Abstract
Understanding the effects of human exploitation on the genetic composition of wild populations is important for predicting species persistence and adaptive potential. We therefore investigated the genetic legacy of large-scale commercial harvesting by reconstructing, on a global scale, the recent demographic history of the Antarctic fur seal (Arctocephalus gazella), a species that was hunted to the brink of extinction by 18th and 19th century sealers. Molecular genetic data from over 2,000 individuals sampled from all eight major breeding locations across the species’ circumpolar geographic distribution, show that at least four relict populations around Antarctica survived commercial hunting. Coalescent simulations suggest that all of these populations experienced severe bottlenecks down to effective population sizes of around 150–200. Nevertheless, comparably high levels of neutral genetic variability were retained as these declines are unlikely to have been strong enough to deplete allelic richness by more than around 15%. These findings suggest that even dramatic short-term declines need not necessarily result in major losses of diversity, and explain the apparent contradiction between the high genetic diversity of this species and its extreme exploitation history.
Subject terms: Population genetics, Genetic variation, Molecular ecology, Conservation biology
Introduction
Anthropogenic exploitation is a major threat to global biodiversity1,2. For example, hunting has decimated many terrestrial species from the plains buffalo to the passenger pigeon3,4, while over-fishing has resulted in the collapse of many fish and marine mammal stocks5,6, reducing the capacity of the world᾿s oceans to provide food and ecosystem services7. This depletion of natural capital has been accelerating in pace as human populations continue to grow2 while animal populations across the planet are declining and being driven to extinction at an unprecedented rate8.
Theoretical and experimental studies suggest that severe declines in the effective population size (Ne) increase extinction risk not only because small populations are more sensitive to demographic and environmental stochasticity9,10 but also because the erosion of genetic diversity that accompanies severe demographic reductions can reduce population viability and adaptive potential11,12. However, the genetic consequences of anthropogenic exploitation are in general poorly understood13 with debate continuing over the contribution of genetic factors to extinction11. For example, comparative studies have shown that threatened species often have lower genetic diversity than non-threatened taxa12,14. However, this pattern does not necessarily imply a causal link between genetic diversity and viability, as many threatened species appear to have small ancestral population sizes, implying that they may not have been very abundant in the first place14.
A compelling approach for investigating the genetic consequences of exploitation on natural populations is to use highly polymorphic genetic markers such as microsatellites to reconstruct the demographic histories of species that were heavily hunted in the past. In particular, recent severe demographic declines or bottlenecks15,16 can be inferred by comparing the observed genetic diversity of a contemporary sample with the diversity expected under alternative historical scenarios simulated based on the coalescent17–19. Applied in a comparative context, this approach has shown that the intensity of recent bottlenecks can be influenced by species-specific traits such as breeding ecology and mating system variation20. However, our understanding of within-species variation in demographic histories remains limited because most studies of individual species are conducted with suboptimal sample sizes of both individuals and loci, and test for the presence or absence of bottlenecks rather than quantifying their intensity21. In order to understand species-wide responses to anthropogenic exploitation, studies are needed that combine exhaustive geographical sampling with modern computational methods capable of deriving quantitative estimates of demographic parameters, such as approximate Bayesian computation (ABC)22,23.
The Antarctic fur seal (Arctocephalus gazella) provides an excellent case to evaluate the genetic consequences of exploitation. This pinniped species is polygynous24, highly site faithful25,26 and has a generation time of around ten years27. Despite breeding on all of the major sub-Antarctic islands (Fig. 1), it was driven to the brink of extinction by the 18th and 19th century sealing industry28,29. Harvesting begun shortly after South Georgia was discovered by Captain James Cook in 1775 and ‘reckless extermination was the only method of seal-hunting (...) so that the first in the field at a new sealing ground was sure of an immense booty, and late-comers as likely as not would go empty away’30. Sealing reached its peak at South Georgia in 1800 when over 100,000 seals were harvested31. Shortly afterwards, with the discovery of the other sub-Antarctic islands, a wave of sealing spread across the entire sub-Antarctic region (Fig. 2), culminating in an estimated 1.7 million Antarctic fur seals being taken32.
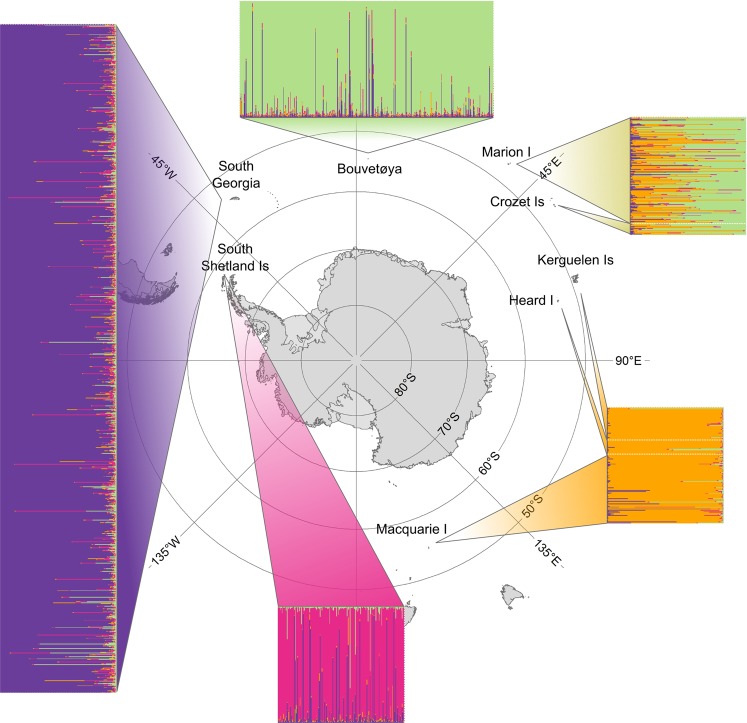
Figure 1.
Global population structure of the Antarctic fur seal inferred by STRUCTURE analysis of 2,000 individuals from eight populations genotyped at 39 microsatellite loci. Each individual is represented by a bar with the proportions of the different colours indicating the estimated membership to each of four inferred genetic populations (see Results for details). The data are plotted separately for each sampling location as indicated on the map. The map was created using ArcMap v. 10.6 https://desktop.arcgis.com/en/arcmap/.
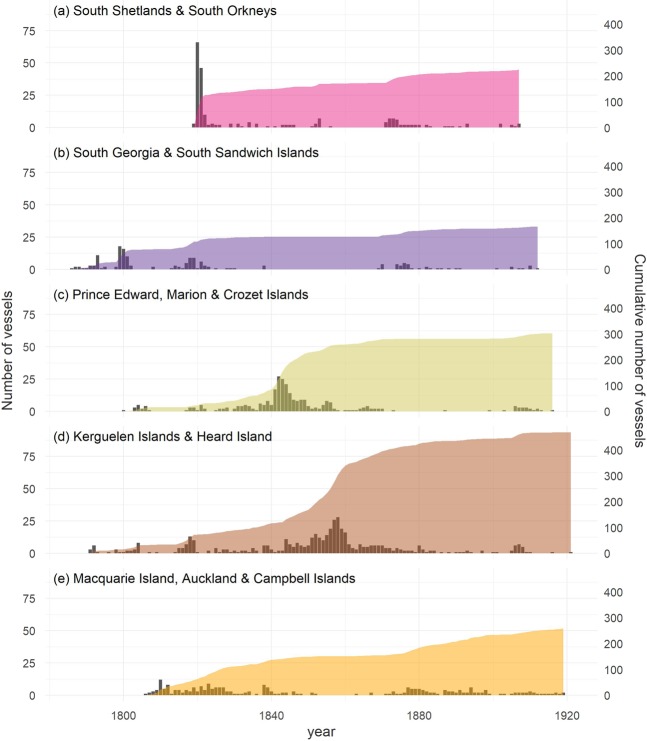
Figure 2.
The spatial and temporal distribution of sealing effort, defined as the number of ships recorded as having visited each of the main sealing grounds. Data are replotted from Headland29 with the permission of the author. The islands are grouped into sealing grounds because individual ships often visited more than one island within a given geographical region; no data were available for Bouvetøya. Annual numbers of ships are shown on the left axis while the cumulative total is shown on the right axis (South Shetlands & South Orkneys = 225, South Georgia & South Sandwich Islands = 167, Prince Edward, Marion Island and Crozet Islands = 303, Kerguelen Islands and Heard Island = 469, Macquarie Island, Auckland & Campbell Islands = 259).
By the turn of the 20th century, Antarctic fur seals were considered all but extinct and the species was placed under legal protection. For the best part of three decades and despite several dedicated sub-Antarctic expeditions, very few individuals were sighted at the traditional breeding grounds28,33–35 with the exception of Bouvetøya where a small population of an uncertain fur seal species was observed in the late 1920s36. However, the Discovery expedition of 1936 observed a small breeding population of Antarctic fur seals with 12 pups at South Georgia37, which grew rapidly in the 1960s and 1970s to reach an estimated five million individuals by 199938. As breeding populations were not observed at the other sub-Antarctic islands until the 1950s to the 1980s39–44, several authors have speculated that the former geographic distribution of this species was likely recolonized from South Georgia28,38,45–47. However, a number of studies have reported genetic differences among some of these populations48–51, pointing towards a more complex picture in which more than one relict population may have survived sealing.
In this study, we genotyped over 2,000 Antarctic fur seals at 39 microsatellite loci to investigate the genetic legacy of extreme exploitation in a marine mammal. In order to determine how the species as a whole was affected by sealing, samples from all major breeding locations were analysed to determine the most likely number of relict populations and ABC was used to reconstruct their recent demographic histories. Contrary to expectations, evidence was found for at least four genetically distinct populations, all of which experienced severe demographic declines. Nevertheless, high levels of neutral genetic variability were retained in comparison to other otariid species. Taken together, our results support the assertion that only the strongest bottlenecks lead to major losses of diversity and highlight the importance of relict populations to species recoveries and the maintenance of genetic variation.
Results
Genetic data were generated from 2,000 Antarctic fur seals sampled from eight populations covering the circumpolar distribution of the species (see Methods for details). The 39 microsatellite loci carried between 2 and 26 alleles (mean = 11.44) and none deviated significantly from Hardy-Weinberg equilibrium in more than two out of eight sampling localities after correction for multiple tests (see Supplementary methods and results). The genotyping error rate determined by re-genotyping 96 samples was low (0.003% per allele or 0.005% per reaction).
Population structure
Bayesian analyses of the microsatellite dataset using STRUCTURE produced consistent results across different clustering solutions (Supplementary Fig. S1). ∆k, which provides an indication of the uppermost hierarchical level of structure, peaked at k = four, while the average log likelihood of the data also gradually increased until k = four, after which a plateau was reached (Supplementary Fig. S2). The clustering solution for k = four resolved the South Shetland Islands, South Georgia and Bouvetøya as three distinct genetic clusters and Kerguelen, Heard and Macquarie Islands as a fourth cluster, while animals from Marion and Crozet Islands showed mixed membership to the third and fourth clusters (Fig. 1). Further increases in k only introduced additional admixture (Supplementary Fig. S1). PCA revealed a similar pattern, with Bouvetøya separating from Kerguelen, Heard and Macquarie Islands along PC1 and PC2, Marion and Crozet Islands occupying an intermediate position, and the South Shetlands and South Georgia separating along PC3 (Supplementary Fig. S3). Pairwise Fst values between populations were significant for all but two comparisons (Supplementary Table S1). Consequently, our genetic data are consistent with four relict populations comprising (i) the South Shetland Islands, (ii) South Georgia, (iii) Bouvetøya; and (iv) Kerguelen, Heard and Macquarie Islands, plus a further genetically admixed population (v) comprising Marion and Crozet Islands. Subsequent analyses therefore focused on these five populations, although similar results were obtained when sampling locations rather than genetic clusters were analysed (Supplementary Fig. S4).
Genetic diversity
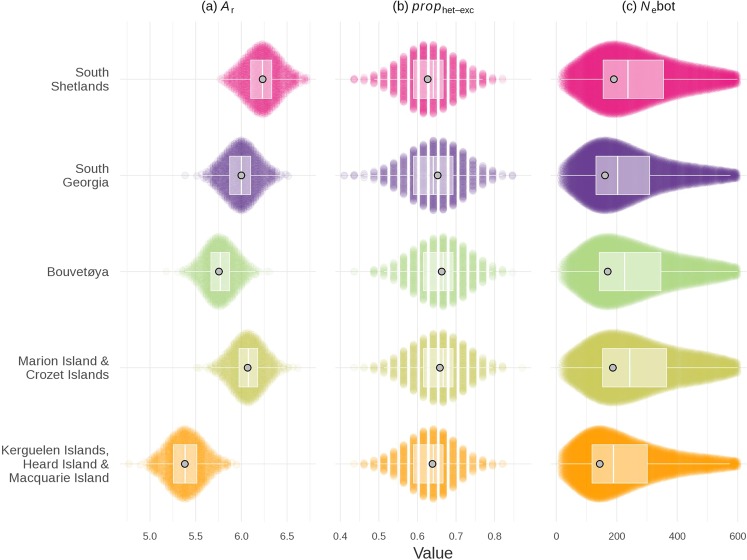
Allelic richness (Ar), standardised across species as the average number of alleles per ten individuals, was high relative to other otariids, with the Antarctic fur seal being ranked fourth highest out of 13 species (Supplementary Fig. S5) for which comparable microsatellite data were available20. In relative terms, genetic diversity did not vary a great deal across the geographical distribution of the Antarctic fur seal (Fig. 3a, Table 1, Supplementary Table S2) with Ar only differing by around one allele per locus between the most diverse (the South Shetland Islands) and the least diverse populations (Kerguelen, Heard and Macquarie Islands).
Figure 3.
Patterns of genetic diversity and bottleneck signatures across the geographic range of the Antarctic fur seal. (a) Genetic diversity summarised as allelic richness (Ar); (b) The proportion of loci in heterozygosity excess (prophet-exc) calculated for the TPM80 model; (c) Estimated bottleneck effective population sizes (Nebot). Data are summarised according to the five main populations identified by the STRUCTURE analysis (see Results for details). To facilitate visual comparisons among populations with different sample sizes while incorporating sampling error, we plotted 1,000 subsets of ten randomly sampled individuals per population as Sinaplots, with the exception of the sinaplot for Nebot, which shows parameter values for 5,000 accepted simulations based on 181 individuals per population. Boxplots (centre line = median, bounds of box = 25th and 75th percentiles, upper and lower whiskers = largest and lowest value but no further than 1.5 * inter-quartile range from the hinge) are superimposed with light grey points representing maximum densities. Bottleneck measures for Marion and Crozet Islands should be interpreted with caution due to admixture.
Table 1.
Genetic diversity and bottleneck signatures per genetic population (as determined by STRUCTURE).
| Population | Sample size | Ar (SD) | Ho (SD) | M-ratio (SD) | Fis (SD) | Private Alleles | Locihet-exc | Sign test p-value | Standardized differences test p-value | Wilcoxon test p-value |
|---|---|---|---|---|---|---|---|---|---|---|
| South Shetlands | 197 | 6.448 (2.555) | 0.719 (0.197) | 0.836 (0.185) | 0.011 (0.046) | 13 | 31 | 0.004 | <0.001 | <0.001 |
| South Georgia | 1042 | 6.235 (2.506) | 0.716 (0.208) | 0.813 (0.200) | 0.010 (0.025) | 19 | 34 | <0.0001 | <0.0001 | <0.0001 |
| Bouvetøya | 396 | 5.963 (2.470) | 0.703 (0.223) | 0.825 (0.195) | 0.014 (0.047) | 8 | 30 | 0.010 | <0.0001 | <0.0001 |
| Marion Island & Crozet Islands | 184 | 6.279 (2.586) | 0.719 (0.215) | 0.830 (0.170) | 0.004 (0.044) | 2 | 31 | 0.004 | <0.001 | <0.001 |
| Kerguelen Islands & Heard Island & Macquarie Island | 181 | 5.578 (2.187) | 0.688 (0.213) | 0.831 (0.187) | 0.006 (0.077) | 7 | 28 | 0.050 | 0.021 | 0.019 |
Rarefied allelic richness (Ar), observed heterozygosity (Ho), M-ratio and inbreeding coefficient Fis given as means and standard deviations (SD) across 39 loci. The numbers of private alleles are summed across loci. The number of loci with heterozygosity excess (locihet-exc) and bottleneck test probabilities (Sign test, standardized differences tests and Wilcoxon test) are given under the two-phase model with 80% single-step mutations (TPM80) based on 1,000 iterations for each population.
Bottleneck inference
Two complementary coalescent-based approaches were used to infer the extent to which these populations experienced recent demographic declines. First, the proportion of loci with heterozygosity-excess (prophet-exc) provides an indicator of recent bottlenecks because rare alleles are preferentially lost during bottlenecks but their loss has little impact on heterozygosity15. Recent bottlenecks therefore generate a transient excess of heterozygosity relative to a population at equilibrium with an equivalent number of alleles, which can be quantified through coalescent simulation. A significant excess of heterozygosity relative to expectations was detected in all five genetic populations identified by STRUCTURE (Table 1 and Supplementary Table S1). The magnitude of prophet-exc was also similar across populations (Fig. 3b) suggestive of more or less equally strong declines.
Second, ABC was used to select between a bottleneck and a non-bottleneck model as well as to estimate posterior distributions of the model parameters. ABC was able to distinguish between the two models, with 87% of the simulations being classified correctly under the bottleneck model and 94% of the simulations being classified correctly under the non-bottleneck model (Supplementary Fig. S6). Posterior probabilities supported the bottleneck model in all five populations and the goodness of fit to the empirical data was consistently better for the bottleneck model (Supplementary Table S3). As another indicator of model quality, posterior predictive checks23,52 showed that the bottleneck model was able to reproduce the relevant observed summary statistics across all populations (Supplementary Fig. S7).
As a further evaluation step, we used cross-validation to estimate prediction errors for all model parameters. Cross-validation in ABC uses randomly selected simulations and treats them as pseudo-observed datasets. For each of these simulations, the ABC model was used to estimate a set of summery statistics. Because we also know the true summary statistics, we can calculate the prediction error as , where is the estimated parameter value and θi is the true parameter value of the ith simulated data set. If the estimated values are closer to the true values than expected by chance (i.e. they contain information about the true values), the prediction error will be below one. Under the bottleneck model, the prediction error was 0.457 for the bottleneck effective population size (Nebot), indicating that posterior estimates for this parameter contain information about the underlying true values (Supplementary Fig. S8). Although in principle the posterior estimates for Nehist also had an acceptable prediction error (0.611), a plot of the cross-validation results (Supplementary Fig. S8) revealed a systematic underestimation of the true values, which is why the posteriors should be interpreted with caution. By contrast, prediction errors for the other parameters were substantially higher (Supplementary Table S4), with visual inspection of the cross-validation results revealing a large amount of scatter and a poor fit of the predicted to the true values. Consequently, subsequent analyses focused on the posteriors for Nebot, while the Nehist estimates are also shown in Supplementary Fig. S9.
The Nebot estimates were similar in magnitude across populations (Fig. 3c), which is again suggestive of recent bottlenecks of comparable strength. However, the posterior distribution of Nebot was skewed towards slightly smaller values (maximum density ~150) in the eastern population comprising Kerguelen, Heard and Macquarie Islands, whereas maximum densities were closer to ~200 elsewhere. Similar results were obtained when the sampling locations were analysed separately (Supplementary Fig. S4). The Nehist posterior estimates were very broad (Fig. S9), suggesting that historical effective population sizes ranging between a few thousand and tens-of-thousands of individuals could have led to similar observed genetic diversities under the bottleneck model.
Expected loss of genetic diversity
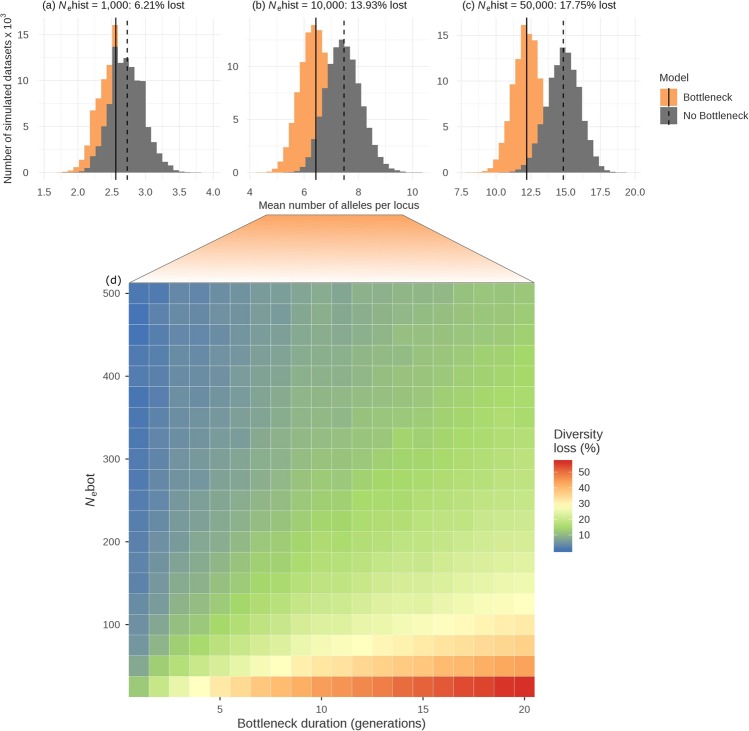
The expected loss of neutral genetic diversity caused by a bottleneck with an effective size Nebot of 200 lasting for ten generations was evaluated using coalescent simulations in fastsimcoal53. As the long-term Ne prior to the bottleneck will affect overall levels of diversity, three scenarios were analysed with Nehist simulated as 1,000, 10,000 and 50,000 respectively. Despite a severe and relatively long-lasting simulated bottleneck, the expected loss of microsatellite alleles was small, ranging from 6% with an Nehist of 1,000 to 18% with an Nehist of 50,000 (Fig. 4). Under the scenario that produced levels of genetic diversity most similar to the empirical values (Nehist = 10,000) only around 14% of alleles were lost. Furthermore, simulated bottlenecks of varying strength and duration, again with an Nehist of 10,000, suggest that only a combination of very strong (i.e. Nebot < 50) and long (i.e. 15–20 generations) bottlenecks would result in genetic diversity losses in excess of 50%. Consequently, the Antarctic fur seal is unlikely to have lost a substantial proportion of neutral genetic diversity despite an inferred steep decline in Ne due to overhunting.
Figure 4.
The expected loss of genetic diversity caused by recent bottlenecks. Panels (a–c) show the distributions of mean allele numbers across loci obtained from 100,000 neutral coalescent simulations of a bottleneck to an Nebot of 200 for ten generations (orange) and a constant population size scenario (grey) with three different pre-bottleneck effective population sizes (Nehist). Panel (d) further explores the effects of bottleneck duration and strength on the expected loss of allelic diversity under the scenario shown in panel (b) as described in the Methods.
Discussion
We investigated the genetic consequences of historical commercial exploitation in the Antarctic fur seal. Information on the number of sealing vessels recorded as having visited each of the main sealing grounds allowed us both to formulate realistic priors for our ABC analysis and to interpret our results in the light of historical evidence. An innovation of this study was to use a comparative approach to shed light on the genetic diversity and recent demographic histories of multiple populations within a single species.
Consistent with previous studies of Antarctic fur seals, we found evidence for strong population structure47–50,54. However, our larger sample size of individuals and exhaustive geographical coverage yields a more comprehensive global picture. On the one hand, the discovery of four genetically distinct clusters builds upon recent studies suggesting that relict populations likely persisted in localities other than South Georgia49,50. This is in contrast to the previously held belief that Antarctic fur seals only survived in one relict population off South Georgia28,38,45,46 and suggests that, at least in some regards, the species as a whole may have been more resilient to commercial exploitation than previously believed. Nevertheless, our data also suggest that at least one and possibly several populations may have gone locally extinct. In particular, Macquarie, Heard and Kerguelen Islands showed little in the way of population structure, consistent with the suggestion that this species was locally extirpated at Macquarie and Heard Islands, leaving these sites to be subsequently recolonised by individuals mainly from the Kerguelen Archipelago47,55. In addition, a strong signal of admixture at Marion and Crozet Islands implies that these two populations may have either gone extinct or been reduced to sufficiently small numbers for the local gene pool to be swamped by emigrants from nearby Bouvetøya and the Kerguelen Islands. This is supported by anecdotal evidence suggesting that Antarctic fur seals were not present after sealing ceased at Marion Island in the early 1930s56.
Two approaches were used to infer the strength of recent population declines. Heterozygosity excess provides a measure of the relative strength of population decline, whereas our ABC analysis specifically tested for an absolute reduction in Ne (i.e. to below 600). By focusing on time priors that reflected the known sealing history, the ABC analysis also evaluated a clear hypothesis‒that 18th and 19th century commercial sealing resulted in strong bottlenecks. Although in principle, similar patterns of genetic diversity might be produced by more ancient bottlenecks, these are unlikely to be detected reliably using microsatellite data when subsequent recovery occurred57. This is in keeping with a previous study in which a simple model incorporating a recent bottleneck was better supported than a more complex model that also included a small population size during the last glacial maximum followed by expansion20.
In practice, heterozygosity excess and ABC produced consistent results, with all of the populations identified by STRUCTURE showing a significant excess of heterozygosity as well as support for recent demographic declines in the ABC analysis. Similar results were also obtained after analyzing each of the sampling localities separately, although Heard Island failed to reach significance in the heterozygosity excess test, possibly due to our relatively small sample size of individuals from this locality. Nebot estimates from the ABC analysis were also quite similar across populations, although the mode of the posterior distribution was slightly lower for the easternmost population. This subtle difference in inferred bottleneck intensity is a reflection of the lower genetic diversity of the Kerguelen Islands, Heard and Macquarie Island populations. It is furthermore consistent with the historical sealing data (Fig. 2), which show that sealing effort was higher in these areas, with Kerguelen and Heard Islands in particular having been visited by more ships in total than any of the other main sealing grounds.
Overall, however, we were struck by the similarity of the Nebot estimates, whose maximal densities consistently fell within the range of around 150 to 200. This provides a good indication that census population sizes on all of the islands may have been similarly diminished, although the relationship between Ne and census size has to be interpreted with caution58. Moreover, mammalian effective population sizes are usually around 2‒3 times smaller than census population sizes59 and this ratio may be even greater in fur seals due to their strongly polygynous mating system24. Our results therefore suggest that the global population of Antarctic fur seals that survived sealing was considerably larger than implied by historical accounts and may have numbered in the thousands.
Several factors may play a role in explaining our results. First, standard economic theory predicts that exploitation will not necessarily lead to extinction due to the escalating costs of harvesting increasingly depleted populations1,60. Consistent with this explanation, historical sealing data show a geographically concordant pattern of early intense exploitation followed by greatly diminished sealing effort during the mid to late 19th century (Fig. 2). This reduction in sealing effort coincides with a crash in the price of fur seal pelts in America and China during the 1850s, which would have substantially decreased profitability61. Consequently, the available historical evidence suggests that sealing probably ceased to become economically viable around the time when the main sealing grounds had been largely depleted of animals.
Having said this, historical accounts suggest that the sealers were extremely thorough in the pursuit of their quarry. Not only did their high mobility allow them to quickly discover and exploit new hunting grounds62, but it was also commonplace for sealing gangs to be left ashore for extended periods to opportunistically harvest any animals that remained on land28,63. It therefore seems unlikely that appreciable numbers of individuals could have survived ashore at the main breeding grounds. This may explain the rarity of sightings of this species during the first few decades of the 20th century28,33–35 and the fact that some breeding populations were not re-established until as late as the 1980s39–44.
It is conceivable that some juveniles, subadults and non-breeders could have escaped the sealers by not coming ashore. However, if this were the case, relict populations should have survived at all of the breeding grounds, whereas our data, together with recent studies and historical accounts, suggest that that Antarctic fur seals may have been locally extirpated at Macquarie, Heard, Crozet and Marion Islands47,50,51. We therefore speculate that tiny remnants of once much larger breeding populations may have persisted in a handful of particularly inaccessible locations that were rarely if ever visited by sealers. Candidates for sites that may have sheltered these relict populations include the Willis Islands off the coast of South Georgia28, the San Telmo Islets in the South Shetlands49 Larsøya and Kapp Norvegia at Bouvetøya36 and any one of a multitude of small islands in the Kerguelen Archipelago48.
This study contributes towards wider debate about the factors shaping genetic diversity and its importance for conservation. In particular, many studies have reported links between known historical bottlenecks and low levels of genetic diversity64–67. However, the implied loss of diversity is often greater than predictions from population genetic theory68,69, suggesting that other mechanisms such as selective sweeps could be involved70. Additionally, the general perception that bottlenecked populations have low genetic variability may also have been influenced by publication bias68. In the current study, it was not possible to directly quantify the amount of genetic diversity lost as a result of sealing due to a lack of historical samples. However, coalescent simulations provided an expectation of the likely magnitude of genetic erosion given the strength of the inferred demographic declines. These simulations suggest that a recent decline from an historical Ne of 10,000 to an Nebot of 200 for ten generations is unlikely to have resulted in the loss of more than around 15% of alleles. Consequently, our study supports the argument of Amos and Balmford68 that even extreme recent bottlenecks need not necessarily lead to major losses of genetic diversity. This may further help to explain why several other pinniped species including northern, Australian and Juan Fernandez fur seals also retained high levels of genetic diversity despite having been harvested in their tens of thousands to millions71–73.
To conclude, demographic reconstruction of an entire species provided insights into the resilience of a marine mammal that was hunted in very large numbers. Our study suggests that, although local extinctions took place, Antarctic fur seals persisted in sufficient numbers in several relict populations to retain a large proportion of the species᾿ pre-sealing genetic diversity. The extreme geographical isolation of some small breeding sites in addition to diminishing economic incentives for sealing likely saved the Antarctic fur seal from extinction. The subsequent recovery of this species is a testament both to its resilience and to the success of protection measures in the 20th century.
Methods
Tissue sampling and microsatellite genotyping
Samples were collected from a total of 2,064 Antarctic fur seal individuals from eight different breeding locations (Fig. 1, for GPS coordinates see Supplementary Table S1). Skin samples were taken from the interdigital margin of the foreflipper with piglet ear notching pliers74 and stored at −20°C in 20% dimethyl sulphoxide saturated with sodium chloride. DNA was extracted using a chloroform/isoamylalcohol extraction protocol75 and genotyped at 39 microsatellite loci as described in the Supplementary methods.
Because Antarctic fur seals are known to hybridise with Subantarctic fur seals (Arctocephalus tropicalis), we additionally sampled 91 pups from a reference population of A. tropicalis at Macquarie Island in order to check the full A. gazella dataset for individuals with a high proportion of A. tropicalis ancestry (see Supplementary methods and results). We found that 49 of the Antarctic fur seal individuals were inadvertently pure A. tropicalis, while five of the A. tropicalis reference individuals were hybrids (see Figs. S11 and S12). A further 15 Antarctic fur seal individuals were also identified as hybrids. After having removed all pure A. tropicalis individuals as well as the hybrids and any additional individuals that failed to genotype at more than four loci from the dataset, we were left with a total of 2,000 Antarctic fur seal individuals, comprising 197 from the South Shetland Islands, 1,042 from South Georgia, 396 from Bouvetøya, 166 from Marion Island, 18 from Crozet Islands, 51 from Kerguelen Islands, 22 from Heard Island and 108 from Macquarie Island.
Population structure and genetic diversity
For the full dataset of 2,000 Antarctic fur seals, we evaluated whether population structure could be detected without prior knowledge of sampling locations using STRUCTURE version 2.3.376. This program distributes individual genotypes into k clusters by subdividing the dataset in a way that maximizes Hardy–Weinberg and linkage equilibrium within the resulting clusters. The membership of each individual to a given cluster is then estimated as q, which varies between 0 and 1, with the latter indicating full cluster membership. STRUCTURE was run using ParallelStructure77 with the following parameters: 500,000 burn-in length, 1,000,000 MCMC replications, 1‒10 assumed clusters (k) and 10 iterations for each k. The most likely number of clusters was evaluated using the maximal average value of Ln P(D) as well as the Evanno method78. Individual admixture proportions were integrated over all simulations using pophelper79. For comparison, we also implemented principle component analysis (PCA) using the R-package adegenet80. Genetic diversity was quantified as allelic richness (Ar) within hierfstat81. Observed heterozygosity (Ho) was calculated with adegenet80, the M-ratio was calculated with StrataG82, the inbreeding coefficient Fis was calculated with diveRsity83 and the number of private alleles was calculated with poppr84. Pairwise Fst values together with their 95% confidence intervals were obtained using hierfstat81.
Bottleneck analyses
We used two coalescent-based approaches to infer recent demographic histories. First, we quantified the proportion of loci in heterozygosity-excess using the program BOTTLENECK v. 1.2.0285. This analysis was based on the two-phase model with 80% single-step mutations (TPM80), consistent with recent estimates from multiple pinniped species20. A total of 1,000 iterations were run for each population and all other parameters were left at default values. Statistical significance was evaluated using sign, standardized differences and Wilcoxon signed ranks tests.
Second, we tested for signatures of recent population bottlenecks using a coalescent-based approximate Bayesian computation (ABC) framework. Support was evaluated for two alternative demographic models (Supplementary Fig. S10). Briefly, to evaluate whether 18th to 20th century commercial exploitation caused severe demographic reductions, we defined a “bottleneck model”, which incorporated a strong decline in population size within strictly bound time priors that fit the time period hunting took place. The priors for pre- and post-bottleneck effective population size were drawn independently from the same distribution to allow the model not only to incorporate a bottleneck, but also longer-term reductions or expansions within realistic bounds. For comparison, we defined a “non-bottleneck model” which was identical to the bottleneck model with the exception of a bottleneck.
For both models, priors for current effective populations size (Ne) and historical pre-bottleneck effective population size (Nehist) were drawn from a log-normal distribution in order to encompass a wide range of values while mainly sampling from the most likely ones. We specified Ne and Nehist ~ lognorm[logmean = 10.5, logsd = 1], which concentrated around 90% of sampling in the range between 3,000 and 15,000 while also occasionally simulating much larger Ne values. This prior range encompassed the estimated current census population sizes across all islands, i.e. from 3,600 at Marion Island86 to 5 × 106 at South Georgia38. For the bottleneck model, the effective population size during the bottleneck (Nebot) was drawn from a uniform distribution between 1 and 600 (Nebot ~ U[1, 600]). Start and end time for the bottleneck were defined by tbotstart and tbotend and drawn from uniform distributions between ten and 40 (tbotstart ~U[10, 40]) and one and 20 (tbotend~ U[1, 20]) generations ago respectively. This places the bottleneck in the last one to four centuries, as the generation time of the Antarctic fur seal is estimated at around nine years87. For the neutral model, the time parameter corresponding to the historical population size (thist) was drawn from a uniform distribution ranging between 10 and 40 generations ago (thist ~U[10, 40]). The microsatellite mutation rate (µ) was drawn from a uniform prior with µ ~ U[10−5, 10−4]. A stepwise mutation model was used with the geometric parameter GSMpar reflecting the proportion of multistep mutations, uniformly distributed from GSMpar ~U[0, 0.3].
Genetic data were simulated under each demographic model for 181 individuals, reflecting the smallest genetic population (as determined by STRUCTURE) in the empirical dataset, and 39 microsatellites using fastsimcoal217 within the R-package strataG82. This resulted in a total of 2 × 107 datasets. We then calculated three summary statistics (as means across all loci) for the simulated data: (1) allelic richness; (2) the proportion of low frequency (<5%) alleles; and (3) the M-ratio88, and retained the 5,000 simulations with summary statistics closest to the empirical data using a tolerance threshold of 5 × 10−4.
To evaluate model specification and fit, we implemented leave-one-out cross-validation using the cv4postpr function of the abc R-package89. Here, simulations are selected at random and their summary statistics are used as pseudo-observed data. The remaining simulations are then used to classify the data into the bottleneck or the neutral model. If ABC is capable of distinguishing between the two models, a large posterior probability will be assigned to the same model under which the pseudo-observed data were generated. This procedure was repeated 100 times and the posterior probabilities for a given model were averaged to derive the rate of misclassification. Furthermore, posterior probabilities were calculated for each model using the rejection method of the postpr function and the fit of the preferred model to the empirical data was evaluated using a formal hypothesis testing procedure implemented by the gfit function of the abc R-package.
Next, we tested the accuracy of the ABC parameter estimates using leave-one-out cross validation in the cv4abc function. For a randomly selected pseudo-observed dataset, parameters were estimated using all remaining simulations via ABC using the rejection method. This procedure was repeated 1,000 times and the mean prediction error was calculated. The smaller the prediction error, the better the fit to the true parameter value. After performing the four checks, we constructed posterior distributions of the parameters using a simple rejection algorithm with the abc function of the abc package.
To provide a measure of the fit of the preferred models to the empirical data, we conducted posterior predictive checks23,52. After generating posterior distributions for each parameter, 1,000 multivariate parameters were sampled from their respective posterior distributions and used to simulate summary statistics a posteriori based on the preferred model. Those summary statistics were then plotted as histograms and superimposed over the observed summary statistics23.
Expected loss of genetic diversity
Finally, we used neutral coalescent simulations in fastsimcoal253 to estimate the expected loss of microsatellite alleles for a bottleneck effective population size (NeBot) of 200 lasting for ten generations. As the historical effective population size Nehist is unknown but will be an important determinant of the overall level of genetic diversity, we simulated three different scenarios with Nehist values of 1,000, 10,000 and 50,000. The post-bottleneck Ne is unlikely to have a major effect on diversity, as only a few generations have passed since the bottleneck, and was thus simulated equal to the Nehist within each scenario. All other model parameters were the same as in the models above. We then compared the resulting allelic richness to the allelic richness generated in equivalent models with constant effective population sizes of 1,000, 10,000 and 50,000 respectively to estimate the proportional loss of alleles due to the bottleneck. To account for uncertainty in building coalescent trees, we simulated 100,000 datasets under both the bottleneck and constant size model and treated the difference in mean allelic richness between the distributions as the average loss of alleles. To further explore the effects of varying bottleneck strength and duration, we conducted a final round of simulations based on the scenario that provided the best fit to the empirical data (Nehist = 10,000, see Results). Specifically, we quantified the expected loss of alleles for Nebot ranging 25 to 600 in steps of 25 and for bottleneck duration ranging from one to 20 generations in steps of one. As hunting ceased by the turn of the 20th century, we fixed tbotend at ten generations ago. 1,000 simulations were conducted for each combination of parameter values.
Animal ethics
Samples were collected from the South Shetlands under Marine Mammal Protection Act Permit no. 774-1847-04 granted by the Office of Protected Resources, National Marine Fisheries Service. Sampling and procedures used on South Georgia were approved by the British Antarctic Survey Animal Welfare and Ethics Review Body (reference no. PEA6) and collected as part of the Polar Science for Planet Earth programme of the British Antarctic Survey. Samples from Bouvetøya were obtained under Permit no. 7001 issued by the Norwegian Department of Plants, Fish, Animals and Food. Samples were collected on Marion Island, Crozet Islands, Kerguelen Islands, Heard Island and Macquarie Island with approval from the Animal Ethics Committee of the Faculty of Veterinary Science, University of Pretoria, South Africa (PN 859; EC077-15), the Prince Edward Islands Management Committee and Department of Environmental Affairs, the Territory of Heard Island and McDonald Islands Environment Protection and Management Ordinance 1987 (Permit no. 00/18) and the Parks and Wildlife Service, Tasmania (Scientific Collecting Permit no. FA 99167). All sampling and procedures were performed in accordance with the relevant guidelines and regulations.
Supplementary information
Acknowledgements
This research was supported by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) in the framework of a Sonderforschungsbereich (project numbers 316099922 and 396774617–TRR 212) and the priority programme “Antarctic Research with Comparative Investigations in Arctic Ice Areas” SPP 1158 (project number 424119118). It was also funded by Norwegian Antarctic Research Expeditions (NARE) programme. This work contributes to the Ecosystems project of the British Antarctic Survey, Natural Environmental Research Council, and is part of the Polar Science for Planet Earth Programme. The Department of Environmental Affairs provided logistical support for research at Marion Island and the Department of Science and Technology of South Africa provided funding through the National Research Foundation (NRF). We are grateful to Caroline Bonin, Debbie Baird-Bower and Iain Staniland together with the seal biologists working within the Marion Island Marine Mammal Programme for sample collection and logistics. We acknowledge support for the Article Processing Charge by the Deutsche Forschungsgemeinschaft and the Open Access Publication Fund of Bielefeld University.
Author contributions
Conceived the study: J.I.H. and A.L. Sample collection and logistics: M.N.B., A.C.C., P.J.N.d.B., J.F., M.E.G., S.D.G., C.G., C.L., K.M.K. Laboratory work: A.J.P., A.C.C. and J.I.H. Analysed data: A.J.P., M.A.S., A.C.C. and J.I.H. Wrote the paper: A.J.P., M.A.S. and J.I.H. All of the authors commented upon and approved the final manuscript.
Data availability
All of the raw data90 are available via the Zenodo repository, doi:10.5281/zenodo.3585717.
Code availability
All data wrangling steps and statistical analyses except for the heterozygosity-excess tests were implemented in R. The complete documented analysis pipeline is available via our GitHub repository: https://github.com/apaijmans/AFS_genetic_legacy.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
5/12/2020
An amendment to this paper has been published and can be accessed via a link at the top of the paper.
Contributor Information
Anneke J. Paijmans, Email: a.paijmans@uni-bielefeld.de
Joseph I. Hoffman, Email: j_i_hoffman@hotmail.com
Supplementary information
is available for this paper at 10.1038/s41598-020-61560-8.
References
- 1.Erickson JD. Endangering the economics of extinction. Wildl. Soc. Bull. 2000;28:34–41. [Google Scholar]
- 2.Peres, C. A. Overharvesting. In Conservation biology for all (eds. Sodhi, N. S. & Ehrlich, P. R.) (Oxford University Press, 2010).
- 3.IUCN red list of threatened species: a global species assessment. (IUCN, 2004). 2004
- 4.Farrow S. Extinction and market forces: two case studies. Ecol. Econ. 1995;13:115–123. doi: 10.1016/0921-8009(94)00075-7. [DOI] [Google Scholar]
- 5.Worm B, et al. Rebuilding global fisheries. Science. 2009;325:578–585. doi: 10.1126/science.1173146. [DOI] [PubMed] [Google Scholar]
- 6.Hutchings JA. Collapse and recovery of marine fishes. Nature. 2000;406:882–885. doi: 10.1038/35022565. [DOI] [PubMed] [Google Scholar]
- 7.Worm B, et al. Impacts of biodiversity loss on ocean ecosystem services. Science. 2006;314:787–790. doi: 10.1126/science.1132294. [DOI] [PubMed] [Google Scholar]
- 8.Ceballos, G., Ehrlich, P. R. & Dirzo, R. Biological annihilation via the ongoing sixth mass extinction signaled by vertebrate population losses and declines. Proc. Natl. Acad. Sci. USA114, E6089–E6096 (2017) [DOI] [PMC free article] [PubMed]
- 9.Lande R. Genetics and demography in biological conservation. Science. 1988;241:1455–1460. doi: 10.1126/science.3420403. [DOI] [PubMed] [Google Scholar]
- 10.Shaffer ML. Minimum population sizes for species conservation. BioScience. 1981;31:131–134. doi: 10.2307/1308256. [DOI] [Google Scholar]
- 11.Frankham R. Genetics and extinction. Biol. Conserv. 2005;126:131–140. doi: 10.1016/j.biocon.2005.05.002. [DOI] [Google Scholar]
- 12.Spielman D, Brook BW, Frankham R. Most species are not driven to extinction before genetic factors impact them. Proc. Natl. Acad. Sci. USA. 2004;101:15261–15264. doi: 10.1073/pnas.0403809101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harris RB, Wall WA, Allendorf FW. Genetic consequences of hunting: what do we know and what should we do? Wildl. Soc. Bull. 2002;30:634–643. [Google Scholar]
- 14.Li H, et al. Large numbers of vertebrates began rapid population decline in the late 19th century. Proc. Natl. Acad. Sci. USA. 2016;113:14079–14084. doi: 10.1073/pnas.1616804113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nei M, Maruyama T, Chakraborty R. The bottleneck effect and genetic variability in populations. Evolution. 1975;29:1–10. doi: 10.1111/j.1558-5646.1975.tb00807.x. [DOI] [PubMed] [Google Scholar]
- 16.Tajima F. The effect of change in population size on DNA polymorphism. Genetics. 1989;123:597–601. doi: 10.1093/genetics/123.3.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Excoffier L, Dupanloup I, Huerta-Sánchez E, Sousa VC, Foll M. Robust demographic inference from genomic and SNP data. Plos Genet. 2013;9:e1003905. doi: 10.1371/journal.pgen.1003905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hudson RR. Generating samples under a Wright–Fisher neutral model of genetic variation. Bioinformatics. 2002;18:337–338. doi: 10.1093/bioinformatics/18.2.337. [DOI] [PubMed] [Google Scholar]
- 19.Kingman JFC. The coalescent. Stoch. Process. Their Appl. 1982;13:235–248. doi: 10.1016/0304-4149(82)90011-4. [DOI] [Google Scholar]
- 20.Stoffel MA, et al. Demographic histories and genetic diversity across pinnipeds are shaped by human exploitation, ecology and life-history. Nat. Commun. 2018;9:4836. doi: 10.1038/s41467-018-06695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peery MZ, et al. Reliability of genetic bottleneck tests for detecting recent population declines. Mol. Ecol. 2012;21:3403–3418. doi: 10.1111/j.1365-294X.2012.05635.x. [DOI] [PubMed] [Google Scholar]
- 22.Beaumont MA, Zhang W, Balding DJ. Approximate Bayesian Computation in population genetics. Genetics. 2002;162:2025–2035. doi: 10.1093/genetics/162.4.2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Csilléry K, Blum MGB, Gaggiotti OE, François O. Approximate Bayesian Computation (ABC) in practice. Trends Ecol. Evol. 2010;25:410–418. doi: 10.1016/j.tree.2010.04.001. [DOI] [PubMed] [Google Scholar]
- 24.Hoffman JI, Boyd IL, Amos W, Ashley M. Male reproductive strategy and the importance of maternal status in the Antarctic fur seal Arctocephalus gazella. Evolution. 2003;57:1917–1930. doi: 10.1111/j.0014-3820.2003.tb00598.x. [DOI] [PubMed] [Google Scholar]
- 25.Hoffman JI, Trathan PN, Amos W. Genetic tagging reveals extreme site fidelity in territorial male Antarctic fur seals Arctocephalus gazella. Mol. Ecol. 2006;15:3841–3847. doi: 10.1111/j.1365-294X.2006.03053.x. [DOI] [PubMed] [Google Scholar]
- 26.Hoffman JI, Forcada J. Extreme natal philopatry in female Antarctic fur seals (Arctocephalus gazella) Mamm. Biol. - Z. Für Säugetierkd. 2012;77:71–73. doi: 10.1016/j.mambio.2011.09.002. [DOI] [Google Scholar]
- 27.Forcada J, Trathan PN, Murphy EJ. Life history buffering in Antarctic mammals and birds against changing patterns of climate and environmental variation. Glob. Change Biol. 2008;14:2473–2488. [Google Scholar]
- 28.Bonner WN. The fur seal of South Georgia. British Antarctic Survey-Scientific reports No. 1968;56:1–81. [Google Scholar]
- 29.Headland, R. K. A chronology of Antarctic exploration: a synopsis of events and activities from the earliest times until the international polar years, 2007-09. (Bernard Quaritch Ltd., 2009).
- 30.Mill, H. R. The siege of the South Pole: the story of Antarctic exploration. (Alston Rivers, Limited, 1905).
- 31.Fanning, E. Voyages & discoveries in the South Seas, 1792–1832. (Marine Research Society, 1833).
- 32.Weddell, J. A voyage towards the South Pole: performed in the years 1822-24. (London: Longman, Hurst, Rees, Orme, Brown, and Green, 1825).
- 33.Rand RW. Notes on the Marion Island fur seal. Proc. Zool. Soc. Lond. 1956;126:65–82. doi: 10.1111/j.1096-3642.1956.tb00425.x. [DOI] [Google Scholar]
- 34.Ingham SE. The status of seals (Pinnipedia) at Australian Antarctic stations. Mammalia. 1960;24:422–430. [Google Scholar]
- 35.Shaughnessy, P. D. & Fletcher, L. Fur seals, Arctocephalus spp., at Macquarie Island. In Status, biology, and ecology of fur seals. Proceedings of an international symposium and workshop, Cambridge, England, 23–27 april 1984 177–188 (NOAA Technical Report NMFS 51, 1987).
- 36.Olstad O. Trekk av Sydishavets dyreliv (Features of the Southern Ocean wildlife) Nor. Geogr. Tidsskr.-Nor. J. Geogr. 1928;2:511–534. doi: 10.1080/00291952808551483. [DOI] [Google Scholar]
- 37.Payne MR. Growth of a fur seal population. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1977;279:67–79. doi: 10.1098/rstb.1977.0072. [DOI] [Google Scholar]
- 38.Boyd IL. Pup production and distribution of breeding Antarctic fur seals (Arctocephalus gazella) at South Georgia. Antarct. Sci. 1993;5:17–24. doi: 10.1017/S0954102093000045. [DOI] [Google Scholar]
- 39.O’Gorman FA. Fur seals breeding in the Falkland Islands Dependencies. Nature. 1961;192:914–916. doi: 10.1038/192914a0. [DOI] [Google Scholar]
- 40.Condy PR. Distribution, abundance and annual cycle for fur seals (Arctocephalus spp.) on the Prince Edward Islands. South Afr. J. Wildl. Res. 1978;8:159–168. [Google Scholar]
- 41.Jouventin, P. & Weimerskirch, H. Long-term changes in seabird and seal populations in the Southern Ocean. In Antarctic ecosystems (eds. Kerry, K. R. & Hempel, G.) (Springer, 1990).
- 42.Jouventin P, Stonehouse B. Biological survey of Ile de Croy, Iles Kerguelen, 1984. Polar Rec. 1985;22:688–691. doi: 10.1017/S0032247400006409. [DOI] [Google Scholar]
- 43.Budd GM, Downes MC. Population increase and breeding in the Kerguelen fur seal, Arctocephalus tropicalis gazella, at Heard Island. Mammalia. 1969;33:58–67. doi: 10.1515/mamm.1969.33.1.58. [DOI] [Google Scholar]
- 44.Goldsworthy SD, et al. Fur seals at Macquarie Island: post-sealing colonisation, trends in abundance and hybridisation of three species. Polar Biol. 2009;32:1473–1486. doi: 10.1007/s00300-009-0645-y. [DOI] [Google Scholar]
- 45.Kerley GIH. Relative population sizes and trends, and hybridization of fur seals Arctocephalus tropicalis and A. gazella at the Prince Edward Islands, Southern Ocean. Afr. Zool. 1983;18:388–392. [Google Scholar]
- 46.Laws RM. Population increase of fur seals at South Georgia. Polar Rec. 1973;16:856–858. doi: 10.1017/S003224740006397X. [DOI] [Google Scholar]
- 47.Cleary AC, et al. Prey differences drive local genetic adaptation in Antarctic fur seals. Mar. Ecol. Prog. Ser. 2019;628:195–209. doi: 10.3354/meps13108. [DOI] [Google Scholar]
- 48.Wynen LP, et al. Postsealing genetic variation and population structure of two species of fur seal (Arctocephalus gazella and A. tropicalis) Mol. Ecol. 2000;9:299–314. doi: 10.1046/j.1365-294x.2000.00856.x. [DOI] [PubMed] [Google Scholar]
- 49.Bonin CA, Goebel ME, Forcada J, Burton RS, Hoffman JI. Unexpected genetic differentiation between recently recolonized populations of a long-lived and highly vagile marine mammal. Ecol. Evol. 2013;3:3701–3712. doi: 10.1002/ece3.732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Humble E, et al. RAD sequencing and a hybrid Antarctic fur seal genome assembly reveal rapidly decaying linkage disequilibrium, global population structure and evidence for inbreeding. G3 Genes Genomes Genet. 2018;8:2709–2722. doi: 10.1534/g3.118.200171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hoffman JI, et al. A global cline in a colour polymorphism suggests a limited contribution of gene flow towards the recovery of a heavily exploited marine mammal. R. Soc. Open Sci. 2018;5:181227. doi: 10.1098/rsos.181227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gelman, A. et al. Bayesian data analysis. (Chapman and Hall/CRC, 2013).
- 53.Excoffier L, Foll M. fastsimcoal: a continuous-time coalescent simulator of genomic diversity under arbitrarily complex evolutionary scenarios. Bioinformatics. 2011;27:1332–1334. doi: 10.1093/bioinformatics/btr124. [DOI] [PubMed] [Google Scholar]
- 54.Hoffman JI, Grant SM, Forcada J, Phillips CD. Bayesian inference of a historical bottleneck in a heavily exploited marine mammal. Mol. Ecol. 2011;20:3989–4008. doi: 10.1111/j.1365-294X.2011.05248.x. [DOI] [PubMed] [Google Scholar]
- 55.Shaughnessy PD, Goldsworthy SD. Population size and breeding season of the Antarctic fur seal Arctocephalus gazella at Heard Island-1987/88. Mar. Mammal Sci. 1990;6:292–304. doi: 10.1111/j.1748-7692.1990.tb00359.x. [DOI] [Google Scholar]
- 56.Wege M, et al. Trend changes in sympatric Subantarctic and Antarctic fur seal pup populations at Marion Island, Southern Ocean. Mar. Mammal Sci. 2016;32:960–982. doi: 10.1111/mms.12306. [DOI] [Google Scholar]
- 57.Hoban SM, Gaggiotti OE, Bertorelle G. The number of markers and samples needed for detecting bottlenecks under realistic scenarios, with and without recovery: a simulation-based study. Mol. Ecol. 2013;22:3444–3450. doi: 10.1111/mec.12258. [DOI] [PubMed] [Google Scholar]
- 58.Hahn, M. W. Molecular population genetics. (Sinauer Associates, 2018).
- 59.Frankham R. Effective population size/adult population size ratios in wildlife: a review. Genet. Res. 1995;66:95–107. doi: 10.1017/S0016672300034455. [DOI] [PubMed] [Google Scholar]
- 60.Grafton RQ, Kompas T, Hilborn RW. Economics of overexploitation revisited. Science. 2007;318:1601–1601. doi: 10.1126/science.1146017. [DOI] [PubMed] [Google Scholar]
- 61.Basberg, B. & Headland, R. K. The 19th century Antarctic sealing industry: sources, data and economic significance. (2008).
- 62.Stackpole, E. A. The Sea-Hunters: the New England whalemen during two centuries, 1635–1835. vol. 23 (J.B. Lippincott & Co., 1954).
- 63.Stackpole, E. A. The voyage of the Huron and the Huntress: the American sealers and the discovery of the continent of Antarctica. (Marine Historical Association, 1955).
- 64.O’Brien SJ. A role for molecular genetics in biological conservation. Proc. Natl. Acad. Sci. USA. 1994;91:5748–5755. doi: 10.1073/pnas.91.13.5748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hoelzel AR. Impact of population bottlenecks on genetic variation and the importance of life-history; a case study of the northern elephant seal. Biol. J. Linn. Soc. 1999;68:23–39. doi: 10.1111/j.1095-8312.1999.tb01156.x. [DOI] [Google Scholar]
- 66.Groombridge JJ, Bruford MW, Jones CG, Nichols RA. Evaluating the severity of the population bottleneck in the Mauritius kestrel Falco punctatus from ringing records using MCMC estimation. J. Anim. Ecol. 2001;70:401–409. doi: 10.1046/j.1365-2656.2001.00502.x. [DOI] [Google Scholar]
- 67.Hoelzel AR, Fleischer RC, Campagna C, Le Boeuf BJ, Alvord G. Impact of a population bottleneck on symmetry and genetic diversity in the northern elephant seal. J. Evol. Biol. 2002;15:567–575. doi: 10.1046/j.1420-9101.2002.00419.x. [DOI] [Google Scholar]
- 68.Amos W, Balmford A. When does conservation genetics matter? Heredity. 2001;87:257–265. doi: 10.1046/j.1365-2540.2001.00940.x. [DOI] [PubMed] [Google Scholar]
- 69.Amos W, Harwood J. Factors affecting levels of genetic diversity in natural populations. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 1998;353:177–186. doi: 10.1098/rstb.1998.0200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Murray GGR, et al. Natural selection shaped the rise and fall of passenger pigeon genomic diversity. Science. 2017;358:951–954. doi: 10.1126/science.aao0960. [DOI] [PubMed] [Google Scholar]
- 71.Dickerson BR, Ream RR, Vignieri SN, Bentzen P. Population structure as revealed by mtDNA and microsatellites in Northern fur seals, Callorhinus ursinus, throughout their range. Plos One. 2010;5:e10671. doi: 10.1371/journal.pone.0010671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lancaster ML, Arnould JPY, Kirkwood R. Genetic status of an endemic marine mammal, the Australian fur seal, following historical harvesting. Anim. Conserv. 2010;13:247–255. doi: 10.1111/j.1469-1795.2009.00325.x. [DOI] [Google Scholar]
- 73.Goldsworthy S, Francis J, Boness D, Fleischer R. Variation in the mitochondrial control region in the Juan Fernández fur seal (Arctocephalus philippii) J. Hered. 2000;91:371–377. doi: 10.1093/jhered/91.5.371. [DOI] [PubMed] [Google Scholar]
- 74.Majluf P, Goebel ME. The capture and handling of female South American fur seals and their pups. Mar. Mammal Sci. 2006;8:187–190. doi: 10.1111/j.1748-7692.1992.tb00382.x. [DOI] [Google Scholar]
- 75.Sambrook, J., Fritsch, E. F. & Maniatis, T. Molecular cloning. vol. 2 (Cold spring harbor laboratory press New York, 1989).
- 76.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Besnier F, Glover KA. ParallelStructure: a R package to distribute parallel runs of the population genetics program STRUCTURE on multi-core computers. Plos One. 2013;8:e70651. doi: 10.1371/journal.pone.0070651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software structure: a simulation study. Mol. Ecol. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- 79.Francis R. M. pophelper: an R package and web app to analyse and visualize population structure. Mol. Ecol. Resour. 2017;17:27–32. doi: 10.1111/1755-0998.12509. [DOI] [PubMed] [Google Scholar]
- 80.Jombart T. adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics. 2008;24:1403–1405. doi: 10.1093/bioinformatics/btn129. [DOI] [PubMed] [Google Scholar]
- 81.Goudet J. hierfstat, a package for r to compute and test hierarchical F-statistics. Mol. Ecol. Notes. 2005;5:184–186. doi: 10.1111/j.1471-8286.2004.00828.x. [DOI] [Google Scholar]
- 82.Archer FI, Adams PE, Schneiders BB. stratag: An R package for manipulating, summarizing and analysing population genetic data. Mol. Ecol. Resour. 2016;17:5–11. doi: 10.1111/1755-0998.12559. [DOI] [PubMed] [Google Scholar]
- 83.Keenan K, McGinnity P, Cross TF, Crozier WW, Prodöhl P. A. diveRsity: An R package for the estimation and exploration of population genetics parameters and their associated errors. Methods Ecol. Evol. 2013;4:782–788. doi: 10.1111/2041-210X.12067. [DOI] [Google Scholar]
- 84.Kamvar ZN, Tabima JF, Grünwald NJ. Poppr: an R package for genetic analysis of populations with clonal, partially clonal, and/or sexual reproduction. PeerJ. 2014;2:e281. doi: 10.7717/peerj.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Piry S, Luikart G, Cornuet JM. BOTTLENECK: a computer program for detecting recent reductions in the effective population size using allele frequency data. J. Hered. 1999;90:502–503. doi: 10.1093/jhered/90.4.502. [DOI] [Google Scholar]
- 86.Hofmeyr GJG, Bester MN, Makhado AB, Pistorius PA. Population changes in Subantarctic and Antarctic fur seals at Marion Island. South Afr. J. Wildl. Res. 2006;36:55–68. [Google Scholar]
- 87.Pacifici M, et al. Generation length for mammals. Nat. Conserv. 2013;5:87–94. [Google Scholar]
- 88.Garza JC, Williamson EG. Detection of reduction in population size using data from microsatellite loci. Mol. Ecol. 2001;10:305–318. doi: 10.1046/j.1365-294x.2001.01190.x. [DOI] [PubMed] [Google Scholar]
- 89.Csilléry K, François O, Blum MG. B. abc: an R package for approximate Bayesian computation (ABC) Methods Ecol. Evol. 2012;3:475–479. doi: 10.1111/j.2041-210X.2011.00179.x. [DOI] [PubMed] [Google Scholar]
- 90.Paijmans AJ, et al. Data from: The genetic legacy of extreme exploitation in a polar vertebrate. Zenodo. 2019 doi: 10.5281/zenodo.3585717. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All of the raw data90 are available via the Zenodo repository, doi:10.5281/zenodo.3585717.
All data wrangling steps and statistical analyses except for the heterozygosity-excess tests were implemented in R. The complete documented analysis pipeline is available via our GitHub repository: https://github.com/apaijmans/AFS_genetic_legacy.