Abstract
Purpose
Hyperpolarized 129Xe MRI characterizes regional lung ventilation in a variety of disease populations, with high sensitivity to airway obstruction in early disease. However, ventilation images are usually limited to a single breath-hold and most-often acquired using gradient-recalled echo sequences with thick slices (~10–15 mm), which increases partial-volume effects, limits ability to observe small defects, and suffers from imperfect slice selection. We demonstrate higher-resolution ventilation images, in shorter breath-holds, using FLORET (Fermat Looped ORthogonally Encoded Trajectories), a center-out 3D-spiral UTE sequence.
Methods
In vivo human adult (N = 4; 2 healthy, 2 with cystic fibrosis) 129Xe images were acquired using 2D gradient-recalled echo, 3D radial, and FLORET. Each sequence was acquired at its highest possible resolution within a 16-second breath-hold with a minimum voxel dimension of 3 mm. Images were compared using 129Xe ventilation defect percentage, SNR, similarity coefficients, and vasculature cross-sections.
Results
The FLORET sequence obtained relative normalized SNR, 40% greater than 2D gradient-recalled echo (P = .012) and 26% greater than 3D radial (P = .067). Moreover, the FLORET images were acquired with 3-fold-higher nominal resolution in a 15% shorter breath-hold. Finally, vasculature was less prominent in FLORET, likely due to diminished susceptibility-induced dephasing at shorter TEs afforded by UTE sequences.
Conclusion
The FLORET sequence yields higher SNR for a given resolution with a shorter breath-hold than traditional ventilation imaging techniques. This sequence more accurately measures ventilation abnormalities and enables reduced scan times in patients with poor compliance and severe lung disease.
Keywords: 129Xe, FLORET, non-Cartesian, spiral, UTE, ventilation
1. INTRODUCTION
Hyperpolarized 129Xe ventilation imaging quantifies regional ventilation and is sensitive to airway obstruction in a wide variety of diseases.1–12 The technique is safe and well-tolerated for many populations, including pediatrics.1,13,14 Ventilation imaging relies on obtaining reliable, spin density–weighted images to quantify xenon regionally in the lung. A standard approach to ventilation imaging incorporates 2D gradient-recalled echo (2D-GRE) sequences limited to a single breath-hold (<16 seconds). Due to the breath-hold and gradient limitations, the lung is typically acquired with thick slices (~10–15 mm), which may suffer from imperfect slice selection.3 Additionally, each slice is acquired at different times during the breath-hold, causing undesired T1 weighting in later slices. These limitations suggest that HP 129Xe imaging using true 3D acquisitions could yield superior performance and quantitative accuracy.
Previously, 129Xe ventilation imaging with 3D-radial was compared with a standard multislice 2D-GRE sequence.7 The 3D-radial acquisitions captured magnetization dynamics and physiologically relevant characteristics such as gravitationally dependent ventilation gradients, but experienced a nearly 2-fold reduction in SNR. The decreased SNR partially resulted from sampling inefficiency, requiring many acquisitions and thus a flip angle approximately 5-fold smaller for the limited total magnetic moment. However, the center-out nature was advantageous, allowing correction/manipulation of the data to obtain a more accurate image of xenon density and dynamic ventilation.15–17
Thus, the ideal acquisition would combine the advantages of center-out 3D radial with efficient k-space encoding, to allow larger flip angles and shorter breath-holds. Relative to 3D radial, center-out spiral acquisitions are more efficient in their sampling of k-space. Higher sampling efficiency permits the use of fewer magnetization-consuming excitations and shorter breath-holds, avoiding 2 main disadvantages of 3D radial.18 One such example, FLORET (Fermat Looped ORthogonally Encoded Trajectories), is a center-out 3D-spiral sequence that has been shown to have a superior performance and efficiency compared with that of 3D radial for greater than approximately 0.8 ms,18,19 much lower than that expected for hyperpolarized gases (>8 ms).20
Here, FLORET is compared with 2D GRE and 3D radial to determine the potential improvements FLORET may provide for 129Xe ventilation imaging. We demonstrate that FLORET provides increased SNR, increased resolution, and shorter breath-holds. Additionally, agreement in identification of regional ventilation defects and overall quantification provides substance to the improvements provided by FLORET 129Xe ventilation.
2. METHODS
2.1. In vivo imaging
Human studies were performed with approval from the internal review board of Cincinnati Children’s Hospital Medical Center and with oversight from the U.S. Food and Drug Administration (USFDA) under an investigational new drug (IND 123,577) protocol. Isotopically enriched xenon (83% 129Xe; Linde Specialty Gases) was polarized to approximately 30% using a Polarean 9820 129Xe hyperpolarizer. By the time of imaging, polarization decayed to 23% ± 7%.
Hyperpolarized 129Xe ventilation images were acquired in 2 healthy volunteers (ages 27 and 33 years) and 2 patients with cystic fibrosis (ages 28 and 48 years) using a Philips 3T Achieva MRI scanner (Best, Netherlands) (maximum gradient amplitude = 80 mT/m; max gradient slew rate = 200 mT/m/ms) and a homebuilt xenon coil.21 Subjects were imaged in the supine position during a breath-hold (maximum of 16 seconds) after inhaling a 1-L dose from functional residual capacity. For each subject, 5 doses of xenon were inhaled (1 calibration dose: 700 mL N2, 300 mL 129Xe; 4 imaging doses: 1 L 129Xe). The calibration dose was used to ascertain the precise center frequency of the xenon signal and to calibrate the flip angle.
Three comparative ventilation sequences were completed for each subject: 300-mm isotropic FOV, shortest possible TE, and a flip angle calculated to maximize the signal of the central acquisition according to θ = atan(√[2/Nex]), where Nex is the number of effective excitations.22 The number of effective excitations for 2D slice-selective scans was equivalent to the number of phase encodings, whereas the 3D scans used a nonselective pulse, making the number of effective excitations equivalent to the number of projections acquired. Because scan duration is the primary limitation, all scans were acquired with identical undersampling factors: 62.5% partial echo and 62.5% half Fourier factors (39% of k-space acquired) for 2D-GRE; 39% of Nyquist radial spokes (equally spaced spokes, maximum undersampling factor, Rmax = 2.56) for 3D-radial; and Rmax value of 2.56 (intertrajectory spacing linearly increasing with k-space radius up to 2.56 times the Nyquist distance18,19) for FLORET. The FLORET sequence was implemented with a readout duration of approximately 8 ms, similar to the global (spectroscopy-based lower limit) of xenon gas in the lung at 3 T23 and with 2 hubs with a maximum cone angle of 45°. The voxel volumes were smaller for FLORET (27 mm3) compared with 3D radial and 2D GRE (86 mm3 and 79 mm3, respectively). Although 2D GRE acquired identical in-plane resolution to FLORET (3 mm), the 2D GRE sequence required almost 3-fold-thicker slices (8.82 mm). The scan time was shortest for FLORET (12.5 seconds) compared with 15.5 seconds for 3D radial and 15.7 seconds for 2D GRE. Additionally, a fully sampled FLORET was acquired using a readout duration of approximately the local (imaging-based) of xenon (18 ms).20 Both FLORET sequences used 30.38 mT/m maximum gradient strengths and 102 mT/m/ms slew rate maximums. The FLORET sampling trajectories and additional information can be found in the Supporting Information. Acquisition parameters can be found in Table 1. A proton 3D-radial image was acquired with the same FOV (300-mm isotropic) and 3.0-mm isotropic resolution during a 1-L breath-hold of room air to obtain an anatomical lung mask (flip angle = 5°, TE = 0.156 ms, bandwidth = 498 Hz/pixel, TR = 2.12 ms, scan duration = 15.6 seconds).
TABLE 1.
Sequence parameters implemented for each ventilation scan
| 2D GRE (comparison) | 3D radial (comparison) | FLORET (comparison) | FLORET (fully sampled) | |
|---|---|---|---|---|
| Resolution (mm) | 3.00 × 3.00 × 8.82 (79 mm3) | 4.41 × 4.41 × 4.41 (86 mm3) | 3.00 × 3.00 × 3.00 (27 mm3) | 3.26 × 3.26 × 3.26 (35 mm3) |
| FOV (mm) | 300 × 300 × 300 | 300 × 300 × 300 | 300 × 300 × 300 | 300 × 300 × 300 |
| Matrix size | 100 × 100 × 34 | 68 × 68 × 68 | 100 × 100 × 100 | 92 × 92 × 92 |
| Effective excitations (Nex) | 63 | 3600 | 1088 | 612 |
| Encoding method | Phase: −ky to +ky Slice: head to foot |
Ten-interleave Archimedean spiral | Two-hub minimal angular difference18 | Two-hub minimal angular difference18 |
| Flip angle | 10.1° | 1.4° | 2.5° | 3.3° |
| TE (ms) | 1.98 | 0.149 | 0.118 | 0.118 |
| Dwell time (us) | 31.5 | 31.6 | 7.3 | 7.4 |
| Readout (ms) | 3.93 | 2.18 | 7.78 | 19.0 |
| TR (ms) | 6.70 | 4.08 | 10.0 | 21.2 |
| Scan duration (seconds) | 15.7 | 15.5 | 12.5 | 14.5 |
2.2. Image reconstruction
Images were reconstructed offline using Graphical Programming Interface (GPI).24 The 2D-GRE reconstruction consisted of Fourier transformation of raw k-space data. For center-out trajectories (3D radial and FLORET), all points in a projection were scaled by the magnitude of their k0 intensities, removing signal decay due to T1 relaxation and flip angle, reducing potential artifacts at the potential cost of SNR.25 The data were corrected for gradient delays but not any other gradient imperfections. The resulting data were formatted into a Cartesian matrix using the default GPI density compensation and gridding settings, including iterative density compensation26,27 and gridding using an exponentially decaying Hanning window with a radius of 2.5 points. The center-out trajectories were corrected for off-resonance frequencies following the method presented by Robison et al,18 which were negligible due to the low gyromagnetic ratio of xenon and an accurate center frequency from the calibration scan.
2.3. Data analysis
Resulting images were imported into MATLAB 2019a (MathWorks, Natick, Massachusetts) for further analysis. Due to the varying matrix dimensions, each ventilation scan was interpolated onto a 100 × 100 × 100 matrix using the nearest-neighbor method to retain the true resolution. Lung masks were generated from the proton image using a combination of intensity-based thresholding and active-contour region-growing segmentation.
The proton image and lung mask were registered to each ventilation image to account for movement. Ventilation defect percentages (VDPs) were determined using a threshold of 60% of the mean 129Xe signal intensity within the lung mask.10 A background mask was generated containing voxels over 10 voxels away from a voxel above the ventilation threshold. The SNR was determined by , which accounts for the Rician nature of the magnitude noise,28 where Slung is mean signal amplitude within the lung mask; SBG is the mean signal amplitude within the background mask; and σBG is the SD of the noise in the background mask. Normalized SNR (SNRn) was calculated according to , where Vvox is the voxel volume, f129 is the isotopic fraction of 129Xe, P129 is the spin polarization, and VXe is the xenon volume.7 Additionally, SNRn was made relative to the subjects’ ventilating lung volume to account for varying amounts of functioning lung despite a fixed xenon volume: relative SNRn = SNRn(mL−2) × Vlung(L).
Lung masks were used to obtain ventilation masks, areas within the lung mask above the defect threshold (60%).10 All images and masks were registered to the 2D-GRE ventilation image. The ventilation masks were compared using Dice scores (for region-based similarities),29 boundary F1 scores (for edge/contour-based similarities),30 and structural similarity index (SSIM, for signal amplitude-based similarities).31 Dice calculates the percent of overlap between the 2 masks, whereas F1 is the percent of boundaries in image 1 that are within x voxels of a boundary in image 2, where x is defined as 0.75% of the image diagonal. The SSIM is more complex, ranging from −1 to 1, and is a measurement of image similarity taking into consideration differences in luminance, contrast, and structure, where a value of 1 is an identical image. For similarity comparisons, the 2D-GRE image was taken as ground truth, as it is the most standard and commonly used technique. Finally, the images were corrected using N4ITK bias correction,32 permitting more accurate cross-section line profiles of areas lacking ventilation due to vasculature. These line profiles were then qualitatively compared among the 3 techniques.
Comparisons among the acquisition groups were compared using a repeated-measures analysis of variance with a Tukey’s honestly significant difference ad hoc test to determine which group differences were significant. Linear regressions were performed among the VDPs of the different acquisition techniques.
3. RESULTS
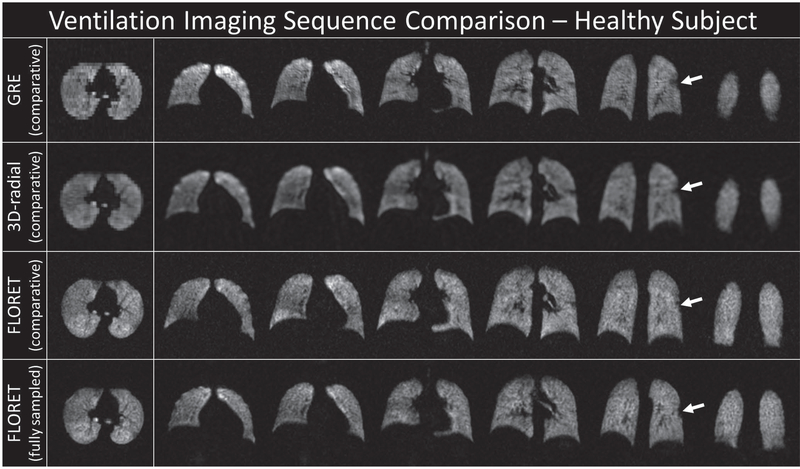
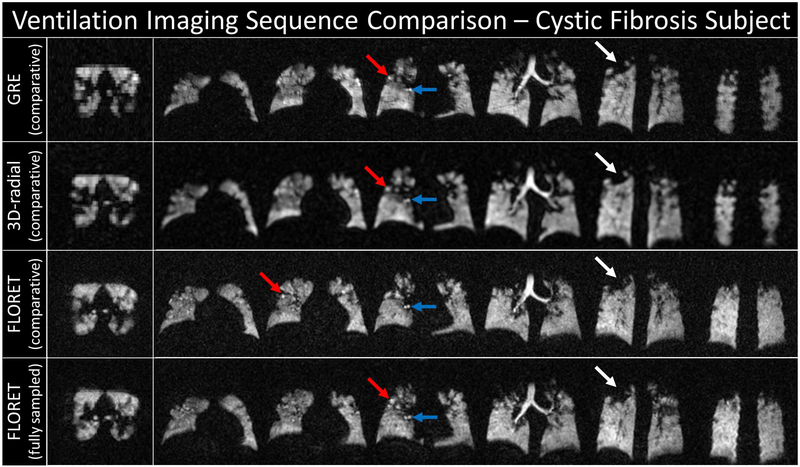
Representative slices from each sequence are shown in Figures 1 and 2. Note that the 2D-GRE sequence is nonisotropic, and while the in-plane resolution is high (3 × 3 mm), the slice thickness is approximately twice as thick as the 3D-radial images and 3 times as thick as the FLORET images. Figure 1 shows the images for a healthy subject. All 4 images (before registration and bias correction) show homogenous ventilation and good correlation of structural features. Figure 2 shows a subject with cystic fibrosis. Many ventilation defects are visible in all images and are predominantly in the middle/upper portions of the lung. Regions of hyperintensity and airway are also observable. This heterogeneity is captured across all sequences (i.e., defects and hyperintensity in the same regions consistently).
FIGURE 1.
Representative slices from the reconstructed images for a healthy subject before registration and bias correction. The white arrows point to the small defect in the left lung. Each row corresponds to 1 of 4 sequences/settings used: top, 2D gradient-recalled echo (GRE) (comparative); upper middle, 3D radial (comparative); lower middle, FLORET (Fermat Looped ORthogonally Encoded Trajectories) (comparative); and bottom, FLORET (fully sampled). Note that the 2D-GRE slices are approximately 3 times thicker than the corresponding in-plane resolution and the resolution in all planes for the comparative FLORET. All other images are fully isotropic
FIGURE 2.
Representative slices from the reconstructed images for a subject with cystic fibrosis before registration and bias correction. White arrows point to a defect; blue arrows point to a hyperintense region of airway; and red arrows point to a hyperintense region likely caused by a defect shunting xenon into other regions of the lung. Each row corresponds to 1 of 4 sequences/settings used: top, 2D GRE (comparative); upper middle, 3D radial (comparative); lower middle, FLORET (comparative); and bottom, FLORET (fully sampled). Note that the 2D-GRE slices are approximately 3 times thicker than the corresponding in-plane resolution and the resolution in all planes for the comparative FLORET. All other images are fully isotropic
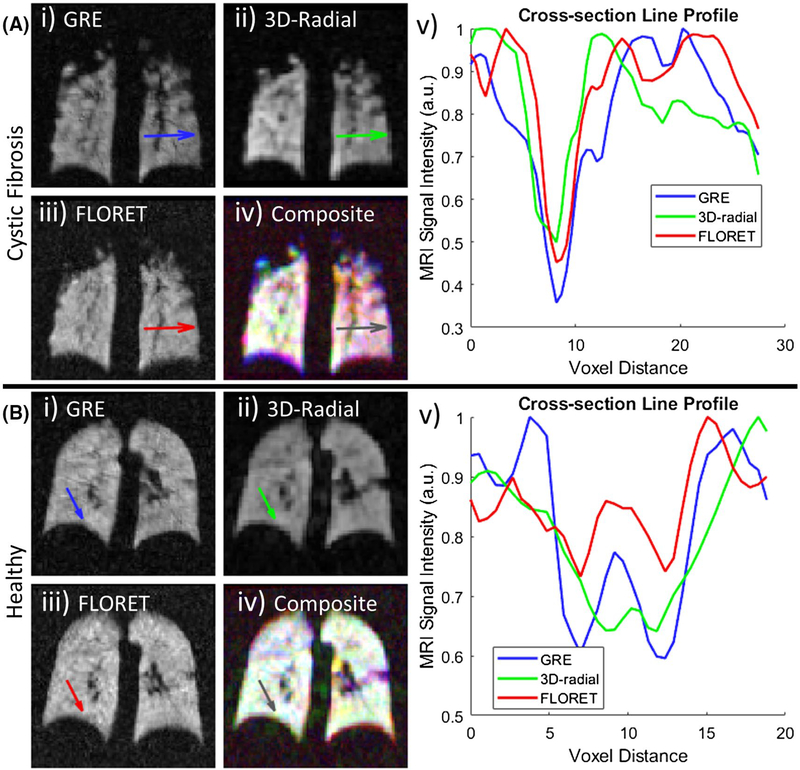
Vasculature cross sections were qualitatively compared among the 3 sequences as shown in Figure 3. Figure 3A illustrates the signal intensity loss due to a larger and more prominent portion of vasculature, whereas Figure 3B illustrates the same for a pair of small vessels, which are difficult to discern by eye, but decrease the signal intensity by about 15%−40%, depending on the sequence and its resolution. The signal amplitude appears to drop more severely and for larger distances in the 2D-GRE images than the FLORET images. This suggests that the longer TE necessary for 2D GRE leads to increased susceptibility-induced signal dephasing in regions adjacent to vasculature, resulting in an overestimation of vessel size. The 3D-radial sequence should benefit from shorter TEs as well, but the effect appears to be lessened by increased partial-volume effects.
FIGURE 3.
Example vasculature cross sections in ventilation images of a subject with cystic fibrosis (CF) (A) and a healthy subject (B). The 3 gray-scale images (i, ii, iii) correspond to 2D GRE, 3D radial, and FLORET, respectively. (iv) Overlay of 3 slices with 2D GRE (blue), 3D radial (green), and FLORET (red) illustrating the excellent agreements among the images following registration. Areas of white indicate relative voxel intensity agreement among all sequences. Areas of color indicate a larger relative intensity for that given sequence. For example, the healthy subject shows excellent agreement within the lung volume with the presence of 3D-radial signal between the lungs. The subject with cystic fibrosis illustrates excellent agreement in the right lung, but a lack of green indicates a lower relative intensity in the 3D-radial sequence. (v) Line profiles (arrows) for each cross section shown in (i)-(iv), illustrating narrower and shallower drops in signal due to vasculature for FLORET, which will reduces the amount of voxels with vasculature classified as ventilation defects
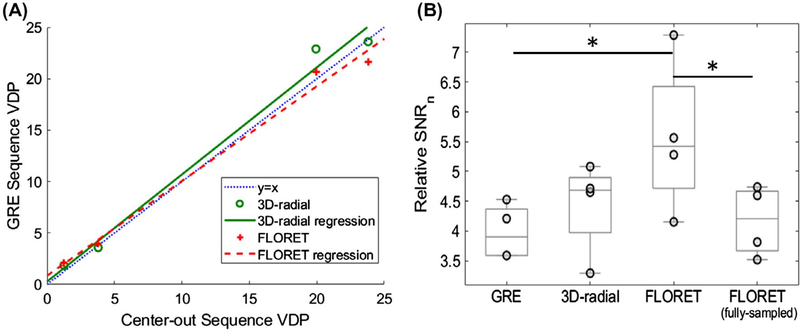
The resulting VDPs for the center-out trajectories were compared with the calculated VDP from the standard 2D-GRE sequence (Figure 4A). This was completed to confirm similar results regardless of the sequence implemented. The 2D-GRE sequence resulted in 1.3% and 3.8% VDPs for the healthy subjects and 19.9% and 23.8% for the subjects with cystic fibrosis. The VDPs from 2D GRE had a linear correlation with 3D radial (y = 1.04x + 0.3, R2 = 0.99) and FLORET (y = 0.92x + 0.85, R2 = 0.99). Thus, all 3 imaging sequences strongly correlate for measuring VDP, as expected. However, future studies will be required to assess any potential bias among the calculated VDPs for each sequence and whether the image-quality improvements result in additional sensitivity to disease severity and progression.
FIGURE 4.
Whole-lung comparisons among the ventilation sequences. A, Comparison of ventilation defect percentage (VDP) derived from center-out sequences and those derived from the standard 2D-GRE sequence. B, Relative SNRn each sequence. The comparable FLORET sequence has a significant 40% increase (P = .012) in SNRn over the 2D-GRE sequence and a significant 34% increase (P = .024) over the fully sampled FLORET
To confirm similar defect locations, quantitative image-space metrics were used, including Dice coefficients, boundary F1 scores, and SSIM indexes. Taking 2D GRE as ground truth, 3D radial had Dice similarity scores of 0.93 ± 0.05, FLORET had scores of 0.94 ± 0.05, and fully sampled FLORET had a score of 0.95 ± 0.03, indicating large overlap of the areas determined as ventilating among all techniques (all P >.72). The 3D-radial sequence had boundary F1 scores of 0.83 ± 0.07, FLORET had scores of 0.885 ± 0.07, and fully sampled FLORET had scores of 0.85 ± 0.06. The boundary F1 score demonstrates that more than 80% of the ventilation boundaries occur in close proximity with each other, regardless of the imaging technique (all P > 0.89). As for SSIM, 3D radial had scores of 0.63 ± 0.08, FLORET had scores of 0.61 ± 0.03, and fully sampled FLORET had scores of 0.66 ± 0.03 (all P > .63). Generally, lower SSIM scores occurred in areas of poorer registration/partial-volume effects and in the anterior portions of the lung. Despite lower SSIM scores, all 3 center-out sequences obtained similar scores, indicating similar intensity discrepancies when compared with 2D GRE. If 3D radial is instead taken as ground truth for the FLORET images, the Dice scores increased by 0.6% and the boundary F1 scores increased 4.3%, while the SSIM scores decreased by 3.4%.
The SNRs were 12.9 ± 1.2 for 2D GRE, 17.8 ± 1.0 for 3D radial, 10.0 ± 1.4 for FLORET, and 12.7 ± 2.2 for the fully sampled FLORET. All images had SNRs much greater than the Rose criterion,33 allowing for accurate analysis. However, because SNR does not account for polarization or voxel volume, SNRn is more informative for comparisons. The value of SNRn was 1.30 ± 0.24 mL−2 for 2D GRE, 1.40 ± 0.24 mL−2 for 3D radial, 1.78 ± 0.50 mL−2 for FLORET, and 1.45 ± 0.28 mL−2 for the fully sampled FLORET. The FLORET sequence provides higher SNRn but with higher SD. This fails to account for functional lung volume: Given the same volume of xenon, a subject with smaller lungs or more ventilation defects will have a larger xenon density in his or her ventilating regions, increasing the signal and SNR. This intersubject variation can be accounted for by calculating a relative SNRn. The resulting relative SNRns is shown in Figure 4B. Using relative SNRn, there are significant differences among the acquisitions (P = 0.011), with comparable FLORET having a 40% larger relative SNRn than 2D GRE (P = .012) and 34% larger relative SNRn than the fully sampled FLORET (P = .024). All other comparisons were not statistically significant.
4. DISCUSSION
The quantitative comparisons (VDP, Dice, F1, and SSIM) illustrate similar amounts of disagreement between the traditional 2D-GRE sequence and the center-out sequences (which show increased ventilation region/boundary agreement with each other). The VDPs were strongly correlated among the techniques, indicative of similar information with regard to ventilation defects. There was strong correlation of ventilating regions using the Dice and F1 boundary scores, and moderate correlation of intensities using SSIM scores. Much of the discrepancies appear to be due to the varying voxel dimensions and separate breath-holds. However, 3D radial and FLORET achieved very similar scores with regard to 2D GRE, indicating a similar amount of discrepancies, independent of the acquisition technique. Additionally, decreased prevalence of vasculature with FLORET would lead to fewer vasculature regions misclassified as ventilation defects, depending on the lung mask. This, and the reduced partial-volume effects, likely lead to more accurate VDP quantifications for FLORET.
Previous results showed approximately half the SNR for 3D-radial ventilation images compared with 2D GRE.7 Here, we observe a slightly higher SNRn (~11%) for 3D radial over 2D GRE. The difference likely is due to the offline reconstruction implemented here for both sequences and removal of the native Fermi filter that, in the previous comparison, had been applied to the 2D-GRE images but not the 3D-radial images. Additionally, kernel/gridding approaches may also play a significant role in the different SNRs we observed compared with the previous 3D-radial results.
Because the FLORET sequence has a higher sampling and SNR efficiency than 3D radial, fewer excitations were necessary, allowing for higher flip angles and thus higher SNR. This is despite the shorter dwell time used in the FLORET sequences. The fully sampled version of FLORET had a significant decrease in SNRn, due to the increased number of excitations, indicating that undersampling could provide noticeable SNR and/or timing gains, as long as undersampling artifacts can be mitigated.
The FLORET sequence efficiency increases with longer readout durations. The comparative sequence used a theoretically optimized readout duration assuming the global of 8 ms at 3 T.18,20 Higher efficiency would be gained if a local of 18 ms is assumed. Additionally, implementation of FLORET has shown little to no effect in image quality/sharpness and only minor decreases in SNR by using longer than optimal readout durations .34 For hyperpolarized imaging, a slight reduction in SNR would likely be overcome with larger flip angles and fewer projections. Thus, the FLORET sequences implemented here may be further.
5. CONCLUSIONS
The use of FLORET for ventilation imaging results in similar ventilation structure to traditional techniques, providing corroboration of FLORET ventilation imaging. Like 3D radial, the center-out nature allows FLORET to more accurately capture physiological gradients and permits advanced reconstruction techniques. The sampling efficiency of FLORET provides fewer excitations and larger flip angles, resulting in higher SNRn when compared with 2D GRE and 3D radial. Perhaps most importantly, the FLORET sampling pattern results in higher-resolution images and/or shorter breath-holds, without the need for high-performance gradients. The FLORET images obtained demonstrate an approximate 3-fold higher resolution and a 40% higher relative SNRn in a 15% shorter breath-hold duration compared with 2D GRE. This should result in higher sensitivity, more accurate quantification of ventilation, and make 129Xe ventilation imaging more feasible for subjects with more advanced lung disease or less compliant subjects who are unable to complete longer-duration breath-holds.
Supplementary Material
FIGURE S1 Base Fermat spirals (kz = 0) for the comparative (left) and fully sampled (right) FLORET sequences. Note that the longer readout samples more of k-space within a single excitation/acquisition
FIGURE S2 Trajectory endpoints for the comparative (left) and fully sampled (right) FLORET sequences. Each hub is colored differently, allowing them to be distinguished. Additionally, endpoints further away in the kx dimension are smaller, to provide depth. Despite the fully sampled nature of the sequence on the right, the left is denser due to its shorter readout duration
FIGURE S3 Complete trajectory sampling for the comparative (left) and fully sampled (right) FLORET sequences. Red to yellow indicates the first hub, with the red indicating the first acquisition of the hub and yellow indicating the final acquisition of the hub. Similarly, the second hub is ordered blue to green. Note that the spirals are acquired relatively randomly in the cone dimension
FIGURE S4 Approximate sampling density maps for the comparative (left) and fully sampled (right) FLORET sequences. Note the cones of increased sampling density due to the overlap of the hubs and the similar amount of undersampling in the cone and spiral dimensions. The minimum undersampling values (at the edges where no hub overlap occurs) is consistent with the reported sampling densities for each sequence
ACKNOWLEDGMENTS
The authors thank the following sources for research funding and support: NIH R01 HL131012, NIH R01 HL143011, NIH R44 HL123299, NIH K99 HL138255, and NIH T32 HL007752.
Funding information
National Heart, Lung, and Blood Institute, Grant/Award Number: K99 HL138255, R01 HL131012, R01 HL143011, R44 HL123299 and T32 HL007752
Footnotes
SUPPORTING INFORMATION
Additional supporting information may be found online in the Supporting Information section.
REFERENCES
- 1.Driehuys B, Martinez-Jimenez S, Cleveland ZI, et al. Chronic obstructive pulmonary disease: safety and tolerability of hyperpolarized 129Xe MR imaging in healthy volunteers and patients. Radiology. 2012;262:279–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mugler JP, Altes TA. Hyperpolarized 129Xe MRI of the human lung. J Magn Reson Imaging. 2013;37:313–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Couch MJ, Thomen R, Kanhere N, et al. A two-center analysis of hyperpolarized 129Xe lung MRI in stable pediatric cystic fibrosis: potential as a biomarker for multi-site trials. J Cyst Fibros. 2019;18:728–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rayment JH, Couch MJ, McDonald N, et al. Hyperpolarised 129 Xe magnetic resonance imaging to monitor treatment response in children with cystic fibrosis. Eur Respir J. 2019;53:1802188. [DOI] [PubMed] [Google Scholar]
- 5.Svenningsen S, Kirby M, Starr D, et al. Hyperpolarized 3He and 129Xe MRI: differences in asthma before bronchodilation. J Magn Reson Imaging. 2013;38:1521–1530. [DOI] [PubMed] [Google Scholar]
- 6.Virgincar RS, Cleveland ZI, Sivaram Kaushik S, et al. Quantitative analysis of hyperpolarized 129Xe ventilation imaging in healthy volunteers and subjects with chronic obstructive pulmonary disease. NMR Biomed. 2013;26:424–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.He MU, Robertson SH, Kaushik SS, et al. Dose and pulse sequence considerations for hyperpolarized 129Xe ventilation MRI. Magn Reson Imaging. 2015;33:877–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roos JE, McAdams HP, Kaushik SS, Driehuys B. Hyperpolarized gas MR imaging: technique and applications. Magn Reson Imaging Clin N Am. 2015;23:217–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.He M, Driehuys B, Que LG, Huang YCT. Using hyperpolarized 129Xe MRI to quantify the pulmonary ventilation distribution. Acad Radiol. 2016;23:1521–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thomen RP, Walkup LL, Roach DJ, Cleveland ZI, Clancy JP, Woods JC. Hyperpolarized 129Xe for investigation of mild cystic fibrosis lung disease in pediatric patients. J Cyst Fibros. 2016;16:275–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ebner L, Kammerman J, Driehuys B, Schiebler ML, Cadman RV, Fain SB. The role of hyperpolarized 129xenon in MR imaging of pulmonary function. Eur J Radiol. 2017;86:343–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stewart NJ, Chan HF, Hughes PJC, et al. Comparison of 3He and 129Xe MRI for evaluation of lung microstructure and ventilation at 1.5T. J Magn Reson Imaging. 2018;48:632–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shukla Y, Wheatley A, Kirby M, et al. Hyperpolarized 129Xe magnetic resonance imaging. Acad Radiol. 2012;19:941–951. [DOI] [PubMed] [Google Scholar]
- 14.Walkup LL, Thomen RP, Akinyi TG, et al. Feasibility, tolerability and safety of pediatric hyperpolarized 129Xe magnetic resonance imaging in healthy volunteers and children with cystic fibrosis. Pediatr Radiol. 2016;46:1651–1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Salerno M, Altes TA, Brookeman JR, De Lange EE, Mugler JP. Dynamic spiral MRI of pulmonary gas flow using hyperpolarized 3He: preliminary studies in healthy and diseased lungs. Magn Reson Med. 2001;46:667–677. [DOI] [PubMed] [Google Scholar]
- 16.De Rochefort L, Maître X, Fodil R, et al. Phase-contrast velocimetry with hyperpolarized 3He for in vitro and in vivo characterization of airflow. Magn Reson Med. 2006;55:1318–1325. [DOI] [PubMed] [Google Scholar]
- 17.Niedbalski PJ, Willmering MM, Robertson SH, et al. Mapping and correcting hyperpolarized magnetization decay with radial keyhole imaging. Magn Reson Med. 2019;82:367–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Robison RK, Anderson AG, Pipe JG. Three-dimensional ultrashort echo-time imaging using a FLORET trajectory. Magn Reson Med. 2017;78:1038–1049. [DOI] [PubMed] [Google Scholar]
- 19.Pipe JG, Zwart NR, Aboussouan EA, Robison RK, Devaraj A, Johnson KO. A new design and rationale for 3D orthogonally oversampled k-space trajectories. Magn Reson Med. 2011;66:1303–1311. [DOI] [PubMed] [Google Scholar]
- 20.Xu X, Norquay G, Parnell SR, et al. Hyperpolarized 129Xe gas lung MRI—SNR and T2* comparisons at 1.5 T and 3 T. Magn Reson Med. 2012;68:1900–1904. [DOI] [PubMed] [Google Scholar]
- 21.Loew W, Thomen RP, Giaquinto R, et al. A dual loop T/R-xenon coil for homogenous excitation with improved comfort and size. In: Proceedings of the 24th Annual Meeting of ISMRM, Singapore, 2016 Abstract 1624. [Google Scholar]
- 22.Miller GW, Altes TA, Brookeman JR, De Lange EE, Mugler JP. Hyperpolarized 3He lung ventilation imaging with B1-inhomogeneity correction in a single breath-hold scan. Magn Reson Mater Phys Biol Med. 2004;16:218–226. [DOI] [PubMed] [Google Scholar]
- 23.Wang Z, He M, Bier E, et al. Hyperpolarized 129Xe gas transfer MRI: the transition from 1.5T to 3T. Magn Reson Med. 2018;80:2374–2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zwart NR, Pipe JG. Graphical programming interface: a development environment for MRI methods. Magn Reson Med. 2015;74:1449–1460. [DOI] [PubMed] [Google Scholar]
- 25.Marshall H, Ajraoui S, Deppe MH, Parra-Robles J, Wild JM. K-space filter deconvolution and flip angle self-calibration in 2D radial hyperpolarised 3He lung MRI. NMR Biomed. 2012;25:389–399. [DOI] [PubMed] [Google Scholar]
- 26.Pipe JG, Menon P. Sampling density compensation in MRI: rationale and an iterative numerical solution. Magn Reson Med. 1999;41:179–186. [DOI] [PubMed] [Google Scholar]
- 27.Zwart NR, Johnson KO, Pipe JG. Efficient sample density estimation by combining gridding and an optimized kernel. Magn Reson Med. 2012;67:701–710. [DOI] [PubMed] [Google Scholar]
- 28.He M, Zha W, Tan F, Rankine L, Fain S, Driehuys B. A comparison of two hyperpolarized 129Xe MRI ventilation quantification pipelines: the effect of signal to noise ratio. Acad Radiol. 2019;26:949–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dice LR. Measures of the amount of ecologic association between species. Ecology. 1945;26:297–302. [Google Scholar]
- 30.Csurka G, Larlus D, Perronnin F. What is a good evaluation measure for semantic segmentation? IEEE Trans Pattern Anal Mach Intell. 2013;26. [Google Scholar]
- 31.Wang Z, Bovik AC, Sheikh HR, Simoncelli EP. Image quality assessment: from error visibility to structural similarity. IEEE Trans Image Process. 2004;13:600–612. [DOI] [PubMed] [Google Scholar]
- 32.Tustison NJ, Avants BB, Cook PA, et al. N4ITK: improved N3 bias correction. IEEE Trans Med Imaging. 2010;29:1310–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rose A The visual process. In: Vision: Human and Electronic. Boston, MA: Springer, US; 1973:1–27. [Google Scholar]
- 34.Willmering MM, Robison RK, Wang H, Pipe JG, Woods JC. Implementation of the FLORET UTE sequence for lung imaging. Magn Reson Med. 2019;82:1091–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
FIGURE S1 Base Fermat spirals (kz = 0) for the comparative (left) and fully sampled (right) FLORET sequences. Note that the longer readout samples more of k-space within a single excitation/acquisition
FIGURE S2 Trajectory endpoints for the comparative (left) and fully sampled (right) FLORET sequences. Each hub is colored differently, allowing them to be distinguished. Additionally, endpoints further away in the kx dimension are smaller, to provide depth. Despite the fully sampled nature of the sequence on the right, the left is denser due to its shorter readout duration
FIGURE S3 Complete trajectory sampling for the comparative (left) and fully sampled (right) FLORET sequences. Red to yellow indicates the first hub, with the red indicating the first acquisition of the hub and yellow indicating the final acquisition of the hub. Similarly, the second hub is ordered blue to green. Note that the spirals are acquired relatively randomly in the cone dimension
FIGURE S4 Approximate sampling density maps for the comparative (left) and fully sampled (right) FLORET sequences. Note the cones of increased sampling density due to the overlap of the hubs and the similar amount of undersampling in the cone and spiral dimensions. The minimum undersampling values (at the edges where no hub overlap occurs) is consistent with the reported sampling densities for each sequence