenov M, Pedamallu CS, Gehlenborg N, et al. Characterization of HPV and host genome interactions in primary head and neck cancers. Proc Natl Acad Sci USA. 2014;111(43):15544‐15549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Inal E, Lacin M, Asal K, et al. The significance of ferritin, lipid‐associated sialic acid, CEA, squamous cell carcinoma (SCC) antigen, and CYFRA 21‐1 levels in SCC of the head and neck. Kulak Burun Bogaz Ihtis Derg. 2004;12(1–2):23‐30. [PubMed] [Google Scholar]
- 32. Ogawa T, Tsurusako Y, Kimura N, et al. Comparison of tumor markers in patients with squamous cell carcinoma of the head and neck. Acta Otolaryngol Suppl. 1999;540:72‐76. [DOI] [PubMed] [Google Scholar]
- 33. Shield KD, Ferlay J, Jemal A, et al. The global incidence of lip, oral cavity, and pharyngeal cancers by subsite in 2012. CA Cancer J Clin. 2017;67(1):51‐64. [DOI] [PubMed] [Google Scholar]
- 34. Chow LQM, Haddad R, Gupta S, et al. Antitumor activity of pembrolizumab in biomarker‐unselected patients with recurrent and/or metastatic head and neck squamous cell carcinoma: results from the phase Ib KEYNOTE‐012 expansion cohort. J Clin Oncol. 2016;34(32):3838‐3845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Shao Y, Ye M, Li Q, et al. LncRNA‐RMRP promotes carcinogenesis by acting as a miR‐206 sponge and is used as a novel biomarker for gastric cancer. Oncotarget. 2016;7(25):37812‐37824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mao L, Hong WK, Papadimitrakopoulou VA. Focus on head and neck cancer. Cancer Cell. 2004;5(4):311‐316. [DOI] [PubMed] [Google Scholar]
- 37. Tawk B, Schwager C, Deffaa O, et al. Comparative analysis of transcriptomics based hypoxia signatures in head‐ and neck squamous cell carcinoma. Radiother Oncol. 2016;118(2):350‐358. [DOI] [PubMed] [Google Scholar]


