Abstract
Streptococcus mutans is an etiologic agent of human dental caries that forms dental plaque biofilms containing functional amyloids. Three amyloidogenic proteins, P1, WapA, and Smu_63c were previously identified. C123 and AgA are naturally occurring amyloid-forming fragments of P1 and WapA, respectively. We determined that four amyloidophilic dyes, ThT, CDy11, BD-oligo, and MK-H4, differentiate C123, AgA, and Smu_63c amyloid from monomers, but non-specific binding to bacterial cells in the absence of amyloid precludes their utility for identifying amyloid in biofilms. Congo red-induced birefringence is a more specific indicator of amyloid formation and differentiates biofilms formed by wild-type S. mutans from a triple ΔP1/WapA/Smu_63c mutant with reduced biofilm forming capabilities. Amyloid accumulation is a late event, appearing in older S. mutans biofilms after 60 hours of growth. Amyloid derived from pure preparations of all three proteins is visualized by electron microscopy as mat-like structures. Typical amyloid fibers become evident following protease digestion to eliminate non-specific aggregates and monomers. Amyloid mats, similar in appearance to those reported in S. mutans biofilm extracellular matrices, are reconstituted by co-incubation of monomers and amyloid fibers. X-ray fiber diffraction of amyloid mats and fibers from all three proteins demonstrate patterns reflective of a cross-β amyloid structure.
Subject terms: Biophysics, Chemical biology, Microbiology, Structural biology
Introduction
Amyloids are ordered protein aggregates with similar quaternary structures and biophysical characteristics. They are formed when soluble proteins change conformation and self-assemble into insoluble fibrillar aggregates1. Amyloids were initially identified in the context of pathology but functional amyloids are now recognized in all kingdoms of life, mediating desirable activities for the organisms that produce them2,3. Microbes use amyloids for their mechanical and biological properties4. Amyloid fibers have a tensile strength comparable to steel5, and are protease and detergent resistant6–8. Multiple bacterial functional amyloids have been identified that can contribute to adhesion, biofilm development, genetic competence, cell density regulation, host interactions, and/or aerial hyphae formation9–12. Microbial amyloid formation is generally recognized as a biofilm-associated process13–15. S. mutans is a major causative agent of human dental caries, a classic biofilm related disease16. Proteins that assemble into amyloid can serve as scaffolds to increase stiffness and unify the extracellular matrix (ECM) and cells within biofilms11,17,18. Bacterial amyloids also participate in other biological processes. For example, Fap amyloid fibrils of Pseudomonas aeruginosa can serve as a functional sink for quorum-sensing molecules19. While bacterial amyloids share many common features, unique species-specific aspects are also being identified.
Amyloids display distinct tinctorial properties and spectral shift upon uptake of dyes such as Thioflavin T (ThT) that increase in fluorescence intensity upon binding. When Congo-red (CR) binds to amyloid it becomes birefringent due to its orientation and optical properties, visualized as anomalous yellow-green or orange colors under crossed-polarizers20,21. Amyloids display a stable assembly of β-sheets packed perpendicular to the fiber axis when examined by X-ray diffraction, Fourier-transform infrared spectroscopy and solid-state nuclear magnetic resonance22,23. Amyloid fibers vary in length but display typical diameters of 4–11 nm6. X-ray fiber diffraction has identified a stacked cross β-sheet quaternary structure of amyloid fibers with a common diffraction pattern of ~4.7–4.8 Å meridional signals, corresponding to the distance between hydrogen-bonded β-strands, and more diffuse ~10–12 Å equatorial signals arising from association of the sheets24. Additionally, the fiber structure of the phenol-soluble modulin α3 peptide of Staphylococcus aureus is reported as an atypical “cross-α“ amyloid-like architecture with amphipathic α-helices stacked perpendicular to the fiber axis in self-associating sheets25. Thus, it is important to characterize individual bacterial amyloids to identify both shared and novel features.
A paradigm is emerging for Gram-positive organisms whereby surface-localized proteins serve dual functions as adhesins, and are also processed into amyloidogenic derivatives that can polymerize depending on prevailing environmental conditions13,17. Streptococcus mutans produces a typical biofilm ECM composed of polysaccharides, proteins, and eDNA. Amyloid formation is a biofilm-related event in S. mutans with three β sheet-rich amyloid-forming proteins identified to date26. P1 (aka AgI/II) and wall-associated protein A (WapA) proteins are surface-localized sortase substrates whose truncation derivatives, C123 (aka, AgII) and AgA, are amyloidogenic. Transpeptidase sortase enzymes covalently link their substrates to the cell wall peptidoglycan27. S. mutans lacking sortase is defective in amyloid production suggesting a potential amyloid nucleation mechanism at the cell surface28, analogous to the process described for E. coli curli amyloid formation15,29,30. Smu_63c, a third amyloidogenic protein identified in S. mutans, is a secreted protein that appears to serve as a negative regulator of genetic competence and biofilm cell density26,31. All three of these S. mutans proteins influence biofilm development, which is inhibited by known inhibitors of amyloid fibrillization such as tannic acid and epigallocatechin gallate26.
The purpose of this study was to evaluate methods to study S. mutans amyloid formation in vitro, to further characterize S. mutans amyloids, and to track the dynamics of amyloid formation in situ within growing biofilms. The new information described herein will facilitate the design of future experiments to evaluate environmental conditions that influence amyloid formation of S. mutans proteins in vitro and within biofilms.
Results
Purification and characterization of amyloid fibers
SDS-Page and Western blot confirmed elimination of protein monomers from each induced amyloid sample by treatment with proteinase K (PK) or PK/Triton. (Fig. S1). All samples showed a different solubility before and after treatment. Prior to amyloid induction, proteins remained in solution. After induction but before protease digestion, amyloid material remained in a homogenous suspension, but once monomers were eliminated the material fell to the bottom of the tubes (Fig. S2). Each protease treated sample was lyophilized and weighed to quantify recovered protease-resistant amyloid material, ~10–20% of the total starting amount of each protein. Insoluble protease-resistant amyloid was suspended in ddH2O by vortexing and small cycles of sonication for further physicochemical characterization.
Transmission electron microscopy
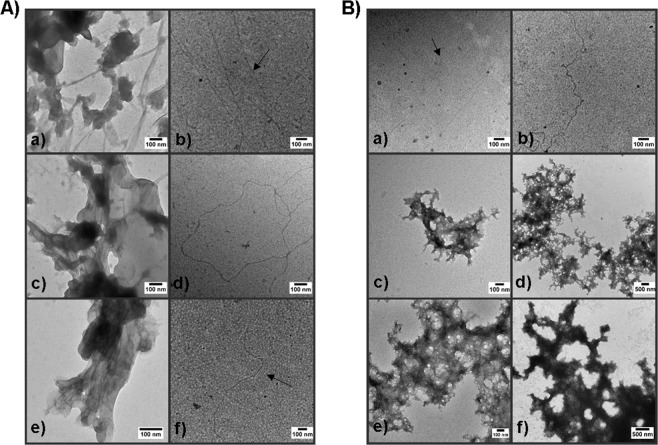
Crude induced amyloid and residual insoluble material harvested after protease digestion were visualized by TEM. Induced amyloid of each protein appeared as mat-like aggregates (Fig. 1A panels a, c, e), consistent with previously published images of purified bacterial amyloid proteins produced by S. mutans as well as by Bacillus subtilis8,26,32, and similar in appearance to nanocomposite structures comprised of amyloid fibers and graphene33. Sample morphologies were notably different following protease digestion in which typical amyloid fibers with diameters of ~4 nm remained (Fig. 1A panels b,d,f). We speculated that the mat-like structures may represent a composite of amyloid fibers and randomly aggregated monomers. Purified fibers of C123 were therefore incubated at 37 °C for 2 or 4 weeks without stirring in the presence of 1 or 10% (w/w) added monomer (Fig. 1B). Fibers without monomer were included as a negative control. The C123 fiber only controls showed no change in appearance over time, but the samples with added monomer reassembled into mat-like structures.
Figure 1.
Transmission electron microscopy (TEM) of amyloid mats and purified fibers (A) TEM images of S. mutans amyloid before and after removal of residual monomer. a, c and e) Induced amyloid produced from purified C123, AgA, and Smu_63c, respectively. b, d and f). Amyloid material produced from C123, AgA, and Smu_63c following proteinase K digestion. (B) TEM of purified C123 amyloid fibers incubated without stirring with and without added monomers. a, c, and e) Purified C123 fibers incubated for 2 weeks alone (a) or with added C123 monomer at final concentrations of 1% (c) or 10% (e). e, d, and f) Purified C123 fibers incubated for 4 weeks alone (b) or with added C123 monomer at final concentrations of 1% (d) or 10% (f).
Fluorescent dye assays
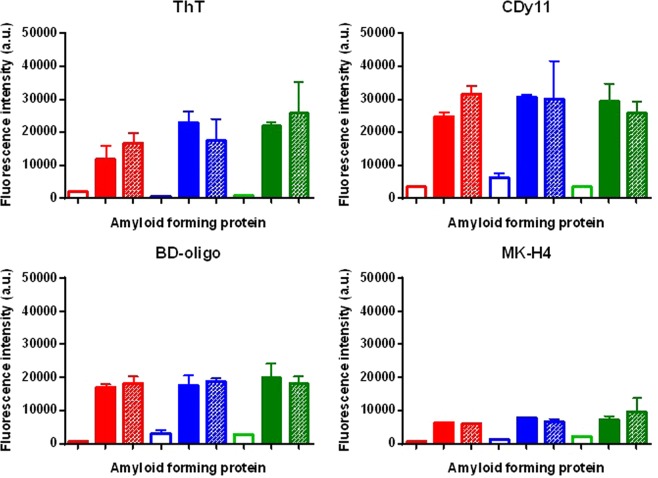
In addition to ThT34, newer amyloidophilic dyes have also been reported. We performed dye uptake assays with ThT, CDy1135, BD-oligo36 and MK-H4 (Young-Tae Chang, personal communication), comparing protein monomers to induced amyloid material and purified protease resistant amyloid fibers (Fig. 2). All dyes readily distinguished between monomeric and both amyloid forms of each protein. Fluorescence values were highest for CDy11, followed by ThT and BD-oligo. MK-H4 was the least sensitive dye tested.
Figure 2.
Amyloidophilic dye uptake assays. Ten micrograms of purified recombinant C123 (red bars), AgA (blue bars), or Smu_63c (green bars) were reacted with ThT, CDy11, BD-oligo, and MK-H4. Empty bars correspond to the purified monomeric proteins, solid bars to the samples after amyloid induction, and diagonal patterned bars to the purified amyloid fibers after treatment of the amyloid material to remove residual monomers. C123 and AgA polypeptides correspond to naturally occurring amyloidogenic derivatives of P1 and WapA, respectively. Arbitrary units (a.u.) ThT (Excitation = 440, Emission = 485), CDy11 and BD-Oligo (Ex = 530, Em = 580), and MK-H4 (Ex = 475, Em = 567). Protein concentration 0.1 mg/mL. Error bars represent standard deviations.
Congo red birefringence
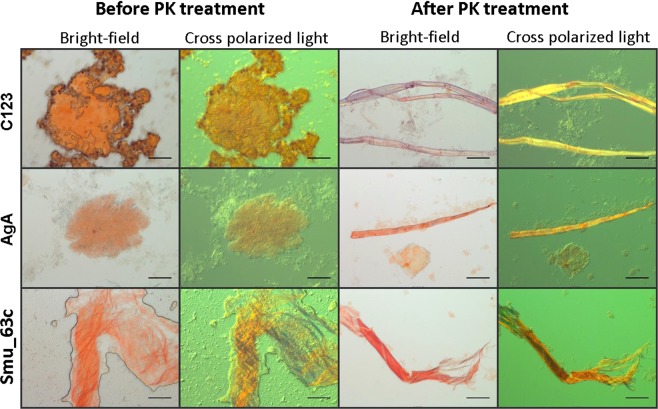
When induced amyloid and purified fibers of each protein were stained with CR, all samples demonstrated typical birefringent properties (Fig. 3). Prior to protease treatment samples looked mat-like and birefringence was more diffuse. After protease treatment birefringence was more pronounced and rope-like structures were visualized. Protein monomers did not stain with CR and were not visible by light microscopy (not shown).
Figure 3.
Congo red-induced birefringence of amyloid material before and after treatment with proteinase K (PK). Purified recombinant Smu_63c, and recombinant C123 and AgA polypeptides, which correspond to naturally occurring amyloidogenic derivatives of P1 and WapA respectively, were evaluated. Samples were visualized by bright-field microscopy and under crossed polarizing light filters. Scale Bar 50 µm.
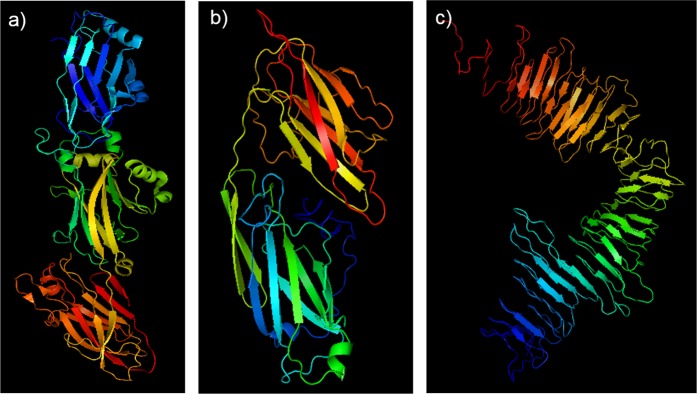
Protein tertiary structure and X-ray fiber diffraction
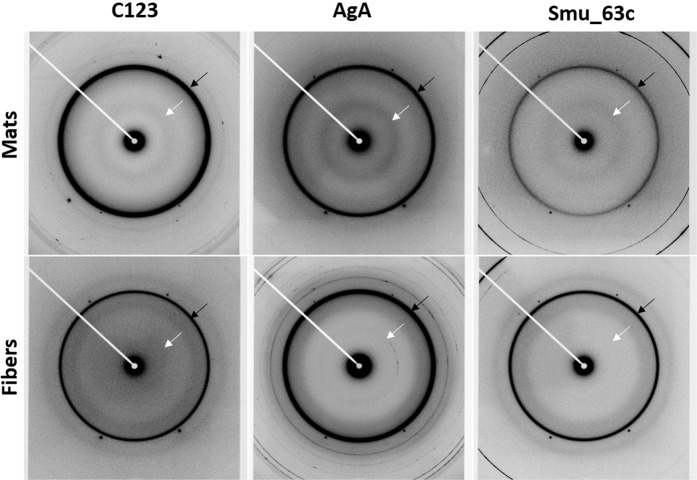
The crystal structure of C123 has been solved and is comprised of three domains (C1–3), each adopting a DE-variant IgG fold with two beta sheets linked through isopeptide bonds37 (Fig. 4, panel a). Crystal structures of AgA and Smu_63c are not known. We utilized I-TASSER38 to predict structures of AgA and Smu_63c (Fig. 4, panels b and c). AgA appears as two beta sheet-rich domains connected by disordered loops. Smu_63c also appears rich in beta sheet structure. While all three proteins exhibit predominantly beta sheet structure, the domain organization of each is distinct and no primary sequence homology is apparent (Fig. S3). We evaluated X-ray diffraction patterns of amyloid mats and purified amyloid fibers of C123, AgA and Smu_63c (Fig. 5). Common features included diffraction rings detected at ~4.8 Å as well as at ~10.5 Å. These signals are in good agreement with reported meridional reflections at ~4.7–4.8 Å that stem from β-strands stacked perpendicular to the fiber axis, and equatorial reflections at ~10–12 Å on the equator that stem from β-sheets running parallel to the fiber axis39. Observed rings, rather than separated reflections, are consistent with diffraction of non-aligned fibers in the samples.
Figure 4.
Schematic representation of the solved or predicted tertiary structures of P1-C123 (a), WapA-AgA (b), and Smu_63c (c). The illustration of C123 is based on the solved crystal structure37 (PDB id:3QE5). The predicted structures of AgA and Smu_63C were determined using I-TASSER38. Blue to red rainbow coloring indicates progression from N- to C-termini.
Figure 5.
X-ray fiber diffraction patterns of amyloid mats and purified amyloid fibers derived from P1-C123, WapA-AgA and, Smu_63c. Black arrows indicate meridonal diffraction at ~4.8 Å. White arrows indicate equatorial diffraction at ~10.5 Å.
Detection of S. mutans amyloid within biofilms in a microfluidics flow cell
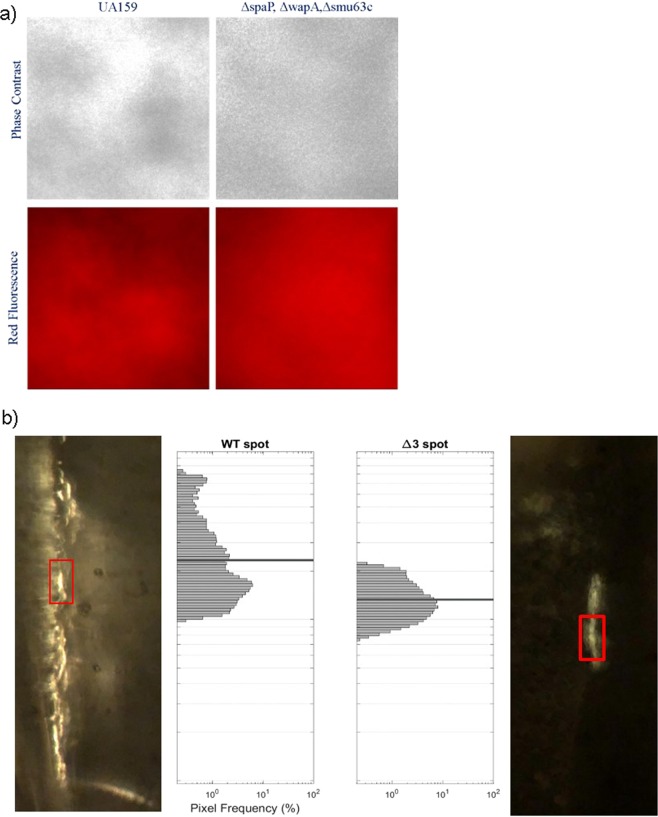
To evaluate amyloid formation in situ within growing biofilms we compared the S. mutans wildtype and a triple deletion (Δ3) mutant26 growing in microfluidic channels under steady flow of medium. We utilized CDy11 since it was the most sensitive dye for detecting amyloids in the in vitro assay (Fig. 2). First, we confirmed that CDy11 could discriminate monomeric from amyloid protein forms by fluorescent microscopy (Fig. S4). Then S. mutans cells were grown in glass capillaries under steady flow for 72 h, exposed to CDy11 and imaged in phase contrast and fluorescence microscopy (Fig. 6, panel a). Bacterial biofilms were evident on the capillary walls of both strains, and CDy11 staining paralleled bacterial accumulation in the chambers over time (not shown), but the strains did not differ in red fluorescence following CDy11 staining. The absence of differential staining between wild-type and mutant strains suggested non-specific background binding of the dye to non-amyloid material in the biofilms. Thus, a potentially more discriminating method for tracking amyloid formation in situ was investigated. S. mutans biofilms were grown in capillaries as described above, but exposed to CR and visualized by bright-field microscopy and under crossed polarizers. CR-induced birefringence was more prevalent and of higher measurable intensity in the wildtype compared to the Δ3 mutant strain (Fig. 6, panel b). Residual CR-induced birefringence in the Δ3 mutant strain biofilm suggests that additional unidentified amyloidogenic proteins may also be present. Further, stronger birefringence was observed with the polarizing axis at 45° with respect to the direction of media flow, than with parallel or perpendicular alignment. The Congo red molecule is believed to bind to amyloid with its electric dipole perpendicular to the long axis of the fibril40. Therefore the maximum Congo red birefringence is expected when the polarizer is aligned at 45° degrees relative to the fibril axis41. Our data therefore suggest that the amyloid fibers are at least partially aligned along the flow direction. We also investigated whether non-specific binding to S. mutans was problematic for ThT. Bacteria were cultured under planktonic conditions in which amyloid production would be minimal8,9. In addition, reactivity of ThT with cells from the wildtype strain was compared to that of the Δ3 and ΔsrtA mutants in which biofilm-associated amyloid formation is impaired26,28. Similar ThT associated fluorescence was observed for all three strains and was directly proportional to cell number (Fig. S5).
Figure 6.
Assessment of amyloid formation during S. mutans biofilm growth in a microfluidics system. (a) Phase contrast and red fluorescence images following staining of 72 h biofilms of S. mutans wild-type and a triple mutant (Δ3) lacking P1 (encoded by spaP), WapA, and Smu_63c with CDy11. Similar patterns of dye binding to both strains suggests CDy11 reactivity with non-amyloid material. (b) CR-induced birefringence of 72 h biofilms of S. mutans wild-type (left) and the Δ3 triple mutant (right). Insets indicate pixel frequency of the brightest visual locations (red boxes) in each image. In contrast to staining with CDy11, biofilms of the WT and mutant strains could be differentiated on the basis of CR-induced birefringence.
Time course of amyloid formation in static biofilms
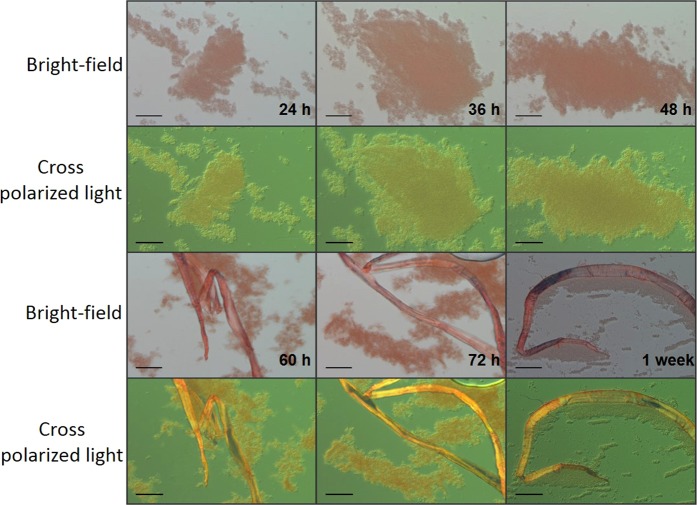
P1, WapA, and Smu_63c are constitutively produced by S. mutans during both planktonic and biofilm growth. Similar to other bacteria14,42–44, amyloid formation by S. mutans is biofilm-associated28. To assess the time course of amyloid formation within S. mutans biofilms, we evaluated static biofilms at 24 h and up to 1 week. CR-induced birefringence was evaluated every 12 h (Fig. 7). Birefringent material was not visible until the biofilms had grown for 60 h.
Figure 7.
Time course of amyloid formation in S. mutans biofilms. Extracellular birefringent rope-like structures were visualized in biofilm material harvested after 60 h of growth as static cultures. Images were captured by bright-field microscopy and under cross-polarizing light filters to visualize Congo Red-induced birefringence at the indicated time points. Scale Bar 50 µm.
Discussion
S. mutans is a highly acidogenic organism that accumulates in dental plaque where it clings to tooth surfaces in a tenacious biofilm encased in an ECM composed of glucan and fructan polymers, proteins, and extracellular DNA (eDNA). While glucosyltransferase enzymes and glucan binding proteins are critical for sucrose-dependent bacterial adhesion, cell surface proteins such as P1 and WapA mediate sucrose-independent adhesion to constituents of the salivary pellicle coating tooth surfaces16,45. In streptococci and staphylococci, truncation derivatives of several adhesins are now known to polymerize into amyloid fibers potentially facilitated by eDNA13,17. The protease resistant amyloid-like material harvested from the ECM of S. mutans biofilms exists in an apparent complex with eDNA8. The C123 and AgA fragments of P1 and WapA are both capable of amyloid formation26. The structure of P1 has been extensively characterized37,46,47. The C123 fragment has been shown to interact with cell wall localized-full-length P148,49. Specific amino acid residues involved in that binding have been characterized by NMR50. While WapA has not been as widely studied as P1, it also contributes to S. mutans cell surface architecture and biofilm formation45,51. The N-terminal amyloidogenic AgA fragment is also reactive with type I collagen and fibronectin52. The third known amyloid forming protein of S. mutans, Smu_63c, is secreted, and a Δsmu_63c single mutant demonstrates increased biofilm cell density and genetic competence26,31. The functional amyloid Fap of Pseudomonas aeruginosa interacts with quorum sensing molecules involved in cell density signaling cascades19; however, it is not yet known whether S. mutans Smu_63c interacts with competence activating peptides53, either in monomeric or amyloid form. Mutants lacking P1, C123, and Smu_63c singly and in combination have been generated, with the triple mutant strain most seriously impaired in biofilm formation26. Two of the three amyloidogenic S. mutans proteins are linked to the cell surface by sortase. A mutant strain lacking this enzyme does not form amyloid despite producing its substrate proteins in secreted form28. Interestingly, three of the eight known amyloidogenic proteins of Streptomyces coelicolor are also sortase substrates54. A common theme that emerges from these studies is that amyloidogenic bacterial proteins are either surface-associated or secreted, and therefore a part of the extracellular matrix. Given the increasing number of bacterial amyloid forming proteins identified, and the potential factors that may contribute to their ability to polymerize, it is of interest to be able to track amyloid formation specifically within the complex milieu of bacterial biofilms.
When amyloid material produced from each of the three S. mutans proteins was visualized by TEM before and after protease digestion, a striking difference in morphology became apparent. Crude amyloid induced by mechanical agitation appeared as mat-like structures, whereas protease digestion of the mats revealed long thin fibers more typical in appearance to that reported for classical disease-related amyloids found in mammals1,39. All four amyloidophilic dyes we tested demonstrated increased fluorescence when reacted with amyloid mats or purified amyloid fibers, but not protein monomers. C123, AgA, and Smu_63c amyloid mats and fibers also displayed CR-induced birefringence. Because of the resemblance of the mats to nanocomposites of graphene and amyloid33, we considered that they might represent a higher order structure composed of fibers held together by protein monomers and/or prefibrillar protease-sensitive oligomeric intermediates. Mat-like structures were indeed reconstituted when fibers and monomers were incubated without agitation, whereas fibers alone were stable over the 4 week time course of the experiment. Similar to engineered amyloid nanocomposites, the fibers “disappeared” when integrated into the supramolecular structure33. The reconstitution of mats in the presence of monomer suggests that the preferred form of S. mutans amyloid is as a supramolecular structure, which may exist in equilibrium with monomer and possibly oligomeric intermediates.
Size and shape of amyloid aggregates and kinetics of their formation are affected by physiochemical factors including the environmental surfaces to which they are exposed55. Extracellular material present in 5 day old S. mutans biofilms was reactive with anti-P1, WapA, and Smu_63c specific antibodies when visualized by immunogold electron microscopy and appeared more mat-like than fibrillar in nature26. This suggests that mats represent a naturally occurring amyloid-containing form produced during S. mutans biofilm growth. Mat-like amyloid aggregates have also been visualized within biofilm ECM of Bacillus subtilis and Staphylococcus aureus17,32. The observed difference in suspension of amyloid mats compared to purified fibers may confer a biological advantage to an organism such as S. mutans that dwells in the aqueous environment of the oral cavity. The encasement of fibrillar amyloid within S. mutans ECM was supported by visualization of thin and thick fibers when extracellular biofilm material was separated from whole cells, treated with DNAse, RNAse and proteinase K, and analyzed by TEM8.
The ability to form amyloid fibers is not directly related to the primary sequence of any given polypeptide backbone, although prediction programs can suggest possible amyloid forming regions. Nonetheless, amyloid fibers that form from different peptides or proteins share the common feature of high β-sheet content that presents as similar X-ray diffraction patterns of the amyloid aggregates56. Analysis of amyloid mats and purified fibers of C123, AgA, and Smu_63c by X-ray fiber diffraction was consistent with a typical stacked cross-β amyloid structure achieved by each protein. Importantly, the observed diffraction patterns of the three different proteins were strikingly similar to one another, as previously reported when amyloids of disparate protein origin have been characterized by this technique57–60. The observed patterns also indicate more conventional β-amyloid assemblies for each of the S. mutans proteins rather than the unusual cross-α amyloid-like assembly that has been reported for the phenol soluble modulins of S. aureus25. The diffraction patterns we observed are similar to those reported for synthetic peptides corresponding to the amyloidogenic chaplin proteins of Streptomyces coelicolor, as well as to a crude mixture of proteins extracted from the bacterial cell wall of that organism54. It is not yet known whether the different chaplin proteins are capable of forming heteroaggregates54. Likewise, it is unknown whether S. mutans proteins form primarily homo or hetero amyloid aggregates during biofilm growth and this question will be the topic of further study.
While, it has been reported that S. mutans amyloid adopts a unique α-sheet structure61, this conclusion was based on indirect evidence in which α-sheet peptides comprised of alternating L- and D-amino acids inhibited ThT uptake and fluorescence by S. mutans biofilms. Our data caution against using fluorescent dye uptake as a surrogate marker of amyloid fibrilization within biofilms of this organism and demonstrate high background binding of dye directly to the bacterial cells in the absence of amyloid (Figs. 6 and S5). Thus in a cellular context uptake of fluorescent dyes is indicative of S. mutans cell number. Because D-amino acids can interfere with bacterial protein synthesis62, as well as with growth of early biofilm foci into larger assemblies of cells after initial bacterial attachment63, it is critical to substantiate conclusions regarding potential amyloid polymerization mechanisms of S. mutans biofilm ECM proteins by biophysical methods other than ThT or CDy11 uptake. CR-induced birefringence appears to be a more specific indicator of amyloid formation in S. mutans biofilms. When a time course experiment was performed to identify amyloid in S. mutans biofilms, CR-induced birefringent material was not evident until biofilms had been cultured for 60 h. This suggests that amyloid is not present or prevalent early in biofilm development during initial adhesion of the bacterial cells, but rather accumulates within the extracellular matrix as the biofilms age. This may aid in cellular consolidation and accumulation into macrocolonies that appear within mature biofilms64.
In addition to amyloid, macromolecules contained within extracellular matrices (ECM) of Streptococcus mutans biofilms include extracellular DNA, as well as soluble and insoluble glucan polymers produced in the presence of sucrose by three different glycosyltransferases, GtfB, C, and D (reviewed in Lemos et al. 2019)16. In S. mutans, sucrose-dependent biofilm formation is severely impaired in a triple mutant lacking gtfBCD, whereas deletion of genes encoding the sucrose-independent adhesins P1 and WapA has a less obvious impact on sucrose-dependent biofilm formation26. In the current study amyloid formation was tracked in biofilms grown without sucrose. However, it was demonstrated previously that amyloid inhibitors impede S. mutans biofilm formation whether sucrose is present or not, and that during growth in sucrose mutant strains lacking genes encoding P1 and/or WapA lose sensitivity to biofilm inhibition by small-molecule amyloid inhibitors compared to wild-type cells26. This suggests functional cooperation of amyloid forms of P1 and WapA-derived polypepetides with adhesive glucans formed during biofilm development in sucrose. Bacterial amyloids present in biofilm extracellular matrices are reported to confer stability by interacting with both extracellular DNA as well as exopolysaccharides13. Taken together, the current results will aid in the design of future studies to investigate amyloid interactions with other ECM components, to evaluate environmental triggers of amyloidogenesis within S. mutans biofilms, and to understand potential mechanisms of action of compounds that inhibit both amyloid and biofilm formation in this organism.
Materials and Methods
Amyloid induction
Recombinant C123, AgA, and Smu_63c were purified and amyloid induced by mechanical agitation (stirring) as described26. Amyloid formation was facilitated in this study by seeding with pre-formed amyloid. Purified proteins were adjusted to 1 mg/mL in 500 µL of 25 mM Tris, 200 mM NaCl, pH 8.0 for C123 and AgA, or 25 mM PO4, 300 mM NaCl, pH 4.0 for Smu_63c, and stirred at 4 °C in a 1.5 ml Eppendorf tube using a 10 ×3 mm micro stir bar (Fisher Scientific) for 3–5 days on a Fisher Scientific Isotemp stir plate at 1200 rpm. Next, 3.5 mL of purified recombinant protein (1 mg/mL) was seeded with the 500 µL of preformed amyloid. Samples were stirred as above in 15 mL falcon tubes using octagon Spinbar magnetic stirring bars (25.4 ×8 mm) for 7 days.
Purification of amyloid fibers
Induced amyloid (1 mg/mL) was treated with proteinase K (PK) at a 10:1 (amyloid:PK) molar ratio. C123 was incubated at 37 °C for 1.5 h, and AgA and Smu_63c were incubated for 3 h. Reactions were stopped by addition of phenylmethylsulfonyl fluoride (PMSF) to 2 mM and incubation for 5 min at room temperature. Samples were ultra-centrifuged at 10 °C for 30 min at 100,000 X g, using a Beckman Optima TLX (TLA-55) rotor. Pellets were resuspended in 1 ml double-distilled water (ddH2O), and ultra-centrifugation repeated with resuspension in 1 mL ddH2O. Samples were monitored for elimination of residual monomers by SDS-polyacrylamide gel electrophoresis and Western blotting using rabbit polyclonal antisera against AgA or Smu_63c, or the C123 specific monoclonal antibody 6–8C48. Residual monomer was eliminated from the C123 sample by additional treatment with 0.1% (v/v) Triton X100 for 15 min at room temperature. To quantify fiber yield, samples were dried overnight under vacuum in tared tubes in a refrigerated vapor trap (Savant RVT 100) followed by weighing with an analytical balance (AL54 Mettler Toledo).
Transmission electron microscopy
One drop of crude amyloid or purified fiber sample (1 mg/mL in water) was placed under a 100-mesh formvar-carbon coated grid FCF100-Cu-UB (Electron Microscopy Sciences, Hatfield, PA) for five minutes. Grids were transferred to one drop of ddH2O for one min and stained by placement over a drop of 2% uranyl acetate for 2 min. Excess uranyl acetate was removed by setting grids in ddH2O for two sec. Imaging was done using a Hitachi H7600 transmission electron microscope (Hitachi High Technologies America, Schaumburg, IL). Digital images were acquired using an AMT digital camera. In the case of mat reconstitution, P1-C123 monomer (1 mg/mL) was added to 100 µL of pure fibers (1 mg/mL) to a final concentration of 1% or 10% (w/w) and incubated at 37 °C for 2 or 4 weeks. The negative control consisted of pure fibers (1 mg/mL) without added monomer.
X-ray fiber diffraction
Induced amyloid and purified fiber samples were prepared in enclosed plastic cuvette chambers as previously reported23, with minor modifications. A 5–10 µL drop of each sample was suspended between two 1 mm glass rods dipped in beeswax held in place 1.5 mm apart. The optimal concentrations needed to suspend the drop and allow the protein samples to dry without breaking were determined in pilot experiments. Amyloid mats were suspended at 20 mg/mL, and purified amyloid fibers at 50 mg/mL, in double distilled water. Cuvettes were placed in closed containers with saturated potassium sulfate for purified fibers, or saturated sodium chloride for amyloid mats, to maintain relative humidity at ~97% or ~75%, respectively. Samples were dried for up to two days. Diffraction data were collected at beamline 4-2 of the Stanford Synchrotron Radiation Lightsource (SSRL) at the SLAC National Accelerator Laboratory. The beam was slit collimated to a size of 200 ×200 µm, and data were recorded using a RayonixMX225HE detector with an energy of 11.5 keV placed at a distance of 334.4 mm from the fiber. The program WCEN65 was used to analyze the data.
Fluorescent dye assays
Thioflavin T (ThT) was from Fluka. CDy11, BoDipy-Oligomer (BD-oligo) and MK-H4 were kind gifts from Dr. Young-Tae Chang, Pohang University of Science and Technology, Korea. A 2 mM stock solution of ThT was prepared in TRIS buffer (25 mM TRIS 200 mM NaCl pH 8). Stock solutions of CDy11, BD-oligo, and MK-H4 were 1 mM each in 10% DMSO. For ThT assays, 10 μL of each protein sample (1 mg/mL) were mixed with 70 μL of ddH2O and 20 μL of 20 μM of ThT stock solution (final [ThT] = 4 μM). For CDy11, BD-oligo, and MK-H4 assays, 10 μL of each protein sample (1 mg/mL) were mixed with 80 μL of ddH2O and 10 μL of 100 μM of dye (final [dye] = 10 μM). Samples were added to 96-well all-black flat-bottom Microfluor plates and incubated in the dark for 15–30 min at room temperature. Fluorescence intensities of dyes were measured using a BioTek Synergy 2 or Synergy H1 spectrophotometer at the following emission and excitation wavelengths: ThT (Ex = 440, Em = 485), CDy11 and BD-Oligo (Ex = 530, Em = 580), and MK-H4 (Ex = 475, Em = 567). All experiments were performed in triplicate with background dye fluorescence subtracted from protein plus dye samples.
Congo red-induced birefringence
Induced amyloid and purified amyloid fibers of each protein were stained with Congo red (CR) and evaluated for birefringence as described26,28. One hundred microliters (1 mg/mL) of each sample were centrifuged at 16,000 x g (Eppendorf centrifuge 5415 R). Pellets were resuspended in 10 μL of 500 µM CR solution in 80% ethanol, and incubated for 30–60 min at room temperature. Visualization was with a Zeiss Scope A1 equipped with a computer-controlled ProgRes C5 Jenoptik inverted camera and crossed polarizing light filters. For static biofilm assays, S. mutans strain UA159 was grown overnight in Todd–Hewitt broth with 0.3% yeast extract, diluted 1:100 in biofilm media66 containing 0.8% of glucose, and 1 mL aliquots placed in wells of a 24 well plate (24 well Costar Flat bottom with lid). Every 12 hours biofilm material was scraped from the bottom of a well and suspended in the culture supernatant, placed in a 1.5 mL Eppendorf tube, and centrifuged at maximum speed in an Eppendorf centrifuge 5415r. Pellets were stained with CR as above. This experiment was performed twice with triplicate wells analyzed for each condition.
Microfluidic experiments
With cells growing in microfluidic flow channels, we compared CDy11 uptake by S. mutans UA159 wild-type to uptake by a triple mutant (Δ3) lacking P1, WapA, and Smu_63c26. Bacteria were grown overnight in brain heart infusion (BHI, BD) at 37 °C in 5% CO2. Cells were harvested by centrifugation (1200 X g for 10 min), washed once in BHI, and resuspended in BHI to an OD600 of 0.1. In order to de-chain the cells, bacterial suspensions were sonicated for 20 seconds using a Fisher Scientific FB120 sonic dismembrator probe at the 20% amplitude setting, and then loaded into a commercial microfluidic device (IBIDI μ-slide VI), which consists of parallel flow channels that are 400 μm deep and 3.8 mm wide. Fresh BHI was flowed through each channel at 0.06 mL/h for 72 hours and then switched to BHI containing 300 nM CDy11 for 4 hours. Phase contrast and red fluorescence images were collected for each strain using an inverted microscope (Nikon TE2000U) equipped with a Photometrics Prime camera and DsRed filter cube. To evaluate CR-induced birefringence, the wild-type and Δ3 mutant strains were grown overnight in Todd-Hewitt broth (THB), harvested by centrifugation and resuspended to an OD600 of 0.1 in THB. Microfluidic channels with low intrinsic birefringence were prepared by washing, drying and autoclaving borosilicate glass capillaries (0.05 mm ×0.05 mm ×50 mm, Wale Apparatus). Capillaries were sealed to clean microbore Tygon tubing with epoxy adhesive, with a Luer-slip connection inserted into the tubing to allow loading of cells into the capillary. Cell cultures were sonicated briefly using a Fisher Scientific FB120 sonic dismembrator probe and then loaded (100 µL of cell culture per channel) into the glass capillaries and allowed to settle for one hour. After that, fresh THB was flowed at 0.06 mLhr−1 for 72 hours and switched to THB with 100 μM CR solution28 for 3 hours. Capillaries were then mounted onto the stage of the Nikon TE2000U inverted microscope and phase contrast and cross-polarized images in visible light were captured.
Supplementary information
Acknowledgements
This work was supported by National Institutes of Health Grant R01 DE21789 to LJB and JRL. Use of the Stanford Synchrotron Radiation Lightsource, SLAC National Accelerator Laboratory, is supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences under Contract No. DE-AC02-76SF00515. The authors thank Dr. Young-Tae Chang, Pohang University of Science and Technology, Korea for the kind gift of CDy11, BD-oligo, and MK-H4 dyes, and Dr. Thomas Weiss at SLAC for technical assistance.
Author contributions
A.L.B.B., J.R.L., S.J.H. and L.J.B. conceived experiments. A.L.B.B., M.H., A.K., G.S., J.R.L., S.J.H. and L.J.B. designed experiments and research methodology. A.L.B.B., S.O., M.H., J.M.-A., G.O. and E.Y. performed the research and data collection. A.L.B.B., A.K., G.S., S.J.H. and L.J.B. analyzed data. S.M. performed protein structure modeling and illustration. A.L.B.B. and L.J.B. wrote the manuscript. All authors reviewed the manuscript drafts and inputted corrections, amendments, and their expertise.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-62115-7.
References
- 1.Johansson J. Amyloid fibrils. FEBS Journal. 2005;272:5941. doi: 10.1111/j.1742-4658.2005.05026.x. [DOI] [PubMed] [Google Scholar]
- 2.Otzen D, Nielsen PH. We find them here, we find them there: Functional bacterial amyloid. Cell. Mol. Life Sci. 2008;65:910–927. doi: 10.1007/s00018-007-7404-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blanco LP, Evans ML, Smith DR, Badtke MP, Chapman MR. Diversity, biogenesis and function of microbial amyloids. Trends in Microbiology. 2012;20:66–73. doi: 10.1016/j.tim.2011.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sawyer EB, Claessen D, Gras SL, Perrett S. Exploiting amyloid: how and why bacteria use cross-β fibrils. Biochem. Soc. Trans. 2012;40:728–734. doi: 10.1042/BST20120013. [DOI] [PubMed] [Google Scholar]
- 5.Smith JF, Knowles TPJ, Dobson CM, MacPhee CE, Welland ME. Characterization of the nanoscale properties of individual amyloid fibrils. Proc. Natl. Acad. Sci. USA. 2006;103:15806–15811. doi: 10.1073/pnas.0604035103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rambaran RN, Serpell LC. Amyloid fibrils: abnormal protein assembly. Prion. 2008;2:112–7. doi: 10.4161/pri.2.3.7488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zurdo J, Guijarro JI, Dobson CM. Preparation and characterization of purified amyloid fibrils. J. Am. Chem. Soc. 2001;123:8141–8142. doi: 10.1021/ja016229b. [DOI] [PubMed] [Google Scholar]
- 8.Chen, D. et al. Characteristics and influencing factors of amyloid fibers in S. mutans biofilm. AMB Express 9 (2019). [DOI] [PMC free article] [PubMed]
- 9.Van Gerven N, Van der Verren SE, Reiter DM, Remaut H. The Role of Functional Amyloids in Bacterial Virulence. J. Mol. Biol. 2018;430:3657–3684. doi: 10.1016/j.jmb.2018.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Larsen P, et al. Amyloid adhesins are abundant in natural biofilms. Environ. Microbiol. 2007;9:3077–3090. doi: 10.1111/j.1462-2920.2007.01418.x. [DOI] [PubMed] [Google Scholar]
- 11.Romero D, Kolter R. Functional amyloids in bacteria. Int. Microbiol. 2014;17:65–73. doi: 10.2436/20.1501.01.208. [DOI] [PubMed] [Google Scholar]
- 12.Fowler DM, Koulov AV, Balch WE, Kelly JW. Functional amyloid – from bacteria to humans. Trends Biochem. Sci. 2007;32:217–224. doi: 10.1016/j.tibs.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 13.Taglialegna A, Lasa I, Valle J. Amyloid structures as biofilm matrix scaffolds. Journal of Bacteriology. 2016;198:2579–2588. doi: 10.1128/JB.00122-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Erskine E, MacPhee CE, Stanley-Wall NR. Functional Amyloid and Other Protein Fibers in the Biofilm Matrix. J. Mol. Biol. 2018;430:3642–3656. doi: 10.1016/j.jmb.2018.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shanmugam N, et al. Microbial functional amyloids serve diverse purposes for structure, adhesion and defence. Biophysical Reviews. 2019;11:287–302. doi: 10.1007/s12551-019-00526-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lemos, J. A. et al. The Biology of Streptococcus mutans. Microbiol. Spectr. 7 (2019). [DOI] [PMC free article] [PubMed]
- 17.Taglialegna A, et al. Staphylococcal Bap Proteins Build Amyloid Scaffold Biofilm Matrices in Response to Environmental Signals. PLOS Pathog. 2016;12:e1005711. doi: 10.1371/journal.ppat.1005711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zeng, G. et al. Functional bacterial amyloid increases Pseudomonas biofilm hydrophobicity and stiffness. Front. Microbiol. 6 (2015). [DOI] [PMC free article] [PubMed]
- 19.Seviour, T. et al. Functional amyloids keep quorum-sensing molecules in check. J. Biol. Chem. 290 (2015). [DOI] [PMC free article] [PubMed]
- 20.Yakupova, E. I., Bobyleva, L. G., Vikhlyantsev, I. M. & Bobylev, A. G. Congo Red and amyloids: History and relationship. Bioscience Reports39 (2019). [DOI] [PMC free article] [PubMed]
- 21.Howie AJ, Brewer DB. Optical properties of amyloid stained by Congo red: History and mechanisms. Micron. 2009;40:285–301. doi: 10.1016/j.micron.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 22.Mettlen M, Chen P-H, Srinivasan S, Danuser G, Schmid SL. Annual Review of Biochemistry Regulation of Clathrin-Mediated Endocytosis. Annu. Rev. Biochem. 2018;86:637–657. doi: 10.1146/annurev-biochem-062917-012644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McDonald M, Kendall A, Tanaka M, Weissman JS, Stubbs G. Enclosed chambers for humidity control and sample containment in fiber diffraction. J. Appl. Crystallogr. 2008;41:206–209. doi: 10.1107/S0021889807060803. [DOI] [Google Scholar]
- 24.Jahn TR, et al. The Common Architecture of Cross-β Amyloid. J. Mol. Biol. 2010;395:717–727. doi: 10.1016/j.jmb.2009.09.039. [DOI] [PubMed] [Google Scholar]
- 25.Tayeb-Fligelman E, et al. The cytotoxic Staphylococcus aureus PSMα3 reveals a cross-α amyloid-like fibril. Science (80-.). 2017;355:831–833. doi: 10.1126/science.aaf4901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Besingi, R. N. et al. Functional amyloids in streptococcus mutans, their use as targets of biofilm inhibition and initial characterization of SMU_63c. Microbiology (United Kingdom)163 (2017). [DOI] [PMC free article] [PubMed]
- 27.Siegel SD, Reardon ME, Ton-That H. Anchoring of LPXTG-like proteins to the gram-positive cell wall envelope. in. Current Topics in Microbiology and Immunology. 2017;404:159–175. doi: 10.1007/82_2016_8. [DOI] [PubMed] [Google Scholar]
- 28.Oli MW, et al. Functional amyloid formation by Streptococcus mutans. Microbiology. 2012;158:2903–16. doi: 10.1099/mic.0.060855-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Taylor JD, Matthews SJ. New insight into the molecular control of bacterial functional amyloids. Front. Cell. Infect. Microbiol. 2015;5:33. doi: 10.3389/fcimb.2015.00033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Evans, M. L. & Chapman, M. R. Curli biogenesis: Order out of disorder. Biochim. Biophys. Acta - Mol. Cell Res. 1843 (2014). [DOI] [PMC free article] [PubMed]
- 31.Binepal G, et al. K+ modulates genetic competence and the stress regulon of Streptococcus mutans. Microbiol. (United Kingdom) 2017;163:719–730. doi: 10.1099/mic.0.000458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Romero D, Sanabria-Valentín E, Vlamakis H, Kolter R. Biofilm inhibitors that target amyloid proteins. Chem. Biol. 2013;20:102–110. doi: 10.1016/j.chembiol.2012.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li C, Adamcik J, Mezzenga R. Biodegradable nanocomposites of amyloid fibrils and graphene with shape-memory and enzyme-sensing properties. Nat. Nanotechnol. 2012;7:421–427. doi: 10.1038/nnano.2012.62. [DOI] [PubMed] [Google Scholar]
- 34.Malmos, K. G. et al. Amyloid The Journal of Protein Folding Disorders ThT 101: a primer on the use of thioflavin T to investigate amyloid formation, 10.1080/13506129.2017.1304905 (2017).
- 35.Kim J-Y, et al. Detection of Pathogenic Biofilms with Bacterial Amyloid Targeting Fluorescent Probe, CDy11. J. Am. Chem. Soc. 2016;138:402–407. doi: 10.1021/jacs.5b11357. [DOI] [PubMed] [Google Scholar]
- 36.Teoh CL, et al. Chemical Fluorescent Probe for Detection of Aβ Oligomers. J. Am. Chem. Soc. 2015;137:13503–13509. doi: 10.1021/jacs.5b06190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Larson MR, et al. Crystal structure of the C-terminal region of Streptococcus mutans antigen I/II and characterization of salivary agglutinin adherence domains. J. Biol. Chem. 2011;286:21657–21666. doi: 10.1074/jbc.M111.231100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang J, Zhang Y. Protein Structure and Function Prediction Using I-TASSER. Curr. Protoc. Bioinforma. 2015;52:5.8.1–5.8.15. doi: 10.1002/0471250953.bi0508s52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morris KL, Serpell LC. X-ray fibre diffraction studies of amyloid fibrils. Methods Mol. Biol. 2012;849:121–135. doi: 10.1007/978-1-61779-551-0_9. [DOI] [PubMed] [Google Scholar]
- 40.Wu C, Scott J, Shea JE. Binding of congo red to amyloid protofibrils of the alzheimer Aβ9-40 peptide probed by molecular dynamics simulations. Biophys. J. 2012;103:550–557. doi: 10.1016/j.bpj.2012.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Howie AJ, Brewer DB, Howell D, Jones AP. Physical basis of colors seen in Congo red-stained amyloid in polarized light. Lab. Investig. 2008;88:232–242. doi: 10.1038/labinvest.3700714. [DOI] [PubMed] [Google Scholar]
- 42.Dueholm MS, et al. Expression of Fap amyloids in Pseudomonas aeruginosa, P. fluorescens, and P. putida results in aggregation and increased biofilm formation. Microbiologyopen. 2013;2:365–382. doi: 10.1002/mbo3.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dragoš A, Kovács ÁT, Claessen D. The Role of Functional Amyloids in Multicellular Growth and Development of Gram-Positive Bacteria. Biomolecules. 2017;7:60. doi: 10.3390/biom7030060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Andreasen, M. et al. Physical determinants of amyloid assembly in biofilm formation. MBio10 (2019). [DOI] [PMC free article] [PubMed]
- 45.Zhu L, et al. Functional characterization of cell-wall-associated protein WapA in Streptococcus mutans. Microbiology. 2006;152:2395–2404. doi: 10.1099/mic.0.28883-0. [DOI] [PubMed] [Google Scholar]
- 46.Larson MR, et al. Elongated fibrillar structure of a streptococcal adhesin assembled by the high-affinity association of α-and PPII-helices. PNAS. 2010;107:5983–5988. doi: 10.1073/pnas.0912293107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Heim KP, et al. An intramolecular lock facilitates folding and stabilizes the tertiary structure of streptococcus mutans adhesin p1. Proc. Natl. Acad. Sci. USA. 2014;111:15711–15716. doi: 10.1073/pnas.1413018111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Heim KP, et al. Identification of a supramolecular functional architecture of Streptococcus mutans adhesin P1 on the bacterial cell surface. J. Biol. Chem. 2015;290:9002–19. doi: 10.1074/jbc.M114.626663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tang W, et al. Specific binding of a naturally occurring amyloidogenic fragment of Streptococcus mutans adhesin P1 to intact P1 on the cell surface characterized by solid state NMR spectroscopy. J. Biomol. NMR. 2016;64:153–164. doi: 10.1007/s10858-016-0017-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rivière, G. et al. Characterization of an intermolecular quaternary interaction between discrete segments of the Streptococcus mutans adhesin P1 by NMR spectroscopy. FEBS J. 15158 (2019). [DOI] [PMC free article] [PubMed]
- 51.Li Y, et al. Live-cell and super-resolution imaging reveal that the distribution of wall-associated protein A is correlated with the cell chain integrity of Streptococcus mutans. Mol. Oral Microbiol. 2015;30:376–383. doi: 10.1111/omi.12100. [DOI] [PubMed] [Google Scholar]
- 52.Han TK, Zhang C, Dao ML. Identification and characterization of collagen-binding activity in Streptococcus mutans wall-associated protein: A possible implication in dental root caries and endocarditis. Biochem. Biophys. Res. Commun. 2006;343:787–792. doi: 10.1016/j.bbrc.2006.03.025. [DOI] [PubMed] [Google Scholar]
- 53.Federle MJ, Morrison DA. One if by land, two if by sea: signalling to the ranks with CSP and XIP. Mol. Microbiol. 2012;86:241–245. doi: 10.1111/mmi.12029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sawyer EB, Claessen D, Haas M, Hurgobin B, Gras SL. The assembly of individual chaplin peptides from streptomyces coelicolor into functional amyloid fibrils. PLoS One. 2011;6:18839. doi: 10.1371/journal.pone.0018839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Moores B, Drolle E, Attwood SJ, Simons J, Leonenko Z. Effect of Surfaces on Amyloid Fibril Formation. PLoS One. 2011;6:25954. doi: 10.1371/journal.pone.0025954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gordon DJ, Balbach JJ, Tycko R, Meredith SC. Increasing the Amphiphilicity of an Amyloidogenic Peptide Changes the β-Sheet Structure in the Fibrils from Antiparallel to Parallel. Biophys. J. 2004;86:428–434. doi: 10.1016/S0006-3495(04)74119-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Diehl A, et al. Structural changes of TasA in biofilm formation of Bacillus subtilis. Proc. Natl. Acad. Sci. USA. 2018;115:3237–3242. doi: 10.1073/pnas.1718102115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wille, H. et al. Natural and synthetic prion structure from X-ray fiber diffraction. [DOI] [PMC free article] [PubMed]
- 59.Dutta A, Bhattacharyya S, Kundu A, Dutta D, Das AK. Macroscopic amyloid fiber formation by staphylococcal biofilm associated SuhB protein. Biophys. Chem. 2016;217:32–41. doi: 10.1016/j.bpc.2016.07.006. [DOI] [PubMed] [Google Scholar]
- 60.Shewmaker F, et al. The functional curli amyloid is not based on in-register parallel β-sheet structure. J. Biol. Chem. 2009;284:25065–25076. doi: 10.1074/jbc.M109.007054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Paranjapye N, Daggett V. De Novo Designed α-Sheet Peptides Inhibit Functional Amyloid Formation of Streptococcus mutans Biofilms. J. Mol. Biol. 2018;430:3764–3773. doi: 10.1016/j.jmb.2018.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Leiman SA, et al. D-Amino acids indirectly inhibit biofilm formation in Bacillus subtilis by interfering with protein synthesis. J. Bacteriol. 2013;195:5391–5395. doi: 10.1128/JB.00975-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hochbaum AI, et al. Inhibitory effects of D-amino acids on staphylococcus aureus biofilm development. J. Bacteriol. 2011;193:5616–5622. doi: 10.1128/JB.05534-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yang L, et al. Current understanding of multispecies biofilms. In International Journal of Oral Science. 2011;3:74–81. doi: 10.4248/IJOS11027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bian W, Wang H, McCullough I, Stubbs G. WCEN: a computer program for initial processing of fiber diffraction patterns. J. Appl. Crystallogr. 2006;39:752–756. doi: 10.1107/S0021889806025386. [DOI] [Google Scholar]
- 66.Bouvet, A., Van De Rijn, I. & Mccarty, M. Nutritionally Variant Streptococci from Patients with Endocarditis: Growth Parameters in a Semisynthetic Medium and Demonstration of a Chromophore. Journal of Bacteriology146 (1981). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.