Abstract
Epithelial-mesenchymal Transition (EMT) is a de-differentiation program that imparts tumor cells with the phenotypic and cellular plasticity required for drug resistance, metastasis, and recurrence. This dynamic and reversible events is governed by a network of EMT-transcription factors (EMT-TFs) through epigenetic regulation. Many chromatin modifying-enzymes utilize metabolic intermediates as cofactors or substrates; this suggests that EMT is subjected to the metabolic regulation. Conversely, EMT rewires metabolic program to accommodate cellular changes during EMT. Here we summarize the latest findings regarding the epigenetic regulation of EMT, and discuss the mutual interactions among metabolism, epigenetic regulation, and EMT. Finally, we provide perspectives of how this interplay contributes to cellular plasticity, which may result in the clinical manifestation of tumor heterogeneity.
Keywords: Epigenetics, Epithelial-mesenchymal transition, Heterogeneity, Metabolism, Plasticity
Introduction
Epithelial-mesenchymal transition (EMT) is a phenotypic conversion that occurs during embryonic development when tissue remodeling and cell migration shape the future organism, or during wound healing to effect tissue repair. Because the genes implicated in EMT also control invasion and metastasis in many solid tumors,1,2 this leads to a prevailing idea that tumor cells hijack this developmental program for their own benefit during metastasis. Activation of EMT provides cells with the increased plasticity required for invasion and metastasis to distant organs.3, 4, 5, 6, 7, 8 Morphologically, EMT confers tumor cells with fibroblast-like properties that result in reduced intercellular adhesion and increased motility. Functionally, EMT endows tumor cells with cancer stem cell (CSC)-like traits and renders them resistant to therapeutics, with a proclivity for recurrence after treatment.9, 10, 11, 12 Consistent with this notion, circulating tumor cells (CTCs), isolated from the blood of breast cancer patients, are enriched in EMT and CSC markers.13, 14, 15, 16, 17, 18, 19, 20, 21
Two distinct phenomena are commonly associated with EMT. First, EMT is provoked by signals that cells receive from their microenvironment.8,22 For pathological conditions, EMT occurs at the edges of wounds during healing and at the invasive fronts of metastatic tumors.23,24 These observations suggest that the migratory and invasive features tumor cells exhibit at the tumor-stromal boundary are initiated and propelled by extrinsic microenvironment signals through EMT induction. A second phenomenon associated with EMT is that it is a reversible process.22 When cancer cells disseminate to distant sites of the body, they no longer encounter the signals that they experienced in the primary tumor, and revert to an epithelial state via a MET.22 Epigenetic regulation provides a reversible and rapid switch in gene expression during transition, while retaining cellular plasticity in response to developmental and microenvironmental signals.25,26 In the eukaryotic nucleus, core histones and other chromosomal proteins package DNA to form chromatin, which carries not only genetic information encoded in the DNA but also epigenetic information carried by the post-translational modifications (PTM) at the N-termini of histones. The reversible PTM of histones facilitates rapid changes in gene expression required for these transitions.25,26 Intriguingly, many of these chromatin modifiers, such as histone deacetylases (HDACs), SirTs, Jumonji C (JmjC)-family demethylases, and ten-eleven translocation (TET) family enzymes, require co-factors, such as NAD+, FAD+, and 2-hydoxyl-glutarate (2-KG), for their enzymatic activities. These observations spurred great interest and immense investigation in recent years; the resultant experimental evidence demonstrates that cellular metabolism has intertwined with the epigenetic program associated with organ development, regeneration, stem cell biology, and EMT.
In this article, we first summarize the latest findings regarding the expanding epigenetic regulation in EMT, we then discuss the contribution of metabolism to the epigenetic regulation. Because recent studies indicate that EMT directs a metabolic reprogramming, we will outline the possible effects of metabolic alternations needed in EMT. Finally, we will touch the interplay among metabolism, epigenetic regulation and EMT in the development of heterogeneity and cellular plasticity of tumor cells.
EMT is regulated by EMT-transcription factors (EMT-TFs) through epigenetic mechanisms
In response to extracellular signals, a network of EMT-TFs orchestrate the EMT process; these include the zinc finger proteins of the SNAIL family, such as Snail1 (Snail), Snail2 (Slug), and Snail3 (Smuc); zinc finger and E-box binding proteins of the ZEB family, such as ZEB1 and ZEB2 (SIP1); bHLH proteins Twist1, Twist2, E47, and E2.2; the homeobox proteins goosecoid (GSC) and Six1; the forkhead-box protein FoxC2 and the Krüppel-like factor KLF8. However, these EMT-TFs cannot functions alone, they require to interact with different chromatin modifying enzymes to control gene expression.
Snail functions as a transcriptional repressor
Snail, first described in Drosophila melanogaster and shown to be essential for the formation of the mesoderm,27,28 contains four C2H2 type zinc fingers that bind to the E-box motif (5′-CANNTG-3′) in target gene promoters.29 Similar to p53 and HIF1α, PTMs are the predominant regulators of Snail activity (Fig. 1B). In the central region of Snail, a serine-rich domain (SRD) and a nuclear export sequence (NES) function to regulate protein stability and subcellular localization of Snail, respectively.30 For example, in human breast cancer cells, canonical Wnt signaling activates EMT by inducing expression of intracellular protein Axin2, and inactivating GSK-3β. Without the phosphorylations provided by GSK-3β, Snail is stabilized and remains in the nucleus.31,32 By blocking the activity of GSK-3β, Wnt can stabilize the level of Snail and β-catenin to generate a synergistic effect for invasion and survival, both of which are required for successful metastasis. Receptor tyrosine kinase (RTKs) signaling activated by HGF, FGF, or EGF, acts through the RAS-MAPK or PI3K-Akt pathway, and inflammation, can also stabilize Snail through a similar mechanism by inactivating GSK-3β.
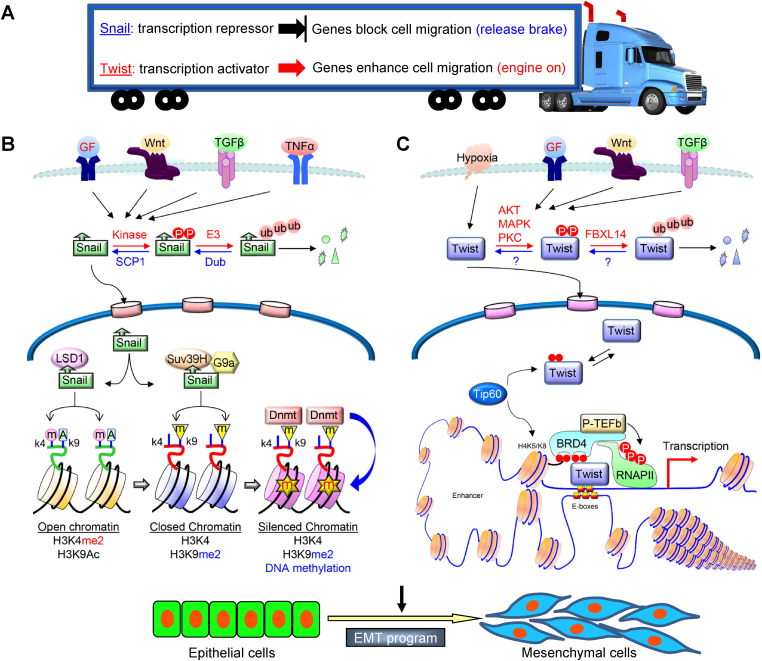
Figure 1.
A schematic diagram showing a distinct but complementary role of Snail and Twist in EMT. (A) A plethora of signaling pathways, including RTKs, TGF-β, Notch, Wnt, TNF-α, and BMPs, transduce signals from tumor microenvironment to activate Snail and Twist, resulting in the EMT induction. Snail functions as a transcriptional repressor to suppress genes that prevent cell migration and growth (such as E-cadherin and FBP1); whereas Twist acts as a transcriptional activator to induce genes that favor cell migration and proliferation (Wnt5A), in an analogy of a moving truck that has disabled brakes and an accelerating engine (B) A Schematic diagram to show the regulation of Snail and the interaction of Snail with chromatin modifiers in EMT. (C) A Schematic diagram to show the regulation of Twist and the interaction of Twist with BRD4 in transcriptional activation.
Once stabilize in the nucleus, the N-terminal of Snail, which contains the evolutionarily conserved SNAG (Snail/Gfi) domain, starts to recruit several transcriptional co-repressor complexes. For instance, Snail interacts with the Sin3A-HDAC1/2 complex through the SNAG domain to deacetylate histones H3 and H4, which leads to E-cadherin repression.33 In addition, Snail has been shown to interact with EZH2 and Suz12 of PRC2 and induce the subsequent trimethylation of H3K27 on the E-cadherin promoter.34 Furthermore, PRMT5 interacts with Snail indirectly through association with Ajuba, which binds to the SNAG domain of Snail. The Snail-Ajuba-PRMT5 ternary complex is found at the proximal promoter region of the E-cadherin gene, and increases arginine methylation of H4R3.35 Our studies identified LSD1 as a partner of Snail36 (Fig. 1B). LSD1 is the first identified histone demethylase that specifically removes methylations from H3K4, a transcriptional mark associated with gene activation.37 The SNAG domain of Snail resembles a histone H3-like structure, and functions as a molecular hook for recruiting LSD1 to the E-cadherin promoter to demethylate H3K4.36 H3K4 demethylation is known to be an initial step in gene repression,38 suggesting that an intermediate step is required to bridge H3K4 demethylation to the DNA methylation on the E-cadherin promoter. Snail interacts with G9a and Suv39H1, two major methyltransferases responsible for H3K9 methylation that intimately link to DNA methylation. G9a interacts with DNA methyltransferase (DNMT) directly and recruits it to target gene promoters for DNA methylation; whereas Suv39H1 creates an H3K9me3 docking site for the adapter molecule HP1, which in turn recruits DNMT and HDAC to catalyze DNA methylation and histone deacetylation, respectively.39, 40, 41 Therefore, demethylation on H3K4 (by LSD1), together with dimethylation and trimethylation on H3K9 (by G9a and Suv39H1), provide a synergistic binary switch in gene repression.
Twist is a transcriptional activator
Twist has multiple roles in cancer progression and metastasis, including the induction of EMT and endowment of CSC-like properties.11,42,43 Twist also plays an important role in the acquisition of resistance to chemotherapy, and antagonizes tumor suppressor-induced apoptosis and oncogene-induced pre-mature senescence.44, 45, 46, 47, 48 Similar to Snail, Twist is also subjected to PTMs. Protein Kinase A (PKA) phosphorylates two residues in the bHLH domain of Twist, Thr125 and Ser127.49 Protein kinase B (PKB/AKT) allows the Ser42-phosphorylated Twist to confer resistance to p53-mediated cell apoptosis in response to DNA damage, and promotes EMT and breast cancer metastasis by enhancing TGF-β signaling.50,51 Three mitogen-activated protein kinases, p38, c-Jun N-terminal kinases (JNK) and extracellular signal-regulated kinases1/2 (ERK1/2) can phosphorylate Twist at Ser68 in vitro and prevent its protein degradation; this phosphorylation also promotes breast cancer cell invasiveness.52 IL-6 activates casein kinase 2, which phosphorylates Twist at Ser18 and Ser20, resulting in Twist stabilization and increase of tumor cell motility.53 Recent studies also show that the stability of Twist is negatively regulated by GSK-3β-mediated phosphorylation on the WR domain (also known as Twist box) of Twist.54
The C-terminus of Twist contains a bHLH motif that is required for recognition of an E-box DNA sequence. A majority of mutations which cause Saethre-Chotzen syndrome are located in the bHLH domain, revealing its critical function in DNA binding.55 The WR domain is located at the end of C-terminus of Twist and modules the dimerization of Twist.56 Several point mutations in the WR domain are also linked to Saethre-Chotzen syndrome.57, 58, 59 Although Twist induces EMT by activating several signaling pathways such as Mir-10b and Platelet-derived growth factor receptor (PDGFR),60, 61, 62 the underlying molecular mechanisms remain obscure. Through unbiased protein purification coupled with mass spectrometric analysis, we found that Twist interacts with BRD4,63 which is a key transcriptional activator that associates p-TEFb and activates RNA polymerase II during transcription in many diverse biological processes.64, 65, 66 The interaction of Twist with BRD4 is dependent on the acetylation of Twist, which is di-acetylated at K73 and K76. The di-acetylated Twist adopts a “histone H4 mimic” GK-X-GK motif that can interact with the second bromodomain of BRD4, while the first bromodomain interacts with acetylated H4, and thereby assemble an activated Twist/BRD4/P-TEFb/RNA-PolII complex at the target promoter and enhancer (Fig. 1C). Although Twist does not directly induce promoter DNA methylation on the E-cadherin promoter, Twist induces Slug expression directly,67 which silences E-cadherin expression in a way similar to that of Snail.
A hallmark of EMT is loss of E-cadherin expression,68,69 which is commonly caused by DNA methylation at the E-cadherin promoter.70,71 Although Snail and Twist have been reported to repress E-cadherin expression either directly or indirectly, we propose that Snail functions as a transcription repressor to repress genes that prevent EMT (such as E-cadherin and FBP1); whereas Twist serves as a transcription activator to induce genes that enhance EMT (such as Slug and Wnt5A). EMT induction mediated by overexpression of Snail or Twist is analogous to a move truck that has disable brakes and an accelerating engine (Fig. 1A).
Bivalent chromatin modification mediated by ZEB1
In embryonic stem cells (ESCs), the promoters of developmental regulatory genes are simultaneously poised with bivalent marks, H3K4me3 and H3K27me3, representing both active and repressive modifications controlled by trithorax and polycomb (PcG) complexes, respectively. During differentiation, pluripotent genes lose active (H3K4me3) modification while maintaining the repressor mark (H3K27me3); whereas the lineage-specific developmental genes, silent in pluripotent ESCs, retain the activation modification and lose the repressive mark upon differentiation. The bivalent configuration of certain EMT-associated genes likely permits a dynamic regulation of differential gene expression, leading to the plastic nature of cells during EMT. For example, Weinberg's group showed that cancer cells that are phenotypically plastic seem to contain bivalent modifications at the ZEB1 promoter, which facilitate their rapid de-differentiation to a stem-like state. Bivalent promoters are also observed in certain cancer cells that exhibit stem cell-like properties. Within the CD44+ stem cell-enriched populations of primary human mammary epithelial tissues, the CDH1 promoter bears the bivalent H3K4me3 and H3K27me3 modifications while being silenced, whereas the more differentiated CD24+ cells that express abundant E-cadherin carry only the active H3K4me3 mark. This chromatin configuration in CD44+ stem cells should logically permit their rapid differentiation into a CD24+ epithelial state through loss of the repressive H3K27me3 mark on the CDH1 promoter and, quite possibly, other epithelial-specific genes.
Using a classic TGF-β-induced EMT model in NMuMG cells,72 Derynck and colleagues was the first to notice that EMT is a dynamic and reversible process: mesenchymal cells can revert to the epithelial state when TGF-β was removed from the medium. However, extended TGF-β exposure produced mesenchymal cells that are difficult to revert to the epithelial state, with concomitant E-cadherin promoter methylation. Genes with bivalent chromatin modifications are not associated with long-term silencing, as their expressions change rapidly and reversibly in response to extracellular signals. However, during the invasion-metastasis cascade, epithelial genes must be repressed for extended periods of time, as in the case of CTCs. Indeed, the majority of the basal-like breast cancer cells are “locked” in the mesenchymal state by DNA methylation at the E-cadherin promoter,70,71 which provides a relative stable “memory” mark for gene silencing. The signals and mechanisms that determine the switch between bivalent chromatin modification and stable DNA methylation in genes involved in the EMT process are an interesting area of continued investigation.
Metabolic influence on chromatin modifications
During EMT, multiple cellular processes, including metabolism, often act synergistically and sequentially. Cellular metabolism directly influences the epigenetic landscape of a cell by modulating the level of metabolites and cofactors, such as oxygen, ATP, acetyl-CoA, S-adenosyl methionine (SAM), NAD+, FAD, and α-KG. Many of these serve as a co-factors or substrates of chromatin modifying enzymes, leading to the change in chromatin modification and gene expression73 (Fig. 2).
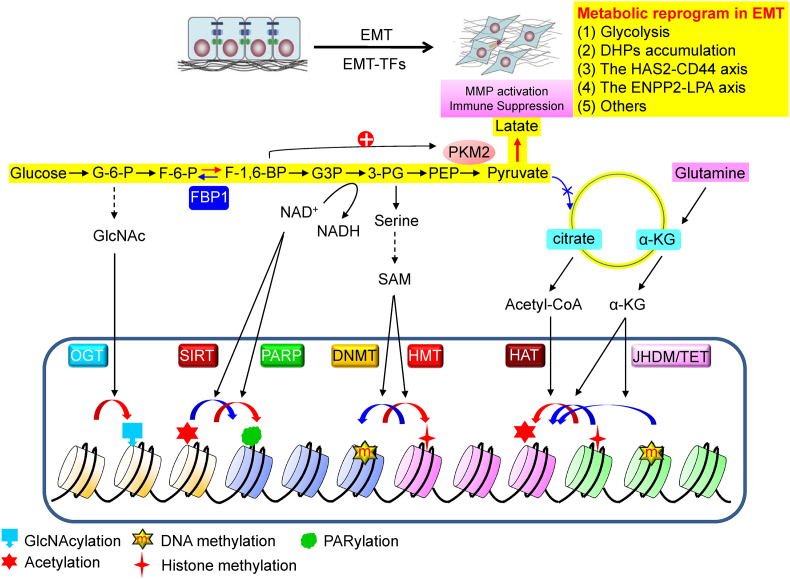
Figure 2.
A schematic diagram of the interaction of metabolism with epigenetic regulation. Numerous metabolic intermediates, showing from glucose metabolism, function as a substrate or co-factor of various chromatin modifying enzymes in controlling the post-translational modifications of histone and DNA methylation/demethylation.
Histone acetylation
Histone acetylation is generally associated with gene activation and often antagonize histone and DNA methylation. Histone acetylation is catalyzed by histone acetyltransferases (HATs), which transfer an acetyl group from acetyl-CoA to lysine residues on histones. Acetyl-CoA is generated from pyruvate by pyruvate dehydrogenase inside mitochondria, and subsequently converted to citrate by citrate synthase through addition of oxaloacetate in the TCA cycle.74,75 Citrate is shuttled to the cytoplasm by a citrate carrier, where ATP-citrate lyase (ACLY) releases acetyl CoA from citrate. In addition to pyruvate, citrate can also be generated from acetate, ketone bodies and the amino acid glutamine.74,75 Citrate is a key metabolite, as it provides acetyl-CoA for fatty acid and cholesterol syntheses in cytosol as well as histone acetylation in nuclei, and ACLY is an important enzyme bridging glucose metabolism to histone acetylation.76 Activation of ACLY by citrate controls histone acetylation in a glucose-dependent manner. Intriguingly, ACLY-mediated histone acetylation is specific, as it regulates genes involved in energy metabolism, including glucose uptake and adipocyte differentiation, but does not cause tubulin acetylation. Significant fluctuation in the acetyl-CoA levels has been noticed in eukaryotic cells; the level of histone acetylation correlates with the peak levels of intracellular acetyl-CoA in yeast and mouse embryonic stem cells (ESCs). For example, mouse ESCs contain high level of acetyl-CoA compared to differentiated cells induced by leukemia-inhibitory factor (LIF) and retinoic acid. It is unclear whether the level of acetyl-CoA fluctuates during the dynamic conversion between epithelial and mesenchymal states in EMT/MET. In addition, it would be interesting to test whether the fluctuation of acetyl-CoA abundance also affect the acetylation of non-histone protein, such as the di-acetylation of Twist in EMT.
Histone de-acetylation
Butyrate, a short-chain fatty acid, is produced in high concentrations from dietary fiber in the lumen of the colon, and is a potent inhibitor of class I/II HDACs. Foods rich in dietary fiber regulate transcriptional programs involved in proliferation of colon cells and prevent colitis and colon cancer in humans.77,78 Ketone body β-hydroxybutyrate (β-OHB) is structurally similar to butyrate. The serum level of ketone bodies, produced mainly in the liver, is elevated during prolonged fasting, calorie restriction, strenuous exercise or with ketogenic diets. Ketone bodies can serve as alternative energy substrates for ATP production by neurons and other peripheral tissues. β-OHB can function as a endogenous inhibitor of HDAC1, 3 and 4, leading to increased acetylation of histone H3K9 and K14, and thus increases gene expression associated with fasting and exercise.79
Class III HDACs (sirtuin 1-7) are homologous to yeast scSir2 that mediates the effect of caloric restriction on longevity. Sirtuin enzymes are proposed as the prime sensors of the cellular NAD+/NADH, because their catalytic activities require NAD+ as a cofactor. During the de-acetylation reaction, NAD+ is cleaved and generates ADP-ribose, which serves as an acyl acceptor to form an acyl-ADP-ribose product. The level of NAD+ is believed to fluctuate during calorie restriction and other metabolic conditions. Intriguingly, NAD+ levels fluctuate substantially in a circadian manner, linking the peripheral clock to transcriptional regulation of metabolism by epigenetic mechanisms involving SIRT1.80 Interesting, resveratrol from red wine mimics an endogenous allosteric metabolite of SIRT1 and thus activates SIRT1,81 suggesting that the activities of sirtuin enzymes are modulated by NAD+ fluctuations and metabolites from some foods.
Histone and DNA methylation
Both histone and DNA methylation play an equal important role in regulating gene expression and chromatin remodeling.82 The balance of methylation is maintained by a group of methyltransferases and demethylases. Methyltransferase utilizes SAM as a methyl donor to catalyze the transfer of methyl group to histones and DNA. Sufficient methionine in culture media is required to maintain SAM levels and global DNA and histone methylation, because long-term methionine deprivation leads to apoptosis through a p53/p38-mediated stress signaling response.83
Histone methylation is a crucial epigenetic mark closely associated with DNA methylation.84 Histone methylation at lysine or arginine residues is part of the histone code, though lysine methylation is more common than arginine methylation in epigenetics. Methylations of H3K4 and H3K36 are associated with gene activation, whereas methylations of H3K9, H3K27 and H4K20 are repressive marks for transcription. There are 33 lysine methyltransferases and 21 lysine demethylases in the human genome; they have a major role in the regulation of chromatin structure and thus control gene expression and maintenance of the genome.84
Histone and DNA demethylation
Histone demethylation is catalyzed by two classes of lysine demethylases.85 LSD1 (KDM1A) belongs to the first class of demethylases that use an amine oxidation reaction with FAD as a cofactor. Thus, the cellular level of FAD, which also serve as a concentration-dependent cofactor for fatty acid oxidation and respiration, can influence the activity of LSD1. LSD1 removes only the mono or di-methylation from H3K4, because it requires a protonated nitrogen to initiate demethylation. The second and more expansive class of enzymes is broadly referred to as the Jumonji demethylases. Members of this large family of 2-oxoglutarate-dependent dioxygenases (2OGD) possess a conserved JmjC domain.86, 87, 88 Unlike LSD1, JmjC enzymes can demethylate all three methyl lysine states, and require α-KG, O2, and Fe(II) as obligatory cofactors for their enzymatic activities.86, 87, 88 Vitamin C (ascorbate) induces reduction of the oxidized Fe(IV) to Fe(II) and thereby actives these enzymes. These 2OGDs have an eight-fold β-sheet core domain, known as the jelly-roll fold, which contains two binding sites for α-KG. Although α-KG is a TCA cycle intermediate, it can also be produced from glutamate by aminotransferases involved in other metabolic pathways in the cytosol. In some cancers, the majority of α-KG is produced by phosphoserine aminotransferase 1 (PSAT1).89 Although the binding domain is highly specific for α-KG, structural analogs, such as succinate and fumarate from TCA cycle can also bind to this domain and thus function as potent inhibitors of 2OGDs. Loss-of-function mutations in succinate dehydrogenase (SDH) and fumarate hydratase (FH) produce high levels of succinate and fumarate, respectively, and result in the suppression of 2OGDs during oncogenesis for several types of cancers.90, 91, 92 Isocitrate dehydrogenases 1 and 2 (IDH1/2) are involved in α-KG catabolism, and mutations in this enzyme generate oncometabolite R(-)-2-hydroxyglutarate (R-2HG), which is a competitive inhibitor of the 2OGDs.93 These studies demonstrate that succinate, fumarate and R-2HG can facilitate tumor development by enhancing histone and DNA hypermethylation. In addition, these 2OGD inhibitors can suppress the activity of PHD1-3 and thus generates a pseudo-hypoxic condition by stabilizing HIF1α during tumor progression and metastasis.
DNA demethylation is carried out by the TET methylcytosine dioxygenases (TET1-3), which also belong to the 2OGD family.86, 87, 88 TETs oxidize the 5 mC in DNA to form 5-hydroxymethylcytosine (5hmC)94; which is an initial modification and subsequently removed by thymine DNA glycosylase (TDG) and the base excision repair (BER) system.95 Genome-wide studies reveal an increased content of 5hmC at active enhancers and promoters, as well as the gene bodies, of actively transcribed genes.96 This indicates that TETs have an important role in the regulation of chromatin structure and gene expression. The level of 5hmC is down-regulated in tumors and mutations in TET2 gene have been linked to human myeloid leukemia.97 Given that TETs are members of the 2OGD family, hypoxia and alterations in mitochondrial metabolism can modulate the enzymatic activity of TETs and thus control DNA methylation.
O-GlcNAcylation
O-GlcNAcylation is a process of adding a GlcNAc moiety to a serine or threonine residue of target proteins, including transcription factors, epigenetic modulators, and RNA polymerase II. This reaction uses UDP-GlcNAc, which is the end product of the hexosamine biosynthetic pathway (HSP), a regulated process requiring precursors derived from glucose, glutamine, acetyl-CoA, ATP, and uridine. The pool of UDP-GlcNAc available for O-GlcNAcylation is influenced by nutritional flux, circadian rhythms, infection, and stress. Thus, O-GlcNAcylation represents a sensitive integration point between metabolism and chromatin structure to control gene expression for homeostasis. Addition and removal of O-GlcNAcylation is catalyzed by O-GlcNAc transferase (OGT) and O-GlcNAcase (OGA; MGEA5), respectively. O-GlcNAc has been shown to directly modify histones H3S10 in mitosis,98 and H2A and H2B in histone exchange.99 The best established role for OGT in transcriptional regulation is involvement in polycomb repression.100 The trithorax protein is associated with H3K4 methylation and it may require O-GlcNAcylation as part of the regulatory network.101 In addition, OGT has been shown to interact with mSin3A and HDAC1/2102 for transcription repression. OGT binds to the TET in transcription.103,104 Intriguingly, OGT also interacts with and confers O-GlcNAcylation on Snail, resulting in the stabilization of Snail and EMT induction under hyperglycemic condition.105
Metabolic reprogramming contributes to EMT induction
EMT provides phenotypic and cellular plasticity and confers CSC-like characteristics to tumor cells, numerous cellular changes are required to accomplish these events, these include changes in the cytoskeleton, the loss of adhesion, and gain of invasion. It is now understood that tumor cells also rewire their metabolic program during EMT, suggesting that metabolic reprogramming is inextricably intertwined with EMT, and that metabolism is not an “accompanied” bystander effect but rather an “actionable” event in EMT.106 Using both functional analysis and bioinformatics to investigate metabolic gene expression in both epithelial and mesenchymal cells, several important signaling pathways essential for EMT are identified.107,108 Below we discuss recent developments in this area and provide new perspectives on the metabolic reprogramming associated with EMT.
Enhanced Warburg effect by suppressing FBP1 expression
Based on gene expression profiling, breast cancer can be divided into five subtypes: luminal A, luminal B, HER2, normal-like and basal-like. Basal-like breast cancer (BLBC) is associated with an aggressive clinical history, development of recurrence, distant metastasis and shorter survival. BLBC cells possess many CSC-like traits and EMT characteristics.109,110 Interestingly, FBP1, a rate-limiting enzyme in gluconeogenesis, is specifically lost in BLBC but not in other subtypes of breast cancer. FBP1 deficiency due to gene mutation results in hypoglycemia and acidosis in adults and causes sudden infant death in infants. FBP1 is also a therapeutic target to restore glucose uptake in the type 2 diabetic model. Loss of FBP1 expression is due to promoter DNA methylation; the Snail-G9a-Dnmt1 complex, which is required for E-cadherin silencing, is also responsible for FBP1 silencing through promoter DNA methylation in BLBC.107 These findings indicate that metabolic reprogramming (resulting from the loss of FBP1) synergizes with the loss of E-cadherin in EMT during dissemination and metastasis. Intriguingly, loss of FBP1 expression is also observed in almost 100% clear cell renal carcinoma, and a majority of malignant hepatomas and lung cancers.111
Loss of FBP1 expression increases the Warburg effect and provides several metabolic benefits for BLBC: (1) increased glucose uptake and sensitivity; (2) increased the production of glycolytic intermediates for biosynthesis (such as PPP, glycerol-3-phosphat and serine); (3) maintenance of ATP production under hypoxia; and (4) reduced oxygen dependence with concomitant less ROS production. Approximately 90% of the ROS is generated from oxidative phosphorylation (OXPHOS) in mitochondria, and protection from ROS is critical for the maintenance of self-renewal of hematopoietic stem cell (HSC) and human and mouse breast CSCs.112 This observation suggests that the self-renewal potential of CSCs is exquisitely sensitive to the level of ROS. Indeed, loss of FBP1 expression in BLBC enhances tumorsphere-formation, increases CSC populations, and promotes tumor growth in vivo. Mechanistically, reduced ROS shifts the interaction of β-catenin from FOXO3a to TCF4 and thus elevates β-catenin activity, which is essential for the maintenance of pluripotency. This study elucidates a crosstalk among EMT, metabolic reprogramming and CSC traits in BLBC.
Accumulation of dihydropyrimidines (DHPs) in EMT
Using a bioinformatics approach in analyzing the expression pattern of 1,704 metabolic genes in 978 various human tumor cell lines, Shaul et al identified a “mesenchymal metabolic signature (MMS)” consisting of 44 genes.108 This MMS includes signaling pathways involved glycan biosynthesis, hyaluronan acid (HA) biosynthesis, and lysophosphatidic acid (LPA) signaling (please see discussion below). The MMS genes are enriched in the BLBC subtype and correlate with known mesenchymal markers in both cancer cell lines and tumors. Unlike the majority of metabolic genes, MMS genes are upregulated upon EMT induction, and knockdown 16 of these 44 genes blocked Twist-mediated EMT induction in HMLE cells. Dihydropyrimidine dehydrogenase (DPYD), a pyrimidine-degrading enzyme, was the leading candidate among these 16 genes. Knockdown of DPYD expression did not affect the growth of tumor cells but inhibited migration and invasion in vitro and metastasis in vivo. Consistent with the elevation of DPYD in EMT, the direct products of DPYD (DHPs), but not the final end product in the pyrimidine-degrading pathway, were significantly increased in EMT cells. These findings indicate that the products of DPYD, but not the pyrimidine-degrading pathway per se, is critical for EMT induction. Although no biological function has been ascribed the DHPs, it is likely that these metabolites may serve as a ligand for other unknown proteins; analogous to kynurenine, an inter-metabolite from the breakdown of tryptophan pathway. Kynurenine generated by the tumor cells binds to the aryl hydrocarbon receptor (AHR) in immune cells. This binding causes suppression of effector T cells (Teff) and inhibition of regulatory B cells (Breg) and dendritic cells, and thus is responsible for immune suppression and the pro-survival effects on tumors.113 Identification of the target and function of DHPs will provide new excitement to the delicate regulation of EMT.
The ATX (autotaxin)-LPA (lysophosphatidate) axis in EMT
This signaling axis, owing to its critical role in development, inflammation and fibrosis, is gaining appreciation as a key mediator of tumor progression and metastasis. LPA is the simplest bioactive phospholipid composed of a glycerol backbone, one fatty acid and a phosphate head group. LPA in plasma is mainly produced from LPC (lysophosphatidylcholine) by ATX that possesses lysophospholipase D activity. ATX, an ecto-nucleotide pyrophosphatase and phosphodiesterase family member 2 (ENPP2), is known as “autocrine motility factor” that can increase cell migration, invasion and metastasis. ATX heterozygote knockout mice have a 50% reduction in circulating LPA levels compared with wild-type mice,114,115 indicating that ATX is a major enzyme responsible for LPA production. LPA binds to specific G-protein-coupled receptors (GPCRs) to exhibit a plethora of actions in developmental, physiological and pathophysiological processes. LPA binds to and activates at least six GPCRs (LPA receptors; LPAR1-6), which in turn couple with and activate three G-protein subfamilies: Gαq/11, Gi/o and Gα12/13. The classic receptors, LPAR1-3, belong to the EDG subfamily of GPCRs, whereas LPAR4-6 are structurally more similar to P2Y nucleotide receptors. Although the LPARs exhibit distinct tissue distribution patterns, they are associated with both common and divergent downstream signaling pathways, including Gαq-mediated stimulation of phospholipase C, Gαi-mediated activation of the Ras-MAPK and PI3K pathways; and Gα12/13-mediated activation of RhoA. Overexpression of ATX and LPARs has also been observed in numerous human malignancies, ranging from neuroblastoma, hepatocellular carcinoma and non-small cell lung cancer, to prostate, ovarian and breast cancers. Mice that overexpress ATX, LPAR1, LPAR2 or LPAR3 in mammary epithelia develop spontaneous metastatic breast tumors.116 In addition, ATX is one of the most commonly up-regulated genes among metastatic cancers.117
Consistent with the upregulation of ATX (ENPP2) in MMS, increased LPA levels are found in cells that undergo EMT.118,119 High level of LPA activate the AKT/GSK-3β signaling pathway and thus fosters the EMT process.120 Clinically, breast cancer cells express more ATX transcripts than normal breast epithelium, and aberrant expression of ATX markedly enhances the aggressiveness of breast cancer cells.121 These results support the finding that elevation of ATX plays a critical role in EMT induction and cancer metastasis.
The HAS2-HA-CD44 axis in EMT
The growth and aggressiveness of tumors rely on a supportive microenvironment characterized by a bioactive extracellular matrix (ECM) network, composed of structural proteins, glycoproteins, proteoglycans, and glycosaminoglycans. The ECM also contains fibroblasts, vascular and lymphatic endothelial cells, and infiltrated immune cells.122 HA is the simplest glycosaminoglycan composed of repeating disaccharide units of d-glucuronic acid and N-acetyl-d-glucosamine, and possesses unique hygroscopic and viscoelastic properties to trap water inside this structure, resulting in soft and elastic tissues.123 HA accumulates in sites of cell division and the rapid matrix remodeling that occur during embryonic morphogenesis, inflammation and tumorigenesis.124 Over production of HA has been correlated to severity of tumors; HA is aberrantly accumulated due to deregulated functions of HA synthases (HAS1-3), HA-degrading enzymes (HYALs and PH-20), or altered clearance from the tissues via lymphatics.125 HA transmits extracellular signals through receptor binding to CD44 and RHAMM, thereby triggering downstream signaling on its own and/or in cooperation with growth factor receptors, such as the receptors for PDGF-BB, TGFβ, and ErbB2. These signaling pathways promote migration, differentiation, and proliferation. CD44 is a transmembrane protein, with a short cytoplasmic tail that interacts with ankyrin and ezrin–radixin–moesin proteins, and provides a link to the cytoskeleton. CD44 can also interact with other molecules, including collagen, fibronectin, osteopontin, growth factors and matrix metalloproteinases (MMPs). Due to alternative splicing and posttranslational modifications, CD44 exhibits wide structural diversity, including standard form of CD44 (CD44s) and CD44 variant forms (CD44v1–10), which all exhibit different affinities for HA. A high correlation between malignancy and HA-rich ECM, as well as with CD44s and CD44v expression have been observed. For example, expression of CD44v is switched to CD44s in EMT and breast cancer metastasis and thus suppresses E-cadherin through activated PI3K/Akt pathway.126 Interestingly, breast CSCs are enriched in a population with the CD44+/CD24low signature.127 Similarly, cells that undergo EMT (such as BLBC cells) express this same signature.
Among HAS1-3, HAS2 is particularly important. HAS2-knockout mice are lethal, because HA deposition in the ECM is blocked and the endocardial cushion cells fail to undergo EMT and migrate to the cardiac jelly; these events result in defects in embryonic heart valve morphogenesis.128,129 Consistently, knockdown of HAS2 inhibits TGFβ-induced EMT.130 HAS2 mRNA is abundantly expressed in the hormone-negative breast cancer cell lines that exhibit aggressive and invasive mesenchymal-like traits. HAS2-mediated HA polymerization increases the growth and self-renewal of the CD44+/CD24low breast CSCs that metastasize to bone.131 The identification of HAS2 in MMS re-enforces the notion that altered metabolic pathways are intertwined with other cellular events directing EMT.
A circuitry of epigenetic-EMT-metabolism in regulating the plasticity of epithelial cells
Tumors are inherently heterogeneous in composition, with cell populations contain different genetic and epigenetic makeup. Of critical importance is the fact that the heterogeneity of tumor, as well as the tumor microenvironment, undergo constant changes as the tumor develops. Tumor heterogeneity is commonly manifested in the following three areas: (1) CSCs vs non-CSCs; (2) migratory and non-migratory tumor cells; and (3) drug resistance. The CSC theory proposes that malignant tumors are hierarchically organized tissues, similar to stem cells in normal tissues/organs, and that the bulk of the tumor mass is composed of the differentiated progeny of CSCs. Only a small population of CSCs have self-renewal and differentiation capabilities and can therefore propagate to give rise to all other cells that form part of the tumor mass. CSCs are also responsible for metastatic growth in breast cancer, which accounts for more than 90% of cancer death. For example, a population of quiescent and migratory, mesenchymal-like CSCs was recently described at the invasive front of breast tumors, whereas a more proliferative, epithelial-like population of CSCs was located toward the center of the tumor.132 In addition, CSCs cause treatment relapse as they have higher chemo resistance than differentiated, non-tumorigenic, tumor cells. The heterogeneity observed in these three processes, CSCs/non-CSCs, invasive/non-invasive, and drug sensitive/resistant cells, can be attributed to the activation of EMT in tumors. In vivo, EMT does not exhibit the two extreme states, the epithelial morphology or the mesenchymal phenotype; instead it displays a full spectrum of cells ranging from epithelial to mesenchymal because some cells undergo partial EMT.133 The mesenchymal cells are more stem cell-like and permit continued self-renewal, as well as possess migratory capabilities. This process is similar to the tissue regeneration that occurs during development and wound healing. Cells, based on the extracellular signals encountered, can bi-directionally shift along this spectrum going from epithelial morphology to mesenchymal phenotype and reverse this course as needed. This cellular plasticity, generated by the activation of EMT, not only occurs in normal cells but also appears more efficiently in tumor cells. Different from the classic CSC model, in which CSCs are hierarchically organized and differentiated tumor cells are unable to de-differentiate back to CSCs, EMT provides tumor cells an opportunity to reversibly shift between two functional states: a more committed epithelial cell versus a mesenchymal stem-like cell with migratory potential depending on the context signals. The reversibility of the differentiated state is supported by a Nobel Prize winning discovery: the generation of induced pluripotent stem (iPS) cells does not require genetic changes and it can be induced by the ectopic expression of a small number of transcription factors. Environmental stimuli, such as vitamin C, kinase inhibitors, α-KG, can modulate the efficiency of iPS cells.
EMT undoubtedly provides cellular plasticity to tumors and thus leads to a mixed tumor cell population with respect to morphology (epithelial or mesenchymal), stem-cell characteristics (more or less), and potential for drug resistance (sensitive/resistant). Fifty years ago, Conrad Waddington conceptualized the directionality of cellular differentiation as a ball falling down a valley (Fig. 3). The shape of the landscape determines the path of a falling ball. This concept is widely used to describe the processes of trans-differentiation, de-differentiation or pluripotent reprogramming, with “energy demanding” processes that must overcome developmental barriers for completion. By analyzing the interactions among all the genes that are expressed in a cell, a recent study indicates that a geometric landscape can be generated to describe the developmental potential of a particular cell, in a graphical manner, similar to Waddington's model134 (Fig. 3). The “valleys” represent lower points of developmental energy (i.e. the final, stable, differentiated cells), whereas the “slopes” are the differentiation routes. The shape of the landscape is determined by the “pulling strings” of the genes, through transcriptional and epigenetic control, located beneath it. This landscape determines the developmental potential and fates of a particular cell. As discussed previously, EMT is mainly regulated through an epigenetic mechanism controlled by a few EMT-TFs and their associated chromatin enzymes. Cellular metabolism, by providing metabolic intermediate for chromatin modifiers, impinges on the epigenetic regulation and gene expression of EMT by modulating the activities of these enzymes. Metabolic reprogramming re-shapes the landscape by disrupting the gene expression profile and thus leads to the opening of new “valleys” that drive tumor cells towards new pathological states. In return, activation of EMT also rewires metabolism to provide a reinforcement to sustain or foster the functionalities of EMT. This feed-forward bi-directional interaction between metabolism and epigenetic modifications provides a dynamic and reversible platform for EMT and generates a vicious cycle for the establishment of the cellular plasticity and heterogeneity. We envision that target the cellular metabolism in addition to the epigenetic regulation may yield a new and effective approach to suppress the plasticity employed by tumor cells in proliferation, metastasis and recurrence.
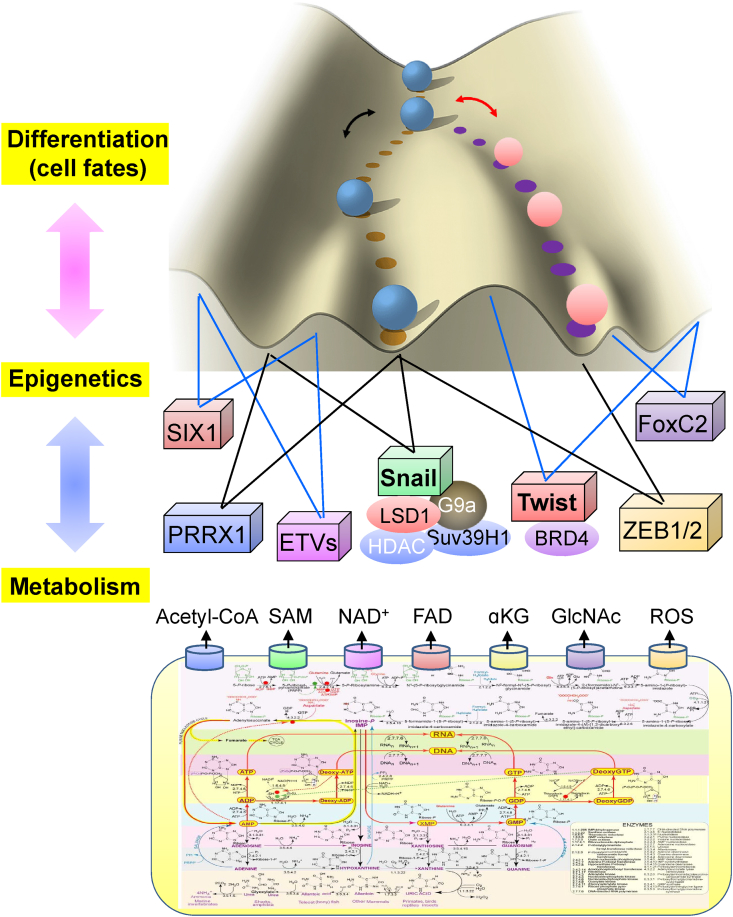
Figure 3.
A schematic diagram showing the relationship of metabolism and epigenetic in contribution to the cell fate determination and tumor heterogeneity. EMT is governed by a set of transcription factors (EMT-TFs); the function and activities of these EMT-TFs are tightly regulated by the corresponding associated chromatin modifying enzymes; the activities of which are subjected to the regulation of metabolic input/flux. The dynamic and reversible interaction between metabolism and epigenetic contributes the cellular plasticity seen in EMT, leading to the manifestation of tumor heterogeneity.
Concluding remarks
In the last several years, we have experienced an information explosion in EMT-related cellular metabolism and epigenetic regulation. New ideas and concepts are being formulated to account for the mechanisms underlying tumor development and metastasis. However, many questions remain. For example, in addition to Snail and Twist, there are several other EMT-TFs that can induce EMT in various cellular contexts; what chromatin modifiers associate with these EMT-TFs and what signals can trigger their activation? Unveiling the transcriptional machinery utilized by different EMT-TFs, and building an epigenome associated with gene expression during EMT will greatly advance our understanding of metastasis. In addition, identification of the contribution that metabolism conveys to epigenetic regulation and the metabolic rewiring mediated by EMT will certainly provide a comprehensive knowledge of the dynamic events in EMT. Lastly, the signaling cross-talk among metabolism, chromatin modifications, and gene expression creates enormous plasticity and differentiation potentials to tumor cells, and results in the clinical manifestations of tumor heterogeneity in metastasis, drug resistance, and tumor recurrence (Fig. 3). Understanding of the contribution and interplay of these processes (metabolism, epigenetics, and EMT) in the development of tumor cell plasticity and heterogeneity unleashes exciting potential for the development of new and effective approaches against metastatic diseases.
Conflict of interest
The authors declare no conflict of interest.
Acknowledgements
We apologize to the many contributors to this field whose work are important while we were unable to cite due to space limitation. Our study is supported by the grants from National Institutes of Health (NIH) (RO1s CA125454 and CA188118), Department of Defense (DOD) Breakthrough Award (BC140733P1), Mary Kay Ash Foundation (to B.P. Zhou), and the Basic Public Welfare Research Program of Zhejiang Province (LGF18H290003 to Y. Wang).
Footnotes
Peer review under responsibility of Chongqing Medical University.
References
- 1.Huber M.A., Kraut N., Beug H. Molecular requirements for epithelial-mesenchymal transition during tumor progression. Curr Opin Cell Biol. 2005;17:548–558. doi: 10.1016/j.ceb.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 2.Thiery J.P. Epithelial-mesenchymal transitions in development and pathologies. Curr Opin Cell Biol. 2003;15:740–746. doi: 10.1016/j.ceb.2003.10.006. [DOI] [PubMed] [Google Scholar]
- 3.Drasin D.J., Robin T.P., Ford H.L. Breast cancer epithelial-to-mesenchymal transition: examining the functional consequences of plasticity. Breast Cancer Res. 2011;13:226. doi: 10.1186/bcr3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Micalizzi D.S., Farabaugh S.M., Ford H.L. Epithelial-mesenchymal transition in cancer: parallels between normal development and tumor progression. J Mammary Gland Biol Neoplasia. 2010;15:117–134. doi: 10.1007/s10911-010-9178-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Polyak K., Weinberg R.A. Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nat Rev Cancer. 2009;9:265–273. doi: 10.1038/nrc2620. [DOI] [PubMed] [Google Scholar]
- 6.Thiery J.P., Acloque H., Huang R.Y., Nieto M.A. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 7.Tsai J.H., Yang J. Epithelial-mesenchymal plasticity in carcinoma metastasis. Genes Dev. 2013;27:2192–2206. doi: 10.1101/gad.225334.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang J., Weinberg R.A. Epithelial-mesenchymal transition: at the crossroads of development and tumor metastasis. Dev Cell. 2008;14:818–829. doi: 10.1016/j.devcel.2008.05.009. [DOI] [PubMed] [Google Scholar]
- 9.Creighton C.J., Li X., Landis M. Residual breast cancers after conventional therapy display mesenchymal as well as tumor-initiating features. Proc Natl Acad Sci USA. 2009;106:13820–13825. doi: 10.1073/pnas.0905718106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li X., Lewis M.T., Huang J. Intrinsic resistance of tumorigenic breast cancer cells to chemotherapy. J Natl Cancer Inst. 2008;100:672–679. doi: 10.1093/jnci/djn123. [DOI] [PubMed] [Google Scholar]
- 11.Mani S.A., Guo W., Liao M.J. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moody S.E., Perez D., Pan T.C. The transcriptional repressor Snail promotes mammary tumor recurrence. Cancer Cell. 2005;8:197–209. doi: 10.1016/j.ccr.2005.07.009. [DOI] [PubMed] [Google Scholar]
- 13.Aktas B., Tewes M., Fehm T., Hauch S., Kimmig R., Kasimir-Bauer S. Stem cell and epithelial-mesenchymal transition markers are frequently overexpressed in circulating tumor cells of metastatic breast cancer patients. Breast Cancer Res. 2009;11:R46. doi: 10.1186/bcr2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Armstrong A.J., Marengo M.S., Oltean S. Circulating tumor cells from patients with advanced prostate and breast cancer display both epithelial and mesenchymal markers. Mol Cancer Res. 2011;9:997–1007. doi: 10.1158/1541-7786.MCR-10-0490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bonnomet A., Brysse A., Tachsidis A. Epithelial-to-mesenchymal transitions and circulating tumor cells. J Mammary Gland Biol Neoplasia. 2010;15:261–273. doi: 10.1007/s10911-010-9174-0. [DOI] [PubMed] [Google Scholar]
- 16.Kallergi G., Papadaki M.A., Politaki E., Mavroudis D., Georgoulias V., Agelaki S. Epithelial to mesenchymal transition markers expressed in circulating tumour cells of early and metastatic breast cancer patients. Breast Cancer Res. 2011;13:R59. doi: 10.1186/bcr2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kasimir-Bauer S., Hoffmann O., Wallwiener D., Kimmig R., Fehm T. Expression of stem cell and epithelial-mesenchymal transition markers in primary breast cancer patients with circulating tumor cells. Breast Cancer Res. 2012;14:R15. doi: 10.1186/bcr3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu J., Fan T., Zhao Q. Isolation of circulating epithelial and tumor progenitor cells with an invasive phenotype from breast cancer patients. Int J Cancer. 2010;126:669–683. doi: 10.1002/ijc.24814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mego M., Mani S.A., Lee B.N. Expression of epithelial-mesenchymal transition-inducing transcription factors in primary breast cancer: the effect of neoadjuvant therapy. Int J Cancer. 2012;130:808–816. doi: 10.1002/ijc.26037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Raimondi C., Gradilone A., Naso G. Epithelial-mesenchymal transition and stemness features in circulating tumor cells from breast cancer patients. Breast Canc Res Treat. 2011;130:449–455. doi: 10.1007/s10549-011-1373-x. [DOI] [PubMed] [Google Scholar]
- 21.Yu M., Bardia A., Wittner B.S. Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science. 2013;339:580–584. doi: 10.1126/science.1228522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thiery J.P., Sleeman J.P. Complex networks orchestrate epithelial-mesenchymal transitions. Nat Rev Mol Cell Biol. 2006;7:131–142. doi: 10.1038/nrm1835. [DOI] [PubMed] [Google Scholar]
- 23.Christofori G. New signals from the invasive front. Nature. 2006;441:444–450. doi: 10.1038/nature04872. [DOI] [PubMed] [Google Scholar]
- 24.Franci C., Takkunen M., Dave N. Expression of Snail protein in tumor-stroma interface. Oncogene. 2006;25:5134–5144. doi: 10.1038/sj.onc.1209519. [DOI] [PubMed] [Google Scholar]
- 25.Feinberg A.P. Phenotypic plasticity and the epigenetics of human disease. Nature. 2007;447:433–440. doi: 10.1038/nature05919. [DOI] [PubMed] [Google Scholar]
- 26.Shi Y. Histone lysine demethylases: emerging roles in development, physiology and disease. Nat Rev Genet. 2007;8:829–833. doi: 10.1038/nrg2218. [DOI] [PubMed] [Google Scholar]
- 27.Grau Y., Carteret C., Simpson P. Mutations and chromosomal rearrangements affecting the expression of snail, a gene involved in embryonic patterning in Drosophila melanogaster. Genetics. 1984;108:347–360. doi: 10.1093/genetics/108.2.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alberga A., Boulay J.L., Kempe E., Dennefeld C., Haenlin M. The snail gene required for mesoderm formation in Drosophila is expressed dynamically in derivatives of all three germ layers. Development. 1991;111:983–992. doi: 10.1242/dev.111.4.983. [DOI] [PubMed] [Google Scholar]
- 29.Nieto M.A. The snail superfamily of zinc-finger transcription factors. Nat Rev Mol Cell Biol. 2002;3:155–166. doi: 10.1038/nrm757. [DOI] [PubMed] [Google Scholar]
- 30.Dominguez D., Montserrat-Sentis B., Virgos-Soler A. Phosphorylation regulates the subcellular location and activity of the snail transcriptional repressor. Mol Cell Biol. 2003;23:5078–5089. doi: 10.1128/MCB.23.14.5078-5089.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yook J.I., Li X.Y., Ota I. A Wnt-Axin2-GSK3beta cascade regulates Snail1 activity in breast cancer cells. Nat Cell Biol. 2006;8:1398–1406. doi: 10.1038/ncb1508. [DOI] [PubMed] [Google Scholar]
- 32.Zhou B.P., Deng J., Xia W. Dual regulation of Snail by GSK-3beta-mediated phosphorylation in control of epithelial-mesenchymal transition. Nat Cell Biol. 2004;6:931–940. doi: 10.1038/ncb1173. [DOI] [PubMed] [Google Scholar]
- 33.Peinado H., Ballestar E., Esteller M., Cano A. Snail mediates E-cadherin repression by the recruitment of the Sin3A/histone deacetylase 1 (HDAC1)/HDAC2 complex. Mol Cell Biol. 2004;24:306–319. doi: 10.1128/MCB.24.1.306-319.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Herranz N., Pasini D., Diaz V.M. Polycomb complex 2 is required for E-cadherin repression by the Snail1 transcription factor. Mol Cell Biol. 2008;28:4772–4781. doi: 10.1128/MCB.00323-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hou Z., Peng H., Ayyanathan K. The LIM protein AJUBA recruits protein arginine methyltransferase 5 to mediate SNAIL-dependent transcriptional repression. Mol Cell Biol. 2008;28:3198–3207. doi: 10.1128/MCB.01435-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin Y., Wu Y., Li J. The SNAG domain of Snail1 functions as a molecular hook for recruiting lysine-specific demethylase 1. EMBO J. 2010;29:1803–1816. doi: 10.1038/emboj.2010.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shi Y., Lan F., Matson C. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 38.Rudolph T., Yonezawa M., Lein S. Heterochromatin formation in Drosophila is initiated through active removal of H3K4 methylation by the LSD1 homolog SU(VAR)3-3. Mol Cell. 2007;26:103–115. doi: 10.1016/j.molcel.2007.02.025. [DOI] [PubMed] [Google Scholar]
- 39.Fuks F. DNA methylation and histone modifications: teaming up to silence genes. Curr Opin Genet Dev. 2005;15:490–495. doi: 10.1016/j.gde.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 40.Dong C., Wu Y., Yao J. G9a interacts with Snail and is critical for Snail-mediated E-cadherin repression in human breast cancer. J Clin Investig. 2012;122:1469–1486. doi: 10.1172/JCI57349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dong C., Wu Y., Wang Y. Interaction with Suv39H1 is critical for Snail-mediated E-cadherin repression in breast cancer. Oncogene. 2012;32:1351–1362. doi: 10.1038/onc.2012.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vesuna F., Lisok A., Kimble B., Raman V. Twist modulates breast cancer stem cells by transcriptional regulation of CD24 expression. Neoplasia. 2009;11:1318–1328. doi: 10.1593/neo.91084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li J., Zhou B.P. Activation of beta-catenin and Akt pathways by Twist are critical for the maintenance of EMT associated cancer stem cell-like characters. BMC Canc. 2011;11:49. doi: 10.1186/1471-2407-11-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maestro R., Dei Tos A.P., Hamamori Y. Twist is a potential oncogene that inhibits apoptosis. Genes Dev. 1999;13:2207–2217. doi: 10.1101/gad.13.17.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Valsesia-Wittmann S., Magdeleine M., Dupasquier S. Oncogenic cooperation between H-Twist and N-Myc overrides failsafe programs in cancer cells. Cancer Cell. 2004;6:625–630. doi: 10.1016/j.ccr.2004.09.033. [DOI] [PubMed] [Google Scholar]
- 46.Ansieau S., Bastid J., Doreau A. Induction of EMT by twist proteins as a collateral effect of tumor-promoting inactivation of premature senescence. Cancer Cell. 2008;14:79–89. doi: 10.1016/j.ccr.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 47.Li Q.Q., Xu J.D., Wang W.J. Twist1-mediated adriamycin-induced epithelial-mesenchymal transition relates to multidrug resistance and invasive potential in breast cancer cells. Clin Cancer Res: Off J Am Assoc Cancer Res. 2009;15:2657–2665. doi: 10.1158/1078-0432.CCR-08-2372. [DOI] [PubMed] [Google Scholar]
- 48.Cheng G.Z., Chan J., Wang Q., Zhang W., Sun C.D., Wang L.H. Twist transcriptionally up-regulates AKT2 in breast cancer cells leading to increased migration, invasion, and resistance to paclitaxel. Cancer Res. 2007;67:1979–1987. doi: 10.1158/0008-5472.CAN-06-1479. [DOI] [PubMed] [Google Scholar]
- 49.Firulli B.A., Krawchuk D., Centonze V.E. Altered Twist1 and Hand2 dimerization is associated with Saethre-Chotzen syndrome and limb abnormalities. Nat Genet. 2005;37:373–381. doi: 10.1038/ng1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vichalkovski A., Gresko E., Hess D., Restuccia D.F., Hemmings B.A. PKB/AKT phosphorylation of the transcription factor Twist-1 at Ser42 inhibits p53 activity in response to DNA damage. Oncogene. 2010;29:3554–3565. doi: 10.1038/onc.2010.115. [DOI] [PubMed] [Google Scholar]
- 51.Xue G., Restuccia D.F., Lan Q. Akt/PKB-mediated phosphorylation of Twist1 promotes tumor metastasis via mediating cross-talk between PI3K/Akt and TGF-beta signaling axes. Cancer Discov. 2012;2:248–259. doi: 10.1158/2159-8290.CD-11-0270. [DOI] [PubMed] [Google Scholar]
- 52.Hong J., Zhou J., Fu J. Phosphorylation of serine 68 of Twist1 by MAPKs stabilizes Twist1 protein and promotes breast cancer cell invasiveness. Cancer Res. 2011;71:3980–3990. doi: 10.1158/0008-5472.CAN-10-2914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Su Y.W., Xie T.X., Sano D., Myers J.N. IL-6 stabilizes Twist and enhances tumor cell motility in head and neck cancer cells through activation of casein kinase 2. PLoS One. 2011;6 doi: 10.1371/journal.pone.0019412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lander R., Nasr T., Ochoa S.D., Nordin K., Prasad M.S., Labonne C. Interactions between Twist and other core epithelial-mesenchymal transition factors are controlled by GSK3-mediated phosphorylation. Nat Commun. 2013;4:1542. doi: 10.1038/ncomms2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kress W., Schropp C., Lieb G. Saethre-Chotzen syndrome caused by TWIST 1 gene mutations: functional differentiation from Muenke coronal synostosis syndrome. Eur J Hum Genet. 2006;14:39–48. doi: 10.1038/sj.ejhg.5201507. [DOI] [PubMed] [Google Scholar]
- 56.Chang A.T., Liu Y., Ayyanathan K. An evolutionarily conserved DNA architecture determines target specificity of the TWIST family bHLH transcription factors. Genes Dev. 2015;29:603–616. doi: 10.1101/gad.242842.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Piccinin S., Tonin E., Sessa S. A “twist box” code of p53 inactivation: twist box: p53 interaction promotes p53 degradation. Cancer Cell. 2012;22:404–415. doi: 10.1016/j.ccr.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 58.Gu S., Boyer T.G., Naski M.C. Basic helix-loop-helix transcription factor Twist1 inhibits transactivator function of master chondrogenic regulator Sox9. J Biol Chem. 2012;287:21082–21092. doi: 10.1074/jbc.M111.328567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Qin Q., Xu Y., He T., Qin C., Xu J. Normal and disease-related biological functions of Twist1 and underlying molecular mechanisms. Cell Res. 2012;22:90–106. doi: 10.1038/cr.2011.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yang J., Mani S.A., Donaher J.L. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117:927–939. doi: 10.1016/j.cell.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 61.Ma L., Teruya-Feldstein J., Weinberg R.A. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449:682–688. doi: 10.1038/nature06174. [DOI] [PubMed] [Google Scholar]
- 62.Eckert M.A., Lwin T.M., Chang A.T. Twist1-induced invadopodia formation promotes tumor metastasis. Cancer Cell. 2011;19:372–386. doi: 10.1016/j.ccr.2011.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Houzelstein D., Bullock S.L., Lynch D.E., Grigorieva E.F., Wilson V.A., Beddington R.S. Growth and early postimplantation defects in mice deficient for the bromodomain-containing protein Brd4. Mol Cell Biol. 2002;22:3794–3802. doi: 10.1128/MCB.22.11.3794-3802.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jang M.K., Mochizuki K., Zhou M., Jeong H.S., Brady J.N., Ozato K. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol Cell. 2005;19:523–534. doi: 10.1016/j.molcel.2005.06.027. [DOI] [PubMed] [Google Scholar]
- 65.Yang Z., Yik J.H., Chen R. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol Cell. 2005;19:535–545. doi: 10.1016/j.molcel.2005.06.029. [DOI] [PubMed] [Google Scholar]
- 66.Devaiah B.N., Lewis B.A., Cherman N. BRD4 is an atypical kinase that phosphorylates serine2 of the RNA polymerase II carboxy-terminal domain. Proc Natl Acad Sci USA. 2012;109:6927–6932. doi: 10.1073/pnas.1120422109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Casas E., Kim J., Bendesky A., Ohno-Machado L., Wolfe C.J., Yang J. Snail2 is an essential mediator of Twist1-induced epithelial mesenchymal transition and metastasis. Cancer Res. 2011;71:245–254. doi: 10.1158/0008-5472.CAN-10-2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kang Y., Massague J. Epithelial-mesenchymal transitions: twist in development and metastasis. Cell. 2004;118:277–279. doi: 10.1016/j.cell.2004.07.011. [DOI] [PubMed] [Google Scholar]
- 69.Thiery J.P. Epithelial-mesenchymal transitions in tumour progression. Nat Rev Cancer. 2002;2:442–454. doi: 10.1038/nrc822. [DOI] [PubMed] [Google Scholar]
- 70.Caldeira J.R., Prando E.C., Quevedo F.C., Neto F.A., Rainho C.A., Rogatto S.R. CDH1 promoter hypermethylation and E-cadherin protein expression in infiltrating breast cancer. BMC Canc. 2006;6:48. doi: 10.1186/1471-2407-6-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lombaerts M., van Wezel T., Philippo K. E-cadherin transcriptional downregulation by promoter methylation but not mutation is related to epithelial-to-mesenchymal transition in breast cancer cell lines. Br J Canc. 2006;94:661–671. doi: 10.1038/sj.bjc.6602996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Miettinen P.J., Ebner R., Lopez A.R., Derynck R. TGF-beta induced transdifferentiation of mammary epithelial cells to mesenchymal cells: involvement of type I receptors. J Cell Biol. 1994;127:2021–2036. doi: 10.1083/jcb.127.6.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kaelin W.G., Jr., McKnight S.L. Influence of metabolism on epigenetics and disease. Cell. 2013;153:56–69. doi: 10.1016/j.cell.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Newman J.C., Verdin E. Ketone bodies as signaling metabolites. Trends Endocrinol Metab. 2014;25:42–52. doi: 10.1016/j.tem.2013.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zaidi N., Swinnen J.V., Smans K. ATP-citrate lyase: a key player in cancer metabolism. Cancer Res. 2012;72:3709–3714. doi: 10.1158/0008-5472.CAN-11-4112. [DOI] [PubMed] [Google Scholar]
- 76.Wellen K.E., Hatzivassiliou G., Sachdeva U.M., Bui T.V., Cross J.R., Thompson C.B. ATP-citrate lyase links cellular metabolism to histone acetylation. Science. 2009;324:1076–1080. doi: 10.1126/science.1164097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Donohoe D.R., Collins L.B., Wali A., Bigler R., Sun W., Bultman S.J. The Warburg effect dictates the mechanism of butyrate-mediated histone acetylation and cell proliferation. Mol Cell. 2012;48:612–626. doi: 10.1016/j.molcel.2012.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kim Y.S., Milner J.A. Dietary modulation of colon cancer risk. J Nutr. 2007;137:2576S–2579S. doi: 10.1093/jn/137.11.2576S. [DOI] [PubMed] [Google Scholar]
- 79.Abdel-Wahab O., Mullally A., Hedvat C. Genetic characterization of TET1, TET2, and TET3 alterations in myeloid malignancies. Blood. 2009;114:144–147. doi: 10.1182/blood-2009-03-210039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Orozco-Solis R., Sassone-Corsi P. Circadian clock: linking epigenetics to aging. Curr Opin Genet Dev. 2014;26:66–72. doi: 10.1016/j.gde.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hubbard B.P., Gomes A.P., Dai H. Evidence for a common mechanism of SIRT1 regulation by allosteric activators. Science. 2013;339:1216–1219. doi: 10.1126/science.1231097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cedar H., Bergman Y. Linking DNA methylation and histone modification: patterns and paradigms. Nat Rev Genet. 2009;10:295–304. doi: 10.1038/nrg2540. [DOI] [PubMed] [Google Scholar]
- 83.Shiraki N., Shiraki Y., Tsuyama T. Methionine metabolism regulates maintenance and differentiation of human pluripotent stem cells. Cell Metabol. 2014;19:780–794. doi: 10.1016/j.cmet.2014.03.017. [DOI] [PubMed] [Google Scholar]
- 84.Black J.C., Van Rechem C., Whetstine J.R. Histone lysine methylation dynamics: establishment, regulation, and biological impact. Mol Cell. 2012;48:491–507. doi: 10.1016/j.molcel.2012.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mosammaparast N., Shi Y. Reversal of histone methylation: biochemical and molecular mechanisms of histone demethylases. Annu Rev Biochem. 2010;79:155–179. doi: 10.1146/annurev.biochem.78.070907.103946. [DOI] [PubMed] [Google Scholar]
- 86.Hausinger R.P. FeII/alpha-ketoglutarate-dependent hydroxylases and related enzymes. Crit Rev Biochem Mol Biol. 2004;39:21–68. doi: 10.1080/10409230490440541. [DOI] [PubMed] [Google Scholar]
- 87.Loenarz C., Schofield C.J. Physiological and biochemical aspects of hydroxylations and demethylations catalyzed by human 2-oxoglutarate oxygenases. Trends Biochem Sci. 2011;36:7–18. doi: 10.1016/j.tibs.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 88.McDonough M.A., Loenarz C., Chowdhury R., Clifton I.J., Schofield C.J. Structural studies on human 2-oxoglutarate dependent oxygenases. Curr Opin Struct Biol. 2010;20:659–672. doi: 10.1016/j.sbi.2010.08.006. [DOI] [PubMed] [Google Scholar]
- 89.Possemato R., Marks K.M., Shaul Y.D. Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature. 2011;476:346–350. doi: 10.1038/nature10350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Adam J., Yang M., Soga T., Pollard P.J. Rare insights into cancer biology. Oncogene. 2014;33:2547–2556. doi: 10.1038/onc.2013.222. [DOI] [PubMed] [Google Scholar]
- 91.Letouze E., Martinelli C., Loriot C. SDH mutations establish a hypermethylator phenotype in paraganglioma. Cancer Cell. 2013;23:739–752. doi: 10.1016/j.ccr.2013.04.018. [DOI] [PubMed] [Google Scholar]
- 92.Xiao M., Yang H., Xu W. Inhibition of alpha-KG-dependent histone and DNA demethylases by fumarate and succinate that are accumulated in mutations of FH and SDH tumor suppressors. Genes Dev. 2012;26:1326–1338. doi: 10.1101/gad.191056.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Xu W., Yang H., Liu Y. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of alpha-ketoglutarate-dependent dioxygenases. Cancer Cell. 2011;19:17–30. doi: 10.1016/j.ccr.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Tahiliani M., Koh K.P., Shen Y. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Delatte B., Deplus R., Fuks F. Playing TETris with DNA modifications. EMBO J. 2014;33:1198–1211. doi: 10.15252/embj.201488290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Pastor W.A., Aravind L., Rao A. TETonic shift: biological roles of TET proteins in DNA demethylation and transcription. Nat Rev Mol Cell Biol. 2013;14:341–356. doi: 10.1038/nrm3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pfeifer G.P., Kadam S., Jin S.G. 5-hydroxymethylcytosine and its potential roles in development and cancer. Epigenet Chromatin. 2013;6:10. doi: 10.1186/1756-8935-6-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Fong J.J., Nguyen S.L., Bridger R. beta-N-Acetylglucosamine (O-GlcNAc) is a novel regulator of mitosis-specific phosphorylations on histone H3. J Biol Chem. 2012;287:12195–12203. doi: 10.1074/jbc.M111.315804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sakabe K., Wang Z., Hart G.W. Beta-N-acetylglucosamine (O-GlcNAc) is part of the histone code. Proc Natl Acad Sci USA. 2010;107:19915–19920. doi: 10.1073/pnas.1009023107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sinclair D.A., Syrzycka M., Macauley M.S. Drosophila O-GlcNAc transferase (OGT) is encoded by the Polycomb group (PcG) gene, super sex combs (sxc) Proc Natl Acad Sci USA. 2009;106:13427–13432. doi: 10.1073/pnas.0904638106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Love D.C., Krause M.W., Hanover J.A. O-GlcNAc cycling: emerging roles in development and epigenetics. Semin Cell Dev Biol. 2010;21:646–654. doi: 10.1016/j.semcdb.2010.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Yang X., Zhang F., Kudlow J.E. Recruitment of O-GlcNAc transferase to promoters by corepressor mSin3A: coupling protein O-GlcNAcylation to transcriptional repression. Cell. 2002;110:69–80. doi: 10.1016/s0092-8674(02)00810-3. [DOI] [PubMed] [Google Scholar]
- 103.Chen Q., Chen Y., Bian C., Fujiki R., Yu X. TET2 promotes histone O-GlcNAcylation during gene transcription. Nature. 2013;493:561–564. doi: 10.1038/nature11742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Deplus R., Delatte B., Schwinn M.K. TET2 and TET3 regulate GlcNAcylation and H3K4 methylation through OGT and SET1/COMPASS. EMBO J. 2013;32:645–655. doi: 10.1038/emboj.2012.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Park S.Y., Kim H.S., Kim N.H. Snail1 is stabilized by O-GlcNAc modification in hyperglycaemic condition. EMBO J. 2010;29:3787–3796. doi: 10.1038/emboj.2010.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ward P.S., Thompson C.B. Metabolic reprogramming: a cancer hallmark even warburg did not anticipate. Cancer Cell. 2012;21:297–308. doi: 10.1016/j.ccr.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Dong C., Yuan T., Wu Y. Loss of FBP1 by Snail-mediated repression provides metabolic advantages in basal-like breast cancer. Cancer Cell. 2013;23:316–331. doi: 10.1016/j.ccr.2013.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Shaul Y.D., Freinkman E., Comb W.C. Dihydropyrimidine accumulation is required for the epithelial-mesenchymal transition. Cell. 2014;158:1094–1109. doi: 10.1016/j.cell.2014.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bergamaschi A., Hjortland G.O., Triulzi T. Molecular profiling and characterization of luminal-like and basal-like in vivo breast cancer xenograft models. Mol Oncol. 2009;3:469–482. doi: 10.1016/j.molonc.2009.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Honeth G., Bendahl P.O., Ringner M. The CD44+/CD24- phenotype is enriched in basal-like breast tumors. Breast Cancer Res. 2008;10:R53. doi: 10.1186/bcr2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Li B., Qiu B., Lee D.S. Fructose-1,6-bisphosphatase opposes renal carcinoma progression. Nature. 2014;513:251–255. doi: 10.1038/nature13557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Diehn M., Cho R.W., Lobo N.A. Association of reactive oxygen species levels and radioresistance in cancer stem cells. Nature. 2009;458:780–783. doi: 10.1038/nature07733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Opitz C.A., Litzenburger U.M., Sahm F. An endogenous tumour-promoting ligand of the human aryl hydrocarbon receptor. Nature. 2011;478:197–203. doi: 10.1038/nature10491. [DOI] [PubMed] [Google Scholar]
- 114.Tanaka M., Okudaira S., Kishi Y. Autotaxin stabilizes blood vessels and is required for embryonic vasculature by producing lysophosphatidic acid. J Biol Chem. 2006;281:25822–25830. doi: 10.1074/jbc.M605142200. [DOI] [PubMed] [Google Scholar]
- 115.van Meeteren L.A., Ruurs P., Stortelers C. Autotaxin, a secreted lysophospholipase D, is essential for blood vessel formation during development. Mol Cell Biol. 2006;26:5015–5022. doi: 10.1128/MCB.02419-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Liu S., Umezu-Goto M., Murph M. Expression of autotaxin and lysophosphatidic acid receptors increases mammary tumorigenesis, invasion, and metastases. Cancer Cell. 2009;15:539–550. doi: 10.1016/j.ccr.2009.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Euer N., Schwirzke M., Evtimova V. Identification of genes associated with metastasis of mammary carcinoma in metastatic versus non-metastatic cell lines. Anticancer Res. 2002;22:733–740. [PubMed] [Google Scholar]
- 118.Jahn S.C., Law M.E., Corsino P.E. An in vivo model of epithelial to mesenchymal transition reveals a mitogenic switch. Cancer Lett. 2012;326:183–190. doi: 10.1016/j.canlet.2012.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Sun C.K., Ng K.T., Lim Z.X. Proline-rich tyrosine kinase 2 (Pyk2) promotes cell motility of hepatocellular carcinoma through induction of epithelial to mesenchymal transition. PLoS One. 2011;6 doi: 10.1371/journal.pone.0018878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Jiang Y., Xie X., Li Z. Functional cooperation of RKTG with p53 in tumorigenesis and epithelial-mesenchymal transition. Cancer Res. 2011;71:2959–2968. doi: 10.1158/0008-5472.CAN-10-4077. [DOI] [PubMed] [Google Scholar]
- 121.Yang S.Y., Lee J., Park C.G. Expression of autotaxin (NPP-2) is closely linked to invasiveness of breast cancer cells. Clin Exp Metastasis. 2002;19:603–608. doi: 10.1023/a:1020950420196. [DOI] [PubMed] [Google Scholar]
- 122.Bissell M.J., Radisky D. Putting tumours in context. Nat Rev Cancer. 2001;1:46–54. doi: 10.1038/35094059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Laurent T.C., Fraser J.R. Hyaluronan. FASEB J: Off Publ Fed Am Soc Exp Biol. 1992;6:2397–2404. [PubMed] [Google Scholar]
- 124.Toole B.P. Hyaluronan: from extracellular glue to pericellular cue. Nat Rev Cancer. 2004;4:528–539. doi: 10.1038/nrc1391. [DOI] [PubMed] [Google Scholar]
- 125.Heldin P., Basu K., Kozlova I., Porsch H. HAS2 and CD44 in breast tumorigenesis. Adv Cancer Res. 2014;123:211–229. doi: 10.1016/B978-0-12-800092-2.00008-3. [DOI] [PubMed] [Google Scholar]
- 126.Brown R.L., Reinke L.M., Damerow M.S. CD44 splice isoform switching in human and mouse epithelium is essential for epithelial-mesenchymal transition and breast cancer progression. J Clin Investig. 2011;121:1064–1074. doi: 10.1172/JCI44540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Marotta L.L., Almendro V., Marusyk A. The JAK2/STAT3 signaling pathway is required for growth of CD44CD24 stem cell-like breast cancer cells in human tumors. J Clin Investig. 2011;121:2723–2735. doi: 10.1172/JCI44745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Camenisch T.D., Schroeder J.A., Bradley J., Klewer S.E., McDonald J.A. Heart-valve mesenchyme formation is dependent on hyaluronan-augmented activation of ErbB2-ErbB3 receptors. Nat Med. 2002;8:850–855. doi: 10.1038/nm742. [DOI] [PubMed] [Google Scholar]
- 129.Camenisch T.D., Spicer A.P., Brehm-Gibson T. Disruption of hyaluronan synthase-2 abrogates normal cardiac morphogenesis and hyaluronan-mediated transformation of epithelium to mesenchyme. J Clin Investig. 2000;106:349–360. doi: 10.1172/JCI10272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Porsch H., Bernert B., Mehic M., Theocharis A.D., Heldin C.H., Heldin P. Efficient TGFbeta-induced epithelial-mesenchymal transition depends on hyaluronan synthase HAS2. Oncogene. 2013;32:4355–4365. doi: 10.1038/onc.2012.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Okuda H., Kobayashi A., Xia B. Hyaluronan synthase HAS2 promotes tumor progression in bone by stimulating the interaction of breast cancer stem-like cells with macrophages and stromal cells. Cancer Res. 2012;72:537–547. doi: 10.1158/0008-5472.CAN-11-1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Liu S., Cong Y., Wang D. Breast cancer stem cells transition between epithelial and mesenchymal states reflective of their normal counterparts. Stem cell Rep. 2014;2:78–91. doi: 10.1016/j.stemcr.2013.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Tan T.Z., Miow Q.H., Miki Y. Epithelial-mesenchymal transition spectrum quantification and its efficacy in deciphering survival and drug responses of cancer patients. EMBO Mol Med. 2014;6:1279–1293. doi: 10.15252/emmm.201404208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Garber M., Yosef N., Goren A. A high-throughput chromatin immunoprecipitation approach reveals principles of dynamic gene regulation in mammals. Mol Cell. 2012;47:810–822. doi: 10.1016/j.molcel.2012.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]