Abstract
Intervertebral disc degeneration (IDD) is the main contributor to low back pain, which is a leading cause of disability worldwide. Although substantial progress has been made in elucidating the molecular mechanisms of IDD, fundamental and long‐lasting treatments for IDD are still lacking. With increased understanding of the complex pathomechanism of IDD, alternative strategies for treating IDD can be discovered. A brief overview of the prevalence and epidemiologic risk factors of IDD is provided in this review, followed by the descriptions of anatomic, cellular, and molecular structure of the intervertebral disc as well as the molecular pathophysiology of IDD. Finally, the recent findings of intervertebral disc progenitors are reviewed and the future perspectives are discussed.
Keywords: epidemiology, genetic, intervertebral disc degeneration, prevalence, progenitors
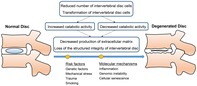
Pathomechanism of intervertebral disc degeneration. Several risk factors including genetic factors as well as mechanical stress, trauma and smoking, attributed to the development of intervertebral disc degeneration. These biological and environmental factors induce the reduction of cell numbers and transformation of intervertebral disc cells through several molecular mechanisms, resulting in decreased production of extracellular matrix due to increased catabolic activity and decreased anabolic activities. Thereafter, the structured integrity of intervertebral disc is lost and intervertebral disc degeneration is further accelerated.
1. INTRODUCTION
Low back pain (LBP) is a common condition affecting approximately 637 million individuals worldwide.1 The high morbidity of LBP is associated with lower health‐related quality of life2 and high medical expenses,3 resulting in increased suffering and high socioeconomic costs. Intervertebral disc degeneration (IDD) is a major contributor to LBP,4 and it also precedes other spinal disorders such as disc herniation, spondylosis, and lumbar spinal stenosis.5 Herein, we will discuss the prevalence of and risk factors for IDD, structure of intervertebral disc (IVD), pathomechanism and histological features of IDD, and IVD progenitors.
2. PREVALENCE OF INTERVERTEBRAL DISC DEGENERATION
IDD develops during adolescence and progresses with age. In 1995, the prevalence of IDD, based on magnetic resonance imaging (MRI) findings, was described in a population‐based cohort in Finland.6 Among 232 men with a mean age of 49.3 (range, 35‐69) years, reduced signal intensity on MRI was observed in 41.6% and 86.0% of the participants at L1/2 and L5/S1, respectively. Reduced disc height was observed in 9.3% and 55.6% of the participants at L1/2 and L5/S1, respectively, suggesting that IDD is more frequent and severe at the lower lumbar disc than at the upper lumbar disc. In 2009, the prevalence of radiographic spondylosis was investigated in a large‐scale nationwide cohort study (Research on Osteoarthritis Against Disability; ROAD) performed in Japan.7 Among 2288 participants (818 men and 1470 women) aged ≥60 years, the prevalence of radiographic spondylosis with Kellgren‐Lawrence grade ≥2 was 75.8% in total, 84.1% in men, and 70.7% in women. Later, the prevalence of IDD based on MRI findings of the entire spine was reported in a population‐based cohort study in Japan,8 in which the presence of IDD was defined by Pfirrmann's grading system9 (where grade 4 and 5 indicated IDD). Among 975 participants (324 men and 651 women) aged 21 to 97 years, the prevalence of IDD was 71% in men and 77% in women aged <50 years, and >90% in both men and women aged >50 years. The prevalence of an intervertebral space with IDD was the highest at C5/6 (men: 51.5%, women: 46%), T6/7 (men: 32.4%, women: 37.7%), and L4/5 (men: 69.1%, women: 75.8%). LBP was associated with the presence of IDD in the lumbar region. Using the same cohort, Teraguchi et al. also examined the association between IDD and LBP, taking endplate signal change and/or Schmorl's node on MRI into consideration.10 Although IDD alone is not associated with the presence of LBP, the combination of IDD and endplate signal change was highly associated with the presence of LBP. More recently, the prevalence of Modic changes based on MRI findings of the lumbar spine was reported in a population‐based cohort study in China.11 Among 2449 participants with a mean age of 40.4 years, the prevalence of Modic changes was 5.8%, and the presence of Modic changes was significantly associated with the presence of IDD and correlated with the presence of LBP. Considering these results, endplate degeneration possibly plays an important role in the mechanism by which IDD causes LBP.
3. RISK FACTORS FOR IDD FROM EPIDEMIOLOGIC STUDIES
Before mid‐ to late 1990s, repetitive mechanical loading or wear and tear were believed to cause IDD. However, recent family and twin studies have suggested that the occurrence of IDD is determined largely by genetic factors, with environmental factors having an important role.12
3.1. Genetic factors
Genetic influences predominate among the reported risk factors for IDD. In 1995, a population‐based cohort study was performed using 115 male monozygotic twin pairs, in which IDD was evaluated by MRI.13 Familial aggregation explained 61% and 34% of IDD scores in the upper and lower lumbar spine, respectively, in multivariate analyses, suggesting that IDD is substantially affected by genetic factors and that compared with IDDs in the lower lumbar spines, those in the upper lumbar spines were more significantly influenced by genetic factors. The predominance of genetic factors was further confirmed by other twin studies, estimating that genetic factors account for up to three‐quarters of susceptibility to lumbar IDD.14, 15 Several candidate genes that may play a role in the onset of IDD have been reported by affected sib‐pair linkage studies or candidate‐gene association studies, such as ACAN,16 CLIP,17 COL1A1,18 COL9A2,19 COL11A1,20 GDF5,21 IGF1R,22 IL‐1,23 IL‐6,24 MMP2,25 MMP3,26 MMP9,27 SKT,28 THBS2,29 and VDR.30, 31 Further, recent genome‐wide association meta‐analyses identified novel candidate genes, including PARK2 32 and CHST3.33 An excellent review focusing on the extensive candidate genes was published recently.34 Many of these candidate genes are known to constitute the extracellular matrix (ECM) of IVD or be involved in ECM turnover, and, thus, they determine the size and mechanical property of the IVD by nature. Genetic defects in these genes presumably render the IVD more vulnerable against external force, leading to early onset of IDD. Further studies are required to elucidate the actual molecular mechanism through which each gene polymorphism causes IDD.
3.2. Mechanical stress
Excessive mechanical stress is thought to induce IDD, considering that IDD is more frequently observed in the lower lumbar spine, where IVDs suffer higher mechanical stress,35 and that IVDs adjacent to vertebral fusion are more likely to suffer IDD.36 On investigating a cohort of monozygotic twin pairs with different physical activities, Battie et al. found that physical activities explained only 2% to 7% of IDD scores in multivariate analyses.13 Similarly, in the aforementioned ROAD study, Muraki et al. examined the association between knee osteoarthritis/lumbar spondylosis and occupation/physical activity of the participants.7 Interestingly, the association between physical activity and lumbar spondylosis was weak, whereas the degree of physical activity was strongly associated with the presence of knee osteoarthritis, indicating that compared with knee joints, IVDs are less likely to be affected by mechanical stress.
3.3. Trauma
A study involving well‐matched cohorts, including 50 subjects who underwent discography and 52 control subjects, revealed that compared to matched controls, subjects who underwent discography showed accelerated IDD at 7 to 10 years of follow‐up; the progression of disc degeneration assessed by MRI was observed in 54 discs (35%) in the discography group compared to 21 discs (14%) in the control group.37 Further, retrospective clinical studies on 14 young patients with previous vertebral fracture and 14 healthy controls showed that IDD was more frequently observed in patients with previous vertebral fracture than in controls (57% and 8%, respectively).38 Hence, trauma is thought to be a risk factor for the onset of IDD.
3.4. Smoking
Smoking is the only chemical exposure known to be associated with the onset of IDD. On investigating the cohort of monozygotic twin pairs with different smoking exposures, Battie et al. found slightly greater IDD scores in the lumbar spine of smokers than in the lumbar spine of nonsmokers.39
4. STRUCTURE OF THE INTERVERTEBRAL DISC
The IVD is composed of different but interrelated tissues, including the central highly hydrated gelatinous nucleus pulposus (NP), surrounding elastic and fibrous annulus fibrosus (AF), and cartilaginous end plates (CEP), which provides connection to the vertebral bodies (Figure 1).40 The NP is derived from the notochord, and notochordal cells remain in the tissue after birth and until around 10 years of age in humans. These cells are thereafter replaced by small chondrocyte‐like cells with lower metabolic activities.41 The ECM of NP consists of type II collagen fibers and elastin that contain proteoglycans such as aggrecan and versican. The presence of proteoglycans with negatively charged side chains makes the NP highly hydrated with high osmolarity, enabling the IVD to resist compressive loads and to deform reversibly.
Figure 1.
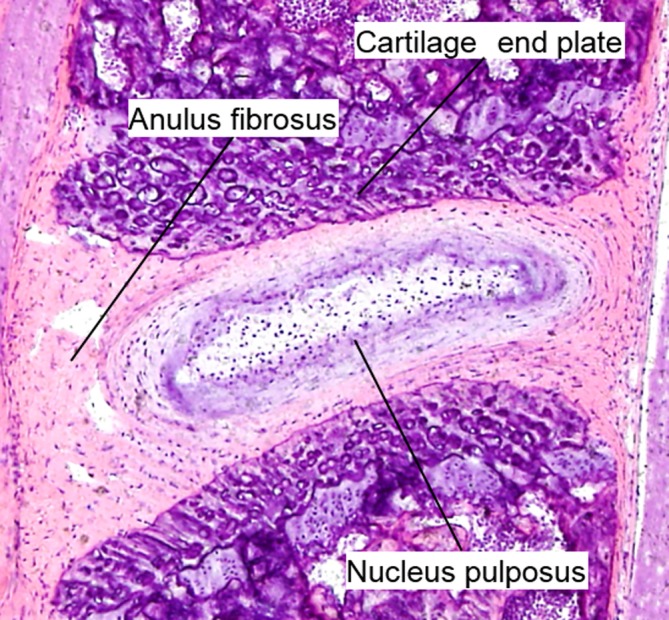
Hematoxylin and eosin staining of mouse lumbar intervertebral disc at 8 weeks of age
The AF consists of a series of concentric rings, or lamellae, with collagen fibers lying parallel within each lamellae, providing tensile strength and the ability to withstand forces applied from any direction.42 The inner AF consists of several layers of fibrocartilage, while the outer AF is a fibrous tissue containing highly organized fibers composed mainly of type I collagen, allowing it to resist tensile loads.43 Proteoglycans and type II collagen fibers decrease gradually closer to the outer AF, while the content of type I collagen fibers increases.44
The CEP is a layer of hyaline cartilage that covers the caudal and cephalic ends of the disc, which plays an important role in the transport of fluids and solutes in/out of the disc.45 Similar to the articular cartilage, the ECM of the CEP consists of type II collagen embedded with chondrocytes.
5. PATHOMECHANISM AND HISTOLOGICAL FEATURES OF HUMAN IDD, AND LESSONS FROM ANIMAL MODELS
IDD can be attributed to several factors, including genetic factors as well as aging, mechanical stress, and injury. These biological and environmental factors induce the reduction of cell number and transformation of IVD cells, resulting in decreased production of ECM of IVD owing to increased catabolic activity and decreased anabolic activities. Thereafter, the structural integrity of IVD is lost and IDD is further accelerated.46, 47 One of the features of the pathomechanism of IDD in humans is increased catabolic and decreased anabolic activities, and the changes in ECM during the IDD process, characterized by the changes in the expression/structure of collagens/proteoglycans. Owing to the difficulty in obtaining human IVD samples, especially normal human tissue, several animal models that mimic these features have been developed to elucidate the pathomechanism of IDD.
5.1. Expression and structural changes of collagens
In humans, a general decrease in type II collagen production and a shift to type I collagen synthesis by NP cells or inner AF cells is observed as IDD progresses.48 In addition, localization of type X collagen has been observed in degenerated IVD, which is associated with the formation of cell clusters and clefts.49 During the process of development of IDD in humans, an increase in nonenzymatic glycosylation of collagen fibers is observed, leading to an accumulation of advanced glycation end‐products. As a result, cross‐linking of collagen fibers increases, causing tissue stiffness and rendering the IVDs more susceptible to mechanical damage during degeneration.50
5.2. Expression and structural changes in proteoglycans
Similar to the changes in the expression of collagens, a decrease in the proteoglycan content of human IVD is observed during degeneration.51, 52, 53, 54 In addition, the composition of glycosaminoglycan chains shifts from chondroitin sulfate to keratin sulfate,52 reducing the water content in the IVD. In synergy with the increased expression of type I collagen, the IVD becomes more fibrotic and less capable of withstanding mechanical stress.
5.3. Histological features of IDD
During the process of development of IDD in humans, several histological findings are observed in NP, including loss of demarcation between NP and AF owing to the shift of synthesis from type II collagen to type I collagen, dehydration caused by the decrease in proteoglycan production, presence of fissure, and cell cluster formation (Table 1).55 With regard to AF, disruption of the lamellar structure of collagen fibers, presence of fissure, and increased degree of vascularization and innervation are observed.56 Structural disorganization of the CEP is observed, including cracks, thinning, mineralization, microfracture in the adjacent subchondral bone, and bone sclerosis.57
Table 1.
Histological findings of intervertebral disc degeneration
| Nucleus pulposus | Anulus fibrosus | Cartilage end plate | |
|---|---|---|---|
| Changes at the molecular level |
Decrease of proteoglycan Decrease of type II collagen Increase of type I collagen |
Cross‐link of collagen fibers | Decrease of proteoglycan |
| Histological changes |
Fissure Fibrosis Appearance of cell cluster Loss of notochordal cells and appearance of chondrocyte‐like cells |
Disruption of lamella Fissure Vascularization and innervation |
Microfracture and sclerosis of subchondral bone Thinning Reduction in the number of vascular channel |
| Biomechanical changes | Decrease of expansive force | Vulnerable against mechanical stress |
5.4. Pathological IDD and disc aging
Organismal aging results from time‐dependent accumulation of molecular and cellular damage, which leads to impaired tissue homeostasis, and eventual physiological and functional decline.58 Compared with other tissues, IVDs appear to undergo age‐related degenerative changes earlier in life.35, 41 On analyzing 44 human lumbar spines from deceased individuals without any spinal disorders, Boos et al. reported that age‐related histological changes of IVD include increased number and size of fissures, presence of granular debris, and neovascularization of the outer AF.41 On investigating 450 skeletons, Edelson et al. reported that the histological features of the CEP associated with aging include ossification and thinning of the CEP, microfractures in the subchondral bone, bone sclerosis, and reduction in the number of vascular channels in the CEP.57 As pathological IDD is caused by factors other than aging, such as genetic predisposition, trauma, and environmental factors, pathological IDD can occur in younger individuals and at a single intervertebral level, whereas disc aging is more systemic and is observed in older individuals in all spinal discs.58 However, we could not precisely distinguish between pathological IDD and IDD associated with aging owing to the almost similar histological features of the two conditions.41
With recent progress in aging research, some of the molecular pathways leading to disc aging have been elucidated. In particular, genomic instability and resulting cellular senescence have been determined to be important drivers of IDD.58 Each cell is constantly subjected to the risk of DNA damage due to the chemical instability of DNA structure, metabolic byproducts, and environmental mutagens and genotoxins.59 Despite several inherent DNA repair mechanisms of cells, the frequency of DNA damage becomes greater than that of DNA repair with aging, resulting in the accumulation of damaged DNA. It has been shown that accumulated genomic damage can lead to disc aging. For example, Ercc1 −/Δ mice, in which ERCC1‐XPF is deficient (involved in DNA damage repair), showed typical features of disc aging, including loss of proteoglycan, decreased IVD height, and an increase in the number of senescent cells.60 The evidence that genotoxic stresses such as tobacco smoking or radiation cause disc aging further supports that DNA damage contributes to disc aging.61, 62 Other possible causes of DNA damage include oxidative stress induced by inflammation. Interleukin‐1 (IL‐1), a predominant cytokine involved in the pathogenesis of IDD,63, 64 has been demonstrated to induce cellular senescence in NP cells. Furthermore, IL‐1 receptor antagonist (IL1‐Ra) knockout mice showed typical features of human IDD, and NP cells from these mice showed the senescent phenotype.65
There are two types of cellular senescence, namely replicative senescence and stress‐induced premature senescence (SIPS). Replicative senescence is characterized by cessation of cell proliferation due to critical telomere shortening after consecutive replicative cell cycles.66 In contrast, SIPS is caused by the accumulation of genomic and mitochondrial damage. SIPS cells acquire a senescence‐associated secretory phenotype (SASP), which is characterized by the secretion of several inflammatory cytokines and matrix proteases that have profound catabolic effects on neighboring cells and ECM, promoting tissue degeneration.67, 68, 69 This pathomechanism of IDD is supported by previous studies that revealed that the number of senescent cells, which were assessed by senescence markers such as senescence‐associated β‐galactosidase and p16INK4A, were increased in human IVD samples. These markers were positively correlated with the expression of matrix metalloproteases including matrix metalloproteinase 13 (MMP13) and a disintegrin and metalloproteinase with thrombospondin motifs 5 (ADAMTS5).70, 71, 72, 73, 74 Recently, Patil et al. demonstrated the causal relationship between cellular senescence and age‐related IDD using the p16‐3MR transgenic mouse model in which p16‐positive senescent cells can be selectively eliminated by treatment with ganciclovir.75 The aging mice (age: 1 year) treated with ganciclovir showed decreased levels of catabolic factors along with improved histological features of IDD at age 2 years compared with control mice, indicating that cellular senescence has a direct impact on IDD development.
5.5. Lessons from animal models
Various kinds of inducers have been used to reproduce IDD in experimental animals, including compression,76, 77, 78 injury,79, 80, 81 instability,82, 83, 84, 85, 86 postural bipedality,87, 88 chemical,89 genetic,90 vibration,91 spontaneous,92 and smoking.93 Many of these models recapitulate the radiological and histological features of human IDD. Recently, we developed a mouse IDD model in which instability was induced without direct injury to IVDs, by surgical resection of posterior elements of the mouse lumbar spine.94 Radiological decrease in IVD height and histological findings compatible with human IDD were observed in this model. It is noteworthy that hypertrophic‐like chondrocytes appeared in the inner AF during degeneration, the morphologies of which apparently differed from those of normal inner AF, and these cells expressed type X collagen and MMP13,94 suggesting these cells contribute to IDD phenotypes as is the case in osteoarthritis.95 Thus, we speculate that the appearance of these cells in the inner AF may be a key event in the development of IDD. Further studies are warranted to investigate whether these cells are activated‐local progenitors or are recruited from other tissues, and whether prevention of these cells from expressing type collagen X and MMP13 is a potential intervention for preventing the progression of IDD.
6. INTERVERTEBRAL DISC PROGENITORS
The presence of local progenitors or the recruitment of appropriate cells into the damaged sites is required for tissue maintenance and repair.96 Despite preliminary results showing the positive effects of cell therapies in regeneration of the IVD, detailed basic research on IVD cells and their niche indicates that transplanted cells are unable to survive and adapt in the avascular niche of the IVD.97 It is imperative to identify the IVD progenitors and understand their niche to succeed in cell therapies for IDD. The IVD niche, which represents the unique microenvironment and communication network within the IVD cells, has been investigated by several researchers.97, 98, 99 The intervertebral disc is avascular because the capillaries terminate at the vertebral endplates and outermost AF, and the nutrition reaches the nucleus pulposus by diffusion through the CEP and outer AF.100, 101, 102, 103 As the NP, which is anatomically farthest from the vascular supply, is exposed to hypoxia, most energy for the NP is derived from anaerobic glycolysis.104 Anaerobic glycolysis in NP cells generates lactic acid and lowers the pH within the IVD. Other features of the IVD niche include low cellular density,56 high osmotic pressure,105 and high mechanical stress.97 IVD cells acquire specific adaptation mechanisms to survive in these harsh microenvironments.
6.1. IVD specific progenitors in vitro
Considering the harsh microenvironments in IVD, activation of endogenous progenitor cells could be a promising therapeutic strategy for IDD. In 2007, using an explant culture to isolate progenitors from degenerate human discs, Risbud et al. identified cells from both NP and AF, expressing typical marrow mesenchymal stem cell markers such as CD105, CD166, CD63, CD49a, CD90, CD73, p75 low‐affinity growth factor receptor, and CD133/1, and these results were also confirmed in rat IVDs.106 Thereafter, several researchers have reported the presence of cells compatible with MSCs from NP, AF, and CEP of normal or degenerated IVDs.107, 108, 109, 110, 111, 112, 113 Further, several microenvironments of IVDs, such as extracellular matrix stiffness, pH, and osmotic pressure, have been shown to affect the properties of these progenitors.114, 115, 116
Sakai et al. identified NP progenitors with novel NP‐specific cell markers, and demonstrated that these progenitors are exhausted with aging and degeneration.117 Using colony‐forming assay with methylcellulose semi‐solid medium, they identified progenitors from human and mouse NPs that express tyrosine‐protein kinase receptor (Tie2) and disialoganglioside 2 (GD2). These cells formed spheroid colonies that highly produced type II collagen and aggrecan and had multipotent and self‐renewal abilities both in vitro and in vivo. Tie2+GD2− cells were found to be precursors of Tie2+GD2+ cells, and CD24, which was previously reported to be a specific marker of the NP,118 was found to be a specific marker of more mature NP cells, which differentiated from Tie2+GD2+ cells. Using these markers, NP cells were classified into four subtypes: dormant stem cells (Tie2+GD2−CD24−), self‐renewing stem cells (Tie2+GD2+CD24−), committed NP progenitor cells (Tie2−GD2+CD24+), and mature NP cells (Tie2−GD2−CD24+).117 Identification of these cell surface markers is epoch‐making in that it enables to evaluate the severity of IDD by quantifying the cell number and function, and in that it makes an index of induction of differentiation from other sources to become NP progenitor cells. Sakai et al. further revealed that angiopoietin‐1, a ligand of Tie2, suppressed apoptosis and promoted the proliferation of Tie2+ cells, enabling the development of a strategy to stimulate ANG‐1 to enhance Tie2+ progenitor cells for prevention of IDD.117 Later, the usefulness of the surface marker Tie2 was validated in a bovine coccygeal model.119
6.2. IVD‐specific progenitors in vivo
Some of the aforementioned progenitor cells maintain their potential multipotent differentiation and self‐renewal in vitro; however, knowledge regarding their in vivo characteristics, such as development, localization, or functional role in the maintenance of IVD homeostasis, is lacking.97
The methods to identify progenitors in vivo include label retention assay and lineage‐tracing experiments. In label retention assay, synthetic nucleic acid analogs, such as 5‐bromo‐2‐deoxyuridine (BrdU) or 5‐ethynyl‐2′‐deoxyuridine (EdU), are used to detect slow‐cycling cells, which are thought to be potential progenitor cells. A prolonged chase period results in dilution of the incorporated nucleic acid analogs, although the slow‐cycling cells remain labeled.120 In knee joints, slow‐cycling cells have been identified in the superficial zone, synovium, fat pad, top narrow reserve zone of the growth plate, and perichondrium (groove of Ranvier).120, 121, 122, 123 With regard to the IVD, Henriksson et al. identified slow‐cycling cells in a region close to the perichondrium, at the junction of the outer AF and the vertebral growth plate, suggesting the presence of stem cell niche.124 This region is analogous to the region known as the groove of Ranvier in the long bone.122 The same group proposed the migration routes of the progenitors from this stem cell niche to the outer AF by analyzing a cell adhesion and migration marker (β1 integrin), and EMT markers (Snail‐1 and ‐2).125
Lineage tracing provides more direct evidences, allowing us to follow cell fate decisions of progenitor cells and their descendants within a living organism without any perturbation.126 Lineage tracing typically uses the Cre‐loxP system to permanently mark the cells of interest. Cre recombinase is expressed under the control of a tissue‐ or cell‐specific promoter in one mouse line.126 That line is crossed with a second mouse line, in which a reporter is flanked by a loxP‐STOP‐loxP sequence.126 In animals expressing both constructs, Cre specifically activates the reporter in cells that express the promoter, by excising the STOP sequence.126 In the knee joint, several novel progenitors have recently been identified by lineage tracing. For example, Prg4‐creERT2 labels the progenitors of the superficial zone of the articular cartilage,127 PTHrP‐creERT2 labels the progenitors in the reserve zone of the growth plate,128 and Axin2‐creERT2 labels the progenitors in and around the groove of Ranvier.129 In studies on IVD, lineage tracing experiments by Choi et al. employing Shh‐cre and Shh‐creERT2 alleles showed that the notochord is the sole source of cells that form the entire NP.130 This was further confirmed by the lineage tracing experiments using the Noto‐cre allele.131 The AF and the CEP were devoid of Shh‐cre or Noto‐cre descendent cells, indicating that the progenitors of AF and CEP never reside in the NP. Although these studies provided important developmental findings, no creERT2 lines have been reported that can specifically mark putative progenitors in IVD tissues.
7. CONCLUSION AND FUTURE PERSPECTIVES
Recent population‐based cohort studies showed that IDD is very common and is associated with the presence of LBP and suggested that the degeneration of CEP has an important role in the pain associated with IDD. Previous epidemiologic studies have clearly shown that IDD is highly heritable. Although candidate gene approaches, family linkage analyses, and recent genome‐wide association studies have revealed several IDD‐sensitive genes, further studies are warranted to elucidate the molecular mechanisms through which each candidate gene causes IDD. The pathomechanisms of IDD have been elucidated using several animal models. However, caution should be exercised while interpreting the information obtained from animal models, as there are many differences between species, including disc size, cell type, nutrition, and mechanical forces.132 Several researchers have reported the presence of progenitors in IVD that behave like mesenchymal stem cells in vitro. However, the actual role and properties of the progenitors in vivo remain unknown owing to the lack of markers that can specifically mark the progenitors in vivo. Recent availability of single‐cell transcriptomic analyses possibly facilitates the identification of such marker genes that can track and localize potential progenitors.133 Establishment of the mouse line where Cre recombinase is expressed under the control of such marker genes will enable the purification of progenitors (eg, using fluorescence‐activated cell sorting), after knowing the properties of these cells in detail through comprehensive expression analyses such as RNA‐seq. Investigation of the signals that promote the function of progenitors will provide essential information for the development of drugs that can activate resident progenitors and, possibly, prevent IDD. In addition, identification of cell surface markers of IVD progenitors will contribute to not only the research of human IVD progenitors but also the development of exogenous cell therapies for IDD.
CONFLICT OF INTEREST
The authors declare no conflicts of interest.
AUTHOR CONTRIBUTIONS
T.O., Y.T., Y.O., S.T., and T.S. contributed the concept of the paper and wrote the manuscript. All authors have read and approved the final submitted manuscript.
Oichi T, Taniguchi Y, Oshima Y, Tanaka S, Saito T. Pathomechanism of intervertebral disc degeneration. JOR Spine. 2020;3:e1076 10.1002/jsp2.1076
REFERENCES
- 1. Global, regional, and national incidence, prevalence, and years lived with disability for 354 diseases and injuries for 195 countries and territories . 1990–2017: a systematic analysis for the global burden of disease study 2017. Lancet. 2018;392:1789‐1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ekman M, Jonhagen S, Hunsche E, Jonsson L. Burden of illness of chronic low back pain in Sweden: a cross‐sectional, retrospective study in primary care setting. Spine. 2005;30:1777‐1785. [DOI] [PubMed] [Google Scholar]
- 3. Katz JN. Lumbar disc disorders and low‐back pain: socioeconomic factors and consequences. J Bone Joint Surg Am. 2006;88(Suppl 2):21‐24. [DOI] [PubMed] [Google Scholar]
- 4. Cheung KM, Karppinen J, Chan D, et al. Prevalence and pattern of lumbar magnetic resonance imaging changes in a population study of one thousand forty‐three individuals. Spine. 2009;34:934‐940. [DOI] [PubMed] [Google Scholar]
- 5. Sakai D. Future perspectives of cell‐based therapy for intervertebral disc disease. Eur Spine J. 2008;17(Suppl 4):452‐458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Videman T, Battie MC, Gill K, Manninen H, Gibbons LE, Fisher LD. Magnetic resonance imaging findings and their relationships in the thoracic and lumbar spine. Insights into the etiopathogenesis of spinal degeneration. Spine. 1995;20:928‐935. [DOI] [PubMed] [Google Scholar]
- 7. Muraki S, Oka H, Akune T, et al. Prevalence of radiographic lumbar spondylosis and its association with low back pain in elderly subjects of population‐based cohorts: the ROAD study. Ann Rheum Dis. 2009;68:1401‐1406. [DOI] [PubMed] [Google Scholar]
- 8. Teraguchi M, Yoshimura N, Hashizume H, et al. Prevalence and distribution of intervertebral disc degeneration over the entire spine in a population‐based cohort: the Wakayama spine study. Osteoarthr Cartil. 2014;22:104‐110. [DOI] [PubMed] [Google Scholar]
- 9. Pfirrmann CW, Metzdorf A, Zanetti M, Hodler J, Boos N. Magnetic resonance classification of lumbar intervertebral disc degeneration. Spine. 2001;26:1873‐1878. [DOI] [PubMed] [Google Scholar]
- 10. Teraguchi M, Yoshimura N, Hashizume H, et al. The association of combination of disc degeneration, end plate signal change, and Schmorl node with low back pain in a large population study: the Wakayama spine study. Spine J. 2015;15:622‐628. [DOI] [PubMed] [Google Scholar]
- 11. Mok FP, Samartzis D, Karppinen J, Fong DY, Luk KD, Cheung KM. Modic changes of the lumbar spine: prevalence, risk factors, and association with disc degeneration and low back pain in a large‐scale population‐based cohort. Spine J. 2016;16:32‐41. [DOI] [PubMed] [Google Scholar]
- 12. Battie MC, Videman T, Kaprio J, et al. The twin spine study: contributions to a changing view of disc degeneration. Spine J. 2009;9:47‐59. [DOI] [PubMed] [Google Scholar]
- 13. Battie MC, Videman T, Gibbons LE, Fisher LD, Manninen H, Gill K. 1995 Volvo award in clinical sciences. Determinants of lumbar disc degeneration. A study relating lifetime exposures and magnetic resonance imaging findings in identical twins. Spine. 1995;20:2601‐2612. [PubMed] [Google Scholar]
- 14. Livshits G, Popham M, Malkin I, et al. Lumbar disc degeneration and genetic factors are the main risk factors for low back pain in women: the UKtwin spine study. Ann Rheum Dis. 2011;70:1740‐1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sambrook PN, MacGregor AJ, Spector TD. Genetic influences on cervical and lumbar disc degeneration: a magnetic resonance imaging study in twins. Arthritis Rheum. 1999;42:366‐372. [DOI] [PubMed] [Google Scholar]
- 16. Kawaguchi Y, Osada R, Kanamori M, et al. Association between an aggrecan gene polymorphism and lumbar disc degeneration. Spine. 1999;24:2456‐2460. [DOI] [PubMed] [Google Scholar]
- 17. Seki S, Kawaguchi Y, Chiba K, et al. A functional SNP in CILP, encoding cartilage intermediate layer protein, is associated with susceptibility to lumbar disc disease. Nat Genet. 2005;37:607‐612. [DOI] [PubMed] [Google Scholar]
- 18. Pluijm SM, van Essen HW, Bravenboer N, et al. Collagen type I alpha1 Sp1 polymorphism, osteoporosis, and intervertebral disc degeneration in older men and women. Ann Rheum Dis. 2004;63:71‐77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Annunen S, Paassilta P, Lohiniva J, et al. An allele of COL9A2 associated with intervertebral disc disease. Science. 1999;285:409‐412. [DOI] [PubMed] [Google Scholar]
- 20. Mio F, Chiba K, Hirose Y, et al. A functional polymorphism in COL11A1, which encodes the alpha 1 chain of type XI collagen, is associated with susceptibility to lumbar disc herniation. Am J Hum Genet. 2007;81:1271‐1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Williams FM, Popham M, Hart DJ, et al. GDF5 single‐nucleotide polymorphism rs143383 is associated with lumbar disc degeneration in northern European women. Arthritis Rheum. 2011;63:708‐712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Urano T, Narusawa K, Shiraki M, et al. Association of a single nucleotide polymorphism in the insulin‐like growth factor‐1 receptor gene with spinal disc degeneration in postmenopausal Japanese women. Spine. 2008;33:1256‐1261. [DOI] [PubMed] [Google Scholar]
- 23. Solovieva S, Kouhia S, Leino‐Arjas P, et al. Interleukin 1 polymorphisms and intervertebral disc degeneration. Epidemiology. 2004;15:626‐633. [DOI] [PubMed] [Google Scholar]
- 24. Noponen‐Hietala N, Virtanen I, Karttunen R, et al. Genetic variations in IL6 associate with intervertebral disc disease characterized by sciatica. Pain. 2005;114:186‐194. [DOI] [PubMed] [Google Scholar]
- 25. Dong DM, Yao M, Liu B, Sun CY, Jiang YQ, Wang YS. Association between the ‐1306C/T polymorphism of matrix metalloproteinase‐2 gene and lumbar disc disease in Chinese young adults. Eur Spine J. 2007;16:1958‐1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Takahashi M, Haro H, Wakabayashi Y, Kawa‐uchi T, Komori H, Shinomiya K. The association of degeneration of the intervertebral disc with 5a/6a polymorphism in the promoter of the human matrix metalloproteinase‐3 gene. J Bone Joint Surg Br. 2001;83:491‐495. [DOI] [PubMed] [Google Scholar]
- 27. Sun ZM, Miao L, Zhang YG, Ming L. Association between the −1562 C/T polymorphism of matrix metalloproteinase‐9 gene and lumbar disc disease in the young adult population in North China. Connect Tissue Res. 2009;50:181‐185. [DOI] [PubMed] [Google Scholar]
- 28. Karasugi T, Semba K, Hirose Y, et al. Association of the tag SNPs in the human SKT gene (KIAA1217) with lumbar disc herniation. J Bone Miner Res. 2009;24:1537‐1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hirose Y, Chiba K, Karasugi T, et al. A functional polymorphism in THBS2 that affects alternative splicing and MMP binding is associated with lumbar‐disc herniation. Am J Hum Genet. 2008;82:1122‐1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kawaguchi Y, Kanamori M, Ishihara H, Ohmori K, Matsui H, Kimura T. The association of lumbar disc disease with vitamin‐D receptor gene polymorphism. J Bone Joint Surg Am. 2002;84:2022‐2028. [DOI] [PubMed] [Google Scholar]
- 31. Videman T, Leppavuori J, Kaprio J, et al. Intragenic polymorphisms of the vitamin D receptor gene associated with intervertebral disc degeneration. Spine. 1998;23:2477‐2485. [DOI] [PubMed] [Google Scholar]
- 32. Williams FM, Bansal AT, van Meurs JB, et al. Novel genetic variants associated with lumbar disc degeneration in northern Europeans: a meta‐analysis of 4600 subjects. Ann Rheum Dis. 2013;72:1141‐1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Song YQ, Karasugi T, Cheung KM, et al. Lumbar disc degeneration is linked to a carbohydrate sulfotransferase 3 variant. J Clin Invest. 2013;123:4909‐4917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kawaguchi Y. Genetic background of degenerative disc disease in the lumbar spine. Spine Surg Relat Res. 2018;2:98‐112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Miller JA, Schmatz C, Schultz AB. Lumbar disc degeneration: correlation with age, sex, and spine level in 600 autopsy specimens. Spine. 1988;13:173‐178. [PubMed] [Google Scholar]
- 36. Lee CK. Accelerated degeneration of the segment adjacent to a lumbar fusion. Spine. 1988;13:375‐377. [DOI] [PubMed] [Google Scholar]
- 37. Carragee EJ, Don AS, Hurwitz EL, Cuellar JM, Carrino JA, Herzog R. 2009 ISSLS prize winner: does discography cause accelerated progression of degeneration changes in the lumbar disc: a ten‐year matched cohort study. Spine. 2009;34:2338‐2345. [DOI] [PubMed] [Google Scholar]
- 38. Kerttula LI, Serlo WS, Tervonen OA, Paakko EL, Vanharanta HV. Post‐traumatic findings of the spine after earlier vertebral fracture in young patients: clinical and MRI study. Spine. 2000;25:1104‐1108. [DOI] [PubMed] [Google Scholar]
- 39. Battie MC, Videman T, Gill K, et al. 1991 Volvo award in clinical sciences. Smoking and lumbar intervertebral disc degeneration: an MRI study of identical twins. Spine. 1991;16:1015‐1021. [PubMed] [Google Scholar]
- 40. Pattappa G, Li Z, Peroglio M, Wismer N, Alini M, Grad S. Diversity of intervertebral disc cells: phenotype and function. J Anat. 2012;221:480‐496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Boos N, Weissbach S, Rohrbach H, Weiler C, Spratt KF, Nerlich AG. Classification of age‐related changes in lumbar intervertebral discs: 2002 Volvo award in basic science. Spine. 2002;27:2631‐2644. [DOI] [PubMed] [Google Scholar]
- 42. Pezowicz CA, Robertson PA, Broom ND. Intralamellar relationships within the collagenous architecture of the annulus fibrosus imaged in its fully hydrated state. J Anat. 2005;207:299‐312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Takimoto A, Kokubu C, Watanabe H, et al. Differential transactivation of the upstream aggrecan enhancer regulated by PAX1/9 depends on SOX9‐driven transactivation. Sci Rep. 2019;9:4605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Roughley PJ. Biology of intervertebral disc aging and degeneration: involvement of the extracellular matrix. Spine. 2004;29:2691‐2699. [DOI] [PubMed] [Google Scholar]
- 45. Wu Y, Cisewski S, Sachs BL, Yao H. Effect of cartilage endplate on cell based disc regeneration: a finite element analysis. Mol Cell Biomech. 2013;10:159‐182. [PMC free article] [PubMed] [Google Scholar]
- 46. Kepler CK, Ponnappan RK, Tannoury CA, Risbud MV, Anderson DG. The molecular basis of intervertebral disc degeneration. Spine J. 2013;13:318‐330. [DOI] [PubMed] [Google Scholar]
- 47. Feng Y, Egan B, Wang J. Genetic factors in intervertebral disc degeneration. Genes Dis. 2016;3:178‐185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Le Maitre CL, Pockert A, Buttle DJ, Freemont AJ, Hoyland JA. Matrix synthesis and degradation in human intervertebral disc degeneration. Biochem Soc Trans. 2007;35:652‐655. [DOI] [PubMed] [Google Scholar]
- 49. Boos N, Nerlich AG, Wiest I, von der Mark K, Aebi M. Immunolocalization of type X collagen in human lumbar intervertebral discs during ageing and degeneration. Histochem Cell Biol. 1997;108:471‐480. [DOI] [PubMed] [Google Scholar]
- 50. Wagner DR, Reiser KM, Lotz JC. Glycation increases human annulus fibrosus stiffness in both experimental measurements and theoretical predictions. J Biomech. 2006;39:1021‐1029. [DOI] [PubMed] [Google Scholar]
- 51. Cs‐Szabo G, Ragasa‐San Juan D, Turumella V, Masuda K, Thonar EJ, An HS. Changes in mRNA and protein levels of proteoglycans of the anulus fibrosus and nucleus pulposus during intervertebral disc degeneration. Spine. 2002;27:2212‐2219. [DOI] [PubMed] [Google Scholar]
- 52. Buckwalter JA, Roughley PJ, Rosenberg LC. Age‐related changes in cartilage proteoglycans: quantitative electron microscopic studies. Microsc Res Tech. 1994;28:398‐408. [DOI] [PubMed] [Google Scholar]
- 53. Pearce RH, Grimmer BJ, Adams ME. Degeneration and the chemical composition of the human lumbar intervertebral disc. J Orthop Res. 1987;5:198‐205. [DOI] [PubMed] [Google Scholar]
- 54. Antoniou J, Steffen T, Nelson F, et al. The human lumbar intervertebral disc: evidence for changes in the biosynthesis and denaturation of the extracellular matrix with growth, maturation, ageing, and degeneration. J Clin Invest. 1996;98:996‐1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Sive JI, Baird P, Jeziorsk M, Watkins A, Hoyland JA, Freemont AJ. Expression of chondrocyte markers by cells of normal and degenerate intervertebral discs. Mol Pathol. 2002;55:91‐97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Roberts S, Evans H, Trivedi J, Menage J. Histology and pathology of the human intervertebral disc. J Bone Joint Surg Am. 2006;88(Suppl 2):10‐14. [DOI] [PubMed] [Google Scholar]
- 57. Edelson JG, Nathan H. Stages in the natural history of the vertebral end‐plates. Spine. 1988;13:21‐26. [DOI] [PubMed] [Google Scholar]
- 58. Vo NV, Hartman RA, Patil PR, et al. Molecular mechanisms of biological aging in intervertebral discs. J Orthop Res. 2016;34:1289‐1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Niedernhofer LJ, Robbins PD. Signaling mechanisms involved in the response to genotoxic stress and regulating lifespan. Int J Biochem Cell Biol. 2008;40:176‐180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Vo N, Seo HY, Robinson A, et al. Accelerated aging of intervertebral discs in a mouse model of progeria. J Orthop Res. 2010;28:1600‐1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Nasto LA, Wang D, Robinson AR, et al. Genotoxic stress accelerates age‐associated degenerative changes in intervertebral discs. Mech Ageing Dev. 2013;134:35‐42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wang D, Nasto LA, Roughley P, et al. Spine degeneration in a murine model of chronic human tobacco smokers. Osteoarthr Cartil. 2012;20:896‐905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Phillips KL, Cullen K, Chiverton N, et al. Potential roles of cytokines and chemokines in human intervertebral disc degeneration: interleukin‐1 is a master regulator of catabolic processes. Osteoarthr Cartil. 2015;23:1165‐1177. [DOI] [PubMed] [Google Scholar]
- 64. Le Maitre CL, Freemont AJ, Hoyland JA. The role of interleukin‐1 in the pathogenesis of human intervertebral disc degeneration. Arthritis Res Ther. 2005;7:R732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Phillips KL, Jordan‐Mahy N, Nicklin MJ, Le Maitre CL. Interleukin‐1 receptor antagonist deficient mice provide insights into pathogenesis of human intervertebral disc degeneration. Ann Rheum Dis. 2013;72:1860‐1867. [DOI] [PubMed] [Google Scholar]
- 66. van Deursen JM. The role of senescent cells in ageing. Nature. 2014;509:439‐446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Campisi J. Senescent cells, tumor suppression, and organismal aging: good citizens, bad neighbors. Cell. 2005;120:513‐522. [DOI] [PubMed] [Google Scholar]
- 68. Coppe JP, Patil CK, Rodier F, et al. Senescence‐associated secretory phenotypes reveal cell‐nonautonomous functions of oncogenic RAS and the p53 tumor suppressor. PLoS Biol. 2008;6:2853‐2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Rodier F, Coppe JP, Patil CK, et al. Persistent DNA damage signalling triggers senescence‐associated inflammatory cytokine secretion. Nat Cell Biol. 2009;11:973‐979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Roberts S, Evans EH, Kletsas D, Jaffray DC, Eisenstein SM. Senescence in human intervertebral discs. Eur Spine J. 2006;15(Suppl 3):S312‐S316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Le Maitre CL, Freemont AJ, Hoyland JA. Accelerated cellular senescence in degenerate intervertebral discs: a possible role in the pathogenesis of intervertebral disc degeneration. Arthritis Res Ther. 2007;9:R45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Heathfield SK, Le Maitre CL, Hoyland JA. Caveolin‐1 expression and stress‐induced premature senescence in human intervertebral disc degeneration. Arthritis Res Ther. 2008;10:R87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Gruber HE, Ingram JA, Davis DE, Hanley EN Jr. Increased cell senescence is associated with decreased cell proliferation in vivo in the degenerating human annulus. Spine J. 2009;9:210‐215. [DOI] [PubMed] [Google Scholar]
- 74. Kim KW, Chung HN, Ha KY, Lee JS, Kim YY. Senescence mechanisms of nucleus pulposus chondrocytes in human intervertebral discs. Spine J. 2009;9:658‐666. [DOI] [PubMed] [Google Scholar]
- 75. Patil P, Dong Q, Wang D, et al. Systemic clearance of p16(INK4a) ‐positive senescent cells mitigates age‐associated intervertebral disc degeneration. Aging Cell. 2019;18:e12927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Yurube T, Nishida K, Suzuki T, et al. Matrix metalloproteinase (MMP)‐3 gene up‐regulation in a rat tail compression loading‐induced disc degeneration model. J Orthop Res. 2010;28:1026‐1032. [DOI] [PubMed] [Google Scholar]
- 77. Lotz JC, Colliou OK, Chin JR, Duncan NA, Liebenberg E. Compression‐induced degeneration of the intervertebral disc: an in vivo mouse model and finite‐element study. Spine. 1998;23:2493‐2506. [DOI] [PubMed] [Google Scholar]
- 78. Sakai D, Nishimura K, Tanaka M, et al. Migration of bone marrow‐derived cells for endogenous repair in a new tail‐looping disc degeneration model in the mouse: a pilot study. Spine J. 2015;15:1356‐1365. [DOI] [PubMed] [Google Scholar]
- 79. Yang F, Leung VY, Luk KD, Chan D, Cheung KM. Injury‐induced sequential transformation of notochordal nucleus pulposus to chondrogenic and fibrocartilaginous phenotype in the mouse. J Pathol. 2009;218:113‐121. [DOI] [PubMed] [Google Scholar]
- 80. Lipson SJ, Muir H. 1980 Volvo award in basic science. Proteoglycans in experimental intervertebral disc degeneration. Spine. 1981;6:194‐210. [DOI] [PubMed] [Google Scholar]
- 81. Masuda K, Aota Y, Muehleman C, et al. A novel rabbit model of mild, reproducible disc degeneration by an anulus needle puncture: correlation between the degree of disc injury and radiological and histological appearances of disc degeneration. Spine. 2005;30:5‐14. [DOI] [PubMed] [Google Scholar]
- 82. Sullivan JD, Farfan HF, Kahn DS. Pathologic changes with intervertebral joint rotational instability in the rabbit. Can J Surg. 1971;14:71‐79. [PubMed] [Google Scholar]
- 83. Fukui D, Kawakami M, Yoshida M, Nakao S, Matsuoka T, Yamada H. Gait abnormality due to spinal instability after lumbar facetectomy in the rat. Eur Spine J. 2015;24:2085‐2094. [DOI] [PubMed] [Google Scholar]
- 84. Hadjipavlou AG, Simmons JW, Yang JP, et al. Torsional injury resulting in disc degeneration: I. An in vivo rabbit model. J Spinal Disord. 1998;11:312‐317. [PubMed] [Google Scholar]
- 85. Miyamoto S, Yonenobu K, Ono K. Experimental cervical spondylosis in the mouse. Spine. 1991;16:S495‐S500. [DOI] [PubMed] [Google Scholar]
- 86. Stokes IA, Counts DF, Frymoyer JW. Experimental instability in the rabbit lumbar spine. Spine. 1989;14:68‐72. [DOI] [PubMed] [Google Scholar]
- 87. Cassidy JD, Yong‐Hing K, Kirkaldy‐Willis WH, Wilkinson AA. A study of the effects of bipedism and upright posture on the lumbosacral spine and paravertebral muscles of the Wistar rat. Spine. 1988;13:301‐308. [DOI] [PubMed] [Google Scholar]
- 88. Liang QQ, Zhou Q, Zhang M, et al. Prolonged upright posture induces degenerative changes in intervertebral discs in rat lumbar spine. Spine. 2008;33:2052‐2058. [DOI] [PubMed] [Google Scholar]
- 89. Norcross JP, Lester GE, Weinhold P, Dahners LE. An in vivo model of degenerative disc disease. J Orthop Res. 2003;21:183‐188. [DOI] [PubMed] [Google Scholar]
- 90. Sahlman J, Inkinen R, Hirvonen T, et al. Premature vertebral endplate ossification and mild disc degeneration in mice after inactivation of one allele belonging to the Col2a1 gene for type II collagen. Spine. 2001;26:2558‐2565. [DOI] [PubMed] [Google Scholar]
- 91. McCann MR, Patel P, Pest MA, et al. Repeated exposure to high‐frequency low‐amplitude vibration induces degeneration of murine intervertebral discs and knee joints. Arthritis Rheumatol. 2015;67:2164‐2175. [DOI] [PubMed] [Google Scholar]
- 92. Gruber HE, Johnson T, Norton HJ, Hanley EN Jr. The sand rat model for disc degeneration: radiologic characterization of age‐related changes: cross‐sectional and prospective analyses. Spine. 2002;27:230‐234. [DOI] [PubMed] [Google Scholar]
- 93. Oda H, Matsuzaki H, Tokuhashi Y, Wakabayashi K, Uematsu Y, Iwahashi M. Degeneration of intervertebral discs due to smoking: experimental assessment in a rat‐smoking model. J Orthop Sci. 2004;9:135‐141. [DOI] [PubMed] [Google Scholar]
- 94. Oichi T, Taniguchi Y, Soma K, et al. A mouse intervertebral disc degeneration model by surgically induced instability. Spine. 2018;43:E557. [DOI] [PubMed] [Google Scholar]
- 95. Jayasuriya CT, Hu N, Li J, et al. Molecular characterization of mesenchymal stem cells in human osteoarthritis cartilage reveals contribution to the OA phenotype. Sci Rep. 2018;8:7044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Weissman IL. Stem cells: units of development, units of regeneration, and units in evolution. Cell. 2000;100:157‐168. [DOI] [PubMed] [Google Scholar]
- 97. Sakai D, Andersson GB. Stem cell therapy for intervertebral disc regeneration: obstacles and solutions. Nat Rev Rheumatol. 2015;11:243‐256. [DOI] [PubMed] [Google Scholar]
- 98. Jiang L, Yuan F, Yin X, Dong J. Responses and adaptations of intervertebral disc cells to microenvironmental stress: a possible central role of autophagy in the adaptive mechanism. Connect Tissue Res. 2014;55:311‐321. [DOI] [PubMed] [Google Scholar]
- 99. Risbud MV, Schipani E, Shapiro IM. Hypoxic regulation of nucleus pulposus cell survival: from niche to notch. Am J Pathol. 2010;176:1577‐1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Urban JP, Holm S, Maroudas A, Nachemson A. Nutrition of the intervertebral disk. An in vivo study of solute transport. Clin Orthop Relat Res. 1977;129:101‐114. [PubMed] [Google Scholar]
- 101. Urban JP, Holm S, Maroudas A, Nachemson A. Nutrition of the intervertebral disc: effect of fluid flow on solute transport. Clin Orthop Relat Res. 1982;170:296‐302. [PubMed] [Google Scholar]
- 102. Grunhagen T, Wilde G, Soukane DM, Shirazi‐Adl SA, Urban JP. Nutrient supply and intervertebral disc metabolism. J Bone Joint Surg Am. 2006;88(Suppl 2):30‐35. [DOI] [PubMed] [Google Scholar]
- 103. Urban JP, Smith S, Fairbank JC. Nutrition of the intervertebral disc. Spine. 2004;29:2700‐2709. [DOI] [PubMed] [Google Scholar]
- 104. Grunhagen T, Shirazi‐Adl A, Fairbank JC, Urban JP. Intervertebral disk nutrition: a review of factors influencing concentrations of nutrients and metabolites. Orthop Clin North Am. 2011;42:465‐477. vii. [DOI] [PubMed] [Google Scholar]
- 105. Urban JP. The role of the physicochemical environment in determining disc cell behaviour. Biochem Soc Trans. 2002;30:858‐864. [DOI] [PubMed] [Google Scholar]
- 106. Risbud MV, Guttapalli A, Tsai TT, et al. Evidence for skeletal progenitor cells in the degenerate human intervertebral disc. Spine. 2007;32:2537‐2544. [DOI] [PubMed] [Google Scholar]
- 107. Blanco JF, Graciani IF, Sanchez‐Guijo FM, et al. Isolation and characterization of mesenchymal stromal cells from human degenerated nucleus pulposus: comparison with bone marrow mesenchymal stromal cells from the same subjects. Spine. 2010;35:2259‐2265. [DOI] [PubMed] [Google Scholar]
- 108. Feng G, Yang X, Shang H, et al. Multipotential differentiation of human anulus fibrosus cells: an in vitro study. J Bone Joint Surg Am. 2010;92:675‐685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Liu LT, Huang B, Li CQ, Zhuang Y, Wang J, Zhou Y. Characteristics of stem cells derived from the degenerated human intervertebral disc cartilage endplate. PLoS One. 2011;6:e26285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Erwin WM, Islam D, Eftekarpour E, Inman RD, Karim MZ, Fehlings MG. Intervertebral disc‐derived stem cells: implications for regenerative medicine and neural repair. Spine. 2013;38:211‐216. [DOI] [PubMed] [Google Scholar]
- 111. Brisby H, Papadimitriou N, Brantsing C, Bergh P, Lindahl A, Barreto HH. The presence of local mesenchymal progenitor cells in human degenerated intervertebral discs and possibilities to influence these in vitro: a descriptive study in humans. Stem Cells Dev. 2013;22:804‐814. [DOI] [PubMed] [Google Scholar]
- 112. Mizrahi O, Sheyn D, Tawackoli W, et al. Nucleus pulposus degeneration alters properties of resident progenitor cells. Spine J. 2013;13:803‐814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Liang L, Li X, Li D, et al. The characteristics of stem cells in human degenerative intervertebral disc. Medicine. 2017;96:e7178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Navaro Y, Bleich‐Kimelman N, Hazanov L, et al. Matrix stiffness determines the fate of nucleus pulposus‐derived stem cells. Biomaterials. 2015;49:68‐76. [DOI] [PubMed] [Google Scholar]
- 115. Li H, Wang J, Li F, Chen G, Chen Q. The influence of hyperosmolarity in the intervertebral disc on the proliferation and chondrogenic differentiation of nucleus pulposus‐derived mesenchymal stem cells. Cells Tissues Organs. 2018;205:178‐188. [DOI] [PubMed] [Google Scholar]
- 116. Wuertz K, Godburn K, Iatridis JC. MSC response to pH levels found in degenerating intervertebral discs. Biochem Biophys Res Commun. 2009;379:824‐829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Sakai D, Nakamura Y, Nakai T, et al. Exhaustion of nucleus pulposus progenitor cells with ageing and degeneration of the intervertebral disc. Nat Commun. 2012;3:1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Fujita N, Miyamoto T, Imai J, et al. CD24 is expressed specifically in the nucleus pulposus of intervertebral discs. Biochem Biophys Res Commun. 2005;338:1890‐1896. [DOI] [PubMed] [Google Scholar]
- 119. Tekari A, Chan SCW, Sakai D, Grad S, Gantenbein B. Angiopoietin‐1 receptor Tie2 distinguishes multipotent differentiation capability in bovine coccygeal nucleus pulposus cells. Stem Cell Res Ther. 2016;7:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Candela ME, Cantley L, Yasuaha R, Iwamoto M, Pacifici M, Enomoto‐Iwamoto M. Distribution of slow‐cycling cells in epiphyseal cartilage and requirement of beta‐catenin signaling for their maintenance in growth plate. J Orthop Res. 2014;32:661‐668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Ohlsson C, Nilsson A, Isaksson O, Lindahl A. Growth hormone induces multiplication of the slowly cycling germinal cells of the rat tibial growth plate. Proc Natl Acad Sci U S A. 1992;89:9826‐9830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Karlsson C, Thornemo M, Henriksson HB, Lindahl A. Identification of a stem cell niche in the zone of Ranvier within the knee joint. J Anat. 2009;215:355‐363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Kurth TB, Dell'accio F, Crouch V, Augello A, Sharpe PT, De Bari C. Functional mesenchymal stem cell niches in adult mouse knee joint synovium in vivo. Arthritis Rheum. 2011;63:1289‐1300. [DOI] [PubMed] [Google Scholar]
- 124. Henriksson H, Thornemo M, Karlsson C, et al. Identification of cell proliferation zones, progenitor cells and a potential stem cell niche in the intervertebral disc region: a study in four species. Spine. 2009;34:2278‐2287. [DOI] [PubMed] [Google Scholar]
- 125. Henriksson HB, Svala E, Skioldebrand E, Lindahl A, Brisby H. Support of concept that migrating progenitor cells from stem cell niches contribute to normal regeneration of the adult mammal intervertebral disc: a descriptive study in the New Zealand white rabbit. Spine. 2012;37:722‐732. [DOI] [PubMed] [Google Scholar]
- 126. Kretzschmar K, Watt FM. Lineage tracing. Cell. 2012;148:33‐45. [DOI] [PubMed] [Google Scholar]
- 127. Kozhemyakina E, Zhang M, Ionescu A, et al. Identification of a Prg4‐expressing articular cartilage progenitor cell population in mice. Arthritis Rheumatol. 2015;67:1261‐1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Mizuhashi K, Ono W, Matsushita Y, et al. Resting zone of the growth plate houses a unique class of skeletal stem cells. Nature. 2018;563:254‐258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Usami Y, Gunawardena AT, Francois NB, et al. Possible contribution of Wnt‐responsive chondroprogenitors to the postnatal murine growth plate. J Bone Miner Res. 2019;34:964‐974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Choi KS, Cohn MJ, Harfe BD. Identification of nucleus pulposus precursor cells and notochordal remnants in the mouse: implications for disk degeneration and chordoma formation. Dev Dyn. 2008;237:3953‐3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. McCann MR, Tamplin OJ, Rossant J, Seguin CA. Tracing notochord‐derived cells using a Noto‐cre mouse: implications for intervertebral disc development. Dis Model Mech. 2012;5:73‐82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Jin L, Balian G, Li XJ. Animal models for disc degeneration‐an update. Histol Histopathol. 2018;33:543‐554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Seguin CA, Chan D, Dahia CL, Gazit Z. Latest advances in intervertebral disc development and progenitor cells. JOR Spine. 2018;1:e1030. [DOI] [PMC free article] [PubMed] [Google Scholar]


