Abstract
Background
Biodegradation of toxic organic dye using nanomaterial-based microbial biocatalyst is an ecofriendly and promising technique.
Materials and Methods
Here, we have investigated the novel properties of functionalized Au-Ag bimetallic nanoparticles using extremophilic Deinococcus radiodurans proteins (Drp-Au-AgNPs) and their degradation efficiency on the toxic triphenylmethane dye malachite green (MG).
Results and Discussion
The prepared Drp-Au-AgNPs with an average particle size of 149.8 nm were capped by proteins through groups including hydroxyl and amide. Drp-Au-AgNPs demonstrated greater degradation ability (83.68%) of MG than D. radiodurans cells and monometallic AuNPs. The major degradation product was identified as 4-(dimethylamino) benzophenone, which is less toxic than MG. The degradation of MG was mainly attributed to the capping proteins on Drp-Au-AgNPs. The bimetallic NPs could be reused and maintained MG degradation ability (>64%) after 2 cycles.
Conclusion
These results suggest that the easily prepared Drp-Au-AgNPs have potential applications as novel nanomedicine for MG detoxification, and nanomaterial for biotreatment of a toxic polyphenyl dye-containing wastewater.
Keywords: bimetallic nanoparticles, Deinococcus radiodurans, biodegradation, toxic triphenylmethane dye, malachite green, detoxification
Introduction
Biomaterials, such as nucleotides,1 amino acids,2 polypeptides and proteins,3–5 cell extracts,6–10 could be used as raw materials for reducing metal ions and producing metal nanoparticles (NPs). Metal NPs have demonstrated catalytic, mechanical, magnetic, thermal, optical, photoelectrochemical and biological properties. Functionalized metal NPs using biomaterials have inestimable merit because of biocompatible and specific properties of the natural biomaterials.11 Various applications of metal NPs have been explored including bacteriostatic agents,12–15 drug carriers16,17 and DNA delivery,18 cancer treatment,19,20 biosensing21,22 and catalysis.23,24 For example, the assay using DNA-gold nanoparticle probes allows absolute and direct microRNA quantification.25 AgNPs, AuNPs and Ag-AuNPs synthesized using plant extract from Plumbago zeylanica exhibited inhibiting and disruptive effects on bacterial biofilms.26
Polyphenyl dyes are chemical products having persistent and toxic effects to human beings. Malachite green (4-[(4-dimethylaminophenyl)-phenyl-methyl]- N,N-dimethyl aniline; MG) is a triphenylmethane dye that has been used as leather and silk dye, aquaculture insect repellent, and cell staining agent. However, residues of malachite green in nature are hazardous compounds that can cause mutagenesis, teratogenesis and chromosomal fractures, leading to serious human health hazards including carcinogenesis, multi-organ tissue injury and developmental abnormalities.27 Although its manufacture has been banned worldwide,27,28 MG is still produced and used in some developing countries because of its low price and lack of substitutes.29 Investigation on biotreatment and detoxification of MG using bio-NPs may provide a new efficient technique to prevent MG-derived health hazards and has attracted increasing attention in recent years.30–32 Generally, bio-NPs mediated decolorization of polyphenyl dyes with two mechanisms, ie, adsorption and degradation, and is more efficient and eco-friendly than physical and chemical methods as well as microbiological degradation method.33–35 Biosynthetic bimetallic gold and silver nanoparticles had catalytic ability to reduce a common waste water pollutant 4-nitrophenol, and demonstrated combinatorial and enhanced functions in comparison with their monometallic counterparts.36
Deinococcus radiodurans is an extreme bacterium known for its resistance to stresses including ionizing radiation (IR), ultraviolet (UV) radiation and oxidative stress. D. radiodurans is particularly attractive for toxic compound remediation, due to its remarkable resistance to oxidative stress incurred from reactive oxygen species (ROS), which are generated upon exposure to oxidant and cellular toxic reagents.37–41 Cell cultures of D. radiodurans were previously used to directly decolorize MG.42 Intracellular reductive proteins, including antioxidant enzymes and oxidoreductases, may provide a reducing microenvironment for transformation and for the synthesis of metal nanomaterials. They might also be involved in the process of MG biodegradation. We have previously synthesized AuNPs and Au-AgNPs using D. radiodurans proteins, which exhibit non-significant cytotoxicity.43,44 Further characterizations of these nanoparticles and their potential use for detoxification of MG are required.
In this study, we characterized the Au-AgNPs produced by D. radiodurans protein extracts (Drp-Au-AgNPs) using X-ray diffraction (XRD) and dynamic light scattering (DLS). The mechanism of formation of Drp-Au-AgNPs was investigated by Fourier-transform infrared spectroscopy (FT-IR) and X-ray photoelectron spectroscopy (XPS). Degradation ability of Drp-Au-AgNPs on MG was measured by decolorization tests using ultraviolet and visible (UV/Vis) spectrum, and by MG product analysis using gas chromatography/mass spectroscopy (GC-MS). The MG degradation ability of recycled Drp-Au-AgNPs was also analyzed. To the best of our knowledge, this is the first study on detoxification of MG using the functionalized Au-Ag bimetallic NPs by extremophilic D. radiodurans proteins.
Materials and Methods
Bacterial Cultures and Chemicals
Deinococcus radiodurans R1 (ATCC13939) was cultured aerobically in tryptone glucose yeast (TGY) (0.5% Bacto tryptone, 0.1% glucose and 0.3% Bacto yeast extract) broth at 30°C with agitation at 220 rpm. Bacterial growth was measured with optical density (OD) at 600 nm (OD600).
Chloroauric acid (HAuCl4·3H2O) and silver nitrate (AgNO3) were purchased from Sigma-Aldrich Co. (St Louis, MO, USA). The Au(III) and Ag(I) solutions used were prepared by dissolving HAuCl4·3H2O and AgNO3 in ultra-pure water, respectively. Malachite green (MG, MW=364.92) was purchased from Aladdin Reagent Co. Ltd., Shanghai, China. AuNPs synthesized using sodium citrate (SC-AuNPs) with an average size of 50 nm were purchased from Shanghai So-Fe Biomedical Co. (Shanghai, China). All other inorganic and organic chemicals used in this study were of analytical grade. The pH of each working solution was adjusted using hydrochloric acid or sodium hydroxide solution.
Preparation of Protein Extracts
A culture of D. radiodurans was grown in TGY medium until its OD600 reached ~1.0. Then, cells were pelleted by centrifugation and re-suspended in phosphate buffer solution (0.01 M, pH 7.2). Cells were disrupted at 4°C using an ultra-high pressure continuous flow cell disrupter followed by centrifugation at 15,000 × g for 30 mins at 4°C to remove cell debris. The supernatant was collected and treated with 80% (w/v) solid ammonium sulfate for 24 hrs at 4°C. Then, the mixture was centrifuged to collect the protein precipitate at 10,000 × g for 20 mins at 4°C. The precipitate was dissolved in ultra-pure water and dialyzed in cellulose acetate membrane (molecular weight [MW] cut-off 14,000 Da) for 12 hrs at 4°C. Protein concentration was quantitated using a BCA Assay Kit (Thermo Fisher Scientific, Waltham, MA, USA).
Biosynthesis of Drp-Au-AgNPs
Metal NPs were prepared according to previously described methods with some modification.44 2mg/mL protein extracts were incubated with 1 mM silver nitrate firstly at room temperature. The mixture turned into brownish yellow after 18 hrs reaction, indicating the formation of AgNPs, and measured by absorption spectrum using a UV/Vis absorption spectrometer (SpectraMax M5; Molecular Devices LLC, Sunnyvale, CA, USA). The AgNPs were centrifuged and washed repeatedly to remove the unreacted substance and then incubated with 1 mM HAuCl4 in an aqueous solution. The formation of Au-AgNPs was monitored by the color change and UV/Vis absorbance profile measured with a spectrophotometer. The bimetallic NPs were centrifuged and washed several times and then freeze-dried for later use.
Characterization of Drp-Au-AgNPs
The elemental composition of metal NPs was analyzed by SEM-energy-dispersive X-ray spectroscopy (SEM-EDS; Hitachi Model SU8010; Hitachi Ltd., Tokyo, Japan). The solid NPs were fixed on an Al carrier. EDS in area scan mode focused on a region of the sample surface.
The crystal structure of the prepared Drp-Au-AgNPs was characterized using an X-ray diffractometer (X’Pert PRO; PANalytical Ltd., Almelo, the Netherlands) with a Cu Kαl radiation source at 1540 Å wavelength. The diffraction angle 2θ was 20° to 90° with a step of 0.02° and 2 s for each step.
Size distribution of NPs was confirmed using a laser Doppler anemometer (Zetasizer Nano ZS; Malvern Instruments, Malvern, UK) with a He-Ne laser beam at wavelength (λ) of 632.8 nm (pH 7.0, 25°C). A 150mV electric field was added to the nanoparticle solution to observe the electrophoretic velocity.
For FT-IR analysis, freeze-dried protein extract or prepared NPs were mixed with KBr in a mortar at the ratio of 1:100 (M/M) and the mixtures were pressed into tablets. The samples were analyzed by the Nicolet 5700 FT-IR spectrometer (Thermo Fisher Scientific, Waltham, MA, USA) in 4000–400 cm−1 frequency range at a resolution of 4 cm−1, over 900 scans.
For XPS analysis, the freeze-dried protein extract or prepared NPs were fixed on a stainless steel holder. Valence state changes and surface functional groups of the samples were measured by a high-performance X-ray photoelectron spectrometer (Escalab 250Xi; Thermo Fisher Scientific, Waltham, MA, USA) with monochromatic Al Kα radiation source at 1486.6 eV. C1s peak at 284.6 eV was used as the charge reference. The XPS spectra were analyzed by XPSPEAK41 software.
Release Kinetics of Drp-Au-AgNPs
NPs solution (15 mL) was placed in a dialysis bag. Continuous measurement of the metal release of NPs over 48 hrs was performed by an inductively coupled plasma-optical emission spectroscopy (ICP-OES; ICP6000; Thermo Fisher Scientific, Waltham, MA, USA). The metal release rate was calculated using the equation as previously described:12 % of cumulative metal released=(Mw/Md) × 100, where Mw is the metal content in the water and Md is the total metal content in the dialysis bag.
Decolorization Assay
To analyze the decolorization capacity of NPs on MG, a certain amount of samples was mixed with 100 mg/L MG solution in a constant volume with ultra-pure water. Each experiment was conducted in parallel in triplicate. The MG decolorization test in the absence of NPs was used as a control. After reaction for 30 mins, the samples were taken and centrifuged at 20,000 × g for 15 mins. To quantify the remaining MG in the mixture, the absorbance of the supernatant was determined by using a spectrophotometer at a wavelength of 617 nm. The decolorization percentage was calculated using the equation: % decolorization= (1- Af/Ai) × 100, where Af is the final absorbance and Ai is the initial absorbance.
HPLC Analysis
High-performance liquid chromatography (HPLC; Alliance 2695, Waters, USA) was used for the analysis of MG. Reverse-phase HPLC analysis was performed with a C18 column (4.66 mm internal diameter/250 mm long/5 μm particle size) using binary elution conditions (20 mM sodium acetate and 100% acetonitrile). For gradient elution, the acetonitrile concentrations were set as follows: 60% (0–10 mins), 60–95% (10–20 mins) and 95% (20–30 mins). The flow rate was maintained at 0.3 mL min−1.
GC-MS Analysis
To determine the products of MG degradation, supernatant from the MG decolorization tests were collected by centrifuge in Amicon-ultracs and extracted twice with ethyl acetate. The extracts were dried and concentrated to about 1 mL for GC-MS analysis. The triple quadrupole gas chromatography-mass spectrometer (GC-MS; 7890B/7000C, Agilent, USA) was used with a DB-5 fused silica capillary column (30 m × 0.25 mm × 0.25 μm). The experimental conditions were as described in previous reports.45–47 The column temperature was increased from 100°C to 290°C at 8°C min−1, and held for 15 mins. The flow of gas was maintained at 1 mL min−1. The products were fully scanned in electron-impact (EI) mode. Agilent mass hunter software was used to match corresponding products in the built-in NIST 17 library.
Reusability Assay of NPs
After the incubation of NPs with MG, the mixture of NPs and MG was centrifuged (20,000 × g for 10 mins at 4°C). The NPs remained in the sediment were resuspended and washed with ultra-pure water. Then, the obtained NPs were repeatedly suspended with water and centrifuged (20,000 × g for 10 mins at 4°C) to remove dye until the supernatant was colorless. The pH value of the NPs solution was adjusted to neutral. The recycled NPs were tested for their properties including hydrodynamic diameter, capping protein groups using DLS, FT-IR and XPS analyses as described above. For investigation on the reusability of NPs, MG decolorization test using the recycled NPs was performed for 5 cycles.
Statistical analysis
Data were processed by OriginPro 9.0 (OriginLab Co., Northampton, MA, USA) and expressed as mean ± standard deviation. The analysis of variance was followed by Student’s t-test. P-value <0.05 was considered as statistically significant.
Results and Discussion
Characterization of Drp-Au-AgNPs
Drp-Au-AgNPs was prepared according to the method described in our previous paper.44 D. radiodurans protein extracts were first incubated with 1 mM Ag(I) to form AgNPs, which showed a characteristic UV/Vis absorption peak at 414 nm. Following purification, the AgNPs were then incubated with 1 mM Au(III). The mixture turned into purple within 1 hrs and its absorption peak changed from 414 nm to 545 nm, indicating that Au-AgNPs were generated (Figure S1a). SEM-EDS of the obtained Au-AgNPs exhibited both Au and Ag element peaks (Figure S1b), confirming the formation of Au-Ag bimetallic NPs (Drp-Au-AgNPs). Au/Ag molar ratio in the prepared Drp-Au-AgNPs was approximately 1:1.2 (Table S1). The metal ion ratio has effects on the size and characteristic absorption wavelength of formed metal nanoparticles.48 The absorption wavelength maximum of bimetallic NPs with a ratio of Ag: Au (3:1) was lower than that of Ag: Au (1:1).19
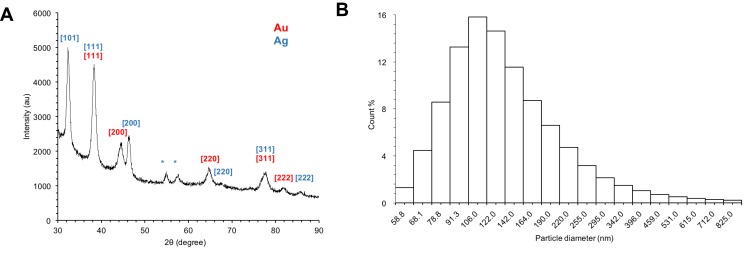
Crystal structures of lyophilized Drp-Au-AgNPs were analyzed by XRD. The characteristic peaks in XRD spectra correspond to different planes of metal crystal face-centered cubic. As shown in Figure 1A, peaks at 32.32°, 38.3°, 46.38°, 66.12°, 77.7°, 85.66° referred to the characteristic diffractions of the [101], [111], [200], [220], [311] and [222] planes of face-centered cubic Ag, respectively. Among them, the peak at 32.32° is the main reflection of AgNPs. Meanwhile, peaks at 38.3°, 44.56°, 64.66°, 77.7° and 81.9° were corresponding to [111], [200], [220], [311] and [222] planes of face-centered cubic Au, respectively. Peaks at 38.3° and 77.7° were corresponding to the planes of face-centered cubic of both Ag and Au. The small peaks at 54.9° and 57.74° might arise from impurities produced during the synthesis of AgNPs.
Figure 1.
XRD and DLS analyses of Drp-Au-AgNPs.
Notes: (A) XRD patterns of the purified Drp-Au-AgNPs. XRD patterns demonstrated the crystal structures of Drp-Au-AgNPs. The numbers in brackets represent the planes of Au (red) and Ag (blue) face-centered cubic crystal structures, respectively. Asterisks indicate the impure signal. (B) The distribution of hydrodynamic diameter of Drp-Au-AgNPs measured by DLS at 25°C, pH 7.0. The average hydrate particle size of the Drp-Au-AgNPs was 149.8 nm with a polydispersity index (PDI) of 0.397±0.021.
Abbreviations: XRD, X-ray diffraction; DLS, dynamic light scattering; Drp-Au-AgNPs, D. radiodurans protein extract-mediated gold-silver bimetallic nanoparticles; D. radiodurans, Deinococcus radiodurans.
The average hydrate particle size of the prepared Drp-Au-AgNPs was 149.8 nm with a polydispersity index (PDI) of 0.397±0.021, indicating that the NPs were evenly distributed (Figure 1B). The highest percentage of the NPs size was 106 nm (15.83%). The size of bimetallic nanoparticles was slightly larger than single metal nanoparticles.49,50
Synthetic Mechanisms of Drp-Au-AgNPs
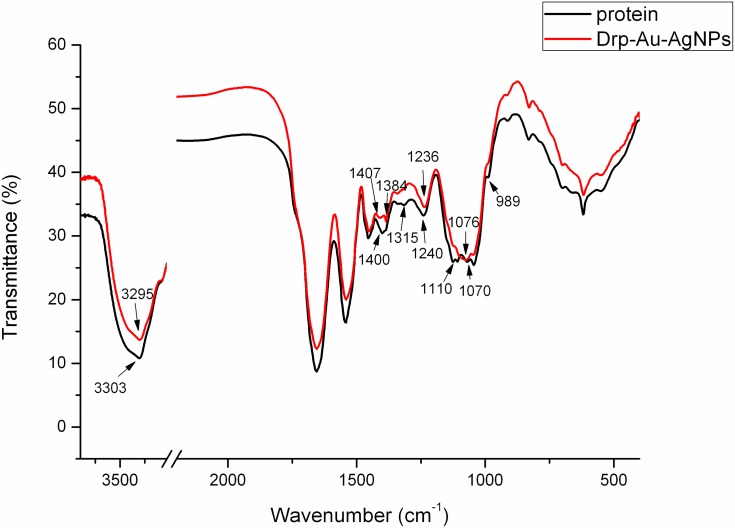
During biosynthesis of metal NPs using proteins, functional groups of proteins might react with metal ions. FT-IR analysis was used to identify the functional groups of D. radiodurans proteins involved in the formation of Au-AgNPs (Figure 2). The strong band at 3303 cm−1 of protein extracts referred to the stretching and out-of-plane bending vibration of O-H groups, and it shifted to 3295 cm−1 in Drp-Au-AgNPs. The band at 1400 cm−1 due to the symmetry extension of COO− groups was changed to 1407 cm−1 during the synthesis of NPs. The band at 1240 cm−1 referred to amide III of proteins shifted to 1236 cm−1 in Drp-Au-AgNPs. These results suggested that O-H, COO−, amide III groups are involved directly in the redox reaction of noble metal ions and formation of Au-AgNPs. The bands at 1110 cm−1 and 1070 cm−1 referred to P-OH and P-O-C, respectively. The P-O-C band shifted to 1076 cm−1 and the P-OH band disappeared. The bands at 1384 cm−1, 1315 cm−1, and 989 cm−1 referred to -CO-CH3, -CH-OH and -COOH, respectively, changed in the synthesis process. These indicated that the groups containing oxygen and phosphorus participated in the constitution of Drp-Au-AgNPs.
Figure 2.
FT-IR analyses of the prepared Drp-Au-AgNPs.
Notes: FT-IR spectra of protein extracts from D. radiodurans (black line) and Drp-Au-AgNPs (red line) were compared. Changes in the corresponding bands’ shape and shift are indicated by wavenumbers and arrows. The changes indicated the functional groups of D. radiodurans proteins involved in the formation of Au-AgNPs.
Abbreviations: FT-IR, Fourier-transform infrared spectroscopy; Drp-Au-AgNPs, D. radiodurans protein extract-mediated gold-silver bimetallic nanoparticles; D. radiodurans, Deinococcus radiodurans.
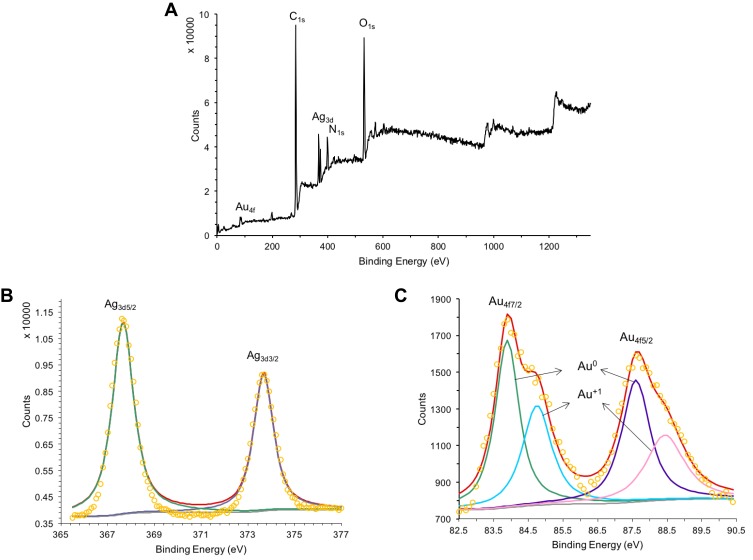
The reduction process of Au and Ag ions was investigated using XPS analysis of Drp-Au-AgNPs compared with D. radiodurans protein extract (Figures 3 and S2). During synthesis of the NPs, the valence state of metal ions changed (Figure 3). Taking the C1s peak at 284.6 eV as a reference, Ag 3d5/2 and Ag 3d3/2 peaks at 367.7 eV and 373.7 eV were the spectra of Ag(0) in the NPs (Figure 3B). Au 4f7/2 and Au4f5/2 peaks at 83.9 eV and 87.6 eV were the dipole peaks of Au(0) in the core-level spectra of Au4f, while the Au(I) characteristic peaks were at 84.8 eV and 88.4 eV, respectively (Figure 3C). This indicated that during the formation of Au-AgNPs from Drp-AgNPs,44 Au(III) was first reduced into Au(I) by hydrogen donation from protein functional groups capped on the AgNPs and then was reduced to Au(0). Compared with the core-level of the Au4f and Ag3d spectrum previously reported,51 the spectra of Drp-Au-AgNPs had ~0.8 eV and ~0.6 eV deviations that possibly were due to the protein coating effects.
Figure 3.
XPS analyses of the Drp-Au-AgNPs.
Notes: Full-scan spectrum of Drp-Au-AgNPs (A) and core level of Au4f (B) and Ag3d (C). Dipole peaks for metal ions of different valence states are indicated by banding energy and arrows. The peak with red line is the fitting peak of dipole peaks and the other colored peaks are the characteristic peaks of metal ion with different valence state. The peak with yellow circle represents the signal value at each binding energy position. XPS analysis of Drp-Au-AgNPs provided evidence for the reduction process of Au and Ag ions.
Abbreviations: XPS, X-ray photoelectron spectroscopy; Drp-Au-AgNPs, D. radiodurans protein extract-mediated gold-silver bimetallic nanoparticles; D. radiodurans, Deinococcus radiodurans.
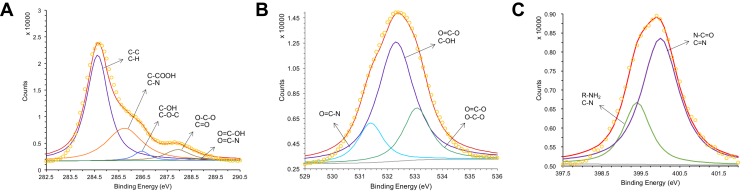
As shown in the XPS full-scan spectra of Drp-Au-AgNPs (Figure 3A), peaks of C1s, O1s, and N1s were similar to the spectra of the protein extract (Figure S2a). It confirmed that the protein functional groups were involved in the synthesis and coating of Au-AgNPs. Comparing C1s core-level spectrum of Drp-Au-AgNPs with that of protein extract (Figures 4A and S2b), we observed that the hydroxyl group (C-OH, generally present in Ser, Thr, Tyr) and C-O-C peaks shifted from 286.2 eV in protein extract to 286.5 eV in Drp-Au-AgNPs. Amide (O=C-N) peak changed from 288.5 eV to 288.7 eV. These peak shifts were also seen in the O1s (Figures S2c and 4B) or N1s (Figures S2d and 4C) core-level spectra, indicating that the hydroxyl groups and amides of proteins took part in the constitution of the NPs.
Figure 4.
XPS analyses of common elements in Drp-Au-AgNPs.
Notes: XPS analyses of the core level of (A) C1s, (B) O1s and (C) N1s of Drp-Au-AgNPs. Dipole peaks for different functional groups are indicated by banding energy and arrows. The peak with red line is the fitting peak of dipole peaks and the other colored peaks are the characteristic peaks of functional groups. The peak with yellow circle represents the signal value at each binding energy position. These XPS results confirmed the involvement of functional groups of protein in the synthesis of Au-AgNPs.
Abbreviations: XPS, X-ray photoelectron spectroscopy; Drp-Au-AgNPs, D. radiodurans protein extract-mediated gold-silver bimetallic nanoparticles; D. radiodurans, Deinococcus radiodurans.
Based on the N1s core-level spectrum (Figures 4C and S2d), the peaks of protonated amino (NH3+) and ammonium (NH4+) disappeared in the N1s core-level spectrum of NPs. The protonated amine groups have a strong reducing capacity by hydrogen donation and might contribute to metal reduction. All the functional groups and related amino acids involved in the reaction were listed in Table S2. These results are consistent with previous studies of amino acid and peptide-mediated synthesis of metal nanoparticles.2–4 Our previous results demonstrated that Drp-AgNPs formed into spherical Au-Ag bimetallic nanoparticles via the oxidation of Drp-AgNPs by the additional Au(III) ions, while as-synthesized Drp-AuNPs could not transform into Drp-Au-AgNPs in the presence of Ag(I).44 The functional groups of D. radiodurans proteins might participate in reducing Ag(I) to Ag(0). The Drp-AgNPs may be acting as a nucleation seed for the synthesis of bimetallic NPs. The coating proteins on Drp-AgNPs could reduce Au(III) into Au(I) and further into Au(0) to form alloy NPs of Au and Ag.
The metal release of Drp-Au-AgNPs was measured over a period of 48 hrs. 3.69% of Au was released and 16.17% of Ag was released was detected at 48 hrs (data not shown), similar to the results obtained in previous works.44,48 The Ag release of Drp-Au-AgNPs was lower than previously reported AgNPs,52 indicating a stronger binding affinity of metals to proteins.53 The characterized Drp-Au-AgNPs were used in the following MG detoxification.
Decolorization of MG by Drp-Au-AgNPs
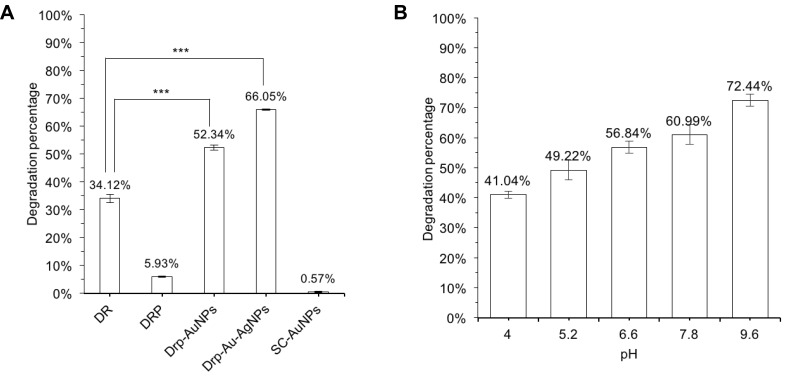
Degradation capability of a biocatalyst on dyes is generally measured by dye decolorization assays and indicated as decolorization percentage. Decolorization capability of Drp-Au-AgNPs on MG was examined in aqueous solution and compared with that of D. radiodurans cell culture, D. radiodurans protein extract, Drp-AuNPs and sodium citrate-AuNPs (SC-AuNPs). The initial concentration of MG was 100 mg/L. Compared with D. radiodurans cells, Drp-AuNPs and Drp-Au-AgNPs demonstrated higher MG degradation capability (Figure 5A). The protein extracts (4 mg/mL) from D. radiodurans showed a relatively low decolorization percentage on MG (5.93%), which might be due to protein precipitation under an acidic condition caused by the MG dye. According to the results of previous literature,42 D. radiodurans cell culture has a strong ability to decolorize MG, which might be attributed to the biosorption by cells and biodegradation by proteins (enzymes) in the microbial cells during growth. Nanoscale Au-Ag particles can promote the solubility of ligand proteins and keep them stable in the acidic condition. Moreover, the protein-capping Au-AgNPs can be easily recovered by centrifugation and serve as recyclable biocatalysts as detailed below.
Figure 5.
Decolorization of MG dye by bimetallic NPs.
Notes: (A) Decolorization percentage of MG at treatment of DR, Drp, Drp-AuNPs, Drp-Au-AgNPs and SC-AuNPs, respectively. The mixture was incubated for 30 mins at room temperature. Initial MG concentration was 100 mg/L. The concentration of samples was 4 mg/mL. ***P˂0.001, indicates statistical significance compared with the treatment by DR cells using Student’s t-test. (B) Decolorization percentage of MG at treatment of the bimetallic NPs at various pH conditions. Values are the mean and standard deviation of three independent experiments.
Abbreviations: MG, malachite green; NPs, nanoparticles; DR, Deinococcus radiodurans; Drp, D. radiodurans protein extract; Drp-AuNPs, D. radiodurans protein extract-mediated gold nanoparticles; Drp-Au-AgNPs, D. radiodurans protein extract-mediated gold-silver bimetallic nanoparticles; SC-NPs, sodium citrate-mediated gold nanoparticles; D. radiodurans, Deinococcus radiodurans.
The activity of the bimetallic NPs (66.05%) was stronger than the single metal NPs (52.34%) (Figure 5A), which might be due to their enhanced surface areas and decreased densities.54 Chen and co-workers reported that silver and gold bimetallic NPs produced using a D. radiodurans cell-free supernatant could degrade a common waste water pollutant 4-nitrophenol.36 The improved and combinatorial properties of the bimetallic NPs compared to their monometallic counterparts might be attributed to the higher surface areas of alloys NPs for enzyme-substrate interaction than that of core-shell forms of single metal nanoparticles. In addition, metal and protein ratio in the nanoparticles might affect the physical and chemical properties of bimetallic NPs.19,48
On the other hand, chemically synthesized AuNPs using sodium citrate (SC-AuNPs) had no substantial degradation ability on MG (Figure 5A), suggesting that the degradation of MG by Drp-Au-AgNPs is attributed to redox reaction between MG and the coating proteins on Au-AgNPs. Degradation capability of Drp-Au-AgNPs increased with the increasing pH of the solution (Figure 5B), demonstrating the highest activity at alkaline condition (pH 9.6). Substantial increase in the decolorization percentage of MG under strong alkaline can be accounted for by an electrostatic force between the biocatalyst and the positively charged MG molecules.55 Besides, the decolorization of MG under strong alkaline conditions can be partially attributed to chemical transformation of MG into its leuco form.
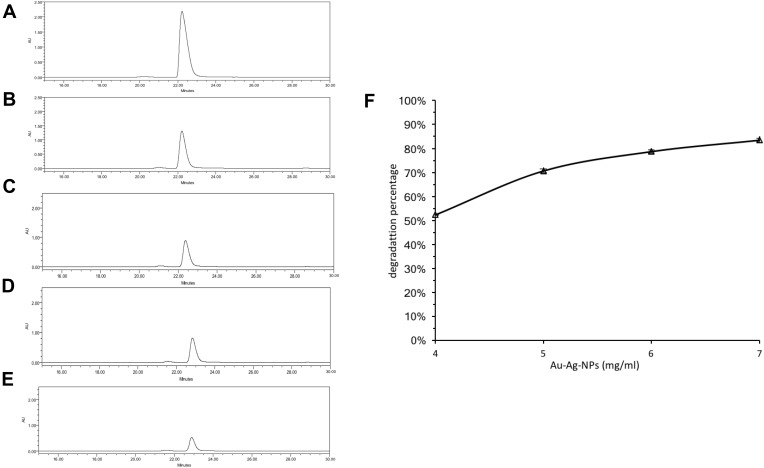
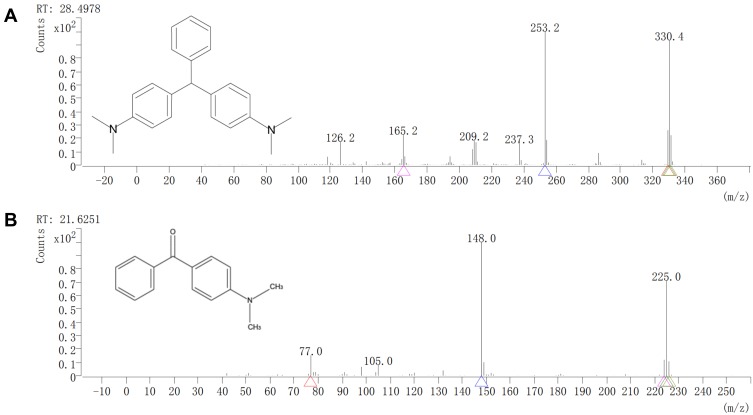
The decolorization ability of bimetallic NPs substantiated by HPLC results, which exhibited up to 83.68% of MG degradation upon treatment with NPs (7 mg/mL) (Figure 6). After the decolorization experiment, the MG products in the mixture were analyzed by GC-MS. The composition of MG degradation by Drp-Au-AgNPs was identified according to the retention times (RT) and m/z of ionized product components. MG molecule losing a Cl− due to the ionization process was detected with an m/z 330.4 at 28.50 mins (Figure 7A). A product fragment with an m/z 225.0 at RT of 21.63 mins, matching 4-(dimethylamino) benzophenone (4-DMABP), was identified as a degradation product of MG (Figure 7B). 4-DMABP has lower toxicity than MG,42 indicating that the toxic MG was degraded into less toxic product by Drp-Au-AgNPs. Therefore, Drp-Au-AgNPs can be used as potential MG detoxification nanomedicine. The detoxification mechanism of MG by Drp-Au-AgNPs might be attributed to the cleavage between benzene and the central carbon, which is the main position in MG degradation reaction.29 There were some unidentified products with characteristic spectra of aromatic aldehydes that had no matches in the GC-MS library. Further study on the unidentified products will elucidate the detailed detoxification mechanism. Our previous work had demonstrated that the Drp-Au-AgNPs have non-significant cytotoxicity to normal human cells,44 suggesting their biocompatible properties and applicability as a safe biomaterial and in biomedicine. Negligible toxicity of AgNPs was reported in a previous study using human cell lines.56
Figure 6.
Degradation of MG dye by bimetallic NPs.
Notes: (A–E) HPLC spectra of MG degradation by (A) ultra-pure water (control); (B) 4 mg/mL Drp-Au-AgNPs; (C) 5 mg/mL Drp-Au-AgNPs; (D) 6 mg/mL Drp-Au-AgNPs; (E) 7 mg/mL Drp-Au-AgNPs. The main peaks were corresponding to MG. (F) Degradation percentage of 100 mg/mL MG at treatment of different concentrations of Drp-Au-AgNPs. Values are the mean and standard deviation of three independent experiments. The bimetallic NPs exhibited up to 83.68% of MG degradation at the treatment of NPs (7 mg/mL).
Abbreviations: NPs, nanoparticles; MG, malachite green; Drp-Au-AgNPs, D. radiodurans protein extract-mediated gold-silver bimetallic nanoparticles; D. radiodurans, Deinococcus radiodurans.
Figure 7.
GC-MS analyses of MG degraded products by bimetallic NPs.
Notes: GC-MS spectra of (A) MG and (B) its degraded product by bimetallic NPs. The corresponding chemical formula is indicated in each panel. The MG was degraded by Drp-Au-AgNPs into less toxic product 4-(dimethylamino) benzophenone with an m/z 225.0 at RT of 21.6251 mins.
Abbreviations: GC-MS, gas chromatography-mass spectrometer; MG, malachite green; NPs, nanoparticles; RT, retention time.
Regeneration of Drp-Au-AgNPs
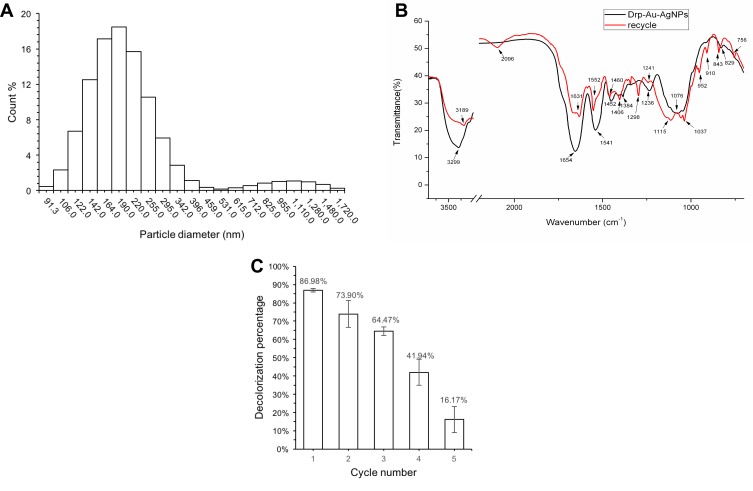
Drp-Au-AgNPs were recycled from MG degradation solution and freeze-dried. SEM-EDS and XRD patterns of the recycled NPS were similar to the original Drp-Au-AgNPs, indicating that their composition and crystal structure were relatively stable. The average hydrate particle size of the recycled Drp-Au-AgNPs as analyzed by DLS was 249.7 nm with a polydispersity index (PDI) of 0.829±0.148 (Figure 8A). The highest percentage of NPs size was 190 nm (18.47%). The distribution sizes were larger than the original Drp-Au-AgNPs, perhaps resulting from the adsorption of MG or its degradation products on the surface of NPs.
Figure 8.
Characterization and reusability analyses of the recycled Drp-Au-AgNPs.
Notes: (A) Distribution of hydrodynamic diameter of recycled NPs measured by DLS at 25°C, pH 7.0. (B) FT-IR spectra of protein from Drp-Au-AgNPs (black line) and recycled bimetallic NPs (red line). Changes in the corresponding bands’ shape and shift are indicated by wavenumbers and arrows. (C) MG decolorization percentage by the recycled bimetallic NPs at room temperature. The decolorization ability of Drp-Au-AgNPs remained above 60% during the first three cycles and decreased at the fourth cycle. Initial MG concentration was 100 mg/L; bimetallic NPs concentration was 7 mg/mL. Values are the mean and standard deviation of three independent experiments.
Abbreviations: Drp-Au-AgNPs, D. radiodurans protein extract-mediated gold-silver bimetallic nanoparticles; D. radiodurans, Deinococcus radiodurans; DLS, dynamic light scattering; FT-IR, Fourier-transform infrared spectroscopy; NPs, nanoparticles.
Degradation of MG by Drp-Au-AgNPs was a redox reaction between MG and the functional groups of the surface proteins on Drp-Au-AgNPs. The participating functional groups involved in the degradation process were confirmed by FTIR analysis of original and recycled Drp-Au-AgNPs (Figure 8B). The O-H groups with a strong band shifted from 3299 cm−1 to 3189 cm−1, and the band of COO− at 1384 cm−1 turned into 1406 cm−1. The amide I, amide II, amide III bands changed following the MG degradation. In the meantime, the bands at 1076 cm−1 and 1044 cm−1 assigning to phosphorus functional groups (P-OH and P-O-C) shifted to 1115 cm−1 and 1057 cm−1, respectively. Besides, the bands at 2096 cm−1 and 952 cm−1 appeared after the reaction might be ascribed to the protonated amino and carboxyl in reclaimed NPs. The hydroxyl, amide, phosphorus and carboxyl functional groups capping on the NPs were important participants in the MG degradation. Of particular, the band for methyl-benzene at 1037 cm−1 appeared, suggesting that the NPs had adsorption effects on degradation products of MG. This result was consistent with the increasing distribution sizes of recycled NPs (Figure 8A). The results provide evidence that proteins took part in the decolorization of MG through degradation and absorption effects.
The XPS spectra of recycled Drp-Au-AgNPs demonstrated the stability of the bimetallic NPs (Figure S3). The full-scan spectrum is shown in Figure S3a. The Au 4f7/2 and Au4f5/2 peaks at 83.7 eV and 87.2 eV in the core-level spectra of Au4f (Figure S3b) were for the dipole peaks of Au(0), and the Ag 3d5/2 and Ag 3d3/2 peaks at 367.5 eV and 373.6 eV were spectrum for Ag(0) (Figure S3c). The peaks of C1s, O1s, and N1s detected in recycled Drp-Au-AgNPs (Figure S3d–f) were similar to those of original Au-AgNPs (Figure 4), confirming that protein was retained coating on the surface of the NPs.
Reusability of Drp-Au-AgNPs
Following the regeneration of Drp-Au-AgNPs, MG decolorization of the recycled bimetallic NPs was tested for 5 cycles to evaluate their reusability (Figure 8C). The decolorization ability of Drp-Au-AgNPs remained above 60% during the first three cycles and decreased sharply at the fourth cycle. The physical and chemical changes in the bimetallic NPs, including aggregation of NPs and deactivation of capping proteins following several cycles of degradation process, may lead to the decrease of degradation ability.
Here, we focus on degradation and detoxification capability of the biosynthesized bimetallic nanoparticles on toxic polyphenyl dye. The protein functional groups on the Au-AgNPs, cooperating with the physical and chemical properties of metal nanoparticles23 contribute to their strong degradation on toxic polyphenyl dye. Comparing with bacterial cells, freeze-dried metal nanoparticles can be easily immobilized and stored.57 Combining with the above advantages, the bimetallic Au-AgNPs with high efficiency of MG detoxification, we believe, have considerable application prospect in bioremediation.
Conclusions
We investigated the functionalized bimetallic nanoparticles using D. radiodurans protein extract and their detoxification ability of MG. Although further study on the species of capping proteins (enzymes) and the kinetics of MG detoxification by Drp-Au-AgNPs are required, our results suggest that Drp-Au-AgNPs have the application potential as MG detoxification nanomedicine. Moreover, Drp-Au-AgNPs can be developed as novel recyclable nanomaterial for biotreatment of toxic MG-containing wastewater to prevent the MG-derived health hazard.
Acknowledgments
This study was supported by grants from the National Natural Science Foundation of China (31670083, 31870025 and 31370119) and the fundamental research funds for the central universities.
Disclosure
The authors declare no conflicts of interest.
References
- 1.Berti L, Burley GA. Nucleic acid and nucleotide-mediated synthesis of inorganic nanoparticles. Nat Nanotechnol. 2008;3(2):81–87. doi: 10.1038/nnano.2007.460 [DOI] [PubMed] [Google Scholar]
- 2.Si S, Mandal TK. Tryptophan-based peptides to synthesize gold and silver nanoparticles: a mechanistic and kinetic study. Chem Eur J. 2007;13(11):3160–3168. doi: 10.1002/(ISSN)1521-3765 [DOI] [PubMed] [Google Scholar]
- 3.Li Y, Tang Z, Prasad PN, Knecht MR, Swihart MT. Peptide-mediated synthesis of gold nanoparticles: effects of peptide sequence and nature of binding on physicochemical properties. Nanoscale. 2014;6(6):3165–3172. doi: 10.1039/C3NR06201E [DOI] [PubMed] [Google Scholar]
- 4.Tan YN, Lee JY, Wang DI. Uncovering the design rules for peptide synthesis of metal nanoparticles. J Am Chem Soc. 2010;132(16):5677–5686. doi: 10.1021/ja907454f [DOI] [PubMed] [Google Scholar]
- 5.Nam KT, Lee YJ, Krauland EM, Kottmann ST, Belcher AM. Peptide-mediated reduction of silver ions on engineered biological scaffolds. ACS Nano. 2008;2(7):1480–1486. doi: 10.1021/nn800018n [DOI] [PubMed] [Google Scholar]
- 6.Chandran SP, Chaudhary M, Pasricha R, Ahmad A, Sastry M. Synthesis of gold nanotriangles and silver nanoparticles using Aloe vera plant extract. Biotechnol Prog. 2006;22(2):577–583. doi: 10.1021/bp0501423 [DOI] [PubMed] [Google Scholar]
- 7.Mabey T, Andrea Cristaldi D, Oyston P, et al. Bacteria and nanosilver: the quest for optimal production. Crit Rev Biotechnol. 2019;39(2):272–287. doi: 10.1080/07388551.2018.1555130 [DOI] [PubMed] [Google Scholar]
- 8.Ahmad N, Sharma AK, Sharma S, et al. Biosynthesized composites of Au-Ag nanoparticles using Trapapeel extract induced ROS-mediated p53 independent apoptosis in cancer cells. Drug Chem Toxicol. 2018;42(1):43–53. doi: 10.1080/01480545.2018.1463241 [DOI] [PubMed] [Google Scholar]
- 9.Das SK, Liang J, Schmidt M, Laffir F, Marsili E. Biomineralization mechanism of gold by zygomycete fungi Rhizopous oryzae. ACS Nano. 2012;6(7):6165–6173. doi: 10.1021/nn301502s [DOI] [PubMed] [Google Scholar]
- 10.Tian B, Li J, Pang R, et al. Gold nanoparticles biosynthesized and functionalized using a hydroxylated tetraterpenoid trigger gene expression changes and apoptosis in cancer cells. ACS Appl Mater Interfaces. 2018;10(43):37353–37363. doi: 10.1021/acsami.8b09206 [DOI] [PubMed] [Google Scholar]
- 11.Chellapandian C, Ramkumar B, Puja P, Shanmuganathan R, Pugazhendhi A, Kumar P. Gold nanoparticles using red seaweed Gracilaria verrucosa: green synthesis, characterization and biocompatibility studies. Process Biochem. 2019;80:58–63. doi: 10.1016/j.procbio.2019.02.009 [DOI] [Google Scholar]
- 12.Oves M, Khan MS, Zaidi A, et al. Antibacterial and cytotoxic efficacy of extracellular silver nanoparticles biofabricated from chromium reducing novel OS4 strain of Stenotrophomonas maltophilia. PLoS One. 2013;8(3):e59140. doi: 10.1371/journal.pone.0059140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saravanan M, Barik SK, MubarakAli D, Prakash P, Pugazhendhi A. Synthesis of silver nanoparticles from Bacillus brevis (NCIM 2533) and their antibacterial activity against pathogenic bacteria. Microb Pathog. 2018;116:221–226. doi: 10.1016/j.micpath.2018.01.038 [DOI] [PubMed] [Google Scholar]
- 14.Shanmuganathan R, Karuppusamy I, Saravanan M, et al. Synthesis of silver nanoparticles and their biomedical applications - A comprehensive review. Curr Pharm Des. 2019;25(24):2650–2660. doi: 10.2174/1381612825666190708185506 [DOI] [PubMed] [Google Scholar]
- 15.Shanmuganathan R, MubarakAli D, Prabakar D, et al. An enhancement of antimicrobial efficacy of biogenic and ceftriaxone-conjugated silver nanoparticles: green approach. Environ Sci Pollut Res. 2018;25(11):10362–10370. doi: 10.1007/s11356-017-9367-9 [DOI] [PubMed] [Google Scholar]
- 16.Chen G, Wen L, Zeng P, Zhang L, Huang W, Wang H. Symbiosis theory-directed green synthesis of silver nanoparticles and their application in infected wound healing. Int J Nanomedicine. 2016;11:2757–2767. doi: 10.2147/IJN [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Prabhu R, Anjali R, Archunan G, NM P, Pugazhendhi A, Suganthy N. Ecofriendly one pot fabrication of methyl gallate@ZIF-L nanoscale hybrid as pH responsive drug delivery system for lung cancer therapy. Process Biochem. 2019;84:39–52. doi: 10.1016/j.procbio.2019.06.015 [DOI] [Google Scholar]
- 18.Ma J-L, Yin B-C, Wu X, Ye B-C. Copper-mediated DNA-scaffolded silver nanocluster on–off switch for detection of pyrophosphate and alkaline phosphatase. Anal Chem. 2016;88(18):9219–9225. doi: 10.1021/acs.analchem.6b02465 [DOI] [PubMed] [Google Scholar]
- 19.Katifelis H, Lyberopoulou A, Mukha I, et al. Ag/Au bimetallic nanoparticles induce apoptosis in human cancer cell lines via P53, CASPASE-3 and BAX/BCL-2 pathways. Artif Cells Nanomed Biotechnol. 2018;46(sup3):S389–S398. doi: 10.1080/21691401.2018.1495645 [DOI] [PubMed] [Google Scholar]
- 20.Pugazhendhi A, Edison TNJI, Karuppusamy I, Kathirvel B. Inorganic nanoparticles: a potential cancer therapy for human welfare. Int J Pharm. 2018;539(1–2):104–111. doi: 10.1016/j.ijpharm.2018.01.034 [DOI] [PubMed] [Google Scholar]
- 21.You -C-C, Miranda OR, Gider B, et al. Detection and identification of proteins using nanoparticle–fluorescent polymer ‘chemical nose’ sensors. Nat Nanotechnol. 2007;2(5):318–323. doi: 10.1038/nnano.2007.99 [DOI] [PubMed] [Google Scholar]
- 22.Wang D, Huang B, Liu J, et al. A novel electrochemical sensor based on Cu@Ni/MWCNTs nanocomposite for simultaneous determination of guanine and adenine. Biosens Bioelectron. 2018;102:389–395. doi: 10.1016/j.bios.2017.11.051 [DOI] [PubMed] [Google Scholar]
- 23.Zhang H, Dai B, Wang X, et al. Non-mercury catalytic acetylene hydrochlorination over bimetallic Au–Co(iii)/SAC catalysts for vinyl chloride monomer production. Green Chem. 2013;15(3):829–836. doi: 10.1039/c3gc36840h [DOI] [Google Scholar]
- 24.Samuel MS, Jose S, Selvarajan E, Mathimani T, Pugazhendhi A. Biosynthesized silver nanoparticles using Bacillus amyloliquefaciens; Application for cytotoxicity effect on A549 cell line and photocatalytic degradation of p-nitrophenol. J Photochem Photobiol B. 2020;202:111642. doi: 10.1016/j.jphotobiol.2019.111642 [DOI] [PubMed] [Google Scholar]
- 25.Degliangeli F, Kshirsagar P, Brunetti V, Pompa PP, Fiammengo R. Absolute and direct microRNA quantification using DNA–gold nanoparticle probes. J Am Chem Soc. 2014;136(6):2264–2267. doi: 10.1021/ja412152x [DOI] [PubMed] [Google Scholar]
- 26.Chopade BA, Salunke GR, Ghosh S, et al. Rapid efficient synthesis and characterization of silver, gold, and bimetallic nanoparticles from the medicinal plant Plumbago zeylanica and their application in biofilm control. Int J Nanomedicine. 2014;9:2635–2653. doi: 10.2147/IJN [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Srivastava S, Sinha R, Roy D. Toxicological effects of malachite green. Aquat Toxicol. 2004;66(3):319–329. doi: 10.1016/j.aquatox.2003.09.008 [DOI] [PubMed] [Google Scholar]
- 28.Lee JB, Kim M. Photo-degradation of malachite green in mudfish tissues — investigation of UV-induced photo-degradation. Food Sci Biotechnol. 2012;21(2):519–524. doi: 10.1007/s10068-012-0066-5 [DOI] [Google Scholar]
- 29.Du L-N, Zhao M, Li G, Xu F-C, Chen W-H, Zhao Y-H. Biodegradation of malachite green by Micrococcus sp. strain BD15: biodegradation pathway and enzyme analysis. Int Biodeterior Biodegradation. 2013;78:108–116. doi: 10.1016/j.ibiod.2012.12.011 [DOI] [Google Scholar]
- 30.Saha S, Wang JM, Pal A. Nano silver impregnation on commercial TiO2 and a comparative photocatalytic account to degrade malachite green. Sep Purif Technol. 2012;89:147–159. doi: 10.1016/j.seppur.2012.01.012 [DOI] [Google Scholar]
- 31.Midya L, Patra AS, Banerjee C, Panda AB, Pal S. Novel nanocomposite derived from ZnO/CdS QDs embedded crosslinked chitosan: an efficient photocatalyst and effective antibacterial agent. J Hazard Mater. 2019;369:398–407. doi: 10.1016/j.jhazmat.2019.02.022 [DOI] [PubMed] [Google Scholar]
- 32.Li B, Gan L, Owens G, Chen Z. New nano-biomaterials for the removal of malachite green from aqueous solution via a response surface methodology. Water Res. 2018;146:55–66. doi: 10.1016/j.watres.2018.09.006 [DOI] [PubMed] [Google Scholar]
- 33.Zhou X-J, Guo W-Q, Yang -S-S, Ren N-Q. A rapid and low energy consumption method to decolorize the high concentration triphenylmethane dye wastewater: operational parameters optimization for the ultrasonic-assisted ozone oxidation process. Bioresour Technol. 2012;105:40–47. doi: 10.1016/j.biortech.2011.11.089 [DOI] [PubMed] [Google Scholar]
- 34.Shang N, Ding M, Dai M, Si H, Li S, Zhao G. Biodegradation of malachite green by an endophytic bacterium Klebsiella aerogenes S27 involving a novel oxidoreductase. Appl Microbiol Biotechnol. 2019;103(5):2141–2153. doi: 10.1007/s00253-018-09583-0 [DOI] [PubMed] [Google Scholar]
- 35.Shedbalkar U, Jadhav JP. Detoxification of malachite green and textile industrial effluent by Penicillium ochrochloron. Biotechnol Bioproc Eng. 2011;16(1):196–204. doi: 10.1007/s12257-010-0069-0 [DOI] [Google Scholar]
- 36.Chen A, Contreras LM, Keitz BK, Liu S-J. Imposed environmental stresses facilitate cell-free nanoparticle formation by Deinococcus radiodurans. Appl Environ Microbiol. 2017;83(18):e00798–17. doi: 10.1128/AEM.00798-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cheng K, Xu H, Chen X, et al. Structural basis for DNA 5´-end resection by RecJ. eLife. 2016;5:e14294. doi: 10.7554/eLife.14294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Daly MJ. Engineering radiation-resistant bacteria for environmental biotechnology. Curr Opin Biotechnol. 2000;11(3):280–285. doi: 10.1016/S0958-1669(00)00096-3 [DOI] [PubMed] [Google Scholar]
- 39.Cox MM, Battista JR. Deinococcus radiodurans — the consummate survivor. Nat Rev Microbiol. 2005;3(11):882–892. doi: 10.1038/nrmicro1264 [DOI] [PubMed] [Google Scholar]
- 40.Slade D, Radman M. Oxidative stress resistance in Deinococcus radiodurans. Microbiol Mol Biol Rev. 2011;75(1):133–191. doi: 10.1128/MMBR.00015-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tian B, Sun Z, Xu Z, Shen S, Wang H, Hua Y. Carotenoid 3′,4′-desaturase is involved in carotenoid biosynthesis in the radioresistant bacterium Deinococcus radiodurans. Microbiology. 2008;154(12):3697–3706. doi: 10.1099/mic.0.2008/021071-0 [DOI] [PubMed] [Google Scholar]
- 42.Lv G-Y, Cheng J-H, Chen X-Y, Zhang Z-F, Fan L-F. Biological decolorization of malachite green by Deinococcus radiodurans R1. Bioresour Technol. 2013;144:275–280. doi: 10.1016/j.biortech.2013.07.003 [DOI] [PubMed] [Google Scholar]
- 43.Li J, Li Q, Ma X, et al. Biosynthesis of gold nanoparticles by the extreme bacterium Deinococcus radiodurans and an evaluation of their antibacterial properties. Int J Nanomedicine. 2016;11:5931–5944. doi: 10.2147/IJN.S119618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li J, Tian B, Li T, et al. Biosynthesis of Au, Ag and Au-Ag bimetallic nanoparticles using protein extracts of Deinococcus radiodurans and evaluation of their cytotoxicity. Int J Nanomedicine. 2018;13:1411–1424. doi: 10.2147/IJN.S149079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ray SK, Dhakal D, Lee SW. Insight into malachite green degradation, mechanism and pathways by morphology-tuned α-NiMoO4 photocatalyst. Photochem Photobiol. 2018;94(3):552–563. doi: 10.1111/php.2018.94.issue-3 [DOI] [PubMed] [Google Scholar]
- 46.Ray SK, Dhakal D, Kshetri YK, Lee SW. Cu-α-NiMoO4 photocatalyst for degradation of methylene blue with pathways and antibacterial performance. J Photochem Photobiol A. 2017;348:18–32. doi: 10.1016/j.jphotochem.2017.08.004 [DOI] [Google Scholar]
- 47.Si S, Bhattacharjee RR, Banerjee A, Tk M. A mechanistic and kinetic study of the formation of metal nanoparticles by using synthetic tyrosine-based oligopeptides. Chem Eur J. 2006;12(4):1256–1265. doi: 10.1002/chem.200500834 [DOI] [PubMed] [Google Scholar]
- 48.Li T, Albee B, Alemayehu M, et al. Comparative toxicity study of Ag, Au, and Ag–Au bimetallic nanoparticles on Daphnia magna. Anal Bioanal Chem. 2010;398(2):689–700. doi: 10.1007/s00216-010-3915-1 [DOI] [PubMed] [Google Scholar]
- 49.Suganthy N, Sri Ramkumar V, Pugazhendhi A, Benelli G, Archunan G. Biogenic synthesis of gold nanoparticles from Terminalia arjuna bark extract: assessment of safety aspects and neuroprotective potential via antioxidant, anticholinesterase, and antiamyloidogenic effects. Environ Sci Pollut Res. 2017;25(11):10418–10433. doi: 10.1007/s11356-017-9789-4 [DOI] [PubMed] [Google Scholar]
- 50.Oves M, Aslam M, Rauf MA, et al. Antimicrobial and anticancer activities of silver nanoparticles synthesized from the root hair extract of Phoenix dactylifera. Mat Sci Eng C. 2018;89:429–443. doi: 10.1016/j.msec.2018.03.035 [DOI] [PubMed] [Google Scholar]
- 51.Luo Y, Shen S, Luo J, Wang X, Sun R. Green synthesis of silver nanoparticles in xylan solution via Tollens reaction and their detection for Hg2+. Nanoscale. 2015;7(2):690–700. doi: 10.1039/C4NR05999A [DOI] [PubMed] [Google Scholar]
- 52.Lu D, Liu Q, Zhang T, Cai Y, Yin Y, Jiang G. Stable silver isotope fractionation in the natural transformation process of silver nanoparticles. Nat Nanotechnol. 2016;11(8):682–686. doi: 10.1038/nnano.2016.93 [DOI] [PubMed] [Google Scholar]
- 53.Levak M, Buric P, Dutour Sikiric M, et al. Effect of protein corona on silver nanoparticle stabilization and ion release kinetics in artificial seawater. Environ Sci Technol. 2017;51(3):1259–1266. doi: 10.1021/acs.est.6b03161 [DOI] [PubMed] [Google Scholar]
- 54.Shin K-S, Kim J-H, Kim I-H KK. Poly(ethylenimine)-stabilized hollow gold-silver bimetallic nanoparticles: fabrication and catalytic application. Bull Korean Chem Soc. 2012;33(3):906–910. doi: 10.5012/bkcs.2012.33.3.906 [DOI] [Google Scholar]
- 55.Chen C-Y, Kuo J-T, Cheng C-Y, Huang Y-T, Ho IH, Chung Y-C. Biological decolorization of dye solution containing malachite green by Pandoraea pulmonicola YC32 using a batch and continuous system. J Hazard Mater. 2009;172(2–3):1439–1445. doi: 10.1016/j.jhazmat.2009.08.009 [DOI] [PubMed] [Google Scholar]
- 56.De Matteis V, Malvindi MA, Galeone A, et al. Negligible particle-specific toxicity mechanism of silver nanoparticles: the role of Ag+ ion release in the cytosol. Nanomed Nanotechnol Biol Med. 2015;11(3):731–739. doi: 10.1016/j.nano.2014.11.002 [DOI] [PubMed] [Google Scholar]
- 57.Jacob JM, John MS, Jacob A, et al. Bactericidal coating of paper towels via sustainable biosynthesis of silver nanoparticles using Ocimum sanctum leaf extract. Mater Res Express. 2019;6(4):045401. doi: 10.1088/2053-1591/aafaed [DOI] [Google Scholar]