Significance
Many RNA-binding proteins contain aggregation-prone prion-like domains (PrLDs), and mutations in several of these have been linked to degenerative diseases. Additionally, many of these proteins are associated with stress granules, which are membraneless organelles that form under stress, in part through reversible protein assembly. Although the mechanisms of stress granule assembly are unclear, PrLDs can play a role in this process. In order to further understand how PrLDs respond to stress, we analyzed the assembly propensities of a selection of different PrLDs from yeast. We find that many PrLDs are efficiently recruited to stress-induced assemblies and that this recruitment is predominantly dependent on the amino acid composition of the PrLDs.
Keywords: prion, prion-like, stress granule, yeast
Abstract
Mutations in a number of stress granule-associated proteins have been linked to various neurodegenerative diseases. Several of these mutations are found in aggregation-prone prion-like domains (PrLDs) within these proteins. In this work, we examine the sequence features governing PrLD localization to stress granules upon stress. We demonstrate that many yeast PrLDs are sufficient for stress-induced assembly into microscopically visible foci that colocalize with stress granule markers. Additionally, compositional biases exist among PrLDs that assemble upon stress, and these biases are consistent across different stressors. Using these biases, we have developed a composition-based prediction method that accurately predicts PrLD assembly into foci upon heat shock. We show that compositional changes alter PrLD assembly behavior in a predictable manner, while scrambling primary sequence has little effect on PrLD assembly and recruitment to stress granules. Furthermore, we were able to design synthetic PrLDs that were efficiently recruited to stress granules, and found that aromatic amino acids, which have previously been linked to PrLD phase separation, were dispensable for this recruitment. These results highlight the flexible sequence requirements for stress granule recruitment and suggest that PrLD localization to stress granules is driven primarily by amino acid composition, rather than primary sequence.
Stress granules are cytoplasmic, membraneless ribonucleoprotein (RNP) assemblies containing mRNAs stalled in translation initiation (1, 2). Stress granules form in response to various stresses and dissipate once the stress-inducing conditions are eliminated (3). Many of the RNA-binding proteins found in stress granules contain prion-like domains (PrLDs), which are glutamine/asparagine (Q/N)-rich domains that compositionally resemble yeast prion domains (4). Recently, mutations in a number of these PrLD-containing RNA-binding proteins have been implicated in various neurodegenerative diseases, including amyotrophic lateral sclerosis and frontotemporal dementia (5, 6).
The relationship between PrLDs and stress granules has attracted research interest for a few reasons. First, some of these domains have been shown to help target proteins to stress granules (7, 8). Second, intrinsically disordered regions (IDRs), of which PrLDs are a subclass, are thought to provide promiscuous, but potentially stabilizing interactions in RNP granules (9). Finally, several of the disease-causing mutations in PrLD-containing stress granule-associated proteins occur within the PrLDs and are associated with irreversible aggregation of the protein in affected individuals (6, 10); these proteins include FUS (11–13), TDP-43 (14, 15), hnRNPA1 (16, 17), and hnRNPA2 (16). These observations have led to the idea that these PrLDs contribute to the functional, reversible assembly of stress granules, but that aggregation-promoting mutations in these domains can negatively affect their normal dynamic behavior (5, 6, 10).
Some constituent proteins of stress granules are able to undergo liquid–liquid phase separation (LLPS) in vitro (11, 17–20), and these proteins seem to retain liquid-like qualities when they assemble in vivo (11, 17, 21). These results have led to a model proposing that stress granules are liquid-like RNP compartments resulting from LLPS of their protein and RNA components. Many PrLDs can also phase separate in vitro, with amino acid sequence and composition affecting LLPS ability (18, 20, 22–24). Interestingly, upon introduction of some disease-associated mutations, PrLDs become more prone to form stable, solid-phase aggregates (11, 17, 21, 25). Other disease-associated mutations alter the propensity of PrLDs to undergo LLPS (26). The sequence features that promote transitions to amorphous aggregates and well-structured amyloids have been extensively studied, and various methods now exist to predict the aggregation propensity of PrLDs (27–32). However, while a number of laboratories have made exciting progress in identifying specific sequence features that promote the formation of reversible, liquid-like assemblies in vivo (20, 24, 33–35), these sequence features have not yet been comprehensively defined; consequently, our ability to predict the propensity of PrLDs to form these assemblies is more limited. For example, cation–pi interactions between positively charged amino acids in RNA-binding domains and aromatic amino acids in PrLDs appear to be a critical driver of phase separation by the FUS family of proteins (24), and have been linked to phase separation of other IDRs (22), but it is not clear whether this is a universal feature of phase-separating PrLDs.
To gain more insight into the sequence requirements for stress granule recruitment, we examined the response of various PrLDs to stress. Using the prion prediction algorithms PAPA and PLAAC (36, 37), we identified a variety of PrLDs in Saccharomyces cerevisiae and tested these PrLDs for assembly into foci in response to heat stress. We found that many of these PrLDs reversibly assemble into foci that colocalize with stress granule markers. Additionally, most PrLDs showed similar assembly activity under two other stresses: sodium azide (NaN3) treatment (oxidative stress) and sorbic acid treatment (pH stress). PrLDs that assembled into foci in response to stress showed substantial compositional biases, including a significant overrepresentation of both charged and hydrophobic amino acids. These compositional biases were sufficient 1) to predict with reasonable accuracy which PrLDs would localize to stress granules in response to heat shock; 2) to design mutations to modulate assembly activity; and 3) to design synthetic PrLDs (sPrLDs) that reversibly assemble in response to stress. Collectively, these results indicate that PrLD composition, rather than primary sequence, is the determining factor dictating recruitment to stress granules.
Results
PrLDs Form Reversible Assemblies upon Heat Shock.
We recently showed that the PrLDs of Sky1, Cdc39, and Ded1 are sufficient for recruitment to stress granules upon heat stress (8). To determine how common this feature is among PrLDs, and to gain a greater understanding of how PrLDs might promote stress granule recruitment, we investigated the behavior of other isolated PrLDs in response to stress. We chose 53 additional PrLDs (SI Appendix, Table S1) from two preexisting yeast datasets. The first dataset was derived from a study by Wallace et al. (38), who utilized mass spectrometry to identify proteins that reversibly assemble in response to heat stress; we screened this dataset with the prion prediction algorithms PAPA and PLAAC (36, 37), and found that 19 of the proteins contained PrLDs. The second dataset was from a previous study in which the yeast proteome was screened with PAPA to identify proteins with high-scoring PrLDs (39).
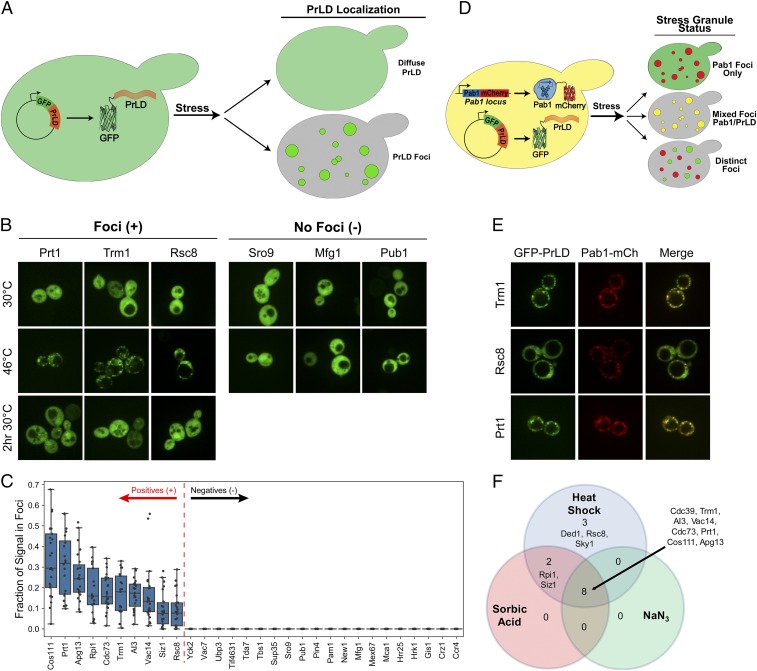
To examine PrLD assembly upon stress, each PrLD was fused to the C terminus of GFP and expressed in S. cerevisiae cells from the constitutive, intermediate-strength SUP35 promoter. Localization of these PrLD fusions was examined before and after 30 min of heat shock at 46 °C (Fig. 1A). About one-fifth of the GFP-PrLD fusions were not detectable on a Western blot and were therefore excluded from further analysis (SI Appendix, Fig. S1 and Table S1). Additionally, PrLDs that were membrane-associated or localized to the nucleus were also excluded (SI Appendix, Table S1). Of the remaining 35 PrLDs, 10 assembled into distinct foci in >60% of cells upon heat stress, 20 rarely or never assembled into foci, and 5 formed foci in a moderate fraction of cells (Fig. 1 B and C and SI Appendix, Fig. S2 and Table S1). Although a few of the foci-forming PrLDs showed a small number of foci during normal growth at 30 °C (including AI3, Vac14, and Cdc73), each had striking increase in foci upon stress. It should be noted that many of the PrLDs expressed poorly or showed multiple bands by Western blot analysis (SI Appendix, Fig. S1); however, despite degradation and/or low protein levels, many PrLDs were still capable of assembling into foci in response to stress.
Fig. 1.
A subset of PrLDs are sufficient to reversibly assemble upon stress. (A) Schematic of heat shock experiments. PrLDs were fused to the C terminus of GFP, and expressed under control of a constitutive promoter. PrLDs that showed diffuse GFP fluorescence under normal growth conditions were visualized after heat shock to test for foci formation. (B) Representative images of three foci-forming PrLDs and three PrLDs that remained diffuse upon heat shock. Cells were imaged under normal growth conditions and after 30 min of heat shock at 46 °C. PrLDs that formed foci upon heat shock were also imaged after 2 h of recovery post heat shock at 30 °C. (C) Box plot depicting the fraction of GFP signal in foci for at least 20 individual cells after heat shock. (D) Schematic of colocalization experiments. Foci-forming PrLDs were coexpressed with Pab1-mCherry as a stress granule marker to assess colocalization with stress granules. Cells were exposed to 30 min of heat shock at 46 °C prior to imaging. (E) Representative examples of colocalization of three positive PrLDs with Pab1. Colocalization is quantified in SI Appendix, Fig. S3. (F) Venn diagram showing PrLDs that assembled in response to heat stress, pH stress upon treatment with 6 mM sorbic acid, or oxidative stress upon treatment with 0.5% NaN3.
Many assemblies induced by heat stress are reversible, disassembling upon stress relief (38). We therefore assessed the ability of foci formed by each assembly-prone PrLD to dissipate post stress. After heat shock, cells were incubated at 30 °C for 2 h to allow for recovery. Of the PrLDs that were able to assemble upon heat shock, most reverted back to their diffuse state during recovery (Fig. 1B and SI Appendix, Fig. S2A). Only AI3 and Cdc73 showed incomplete dissolution of stress-induced foci. These data suggest that most PrLDs are not forming insoluble, irreversible aggregates, but rather dynamic, reversible assemblies that are more analogous to RNP granules.
We next asked whether these PrLDs were localizing to stress granules (Fig. 1D). Each PrLD that assembled upon stress was coexpressed with mCherry-tagged Pab1, a known stress granule protein (40). All of the assembling PrLDs colocalized with Pab1-mCh (Fig. 1E and SI Appendix, Fig. S3) upon heat shock; all except Rsc8 had median Pearson correlation coefficients between the GFP and mCherry signals of greater than 0.80 (SI Appendix, Fig. S3B), indicating that these PrLDs alone are sufficient for localization to stress granules.
Different stresses result in stress granules with overlapping but distinct compositions (41). We therefore tested whether our PrLDs would show similar responses to other stresses. We exposed the same PrLDs, plus the previously identified Sky1, Ded1, and Cdc39 PrLDs (8), to 30-min treatments with either 0.5% NaN3, which results in oxidative stress (41), or 6 mM sorbic acid, which causes pH stress (42). Interestingly, most PrLDs showed similar behavior across the various stresses; although slightly fewer PrLDs assembled in response to oxidative and pH stress (8 and 10 PrLDs, respectively), every PrLD that assembled under either of these two stresses also assembled in response to heat stress (Fig. 1F and SI Appendix, Figs. S4 and S5 and Table S1).
Composition Is the Primary Determinant of Stress-Induced Assembly.
The tested PrLDs provide a useful dataset to examine the sequence features that promote stress-induced assembly. Because there is evidence that amino acid composition contributes significantly to LLPS of PrLDs (22–24), we first examined whether each amino acid was overrepresented or underrepresented among the PrLDs that assembled under each stress relative to those PrLDs that did not assemble. Strikingly, the major compositional biases were consistent across all three stresses. Charged and hydrophobic residues were overrepresented in PrLDs that assembled in response to each stress, whereas glutamine, asparagine, proline, and alanine were overrepresented in PrLDs that did not show stress-dependent assembly (Fig. 2 and SI Appendix, Fig. S6 and Table S2). These compositional biases are surprising since they contradict the features that make PrLDs prion-like. Classical yeast prion domains are generally enriched in Q/N residues and depleted in charged and hydrophobic residues (43). These results strongly suggest that, among PrLDs, distinct sequence features maximize reversible stress-induced assembly versus stable prion aggregate formation.
Fig. 2.
Box plots depicting compositional biases observed among assembly-forming (blue boxes) and non–assembly-forming (orange boxes) PrLDs for each stress. HS, heat shock; NaN3, sodium azide stress; SA, sorbic acid stress. The left-most plot depicts the percent composition of charged amino acids (H, D, E, K, and R) among the assembly- and non–assembly-forming PrLDs, the middle plot depicts the percent composition of hydrophobic amino acids (I, L, V, and M), and the right-most plot depicts the percent composition of polar amino acids (Q, N, S, and T). Gray violin plots (with overlaid miniature boxplots) on the right of each graph indicate the distribution derived from scanning the entire yeast proteome with a 100-aa window and calculating the percent composition for the indicated amino acid groups in each window. A two-sided Mann–Whitney U test was used to calculate P values.
Interestingly, various other features that have previously been linked to the formation of membraneless organelles did not show obvious biases in our dataset. For example, asymmetric charge distribution has been shown to promote complex coacervation (23). However, although charged amino acids were enriched among PrLDs that assembled in response to heat stress (SI Appendix, Fig. S7A), the nonassembling PrLDs actually showed slightly higher charge asymmetry (SI Appendix, Fig. S7B), although this difference was not statistically significant. There was also no significant difference in the predicted isoelectric points or net charge per residue for the assembling and nonassembling PrLDs (SI Appendix, Fig. S7B). Likewise, IDRs are enriched among proteins that are recruited to heat-induced stress granules in yeast (44); however, while almost every PrLD in our dataset contains regions that are predicted to be disordered by multiple prediction methods [IUPred (45), FoldUnfold (46), and FoldIndex (47)], the PrLDs that failed to assemble in response to heat stress actually tended to show a higher degree of predicted disorder (SI Appendix, Fig. S8).
Given the compositional biases observed among assembly-prone PrLDs, we examined whether these biases were sufficient to predict which PrLDs would assemble in response to stress. Because the compositional biases were similar across all three stresses, we focused our follow-up investigations on heat stress. We calculated the mean frequency of occurrence of each amino acid among assembly-forming (positive) PrLDs and non–assembly-forming (negative) PrLDs (Table 1). These values were used to calculate a log-odds ratio for each amino acid, representing the degree of overrepresentation or underrepresentation of that amino acid in PrLDs that form assemblies (Table 1). To score the predicted assembly activity of each PrLD, for each amino acid we multiplied the frequency of occurrence of the amino acid in the PrLD by the log-odds ratio for that amino acid, and then summed these values (SI Appendix, Table S1).
Table 1.
Log-odds ratio scores for each amino acid
| Amino acid | Mean frequency in nonassembling PrLDs | Mean frequency in assembly-prone PrLDs | Log-odds ratio |
| A | 5.38 | 2.79 | −0.68 |
| C | 0.23 | 1.22 | 1.67 |
| D | 4.33 | 4.58 | 0.06 |
| E | 2.44 | 4.11 | 0.54 |
| F | 2.41 | 4.71 | 0.69 |
| G | 6.63 | 5.27 | −0.24 |
| H | 2.06 | 2.94 | 0.37 |
| I | 2.78 | 5.23 | 0.66 |
| K | 3.12 | 4.83 | 0.45 |
| L | 4.26 | 6.07 | 0.37 |
| M | 2.98 | 1.67 | −0.59 |
| N | 16.15 | 13.06 | −0.25 |
| P | 8.30 | 3.94 | −0.79 |
| Q | 13.91 | 6.79 | −0.8 |
| R | 3.07 | 6.04 | 0.71 |
| S | 9.57 | 12.57 | 0.31 |
| T | 4.54 | 4.46 | −0.02 |
| V | 2.72 | 5.19 | 0.67 |
| W | 0.30 | 1.22 | 1.42 |
| Y | 4.83 | 3.31 | −0.39 |
The frequency of each amino acid within each PrLD was determined. A log-odds ratio was then calculated, comparing the mean frequency of occurrence among those PrLDs that assembled in response to heat versus those that did not. Positive log-odds ratios indicate that an amino acid was more common among assembly-prone PrLDs.
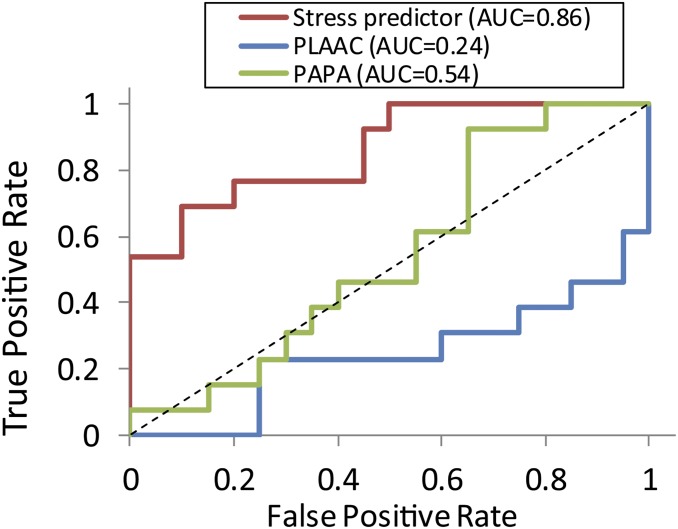
The 9 highest scoring PrLDs all formed stress-induced assemblies, while the 11 lowest scoring all failed to assemble. To evaluate the accuracy of the predictor, we performed an iterative leave-one-out analysis for all of the PrLDs and plotted the resulting receiver operating characteristic (ROC) curve (Fig. 3). The area under the curve (AUC) of 0.86 for our predictor indicates a reasonably accurate predictive ability. By contrast, traditional prion prediction algorithms were not effective at predicting stress-induced assembly of PrLDs, with PAPA (37) yielding an AUC of 0.54, and PLAAC (36) yielding an AUC of 0.24. This further confirms that the compositional requirements for prion formation and those for reversible stress-induced assemblies are distinct.
Fig. 3.
Amino acid composition predicts PrLD assembly. The ROC plot depicts the true-positive rate versus the false-positive rate for the composition-based stress-induced assembly predictor, and for the prion prediction algorithms PAPA and PLAAC. Area under the curve (AUC) values are indicated. For the stress predictor, PrLDs from the full PrLD dataset (n = 33) were scored using an iterative leave-one-out procedure.
To test the robustness of our predictor, we utilized the predictor to identify eight additional PrLDs with a range of predicted stress-induced assembly propensities (SI Appendix, Fig. S9A) and evaluated these for assembly upon heat shock. One of these, Tif4632, was poorly expressed and predominantly in foci under normal growth conditions (SI Appendix, Fig. S9 B and C), so was excluded from further analysis. The three highest-scoring PrLDs all reversibly formed stress-induced foci (SI Appendix, Fig. S9D), and these foci all colocalized with Pab1-mCh (SI Appendix, Fig. S9E). By contrast, the four lowest-scoring PrLDs (excluding Tif4632) failed to assemble (SI Appendix, Fig. S9F). However, our predictor was not effective when applied to IDRs that do not have prion-like composition. Specifically, we identified six IDRs that were predicted by our algorithm to have high stress-induced assembly propensities but that, unlike classical yeast prion domains, had low Q/N content (SI Appendix, Fig. S10A). Only one of these assembled in response to heat stress (SI Appendix, Fig. S10B). Together, these results indicate that the strong compositional biases observed among the PrLDs that form stress-induced assemblies are sufficient to predict with reasonable accuracy whether a PrLD will assemble into stress-induced foci under heat stress, but that this predictive accuracy is specific to PrLDs.
Modulating Stress-Induced Assembly.
The fact that we were able to predict stress-induced assembly of PrLDs based on composition suggests that we should similarly be able to modulate assembly propensity by rationally changing a PrLD’s amino acid composition. To test this, we selected three PrLDs that formed stress induced assemblies (Prt1, Trm1, and Rsc8). These PrLDs were selected for two reasons. First, they are all relatively short and therefore should require fewer mutations to change their ability to assemble. Second, they all had composition scores within the range that our predictor was highly accurate (>0.10; SI Appendix, Table S1 and Fig. 3), suggesting that their compositions are sufficient to explain their assembly propensities.
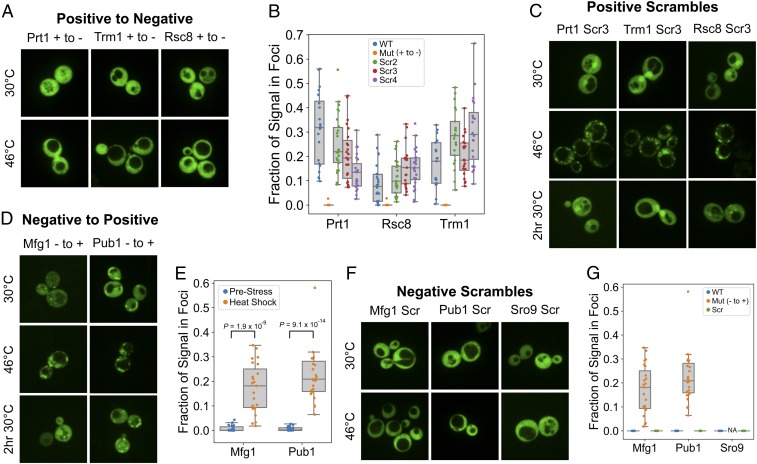
None of the 10 PrLDs that had assembly scores below −0.14 formed stress-induced foci. Therefore, to prevent assembly of the Prt1, Trm1, and Rsc8 PrLDs, we randomly selected amino acids that were overrepresented among assembly-prone PrLDs (C, D, E, F, I, K, L, R, V, and W), and serially replaced them with amino acids that were underrepresented among assembly-prone PrLDs (Q, N, A, and P) until the predicted assembly score of each PrLD was below −0.14 (SI Appendix, Table S3). It should be noted that although C, D, E, F, I, K, L, R, V, and W were all more common among assembly-prone PrLDs, not all of these biases were statistically significant; however, we wanted to be conservative in targeting a range of amino acids that might contribute to assembly. Q, A, N, and P were chosen as replacements because each showed a reasonably strong underrepresentation among assembly-prone PrLDs. All three mutated PrLDs remained diffuse upon heat shock (Fig. 4 A and B), demonstrating that composition alone is sufficient to design substitutions that modulate assembly behavior.
Fig. 4.
Effects of rational mutation or scrambling on PrLD behavior. Mutated and scrambled PrLDs were fused to GFP and imaged under normal growth conditions and after 30 min of heat shock at 46 °C. PrLDs that formed foci upon heat shock were also imaged after 2 h of recovery post heat shock at 30 °C. (A) Fluorescence microscopy of assembly-prone PrLDs that were rationally mutated to inhibit stress-induced assembly. (B) Box plot depicting the fraction of GFP signal in foci after heat shock for at least 20 individual cells for wild-type (WT), mutated, and scrambled versions of the assembly-prone PrLDs. (C) The primary sequence of assembly-prone PrLDs was scrambled to determine the effects on localization. Four scrambled versions were made for each. One representative version for each is shown; the others are shown in SI Appendix, Fig. S11. (D) Nonassembling PrLDs (from Mfg1, Pub1, and Sro9) were rationally mutated to promote assembly; the mutated version of Sro9 did not express, so was not analyzed. (E) Box plot depicting the fraction of GFP signal in foci before and after heat shock for at least 20 individual cells for mutated versions of the nonassembling PrLDs. Two-sided t tests were used to calculate P values. (F) Scrambled versions of nonassembling PrLDs were tested for assembly before and after heat stress. (G) Box plot comparing the fraction of GFP signal in foci after heat stress for the rationally mutated and scrambled versions of Mfg1, Pub1, and Sro9. The nonexpressing Sro9 mutant was not analyzed (NA).
Previous work has shown that scrambling the primary sequence of classical yeast prion domains does not prevent prion formation (48, 49). Because we were able to predict and alter assembly properties based on amino acid composition, we tested whether stress-induced assembly would show similar insensitivity to scrambling. We scrambled the same three PrLDs that were rationally mutated in Fig. 4A. Initial scrambled versions of the three assembly-prone PrLDs were not detectable by Western blot (SI Appendix, Fig. S11A), so multiple scrambled versions were constructed and tested (SI Appendix, Table S3). Strikingly, each additional scrambled version formed foci upon heat shock (Fig. 4C and SI Appendix, Fig. S11B), and these foci colocalized with Pab1-mCherry (SI Appendix, Fig. S12). These results suggest that amino acid composition is the predominant determinant of a PrLD’s recruitment to stress granules.
We then tested whether nonassembling PrLDs could likewise be rationally mutated to form stress-induced assemblies. We mutated Mfg1, Pub1, and Sro9 by serially replacing randomly selected amino acids that were underrepresented among assembly-prone PrLDs (Q, N, A, P, M, and T) with amino acids that were overrepresented (E, V, F, I, R, K, L, and D) (Table 1 and SI Appendix, Fig. S3). The mutated version of Sro9 did not express, and so could not be evaluated for stress-induced assembly (SI Appendix, Fig. S13A). However, while the mutated versions of both Mfg1 and Pub1 formed small foci in some cells prior to stress (Fig. 4 D and E), both showed a substantial, statistically significant increase in foci formation upon heat stress (Fig. 4 D and E), and these foci partially colocalized with Pab-mCherry (SI Appendix, Fig. S13B). By contrast, all three nonassembling PrLDs maintained their nonassembling phenotypes upon scrambling (Fig. 4 F and G).
Thus, for five of the six PrLDs, we were able to successfully reverse the stress-induced assembly or nonassembly phenotype through rational mutation. By contrast, in all cases the assembling or nonassembling phenotype was retained upon scrambling, indicating that amino acid composition, rather than primary sequence, is the predominant determinant of stress-induced assembly by PrLDs. Additionally, the same sequence features that promote assembly also appear to promote recruitment of these assemblies to bona fide stress granules.
Designing sPrLDs.
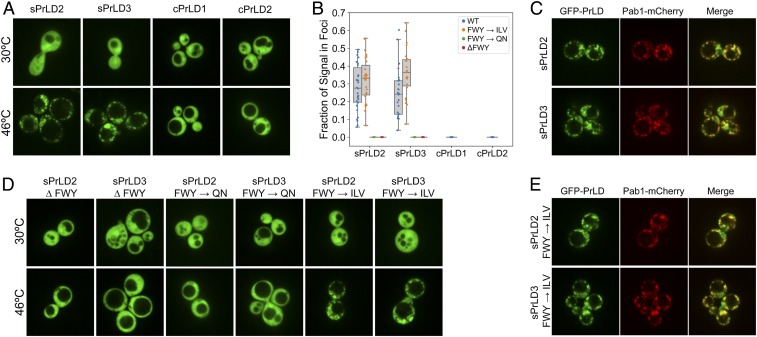
If PrLD stress-induced assembly is based primarily on amino acid composition, we reasoned that it should be possible to design sPrLDs that would assemble in response to stress. We used the average frequencies of each amino acid among the assembly-prone PrLDs to design three sPrLDs predicted to have high assembly propensity (see Materials and Methods for design details); one of these three (sPrLD1) failed to show detectable expression (SI Appendix, Fig. S14A). As a negative control, we used the average frequencies of each amino acid among the nonassembling PrLDs to design two control PrLDs (cPrLDs). Both the sPrLDs and cPrLDs showed diffuse localization prior to stress (Fig. 5A). Upon heat shock, the sPrLDs both efficiently formed reversible foci that colocalized with Pab1-mCherry, while the cPrLDs remained diffuse (Fig. 5 A–C and SI Appendix, Fig. S14 B and C).
Fig. 5.
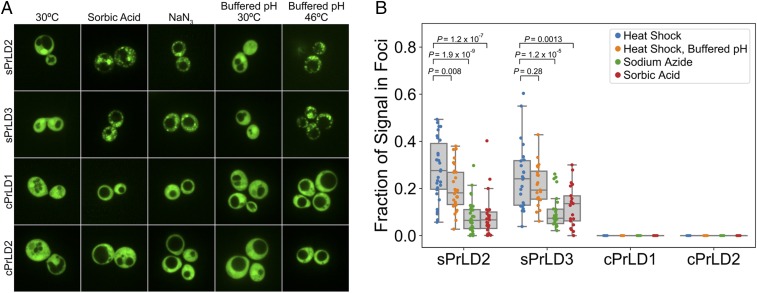
sPrLDs localize to stress granules. (A) sPrLDs and cPrLDs were fused to GFP and imaged under normal growth conditions and after 30 min of heat shock at 46 °C. (B) Box plot depicting the fraction of GFP signal in foci for at least 20 individual cells under heat shock for wild-type (WT) and mutant PrLDs. (C) sPrLDs colocalized with Pab1 upon heat shock. (D) Replacement of aromatic amino acids within the sPrLD with hydrophobic amino acids (I/L/V) maintained stress-induced assembly capacity, whereas deletion or substitution with Q/N prevented stress-induced assembly. (E) sPrLD mutants in which the aromatic amino acids were replaced with I/L/V colocalize with Pab1-mCherry.
These sPrLDs provide a useful tool for examining the features that promote PrLD assembly. Various studies have pointed to an important role for aromatic amino acids in driving LLPS or recruitment into phase-separated organelles (18, 22–24, 50). We therefore examined whether aromatic amino acids were required for stress-induced assembly. Either deletion of the aromatic amino acids in the sPrLDs or replacement of these amino acids with glutamine and asparagine prevented stress-induced assembly (Fig. 5 B and D), supporting a role for aromatic amino acids in promoting assembly. However, when all of the aromatic amino acids in the sPrLDs were replaced with nonaromatic hydrophobic amino acids (Val, Leu, and Ile), reversible localization to stress granules was restored (Fig. 5 B, D, and E and SI Appendix, Fig. S14 B and C). Thus, while aromatic amino acids appear to play an important role for stress granule recruitment or LLPS by some PrLDs, aromatic amino acids are not required for PrLD stress granule recruitment.
Finally, we tested whether these sPrLDs and cPrLDs, designed based on the heat shock dataset, would show similar behavior under other stresses. Indeed, both sPrLDs assembled in response to oxidative and pH stress, while the cPrLDs failed to assemble under each stress condition (Fig. 6). Standard yeast growth media is acidic, but yeast utilize ATPases to prevent cellular acidification; however, heat stress results in cellular acidification (51), potentially explaining why our PrLDs show similar response to heat and pH stress. To test the role of cellular acidification on heat-induced assembly of the sPrLD, we repeated the heat shock experiments in media buffered to pH 7.5. Previous work has shown that buffered media modestly reduces, but does not prevent, stress granule formation (52). Consistent with these results, buffered media slightly reduced assembly by both sPrLDs, although this difference was only statistically significant for sPrLD2 (Fig. 6). Thus, assembly does not require cellular acidification.
Fig. 6.
sPrLDs assemble under pH and oxidative stress. (A) Cells expressing sPrLDs or cPrLDs fused to GFP imaged under normal growth conditions; after 30 min of incubation with 0.5% NaN3; after 30 min of incubation with 6 mM sorbic acid; or before and after 30 min of heat shock at 46 °C in media buffered to pH 7.5. (B) Box plots depicting the fraction of GFP signal in foci for at least 20 individual cells under each stress. Two-sided t tests were used to calculate P values.
Discussion
PrLDs are relatively common among RNA-binding proteins, particularly those that are recruited to RNP granules (10). Although there have been efforts to understand how these domains affect RNP granule dynamics, their role within these assemblies remains unclear. While various studies have examined specific sequence and composition features that promote phase separation in vitro (18, 20, 22–24), or that affect stress-induced assembly of individual proteins (20, 53), this work systematically examines the range of PrLDs that can assemble in response to stress in vivo. We have demonstrated that many PrLDs are sufficient to be reversibly recruited to stress granules in yeast and that this recruitment occurs in a composition-dependent manner.
We were able to discern clear compositional biases among PrLDs that were sufficient to form stress-induced assemblies. Four pieces of data strongly argue that composition is the dominant feature driving stress-induced assembly: the ability of our composition-based predictor to predict which of the PrLDs will assemble into foci; the ability to predict new assembly-prone PrLDs and modulate the assembly propensity of existing PrLDs based solely on composition; the relative insensitivity of high- and low-scoring PrLD assembly propensity to scrambling; and the ability to design sPrLDs that are efficiently recruited to stress granules, based only amino acid composition. The fact that amino acid composition was the dominant determinant of reversible assembly is consistent with emerging results from studies of biological LLPS (23, 34); this result suggests that the basic physical properties of a PrLD determine whether it will reversibly partition into the cytosolic or stress granule phase upon stress.
Nevertheless, our data do not rule out modest effects of primary sequence. Although PrLDs with very high or low assembly scores were accurately predicted solely based on composition, our predictor was less accurate across the middle of the assembly score range. Some of this inaccuracy may simply reflect imperfections in our prediction algorithm. The assembly propensities for each amino acid have large confidence intervals, creating uncertainty in the final predictions. Alternatively, the inaccuracy of our predictions across the middle of the range may suggest that primary sequence can modulate assembly propensity. Primary sequence features could affect the intrinsic assembly propensity of PrLDs or could promote stress granule recruitment through interactions with binding partners. Such primary sequence effects appear insufficient to overcome strong compositional effects, but may be enough to nudge moderately scored domains across the boundary of assembly, in either direction. By defining the compositional contributions to assembly into stress-induced foci, our work should facilitate the identification of contributing primary sequence motifs.
While some of the biases that we observe are consistent with previous work examining either protein aggregation or LLPS, other results were unexpected. For example, the overrepresentation of charged residues among assembly-forming PrLDs (Fig. 2) is consistent with previous work analyzing sequence features that promote LLPS (22, 23, 54). However, charge patterning, particularly asymmetric charge distribution, has also been suggested to affect phase separation (22, 23), yet we found that the nonassembling PrLDs actually had slightly higher degrees of charge asymmetry.
A variety of evidence suggests that cation–pi interactions, particularly between arginine and tyrosine, are a key driver of phase separation by PrLDs and IDRs (18, 23, 24, 34, 55). However, we found that phenylalanine, but not tyrosine, was significantly overrepresented among assembly-forming PrLDs (SI Appendix, Fig. S6 and Table 1), although these data do not exclude a positive role for tyrosine in some sequences. It is worth noting that phenylalanine can also participate in cation–pi interactions and is capable of promoting LLPS, but tends to do so less efficiently than tyrosine in otherwise equivalent PrLD sequences. It is possible that phenylalanine is favored in our assays because it provides the proper balance of solubility and assembly propensity under our experimental conditions, as suggested in principle previously (24). Additionally, although aromatic amino acids overall promoted stress-induced assembly by our sPrLDs, aromatic amino acids were not required for assembly.
Another unexpected compositional bias was the overrepresentation of Q/N residues in PrLDs that do not form assemblies. This result was surprising because PrLDs are defined in part by their high Q/N content (43). This bias against Q/N residues highlights an apparent contradiction in our data. The fact that many of the tested PrLDs formed stress-induced assemblies clearly suggests that PrLDs are prone to form these assemblies, and indicates that some degree of Q/N enrichment may facilitate PrLD assembly. However, sequence features that made PrLDs maximally “prion-like” actually reduced assembly; although PrLDs tend to contain an overrepresentation of Q/N residues and an underrepresentation of charged and hydrophobic amino acids (43), PrLDs within our set with highest Q/N content and lowest hydrophobic content tended not to form stress-induced assemblies. These results are analogous to previous studies examining bona fide prion formation (56), which showed that while compositional similarity to known prion domains is an excellent way to identify prion candidates, such methods are relatively ineffective at ranking these candidates (56) or predicting the effects of mutations (57, 58). For stress-induced assembly, our results go one step further; although the prediction algorithm PLAAC was good at identifying candidates for stress-induced assembly, the rankings of the highest-scoring prion candidates were actually anticorrelated with the propensity to form stress-induced assemblies. These results suggest that the sequence features that promote stress-induced assembly partially, but incompletely, overlap with features that make a sequence prion-like. For example, while extremely elevated Q/N content was associated with lower assembly propensity, modestly elevated Q/N content may facilitate assembly, perhaps by increasing disorder propensity. This “Goldilocks” idea (that either too many or too few Q/N residues could inhibit assembly) might explain why our predictor was not effective for non–prion-like IDRs. Indeed, although the assembly-prone PrLDs tended to have more charged and hydrophobic amino acids and fewer polar amino acids compared to the nonassembling PrLDs, they still on average were enriched in polar amino acids and depleted in charged and hydrophobic amino acids compared to the yeast proteome (Fig. 2).
We were also surprised to find no substantial differences in compositional bias among three different stresses. Because different stresses likely result in different changes to the cellular environment, we expected that each stress would select for distinct compositional features, potentially explaining why the composition of stress granules differs depending on the stress (41). Instead, this result suggests that common features of the stress response contribute to PrLD assembly. We examined whether cellular acidification could be a common cause of assembly, as exposure of yeast cells to various stresses results in acidification (59). However, the fact that assembly of our sPrLDs occurred even in buffered media suggests that this acidification is not required for PrLD assembly, and that some other feature of the stress response drives PrLD assembly. Our results may also suggest that PrLDs generally provide a consistent contribution to stress-induced assembly across a variety of stresses, while the remainder of each PrLD-containing protein might dictate localization to granules in a stress type-dependent manner.
Strikingly, all of the assembly-prone PrLDs reversibly localized to stress granules in response to heat shock, suggesting that similar sequence features drive PrLD stress-induced assembly and stress granule localization. This allowed us to design sPrLDs that localize to stress granules. These sPrLDs may provide a useful tool for future experiments, as nonnatural sequences are less likely to engage in specific cellular interactions that could confound experiments. Additionally, these sPrLDs could potentially be used to artificially target proteins to stress granules, as well as to further dissect the sequence features that drive assembly.
While we have made significant strides toward understanding the sequence features of PrLDs that drive stress granule recruitment, there are likely many mechanisms of stress granule targeting; these may range from composition-based LLPS to very specific binding interactions. Thus, the behavior of a PrLD in isolation does not necessarily dictate the behavior of its respective full-length protein, as other regions of PrLD-containing proteins may either enhance or inhibit stress granule recruitment. The PrLD-containing protein kinase Sky1 is a good example of this concept. We recently identified Sky1 as a stress granule component and regulator of stress granule dynamics (8). However, while the Sky1 PrLD is sufficient for stress granule recruitment, deletion of the PrLD diminishes, but does not prevent, recruitment, highlighting a role for regions outside the PrLD (8). Like Sky1, some other full-length proteins containing assembly-prone PrLDs from our screen appear to localize to stress granules (SI Appendix, Fig. S15A). However, in other cases it appears that regions outside the PrLD restrict localization, preventing recruitment to stress granules. For example, while the PrLDs of Cdc73 and Trm1 are recruited to stress granules, their respective full-length proteins are not, but instead remain in the nucleus or at the nuclear periphery upon heat stress (SI Appendix, Fig. S15B). Therefore, while many of the proteins identified in proteomic screens of stress-induced assembly contain PrLDs (SI Appendix, Table S4 and refs. 38, 44, and 60–62), future experiments will be required to determine the exact contribution of the PrLD in each case.
Finally, the variable expression levels of the tested PrLDs conceivably could influence assembly behavior, as aggregation is a concentration-dependent process. However, the fact that we were able to build an effective prediction algorithm from this dataset, despite the possibility of variable expression creating noise in the analysis, only further highlights the dominant effects of composition. Furthermore, the assembly-prone sequences showed a diverse range of expression levels; indeed, while there were both poorly expressing and strongly expressing PrLDs among both the assembly-prone and nonassembling sequences, the poorly expressing sequences were actually more likely to assemble. This may simply be because assembly-prone PrLDs tended to have more hydrophobic amino acids and fewer Q/N residues; the presence of hydrophobic amino acids in PrLDs can promote protein degradation, while more Q/N-rich PrLDs seem to be more resistant to this degradation (63, 64). The effects of concentration will also depend on the mechanism of recruitment. In vitro, LLPS by an isolated purified protein tends to show a strong concentration dependence, in which LLPS occurs above a defined saturation concentration; however, in multicomponent systems where assembly is driven by heterotypic interactions, simple saturations concentrations are unlikely to apply (65).
With these results, we have established a groundwork for understanding the interactions that PrLDs contribute toward RNP granule dynamics. Using our foundational understanding of the effects of composition on PrLD localization, we can now begin to identify additional layers of sequence features that dictate assembly and localization upon stress.
Materials and Methods
Strains and Growth Conditions.
All experiments were performed in Saccharomyces cerevisiae using standard yeast media and growth conditions (66). All plasmids were transformed into either YER1405 (MATa his3D1 leu2D0 met15D0 ura3D0 PAB1-mCherry::URA3) or YER1997 (MATa his3D1 leu2D0 met15D0 ura3D0 PBP1-HA-mCherry) using standard yeast transformation methods (66). Both strains are derivatives of BY4741 (67). Yeast were grown at 30 °C, unless otherwise specified.
Cloning PrLDs.
Amino acid coordinates for each PrLD are listed in SI Appendix, Table S1. Var1, AI3, YML053C, Cdc39, and Fab1 all have slight sequence changes relative to the reference strains in the Saccharomyces Genome Database, as described in ref. 39. PrLDs were first amplified from yeast genomic DNA and then amplified a second time with primers containing tails for cloning into pER843. pER843 was built from pJ526 (49) by inserting GFP between the HindIII and BamHI restriction sites following the SUP35 promoter. PrLDs were cloned into this plasmid after GFP (in-frame) between the BamHI and BglII sites.
Mutation Design.
For each PrLD that was mutated from a negative to a positive (Mfg1, Pub1, and Sro9), Q, N, A, P, M, and T residues within the PrLD were randomly selected using an Excel random number generator; these residues were replaced with randomly selected E, V, F, I, R, K, L, and D residues until the score rose above 0.14. For PrLDs that were mutated from positive to negative (Prt1, Trm1, and Rsc8), C, D, E, F, I, K, L, R, V, and W were randomly chosen, and replaced with a random mixture of Q, N, A, and P (in a 2:2:1:1 ratio of Q:N:A:P) until the score dropped below −0.14. To design the scrambled PrLDs, sequences were randomly shuffled using the Excel random number generator. Both mutant and scrambled PrLDs were built synthetically using overlapping primers and then cloned into pER843 as described above.
Designing sPrLDs and cPrLDs.
Each sPrLD and cPrLD is 100 amino acids long. The average frequency of each amino acid among the assembling and nonassembling PrLDs (Table 1) were used as a starting point for design of the sPrLDs and cPrLDs, respectively. To achieve whole-number values for each amino acid, frequencies were rounded in the direction predicted to promote or inhibit assembly for the sPrLDs and cPrLDs, respectively; the numbers were then modestly adjusted such that the final values added up to 100. An Excel random number generator was used to randomly order the amino acids. Sequences were built synthetically using overlapping primers and then cloned into pER843 as described above.
Stress Conditions.
Cells were grown to midlog phase at 30 °C in SC-Leu media (to select for the plasmids) prior to stress induction. For heat shock, 1 mL of cells was concentrated to 50 μL and incubated in a 46 °C water bath for 30 min prior to imaging. For oxidative stress, NaN3 was added to cells to a final concentration of 0.5% (vol/vol), and cells were incubated at 30 °C with shaking for 30 min. Following NaN3 treatment, cells were then concentrated to 50 μL and imaged. For pH stress, 1 mL of cells was harvested and media exchanged for 6 mM sorbic acid in SC-Leu. Cells were incubated at 30 °C for 30 min with rotation or shaking and then concentrated to 50 μL prior to imaging. For the heat shock experiment in buffered media, 10 mM Tris was added to the media, and the media was adjusted to pH 7.5.
Confocal Fluorescence Microscopy.
Imaging was performed on an Olympus (IX83) Inverted Spinning Disk Confocal Microscope using a 100× objective. Images were captured as single planes. Quantification was performed using Slidebook (Intelligent Imaging Innovations, 3i). All quantification was performed blinded. To determine the fraction of protein in foci, region masks were applied to cells by tracing the cell outline. A minimum of three positions per cell were sampled to estimate the average background fluorescence in each cell. Regions within each cell with fluorescence at least twofold above background were defined as foci. A minimum of 20 cells was analyzed for each PrLD.
Western Blotting.
Cells were grown to midlog phase at 30 °C in SC-Leu media (to select for the plasmids) before harvesting. Volumes collected for each culture were normalized to the lowest density culture, as assessed by OD600 measurements. Cells were harvested by centrifugation at 3,000 rpm for 5 min at 4 °C. Cell lysis was performed as described previously (68). A volume of 15 μL of each sample was run on a polyacrylamide gel and then transferred onto a PVDF membrane. GFP-PrLD fusions were probed using an anti-GFP antibody (Santa Cruz Biotechnology). Images were acquired using Li-Cor Odyssey Clx imaging system.
PrLD Composition Analyses.
PrLD compositions were analyzed using in-house Python scripts. Briefly, the percent compositions of individual amino acids (SI Appendix, Fig. S6) or amino acid groups (Fig. 2) were calculated for each PrLD. For each amino acid or amino acid group, the composition distribution associated with PrLDs localizing to stress granules was statistically compared (two-sided Mann–Whitney U test) to the composition distribution associated with PrLDs that did not localize to stress granules, and plotted as adjacent boxplots. Composition analyses were performed independently for each stress condition. Plotting and statistical tests were performed using the Matplotlib/Seaborn and Scipy packages, respectively.
Algorithm Generation.
For each amino acid, an odds ratio (ORaa) was determined as follows:
| [1] |
where fa is the mean frequency of occurrence of the amino acid among assembly-prone PrLDs, and fn is the mean frequency of occurrence among nonassembling PrLDs. The assembly propensity of each amino acid was defined based on its log-odds ratio. To predict the assembly propensity of a PrLD, the frequency of occurrence of each amino acid in the PrLD was multiplied by the amino acid’s log-odds ratio, and these values were summed.
Data Availability Statement.
All data are included in the figures and SI Appendix.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grants GM105991 and R35GM130352 (to E.D.R.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. M.K.R. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1912723117/-/DCSupplemental.
References
- 1.Ivanov P., Kedersha N., Anderson P., Stress granules and processing bodies in translational control. Cold Spring Harb. Perspect. Biol. 11, a032813 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Protter D. S. W., Parker R., Principles and properties of stress granules. Trends Cell Biol. 26, 668–679 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kedersha N., Ivanov P., Anderson P., Stress granules and cell signaling: More than just a passing phase? Trends Biochem. Sci. 38, 494–506 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li L., McGinnis J. P., Si K., Translational control by prion-like proteins. Trends Cell Biol. 28, 494–505 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ramaswami M., Taylor J. P., Parker R., Altered ribostasis: RNA-protein granules in degenerative disorders. Cell 154, 727–736 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baradaran-Heravi Y., Van Broeckhoven C., van der Zee J., Stress granule mediated protein aggregation and underlying gene defects in the FTD-ALS spectrum. Neurobiol. Dis. 134, 104639 (2020). [DOI] [PubMed] [Google Scholar]
- 7.Gilks N., et al. , Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol. Biol. Cell 15, 5383–5398 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shattuck J. E., Paul K. R., Cascarina S. M., Ross E. D., The prion-like protein kinase Sky1 is required for efficient stress granule disassembly. Nat. Commun. 10, 3614 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Protter D. S. W., et al. , Intrinsically disordered regions can contribute promiscuous interactions to RNP granule assembly. Cell Rep. 22, 1401–1412 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.March Z. M., King O. D., Shorter J., Prion-like domains as epigenetic regulators, scaffolds for subcellular organization, and drivers of neurodegenerative disease. Brain Res. 1647, 9–18 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Patel A., et al. , A liquid-to-solid phase transition of the ALS protein FUS accelerated by disease mutation. Cell 162, 1066–1077 (2015). [DOI] [PubMed] [Google Scholar]
- 12.Vance C., et al. , Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science 323, 1208–1211 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kwiatkowski T. J. Jr, et al. , Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science 323, 1205–1208 (2009). [DOI] [PubMed] [Google Scholar]
- 14.Guo W., et al. , An ALS-associated mutation affecting TDP-43 enhances protein aggregation, fibril formation and neurotoxicity. Nat. Struct. Mol. Biol. 18, 822–830 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Neumann M., et al. , Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science 314, 130–133 (2006). [DOI] [PubMed] [Google Scholar]
- 16.Kim H. J., et al. , Mutations in prion-like domains in hnRNPA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS. Nature 495, 467–473 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Molliex A., et al. , Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell 163, 123–133 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kato M., et al. , Cell-free formation of RNA granules: Low complexity sequence domains form dynamic fibers within hydrogels. Cell 149, 753–767 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin Y., Protter D. S., Rosen M. K., Parker R., Formation and maturation of phase-separated liquid droplets by RNA-binding proteins. Mol. Cell 60, 208–219 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Riback J. A., et al. , Stress-triggered phase separation is an adaptive, evolutionarily tuned response. Cell 168, 1028–1040.e19 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Murakami T., et al. , ALS/FTD mutation-induced phase transition of FUS liquid droplets and reversible hydrogels into irreversible hydrogels impairs RNP granule function. Neuron 88, 678–690 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nott T. J., et al. , Phase transition of a disordered nuage protein generates environmentally responsive membraneless organelles. Mol. Cell 57, 936–947 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pak C. W., et al. , Sequence determinants of intracellular phase separation by complex coacervation of a disordered protein. Mol. Cell 63, 72–85 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang J., et al. , A molecular grammar governing the driving forces for phase separation of prion-like RNA binding proteins. Cell 174, 688–699.e16 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Conicella A. E., Zerze G. H., Mittal J., Fawzi N. L., ALS mutations disrupt phase separation mediated by α-helical structure in the TDP-43 low-complexity C-terminal domain. Structure 24, 1537–1549 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mackenzie I. R., et al. , TIA1 mutations in amyotrophic lateral sclerosis and frontotemporal dementia promote phase separation and alter stress granule dynamics. Neuron 95, 808–816.e9 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cascarina S. M., Ross E. D., Yeast prions and human prion-like proteins: Sequence features and prediction methods. Cell. Mol. Life Sci. 71, 2047–2063 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Iglesias V., Conchillo-Sole O., Batlle C., Ventura S., AMYCO: Evaluation of mutational impact on prion-like proteins aggregation propensity. BMC Bioinformatics 20, 24 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paul K. R., et al. , Effects of mutations on the aggregation propensity of the human prion-like protein hnRNPA2B1. Mol. Cell. Biol. 37, e00652-16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zambrano R., et al. , PrionW: A server to identify proteins containing glutamine/asparagine rich prion-like domains and their amyloid cores. Nucleic Acids Res. 43, W331-7 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goldschmidt L., Teng P. K., Riek R., Eisenberg D., Identifying the amylome, proteins capable of forming amyloid-like fibrils. Proc. Natl. Acad. Sci. U.S.A. 107, 3487–3492 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Afsar Minhas F. U. A., Ross E. D., Ben-Hur A., Amino acid composition predicts prion activity. PLoS Comput. Biol. 13, e1005465 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hughes M. P., et al. , Atomic structures of low-complexity protein segments reveal kinked β sheets that assemble networks. Science 359, 698–701 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin Y., Currie S. L., Rosen M. K., Intrinsically disordered sequences enable modulation of protein phase separation through distributed tyrosine motifs. J. Biol. Chem. 292, 19110–19120 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maharana S., et al. , RNA buffers the phase separation behavior of prion-like RNA binding proteins. Science 360, 918–921 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lancaster A. K., Nutter-Upham A., Lindquist S., King O. D., PLAAC: A web and command-line application to identify proteins with prion-like amino acid composition. Bioinformatics 30, 2501–2502 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ross E. D., Maclea K. S., Anderson C., Ben-Hur A., A bioinformatics method for identifying Q/N-rich prion-like domains in proteins. Methods Mol. Biol. 1017, 219–228 (2013). [DOI] [PubMed] [Google Scholar]
- 38.Wallace E. W., et al. , Reversible, specific, active aggregates of endogenous proteins assemble upon heat stress. Cell 162, 1286–1298 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shattuck J. E., Waechter A. C., Ross E. D., The effects of glutamine/asparagine content on aggregation and heterologous prion induction by yeast prion-like domains. Prion 11, 249–264 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Buchan J. R., Muhlrad D., Parker R., P bodies promote stress granule assembly in Saccharomyces cerevisiae. J. Cell Biol. 183, 441–455 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Buchan J. R., Yoon J. H., Parker R., Stress-specific composition, assembly and kinetics of stress granules in Saccharomyces cerevisiae. J. Cell Sci. 124, 228–239 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Munder M. C., et al. , A pH-driven transition of the cytoplasm from a fluid- to a solid-like state promotes entry into dormancy. eLife 5, e09347 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Harrison P. M., Gerstein M., A method to assess compositional bias in biological sequences and its application to prion-like glutamine/asparagine-rich domains in eukaryotic proteomes. Genome Biol. 4, R40 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhu M., et al. , Proteomic analysis reveals the recruitment of intrinsically disordered regions to stress granules. bioRxiv:10.1101/758599 (5 September 2019). [DOI] [PubMed]
- 45.Dosztányi Z., Csizmok V., Tompa P., Simon I., IUPred: Web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics 21, 3433–3434 (2005). [DOI] [PubMed] [Google Scholar]
- 46.Galzitskaya O. V., Garbuzynskiy S. O., Lobanov M. Y., FoldUnfold: Web server for the prediction of disordered regions in protein chain. Bioinformatics 22, 2948–2949 (2006). [DOI] [PubMed] [Google Scholar]
- 47.Prilusky J., et al. , FoldIndex: A simple tool to predict whether a given protein sequence is intrinsically unfolded. Bioinformatics 21, 3435–3438 (2005). [DOI] [PubMed] [Google Scholar]
- 48.Ross E. D., Baxa U., Wickner R. B., Scrambled prion domains form prions and amyloid. Mol. Cell. Biol. 24, 7206–7213 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ross E. D., Edskes H. K., Terry M. J., Wickner R. B., Primary sequence independence for prion formation. Proc. Natl. Acad. Sci. U.S.A. 102, 12825–12830 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xiang S., et al. , The LC domain of hnRNPA2 adopts similar conformations in hydrogel polymers, liquid-like droplets, and nuclei. Cell 163, 829–839 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weitzel G., Pilatus U., Rensing L., The cytoplasmic pH, ATP content and total protein synthesis rate during heat-shock protein inducing treatments in yeast. Exp. Cell Res. 170, 64–79 (1987). [DOI] [PubMed] [Google Scholar]
- 52.Kroschwald S., et al. , Different material states of Pub1 condensates define distinct modes of stress adaptation and recovery. Cell Rep. 23, 3327–3339 (2018). [DOI] [PubMed] [Google Scholar]
- 53.Franzmann T. M., et al. , Phase separation of a yeast prion protein promotes cellular fitness. Science 359, eaao5654 (2018). [DOI] [PubMed] [Google Scholar]
- 54.Elbaum-Garfinkle S., et al. , The disordered P granule protein LAF-1 drives phase separation into droplets with tunable viscosity and dynamics. Proc. Natl. Acad. Sci. U.S.A. 112, 7189–7194 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Qamar S., et al. , FUS phase separation is modulated by a molecular chaperone and methylation of arginine cation-π interactions. Cell 173, 720–734.e15 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Toombs J. A., McCarty B. R., Ross E. D., Compositional determinants of prion formation in yeast. Mol. Cell. Biol. 30, 319–332 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gonzalez Nelson A. C., et al. , Increasing prion propensity by hydrophobic insertion. PLoS One 9, e89286 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Paul K. R., Hendrich C. G., Waechter A., Harman M. R., Ross E. D., Generating new prions by targeted mutation or segment duplication. Proc. Natl. Acad. Sci. U.S.A. 112, 8584–8589 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yoo H., Triandafillou C., Drummond D. A., Cellular sensing by phase separation: Using the process, not just the products. J. Biol. Chem. 294, 7151–7159 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cherkasov V., et al. , Systemic control of protein synthesis through sequestration of translation and ribosome biogenesis factors during severe heat stress. FEBS Lett. 589, 3654–3664 (2015). [DOI] [PubMed] [Google Scholar]
- 61.Jain S., et al. , ATPase-modulated stress granules contain a diverse proteome and substructure. Cell 164, 487–498 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cary G. A., Vinh D. B., May P., Kuestner R., Dudley A. M., Proteomic analysis of Dhh1 complexes reveals a role for Hsp40 chaperone Ydj1 in yeast P-body assembly. G3 (Bethesda) 5, 2497–2511 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cascarina S. M., Paul K. R., Machihara S., Ross E. D., Sequence features governing aggregation or degradation of prion-like proteins. PLoS Genet. 14, e1007517 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cascarina S. M., Ross E. D., Aggregation and degradation scales for prion-like domains: Sequence features and context weigh in. Curr. Genet. 65, 387–392 (2019). [DOI] [PubMed] [Google Scholar]
- 65.Choi J. M., Dar F., Pappu R. V., LASSI: A lattice model for simulating phase transitions of multivalent proteins. PLoS Comput. Biol. 15, e1007028 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sherman F., Getting started with yeast. Methods Enzymol. 194, 3–21 (1991). [DOI] [PubMed] [Google Scholar]
- 67.Brachmann C. B., et al. , Designer deletion strains derived from Saccharomyces cerevisiae S288C: A useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14, 115–132 (1998). [DOI] [PubMed] [Google Scholar]
- 68.Fredrickson E. K., Gallagher P. S., Clowes Candadai S. V., Gardner R. G., Substrate recognition in nuclear protein quality control degradation is governed by exposed hydrophobicity that correlates with aggregation and insolubility. J. Biol. Chem. 288, 6130–6139 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are included in the figures and SI Appendix.