Significance
Here, we show that expression of key circadian clock genes in the skin is altered by acute inflammation in mice treated topically with the immune activator imiquimod and by chronic inflammation in human psoriatic lesions. We show time-of-day–dependent activation of the interferon pathway, a key pathway involved in the host defense response. Mice lacking circadian rhythms have greater epidermal hyperplasia and more robust activation of the interferon pathway. Furthermore, we show that daytime-restricted feeding shifts the phase of interferon-sensitive gene expression in mouse skin. These findings demonstrate a role for the circadian clock in a major defense pathway in the skin’s response to microbes and cancer and suggest that timing of feeding can affect this response in the skin.
Keywords: Bmal1, interferon, immune, circadian, antiviral
Abstract
The circadian clock coordinates a variety of immune responses with signals from the external environment to promote survival. We investigated the potential reciprocal relationship between the circadian clock and skin inflammation. We treated mice topically with the Toll-like receptor 7 (TLR7) agonist imiquimod (IMQ) to activate IFN-sensitive gene (ISG) pathways and induce psoriasiform inflammation. IMQ transiently altered core clock gene expression, an effect mirrored in human patient psoriatic lesions. In mouse skin 1 d after IMQ treatment, ISGs, including the key ISG transcription factor IFN regulatory factor 7 (Irf7), were more highly induced after treatment during the day than the night. Nuclear localization of phosphorylated-IRF7 was most prominently time-of-day dependent in epidermal leukocytes, suggesting that these cell types play an important role in the diurnal ISG response to IMQ. Mice lacking Bmal1 systemically had exacerbated and arrhythmic ISG/Irf7 expression after IMQ. Furthermore, daytime-restricted feeding, which affects the phase of the skin circadian clock, reverses the diurnal rhythm of IMQ-induced ISG expression in the skin. These results suggest a role for the circadian clock, driven by BMAL1, as a negative regulator of the ISG response, and highlight the finding that feeding time can modulate the skin immune response. Since the IFN response is essential for the antiviral and antitumor effects of TLR activation, these findings are consistent with the time-of-day–dependent variability in the ability to fight microbial pathogens and tumor initiation and offer support for the use of chronotherapy for their treatment.
The skin contains a circadian clock that is under the influence of the suprachiasmatic nucleus (1, 2) and, surprisingly, feeding time (3). Coordination of intermediary metabolism with the cell cycle in epidermal stem cells (4, 5) is one of the key roles of the skin clock, and there is also strong evidence for clock regulation of skin resident and migratory immune cells (1, 6). Circadian regulation of mast cells, for example, plays a role in the diurnal variation of allergic symptoms in the skin (7), and mice mutated for core circadian clock gene CLOCK have severe skin allergic reactions (8). Antigen-presenting dendritic cells (DCs) are recruited to the skin in a diurnal fashion; loss of circadian control dampens trafficking of these cells to the skin during delayed-type hypersensitivity reactions (9). The circadian clock also attenuates imiquimod (IMQ)-induced skin inflammation (10). The circadian clock, then, influences skin immune cells and modulates the skin’s inflammatory response, but the reciprocal interactions between the skin clock and the immune system are not fully understood, and it is unknown if altered feeding times, which shifts the phase of the skin clock, affect the skin immune response.
As the first line of defense against pathogens, the skin is paramount in preventing and responding to infections. While the influence of the circadian clock on skin viral infections is incompletely understood, recent evidence suggests that the circadian clock may regulate the defense against viral infections in other organs. Mice intranasally infected with herpes or influenza A viruses during the daytime have greater infection rates and mortality than mice infected at night. This diurnal effect may be mediated by the circadian clock as deletion of the core clock gene Bmal1 exacerbates viral infections (11). Furthermore, disruption of circadian function by jet lag or Bmal1 deletion leads to increased acute viral bronchiolitis after Sendai virus and influenza A viral infections (12). The mechanism by which the circadian clock and BMAL1 contribute to host defense response against viruses is currently unclear. The complex relationship between the circadian clock and the skin immune system is important not only for defense against viral infections, but also for tumorigenesis and antitumor actions, which rely on activation of antiviral pathways, including type I interferons (IFNs) (13).
IFN mediates greater antiviral effects during the night compared to the day (14), and circadian and feeding time regulation of IFN in the skin has not been previously described. We used the therapeutic drug and single-stranded RNA virus mimic IMQ as a model system to investigate the circadian clock mechanisms that may alter the skin’s immune response against tumors or viral infections. IMQ was originally developed as a topical treatment for human papilloma virus-associated anogenital warts, and it has since been approved for the treatment of nonviral tumors such as actinic keratosis and superficial basal cell carcinomas. IMQ causes immune activation through induction of antiviral proteins, proinflammatory cytokines, and chemokines (15).
When IMQ is applied repetitively to mouse skin, it induces robust inflammation affected by sensory neurons containing TRPV1 and NaV1.8 ion channels (16). This causes epidermal hyperproliferation, parakaratosis (nuclei retained in the stratum corneum), acanthosis (thickening of the epidermis), and Munro’s microabscesses—features that are similar to human psoriasis (15, 17). In humans, there is a higher incidence of psoriatic lesions in night-shift workers with misaligned circadian clocks (18). Consistently, the response to IMQ in mice has diurnal features and is affected by clock gene deletions (10).
IMQ activates TLR7 and TLR8 (in humans) receptors located on endosomes within a variety of different cell types, including macrophages (19), keratinocytes (KCs) (20), mast cells (21), monocytes (22), classic dendritic cells (23), and plasmacytoid dendritic cells (24). Upon IMQ binding, TLR7 dimerizes and recruits the adaptor molecule MyD88, leading to a signaling cascade involving PI3K (13), IRAK1-, IKKα-, and/or LMP1-mediated phosphorylation, and nuclear translocation of IRF7. Once in the nucleus, IRF7 binds to IFN-responsive elements (IREs) within promoters of type I IFN-encoding genes and other IFN-sensitive genes (ISGs), including Irf7. IRF7, then, is the key transcription factor induced by this pathway, acting in a feed-forward manner by facilitating the production of ISGs and type I IFNs through the MyD88 pathway. This feed-forward loop induces robust expression of ISGs which activate the immune system to kill cancer cells, bacteria, and viruses.
The goal of our study was to investigate the reciprocal relationship between the circadian clock and TLR7-induced inflammatory responses in skin. Inflammatory parameters, including spleen weight, epidermal thickness, and epidermal cell proliferation, were diurnal under homeostasis and/or after one dose of topical 1% IMQ, but became arrhythmic and elevated with five consecutive daily doses of 1% IMQ. Mice lacking Bmal1 systemically had greater epidermal thickness at 5 d of IMQ compared to wild type (Wt). Certain circadian clock genes are also perturbed in human patient psoriatic skin samples. We show that IMQ alters core clock gene expression in mouse skin in an acute manner that recovers after 1 d. Likewise, the early IMQ-induced ISG response is diurnal in mouse skin, but this diurnal response is lost after repeated daily IMQ treatments, presumably because of IMQ’s effect on the clock (25). Consistent with this idea, the ISG response is enhanced and occurs without diurnal variation in Bmal1-deleted mice. Through cell sorting and single-cell RNA sequencing (RNA-seq) we determined that IMQ up-regulates Irf7 mRNA more potently in epidermal immune cells (Langerhans cells [LCs], T cells, and monocytes) than in keratinocytes (26). Intriguingly, shifting the phase of the core circadian clock through daytime-restricted feeding alters the rhythm of IMQ-induced ISG induction. Together, these studies indicate that BMAL1 is a negative regulator of ISG gene expression in skin immune cells and that the skin’s antiviral and antitumor response can be modulated by feeding time.
Results
Repeated Skin Applications of IMQ Dampen Diurnal Rhythms of Inflammation.
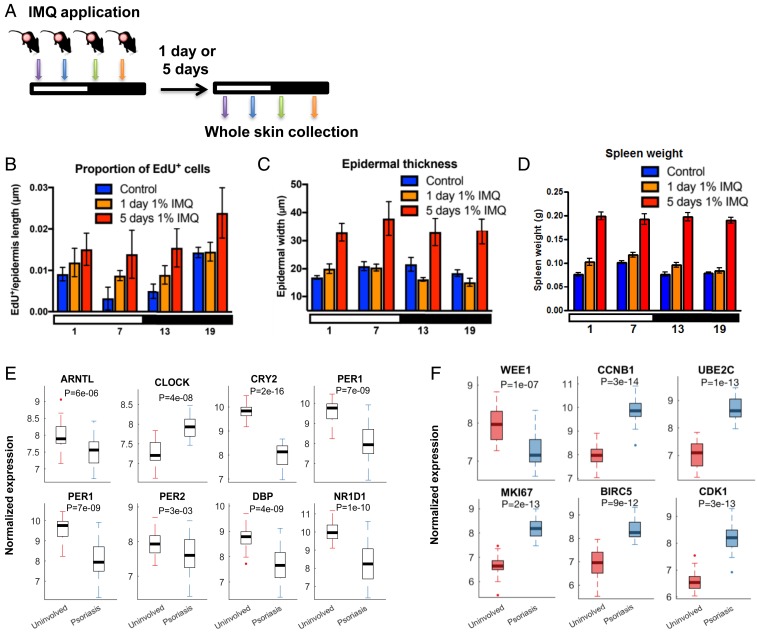
We first tested whether there are time-of-day–dependent inflammatory effects of topically applied IMQ by treating mice with daily 1% IMQ at early morning (ZT01), afternoon (ZT07), early evening (ZT13), or late night (ZT19) for 1 or 5 d and harvesting skin 24 h after IMQ applications (Fig. 1A). The circadian clock-dependent diurnal variation in interfollicular epidermal (IFE) stem cell DNA replication, where most cells are in S phase late at night (4), is lost after 1 and 5 d of IMQ; there is also increased IFE cell proliferation after both treatment durations (Fig. 1B). Interestingly, we observed a trend for diurnal rhythm in epidermal thickness in homeostasis. This diurnal variation was significant after 1 d of IMQ, but lost after 5 d of IMQ (Fig. 1C). Furthermore, spleen weight, a measurement of systemic inflammation, differed significantly across the day/night cycle under homeostasis and after 1 d of 1% IMQ but was constitutively elevated by 5 d of IMQ (Fig. 1D).
Fig. 1.
IMQ effects on inflammation and core clock gene expression. (A) IMQ treatment protocol. Mice were treated topically with 1% IMQ (colored arrows) at 6-h intervals throughout the day/night cycle (indicated by white and black bars) for either 1 d, or each day for 5 d, and collected 1 d after the last treatment. (B) Epidermal cell proliferation (EdU+ cells). Two-way ANOVA test shows significance between treatments (P = 0.02) and time points (P = 0.005). Tukey’s post hoc test for control vs. 5 d 1% IMQ, P = 0.01. One-way ANOVA for control across ZT times was P < 0.003. (C) Epidermal thickness. Two-way ANOVA shows significance between treatments (P < 0.0001), for both control vs. 5 d IMQ, and 1 d IMQ vs. 5 d IMQ (Tukey’s post hoc test P < 0.0001 for both). Epidermal thickness after 1 d IMQ differed across ZT time (one-way ANOVA P = 0.01). (D) Spleen weights. Spleen weight differed across ZT time and treatments (two-way ANOVA P = 0.03 and P < 0.0001). Spleens weighed more after 1 d of 1% IMQ and 5 d of 1% IMQ compared to controls (Tukey’s post hoc test, P = 0.004 and P < 0.0001). Spleen weight changed across ZT time in controls, and after 1 d of 1% IMQ (one-way ANOVA P = 0.0002 and P = 0.003, respectively). (B–D) Control (blue, n = 5), 1 d of 1% IMQ (orange, n = 7), and 5 d of 1% IMQ (red, n = 5 to 7). Data are presented as mean ± SEM. Comparison of gene expression of core clock genes (E) and cell cycle genes (F) between uninvolved and psoriatic biopsies using RNA-seq data from human skin biopsies. Uninvolved and psoriatic biopsies were collected from the same patient (n = 27) during the time between 9 AM and 4 PM (E and F). (Data in E and F from ref. 47.) Statistical significance was determined by Student’s paired t test and significant P values are shown.
We also performed experiments in Bmal1 knockout (KO) mice, and found that, while their spleens weigh less under homeostasis and after 24 h of nighttime IMQ treatment compared to Wt (SI Appendix, Fig. S1A), they did not differ significantly by 5 d of IMQ (SI Appendix, Fig. S1B). On the other hand, epidermal thickness was increased in Bmal1 KO mice compared to Wt after 5 d of IMQ (SI Appendix, Fig. S1C). Although Bmal1 KOs had greater epidermal proliferation under homeostasis as previously observed (4), the proportion of proliferating cells in the epidermis of Bmal1 KO and Wt mice did not differ significantly after 5 d of repeated IMQ treatment at the timepoint measured (ZT13) (SI Appendix, Fig. S1 D and E). We also treated ears with IMQ, measured ear width, and found a trend for increased ear width in Bmal1 KO mice compared to Wt at days 4 and 5 but not at 6 d of IMQ treatment (SI Appendix, Fig. S1F).
These data demonstrate that: 1) systemic and skin-specific inflammation progressively increased from short-term (1 d) to repeated (5 d) treatment with IMQ; 2) increased inflammation associated with repetitive doses of IMQ obliterates diurnal rhythms in IFE stem cell proliferation and thickness and in spleen weight; and 3) in the absence of Bmal1, IMQ-induced epidermal hyperplasia is exacerbated.
To test whether the IMQ-induced obliteration of diurnal rhythms in IFE stem cell proliferation and inflammatory parameters was associated with altered clock gene expression, we measured the expression of clock output genes in mouse back skin after 6 h, 24 h, and/or 5 d of 1% IMQ treatment. Rhythmic expression of core clock genes Bmal1 and Dbp measured by qPCR (SI Appendix, Fig. S1 G and H) and microarray (SI Appendix, Fig. S1I) (25) after 1 and 5 d of IMQ matches control when it is measured 24 h after the last dose of IMQ. However, if we look at a more immediate timepoint (6 h) after a single IMQ treatment, IMQ dampens rhythmic Bmal1 expression (SI Appendix, Fig. S1J) and down-regulates both Rev-erbα and Dbp to similar levels as seen in Bmal1 KO (SI Appendix, Fig. S1 K and L). These qPCR results are confirmed by our microarray studies (25), which also show abnormal up-regulation in Cry1/2 and Per2 (SI Appendix, Fig. S1M).
To explore the mechanism by which the circadian clock modulates IMQ-induced inflammation, we assessed the expression of Il23r, since a previous study (10) showed that IL-23R expression in γ/δ+ T cells from the spleen or lymph node was altered by Clock mutations and correlated with IMQ-induced inflammation. Interestingly, we found no diurnal rhythm of Il23r expression in the skin at the timepoints assessed, and no difference between Bmal1 KO and Wt (SI Appendix, Fig. S1N).
Consistent with IMQ-induced inflammation altering clock gene expression in murine skin (SI Appendix, Fig. S1 J–M), we found that the expression of core clock genes (ARNTL, CRY2, PER1, PER2, DBP, and NR1D1) is altered in human psoriatic lesions compared to uninvolved skin from the same patient collected at the same time (Fig. 1E and SI Appendix, Fig. S1 O and Q). Diurnal cell cycle genes (WEE1, CCNB1, UBE2C, MK167, BIRC5, and CDK1) also showed altered expression in human psoriatic lesions (Fig. 1F and SI Appendix, Fig. S1 P and R). WEE1, a gene which acts to inhibit cell entry into mitosis, was down-regulated in the psoriatic lesions, whereas other cell cycle genes were up-regulated. This is consistent with the increase in proliferation that we found in mice treated for 5 d with 1% IMQ. We did not observe significant differences in gene expression between male and female patients (SI Appendix, Fig. S1 Q and R). Together, these findings suggest that cutaneous inflammation disrupts the skin circadian clock and alters the expression of diurnal cell cycle genes.
IMQ Alters Gene Expression in Skin.
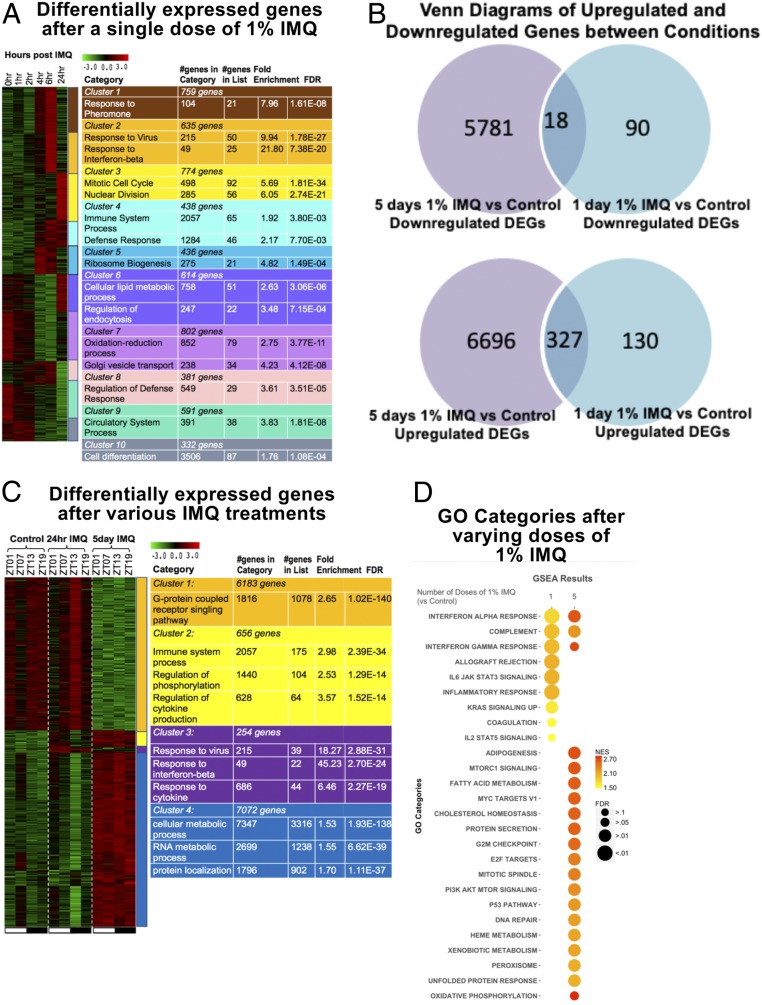
To identify inflammatory pathways induced by IMQ that might be subject to clock regulation, we first assessed IMQ-induced gene expression changes in skin by measuring global gene expression before (0 h) and 1, 2, 4, 6, and 24 h after a single application of IMQ at ZT09. K-means clustering followed by gene ontology (GO) analysis was used to identify gene pathways induced or repressed by IMQ. We found that 5,762 genes were significantly altered across the 24-h time course. These genes could be grouped into 10 temporal clusters, each with enrichment of specific functional categories, including cluster 2 that was markedly up-regulated by 4 h and enriched for IFN-β–associated genes (Fig. 2A).
Fig. 2.
Summary of differentially expressed genes (DEGs) and ISGs following single and multiple treatments of 1% IMQ. (A) Heatmap of expression of differentially expressed genes across time, normalized across genes. (B) Venn diagrams of up-regulated and down-regulated genes between 5 d 1% IMQ vs. control and 1 d 1% IMQ vs. control. Scale of circle to other circles within the same diagram do not correspond with number of genes in category. DEGs were identified using Tukey’s post hoc test with a P < 0.05. (C) Heatmap of expression of DEGs between control, 1 d 1% IMQ, and 5 d 1% IMQ timepoints normalized across genes. (A and C) K-means clustering was performed, and clusters were color coded. Color-coded tables on Right list relevant gene ontologies for cluster associated with color. Tables also list the number of genes in each cluster and the number of genes in GO category, number of genes in GO category in the cluster, fold enrichment of GO category in the cluster, and false discovery rate (FDR) value. DEGs were identified using one-way ANOVA and Tukey’s post hoc test. For the heatmap, green, low expression and red, high expression. (D) Two-dimensional (2D) heatmap of GO categories determined using GSEA for 1 or 5 doses of 1% IMQ. Size of circle corresponds to significance of GO category for the treatment and condition. Color of the circle corresponds to enrichment of category.
We also compared genes that were up-regulated and down-regulated after 1 and 5 d of IMQ (Fig. 2B and SI Appendix, Fig. S2). To do this, we treated mice with IMQ at ZT01, ZT07, ZT13, or ZT19 and collected back skin 24 h after single doses, or 24 h after the last dose of 5 consecutive days of treatment (as described in Fig. 1A). Of the 7,153 genes that were up-regulated after 1 or 5 d of IMQ, 327 were up-regulated in both treatments, leaving 130 uniquely up-regulated after 1 d of IMQ and 6,696 uniquely up-regulated after 5 d of IMQ (Fig. 2B). In addition, 6,889 genes were down-regulated after 1 or 5 d of IMQ. Of the 6,889 genes, 90 were uniquely down-regulated after 1 d of IMQ, 5,781 were uniquely down-regulated after 5 d of IMQ, and only 18 were down-regulated after both 1 and 5 d of treatment (Fig. 2B). The number of genes up-regulated or down-regulated was also time-of-day specific (SI Appendix, Fig. S2).
After 1 and 5 d of IMQ, 566 and 12,871 genes, respectively, were significantly altered compared to control (Fig. 2C). To explore what the physiological significance of these gene expression changes may be, we performed gene set enrichment analysis (GSEA) to identify gene ontologies that were enriched based on our differential gene expression analysis (Fig. 2D). After a single treatment with IMQ, enriched hallmark pathways associated with the immune system included allograft rejection, IL-6 Jak/Stat3 signaling, inflammatory response (including IFN-α/IFN-γ response), KRAS signaling down-regulated, and IL-2/Stat5 signaling (Fig. 2 C and D). These data also point to IFN signaling being prominent after a single dose of IMQ. Pathways enriched after five daily applications of IMQ included protein secretion, G2M checkpoint, and metabolic-associated pathways (Fig. 2 C and D). These findings are supported by the earlier results that IFE cell proliferation is markedly increased after 5 d of treatment.
Overall, these data point to the IFN pathway as a mediator of the early response to a single dose of IMQ (14). In addition, while some diurnal gene expression changes persist over 5 d of IMQ treatment, a much larger set of genes becomes dysregulated with repeated treatment over multiple days.
Diurnal Gene Expression Responses to IMQ Skin Treatment.
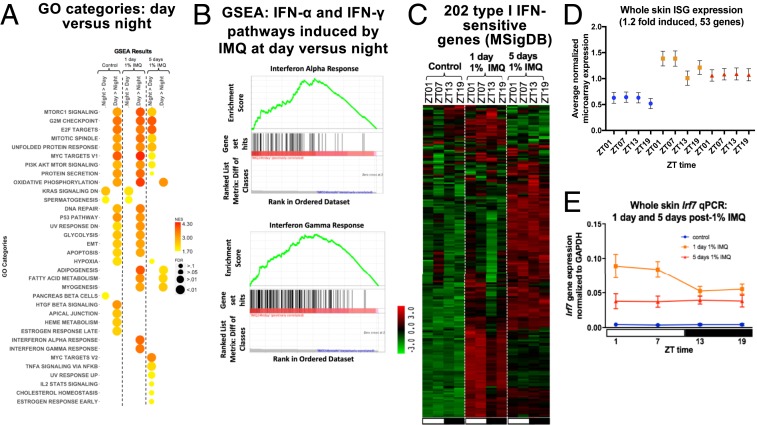
To identify genes and gene ontology categories that respond to IMQ in a time-of-day–dependent manner, we averaged the gene expression for daytime timepoints (ZT01 and ZT07) and nighttime timepoints (ZT13 and ZT19). We then used a fold cutoff of 1.2 or greater to define genes differentially expressed between day and night. Out of the 566 genes significantly altered after 1 d of IMQ, 17 genes showed greater up-regulation when IMQ was applied during the day compared to the night and 78 genes showed greater up-regulation when IMQ was applied during the night compared to the day. The genes with greater induction after daytime IMQ were enriched for IFN-α response, IFN-γ response, adipogenesis, fatty acid metabolism, and myogenesis (Fig. 3A). The genes with greater up-regulation when IMQ was applied during the night did not form specific functional categories. After five daily IMQ treatments at night, enriched pathways were associated with cell cycle regulation, E2F and Myc targets, TNFα signaling via NF-κB, and IL-2/STAT5 signaling. However, there were few enriched pathways after 5 d of daytime IMQ treatment, and these pathways were associated mainly with metabolic functions (Fig. 3A). These findings suggest distinct time-of-day–dependent gene expression changes after single and repeated IMQ doses, with greater IFN responses after one dose of IMQ during the day, and greater cell cycle pathway enrichment after nighttime treatment for 5 d with IMQ.
Fig. 3.
Analysis of genes and ISGs following day or night 1% IMQ treatment. (A) Two-dimensional (2D) heatmap of GO categories determined using GSEA in two different treatment groups (night vs. day and day vs. night) across three conditions (control, 1 d 1% IMQ, and 5 d 1% IMQ). Size of circle corresponds to significance of GO category for the treatment and condition. Color of the circle corresponds to enrichment of category. (B) Enrichment plot for IFN-α and IFN-γ hallmark pathways shows a significant enrichment of these pathways after 1 d of 1% IMQ during the day vs. night. (C) Heatmap of 202 ISGs (from MSigDB) from the samples collected in Fig. 2C. Green, low expression and red, high expression. (D) Average normalized whole skin microarray expression (25) of ISGs induced by 1.2-fold or more after IMQ, under homeostasis (blue), 1 d 1% IMQ (orange), and 5 d 1% IMQ (red). One-way ANOVA, control P = 0.001, 1 d 1% IMQ P < 0.0001, 5 d 1% IMQ P = 0.56. (E) qPCR for Irf7 on individual samples collected in Fig. 1A. Student’s unpaired t test comparing daytime vs. nighttime under control [not significant (N.S.)], 1 d (P = 0.02), and 5 d (N.S.). Data represents mean ± SEM of n = 5 to 7.
As many of IMQ’s immunogenic effects are mediated through induction of IFN pathways (27), we further investigated genes associated with the IFN response. By GSEA, the IFN-α and IFN-γ response genes were markedly enriched among genes that were expressed higher after a single dose of IMQ during the day than night (Fig. 3B). Using the Molecular Signatures database (MSigDB), we identified 202 ISGs, some of which were up-regulated after 1 d of IMQ, and others were up-regulated by 5 d of IMQ (Fig. 3C). We further analyzed the 202 ISGs by identifying which were up-regulated, on average, by 1.2-fold or greater after 1 d of IMQ (53 genes); there is a clear diurnal difference with greater expression of these up-regulated genes during the day compared to the night after 1 d of IMQ (Fig. 3D). The expression pattern of Irf7, a master regulator of ISG transcription, when measured by qPCR, had greater induction after IMQ treatment during the day than night after 1 d (Fig. 3E), consistent with the results of the microarray study (25).
To more specifically define the biological processes that the 53 up-regulated ISGs could be regulating, we performed gene ontology analysis using the online database DAVID (28). The majority of these genes were associated with antiviral activities; examples include genes encoding antiviral proteins that inhibit replication of RNA viruses, such as Isg15, Ifit1, Mx1, and Oas1a, as well as negative feedback factors that dampen proinflammatory responses, such as Irgm1, Usp18, and Trim30.
In sum, IMQ treatment alters skin gene expression which differs depending on both the treatment time during the circadian cycle and length of treatment (1 d or 5 d). There is a time-of-day–dependent induction of ISG-associated pathways (IFN-α and IFN-γ response) 1 d after a single dose of IMQ, whereas few pathways, mainly associated with cell cycle or metabolism, exhibit diurnal regulation after 5 d of daily IMQ treatment (Fig. 3A).
IMQ Up-Regulates ISG Expression in Epidermal Cell Subsets.
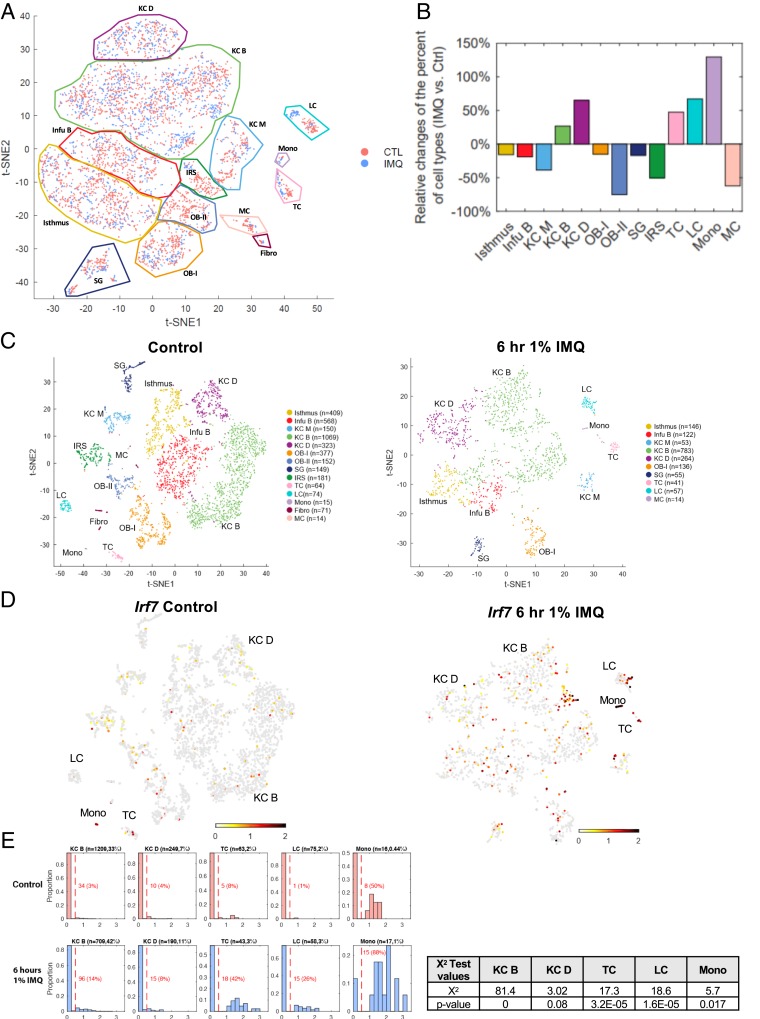
To define which epidermal cell types induce ISG expression in response to IMQ, we performed single-cell RNA-seq on epidermis isolated from adult mice under homeostasis and 6 h after a single dose of IMQ (26). We used Seurat (29) to cluster cells based on their gene expression profiles and t-distributed stochastic neighbor embedding (t-SNE) to display the cell clusters, which were annotated manually based on markers from previous studies (30). To assess any compositional differences between the control and treated samples, we pooled the control and IMQ-treated samples onto one t-SNE plot (Fig. 4A). By analyzing each of the 13 clusters (we also had a small cluster of contaminating fibroblasts), we determined the relative change in the percent of cell types in IMQ-treated samples over the control (Fig. 4B). We noticed a small cluster of immune cells in normal epidermis, which we classified as monocytes (labeled “mono”) by searching the top 50 genes that were differentially expressed in this cluster compared to other epidermal populations in the Immunological Genome Project (Immgen) database (31) (SI Appendix, Fig. S3A). Genes enriched in the monocyte population include myeloid-associated genes (Ly6e, CD14, and Lyz2), M2 polarization markers (CD16 and Adgre1 [F4/80]), phagolysosomes/oxidative burst (Coro1a, Ctss, Cyba, Cybb, and fcgr3), and chemokines (Ccl6, Ccl4, Ccl3, and Ccl9). Interestingly, the percent of monocytes increased by greater than 100% following treatment with IMQ compared to control, indicating that IMQ plays a role in the recruitment of monocytes. On the other hand, some cell populations were absent in the IMQ sample: fibroblasts, melanocytes, outer bulge group 2 cells, and inner root sheath cells.
Fig. 4.
IMQ up-regulates ISG expression in keratinocytes, Langerhans cells, T cells, and monocytes within the epidermis. scRNA-seq was performed on epidermal cells isolated from control and 6 h of 1% IMQ-treated mice (26). (A) Cell from both control (pink) and 6 h 1% IMQ (blue) samples were jointly projected on the same t-SNE plot to better compare different cell subtypes. (B) Histogram representing the relative changes of each cell type in control and IMQ samples. Positive value bars represent percent of cells that were more present in IMQ-treated samples over control and negative value bars the opposite. (C) t-SNE plots of cells isolated from adult male mice, untreated (Left) or treated with 1% IMQ for 6 h (Right). Differentiated keratinocytes (KC D), basal keratinocytes (KC B), basal infundibulum (Infu B), mitotic keratinocytes (KC M), Langerhans cells (LCs), sebaceous gland (SG), melanocytes (MCs), fibroblasts (fibro), monocytes (mono), T cells (TC), Isthmus cells, outer bulge cells group 1 (OB-I), outer bulge cells group 2 (OB-II), and inner root sheath cells (IRS). N refers to the number of cells per population. (D) Irf7 mRNA expression in single-cell data (26). Gray, no expression; red, intensity of expression. (E) Proportion of differentiated KCs (KC D; Krt1+), basal KCs (KC B; Col17a1+), T cells (TC; Cd3g+), LCs (Cd207+), and monocytes (mono; Cybb+) expressing Irf7 in control and IMQ-treated samples. Normalized Irf7 expression is on the x axis and the proportion of cells expressing Irf7 mRNA out of each population is on the y axis. Number of cells analyzed and percent of each population out of total cells analyzed is listed above each graph. Number and percent of cells expressing Irf7 out of each population is listed in red. χ2 test results (Right) of the portion of positive Irf7 between control and treated samples of selected subpopulations and the corresponding P values.
We also studied the control and IMQ-treated samples separately (Fig. 4C), identifying 14 clusters for control and 10 clusters for IMQ treated. Under homeostasis, we detected few Irf7 positive cells within the epidermis (Fig. 4D), with highest expression found in the monocyte population. After IMQ, we identified four main cell populations that increased expression of Irf7: a small group of KCs, LCs, T cells, and monocytes (Fig. 4D). Irf7 expression was consistent with the expression of the 53 ISGs identified in Fig. 3D (SI Appendix, Fig. S3A).
Out of all cell types, the small group of monocytes expressed Irf7 and ISGs most robustly in response to IMQ (Fig. 4E and SI Appendix, Fig. S3B). Fifty percent of monocytes expressed Irf7 under homeostasis, while 88% expressed Irf7 after IMQ (Fig. 4E). Out of the differentiated KC (Krt1+) population, 4% of cells express Irf7 under homeostasis, and this increases to 8% after IMQ. The same trend was observed in the basal KC population (Col17a1+) (Fig. 4E). In T cells (Cd3g+) and LCs (Cd207+), 8% and 1% of cells expressed Irf7 under homeostasis, and this increased to 42% and 26%, respectively, after IMQ (Fig. 4E). Interestingly, only the monocytes from the control sample expressed Tlr7 (Tlr7 was not detected in the IMQ sample) (SI Appendix, Fig. S3C).
We found that, at the single-cell level, core clock genes Rev-erbα (Nr1d1) and Dbp were suppressed by 6 h of IMQ (SI Appendix, Fig. S3D) in the major cell populations of keratinocytes (Isthmus, KC B, and KC D) (26). In addition, we fluorescence-activated cell sorting (FACS) sorted epidermal cell subsets (T cells, DCs, and CD49f+ KCs) after 6 h of IMQ during the day (ZT0) or night (ZT12) and performed qPCR for Dbp and found similar down-regulation (SI Appendix, Fig. S3E). These results support the data presented in SI Appendix, Fig. S1 D–G showing 6 h of IMQ down-regulates the expression of certain core clock genes.
Taken together, these results show that multiple cell types within the epidermis respond to IMQ by up-regulating Irf7 and ISG expression and down-regulating Rev-erbα (Nr1d1) and Dbp expression. In terms of ISG expression, immune cells, mainly monocytes and T cells, are the most potent responders, followed by LCs and a small population of KCs.
IMQ Induces Total and Phosphorylated-IRF7 Protein Nuclear Translocation in Epidermal Cell Subsets.
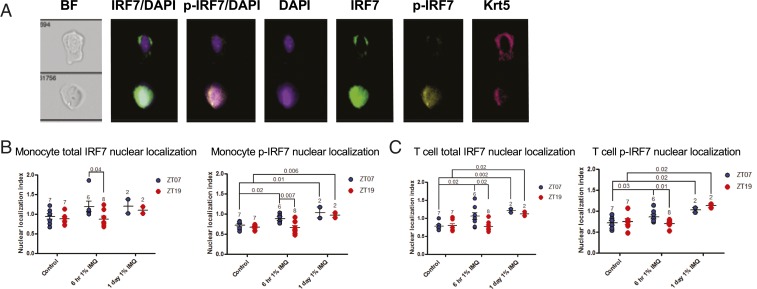
To activate ISG expression, IRF7 must be phosphorylated and translocated into the nucleus (13). To test whether IRF7 nuclear translocation differed depending on the time of day of IMQ application, we treated mice topically with IMQ during the day (ZT07) or night (ZT19) and collected epidermal cells 6 h and 1 d later and analyzed IRF7 and phosphorylated IRF7 (p-IRF7) nuclear localization with the ImageStream Flow Cytometer (Fig. 5A) (32). We measured nuclear similarity index (a measurement of the relative nuclear localization of a given marker) of total IRF7 and p-IRF7, finding that monocytes (CD11c−CD3e−CD45+) had greater nuclear localization of total and p-IRF7 after 6 h of IMQ during the day than night. At 1 d, p-IRF7 nuclear localization was significantly increased and similar at both treatment times (Fig. 5B). Similar results were seen for T cells (Fig. 5C). Nuclear localization of total IRF7 and p-IRF7 was significantly increased in KCs 1 d after IMQ compared to control, with greater nuclear localization of total IRF7 seen after nighttime IMQ treatment compared to daytime treatment (SI Appendix, Fig. S4A). In DCs, no significant increase in nuclear localization of total or p-IRF7 6 h after IMQ was observed, but by 1 d, nuclear localization of total and p-IRF7 was significantly increased with no time-of-day dependence (SI Appendix, Fig. S4B).
Fig. 5.
IMQ-induced nuclear localization of total and phosphorylated IRF7 is diurnal in epidermal T cells and monocytes. Male Wt mice were treated with 1% IMQ for 6 h or 1 d during the day (ZT07) or night (ZT19) and back epidermis was dissociated, stained for cell surface markers and intracellular IRF7 and p-IRF7, and analyzed on the ImageStream Flow Cytometer. Nuclear localization index was calculated using IDEAS software. (A) Example images (60x) taken during ImageStream fluorescence imaging, showing a cell with no p-IRF7 nuclear localization (Top) and one with p-IRF7 nuclear localization (Bottom). (B) Average nuclear translocation index for total IRF7 (Left) and p-IRF7 (Right) in monocytes (CD45+CD11c−CD3e−). (C) Average nuclear translocation index for total IRF7 (Left) and p-IRF7 (Right) in T cells (CD45+CD3e+CD11c−). (A–C) Each data point represents one mouse, and mean ± SEM is indicated. Statistical significance was determined by Student’s paired t test and significant or near-significant P values are shown.
In summary, the activity of IRF7, indicated by its phosphorylation and nuclear localization, is induced after IMQ in epidermal KCs, T cells, dendritic cells, and monocytes, but is time-of-day dependent only in T cells and monocytes.
Systemic Deletion of Bmal1 Increases IMQ-Induced ISG Expression in the Skin.
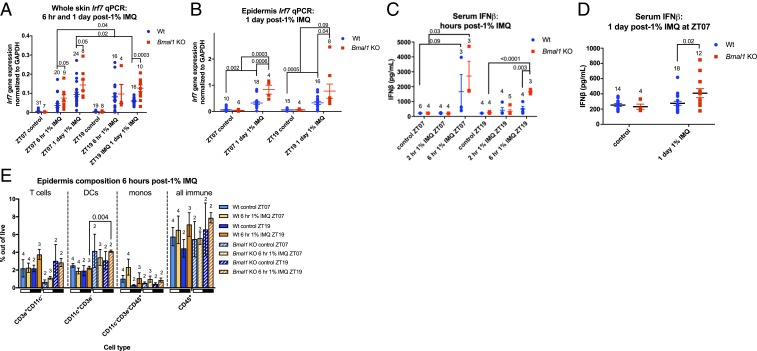
We tested how IMQ treatment length affected the diurnal ISG response and whether the circadian clock plays a direct role in the rhythmic IMQ-induced ISG response. We treated Wt and Bmal1 KO mice topically with IMQ and collected dorsal skin 6 h or 1 d after IMQ treatment during the day (ZT07) or night (ZT19). Wt mice had greater induction of Irf7 after 6 h of IMQ during the night than the day (Fig. 6A), which contrasts with that observed with 1 d after treatment in which Irf7 is more highly induced after daytime than nighttime IMQ treatment (Figs. 3E and 6A). These findings suggest that the diurnal ISG response elicited by IMQ is dependent on treatment duration; the shorter treatment duration (6 h) leads to greater response at night, whereas the longer treatment duration (1 d), the time at which Irf7 expression peaks after IMQ, leads to greater response during the day (Fig. 6A).
Fig. 6.
Systemic Bmal1 deletion results in exacerbated IMQ-induced serum IFN-β and ISG expression in the skin. (A) Irf7 qPCR on whole back skin from Wt (blue, n = 17 to 30) and Bmal1 KO mice (red, n = 6 to 9) after 6 h or 1 d of 1% IMQ during the day (ZT07) or night (ZT19). (B) Irf7 qPCR on isolated epidermis from Wt (blue, n = 10 to 18) and Bmal1 KO mice (red, n = 4 to 8) after 1% IMQ during the day (ZT07) or night (ZT19) for 1 d. (C) Wt serum IFN-β after 2 or 6 h of 1% IMQ during the day (ZT07) or night (ZT19). (D) Wt (blue) vs. Bmal1 KO (red) serum IFN-β after 1 d of 1% IMQ at ZT07. (E) Flow cytometry quantification of immune cell populations within the epidermis in Wt and Bmal1 KO mice treated with 1% IMQ for 6 h during the day or night (indicated by white and black bars). (A–E) Each data point represents one mouse, and mean ± SEM is indicated. The numbers above each group indicate the number of samples analyzed. Statistical significance was determined by Student’s paired t test and significant or near-significant P values are shown.
We then tested the IMQ effect in mice deleted for Bmal1. Irf7 gene expression was increased in the skin of Bmal1 KO mice compared to Wt after 6 h of IMQ treatment during the day, whereas there was no difference between the genotypes after 6 h of IMQ at night; the 1 d of IMQ treatment of Bmal1 KO mice induced significantly greater Irf7 expression compared to Wt, irrespective of whether IMQ was applied during the day or night (Fig. 6A). These findings are consistent with the circadian clock controlling the diurnal response to IMQ. Also, in contrast to epidermis isolated from Wt mice, Irf7 mRNA was induced by approximately fourfold 1 d after IMQ, with no diurnal difference (Fig. 6B). Interestingly, Irf7 mRNA in the epidermis of Bmal1 KO mice was exacerbated almost fourfold after IMQ compared to Wt (Fig. 6B).
To determine whether systemic type I IFN activity was affected by Bmal1 deletion, we measured serum IFN-β levels in Wt and Bmal1 KO mice treated topically with IMQ for 2 or 6 h during the day or night. There was no significant IFN-β induction after 2 h of IMQ in either genotype, whereas IFN-β levels were significantly more up-regulated after 6 h of IMQ in the Bmal1 KO mice compared to control, both when IMQ was applied during the day or the night (Fig. 6C). After 1 d of IMQ, IFN-β levels were similar to control in the Wt, while Bmal1 KO mice maintained elevated IFN-β levels (Fig. 6D).
To decipher whether the diurnal and BMAL1-affected ISG response to IMQ was a result of differential recruitment of immune cells (which express ISGs more robustly compared to KCs) (Fig. 4), we measured the immune cell composition in the epidermis (Fig. 6E). We observed no major significant difference in composition of immune cells, including T cells, DCs, monocytes, and after 6 h of IMQ during the day vs. night in Wt and Bmal1 KO mice (Fig. 6E).
These results show that the antiviral (ISG) response to IMQ not only depends on the duration of treatment and the time of day, but is also greatly impacted by deletion of Bmal1. Furthermore, this effect is mediated at the transcript level, as the cellular composition of immune cells within the skin is unaffected by time of day or Bmal1 deletion. Based on these data, we hypothesize that Bmal1 plays a regulatory role in suppressing type I IFN levels and ISG expression.
Feeding Schedules Alter the Skin’s Type I IFN Response after IMQ.
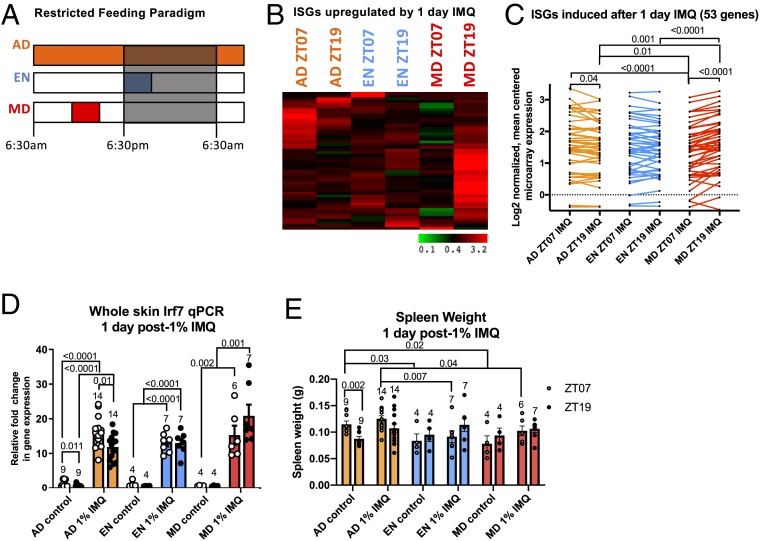
Previously we found that the rhythmic skin expression of many immune-associated genes was shifted after daytime-restricted feeding (3). Some of these genes encode for proteins that mediate the IMQ-induced ISG response, including Tlr7, Ifnar2, and Irf7. Therefore, we sought to determine if shifting the skin clock with time-restricted feeding could affect the rhythm of the IMQ-induced ISG response. We subjected mice to time-restricted feeding with EN (early night; ZT12–ZT16) and MD (midday; ZT05–ZT09) feeding schedules as well as an AD (ad libitum) feeding schedule as a control (Fig. 7A). After 21 d, these mice were treated topically with IMQ during the day (ZT07) or night (ZT19), and skin was harvested for RNA analysis 1 d later (Fig. 7A). Consistent with our previous findings (Fig. 3D), AD mice exhibited a diurnal rhythm of ISG induction after IMQ, as shown by the expression of the 53 induced ISGs identified in Fig. 3D (Fig. 7 B and C). Interestingly, in MD mice this rhythm was reversed with greater ISG induction after IMQ treatment during the night than during the day (Fig. 7 B and C). qPCR for Irf7 expression in individual mice (Fig. 7D) confirmed the results from the microarray study (Fig. 7C) (25). To assess whether restricted feeding impacts the systemic inflammatory response to IMQ, we measured spleen weight in these mice and found that in AD mice, spleen weight is higher during the day than at night under homeostasis, and that a similar trend is observed after 1 d of 1% IMQ. The EN and MD feeding groups generally exhibited decreased spleen weight compared to AD and no diurnal differences (Fig. 7E). These results demonstrate that time-restricted feeding, which shifts the phase of the skin circadian clock (3), shifts the rhythmic expression of ISGs after IMQ, suggesting that meal timing may directly affect our susceptibility to insults protected against by the ISG response.
Fig. 7.
Shifting the clock by daytime-restricted feeding (RF) alters the diurnal rhythm of IMQ-induced ISG expression. (A) Restricted feeding followed by IMQ treatment protocol. Mice were subjected to RF feeding schedules and then shaved and treated with 1% IMQ during the day (ZT07) or night (ZT19) for 1 d and then whole skin was collected for qPCR and pooled for microarray analysis (25). AD, ad libitum fed; EN, early night fed; MD, midday fed. (B) Heatmap of 202 ISGs (from MSigDB) from the samples collected in A. Green, lowly expressed genes; red, highly expressed genes. (C) Whole skin microarray expression of ISGs identified by MSig database (25), up-regulated by 1.2-fold or more after 1 d of treatment, is plotted. Data points indicate individual ISG expression values after IMQ, with average ISG expression per group indicated by colored lines (AD, orange; EN, blue; MD, red). Statistical significance was measured by one-way ANOVA and significant P values are shown. (D) Whole skin RNA samples from A were subject to qPCR for Irf7 (n = 5 to 14). Data are presented as mean ± SEM. (E) Spleen weights of mice 1 d post-1% IMQ treatment. (D and E) Data presented with points representing individual mice at ZT07 (white) and ZT19 (black), and colored bars representing the mean with error bars indicating ± SEM. Statistical significance was determined by Student’s paired t test, and significant P values are shown.
Discussion
Our study sheds light on the underappreciated role of the circadian clock in regulating IFN responses in skin, a response important for defenses against skin cancer and viral infections and a pathogenetic mechanism in chronic autoimmune diseases such as psoriasis. The study also suggests that the response to IMQ—a drug used to treat actinic keratosis, superficial basal cell carcinoma, and other dermatological diseases—may be time-of-day dependent. Furthermore, we demonstrate that the IFN response to IMQ is modulated by time of feeding, suggesting that feeding has a previously unrecognized influence over the skin immune response.
An important barrier against infections and injury, the epidermis constantly renews through the proliferation of IFE stem cells (1). In these stem cells, the circadian clock controls diurnal rhythms in DNA replication and repair (4, 33–35); deletion of Bmal1 leads to higher and constant IFE cell proliferation (4). We propose that these daily cell proliferation rhythms allow coordination with diurnal cycles in oxidative phosphorylation and glycolysis to minimize ROS-induced genotoxicity (1, 4, 5). Here we find that IMQ-induced inflammation overrides the circadian control of the cell cycle, causing increased and constant IFE stem cell proliferation (Fig. 1B), similar to that found in the absence of Bmal1 (SI Appendix, Fig. S1D). This is especially pronounced after repeated IMQ doses. We speculate that it is advantageous to suspend the clock control of the cell cycle in stem cells to maximize the proliferative response to inflammation-induced tissue injury. Consistent with this idea, we found that topical IMQ application acutely down-regulates clock output genes Bmal1, Dbp, and Rev-erbα 6 h after treatment and up-regulates negative BMAL1:CLOCK regulators, Cry1/2 and Per2 (SI Appendix, Fig. S1 J–M). While core clock gene expression was restored 24 h after a single treatment in mice, we observed that human psoriatic skin biopsies had constitutively dampened core clock gene expression (Fig. 1 and SI Appendix, Fig. S1). We also found that 5-d treatment with IMQ markedly increased and obliterated diurnal rhythms in spleen weight (Fig. 1D), suggesting that IMQ-induced inflammation may suspend diurnal rhythms in the systemic immune response.
A previous study found greater skin thickening when IMQ was applied at night than during the day, suggesting diurnal rhythms in inflammatory activity, even after repeated daily treatments with IMQ (10). In contrast, we demonstrate that repeated IMQ administration disrupts circadian control of the epidermal proliferation and thickness (Fig. 1). Differences between the two studies may account for these results: they used 2.5% IMQ cream, we used 1% IMQ; they analyzed ear skin, we analyzed back skin; they used ICR albino female mice, we used male C57/B6J mice. The two mouse strains have been shown to have different inflammatory responses to repetitive IMQ treatment (36); ICR mice have greater IMQ-induced splenomegaly compared to C57/B6J, but C57/B6J have greater epidermal thickness and weight loss in response to IMQ than ICR mice (36).
In the skin, topical IMQ treatment induces the production of ISGs which facilitate proinflammatory, antiviral immunity and help steer the host’s innate and adaptive immune system to defend against pathogens and cancers. Our results show that the acute ISG response in the skin elicited by IMQ is time-of-day dependent, while chronic treatment (5 d) results in ablation of rhythmic ISG expression (Fig. 3). We observe greater ISG induction after 6 h of IMQ treatment during the night compared to the day, while by 1 d after treatment, ISGs are higher during the day compared to the night. This temporally dynamic rhythm in ISG expression is supported by a previous study in which systemic injection of IFN produced greater IFN-induced myelosuppressive activity during the night at early (12 h) timepoints, whereas by 24 to 48 h myelosuppressive activity was greater during the day (37). Moreover, IFN administration during the day yielded higher antitumor activity than IFN administered at night, highlighting the functional significance of circadian IFN responsiveness (38).
This study identifies a population of IRF7-expressing monocytes within the mouse epidermis under homeostasis. In response to IMQ, this population turns on ISG expression most robustly out of all cell types (Fig. 4); it is tempting to posit that these cells are key initiators of IFN-mediated defense responses in the skin. Consistent with that notion, under homeostasis this is the only cell population that expresses Tlr7 to detectable levels in the scRNA-seq experiments (SI Appendix, Fig. S3C). In epidermal T cells and monocytes, p-IRF7 exhibits greater nuclear localization after 6 h of IMQ during the day than night (Fig. 5), supporting the idea that IRF7 regulates the activation of ISG expression after phosphorylation and translocation into the nucleus in a time-of-day–dependent manner.
We found that systemic Bmal1 KO mice have an exacerbated IMQ-induced ISG response in skin and isolated epidermis. Considering that the Bmal1 KO phenotype is observed in isolated epidermis (Fig. 6), and mainly epidermal immune cells (few KCs) up-regulate Irf7/ISG expression after IMQ (Fig. 4), we propose that BMAL1 acts as a negative transcriptional regulator of ISGs in a cell-type–specific manner, mainly in skin-residing or infiltrating leukocytes. This hypothesis does not exclude the possibility that BMAL1 also regulates the ISG response in other cells; considering that we observe elevated serum IFN-β levels after IMQ in Bmal1 KO mice, it is likely that BMAL1 plays this regulatory role in other immune populations throughout the body.
BMAL1 has been described as a positive regulator of antiviral responses. Intranasal influenza A, herpesvirus, and Sendai virus infections in mice exhibit greater virulence during the day than night, with deletion of core clock genes causing increased viral titers and mortality (11, 12). Furthermore, BMAL1 inhibits replication of respiratory syncytial virus and parainfluenza virus type 3 viruses in a cell-intrinsic manner (39). Here, we demonstrate that Bmal1 deletion increases the expression of ISGs in response to a TLR7 agonist. The increased susceptibility to viral infections in Bmal1 KO mice despite a more robust ISG response (also shown by others) (12) can be reconciled by multiple explanations. First, excessive IFN responses worsens pathologies by reducing the effectiveness of the adaptive immune system, such as in chronic IFN signaling due to a lymphocytic choriomeningitis virus infection (40), indicating that the antiviral power of the IFN-activated pathways requires a balance between activating signals and inhibitory mechanisms. Additionally, recombinant IFN-β treatment in patients with relapsing-remitting multiple sclerosis suppress T cell activity, leading to better prognoses (41). Second, Bmal1 deletion results in up-regulation of the core clock gene CLOCK (4, 42), which associates with the viral transcriptional complex of the herpesvirus to mediate viral gene expression (43, 44). Third, cells lacking circadian clocks have elevated expression of certain protein biosynthesis enzymes, some of which are controlled by the circadian clock, and may facilitate greater production of viral gene products in cells whose transcriptional machinery has been hijacked by viral effectors (11).
Timing of food intake alters the circadian clock in peripheral organs (3, 45). We previously showed that shifting the skin clock via time-restricted feeding affects the amount of UVB-induced DNA damage (3). In relation to our study, time-restricted feeding affects adrenal gland glucocorticoid secretion, which in turn regulates IFN-α/β receptor 1 expression in the liver (46). We expand on these results and demonstrate that the rhythmic expression of many ISGs was shifted after IMQ treatment, suggesting that susceptibility to viral infections and tumorigenesis may be affected by meal timing (Fig. 7).
In conclusion, we show: 1) Core clock gene expression is altered by cutaneous inflammation; 2) a requirement for core clock gene Bmal1 for diurnal rhythms in the skin’s ISG response, suggesting the circadian clock modulates the ISG response in skin; 3) that a population of epidermal leukocytes, rather than the bulk population of keratinocytes, mediates the most robust IFN response to IMQ; and 4) that shifting the clock by daytime-restricted feeding reverses the phase in ISG pathway activation in the skin, suggesting that meal timing may be a previously unknown modulator of skin immune responses.
Materials and Methods
Detailed descriptions of all materials and methods are available in SI Appendix.
Animals.
Mice were maintained according to NIH guidelines and approved by the Institutional Animal Care and Use Committee of the University of California, Irvine (protocol number 2001-2239).
IMQ Treatment.
All mice (including controls) were anesthetized by i.p. injection with ketamine (100 mg/kg) and xylazine (10 mg/kg) and shaved 2 d prior to experiment initiation. To make 1% IMQ cream, 5% pharmaceutical IMQ (Perrigo brand) was diluted in a ratio of 1:4 with CVS moisturizing cream. The 1% of the stock 5% cream was weighed out in 0.0625 g aliquots to apply to the back and 0.031 g aliquots to apply to the ears. For some experiments, mice were injected with EdU at 5 mg/kg of body weight 2 h prior to collection. For specimen collection, mice were killed by CO2 followed by cervical dislocation. Whole back skins were collected and preserved in RNAlater (Thermo Fisher Scientific).
Microarray Analysis of Whole Skin RNA.
Whole skin RNA from four to seven mice with RNA integrity numbers (RINs) above 7 were pooled. A total of 200 ng of total RNA was hybridized to Affymetrix Mouse Gene 1.0 ST arrays. All reads were Plier normalized, and duplicate reads and genes with expression level below 50 were removed. Expression was Log2 transformed and mean centered across all arrays being compared.
Analysis of RNA-Seq Data of Human Skin Biopsies.
Uninvolved and psoriatic biopsies were collected from the same patient at the same time (between 9 AM and 4 PM). The Log2-transformed data were used for downstream analysis. Human data were acquired from a published dataset (47). All subjects involved in this study provided written informed consent under a protocol adherent to the Helsinki Guidelines and approved by the Institutional Review Board of the University of Michigan Medical School.
Single-Cell RNA-Seq Analysis: Data Processing and Dimension Reduction and Clustering Analysis.
For downstream analyses, low-quality cells were removed based on the outliers of a distribution of several quality control metrics. Cells with a number of expressed genes greater than 5,000 (control samples) or greater than 4,500 (IMQ samples), or the proportion of counts in mitochondrial genes greater than 10%, were removed. In sum, 132 and 78 cells were removed in control and IMQ samples, respectively, leading to 3,616 and 1,673 cells for downstream analyses. Clustering of cells was performed using the Seurat R package (29). The control and IMQ samples were analyzed separately as well as in a pooled fashion in order to perform integrative analysis using canonical correlation analysis (CCA) (48), as described in SI Appendix, Materials and Methods.
Preparation of Epidermal Cells for ImageStream Analysis.
Cells were stained as described in SI Appendix, Materials and Methods. Immediately prior to acquisition on the ImageStream Flow Cytometer (Amnis Corporation), cells were incubated with 4′,6-diamidino-2-phenylindole (DAPI) to label nuclei. IDEAS analysis software was used to calculate the nuclear translocation index of total IRF7 and p-IRF7.
Statistical Methods.
Statistics are described in the figure legends.
Data Availability.
The scRNA-seq, mouse skin microarray, and human psoriasis data discussed herein were deposited in GEO (accession nos. GSE142345, GSE142165, and GSE63980 respectively).
Supplementary Material
Acknowledgments
This study was supported by the Irving Weinstein Foundation and NIH Grants AR56439 and AR075047 (to B.A.); NSF Graduate Research Fellowship DGE-1321846 (to E.N.G.); NIH Grants U01AR073159 and R01GM123731 (to Q.N.); NSF Grants DMS1562176 and DMS1763272 (to Q.N.); a Simons Foundation Grant (594598, Q.N.); NIH Grants AR069071, AI130025, and AR075043 (J.E.G.); and the A. Alfred Taubman Medical Research Institute (J.E.G.). L.C.T. is supported by the Dermatology Foundation, the National Psoriasis Foundation, and the Arthritis National Research Foundation. J.S.T. is an Investigator in the Howard Hughes Medical Institute.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: The mouse epidermis scRNAseq, mouse skin microarray, and human skin psoriasis data are deposited in Gene Expression Omnibus (GEO) (accession nos. GSE142165, GSE142345, GSE63980, respectively).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1915773117/-/DCSupplemental.
References
- 1.Plikus M. V., et al. , The circadian clock in skin: Implications for adult stem cells, tissue regeneration, cancer, aging, and immunity. J. Biol. Rhythms 30, 163–182 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tanioka M., et al. , Molecular clocks in mouse skin. J. Invest. Dermatol. 129, 1225–1231 (2009). [DOI] [PubMed] [Google Scholar]
- 3.Wang H., et al. , Time-restricted feeding shifts the skin circadian clock and alters UVB-induced DNA damage. Cell Rep. 20, 1061–1072 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Geyfman M., et al. , Brain and muscle Arnt-like protein-1 (BMAL1) controls circadian cell proliferation and susceptibility to UVB-induced DNA damage in the epidermis. Proc. Natl. Acad. Sci. U.S.A. 109, 11758–11763 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stringari C., et al. , In vivo single-cell detection of metabolic oscillations in stem cells. Cell Rep. 10, 1–7 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pasparakis M., Haase I., Nestle F. O., Mechanisms regulating skin immunity and inflammation. Nat. Rev. Immunol. 14, 289–301 (2014). [DOI] [PubMed] [Google Scholar]
- 7.Nakamura Y., et al. , Circadian regulation of allergic reactions by the mast cell clock in mice. J. Allergy Clin. Immunol. 133, 568–575 (2014). [DOI] [PubMed] [Google Scholar]
- 8.Takita E., et al. , Biological clock dysfunction exacerbates contact hypersensitivity in mice. Br. J. Dermatol. 168, 39–46 (2013). [DOI] [PubMed] [Google Scholar]
- 9.Prendergast B. J., et al. , Impaired leukocyte trafficking and skin inflammatory responses in hamsters lacking a functional circadian system. Brain Behav. Immun. 32, 94–104 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ando N., et al. , Circadian gene clock regulates psoriasis-like skin inflammation in mice. J. Invest. Dermatol. 135, 3001–3008 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Edgar R. S., et al. , Cell autonomous regulation of herpes and influenza virus infection by the circadian clock. Proc. Natl. Acad. Sci. U.S.A. 113, 10085–10090 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ehlers A., et al. , BMAL1 links the circadian clock to viral airway pathology and asthma phenotypes. Mucosal Immunol. 11, 97–111 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guiducci C., et al. , PI3K is critical for the nuclear translocation of IRF-7 and type I IFN production by human plasmacytoid predendritic cells in response to TLR activation. J. Exp. Med. 205, 315–322 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takane H., et al. , Relationship between 24-hour rhythm in antiviral effect of interferon-beta and interferon-alpha/beta receptor expression in mice. Jpn. J. Pharmacol. 90, 304–312 (2002). [DOI] [PubMed] [Google Scholar]
- 15.van der Fits L., et al. , Imiquimod-induced psoriasis-like skin inflammation in mice is mediated via the IL-23/IL-17 axis. J. Immunol. 182, 5836–5845 (2009). [DOI] [PubMed] [Google Scholar]
- 16.Riol-Blanco L., et al. , Nociceptive sensory neurons drive interleukin-23-mediated psoriasiform skin inflammation. Nature 510, 157–161 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Suzuki H., et al. , Imiquimod, a topical immune response modifier, induces migration of Langerhans cells. J. Invest. Dermatol. 114, 135–141 (2000). [DOI] [PubMed] [Google Scholar]
- 18.Li W. Q., Qureshi A. A., Schernhammer E. S., Han J., Rotating night-shift work and risk of psoriasis in US women. J. Invest. Dermatol. 133, 565–567 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hemmi H., et al. , Small anti-viral compounds activate immune cells via the TLR7 MyD88-dependent signaling pathway. Nat. Immunol. 3, 196–200 (2002). [DOI] [PubMed] [Google Scholar]
- 20.Yin C., et al. , TLR7-expressing cells comprise an interfollicular epidermal stem cell population in murine epidermis. Sci. Rep. 4, 5831 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heib V., et al. , Mast cells are crucial for early inflammation, migration of Langerhans cells, and CTL responses following topical application of TLR7 ligand in mice. Blood 110, 946–953 (2007). [DOI] [PubMed] [Google Scholar]
- 22.Hornung V., et al. , Quantitative expression of toll-like receptor 1-10 mRNA in cellular subsets of human peripheral blood mononuclear cells and sensitivity to CpG oligodeoxynucleotides. J. Immunol. 168, 4531–4537 (2002). [DOI] [PubMed] [Google Scholar]
- 23.Wohn C., et al. , Langerin(neg) conventional dendritic cells produce IL-23 to drive psoriatic plaque formation in mice. Proc. Natl. Acad. Sci. U.S.A. 110, 10723–10728 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Di Domizio J., et al. , TLR7 stimulation in human plasmacytoid dendritic cells leads to the induction of early IFN-inducible genes in the absence of type I IFN. Blood 114, 1794–1802 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Greenberg E., Microarray expression data from P50 mouse full thickness back skin of C57BL/6 mice - various durations of IMQ treatment and timepoints. Gene Expression Omnibus (GEO). https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE142165. Deposited 17 December 2019.
- 26.Greenberg E., Andersen B., scRNAseq of mouse epidermis with and without 6h of IMQ treatment. Gene Expression Omnibus (GEO). https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE142345. Deposited 19 December 2019.
- 27.Takagi H., et al. , Plasmacytoid dendritic cells orchestrate TLR7-mediated innate and adaptive immunity for the initiation of autoimmune inflammation. Sci. Rep. 6, 24477 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sherman B. T., et al. , DAVID knowledgebase: A gene-centered database integrating heterogeneous gene annotation resources to facilitate high-throughput gene functional analysis. BMC Bioinformatics 8, 426 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Satija R., Farrell J. A., Gennert D., Schier A. F., Regev A., Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 33, 495–502 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Joost S., et al. , Single-cell transcriptomics reveals that differentiation and spatial signatures shape epidermal and hair follicle heterogeneity. Cell Syst. 3, 221–237.e9 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Heng T. S., Painter M. W.; Immunological Genome Project Consortium , The immunological genome project: Networks of gene expression in immune cells. Nat. Immunol. 9, 1091–1094 (2008). [DOI] [PubMed] [Google Scholar]
- 32.George T. C., et al. , Quantitative measurement of nuclear translocation events using similarity analysis of multispectral cellular images obtained in flow. J. Immunol. Methods 311, 117–129 (2006). [DOI] [PubMed] [Google Scholar]
- 33.Plikus M. V., et al. , Local circadian clock gates cell cycle progression of transient amplifying cells during regenerative hair cycling. Proc. Natl. Acad. Sci. U.S.A. 110, E2106–E2115 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sancar A., et al. , Circadian clock, cancer, and chemotherapy. Biochemistry 54, 110–123 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van Spyk E., Greenberg M., Mourad F., Andersen B., Regulation of Cutaneous Stress Response Pathways by the Circadian Clock: From Molecular Pathways to Therapeutic Opportunities (Springer International Publishing, Cham, 2016), pp. 281–300. [Google Scholar]
- 36.Swindell W. R., et al. , Imiquimod has strain-dependent effects in mice and does not uniquely model human psoriasis. Genome Med. 9, 24 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koren S., Fleischmann W. R. Jr, Optimal circadian timing reduces the myelosuppressive activity of recombinant murine interferon-gamma administered to mice. J. Interferon Res. 13, 187–195 (1993). [DOI] [PubMed] [Google Scholar]
- 38.Koren S., Whorton E. B. Jr., Fleischmann W. R. Jr, Circadian dependence of interferon antitumor activity in mice. J. Natl. Cancer Inst. 85, 1927–1932 (1993). [DOI] [PubMed] [Google Scholar]
- 39.Majumdar T., Dhar J., Patel S., Kondratov R., Barik S., Circadian transcription factor BMAL1 regulates innate immunity against select RNA viruses. Innate Immun. 23, 147–154 (2017). [DOI] [PubMed] [Google Scholar]
- 40.Teijaro J. R., et al. , Persistent LCMV infection is controlled by blockade of type I interferon signaling. Science 340, 207–211 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dhib-Jalbut S., Marks S., Interferon-beta mechanisms of action in multiple sclerosis. Neurology 74 (suppl. 1), S17–S24 (2010). [DOI] [PubMed] [Google Scholar]
- 42.Lin K. K., et al. , Circadian clock genes contribute to the regulation of hair follicle cycling. PLoS Genet. 5, e1000573 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kalamvoki M., Roizman B., Circadian CLOCK histone acetyl transferase localizes at ND10 nuclear bodies and enables herpes simplex virus gene expression. Proc. Natl. Acad. Sci. U.S.A. 107, 17721–17726 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kalamvoki M., Roizman B., The histone acetyltransferase CLOCK is an essential component of the herpes simplex virus 1 transcriptome that includes TFIID, ICP4, ICP27, and ICP22. J. Virol. 85, 9472–9477 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reinke H., Asher G., Crosstalk between metabolism and circadian clocks. Nat. Rev. Mol. Cell Biol. 20, 227–241 (2019). [DOI] [PubMed] [Google Scholar]
- 46.Koyanagi S., et al. , Glucocorticoid regulation of 24-hour oscillation in interferon receptor gene expression in mouse liver. Endocrinology 147, 5034–5040 (2006). [DOI] [PubMed] [Google Scholar]
- 47.Li B., Tsoi L. C., Swindell W. R., Gudjonsson J. E. et al. , Transcriptome analysis of psoriasis in a large case-control sample: RNA-seq provides insights into disease mechanisms. J. Invest. Dermatol. 134, 1828–1838 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Butler A., Satija R., Integrated analysis of single cell transcriptomic data across conditions, technologies, and species. bioRxiv:10.1101/164889 (18 July 2017). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The scRNA-seq, mouse skin microarray, and human psoriasis data discussed herein were deposited in GEO (accession nos. GSE142345, GSE142165, and GSE63980 respectively).