Abstract
As major fuels for the small intestinal mucosa, dietary amino acids (AA) are catabolized in the mitochondria and serve as sources of energy production. The present study was conducted to investigate AA metabolism that supply cell energy and the underlying signaling pathways in porcine enterocytes. Intestinal porcine epithelial cells (IPEC-J2) were treated with different concentrations of AA, inhibitor, or agonist of mammalian target of rapamycin complex 1 (mTORC1) and adenosine monophosphate activated protein kinase (AMPK), and mitochondrial respiration was monitored. The results showed that AA treatments resulted in enhanced mitochondrial respiration, increased intracellular content of pyruvic acid and lactic acid, and increased hormone-sensitive lipase mRNA expression. Meanwhile, decreased citrate synthase, isocitrate dehydrogenase alpha, and carnitine palmitoyltransferase 1 mRNA expression were also observed. We found that AA treatments increased the protein levels of phosphorylated mammalian target of rapamycin (p-mTOR), phosphorylated-p70 ribosomal protein S6 kinase, and phosphorylated-4E-binding protein 1. What is more, the protein levels of phosphorylated AMPK α (p-AMPKα) and nicotinamide adenine dinucleotide (NAD)-dependent protein deacetylase sirtuin-1 (SIRT1) were decreased by AA treatments in a time depending manner. Mitochondrial bioenergetics and the production of tricarboxylic acid cycle intermediates were decreased upon inhibition of mTORC1 or AMPK. Moreover, AMPK activation could up-regulate the mRNA expressions of inhibitor of nuclear factor kappa-B kinase subunit beta (Ikbkβ), integrin-linked protein kinase (ILK), unconventional myosin-Ic (Myo1c), ribosomal protein S6 kinase beta-2 (RPS6Kβ2), and vascular endothelial growth factor (VEGF)-β, which are downstream effectors of mammalian target of rapamycin (mTOR). The mRNA expressions of phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform (PIK3CD) and 5′-AMP-activated protein kinase subunit gamma-1 (PRKAG1), which are upstream regulators of mTOR, were also up-regulated by AMPK activation. On the other hand, AMPK activation also down-regulated FK506-binding protein 1A (FKBP1A), serine/threonine-protein phosphatase 2A 55 kDa regulatory subunit B beta isoform, phosphatase and tensin homolog (PTEN), and unc-51 like autophagy activating kinase 1 (Ulk1), which are up-stream regulators of mTORC1. Taken together, these data indicated that AA regulated cellular energy metabolism through mTOR and AMPK pathway in porcine enterocytes. These results demonstrated interactions of AMPK and mTORC1 pathways in AA catabolism and energy metabolism in intestinal mucosa cells of piglets, and also provided reference for using AA to remedy human intestinal diseases.
Keywords: Amino acids, Mammalian target of rapamycin, Adenosine monophosphate activated protein kinase, Mitochondrial respiration, Energy utilization
1. Introduction
Diets changing from sow milk that is highly digestible and high in protein, fat and lactose to a dry and less digestible starch-based diet cause significantly reduced energy intake for maintenance of epithelial structure in weaning piglets. Enterocytes act as an energy flow sensor, which account for 90% of the mucosal epithelial cell population in the small intestine. Dietary amino acids (AA) are major fuels for the small intestinal mucosa, such as glutamine, glutamate, and aspartate which are the major energy substrates for the small intestine. Mitochondria are energy-producing organelles that produce adenosine triphosphate for cell metabolism, and mitochondrial dysfunctions play relevant roles in the pathophysiology of metabolic diseases, including insulin resistance syndrome, obesity, diabetes mellitus, and cardiovascular diseases (Nisoli et al., 2008, Johnson et al., 2014). Mitochondria utilize nutrients, including AA, for cell energy production and modulate AA homeostasis according to the particular requirements and resources for a cell (Johnson et al., 2014). There is increasing evidence showing the functional role of AA in mitochondrial biogenesis. Branched-chain AA-enriched mixture (BCAAem) promoted mitochondrial biogenesis and increased survival rate of middle-aged mice (D'Antona et al., 2010). Similarly, leucine significantly increased mitochondrial content, mitochondrial biogenesis-related genes expression through nicotinamide adenine dinucleotide (NAD)-dependent protein deacetylase sirtuin-1 (SIRT1)-adenosine monophosphate activated protein kinase (AMPK) signaling in C2C12 myotubes (Liang et al., 2014). Our previous study also found that arginine improved DNA synthesis and mitochondrial bioenergetics of LPS-treated porcine enterocytes (Tan et al., 2015). However, the underlying signaling pathway of AA-regulated cell energy metabolism in enterocytes has not been well characterized.
Inhibition of mammalian target of rapamycin (mTOR) could decrease the mRNA expressions of genes involved in mitochondrial oxidative phosphorylation by disrupting the protein complex containing target of rapamycin complex 1 (TORC1), yin-yang 1 (YY1), and peroxisome proliferator-activated receptor coactivator-1α (PGC-1α), thereby preventing the coactivation of YY1 by PGC-1α (Cunningham et al., 2007). Mammalian target of rapamycin was also reported to regulate the transcription of nuclear encoded mitochondrial genes (Chaveroux et al., 2013). Furthermore, mTOR is also believed to regulate glycolytic and respiratory metabolism, and govern the transcription of estrogen-related receptor α (ERRα)-target genes involved in citric acid cycle and lipogenesis (Chaveroux et al., 2013, Ramanathan and Schreiber, 2009). However, the role of mTOR and AMPK signaling pathway on regulating AA metabolism for cell energy supply in the enterocyte has not been well characterized. Therefore, the present study was designed to test the effects of AA on cell energy metabolism, as well as signaling pathways including mTOR and AMPK using IPEC-J2 cells model.
2. Materials and methods
2.1. Cell culture and treatment
The IPEC-J2 cells were purchased from Guangzhou Genio Biotechnology Co., Ltd. (Guangzhou, China). The cell culture was referred to our previous study (Xiao et al., 2016). Dulbecco's modified Eagle's medium (high glucose, 25 mmol/L) (DMEM-H), fetal bovine serum (FBS), and antibiotics were procured from Invitrogen (Grand Island, NY, USA). Plastic culture plates were manufactured by Corning Inc. (Corning, NY, USA). Unless indicated, all other chemicals were purchased from Sigma–Aldrich (St. Louis, MO, USA). IPEC-J2 cells were seeded and cultured with DMEM-H medium containing 10% FBS, 5 mmol/L L-glutamine, 100 U/mL penicillin, and 100 μg/mL streptomycin at 37 °C in a 5% CO2 incubator. After an overnight incubation, 1) the cells were starved for 6 h, and then treated by a basal medium (Table 1) without AA or containing 0.5-, 1- or 2-fold AA for 30 min, 2, 4 or 8 h, respectively; 2) the cells were cultured in the basal medium (control), the basal medium + 10 nmol/L rapamycin, the basal medium + 0.5 mmol/L AICAR (an activator for AMPK) or the basal medium + 1 mmol/L compound C for 24 h. Cells were treated and collected for the analysis of extracellular flux, Western blot, gas chromatography-mass spectrometry (GC–MS) and RNA extraction.
Table 1.
The composition of Dulbecco's modified Eagle's medium - high glucose (DMEM-H).
| Composition | Molecular weight | Content, mg/L |
|---|---|---|
| Inorganic salts | ||
| CaCl2 | 111 | 200.00 |
| Fe(NO3)3•9H2O | 404 | 0.10 |
| KCl | 75 | 400.00 |
| MgSO4 | 120 | 97.67 |
| Salt | 58 | 6,400.00 |
| NaH2PO4 | 138 | 125.00 |
| NaHCO3 | 84.0 | 3700.0 |
| Vitamins | ||
| Choline chloride | 140 | 4.00 |
| D-calcium pantothenate | 477 | 4.00 |
| Folic acid | 441 | 4.00 |
| Niacinamide | 122 | 4.00 |
| Riboflavin | 376 | 0.40 |
| Thiamine hydrochloride | 337 | 4.00 |
| i-inositol | 180 | 7.20 |
| Pyridoxine hydrochloride | 206 | 4.00 |
| Others | ||
| D-Glucose | 180 | 4,500.00 |
| Sodium pyruvate | 110 | 110.00 |
| Phenol red | 376 | 15.00 |
| Amino acids1 | ||
| Glycine | 75 | 30.00 |
| L-arginine hydrochloride | 211 | 84.00 |
| L-cystine | 240 | 48.00 |
| L-glutamine | 146 | 584.00 |
| L-histidine hydrochloride-H2O | 210 | 42.00 |
| L-isoleucine | 131 | 105.00 |
| L-leucine | 131 | 105.00 |
| L-lysine hydrochloride | 183 | 146.00 |
| L-methionine | 149 | 30.00 |
| L-phenylalanine | 165 | 66.00 |
| L-serine | 105 | 42.00 |
| L-threonine | 119 | 95.00 |
| L-tryptophan | 204 | 16.00 |
| L-tyrosine disodium salt hydrate | 225 | 90.00 |
| L-valine | 117 | 94.00 |
The content of amino acids is for 1AA group (1-fold AA medium).
2.2. Real-time PCR
IPEC-J2 cells (3 × 104 cells per well) were seeded in a 6-well plate. After the test, the cells were collected by Trizol. The protocol of total RNA extraction, quantification, cDNA synthesis and real-time PCR was adapted from the method of Li et al. (2015). Briefly, total RNA was isolated from cell samples by using the Trizol method. Real-time PCR of mRNA in this study was carried out using SYBR Green Realtime PCR Master Mix (Takara biotechnology Co., Ltd, Dalian, China) as described by the manufacturer. Forward and reverse primers (Table 2) were used to amplify the target genes. For quantification, the following conditions of PCR were used: incubation for 10 min at 95 °C, followed by 40 cycles of denaturation for 15 s at 95 °C, and annealing and extension for 60 s at 56 to 64 °C. The mRNA expressions of target genes were calculated by the value of the threshold cycle (Ct) of the real-time PCR as related to that of β-actin using the method (Duan et al., 2017), in which
Table 2.
Forward (F) and reverse (R) primers of relative kinases involved in energy metabolism.
| Genes | Primer sequence (5′ to 3′) |
|---|---|
| β-actin | F: GGACCTGACCGACTACCTCA |
| R: CACAGCTTCTCCTTGATGTCC | |
| Hexokinase (Hexok) | F: CTCATCACAACCGTTACCA |
| R: TGTCATTAGTGTCCTCATCC | |
| Phosphoenol pyruvate kinase 1 (PCK-1) | F: GATGGTTCAGCCTAGGGACA |
| R: GCTACAGCAGAAGGGCAAAA | |
| Pyruvate dehydrogenase (PDH) | F: GCAGACTTACCGTTACCAT |
| R: GATAGCCGAGTTCTTCCAA | |
| Citrate synthase (CS) | F: TCTCAGCTCAGTGCAGCCATTACA |
| R: CTGCAACACAAGGTAGCTTTGCGA | |
| Isocitrate dehydrogenase alpha (ICDHα) | F: TGTGGTTCCTGGTGAGAG |
| R: CGAGATTGAGATGCCGTAG | |
| Isocitrate dehydrogenase beta (ICDHβ) | F: GGTGGAGAGCCTCAAGAT |
| R: TGGTGGTGTTGTCTACGA | |
| Phosphorylated acetyl-CoA carboxylase (p-ACC) | F: ATGTTTCGGCAGTCCCTGAT |
| R: TGTGGACCAGCTGACCTTGA | |
| Fatty acid synthase (FAS) | F: CTGCTGAAGCCTAACTCCTCG |
| R: TTGCTCCTTGGAACCGTCTG | |
| Carnitine palmitoyltransferase 1 (CPT-1) | F: TATCTGTGTCCGCCTTCTGTC |
| R: GGCTGTATTCCTCGTCATCC | |
| Hormone sensitive lipase (HSL) | F: AGCTCCTTCTTCGAGGGTGAC |
| R: AGGCCTGTTTCATTGCGTTT |
2.3. Western blotting analysis
IPEC-J2 cells (3 × 104 cells per well) were seeded in a 6-well plate for Western blotting analysis. Cell samples were collected as described by Tan et al. (2015). Protein concentrations of cell homogenates were measured by using the bicinchoninic acid (BCA) method and bovine serum albumin as standard (Beyotime Institute of Biotechnology, Shanghai, China). All samples were adjusted to an equal concentration, and the loading amount of each sample was 50 μg protein. The supernatant fluid (containing tissue proteins) was then diluted with 4 × sodium dodecyl sulfate (SDS) sample buffer (Beyotime Institute of Biotechnology, Shanghai, China) and heated in boiling water for 5 min. The Western blotting was conducted based on previous description (Xiao et al., 2013). Briefly, aliquots of samples were loaded onto SDS-polyacrylamide gels (Invitrogen, Carlsbad, CA, USA). After separation on 4% to 12% gels, proteins were transferred to a polyvinylidene difluoride (PVDF) membrane (Millipore, Bedford, MA, USA) under 200 mA for 1.5 to 2.5 h based on the molecular weight of proteins using an apparatus (Bio-Rad Transblot; Bio-Rad, Hercules, CA, USA). The membranes were blocked in 5% BCA in Tris-Tween buffered saline (TTBS; Applygen, Beijing, China) for 1.5 to 2 h and then incubated with these primary antibodies 2 h or overnight at 4 °C with gentle rocking. The primary antibodies (1:1,000) for mTOR, phosphorylated mammalian target of rapamycin (p-mTOR)-Ser2448, SIRT1, phosphorylated-p70 ribosomal protein S6 kinase (p-P70S6K)-Thr389, phosphorylated acetyl-CoA carboxylase (p-ACC)-Ser79, phosphorylated AMPK α (p-AMPKα), phosphorylated-4E-binding protein 1 (p-4EBP1)-Thr37/46 were purchased from Cell Signaling Technology (Danvers, MA, USA), and that (1:400) for β-actin was purchased from Santa Cruz Biotechnology (Dallas, TX, USA). After being washed 4 times with TTBS, the membranes were incubated at room temperature for 2 h with secondary antibodies at 1:2,000 (horseradish peroxidase-conjugated goat anti-rabbit IgG or peroxidase-labeled goat anti-mouse IgG, Santa Cruz Biotechnology, Dallas, TX, USA). Finally, the membranes were washed 4 times with TTBS and then developed using a substrate (Supersignal West Dura Extended Duration Substrate; Pierce, Rockford, IL, USA). The images were detected by chemiluminescence (Millipore, Billerica, MA, USA). Western blots were quantified by measuring the intensity of correctly sized bands using software (AlphaImager 2200 Software; Alpha Innotech Corporation, San Leandro, CA, USA). All protein measurements were normalized to β-actin.
2.4. Extracellular flux assays
IPEC-J2 cells (1 × 104 cells per well) were seeded in a 24-well plate for extracellular flux assays. The XF-24 Extracellular Flux Analyzer and Cell Mito Stress Test Kit from Seahorse Biosciences was used to examine the effects of addition of different concentrations of AA, inhibition of mTOR and activation of AMPK pathway on mitochondrial respiration in IPEC-J2 cells as described by Tan et al. (2015) and Xiao et al. (2016). Briefly, the base medium was changed prior to the bioenergetic measurements to serum-free unbuffered (without sodium bicarbonate) DMEM-H medium base supplemented with 2 mmol/L L-glutamine, 25 mmol/L D-glucose and 1 mmol/L sodium pyruvate, at pH 7.4 ± 0.1 at 37 °C. To measure indices of mitochondrial function, oligomycin (an inhibitor of the ATP synthesis), carbonyl cyanide-p-trifluoromethoxyphenylhydrazone (FCCP) (an inhibitor of the ATP synthesis), as well as rotenone and antimycin A (an inhibitor of the electron transport chain) were injected sequentially at the final concentrations of 0.5, 4, and 1 μmol/L, respectively. This allowed for an estimation of the contribution of non-ATP–linked oxygen consumption (proton leak) and ATP–linked mitochondrial oxygen consumption (ATP production). The maximal respiration capacity was determined using the FCCP-stimulated rate. The spare respiratory capacity was represented by the maximal respiratory capacity subtracted from the baseline oxygen consumption rate (OCR). The residual oxygen consumption that occurred after addition of rotenone and antimycin A was ascribed to non-mitochondrial respiration and was subtracted from all measured values in the analysis. Owing to the effects of AA, rapamycin, AICAR and compound C on IPEC-J2 cell proliferation, total cellular protein was determined and used to normalize mitochondrial respiration rates.
2.5. Detection of tricarboxylic acid (TCA) cycle intermediates by gas chromatography-mass spectrometer
IPEC-J2 cells (5 × 105 cells per dish) were seeded in 10 cm dishes for GC–MS analysis as described by our previous study (Xiao et al., 2016). Briefly, cells were washed with phosphate buffer saline (PBS) and treated by 0.25% trypsin. And then, cells were collected and pelleted at 1,000 × g for 5 min. After being quenched using 500 L of prechilled 50% (vol/vol) methanol, cells were centrifuged at 1,000 × g for 5 min and then removed and added 500 L of prechilled 100% (vol/vol) methanol. Cells were measured by an Agilent 7890B-5977A GC–MS equipped with HP-5 ms (30 m × 250 m × 0.25 m) capillary column (Agilent J&W, Santa Clara, CA, USA). All metabolites were previously validated using authentic standards (Sigma, St. Louis, MO, USA). The data are expressed relative to the control cells.
2.6. PCR array test
IPEC-J2 cells (3 × 104 cells per well) were seeded in a 6-well plate. Total RNA was extracted and cDNA was synthesized following manufacturer's instructions for RT2 First Strand Assay Kit (QIAGEN, Germany). The protocol for real time-PCR was performed following manufacturer's instructions for RT2 SYBR Green MasterMix (QIAGEN, Germany) and RT2 Profiler PCR Array (QIAGEN, Germany). Real-time PCR was performed by using Bio-Rad Real-Time PCR (CFX96). Data analysis was performed by using RT2 Profiler PCR Array Data Analysis (QIAGEN, Germany).
2.7. Statistical analysis
Results are expressed as means ± SD. All statistical analyses were performed using SPSS software (SPSS Inc., Chicago, IL, USA). The differences among treatments were evaluated using Tukey's test. Probability values < 0.05 were considered statistically significant.
3. Results
3.1. Amino acids regulate mitochondrial bioenergetics and energy metabolism in a dose depending manner
The cell viability after AA treatments is illustrated in Fig. 1. Amino acid treatments significantly increased the cell proliferation of IPEC-J2 cells compared with 0 AA group. To detect the effects of different concentrations of AA on mitochondrial bioenergetics, OCR was measured in IPEC-J2 cells for 4 h (Fig. 1B and C). The individual parameters for basal respiration, proton leak, maximal respiration, and spare respiratory capacity were gradually increased by AA treatments (P < 0.05), and no effect on non-mitochondrial oxygen consumption was found. The individual parameter for proton leak in the 2 AA group was significantly increased (P < 0.05) compared with the 0.5 AA group. As shown in Fig. 1D, the content of pyruvic acid in 0.5 AA and 1 AA groups was significantly higher than that in 0 AA group (P < 0.05), and significantly lower than that in 2 AA group (P < 0.05), but the content of lactic acid in 0.5 AA and 1 AA groups was significantly lower than that in 0 AA and 2 AA groups. Increasing concentrations of AA decreased (P < 0.05) the content of citric acid. The treatment of 1 AA increased (P < 0.05) the content of malic acid, however, 0.5 AA group decreased (P < 0.05) the content of malic acid compared with the other groups. There were no differences in the content of succinic acid and fumaric acid among the 4 groups (Fig. 1D).
Fig. 1.
The concentrations of amino acids (AA) affected energy metabolism. (A) cell viability of AA treatment; (B) oxygen consumption rate (OCR) assessed by extracellular flux analysis; (C) basal respiration, proton leak, maximal respiration, and ATP production; (D) the content of pyruvic acid, lactic acid, and the tricarboxylic acid (TCA) cycle intermediates (succinic acid, alpha-ketoglutarate, fumaric acid, malic acid, and citric acid), and the data are expressed relative to the control cells; (E) the mRNA expressions of relative kinases of glycolysis; (F) the mRNA expressions of relative kinases of fatty acid metabolism. Cells were cultured for 4 h in AA-free Dulbecco's modified Eagle's medium (high glucose) (DMEM-H) containing 0-, 0.5-, 1-, and 2-fold AA. Data are expressed as means ± SD of at least 3 independent experiments (n = 5 for extracellular flux analysis, n = 6 in other studies). Hexok = hexokinase; PDH = pyruvate dehydrogenase; PCK-1 = phosphoenol pyruvate kinase 1; CS = citrate synthase; ICDHα = isocitrate dehydrogenase alpha; ICDHβ = isocitrate dehydrogenase beta; ACC = acetyl-CoA carboxylase; FAS = fatty acid synthase; CPT-1 = carnitine palmitoyltransferase 1; HSL = hormone sensitive lipase. a–d Value columns with different letters are significantly different (P < 0.05).
Based on the knowledge of energy metabolism, we next determined the effect of AA on the mRNA expressions of relative kinases of glycolysis and fatty acid metabolism. As shown in Fig. 1E and F, increasing concentrations of AA decreased (P < 0.05) mRNA expressions of citrate synthase (CS), isocitrate dehydrogenase alpha (ICDHα) and carnitine palmitoyltransferase 1 (CPT-1), and increased (P < 0.05) the mRNA expression of hormone sensitive lipase (HSL). Moreover, 1 AA treatment increased (P < 0.05) the mRNA expressions of hexokinase (Hexok) and phosphoenol pyruvate kinase 1 (PCK-1) compared with the other groups, where as 0.5 AA group significantly decreased (P < 0.05) the mRNA expressions of Hexok and pyruvate dehydrogenase (PDH) compared with 2 AA group. There was no significant difference in other measured genes among the 4 groups.
3.2. Amino acids affected energy metabolism via mTOR and AMPK pathways
The 2 pathways, mTOR and AMPK pathways, have been reported to regulate energy metabolism, and AA would activate mTOR pathway to regulate cell metabolism. Therefore, we further determined the effects of different concentrations of AA on the 2 pathways. As shown in Fig. 2, AA treatments increased (P < 0.05) the protein levels of p-mTOR, p-P70S6K and p-4EBP1, compared with the 0 AA group, except for p-mTOR in 0.5 AA group at 4 h. The protein levels of p-AMPKα were gradually increased (P < 0.05) with increasing concentrations of AA in the medium after 30 min and 2 h, but were reduced (P < 0.05) in 2 AA group after 4 h and in all AA groups after 8 h compared with 0 AA group. Moreover, 2 AA treatment decreased (P < 0.05) the protein level of SIRT1 after 2, 4 and 8 h, and 0.5 AA increased the protein level of SIRT1 after 2 h compared with 0 AA group. Amino acid treatments increased the protein levels of p-ACC after 30 min, 4 h, and 8 h, except for the data of 0.5 AA after 4 h. The treatment of 1 AA increased (P < 0.05) the protein level of p-ACC after 2 h compared with 0 AA group. Collectively, these results indicate that mTOR and AMPK pathways are involved in the regulation of cell metabolism of AA.
Fig. 2.
Amino acids (AA) affected energy metabolism through mammalian target of rapamycin complex 1 (mTORC1) and adenosine monophosphate activated protein kinase (AMPK) pathways. (A) representative Western blots; (B) to (G) relative protein levels of mTOR and AMPK pathways. Cells were cultured for 30 min, 2, 4 and 8 h in AA-free Dulbecco's modified Eagle's medium (high glucose) (DMEM-H) containing 0-, 0.5-, 1-, and 2-fold AA. Data are expressed as means ± SD of at least 3 independent experiments (n = 4). p-p70S6K = phosphorylated-p70 ribosomal protein S6 kinase; p-ACC = phosphorylated acetyl-CoA carboxylase; p-AMPKα = phosphorylated adenosine monophosphate activated protein kinase α; p-mTOR = phosphorylated mammalian target of rapamycin; SIRT1 = nicotinamide adenine dinucleotide (NAD)-dependent protein deacetylase sirtuin-1; p-4EBP1 = phosphorylated-4E-binding protein 1. a–d Value columns with different letters are significantly different (P < 0.05).
3.3. Inhibition of mTOR and AMPK suppressed cell respiratory metabolism
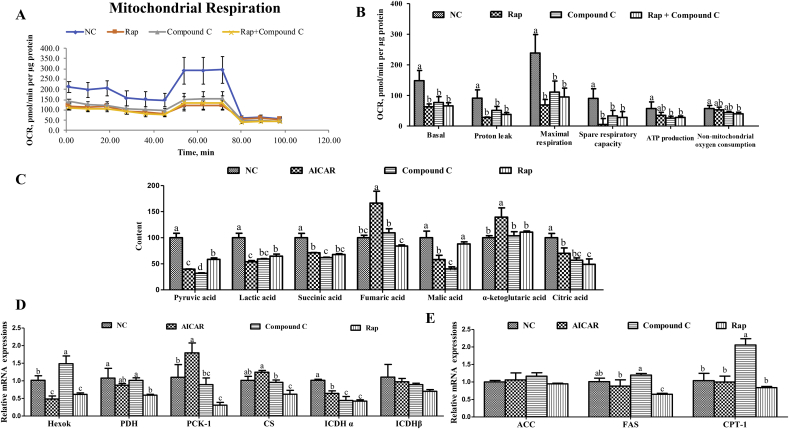
To assess the effects of mTOR and AMPK on cell respiratory metabolism, mitochondrial respiration, relative content of pyruvic acid, lactic acid and TCA cycle intermediates and mRNA expressions of relative kinases of glycolysis and fatty acid metabolism are illustrated in Fig. 3. Compared with the control group, addition of rapamycin decreased (P < 0.05) individual parameters for basal respiration, proton leak, maximal respiration, and spare respiratory capacity, and addition of compound C decreased the result of basal respiration, proton leak, maximal respiration, spare respiratory capacity, and ATP production (Fig. 3A and B). Addition of both rapamycin and compound C decreased the individual parameter of basal respiration, proton leak, maximal respiration, spare respiratory capacity, ATP production, and non-mitochondrial oxygen consumption (Fig. 3A and B). The content of pyruvic acid, lactic acid, citric acid, and succinic acid were reduced (P < 0.05) by addition of rapamycin, AICAR and compound C and AMPK activation increased the content of apha-ketoglutarate and fumaric acid, and AICAR and compound C groups reduced the content of malic acid, compared with control group (Fig. 3C). According to the results of Fig. 3D and E, inhibition of mTOR significantly decreased (P < 0.05) the mRNA expressions of Hexok, PDH, PCK-1, CS, ICDHα, and fatty acid synthase (FAS), and inhibition of AMPK decreased (P < 0.05) the mRNA expression of ICDHα, and increased (P < 0.05) those of Hexok and CPT-1, compared with control group. Moreover, activation of AMPK promoted (P < 0.05) the mRNA expression of PCK-1 and decreased (P < 0.05) those of Hexok and ICDHα, compared with control group.
Fig. 3.
Inhibition of mammalian target of rapamycin complex 1 (mTORC1) and adenosine monophosphate-activated protein kinase (AMPK) suppressed cell respiratory metabolism. (A) oxygen consumption rate (OCR) assessed by extracellular flux analysis; (B) basal respiration, proton leak, maximal respiration, and ATP production; (C) the content of pyruvic acid, lactic acid, and the TCA cycle intermediates (succinic acid, alpha-ketoglutarate, fumaric acid, malic acid, and citric acid) were determined; (D) the mRNA expressions of relative kinases of glycolysis; (E) the mRNA expressions of relative kinases of fatty acid metabolism. Cells were cultured for 24 h in a basal medium (control), the basal medium + 10 nmol/L rapamycin (an inhibitor of mTORC1) (Rap), the basal medium + 1 mmol/L compound C (Compound C), and the basal medium + 0.5 mmol/L AICAR (an agonist of AMPK). Data are expressed as means ± SD of at least 3 independent experiments (n = 5 for extracellular flux analysis, n = 6 in other study). Hexok = hexokinase; PDH = pyruvate dehydrogenase; PCK-1 = phosphoenol pyruvate kinase 1; CS = citrate synthase; ICDHα = isocitrate dehydrogenase alpha; ICDHβ = isocitrate dehydrogenase beta; ACC = acetyl-CoA carboxylase; FAS = fatty acid synthase; CPT-1 = carnitine palmitoyltransferase 1. a–d Value columns with different letters are significantly different (P < 0.05).
3.4. The relationship of AMPK and mTOR in porcine enterocytes
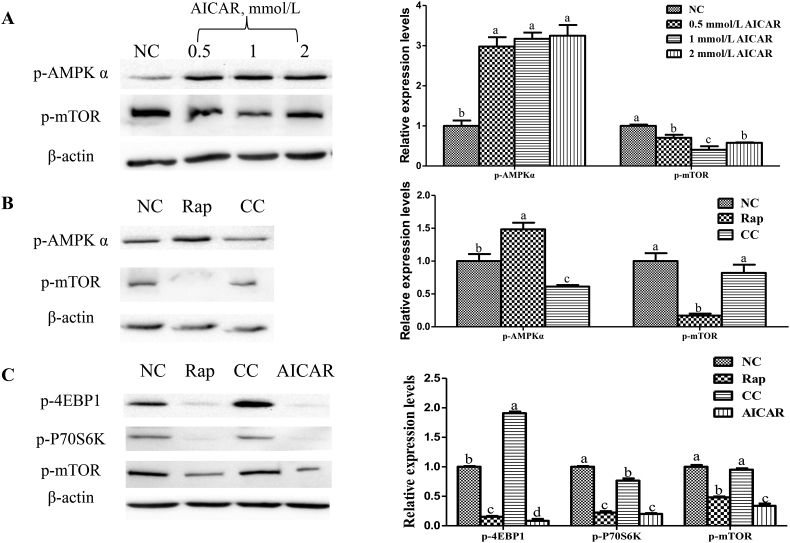
Relative protein levels for p-AMPKα and p-mTOR are illustrated in Fig. 4. Addition of different dosages of AICAR significantly increased (P < 0.05) the protein level for p-AMPKα, and decreased (P < 0.05) the protein level for p-mTOR. Addition of rapamycin decreased (P < 0.05) the protein levels for p-4EBP1, p-p70S6K and p-mTOR, and increased (P < 0.05) the protein level of p-AMPKα compared with the control group. Inhibition of AMPK significantly decreased the protein levels of p-AMPKα and p-p70S6K, and increased (P < 0.05) the protein level of p-4EBP1, but has no effects on the protein level for p-mTOR. AICAR decreased the protein levels of p-4EBP1, p-p70S6K and p-mTOR compared with control group.
Fig. 4.
Interaction of mammalian target of rapamycin complex 1 (mTORC1) and adenosine monophosphate activated protein kinase (AMPK) pathways. (A) the effect of AMPK activation on phosphorylated mammalian target of rapamycin (p-mTOR), and phosphorylated AMPK α (p-AMPKα); (B) the effect of mTOR and AMPK inhibition on p-mTOR and p-AMPKα; (C) the effect of mTOR and AMPK inhibition on mTOR pathway. Cells were cultured for 24 h in a basal medium (control), the basal medium + 10 nmol/L Rapamycin (Rap), the basal medium + 1 mmol/L compound C (Compound C), and the basal medium + 0.5 mmol/L AICAR (an agonist of AMPK). Data are expressed as means ± SD of at least 3 independent experiments (n = 4). p-4EBP1 = phosphorylated-4E-binding protein 1; p-p70S6K = phosphorylated-p70 ribosomal protein S6 kinase. a–c Value columns with different letters are significantly different (P < 0.05).
According to the results of RT2 Profiler PCR Array (Fig. 5 and Table 3), activated AMPK up-regulated (P < 0.05) 8 genes (RAC-beta serine/threonine-protein kinase [Akt2], inhibitor of nuclear factor kappa-B kinase subunit beta [Ikbkβ], integrin-linked protein kinase [ILK], unconventional myosin-Ic [Myo1c], vascular endothelial growth factor [VEGF]-β, phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform [PIK3CD], 5′-AMP-activated protein kinase subunit gamma-1 [PRKAG1], ribosomal protein S6 kinase beta-2 [RPS6Kβ2]) and down-regulated (P < 0.05) 9 genes (heat shock 70 kDa protein 4 [HSPA4], GTPase KRas, FK506-binding protein 1A [FKBP1A], GTPase NRas, serine/threonine-protein phosphatase 2A 55 kDa regulatory subunit B beta isoform [PPP2R2-β], phosphatase and tensin homolog [PTEN], ribosomal protein S6 kinase alpha-2 [RPS6Kα2], telomere length regulation protein TEL2 homolog [TELO2], unc-51 like autophagy activating kinase 1 [Ulk1]) for mTOR pathway.
Fig. 5.
Effects of AMPK activation on mTOR pathway. (A) heatmap of genes from mTOR pathway after AICAR treatment (Green to red gradation in color represent lower to higher gene expression); (B). scatter diagram from mTOR pathway after AICAR treatment; (C) a diagram for possible interaction pathways between mTOR and AMPK pathways. Cells were cultured for 24 h in a basal medium (control), and the basal medium + 0.5 mmol/L AICAR (an agonist of AMPK). Data are expressed as means ± SD of at least 3 independent experiments (n = 6). AICAR = an AMPK activator; AMPK = adenosine monophosphate activated protein kinase; PI3K = phosphatidylinositol 4,5-bisphosphate 3-kinase; PTEN = phosphatase and tensin homolog; Ulk = unc-51 like autophagy activating kinase; Akt = RAC-beta serine/threonine-protein kinase; MAPK = p53 and K-Ras-mitogen-activated protein kinase; mTOR = mammalian target of rapamycin.
Table 3.
The change of gene expressions of mTOR signaling pathway in enterocytes.
| Genes | Fold regulation | P-value |
|---|---|---|
| RAC-beta serine/threonine-protein kinase (Akt2) | 1.9033 | 0.002695 |
| Inhibitor of nuclear factor kappa-B kinase subunit beta (Ikbkβ) | 1.8781 | 0.002409 |
| Integrin-linked protein kinase (ILK) | 4.3749 | 0.001899 |
| Unconventional myosin-Ic (Myo1c) | 1.7143 | 0.001645 |
| Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform (PIK3CD) | 1.5053 | 0.034384 |
| 5′-AMP-activated protein kinase subunit gamma-1 (PRKAG1) | 2.0126 | 0.013545 |
| Ribosomal protein S6 kinase beta-2 (RPS6Kβ2) | 1.5272 | 0.003714 |
| Vascular endothelial growth factor beta (VEGF-β) | 2.0859 | 0.0436 |
| Heat shock 70 kDa protein 4 (HSPA4) | −1.6393 | 0.00758 |
| GTPase KRas | −1.631 | 0.005048 |
| FK506-binding protein 1A (FKBP1A) | −1.9455 | 0.004166 |
| GTPase NRas | −1.9945 | 0.008077 |
| Serine/threonine-protein phosphatase 2A 55 kDa regulatory subunit B beta isoform (PPP2R2-β) | −1.5542 | 0.009749 |
| Phosphatase and tensin homolog (PTEN) | −1.5962 | 0.017123 |
| Ribosomal protein S6 kinase alpha-2 (RPS6Kα2) | −2.0988 | 0.002204 |
| Telomere length regulation protein TEL2 homolog (TELO2) | −1.833 | 0.031739 |
| Unc-51 like autophagy activating kinase 1 (Ulk1) | −4.0356 | 0.019955 |
4. Discussion
In the present study, we demonstrated that the concentrations of total AA affected mitochondrial bioenergetics and energy metabolism. This is in line with the previous studies that different concentrations of glutamine, leucine, BCAAem and arginine affected mitochondrial functions in vitro and in vivo (Wu et al., 1994, Liang et al., 2014, Tan et al., 2015, D'Antona et al., 2010, Wu et al., 1995). Amino acids, such as glutamine, glutamic acid and aspartic acid, are utilized by the enterocytes of the small intestine as major energy substrates and could contribute more ATP to enterocytes of pig (Wu et al., 1994, Wu et al., 1995, Rezaei et al., 2013). Moreover, proteolysis regulated AA balance, and autophagy would supply AA to replenish TCA intermediates during AA starvation (Tan et al., 2017). Normally, addition of AA increased the contents of pyruvic acid, malic acid, a-ketoglutaric acid, and the expressions of PCK-1 and HSL. Amino acid supplementation decreased the expressions of CS and ICDH, therefore decreased the content of citric acid. Moreover, AA addition decreased the mRNA expression of CPT-1, which indicates AA affect fatty acid oxidation, which is in line with the previous study (Cheng et al., 2010), but it requires further investigation.
Mitochondrial biogenesis is regulated by a series of transcriptional and signaling networks: 1) transcriptional programs that coordinate induction of both mitochondrial- and nuclear-localized genes that encode mitochondrial proteins, such as the transcriptional coactivator peroxisome proliferator-activated receptor gamma coactivator-1 alpha (PGC-1α) (Tan et al., 2016); 2) key mechanisms of signaling pathways, c-Myc, phosphatidylinositol 4,5-bisphosphate 3-kinase (PI3K)/Akt/mTOR, p53 and K-Ras-mitogen-activated protein kinase (MAPK) signaling pathways (Morita et al., 2015, Li et al., 2005, Pylayeva-Gupta et al., 2011, Vyas et al., 2016). Amino acid availability is a major determinant of growth and is sensed through central signaling pathways, and one central nutrient sensing pathway is mTOR pathway (Bar-Peled and Sabatini, 2014). Low nutrient conditions such as lack of AA result in a high AMP-to-ATP ratio that activates AMPK, which opposes the mTOR pathway (Vyas et al., 2016). Recent studies have found that mTOR played an important role in regulating mitochondrial energy metabolism. Mammalian target of rapamycin regulated RNA translation mitochondrial activity and biosynthesis by modulating activity of 4EBP1 (Morita et al., 2013). Inhibition of mTOR by rapamycin reduced the expressions of genes related to mitochondrial oxidation by damaging mTORC1, YY1 and PGC-1 complex, and then inhibited the interaction between YY1 and PGC-1 (Ramanathan and Schreiber, 2009). Therefore, in the present study, addition of different concentrations of AA for different times could influence activities of mTOR and AMPK pathways. Increasing concentrations of AA almost activated mTOR pathway in all groups, and activated AMPK pathway at 30 min and 2 h, and inhibited AMPK pathway at 4 and 8 h, compared with the control. It indicated that cells need time to feedback regulation on AA supplementation.
In the present study, the respiratory metabolism of IPEC-J2 cells could be significantly inhibited after the mTOR and AMPK pathways were inhibited, indicating that the energy balance in cells required both mTOR and AMPK pathways. Schieke et al. (2006) found that mTORC1 activity was positively correlated with mitochondrial membrane potential, maximum oxidation capacity, and intracellular ATP content. Ramanathan and Schreiber (2009) found that inhibition of mTOR by rapamycin decreased oxygen consumption and mitochondrial capacity, and inducted oxidative phosphorylation of glycolysis (Ramanathan and Schreiber, 2009), which is consistent with our findings. The researchers found that the lysosomal v-ATPase-ragulator complex is a common activator for AMPK and mTORC1 in energy metabolism acting as a switch between catabolism and anabolism. During glucose starvation, the v-ATPase-ragulator complex is accessible to AXIN/liver kinase B1 (LKB1) for AMPK activation. Concurrently, the guanine nucleotide exchange factor activity of ragulators toward RAG is inhibited by AXIN, causing dissociation from endosome and inactivation of mTORC1 (Zhang et al., 2014). When the intracellular energy level is high, mTORC1 is activated, opening the synthetic metabolic pathway (Zoncu et al., 2011, Zhang et al., 2014). Supplication of cellular energy regulates mTOR and AMPK pathways, and meantime energy metabolism requires a combination of these 2 pathways.
In this experiment, the effect of activated AMPK pathway for respiratory metabolism is not very clear, it might involve the feedback regulation of other pathways. Therefore, we used PCR array to detect mRNA expressions of genes related to mTOR pathways by activating AMPK pathway. The results showed that the PI3K-Akt pathway is mainly involved in up-regulated genes, and the suppressor gene PTEN is present in the down-regulated genes, indicating that activation of AMPK pathway increased the PI3K-Akt pathway, and inhibited the mTOR-S6K pathway. However, both KRAS and NRAS in the down-regulated genes were decreased, indicating that the MAPK pathway might be inhibited, then with reducing the transcription of related protein genes, leading to the reduction of the energy consumption. The interaction between AMPK activation and mTOR inhibition activated Ulk1 (Hardie, 2011, Moscat and Diaz-Meco, 2011, Kim et al., 2011). However, the recent study showed that the expression of Ulk1 was decreased, probably because AMPK was activated for a long time and cell energy consumption was serious, so it could activate cell apoptosis instead of autophagy.
5. Conclusion
In summary, AA regulate energy metabolism through mTOR and AMPK pathways, and mTOR negatively interacts with AMPK to regulate energy balance in porcine enterocytes. The balance of cell energy requires the interaction of AMPK and mTOR, and AMPK mainly regulates the expression of related upstream genes of the mTOR pathway in porcine enterocytes. These results elucidate that interactive mechanism of AMPK and mTORC1 on energy utilization by AA in the intestinal mucosa cells of piglets, and also provide reference for using AA to remedy human intestinal diseases.
Conflict of Interest
We declare that we have no financial and personal relationships with other people or organizations that can inappropriately influence our work, there is no professional or other personal interest of any nature or kind in any product, service and/or company that could be construed as influencing the content of this paper.
Acknowledgments
This project was funded by the National Natural Science Foundation of China (31672433, 31560640), China; Special fund for scientific innovation strategy-construction of high level Academy of Agriculture Science -the Outstanding Talents Training Program of Guangdong Academy of Agricultural Sciences (R2018PY-JC001), China; Special fund for scientific innovation strategy-construction of high level Academy Of Agriculture Science -The Talents Training Program Of Guangdong Academy Of Agricultural Sciences-Young Associate Researcher (R2018PY-QF001), China.
Footnotes
Peer review under responsibility of Chinese Association of Animal Science and Veterinary Medicine.
References
- Bar-Peled L., Sabatini D.M. Regulation of mTORC1 by amino acids. Trends Cell Biol. 2014;24:400–406. doi: 10.1016/j.tcb.2014.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaveroux C., Eichner L.J., Dufour C.R., Shatnawi A., Khoutorsky A., Bourque G., Sonenberg N., Giguere V. Molecular and genetic crosstalks between mTOR and ERRalpha are key determinants of rapamycin-induced nonalcoholic fatty liver. Cell Metabol. 2013;17:586–598. doi: 10.1016/j.cmet.2013.03.003. [DOI] [PubMed] [Google Scholar]
- Cheng Y., Meng Q., Wang C., Li H., Huang Z., Chen S., Xiao F., Guo F. Leucine deprivation decreases fat mass by stimulation of lipolysis in white adipose tissue and upregulation of uncoupling protein 1 (UCP1) in brown adipose tissue. Diabetes. 2010;59:17–25. doi: 10.2337/db09-0929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham J.T., Rodgers J.T., Arlow D.H., Vazquez F., Mootha V.K., Puigserver P. mTOR controls mitochondrial oxidative function through a YY1-PGC-1alpha transcriptional complex. Nature. 2007;450:736–740. doi: 10.1038/nature06322. [DOI] [PubMed] [Google Scholar]
- D'Antona G., Ragni M., Cardile A., Tedesco L., Dossena M., Bruttini F., Caliaro F., Corsetti G., Bottinelli R., Carruba M.O., Valerio A., Nisoli E. Branched-chain amino acid supplementation promotes survival and supports cardiac and skeletal muscle mitochondrial biogenesis in middle-aged mice. Cell Metabol. 2010;12:362–372. doi: 10.1016/j.cmet.2010.08.016. [DOI] [PubMed] [Google Scholar]
- Duan Y., Zeng L., Li F., Wang W., Li Y., Guo Q., Ji Y., Tan B., Yin Y. Effect of branched-chain amino acid ratio on the proliferation, differentiation, and expression levels of key regulators involved in protein metabolism of myocytes. Nutrition. 2017;36:8–16. doi: 10.1016/j.nut.2016.10.016. [DOI] [PubMed] [Google Scholar]
- Hardie D.G. AMPK and autophagy get connected. EMBO J. 2011;30:634–635. doi: 10.1038/emboj.2011.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson M.A., Vidoni S., Durigon R., Pearce S.F., Rorbach J., He J., Brea-Calvo G., Minczuk M., Reyes A., Holt I.J., Spinazzola A. Amino acid starvation has opposite effects on mitochondrial and cytosolic protein synthesis. PLoS One. 2014;9 doi: 10.1371/journal.pone.0093597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Kundu M., Viollet B., Guan K.L. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13:132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Wang Y., Zeller K.I., Potter J.J., Wonsey D.R., O'Donnell K.A., Kim J.W., Yustein J.T., Lee L.A., Dang C.V. Myc stimulates nuclearly encoded mitochondrial genes and mitochondrial biogenesis. Mol Cell Biol. 2005;25:6225–6234. doi: 10.1128/MCB.25.14.6225-6234.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G., Li J., Tan B., Wang J., Kong X., Guan G., Li F., Yin Y. Characterization and regulation of the amino acid transporter SNAT2 in the small intestine of piglets. PLoS One. 2015;10 doi: 10.1371/journal.pone.0128207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang C., Curry B.J., Brown P.L., Zemel M.B. Leucine modulates mitochondrial biogenesis and SIRT1-AMPK signaling in C2C12 myotubes. J Nutr Metab. 2014;2014 doi: 10.1155/2014/239750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita M., Gravel S.P., Chenard V., Sikstrom K., Zheng L., Alain T., Gandin V., Avizonis D., Arguello M., Zakaria C., McLaughlan S., Nouet Y., Pause A., Pollak M., Gottlieb E., Larsson O., St-Pierre J., Topisirovic I., Sonenberg N. mTORC1 controls mitochondrial activity and biogenesis through 4E-BP-dependent translational regulation. Cell Metabol. 2013;18:698–711. doi: 10.1016/j.cmet.2013.10.001. [DOI] [PubMed] [Google Scholar]
- Morita M., Gravel S.P., Hulea L., Larsson O., Pollak M., St-Pierre J., Topisirovic I. mTOR coordinates protein synthesis, mitochondrial activity and proliferation. Cell Cycle. 2015;14:473–480. doi: 10.4161/15384101.2014.991572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moscat J., Diaz-Meco M.T. Feedback on fat: p62-mTORC1-autophagy connections. Cell. 2011;147:724–727. doi: 10.1016/j.cell.2011.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nisoli E., Cozzi V., Carruba M.O. Amino acids and mitochondrial biogenesis. Am J Cardiol. 2008;101:22E–25E. doi: 10.1016/j.amjcard.2008.02.077. [DOI] [PubMed] [Google Scholar]
- Pylayeva-Gupta Y., Grabocka E., Bar-Sagi D. RAS oncogenes: weaving a tumorigenic web. Nat Rev Cancer. 2011;11:761–774. doi: 10.1038/nrc3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanathan A., Schreiber S.L. Direct control of mitochondrial function by mTOR. Proc Natl Acad Sci U S A. 2009;106:22229–22232. doi: 10.1073/pnas.0912074106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezaei R., Wang W., Wu Z., Dai Z., Wang J., Wu G. Biochemical and physiological bases for utilization of dietary amino acids by young pigs. J Anim Sci Biotechnol. 2013;4:7. doi: 10.1186/2049-1891-4-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schieke S.M., Phillips D., McCoy J.P., Jr., Aponte A.M., Shen R.F., Balaban R.S., Finkel T. The mammalian target of rapamycin (mTOR) pathway regulates mitochondrial oxygen consumption and oxidative capacity. J Biol Chem. 2006;281:27643–27652. doi: 10.1074/jbc.M603536200. [DOI] [PubMed] [Google Scholar]
- Tan B., Xiao H., Xiong X., Wang J., Li G., Yin Y., Huang B., Hou Y., Wu G. L-arginine improves DNA synthesis in LPS-challenged enterocytes. Front Biosci (Landmark Ed) 2015;20:989–1003. doi: 10.2741/4352. [DOI] [PubMed] [Google Scholar]
- Tan H.W.S., Sim A.Y.L., Long Y.C. Glutamine metabolism regulates autophagy-dependent mTORC1 reactivation during amino acid starvation. Nat Commun. 2017;8:338. doi: 10.1038/s41467-017-00369-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Z., Luo X., Xiao L., Tang M., Bode A.M., Dong Z., Cao Y. The role of PGC1alpha in cancer metabolism and its therapeutic implications. Mol Cancer Ther. 2016;15:774–782. doi: 10.1158/1535-7163.MCT-15-0621. [DOI] [PubMed] [Google Scholar]
- Vyas S., Zaganjor E., Haigis M.C. Mitochondria and cancer. Cell. 2016;166:555–566. doi: 10.1016/j.cell.2016.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G., Knabe D.A., Flynn N.E. Synthesis of citrulline from glutamine in pig enterocytes. Biochem J. 1994;299(Pt 1):115–121. doi: 10.1042/bj2990115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G., Knabe D.A., Yan W., Flynn N.E. Glutamine and glucose metabolism in enterocytes of the neonatal pig. Am J Physiol. 1995;268:R334–R342. doi: 10.1152/ajpregu.1995.268.2.R334. [DOI] [PubMed] [Google Scholar]
- Xiao H., Tan B.E., Wu M.M., Yin Y.L., Li T.J., Yuan D.X., Li L. Effects of composite antimicrobial peptides in weanling piglets challenged with deoxynivalenol: II. Intestinal morphology and function. J Anim Sci. 2013;91:4750–4756. doi: 10.2527/jas.2013-6427. [DOI] [PubMed] [Google Scholar]
- Xiao H., Wu M., Shao F., Guan G., Huang B., Tan B., Yin Y. N-Acetyl-L-cysteine protects the enterocyte against oxidative damage by modulation of mitochondrial function. Mediat Inflamm. 2016;2016 doi: 10.1155/2016/8364279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C.S., Jiang B., Li M., Zhu M., Peng Y., Zhang Y.L., Wu Y.Q., Li T.Y., Liang Y., Lu Z., Lian G., Liu Q., Guo H., Yin Z., Ye Z., Han J., Wu J.W., Yin H., Lin S.Y., Lin S.C. The lysosomal v-ATPase-ragulator complex is a common activator for AMPK and mTORC1, acting as a switch between catabolism and anabolism. Cell Metabol. 2014;20:526–540. doi: 10.1016/j.cmet.2014.06.014. [DOI] [PubMed] [Google Scholar]
- Zoncu R., Efeyan A., Sabatini D.M. mTOR: from growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol. 2011;12:21–35. doi: 10.1038/nrm3025. [DOI] [PMC free article] [PubMed] [Google Scholar]