Abstract
Over the last decade, multiple studies have shown that signaling proteins activated in different temporal patterns—such as oscillatory, transient, and sustained—can result in distinct gene expression patterns or cell fates. However, the molecular events that ensure appropriate stimulus- and dose-dependent dynamics are not often understood and are difficult to investigate. Here, we used single-cell analysis to dissect the mechanisms underlying the stimulus- and dose-encoding patterns in the innate immune signaling network. We found that Toll-like receptor (TLR) and interleukin-1 receptor (IL-1R) signaling dynamics relied on a dose-dependent, auto-inhibitory loop that rendered cells refractory to further stimulation. Using inducible gene expression and optogenetics to perturb the network at different levels, we identified the IL-1R–associated kinase1 (IRAK1) as the dose-sensing node responsible for limiting signal flow during the innate immune response. Although the kinase activity of IRAK1 was not required for signal propagation, it played a critical role in inhibiting the nucleocytoplasmic oscillations of the transcription factor NF-κB. Thus, protein activities that may be “dispensable” from a topological perspective can nevertheless be essential in shaping the dynamic response to the external environment.
Introduction
Living cells encounter an overwhelming amount of biological, chemical, and physical information that must be identified and quantified appropriately to elicit an appropriate response (1). Understanding how cells process all of this information requires uncovering the link between the organization of signaling components (that is, the topology of the pathway) and their temporal patterns of activity (that is, their dynamics) (2). Although cell population-based studies have been critical to map the topology of signaling networks, the complexity of the temporal patterns of signaling was underappreciated before the advent of live, single-cell studies. Unsynchronized oscillatory patterns of activity have been found in various pathways including those involving the extracellular signal-regulated kinase (ERK) (3–5), the tumor suppressor p53 (6–8), and the innate immunity regulator and transcription factor nuclear factor κB (NF-κB) (9–12). These pathways respond to a broad spectrum of stimuli and execute specific gene expression programs by precisely regulating the stimulus- and dose-dependent dynamics of kinase and transcription factor activity (11, 13–15). However, these dynamics cannot be predicted solely based on the topological organization of signaling components. Indeed, the signaling nodes and circuit patterns that ensure appropriate dynamics are not often understood.
The innate immune signaling network displays oscillatory dynamics in response to a wide range of cytokines and pathogen associated molecular patterns (PAMPs) (16, 17). These molecules activate distinct cell surface receptors, including the tumor necrosis factor receptor (TNFR), the interleukin-1 receptor (IL-1R), and Toll-like receptors (TLRs). However, they signal through the same pathways, the stress activated protein kinases (SAPKs) cJun N-terminal kinase (JNK) and p38, and the transcription factor NF-κB (18, 19), to name a few. Mechanistically, NF-κB oscillations from the nucleus to the cytoplasm in response to tumor necrosis factor alpha (TNFα) are due to transcriptionally induced negative regulators, such as IκB-α or A20 (20). The oscillation frequency is highly variable between isogenic cells and heavily influenced by both extrinsic noise (for example, the availability of transcriptional and translational machinery) and intrinsic noise (for example the probability of transcription bursting) (17). This heterogeneity is important to mitigate biological noise and increase robustness at the population level (21, 22).
Differences in dynamic signaling patterns also lead to distinct phenotypic outcomes (23). Moreover, multiple studies have suggested that gene expression can be quantitatively and qualitatively regulated by transcription factor dynamics (13, 24–26). We previously reported a combined measurement of both signaling dynamics and global transcription output in the same individual cells. This study demonstrated that cells displaying different temporal patterns of NF-κB activity execute distinct cytokine expression patterns (15).
It is therefore clear that developing an accurate, fundamental understanding of the innate immune response will depend on elucidating the factors that shape the dynamic response. However, the molecular mechanisms dedicated to fine-tune NF-κB signaling dynamics remain elusive. Here, we used live single-cell analysis, new optogenetic tools, and CRISPR to describe the mechanistic basis of NF-κB signaling dynamics. Previous studies have used optical strategies to model dynamic behaviors (14, 27, 28), and others have used population-based studies to investigate mechanisms (29–33); here, we bridged these studies by using optical tools to identify the mechanistic basis of signaling dynamics in single cells.
Results
TLR4 elicits non-monotonic dose-encoding NF-κB dynamics
The innate immune network is activated by multiple cytokines and PAMPs, such as TNFα, lipopolysacharide (LPS), and IL-1β. These three molecules bind to different cell surface receptors (TNFR, TLR4, and IL-1R respectively) but signal through the same mitogen activated protein kinase kinase kinase (MAP3K) TAK1 [transforming growth factor- β (TGF-β) activated kinase 1]. TAK1 stimulates the nuclear translocation of NF-κB through IκB kinase (IKK)-mediated degradation of IκBα and the activation of the MAPKs JNK and p38. However, whereas LPS and IL-1β activate TAK1 through the same complex, which involves the adaptor MyD88, IRAK4, IRAK1 or IRAK2 and the E3 ubiquitin ligase TRAF6, TNFα uses a different set of upstream components, which include the adaptor TNF receptor associated death domain (TRADD), TRAF2 or TRAF5 and receptor interacting protein 1 kinase (RIP1) (Fig. 1A) (34, 35).
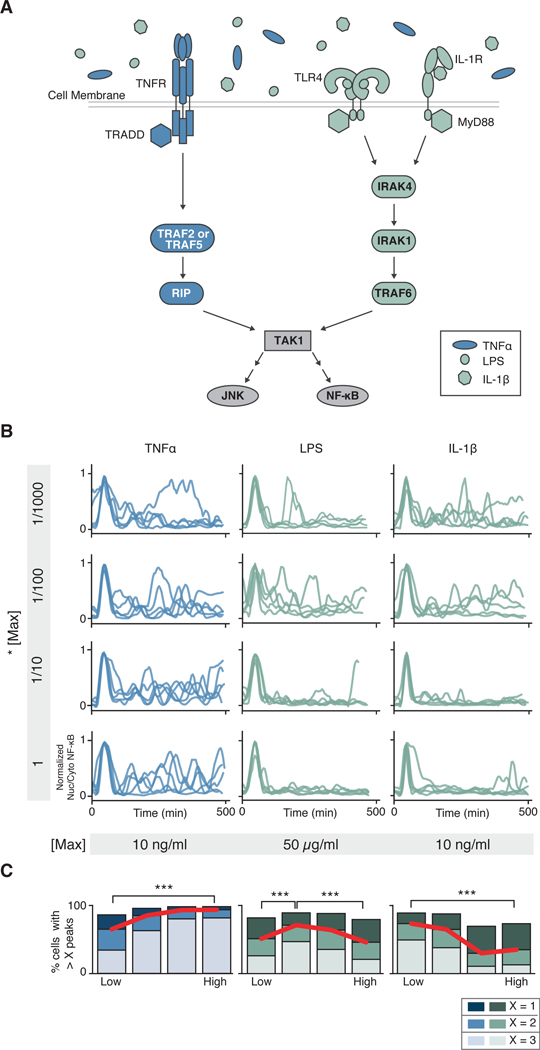
Fig. 1. The TNFR and TLR or IL-1R elicit qualitatively different dose-dependent NF-κB dynamics.
(A) Schematic representation of the innate immune signaling network. TNFα, LPS, and IL-1β activate innate immune signaling through different receptors and signaling proteins. While TLR4 (LPS) and IL-1R (IL-1β) share most signaling components, TNFR (TNFα) signals through different proteins. Both cascades lead to activation of NF-κB and MAPK signaling via TAK1. (B) Parental strain (PS) cells (NIH 3T3, RelA−/−, p65-dsRed, H2B-EGFP) were treated with increasing concentrations (top to bottom) of indicated stimuli, imaged, and quantified as described in the Methods (see sections on live cell imaging and segmentation and tracking). Five randomly selected traces from > 2,000 are shown per condition. Traces have been aligned to their first peak for clarity. (C) Peak counting quantification (see Methods for details) of the data presented in (B). Fractions of cells with more than 1, 2, or 3 peaks are shown to highlight population distribution. Data are representative of three independent experiments. High vs. Low, ***P< 0.001 by chi-square test. For TNFα, LPS and IL-1β, > 6,700, 9,600 and 11,600 cells respectively were analyzed across concentration.
To determine the dose-response kinetics of NF-κB signaling in response to different stimuli, we stimulated RelA (also known as p65) deficient mouse embryonic fibroblasts (RelA−/−) expressing p65-DsRed with TNFα, LPS or IL-1β, and measured the dynamics of p65 nuclear translocation at the single-cell level (Fig. 1B and fig. S1). Our data showed that whereas increasing concentrations of TNFα monotonically increased the number of NF-κB oscillations per cell, increasing concentrations of either LPS or IL-1β decreased NF-κB oscillations (Fig. 1C). Additional analysis of NF-κB signaling dynamics showed that other parameters such as area under the curve, percentage of responding cells or amplitude of the first peak monotonically increased with both TNFα or IL-1β concentration (fig. S2). Furthermore, the LPS dose-encoding pattern showed a statistically significant non-monotonic behavior by which low and high concentrations stimulated fewer oscillations compared to intermediate concentrations. These data suggest that a dose-dependent inhibition of signal flow occurs during TLR and IL-1R signaling.
TLR and IL-1R stimuli render cells cross-tolerant to further stimulation in a dose-dependent manner
Previous studies have shown that refractory periods within the NF-κB signaling network exist (36). In addition, the qualitative difference observed between the dose-encoding patterns of different innate immune signaling molecules suggested that NF-κB activation may be temporarily inhibited after TLR and IL-1R signaling. Thus, we performed sequential stimulation with all combinations of TNFα, LPS and IL-1β. Briefly, we stimulated cells with a primary input for 30 min which was followed by a three-hour recovery period and a secondary stimulation (Fig. 2A). Individual cells were tracked and NF-κB translocation dynamics were recorded throughout the experiment.
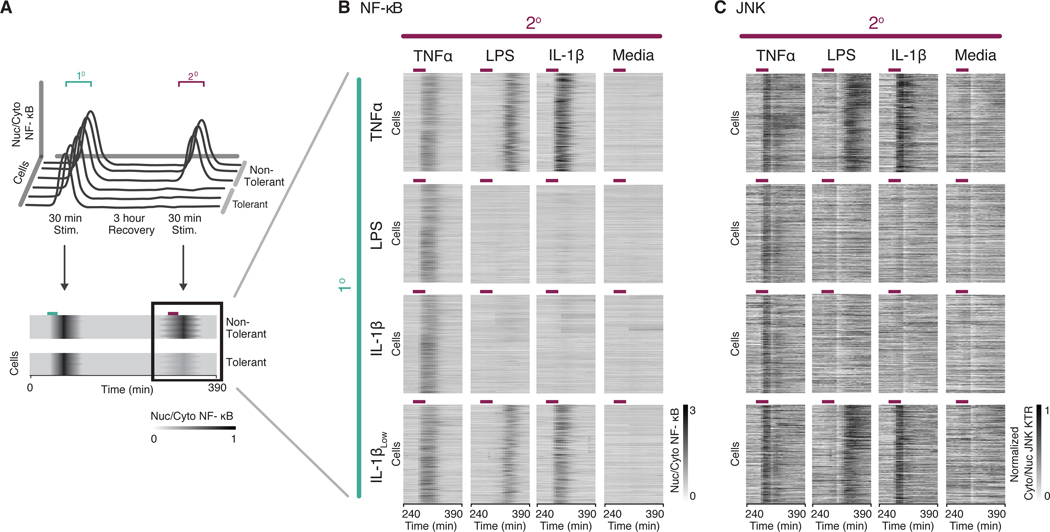
Fig. 2. TLR and IL-1R stimuli render cells cross-tolerant to further stimulation.
(A) Schematic representation of the experimental timeline. During tolerance experiments, cells were stimulated with a primary (1°) and secondary (2°) stimulation for 30 minutes with an intervening rest period of 3 hours. Traces of NF-κB activity during the secondary response were converted to heatmaps wherein rows indicate individual cells, columns indicate time, and the grayscale colormap represents nuclear to cytoplasmic median intensity ratio of p65-DsRed. (B and C) Parental Strain (PS) cells (RelA−/−, p65-DsRed, H2B-EGFP) expressing JNK-KTR-mCerulean3 were stimulated with different combinations of primary and secondary inputs as indicated (10 ng/ml TNFα, 5 μg/ml LPS, 1 ng/ml IL-1β, 0.1 ng/ml IL-1βLow), and monitored for NF-κB (B) and JNK (C) activity. Time period for secondary stimulation only is shown for clarity. Purple lines over heatmaps indicate time period when cells were in secondary stimulus. Cells were filtered to include only those responding to the primary stimulus. Data represents two independent experiments (n > 300 cells per condition, > 9,000 cells total).
We observed that cells that were stimulated first with TNFα responded to any secondary stimulus. However, cells that were stimulated first with high doses of LPS or IL-1β were refractory (or “cross-tolerant”) to a secondary stimulus through TLR or IL-1R. Furthermore, these same cells responded normally to a secondary TNFα stimulus, indicating that the TAK1-NF-κB signaling axis was fully functional (Fig. 2B). Whereas receptor inactivation (for example, by endocytosis) might explain tolerance to the same stimulus, the fact that cells were also refractory to other TLR or IL-1R ligands suggests a common mechanism downstream of the receptor. Indeed, the cross-tolerance effect was dose-dependent: cells stimulated first with a low dose of IL-1β or LPS were responsive to any secondary stimulus (Fig. 2B and fig. S3A). These results confirm that a high dose of TLR or IL-1R ligand renders the cells insensitive to a subsequent stimulus through these receptors.
To test whether this negative feedback affected signaling downstream of TAK1, we used our previously described Kinase Translocation Reporter (KTR) technology to measure JNK activity (37) in the same sequential stimulation experiments. Specifically, JNK KTR transforms JNK kinase activity into the nucleocytoplasmic translocation of a single fluorescent protein that can be easily measured by epifluorescence. Our results showed that TLR and IL-1R-dependent JNK activation was also abolished in cells previously stimulated with high doses of LPS or IL-1β (Fig. 2C). In summary, these results suggest that the mechanism responsible for the cross-tolerance effect targets signaling upstream of TAK1 and downstream of the receptors, TLR4 and IL-1R.
Optogenetic control of signaling at the MyD88 and TRAF6 nodes identifies IRAK1 as the mechanism of dose-dependent cross tolerance
In order to identify the molecular mechanism leading to dose-dependent cross-tolerance, we aimed to develop molecular tools to control signaling at different nodes in the network. TLR and IL-1R signaling is initiated by receptor-mediated nucleation of the Myddosome, a macromolecular complex composed of multiple subunits of MyD88, IRAK4 and IRAK1/2 (38). Within this complex, IRAK4 activates IRAK1 to recruit the RING finger ubiquitin ligase TRAF6 that mediates TAK1 activation (33, 39). The fact that multimerization is a recurrent mechanism to activate signaling in this pathway suggested that a light-induced aggregation of key pathway intermediates may be sufficient to elicit signaling. The flavoprotein cryptochrome 2 mutant (Cry2o) has previously been shown to aggregate in response to blue wavelength light (40). We fused this protein to MyD88 (OptoMyD88) (Fig. 3A) as well as the N-terminal ubiquitin ligase domain of TRAF6 (OptoTRAF6) (Fig. 3B), and expressed these in NF-κB reporter cells. We found that a brief pulse of blue light (470/24 nm excitation coupled to a FITC filter) caused robust nuclear translocation of NF-κB, comparable to cytokine induced responses (Fig. 3C and 3D).
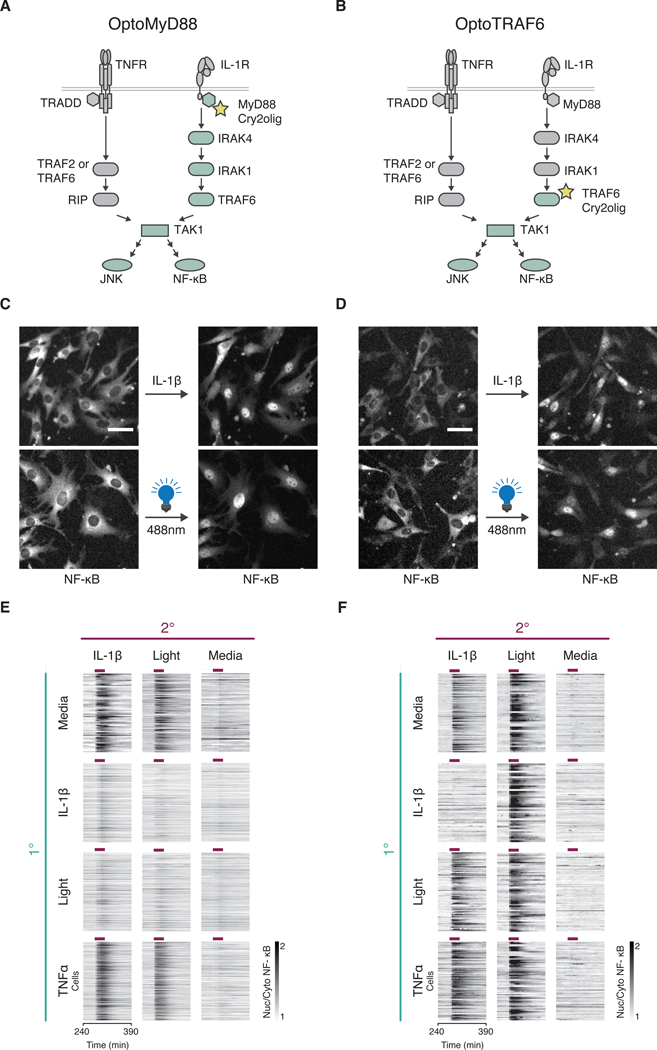
Fig. 3. Optogenetic control of signaling at MyD88 and TRAF6 nodes maps the cross-tolerance mechanism to IRAK proteins.
(A and B) Schematic representation of OptoMyD88 (A) or OptoTRAF6 (B) activation of innate immune signaling. Upon light stimulation, Opto tools activate their respective downstream proteins labeled in light green. (C and D) Cells (NIH 3T3, p65-mRuby, H2B-iRFP) expressing OptoMyD88 (C) or OptoTRAF6 (D) under the Tet-Responsive Element Third Generation (TRE3G) promoter were incubated in doxycycline (dox, 2 μg/ml) overnight and stimulated with either IL-1β (1 ng/ml) or light (470/24 nm, five 250 ms pulses with 5 min intervals). Representative NF-κB localization images before and after indicated stimuli are shown. Experiment was performed with a 10x objective. Scale bar, 50 μm. (E and F) Secondary response heatmaps of NF-κB nuclear translocation in OptoMyD88 (E) or OptoTRAF6 (F) cells with varied primary (1°) and secondary (2°) stimulations (experimental timeline as in Fig. 2A). Cells were incubated in dox (2 μg/ml) overnight and treated with TNFα (10 ng/ml), IL-1β (1 ng/ml), light (470/24 nm, 250 ms pulses), or media for 30 minutes (purple bars). Data represents three independent experiments (n > 100 cells per condition). Cells were filtered to include only those responding to the primary stimulus, except for those with no primary stimulus.
Next, we tested whether optically induced signaling with OptoTRAF6 or OptoMyD88 renders cells cross tolerant to further TLR or IL-1R stimulation, and vice versa. Our results showed that activation of signaling at the MyD88 level with OptoMyD88 strongly inhibited secondary TLR or IL-1R signaling (Fig. 3E), but not TNFR signaling (fig. S3B). However, signaling at the TRAF6 level with OptoTRAF6 did not inhibit further signaling (Fig. 3F). Conversely, OptoTRAF6 was able to bypass TLR and IL-1R induced tolerance, and OptoMyD88 was not (Fig. 3, E and F).
These data suggest that the mechanism of tolerance originates and acts upstream of TRAF6 and downstream of MyD88, leaving only the IRAK proteins as potential candidates for the cross-tolerance mechanism. To determine whether tolerant cells activate IRAK4 and/or IRAK1 in response to TLR or IL-1R stimulation, we performed immunoblotting assays against the active phosphorylated form of IRAK4 and total IRAK1 (Fig. 4A). These experiments showed that IL-1β induced IRAK4 phosphorylation in both tolerant cells primed with cytokine stimulation, and non-tolerant cells primed with OptoTRAF6 stimulation (Fig. 4B and fig. S4). As previously described, IRAK1 levels appear to decrease upon TLR or IL-1R stimulation (30), and these levels remain low at the time of secondary stimulation (Fig. 4C). In contrast, cells stimulated with OptoTRAF6 (i.e., non-tolerant cells) did not show any decrease in IRAK1 abundance (Fig. 4C). Together these data suggest that when cells are in a tolerant state, subsequent signaling can still activate IRAK4 but not IRAK1. Previous studies have shown that the apparent decrease of IRAK1 abundance in response to stimulation is reversible by treating protein extracts with phosphatases and deubiquitinating enzymes, indicating that IRAK1 is not degraded but heavily modified under stimulation (41). In fact, endogenous IRAK1 is heavily modified upon IL-1β treatment (fig. S5A). Thus, one possibility is that IRAK1 posttranslational modification is negatively regulating IRAK1 function and rendering cells cross tolerant to further stimulation.
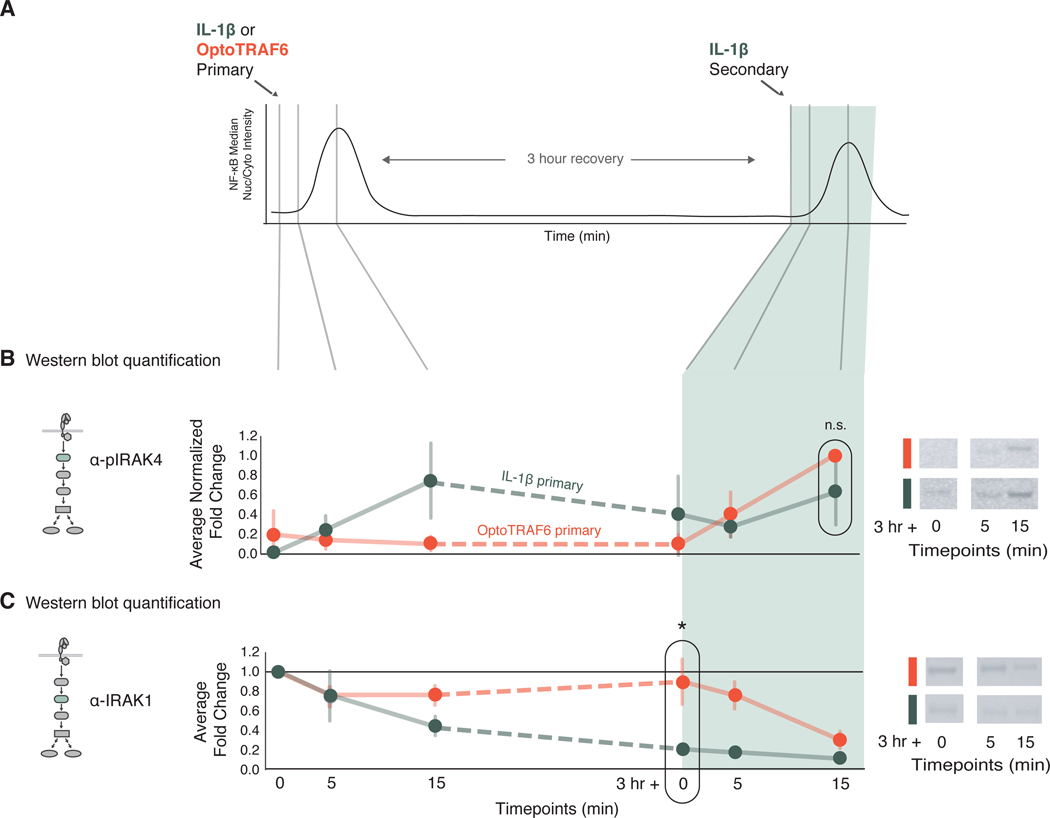
Fig. 4. Low IRAK1 abundance correlates with the cross-tolerant state.
(A) Schematic detailing of stimulation and sample collection timeline for western blotting. Cells were stimulated with primary stimuli as indicated [1 ng/ml IL-1β, green; OptoTRAF6 (light, 488nm), orange] and sampled at 0, 5, and 15 min after primary stimulation. Cells were washed after 30 min of primary stimulation, allowed to recover for 3 hours, then challenged with secondary stimulation of IL-1β, and sampled at 0, 5 and 15 min. (B) Quantification of immunoblots for phosphorylated IRAK4 (pIRAK4) in cells treated with light (orange) or IL-1β (green) and subjected to a secondary IL-1β stimulation. pIRAK4 abundance was first normalized to a β-actin loading control, then fold-change compared to the unstimulated condition (first time point) was calculated for each sample, then normalized between 0 and 1 across all time points. Data are means ± SD of three independent experiments, n.s. = not significant by a t-test. Representative blots are shown for the last three timepoints, for both IL-1β and OptoTRAF6 primary conditions. Full blots and additional stimulation combinations are presented in fig. S4. (C) Quantification of Western blots for IRAK1, as described in (B). IRAK1 expression was first normalized to a β-actin loading control, then fold-change to the unstimulated condition was calculated for each sample. Data are mean ± SD of three independent experiments, *p<0.05 by a t-test. Representative blots are shown for the last three timepoints, for both IL-1β and OptoTRAF6 primary conditions. As in (B), full blots and additional stimulation combinations are shown in fig. S4.
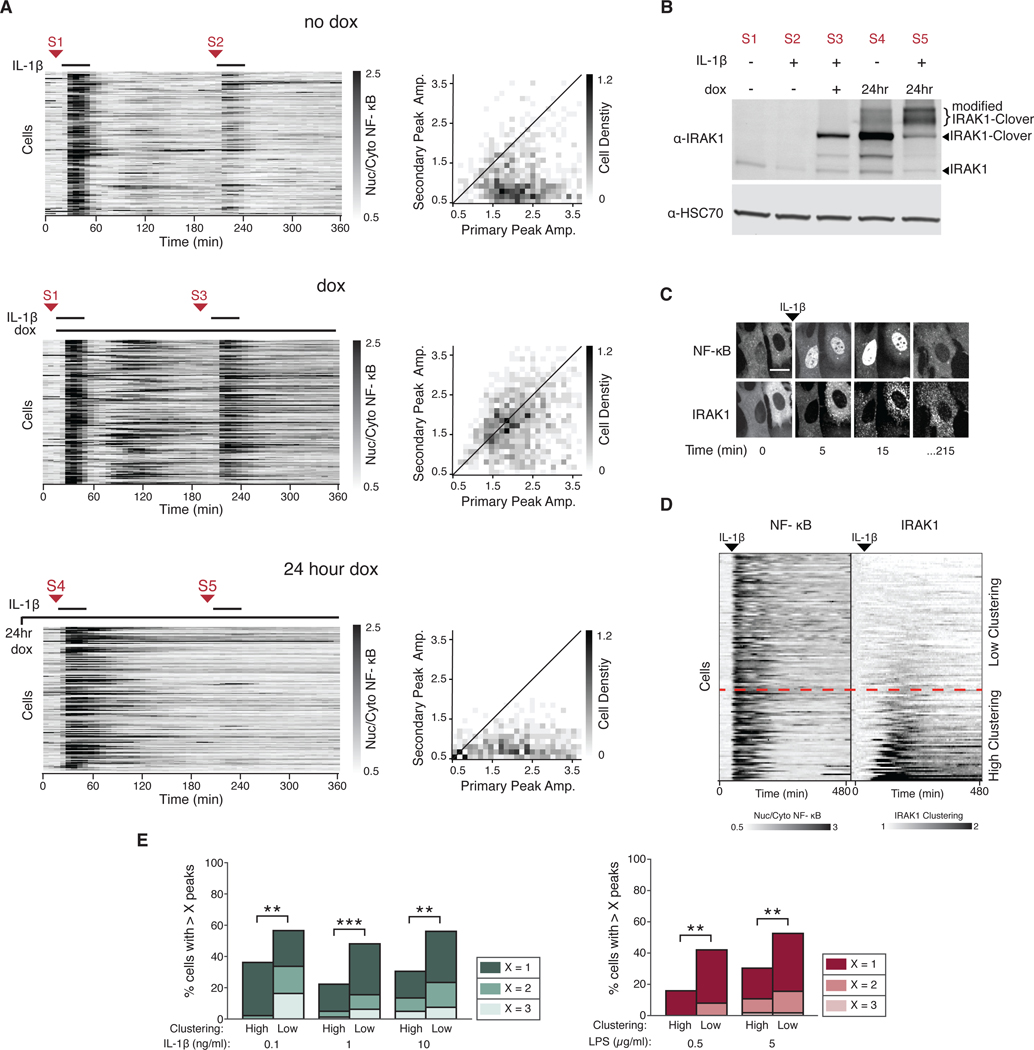
To more directly test whether unmodified IRAK1 was able to bypass the tolerant state, we infected a wild type NF-κB reporter-expressing cell line with a lentiviral vector containing IRAK1 under the control of a doxycycline inducible promoter. This strategy enabled us to test if expression of IRAK1 before or after the primary stimulation is enough to bypass tolerance. We found that doxycycline-induced expression of IRAK1 after the primary stimulation enabled cells to respond to a secondary stimulation (Fig. 5A). However, overexpression of IRAK1 before primary stimulation resulted in increased duration of the NF-κB primary response but did not bypass tolerance (Fig. 5A). These data suggest that IRAK1 levels are limiting the primary response which is in turn coupled to IRAK1 inactivation by posttranslational modification. Thus, newly synthetized (i.e. unmodified) IRAK1 after the primary stimulus allows cells to respond to the secondary IL-1β stimulation (Fig. 5B). The ability of IRAK1 to bypass tolerance is specific because cells overexpressing either MyD88 or TRAF6 remained insensitive to a second stimulus (fig. S5, B and C). Together our data suggests that IRAK1 has a dual role in controlling signal flow: IRAK1 abundance is required for signal propagation (30, 39) and at the same time its posttranslational modification inhibits signaling after a primary stimulation.
Fig. 5. Expression of unmodified IRAK1 protein bypasses tolerance.
(A) Induced expression of IRAK1 after primary stimulation. Cells containing TRE3G::IRAK1-Clover were incubated without doxycycline (no dox), with dox at the time of primary stimulation (dox), or 24 hours overnight (24 hour dox). Cells were stimulated with IL-1β (1 ng/ml) and imaged as described in the Methods. After 30 min, cells were washed and allowed to recover for 3 hours and stimulated again with IL-1β (1 ng/ml). Red arrows indicate sample collection points for the Western blot shown in (B). 2D histograms show the distribution of peak amplitudes of nuclear/cytoplasmic NF-κB median intensity during the primary vs secondary response in each cell. Black line indicates primary equals secondary NF-κB amplitude. Data represents three independent experiments with n>100 cells. (B) Western blotting for IRAK1 protein abundance between primary and secondary stimuli in cells treated as in (A) were harvested at indicated times [S1-S5, corresponding to those in A)]. HSC70 was used as a loading control. Blot is representative of three independent experiments. (C) Representative confocal images of IRAK1-Clover cluster formation following NF-kB activation. Cells stably expressing IRAK1-Clover were imaged before and after IL-1β stimulation (1 ng/ml). Scale bar, 50 μm.(D) IRAK1-Clover cells were imaged for 8 hours and stimulated with IL-1β (0.1 ng/ml) 45 min into the time course. NF-κB nuclear/cytoplasmic intensity ratio (left) and IRAK1-Clover clustering dynamics (right) are displayed in tandem. Heatmap rows are ordered top to bottom based on increasing IRAK1 clustering (see Materials and Methods). Dashed red line represents an arbitrary IRAK1 clustering threshold (1.3-fold change). (E) Grouping of cells by IRAK1 clustering separates oscillatory vs. non-oscillatory cells. Irak1-KO cells expressing IRAK1-Clover were stimulated with IL-1β (0.1, 1, and 10 ng/ml) or LPS (0.5 and 5 μg/ml). Peaks of NF-κB activity and IRAK1 clustering from single cell traces obtained in (D) and fig. S6B were measured as described in the Methods. An arbitrary threshold of 1.3-fold increase in IRAK1 clustering was used in all conditions to group high vs low IRAK1 clustering cells. Within each group, fractions of cells with more than 1, 2, or 3 peaks are shown to highlight population distribution. (n >100 cells; **P<0.01 and ***P<0.001 by chi-square test). Additional clustering quantification is provided in fig. S8A, and heatmaps of additional concentrations of IL-1β are provided in fig. S6B.
We observed that IRAK1 formed aggregates in response to either LPS or IL-1β (Fig. 5C). This IRAK1 aggregation was highly heterogeneous between isogenic cells and depended on TLR or IL-1R signaling, because TNFα failed to aggregate IRAK1 (fig. S6A). Because IRAK1 clustering occurred after NF-κB activation and persisted long after stimulation, we hypothesized that it may be correlated with the inhibitory function of IRAK1 rather than the signal propagation function. Therefore, we further explored the relationship between IRAK1 clustering and signaling dynamics by stimulating cells expressing fluorescently tagged IRAK1 with IL-1β or LPS and measuring both NF-κB dynamics and IRAK1 clustering in each individual cell. Our results showed a negative correlation between IRAK1 clustering and NF-κB oscillations. In fact, grouping of cells based on the degree of IRAK1 clustering was able to predict oscillatory behavior (Fig. 5, D and E, and fig. S6B). Moreover, the percentage of IRAK1 clustered cells increased with IL-1β concentration as expected (fig. S6C). In addition, varying the resting period between primary and secondary stimulation showed that the percentage of responding cells inversely correlated with IRAK1 posttranslational modification and clustering (fig. S6, D and E). These data suggest that IRAK1 cluster formation regulates signaling dynamics in the innate immune signaling network.
IRAK1 kinase activity is critical to regulate signaling dynamics in the innate immune signaling network
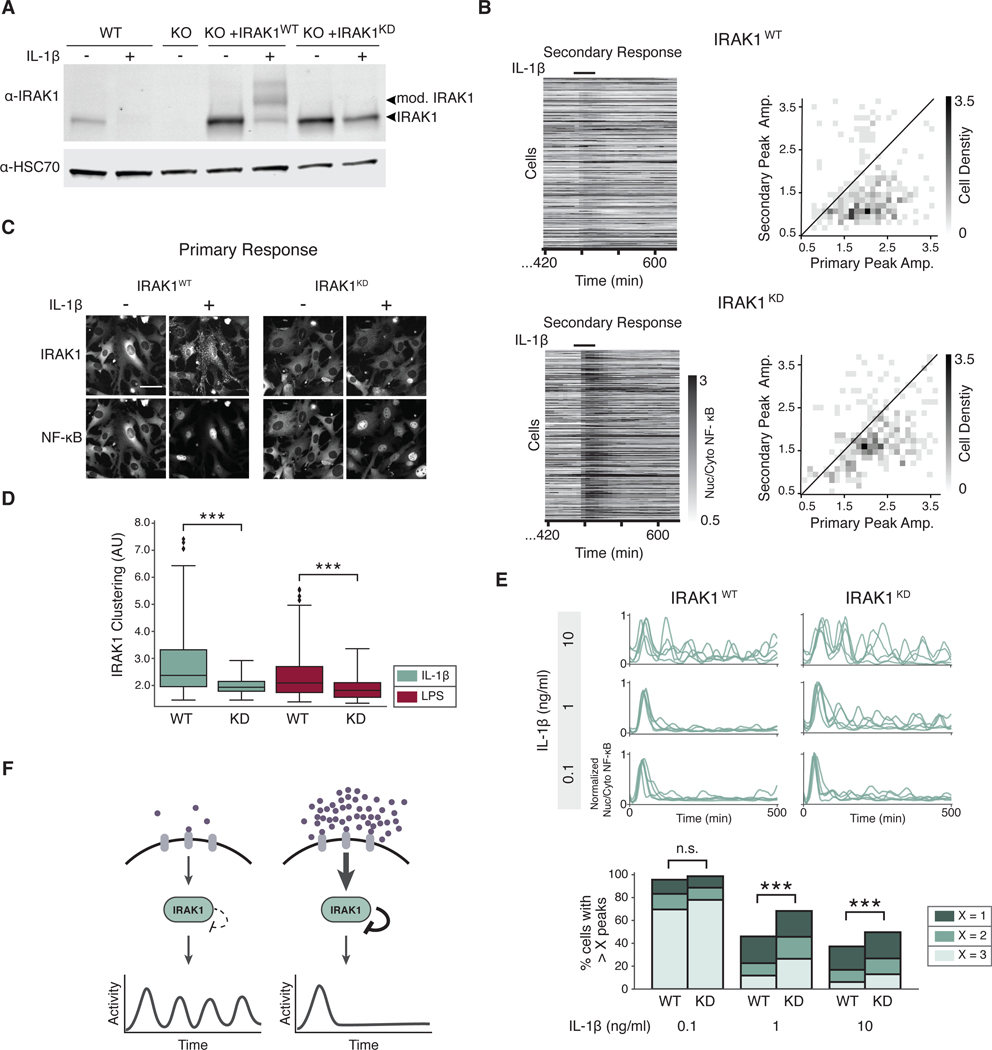
To mechanistically understand the dual role of IRAK1 in regulating NF-κB signaling dynamics, we aimed to uncouple the positive and negative activities of IRAK1 using specific point mutations. First, we generated an NIH3T3 Irak1 knockout cell line with CRISPR Cas9 (Irak1-KO) and complemented it with wild-type (WT) IRAK1 or one of several IRAK1 mutants, including the non-phosphorylatable IRAK1T209A and the kinase-deficient IRAK1K239S (IRAK1KD) (29, 42). Of note, Irak1-KO cells were not responsive to IL-1β or LPS, confirming that signaling in the complemented cell lines was exclusively due to reconstituted IRAK1 (fig. S7A). Both NF-κB dynamics and IRAK1 post-translational modification in response to IL-1β stimulation were determined for each cell line (fig. S7, B and C). Our results showed that only the IRAK1KD mutant was able to propagate the signal but unable to undergo post-translational modification (Fig. 6A). Previous studies have suggested that IRAK1 kinase activity is dispensable for signaling, however the lack of post-translational modification prompted us to ask whether these cells could still activate tolerance or not. Because of the increased duration of NF-κB response between the IRAK1 reconstituted and WT cells, we increased the recovery period between primary and secondary stimulation to 8 hours. Whereas IRAK1WT cells were still tolerant, IRAK1KD cells were responsive to a secondary stimulation of TLR or IL-1R (Fig. 6B), suggesting that IRAK1KD cells are capable of signaling but show significantly impaired tolerance. Together, these data indicate that the inhibitory role of IRAK1 depends on its own kinase activity constituting a negative autoregulation loop. Although these two roles appear to oppose, the inhibitory function of IRAK1 is only present at high ligand concentrations, allowing cells to tightly control the dose response of the pathway in a single node.
Fig. 6. IRAK1 kinase activity is critical to regulate NF-κB signaling dynamics.
(A) WT cells and Irak1-KO cells reconstituted with IRAK1WT or IRAK1KD were incubated with or without IL-1β (1 ng/ml) for 3 hours. Lysate was collected and immunoblotted against IRAK1. Arrows indicate posttranslationally modified and unmodified IRAK1 protein. HSC70 was used as a loading control. Blot is representative of 3 experiments. (B) Secondary response heatmaps of IRAK1WT or IRAK1KD cells show reduced tolerance in IRAK1KD. Irak1-KO cells reconstituted with IRAK1WT and IRAK1KD were stimulated with a 30-min pulse of IL-1β (1 ng/ml), washed, allowed to recover for 8 hours, and stimulated again with a secondary pulse of IL-1β (1 ng/ml). 2D histograms show the distribution of peak amplitudes of nuclear/cytoplasmic NF-κB median intensity during the primary vs. secondary response in each cell. Black line indicates primary equals secondary NF-κB amplitude. Data represents three independent experiments with n>100 cells. (C) Irak1-KO cells expressing IRAK1WT-Clover or IRAK1KD-Clover were imaged prior to and 20 min after stimulation with IL-1β (1 ng/ml). Representative images are shown. Scale bar, 50 μm. (D) IRAK1 clustering was quantified as described in methods in IRAK1WT-Clover or IRAK1KD-Clover cells stimulated with IL-1β (0.1 ng/ml) or LPS (0.5 μg/ml). Data represent n >100 cells; ***P<0.001 by a Kolmogorov–Smirnov test. (E) IRAK1 kinase activity regulates oscillatory dynamics. PS cells expressing IRAK1WT-Clover or IRAK1KD -Clover were stimulated with IL-1β (0.1, 1, or 10 ng/ml) and imaged for 8 hours. Five randomly selected single cell traces are presented for each condition. Peak counting of NF-κB oscillations were counted as described in methods. Fractions of cells with more than 1, 2, or 3 peaks are shown to highlight population distribution. Data represents three independent experiments with n >100 cells (n.s.= not significant; ***p < 0.001; chi-square test). See fig. S8B for TNFα data. (F) Schematic model of the effects of IRAK1-dependent autoinhibitory loop in NF-κB signaling dynamics. When ligand is in low abundance, TLR and IL-1R signaling is not inhibited after the initial activation, and continues to signal in an oscillatory pattern. When ligand concentration is high, IRAK1 kinase activity strongly inhibits signaling following the initial activation, and oscillations are not detected.
Because IRAK1 clustering is correlated with inhibition of NF-κB oscillations and IRAK1 kinase activity appears to be important for the inhibitory role of IRAK1, we hypothesized that IRAK1 kinase activity may be necessary to induce clustering. To address this question, we expressed a fluorescent IRAK1KD and stimulated cells with TLR or IL-1R ligands. As expected, IRAK1KD was unable to aggregate after stimulation (Fig. 6, C and D, and fig. S8A). Together, our data suggest that both IRAK1 posttranslational modification and clustering depend on IRAK1 kinase activity and correlate with the inhibitory function of IRAK1.
The ability to uncouple the positive and negative functions of IRAK1 further enabled us to interrogate the role of IRAK1 autoinhibition in regulating signaling dynamics. Thus, we expressed IRAK1WT or IRAK1KD in our reporter cell line (RelA−/−, p65-DsRed) and recorded single cell NF-κB dynamics under multiple concentrations of IL-1β and TNFα. Results showed that IRAK1KD expressing cells had significantly more NF-κB oscillations than IRAK1WT cells at high concentrations of IL-1β (Fig. 6E), but not with high TNFα concentrations (fig. S8B). Therefore, we conclude that the dual role of IRAK1 in innate immune signaling is dose-dependent; low ligand concentrations trigger signaling dependent on IRAK1 abundance whereas high concentrations trigger IRAK1 autoinhibition, which is critical to inhibit further signaling (Fig. 6F). We note that even in IRAK1KD cells, the oscillatory dynamics is reduced at high IL-1β concentrations, suggesting that other mechanisms may be in place to terminate the signal. In addition, these data suggest that although IRAK1 kinase activity is not necessary for signal propagation, it is important to terminate NF-κB pathway activity.
Discussion
In higher eukaryotes, the innate immune network is activated by a broad spectrum of cytokines and PAMPs. These stimuli must be precisely identified, quantified and integrated because insufficient or exaggerated immune responses can have devastating consequences. Although the main components of the innate immune signaling network have been identified, the mechanisms and circuit topologies that ensure appropriate stimuli-dependent and dose-dependent signaling are unclear. Here, we used live-cell biosensors combined with optogenetics to identify molecular determinants of innate immune signaling dynamics.
Our data shows that NF-κB oscillatory dynamics depend on the nature of the stimulation. Whereas increasing doses of TNFα monotonically augmented the number of oscillations per cell, increasing doses of either LPS or IL-1β decreased NF-κB oscillations. Indeed, high concentrations of TLR or IL-1R ligands led to an abbreviated response and rendered the cells insensitive to further stimulation. Moreover, the effect of LPS concentrations on NF-κB dynamics was non-monotonic; the signaling responses of greatest duration were elicited at intermediate concentrations. Non-monotonic behaviors have been observed in transcriptional networks, but are rarely found in signaling networks (43–45). This innate immune non-monotonic signaling may help prevent the harmful effects of enhanced inflammation.
By using sequential stimulation with multiple inputs, we demonstrated that TLR and IL-1R signaling were strongly inhibited after stimulation, rendering cells cross-tolerant to further stimulation. Although we did not observe any tolerance with TNFα at three hours after stimulation, a previous study described a refractory period lasting for one hour after TNFα treatment (36). Thus, the presence of heterogeneous tolerant states upon multiple activators of the innate immune signaling network emphasizes the importance of balancing innate immune signaling.
The discovery of light-regulated domains to manipulate signaling has opened up the exciting possibility of simultaneously measuring and perturbing signaling dynamics in single cells (27, 46). To identify the molecular mechanisms leading to TLR and IL-1R tolerance and dose-response function, we used two new optogenetic tools, OptoTRAF6 and OptoMyD88, to stimulate signaling at different nodes in the network. These tools hold two main promises: (i) enabling future research regarding the physiological consequences of signaling dynamics by enabling fine temporal control of signaling, and (ii) opening the possibility of interrogating paracrine communication dynamics between immune cells by enabling spatially restricted control of NF-κB signaling.
In this study, the use of optogenetic tools enabled us to control signaling dynamics to identify IRAK1 as the dose-sensing node responsible for TLR and IL-1R signaling dynamics and that IRAK1 has a dual role in controlling signal flow. On the one hand, IRAK1 abundance was important to for signal propagation, but on the other hand, its kinase activity was responsible for an autophosphorylation event that eventually switched the cell between active and tolerant states. Whereas the activating role of IRAK1 was present at all concentrations, the effects of the autoinhibitory phosphorylation were only seen at high concentrations, when the pool of active IRAK1 was depleted. In addition, we showed that the autoinhibitory loop of IRAK1 is critical to regulate NF-κB oscillatory dynamics.
Our results showed that NF-κB signaling is extremely sensitive to IRAK1 abundance, suggesting that alternative mechanisms may exist to regulate IRAK1 expression. Indeed, previous studies showed that IRAK1 mRNA translation is regulated by the micro-RNA miR-146a, which can bind to the 3’UTR of the mRNA and inhibit its translation (47, 48). Moreover, patients carrying mutations in the 3’UTR region of IRAK1 mRNA have an increased risk of rheumatoid arthritis (49). This extra layer of regulation reinforces the notion that IRAK1 is responsible for controlling innate immune signal flow.
In addition to being linked to autoimmunity, IRAK1 mutations have been linked to multiple cancers (50, 51). However, efforts to use IRAK1 as a therapeutic target are based on dual specificity IRAK4 and IRAK1 inhibitors. Our analysis of IRAK1 mutants demonstrates that IRAK1 kinase activity is dispensable for signaling but critical to regulate NF-κB dynamics. Thus, the assumption that dual specificity inhibitors against IRAK4 and IRAK1 cause similar effects to specific IRAK1 inhibitor may be incorrect. Indeed, because IRAK1 activity decreased NF-κB oscillations, a specific IRAK1 inhibitor may cause paradoxical effects. Further investigation into the nature of cancer-driving mutations in IRAK1 will determine whether the signaling or the tolerance role of IRAK1 is linked to tumorigenesis.
In clinical immunology, the phenomenon of LPS tolerance has been extensively studied (52). This phenomenon consists of a substantial inhibition of the immune function after a systemic exposure to endotoxin, to the point where sepsis patients ultimately undergo fatal secondary infections. Although multiple mechanisms have been connected to long-term tolerance, including epigenetic regulation and cytokine production, the IRAK1 autoinhibition mechanism shown here likely contributes to acute tolerance. We speculate that further characterization of human IRAK1 homologs and the development of specific inhibitors may help to relieve acute tolerance.
The identification of molecular mechanisms that serve to regulate signaling dynamics reinforces the idea that signaling should be understood as a continuous process composed of fluxes, equilibria, and feedbacks loops, rather than discrete events. Moreover, the complexity of signaling dynamics and its role in regulating cell physiology require new approaches involving live single-cell analysis. These approaches will be critical to define the molecular mechanisms responsible for controlling signaling dynamics and quantitatively encode complex environmental information. We anticipate that such understanding will be critical to design efficient pharmacological intervention.
MATERIALS AND METHODS
Cell lines and cell culture
Cells were maintained in Dulbecco’s Modified Eagle Medium (DMEM) (Gibco) supplemented with 10% Fetal Bovine Serum (FBS) (Omega Scientific), 2mM L-Glutamine (Gibco), 1x Penicillin/Streptomycin (Gibco)) at 37°C and 5% CO2. OptoTRAF6 and OptoMyD88 expressing cells were handled in minimal ambient light to avoid risk of pathway activation.
Plasmid generation
All plasmids were generated using Gibson assembly (53) into a pENTR vector, sequenced, then recombined into a pLenti vector using Gateway cloning. The OptoTRAF6 construct is based on the previously described TRAF6-GryB fusion (31) and consists of the N-terminal effector domain of TRAF6 (AA 1–359) fused to the PHR domain of Cry2olig (Cry2o) (40) followed by a 2A peptide and nuclear reporter H2B-iRFP. The OptoMyD88 construct is full-length MyD88 fused to the PHR domain of Cry2olig (40).
Cell line generation
All cell lines were established with lentiviral infection. Lentivirus was generated with lipofectamine transfection of third generation viral packaging plasmids and lentiviral construct into HEK293FT cells. Cells were incubated in viral supernatant with Polybrene (10 μg/ml) (EMD Millipore) for 48 hours before selection. Parental Strain (PS) cell line is RelA−/− NIH3T3 fibroblasts transduced with p65-DsRed and H2B-EGFP nuclear marker (10). To construct PS-JNK-KTR-mCer cells, PS cells were infected with lentiviral JNK-KTR-mCerulean3 and selected with puromycin (Invivogen) (2 μg/ml). To construct OptoTRAF6 and OptoMyD88 cell lines, NIH3T3 fibroblasts expressing p65-mRuby and H2B-iRFP were transduced with a Tet Responsive Element 3rd Generation promoter (TRE3G), inducible lentiviral construct. Tet inducible IRAK1, MyD88, and TRAF6 cell lines were made in the NIH3T3 p65-mRuby, H2B-iRFP background with a TRE3G promoter followed by IRAK1 (or MyD88 or TRAF6) and Clover (54). Irak1 knockout (Irak1-KO) was achieved by ligation of guide RNA targeting exon 1 (target sequence: CTGGAACCACAGGCTCCC) of the mouse Irak1 gene into a CRISPR V2 Neomycin resistant vector (provided by Dr. Andrew Holland, Johns Hopkins University School of Medicine) and transduced into NIH3T3 cells stably expressing H2B-iRFP and p65-mRuby. Cells were selected for neomycin resistance (0.5 mg/ml) and clonally expanded. Genomic DNA was amplified by PCR across the target site and sequenced to verify a successful frame shift mutation (55). IRAK1-WT and IRAK1KD constructs were expressed in three backgrounds: the Irak1-KO, NIH3T3 p65-mRuby H2B-iRFP, and the PS cell line. Cells were validated for expression and activity using live cell imaging or Western blotting.
Live cell imaging
Cells were seeded on glass bottom 96-well plates (Thermo Fisher) coated with fibronectin (10 μg/ml) (Sigma Aldrich), and left to adhere overnight. The next day, the cells were washed and media was changed to imaging media (Fluorobrite) (Thermo Fisher Scientific) (10 mM Hepes, 1% FBS, 1 x Penicillin/Streptomycin (Gibco), 2mM L-Glutamine (Gibco)). Cells were imaged with a Nikon Eclipse Ti-E inverted fluorescence microscope with Andor Neo 5.5 or Hamamatsu sCMOS camera and a Prime 95B camera for confocal images, controlled by Micromanager or Metamorph software. LED excitation light source (SPECTRA X) was used at 430/24 nm (mCerulean3), 470/24 (mClover or GFP), 500/20 (mClover, used when imaged with mCerulean3), 550/15 nm (mRuby2 or dsRed), and 640/30 nm (iRFP670). Unless otherwise stated, experiments were imaged with a 20x objective. Exposure times for each light channel were limited to 250 ms and frequency of imaging acquisition was no higher than 5 minutes (unless otherwise stated) to preserve cell health. Temperature (37°C), CO2 (5%), and humidity were controlled throughout the experiments.
Imaging of cells for peak counting experiments
Cells were imaged every five minutes with controlled temperature (37°C), CO2 (5%) and humidity over an 8-hour time course. Cells were pre-imaged to establish a baseline then stimulated with the indicated concentration of TNFα (Roche), LPS (Enzo Life Sciences or Thermofisher), or IL-1β (R & D).
Tolerance experiment imaging
For tolerance experiments, cells were pre-imaged to establish a baseline, then stimulated with the indicated primary stimulus for 30 min, washed, imaged for 3 hours, then stimulated with secondary stimulus for 30 min, washed, and imaged for another 1.5 hours. For OptoTRAF6 and OptoMyD88 experiments, cells were incubated in doxycycline (2 μg/ml) (Thermo Scientific) for 15 hours prior to the experiment and all imaging media and stimuli thereafter contained doxycycline. Cells were imaged every 5 minutes and received 30 minutes of primary stimulations of either TNFα (10 ng/ml), IL-1β (1 ng/ml), or Light (470/24 nm excitation coupled with a FITC filter, five 250 ms pulses with 5 min intervals), washed, allowed a 3 hour resting period, then stimulated again with secondary stimulations as indicated.
Segmentation and tracking
Image correction, segmentation and cell tracking were implemented by custom python scripts that combine existing packages such as SimpleITK (56) using the image analysis pipeline outlined in Kudo et. al. (57). The nuclear images were preprocessed using the histogram-matching standardization. This accounts for an overall intensity change during time-lapse experiments and helps consistent segmentation and tracking results. After the illumination bias correction based on the improved N3 bias correction (58), the background of the nuclear images and NF-κB images were estimated using an iterative wavelet transform (59) algorithm and a block-based estimation, respectively. In the latter method, the image was divided into 100 blocks (10 grids for both x and y), and 0.1 percentile of each block pixels was calculated to produce 10×10 matrix. This matrix was then resized using bilinear interpolation, which produces the estimated background generally more conservative than those produced by the iterative wavelet transform.
For the segmentation, the high-pass filtered laplacian of gaussian images were then computed and added to the log-transform of nuclear images to enhance separation of nuclei, and nuclei were segmented by global thresholding followed by smoothing with morphological operations including opening. Nuclei areas averaged 145 pixels.
For tracking, first, each nuclei in consecutive frames were subjected for linking with a linear assignment problem (60) framework that minimizes a global linking cost based on squared distances between objects. Unlinked objects were then attempted to be separated by a marker-controlled watershed segmentation until one object is linked to a nucleus in a previous frame based on changes in intensity and distance. This enables an adaptive separation of two touching nuclei. Additionally, a gap closing algorithm was used to connect cells that disappear for less than 6 frames, and cells that do not appear for more than 20 frames were discarded from analysis. Once tracked nuclei were obtained, cytoplasmic region is identified by making a ring around each nuclei.
Analysis
For all NF-κB nuclear translocation analysis, NF-κB nuclear/cytoplasmic median intensities were used. Outlier cells with NF-κB expression outside a 95% range, were rejected. Sharp increases in nuclear/cytoplasmic intensity indicate division events. Thresholding was applied to detect division events, the spikes were removed, and the space between interpolated. When indicated a basic python peak finder, PeakUtils (https://bitbucket.org/lucashnegri/peakutils) was used to identify cells responding to a stimulus. For peak counting quantification, data was exported to Matlab, a Savitzky-Golay filter was applied to reduce the chance of false positive peak identification and a more robust peak counter, findPeaks (T.C. O’Haver 2014, Mathworks.com) was used. Additional image analysis was performed using custom python and Matlab scripts.
IRAK1 clustering analysis
For experiments involving IRAK1-Clover clustering measurements, a local standard deviation filter (MathWorks, stdfilt) that calculated the standard deviation divided by the mean intensity in each 3×3 pixel square was applied to IRAK1-Clover images. The mean intensity of the cytoring object in filtered images was used to measure IRAK1 texture and displayed in boxplots (Fig. 6 and fig. S8A). IRAK1 traces were converted into a fold change over the average of the first 5 (pre-stimulation) time points. To threshold heatmaps, the 90th percentile of the fold change stdfilt intensity trace was calculated to assign a value of IRAK1 clustering per cell and avoid outlier timepoints. Cells with an IRAK1 clustering value lower than 1.3 fold were assigned “low clustering” while cells higher than 1.3 were labeled “high clustering”.
Immunoblotting
Cells were seeded at 80% confluence and left to adhere overnight. The next morning cells were stimulated as indicated, washed with PBS, fully aspirated, then flash frozen at −80°C. Cells were harvested and proteins were extracted in Lysis Buffer [50 mM Tris pH 7.4, 150 mM NaCl, 1% Triton, 0.5% Na-Deoxycholate, 1% SDS, 2 mM EDTA, 1x fresh Halt Protease and Phosphatase Inhibitor Cocktail (Thermo Fisher Scientific), 1mM DTT]. Protein concentration was determined using a BCA Protein Assay Kit (Thermo Fisher Scientific). Protein was boiled for 5 min in Laemmli sample buffer (Thermo Fisher or Bio-Rad), resolved by SDS-PAGE, and transferred to a PVDF membrane (Milipore-Sigma). Blocking and antibody dilutions were done in Odyssey Blocking Buffer (Li-COR) and washes in PBS/Tween-20 (0.1%). IRAK1 (CST 4505), actin (CST 3700), pIRAK4 (CST 11927), pJNK (CST 4668), and JNK (CST 9258) antibodies were from Cell Signaling Technology; HSC70 (Sc-7298) antibody was from Santa Cruz Biotechnology; GFP (ab290) antibody was from Abcam; and all secondary antibodies were from Li-COR. Membranes were analyzed with a Li-COR scanner and quantified using ImageStudioLite software and ImageJ.
Light board construction
Western blot sample light stimulation was carried out on a customized programmable LED board (Evil Mad Scientist), with blue 470 nm LEDs (Evil Mad Scientist). To make sure the spread of light was even, a diffuser (Bright View Technologies) was applied 2 inches above the board attached to a custom 3D printed stage. Diffused light intensity was measured at 500 lux with a light meter. Cell stimulation was confirmed with imaging.
Statistical analysis
Several statistical methods were used to test for significance in different experiments. For discrete distribution data (such as total number of peaks counted in single cell traces), the chi-square test was used. For continuous distribution data (such as degree of clustering or Western blot quantification) the Kolmogorov-Smirnov or t-test was used.
Supplementary Material
Fig. S1. Graphical summary of the image analysis pipeline.
Fig. S2. Additional analysis and TNF concentration for data presented in Fig. 1.
Fig. S3. Additional stimulus combinations for Figs. 2 and 3.
Fig. S4. Full gels and quantification of Western blots in Fig. 4
Fig. S5. Increased abundance of MyD88 or TRAF6 after primary stimulation cannot bypass tolerance.
Fig. S6. IRAK1 modification and clustering with varied recovery periods.
Fig. S7. IRAK1 mutant screen and knockout characterization.
Fig. S8. Additional IRAK1 WT and KD-Clover characterization.
Acknowledgements:
We thank all members of the M.W.C. and S.R. laboratories for technical assistance and critical commentary on the manuscript. Dr. Holland, Dr. Seydoux, Dr. Urban and their laboratory members for providing us with reagents, equipment and technical assistance.
Funding: We gratefully acknowledge funding from several sources, including an NSF Graduate Research Fellowship and NIH training grant (T32GM007445) to H.R.C.; a Distinguished Investigator award and a Discovery Center grant from the Paul Allen Family Foundation, an NIH Pioneer Award (5DP1LM01150–05) and a Systems Biology Center grant (P50 GM107615) to M.W.C.; an NSF CAREER award (1844994), a Kimmel Scholar Award and Jerome L. Green Foundation award to S.R.
Footnotes
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions of this study are available in the paper or the supplementary materials.
References and Notes
- 1.Kholodenko BN, Cell-signalling dynamics in time and space. Nature reviews. Molecular cell biology 7, 165–176 (2006); published online EpubMar ( 10.1038/nrm1838). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Purvis JE, Lahav G, Encoding and decoding cellular information through signaling dynamics. Cell 152, 945–956 (2013); published online EpubFeb 28 ( 10.1016/j.cell.2013.02.005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Albeck JG, Mills GB, Brugge JS, Frequency-modulated pulses of ERK activity transmit quantitative proliferation signals. Molecular cell 49, 249–261 (2013); published online EpubJan 24 ( 10.1016/j.molcel.2012.11.002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shankaran H, Ippolito DL, Chrisler WB, Resat H, Bollinger N, Opresko LK, Wiley HS, Rapid and sustained nuclear-cytoplasmic ERK oscillations induced by epidermal growth factor. Molecular systems biology 5, 332 (2009) 10.1038/msb.2009.90). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sparta B, Pargett M, Minguet M, Distor K, Bell G, Albeck JG, Receptor Level Mechanisms Are Required for Epidermal Growth Factor (EGF)-stimulated Extracellular Signal-regulated Kinase (ERK) Activity Pulses. The Journal of biological chemistry 290, 24784–24792 (2015); published online EpubOct 09 ( 10.1074/jbc.M115.662247). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lahav G, Rosenfeld N, Sigal A, Geva-Zatorsky N, Levine AJ, Elowitz MB, Alon U, Dynamics of the p53-Mdm2 feedback loop in individual cells. Nature genetics 36, 147–150 (2004); published online EpubFeb ( 10.1038/ng1293). [DOI] [PubMed] [Google Scholar]
- 7.Purvis JE, Karhohs KW, Mock C, Batchelor E, Loewer A, Lahav G, p53 dynamics control cell fate. Science 336, 1440–1444 (2012); published online EpubJun 15 ( 10.1126/science.1218351). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stewart-Ornstein J, Cheng HWJ, Lahav G, Conservation and Divergence of p53 Oscillation Dynamics across Species. Cell Syst 5, 410–417 e414 (2017); published online EpubOct 25 ( 10.1016/j.cels.2017.09.012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee TK, Denny EM, Sanghvi JC, Gaston JE, Maynard ND, Hughey JJ, Covert MW, A noisy paracrine signal determines the cellular NF-kappaB response to lipopolysaccharide. Science signaling 2, ra65 (2009) 10.1126/scisignal.2000599). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tay S, Hughey JJ, Lee TK, Lipniacki T, Quake SR, Covert MW, Single-cell NF-kappaB dynamics reveal digital activation and analogue information processing. Nature 466, 267–271 (2010); published online EpubJul 8 ( 10.1038/nature09145). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nelson DE, Ihekwaba AE, Elliott M, Johnson JR, Gibney CA, Foreman BE, Nelson G, See V, Horton CA, Spiller DG, Edwards SW, McDowell HP, Unitt JF, Sullivan E, Grimley R, Benson N, Broomhead D, Kell DB, White MR, Oscillations in NF-kappaB signaling control the dynamics of gene expression. Science 306, 704–708 (2004); published online EpubOct 22 ( 10.1126/science.1099962). [DOI] [PubMed] [Google Scholar]
- 12.Sung MH, Salvatore L, De Lorenzi R, Indrawan A, Pasparakis M, Hager GL, Bianchi ME, Agresti A, Sustained oscillations of NF-kappaB produce distinct genome scanning and gene expression profiles. PloS one 4, e7163 (2009); published online EpubSep 29 ( 10.1371/journal.pone.0007163). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hafner A, Stewart-Ornstein J, Purvis JE, Forrester WC, Bulyk ML, Lahav G, p53 pulses lead to distinct patterns of gene expression albeit similar DNA-binding dynamics. Nat Struct Mol Biol 24, 840–847 (2017); published online EpubOct ( 10.1038/nsmb.3452). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wilson MZ, Ravindran PT, Lim WA, Toettcher JE, Tracing Information Flow from Erk to Target Gene Induction Reveals Mechanisms of Dynamic and Combinatorial Control. Molecular cell 67, 757–769 e755 (2017); published online EpubSep 07 ( 10.1016/j.molcel.2017.07.016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lane K, Van Valen D, DeFelice MM, Macklin DN, Kudo T, Jaimovich A, Carr A, Meyer T, Pe’er D, Boutet SC, Covert MW, Measuring Signaling and RNA-Seq in the Same Cell Links Gene Expression to Dynamic Patterns of NF-kappaB Activation. Cell Syst 4, 458–469 e455 (2017); published online EpubApr 26 ( 10.1016/j.cels.2017.03.010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoffmann A, Levchenko A, Scott ML, Baltimore D, The IkappaB-NF-kappaB signaling module: temporal control and selective gene activation. Science 298, 1241–1245 (2002); published online EpubNov 08 ( 10.1126/science.1071914). [DOI] [PubMed] [Google Scholar]
- 17.Hughey JJ, Gutschow MV, Bajar BT, Covert MW, Single-cell variation leads to population invariance in NF-kappaB signaling dynamics. Molecular biology of the cell 26, 583–590 (2015); published online EpubFeb 01 ( 10.1091/mbc.E14-08-1267). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kyriakis JM, Avruch J, Mammalian MAPK signal transduction pathways activated by stress and inflammation: a 10-year update. Physiol Rev 92, 689–737 (2012); published online EpubApr ( 10.1152/physrev.00028.2011). [DOI] [PubMed] [Google Scholar]
- 19.Newton K, Dixit VM, Signaling in innate immunity and inflammation. Cold Spring Harb Perspect Biol 4, (2012); published online EpubMar 01 ( 10.1101/cshperspect.a006049). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Werner SL, Kearns JD, Zadorozhnaya V, Lynch C, O’Dea E, Boldin MP, Ma A, Baltimore D, Hoffmann A, Encoding NF-kappaB temporal control in response to TNF: distinct roles for the negative regulators IkappaBalpha and A20. Genes & development 22, 2093–2101 (2008); published online EpubAug 01 ( 10.1101/gad.1680708). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Selimkhanov J, Taylor B, Yao J, Pilko A, Albeck J, Hoffmann A, Tsimring L, Wollman R, Systems biology. Accurate information transmission through dynamic biochemical signaling networks. Science 346, 1370–1373 (2014); published online EpubDec 12 ( 10.1126/science.1254933). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kellogg RA, Tay S, Noise facilitates transcriptional control under dynamic inputs. Cell 160, 381–392 (2015); published online EpubJan 29 ( 10.1016/j.cell.2015.01.013). [DOI] [PubMed] [Google Scholar]
- 23.Santos SD, Verveer PJ, Bastiaens PI, Growth factor-induced MAPK network topology shapes Erk response determining PC-12 cell fate. Nature cell biology 9, 324–330 (2007); published online EpubMar ( 10.1038/ncb1543). [DOI] [PubMed] [Google Scholar]
- 24.Hao N, O’Shea EK, Signal-dependent dynamics of transcription factor translocation controls gene expression. Nat Struct Mol Biol 19, 31–39 (2012); published online EpubJan ( 10.1038/nsmb.2192). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Porter JR, Fisher BE, Batchelor E, p53 Pulses Diversify Target Gene Expression Dynamics in an mRNA Half-Life-Dependent Manner and Delineate Co-regulated Target Gene Subnetworks. Cell Syst 2, 272–282 (2016); published online EpubApr 27 ( 10.1016/j.cels.2016.03.006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tian B, Nowak DE, Brasier AR, A TNF-induced gene expression program under oscillatory NF-kappaB control. BMC Genomics 6, 137 (2005); published online EpubSep 28 ( 10.1186/1471-2164-6-137). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Toettcher JE, Voigt CA, Weiner OD, Lim WA, The promise of optogenetics in cell biology: interrogating molecular circuits in space and time. Nature methods 8, 35–38 (2011); published online EpubJan ( 10.1038/nmeth.f.326). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Toettcher JE, Weiner OD, Lim WA, Using optogenetics to interrogate the dynamic control of signal transmission by the Ras/Erk module. Cell 155, 1422–1434 (2013); published online EpubDec 5 ( 10.1016/j.cell.2013.11.004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dunne A, Carpenter S, Brikos C, Gray P, Strelow A, Wesche H, Morrice N, O’Neill LA, IRAK1 and IRAK4 promote phosphorylation, ubiquitination, and degradation of MyD88 adaptor-like (Mal). The Journal of biological chemistry 285, 18276–18282 (2010); published online EpubJun 11 ( 10.1074/jbc.M109.098137). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goh ET, Arthur JS, Cheung PC, Akira S, Toth R, Cohen P, Identification of the protein kinases that activate the E3 ubiquitin ligase Pellino 1 in the innate immune system. Biochem J 441, 339–346 (2012); published online EpubJan 01 ( 10.1042/BJ20111415). [DOI] [PubMed] [Google Scholar]
- 31.Hacker H, Redecke V, Blagoev B, Kratchmarova I, Hsu LC, Wang GG, Kamps MP, Raz E, Wagner H, Hacker G, Mann M, Karin M, Specificity in Toll-like receptor signalling through distinct effector functions of TRAF3 and TRAF6. Nature 439, 204–207 (2006); published online EpubJan 12 ( 10.1038/nature04369). [DOI] [PubMed] [Google Scholar]
- 32.Koziczak-Holbro M, Joyce C, Gluck A, Kinzel B, Muller M, Tschopp C, Mathison JC, Davis CN, Gram H, IRAK-4 kinase activity is required for interleukin-1 (IL-1) receptor- and toll-like receptor 7-mediated signaling and gene expression. The Journal of biological chemistry 282, 13552–13560 (2007); published online EpubMay 04 ( 10.1074/jbc.M700548200). [DOI] [PubMed] [Google Scholar]
- 33.Walsh MC, Kim GK, Maurizio PL, Molnar EE, Choi Y, TRAF6 autoubiquitination-independent activation of the NFkappaB and MAPK pathways in response to IL-1 and RANKL. PloS one 3, e4064 (2008) 10.1371/journal.pone.0004064). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.O’Neill LA, Golenbock D, Bowie AG, The history of Toll-like receptors - redefining innate immunity. Nat Rev Immunol 13, 453–460 (2013); published online EpubJun ( 10.1038/nri3446). [DOI] [PubMed] [Google Scholar]
- 35.Brenner D, Blaser H, Mak TW, Regulation of tumour necrosis factor signalling: live or let die. Nat Rev Immunol 15, 362–374 (2015); published online EpubJun ( 10.1038/nri3834). [DOI] [PubMed] [Google Scholar]
- 36.Adamson A, Boddington C, Downton P, Rowe W, Bagnall J, Lam C, Maya-Mendoza A, Schmidt L, Harper CV, Spiller DG, Rand DA, Jackson DA, White MR, Paszek P, Signal transduction controls heterogeneous NF-kappaB dynamics and target gene expression through cytokine-specific refractory states. Nature communications 7, 12057 (2016); published online EpubJul 06 ( 10.1038/ncomms12057). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Regot S, Hughey JJ, Bajar BT, Carrasco S, Covert MW, High-sensitivity measurements of multiple kinase activities in live single cells. Cell 157, 1724–1734 (2014); published online EpubJun 19 ( 10.1016/j.cell.2014.04.039). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gay NJ, Gangloff M, O’Neill LA, What the Myddosome structure tells us about the initiation of innate immunity. Trends Immunol 32, 104–109 (2011); published online EpubMar ( 10.1016/j.it.2010.12.005). [DOI] [PubMed] [Google Scholar]
- 39.Vollmer S, Strickson S, Zhang T, Gray N, Lee KL, Rao VR, Cohen P, The mechanism of activation of IRAK1 and IRAK4 by interleukin-1 and Toll-like receptor agonists. Biochem J 474, 2027–2038 (2017); published online EpubJun 06 ( 10.1042/BCJ20170097). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Taslimi A, Vrana JD, Chen D, Borinskaya S, Mayer BJ, Kennedy MJ, Tucker CL, An optimized optogenetic clustering tool for probing protein interaction and function. Nature communications 5, 4925 (2014) 10.1038/ncomms5925). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Emmerich CH, Cohen P, Optimising methods for the preservation, capture and identification of ubiquitin chains and ubiquitylated proteins by immunoblotting. Biochem Biophys Res Commun 466, 1–14 (2015); published online EpubOct 09 ( 10.1016/j.bbrc.2015.08.109). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kollewe C, Mackensen AC, Neumann D, Knop J, Cao P, Li S, Wesche H, Martin MU, Sequential autophosphorylation steps in the interleukin-1 receptor-associated kinase-1 regulate its availability as an adapter in interleukin-1 signaling. The Journal of biological chemistry 279, 5227–5236 (2004); published online EpubFeb 13 ( 10.1074/jbc.M309251200). [DOI] [PubMed] [Google Scholar]
- 43.Ascensao JA, Datta P, Hancioglu B, Sontag E, Gennaro ML, Igoshin OA, Non-monotonic Response to Monotonic Stimulus: Regulation of Glyoxylate Shunt Gene-Expression Dynamics in Mycobacterium tuberculosis. PLoS Comput Biol 12, e1004741 (2016); published online EpubFeb ( 10.1371/journal.pcbi.1004741). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kaplan S, Bren A, Dekel E, Alon U, The incoherent feed-forward loop can generate non-monotonic input functions for genes. Molecular systems biology 4, 203 (2008) 10.1038/msb.2008.43). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li L, Andersen ME, Heber S, Zhang Q, Non-monotonic dose-response relationship in steroid hormone receptor-mediated gene expression. J Mol Endocrinol 38, 569–585 (2007); published online EpubMay ( 10.1677/JME-07-0003). [DOI] [PubMed] [Google Scholar]
- 46.Zhou XX, Fan LZ, Li P, Shen K, Lin MZ, Optical control of cell signaling by single-chain photoswitchable kinases. Science 355, 836–842 (2017); published online EpubFeb 24 ( 10.1126/science.aah3605). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schmiedel JM, Klemm SL, Zheng Y, Sahay A, Bluthgen N, Marks DS, van Oudenaarden A, Gene expression. MicroRNA control of protein expression noise. Science 348, 128–132 (2015); published online EpubApr 3 ( 10.1126/science.aaa1738). [DOI] [PubMed] [Google Scholar]
- 48.Taganov KD, Boldin MP, Chang KJ, Baltimore D, NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proceedings of the National Academy of Sciences of the United States of America 103, 12481–12486 (2006); published online EpubAug 15 ( 10.1073/pnas.0605298103). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chatzikyriakidou A, Voulgari PV, Georgiou I, Drosos AA, A polymorphism in the 3’-UTR of interleukin-1 receptor-associated kinase (IRAK1), a target gene of miR-146a, is associated with rheumatoid arthritis susceptibility. Joint Bone Spine 77, 411–413 (2010); published online EpubOct ( 10.1016/j.jbspin.2010.05.013). [DOI] [PubMed] [Google Scholar]
- 50.Yang D, Chen W, Xiong J, Sherrod CJ, Henry DH, Dittmer DP, Interleukin 1 receptor-associated kinase 1 (IRAK1) mutation is a common, essential driver for Kaposi sarcoma herpesvirus lymphoma. Proceedings of the National Academy of Sciences of the United States of America 111, E4762–4768 (2014); published online EpubNov 04 ( 10.1073/pnas.1405423111). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wee ZN, Yatim SM, Kohlbauer VK, Feng M, Goh JY, Bao Y, Lee PL, Zhang S, Wang PP, Lim E, Tam WL, Cai Y, Ditzel HJ, Hoon DS, Tan EY, Yu Q, IRAK1 is a therapeutic target that drives breast cancer metastasis and resistance to paclitaxel. Nature communications 6, 8746 (2015); published online EpubOct 27 ( 10.1038/ncomms9746). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Biswas SK, Lopez-Collazo E, Endotoxin tolerance: new mechanisms, molecules and clinical significance. Trends Immunol 30, 475–487 (2009); published online EpubOct ( 10.1016/j.it.2009.07.009). [DOI] [PubMed] [Google Scholar]
- 53.Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA 3rd, Smith HO, Enzymatic assembly of DNA molecules up to several hundred kilobases. Nature methods 6, 343–345 (2009); published online EpubMay ( 10.1038/nmeth.1318). [DOI] [PubMed] [Google Scholar]
- 54.Lam AJ, St-Pierre F, Gong Y, Marshall JD, Cranfill PJ, Baird MA, McKeown MR, Wiedenmann J, Davidson MW, Schnitzer MJ, Tsien RY, Lin MZ, Improving FRET dynamic range with bright green and red fluorescent proteins. Nature methods 9, 1005–1012 (2012); published online EpubOct ( 10.1038/nmeth.2171). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Moyer TC, Holland AJ, Generation of a conditional analog-sensitive kinase in human cells using CRISPR/Cas9-mediated genome engineering. Methods Cell Biol 129, 19–36 (2015) 10.1016/bs.mcb.2015.03.017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lowekamp BC, Chen DT, Ibanez L, Blezek D, The Design of SimpleITK. Front Neuroinform 7, 45 (2013) 10.3389/fninf.2013.00045). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kudo T, Jeknic S, Macklin DN, Akhter S, Hughey JJ, Regot S, Covert MW, Live-cell measurements of kinase activity in single cells using translocation reporters. Nat Protoc 13, 155–169 (2018); published online EpubJan ( 10.1038/nprot.2017.128). [DOI] [PubMed] [Google Scholar]
- 58.Tustison NJ, Avants BB, Cook PA, Zheng Y, Egan A, Yushkevich PA, Gee JC, N4ITK: improved N3 bias correction. IEEE Trans Med Imaging 29, 1310–1320 (2010); published online EpubJun ( 10.1109/TMI.2010.2046908). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Galloway CM, Le Ru EC, Etchegoin PG, An iterative algorithm for background removal in spectroscopy by wavelet transforms. Appl Spectrosc 63, 1370–1376 (2009); published online EpubDec ( 10.1366/000370209790108905). [DOI] [PubMed] [Google Scholar]
- 60.Jaqaman K, Loerke D, Mettlen M, Kuwata H, Grinstein S, Schmid SL, Danuser G, Robust single-particle tracking in live-cell time-lapse sequences. Nature methods 5, 695–702 (2008); published online EpubAug ( 10.1038/nmeth.1237). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Graphical summary of the image analysis pipeline.
Fig. S2. Additional analysis and TNF concentration for data presented in Fig. 1.
Fig. S3. Additional stimulus combinations for Figs. 2 and 3.
Fig. S4. Full gels and quantification of Western blots in Fig. 4
Fig. S5. Increased abundance of MyD88 or TRAF6 after primary stimulation cannot bypass tolerance.
Fig. S6. IRAK1 modification and clustering with varied recovery periods.
Fig. S7. IRAK1 mutant screen and knockout characterization.
Fig. S8. Additional IRAK1 WT and KD-Clover characterization.