Abstract
As a key technique for the CRISPR-Cas9 system, identification of single-guide RNAs (sgRNAs) on-target activity is critical for both theoretical research (investigation of RNA functions) and real-world applications (genome editing and synthetic biology). Because of its importance, several computational predictors have been proposed to predict sgRNAs on-target activity. All of these methods have clearly contributed to the developments of this very important field. However, they are suffering from certain limitations. We proposed two new methods called “sgRNA-PSM” and “sgRNA-ExPSM” for sgRNAs on-target activity prediction via capturing the long-range sequence information and evolutionary information using a new way to reduce the dimension of the feature vector to avoid the risk of overfitting. Rigorous leave-one-gene-out cross-validation on a benchmark dataset with 11 human genes and 6 mouse genes, as well as an independent dataset, indicated that the two new methods outperformed other competing methods. To make it easier for users to use the proposed sgRNA-PSM predictor, we have established a corresponding web server, which is available at http://bliulab.net/sgRNA-PSM/.
Keywords: sgRNAs on-target activity, position-specific mismatch, XGBoost
Introduction
Three main genome editing tools, including zinc-finger nucleases (ZFNs),1 transcription activator-like effector nucleases (TALENs),2 and CRISPR-Cas9 RNA-guided technologies,3,4, can be used to recognize and cleave specific DNA sequences.5 Compared with ZFNs and TALENs, CRISPR-Cas9 has been widely applied in various cell types and organisms in recent years. In the type II CRISPR-Cas9 system, single-guide RNA (sgRNA) directs Cas9 protein to the target site to cleave the DNA target sequences, and sgRNA should be designed to have around a 20-nt sequence to be complementary to the guide sequence in the DNA target sequences.6,7. Rational design of sgRNA is a crucial part for CRISPR-Cas9. Therefore, the prediction of sgRNAs on-target activity is very important for CRISPR-Cas9.
Researchers have proposed several computational methods for sgRNAs on-target activity prediction. Most of them treat the prediction problem of sgRNA as a binary classification task or a regression task, and the computational predictors were constructed based on machine learning algorithms. The differences between these approaches are feature extraction methods and machine learning techniques, such as gradient boosting regression (GBR),8 support vector machines (SVMs),9, 10, 11, 12, 13, 14, 15, 16, 17, 18 ensemble classifiers19, 20, 21, 22, 23, 24, and deep learning,25, 26, 27, 28, 29, 30, 31, 32 among others. As shown in the aforementioned studies,33,34 discriminative features are critical for constructing the computational predictors. Accordingly, some features have been proposed to capture the characteristics of sgRNAs, for example, because the position of a nucleotide in sgRNA will affect its activity, and thus the position-specific (PS)35 feature was proposed to incorporate these sequence patterns, which has been used in ge-CRISPR,36, Azimuth,37 and CRISPRpred.38 Kaur et al.36 proposed an integrated pipeline called ge-CRISPR to predict and analyze the genome editing efficiency of sgRNAs. Azimuth37 employed GBR to train the model, achieving state-of-the-art performance. CRISPRpred38 is another efficient predictor, combining the discriminative features selected by random forest (RF)39 and the SVM regression.
All of the aforementioned predictors have obtained encouraging results and played a role in the development of computational predictors for sgRNAs on-target activity prediction, but they are also suffering from some problems or limitations. Further work is required for the following reasons: (1) these predictors are only able to consider the short-range sequence information of the DNA sequences, otherwise they will cause “high-dimension disaster”;40,41 and (2) these predictors failed to incorporate the evolutionary information, ignoring information between non-consecutive nucleotides.
In order to solve these aforementioned problems, we proposed a novel feature, PS mismatch (PSM), sharing the advantages of both PS35 and mismatch features.41 RNA sequence evolution involves single nucleotides, insertions and deletions of several nucleotides, and other factors. With the long-term accumulation of these changes in evolution, although the similarities between the initial and the final RNA sequences are gradually reduced, these RNA sequences still have many features in common. PSM is such a method for extracting the evolutionary information from RNA sequences by allowing mismatches occurring in k-mers from specific positions.41 PSM has been applied to predict sgRNAs on-target activity, and two predictors were established called “sgRNA-PSM” and “sgRNA-ExPSM” (sgRNA-extended PSM). Finally, a corresponding web server has been constructed (http://bliulab.net/sgRNA-PSM/).
Results and Discussion
Parameter Optimization
According to Equations 9 and 10, there are two parameters in PSM, k and m, and three parameters in the XGBoost algorithm, C, R, and F. These parameters were optimized according to AUC (area under the curve) by using leave-one-gene-out cross-validation on the benchmark dataset S (cf. Equation 3). In this study, these parameters were optimized in the ranges listed in the following:
| (Equation 1) |
The final optimal values of the five parameters (cf. Equation 1) were optimized based on the AUC on the benchmark dataset S (cf. Equation 3), as given by
| (Equation 2) |
Feature Selection and Analysis
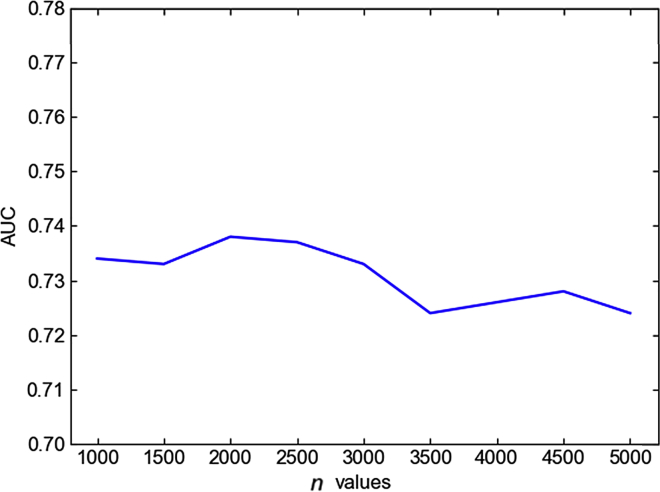
In order to remove the redundant features and reduce the dimension of the resulting feature vectors, here we used SelectKBest in scikit-learn42 to select the top number of features with the highest scores based on the scoring function f_regression, which can avoid the overfitting risk with low computational cost.43 We investigated the influence of the value n (number of selected features) in SelectKBest on the predictive performance of sgRNA-PSM, and the results are shown in Figure 1, from which we can see that the values of n have little impact on the performance, and sgRNA-PSM can achieve the best performance when n is equal to 2,000.
Figure 1.
Graph Showing AUC Scores of the sgRNA-PSM Predictors with Different n Values, where n Denotes the Number of Selected Features
The importance of each feature can be analyzed based on F_score. To explore the reason why the proposed sgRNA-PSM predictor works so well, we analyzed the contribution of each feature. Table 1 lists the 10 most important features, from which we can see that (1) the top 9 most important features belong to the features generated in the sequence positions from 23 to 30. In the CRISPR-Cas9 system, the DNA target sequences are composed of two parts:44 one is the guide sequence, and the other is the protospacer adjacent motif (PAM). The guide sequence is complementary to around a 20-bp sequence in sgRNA, and PAM is the downstream short sequence of the guide sequence6 and is recognized by the Cas9 protein.45 In the benchmark dataset S (cf. Equation 3), the guide sequence is in the sequence positions from 5 to 24, and PAM is the short sequence in the sequence positions from 25 to 27.37 Therefore, the top 9 most important features all cover PAM, indicating that the proposed PSM is able to incorporate this important sequence pattern. (2) PAM is composed of any nucleotide in sequence position 25 followed by GG in positions 26 and 27.6,37 7 of the 10 most important features capture this sequence pattern.
Table 1.
The 10 Most Important Features in the sgRNA-PSM Predictor
| No. | PSM Featurea | Sequence Positionb | F_scorec |
|---|---|---|---|
| 1 | *G*GG | 23–27 | 185.6 |
| 2 | G*GG* | 24–28 | 185.6 |
| 3 | C*G*G | 24–28 | 136.2 |
| 4 | C**GG | 24–28 | 136.2 |
| 5 | *C*GG | 23–27 | 129.0 |
| 6 | C*GG* | 24–28 | 129.0 |
| 7 | **GGG | 24–28 | 128.0 |
| 8 | *GGG* | 25–29 | 128.0 |
| 9 | GGG** | 26–30 | 128.0 |
| 10 | **TTC | 20–24 | 113.0 |
Parameters were k = 5, m = 2.
The sequence position of mismatches.
Calculated by F regression.
Comparison with Other Methods
The results obtained by sgRNA-PSM and sgRNA-ExPSM on the benchmark dataset S are listed in Table 2, from which we can see that the AUC achieved by sgRNA-PSM was 73.8%. The corresponding AUC achieved by sgRNA-ExPSM was even better, which was 74.4%. This is reasonable because the acid cut position and percent peptide features referred to in Equation 11 are complementary with the PSM features in Equation 9. The PSM feature vector reflects long-range sequence information, while the amino acid cut position and percent peptide are guide-positional features corresponding to the start distance of the protein coding region of the gene where the cleavage site of the sgRNA is positioned.37
Table 2.
List of AUC Scores Obtained by Various Methods via the Leave-One-Gene-Out Cross-Validation on the Same Benchmark Dataset S (cf. Equation 3)
AUC means the area under the ROC curve;56,57 the better predictor corresponds to larger AUC values.
Results obtained by in-house implementation from Doench et al.37
Results obtained by in-house implementation from Kaur et al.36
Results obtained by in-house implementation from Rahman and Rahman.38
For the proposed predictor in this article, see Equations 9 and 10 with k = 5, m = 2, = 3, R = 0.1, F = 800.
For the proposed predictor in this article, see Equations 10 and 11 with k = 5, m = 2, = 3, R = 0.1, F = 800.
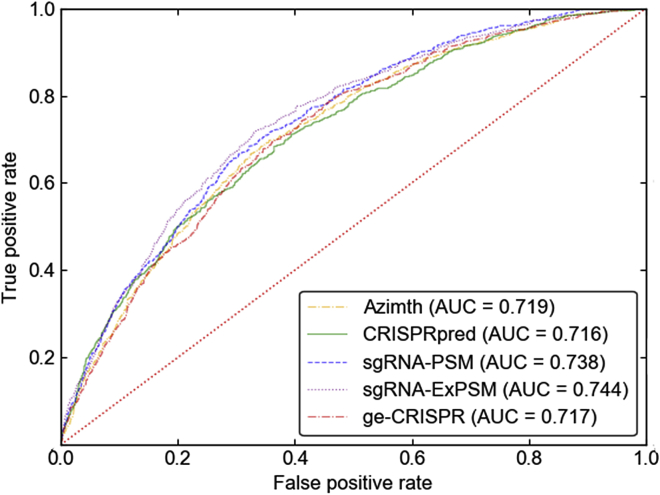
Then, we made a comparison of the sgRNA-PSM and sgRNA-ExPSM with ge-CRISPR,36 Azimth,37 and CRISPRpred.38 All of these predictors were examined by the leave-one-gene-out cross-validation on the benchmark dataset S (cf. Equation 3). For facilitating comparison, the corresponding results obtained by the ge-CRISPR predictor, the Azimth predictor, and the CRISPRpred predictor are also given in Table 2 and Figure 2. Here, Figure 2 includes the corresponding receiver operating characteristic (ROC) curves showing the performance of the five predictors. A diagonal from the point (0,0) to (1,1) means a random guess. The better performance of the predictor corresponds to a larger AUC.
Figure 2.
Graph Showing the Predictive Quality of the Aforementioned Predictors via the ROC Curves
The corresponding AUC scores are 0.717, 0.716, 0.719, 0.738, and 0.744 for ge-CRISPR, CRISPRpred, Azimth, sgRNA-PSM, and sgRNA-ExPSM predictors via the leave-one-gene-out cross-validation on the same benchmark dataset S, respectively.
The following conclusions can be drawn from Table 2 and Figure 2: (1) the AUC score achieved by the proposed sgRNA-PSM predictor is higher than that of ge-CRISPR, and even higher than those of Azimth and CRISPRpred based on the wet experiment features, such as amino acid cut position and percent peptide. Please note that these two features are not sequence-based features, and they are often unavailable. (2) The sgRNA-ExPSM predictor outperforms the sgRNA-PSM predictor by incorporating the amino acid cut position feature and percent peptide feature.
In addition, the sgRNA-PSM predictor was further compared with Azimuth37 and DeepCRISPR (pt+aug CNN)46 on the on-target dataset.46,47. In order to make a fair comparison, the sgRNA-PSM predictor was trained on the training set of on-target dataset reported in Chuai et al.46 and tested on the independent test dataset46 for the hct116, hela, and hl60 cell types. The hek293t dataset reported in Doench et al.37 is a subset of our benchmark dataset S (cf. Equation 3). Therefore, our method was not tested on the hek293t dataset again. For sgRNA-PSM, SelectKBest with the scoring function chi2 in scikit-learn was used to select 1,100 dimensions of the PSM features and fed into XGBoost for classification. The predictive results of sgRNA-PSM, DeepCRISPR (pt+aug CNN), and Azimuth are shown in Table 3. As shown in this table, our method outperformed Azimuth and DeepCRISPR (pt+aug CNN) on the hct116 and hela cell types, and it is highly comparable to DeepCRISPR (pt+aug CNN) on the hl60 cell type.
Table 3.
List of the AUC Scores Obtained by Various Methods on the On-Target Dataset Reported in Chuai et al.46
| Cell Typea | Methods | AUC (%) |
|---|---|---|
| hct116 | Azimuthb | 74.1 |
| DeepCRISPR (pt+aug CNN)c | 87.4 | |
| sgRNA-PSMd | 91.7 | |
| Retrained sgRNA-PSMe | 74.0 | |
| Hela | Azimuthb | 67.5 |
| DeepCRISPR (pt+aug CNN)c | 78.2 | |
| sgRNA-PSMd | 82.8 | |
| Retrained sgRNA-PSMe | 72.1 | |
| hl60 | Azimuthb | 79.2 |
| DeepCRISPR (pt+aug CNN)c | 73.9 | |
| sgRNA-PSMd | 77.6 | |
| Retrained sgRNA-PSMe | 83.7 |
The cell type of the independent test dataset.
Results reported in Chuai et al.46
Results reported in Chuai et al.46
The sgRNA-PSM predictor trained with the dataset reported in Chuai et al.;46 see Equations 9 and 10 with k = 4, m = 2, = 9, R = 0.05, F = 2,300.
The sgRNA-PSM predictor trained with each of the three datasets (hct116, hela, and hl60).
To further explore the reasons why our method cannot perform well on the hl60 cell type, we retrained the sgRNA-PSM classifier with each of the three datasets (hct116, hela, and hl60). For each dataset, 20% of the samples were used as the test dataset, which were stratified by labels following Chuai et al.,46 and the remaining 80% of the samples were used as the training dataset. The results are also listed in Table 3, from which we can see that the sgRNA-PSM trained with the hl60 dataset outperformed the corresponding classifier trained with the training data consisting of all four cell types, and it even outperformed Azimuth. The results are not surprising because the four different cell types have different data distributions. Noise information was introduced when combining all four cell types to train a computational predictor. Therefore, the overall performance of sgRNA-PSM is better than that of all of the other competing methods.
Web Server and User Guide
Providing a user-friendly and freely accessible web server for a new predictor can obviously improve its impact.48 To make it easier for users to use the proposed predictor, we established the corresponding sgRNA-PSM web server. Because the sgRNA-ExPSM predictor requires two features obtained from wet experiments, which are often unavailable, its corresponding web server is not able to be constructed. The web server has the following functions: (1) it allows users to input sgRNA sequences in reverse-complementary order, and (2) it allows users to input longer sequences (30–1,000 bp). The web server will detect all of the possible sgRNAs and predict their on-target activities. The steps for using the sgRNA-PSM web server are as follows:
Step 1. Click on the website address http://bliulab.net/sgRNA-PSM/ to open the sgRNA-PSM web server, at which point the homepage of the website will appear as shown in Figure 3. The detailed introduction to the web server can be obtained by clicking on the “Read Me” button.
Step 2. Click on the “Browse” button to upload the input file or type your query DNA sequences in FASTA format.
Step 3. Click on the “Submit” button to get the final predictive results. When inputting the four DNA sequences in the “Example” window, you will see that the first and second are predicted as high on-target activity sgRNAs, while the third is the sequence in reverse-complementary order, which is predicted as low on-target activity sgRNA, and the fourth has four low on-target activity sgRNAs and one high on-target activity sgRNA. These results are consistent with the experimental results. In order to help the users to solve the problems when using the web server, the Frequently Questioned Answers (FQA) are provided by clicking on the FQA button.
Figure 3.
Graphic of the Homepage of the Web Server http://bliulab.net/sgRNA-PSM/
Materials and Methods
Benchmark Datasets
In this study, a widely used benchmark dataset37 constructed by the FC dataset35 and the RES dataset37 was employed to evaluate the performance of different methods. The benchmark dataset consists of 5,310 sequences from 11 human genes (CD33, MED12, NF2, CD13, TADA2B, CUL3, TADA1, HPRT, NF1, CD15, CCDC101) and 6 mouse genes (Cd45, Cd43, Cd28,H2-K, Cd5, Thy1). There are 1,059 high on-target activity sgRNAs and 4,251 low on-target activity sgRNAs. The benchmark dataset S is as follows:
| (Equation 3) |
where
| (Equation 4) |
with
| (Equation 5) |
where represents the union symbol between two sets, denotes the subset whose sgRNAs are from the ith targeting gene, the positive subset contains high on-target activity sgRNAs, the negative subset contains the low on-target activity sgRNAs, represents the number of sgRNAs in , represents the number of sgRNAs in , and denotes the number of sgRNAs in and in a ratio of about 1:4. The corresponding detailed sequences can be found in Data S1.
The most updated on-target dataset established in Chuai et al.46 was employed to further evaluate the performance of the proposed method. This on-target dataset was constructed based on hct116,49 hek293t,37 hela,49 and hl60.50 Those datasets were also employed by Haeussler et al.47
PSM
Feature extraction is very important for constructing a computational predictor.51 Inspired by the PS35 and mismatch features,41 here a novel feature extraction method, PSM, was proposed to capture the long-range sequence information and evolutionary information. Furthermore, PSM is able to efficiently reduce the dimension of the feature vectors. The detailed procedures of generating PSM are described as follows.
A DNA sample D can be represented as follows:
| (Equation 6) |
where
| (Equation 7) |
represents the ith nucleobase in the sequence, the symbol denotes ‘‘member of’’ in the set, and L represents the length of D.
The PS feature is an important and useful feature extraction method widely used in previous studies.35, 36, 37, 38 Because the position of nucleotide in a sgRNA affects its activity, the PS feature incorporates the local sequence position information by representing the k-mers41,52 along a DNA sample D (cf. Equation 6) by “one-hot” encoding.53 By using the PS feature, D can be represented as follows:35, 36, 37, 38
| (Equation 8) |
where T represents the transpose symbol, denotes the jth feature in the one-hot encoding at the ith position in D, whose value is 0 or 1, and k is the number of adjacent nucleotides in a k-mer.
From Equation 8, we can see that the dimension of the PS vector will increase rapidly with the incensement of k values. For example, when k is equal to 6, the dimension of the PS feature vector will be 46 × (30 − 6 + 1) = 1.024 × 105, which will cause high-dimension disaster.40,41,54 Therefore, Equation 8 is useful only when k is small, and it ignores the information of non-consecutive nucleotides. As a result, it can only incorporate the short-range and consecutive nucleotide information without considering the long-range and non-consecutive nucleotide information.
The mismatch feature considers the evolutionary process and allows mismatches occurring in k-mers. Therefore, the dimension of the corresponding feature vectors can be obviously decreased compared with those of k-mers. In this study, we combined the mismatch with the PS feature and proposed a novel feature, i.e., PSM, which is defined as follows:
| (Equation 9) |
where represents the jth feature in one-hot encoding at the ith position in D, whose value is 0 or 1, and α denotes the number of mismatch features considering the one-hot encoding, which can be defined as follows:
| (Equation 10) |
where m is the number of mismatches in k-mers.
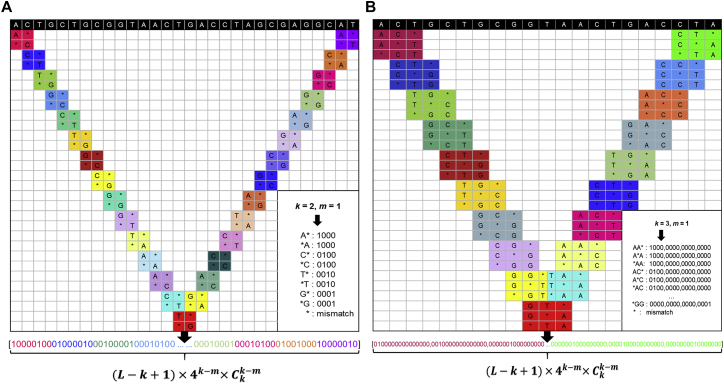
As shown in Equations 9 and 10, the first components reflect the one-hot-encoded feature vector corresponding to the first sequence position, whereas the components from to reflect the one-hot-encoded feature vector corresponding to the second sequence position, and so forth. A feature vector formed with components is called the PSM vector for D as defined in Equation 9. A schematic diagram illustrating how to generate the PSM vector for D is shown in Figure 4. Compared to the PS vector defined in Equation 8, the dimension of the PSM vector will be significantly reduced. For example, when k = 6, the PS feature vector’s dimension (cf. Equation 8) is 1.024 × 105, while the PSM feature vector’s dimension is as defined in Equations 9 and 10. Now, when we assume m =5, the dimension will be . The size of the latter is around 1/170th that of the former. Namely, PSM can obviously reduce the dimension of the feature vector compared with PS. It is especially true for larger k values (see Table 4).
Figure 4.
Schematic Diagram Illustrating How to Generate the PSM Vector for a DNA Sequence
(A) Example of PSM with parameters of k = 2, m = 1. (B) Example of PSM with parameters of k = 3, m = 1.
Table 4.
Comparison between the PS Feature Vector’s dimension (cf. Equation 8) and the PSM Feature Vector’s Dimension (cf. Equation 9)
| k | Dimension of PS Vectora | m | Dimension of PSM Vectorb | Ratio γc |
|---|---|---|---|---|
| 2 | 464 | 1 | 232 | ∼2 |
| 3 | 1,792 | 1 | 1,344 | ∼1.3 |
| 2 | 336 | ∼5.3 | ||
| 4 | 6,912 | 1 | 6,912 | 1 |
| 2 | 2,592 | ∼2.7 | ||
| 3 | 432 | ∼16 | ||
| 5 | 26,624 | 2 | 16,640 | ∼1.6 |
| 3 | 4,160 | ∼6.4 | ||
| 4 | 520 | ∼51.2 | ||
| 6 | 102,400 | 4 | 6,000 | ∼17.07 |
| 5 | 600 | ∼170.67 | ||
Calculated by Equation 8.
Calculated by Equation 9.
Ratio of the number of column 2 and the number of column 4; it is the same with , where m is given in column 3.
Therefore, the PSM vector (cf. Equation 9) should be used to represent the DNA samples, because PSM can overcome the aforementioned limitations for large values of k, while avoiding the high-dimension disaster problem.
Finally, we can augment the PSM vector (cf. Equation 9) to
| (Equation 11) |
where is the augmented PSM, a is the amino acid cut position, and b is the percent peptide given in Doench et al.37 Both of these two features were obtained by wet experiments, which are often unavailable. The feature vector formed with components is the ExPSM vector for D.
XGBoost Algorithm
The XGBoost algorithm55 is a technique for classification and regression tasks, which is based on tree boosting.8 The most important advantage of XGBoost is its scalability in all scenarios. For more detailed information on XGBoost, please refer to Chen and Guestrin.55
In this study, the regression model of the XGBoost algorithm was employed. We used the scikit-learn package42 to implement the XGBoost algorithm. The values of its three main parameters (maximum depth of a tree C, boosting learning rate R, and number of boosted trees F) are given in the following sections, and all the other parameters were set as default values.
Finally, according to Equations 9 and 11, two predictors have been proposed as follows:
| (Equation 12) |
Evaluation Method of Performance
The AUC, as it pertains to the ROC curve,56, 57, 58 is a widely used measure for evaluating the performance of the predictors. The better predictor corresponds to larger AUC values.
Cross-Validation
The cross-validation method is an important step for evaluating the performance of a predictor.59 In this study, in order to ensure that a predictor can be generalized across genes, the leave-one-gene-out cross-validation35,37 was used, where each of the 17 subsets of Si (cf. Equation 3) was selected one by one as the test set, while the other 16 subsets were used to construct the training set to train the predictor. This process was repeated for 17 times, and each subset was selected as the test set once.
Implementation of the Competing Methods
In this study, we compared the proposed methods with three state-of-the-art methods, including ge-CRISPR,36 Azimuth,37 and CRISPRpred.38 The detailed processes of these three approaches were introduced as follows: for ge-CRISPR, the 464 dinucleotide (1-degree) binary features were finally fed into SVM regressor with a radial basis function (RBF) kernel with a c value of 25 for regression. For Azimuth, seven features were used to represent the samples, including position-independent, position-specific, GC count, NGGN, thermodynamic features, amino acid cut position, and percent peptide. These features were combined with GBR with the parameters learning_rate = 0.1, max_depth = 3, and n_estimators = 100 to construct the predictor. For CRISPRpred, five different feature extraction methods were employed, including position-independent, position-specific, thermodynamic features, amino acid cut position, and percent peptide. Please note that ViennaRNA package version 2.060 was used to generate thermodynamic features. RF39 was then performed on these features to select 2,899 relevant features according to the importance scores (Mean Decrease Gini) with the maximum number of trees of 500. These features were finally fed into the SVM regressor with linear kernel function with a c value of 2−2 for regression.
Acknowledgments
This work was supported by the Beijing Natural Science Foundation (JQ19019); the National Natural Science Foundation of China (61822306 and 61672184); the Fok Ying-Tung Education Foundation for Young Teachers in the Higher Education Institutions of China (161063); and the Scientific Research Foundation in Shenzhen (JCYJ20180306172207178, JCYJ20180306172156841, and JCYJ20180507183608379).
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2020.01.029.
Supplemental Information
There are 1,059 high on-target activity sgRNAs and 4,251 low on-target activity sgRNAs. See the text of the paper for further explanation.
References
- 1.Urnov F.D., Miller J.C., Lee Y.L., Beausejour C.M., Rock J.M., Augustus S., Jamieson A.C., Porteus M.H., Gregory P.D., Holmes M.C. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature. 2005;435:646–651. doi: 10.1038/nature03556. [DOI] [PubMed] [Google Scholar]
- 2.Mussolino C., Morbitzer R., Lütge F., Dannemann N., Lahaye T., Cathomen T. A novel TALE nuclease scaffold enables high genome editing activity in combination with low toxicity. Nucleic Acids Res. 2011;39:9283–9293. doi: 10.1093/nar/gkr597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A., Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mali P., Yang L., Esvelt K.M., Aach J., Guell M., DiCarlo J.E., Norville J.E., Church G.M. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lander E.S. The heroes of CRISPR. Cell. 2016;164:18–28. doi: 10.1016/j.cell.2015.12.041. [DOI] [PubMed] [Google Scholar]
- 6.Hartenian E., Doench J.G. Genetic screens and functional genomics using CRISPR/Cas9 technology. FEBS J. 2015;282:1383–1393. doi: 10.1111/febs.13248. [DOI] [PubMed] [Google Scholar]
- 7.Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Friedman J.H. Greedy function approximation: a gradient boosting machine. Ann. Stat. 2001;29:1189–1232. [Google Scholar]
- 9.Fu X., Zhu W., Cai L., Liao B., Peng L., Chen Y., Yang J. Improved pre-miRNAs identification through mutual information of pre-miRNA sequences and structures. Front. Genet. 2019;10:119. doi: 10.3389/fgene.2019.00119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cai Y.D., Zhou G.P., Chou K.C. Support vector machines for predicting membrane protein types by using functional domain composition. Biophys. J. 2003;84:3257–3263. doi: 10.1016/S0006-3495(03)70050-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Suykens J.A.K., Vandewalle J. Least squares support vector machine classifiers. Neural Process. Lett. 1999;9:293–300. [Google Scholar]
- 12.Li D., Ju Y., Zou Q. Protein folds prediction with hierarchical structured SVM. Curr. Proteomics. 2016;13:79–85. [Google Scholar]
- 13.Liu B., Li C.C., Yan K. DeepSVM-fold: protein fold recognition by combining support vector machines and pairwise sequence similarity scores generated by deep learning networks. Brief. Bioinform. 2019 doi: 10.1093/bib/bbz098. Published online October 28, 2019. [DOI] [PubMed] [Google Scholar]
- 14.Fu X., Ke L., Cai L., Chen X., Ren X., Gao M. Improved prediction of cell-penetrating peptides via effective orchestrating amino acid composition feature representation. IEEE Access. 2019;7:163547–163555. [Google Scholar]
- 15.Lu X., Qian X., Li X., Miao Q., Peng S. DMCM: a data-adaptive mutation clustering method to identify cancer-related mutation clusters. Bioinformatics. 2019;35:389–397. doi: 10.1093/bioinformatics/bty624. [DOI] [PubMed] [Google Scholar]
- 16.Lu X., Li X., Liu P., Qian X., Miao Q., Peng S. The integrative method based on the module-network for identifying driver genes in cancer subtypes. Molecules. 2018;23:183. doi: 10.3390/molecules23020183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zeng X., Liu L., Lü L., Zou Q. Prediction of potential disease-associated microRNAs using structural perturbation method. Bioinformatics. 2018;34:2425–2432. doi: 10.1093/bioinformatics/bty112. [DOI] [PubMed] [Google Scholar]
- 18.Fu X., Zhu W., Liao B., Cai L., Peng L., Yang J. Improved DNA-binding protein identification by incorporating evolutionary information into the Chou’s PseAAC. IEEE Access. 2018;6:66545–66556. [Google Scholar]
- 19.Lin C., Chen W., Qiu C., Wu Y., Krishnan S., Zou Q. LibD3C: ensemble classifiers with a clustering and dynamic selection strategy. Neurocomputing. 2014;123:424–435. [Google Scholar]
- 20.Zou Q., Guo J., Ju Y., Wu M., Zeng X., Hong Z. Improving tRNAscan-SE annotation results via ensemble classifiers. Mol. Inform. 2015;34:761–770. doi: 10.1002/minf.201500031. [DOI] [PubMed] [Google Scholar]
- 21.Zeng X., Wang W., Chen C., Yen G.G. A Consensus Community-Based Particle Swarm Optimization for Dynamic Community Detection. IEEE Trans. Cybern. 2019 doi: 10.1109/TCYB.2019.2938895. Published online September 23, 2019. [DOI] [PubMed] [Google Scholar]
- 22.Wei L., Chen H., Su R. M6APred-EL: a sequence-based predictor for identifying N6-methyladenosine sites using ensemble learning. Mol. Ther. Nucleic Acids. 2018;12:635–644. doi: 10.1016/j.omtn.2018.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wei L., Wan S., Guo J., Wong K.K. A novel hierarchical selective ensemble classifier with bioinformatics application. Artif. Intell. Med. 2017;83:82–90. doi: 10.1016/j.artmed.2017.02.005. [DOI] [PubMed] [Google Scholar]
- 24.Wei L., Xing P., Zeng J., Chen J., Su R., Guo F. Improved prediction of protein-protein interactions using novel negative samples, features, and an ensemble classifier. Artif. Intell. Med. 2017;83:67–74. doi: 10.1016/j.artmed.2017.03.001. [DOI] [PubMed] [Google Scholar]
- 25.Zeng X., Lin Y., He Y., Lv L., Min X., Rodriguez-Paton A. 2019. Deep collaborative filtering for prediction of disease genes. IEEE/ACM Trans. Comput. Biol. Bioinformat. Published online March 26, 2019. [DOI] [PubMed] [Google Scholar]
- 26.Wei L., Ding Y., Su R., Tang J., Zou Q. Prediction of human protein subcellular localization using deep learning. J. Parallel Distrib. Comput. 2018;117:212–217. [Google Scholar]
- 27.Lin X., Quan Z., Wang Z.-J., Huang H., Zeng X. A novel molecular representation with BiGRU neural networks for learning atom. Brief. Bioinform. 2019 doi: 10.1093/bib/bbz125. Published online November 15, 2019. doi. [DOI] [PubMed] [Google Scholar]
- 28.Yu L., Sun X., Tian S.W., Shi X.Y., Yan Y.L. Drug and nondrug classification based on deep learning with various feature selection strategies. Curr. Bioinform. 2018;13:253–259. [Google Scholar]
- 29.Song T., Rodríguez-Patón A., Zheng P., Zeng X. Spiking neural P systems with colored spikes. IEEE Trans. Cogn. Dev. Syst. 2018;10:1106–1115. doi: 10.1109/TNB.2018.2873221. [DOI] [PubMed] [Google Scholar]
- 30.Wei L., Su R., Wang B., Li X., Zou Q. Integration of deep feature representations and handcrafted features to improve the prediction of N6-methyladenosine sites. Neurocomputing. 2019;324:3–9. [Google Scholar]
- 31.Hong Z., Zeng X., Wei L., Liu X. Identifying enhancer-promoter interactions with neural network based on pre-trained DNA vectors and attention mechanism. Bioinformatics. 2019 doi: 10.1093/bioinformatics/btz694. Published online September 6, 2019. [DOI] [PubMed] [Google Scholar]
- 32.Liu X., Hong Z., Liu J., Lin Y., Rodríguez-Patón A., Zou Q., Zeng X. Computational methods for identifying the critical nodes in biological networks. Brief. Bioinform. 2019 doi: 10.1093/bib/bbz011. Published online February 12, 2019. [DOI] [PubMed] [Google Scholar]
- 33.Yan K., Xu Y., Fang X., Zheng C., Liu B. Protein fold recognition based on sparse representation based classification. Artif. Intell. Med. 2017;79:1–8. doi: 10.1016/j.artmed.2017.03.006. [DOI] [PubMed] [Google Scholar]
- 34.Liu B., Weng F., Huang D.-S., Chou K.-C. iRO-3wPseKNC: identify DNA replication origins by three-window-based PseKNC. Bioinformatics. 2018;34:3086–3093. doi: 10.1093/bioinformatics/bty312. [DOI] [PubMed] [Google Scholar]
- 35.Doench J.G., Hartenian E., Graham D.B., Tothova Z., Hegde M., Smith I., Sullender M., Ebert B.L., Xavier R.J., Root D.E. Rational design of highly active sgRNAs for CRISPR-Cas9-mediated gene inactivation. Nat. Biotechnol. 2014;32:1262–1267. doi: 10.1038/nbt.3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kaur K., Gupta A.K., Rajput A., Kumar M. ge-CRISPR—an integrated pipeline for the prediction and analysis of sgRNAs genome editing efficiency for CRISPR/Cas system. Sci. Rep. 2016;6:30870. doi: 10.1038/srep30870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Doench J.G., Fusi N., Sullender M., Hegde M., Vaimberg E.W., Donovan K.F., Smith I., Tothova Z., Wilen C., Orchard R. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat. Biotechnol. 2016;34:184–191. doi: 10.1038/nbt.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rahman M.K., Rahman M.S. CRISPRpred: a flexible and efficient tool for sgRNAs on-target activity prediction in CRISPR/Cas9 systems. PLoS ONE. 2017;12:e0181943. doi: 10.1371/journal.pone.0181943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ho T.K. The random subspace method for constructing decision forests. IEEE Trans. Pattern Anal. 1998;20:832–844. [Google Scholar]
- 40.Wang T., Yang J., Shen H.B., Chou K.C. Predicting membrane protein types by the LLDA algorithm. Protein Pept. Lett. 2008;15:915–921. doi: 10.2174/092986608785849308. [DOI] [PubMed] [Google Scholar]
- 41.Liu B., Fang L., Wang S., Wang X., Li H., Chou K.C. Identification of microRNA precursor with the degenerate K-tuple or Kmer strategy. J. Theor. Biol. 2015;385:153–159. doi: 10.1016/j.jtbi.2015.08.025. [DOI] [PubMed] [Google Scholar]
- 42.Pedregosa F., Varoquaux G., Gramfort A., Michel V., Thirion B., Grisel O., Blondel M., Müller A., Nothman J., Louppe G. scikit-learn: machine learning in Python. J. Mach. Learn. Res. 2011;12:2825–2830. [Google Scholar]
- 43.Chandrashekar G., Sahin F. A survey on feature selection methods. Comput. Electr. Eng. 2014;40:16–28. [Google Scholar]
- 44.Zhu L.J., Holmes B.R., Aronin N., Brodsky M.H. CRISPRseek: a bioconductor package to identify target-specific guide RNAs for CRISPR-Cas9 genome-editing systems. PLoS ONE. 2014;9:e108424. doi: 10.1371/journal.pone.0108424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nishimasu H., Ran F.A., Hsu P.D., Konermann S., Shehata S.I., Dohmae N., Ishitani R., Zhang F., Nureki O. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell. 2014;156:935–949. doi: 10.1016/j.cell.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chuai G., Ma H., Yan J., Chen M., Hong N., Xue D., Zhou C., Zhu C., Chen K., Duan B. DeepCRISPR: optimized CRISPR guide RNA design by deep learning. Genome Biol. 2018;19:80. doi: 10.1186/s13059-018-1459-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Haeussler M., Schönig K., Eckert H., Eschstruth A., Mianné J., Renaud J.B., Schneider-Maunoury S., Shkumatava A., Teboul L., Kent J. Evaluation of off-target and on-target scoring algorithms and integration into the guide RNA selection tool CRISPOR. Genome Biol. 2016;17:148. doi: 10.1186/s13059-016-1012-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu B., Gao X., Zhang H. BioSeq-Analysis2.0: an updated platform for analyzing DNA, RNA and protein sequences at sequence level and residue level based on machine learning approaches. Nucleic Acids Res. 2019;47:e127. doi: 10.1093/nar/gkz740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hart T., Chandrashekhar M., Aregger M., Steinhart Z., Brown K.R., MacLeod G., Mis M., Zimmermann M., Fradet-Turcotte A., Sun S. High-resolution CRISPR screens reveal fitness genes and genotype-specific cancer liabilities. Cell. 2015;163:1515–1526. doi: 10.1016/j.cell.2015.11.015. [DOI] [PubMed] [Google Scholar]
- 50.Wang T., Wei J.J., Sabatini D.M., Lander E.S. Genetic screens in human cells using the CRISPR-Cas9 system. Science. 2014;343:80–84. doi: 10.1126/science.1246981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu B., Li K. iPromoter-2L2.0: identifying promoters and their types by combining smoothing cutting window algorithm and sequence-based features. Mol. Ther. Nucleic Acids. 2019;18:80–87. doi: 10.1016/j.omtn.2019.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu B. BioSeq-Analysis: a platform for DNA, RNA and protein sequence analysis based on machine learning approaches. Brief. Bioinform. 2019;20:1280–1294. doi: 10.1093/bib/bbx165. [DOI] [PubMed] [Google Scholar]
- 53.Harris D.M., Harris S. IEEE; 2013. Introductory digital design & computer architecture curriculum. Proceedings of the 2013 IEEE International Conference on Microelectronic Systems Education; pp. 14–16. [Google Scholar]
- 54.Li C.-C., Liu B. MotifCNN-fold: protein fold recognition based on fold-specific features extracted by motif-based convolutional neural networks. Brief. Bioinform. 2019 doi: 10.1093/bib/bbz133. Published online November 28, 2019. [DOI] [PubMed] [Google Scholar]
- 55.Chen T., Guestrin C. ACM; 2016. Xgboost: a scalable tree boosting system. Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining; pp. 785–794. [Google Scholar]
- 56.Fawcett T. An introduction to ROC analysis. Pattern Recognit. Lett. 2006;27:861–874. [Google Scholar]
- 57.Hanley J.A., McNeil B.J. The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology. 1982;143:29–36. doi: 10.1148/radiology.143.1.7063747. [DOI] [PubMed] [Google Scholar]
- 58.Liu B., Zhu Y. ProtDec-LTR3.0: protein remote homology detection by incorporating profile-based features into Learning to Rank. IEEE Access. 2019;7:102499–102507. [Google Scholar]
- 59.Liu B., Zhu Y., Yan K. Fold-LTR-TCP: protein fold recognition based on triadic closure principle. Brief. Bioinform. 2019 doi: 10.1093/bib/bbz139. Published online December 8, 2019. [DOI] [PubMed] [Google Scholar]
- 60.Lorenz R., Bernhart S.H., Höner Zu Siederdissen C., Tafer H., Flamm C., Stadler P.F., Hofacker I.L. ViennaRNA package 2.0. Algorithms Mol. Biol. 2011;6:26. doi: 10.1186/1748-7188-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
There are 1,059 high on-target activity sgRNAs and 4,251 low on-target activity sgRNAs. See the text of the paper for further explanation.