Significance
Mathematical models of evolution help us understand mechanisms driving protein-sequence change. Previous models recapitulate a disjoint subset of statistical features of natural sequences. We present a neutral evolution model that unifies features including extreme variance of the molecular clock’s tick rate and the observation of an evolutionary Stokes shift, an irreversible effect of mutations in the fitness landscape during sequence evolution. We show that interactions between amino acid sites, which inform our fitness metric, are required to observe these features. These interactions are inferred by using direct coupling analysis, which has been successfully utilized to predict protein structures, dynamics, and complexes from coevolutionary information. We anticipate our model will have applications in phylogenetics, ancestral reconstruction of sequences, and protein design.
Keywords: amino acid evolution, epistasis, generative models, substitution models, direct couplings
Abstract
We introduce a model of amino acid sequence evolution that accounts for the statistical behavior of real sequences induced by epistatic interactions. We base the model dynamics on parameters derived from multiple sequence alignments analyzed by using direct coupling analysis methodology. Known statistical properties such as overdispersion, heterotachy, and gamma-distributed rate-across-sites are shown to be emergent properties of this model while being consistent with neutral evolution theory, thereby unifying observations from previously disjointed evolutionary models of sequences. The relationship between site restriction and heterotachy is characterized by tracking the effective alphabet dynamics of sites. We also observe an evolutionary Stokes shift in the fitness of sequences that have undergone evolution under our simulation. By analyzing the structural information of some proteins, we corroborate that the strongest Stokes shifts derive from sites that physically interact in networks near biochemically important regions. Perspectives on the implementation of our model in the context of the molecular clock are discussed.
Over the last few decades, several phylogenetic and evolutionary models have been proposed to account for observed differences in homologous DNA and protein sequences across species. These models infer, based on interspecies sequence or structural homology, that these differences arose as polymorphisms in a population that then became fixed substitutions over time via a combination of neutral and selective processes as the population split into new species. Besides having theoretical significance, such models are key to applications such as the calibration of the molecular clock and phylogenetics. Despite such usefulness, current models are yet to capture important properties and patterns observed in real sequence data.
With the founding of population genetics, it became possible to calculate that, in order to fix substitutions in a population via natural selection, an untenable number of individuals must be born and die without reproducing (1). In response to Haldane’s upper limit on selective forces (1), Kimura proposed that, rather than being beneficial or deleterious to fitness, most mutations are perfectly neutral (2–4). Using molecular evolution data and population genetics theory, he proposed that changes to allele frequencies and fixation occur mainly through drift. The Neutral Theory of Evolution has been a successful assumption that accounts for the relative constancy of the substitution rate observed in the molecular clock as well as how allele diversity is produced by genetic drift (5–7).
Zuckerkandl and Pauling proposed that amino acid substitutions occur at regular rates, giving rise to the idea of a molecular clock (8). Later, statistical corrections to this model were considered by Ohta and Kimura, whereby they assumed a Poisson distribution for the substitution rate (6, 9). Importantly, this assumption could only be valid if substitutions are indeed independent events. This has been shown not to be the case, however, since some changes can induce selective pressure that impacts the substitution rates of other sites (2, 10, 11). As a measure of divergence from Poissonian behavior, the index of dispersion was introduced, defined as the ratio of the variance in substitution counts across branches of a phylogeny to the mean number of substitutions across branches (2, 12). A large index of dispersion, or overdispersion, suggests significant deviation from independence in the substitution rates across sites.
Additional models were developed, each one designed to account for some of the statistical behaviors observed in phylogenies and biochemical data. For example, proteins were shown to have nonuniform substitution rates across sites, in accordance with the knowledge that positions in a sequence vary in their contribution to the protein function, resulting in different selective constraints across the sequence. The distribution of these various rates among sites could generally be fit to a gamma distribution (13). Taking this observation into the phylogenetics field, the Rate-Across-Site (RAS) model assumed that each site has a unique, but constant, substitution rate along the amino acid sequence history and that these rates are sampled from a gamma distribution (14–16).
Lopez and coworkers subsequently discovered, via the particularly well-represented Multiple Sequence Alignment (MSA) of mitochondrial cytochrome b, that the evolutionary rate of a given site changed between taxonomic groups (17, 18). They coined the term “heterotachy” to name this property, based on the idea that per-site substitution rate or movement (“tachy”) differed (“hetero”) over time (or across phylogenetic branches). The RAS model did not display significant heterotachy at sites, but this was by construction and, therefore, expected. Fitch and Markowitz (19) observed the fixation of codons in cytochrome c in its phylogenetic tree. This observation was consistent with the idea that, at a given moment, only certain sites are allowed to mutate, but, as time passes by, the fixed sites change. Based on these observations, the covarion models were proposed. In such an approach, a fraction of sites are allowed to mutate, referred to as covarions, and, after a round of mutational events, some of the designated covarions are reassigned as fixed residues, while some fixed sites become covarions. By adjusting the mutation rates and the ratio of covarions, this model replicated both the gamma distribution across sites and the heterotachy of sites. However, these properties manifest by design instead of being emergent properties of the model.
Not only are substitution rates overdispersed, and not only do substitution rates of a particular site vary significantly from one branch of a lineage to the next (heterotachy), but, indeed, the acceptability of specific substitutions at a site is also not constant. Pollock et al. (11) performed simulations from a thermodynamical point of view showing that the acceptance probability for an amino acid mutation at a specific site increased over evolutionary time due to compensatory changes at the rest of the sequence. This shift in acceptance probability was dubbed a “Stokes shift,” after the spectroscopic effect in which an excited molecule adjusts to the higher-energy state so that a smaller quantum of energy is released when it relaxes back to the ground state. The magnitude of the Stokes shift a site displays over evolution depends on a combination of its substitution rate and its degree of involvement in coevolutionary interactions.
Also taking a thermodynamics approach, the Structurally Constrained Neutral (SCN) Model, put forth by Bastolla et al. (12), divides mutations into two classes: One set inactivates the protein or abrogates function, and the other allows the protein to remain active. These neutral mutations are the only changes that can be fixed according to this model. Rather than assuming that the rate of appearance of neutral mutations is constant, as did Kimura in his neutral model, Bastolla et al. allow the rate of neutral mutation to be a free parameter and then calculate the effect of mutations on protein-fold stability. This approach provides a genotype-to-phenotype mapping that allows the rate of occurrence of neutral mutations to be an outcome of the model. While they did not report capturing heterotachy or a gamma distribution, they did find that the rate of neutral substitutions fluctuates significantly across sites; i.e., overdispersion was captured by this model.
In recent years, new methods have been developed to analyze MSAs of protein families using a joint probability model that takes into account pairwise and single-site interactions. Particularly, direct coupling analysis (DCA) (20, 21) has been shown to be a powerful tool for predicting sites that are coupled during evolution and have been utilized as guides in inferring protein structures (22–26), understanding the thermodynamics of folding (27, 28), predicting protein–protein interactions (29–37), conformational dynamics (38), and uncovering mutational landscapes (39–42), as well as possible biomedical applications (43–50).
Here, we present a model of neutral evolution that incorporates coevolutionary information estimated from the statistical features of the MSAs of domain families as epistatic contributions of the sequence composition of proteins (51). Our model produces new members of the family with each step of the simulation, and it is neutral in that it does not innovate proteins with new functions, but preserves the functions typical to the family. A key result is that our model displays all of the features of previous neutral evolution models, not by construction, but as an emergent property, including overdispersion, gamma distribution of rates across sites, and heterotachous sites. We are also able to detect significant evolutionary Stokes shifts at many sites and to show that our fitness metric correlates with divergence from the root in a phylogenetic tree. Overall, the use of coevolutionary information could integrate statistical features of evolution to develop new models that display more realistic behavior based not only on sequence composition but also on evolutionary constraints imposed by structure and function. We refer to this model as Sequence Evolution with Epistatic Contributions (SEEC). Previous evolutionary simulations have considered epistasis in the fitness function and based the definition of fitness on predicted protein stability (52–55) or a statistical physics-based concept of energy (49). In the past, DCA has been used to identify effects of evolution in sequences (56–58), but here, we are using it to model neutral evolution and, furthermore, to unify existing models.
Results
Model Construction.
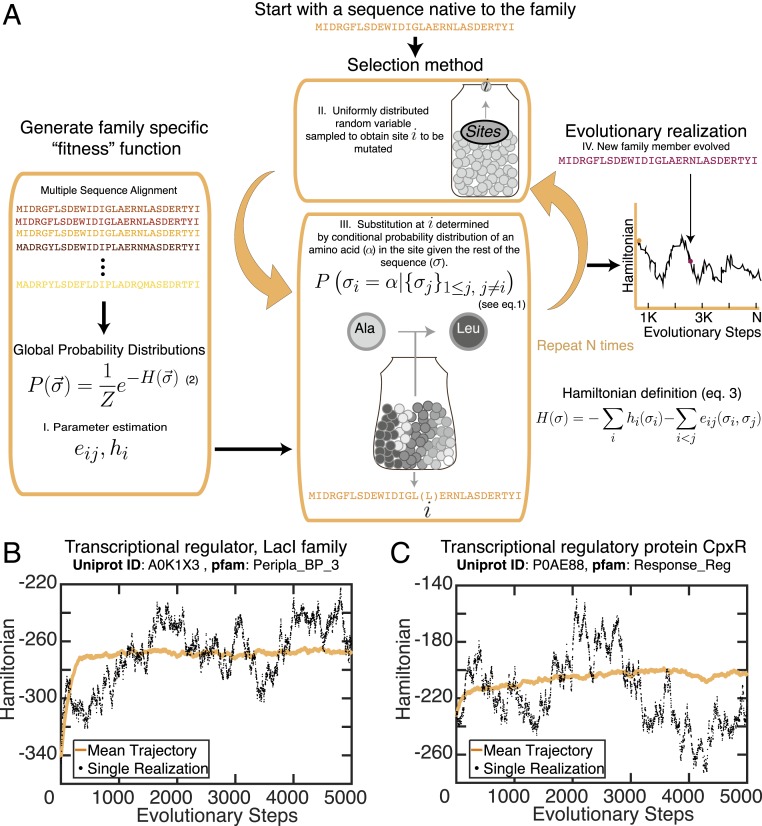
To demonstrate the properties of the SEEC model, we performed several analyses on the sequences of different protein families: 1) the Lac repressor protein (Uniprot ID A0K1X3; Protein Data Bank [PDB] ID code 4RKR) from the periplasmic binding protein family (PF13377) and 2) the transcriptional regulatory protein CPxR (Uniprot ID P0AE88; PDB ID code 4UHJ) from the Response regulator (RR) family (PF00072), as classified by Pfam (51). We also provide analyses of eight additional families, listed in SI Appendix, Table S1. Simulating evolution for a particular sequence under our model starts by creating a fitness metric related to the probability of a sequence to belong to a given family.
The MSA of the family to which that sequence belongs is used to compile a sample of the sequence space sampled by evolution (Fig. 1A). We used the MSA to infer parameters of a global probability distribution of sequences in this family using DCA (20, 59); specifically, we estimated pairwise coupling parameters, , and the single-site propensities, , called “local fields” (Fig. 1 A, Left). Both sets of parameters arise from a maximum-entropy inference approach, yielding a global probability distribution (Materials and Methods) whose marginals replicate the single-site and pairwise empirical frequencies of the MSA. These parameters have been used extensively for problems in structural biology, as well as for generative models of sequences (48, 49, 60–63). The proper inference of the coupling and local fields allowed us to build a Hamiltonian function that associates a statistical energy to each sequence as a score and, correspondingly, a probability of realization similar to a Boltzmann distribution (20, 21, 64, 65). As discussed in more detail below, this Hamiltonian acts as a fitness function for a given sequence, where fitness increases the more negative its statistical energy becomes (Materials and Methods). Such a fitness function has recently been used to predict the effects of mutations on histidine kinase (HK)–RR domain–domain interactions and specificity (41).
Fig. 1.
(A) Schematic of the SEEC model. The main steps are: I) statistical estimation of parameters from the MSA, II) site selection, and III) amino acid selection for the chosen site (steps II and III are iterated for each evolutionary step). Finally, IV) the Hamiltonian of the mutated sequence is calculated based on the conditional probability function (Eq. 3) and can be analyzed as a trajectory with respect to evolutionary step. (B and C) Hamiltonian evolutionary trajectories are shown for Pfam domains Peripla_BP_3 (PF13377) (B) and Response_reg (PF00072) (C). See SI Appendix, Fig. S1 for longer trajectories for all 10 families considered.
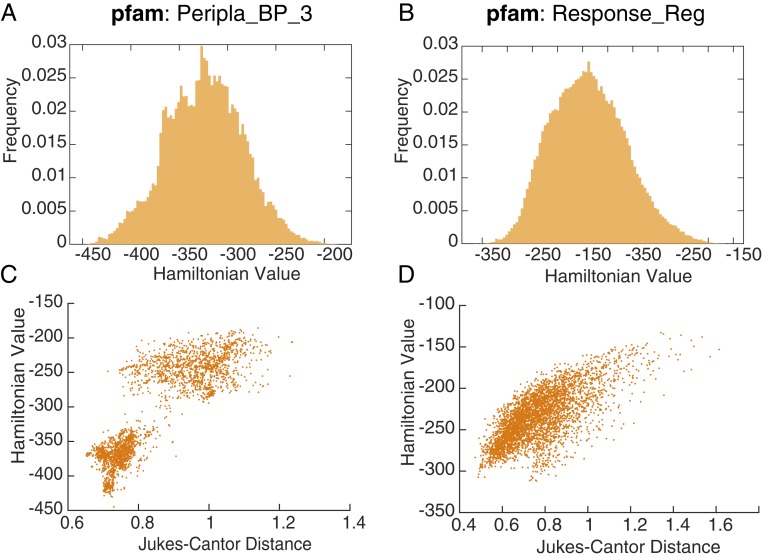
The sequences native to these families are diverse, with a broad distribution of Hamiltonian values (Fig. 2 A and B). In addition, the Jukes–Cantor distance between each sequence and the tree’s root is positively correlated with the Hamiltonian (Pearson correlation coefficient [CC] of 0.85 (Fig. 2C) and 0.70 (Fig. 2D), as defined in Materials and Methods concerning effective size) (Fig. 2 C and D), which hints that phylogenetic relationships among these sequences are also captured with the Hamiltonian. The statistical energy also tends to decrease as sequences become more derived, or distal from the tree root (Fig. 2 C and D), so that fitter sequences appear “older” or more basal to the lineage. While the relationship is compelling, we cannot rule out the possibility that the reason basal sequences are observed to have lower energy is due to the Hamiltonian parameters being inferred from phylogenetically correlated data, rather than it reflecting an accurate historical narrative.
Fig. 2.
A broad range of Hamiltonian values applies to proteins that are native to the family. (A) Hamiltonian distribution for 93,955 sequences in the family of periplasmic binding proteins. (B) Hamiltonian distribution for 155,996 sequences in the family of response regulators. (C and D) Relationship between Hamiltonian and the phylogenetic distance from tree root. There is a strong correlation between the change in Hamiltonian value and the Jukes–Cantor distance change.
The exact meaning of “fitness” varies for each evolutionary model. Here, fitness is defined by the inferred statistical energy: A sequence with a low energy optimizes both the local fields (determined by the single-site amino acid frequencies in an MSA position) and the pairwise coupling constraints or epistatic relationships that are common to the family (40), yielding a sequence that is representative according to the statistics of the MSA used to generate such parameters. The Hamiltonian histograms illustrate that, for the specific family members, fitness is distinguished from popularity within the family; because it is difficult to fully satisfy all of the coupling and frequency constraints of the family, the average sequence will not do so.
With this fitness parameter in hand, we then use this information encoded in the Hamiltonian to model an evolutionary process for sequence change over generations (Fig. 1 A, Center). We start with a sequence native to the family, and, then, at each step, a site along the protein is chosen by using a uniformly distributed random variable. An amino acid is then either retained or substituted at that site based on its associated conditional probability distribution , which describes the probability of finding each amino acid in that position, given that the rest of the sequence remains unchanged. The exact form of this conditional probability is described by equation Eq. 1. For further details, see Selection Method.
| [1] |
Importantly, we consider that each time a site is sampled, a nucleotide mutation occurs. This allows us to incorporate the concept of a synonymous mutation into our model and analyses, even though the simulation is following changes to the protein sequence. These two operations—i.e., a base-substitution event—are iterated for each evolutionary step. As expected, the majority of the time, a conserved amino acid remains in that position due to its high probability in the conditional probability distribution. This feature enables the study of the persistence of certain residues in specific sites during the evolutionary realization. Such sites are conserved because of the importance of their roles in the structure and function of the domain.
Our model generates sequences for which the Hamiltonian function levels off after a transient period, while still presenting fluctuations around a center value. In individual trajectories, we observed that if the Hamiltonian increases in value, it tends to collapse back to values closer to the average of the trajectories (Fig. 1 B and C and SI Appendix, Fig. S1) (66). The conditional probability takes into account how this site is coupled with other sites and not only the statistics of the local site. Because of this, the oscillations of the Hamiltonian around a given value could be understood as the increment of the space of allowed changes for a sequence to undergo to reduce its Hamiltonian; as the fitness of the function decreases, the allowed changes are those that will correct for the previous deviations. Importantly, when couplings are set to zero during the selection process, this smooth oscillation gets more noisy compared to the trajectories evolved under the influence of couplings (SI Appendix, Fig. S2). This result reveals that the progression of the Hamiltonian during evolutionary simulations is largely dependent on site–site coupling and hints at the critical nature of epistatic interactions in evolutionary processes.
The Substitution Rate of SEEC Displays Overdispersion That Is Enhanced by Epistatic Contributions.
Early molecular-clock models predicted that the substitutions at sites across protein sequences would be fixed at constant rates following a Poissonian distribution, characterized by a variance to mean ratio of 1 (5, 6, 10, 12). Simulations and phylogenetic analysis of natural protein sequences, however, consistently revealed overdispersion, wherein the variance of the fixation rates exceeded their mean (10, 12, 67).
In order to test if the SEEC model based on global couplings displays overdispersion, a total of 100 simulations of 30 K-steps were performed. After every 50 steps, the number of steps between consecutive substitutions was recorded, and the average number of substitutions and its variance were calculated for each round to obtain the index of dispersion as a function of the accumulated number of evolutionary steps (Materials and Methods). The corresponding results are displayed in Fig. 3 A and B (see also SI Appendix, Fig. S3).
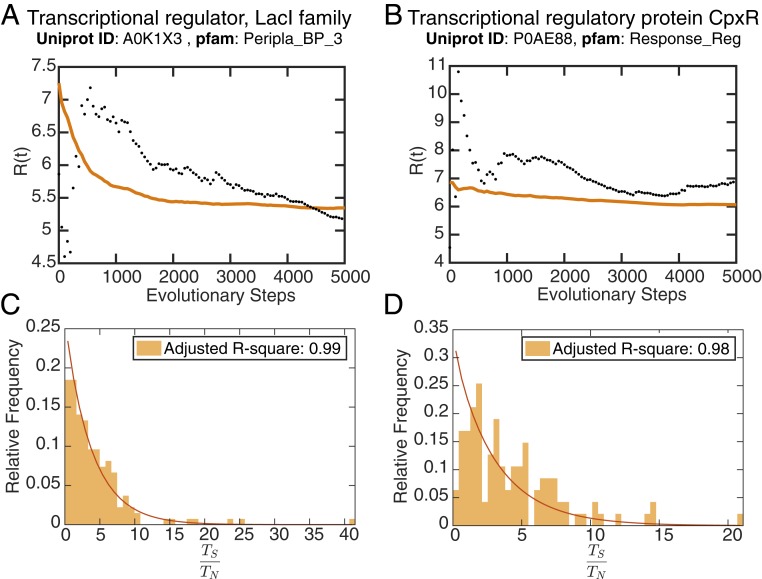
Fig. 3.
(A and B) Dispersion index, R(t), of the fixation rates across all sites over 3,000 evolutionary steps is displayed. The mean trajectory of 100 simulations is shown as an orange line. Fixation rates have dispersion indexes >1, indicative of a non-Poissonian process. (C and D) The substitution rates across the sequence, as defined by , follow a gamma distribution, an emergent property of our model. (A and C) Peripla_BP_3 (PF13377). (B and D) Response_reg (PF00072). See SI Appendix, Figs. S3 and S5 for dispersion-index trajectories and substitution-rate distributions of all 10 families considered, respectively.
By 1,000 evolutionary steps, the threshold for Poissonian processes has been exceeded, finally capping out at R(t) values ranging from 4 to 25, in agreement with reported overdispersion measurements (9, 10, 12, 68–70). Of note, in the absence of global couplings, overdispersion was significantly reduced (SI Appendix, Fig. S4), signifying that this effect was enhanced by epistatic relationships within the protein.
Substitution Rates across Sites Follow a Gamma Distribution, and Sites Display Heterotachy.
The notion that fixations across gene sequences occur at rates that follow a Poisson distribution was rejected by Uzzell and Corbin (13) based on several lines of evidence. Their analysis suggested that the distribution of fixations of nucleotide base-pair substitutions be described by the negative binomial distribution. This distribution is based on the assumption that the probability of fixing an additional nucleotide base-pair substitution at any given position is randomly drawn from a gamma-distributed probability density , where the mean probability is equal to the constant, fixed probability of the Poisson distribution that best describes the data (13). The RAS model also assumed that each site has a unique, but constant, substitution rate along the amino acid sequence history and that these rates are sampled from a gamma distribution (14–16).
Rather than evaluating patterns of inferred fixation across natural sequences, our model generates sequences that fit the characteristics of the natural family. To determine if the statistical properties of inferred fixations across natural sequences are also found in the sequences generated by our model, we measured the distribution of substitution rates over 1,000 realizations of our simulation (Materials and Methods). Fig. 3 C and D show that the distribution of fixation rates indeed fits into the space of gamma distributions (see also SI Appendix, Fig. S5 for other families). Of note is the observation of outliers that represent a deviation from the exponential decrease of a typical gamma distribution. These outliers might be the result of sampling effects or other phenomena not captured by the gamma distribution.
We then ran independent simulations starting from the same native sequence and quantified the rates of each site at each simulation within algorithmic constraints (Materials and Methods) by quantifying the percentage of realizations at which a site had an atypical rate (median absolute deviation [MAD] criterion), and we assigned a heterotachy degree (17). Fig. 4 A and B and SI Appendix, Fig. S6 show that highly heterotachous sites are found in a significant fraction of sites; they are spread across the sequence and are ubiquitous across protein families, as observed in phylogenetic data (17). Of significance here is that the findings of gamma-distributed fixation rates and heterotachy were not imposed on the model a priori, but they nevertheless emerged as properties of the evolved sequences. One clue as to why heterotachy happens was provided within the evidence for rejecting the Poisson distribution of fixation rates, which was “contagion and interaction,” or coevolution, between sites (13). This suggests that coevolutionary couplings, which are connected to structure, influence single-site rates via the evolutionary pressures imposed by the interaction with other sites and the constraints to maintain structure and function.
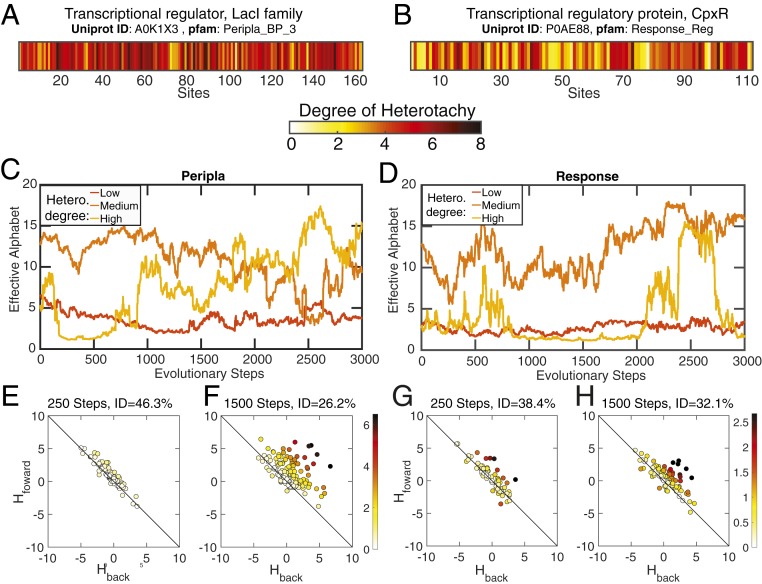
Fig. 4.
(A and B) The degree of heterotachy at each site in the sequence for LacI and CpxR regulators. Nearly every site displays some degree of heterotachy. (C and D) Trajectories of the effective alphabet for three sites over the course of the evolutionary realization. Sites were chosen based on having the lowest, medium (i.e., average), or highest degrees of heterotachy (Hetero), excluding sites containing gaps in the native sequence. (E–H) Relationship between the Hamiltonian resulting from forward and backward mutations in original and evolved sequences. Forward Hamiltonians (analogous to ) and back Hamiltonians (analogous to ) are the result of changing Y back to X after completing 250 (E and G) or 1,500 (F and H) steps along the simulation. Identity (ID) of the evolved sequence with respect to the original sequence and CCs is shown with each plot. Color in data points represents the absolute distance of the points to the line .
Heterotachy is an indication that a site’s degree of restriction is changing over time; this is largely explained by the fact that the connectivities, or coevolutionary forces, among sequence positions are changing as the sequence changes. To quantify how restricted a site is throughout the evolutionary simulation, we calculated the effective sequence alphabet, as defined in Materials and Methods. A larger effective alphabet implies fewer restrictions imposed on the mutation and substitution process, allowing for a larger variability of the site’s residues. We evaluated the effective alphabet at each evolutionary step of a simulation using a native sequence as starting point. We then compared the trajectories of three sites in the sequence having either low, medium (average), or high degrees of heterotachy (Fig. 4 C and D and SI Appendix, Fig. S7). Having a low degree of heterotachy corresponds with having small fluctuations of the effective alphabet over the course of the simulation, an indication of the site restriction remaining constant. As heterotachy increases, the variability of the effective alphabet also tends to increase, due to strong fluctuations in the site restrictions. Different effective alphabets imply different probability distributions for each site, being more or less restricted. As a comparison, simulations run with a Hamiltonian based on site–site independence (Materials and Methods) yield varying effective alphabets across sites that nevertheless remain fixed throughout the simulation.
Epistasis Induces Evolutionary Stokes Shifts at Residues Clustered in Loops.
Pollock et al. (11) observed, using an energetic model, that when a change is made to a site, the compensatory changes to the rest of the sequence tend to make it favorable for that particular amino acid to remain in that position. They found that the for a mutation in a sequence has the opposite sign, but does not have the same magnitude when the inverse change is made to the sequence once it has evolved. This effect was named an evolutionary Stokes shift (11). We quantified a similar effect in our evolved sequences using the SEEC Hamiltonian instead of . Briefly, native sequences were evolved for different numbers of evolutionary steps (250 and 1,500) and then compared to their diverged counterparts at these time points. The Hamiltonian cost of exchanging residues at each site was measured for both the native sequence () and the diverged sequence (). Early in the simulation (250 steps), when sites have not yet experienced a substitution, the , value is at the origin. As substitutions accumulate under the model, these values spread along the diagonal (indicating that the for mutations and for the reverse mutations have Hamiltonian effects of the same magnitude and opposite sign). Finally, the , values migrate into the upper right half of the plot, indicating that the cost of going back after substitutions at other sites have accumulated is higher than the cost of the initial mutation (Fig. 4 E and G and SI Appendix, Fig. S8).
Later in the simulation (1,500 steps), the sequence divergence increases, and the association moves even further from the diagonal (Fig. 4 F and H). There is a Hamiltonian cost for mutating each position that differs based on the changes to the rest of the sequence, showcasing the importance of coevolutionary information for producing the evolutionary Stokes shift. As observed by Pollock et al. (11), the inverse mutations are almost always in the upper right quadrant, which shows that mutations away from the original amino acid are almost always detrimental (i.e., positive ). In our case, the inverse mutations cause the Hamiltonian to increase, which indicates a loss of fitness relative to the original sequence.
In ref. 11, authors measure the propensity of a site to contain a given residue based on the probability distribution induced by a thermodynamic equivalent of the SEEC Hamiltonian, i.e., an empirical Gibbs free-energy model. Even though the approach is similar, we use the sequence Hamiltonian equivalent to directly simulate the evolutionary process.
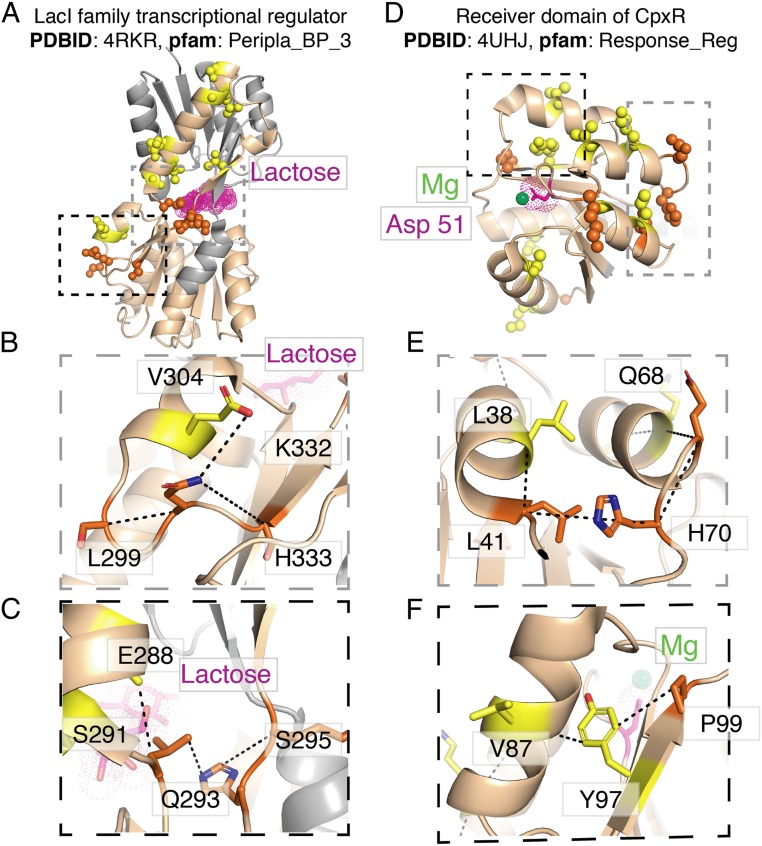
In order to map the Stokes shifts to structural and functional elements of the domain, we highlighted the residues possessing the largest 15 Stokes shifts on the three-dimensional (3D) structures of LacI family transcriptional regulator (Fig. 5 A–C) and the receiver domain of CpxR (Fig. 5 D–F). We observed that many of these highly Stokes-shifted residues were found near the biochemically important region of the protein, i.e., the bound lactose (Fig. 5A) or the catalytic Asp51 and magnesium cation (Fig. 5D). In addition, several of these residues fell either within a loop region of the structure or at the junction between a loop and a secondary structural element (colored orange in Fig. 5). Moreover, clusters of highly Stokes-shifted residues featured loop positions. For example, the black dashed lines in Fig. 5 B and C represent distances that are <8 Å, revealing that H333, K332, L299, and V304 (Fig. 5B); and E288, S291, Q293, and S295 (Fig. 5C) also form a physically contacted network of positions, most of which are in loops, suggesting that their cooperation is important for structure and function of the protein.
Fig. 5.
(A) Structural significance of the top 15 Stokes-shifted residues. Side chains are shown as spheres in A and D and as sticks in B, C, E, and F. These residues are colored orange when located in loops or at the end of helices or beta strands and colored yellow when in the middle of a secondary structural element. Areas from A and D that are magnified in B, C, E, and F are distinguished by gray or black dashed boxes. Portions of the structure that are not part of the Pfam domain family are colored gray. (A–C) X-ray crystal structure LacI family transcriptional regulator from Arthrobacter complexed with lactose (magenta dots). Of the top 15 Stokes-shifted residues, five are in loops or at the junction between helices or beta strands. (D–F) X-ray crystal structure of the receiver domain of CpxR from Escherichia coli. The catalytic Asp-51 (magenta dots) and magnesium ion (lime green sphere) are highlighted. Six of the top 15 Stokes-shifted residues are in loops or the ends of secondary structural elements. B, C, E, and F depict the physical contacts made between clustered residues. Residues that are <8 Å apart are indicated with a dashed black line, revealing networks of interactions among these residues.
In the case of the receiver domain of CpxR, clusters also form near biochemically relevant sites; Leu38, Leu31, His70, and Gln68 form close contacts of <8 Å (Fig. 5E). Val87, Tyr97, and Pro99 form close contacts in addition to being near to the catalytic magnesium ion, which interacts with Asp51 during phospho-transfer from a partner HK protein (Fig. 5F). Both of these groups feature one or more loop residues. Taken together, these results corroborate that the coevolutionary signals that produce evolutionary Stokes shifts are derived from the networking and coparticipation of residues in generating structural elements and biochemical activities and that loop regions have biophysical capacities enabling them to play important roles in facilitating those functions. Others have noticed that hinge neighboring residues are also important for function, perhaps even involved in disease (71).
Discussion
We introduce a model that recapitulates the properties found in natural phylogenies without having to impose these properties as an assumption of the model. Our model reconciles several models, including the RAS, covarion, and SCN, as well as displaying an evolutionary Stokes shift. These models measure or observe only some of the statistical properties of natural sequences. We find that the main driving force for these observations within our model is the effect of epistatic contributions that arose via amino acid coevolution. Coevolutionary relationships connect positions to each other in a sequence which leads to non-Poissonian fixation rates and wild variance in the fixation rates on different branches of a phylogeny (heterotachy), as well as overdispersion. Deviations from a Poissonian behavior in the overall mutation rate and differences at the site-mutation rate between different evolutionary trajectories can be attributed to different site restrictions due to coevolution along the evolutionary process. These conditions were implicitly included in previous models by fitness criteria containing structural information, as in refs. 12, 72, and 73, or by an explicit constraint, as in covarion simulations. For our model, this is an emergent property derived from the fact that coevolutionary constraints are encoded in the couplings fields used in the calculation of the global probability distribution from a family of sequences. The SEEC model includes structural constraints, but relies only on sequence information (20, 27, 28). For this study, the inference of the fields for the main text was based on Boltzmann-machine learning (bmDCA), as described in Materials and Methods. These same overall effects reported here were also seen with a mean-field DCA (mfDCA) approach, except that evolutionary energetic changes were more pronounced at the beginning of the trajectory (SI Appendix, Fig. S9). This highlights the importance of couplings to produce these statistical properties, as mfDCA seems to overestimate the contribution of the coupling parameters.
As shown in Fig. 4 C and D and SI Appendix, Fig. S7, the effective alphabet of a given site may vary along a simulation. Particularly, some sites can change from an alphabet of one to higher values. This could be interpreted as the site undergoing strong restrictions on substitutions and then suddenly being allowed to mutate after modifying other sites of the sequence that are coevolving with it. Similarly, sites with low restrictions can become fixed as their effective alphabet reduces to one during the simulation, due to changes in the rest of the sequence. This is similar to the behavior in the covarion models in which certain sites are allowed to mutate, referred as covarions, while others are completely fixed, changing between one another randomly.
The degree to which sites coevolve is connected with the degree to which they are under selection. Nevertheless, we present this model as a model of neutral evolution, because it maintains the fitness function in accordance with that which is typical of the family. It is not meant to show how adaptation to new evolutionary constraints occurs.
We foresee an impact of SEEC in several areas, including molecular-clock calibration, phylogenetic-tree constructions, ancestral reconstruction, and protein design. For example, the molecular clock has well-known issues with calibration (74), as well as consistency over time (75), where, due to reversions and other mutations to the same site, divergence times between sequences appear younger and younger the more taxonomically separated the species are. We found that the cumulative substitution times were linear with respect to evolutionary time, signaling that the substitution process under our model displays clock-like behavior at long timescales. We hope that future studies using SEEC will reveal a way to understand the underlying processes driving these observed phenomena.
Traditional distance-based and character-based phylogenetic-tree construction methods, such as the unweighted pair-group method with arithmetic mean, neighbor joining vs. maximum parsimony, and maximum likelihood, respectively, each assume that positions in the sequence alignment are statistically independent. A SEEC-based approach, however, would measure sequence distances by using both local propensities of amino acids and the coupling information to construct trees. We could use this distance to check the conservation of function, or family typicality, as sequences evolve, effectively using the Hamiltonian as a factor to evaluate the likelihood of trees. Moreover, a common observation found among ancestrally reconstructed proteins is that they are far more thermostable than their extant descendants, regardless of whether or not the ancestors emerged during the period when the earth was still hot (76). Likely an artifact of the current procedures and assumptions used for ancestral reconstruction, we envision that SEEC-based approaches may reduce such artifacts. For example, by assuming that the distribution of possible ancestors remains consistent over evolutionary time, pools of ancestral sequences can be constructed such that their Hamiltonians match the extant distribution of family members. Lastly, our model can direct protein-design approaches that utilize the Hamiltonian and conditional probability to design and optimize novel protein functions into existing protein scaffolds (77).
Materials and Methods
Global Probability Distributions.
Each sequence was considered as a realization of a multivariate random variable with a Boltzmann-type probability distribution. This probability distribution was derived following a maximumentropy treatment for a probability distribution constrained by local and pairwise marginal distributions empirically estimated from a MSA of amino acid sequences (20, 78). From this, the probability distribution for a given sequence to be inferred and observed within an MSA can be written as
| [2] |
where we introduced a Hamiltonian function parameterized by couplings and local fields , which account for pairwise and local interaction of sites, respectively, according to its identity. In our notation, indices refer to position along the sequence, and Greek-letter indices refer to the amino acid identity. For a given sequence , the corresponding Hamiltonian is given by:
| [3] |
This function assigns a statistical energy to every sequence, such that a sampling of the distribution (2) would replicate the single-site and pairwise statistics of the MSA, and its parameters would reflect coevolutionary direct relationships across sites in the sequence. Such couplings are typically related to physically interacting amino acids in the 3D structure of a protein in a family, as well as functional relationships (20, 21, 64, 65).
Parameter Inference.
The inference of couplings and local fields corresponding to each family is a computationally complex task. In the past, this issue has been approximated by using a mean field approach termed mfDCA (20) and pseudo-likelihood methods (79). However, since our goal is to produce a neutral model of sequence evolution, we have used a more computationally complex, but generative, version of DCA that uses a Boltzmann machine learning implementation based on Markov-chain Monte Carlo sampling (MCMC), as described by Ackley et al. (80). The specific implementation used in this study is described by ref. 59 and is available online (bmDCA).
In this approach, the marginal single-site and pairwise frequencies are approximated according to the reweighted frequencies described in ref. 59 from the MSA. Then, for a set of couplings and local fields, the single and pairwise marginals of the model are estimated by using MCMC. Finally, parameters are adjusted to correct for deviations between the estimated and the empirical frequencies.
Alternatively, for comparative purposes (SI Appendix, Fig. S9), we also use mfDCA, where an inverse of the cross-correlation matrix leads to a reliable approximation of the couplings and the local fields are fit to the marginalized two-site distribution of every pair of sites to satisfy the constraint of the single-site reweighted frequency using a message passing implementation (20).
Selection Method.
We refer to every event of the simulation as an evolutionary step. At each of those steps, one position of the sequence is chosen by sampling an uniform distribution over all of the sites. Once chosen, we calculate the probability distribution of the amino acid identity, as defined in Eq. 1, given that the rest of the sequence is held fixed and a residue is sampled from it.
Once the distribution is sampled, a new sequence is produced, with the corresponding site having the amino acid resulting from the sampling. This sequence is then used for the next evolutionary step. In the case that the selected amino acid is different from the previous step, we count it as a nonsynonymous substitution; else, we consider that the site underwent a synonymous substitution. The selection process is not performed explicitly in this model, but it, rather, occurs as a consequence of the sampling derived from the coevolutionary parameters. Simultaneously, at every evolutionary step, we sample a Poisson random variable with rate parameter to set some arbitrary timescale on the evolutionary events.
Dispersion Measurements.
The times at which a mutation is fixed were recorded from the timescale given by the Poisson sampling of the evolutionary step; every 50 steps, we calculated the average and variance on the number of evolutionary steps between two consecutive fixations up to that stage of the simulation. From this, we evaluated the index of dispersion for the fixation of mutation on along the simulations, defined as:
| [4] |
If fixation events were independent, they could be modeled as a Poissonian process, leading to a dispersion of one. However, once correlations are present between events, either by single or pairwise restrictions, the dispersion may vary to values below unity in underdispersed realizations or to larger, overdispersed, values, as in negative-binomial samplings. In the case of our model, from the law of total expectation and the law of total variance, one can see that the index of dispersion tends to , where is the random variable modeling the average number of evolutionary steps between nonsynonymous substitutions. Since this implies , regardless of the value of , we fixed the rate parameter to 10, just large enough to avoid sampling a 0 interval in 10,000 steps, i.e., PPoisson(X = 0) < 1/10,000.
Evolutionary Trajectories for Proteins.
A number of 10 protein families, including the Lac repressor protein (PDB ID codes 1EFA and 4RKR) and the transcriptional regulatory protein CpxR (PDB ID code 4UHJ), were retrieved and aligned with a hidden Markov model from Pfam (51). A representative sequence was chosen from the alignment and used as a starting point for the simulations.
Two kinds of evolutionary trajectories were performed. For representative measurements, as in Fig. 1 B and C, Fig. 4 E and F, and SI Appendix, Figs. S1 and S8, a single base simulation was performed with 30,000 steps, 5,000 of which were shown, storing the whole set of statistics and the actual trajectory. For average measurements, 100 simulations were performed, storing only the ensemble of the statistic of interest. All averaged quantities were calculated from the same ensemble of simulations.
Stokes Shift.
We proceeded in a similar manner to Pollock et al. (11). We took a test-evolved sequence generated along the base simulation, together with the native one. We generated multiple copies of both of them, one per each site in the sequence. For each copy of the native, we replaced one residue by the amino acid in the corresponding position at the test sequence and evaluated the difference in Hamiltonian for this new sequences with respect to the native to obtain the values as an analogous measure of . Similarly, for each copy of the test sequence, we replaced one residue by the amino acid in the corresponding position at the native sequence and evaluated the difference in Hamiltonian for this new sequence with respect to the test one to obtain the values as an analogous measure of . Hamiltonian shift points for the same position are shown in scatter plots (Fig. 4 E–H and SI Appendix, Fig. S8), and the line is shown as reference of the noninteracting scenario.
Site-Rate Calculation.
For a given evolutionary trajectory, we identified those sites that were chosen for substitution at least twice along the simulation. We measured the recurrence interval between consecutive synonymous substitutions of the same site that passed between sampling the same position twice without changing its identity, as defined in the selection method. Similarly, we obtained the recurrence interval between consecutive nonsynonymous substitutions of the same site , as defined in the selection method. From these quantities, an empirical mutation rate was assigned to the site, defined as , for the particular trajectory.
Heterotachy Tests.
In order to measure heterotachy, we ran 1,000 independent simulations per family with 10,000 steps starting from the same native sequences, such that the Hamiltonian has already reached a steady state. We identified the sites that changed in identity at least twice and obtained the empirical mutation rate, as described above.
Then, we compared between simulations those sites that were identified in every one of the 1,000. Those sites whose rate was more than three scaled MADs away from the median along the simulations were counted as anomalous realizations. We defined the heterotachy degree as the percentage of anomalous realizations of the site within the 1,000 realizations.
Gamma Distribution Across Sites.
To study the gamma distribution of the rate across sites for each independent trajectory, we took the same rates calculated for the heterotachy test and grouped them in 50 bins of the same size, spanning the range of rates of each individual simulation. From the grouped data, we performed a nonlinear least-squares fitting with a gamma distribution with the scale and shape parameters as fitting coefficients. We performed the fit using the Trust-Region algorithm from the Curve Fitting Toolbox (MathWorks, Inc.). Most shape parameters were between 0.5 and 1.9 values (SI Appendix, Table S3). The broad variation of the shape parameter cannot be interpreted solely on sampling error, since different trajectories can yield disjointed CIs (95% confidence), implying that the rates of sites were being sampled from a different gamma distribution, due to heterotachy effects. To evaluate the fitting, the degree-of-freedom adjusted coefficient of determination was used, defined by adj- for two-parameter estimation, where SSE and SST stand for the summed square of residuals and the sum of squares about the mean, respectively.
Statistical Entropy and Effective Alphabet.
It is possible to quantify how restricted a site is in a given an evolutionary step by using the statistical entropy associated to the conditional probability of such a site at each step. This quantity is always nonnegative, and it is only equal to zero whenever the distribution is certain; i.e., there is only one residue with probability one of occurrence, implying a highly restricted site with no mutations allowed. Entropy is maximal if the distribution is uniform along the possible residues, yielding a value of (78). Further, we can introduce an effective alphabet analogous to the one utilized by Pollock et al. (11) as a more tangible measure of this restriction derived from the site entropy. The effective alphabet per site is given by
| [5] |
Given the properties of Eq. 5, ranges from one whenever a site is restricted to a single amino acid up to when it is completely free to undergo substitutions to any of the amino acids. If a site is correlated with another position through a nonvanishing coupling matrix , changing the identity of the residue at site could influence the conditional probability of site , which could be measure as a change of the effective alphabet, getting reduced if the site’s restrictions are more severe now, or augmented if the site is in the opposite case. By definition, this quantity does not depend on the current amino acid in site , but on the state of the rest of the sequence; this allows us to directly quantify how the ensemble of coevolving sites in the protein restrict the amino acid identity at a given position.
Effect Size.
CCs and r values were calculated by using the following expression:
| [6] |
where and represent the analyzed quantities, is the SD as calculated from the data, and is the covariance between the two variables.
Tree Reconstruction.
The trees were constructed by using subsamples of 4,000 sequences from the original MSAs. The Jukes–Cantor pairwise distance was calculated for the subsamples; this is defined as a maximum-likelihood estimate of the number of substitutions based on the Hamming distance between two sequences. The phylogenetic-tree construction was done by using the neighbor-joining method, assuming equal variance and independence of evolutionary distance estimates, as in refs. 81 and 82. No ancestral sequences were reconstructed. Both the Jukes–Cantor distance and the neighbor-joining implementations are provided in the Bioinformatics Toolbox in MATLAB (MathWorks, Inc.).
Data Availability.
Data related to this study can be accessed in Datadryad.org (https://doi.org/10.5061/dryad.2ngf1vhj8). Scripts and model details are accessible in a GitHub repository (https://github.com/AlbertodelaPaz/SEEC) and at http://morcoslab.org.
Supplementary Material
Acknowledgments
This work was supported by the University of Texas at Dallas (J.A.d.l.P., C.M.N., and F.M.); NIH Grant R35GM133631 (to F.M.); and NSF Grant MCB-1943442 (to F.M.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: Data related to this study can be accessed in Datadryad.org (https://doi.org/10.5061/dryad.2ngf1vhj8). Scripts and model details are accessible in a GitHub repository (https://github.com/AlbertodelaPaz/SEEC) and at http://morcoslab.org.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1913071117/-/DCSupplemental.
References
- 1.Haldane J. B. S., The cost of natural selection. J. Genet. 55, 511–524 (1957). [Google Scholar]
- 2.Ohta T., Gillespie J. H., Development of neutral and nearly neutral theories. Theor. Popul. Biol. 49, 128–142 (1996). [DOI] [PubMed] [Google Scholar]
- 3.Nei M., Suzuki Y., Nozawa M., The neutral theory of molecular evolution in the genomic era. Annu. Rev. Genom. Hum. Genet. 11, 265–289 (2010). [DOI] [PubMed] [Google Scholar]
- 4.Kimura M., On the probability of fixation of mutant genes in a population. Genetics 47, 713–719 (1962). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bromham L., Penny D., The modern molecular clock. Nat. Rev. Genet. 4, 216–224 (2003). [DOI] [PubMed] [Google Scholar]
- 6.Kumar S., Molecular clocks: Four decades of evolution. Nat. Rev. Genet. 6, 654–662 (2005). [DOI] [PubMed] [Google Scholar]
- 7.Kimura M., Ohta T., Protein polymorphism as a phase of molecular evolution. Nature 229, 467–469 (1971). [DOI] [PubMed] [Google Scholar]
- 8.Pauling L., Molecular disease and evolution. Bull. N. Y. Acad. Med. 40, 334–342 (1964). [PMC free article] [PubMed] [Google Scholar]
- 9.Ohta T., Kimura M., On the constancy of the evolutionary rate of cistrons. J. Mol. Evol. 1, 18–25 (1971). [DOI] [PubMed] [Google Scholar]
- 10.Bedford T., Hartl D. L., Overdispersion of the molecular clock: Temporal variation of gene-specific substitution rates in Drosophila. Mol. Biol. Evol. 25, 1631–1638 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pollock D. D., Thiltgen G., Goldstein R. A., Amino acid coevolution induces an evolutionary Stokes shift. Proc. Natl. Acad. Sci. U.S.A. 109, E1352–E1359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bastolla U., Porto M., Roman E. H., Vendruscolo M., Connectivity of neutral networks, overdispersion, and structural conservation in protein evolution. J. Mol. Evol. 56, 243–254 (2003). [DOI] [PubMed] [Google Scholar]
- 13.Uzzell T., Corbin K. W., Fitting discrete probability distributions to evolutionary events. Science 172, 1089–1096 (1971). [DOI] [PubMed] [Google Scholar]
- 14.Rzhetsky A., Nei M., Unbiased estimates of the number of nucleotide substitutions when substitution rate varies among different sites. J. Mol. Evol. 38, 295–299 (1994). [DOI] [PubMed] [Google Scholar]
- 15.Strimmer K., Von Haeseler A., Quartet puzzling: A quartet maximum likelihood method for reconstructing tree topologies. Mol. Biol. Evol. 13, 964–969 (1996). [Google Scholar]
- 16.Yang Z., Paml: Phylogenetic analysis by maximum-likelihood. Comput. Appl. Biosci. 13, 555–556 (1997). [DOI] [PubMed] [Google Scholar]
- 17.Lopez P., Casane D., Philippe H., Heterotachy, an important process of protein evolution. Mol. Biol. Evol. 19, 1–7 (2002). [DOI] [PubMed] [Google Scholar]
- 18.Lopez P., Forterre P., Philippe H., The root of the tree of life in the light of the covarion model. J. Mol. Evol. 49, 496–508 (1999). [DOI] [PubMed] [Google Scholar]
- 19.Fitch W. M., Markowitz E., An improved method for determining codon variability in a gene and its application to the rate of fixation of mutations in evolution. Biochem. Genet. 4, 579–593 (1970). [DOI] [PubMed] [Google Scholar]
- 20.Morcos F., et al. , Direct-coupling analysis of residue coevolution captures native contacts across many protein families. Proc. Natl. Acad. Sci. U.S.A. 108, E1293–E1301 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weigt M., White R. A., Szurmant H., Hoch J. A., Hwa T., Identification of direct residue contacts in protein-protein interaction by message passing. Proc. Natl. Acad. Sci. U.S.A. 106, 67–72 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu Qi., et al. , Protein contact prediction using metagenome sequence data and residual neural networks. Bioinformatics 36, 41–48 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marks D. S., et al. , Protein 3D structure computed from evolutionary sequence variation. PLoS One 6, 1–20 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schaarschmidt J., Monastyrskyy B., Kryshtafovych A., Bonvin A. M. J. J., Assessment of contact predictions in CASP12: Co-evolution and deep learning coming of age. Proteins Struct. Funct. Bioinf. 86, 51–66 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cocco S., Monasson R., Weigt M., From principal component to direct coupling analysis of coevolution in proteins: Low-eigenvalue modes are needed for structure prediction. PLoS Comput. Biol. 9, 1–17 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sułkowska J. I., Morcos F., Weigt M., Hwa T., Onuchic J. N., Genomics-aided structure prediction. Proc. Natl. Acad. Sci. U.S.A. 109, 10340–10345 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jana B., Morcos F., Onuchic J. N., From structure to function: The convergence of structure based models and co-evolutionary information. Phys. Chem. Chem. Phys. 16, 6496–6507 (2014). [DOI] [PubMed] [Google Scholar]
- 28.Morcos F., Schafer N. P., Cheng R. R., Onuchic J. N., Wolynes P. G., Coevolutionary information, protein folding landscapes, and the thermodynamics of natural selection. Proc. Natl. Acad. Sci. U.S.A. 111, 12408–12413 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ovchinnikov S., Kamisetty H., Baker D., Robust and accurate prediction of residue–residue interactions across protein interfaces using evolutionary information. eLife 3, e02030 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.dos Santos R. N., Morcos F., Jana B., Andricopulo A. D., Onuchic J. N., Dimeric interactions and complex formation using direct coevolutionary couplings. Sci. Rep. 5, 13652 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cheng R. R., Morcos F., Levine H., Onuchic J. N., Toward rationally redesigning bacterial two-component signaling systems using coevolutionary information. Proc. Natl. Acad. Sci. U.S.A. 111, E563–E571 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tamir S., et al. , Integrated strategy reveals the protein interface between cancer targets Bcl-2 and NAF-1. Proc. Natl. Acad. Sci. U.S.A. 111, 5177–5182 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schug A., Weigt M., Onuchic J. N., Hwa T., Szurmant H., High-resolution protein complexes from integrating genomic information with molecular simulation. Proc. Natl. Acad. Sci. U.S.A. 106, 22124–22129 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hopf T. A., et al. , Sequence co-evolution gives 3D contacts and structures of protein complexes. eLife 3, e03430 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Uguzzoni G., et al. , Large-scale identification of coevolution signals across homo-oligomeric protein interfaces by direct coupling analysis. Proc. Natl. Acad. Sci. U.S.A. 114, E2662–E2671 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Podgornaia A. I., Laub M. T., Protein evolution. pervasive degeneracy and epistasis in a protein-protein interface. Science 347, 673–677 (2015). [DOI] [PubMed] [Google Scholar]
- 37.Bitbol A.-F., Dwyer R. S., Colwell L. J., Wingreen N. S., Inferring interaction partners from protein sequences. Proc. Natl. Acad. Sci. U.S.A. 113, 12180–12185 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Malinverni D., Marsili S., A. Barducci, P. De Los Rios, Large-scale conformational transitions and dimerization are encoded in the amino-acid sequences of Hsp70 chaperones. PLoS Comput. Biol. 11, e1004262 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou Q., et al. , Global pairwise RNA interaction landscapes reveal core features of protein recognition. Nat. Commun. 9, 2511 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Figliuzzi M., Jacquier H., Schug A., Tenaillon O., Weigt M., Coevolutionary landscape inference and the context-dependence of mutations in beta-lactamase TEM-1. Mol. Biol. Evol. 33, 268–280 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cheng R. R., et al. , Connecting the sequence-space of bacterial signaling proteins to phenotypes using coevolutionary landscapes. Mol. Biol. Evol. 33, 3054–3064 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hopf T. A., et al. , Sequence co-evolution gives 3D contacts and structures of protein complexes. eLife 3, e03430 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bai F., Morcos F., Cheng R. R., Jiang H., Onuchic J. N., Elucidating the druggable interface of protein-protein interactions using fragment docking and coevolutionary analysis. Proc. Natl. Acad. Sci. U.S.A. 113, E8051–E8058 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jiang X.-L., Martinez-Ledesma E., Morcos F., Revealing protein networks and gene-drug connectivity in cancer from direct information. Sci. Rep. 7, 3739 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mann J. K., et al. , The fitness landscape of HIV-1 Gag: Advanced modeling approaches and validation of model predictions by in vitro testing. PLoS Comput. Biol. 10, e1003776 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chakraborty A. K., Barton J., Rational design of vaccine targets and strategies for HIV: A crossroad of statistical physics, biology, and medicine. Rep. Prog. Phys. 80, 032601 (2017). [DOI] [PubMed] [Google Scholar]
- 47.Butler T., Barton J., Kardar M., Chakraborty A. K., Identification of drug resistance mutations in HIV from constraints on natural evolution. Phys. Rev. E 93 022412 (2015). [DOI] [PubMed] [Google Scholar]
- 48.Ferguson A., et al. , Translating HIV sequences into quantitative fitness landscapes predicts viral vulnerabilities for rational immunogen design. Immunity 38, 606–617 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Barton J., et al. , Relative rate and location of intra-host HIV evolution to evade cellular immunity are predictable. Nat. Commun. 7, 11660 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hart G. R., Ferguson A. L., Computational design of hepatitis C virus immunogens from host-pathogen dynamics over empirical viral fitness landscapes. Phys. Biol. 16, 016004 (2018). [DOI] [PubMed] [Google Scholar]
- 51.Finn R. D., et al. , Pfam: The protein families database. Nucleic Acids Res. 42, D222–D230 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shah P., McCandlish D. M., Plotkin J. B., Contingency and entrenchment in protein evolution under purifying selection. Proc. Natl. Acad. Sci. U.S.A. 112, E3226–E3235 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Starr T. N., Thornton J. W., Epistasis in protein evolution. Protein Sci. 25, 1204–1218 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Goldstein R. A., Pollock D. D., Sequence entropy of folding and the absolute rate of amino acid substitutions. Nat. Ecol. Evol. 1, 1923–1930 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kryazhimskiy S., Rice D. P., Jerison E. R., Desai M. M., Global epistasis makes adaptation predictable despite sequence-level stochasticity. Science 344, 1519–1522 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shekhar K., et al. , Spin models inferred from patient-derived viral sequence data faithfully describe HIV fitness landscapes. Phys. Rev. E 88, 062705 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Couce A., et al. , Mutator genomes decay, despite sustained fitness gains, in a long-term experiment with bacteria. Proc. Natl. Acad. Sci. U.S.A. 114, E9026–E9035 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gao C.-Y., Cecconi F., Vulpiani A., Zhou H.-J., Aurell E., DCA for genome-wide epistasis analysis: The statistical genetics perspective. Phys. Biol. 16, 026002 (2019). [DOI] [PubMed] [Google Scholar]
- 59.Figliuzzi M., Barrat-Charlaix P., Weigt M., How pairwise coevolutionary models capture the collective residue variability in proteins? Mol. Biol. Evol. 35, 1018–1027 (2018). [DOI] [PubMed] [Google Scholar]
- 60.Baldassi C., et al. , Fast and accurate multivariate Gaussian modeling of protein families: Predicting residue contacts and protein-interaction partners. PLoS One 9, e92721 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Balakrishnan S., Kamisetty H., Carbonell J. G., Lee S.-I., Langmead C. J., Learning generative models for protein fold families. Proteins 79, 1061–1078 (2011). [DOI] [PubMed] [Google Scholar]
- 62.Flynn W. F., Haldane A., Torbett B. E., Levy R. M., Inference of epistatic effects leading to entrenchment and drug resistance in HIV-1 protease. Mol. Biol. Evol. 34, 1291–1306 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mann J. K., et al. , The fitness landscape of HIV-1 Gag: Advanced modeling approaches and validation of model predictions by in vitro testing. PLoS Comput. Biol. 10, e1003776 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Haldane A., Flynn W. F., He P., Levy R. M., Coevolutionary landscape of kinase family proteins: Sequence probabilities and functional motifs. Biophys. J. 114, 21–31 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Haldane A., Flynn W. F., He P., Vijayan R. S. K., Levy R. M., Structural propensities of kinase family proteins from a Potts model of residue co-variation. Protein Sci. 25, 1378–1384 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nelson Onuchic Z. L.-S. J., Wolynes P. G., Theory of protein folding: The energy landscape perspective. Annu. Rev. Phys. Chem. 48, 545–600 (1997). [DOI] [PubMed] [Google Scholar]
- 67.Bedford T., Wapinski I., Hartl D. L., Overdispersion of the molecular clock varies between yeast, Drosophila and mammals. Genetics 179, 977–984 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gillespie J. H., The Causes of Molecular Evolution (Oxford University Press, New York, NY, 1991). [Google Scholar]
- 69.Langley C. H., Fitch W. M., An examination of the constancy of the rate of molecular evolution. J. Mol. Evol. 3, 162–177 (1974). [DOI] [PubMed] [Google Scholar]
- 70.Wilson A. C., Carlson S. S., White T. J., Biochemical evolution. Annu. Rev. Biochem. 46, 573–639 (1977). [DOI] [PubMed] [Google Scholar]
- 71.Sayılgan J. F., Haliloğlu T., Gönen M., Protein dynamics analysis reveals that missense mutations in cancer-related genes appear frequently on hinge-neighboring residues. Proteins 87, 512–519 (2019). [DOI] [PubMed] [Google Scholar]
- 72.Fitch W. M., Markowitz E., An improved method for determining codon variability in a gene and its application to the rate of fixation of mutations in evolution. Biochem. Genet. 4, 579–593 (1970). [DOI] [PubMed] [Google Scholar]
- 73.Uzzell T., Corbin K. W., Fitting discrete probability distributions to evolutionary events. Science 172, 1089–1096 (1971). [DOI] [PubMed] [Google Scholar]
- 74.Ayala F. J., Vagaries of the molecular clock. Proc. Natl. Acad. Sci. U.S.A. 94, 7776–7783 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ho S. Y. W., Phillips M. J., Cooper A., Drummond A. J., Time dependency of molecular rate estimates and systematic overestimation of recent divergence times. Mol. Biol. Evol. 22, 1561–1568 (2005). [DOI] [PubMed] [Google Scholar]
- 76.Trudeau D. L., Kaltenbach M., Tawfik D. S., On the potential origins of the high stability of reconstructed ancestral proteins. Mol. Biol. Evol. 33, 2633–2641 (2016). [DOI] [PubMed] [Google Scholar]
- 77.Dimas R. P., Jiang X.-L., Alberto de la Paz J., Morcos F., Chan C. T. Y., Engineering repressors with coevolutionary cues facilitates toggle switches with a master reset. Nucleic Acids Res. 47, 5449–5463 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mézard M., Montanari A., Information, Physics, and Computation (Oxford University Press, Oxford, UK, 2012). [Google Scholar]
- 79.Ekeberg M., Lövkvist C., Lan Y., Weigt M., Aurell E., Improved contact prediction in proteins: Using pseudolikelihoods to infer Potts models. Phys. Rev. E 87, 012707 (2013). [DOI] [PubMed] [Google Scholar]
- 80.Ackley D. H., Hinton G. E., Sejnowski T. J., A learning algorithm for Boltzmann machines. Cognit. Sci. 9, 147–169 (1985). [Google Scholar]
- 81.Saitou N., Nei M., The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425 (1987). [DOI] [PubMed] [Google Scholar]
- 82.Studier J. A., Keppler K. J., A note on the neighbor-joining algorithm of Saitou and Nei. Mol. Biol. Evol. 5, 729–731 (1988). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data related to this study can be accessed in Datadryad.org (https://doi.org/10.5061/dryad.2ngf1vhj8). Scripts and model details are accessible in a GitHub repository (https://github.com/AlbertodelaPaz/SEEC) and at http://morcoslab.org.