Abstract
The genetic alterations associated with cell transformation are in large measure expressed in the metabolic phenotype as cancer cells proliferate and change their local environment, and prepare for metastasis. Qualitatively, the fundamental biochemistry of cancer cells is generally the same as in the untransformed cells, but the cancer cells produce a local environment, the TME, that is hostile to the stromal cells, and compete for nutrients. In order to proliferate, cells need sufficient nutrients, either those that cannot be made by the cells themselves, or must be made from simpler precursors. However, in solid tumors, the nutrient supply is often limiting given the potential for rapid proliferation, and the poor quality of the vasculature. Thus, cancer cells may employ a variety of strategies to obtain nutrients for survival, growth and metastasis. Although much has been learned using established cell lines in standard culture conditions, it is becoming clear from in vivo metabolic studies that this can also be misleading, and which nutrients are used for energy production versus building blocks for synthesis of macromolecules can vary greatly from tumor to tumor, and even within the same tumor. Here we review the operation of metabolic networks, and how recent understanding of nutrient supply in the TME and utilization are being revealed using stable isotope tracers in vivo as well as in vitro.
Keywords: Cancer metabolism, Nutrient supply, Stable isotope resolved metabolomics, Tumor microenvironment, Metabolic flux
Abbreviations: ACO1,2, aconitase 1,2; CP-MAS, Cross polarization Magic Angle Spinning; DMEM, Dulbeccos Modified Eagles Medium; ECAR, extracellular acidification rate; ECM, extracellular matrix; EMP, Embden-Meyerhof Pathway; IDH1,2, isocitrate dehydrogenase 1,2 (NADP+dependent); IF, interstitial fluid; 2OG, 2-oxoglutarate; ME, malic enzyme; RPMI, Roswell Park Memorial Institute; SIRM, Stable Isotope Resolved Metabolomics; TIL, tumor infiltrating lymphocyte; TIM/TPI, triose phosphate isomerase; TME, Tumor Micro Environment
Introduction
Apart from mutations, all somatic cells in an organism nominally contain the same genetic instruction set.1 However, early in differentiation, gene expression varies according to position and nascent tissue morphogenesis, even for cells of the same nominal lineage. For example, although epithelial cells are recognizable morphologically and functionally, there are substantial differences in behavior between epithelial cells lining the upper airways and those lining the alveolae.2 Furthermore, these different parts of the tissue not only differentially express proteins, but are also contact and interact with different cell types. This is critically important for carrying out the specialized functions of differentiated tissue.
Cellular metabolism
Metabolism is the essence of all live cells, which includes the basal metabolism needed to maintain basic critical cell functions, cell division proliferating, migration, and tissue-specific functions. These processes all require input of oxidizable matter from external sources both to generate energy (catabolism) and to provide building blocks for macromolecular synthesis (anabolism). Mammals are capable of only a limited subset of all metabolic processes, and rely to a significant extent on absorbing nutrients needed for biosynthesis, for example the essential amino acids and fatty acids. Even where the genes are present for synthesizing precursors, cells often will utilize exogenous molecules rather than synthesize them, as the energy costs are much lower, but which requires regulation of modes of transport, including specific transporters as well as pinocytosis.
The hydrolysis of ATP, and to a lesser extent GTP, is needed to drive thermodynamically unfavorable reactions, including maintaining membrane potentials and synthesis of macromolecules and their precursors, as well as for activating the small metabolites which are otherwise (fortunately) quite unreactive. The latter includes ATP + X- > AMP-X (e.g. aminoacylation in protein synthesis) + PPi, A/G/U/CTP + Y - > Y - A/G/U/CDP + PPi (production of activated sugars for glycosylation).
Cancer cells in a tumor need to survive, divide, and eventually metastasize. Survival, i.e. remaining viable without cell division still requires energy. Ultimately, this energy derives from the uptake and oxidation of exogenous nutrients, which can be used to replenish internal stores. Some cells are able to store molecules that can provide energy and/or carbon for precursors, which include fats in the form of lipid droplets3, 4, 5, 6, 7 and glycogen.8, 9, 10, 11, 12 Cells are also constantly recycling macromolecules, but such repair processes cannot be used for net growth, and also require energy both for breaking down proteins (cf proteasomal degradation) and for (re)making new bonds.
Autophagy has been of interest as a recycling mechanism that can be help cancer cells survive under conditions of extreme nutrient deprivation,13,14 but net proliferation cannot be supported as the total biomass does not increase. That is, in the absence of exogenous nutrients, any internal stores will eventually deplete, and the cells will be unable to support cell maintenance and membrane potentials, and thus undergo necrosis. The same considerations apply to cell division, as without exogenous nutrients to produce biomass, the cells would have to become smaller on each division until eventually they become non-functional, a kind of metabolic “black holes”. As part of an ecosystem, cancer cells may temporarily outcompete other stromal cells, which may die and release a balanced array of nutrients into the interstitial fluid, but absent the net input of exogenous nutrients to the tumor ecosystem as a whole, the system becomes self-limiting.
A longer term strategy for cancer cells instead, is to metastasize to other locations that have a richer source of nutrients, or to send out signals that induce cell breakdown elsewhere (such as muscle and adipose) and replenish the system with essential supplies (cf. cachexia5,6,15, 16, 17). This is also ultimately self-limiting when the supply of whole organismal nutrients cannot keep up with the demands of the proliferating cancer.
What is needed to produce a new cell?
Eukaryotic cells are roughly 70% water, and the remaining 30% of solutes comprises primarily organic compounds-the various ions such as chloride, phosphate, sulfate, potassium calcium and magnesium account for 1–2% of the mass. The largest component of the biomass is protein, which is present at roughly 200 mg/mL in cells depending on size,18 or ca. 1.8 M in peptide units, and accounts for ca. 60–65% of the biomass by weight. The remainder of the biomass by weight comprises lipids, nucleic acids and complex carbohydrates, which vary from cell type to cell type and according to their metabolic state.19,20 A diploid human cell contains 6.4 109 nucleotides or 3.2 pg and ca. 15 pg ribonucleotides as RNA. For an epithelial cell of volume 2 pL, there would be 350–400 pg protein and ca. 100 pg of lipids and complex carbohydrates. Nucleic acids then account for only about 3% of the biomass. Small metabolites comprise ca. 3% of the biomass.19 Thus the macromolecular biomass collectively substantially outweighs the total amount of “small molecule” metabolites.
30 cell divisions will increase the cancer cell number 109 fold, producing a new mass of 1–2 gm, with the synthesis of 0.3–0.6 g biomass. This is equivalent to approximately 3–4 mmol peptides. As 4 ATP equivalents are needed to form each peptide bond, the energy requirement for the protein component would be 12–16 mmol ATP, assuming all of the amino acids are transported the medium. Roughly half of the amino acids are essential so must be transported; the other 50% may be transported or synthesized de novo (see below). De novo synthesis requires uptake of carbon and nitrogen. Although the nucleotide content is low, the ATP requirement for complete synthesis of the nucleotides is relatively high.21 Lipids are also demanding in their carbon and energy requirements, and under conditions of poor availability of oxidizable substrates needed to generate energy, scavenging from the local environment is an alternative (see below).
Although it is common practice in areas such as microbial metabolism to measure the biomass, this is rarely done for cancer cells or tissues, which is puzzling given that the mass balance of carbon (and nitrogen) mass from different sources that end up in these different components is an important aspect of cell function and dynamics. Recently solid state NMR was applied to extracts of cells grown on 13C glucose, which enabled discrimination into generalized groups of molecules (e.g. protein, lipid, carbohydrate) in HeLa cells.22 This approach has the advantage of not requiring extensive separations into fine biochemical classes, at the cost of biochemical resolution, and there are some technical issues with quantification in CP-MAS NMR. Alternatives are to separate the major classes of macromolecules in cells, such as the nucleic acids,23 proteins24 and complex carbohydrates, and analyze the subunits after hydrolysis for total 13C (or 15N) as well as enrichment using high resolution NMR and/or mass spectrometry. Crucially, important volatile metabolites such as CO2 and NH3 are not often accounted for, which might be important on a molar basis via oxidative decarboxylation and deamidation reactions.
In this review we will briefly cover background basics of nutrient supply and transformations through metabolic networks, kinetics, and how pathways can be probed and quantified using Stable Isotope Resolved Metabolomics (SIRM), with recent examples relevant to the altered metabolism found inside the tumor microenvironment. We have recently reviewed labeling schemes and the MS and NMR-based analytical approaches for determining isotopomer and isotopologue distributions by stable isotope resolved metabolomics (SIRM) studies in different systems25, 26, 27, 28
Metabolic networks
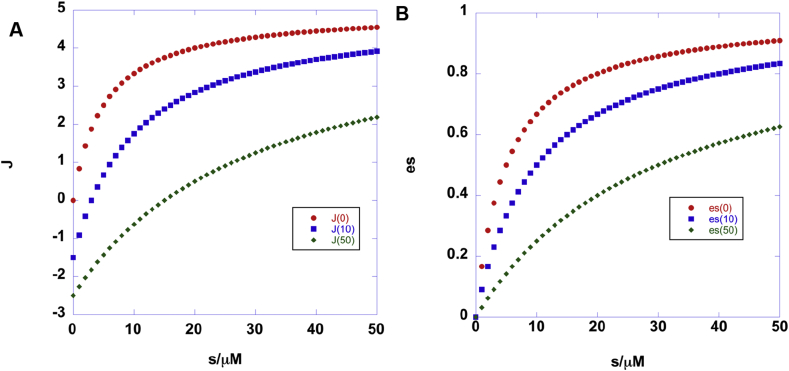
Although pathways such as glycolysis are often drawn in isolation on metabolic charts, they are in practice embedded within complex networks, such that even local changes in activity can greatly impact flow through different parts of an extensive network. Consider a simple subnetwork as in Fig. 1. Glucose enters the cell via the glucose transporters, and is then phosphorylated to G6P, which has several possible immediate fates, e.g. the oxidative branch of the pentose phosphate pathway, glycogen synthesis and glycolysis. Many other intermediates of the Embden-Meyerhof pathway have several possible fates, including the non-oxidative branch of the PPP (F6P, GAP), hexosamine pathway (F6P), lipid synthesis (DHAP) serine/glycine synthesis (3PGA) and lactic fermentation/Ala or Krebs cycle (pyruvate), involving separate compartments. Other nutrients can also enter the glycolytic pathway via glycogenolysis, and other hexoses such as galactose29 and fructose.30, 31, 32
Figure 1.
Metabolic subnetwork centered on glucose metabolism. Double headed arrows represent reversible reactions catalyzed by the same enzyme; two arrows represent reactions catalyzed by different enzymes. Enzyme names are in red. G6P: glucose-6-phosphate; F6P fructose-6-phosphate; F1,6BP fructose-1,6-bisphosphate; GAP glyceraldehyde-3-phosphate; DHAP dihydroxyacetone phosphate; 1,3bisPG 1,3-bisphosphoglycerate; 2PGA 2-phosphogycerate; PEP phosphoenolpyruvate; Pyr pyruvate; OAA oxaloacetate; AcCoA acetyl CoA; Lac lactate; Ru5P ribose-5-phosphate; GSH reduced glutathione; GALK galactose 1 kinase; GALT galcaose-1-phosphate uridyltransferase; HK hexokinase; G6Pase glucose-6-phosphatase; PGI phosphoglucose isomerase; PFK1 phosphofructokinase 1; FBPase fructose 1,6 bisphosphatase; ALD aldolase; GAPDH glyceraldehyde-3- phosphate dehydrogenase; PGK phosphoglycerate kinase; PGM phosphoglycerate mutase; ENO enolase; PK pyruvate kinase; LDH lactate dehydrogenase; ALT alanine aminotransferase; PC pyruvate carboxylase; PDH pyruvate dehydrogenase; GOT glutamate oxaloacetate aminotransferase; PPPox oxidative branch of the pentose phosphate pathway; PPPnx non-oxidative branch of the pentose phosphate pathway.
Metabolic flux
The flux through glycolysis is determined by multiple factors, which include the active protein expression levels of the various enzymes, substrate availability, product levels, allosteric effector levels, and posttranslational modifications that modulate the activity.
Consider a simple Michaelis–Menten enzyme that catalyzes the interconversion of substrate S and product P (a so-called “uni–uni” reaction):
| (1A) |
Ks and Kp are the apparent dissociation constants for the substrate and product, respectively and k2, k-2 represent the rate constants for the chemical reaction in the enzyme active site. At steady state, the instantaneous flux, J, is given by:
| J = k2ets[1-p/sKeq]/[s + Ks(1 + p/Kp)] | (1B) |
here et is the total enzyme concentration = e + es + ep and s, p, e, es, ep are the free concentrations of S, P, E, ES and EP. k2et may also be written as Vmax.
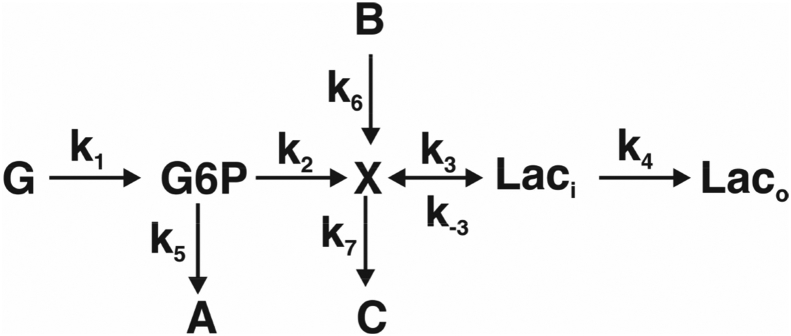
Eq. (1B) and Fig. 2A shows that the flux is a hyperbolic function of the free concentration of the substrate, s but is also subject to inhibition by the product (increases the apparent Km for S) and also demonstrates how the sign of the flux changes as the ratio of s/p varies with respect to the overall equilibrium constant Keq = <p>/<s>, as expressed via the term [1-p/sKeq]. As Fig. 2 shows, the net flux, J, can be positive, negative or zero (i.e. at equilibrium).
Figure 2.
Flux simulations for a reversible enzyme. J is the flux calculated as a function of substrate concentration s from different concentrations of product p (0, 10, 50 μM) from Eq. (1A), (1B), (1C), (1D) using the following values: KS = 5 μM, Kp = 10 μM, k2 = 5 s−1 k-2 = 3 s−1. Keq = 3.33333. es is the corresponding concentration of the ES complex calculated according to Eq. (1D). (A). Net flux for p = 0,10,20 μM (et = 1). (B). es for p = 0,10,20 μM (et = 1).
The net flux is linearly dependent on the total active enzyme concentration, and further on the intrinsic catalytic properties of the enzyme, e.g. kcat and the Km values. The enzyme concentration is determined by the balance between the rates of synthesis and degradation of the protein, which are themselves subject to regulation by numerous factors including gene expression, mRNA stability, oxidative stress, aminoacyl tRNA availability and ribosome number, as well as many others. Furthermore, the enzyme concentration can vary greatly in different parts of the cell (“compartmentation”), as can substrate and product concentrations. An allosteric effector (i.e. a compound that does not directly compete for S or P) may alter either or both Km and kcat, positively or negatively, unlike a competitive inhibitor, which can only increase the Km.
The majority of metabolic studies allow the reactions to occur in vivo, followed by quenching and extraction of the metabolites for quantitative analysis. This immediately destroys all information about compartmentation and relevant pool sizes, posing significant difficulties for flux analysis in eukaryotic cells or tissues. Furthermore, the form of Eq. (1B) is deceptively simple, because it is written in terms of free substrate and product concentration. However, neither “free” nor “concentration” is usually measured in extracts, rather what is obtained is the total substrate abundance, which is the sum of the free substrate and that bound to all the proteins that bind it, or st = s+Σesi. The actual substrate concentration in the relevant compartment that contains the enzymes is generally unknown. For low abundance enzymes and high abundance substrate, st ≈ s. However, for some central pathways, the enzyme concentrations can be high and even comparable to the substrate concentration.33,34 In that case, the flux in terms of total substrate concentration and free product concentration is given by:
| J = k2es(1-p/sKeq) | (1C) |
| 2es = et + st + Ks(1 + p/Kp) + {[et + st + Ks(1 + p/Kp)]2–4 etst}0.5 | (1D) |
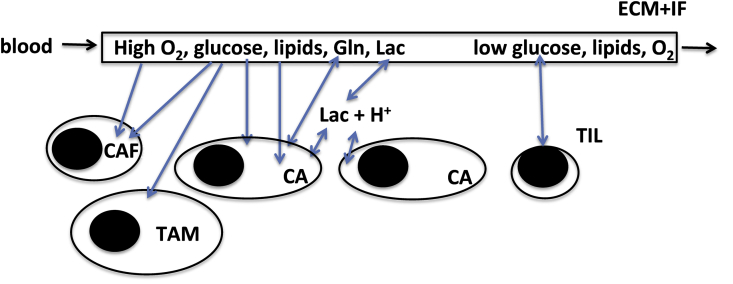
At steady state, the flux through a linear pathway is identical at each point in the pathway. However the reality is more complicated as there are usually multiple branch points in metabolic networks. Furthermore, pathway flux is more likely to be measured as consumption at the start of the pathway and/or production at the end. The following simple scheme (Scheme 1) shows how this can be misleading.35
Scheme 1.
Simple branched kinetic pathway. ki are rate constants for the interconversions. The rate of G consumption then is –dg/dt = k1 g where lower case g genotes concentration of the metabolite G. Expressions for the rates of production of A, C and Lac are developed in the text.
This branched pathway shows that there are two sources of Lac, namely G and B, and not all of G or B ends up in Lac as some is converted to A and C. To determine the various contributing sources and sinks, tracer methodology is needed, such as via the use of stable isotope-enriched precursors,.28 For this scheme, if G is isotopically enriched, the enrichment in Lac will determined by the relative rate of utilization of G and B, as described in Scheme 1, as follows:
| dlo(t)/dt = k1k2k3k4g/[(k2 + k5)(k3k4 + k7(k-3 + k4)] + k3k4k6b/[k3 k4 + k7(k-3 + k4)] | (2A) |
| da(t)/dt = k1k5g/(k2 + k5) | (2B) |
| dc(t)/dt = k1k2k7(k-3 + k4)g/[(k2 + k5)(k3k4 + k7(k-3 + k4))] + k6k7(k-3 + k4)b/[k3k4 + k7(k-3 + k4)] | (2C) |
| ki = kcat,i ei/Km,i |
Steady state is maintained if g exceeds the transporter Km value, or is supplied at a rate equal to that of its utilization.
Here lo represents the extracellular concentration of Lac, that might be easily measured, and Eq. (2A) describes the total rate of extracellular lactate production. Suppose the system were supplied with isotope enriched G, i.e. G*, then the rate of production of Laco* would be:
| dlo*(t)/dt = k1k2k3k4g*/[(k2 + k5)(k3k4 + k7(k-3 + k4))] | (2D) |
The fraction enrichment, f in the extracellular lactate (Laco) would then be
| f(t) = lo*(t)/[lo*t) + lo(t)] | (3A) |
For changes in lo between two time points, f = 1/{1 + k6 (k2 + k5)b/k1k2g*}. This function approaches unity when k1g*>>k6b.
The fraction of lactate produced from G that is consumed, using a tracer, would be
| F(t) = dlo*/dt/-dg*/dt | (3B) |
In the limit that the glucose utilization is a linear function of time
| F = k2k3k4/[(k2 + k5)(k3k4 + k7(k-3 + k4))] | (3C) |
This function approaches unity only when both k5 and k7 are small.
Relationship to glycolytic flux
Despite the above difficulties, the rate of glycolysis is important to determine in the analysis of carbohydrate metabolism, and there are several approaches to determining this flux.36 The Embden-Meyerhof pathway (EMP) describes the conversion of intracellular glucose to pyruvate, which has numerous possible fates depending on the cellular state and conditions. Although the EMP is usually written as a linear pathway, in practice it is embedded in a more complex network of reactions, such that many of the intermediates have alternative fates (Fig. 1). For example, G6P is also the entry point into the oxidative branch of the pentose phosphate pathway as well as glycogenesis after isomerization to G1P. Furthermore, the Leloir pathway may convert lactose-derived galactose to G1P and G6P.29 F6P is the entry point into hexosamine synthesis, as well as being an intermediate in the non-oxidative branch of the PPP. DHAP, a product of the aldolase reaction, generally equilibrates with GAP via TIM (TPI), but is also the precursor to the glycerol subunit of the glycerolipids. Fructokinase converts fructose to F1P which via aldolase B produces DHAP and glyceraldehyde30,31 and thence to lactate and/or glycogen.37 3PGA is the starting point for the serine–glycine pathway, and pyruvate may be transaminated to Ala, reduced to lactate (fermentation), oxidatively decarboxylated and condensed with CoA to make AcCoA via pyruvate dehydrogenase (PDH), or carboxylated via pyruvate carboxylase (PC). The key concept is that is rare for all of the glucose to be converted to lactate, so measurement of lactate production alone is not a direct measurement of glycolysis. Indeed, pyruvate can and is also made from other carbon sources via malic enzymes (which has three isoforms), such that glycolysis will be overestimated. Similarly, glucose uptake rates alone will also overestimate the glycolytic flux or -dg/dt > FEMP > dlac/dt.
Glycolytic flux is also estimated from the rate of proton extrusion, or ECAR.38 This is usually equated with the release of lactate, which generally occurs as a lactate-proton symport via the monocarboxylate transporters.38 If – and only if – the symport of lactate and protons has a stoichiometry of 1:1, then the ECAR is a measure of the lactate release. However, as stated above the rate of lactate release is not a direct measure of the EMP. Furthermore the source of protons in the cytosol and those extruded into the extracellular space (which is the ECAR) is complex. Even simple considerations of lactic fermentation indicate that the stoichiometry of proton production by the EMP and export is not necessarily integral. The numbers of protons produced by the EMP is pH-dependent, and the LDH reaction itself consumes protons.39 This is further complicated if glycogenolysis is also occurring.40 Indeed, lactate/proton symport is not the only source of extracellular acidification, as other transporters and carbonic anhydrases among others can also contribute.41,42 To determine the absolute rate of proton release (rather than pH changes) requires accounting for the buffer capacity of the medium. An alternative approach is to measure the release of 3H to water from [5-3H] glucose which occurs late in glycolysis (via the enolase-catalyzed reaction).36 However, 3H would also be lost at the first triose step as GAP and DHAP are interconverted via TIM, and the stoichiometry will also depend on how much DHAP is also removed from glycolysis as glycerol-3-phosphate (cf. Fig. 1).
In summary, determining absolute, or even relative intracellular fluxes, requires a model for analyzing the experimental data. Scheme 1 is an example of a very simplified model, which shows that in the presence of reversible reaction and multiple inputs and outputs, as in glycolysis, pathway fluxes can be difficult to determine without appropriate network modeling using tracers.
Determining enzyme, substrate and modulator concentrations-compartmentation
It is common for different isoforms of enzymes to be located in different compartments, which means that they will be exposed to different concentrations of substrate, products, and allosteric modulators, as well as potentially having different levels of expression and intrinsic catalytic properties (cf Eq. (1A), (1B), (1C), (1D)). As previously stated, the absolute amounts of a given enzyme present in a given compartment depend on the rate of gene expression, mRNA stability, rate of translation, protein stability, and transport into the destined compartment. Substrate and product levels then depend on the activity of the enzymes and any transport processes between compartments.
Examples that are important in cancer metabolism are GOT1,2, IDH1,2,3, ACO1,2 (thus 2OG and citrate can be interconverted in the cytoplasm as well as in the mitochondrial matrix); ME1,2,3; which are found in either the cytosol or in the mitochondria, and may be involved in cycling metabolites across the mitochondrial membrane for which there are no direct transporters (cf. NADH and the Asp/malate shuttle)43 or NADPH-dependent hydride shuttle across the mitochondrial membranes via IDH1 and 2.44
The amounts of substrates and enzymes that are determined in most metabolic studies however result from extraction of cells or tissues and therefore are averaged over all compartments.
The tumor microenvironment and nutrient sources
Although much of our understanding of cancer cell proliferation and survival has been learned from cell culture,45, 46, 47, 48, 49, 50, 51 there are many limitations of this model.35,52, 53, 54 In addition to the lack of cell–cell interactions, the absence of stroma, and a full extracellular matrix, the nutrient supply is also usually very different from what is found in actual diseased tissues (cf. Fig. 3).
Figure 3.
Tumor microenvironment and metabolism. Cancer cells compete with stromal cells for nutrients supplied by the blood, especially glucose, oxygen and lipids, and excrete compounds such as PD-L1 lactate and protons that are immunosuppressive, and which induce changes in macrophage polarization and thence TILs. Cancer cells may also engulf ECM and debris by macropinocyosis to fuel growth and survival (see text). CAF cancer associated fibroblast; TAM tumor associated macrophage; TIL tumor infiltrating lymphocyte; CA cancer cell. ECM + IF extracellular matrix + interstitial fluid.
Table 1 summarizes the metabolite composition of three commonly used cell culture media, in comparison with adult humans.
Table 1.
Composition of common cell culture media and human blood.
| Compound | RPMI1640a | DMEMb | Hams F12c | Adult bloodd |
|---|---|---|---|---|
| Glycine | 0.133 | 0.40 | 0.1 | 0.25 ± 0.06 |
| l-Alanine | 0 | 0 | 0.1 | 0.2–0.6 |
| l-Argininef | 1.15 | 0.4 | 1.0 | 0.11 ± 0.03 |
| l-Asparagine | 0.379 | 0 | 0.1 | 0.04 ± 0.02 |
| l-Aspartic acid | 0.150 | 0 | 0.01 | 0.02 ± 0.01 |
| l-Cystine | 0.208 | 0.2 | 0 | 0.1 ± 0.03 |
| l-Cysteine | 0 | 0 | 0.2 | 0.03–0.3 |
| l-Glutamic Acid | 0.136 | 0 | 0.1 | 0.05–0.15 |
| l-Glutamine | 2.05 | 4 | 1 | 0.6 ± 0.1 |
| l-Histidinee | 0.0968 | 0.2 | 0.074 | 0.09 ± 0.01 |
| l-Hydroxyproline | 0.153 | 0 | 0 | 0.015 ± 0.01 |
| l-Isoleucinee | 0.382 | 0.8 | 0.03 | 0.07 ± 0.01 |
| l-Leucinee | 0.382 | 0.8 | 0.1 | 0.16 ± 0.03 |
| l-Lysinee | 0.274 | 0.8 | 0.2 | 0.18 ± 0.03 |
| l-Methioninee | 0.101 | 0.2 | 0.032 | 0.03 ± 0.06 |
| l-Phenylalaninee | 0.0909 | 0.4 | 0.03 | 0.06 ± 0.02 |
| l-Proline | 0.174 | 0 | 0.03 | 0.18 ± 0.05 |
| l-Serine | 0.286 | 0.4 | 0.1 | 0.15 ± 0.03 |
| l-Threoninee | 0.168 | 0.8 | 0.1 | 0.13 ± 0.03 |
| l-Tryptophane | 0.0245 | 0.078 | 0.01 | 0.055 ± 0.03 |
| l-Tyrosinef | 0.111 | 0.4 | 0.03 | 0.07 ± 0.04 |
| l-Valinee | 0.171 | 0.8 | 0.1 | 0.2 ± 0.06 |
| d Glucose | 11.1 | 25 | 10 | 4 (3–6) |
| d-Fructose | 0 | 0 | 0 | 0.03–0.06g |
| d-Galactose | 0 | 0 | 0 | 0.03–0.12 |
| l-lactate | 0 | 0 | 0 | 1–2 |
| pyruvate | 0/0.1 | 0/ | 0.1 | 0.02–0.25 |
| Acetate | 0 | 0 | 0 | 0.04 ± 0.02 |
| 3-OHbutyrate | 0 | 0 | 0 | 0.04 ± 0.03 |
| GSH | 0.00326 | 0 | 0 | 0.001–1 |
| HCO3- | 23.81 | 44 | 14 | 25 |
% arginine can be synthesized, but generally not at a sufficient rate.
Invitrogen.
Thermo-Fisher 11965.
Sigma Aldrich.
Fasted normal human adult blood.55
Essential amino acid.
Tyrosine is contingent on its direct precursor, Phe.
Much higher after meals containing high fructose corn syrup.
In many cancer cell studies, the media are supplemented with 10% FBS, which is the only source of exogenous lipids to the cells, unless explicitly stripped and replaced by defined lipid composition. As FBS is variable in composition, some studies have used dialyzed serum, which depletes all of the small metabolites, so that the concentrations in the culture medium are 10% lower. However, the concentrations of amino acids, glucose and bicarbonate differ greatly from one medium to another, and differ also from the concentrations observed in adult human blood, which is itself variable (Table 1). The media do not contain lipids, most of which are insoluble in water, so are supplied by the FBS, mainly as fatty acids bound to serum albumin. It has been reported that some proliferating cells obtain most of their fatty acids from exogenous sources56,57 rather than from synthesis, though other cancer cells upregulate fatty acid synthase.58,59 The cholesterol concentration in human adult blood is 4–6 mM (bound to lipoproteins), and total palmitate is around 1 mM.55 To control the levels of these important nutrients, charcoal and solvent stripping may be needed.60 Further, many cell culture experiments are carried out at ca. 20% oxygen and 5% CO2, neither of which are typical for most tissues. The oxygen concentration in solid tumors can drop below 1% in some parts. It is perhaps not surprising that differences are observed between different culture systems, both at the nutrient level,61 and the responses engendered by a 3D environment.62
It is also crucial to keep in mind, that although some nutrients are present at low concentrations in blood, the blood represents a very large constantly replenishable reservoir-even at 20 μM Asp the total amount in 5 L blood is 100 μmol, which is enough to make protein and nucleic acids for ca. 109 cells, or a >1 cm3 tumor. The major issue is more likely the rate of delivery within a solid tumor, which may be a significant factor in the rate of tumor growth.
Sources of lactate and utilization
It has been known for decades that lactate produced by various tissues in an organism can be taken up by other tissues for gluconeogenesis (e.g. liver and kidney via the Cori cycle63) or for oxidation as an energy source under appropriate conditions (e.g. cardiomyocytes64,65 and neurons66,67). More recently, lactate has been shown to be an important nutrient source for tumors.54,68, 69, 70, 71 The concentration of lactate in adult human blood is in the range 1–3 mM under resting conditions,72 and a substantial fraction of this lactate originates from lactic fermentation by the erythrocytes,73 which convert most of the readily available glucose to lactate via glycolysis, with loss of a small fraction of carbon as CO2 via the oxidative branch of the PPP (the rest is returned to glycolysis via the non oxidative branch) and is found in abundance in most tissues. The monocarboxylate transporters (MCT) are freely reversible, and lactate is symported with a proton.74,75 Recently it has been suggested that this proton dependence may be important for determining the net flow of lactate in and out of tumors.76 Similarly, the interconversion of lactate and pyruvate is readily reversible via the activity of LDH, so the net production of extracellular lactate or intracellular pyruvate is governed by the disequilibrium activity ratios (cf Eq. (1B)):
| ΓMCT = laci10−pHi/laco10−pHo ΓLDH = lac.nad+/pyr. nadh10−pH | (4) |
There would be a net inward flow of lactate from the extracellular environment if ΓMCT < 1 which will occur if the extracellular space is more acidic than the cytoplasm (generally the case in tumors41,77) and/or the intracellular lactate concentration is lower than the (local) extracellular lactate. For example, a pH difference of 0.5 across the plasma membrane is equivalent to a proton ratio of 3.16 fold, which would support a net influx of lactate plus a proton, at an intracellular to extracellular lactate ratio of exactly 3.16. Similarly, there will be a net production of pyruvate from lactate if ΓLDH > Keq, which is ca. 2.5 E4 at pH 778 which corresponds to a pH-independent equilibrium constant of 2.5 E11 M−1 favoring lactate. As the concentration ratio of free NAD+/NADH in cancer cells is maintained in the 700–1500 depending on cell type and presumably conditions79 there would be net net intracellular flux for pyruvate to lactate for lac/pyr ratios up to 11 to 5 at pH 7.5 or 36 to 17 at pH 7. Given the uncertainties in Keq and compartmentation of NAD+/NADH ratios, this can be regarded as a poised system, and the net flux would need only modest changes in the lac/pyr ratio. For example, where thin tissue slices are incubated in the presence of physiological glucose concentrations, there is certainly a net excretion of glucose-derived lactate into the medium,80,81 but this does not show which cells are the net exporter, nor whether this would remain so under continuing experimental conditions of low extracellular glucose and high lactate concentrations. Eventually, there will be a net influx of lactate plus H+ that will acidify the intracellular space, and for lactate to be used either for oxidation or for gluconeogenesis, must first be converted to pyruvate, which will consume NAD+ and generate NADH. The resulting pyruvate can also be transaminated to Ala, or once in the mitochondria, carboxylated to OAA via the ATP-dependent pyruvate carboxylase, which is upregulated in at least some tumors.54,80,82,83 If the LDH activity is exclusively cytoplasmic, then the NADH would need to be reoxidized via the malate-aspartate shuttles even if the pyruvate were imported into the mitochondria for oxidation or for anaplerosis. The trinucleotide yield would be 14 ATP +1 GTP (cf. Table 2).
Table 2.
Metabolic energy yields from different substrates. It is assumed that mitochondria produces 2.5 ATP/NADH and FADH2 produces 1.5 ATP. GTP is made in the substrate level phosphorylation via succinyl CoA synthetase.
| Compound | Pathway | ATP yield | Comments |
|---|---|---|---|
| Glycogen | Glycogenolysis + lactic fermentation | 3/glucose subunit | Phosphorolysis generates G1P |
| Glucose | Lactic fermentation | 2/glucose subunit | |
| Glycolysis | 2 + 3 | 3 for shuttles to respiring mitochondria | |
| Fructose | Fructose cleavage to lactate | 2 | Fructokinase + glyceraldehyde kinase |
| Galactose | Lactic fermentation | 2 | |
| Glycolysis | 2 + 3 | 3 for shuttles to respiring mitochondria | |
| Pyruvate | Krebs Cycle | 11.5 ATP + 1 GTP | |
| Lactate | Krebs Cycle | 14 ATP + 1 GTP | NADH produced reoxidized via shuttle |
| Acetate | Krebs Cycle | 8 ATP + 1 GTP | 1 ATP used for activation of acetate to AcCoA |
| Palmitate | Beta-oxidation/Krebs Cycle | 106 ATP + 7 GTP | |
| Glutamine- > OAA | Krebs Cycle | 6.5 ATP + 1 GTP | Transamination of Glu via GOT |
| 9 ATP + 1 GTP | Oxidative deamination of Glu via glutamate dehydrogenase |
Alternatively, it has been suggested that there is a mitochondrial form of LDH (LDH B), which reoxidizes lactate intramitochondrially in cancer, that is used for making AcCoA for lipid biosynthesis71 (after shuttling out to the cytoplasm as citrate). The existence of a mitochondrial LDH in healthy cells has been a controversial suggestion for more than 60 years, as reviewed recently.84 In this scenario, the NADH produced by LDH would be reoxidized by the respiratory chain, again with an overall energy yield of 14 ATP +1 GTP.
Note that as these reactions are freely reversible, there will always be an exchange flux associated with the reactions, which appears to be very extensive in vivo according to DNP studies.85,86
Acetate
There has been considerable interest recently in acetate both as a nutrient source, and for its involvement in protein acetylation.87, 88, 89, 90, 91, 92, 93, 94, 95, 96 The abundance of acetate in adult plasma is approximately 20–60 μM (Table 1). Acetate is the shortest chain fatty acid that can undergo beta oxidation. To be utilized as an energy source, it must first be converted to AcCoA via the acetate thiokinases (or Acetate CoA synthetase, ACS1,2) in an ATP-consuming reaction. The energy yield from beta oxidation then is 8 ATP + 1 GTP. Alternatively, this is a cytoplasmic (and also nuclear91) source of AcCoA that might be used for either fatty acid synthesis or protein acetylation.93 Recently it was shown that acetate enters the Krebs cycle in clear cell renal carcinomas in situ, whereas glucose was preferentially shunted to lactate, which is different to what was observed in NSCLC with 13C glucose tracing.97 Furthermore, acetate is also oxidized to a high extent in some tissues,98,99 suggesting that this simple metabolite has multiple context-dependent fates.
Utilization and production of non-essential amino acids
Alanine
Alanine is readily available from blood, is an abundant amino acid (on average 200–500 μM in healthy adults, Table 1), and is readily converted to pyruvate by alanine aminotransferase, ALT (AST) using 2-oxoglutarate as the keto acid. This enzyme is highly expressed in gluconeogenic tissues (liver and kidney) though its isoforms are also expressed in many other tissues,100 including cancers, and in concert with GOT is able to shuttle amino groups between Glu/Asp/Ala. It behaves analogously to lactate in the Cori cycle, as well as being a potential intracellular reservoir for pyruvate, which has implications for in vivo studies using SIRM.
Glutamine
Glutamine has received considerable attention, as many cancer cells in vitro show a very strong dependence on exogenous Gln for proliferation and survival.50,101, 102, 103 In addition to being a proteinaceous amino acid, via glutaminolysis it is a major source of anaplerotic carbon in the Krebs cycle and also an important nitrogen donor for nucleotide23,102 and amino sugar production104 as well as a major source of glutamate used for glutathione synthesis. 50 Although Gln is the most abundant amino acid in blood (Table 1), recent studies have indicated that the degree of “addiction” to Gln in vivo for some tumors (e.g. KRas-driven lung adenocarcinomas) may be less than observed in cell culture52; but this is not readily generalizable as several tissue types express glutamine synthetase (reviewed in105). We have shown that glutaminase expression in human lung tumors is highly variable, though tissues do metabolize glutamine extensively.80,106 Furthermore, the level of glutamine and glucose labeling among different breast cancer cell type, both in vitro and in mouse xenografts,45 suggesting again that metabolic phenotypes are cell and environment dependent.
Serine and glycine
Serine and Gly are proteinaceous amino acids that are also used for making numerous other compounds, including the serine-containing lipids (e.g. PS and the sphingolipids) and Gly, which can be readily made from Ser, is used for purine biosynthesis, glutathione production and in 1-carbon metabolism (Fig. 1). Both are abundant in blood as well as common cell culture media (Table 1). However, some cells will synthesize their own serine and glycine whereas others will prefer to use exogenous sources, or both, which appears to depend on cell type and growth conditions and some cells show net efflux of glycine18,48,107, 108, 109, 110, 111, 112, 113, 114, 115
Aspartate and asparagine
Asp and Asn are non-essential amino acids needed for protein biosynthesis, and Asp is also the direct precursor of uracil, as well as being a nitrogen donor in nucleotide synthesis.21 Although Asp and Asn are non-essential amino acids, their concentration in blood is low (Table 1). In cell culture studies, where exogenous Asp is sometimes zero (e.g. in DMEM), cells must synthesize these amino acids to maintain proliferation. However it appears that cells make Asp even when there is an exogenous source, and do so using both glucose and glutamine carbon.23 Furthermore, production of Asp is essential for proliferation when mitochondria are fully functional.116 A corollary is that under hypoxia, cancer cells cannot make Asp (as the enzyme dihydroorotate dehydrogenase, DHODH is mitochondrial), and under those conditions must import exogenous Asp,117 which in tumors may be limiting.116
Asp is also the precursor of Asn, via an ATP-dependent amidation reaction (the reverse reaction is catalyzed by asparaginase, which is not expressed in mammals). When mammalian cells are deprived of glutamine, exogenous Asn is used for protein synthesis and rescues cell survival and growth.118,119 Similarly, the expression level of asparagine synthetase (ASNS) correlates with efficacy of asparaginase treatment in acute lymphoblastic leukemia.120 However, although Asn is usually not metabolized extensively, it does have other critical functions, including as an exchange factor for amino acid transport, especially Ser, Arg, and His121 which would be needed to support protein synthesis in proliferating cells.
Nutrients in the TME
Tumor microenvironments are heterogeneous and variable in many ways.54,77,122, 123, 124, 125, 126, 127 As a tumor grows, its internal vasculature and blood supply changes non-uniformly such that some parts of the tumor are very poorly vascularized whereas others may be relatively close to blood vessels. It must also be kept in mind that the microvasculature within tumors is generally of poor quality, with many bends and blind endings, leading to poor blood flow,128,129 hypoxia130 and a depletion of essential nutrients for supporting cell growth and survival, such as glucose, Gln, lipids,127,131, 132, 133 and essential amino acids (which by definition, must be taken up from the blood or extracellular space). Furthermore, the cancer cells excrete a variety of metabolites that may influence their local environment, such as protons,77 lactate which may be used by other cells in the tumor,127,134,135 glutamate (antiported with Cystine via xc−, needed for protein and glutathione synthesis)136 among other amino acids. This altered environment impacts the activities and responses of other cells in the tumor. The cancer cells may excrete PD-L1 which prevents T cell activation,137, 138, 139 the hypoxic and acidic environment140,141 and lactate also immunosuppressive, and further promote pro-tumorigenic states of immune cells, such as the tumor associated macrophages (TAMs)142,143 and fibroblasts (CAFs) (cf Fig. 3) which is associated with alerted metabolism.144,145 For these reasons there has been increasing interest in establishing what are the most significant nutrient sources for different cancers under various conditions.
In addition to blood, other possible sources of nutrients are via the lymphatic system, the interstitial fluid (IF) and the extracellular matrix (ECM). The ECM not only provides a matrix for cell attachment but also determines cell migration. Its structure and composition vary from tissue to tissue, and may become modified in tumors.146 However, the major components are the fibers of collagen, which is associated with numerous other proteins including fibronectin, elastin and laminins. These are complemented by proteoglycans and hyaluronate.133 The ECM is thus a store of amino acids and sugars. Metabolic scavenging via e.g. macropinocytosis of the ECM may be an important mechanism for survival and growth of cancer cells.131,147, 148, 149, 150 In addition to macromolecules that can be engulfed, the interstitial fluid (IF), which derives from serum, also contains abundant small molecules that can be transported into tumor cells.132 However, the composition of the IF within tumors differs in specific ways from that in non-cancerous tissue or serum itself. Most notable was that the tumors studied not only reduced the glucose content of the blood flowing through the tumor, but also depleted it in the IF to very low values. Similarly the cholesterol levels were much lower in the IF than in serum. Cancer cells clearly deplete certain critical metabolites in the tumor microenvironment, which can impact the function of other cells within the tumor. It was recently reported that ovarian cancer cells promote membrane-cholesterol efflux and depletion of lipid rafts from macrophages, making them protumorigenic.151 The low concentration of glucose in parts of solid tumors poses an obvious problem for the Warburg effect, which has so dominated discussion of tumor metabolism in recent years. Nevertheless, 13C enriched metabolites do enter solid tumors and are transformed in both human subjects and animal models45,52,54,68,80,97,152, 153, 154
In larger tumors, these and other factors also lead to regions of necrotic tissue, the remnants of which could be used as rich sources of nutrients by active cancer cells, such as via macropinocytosis131,147, 148, 149, 150 The cancer cells are also competing with other cells within the tumor including remaining untransformed epithelial cells, the endothelial cells of the vasculature, fibroblasts and several kinds of immune cells. The cancer cellularity of solid tumors can be very variable, ranging from a few percent of the total to >90%, with consequences for the physical architecture of the tumor and the microenvironment.
Concluding remarks
Metabolic adaptations to changing available nutrient sources in the context of competition is vital for cancer cell growth, survival, and metastasis. These processes have vastly different metabolic requirements, and cancer cells not only have a high demand for glucose and glutamine, but also excrete molecules that affect the phenotype of stromal cells. For example infiltrating macrophages and T lymphocytes inside solid tumors are in a different state than those in the non-diseased tissue.62,144,155, 156, 157, 158 Furthermore, TILs159 and macrophages160 are not uniformly distributed throughout the tumor mass, and in some instances may be excluded from nests or islets of cancer cells.161 Given the different metabolic adaptations of the various cell types, it follows that the metabolic profiles must also be spatially heterogeneous.
Conclusions based on cell culture with established cell lines under standard growth conditions are likely to be misleading as metabolism by its very nature is plastic and condition-sensitive. Solid tumors are often highly heterogeneous both in terms of the distribution of cells, and the high variability in the relative number of cancer and immune cells from tumor to tumor and within a tumor. This is associated, especially in larger tumors, with poor vascularization and thus poor nutrient availability, which may lead to alternative strategies for survival within the hostile environment (largely of the cancer's own making). Conversely, such larger tumors may also be locally nutrient-rich due to sources from necrotic tissues. These mechanisms include selection of cells that have acquired metabolic advantages via substantial quantitative reprogramming and upregulation of various scavenging mechanisms. As the particular adaptations are both cell type genetic lesion dependent there may be several ways for cells to survive and ultimately metastasize. The particular nutrient requirements for cancer cell survival and proliferation under different conditions is still being determined. How they achieve this in vivo remains a challenging problem, that will need a combination of more realistic models as well as actual in vivo analyses.35
Conflict of interest
The authors declare no conflict of interest.
Acknowledgments
This work was supported in part by the Carmen L Buck Chair in Oncology (to ANL), the Edith D. Gardner Chair in Cancer Research (to TWMF) and funding from NIH 1P01CA163223-01A1, 5P20GM121327 and P30CA177558.
Footnotes
Peer review under responsibility of Chongqing Medical University.
References
- 1.Weinberg R.A. Garland Science; New York: 2014. The Biology of Cancer. [Google Scholar]
- 2.Crystal R.G., Randell S.H., Engelhardt J.F., Voynow J., Sunday M.E. Airway epithelial cells current concepts and challenges. Proc Am Thorac Soc. 2008;5:772–777. doi: 10.1513/pats.200805-041HR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Farese R.V., Jr., Walther T.C. Lipid droplets finally get a little R-E-S-P-E-C-T. Cell. 2009;139:855–860. doi: 10.1016/j.cell.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crooks D., Maio N., Lane A. Acute loss of iron-sulfur clusters results in metabolic reprogramming and generation of lipid droplets in mammalian cells. J Biol Chem. 2018;293:8297–8311. doi: 10.1074/jbc.RA118.001885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tirinato L., Pagliari F., Limongi T. An overview of lipid droplets in cancer and cancer stem cells. Stem Cell Int. 2017 doi: 10.1155/2017/1656053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Krahmer N., Farese R.V., Walther T.C. Balancing the fat: lipid droplets and human disease. EMBO Mol Med. 2013;5:973–983. doi: 10.1002/emmm.201100671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yue S., Li J., Lee S.-Y. Cholesteryl ester accumulation induced by PTEN loss and PI3K/AKT activation underlies human prostate cancer aggressiveness. Cell Metabol. 2014;19:393–406. doi: 10.1016/j.cmet.2014.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zois C.E., Harris A.L. Glycogen metabolism has a key role in the cancer microenvironment and provides new targets for cancer therapy. J Mol Med. 2016;94:137–154. doi: 10.1007/s00109-015-1377-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ritterson Lew C., Guin S., Theodorescu D. Targeting glycogen metabolism in bladder cancer. Nat Rev Urol. 2015;12:383–391. doi: 10.1038/nrurol.2015.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ros S., Schulze A. Linking glycogen and senescence in cancer cells. Cell Metabol. 2012;16:687–688. doi: 10.1016/j.cmet.2012.11.010. [DOI] [PubMed] [Google Scholar]
- 11.Favaro E., Bensaad K., Chong M.G. Glucose utilization via glycogen phosphorylase sustains proliferation and prevents premature senescence in cancer cells. Cell Metabol. 2012;16:751–764. doi: 10.1016/j.cmet.2012.10.017. [DOI] [PubMed] [Google Scholar]
- 12.Lee W.N.P., Guo P., Lim S. Metabolic sensitivity of pancreatic tumour cell apoptosis to glycogen phosphorylase inhibitor treatment. Br J Canc. 2004;91:2094–2100. doi: 10.1038/sj.bjc.6602243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mathew R., Karantza-Wadsworth V., White E. Role of autophagy in cancer. Nat Rev Cancer. 2007;7 doi: 10.1038/nrc2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kimmelman A.C., White E. Autophagy and tumor metabolism. Cell Metabol. 2017;25:1037–1043. doi: 10.1016/j.cmet.2017.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tisdale M.J. Cancer anorexia and cachexia. Nutrition. 2001;17:438–442. doi: 10.1016/s0899-9007(01)00506-8. [DOI] [PubMed] [Google Scholar]
- 16.Martinelli G.B., Olivari D., Cecconi A.D.R. Activation of the SDF1/CXCR4 pathway retards muscle atrophy during cancer cachexia. Oncogene. 2016;35:6212–6222. doi: 10.1038/onc.2016.153. [DOI] [PubMed] [Google Scholar]
- 17.Lecker S.H., Jagoe R.T., Gilbert A. Multiple types of skeletal muscle atrophy involve a common program of changes in gene expression. FASEB J. 2004;18:39–51. doi: 10.1096/fj.03-0610com. [DOI] [PubMed] [Google Scholar]
- 18.Dolfi S.C., Chan L.L.-Y., Qiu J. The metabolic demands of cancer cells are coupled to their size and protein synthesis rates. Cancer Metabol. 2013;1:20. doi: 10.1186/2049-3002-1-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alberts B., Johnson A., Lewis J., Raff M., Roberts K., Walter P. Garland Science; New York: 2002. Molecular Biology of the Cell. [Google Scholar]
- 20.Feijo´ Delgado F., Cermak N., Hecht V.C. Intracellular water exchange for measuring the dry mass, water mass and changes in chemical composition of living cells. PLoS One. 2013;8:e67590. doi: 10.1371/journal.pone.0067590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lane A.N., Fan T.W.-M. Regulation of mammalian nucleotide metabolism and biosynthesis. Nucleic Acids Res. 2015;43:2466–2485. doi: 10.1093/nar/gkv047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen Y., Jr., Huang X., Mahieu N.G., Cho K., Schaefer J., Patti G.J. Differential incorporation of glucose into biomass during Warburg metabolism. Biochemistry. 2014;53:4755–4757. doi: 10.1021/bi500763u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fan T.W.-M., Tan J.L., McKinney M.M., Lane A.N. Stable isotope resolved metabolomics analysis of ribonucleotide and RNA metabolism in human lung cancer cells. Metabolomics. 2012;8:517–527. doi: 10.1007/s11306-011-0337-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang Y., Fan W.W.-M., Lane A.N., Higashi R.M. Chloroformate derivatization for tracing the fate of amino acids in cells by multiple stable isotope resolved metabolomics (mSIRM) Anal Chim Acta. 2017;976:63–73. doi: 10.1016/j.aca.2017.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lane A.N., Higashi R.M., Fan T.W.-M. NMR and MS-based stable isotope-resolved metabolomics and applications in cancer metabolism. Trends Anal Chem. 2019 doi: 10.1016/j.trac.2018.11.020. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lane A.N., Fan T.W.-M. NMR-based stable isotope resolved metabolomics in systems biochemistry. Arch Biochem Biophys. 2017;628:123–131. doi: 10.1016/j.abb.2017.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fan T.W.-M., Lane A.N. Applications of NMR to systems biochemistry. Prog Nucl Magn Reson Spectrosc. 2016;92:18–53. doi: 10.1016/j.pnmrs.2016.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bruntz R.C., Higashi R.M., Lane A.N., Fan T.W.-M. Exploring cancer metabolism using stable isotope resolved metabolomics (SIRM) J Biol Chem. 2017;292:11601–11609. doi: 10.1074/jbc.R117.776054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Frey P.A. The Leloir pathway: a mechanistic imperative for three enzymes to change the stereochemical configuration of a single carbon in galactose. FASEB J. 1996;10:461–470. [PubMed] [Google Scholar]
- 30.Jang C., Hui S., Lu W. The small intestine converts dietary fructose into glucose and organic acids. Cell Metabol. 2018;27:351. doi: 10.1016/j.cmet.2017.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bu P., Chen K.-Y., Xiang K. Aldolase B-mediated fructose metabolism drives metabolic reprogramming of colon cancer liver metastasis. Cell Metabol. 2018;27:1249. doi: 10.1016/j.cmet.2018.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fan X., Liu H., Liu M., Wang Y., Qiu L., Cui Y. Increased utilization of fructose has a positive effect on the development of breast cancer. PeerJ. 2017;5:e3804. doi: 10.7717/peerj.3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maughan D.W., Henkins J.A., Vigoreaux J.O. Concentrations of glycolytic enzymes and other cytosolic proteins in the diffusible fraction of a vertebrate muscle proteome. Mol Cell Proteom. 2005;4:1541–1549. doi: 10.1074/mcp.M500053-MCP200. [DOI] [PubMed] [Google Scholar]
- 34.Srere P.A. The infrastructure of the mitochondrial matrix. Trends Biochem Sci. 1980;5:120–121. [Google Scholar]
- 35.Lane A.N., Higashi R.M., Fan T.W.-M. Preclinical models for interrogating drug action in human cancers using Stable Isotope Resolved Metabolomics. Metabolomics. 2016;12:1–15. doi: 10.1007/s11306-016-1065-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.TeSlaa T., Teitell M.A. Techniques to monitor glycolysis. Methods Enzymol. 2014;542:91–114. doi: 10.1016/B978-0-12-416618-9.00005-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sun S.Z., Empie M.W. Fructose metabolism in humans – what isotopic tracer studies tell us. Nutr Metab. 2012;9:89. doi: 10.1186/1743-7075-9-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu M., Neilson A., Swift A.L. Multiparameter metabolic analysis reveals a close link between attenuated mitochondrial bioenergetic function and enhanced glycolysis dependency in human tumor cells. Am J Physiol Cell Physiol. 2007;292:C125–C136. doi: 10.1152/ajpcell.00247.2006. [DOI] [PubMed] [Google Scholar]
- 39.Lane A.N., Fan T.W.-M., Higashi R.M. Metabolic acidosis and the importance of balanced equations. Metabolomics. 2009;5:163–165. [Google Scholar]
- 40.Ipata P.L., Balestri F. Glycogen as a fuel: metabolic interaction between glycogen and ATP catabolism in oxygen-independent muscle contraction. Metabolomics. 2012;8:736–741. [Google Scholar]
- 41.Mehdi Damaghi J.W.G. pH sensing and regulation in cancer. Front Physiol. 2013;4 doi: 10.3389/fphys.2013.00370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mboge M.Y., Mahon B.P., McKenna R., Frost S.C. Carbonic anhydrases: role in pH control and cancer. Metabolites. 2018;8:19. doi: 10.3390/metabo8010019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang Y.P., Zhou W., Wang J. Arginine methylation of MDH1 by CARM1 inhibits glutamine metabolism and suppresses pancreatic cancer. Mol Cell. 2016;64:673–687. doi: 10.1016/j.molcel.2016.09.028. [DOI] [PubMed] [Google Scholar]
- 44.Jiang L., Shestov A.A., Swain P. Reductive carboxylation supports redox homeostasis during anchorage-independent growth. Nature. 2016;532:255–258. doi: 10.1038/nature17393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lane A.N., Tan J., Wang Y., Yan J., Higashi R.M., Fan T.W.-M. Probing the metabolic phenotype of breast cancer cells by multiple tracer Stable Isotope Resolved Metabolomics. Metab Eng. 2017;43:125–136. doi: 10.1016/j.ymben.2017.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.DeBerardinis R.J., Mancuso A., Daikhin E. Beyond aerobic glycolysis: transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc Natl Acad Sci USA. 2007;104:19345–19350. doi: 10.1073/pnas.0709747104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mullen A.R., Wheaton W.W., Jin E.S. Reductive carboxylation supports growth in tumour cells with defective mitochondria. Nature. 2011;481:385–388. doi: 10.1038/nature10642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tedeschi P.M., Markert E.K., Gounder M. Contribution of serine, folate and glycine metabolism to the ATP, NADPH and purine requirements of cancer cells. Cell Death Dis. 2013;4 doi: 10.1038/cddis.2013.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Centelles J.J., Ramos-Montoya A., Lim S. Metabolic profile and quantification of deoxyribose synthesis pathways in HepG2 cells. Metabolomics. 2007;3:105–111. [Google Scholar]
- 50.Le A., Lane A.N., Hamaker M. Myc induction of hypoxic glutamine metabolism and a glucose-independent TCA cycle in human B lymphocytes. Cell Metabol. 2012;15:110–121. doi: 10.1016/j.cmet.2011.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vander Heiden M.G., Locasale J.W., Swanson K.D. Evidence for an alternative glycolytic pathway in rapidly proliferating cells. Science. 2010;329:1492–1499. doi: 10.1126/science.1188015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Davidson S.M., Papagiannakopoulos T., Olenchock B.A. Environment impacts the metabolic dependencies of ras-driven non-small cell lung cancer. Cell Metabol. 2016;23:517–528. doi: 10.1016/j.cmet.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xie J., Wu H., Dai C. Beyond Warburg effect – dual metabolic nature of cancer cells. Sci Rep. 2014;4:4927. doi: 10.1038/srep04927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hensley C.T., Faubert B., Yuan Q. Metabolic heterogeneity in human lung tumors. Cell. 2016;164:681–694. doi: 10.1016/j.cell.2015.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wishart D.S., Guo A.C., Wilson M. HMDB 3.0-the human metabolome database in 2013. Nucleic Acids Res. 2013;41:D801–D807. doi: 10.1093/nar/gks1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yao C.-H., Fowle-Grider R., Mahieu N.G. Exogenous fatty acids are the preferred source of membrane lipids in proliferating fibroblasts. Cell Chem Biol. 2016;23:483–493. doi: 10.1016/j.chembiol.2016.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mackenzie C.G., Mackenzie J.B., Reiss O.K., Wisneski J.A. Identification of albumin-bound fatty acids as major factor in serum-induced lipid accumulation by cultured cells. JLR (J Lipid Res) 1970;11:571. [PubMed] [Google Scholar]
- 58.Zaytseva Y.Y., Rychahou P.G., Le A.-T. Preclinical evaluation of novel fatty acid synthase inhibitors in primary colorectal cancer cells and a patient-derived xenograft model of colorectal cancer. Oncotarget. 2018;9:24787–24800. doi: 10.18632/oncotarget.25361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zaytseva Y.Y., Harris J.W., Mitov M.I. Increased expression of fatty acid synthase provides a survival advantage to colorectal cancer cells via upregulation of cellular respiration. Oncotarget. 2015;6:18891–18904. doi: 10.18632/oncotarget.3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hosios A.M., Li Z., Lien E.C., Vander Heiden M.G. Preparation of lipid-stripped serum for the study of lipid metabolism in cell culture. Dev Cell. 2018;8:e2876. doi: 10.21769/BioProtoc.2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chihanga T., Hausmann S.M., Ni S., Kennedy M.A. Influence of media selection on NMR based metabolic profiling of human cell lines. Metabolomics. 2018;14:28. doi: 10.1007/s11306-018-1323-2. [DOI] [PubMed] [Google Scholar]
- 62.Fan T.W.-M., El-Amouri S.S., Macedo J.K.A., Wang Q.J., Cassel T.A., Lane A.N. Mapping metabolic networks in 3D spheroids using stable isotope-resolved metabolomics. Metabolites. 2018;8:40. doi: 10.3390/metabo8030040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tayek J.A., Katz J. Glucose production, recycling, Cori cycle, and gluconeogenesis in humans: relationship to serum cortisol. Am J Physiol. 1997;272:E476–E484. doi: 10.1152/ajpendo.1997.272.3.E476. [DOI] [PubMed] [Google Scholar]
- 64.Chatham J.C. Lactate – the forgotten fuel! J Physiol. 2002;542:333. doi: 10.1113/jphysiol.2002.020974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Khairallah M., Labarthe F., Bouchard B., Danialou G.T., Petrof B.J., Des Rosiers C. Profiling substrate fluxes in the isolated working mouse heart using C-13-labeled substrates: focusing on the origin and fate of pyruvate and citrate carbons. Am J Physiol Heart Circ Physiol. 2004;286:H1461–H1470. doi: 10.1152/ajpheart.00942.2003. [DOI] [PubMed] [Google Scholar]
- 66.van Hall G., Strømstad M., Rasmussen P. Blood lactate is an important energy source for the human brain. J Cereb Blood Flow Metab. 2009;29:1121–1129. doi: 10.1038/jcbfm.2009.35. [DOI] [PubMed] [Google Scholar]
- 67.Costalat R., Aubert A., Magistretti P., Pellerin L. Is the lactate a major energy substrate for the neurons? M-S (Med Sci) 2006;22:356–357. doi: 10.1051/medsci/2006224356. [DOI] [PubMed] [Google Scholar]
- 68.Hui S., Ghergurovich J.M., Morscher R.J. Glucose feeds the TCA cycle via circulating lactate. Nature. 2017;551:115. doi: 10.1038/nature24057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Faubert B., Li K.Y., Cai L. Lactate metabolism in human lung tumors. Cell. 2017;171:358. doi: 10.1016/j.cell.2017.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Updegraff B.L., Zhou X., Guo Y. Transmembrane protease TMPRSS11B promotes lung cancer growth by enhancing lactate export and glycolytic metabolism. Cell Rep. 2018;25:2223. doi: 10.1016/j.celrep.2018.10.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chen Y., Jr., Mahieu N.G., Huang X. Lactate metabolism is associated with mammalian mitochondria. Nat Chem Biol. 2016;12:937. doi: 10.1038/nchembio.2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wishart D.S., Jewison T., Guo A.C. HMDB 3.0-the human metabolome database in 2013. Nucleic Acids Res. 2013;41:D801–D807. doi: 10.1093/nar/gks1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lieberman M., Marks A.D. Wolters Kluwer; 2009. In Mark's Basic Medical Biochemistry: A Clinical Approach 410. [Google Scholar]
- 74.Halestrap A.P., Meredith D. The SLC16 gene family—from monocarboxylate transporters (MCTs) to aromatic amino acid transporters and beyond. Pflüg Arch. 2004;447:619–628. doi: 10.1007/s00424-003-1067-2. [DOI] [PubMed] [Google Scholar]
- 75.Halestrap A.P., Wilson M.C. The monocarboxylate transporter family--role and regulation. IUBMB Life. 2012;64:109–119. doi: 10.1002/iub.572. [DOI] [PubMed] [Google Scholar]
- 76.García-Cañaveras J.C., Chen L., Joshua D., Rabinpwitz The tumor metabolic microenvironment: lessons from lactate. Cancer Res. 2019;79:3155–3162. doi: 10.1158/0008-5472.CAN-18-3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gatenby R.A., Gillies R.J. Hypoxia and metabolism - opinion - A microenvironmental model of carcinogenesis. Nat Rev Cancer. 2008;8:56–61. doi: 10.1038/nrc2255. [DOI] [PubMed] [Google Scholar]
- 78.Schwert G.W. The estimation of kinetic constants for the lactate dehydrogenase system by the use of integrated rate equations. J Biol Chem. 1969;244:1285–1290. [PubMed] [Google Scholar]
- 79.Lerche M.H., Jensen P.R., Karlsson M., Meier S. NMR insights into the inner workings of living cells. Anal Chem. 2015;87:119–132. doi: 10.1021/ac501467x. [DOI] [PubMed] [Google Scholar]
- 80.Sellers K., Fox M.P., Bousamra M. Pyruvate carboxylase is critical for non-small-cell lung cancer proliferation. J Clin Investig. 2015;125:687–698. doi: 10.1172/JCI72873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Xie H., Hanai J., Ren J.-G. Targeting lactate dehydrogenase-A (LDH-A) inhibits tumorigenesis and tumor progression in mouse models of lung cancer and impacts tumor initiating cells. Cell Metabol. 2014;19:795–809. doi: 10.1016/j.cmet.2014.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Fan T.W., Lane A.N., Higashi R.M. Altered regulation of metabolic pathways in human lung cancer discerned by 13C stable isotope-resolved metabolomics (SIRM) Mol Cancer. 2009;8:41. doi: 10.1186/1476-4598-8-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Christen S., Lorendeau D., Schmieder R. Breast cancer-derived lung metastases show increased pyruvate carboxylase-dependent anaplerosis. Cell Rep. 2016;17:837–848. doi: 10.1016/j.celrep.2016.09.042. [DOI] [PubMed] [Google Scholar]
- 84.Passarella S., Paventi G., Pizzuto R. The mitochondrial L-lactate dehydrogenase affair. Front Neurosci. 2014;8 doi: 10.3389/fnins.2014.00407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kennedy B.W.C., Kettunen M.I., Hu D.E., Brindle K.M. Probing lactate dehydrogenase activity in tumors by measuring hydrogen/deuterium exchange in hyperpolarized L- 1-C-13,U-H-2 lactate. J Am Chem Soc. 2012;134:4969–4977. doi: 10.1021/ja300222e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Brindle K.M. Imaging metabolism with hyperpolarized C-13-Labeled cell substrates. J Am Chem Soc. 2015;137:6418–6427. doi: 10.1021/jacs.5b03300. [DOI] [PubMed] [Google Scholar]
- 87.Schug Z.T., Peck B., Jones D.T. Acetyl-CoA synthetase 2 promotes acetate utilization and maintains cancer cell growth under metabolic stress. Cancer Cell. 2015;27:57–71. doi: 10.1016/j.ccell.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sutendra G., Kinnaird A., Dromparis P. A nuclear pyruvate dehydrogenase complex is important for the generation of acetyl-CoA and histone acetylation. Cell. 2014;158:84–97. doi: 10.1016/j.cell.2014.04.046. [DOI] [PubMed] [Google Scholar]
- 89.Lyssiotis C.A., Cantley L.C. Acetate fuels the cancer engine. Cell. 2014;159:1492–1494. doi: 10.1016/j.cell.2014.12.009. [DOI] [PubMed] [Google Scholar]
- 90.Comerford S.A., Huang Z., Du X. Acetate dependence of tumors. Cell. 2014;159:1591–1602. doi: 10.1016/j.cell.2014.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bulusu V., Tumanov S., Michalopoulou E. Acetate recapturing by nuclear acetyl-CoA synthetase 2 prevents loss of histone acetylation during oxygen and serum limitation. Cell Rep. 2017;18:647–658. doi: 10.1016/j.celrep.2016.12.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhao S., Torres A., Henry R.A. ATP-citrate lyase controls a glucose-to-acetate metabolic switch. Cell Rep. 2016;17:1037–1052. doi: 10.1016/j.celrep.2016.09.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Schug Z.T., Vande Voorde J., Gottlieb E. The metabolic fate of acetate in cancer. Nat Rev Cancer. 2016;16:708–717. doi: 10.1038/nrc.2016.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Carrer A., Trefely S., Zhao S. Acetyl-CoA metabolism supports multi-step pancreatic tumorigenesis. Cancer Discov. 2019;9:416–435. doi: 10.1158/2159-8290.CD-18-0567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Liu X., Cooper D.E., Cluntun A.A. Acetate production from glucose and coupling to mitochondrial metabolism in mammals. Cell. 2018;175:502. doi: 10.1016/j.cell.2018.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lee J.V., Berry C.T., Kim K. Acetyl-CoA promotes glioblastoma cell adhesion and migration through Ca2+-NFAT signaling. Genes Dev. 2018;32:497–511. doi: 10.1101/gad.311027.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Courtney K.D., Bezwada D., Mashimo T. Isotope tracing of human clear cell renal cell carcinomas demonstrates suppressed glucose oxidation in vivo. Cell Metabol. 2018;28:793. doi: 10.1016/j.cmet.2018.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Patel A.B., De Graaf R.A., Rothman D.L., Behar K.L., Mason G.F. Evaluation of cerebral acetate transport and metabolic rates in the rat brain in vivo using H-1- C-13 -NMR. J Cereb Blood Flow Metab. 2010;30:1200–1213. doi: 10.1038/jcbfm.2010.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Marin-Valencia I., Ali Hooshyar M., Pichumani K., Sherry A.D., Malloy C.R. The ratio of acetate-to-glucose oxidation in astrocytes from a single 13C NMR spectrum of cerebral cortex. J Neurochem. 2014;132:99–109. doi: 10.1111/jnc.12948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Liu L., Zhong S., Yang R. Expression, purification, and initial characterization of human alanine aminotransferase (ALT) isoenzyme 1 and 2 in High-five insect cells. Protein Expr Purif. 2008 Aug;60(2):225–231. doi: 10.1016/j.pep.2008.04.006. 60, 225-231 (2008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yuneva M., Zamboni N., Oefner P., Sachidanandam R., Lazebnik Y. Deficiency in glutamine but not glucose induces MYC-dependent apoptosis in human cells. JCB (J Cell Biol) 2007;178:93–105. doi: 10.1083/jcb.200703099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Yuneva M.O., Fan T.W.-M., Allen T.D. The metabolic profile of tumors depends on both the responsible genetic lesion and tissue type. Cell Metabol. 2012;15:157–170. doi: 10.1016/j.cmet.2011.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Shanware N.P., Mullen A.R., DeBerardinis R.J., Abraham R.T. Glutamine: pleiotropic roles in tumor growth and stress resistance. J Mol Med. 2011;89:229–236. doi: 10.1007/s00109-011-0731-9. [DOI] [PubMed] [Google Scholar]
- 104.DeBerardinis R.J., Cheng T. Q's next: the diverse functions of glutamine in metabolism, cell biology and cancer. Oncogene. 2010;29:313–324. doi: 10.1038/onc.2009.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Cluntun A.A., Lukey M.J., Cerione R.A., Locasale J.W. Glutamine metabolism in cancer: understanding the heterogeneity. Trends Cancer. 2017;3:169–180. doi: 10.1016/j.trecan.2017.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sellers K., Allen T., Bousamra M. Metabolic reprogramming and Notch activity distinguish between non-small cell lung cancer subtypes. Br J Canc. 2019;121:51–64. doi: 10.1038/s41416-019-0464-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Labuschagne C., van den Broek N., Mackay G., Vousden K., Maddocks O.D. Serine, but not glycine, supports one-carbon metabolism and proliferation of cancer cells. Cell Rep. 2014;7:1248–1258. doi: 10.1016/j.celrep.2014.04.045. [DOI] [PubMed] [Google Scholar]
- 108.Ben-Sahra I., Hoxhaj G., Ricoult S.J.H., Asara J.M., Manning B.D. mTORC1 induces purine synthesis through control of the mitochondrial tetrahydrofolate cycle. Science. 2016;351:728–733. doi: 10.1126/science.aad0489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Jain M., Nilsson R., Sharma S. Metabolite profiling identifies a key role for glycine in rapid cancer cell proliferation. Science. 2012;366:1040–1044. doi: 10.1126/science.1218595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Locasale J.W., Grassian A.R., Melman T. Phosphoglycerate dehydrogenase diverts glycolytic flux and contributes to oncogenesis. Nat Genet. 2011;43:869–879. doi: 10.1038/ng.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Reid M.A., Allen A.E., Liu S. Serine synthesis through PHGDH coordinates nucleotide levels by maintaining central carbon metabolism. Nat Commun. 2018;9 doi: 10.1038/s41467-018-07868-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Possemato R., Marks K.M., Shaul Y.D. Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature. 2011;476:346–350. doi: 10.1038/nature10350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.DeNicola G.M., Chen P.-H., Mullarky E. NRF2 regulates serine biosynthesis in non-small cell lung cancer. Nat Genet. 2015;47:1475. doi: 10.1038/ng.3421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Maddocks O.D.K., Athineos D., Cheung E.C. Modulating the therapeutic response of tumours to dietary serine and glycine starvation. Nature. 2017;544:372–376. doi: 10.1038/nature22056. [DOI] [PubMed] [Google Scholar]
- 115.Fan T.W.-M., Bruntz R.C., Yang Y. De novo synthesis of serine and glycine fuels purine nucleotide biosynthesis in human lung cancer tissues. J Biol Chem. 2019;294(294):13464–13477. doi: 10.1074/jbc.RA119.008743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Sullivan L.B., Gui D.Y., Hosios A.M. Supporting aspartate biosynthesis is an essential function of respiration in proliferating cells. Cell. 2015;162:552–563. doi: 10.1016/j.cell.2015.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Garcia-Bermudez J., Baudrier L., La K. Aspartate is a limiting metabolite for cancer cell proliferation under hypoxia and in tumours. Nat Cell Biol. 2018;20:775–781. doi: 10.1038/s41556-018-0118-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Pavlova N.N., Hui S., Ghergurovich J.M. As extracellular glutamine levels decline, asparagine becomes an essential amino acid. Cell Metabol. 2018;27:428. doi: 10.1016/j.cmet.2017.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Jiang J., Pavlova N.N., Zhang J. Asparagine, a critical limiting metabolite during glutamine starvation. Mol Cell Oncol. 2018;5 doi: 10.1080/23723556.2018.1441633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Nakamura Akito, Nambu Tadahiro, Ebara Shunsuke. Inhibition of GCN2 sensitizes ASNS-low cancer cells to asparaginase by disrupting the amino acid response. Proc Natl Acad Sci. 2018;115:E7776–E7785. doi: 10.1073/pnas.1805523115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Krall A.S., Xu S., Graeber T.G., Braas D., Christofk H.R. Asparagine promotes cancer cell proliferation through use as an amino acid exchange factor. Nat Commun. 2016;7:11457. doi: 10.1038/ncomms11457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Skoulidis F., Byers L.A., Diao L.X. Co-occurring genomic alterations define major subsets of KRAS-mutant lung adenocarcinoma with distinct biology, immune profiles, and therapeutic vulnerabilities. Cancer Discov. 2015;5:860–877. doi: 10.1158/2159-8290.CD-14-1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Cassidy J.W., Caldas C., Bruna A. Maintaining tumor heterogeneity in patient-derived tumor xenografts. Cancer Res. 2015;75:2963–2968. doi: 10.1158/0008-5472.CAN-15-0727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Fan T.W.-M., Warmoes M.O., Sun Q. Distinctly perturbed metabolic networks underlie differential tumor tissue damages induced by immune modulator β-glucan in a two-case ex vivo non-small cell lung cancer study. Molec Case Stud. 2016;2:a000893. doi: 10.1101/mcs.a000893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Torres L., Ribeiro F.R., Pandis N., Andersen J.A., Heim S., Teixeira M.R. Intratumor genomic heterogeneity in breast cancer with clonal divergence between primary carcinomas and lymph node metastases. Breast Canc Res Treat. 2007;102:143–155. doi: 10.1007/s10549-006-9317-6. [DOI] [PubMed] [Google Scholar]
- 126.Dang C.V., Kim J.W., Gao P., Yustein J. Hypoxia and metabolism - opinion - the interplay between MYC and HIF in cancer. Nat Rev Cancer. 2008;8:51–56. doi: 10.1038/nrc2274. [DOI] [PubMed] [Google Scholar]
- 127.Schroeder T., Yuan H., Viglianti B.L. Spatial heterogeneity and oxygen dependence of glucose consumption in R3230Ac and fibrosarcomas of the fischer 344 rat. Cancer Res. 2005;65:5163–5171. doi: 10.1158/0008-5472.CAN-04-3900. [DOI] [PubMed] [Google Scholar]
- 128.Fukumura D., Duda D.G., Munn L.L., Jain R.K. Tumor microvasculature and microenvironment: novel insights through intravital imaging in pre-clinical models. Microcirculation. 2010;17:206–225. doi: 10.1111/j.1549-8719.2010.00029.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Fukumura D., Jain R.K. Tumor microvasculature and microenvironment: targets for anti-angiogenesis and normalization. Microvasc Res. 2007;74:72–84. doi: 10.1016/j.mvr.2007.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Cardenas-Navia L.I., Mace D., Richardson R.A., Wilson D.F., Shan S., Dewhirst M.W. The pervasive presence of fluctuating oxygenation in tumors. Cancer Res. 2008;68:5812–5819. doi: 10.1158/0008-5472.CAN-07-6387. [DOI] [PubMed] [Google Scholar]
- 131.Kamphorst J.J., Nofal M., Commisso C. Human pancreatic cancer tumors are nutrient poor and tumor cells actively scavenge extracellular protein. Cancer Res. 2015;75:544–553. doi: 10.1158/0008-5472.CAN-14-2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Gullino P.M., Clark S.H., Grantham F.H. The interstial fluid of solid tumors. Cancer Res. 1964;24:780–797. [PubMed] [Google Scholar]
- 133.Jain R.K. Transport of molecules in the tumor interstitium: a review. Cancer Res. 1987;47:3039–3051. [PubMed] [Google Scholar]
- 134.Sonveaux P., Vegran F., Schroeder T. Targeting lactate-fueled respiration selectively kills hypoxic tumor cells in mice. J Clin Investig. 2008;118:3930–3942. doi: 10.1172/JCI36843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Kelly M., Kennedy B., MT (ASCP), Dewhirst Mark W. Tumor metabolism of lactate: the influence and therapeutic potential for MCT and CD147 regulation. Future Oncol. 2010;6:127. doi: 10.2217/fon.09.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Lewerenz J., Hewett S.J., Huang Y. The cystine/glutamate antiporter system x(c)(-) in health and disease: from molecular mechanisms to novel therapeutic opportunities. Antioxidants Redox Signal. 2013;18:522–555. doi: 10.1089/ars.2011.4391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Binnewies M., Roberts E.W., Kersten K. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat Med. 2018;24:541–550. doi: 10.1038/s41591-018-0014-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Scharping N.E., Delgoffe G.M. Tumor microenvironment metabolism: a new checkpoint for anti-tumor immunity. Vaccines. 2016;4 doi: 10.3390/vaccines4040046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Sun C., Mezzadra R., Schumacher T.N. Regulation and function of the PD-L1 checkpoint. Immunity. 2018;48:434–452. doi: 10.1016/j.immuni.2018.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Henze A.T., Mazzone M. The impact of hypoxia on tumor-associated macrophages. J Clin Investig. 2016;126:3672–3679. doi: 10.1172/JCI84427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Li Y., Patel S.P., Roszik J., Qin Y. Hypoxia-driven immunosuppressive metabolites in the tumor microenvironment: new approaches for combinational immunotherapy. Front Immunol. 2018;9 doi: 10.3389/fimmu.2018.01591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Yang M., McKay D., Pollard J.W., Lewis C.E. Diverse functions of macrophages in different tumor microenvironments. Cancer Res. 2018;78:5492–5503. doi: 10.1158/0008-5472.CAN-18-1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Shapouri-Moghaddam A., Mohammadian S., Vazini H. Macrophage plasticity, polarization, and function in health and disease. J Cell Physiol. 2018;233:6425–6440. doi: 10.1002/jcp.26429. [DOI] [PubMed] [Google Scholar]
- 144.Liu M., Luo F., Ding C. Dectin-1 activation by a natural product beta-glucan converts immunosuppressive macrophages into an M1-like phenotype. J Immunol. 2015;195:5055–5065. doi: 10.4049/jimmunol.1501158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Mills E.L., Ryan D.G., Prag H.A. Itaconate is an anti-inflammatory metabolite that activates Nrf 2 via alkylation of KEAP1. Nature. 2018;556:113. doi: 10.1038/nature25986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Frantz C., Stewart K.M., Weaver V.M. The extracellular matrix at a glance. J Cell Sci. 2010;123:4195–4200. doi: 10.1242/jcs.023820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Kamphorst J.J., Cross J.R., Fan J. Hypoxic and Ras-transformed cells support growth by scavenging unsaturated fatty acids from lysophospholipids. Proc Natl Acad Sci USA. 2013;110:8882–8887. doi: 10.1073/pnas.1307237110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Commisso C., Davidson S.M., Soydaner-Azeloglu R.G. Macropinocytosis of protein is an amino acid supply route in Ras-transformed cells. Nature. 2013;497:633. doi: 10.1038/nature12138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Olivares O., Mayers J.R., Gouirand V. Collagen-derived proline promotes pancreatic ductal adenocarcinoma cell survival under nutrient limited conditions. Nat Commun. 2017;8 doi: 10.1038/ncomms16031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Michalopoulou E., Bulusu V., Kamphorst J.J. Metabolic scavenging by cancer cells: when the going gets tough, the tough keep eating. Br J Canc. 2016;115:635–640. doi: 10.1038/bjc.2016.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Goossens P., Rodriguez-Vita J., Etzerodt A. Membrane cholesterol efflux drives tumor-associated macrophage reprogramming and tumor progression. Cell Metabol. 2019;29:1376–1389. doi: 10.1016/j.cmet.2019.02.016. [DOI] [PubMed] [Google Scholar]
- 152.Sun R.C., Fan T.W.-M., Deng P. Noninvasive liquid diet delivery of stable isotopes into mouse models for deep metabolic network tracing. Nat Commun. 2017;8:1646. doi: 10.1038/s41467-017-01518-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Cheshkov S.1, Dimitrov I.E.1, Jakkamsetti V.4. Oxidation of [U-13 C]glucose in the human brain at 7T under steady state conditions. Magn Reson Med. 2017;73:2065–2071. doi: 10.1002/mrm.26603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Maher E.A., Marin-Valencia I., Bachoo R.M. Metabolism of U-13C glucose in human brain tumors in vivo. NMR Biomed. 2012;25:1234–1244. doi: 10.1002/nbm.2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Gordon S.R.1., Maute R.L., Dulken B.W. PD-1 expression by tumour-associated macrophages inhibits phagocytosis and tumour immunity. Nature. 2017;545:495–499. doi: 10.1038/nature22396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Almatroodi S.A., McDonald C.F., Pouniotis D.S. Alveolar macrophage polarisation in lung cancer. Lung Cancer Int. 2014;2014 doi: 10.1155/2014/721087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Loose D., Van de Wiele C. The immune system and cancer. Cancer Biother Radiopharm. 2009;24:369–376. doi: 10.1089/cbr.2008.0593. [DOI] [PubMed] [Google Scholar]
- 158.Wang R., Dillon C.P., Shi L.Z. The transcription factor Myc controls metabolic reprogramming upon T lymphocyte activation. Immunity. 2011;35:871–882. doi: 10.1016/j.immuni.2011.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Rosenthal R., Cadieux E.L., Salgado R. Nature. 2019:479–485. doi: 10.1038/s41586-019-1032-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Edin S., Wikberg M.L., Dahlin A.M. The distribution of macrophages with a M1 or M2 phenotype in relation to prognosis and the molecular characteristics of colorectal cancer. PLoS One. 2012;7:e47045. doi: 10.1371/journal.pone.0047045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Jackute J., Zemaitis M., Pranys D. Distribution of M1 and M2 macrophages in tumor islets and stroma in relation to prognosis of non-small cell lung cancer. BMC Immunol. 2018;19:3. doi: 10.1186/s12865-018-0241-4. [DOI] [PMC free article] [PubMed] [Google Scholar]